-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
Genetic studies often collect family histories from diagnosed individuals. Some diseases exhibit inter-lineage asymmetry: mothers and their progenitors have higher (or lower) risk than fathers and their progenitors, and descendants of female cases have higher (or lower) risk than descendants of male cases. We describe how certain non-standard genetic mechanisms might underlie that asymmetry and make substantial contributions to disease susceptibility. Besides variants on sex chromosomes, these mechanisms include variants in the mother's genome that influence fetal development and hence later risk, variants in the mitochondria that modulate risk, and susceptibility variants in particular inherited genes whose expression depends on whether the variant came from the mother or the father. Applying our ideas to a study of more than 30,000 families with breast cancer, we found that more maternal grandmothers of cases than paternal grandmothers of cases had breast cancer, giving evidence that such non-standard mechanisms may be important contributors to breast cancer risk.
Published in the journal: Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer. PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004174
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004174Summary
Genetic studies often collect family histories from diagnosed individuals. Some diseases exhibit inter-lineage asymmetry: mothers and their progenitors have higher (or lower) risk than fathers and their progenitors, and descendants of female cases have higher (or lower) risk than descendants of male cases. We describe how certain non-standard genetic mechanisms might underlie that asymmetry and make substantial contributions to disease susceptibility. Besides variants on sex chromosomes, these mechanisms include variants in the mother's genome that influence fetal development and hence later risk, variants in the mitochondria that modulate risk, and susceptibility variants in particular inherited genes whose expression depends on whether the variant came from the mother or the father. Applying our ideas to a study of more than 30,000 families with breast cancer, we found that more maternal grandmothers of cases than paternal grandmothers of cases had breast cancer, giving evidence that such non-standard mechanisms may be important contributors to breast cancer risk.
Introduction
Genome-wide association studies (GWASs) that compare affected individuals and controls have identified many inherited genetic variants associated with complex diseases [1]. Nevertheless, effects of single nucleotide polymorphisms (SNPs) tend to be small [2] and much of the heritability for major diseases remains unexplained. For example, the most important GWAS-derived SNPs for breast cancer explain little of the risk [3].
Four other genetic mechanisms (henceforth referred to as “nonstandard”) are overlooked in a typical GWAS. Sex-linked genetic variants on the Y or the X chromosome are often not considered, although polymorphic X loci may be relevant to breast cancer [4].
The mother's genome can also exert effects on the developing fetus, of consequence for both birth outcomes and adult phenotypes [5], [6], [7]. Such maternally-mediated prenatal effects remain unexplored for breast cancer, though a prenatal influence on adult risk is suggested by the fact that birth weight is a risk factor [8]. A third mechanism involves variants in mitochondrial DNA, as reported for breast cancer in African-American women [9]. Finally, parent-of-origin effects (e.g. due to imprinted polymorphic autosomal genes) can also influence risk, as exemplified by a report based on Icelandic families [10], where the effect of an allele related to breast cancer differed depending on whether its origin was maternal or paternal.
Each of these nonstandard mechanisms produces asymmetry in family history data. We define inter-lineage asymmetry as the presence of a higher (or lower) risk either for the mother and her progenitors compared to the father and his progenitors, or for descendants of female cases compared to descendants of male cases. Although Table 1 includes sex-linked effects, we will not consider them further here. A maternally-mediated prenatal effect should produce increased risk in an affected individual's mother's (but not father's) progenitors, in a pattern where risk diminishes toward earlier generations. By contrast, a parent-of-origin effect could show a diminishing pattern of increased risk in the affected individual's father's progenitors if only the paternally inherited copy is expressed. The action of these understudied mechanisms can be discerned by studying family histories.
Tab. 1. Qualitative asymmetries produced by non-autosomal genetic mechanisms. 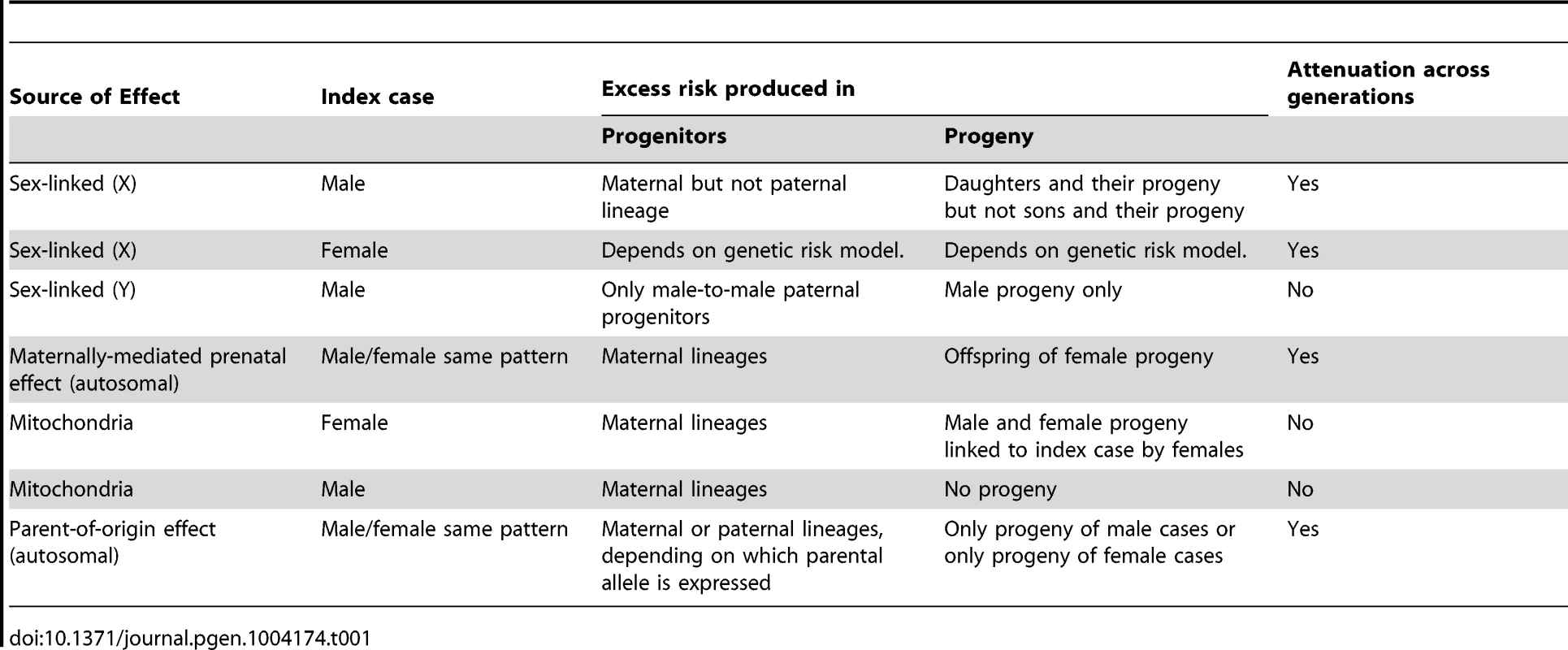
The presence of disease in a proband statistically induces enrichment in their progenitors and progeny for risk-related alleles. For the mechanisms in Table 1, that enrichment manifests as inter-lineage asymmetry. To quantify that asymmetry, we define several inter-lineage relative risks, whose exact definitions (and magnitudes) depend on the familial relationship to the affected proband of the individuals whose risks are being compared. We denote mother, father and child by M, F, and C, and extend the same notation for progenitors, e.g., as MM for the mother's mother and MMF for the mother's mother's father. C will denote the grandchild when considering parents of parents. Let , , denote the events that the child, mother, father has the disease, respectively, with analogous notation for other relatives. Let , denote the events that a female (girl), male (boy) in the population has the disease, respectively. We can compare risk in the proband's grandparental generation directly by comparing two sex-matched grandparents, either grandmothers (MM vs. FM as ) or grandfathers (MF vs. FF as ), without normalizing to the population. By contrast, assessing asymmetry in risk between mothers and fathers of affected individuals requires normalizing those risks to risk for females versus males in the general population. Thus, the inter-lineage parent relative risk is the risk for the proband's mother compared to that for females in the population divided by the risk for the proband's father compared to that for males in the population, expressed symbolically as .
Investigators have also looked prospectively for asymmetry, by comparing risk in the offspring of male versus female affected individuals [11]. The inter-lineage son (daughter) relative risk is the risk for sons (daughters) of affected mothers divided by the risk for sons (daughters) of affected fathers. Let and denote the events that a son or daughter, respectively, has the disease. We express the inter-lineage relative risk for sons as and that for daughters as .
Epidemiologic studies sometimes assemble very large case-control samples or cohorts at elevated risk [12], [13] and ascertain extensive family history data for affected families, enabling powerful comparisons of disease rates for maternal versus paternal lineages. Although studies related to birth defects have made use of multigenerational data [11] [14], [15], the huge consortia assembled for case-control and cohort studies of diseases like cancer [16] have thus far not probed for these less accessible genetic mechanisms.
The NIEHS Sister Study enrolled 50,884 women who each had a sister diagnosed with breast cancer. We here use data from that cohort to compare rates of breast cancer in maternal versus paternal grandmothers. We develop general results to relate the inter-lineage relative risks in progenitors to the inter-lineage relative risks in descendants. Under simplifying assumptions, we calculate how large a maternally-mediated prenatal effect or a parent-of-origin effect would have to be to explain any particular inter-lineage asymmetry and conclude that those causative relative risks would have to be substantial to produce the inter-lineage asymmetry evident in the Sister Study.
Materials and Methods
We analyzed family history data for a large number of cases of breast cancer, to compare the risk of breast cancer in the maternal versus the paternal grandmother. The Sister Study [17] enrolled 50,884 women aged 35 to 74 in the United States and Puerto Rico between 2004 and 2009; each had a sister diagnosed with breast cancer. The unaffected sisters are being followed for newly incident breast cancer and other conditions. Participants completed a detailed family history questionnaire and were old enough that the data effectively encompass their grandmothers' lifetime risks. The Sister Study secured informed consent and was carried out with human-subjects approval and oversight from the NIEHS Institutional Review Board and the Copernicus Group Institutional Review Board.
To assess asymmetry, we calculated odds ratios (which approximate the relative risks for maternal versus paternal grandmothers) using families where exactly one of the two grandmothers had breast cancer, i.e., discordant grandmother pairs.
Using algebra we then derived formulae to assess the likely strength of the mechanisms that could underlie an observed asymmetry in family history. Foreseeing applications beyond breast cancer, we similarly derived expressions to assess the degree of asymmetry that those nonstandard mechanisms would produce in maternal versus paternal progenitors, and the degree of asymmetry produced in the offspring of affected male versus female individuals.
Results
The Sister Study
Our analysis used 32,929 women, each from a distinct family, where each woman was the full sister of a case and could report breast cancer history for both grandmothers. Of these grandmothers, 3046 on the maternal side (9%) and 2639 (8%) on the paternal side had developed breast cancer. These reported rates are in general agreement with expected rates for their birth cohort [18]. However, presumably reflecting heritability, the probability that at least one of the two grandmothers had developed breast cancer decreased as a function of the youngest age at diagnosis of a sister in the participating family (Figure 1, Table 2).
Fig. 1. Risk of breast cancer in either grandmother as related to the youngest age at diagnosis of a granddaughter in the family studied (data taken from <em class="ref">Table 1</em>). 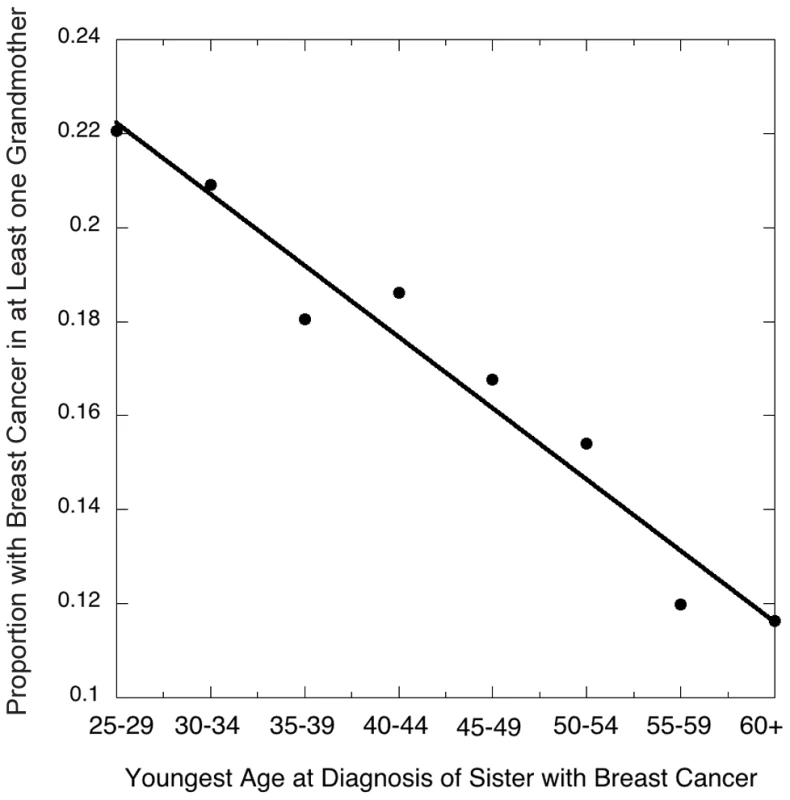
Tab. 2. Grandmothers' breast cancer history by age at breast cancer diagnosis of the youngest-onset grand-daughter. 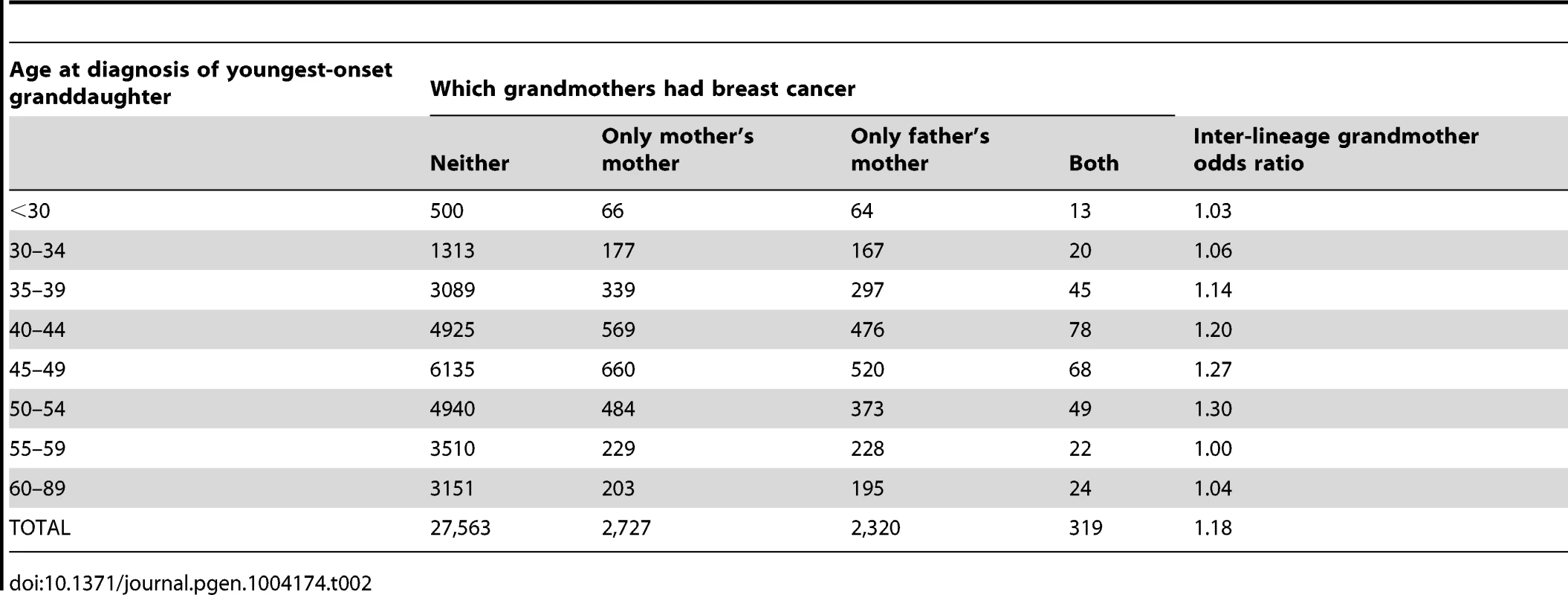
Figure 2 shows the inter-lineage odds ratios (which approximate the relative risks, ) for maternal versus paternal grandmothers, using families where exactly one of the two grandmothers had breast cancer, i.e., discordant grandmother pairs. That is, we calculated the ratio of maternal to paternal grandmothers among the discordant pairs. The estimated overall odds ratio for a positive family history on the maternal versus paternal side was 1.18 (95% CI 1.11, 1.24, p<0.0001).
Fig. 2. Grandmothers' odds ratio (maternal versus paternal) in the Sister Study as a function of youngest age at diagnosis of a granddaughter in the family studied. 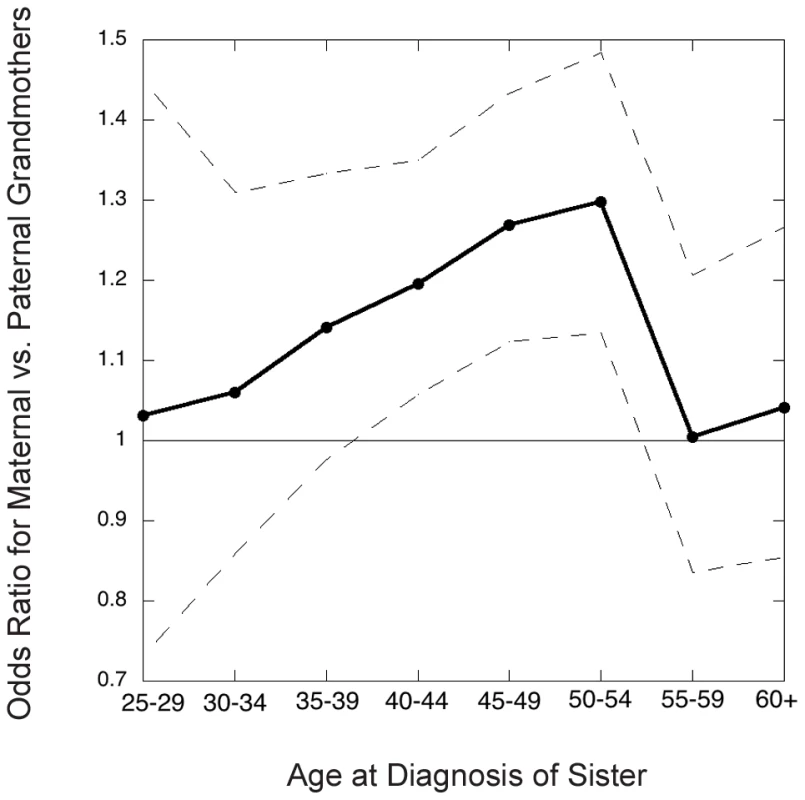
Dots connected by solid line segments are estimated odds ratios (approximately the relative risks); dashed lines connect 95% point-wise confidence limits. The inter-lineage odds ratio depended on the granddaughter's age at diagnosis (Figure 2), being most pronounced for cancers diagnosed in the age decade 45–54, i.e. in the perimenopausal years, and much less pronounced for later-diagnosed or very young onset cancers. We next wanted to relate the magnitude of the excess in maternal grandmothers to possible nonstandard genetic mechanisms.
Quantifying Mechanisms That Can Underlie Asymmetry in Family History
An analytic result related to asymmetry in family histories
The degree of asymmetry in progenitors of affected individuals and the degree of asymmetry in offspring of affected individuals are closely related. Assume that any negative effect the disease has on reproductive success is equivalent in males and females and also that the relative risk for mothers versus fathers is the same as that for women versus men, and the risk for maternal grandmothers (grandfathers) is the same as that for paternal grandmothers (grandfathers). Straightforward manipulation of conditional probabilities proves the following result (Text S1).
Result 1: Under the above-stated assumptions, a) the inter-lineage relative risk for mothers with an affected offspring versus fathers with an affected offspring is the same as the inter-lineage relative risk for offspring with an affected mother versus those with an affected father, that is,
b), the inter-lineage relative risk for maternal grandmothers (grandfathers) with an affected grandchild versus paternal grandmothers (grandfathers) with an affected grandchild is the same as the inter-lineage relative risk for grandchildren with an affected maternal grandmother (grandfather) versus those with an affected paternal grandmother (grandfather). That is,
c) Similarly, provided the inter-lineage relative risk for sons equals that for daughters, the inter-lineage son (daughter) relative risk is the same as the inter-lineage parental relative risk for mothers versus fathers of affected offspring. That is,
Note that Result 1 refers to a particular offspring or grandchild (for example, first-born or randomly selected) and thus the result does not apply to families selected because one out of some large number of progeny developed the disease.
Calculating genotype distributions across generations
To quantify inter-lineage relative risks, we adopt some additional simplifying assumptions: a rare outcome, random mating (relative to the di-allelic locus under study), Mendelian inheritance, Hardy-Weinberg equilibrium (HWE), and that the locus is not in linkage disequilibrium with and does not interact with another risk-related locus.
Let P be the 1×3 row vector containing an individual's autosomal genotype probabilities for minor allele counts 0, 1, 2 for a particular locus. Under Hardy-Weinberg Equilibrium (HWE), for a randomly sampled individual, where is the minor allele frequency. Now suppose sampling is instead based on the disease status of a designated relative, the proband case.
We need to introduce the concept of “risk relevant” genotypes. For maternally-mediated prenatal effects, those where the maternal genome affects the offspring's risk through prenatal influences on fetal development, the risk-relevant genotype at that locus is that of the affected proband's mother; and for autosomal genes with parent-of-origin effects the risk-relevant genotypes are those of both the proband and the proband's parents. Those risk-relevant genotypes will be distorted away from HWE for a locus related to risk. Other family members who are the progenitors or descendants of risk-relevant individuals are not risk-relevant to the proband, in that their genotypes at that locus are unrelated to the proband's risk conditional on the genotypes of risk-relevant individuals. However, their genotype distributions may still be distorted away from HWE, due to their relationship with the proband. The following result, proven in Text S2 (Table S1), facilitates calculation of autosomal genotype distributions for individuals who are not risk-relevant in this sense. We define the “index person” as the person whose parent's or whose offspring's genotype distribution is desired.
Result 2: Assume that the population meets the assumptions stated earlier. Let be the genotype distribution for an index person's parent (either mother or father), be the genotype distribution for an index person's offspring, and be the genotype distribution for the index person. Let be the 3×3 matrix
If is not risk-relevant to the proband, then . Similarly, if P+ is not risk-relevant to the proband, then .
The elements of the matrix V are the conditional probabilities for the offspring or parent to have a certain allele count, conditional on the index person having a certain allele count. Note that Result 2 holds under our assumptions even if the locus under study is not related to risk. This result allows the extrapolation of enrichment of the risk allele back to progenitors and forward to descendants.
Maternal Effects
Suppose a genetic variant carried by the mother has a prenatal effect on fetal development, hence on later risk to her offspring. We quantify the asymmetry induced by such a maternal effect (Text S3).
Let the relative risk for the offspring of a mother with one (two) copies of the variant allele be relative to the risk for the offspring of a mother with no copies. For simplicity we consider an outcome where gender itself does not influence risk and take (log-additive risk model). Then mothers of affected offspring are enriched for the risk allele and so are their mothers, that is the maternal grandmothers. Consequently, the mothers themselves have greater risk than fathers whenever (Figure 3). Note that both causative and protective maternal effects produce increased risk in the maternal lineage, reflecting the fact that whichever of the two alleles confers higher risk will tend to be over-represented in the maternal lineage of a proband. Also, note that a given small inter-lineage relative risk is induced by a more extreme maternally-mediated relative risk .
Fig. 3. Progenitors relative risk (mothers versus fathers or maternal grandmothers versus paternal grandmothers) as a function of maternally mediated relative risk () under a log-additive risk model (), or the imprinting relative risk, I, for allele frequency 0.2 for a locus for which only a specific parental copy is expressed. 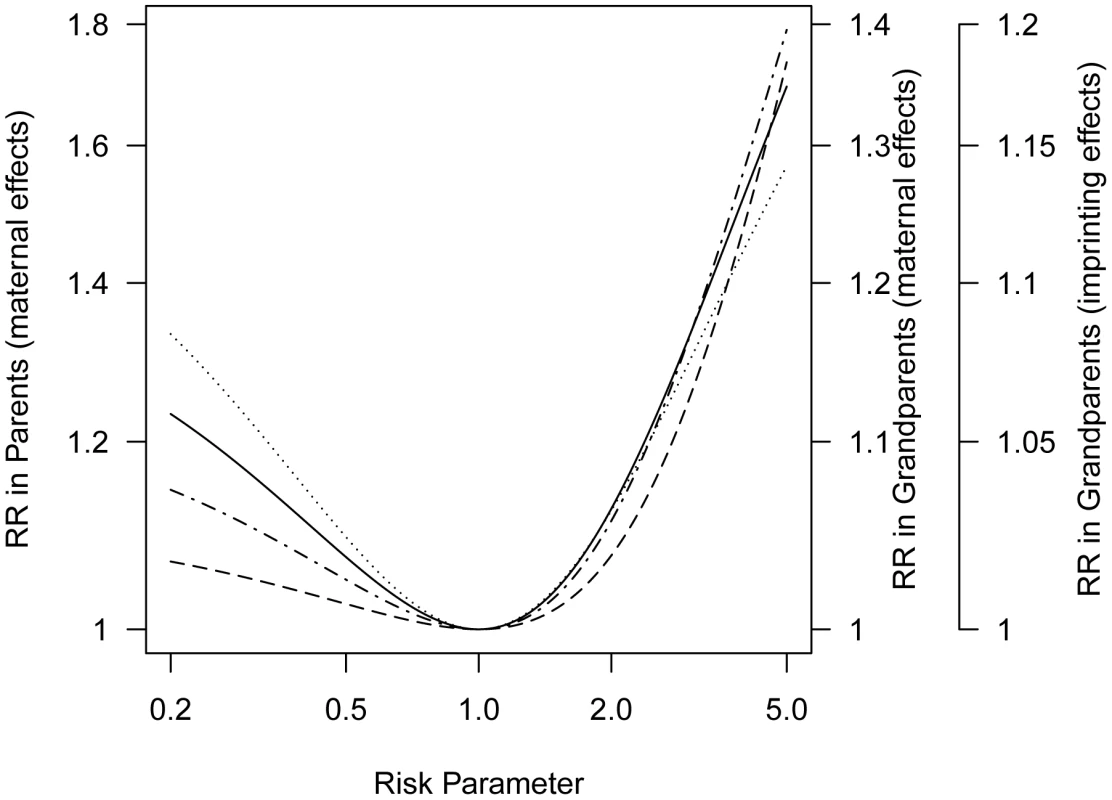
The curve for parents for imprinting would overlay the curve for grandparents for a maternal effect. This attenuation of seen in the inter-lineage parent relative risk reflects the fact that the mother of a case could have inherited the susceptibility allele that influenced her offspring's risk from either her mother or her father and in the event it came from her father it would not have affected her own risk.
In general, we expect weaker asymmetry in the grandparental generation than in the parental generation. Assessment of risk to grandparents requires repeated use of the V matrix to calculate conditional genotype probabilities for their mothers, i.e., the sampled proband case's great-grandmothers (Text S3, Table S2). Because the relative risk for grandparents is a linear function of the relative risk for parents (Text S4), both sets of curves can be displayed in one figure with a simple scale change (Figure 3).
Returning to breast cancer, we see that if the modest perimenopausal asymmetry we saw (about 1.3) were due entirely to a maternally-acting SNP with a frequency of 10% and the effect obeyed a log-additive risk model, the relative risk for an offspring whose mother carries one copy of that SNP would be about 4.0.
Most diseases affect both males and females, so that, even if risk is maternally mediated, both grandmothers and grandfathers can contribute to the asymmetry analysis. In that event, the inter-lineage grandparental odds ratio (relative risk) is estimated by dividing the number of discordant pairs where the affected grandparent (either grandmother or grandfather) is on the maternal side by the number of discordant pairs where the affected grandparent is on the paternal side. If the condition is not rare so that some families contribute two discordant pairs, a within-cluster resampling approach [19] or generalized estimating equations approach [20] can accommodate family-based dependencies.
Parent-of-Origin Effects
Another plausible source of asymmetry in family history involves parent-of-origin effects such as genetic imprinting. Such an effect was reported for breast cancer by Kong, et al [10] based on Icelandic family data. For simplicity we consider a situation where only the maternally-inherited copy at a risk-related locus is expressed (Table S2). Suppose the relative risk is I for offspring who carry a maternally-inherited copy of the variant compared to a risk of in individuals who do not (although for simplicity we assume and I are the same for both sexes, they could be sex-specific) (Text S5, Table S3). For a parent-of-origin effect, here based on imprinting, maternal grandmothers of affected children show greater risk than paternal grandmothers whenever (Figure 3). Again the same set of curves can serve, as there is a simple scale change involved in moving from asymmetry induced by log-additive maternal effects to asymmetry induced by a parent-of-origin effect (Text S6). The observed grandparental relative risk of approximately 1.2 could reflect a polymorphic imprinted gene if the risk associated with a maternally-inherited allele (I) is 5.
Inherited Mitochondrial Variants
Variants in mitochondrial DNA (mtDNA) can also produce asymmetry in families. A recent report found such a variant to be related to breast cancer risk in African-American women [9]. Since each person inherits virtually all their mitochondria from their mother, the asymmetry produced by this genetic mode of effect could show little or no diminution across generations of females, unless the mitochondria become heteroplasmic. The chain of mitochondrial inheritance is broken, however, by males: risk for the mother's father would on average return to the population risk because his mitochondria came from a separate maternal line. Returning to breast cancer, for a mitochondrial effect to explain the observed asymmetry it would have to confer about the same relative risk as that seen in the grandmothers, i.e., on the order of 1.2–1.3.
Discussion
While case-control GWASs have revealed many SNPs related to susceptibility to complex diseases, nonstandard genetic mechanisms may also play a role. We considered three such mechanisms that can produce asymmetry in family histories: maternal genetic effects that influence risk via the prenatal environment; parent-of-origin effects, for example where the expression of an imprinted polymorphic gene variant depends on parental source; and effects of variants in the mitochondrial DNA, which are exclusively inherited from mothers. Our algebraic quantification of the relationship between inter-lineage asymmetry and its driving cause led us to conclude that, although the driving effect would be small if due to a mitochondrial variant, if a single variant acted through maternally-mediated prenatal effects or was subject to parent-of-origin effects, then that cause would be associated with a large relative risk, at least on the order of 4 or 5, for breast cancer.
Although a single-allele scenario seems unlikely, we wondered whether a single allele acting through a maternally-mediated genetic mechanism could explain the known increased risk seen in sisters. Under our simplifying homogeneity assumptions, one can show with a little added algebra that a single, log-additive maternal effect with a single-copy relative risk of 4.0 involving an allele with frequency about 0.07 would produce about the observed two-fold increased risk for sisters of cases.
Although we have focused our methods and analysis on progenitors and descendants, siblings, aunts and uncles would also be informative. A prenatal maternal effect and a parent-of-origin effect where only the maternal allele is expressed share an interesting feature: A half sibling of a case would have risk similar to that of a full sibling if the shared parent is the mother, but no increased risk if the shared parent is the father. A Danish study reported that pattern of asymmetry in half-brothers of cases with the birth defect cryptorchidism [21]. Under a maternally-mediated prenatal effect, because siblings share the same mother, the relative risk for the siblings of a mother with an affected child versus siblings of that child's father should be greater than 1. Thus, one could glean even more insight by comparing histories of maternal versus paternal blood-relative aunts and uncles, in addition to parents.
Several small studies have compared rates of breast cancer in maternal and paternal relatives. A registry-based Swedish study found no difference in maternal and paternal grandmothers but included fewer than a thousand breast cancer cases [22]. Two studies of healthy adults found an excess of cancer reported for female relatives [23], [24], but presumably more fathers than mothers were estranged and participants were not asked if they knew about particular relatives' disease histories. In the Sister Study, out of 44,307 families 1,843 reported about their paternal grandmother but not their maternal grandmother, while 4,895 reported about their maternal grandmother but not their paternal grandmother, suggesting that knowledge about maternal versus paternal grandmothers is differential. Unlike other studies, however, we restricted our analysis to families where the status of both was reported to minimize information bias.
Although our sample is large, with almost 33,000 families represented, cases reported among sisters and grandmothers were not generally validated clinically. Nevertheless, our participants are sisters of women with breast cancer; and they have proven themselves an informed and dedicated cohort, providing bio-samples, completing lengthy questionnaires and maintaining commitment to follow-up with a very low dropout rate. Moreover, though the study staff did not obtain medical records for cases whose sister enrolled in the Sister Study, we did request medical records for 1422 affected sisters who joined our family-based add-on “Two Sister Study” [25]. Their diagnosis of breast cancer was confirmed by medical records for all but 3 (who had lobular carcinoma in situ) of 1251.
We made some simplifying assumptions (HWE, Mendelian transmission, random mating, rare disease, effect of only a single locus) and thus our figures depict idealized settings, which are not fully appropriate for the Sister Study. There are also effects secondary to ascertainment. The Sister Study is more likely to enroll unaffected women from larger families who have lower genetic risks. Consider two families, each with a single daughter with breast cancer and suppose one family has 10 daughters and the second has only two daughters. The first family is more likely to be in our study because any one of the 9 unaffected daughters can join, but the same large family has demonstrated lower genetic risk with only one of 10 affected, compared to the second family with 1 of 2 affected. The Sister Study consequently would have sampled families with less genetic enrichment for the allele under study than the two-fold increase presumed. Because this ascertainment effect should distort the inter-lineage asymmetry toward the null, the estimate of 4 for a maternally-mediated prenatal effect may be too low.
We implicitly assumed that the reported father is the biological father, but reported paternity can be incorrect. However, out of a subset of 602 families in the Two Sister Study where DNA was acquired, only 5 fathers failed the paternity test. Moreover, the observed strong pattern where asymmetry was related to the age at diagnosis of the granddaughter could not be explained by misidentified paternity. Another issue is that in a large series of cases such as the Sister Study, some affected sisters may unknowingly have been adopted; however, we expect that proportion to be small. Also, unaware adoptees would report the wrong history on both the maternal and the paternal side, driving estimates toward symmetry.
Reporting bias and self-selection may be at work. One might expect a woman with both a sister and a mother with breast cancer to be more likely to join the Sister Study than a woman with only an affected sister. Also, women whose mother had breast cancer may undergo more regular screening. However, the rate of breast cancer reported for mothers was about 18%, which does not exceed expectation based on having a first degree relative with breast cancer.
With combined data including as many as 45,000 cases, large consortial efforts are underway [16] to study the genetics of cancers and other complex diseases. We believe that important clues could be elicited by also studying asymmetries in the reported family histories for those cases.
Although we saw evidence for inter-lineage asymmetry based on the family histories from the Sister Study participants, with more breast cancer in the maternal lineage, one cannot differentiate among the three nonstandard mechanisms using only phenotypic family histories of affected individuals. Only family-based genotype data will enable an investigator to identify the genetic mechanism and identify relevant variants [26], [27].
In summary, susceptibility to complex disease can be influenced by inherited autosomal gene variants, as most GWASs assume, but can also be influenced by sex-linked genes, maternally-mediated prenatal effects, parent-of-origin effects, and mitochondrial variants. These under-studied genetic mechanisms are best explored through family studies. Yet even without access to genetic data on family members, evidence that those phenomena play a role can be adduced through careful analyses of family history data from large assemblages of cases. Breast cancer appears to be subject to genetic mechanisms that produce family history asymmetry, particularly when diagnosed in the perimenopausal years.
Supporting Information
Zdroje
1. National Human Genome Research Institute, National Institutes of Health, “A Catalog of Published Genome-wide Association Studies,” Available:, http://www.genome.gov/GWAStudies, last updated 10 January 2014.
2. ParkJH, GailMH, WeinbergCR, CarrollRJ, ChungCC, et al. (2011) Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci U S A 108 : 18026–18031.
3. WacholderS, HartgeP, PrenticeR, Garcia-ClosasM, FeigelsonHS, et al. (2010) Performance of common genetic variants in breast-cancer risk models. N Engl J Med 362 : 986–993.
4. RaskinL, RennertG, GruberSB (2009) FOXP3 germline polymorphisms are not associated with risk of breast cancer. Cancer Genet Cytogenet 190 : 40–42.
5. Hill, M.A. Abnormal Development - Fetal Origins Hypothesis. Available: http://embryology.med.unsw.edu.au/embryology/index.php?title=Abnormal_Development_-_Fetal_Origins_Hypothesis, last updated 3 April, 2012.
6. WilcoxAJ, WeinbergCR, LieRT (1998) Distinguishing the effects of maternal and offspring genes through studies of “case-parent triads”. Am J Epidemiol 148 : 893–901.
7. LupoPJ, NousomeD, OkcuMF, ChintagumpalaM, ScheurerME (2012) Maternal variation in EPHX1, a xenobiotic metabolism gene, is associated with childhood medulloblastoma: an exploratory case-parent triad study. Pediatr Hematol Oncol 29 : 679–685.
8. MichelsKB, TrichopoulosD, RobinsJM, RosnerBA, MansonJE, et al. (1996) Birthweight as a risk factor for breast cancer. Lancet 348 : 1542–1546.
9. CanterJA, KallianpurAR, ParlFF, MillikanRC (2005) Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res 65 : 8028–8033.
10. KongA, SteinthorsdottirV, MassonG, ThorleifssonG, SulemP, et al. (2009) Parental origin of sequence variants associated with complex diseases. Nature 462 : 868–874.
11. Romano-ZelekhaO, HirshR, BliedenL, GreenM, ShohatT (2001) The risk for congenital heart defects in offspring of individuals with congenital heart defects. Clin Genet 59 : 325–329.
12. WeinbergCR, ShoreDL, UmbachDM, SandlerDP (2007) Using risk-based sampling to enrich cohorts for endpoints, genes, and exposures. Am J Epidemiol 166 : 447–455.
13. NewschafferCJ, CroenLA, FallinMD, Hertz-PicciottoI, NguyenDV, et al. (2012) Infant siblings and the investigation of autism risk factors. J Neurodev Disord 4 : 7.
14. VikanesA, SkjaervenR, GrjibovskiAM, GunnesN, VangenS, et al. (2010) Recurrence of hyperemesis gravidarum across generations: population based cohort study. BMJ 340: c2050.
15. CarterCO, EvansK, CoffeyR, RobertsJA, BuckA, et al. (1982) A three generation family study of cleft lip with or without cleft palate. J Med Genet 19 : 246–261.
16. Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, et al.. (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45: : 353–361, 361e351–352.
17. National Institute of Environmental Health Sciences, National Institutes of Health, The Sister Study. Available: http://sisterstudy.niehs.nih.gov/English/index1.htm.
18. CampbellMK, FeuerEJ, WunLM (1994) Cohort-specific risks of developing breast cancer to age 85 in Connecticut. Epidemiology 5 : 290–296.
19. HoffmanE, SenP, WeinbergC (2001) Within-cluster resampling. Biometrika 88 : 1121–1134.
20. LipsitzSR, LairdNM, HarringtonDP (1991) Generalized estimating equations for correlated binary data: Using the odds ratio as a measure of association. Biometrika 78 : 153–160.
21. JensenMS, ToftG, ThulstrupAM, HenriksenTB, OlsenJ, et al. (2010) Cryptorchidism concordance in monozygotic and dizygotic twin brothers, full brothers, and half-brothers. Fertil Steril 93 : 124–129.
22. CoutoE, HemminkiK (2007) Estimates of heritable and environmental components of familial breast cancer using family history information. Br J Cancer 96 : 1740–1742.
23. BevierM, SundquistK, HemminkiK (2012) Risk of breast cancer in families of multiple affected women and men. Breast Cancer Res Treat 132 : 723–728.
24. OzanneEM, O'ConnellA, BouzanC, BosinoffP, RourkeT, et al. (2012) Bias in the reporting of family history: implications for clinical care. J Genet Couns 21 : 547–556.
25. National Institute of Environmental Health Sciences, National Institutes of Health, The Two Sister Study, Available: http://sisterstudy.niehs.nih.gov/English/2sis.htm.
26. WeinbergCR, WilcoxAJ, LieRT (1998) A log-linear approach to case-parent triad data: Assessing effects of disease genes that act directly or through maternal effects, and may be subject to parental imprinting. Am J Hum Gen 62 : 969–978.
27. WilcoxA, WeinbergC, LieR (1998) Distinguishing the effects of maternal and offspring genes through studies of “case-parent triads”. Am J Epid 148 : 893–901.
Štítky
Genetika Reprodukčná medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy







