-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Coming of Age—Sexual Reproduction in Species
article has not abstract
Published in the journal: Coming of Age—Sexual Reproduction in Species. PLoS Pathog 6(12): e32767. doi:10.1371/journal.ppat.1001155
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1001155Summary
article has not abstract
What Are Candida Species and Why Are They Important?
Historically, Candida species represented a catch-all taxonomic grouping for yeasts that exhibited hyphal or pseudohyphal branching and did not form sexual spores. However, with the advent of molecular typing of DNA sequences, it is now apparent that Candida represent a diverse range of species within the Hemiascomycetes, which also includes the model yeast Saccharomyces cerevisiae. Several Candida species are prominent human pathogens, including Candida glabrata, Candida parapsilosis, and Candida albicans, the latter being the most common human pathogenic fungus. While C. albicans is a natural commensal, typically found amongst the microbiota inhabiting the gastrointestinal tract, it is also the cause of debilitating mucosal infections as well as life-threatening systemic infections. Taken together, Candida species are the fourth most common cause of nosocomial bloodstream infections, where they account for 8%–10% of such infections [1].
The most clinically important Candida species, with the exception of C. glabrata and Candida krusei, cluster together in a single clade [2]. The species within this clade share an altered genetic code in which CUG codons encode serine rather than the universal leucine [3]. The Candida clade species can be further subdivided into two separate sub-clades; one contains haploid species that are relatively rare pathogens (e.g., Candida lusitaniae), while the other contains diploid species such as C. albicans and C. parapsilosis that are frequent pathogens (Figure 1A).
Fig. 1. Phylogeny of the Candida clade and the parasexual mating cycle of Candida albicans. 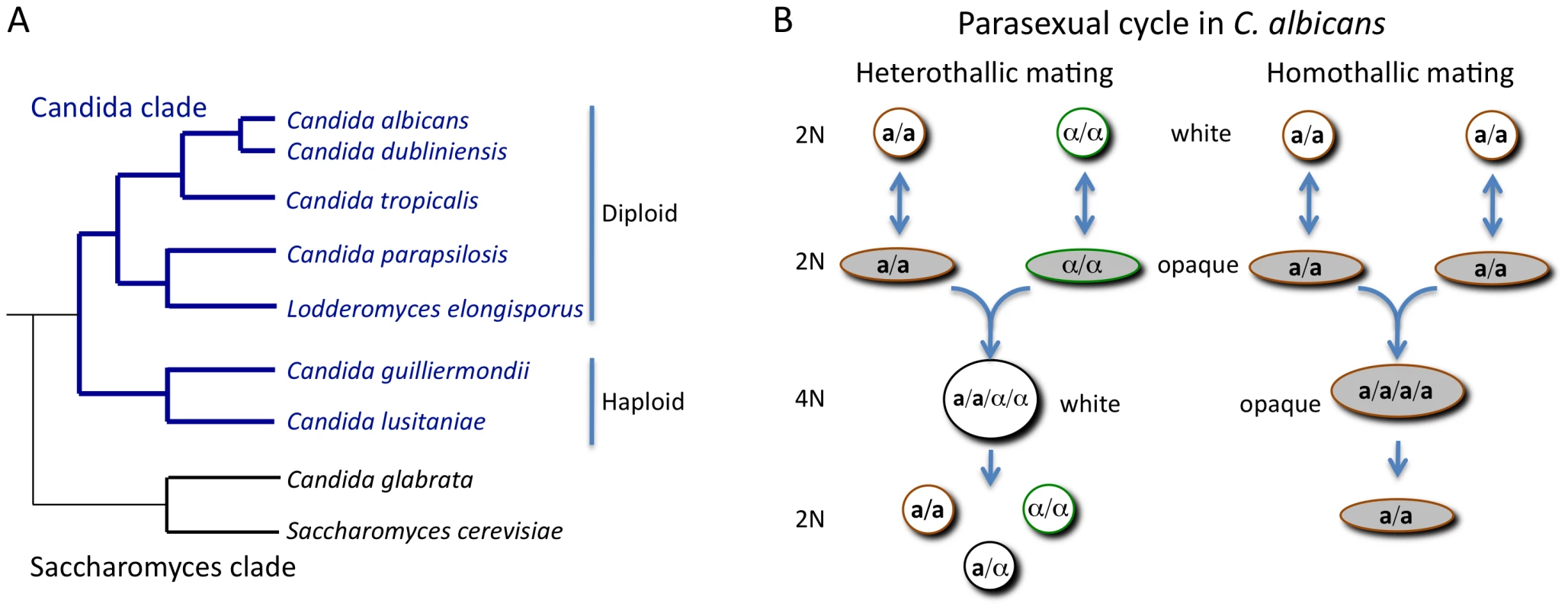
(A) Phylogenetic organization of the Candida clade of species and its relationship to the Saccharomyces clade. Most Candida species belong to the Candida clade, including the most frequently isolated species, C. albicans. A notable exception is the pathogen C. glabrata, which is more closely related to S. cerevisiae than to other Candida species. (B) Heterothallic and homothallic mating cycles of C. albicans. In both mating cycles cells must switch from the white state to the opaque state to become mating competent. Heterothallic mating involves fusion of diploid a and α cells to form a white tetraploid a/α cell. Under some conditions, homothallic mating can occur with between two opaque a cells (or two α cells) to form an opaque tetraploid cell. How Was Mating Discovered in C. albicans?
Sexual reproduction in fungi is often regulated by genes encoded at a mating-type locus, as exemplified by the MAT locus of S. cerevisiae. MATa-containing cells mate with MATα-containing cells in a program choreographed by MAT-encoded transcription factors. The discovery of a mating-type–like (MTL) locus in C. albicans [4] provided the impetus for re-examining the potential for sexual reproduction in this species. Subsequent experiments in both the Johnson and Magee groups revealed that diploid MTLa and MTLα strains could indeed be made to mate and form stable tetraploids, either on laboratory media or during bloodstream infection of a mammalian host [5], [6]. Despite uncovering mating in the “asexual” C. albicans, the frequency of cell–cell conjugation observed was extremely low. This conundrum was solved when it was discovered that C. albicans mating is regulated by a unique mechanism of phenotypic switching. First described by Soll and colleagues, some isolates of C. albicans had been noted for their ability to undergo switching between “white” and “opaque” states [7]. White cells are round and give rise to shiny, domed-shaped colonies while opaque cells are more elongated and produce darker, flatter colonies. Miller and Johnson noted that only a or α strains (and not a/α strains) could undergo switching to the opaque form. Furthermore, opaque cells were demonstrated to mate a million times more efficiently than white cells (Figure 1B) [8].
Completion of fungal sexual cycles is usually accomplished via meiosis, in which one round of DNA replication precedes two rounds of DNA division. In the case of C. albicans, many potential meiotic genes were found in the sequenced genome but no experimental evidence for a conventional meiosis exists [9]. Instead, a genetic screen showed that a parasexual mechanism of chromosome loss could complete the mating cycle. Growth of C. albicans tetraploids on selective media caused random, but concerted, chromosome loss resulting in diploid (and aneuploid) products [10]. A subset of parasexual cells was analyzed and shown to have undergone inter-chromosomal recombination, indicating that the parasexual cycle generates recombinant forms of the species with potentially novel properties [11].
What Is the Mechanism of White–Opaque Phenotypic Switching?
Recent studies have begun to shed light on the molecular mechanism underlying this bistable switch that regulates pleiotropic aspects of C. albicans biology. In addition to mating, the white–opaque switch influences the expression of a number of metabolic genes, determines how C. albicans cells interact with host immune cells, and also modulates the virulence of strains during host infection [12]–[14]. It is now established that the master regulator of the opaque state is the transcription factor Wor1p. High levels of Wor1p cause switching to opaque, and Wor1p binding to its own promoter drives a positive feedback loop that stabilizes the opaque form [15]–[17]. Wor1p appears to be representative of a novel superfamily of DNA-binding proteins that is conserved across all fungi [18]. Three additional transcription factors (Efg1p, Czf1p, and Wor2p) complete a network of complementary transcriptional feedback loops with Wor1p that generates the two alternative phenotypic states [19].
In addition to new mechanistic insights into switching, several studies have illustrated the sensitivity of the white–opaque switch to multiple external stimuli. A growing list of conditions that promote switching to opaque includes anaerobic culture, high levels of CO2, N-acetyl glucosamine (also a product of commensal bacteria), oxidative stress, and slower growth of the cell [20]–[23]. Significantly, some environmental conditions can stabilize the opaque form at 37°C, a temperature that normally results in conversion en masse of opaque cells to white cells [21]. The fact that opaque cells can persist in certain host niches is supported by observations of white-to-opaque switching in the gastrointestinal tract by some strains of C. albicans [23].
The central question remains as to why the white–opaque switch evolved to regulate mating in C. albicans. It is possible that precise regulation of mating is required to limit it to specific niches in the host. Opaque cells are also targeted less efficiently by host immune cells than white cells, perhaps shielding mating events from destruction [24]. In addition, it is now apparent that white cells, although incapable of mating, could play a key role in sexual reproduction. While opaque cells respond to secreted pheromones by forming mating projections and undergoing cell–cell fusion, white cells exhibit a very different response in which they display increased adhesion to each other and to synthetic surfaces [25]. The ability of white cells to form such pheromone-induced biofilms could promote mating of opaque cells by allowing stable diffusion of pheromone gradients across relatively large distances. Strikingly, the differential response of white and opaque cells involves distinct transcription factors activated by the pheromone mitogen-activated protein (MAP) kinase cascade; this conserved signaling pathway activates the Ste12/Cph1 transcription factor in opaque cells, yet acts via the Tec1 transcription factor in white cells (Figure 2) [26]. It remains to be seen how these two different transcription factors are targeted by the same MAP kinase cascade, although multiple mechanisms help prevent cross-talk between related MAP kinase signaling pathways in S. cerevisiae [27].
Fig. 2. Differential response of C. albicans white and opaque cells to pheromone. 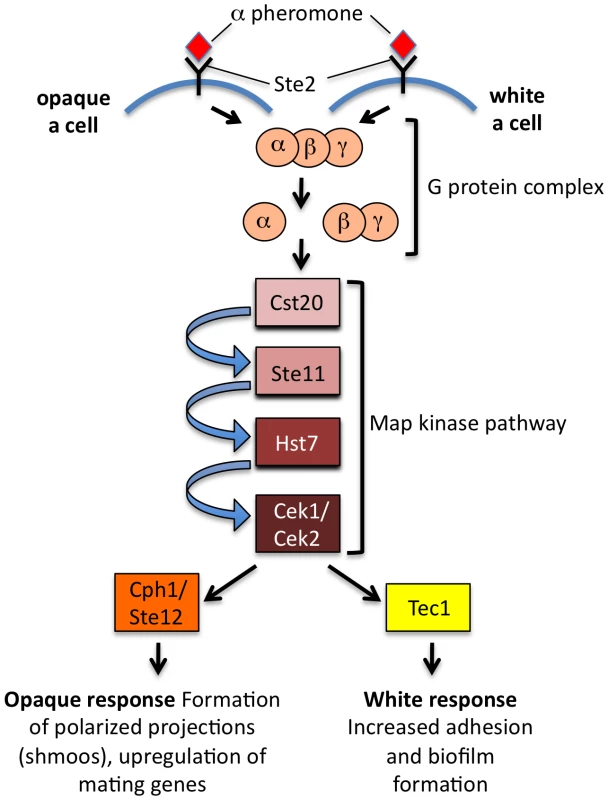
Both cell types respond to pheromone using the conserved pheromone MAP kinase cascade, but opaque cells use the Ste12/Cph1 transcription factor to turn on genes necessary for cell–cell conjugation, whereas white cells use the Tec1 transcription factor to turn on genes involved in adherence and biofilm formation. Coordination of these pathways may promote mating in vivo. Adapted from Sahni et al. [26]. How Does C. albicans Undergo Homothallic Mating?
Heterothallic mating between opaque a and α cells is now well established in C. albicans, but recent studies demonstrate that homothallic mating, or self-fertilization, can also occur. Self-mating between a cells was first detected upon deletion of BAR1, a gene encoding a protease activity that degrades α pheromone [28]. In conventional a-α mating in yeast, a cells secrete a pheromone while α cells secrete α pheromone to attract partners of the opposite sex. However, it is now apparent that C. albicans a cells secrete both a and α pheromones; α pheromone is normally degraded by Bar1p, but in the absence of this protease α pheromone accumulates, leading to autocrine signaling and induction of self-mating in unisexual populations of (opaque) a cells. The products of same-sex mating are a-a tetraploid cells that can still undergo the parasexual program of chromosome loss to return to the diploid state [28]. The discovery of same-sex mating in C. albicans shows interesting parallels with that previously made in a distantly related fungal pathogen, Cryptococcus neoformans. In C. neoformans, efficient mating between a and α cells can take place, but the predominant mode of mating in nature appears to be unisexual, with α cells self-mating to form α-α cells that can still undergo meiosis and sporulation [29], [30].
Why should such a mechanism of self-fertilization evolve in C. albicans and C. neoformans, two such diverse species? Several possibilities have been considered, including the fact that self-mating could help maintain the sexual/parasexual machinery for future (rare) heterothallic mating events [30]. In addition, same-sex mating events have the potential to increase genetic diversity as well as generate changes in ploidy that may increase fitness [29]. In the case of C. albicans, same-sex mating products can also exist in the opaque state (whereas conventional a-α products are obligate white cells), and this may provide additional, yet to be discovered, benefits for the species (see Figure 1B) [28].
What About Sexual Reproduction in Other Candida Species?
With the recent release of genome sequences for multiple Candida species [2], new insights into mechanisms of fungal sex are emerging. A conserved MTL locus is present in the majority of strains from the Candida clade, although surprises have been found in species such as Lodderomyces elongisporus. This yeast is thought to be homothallic due to the production of asci-like spores, and yet it has lost all of the conserved transcriptional regulators of sexual identity at the MTL [2]. If a sexual cycle does take place in L. elongisporus it must therefore be under novel regulatory control. In this regard, it is interesting to note that mating in the basidiomycete C. neoformans was recently shown to be regulated by transcription factors that are not encoded at the mating locus [31]. In addition, the re-wiring of sexual transcriptional circuits has occurred multiple times during the evolution of the Candida clade. An example of this is seen in C. lusitaniae, which exhibits a complete sexual cycle culminating in meiosis and sporulation despite lacking the MTL transcription factor α2. Furthermore, C. lusitaniae has apparently lost several conserved meiosis factors, including the strand exchange protein Dmc1 and the synaptonemal complex proteins Zip1-4 and Hop1 [32]. Conversely, the asexual species C. tropicalis and C. parapsilosis do not appear to undergo mating or asci formation, yet contain many of the genes required for mating and meiosis [2]. These observations emphasize that sequence analysis alone is not sufficient to determine whether a species is able to complete a sexual (or parasexual) cycle. It is likely that cryptic mating cycles are still to be discovered in some Candida species, while in others, the sexual machinery is under different selective pressures and components have been retained for uses other than mating and recombination. What is certain is that defining the roles of mating and mating-related mechanisms in Candida, and their potential function in commensal and infectious growth, will provide very fertile ground in the years to come.
Zdroje
1. WisplinghoffH
BischoffT
TallentSM
SeifertH
WenzelRP
2004
Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study.
Clin Infect Dis
39
309
317
2. ButlerG
RasmussenMD
LinMF
SantosMA
SakthikumarS
2009
Evolution of pathogenicity and sexual reproduction in eight Candida genomes.
Nature
459
657
662
3. MouraGR
ParedesJA
SantosMA
2010
Development of the genetic code: insights from a fungal codon reassignment.
FEBS Lett
584
334
341
4. HullCM
JohnsonAD
1999
Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans.
Science
285
1271
1275
5. HullCM
RaisnerRM
JohnsonAD
2000
Evidence for mating of the "asexual" yeast Candida albicans in a mammalian host.
Science
289
307
310
6. MageeBB
MageePT
2000
Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains.
Science
289
310
313
7. SlutskyB
StaebellM
AndersonJ
RisenL
PfallerM
1987
"White-opaque transition": a second high-frequency switching system in Candida albicans.
J Bacteriol
169
189
197
8. MillerMG
JohnsonAD
2002
White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating.
Cell
110
293
302
9. BennettRJ
JohnsonAD
2005
Mating in Candida albicans and the search for a sexual cycle.
Annu Rev Microbiol
59
233
255
10. BennettRJ
UhlMA
MillerMG
JohnsonAD
2003
Identification and characterization of a Candida albicans mating pheromone.
Mol Cell Biol
23
8189
8201
11. ForcheA
AlbyK
SchaeferD
JohnsonAD
BermanJ
2008
The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains.
PLoS Biol
6
e110
doi:10.1371/journal.pbio.0060110
12. LohseMB
JohnsonAD
2009
White-opaque switching in Candida albicans.
Curr Opin Microbiol
12
650
654
13. SollDR
2009
Why does Candida albicans switch?
FEMS Yeast Res
9
973
989
14. MorschhauserJ
2010
Regulation of white-opaque switching in Candida albicans.
Med Microbiol Immunol
15. HuangG
WangH
ChouS
NieX
ChenJ
2006
Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans.
Proc Natl Acad Sci U S A
103
12813
12818
16. ZordanRE
GalgoczyDJ
JohnsonAD
2006
Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop.
Proc Natl Acad Sci U S A
103
12807
12812
17. SrikanthaT
BornemanAR
DanielsKJ
PujolC
WuW
2006
TOS9 regulates white-opaque switching in Candida albicans.
Eukaryot Cell
5
1674
1687
18. LohseMB
ZordanRE
CainCW
JohnsonAD
2010
Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans.
Proc Natl Acad Sci U S A
107
14105
14110
19. ZordanRE
MillerMG
GalgoczyDJ
TuchBB
JohnsonAD
2007
Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans.
PLoS Biol
5
e256
doi:10.1371/journal.pbio.0050256
20. AlbyK
BennettRJ
2009
Stress-induced phenotypic switching in Candida albicans.
Mol Biol Cell
20
3178
3191
21. HuangG
SrikanthaT
SahniN
YiS
SollDR
2009
CO(2) regulates white-to-opaque switching in Candida albicans.
Curr Biol
19
330
334
22. HuangG
YiS
SahniN
DanielsKJ
SrikanthaT
2010
N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans.
PLoS Pathog
6
e1000806
doi:10.1371/journal.ppat.1000806
23. Ramirez-ZavalaB
ReussO
ParkYN
OhlsenK
MorschhauserJ
2008
Environmental induction of white-opaque switching in Candida albicans.
PLoS Pathog
4
e1000089
doi:10.1371/journal.ppat.1000089
24. LohseMB
JohnsonAD
2008
Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes.
PLoS ONE
3
e1473
doi:10.1371/journal.pone.0001473
25. DanielsKJ
SrikanthaT
LockhartSR
PujolC
SollDR
2006
Opaque cells signal white cells to form biofilms in Candida albicans.
Embo J
25
2240
2252
26. SahniN
YiS
DanielsKJ
HuangG
SrikanthaT
2010
Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways.
PLoS Biol
8
e1000363
doi:10.1371/journal.pbio.1000363
27. SchwartzMA
MadhaniHD
2004
Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae.
Annu Rev Genet
38
725
748
28. AlbyK
SchaeferD
BennettRJ
2009
Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans.
Nature
460
890
893
29. LinX
HullCM
HeitmanJ
2005
Sexual reproduction between partners of the same mating type in Cryptococcus neoformans.
Nature
434
1017
1021
30. LeeSC
NiM
LiW
ShertzC
HeitmanJ
2010
The evolution of sex: a perspective from the fungal kingdom.
Microbiol Mol Biol Rev
74
298
340
31. LinX
JacksonJC
FeretzakiM
XueC
HeitmanJ
2010
Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite - and same-sex mating in Cryptococcus neoformans.
PLoS Genet
6
e1000953
doi:10.1371/journal.pgen.1000953
32. ReedyJL
FloydAM
HeitmanJ
2009
Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex.
Curr Biol
19
891
899
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 12- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Is Adherence to Erythrocytes a Factor in Extrapulmonary Dissemination?
- Identifying the Age Cohort Responsible for Transmission in a Natural Outbreak of
- Blockade of Immunosuppressive Cytokines Restores NK Cell Antiviral Function in Chronic Hepatitis B Virus Infection
- Metaeffector Exploits Host Proteasome to Temporally Regulate Cognate Effector
- The p53-Target Gene Drives Neutrophil-Mediated Protection against Lethal Bacterial Sepsis
- Development of an RNAi Protocol to Investigate Gene Function in the Filarial Nematode,
- Lysine Acetyltransferase GCN5-A Functions in the Cellular Response to Alkaline Stress and Expression of Cyst Genes
- Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus BHRF1
- Molecular Architectures of Trimeric SIV and HIV-1 Envelope Glycoproteins on Intact Viruses: Strain-Dependent Variation in Quaternary Structure
- Interaction of c-Cbl with Myosin IIA Regulates Bleb Associated Macropinocytosis of Kaposi's Sarcoma-Associated Herpesvirus
- Glacial Refugia in Pathogens: European Genetic Structure of Anther Smut Pathogens on and
- Role for Sumoylation in Systemic Inflammation and Immune Homeostasis in Larvae
- Inflammasome Sensor Nlrp1b-Dependent Resistance to Anthrax Is Mediated by Caspase-1, IL-1 Signaling and Neutrophil Recruitment
- Noise Cancellation: Viral Fine Tuning of the Cellular Environment for Its Own Genome Replication
- Infectious Speciation Revisited: Impact of Symbiont-Depletion on Female Fitness and Mating Behavior of
- Eis Regulates Autophagy, Inflammation, and Cell Death through Redox-dependent Signaling
- NleC, a Type III Secretion Protease, Compromises NF-κB Activation by Targeting p65/RelA
- Early Myeloid Dendritic Cell Dysregulation is Predictive of Disease Progression in Simian Immunodeficiency Virus Infection
- HIV Capsid is a Tractable Target for Small Molecule Therapeutic Intervention
- Structural and Functional Studies of Nonstructural Protein 2 of the Hepatitis C Virus Reveal Its Key Role as Organizer of Virion Assembly
- Coming of Age—Sexual Reproduction in Species
- HIV-1 Envelope Subregion Length Variation during Disease Progression
- Compartmentation of Redox Metabolism in Malaria Parasites
- Evidence That Intracellular Stages of Utilize Amino Sugars as a Major Carbon Source
- Rapid End-Point Quantitation of Prion Seeding Activity with Sensitivity Comparable to Bioassays
- Dimeric 2G12 as a Potent Protection against HIV-1
- Hypoxia Induces an Immunodominant Target of Tuberculosis Specific T Cells Absent from Common BCG Vaccines
- CD4 Natural Regulatory T Cells Prevent Experimental Cerebral Malaria via CTLA-4 When Expanded In Vivo
- The Killing of African Trypanosomes by Ethidium Bromide
- H2A.Z Demarcates Intergenic Regions of the Epigenome That Are Dynamically Marked by H3K9ac and H3K4me3
- Large-Scale Field Application of RNAi Technology Reducing Israeli Acute Paralysis Virus Disease in Honey Bees (, Hymenoptera: Apidae)
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- HIV-1 Envelope Subregion Length Variation during Disease Progression
- Coming of Age—Sexual Reproduction in Species
- Evidence That Intracellular Stages of Utilize Amino Sugars as a Major Carbon Source
- Compartmentation of Redox Metabolism in Malaria Parasites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



