-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
The Escherichia coli chromosome is organized into four macrodomains, the function and organisation of which are poorly understood. In this review we focus on the MatP, SeqA, and SlmA proteins that have recently been identified as the first examples of factors with macrodomain-specific DNA-binding properties. In particular, we review the evidence that these factors contribute towards the control of chromosome replication and segregation by specifically targeting subregions of the genome and contributing towards their unique properties. Genome sequence analysis of multiple related bacteria, including pathogenic species, reveals that macrodomain-specific distribution of SeqA, SlmA, and MatP is conserved, suggesting common principles of chromosome organisation in these organisms. This discovery of proteins with macrodomain-specific binding properties hints that there are other proteins with similar specificity yet to be unveiled. We discuss the roles of the proteins identified to date as well as strategies that may be employed to discover new factors.
Published in the journal: Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria. PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002123
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1002123Summary
The Escherichia coli chromosome is organized into four macrodomains, the function and organisation of which are poorly understood. In this review we focus on the MatP, SeqA, and SlmA proteins that have recently been identified as the first examples of factors with macrodomain-specific DNA-binding properties. In particular, we review the evidence that these factors contribute towards the control of chromosome replication and segregation by specifically targeting subregions of the genome and contributing towards their unique properties. Genome sequence analysis of multiple related bacteria, including pathogenic species, reveals that macrodomain-specific distribution of SeqA, SlmA, and MatP is conserved, suggesting common principles of chromosome organisation in these organisms. This discovery of proteins with macrodomain-specific binding properties hints that there are other proteins with similar specificity yet to be unveiled. We discuss the roles of the proteins identified to date as well as strategies that may be employed to discover new factors.
Introduction
All organisms are faced with the challenge of organising their genetic content within the confines of the cell or its compartments. In eukaryotes, DNA is packed inside the nucleus and histone proteins are known to wrap DNA into nucleosomes. Nucleosomal arrays are folded into chromatin fibers, which are themselves folded into higher order structures. Whilst our understanding of this process at the nucleosomal level is well developed, higher levels of organization are poorly understood [1], [2]. Similarly, mechanisms of chromosome organisation in bacteria are poorly defined. The folded bacterial genome, or nucleoid, is known to be organized by “nucleoid-associated” DNA-binding proteins (NAPs), DNA supercoiling, and transcription [3]. Nucleoid-associated proteins are abundant, often bind DNA with a low degree of sequence specificity, and impose constraints on DNA topology that are best understood at the nm scale (Figure 1A). For example histone-like nucleoid structuring protein (H-NS) can stimulate DNA-bridging events, the integration host factor (IHF) can introduce hair-pin bends into the double helix and curved DNA binding protein A (CbpA) forms aggregates with DNA [4]–[6]. It is likely that some of these nucleoid-associated proteins contribute to the formation of structures at larger scales, such as topologically isolated supercoiled domains and transcription foci (Figure 1B), but fine molecular details remain to be elucidated [7], [8]. In this review, we focus on recent observations concerning organisation of bacterial chromosomes into even larger organisational units at the µm scale: macrodomains (Figure 1C) [9]–[12]. In particular we focus on the implications of recent findings regarding three proteins—SeqA, SlmA, and macrodomain Ter protein (MatP)—with macrodomain-specific DNA-binding properties.
Fig. 1. Hierarchical levels of organization in bacterial chromosomes. 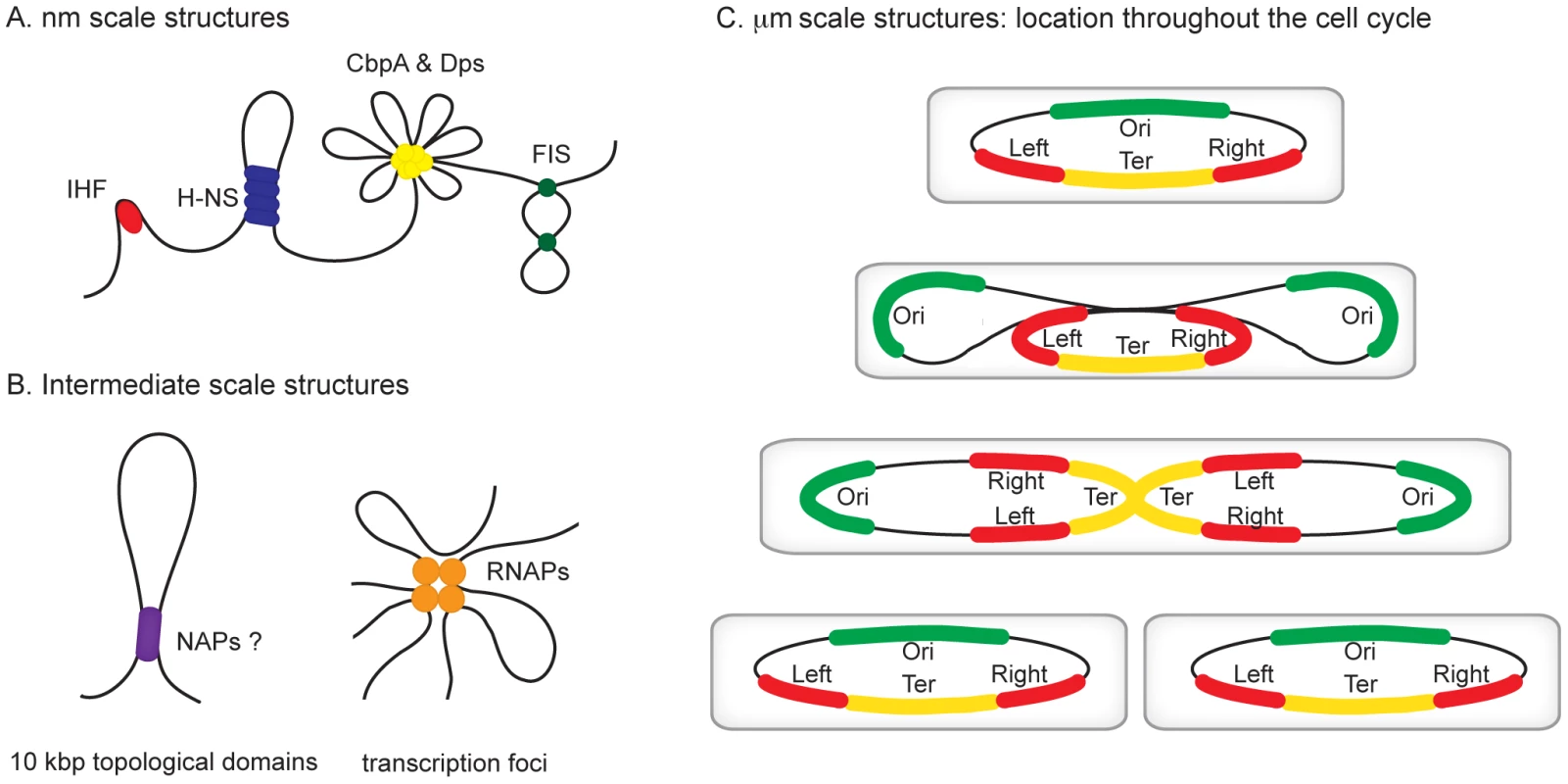
Different levels of organization exist within bacterial chromosomes. (A) At the nm scale nucleoid proteins such as HU, H-NS, CbpA, Dps, and Fis organize the genome by driving events such as DNA bending, bridging, and aggregation. (B) Structures such as seen in (A) likely exist within, and may contribute towards the formation of looped topological domains (on average each ∼10 kbp in size) and transcription foci, where multiple transcribing RNA polymerase molecules are clustered potentially also yielding loops along the genome. (C) All of the above could add to the complexity of the organization within individual macrodomains. The individual macrodomains have a defined localization within the cell throughout the cell cycle. In newborn cells ori and ter are located at mid-cell positions. These sites are located centrally within the Ori and Ter macrodomains. The Left and Right macrodomains occupy positions close to the cell poles. Upon replication, the Ori domains move towards the cell poles. Right before cell division the replicated Ter domains segregate. The chromosome in the daughter cells has again the same Left-Right orientation. MatP preferentially occupies sites in the Ter domain, whereas SlmA and SeqA are absent from this domain. Identification of the Chromosomal Macrodomains
Evidence for the existence of chromosomal “macrodomains” in E. coli has been established during the last 5 years by Boccard and coworkers [9], [13]–[15], building on the ideas of Niki et al. [10]. The existence and positioning of the four macrodomains was first determined in assays aimed at resolving spatial proximity of genomic regions by measuring the frequency of recombination between phage λ att sites scattered throughout the E. coli chromosome [13]. This analysis revealed a clear bias in the positioning of pairs of att sites that supported efficient recombination and thus were spatially close. On the basis of these observations, it was concluded that the E. coli chromosome is organized into four discrete structured subdomains and that att sites in each domain interact primarily with the att sites in the same domain. Each of these domains (Ori, Right, Left, and Ter) contains approximately 1 Mbp of DNA. The localization of the macrodomains is subject to changes during the cell cycle, but is fairly well defined (Figure 1C). The degree of linear DNA compaction as measured in vivo using genomic markers varies among domains. The 800-kb domain around Ter is on average five times less compact than the rest of the genome and extends between two opposing ends of the nucleoid [16]. The highly abundant nucleoid-associated proteins are obvious candidates for bestowing unique properties on the individual macrodomains. However, available evidence suggests that this is unlikely; well-characterised nucleoid-associated proteins such as H-NS and IHF are found to bind with all of the macrodomains in chromatin immunoprecipitation (ChIP) experiments (Figure 2A). Indeed, amongst the known drivers of chromosome structure, only RNA polymerase displays any domain-specific binding behaviour; its primary targets, the seven rRNA operons, are all in the oriC half of the chromosome (Figure 2A).
Fig. 2. Distribution of nucleoid-associated proteins across the E. coli chromosome. 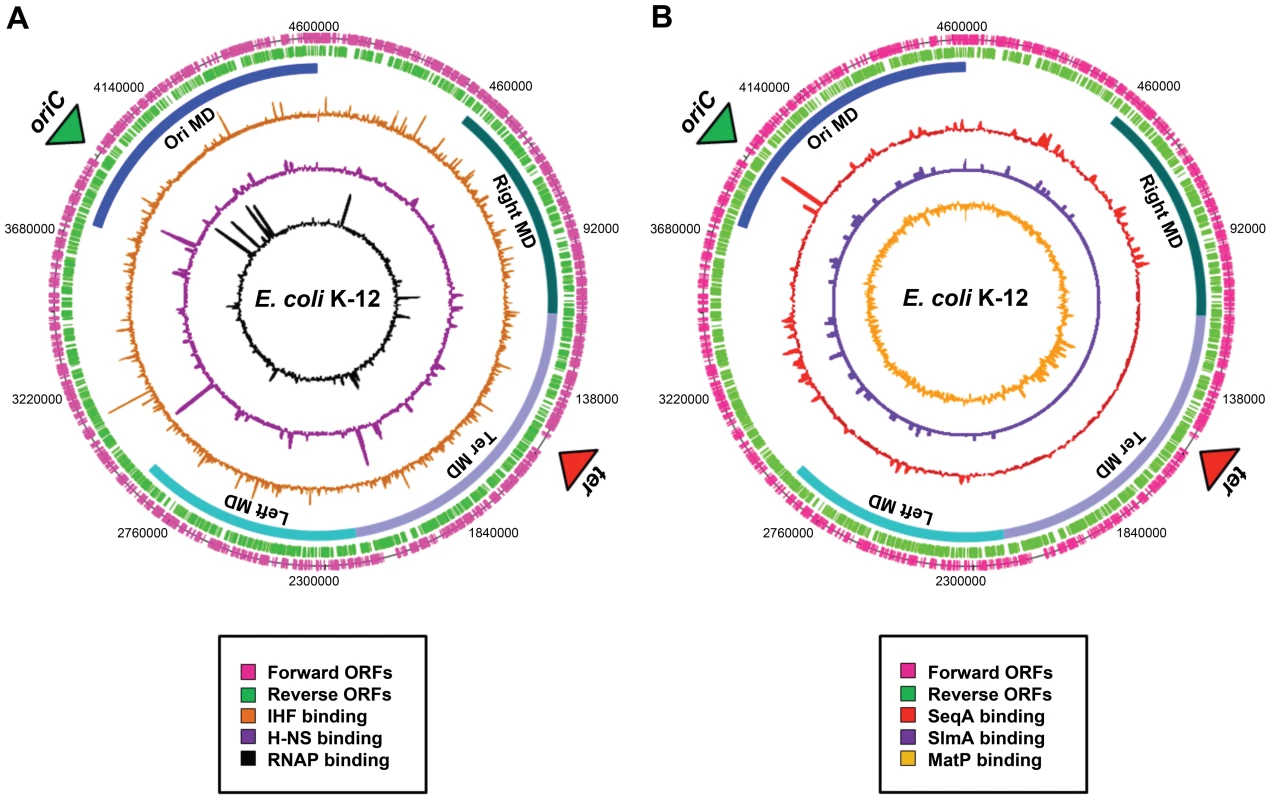
(A) A genome atlas where ChIP-chip datasets [41] for IHF (orange), H-NS (purple), and RNA polymerase (black) are plotted against the features of the E. coli chromosome. (B) A genome atlas where ChIP-chip or ChIP-Seq datasets for SeqA (red) [17], SlmA [purple] (19) and MatP [orange] (20) are plotted against the features of the E. coli chromosome. The locations of ORFs are shown as pink and green lines. The positions of the four macrodomains (MDs) are shown as blue bars and are labelled. Proteins with Macrodomain-Specific DNA-Binding Properties
High-throughput analysis of DNA-binding events across bacterial genomes using ChIP has revealed that some major regulators of the cell cycle have macrodomain-specific DNA-binding profiles [17]–[21]. MatP binds exclusively to the Ter macrodomain [20], whilst both SeqA and SlmA are excluded from this region of the chromosome [17]–[19], [21]. The fact that SeqA, SlmA, and MatP bind to nondegenerate DNA target sites with a high degree of specificity, sets them apart from the classical nucleoid-associated proteins [16], [19]–[21]. However, since the term “nucleoid-associated protein” is clearly ambiguous (discussed in [3]), we argue that it can be applied to any protein that plays a role in organising the chromosome. Thus, below we discuss the known properties of SeqA, SlmA, and MatP in light of their recently discovered macrodomain-specific chromosome-binding properties.
SeqA
The SeqA protein was originally discovered as the factor responsible for sequestration of chromosome replication origins in bacteria [22]. It has subsequently been shown that SeqA plays a key role in preventing the over-initiation of chromosome replication [23] and delays the separation of new chromosomes [24]. SeqA recognises pairs of hemi-methylated GATC motifs that are found in newly replicated DNA. Whilst these motifs are most densely concentrated near oriC, many other potential SeqA targets are distributed across the chromosome. It has long been assumed that SeqA might bind hundreds of sites distal to oriC, and two ChIP studies recently confirmed these suspicions [17], [18]. Surprisingly, these studies also demonstrated that SeqA is excluded from the Ter macrodomain except under artificial conditions where chromosome replication is blocked (Figure 2B) [17]. This exclusion is most likely due to a lack of high affinity SeqA binding sites in the Ter macrodomain [17]. SeqA is known to associate with the cell membrane and, given the skewed binding of SeqA across the genome, SeqA may play a role to properly orientate the chromosome during cell division. Due to changes in the methylation state of the DNA as the chromosome is replicated, the SeqA distribution across the genome is dynamic. These changes may influence the structure and/or cellular position of the Ori, Right, and Left macrodomains as the chromosome is copied. It is unknown if the process of DNA replication affects SlmA or MatP binding but, as outlined below, all three proteins are known to play key roles in controlling chromosome replication and separation.
SlmA
The SlmA protein was identified in genetic screens as a “nucleoid occlusion” factor, i.e., as a protein involved in coordinating positioning and proper assembly of the so-called Z-ring at mid-cell prior to cell division [25]. The assembly of the Z-ring relies on the multimerization of the tubulin-like FtsZ protein, to which subsequently other septal ring components are recruited. The molecular basis underlying the action of SlmA was recently investigated in two parallel studies [19], [21]. These studies showed that SlmA can bind DNA and simultaneously interact with FtsZ, interfering with Z-ring assembly [19], [21]. Genome-wide ChIP showed that SlmA binds to a 12-bp palindromic consensus sequence (GTGAGTACTCAC), which is found 50 times along the E. coli K-12 genome. Strikingly, none of these sites are found in the Ter macrodomain and they are underrepresented in the Left and Right macrodomains (Figure 2B). Sequence analysis reveals that putative SlmA binding sites are also excluded from the Ter macrodomain of pathogenic E. coli strains, Salmonella Typhimurium, and Klebsiella pneumoniae [19]. The unique presence of SlmA binding sites in non-Ter domains suggests a model in which SlmA bound in these genomic regions prevents undesired Z-ring formation, whilst permitting Z-ring formation at Ter-sites that prior to cell division are located at mid-cell (Figure 3) [26]. One might speculate that the FtsZ-SlmA structures that are nonproductive for Z-ring formation act in contributing to a structural framework to which the nucleoid is tethered. SlmA works together with the MinCDE system in ensuring that the cytokinetic ring is properly positioned. MinCDE prevents cells from dividing near the poles and promotes the positioning of the cytokinetic ring near midcell, while SlmA prevents the premature assembly of the cytokinetic ring over unsegregated chromosomes [21], [27]. Although this review is focused on the E. coli system, it is pertinent to note that proteins similar in function to SlmA have been identified in other bacteria. Thus, the nucleoid occlusion protein Noc of Bacillus subtilis also acts as a spatial regulator of cell division by binding to sites outside the terC region of the chromosome [28]. The MipZ protein appears to play a similar role in Caulobacter. Owing to its interaction with ParB, which binds specifically to the origin region, upon origin segregation MipZ localizes to the poles where it destabilizes the polar FtsZ complex and directs FtsZ polymerization towards midcell [29].
Fig. 3. Localization of MatP and SlmA on the E. coli chromosome. 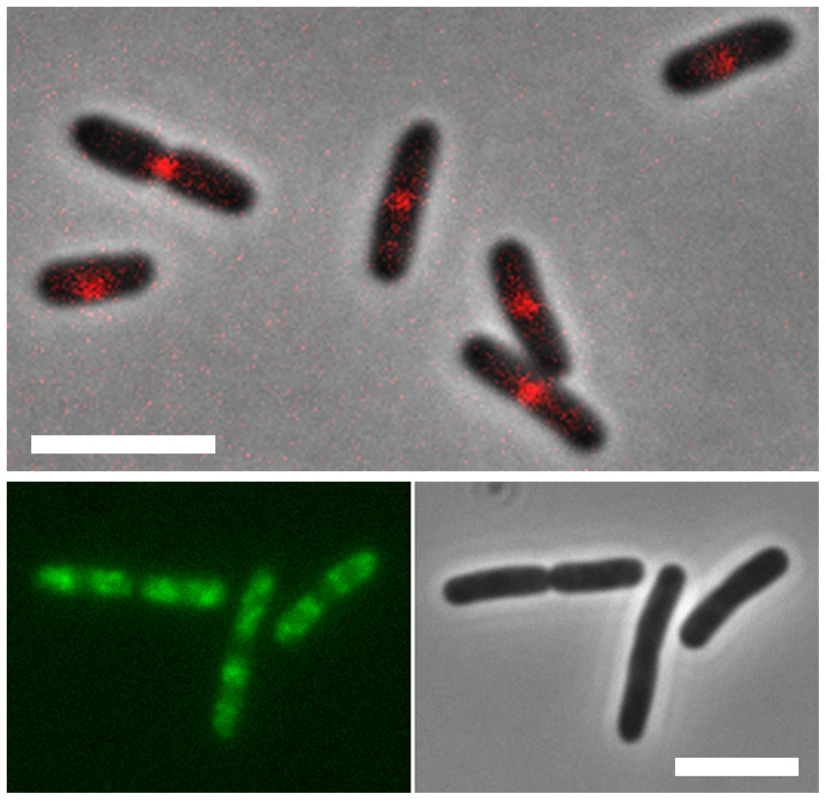
E. coli cells expressing fluorescent derivatives of matP (matP-Cherry) (top panel) and SlmA (GFP-SlmA) (bottom panel). An overlay of phase contrast and fluorescence images is shown for matP, whereas separate fluorescence and DIC images are shown for SlmA. Scale bar, 4 µm. MatP predominantly localizes to the Ter macrodomain, whereas SlmA is absent from this domain. MatP
MatP is a small DNA-binding protein that—unlike SeqA and SlmA—is associated exclusively with the Ter domain of the E. coli genome (Figure 3) [20]. It binds specifically to a signature motif of 13 bps (GTGACA/GNT/CGTCAC) repeated 23 times within the Ter region. It is intriguing to note that the flanking four bps of the binding site of MatP and that of SlmA are identical. The MatP binding motif (matS), was discovered in silico by searching for scattered domain-specific targets of nucleoid-associated proteins. The factor specifically binding to this site (MatP) was identified in DNA-binding assays using crude E. coli extracts [20] as the product of the ycbG gene. The high affinity binding of MatP within the Ter domain was visualized in vivo using fluorescent microscopy. These experiments showed that MatP prevents premature chromosome segregation early during the cell cycle by keeping the Ter regions of two chromosomes together. In MatP knock-out cells this prolonged colocalization of the Ter domains is not observed. Fast growing cells deficient in MatP display a filament-like or anucleate phenotype. A delay in segregation of the daughter chromosomes due to the binding of MatP to the Ter region thus appears essential in coordinating chromosome segregation and cell division. Also, without MatP, the Ter domain displays higher mobility and a lower degree of compaction. Surprisingly the effects of MatP-DNA binding stretch over long distances. The deletion of a matS site increases the mobility of regions even several tens of kb away. While the role of this protein in the cell cycle and the organization of the Ter domain is apparent, the mechanism of MatP action is still unknown. Two models have been proposed for how MatP organizes the Ter domain. According to the first model MatP dimers bridge two matS sites located on either separate chromosomes or within one chromosome. It is possible, that bridging nucleates at matS sites and that flanking regions are zipped up by additional nonspecific binding (and bridging) of MatP. The second model invokes an as yet unknown cofactor. After the binding of MatP, this factor would be recruited to regions surrounding matS sites and spread over distances up to several kb. An obvious candidate for such binding would be the H-NS protein [4] or any other NAP exhibiting cooperative binding (and bridging), but ChIP data on known NAPs do not show any evident overlap in binding patterns.
SeqA, SlmA, MatP, and the Control of Gene Expression
As mentioned above, SeqA, SlmA, and MatP are distinct from the classical nucleoid-associated proteins in that they recognise DNA with a high degree of sequence specificity. In this respect the DNA-binding properties of SeqA, SlmA, and MatP are more akin to those of transcription factors. Intriguingly, many SeqA binding sites are located at promoters and within coding regions of genes involved in DNA replication and repair [17], and it is tempting to speculate that SeqA might regulate expression of these genes. Indeed, at some such targets (for example mioC, dnaA, ftsZ, and mukB), SeqA binding is thought to exert cell cycle–dependent control on gene expression [17], [30]–[32]. However, in other instances, SeqA binding was found to have no effect [17]. Moreover, there is little correlation between SeqA binding and changes in gene expression observed in a seqA mutant [17], [33]. SlmA binding sites were found mainly in coding regions of the chromosome, consistent with observations that SlmA does not appear to function as a regulator of gene expression [19], [21]. This is despite the fact that SlmA is structurally related to the TetR family of transcription factors. Similarly, whilst some MatP targets were located in intergenic regions, MatP was found to have no effect on the expression of genes in the Ter macrodomain [20]. Thus, the available data suggest that a significant proportion of binding sites for SeqA, SlmA, and MatP are not directly involved in the regulation of gene expression. Since evolution has clearly dictated that these proteins bind to specific subregions of the chromosome, we postulate that the relative positioning of SeqA, SlmA, and MatP binding sites across the genome, rather than genes targeted, is crucial. SeqA, SlmA. and MatP may act as “markers” that permit the cell to orientate chromosomes correctly, for instance, to ensure that cell division occurs where genome replication has just finished. Ultimately, detailed studies of individual SeqA, SlmA, and MatP binding loci will be required to determine the precise role of these proteins.
Perspectives for the Future
The pattern of SeqA, SlmA, and MatP binding is probably similar among Gram negative bacteria, including the many pathogenic organisms, related to E. coli [17], [19], [20]. We anticipate that other proteins with macrodomain-specific DNA-binding profiles will be unearthed in the coming years. The discovery of such factors will provide new mechanistic insights into chromosome organisation, replication, and separation inside cells. The rapid detection of such proteins will require an integrated experimental approach utilizing a combination of bioinformatic, genomic, and imaging technologies. Mercier and colleagues demonstrated that careful analysis of DNA sequence can quickly pinpoint potential binding sites for proteins with macrodomain-specific DNA-binding properties [20]. Once identified such DNA sequences can be used to isolate the cognate binding factor. In this respect, recently developed “DNA-sampling” technologies, which allow the proteins bound to a specific portion of the genome to be defined, may be of particular use [34]. Currently, this approach is limited to DNA fragments a few thousand base pairs in length. However, we speculate that it may be possible to isolate individual macrodomains and apply biophysical approaches to probe their structure and protein content. Indeed, the intact nucleoid has already been purified and crudely analyzed in this way [35]. Once detected, it is essential to probe the specific role of macrodomain-associated proteins using state-of-the-art techniques, common ground already in the field of eukaryotic chromatin organisation. Specifically, detailed knowledge can be obtained using 3C-based techniques [36] that map at high resolution the spatial interaction frequencies between genomic sites. Super-resolution imaging techniques [37], [38] can provide single-cell information on the position and function of these proteins within the nucleoidal framework, as well as on spatial distance of genomic sites of interest. Finally, it is not known if macrodomains are maintained under different physiological conditions. For instance, in starved cells, the chromosome undergoes a process of super-compaction attributed to stationary phase-specific proteins Dps and CbpA [6], [39]. Drug treatment can also trigger changes in chromosome morphology [40] and this process may be particularly important for understanding the response of pathogenic bacteria to antibiotics.
Zdroje
1. MisteliT 2010 Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol 2 a000794
2. LugerKMäderAWRichmondRKSargentDFRichmondTJ 1997 Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389 251 260
3. DillonSCDormanCJ 2010 Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8 185 195
4. DameRTNoomMCWuiteGJ 2006 Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 444 387 390
5. van NoortJVerbruggeSGoosenNDekkerCDameRT 2004 Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc Natl Acad Sci U S A 101 6969 6974
6. CosgriffSChintakayalaKChimYTChenXAllenS 2010 Dimerization and DNA-dependent aggregation of the Escherichia coli nucleoid protein and chaperone CbpA. Mol Microbiol 77 1289 1300
7. ChoBKKnightEMBarrettCLPalssonBØ 2008 Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res 18 900 910
8. NoomMCNavarreWWOshimaTWuiteGJDameRT 2007 H-NS promotes looped domain formation in the bacterial chromosome. Curr Biol 17 R913 R914
9. BoccardFEsnaultEValensM 2005 Spatial arrangement and macrodomain organization of bacterial chromosomes. Mol Microbiol 57 9 16
10. NikiHYamaichiYHiragaS 2000 Dynamic organisation of chromosomal DNA in Escherichia coli. Genes Dev 14 212 223
11. NielsenHJOttensenJRYoungrenBAustinSJHansenFG 2006 The Escherichia coli chromosome is organised with the left and right chromosome arms in separate cell halves. Mol Microbiol 62 331 338
12. WangXLiuXPossozCSherattDJ 2006 The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev 20 1727 1731
13. ValensMPenaudSRossignolMCornetFBoccardF 2004 Macrodomain organization of the Escherichia coli chromosome. EMBO J 23 4330 4341
14. EspeliOMercierRBoccardF 2008 DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68 1418 1427
15. LovettSTSegallAM 2004 New views of the bacterial chromosome. EMBO Rep 5 860 864
16. WigginsPACheverallsKCMartinJSLintnerRKondevJ 2010 Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci 107 4991 4995
17. Sánchez-RomeroMABusbySJDyerNPOttSMillardAD 2010 Dynamic distribution of SeqA protein across the chromosome of Escherichia coli K-12. mBio 1 e00012 10
18. WaldminghausTSkarstadK 2010 ChIP on Chip: surprising results are often artefacts. BMC Genomics 11 414
19. TonthatNKAroldSTPickeringBFVan DykeMWLiangS 2011 Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J 30 154 164
20. MercierRPetitMASchbathSRobinSEl KarouiM 2008 The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 475 485
21. ChoHMcManusHRDoveSLBernhardtTG 2011 Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerisation antagonist. Proc Natl Acad Sci U S A 108 3773 3778
22. LuMCampbellJLBoyeEKlecknerN 1994 SeqA: a negative modulator of replication initiation in E. coli. Cell 77 413 426
23. von FreieslebenURasmussenKVSchaechterM 1994 SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol 14 763 772
24. BachTKreklingMASkarstadK 2003 Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. EMBO J 22 315 323
25. BernhardtTGde BoerPA 2005 SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol. Cell 18 555 564
26. LiYYoungrenBSergueevKAustinS 2003 Segregation of the Escherichia coli chromosome terminus. Mol Microbiol 50 825 834
27. MargolinW 2005 FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6 862 871
28. WuLJIshiwakaSKawaiYOshimaTOgasawaraN 2009 Noc protein binds to specific DNA sequences to co-ordinate cell division with chromosome segregation. EMBO J 28 1940 1952
29. ThanbichlerMShapiroL 2006 MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126 147 162
30. ZhouPBoganJAWelchKPickettSRWangHJ 1997 Gene transcription and chromosome replication in Escherichia coli. J Bacteriol 179 163 169
31. BoganJAHelmstetterCE 1996 mioC transcription, initiation of replication, and the eclipse in Escherichia coli. J Bacteriol 178 3201 3206
32. ZhouPHelmstetterCE 1994 Relationship between ftsZ gene expression and chromosome replication in Escherichia coli. J Bacteriol 176 6100 6106
33. Løbner-OlesenAMarinusMGHansenFG 2003 Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc Natl Acad Sci U S A 100 4672 4677
34. ButalaMBusbySJLeeDJ 2009 DNA sampling: a method for probing protein binding at specific loci on bacterial chromosomes. Nucleic Acids Res 37 e37
35. ZimmermanSB 2006 Cooperative transitions of isolated Escherichia coli nucleoids: implications for the nucleoid as a cellular phase. J Struct Biol 153 160 175
36. van BerkumNLDekkerJ 2009 Determining spatial chromatin organization of large genomic regions using 5C technology. Methods Mol Biol 567 189 213
37. GitaiZ 2009 New fluorescence microscopy methods for microbiology: sharper, faster, and quantitative. Curr Opin Microbiol 12 341 346
38. XieXSChoiPJLiG-WLeeNKLiaG 2008 Single-molecule approach to molecular biology in living bacterial cells. Ann Rev Biophys 37 417 444
39. OhniwaRLMorikawaKKimJOhtaTIshihamaA 2006 Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J 25 5591 5602
40. CabreraJECaglieroCQuanSSquiresCLJinDJ 2009 Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: examining the effect of transcription on nucleoid structure in the absence of transertion. J Bacteriol 191 4180 4185
41. GraingerDCHurdDGoldbergMDBusbySJ 2006 Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res 34 4642 4652
Štítky
Genetika Reprodukčná medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



