-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Impaired mitochondrial oxidative phosphorylation (OXPHOS) has been proposed as an etiological mechanism underlying insulin resistance. However, the initiating organ of OXPHOS dysfunction during the development of systemic insulin resistance has yet to be identified. To determine whether adipose OXPHOS deficiency plays an etiological role in systemic insulin resistance, the metabolic phenotype of mice with OXPHOS–deficient adipose tissue was examined. Crif1 is a protein required for the intramitochondrial production of mtDNA–encoded OXPHOS subunits; therefore, Crif1 haploinsufficient deficiency in mice results in a mild, but specific, failure of OXPHOS capacity in vivo. Although adipose-specific Crif1-haploinsufficient mice showed normal growth and development, they became insulin-resistant. Crif1-silenced adipocytes showed higher expression of chemokines, the expression of which is dependent upon stress kinases and antioxidant. Accordingly, examination of adipose tissue from Crif1-haploinsufficient mice revealed increased secretion of MCP1 and TNFα, as well as marked infiltration by macrophages. These findings indicate that the OXPHOS status of adipose tissue determines its metabolic and inflammatory responses, and may cause systemic inflammation and insulin resistance.
Published in the journal: Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice. PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003356
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003356Summary
Impaired mitochondrial oxidative phosphorylation (OXPHOS) has been proposed as an etiological mechanism underlying insulin resistance. However, the initiating organ of OXPHOS dysfunction during the development of systemic insulin resistance has yet to be identified. To determine whether adipose OXPHOS deficiency plays an etiological role in systemic insulin resistance, the metabolic phenotype of mice with OXPHOS–deficient adipose tissue was examined. Crif1 is a protein required for the intramitochondrial production of mtDNA–encoded OXPHOS subunits; therefore, Crif1 haploinsufficient deficiency in mice results in a mild, but specific, failure of OXPHOS capacity in vivo. Although adipose-specific Crif1-haploinsufficient mice showed normal growth and development, they became insulin-resistant. Crif1-silenced adipocytes showed higher expression of chemokines, the expression of which is dependent upon stress kinases and antioxidant. Accordingly, examination of adipose tissue from Crif1-haploinsufficient mice revealed increased secretion of MCP1 and TNFα, as well as marked infiltration by macrophages. These findings indicate that the OXPHOS status of adipose tissue determines its metabolic and inflammatory responses, and may cause systemic inflammation and insulin resistance.
Introduction
White adipose tissue (WAT) determines whole-body energy metabolism by controlling lipid storage and by releasing adipokines, which may directly or indirectly affect the physiological functions of almost all cell types (for a review, see [1], [2]). These adipocyte functions are perturbed by genetic and environmental factors, which lead to adipocyte dysfunction characterized by hypertrophy, hypoxia and inflammatory process within adipose tissue [3]. Adipocyte dysfunction is further characterized by impaired insulin sensitivity, which is associated with changes in cellular composition or organelle dysfunction, particularly of the endoplasmic reticulum (ER) and mitochondria. An emerging concept to explain insulin resistance in obese individuals is maladaptive responses within the ER, which are prominent in adipose tissue (for a review, see [4]–[6]).
Besides the ER, the mitochondria in white adipocytes are linked with adipocyte differentiation and with the function of mature adipocytes. Recent studies show that drastic increases in mitochondrial biogenesis and reactive oxygen species (ROS) production via the OXPHOS complex play a crucial role in adipocyte differentiation. In addition, the mitochondria in differentiating adipocytes support high energy-consuming lipogenic processes to maintain mature adipocyte function [5], [7]. Therefore, it is suggested that the contribution of adipocyte mitochondria to whole-body energy metabolism or adipocyte plasticity may depend on the mitochondrial OXPHOS capacity of the adipose tissue [6]. Consistent with this, decreased mitochondrial capacity in adipocytes may also alter their insulin sensitivity and/or function due to the high energy requirements of fatty acid storage, adipokine secretion, insulin signaling, and glucose uptake [8], [9].
It is interesting that a marked decrease in the level of transcripts for nuclear-encoded mitochondrial genes in cells derived from the epididymal fat pads of ob/ob mice accompanies the onset of obesity [10]. In db/db and diet-induced obese mice, the expression of OXPHOS genes was markedly reduced compared with that in db/+ mice and control mice fed a standard-fat diet, respectively [11]. In humans, the mtDNA copy number is enriched in adipocytes in adipose tissue, but it decreases slightly with age and increasing BMI, and shows a strong positive correlation with basal and insulin-stimulated lipogenesis in fat cells [12]. More interestingly, suppression of OXPHOS genes is prominent in the visceral adipose tissue of humans with type 2 diabetes independent of obesity [13]. Agonists of peroxisome proliferator-activated receptor-gamma (PPARγ) increase the number of mitochondria and induce mitochondrial remodeling in adipocytes [10], [11], [14], and significantly increase the mitochondrial copy number and expression of factors involved in mitochondrial biogenesis, including PPARγ coactivator-1alpha (PGC1α) and mitochondrial transcription factor A (TFAM), which are required for mitochondrial transcription of OXPHOS genes in humans [15]. These observations in rodent models and human subjects suggest that the OXPHOS capacity of adipose tissue may affect the changes in adipocyte plasticity, which controls insulin sensitivity and may determine the therapeutic responsiveness to antidiabetic agents such as thiazolidinediones and CB1 receptor blockers that affect the mitochondrial content of adipocytes [10], [16].
Here, we demonstrate that primary OXPHOS dysfunction in adipose tissue causes insulin resistance and a diabetic phenotype in mice with a Crif1 loss-of-function mutation. Crif1 is a mitochondrial protein that associates with large mitoribosomal subunits, which are located close to the polypeptide exit tunnel, and the elimination of Crif1 led to both aberrant synthesis and defective insertion of mtDNA-encoded nascent OXPHOS polypeptides into the inner membrane [17]. Targeted elimination of the Crif1 gene resulted in a phenotype characterized by organ-specific failure of OXPHOS function; therefore, we attempted to identify the adipose tissue phenotypes of adipose-specific Crif1-knockout mice using Fabp4-Cre and Adiponectin-Cre mice models. Reduced OXPHOS capacity in the WAT of Crif1-deficient mice triggered spontaneous adipose inflammation, which was characterized by macrophage infiltration and systemic insulin resistance. Therefore, the OXPHOS reserve may be the critical determinant controlling the metabolic and inflammatory responses of adipose tissue, which are closely related to systemic changes in insulin sensitivity.
Results
Homozygotic loss of Crif1 causes marked impairment of WAT development
Crif1 is a mitochondrial protein that specifically interacts with the protein components of the large subunit of the mitochondrial ribosome [17]. It specifically regulates the translation and insertion of the 13 polypeptide subunits that comprise mitochondrial OXPHOS complexes I, III, IV and V. Homozygous Crif1-null mouse embryonic fibroblasts (MEFs) showed a profound failure in translation and expression of these subunits, along with markedly low levels of basal and stimulated (CCCP-treated) mitochondrial oxygen consumption [17]. Disruption of the mouse Crif1 gene consistently resulted in a profound OXPHOS deficiency characterized by the loss of OXPHOS complex subunits and respiratory complexes in vivo.
Crif1 mRNA is ubiquitously expressed, and it is highly expressed in brain, heart, liver kidney and skeletal muscle (Figure S1A). Two types of adipose tissues, brown (BAT) and white (WAT), contained substantial amounts of Crif1 mRNA (Figure S1A). Crif1 mRNA levels were decreased in the WAT, BAT and liver of db/db and ob/ob mice compared to db/+ and ob/+ mice, respectively (Figure S1B). Interestingly, Crif1 mRNA expression in WAT of C57BL/6 mice was downregulated when they were fed a high fat diet (HFD) for 8 weeks (Figure S1C). These findings indicate that Crif1 expression correlates with the nutritional status in adipose tissue.
To identify the roles of Crif1 and mitochondrial OXPHOS in adipose tissue, we tried to induce primary OXPHOS deficiency in adipose tissue in vivo using conditional Crif1 knockout mice. We crossed conditional Crif1 mice (Crif1flox/flox) [18] with mice expressing a Cre recombinase gene under the control of the fatty acid binding protein-4 (Fabp4) promoter (Fabp4-Cre) and the adiponectin promoter (Adipoq-Cre). The resulting pups were born healthy and viable, and showed a normal Mendelian ratio. However, these homozygous Crif1f/f,Fabp4 mice showed delayed weight gain and poor development of adipose tissue (Figure 1A–1C). Unlike the control (Crif1+/+,Fabp4) and Crif1 heterozygous (Crif1f/+Fabp4) mice, Crif1f/f,Fabp4 mice showed uniform lethality within 24 days of birth (median survival = 19.4 days) (Figure 1D).
Fig. 1. Marked failure of adipose tissue development in Crif1f/f,Fabp4 mice. 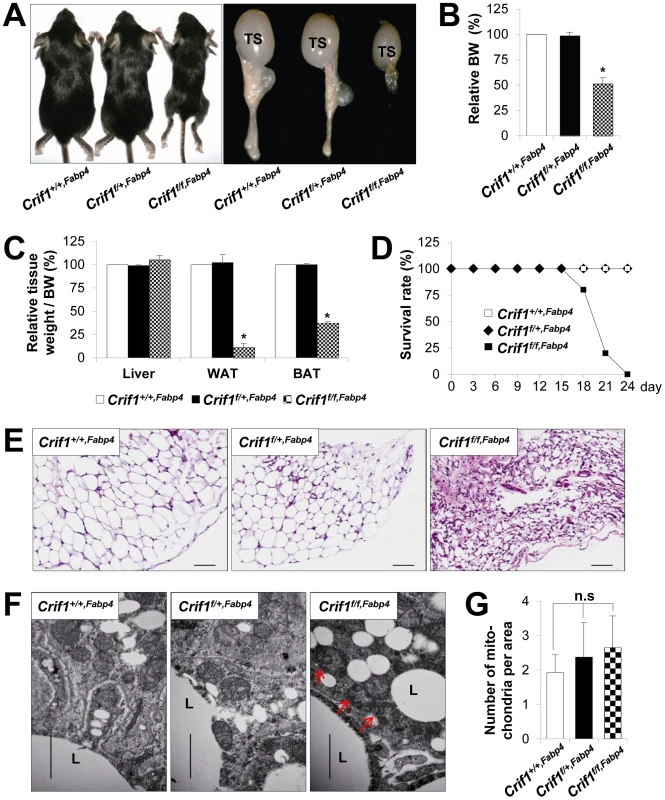
Mice were generated by breeding two mouse lines, a Crif1flox/flox transgenic mouse line and a Fabp4-Cre recombinase transgenic mouse line. Data from Crif1f/f,Fabp4 mice were obtained at 3 weeks-of-age. (A) Gross characteristics of control mice (Crif1+/+,Fabp4), adipose-specific Crif1 heterozygous mice (Crif1f/+,Fabp4), and adipose-specific Crif1 homozygous knockout mice (Crif1f/f,Fabp4). TS, testis. (B and C) Body weight and tissue weight relative to control mice (n = 10). Values are means + SD, *p<0.05 versus control mice. BAT, brown adipose tissue; WAT, white adipose tissue. (D) Crif1f/f,Fabp4 mice showed markedly reduced survival rates after 2 weeks-of-age (n = 20). (E) Hematoxylin & eosin (H&E) staining of perirenal WAT (pWAT). Scale: 100 µm. (F) Transmission electron microscopy (TEM) of subcutaneous WAT (sWAT) revealed that the mitochondria of Crif1f/f,Fabp4 mice developed swollen cristae (red arrows). L, lipid droplet. Scale: 6,000 nm. (G) Mitochondria number per area in sWAT (n = 10). Values are means + SD. n.s, not significant. The perirenal, subcutaneous and epididymal fat pads of Crif1f/f,Fabp4 mice comprised small adipocytes with dystrophic changes (Figure 1E). To verify any mitochondrial abnormalities, the adipose tissues of Crif1f/f,Fabp4 mice were examined by transmission electron microscopy (TEM). The adipocytes of these mice contained mitochondria with ultrastructural abnormalities, such as swollen and distorted cristae, but mitochondrial number was unaffected (Figure 1F and 1G). In heterozygous Crif1f/+,Fabp4 mice, hematoxylin and eosin (H&E) staining of adipose tissue showed no evidence of histological abnormalities compared with the controls (Figure 1E). Consistent with the results of H&E staining, the mitochondria of Crif1f/+,Fabp4 mice showed no morphological or numerical abnormalities of mitochondria in TEM (Figure 1F and 1G). Collectively, this comprehensive analysis of the adipose tissues in Crif1f/f,Fabp4 mice indicated that loss of Crif1 results in a marked failure of WAT and BAT development.
Characterization of BAT in Crif1f/f,Fabp4 mice
The Fabp4-Cre transgene is expressed and localized within the dorsal root ganglion, centrum of the vertebra and the carpals of the embryo from the mid-gestation stage [19]. Neonatal Crif1f/+,Fabp4 or Crif1f/f,Fabp4 mice did not show developmental abnormalities when compared with control mice. Therefore, embryonic expression of the Fabp4-Cre transgene may not affect the development of Crif1f/+,Fabp4 and Crif1f/f,Fabp4 mice, and may not be a plausible reason for observed lethality at around post-natal Week 3. Mice are normally weaned at post-natal Week 3, at which point the rate of lipogenesis and UCP1 expression in the BAT rises sharply and reaches maximal levels to enhance thermogenesis [20]. The Fabp4-Cre transgene was uniformly detected in BAT from the early post-natal period (Day 7), the Crif1 protein and OXPHOS complex subunits are downregulated in the BAT of 3-week-old mice (Table S1). As shown in Figure 1C, Crif1f/f,Fabp4 mice had less BAT at Day 21, but histological examination of inter-scapular BAT showed normal histological findings (Figure S2A). Crif1f/f,Fabp4 mice had fewer mitochondria than control mice, but these were larger in size and were characteristically disorganized and swollen, suggesting OXPHOS defects (Figure S2B and S2C). Consistent with these findings, Crif1f/f,Fabp4 mice showed a low body temperature under ambient conditions (23°C) and rapidly reached a fatally low rectal temperature within 5 minutes of immersion in cold water (4°C) (Figure S2D). However, although the mass of BAT was reduced, the level of UCP1 expression was not altered in Crif1f/f,Fabp4 mice (data not shown). When Crif1f/f,Fabp4 mice were housed at thermoneutrality (30°C), the median survival rate was increased and mortality was reduced (Figure S2E). This indicates that thermal stress caused by mitochondrial OXPHOS dysfunction in BAT following Crif1 ablation may be a critical factor in the premature death of Crif1f/f,Fabp4 mice.
By contrast, the BAT of Crif1f/+,Fabp4 mice showed normal development and histological and ultrastructural findings (Figure S2A–S2C). Furthermore, the response of Crif1f/+,Fabp4 mice (in terms of core temperature) to a cold environment were identical to those of control mice (Figure S2D). These results showed that Fabp4-Cre driven haploinsufficiency of Crif1 may not affect the physiological function of BAT.
Characterization of mitochondrial OXPHOS function in Crif1f/+,Fabp4 mice
A previous study revealed that Crif1-deficient (−/Δ) MEFs prepared from Crif1−/flox mice showed marked OXPHOS defects due to a profound failure of translation and insertion of the newly-synthesized OXPHOS polypeptides encoded by the mtDNA. Also, Crif1 −/Δ MEFs showed increased anaerobic glycolysis, which eventually led to accelerated cell death [17]. Similar to Crif1 −/Δ MEFs, loss of Crif1 in adipose-derived stem cells (ADSCs) (Crif1M−/−) resulted in marked impairment of differentiation and accelerated cell death, which prevented functional analysis of the mitochondria (data not shown). However, control (Crif1+/+) and Crif1-haploinsufficient ADSCs (Crif1+/−) prepared from Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice showed identical levels of cell viability and differentiation to those of control cells (Figure 2A and 2B). Interestingly, Crif1+/− ADSCs showed lower expression of OXPHOS subunits (ND1, NDUFA9, UQCRC2 and COX4) and assembled OXPHOS complex I on Western blot and Blue Native PAGE (BN-PAGE) analysis, respectively (Figure 2C and 2D). Crif1+/− ADSCs consumed less oxygen under basal conditions and showed reduced maximal OXPHOS capacity (Figure 2E). Taken together, Crif1 haploinsufficiency in adipocytes resulted in normal differentiation but reduced genetically-determined OXPHOS capacity.
Fig. 2. Dysfunctional mitochondria in Crif1 haploinsufficient adipocytes. 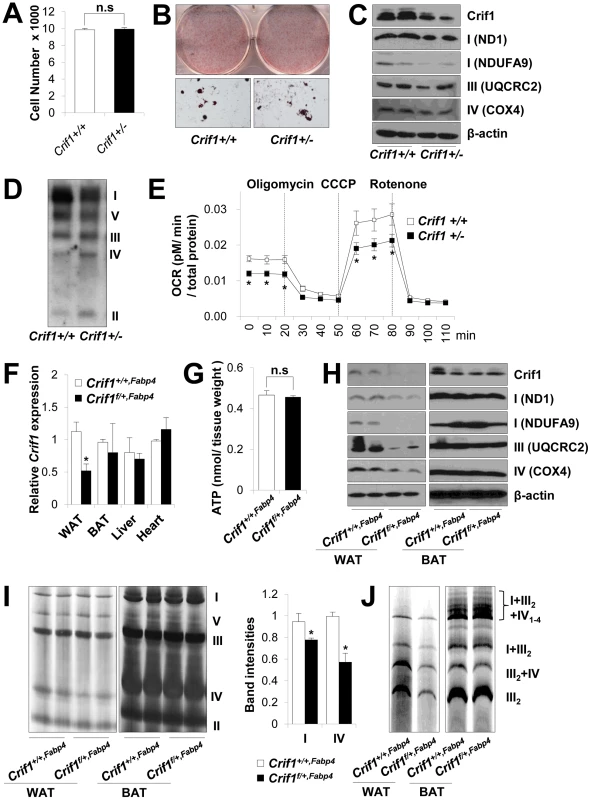
(A) Adipose derived stem cells (ADSCs) were isolated from eWAT of Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice. The number of Crif1+/+ and Crif1+/− ADSCs was counted in 96 well plates after 10 days of differentiation (n = 6). Values are means + SD. n.s, not significant. (B) Oil red O staining of Crif1+/+ and Crif1+/− ADSCs. (C) OXPHOS complex subunits were detected by western blotting with the appropriate antibodies. Anti-ND1 and anti-NDUFA9 were used to detect OXPHOS complex I, anti-UQCRC2 was used for complex III, and anti-COX4 was used for complex IV. (D) BN-PAGE analysis using a mixture of anti-OXPHOS antibodies to detect assembled OXPHOS complexes. (E) Oxygen consumption rates (OCR) measured by Seahorse XF-24 flux analyzer in Crif1+/+ and Crif1+/− ADSCs (n = 6). Values are means ± SD, *p<0.05. CCCP, carbonyl cyanide m-chloro phenyl hydrazine. (F) Crif1 mRNA expression in eWAT, BAT, liver and heart of Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice (n = 8). Values are means + SD, *p<0.05. (G) Level of ATP in eWAT of Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice (n = 8). Values are means + SD. n.s, not significant. (H) Western blots analysis of Crif1 and OXPHOS subunits from eWAT and BAT of mice. ND1 and NDUFA9, subunit of OXPHOS complex I; UQCRC2, subunit of OXPHOS complex III; COX4, subunit of OXPHOS complex IV. (I) BN-PAGE analysis of assembled OXPHOS complexes (I, II, III, IV and V). Percentage of band intensities are presented in the graph (n = 4). Values are means + SD, *p<0.05. (J) BN-PAGE analysis of supercomplex (I+III2+IV1–4, I+III2, III2+IV and III2). Several experimental criteria have been proposed to test whether a primary in vivo OXPHOS deficiency plays a causal role in insulin resistance [21]. One of these criteria is that perturbations in OXPHOS gene expression and function must be as modest as possible [21]. Thus, we analyzed Crif1 and OXPHOS gene expression to test whether Crif1f/+,Fabp4 mice were suitable for our proposed experiments. Compared with Crif1+/+,Fabp4 mice, Crif1f/+,Fabp4 mice showed about ∼50% of the Crif1 mRNA and protein expression in epididymal WAT (eWAT) (Figure 2F and 2H). Although basal ATP levels in eWAT were not affected by Crif1 haploinsufficiency (Figure 2G), the expression levels of OXPHOS complex I, III and IV subunits were reduced in the epididymal fat pads of Crif1f/+,Fabp4 mice (Figure 2H). BN-PAGE analysis showed that the levels of Complex I and IV and supercomplex in WAT were approximately 20%, 40% and 50% lower, respectively, in Crif1f/+,Fabp4 mice compared to control mice (Figure 2I and 2J). However, normal levels of Crif1 and OXPHOS complexes were expressed in the liver and heart of Crif1f/+,Fabp4 mice (Figure S3A–S3C). In contrast to homozygous Crif1f/f,Fabp4 mice, heterozygous Crif1f/+,Fabp4 mice exhibited normal levels of OXPHOS subunits in BAT and mitochondrial morphology was normal (Figure 2H–2J and Figure S2B).
Food intake and weight gain were comparable in Crif1f/+,Fabp4 and Crif1+/+,Fabp4 mice when fed a normal chow diet (NCD) (Figure S4A and S4B). MR images of control and Crif1f/+,Fabp4 mice fed a NCD or a HFD showed a similar pattern of adipose distribution (Figure S4C). Triglyceride levels in the liver and plasma of Crif1f/+,Fabp4 mice were the same as those in control mice, regardless of whether they were fed a NCD or a HFD. Serum free fatty acid (FFA) levels tended to be higher in Crif1f/+,Fabp4 mice, but were not significantly different from those in control mice (Figure S4D–S4F). Taken together, these results show that Crif1f/+,Fabp4 mice have mildly reduced primary OXPHOS deficiency in adipose tissue but, unlike the lipodystrophic model, they show no defects in adipose tissue development, and no hyperlipidemia or ectopic lipid accumulation.
Insulin resistance in Crif1f/+,Fabp4 mice
To identify the relationship between insulin resistance and reduced OXPHOS capacity in adipocytes in vivo, control and Crif1f/+,Fabp4 mice were subjected to glucose tolerance tests after 8 weeks or 14 weeks on a NCD or HFD. Neither control nor Crif1f/+,Fabp4 mice fed a NCD for 8 weeks showed any differences in glucose tolerance following an intraperitoneal injection of glucose (IPGTT, 2 g/kg body weight) (Figure 3A). However, Crif1f/+,Fabp4 mice fed a NCD for 14 weeks developed glucose intolerance (Figure 3B). More impressively, Crif1f/+,Fabp4 mice fed a HFD for 8 weeks showed an earlier onset of glucose intolerance, which was characterized by higher peak glucose levels than those measured in control mice in the intraperitoneal glucose tolerance tests (Figure 3C). Crif1f/+,Fabp4 mice fed a HFD for 14 weeks showed more advanced glucose intolerance, with higher basal (168.8±13.2 mg/dL vs 131.3±8 mg/dL) and peak (516.8±34.8 mg/dL vs 420.4±52.3 mg/dL) plasma glucose levels (Figure 3D). Therefore, regardless of the caloric state, mice with Crif1 haploinsufficiency showed reduced glucose tolerance. Crif1f/+,Fabp4 mice fed a HFD for 14 weeks showed decreased Akt phosphorylation in the liver and muscle and a reduced glucose disposal rate after an intraperitoneal insulin challenge (Figure 3E and 3F). Furthermore, suppression of hepatic glucose production (HGP) by insulin was not different between the two groups, but the glucose infusion rate (GIR) and glucose uptake rate decreased by approximately 18.6% and 14.7%, respectively, during hyperinsulinemic euglycemic clamping after 14 weeks on a HFD (Figure 3G); these data supported the insulin tolerance tests (ITT) results. These findings indicate that Crif1f/+,Fabp4 mice, which have limited OXPHOS capacity in their adipose tissue, may show exacerbated diabetic mechanisms, which are characterized by insulin resistance.
Fig. 3. Metabolic phenotypes and insulin resistance in Crif1f/+,Fabp4 mice. 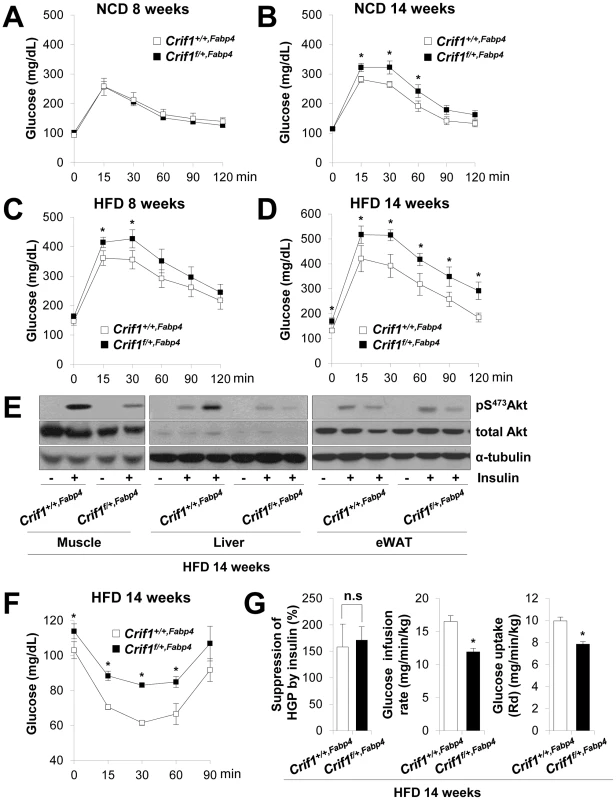
(A–D) An intraperitoneal glucose tolerance test (IPGTT) was performed with 1 g/kg glucose, after a 16 h fast in NCD or HFD mice (n = 8). Values are means ± SD, *p<0.05 versus control mice. (E) Western blot analysis of p-Akt in the gastrocnemius muscle and liver after injecting 1 U/kg insulin in control and Crif1f/+,Fabp4 mice with 14 weeks of HFD. Data are representative of three independent experiments. p-Akt, phospho-Akt; t-Akt, total-Akt. (F) Insulin (0.75 U/kg) tolerance tests (ITTs) (n = 8). Values are means ± SD, *p<0.05 versus control mice. (G) Hyperinsulinemic euglycemic clamp analysis in Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice with 14 weeks of HFD. HGP, hepatic glucose production (n = 8). Values are means + SD, *p<0.05, n.s, not significant. The levels of saturated fatty acids and ceramides in WAT, muscle and liver were not significantly altered in Crif1f/+,Fabp4 mice (Figure S4G and S4H). Thus, abnormal accumulation of ceramides and saturated fatty acids in insulin sensitive tissues does not appear to underlie the insulin resistance of Crif1f/+,Fabp4 mice (Figure S4G and S4H).
Dysregulation of chemokines in Crif1-deficient differentiated adipocytes in vitro
To determine the molecular pathways that are dysregulated by mitochondrial OXPHOS dysfunction following Crif1 knockdown by siRNA in adipocytes, we introduced Crif1 siRNA into fully-differentiated 3T3-L1 cells. Crif1 knockdown in differentiated 3T3-L1 cells resulted in decreased expression of the OXPHOS subunits, ND1, NDUFA9, UQCRC2 and ATP5A1, but did not affect the expression of Ppar-gamma, adiponectin, and Cd36 (Figure 4A). A complementary DNA (cDNA) microarray analysis showed prominent increases in the expression levels of inflammatory cytokine and chemokine genes in adipocytes following knockdown of Crif1 (Figure S5). In particular, the chemokines monocyte chemotactic protein 1 (Mcp1/Ccl2), IFN-γ-inducible protein (Ip10/Cxcl10), Regulated upon Activation, Normal T cell Expressed and Secreted (Rantes/Ccl5) and Mig/Cxcl9, which are important for the recruitment of macrophages and T cells to WAT, were elevated in Crif1-silenced 3T3-L1 adipocytes [22]. The elevation of Mcp1, Ip10, and Rantes expression observed in cDNA microarrays was confirmed by real-time PCR experiments with Crif1-silenced 3T3-L1 adipocytes (Figure 4B). In parallel experiments, levels of mitochondrial and cytoplasmic superoxide anions were increased in Crif1-silenced 3T3-L1 adipocytes compared to control cells (Figure 4C). Treatment with the antioxidant N-acetylcysteine (NAC) suppressed the expression of Mcp1, Ip10 and Rantes in Crif1-silenced 3T3-L1 adipocytes (Figure 4D).
Fig. 4. Dysregulation of chemokines and activation of stress kinases in Crif1-deficient 3T3-L1 adipocytes. 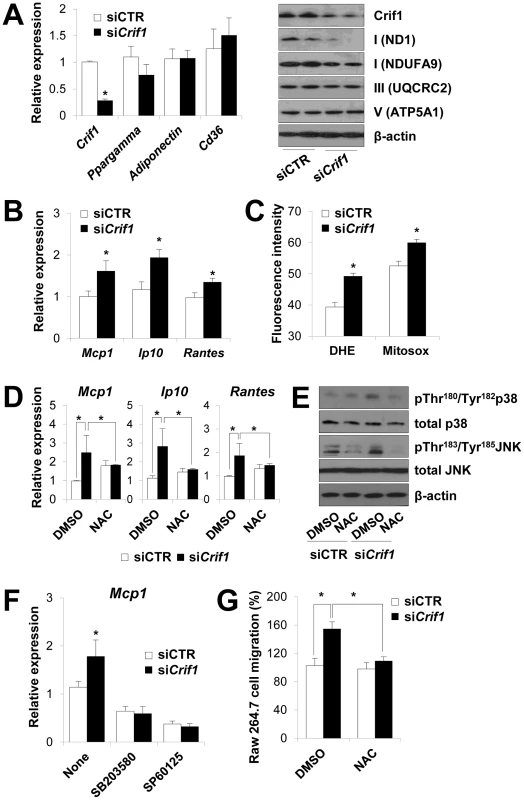
(A) Silencing of Crif1 and expression of adipogenic genes, Ppargamma, adiponectin and Cd36 in 3T3-L1 cells. Western blot analysis with anti-ND1 and NDUFA9 (Complex I), anti-UQCRC2 (Complex III) and anti-ATP5A1 (Complex V) after 4 days of Crif1 silencing (n = 10). siCTR, control siRNA; siCrif1, Crif1 siRNA. Values are means + SD, *p<0.05. (B) Expression of macrophage recruiting chemokines (Mcp1, monocyte attractant chemokine; Ip10, IFN-γ–inducible protein 10; Rantes, Regulated upon Activation, Normal T cell Expressed and Secreted) by 3T3-L1 cells subjected to Crif1 siRNA (n = 10). Values are means + SD, *p<0.05. (C) Measurement of MitoSox- or DHE-stained cells by flow cytometry. DHE, di-hydroethidium (n = 10). Values are means + SD, *p<0.05. (D) Mcp1, Ip10 and Rantes mRNA levels with/without NAC treatment (1 mM) for 24 h. NAC, N-acetyl-L-cysteine (n = 10). Values are means + SD, *p<0.05. (E) p-p38 MAPK and p-JNK levels in Crif1-deficient 3T3-L1 adipocytes with/without NAC treatment (1 mM) for 24 h. p38 MAPK, p38 mitogen-activated protein kinases; JNK, c-Jun N-terminal kinases. (F) Inhibition of Mcp1 expression after treatment with p-p38 (SB203580; 10 µM for 24 h) or p-JNK (SP60125; 20 µM for 24 h) inhibitors, respectively (n = 10). Values are means + SD, *p<0.05. (G) Migration of Raw264.7 macrophages examined using culture supernatants from control siRNA- or Crif1 siRNA-treated 3T3-L1 adipocytes with/without NAC (n = 10). Values are means + SD, *p<0.05. Adipose inflammation links adipocyte dysfunction to insulin resistance, which are frequently observed in excessive adiposity (for a review, see). The inflammatory process in adipose tissue is provoked by the activation of stress kinases, e.g., c-Jun N-terminal kinase (JNK), which inhibit insulin signaling and activate transcription factors that mediate the expression of chemokine genes [24], [25]. Intracellular stress signals including mitochondrial ROS, FFA, ceramide and ER stress activates the stress kinases, JNK, p38 MAPK and NF-κB in adipocytes [4], [26], [27]. JNK mediates macrophage activation and expression of proinflammatory cytokines and inhibits insulin receptor substrate 1 (IRS-1)-mediated insulin signaling pathways (for a review, see [28], [29]). To identify the roles of stress kinases in the expression of chemokines in Crif1-silenced 3T3-L1 adipocytes, p38 MAPK and JNK phosphorylation were observed by Western blot analysis. Levels of phosphorylated p38 MAPK and JNK were elevated in Crif1-silenced 3T3-L1 adipocytes compared to control cells; however, this activation was suppressed by NAC treatment (Figure 4E). These results indicate that chemokine dysregulation is associated with increased ROS generation and inappropriate activation of p38 MAPK and JNK. To confirm these results, 3T3-L1 cells were treated with inhibitors of p38 MAPK and JNK (SB203580 and SP60125, respectively). Two inhibitors effectively inhibited the expression of Mcp1 in Crif1 silenced 3T3-L1 cells (Figure 4F). Crif1 deficiency in MEFs results in increased ROS production [17] and induces phosphorylation of p38 (Figure S6A). However, Crif1 -/Δ MEFs did not show increased Mcp1 and Ip10 expression (Figure S6B). Taken together, these results suggest that limited mitochondrial OXPHOS function in fully-differentiated adipocytes triggers the expression of chemokines (Mcp1, Ip10 and Rantes) in a cell-specific manner.
The chemokines (Mcp1, Ip10 and Rantes) upregulated in Crif1 siRNA-treated adipocytes are thought to be critical for attracting macrophages and T lymphocytes into adipose tissue in obese subjects [30]. Therefore, we wondered whether Crif1-silenced 3T3-L1 cells would trigger the migration of macrophages. As shown in Figure 4G, Crif1-silenced 3T3-L1 cells enhanced the migration of RAW 264.7 cells and NAC treatment inhibited the migration of RAW 264.7 cells. Thus, our in vitro studies show that OXPHOS deficiency induced in differentiated cultured 3T3-L1 adipocytes by Crif1 silencing results in the upregulated expression of chemokines, which then recruit or activate macrophages, ROS and stress kinase dependently.
Macrophage infiltration and systemic inflammatory responses in Crif1f/+,Fabp4 mice
To observe ROS stress associated with abnormal chemokine responses in adipose tissues in vivo, we measured lipid peroxidation (TBAR assays), stress kinase activation and cytokine expression in WAT of Crif1f/+,Fabp4 mice fed a NCD or a HFD for 8 weeks. Consistent with the in vitro studies, lipid peroxidation in WAT and plasma was increased in Crif1f/+,Fabp4 mice fed a HFD for 8 weeks compared to control mice (Figure S7A). Levels of p38 MAPK and JNK phosphorylation were higher in WAT of Crif1f/+,Fabp4 mice fed a HFD for 8 weeks than in control mice (Figure 5A). Furthermore, the expression of Mcp1, Ip10 and Rantes was higher in adipose tissue from Crif1f/+,Fabp4 mice than in control mice (Figure 5B). In addition, the level of secreted MCP1, but not IP10, were higher in the serum of Crif1f/+,Fabp4 mice than in control mice (Figure S7B). The results showing dysregulation of chemokines in the absence of Crif1 suggest that mitochondrial OXPHOS dysfunction may trigger immune cell recruitment in adipose tissue.
Fig. 5. Macrophage infiltration and inflammation in adipose tissue of Crif1f/+,Fabp4 mice. 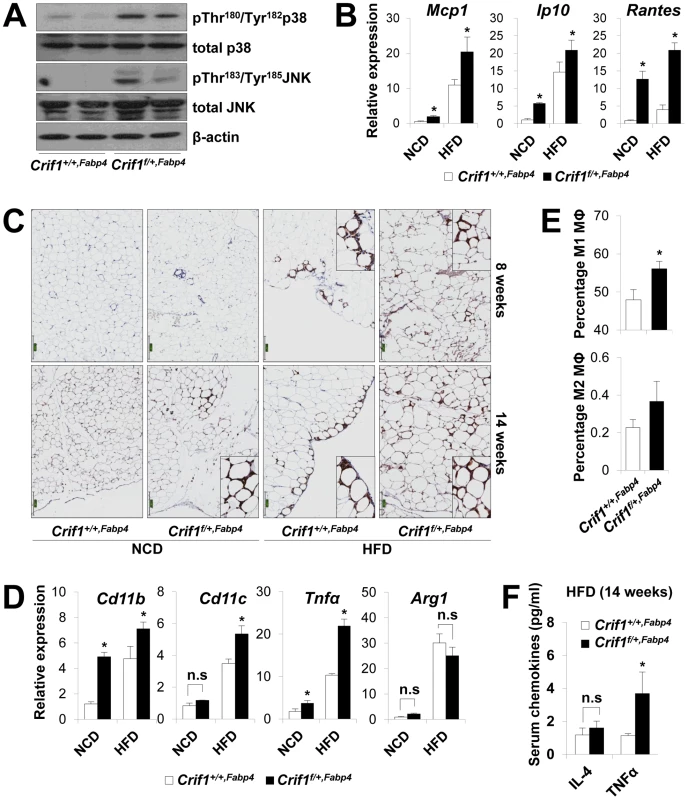
(A) Western blotting for p38 MAPK and JNK phosphorylation in WAT of Crif1f/+,Fabp4 mice fed a HFD for 8 weeks. p38 MAPK, p38 mitogen-activated protein kinase; JNK, c-Jun N-terminal kinase. (B) Mcp1, Ip10 and Rantes mRNA levels in eWAT from control mice fed a NCD or a HFD for 8 weeks (n = 8). Values are means + SD, *p<0.05 versus control mice. (C) Immunohistochemistry with anti-F4/80 and hematoxylin staining reveals macrophage accumulation in eWAT. Scale: 200 µm. (D) Expression of the M1 macrophage and M2 macrophage-specific genes (Cd11c, Cd11b, Tnfα and arginase1) in eWAT (n = 8). Values are means + SD, *p<0.05 versus control mice, n.s, not significant. (E) Percentage of proinflammatory M1 macrophages and anti-inflammatory M2 macrophages assessed by counting CD11c-PE-A- and CD206-FITC-stained cells in the SVF in eWAT of 14 weeks HFD feeding mice (n = 8). Values are means + SD, *p<0.05 versus control mice. MΦ, Macrophage. (F) Serum IL4 and TNFα levels in mice fed HFD, as measured by Multiplex (n = 8). Values are means + SD, *p<0.05 versus control mice, n.s, not significant. To observe inflammation in the adipose tissue of Crif1f/+,Fabp4 mice directly, the eWAT were stained with anti-F4/80, an antibody that detects macrophages. Increased F4/80 reactivity was observed in the eWAT of Crif1f/+,Fabp4 mice fed a NCD for 8 weeks. Aging and a HFD had an even more pronounced effect (Figure 5C). Based on the quantitative real-time PCR results, the relative expression of proinflammatory M1 macrophage markers (Cd11c, Cd11b and Tnfα) increased significantly; however, the relative gene expression of an anti-inflammatory M2 macrophage marker (arginase 1) did not change (Figure 5D). To quantify the number of macrophages in the adipose tissue, multi-parameter flow cytometry was performed with anti-F4/80, anti-CD11c and anti-CD206 antibodies using isolated stromal vascular fractions (SVF). F4/80+/CD11c+/CD206 - M1 macrophages were predominant in Crif1f/+,Fabp4 mice compared with control mice. The proportion of F4/80+/CD11c-/CD206+ M2 macrophages tended to be higher in Crif1f/+,Fabp4 mice, but this did not reach statistical significance (Figure 5E). Taken together, the results suggested that the infiltrating macrophages were skewed towards the M1 phenotype in Crif1f/+,Fabp4 mice. Recent studies show that B cell-mediated CD4+ and CD8+ T cell activation is required to induce inflammation and insulin resistance [31], [32]. The present study found no difference between the numbers of CD4+ and CD8+ T cells in Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice (data not shown).
Adipocytes in adipose tissue secretes adipokines, such as adiponectin, leptin, IL-6 and TNFα, which are involved in the control of whole-body insulin sensitivity. However, proinflammatory TNFα is released by dysfunctional adipocytes and amplifies local immune responses by recruiting macrophages to WAT [33], [34]. Serum levels of TNFα were consistently higher in Crif1f/+,Fabp4 mice fed a HFD than in control mice (Figure 5F). This indicates that TNFα may be a crucial mediator of inflammation in WAT and whole-body insulin resistance of Crif1f/+,Fabp4 mice.
The Fabp4 gene is expressed in macrophages [35], but no Cre expression or activity was detected in macrophages isolated from Crif1f/+,Fabp4 mice. As shown in Figure S8A, the expression levels of Crif1 and macrophage markers (Cd11c, Tnfα, Cd11b, and Arg1) were not reduced in peritoneal macrophages obtained from Crif1f/+,Fabp4 mice (Figure S8A). Homologous recombination using PCR [36] identified Cre recombinase activity in WAT and BAT, but not in peritoneal macrophages in Crif1f/+,Fabp4 mice at 20 weeks-of-age (Figure S8B and S8C).
Adipose inflammation and glucose intolerance in Crif1f/f,Adipoq mice
To verify the adipose inflammation characterized by macrophage infiltration in Crif1-null mice, we generated another adipose tissue-specific Crif1 knockout mouse by crossing floxed Crif1 mice with Adipoq-Cre transgenic mice. Adipoq-Cre transgenic mice expressed Cre recombinase in WAT and BAT, but not in macrophages (including adipose tissue resident macrophages, alveolar macrophages, or thioglycolate-stimulated peritoneal macrophages) [37].
The homozygous Crif1 knockout mice (Crif1f/f,Adipoq) showed about ∼30% of the Crif1 expression observed in the eWAT of controls (Figure S9A). They showed decreased expression of OXPHOS subunits (ND1, NDUFA9, UQCRC2 and COX4) in eWAT and BAT, not in liver and heart (Figure S9B). We compared adipocyte development in the adipocyte-specific Crif1 knockout mouse with that of Adipoq-Cre mice. H&E staining of adipose tissues indicated that the adipocytes of Crif1f/f,Adipoq mice were relatively smaller and irregularly shaped in comparison to those of Crif1+/+,Adipoq mice (data not shown).
Consistent with the Crif1f/+,Fabp4 mouse model, Crif1f/f,Adipoq mice showed higher plasma levels of MCP1 (1.9-fold higher), IP10 (2.5-fold higher) and marked F4/80 immuno-reactivities in eWAT, suggesting inflammation in WAT (Figure 6A and 6B). The nature of the macrophage phenotypes was further identified by flow cytometry using fluorescently-labeled anti-F4/80, anti-CD11c, and anti-CD206 antibodies. In addition, the T cell population was also analyzed using anti-CD3, anti-CD8, and anti-CD4 antibodies. Crif1f/f,Adipoq mice had a higher level of M1 macrophages and a lower level of M2 macrophages in eWAT compared to Crif1+/+,Adipoq mice (Figure 6C). The level of cytotoxic CD8-positive T cells was increased 5.5-fold and the level of CD4-positive helper T cells was decreased 0.5-fold in Crif1f/f,Adipoq in comparison to control mice; however, the levels of these cells in Crif1f/+,Fabp4 mice were not significantly different from control mice (Figure 6D). This reflect differences in the severity of the defect in the mitochondrial OXPHOS complex in Crif1f/f,Adipoq and Crif1f/+,Fabp4mice. Similar to Crif1f/+,Fabp4 mice, Crif1f/f,Adipoq mice developed glucose intolerance even in being fed a NCD, at 8 weeks-of-age (Figure 6E).
Fig. 6. Recruitment of macrophages and T cells to adipose tissue in Crif1f/f,Adipoq mice. 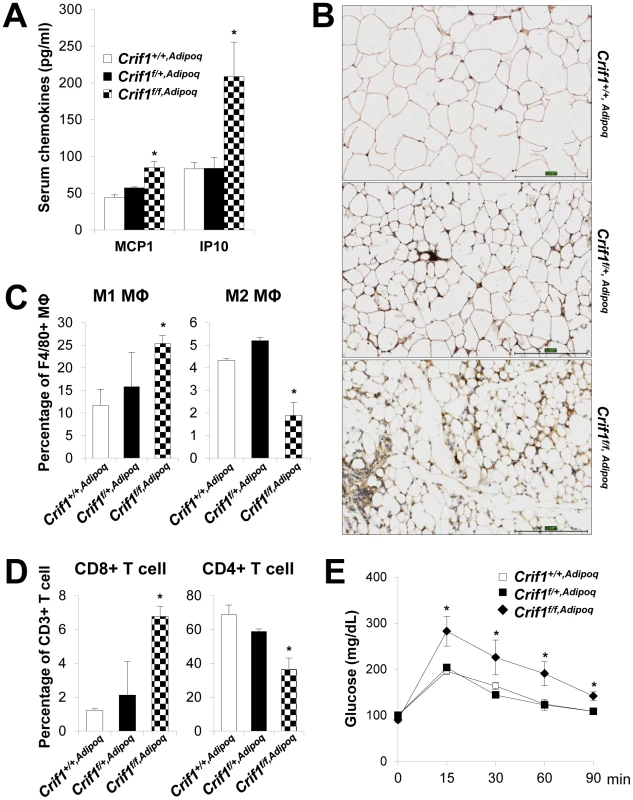
Control (Crif1+/+,Adipoq), adipose-specific Crif1 heterozygous (Crif1f/+,Adipoq), and homozygous (Crif1f/f,Adipoq) knockout mice were generated by cross-breeding with Adipoq-Cre recombinase transgenic mice. Phenotypic analysis all of these mice fed with a NCD was performed at 8 weeks-of-age. (A) Secreted MCP1 and IP10 levels in the serum in the three strains of mice (n = 6). Values are means + SD, *p<0.05 versus control mice. (B) Immunohistochemical staining with an anti-F4/80 antibody shows severe macrophage accumulation in eWAT of Crif1f/f,Adipoq mice. Scale: 200 µm. (C) Percentage of proinflammatory M1 macrophages (CD11c-PE-A-) and anti-inflammatory M2 macrophages (CD206-FITC-) (n = 6). Values are means + SD, *p<0.05. MΦ, Macrophage. (D) Percentage of CD8 positive T cells and CD4 positive T cells in the SVF in eWAT. Quantification of the CD8/CD4 ratio by counting cells stained with CD8-FITC and CD-PE-cy7 (n = 6). Values are means + SD, *p<0.05. (E) IPGTT was performed in NCD mice after a 16 h fast with 1 g/kg of glucose (n = 6). Values are means ± SD, *p<0.05 versus control. Unlike Crif1f/f,Fabp4 mice, Crif1f/f,Adipoq mice were viable. This discrepancy could be due to the inherent differences in the activities of Cre-recombinase driven by Fabp4 and adiponectin promoter (Table S1). Loss of Crif1 was consistently observed in WAT in both mouse lines; however, the degree of Crif1 loss was more severe in Crif1f/f,Fabp4 mice than in Crif1f/f,Adipoq mice. Also, Crif1f/f,Fabp4 mice exhibited a severe loss of BAT and WAT mass, whereas the mass of these tissues was only mildly reduced in Crif1f/f,Adipoq mice. Consistent with these findings, homozygous Crif1f/f,Fabp4 mice rapidly reached a fatal low rectal temperature of 22.6+1.9°C (Figure S2D), whereas homozygous Crif1f/f,Adipoq mice reached a milder rectal temperature of 29.5+0.7°C within 5 minutes of immersion in cold water (4°C) (Figure S9C). These results indicated that thermal stress caused by mitochondrial OXPHOS dysfunction in BAT following Crif1 ablation may be a causative factor of the premature death of adipocyte specific Crif1 knockout mice, and that BAT dysfunction may be partially involved in systemic glucose intolerance.
Reversal of insulin resistance in adipose-specific Crif1-deficient mice by clodronate treatment
To determine whether macrophages in the adipose tissue of Crif1f/+,Fabp4 mice play a role in insulin resistance, we depleted macrophages from adipose tissue by intraperitoneal treatment with clodronate liposomes [38]. Clodronate is an apoptosis-inducing drug; therefore, injection of liposome-encapsulated clodronate into the intraperitoneal cavity can deplete phagocytic cells, such as macrophages. Control and Crif1f/+,Fabp4 mice fed a HFD for 8 weeks were intraperitoneally administered two rounds of clodronate liposomes with an interval of 3 days. The accumulation of macrophages positively stained with an anti-F4/80 antibody was decreased in the eWAT following injection of clodronate liposomes (Figure 7A). Furthermore, the level of Cd68 mRNA was significantly lower in the eWAT of mice injected with clodronate than in untreated mice (Figure 7B). Administration of clodronate liposomes dramatically improved the insulin and glucose tolerance of Crif1f/+,Fabp4 mice fed a HFD for 10 weeks (Figure 7C and 7D). These findings indicate that suboptimal reserves of mitochondrial OXPHOS in the adipose tissue of Crif1-deficient mice induce macrophage recruitment, which may trigger systemic insulin resistance (Figure 8).
Fig. 7. Depletion of macrophages in adipose tissue by clodronate. 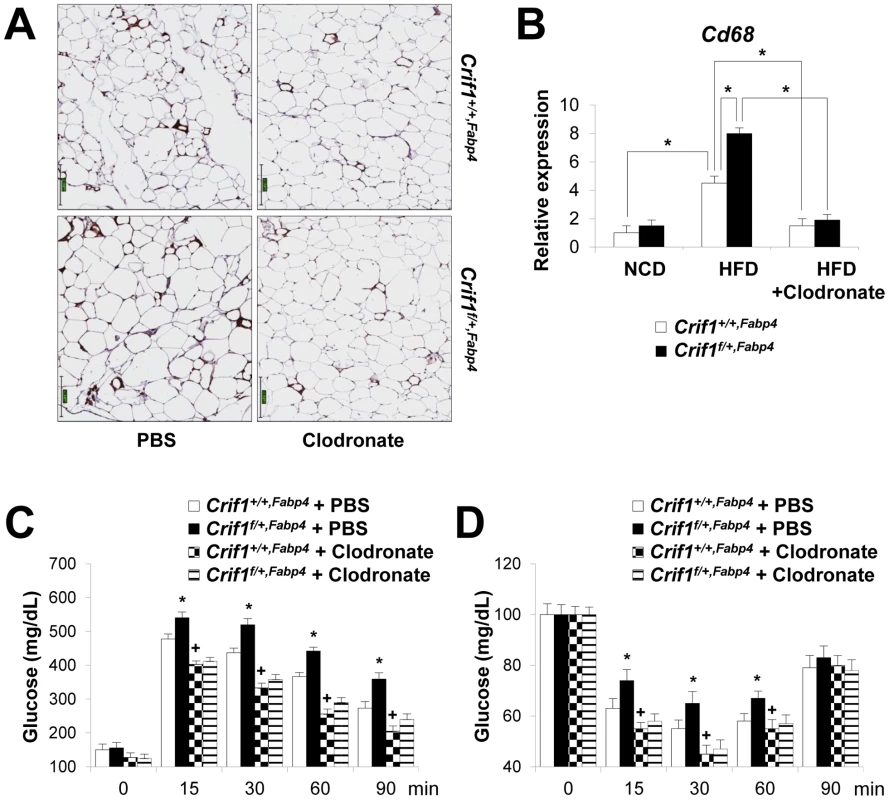
The clodronate study was performed after feeding Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice a HFD for 8 weeks. Two intraperitoneal injections of clodronate were given with a 3 day interval between each. IPGTT and ITT were performed 6 days after the first injection. (A) Immunohistochemistry with anti-F4/80 after macrophage depletion by liposomal clodronate in Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice fed HFD. Scale: 200 µm. (B) Real-time PCR using primers for the macrophage marker, Cd68 (n = 8). Values are means + SD, *p<0.05. (C and D) IPGTT and ITT after macrophage depletion by intraperitoneal injection of liposomal clodronate or PBS control (n = 8). Values are means + SD, *p<0.05, Crif1f/+,Fabp4 mice versus control mice; + p<0.05, clodronate versus PBS in control mice. Fig. 8. A model of the systemic insulin resistance developed by <i>Crif1</i>-haploinsufficient mice, which shows limited adipose OXPHOS capacity. 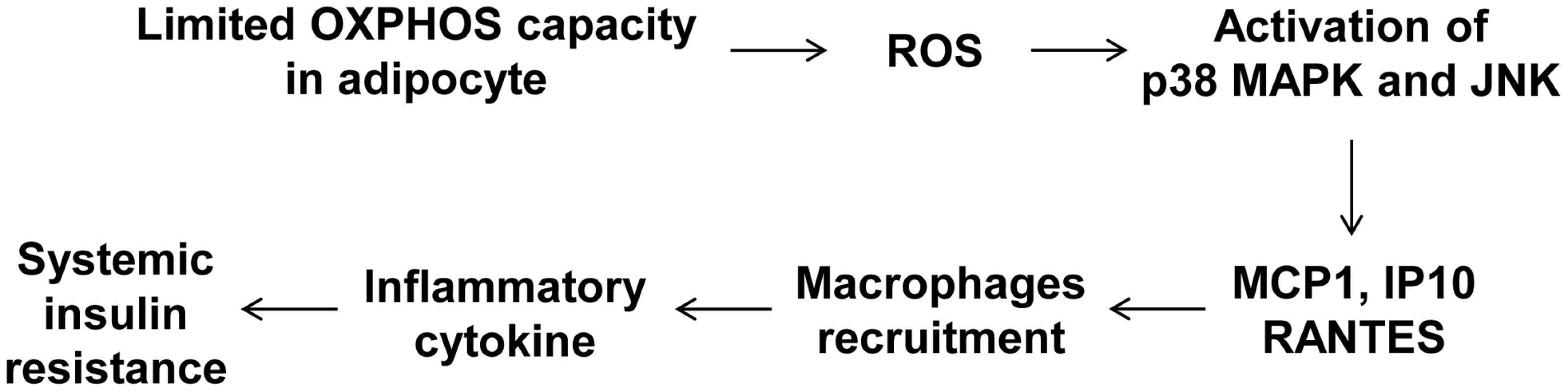
Discussion
Mitochondrial dysfunction, characterized by reduced OXPHOS function in liver and skeletal muscle, is thought to be one of the underlying causes of insulin resistance and type 2 diabetes (for a review, see [39], [40]). In addition, reduced hepatic OXPHOS function is closely related to hepatic lipid accumulation and insulin resistance [41]. Collectively, these studies provide evidence of a role for mitochondrial OXPHOS dysfunction in the development of human insulin resistance and type 2 diabetes. However, animal models of OXPHOS dysfunction in skeletal muscle and liver do not exhibit the human insulin resistance and type 2 diabetes phenotype [21], [42], [43]. The absence of insulin resistance in mice with homozygous or heterozygous Crif1 deletion in the liver (Crif1f/f,Alb, Albumin-Cre) or skeletal muscle (Crif1f/+,MLC, MLC-Cre) is in agreement with previous findings that hepatic and skeletal mitochondrial dysfunction does not cause insulin resistance (Figure S10). Therefore, whether or how mitochondrial OXPHOS contributes to the pathogenesis of insulin resistance remains to be resolved. It is reported that adipose OXPHOS capacity is controlled by both genetic and diet-induced obesity [10], [44], [45], which potentially contribute to adipose tissue dysfunction and exacerbation of insulin resistance. However, whether changes in adipose OXPHOS capacity are a cause or a consequence of complications associated with insulin resistance has not been clarified in vivo.
In this study, we have shown an association between limited mitochondrial OXPHOS capacity and adipose tissue inflammation and insulin resistance in a Crif1 haploinsufficiency animal model. Mitochondria play a key role in the differentiation and maturation of adipocytes. It is reported that marked mitochondrial biogenesis is observed during the adipocyte differentiation process in vitro. In fact, the concentration of mitochondrial proteins in differentiated 3T3-L1 adipocytes showed a 20 - or 30-fold increase compared with that in pre-adipocytes [14], [46]. Notably, chemical inhibition of respiratory chain function, for example by rotenone treatment, suppresses adipogenesis and induces changes in the expression levels of the key transcription factors, C/EBPα, PPARγ, and SREBP-1c [47]. However, the role played by mitochondria during adipogenesis has mostly been investigated in vitro by inhibiting or knocking down the genes encoding the OXPHOS complex. This study showed that homozygous Crif1 null mice generated by both Fabp4-Cre and Adipoq-Cre recombinase have defects in WAT and BAT development. These observations support that notion that intact OXPHOS function is critical for adipogenesis in vivo. By contrast, our own observations show that heterozygous Crif1 knockout mice do not have defects in adipogenesis and maturation under NCD and HFD conditions. This finding indicates that Crif1 haploinsufficiency and mildly reduced OXPHOS capacity do not cause the apparent failure of adipogenesis in WAT and BAT. Consistently, plasma and liver lipid levels were not increased in heterozygous Crif1 knockout mice, suggesting that the mice do not have the lipodystrophy phenotype. Furthermore, it is reported that insulin resistance in a mouse model of lipodystrophy was not relieved by controlling inflammation [48]. Therefore, insulin resistance in Crif1 haploinsufficient knockout mice may not be related to lipodystrophic changes.
Similar to WAT, the development of BAT was severely perturbed in homozygous Crif1-null mice (Crif1f/f,Fabp4), which may be a critical factor in the early mortality of these mice. By contrast, BAT development was normal in heterozygous Crif1f/+,Fabp4 mice, and the histology and ultrastructure of mitochondria were normal. Furthermore, core temperature responses to a cold environment suggest that Fabp4-Cre-driven haploinsufficiency of Crif1 may not affect the physiological function of BAT. Therefore, decreased BAT function and impaired energy expenditure may not be principal cause of the development of insulin resistance in Crif1-haploinsufficient mice.
The OXPHOS capacity in adipose tissue may be controlled by tissue-specific, genetic and environmental factors. In fact, it is well known that each cell type develops and maintains a specific OXPHOS capacity to satisfy its metabolic and energetic demands. In addition, individual OXPHOS capacity is genetically determined by specific tissues [49]. The cellular and genetic factors that control adipose-specific OXPHOS capacity are not fully understood. Therefore, white adipocyte responses to marginal or limited OXPHOS capacity in vitro and in vivo remain to be elucidated. In the present study, we characterized the enhanced secretory chemokine responses in Crif1-silenced mature adipocytes. Chemokine production in WAT is physiological, but enhanced production is linked to adipose inflammation, which is usually observed in cases of excessive adiposity (for a review, see [50]). Therefore, the earliest events that trigger the process of enhanced chemokine secretion are of great interest. Studies on signals that initiate adipose inflammation are mainly based on the model of ER homeostasis, lipolysis, and fatty acid signals in obese individuals (for a review, see [4]). It is not known how mitochondria or OXPHOS dysfunction modify the chemokine responses in WAT under physiological and pathological conditions. We found that abnormal increases in ROS production and activation of p38 and JNK were associated with increased expression of Mcp1, Ip10 and Rantes. The increase in ROS and the activation of p38 and JNK in adipose tissue are common denominators that respond to cellular stresses. Unexpectedly, haploinsufficient heterozygous Crif1f/+,Fabp4 mice and control mice exhibited similar levels of adipose Akt phosphorylation in response to insulin injection. This indicates that insulin signaling in adipose tissue may not be the principal cause of the systemic glucose intolerance of Crif1f/+,Fabp4 mice. Therefore, the relative importance of these factors (increased ROS, p38 and JNK activation) need to be addressed by suppressing or eliminating these events while studying abnormal chemokine responses and systemic insulin resistance.
We showed that the WAT in Crif1-deficient mice is predominantly infiltrated by macrophages, regardless of excessive adiposity. An increase in the number of adipose tissue macrophages (ATM) is a prominent feature associated with excessive adiposity [51], [52]. Increased expression of chemokines, especially MCP1, is responsible for recruiting macrophages into the WAT [30]. The ATM infiltration of epididymal fat pads in Crif1-deficient mice showed several characteristic features. First, it was present in fat pads with normal adiposity. This finding suggests that macrophage recruitment to adipose tissue caused by impaired OXPHOS capacity may also develop independently of excessive adiposity, but is accentuated in cases of increased adiposity. Mitochondrial OXPHOS dysfunction in the adipose tissue of Crif1f/+,Fabp4 mice fed a NCD for 8 weeks resulted in macrophage recruitment; however, the mice showed normal glucose tolerance. This suggests that a threshold level of macrophage recruitment or activation is required for the development of insulin resistance. Phenotypic analysis of ATM in Crif1-deficient mice demonstrated that the proportion of both M1 and M2 macrophages tended to be increased under NCD and HFD conditions. However, a phenotypic shift toward M1 macrophages was observed in the adipose tissue of Crif1-deficient mice. Thus, these features of macrophage recruitment in WAT were similar to those observed in a mouse model of diet-induced obesity [53].
Our data provide novel insights into the relationship between adipose inflammation and insulin resistance. This study supports the idea that adiposity overwhelms the genetically-determined OXPHOS capacity in adipose tissue, provoking an inflammatory response and insulin resistance. Therefore, it is possible that adipose mitochondrial OXPHOS capacity is an independent factor determining the risk of adipose inflammation and systemic insulin resistance in obese and even in non-obese subjects.
Materials and Methods
Cell culture
3T3-L1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine calf serum (Gibco BRL). Forty-eight hours post-confluence, the cells were differentiated with IBMX (0.5 mM), dexamethasone (1 µM), insulin (10 µg/ml) and 10% fetal bovine serum (Gibco BRL) [54]. Crif1 siRNA (GGA GUG CUC GCU UCC AGG AAC UAU U) was transfected by using Lipofectamine RNAiMAX reagent (Invitrogen) into 3T3-L1 adipocytes on day 4 of differentiation. Migration of Raw264.7 cells was examined in 8.0 µm Transwell filters (Corning Corp). Raw 264.7 cells were maintained on the top well, with the media from 3T3-L1 adipocytes in the bottom well. After twenty-four hours, the Raw 264.7 cells that had not migrated to the filter were removed, and the cells that had migrated through the filter were collected and stained with trypan-blue. ADSCs were cultured as previously described [55]. ADSCs were differentiated into adipocytes using IBMX (0.5 mM), dexamethasone (1 µM), insulin (10 µg/ml) and rosiglitazone (0.5 µM) in M199 medium (Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL). After induction of differentiation, lipid accumulation was detected with Oil red O staining. ADSCs were fixed with 10% neutralized formalin, washed with water, and then stained with freshly prepared 0.2% Oil red O solution.
Western blot analysis
Primary antibodies against OXPHOS complex subunits (NDUFA9, SDHA, UQCRC2, and ATP5A1) were purchased from Invitrogen. Anti-COX4 (#4844) antibody was purchased from Cell Signaling. Anti-ND1 antibody (sc-65237) was purchased from Santa Cruz Biotechnology. Secondary antibodies (goat anti-mouse and goat anti-rabbit) were obtained from Cell Signaling. Anti-p38 antibody, anti-phospho-p38 antibody, anti-JNK-antibody, anti-phospho-JNK antibody, anti-phospho-Akt and total-Akt antibodies were obtained from Cell Signaling and anti-β-actin, α-tubulin antibody was obtained from Sigma-Aldrich. Anti-UCP1 antibody was obtained from Abcam.
Northern blot analysis and real-time PCR
Total RNA was isolated using Trizol (Invitrogen). For Northern blot analysis, 10–20 µg of total RNA was loaded onto a 1.5% agarose-formaldehyde gel. A Crif1 probe was constructed using the mouse Crif1 gene digested with KpnI enzyme. The relative intensity of the Crif1/β-actin bands was normalized against that in the brain. Complementary DNA (cDNA) was prepared from total RNA using M-MLV Reverse Transcriptase and oligo-dT primers (Invitrogen). Real-time PCR was performed using cDNA, QuantiTect SYBR Green PCR Master Mix (QIAGEN), and specific primers. The primers used are described in Table S2. Relative expressions were calculated normalized with 18s ribosomal RNA, using Rotor-Gene 6000 real-time rotary analyzer Software (Version 1.7, Corbett Life Science).
Complementary DNA microarray analysis
Total RNA was prepared from fully-differentiated 3T3-L1 adipocytes transfected with control or Crif1 siRNA. RNA amplification and labeling were performed with the Low RNA Input Linear Amplification kit PLUS (Agilent Technologies). Array hybridization and scanning were performed with a DNA microarray Chip and scanner (Agilent Technologies). Array data was analyzed using the Feature Extraction and GeneSpring Software (Agilent Technologies).
ROS staining
Dihydroethidium (DHE) or MitoSOX were used to detect intracellular superoxide. Fully-differentiated 3T3-L1 cells were incubated with 10 µM DHE or 5 µM MitoSOX at 37°C for 15 min. Fully-differentiated 3T3-L1 cells were washed with Krebs-HEPES buffer (pH 7.4) or HBSS. Images of cells stained with DHE or MitoSOX were obtained by fluorescence microscopy (Olympus, Japan). Cells were trypsinized and analyzed using a FACScan flow cytometer (BD Bioscience) and data analysis was performed using BD FACSDiva software (BD Bioscience).
Blue native-polyacrylamide gel electrophoresis (BN-PAGE)
Before BN-PAGE, mitochondrial isolation was performed as previously described [56] with modifications. Pellets of ADSCs or tissues from mice were resuspended in buffer B (210 mM mannitol, 70 mM sucrose, 1 mM EGTA, and 5 mM HEPES, pH 7.2) and incubated for 5 min at 4°C. After centrifugation at 600× g for 10 min, the supernatant was re-centrifuged at 17,000× g for 10 min. The pellet containing the mitochondrial fraction was used in the Native PAGE Novex Bis-Tris Gel system (Invitrogen) to determine the content of the OXPHOS complex. A total of 20 µg of the mitochondrial fraction in Native PAGE sample buffer supplemented with 0.5% n-dodecyl-β-D-maltoside was loaded onto a Native PAGE Novex 3–12% Bis-Tris gel. The mitochondrial fraction was mixed with Native PAGE sample buffer containing 1% of digitonin to detect the supercomplexes. After electrophoresis, the separated proteins in the gel were transferred to a PVDF membrane, which was then incubated with an anti-OXPHOS antibody mixture (Invitrogen).
Oxygen consumption rate (OCR)
OCR was measured using a Seahorse XF-24 analyzer (Seahorse Bioscience). Control Crif1+/+ and Crif1+/− ADSCs were prepared from the eWAT of Crif1+/+,Fabp4 and Crif1f/+,Fabp4 mice. After seeding ADSCs on an XF-24 plate, cells were incubated in differentiation M199 media contained with FBS, IBMX, dexamethasone, insulin and rosiglitazone. After 2 days later, Crif1+/+ and Crif1+/− ADSCs maintained M199 media with insulin for 8 days. The day before OCR measurement, the sensor cartridge was calibrated with calibration buffer (Seahorse Bioscience) at 37°C. Fully-differentiated ADSCs were washed and incubated with M199 (Gibco BRL) without sodium bicarbonate at 37°C in an incubator. Three readings were taken after each addition of mitochondrial inhibitor before injection of the subsequent inhibitors. The mitochondrial inhibitors used were oligomycin (2 µg/ml), carbonyl cyanide m-chloro phenyl hydrazine (CCCP, 10 µM), and rotenone (1 µM). OCR was automatically calculated and recorded by the sensor cartridge and Seahorse XF-24 software. The plates were saved and the protein concentration was calculated to confirm that there were an approximately equal number of cells in each well.
Mice
Floxed Crif1 (Crif1flox/flox) mice were generated as previously described [18]. Fabp4-Cre, Albumin-Cre transgenic mice (C57BL/6J) were purchased from the Jackson Laboratory. Adiponectin-Cre transgenic mice were kindly provided by Dr. Evan Rosen. Dr. Steven J Burden provided the MLC-Cre mouse strain. The HFD, which contained 60% fat, was purchased from Research Diets Inc. (D12492). Mice were maintained in a controlled environment (12 h light/12 h dark cycle; humidity 50–60%; ambient temperature 23°C±1°C) and fed ad libitum. For the cold challenge experiments, mice were individually housed in cages pre-chilled to 4°C. Body temperature was monitored using a rectal probe attached to a digital thermometer (TD-300, Shibaura Denshi. Japan) with/without cold stress. For the thermoneutrality experiments, 2-week-old mice were housed with their mothers at a temperature of 30°C±1°C. All mouse experiments were performed in the animal facility according to institutional guidelines, and the experimental protocols were approved by the institutional review board of Korean Research Institute of Biotechnology and Bioscience, and Chungnam National University.
Genomic PCR
To measure the activity of Cre recombinase, PCR was performed as previously reported [36]. Briefly, after isolation of genomic DNA from WAT, BAT, and thioglycolate-induced peritoneal macrophages, PCR was performed with a combination of three primers: forward primer 1, GGGCTGGTGA AATGTGTTG; reverse primer 2, TCAGCTAGGG TGGGACAGA; and reverse primer 3, TATCAGTCCG AGAAGACCTG. To ensure product specificity from PCR, the extension time was limited to 30 sec.
Histological and morphometric analysis
WAT was fixed in 10% neutralized formalin for 16 h, washed, and then embedded in paraffin. Tissue sections of 5 µm thickness were deparaffinized, rehydrated, and heated in a microwave for 10 min in citrate buffer. The tissue sections were then incubated with primary antibodies (anti-F4/80 (diluted 1∶100; Abcam)) for 16 h at 4°C. Immunohistochemistry was performed using a Polink-1 HRP Rat-NM DAB Detection System (GBI Inc).
Transmission electron microscopy (TEM)
WAT and BAT were fixed in 1% glutaraldehyde at 4°C and then washed with 0.1 M cacodylate buffer at 4°C. After washing five times, the tissue was post-fixed with 1% OsO4 in an 0.1 M cacodylate buffer (pH 7.2) containing 0.1% CaCl2 for 1 h at 4°C. Samples were dehydrated by serial ethanol and propylene oxide treatment and embedded in Embed-812 (EMS). The resin was then polymerized at 60°C for 36 h. Tissue was sectioned using an EM UC6 ultramicrotome (LEICA) and stained with 4% uranyl acetate and citrate. Observation was performed using a Tecnai G2 Spirit Twin transmission electron microscope (FEI Company, USA) and a JEM ARM 1300S high-voltage electron microscope (JEOL, Japan).
Intraperitoneal glucose tolerance (IPGTT) and insulin tolerance tests (ITT)
For IPGTT, mice were fasted for 16 h and then 2 g/kg or 1 g/kg glucose was injected into the intraperitoneal cavity of each mouse. Blood glucose levels were measured at 0, 15, 30, 60, and 90 min using a glucometer (Bayer breeze). ITT was performed by measuring blood glucose after 6 h of fasting followed by intraperitoneal insulin injection (0.75 U/kg; Humalog).
Hyperinsulinemic euglycemic clamping
Hyperinsulinemic euglycemic clamping was performed as previously described [57]. Briefly, after an overnight fast, a 2 h hyperinsulinemic euglycemic clamping was performed in Crif1f/+,Fabp4 and control littermates (n = 8). The insulin clamp began with a primed-continuous infusion of insulin (0.3 U/kg bolus followed by 2.5 mU/kg/min). Blood samples (20 µl) were collected at 10 to 20 min intervals for immediate measurement of plasma glucose concentrations, and 20% glucose was infused at variable rates to maintain glucose at basal concentrations (∼120 mg/dL). Basal and insulin-stimulated whole-body glucose uptake was estimated with a continuous infusion of 3H glucose (Perkin Elmer Life and Analytical Sciences) for 2 h before clamping (0.05 µCi/min) and throughout the clamping (0.1 µCi/min), respectively. At 75 min after the start of the clamp, 2-deoxy-d-1-14C glucose (PerkinElmer Life and Analytical Sciences) was injected with a Hamilton syringe to measure insulin-stimulated glucose transport activity and metabolism in skeletal muscle. Blood samples were taken before, during, and at the end of the clamps for measurement of plasma 3H glucose and 2-deoxy-d-1-14C glucose concentrations, and/or insulin concentrations. At the end of the clamps, tissue samples (gastrocnemius, eWAT, and liver) were rapidly taken and stored at −70°C prior to biochemical and molecular analysis.
Flow cytometry
To quantified M1 macrophages, M2 macrophages, and CD4+ and CD8+ T cell populations, the stromal vascular fractions (SVF) was isolated from mouse eWAT. The SVF was prepared by the lysis of eWAT with type 1 collagenase (Gibco BRL) in collagenase buffer at 37°C in a shaking water bath for 40 min, followed by centrifuging at 2000 rpm for 5 min. The suspended solid matter comprised adipocytes and the cell pellet comprised T cells, B cells and macrophages. The cell pellet was then incubated with RBC lysis buffer and the remaining cells were stained with specific antibodies. Anti-CD3 (BD bioscience), anti-CD4 (BD Bioscience) and anti-CD8 (eBioscience) were used to stain the T cell population [58], and F4/80 (eBioscience), CD206 (eBioscience) and CD11c (eBioscience) were used to stain M1/M2 macrophages. The stained SVF cells were analyzed using a FACScan flow cytometer (BD Bioscience) and data analysis was performed using BD FACSDiva software (BD Bioscience).
Thiobarbituric acid reactive substance (TBAR) assay
The TBAR assay kit (Cayman Chemicals) was used to measure lipid peroxidation in the WAT and plasma of mice. WAT (25 mg) suspended in RIPA buffer was sonicated, centrifuged at 1,600 g for 10 min at 4°C, and the supernatant was collected. The SDS solution was added to the supernatant, which was then mixed with the Color reagent according to the manufacturer's instructions. The sample was boiled for 1 h, centrifuged, and the supernatant was collected. Fluorescence at the excitation wavelength of 530 nm and emission wavelength of 550 nm was measured.
Macrophage depletion by clodronate
The generation of liposome-encapsulated clodronate was performed as previously described [38]. Cholesterol (10 mg/ml; Sigma-Aldrich) was dissolved in 100% ethanol, and 100 mg/ml phosphatidylcholine in 100% ethanol (Sigma-Aldrich) was made into a phospho-lipid film by drying with a low-vacuum rotary. Clodronate (0.6 M) (Sigma-Aldrich) was dissolved in purified water and incubated with the phospho-lipid film by gentle rotation at room temperature and sonication in a water bath for 3 min at 55 kHz. After removing the non-encapsulated clodronate, liposome-encapsulated clodronate was resuspended in 1X PBS. Two intraperitoneal injections (3 days apart) of clodronate were administered to mice fed a HFD for 8 weeks. IPGTT and ITT were performed 6 days after the first injection.
Analysis of triglyceride, ceramide, and saturated fatty acids in tissues
Measurement of hepatic triglycerides: Liver triglycerides were extracted with chloroform and methanol, dissolved in 1× PBS, and measured in a Hitachi 7150 chemistry analyzer (Hitachi, Japan).
Measurement of ceramides in WAT, liver and muscle: Prior to extraction of total lipids, C17 ceramide was added as an internal standard. Ceramides were measured as previously described [59]. All liquid chromatography-mass spectrometry (LC-MS/MS) experiments were performed using an Agilent 1200 HPLC system (Agilent Technologies, Santa Clara, CA, USA) coupled to a Thermo LTQ linear ion trap mass spectrometer (Thermo Scientific, San Jose, CA) equipped with an electro spray ionization (ESI) source. Briefly, LC separation was achieved using a LunaC18 RP column (150 mm×2 mm I.D., 5 µm 100 Å particles; Phenomenex, Torrance, CA) with gradient elution. Lipid molecules separated by LC were detected by the mass spectrometer in Positive ESI mode using selected reaction monitoring (SRM). The SRM channels were arranged as follows: 538→264 for C16, 552→264 for C17, 566→264 for C18, 594→264 for C20, 648→264 for C24 : 1, and 650→264 for C24. The peak area was normalized according to the internal standard and tissue weight. All values are presented as relative differences in the ratio of the extracted lipids to the internal standard.
To measure the level of saturated fatty acids, tissues were homogenized in ice-cold methanol, and 1 µg of pentadecanoic acid (C15:0) was added as an internal standard. Samples were incubated at 45°C overnight, then cooled to room temperature. Hexane and 1 mL of H2O were added, samples were vortexed and centrifuged, and fatty acid methyl esters were collected from the upper hexane layer. Samples were analyzed by gas chromatography–mass spectrometry (GC-MS) using an Agilent HP6890 GC interfaced with an HP5973N MSD. A DB-5 column was used. The GC oven temperature was initially 150°C and then increased to 280°C for 52 min. Full MS scans over a m/z range of 60 to 800 were obtained and the peaks of the characteristic ion chromatogram for each fatty acid methyl ester were used for quantification. All samples were normalized against the internal standard.
Serum analysis
Whole cardiac blood from the mice was incubated at room temperature for 2 h. The blood was centrifuged at 2,500 rpm for 5 min, and the supernatant was collected. TNFα and IL-4 were measured using a mouse cytokine/chemokine multiplex panel (Millipore). MCP1 and IP10 were measured using an ELISA kit (R&D Systems). Serum triglycerides and FFAs were measured with a Hitachi 7150 chemistry analyzer (Hitachi, Japan).
Statistical analyses
Data are presented as means ± or + standard deviation (SD). Statistical significance for comparisons was determined using the Student's two-tailed T-test. A p value less than 0.05 was considered statistically significant.
Supporting Information
Zdroje
1. RosenED, SpiegelmanBM (2006) Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444 (7121)
847–53.
2. FlierJS (2004) Obesity wars: molecular progress confronts an expanding epidemic. Cell 116 (2)
337–50.
3. KahnBB, FlierJS (2000) Obesity and insulin resistance. J Clin Invest 106 (4)
473–81.
4. HotamisligilGS (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140 (6)
900–17.
5. KusminskiCM, SchererPE (2012) Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab 23 (9)
435–43.
6. PattiME, CorveraS (2010) The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev 31 (3)
364–95.
7. TormosKV, et al. (2011) Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab 14 (4)
537–44.
8. ShiX, et al. (2008) Paradoxical effect of mitochondrial respiratory chain impairment on insulin signaling and glucose transport in adipose cells. J Biol Chem 283 (45)
30658–67.
9. KohEH, et al. (2007) Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes 56 (12)
2973–81.
10. Wilson-FritchL, et al. (2004) Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest 114 (9)
1281–9.
11. RongJX, et al. (2007) Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes 56 (7)
1751–60.
12. KaamanM, et al. (2007) Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia 50 (12)
2526–33.
13. DahlmanI, et al. (2006) Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-alpha. Diabetes 55 (6)
1792–9.
14. Wilson-FritchL, et al. (2003) Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol 23 (3)
1085–94.
15. BogackaI, et al. (2005) Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 54 (5)
1392–9.
16. TedescoL, et al. (2008) Cannabinoid type 1 receptor blockade promotes mitochondrial biogenesis through endothelial nitric oxide synthase expression in white adipocytes. Diabetes 57 (8)
2028–36.
17. KimSJ, et al. (2012) CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab 16 (2)
274–83.
18. KwonMC, et al. (2008) Crif1 is a novel transcriptional coactivator of STAT3. EMBO J 27 (4)
642–53.
19. UrsS, et al. (2006) Selective expression of an aP2/Fatty Acid Binding Protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res 15 (5)
647–53.
20. XueB, et al. (2007) Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. J Lipid Res 48 (1)
41–51.
21. PospisilikJA, et al. (2007) Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131 (3)
476–91.
22. RochaVZ, Libby (2009) Obesity, inflammation, and atherosclerosis. Nat Rev Cardiol 6 (6)
399–409.
23. SunK, KusminskiCM, SchererPE (2011) Adipose tissue remodeling and obesity. J Clin Invest 121 (6)
2094–101.
24. GaoYJ, et al. (2009) JNK-induced MCP-1 production in spinal cord astrocytes contributes to central sensitization and neuropathic pain. J Neurosci 29 (13)
4096–108.
25. ShenQ, ZhangR, BhatNR (2006) MAP kinase regulation of IP10/CXCL10 chemokine gene expression in microglial cells. Brain Research 1086 : 9–16.
26. ShoelsonSE, LeeJ, GoldfineAB (2006) Inflammation and insulin resistance. J Clin Invest 116 (7)
1793–801.
27. ChenCL, et al. (2008) Ceramide induces p38 MAPK and JNK activation through a mechanism involving a thioredoxin-interacting protein-mediated pathway. Blood 111 (8)
4365–74.
28. OlefskyJM, GlassCK (2010) Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 72 : 219–46.
29. SamuelVT, ShulmanGI (2012) Mechanisms for insulin resistance: common threads and missing links. Cell 148 (5)
852–71.
30. KandaH, et al. (2006) MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest 116 (6)
1494–505.
31. LumengCN, MaillardI, SaltielAR (2009) T-ing up inflammation in fat. Nat Med 15 (8)
846–7.
32. NishimuraS, et al. (2009) CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med 15 (8)
914–20.
33. HotamisligilGS, et al. (1995) Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest 95 (5)
2409–15.
34. SchenkS, SaberiM, OlefskyJM (2008) Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest 118 (9)
2992–3002.
35. MakowskiL, et al. (2001) Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med 7 (6)
699–705.
36. GareusR, et al. (2008) Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metab 8 (5)
372–83.
37. EguchiJ, et al. (2011) Transcriptional control of adipose lipid handling by IRF4. Cell Metab 13 (3)
249–59.
38. van RooijenN, HendrikxE (2010) Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol Biol 605 : 189–203.
39. LowellBB, ShulmanGI (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307 (5708)
384–7.
40. MorinoK, PetersenKF, ShulmanGI (2006) Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes 55 Suppl 2: S9–S15.
41. SchmidAI, et al. (2011) Liver ATP synthesis is lower and relates to insulin sensitivity in patients with type 2 diabetes. Diabetes Care 34 (2)
448–53.
42. FreyerC, LarssonNG (2007) Is energy deficiency good in moderation?. Cell 131 (3)
448–50.
43. WredenbergA, et al. (2006) Respiratory chain dysfunction in skeletal muscle does not cause insulin resistance. Biochem Biophys Res Commun 350 (1)
202–7.
44. CrunkhornS, et al. (2007) Peroxisome proliferator activator receptor gamma coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem 282 (21)
15439–50.
45. SutherlandLN, et al. (2008) Time course of high-fat diet-induced reductions in adipose tissue mitochondrial proteins: potential mechanisms and the relationship to glucose intolerance. Am J Physiol Endocrinol Metab 295 (5)
E1076–83.
46. NewtonBW, et al. (2011) Proteomic analysis of 3T3-L1 adipocyte mitochondria during differentiation and enlargement. J Proteome Res 10 (10)
4692–702.
47. LuRH, et al. (2010) Mitochondrial development and the influence of its dysfunction during rat adipocyte differentiation. Mol Biol Rep 37 (5)
2173–82.
48. HerreroL, et al. (2010) Inflammation and adipose tissue macrophages in lipodystrophic mice. Proc Natl Acad Sci U S A 107 (1)
240–5.
49. PetersenKF, et al. (2004) Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350 (7)
664–71.
50. OsbornO, OlefskyJM (2012) The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med 18 (3)
363–74.
51. XuH, et al. (2003) Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 112 (12)
1821–30.
52. WeisbergSP, et al. (2003) Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112 (12)
1796–808.
53. LumengCN, BodzinJL, SaltielAR (2007) Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 117 (1)
175–84.
54. LeeKW, et al. (2011) Effects of Sulfonylureas on Peroxisome Proliferator-Activated Receptor gamma Activity and on Glucose Uptake by Thiazolidinediones. Diabetes Metab J 35 (4)
340–7.
55. EstesBT, et al. (2010) Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat Protoc 5 (7)
1294–311.
56. BoldoghIR, PonLA (2007) Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol 80 : 45–64.
57. ParkSY, et al. (2005) Cardiac-specific overexpression of peroxisome proliferator-activated receptor-alpha causes insulin resistance in heart and liver. Diabetes 54 (9)
2514–24.
58. KangSM, et al. (2008) CD4+CD25+ regulatory T cells selectively diminish systemic autoreactivity in arthritic K/BxN mice. Mol Cells 25 (1)
64–9.
59. KasumovT, et al. (2010) Quantification of ceramide species in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Anal Biochem 401 (1)
154–61.
Štítky
Genetika Reprodukčná medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



