-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
Due to their deregulation in cancer and their potential to be inhibited by small chemical compounds, tyrosine kinases are among the most important targets under consideration for cancer therapeutics. One such oncogenic tyrosine kinase is FAK, which is known to regulate cellular signalling downstream of Integrins and Receptor Tyrosine Kinases (RTK) at the cell surface. In this study, however, we report that FAK can act as a suppressor of oncogenic Receptor Tyrosine Kinases. This mechanism was observed in fruit fly tissues in vivo and human cancer-derived cells in vitro, which additionally suggests it is an evolutionary conserved mechanism in humans. FAK mediated this inhibition by controlling the sub-cellular localisation of receptors, via suppression of receptor recycling to the cell surface. These results suggest that in some particular cancer contexts such as RTK-driven tumours, FAK may act as a tumour suppressor and therefore, may not be a valid drug target.
Published in the journal: FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells. PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004262
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004262Summary
Due to their deregulation in cancer and their potential to be inhibited by small chemical compounds, tyrosine kinases are among the most important targets under consideration for cancer therapeutics. One such oncogenic tyrosine kinase is FAK, which is known to regulate cellular signalling downstream of Integrins and Receptor Tyrosine Kinases (RTK) at the cell surface. In this study, however, we report that FAK can act as a suppressor of oncogenic Receptor Tyrosine Kinases. This mechanism was observed in fruit fly tissues in vivo and human cancer-derived cells in vitro, which additionally suggests it is an evolutionary conserved mechanism in humans. FAK mediated this inhibition by controlling the sub-cellular localisation of receptors, via suppression of receptor recycling to the cell surface. These results suggest that in some particular cancer contexts such as RTK-driven tumours, FAK may act as a tumour suppressor and therefore, may not be a valid drug target.
Introduction
Research in model organisms can provide important insights on the effects of oncogenic pathways in different in vivo environments [1], [2]. Particularly, Drosophila melanogaster has made numerous contributions to cancer biology, e.g. by identifying components of several signalling pathways such as the Hippo [3] and RTK/Ras/MAPK signalling pathways [4]–[7].
FAK is a cytoplasmic non-receptor tyrosine kinase that interacts primarily with Integrins at the focal adhesion sub-domains of the plasma membrane (reviewed in [8]). FAK belongs to a hub where phosphorylation signals are regulated and transferred into the cell, therefore it is implicated in many cellular processes such as adhesion, migration, survival and differentiation [8], [9], and is normally found over-expressed in migrating and invasive tumour cells [10]. The current knowledge suggests that abnormal FAK activation is a key driver of tumour cell motility and survival in conditions that would trigger anoikis (detachment-dependent apoptosis) (reviewed in [10], [11]). Thus, FAK has been regarded as a potential target for cancer therapeutics.
In Drosophila, there is a single FAK homologue (FAK56D, here aftercalled dFAK [12]–[14]); dFAK is ubiquitously expressed, with particularly high levels in the developing Central Nervous System (CNS) and muscle [14]. Consistently, dFAK mutants have abnormal neuromuscular junction growth and optic stalk structure [15], [16]. Nevertheless, dFAK mutants are viable and fertile [17], proving it is dispensable for general development. This suggests the role of dFAK may become apparent only under conditions of stress. In fact, dFAK mutants display sensitivity to mechanical stimuli, suffering seizure and temporal paralysis [18].
As oncogene activation is a condition of stress, we examined the role of dFAK within a context of a Drosophila model of cancer [19]–[21] achieved by the expression of the receptor tyrosine kinase RET (Rearranged during transformation). Activating mutations in RET cause the familial cancer syndrome Multiple Endocrine Neoplasia type 2 (MEN2) (reviewed in [22], [23]). Furthermore, chromosomal translocations implicating ectopic expression of RET are frequent in Papillary thyroid Carcinoma (PTC), the most common type of thyroid cancer [24], [25], pheochromocytomas [26] and breast carcinoma [27].
Several RTKs were described to directly phosphorylate and activate FAK [28], [29]. Interestingly, direct interaction and mutual phosphorylation between FAK and RET has also been reported [30], [31]. Nevertheless, the functional importance of FAK in RTK signalling in vivo, particularly in the context of tumour development, is not yet clear [29] (reviewed in [11]). Therefore, dFAK is a likely candidate to be activated by Drosophila RET (here after called dRET) and mediate its signalling cascade in Drosophila.
In this study, we investigated the regulatory role of FAK downstream of RTKs in epithelial tissues. Unexpectedly, our findings demonstrate that FAK constitutes a negative regulator of RET and also, the RTK epithelial growth factor receptor (EGFR). FAK impairs RTK signalling specifically via the Ras/MAPK pathway, which in turn impacts on the survival and differentiation outcomes of RTK-expressing tissues. Because FAK is currently considered an oncogenic target, our study may have implications in future cancer therapies. Our results suggest that targeting FAK in the context of some RTK-driven tumours might be detrimental rather than beneficial for the host.
Results
RET activates FAK and MAPK
We expressed a constitutively activated form of dRET (hereafter called dRETCA) with the ptc-gal4 driver, which is active in a stripe of cells immediately anterior to the Anterior/Posterior compartmental boundary of the developing wing imaginal discs (Figure 1A and 1D). GFP expression by itself did not affect this cell population nor produced activation of the cytoplasmic kinases Src, FAK or MAPK (Figure 1A–C). As expected from previous studies [19], expression of dRETCA (ptc>dRETCA) led to phosphorylation of Src and MAPK on residues that report their activation (Figure 1D–E, see methods). Interestingly, we also observed increased phosphorylation of FAK (Figure 1F).
Fig. 1. Genetic interactions between RET and FAK. 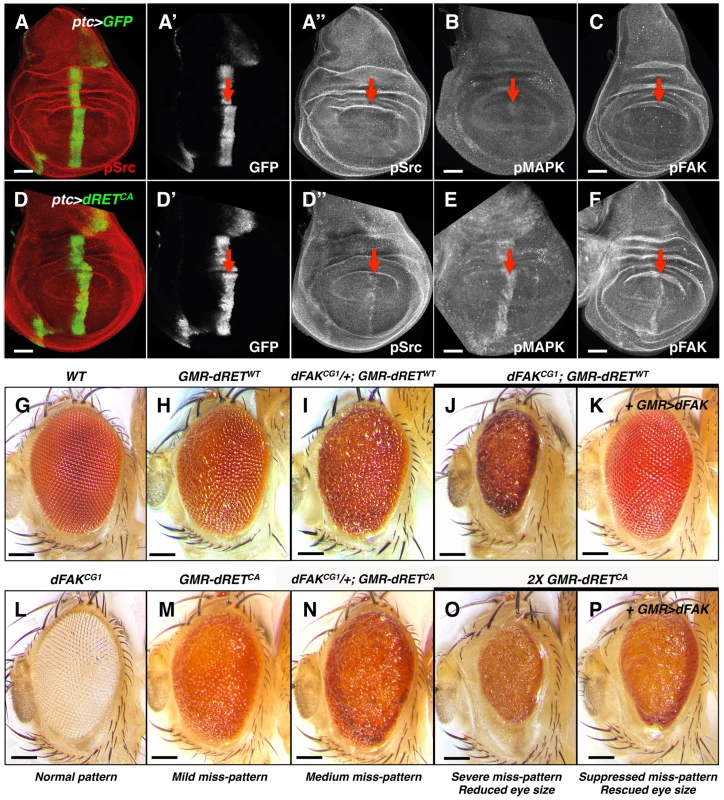
(A–F) Confocal images of wing disc epithelia. Control tissues (A–C) and experimental tissues (D–F) expressed GFP driven by ptc-gal4, shown in A′ and D′. Experimental tissues (D–F) also expressed dRETCA. Immunostaining against pSrc (A″ and D″), pMAPK (B and E) and pFAK proteins (C and F; see methods), as a proxy for probing their activation levels, are shown in grayscale panels. Note increased phosphorylation of Src, MAPK and FAK after dRETCA expression within the ptc domain, indicated by red arrows. Scale bars, 50 µm. (G–P) Images of adult eyes with indicated relevant genotypes; full genotypes are listed in supplemental material. GMR (glass multimer reporter) is an eye specific promoter. GMR-gal4 was used to drive UAS-dFAK transgene expression. GMR-dRETWT and GMR-dRETCA are fusion recombinant constructs. Wild type (G) and dFAKCG1 (L) animals displayed a normal eye pattern; note that the dFAKCG1 is in a white background (see Figure S1B). Expression of dRETWT caused a mild eye miss-patterning phenotype (H), and lowering the genetic dose of dFAK gene in these animals either enhanced eye roughness (I–J) or completely disrupted patterning and decreased eye size (J). Reciprocally, suppression of both effects was observed by co-expression of dFAK (K). (M–N) A similar enhancement was observed by halving the dose of dFAK gene after expression of dRETCA. (O) Doubling dRETCA dose caused a very rough and small eye, comparable to (J), which was partially suppressed when dFAK was also expressed (P). Eye size quantifications of panels G, H, J, K, O and P are shown in Figure S1D. Scale bars, 100 µm. Next, we tested genetically the importance of Src, Ras/MAPK and FAK downstream of RET. The Drosophila compound eye is an elegant structure composed of about 750 hexagonal units called ommatidia, which pattern in a honeycomb-shaped array (Figure 1G) [32]. This repetitive array makes the eye very sensitive to perturbations in signalling pathways. The ectopic expression of a single wild type copy of the Drosophila RET gene (hereafter called dRETWT) under the direct control of the eye-specific GMR promoter (GMR-dRETWT) disturbed the normal array of ommatidia, creating a ‘rough’ eye phenotype (Figure 1H; [19]). In a search of genes involved in dRET signalling, previously known members of the Ras/MAPK and Src signalling pathways were identified (drk (Grb2), Sos, Ras85D, ksr, Gap1 (RasGAP), Src42A, Src64B, Jra (c-Jun) and basket (JNK) [19]. As expected, we observed that the dRET-induced phenotype was suppressed when Src42A or Ras85D proteins were knocked down by RNA interference (Figure S2A), confirming that they play key roles downstream of dRET signalling.
Eyes from dFAKCG1 null mutants displayed normal patterning (Figure 1L). Interestingly, and contrary to expectations of a potential dRET-signalling effector, loss of FAK did not suppress the rough eye phenotype. In fact, hetero - and homozygosis of dFAKCG1 either enhanced the miss-patterning or caused a smaller eye due to a completely disrupted ommatidial pattern, respectively (Figure 1I and 1J).
To prove that only the loss of dFAK was responsible for the phenotype observed in dFAKCG1; GMR-dRETWT animals (Figure 1J) and no other genetic background mutation was influencing the results, we next used two additional dFAK mutant strains (Figure S1A). These strains bear a P-element insertion within dFAK and by measuring the expression levels of dFAK mRNA transcripts were confirmed to be hypomorphic lines (Figure S1B). Different dFAK allelic combinations showed a similar genetic interaction with GMR-dRETWT (Figures S1C).
The latter phenotype was comparable to the effect mediated by high expression of dRETCA in the eye (Figure 1O). dFAK overexpression rescued the effects of dFAK null mutation (Figure 1K) and partially restored the small size induced by the constitutively active RET isoform (compare Figure 1O with 1P; eye sizes are quantified in S1D).
Overall, these data suggest that dRET activates dFAK, which in turn may have unanticipated suppressive effects on RET signalling.
FAK suppresses RET in different fly epithelia
To further test whether FAK can inhibit RET signalling we combined dRET and dFAK in different imaginal discs using the GMR-gal4, dpp-gal4 and ptc-gal4 drivers. dFAK overexpression by itself had no apparent phenotype on adult survival and tissue patterning (Figure 2A, B, E, F, I, J, M).
Fig. 2. FAK suppresses RET-driven effects in different fly tissues. 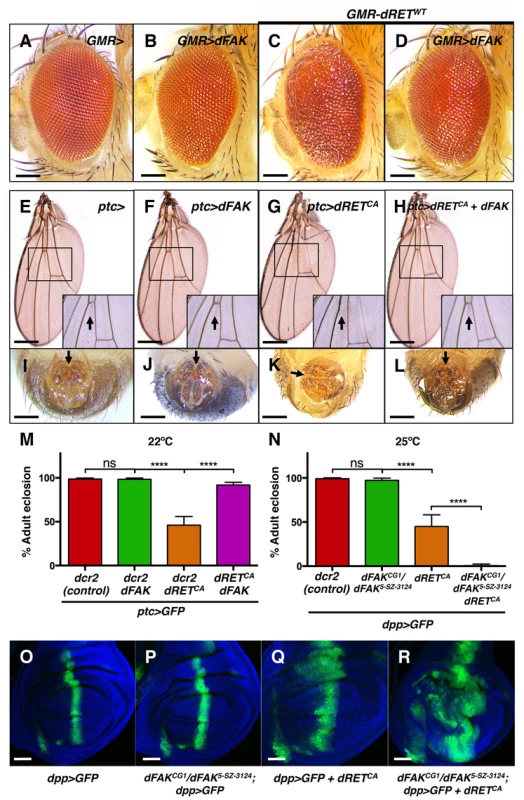
(A–D) Eyes expressing dFAK displayed a normal adult eye phenotype, while dRETWT expression perturbed the normal pattern. Co-expression of dRETWT and dFAK supressed dRET-driven mis-patterning defects. Scale bar, 100 µm. (E–H) dFAK-expressing wings via ptc-gal4 showed no detectable defects similarly to control wings. Expression of dRETCA led to disappearance of anterior cross veins in all adult escapers (arrow in inset box), which was rescued by simultaneous expression of dFAK with full genetic penetrance. Scale bar, 500 µm. (I–L) ptc-driven dRETCA expression also led to incomplete rotation of the male genitalia in all adult escapers (arrows). dFAK co-expression rescued this phenotype with full penetrance and it did not affect the normal development of the genitalia by itself. Scale bar, 100 µm. (M) Quantification of the penetrance of adult eclosion for the indicated genotypes, note that dFAK co-expression rescued significantly the developmental lethality of ptc>dRETCA animals. Error bars are standard deviation in this and all plots; ‘ns’ stands for non-statistically significant, **** means p<0.0001 in this and all plots (see methods). (N) Conversely, dFAK loss, which by itself had no effect in viability, enhanced to almost full penetrance the developmental lethality of dpp>dRETCA animals. (O–R) Confocal images from wing discs with the indicated genotypes. Note that dFAK mutation enhanced the size and shape defects associated with ectopic expression of dRETCA within the dpp stripe. For a detailed characterisation of the dFAK mutant alleles used here, please see Figure S1A–B. Scale bars, 50 µm. However, dRET expression did affect several tissues of the adult fly. As mentioned above, dRETWT expression in the eye altered the normal pattern of ommatidia (Figure 2C, see also 1H), which was prevented when dFAK was simultaneously expressed (Figure 2D). All escaper ptc>dRETCA adults showed wing vein defects [33], with absence of the anterior cross vein, which lies within the ptc domain of the wing anlage (Figure 2G). Remarkably, this phenotype was rescued by the co-expression of dFAK and dRETCA (Figure 2H). All escaper ptc>dRETCA adult males also displayed rotation defects in the epandrium (Figure 2K). The patched gene is expressed in a compartment-specific manner across most imaginal discs in the larva, including the genital disc [34]. Therefore, dRET expression in this tissue perturbed normal development and rotation of the male genital organ of adult escapers [35], [36]. Importantly, dFAK also suppressed this dRET-induced phenotype and restored the proper orientation of the male genitalia (Figure 2L).
Additionally, dRET expression also resulted in developmental toxicity (with a penetrance that depended on the temperature and other factors [33]). Correspondingly, 46% of ptc>dRETCA-expressing flies made it to adulthood (n = 187) in our experimental conditions. Co-expression of dRETCA and dFAK increased survival up to 92% (n = 106) (Figure 2M). Conversely, loss of dFAK enhanced the morphological defects induced by dRET in the imaginal discs (Figure 2O–R) and reduced the survival rate of dpp>dRETCA animals (Figure 2N).
Together, these observations demonstrate that dFAK inhibits dRET-induced phenotypic effects in multiple imaginal disc epithelia.
The N-terminal domain of FAK, but not its kinase activity, is required to suppress RET
To gain insight into the mechanism by which FAK suppresses RET signalling, we co-expressed dRETCA and different dFAK mutant isoforms in the eye (schematised in Figure 3A). When driven by GMR-gal4, the transcript levels from these UAS-dFAK transgenes ranged from 20 - to 40-fold over the endogenous levels of dFAK transcript (Figure 3B). When expressed simultaneously with dRETCA and by measuring the eye areas of the consequent phenotypes (Figure 3C) we observed that a wild type allele of dFAK significantly restored the eye size when compared to control GMR>dRETCA eyes (Figure 3D and 3E). Interestingly, and in correlation with observations made in different systems [30], [31], the N-terminal FERM domain of dFAK appears important for this functional interaction between dFAK and dRET: expression of a N-terminal deletion mutant (dFAKΔN) failed to modify the eye size of GMR>dRETCA flies (Figure 3C and 3F). On the other hand, a point mutant isoform in dFAK major auto-phosphorylation site (FAKY430F, equivalent to tyrosine Y-397 in human FAK), did significantly rescue the eye size of GMR>dRETCA flies and the patterning of FAKCG1/+; GMR>dRETWT eyes (Figure 3G and S2C). These results suggest that the FERM domain of dFAK is necessary to inhibit RET signalling, while the major autophosphorylation site — required for full FAK kinase activity [37] — may not be essential. Some systems require FAK as a scaffold protein but not as a kinase [38] and our results suggest that this is the case in the functional interaction with RET.
Fig. 3. Requirement of the N-terminal FAK FERM domain. 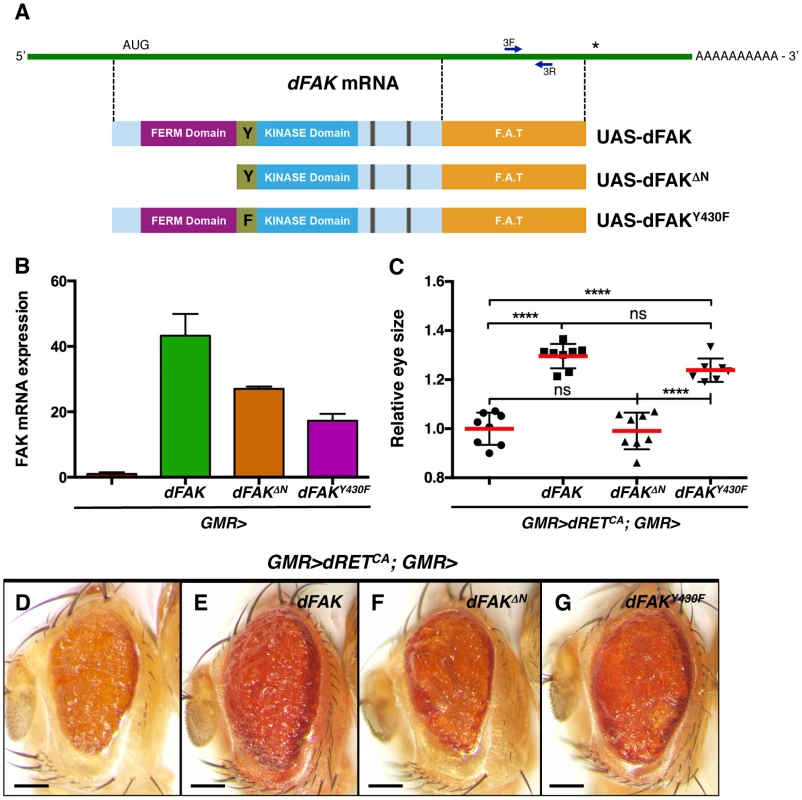
(A) Linear representation of dFAK mRNA, its derivatives UAS-transgenes and their resultant protein isoforms: a full-length dFAK isoform; an N-terminal deletion mutant that lacks the first 400 amino acids residues of dFAK including its FERM domain; and a point mutant isoform that bears a replacement of the Tyrosine430 residue for a Phenylalanine residue, which impairs the auto-phosphorylation site and consequently the kinase activity of dFAK. (B) Expression profiles of each UAS-dFAK transgene in the eye (driven by GMR-gal4) as determined by quantitative (q) PCR of RNA samples (see methods). We used a pair of primers (3F and 3L) flanking a 200 bp region corresponding to the C-terminal domain (FAT: Focal adhesion targeting domain), which is a common region to all the isoforms. (C) Eye size quantification of the indicated genotypes, shown in D-G. Eye sizes on the Y-axis are represented as relative values to the mean of GMR>dRETCA (‘ns’: not statistically significant; **** = p<0.0001; n = 8–10 for each genotype). (D–G) Eye micrographs correspond to the indicated genotypes. Note that while the auto-phosphorylation mutant version of dFAK was expressed at lower levels than the N-terminal mutant isoform (B), it was still able to rescue the size of dRETCA-expressing eyes (G), to a similar extent as the full-length dFAK isoform (E). However, the N-terminal deletion mutant isoform did not suppress the small eye size of dRETCA animals (C, D and F). Scale bar, 100 µm. Moderate RET/FAK ratios suppress apoptosis
Next, we further characterized the RET/FAK regulatory loop and its signalling output; specifically, we analysed how different experimental conditions that altered relative RET/FAK levels affected eye patterning. To gain further insights on how different RET/FAK ratios influence tissue cell fate in vivo, we examined the cell composition of the eye tissue. Each ommatidial unit consists of eight photoreceptor and six supporting cells (four cone cells and two primary pigment cells) (Figure 4A) [32] while a hexagonal lattice of secondary and tertiary pigment cells surrounds the units (white coloured in Figure 4A′–C′). Photoreceptor cell clusters are specified first; they constitute ‘organizing centres’ that instruct neighbouring cells to differentiate into cone cells and primary pigment cells. The hexagonal lattice patterns by local cell reorganization and elimination of surplus cells via a wave of developmental programmed cell death.
Fig. 4. Moderate relative RET/FAK levels lead to inhibition of programmed cell death. 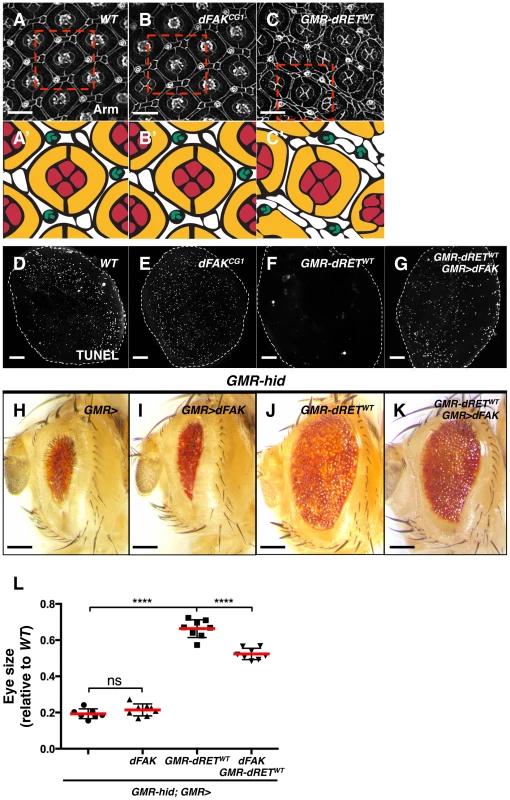
(A–C) Armadillo immunostaining revealed cell outlines of wild type (A), dFAKCG1 (B), and GMR-dRETWT (C) retinas at 42 hs after puparium formation (APF). The boxed areas were traced to highlight their cellular composition (A′–C′). Each ommatidium is composed of 4 cone cells (red), 2 primary pigments cells (yellow), 6 secondary and three tertiary cells (white), and three-bristle cells (green) make the hexagonal lattice. Note that dFAKCG1 eyes display normal patterning (B′) while GMR-dRETWT retinas displayed normal ommatidial cores but additional interommatidial cells (white cells in C′). Scale bars, 10 µm. (D–G) TUNEL labelling of retinas at 28 h APF. Note that the developmental programmed cell death observed in wild type and dFAKCG1 retinas were suppressed in GMR-dRETWT retinas. Co-expression of dFAK rescued this inhibition of cell death (G). Scale bar, 50 µm. (H–K) Hid overexpression (GMR-hid) gave a small eye phenotype, which was suppressed by dRETWT co-expression (J). This dRET-dependent inhibition was also suppressed by dFAK co-expression (K) while dFAK itself did not suppress Hid-mediated effects in the eye (I). Scale bar, 100 µm. (L) Eye size quantification of the indicated genotypes (as depicted in panels H–K) represented as relative values to the wild type mean value (‘ns’: not statistically significant; **** = p<0.0001; n = 8–10 for each genotype). We visualized the final pattern of retinas at 42 hs after puparium formation (APF). dFAK mutant retinas were indistinguishable from their wild type counterparts (Figure 4A and 4B). When dRETWT was expressed by itself, the ommatidial cores remained normal, with four cone cells surrounded by two primary pigment cells. Nevertheless, eye patterning was altered, displaying supernumerary interommatidial cells (Figure 4C). This suggested that developmental programmed cell death might be suppressed during eye development. Indeed, while wild type or FAKCG1 retinas displayed a large number of apoptotic cells at 28 h APF —a time of high levels of developmental apoptotic cell death — there were virtually no apoptotic cells in GMR-dRETWT retinas (Figures 4D–F). Remarkably, developmental cell death was rescued by co-expression of dRETWT and dFAK, further demonstrating the ability of dFAK to suppress dRET signalling (Figure 4G).
Programmed cell death during eye development is mainly dependent on the pro-apoptotic protein Hid (Head Involution Defective) [39]–[41]. Hid over-expression in the developing eye (GMR-hid) triggers apoptotic cell death throughout the tissue leading to a small eye phenotype (Figure 4H) [42]. When dRETWT was simultaneously expressed with Hid, the eye size increased significantly (Figure 4J and 4L), indicating that RET signalling could block Hid or its downstream effectors. The ability of dRET to suppress Hid-induced apoptosis depended on its known effectors Src and Ras: down-regulation of Ras85D or Src42A by RNA interference prevented dRET-mediated suppression of GMR-hid small eye phenotype (Figure S2B). In contrast, while dFAK expression by itself had no significant effects on the GMR-hid eye phenotype (Figure 4I), it did suppress dRET inhibitory effect (Figure 4K and 4L).
Taken together, our data suggest that dRET expression suppresses both ectopic (Hid-induced) and developmental cell death via its known effectors Src and Ras, and this effect can be suppressed by dFAK co-expression. Therefore, in the retina, the output of dRET overexpression in a dFAK wild type background—genetically defined here as a moderate RET/FAK ratio and expected to produce a moderate level of Ras/MAPK signalling — is the inhibition of cell death.
High RET/FAK ratios drive ectopic differentiation
While the output of moderate RET/FAK ratios resulted in suppression of cell death, the small eye phenotypes observed under conditions of higher RET/FAK ratios suggested different cell fate outcomes. We then further analysed two different experimental conditions expected to produce high relative levels between RET and FAK, namely (i) the expression of one copy of dRETWT in a dFAK mutant background (dFAKCG1; GMR-dRETWT), and (ii) the expression of two copies of dRETCA in a dFAK wild type background (2X GMR-dRETCA), as these displayed the roughest, reduced eye size phenotypes (Figure 5B and 5C, see also Figure 1J and O). dFAKCG1; GMR-dRETWT pupal retinas lacked the hexagonal array and identifiable ommatidial units (Figure 5F–F′). In this case, there were clusters of numerous cone-like cells. Some bristle cells and a few cells recognisable as primary pigment-like remained, but there were no detectable cells with the appearance of normal interommatidial cells (Figure 5F′). In order to confirm the identity of those cells, we stained for the transcription factor Cut, a well-known cone cell marker [43]. In control retinas, Cut localised constitutively to the nucleus of the four cone cells from each ommatidium (Figure 5E″) [43]. In contrast, in dFAKCG1; GMR-dRETWT retinas (Figure 5F″), the cell clusters were indeed made of numerous Cut-expressing cone-like cells. Thus, in these experimental conditions, dRET signalling drives ectopic differentiation into the cone cell fate.
Fig. 5. High relative levels between RET and FAK induce ectopic cone cell differentiation in the eye. 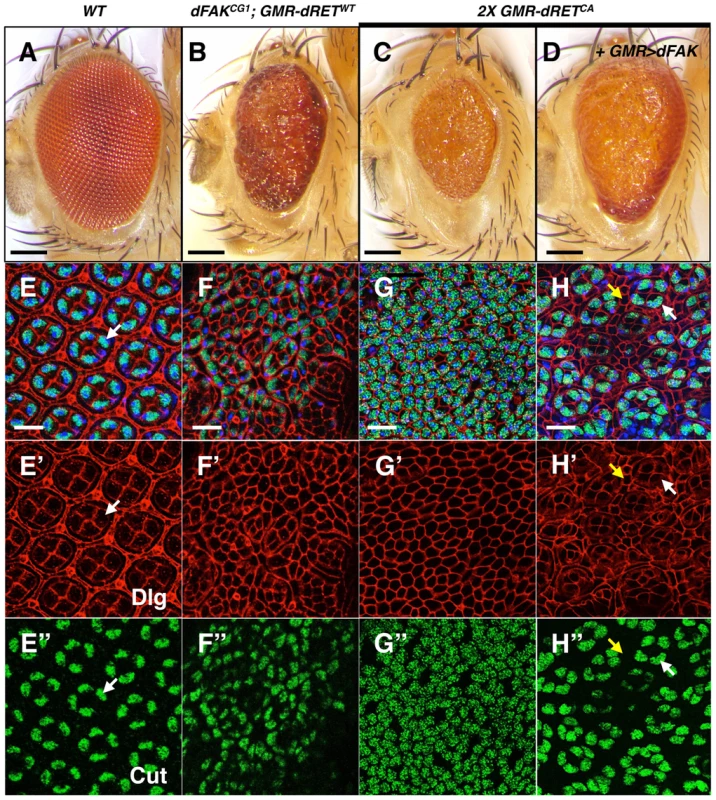
We examined the cellular patterning of the pupal retinas in correspondence to the adult eye phenotypes shown in panels A–D. Scale bars, 100 µm. (E–H) Merged images of stainings for nuclei (DAPI, blue), Dlg (cell outlines, red) and Cut (cone cells, green), from retinas at 42 h APF. Bottom panels show Dlg (E′–H′) and Cut (E″–H″) immunostainings individually. (E) Note the symmetric hexagonal array, and four Cut+ cone cells per ommatidium (white arrows) in control retinas. (F and G) Note the change in cellular composition of these retinas with high RET/FAK ratios, primarily composed of Cut+ cone-like cells. (H) dFAK expression within a 2X GMR-dRETCA background suppressed this phenotype (also see S1D); some normal four-cone cell clusters (white arrows) can be identified and interommatidial cells reappeared (yellow arrows). Scale bars, 10 µm. We next tested whether the supernumerary cone cells could be a consequence of ectopic differentiation during the larval stage. Since photoreceptor cells induce cone cell differentiation, one possibility is that aberrant photoreceptor cell numbers trigger ectopic cone cell differentiation. We observed normal numbers of photoreceptors (i.e.; clusters of eight) in dFAKCG1; GMR-dRETWT eye discs, albeit some clusters had rotation defects (Figure S3A–C). We also observe ommatidial units with extranumerary cone cells at early stages of eye development (4 h APF; Figure S3E) that suggests these ectopic clusters might accumulate during development until a mesh of mostly cone cells is observed by 42 h APF.
Similarly, 2X GMR-dRETCA pupal retinas displayed a mesh of cone-like cells as shown by Cut staining (Figure 5C and 5G–G″), where the few remaining cells displayed bristle cell morphology. The expression of dFAK within this context rescued the small eye size phenotype (Figure 5D; quantified in S1D). Most remarkably, it also reduced the number of ectopic cone cells and resulted in the re-appearance of normal ommatidial cores and surrounding interommatidial cells in the pupal retinas (Figure 5H–H″). Moreover, the down-regulation of Ras85D also prevented this ectopic differentiation, restoring the eye size and suppressing patterning defects (Figure S3D).
Together, these results indicate that in a genetically defined high RET/FAK ratio, expected to produce high Ras/MAPK signalling, most of non-neuronal eye cell types ectopically differentiate into cone cells.
FAK impairs MAPK signalling downstream of RET
Next, in order to gain insights into the mechanism of dRET signalling inhibition, we assessed the role of dFAK in influencing dRET-signalling effectors.
dRET was reported to activate the PI3K/Akt pathway [33], therefore we assessed whether there was also an ectopic activation of Drosophila Akt1 in our experimental conditions. Overexpression of Drosophila Insulin receptor (dInR), an RTK known to signal primarily via the PI3K/Akt pathway [44], resulted in increased immunostaining for phosphorylated-Akt1 (pAkt) (Figure S4E and S4G); nevertheless, that was not the case after expression of dRETCA (Figure S4C and S4G). This suggests that dRET signals independently of PI3K/Akt in the imaginal disc domains we utilized.
We showed above that Src and MAPK were activated upon RET expression (Figure 1D–E). dFAK overexpression on its own within the ptc domain of the wing disc also resulted in increased levels of pSrc (Figure S4I). However, the co-expression of dRET and dFAK did not affect notably the levels of pSrc (Figure S4K and S4L). These results indicate that dFAK does not suppress dRET signalling via Src.
Regarding MAPK, dFAK expression did not modulate MAPK phosphorylation when expressed by itself (ptc>dFAK) (Figure 6B) but did reduce MAPK phosphorylation within the dRETCA-overexpressing ptc compartment of the wing disc (Figure 6C–E). This indicates that dFAK is able to inhibit dRET signalling via the MAPK pathway.
Fig. 6. FAK inhibits RTK signalling by impairing Ras/MAPK pathway. 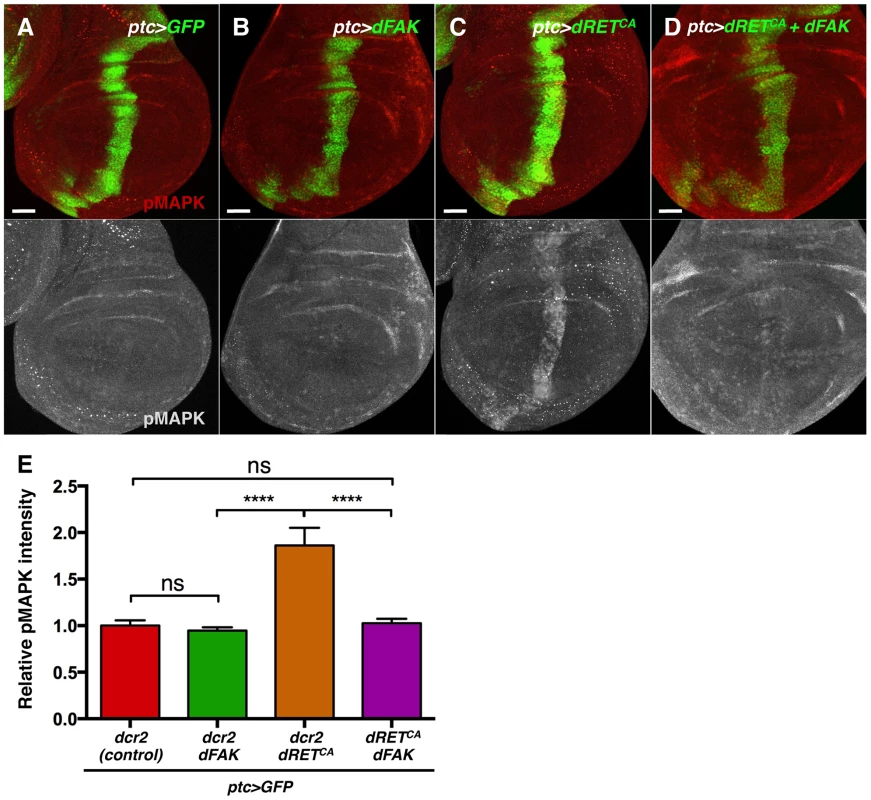
(A–D) Phosphorylated (active) MAPK immunostainings from wing discs with the indicated genotypes. (A–B) When dFAK was expressed in the ptc-compartment (green), pMAPK staining was unchanged compared to GFP-only expressing cells. (C–D) dRETCA expression increased pMAPK staining in the ptc domain but co-expression with dFAK suppressed this dRETCA-induced activation of MAPK. Scale bars, 50 µm. (E) Quantification of pMAPK immunostaining within the ptc stripe (see methods). Intensity of pMAPK signal is represented as relative values to the mean intensity of control tissues (A) (‘ns’: not statistically significant; **** = p<0.0001; n = 4–6 for each genotype). Interestingly, the eye phenotypes mediated by activated isoforms of Ras and Raf, two components of the MAPK kinase pathway, were not suppressed by dFAK co-expression (Figure S5), suggesting that dFAK-mediated inhibition of dRET/MAPK signalling occurs upstream of Ras.
FAK suppresses EGFR signalling
Next, we evaluated whether the ability of FAK to inhibit RTK/MAPK signalling was specific to RET. The epithelial growth factor receptor (EGFR) is known to bind to FAK in mammals [29], [45] and to activate MAPK in Drosophila [46]. Therefore, we took a similar approach and co-expressed the dEGFR with dFAK in the eye or in the ptc domains through the GMR-gal4 and ptc-gal4 drivers, respectively. As it happened with dRET, dEGFR also induced dFAK (Figure S6A) but not Akt1 activation (Figure S6B–B′). Co-expression with dFAK resulted in a significant rescue of the GMR>dEGFR phenotype (Figure 7A and 7B) and inhibition of MAPK activation (Figure 7C–E). Moreover, dFAK allowed a remarkable increase in survival of ptc>dEGFR flies (Figure 7F).
Fig. 7. FAK suppression of RTK signalling is conserved. 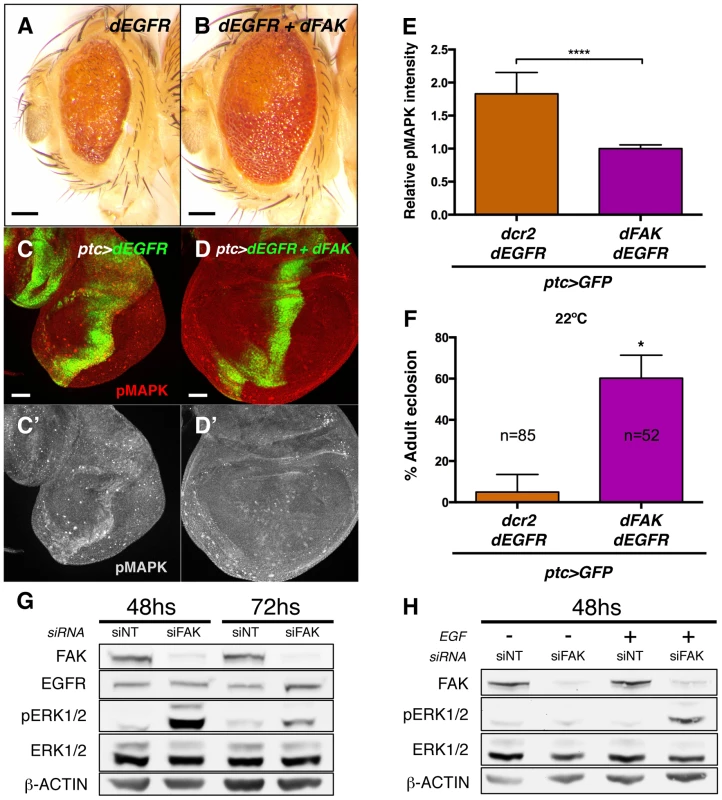
(A–B) Adult eyes images of animals expressing Drosophila EGFR (dEGFR) alone or in combination with dFAK. Note that dFAK expression suppressed the rough, small eye phenotype driven by dEGFR. Scale bar, 100 µm. (C–C′) Expression of dEGFR within the ptc domain resulted in increased MAPK phosphorylation, and co-expression of dFAK rescued the ectopic pMAPK staining within the ptc stripe (D–D′). Scale bar, 50 µm. (E) Quantification of pMAPK immunostaining within the ptc stripe (see methods). Intensity of pMAPK signal is represented as relative values to the mean intensity of control tissues (Figure 6A; **** = p<0.0001; n = 4–6 for each genotype). (F) Quantification of the penetrance on adult eclosion for the indicated genotypes. Note that dFAK co-expression significantly rescued the developmental lethality associated to ptc-driven dEGFR expression (* = p<0.05). (G) Western blots from protein extracts from MDA-MB-231 cells after 48 or 72 h transfection with FAK siRNA. FAK protein levels were effectively knocked down. While total levels of EGFR and ERK were not changed at 48 h, there was a marked upregulation in phosphorylated ERK1/2 upon FAK knockdown, which was more apparent at 48 h after siRNA transfection. Actin levels were probed as an additional loading control. (H) MDA-MB-231 cells were transfected with either non-targeting (siNT) or FAK-specific siRNA (siFAK) and serum starved prior to addition of EGF. Note that FAK knockdown resulted in increased phosphorylation of ERK1/2 in response to EGF treatment (30 µM, 15 minutes). Mechanistically, dFAK seems to act in a similar fashion with both RTKs as the mutant isoforms of dFAK showed the same pattern of suppression when co-expressed within a GMR>dEGFR context: the auto-phosphorylation mutant was still capable of rescuing (Figure S6F), while the amino-terminal domain mutant failed to modify the phenotype of dEGFR overexpression (Figure S6E). Thus, the FERM domain of dFAK appears essential to initiate the negative regulation over RTKs.
This implies that dFAK not only suppressed dRET but can also suppress other RTKs, namely dEGFR.
Phylogenetic conservation role for FAK downstream of RTK signalling
We next tested whether the negative role of FAK downstream of RTK signalling was evolutionary conserved. Since FAK inhibited dEGFR signalling, we utilized MDA-MB-231 cells, derived from a human breast adenocarcinoma, which express high levels of EGFR and FAK [47]–[50]. An efficient knockdown of the FAK protein was achieved using small interfering RNAs by 48 hours after transfection (Figure 7G and S7A). Remarkably, when cells were grown in presence of serum we observed an increase of ERK1/2 (MAPK) phosphorylation (pERK1/2) after FAK knockdown, while the total ERK1/2 and EGFR levels remained constant. ERK1/2 phosphorylation levels were also increased at 72 hs after transfection but at this later time point EGFR levels were mildly upregulated (Figure 7G and S7A).
When serum-starved FAK-siRNA MDA-MB-231 cells were treated with EGF in order to selectively stimulate EGFR receptor, the same dramatic increase of ERK (MAPK) activation was observed (Figure 7H and S7B). These results indicate that the suppressive role of FAK on RTK/MAPK signalling is evolutionary conserved through EGF/EGFR signalling in human breast cancer cells.
FAK reduces the fraction of EGFR located at the cell surface
To gain mechanistic insights into how FAK suppresses RTK/MAPK signalling, we next examined the cellular distribution of EGFR in MDA-MB-231 cells. Previous work indicated that growth factor receptors regulate cell signalling differently depending on its localization at the plasma membrane or within internalized vesicles [51]–[53]. In most contexts, EGFR signals to MAPK preferentially when located at the cell surface [54], [55]. Since knockdown of FAK at 48 h did not affect total levels of EGFR, we hypothesised that a change of receptor subcellular localization could explain the enhanced ERK signalling.
To assess whether FAK affects receptor subcellular localization, we performed immunofluorescence stainings for EGFR in control and FAK knockdown cells (Figure 8A and 8B). A 27% increase in the fraction of total EGFR located at the plasma membrane was observed in siFAK-treated cells, at the expense of the intracellular pool (Figure 8C). This suggested that the increase of EGFR at the cell surface could account for the increased ERK signalling in cells with reduced FAK levels. To further test this hypothesis, we experimentally increased the fraction of EGFR at the plasma membrane using the Dynamin GTPase inhibitor Dynasore, widely used to retain receptors at the cell surface [56]–[59]. This treatment phenocopied FAK knockdown as it led to increased pERK1/2 levels without affecting ERK1/2 or EGFR total levels (Figure 8D and S8).
Fig. 8. FAK decreases EGFR at the plasma membrane via reduced recycling. 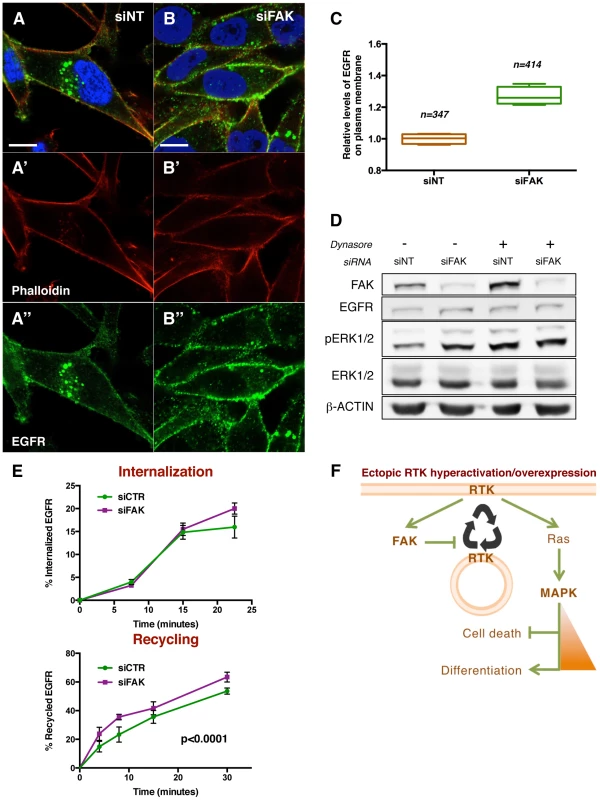
(A–B) MDA-MB-231 cells transfected with non-targeting (NT) siRNA or FAK siRNA were immunostained with anti-EGFR antibody (green, A″ and B″), Rhodamine-phalloidin (red, A′ and B′) and DAPI (blue). Note the differential localisation of EGFR; while in siNT cells the receptor is distributed in plasma membrane and internal vesicles, FAK downregulation leads to an increase of EGFR levels at the cellular membrane. Scale bar, 10 µm. (C) Quantification of relative EGFR membrane levels, values are expressed as relative levels of the receptor against the mean value of siNT cells; four confocal fields for each condition were analysed: n = 347 cells (siNT), and n = 414 cells (siFAK). p<0.0286 in a Mann-Whitney test. (D) MDA-MB-231 cells were transfected with either non-targeting (siNT) or FAK-specific siRNA (siFAK) and deprived of serum prior to addition of 80 µM Dynasore. siNT-transfected cells showed an increased pERK1/2 level in response to both Dynasore treatment (80 µM, 30 minutes) and FAK knockdown. Total levels of EGFR and ERK were not changed and actin levels were probed as an additional loading control. (E) The internalization of membrane EGFR (top panel) and recycling of internalised EGFR (bottom panel) were determined in MDA-MB-231 cells transfected with non-targeting siRNA (siCTR) or FAK siRNA (siFAK). Values are means ± Standard Deviation (SD) of two independent experiments with four to eight replicates of each time point per genotype. See materials and methods for more details. FAK knockdown did not affect receptor internalization but increased the recycling of the internalised EGFR pool. (F) A working model for the regulatory mechanism of FAK. Ectopic expression and/or hyperactivation of RTKs activate FAK and Ras among other signalling pathways. FAK mediates a negative regulation of receptor recycling; when FAK is reduced or absent, there are more RTKs molecules at the plasma membrane, thus enabling a higher flux of signalling through Ras/MAPK pathway. See the text for more details. Taken together, these results indicate that FAK disfavours localisation of EGFR at the plasma membrane thus reducing receptor signalling into the MAPK pathway.
FAK reduces EGFR recycling to the cell surface
Finally, we asked how FAK might affect the receptor sub-cellular localisation. Receptors are normally internalised into the endosomal/lysosomal compartment, when they can be either degraded or spared from degradation and returned to the cell surface [60].
We performed pulse-chase biochemical assays [61] to quantify the rates of EGFR internalisation and recycling. We did not detect significant differences in the rate of EGFR internalization (Figure 8E). Instead, the recycling of the internal EGFR pool was significantly higher (36%) in cells with reduced FAK versus control cells (Figure 8E). These results correlate with the percentages of membrane EGFR observed in Figure 8A–C and hence indicate that FAK suppresses EGFR localisation at the plasma membrane by suppressing receptor recycling but not internalization.
Discussion
This study provides evidence that RTK signalling can be moderated by FAK through the regulation of RTK recycling. This mechanism appears specific for the case of RTK over-expression/hyper-activation. We found that ectopic dRET and dEGFR signalling were able to activate dFAK and the MAPK pathway. Nevertheless, dFAK negatively regulated RTK-induced signalling to the MAPK pathway. The highly conserved FERM domain of dFAK appeared necessary for its functional suppression of Drosophila RTK signalling, which is consistent with the physical interactions of FAK and several RTKs described in mammals [28], [30], [62]. This negative feedback regulation indicates that the balance between RTK and FAK is what determines the intensity of MAPK pathway activity, which ultimately dictates cellular fate (Figure 8F).
We characterized cell fate outcomes in detail in the patterning eye anlage. In this tissue, it is known that different thresholds of MAPK pathway activation result in different outcomes: moderate levels of activation promote survival of cells during the wave of developmental apoptosis [63], while high levels of activation result in ectopic differentiation into photoreceptors [63] or the cone cell fate [64]. Correspondingly, we observed that experimental genetic manipulations expected to produce moderate RET/FAK ratios —such as the expression of one copy of wild type dRET in a dFAK wild type background — resulted in reduced developmental apoptosis and supernumerary interommatidial cells. On the other hand, genetically defined high RET/FAK ratios —such as the expression of one copy of wild type dRET in a dFAK mutant background, or the expression of two copies of constitutively activated dRET in a dFAK wild type background — resulted in ectopic differentiation into cone cells. Importantly, all dRET-driven phenotypes were suppressed by the co-overexpression of dFAK, which lowered RET/FAK ratios.
The initial characterization of the dRET-driven eye model identified several components of the Src and Ras/MAPK pathways [19]. These authors further observed that high levels of dRET signalling, achieved by the expression of two copies of activated RET, resulted in non-patterned retinas composed of identical cells with cuboidal morphology, which were proposed to be undifferentiated precursor cells. This led to the conclusion that high dRET signalling could block differentiation in this tissue. We report here that under similar experimental conditions, such cuboidal cells express the cone cell marker Cut. Thus, we conclude that high relative levels between dRET and dFAK result in a proportional activation of the MAPK pathway, which force differentiation into the cone cell fate.
Previous work reported that in dFAK mutant embryos, the activity of the MAPK pathway is normal during development [16]. This indicates that endogenous MAPK signalling does not normally became hyper-activated simply as a result of dFAK loss and is in sharp contrast to the case of over-expression of RET or EGFR as reported here. We postulate that in imaginal disc epithelia, dFAK constitutes a signalling “fuse” that can act in a negative feedback loop over RET and other RTKs, specifically in conditions of ectopic RTK activation or expression. Relevant to this, we recently reported that mammalian FAK can protect epithelial cells from deregulated RET or Src signalling; when FAK is absent, some epithelial cancer cells specifically respond by targeting these promiscuous oncogenic kinases for autophagic degradation [31], [65]. It therefore seems that a common feature of FAK regulatory function is to ‘buffer’ cells against potentially hazardous tyrosine kinases-mediated oncogenic signalling by promoting their internalization [31], [65].
In Drosophila, negative regulation of MAPK by dFAK has been previously observed in neuromuscular junction (NMJ) growth [16]; in fact, this was one of the few developmental defects detectable in dFAK mutant animals. Interestingly, it was suggested that this ‘non-canonical’ negative regulation of MAPK by dFAK was specific to the process of Integrin-dependent NMJ growth [16]. Importantly, our data imply instead that this is a more commonly used mechanism that occurs also in epithelial tissues, downstream of ectopic dRET and dEGFR signalling. Thus, the negative regulation of RTK/MAPK signalling by dFAK is more widespread across Drosophila tissues, and we show that this has important consequences for cell and tissue fate. Most importantly, we observed that the ability dFAK to restrain signalling through the MAPK pathway is evolutionary conserved in a human breast carcinoma cell line downstream of EGFR signalling. We demonstrate that this novel role of FAK relies on its ability to affect receptor sub-cellular localisation. RTKs normally reside at the plasma membrane or within internalised cytosolic vesicles. There is a constant transport of vesicles between these two pools, which allow cells to keep a healthy RTK signalling by degrading old receptor molecules or resetting their activity and sending them back to the cell surface. RTKs can activate a certain signalling pathway either from the plasma membrane or endocytic vesicles; here we showed that EGFR signals to Ras/MAPK pathway preferentially from the cell surface, and FAK is able to regulate its localisation by reducing recycling, while the internalization rates are unaffected. Thus, in a context where MAPK signalling is activated preferentially by the plasma membrane pool of EGFR [51]–[53], an increase of receptor located at the cell surface results in enhanced MAPK signalling.
We also present evidence that the N-terminal FERM domain of dFAK may be essential in the regulation of RTKs, while its kinase activity appears dispensable. These results highlighted the important regulatory roles of the FERM domain, likely by mediating interactions with RET and EGFR. Future studies should elucidate how FAK, presumably acting as a scaffold through its FERM domain, interacts with the recycling endosome machinery to control RTK recycling.
The attenuation of the MAPK signal transduction pathway by FAK is in stark contrast to the well-established role of FAK linking integrin engagement to the activation of Ras/MAPK [66]. Therefore, the regulation of MAPK by FAK may be context-dependent. It is worth noting that most previous studies reporting the activation of MAPK by FAK utilized immortalized cultured fibroblasts [66]; it is possible that FAK negative regulation of MAPK applies to epithelial cells in situ and acts downstream of RTKs but Integrins.
High expression or activation of FAK in a range of human carcinomas [49] and its role promoting migration and survival of malignant cells, make it an attractive therapeutic target. In fact, many small molecule inhibitors have been developed to target FAK kinase activity or its FERM domain, and clinical trials are in progress [67] [68]. However, in some tumors FAK downregulation has also been related with malignancy [69]–[72]. For instance, in glioblastoma, a malignancy that is associated with EGFR activation, there is an inverse correlation between pFAK and pMAPK [73], as predicted in our model.
Therefore, the signaling crosstalk between FAK and RTK/MAPK is complex and context-dependent [73], and a better understanding of such roles of FAK will be useful as FAK inhibitors move towards potential clinical use.
Materials and Methods
Drosophila stocks and culture
The following fly stocks were used: w1118 and Canton-S as a wild-type reference; GMR-gal4 and sev-gal4 as expression drivers in the eye [74]; patched (ptc)-gal4 and decapentaplegic (dpp)-gal4 as expression drivers in a stripe of cells in the wing and genital discs [34]; the dFAKCG1 mutant line, UAS-dFAK, UAS-dFAKΔN and UAS-dFAKY430F lines were made by R. Palmer [13], [17]; GMR-dRETWT, UAS-dRETC695R(MEN2A) and GMR-dRETC695R(MEN2A) were a gift from R. Cagan (Mount Sinai Medical School, New York, USA) [19]; dFAKKG00304, Ras85DRNAi, UAS-Ras85DV12, UAS-RafF179, GMR-hid, and UAS-dInR lines were obtained from the Bloomington Drosophila Stock Centre. Src42ARNAi line was obtained from the Vienna Drosophila RNAi Centre. dFAK5-SZ-3124 was obtained from the Drosophila Genetic Resource Centre. The UAS-dEGFR line was obtained from Matthew Freeman [75]. All flies were cultured at 25°C on standard molasses diet unless otherwise stated. Please find in Text S1 the detailed genotypes of animals used in all figures.
Overexpression using GAL4/UAS system
The GMR-gal4 line drives expression of UAS-linked transgenes in differentiating and post-mitotic cells of the developing eye, posterior to the morphogenetic furrow; the sevenless-gal4 line is active in R7 photoreceptor cells; the patched-gal4 and decapentaplegic-gal4 lines drive expression of UAS-linked transgenes within compartments of cells in imaginal discs.
Immunofluorescence assays
Eye and wing imaginal discs or pupal retinas were dissected at the indicated time points, in 1X PBS, fixed in 4% formaldehyde for 30 minutes at room temperature, and rinsed in PBST (PBS, 0.1% Triton X-100). Tissues were incubated in primary antibody at 4°C overnight. After PBST washing (3 times, 10 minutes each), tissues were incubated for 2 hours in secondary antibody. Tissues were rinsed in PBST and counterstained with DAPI (1 µg/ml, SIGMA) for 5 min at RT and then mounted in Vectashield. Primary antibodies used were anti-Armadillo (1∶3, DSHB) and anti-Cut (1∶50, DSHB), anti-phosphorylated (Y418)-Src (1∶100, Cell Signalling), anti-phosphorylated (Y397)-FAK (1∶100, Cell Signalling), anti-phosphorylated (T202/Y204)-MAPK (1∶200, Cell signalling), anti-Dlg (1∶50, DSHB), anti-Prospero (1∶30, DSHB), anti-Elav (1∶500, DSHB), anti-phosphorylated (S-473)-Akt1 (1∶100, Cell Signalling). Secondary antibodies were Alexa-conjugated 488, 594, or 633 (Molecular Probes). All preparations were analysed on a Zeiss 710 upright confocal microscope and images were processed with Fiji (ImageJ) program (NIH).
Quantification of immunofluorescence stainings
In order to quantify the intensity of immunostainings within the ptc compartment of the wing discs, sum projections were created from the original multi-slice files. Two identical areas were defined, within the ptc domain (A1) and posterior to it (A2). The ‘mean grayscale value’ of each area was measured with Image J, and the ratio between A1 and A2 was taken as the normalised intensity of signal.
TUNEL (Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP Nick End Labelling)
Eye discs or retinas were dissected and fixed in 4% formaldehyde in PBT for 20 min at RT. Samples were permeabilized in 100 mM Sodium Citrate in PBST (PBS+0.1% Triton X-100) at 65°C for 30 min followed by the addition of TUNEL reagent according to the manufacturer's instructions (In Situ Cell Death Detection Kit, Roche) and incubated at 37°C for 2 h on a rotating platform.
Light microscopy
Adult eye and male genitalia images were taken with a Leica M205 FA stereomicroscope equipped with Montage software. Wing blades images were taken with an Olympus BX51 FL Microscope.
RNA quantification
Total RNA was extracted from 10–15 heads or whole bodies per biological replicate using RNeasy kit (Qiagen) and converted into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). MAXIMA SYBR Green Master Mix (Fermentas) was used for quantitative (q) PCR. Data from three biological replicates were analyzed using Applied Biosystems 7500 software version 2.0 and GraphPad Prism 6 software. Data are presented as the mean fold change relative to wild type with standard deviations. Primers are listed in Text S1.
Statistical analysis
To statistically analyse eye size measurements, adult eclosion and immunofluorescence signal we used Student's unpaired t-test to compare two groups of data or One-way ANOVA followed by Bonferroni's or Tukey's post-test corrections to compare more than two groups of data. Error bars are standard deviation in all plots.
Cell culture and transfection
MDA-MB-231 cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 2 mM glutamine and 10% FBS (Fetal Bovine Serum) at 37°C in a humidified 5% CO2 atmosphere. siGENOME non-targeting (NT) siRNA pool (D-001206-13-05) and Smartpool siRNAs targeting FAK (L-003164-00) were obtained from Dharmacon, 10 ul of the 20 uM stock was used in each transfection. Non-targeting and FAK siRNAs were transfected into cells using Nucleofector Technology (Nucleofector Solution V, program X-013; Lonza) and Nucleofector II, Amaxa Biosystems.
Human cell imaging and analysis
MDA-MB-231 transfected cells were washed in ice cold PBS, fixed in 4% paraformaldehyde for 10 minutes at room temperature (RT), and permeabilised during 5 minutes in PBS+0.2% Triton X-100. Then, cells were blocked in 1% BSA/PBS solution for 30 minutes and incubated with primary antibody overnight. Secondary antibody was added to cells for 1 hour at RT and then cells were washed, incubated with FITC-Phalloidin for 10 minutes and finally mounted in Vectashield with DAPI.
Imaging was done with an inverted confocal microscope (FluoView FV1000; Olympus) with FluoView software (Olympus) and processed with Fiji (ImageJ) software (National Institute of Health). Immunofluorescence intensity values of EGFR were obtained by creating a mask of the cell outline, defining a threshold and measuring fluorescent intensity. Data analysis and Mann-Whitney statistical tests were performed and plotted in GraphPad Prism 6 software.
Western blotting
Assays were set up 48 or 72 hours after transfection. EGF treatment (30 uM, 15 minutes - Millipore) was performed on serum-starved cells, whereas all other experiments were conducted in the presence of serum. Each assay was independently repeated three times with similar results; the blots with the most equalized loading as judged by β-actin labelling are shown.
Cells were washed in PBS (Phosphate buffered saline) and lysed in lysis buffer (2% SDS, 100 mM Tris-HCl, pH 7.4). Cell protein extracts were incubated at 95°C for 5 minutes, sonicated and clarified by centrifugation at 10,000 g for 15 min. Protein concentration was determined by Bradford method [76]. Proteins were resolved by SDS-PAGE and analysed by Western blotting. The following antibodies were used for immunoblotting: anti-β-actin (1∶10000, Abcam), anti-FAK (1∶1000, C-20, Santa Cruz Biotechnology), anti-phosphorylated (T202/Y204)-MAPK (pERK1/2) (1∶200, Cell signalling), anti-ERK1/2 (1∶10000, Promega), anti-EGFR (1∶2000, BD Transduction Laboratories). The Nitrocellulose membranes were incubated with secondary antibodies IRDye 680RD anti-mouse (1∶10000, LI-COR) and IRDye 800CW anti-rabbit (1∶10000, LI-COR) imaged with Odyssey Imager (LI-COR Biosciences) and analysed with Image Studio Lite software.
Receptor internalization and recycling assays
EGFR internalization and recycling assays were performed as described previously for Integrins [61] [77], but without re-feeding or serum-starvation. For internalization assays: MDA-MD-231 cells were transferred to ice, washed twice in cold PBS, and their surface proteins labelled with 0.13 mg/ml NHS-SS-Biotin in PBS at 4°C for 1 h. Labelled cells were washed in cold PBS and transferred to serum-containing DMEM at 37°C for the indicated times to allow internalization. After internalization, dishes were rapidly transferred to ice and washed. Biotin remaining at cell surface was removed by incubation with 20 mM MesNa (sodium 2-mercaptoethanesulphonate), 50 mM Tris pH 8.6, 100 mM NaCl for 60 min at 4°C. MesNa was quenched by addition of 20 mM iodoacetamide for 20 min. Then, cells were washed and lysed in 200 mM NaCl, 75 mM Tris, 15 mM NaF, 1.5 mM Na3VO4, 7.5 mM EDTA, 7.5 mM EGTA, 1.5% Triton X-100, 0.75% IGEPAL CA-630, 50 µg/ml leupeptin, 50 µg/ml aprotinin, and 1 mM 4-(2-aminoethyl) benzynesulphonyl fluoride. Lysates were clarified by centrifugation at 10,000 g for 10 min, and biotinylated EGFR was determined by ELISA. Briefly, 96-well plates were coated overnight with monoclonal mouse anti–EGFR (clone EGFR.1; BD Biosciences) at 5 µg/ml at 4C° in 0.05 M Na2CO3 pH 9.6, and then blocked in PBS, 0.05% Tween 20, 5% BSA for 1 h at room temperature. EGF Receptors were captured by overnight incubation of 50 µl cell lysate at 4°C. After extensive washing with PBS-T to remove unbound material, wells were incubated with streptavidin-conjugated horseradish peroxidase for 1 h at 4°C. After further washing, biotinylated EGFR molecules were detected by chromogenic reaction with orthophenylenediamine.
For recycling assays, after surface labelling with Biotin, cells were transferred to serum-containing DMEM and incubated at 37°C for 30 min to allow internalization. Then, cells were returned to ice and washed in ice-cold PBS, and biotin was removed from cell surface proteins by reduction with MesNa. The internalized fraction was then chased by returning cells to 37°C in serum-containing DMEM. After recycling, cells were returned to ice, and biotin was removed from recycled proteins at the cell surface by a second reduction with MesNa. Biotinylated EGFR molecules were then determined by capture ELISA as described above. Two independent recycling experiments were performed with 4 to 8 independent measures of every time point and genotype. Two-way ANOVA with Sidak's multiple comparison test was performed to obtain statistical significance between time points.
Supporting Information
Zdroje
1. VidalM, CaganRL (2006) Drosophila models for cancer research. Curr Opin Genet Dev 16 : 10–16.
2. TenenbaumD (2003) What's All the Buzz? Fruit Flies Provide Unique Model for Cancer Research. J Natl Cancer Inst 95 : 1742–1744.
3. EdgarBA (2006) From cell structure to transcription: Hippo forges a new path. Cell 124 : 267–273.
4. KarimFD, ChangHC, TherrienM, WassarmanDA, LavertyT, et al. (1996) A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143 : 315–329.
5. DicksonBJ, van der StratenA, DominguezM, HafenE (1996) Mutations Modulating Raf signaling in Drosophila eye development. Genetics 142 : 163–171.
6. TherrienM, MorrisonDK, WongAM, RubinGM (2000) A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics 156 : 1231–1242.
7. RebayI, ChenF, HsiaoF, KolodziejPA, KuangBH, et al. (2000) A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154 : 695–712.
8. ParsonsJT (2003) Focal adhesion kinase: the first ten years. J Cell Sci 116 : 1409–1416.
9. GelmanIH (2003) Pyk 2 FAKs, any two FAKs. Cell Biol Int 27 : 507–510.
10. SiesserPM, HanksSK (2006) The signaling and biological implications of FAK overexpression in cancer. Clin Cancer Res 12 : 3233–3237.
11. ZhaoJ, GuanJL (2009) Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev 28 : 35–49.
12. FujimotoJ, SawamotoK, OkabeM, TakagiY, TezukaT, et al. (1999) Cloning and characterization of Dfak56, a homolog of focal adhesion kinase, in Drosophila melanogaster. J Biol Chem 274 : 29196–29201.
13. PalmerRH, FesslerLI, EdeenPT, MadiganSJ, McKeownM, et al. (1999) DFak56 is a novel Drosophila melanogaster focal adhesion kinase. J Biol Chem 274 : 35621–35629.
14. FoxGL, RebayI, HynesRO (1999) Expression of DFak56, a Drosophila homolog of vertebrate focal adhesion kinase, supports a role in cell migration in vivo. Proc Natl Acad Sci USA 96 : 14978–14983.
15. MurakamiS, UmetsuD, MaeyamaY, SatoM, YoshidaS, et al. (2007) Focal adhesion kinase controls morphogenesis of the Drosophila optic stalk. Development 134 : 1539–1548.
16. TsaiP-I, KaoH-H, GrabbeC, LeeY-T, GhoseA, et al. (2008) Fak56 functions downstream of integrin alphaPS3betanu and suppresses MAPK activation in neuromuscular junction growth. Neural development 3 : 26.
17. GrabbeC, ZervasCG, HunterT, BrownNH, PalmerRH (2004) Focal adhesion kinase is not required for integrin function or viability in Drosophila. Development 131 : 5795–5805.
18. UedaA, GrabbeC, LeeJ, LeeJ, PalmerRH, et al. (2008) Mutation of Drosophila focal adhesion kinase induces bang-sensitive behavior and disrupts glial function, axonal conduction and synaptic transmission. Eur J Neurosci 27 : 2860–2870.
19. ReadRD, GoodfellowPJ, MardisER, NovakN, ArmstrongJR, et al. (2005) A Drosophila model of multiple endocrine neoplasia type 2. Genetics 171 : 1057–1081.
20. VidalM, WellsS, RyanA, CaganR (2005) ZD6474 suppresses oncogenic RET isoforms in a Drosophila model for type 2 multiple endocrine neoplasia syndromes and papillary thyroid carcinoma. Cancer Res 65 : 3538–3541.
21. DasT, CaganR (2010) Drosophila as a novel therapeutic discovery tool for thyroid cancer. Thyroid 20 : 689–695.
22. JhiangSM (2000) The RET proto-oncogene in human cancers. Oncogene 19 : 5590–5597.
23. LeboulleuxS, BaudinE, TravagliJP, SchlumbergerM (2004) Medullary thyroid carcinoma. Clin Endocrinol (Oxf) 61 : 299–310.
24. GriecoM, SantoroM, BerlingieriMT, MelilloRM, DonghiR, et al. (1990) PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell 60 : 557–563.
25. BongarzoneI, ButtiMG, CoronelliS, BorrelloMG, SantoroM, et al. (1994) Frequent activation of ret protooncogene by fusion with a new activating gene in papillary thyroid carcinomas. Cancer Res 54 : 2979–2985.
26. SantoroM, RosatiR, GriecoM, BerlingieriMT, D'AmatoGL, et al. (1990) The ret proto-oncogene is consistently expressed in human pheochromocytomas and thyroid medullary carcinomas. Oncogene 5 : 1595–1598.
27. BoulayA, BreuleuxM, StephanC, FuxC, BriskenC, et al. (2008) The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res 68 : 3743–3751.
28. ChenSY, ChenHC (2006) Direct interaction of focal adhesion kinase (FAK) with Met is required for FAK to promote hepatocyte growth factor-induced cell invasion. Mol Cell Biol 26 : 5155–5167.
29. SiegDJ, HauckCR, IlicD, KlingbeilCK, SchaeferE, et al. (2000) FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol 2 : 249–256.
30. Plaza-MenachoI, MorandiA, MologniL, BoenderP, Gambacorti-PasseriniC, et al. (2011) Focal adhesion kinase (FAK) binds RET kinase via its FERM domain, priming a direct and reciprocal RET-FAK transactivation mechanism. J Biol Chem 286 : 17292–17302.
31. SandilandsE, SerrelsB, WilkinsonS, FrameMC (2012) Src-dependent autophagic degradation of Ret in FAK-signalling-defective cancer cells. EMBO Rep 13 : 733–740.
32. CorderoJ, JassimO, BaoS, CaganR (2004) A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech Dev 121 : 1523–1530.
33. DarAC, DasTK, ShokatKM, CaganRL (2012) Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486 : 80–84.
34. SpeicherSA, ThomasU, HinzU, KnustE (1994) The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development 120 : 535–544.
35. GleichaufR (1936) Anatomie und Variabilitat des Geschlechtapparates von Drosophila melanogaster (Meigen).. ZWissZool 148 : 1–66.
36. AdamG, PerrimonN, NoselliS (2003) The retinoic-like juvenile hormone controls the looping of left-right asymmetric organs in Drosophila. Development 130 : 2397–2406.
37. FrameMC, PatelH, SerrelsB, LiethaD, EckMJ (2010) The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol 11 : 802–814.
38. ZhaoX, PengX, SunS, ParkAY, GuanJL (2010) Role of kinase-independent and -dependent functions of FAK in endothelial cell survival and barrier function during embryonic development. J Cell Biol 189 : 955–965.
39. SawamotoK, TaguchiA, HirotaY, YamadaC, JinMH, et al. (1998) Argos induces programmed cell death in the developing Drosophila eye by inhibition of the Ras pathway. Cell Death Differ 5 : 262–270.
40. BergmannA, AgapiteJ, McCallK, StellerH (1998) The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95 : 331–341.
41. KuradaP, WhiteK (1998) Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95 : 319–329.
42. GretherME, AbramsJM, AgapiteJ, WhiteK, StellerH (1995) The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 9 : 1694–1708.
43. BlochlingerK, JanLY, JanYN (1993) Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development 117 : 441–450.
44. VerduJ, BuratovichMA, WilderEL, BirnbaumMJ (1999) Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol 1 : 500–506.
45. LongW, YiP, AmazitL, LamarcaHL, AshcroftF, et al. (2010) SRC-3Delta4 Mediates the Interaction of EGFR with FAK to Promote Cell Migration. Molecular cell 37 : 321–332.
46. Diaz-BenjumeaFJ, HafenE (1994) The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development 120 : 569–578.
47. CorkeryB, CrownJ, ClynesM, O'DonovanN (2009) Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol 20 : 862–867.
48. PriceJT, TiganisT, AgarwalA, DjakiewD, ThompsonEW (1999) Epidermal growth factor promotes MDA-MB-231 breast cancer cell migration through a phosphatidylinositol 3′-kinase and phospholipase C-dependent mechanism. Cancer Res 59 : 5475–5478.
49. OwensLV, XuL, CravenRJ, DentGA, WeinerTM, et al. (1995) Overexpression of the focal adhesion kinase (p125FAK) in invasive human tumors. Cancer Res 55 : 2752–2755.
50. AgochiyaM, BruntonVG, OwensDW, ParkinsonEK, ParaskevaC, et al. (1999) Increased dosage and amplification of the focal adhesion kinase gene in human cancer cells. Oncogene 18 : 5646–5653.
51. SigismundS, ArgenzioE, TosoniD, CavallaroE, PoloS, et al. (2008) Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell 15 : 209–219.
52. MiaczynskaM, ChristoforidisS, GinerA, ShevchenkoA, Uttenweiler-JosephS, et al. (2004) APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116 : 445–456.
53. SadowskiL, PileckaI, MiaczynskaM (2009) Signaling from endosomes: location makes a difference. Exp Cell Res 315 : 1601–1609.
54. MarshallCJ (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80 : 179–185.
55. IrwinME, MuellerKL, BohinN, GeY, BoernerJL (2011) Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol 226 : 2316–2328.
56. MaciaE, EhrlichM, MassolR, BoucrotE, BrunnerC, et al. (2006) Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10 : 839–850.
57. MesakiK, TanabeK, ObayashiM, OeN, TakeiK (2011) Fission of tubular endosomes triggers endosomal acidification and movement. PLoS One 6: e19764.
58. HenriksenL, GrandalMV, KnudsenSL, van DeursB, GrovdalLM (2013) Internalization mechanisms of the epidermal growth factor receptor after activation with different ligands. PLoS One 8: e58148.
59. RizzolioS, RabinowiczN, RaineroE, LanzettiL, SeriniG, et al. (2012) Neuropilin-1-dependent regulation of EGF-receptor signaling. Cancer Res 72 : 5801–5811.
60. RaineroE, NormanJC (2013) Late endosomal and lysosomal trafficking during integrin-mediated cell migration and invasion: cell matrix receptors are trafficked through the late endosomal pathway in a way that dictates how cells migrate. Bioessays 35 : 523–532.
61. RobertsM, BarryS, WoodsA, van der SluijsP, NormanJ (2001) PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol 11 : 1392–1402.
62. ChenT-H, ChanP-C, ChenC-L, ChenH-C (2011) Phosphorylation of focal adhesion kinase on tyrosine 194 by Met leads to its activation through relief of autoinhibition. Oncogene 30 : 153–166.
63. HalfarK, RommelC, StockerH, HafenE (2001) Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development 128 : 1687–1696.
64. MatsuoT, TakahashiK, KondoS, KaibuchiK, YamamotoD (1997) Regulation of cone cell formation by Canoe and Ras in the developing Drosophila eye. Development 124 : 2671–2680.
65. SandilandsE, SerrelsB, McEwanDG, MortonJP, MacagnoJP, et al. (2012) Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol 14 : 51–60.
66. SchlaepferDD, HanksSK, HunterT, van der GeerP (1994) Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature 372 : 786–791.
67. CanceWG, KurenovaE, MarloweT, GolubovskayaV (2013) Disrupting the scaffold to improve focal adhesion kinase-targeted cancer therapeutics. Sci Signal 6: pe10.
68. InfanteJR, CamidgeDR, MileshkinLR, ChenEX, HicksRJ, et al. (2012) Safety, pharmacokinetic, and pharmacodynamic phase I dose-escalation trial of PF-00562271, an inhibitor of focal adhesion kinase, in advanced solid tumors. J Clin Oncol 30 : 1527–1533.
69. GabrielB, zur HausenA, StickelerE, DietzC, GitschG, et al. (2006) Weak expression of focal adhesion kinase (pp125FAK) in patients with cervical cancer is associated with poor disease outcome. Clin Cancer Res 12 : 2476–2483.
70. AyakiM, KomatsuK, MukaiM, MurataK, KameyamaM, et al. (2001) Reduced expression of focal adhesion kinase in liver metastases compared with matched primary human colorectal adenocarcinomas. Clin Cancer Res 7 : 3106–3112.
71. OhtaR, YamashitaY, TaketomiA, KitagawaD, KurodaY, et al. (2006) Reduced expression of focal adhesion kinase in intrahepatic cholangiocarcinoma is associated with poor tumor differentiation. Oncology 71 : 417–422.
72. LuZ, JiangG, Blume-JensenP, HunterT (2001) Epidermal growth factor-induced tumor cell invasion and metastasis initiated by dephosphorylation and downregulation of focal adhesion kinase. Mol Cell Biol 21 : 4016–4031.
73. ZhengY, LuZ (2009) Paradoxical roles of FAK in tumor cell migration and metastasis. Cell Cycle 8 : 3474–3479.
74. HayBA, MaileR, RubinGM (1997) P element insertion-dependent gene activation in the Drosophila eye. Proc Natl Acad Sci U S A 94 : 5195–5200.
75. FreemanM (1996) Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87 : 651–660.
76. BradfordMM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 : 248–254.
77. RaineroE, CaswellPT, MullerPA, GrindlayJ, McCaffreyMW, et al. (2012) Diacylglycerol kinase alpha controls RCP-dependent integrin trafficking to promote invasive migration. J Cell Biol 196 : 277–295.
Štítky
Genetika Reprodukčná medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



