-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
Danforth's short tail mutant (Sd) mouse, first described in 1930, is a classic spontaneous mutant exhibiting defects of the axial skeleton, hindgut, and urogenital system. We used meiotic mapping in 1,497 segregants to localize the mutation to a 42.8-kb intergenic segment on chromosome 2. Resequencing of this region identified an 8.5-kb early retrotransposon (ETn) insertion within the highly conserved regulatory sequences upstream of Pancreas Specific Transcription Factor, 1a (Ptf1a). This mutation resulted in up to tenfold increased expression of Ptf1a as compared to wild-type embryos at E9.5 but no detectable changes in the expression levels of other neighboring genes. At E9.5, Sd mutants exhibit ectopic Ptf1a expression in embryonic progenitors of every organ that will manifest a developmental defect: the notochord, the hindgut, and the mesonephric ducts. Moreover, at E 8.5, Sd mutant mice exhibit ectopic Ptf1a expression in the lateral plate mesoderm, tail bud mesenchyme, and in the notochord, preceding the onset of visible defects such as notochord degeneration. The Sd heterozygote phenotype was not ameliorated by Ptf1a haploinsufficiency, further suggesting that the developmental defects result from ectopic expression of Ptf1a. These data identify disruption of the spatio-temporal pattern of Ptf1a expression as the unifying mechanism underlying the multiple congenital defects in Danforth's short tail mouse. This striking example of an enhancer mutation resulting in profound developmental defects suggests that disruption of conserved regulatory elements may also contribute to human malformation syndromes.
Published in the journal: A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse. PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003206
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003206Summary
Danforth's short tail mutant (Sd) mouse, first described in 1930, is a classic spontaneous mutant exhibiting defects of the axial skeleton, hindgut, and urogenital system. We used meiotic mapping in 1,497 segregants to localize the mutation to a 42.8-kb intergenic segment on chromosome 2. Resequencing of this region identified an 8.5-kb early retrotransposon (ETn) insertion within the highly conserved regulatory sequences upstream of Pancreas Specific Transcription Factor, 1a (Ptf1a). This mutation resulted in up to tenfold increased expression of Ptf1a as compared to wild-type embryos at E9.5 but no detectable changes in the expression levels of other neighboring genes. At E9.5, Sd mutants exhibit ectopic Ptf1a expression in embryonic progenitors of every organ that will manifest a developmental defect: the notochord, the hindgut, and the mesonephric ducts. Moreover, at E 8.5, Sd mutant mice exhibit ectopic Ptf1a expression in the lateral plate mesoderm, tail bud mesenchyme, and in the notochord, preceding the onset of visible defects such as notochord degeneration. The Sd heterozygote phenotype was not ameliorated by Ptf1a haploinsufficiency, further suggesting that the developmental defects result from ectopic expression of Ptf1a. These data identify disruption of the spatio-temporal pattern of Ptf1a expression as the unifying mechanism underlying the multiple congenital defects in Danforth's short tail mouse. This striking example of an enhancer mutation resulting in profound developmental defects suggests that disruption of conserved regulatory elements may also contribute to human malformation syndromes.
Introduction
Congenital malformations occur in about 3% of all live births and are a major cause of childhood morbidity and mortality [1], [2]. These defects can affect multiple systems, defining syndromes. Caudal regression syndrome (or caudal dysgenesis) is a major developmental syndrome characterized by malformation of the neural tube, caudal spine, the hindgut and lower limbs [3], [4]. Caudal regression can also occur with bilateral renal agenesis, and this form is usually fatal because it produces secondary pulmonary hypoplasia [3], [4]. Caudal regression most often occurs as sporadic disease; mutations in HLXB9 cause Currarino syndrome (sacral agenesis, OMIM 176450), accounting for a minority of cases [5]. On the milder side of the clinical spectrum, isolated defects, such as renal hypoplasia or unilateral renal agenesis, are common but often remain subclinical [6], [7]. Kidney malformations are highly genetically heterogeneous. Mutations in the PAX2 and HNF1B genes may account for up to 15–20% of pediatric renal hypoplasia [8]–[10]. However, the molecular basis for the majority of severe syndromes, such as bilateral renal agenesis and caudal regression are not well understood.
The Danforth's short tail mutant mouse is a classic Mendelian model of caudal malformations [11]. First described in 1930, this spontaneous mutation (symbol Sd) produces combined defects of the axial skeleton, urogenital system and distal gut [11], [12]. The homozygous mutant mice have fully penetrant abnormalities including truncation of the caudal vertebral column - resulting in a short or absent tail - as well as bilateral renal agenesis, colonic aganglionosis and absence of anorectal opening [12]. The Sd/Sd mice die shortly after birth due to these multiple organ malformations. The heterozygous (Sd/+) mutant mice exhibit short tails with complete penetrance and 30–40% incidence of unilateral renal agenesis; the solitary kidneys in the Sd/+ mice are devoid of major structural defects and consequently, these mice have a normal lifespan [12], [13]. The Sd homozygote and heterozygote mutants thus represent excellent models of human caudal regression syndrome and isolated unilateral renal agenesis, respectively.
The earliest defect detected in Sd/Sd mice is the progressive disintegration of the notochord and the floor plate starting at E9.5, causing patterning defects in both neural tube and somites, leading to vertebral defects; the ureteric bud, derived from the mesonephric duct, either fails to reach or fails to induce the metanephric blastema, resulting in renal hypoplasia or aplasia; abnormalities in the development of the hindgut and cloaca result in the absence of an anorectal opening and could also be the cause of aganglionosis of the rectal pouch [12]–[16]. Studies of chimeric embryos have shown that the Sd cells are selectively lost from the notochord and ventral hindgut endoderm starting at E9.5, implicating a cell-autonomous defect in these tissues [17]; in rare instances that metanephric kidneys develop, however, Sd cells are robustly incorporated into chimeric kidneys, suggesting that the urogenital defect may be cell-nonautonomous or due to specific impairment of signaling for the mesonephric to metanephric transition [17]. More recent studies have shown that genetic ablation of the notochord with Diphtheria toxin recapitulates the axial defects observed in the Sd mutants, but notochord-ablated mice exhibited only kidney fusion and no noticeable defects in nephrogenesis, suggesting that an additional mechanism accounts for the renal agenesis in the Sd/Sd mouse [18]. Thus, identification of the genetic basis of the Sd mutation will provide insight into mechanisms of axial skeletal development and reconcile potentially contradictory findings about the origin of visceral defects in this mutant strain.
Previous studies have assigned the Sd locus to Chr. 2A3 but the mutation was not known [19]. Here we refined the Sd locus to a 42.8 kb interval and identified the Sd mutation as an insertion in the 5′ regulatory domain of Pancreas Specific Transcription Factor 1a gene (Ptf1a). This mutation results in ectopic expression of Ptf1a in the notochord, mesonephros and gut providing an explanation for the complete spectrum of abnormalities seen in Danforth's short tail mutants.
Results
Mapping of the Sd mutation to a 42.8-kb intergenic region on Chr. 2
The Sd mutation arose in 1930 on an outbred stock prior to generation of classical inbred strains. Mice carrying the Sd mutations were transferred in 1950 to Jackson laboratories in a linkage testing stock called E1, which was sequentially outcrossed to C57BL/6J, C3H/He and CBA and then maintained as closed colony until 1970 (Jackson laboratories). This colony, segregating the Sd mutation, was subsequently sibling mated at Jackson laboratories and named RSV/LeJ. We obtained RSV/LeJ-Sd/+ mice and confirmed that Sd/Sd, Sd/+ and +/+ mice are obtained in an expected Mendelian ratio. RSV/LeJ-Sd/Sd mice were readily recognizable at birth because they manifested all the organ malformations defects originally described [12]–[14]. RSV/LeJ-Sd/+ exhibited short tails with no other outwardly visible defects; on dissection, unilateral renal agenesis was detected in 10/28 (35%) of RSV/LeJ-Sd/+ mice, consistent with previous reports [12]–[14]. RSV/LeJ - +/+ exhibited long tails and were anatomically indistinguishable from other inbred strains.
Previous studies had mapped the Sd locus to a 5 cM interval on Chr. 2 [19]. To confirm and refine this interval, we generated 174 backcross (BC) and 120 F2 intercross progeny (using C57BL/6J as the counterstrain). Analysis of linkage with 10 informative markers confirmed the mapping of the Sd locus to this region, yielding a peak lod score of 39 (p<6×10−41, Figure 1 and Figure S1). Three informative recombinants refined the locus to 1.3 Mb interval containing nine transcriptional units, including five genes with orthologs in other mammalian species (Pip4k2a, Armc3, Mrsb2, Ptf1a, Otud1, Figure 1A and 1B).
Fig. 1. Fine mapping and identification of the Sd mutation. 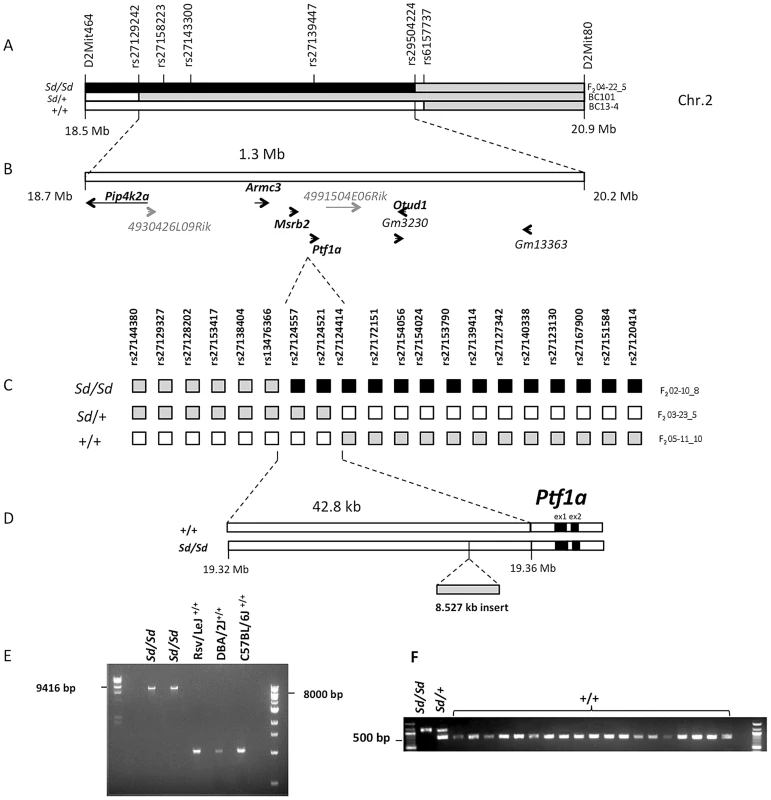
(A) Meiotic mapping on mouse Chr.2 on 174 backcross (BC) and 120 F2 intercross mice refines the Sd locus to a 1.3 Mb interval. The critical recombinants in 2 BC and 1 F2 mice are shown. The phenotype of each mouse (Sd/Sd, Sd/+ and +/+) is indicated on the left. The genotypes are indicated by the color of each haplotype: black = RSV/RSV genotype; gray = RSV/B6 genotype; white = B6/B6 genotype. The location of genotyped markers is indicated above the haplotypes. (B) The initial mapping localized the Sd mutation to a 1.3 Mb interval on Chr. 2 between rs27129242 and rs29504224. This region contains 9 genes, of which five have mammalian orthologs (shown bold black). Predicted genes without mammalian orthologs are indicated in gray. (C) High resolution mapping in 1203 F2 mice. The critical recombinants in 3 F2 mice are shown. The phenotype and genotypes are indicated as in (A). (D) Localization of the Sd mutation to a 42.8 kb interval between rs13476366 and rs27124414, 12.2 kb upstream of the Ptf1a start site and identification of an insertion of 8.5 kb in the Sd/Sd mouse. (E) Long-range PCR identifies the insertion in 2 different mice Sd/Sd mutants but not in the RSV/LeJ strain and the two counterstrains used in the mapping study (C57BL/6J and DBA/2J). (F) Genotyping identified the insertion in Sd/Sd, and Sd/+ samples but not in any additional mouse strain tested, including 4 wild-derived and 15 other laboratory inbred strains (listed in methods). However, sequence analysis of exons and flanking regions as well as copy number analysis of all nine positional candidates did not identify any coding variants or intragenic copy number changes that distinguished RSV/LeJ-Sd/Sd mice from the background strain. Because the mapping was based on multiple critical recombinants in affected mice, this excluded the possibility of mismapping due to incomplete penetrance. Taken together these data indicated that the Sd mutation occurs in a noncoding region within this interval.
We therefore undertook further refinement of the Sd locus by meiotic mapping in additional F2 intercross mice. We did not identify informative markers between B6 and RSV/LeJ-Sd/Sd, or RSV/LeJ - +/+ across this minimal interval, suggesting that these strains share the same ancestral haplotype in this region. However, DBA/2J harbored informative SNPs and was therefore used as the counterstrain for fine mapping. We generated 1203 F2 intercross mice and genotyped 29 informative markers across the minimal 1.3 Mb interval. We identified 14 F2 progeny with recombinants within this interval, with 4, 7 and 3 mice exhibiting the wildtype, heterozygote and homozygote phenotypes, respectively. Among these, three critical recombinants localized the Sd locus to a 42.8 Kb intergenic region between rs13476366 and rs27124414, proximal to Ptf1a (Figure 1C, 1D). These mapping results were highly reliable because two of these critical recombinants occurred in affected mice exhibiting the homozygous and heterozygous mutant phenotype (Figure 1C).
The Sd mutation is an insertion 12.2 kb upstream of the Ptf1a gene
We performed Sanger sequencing of the 42.8 Kb segment spanning the minimal meiotic interval in an RSV/LeJ-Sd/Sd and an RSV/LeJ-+/+ mouse, achieving 100% coverage of the interval with an average base call accuracy of 99%. We identified an 8.53 Kb insertion within this region in Sd homozygotes (located at nucleotide position 19,355,026 on Chr. 2, genome build 37.2). The insertion was present in homozygosity in RSV/LeJ-Sd/Sd mice, in heterozygosity in RSV/LeJ-Sd/+ mice, and was absent in the background RSV/LeJ-+/+ mice and in the two counterstrains used for the mapping study (DBA/2J, C57BL/6J) (Figure 1E). We also genotyped this insertion in a random sample of 142 mice from the F2 mapping progeny and found that it perfectly segregated with the heterozygote and homozygote Sd phenotypes. There were no other variants distinguishing RSV/LeJ-Sd/Sd from RSV/LeJ-+/+ within the 42.8 kb minimal recombinant intervals, and the RSV/LeJ-Sd/Sd sequence otherwise shows identity with the C57BL/6J reference sequence (Figure 1F).
Sequence comparison with Repbase data indicated that the insertion is an endogenous retroviral element, with the closest alignment with the early retrotransposon (ETn) subtype (Figure 2A and 2B, Table S1). The insertion results in duplication of 6 bp at its flanking sites, without loss of reference sequence, which is typical of ETn sequences (Figure 2C) [20], [21]. Many copies of this mobile element are interspersed across the mouse genome, and transposition of ETn sequences into genic regions is responsible for many spontaneous mutant mouse phenotypes [20]–[23]. We further determined the haplotype on which this mutation occurred by genotyping 37 SNPs in the 95.5 kb interval surrounding Ptf1a (2.6 kb spacing, Table 1). This analysis demonstrated that in the RSV/LeJ-Sd/Sd and RSV/LeJ-+/+ strains shared the same ancestral haplotype at the Ptf1a locus and therefore RSV/LeJ-+/+ provides a valid reference haplotype for RSV/LeJ-Sd/Sd. This same haplotype is also present in ten other inbred strains (C57BL/6J, CBA/J, BALB/cJ, C3H/HeJ, C57BL/6NJ, LP/J, 129P2/OlaHsd, 129S1/SvImJ, 129S5SvEv, and NZO/HlLtJ, Figure 3). These data are consistent with the lack of polymorphism between C57BL/6J, RSV/LeJ-Sd/Sd and RSV/LeJ-+/+ based on sequencing of the 42.8 kb minimal recombinant interval. Genotyping and Sanger Sequencing verified that the ETn insertion was absent in these ten strains as well as fifteen other strains with different haplotypes at the Ptf1a locus (Figure 1F). These data demonstrate that the ETn insertion is not an old polymorphism but is a new variant that arose on ancestral haplotype still commonly represented among laboratory inbred strains. Altogether, the precise mapping of the Sd locus to a 42.8 kb interval, the detection of an ETn insertion in the Sd mice and its absence in the RVS/LeJ background strain and twenty-five additional inbred strains, including strains with the same ancestral haplotype, demonstrated that we successfully identified the mutation responsible for the Danforth mouse phenotype.
Fig. 2. The Sd mutation is an insertion in the 5′ regulatory of Ptf1a. 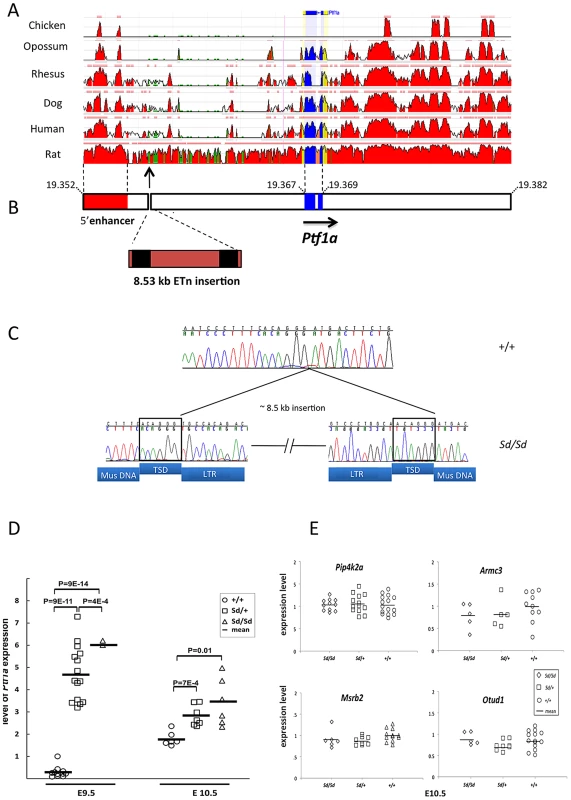
(A) Conservation of the Ptf1a genomic region across vertebrate species (ECR Browser at dcode.org). The height of the red peaks indicates the level of conservation. The Ptf1a exons are shown in blue. The Sd insertion is shown by the arrow. There is a highly conserved 5′ enhancer upstream of the Sd mutation (shown in red, located between positions 13.4–15.6 kb upstream of the Ptf1a start site). (B) Schematic representation of the Ptf1a locus. Sequence coordinates are shown in Mb (build 37.2). The Ptf1a exons are shown in blue. The location of the highly conserved 5′ enhancer element is shown in red. The Sd mutation is an insertion of an 8.527 kb ETn at nucleotide 19,355,020 on Chr. 2. The LTRs are indicated in black. The full sequence of the element is in Table 1. (C) Electropherogram of the site of the insertion in wild-type and Sd/Sd mutant mice. The target site duplications (TSD) are boxed. The sequences corresponding to LTRs, TSD and mouse genomic DNA (Mus DNA) are indicated below the nucleotide sequence. (D) Quantitative PCR of whole embryo shows up to 10-fold increased expression of Ptf1a at E9.5 in Sd/Sd mutants. Ptf1a expression remains significantly at E10.5. P-values for comparisons are indicated (t-test). (E) Quantitative PCR of whole embryo of neighboring genes with mammalian orthologs did not reveal any differences in expression between Sd mutants and wildtype mice (p>0.2 for all comparisons). Fig. 3. Haplotype map of the Ptf1a locus. 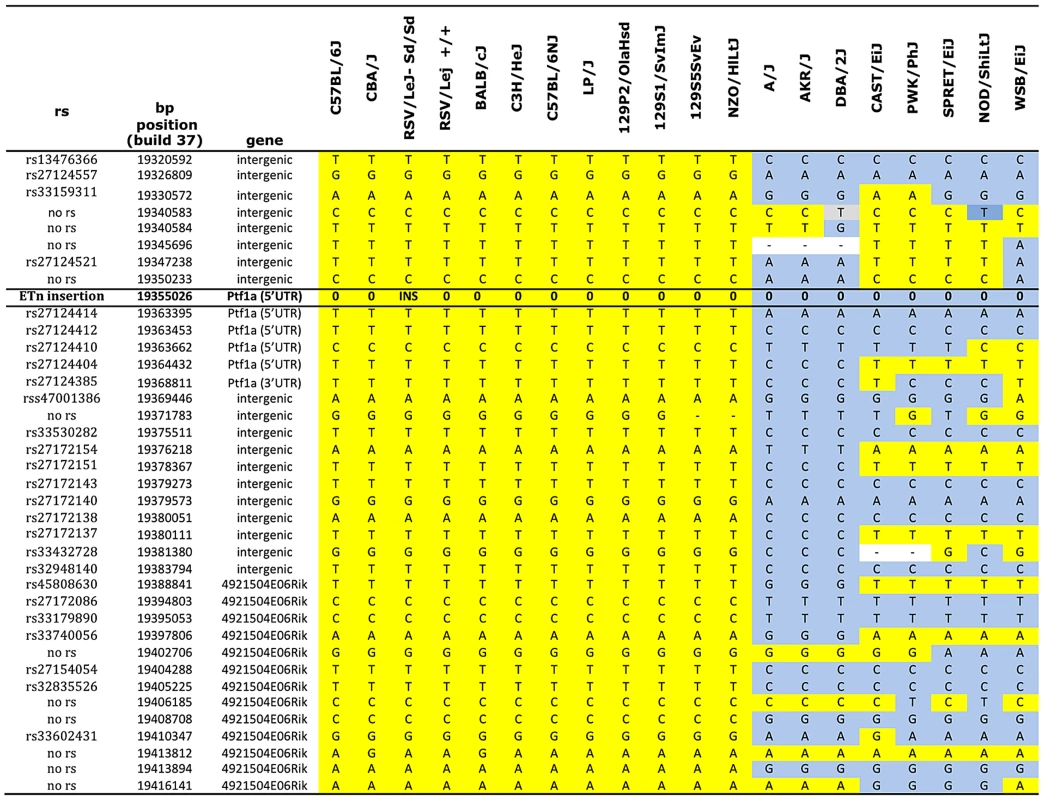
Genotype data were obtained from the Mouse Phenome Database (Sanger data). All alleles identical to C57BL/6J are highlighted in yellow. Non- C57BL/6J alleles are highlighted in gray. The ETn insertion is indicated (INS). Missing genotypes are denoted by (-). Tab. 1. Phenotypes of (Sd/+ X Ptf1a-cre/+) F1 mice. 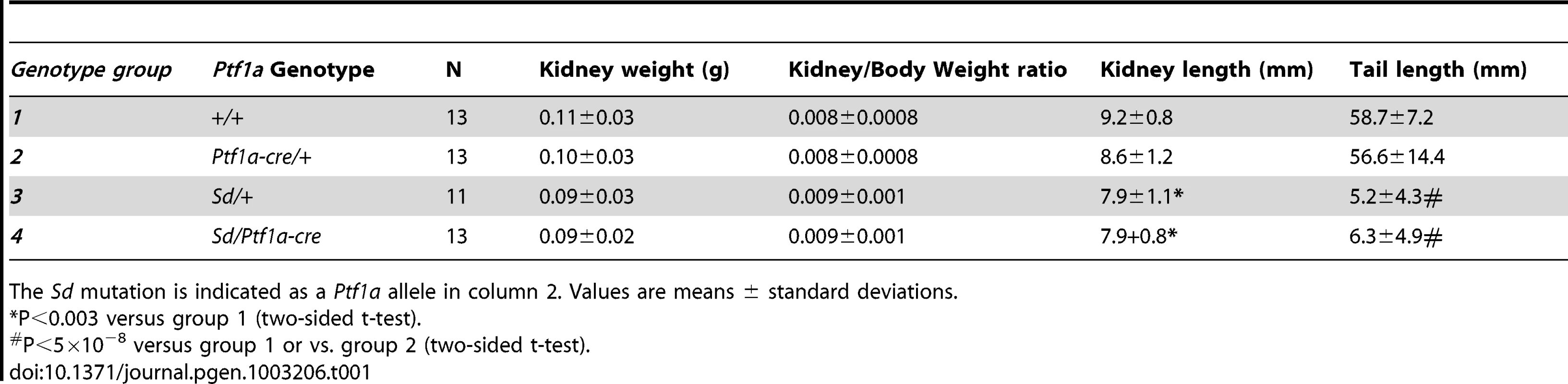
The Sd mutation is indicated as a Ptf1a allele in column 2. Values are means ± standard deviations. The ETn is inserted 12.2 kb upstream of the Pancreas Specific Transcription Factor 1a (Ptf1a) start site (Figure 2A). Ptf1a encodes a subunit of a trimeric Pancreas Specific Transcription Factor Complex (PTF1) which regulates cerebellar, retinal, pancreatic and spinal cord development [24]. The 5′ regulatory region of Ptf1a contains highly conserved elements, including a 2.3 kb autoregulatory enhancer domain located 13.4 kb upstream of the Ptf1a start site (Figure 2B), which normally directs Ptf1a expression to the dorsal spinal cord [25], [26]. Deletion of this 2.3 kb enhancer results in mislocalization of reporter constructs to the ventral spinal cord [26]. The Sd insertion occurs downstream of this highly conserved tissue enhancer, and displaces this element 8.5 kb upstream from its conserved position in the Ptf1a regulatory region (Figure 2A and 2B). The next closest gene Mrsb2, is located 64 kb proximal to the insertion, and has very low sequence conservation around its 3′ untranslated region. The data suggested that the ETn insertion is most likely to affect Ptf1a expression.
The Sd mutation results in dysregulated Ptf1a expression
Analysis with GENSCAN predicted a few low probability open reading frames within the transposon sequence, but quantitative PCR analysis of whole embryo found no evidence that these sequences are transcribed, consistent with the known lack of transcriptional activity of ETn elements (data not shown) [20]. We examined expression of Ptf1a as well as other genes that were located 500,000 bp upstream or downstream of the Sd mutation. Quantitative PCR expression analysis of whole embryos at E9.5 and E10.5 revealed a consistent four-to ten fold increased expression of Ptf1a in Sd mutants (p<9×10−14 for comparison of Sd/Sd vs. +/+ mice, Figure 2D). There were no detectable differences in expression of neighboring genes with mammalian orthologs at these two time points (Pip4k2a, Armc3, OtuD1 and Mrsb2, Figure 2E). The other predicted genes in the region did not have mammalian orthologs and did not have detectable expression at these time points. These data rule out an effect on other genes located 500 kb upstream or downstream of Ptf1a.
To follow-up these findings, we performed in situ hybridization for Ptf1a in developing embryos. Consistent with a previous study [27], Ptf1a was robustly expressed throughout most of the length of the dorsal neural tube by E10.5 in wild type and was also seen in the same location in Sd/+ and Sd/Sd embryos (Figure 4A–4F). In addition to this normal expression pattern, Sd/+ and Sd/Sd embryos showed ectopic expression of Ptf1a in the hindgut, around the cloaca and in the hindgut diverticulum extending into the tail (Figure 4H, 4I, 4K, 4L). Consistent with E10.5 qPCR data, they're appeared to be a higher level of expression in the Sd/Sd embryos compared to Sd/+ (Figure 4B, 4C, 4E, 4F), and also a higher level of endogenous expression in Sd/+ compared to wild type (Figure 4A, 4B, 4D, 4E).
Fig. 4. Ptf1a expression in wild-type, Sd/+, and Sd/Sd embryos at E10.5. 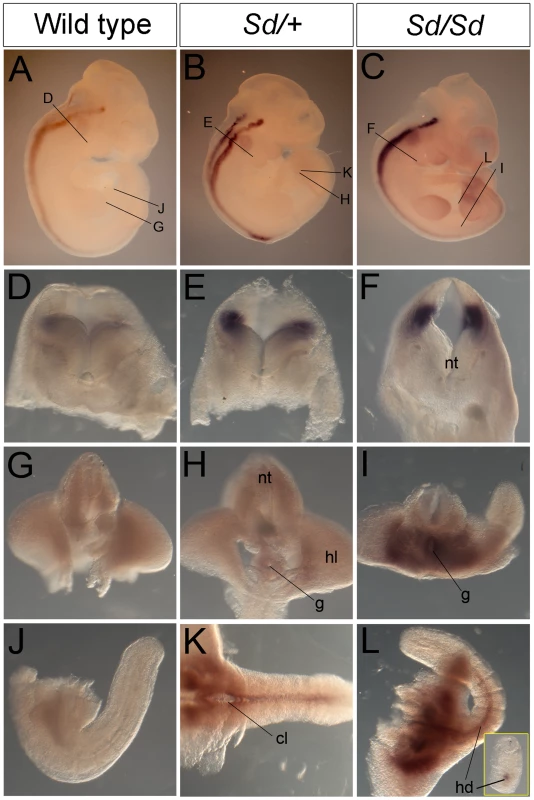
(A–C) Whole mount ISH showing Ptf1a expression in wild type, Sd/+ and Sd/Sd. embryos at E10.5. Ptf1a expression is seen in the hindbrain and throughout most of the neural tube. Lines indicate corresponding slices in D–I. (D–I) Slices from embryos in A–C showing Ptf1a expression at the level of hindbrain (D–F), hindlimb (G–I). (J–L) Isolated tails show ectopic expression of Ptf1a in the hindgut diverticulum (hd) in K,L. Inset in L is a slice close to the end of the tail showing expression in the hindgut diverticulum. Ectopic expression of Ptf1a is seen around the cloacal opening (cl) in K. nt-neural tube, g-gut, hl-hind limb. A day earlier at E9.5, extensive ectopic Ptf1a expression was evident in Sd/+ and Sd/Sd embryos in the tailbud mesenchyme (Figure 5B, 5C, 5E, 5F, 5N, 5O), the notochord and hindgut (Figure 5K, 5L, 5N, 5O), and throughout the length of the developing mesonephros and mesonephric duct (Figure 5B, 5C, 5K, 5L). Ectopic expression was evident even at E8.5 in the lateral plate mesoderm and tail bud mesenchyme of Sd/+ and Sd/Sd embryos (Figure 6A, 6B, 6C, 6E, 6F) and the notochord of Sd/Sd embryos (Figure 6C, 6F). In summary, we detected ectopic Ptf1a expression in every organ that will manifest a developmental defect in Sd/+ and Sd/Sd mice - the notochord, the mesonephros and the hindgut.
Fig. 5. Ectopic expression of Ptf1a in Sd/+ and Sd/Sd embryos at E9.5. 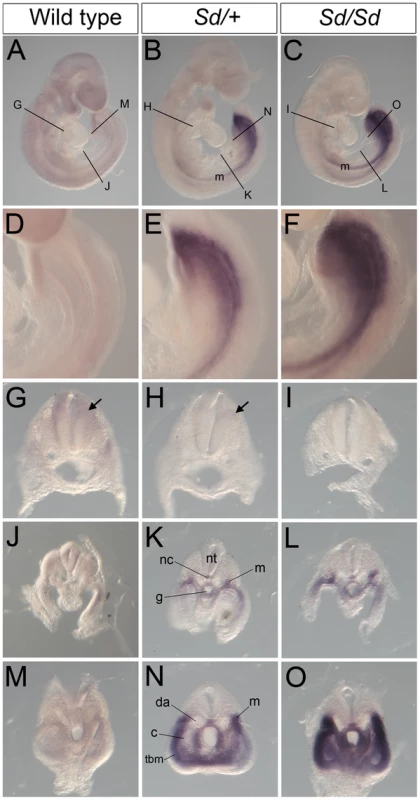
(A–C) Whole mount ISH showing Ptf1a expression in wild type, Sd/+ and Sd/Sd embryos at E9.5. Lines indicate corresponding slices in G–O. (D–F) High magnification views of the caudal end of the embryos shown in A,B,C. (G–O) Slices from embryos in A, B and C showing Ptf1a expression at the level of foregut (G–I), midgut (J–L) and the tail bud (M–O) of the embryo. Ptf1a is ectopically expressed in the posterior notochord (nc), mesonephros (m), and tail bud mesenchyme surrounding the gut tube (g) and the coelomic cavity (c) in the Sd/+ and Sd/Sd embryos. da-dorsal aortae, nt-neural tube, tbm-tail bud mesenchyme. Arrows point to expression in the dorsal neural tube. Fig. 6. Ectopic expression of Ptf1a in Sd/+ and Sd/Sd embryos at E8.5. 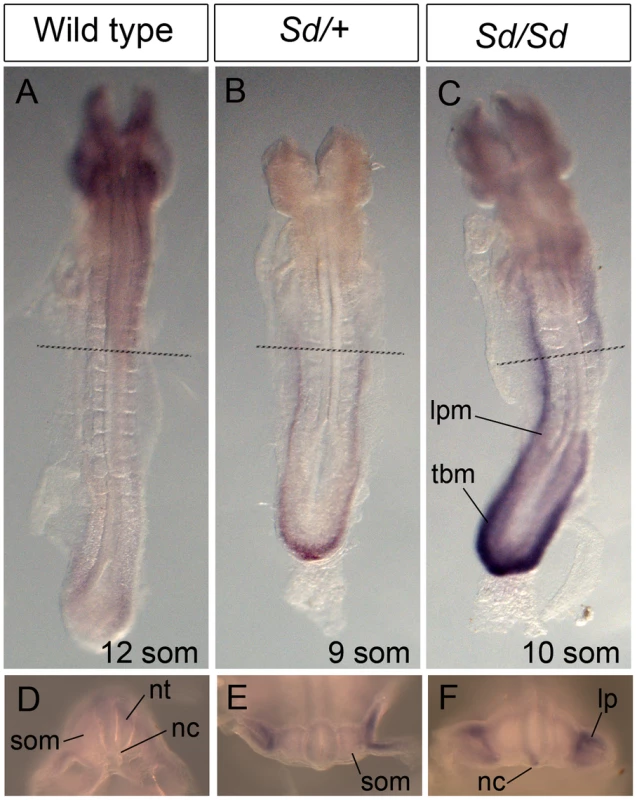
(A–C) Dorsal view of whole mount ISH showing Ptf1a expression in wild type, Sd/+ and Sd/Sd embryos at E8.5. Lines indicate corresponding slices in D–F. (D–F) Slices from embryos in A–C at the level of the fourth somite. Ptf1a is ectopically expressed in the lateral plate mesoderm (lpm) and tail bud mesenchyme (tbm) of Sd/+ and Sd/Sd embryos and in the notochord (nc) of Sd/Sd embryos. nt-neural tube, som-somite. These data suggested that dysregulated timing and localization of Ptf1a expression is responsible for the Danforth phenotype. Nonetheless, because increased Ptf1a expression was also detected in mutant embryos, we attempted to distinguish hypermorphic from neomorphic effects of the Sd allele by crossbreeding Sd/+ mice to Ptf1a haploinsufficient mice (Ptf1a-cre mice, reference [28]). If the Danforth phenotype was solely due to increased expression of Ptf1a, then reduction of Ptf1a gene dosage should attenuate the organ malformations. While the Sd mutants had significantly reduced kidney and tail lengths, there were no phenotypic differences between Sd/+, and Sd/Ptf1a-cre mice at weaning (Table 1). Histologic analysis of the kidneys also did not reveal morphological defects, consistent with prior reports of normal histology in Sd heterozygotes [12]–[16]. The absence of rescue by Ptf1a haploinsufficiency suggests that inactivation of one wild-type allele could not compensate for the increased expression from the Sd allele or more likely, that the neomorphic effects of the Sd allele predominate in the pathogenesis of malformations in the Danforth mice.
Only a few PTF1a targets are known. Recent data suggest that PTF1a regulates Mnx1, Nkx6-1, Bmp7, Dll1 and Onecut1 expression [29]. However, we did not detect any differences in expression of these genes between Sd/+, Sd/Sd and +/+ mice at E 9.5 (Figure S2), suggesting that increased Ptf1a expression is not sufficient to activate these particular targets in this tissue context and this time point.
Discussion
Danforth first described his spontaneous mutant strain over 80 years ago [11]. Since that time, this strain has been studied as a classical model of developmental defects of the spinal cord, hindgut and the urogenital tract [12]–[19]. Although the Sd locus was assigned to Chr. 2A3 in 1980, the underlying mutation had not been identified. We used meiotic mapping in 1203 F2 mice to precisely map the mutation to a 42.8 kb intergenic region. This segment contained an ETn insertion upstream of Ptf1a in the mutant strain, which was absent in the RSV/LeJ background strain and twenty-five wild-derived or classical inbred strains. SNP analysis of the Ptf1a locus suggests that the mutation arose on an ancestral haplotype that is shared by at least eleven laboratory strains, including RSV/LeJ and three strains used in the derivation of this laboratory strain (C57BL/6J, CBA/J and C3H/HeJ). Our findings are independently supported by accompanying papers (by Vlangos et al. and Semba et al.) who used a alternatives approaches to identify the same mutation. Taken together, these data establish that we have identified the genetic basis of the Sd mutation. The presence of a noncoding mutation explains the difficulties in identifying the genetic lesion in the Danforth strain since its initial description in 1930. We observed perfect co-segregation of all of the skeletal, urogenital and gastroenterological phenotypes in all affected F2 and BC mice generated in this study, indicating that the noncoding mutation is at the origin of the entire spectrum of defects. The ETn insertion occurs within the regulatory sequences upstream of Ptf1a, and is associated with dysregulated dosage, timing and localization of expression. These data implicate dysregulated Ptf1a expression as the cause of developmental defects in the Danforth mouse and indicate that Sd should be recognized as a neomorphic Ptf1a allele.
Ptf1a is a member of the trimeric transcriptional complex PTF1, and loss of function mutations result in recessive cerebellar and pancreatic hypoplasia in humans and mice [24]–[26], [30]–[34]. Ptf1a has highly restricted temporo-spatial expression during embryogenesis [24]–[26], [30]–[33]. Transient Ptf1a expression between E10 to E13 initiates the genetic programs required for specification of dorsal horn neurons, the cerebellum and the pancreas; Ptf1a expression subsequently declines to undetectable levels, with postnatal expression present only in pancreatic acinar cells [24], [31], [32], [35]–[42]. This tightly regulated temporal and spatial expression pattern is controlled by highly conserved regulatory elements upstream and downstream of Ptf1a but the consequences of regulatory mutations have not been reported. The Sd mutation occurs in the vicinity of a highly conserved enhancer sequence (positioned from 13.4 to 15.6 kb upstream of the Ptf1a start site), which contains multiple autoregulatory domains required for restricting expression to the dorsal spinal cord and maintaining expression in the adult acinar cells (Figure 2B) [25], [26]. Reporter constructs lacking this enhancer are ectopically expressed in the ventral spinal cord [26] and although not specifically reported, are also evident in the hindgut at E10.5 (see reference [26], Figure 1B), consistent with the mislocalized expression pattern in Sd/+ and Sd/Sd mutants (Figure 3, Figure 4). The retrotransposon insertion may disrupt this enhancer or a neighboring negative cis-acting element, or may act as a broadly-acting positive regulator, resulting in ectopic expression of Ptf1a in Sd mutants.
We show that each tissue that has been implicated as being primarily affected in Sd mutant mice – the notochord, the mesonephric duct and the hindgut – shows ectopic Ptf1a expression at a critical stage in its early development, indicating that dysregulated Ptf1a expression is at the origin of the developmental defects arising from these compartments. Notably, ectopic expression of Ptf1a is present in the notochord at E8.5, prior to or coincident with the earliest notochord defects and prior to the start of notochord disintegration [12]–[16]. Our data also suggest that the mouse embryo is very sensitive to Ptf1a gene dosage as Sd/Sd mice, which have a higher level of ectopic expression, consistently manifest more severe axial defects and near complete penetrance of bilateral renal agenesis resulting in death, whereas Sd/+ animals have less severe defects compatible with survival.
PTF1a requires two cofactors to form an active transcriptional complex [34], [37]. If its canonical partners were present, its misexpression may activate its direct downstream targets, ectopically initiating neural or pancreatic developmental programs [43]. Alternatively, PTF1a may form transcriptional complexes inappropriately in regions where it is ectopically expressed, interfering with normal developmental processes and impairing notochord, urogenital and hindgut development. At present, the downstream targets of Ptf1a have not been comprehensively identified. We examined five known Ptf1a targets but did not detect increased expression in Sd mutant mice, indicating that unbiased genome-wide approaches will be required to discover dysregulated developmental programs downstream of ectopic Ptf1a expression.
The present findings provide a unifying mechanism for the multiple developmental defects in the Danforth mouse and reconcile several prior observations. For example, our data confirm a previous study that used genetic interaction to infer that Sd is a gain-in-function mutation [44]. The present data also explain why the Danforth mutation was not fully recapitulated by ablation of the notochord [18] and could not be explained based on dysregulation of any single embryonic compartment [17], as ectopic Ptf1a expression likely misdirects developmental programs independently in each affected tissue. Now that we identified the initiating genetic lesion, future studies can determine the precise molecular cascade leading to the developmental defects in each compartment.
To date, the vast majority of developmental defects reported in mice and humans are produced by coding mutations that result in loss of function of the encoded protein; developmental defects arising from disruption of conserved regulatory elements have been less frequently described [45]. Comparative genomics studies of bony vertebrates have identified highly conserved sequences that are enriched around genes that have tissue-specific enhancer activity, acting as developmental regulators [45]–[47]. The Sd mutation provides a striking example of a cis-regulatory mutation that produces profound developmental defects that are quite distinct from phenotypes resulting from simple loss/gain of function mutations. The Danforth mouse can thus serve as an excellent model for dissecting the role of enhancer elements on the temporo-spatial regulation of gene expression in vertebrate development. These data also suggest that mutations in conserved regulatory elements may contribute to human malformation syndromes. For example, we recently studied 522 children with kidney malformations and identified 72 rare copy number disorders that disrupt coding segments, accounting for up to 16.6% of cases [48]. However, we also identified many rare or unique intergenic CNVs in this population, suggesting that disruption of noncoding elements may also play a pathogenic role in this phenotype [48]. Given the high sequence conservation in the Ptf1a 5′ regulatory region, this segment is a good candidate for mutational screening in larger patient populations with caudal regression, axial or urogenital defects.
Methods
Mice
The Association for Assessment and Accreditation of Laboratory Animal Care guidelines were followed for all animal procedures, and all procedures were approved by the Institutional Animal Care and Use Committee of Columbia University. All inbred strains including the strain carrying the Sd mutant allele (RSV/LeJ-Sd/+) mice were purchased from The Jackson Laboratory. To refine the Sd locus, we generated 174 backcross and 120 intercross mice between RSV/LeJ-Sd/+ mice and C57BL/6J mice and also a second mapping F2 intercross cohort of 1203 mice by intercrossing (RSV/LeJ-Sd/+ x DBA/2J) F1 mice. Mice were phenotyped at birth by visual inspection. Affection status was assigned based on presence of the short tail in homozygotes and heterozygotes. While heterozygotes have no other outwardly visible phenotypes, the homozygote mutants are readily recognizable based on the presence of caudal agenesis, and on dissection have major gut and urogenital malformations as previously described [11], [13], [14], [16]–[19]. The Ptf1atm1(cre)Wri or Ptf1a–cre mice (carrying a Cre recombinase replacing the Ptf1a protein-coding region, resulting in a Ptf1a null allele) were obtained from Wright and colleagues [28]. To examine the effect of Ptf1a haploinsufficiency on the heterozygote phenotypes, we generated F1 hybrids between Ptf1a–cre/+mice and RSV/LeJ-Sd/+ mice. Phenotypes were determined at weaning (postnatal day 22), by measurement of tail length, kidney length and kidney weight. H&E staining of kidneys were also performed. Genomic DNA was isolated using the Genomic DNA Isolation kit (Lamda Biotech).
Genotyping and analysis of linkage
Marker loci for inbred strains were obtained from the Mouse Phenome Database (http://phenome.jax.org/); we filtered these for SNPs that were polymorphic between C57Bl/6J, DBA/2J and RSV/LeJ. The BC group was genotyped using microsatellite markers across the Sd locus. Multipoint lod score was calculated using the R/QTL package, utilizing the discrete trait analytic model. The F2 cohort was genotyped with 29 informative SNPs distributed across the 1.3 cM region delimited by rs27129240 and rs29504224 (Sequenom Mass Array system, Columbia University Genotyping facility). To define the haplotype at the Ptf1a locus, we searched SNPs in the Mouse Phenome Database (MPD) and genotyped 37 SNPs that differentiated common haplotypes between 17 inbred strains sequenced by the Sanger Center [49]. Haplotype analysis was performed by Sanger sequencing of RSV/LeJ-+/+, RSV/LeJ-Sd/Sd, C57BL/6J and DBA/2J The genotypes in C57BL/6J and DBA/2J mice demonstrated a 100% concordance with genotypes from dbSNP, confirming accuracy of Sanger sequencing.
Sequencing
Gene annotation was performed using NCBI database (http://www.ncbi.nlm.nih.gov/) and UC Santa Cruz genome browser (http://genome.ucsc.edu/). Mutational screening of the positional candidate genes was performed by Sanger sequencing of exons and flanking introns, comparing RSV/LeJ-Sd/Sd mouse to a RSV/LeJ - +/+ mouse. Long range-PCR was performed in order to amplify the insertion in the Sd/Sd mutant (TaKaRa long range PCR). We performed Sanger sequencing of the 42.8 kb minimal recombinant interval in an RSV/LeJ -Sd/Sd and RSV/LeJ −+/+ mouse. We achieved 100% coverage of this interval with an average base call accuracy of 99% (Phred scores of 20); there were no gaps or ambiguity in the interval. Bidirectional sequencing was obtained for 76% of the region. The sequence of the insertion was analyzed by alignment with the Repbase data, the most commonly used database of repetitive DNA elements (http://www.girinst.org/repbase/index.html ref [50]). From The Jackson Laboratory, we obtained genomic DNA from 25 strains of mice which comprehensively represent all Mus musculus sub-species, including 4 wild-derived (PWD/PhJ, MOLF/EiJ, WSB/EiJ, CAST/EiJ) and 21 inbred strains (DBA/2J, DBA/1J, C57BL/6J, C57BL/6NJ, BALB/cJ, FVB/NJ, RIIIS/J, C3H/HeJ, AKR/J, NOD/LtJ, SJL/J, 129/SvimJ, 129/SvEv, 129P2/OlaHsd, CBA/J, CFW, SWR/J, BTBR T<+>tfJ, NZW/LacJ, KK/HlJ, and A/J).
Quantitative analysis of cDNA
Timed embryos were collected at embryonic day (E) 9.5, 10.5 and E12.5 (where E0.5 is the day of detection of the vaginal plug). Comparisons were made between littermates with differing genotypes. RNA was isolated from whole embryos with TRIzol (Invitrogen) followed by DNaseI treatment and clean-up using the RNeasy mini kit (Qiagen). cDNA was generated with the Omni-Script kit (Qiagen). cDNA levels were quantified in duplicate by qPCR using SYBR Green Mix on an IQ thermal cycler (Bio-Rad). The same internal control was included in each run to standardize each qPCR run, and β-actin was used as reference gene (using Pfaffl algorithm). Expression levels were standardized to the same E10.5 wild-type mouse embryo.
Embryo dissections and in situ hybridization
Embryos were collected at E8.5, E9.5 and E10.5 and yolk sacs were removed for PCR genotyping. Embryos were fixed in 4% paraformaldehyde (PFA) (Fisher), washed in PBT, dehydrated in methanol and processed for whole mount in situ hybridization (ISH) as described previously [51]. Samples at each developmental stage were processed together and images captured under identical settings to allow qualitative comparison of staining intensity between genotypes. The Ptf1a probe was generated by cloning the full-length Ptf1a cDNA from mouse embryonic pancreas into the TOPO blunt PCR cloning vector (Invitrogen). Briefly, embryos were bleached in hydrogen peroxide for an hour, treated with proteinase K (10 µg/ml) (Roche) and fixed in PFA. Further the embryos were incubated at 65°C in hybridization buffer for an hour and then digoxygenin labeled Ptf1a antisense RNA probe was added to hybridization buffer overnight. The following day the embryos were washed with solutions of decreasing stringency of saline sodium citrate salt solutions, washed in Tris-buffered saline with tween (TBST) and incubated in alkaline-phosphatase labeled anti-digoxygenin (Roche) in TBST overnight at 4°C. On the third day the embryos were washed in TBST followed by an overnight wash. On day four the embryos were developed using the BM purple (Roche) solution in dark. The embryos were washed in PBT after developing, post fixed with PFA and stored in PBT until photographed on a Nikon SMZ1500 dissecting microscope.
Supporting Information
Zdroje
1. ParkerSE, MaiCT, CanfieldMA, RickardR, WangY, et al. (2010) Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol 88 : 1008–1016.
2. Population-based birth defects surveillance data from selected states, 2000–2004. Birth Defects Res A Clin Mol Teratol 79 : 874–942.
3. BoulasMM (2009) Recognition of caudal regression syndrome. Adv Neonatal Care 9 : 61–69 quiz 70–61.
4. BohringA, LewinSO, ReynoldsJF, VoigtlanderT, RittingerO, et al. (1999) Polytopic anomalies with agenesis of the lower vertebral column. Am J Med Genet 87 : 99–114.
5. RossAJ, Ruiz-PerezV, WangY, HaganDM, SchererS, et al. (1998) A homeobox gene, HLXB9, is the major locus for dominantly inherited sacral agenesis. Nat Genet 20 : 358–361.
6. WoolfAS, PriceKL, ScamblerPJ, WinyardPJ (2004) Evolving concepts in human renal dysplasia. J Am Soc Nephrol 15 : 998–1007.
7. SchedlA (2007) Renal abnormalities and their developmental origin. Nat Rev Genet 8 : 791–802.
8. WeberS, MoriniereV, KnuppelT, CharbitM, DusekJ, et al. (2006) Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol 17 : 2864–2870.
9. UlinskiT, LescureS, BeaufilsS, GuigonisV, DecramerS, et al. (2006) Renal phenotypes related to hepatocyte nuclear factor-1beta (TCF2) mutations in a pediatric cohort. J Am Soc Nephrol 17 : 497–503.
10. ThomasR, Sanna-CherchiS, WaradyBA, FurthSL, KaskelFJ, et al. (2011) HNF1B and PAX2 mutations are a common cause of renal hypodysplasia in the CKiD cohort. Pediatr Nephrol 26 : 897–903.
11. DanforthCH (1930) Developmental Anomalies in a Special Strain of Mice. Amer Jour Anat 45 : 275–287.
12. DunnLC, Gluecksohn-SchoenheimerS, BrysonV (1940) A New Mutation in the Mouse: Affecting Spinal Column and Urogenital System J Heredity. 31 : 343–348.
13. Gluecksohn-SchoenheimerS (1943) The Morphological Manifestations of a Dominant Mutation in Mice Affecting Tail and Urogenital System. Genetics 28 : 341–348.
14. Gluecksohn-SchoenheimerS (1945) The Embryonic Development of Mutants of the Sd-Strain in Mice. Genetics 30 : 29–38.
15. GrunebergH (1958) Genetical studies on the skeleton of the mouse. XXII. The development of Danforth's short-tail. J Embryol Exp Morphol 6 : 124–148.
16. AsakuraA, TapscottSJ (1998) Apoptosis of epaxial myotome in Danforth's short-tail (Sd) mice in somites that form following notochord degeneration. Dev Biol 203 : 276–289.
17. MaatmanR, ZachgoJ, GosslerA (1997) The Danforth's short tail mutation acts cell autonomously in notochord cells and ventral hindgut endoderm. Development 124 : 4019–4028.
18. TripathiP, GuoQ, WangY, CoussensM, LiapisH, et al. (2010) Midline signaling regulates kidney positioning but not nephrogenesis through Shh. Dev Biol 340 : 518–527.
19. AlfredJB, RanceK, TaylorBA, PhillipsSJ, AbbottCM, et al. (1997) Mapping in the region of Danforth's short tail and the localization of tail length modifiers. Genome Res 7 : 108–117.
20. MaksakovaIA, RomanishMT, GagnierL, DunnCA, van de LagemaatLN, et al. (2006) Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2: e2 doi:10.1371/journal.pgen.0020002
21. NellakerC, KeaneTM, YalcinB, WongK, AgamA, et al. (2012) The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol 13: R45.
22. Fernandez-GonzalezA, La SpadaAR, TreadawayJ, HigdonJC, HarrisBS, et al. (2002) Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science 295 : 1904–1906.
23. MaJ, NortonJC, AllenAC, BurnsJB, HaselKW, et al. (1995) Retinal degeneration slow (rds) in mouse results from simple insertion of a t haplotype-specific element into protein-coding exon II. Genomics 28 : 212–219.
24. GittesGK (2009) Developmental biology of the pancreas: a comprehensive review. Dev Biol 326 : 4–35.
25. MasuiT, SwiftGH, HaleMA, MeredithDM, JohnsonJE, et al. (2008) Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol 28 : 5458–5468.
26. MeredithDM, MasuiT, SwiftGH, MacDonaldRJ, JohnsonJE (2009) Multiple transcriptional mechanisms control Ptf1a levels during neural development including autoregulation by the PTF1-J complex. J Neurosci 29 : 11139–11148.
27. ObataJ, YanoM, MimuraH, GotoT, NakayamaR, et al. (2001) p48 subunit of mouse PTF1 binds to RBP-Jkappa/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes to cells : devoted to molecular & cellular mechanisms 6 : 345–360.
28. KawaguchiY, CooperB, GannonM, RayM, MacDonaldRJ, et al. (2002) The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nature genetics 32 : 128–134.
29. ThompsonN, GesinaE, ScheinertP, BucherP, Grapin-BottonA (2012) RNA profiling and chromatin immunoprecipitation-sequencing reveal that PTF1a stabilizes pancreas progenitor identity via the control of MNX1/HLXB9 and a network of other transcription factors. Molecular and cellular biology 32 : 1189–1199.
30. HenkeRM, SavageTK, MeredithDM, GlasgowSM, HoriK, et al. (2009) Neurog2 is a direct downstream target of the Ptf1a-Rbpj transcription complex in dorsal spinal cord. Development 136 : 2945–2954.
31. HoriK, Cholewa-WaclawJ, NakadaY, GlasgowSM, MasuiT, et al. (2008) A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev 22 : 166–178.
32. GlasgowSM, HenkeRM, MacdonaldRJ, WrightCV, JohnsonJE (2005) Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development 132 : 5461–5469.
33. ObataJ, YanoM, MimuraH, GotoT, NakayamaR, et al. (2001) p48 subunit of mouse PTF1 binds to RBP-Jkappa/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells 6 : 345–360.
34. RouxE, StrubinM, HagenbuchleO, WellauerPK (1989) The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes Dev 3 : 1613–1624.
35. KawaguchiY, CooperB, GannonM, RayM, MacDonaldRJ, et al. (2002) The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 32 : 128–134.
36. MasuiT, LongQ, BeresTM, MagnusonMA, MacDonaldRJ (2007) Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev 21 : 2629–2643.
37. BeresTM, MasuiT, SwiftGH, ShiL, HenkeRM, et al. (2006) PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol 26 : 117–130.
38. FujitaniY, FujitaniS, LuoH, QiuF, BurlisonJ, et al. (2006) Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 133 : 4439–4450.
39. SellickGS, BarkerKT, Stolte-DijkstraI, FleischmannC, ColemanRJ, et al. (2004) Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet 36 : 1301–1305.
40. MizuguchiR, KriksS, CordesR, GosslerA, MaQ, et al. (2006) Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci 9 : 770–778.
41. HoshinoM, NakamuraS, MoriK, KawauchiT, TeraoM, et al. (2005) Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47 : 201–213.
42. PascualM, AbasoloI, Mingorance-Le MeurA, MartinezA, Del RioJA, et al. (2007) Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci U S A 104 : 5193–5198.
43. FukudaA, KawaguchiY, FuruyamaK, KodamaS, HoriguchiM, et al. (2006) Ectopic pancreas formation in Hes1 -knockout mice reveals plasticity of endodermal progenitors of the gut, bile duct, and pancreas. J Clin Invest 116 : 1484–1493.
44. ZachgoJ, KornR, GosslerA (1998) Genetic interactions suggest that Danforth's short tail (Sd) is a gain-of-function mutation. Dev Genet 23 : 86–96.
45. KleinjanDA, van HeyningenV (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76 : 8–32.
46. WoolfeA, GoodsonM, GoodeDK, SnellP, McEwenGK, et al. (2005) Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7 doi:10.1371/journal.pbio.0030007
47. WoolfeA, ElgarG (2008) Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv Genet 61 : 307–338.
48. Sanna-CherchiS, KirylukK, BurgessK, BodriaM, SampsonM, et al. (2012) Copy Number Disorders are a Common Cause of Congenital Kidney Malformations. Am J Hum Genet (in press).
49. KeaneTM, GoodstadtL, DanecekP, WhiteMA, WongK, et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477 : 289–294.
50. KohanyO, GentlesAJ, HankusL, JurkaJ (2006) Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7 : 474.
51. WilkinsonDG, NietoMA (1993) Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol 225 : 361–373.
Štítky
Genetika Reprodukčná medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



