-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
Higher plants are able to sense and interpret diverse light signals to modulate their growth. In response to long-wavelength and low-intensity ultraviolet-B (UV-B) light, plants establish photomorphogenic development and stress acclimation. UV RESISTANCE LOCUS 8 (UVR8) is a unique UV-B photoreceptor that triggers photomorphogenesis in Arabidopsis thaliana. However, the signaling process following UV-B light perception by plants is not fully understood. In this study, by generating transgenic UVR8 variants in Arabidopsis, we have extensively analyzed the biological significance of key residues in UVR8 for UV-B-induced photomorphogenesis. Furthermore, by engineering and characterizing two constitutively active UVR8 variants, we have provided the biochemical insight that the in vivo association between UVR8 and CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) critically determines the photomorphogenic UV-B signaling output.
Published in the journal: Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in. PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004218
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004218Summary
Higher plants are able to sense and interpret diverse light signals to modulate their growth. In response to long-wavelength and low-intensity ultraviolet-B (UV-B) light, plants establish photomorphogenic development and stress acclimation. UV RESISTANCE LOCUS 8 (UVR8) is a unique UV-B photoreceptor that triggers photomorphogenesis in Arabidopsis thaliana. However, the signaling process following UV-B light perception by plants is not fully understood. In this study, by generating transgenic UVR8 variants in Arabidopsis, we have extensively analyzed the biological significance of key residues in UVR8 for UV-B-induced photomorphogenesis. Furthermore, by engineering and characterizing two constitutively active UVR8 variants, we have provided the biochemical insight that the in vivo association between UVR8 and CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) critically determines the photomorphogenic UV-B signaling output.
Introduction
Light is a critical environmental stimulus that regulates a number of developmental and physiological processes of living organisms. In sessile plants, perception of light is the initial and decisive step in light signaling transduction, and is achieved by several groups of photosensory receptor proteins. Phytochromes sense far-red and red light [1], [2]. Cryptochromes and phototropins perceive blue and ultraviolet (UV)-A light [3], [4], [5], [6]. In Arabidopsis thaliana, UV RESISTANCE LOCUS 8 (UVR8) has recently been identified as a photoreceptor that detects UV-B (280 to 320 nm) light [7]. Long-wavelength and low-fluence UV-B induces plant photomorphogenic development that is physically characterized by the inhibition of hypocotyl elongation, flavonoid accumulation, and UV-B stress tolerance [8], [9], [10], [11].
UVR8 was originally isolated as a UV-resistance gene, having been shown to contribute to the UV-B-induced flavonoid accumulation and UV-B protection [12]. Transcriptomic analyses have revealed that UVR8 positively orchestrates UV-B signaling specifically under photomorphogenic UV-B [13]. Later, a series of functional studies have disclosed that UVR8 exhibits a number of features characteristic of photoreceptors, including a broad-range loss of UV-B responsive gene expression in the uvr8 null mutant [13], [14], the enrichment of aromatic residues in UVR8 protein and UV-B-induced conformational change of UVR8 [7].
Despite these insights, however, the exact process by which UVR8 mediates UV-B light perception remained unclear until two research groups independently reported the structure of UVR8 [15], [16]. Without the presence of UV-B light, UVR8 appears as a symmetric seven-bladed-β-propeller homodimer that is stabilized by arginines primarily Arg 286 and Arg 338. These arginine residues shape intramolecular cation-π interactions with their surrounding tryptophan residues, among which Trp 285 and Trp 233 act as the internal UV-B chromophore. Consequently, unlike phytochromes and cryptochromes, UVR8 is devoid of external small molecules as chromophore. Upon UV-B irradiation, the dark-state dimer of UVR8 is monomerized as a result of the disruption of the intramolecular cation-π interactions and the intermolecular hydrogen bonds mediated by Arg 286 and Arg 338 [7], [15], [16]. This structural conversion, which takes place in seconds, is a major determinant for UVR8 to sequester CONSTITUTIVELY PHOTOMORPHOGENIC 1-SUPPRESSOR OF PHYA (COP1-SPA) core complex(es) from the CULLIN 4-DAMAGED DNA BINDING PROTEIN 1 (CUL4-DDB1) E3 apparatus. Ultimately, this complex reorganization enables COP1 to act as a positive regulator in the UV-B-induced photomorphogenesis by facilitating the stability and activity of a photomorphogenesis-promoting transcription factor ELONGATED HYPOCOTYL 5 (HY5) [7], [17]. Reversibly, upon the elimination of UV-B irradiation, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2, two UVR8-interacting proteins, might disrupt the physical contact of UVR8 and COP1, so that UVR8 dimerization can be regenerated [7], [15], [16], [18], [19], [20]. However, the exact biological significance of key residues in UVR8 has not been fully determined to date.
Here we take advantage of site-directed mutagenesis to generate UVR8 variant proteins in Arabidopsis, and demonstrate the pivotal roles of two light-absorbing tryptophans, W233 and W285, and two dimer-stabilizing arginines, R286 and R338 in UVR8-initiated UV-B-induced photomorphogenesis. We also characterize two constitutively active forms of UVR8, UVR8W285A and UVR8R338A, whose photobiological activity is enhanced when the repressor CUL4 is suppressed. Overall, our molecular and biochemical evidence has supported that the intrinsic affinity of UVR8-COP1 critically determines the efficiency of photomorphogenic UV-B signal transduction coupling with UVR8-mediated UV-B light perception.
Results
UVR8 variants differentially interact with COP1 in yeast
In order to assess the roles of key residues in UVR8, we generated six UVR8 variants in two groups based on their functional classification via site-directed mutagenesis. The first group, which included UVR8W233A, UVR8W233F, UVR8W285A and UVR8W285F, and the second group, which included UVR8R286A and UVR8R338A, were designed to interrupt UVR8's perception of UV-B light and dimer stabilization respectively (Figure S1A). As yeast has been widely used as an efficient system to determine the conformational status of UVR8 and its interaction with other proteins in response to UV-B [7], [21], [22], we first introduced wild-type and these six mutated UVR8 (mUVR8) into yeast. Using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblot analyses, we found that UVR8WT was dimeric under −UV-B and monomeric under +UV-B (Figure 1A), while UVR8W233F, UVR8W285A and UVR8W285F were constitutively monomeric, monomeric and dimeric respectively (Figure 1A). This is consistent with the results reported previously [7]. The other UVR8 variants, UVR8W233A, UVR8R286A and UVR8W338A appeared as monomers irrespective of UV-B treatment (Figure 1A). These results demonstrate that in yeast, in addition to the dimer-stabilizing arginines, R286 and R338, the light-absorbing tryptophans, W233 and W285, critically contribute to the dimer-to-monomer switch of UVR8 upon UV-B irradiation.
Fig. 1. UVR8 variants display altered interaction with COP1 in yeast. 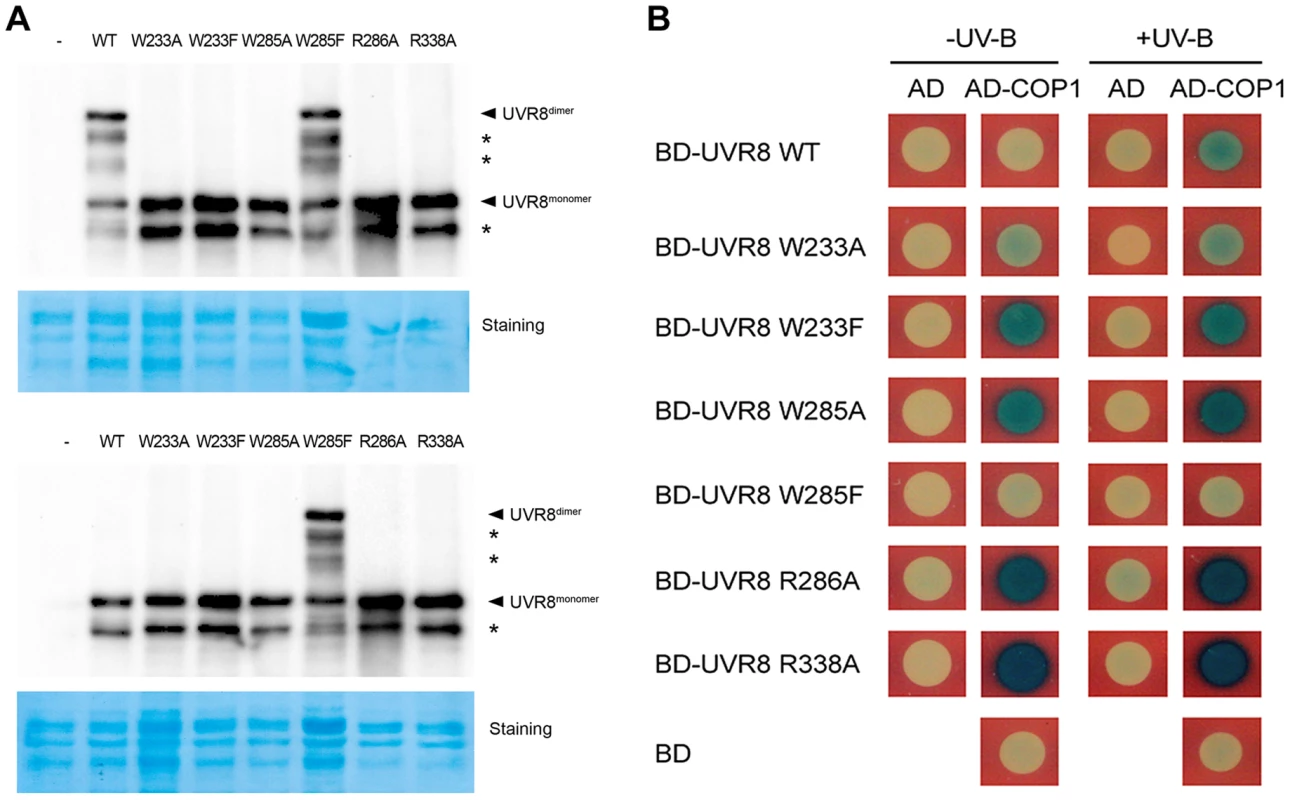
(A) Conformational status of wild-type and mutated UVR8 proteins in yeast. Total proteins of yeast expressing LexA fused wild-type and mutated UVR8 were extracted and incubated under −UV-B and +UV-B for 20 min. Protein samples without heat denaturation were assayed in SDS-PAGE and immunoblot analysis by anti-LexA antibody. Staining by Coomassie brilliant blue (CBB) is shown as a loading control. The asterisks indicate unspecific degradation products. (B) Interaction of wild-type and mutated UVR8 proteins with COP1 in yeast two-hybrid assays. Transformants in the respective combinations were incubated under −UV-B and +UV-B for 16 h. We next performed a series of yeast two-hybrid assays in order to examine the effects of the UVR8 mutations on the interaction between UVR8 and COP1. While COP1 only interacted with UVR8WT under +UV-B, it interacted with UVR8W233A, UVR8W233F, UVR8W285A, UVR8R286A and UVR8R338A under both −UV-B and +UV-B. The interaction between UVR8W285F and COP1 was barely observed (Figure 1B). These results suggest that the UVR8-COP1 interaction in yeast requires UVR8 to be in its monomeric form. Furthermore, previous studies have also proposed that RUP1 and RUP2 interact with UVR8 independent of UV-B irradiation [19]. Specifically, it has been suggested that these two proteins mediate UVR8 redimerization and disrupt UVR8-COP1 interaction, so as to facilitate the inactivation of the photoreceptor [18]. In yeast, RUP1 was found to constitutively interact with UVR8W233A, UVR8W233F, UVR8W285A and UVR8R286A, but was not observed to interact with UVR8WT, UVR8W285F and UVR8R338A. Differently, RUP2 was observed to interact with the wild-type UVR8 and all the mutant UVR8 proteins (Figure S1B). These results collectively suggest that light perception and conformational change of UVR8 are both major determinants of its interaction with key players of UV-B-induced photomorphogenesis.
UVR8 variants alter physiological responses to photomorphogenic UV-B in Arabidopsis
To investigate the biological activity of these UVR8 variants in Arabidopsis, we introduced wild-type and mutated UVR8 fused with yellow fluorescent protein (YFP) driven by the native UVR8 promoter into the uvr8 null mutant uvr8-6 background. As expected, proUVR8-YFP-UVR8/uvr8-6 (YFP-UVR8WT) was found to express the transgenic UVR8 protein at a level comparable to the endogenous UVR8 protein in Col under −UV-B and +UV-B (Figure S2A). It was able to rescue uvr8-6 in UV-B-induced hypocotyl growth and anthocyanin accumulation (Figures 2A, 2B and 2C) which are characteristic physiological responses to photomorphogenic UV-B [14], [23]. We next examined these two responses in the six transgenic UVR8 variants expressing YFP-UVR8 proteins at equivalent levels to that in YFP-UVR8WT (Figure 2D). The hypocotyl shortening in all the variants failed to reach the shortening degree detected in YFP-UVR8WT. Interestingly, YFP-UVR8W285A and YFP-UVR8R338A displayed shorter hypocotyl than YFP-UVR8WT under −UV-B, while YFP-UVR8R338A instead of YFP-UVR8W285A showed further shortened hypocotyl under +UV-B (Figures 2A and 2B). Furthermore, compared with YFP-UVR8WT, reduced anthocyanin accumulation was found in YFP-UVR8W233A, YFP-UVR8W233F, YFP-UVR8W285F and YFP-UVR8R286A under both −UV-B and +UV-B. In contrast, enhanced anthocyanin accumulation was detected in YFP-UVR8W285A and YFP-UVR8R338A under −UV-B. YFP-UVR8R338A was even able to accumulate anthocyanin under +UV-B, though at a level lower than that in YFP-UVR8WT (Figure 2C).
Fig. 2. UVR8 variants result in impaired UV-B-induced photomorphogenesis. 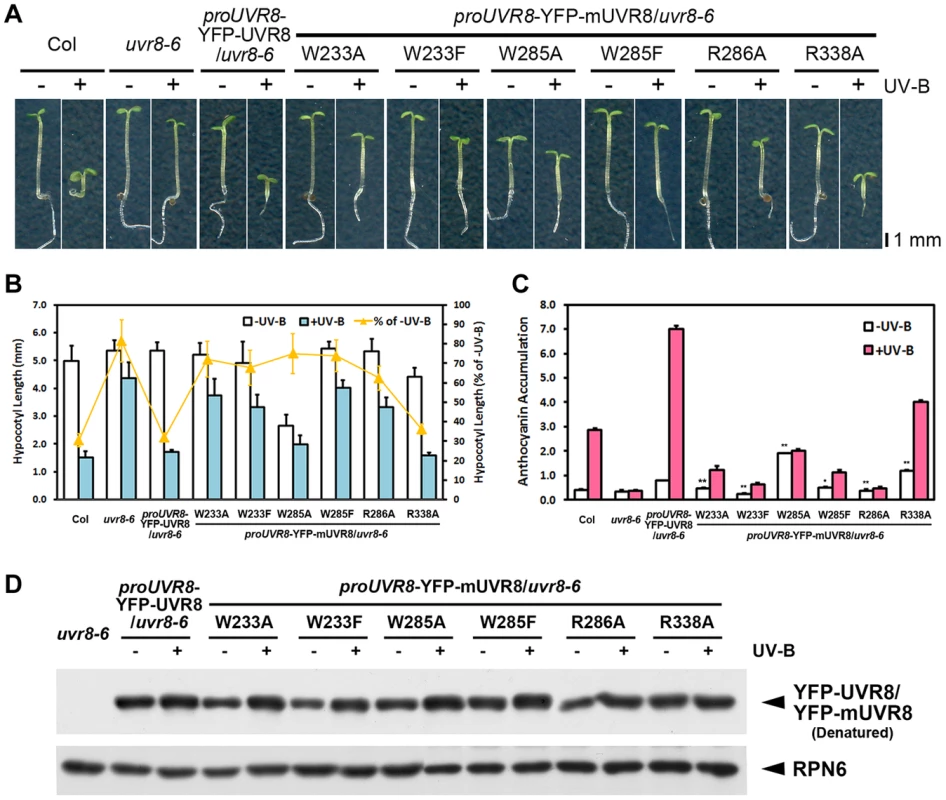
(A) Phenotypes of 4-day-old seedlings of transgenic UVR8 variant lines grown under −UV-B and +UV-B. (B) Hypocotyl length of the seedlings shown in (A). Data are shown as mean ± SD; n>30. (C) Anthocyanin content of the seedlings shown in (A). Data are shown as mean ± SD; n = 3. The difference significance of UVR8 variants from wild-type UVR8 in anthocyanin content under −UV-B was analyzed by Student's t test. *, p<0.05. **, p<0.01. (D) Immunoblot assay of wild-type and mutated UVR8 proteins (by anti-GFP antibody) in 4-day-old transgenic seedlings grown under −UV-B and +UV-B. Anti-RPN6 was used as a loading control. These observations showed that all the mutations interfered with UV-B-induced photomorphogenesis, though to varying degrees. The mutations in UV-B-absorbing residues resulted in more severe phenotypic defects than those in UVR8-dimerizing residues, suggesting the importance of the sequential action of UV-B light perception before UVR8 monomerization. Meanwhile, a specific activity of UVR8W285A and UVR8R338A is indicated by the constitutive short hypocotyl and high anthocyanin content found in YFP-UVR8W285A and YFP-UVR8R338A (Figures 2B and 2C).
UVR8 mutations affect UV-B-responsive gene expression
It is well known that differential transcriptomic regulation is orchestrated by photomorphogenic UV-B in Arabidopsis [9], [13]. Using 4-day-old seedlings grown under −UV-B and +UV-B, we examined the expression pattern of several UV-B-responsive marker genes, EARLY LIGHT-INDUCIBLE PROTEIN 2 (ELIP2), UDP-GLYCOSYLTRANSFERASE 84A1 (UGT84A1) and CHALCONE SYNTHASE (CHS). The UV-B-induced activation of these genes observed in YFP-UVR8WT was largely impaired in YFP-UVR8W233A, YFP-UVR8W233F, YFP-UVR8W285A, YFP-UVR8W285F and YFP-UVR8R286A, while the activation was readily detected in YFP-UVR8R338A though it showed a reduced induction of UGT84A1. Again, we noticed YFP-UVR8W285A and YFP-UVR8R338A as these two variants accumulated higher transcript levels of these genes than YFP-UVR8WT under −UV-B (Figures 3A, 3B and 3C).
Fig. 3. UVR8 mutations lead to abnormal UV-B-responsive gene expression. 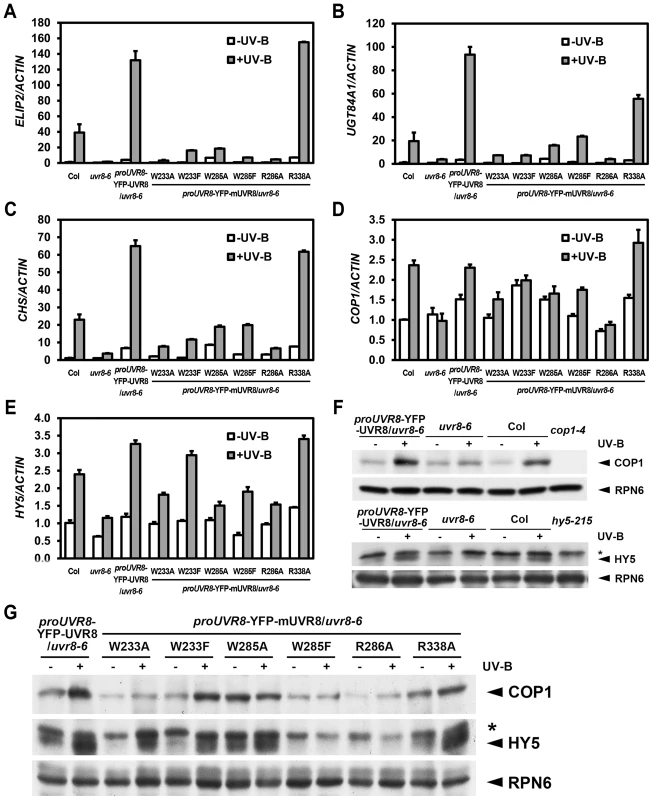
(A–E) qRT-PCR analysis of UV-B-responsive gene expression ELIP2 (A), UGT84A1 (B), CHS (C), COP1 (D) and HY5 (E) in 4-day-old seedlings grown under −UV-B and +UV-B. Data are shown as mean ± SD; n = 3. (F–G) Immunoblot assay of COP1 and HY5 proteins (by anti-COP1 and anti-HY5 antibodies) in 4-day-old seedlings grown under −UV-B and +UV-B. Anti-RPN6 was used as a loading control. The asterisk indicates an unspecific cross-reactive band. In addition to these marker genes, key regulators of UV-B specific signaling, including the positive regulators COP1 and ELONGATED HYPOCOTYL 5 (HY5) are also transcriptionally governed by photomorphogenic UV-B [24], [25]. We found that the accumulation of COP1 mRNA was diminished in all UVR8 variants with the exception of YFP-UVR8R338A (Figure 3D). Similarly, though the accumulation of HY5 mRNA was observed in YFP-UVR8W233F and YFP-UVR8R338A, it was limited in YFP-UVR8W233A, YFP-UVR8W285A, YFP-UVR8W285F and YFP-UVR8R286A (Figure 3E). At the post-transcriptional level, COP1 protein was clearly induced in YFP-UVR8WT, YFP-UVR8W233F and YFP-UVR8R338A, slightly increased in YFP-UVR8W233A under +UV-B, and relatively high under both −UV-B and +UV-B in YFP-UVR8W285A. However, the accumulation of COP1 protein mediated by photomorphogenic UV-B was scarcely detected in YFP-UVR8W285F and YFP-UVR8R286A (Figures 3F and 3G). A similar pattern was observed for the case of HY5 protein accumulation (Figures 3F and 3G). Taken together, the altered expression profiles of these UV-B responsive genes suggest that these residues responsible for UV-B light perception and UVR8 dimerization are crucial for UVR8 activity in the transcriptional control of UV-B signaling at a molecular level.
Downstream of UVR8-COP1, RUP1 and RUP2 contribute to the negative feedback regulation of UV-B-induced photomorphogenesis. Given that RUP1 and RUP2 are known to be induced by photomorphogenic UV-B dependent on UVR8, COP1 and HY5 [19], it was noteworthy that the accumulation of RUP1 and RUP2 mRNA was apparently reduced in all UVR8 variants except UVR8R338A (Figures 4A and 4B). As early UV-B responsive genes, RUP1 and RUP2 were activated in a temporal manner in response to photomorphogenic UV-B to balance UV-B specific signaling [19]. Using 4-day-old seedlings grown under −UV-B and then transferred to +UV-B for various periods of time, we found that the temporal induction of RUP1 and RUP2 by photomorphogenic UV-B was apparently observed in YFP-UVR8WT, but was retarded in all the UVR8 variant lines (Figures 4C and 4D). It is worth pointing out that the transcript levels of RUP1 and RUP2 were elevated within 1 hours of UV-B irradiation to a peak and then fell back in YFP-UVR8WT, whereas they continued to rise to a lower peak particularly in YFP-UVR8W285A, YFP-UVR8R286A and YFP-UVR8R338A over the 12-hour UV-B treatment (Figures 4C and 4D). These results suggest that RUP1 and RUP2 fail to be activated in our UVR8 variants, and thus do not establish the repressive transcriptional modules required for balanced UV-B signaling. Overall, none of the UVR8 variants are functionally equivalent to YFP-UVR8WT, which was consistent with our phenotypic observations.
Fig. 4. UVR8 variants show defective transcriptional regulation of RUP1 and RUP2 by photomorphogenic UV-B. 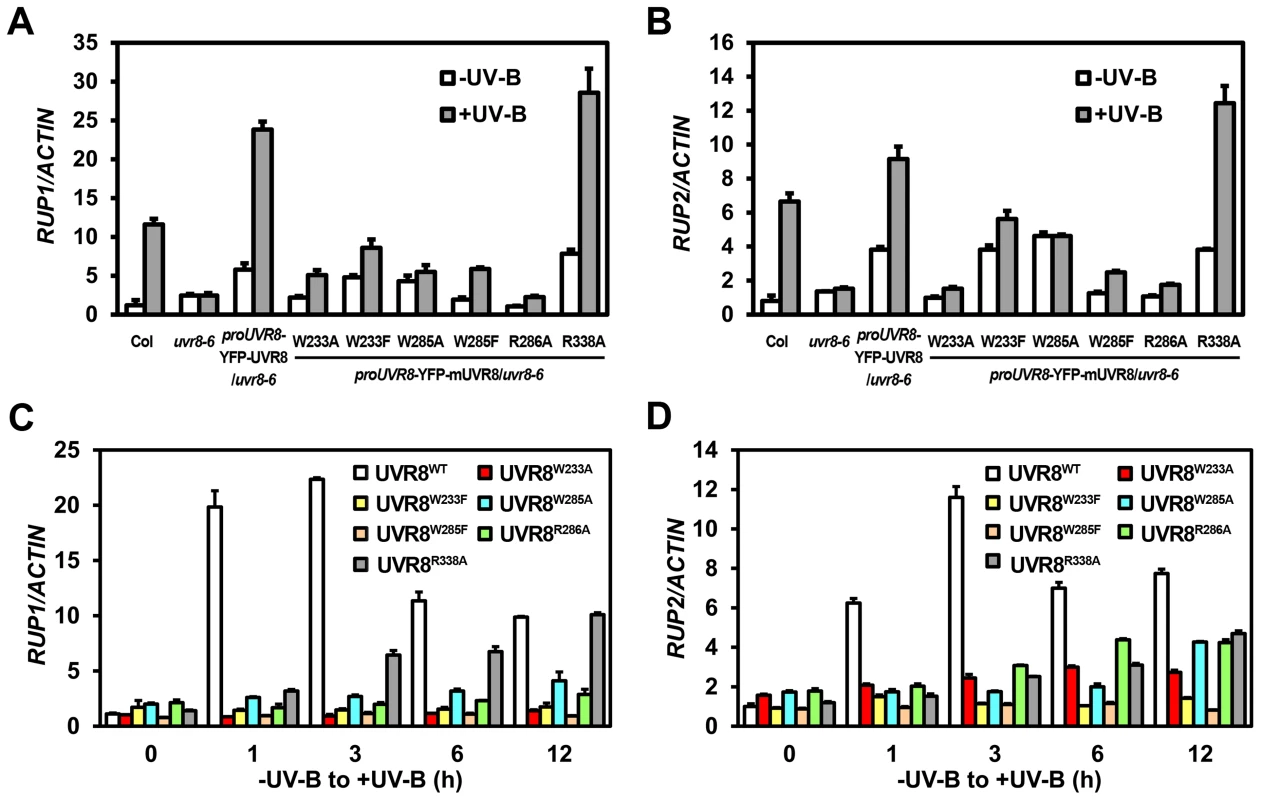
(A–B) qRT-PCR analysis of RUP1 (A) and RUP2 (B) in 4-day-old seedlings grown under −UV-B and +UV-B. Data are shown as mean ± SD; n = 3. (C–D) qRT-PCR analysis of RUP1 (C) and RUP2 (D) in 4-day-old seedlings transferred from −UV-B to +UV-B and harvested at indicated time points. Data are shown as mean ± SD; n = 3. UVR8 variants impair UV-B light perception, UVR8 monomerization and UVR8-COP1 association
In order to better understand the biochemical activity of these residues in vivo, we investigated the efficiency of UV-B light perception, UVR8 monomerization and the formation of UVR8-containing complex in all of our UVR8 variant plants. By measuring UV-B absorbance at 310 nm, the central wavelength of our photomorphogenic UV-B condition, we found that Col and YFP-UVR8WT exhibited a strong ability to sense UV-B. In contrast, mutations in either residue of the internal chromophore, W233 or W285, led to a complete loss of UV-B absorption. YFP-UVR8R286A and YFP-UVR8R338A retained the ability to sense UV-B, but at significantly diminished levels, which suggests that the disruption of the homodimeric interface has a negative impact on the full activity of UVR8 to perceive UV-B light (Figure 5A).
Fig. 5. UVR8 mutations affect UV-B light perception, UVR8 monomerization and UVR8-COP1 association. 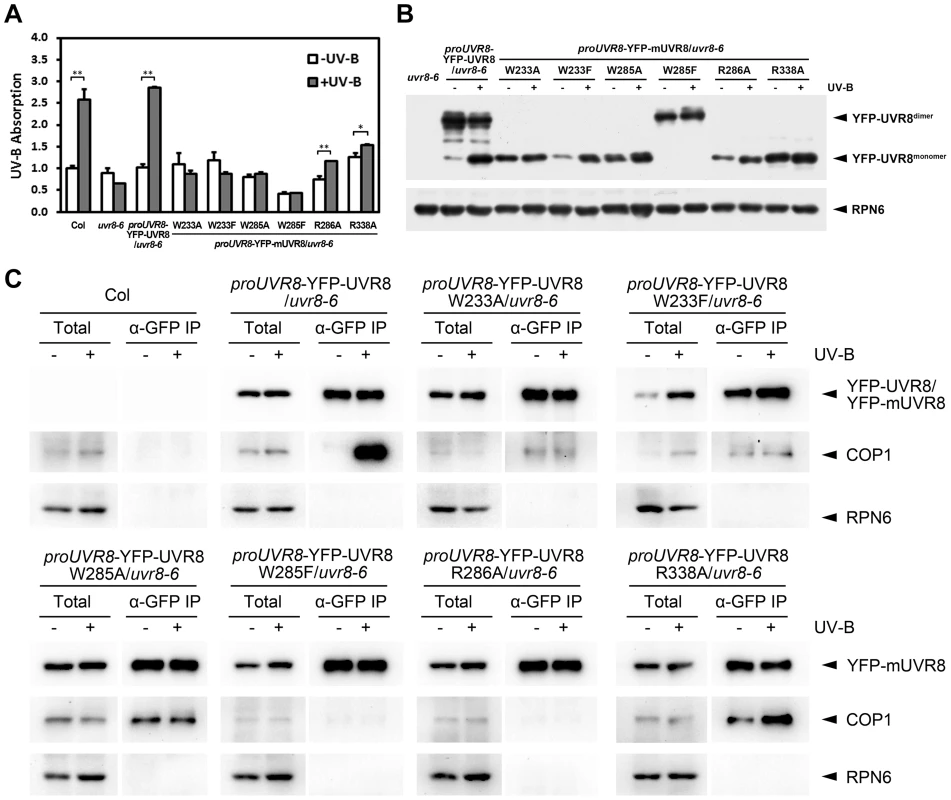
(A) Absorbance at 310 nm of plant total proteins extracted from 4-day-old seedlings grown under −UV-B and +UV-B. Data are shown as mean ± SD; n = 3. *, p<0.05. **, p<0.01. Student's t test. (B) Conformational status of wild-type and mutated UVR8 proteins in 4-day-old seedlings grown under −UV-B and +UV-B. Total plant proteins without heat denaturation were assayed in SDS-PAGE and immunoblot analysis by anti-GFP antibody. Anti-RPN6 was used as a loading control. The asterisks indicate unspecific degradation products. (C) In vivo co-immunoprecipitation (co-IP) assays using 4-day-old seedlings grown under −UV-B and +UV-B by anti-GFP antibody. Immunoblot analysis was performed by anti-GFP and anti-COP1 antibodies. Anti-RPN6 was used as a un-immunoprecipitated and loading control. In SDS-PAGE analysis, we found that in response to photomorphogenic UV-B, while YFP-UVR8WT switched from dimer to monomer, none of YFP-UVR8 variant proteins showed a comparable conformational change. YFP-UVR8W233A, YFP-UVR8W233F, YFP-UVR8W285A, YFP-UVR8R286A and YFP-UVR8R338A were monomeric, while YFP - UVR8W285F was dimeric (Figure 5B). Thus the conformational profiles of wild-type and variant UVR8 observed in Arabidopsis were consistent with those found in yeast.
We next examined the endogenous association of UVR8 variants and COP1 by using in vivo co-immunoprecipitation (co-IP) assays. In agreement with previous studies [7], [14], YFP-UVR8WT co-immunoprecipitated a high level of COP1 specifically under +UV-B. However, independent of UV-B, YFP-UVR8W285F and YFP-UVR8R286A scarcely co-immunoprecipitated COP1, while YFP-UVR8W233F and YFP-UVR8W233A co-immunoprecipitated very low levels of COP1. YFP-UVR8W285A and YFP-UVR8R338A co-immunoprecipitated medium levels of COP1 under −UV-B, while the latter was also observed to co-immunoprecipitate more COP1 under +UV-B (Figure 5C). These results reveal that the monomeric conformation is not sufficient for UVR8 to associate with COP1 in vivo. The relatively close association of YFP-UVR8W285A and YFP-UVR8R338A with COP1 without UV-B treatment indicates that a specific activity of UVR8 might be produced by W285A or R338A mutation.
Constitutive activity of UVR8 is dependent on constant UVR8-COP1 interaction
As both cop1-4 and uvr8-6 suffer from diminished gene expression, hypocotyl growth, anthocyanin accumulation and acclimation in response to photomorphogenic UV-B, COP1 and UVR8 share a high degree of functional similarity in photomorphogenic UV-B signaling [14], [25]. We found that YFP-UVR8WT/cop1-4 phenocopied cop1-4 (Figure 6A), clearly demonstrating that COP1 acts genetically downstream of UVR8. The observation that YFP-UVR8WT failed to rescue cop1-4 (Figure 6A) also suggests that the function of UVR8 is dependent on COP1. Though CUL4 works in concert with COP1 in darkness, it functionally disassociates from COP1 and plays a negative role in UV-B-induced photomorphogenesis [17]. Since cul4cs exhibited no obvious defect in hypocotyl growth under UV-B [17], we consistently found that YFP-UVR8WT/cul4cs phenocopied YFP-UVR8WT and cul4cs (Figure 6A).
Fig. 6. UVR8W285A and UVR8R338A display constitutive activity in darkness. 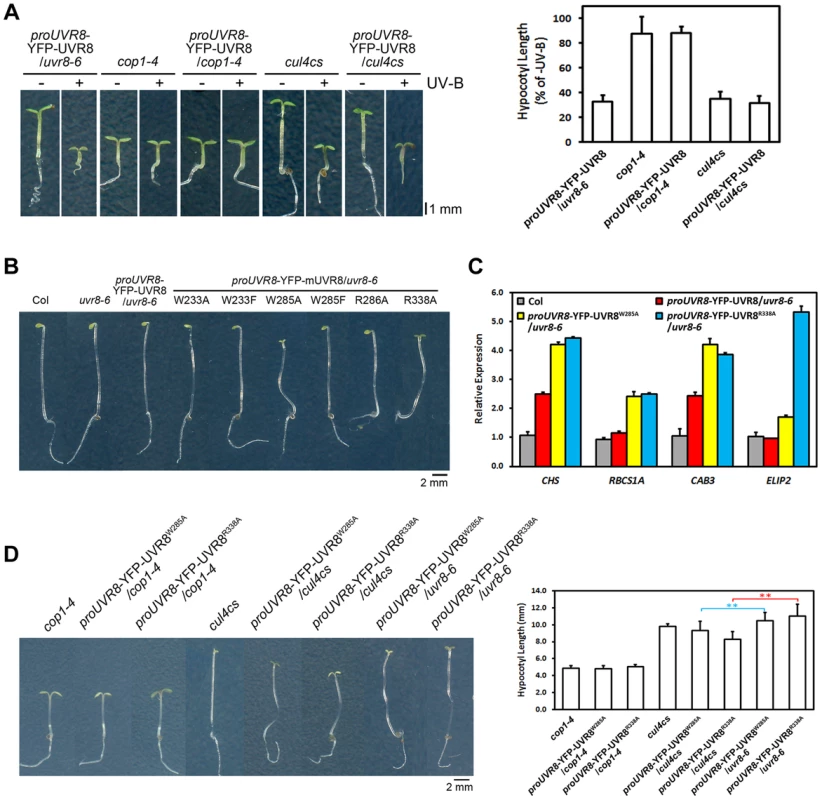
(A) Phenotypes and relative hypocotyl length of 4-day-old seedlings of indicated genotypes grown under −UV-B and +UV-B. Data are shown as mean ± SD; n>30. (B) Phenotypes of 4-day-old dark-grown seedlings. Data are shown as mean ± SD; n>30. (C) qRT-PCR analysis of light-regulated gene expression in 4-day-old dark-grown seedlings of indicated genotypes. Data are shown as mean ± SD; n = 3. (D) Phenotypes and hypocotyl length of 4-day-old dark-grown seedlings of indicated genotypes. Data are shown as mean ± SD; n>30. **, p<0.01. Student's t test. We have previously proposed that the UVR8-COP1 interaction mediated by UV-B enables a reorganization of COP1 complexes and eventually results in a functional switch of COP1 from a repressor to a promoter of photomorphogenesis [17]. Both the enhanced photomorphogenesis of YFP-UVR8W285A and YFP-UVR8R338A under −UV-B (Figures 2A, 2B and 2C) and the persistent binding of UVR8W285A and UVR8R338A (Figures 1B and 5C) to COP1 prompted us to examine the development of all the UVR8 variants in darkness. In addition to Col, uvr8-6 and YFP-UVR8WT, YFP-UVR8W233A, YFP-UVR8W233F, YFP-UVR8W285F and YFP-UVR8R286A all showed typical skotomorphogenic responses, demonstrating long hypocotyls, closed cotyledons and apical hooks. In contrast, YFP-UVR8W285A and YFP-UVR8R338A displayed open cotyledons, suggesting that these two UVR8 variants were capable of inducing constitutive photomorphogenesis irrespective of their exposure to light (Figure 6B). We then examined light-regulated gene expression, and found highly accumulated transcripts of CHS, RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN 1A (RBCS1A), CHLOROPHYLL A/B BINDING PROTEIN 3 (CAB3) and ELIP2, in YFP-UVR8W285A and YFP-UVR8R338A (Figure 6C). Though UVR8W233A, UVR8W233F and UVR8R286A had constitutive physical interactions with COP1 in yeast (Figure 1B), their affinity with COP1 in vivo was much weaker than that of UVR8W285A and UVR8R338A (Figure 5C), indicating that conversion of COP1's function might require a threshold level of UVR8-COP1 interaction.
We have also pointed out that in response to UV-B, monomerized UVR8 might sequester COP1 from the CUL4-DDB1 based E3 apparatus [17]. In darkness, compared with YFP-UVR8W285A/uvr8-6 and YFP-UVR8R338A/uvr8-6, YFP-UVR8W285A/cop1-4 and YFP-UVR8R338A/cop1-4 mimicked cop1-4 (Figure 6D), which is consistent with our conclusion that UVR8 functions in a COP1-dependent manner. YFP-UVR8W285A/cul4cs and YFP-UVR8R338A/cul4cs exhibited enhanced constitutive photomorphogenesis with decreased hypocotyl length (Figure 6D). These results indicate that the reduced CUL4 protein abundance might facilitate the release of an increased amount of COP1 from CUL4-DDB1 to allow an improved association between UVR8 and COP1, and in turn achieve a highly switched function of COP1 in promoting photomorphogenesis.
Discussion
Light-absorbing tryptophans and dimer-stabilizing arginines are intrinsically coordinated to fulfill UVR8 function
The molecular framework of UV-B-induced photomorphogenesis has been gradually established over the past ten years. For example, both the identification of UVR8 as a UV-B photoreceptor and the subsequent structural analysis of recombinant UVR8 have recently led to a proposed mechanism of UVR8-dependent UV-B signaling initiation [7], [15], [16]. A series of UVR8 variant proteins have been generated to further demonstrate that UVR8 exploits its own light-absorbing tryptophans and dimer-stabilizing arginines to perceive light and initiate protein conformational changes, respectively [15], [16]. UVR8W233F, UVR8W285A and UVR8W285F are lack of the ability to perceive UV-B in vitro [16]. UVR8W285A is at least partially monomeric and interacts with COP1. UVR8W285F, on the other hand, is dimeric and unable to interact with COP1 in yeast, plant and mammalian cells [7], [15], [16], [22], [26]. UVR8R286A and UVR8R338A are monomeric and display an obviously diminished perception of UV-B light in vitro [16]. In Arabidopsis, mutations in UVR8's tryptophan residues have been pointed out to result in the physical impairment of photomorphogenic UV-B responses [22]. However, the exact mechanism driving this hindered response remains unknown. Moreover, it is far less understood concerning the biological significance of light-absorbing and dimer-stabilizing residues in UVR8, as well as the signaling process that connects UV-B light perception, UVR8 monomerization and subsequent signaling events including the organization of UVR8-COP1-SPA complex(es).
In our study, we generated transgenic plants expressing UVR8 variant proteins under UVR8's native promoter, based on the site-directed mutagenesis of two light-absorbing tryptophans and two dimer-stabilizing arginines. These variant proteins drove varied levels of UV-B-induced photomorphogenesis. All the variants were observed to impair both UVR8's activity to perceive UV-B light and its ability to undergo a dimer-to-monomer transition (Figures 5A and 5B), suggesting the reciprocal impacts by these residues involved in the same intramolecular interaction network. More severe phenotypic defects were observed in UV-B-absorbing variants than those in UVR8-dimerizing variants (Figures 2A, 2B and 2C), confirming that UV-B light perception precedes UVR8 monomerization to launch UV-B signaling. In addition, the unchangeable conformational status of the UVR8 variants (Figure 5B) abolished the dimer-monomer-dimer cycling of UVR8, and further disturbed the balance in UV-B signaling. Molecularly, all the mutations caused an altered hierarchy of UV-B responsive gene expression (Figures 3 and 4). They failed to accurately establish the promotive module formed by the UVR8-COP1-HY5 core pathway and the negative transcriptional feedback mediated by RUP1 and RUP2, leading to inadequate and unbalanced photomorphogenic UV-B responses. Taken together, the roles of light-absorbing tryptophans and dimer-stabilizing arginines in UVR8 are intrinsically coordinated for UVR8 activity in UV-B-induced photomorphogenesis.
A threshold UVR8-COP1 interaction plays a critical role in coupling with UV-B light perception to signal transduction
The direct interaction between UVR8 and COP1 takes place rapidly following UV-B light perception and UVR8 monomerization [7]. It requires UVR8 to be in its monomeric form [7] whereas physically monomeric UVR8 is not sufficient for the formation of the UV-B-dependent COP1 complex and to mediate photomorphogenesis in response to UV-B in plants (Figures 2A and 5C). Though this point has been previously articulated [22], the molecular mechanism underlying this empirical observation is still unknown. This discrepancy serves to indicate that factors downstream of UV-B light perception and UVR8 monomerization might essentially govern the progression of UV-B signaling.
In terms of the transcriptome reprogramming induced by photomorphogenic UV-B, cop1-4 resulted in the loss of transcriptional responses of a broader range of genes than uvr8-6 [14]. While UVR8's ability to mediate UV-B-induced photomorphogenesis was observed to be dependent on COP1 (Figure 6A), COP1 did not appear to significantly influence UVR8 conformation [7], [20]. These data collectively indicate that COP1 is at least as essential as UVR8 in photomorphogenic UV-B signaling, if not superior to UVR8, due to its role in the formation of UVR8-COP1-SPA complex(es). By using the native promoter of UVR8 to drive UVR8 variants in plants, we selected those transgenic lines expressing comparable levels of UVR8 variant proteins (Figure 2D), in order to stringently analyze each variant in parallel, particularly to examine the in vivo association intensity between UVR8 and COP1 (Figure 5C). As a result, we propose that a threshold level of the in vivo association between UVR8 and COP1 is critically required for photomorphogenic UV-B signaling output, founded on the following evidence. Firstly, UVR8 monomers, rather than UVR8-COP1 interaction, were detected in −UV-B-treated plant and yeast cells expressing wild-type UVR8 (Figures 1, 5B and 5C). Secondly, the in vivo levels of UVR8-COP1 association correspond with the physiological and molecular features of the transgenic UVR8 variant lines (Figures 2A and 5C). Only those UVR8 variants that possess high affinity with COP1 in plants, namely YFP-UVR8WT under +UV-B, and YFP-UVR8W285A and YFP-UVR8R338A under −UV-B and +UV-B in our study, result in photobiological activity in specific light contexts (Figures 5C, 2A and 6B). Thirdly, once the affinity between UVR8 and COP1 is conditionally increased, such as the situation in cul4cs that might release more COP1 for UVR8 to interact with, the photobiological activity of UVR8 is ultimately enhanced (Figure 6D). In agreement with our hypothesis, a most recent report has presented that comparing with the overexpressed wild-type UVR8 (UVR8-OX), UVR8W285A leads to improved photomorphogenesis and UV-B tolerance by increased COP1 binding affinity, though UVR8W285A and UVR8-OX express equivalent UVR8 proteins [27].
Though our UVR8 variants do not demonstrate equivalent patterns of interaction with COP1 in yeast and plants (Figures 1B and 5C), this discrepancy can most likely be explained by the fact that the yeast two-hybrid system is devoid of any other factors that might influence the UVR8-COP1 interaction. The variation in the UVR8 variants' affinity for COP1 observed in plants may be due to a wide variety of factors. For example, UVR8 monomer variants might undergo protein folding processes in plants that are distinct from those in yeast, given that each mutated residue locates specifically in the intramolecular interaction network (Figure S1A). Similarly, the protein levels of endogenous COP1 vary amongst the different UVR8 monomer variants (Figure 3G). Finally, a number of the other proteins present in vivo might interact with COP1 and/or UVR8 in manner that either enhances or hinders the in vivo contact of UVR8 and COP1. Further research is required, however, in order to fully disentangle and elucidate the impact of the great complexity in vivo.
Constitutive light signaling is mediated by UVR8W285A and UVR8R338A
Two constitutively active forms of UVR8, UVR8W285A and UVR8R338A have been uncovered in our study. Though they differed in the phenotypic features of UV-B-induced photomorphogenesis, they both displayed constitutive interaction with COP1 (Figure 5C) and photomorphogenic development in darkness (Figure 6B). It is worth pointing out that a previous report did not find constitutive photomorphogenesis in GFP-UVR8W285A [22] whereas a recent independent study reached a consensus with ours by showing the constitutive photomorphogenesis in UVR8W285A [27].
Structure of dimeric UVR8 has revealed that W285, the UV-B chromophore, is located at the center of the strong cation-π and π-π interaction network at the dimer interface, while R338, responsible for dimer stabilization, is located at the edge of the interaction network [16]. It is suggested that W285 is more essential to the function of UVR8 than R338. Hence it is reasonable that YFP-UVR8W285A failed to respond to photomorphogenic UV-B while YFP-UVR8R338A was still able to, in terms of UV-B light perception, UV-B-induced UVR8-COP1 association and gene expression, and eventually photomorphogenic development (Figures 2A–C, 3A–C, 5A and 5C). On the other hand, in the absence of UV-B, YFP-UVR8W285A and YFP-UVR8R338A associated with a sufficient amount of COP1, and thus promoted hypocotyl growth, anthocyanin accumulation and gene activation under −UV-B (Figures 2A–C, 3A–C and 5C), and even photomorphogenesis in darkness (Figures 6B and 6C).
As reported, other gain-of-function alleles of photoreceptors were also produced via point mutations in their photosensory regions, such as the GAF domain tyrosine mutants of phytochromes (PHY), PHYAY242H and PHYBY276H [28], and the photolyase-related (PHR) domain glycine mutants of cryptochromes (CRY), CRY1G380R, CRY2G377R [29]. PHYBY276H was proposed to interact with COP1 in a light-independent manner to diminish the degradation of HY5 by COP1 [28]. CRY1G380R was found to co-localize in nucleus with COP1 to promote the nuclear exclusion of COP1 in darkness [29]. In contrast to its role in the repression of photomorphogenesis induced by far-red and visible light [30], COP1 is known to promote UV-B-induced photomorphogenesis [25]. Upon UV-B irradiation, COP1 rapidly interacts with UVR8 in nucleus [7], and switches from degrading to stabilizing HY5 [17], [31]. The constitutive photomorphogenesis in YFP-UVR8W285A and YFP-UVR8R338A is not resulted from a loss in COP1 protein abundance that was observed in cop1-4 (Figure S3). Therefore, the way UVR8 modulates COP1 dramatically differs from the activity repression of COP1 by cryptochromes or phytochromes. Furthermore, UVR8 is involved in diverse developmental processes in plants. Photomorphogenic UV-B signaling displays crosstalk with circadian regulation [32], and it also controls leaf morphogenesis [33], [34], drought tolerance [35] and plant immune response [36]. Beyond the area of plant researches, UVR8 has also been implemented in optical control of protein interactions [26], [37] and multi-chromatic expression regulation [38] in mammalian cells. Thus, the characterization of constitutively active UVR8 variants should serve to elucidate the mechanism of UV-B specific signaling in plants and advance protein engineering pertinent to a variety of medical applications.
Materials and Methods
Plant materials and growth conditions
The wild-type Arabidopsis thaliana used in this study is of the Columbia (Col) ecotype. Some of the mutants and transgenic lines used in this study were described previously: cop1-4 [39], uvr8-6 [14], cul4cs [40], proUVR8-YFP-UVR8/uvr8-6, proUVR8-YFP-UVR8W285A/uvr8-6 and proUVR8-YFP-UVR8W285F/uvr8-6 [17].
The vectors for UVR8 variant transgenic lines, proUVR8-YFP-mUVR8/uvr8-6, were generated using the QuikChange site-directed mutagenesis kit (Stratagene). The primers are listed in Table S1. These transgenic lines were prepared using floral dipping method [41].
The Arabidopsis materials were grown as described previously [24]. The seeds were surface-sterilized and sown on solid Murashige and Skoog medium supplemented with 1% sucrose for biochemical assays or with 0.3% sucrose for phenotypic analysis, and cold treated at 4°C for 4 days. For photomorphogenic UV-B treatment, seedlings were grown at 22°C under continuous white light (3 µmol·m−2·s−1, measured by LI-250 Light Meter, LI-COR Biosciences) supplemented with Philips TL20W/01RS narrowband UV-B tubes (1.5 µmol·m−2·s−1, measured by TN-340 UV-B Light Meter, China) under a 350-nm cutoff (half-maximal transmission at 350 nm) filter ZUL0350 (−UV-B; Asahi spectra, USA) or a 300-nm cutoff (half-maximal transmission at 300 nm) filter ZUL0300 (+UV-B; Asahi spectra, USA).
UVR8 dimer/monomer assay
For the assay in yeast, the vectors of LexA fused wild-type and mutated UVR8 were transformed into the yeast strain EGY48 (Clontech). Total proteins were extracted from transformants in Yeast Protein Extraction Reagent (Thermo), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× complete protease inhibitor cocktail (Roche), and then kept on ice under −UV-B (3 µmol·m−2·s−1 of white light) or +UV-B (3 µmol·m−2·s−1 of white light and 1.5 µmol·m−2·s−1 of UV-B) for 20 min. Added with 4× loading buffer containing 250 mM Tris-HCl (pH 6.8), 2% SDS, 20% β-mercaptoethanol, 40% glycerol, and 0.5% bromophenol blue, the samples were subjected to immunoblot analysis without boiling.
The assay in plants was performed as previously described [17]. Total proteins were extracted from 4-day-old Arabidopsis seedlings grown under −UV-B or +UV-B in protein extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× complete protease inhibitor cocktail (Roche). Then the cell extracts were kept on ice under exactly the same condition (−UV-B or +UV-B) as where the seedlings were grown for 30 min. Added with 4× loading buffer containing 250 mM Tris-HCl (pH 6.8), 2% SDS, 20% β-mercaptoethanol, 40% glycerol, and 0.5% bromophenol blue, the samples were subjected to immunoblot analysis without boiling.
Yeast two-hybrid assay
The respective combinations of vectors were cotransformed into the yeast strain EGY48 (Clontech) containing the reporter plasmid p8op::LacZ. Transformants were grown under −UV-B (3 µmol·m−2·s−1 of white light) and +UV-B (3 µmol·m−2·s−1 of white light and 1.5 µmol·m−2·s−1 of UV-B) on proper dropout plates containing X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) for blue color development.
Hypocotyl and anthocyanin measurement
Hypocotyl length was measured as previously described [24]. For each line grown under −UV-B or +UV-B for 4 days, hypocotyl length was analyzed in three biological replicates. In each replicate, at least 30 Arabidopsis seedlings were measured. The relative hypocotyl length was presented as the percentage of the hypocotyl length under +UV-B with respect to that under −UV-B (% of −UV-B). For each line grown in darkness, hypocotyl length was analyzed using at least 30 Arabidopsis seedlings. The quantification of hypocotyl length was performed by ImageJ (http://rsb.info.nih.gov/ij/).
Anthocyanin was extracted and quantified as previously described [42]. Briefly, Arabidopsis seedlings were harvested and placed into extraction solution (18% 1-propanol and 1% HCl), and boiled for 3 minutes. Then the mixture was left in darkness for at least 3 hours at room temperature. After a brief centrifugation to pellet the tissue debris, the supernatant was removed and diluted with the extraction solution. The anthocyanin content was presented as A535−2(A650) g−1 fresh weight.
Quantitative real-time PCR
Total RNA was extracted from 4-day-old Arabidopsis seedlings grown under −UV-B or +UV-B using the RNeasy plant mini kit (Qiagen). Reverse transcription was performed using SuperScript II first-strand cDNA synthesis system (Invitrogen) according to the manufacturer's instructions. Real-time qPCR analysis was performed using SYBR Premix Ex Taq (Takara) with Applied Biosystems 7500 Real-Time PCR System. Each experiment was repeated with three independent samples, and RT-PCR reactions were performed in three technical replicates for each sample. The primers are listed in Table S1.
Measurement of UV-B absorbance
Total proteins was extracted from 4-day-old Arabidopsis seedlings grown under −UV-B and +UV-B in protein extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM PMSF, and 1× complete protease inhibitor cocktail (Roche). Absorbance at 310 nm of plant total proteins adjusted to equal concentration and total amount were measured. Each experiment was repeated with three independent samples.
Co-immunoprecipitation assays and immunoblot analysis
1 mg of total proteins was extracted from 4-day-old Arabidopsis seedlings in protein extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween 20, 1 mM PMSF, and 1× complete protease inhibitor cocktail (Roche). The extracts were incubated with 8 µl anti-GFP antibodies (Invitrogen) coupled with 25 µl Dynabeads Protein G (Invitrogen) for 3 hours at 4°C under the same condition (−UV-B or +UV-B) as where the seedlings were grown. Then the dynabeads were washed three times by protein extraction buffer. Next the precipitates were eluted into 100 mM Glycine (pH 2.5) and 100 mM NaCl, and immediately neutralized by 2 M Tris-HCl (pH 9.0) and 100 mM NaCl, and finally concentrated using Strataresin (Stratagene) before immunoblot analysis. Primary antibodies used in this study were anti-COP1 and anti-RPN6 [40], anti-HY5 [31], anti-GFP (Invitrogen) and anti-UVR8 [17] antibodies.
Supporting Information
Zdroje
1. QuailPH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3 : 85–93.
2. ChenM, ChoryJ (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21 : 664–671.
3. CashmoreAR, JarilloJA, WuYJ, LiuD (1999) Cryptochromes: blue light receptors for plants and animals. Science 284 : 760–765.
4. BriggsWR, ChristieJM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7 : 204–210.
5. LiuH, LiuB, ZhaoC, PepperM, LinC (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16 : 684–691.
6. ChristieJM (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58 : 21–45.
7. RizziniL, FavoryJJ, CloixC, FaggionatoD, O'HaraA, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332 : 103–106.
8. FrohnmeyerH, StaigerD (2003) Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol 133 : 1420–1428.
9. UlmR, NagyF (2005) Signalling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol 8 : 477–482.
10. HectorsK, PrinsenE, De CoenW, JansenMA, GuisezY (2007) Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol 175 : 255–270.
11. JenkinsGI (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60 : 407–431.
12. KliebensteinDJ, LimJE, LandryLG, LastRL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130 : 234–243.
13. BrownBA, CloixC, JiangGH, KaiserliE, HerzykP, et al. (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A 102 : 18225–18230.
14. FavoryJJ, StecA, GruberH, RizziniL, OraveczA, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28 : 591–601.
15. ChristieJM, ArvaiAS, BaxterKJ, HeilmannM, PrattAJ, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335 : 1492–1496.
16. WuD, HuQ, YanZ, ChenW, YanC, et al. (2012) Structural basis of ultraviolet-B perception by UVR8. Nature 484 : 214–219.
17. HuangX, OuyangX, YangP, LauOS, ChenL, et al. (2013) Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci U S A 110 : 16669–16674.
18. HeijdeM, UlmR (2013) Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci U S A 110 : 1113–1118.
19. GruberH, HeijdeM, HellerW, AlbertA, SeidlitzHK, et al. (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci U S A 107 : 20132–20137.
20. HeilmannM, JenkinsGI (2013) Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol 161 : 547–555.
21. CloixC, KaiserliE, HeilmannM, BaxterKJ, BrownBA, et al. (2012) C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci U S A 109 : 16366–16370.
22. O'HaraA, JenkinsGI (2012) In vivo function of tryptophans in the Arabidopsis UV-B photoreceptor UVR8. Plant Cell 24 : 3755–3766.
23. GardnerG, LinC, TobinEM, LoehrerH, BrinkmanD (2009) Photobiological properties of the inhibition of etiolated Arabidopsis seedling growth by ultraviolet-B irradiation. Plant Cell Environ 32 : 1573–1583.
24. HuangX, OuyangX, YangP, LauOS, LiG, et al. (2012) Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24 : 4590–4606.
25. OraveczA, BaumannA, MateZ, BrzezinskaA, MolinierJ, et al. (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18 : 1975–1990.
26. CrefcoeurRP, YinR, UlmR, HalazonetisTD (2013) Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat Commun 4 : 1779.
27. HeijdeM, BinkertM, YinR, Ares-OrpelF, RizziniL, et al. (2013) Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc Natl Acad Sci USA 110 : 20326–20331.
28. SuYS, LagariasJC (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19 : 2124–2139.
29. GuNN, ZhangYC, YangHQ (2012) Substitution of a conserved glycine in the PHR domain of Arabidopsis cryptochrome 1 confers a constitutive light response. Mol Plant 5 : 85–97.
30. OsterlundMT, AngLH, DengXW (1999) The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol 9 : 113–118.
31. OsterlundMT, HardtkeCS, WeiN, DengXW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 : 462–466.
32. FeherB, Kozma-BognarL, KeveiE, HajduA, BinkertM, et al. (2011) Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J 67 : 37–48.
33. BrownBA, JenkinsGI (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146 : 576–588.
34. WargentJJ, GegasVC, JenkinsGI, DoonanJH, PaulND (2009) UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol 183 : 315–326.
35. GitzDC, Liu-GitzL (2003) How do UV photomorphogenic responses confer water stress tolerance? Photochem Photobiol 78 : 529–534.
36. DemkuraPV, BallareCL (2012) UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol Plant 5 : 642–652.
37. ChenD, GibsonES, KennedyMJ (2013) A light-triggered protein secretion system. J Cell Biol 201 : 631–640.
38. MullerK, EngesserR, SchulzS, SteinbergT, TomakidiP, et al. (2013) Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res 41: e124.
39. McNellisTW, von ArnimAG, ArakiT, KomedaY, MiseraS, et al. (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 : 487–500.
40. ChenH, ShenY, TangX, YuL, WangJ, et al. (2006) Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell 18 : 1991–2004.
41. WeigelD, AhnJH, BlazquezMA, BorevitzJO, ChristensenSK, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122 : 1003–1013.
42. NohB, SpaldingEP (1998) Anion channels and the stimulation of anthocyanin accumulation by blue light in Arabidopsis seedlings. Plant Physiol 116 : 503–509.
Štítky
Genetika Reprodukčná medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



