-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Kinase-Independent Role for the Rad3-Rad26 Complex in Recruitment of Tel1 to Telomeres in Fission Yeast
ATM and ATR are two redundant checkpoint kinases essential for the stable maintenance of telomeres in eukaryotes. Previous studies have established that MRN (Mre11-Rad50-Nbs1) and ATRIP (ATR Interacting Protein) interact with ATM and ATR, respectively, and recruit their partner kinases to sites of DNA damage. Here, we investigated how Tel1ATM and Rad3ATR recruitment to telomeres is regulated in fission yeast. Quantitative chromatin immunoprecipitation (ChIP) assays unexpectedly revealed that the MRN complex could also contribute to the recruitment of Tel1ATM to telomeres independently of the previously established Nbs1 C-terminal Tel1ATM interaction domain. Recruitment of Tel1ATM to telomeres in nbs1-c60Δ cells, which lack the C-terminal 60 amino acid Tel1ATM interaction domain of Nbs1, was dependent on Rad3ATR-Rad26ATRIP, but the kinase domain of Rad3ATR was dispensable. Thus, our results establish that the Rad3ATR-Rad26ATRIP complex contributes to the recruitment of Tel1ATM independently of Rad3ATR kinase activity, by a mechanism redundant with the Tel1ATM interaction domain of Nbs1. Furthermore, we found that the N-terminus of Nbs1 contributes to the recruitment of Rad3ATR-Rad26ATRIP to telomeres. In response to replication stress, mammalian ATR–ATRIP also contributes to ATM activation by a mechanism that is dependent on the MRN complex but independent of the C-terminal ATM interaction domain of Nbs1. Since telomere protection and DNA damage response mechanisms are very well conserved between fission yeast and mammalian cells, mammalian ATR–ATRIP may also contribute to the recruitment of ATM to telomeres and to sites of DNA damage independently of ATR kinase activity.
Published in the journal: A Kinase-Independent Role for the Rad3-Rad26 Complex in Recruitment of Tel1 to Telomeres in Fission Yeast. PLoS Genet 6(2): e32767. doi:10.1371/journal.pgen.1000839
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000839Summary
ATM and ATR are two redundant checkpoint kinases essential for the stable maintenance of telomeres in eukaryotes. Previous studies have established that MRN (Mre11-Rad50-Nbs1) and ATRIP (ATR Interacting Protein) interact with ATM and ATR, respectively, and recruit their partner kinases to sites of DNA damage. Here, we investigated how Tel1ATM and Rad3ATR recruitment to telomeres is regulated in fission yeast. Quantitative chromatin immunoprecipitation (ChIP) assays unexpectedly revealed that the MRN complex could also contribute to the recruitment of Tel1ATM to telomeres independently of the previously established Nbs1 C-terminal Tel1ATM interaction domain. Recruitment of Tel1ATM to telomeres in nbs1-c60Δ cells, which lack the C-terminal 60 amino acid Tel1ATM interaction domain of Nbs1, was dependent on Rad3ATR-Rad26ATRIP, but the kinase domain of Rad3ATR was dispensable. Thus, our results establish that the Rad3ATR-Rad26ATRIP complex contributes to the recruitment of Tel1ATM independently of Rad3ATR kinase activity, by a mechanism redundant with the Tel1ATM interaction domain of Nbs1. Furthermore, we found that the N-terminus of Nbs1 contributes to the recruitment of Rad3ATR-Rad26ATRIP to telomeres. In response to replication stress, mammalian ATR–ATRIP also contributes to ATM activation by a mechanism that is dependent on the MRN complex but independent of the C-terminal ATM interaction domain of Nbs1. Since telomere protection and DNA damage response mechanisms are very well conserved between fission yeast and mammalian cells, mammalian ATR–ATRIP may also contribute to the recruitment of ATM to telomeres and to sites of DNA damage independently of ATR kinase activity.
Introduction
ATM (Ataxia Telangiectasia Mutated) and ATR (ATM and Rad3-related), members of the phosphoinositol-3-kinase like kinase (PIKK) family, are central players in coordinating cellular responses to various forms of DNA damage, such as DNA double-stranded breaks (DSBs) and problems that are encountered by DNA replication forks, in eukaryotic cells [1],[2]. ATM and ATR both preferentially recognize and phosphorylate Serine (S) or Threonine (T) amino acid residues followed by Glutamate (Q), and over 900 sites in more than 700 proteins have been identified as potential phosphorylation sites for these two kinases in mammalian cells [3].
Previous studies have identified the Mre11-Rad50-Nbs1 (MRN) DNA repair complex as a key player in the activation of ATM kinase in response to DSBs [1],[4]. The MRN complex interacts with ATM through an evolutionarily conserved C-terminal motif in its Nbs1 subunit, and this interaction is critical for recruitment of ATM to DSBs and phosphorylation of downstream targets by ATM [5],[6]. Likewise, ATRIP (ATR-Interacting Protein) interacts with ATR through its evolutionarily conserved extreme C-terminal motif and promotes recruitment of ATR to sites of DNA damage [6]. RPA (Replication Protein A)-coated single-stranded DNA (ssDNA) serves as a platform for recruitment of the ATR-ATRIP complex, where phosphorylation of various downstream targets can take place [7]. Besides its role in activation of ATM, the MRN complex has also been shown to contribute to ATR signaling in mammalian cells [8].
Initial studies have suggested that ATM is particularly important for recognition of DSBs, while ATR is more important for recognition of replication stress and ultraviolet radiation (UV)-induced DNA damage [1],[2]. However, recent studies have uncovered an intimate crosstalk between the ATM and ATR signaling pathways in response to DNA damage. For example, ATM-MRN can act upstream of ATR-ATRIP in cellular responses to DSBs by promoting recruitment of ATR-ATRIP through its contribution to generate RPA-coated ssDNA at DSBs [9]–[11]. Conversely, ATR contributes to the activation of ATM in response to DNA replication stress by converting inactive ATM dimers into active monomers through direct phosphorylation of ATM [12]. However, it is currently unknown if ATR-ATRIP may also contribute to the recruitment of ATM to stalled replication forks. In contrast to DSB-induced ATM activation, the C-terminal ATM interaction domain of Nbs1 is not required for ATR-dependent activation of ATM in response to DNA replication stress; however, the Nbs1 N-terminus is essential for ATR-dependent activation of ATM [12].
ATM-MRN and ATR-ATRIP are also redundantly required for the maintenance of telomeres, stable DSBs at ends of chromosomes, in a wide variety of eukaryotic species [13] (Figure 1A). Studies in yeasts and mammalian cells have shown that ATM-MRN and ATR-ATRIP are recruited to functional telomeres during S/G2 phases of the cell cycle [14]–[16]. In budding yeast, Tel1ATM and Mec1ATR have been shown to phosphorylate several Serine residues within the telomerase-recruitment domain of the telomere capping protein Cdc13, and these phosphorylation events have been proposed to promote efficient telomerase recruitment to telomeres [14],[17]. In fission yeast, we have recently shown that Tel1ATM and Rad3ATR redundantly promote interaction between the Pot1 telomere capping complex (consisting of Pot1, Tpz1, Poz1 and Ccq1 subunits) and telomerase, and thereby help to recruit telomerase to telomeres [18]. On the other hand, the telomeric GT-rich repeat DNA binding factors TRF2 and POT1, essential for protection of telomeres against degradation and recombination, play critical roles in attenuating DNA damage checkpoint activation mediated by ATM and ATR in mammalian cells [19]–[21]. Therefore, it has been suggested that telomeres transiently become de-protected during S - and G2-phases, and can thus be recognized as DSBs by ATM/ATR to allow the timely recruitment of telomerase [22].
Fig. 1. Kinase-independent function of Rad3ATR-Rad26ATRIP contributes to telomere maintenance in the absence of the Nbs1-Tel1ATM interaction domain of Nbs1. 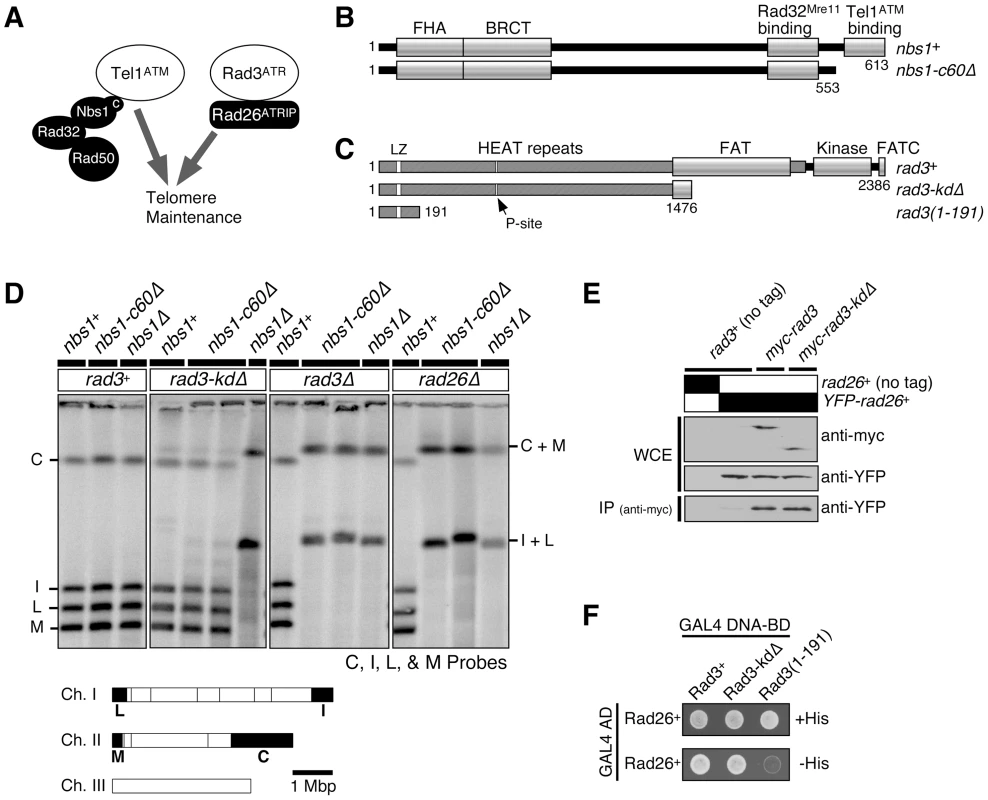
(A) Tel1ATM-MRN (Rad32Mre11-Rad50-Nbs1) and Rad3ATR-Rad26ATRIP represent two redundant pathways essential for telomere maintenance in fission yeast. (B) Schematic representation of Nbs1 protein expressed in nbs1+ and nbs1-c60Δ. Conserved motifs and functional domains [5],[27] are indicated. (C) Schematic representation of various Rad3 constructs tested. Amino acids 1477–2386 are deleted in rad3-kdΔ, while amino acids 1–2363 are deleted in rad3Δ. Conserved motifs and functional domains are indicated [26],[58]. The Leucine zipper (LZ) and putative PCNA-interaction site (P-site) were previously implicated in protein-protein interaction [59]. (D) Characterization of telomere status by pulsed-field gel electrophoresis (PFGE) for indicated strains. Cells with defects in telomere maintenance show C+M and I+L bands, corresponding to the fused telomeric fragments. Bottom, NotI restriction map of fission yeast chromosomes, shown with telomeric C, I, L, and M fragments marked as black boxes. (E) Rad3-kdΔ forms a stable complex with Rad26 in vivo. Anti-myc immunoprecipitation was performed using extracts from strains expressing YFP-Rad26 and either wild-type myc-Rad3 or myc-Rad3-kdΔ, and co-immunoprecipitated Rad26 was visualized by anti-GFP western blot. (F) Two-hybrid assays of S. pombe Rad26 with full-length Rad3 (Rad3+), C-terminal truncation of Rad3 (Rad3-kdΔ), or the N-terminal 191 amino acids of Rad3. A positive two-hybrid interaction is identified by growth on -His plates. On the other hand, we have recently found that the arrival of lagging strand DNA polymerases (α and δ) to telomeres is significantly delayed compared to the arrival of the leading strand DNA polymerase ε in fission yeast, and that significant quantities of RPA and Rad26ATRIP transiently accumulate at replicating telomeres [15]. These observations thus raised the possibility that replicating telomeres may be primarily recognized as unusual/stressed replication forks by Rad3ATR-Rad26ATRIP rather than being recognized as DSBs in fission yeast [15]. In mammalian cells, recruitment of ATR and MRN to telomeres precedes recruitment of ATM to telomeres [16], and replication of lagging-strand telomeres also appears to be significantly delayed compared to leading-strand telomeres [23]. Thus, replicating mammalian telomeres might also accumulate unusually high levels of RPA on the lagging strand and activate ATR, and ATR might subsequently contribute to the activation of ATM signaling at telomeres to promote stable telomere maintenance.
Here, we investigated inter-dependencies among Tel1ATM, Nbs1, and Rad3ATR-Rad26ATRIP for their recruitment to telomeres in fission yeast. Our studies uncovered a surprising kinase-independent role of the Rad3ATR-Rad26ATRIP complex, which is redundant with the C-terminal Tel1ATM-interacting domain of Nbs1, for the recruitment of Tel1ATM to telomeres. We also demonstrate that the N-terminal domain of Nbs1 contributes to Rad3ATR-Rad26ATRIP function by promoting recruitment of Rad3ATR-Rad26ATRIP to telomeres. Thus, our findings provide mechanistic insights into how recruitment of Tel1ATM and Rad3ATR to telomeres is regulated, and define a novel molecular crosstalk between Rad3ATR and Tel1ATM in recognition of fission yeast telomeres, potentially resembling the ATR-ATM crosstalk in response to DNA replication stress in mammalian cells.
Results
Tel1ATM interaction domain of Nbs1 is dispensable for telomere maintenance in rad3-kdΔ cells
The C-terminus of Nbs1 is important for its interaction with Tel1ATM in fission yeast Schizosaccharomyces pombe [5] (Figure 1B). This interaction is evolutionarily conserved, since the corresponding region of Nbs1 is also required for its interaction with ATM in Xenopus and humans [5],[6]. Mutating or truncating the Nbs1 C-terminus in fission yeast cells lacking functional Rad3ATR eliminated DNA damage-induced histone H2A phosphorylation (γ-H2A) and foci formation of the checkpoint mediator protein Crb2/Rhp9 [5], since Tel1ATM and Rad3ATR are redundantly required for DNA damage-induced histone H2A phosphorylation and Crb2 foci formation [24]. Tel1ATM-MRN (Rad32Mre11-Rad50-Nbs1) and Rad3ATR-Rad26ATRIP pathways are also redundantly required for stable telomere maintenance, and simultaneous inactivation of these two pathways results in chromosome circularization in fission yeast [25] (Figure 1A). Thus, it was previously hypothesized that disruption of Tel1ATM-Nbs1 interaction in fission yeast cells lacking functional Rad3ATR would lead to telomere dysfunction and chromosome circularization [5].
We tested this hypothesis by combining the most commonly used rad3 deletion allele (rad3Δ::ura4+, hereafter referred to as rad3-kdΔ) with the Nbs1 C-terminal 60 amino acid deletion allele nbs1-c60Δ that disrupts Tel1ATM-Nbs1 interaction [5],[26] (Figure 1B and 1C). After extensive restreaking of fission yeast cells on agar plates, we examined the status of telomeres by separating NotI-digested chromosomal DNA by pulsed-field gel electrophoresis (PFGE), and then performing Southern blot analysis with telomeric NotI fragment-specific probes C, I, L and M (Figure 1D). In this assay, cells carrying circular chromosomes lose individual telomeric NotI fragments and show I+L and C+M bands, corresponding to the fused telomeric fragments from chromosomes I and II.
We were surprised to find that the double mutant nbs1-c60Δ rad3-kdΔ cells stably maintained telomeres (Figure 1D). By contrast, combining tel1Δ or nbs1Δ with rad3-kdΔ caused complete loss of telomeres and chromosome circularization, as previously shown [25],[27] (Figure 1D). Thus, while Nbs1 and Tel1ATM are both essential for telomere maintenance in rad3-kdΔ cells, the Nbs1-Tel1ATM interaction is dispensable for telomere maintenance in rad3-kdΔ cells. The mutant Nbs1-c60Δ protein retains its ability to interact with Rad32Mre11, and both rad32Δ rad3-kdΔ and rad50Δ rad3-kdΔ cells are unable to maintain telomeres [5],[27]. Thus, the N-terminus of Nbs1 is required for the MRN (Rad32Mre11-Rad50-Nbs1) complex to fulfill its essential telomere maintenance function(s) in rad3-kdΔ cells.
The N-terminal non-kinase domain of Rad3ATR and its regulatory subunit Rad26ATRIP are both required for telomere maintenance in the absence of Nbs1-Tel1ATM interaction domain
The rad3-kdΔ allele does not completely delete the rad3+ coding region and only removes a C-terminal portion of the Rad3ATR protein including its kinase domain [26] (Figure 1C). We found that the truncated Rad3ATR protein (Rad3-kdΔATR) expressed stably, and retained its ability to interact with its regulatory subunit Rad26ATRIP in co-immunoprecipitation (co-IP) experiments and yeast 2-hybrid assays (Figure 1E and 1F). Similarly, the N-terminal regions of mammalian ATR and budding yeast Mec1, outside of their C-terminal kinase domains, interact with their respective regulatory subunits ATRIP and Ddc2 [28],[29]. Furthermore, when the fission yeast rad3+ gene was originally cloned, a truncated Rad3ATR protein lacking its kinase domain was found to fully suppress UV, IR and HU hypersensitivities and a G2-checkpoint defect of rad3-136 cells [30]. In addition, over-expression of the N-terminal fragments of Xenopus ATR or budding yeast Mec1ATR lacking the kinase domain has been shown to partially suppress the DNA damage sensitivity of mec1-1 budding yeast cells by activating the spindle checkpoint [31]. Therefore, we next examined if a complete deletion allele of rad3+ (rad3Δ::LEU2, hereafter referred to as rad3Δ, deletes genomic DNA corresponding to amino acids 1-2363 of Rad3ATR [25]) would show a different genetic interaction with nbs1-c60Δ with respect to DNA damage sensitivity and telomere maintenance, even though we saw no difference between rad3-kdΔ and rad3Δ cells for their average telomere lengths in nbs1+ background (Figure S1A).
While we detected no differences in sensitivity towards various DNA damaging agents between rad3-kdΔ and rad3Δ alleles in nbs1+, nbs1-c60Δ or nbs1Δ backgrounds (Figure S1), we found that rad3Δ nbs1-c60Δ cells cannot maintain stable telomeres, unlike rad3-kdΔ nbs1-c60Δ cells (Figure 1D). Thus, the N-terminal domain of Rad3ATR contributes to Tel1ATM-dependent telomere maintenance in nbs1-c60Δ cells. Additionally, we found that rad26Δ nbs1-c60Δ cells cannot maintain telomeres (Figure 1D), establishing that Rad26ATRIP is essential for telomere maintenance in nbs1-c60Δ cells. Thus, these results indicated that the Rad3-kdΔATR-Rad26ATRIP complex has a kinase-independent function for telomere maintenance in cells lacking the C-terminal Tel1ATM interaction domain of Nbs1. The apparent discrepancy in absolute requirement for the Rad3ATR kinase domain for DNA damage sensitivity versus telomere maintenance is plausible, since previous studies have established that the downstream checkpoint kinases Chk1 and Cds1CHK2 are not involved in telomere maintenance in fission yeast, while these kinases play critical roles in cell survival in response to DNA damage [25],[32].
The N-terminal domain of Rad3ATR is required for recruitment of Rad26ATRIP to telomeres
To gain insights into how the Rad3-kdΔATR-Rad26ATRIP complex contributes to telomere maintenance in nbs1-c60Δ cells, we next examined the association of the Rad3ATR-Rad26ATRIP complex with fission yeast telomeres by quantitative chromatin immunoprecipitation (ChIP) assays. While a significant amount of telomeric DNA was found to associate with Rad26ATRIP in rad3+ and rad3-kdΔ cells, the amount of telomeric DNA precipitated by Rad26ATRIP in rad3Δ cells was reduced to background levels (Figure 2A). Conversely, both wild-type Rad3ATR and Rad3-kdΔATR proteins associate with telomeric DNA in a Rad26-dependent manner, although the precipitation efficiency for Rad3-kdΔATR was reduced compared to wild-type Rad3ATR (Figure 2B). Taken together, these results are consistent with the notion that Rad26ATRIP is essential for recruitment of Rad3ATR to telomeres and that the N-terminal domain of Rad3ATR contributes to the recruitment of Rad26ATRIP to telomeres.
Fig. 2. The C-terminal Tel1ATM interaction domain of Nbs1 and the non-kinase domain of Rad3ATR redundantly contribute to recruitment of Tel1ATM to telomeres. 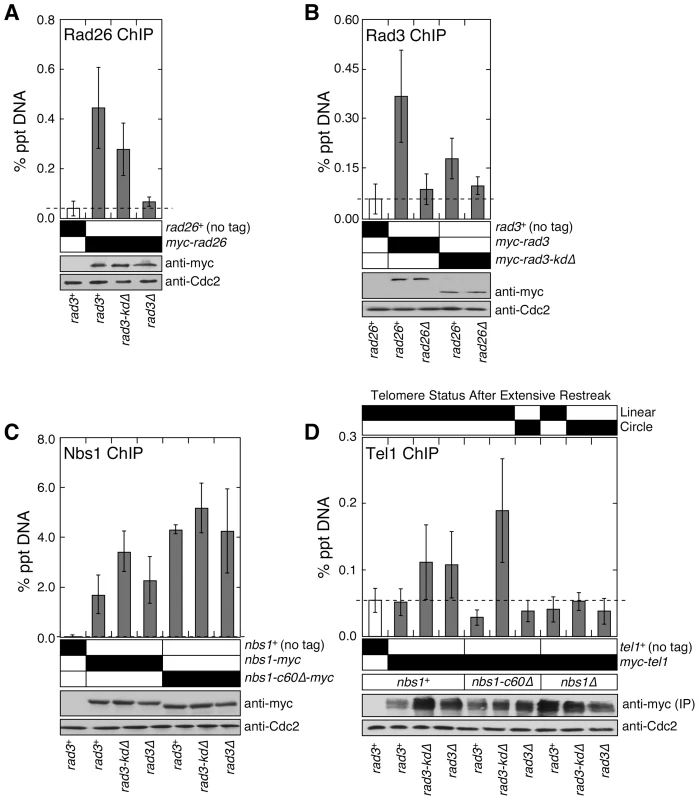
(A) Recruitment of Rad26ATRIP to telomeres in rad3+, rad3-kdΔ, and rad3Δ, monitored by quantitative ChIP assays. Mean values plus or minus one standard deviation for two to nine independent experiments are plotted. Compared to untagged control, Rad26ATRIP showed significant telomere binding in rad3+ and rad3-kdΔ (P values≤0.00003), but not in rad3Δ (P = 0.282). Based on anti-myc western blot, comparable level of Rad26 was expressed. Anti-Cdc2 western blots were used as loading controls. (B) Recruitment of wild-type Rad3ATR or Rad3-kdΔ to telomeres in rad26+ or rad26Δ, monitored by quantitative ChIP assays. Mean values plus or minus one standard deviation for three to eleven independent experiments are plotted. Compared to untagged control, both wild-type Rad3 and Rad3-kdΔ showed significant telomere binding in rad26+ (P values≤0.0003) but not in rad26Δ (P values≥0.154). Based on anti-myc western blot, expression levels of wild-type or truncated Rad3ATR are not affected by deletion of rad26. Anti-Cdc2 western blots were used as loading controls. (C) Recruitment of wild-type Nbs1 or Nbs1-c60Δ to telomeres in rad3+, rad3-kdΔ, or rad3Δ, monitored by quantitative ChIP assays. Mean values plus or minus one standard deviation for three to seven independent experiments are plotted. Compared to untagged control, both wild-type Nbs1 and Nbs1-c60Δ showed significant telomere binding in all genetic backgrounds tested (P values≤0.0001). Telomere binding of wild-type Nbs1 was significantly increased in rad3-kdΔ (P = 0.004) but not rad3Δ cells (P = 0.295), compared to rad3+ cells. For Nbs1-c60Δ, there was no significant change in telomere association in rad3-kdΔ (P = 0.214) or rad3Δ cells (P = 0.952), compared to rad3+ cells. Based on anti-myc western blot, expression levels of wild-type or truncated Nbs1 are not affected by mutations in Rad3. Anti-Cdc2 western blots were used as loading controls. (D) Recruitment of Tel1ATM to telomeres in various mutant combinations among nbs1 and rad3, monitored by quantitative ChIP assays. Mean values plus or minus one standard deviation for two to four independent experiments are plotted. Only rad3-kdΔ, rad3Δ and rad3-kdΔ nbs1-c60Δ showed higher mean % precipitated DNA (% ppt DNA) values compared to untagged control. Tel1 binding to telomeres in rad3-kdΔ and rad3Δ cells was deemed statistically insignificant (P values 0.155 and 0.139, respectively) compared to untagged control, due to large standard deviation in % ppt DNA values among four independent ChIP experiments. However, we consistently found that % ppt DNA values for rad3-kdΔ and rad3Δ cells are higher than untagged control or rad3+ cells for a given set of ChIP experiment performed the same day. Tel1 binding to telomeres in rad3-kdΔ nbs1-c60Δ cells, compared to untagged control, was statistically significant (P = 0.016). Since myc-Tel1 expressed from its endogenous promoter could not be detected in whole cell extracts by anti-myc western blot, anti-myc immunoprecipitation (IP) was performed to enrich for myc-Tel1. Based on anti-myc western blot, comparable amounts of Tel1 were pulled down in different mutant backgrounds. Anti-Cdc2 western blots were used as loading controls. We also examined the recruitment of wild-type Nbs1 and Nbs1-c60Δ to telomeres in rad3+, rad3-kdΔ, and rad3Δ background by ChIP assays (Figure 2C). Since we were able to still detect a robust association of Nbs1-c60Δ with telomeres even in rad3Δ cells, we could exclude the possibility that the Rad3-kdΔATR-Rad26ATRIP complex contributes to telomere maintenance by promoting recruitment of Nbs1-c60Δ to telomeres.
The Rad3-kdΔATR-Rad26ATRIP complex and the N-terminal domain of Nbs1 are both required for recruitment of Tel1ATM to telomeres in the absence of Nbs1-Tel1ATM interaction
Since expression of Rad3-kdΔATR allowed cells to maintain stable telomeres in the absence of Nbs1-Tel1ATM interaction but not in tel1Δ and nbs1Δ cells (Figure 1D), we hypothesized that Nbs1-c60Δ protein and the Rad3-kdΔATR-Rad26ATRIP complex may function collaboratively to recruit Tel1ATM to telomeres and maintain telomeres. Indeed, we detected robust Tel1ATM recruitment to telomeres in nbs1-c60Δ rad3-kdΔ cells, but failed to detect Tel1ATM at telomeres in nbs1-c60Δ rad3Δ, nbs1Δ rad3-kdΔ, and nbs1Δ rad3Δ cells (Figure 2D). The ability to recruit Tel1ATM to telomeres in rad3 mutant backgrounds (either rad3-kdΔ or rad3Δ) correlated perfectly with the telomere maintenance phenotypes of these mutant cells (Figure 1D and Figure 2D). Thus, our ChIP data establish that the N-terminal domain of Nbs1 and the telomere-bound Rad3-kdΔATR-Rad26ATRIP complex must both be present, in order for Tel1ATM to be recruited independently of the C-terminal Tel1ATM-interaction domain of Nbs1.
It should be noted that for those mutant strains that are defective in telomere maintenance and ultimately circularize their chromosomes, ChIP assays were performed with early generation cells prior to chromosome circularization by utilizing our recently described Rad3 plasmid loss system [18]. This system allows us to harvest cells immediately after the loss of Rad3ATR, when the average telomere length is comparable or slightly longer than in rad3Δ or rad3-kdΔ cells. Furthermore, by monitoring real-time PCR amplification cycle numbers for all ChIP input samples, we have ensured that mutant cells utilized in our experiments have not yet circularized their chromosomes, since the PCR primer-annealing sites would be completely lost upon chromosome circularization, resulting in a significant delay in PCR amplification.
In contrast to nbs1-c60Δ cells, nbs1+ cells were able to recruit Tel1ATM to telomeres in both rad3-kdΔ and rad3Δ backgrounds (Figure 2D). Thus, our ChIP data also support the notion that the C-terminal Tel1ATM interaction domain of Nbs1 and the Rad3-kdΔATR-Rad26ATRIP complex represent two pathways that are redundantly required for recruitment of Tel1ATM to telomeres. Since we detect Tel1ATM at telomeres only in cells carrying very short telomere-repeats (∼180 bp in rad3-kdΔ and rad3Δ cells), but not in cells with wild-type telomere-repeat length (∼280 bp in rad3+ cells) (Figure 2D, Figure S1A), fission yeast Tel1ATM may be preferentially recruited to short telomeres. Alternatively, active Rad3ATR kinase might prevent association of Tel1ATM with telomeres. In any case, our results establish that the C-terminal 60 amino acids of fission yeast Nbs1 are not essential for the recruitment of Tel1ATM to critically short telomeres, due to a redundant non-kinase contribution of Rad3ATR-Rad26ATRIP to Tel1ATM recruitment.
The N-terminal domain of Nbs1 contributes to the recruitment of Rad3ATR-Rad26ATRIP to telomeres
In order to better understand how the N-terminal domain of Nbs1 contributes to the Rad3-kdΔATR-dependent maintenance of telomeres in nbs1-c60Δ cells, we next examined the recruitment of Rad3ATR or Rad3-kdΔATR to telomeres in nbs1+, nbs1-c60Δ or nbs1Δ backgrounds by ChIP assays. As shown in Figure 3A, we found that nbs1Δ cells recruit significantly less wild-type Rad3 or Rad3-kdΔATR proteins to telomeres compared to nbs1+ or nbs1-c60Δ cells, suggesting that Nbs1-c60Δ protein contributes to the efficient recruitment of the Rad3ATR-Rad26ATRIP complex to telomeres. Accordingly, the loss of the Rad3-kdΔATR-Rad26ATRIP complex from telomeres in nbs1Δ cells might explain why rad3-kdΔ nbs1Δ cells fail to recruit Tel1ATM to telomeres and circularize chromosomes (Figure 1D, Figure 2D, and Figure 3A). It should be noted that a substantial amount of wild-type Rad3ATR could still be recruited to telomeres in nbs1Δ cells. In fact, since nbs1Δ, rad50Δ, rad32Δmre11 and tel1Δ cells all maintain essentially wild-type length or only slightly shorter telomeres in fission yeast [27],[33], it appears that the MRN complex and Tel1ATM do not make significant contributions to telomere maintenance as long as cells express the wild-type Rad3ATR-Rad26ATRIP complex.
Fig. 3. Nbs1 N-terminal domain is required to promote recruitment of the Rad3ATR-Rad26ATRIP complex to telomeres. 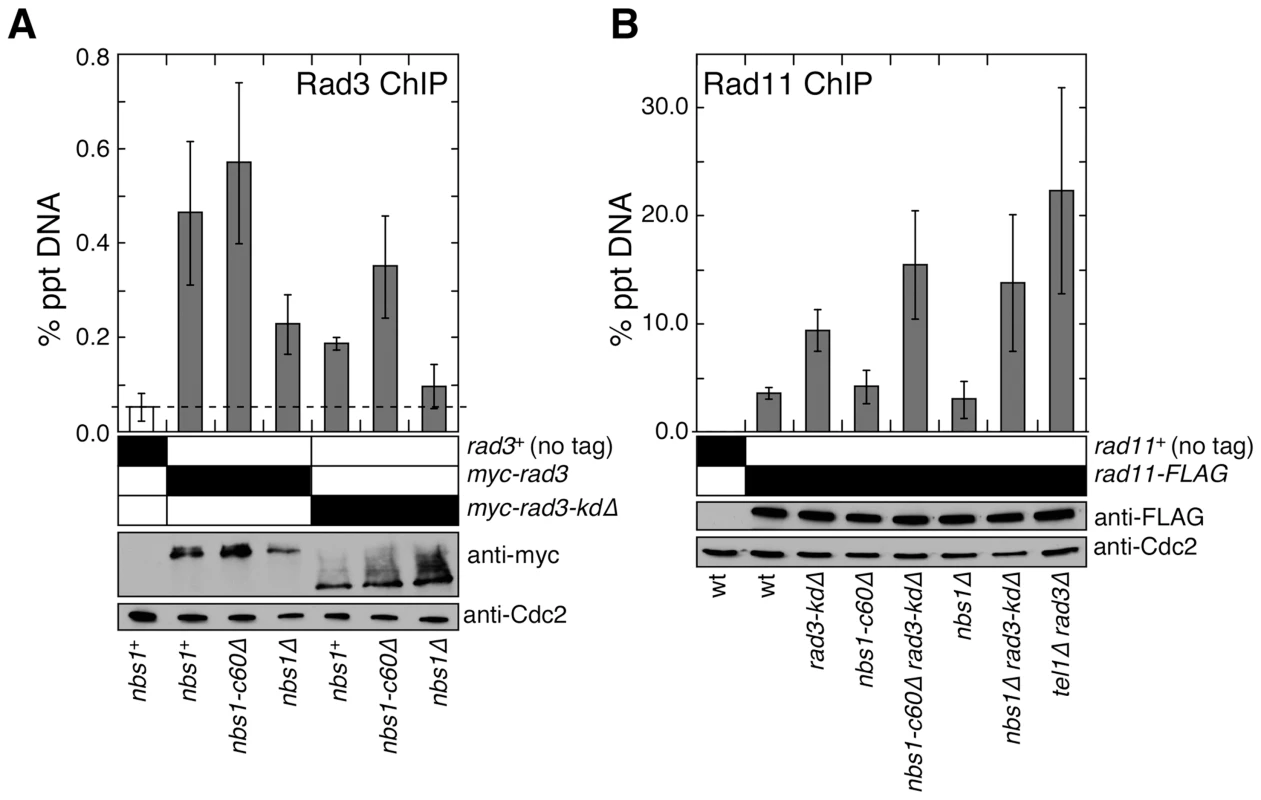
(A) Recruitment of wild-type Rad3ATR or Rad3-kdΔ to telomeres in nbs1+, nbs1-c60Δ or nbs1Δ was monitored by quantitative ChIP assays. Averages and standard deviations from three to five independent ChIP experiments are plotted. Compared to untagged control, wild-type Rad3 was detected at telomeres in all genetic backgrounds tested (P values≤0.0003). Compared to nbs1+ cells, wild-type Rad3 binding to telomeres was significantly reduced in nbs1Δ (P = 0.011) but not in nbs1-c60Δ (P = 0.316). Compared to untagged control, Rad3-kdΔ showed telomere recruitment in nbs1+ and nbs1-c60Δ (P values≤0.0002) but not in nbs1Δ (P = 0.118). Based on anti-myc western blot, expression levels of wild-type or truncated Rad3ATR are not affected by mutations in nbs1. Anti-Cdc2 western blots were used as loading controls. (B) Recruitment of RPA largest subunit Rad11 to telomeres, monitored by quantitative ChIP assays, did not change greatly among strains mutated for nbs1 and rad3. Averages and standard deviations from three to six independent experiments are plotted. Compared to untagged control, Rad11 binding to telomeres was significant in all genetic backgrounds tested (P values≤0.01). Strains carrying rad3-kdΔ showed significantly higher telomere recruitment of Rad11 compared to rad3+ cells for all nbs1 alleles tested (P values≤0.01). No significant change in recruitment of Rad11 to telomeres among rad3-kdΔ, nbs1-c60Δ rad3-kdΔ, and nbs1Δ rad3-kdΔ cells was detected (P values≥0.06). Based on anti–FLAG western blot, expression levels of Rad11 are not affected by mutations in nbs1 or rad3. Western blots with anti-Cdc2 were used as loading controls. The ATR-ATRIP complex preferentially binds to RPA-coated ssDNA, and the MRN complex promotes formation of 3′ ssDNA at DSBs and telomeres [10], [34]–[36]. Thus, we considered the possibility that the fission yeast MRN complex, independently of its Nbs1 C-terminal Tel1ATM interaction domain, might promote recruitment of Rad3ATR-Rad26ATRIP to telomeres by promoting accumulation of RPA-coated ssDNA at telomeres. However, we found comparable level of RPA (Rad11) associated with telomeric DNA in rad3-kdΔ nbs1-c60Δ and rad3-kdΔ nbs1Δ cells (Figure 3B). We also considered the possibility that the N-terminus of Nbs1 might contribute to the recruitment of Rad3ATR-Rad26ATRIP to telomeres by promoting association between the MRN complex and the Rad3ATR-Rad26ATRIP complex, since it was previously shown that mammalian MRN interacts with ATR-ATRIP [37]. However, we could not detect a direct interaction between Nbs1 and Rad3ATR or between Nbs1 and Rad26ATRIP by yeast 2-hybrid assays, and failed to detect an interaction between Nbs1 and Rad26ATRIP by co-IP experiments (data not shown). Thus, further investigations are necessary to fully understand how the N-terminal domain of Nbs1 contributes to efficient recruitment of the Rad3ATR-Rad26ATRIP complex to telomeres.
Over-expression of Tel1ATM can bypass the requirement for the Nbs1 C-terminal domain and Rad3ATR-Rad26ATRIP complex in telomere maintenance
Over-expression of Tel1ATM results in MRX (Mre11-Rad50-Xrs2) complex-dependent elongation of telomeres in budding yeast [14],[38],[39], consistent with the notion that recruitment of Tel1ATM to telomeres represents the critical rate limiting step in regulating telomere length in budding yeast. By contrast, over-expression of Tel1ATM was unable to increase telomere length in wild-type fission yeast cells (Figure 4A), even though over-expressed Tel1ATM can be detected at telomeres by ChIP assays (Figure 4D). However, we did observe a partial suppression of telomere shortening in rad3-kdΔ and rad3Δ cells upon Tel1ATM over-expression (Figure 4A), suggesting that the level of Tel1ATM recruitment to short telomeres becomes a critical limiting factor in telomere length determination in the absence of the functional Rad3ATR-Rad26ATRIP complex. Interestingly, we also found that the Nbs1 C-terminal Tel1ATM interaction domain is essential for over-expressed Tel1ATM to associate with telomeres in rad3+ cells, but dispensable in rad3-kdΔ and rad3Δ cells (Figure 4D). Thus, it appears that the presence of the kinase active Rad3ATR-Rad26ATRIP complex can prevent recruitment of over-expressed Tel1ATM to telomeres if cells are missing the C-terminal Tel1ATM interaction domain of Nbs1.
Fig. 4. Over-expression of Tel1ATM bypasses the Tel1ATM recruitment function redundantly provided by the Nbs1 C-terminus and the Rad3ATR-Rad26ATRIP complex, but does not bypass the complete deletion of Nbs1. 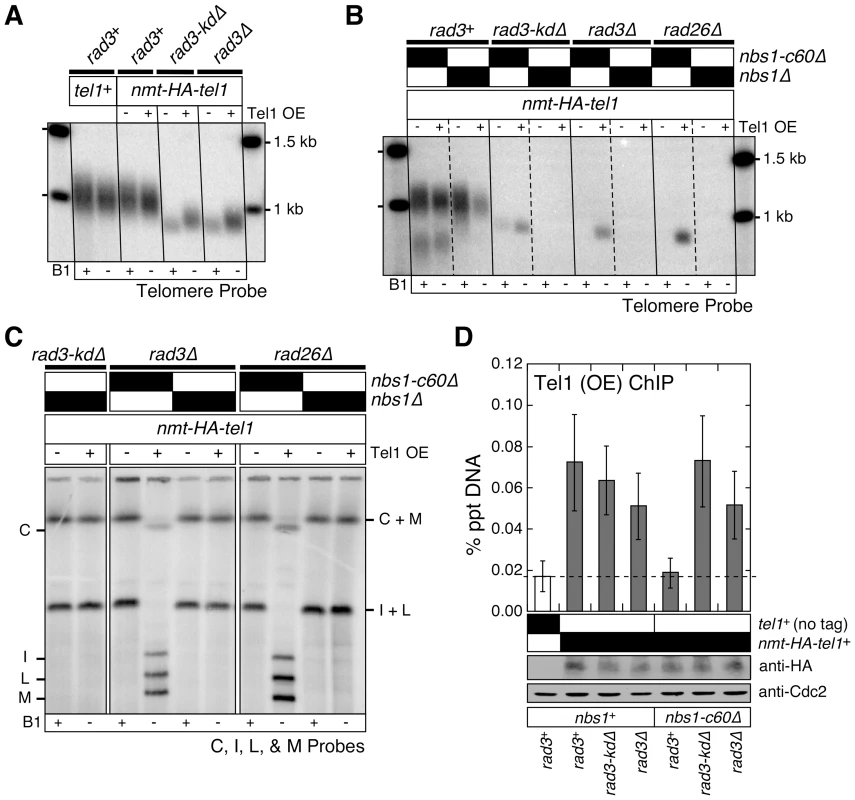
(A, B) Southern blot analysis of EcoRI digested genomic DNA probed with a telomeric repeat specific probe for indicated strains. See Figure S1A for a schematic drawing of the fission yeast telomere restriction map. (C) Characterization of telomere status by PFGE for indicated strains. For all panels, fission yeast cells were grown in minimal media in the presence or absence of thiamine (B1) to repress (−) or induce (+) expression of Tel1 under the control of the nmt1+ promoter respectively, for at least 150 cell divisions. (D) Recruitment of over-expressed Tel1ATM to telomeres in various mutant combinations among nbs1 and rad3, monitored by quantitative ChIP assays. Averages and standard deviations from three to four independent ChIP experiments are plotted. Compared to untagged control, over-expressed Tel1ATM was detected at telomeres in all genetic backgrounds tested (P values ≤0.019), except in rad3+ nbs1-c60Δ (P = 0.81). Tel1ATM was transiently over-expressed by growing cells in minimal media lacking thiamine for 21 hr prior to harvest. Based on anti-HA western blot, expression levels of over-expressed Tel1ATM are comparable among all genetic backgrounds tested. Anti-Cdc2 western blots were used as loading controls. We reasoned that over-expression of Tel1ATM might be able to suppress chromosome circularization caused by simultaneous mutations in Nbs1 and Rad3ATR-Rad26ATRIP, if the essential telomere maintenance function, redundantly provided by the Nbs1 C-terminal Tel1ATM interaction domain and the kinase-inactive Rad3ATR-Rad26ATRIP complex, were to promote efficient recruitment of Tel1ATM to telomeres. Indeed, over-expression of Tel1ATM allowed recruitment of Tel1ATM in nbs1-c60Δ rad3Δ cells (Figure 4D) and suppressed chromosome circularization in nbs1-c60Δ rad3Δ and nbs1-c60Δ rad26Δ cells (Figure 4B and 4C). However, over-expression of Tel1ATM was unable to suppress chromosome circularization in nbs1Δ rad3-kdΔ, nbs1Δ rad3Δ and nbs1Δ rad26Δ cells, indicating that the expression of Nbs1 (and likely other subunits of the MRN complex) is essential for telomere maintenance in the absence of kinase-active Rad3ATR-Rad26ATRIP even when Tel1ATM is over-expressed. Taken together, our results indicate that the Nbs1 C-terminal Tel1ATM interaction domain and the kinase-independent function of the Rad3-kdΔATR-Rad26ATRIP complex redundantly contribute to efficient recruitment of Tel1ATM to telomeres, when Tel1ATM is expressed at endogenous level. Our results further demonstrate that when Tel1ATM is over-expressed, the MRN complex can contribute to telomere maintenance even in the absence of both the C-terminal Tel1ATM interaction domain of Nbs1 and the Rad3ATR-Rad26ATRIP complex.
Over-expression of Rad3ATR can bypass the requirement for Rad26ATRIP and Nbs1, but not Tel1ATM in telomere maintenance
Our ChIP data indicated that both the Rad26ATRIP regulatory subunit and the N-terminal domain of Nbs1 play critical roles in promoting recruitment of Rad3ATR to telomeres (Figure 2B, Figure 3A). Therefore, we next investigated if over-expression of Rad3ATR might be able to bypass the requirement for Rad26ATR and/or Nbs1 in telomere maintenance. Based on ChIP analysis, over-expression of Rad3ATR bypassed the requirement for Rad26ATRIP in recruitment of Rad3ATR to telomeres (Figure 5C). However, over-expression of Rad3ATR was not able to suppress telomere shortening observed in rad26Δ cells (Figure 5A). Thus, in the presence of wild-type Nbs1 (and thus intact Nbs1-Tel1ATM interaction), forced recruitment of Rad3ATR to telomeres by over-expression is not sufficient to elongate telomeres in rad26Δ cells. On the other hand, we found that Rad3ATR over-expression completely suppressed chromosome circularization in rad26Δ nbs1-c60Δ cells (Figure 5B), consistent with the notion that the essential telomere maintenance function contributed by Rad26ATRIP in nbs1-c60Δ is to promote recruitment of Rad3ATR to telomeres.
Fig. 5. Over-expression of Rad3ATR can bypass the Rad3ATR recruitment function provided by Rad26ATRIP and Nbs1. 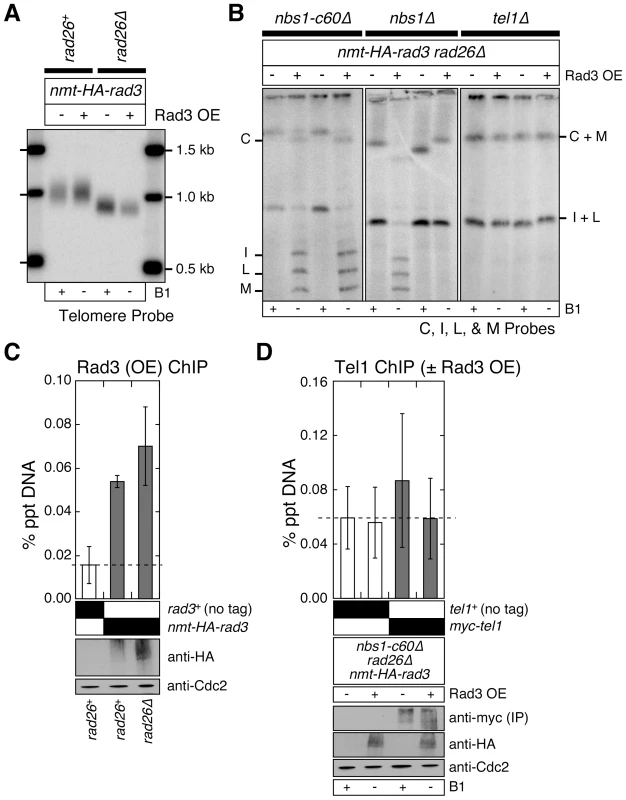
(A) Southern blot analysis of EcoRI digested genomic DNA, hybridized to telomere repeat specific probe. Fission yeast cells were grown in minimal media in the presence or absence of thiamine (B1) to repress (−) or induce (+) expression of Rad3 under the control of the nmt1+ promoter, respectively, for at least 150 cell divisions. See Figure S1A for a schematic drawing of the fission yeast telomere restriction map. (B) Characterization of telomere status by PFGE for indicated strains. Fission yeast cells were grown as in (A). Two independent clones of each genotype are shown. Rad3 over-expression in nbs1Δ rad26Δ cells resulted in mixed telomere phenotypes, suggesting partial suppression of telomere dysfunction. (C) Recruitment of over-expressed Rad3ATR to telomeres in rad26+ or rad26Δ cells, monitored by quantitative ChIP assays. Averages and standard deviations from three independent ChIP experiments are plotted. Compared to untagged control, over-expressed Rad3ATR was detected at telomeres in both rad26+ (P = 0.0016) and rad26Δ (P = 0.0087) cells. Rad3ATR was transiently over-expressed by growing cells in minimal media lacking thiamine for 21 hr prior to harvest. Expression levels of over-expressed Rad3ATR are comparable, based on anti-HA western blot. Anti-Cdc2 western blots were used as loading controls. (D) Recruitment of endogenous Tel1ATM to telomeres in nbs1-c60Δ rad26Δ cells, with or without over-expressed Rad3ATR, was monitored by quantitative ChIP assays. Averages and standard deviations from three independent ChIP experiments are plotted. No significant binding of Tel1ATM to telomeres above untagged control was observed. Rad3ATR was transiently over-expressed as in (C). Anti-HA western blots were used to monitor expression levels of over-expressed Rad3ATR. Since myc-Tel1 expressed from its endogenous promoter could not be detected in whole cell extracts by anti-myc western blot, anti-myc immunoprecipitation (IP) was performed to enrich for myc-Tel1. Anti-Cdc2 western blots were used as loading controls. Even when Rad3ATR is over-expressed, the N-terminus of Nbs1 still appears to contribute to telomere maintenance in rad26Δ cells, since suppression of chromosome circularization by over-expression of Rad3ATR was much more complete in rad26Δ nbs1-c60Δ cells than in rad26Δ nbs1Δ cells (Figure 5B). Since we observed a mixed telomere phenotype among survivor cells after extensive restreaking of nbs1Δ rad26Δ cells over-expressing Rad3ATR on agar plates, we concluded that over-expression of Rad3ATR can partially bypass the essential telomere function of Nbs1 in the absence of Rad26ATRIP (Figure 5B). The observed partial suppression of chromosome circularization in nbs1Δ rad26Δ by over-expression of Rad3ATR is consistent with the notion that the N-terminal domain of Nbs1 contributes to telomere maintenance by promoting recruitment of Rad3ATR to telomeres.
By contrast, Rad3ATR over-expression was unable to suppress chromosome circularization observed in rad26Δ tel1Δ cells (Figure 5B). Thus, it appears that over-expression of Rad3ATR allows the Tel1ATM-dependent mechanism to maintain telomeres in rad26Δ nbs1-c60Δ or rad26Δ nbs1Δ cells. One possible mechanism by which Rad3ATR over-expression might promote Tel1ATM-dependent telomere maintenance in rad26Δ nbs1-c60Δ or rad26Δ nbs1Δ cells is to promote recruitment of Tel1ATM to telomeres. Therefore, we examined changes in Tel1ATM recruitment to telomeres in nbs1-c60Δ rad26Δ cells with or without over-expression of Rad3ATR by ChIP analyses. However, we were unable to detect Rad3ATR over-expression dependent recruitment of Tel1ATM to telomeres (Figure 5D). Thus, further investigations are necessary to fully understand how over-expression of Rad3ATR contributes to the Tel1ATM-dependent suppression of chromosome circularization in fission yeast cells simultaneously lacking functional Rad26ATRIP and Nbs1.
Discussion
In the current study, we investigated how two PIKK-containing complexes, Tel1ATM-MRN and Rad3ATR-Rad26ATRIP, contribute to telomere length regulation in fission yeast (summarized in Figure 6). We have demonstrated that fission yeast Tel1ATM can be recruited to telomeres by two alternative and redundant mechanisms, which are either dependent or independent of the Nbs1 C-terminal Tel1ATM interaction domain (Figure 6A and 6B). Our analyses indicated that the Nbs1 C-terminus dependent mode of Tel1ATM recruitment to telomeres does not require the Rad3ATR-Rad26ATRIP complex, and in fact, it might be inhibited by the presence of wild-type Rad3ATR-Rad26ATRIP (Figure 2D). On the other hand, the Nbs1 C-terminus independent mode of Tel1ATM recruitment requires the presence of telomere bound kinase-inactive Rad3-kdΔATR-Rad26ATRIP complex (Figure 2A, 2B, and 2D). Since the N-terminal domain of Nbs1 is essential for recruitment of the Rad3-kdΔATR-Rad26ATRIP complex to telomeres (Figure 3A), all our results are consistent with the notion that association of Rad3-kdΔATR-Rad26ATRIP to telomeres is the crucial determinant that allows recruitment of Tel1ATM to telomeres in the absence of the Nbs1 C-terminal Tel1ATM interaction domain. The notion that the C-terminus of Nbs1 and the Rad3-kdΔATR-Rad26ATRIP complex redundantly contribute to the recruitment of Tel1ATM is also supported by our finding that over-expression of Tel1ATM can entirely bypass the requirement for Rad3ATR-Rad26ATRIP for telomere maintenance and Tel1ATM recruitment to telomeres in nbs1-c60Δ cells (Figure 4, Figure 6C). Moreover, the finding that over-expression of Rad3ATR was able to at least partially suppress the loss of Nbs1 protein (Figure 5B, Figure 6D) provided further support for the notion that the N-terminal domain of Nbs1 also contributes to the recruitment of Rad3ATR to telomeres.
Fig. 6. Summary of mechanisms controlling the recruitment of Rad3ATR-Rad26ATRIP and Tel1ATM to telomeres in fission yeast. 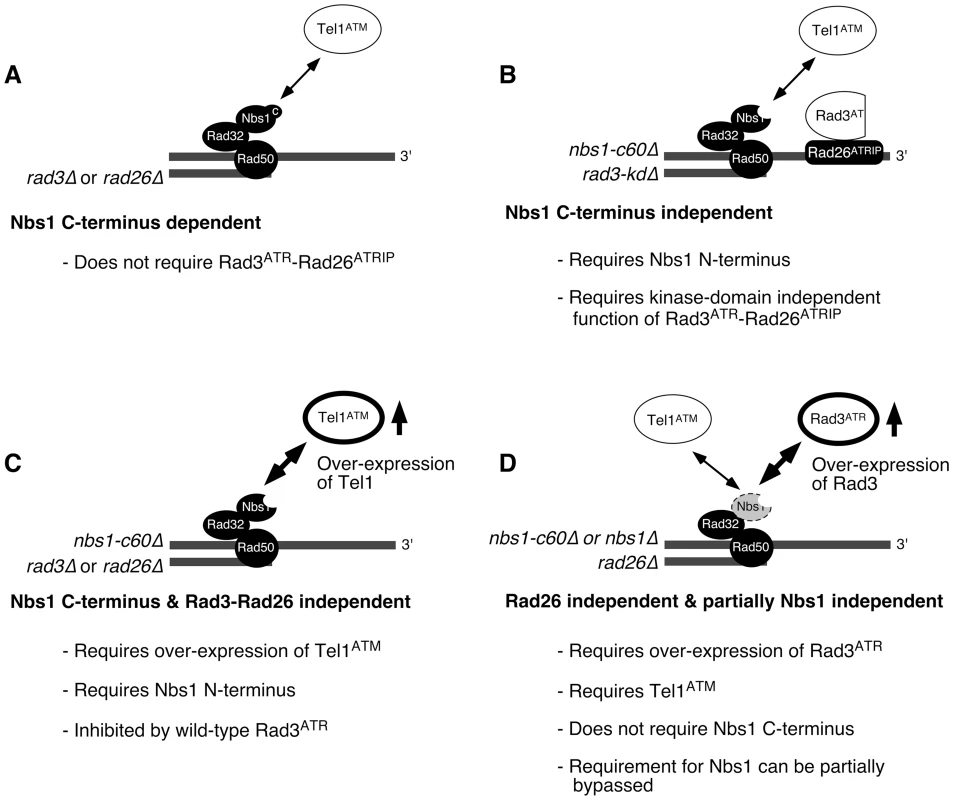
See main text for details. Over-expressed Tel1ATM (C) and Rad3ATR (D) are emphasized with thicker lines for their respective ovals. Thus, our findings are generally consistent with the model that at least one PIKK activity needs to be localized at telomeres, in order to stably maintain telomeres in fission yeast, although we have not yet tested if Tel1ATM kinase activity is indeed required for telomere maintenance in nbs1-c60Δ rad3-kdΔ cells. It is currently unknown which telomere-associated protein(s) represent critical substrate(s) of Tel1/Rad3 that are essential for telomere maintenance. However, we were able to demonstrate recently that Tel1ATM and Rad3ATR are redundantly required to prevent accumulation of homologous recombination DNA repair factors and RPA at telomeres, to promote efficient telomere recruitment of Tpz1 and Ccq1 (subunits of the Pot1 telomere capping complex), and to recruit telomerase to telomeres [18].
Our results also indicate that fission yeast Tel1ATM is recruited preferentially to short telomeres (Figure 2D). Due to the kinase-independent function of the Rad3ATR-Rad26ATRIP complex, the Nbs1 C-terminal 60 amino acid Tel1ATM interaction domain is dispensable for recruitment of Tel1ATM to short telomeres. Tel1ATM is also recruited preferentially to short telomeres in budding yeast [14],[38],[40]. Based on studies utilizing xrs2-664 mutant cells, which express the truncated Xrs2Nbs1 protein lacking the C-terminal 190 amino acids (full length 884 amino acids), it was suggested that the Xrs2Nbs1 C-terminal Tel1ATM interaction domain is essential for recruitment of Tel1ATM to critically short telomeres [14]. However, a recent study has shown that, while deleting as little as 20 amino acids from C-terminus of Xrs2Nbs1 is sufficient to disrupt the interaction between Tel1ATM and Xrs2Nbs1, xrs2 mutant cells lacking the C-terminal 20 amino acids maintain significantly longer telomeres than xrs2-664 or xrs2Δ cells [41]. Therefore, budding yeast Xrs2Nbs1 also appears to contribute to telomere maintenance independently of its C-terminal Tel1ATM interaction domain. Thus, it would be interesting to test if the N-terminal domain of Xrs2Nbs1 might also collaborate with the Mec1ATR-Ddc2ATRIP complex, perhaps independently of Mec1ATR kinase activity, in regulating Tel1ATM recruitment to telomeres in budding yeast.
Our ChIP data indicate that very little or no Tel1ATM is recruited to telomeres in wild-type fission yeast cells. On the other hand, Rad3ATR-Rad26ATRIP is specifically recruited to replicating telomeres and appears to act as the primary sensor of transiently “open” telomeres during S-phase [15]. Therefore, it makes sense that rad3Δ or rad26Δ cause a more severe shortening of telomeres than tel1Δ in fission yeast [25],[27],[42]. Thus, Tel1ATM is likely to function as a back-up mechanism to extend critically short telomeres in fission yeast. In contrast, budding yeast mec1Δ cells show very little shortening of telomeres [43], while tel1Δ or xrs2Δ cells carry extremely short telomeres [44],[45]. It has also been reported that budding yeast Mec1ATR cannot be detected at telomeres by ChIP in wild-type cells [13]. However, simultaneous loss of Mec1ATR-Ddc2ATRIP and Tel1ATM-MRX pathways results in a severe defect in telomere maintenance [43],[46], much like in fission yeast cells. Thus, available data suggest that budding yeast telomeres are primarily regulated by Tel1ATM-MRX, and Mec1ATR-Ddc2ATRIP fulfills a back-up role for maintaining telomeres.
There are intriguing similarities between our current findings and findings in mammalian cells, where ATR-ATRIP was found to act upstream of ATM. In response to replication stress or UV irradiation during S-phase, the Nbs1 C-terminal ATM interaction domain is dispensable, but the N-terminus of Nbs1 is essential, for ATR to activate ATM in mammalian cells [12]. However, it is currently unknown whether ATR-ATRIP might also contribute to the recruitment of ATM to sites of stalled or distressed DNA replication forks in a kinase-independent manner, besides the previously established kinase-dependent role of ATR in converting inactive ATM dimers into active monomers [12]. However, the similar requirement for the N-terminus, but not the C-terminus, of Nbs1 for the ATR-ATM crosstalk in response to DNA replication stress in mammalian cells and telomere maintenance in fission yeast might suggest that fission yeast telomeres may be recognized primarily as stressed and/or abnormal replication forks by ATM/ATR kinases.
Studies in mammalian cells have also established the existence of an ATM-ATR crosstalk in response to DNA DSBs, where ATM and the MRN complex contribute to the recruitment of ATR-ATRIP to DSBs by promoting the generation of RPA-coated ssDNA at DSB sites [9]–[11],[47],[48]. The Nbs1 C-terminal ATM interaction domain is required to recruit ATM to DSBs induced by ionizing radiation (IR) and to promote ATM-dependent phosphorylation events [5],[6],[49], and it is thus critical for the ATM-ATR crosstalk in response to DSBs. While the critical role of Tel1ATM in preferentially extending short telomeres in budding yeast has been well established [14],[38], mouse ATM was found not to be critical for extending short telomeres [50]. This difference might indicate that mouse ATR-ATRIP and ATM-MRN pathways work redundantly in extending critically short telomeres. Alternatively, since recruitment of human ATR to telomeres was found to occur earlier in S-phase than recruitment of ATM to telomeres [16], mammalian ATR-ATRIP may function upstream of ATM in telomere length maintenance, and perhaps also possess kinase-independent functions in recruitment of ATM to telomeres. While further analyses are clearly needed to test these speculations, our current findings highlight a complex molecular crosstalk between ATM-MRN and ATR-ATRIP pathways in recognizing an “open” configuration of telomeres to allow their stable maintenance.
Materials and Methods
Reagents and general methods
Fission yeast strains used in this study were constructed by standard techniques [51] and are listed in Table S1. Primers listed in Table S2 were used to construct new strains. For nbs1-myc, nmt-HA-rad3, myc-rad26 and YFP-rad26, original strains were described previously [15],[24],[27],[52]. For myc-rad3, nbs1-c60Δ-myc, myc-tel1 and nmt-HA-tel1, PCR-based methods [53],[54] were used to generate tagged strains. For nbs1Δ::kanMX, nbs1-c60Δ::kanMX6, rad3-kdΔ::ura4+, rad3Δ::LEU2 and rad26Δ::ura4+, original strains were described previously [5], [25]–[27],[55]. For nbs1Δ::natMX, PCR-based methods [53],[54] were used to generate deletion strains. Budding yeast strains used in yeast two-hybrid assays are also listed in Table S1. Plasmids used in this study are listed in Table S3. Yeast two hybrid assays were performed by mating S. cerevisiae MATa strains harboring GAL4-DBD plasmids with MATα strains harboring GAL4-AD plasmids, as described in the MATCHMAKER system manual (Clonetech). Positive two-hybrid interactions were identified by spotting mated cells onto SD-HTL plates. Sensitivities of fission yeast cells to IR, UV, HU (hydroxyurea) and CPT (camptothecin) were assayed as previously described [24].
Co-immunoprecipitation and western blot analyses
For most strains, cell extracts were prepared in lysis buffer 1 [50mM Tris pH8.0, 150mM NaCl, 10% glycerol, 5mM EDTA, 0.5% NP40, 50mM NaF, 1mM DTT, 1mM PMSF, 1mM Na3VO4, ‘Complete’ protease inhibitor cocktail (Roche)], either by glass bead disruption using FastPrep homogenizer (MP Biomedical) or by cryogenic disruption using MM301 Ball Mill (Retsch). For strains expressing myc-Rad3, myc-Rad3-kdΔ or myc-Tel1, lysis buffer 2 [25mM Tris pH7.5, 100mM NaCl, 10% glycerol, 15mM EDTA, 0.1% NP40, 1% Triton, 15mM MgCl2, 0.1mM NaF, 0.5mM DTT, 1mM PMSF, 1mM Na3VO4, ‘Complete’ protease inhibitor cocktail] was used. For co-immunoprecipitation analyses, proteins were immunoprecipitated using either monoclonal anti-myc antibody (9B11, Cell Signaling) or monoclonal anti-HA antibody (12CA5, Roche), and protein G Dynabeads (Invitrogen) or protein G sepharose beads (GE) respectively. Proteins in whole cell extract or from immunoprecipitations were analyzed by western blots using monoclonal anti-HA antibody (12CA5), monoclonal anti-myc antibody (9B11), monoclonal anti-FLAG antibody (M2, F1804, Sigma) or monoclonal anti-GFP antibody (7.1/B.1, Roche). Anti-Cdc2 antibody (y100.4, Abcam) was used for loading control.
Pulsed-field gel electrophoresis (PFGE)
Chromosomal DNA samples were prepared in agarose plugs, digested with NotI restriction enzyme, and fractionated in 1% agarose gels using the CHEF-DR III system (Bio-Rad) as previously described [25]. C, I, L, and M probes specific for telomeric NotI fragments were prepared as previously described [56]. Except for Tel1 or Rad3 over-expression experiments, cells were extensively restreaked on YES agar plates to achieve terminal telomere states prior to harvesting. For over-expression experiments, minimal media was used for nmt1+ promoter-controlled over-expression.
Southern blot analyses
S. pombe genomic DNA samples were digested with EcoRI or ApaI, separated on 1.2% (EcoRI) or 2% (ApaI) agarose, transferred to Hybond-XL membrane (GE), and hybridized to telomere probe [57] in Church Buffer [0.25M sodium phosphate buffer pH7.2, 1mM EDTA, 1% BSA, 7% SDS] at 65°C overnight to monitor telomere length.
ChIP assays
Cells were processed for ChIP and analyzed as previously described [15], using either monoclonal anti-myc (9B11; Cell Signaling), anti-FLAG (M2, F1804, Sigma), or anti-HA (12CA5) antibodies. Percent precipitated DNA values (% ppt DNA) were calculated based on ΔCt between Input and IP samples after performing several independent triplicate SYBR Green-based real-time PCR (Bio-Rad) using telomere primers jk380 and jk381 [15]. For genetic backgrounds that cause eventual circularization of chromosomes due to a telomere maintenance defect, a Rad3 plasmid (pREP41H-rad3) was utilized to maintain linear chromosomes during strain construction [18]. Prior to ChIP experiments, single colonies that had lost the Rad3 plasmid were selected based on lack of growth on media lacking histidine and sensitivity to HU, and immediately utilized in ChIP experiments. Based on Southern blot analysis, the early generation strains that have just lost the Rad3 plasmid carry comparable or slightly longer telomeres than rad3Δ cells [18] (data not shown).
Statistical analysis
In order to determine statistical significance of our data, two-tailed Student's t-tests were performed, and P values ≤0.05 were considered as statistically significant differences.
Supporting Information
Zdroje
1. LeeJH
PaullTT
2007 Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26 7741 7748
2. CimprichKA
CortezD
2008 ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9 616 627
3. MatsuokaS
BallifBA
SmogorzewskaA
McDonaldER3rd
HurovKE
2007 ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316 1160 1166
4. WilliamsRS
WilliamsJS
TainerJA
2007 Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 85 509 520
5. YouZ
ChahwanC
BailisJ
HunterT
RussellP
2005 ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25 5363 5379
6. FalckJ
CoatesJ
JacksonSP
2005 Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434 605 611
7. ZouL
ElledgeSJ
2003 Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300 1542 1548
8. StiffT
CerosalettiK
ConcannonP
O'DriscollM
JeggoPA
2008 Replication independent ATR signalling leads to G2/M arrest requiring Nbs1, 53BP1 and MDC1. Hum Mol Genet 17 3247 3253
9. JazayeriA
FalckJ
LukasC
BartekJ
SmithGC
2006 ATM - and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8 37 45
10. ShiotaniB
ZouL
2009 Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33 547 558
11. MyersJS
CortezD
2006 Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 281 9346 9350
12. StiffT
WalkerSA
CerosalettiK
GoodarziAA
PetermannE
2006 ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J 25 5775 5782
13. SabourinM
ZakianVA
2008 ATM-like kinases and regulation of telomerase: lessons from yeast and mammals. Trends Cell Biol 18 337 346
14. SabourinM
TuzonCT
ZakianVA
2007 Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27 550 561
15. MoserBA
SubramanianL
ChangYT
NoguchiC
NoguchiE
2009 Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J 28 810 820
16. VerdunRE
KarlsederJ
2006 The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127 709 720
17. TsengSF
LinJJ
TengSC
2006 The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res 34 6327 6336
18. MoserBA
SubramanianL
KhairL
ChangYT
NakamuraTM
2009 Fission yeast Tel1ATM and Rad3ATR promote telomere protection and telomerase recruitment. PLoS Genet 5 e1000622 doi:10.1371/journal.pgen.1000622
19. DenchiEL
de LangeT
2007 Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448 1068 1071
20. NegriniS
RibaudV
BianchiA
ShoreD
2007 DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev 21 292 302
21. GasparyanHJ
XuL
PetreacaRC
RexAE
SmallVY
2009 Yeast telomere capping protein Stn1 overrides DNA replication control through the S phase checkpoint. Proc Natl Acad Sci U S A 106 2206 2211
22. LongheseMP
2008 DNA damage response at functional and dysfunctional telomeres. Genes Dev 22 125 140
23. ZhaoY
SfeirAJ
ZouY
BusemanCM
ChowTT
2009 Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138 463 475
24. NakamuraTM
DuLL
RedonC
RussellP
2004 Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24 6215 6230
25. NakamuraTM
MoserBA
RussellP
2002 Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161 1437 1452
26. BentleyNJ
HoltzmanDA
FlaggsG
KeeganKS
DeMaggioA
1996 The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J 15 6641 6651
27. ChahwanC
NakamuraTM
SivakumarS
RussellP
RhindN
2003 The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol Cell Biol 23 6564 6573
28. BallHL
MyersJS
CortezD
2005 ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 16 2372 2381
29. WakayamaT
KondoT
AndoS
MatsumotoK
SugimotoK
2001 Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol 21 755 764
30. SeatonBL
YucelJ
SunnerhagenP
SubramaniS
1992 Isolation and characterization of the Schizosaccharomyces pombe rad3 gene, involved in the DNA damage and DNA synthesis checkpoints. Gene 119 83 89
31. McSherryTD
KitazonoAA
JavaheriA
KronSJ
MuellerPR
2007 Non-catalytic function for ATR in the checkpoint response. Cell Cycle 6 2019 2030
32. MatsuuraA
NaitoT
IshikawaF
1999 Genetic control of telomere integrity in Schizosaccharomyces pombe: rad3+ and tel1+ are parts of two regulatory networks independent of the downstream protein kinases chk1+ and cds1+. Genetics 152 1501 1512
33. UenoM
NakazakiT
AkamatsuY
WatanabeK
TomitaK
2003 Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol Cell Biol 23 6553 6563
34. ZouL
ElledgeSJ
2001 Sensing and signaling DNA damage: roles of Rad17 and Rad9 complexes in the cellular response to DNA damage. Harvey Lect 97 1 15
35. LarriveeM
LeBelC
WellingerRJ
2004 The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18 1391 1396
36. TomitaK
MatsuuraA
CaspariT
CarrAM
AkamatsuY
2003 Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23 5186 5197
37. OlsonE
NieveraCJ
LeeAY
ChenL
WuX
2007 The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem 282 22939 22952
38. HectorRE
ShtofmanRL
RayA
ChenBR
NyunT
2007 Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27 851 858
39. ViscardiV
BonettiD
Cartagena-LirolaH
LucchiniG
LongheseMP
2007 MRX-dependent DNA damage response to short telomeres. Mol Biol Cell 18 3047 3058
40. BianchiA
ShoreD
2007 Increased association of telomerase with short telomeres in yeast. Genes Dev 21 1726 1730
41. MaY
GreiderCW
2009 Kinase-independent functions of TEL1 in telomere maintenance. Mol Cell Biol 29 5193 5202
42. NaitoT
MatsuuraA
IshikawaF
1998 Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet 20 203 206
43. RitchieKB
MalloryJC
PetesTD
1999 Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol 19 6065 6075
44. LustigAJ
PetesTD
1986 Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci U S A 83 1398 1402
45. NugentCI
BoscoG
RossLO
EvansSK
SalingerAP
1998 Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol 8 657 660
46. RitchieKB
PetesTD
2000 The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155 475 479
47. AdamsKE
MedhurstAL
DartDA
LakinND
2006 Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 25 3894 3904
48. CuadradoM
Martinez-PastorB
MurgaM
ToledoLI
Gutierrez-MartinezP
2006 ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med 203 297 303
49. CerosalettiK
WrightJ
ConcannonP
2006 Active role for nibrin in the kinetics of Atm activation. Mol Cell Biol 26 1691 1699
50. FeldserD
StrongMA
GreiderCW
2006 Ataxia telangiectasia mutated (Atm) is not required for telomerase-mediated elongation of short telomeres. Proc Natl Acad Sci U S A 103 2249 2251
51. AlfaC
FantesP
HyamsJ
McLoedM
WarbrickE
1993 Experiments with Fission Yeast Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press
52. MoserBA
BrondelloJM
Baber-FurnariB
RussellP
2000 Mechanism of caffeine-induced checkpoint override in fission yeast. Mol Cell Biol 20 4288 4294
53. BählerJ
WuJQ
LongtineMS
ShahNG
McKenzieA3rd
1998 Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14 943 951
54. KrawchukMD
WahlsWP
1999 High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15 1419 1427
55. al-KhodairyF
FotouE
SheldrickKS
GriffithsDJ
LehmannAR
1994 Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell 5 147 160
56. NakamuraTM
CooperJP
CechTR
1998 Two modes of survival of fission yeast without telomerase. Science 282 493 496
57. NakamuraTM
MorinGB
ChapmanKB
WeinrichSL
AndrewsWH
1997 Telomerase catalytic subunit homologs from fission yeast and human. Science 277 955 959
58. PerryJ
KlecknerN
2003 The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112 151 155
59. ChapmanCR
EvansST
CarrAM
EnochT
1999 Requirement of sequences outside the conserved kinase domain of fission yeast Rad3p for checkpoint control. Mol Biol Cell 10 3223 3238
Štítky
Genetika Reprodukčná medicína
Článek Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the GenomeČlánek Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in MiceČlánek Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' DiseaseČlánek Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD TagsČlánek Wing Patterns in the Mist
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2010 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Scale of Population Structure in
- Allelic Exchange of Pheromones and Their Receptors Reprograms Sexual Identity in
- Genetic and Functional Dissection of and in Age-Related Macular Degeneration
- A Single Nucleotide Polymorphism within the Acetyl-Coenzyme A Carboxylase Beta Gene Is Associated with Proteinuria in Patients with Type 2 Diabetes
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Cdk2 Is Required for p53-Independent G/M Checkpoint Control
- Uncoupling of Satellite DNA and Centromeric Function in the Genus
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in the Clade
- Use of DNA–Damaging Agents and RNA Pooling to Assess Expression Profiles Associated with and Mutation Status in Familial Breast Cancer Patients
- Cheating by Exploitation of Developmental Prestalk Patterning in
- Replication and Active Demethylation Represent Partially Overlapping Mechanisms for Erasure of H3K4me3 in Budding Yeast
- Cdk1 Targets Srs2 to Complete Synthesis-Dependent Strand Annealing and to Promote Recombinational Repair
- A Genome-Wide Association Study Identifies Susceptibility Variants for Type 2 Diabetes in Han Chinese
- Genome-Wide Identification of Susceptibility Alleles for Viral Infections through a Population Genetics Approach
- Transcriptional Rewiring of the Sex Determining Gene Duplicate by Transposable Elements
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in
- Proteasome Nuclear Activity Affects Chromosome Stability by Controlling the Turnover of Mms22, a Protein Important for DNA Repair
- Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in Mice
- Structure, Function, and Evolution of the spp. Genome
- Human and Non-Human Primate Genomes Share Hotspots of Positive Selection
- A Kinase-Independent Role for the Rad3-Rad26 Complex in Recruitment of Tel1 to Telomeres in Fission Yeast
- Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' Disease
- Molecular Evolution and Functional Characterization of Insulin-Like Peptides
- Molecular Poltergeists: Mitochondrial DNA Copies () in Sequenced Nuclear Genomes
- Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD Tags
- Wing Patterns in the Mist
- DNA Binding of Centromere Protein C (CENPC) Is Stabilized by Single-Stranded RNA
- Genome-Wide Association Study Reveals Multiple Loci Associated with Primary Tooth Development during Infancy
- Mutations in , Encoding an Equilibrative Nucleoside Transporter ENT3, Cause a Familial Histiocytosis Syndrome (Faisalabad Histiocytosis) and Familial Rosai-Dorfman Disease
- Genome-Wide Identification of Binding Sites Defines Distinct Functions for PHA-4/FOXA in Development and Environmental Response
- Ku Regulates the Non-Homologous End Joining Pathway Choice of DNA Double-Strand Break Repair in Human Somatic Cells
- Nucleoporins and Transcription: New Connections, New Questions
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Nucleoporins and Transcription: New Connections, New Questions
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



