-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
Centromere behavior is specialized in meiosis I, so that sister chromatids of homologous chromosomes are pulled toward the same side of the spindle (through kinetochore mono-orientation) and chromosome number is reduced. Factors required for mono-orientation have been identified in yeast. However, comparatively little is known about how meiotic centromere behavior is specialized in animals and plants that typically have large tandem repeat centromeres. Kinetochores are nucleated by the centromere-specific histone CENH3. Unlike conventional histone H3s, CENH3 is rapidly evolving, particularly in its N-terminal tail domain. Here we describe chimeric variants of CENH3 with alterations in the N-terminal tail that are specifically defective in meiosis. Arabidopsis thaliana cenh3 mutants expressing a GFP-tagged chimeric protein containing the H3 N-terminal tail and the CENH3 C-terminus (termed GFP-tailswap) are sterile because of random meiotic chromosome segregation. These defects result from the specific depletion of GFP-tailswap protein from meiotic kinetochores, which contrasts with its normal localization in mitotic cells. Loss of the GFP-tailswap CENH3 variant in meiosis affects recruitment of the essential kinetochore protein MIS12. Our findings suggest that CENH3 loading dynamics might be regulated differently in mitosis and meiosis. As further support for our hypothesis, we show that GFP-tailswap protein is recruited back to centromeres in a subset of pollen grains in GFP-tailswap once they resume haploid mitosis. Meiotic recruitment of the GFP-tailswap CENH3 variant is not restored by removal of the meiosis-specific cohesin subunit REC8. Our results reveal the existence of a specialized loading pathway for CENH3 during meiosis that is likely to involve the hypervariable N-terminal tail. Meiosis-specific CENH3 dynamics may play a role in modulating meiotic centromere behavior.
Published in the journal: Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in. PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002121
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002121Summary
Centromere behavior is specialized in meiosis I, so that sister chromatids of homologous chromosomes are pulled toward the same side of the spindle (through kinetochore mono-orientation) and chromosome number is reduced. Factors required for mono-orientation have been identified in yeast. However, comparatively little is known about how meiotic centromere behavior is specialized in animals and plants that typically have large tandem repeat centromeres. Kinetochores are nucleated by the centromere-specific histone CENH3. Unlike conventional histone H3s, CENH3 is rapidly evolving, particularly in its N-terminal tail domain. Here we describe chimeric variants of CENH3 with alterations in the N-terminal tail that are specifically defective in meiosis. Arabidopsis thaliana cenh3 mutants expressing a GFP-tagged chimeric protein containing the H3 N-terminal tail and the CENH3 C-terminus (termed GFP-tailswap) are sterile because of random meiotic chromosome segregation. These defects result from the specific depletion of GFP-tailswap protein from meiotic kinetochores, which contrasts with its normal localization in mitotic cells. Loss of the GFP-tailswap CENH3 variant in meiosis affects recruitment of the essential kinetochore protein MIS12. Our findings suggest that CENH3 loading dynamics might be regulated differently in mitosis and meiosis. As further support for our hypothesis, we show that GFP-tailswap protein is recruited back to centromeres in a subset of pollen grains in GFP-tailswap once they resume haploid mitosis. Meiotic recruitment of the GFP-tailswap CENH3 variant is not restored by removal of the meiosis-specific cohesin subunit REC8. Our results reveal the existence of a specialized loading pathway for CENH3 during meiosis that is likely to involve the hypervariable N-terminal tail. Meiosis-specific CENH3 dynamics may play a role in modulating meiotic centromere behavior.
Introduction
Centromeres are loci that direct faithful segregation of chromosomes during eukaryote cell division. They provide a platform for the assembly of kinetochores, structures that bind to spindle microtubules and coordinate chromosome movement. Centromere behavior must be regulated differently in mitosis and meiosis [1]. In mitosis, centromeres from sister chromatids face in opposite directions (bi-orientation), allowing the spindle to pull the replicated sisters apart at anaphase. In meiosis I, sister centromeres face in the same direction (mono-orientation). This allows sister chromatids to move to the same side of the spindle in anaphase I, when homologous chromosomes are segregated apart. Chromosome segregation errors in meiosis I are a primary cause of spontaneous abortion and birth defects, highlighting the importance of studying meiotic centromere behavior [1].
The mechanism of mono-orientation has been illuminated by yeast studies. In Schizosaccharomyces pombe, the meiosis-specific cohesin subunit Rec8 fuses sister kinetochores together in a geometry that favors attachment to microtubules from the same side of the spindle [2]. This appears to be a conserved mechanism, because rec8 mutants in the plant Arabidopsis thaliana also show bi-oriented sister kinetochores in meiosis I [3]. Furthermore, fused sister kinetochores have been observed in maize meiosis I [4]. Other proteins required for mono-orientation in yeast, such as S. pombe Moa1p or the monopolin complex of Saccharomyces cerevisiae, are not found in animals or in plants [5], [6]. These proteins may evolve rapidly. However it is also possible that the mechanism of bi-orientation has different features between yeast kinetochores that are nucleated by small DNA sequences and animal and plant kinetochores that assemble on megabase-scale tandem repeat arrays [7].
Centromere function requires the centromere specific histone H3 variant (CENH3, called CENP-A in human), which replaces histone H3 in centromeric nucleosomes and recruits many essential kinetochore proteins [8]. Unlike conventional histones, which are extremely well conserved, CENH3s are fast evolving [9]. Genetic experiments in A. thaliana and in S. cerevisiae, as well as localization studies in Drosophila melanogaster have shown that evolutionarily divergent CENH3s cannot substitute for one another (although gene silencing experiments in human cells suggest greater promiscuity) [10]–[13]. The N-terminal tail domain of CENH3s is even more hypervariable than the C-terminal histone-fold domain, and shares almost no similarity between plant species such as A. thaliana and maize (Zea mays), let alone between plants and other eukaryotes [9], [13].
As the histone-fold domain of CENH3 is sufficient for kinetochore localization, the role of the tail domain is enigmatic. We have shown that a CENH3 protein lacking the tail is targeted to kinetochores, but fails to complement an A. thaliana cenh3 null mutant [13]. However, replacing the CENH3 tail with the tail of conventional H3.3 in a GFP-tagged protein gives rise to viable but sterile plants (cenh3 plants complemented with this transgene are referred to as GFP-tailswap) [14]. This unexpected result suggested that the CENH3 tail might have a specific function in meiosis, even though CENH3 is required for both mitotic and meiotic kinetochore functions. Most studies of CENH3 dynamics and function in eukaryotes with tandem repeat centromeres are limited to mitosis, since the knockout of CENH3 is zygotic lethal. The viability and subsequent sterility of cenh3 GFP-tailswap plants enabled us to investigate the meiosis specific role of CENH3 in A. thaliana.
Here we present a detailed analysis of the meiotic phenotype of A. thaliana plants expressing GFP-tagged CENH3 variants with alterations in the CENH3 tail domain. Sterility in these plants was caused by random chromosome segregation, with the first defects appearing at the onset of metaphase I (the stage at which centromere behavior is expected to differ between mitosis and meiosis). Chromosome segregation defects in meiosis were explained by severe depletion of the GFP-tailswap CENH3 variant at meiotic kinetochores (the same protein is loaded normally in mitosis). Depletion of CENH3 at centromeres also compromised the recruitment of the essential kinetochore protein Mis12. Our results thus reveal that centromeres have a meiosis-specific assembly mechanism which involves the CENH3 tail domain. This previously unsuspected pathway may play a role in modulating kinetochore dynamics to ensure differential centromere behaviour during meiosis.
Results
Viable but sterile cenh3 variants suggest that CENH3 function differs between mitosis and meiosis
We previously observed sterility in GFP-tailswap plants but not in plants expressing GFP-CENH3, suggesting that the N-terminal tail of CENH3 might have a specific role in plant reproduction [14]. The sterile phenotype of GFP-tailswap could result from the absence of the CENH3 tail, or from the presence of the H3.3 tail. To differentiate between these possibilities, we created a chimera in which the A. thaliana CENH3 tail was replaced with an unrelated CENH3 tail domain from maize (Zea mays), and transformed it into cenh3-1 heterozygotes (Figure 1A). This GFP-maizetailswap protein was targeted to kinetochores and rescued the embryo-lethal phenotype of cenh3-1. In contrast, full-length maize CENH3 protein was targeted to A. thaliana kinetochores but failed to rescue cenh3-1 embryo lethality [13]. Complemented GFP-maizetailswap plants showed the vegetative phenotype previously seen in GFP-tailswap plants but were even more sterile than GFP-tailswap plants (although one partially fertile GFP-maizetailswap plant was recovered) (Figure 1A) [13].
Fig. 1. Altering the CENH3 N-terminal tail domain leads to defects in meiotic chromosome segregation. 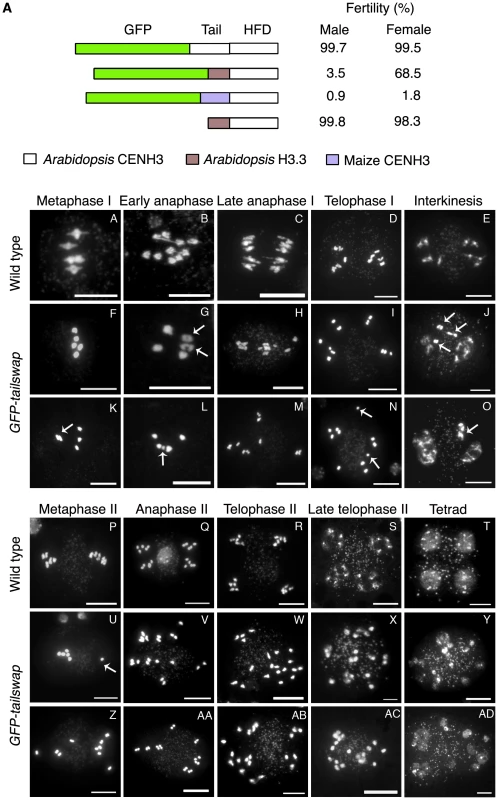
a) CENH3 transgenes tested for fertility in a cenh3-1 homozygous mutant background. The male fertility was examined by Alexander staining. Viable pollen stains pink/red. Female fertility was judged by differential intereference contrast (DIC) microscopy of embryo sacs from at least 100 cleared mature ovules per genotype (Figure S1A). Single cell arrested ovules and ovules without an embryo sac (Figure S1B) were counted as non-viable, and ovules with 7–8 celled embryo sacs (Figure S1B) were counted as viable. Viable ovules may be haploid or aneuploid. b) Male meiotic chromosome spreads from wild type and GFP-tailswap plants. Metaphase I bivalents in the mutant are oval/round in shape, lacking the rhombus shape that indicates tension in wild type (compare A and F). Some metaphase I cells showed chromosomes that failed to congress to the spindle midzone (arrowed in K). Chromosome segregation at anaphase I is random in GFP-tailswap (G to I, L to N). Asynchronous homolog separation was seen at anaphase I (arrowed in G), and premature sister chromatid separation was also seen in meiosis I (arrowed in N). Decondensation at interkinesis was frequently delayed, especially for lagging chromosomes near the spindle midzone (arrowed in J, O). Metaphase II cells in the mutant show random chromosome alignment (U, Z). U shows one univalent (arrowed) and four bivalents plus the remaining univalent on the other side of the cell. Anaphase II chromosome segregation is random (V–X, AA–AC). Tetrad equivalent stages in GFP-tailswap (Y, AD) show several small nuclei instead of the expected four uniform nuclei seen in wild type. Scale bars −1 µm. A GFP tag can have a deleterious effect on CENH3 function [15]. Self-pollinated GFP-CENH3 plants are phenotypically indistinguishable from wild type and fully fertile, indicating that the GFP tag does not interfere with meiosis. We constructed an untagged tailswap transgene to test the role of the CENH3 N-terminus in a protein lacking an N-terminal GFP (Figure 1A). Interestingly, cenh3-1 plants expressing a tailswap transgene without GFP were viable and fertile, indicating that the meiosis-specific role of the CENH3 tail domain is evident only when protein function is compromised by the presence of an N-terminal GFP tag.
Together, these results suggest that either the H3.3 tail or the maize CENH3 tail in the place of the native Arabidopsis CENH3 tail can severely compromise plant reproduction when combined with a GFP tag. This is especially interesting because the CENH3 tail is not required for centromere localization during mitosis, and is extremely fast-evolving [9], [16].
Sterility in tailswap cenh3 variants is caused by meiotic chromosome segregation defects
Sterility in GFP-tailswap plants could be caused by meiotic defects (during sporogenesis), or by later defects in post-meiotic cell divisions (gametogenesis). To investigate the cause of sterility, we analyzed the course of male meiosis in chromosome spreads from anthers. The major early events of meiotic homolog pairing and recombination appeared normal in GFP-tailswap (Figure S2). In prophase I, progressive condensation of chromosomes, homolog pairing, chiasmata formation and subsequent desynapsis of homologs (at diplotene) were similar in GFP-tailswap (n = 317) and wildtype meioses (Figure S2).
The first defects in GFP-tailswap were seen in metaphase I (Figure 1B). In most mutant cells (98/112), bivalent chromosomes congressed normally to the spindle midzone (Figure 1B, panel F). However, a few (14/112) showed alignment defects such as widely spaced metaphase plates, and unaligned bivalent chromosomes (Figure 1B, panel K). Chromosomes can congress to the metaphase plate in the absence of a centromere [17]. Such movements may be driven by chromosome arm-associated kinesins that are independent of the presence of a functional kinetochore [18]. Plant genomes do not contain chromokinesins, the animal proteins that perform this role, but functional counterparts may exist.
A striking defect was seen in the shape of chromosomes during the metaphase I to anaphase I transition. In wild-type meiosis, bivalent chromosomes at this stage assume a rhombus - or linear-shaped configuration caused by tension between spindle microtubules pulling on the kinetochore and chiasmata that hold homologous chromosomes together (Figure 1B, panel A). In GFP-tailswap meiocytes, wild-type metaphase configurations were rarely observed.
Instead, bivalents were oval and irregularly shaped, resembling prometaphase I stage meiocytes (n = 79) (Figure 1B, panels F and K). Prometaphase stage chromosomes are rarely seen in wildtype meiotic spreads (in a total of 392 prophase stage meiocytes, only 4 of this type were observed) because this stage has only a short duration before the onset of metaphase I. Our observations suggest that meiocytes in GFP-tailswap stall at the prometaphase stage and proceed directly to anaphase I without going through a typical metaphase-anaphase transition stage at which tension is exerted by the spindle.
During anaphase I, GFP-tailswap bivalents frequently segregated both homologs to the same side of the spindle, in addition to the normal behavior of resolving homologs and segregating them to opposite poles (Figure 1B, panels G and L). In wild type, pulling forces from the meiosis I spindle help to resolve homologs at anaphase. In GFP-tailswap, homolog separation was more asynchronous than in wild type, yielding cells that contained a mixture of bivalents and univalents (Figure 1B, panels G, H, L and M). The bivalents then separated their homologs at a later stage of anaphase I (Figure 1B, panel M). Some cells contained lagging chromosomes at the spindle midzone, supporting the idea that attachment to the meiotic spindle is impaired (Figure 1B, panels J and O). In rare instances (4/103), sister chromatids separated prematurely during meiosis I (in contrast to normal separation at anaphase II) (Figure 1B, panel N). As a result of random chromosome segregation, meiosis I in GFP-tailswap yielded predominantly unbalanced dyads with 6-4, 7-3 configurations e.t.c. instead of the 5-5 segregation that is universally seen in wild type (reductional segregation to give 5-5 dyads was seen in 6% of mutant cells) (Figure 1B, panels J and O).
At interkinesis, an intermediate stage between meiosis I and II, wild-type chromosomes decondense and recondense. In GFP-tailswap, decondensation and recondensation were delayed in some chromosomes (especially those located at or near the midzone) (Figure 1B, panels J and O). Instead of regrouping and aligning on the metaphase plate during meiosis II, GFP-tailswap chromosomes remained scattered throughout the meiocyte in a manner similar to anaphase I chromosomes in the mutant (Figure 1B, panel Z). Anaphase II in wild type A. thaliana separates sister chromatids to form four groups of 5 chromosomes, each of which contains one haploid genome. In GFP-tailswap, sister chromatids separated while chromosomes were scattered, showing that anaphase II begins without correct chromosome alignment on the metaphase plate (Figure 1B, panels V and AA). Chromosome decondensation also occurred at dispersed locations throughout the cell (Figure 1B, panels X and AC). Instead of the wild type tetrad containing four haploid nuclei, GFP-tailswap meiocytes after meiosis II are polyads containing many small nuclei (in A. thaliana, cytokinesis occurs after the tetrad is formed) (Figure 1B, panels Y and AD). Analysis of male meiosis in GFP-maizetailswap plants revealed meiotic chromosome segregation defects similar to those in GFP-tailswap plants (Figure S3). Based on the chromosome segregation phenotypes described above, it is clear that sterility in GFP-tailswap and GFP-maizetailswap is caused by a severe meiosis-specific defect in centromere function.
Centromere function is required to compact segregating chromosomes into a single nucleus in microspores
Analysis of microspores (pollen grains) from GFP-tailswap plants revealed that each contained 1–8 nuclei instead of the single nucleus that is always seen in wild type (Figure 2A–2F). These micronuclei varied in size, suggesting that they contained different numbers of chromosomes. Fluorescence in situ hybridization (FISH) using a probe that recognizes the 180 bp centromere tandem repeats confirmed that each micronucleus contained from 1–4 chromosomes (Figure 2J–2L). This observation suggests that randomly scattered chromosomes that lie in close proximity reassemble their own nuclear envelope at the end of telophase II, resulting in multiple micronuclei within each microspore. A similar defect has been reported in A. thaliana separase (esp) mutants defective in the enzyme that releases sister chromatid cohesion [19]. In mammalian somatic cells, micronuclei formation is triggered by the depletion of factors required for chromosome segregation [20]–[22]. Formation of micronuclei might be a general feature of perturbations that drastically affect chromosome movement in mitosis or meiosis.
Fig. 2. Lack of centromere function in meiosis causes micronuclei to form in GFP-tailswap pollen. 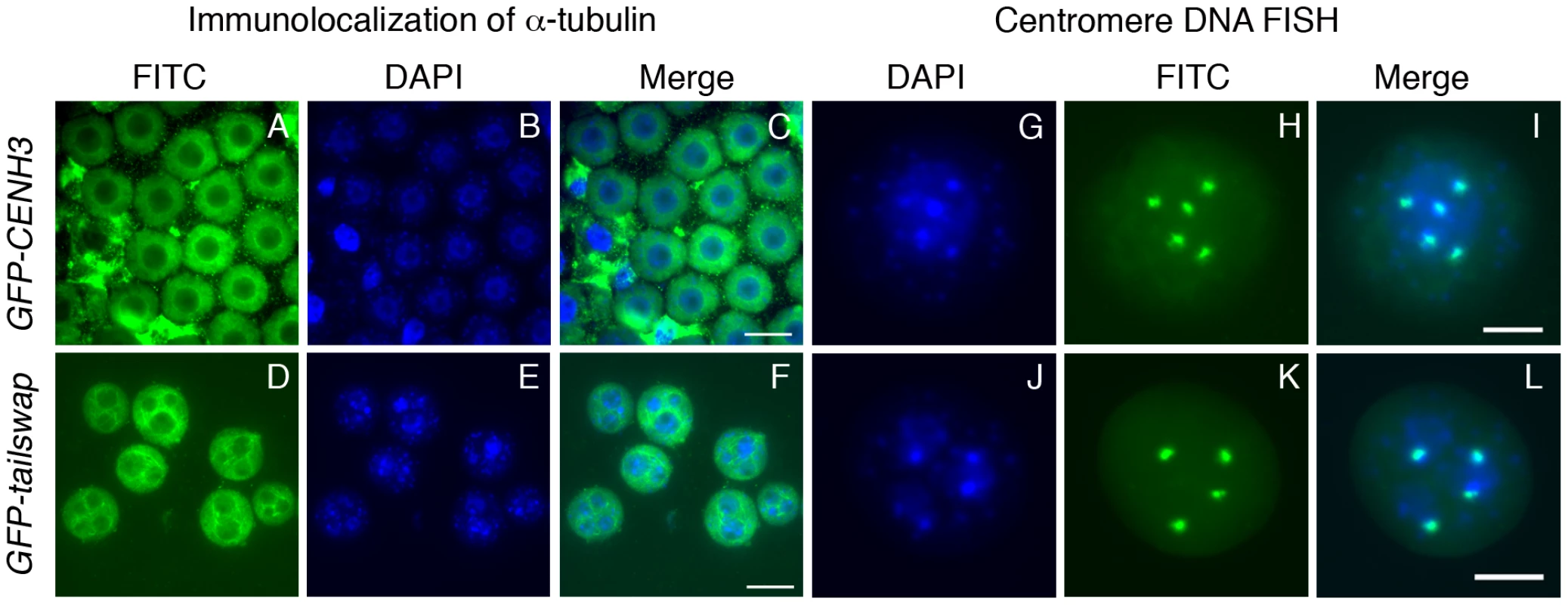
Immunolocalization of alpha-tubulin outlines the nuclear envelope in microspores of GFP-CENH3 and GFP-tailswap pollen (A–F). GFP-tailswap pollen contains multiple micronuclei. Centromere DNA FISH shows that micronuclei contain 1–2 chromosomes each, as opposed to 5 chromosomes in a normal A. thaliana haploid pollen genome (G–L). The pollen grain shown in J–L has three micronuclei. Two contain one chromosome each, while the third contains two chromosomes. Viable male and female gametes in GFP-tailswap are expected to contain a single nucleus with a haploid genome of five chromosomes, because most (95%) of the viable progeny from self fertilized GFP-tailswap are diploid [14], [13]. This can be contrasted with several plant meiotic mutants which show random chromosome segregation during meiosis [23]–[25]. After meiosis II, these mutants often contain more than the normal four nuclei within a single meiocyte. However, the microspores resulting from such meiocytes usually contain a single nucleus, in contrast to the multinucleate microspores of separase mutants and GFP-tailswap. In A. thaliana mutants with general meiotic defects (for example, ask1 and spo11), a high fraction of viable gametes are aneuploid [24], [25]. When such plants are self-fertilized, a high fraction of viable progeny are aneuploid. Formation of micronuclei in the microspore selects against otherwise viable aneuploid pollen in GFP-tailswap, so >90% of progeny obtained by self-fertilization are diploid, despite random chromosome segregation in meiosis. We conclude that functional centromeres are required to package segregating chromosomes into a single pollen nucleus, and are thus necessary for accurate transmission of a haploid genome after meiosis.
Chromosomes in GFP-tailswap meiosis appear to lack tension from the spindle and show abnormal alignment in metaphase I
A defect in kinetochore attachment to spindle microtubules in GFP-tailswap is suggested by the lack of apparent tension in metaphase I chromosomes and by random chromosome segregation (in the absence of pairing or recombination defects). The distance between opposing kinetochores (interkinetochore distance) during metaphase is a more precise measure of tension generated by the spindle during mitosis. To investigate interkinetochore distance in GFP-tailswap meiosis I, we used FISH with a centromere tandem repeat probe (Figure 3A). In meiotic chromosome spreads, the outer limit of centromere DNA staining indicates the likely position of the kinetochore. Wild type cells at the metaphase I to anaphase I transition showed centromere DNA foci whose outer edges were separated by a 405±68 nm distance (n = 15 bivalent chromosomes). Centromere DNA was clearly stretched out on either side of the non-hybridizing DNA representing chromosome arms (Figure 3A, panel D). In GFP-tailswap chromosomes at an equivalent stage, centromere DNA extremities were much closer to each other at 234±30 nm (n = 15 bivalent chromosomes) (Figure 3A, panel H). Furthermore, the centromere DNA stretch characteristic of wild-type chromosomes under tension was not seen in the mutant. We conclude that GFP-tailswap kinetochores may not be efficiently pulled by spindle microtubules.
Fig. 3. Reduced inter-kinetochore distance and meiotic spindle defects suggest lack of kinetochore function in GFP-tailswap. 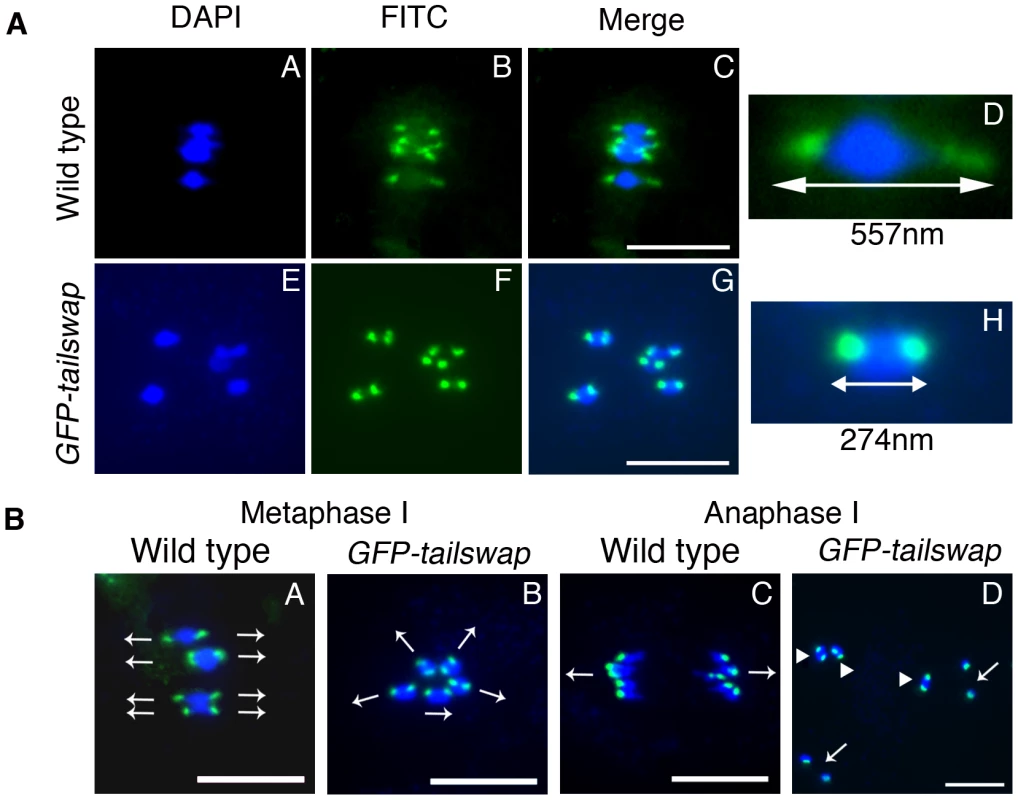
a) Centromere DNA FISH from metaphase I in wild type and in GFP-tailswap. Blue = DNA (DAPI), green = centromere DNA FISH (FITC). GFP-tailswap bivalents lack the centromere stretch exerted by the spindle in wild type, and have reduced inter-kinetochore distance. Representative bivalents are magnified in D and H. Scale bars −1 µm. b) Centromere DNA FISH shows random orientation of bivalent chromosomes in GFP-tailswap meiosis I. Blue = DNA (DAPI), green = centromere DNA FISH (FITC). Metaphase I chromosomes are frequently aligned at unusual angles in the mutant (B). Anaphase I chromosomes show random alignment and premature sister chromatid separation (D). Arrows in A and B show presumed orientation of sister centromeres. Arrows in D indicate separated univalents, while arrowheads show intact bivalents. Scale bars −1 µm. FISH analysis of metaphase I meiocytes in GFP-tailswap also revealed abnormal alignment of centromeres with respect to the cell plate (Figure 3B). In wild-type meiosis I, centromeres from homolog pairs align in a direction perpendicular to the future division plane. In GFP-tailswap they aligned in multiple directions, presaging the random chromosome segregation that occurs at anaphase I. This data further supports the hypothesis that spindle microtubules fail to pull on kinetochores in GFP-tailswap, leading to a lack of tension and incorrect chromosome orientation during meiosis I.
GFP-tailswap protein is loaded normally onto mitotic kinetochores but not onto meiotic kinetochores
As meiotic kinetochores appeared to be non-functional in GFP-tailswap plants, we investigated whether the GFP-tailswap variant of CENH3 was localized to meiotic kinetochores (Figure 4 and Figure 5). We have previously shown that GFP-tailswap protein is localized normally to mitotic kinetochores, and that mitosis is accurate in GFP-tailswap plants [13]. A. thaliana male meiocytes can be extruded as a cell conglomerate by gently squeezing anthers [26]. We imaged meiocytes from GFP-CENH3 plants, and found that the GFP-tagged CENH3 protein was visualized at kinetochores in all stages of meiosis, as well as in haploid pollen grains. However in GFP-tailswap meiocytes, the protein was only faintly visualized at kinetochores during premeiotic and early prophase I stages (Figure 4 and Figure 5) and was not detected in later stages of meiosis I (starting from pachytene) and meiosis II. Depletion of GFP-tailswap from meiotic kinetochores contrasted with somatic cells from the same anther, which showed GFP fluorescence at mitotic kinetochores that appeared identical to wild-type (Figure S4).
Fig. 4. GFP-tailswap protein is depleted from kinetochores during pre-meiotic interphase. 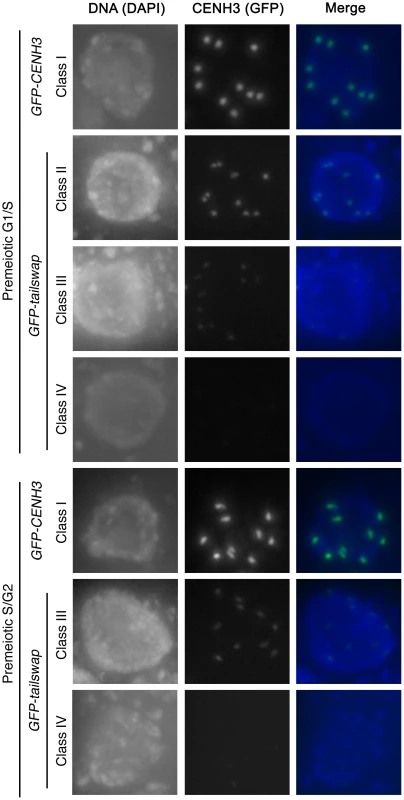
Meiocytes from anthers of GFP-CENH3 and GFP-tailswap were imaged using identical exposure times. Flattened projections of several stacked images are shown. The GFP-CENH3 protein showed bright fluorescence at kinetochores in all meiocytes (class I). GFP-tailswap protein always showed reduced fluorescence relative to GFP-CENH3. Three classes of GFP fluorescence were observed in GFP-tailswap: faintly visible (class II, 7%), barely detectable (class III, 47%), and undetectable (class IV, 46%). Fig. 5. Depletion of GFP-tailswap protein from kinetochores continues progressively during meiosis. 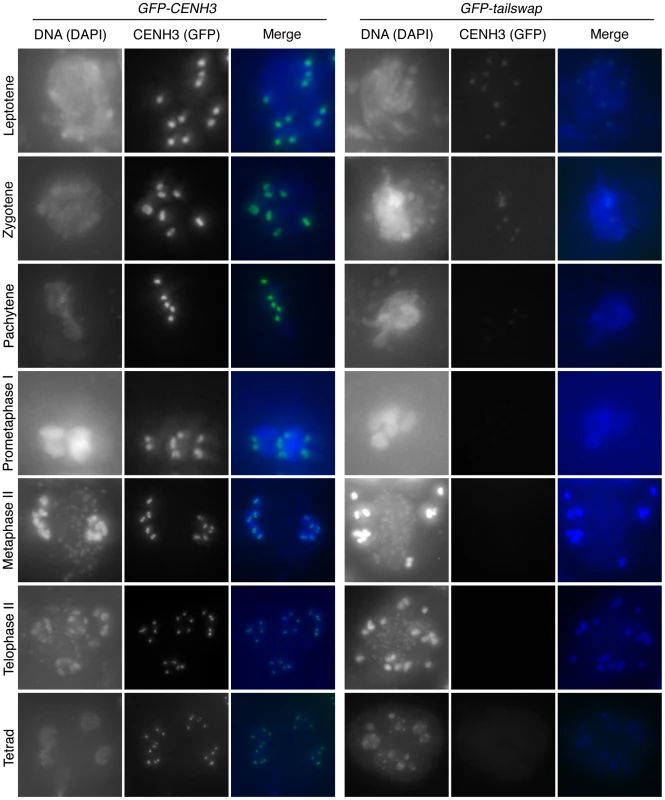
a) Dynamics of kinetochore GFP-CENH3 and GFP-tailswap proteins during meiosis were visualized in anthers. Flattened projections of several stacked images are shown. GFP-CENH3 is visible at uniform intensity at kinetochores throughout meiosis. GFP-tailswap is barely detectable or undetectable in leptotene and zygotene (comparable to class III and class IV in Figure 4). In pachytene and subsequent stages, we did not detect kinetochore GFP fluorescence in GFP-tailswap meiocytes. To further understand the dynamics of the GFP-tailswap protein during meiosis, we analyzed kinetochore GFP fluorescence at all meiotic stages from anther squashes (identification of these stages is described in the Materials and Methods). In premeiotic interphase of GFP-CENH3 meiocytes, kinetochore GFP fluorescence was bright and uniform (Figure 4) (n = 93, 4 plants). GFP-tailswap meiocytes never showed bright kinetochore GFP fluorescence (Figure 4) (n = 119, 5 plants). Instead, we categorized them into three classes of reduced fluorescence, where GFP-CENH3 fluorescence is class I: 7% of meiocytes showed reduced kinetochore signal (class II), 47% had barely detectable fluorescence (class III), and the remaining 46% showed no GFP at kinetochores (class IV). This observation suggests that the GFP-tailswap protein is not replenished during premeiotic interphase, and that GFP-tailswap protein inherited from the somatic precursor cell is gradually removed from the centromere. Depletion of GFP-tailswap protein from meiocytes continued in subsequent stages of meiosis I (Figure 5 and Figure 6). Kinetochore GFP signal gradually disappeared during leptotene and zygotene stages of early prophase I (Figure 5). From late pachytene stage onwards until the completion of meiosis we could not detect GFP signal in any meiocytes (Figure 5 and Figure 6). We also used anti-GFP antibodies to immunolocalize GFP-CENH3 and GFP-tailswap proteins during meiosis, and found similar results (Figure S5). Residual GFP-tailswap protein may remain at kinetochores at a level below the detection limit. To verify that the GFP-tailswap mRNA was correctly spliced during meiosis, we extracted meiocytes from GFP-CENH3 and GFP-tailswap anthers using a capillary-based method [26]. RT-PCR and subsequent sequencing of cDNA showed that the GFP-tailswap mRNA was identically spliced in somatic and meiotic cells (Figure S6). As CENH3 is essential, specific depletion of GFP-tailswap from meiotic kinetochores explains the chromosome missegregation that leads to sterility in GFP-tailswap plants.
Fig. 6. Percentage of <i>GFP-CENH3</i> and <i>GFP-tailswap</i> meiocytes showing particular classes of GFP fluorescence at kinetochores. 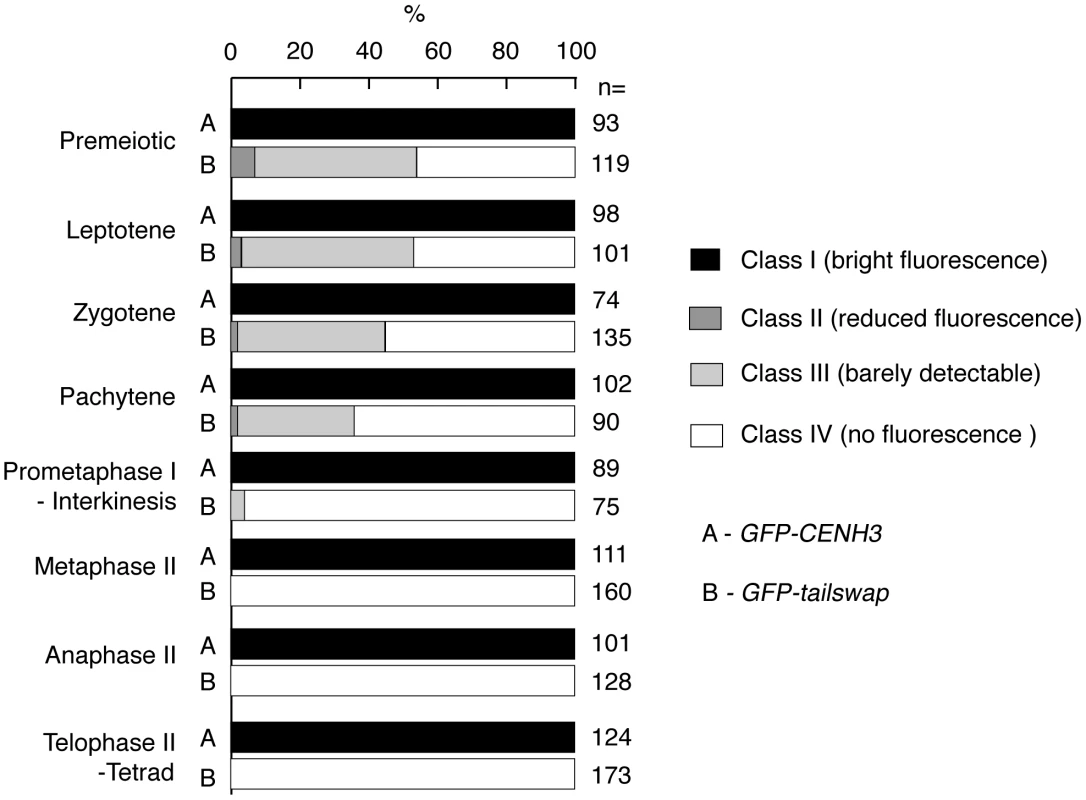
The GFP-maizetailswap protein was also absent from meiotic kinetochores but present normally at mitotic kinetochores. Plants that co-express either GFP-tailswap or GFP-maizetailswap along with a wild-type endogenous CENH3 gene were fully fertile. However, the GFP-tailswap and GFP-maize-tailswap proteins were poorly loaded onto meiotic kinetochores even in the presence of functional endogenous CENH3 (importantly, GFP-CENH3 loads normally in the presence of wild type CENH3) (Figure S7). Thus, the CENH3 tail domain appears to be required specifically for recruitment of the protein to meiotic kinetochores (when protein function is compromised by a GFP tag).
Kinetochore proteins that act only during meiosis have been described [1]. To our knowledge, this is the first example of an alteration in CENH3 that causes a meiosis-specific defect but allows for accurate mitosis. In A. thaliana, CENH3 is recruited to mitotic kinetochores after S phase, to replenish kinetochore CENH3 levels that were diluted by DNA replication [16]. If the GFP-tailswap and GFP-maizetailswap proteins were simply unable to replenish kinetochores after pre-meiotic DNA replication, we would expect to see half the GFP signal found at mitotic kinetochores in meiosis I cells. The fact that these proteins are greatly reduced at almost all meiotic kinetochores suggests that CENH3 chromatin is actively disassembled during meiosis in mutant plants. We believe that the GFP-tailswap and GFP-maizetailswap mutants reveal a meiosis-specific kinetochore assembly pathway whose existence was previously unsuspected.
Depletion of the GFP-tailswap CENH3 variant from meiotic kinetochores causes loss of MIS12
CENH3 is required to recruit a large number of essential kinetochore proteins in other organisms. To further characterize the effects of the GFP-tailswap variant on kinetochore assembly, we performed immunostaining on GFP-tailswap and control GFP-CENH3 anther squashes with antibodies raised against the A. thaliana kinetochore proteins CENP-C and MIS12 [27], [28]. CENP-C antibodies did not yield specific staining of kinetochores in meiocytes from either GFP-CENH3 or GFP-tailswap plants. However, MIS12 staining was observed at kinetochores in GFP-CENH3 meiocytes (n = 44), but not in GFP-tailswap meiocytes (n = 33) (Figure 7C and 7K). Somatic cells from both GFP-CENH3 and GFP-tailswap plants showed kinetochore localization of MIS12 (Figure 7G and 7O). Although MIS12 may be recruited in a CENH3-independent way in human cells [20], our results show that loss of A. thaliana CENH3 in meiosis also depletes MIS12 from the kinetochore. As MIS12 is a component of the KMN network that connects kinetochores to spindle microtubules, we predict that this will compromise kinetochore-microtubule attachment [29]. Furthermore, MIS12 is important for mono-orientation during meiosis in maize [4]. In summary, severe depletion of the GFP-tailswap protein during meiosis and downstream effects on kinetochore assembly can explain the chromosome segregation defects observed in the mutant.
Fig. 7. Depletion of GFP-tailswap protein from meiotic kinetochores causes removal of MIS12. 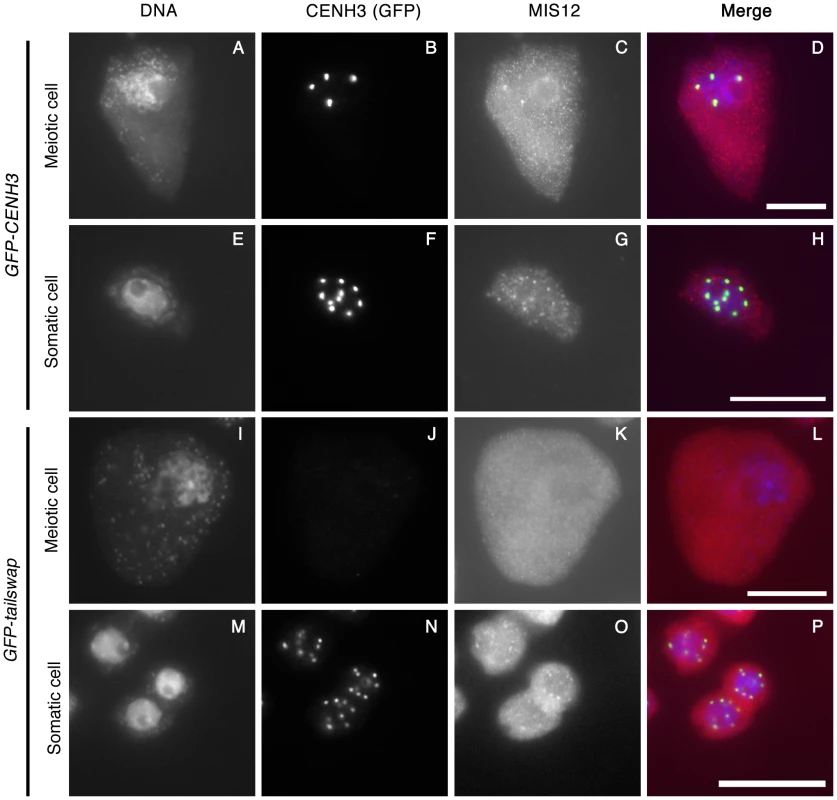
GFP-CENH3, GFP-tailswap and MIS12 proteins were immunolocalized in anthers during the pachytene stage of meiosis with anti-GFP and anti-MIS12 antibodies. Somatic cells from the same anther are shown as a control. GFP-CENH3 and MIS12 were visualized at both meiotic and somatic kinetochores of GFP-CENH3 plants. In GFP-tailswap plants, GFP-tailswap and MIS12 were both undetectable in GFP-tailswap meiotic kinetochores but can be seen in somatic kinetochores. Scale bars −5 µm. Meiotic spindles are disordered in GFP-tailswap
Depletion or removal of CENH3 or other essential kinetochore proteins from the centromere results in compromised kinetochore function, which destabilizes the formation of a normal spindle [30], [31]. To gain insight into kinetochore-spindle microtubule interactions in GFP-tailswap, we visualized microtubules in meiocytes with anti-alpha-tubulin antibodies (Figure 8). A bipolar spindle is formed during metaphase I in mutants, but it was longer and more disorganized than the wild-type meiosis I spindle (Figure 8, panel E and I). Although we cannot conclude that kinetochores in GFP-tailswap are completely non-functional, our data is consistent with previous studies showing that kinetochores are not required to assemble a bipolar spindle in either mitosis or meiosis [32], [33].
Fig. 8. Immunolocalization of α-tubulin in wild type and GFP-tailswap meiocytes. 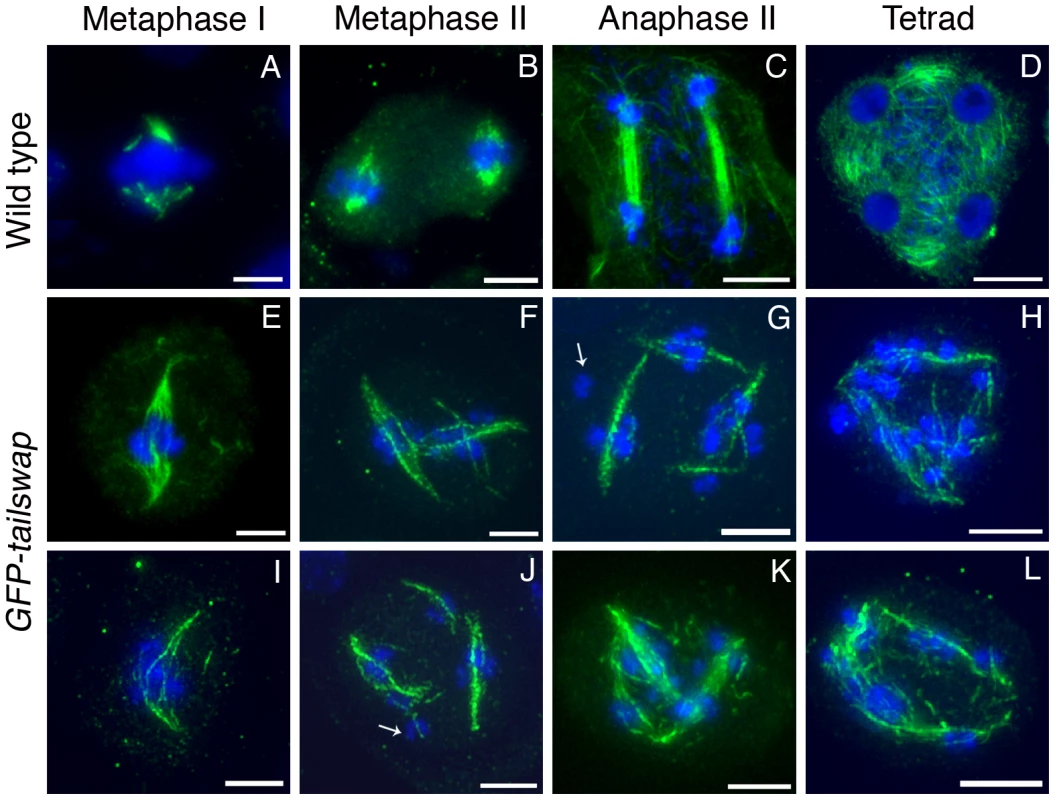
Anther meiocytes from wild type and GFP-tailswap plants were stained with anti-tubulin antibodies. Metaphase I spindles are disorganized and longer in GFP-tailswap, and may contain fewer microtubules (E and I). Meiosis II cells in GFP-tailswap often contain more than two spindles (F–G, J–K). Spindle appearance and orientation are disordered, and may fail to include some chromosomes (arrowed in G). Tetrad equivalent cells (H, L) lack the radial microtubule system that surrounds the four haploid nuclei in wild type. Scale bars −5 µm. It is possible that interactions between spindle microtubules and chromosome arm-binding kinesins can organize a spindle in the absence of fully functional kinetochores. In general, longer spindles are correlated with smaller kinetochores, and with abnormal chromosome movement [20]. This provides further evidence that kinetochores are functionally compromised in GFP-tailswap meiocytes.
Meiosis II spindles were even more disorganized in GFP-tailswap (Figure 8). Many meiocytes at this stage contained more than two spindles (multipolar spindles). Spindles were frequently perpendicular to each other or generally lacking the neat parallel appearance of spindles in wild-type meiosis II (Figure 8, panels F, G, J and K). These phenotypes may explain the inability of chromosomes to align on the metaphase II plate. Some kinetochores in GFP-tailswap meiocytes appeared to lack nearby spindle microtubules (Figure 8, panels G and J). However, it is difficult to conclude from our data that kinetochores fail to bind stably to spindle microtubules in the mutant, because the detection limit of tubulin staining in meiocytes is unknown. Furthermore, we cannot easily distinguish kinetochore microtubules from interpolar microtubules.
GFP-tailswap recruitment to meiotic kinetochores is not restored by imposing mitosis-like chromosome behavior during meiosis I
If GFP-tailswap has a specific defect in recruitment to meiotic centromeres, can we suppress this phenotype by imposing a mitosis-like behavior on kinetochores in meiosis I? REC8 is a meiosis-specific cohesin subunit that functionally replaces its mitotic counterpart RAD21. REC8 is required to hold sister kinetochores together and force them to orient towards the same side of the spindle [3], [2]. In S. pombe rec8 mutants, sister kinetochores in meiosis I show bipolar attachment to the spindle and segregate apart from each other, much as they do in mitosis [34]. The role of REC8 in mono-orientation is conserved in plants, as shown by A. thaliana rec8 spo11 mutants (the spo11 mutation is needed to prevent chromosome fragmentation, as rec8 mutants cannot repair meiotic double-stranded breaks) [3]. To test whether CENH3 recruitment employs the mitotic loading pathway when REC8 is removed from meiotic kinetochores, we generated rec8 spo11-1 cenh3-1 triple mutant plants carrying the GFP-tailswap transgene. If removing REC8 in GFP-tailswap mutants fully converted kinetochores from meiotic to mitotic behavior, we expected to see two experimental readouts. First, GFP-tailswap would be recruited to meiotic kinetochores in a manner similar to GFP-CENH3. Second, chromosomes in meiosis I would show a mitosis-like segregation pattern similar to the rec8 spo11-1 mutant, because functional centromeres would be restored.
GFP-tailswap protein was not loaded onto meiotic kinetochores in rec8 spo11-1 cenh3-1 GFP-tailswap plants, showing that REC8 removal does not restore the mitotic CENH3 loading pathway during meiosis (Figure S8). In rec8 spo11-1 mutants, chromosomes remain as univalents during prophase I and separate their sister chromatids at anaphase I as they do in mitosis (10-10 segregation instead of 5-5 segregation) (Figure 9 and Figure S8) [3]. By contrast, meiosis I in rec8 spo11-1 cenh3-1 GFP-tailswap plants showed random segregation of the unpaired univalent chromosomes, confirming that kinetochores were still non-functional (Figure 9 and Figure S8). The inability of GFP-tailswap to load onto meiotic kinetochores suggests that the meiosis-specific CENH3 loading pathway is still functional even if we impose mitotic chromosome-like behaviour in meiotic cells by removing REC8 from centromeres.
Fig. 9. Removing the meiosis-specific cohesin REC8 does not restore meiotic kinetochore function in GFP-tailswap. 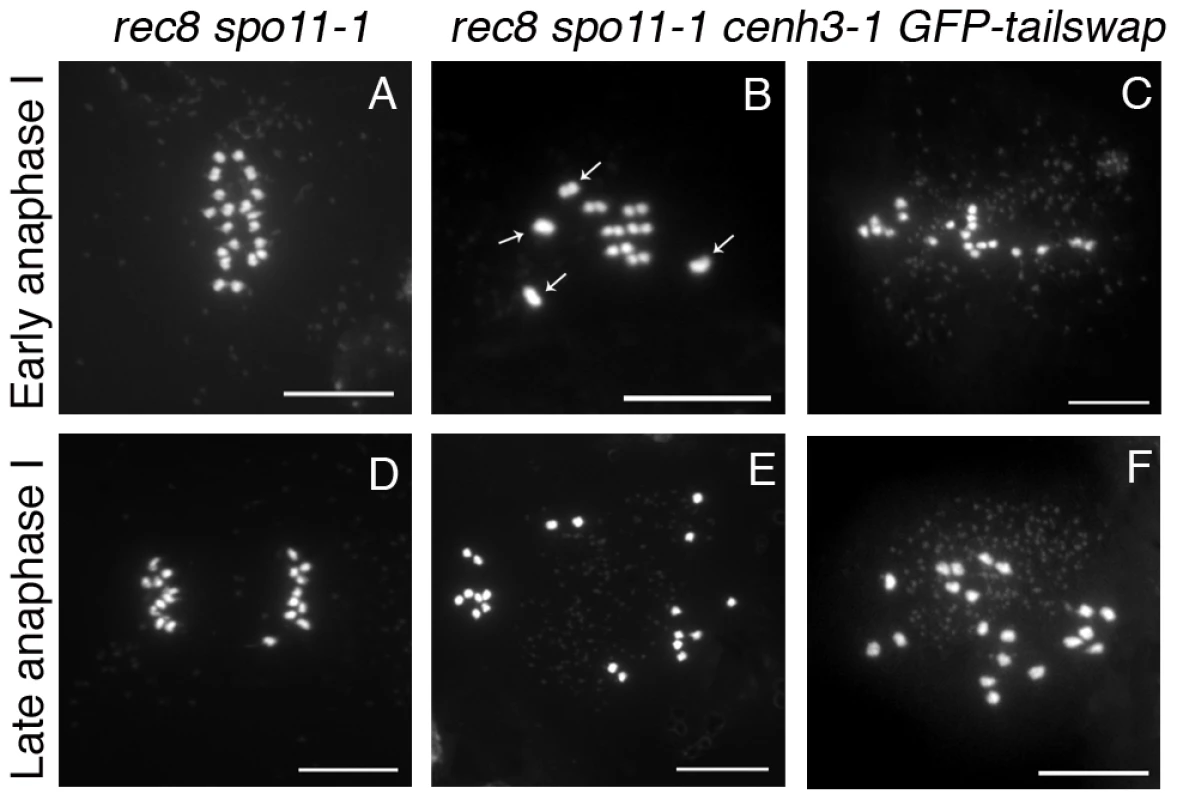
Chromosome spreads from male meiosis in rec8 spo11-1 and rec8 spo11-1 cenh3-1 GFP-tailswap. Anaphase I in rec8 spo11-1 shows orderly separation of sister chromatids that is similar to mitosis, because the rec8 mutation converts kinetochores to a mitosis-like behavior (A, D). Anaphase I in rec8 spo11-1 cenh3-1 GFP-tailswap shows random segregation of univalent chromosomes (B, C, E, F). This is consistent with the observation that removing REC8 does not restore loading of the GFP-CENH3 protein (Figure S6). GFP-tailswap reloads onto mitotic kinetochores after meiosis
Unlike animal gametes, the haploid cells produced by plant meiosis undergo mitotic divisions to generate mature gametes. Male meiosis in GFP-tailswap produces a few viable pollen grains, presumably in those rare cases where a complete haploid genome is clustered into a single microspore nucleus. These haploid microspores (and rarely, viable aneuploid microspores) must undergo mitotic divisions, so we asked whether the GFP-tailswap protein was recruited to kinetochores after meiosis, when mitosis resumes. Although a large majority of GFP-tailswap microspores (407/504 or 81%, n = 6 plants) are dead due to micronuclei formation and did not show GFP fluorescence, remaining microspores (97/504 or 19%) in the mutant showed GFP fluorescence at kinetochores at a level equivalent to wildtype (Figure 10A and 10B).
Fig. 10. GFP-tailswap protein reloads onto centromeres after meiosis, when mitosis resumes. 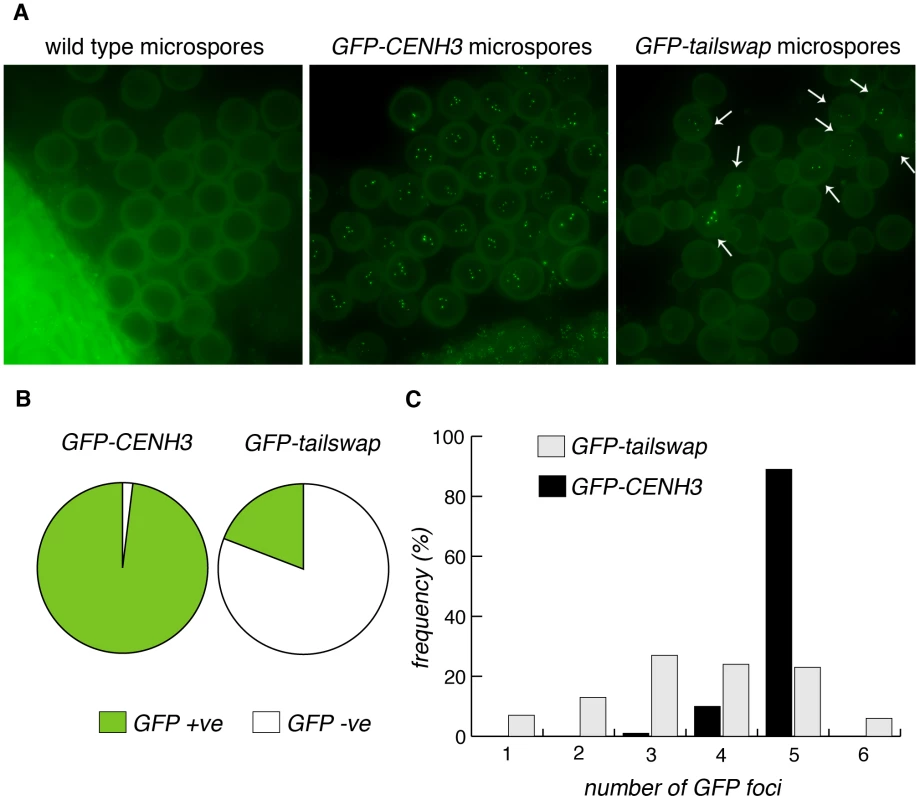
a) Microspores of wild type, GFP-CENH3 and GFP-tailswap. Tetrad equivalent stages in GFP-tailswap do not show any GFP fluorescence at meiotic kinetochores (Figure S4). However, GFP-tailswap protein reloads onto kinetochores in a small fraction of microspores, when haploid mitotic divisions are expected to resume. b) Frequency of GFP-positive and -negative microspores from GFP-CENH3 and GFP-tailswap plants. c) The number of GFP foci in each GFP-positive spore is shown. Some of the microspores that showed kinetochore fluorescence contained fewer than five GFP foci, and are unlikely to contain a full haploid genome (these microspores should lead to inviable pollen) (Figure 10C). Kinetochore localization of GFP-tailswap in haploid spores was brighter than that seen in early stages of meiosis (class II and class III). Furthermore, we did not observe GFP-tailswap at kinetochores from pachytene until telophase II of meiosis. We conclude that GFP-tailswap is loaded afresh onto mitotic kinetochores after meiosis and is not simply carried over from those meiocytes that showed faint localization. This result confirms the existence of two distinct kinetochore assembly pathways, for mitosis and meiosis respectively. It also raises the question of how the GFP-tailswap variant of CENH3 recognizes mitotic kinetochores after meiosis. If GFP-tailswap is truly removed from kinetochores in meiosis, there must be a targeting mechanism that does not require the prior presence of CENH3 in centromeric nucleosomes.
Discussion
Centromeres are differentially configured during mitotic and meiotic cell divisions, resulting in separation of either sister chromatids or homologous chromosomes during anaphase. Segregation of sister chromatids to the same spindle pole in meiosis I depends on meiosis-specific proteins such as monopolin components and Moa1p, but may also involve specialized functions of constitutive kinetochore proteins. The essential kinetochore protein CENP-C recruits Moa1p in S. pombe [35]. In maize, MIS12 has a role in fusing kinetochores, facilitating mono-orientation during meiosis I [4]. In addition, linker histone H1 variants have a meiosis-specific role in plants and are present only at meiotic centromeres in lily [36]–[39]. CENH3 is essential in both mitosis and meiosis, but our viable yet sterile A. thaliana mutants have uncovered a meiosis-specific loading pathway for CENH3 that most likely depends on its fast evolving N-terminal tail domain. Male and female meiosis are differentially affected in GFP-tailswap plants, although the basis for this is unclear [14]. Sex specific meiotic chromosome segregation defects have also been observed in tobacco plants defective for linker histone H1 variants [39].
During DNA replication, CENH3 nucleosomes are randomly partitioned between replicated sister chromatids and voids created in chromatin are filled by fresh loading of free CENH3. The cell cycle loading time of CENH3, and the chaperones that direct it to kinetochores, can vary between organisms [16], [40], [41] [42]–[45]. Our results may indicate that CENH3 has meiosis-specific chaperones that cannot recruit the GFP-tailswap and GFP-maizetailswap variants. In C. elegans, CENH3/HCP-3 is also differentially recruited during mitosis and meiosis [46]. Furthermore, C. elegans CENH3 has a different distribution from outer kinetochore proteins in meiosis, and is dispensable for meiotic chromosome segregation [46], [47]. These properties are probably related to the holocentric nature of C. elegans chromosomes, whereas the meiosis-specific defect we have found confirms that CENH3 is essential for meiotic segregation of monocentric A. thaliana chromosomes.
Severe depletion of GFP-tailswap at meiotic kinetochores suggests that endogenous CENH3 undergoes a removal and reloading process in meiosis. If this is the case, reloading must be closely coupled to removal, as we never observed meiotic cells from GFP-CENH3 plants that lacked kinetochore fluorescence. There are precedents for dynamic reorganization of CENH3 chromatin during particular developmental stages. In C. elegans meiosis, CENH3/HCP-3 is removed, and kinetochores are assembled de novo when mitosis resumes [46]. Parental CENH3 is also greatly depleted in the fertilized zygote of A. thaliana, and replaced by CENH3 synthesized from the zygotic genome [48]. In an alternative model to explain GFP-tailswap dysfunction, CENH3 at meiotic kinetochores might assume a different conformation that is destabilized by the combination of a bulky tag with the absence of the CENH3 N-terminal tail domain. Meiosis specific kinetochore architecture could involve an altered nucleosome structure (such as a conversion between octameric and tetrameric forms), or a change in the arrangement of CENH3 nucleosomes [49]–[51]. CENH3 levels can be regulated by proteolysis [52], and GFP-tailswap and GFP-maizetailswap proteins may be specifically degraded in meiotic cells (possibly because they are not loaded into centromeric chromatin). The fact that A. thaliana has a centromere DNA structure similar to most animals and plants favors the hypothesis that meiosis-specific CENH3 assembly will be a conserved phenomenon.
The involvement of the CENH3 N-terminal tail in meiosis-specific centromere assembly is intriguing because the tail is so fast-evolving, and because it is dispensable for accurate mitosis. This requirement is not absolute, as the H3.3 tail substitution in the absence of GFP is fertile – others have proposed that a GFP tag on CENH3 might disturb higher order chromatin structure [15]. The CENH3 tail (81 amino acids) is longer than the H3 tail (43 amino acids), so GFP is closer to the histone-fold domain in the GFP-tailswap protein. However, we do not think this is the sole cause of sterility in GFP-tailswap, because GFP-maizetailswap plants are even more sterile, and the maize CENH3 tail (61 amino acids) is longer than the H3 tail. The N-terminal tail of histone H3 can contain many functionally important post-translational modifications, and CENH3 can also be post-translationally modified [53], [54]. Meiosis-specific modifications of CENH3 seem unlikely to be significant, because the amino acid sequence of the tail changes so rapidly, and important modified residues would be expected to be conserved. The N-terminal tail may interact with as yet unidentified meiosis-specific histone chaperones, with other proteins important for mono-orientation, or even with centromere DNA directly [55]. If the latter is the case, this binding must be especially critical during meiosis.
The “centromere paradox” refers to the fact that centromere DNAs and CENH3 are remarkably fast evolving, despite their essential function [56]. It has been proposed that differences between the centromeres of two parents cause hybrid defects when they are crossed, leading to speciation [56]. Centromere differences could reduce the fitness of hybrid offspring by affecting either meiosis or mitosis. We previously reported that a cross between two phenotypically indistinguishable parents with CENH3 differences (GFP-CENH3 and wild type) caused mitotic chromosome segregation errors in the fertilized zygote [14]. Now we have found that meiosis (particularly in the male) can be specifically affected by changes in CENH3, albeit in plants that contain only a single altered CENH3 protein. This result is analogous to the observation that a centromere DNA polymorphism in the monkeyflower Mimulus guttatus can cause male meiotic defects when it is homozygous [57]. We speculate that CENH3 interactions with centromere DNA may be altered by the M. guttatus centromere DNA polymorphism, and that A. thaliana GFP-tailswap protein may fail in meiosis because it cannot interact with centromere DNA appropriately. The existence of a meiosis-specific loading pathway for CENH3 further supports the concept that rapid evolution reduces hybrid fertility by weakening kinetochore function in meiosis.
Materials and Methods
Plant growth and materials
Plants were grown under a 16 hr light/8 hr dark regime at 20°C. GFP tailswap plants have been previously described [14], [13]. Plant transformations used the floral dip method. The rec8 (Ler accession) and spo11-1 (Ws-0 accession) mutants have been described [3]. rec8 spo11-1 cenh3-1 triple mutants expressing GFP-tailswap were generated by selfing REC8/rec8 SPO11-1/spo11-1 CENH3/cenh3-1 plants carrying the GFP tailswap transgene. Primer sequences for genotyping are listed in Table S1.
Plasmids and transgenes
The GFP-maizetailswap transgene fuses the Zea mays CENH3 N-terminal tail (amino acids 1–61) and the A. thaliana CENH3 histone fold domain (amino acids 82–179). GFP-maizetailswap was constructed by overlapping PCR and cloned as a SalI-XbaI fragment into the binary vector CP93 [13]. The tailswap transgene without an N-terminal GFP was constructed by overlapping PCR and cloned into CP93 from SalI to XbaI. Primer sequences for overlapping PCR are listed in Table S1.
Differential interference contrast microscopy
Developmental analysis of unfertilized fixed ovules by differential interference contrast microscopy was performed as described [58].
Meiotic chromosome spreads and FISH
Male meiotic spreads were prepared as described [59] except for a modification in the enzyme cocktail for tissue digestion, which contained 0.3% cellulose and 0.3% pectolyase in 10 mM citrate buffer (pH 4.5). FISH analysis on male meiotic chromosome spreads was performed as described [13]. The distance between centromeres at the metaphase I to anaphase I transition was measured using NIH Image J software.
Immunolocalization analysis
Immunolocalization of alpha-tubulin in meiocytes and microspores was carried out as described [60]. The primary antibody was a mouse monoclonal anti-alpha-tubulin (Sigma T6199). The secondary antibody was a goat anti-mouse IgG (Sigma F0257). Images were captured with a Deltavision deconvolution microscope. Immunolocalization of GFP and MIS12 was performed as described previously [28]. We used an anti-GFP antibody from Acris (R1461P).
GFP localization in meiocytes
To visualize GFP fluorescence in meiocytes, anthers were dissected from unfixed, fresh flower buds using insulin needles in a drop of staining solution (50% glycerol, 1% PBS and 1 µg/ml DAPI). After removing other floral tissues, anthers were fully submerged by fresh addition of 10–20 µl of staining solution onto the slide. A thin coverslip was placed on the slide and gently pressed with the plunger end of the insulin syringe until meiocytes were finely extruded out from the anther sacs. After sealing the coverslip with valap wax (vaseline∶lanolin∶paraffin wax in a 1∶1∶1 ratio), slides were imaged using a Deltavision deconvolution microscope. Images were captured at 60× magnification with an exposure time of 0.5 seconds. Z-stacks with a step size of 0.2 µM were captured and further transformed into two-dimensional (2D) flattened projections using SoftWoRx software (Applied Precision). TIF files were edited using Adobe Photoshop and Illustrator.
In live meiocytes stained with DAPI, it was difficult to accurately pinpoint the early stages of meiosis, especially the premeiotic and early prophase I stages. However, we could gauge the approximate stage by looking for certain landmark phenotypes. In the premeiotic stage, the chromosomes are highly decondensed and diffuse, and thus DAPI stains the whole nucleus. Therefore, premeiotic stage meiocytes were identified as being spherical or round in appearance. In premeiotic stage meiocytes, the G1 phase cells were identified as ones that showed round/spherical GFP fluorescence, whereas in S-G2 phase meiocytes, GFP signals are more elongated as a result of chromosome doubling. We never observed the clear separation of replicated sister kinetochores seen in mitotic G2 cells (paired GFP foci). As meiosis proceeds, chromosomes start to condense. When the round appearance of chromosome mass became more irregular, they were identified as leptotene stage meiocytes. During zygotene, pairing of homologous partners starts and hence there were 5–10 GFP signals corresponding to the centromeres. During pachytene, pairing is complete and thus we detected 5 GFP signals corresponding to 5 fused kinetochores. In pachytene, chromosomes threads are much compact than early stages. From metaphase I onwards, meiotic stages were distinguished by their chromosome segregation behaviour using DAPI staining.
Meiocyte extraction
Male meiocytes were extracted with a microcapillary-based method as described previously [26].
Supporting Information
Zdroje
1. BrarGAAmonA 2008 Emerging roles for centromeres in meiosis I chromosome segregation. Nat Rev Genet 9 12 899 910
2. SakunoTTadaKWatanabeY 2009 Kinetochore geometry defined by cohesion within the centromere. Nature 458 7240 852 858
3. ChelyshevaLDialloSVezonDGendrotGVrielynckN 2005 AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118 Pt 20 4621 4632
4. LiXDaweRK 2009 Fused sister kinetochores initiate the reductional division in meiosis I. Nat Cell Biol 11 9 1103 1108
5. YokobayashiSWatanabeY 2005 The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123 5 803 817
6. Monje-CasasFPrabhuVRLeeBHBoselliMAmonA 2007 Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128 3 477 490
7. ClevelandDWMaoYSullivanKF 2003 Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112 4 407 421
8. BlackBEBassettEA 2008 The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20 1 91 100
9. MalikHSHenikoffS 2003 Phylogenomics of the nucleosome. Nat Struct Biol 10 11 882 891
10. VermaakDHaydenHSHenikoffS 2002 Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol 22 21 7553 7561
11. WielandGOrthausSOhndorfSDiekmannSHemmerichP 2004 Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol 24 15 6620 6630
12. BakerRERogersK 2006 Phylogenetic analysis of fungal centromere H3 proteins. Genetics 174 3 1481 1492
13. RaviMKwongPNMenorcaRMValenciaJTRamahiJS 2010 The Rapidly Evolving Centromere-specific Histone Has Stringent Functional Requirements in Arabidopsis thaliana. Genetics 186 461 471
14. RaviMChanSW 2010 Haploid plants produced by centromere-mediated genome elimination. Nature 464 7288 615 618
15. KalitsisPFowlerKJEarleEGriffithsBHowmanE 2003 Partially functional Cenpa-GFP fusion protein causes increased chromosome missegregation and apoptosis during mouse embryogenesis. Chromosome Res 11 4 345 357
16. LermontovaISchubertVFuchsJKlatteSMacasJ 2006 Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18 10 2443 2451
17. KhodjakovARiederCL 1996 Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J Cell Biol 135 2 315 327
18. MazumdarMMisteliT 2005 Chromokinesins: multitalented players in mitosis. Trends Cell Biol 15 7 349 355
19. LiuZMakaroffCA 2006 Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis. Plant Cell 18 5 1213 1225
20. GoshimaGKiyomitsuTYodaKYanagidaM 2003 Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol 160 1 25 39
21. SalinaDEnarsonPRattnerJBBurkeB 2003 Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 162 6 991 1001
22. OhsugiMAdachiKHoraiRKakutaSSudoK 2008 Kid-mediated chromosome compaction ensures proper nuclear envelope formation. Cell 132 5 771 782
23. KoduruPRKRaoMK 1981 Cytogenetics of synaptic mutants in higher plants 59 197 214
24. GrelonMVezonDGendrotGPelletierG 2001 AtSPO11-1 is necessary for efficient meiotic recombination in plants. Embo J 20 3 589 600
25. ZhaoDYangXQuanLTimofejevaLRigelNW 2006 ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol Biol 62 1–2 99 110
26. ChenCFarmerADLangleyRJMudgeJCrowJA 2010 Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol 10 280
27. OguraYShibataFSatoHMurataM 2004 Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet Syst 79 3 139 144
28. SatoHShibataFMurataM 2005 Characterization of a Mis12 homologue in Arabidopsis thaliana. Chromosome Res 13 8 827 834
29. CheesemanIMChappieJSWilson-KubalekEMDesaiA 2006 The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127 5 983 997
30. GoshimaGSaitohSYanagidaM 1999 Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev 13 13 1664 1677
31. OegemaKDesaiARybinaSKirkhamMHymanAA 2001 Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 153 6 1209 1226
32. SorgerPKDohenyKFHieterPKopskiKMHuffakerTC 1995 Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc Natl Acad Sci U S A 92 26 12026 12030
33. HealdRTournebizeRBlankTSandaltzopoulosRBeckerP 1996 Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382 6590 420 425
34. WatanabeYNurseP 1999 Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400 6743 461 464
35. TanakaKChangHLKagamiAWatanabeY 2009 CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell 17 3 334 343
36. SasakiYYasudaHOhbaYHaradaH 1990 Isolation and characterization of a novel nuclear protein from pollen mother cells of lily. Plant Physiol 94 3 1467 1471
37. RiggsCDHasenkampfCA 1991 Antibodies directed against a meiosis-specific, chromatin-associated protein identify conserved meiotic epitopes. Chromosoma 101 2 92 98
38. SuzukiTIdeNTanakaI 1997 Immunocytochemical visualization of the centromeres during male and female meiosis in Lilium longiflorum. Chromosoma 106 7 435 445
39. Prymakowska-BosakMPrzewlokaMRSlusarczykJKurasMLichotaJ 1999 Linker histones play a role in male meiosis and the development of pollen grains in tobacco. Plant Cell 11 12 2317 2329
40. JansenLEBlackBEFoltzDRClevelandDW 2007 Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176 6 795 805
41. SchuhMLehnerCFHeidmannS 2007 Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17 3 237 243
42. MaddoxPSHyndmanFMonenJOegemaKDesaiA 2007 Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176 6 757 763
43. ErhardtSMelloneBGBettsCMZhangWKarpenGH 2008 Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol 183 5 805 818
44. DunleavyEMRocheDTagamiHLacosteNRay-GalletD 2009 HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137 3 485 497
45. FoltzDRJansenLEBaileyAOYatesJR3rdBassettEA 2009 Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137 3 472 484
46. MonenJMaddoxPSHyndmanFOegemaKDesaiA 2005 Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol 7 12 1248 1255
47. DumontJOegemaKDesaiA 2010 A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol
48. IngouffMRademacherSHolecSSoljicLXinN 2010 Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20 23 2137 2143
49. DalalYWangHLindsaySHenikoffS 2007 Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol 5 e218 doi:10.1371/journal.pbio.0050218
50. FuruyamaTHenikoffS 2009 Centromeric nucleosomes induce positive DNA supercoils. Cell 138 1 104 113
51. DalalYBuiM 2010 Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol 22 3 392 402
52. CollinsKAFuruyamaSBigginsS 2004 Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol 14 21 1968 1972
53. ZeitlinSGShelbyRDSullivanKF 2001 CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol 155 7 1147 1157
54. KouzaridesT 2007 Chromatin modifications and their function. Cell 128 4 693 705
55. MalikHSVermaakDHenikoffS 2002 Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci U S A 99 3 1449 1454
56. HenikoffSAhmadKMalikHS 2001 The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293 5532 1098 1102
57. FishmanLSaundersA 2008 Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322 5907 1559 1562
58. SiddiqiIGaneshGGrossniklausUSubbiahV 2000 The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127 1 197 207
59. RossKJFranszPJonesGH 1996 A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4 7 507 516
60. MercierRVezonDBullierEMotamayorJCSellierA 2001 SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev 15 14 1859 1871
Štítky
Genetika Reprodukčná medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



