-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Mechanisms of Chromosome Number Evolution in Yeast
The whole-genome duplication (WGD) that occurred during yeast evolution changed the basal number of chromosomes from 8 to 16. However, the number of chromosomes in post-WGD species now ranges between 10 and 16, and the number in non-WGD species (Zygosaccharomyces, Kluyveromyces, Lachancea, and Ashbya) ranges between 6 and 8. To study the mechanism by which chromosome number changes, we traced the ancestry of centromeres and telomeres in each species. We observe only two mechanisms by which the number of chromosomes has decreased, as indicated by the loss of a centromere. The most frequent mechanism, seen 8 times, is telomere-to-telomere fusion between two chromosomes with the concomitant death of one centromere. The other mechanism, seen once, involves the breakage of a chromosome at its centromere, followed by the fusion of the two arms to the telomeres of two other chromosomes. The only mechanism by which chromosome number has increased in these species is WGD. Translocations and inversions have cycled telomere locations, internalizing some previously telomeric genes and creating novel telomeric locations. Comparison of centromere structures shows that the length of the CDEII region is variable between species but uniform within species. We trace the complete rearrangement history of the Lachancea kluyveri genome since its common ancestor with Saccharomyces and propose that its exceptionally low level of rearrangement is a consequence of the loss of the non-homologous end joining (NHEJ) DNA repair pathway in this species.
Published in the journal: Mechanisms of Chromosome Number Evolution in Yeast. PLoS Genet 7(7): e32767. doi:10.1371/journal.pgen.1002190
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002190Summary
The whole-genome duplication (WGD) that occurred during yeast evolution changed the basal number of chromosomes from 8 to 16. However, the number of chromosomes in post-WGD species now ranges between 10 and 16, and the number in non-WGD species (Zygosaccharomyces, Kluyveromyces, Lachancea, and Ashbya) ranges between 6 and 8. To study the mechanism by which chromosome number changes, we traced the ancestry of centromeres and telomeres in each species. We observe only two mechanisms by which the number of chromosomes has decreased, as indicated by the loss of a centromere. The most frequent mechanism, seen 8 times, is telomere-to-telomere fusion between two chromosomes with the concomitant death of one centromere. The other mechanism, seen once, involves the breakage of a chromosome at its centromere, followed by the fusion of the two arms to the telomeres of two other chromosomes. The only mechanism by which chromosome number has increased in these species is WGD. Translocations and inversions have cycled telomere locations, internalizing some previously telomeric genes and creating novel telomeric locations. Comparison of centromere structures shows that the length of the CDEII region is variable between species but uniform within species. We trace the complete rearrangement history of the Lachancea kluyveri genome since its common ancestor with Saccharomyces and propose that its exceptionally low level of rearrangement is a consequence of the loss of the non-homologous end joining (NHEJ) DNA repair pathway in this species.
Introduction
Centromeres and telomeres are essential genetic and structural elements of eukaryotic chromosomes. To maintain the accurate transmission of the genome to the next generation, each chromosome must have exactly one centromere and two telomeres. Evolutionary changes in an organism's number of chromosomes are caused by, or result in, structural rearrangements at centromeres and telomeres. Some particular chromosome number changes have been studied in detail in other eukaryotes, such as the fusion of two chromosomes in human since the divergence from chimpanzee [1]–[2] and the insertions of whole chromosomes into other centromeres that occurred during grass evolution [3]–[4]. Here we present the first study of this kind in yeast species.
Centromeres in all eukaryotes are the site at which the kinetochore forms and is attached to spindle microtubules, which segregate sister chromosomes to opposite poles of a dividing cell during anaphase I of meiosis, and sister chromatids during mitosis and anaphase II of meiosis. They also play a role in the pairing of homologous chromosomes during meiosis [5]. Centromere malfunction can lead to aneuploidy, resulting in inviable cells or severe genetic conditions. With few exceptions, centromeres are limited to one location per chromosome, because having more than one can lead to differential attachment to opposite spindle pole bodies during cell division, causing chromosome breakage by mechanical shearing during chromosome segregation.
There are several different types of centromeres in eukaryotes [6]. Most species have ‘regional’ centromeres that are defined epigenetically and can range in size from a few kilobases, to hundreds of kilobases. These regions are often heterochromatic and contain repetitive arrays of DNA satellites. Several diverse eukaryotic species have holocentric chromosomes which are thought to have evolved independently, where the centromeric function is spread along the entire chromosome [7]. Yeasts related to Saccharomyces cerevisiae have a unique type of centromere, known as point centromeres [8]–[9]. These are generally less than 200 bases long and are defined by specific sequences, the CDEI, CDEII and CDEIII regions which are bound by CEN DNA-binding proteins [10]–[11]. Point centromeres are probably an evolutionary state derived from epigenetic centromeres, as more divergent fungal lineages have epigenetic centromeres that cannot be identified by sequence [12]–[13]. It has been proposed that point centromeres evolved from the partitioning elements found on selfish plasmids, which supplanted the epigenetic centromeres in the Saccharomycetaceae lineage [6]. The point centromeres in yeast are some of the fastest diverging regions in the genome [11].
Telomeres are also ubiquitous and essential in all eukaryotes. They are heterochromatic regions that serve a protective function for the chromosomes [14]–[17]. Telomeres prevent the degradation of chromosomes from their ends and stop them from being recognized as double strand breaks (DSBs). Wild type telomeres are ‘capped’ with a combination of binding proteins, chromatin structure and DNA secondary structure folding into t-loops or other higher order chromatin structures [18]–[21]. Uncapped telomeres act and are recognized as DSBs, which initiate cell cycle arrest and DSB repair pathways [19], [22]. Telomeres of S. cerevisiae chromosomes consist of a heterogeneous repeating sequence (basic unit TGGGTG(TG)0–3) that is maintained by the enzyme telomerase in an array 325±75 bp long [23]–[24]. Other species such as Naumovozyma castellii and Candida glabrata have a similar organization though the sequence and length can vary [25]. Proximal to the telomere itself is a ‘subtelomeric’ region, which in S. cerevisiae consists of larger repeat sequences such as the Y′ element. Further proximal again are the first genes on the chromosome, which tend to be members of subtelomere-specific repeat families such as the DAN/TIR and FLO gene families in S. cerevisiae.
Many species from the Saccharomycetaceae family [26] have had their genomes sequenced (Figure 1) [27]–[33]. Central in this phylogeny is a whole genome duplication (WGD) event that occurred roughly 100 million years ago and gave rise to several extant paleopolyploids with reduced duplicate gene content [34]. Multiple genome sequences are available representing lineages that arose both before and after the WGD (Figure 1), referred to as non-WGD and post-WGD species, respectively [28], [33], [35].
Fig. 1. Phylogeny of the Saccharomycetaceae species used in this study. 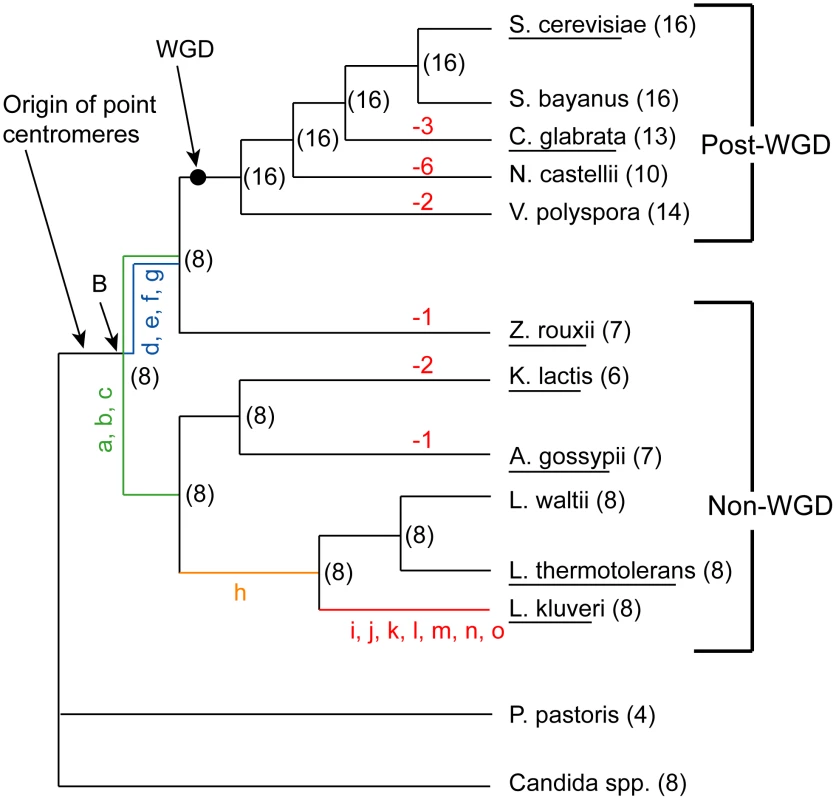
Parentheses show the numbers of chromosomes in extant species, and the inferred numbers at nodes in the tree. Negative numbers in red show chromosome number reductions. The black dot indicates the position of the WGD and the Ancestral genome sequence. Node ‘B’ is an older node that is the common ancestor of all non-WGD and post-WGD species. Lowercase letters represent specific rearrangements that differentiate L. kluyveri from the WGD Ancestor (black dot) as shown in Figure 2. Species whose names are underlined are those for which end-to-end complete chromosome sequences are available. The phylogeny used is that of Hedtke et al [88]. We previously inferred the gene order and core genome structure of the ancestral species that existed immediately before the WGD [36]. This ancestral genome contained a minimum complement of roughly 4,700 genes arranged on 8 chromosomes. The WGD doubled this basal chromosome number from 8 to 16. However, many of the post-WGD species do not have exactly 16 chromosomes; C. glabrata for instance has only 13. Karyotype data from pulse field gel electrophoresis (PFGE) also indicates a chromosome complement that ranges between 8 and 16 chromosomes for a range of post-WGD species [37]–[38]. Similarly, some of the non-WGD species have fewer than 8 chromosomes, such as Kluyveromyces lactis with 6. The ancestral reconstruction has allowed us to trace the genomic rearrangements that gave rise to the genome structures of extant species. Here, we mapped the locations of the ancestral centromeres and telomeres to sites in extant species, and identified the rearrangements that caused the chromosome number to change during the evolution of these species.
Results/Discussion
Mapping ancestral centromere and telomere locations
We previously inferred the structure of the yeast genome as it existed immediately before the WGD occurred [36]. We refer to this genome as the ‘Ancestral genome’, and to the organism that contained it as the ‘Ancestor’. It corresponds to the point marked ‘WGD’ on the phylogenetic tree in Figure 1. The approximate locations of telomeres in this genome are already known [36]. We inferred centromere locations in the Ancestral genome by using the same parsimony approach as in [36] combined with available centromere annotations from sequenced species. The inferred Ancestral centromere locations have been included in YGOB [39]. In summary, if a centromere is present in an orthologous intergenic region in at least one non-WGD and one post-WGD species, or in paralogous ‘sister’ regions of a post-WGD species, then that centromere was inferred to have been present in the Ancestral genome (WGD node in Figure 1). We extended the inferences of centromeres and telomere locations further back along the phylogeny to the common ancestor of the non-WGD and post-WGD species (Node ‘B’ in Figure 1) to allow for inferences about the evolution of centromeres and telomeres in the genera Kluyveromyces, Lachancea and Ashbya.
Lack of rearrangement in Lachancea kluyveri
While inferring node B we found that the genome of the non-WGD species L. kluyveri differs from the Ancestor by only 15 rearrangements (not including inversions within synteny blocks) as shown in Figure 2 (details are given in Table S1). We then assigned these rearrangements to different branches of the tree based on their presence or absence in other non-WGD species and the outgroup Candida and Pichia clades (Figure 1). The centromere and telomere locations are nearly identical between L. kluyveri and the Ancestor, allowing us to infer the centromere and telomere locations in the common ancestor of the non-WGD and post-WGD species (Node ‘B’ in Figure 1).
Fig. 2. Cartoon showing the rearrangements indicated by lowercase letters in Figure 1. 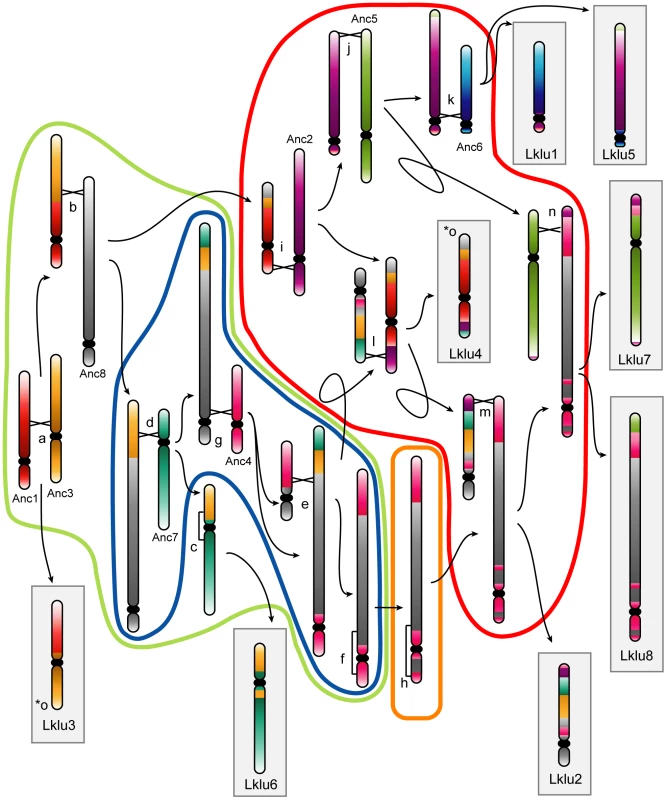
Monocolored chromosomes belong to the WGD Ancestor. Chromosomes in gray boxes are extant L. kluyveri chromosomes. Events encircled by a color correspond to events on branches of the same color in Figure 1. Black crossed lines between chromosomes represent points of interchromosomal translocations, and square brackets along chromosomes (events c, f and h) represent inversions. Arrows point to the products resulting from each rearrangement. The rearrangement for event o (marked with two asterisks) is not shown as it involves a reciprocal translocation located one gene from the edge of the Ancestral inference, which essentially swaps the telomeres of Anc3 and Anc8 at the ends of Lklu3 and Lklu4. Interestingly, by examining which Ancestral genes were not present in L. kluyveri, we noticed that four genes involved in non-homologous end joining (NHEJ) (DNL4, POL4, NEJ1 and LIF1) are missing from the genome of L. kluyveri with only a degraded DNL4 pseudogene and weak traces of an NEJ1 pseudogene remaining in the ancestral locations. These four proteins are part of the end-processing complex which plays a role in NHEJ [40]–[42], and DNL4, NEJ1 and LIF1 are also part of the end-bridging complex [40]–[41]. NHEJ is generally limited to haploid yeast cells because the expression of NEJ1, a major regulator of NHEJ, is down-regulated in MATa/MATα diploid cells [43]–[44]. DNL4 is required for NHEJ, and NEJ1 regulates NHEJ, so it appears that the NHEJ pathway is missing in L. kluyveri. POL4, NEJ1 and DNL4 have also been shown to play roles in the alternative microhomology-mediated end joining (MMEJ) pathway, and deletions of these genes reduce the efficiency of this process several-fold [45]–[47]. We hypothesize that the loss of the NHEJ and MMEJ pathways (or a large reduction in their efficiency) in L. kluyveri may be linked to the low number of genomic rearrangements and lack of telomere-to-telomere fusions in this lineage. It may also be linked to the predominantly diploid lifecycle of this yeast [48], which also suggests that most DSB repair in L. kluyveri is through homologous recombination. Although the NHEJ machinery is not essential, to our knowledge L. kluyveri is the only eukaryote so far identified that lacks it. Genes for all members of the MRX and Ku complexes are still present in L. kluyveri, and the related species L. thermotolerans has a complete set of NHEJ genes.
Mapping centromeres
The locations of centromeres were already inferred bioinformatically by the original sequencing groups for all species except Saccharomyces bayanus, Vanderwaltozyma polyspora (previously called Kluyveromyces polysporus) and Naumovozyma castellii (previously called Saccharomyces castellii or Naumovia castellii). We identified and annotated centromeres in S. bayanus and V. polyspora by extracting the intergenic regions in these species orthologous to the inferred Ancestral centromeres, and used MEME [49] to generate consensus CDEI and CDEIII profiles (full sequences of all centromeric loci are in Table S2). For N. castellii, Cliften et al. [50] were unable to identify any consensus centromere sequence. We too were unable to identify consensus centromere sequences at the Ancestral centromeric locations in N. castellii (Dataset S1). We also searched the whole N. castellii genome using the consensus motif for Saccharomycetaceae point centromeres derived from all identified centromeres in all species, but still could not find any candidates. Inspection of the intergenic regions corresponding to Ancestral centromeres in preliminary genome sequence data from the related species N. dairenensis also failed to locate any candidate point centromeres (data not shown). We hypothesize that these species may represent a novel transition of centromere structure in Naumovozyma which could be analogous to the earlier replacement of epigenetic centromeres by point centromeres in yeasts [6]. The system that has potentially superseded point centromeres in Naumovozyma will require functional characterization in the laboratory.
The correspondence between Ancestral centromere locations and current centromeres for all other extant species in the YGOB species set are shown in Table 1. All but one current centromere mapped in a straightforward manner to a corresponding Ancestral centromere with full or partially conserved syntenic gene content bordering the centromeres relative to the Ancestor. The exceptional case was CEN9 of C. glabrata, which maps to Ancestral CEN6 and has undergone a series of rearrangements with breakpoints on both sides of the centromere which have eliminated all traces of synteny at this locus (Figure S1).
Tab. 1. Mapping between Ancestral centromeres and centromeres in extant species. 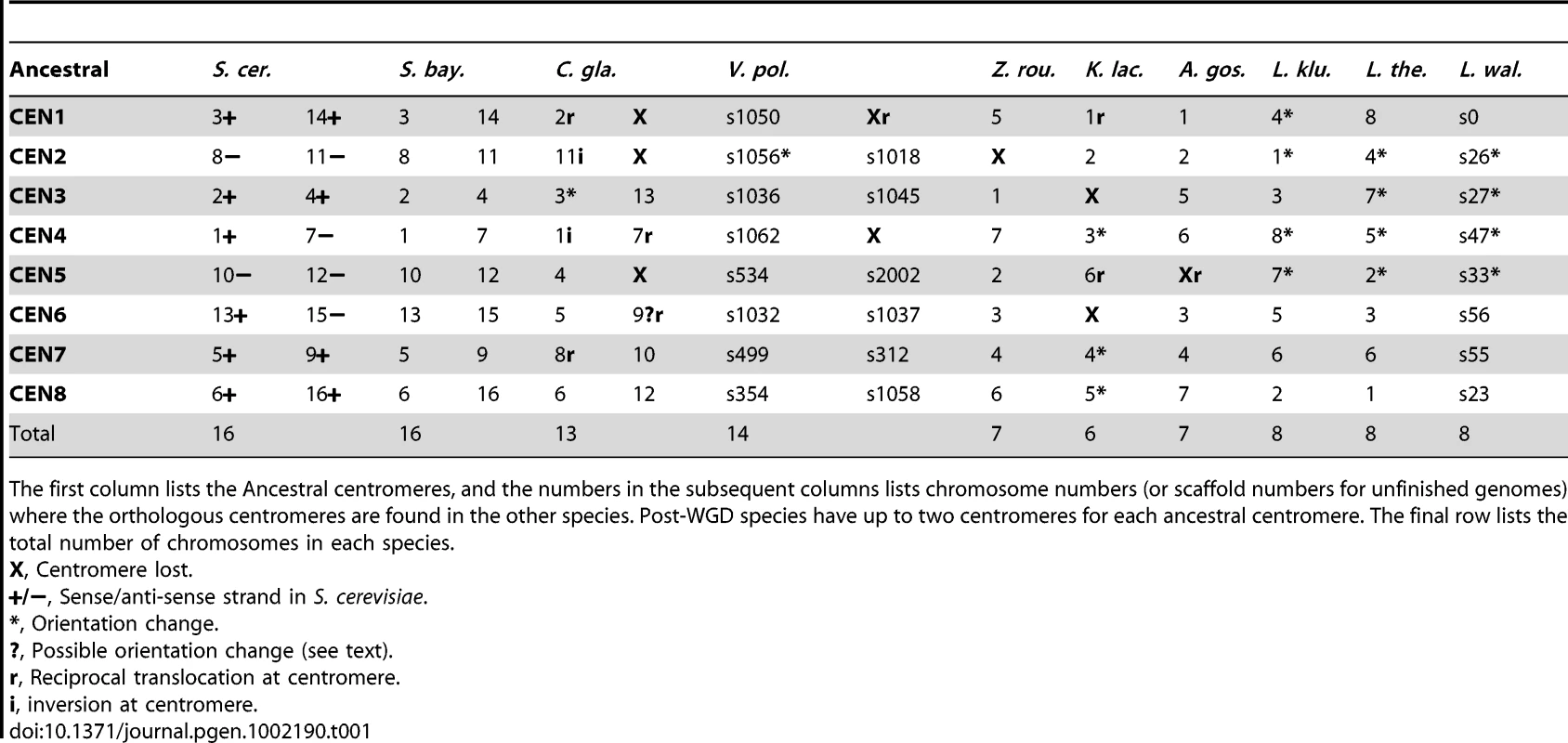
The first column lists the Ancestral centromeres, and the numbers in the subsequent columns lists chromosome numbers (or scaffold numbers for unfinished genomes) where the orthologous centromeres are found in the other species. Post-WGD species have up to two centromeres for each ancestral centromere. The final row lists the total number of chromosomes in each species. Mapping telomeres
We traced the evolution of telomere locations in all the species for which completely finished genome sequences are available, but not for those whose genomes consist of numerous scaffolds, due to the uncertainty in identifying real telomeric regions in scaffold data (Table 2). In most of the genomes, mapping the current telomeres to Ancestral locations is relatively trivial as there is a direct correspondence without genome rearrangements at those locations (Table 2). However in C. glabrata, A. gossypii and K. lactis several telomeres mapped to Ancestral locations through a complex set of rearrangements including breakpoint reuse. The genomes of these species are also the most rearranged of those examined. By contrast, members of the Lachancea clade have had relatively few genomic rearrangements on the evolutionary path between them and the Ancestor. The mapping of telomeres to Ancestral telomeres is more tentative than for the centromeric mapping, due to the inherently unstable nature of telomeres, and the possibility of movement of the telomeric boundaries. For example, if we had genome sequences from more species, it might become possible to extend the Ancestral genome inference further towards the telomeres and so reveal rearrangements that are presently inaccessible that may alter the mapping. The current telomere assignments represent the most parsimonious mappings given the data that is currently available.
Tab. 2. Mapping between Ancestral telomeres and telomeres in extant species with finished genome sequences. 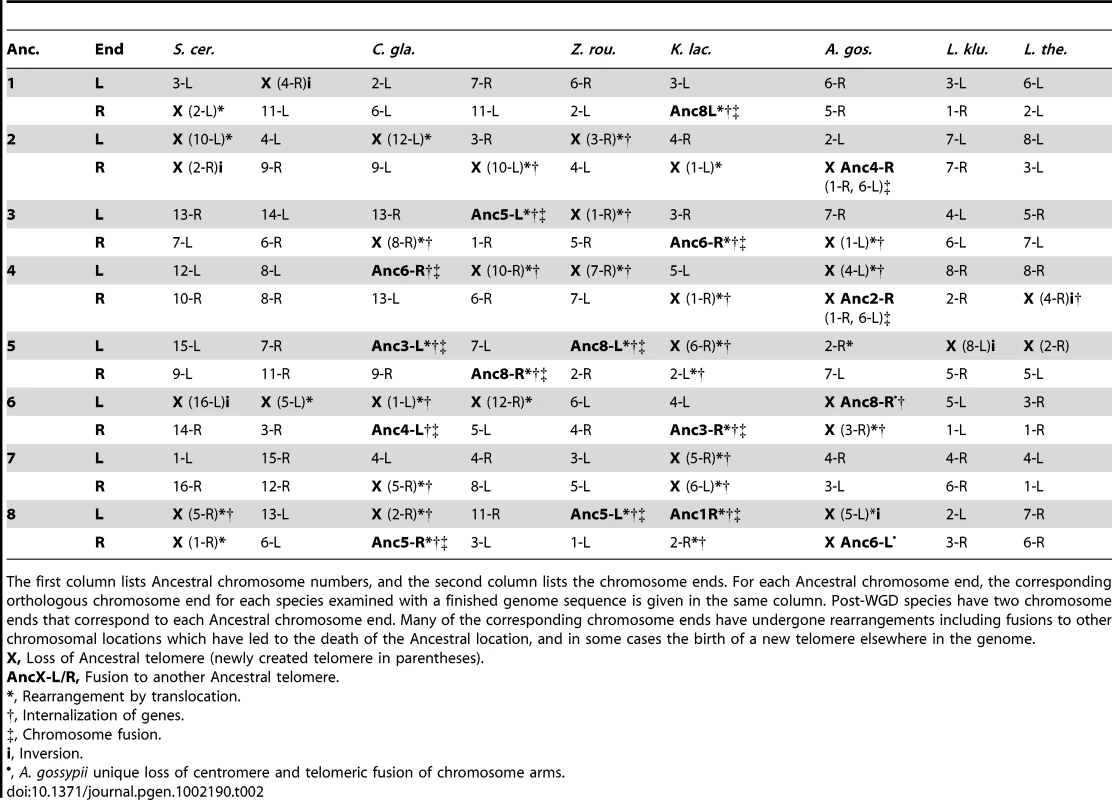
The first column lists Ancestral chromosome numbers, and the second column lists the chromosome ends. For each Ancestral chromosome end, the corresponding orthologous chromosome end for each species examined with a finished genome sequence is given in the same column. Post-WGD species have two chromosome ends that correspond to each Ancestral chromosome end. Many of the corresponding chromosome ends have undergone rearrangements including fusions to other chromosomal locations which have led to the death of the Ancestral location, and in some cases the birth of a new telomere elsewhere in the genome. Centromere losses
We identified nine losses of a centromere, corresponding to nine decreases of chromosome number. Three of these occurred in C. glabrata, two each in V. polyspora and K. lactis, and one each in Z. rouxii and A. gossypii (Figure 1). The major mechanism of centromere loss was associated with the telomere-to-telomere fusion of two chromosomes with the loss of one of the centromeres. This mechanism is illustrated by the chromosome fusion and single centromere loss that occurred in Z. rouxii, whose details are shown in Figure 3. In this example, the process also resulted in the internalization of many genes that were previously located near telomeres. All but perhaps one of the nine centromere losses occurred in this fashion, resulting in the loss of at least 14 of the 112 telomere locations examined. The removal of centromeres appears to have been quite specific, generally leaving adjacent genes intact. In some cases a centromere and some adjacent genes are missing, but all these cases occur in post-WGD species where gene deletion is relatively common due to the redundancy created by the WGD. None of the centromere losses in non-WGD species is accompanied by loss of centromere-adjacent genes.
Fig. 3. Progression of rearrangements and chromosome fusions leading to the loss of a centromere in Z. rouxii. 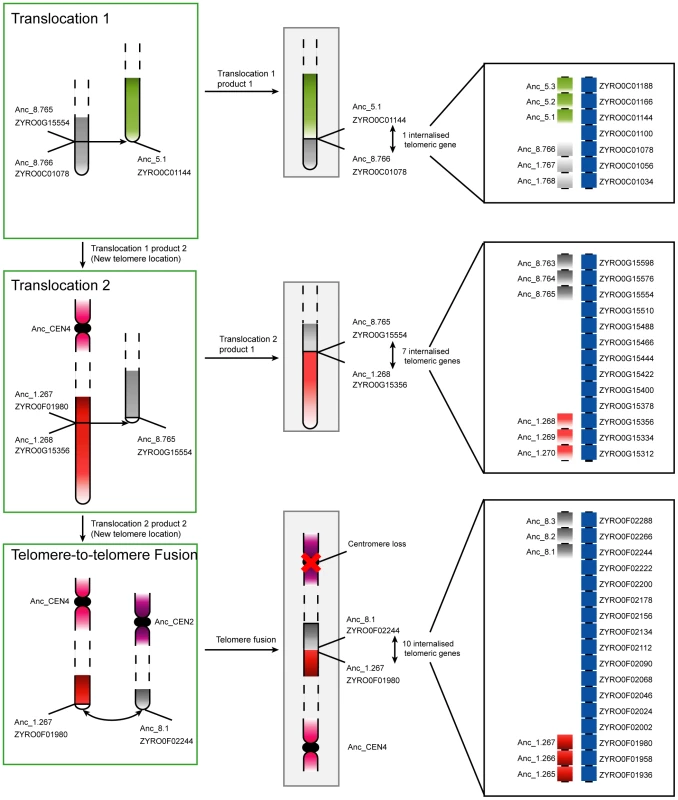
Two non-reciprocal telomeric translocations and a telomere-to-telomere fusion gave rise to the extant chromosome structures in Z. rouxii. Chromosomes in green boxes are those that underwent rearrangements, while those in gray boxes are finished translocation products (i.e., extant regions in Z. rouxii). The edges of the breakpoints are labelled with both the Ancestral and current Z. rouxii gene names. In the bottom step, the loss of a centromere occured contemporaneously with the two chromosomes fusing at their telomeres. All three rearrangements led to the internalisation of previously telomeric genes. The panels on the right show details of the gene orders and internalized telomeric genes at the junctions. The majority of centromere losses in yeast appear to have involved the fusion of whole chromosomes. In these cases, two possible scenarios exist that differ only in the order of events. The first scenario is the initial fusion of the chromosomes at telomeric locations, with subsequent loss of one of the two centromeres. In this case selection would likely act to suppress one of the two centromeres to avoid problems during cell division. The second scenario is that the centromere of a chromosome is first lost or disabled, with the chromosome subsequently being rescued from cellular loss by fusion to another chromosome with a functional centromere. Under the latter scenario, selection acts to maintain the genes contained on the chromosome without a centromere, because cells missing a whole chromosome will certainly be inviable. Chromosome fusions have been generated experimentally in S. cerevisiae by the inactivation of a centromere [51]. Interestingly, if the centromere is reactivated, it often leads to fission of the resulting chromosome at or near the fusion site to reconstitute the parental karyotype [51], indicating that the fusion point may be a fragile site. This fragility might explain the reuse of fission/fusion breakpoints like those shared between Translocations 1, 2 and 3 in Figure 3.
The unique case observed in A. gossypii appears to have occurred by the breakage of a chromosome in the intergenic region that contained Ancestral centromere Anc_CEN5 (Figure 4). The resulting two chromosome arms then fused to two other chromosomes, joining the previously centromere-proximal sequences to the telomeres of the other chromosomes. The exact nature of this fission and fusion is not known, and we cannot tell the difference between chromosome breakage and religation to new locations, or translocation events. It is also not possible to infer whether the centromere was destroyed in the fission event, or whether it was still intact at the end of one of the arms that subsequently fused to another telomere and was lost later due to the constraint of having one centromere per chromosome.
Fig. 4. Loss of a centromere in A. gossypii by the breakage of a chromosome at its centromere. 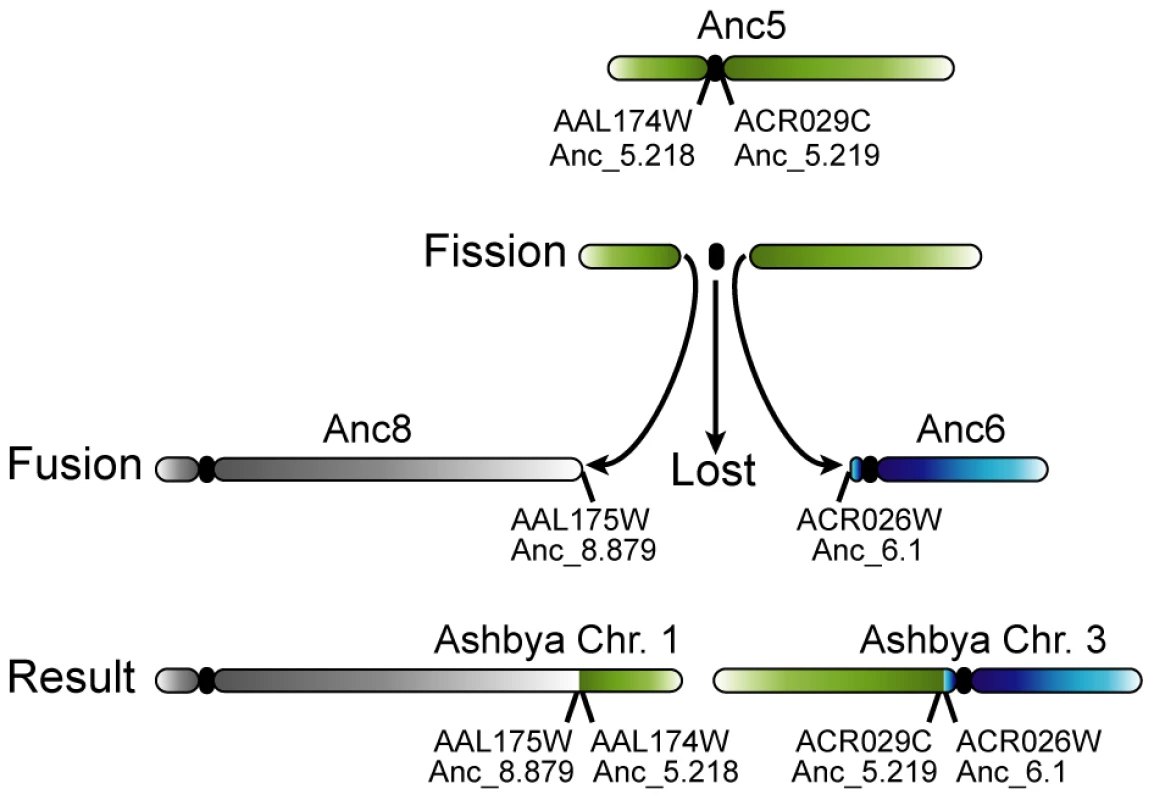
The green chromosome at the top represents chromosome 5 at Node ‘B’ of the tree (Figure 1), which is identical to chromosome 5 of the WGD Ancestor (see Figure 2). After A. gossypii diverged from K. lactis, this chromosome broke in the intergenic region containing its centromere. To avoid losing large numbers of genes during cell division, both arms of the split chromosome fused their broken edges to the telomeres of Ancestral chromosomes 6 and 8, which gave rise to the organisation on the extant A. gossypii chromosomes 1 and 3. The timing of loss of the centromere is unclear: it may have happened as a part of the rearrangement, or the centromere may have been carried on one of the chromosome arms and lost after fusion to the telomere of another centromere-containing chromosome. The mechanism of the fission event is also ambiguous: it may have occurred by the chromosome actually breaking into two, or by two separate translocations to other chromosome ends that separated the centromere from its neighboring genes. We observed no cases of de novo centromere gain. Apparently, the only mechanism by which chromosome number has increased during the evolution of Saccharomycetaceae is WGD (Figure 1). This discovery is quite surprising, because the spontaneous formation of aneuploids with duplications of single centromeres or chromosomes has frequently been reported, both in S. cerevisiae [52]–[53] and C. glabrata [54]. Interestingly, from the sequenced genomes only species in the genus Saccharomyces have retained all 16 centromeres from the WGD, while the other sequenced post-WGD species (V. polyspora, N. castellii and C. glabrata) all have a reduced chromosome complement that arose independently in their respective lineages (Figure 1). Previous PFGE karyotype analyses indicated that some strains of Kazachstania exigua may also have a chromosome complement of 16 [37]–[38], the most likely explanation of which is that this species has also retained all of its centromeres since the WGD.
Consensus centromere sequences
We compiled and compared the CDE consensus sequences for all sequenced yeasts with point centromeres (Figure S3). All the centromeres of S. cerevisiae have been characterized functionally [8]–[9], and a few have been cloned from other yeasts: S. bayanus [55]–[56], C. glabrata [57], Z. rouxii [58] and K. lactis [59]. The genome sequencing groups made bioinformatic predictions about centromere locations for most of the other chromosomes and species, based on matches to the CDEI–III consensus sequences [27]–[28], [30], [33]. We used these in our analysis, though we revised the coordinates of two L. waltii centromeres (Table S4). We identified CDE regions for centromeres in S. bayanus (Table S5) and V. polyspora (Table S6), finding 16 and 14 centromeres respectively. Although the genome sequence of V. polyspora is incomplete [32], there is complete intergenic sequence spanning both of the lost centromeres meaning we are confident of their absence. Our count of 14 centromeres is one more than the previous estimate of chromosome number in this species [60].
With over a hundred yeast centromeres in our dataset we searched for features common to all point centromeres (Figure 5). For consistency with S. cerevisiae, in this analysis we delineated the boundaries of CDEI, CDEII and CDEIII regions in the same way across all genomes disregarding small differences in the boundary choices made by different sequencing groups. The CDEI regions have an 8 bp consensus motif with four invariant sites (NNCAVBTG). The CDEIII regions have an invariant 5 bp motif (CCGAA) and the whole CDEIII consensus is 26 bp. Within a given species there are often further invariant sites in their CDEI or CDEIII regions, for example G at positions 2 and 8 in S. cerevisiae CDEIII. The intervening CDEII regions are always highly AT-rich (76–98%). The length of CDEII varies twofold among species, but there is remarkably little CDEII length variation within each species, and a clear correlation of CDEII lengths among related species (Figure 5C).
Fig. 5. CDE conservation in the Saccharomycetaceae. 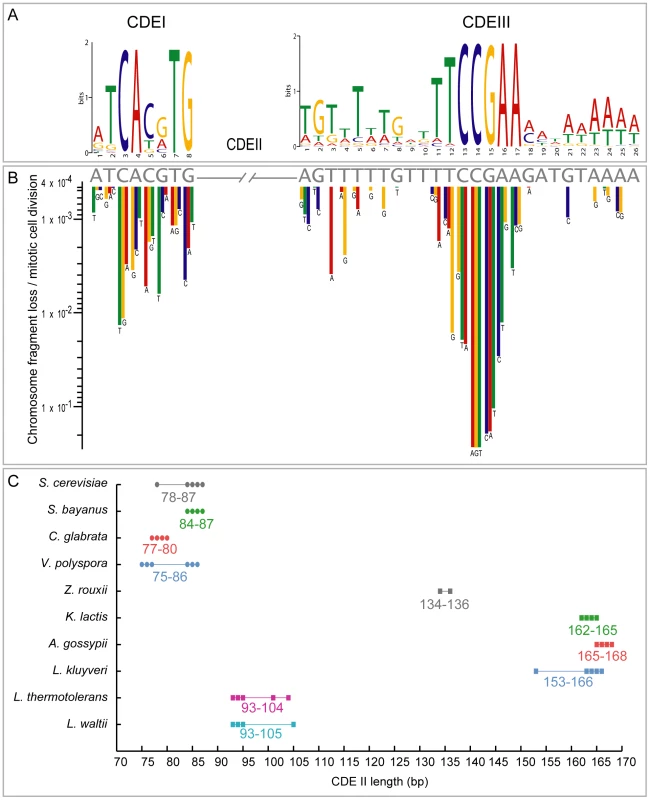
(A) Sequence logo showing base frequencies at each position in all annotated CDEI and CDEIII regions from 10 species. (B) Rate of chromosome loss per mitotic cell division caused by mutagenesis of individual residues in CDEI and CDEIII sequences (gray letters) of S. cerevisiae CEN6 (redrawn from [61]). Sites conserved in the logo tend to have the largest effects on chromosome loss when mutated. (C) Variation of CDEII lengths in species with identifiable point centromeres. The number of points is fewer than the number of chromosomes in each species because some chomosomes have identical CDEII length. Hegemann and Fleig [61] compiled and summarized mutagenesis studies on S. cerevisiae CEN6 [62]–[64], measuring the frequency of chromosome fragment loss resulting from point mutations at many sites in CEN6. There is a strikingly strong correlation between their results and the evolutionary conservation of individual sites in CDEI and CDEIII (Figure 5A, 5B). None of the 13 nucleotide changes with the most severe phenotypes (chromosome fragment loss rates >10−2 per mitotic cell division) at CEN6 occurs as a natural variant in the 102 centromeres we compiled. Thus the evolutionary conservation of these regions over hundreds of millions of years correlates well with the highest impact point mutations from the mutational data. Due to these constraints, we suggest that the de novo formation of a point centromere in these yeast species is much less likely than the de novo creation of regional centromeres in other species such as Candida albicans [65] because heritable epigenetic changes can occur on a much smaller timescale than sequence-based evolution.
Rearrangements at centromeres
Reciprocal translocation and inversion breakpoints were observed adjacent to centromeres in C. glabrata, V. polyspora, A. gossypii and K. lactis, as were orientation changes of the centromeres (Table 1). V. polyspora and A. gossypii each show only one such event, and in both cases the rearrangement breakpoints coincide with the site of a centromere loss in these species. K. lactis has three rearrangement breakpoints adjacent to centromeres, and C. glabrata has six, none of which coincide with centromere loses in either species. Interestingly, the breakpoints adjacent to the three centromeres in K. lactis are all part of one rearrangement cycle (Figure S2), indicating that there have been reciprocal translocations between intergenic locations containing centromeres.
Telomere cycling and internalization of telomeric genes
Translocations causing a terminal segment of one chromosome to be transferred and joined to another chromosome were observed in Z. rouxii (Figure 3), S. cerevisiae, C. glabrata, K. lactis and A. gossypii. As well as physically moving an existing telomere to a new chromosome, this type of rearrangement results in some previously subtelomeric DNA becoming internal to chromosomes where the fusion occurred (Figure 3). These events can be inferred at the level of synteny blocks, but they probably occurred millions of years ago and there is currently no telomere-like DNA sequence at the rearrangement points. Conversely, previously internal regions on the chromosomes located at the breakpoints of telomeric translocations become novel telomere sites (e.g., gene ZYRO0G15554 after Translocation 1 in Figure 3, before it became the join-site of another telomeric translocation). Analogous birth and death of telomere locations can occur by inversions and are found in S. cerevisiae, A. gossypii, Z. rouxii and K. thermotolerans (Table 2). Telomeric translocations and inversions have resulted in the turnover of more than a quarter (33/112) of telomere locations relative to the ancestor. As well as inversions and translocations, the death of telomere locations can be caused by telomere-to-telomere fusions. The gain of novel telomere sites is presumably by telomere capture, a process that has been observed in cells that survive the absence of telomerase or defective telomere capping. Novel telomeres can also be generated at the site of a DSB by telomerase, a process that is enhanced by G-rich telomeric seed sequences lying close to the DSB [66]–[69].
Internal chromosomal positions differ from subtelomeric locations in terms of their chromatin configurations, which in turn affect the expression of nearby genes [70]–[72]. In general, subtelomeric regions tend to have higher nucleosome occupancy and silencing protein association, both of which generally reduce gene expression [70]–[72]. Subtelomeric genes are likely to be under less evolutionary constraint than genes in internal locations, are less essential and have higher variance in their expression profiles [73]. The rate of sequence evolution is negatively correlated with expression and essentiality, but positively correlated with the variance of gene expression [74]–[77]. Thus relocating a gene from telomeric to internal regions is likely to increase the evolutionary constraints on its sequence. Conversely, evolution may proceed at a faster pace at telomeres due to more relaxed selective constraints. If this higher evolutionary rate leads to an advantageous allele at a telomere, we hypothesize that it may be beneficial to relocate the gene to somewhere else in the genome where selection will maintain the advantageous allele under higher constraint. This could potentially constitute an ongoing cycle over evolutionary time, where the telomeres act as the cooking pots of evolution [78], with successful innovations moving to more stable regions.
Rearrangements that internalize genes appear to be more common in genomes that have high rates of genome rearrangement. In S. cerevisiae, which is the least rearranged post-WGD species [36], only two genes (GAL2 and SRL2, which are in the same breakpoint location) were internalized by rearrangement from a telomere (Table S3). In C. glabrata, arguably the most rearranged post-WGD species [36], there are at least 17 internalized genes in 8 locations (Table S3) even though the telomeres of C. glabrata contain many fewer annotated genes than those of S. cerevisiae. Non-WGD genomes that have high levels of rearrangement such as K. lactis and A. gossypii [36] contain high numbers of these genes (at least 48 genes in 19 locations and 15 genes at 8 locations respectively) (Table S3). In Z. rouxii, which is intermediate in terms of rearrangement, there are at least 27 genes at 7 locations, while in the rearrangement poor L. thermotolerans, there are 6 genes at a single location. There are no internalized genes in the L. kluyveri, the least rearranged non-WGD species. These numbers also somewhat reflect the overall numbers of subtelomeric genes annotated in these species.
Large scale genomic rearrangements like the fusions of telomeres to other telomeres or internal chromosomal sections inferred in this work are generally considered to be detrimental to cells although they are not necessarily so. Many cancers involve similar types of rearrangements, and there are several pathways and mechanisms in place in cells to prevent and repair them, including proteins involved in telomere structure and maintenance, cell cycle arrest signalling, homologous recombination (HR) and NHEJ repair pathways [19], [22], [69], [79]–[81]. Interestingly, many of the components of the HR and NHEJ machinery such as the MRX complex, Yku70/80 proteins and Rad17/Mec3/Ddc1 complex also play roles in telomere structure and stability and are associated to telomeres [19], [22], [79]–[81]. Experimental deletions of genes involved in these pathways as well as those involved in telomeric structure have helped to tease apart their functions at telomeres, and many of the deletions result in chromosomal rearrangements such as telomere-to-telomere fusions and non-reciprocal translocations, similar to those inferred in our work [19], [22], [80]–[82]. The gross chromosomal rearrangements observed in these mutants generally manifest through a NHEJ-like mechanism requiring Dnl4 (Lig4), an NHEJ ligase [79]–[81].
Spontaneous rearrangements involving telomere fusions to other telomeres or DSBs occur in wild type S. cerevisiae cells at a rate of 1–6×10−7 events per genome per cell division [80], but have only been fixed a few times throughout Saccharomycetaceae evolution. Together with evidence that S. cerevisiae is capable of rescuing cells from DSBs by telomere capture at the edge of the DSB from the centromere-containing part of the chromosome [66], [68], [83], it appears that telomeric rearrangements such as telomere-to-telomere fusions and non-reciprocal translocations likely represent rare errors in the systems that protect and cap telomeres or repair DSBs that have been fixed over evolutionary time. It is only possible to speculate about the exact causes of the rearrangements, how they became fixed in populations, and whether they were selectively advantageous, neutral or disadvantageous. The observed rearrangements are in the order of millions of years old, and are thus unlikely to contain any sequence information that could provide empirical evidence about their mechanism of formation.
We suggest that the rearrangements probably occurred in haploid cells, as in a diploid it would be expected that DSBs would be repaired via homologous recombination using the homologous chromosome as templates. In the Saccharomycetaceae where mating-type switching occurs [28], [84], rearrangements in haploids would also avoid mating incompatibilities that could arise in a diploid due to meiotic segregation difficulties [85]. A haploid cell could divide, change mating type and then mate with the daughter cell, thus avoiding potential chromosome pairing problems and aneuploidy.
Perspectives
Among the species studied here (the family Saccharomycetaceae) [26], we find that chromosome number has evolved by two very different mechanisms. The only mechanism of increase was polyploidization. We suggest that the lack of any other new centromere formation is a consequence of the sequence-defined nature of point centromeres, but it is unclear why the formation of a new centromere by small-scale DNA duplication of an existing centromere, as seen in C. glabrata drug resistance isolates [54], is not seen during evolution. The mechanism of decrease in chromosome number was by rearrangements involving telomeres, primarily telomere-to-telomere fusions with the loss of a centromere belonging to one of the fused chromosomes. The temporal sequence of the chromosome fusion and centromere loss is ambiguous. Telomeric rearrangements have also frequently moved genes from subtelomeric locations to internal genomic locations. These movements have the potential to change the selective constraints on the genes and could be evolutionarily adaptive.
Materials and Methods
Mapping centromeres and telomeres to the Ancestor
The Ancestral centromere locations were generally trivial to find because numerous comparisons among extant non-WGD and post-WGD species can be made, most centromere locations are in syntenic regions among species, and most rearrangements that might obscure these relationships are species specific. Ancestral centromere loci were added to YGOB following the same parsimony rules as in [36], by using species for which centromere annotations have already been made. These Ancestral centromere locations were then used to guide the search for unannotated centromeres in orthologous intergenic regions by searching for CDEI and CDEIII sequence motifs using MEME [49].
To map the rearrangements that had occurred at a centromere in any particular species, we examined the breakpoints between synteny blocks in that species relative to the Ancestor and tried to locate the reciprocal breakpoint elsewhere in the genome. In some cases, a reciprocal breakpoint did not exist; these cases represent breakpoint reuse [36]. They can be solved by following one edge of the breakpoint (A|B) locating the reciprocal edge at another location (B′|C), then finding the breakpoint partner's reciprocal edge (C′|D) and iterating this process until reaching the original breakpoint's other edge (D′|A′). This process identifies a cycle of breakpoint edges that eventually leads back to the adjacent edge of the centromeric breakpoint.
Telomeric locations were mapped between the Ancestor and extant species in a similar way, except the extant telomere positions were defined as the regions at the ends of chromosomes where it is no longer possible to define Ancestral genes based on synteny across species, i.e. the regions in extant species that lie beyond the edges of the Ancestral chromosome reconstruction. As telomeres have a very high rate of rearrangement, we regard telomeres as locations rather than as any particular genes. Thus the telomere locations of a chromosome were defined as the locations beside the leftmost and rightmost genes on that chromosome that have orthologs in the Ancestral genome. We only analyzed the evolution of telomere locations in species whose genomes are completely sequenced, because for incompletely sequenced species we cannot be sure that there is a telomere at the end of each scaffold.
To trace the evolution of centromere and telomere positional evolution in the non-WGD species, which are not direct descendants of the Ancestor (Figure 1), we mapped the translocational rearrangements between the Ancestor and the non-WGD species L. kluyveri onto the phylogeny by comparing their presence and absence in other extant species in the Saccharomycetaceae and outgroups (Pichia pastoris [86] and the Candida clade of species [87]).
Absence of NHEJ genes in L. kluyveri
The four genes involved in NHEJ that are missing from L. kluyveri were identified by compiling a list of genes in the YGOB database that are present in the Ancestral genome but not in the L. kluyveri genome. We noticed that four genes in the list had a role in NHEJ. We then examined the L. kluyveri intergenic locations where these genes would be expected to reside, to make sure that they were not present but unannotated. No potentially coding ORFs were found in these regions, but pseudogene relics of DNL4 and NEJ1 were identified. Finally, protein sequences from the four genes from the closely related L. thermotolerans were used as TBLASTN queries against the L. kluyveri chromosome sequences to make sure they were not present elsewhere in the genome.
Supporting Information
Zdroje
1. JWIJBaldiniAWardDCReedersSTWellsRA 1991 Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci U S A 88 9051 9055
2. HillierLWGravesTAFultonRSFultonLAPepinKH 2005 Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature 434 724 731
3. LuoMCDealKRAkhunovEDAkhunovaARAndersonOD 2009 Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci U S A
4. International Brachypodium Initiative 2010 Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463 763 768
5. GuerraCEKabackDB 1999 The role of centromere alignment in meiosis I segregation of homologous chromosomes in Saccharomyces cerevisiae. Genetics 153 1547 1560
6. MalikHSHenikoffS 2009 Major evolutionary transitions in centromere complexity. Cell 138 1067 1082
7. DernburgAF 2001 Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol 153 F33 38
8. FleigUBeinhauerJDHegemannJH 1995 Functional selection for the centromere DNA from yeast chromosome VIII. Nucleic Acids Res 23 922 924
9. HieterPPridmoreDHegemannJHThomasMDavisRW 1985 Functional selection and analysis of yeast centromeric DNA. Cell 42 913 921
10. KennaMAmayaEBloomK 1988 Selective excision of the centromere chromatin complex from Saccharomyces cerevisiae. J Cell Biol 107 9 15
11. BensassonDZarowieckiMBurtAKoufopanouV 2008 Rapid evolution of yeast centromeres in the absence of drive. Genetics 178 2161 2167
12. SanyalKBaumMCarbonJ 2004 Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A 101 11374 11379
13. LynchDBLogueMEButlerGWolfeKH 2010 Chromosomal G+C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol Evol 2 572 583
14. BlackburnEHGallJG 1978 A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120 33 53
15. McClintockB 1939 The Behavior in Successive Nuclear Divisions of a Chromosome Broken at Meiosis. Proc Natl Acad Sci U S A 25 405 416
16. BoscoGHaberJE 1998 Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150 1037 1047
17. CechTR 2004 Beginning to understand the end of the chromosome. Cell 116 273 279
18. de BruinDKantrowSMLiberatoreRAZakianVA 2000 Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol 20 7991 8000
19. LydallD 2003 Hiding at the ends of yeast chromosomes: telomeres, nucleases and checkpoint pathways. J Cell Sci 116 4057 4065
20. WeinertT 2005 Do telomeres ask checkpoint proteins: “gimme shelter-in”? Dev Cell 9 725 726
21. de BruinDZamanZLiberatoreRAPtashneM 2001 Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409 109 113
22. ChanSWBlackburnEH 2003 Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol Cell 11 1379 1387
23. RayARungeKW 1999 The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol Cell Biol 19 31 45
24. RungeKWZakianVA 1989 Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol Cell Biol 9 1488 1497
25. CohnMMcEachernMJBlackburnEH 1998 Telomeric sequence diversity within the genus Saccharomyces. Curr Genet 33 83 91
26. KurtzmanCP 2011 Discussion of teleomorphic and anamorphic ascomycetous yeasts and yeast-like taxa. BoekhoutT The Yeasts, a Taxonomic Study. 5 ed Amsterdam Elsevier 293 307
27. DietrichFSVoegeliSBrachatSLerchAGatesK 2004 The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304 304 307
28. DujonBShermanDFischerGDurrensPCasaregolaS 2004 Genome evolution in yeasts. Nature 430 35 44
29. GoffeauABarrellBGBusseyHDavisRWDujonB 1996 Life with 6000 genes. Science 274 546, 563–547
30. KellisMBirrenBWLanderES 2004 Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 617 624
31. KellisMPattersonNEndrizziMBirrenBLanderES 2003 Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423 241 254
32. ScannellDRFrankACConantGCByrneKPWoolfitM 2007 Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci U S A 104 8397 8402
33. SoucietJLDujonBGaillardinCJohnstonMBaretPV 2009 Comparative genomics of protoploid Saccharomycetaceae. Genome Res 19 1696 1709
34. WolfeKHShieldsDC 1997 Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387 708 713
35. WolfeKH 2006 Comparative genomics and genome evolution in yeasts. Philos Trans R Soc Lond B Biol Sci 361 403 412
36. GordonJLByrneKPWolfeKH 2009 Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet 5 e1000485 doi:10.1371/journal.pgen.1000485
37. SpirekMYangJGrothCPetersenRFLangkjaerRB 2003 High-rate evolution of Saccharomyces sensu lato chromosomes. FEMS Yeast Res 3 363 373
38. PetersenRFNilsson-TillgrenTPiskurJ 1999 Karyotypes of Saccharomyces sensu lato species. Int J Syst Bacteriol 49 Pt 4 1925 1931
39. ByrneKPWolfeKH 2005 The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15 1456 1461
40. EllenbergerTTomkinsonAE 2008 Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem 77 313 338
41. TsengHMTomkinsonAE 2004 Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J Biol Chem 279 47580 47588
42. WilsonTELieberMR 1999 Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem 274 23599 23609
43. KegelASjostrandJOAstromSU 2001 Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol 11 1611 1617
44. ValenciaMBenteleMVazeMBHerrmannGKrausE 2001 NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414 666 669
45. DecottigniesA 2007 Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics 176 1403 1415
46. MaJLKimEMHaberJELeeSE 2003 Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol 23 8820 8828
47. LeeKLeeSE 2007 Saccharomyces cerevisiae Sae2 - and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 176 2003 2014
48. de ClareMPirPOliverSG 2011 Haploinsufficiency and the sex chromosomes from yeasts to humans. BMC Biol 9 15
49. BaileyTLElkanC 1994 Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2 28 36
50. CliftenPFFultonRSWilsonRKJohnstonM 2006 After the duplication: gene loss and adaptation in Saccharomyces genomes. Genetics 172 863 872
51. PobiegaSMarcandS 2010 Dicentric breakage at telomere fusions. Genes Dev 24 720 733
52. HughesTRRobertsCJDaiHJonesARMeyerMR 2000 Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25 333 337
53. DelneriDColsonIGrammenoudiSRobertsINLouisEJ 2003 Engineering evolution to study speciation in yeasts. Nature 422 68 72
54. PolakovaSBlumeCZarateJAMentelMJorck-RambergD 2009 Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci U S A 106 2688 2693
55. HubermanJAPridmoreRDJägerDZonneveldBPhilippsenP 1986 Centromeric DNA from Saccharomyces uvarum is functional in Saccharomyces cerevisiae. Chromosoma 94 162 168
56. YamaneSKarashimaHMatsuzakiHHatanoTFukuiS 1999 Isolation of centromeric DNA from Saccharomyces bayanus. J Gen Appl Microbiol 45 89 92
57. KitadaKYamaguchiEHamadaKArisawaM 1997 Structural analysis of a Candida glabrata centromere and its functional homology to the Saccharomyces cerevisiae centromere. Curr Genet 31 122 127
58. PribylovaLStraubM-LSychrovaHde MontignyJ 2007 Characterisation of Zygosaccharomyces rouxii centromeres and construction of first Z. rouxii centromeric vectors. Chromosome Res 15 439 445
59. HeusJJZonneveldBJSteensmaHYvan den BergJA 1993 The consensus sequence of Kluyveromyces lactis centromeres shows homology to functional centromeric DNA from Saccharomyces cerevisiae. Mol Gen Genet 236 355 362
60. BellochCBarrioEGarciaMDQuerolA 1998 Inter - and intraspecific chromosome pattern variation in the yeast genus Kluyveromyces. Yeast 14 1341 1354
61. HegemannJHFleigUN 1993 The centromere of budding yeast. Bioessays 15 451 460
62. HegemannJHSheroJHCottarelGPhilippsenPHieterP 1988 Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol 8 2523 2535
63. NiedenthalRStollRHegemannJH 1991 In vivo characterization of the Saccharomyces cerevisiae centromere DNA element I, a binding site for the helix-loop-helix protein CPF1. Mol Cell Biol 11 3545 3553
64. JehnBNiedenthalRHegemannJH 1991 In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol Cell Biol 11 5212 5221
65. KetelCWangHSMcClellanMBouchonvilleKSelmeckiA 2009 Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5 e1000400 doi:10.1371/journal.pgen.1000400
66. DiedeSJGottschlingDE 1999 Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99 723 733
67. KramerKMHaberJE 1993 New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev 7 2345 2356
68. PutnamCDPennaneachVKolodnerRD 2004 Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101 13262 13267
69. MyungKDattaAKolodnerRD 2001 Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104 397 408
70. WyrickJJHolstegeFCJenningsEGCaustonHCShoreD 1999 Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402 418 421
71. LoneyERInglisPWSharpSPrydeFEKentNA 2009 Repressive and non-repressive chromatin at native telomeres in Saccharomyces cerevisiae. Epigenetics Chromatin 2 18
72. MartinAMPouchnikDJWalkerJLWyrickJJ 2004 Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics 167 1123 1132
73. BatadaNNHurstLD 2007 Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet 39 945 949
74. PalCPappBLercherMJ 2006 An integrated view of protein evolution. Nat Rev Genet 7 337 348
75. DrummondDABloomJDAdamiCWilkeCOArnoldFH 2005 Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A 102 14338 14343
76. PalCPappBHurstLD 2001 Highly expressed genes in yeast evolve slowly. Genetics 158 927 931
77. PalCPappBHurstLD 2003 Genomic function: Rate of evolution and gene dispensability. Nature 421 496 497; discussion 497–498
78. KentWJBaertschRHinrichsAMillerWHausslerD 2003 Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A 100 11484 11489
79. MyungKChenCKolodnerRD 2001 Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411 1073 1076
80. MieczkowskiPAMieczkowskaJODominskaMPetesTD 2003 Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc Natl Acad Sci U S A 100 10854 10859
81. LitiGLouisEJ 2003 NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol Cell 11 1373 1378
82. GreenwoodJCooperJP 2009 Trapping Rap1 at the telomere to prevent chromosome end fusions. EMBO J 28 3277 3278
83. PennaneachVPutnamCDKolodnerRD 2006 Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol 59 1357 1368
84. ButlerGKennyCFaganAKurischkoCGaillardinC 2004 Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc Natl Acad Sci U S A 101 1632 1637
85. DelneriDColsonIGrammenoudiSRobertsINLouisEJ 2003 Engineering evolution to study speciation in yeasts. Nature 422 68 72
86. De SchutterKLinYCTielsPVan HeckeAGlinkaS 2009 Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol 27 561 566
87. ButlerGRasmussenMDLinMFSantosMASakthikumarS 2009 Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459 657 662
88. HedtkeSMTownsendTMHillisDM 2006 Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol 55 522 529
Štítky
Genetika Reprodukčná medicína
Článek Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal MutationsČlánek Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene ExpressionČlánek Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary IntegrityČlánek A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with ExpressionČlánek Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line PedigreeČlánek Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 7- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Gene-Based Tests of Association
- The Demoiselle of X-Inactivation: 50 Years Old and As Trendy and Mesmerising As Ever
- Variants in and Underlie Natural Variation in Translation Termination Efficiency in
- SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
- Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal Mutations
- Genetic Anticipation Is Associated with Telomere Shortening in Hereditary Breast Cancer
- Identification of a Mutation Associated with Fatal Foal Immunodeficiency Syndrome in the Fell and Dales Pony
- Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene Expression
- An Epigenetic Switch Involving Overlapping Fur and DNA Methylation Optimizes Expression of a Type VI Secretion Gene Cluster
- Recombination and Population Structure in
- A Rice Plastidial Nucleotide Sugar Epimerase Is Involved in Galactolipid Biosynthesis and Improves Photosynthetic Efficiency
- A Role for Phosphatidic Acid in the Formation of “Supersized” Lipid Droplets
- Colon Stem Cell and Crypt Dynamics Exposed by Cell Lineage Reconstruction
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Interactions between Glucocorticoid Treatment and Cis-Regulatory Polymorphisms Contribute to Cellular Response Phenotypes
- Translation Reinitiation Relies on the Interaction between eIF3a/TIF32 and Progressively Folded -Acting mRNA Elements Preceding Short uORFs
- DAF-12 Regulates a Connected Network of Genes to Ensure Robust Developmental Decisions
- Adult Circadian Behavior in Requires Developmental Expression of , But Not
- Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary Integrity
- Proteins in the Nutrient-Sensing and DNA Damage Checkpoint Pathways Cooperate to Restrain Mitotic Progression following DNA Damage
- Complex Evolutionary Events at a Tandem Cluster of Genes Resulting in a Single-Locus Genetic Incompatibility
- () and Its Regulated Homeodomain Gene Mediate Abscisic Acid Response in
- A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with Expression
- LGI2 Truncation Causes a Remitting Focal Epilepsy in Dogs
- Adaptations to Endosymbiosis in a Cnidarian-Dinoflagellate Association: Differential Gene Expression and Specific Gene Duplications
- The Translation Initiation Factor eIF4E Regulates the Sex-Specific Expression of the Master Switch Gene in
- Somatic Genetics Empowers the Mouse for Modeling and Interrogating Developmental and Disease Processes
- Molecular Mechanisms Generating and Stabilizing Terminal 22q13 Deletions in 44 Subjects with Phelan/McDermid Syndrome
- Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line Pedigree
- Mechanisms of Chromosome Number Evolution in Yeast
- Regulatory Cross-Talk Links Chromosome II Replication and Segregation
- Ancestral Genes Can Control the Ability of Horizontally Acquired Loci to Confer New Traits
- Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
- Rare and Common Regulatory Variation in Population-Scale Sequenced Human Genomes
- The Epistatic Relationship between BRCA2 and the Other RAD51 Mediators in Homologous Recombination
- Identification of Novel Genetic Markers Associated with Clinical Phenotypes of Systemic Sclerosis through a Genome-Wide Association Strategy
- NatF Contributes to an Evolutionary Shift in Protein N-Terminal Acetylation and Is Important for Normal Chromosome Segregation
- Araucan and Caupolican Integrate Intrinsic and Signalling Inputs for the Acquisition by Muscle Progenitors of the Lateral Transverse Fate
- Pathologic and Phenotypic Alterations in a Mouse Expressing a Connexin47 Missense Mutation That Causes Pelizaeus-Merzbacher–Like Disease in Humans
- Recombinant Inbred Line Genotypes Reveal Inter-Strain Incompatibility and the Evolution of Recombination
- Epistatic Relationships in the BRCA1-BRCA2 Pathway
- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Genetic Loci Associated with Plasma Phospholipid n-3 Fatty Acids: A Meta-Analysis of Genome-Wide Association Studies from the CHARGE Consortium
- Fine Mapping of Five Loci Associated with Low-Density Lipoprotein Cholesterol Detects Variants That Double the Explained Heritability
- CHD1 Remodels Chromatin and Influences Transient DNA Methylation at the Clock Gene
- Nonlinear Fitness Landscape of a Molecular Pathway
- Genome-Wide Scan Identifies , , and as Novel Risk Loci for Systemic Sclerosis
- Quantitative and Qualitative Stem Rust Resistance Factors in Barley Are Associated with Transcriptional Suppression of Defense Regulons
- A Systematic Screen for Tube Morphogenesis and Branching Genes in the Tracheal System
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Gene-Based Tests of Association
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



