-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC
Methylation of DNA and of Lysine 9 on histone H3 (H3K9) is associated with gene silencing in many animals, plants, and fungi. In Neurospora crassa, methylation of H3K9 by DIM-5 directs cytosine methylation by recruiting a complex containing Heterochromatin Protein-1 (HP1) and the DIM-2 DNA methyltransferase. We report genetic, proteomic, and biochemical investigations into how DIM-5 is controlled. These studies revealed DCDC, a previously unknown protein complex including DIM-5, DIM-7, DIM-9, CUL4, and DDB1. Components of DCDC are required for H3K9me3, proper chromosome segregation, and DNA methylation. DCDC-defective strains, but not HP1-defective strains, are hypersensitive to MMS, revealing an HP1-independent function of H3K9 methylation. In addition to DDB1, DIM-7, and the WD40 domain protein DIM-9, other presumptive DCAFs (DDB1/CUL4 associated factors) co-purified with CUL4, suggesting that CUL4/DDB1 forms multiple complexes with distinct functions. This conclusion was supported by results of drug sensitivity tests. CUL4, DDB1, and DIM-9 are not required for localization of DIM-5 to incipient heterochromatin domains, indicating that recruitment of DIM-5 to chromatin is not sufficient to direct H3K9me3. DIM-7 is required for DIM-5 localization and mediates interaction of DIM-5 with DDB1/CUL4 through DIM-9. These data support a two-step mechanism for H3K9 methylation in Neurospora.
Published in the journal: DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC. PLoS Genet 6(11): e32767. doi:10.1371/journal.pgen.1001196
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001196Summary
Methylation of DNA and of Lysine 9 on histone H3 (H3K9) is associated with gene silencing in many animals, plants, and fungi. In Neurospora crassa, methylation of H3K9 by DIM-5 directs cytosine methylation by recruiting a complex containing Heterochromatin Protein-1 (HP1) and the DIM-2 DNA methyltransferase. We report genetic, proteomic, and biochemical investigations into how DIM-5 is controlled. These studies revealed DCDC, a previously unknown protein complex including DIM-5, DIM-7, DIM-9, CUL4, and DDB1. Components of DCDC are required for H3K9me3, proper chromosome segregation, and DNA methylation. DCDC-defective strains, but not HP1-defective strains, are hypersensitive to MMS, revealing an HP1-independent function of H3K9 methylation. In addition to DDB1, DIM-7, and the WD40 domain protein DIM-9, other presumptive DCAFs (DDB1/CUL4 associated factors) co-purified with CUL4, suggesting that CUL4/DDB1 forms multiple complexes with distinct functions. This conclusion was supported by results of drug sensitivity tests. CUL4, DDB1, and DIM-9 are not required for localization of DIM-5 to incipient heterochromatin domains, indicating that recruitment of DIM-5 to chromatin is not sufficient to direct H3K9me3. DIM-7 is required for DIM-5 localization and mediates interaction of DIM-5 with DDB1/CUL4 through DIM-9. These data support a two-step mechanism for H3K9 methylation in Neurospora.
Introduction
Methylation of selected cytosines in DNA is a prototypical epigenetic process found in many eukaryotes. DNA methylation has been implicated in embryonic development, genome imprinting, X chromosome inactivation, transposon silencing and gene regulation [1]–[5]. Conversely, abnormal DNA methylation has been associated with disease in humans, developmental defects in plants and growth defects in Neurospora [6]–[8]. Although some functions of DNA methylation have been identified, its regulation is not completely understood. The filamentous fungus Neurospora crassa has emerged as an excellent model system to elucidate the control of DNA methylation. In this organism, DNA methylation is found almost exclusively associated with relics of a genome defense system, RIP (repeat-induced point mutation) [9], [10]. The RIP machinery detects and mutates duplicate sequences during the sexual cycle, littering each copy with C to T transition mutations [11], [12]. Notably, the resulting A:T-rich sequences tend to be potent signals for de novo DNA methylation [9], [13], [14]. Our previous genetic studies revealed that all DNA methylation in Neurospora is dependent on a single DNA methyltransferase, DIM-2, (named for defective in DNA methylation) [15], an H3K9 methyltransferase (KMT), DIM-5 [16], Heterochromatin Protein-1 (HP1) [17] and DIM-7, a protein that interacts with DIM-5 [18]. The demonstration that DNA methylation depends on H3K9 methylation in Neurospora was followed quickly by findings that histone methylation is also critical for some DNA methylation in both plants and animals [19]–[21], suggesting that components of the DNA methylation pathway of Neurospora may be conserved in higher eukaryotes.
DIM-5 catalyzes tri-methylation of H3K9 (H3K9me3), which is recognized and bound by a complex of HP1 and DIM-2 [17], [22], [23]. Direct interaction of the chromo shadow domain of HP1 with a pair of PXVXL-like motifs in DIM-2 is essential for DNA methylation and does not depend on H3K9me3 [23]. In Neurospora, H3K9me3, HP1 and DNA methylation co-localize at RIP'd sequences and together define domains of heterochromatin at centromeres, telomeres and dispersed RIP'd regions throughout the genome [9]. Notably, the distribution of H3K9me3 is unaffected in the dim-2 mutant and is also independent of HP1 at nearly all heterochromatin domains [9], [23]. Efficient de novo DNA methylation is observed following depletion and subsequent re-introduction of H3K9 methylation [9]. Thus, RIP'd DNA directs H3K9 methylation and subsequent DNA methylation primarily through a unidirectional pathway.
Here we report that purification of DIM-5-associated proteins, in conjunction with genetic studies based on a powerful new selection for mutants defective in DNA methylation [18], revealed a multi-subunit complex, DCDC, that directs histone methylation in Neurospora. All five core members of the complex, DIM-7, DIM-8 (DDB1), DIM-9 and CUL4, are essential for H3K9 and DNA methylation but DIM-7 is uniquely required to target DIM-5 to heterochromatin domains and is also required to connect DIM-5 to the DCAF (DDB1/CUL4 Associated Factor), DIM-9.
Results
Identification of dim genes essential for DNA and H3K9 methylation
Neurospora mutants defective in DNA methylation, such as dim-2 [24] and dim-5 [16], were initially identified by laborious screening, by happenstance or, later, by reverse genetics [17]. Because there was no indication that the genome had been thoroughly searched for non-essential dim genes, we recently developed a dual reporter strain harboring methylated copies of drug-resistance genes (bar and hph, conferring resistance to basta and hygromycin, respectively) that could be used to select for dim mutants [18]. We decided to use this strain for an insertional mutagenesis, reasoning that the insertions could be used as tags to quickly identify the dim genes (see Methods). We identified eleven candidate insertional mutants, which were basta - and hygromycin-resistant, exhibited reduced or no DNA methylation at the normally methylated 8:A6 region [10] and gave rise to Dim− progeny in sexual crosses (Figure 1A, 1B and data not shown). Curiously, genetic analyses revealed that the insertion cassette was not responsible for the Dim− phenotype of 10 of the 11 mutants (data not shown). The single potential insertional mutant strain, which we named dim-8, displayed an apparent complete loss of DNA methylation (Figure 1B, 1C). Using inverse PCR, we found that the insertion cassette had integrated within NCU06605, a gene encoding the Neurospora homolog of DDB1 (Damaged DNA Binding Protein-1; see Text S1). To confirm that the insertion into NCU06605 was indeed responsible for the Dim− phenotype, we tested a NCU06605 knockout strain available from the Neurospora Genome Project [25]. Like our dim-8 strain, the NCU06605 knockout strain displayed an apparent complete loss of DNA methylation (Figure 1C). We next tested for complementation of the methylation defects of the dim-8 and NCU06605 knockout strains by introducing a 3XHA-tagged copy of the gene. DNA methylation was successfully restored in both strains (Figure 1C), confirming that disruption of NCU06605 was responsible for the methylation defect of the dim-8 strain. We therefore refer to NCU06605 as dim-8 and its protein product as DDB1.
Fig. 1. CUL4 and DDB1 are essential for DNA methylation. 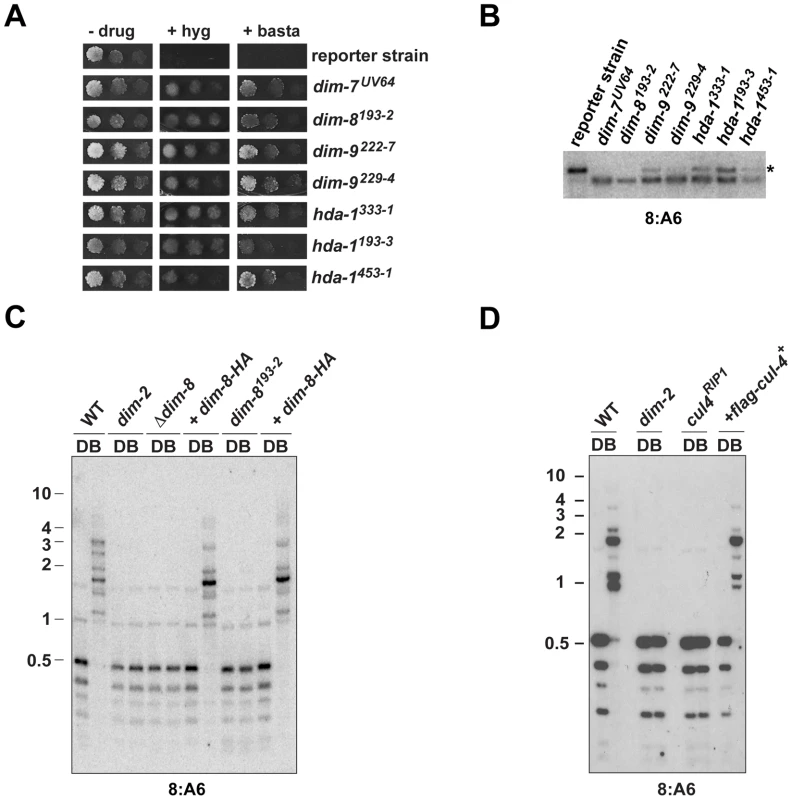
(A) Suspensions of 104, 103 or 102 conidia of the indicated strains were spot-tested on media with or without basta or hygromycin. (B) Southern hybridization with a probe corresponding to the normally methylated 8:A6 region was performed using genomic DNA digested with BamHI and EcoRI from methylation-positive (reporter strain) and -negative (dim-7UV64) control strains, as well as putative dim mutants. The asterisk indicates the expected position of the methylated DNA fragment (C, D) Southern hybridizations with a probe corresponding to the normally methylated 8:A6 region was performed using genomic DNA from the indicated strains digested with the cytosine-methylation-sensitive BfuCI (B) and –insenstive DpnII (D) endonucleases. The numbers at the left of each blot indicate the size, in kilobases, and position of molecular weight markers. Two additional mutants mapped to LGII and comprised a novel complementation group, which defined the dim-9 gene. The identity of dim-9 was revealed following purification and identification of DIM-5-associated proteins (see below). Complementation analyses also revealed that three additional strains represent new alleles of histone deacetylase-1, which we already knew is required for normal levels of DNA methylation [26].
CUL4 is essential for methylation of DNA and H3K9
DDB1 is known to interact with Cullin4 (CUL4) to form the core of an E3 ubiquitin ligase [27]. We utilized RIP to create a cul4 mutant strain and found that DNA methylation was abolished in this strain (Figure 1D, Figure S1). To verify that disruption of cul4 was responsible for the loss of DNA methylation, we introduced a FLAG-tagged copy of CUL4 (FLAG-CUL4; see Text S1). DNA methylation was restored in this strain, demonstrating that like DDB1, CUL4 is essential for DNA methylation (Figure 1D, Figure S1).
In Schizosaccharomyces pombe, CUL4 and the divergent DDB1 homolog Rik1 are essential for H3K9 methylation at heterochromatin domains [28], [29]. Although Neurospora DDB1 is more similar to DDB1 homologues than to Rik1 (49% similar to Arabidopsis DDB1A; 46% similar to S. pombe Ddb1; 45% similar to human DDB1; 39% similar to S. pombe Rik1; determined by BLAST searches queried with Neurospora DDB1), the similarity between these proteins suggested that they could perform similar functions. We therefore tested if CUL4 and DDB1 are required for H3K9 methylation in Neurospora, which we already knew is essential for DNA methylation in this organism [16]. Western blots revealed that H3K9me3 was completely abolished in the cul4RIP1 and Δdim-8 mutant strains (Figure 2A). Recent work with mammalian cells revealed that CUL4 and DDB1 are important for methylation of additional residues on H3, including H3K4 and H3K27 [30]. We therefore examined the levels of H3K4me, H3K27me, H3K36me, H3K79me and H4K20me in these mutant strains. Western blots revealed that only H3K9 methylation was affected in the cul4 and dim-8 strains (Figure 2A). HP1 localization to heterochromatic foci within the nucleus is dependent on H3K9me3 in Neurospora [17]. As expected, HP1 was mislocalized in the cul4 and dim-8 strains, consistent with a complete loss of H3K9 methylation (Figure 2B).
Fig. 2. CUL4 and DDB1 are essential for H3K9 methylation. 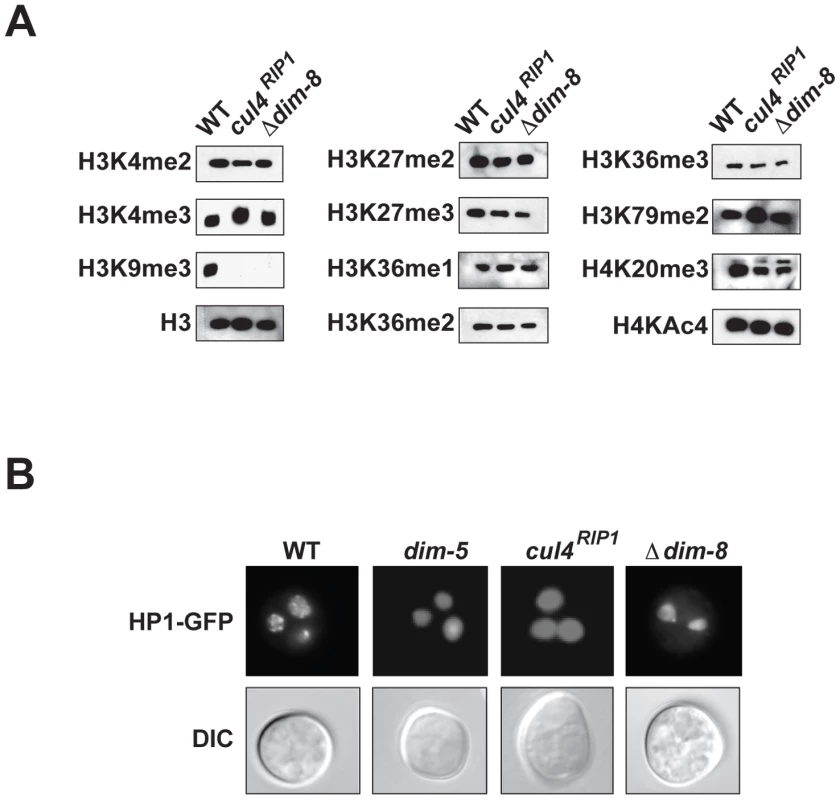
(A) Nuclear extracts from wildtype, dim-8 and cul4 were subjected to western blotting using the indicated antibodies specific for various modified histone proteins or for unmodified H3. (B) The distribution of HP1-GFP is shown in multinucleate conidia for wildtype, dim-5, dim-8 and cul4 strains (DIC; differential interference contrast). Identification of DIM-5–associated proteins
In addition to the genetic approach described above, we also employed biochemical approaches to identify DIM-5-associated proteins. We engineered a strain expressing DIM-5 fused to a HAT-FLAG tandem affinity tag [31] and used this in a two-step purification of DIM-5. The purified material was then analyzed by mass spectrometry. We identified peptides covering 25% of DIM-5, 25% of the previously characterized DIM-5-interacting protein DIM-7 [18], 11% of CUL4 and 13% of DDB1 (Table S1). Other potentially relevant proteins were also identified. CUL4/DDB1 complexes are known to interact with DCAFs that have WD40 domains and serve as substrate specificity factors [32], [33]. We identified peptides covering 8% of a WD40 domain-containing protein encoded by NCU01656 (Table S1). This gene resides on LGII, which raised the possibility that it was the unidentified dim-9 gene revealed in our mutant hunt. To test this possibility, we sequenced NCU01656 from the dim-9222-7 strain. A 120 bp deletion near the C-terminus was found, which would remove amino acids 1178 to 1217 from the predicted protein (XP_956278.2), suggesting that this gene was dim-9. We next introduced a wildtype copy of the NCU01656 gene into the dim-9 strain to test for complementation. DNA methylation was restored (Figure S2), demonstrating that mutations in NCU01656 are indeed responsible for loss of methylation in the dim-9 strains. We therefore refer to NCU01656 as dim-9 and the encoded protein as DIM-9. This gene had been replaced with an hph cassette as part of the Neurospora genome project [25] but homokaryotic strains had not been successfully isolated, suggesting that DIM-9 might be essential for viability or meiosis. To examine these possibilities, we crossed the heterokaryotic dim-9 replacement strain to a Sad-1 strain to prevent meiotic silencing by unpaired DNA [34] and isolated hygromycin-resistant progeny. We were able to obtain homokaryotic dim-9 knock-out progeny, indicating that the gene is not essential for viability. Southern blot analyses revealed that DIM-9 is essential for DNA methylation, like DIM-5, DIM-7, DDB1 and CUL4 (Figure 3A). Similarly, western blots revealed that the dim-9 knock-out strain displayed an apparent complete loss of H3K9me3 (Figure 3B).
Fig. 3. DIM-5 interacts with CUL4, DDB1, and DIM-9. 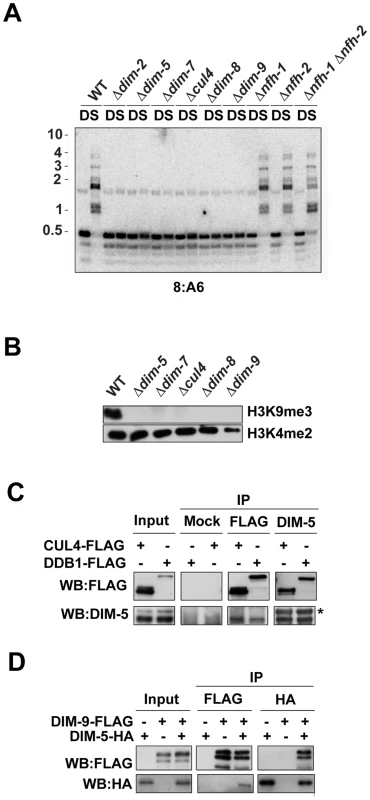
(A) Southern hybridization with a probe corresponding to the normally methylated 8:A6 region was performed using genomic DNA from the indicated strains digested with the cytosine-methylation-sensitive BfuCI (B) and –insensitive DpnII (D) endonucleases. (B) Histones extracted from wildtype, dim-5, dim-7, dim-8, cul4 and dim-9 were subjected to western blotting using antibodies to H3K9me3 and H3K4me2 as indicated. (C) Immunoprecipitation experiments were performed using extracts from strains expressing FLAG-CUL4 or DDB1-FLAG (+). The input fraction, the α-FLAG immunoprecipitate (IP:αFLAG), the α-DIM-5 immunoprecipitate (IP: α-DIM-5) and the mock immunoprecipitate (IP: mock) were subject to western blotting and probed with the α-FLAG or α-DIM-5 antibodies as indicated (WB). The asterisk indicates a non-specific cross-reacting band. (D) Immunoprecipitation experiments were performed using extracts from strains expressing DIM-9-FLAG, DIM-5-HA, or both (indicated by + or −). The input fraction, the α-HA immunoprecipitate fraction (IP:α-HA) and the α-FLAG immunoprecipitate fraction (IP:α-FLAG) were subjected to western blotting and probed with the α-FLAG or α-HA antibodies as indicated (WB). In addition to DIM-9, we identified peptides covering 19% of one Neurospora 14-3-3 domain-containing protein and 12% of another such protein (Table S1), which together represent the only two genes encoding 14-3-3 domain proteins in the N. crassa genome [35]. We refer to these previously uncharacterized genes as Neurospora fourteen-three-three homolog-1 (nfh-1; NCU3300) and nfh-2 (NCU02806). An S. pombe 14-3-3 protein was recently shown to interact with the Clr4 KMT and to function in heterochromatin formation [36]. We were interested to determine if one or both Neurospora 14-3-3 protein(s) is/are required for heterochromatin formation. Because knockout strains lacking nfh-2 were not available, we replaced the nfh-2 gene with the selectable bar gene [37] by targeted gene replacement [38]. Southern analysis of this and an nfh-1 knockout strain obtained from the Neurospora genome project revealed normal DNA methylation in both nfh mutant strains (Figure 3A). The predicted amino acid sequences of NFH-1 and NFH-2 are similar, suggesting that these proteins may perform redundant functions. To test this, we created an nfh-1, nfh-2 double mutant strain. The double mutant exhibited severe growth defects but we were able to obtain enough tissue to assess DNA methylation. In contrast to the results obtained for dim-5, dim-7, dim-8, cul4 and dim-9 strains, Southern blots revealed only a mild loss of DNA methylation in the nfh-1, nfh-2 strain (Figure 3A). Although we were unable to obtain enough tissue from the nfh double mutant to isolate histones, the persistence of DNA methylation predicts that H3K9 methylation is present in this strain.
We previously showed that DIM-7 interacts with DIM-5 in vivo [18]. To verify that DDB1, CUL4 and DIM-9 also interact with DIM-5, we performed coimmunoprecipitation (CoIP) experiments with strains expressing epitope-tagged proteins. We expressed a C-terminal, 3XFLAG-tagged DDB1 (DDB1-FLAG) from its native locus and similarly used the FLAG-CUL4 strain described above. Following immunoprecipitation with anti-FLAG antibodies or anti-DIM-5 antibodies, western blots revealed both DIM-5 and the expressed DDB1-FLAG or FLAG-CUL4 protein in the input, the anti-FLAG immunoprecipitate (IP) and the anti-DIM-5 IP fractions. In contrast, neither FLAG-tagged protein nor DIM-5 was detected in the mock IP (Figure 3C). Similarly, we performed CoIP experiments using a strain expressing 3XFLAG-tagged DIM-9 (DIM-9-FLAG) and 3XHA-tagged DIM-5 (DIM-5-HA). Western blots revealed both proteins in the input, the anti-FLAG IP, and the anti-HA IP fractions, confirming that these proteins interact in vivo (Figure 3D).
Our finding that the products of dim-7, dim-8, dim-9 and cul4 genes co-purified with DIM-5 and are all absolutely required for DIM-5 function, and our confirmation of key interactions by CoIP experiments, led us to conclude that DIM-5 is part of a complex necessary for DNA methylation in Neurospora. We will refer to this complex as DCDC (the DIM-5/-7/-9, CUL4/DDB1 complex). We were interested to learn whether some or all of the identified DCDC proteins would co-purify with CUL4. To investigate this, we engineered a strain expressing CUL4 fused to a tandem HAT-FLAT affinity tag [31], purified the tagged protein, and identified associated proteins by mass spectrometry. We identified peptides corresponding to CUL4 (49% coverage), DDB1 (44% coverage), DIM-7 (30% coverage) and DIM-9 (28% coverage). Interestingly, DIM-5, NFH-1 and NFH-2 were not identified in the purified fraction (Table S2), suggesting that DIM-5 only associates with a fraction of the total CUL4/DDB1 protein complex in the cell, consistent with the expectation that CUL4/DDB1 serves as a scaffold for more than one complex.
Purification of CUL4-associated proteins revealed additional proteins that do not seem to be members of DCDC, but are known to interact with CUL4/DDB1 in other organisms [32], [33] (Table S2). These include several WD40 domain-containing proteins, which presumably correspond to Neurospora DCAFs, plus members of the COP9 signalosome complex. Cullin proteins are typically modified post-translationally by attachment of the small ubiquitin-like protein, NEDD8. We identified peptides corresponding to Neurospora NEDD8 in the band that contained CUL4, suggesting that Neurospora CUL4 is neddylated. We examined DNA methylation levels in mutant strains lacking individual DCAFs or components of the COP9 signalosome complex and found normal DNA methylation in these strains (Table S2). These data suggest that Neurospora CUL4 and DDB1 interact with DCAFs to form distinct complexes that participate in various cellular processes.
DCDC is required for normal chromosome segregation
Mutant strains lacking components of DCDC exhibit growth defects (representative data shown for cul4 in Figure S3), similar to previously reported defects observed for dim-5 and hpo strains [16], [17]. To test heterochromatin-deficient mutants for specific defects in transcription, centromere function, and DNA repair, we tested their sensitivity to diagnostic drugs. Serial dilutions of conidia of wildtype, dim-2, hpo and DCDC-defective strains were spot-tested on unsupplemented medium and media supplemented with hydroxyurea (HU; ribonucleotide reductase inhibitor), methyl methanesulfonate (MMS; alkylating agent), camptothecin (CPT; topoisomerase I inhibitor) or thiabendazole (TBZ; microtubule inhibitor). All strains were able to grow on HU (Figure 4A). In contrast, hpo and the DCDC mutants were hypersensitive to TBZ, whereas the dim-2 and wildtype control strains were not. Interestingly, the DCDC mutants, but not hpo, were hypersensitive to MMS, suggesting that some functions of H3K9me3 are not dependent on HP1. Finally, cul4 and dim-8 mutants were hypersensitive to CPT, whereas all other strains tested grew on this drug, consistent with a role for CUL4/DDB1 in additional cellular processes, presumably mediated by additional DCAFs.
Fig. 4. Heterochromatin is required for normal chromosome segregation. 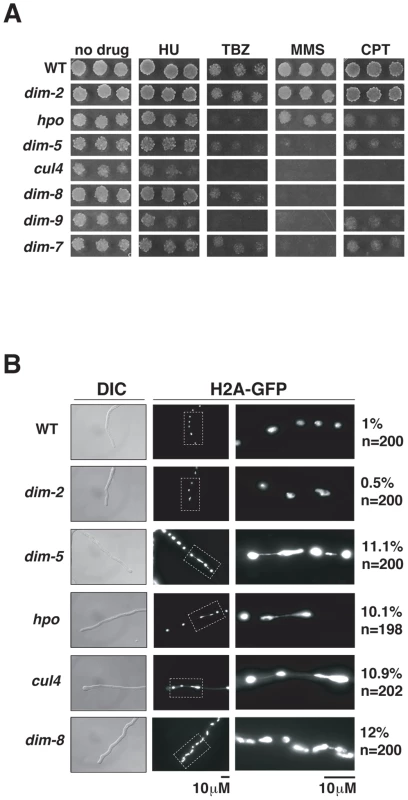
(A) Suspensions of 104, 103 or 102 conidia of the indicated strains were spot-tested on media with or without hydroxyurea (HU), methyl methanesulfonate (MMS), camptothecin (CPT) or thiabendazole (TBZ). (B) The distribution of H2A-GFP in growing hyphal tips in wildtype, dim-2, dim-5, dim-8 and cul4 strains. The frequency of observed chromatin bridges/total number of nuclei is shown beside each micrograph. The sensitivity to the microtubule inhibitor TBZ observed for hpo and DCDC mutants suggests H3K9me3 and HP1 are important for centromere function. To test this possibility, we examined chromosome segregation in live cells using a GFP-tagged H2A to visualize chromatin. Indeed, hpo, dim-5, cul4 and dim-8 mutants displayed high frequencies of lagging chromosomes, indicating that centromere function is impaired in these strains. All of these mutants showed chromosome bridges associated with approximately 10% of the nuclei, whereas evidence of lagging chromosomes was rarely observed in wildtype or dim-2 stains (Figure 4B).
DIM-7 recruits DIM-5 to form DCDC
In an attempt to determine which components of DCDC are responsible for recruiting DIM-5 to the complex, we initially tested for direct interaction between DIM-5 and each DCDC member by the yeast two-hybrid assay but these experiments failed to demonstrate a direct interaction between DIM-5 and any other DCDC component (data not shown). We therefore carried out CoIP experiments to test individual DCDC knockout strains for their ability to support pair-wise interactions between DIM-5 and other members of DCDC. FLAG-tagged versions of DDB1, DIM-9 and DIM-7 were expressed from their native loci. Tagged proteins were precipitated with anti-FLAG antibodies, and the input and IP fractions were interrogated with anti-FLAG and anti-DIM-5 antibodies. Immunoprecipitation of DDB1-FLAG, DIM-9-FLAG and DIM-7-FLAG revealed that all three proteins interact with DIM-5 in both wildtype and cul4 strains (Figure 5A–5C), indicating that CUL4 is dispensable for interaction of DIM-5 with other DCDC components. We note that although the DIM-9-DIM-5 interaction appears reduced in the experiment illustrated (Figure 5B), we observed increased interaction between these two proteins in a replicate experiment (Figure S4). Interestingly, immunoprecipitation of DDB1-FLAG failed to reveal a DDB1-DIM-5 interaction in the dim-9 or dim-7 strains (Figure 5A), suggesting that DIM-9 and DIM-7 mediate the indirect interaction of DIM-5 with DDB1.
Fig. 5. DIM-7 recruits DIM-5 to form DCDC. 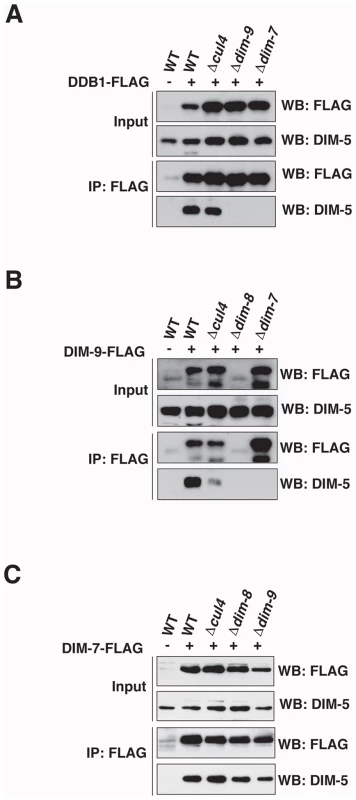
Immunoprecipitation experiments were performed using extracts from the indicated wildtype, cul4, dim-8, dim-9 or dim-7 strains with or without (A) DDB1-FLAG, (B) DIM-9-FLAG or (C) DIM-7-FLAG (indicated by + or −). The input fraction and the α-FLAG immunoprecipitate (IP:α-FLAG) were subjected to western blotting and probed with antibodies to α-FLAG or α-DIM-5 as indicated (WB). Western blots of both the input and IP fractions revealed that DIM-9-FLAG levels were markedly reduced in the dim-8 strain (Figure 5B), suggesting that DIM-9 stability depends on DDB1. Consistent with this, DIM-5 was not found in the DIM-9-FLAG IP fraction from the dim-8 strain (Figure 5B). Yeast two-hybrid assays revealed an interaction between DIM-9 and DDB1 (data not shown), suggesting that these proteins interact directly, as expected. Together, these data suggest that direct interaction of DDB1 and DIM-9 is important for DIM-9 stability.
DIM-9-FLAG was readily detectable in the dim-7 strain, but DIM-5 was not found in the DIM-9-FLAG IP fraction of this strain (Figure 5B). These findings indicate that the DIM-9-DIM-5 interaction depends on DIM-7. In contrast to the situation for DDB1 and DIM-9, the interaction of DIM-5 and DIM-7-FLAG was independent of all other DCDC members. Indeed, DIM-5 was detected in the IP fraction following immunoprecipitation of DIM-7-FLAG from wildtype, cul4, dim-8, and dim-9 strains (Figure 5C). These data suggest that DIM-7 is required to mediate interaction of DIM-5 with DCDC, most likely via DIM-9.
DIM-7 directs DIM-5 to heterochromatin domains
We recently adapted the DamID technique [18], [39] to test for chromatin association of DIM-5 and showed that DIM-7 is required to target DIM-5 to heterochromatin domains. Because DIM-7 is required to recruit DIM-5 to form the DCDC, we tested if the other components of DCDC are also required for association of DIM-5 with chromatin regions destined to be methylated. We introduced a DIM-5-Dam fusion construct into the cul4, dim-8 and dim-9 strains. We then tested for Dam activity in these strains, as well as positive - (wildtype) and negative - (dim-7) control strains by treating genomic DNA with DpnI, which specifically cuts GATC sites containing methylated adenines, but does not digest unmethylated GATC sites (Figure 6A). The digested DNA was fractionated by electrophoresis and probed for the heterochromatin regions 8:G3 and 8:A6, as well as for the euchromatic genes mtr and Sms-2. For the wildtype, cul4, dim-8 and dim-9 strains, the heterochromatin probes detected low molecular weight fragments corresponding to completely digested DNA and some intermediate molecular weight fragments corresponding to partially digested DNA. In contrast, only high molecular weight DNA was detected in the dim-7 background. Importantly, probes for Sms-2 and mtr hybridized to high molecular weight DNA corresponding to largely undigested DNA in all strains. These data suggest that DIM-7 is required to recruit DIM-5 to heterochromatin domains, while the remaining DCDC members are not.
Fig. 6. DIM-7, but not CUL4/DDB1DIM-9, is required for recruitment of DIM-5 to heterochromatin domains. 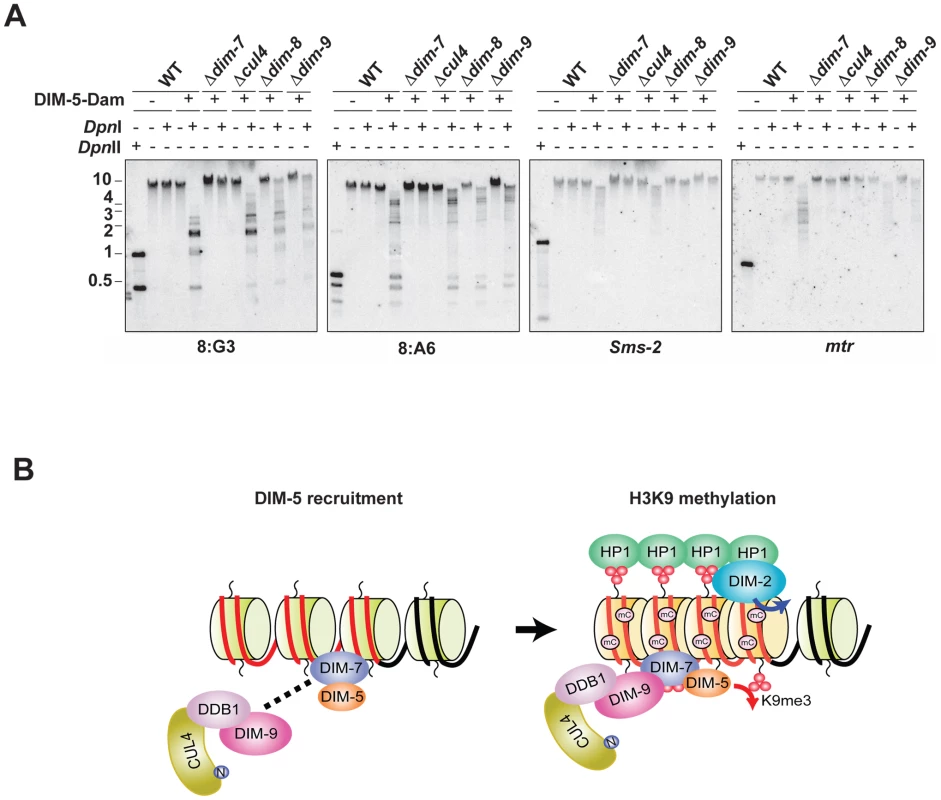
(A) Genomic DNA from wildtype, which does not express DIM-5-Dam, as well as Dim+, dim-7, dim-8, cul4 and dim-9 strains expressing DIM-5-Dam were incubated with or without DpnI, which cuts GATC only when the adenine is methylated. As an indicator of completely digested DNA, genomic DNA from the wildtype strain was incubated with the cytosine methylation-insensitive enzyme DpnII. Digested DNA was used for Southern hybridizations with probes corresponding to the indicated heterochromatic 8:A6 and 8:G3 regions and the euchromatic mtr and Sms-2 genes. (B) A model for H3K9 methylation by DCDC is shown (see text). Discussion
DNA methylation, which is frequently associated with heterochromatin, is essential for development, genome defense, genome imprinting and X-chromosome inactivation [1]–[4], and misregulation of DNA methylation has been implicated in disease [8]. Unfortunately, the mechanisms that direct heterochromatin and DNA methylation are not well understood. To uncover the mechanisms responsible for regulating DNA methylation in Neurospora, we carried out three independent lines of investigation: 1) We selected for mutants that are defective in DNA methylation; 2) we identified DIM-5-associated proteins by mass spectrometry; and 3) we made, and tested the effects of, mutations in candidate genes, such as cul4. These approaches proved complementary, revealing that a complex of DIM-5, DIM-7, CUL4, DDB1 and DIM-9, which we named DCDC, is required for H3K9 methylation and DNA methylation.
CUL4 and DDB1 are conserved from S. pombe to humans and are known to participate in a variety of cellular processes [32], [33]. Our discovery that CUL4 and DDB1 are required for DNA methylation is consistent with a report published while this paper was in preparation [40]. Our more comprehensive analyses revealed additional components of a DIM-5-containing complex and a hierarchy of interactions within the complex. Distinct functions of CUL4/DDB1 complexes are mediated by variable, WD40 domain-containing subunits called DCAFs, which interact directly with DDB1 and are thought to determine the substrate specificities of the various CUL4/DDB1 ubiquitin ligase complexes. DIM-9 is a WD40 domain-containing protein, suggesting that DIM-9 is the DCAF component of DCDC. Consistent with this, yeast two-hybrid analyses revealed that DIM-9 interacts directly with DDB1. In addition, DDB1 is required for stability of DIM-9. Our data also indicate that DIM-9 is required to mediate interaction of DIM-7/DIM-5 with CUL4/DDB1. These findings would be consistent with the possibility that DIM-7 or DIM-5 is a substrate for the CUL4/DDB1DIM-9 ubiquitin ligase; however, several attempts to identify ubiquitylated forms of either protein in Neurospora extracts were unsuccessful (data not shown). In addition, we note that sequence alignments of fungal DIM-7 homologues reveal only a handful of conserved residues, none of which are lysine residues [18], suggesting that ubiquitylation of DIM-7 is unlikely.
Although the putative substrate of CUL4/DDB1DIM-9 is unknown, our results are consistent with the possibility that the complex does serve as a ubiquitin ligase. We found that DIM-5 recruitment to heterochromatin domains is independent of CUL4, DDB1 and DIM-9. These data demonstrate that recruitment of DIM-5 to heterochromatin is not sufficient to direct H3K9 methylation. Similarly, recent work in S. pombe demonstrated that tethering of the Clr4 KMT to chromatin is not sufficient to direct H3K9 methylation in the absence of Rik1 [41]. It is notable that recombinant DIM-5 shows robust and specific methyltransferase activity on naked histones but not on nucleosomal substrates [22]. One possible role for the CUL4/DDB1DIM-9 components of DCDC would be to direct ubiquitination of a histone, thereby making H3 more accessible for methylation by DIM-5.
Interestingly, purification of CUL4/DDB1 complexes from mammalian cells has uncovered several DCAFs that are also components of histone lysine methyltransferase complexes [32], [33]. Furthermore, knock down of cul4 or dim-8 (DDB1) gene expression led to reduced methylation at several histone residues [30], consistent with a general role for CUL4 and DDB1 in histone methylation. Here we observed normal levels of H3K4, H3K27, H3K36, H3K79 and H4K20 methylation in CUL4 - and DDB1-deficent strains, indicating that these proteins are not required for general histone methylation in Neurospora. Rather, they specifically regulate H3K9 methylation. CUL4 - and DDB1-deficient strains exhibited hypersensitivity to the topoisomerase I inhibitor, CPT, whereas mutants deficient in other members of DCDC did not, supporting the expectation that CUL4 and DDB1 perform functions in addition to their function required for heterochromatin formation. Consistent with this, purification of CUL4 revealed additional DCAF proteins, suggesting that CUL4 and DDB1 form multiple ubiquitin ligase complexes as in other organisms.
We observed that DCDC and HP1 mutants are hypersensitive to the microtubule inhibitor TBZ, and that these strains exhibit high frequencies of lagging chromosomes. These data suggest that H3K9 methylation and HP1 are important for chromosome segregation in Neurospora, similar to the case in mammals, Drosophila and S. pombe [42]–[45]. This observation provides an explanation for the poor growth of Neurospora heterochromatin-deficient strains [16], [17].
Although S. pombe lacks DNA methylation, CLRC, a Clr4-containing complex that is essential for H3K9 methylation in this yeast [28], [29], resembles N. crassa DCDC. These complexes exhibit several significant differences, however. First, DCDC includes the conserved CUL4 binding partner DDB1, whereas CLRC utilizes the DDB1-like protein Rik1. In addition, these subunits appear to perform different functions. Rik1 is essential for RNAi-dependent recruitment of CLRC to heterochromatin nucleation sites [46], while DIM-5 recruitment to heterochromatin domains is independent of DDB1. DIM-5 recruitment is also independent of the DCDC components CUL4 and DIM-9, whereas recruitment of S. pombe Clr4 to heterochromatin is dramatically reduced in a Cul4 mutant strain [29], [46]. In contrast, DIM-7 is required to target DIM-5 to heterochromatin domains [18]. Another distinction between S. pombe CLRC and Neurospora DCDC involves the requirement of a 14-3-3 domain-containing subunit. In S. pombe, Rad24 co-purified with CLRC and is required for heterochromatic gene silencing and siRNA production [36]. Purification of Neurospora DIM-5 revealed NFH-1 and -2, but inactivation of the corresponding genes did not markedly effect DNA methylation, indicating that these proteins are not essential for maintenance of heterochromatin in Neurospora. These differences between S. pombe CLRC and Neurospora DCDC are not surprising given that these fungi employ different mechanisms to regulate heterochromatin formation. Indeed, work with S. pombe revealed that CLRC interacts with the Argonaute-containing RITS complex via the protein Stc1 to target H3K9 methylation [46], [47], whereas in Neurospora, H3K9 and DNA methylation do not depend on RNAi, but instead are directed by A:T-rich DNA [9], [13], [14], [48].
Mass spectrometry of DIM-5-associated proteins revealed that DIM-7 was the best represented DIM-5-associated component of DCDC, suggesting that DIM-5 and DIM-7 may interact directly. Consistent with this possibility, we demonstrated that the DIM-5/DIM-7 interaction is independent of other DCDC components, whereas DIM-7 is required for interaction of DIM-5 with DDB1 and DIM-9. Taken together, these data suggest that DIM-7 is required to recruit DIM-5 to form DCDC and lead us to propose a model (Figure 6B) in which DIM-5 and DIM-7 directly interact. We propose a two-step mechanism for H3K9 methylation by DCDC. First, DIM-7 recruits DIM-5 to form DCDC and somehow targets the complex to A:T-rich relics of RIP, by either a direct or indirect interaction with chromatin. We found that histones H3, H2A and H2B co-purify with DIM-7 (unpublished data of Z. Lewis and E. Selker), lending support to this model. In the second step, DIM-5 performs tri-methylation of H3K9 associated with RIP'd DNA in a CUL4/DDB1DIM-9-dependent manner. DIM-7 is not well conserved, but it appears to be a distant homolog of the CLRC component Raf-2. Therefore it would be interesting to know if Raf2 is responsible for recruitment of Clr4 to form the CLRC complex.
It seems quite possible that H3K9 KMTs exist in multi-protein complexes, generally [49], and that KMT-interacting proteins are important for targeting H3K9 methylation to appropriate chromatin domains. Purification of the mammalian H3K9 KMTs, Suv39H1, Suv39H2, G9a and SETDB1, did not reveal an interaction with CUL4 or DDB1 proteins [49] but these results do not rule out a possible role for a mammalian CUL4/DDB1 complex in heterochromatin formation. Moreover, a weak but biologically relevant interaction between mammalian H3K9 KMTs and CUL4/DDB1 proteins could be missed in analyses of affinity-purified proteins. Interestingly, mammalian cells in which DDB1 and CUL4 expression were knocked down showed reduced levels of H3K9 methylation [30], suggesting that these proteins may play a conserved role in heterochromatin formation from fungi to mammals.
Methods
Neurospora growth and molecular analyses
All strains used in this study are listed in Table S3. N. crassa strains were maintained, grown and crossed using previously described procedures [50]. Neurospora transformation [51], DNA isolation [52], Southern blotting [13], isolation of nuclei [53], fluorescence microscopy [17], protein isolation, histone isolation, coimmunoprecipitation [23] and construction of FLAG-tagged strains [31] were performed as described. All primers used in this study are listed in . Detailed descriptions of knock-out and epitope-tagged strain construction and a list of antibodies used for western blot analyses and coimmunoprecipitation experiments are available in the supplementary information.
In Neurospora, transforming DNA is typically integrated into the genome in an apparently random manner [38]. We therefore performed approximately three hundred transformations of our methylation reporter strain (N2977) as an attempt to generate mutations associated with the introduced DNA and selected for basta-resistant transformants as described in Text S1.
Identification of DIM-5–associated proteins
Construction of HAT-FLAG-tandem-affinity-tagged strains and the two-step purification were performed as described (Honda and Selker, 2009). Purified samples were separated by SDS-PAGE. As expected, DIM-5 was resolved with an apparent molecular weight of 38 kD. Gel slices containing bands were excised, washed and in-gel digested with trypsin overnight at 37°C. Tryptic peptides were separated by nano-HPLC (Rheos 2000) coupled to a 3D-ion trap mass spectrometer (LCQ Deca XP, both Thermo Fisher Scientific). The LC system was equipped with a capillary column with an integrated nanospray tip (100 µm i.d. ×100 mm, Swiss BioAnalytics AG) filled with Magic C18 (Michrom Bioresources, Inc.). Samples were loaded on a Peptide CapTrap (Michrom BioResources, Inc.) using a CTC PAL autosampler (CTC Analytics AG). Elution was performed with a gradient of 0 – 45% solvent B in 30 min at a flow rate of 500 nL/min. Solvent A consisted of 0.1% formic acid/2% acetonitrile; solvent B was composed of 0.1% formic acid/80% acetonitrile. In the data-dependent mode, the mass spectrometer cycled through four analyses, one MS full scan followed by MSMS scans for each of the three most intense peaks. Peptides were identified searching UniProt 15.14 using Mascot Distiller 2.3 for data extraction and conversion and Mascot 2.2 (Matrix Science). Results were compiled with Scaffold 2.06.
Phenotypic analyses of heterochromatin mutants
For drug sensitivity assays, serial dilutions of conidia were spot-tested on media with or without HU (8 mM), MMS (0.015%), CPT (0.3 µg/ml), or TBZ (0.5 µg/ml) obtained from Sigma Aldrich. To facilitate tracking chromatin cytologically, H2A-GFP (see Text S1) in growing hyphae was visualized using a Zeiss Axioplan 2 Imaging system with 100X oil immersion lens. Bright field and fluorescence images were collected using Images and processed using Axiovision (4.6.3) and Adobe Photoshop CS (version 8) software. Approximately 200 hyphal tips were counted for each culture and the number of tips that displayed nuclei with lagging chromosome bridges was noted to quantify the chromosome segregation defects.
Supporting Information
Zdroje
1. ReikW
DeanW
WalterJ
2001 Epigenetic reprogramming in mammalian development. Science 293 1089 1093
2. ReikW
WalterJ
2001 Genomic imprinting: parental influence on the genome. Nat Rev Genet 2 21 32
3. MiuraA
YonebayashiS
WatanabeK
ToyamaT
ShimadaH
2001 Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411 212 214
4. HeardE
DistecheCM
2006 Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20 1848 1867
5. WeberM
SchubelerD
2007 Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol 19 273 280
6. SazeH
ShiraishiA
MiuraA
KakutaniT
2008 Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319 462 465
7. HondaS
LewisZA
HuarteM
ChoLY
DavidLL
2010 The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa. Genes Dev 24 443 454
8. JonesPA
BaylinSB
2007 The epigenomics of cancer. Cell 128 683 692
9. LewisZA
HondaS
KhlafallahTK
JeffressJK
FreitagM
2009 Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res 19 427 437
10. SelkerEU
TountasNA
CrossSH
MargolinBS
MurphyJG
2003 The methylated component of the Neurospora crassa genome. Nature 422 893 897
11. CambareriEB
JensenBC
SchabtachE
SelkerEU
1989 Repeat-induced G-C to A-T mutations in Neurospora. Science 244 1571 1575
12. SelkerEU
1990 Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet 24 579 613
13. MiaoVP
FreitagM
SelkerEU
2000 Short TpA-rich segments of the zeta-eta region induce DNA methylation in Neurospora crassa. J Mol Biol 300 249 273
14. TamaruH
SelkerEU
2003 Synthesis of Signals for De Novo DNA Methylation in Neurospora crassa. Mol Cell Biol 23 2379 2394
15. KouzminovaEA
SelkerEU
2001 dim-2 encodes a DNA-methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO Journal 20 4309 4323
16. TamaruH
SelkerEU
2001 A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 277 283
17. FreitagM
HickeyPC
KhlafallahTK
ReadND
SelkerEU
2004 HP1 is essential for DNA methylation in Neurospora. Mol Cell 13 427 434
18. LewisZA
AdhvaryuKK
HondaS
ShiverAL
SelkerEU
2010 Identification of DIM-7, a protein required to target the DIM-5 H3 methyltransferase to chromatin. Proc Natl Acad Sci U S A 107 8310 8315
19. LehnertzB
UedaY
DerijckAA
BraunschweigU
Perez-BurgosL
2003 Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13 1192 1200
20. JacksonJP
LindrothAM
CaoX
JacobsenSE
2002 Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556 560
21. MalagnacF
BarteeL
BenderJ
2002 An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21 6842 6852
22. TamaruH
ZhangX
McMillenD
SinghPB
NakayamaJ
2003 Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet 34 75 79
23. HondaS
SelkerEU
2008 Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol Cell Biol 28 6044 6055
24. FossHM
RobertsCJ
ClaeysKM
SelkerEU
1993 Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science 262 1737 1741
25. ColotHV
ParkG
TurnerGE
RingelbergC
CrewCM
2006 A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103 10352 10357
26. SmithKM
DobosyJR
ReifsnyderJE
RountreeMR
AndersonDC
2010 H2B - and H3-specific histone deacetylases are required for DNA methylation in Neurospora crassa. Genetics(in press)
27. PetroskiMD
DeshaiesRJ
2005 Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 9 20
28. HornPJ
BastieJN
PetersonCL
2005 A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19 1705 1714
29. JiaS
KobayashiR
GrewalSI
2005 Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 7 1007 1013
30. HigaLA
WuM
YeT
KobayashiR
SunH
2006 CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 8 1277 1283
31. HondaS
SelkerEU
2009 Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182 11 23
32. HigaLA
ZhangH
2007 Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div 2 5
33. LeeJ
ZhouP
2007 DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 26 775 780
34. ShiuPK
RajuNB
ZicklerD
MetzenbergRL
2001 Meiotic silencing by unpaired DNA. Cell 107 905 916
35. MorrisonDK
2009 The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 19 16 23
36. HongEJ
VillenJ
GeraceEL
GygiSP
MoazedD
2005 A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol 2 106 111
37. PallML
1993 The use of Ignite (Basta;glufosinate;phosphinothricin) to select transformants of bar-containing plasmids in Neurospora crassa. Fungal Genetics Newsletter 40 58
38. NinomiyaY
SuzukiK
IshiiC
InoueH
2004 Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci U S A 101 12248 12253
39. VogelMJ
Peric-HupkesD
van SteenselB
2007 Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc 2 1467 1478
40. ZhaoY
ShenY
YangS
WangJ
HuQ
2010 Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem 285 4355 4365
41. KaganskyA
FolcoHD
AlmeidaR
PidouxAL
BoukabaA
2009 Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324 1716 1719
42. KellumR
AlbertsBM
1995 Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci 108 1419 1431
43. BernardP
MaureJF
PartridgeJF
GenierS
JaverzatJP
2001 Requirement of heterochromatin for cohesion at centromeres. Science 294 2539 2542
44. NonakaN
KitajimaT
YokobayashiS
XiaoG
YamamotoM
2002 Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4 89 93
45. PetersAH
O'CarrollD
ScherthanH
MechtlerK
SauerS
2001 Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107 323 337
46. ZhangK
MoschK
FischleW
GrewalSI
2008 Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15 381 388
47. BayneEH
WhiteSA
KaganskyA
BijosDA
Sanchez-PulidoL
2010 Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140 666 677
48. FreitagM
LeeDW
KotheGO
PrattRJ
AramayoR
2004 DNA methylation is independent of RNA interference in Neurospora. Science 304 1939
49. FritschL
RobinP
MathieuJR
SouidiM
HinauxH
2010 A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37 46 56
50. DavisRH
2000 Neurospora: Contributions of a Model Organism: Oxford Oxford University Press 333
51. MargolinBS
FreitagM
SelkerEU
1997 Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genetics Newsletter 44 34 36
52. PomraningKR
SmithKM
FreitagM
2009 Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47 142 150
53. RountreeMR
SelkerEU
1997 DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev 11 2383 2395
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2010 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genome-Wide Association Meta-Analysis of Cortical Bone Mineral Density Unravels Allelic Heterogeneity at the Locus and Potential Pleiotropic Effects on Bone
- Beyond QTL Cloning
- An Evolutionary Framework for Association Testing in Resequencing Studies
- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- The Functional Interplay between Protein Kinase CK2 and CCA1 Transcriptional Activity Is Essential for Clock Temperature Compensation in Arabidopsis
- Endogenous Viral Elements in Animal Genomes
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC
- Sarcomere Formation Occurs by the Assembly of Multiple Latent Protein Complexes
- Genetic Basis of Growth Adaptation of after Deletion of , a Major Metabolic Gene
- Nomadic Enhancers: Tissue-Specific -Regulatory Elements of Have Divergent Genomic Positions among Species
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- CTCF-Dependent Chromatin Bias Constitutes Transient Epigenetic Memory of the Mother at the Imprinting Control Region in Prospermatogonia
- Systematic Dissection and Trajectory-Scanning Mutagenesis of the Molecular Interface That Ensures Specificity of Two-Component Signaling Pathways
- Nucleolin Is Required for DNA Methylation State and the Expression of rRNA Gene Variants in
- The Complex Genetic Architecture of the Metabolome
- ATM Limits Incorrect End Utilization during Non-Homologous End Joining of Multiple Chromosome Breaks
- Mutation Disrupts Synaptonemal Complex Formation, Recombination, and Chromosome Segregation in Mammalian Meiosis
- Mismatch Repair–Independent Increase in Spontaneous Mutagenesis in Yeast Lacking Non-Essential Subunits of DNA Polymerase ε
- The Kinesin-3 Motor UNC-104/KIF1A Is Degraded upon Loss of Specific Binding to Cargo
- Epigenetic Silencing of Spermatocyte-Specific and Neuronal Genes by SUMO Modification of the Transcription Factor Sp3
- A Coastal Cline in Sodium Accumulation in Is Driven by Natural Variation of the Sodium Transporter AtHKT1;1
- Cyclin B3 Is Required for Multiple Mitotic Processes Including Alleviation of a Spindle Checkpoint–Dependent Block in Anaphase Chromosome Segregation
- Altered DNA Methylation in Leukocytes with Trisomy 21
- Human-Specific Evolution and Adaptation Led to Major Qualitative Differences in the Variable Receptors of Human and Chimpanzee Natural Killer Cells
- Leptotene/Zygotene Chromosome Movement Via the SUN/KASH Protein Bridge in
- RACK-1 Acts with Rac GTPase Signaling and UNC-115/abLIM in Axon Pathfinding and Cell Migration
- Genome-Wide Effects of Long-Term Divergent Selection
- Endless Forms Most Viral
- Conflict between Noise and Plasticity in Yeast
- Essential Functions of the Histone Demethylase Lid
- The Transcriptional Regulator Rok Binds A+T-Rich DNA and Is Involved in Repression of a Mobile Genetic Element in
- The Cellular Robustness by Genetic Redundancy in Budding Yeast
- Localization of a Guanylyl Cyclase to Chemosensory Cilia Requires the Novel Ciliary MYND Domain Protein DAF-25
- A Buoyancy-Based Screen of Drosophila Larvae for Fat-Storage Mutants Reveals a Role for in Coupling Fat Storage to Nutrient Availability
- A Functional Genomics Approach Identifies Candidate Effectors from the Aphid Species (Green Peach Aphid)
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- Genome-Wide Effects of Long-Term Divergent Selection
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



