-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Endothelial Galectin-1 Binds to Specific Glycans on Nipah Virus Fusion Protein and Inhibits Maturation, Mobility, and Function to Block Syncytia Formation
Nipah virus targets human endothelial cells via NiV-F and NiV-G envelope glycoproteins, resulting in endothelial syncytia formation and vascular compromise. Endothelial cells respond to viral infection by releasing innate immune effectors, including galectins, which are secreted proteins that bind to specific glycan ligands on cell surface glycoproteins. We demonstrate that galectin-1 reduces NiV-F mediated fusion of endothelial cells, and that endogenous galectin-1 in endothelial cells is sufficient to inhibit syncytia formation. Galectin-1 regulates NiV-F mediated cell fusion at three distinct points, including retarding maturation of nascent NiV-F, reducing NiV-F lateral mobility on the plasma membrane, and directly inhibiting the conformational change in NiV-F required for triggering fusion. Characterization of the NiV-F N-glycome showed that the critical site for galectin-1 inhibition is rich in glycan structures known to bind galectin-1. These studies identify a unique set of mechanisms for regulating pathophysiology of NiV infection at the level of the target cell.
Published in the journal: Endothelial Galectin-1 Binds to Specific Glycans on Nipah Virus Fusion Protein and Inhibits Maturation, Mobility, and Function to Block Syncytia Formation. PLoS Pathog 6(7): e32767. doi:10.1371/journal.ppat.1000993
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000993Summary
Nipah virus targets human endothelial cells via NiV-F and NiV-G envelope glycoproteins, resulting in endothelial syncytia formation and vascular compromise. Endothelial cells respond to viral infection by releasing innate immune effectors, including galectins, which are secreted proteins that bind to specific glycan ligands on cell surface glycoproteins. We demonstrate that galectin-1 reduces NiV-F mediated fusion of endothelial cells, and that endogenous galectin-1 in endothelial cells is sufficient to inhibit syncytia formation. Galectin-1 regulates NiV-F mediated cell fusion at three distinct points, including retarding maturation of nascent NiV-F, reducing NiV-F lateral mobility on the plasma membrane, and directly inhibiting the conformational change in NiV-F required for triggering fusion. Characterization of the NiV-F N-glycome showed that the critical site for galectin-1 inhibition is rich in glycan structures known to bind galectin-1. These studies identify a unique set of mechanisms for regulating pathophysiology of NiV infection at the level of the target cell.
Introduction
Nipah virus (NiV) is a lethal emerging virus that infects agricultural livestock and humans. In 1999–2000, NiV infection of agricultural workers in Malaysia and Singapore resulted in a 40% mortality rate, and subsequent outbreaks in Bangladesh resulted in an average case-fatality ratio greater than 70% [1]. In humans, NiV targets endothelial and neural cells, with resulting respiratory and neurologic sequelae; patients infected with Nipah virus often succumb to acute encephalitis with accompanying multi-organ failure due to systemic vasculitis. Autopsy studies of NiV victims identified virus in endothelial cells, with endothelial cell syncytia formation being a pathognomonic hallmark of NiV infection [2], [3], [4].
NiV, a member of a new genus of Paramyxoviridae, is an enveloped virus with two viral envelope glycoproteins, NiV-G and NiV-F, that mediate viral entry [5]. The NiV-G attachment glycoprotein binds to specific receptors, primarily ephrinB2 or alternatively ephrinB3 [6], [7], [8]. While ephrinB2 and ephrinB3 are expressed in neuronal cells, only ephrinB2 is highly expressed on endothelial cells [9]. The NiV-F glycoprotein mediates fusion of bound virus with target cells. After endothelial cell infection, NiV-F and NiV-G glycoproteins are expressed on the surface of infected cells. This triggers cell-cell fusion with neighboring endothelial cells (homologous fusion) or stromal cells (heterologous fusion), resulting in endothelial syncytia formation, endothelial disruption, and subsequent hemorrhage and tissue damage.
The NiV-F fusion glycoprotein, NiV-F0, is initially expressed at the cell surface as a single glycosylated polypeptide precursor, but subsequently undergoes endocytosis and endosomal proteolytic cleavage by cathepsin L into F1 and F2 subunits that are disulfide linked to form the mature fusion protein NiV-F1/2 [10], [11], [12]. Mature NiV-F traffics back to the cell surface, where the protein can initiate cell-cell fusion at neutral pH when it is appropriately triggered by receptor binding to NiV-G. Our labs and others have previously demonstrated that specific N-glycans on NiV-F play important roles in regulating the extent of cell fusion [13], [14].
Infection of endothelial cells by viruses results in the release of innate immune effectors, including galectins, a family of mammalian lectins [15], [16]. Galectins are soluble, secreted proteins that stay associated with the cell surface by binding specific cell surface glycan ligands on specific glycoprotein receptors [17]. All galectins are multivalent or form higher order multimers, and can thus cross-link glycan ligands and the glycoprotein receptors that bear these ligands on the surface of cells. Human endothelial cells express galectin-1, as well as galectin-3, -8, and -9, and in vitro and in vivo activation of human endothelial cells increased synthesis as well as secretion and cell surface localization of galectin-1 [17], [18], [19].
We have previously shown that recombinant human galectin-1 inhibits cell fusion and syncytia formation caused by NiV-F [20]. The mechanism by which galectin-1 inhibits NiV-F mediated cell fusion is not well understood; however, we found that galectin-1 bound directly to NiV-F and caused NiV-F to oligomerize, suggesting that galectin-1 can cross link NiV-F on the surface of infected cells. Moreover, a single N-glycan site in NiV-F, the F3 glycosylation site, appears to be critical for galectin-1 mediated inhibition of cell fusion, as mutation of that site to abolish N-glycan addition, reduced galectin-1 binding to NiV-F and reduced the inhibitory effect of galectin-1 by 50% [20].
In the present study, we demonstrate that galectin-1 regulates NiV-F mediated fusion of endothelial cells and neural cells, the targets of NiV infection in vivo. Since endothelial cell syncytia formation is a pathognomic feature of NiV infection, we investigated the mechanisms involved in galectin-1 modulation of cell-cell fusion. We found that galectin-1 regulates cell fusion at three distinct points in the process; galectin-1 retains immature NiV-F0 on the cell surface to reduce production of the mature NiV-F fusion protein, galectin-1 reduces lateral movement of NiV-F on the plasma membrane that is required for cell-cell fusion, and galectin-1 directly inhibits the fusogenic activity of NiV-F by preventing fusion-peptide exposure and pre-hairpin intermediate (PHI) formation. The biological significance of our results is underscored by our demonstration that endogenous galectin-1 on endothelial cells is sufficient to reduce NiV-F mediated fusion. These studies identify a unique set of mechanisms for regulating the pathophysiology of NiV induced syncytia formation at the target cell level, and contribute to our understanding of the interaction between galectin-1 and glycoproteins of microbial pathogens.
Results
Cell surface galectin-1 inhibits syncytia formation in relevant cell types
We previously found that galectin-1 inhibits syncytia formation in Vero cells mediated by NiV-F and NiV-G [20]. In vivo, endothelial cells and neuronal cells are the main targets of Nipah virus [2], due to the high expression of ephrinB2 and/or ephrinB3 by these cells [6], [8]. To explore the role of galectin-1 in NiV-F and G mediated endothelial and glial cell syncytia formation, we used a heterologous overlay fusion assay. EphrinB2 and ephrinB3-negative cells (PK13) were transfected with NiV-F and NiV-G and plated on a confluent layer of the indicated target cell type (Fig. 1). Cell-cell fusion could be observed as early as 45 min and plateaued after approximately 6 hrs. Addition of recombinant galectin-1 treatment to the co-cultures significantly inhibited syncytia formation in each of the target cell lines. Vero cells, human umbilical endothelial cells (HUVECs), and microvascular endothelial cells (mVECs) all showed a 90% reduction in cell-cell fusion, and U87 glioblastoma cells showed a 60% reduction in fusion, compared to control cultures, in the presence of galectin-1 (Fig. 1A). Representative images from the syncytia assays for each target cell type are shown in Fig. 1B.
Fig. 1. Galectin-1 blocks NiV-F and G mediated syncytia formation of endothelial and glial cells. 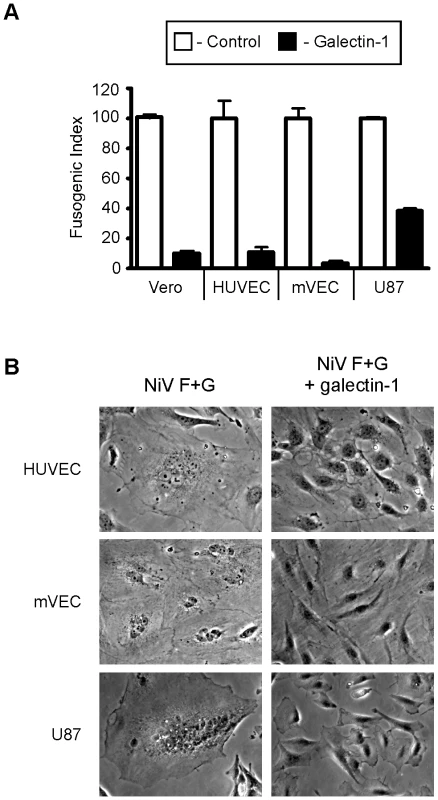
A, Quantification of galectin-1 inhibition. PK-13 (ephrinB2 negative) cells expressing NiV-F and NiV-G were added to monolayers of ephrinB2 positive cells, Vero (control), HUVEC, mVEC, and U87. Heterologous fusion in the absence and presence of 20µM galectin-1 (white and black bars respectively) were quantified as described in Experimental Procedures. Data are mean ± SD of triplicate samples from one of three replicate experiments. B, Representative images of cell fusion in the absence or presence of galectin-1. Left panels show multinucleated syncytia in the indicated cell type. Right panels are cells treated with galectin-1 (20×). Endogenous endothelial galectin-1 inhibits syncytia formation
Nipah virus infection results in extensive damage to endothelial cells as a result of syncytia formation, culminating in multi-organ hemorrhage and death. During viral infection, endothelial cells become activated and release immune mediators, including galectin-1 [18], [21]. We asked if endogenous endothelial galectin-1 could affect NiV-F mediated syncytia formation, using the heterologous fusion assay as in Fig. 1. To activate HUVECs, the cells were cultured in 20% human serum for four days, a process that increased cell surface galectin-1 protein expression in these cells [21]. Galectin-1 expression on the cell surface was quantified by flow cytometry, and activated HUVECs demonstrated a consistent increase in cell surface galectin-1 compared to resting cells (Fig. 2A). Activated HUVECs also showed a significant decrease (40–50%) in cell-cell fusion, which correlated with the increase in cell surface galectin-1 (Fig. 2B).
Fig. 2. Endogenous endothelial galectin-1 inhibits NiV-F and NiV-G mediated syncytia formation. 
A, Activated HUVECs have increased cell surface galectin-1 compared to resting cells. Flow cytometric analysis of cell surface galectin-1 on resting (light grey) and activated (dark grey) HUVECs, and on resting HUVECs plus exogenous galectin-1 (black) Data are mean ± SEM of three independent experiments, each done in triplicate. B, Activated HUVECs are resistant to NiV-F and NiV-G mediated cell fusion. Heterologous cell fusion of resting (light grey) and activated (dark grey) HUVECs, and resting HUVECs plus exogenous galectin-1 (black). Data are mean ± SD of triplicate samples from one of three replicate experiments. C, Reduction of cell surface galectin-1 by siRNA. Flow cytometric analysis of cell surface galectin-1 on resting HUVECs (grey filled), siRNA treated HUVECs (grey line), and siRNA treated HUVECS with 20µM exogenous galectin-1 (black line). D, Reduction of cell surface galectin-1 in HUVECs increases susceptibility to NiV-F and G mediated cell fusion. Heterologous cell fusion of resting cells (light grey), cells with reduced galectin-1 (white), and cells with reduced galectin-1 plus exogenous galectin-1 (dark grey). Data are mean ± SD of triplicate samples from one of three replicate experiments. Conversely, in order to determine if endogenous galectin-1, even on resting HUVECs, was sufficient to affect NiV-F and G mediated syncytia formation, we reduced expression of galectin-1 in HUVECs using lentiviral vectors expressing siRNAs targeted against galectin-1. A combination of three siRNAs reduced galectin-1 protein approximately 70% (data not shown), and reduced cell surface galectin-1 approximately two-fold (Fig. 2C). Reduction of endogenous galectin-1 had a dramatic effect on syncytia formation, as HUVECs treated with siRNA to galectin-1 demonstrated 2.5-fold increase in syncytia formation, compared to cells treated with control siRNA (Fig. 2D). Infection of HUVECs with lentiviral vectors containing no siRNA, or siRNA against an irrelevant protein, had no effect on syncytia formation, which was identical to that observed in uninfected cells (data not shown).
To confirm that cell surface galectin-1 was responsible for the effect on syncytia formation, we added exogenous galectin-1 to HUVECs in which galectin-1 expression was decreased by siRNA. We observed the expected increase of cell surface galectin-1 (Fig. 2C), as well as a decrease in syncytia formation (Figure 2D). Thus, endogenous galectin-1 on the surface of endothelial cells inhibits NiV-F/G mediated syncytia formation, underscoring the biological relevance of the effect of galectin-1 on NiV mediated cell-cell fusion.
Galectin-1 decreases the lateral mobility of NiV-F on the plasma membrane
Galectin-1 regulates the distribution and residence time of cell surface glycoproteins by binding to glycan branches on glycoproteins to create a cell surface lectin-glycoprotein lattice [22]. Lattice formation can have a variety of effects, including decreasing lateral mobility of glycoproteins. Lateral mobility is critical for effective cell-cell fusion mediated by NiV-F and NiV-G, as it is assumed that the F and G glycoproteins must physically separate in order to facilitate cell fusion [13], [23]. To determine the effect of galectin-1 on the lateral mobility of NiV-F, we performed fluorescence recovery after photobleaching (FRAP) analysis, using GFP-tagged NiV-F [24]. NiV - FGFP was expressed on transfected Vero cells (Fig. 3A) and mediated cell-cell fusion when co-transfected with NiV-G (Fig. 3B). Galectin-1 was also efficiently able to inhibit fusion mediated by NiV-FGFP and NiV-G (Fig. 3C).
Fig. 3. Galectin-1 interferes with lateral movement of NiV-F on the plasma membrane. 
A, NiV-FGFP expression on the surface of two individual Vero cells. B, NiV-FGFP promotes cell fusion when transfected into Vero cells with NiV-G. (20×) C, Galectin-1 inhibits fusion mediated by NiV-FGFP. Fusion of Vero cells in the absence (−) or presence (+) of 20µM galectin-1 was measured as in Fig. 1. D, Galectin-1 inhibited fluorescence recovery after photobleaching. NiV-FGFP transfected Vero cells were treated with buffer control (black line with black squares) or 20µM galectin-1 (grey line with black triangles), and a portion of the membrane was bleached and measured for fluorescent recovery (y-axis) as a function of time in seconds (x-axis). Data are mean ± SD of six replicate measurements from one of two independent experiments. To determine the effect of galectin-1 on NiV-F lateral movement, we measured the fluorescence recovery of NiV-FGFP (lateral diffusion) in the presence or absence of galectin-1 (Fig. 3D). We observed 50–60% recovery of NiV-FGFP fluorescence within 20 sec of photobleaching in the absence of galectin-1 (Fig. 3D). Addition of galectin-1 slowed the initial rate of fluorescence recovery, and also reduced overall fluorescence recovery to 25% (Fig. 3D). Thus, the presence of galectin-1 retarded NiV-F lateral movement on the cell surface.
Galectin-1 prevents endocytosis of NiV-F and inhibits maturation
The formation of galectin-glycan lattices on the cell surface has also been shown to reduce the rate of endocytosis of cell surface glycoproteins [25], [26]. As described above, NiV-F is produced as an immature precursor, NiV-F0, which is expressed on the cell surface and undergoes endocytosis and proteolytic processing to produce the fusion competent mature protein NiV-F1/2. We asked if cell surface galectin-1, in addition to reducing lateral mobility of NiV-F, altered endocytosis of NiV-F0. Cells expressing NiV-F were biotinylated to label cell surface proteins and incubated at 37°C to allow for endocytosis in the presence or absence of galectin-1. Following endocytosis, remaining cell surface biotin was removed by reduction with glutathione, and internalized biotinylated NiV-F was quantified in cell lysates. Addition of galectin-1 decreased the amount of internalized NiV-F by approximately 50%, compared to control-treated cells (Fig. 4A). Galectin-1 also decreased the rate of NiV-F internalization by approximately 50%, (Fig. 4A) over the first 30 min, before reaching equilibrium.
Fig. 4. Galectin-1 inhibits NiV-F0 endocytosis and maturation. 
A, Galectin-1 decreases internalization of NiV-F0 from the plasma membrane. Cells transfected with NiV-F were cell surface biotinylated, then incubated in the presence of 20µM galectin-1 (bold line), or buffer control (dashed line), for the indicated times to allow internalization. Internalized biotinylated NiV-F was quantified by ELISA. Percent internalization was determined as the amount of internalized biotinylated NiV-F compared to total biotinylated NiV-F at the initial timepoint. Data are mean ± SEM for seven replicate experiments. B, Galectin-1 inhibits NiV-F0 proteolytic processing. 293T cells expressing NiV-F were pulse-labeled with 35S-methionine, then chased for 4 or 6 hrs in the presence or absence of galectin-1. NiV-F was immunoprecipitated with anti-NiV-F polyclonal sera and proteolytic processing analyzed by immunoblotting. C, Graphic representation of data in B. Cleavage ratio was determined as the amount of processed NiV-F (F1+F2) compared to total NiV-F protein. Data are mean ± SEM of three replicate experiments. As galectin-1 inhibited endocytosis of NiV-F from the cell surface, and NiV-F endocytosis is required for proteolytic processing [11], [27], we investigated the effect of galectin-1 on proteolytic processing of NiV-F. Cells expressing NiV-F were pulse labeled and chased in the presence or absence of galectin-1. As shown in Fig. 4B, galectin-1 treatment decreased the processing of NiV-F0 into NiV-F1/2. At time 0, there was little to no NiV-F0 cleavage, indicated by the absence of F1 and F2 bands. After four to six hrs, a fraction of NiV-F0 was cleaved into NiV-F1 and NiV-F2; however, the presence of galectin-1 significantly reduced processing of NiV-F0 as evidenced by the decrease in NiV-F1 and NiV-F2. The amount of cleavage was quantified by comparing the amount of processed NiV-F (F1+F2) to total NiV-F protein. Addition of galectin-1 decreased NiV-F processing by approximately 50% at the 6 hour time point (Fig. 4C), which is approximately the amount of endocytosis reduction seen in the presence of galectin-1 (Fig. 4A). Taken together, these results demonstrate that galectin-1 reduces NiV-F processing by binding to and decreasing internalization of the NiV-F0 precursor.
Galectin-1 inhibits the function of mature NiV-F1/2 fusion protein
To further explore the interaction between galectin-1 and NiV-F, we used a quantitative heterologous fusion assay [13], [20], which combines ephrinB2 positive cells stably expressing the T7 polymerase (BSRT7) and ephrinB2 negative cells (PK-13) transfected with NiV-F, NiV-G and a luciferase construct with a T7 dependent promoter; in this system, luciferase expression is dependent on fusion of the two cell types. We used this assay to ask if galectin-1 inhibition of fusion involved interaction of galectin-1 with mature NiV-F, as well as inhibition of NiV-F0 endocytosis and maturation, as seen in Fig. 4. We found that, in this assay, addition of galectin-1 inhibited cell fusion by 80%, and galectin-1 mediated inhibition required the dimeric form of galectin-1, as a monomeric galectin-1, N-Gal-1, was not effective at blocking cell fusion, while a covalently linked galectin-1 dimer (GG) was effective at blocking fusion (Supplmentary Fig. S1A). In addition, in this assay, galectin-1 mediated inhibition of fusion was specific and carbohydrate mediated, as the inhibitory effect was abrogated by addition of lactose, a preferred glycan ligand for galectin-1, but not by sucrose (Supplementary Fig. S1B).
Chlorpromazine is an endocytosis inhibitor that reduces NiV-F endocytosis, cleavage, and maturation [27]; cells treated with chlorpromazine at the time of transfection with NiV-G and NiV-F demonstrated virtually no maturation of nascent NiV-F0 (Fig. 5A). We reasoned that, if galectin-1 plus chlorpromazine were added to cells already expressing mature NiV-F, any effect of galectin-1 on cell fusion would indicate that galectin-1 bound to and directly inhibited the fusogenic activity of mature NiV-F, rather than reducing maturation of NiV-F0. Indeed, we found that addition of galectin-1 to chlorpromazine treated cells inhibited syncytia formation above the level of inhibition observed with chlorpromazine alone (Fig. 5B), suggesting that galectin-1 can inhibit the function of the mature fusion protein, in addition to inhibiting endocytosis of NiV-F0.
Fig. 5. Galectin-1 inhibits function of mature NiV-F. 
A, Chlorpromazine inhibits maturation of NiV-F0. PK-13 were transfected with NiV-F in the absence or presence of chlorpromazine (50µM). Cells were incubated overnight and NiV-F0 and NiV-F1 detected by immunoblotting. B, Galectin-1 inhibits heterologous cell fusion in the presence of chlorpromazine. BSRT7 ephrinB2 positive cells were added to a monolayer of PK13 cells transfected with NiV-F, NiV-G and a luciferase construct with a T7 dependent promoter in the presence or absence of chlorpromazine (50µM) and galectin-1 (20µM). * p = 0.0002, calculated using Student's t test. C, Fusion kinetics in the presence or absence of galectin-1. NiV-G and NiV-F were expressed in effector PK13 cells, and the relative rate of fusion assessed on target 293T cells loaded with CCF2 dye. Relative fusion is the ratio of blue to green fluorescence from NiV-G and NiV-F-transfected effector cells minus the ratio of background blue to green fluorescence from empty-vector (pcDNA3)-transfected cells. Each data point is the mean of three independent experiments. D, Galectin-1 inhibits the ability of NiV-F to be triggered for membrane fusion. CHO cells expressing NiV-F and NiV-G were mixed with CHO cells (negative control, grey shaded) or CHOB2 cells (ephrinB2 positive, black line) for 1.5 hr at 4°C. Cell mixtures were brought to 37°C or kept at 4°C for 1.5 hr with 1 µM biotinylated HR2 peptide, in the presence or absence of galectin-1GG; top, 4°C without and with galectin-1; bottom, 37°C without and with galectin-1. E, Inhibition of F triggering at 37°C; data are mean fluorescence intensity of triplicate determinations ± SEM. See also Supplementary Figure S1. To further demonstrate that galectin-1's inhibitory activity can be effected through mature NiV-F, we used a real-time fusion kinetics assay [28] (Fig. 5C). Non-permissive effector cells (receptor-negative) were co-transfected with beta-lactamase, NiV-G, and NiV-F and then added to ephrinB2-expressing 293T target cells labeled with CCF2-AM dye. Cell-cell fusion was detected by analyzing the shift from green to blue fluorescence, indicating cytoplasmic mixing and beta-lactamase cleavage of CCF2-AM. In the absence of galectin-1, cell-cell fusion plateaued at about 100 min after mixing of the cells, while addition of galectin-1 completely inhibited fusion in this assay. As we start to observe an effect of galectin-1 on cell fusion within 25 min, galectin-1 is likely affecting mature fusion protein already on the effector cell surface, rather than solely affecting maturation of nascent NiV-F0.Thus, the ability of galectin-1 to inhibit fusion in this assay further supports a direct interaction between galectin-1 and the mature NiV-F fusion protein.
To more specifically understand the effect of galectin-1 binding to mature NiV-F on the fusion process, we examined triggering of NiV-F in the presence and absence of galectin-1. Current models of Nipah virus membrane fusion suggest that NiV-G binding to the cell surface receptor ephrinB2 triggers a conformational change in NiV-F; this triggering results in exposure of the fusion peptide in the pre-hairpin intermediate (PHI). The PHI then undergoes six-helix bundle formation, which is the conformational change that physically drives fusion of opposing membranes [29]. In NiV-F, the two heptad repeat regions, HR1 and HR2, fold next to each other during six-helix bundle formation. The HR1 region is transiently exposed during PHI formation, but before six-helix bundle formation. We have previously demonstrated [23] that a biotinylated peptide corresponding to the HR2 region can inhibit NiV-F mediated fusion by binding to the exposed HR1 region during PHI formation; HR2 peptide binding serves as a functional assay for formation of the PHI or NiV-F triggering. As Figs. 5B and 5C indicated that galectin-1 can block the fusogenic activity of mature NiV-F, we asked if galectin-1 binding to mature NiV-F could inhibit the conformational change in NiV-F (triggering or PHI formation) necessary to expose the fusion peptide, thereby inhibiting membrane fusion and syncytia formation.
To detect triggering, wild type CHO cells or CHO cells stably expressing ephrinB2 (CHOB2) were mixed with CHO cells transfected with NiV-F and G at 4°C for 1.5 hrs; NiV-G binding to ephrinB2 is an energy independent process. The biotinylated HR2 peptide was added to the cells in the presence or absence of galectin-1, and fusion was induced by incubation at 37°C for an additional 1.5 hours, because PHI formation is an energy dependent process. Binding of the HR2 peptide indicates that triggering has occurred. We observed no triggering of NiV-F at 4°C, regardless of the presence of galectin-1 (Fig. 5D). In the absence of galectin-1, incubation of the cell mixture at 37°C resulted in NiV-F triggering as seen by an increase in HR2 peptide binding. However, addition of galectin-1 reduced HR2 peptide binding, demonstrating that galectin-1 inhibited triggering of mature NiV-F (Fig. 5D). The change in HR2 peptide binding in the presence of galectin-1 is quantified in Fig. 5E. These results clearly show that galectin-1 can directly inhibit NiV-F triggering, so that this effect, in addition to inhibition of NiV-F lateral movement on the cell surface and inhibition of NiV-F0 maturation, contributes to the mechanism of galectin-1 mediated inhibition of NiV-F mediated cell fusion.
Site-specific glycosylation on NiV-F
The NiV-F fusion protein contains 5 consensus sites for N-glycosylation, labeled F1–F5; four of these predicted sites have been found to be glycosylated in vivo (F2–F5) [13]. To establish the types of N-glycans expressed by NiV-F0 and NiV-F1, glycomic screening was performed using MALDI-TOF mass spectrometry. The N-glycans were released by PNGase F and analyzed as their permethylated derivatives. The resulting spectra exhibit a series of singly charged sodiated molecular ions ([M+Na]+) to which putative structures are assigned based on the molecular compositions and knowledge of the N-glycan biosynthetic pathway. The profile for the complete propeptide, NiV-F0, can be seen in Fig. 6A. The assigned structures on NiV-F0 include high mannose (m/z 1988, 2192, 2396; Man7–9GlcNAc2), complex (m/z 2040–3416; Fuc0–1NeuAc0–2Hex0–7HexNAc4–6) and hybrid structures (m/z 1550, 1999; Fuc1Hex4–5HexNAc2–3). The complex and hybrid structures contain lactosamine (Gal-GlcNAc) moieties that can be recognized by galectin-1. The MALDI-TOF mass spectrum of permethylated N-glycans from NiV-F1 is displayed in Supplementary Figure S2. Comparison of the structures released from NiV-F0 and NiV-F1 suggests that the glycosylation sites on the NiV-F2 subunit (F2 and F3 glycans) are modified with larger complex structures than those on NiV-F1 subunit (F4 and F5 glycans).
Fig. 6. The F3 N-glycan on NiV-F is a complex N-glycan containing putative binding sites for galectin-1. 
A, MALDI-TOF mass spectrum of all permethylated N-glycans from NiV-F0. Annotated structures were deduced by taking into account theoretical compositions and knowledge of the biosynthetic pathways (for further information, refer to http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=glyco.TOC&depth=2). All molecular ions are [M+Na]+. Peaks labeled with * represent contaminating hexose polymers. Unlabelled peaks are non-carbohydrate contaminants or permethylation products. B, Glycan component of the F3 glycopeptide, GALEIYKNNTHDLVGDVR, and effect of sialidase S digestion. Top panel – NiV-F0 was digested with trypsin and the peptide/glycopeptide mixture was analysed by LC-ES-MS/MS; the summed MS data for the F3 glycopeptide are shown. Bottom panel – The LC-ES-MS/MS experiment was repeated after treatment of the tryptic digest of NiV-F0 with Sialidase S; summed MS data for the partially desialylated F3 glycopeptide are shown. Unannotated peaks correspond to peptides. Molecular ions attributable to glycopeptides are annotated with m/z values and subscripted charge states. Peaks labeled in bold correspond to molecular ions that have shifted on Sialidase S digestion; these peaks are also assigned a potential glycan structure. Symbol nomenclature is that used by the Consortium of Functional Glycomics (CFG) (see key below). See also Figure S2. Key: Galactose (yellow circle), Mannose (green circle), GlcNAc (blue square), Fucose (red triangle), NeuAc, (purple diamond). Previous research has shown that galectin-1 preferentially binds to the F3 glycan and this contributes significantly to galectin-1 inhibition of NiV-F mediated fusion of Vero cells. The other NiV-F N-glycans can also contribute to galectin-1 binding, as shown by co-immunoprecipitation studies with the NiV-F3 mutant [20]. In order to confirm the differences in site glycosylation suggested in the comparative MS data, and, where possible, deduce which subtypes of N-glycan occur at each glycosylation site, we performed online nano-LC ES-MS and data-dependent MS/MS analyses on tryptic peptides and glycopeptides from purified NiV-F samples. A peptide containing the F3 glycosylation site (GALEIYKNNTHDLVGDVR) was observed at an elution time of ∼50 min (Figure 6B, top panel). Carbohydrate structures were assigned by identifying neighboring ions separated by mass differences corresponding to sugar residues. At the F3 site, a total of 26 different glycan compositions were assigned. The carbohydrate structures are mostly complex and hybrid-type structures. The most intense peak assigned in Figure 6B (top panel) corresponds to a mono-sialylated, core-fucosylated bianntennary carbohydrate structure (m/z 1019.384+). Previous research has shown that alpha 2,6-linked sialic acid caps block galectin-1 binding while alpha 2,3 sialic acid caps do not (Stowell et al. 2008). Treatment of the tryptic glycopeptides prior to the nano-LC ES-MS experiment with Sialidase S (a sialidase that specifically cleaves the alpha 2,3 linked sialic acid) revealed that both alpha 2,3 and alpha 2,6-linked sialic acid are present, with 2,3-linked sialic acid being the more abundant (Fig. 6B, lower panel). These results suggest that the F3 glycan contains abundant putative binding sites for galectin-1. Analysis of the other glycosylation sites revealed that the F5 site carries only high mannose-type glycans (Supplementary Fig. S3), indicating that the F5 glycan is unlikely to contribute substantially to galectin-1 binding. The F4 site carries mostly complex-type structures, and the major peak at F4 is the sialylated biantennary glycan without core fucose (Supplementary Fig. S4), indicating that the F4 glycan may contribute to galectin-1 binding.
Galectin-1 inhibits NiV-F maturation and fusion through binding to the F3 glycan
We previously found that only the F3 glycan on NiV-F, but not the other N-glycans, is critical for optimal galectin-1 binding and inhibition of fusion of Vero cells [20]. Thus, we asked if the F3 glycan was also important for galectin-1 inhibition of fusion of endothelial cells and glial cells, the target cells of NiV. PK13 cells transfected with NiV-G and NiV-F or NiV-F3, lacking the F3 glycan, were added to HUVECs and U87 cells, and syncytia formation was scored in the presence or absence of galectin-1 (Fig. 7A). There was reduced inhibition of fusion by galectin-1 between PK13 cells expressing NiV-F3 and target cells, compared to cells expressing wildtype NiV-F3. These results indicate that galectin-1 partially inhibits fusion in relevant cell types by binding to the F3 glycan on NiV-F.
Fig. 7. The F3 glycan is critical for galectin-1 inhibition of NiV-F maturation and function. 
A, The F3 mutant (NiV-F missing the F3 glycan) is resistant to galectin-1 inhibition of syncytia formation in endothelial and glial cells, using the heterologous cell fusion assay described in Fig. 1. PK13 cells expressing NiV-G and either wildtype NiV-F (white) or NiV-F lacking the F3 glycan (black) were added to indicated cells in the presence of galectin-1 (HUVEC, 10µM; U87, 20µM). The y-axis shows percent inhibition of fusion. Data are mean ± SD of triplicate samples from one of three replicate experiments. * p = 0.0001, calculated using unpaired Student's t test. B, The F3 glycan on NiV-F is critical for galectin-1 inhibition of NiV-F0 internalization. Cells transfected with NiV-F3 (lacking the F3 glycan) were cell surface biotinylated and incubated in the presence of galectin-1 (20µM) (bold line), or buffer control (dashed line), for the indicated times to allow internalization. Internalized NiV-F3 was quantified as in Fig. 4. Data are mean ± SEM for seven replicate experiments. We also asked if the F3 glycan on NiV-F0 is critical for galectin-1 inhibition of endocytosis and maturation, as seen in Fig. 4A. The reduced endocytosis of NiV-F0 in the presence of galectin-1 was essentially abrogated in cells expressing the NiV-F3 mutant (Fig. 7B); in addition, we quantified processed NiV-F and NiV-F3, and found that, as we saw in Fig. 4B, processing of wildtype NiV-F was reduced approximately 50% in the presence of galectin-1, while there was <20% reduction of NiV-F3 processing in the presence of galectin-1 (data not shown). Taken together these data indicate that the F3 glycan on NiV-F is critical for galectin-1 inhibition of endocytosis of immature NiV-F0.
Discussion
The galectins, a family of mammalian carbohydrate binding proteins, interact with a broad range of mammalian cell surface glycoproteins to regulate essential functions in virtually every type of cell, including neural, immune and endothelial cells. Recent reports have shown that galectins can also directly bind glycoproteins on microbial pathogens, including viruses, bacteria and fungi, and that some galectins can participate as innate immune effectors. For example, galectin-3 binding to Candida albicans is fungicidal [30] and galectin-4 and galectin-8 binding to Escherichia coli is bacteriocidal [31]. Conversely, some pathogens exploit endogenous galectins to promote infection or evade host immune responses [32], [33], [34], [35], [36], [37], [38], [39]. Enveloped viruses use glycoproteins as both fusion and attachment molecules, and galectins have been shown to interact with envelope glycoproteins on HIV, HTLV and NiV [20], [33], [35], [37], [40]. Previous work from our laboratories demonstrated that both NiV-G and NiV-F glycoproteins bind galectin-1, and that exogenous galectin-1 reduced syncytia formation triggered by NiV-F; galectin-1 inhibition of syncytia formation was specific for paramyxoviruses, as we did not observe inhibition of cell fusion induced by HTLV-2, vaccinia and MLV [20]. As inflammation and viral infection can stimulate production of galectins by endothelial cells [15], [16], [18], endogenous galectin-1 may contribute to host defense against NiV infection by mitigating the endothelial cell syncytia formation that is a hallmark of Nipah infection, as we have clearly demonstrated that endogenous galectin-1 can attenuate endothelial cell fusion in vitro (Fig. 2). While autopsy studies of patients who succumbed to NiV infection found endothelial cell syncytia in numerous organs including brain [2], [3], variation in galectin-1 expression among infected individuals may contribute to susceptibility or resistance to viral-induced pathophysiology and, in part, explain why some infected individuals do not progress to encephalitis.
We investigated the effect of galectin-1 at each stage of the cell fusion process, and have identified three points at which galectin-1 inhibits NiV-F mediated syncytia formation. First, galectin-1 reduced lateral mobility of NiV-F on the plasma membrane (Fig. 3). Second, galectin-1 retarded endocytosis and maturation of the precursor NiV-F0 (Fig. 4). Third, galectin-1 prevented the triggering of mature NiV-F into the fusion-competent form (Fig. 5). Lateral mobility in the plasma membrane is important for NiV-F mediated cell fusion, as the physical separation of NiV-F and NiV-G is required for the conformational change in NiV-F necessary for membrane fusion [23], [28]. Galectin-1 inhibition of the lateral movement of NiV-F could contribute to reduced ability of mature NiV-F to form the fusion competent PHI on the cell surface, in addition to the direct inhibition of PHI formation that we observed in Fig. 5.
All these effects are congruent with data from prior studies demonstrating that galectin interacts with endogenous mammalian plasma membrane glycoproteins to form galectin-glycoprotein lattices on the cell surface [22], [41]. Formation of these lattices increases the local concentration of galectins at the cell surface, and also has direct effects on the glycoproteins in the lattice, as galectin-glycoprotein lattices can regulate the distribution, residence time, and function of glycoproteins on the plasma membrane [24], [25], [42], [43], [44], [45]. Galectin-glycoprotein lattice formation has been shown to inhibit lateral diffusion of EGF receptors on tumor cells [24]. Galectin-glycoprotein lattices also reduce endocytosis of EGF receptors on tumor cells, Glut-2 receptors on pancreatic cells, and receptor tyrosine phosphatase beta on neural cells [24], [25], [26]. The present study provides the first demonstration that galectin-1 on the cell surface reduces lateral mobility and endocytosis of a viral glycoprotein, rather than an endogenous mammalian glycoprotein. Lattice formation between galectin-1 and NiV-F likely contributes to inhibition of NiV-F maturation, reduced mobility, and reduced triggering that we observed.
Our findings also emphasize the specificity of the interaction between galectin-1 and unique glycans on a viral envelope glycoprotein. Glycoproteomic analysis showed that the most abundant glycan at the F3 site is a monosialylated biantennary glycan with a 2–3 linked sialic acid (Fig. 6). This structure fulfils the known requirement for a galectin-1 ligand [46]. Fig. 7, as well as our previous report, shows that the F3 glycan on NiV-F participates in galectin-1 inhibition of syncytia formation at two distinct points; the F3 glycan appears essential for galectin-1 inhibiting maturation of NiV-F0 and also substantially contributes to the galectin-1 effect on the function of mature NiV-F. This level of specificity for a particular glycan on a glycoprotein counter-receptor is quite novel, in that previous reports on mammalian lectins interacting with viral glycoproteins emphasized simple pattern matching or binding to dense and repetitive arrays of terminal monosaccharide ligands [47], [48], [49]. In contrast, a significant component of the interaction between galectin-1 and NiV-F primarily involves a single glycosylation site on the viral glycoprotein. This interaction emphasizes the site-specific nature of glycan addition and subsequent modification during viral glycoprotein maturation and transport to the cell surface. As shown in Fig. 6, there is significant heterogeneity among the different glycans attached to NiV-F. Similar microheterogeneity has been observed for the glycans attached to HIV gp120 [50]. This microheterogeneity would substantially contribute to selective interaction of viral glycoproteins with endogenous mammalian lectins, such as C-type lectins and galectins, or with antibodies that recognize specific glycans on viral glycoproteins. Interestingly, F3 glycan removal resulted in the highest levels of hyperfusiogenicity compared to the removal of other glycans on NiV-F [13]. This is consistent with F3 glycan removal reducing the inhibitory effects of endogenous galectin-1.
This report defines several molecular mechanisms by which galectin-1 binding to NiV-F interferes with maturation and function of NiV-F to reduce cell fusion, and also demonstrates that endogenous endothelial galectin-1 can influence the extent of syncytia formation. Since syncytia formation is the pathognomonic hallmark of NiV infection, we predict that, in vivo, galectin-1 may reduce pathophysiologic consequences during the course of an infection. An 11kb region containing the gene coding for galectin-1 contains 14 different single nucleotide polymorphisms (SNP) [51], and this genetic variability may partially account for the range of pathophysiological consequences seen in Nipah virus infection, as SNPs in other galectins have been found to be associated with increased disease risk [52]. Our studies focus on cell-cell fusion, as syncytia formation is the event that primarily contributes to the endothelial destruction and hemorrhagic sequelae in NiV infection. However, galectins may also influence attachment of viruses to target cells [33], [35], [37], [40], although galectin mediated cell attachment is fundamentally distinct from the specific fusion inhibitory mechanisms that we have elucidated in this study. Galectins can play multiple distinct roles in complex biologic processes such as pathogen entry, replication and dissemination, so that our goal is to define all the roles played by galectin-1 during the entire course of Nipah virus infection of human host cells.
Materials and Methods
Cell lines and reagents
Vero cells and CHO cells (ATCC) were maintained in MEM alpha (Invitrogen) with 10% FBS (Hyclone) and 2mM Glutamax in 5%CO2 at 37°C. PK-13 porcine fibroblast cells and 293T cells were maintained in DMEM (Invitrogen) with 10% FBS (Hyclone) and 2mM Glutamax. BSR cells stably transfected with T7 polymerase were maintained in DMEM with 10% FBS (Hyclone), 2mM Glutamax, and 0.5mg/ml G418 (Sigma). U87 glioblastoma cells (gift of P. Mischel, UCLA) were maintained in DMEM with 10% FBS (heat inactivated at 55°C for 30 min), 2mM Glutamax, and 50 units/ml penicillin/streptomycin. mVECs [6] and HUVECs (BD Biosciences) were maintained in MDCB-131 Complete media with fetal bovine serum and antibiotics (VEC Technologies, INC.).
Codon-optimized NiV-F and G plasmids tagged with AU1 were previously described [20]. NiV-F3 plasmid (encoding NIV-F lacking the F3 glycan) was previously described [13]. NiV-F-GFP plasmid was created by synthesizing a fusion gene between NiV-F and GFP using overlapping PCR. Oligonucleotide sequences were designed that flank the 5′ region of codon-optimized NiV-F and 3′region of GFP. An additional oligonucleotide that overlaps the 3′ region of NiV-F and the 5′ region of GFP was also designed which did not contain the stop codon at the end of the NiV-F ORF and included a GGG linker between the two genes. NiV-F (1.6 kb) and GFP (0.8kb) genes were amplified by PCR using the appropriate 5′ or 3′ oligonucleotide primers and the overlapping primer. The two PCR products were gel purified and used together as template for another PCR reaction using the original 5′ and 3′ primers. The resulting 2.2kb fusion gene product was subcloned into pcDNA3 (Invitrogen) and sequence verified. Recombinant human galectin-1 was expressed in E. coli and purified by affinity chromatography on lactosyl-Sepharose, as in [53]; in all assays, the buffer control includes 8mM dithiothreitol (DTT), as galectin-1 is prepared and stored in PBS with DTT.
Heterologous fusion assay
PK-13 cells were transfected with codon-optimized, AU1-tagged NiV-F, NiV-F3, or NiV-FGFP and HA-tagged NiV-G at 15ug per plasmid using Lipofectamine 2000 (Invitrogen). Cells were cultured overnight, lifted with 5mM EDTA (ethylenediaminetetraacetic acid) and overlayed in the absence or presence of galectin-1 onto ephrinB2 positive cells (Vero, U87, mVEC, or HUVEC). After 2 hrs (Vero/U87) or 6 hrs (HUVEC/mVEC), cells were fixed with 2% paraformaldehyde (EMS). After 4′,6′-diamidino-2-phenylindole (DAPI) staining (Invitrogen), nuclei inside syncytia per ×100 field were counted by fluorescence microscopy as previously described [20].
Cell surface staining for galectin-1
HUVEC cells were incubated with or without 20µM galectin-1 for 30 min at 37°C. Cells were fixed in DTSSP (Thermo Scientific) at 0.2mg/ml for 10 min at room temperature, and quenched by addition of 100µl of 1M Tris pH 7.5 for 15 min at room temperature. Cells were washed with PBS and lifted with 5mM EDTA at 37°C for 10 min. Cells were stained with a rabbit galectin-1 antibody (Strategic) for 1 hr at 4°C. Cells were washed with PBS and stained with FITC-conjugated AffiniPure goat anti-rabbit IgG (H+L) antibody (Jackson ImmunoResearch) at 8µg/ml for 1 hour at 4°C. Cells were washed in PBS and resuspended in PBS with 1% BSA (Gemini BioProducts) for analysis by flow cytometry. Flow cytometric analysis of HUVEC cell surface galectin-1 was performed on a Becton-Dickinson FACScan, using CellQuest software (Becton-Dickinson).
Galectin-1 knockdown
3 different siRNA constructs directed toward human galectin-1 mRNA in lentivectors (pSIH1-H1-copGFP plasmid) (System Biosciences) were packaged into VSV using pPACKH1 Lentivector Packaging Kit (System Bioscience). Lentivirus was produced in 293T cells; three different viruses corresponding to the 3 different siRNAs were isolated, as well as a virus containing only the GFP infection marker, and a virus containing siRNA to an irrelevant protein, CD43. HUVECs were plated at 1.4×105 cells per well in a 6 well tissue culture dish (Corning). A single well of HUVECs was treated with each virus (MOI of 3.3, total MOI of 10) in PBS with 1% heat inactivated FBS (Hyclone) and cells were spinoculated at 2000 rpm at 37°C for 2 hrs. Infected cells were grown in full media for 4 days and knockdown was assessed by western blot and flow cytometry.
FRAP
Fluorescence Recovery After Photobleaching analysis of Vero cells transfected with NiV-FGFP plasmid was performed in 35mm glass bottom culture dishes (MatTek) on a 37°C heated stage. Cells were treated with 20µM galectin-1 or buffer control for 10 min prior to photobleaching. Images were acquired on a confocal microscope (Leica SP2 1P-FCS) with a HCX PL APO 63.0×1.40 oil objective and fully opened pinhole. Photobleaching of NiV-F-GFP was performed using 5 scans with the 488-nm laser at full power. Recovery data (six cells per time point from each of two independent experiments) was collected every 2.2 seconds for a total of 25 time points. Images were acquired with equivalent acquisition settings including pre-bleach, bleach, and post-bleach measurements. Bleaching and recovery were measured in a fixed area and compared to an area absent of fluorescence outside of the cell.
NiV-F endocytosis assay
PK-13 cells were transfected with NiV-F or NiV-F3. After overnight incubation, cells were washed twice with KRPH buffer (128mM NaCl, 4.7mM KCl, 1.25mM CaCl2, 1.25mM MgSO4, 5mM Na2HPO4, 20mM Hepes pH 7.4) and cell surface biotinylated using the EZ-Link sulfo-NHS-SS-Biotin (Pierce). Biotinylation was quenched in 20mM glycine and cells were washed again in KRPH buffer. Cells were incubated in media with or without galectin-1 or buffer control for the designated times at 37°C to allow endocytosis. After timepoints, cells were washed and remaining biotin cleaved twice with cleavage buffer (90mM NaCl, 1.25mM CaCl2, 1.25mM MgSO4, 2mg/ml BSA, 50mM glutathione pH 8.6) and quenched in 20mM glycine for 15 min. Cells were lysed in lysis buffer (20mM Tris-HCL pH 7.5, 100mM NH2SO4, 0.1% BSA, 0.75% Trition X-100, 0.01% NaN3). Biotinylated NiV-F in cell lysates was quantified by ELISA using mouse anti-AU1 (1∶1000) coated Reacti-Bind goat anti-mouse plates (Pierce) to capture AU1-tagged NiV-F, and detected with streptavidin-HRP (Biorad).
Metabolic labeling and pulse chase
293T cells grown in 6-well plates were transfected with 0.25µg of NiV-F plasmid DNA and 1.75µg pcDNA3 per well. 24 hrs post transfection, cells were incubated in media lacking methionine and cysteine for 45 min followed by labeling with media containing 35S-cysteine and 35S-methionine (100mCi/ml) for 30 min, and then in chase (non-radioactive) media for 4 to 6 hrs, in the presence or absence of 20µM galectin-1. Cells were lysed in 200µl RIPA buffer (20mM Tris-HCl pH 7.4, 137mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP-40, 2mM EDTA). NiV-F was immunoprecipitated from cleared cell lysates using a combination of anti-NiV-F polysera [13] and anti-AU1 antibody each at 1∶100 dilution, and Protein G agarose (Pierce). Precipitates were washed twice in RIPA buffer and twice with RIPA buffer plus 0.5M NaCl. Samples were separated on a 14% polyacrylamide gel and data were quantified and analyzed using a phosphoimager (Molecular Dynamics 445SI) and ImageQuant (v5.2).
Fusion reporter assay
Fusion-nonpermissive PK-13 target cells were transfected with codon optimized NiV-F, NiV-G, and a plasmid containing a T7 promoter driven luciferase at a 3∶3∶1 ratio with 30µg of total plasmid DNA and grown overnight. BSR cells stably transfected with a T7 polymerase (BSRT7) were lifted with 5mM EDTA at 37°C for 10 min. BSRT7 cells were co-cultured with transfected PK-13 cells for 6 hrs with or without galectin-1. After 6 hrs, the cells were lysed in 0.3% Triton-X 100 by two rounds of freeze/thaw at −80°C. Luciferase expression was quantified using a Luciferase assay system (Promega). Briefly, lysates were transferred to a 96 well opaque black plate, luciferase assay substrate was added, and light production was measured by luminometry (Turner Biosystems).
Chlorpromazine treatment
PK-13 cells were transfected with NiV-F and simultaneously treated with chlorpromazine (Sigma) for 16 hrs. Cells were lysed in 50mM Tris-HCl (pH 7.4), 1% Nonidet P-40, 5mM EDTA, 150mM NaCl, 1mM PMSF, 10mg/ml aprotinin, 10mg/ml leupeptin, and 10mM sodium orthovanadate with scraping. Lysates were microfuged for 15 min at 10,000 rpm. Samples were denatured in NuPAGE reducing agent and NuPAGE Sample Buffer (Invitrogen) before loading. Lysates (10 µg) were separated on a 12% Bis-Tris gel (Invitrogen NuPAGE Electrophoresis System) and electroblotted onto nitrocellulose (Whatman). The membrane was blocked and probed as previously described [53] using an AU1 antibody (Covance) and NiV-F proteins were visualized by ECL.
Triggering of NiV-F by NiV-G in the presence or absence of galectin-1
NiV-F triggering was measured essentially as in [23] except in the presence or absence of 0.4 µM galectin-1GG [54]. Briefly, CHO cells were transfected with NIV-F and NiV-G expression plasmids, plus GFP expression plasmid, at a 13∶6∶1 ratio, respectively. 18 hrs post-transfection, a 1∶1 ratio of transfected cells and either CHO (negative control) or CHOB2 (CHO cells transfected with ephrinB2 [23]) cells were mixed and incubated for 2 hr at 4°C, followed by a 90 min incubation at either 4°C or 37°C in the presence of excess biotinylated HR2 peptide and in the presence or absence of 0.4 µM galectin-1GG, as indicated. Cells were washed with wash buffer (1% FBS in PBS), fixed in 0.5% paraformaldehyde in wash buffer, and washed twice with wash buffer. Biotinylated HR2 peptide bound to F was detected using streptavidin-APC (ebioscience). GFP-positive cells were gated and analyzed for HR2-biotin binding.
Fusion kinetics in the presence or absence of galectin-1
Fusion kinetics were determined in a beta-lactamase reporter cell-cell fusion assay, as previously described [13], [28], [55], [56], using a catalytically enhanced and codon optimized beta-lactamase gene [57], [58]. Fusion-nonpermissive PK13 effector cells were co-transfected with beta-lactamase, NiV-G, and NiV-F expression plasmids, and mixed with 293T target cells labeled with CCF2-AM dye for 30 min at 4°C. Galectin-1GG (0.4 µM) or buffer control was added and the cells moved immediately to 37°C. Cell-cell fusion was detected by analyzing the shift from green to blue fluorescence, indicating beta-lactamase cleavage of CCF2-AM. Fluorescence was quantified every 3 min with a Synergy 2 Multi-mode microplate reader (BioTek Instruments, Winooski, VT). Results are expressed as the ratio of blue to green fluorescence obtained with NiV-G - and NiV-F-transfected effectors minus the background blue to green fluorescence ratios obtained with NiV-G - and empty-vector-transfected cells.
Preparation of NiV-F sample for mass spectrometric analysis
Glycan analysis was performed on NiV-F protein pseudotyped onto VSV viral-like particles produced in 293T cells. Gel bands containing purified NiV-F0 and NiV-F1 were destained using 100% acetonitrile, incubated with 10mM DTT for 30 min at 56°C, followed by 55 mM iodoacetic acid for 30 min at RT. Reduced and carboxymethylated NiV-F was digested with 0.5µg sequencing grade-modified trypsin (Promega, UK) at 37°C for 14 hrs. Tryptic peptides and glycopeptides were extracted from the gel pieces by incubating with 0.1% trifluoroacetic acid (TFA) and 100% acetonitrile; the supernatant was pooled and reduced in volume on a rotary evaporator. For glycomic screening the supernatant was lyophilised before being dissolved in 200µl ammonium bicarbonate (50mM, pH 8.4) and incubated with 5U of Peptide-N-glycosidase F (PNGase F) (Roche Applied Science, UK) for 24 hrs at 37°C to release the N-glycans. The reaction was terminated by lyophilisation. Glycans were separated from peptides by C18 Sep-Pak purification and permethylated as previously described [59].
MALDI-TOF and online nano-LC-ES-MS/MS analysis
MALDI-TOF MS and MS/MS data on permethylated N-glycan samples were acquired in positive ion mode [M+Na]+ using a 4800 MALDI-TOF/TOF (Applied Biosystems, UK) mass spectrometer as previously described [60]. The MALDI data were processed using Data Explorer 4.9 Software (Applied Biosystems, UK). Tryptic digests were analysed by nano-LC-ES-MS/MS using a reverse-phase nano-HPLC system (Dionex (UK) Ltd, Camberley) connected to a quadrupole TOF mass spectrometer (API Q-STAR Pulsar I, Applied Biosystems, UK) as previously described [61]. Analysis of the ES-MS and MS/MS data was aided by use of the Peptoonist algorithm [62].
Sialidase S digestion of NiV-F glycopeptides
Sample was dried, resuspended in 50µl of 50mM ammonium acetate (pH 5.5) and incubated with 10mU of Sialidase S at 37°C for 14 hrs. After digestion, NiV-F peptides and desialylated glycopeptides were desalted and separated using a C18-microtrap peptide cartridge (Presearch, Basingstoke). The sample was loaded directly onto the microtrap cartridge with a 25µl gastight syringe. The microtrap was first solvated with methanol, washed off with acetonitrile and conditioned with 0.1% TFA. The sample was loaded onto the column and washed with 0.1% TFA prior to eluting with 15µl 30 and 60% acetonitrile in 0.1% TFA, respectively. Eluted fractions were combined and dried down gently under nitrogen before online LC-ES-MS and MS/MS analysis.
Supporting Information
Zdroje
1. BlumLS
KhanR
NaharN
BreimanRF
2009 In-depth assessment of an outbreak of Nipah encephalitis with person-to-person transmission in Bangladesh: implications for prevention and control strategies. Am J Trop Med Hyg 80 96 102
2. WongKT
ShiehWJ
KumarS
NorainK
AbdullahW
2002 Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol 161 2153 2167
3. HooperP
ZakiS
DanielsP
MiddletonD
2001 Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect 3 315 322
4. MaisnerA
NeufeldJ
WeingartlH
2009 Organ - and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb Haemost 102 1014 1023
5. EatonBT
BroderCC
MiddletonD
WangLF
2006 Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol 4 23 35
6. NegreteOA
LevroneyEL
AguilarHC
Bertolotti-CiarletA
NazarianR
2005 EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436 401 405
7. BonaparteMI
DimitrovAS
BossartKN
CrameriG
MungallBA
2005 Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A 102 10652 10657
8. NegreteOA
WolfMC
AguilarHC
EnterleinS
WangW
2006 Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog 2 e7
9. LeeB
2007 Envelope-receptor interactions in Nipah virus pathobiology. Ann N Y Acad Sci 1102 51 65
10. VogtC
EickmannM
DiederichS
MollM
MaisnerA
2005 Endocytosis of the Nipah virus glycoproteins. J Virol 79 3865 3872
11. DiederichS
MollM
KlenkHD
MaisnerA
2005 The nipah virus fusion protein is cleaved within the endosomal compartment. J Biol Chem 280 29899 29903
12. PagerCT
CraftWWJr
PatchJ
DutchRE
2006 A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346 251 257
13. AguilarHC
MatreyekKA
FiloneCM
HashimiST
LevroneyEL
2006 N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80 4878 4889
14. MollM
KaufmannA
MaisnerA
2004 Influence of N-glycans on processing and biological activity of the nipah virus fusion protein. J Virol 78 7274 7278
15. RabinovichGA
BaumLG
TinariN
PaganelliR
NatoliC
2002 Galectins and their ligands: amplifiers, silencers or tuners of the inflammatory response? Trends Immunol 23 313 320
16. WarkeRV
XhajaK
MartinKJ
FournierMF
ShawSK
2003 Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol 77 11822 11832
17. RamasamyS
DuraisamyS
BarbashovS
KawanoT
KharbandaS
2007 The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell 27 992 1004
18. BaumLG
SeilhamerJJ
PangM
LevineWB
BeynonD
1995 Synthesis of an endogeneous lectin, galectin-1, by human endothelial cells is up-regulated by endothelial cell activation. Glycoconj J 12 63 68
19. HeJ
BaumLG
2006 Endothelial cell expression of galectin-1 induced by prostate cancer cells inhibits T-cell transendothelial migration. Lab Invest 86 578 590
20. LevroneyEL
AguilarHC
FulcherJA
KohatsuL
PaceKE
2005 Novel innate immune functions for galectin-1: galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J Immunol 175 413 420
21. ThijssenVL
HulsmansS
GriffioenAW
2008 The galectin profile of the endothelium: altered expression and localization in activated and tumor endothelial cells. Am J Pathol 172 545 553
22. GarnerOB
BaumLG
2008 Galectin-glycan lattices regulate cell-surface glycoprotein organization and signalling. Biochem Soc Trans 36 1472 1477
23. AguilarHC
AtamanZA
AspericuetaV
FangAQ
StroudM
2009 A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F). J Biol Chem 284 1628 1635
24. LajoieP
PartridgeEA
GuayG
GoetzJG
PawlingJ
2007 Plasma membrane domain organization regulates EGFR signaling in tumor cells. J Cell Biol 179 341 356
25. OhtsuboK
TakamatsuS
MinowaMT
YoshidaA
TakeuchiM
2005 Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 123 1307 1321
26. AbbottKL
MatthewsRT
PierceM
2008 Receptor tyrosine phosphatase beta (RPTPbeta) activity and signaling are attenuated by glycosylation and subsequent cell surface galectin-1 binding. J Biol Chem 283 33026 33035
27. DiederichS
ThielL
MaisnerA
2008 Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology 375 391 400
28. AguilarHC
MatreyekKA
ChoiDY
FiloneCM
YoungS
2007 Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol 81 4520 4532
29. LambRA
JardetzkyTS
2007 Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol 17 427 436
30. KohatsuL
HsuDK
JegalianAG
LiuFT
BaumLG
2006 Galectin-3 induces death of Candida species expressing specific beta-1,2-linked mannans. J Immunol 177 4718 4726
31. StowellSR
ArthurCM
Dias-BaruffiM
RodriguesLC
GourdineJP
Innate immune lectins kill bacteria expressing blood group antigen. Nat Med
32. MoodyTN
OchiengJ
VillaltaF
2000 Novel mechanism that Trypanosoma cruzi uses to adhere to the extracellular matrix mediated by human galectin-3. FEBS Lett 470 305 308
33. OuelletM
MercierS
PelletierI
BounouS
RoyJ
2005 Galectin-1 acts as a soluble host factor that promotes HIV-1 infectivity through stabilization of virus attachment to host cells. J Immunol 174 4120 4126
34. FowlerM
ThomasRJ
AthertonJ
RobertsIS
HighNJ
2006 Galectin-3 binds to Helicobacter pylori O-antigen: it is upregulated and rapidly secreted by gastric epithelial cells in response to H. pylori adhesion. Cell Microbiol 8 44 54
35. MercierS
St-PierreC
PelletierI
OuelletM
TremblayMJ
2008 Galectin-1 promotes HIV-1 infectivity in macrophages through stabilization of viral adsorption. Virology 371 121 129
36. OkumuraCY
BaumLG
JohnsonPJ
2008 Galectin-1 on cervical epithelial cells is a receptor for the sexually transmitted human parasite Trichomonas vaginalis. Cell Microbiol 10 2078 2090
37. GauthierS
PelletierI
OuelletM
VargasA
TremblayMJ
2008 Induction of galectin-1 expression by HTLV-I Tax and its impact on HTLV-I infectivity. Retrovirology 5 105
38. VastaGR
2009 Roles of galectins in infection. Nat Rev Microbiol 7 424 438
39. SatoS
St-PierreC
BhaumikP
NieminenJ
2009 Galectins in innate immunity: dual functions of host soluble beta-galactoside-binding lectins as damage-associated molecular patterns (DAMPs) and as receptors for pathogen-associated molecular patterns (PAMPs). Immunol Rev 230 172 187
40. Pais-CorreiaAM
SachseM
GuadagniniS
RobbiatiV
LasserreR
Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med 16 83 89
41. BrewerCF
MiceliMC
BaumLG
2002 Clusters, bundles, arrays and lattices: novel mechanisms for lectin-saccharide-mediated cellular interactions. Curr Opin Struct Biol 12 616 623
42. BracciaA
VillaniM
ImmerdalL
Niels-ChristiansenLL
NystromBT
2003 Microvillar membrane microdomains exist at physiological temperature. Role of galectin-4 as lipid raft stabilizer revealed by “superrafts”. J Biol Chem 278 15679 15684
43. NguyenJT
EvansDP
GalvanM
PaceKE
LeitenbergD
2001 CD45 modulates galectin-1-induced T cell death: regulation by expression of core 2 O-glycans. J Immunol 167 5697 5707
44. LiuSD
TomassianT
BruhnKW
MillerJF
PoirierF
2009 Galectin-1 tunes TCR binding and signal transduction to regulate CD8 burst size. J Immunol 182 5283 5295
45. LauKS
PartridgeEA
GrigorianA
SilvescuCI
ReinholdVN
2007 Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129 123 134
46. StowellSR
ArthurCM
MehtaP
SlaninaKA
BlixtO
2008 Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem 283 10109 10123
47. TanakaH
ChibaH
InokoshiJ
KunoA
SugaiT
2009 Mechanism by which the lectin actinohivin blocks HIV infection of target cells. Proc Natl Acad Sci U S A
48. MenonS
RosenbergK
GrahamSA
WardEM
TaylorME
2009 Binding-site geometry and flexibility in DC-SIGN demonstrated with surface force measurements. Proc Natl Acad Sci U S A 106 11524 11529
49. DamTK
BrewerCF
2009 Lectins as Pattern Recognition Molecules: The Effects of Epitope Density in Innate Immunity#. Glycobiology
50. LiY
ClevelandB
KlotsI
TravisB
RichardsonBA
2008 Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol 82 638 651
51. IidaA
OzakiK
TanakaT
NakamuraY
2005 Fine-scale SNP map of an 11-kb genomic region at 22q13.1 containing the galectin-1 gene. J Hum Genet 50 42 45
52. OzakiK
InoueK
SatoH
IidaA
OhnishiY
2004 Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-alpha secretion in vitro. Nature 429 72 75
53. PaceKE
LeeC
StewartPL
BaumLG
1999 Restricted receptor segregation into membrane microdomains occurs on human T cells during apoptosis induced by galectin-1. J Immunol 163 3801 3811
54. BiS
EarlLA
JacobsL
BaumLG
2008 Structural features of galectin-9 and galectin-1 that determine distinct T cell death pathways. J Biol Chem 283 12248 12258
55. LinebergerJE
DanzeisenR
HazudaDJ
SimonAJ
MillerMD
2002 Altering expression levels of human immunodeficiency virus type 1 gp120-gp41 affects efficiency but not kinetics of cell-cell fusion. J Virol 76 3522 3533
56. ReevesJD
MiamidianJL
BisconeMJ
LeeFH
AhmadN
2004 Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J Virol 78 5476 5485
57. DoucetN
De WalsPY
PelletierJN
2004 Site-saturation mutagenesis of Tyr-105 reveals its importance in substrate stabilization and discrimination in TEM-1 beta-lactamase. J Biol Chem 279 46295 46303
58. WolfMC
WangY
FreibergAN
AguilarHC
HolbrookMR
2009 A catalytically and genetically optimized beta-lactamase-matrix based assay for sensitive, specific, and higher throughput analysis of native henipavirus entry characteristics. Virol J 6 119
59. Jang-LeeJ
NorthSJ
Sutton-SmithM
GoldbergD
PanicoM
2006 Glycomic profiling of cells and tissues by mass spectrometry: fingerprinting and sequencing methodologies. Methods Enzymol 415 59 86
60. PangPC
TissotB
DrobnisEZ
MorrisHR
DellA
2009 Analysis of the human seminal plasma glycome reveals the presence of immunomodulatory carbohydrate functional groups. J Proteome Res 8 4906 4915
61. TissotB
NorthSJ
CeroniA
PangPC
PanicoM
2009 Glycoproteomics: past, present and future. FEBS Lett 583 1728 1735
62. GoldbergD
BernM
ParryS
Sutton-SmithM
PanicoM
2007 Automated N-glycopeptide identification using a combination of single - and tandem-MS. J Proteome Res 6 3995 4005
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System inČlánek A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 7- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Mouse Resistance Protein Irgm1 (LRG-47): A Regulator or an Effector of Pathogen Defense?
- Leprosy and the Adaptation of Human Toll-Like Receptor 1
- Intergenomic Arms Races: Detection of a Nuclear Rescue Gene of Male-Killing in a Ladybird
- The Role of Chemokines during Viral Infection of the CNS
- Bottlenecks and the Maintenance of Minor Genotypes during the Life Cycle of
- DNA Damage Triggers Genetic Exchange in
- The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System in
- Uropathogenic Modulates Immune Responses and Its Curli Fimbriae Interact with the Antimicrobial Peptide LL-37
- Biogenesis of the Inner Membrane Complex Is Dependent on Vesicular Transport by the Alveolate Specific GTPase Rab11B
- A Spatio-Temporal Analysis of Matrix Protein and Nucleocapsid Trafficking during Vesicular Stomatitis Virus Uncoating
- Hepatitis B Virus Polymerase Blocks Pattern Recognition Receptor Signaling via Interaction with DDX3: Implications for Immune Evasion
- Quasispecies Theory and the Behavior of RNA Viruses
- Bid Regulates the Pathogenesis of Neurotropic Reovirus
- Distinct Roles for Dectin-1 and TLR4 in the Pathogenesis of Keratitis
- Unexpected Inheritance: Multiple Integrations of Ancient Bornavirus and Ebolavirus/Marburgvirus Sequences in Vertebrate Genomes
- Balanced Nuclear and Cytoplasmic Activities of EDS1 Are Required for a Complete Plant Innate Immune Response
- Adaptation of Hepatitis C Virus to Mouse CD81 Permits Infection of Mouse Cells in the Absence of Human Entry Factors
- An Outer Membrane Receptor of Involved in Zinc Acquisition with Vaccine Potential
- Inositol Hexakisphosphate-Induced Autoprocessing of Large Bacterial Protein Toxins
- Plus- and Minus-End Directed Microtubule Motors Bind Simultaneously to Herpes Simplex Virus Capsids Using Different Inner Tegument Structures
- Distinct Pathogenesis and Host Responses during Infection of by and
- Can Bacteria Evolve Resistance to Quorum Sensing Disruption?
- RNA Virus Replication Complexes
- PPARγ and LXR Signaling Inhibit Dendritic Cell-Mediated HIV-1 Capture and -Infection
- The Virulence Protein SopD2 Regulates Membrane Dynamics of -Containing Vacuoles
- Genome-Wide Mutagenesis Reveals That ORF7 Is a Novel VZV Skin-Tropic Factor
- Adaptive Evolution of Includes Retroviral Insertion and Positive Selection at Two Clusters of Residues Flanking the Substrate Groove
- A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
- Transduction of Human T Cells with a Novel T-Cell Receptor Confers Anti-HCV Reactivity
- Identification of GBV-D, a Novel GB-like Flavivirus from Old World Frugivorous Bats () in Bangladesh
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- HIV gp41 Engages gC1qR on CD4+ T Cells to Induce the Expression of an NK Ligand through the PIP3/H2O2 Pathway
- Oxidation of Helix-3 Methionines Precedes the Formation of PK Resistant PrP
- Protection from the 2009 H1N1 Pandemic Influenza by an Antibody from Combinatorial Survivor-Based Libraries
- Murine Gamma-Herpesvirus 68 Hijacks MAVS and IKKβ to Initiate Lytic Replication
- Viral Protein Inhibits RISC Activity by Argonaute Binding through Conserved WG/GW Motifs
- TOPO3α Influences Antigenic Variation by Monitoring Expression-Site-Associated Switching in
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- The Meningococcal Vaccine Candidate Neisserial Surface Protein A (NspA) Binds to Factor H and Enhances Meningococcal Resistance to Complement
- Network Modeling Reveals Prevalent Negative Regulatory Relationships between Signaling Sectors in Arabidopsis Immune Signaling
- Endothelial Galectin-1 Binds to Specific Glycans on Nipah Virus Fusion Protein and Inhibits Maturation, Mobility, and Function to Block Syncytia Formation
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
- Integration Preferences of Wildtype AAV-2 for Consensus Rep-Binding Sites at Numerous Loci in the Human Genome
- Epigenetic Analysis of KSHV Latent and Lytic Genomes
- Vaccinia Virus–Encoded Ribonucleotide Reductase Subunits Are Differentially Required for Replication and Pathogenesis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- RNA Virus Replication Complexes
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



