-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
The extracellular matrix (ECM) supports vascular integrity during embryonic development. Proteolytic degradation of ECM components is required for angiogenesis, but excessive ECM proteolysis causes blood vessel fragility and hemorrhage. Little is understood about how ECM proteolysis is transcriptionally regulated during embryonic vascular development. We now show that the NuRD ATP-dependent chromatin-remodeling complex promotes vascular integrity by preventing excessive ECM proteolysis in vivo. Mice lacking endothelial CHD4—a catalytic subunit of NuRD complexes—died at midgestation from vascular rupture. ECM components surrounding rupture-prone vessels in Chd4 mutants were significantly downregulated prior to embryonic lethality. Using qPCR arrays, we found two critical mediators of ECM stability misregulated in mutant endothelial cells: the urokinase-type plasminogen activator receptor (uPAR or Plaur) was upregulated, and thrombospondin-1 (Thbs1) was downregulated. Chromatin immunoprecipitation assays showed that CHD4-containing NuRD complexes directly bound the promoters of these genes in endothelial cells. uPAR and THBS1 respectively promote and inhibit activation of the potent ECM protease plasmin, and we detected increased plasmin activity around rupture-prone vessels in Chd4 mutants. We rescued ECM components and vascular rupture in Chd4 mutants by genetically reducing urokinase (uPA or Plau), which cooperates with uPAR to activate plasmin. Our findings provide a novel mechanism by which a chromatin-remodeling enzyme regulates ECM stability to maintain vascular integrity during embryonic development.
Published in the journal: The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis. PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004031
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004031Summary
The extracellular matrix (ECM) supports vascular integrity during embryonic development. Proteolytic degradation of ECM components is required for angiogenesis, but excessive ECM proteolysis causes blood vessel fragility and hemorrhage. Little is understood about how ECM proteolysis is transcriptionally regulated during embryonic vascular development. We now show that the NuRD ATP-dependent chromatin-remodeling complex promotes vascular integrity by preventing excessive ECM proteolysis in vivo. Mice lacking endothelial CHD4—a catalytic subunit of NuRD complexes—died at midgestation from vascular rupture. ECM components surrounding rupture-prone vessels in Chd4 mutants were significantly downregulated prior to embryonic lethality. Using qPCR arrays, we found two critical mediators of ECM stability misregulated in mutant endothelial cells: the urokinase-type plasminogen activator receptor (uPAR or Plaur) was upregulated, and thrombospondin-1 (Thbs1) was downregulated. Chromatin immunoprecipitation assays showed that CHD4-containing NuRD complexes directly bound the promoters of these genes in endothelial cells. uPAR and THBS1 respectively promote and inhibit activation of the potent ECM protease plasmin, and we detected increased plasmin activity around rupture-prone vessels in Chd4 mutants. We rescued ECM components and vascular rupture in Chd4 mutants by genetically reducing urokinase (uPA or Plau), which cooperates with uPAR to activate plasmin. Our findings provide a novel mechanism by which a chromatin-remodeling enzyme regulates ECM stability to maintain vascular integrity during embryonic development.
Introduction
The extracellular matrix (ECM) plays a critical role in maintaining blood vessel integrity by serving as a scaffold to which endothelial cells and vascular support cells can adhere [1], [2]. Signals between ECM components and endothelial cells likewise promote vessel stabilization by influencing endothelial cell proliferation, morphology, and survival. Aberrant degradation of ECM components is associated with life-threatening aortic and cerebral aneurysms, which are characterized by vascular fragility or rupture [3], [4].
ECM composition and stability also impact vascular integrity during embryonic development. Mice with mutations in ECM components or in regulators of ECM deposition die during midgestation with vascular rupture and hemorrhage [5], [6]. The timing of these lethal phenotypes is likely influenced by the combination of weakened vascular ECM and increasing intravascular pressure that results from embryonic heart rate acceleration during midgestation [7]. Mice with mutations that cause excessive vascular ECM degradation also succumb to vascular rupture at midgestation [8], [9]. ECM proteases are produced by activated endothelial cells during vascular development and are necessary for sprouting angiogenesis [10]. However, ECM proteases must be tightly regulated to prevent excessive proteolysis of the vascular matrix and blood vessel fragility. Currently, the transcriptional regulation and coordination of ECM proteolysis during vascular development is poorly understood.
Epigenetic factors are important transcriptional regulators of developmental processes, and increasing evidence supports their roles in cardiovascular development. Enzymes that add or subtract covalent modifications from histone tails affect transcription of target genes that impact heart and blood vessel development [11], [12]. Likewise, ATP-dependent chromatin-remodeling complexes, which transiently displace nucleosomes at gene regulatory regions, mediate transcriptional regulation of genes involved in cardiovascular development [13], [14]. Mammalian Nucleosome-Remodeling and Histone Deacetylase (NuRD) chromatin-remodeling complexes contain both histone-modifying and chromatin-remodeling enzymes [15]–[17]. These multi-subunit complexes incorporate histone deacetylases 1 and 2 (HDAC1 and HDAC2), which remove acetyl groups from histone tails. NuRD complexes also contain ATPase catalytic subunits belonging to the SNF2 superfamily: the Chromodomain-Helicase-DNA-binding proteins CHD3 and CHD4 (also called Mi-2α and Mi-2β, respectively). CHD3 and CHD4 contain protein - and DNA-binding domains in addition to an ATPase domain that provides energy for nucleosome remodeling [18], [19]. NuRD complexes were traditionally thought to act in a transcriptionally repressive manner, since histone deacetylation is associated with gene silencing. However, evidence from developing blood and nerve cell lineages indicates that NuRD complexes can facilitate target gene activation in addition to silencing [20]–[22]. Although our lab previously showed NuRD negatively regulates vascular Wnt signaling in the developing extraembryonic yolk sac [23], a role for NuRD in the embryonic vasculature has not yet been described.
We now report that embryos depleted of the NuRD ATPase CHD4 in vascular endothelial cells died by embryonic day 11.5 (E11.5) with blood vessel rupture and massive hemorrhage. Our studies reveal coordinated chromatin-based mechanisms by which CHD4-containing NuRD complexes transcriptionally regulate ECM proteolysis to maintain vascular integrity during embryonic development.
Results
Chd4fl/fl;Tie2-Cre+ Embryos Die at Midgestation
We previously deleted Chd4 from embryonic endothelial cells with a Tie2-Cre transgenic mouse line in order to assess the role of CHD4-containing NuRD complexes in vascular development. We found Wnt signaling was upregulated in Chd4fl/fl;Tie2-Cre+ yolk sac vessels at E10.5, but embryonic vascular phenotypes were not apparent at this developmental time point [23]. In order to determine whether Chd4fl/fl;Tie2-Cre+ embryos could survive development, we mated Chd4fl/+ and Chd4fl/+;Tie2-Cre+ mice together and expected to obtain 12.5% Chd4fl/fl;Tie2-Cre+ offspring. However, no Chd4fl/fl;Tie2-Cre+ animals were detected at weaning (Figure 1A). These results indicated that expression of Chd4 on developing endothelial cells was important for embryonic survival.
Fig. 1. Chd4fl/fl;Tie2-Cre+ embryos undergo vascular rupture by E11.25. 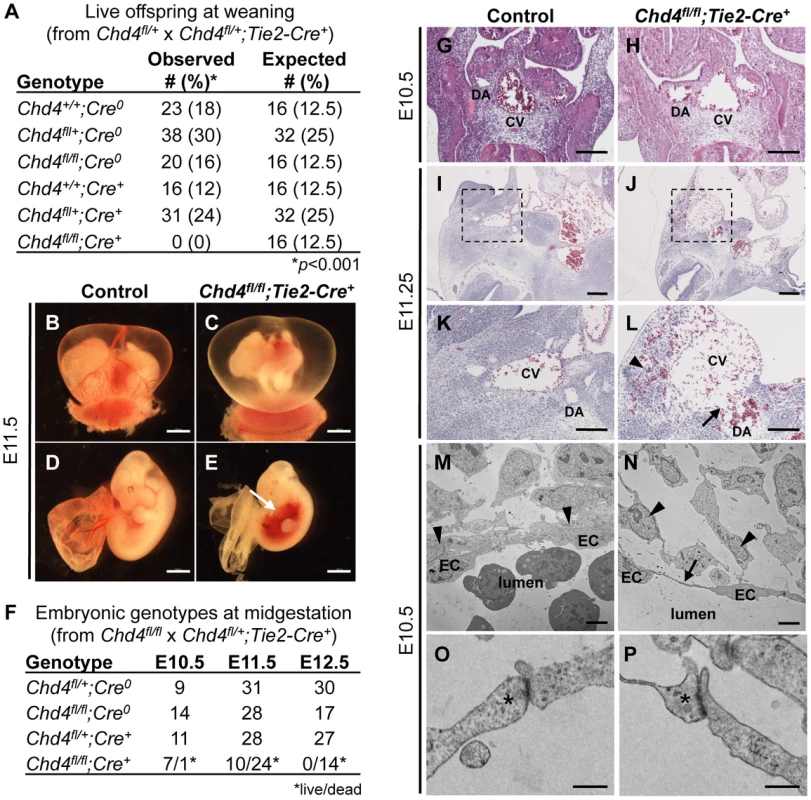
(A) Chd4fl/+ females were mated with Chd4fl/+;Tie2-Cre+ males, and live progeny from 22 litters were genotyped and scored at weaning. No live Chd4fl/fl;Tie2-Cre+ mice were recovered [χ2(5dof): p<0.001]. (B–E) Gross images of E11.5 littermate control (B,D) and Chd4fl/fl;Tie2-Cre+ (C,E) embryos. Arrow in panel E indicates massive hemorrhage within the ventral trunk region of a Chd4fl/fl;Tie2-Cre+ embryo. (F) Chd4fl/fl females were mated with Chd4fl/+;Tie2-Cre+ males, and dissections were performed on E10.5–12.5 embryos. Dead embryos were characterized by absence of heartbeat and onset of necrosis. No surviving Chd4fl/fl;Tie2-Cre+ embryos were found at E12.5 [χ2(3dof): p<0.01]. (G–L) Hematoxylin and eosin (H&E) staining of E10.5 (G,H) or E11.2±5 (I–L) littermate control (G,I,K) and Chd4fl/fl;Tie2-Cre+ (H,J,L) embryos. Boxed regions in panels I and J are shown at higher magnification in panels K and L respectively. Arrow in panel L reveals site of vascular rupture between the mutant dorsal aorta and cardinal vein; arrowhead indicates blood in extravascular tissues. DA, dorsal aorta; CV, cardinal vein. (M–P) Transmission electron micrographs from vessel walls of E10.5 littermate control (M,O) and Chd4fl/fl;Tie2-Cre+ (N,P) embryos. Arrowheads in panels M and N indicate smooth muscle cells adjacent to endothelial cells (ECs). Arrow in panel N points to a long, thin EC extension. (O,P) Endothelial cell junctions (*) are intact in both control and mutant sections. Scale bars: 1 mm (B–E); 100 µm (G–L); 2 µm (M,N); 500 nm (O,P). Since many mutants with vascular defects die during midgestation [24], we focused on this time period for preliminary analysis of our Chd4fl/fl;Tie2-Cre+ embryos. CHD4 is broadly expressed at E10.5, and we detected it by immunostaining in endothelial cells of both large and small vessels (Figure S1). We performed dissections on E10.5–12.5 embryos and consistently found Chd4fl/fl;Tie2-Cre+ embryos with massive hemorrhage in the trunk region at E11.5 (Figure 1E). Chd4fl/fl;Tie2-Cre+ embryos and yolk sacs appeared pale in comparison to littermate controls due to pooling of embryonic blood (Figure 1B–1E). Prior to hemorrhage, mutant embryos were visibly normal and displayed minimal developmental delay compared to E10.5 littermate controls. Thus, we determined that Chd4fl/fl;Tie2-Cre+ embryos died at E11.0–11.75 from sudden and massive hemorrhage, since embryos appeared normal at E10.5 and were found dead by E12.5 (Figure 1F).
Chd4fl/fl;Tie2-Cre+ Dorsal Aortae and Cardinal Veins Are Prone to Rupture
The localized hemorrhage observed at E11.5 suggested vascular rupture in the trunk region of Chd4fl/fl;Tie2-Cre+ embryos. Histological analysis revealed rupture of the dorsal aortae and cardinal veins of Chd4fl/fl;Tie2-Cre+ embryos at E11.25 (Figure 1J and 1L), corresponding closely with the time of death. However, there was no indication of impending vascular rupture or evidence of blood leakage from Chd4fl/fl;Tie2-Cre+ dorsal aortae or cardinal veins at E10.5 (Figure 1H). Vascular patterning within Chd4fl/fl;Tie2-Cre+ embryos was largely normal at E10.5 (Figure S2A–S2F). Likewise, vascular patterning was comparable in control and Chd4fl/fl;Tie2-Cre+ yolk sacs at E10.5 (Figure S2G–S2K), as we previously reported [23]. Chd4fl/fl;Tie2-Cre+ hearts showed slight evidence of hypotrabeculation compared to littermate controls at E10.5 (Figure S3D). This hypotrabeculation was accompanied by a subtle decrease in cardiac ECM, as assessed by Alcian blue staining (Figure S3B). Since cardiac ECM is critical for supporting trabeculation [25], we suspect that Chd4fl/fl;Tie2-Cre+ hypotrabeculation is secondary to reduced cardiac ECM. Nevertheless, hypotrabeculation and diminished cardiac ECM are not associated with vascular rupture in other mutants [25]–[30]. Therefore the sudden vascular rupture and lethal hemorrhage in Chd4fl/fl;Tie2-Cre+ embryos did not likely result from grossly observable cardiovascular anomalies.
Chd4fl/fl;Tie2-Cre+ Endothelial Cells Have Long Cytoplasmic Extensions but Intact Intercellular Junctions Prior to Vascular Rupture
In order to evaluate the morphology of Chd4fl/fl;Tie2-Cre+ endothelial cells prior to vascular rupture, we processed E10.5 control and mutant littermate embryos for transmission electron microscopy (TEM). We found that endothelial cells lining Chd4fl/fl;Tie2-Cre+ vessels had long, thin extensions between cells that were not seen in control endothelial cells (Figure 1M and 1N). In addition, we observed smooth muscle cells attached tightly to endothelial cells outside control dorsal aortae (Figure 1M, arrowheads), but these attachments were disrupted outside Chd4fl/fl;Tie2-Cre+ dorsal aortae (Figure 1N). These phenotypes indicated that Chd4fl/fl;Tie2-Cre+ vessels were fragile and lacked closely apposed supporting smooth muscle cells prior to rupture.
We also evaluated Chd4fl/fl;Tie2-Cre+ endothelial cell proliferation and viability prior to vascular rupture. At E10.5, no differences were seen in numbers of proliferating endothelial cells within dorsal aortae of control and Chd4fl/fl;Tie2-Cre+ embryos, as assessed by immunostaining with antibodies against the proliferation markers phosphorylated histone H3 (PPH3) or Ki67 (Figure S4A–S4D). Likewise no differences were seen in numbers of apoptotic endothelial cells in control and Chd4fl/fl;Tie2-Cre+ embryos following TUNEL staining or immunostaining with an antibody against active Caspase 3 (Figure S4E–S4H). We concluded that the long endothelial cell extensions and loosely connected smooth muscle cells lining Chd4fl/fl;Tie2-Cre+ vessels at E10.5 were not influenced by aberrant endothelial cell apoptosis or deficient proliferation.
Since mutant embryos with defective endothelial cell junctions can display thin, extended endothelial cells prior to hemorrhagic events [31], we analyzed endothelial junctional components and morphology in control and Chd4fl/fl;Tie2-Cre+ embryos prior to vascular rupture. We confirmed that intercellular junctions appeared morphologically similar by TEM in control and Chd4fl/fl;Tie2-Cre+ endothelial cells at E10.5 (Figure 1O and 1P). Likewise, E10.5 control and Chd4fl/fl;Tie2-Cre+ dorsal aortae immunostained with antibodies against the adherens junction protein vascular endothelial cadherin (VE-cadherin; Figure S5A and S5B) or the tight junction protein zonula occludens-1 (ZO-1; Figure S5C and S5D) revealed comparable levels and patterns of expression. Therefore, our data suggested that defective cell junctions did not contribute to the thin endothelial cell extensions we saw at E10.5 or to vascular rupture at E11.5.
Extracellular Matrix Components Are Diminished around Chd4fl/fl;Tie2-Cre+ Rupture-Prone Vessels
In addition to endothelial cell junctions, the extracellular matrix plays an important role in the maintenance of vascular integrity [2]. One element of the ECM, the basement membrane, is a sheet-like structure that lines and supports endothelial cells. We immunostained sections of control and Chd4fl/fl;Tie2-Cre+ embryos with an antibody against type IV collagen—a major component of the basement membrane—to assess its expression around rupture-prone dorsal aortae. Type IV collagen was diminished around Chd4fl/fl;Tie2-Cre+ versus control dorsal aortae at E10.5 (Figure 2A and 2B). Likewise, fibronectin, another ECM component that is abundantly expressed around dorsal aortae at midgestation [32], was downregulated around Chd4fl/fl;Tie2-Cre+ versus control dorsal aortae at E10.5 (Figure 2C and 2D). Therefore ECM components were diminished around Chd4fl/fl;Tie2-Cre+ dorsal aortae prior to the vascular rupture we consistently saw in mutant embryos at E11.5. Interestingly, type IV collagen and fibronectin were not significantly diminished around small vessels we examined in mutant limb buds (data not shown), and these microvessels were not prone to rupture in Chd4fl/fl;Tie2-Cre+ mutants.
Fig. 2. Extracellular matrix (ECM) components are diminished around Chd4fl/fl;Tie2-Cre+ vessels prior to rupture. 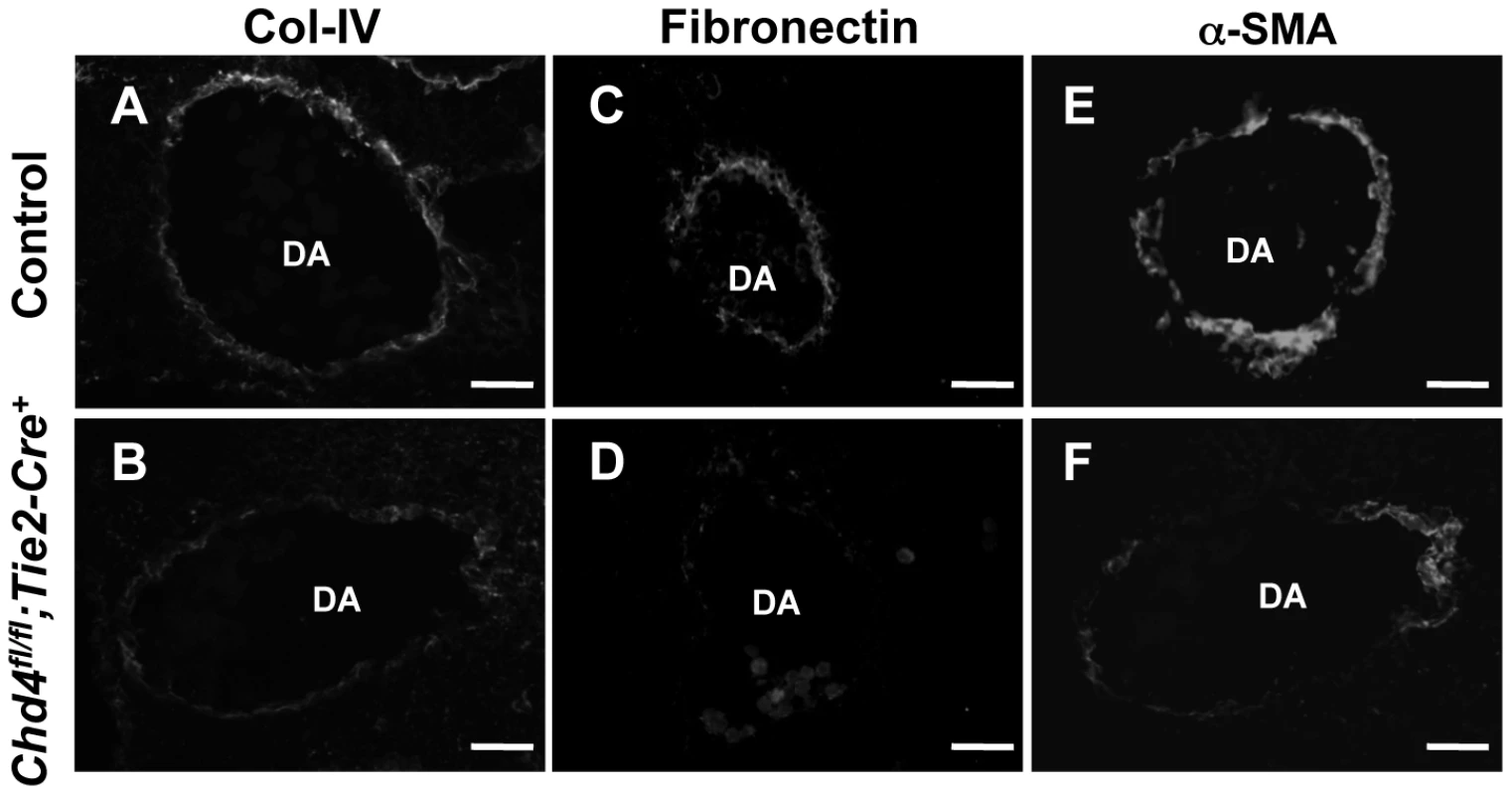
Histological sections of dorsal aortae (DA) from E10.5 littermate control (A,C,E) and Chd4fl/fl;Tie2-Cre+ (B,D,F) embryos were stained with antibodies against the ECM basement membrane component type IV collagen (Col-IV; A,B) or the ECM component fibronectin (C,D) or against the smooth muscle cell differentiation marker α-SMA (E,F). Representative images from three independent experiments on separate sets of embryos are shown. Scale bars: 50 µm (A–F). Because smooth muscle cells (SMCs) provide mechanical support for developing arteries, we also assessed E10.5 Chd4fl/fl;Tie2-Cre+ dorsal aortae for expression of the SMC differentiation marker alpha-smooth muscle actin (α-SMA). Cells surrounding Chd4fl/fl;Tie2-Cre+ dorsal aortae expressed less α-SMA than those surrounding control dorsal aortae (Figure 2E and 2F). Since α-SMA expression is influenced by ECM components such as type IV collagen [33], we suspected the reduced type IV collagen around rupture-prone Chd4fl/fl;Tie2-Cre+ dorsal aortae contributed to diminished α-SMA levels on SMCs around those vessels. Alternatively, the reduced α-SMA expression and loose attachments between endothelial cells and SMCs observed by TEM (Figure 1N) could indicate that mutant endothelial cells failed to recruit SMCs effectively. To address this possibility, we examined expression of several genes implicated in SMC recruitment toward endothelium in E10.5 control versus Chd4fl/fl;Tie2-Cre+ endothelial cells (Tgfb1, Tgfbr1, Eng, Angpt1, Angpt2, and S1pr1; Table S1) or in an endothelial cell line depleted of CHD4 (Hbegf and Tgfbr2; Figure S6). We did not see significant expression changes in these genes, although we did see downregulation of Tie2 (Tek; Table S2), which plays important roles in communication between endothelial cells and SMCs [34]. Additionally, we saw significant upregulation of Pdgfb in CHD4-depleted endothelial cells (Figure S6), which reduces α-SMA expression on vascular SMCs [35], [36]. Therefore, α-SMA expression on cells surrounding rupture-prone Chd4fl/fl;Tie2-Cre+ vessels could be diminished due to multiple causes. Overall, we hypothesized that Chd4fl/fl;Tie2-Cre+ vessels were susceptible to rupture because their walls were weakened from a paucity of ECM components and differentiated SMCs.
CHD4 Directly Inhibits Expression of uPAR
Since CHD4 typically exerts its cellular actions through transcriptional regulation of target genes, we sought to determine which endothelial CHD4 target genes could impact ECM composition. We isolated endothelial cells from E10.5 littermate control and Chd4fl/fl;Tie2-Cre+ and embryos and identified significant gene expression changes using two commercially available quantitative real-time PCR (qPCR) arrays containing a total of 157 genes. We chose these arrays because they contained many genes with important roles in ECM composition or proteolysis. Notably, we did not see significant downregulation of genes encoding midgestational ECM components, such as collagens, laminins, and fibronectin (Table S1). Nor did we see upregulation of matrix metalloproteinases that could contribute to ECM proteolysis. However, the arrays revealed the urokinase-type plasminogen activator (uPA, encoded by the gene Plau) was significantly upregulated in Chd4fl/fl;Tie2-Cre+ endothelial cells (Table S2). This result was notable since we had previously identified the uPA receptor (uPAR, encoded by the gene Plaur) as a CHD4 target gene in yolk sac vessels but had not determined the functional relevance of its upregulation in Chd4fl/fl;Tie2-Cre+ endothelial cells [23]. uPAR regulates ECM proteolysis by binding and localizing uPA to cell surfaces where it activates the serine protease plasmin [37]. Therefore, we hypothesized that upregulation of uPA/uPAR and subsequent plasmin activity might contribute to ECM fragility, vascular rupture, and lethality in Chd4fl/fl;Tie2-Cre+ embryos.
In order to validate the qPCR array results and determine whether Plau expression was altered in our mutant embryos prior to vascular rupture, we isolated endothelial cells from E10.5 control and Chd4fl/fl;Tie2-Cre+ embryos and performed qPCR using primers specific for Plau and Plaur. Indeed, we found both Plau and Plaur were significantly upregulated in Chd4fl/fl;Tie2-Cre+ endothelial cells (Figure 3A). We next performed chromatin immunoprecipitation (ChIP) assays to determine whether Plau—like Plaur—is a direct CHD4 target gene. In order to obtain the number of cells required for ChIP, we used the C166 yolk sac-derived murine endothelial cell line [38]. We chose this cell line for ChIP because knockdown of CHD4 in C166 cells resulted in upregulation of Plaur mRNA (Figure 3C), as we saw in primary Chd4fl/fl;Tie2-Cre+ endothelial cells. Plau expression trended upward as well in CHD4 knockdown C166 cells, but it was not significantly upregulated like Plaur (Figure 3C). We found no evidence that CHD4 bound the Plau promoter in wildtype C166 cells by ChIP but confirmed that CHD4 bound the Plaur promoter in a region approximately 900 bp upstream of the transcription start site (TSS) (Figure 3D). Therefore, our data indicated that CHD4 directly inhibited Plaur—but not Plau—expression in endothelial cells. Since Plau gene expression is influenced by uPAR signaling [39], [40], we suspected that the upregulated Plau expression we saw in Chd4fl/fl;Tie2-Cre+ endothelial cells was secondary to upregulated Plaur. Importantly, we also detected binding of HDAC1 to the same region of the Plaur promoter at which we detected CHD4 binding by ChIP (Figure 3D). Since HDAC1 can participate in NuRD complexes [15], these data indicated that CHD4/HDAC1-containing NuRD complexes repressed Plaur transcription in endothelial cells.
Fig. 3. CHD4 differentially regulates Plaur and Thbs1 expression in endothelial cells. 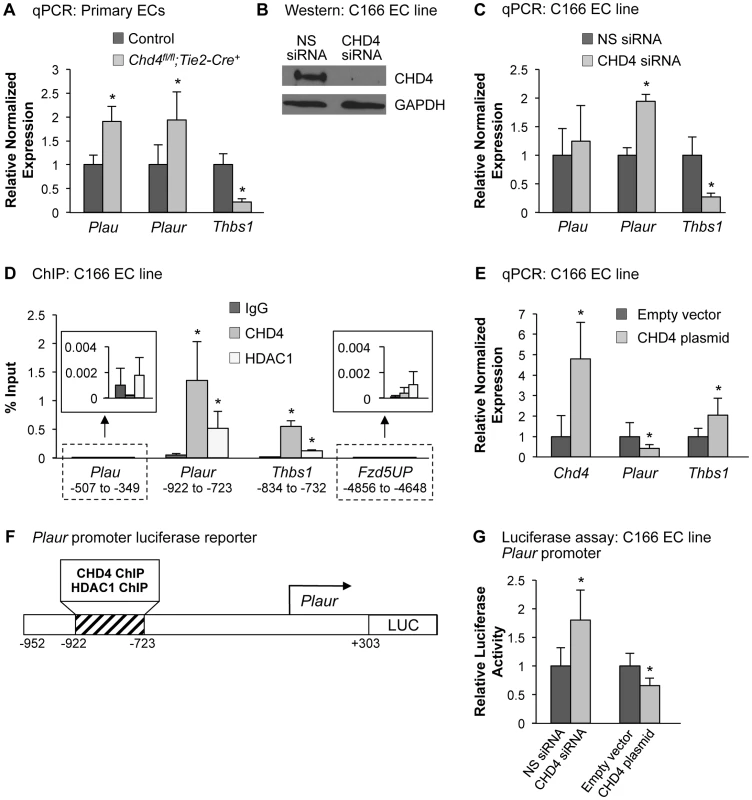
(A) qPCR with gene-specific primers for Plau, Plaur, and Thbs1 was performed on endothelial cells isolated from E10.5 littermate control and Chd4fl/fl;Tie2-Cre+ embryos. Data were normalized to the relative expression of control samples. Error bars represent SD of results from three independent experiments. (B and C) C166 endothelial cells were transfected with nonspecific (NS) or CHD4-specific siRNA oligonucleotides for 24 h. (B) Western blot analysis was performed on cell lysates using antibodies that recognize CHD4 or GAPDH. A representative blot from 3 independent experiments is shown. (C) RNA was isolated, cDNA was synthesized, and qPCR was performed using Plau-, Plaur-, or Thbs1-specific primers. Data were normalized to the relative expression of NS siRNA-treated samples. Error bars represent SD of results from three to four independent experiments. (D) Chromatin immunoprecipitation (ChIP) assays were carried out in C166 endothelial cells using antibodies against normal mouse IgG (negative control), CHD4, or HDAC1. Immunoprecipitated DNA was analyzed by qPCR to examine CHD4 and HDAC1 enrichment at the Plau, Plaur, and Thbs1 promoters. A transcriptionally inactive region approximately 5 kb upstream of the Fzd5 transcription start site (Fzd5UP) was assessed as a negative control for CHD4 and HDAC1 binding. Data are represented as a percent of total input chromatin. Error bars represent SD of results from three independent experiments. Results for Plau and Fzd5UP ChIP experiments are magnified in the insets. For the Plaur and Thbs1 ChIPs, CHD4 and HDAC1 binding were statistically compared against IgG binding at the respective loci or against CHD4 and HDAC1 binding at the Fzd5UP locus; both sets of comparisons revealed significant enrichment. (E) qPCR with gene-specific primers for Chd4, Plaur, and Thbs1 was performed on C166 endothelial cells transfected with 0.02 ng of a CHD4 expression plasmid or the analogous empty vector backbone (control). Data were normalized to the relative expression of control samples. Error bars represent SD of results from three independent experiments. (F) Schematic of the region of the murine Plaur promoter that was cloned into a luciferase (LUC) reporter plasmid for use in G. The Plaur promoter fragment encompasses the region to which CHD4 and HDAC1 were shown to bind by ChIP in D. (G) Luciferase assays were performed in C166 cells co-transfected with 250 ng of the reporter schematized in F and 10 ng of a constitutive Renilla luciferase control plasmid. Cells were also transfected with either 10 pmol of non-specific (NS) siRNA or CHD4 siRNA oligonucleotides to knock down endogenous CHD4 or with the CHD4 expression plasmid or its relevant control (empty vector) described in E. All transfections were performed for 24 h. Ratios of relative luciferase∶renilla activity were normalized to results from the control samples. Error bars represent SD of results from four independent experiments (with triplicate samples) for the siRNA-transfected samples and from five independent experiments (with triplicate samples) for the CHD4/control plasmid-transfected samples. All statistical calculations for Figure 3 were performed using a two-tailed Student's t test (*, p<0.05). To further assess the impact of CHD4 on Plaur transcription, we transfected a CHD4 expression plasmid into C166 cells and assessed Plaur transcription by qPCR. We detected significant downregulation of Plaur with the overexpression of CHD4 (Figure 3E), which complemented our genetic and knockdown data and suggested that CHD4 inhibited Plaur transcription. Finally, we generated a luciferase reporter containing a fragment of the Plaur promoter that included the region in which we detected positive CHD4 and HDAC1 binding by ChIP (Figure 3F), and examined reporter activity in C166 endothelial cells. We saw a significant increase in reporter activity upon knockdown of endogenous CHD4, and overexpression of CHD4 significantly decreased reporter activity (Figure 3G). Altogether, these data indicated that CHD4 directly and negatively regulated Plaur transcription in endothelial cells.
To assess how CHD4 mediated negative regulation of Plaur transcription, we analyzed the acetylation status of H3K9 and H3K27, two covalent histone marks associated with transcriptional activation that can be influenced by NuRD deacetylase activity [41], [42]. Following CHD4 knockdown we saw no significant changes in H3K9Ac or H3K27Ac marks in the region of the Plaur promoter that bound CHD4 and HDAC1 (Figure S7). Therefore CHD4/HDAC1-containing NuRD complexes do not appear to repress Plaur transcription through deacetylation of H3K9 or H3K27 in the promoter region we analyzed. Further analysis of the Plaur promoter in CHD4 knockdown cells will be required to discern the precise mechanism by which NuRD negatively regulates Plaur transcription in endothelium.
CHD4 Directly Promotes Expression of Thrombospondin-1
In addition to Plau, our commercial qPCR arrays also revealed that Thrombospondin-1 (Thbs1) was misregulated in Chd4fl/fl;Tie2-Cre+ endothelial cells (Table S2). However, unlike Plau, which was upregulated in the absence of CHD4, Thbs1 was significantly downregulated. This result was noteworthy because THBS1 inhibits the ECM protease plasmin as well as matrix metalloproteinases involved in ECM degradation [43], [44]. Therefore, downregulation of Thbs1 could contribute to the excessive degradation of ECM components seen in our Chd4fl/fl;Tie2-Cre+ embryos.
To validate our array data, we performed direct qPCR with Thbs1-specific primers and found a significant decrease in the transcript level of Thbs1 in endothelial cells isolated from Chd4fl/fl;Tie2-Cre+ embryos (Figure 3A). Thbs1 transcripts were similarly decreased in CHD4 knockdown C166 endothelial cells (Figure 3C). Immunostaining confirmed that THBS1 protein expression was diminished around the dorsal aortae of E9.5 Chd4fl/fl;Tie2-Cre+ embryos (Figure S8D and S8F). Furthermore, the matrix metalloproteinase MMP9—which is normally inhibited by THBS1 [44]—was upregulated around E10.5 Chd4fl/fl;Tie2-Cre+ dorsal aortae (Figures S8H and S8J). ChIP assays revealed that CHD4 and HDAC1 associated with the Thbs1 promoter approximately 800 bp upstream of the transcription start site (Figure 3D). Furthermore, overexpression of CHD4 in C166 endothelial cells resulted in significantly increased Thbs1 transcription, as measured by qPCR (Figure 3E). Altogether our data indicated that CHD4 directly promoted Thbs1 expression in endothelial cells.
To examine the mechanism by which CHD4 promoted Thbs1 expression in endothelial cells, we assessed histone acetylation by ChIP in the region of the Thbs1 promoter where CHD4 and HDAC1 bound. As with the Plaur promoter, we saw no changes in H3K9 or H3K27 acetylation upon CHD4 knockdown (Figure S7). Therefore, the mechanism by which CHD4-containing NuRD complexes promote Thbs1 expression in endothelial cells remains unknown.
Plasmin Activity Is High around Rupture-Prone Chd4fl/fl;Tie2-Cre+ Vessels
Since uPAR and THBS1 respectively promote and inhibit activation of the ECM protease plasmin [43], [45], we hypothesized that upregulated uPAR/uPA and downregulated THBS1 activities led to excessive plasmin activation around rupture-prone Chd4fl/fl;Tie2-Cre+ vessels. To test this hypothesis, we conducted in situ zymography assays on sections of E10.5 control and Chd4fl/fl;Tie2-Cre+ embryos (Figure 4A–4D). Plasmin activity within tissue sections was detected with the quenched fluorescent plasmin substrate casein, which fluoresces upon cleavage. After addition of the plasmin zymogen plasminogen, which normally circulates in plasma but must be added exogenously to tissue sections, plasmin activity was significantly elevated around Chd4fl/fl;Tie2-Cre+ versus control dorsal aortae (Figure 4D versus Figure 4B; quantification in Figure 4E). These results indicated that proteolytic activators of plasmin were upregulated around rupture-prone Chd4fl/fl;Tie2-Cre+ blood vessels. Elevated plasmin activity was also detected in E10.5 Chd4fl/fl;Tie2-Cre+ hearts (Figure S9), which may explain the slight hypotrabeculation seen in mutant hearts (Figure S3). We did not see elevated plasmin activity around microvessels in mutant limb buds (data not shown), which was consistent with the undiminished ECM components we observed around these vessels and our finding that they were not prone to rupture. Therefore, although CHD4 is expressed in endothelium lining both large and small vessels (Figure S1), it appears to regulate plasmin production preferentially around large truncal vessels that are subject to higher hemodynamic forces at midgestation.
Fig. 4. Excessive plasmin activity is detected around rupture-prone Chd4fl/fl;Tie2-Cre+ blood vessels. 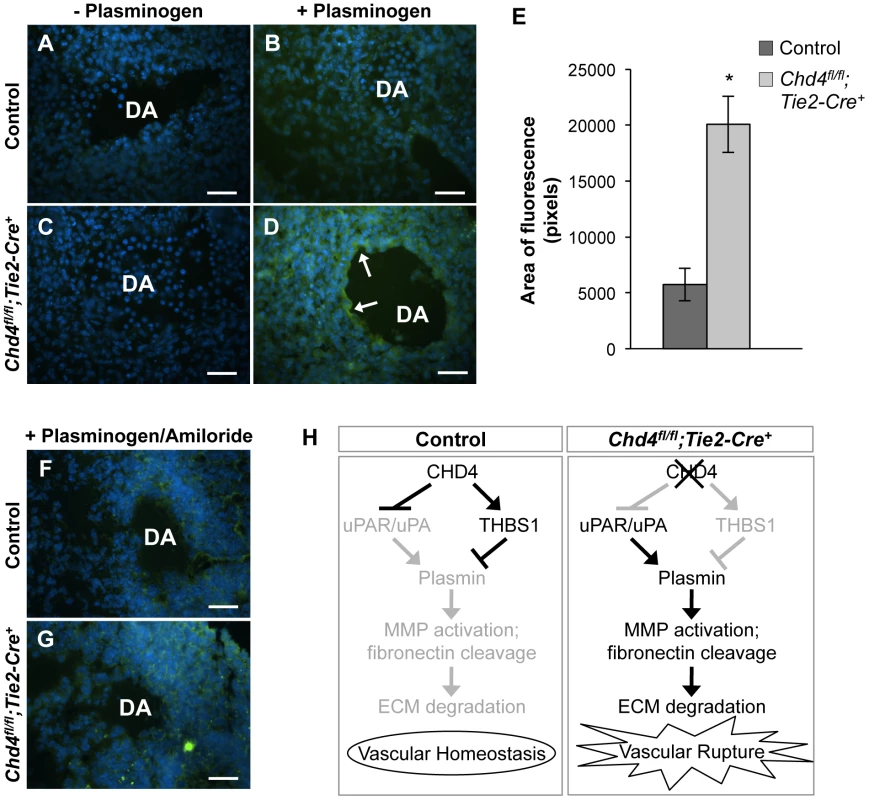
(A–D) Representative images of E10.5 littermate control (A,B) and Chd4fl/fl;Tie2-Cre+ (C,D) dorsal aortae (DA) subjected to in situ zymography for detection of plasmin activity. Unfixed sections were overlaid with quenched fluorescent casein, a plasmin substrate that fluoresces upon cleavage. In the presence of exogenous plasminogen, casein cleavage (green fluorescence) was substantially higher around Chd4fl/fl;Tie2-Cre+ versus control dorsal aortae (arrows in D versus B). The lack of significant casein cleavage in the absence of exogenous plasminogen (A and C) indicates that the casein cleavage seen in D resulted from plasmin activity. Hoechst (blue) was used for counterstaining. (E) Quantification of three independent in situ zymography experiments such as those shown in B and D from three sets of control and Chd4fl/fl;Tie2-Cre+ embryos. Fluorescence generated by casein cleavage was quantified and measured in pixels, comparing comparably sized control and Chd4fl/fl;Tie2-Cre+ dorsal aortae. Data are presented as mean ± SD. (F–G) In situ zymography for plasmin activity was performed as described in A for control and Chd4fl/fl;Tie2-Cre+ embryonic sections in the presence of the uPA inhibitor amiloride. Amiloride decreased the level of plasmin activity surrounding the Chd4fl/fl;Tie2-Cre+ DA (G) to that seen surrounding the control DA (F). (H) Model for how CHD4 impacts ECM proteolysis and vascular integrity through transcriptional regulation of Thbs1 and Plaur. (Left panel) In control endothelial cells, plasmin production and ECM degradation are curbed by CHD4-mediated inhibition of the genes encoding uPA/uPAR and activation of the gene encoding THBS1, resulting in vascular homeostasis. (Right panel) In Chd4fl/fl;Tie2-Cre+ endothelial cells, loss of CHD4 leads to increased plasmin activation, which enhances MMP activation and fibronectin cleavage. The net result is excessive ECM degradation and vascular rupture. Scale bars: 100 µm. Plasmin can be activated by two independent triggers: uPA or the tissue-type plasminogen activator (tPA). We assessed expression of tPA (Plat) by qPCR in CHD4 knockdown C166 endothelial cells to determine if tPA—like uPA—was aberrantly upregulated in the absence of CHD4. We did not see significant chances in Plat transcription (Figure S10). To further confirm that elevated uPA triggered the excessive plasmin activity seen around rupture-prone Chd4fl/fl;Tie2-Cre+ dorsal aortae, we performed in situ zymography assays on sections of control and mutant embryos in the presence of the uPA-specific inhibitor amiloride, which does not inhibit tPA activity [46]. Amiloride treatment caused a dramatic decrease in the amount of plasmin activity around Chd4fl/fl;Tie2-Cre+ dorsal aortae (Figure 4G versus 4D). These data indicated that elevated plasmin activity around rupture-prone Chd4fl/fl;Tie2-Cre+ vessels resulted from upregulated uPA. We suspected that this elevated plasmin activity was responsible for the ECM degradation and vascular rupture seen in Chd4fl/fl;Tie2-Cre+ embryos (Figure 4H), since excessive plasmin production is associated with vessel wall destruction and aneurysm formation in adult mice [47].
Genetic Reduction of Urokinase Partially Rescues Chd4fl/fl;Tie2-Cre+ Vascular Rupture at E11.5
Since urokinase (uPA) is a potent activator of plasmin and is upregulated in Chd4fl/fl;Tie2-Cre+ endothelial cells, we predicted that genetic reduction of uPA (Plau) could rescue Chd4fl/fl;Tie2-Cre+ embryos from vascular rupture. Plau−/− mice bred poorly in our colony, so we crossed Chd4fl/fl;Tie2-Cre+ embryos onto a Plau+/− background. We dissected embryos at E12.5 to determine whether urokinase reduction could rescue the vascular rupture typically seen in Chd4fl/fl;Tie2-Cre+ embryos at E11.5. At E12.5, all the Chd4fl/fl;Tie2-Cre+ embryos we assessed (N = 20) were severely necrotic and partially resorbed (Figure 5B). By contrast, 60% of Chd4fl/fl;Plau+/−;Tie2-Cre+ embryos (9 out of 15) were comparable in size to littermate controls and showed no evidence of necrosis (Figure 5A and 5C). Additionally, dorsal aortae within these visibly rescued Chd4fl/fl;Plau+/−;Tie2-Cre+ embryos were intact and appeared histologically comparable to control dorsal aortae at E12.5 (Figure S11A and S11B). However, although Chd4fl/fl;Plau+/−;Tie2-Cre+ embryos bypassed the dorsal aorta/cardinal vein rupture consistently seen in Chd4fl/fl;Tie2-Cre+ embryos at E11.5, many Chd4fl/fl;Plau+/−;Tie2-Cre+ embryos displayed blood in their brain ventricles and in the central canal of their spinal cords (Figure 5C and Figure S11D). Therefore CHD4 may play a role in central nervous system vascular formation or maintenance at E12.5. Nevertheless, these rescue data demonstrated that urokinase activity contributed to Chd4fl/fl;Tie2-Cre+ vascular rupture and lethality at E11.5.
Fig. 5. Genetic reduction of urokinase restores ECM components and partially rescues vascular rupture in Chd4fl/fl;Tie2-Cre+ embryos. 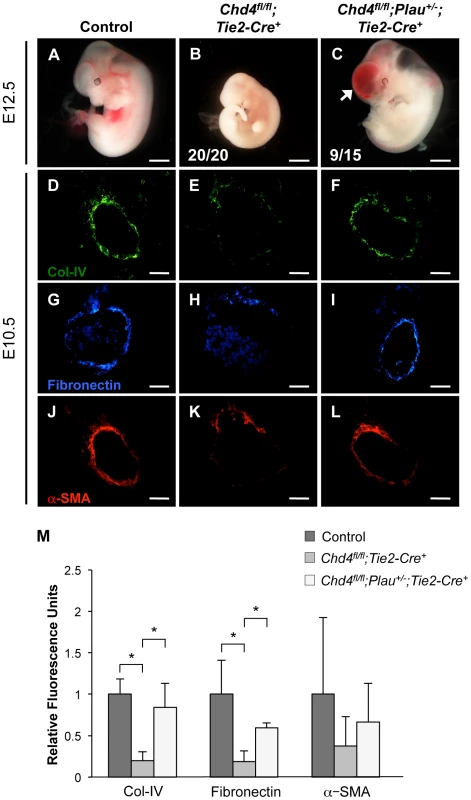
(A–C) Representative images of littermate control (A), Chd4fl/fl;Tie2-Cre+ (B), and Chd4fl/fl;Plau+/−;Tie2-Cre+ (C) embryos at E12.5. All Chd4fl/fl;Tie2-Cre+ embryos examined (20/20) were pale and necrotic (B). 60% of Chd4fl/fl;Plau+/−;Tie2-Cre+ embryos (9 out of 15) were comparable in size to littermate control embryos, although they displayed blood in the brain (arrow) and/or spinal cord (C). (D–L) Histological sections of dorsal aortae from E10.5 control, Chd4fl/fl;Tie2-Cre+, and Chd4fl/fl;Plau+/−;Tie2-Cre+ embryos were stained with antibodies against type IV collagen (Col-IV; D–F), fibronectin (G–I), or α-SMA (J–L). Scale bars: 1 mm (A–C); 50 µm (D–L). (M) Quantification of immunostained ECM components, such as those shown in panels D–L. Relative fluorescent intensity was measured and normalized to fluorescence in the control sections. Error bars represent SD of results from three independent experiments using three different sets of littermate embryos. Significant differences were calculated using a two-tailed Student's t test (*, p<0.05). Genetic Reduction of Urokinase Rescues Expression of ECM Components around Chd4fl/fl;Tie2-Cre+ Vessels
Since urokinase reduction partially rescued vascular rupture and lethality in E11.5 Chd4fl/fl;Tie2-Cre+ embryos, we questioned whether it also rescued ECM components around rupture-prone vessels. By immunostaining, we saw a significant rescue of type IV collagen and fibronectin expression levels around E10.5 Chd4fl/fl;Plau+/−;Tie2-Cre+ versus Chd4fl/fl;Tie2-Cre+ dorsal aortae (Figure 5D–5I and Figure 5M). We likewise saw an increase in α-SMA-positive cells around Chd4fl/fl;Plau+/−;Tie2-Cre+ dorsal aortae compared to Chd4fl/fl;Tie2-Cre+ dorsal aortae (Figure 5J–5M). These findings supported our central hypothesis that misregulation of uPA/uPAR activity resulted in excessive ECM proteolysis that contributed to Chd4fl/fl;Tie2-Cre+ vascular rupture and lethality at E11.5.
Discussion
The extracellular matrix plays a key role in supporting vascular integrity, however little is known about how ECM production and degradation are regulated and coordinated during embryonic blood vessel development. The first evidence that epigenetic factors could impact embryonic vascular integrity through regulation of ECM composition came from genetic deletion of the histone deacetylase HDAC7. Conditional deletion of Hdac7 in endothelial cells results in lethal vascular rupture and hemorrhage by E11.5 associated with upregulated expression of the matrix metalloproteinase Mmp10 [8]. The ATP-dependent chromatin-remodeling enzyme BRG1 similarly regulates endocardial expression of the secreted matrix metalloproteinase Adamts1, although vascular Brg1 mutants suffer from defective cardiac trabeculation rather than vascular rupture due to increased ADAMTS1 activity [25]. Our work now builds on these precedents by demonstrating that ATP-dependent chromatin-remodeling complexes also contribute to the regulation of embryonic vascular integrity by modulating expression of ECM regulators in endothelial cells. We show that NuRD coordinates differential expression of at least two genes to achieve a common outcome of ECM homeostasis. Importantly, genetic reduction of ECM proteolysis partially rescues vascular rupture in our NuRD-deficient mutants, providing convincing evidence that NuRD promotes ECM stability to maintain vascular integrity.
We hypothesize that excessive plasmin production contributes significantly to Chd4fl/fl;Tie2-Cre+ vascular rupture. Importantly, we did not see evidence of significant plasmin generation around large or small vessels in control embryos by in situ zymography, suggesting that plasmin levels are normally low at midgestation. Plasmin mediates basement membrane degradation required for sprouting angiogenesis in maternal vessels during embryonic implantation and in tumor vessels during neovascularization [48], [49]. However, it is questionable whether plasmin plays a comparable role during embryonic angiogenesis. Knockouts for various plasmin-generating components such as plasminogen (Plg), uPA (Plau), and uPAR (Plaur) all survive development without notable angiogenesis defects [50]–[53]. Moreover, plasmin-mediated activation of the matrix metalloproteinases MMP1 and MMP10 causes vascular regression rather than sprouting in vitro [54]–[56]. These findings are supported by the report that excessive activation of MMP10 in vivo results in vascular rupture similar to that seen in our Chd4fl/fl;Tie2-Cre+ embryos [8]. Therefore, plasmin may not promote vessel formation in the midgestation embryo but rather may trigger detrimental vessel breakdown by activation of pro-regressive proteinases. If true, minimization of plasmin production or activity—particularly in large vessels that are subject to high hemodynamic forces and increased fragility—would be desirable during embryonic development.
In adult tissues excessive plasmin generation and activity are curtailed by an anti-plasmin system that includes the plasminogen activator inhibitors 1 and 2 (PAI1/2) and α2-antiplasmin. However, expression levels and activities of these inhibitors in the midgestation embryo are unclear. Mice genetically depleted of PAI-1 (Serpine1), PAI-2 (Serpinb2), or α2-antiplasmin (Serpinf2)—either alone or in combination—survive development without hemorrhagic events [57]–[59]. These findings suggest that the traditional anti-plasmin system is not required for suppressing excessive plasmin production and protecting vascular integrity during embryonic development. Instead, we propose that a chromatin-based mechanism for regulating transcription of plasminogen activators may have evolved to limit plasmin production prior to the developmental time point at which a robust anti-plasmin system is established.
Plasmin directly cleaves the ECM components laminin and fibronectin and activates multiple matrix metalloproteinases (MMPs) that degrade the ECM components collagen and elastin [60]. One of the MMPs activated by plasmin, MMP9, is directly inhibited by THBS1 [44]. Therefore, THBS1 may influence ECM proteolysis and vascular integrity during development by inhibiting both plasmin and MMP9. In addition, THBS1 is a chemoattractant for vascular smooth muscle cells [61], so diminished Thbs1 expression in Chd4fl/fl;Tie2-Cre+ endothelial cells may contribute to the reduced α-SMA staining and the disrupted attachments we saw between smooth muscle cells and endothelial cells around mutant vessels. Given the multiple ways that THBS1 can impact ECM stability and vascular integrity, it is possible that the unexplained 25% embryonic lethality observed in Thbs1−/− mice [62] may be due to vascular rupture and lethal hemorrhage such as that seen in Chd4fl/fl;Tie2-Cre+ embryos. We suspect the partial rescue of vascular rupture seen in Chd4fl/fl;Plau+/−;Tie2-Cre+ embryos would be more penetrant if this cross were able to rescue additional functions of THBS1 beyond plasmin inhibition.
It is also likely that CHD4/NuRD has other primary or secondary genomic targets beyond Thbs1 and Plaur whose misregulation contribute to the vascular rupture phenotype seen in Chd4fl/fl;Tie2-Cre+ embryos. For example, integrin β3 (Itgb3) was significantly downregulated in Chd4fl/fl;Tie2-Cre+ endothelial cells (Table S2). Integrin β3 mediates interactions between the ECM and endothelial cells [63], and vascular integrity is compromised in Itgb3−/− mice, which experience postnatal hemorrhage [64]. We could not detect binding of CHD4 to the Itgb3 promoter by chromatin immunoprecipitation, so we suspect Itgb3 downregulation in Chd4fl/fl;Tie2-Cre+ endothelial cells was a secondary consequence of Chd4 deletion. Nevertheless, decreased Itgb3 in Chd4fl/fl;Tie2-Cre+ endothelial cells may partially contribute to vascular fragility at midgestation. Likewise, it is possible that misregulated Wnt signaling contributes to vascular fragility in Chd4fl/fl;Tie2-Cre+ embryos. We had previously determined that the Wnt-responsive transcription factor Tcf7 and a subset of Wnt target genes—including Plaur—are upregulated in Chd4fl/fl;Tie2-Cre+ yolk sac endothelial cells [23]. However, this upregulation of Wnt signaling does not appear to impact vascular patterning, as is seen in vascular gain-of-function mutants for the central Wnt signaling component β-catenin (β-cateninlox(ex3)/lox(ex3);Tie2-Cre+) [65]. This phenotypic dissimilarity may be due to the fact that fewer Wnt target genes are misregulated in Chd4fl/fl;Tie2-Cre+ vasculature than in β-cateninlox(ex3)/lox(ex3);Tie2-Cre+ blood vessels. Although β-cateninlox(ex3)/lox(ex3);Tie2-Cre+ embryos show no evidence of vascular rupture or hemorrhage prior to lethality at E11.5 [65], we cannot exclude the possibility that upregulated vascular Wnt signaling might contribute to blood vessel fragility in Chd4fl/fl;Tie2-Cre+ embryos.
Finally, it will be important to determine if the increased ECM proteolysis and vascular rupture seen in Chd4fl/fl;Tie2-Cre+ embryos are recapitulated in postnatal mice with induced vascular Chd4 excision. It is possible that plasminogen/plasmin inhibitors such as PAI-1 and α2-antiplasmin reduce the requirement for CHD4 in adult vasculature. However, if CHD4 limits plasmin production in adult endothelium as it does in the embryo, it may play an important preventative role in vascular pathologies such as abdominal aortic aneurysms, which are associated with excessive ECM degradation around vessel walls [66]. uPA/uPAR and plasmin have previously been recognized as promising therapeutic targets for combating aneurysm progression [67], and we now present a novel chromatin-based mechanism by which the activity of these proteins can be regulated during embryonic vascular development. Future studies will determine if NuRD influences proteolytically triggered aneurysms in adults.
Materials and Methods
Ethics Statement
The Oklahoma Medical Research Foundation (OMRF) is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the Guide for Care and Use of Laboratory Animals (National Academies Press; Washington, D.C.; 1996). The OMRF Institutional Animal Care and Use Committee approved all animal use protocols.
Mice
Chd4-floxed mice (Chd4fl/fl), uPA+/− mice, and Tie2-Cre transgenic mice have been described [20], [52], [68]. All mice were maintained on a mixed genetic background at the Oklahoma Medical Research Foundation animal facility. Tie2-Cre transgenic and Chd4-floxed embryos and mice were genotyped as described [23], [69]. uPA+/− mice and embryos were genotyped by PCR, using primer sequences obtained from Jackson Laboratory. The wildtype uPA allele was amplified with a forward primer (5′-TCTGGAGGACCGCTTATCTG-3′) and a reverse primer (5′-CTCTTCTCCAATGTGGGATTG-3′) that yielded a 153 bp amplicon. The targeted uPA allele was amplified by a neomycin-specific forward primer (5′-CACGAGACTAGTGAGACGTG-3′) and the same reverse primer used for detecting the wildtype allele, to yield a 337 bp amplicon.
Histological Staining
Hematoxylin and eosin (H&E) staining was performed on embryo cryosections as described previously [70]. Alcian blue staining was performed on paraffin sections of embryonic hearts. Sections were dewaxed, rehydrated, and stained with a 1% solution of Alcian blue 8 GX (Sigma) in acetic acid for 30 min. After Alcian blue staining, sections were counterstained with Nuclear Fast Red (Vector Laboratories) for 5 min.
Transmission Electron Microscopy
Tissue samples were fixed by immersion in a mixture of 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate for 1 h, followed by postfixation in 1% osmium tetroxide for 90 min and 1% tannic acid for 1 h. The samples were subsequently dehydrated in a graded ethanol series and embedded in epoxy resin (Electron Microscopy Sciences). Ultrathin sections (70 nm) were obtained using an RMC 7000 ultramicrotome (Boeckeler Instruments) equipped with a diamond knife. Sections were stained with uranyl acetate and lead citrate before being viewed with a Hitachi H-7600 electron microscope equipped with a 4 megapixel digital monochrome camera and AMT-EM image acquisition software (Advanced Microscopy Techniques).
Immunohistochemistry
Chd4fl/fl;Tie2-Cre+ and littermate control embryos were dissected, fixed in 4% PFA overnight, and passed through 10% sucrose for 10 min, 15% sucrose for 10 min, 20% sucrose for 1 hr and a 1∶1 mixture of 20% sucrose and Optimal Cutting Temperature compound (O.C.T.; Sakura Finetek) overnight. Embryos were embedded in O.C.T. the following morning. Yolk sac tissue was used for genotyping. 8 µm sections were cut with a Microm HM 505 E cryotome (Microm International) and adhered to Superfrost Plus slides (Fisher Scientific). Histological sections were blocked in blocking solution [5% normal goat serum/5% normal donkey serum/0.3% bovine serum albumin/0.1% Triton X-100 in phosphate buffered saline (PBS)] for 2 h at room temperature. Sections were incubated in primary antibody at a 1∶100–1∶500 concentration in blocking solution for 1 h at room temperature, washed three times (3 min each) in 0.1% Triton X-100/PBS, then incubated for 1 h at room temperature in secondary antibody at a 1∶500 concentration in blocking solution along with 20 ug/mL Hoechst stain. Sections were then washed three times (3 min each) in 0.1% Triton X-100/PBS and coverslipped with 2.5% DABCO/90% glycerol/PBS, pH 8.6. Primary antibodies used for immunohistochemistry were rat-anti-PECAM-1 (BD Pharmingen, 553370); rabbit-anti-phospho-histone H3 (Millipore, 06-570); rabbit-anti-Ki67 (Thermo Scientific, RM-9106-SO); Cy3-labeled mouse-anti-α-SMA (Sigma-Aldrich, C6198); rabbit-anti-active-Caspase 3 (Abcam, ab13847); mouse-anti-THBS-1 (Abcam, ab1823); rat-anti-VE-Cadherin (BD Pharmingen, 550548); rabbit-anti-Col-IV (Abcam, ab19808); mouse-anti-Fibronectin (Sigma, F6140); rabbit-anti-MMP9 (Abcam, ab38898); rabbit-anti-CHD4 (Active Motif, 39289); and rabbit-anti-ZO-1 [71]. Secondary antibodies used for immunohistochemistry were Cy3-donkey-anti-rat IgG (Jackson ImmunoResearch), Alexa 488-goat-anti-rabbit IgG (Invitrogen) and Alexa 488-goat-anti-mouse IgG (Invitrogen).
Cell Culture and Transfections
The C166 murine yolk sac-derived endothelial cell line (ATCC, CRL-2581) was maintained as described [69]. For CHD4 knockdown, cells were transfected with 100 nM CHD4 siGENOME SMARTpool or nontargeting control small interfering RNA (siRNA) oligonucleotides (Dharmacon catalog numbers M-052142-01 or D-001210-01, respectively) or with 100 nM CHD4 Silencer Select or nontargeting control siRNA oligonucleotides (Life Technologies, AM4637 and 4390816, respectively) using Lipofectamine 2000 (Invitrogen) in serum-free OptiMEM (Invitrogen). After 24 h, cells were harvested in Laemmli buffer (62.5 mM Tris, pH 6.8, 10% glycerol, 5% SDS, 0.01% bromophenol blue) for Western blot analysis or TRIzol (Invitrogen) for transcript analysis. For transfection with the CHD4 expression plasmid (Fisher, MMM1013-202770503) or with its analogous empty vector backbone, which was generated by removing CHD4 with SalI and NotI restriction enzymes, C166 cells were transfected as above with 0.02 ng of plasmid for 24 h.
Western Blot
Total protein was harvested from siRNA-transfected C166 cells, fractionated in a 9% SDS polyacrylamide gel, and transferred to a PVDF membrane for Western blot analysis with antibodies to CHD4 (Abcam, ab72418) and GAPDH (Sigma, G9545).
Luciferase Assays
A fragment of the murine Plaur promoter spanning the transcription start site (−952 bp to +303 bp) was amplified by PCR and cloned into a modified chromatinizable pCEP9 vector (gift of Jesse Raab). The luciferase gene from a pGL3 luciferase reporter vector was also cloned into the pCEP9 vector. The complete Plaur promoter reporter was transfected into C166 endothelial cells (250 ng) using Lipofectamine 2000. A renilla luciferase plasmid (pRL-TK) was co-transfected (10 ng) as a control. Reporter activity was assayed by transfection of 10 pmol NS siRNA or CHD4 siRNA oligonucleotides (Life Technologies, AM4637 and 4390816, respectively) or with 20 ng of the CHD4 expression plasmid (or analogous control plasmid) described above in “cell culture and transfections.” Twenty-four hours after transfection, cells were harvested and analyzed with the Dual-Luciferase Reporter Assay System (Promega) and a Glomax 20/20 Luminometer (Promega).
Fluorescence Microscopy and Image Acquisition
Gross embryonic images were obtained with a Nikon SMZ800 stereomicroscope and Nikon DS-Fi1 camera and monitor. Brightfield histological images were obtained with a Nikon Eclipse 80i microscope using a 10× (NA 0.3) and 40× (NA 0.75) objective and a Nikon DS-Fi1 camera. Fluorescent images were obtained with a Nikon Eclipse 80i microscope using 10× (NA 0.3), 20× (NA 0.5), and 40× (NA 0.75) objectives, an X-cite 120Q light source, and a Nikon DS-Qi1Mc camera. NIS-Elements AR3.0 (Nikon) software was used for all brightfield and fluorescent image acquisition and assembly. Relative fluorescence was determined using ImageJ software (National Institutes of Health).
Confocal Microscopy and Image Acquisition
Confocal images for VE-Cadherin and ZO-1 immunostained sections were obtained on an Olympus IX81 motorized inverted microscope with a Lumens 200 light source. Z stacks were acquired using an Olympus DSU spinning disc confocal system and analyzed using Slidebook 5.0 imaging software (Intelligent Imaging Innovations).
In Situ Zymography
E10.5 embryos were cryosectioned without prior fixation. 20 µm sections of control and Chd4fl/fl;Tie2-Cre+ embryos were affixed to the same slide and were overlaid with in situ zymography solution (ISZS) consisting of 1% low melting point agarose (Invitrogen), 0.1 mg/mL quenched BODIPY FL casein from the EnzChek Protease Assay Kit (Invitrogen), 1 U/mL human Glu-plasminogen (Sigma), and Hoechst (20 µg/mL). Overlaid sections were coverslipped and incubated for 1 h at 37°C. Casein cleavage was detected by fluorescence microscopy. Plasminogen-free ISZS was used as a negative control to verify that casein cleavage was plasmin-dependent. For amiloride treatment, 0.1 mM amiloride (Sigma, A7410) was mixed with the ISZS prior to overlaying embryo sections.
Primary Endothelial Cell Isolation
E10.5 embryos were digested with collagenase-DNase solution [1.5 mg/mL collagenase, 25 mg/mL DNase, 25 mM HEPES in Dulbecco's Modified Eagle Medium (DMEM)] for 30 min at 37°C. Cells were washed once with PBS/0.1% BSA and resuspended in DMEM. Dynabeads (Life Technologies) conjugated to PECAM-1 antibody (BD Pharmingen, 557355) were added, and samples were incubated for 30 min at 4°C with rotation. Immunoprecipitated cells were washed once with PBS/0.1% BSA and eluted in Trizol (Life Technologies).
qPCR
To analyze transcript levels, total RNA isolated from primary vascular endothelial cells was purified using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. cDNA was prepared using the MultiScribe Reverse Transcriptase kit (Applied Biosystems), and real-time quantitative PCR was performed using 2× SYBR green qPCR master mix (Life Technologies) and the CFX96 Real-Time System (Bio-Rad) with gene-specific primers. The following primers were used for qPCR: Chd4 (5′-TCCTCTGTCCACCATCATCA-3′ and 5′-ACCCAAGATGGCCATATCAA-3′); Plau (5′-GCGCCTTGGTGGTGAAAAAC-3′ and 5′-TTGTAGGACACGCATACACCT-3′); Plaur (5′-GGCTTAGATGTGCTGGGAAA-3′ and 5′-CAATGAGGCTGAGTTGAGCA-3′); Thbs1 (5′-CCAAAGCCTGCAAGAAAGAC -3′ and 5′-CCTGCTTGTTGCAAACTTGA-3′); Hbegf (5′-GACCCATGCCTCAGGAAATA-3′ and 5′-GGCATTTGCAAGAGGGAGTA-3′); Pdgfb (5′-CTGCTGCAATAACCGCAAT-3′ and 5′-CCGAGGGGTCACTACTGTCT-3′); Tgfbr2 (5′-GCAAGTTTTGCGATGTGAGA-3′ and 5′-GGCATCTTCCAGAGTGAAGC-3′); Plat (5′-CTGAGGTCACAGTCCAAGCA-3′ and 5′-TCAGCCGGTCAGAGAAGAAT-3′); Gapdh (5′-TCAACGGCACAGTCAAGG-3′ and 5′-ACTCCACGACATACTCAGC-3′); and β-actin (5′-TGTTACCAACTGGGACGACA-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′).
qPCR Analysis
The relative fold change in transcription was determined using the comparative CT method and the housekeeping genes, Gapdh and β-actin, as internal controls. Data from at least 3 independent sets of littermate control and mutant embryos or from 3 independent siRNA knockdown experiments or overexpression experiments were combined and presented as the mean plus standard deviation (SD). Statistical differences were detected using a two-tailed unpaired Student's t test.
Angiogenesis and Extracellular Matrix qPCR Arrays
Total RNA from primary vascular endothelial cells was isolated using Trizol (Life Technologies) according to the manufacturer's instructions. The DNA-free kit (Life Technologies) was used to digest any contaminating DNA, and cDNA was prepared using the RT2 First Strand kit (SABiosciences/QIAGEN). Three independent experiments were performed using Mouse Angiogenesis RT2 Profiler PCR Array and Mouse Extracellular Matrix and Adhesion Molecules RT2 Profiler PCR Array (SABiosciences/QIAGEN) according to the manufacturer's instructions. Data analysis was performed using the web-based PCR array data analysis tool available through the SABiosciences/QIAGEN website.
Chromatin Immunoprecipitation (ChIP)
Sub-confluent C166 yolk sac endothelial cells were crosslinked with 1% formaldehyde for 10 min and processed for ChIP with the MAGnify Chromatin Immunoprecipitation System (Invitrogen) according to the manufacturer's instructions. A CHD4-specific antibody (Abcam; ab70469) was used to immunoprecipitate protein-DNA complexes, and a normal mouse IgG antibody (Invitrogen; 100005291) was used as a negative control for CHD4 ChIP assays. An HDAC1-specific antibody (Abcam, ab46985) and normal mouse IgG antibody were used for HDAC1 ChIP assays. For H3K9Ac and H3K27Ac ChIP assays, chromatin was harvested from C166 cells that were transfected for 24 h with nonspecific or CHD4-specific siRNAs. Sonicated chromatin was immunoprecipitated using an anti-histone H3K9Ac antibody (Active Motif; 39137) or an anti-histone H3K27Ac antibody (Abcam; ab4729), and isotype matched IgG antibodies (Invitrogen) were used as negative controls. All ChIP assays were performed with 10 µg of experimental or control antibodies. Real-time quantitative PCR was performed using RT2 Fast SYBR green qPCR master mix (SABiosciences) and the CFX96 detection system (Bio-Rad) with specific primers. For the Thbs1 gene, the primers 5′-AGCCAGATGGTTCAGCAAAT-3′ and 5′-CCCTCAAGGCAAAATTGAAA-3′ (located 834 bp and 732 bp upstream of the TSS, respectively) were used for CHD4, HDAC1, H3K9Ac and H3K27Ac ChIP assays. For the Plaur gene, the primers 5′-ACTGAGCCGCTCTGAGTGAT-3′ and 5′-CCAGGGGAAAAACAAGTTGA-3′ (located 922 bp and 723 bp upstream of the TSS, respectively) were used for the ChIP assays described above. For the Plau gene, the primers 5′-GACATGTGGGAGCCTTTGTT-3′ and 5′-CTCGCTGATCTCAAGCCTCT-3′ (located 507 bp and 349 bp upstream of the TSS, respectively) were used for CHD4 and HDAC1 ChIP assays. Primers designed for a transcriptionally inactive region far upstream of the Fzd5 TSS were used to amplify a negative control region for CHD4 binding and activity. These Fzd5UP primers, 5′-GGTGACTTAGGGCAAAACCA-3′ and 5′-AGGCCACCATACCAGGTTCT-3′, were located 4856 bp and 4648 bp upstream of the TSS, respectively.
ChIP Analysis
A defined fraction of input chromatin that was subjected to sonication was set aside prior to immunoprecipitation but was otherwise treated identically to immunoprecipitated samples through reverse crosslinking and DNA purification steps. DNA from input chromatin was diluted and analyzed by qPCR along with DNA from immunoprecipitated samples. Input CT values were adjusted for dilution and used to calculate % input values for immunoprecipitated samples. A two-tailed Student's t test was used for statistical comparisons between samples immunoprecipitated with negative control (i.e. IgG) or experimental antibodies at a particular locus. Comparisons between immunoprecipitated samples at an experimental locus (i.e. Plaur or Thbs1) versus a negative control locus (i.e. Fzd5UP) were also analyzed.
Whole-Mount Yolk Sac Staining
Embryos with attached yolk sacs were dissected from maternal tissue, fixed, permeabilized and immunostained with rat anti-mouse PECAM-1 antibody (BD Pharmingen, 557355) as described [69]. Vitelline vessel diameters were measured using NIS-Elements AR3.0 (Nikon) software.
Whole-Mount Embryo Staining
Whole-mount immunostaining for PECAM-1 was performed using a rat anti-mouse PECAM-1 antibody (BD Pharmingen, 557355) as described [72].
Supporting Information
Zdroje
1. DavisGE, SengerDR (2005) Endothelial extracellular matrix: biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ Res 97 : 1093–1107.
2. MurakamiM, SimonsM (2009) Regulation of vascular integrity. J Mol Med 87 : 571–582.
3. SakalihasanN, LimetR, DefaweOD (2005) Abdominal aortic aneurysm. Lancet 365 : 1577–1589.
4. RuigrokYM, RinkelGJ, WijmengaC (2005) Genetics of intracranial aneurysms. Lancet Neurol 4 : 179–189.
5. PoschlE, Schlotzer-SchrehardtU, BrachvogelB, SaitoK, NinomiyaY, et al. (2004) Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 131 : 1619–1628.
6. TangN, MackF, HaaseVH, SimonMC, JohnsonRS (2006) pVHL function is essential for endothelial extracellular matrix deposition. Mol Cell Biol 26 : 2519–2530.
7. PhoonCK, AristizabalO, TurnbullDH (2000) 40 MHz Doppler characterization of umbilical and dorsal aortic blood flow in the early mouse embryo. Ultrasound Med Biol 26 : 1275–1283.
8. ChangS, YoungBD, LiS, QiX, RichardsonJA, et al. (2006) Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 126 : 321–334.
9. OhJ, TakahashiR, KondoS, MizoguchiA, AdachiE, et al. (2001) The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 107 : 789–800.
10. van HinsberghVW, KoolwijkP (2008) Endothelial sprouting and angiogenesis: matrix metalloproteinases in the lead. Cardiovasc Res 78 : 203–212.
11. ChangCP, BruneauB (2011) Epigenetics and Cardiovascular Development. Annu Rev Physiol 74 : 41–68.
12. OhtaniK, DimmelerS (2011) Epigenetic regulation of cardiovascular differentiation. Cardiovasc Res 90 : 404–412.
13. CurtisCD, DavisRB, IngramKG, GriffinCT (2012) Chromatin-remodeling complex specificity and embryonic vascular development. Cell Mol Life Sci 69 : 3921–3931.
14. van WeerdJH, Koshiba-TakeuchiK, KwonC, TakeuchiJK (2011) Epigenetic factors and cardiac development. Cardiovasc Res 91 : 203–211.
15. TongJK, HassigCA, SchnitzlerGR, KingstonRE, SchreiberSL (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395 : 917–921.
16. WadePA, JonesPL, VermaakD, WolffeAP (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol 8 : 843–846.
17. XueY, WongJ, MorenoGT, YoungMK, CoteJ, et al. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2 : 851–861.
18. ZhangY, LeRoyG, SeeligHP, LaneWS, ReinbergD (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95 : 279–289.
19. LaiAY, WadePA (2011) Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 11 : 588–596.
20. WilliamsCJ, NaitoT, ArcoPG, SeavittJR, CashmanSM, et al. (2004) The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20 : 719–733.
21. MiccioA, WangY, HongW, GregoryGD, WangH, et al. (2010) NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. Embo J 29 : 442–456.
22. HungH, KohnkenR, SvarenJ (2012) The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci 32 : 1517–1527.
23. CurtisCD, GriffinCT (2012) The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Mol Cell Biol 32 : 1312–1320.
24. CoultasL, ChawengsaksophakK, RossantJ (2005) Endothelial cells and VEGF in vascular development. Nature 438 : 937–945.
25. StankunasK, HangCT, TsunZY, ChenH, LeeNV, et al. (2008) Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell 14 : 298–311.
26. MeyerD, BirchmeierC (1995) Multiple essential functions of neuregulin in development. Nature 378 : 386–390.
27. GassmannM, CasagrandaF, OrioliD, SimonH, LaiC, et al. (1995) Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378 : 390–394.
28. LeeKF, SimonH, ChenH, BatesB, HungMC, et al. (1995) Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378 : 394–398.
29. SuriC, JonesPF, PatanS, BartunkovaS, MaisonpierrePC, et al. (1996) Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell 87 : 1171–1180.
30. CamenischTD, SpicerAP, Brehm-GibsonT, BiesterfeldtJ, AugustineML, et al. (2000) Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest 106 : 349–360.
31. CattelinoA, LiebnerS, GalliniR, ZanettiA, BalconiG, et al. (2003) The conditional inactivation of the beta-catenin gene in endothelial cells causes a defective vascular pattern and increased vascular fragility. J Cell Biol 162 : 1111–1122.
32. GeorgeEL, Georges-LabouesseEN, Patel-KingRS, RayburnH, HynesRO (1993) Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 119 : 1079–1091.
33. OrrAW, LeeMY, LemmonJA, YurdagulAJr, GomezMF, et al. (2009) Molecular mechanisms of collagen isotype-specific modulation of smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol 29 : 225–231.
34. ArmulikA, AbramssonA, BetsholtzC (2005) Endothelial/pericyte interactions. Circ Res 97 : 512–523.
35. HolycrossBJ, BlankRS, ThompsonMM, PeachMJ, OwensGK (1992) Platelet-derived growth factor-BB-induced suppression of smooth muscle cell differentiation. Circ Res 71 : 1525–1532.
36. CorjayMH, ThompsonMM, LynchKR, OwensGK (1989) Differential effect of platelet-derived growth factor - versus serum-induced growth on smooth muscle alpha-actin and nonmuscle beta-actin mRNA expression in cultured rat aortic smooth muscle cells. J Biol Chem 264 : 10501–10506.
37. BlasiF, SideniusN (2010) The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett 584 : 1923–1930.
38. WangSJ, GreerP, AuerbachR (1996) Isolation and propagation of yolk-sac-derived endothelial cells from a hypervascular transgenic mouse expressing a gain-of-function fps/fes proto-oncogene. In Vitro Cell Dev Biol Anim 32 : 292–299.
39. GhoshS, JohnsonJJ, SenR, MukhopadhyayS, LiuY, et al. (2006) Functional relevance of urinary-type plasminogen activator receptor-alpha3beta1 integrin association in proteinase regulatory pathways. J Biol Chem 281 : 13021–13029.
40. SmithHW, MarshallCJ (2010) Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol 11 : 23–36.
41. ReynoldsN, Salmon-DivonM, DvingeH, Hynes-AllenA, BalasooriyaG, et al. (2012) NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. Embo J 31 : 593–605.
42. ZhangJ, JacksonAF, NaitoT, DoseM, SeavittJ, et al. (2012) Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13 : 86–94.
43. HoggPJ, StenfloJ, MosherDF (1992) Thrombospondin is a slow tight-binding inhibitor of plasmin. Biochemistry 31 : 265–269.
44. Rodriguez-ManzanequeJC, LaneTF, OrtegaMA, HynesRO, LawlerJ, et al. (2001) Thrombospondin-1 suppresses spontaneous tumor growth and inhibits activation of matrix metalloproteinase-9 and mobilization of vascular endothelial growth factor. Proc Natl acad Sci U S A 98 : 12485–12490.
45. VassalliJD, SappinoAP, BelinD (1991) The plasminogen activator/plasmin system. J Clin Invest 88 : 1067–1072.
46. VassalliJD, BelinD (1987) Amiloride selectively inhibits the urokinase-type plasminogen activator. FEBS Lett 214 : 187–191.
47. CarmelietP, MoonsL, LijnenR, BaesM, LemaitreV, et al. (1997) Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat Genet 17 : 439–444.
48. BacharachE, ItinA, KeshetE (1992) In vivo patterns of expression of urokinase and its inhibitor PAI-1 suggest a concerted role in regulating physiological angiogenesis. Proc Natl Acad Sci U S A 89 : 10686–10690.
49. MazarAP, HenkinJ, GoldfarbRH (1999) The urokinase plasminogen activator system in cancer: implications for tumor angiogenesis and metastasis. Angiogenesis 3 : 15–32.
50. BuggeTH, FlickMJ, DaughertyCC, DegenJL (1995) Plasminogen deficiency causes severe thrombosis but is compatible with development and reproduction. Genes Dev 9 : 794–807.
51. PloplisVA, CarmelietP, VazirzadehS, Van VlaenderenI, MoonsL, et al. (1995) Effects of disruption of the plasminogen gene on thrombosis, growth, and health in mice. Circulation 92 : 2585–2593.
52. CarmelietP, SchoonjansL, KieckensL, ReamB, DegenJ, et al. (1994) Physiological consequences of loss of plasminogen activator gene function in mice. Nature 368 : 419–424.
53. BuggeTH, SuhTT, FlickMJ, DaughertyCC, RomerJ, et al. (1995) The receptor for urokinase-type plasminogen activator is not essential for mouse development or fertility. J Biol Chem 270 : 16886–16894.
54. DavisGE, Pintar AllenKA, SalazarR, MaxwellSA (2001) Matrix metalloproteinase-1 and -9 activation by plasmin regulates a novel endothelial cell-mediated mechanism of collagen gel contraction and capillary tube regression in three-dimensional collagen matrices. J Cell Sci 114 : 917–930.
55. DavisGE, SaundersWB (2006) Molecular balance of capillary tube formation versus regression in wound repair: role of matrix metalloproteinases and their inhibitors. J Investig Dermatol Symp Proc 11 : 44–56.
56. SaundersWB, BaylessKJ, DavisGE (2005) MMP-1 activation by serine proteases and MMP-10 induces human capillary tubular network collapse and regression in 3D collagen matrices. J Cell Sci 118 : 2325–2340.
57. CarmelietP, StassenJM, SchoonjansL, ReamB, van den OordJJ, et al. (1993) Plasminogen activator inhibitor-1 gene-deficient mice. II. Effects on hemostasis, thrombosis, and thrombolysis. J Clin Invest 92 : 2756–2760.
58. DoughertyKM, PearsonJM, YangAY, WestrickRJ, BakerMS, et al. (1999) The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival. Proc Natl Acad Sci U S A 96 : 686–691.
59. DewerchinM, CollenD, LijnenHR (2001) Enhanced fibrinolytic potential in mice with combined homozygous deficiency of alpha2-antiplasmin and PAI-1. Thromb Haemost 86 : 640–646.
60. LijnenHR (2001) Plasmin and matrix metalloproteinases in vascular remodeling. Thromb Haemost 86 : 324–333.
61. PatelMK, LymnJS, ClunnGF, HughesAD (1997) Thrombospondin-1 is a potent mitogen and chemoattractant for human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 17 : 2107–2114.
62. LawlerJ, SundayM, ThibertV, DuquetteM, GeorgeEL, et al. (1998) Thrombospondin-1 is required for normal murine pulmonary homeostasis and its absence causes pneumonia. J Clin Invest 101 : 982–992.
63. ShattilSJ (1995) Function and regulation of the beta 3 integrins in hemostasis and vascular biology. Thromb Haemost 74 : 149–155.
64. Hodivala-DilkeKM, McHughKP, TsakirisDA, RayburnH, CrowleyD, et al. (1999) Beta3-integrin-deficient mice are a model for Glanzmann thrombasthenia showing placental defects and reduced survival. J Clin Invest 103 : 229–238.
65. CoradaM, NyqvistD, OrsenigoF, CapriniA, GiampietroC, et al. (2010) The Wnt/beta-catenin pathway modulates vascular remodeling and specification by upregulating Dll4/Notch signaling. Dev Cell 18 : 938–949.
66. MacSweeneyST, PowellJT, GreenhalghRM (1994) Pathogenesis of abdominal aortic aneurysm. Br J Surg 81 : 935–941.
67. CarmelietP (2000) Proteinases in cardiovascular aneurysms and rupture: targets for therapy? J Clin Invest 105 : 1519–1520.
68. KisanukiYY, HammerRE, MiyazakiJ, WilliamsSC, RichardsonJA, et al. (2001) Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol 230 : 230–242.
69. GriffinCT, CurtisCD, DavisRB, MuthukumarV, MagnusonT (2011) The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc Natl Acad Sci U S A 108 : 2282–2287.
70. GriffinCT, BrennanJ, MagnusonT (2008) The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development 135 : 493–500.
71. CarmelietP, LampugnaniMG, MoonsL, BreviarioF, CompernolleV, et al. (1999) Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell 98 : 147–157.
72. SchlaegerTM, QinY, FujiwaraY, MagramJ, SatoTN (1995) Vascular endothelial cell lineage-specific promoter in transgenic mice. Development 121 : 1089–1098.
Štítky
Genetika Reprodukčná medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



