-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Bottlenecks and the Maintenance of Minor Genotypes during the Life Cycle of
African trypanosomes are digenetic parasites that undergo part of their developmental cycle in mammals and part in tsetse flies. We established a novel technique to monitor the population dynamics of Trypanosoma brucei throughout its life cycle while minimising the confounding factors of strain differences or variation in fitness. Clones derived from a single trypanosome were tagged with short synthetic DNA sequences in a non-transcribed region of the genome. Infections were initiated with mixtures of tagged parasites and a combination of polymerase chain reaction and deep sequencing were used to monitor the composition of populations throughout the life cycle. This revealed that a minimum of several hundred parasites survived transmission from a tsetse fly to a mouse, or vice versa, and contributed to the infection in the new host. In contrast, the parasites experienced a pronounced bottleneck during differentiation and migration from the midgut to the salivary glands of tsetse. In two cases a single tag accounted for ≥99% of the population in the glands, although minor tags could be also detected. Minor tags were transmitted to mice together with the dominant tag(s), persisted during a chronic infection, and survived transmission to a new insect host. An important outcome of the bottleneck within the tsetse is that rare variants can be amplified in individual flies and disseminated by them. This is compatible with the epidemic population structure of T. brucei, in which clonal expansion of a few genotypes in a region occurs against a background of frequent recombination between strains.
Published in the journal: Bottlenecks and the Maintenance of Minor Genotypes during the Life Cycle of. PLoS Pathog 6(7): e32767. doi:10.1371/journal.ppat.1001023
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001023Summary
African trypanosomes are digenetic parasites that undergo part of their developmental cycle in mammals and part in tsetse flies. We established a novel technique to monitor the population dynamics of Trypanosoma brucei throughout its life cycle while minimising the confounding factors of strain differences or variation in fitness. Clones derived from a single trypanosome were tagged with short synthetic DNA sequences in a non-transcribed region of the genome. Infections were initiated with mixtures of tagged parasites and a combination of polymerase chain reaction and deep sequencing were used to monitor the composition of populations throughout the life cycle. This revealed that a minimum of several hundred parasites survived transmission from a tsetse fly to a mouse, or vice versa, and contributed to the infection in the new host. In contrast, the parasites experienced a pronounced bottleneck during differentiation and migration from the midgut to the salivary glands of tsetse. In two cases a single tag accounted for ≥99% of the population in the glands, although minor tags could be also detected. Minor tags were transmitted to mice together with the dominant tag(s), persisted during a chronic infection, and survived transmission to a new insect host. An important outcome of the bottleneck within the tsetse is that rare variants can be amplified in individual flies and disseminated by them. This is compatible with the epidemic population structure of T. brucei, in which clonal expansion of a few genotypes in a region occurs against a background of frequent recombination between strains.
Introduction
A bottleneck is an event in which the population size of a species is temporarily severely reduced. Bottlenecks can have strong evolutionary effects because limited population sizes can lead to dramatic shifts that favour certain genotypes (founder effects) [1], [2], [3] and to the stochastic loss of others [4], with rare genotypes being especially prone to being lost. Many digenetic parasites are presumed to experience bottlenecks because their population sizes are reduced during transmission between their two hosts, but there is little information on the size of such bottlenecks or the impact that this may have on genetic diversity. In addition, parasites may also encounter bottlenecks within a host as they differentiate and migrate from one tissue to another or infect different cell types.
The protozoan parasite Trypanosoma brucei brucei causes Nagana in cattle, while its close relatives T. b. rhodesiense and T. b. gambiense cause human sleeping sickness. All three sub-species undergo part of their developmental cycle in their insect vector, the tsetse fly (Glossina spp.), and part in their mammalian host. Within their life cycles, there are several phases where parasite numbers are severely reduced (shown schematically in Figure 1). When a fly feeds on an infected mammal, the parasites that are taken up reach the midgut together with the blood meal. Depending on the trypanosome density in the mammalian host, the fly may ingest anywhere from a few hundred to several hundred thousand organisms. Many species of tsetse are completely refractory to infection by a particular species of trypanosome (reviewed in [5]). Even when a fly species is susceptible, the number of parasites in the midgut can decrease by three orders of magnitude after 3–5 days [6] (Figure 1; A). Attrition of the parasite population also occurs when an infection is initiated with procyclic forms fed to flies through a silicon membrane [7] indicating that the drop in numbers is not solely due to parasites failing to differentiate. In many flies the infection is eradicated at this point; in flies that sustain an infection, the surviving parasites multiply as procyclic forms and colonise the ectoperitrophic space, reaching densities of up to 5×105 parasites per midgut [6].
Fig. 1. Schematic depiction of population bottlenecks during the life cycle of Trypanosoma brucei. 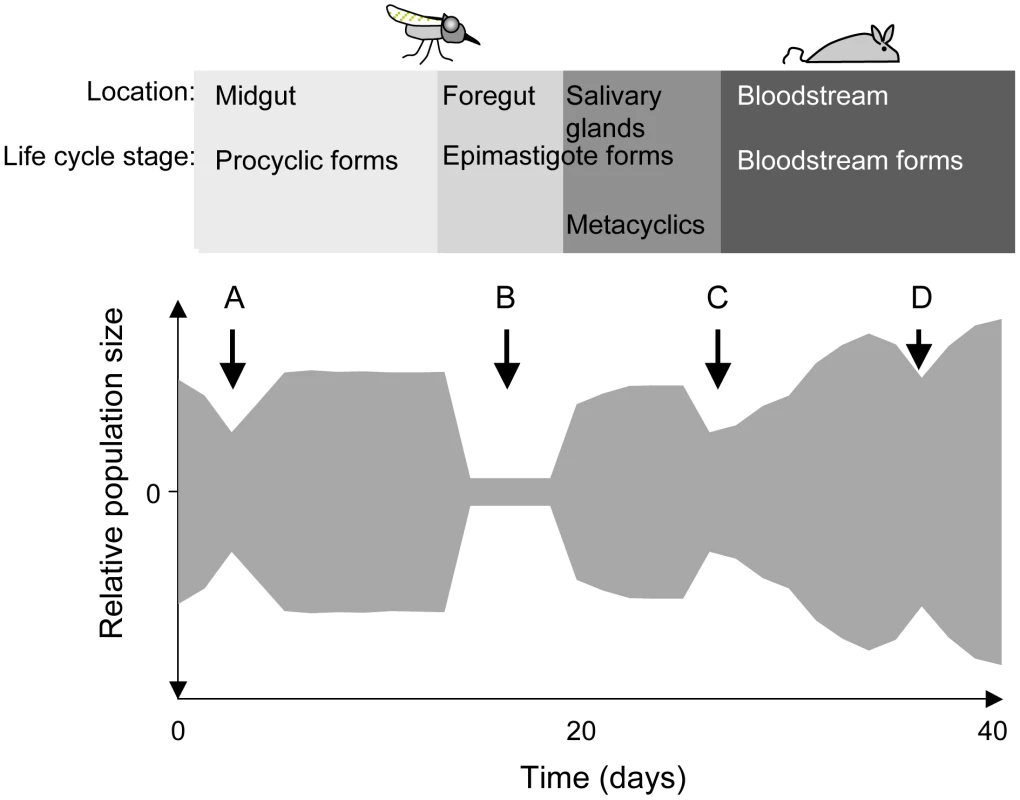
The relative population size is shown on the y-axis. When trypanosomes are taken up by a tsetse fly, the population collapses during the adaptation to the midgut (A) and recovers thereafter. Only a few trypanosomes at a time are presumed to migrate to the salivary glands (B). Migration might take place during a defined period [6] or continuously [8]. Long epimastigote forms can reach the salivary glands where they deposit the short epimastigote forms that colonise the epithelia and give rise to metacyclic forms. During a blood meal, metacyclic forms are injected into a mammalian host. The injection and the relocation of trypanosomes from the site of injection into the bloodstream may reduce its number (C). In the mammalian host, the trypanosome population is periodically reduced in size owing to the adaptive immune response and to the differentiation of long slender bloodstream forms into non-dividing short stumpy forms (D). To complete the life cycle, trypanosomes must migrate to the salivary glands via the foregut and the proboscis [6], [8]. In a large proportion of tsetse flies with infected midguts, trypanosomes fail to infect the salivary glands [9], [10] (Figure 1; B). The factors that promote or hinder colonisation of the salivary glands are not known and it is under debate whether migration is continuous [8] or restricted to a defined period [6]. It has been proposed that only a few trypanosomes undertake this journey and that asymmetrically dividing epimastigotes are the only forms capable of colonising the salivary glands [6]. Two lines of evidence support the notion of a limited founder population in the glands: first, fewer than ten epimastigote forms could be detected in the salivary gland ducts of individual flies [6] and second, in mixed infections with two strains of trypanosomes, each tagged with a different fluorescent protein, colonisation of a gland by only one strain was observed on several occasions [11], [12].
Epimastigotes in the salivary glands attach to the epithelium and proliferate, giving rise to the mammalian-infective metacyclic forms [13] that are transmitted to a susceptible mammal during a blood meal. It has been estimated that flies can inject up to several thousand trypanosomes when they feed on a new host [14], [15], but it is not known how many of these differentiate into bloodstream forms and establish an infection (Figure 1; C). Within the mammalian host, a chronic infection is characterised by repeated waves of parasitaemia (Figure 1; D). These are due to the interplay between three phenomena: the host immune response to the parasite's variant surface glycoprotein (VSG) coat, resulting in elimination of the population that expresses this particular variant, outgrowth of minor populations that have switched to a different VSG, and differentiation of proliferating slender bloodstream forms to non-dividing stumpy forms at high parasite densities. Stumpy forms are preadapted for further differentiation in the fly and have a lifespan of only a few days in the mammalian bloodstream [16].
Given the right conditions, trypanosomes can infect their mammalian and insect hosts very efficiently: a single parasite is sufficient to infect a tsetse fly [17] and one bite of an infected fly is sufficient to infect a mammal [18] with a minimal infective dose of one metacyclic trypanosome [19]. This high infectivity implies that trypanosomes can cope with very narrow bottlenecks. If transmission bottlenecks are so small and so frequent, however, trypanosomes might risk a loss of fitness and the accumulation of deleterious mutations [20]. In addition, any acquired mutations (such as drug resistance) that are beneficial to the parasite in one host might be lost during transmission through the second host.
In the case of endoparasites, the quantification of bottlenecks can be difficult because populations are not easily observed over time. Furthermore, it is not straightforward to distinguish between random and selective population reduction. To resolve these problems we used a novel methodology to monitor the population dynamics of T. b. brucei in tsetse. This was subsequently extended to the rest of the life cycle, including transmission from the fly to the mammalian host and vice versa. Different strains of trypanosomes can vary greatly in their ability to be transmitted by tsetse [21], [22]. Genetic differences were minimised by tagging the progeny of a single trypanosome with short unique DNA sequences that were integrated into a non-transcribed region of the genome. These tags were subsequently used to identify the different populations by amplifying them by polymerase chain reaction and subjecting them to deep sequencing. This approach has the advantage that it yields quantitative data about the different populations that co-exist, as well as allowing an estimate of the population size after a bottleneck.
Results
Repeated syringe passage of bloodstream forms in rodents or prolonged culture of procyclic forms can reduce infectivity for flies. We therefore used the following protocol (Figure 2A) to obtain trypanosomes that were genetically homogeneous and capable of completing the life cycle: procyclic forms of T. b. brucei were cloned and a single clone was transmitted through a fly and a mouse. Bloodstream forms isolated from the mouse were triggered to differentiate to procyclic forms in culture. To generate parasites that were distinguishable from each other, aliquots of the culture were transfected with plasmids containing a unique 40bp tag (Figure 2B and Table S1). The tag in each plasmid lies upstream of the promoter and should not be transcribed, and therefore not influence the fitness of the parasite.
Fig. 2. Cloning procedure and generation of tagged trypanosomes. 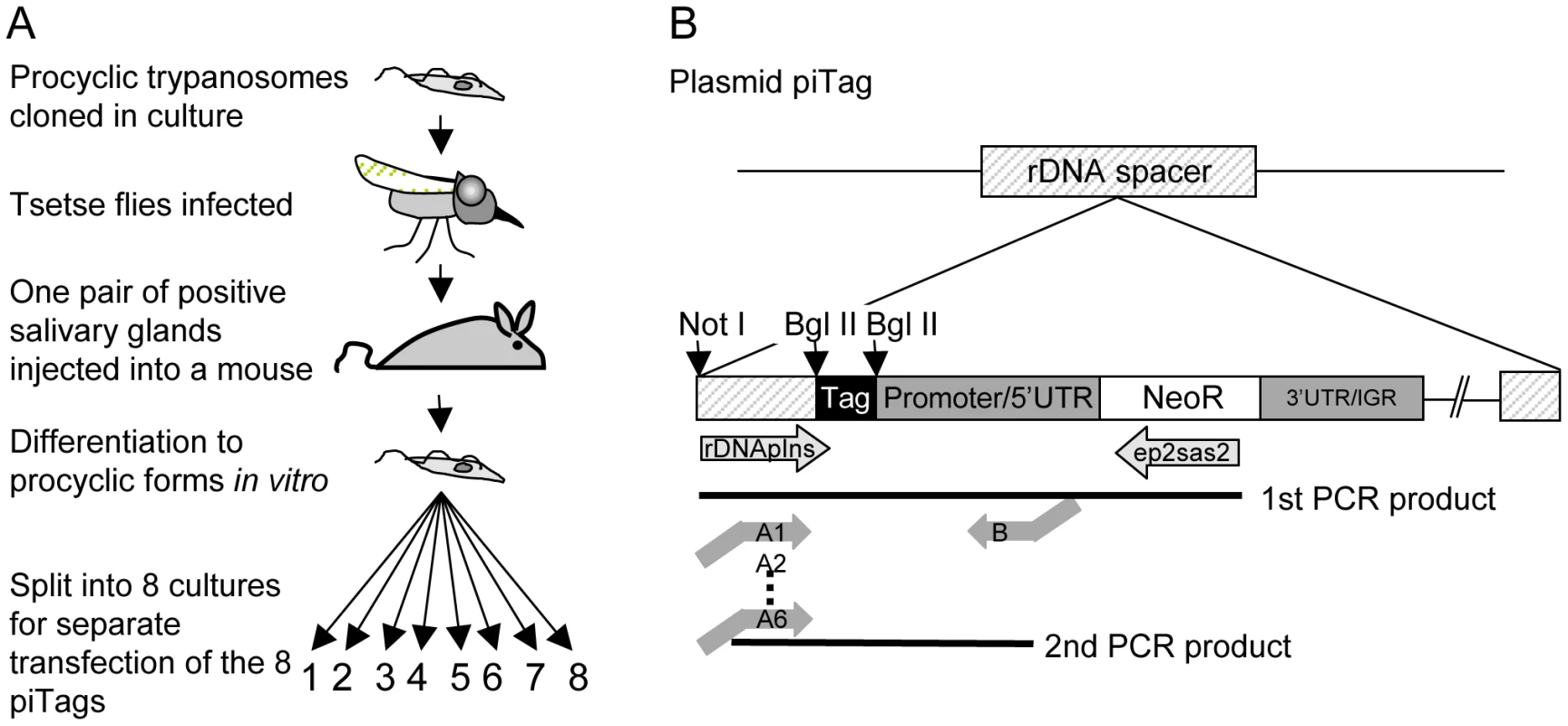
A. Cloned procyclic trypanosomes were passaged through a tsetse fly and a mouse and bloodstream forms were triggered to differentiate to procyclic forms in vitro. The plasmids piTag 1–8 were then transfected separately into T. brucei. Cloned stable transformants containing each of the eight tags were isolated. B. Construction of the plasmid piTag. Eight different 40mers were integrated into the plasmid upstream of the procyclin promoter. Expression of the neomycin resistance gene (NeoR) in trypanosomes is controlled by the EP1 procyclin promoter and 5′ untranslated region (UTR) and the last 19 bases of the 3′ untranslated region and intergenic region (IGR) of EP2 procyclin. The linearised plasmid integrates into an rDNA spacer in the genome. Tags were amplified from genomic DNA by nested PCR. Trypanosome clones (one for each of eight tags) were isolated and tested for growth in culture. All grew at similar rates (Figure S1). Cultures of the eight clones were mixed and used to infect tsetse. Three flies (A, B, and C) that were positive for metacyclic forms were selected and allowed to infect mice. Parasites were first detected in the corresponding mice 6, 7 and 4 days, respectively, after the infective bite. Subsequently, the salivary glands and midguts of the flies were isolated by dissection and DNA was extracted. Tail blood samples were collected from each mouse to monitor the parasitaemia and DNA was prepared from samples in weeks 1, 2, 3 and 4 and at the termination of the experiment after 7–10 weeks. Two batches of ten flies were fed on mouse C 18 and 30 days post infection. Midguts were dissected after 10 and 12 days and one positive midgut from each batch was taken for tag analysis (fly D and fly E in Figure 3). Tags were amplified by PCR, sequenced by 454 massively parallel pyrosequencing and analysed for their frequency and distribution. In total, we identified 30,592 sequences, an average of 1330 per sample (Figure S2). Control experiments confirmed that the barcoded primers did not affect the frequency with which individual sequences were detected (Figure S2).
Fig. 3. Diversity and frequency of tagged trypanosomes in three experiments. 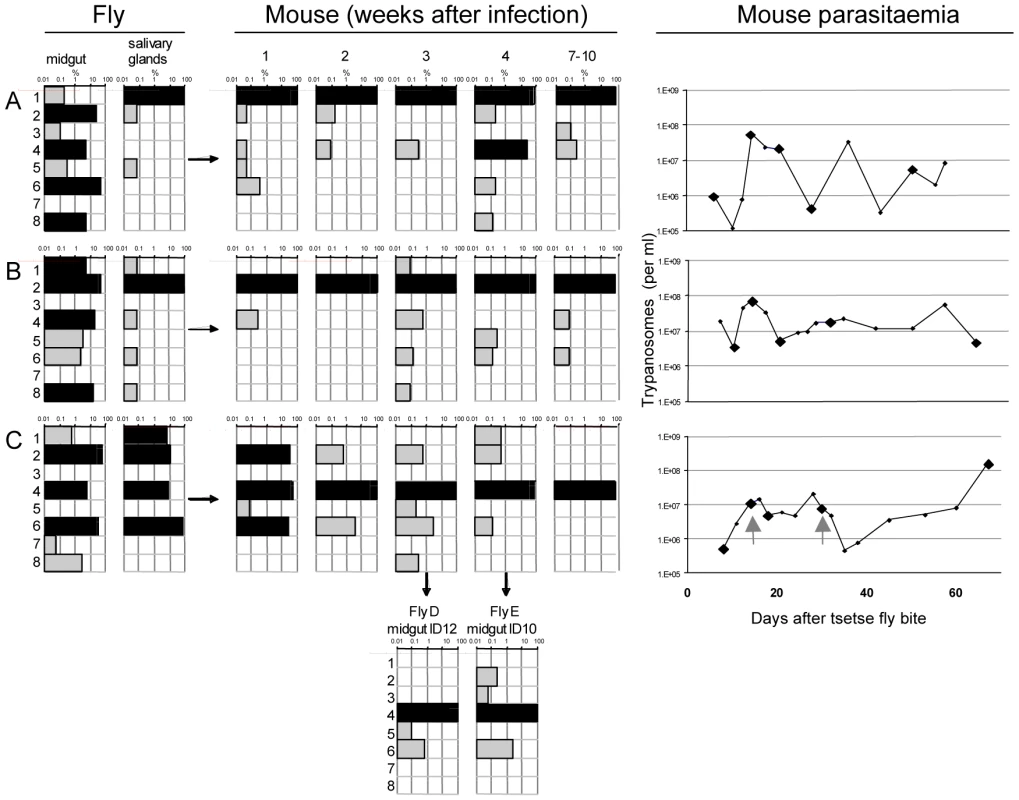
The bars in each chart represent the 8 tags. Frequency is shown on a log scale ranging from 0.01–100 per cent with dominant tags (>5%) highlighted in black. Tsetse flies were infected with procyclic trypanosomes with an even distribution of the tags. Three flies (A, B, and C) had infected midguts (mg) and salivary glands (sg). These flies each infected one mouse whose parasitaemia was monitored over a period of 2–3 months. Since the mice are immunocompetent, antigenic variation occurs, with each peak of parasitaemia containing trypanosomes expressing new VSGs compared to the preceding peaks. Five blood samples were taken from each mouse for analysis of the tags (indicated by large diamonds in the right panel) after 1, 2, 3, and 4 weeks and at the end of the experiment after 7–10 weeks. Flies D and E were infected on mouse C 18 and 30 days after infection (indicated with the grey arrows in the right panel). Midguts were dissected after 12 days (ID12) and 10 days (ID10). The number of sequences obtained from each sample is shown in Figure S2A. A common pattern in all three experiments (Figure 3) was the large number of different tags detected in the midgut, many of them at high frequency (>5%). Each midgut contained at least 6 different tags and, taken together, all 8 tags could be detected in the three flies. This demonstrates that the procyclic culture forms used to infect the flies maintained their diversity in the gut lumen. The frequency of individual tags in the salivary glands changed compared to the midgut (Figure 3). This was most striking in fly A, in which tag 1 was minor (0.2%) and tag 6 dominant (52%) in the midgut, whereas in the salivary glands tag 1 was dominant (99.9%) and tag 6 undetectable. In fly B, four tags were dominant in the midgut, but only one of these (tag 2) was dominant in the salivary glands. In fly C, the three tags that were dominant in the midgut (tags 2, 4 and 6) were also dominant in the salivary glands, with tag 6 constituting 74% of the population. In addition, tag 1, which was present at <1% in the midgut, accounted for 6.9% of the parasites in the salivary glands. This analysis demonstrates that tags that are dominant in the midgut are not necessarily so in the salivary glands, and that their relative frequencies can be altered. This is reflected by the diverse correlation coefficients: r2 = 0.08, 0.86 and 0.25 for flies A, B, and C respectively, and implies that when trypanosomes are equally fit, any of the parasites from the midgut is capable of migrating to the salivary glands and founding the dominant population.
At the beginning of the infection in mice, the distribution and frequency of tags was very similar to the parasite populations in the salivary glands of the corresponding tsetse fly. The dominant tags in the salivary glands retained their dominance in mouse A and mouse B from the first sample onwards. In mouse C, tags 2 (32%), 4 (43%), and 6 (25%) were present in the first week of infection. Tag 4 was the only dominant tag from the second week onwards and finally the only one detectable after ten weeks. Tag 1, with a frequency of 6.9% in the salivary glands of fly C, became very minor in the following mouse infection and was detected only once after four weeks. The presence of one dominant tag and a few minor tags during mouse infections led to a very uneven distribution of individual tags, which showed up to a thousand-fold variation in frequency within a single sample. This uneven distribution is also reflected in a low Simpson's diversity index [23] during the course of infection in the mice (Table 1). Based on the parasite density and the frequency with which each tag occurred at the different time points sampled, the parasitaemia of individual populations could be extrapolated from the data (Figure S3). This revealed that the dominant and minor populations showed similar fluctuations in parasitaemia, although their titres differed by several orders of magnitude.
Tab. 1. Diversity index (Simpson's index). 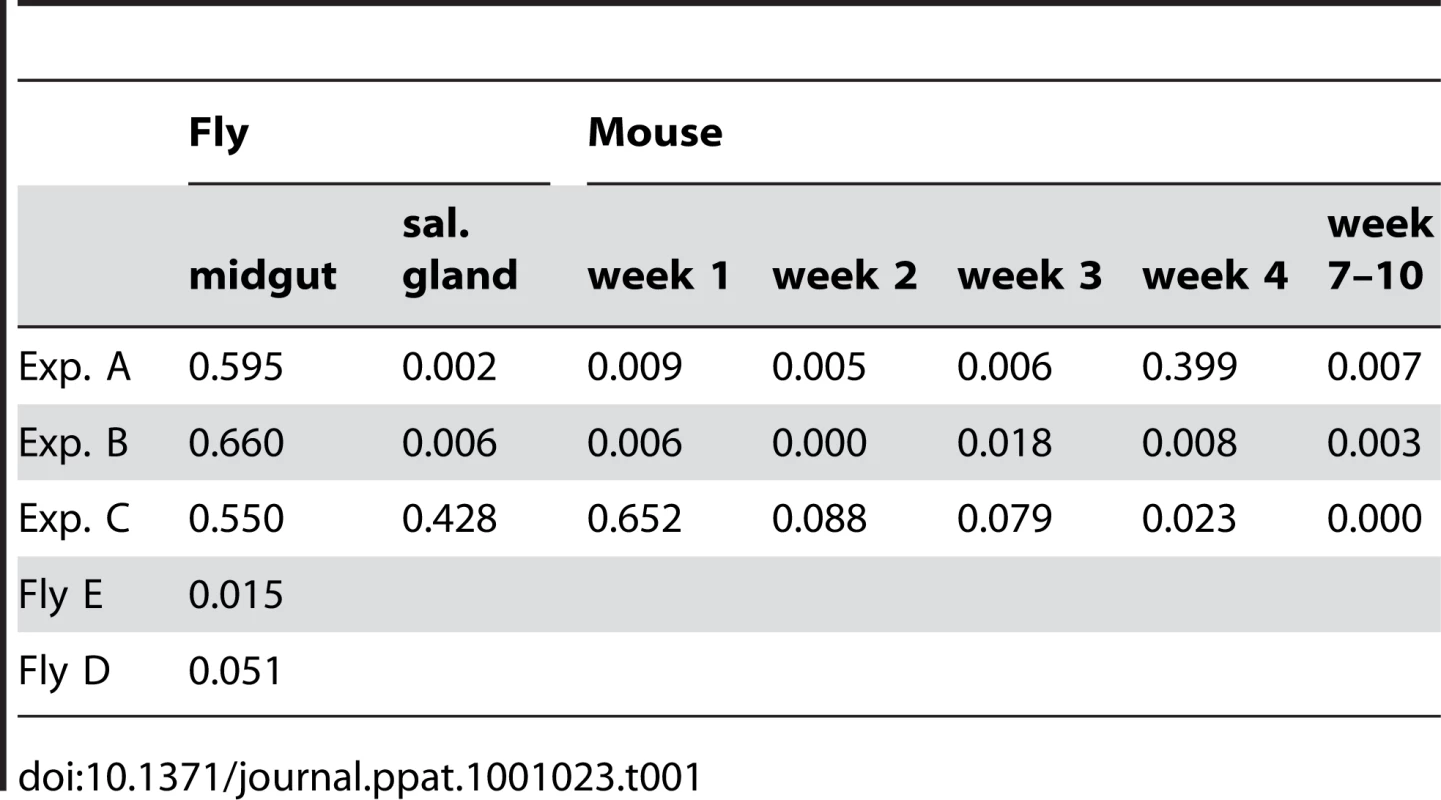
Analysis of the tags present in the midguts of flies D and E, which became infected after feeding on mouse C, revealed two interesting outcomes. First, the tag that was dominant in the mouse remained so in the midgut of both flies (Figure 3). Second, minor tags in the bloodstream form population were also present. For example tags with frequencies of 0.5–3% in the bloodstream form population (tags 2, 5, and 6) were detectable in the midgut. Interestingly, tag 3 was detectable in fly E even though it was under the detection limit in all the samples from mouse C. Together, the two flies took up five different tags. This mouse had a parasitaemia of 4.9×106 and 7.7×106 per ml on days 18 and 30, respectively. Assuming the flies imbibed approximately 20µl of blood, about 1–1.5×105 trypanosomes might reach the midgut of each fly. If approximately 1% survived [6] even very minor tags would be represented by 2–30 individual trypanosomes that could contribute to establishing the midgut infection.
Discussion
By using tagged trypanosomes originating from a single clone, we have been able to monitor the dynamics of a parasite population throughout the life cycle without the confounding factor of strain differences. This analysis revealed that a major bottleneck in the life cycle occurs during migration of parasites from the midgut to the salivary glands, leading to the establishment of one or a few dominant genotypes in each fly. Minor genotypes constituting <1% of the population could also be detected in the glands, however. These were transmitted to mice together with the dominant genotype(s), and were found to persist during a chronic infection and survive transmission to a new insect vector.
The frequency and diversity of tags enabled us to extrapolate the minimum number of parasites transferred between hosts and provide an estimate of the size of the bottleneck (Figure 4). We estimate that at least 500–1000 trypanosomes must have survived the transfer from mouse C to flies D and E and colonised their midguts. The population structure was similar to that in the mouse blood at the time of the blood meal, indicating that despite the reduction in parasite numbers, transmission from the mammal to the insect does not represent a severe bottleneck.
Fig. 4. Population size through the life cycle of T. brucei based on data from this study. 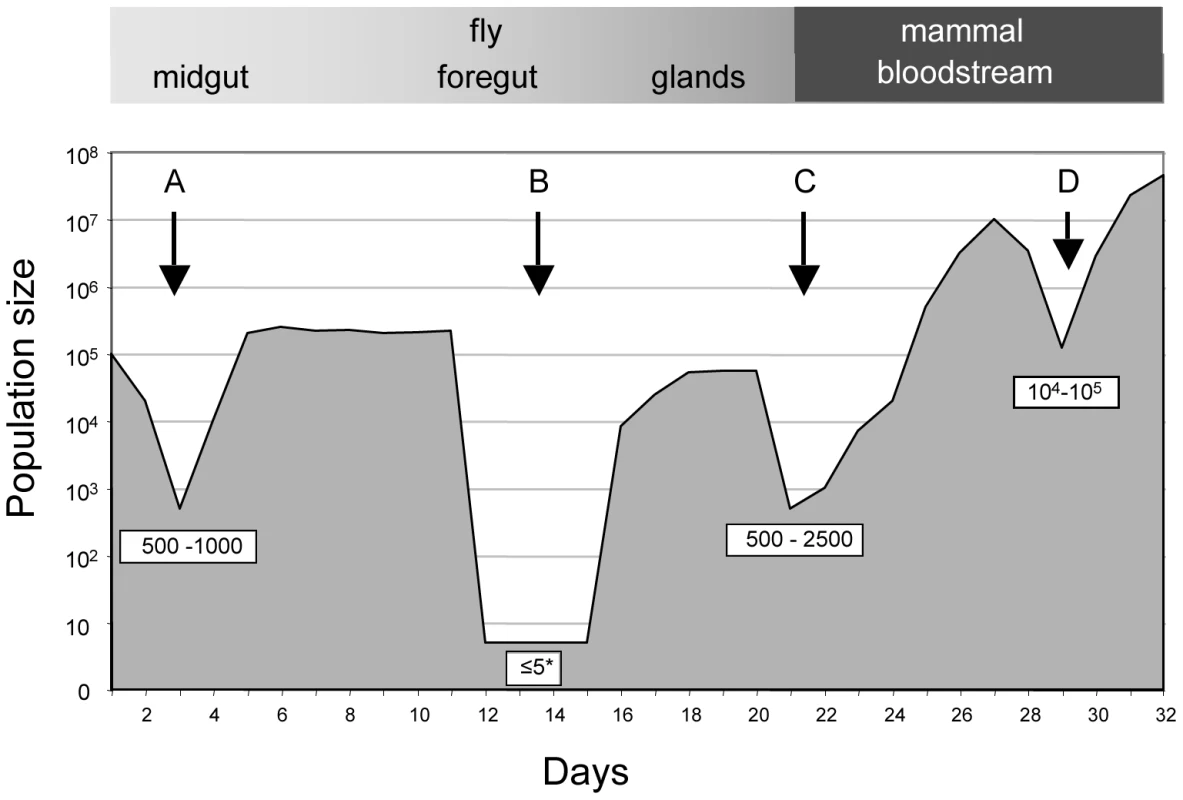
A: establishment of infection in the fly midgut following a blood meal; B: trypanosomes migrating to the salivary glands; C: establishment of infection in the mammal following fly transmission; D: fluctuations in parasite numbers in the mammalian bloodstream. Numbers in boxes refer to minimum numbers of trypanosomes surviving the transition between different tissues and hosts. The asterisk depicts the possibility that there might be waves of migration of a few parasites at a time from the midgut to the salivary glands. Likewise, the minimum number of trypanosomes that survived the transfer from infected flies and initiated the infection in the 3 mice was estimated by drawing simulation to range from 500–2500 (Figures 4 and S4). This is similar to the number of metacyclic forms extruded by infected flies [14], [15], implying that most of the parasites that are inoculated can contribute to an infection. With the exception of a single major tag that was present in the salivary glands of fly C, parasites isolated from mice during the first week of infection reflected the diversity and distribution of parasites in salivary glands of the infecting fly. A large number of tags were detected during the chronic infections, but their frequency was highly variable. Some tags were detected only once in a total of 6800 sequences obtained from five blood samples (e.g. tag 1 or tag 8 in mouse B). Considering that the parasitaemia never fell below 105 ml−1, we consider it likely that most tags transmitted by the tsetse were present continuously during mouse infection although they were under the detection limit in individual samples. Antigenic variation must have occurred several times during the course of these infections, since the mice were immunocompetent. Ordered (hierarchical) VSG expression is a widely accepted model for antigenic variation [24], [25], [26]. Since the tagged trypanosomes derive from a clone, it is possible that they obeyed the same hierarchy, which would result in synchronised VSG expression by trypanosomes with different tags. This is not mandatory, however, as simultaneous expression of several variants is both predicted by the model of Lythgoe et al. [26] and can occur in vivo [27].
In contrast to transmission between hosts, migration from the midgut to the salivary glands of tsetse caused profound changes in relative frequencies of different tags, with up to five tags per experiment changing from dominant to minor, or vice versa. Two flies had a single dominant tag accounting for more than 99% of the population in the glands, while the remaining fly had 4 dominant tags (one of which accounted for 74% of the population). When minor populations from the glands and the corresponding mice were taken into account, all the tags that were found in the midgut were represented, meaning that at least 6 trypanosomes must have reached the salivary glands in each fly. The strong dominance of one tag each in flies A and B, and to a slightly lesser extent in Fly C, could best be explained by waves of migration by very few parasites at a time. Trypanosomes are tightly packed in infected glands and have to compete for space. Early migration might be a more important factor than dominance in the midgut if parasites that arrive first can disperse and colonise the glands more readily than latecomers. This “race for space” would account for the shift in frequencies between the midgut and salivary glands of a single fly and also explain why a single tag can dominate the population while others remain very minor. It would also be compatible with publications that have reported changes in the relative frequencies of two strains of trypanosomes between the midgut and glands, as well as glands colonised by only one of the two strains [11], [12]. The latter study catalogued whether each of the pair of salivary glands contained one or both strains, thus allowing the minimum number of founder trypanosomes in each fly to be extrapolated from the data. In these experiments the salivary gland infections of approximately two-thirds of the flies could have been established by as few as one or two trypanosomes, while the remaining third would have required at least three or four. It could not be excluded, however, that several trypanosomes of one strain migrated to the same gland, or that very minor populations might have been overlooked.
An important outcome of the extreme bottleneck that can occur between the midgut and the salivary glands is that rare variants can be amplified in individual flies and be disseminated by them. If a variant has a selective advantage in mammals, such as altered host range or increased resistance to drugs, this might cause it to become the major species circulating locally [28], [29]. Such a phenomenon could explain the epidemic population structure of T. brucei documented by MacLeod and coworkers, in which clonal expansion of a few genotypes in a region occurs against a background of frequent recombination between strains [30].
Our data indicate that both mammals and tsetse can readily acquire and transmit more than one genotype in the course of a single blood meal. Mixed genotypes have been detected fairly frequently in field isolates from cattle, humans and tsetse [30], [31], [32], [33], and it is possible that they might be even more widespread since minor populations would escape detection. Co-infection of susceptible flies with different strains of trypanosomes might affect parasite population dynamics [34] and also increase the chances of genetic exchange [35], [36] if the parasites develop and migrate in parallel.
The approach that we have used here can be extended to study other facets of infection with trypanosomes, for example the population found in the central nervous system during the late stage of sleeping sickness or the parasites causing relapse infections after drug treatment. It can also be applied to the analysis of population dynamics of any other parasite that is amenable to transfection, including other African trypanosomes that do not infect the salivary glands and may therefore show different dynamics in the tsetse fly.
Materials and Methods
Ethics statement
Animal experiments were approved by the local veterinary authorities (Veterinäramt Basel-Stadt) in compliance with Swiss federal law (TSchG) and cantonal by-laws (TSchV Basel-Stadt).
Trypanosomes
Trypanosoma brucei brucei AnTat 1.1 [37] procyclic forms were cloned by the micro-drop method [38] and stabilates were made after 22 days. Tsetse flies (see below) were infected with one clone, and the infected salivary glands of one fly were dissected at day 27 and inoculated intraperitoneally into a female NMRI mouse (RCC, Ittingen, Switzerland), which developed a parasitaemia of 108 trypanosomes ml−1 at day 5 post infection. Bloodstream forms, obtained by heart puncture, were triggered to differentiate into procyclic forms in SDM-79 supplemented with 10% foetal bovine serum, 3 mM sodium citrate and cis-aconitate (CCA) [39], and 20mM glycerol at 27°C for 3 days [40]. Procyclic forms were cultured thereafter in the same medium without CCA.
Infection of tsetse flies
Pupae of Glossina morsitans morsitans were obtained from the Institute of Zoology, Bratislava, Slovakia. The flies were maintained at 25°C and 70% relative humidity with 12 hours of light per day. Teneral tsetse flies (under the age of 72 hours) were infected with procyclic forms as described previously [22]. Starting twenty days post-infection, tsetse flies were examined for the presence of metacyclic forms in their saliva. Tsetse flies with a mature infection were allowed to feed on NMRI mice 2 to 4 days after the appearance of first metacyclic forms. Subsequently the paired ducts of the salivary glands were extracted from the neck of the tsetse flies. This prevented contamination with midgut forms. The midgut (including the proventriculus) was then dissected out of the abdomen. The tissues were dissected on separate slides in a drop of PBS and then transferred to an Eppendorf tube containing 200µl lysis buffer (see below) and stored at −20°C prior to DNA extraction.
Infection of mice
Female NMRI mice (RCC, Ittingen) were kept at 22°C, 70% relative humidity and with 12 hours of light per day. To determine the parasitaemia, 10 µl tail blood were mixed with 40 µl 3.2% sodium citrate, and 4 µl were uniformly distributed under a 20 mm2 cover slip. For each sample, 10–15 fields were counted. The parasitaemia is given as the number of trypanosomes per ml mouse blood. For analysis of the tags 50 µl of tail blood was processed.
Plasmid constructs and stable transfection of trypanosomes
The insert from the plasmid pKON [21], was amplified by PCR using the primers Bgl II-promotor (ATAGATCTCGAAAACTCTTCGGGA) and KO 2 (TATCTAGAGGGCACTGCAGT). Bgl II and Xba I sites, respectively, are underlined. The PCR product encompassing the EP1 promoter, the neomycin resistance gene and 19bp of the EP2 3′ untranslated region (UTR) was digested with Bgl II and Xba I. The plasmid pLew111 (http://tryps.rockefeller.edu/) was digested with Bgl II and Nhe I to provide the plasmid backbone with the rDNA spacer. The digested PCR product was ligated to the backbone (Xba I and Nhe I have compatible ends). The resulting construct, pIns has a single Bgl II site between the rDNA spacer and the procyclin promoter that was used for the insertion of unique tags (Figure 2B).
An oligonucleotide (ATCACGGCCGGGAGATCT(N)40AGATCTGTGAGACCCATTAAGCTTCC) containing a variable 40mer flanked by two constant regions with Bgl II sites (underlined), was purchased from Microsynth AG, Balgach, Switzerland. Double-stranded DNA was produced by amplification with the constant flanking sequences: iTag-oligo (ATCACGGCCGGGAGATCT) and BIL-4A (GGAAGCTTAATGGGTCTCAC). The PCR product was inserted into pCR2.1 TOPO (Invitrogen, Carlsbad Ca, USA) according to the manufacturer's protocol and used to transform E. coli XL-1 blue. Purified plasmids were sequenced using standard methods. Eight tags were selected (Table S1a), the fragments released with Bgl II and ligated into pIns to generate the plasmid series piTag1–8.
Transfection was performed with 10µg of each plasmid (piTag1–8) linearised with Not I. Plasmids were electroporated separately into 2.5×107 procyclic trypanosomes. Transfection and cloning by limiting dilution were carried out as described elsewhere [40]. G418 (25µg ml−1) was used to select stable transformants.
DNA extraction, nested PCR and sequencing
Samples were resuspended in 200µl lysis buffer (100 mM NaCl, 5 mM sodium EDTA, 10 mM tris-HCl, pH 8), supplemented with 20 µl RNAse A (1 mg/ml) and incubated for 1 h at 37°C, followed by the addition of 10µl Pronase (20mg/ml) and incubation for a further 2 h. Genomic DNA was isolated by phenol/chloroform extraction, precipitated with ethanol and resuspended in 50 µl water. Genomic DNA obtained from blood samples was subjected to an additional precipitation using 0.5 volumes of 7.5M ammonium acetate and 5 volumes ethanol.
Nested PCR was performed on 1 µl of each DNA sample. The oligonucleotides rDNAsense/ep2sas2 (Table S1b) were used to generate a product of ∼700 bp using the following conditions: 3 min at 96°C, 30 cycles of 1 min 94°C, 1 min 45°C, and 45 sec 72°C, followed by 10 min extension at 72°C. The second PCR performed with 1 µl from the first reaction as template and the fusion primers A and B, these consist of two regions: a template specific region for PCR amplification and a fusion region for 454 sequencing (Microsynth, Balgach, Switzerland). The primers A1–A6 can be distinguished by a variable 6mer barcode that connects the two regions (Table S1b). This barcode was used to allocate samples in the same region on the pyrosequencing plate (see below). The PCR conditions with the primers A and B were: 3 min at 96°C, 30 cycles of 1 min 94°C, 50 sec 52°C and 30 sec 72°C followed by 10 min extension at 72°C, yielding products of 177 bp. 454 picotiter plate pyrosequencing was performed by Microsynth, Balgach, Switzerland, with the Roche Genome Sequencer FLX System as described elsewhere [41], [42]. Five regions of a picotiter plate were used for this study: one (I) for the control DNA and four regions (II–V) for the samples (Figure S2, panel A). For the control sequencing reactions, cultures of individual clones were mixed, genomic DNA extracted, and split into 6 aliquots. DNA from each aliquot was amplified with a different barcoded fusion primer A1–A6 together with the primer B. The samples collected from the flies and mice were amplified with the primers A1/B–A6/B as indicated in Figure 2. The barcodes were identified with the SFF file program sfffile (included in the 454 software package), allowing one mismatch. The distribution of the tags (Figure S2, panel B) was very similar among all control samples (two-sided paired T-test; p>0.3 for all combinations).
Supporting Information
Zdroje
1. BartonN
CharlesworthB
1984 Genetic Revolutions, Founder Effects, and Speciation. Annual Review of Ecology and Systematics 15 133.164
2. RichSM
LichtMC
HudsonRR
AyalaFJ
1998 Malaria's Eve: evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum. Proc Natl Acad Sci U S A 95 4425 4430
3. MackinnonMJ
BellA
ReadAF
2005 The effects of mosquito transmission and population bottlenecking on virulence, multiplication rate and rosetting in rodent malaria. Int J Parasitol 35 145 153
4. NeiM
MaruyamaT
ChakrabortyR
1975 The bottleneck effect and genetic variability in populations. Evolution 29 1 10
5. RoditiI
LehaneMJ
2008 Interactions between trypanosomes and tsetse flies. Curr Opin Microbiol 11 345 351
6. Van Den AbbeeleJ
ClaesY
van BockstaeleD
Le RayD
CoosemansM
1999 Tryanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology 118 469 478
7. MolooSK
KabataJM
SabwaCL
1994 A study on the maturation of procyclic Trypanosoma brucei brucei in Glossina morsitans centralis and G. brevipalpis. Med Vet Entomol 8 369 374
8. SharmaR
PeacockL
GluenzE
GullK
GibsonW
2008 Asymmetric cell division as a route to reduction in cell length and change in cell morphology in trypanosomes. Protist 159 137 151
9. BruceD
HamertonAE
BatemanHR
MackieFP
1911 Further researches on the development of Trypanosoma gambiense in Glossina palpalis. Proc Roy Soc B 83 513 527
10. HarmsenR
1973 The nature of the establishment barrier for Trypanosoma brucei in the gut of Glossina pallidipes. Trans R Soc Trop Med Hyg 67 364 373
11. PeacockL
FerrisV
BaileyM
GibsonW
2007 Dynamics of infection and competition between two strains of Trypanosoma brucei brucei in the tsetse fly observed using fluorescent markers. Kinetoplastid Biol Dis 6 4
12. GibsonW
PeacockL
FerrisV
WilliamsK
BaileyM
2008 The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. Parasit Vectors 1 4
13. VickermanK
1985 Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull 41 105 114
14. FairbairnH
BurttE
1946 The infectivity to man of a strain of Trypanosoma rhodesiense transmitted cyclically by Glossina morsitans through sheep and antelope: evidence that man requires a minimum infective dose of metacyclic trypanosomes. Annals of Tropical Medicine and Parasitology 40 270 313
15. BrunR
JenniL
1987 Human serum resistance of metacyclic forms of Trypanosoma brucei brucei, T. brucei rhodesiense and T. brucei gambiense. Parasitol Res 73 218 223
16. VickermanK
TetleyL
HendryKA
TurnerCM
1988 Biology of African trypanosomes in the tsetse fly. Biol Cell 64 109 119
17. MaudlinI
WelburnSC
1989 A single trypanosome is sufficient to infect a tsetse fly. Ann Trop Med Parasitol 83 431 433
18. ThuitaJK
KagiraJM
MwangangiD
MatovuE
TurnerCM
2008 Trypanosoma brucei rhodesiense Transmitted by a Single Tsetse Fly Bite in Vervet Monkeys as a Model of Human African Trypanosomiasis. PLoS Negl Trop Dis 2 e238
19. GingrichJB
WardRA
MackenLM
EsserKM
1981 Some phenomena associated with the development of Trypanosoma brucei rhodesiense infections in the tsetse fly, Glossina morsitans. Am J Trop Med Hyg 30 570 574
20. EbertD
1998 Experimental evolution of parasites. Science 282 1432 1435
21. RueppS
FurgerA
KurathU
RenggliCK
HemphillA
1997 Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J Cell Biol 137 1369 1379
22. VassellaE
OberleM
UrwylerS
RenggliCK
StuderE
2009 Major surface glycoproteins of insect forms of Trypanosoma brucei are not essential for cyclical transmission by tsetse. PLoS One 4 e4493
23. SimpsonEH
1949 Measurement of diversity. Nature 163 688
24. AitchesonN
TalbotS
ShapiroJ
HughesK
AdkinC
2005 VSG switching in Trypanosoma brucei: antigenic variation analysed using RNAi in the absence of immune selection. Mol Microbiol 57 1608 1622
25. MorrisonLJ
MajiwaP
ReadAF
BarryJD
2005 Probabilistic order in antigenic variation of Trypanosoma brucei. Int J Parasitol
26. LythgoeKA
MorrisonLJ
ReadAF
BarryJD
2007 Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc Natl Acad Sci U S A 104 8095 8100
27. CapbernA
GiroudC
BaltzT
MatternP
1977 Trypanosoma equiperdum: antigenic variations in experimental trypanosomiasis of rabbits. Exp Parasitol 42 6 13
28. WoottonJC
FengX
FerdigMT
CooperRA
MuJ
2002 Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418 320 323
29. de RoodeJC
CulletonR
BellAS
ReadAF
2004 Competitive release of drug resistance following drug treatment of mixed Plasmodium chabaudi infections. Malar J 3 33
30. MacLeodA
TweedieA
WelburnSC
MaudlinI
TurnerCM
2000 Minisatellite marker analysis of Trypanosoma brucei: reconciliation of clonal, panmictic, and epidemic population genetic structures. Proc Natl Acad Sci U S A 97 13442 13447
31. StevensJR
Mathieu DaudeF
McNamaraJJ
MizenVH
NzilaA
1994 Mixed populations of Trypanosoma brucei in wild Glossina palpalis palpalis. Trop Med Parasitol 45 313 318
32. MacLeodA
TurnerCM
TaitA
1999 A high level of mixed Trypanosoma brucei infections in tsetse flies detected by three hypervariable minisatellites. Mol Biochem Parasitol 102 237 248
33. BalmerO
CacconeA
2008 Multiple-strain infections of Trypanosoma brucei across Africa. Acta Trop 107 275 279
34. BalmerO
StearnsSC
SchotzauA
BrunR
2009 Intraspecific competition between co-infecting parasite strains enhances host survival in African trypanosomes. Ecology 90 3367 3378
35. TaitA
1980 Evidence for diploidy and mating in trypanosomes. Nature 287 536 538
36. JenniL
MartiS
SchweizerJ
BetschartB
Le PageRW
1986 Hybrid formation between African trypanosomes during cyclical transmission. Nature 322 173 175
37. Le RayD
BarryJD
EastonC
VickermanK
1977 First tsetse fly transmission of the “AnTat” serodeme of Trypanosoma brucei. Ann Soc Belg Med Trop 57 369 381
38. BrunR
SchoenenbergerM
1979 Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Tropica 36 289 292
39. BrunR
SchoenenbergerM
1981 Stimulating effect of citrate and cis-aconitate on the transformation of Trypanosoma brucei bloodstream forms to procyclic forms in vitro. Z Parasitenkd 66 17 24
40. VassellaE
Den AbbeeleJV
BütikoferP
RenggliCK
FurgerA
2000 A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post - transcriptionally by glycerol or hypoxia. Genes Dev 14 615 626
41. ThomasRK
NickersonE
SimonsJF
JannePA
TengsT
2006 Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med 12 852 855
42. MarguliesM
EgholmM
AltmanWE
AttiyaS
BaderJS
2005 Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376 380
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System inČlánek A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 7- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Mouse Resistance Protein Irgm1 (LRG-47): A Regulator or an Effector of Pathogen Defense?
- Leprosy and the Adaptation of Human Toll-Like Receptor 1
- Intergenomic Arms Races: Detection of a Nuclear Rescue Gene of Male-Killing in a Ladybird
- The Role of Chemokines during Viral Infection of the CNS
- Bottlenecks and the Maintenance of Minor Genotypes during the Life Cycle of
- DNA Damage Triggers Genetic Exchange in
- The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System in
- Uropathogenic Modulates Immune Responses and Its Curli Fimbriae Interact with the Antimicrobial Peptide LL-37
- Biogenesis of the Inner Membrane Complex Is Dependent on Vesicular Transport by the Alveolate Specific GTPase Rab11B
- A Spatio-Temporal Analysis of Matrix Protein and Nucleocapsid Trafficking during Vesicular Stomatitis Virus Uncoating
- Hepatitis B Virus Polymerase Blocks Pattern Recognition Receptor Signaling via Interaction with DDX3: Implications for Immune Evasion
- Quasispecies Theory and the Behavior of RNA Viruses
- Bid Regulates the Pathogenesis of Neurotropic Reovirus
- Distinct Roles for Dectin-1 and TLR4 in the Pathogenesis of Keratitis
- Unexpected Inheritance: Multiple Integrations of Ancient Bornavirus and Ebolavirus/Marburgvirus Sequences in Vertebrate Genomes
- Balanced Nuclear and Cytoplasmic Activities of EDS1 Are Required for a Complete Plant Innate Immune Response
- Adaptation of Hepatitis C Virus to Mouse CD81 Permits Infection of Mouse Cells in the Absence of Human Entry Factors
- An Outer Membrane Receptor of Involved in Zinc Acquisition with Vaccine Potential
- Inositol Hexakisphosphate-Induced Autoprocessing of Large Bacterial Protein Toxins
- Plus- and Minus-End Directed Microtubule Motors Bind Simultaneously to Herpes Simplex Virus Capsids Using Different Inner Tegument Structures
- Distinct Pathogenesis and Host Responses during Infection of by and
- Can Bacteria Evolve Resistance to Quorum Sensing Disruption?
- RNA Virus Replication Complexes
- PPARγ and LXR Signaling Inhibit Dendritic Cell-Mediated HIV-1 Capture and -Infection
- The Virulence Protein SopD2 Regulates Membrane Dynamics of -Containing Vacuoles
- Genome-Wide Mutagenesis Reveals That ORF7 Is a Novel VZV Skin-Tropic Factor
- Adaptive Evolution of Includes Retroviral Insertion and Positive Selection at Two Clusters of Residues Flanking the Substrate Groove
- A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
- Transduction of Human T Cells with a Novel T-Cell Receptor Confers Anti-HCV Reactivity
- Identification of GBV-D, a Novel GB-like Flavivirus from Old World Frugivorous Bats () in Bangladesh
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- HIV gp41 Engages gC1qR on CD4+ T Cells to Induce the Expression of an NK Ligand through the PIP3/H2O2 Pathway
- Oxidation of Helix-3 Methionines Precedes the Formation of PK Resistant PrP
- Protection from the 2009 H1N1 Pandemic Influenza by an Antibody from Combinatorial Survivor-Based Libraries
- Murine Gamma-Herpesvirus 68 Hijacks MAVS and IKKβ to Initiate Lytic Replication
- Viral Protein Inhibits RISC Activity by Argonaute Binding through Conserved WG/GW Motifs
- TOPO3α Influences Antigenic Variation by Monitoring Expression-Site-Associated Switching in
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- The Meningococcal Vaccine Candidate Neisserial Surface Protein A (NspA) Binds to Factor H and Enhances Meningococcal Resistance to Complement
- Network Modeling Reveals Prevalent Negative Regulatory Relationships between Signaling Sectors in Arabidopsis Immune Signaling
- Endothelial Galectin-1 Binds to Specific Glycans on Nipah Virus Fusion Protein and Inhibits Maturation, Mobility, and Function to Block Syncytia Formation
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
- Integration Preferences of Wildtype AAV-2 for Consensus Rep-Binding Sites at Numerous Loci in the Human Genome
- Epigenetic Analysis of KSHV Latent and Lytic Genomes
- Vaccinia Virus–Encoded Ribonucleotide Reductase Subunits Are Differentially Required for Replication and Pathogenesis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- RNA Virus Replication Complexes
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



