-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
Small non-coding RNAs (sRNAs) that act as regulators of gene expression have been identified in all kingdoms of life, including microRNA (miRNA) and small interfering RNA (siRNA) in eukaryotic cells. Numerous sRNAs identified in Salmonella are encoded by genes located at Salmonella pathogenicity islands (SPIs) that are commonly found in pathogenic strains. Whether these sRNAs are important for Salmonella pathogenesis and virulence in animals has not been reported. In this study, we provide the first direct evidence that a pathogenicity island-encoded sRNA, IsrM, is important for Salmonella invasion of epithelial cells, intracellular replication inside macrophages, and virulence and colonization in mice. IsrM RNA is expressed in vitro under conditions resembling those during infection in the gastrointestinal tract. Furthermore, IsrM is found to be differentially expressed in vivo, with higher expression in the ileum than in the spleen. IsrM targets the mRNAs coding for SopA, a SPI-1 effector, and HilE, a global regulator of the expression of SPI-1 proteins, which are major virulence factors essential for bacterial invasion. Mutations in IsrM result in disregulation of expression of HilE and SopA, as well as other SPI-1 genes whose expression is regulated by HilE. Salmonella with deletion of isrM is defective in bacteria invasion of epithelial cells and intracellular replication/survival in macrophages. Moreover, Salmonella with mutations in isrM is attenuated in killing animals and defective in growth in the ileum and spleen in mice. Our study has shown that IsrM sRNA functions as a pathogenicity island-encoded sRNA directly involved in Salmonella pathogenesis in animals. Our results also suggest that sRNAs may represent a distinct class of virulence factors that are important for bacterial infection in vivo.
Published in the journal: A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors. PLoS Pathog 7(9): e32767. doi:10.1371/journal.ppat.1002120
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002120Summary
Small non-coding RNAs (sRNAs) that act as regulators of gene expression have been identified in all kingdoms of life, including microRNA (miRNA) and small interfering RNA (siRNA) in eukaryotic cells. Numerous sRNAs identified in Salmonella are encoded by genes located at Salmonella pathogenicity islands (SPIs) that are commonly found in pathogenic strains. Whether these sRNAs are important for Salmonella pathogenesis and virulence in animals has not been reported. In this study, we provide the first direct evidence that a pathogenicity island-encoded sRNA, IsrM, is important for Salmonella invasion of epithelial cells, intracellular replication inside macrophages, and virulence and colonization in mice. IsrM RNA is expressed in vitro under conditions resembling those during infection in the gastrointestinal tract. Furthermore, IsrM is found to be differentially expressed in vivo, with higher expression in the ileum than in the spleen. IsrM targets the mRNAs coding for SopA, a SPI-1 effector, and HilE, a global regulator of the expression of SPI-1 proteins, which are major virulence factors essential for bacterial invasion. Mutations in IsrM result in disregulation of expression of HilE and SopA, as well as other SPI-1 genes whose expression is regulated by HilE. Salmonella with deletion of isrM is defective in bacteria invasion of epithelial cells and intracellular replication/survival in macrophages. Moreover, Salmonella with mutations in isrM is attenuated in killing animals and defective in growth in the ileum and spleen in mice. Our study has shown that IsrM sRNA functions as a pathogenicity island-encoded sRNA directly involved in Salmonella pathogenesis in animals. Our results also suggest that sRNAs may represent a distinct class of virulence factors that are important for bacterial infection in vivo.
Introduction
Salmonella (e.g. S. enterica serovars Typhimurium and Enteritidis) is the leading cause of food-borne illnesses in the United States, causing diverse diseases ranging from mild, self-limiting gastroenteritis to life-threatening systemic infection [1]. As a facultative intracellular pathogen, Salmonella invades non-phagocytic cells such as intestinal epithelial cells and replicates in phagocytes during systemic infection. Two hallmarks of Salmonella pathogenesis, i.e. host invasion and intracellular proliferation, correlate with the genes in Salmonella pathogenicity islands (SPIs), which are distinct, relatively large chromosomal regions harboring virulence genes and are commonly found in pathogenic strains [2], [3]. For example, Salmonella pathogenicity island 1 (SPI-1) contains invasion genes, while Salmonella pathogenicity island 2 (SPI-2) contains genes required for intracellular survival and replication [4], [5], [6]. Both SPI-1 and SPI-2 encode type III secretion systems (T3SS), which are specialized organelles that deliver effector proteins to the cytosol of host cells [3], [7]. The T3SS apparatus is a needle-like structure that spans the inner and outer membranes of the bacterial envelope and penetrates host cell membranes. Through T3SS Salmonella secretes translocon proteins that allow the delivery of effector proteins into eukaryotic cells [3], [7], leading to modulation of host cells and immune responses, and promotion of bacterial pathogenesis [4], [5], [6]. Highly regulated expression of the genes in SPIs and those encoding their effector proteins is observed both in vitro and in vivo and is required for bacterial pathogenesis and infection [6], [8].
The regulation of expression of genes coding for SPI-1 proteins and its effectors is remarkably complex and is yet to be fully characterized [8]. For example, the expression of some genes coding for SPI-1 proteins, including SopA, has been shown to be induced upon invasion of both macrophages and epithelial cells and at the late stage of Salmonella infection in animals [9], [10], [11], [12], [13]. These results suggest that in addition to their generally recognized roles in invasion, the SPI-1 factors may play an important role post-invasion, possibly in intracellular replication/survival.
Small non-coding RNAs (sRNAs) that act as regulators of gene expression have been identified in all kingdoms of life, including microRNA (miRNA) and small interfering RNA (siRNA) in eukaryotic cells [14], [15], [16], [17]. The bacterial sRNAs are generally untranslated, and range from 50–300 nucleotides in length. They play diverse physiological roles such as those in the regulation of stress responses and metabolism, and control of bacterial envelope composition [16], [18], [19]. The mechanisms by which bacterial sRNAs modulate gene expression are diverse, and two general modes of action have been established, dividing regulatory sRNAs into two classes [16], [18], [19], [20]. One class of sRNAs acts by interacting with a protein target to modify its activity [21], [22]. The other class of sRNAs acts by base-pairing with one or more target RNAs. Most of these antisense RNAs base-pair with target mRNAs in trans with partial complementarity to modulate their translation and/or stability [16], [17], [20].
While many protein factors (e.g. SPI-1 and SPI-2 factors) are shown to be important for bacterial pathogenesis, little is known about the functions of sRNAs in Salmonella virulence. Recently novel sRNA genes have been identified within the SPIs [23], [24], [25]. The expression of many of these sRNA genes is associated with stress conditions and the stationary phase [25]. In addition, several SPI-encoded sRNAs were found to be expressed in Salmonella that resides within macrophages [23]. The location of these sRNA genes in the SPIs suggests that these sRNAs may play important roles in bacterial pathogenesis and virulence.
Here we demonstrate that a SPI-encoded sRNA, IsrM sRNA, modulates the differential expression of SPI-1 genes by independently targeting the mRNAs of both the global SPI-1 regulator HilE and specific SPI-1 effector SopA. Salmonella with mutations in isrM is defective in bacterial invasion and intracellular replication in vitro, and in virulence and colonization in infected mice. To our knowledge, IsrM is the first pathogenicity island-encoded RNA known to be important in Salmonella pathogenesis in vivo. Our study suggests that sRNAs may represent a distinct class of virulence factors that significantly contribute to bacterial pathogenesis in vivo.
Results
Expression of IsrM RNA in vitro under conditions resembling those of the gastrointestinal tract
Because IsrM is a pathogenicity island-specific sRNA of Salmonella and is not found in E.coli [23], it is conceivable that IsrM plays a role in pathogenesis of Salmonella. The gene encoding IsrM can be found in several serovars of Salmonella such as Salmonella typhimurium but not other serovars such as Salmonella typhi or Samonella bongori (Figure 1). It is currently unclear why IsrM is found in some serovars but not other serovars of Salmonella, although it is possible that the presence of IsrM in a specific serovar may be related to the unique pathogenesis of the serovar in a particular host. IsrM is expressed as a 329-nucleotide RNA at both the logarithm (log) and stationary growth phases, and is expressed at a moderate level in Salmonella isolated from J774 macrophages [23]. However, little is currently known about the expression of IsrM in vitro under conditions resembling those at early stages of infection, and its expression during Salmonella infection in vivo has not been reported.
Fig. 1. Alignment of isrM sequences of different Salmonella strains including the upstream promoter region, based on BLAST searches of the IsrM sequences in GenBank. 
The transcription start sites, −10 boxes, −35 boxes, and transcription terminators of isrM genes are boxed and in red [23]. The targeting sequences for SopA and HilE mRNAs that were identified in this study are boxed and in green. During natural infection via the oral route, Salmonella encounters a series of environmental changes in the gastrointestinal track that prime Salmonella for invasion of the intestinal epithelium [26], [27], [28], [29], [30], [31], [32]. To study the role of IsrM in Salmonella invasion, we exposed Salmonella to three different stimuli that resembled those Salmonella likely encounters during the early stages of its oral infection: pH, osmolarity, and oxygen limitation. The expression of IsrM was examined using quantitative RT-PCR (qRT-PCR) assays with the level of 16S rRNA as the internal control (Figure 2A–C).
Fig. 2. Expression of IsrM RNA in Salmonella. 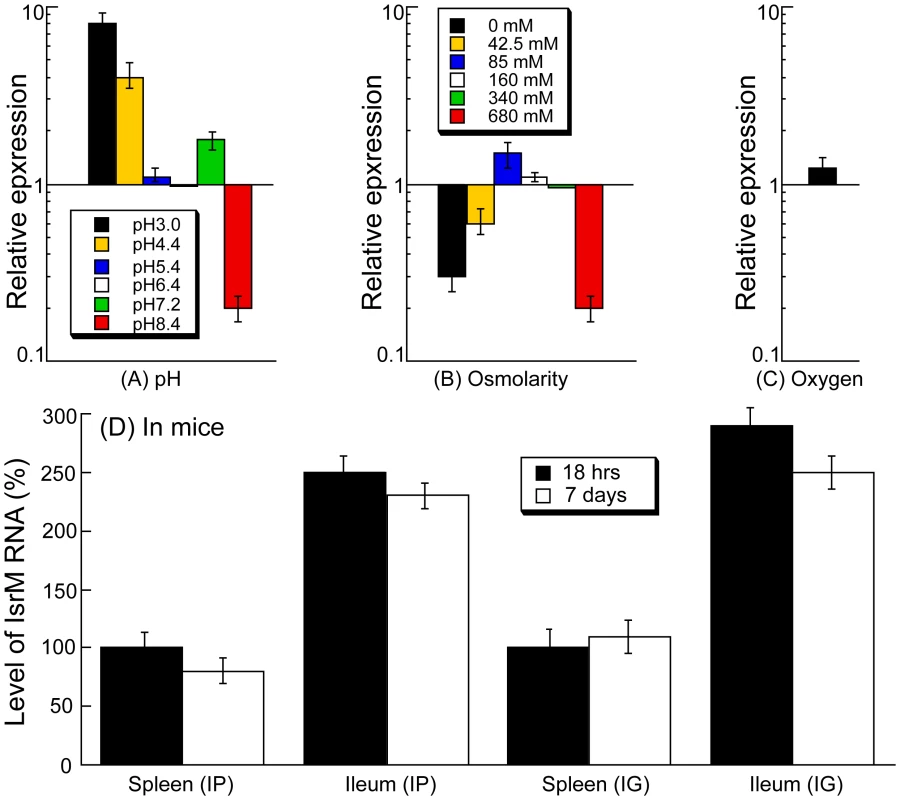
Salmonella was grown either in vitro (A–C) or in mice (D). Effect of pH values (A), osmolarity (B), and limitation of oxygen (C). Cultures of ST14028s were grown in culture media at different pH, at various concentrations of NaCl, or in the presence and limitation of oxygen. The values of the relative expression, which are the means from triplicate experiments, represent the ratios of the levels of IsrM under each pH condition to that under the control pH7.0 condition (A), the ratios of the levels of IsrM under different osmolarity conditions to that under the control condition of 170 mM NaCl as in regular LB broth (B), or the ratios of the level of IsrM under oxygen limitation to that under the control condition (i.e. in the presence of oxygen) (C). The analyses were repeated three times. The standard deviation is indicated by the error bars. (D) Level of IsrM from Salmonella recovered from the spleen and ileum of infected mice. SCID mice were intraperitoneally (IP) or intragastrically (IG) infected with ST14028s, and bacteria were recovered from the organs at 18 hours or 7 days post inoculation, respectively. The values in the IP and IG experiments, which are the means from triplicate experiments, represent the relative level of IsrM in ST14028s recovered from the spleen and ileum, as compared to the level of IsrM in Salmonella recovered from the spleen of the IP- and IG-infected mice at 18 hours post inoculation, respectively. During enteric infection, Salmonella needs to survive in the stomach, which is acidic, before establishing colonization in the intestine, which is relatively basic. Quantitative RT-PCR assays showed that IsrM had higher expression at pH3.0, pH4.4, and pH7.2, with the highest expression at pH3.0, compared to that at pH7.0 (Figure 2A). These results were further confirmed using Northern blot analyses. For example, using the level of 5S RNA as the internal control, Northern analyses showed that the level of IsrM was higher at pH4.4 than at pH7.0 (Figure S1)(Supporting Information). These observations suggest that this sRNA may be expressed as early as in the stomach during Salmonella infection in vivo. High osmolarity is an environmental stress that bacteria encounters in the intestine, and is believed to promote Salmonella adhesion and invasion of intestinal epithelial cells, contributing to its virulence [28]. Increasing osmolarity up to 85 mM NaCl favored the expression of IsrM (Figure 2B). Oxygen limitation, an important characteristic of the environment in the intestine, can induce Salmonella invasiveness of intestinal mucosa while aerobic conditions render Salmonella less invasive [29], [30], [31], [32]. In our experiments, IsrM expression was moderately induced by oxygen limitation (Figure 2C). Thus, IsrM RNA was apparently expressed under conditions that resembled the early stages of its oral infection in the gastrointestinal tract.
Expression of IsrM during Salmonella infection in vivo
Both immunocompetent BALB/c mice and immunodeficient SCID mice were infected intraperitoneally to study IsrM expression during systemic bacterial infection. Immunodeficient animals, such as the CB17 SCID mice that lack functional T and B lymphocytes, are extremely susceptible to Salmonella infection [12], [13]. Analysis of bacterial growth in these mice can be used for comparing the virulence and studying the pathogenesis of different bacterial strains and mutants [12], [13]. At different time points postinfection, the spleen and ileum were harvested. Quantitative RT-PCR was carried out to determine the expression of IsrM RNA in Salmonella isolated from the tissues, using the expression of bacterial 16S rRNA as the internal control (Figure 2D). Normalization of samples was also carried out by using total RNAs extracted from the same colony forming unit (CFU) (e.g. 5×104 CFU) of bacteria. Similar amounts of 16S rRNA were detected from same CFU Salmonella regardless of infection route (intraperitoneally or intragastrically) or time point postinfection (12–24 hours or 5–7 days) (data not shown), suggesting that the level of 16S rRNA was not significantly different in bacteria from the spleen and ileum. Salmonella presumably trafficked to the ileum tissue via the lymphatic system in intraperitoneally-infected mice, while in intragastrically-infected mice, the bacteria could directly invade and infect the ileum tissue.
Salmonella isolated from both the spleen and ileum of SCID mice at 18 hours postinfection were found to express IsrM, with its expression in the ileum approximately 2.5-fold higher than that in the spleen (Figure 2D). To rule out the residual expression of IsrM from in vitro bacterial growth, mice were intraperitoneally infected with bacteria for a longer period and tissues were harvested at 7 days postinfection. We also found that IsrM was expressed in both organs, with its expression in the ileum approximately 2.5-fold higher than that in the spleen. Similar results were also observed in bacteria isolated from the organs of the intraperitoneally-infected BALB/c mice (data not shown).
SCID mice were also infected intragastrically with bacteria in order to study whether IsrM is expressed during Salmonella infection acquired by the oral route. Spleens and ileums were collected and the bacteria were recovered at 18 hrs and 7 days postinfection. IsrM was detected in Salmonella isolated from both the spleen and ileum, with its level in the ileum approximately 2.5-fold higher than that in the spleen (Figure 2D). Similar results were also observed in Salmonella isolated from the organs of infected BALB/c mice. Taken together, these results suggest that IsrM is expressed during the early and late stages of Salmonella infection, and that it is differentially expressed, with higher expression in the ileum than in the spleen.
IsrM is dispensable for Salmonella growth in vitro but important for the expression of Salmonella SPI-1 proteins
To investigate the role of IsrM in Salmonella growth, a mutant, ΔIsrM, was derived from the wild type S. typhimurium 14028s strain by deleting the IsrM sequence. Mutant ΔIsrM grew as well as ST14028s in LB broth (Figure S2A)(Supporting Information), indicating that IsrM is dispensable for bacterial growth in vitro.
The unique presence of IsrM in a SPI region and its induced expression in the ileum suggests that IsrM may play a role in the pathogenesis of Salmonella in the intestine. Since SPI-1 proteins are major virulence determinants essential for Salmonella invasion [3], [7], we reasoned that IsrM may function to regulate the expression of SPI-1 factors, facilitating Salmonella infection and pathogenesis. To investigate this possibility, we examined the effect of a deletion of the IsrM sequence on the expression of SPI-1 factors. Bacterial strains T-invJ, T-sopE2, T-sptP, T-spaO, T-sipD, T-sipC, T-sipB, T-sipA, T-sopB, and T-sopA were previously derived from the wild type Salmonella strain ST14028s by inserting the FLAG epitope tag sequence into SPI-1 ORFs invJ, sopE2, sptP, spaO, sipD, sipC, sipB, sipA, sopB, and sopA, respectively [12], [13]. Tagging of the SPI-1 ORFs in these mutants did not impair the invasiveness, growth, or the virulence of the bacteria, and the tagged strains can be used as model strains to study the infection of Salmonella in vitro and in vivo, including the expression of SPI-1 proteins and effectors [12], [13]. We constructed mutants ΔM-T-invJ, ΔM-T-sopE2, ΔM-T-sptP, ΔM-T-spaO, ΔM-T-sipD, ΔM-T-sipC, ΔM-T-sipB, ΔM-T-sipA, ΔM-T-sopB, and ΔM-T-sopA from T-invJ, T-sopE2, T-sptP, T-spaO, T-sipD, T-sipC, T-sipB, T-sipA, T-sopB, and T-sopA, respectively, by deleting the IsrM sequence in each SPI-1 tagged strain (Table 1). These newly constructed isrM-deletion mutant strains grew as well as the parental strains and the wild type ST14028s in LB broth (Figure S2B)(Supporting Information), consistent with our results (Figure S2A)(Supporting Information) that IsrM is not essential for bacterial growth in vitro.
Tab. 1. Bacterial strains used in this study. 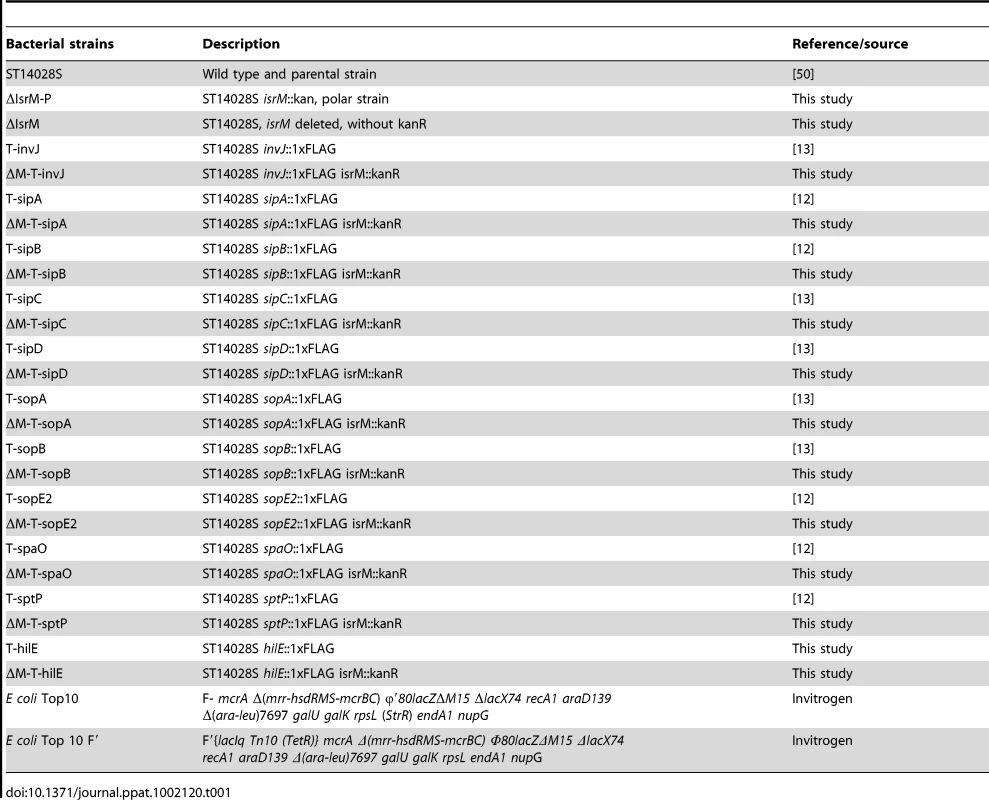
We performed Western analyses to determine the expression of the tagged SPI-1 proteins with an anti-FLAG antibody, using the expression of bacterial DnaK protein as the internal control (Figure 3A). Normalization of samples was also carried out by loading total protein extracted from the same CFU (e.g. 5×107 CFU) of bacteria in each lane. The levels of the tagged InvJ, SpoE2, SptP, SpaO, SipD, SipC, SipB, SipA, and SopB proteins in the IsrM deletion strains were found to be lower than those in the parental strains (Figure 3). In contrast, the protein level of SopA in the IsrM-deletion strain (i.e. ΔM-T-sopA) was found to be approximately 2.5-fold higher than that in the parental T-sopA strain (Figure 3A, compare lanes 1 and 2). Transformation of plasmid pIsrM, which contained the sequence of IsrM under its native promoter and transcriptional terminator sequence, in the deletion mutants restored the levels of the tagged proteins to the wildtype levels (Figure 3, lanes 3 and 6). Using 16S rRNA as the internal control, qRT-PCR assays showed that the levels of IsrM were similar in the parental strain T-sopA and the deletion mutant ΔM-T-SpoA that contained the complementation construct pIsrM (data not shown). Taken together, these results suggest that deletion of IsrM may lead to a global disregulation of expression of the SPI-1 proteins.
Fig. 3. Expression of the tagged proteins in Salmonella. 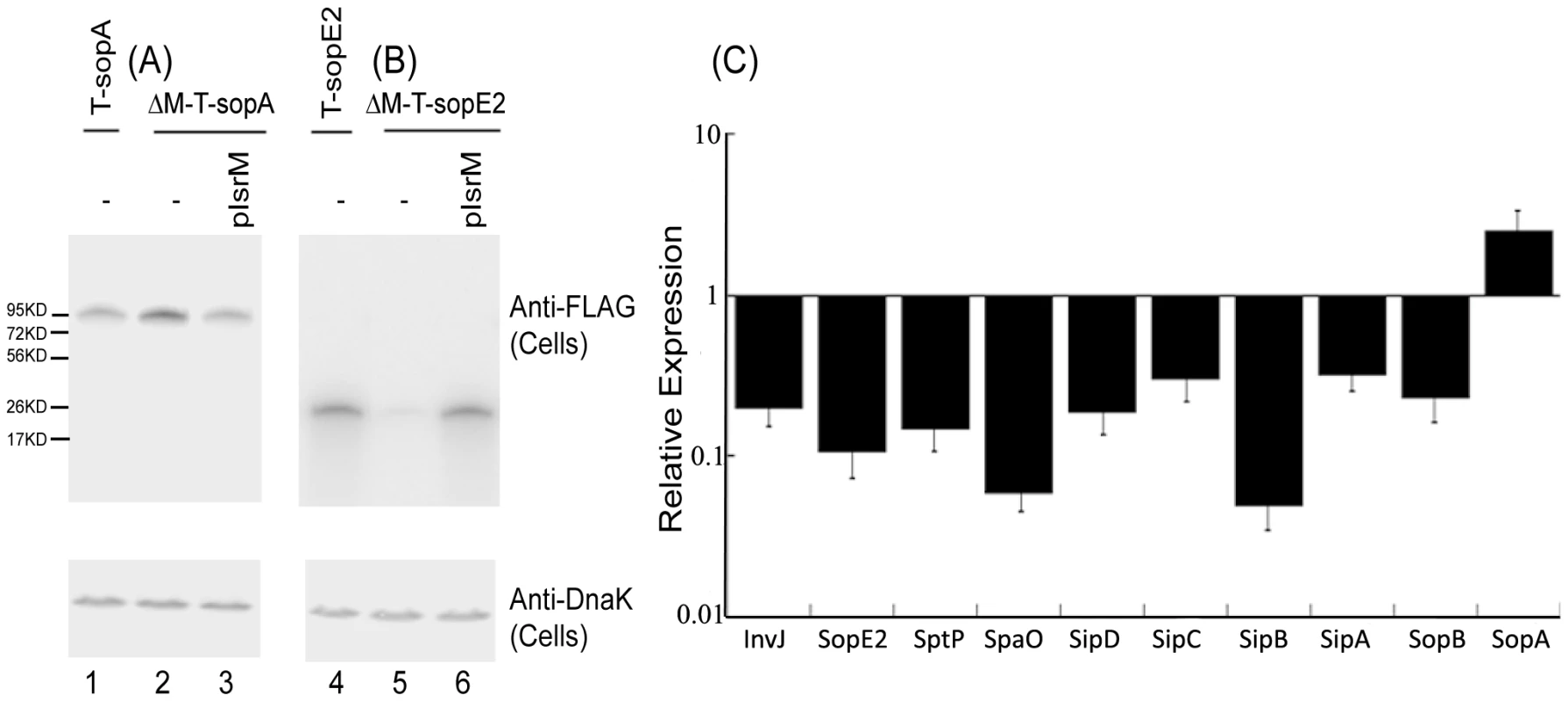
(A–B) Western blot analyses of the expression of the tagged proteins from bacterial strains T-sopA and ΔM-T-sopA (A), and T-sopE2 and ΔM-T-sopE2 (B). The expression of bacterial DnaK was used as the internal control. Protein samples were separated in SDS-polyacrylamide gels and reacted with antibodies against the FLAG sequence and DnaK. Each lane was loaded with lysate prepared from 5×107 CFU bacteria. (C) The effect of the deletion of the isrM sequence on the expression of SPI-1 proteins. The values of the relative expression, which are the means of triplicate experiments, represent the ratios of the levels of the tagged proteins in ΔM-T-invJ, ΔM-T-sopE2, ΔM-T-sptP, ΔM-T-spaO, ΔM-T-sipD, ΔM-T-sipC, ΔM-T-sipB, ΔM-T-sipA, ΔM-T-sopB, and ΔM-T-sopA to those in T-invJ, T-sopE2, T-sptP, T-spaO, T-sipD, T-sipC, T-sipB, T-sipA, T-sopB, and T-sopA, respectively. Targeting the 5′ untranslated region (UTR) of the HilE mRNA by IsrM
Bacterial sRNAs generally function to modulate gene expression by hybridizing to the target mRNAs [18], [19], [20]. No targets for IsrM have been reported. Using the TargetRNA and RNAhybrid algorithms, we searched the regions of Salmonella mRNAs for potential RNA duplex formation with IsrM [33], [34]. This analysis predicted stable pairing of two distinct and non-overlapping regions of IsrM with the entire Shine-Dalgarno (SD) sequence of hilE and sopA mRNAs, respectively (Figure 4A–B), which may cause translational repression [18], [19], [20].
Fig. 4. Post-transcriptional targeting of hilE and sopA by IsrM. 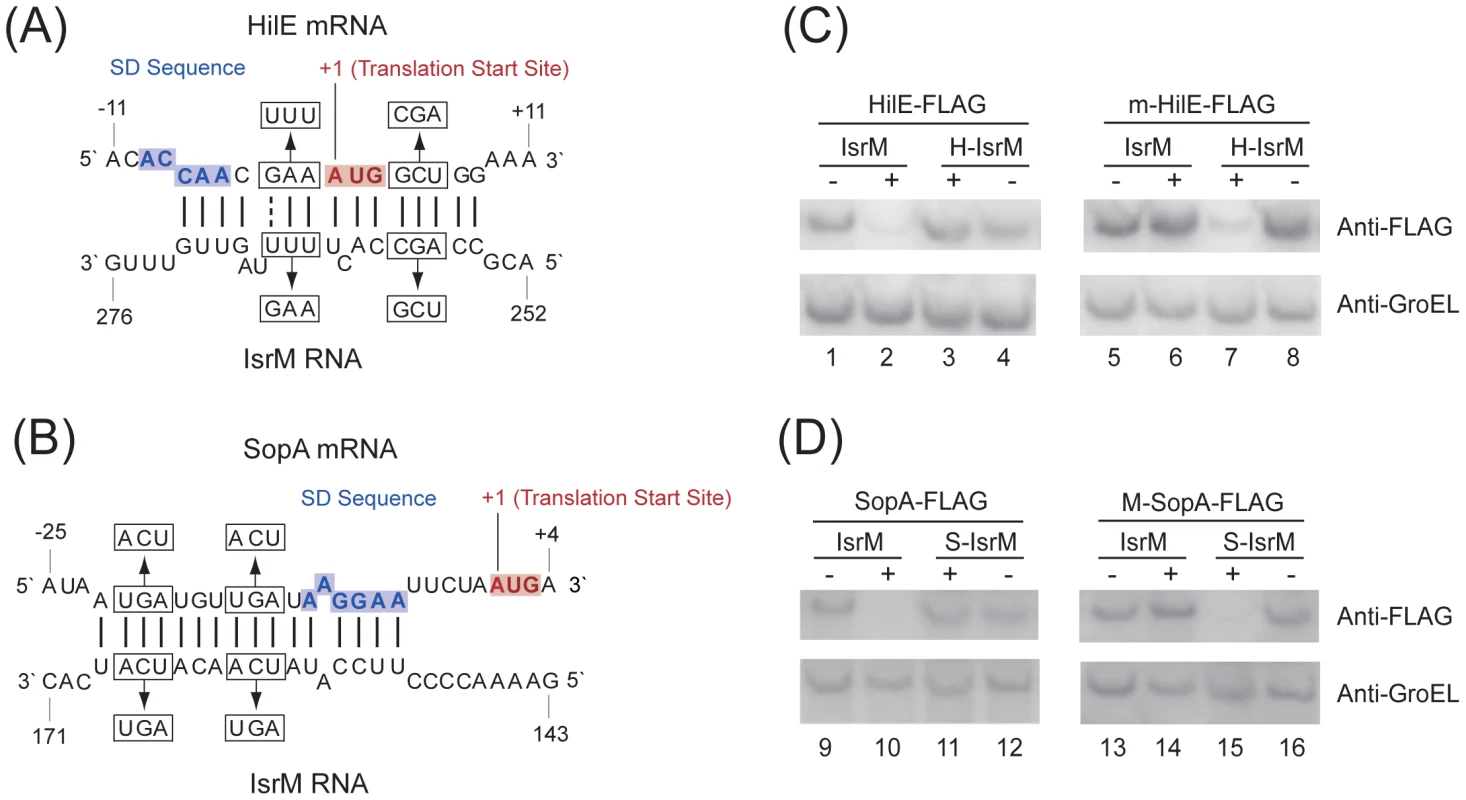
(A–B) Schematic representation of the proposed interactions of IsrM sRNA to hilE (A) and sopA mRNAs (B), and of compensatory base-pair changes. Numbers indicate relative position to the translational start site of hilE and sopA or position downstream of the transcriptional start site of isrM. Arrows denote nucleotide substitutions (in box) introduced to IsrM, and hilE and sopA mRNAs. The SD and AUG sequences are highlighted in blue and red, respectively. (C–D) Western blot analysis of E. coli carrying pLaco-IsrM, pLaco-H-IsrM and pLaco-S-IsrM, in combination with either wild type or mutant target plasmids, as indicated. The expression of the tagged HilE (C) and SopA (D) proteins was determined using that of GroEL as the internal control. HilE is a negative global regulator of the expression of many SPI-1 genes by sequestering HilD, a major SPI-1 transcriptional activator [8], [35]. To determine if IsrM regulates hilE, we constructed a tagged hilE strain T-hilE by inserting the FLAG epitope tag sequence into hilE of ST14028s and furthermore, a corresponding isrM deletion mutant of T-hilE, ΔM-T-hilE (Table 1). Both T-hilE and ΔM-T-hilE grew as well as ST14028s in LB broth, and tagging of hilE did not impair the invasiveness, growth, or the virulence of the bacteria in mice (data not shown).
Using the expression level of DnaK as the internal control, Western blot analyses with an anti-FLAG antibody indicated that deletion of IsrM increased the level of HilE (Figure 5, compare lane 1 and 4). Transformation of plasmid pIsrM carrying the IsrM sequence with the native promoter and transcriptional terminator reduced the HilE expression to the wildtype level in ΔM-T-hilE (Figure 5, lane 2), suggesting that IsrM specifically modulates the protein level of HilE.
Fig. 5. Western blot analysis of the expression of the tagged proteins from Salmonella. 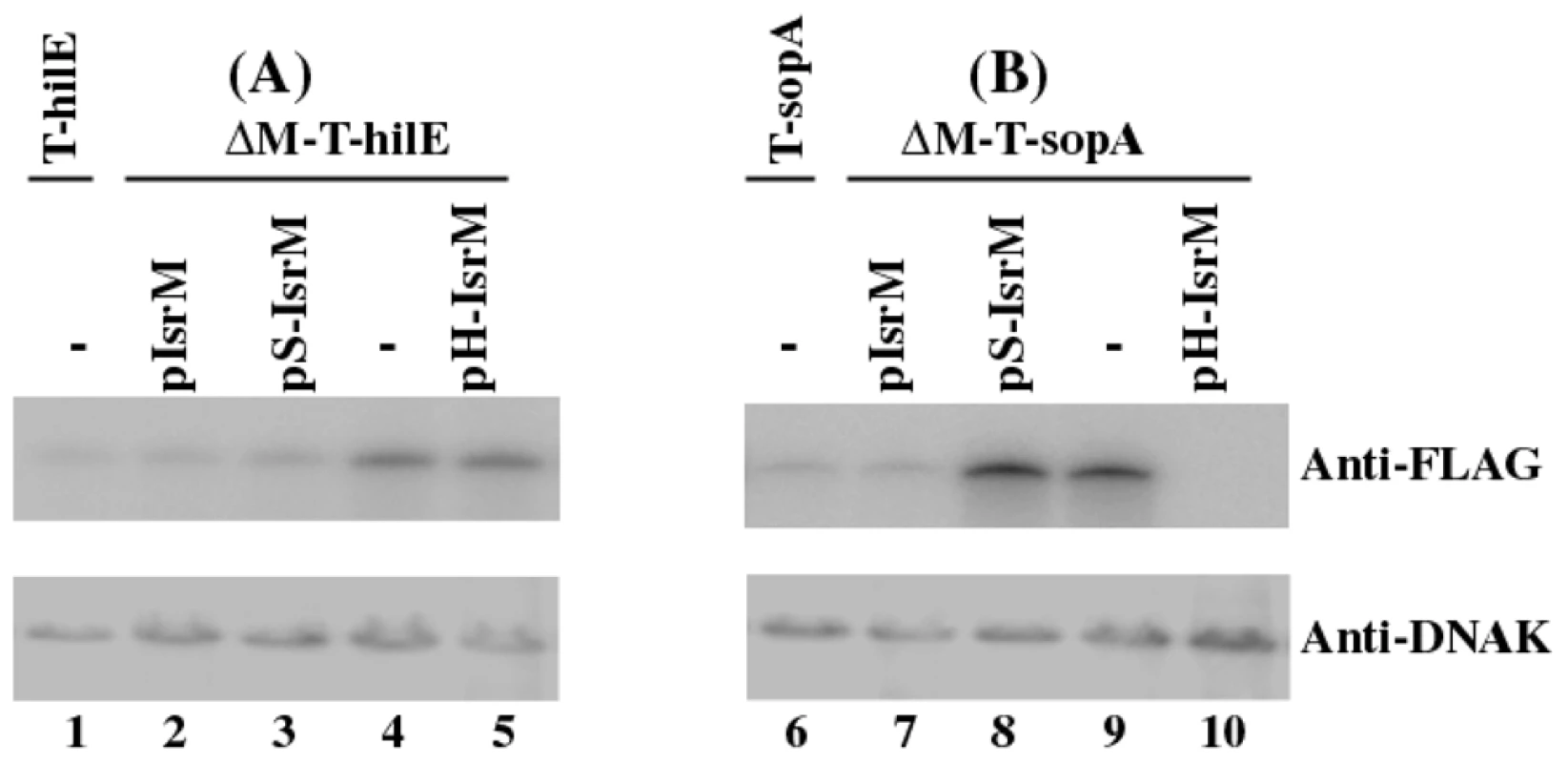
The tagged proteins were expressed from strains T-hilE and ΔM-T-hilE (A), and T-sopA and ΔM-T-sopA (B). The expression of bacterial DnaK was used as the internal control. Protein samples were separated in SDS-polyacrylamide gels and reacted with antibodies against the FLAG sequence and DnaK. Each lane was loaded with lysate prepared from 5×107 CFU bacteria. To investigate the potential interactions between IsrM and hilE mRNA, we used a two plasmid system [36] that involved co-expression of the sRNA and a translational target gene with the FLAG fusion at its carboxyl terminus. By transforming both plasmids in E.coli, we tested whether the sRNA expression directly affects the protein level of the FLAG-tagged target gene. The 5′ UTR region and the entire coding sequence of hilE were cloned into a FLAG fusion vector. Transcription of the HilE-FLAG fusion was driven by the constitutive PLtetO-1 promoter to specifically assay post-transcriptional regulation. The IsrM sequence was cloned under the control of the IPTG-inducible PLaco promoter, yielding construct pLaco-IsrM. E.coli transformed with both the hilE::flag fusion and pLaco-IsrM plasmid was grown and split into two cultures, only one of which was treated with IPTG. IsrM-dependent regulation was determined by Western blot analysis of the HilE-FLAG expression (Figure 4C). Our results showed that the expression of IsrM significantly reduced HilE-FLAG expression (Figure 4C, lanes 1 and 2).
To confirm the predicted base-pairing interactions between hilE mRNA and IsrM, point mutations were introduced to isrM (U264U265U266 ->A264A265G266) and hilE (G-3A-2A-1 ->U-3U-2U-1)to generate mutant H-IsrM and m-HilE (Figure 4A). The expression of HilE-FLAG was not repressed by mutant H-IsrM (Figure 4C, lanes 3 and 4), while IsrM did not significantly affect the expression of mutant m-HilE-FLAG (lanes 5 and 6). Repression of HilE expression by IsrM was restored using m-HilE-FLAG and H-IsrM, which contained the compensatory mutations (lanes 7 and 8). To determine the degree of IsrM over-expression from the plasmids used and to correlate the level of IsrM expression with the levels of HilE protein, qRT-PCR assays were performed to determine the level of IsrM. Table S1 (Supporting Information) shows the relative levels of IsrM and the respective levels of HilE protein in the experiments. These results suggest that no significant difference in the expression levels of IsrM in the presence of IPTG was found in these experiments, and that base-pairing interactions between the specific sequences of IsrM (U264U265U266) and HilE mRNA (G-3A-2A-1) were required for IsrM-mediated downregulation of HilE protein expression.
To further validate the interactions between IsrM and the hilE mRNA, an additional pair of compensatory mutations was introduced to isrM (A257G258C259 ->U257C258G259) and hilE (G4C5U6 ->C4G5A6) to generate mutants H2-IsrM and m2-HilE, respectively (Figure 4A). Repression of the m2-HilE-FLAG protein was restored only by using H2-IsrM, which contained compensatory mutations (data not shown). These results suggest that IsrM inhibits HilE expression by binding to its mRNA around the SD sequence.
Targeting the 5′ UTR of the SopA mRNA by IsrM
Similar experiments were performed to validate the potential interactions between IsrM and the sopA mRNA. Since the transcription initiation site of the sopA mRNA has not definitively been determined, the sequence between the position 61 nt upstream the translation initiation codon and the position coding for the 258th amino acid residue of SopA, which covered the sequence predicted to interact with IsrM, was cloned into the PLtetO-1-FLAG fusion vector. E.coli transformed with both the sopA::flag fusion and pLaco-IsrM plasmid was grown and split into two cultures, only one of which was treated with IPTG. Western blot analysis indicated that the expression of IsrM significantly reduced SopA-FLAG expression (Figure 4D; lanes 9 and 10). To confirm the predicted potential base-pairing interactions between IsrM and the sopA mRNA, point mutations were introduced to isrM (U165C166A167 ->A165G166U167) and sopA (U-21G-20A-19 ->A-21C-20U-19) to generate mutant S-IsrM and M-SopA (Figure 4B). The expression of SopA-FLAG was not repressed by S-IsrM (Figure 4D, lanes 11 and 12), while IsrM did not significantly affect the expression of M-SopA-FLAG (lanes 13 and 14). Repression of M-SopA-FLAG expression was restored by using S-IsrM, which contained the compensatory mutations (lanes 15 and 16). To determine the degree of IsrM over-expression from the plasmids used and to correlate the level of IsrM expression with the levels of SopA protein, qRT-PCR assays were performed to determine the level of IsrM. Table S2 (Supporting Information) shows the relative levels of IsrM and the respective levels of SopA protein in the experiments. These results suggest that no significant difference in the expression levels of IsrM in the presence of IPTG was found in these experiments, and that base-pairing interactions between the specific sequences of IsrM (U165C166A167) and SopA mRNA (U-21G-20A-19) were required for IsrM-mediated downregulation of SopA protein expression.
To further validate the interactions between IsrM and sopA mRNA, an additional pair of compensatory mutations was introduced to isrM (U159C160A161 ->A159G160U161) and sopA (U-15G-14A-13 ->A-15C-14U-13) to generate mutants S2-IsrM and M2-sopA, respectively (Figure 4B). Repression of the M2-SopA-FLAG protein was restored only by using S2-IsrM, which contained compensatory mutations (data not shown). Taken together, these results suggest that IsrM inhibits SopA expression by binding to its mRNA around the SD sequence.
Independent targeting of hilE and sopA mRNAs by IsrM in Salmonella
Since HilE globally regulates the expression of most of SPI-1 proteins including SopA [8], targeting of either hilE or sopA mRNAs by IsrM is expected to modulate the SopA protein expression. Consistent with this notion, the expression of SopA in the isrM deletion mutant ΔM-T-SopA was suppressed to the wildtype level only by transformation of plasmid pIsrM (Figure 5, lane 7), which contained the wild type full length IsrM sequence, but not by pS-IsrM, which contained mutated IsrM sequence at the SopA binding site with nucleotide substitutions (U165C166A167 ->A165G166U167) (Figure 5, lane 8). The over-expression of SopA in ΔM-T-SopA was also suppressed by transformation of pH-IsrM (Figure 5, lane 10), which contained mutations in IsrM sequence that disrupted the binding of IsrM to hilE mRNA (U264U265U266 ->A264A265G266). Interestingly, the SopA level was lower in the pH-IsrM transformed ΔM-T-SopA than the pIsrM transformed mutant (compare lane 10 to lane 7 of Figure 5). One possible explanation for the observation is that pIsrM suppresses HilE while pH-IsrM does not due to mutations in the hilE mRNA binding site, and therefore the HilE level in the pH-IsrM transformed mutant is likely higher than in the pIsrM transformed mutant (compared lane 5 and 2 of Figure 5). Since HilE negatively regulates SopA and other SPI-1 factors, the higher HilE level in the pH-IsrM transformed mutant may lead to further suppression of SopA as compared to the pIsrM transformed mutant. The expression of tagged HilE protein could also be reduced to the wildtype level in the isrM deletion mutant ΔM-T-hilE by construct pIsrM or pS-IsrM with mutations in the binding site to sopA mRNA, but not with construct pH-IsrM that contained mutations in the binding site to hilE mRNA (Figure 5A). Quantitative RT-PCR assays showed no significant differences in the expression levels of the wild type and mutant IsrM between Salmonella carrying complementation constructs pIsrM, pH-IsrM, or pS-IsrM (data not shown). These observations suggest that IsrM targets HilE and SopA independently by using different regions to bind to these mRNAs separately.
Role of IsrM in Salmonella invasion
It is expected that mutations at IsrM, which result in altered expression levels of HilE, SopA, and other SPI-1 proteins, affect Salmonella invasion since proper expression of SPI-1 factors is required for Salmonella entry to nonphagocytosed cells. To determine whether this is the case, we tested the ability of various mutant Salmonella strains to invade cultured epithelial cells. A reduction of about 90% in invasion of HeLa cells was observed in ΔIsrM, compared to the wild type ST14028s (Figure 6). Transformation with the wild type complementation construct pIsrM restored the ability of ΔIsrM to invade (Figure 6). Furthermore, the level of invasion of ΔIsrM could be restored by more than 90% with transformation of construct pS-IsrM, which expressed a mutated IsrM (S-IsrM) with mutations in the binding site to the sopA mRNA, but not with construct pH-IsrM, which expressed a mutated IsrM (H-IsrM) with mutations in the binding site to the hilE mRNA (Figure 6). These results suggest that IsrM is important for bacterial invasion by modulating the expression of HilE protein.
Fig. 6. Epithelial cell invasion and intracellular replication in macrophages. 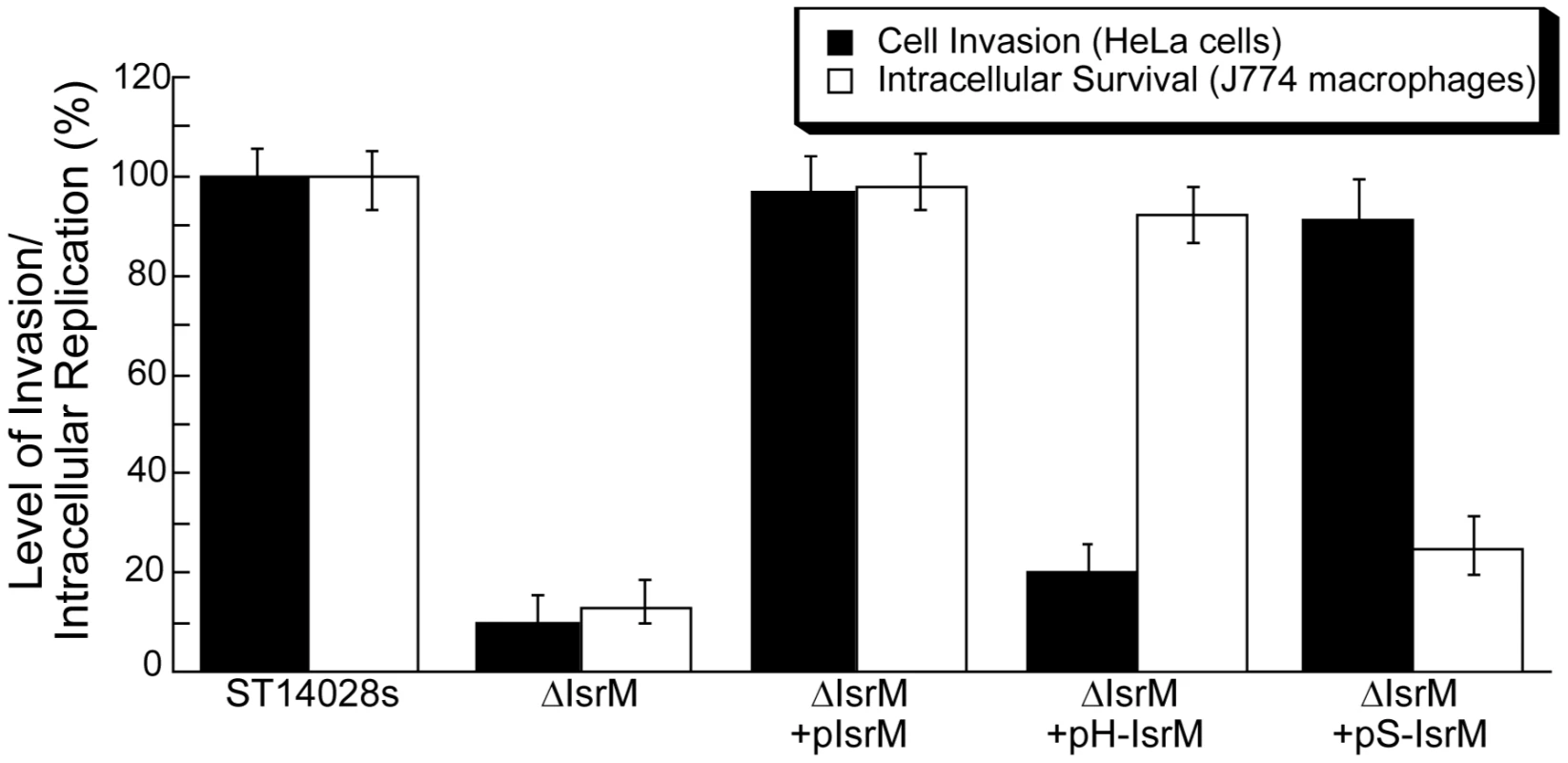
Isogenic Salmonella strains carrying different constructs were added to HeLa cells at a MOI of 10, and cell invasion was assayed by determining the ratio of the number of intracellular bacteria at 1 hour postinfection to the number of input bacteria. The ratio for wild-type strain ST14028s at 1 h postinfection was arbitrarily defined as 100%, and the ratios for other samples were expressed as relative values. For the intracellular replication assay, Salmonella was added to J774 macrophages at a MOI of 10. Intracellular growth was assayed by determining the ratio of the number of intracellular bacteria at 8 hour postinfection to the initial number of bacteria at time zero. The ratio for wild-type strain ST14028s at 8 h postinfection was arbitrarily defined as 100%, and the ratios for other samples were expressed as relative values. The data are the averages of three experiments performed in triplicate. The error bars indicate standard deviations. Role of IsrM in intracellular survival in macrophages
Intracellular replication in macrophages represents a major aspect of Salmonella pathogenesis [4], [5], [6]. The ability of various strains to proliferate intracellularly was assayed in mouse J774 macrophages [37]. A reduction of about 85% in intracellular replication was observed in ΔIsrM, compared to the wild type ST14028s (Figure 6). Transformation with the wild type complementation construct pIsrM fully restored the ability of ΔIsrM to replicate in macrophages, while the ability of ΔIsrM to replicate in J774 cells was also restored by about 80% by transformation with construct pH-IsrM that expressed the hilE mRNA binding-defective IsrM mutant (H-IsrM), but not with construct pS-IsrM that expressed the sopA mRNA binding-defective IsrM mutant (S-IsrM) (Figure 6). Meanwhile, these strains, which carried different complementation constructs, appeared to have no growth defect in vitro as they replicated as well as the wild type ST14028s strain in LB (Figure S2)(Supporting Information). These results suggest that IsrM is important for intracellular proliferation of Salmonella in macrophages by modulating the expression of SopA protein.
Role of IsrM in bacterial pathogenesis and virulence in mice
To determine if IsrM is important for pathogenesis in vivo, BALB/c mice were infected intragastrically with the constructed isrM mutants. To study the virulence of Salmonella, the survival rates of the infected animals were determined. When mice were infected intragastrically with 5×106 CFU bacteria, all animals inoculated with the wild type ST14028s strain and ΔIsrM carrying complementation construct pIsrM died within 5 days postinfection (Figure 7A). In contrast, mice infected with ΔIsrM, and ΔIsrM carrying construct pH-IsrM and pS-IsrM remained alive until 15, 12, and 10 days postinfection, respectively. Similar results were also observed in SCID mice that were intragastrically infected with these Salmonella strains (Figure S3)(Supporting Information). These results suggest that the reduced virulence of ΔIsrM is due to the deletion of IsrM and that IsrM is important for Salmonella virulence in vivo.
Fig. 7. Virulence and colonization of Salmonella in mice. 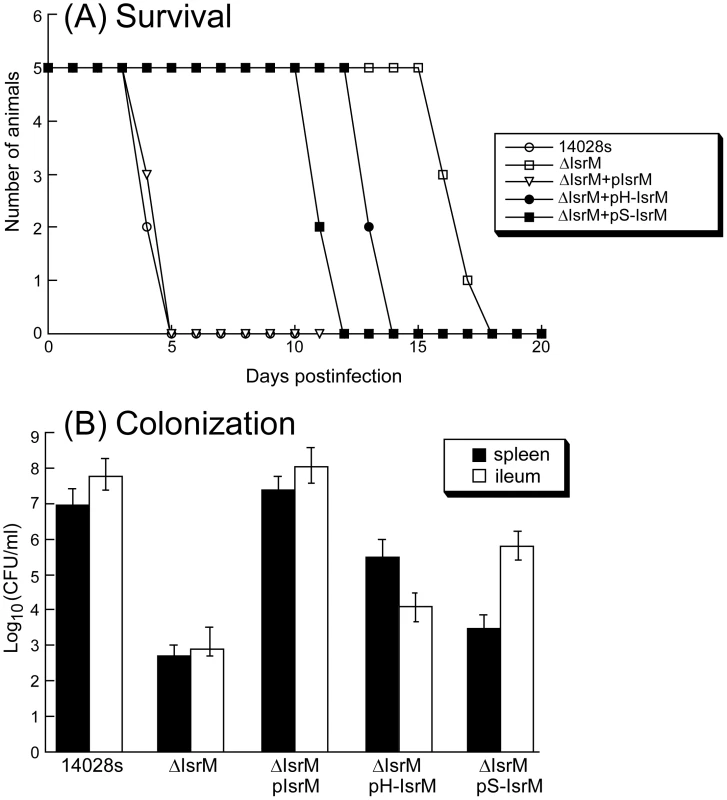
(A) Mortality of the BALB/c mice infected with isogenic strains carrying different constructs. BALB/c mice (5 animals per group) were infected intragastrically with Salmonella (5×106 CFU). (B) The numbers of bacteria (CFU) in spleen and ileum of the infected animals. Groups of BALB/c (5 animals per group) mice were infected intragastrically (IG) with 1×105 CFU of isogenic strains carrying different constructs, and bacteria were recovered from the organs at 7 days post inoculation. Each sample was analyzed in triplicate and the analysis was repeated at least three times. The CFU of the sample was expressed as the average of the values obtained. The concentrations of bacteria were recorded as CFU/ml of organ homogenate. The limit of bacteria detection in the organ homogenates was 10 CFU/ml. To further study the pathogenesis of Salmonella mutants in these animals, colonization of the mutants in the spleen and ileum was studied during a 7-day infection period before the onset of mortality of animals infected with the wild type ST14028s strain. At 7 days postinfection, the level of colonization of ΔIsrM carrying construct pIsrM was similar to that of ST14028s in the organs examined (Figure 7B). In contrast, the counts of ΔIsrM in spleens and ileums of the infected animals were lower than those of the ST14028s strain by approximately 15,000 and 70,000 fold, respectively (Figure 7B). The presence of construct pH-IsrM and pS-IsrM, which contained the mutated IsrM sequences with mutations that disrupted the interactions of IsrM RNA with the hilE and sopA mRNAs, respectively, did not completely restore the ability of mutant ΔIsrM to colonize these mice. Similar results were also observed in SCID mice that were intragastrically infected with these Salmonella strains (Figure S3)(Supporting Information). These results suggest that the attenuated colonization of ΔIsrM in these organs is probably due to the disruption of isrM and that IsrM sRNA may be required for optimal colonization and growth of Salmonella in these organs in mice.
Discussion
Pathogenicity island-encoded sRNA as a virulence factor
In this report, we presented the first direct evidence that IsrM modulates the expression of HilE and SopA proteins, and that its function is important for Salmonella invasion of epithelial cells, intracellular replication inside macrophages, and virulence and colonization in mice. A significant number of the sRNAs identified in Salmonella are encoded in the pathogenicity islands, which are not present in commensal bacteria or E.coli [23], [24], [25]. Only two of these sRNAs, InvR and IsrJ, have been characterized in detail. InvR, encoded in the SPI-1 region, represses outer membrane protein synthesis of Salmonella by regulating chromosomal genes and does not target any genes in the SPIs [24]. IsrJ, whose gene is located between STM2614 and STM2616, is expressed in the presence of the major SPI-1 transcription factor, HilA, and an isrJ-deletion mutant is defective in host cell invasion and effector translocation in vitro [23]. However, the targets of IsrJ sRNA have not been identified. Characterization of the mutants with inactivating InvR and IsrJ RNAs in animals has not been reported. The roles of either InvR or IsrJ sRNAs in bacterial pathogenesis and virulence in mice have not been determined.
Our results showed that IsrM sRNA targets the mRNAs of HilE and SopA independently, leading to down-regulation of expression of these two proteins. IsrM was expressed in vitro under conditions resembling those during the initiation of infection in the gastrointestinal tract (e. g. low pH, oxygen limitation, and high osmolarity). Furthermore, IsrM was found to be differentially expressed in vivo, with higher expression in the ileum than in the spleen. Mutations in IsrM resulted in disregulation of expression of HilE and SopA, as well as other SPI-1 genes whose expression is regulated by HilE. Salmonella with a deletion of isrM was defective in invasion of epithelial cells and intracellular replication/survival in macrophages, and the defect could be corrected by introduction of a complementation construct containing the full length isrM sequence. Moreover, Salmonella with mutations in isrM was attenuated in its virulence in killing animals and was defective in colonization in the ileum and spleen in vivo. These results suggest that IsrM sRNA encodes a virulence factor important for bacterial infection and demonstrate that a pathogenicity island-encoded sRNA is directly involved in Salmonella pathogenesis in mice. Our study raises the possibility that additional pathogenicity island-encoded sRNA may function in different aspects of Salmonella pathogenesis, including invasion of epithelium and intracellular replication in phagocytes.
Differential expression of SPI-1 proteins regulated by sRNA
The regulation of expression of genes encoding SPI-1 proteins and its effectors is remarkably complex. For example, the transcription of many SPI-1 genes can be regulated by HilA, HilC, HilD, HilE, and PhoP/Q [8]. HilE encodes a global regulator that negatively modulates the expression of SPI-1 genes by sequestering HilD, which is a major transcriptional activator required for expression of most of the SPI-1 genes [35], [38]. Our results indicated that disruption of IsrM-mediated targeting of hilE mRNA results in increased expression of HilE protein and disregulated expression of SPI-1 proteins, leading to reduced invasion efficiency in cultured cells and attenuation of bacterial virulence and pathogenesis in vivo. Thus, bacterial sRNA presents another mechanism for global regulation of SPI-1 gene expression.
In addition to globally modulating the expression of SPI-1 proteins through HilE, IsrM also interacts with the mRNA of a specific SPI-1 effector, SopA. This interaction is likely to be important as IsrM-mediated modulation of SopA protein expression affects bacterial intracellular replication in macrophages (Figure 6). SopA has been shown to function as an E3 ubiquitin ligase regulating host inflammatory responses [39], [40]. The level of this protein is highly regulated in vivo as SopA is found to be expressed during the early and late stage of Salmonella infection in vivo, consistent with its role in invasion as well as in intracellular replication/survival [9], [12], [13]. Appropriate level of expression of SopA protein in different cells and at specific time points may contribute to defined consequences of pathogenesis. This is consistent with the recent observations that SopA exerts its functions in concert with other effector proteins during Salmonella infection of non-phagocytic cells; however, in the context of systemic infection, its ubiquitin ligase activity may facilitate bacterial replication in phagocytes [39], [41], [42]. IsrM can potentially negatively regulate SopA protein level at the translation level through its direct binding of the sopA mRNA and positively at the transcriptional level simultaneously through its down-regulation of the negative regulator HilE. This scenario may represent a novel mechanism of precise control of bacterial protein levels in a temporal and tissue/cell specific manner as required for pathogenesis.
It is currently unclear why IsrM is found in some serovars of Salmonella but not others. It is possible that the presence of IsrM in a specific serovar contributes to host specificity in a particular host. This issue can be investigated further by studying the presence of IsrM in different serovars and its role in the pathogenesis of different Salmonella strains in specific hosts in vivo. Our results indicated that IsrM binds to the 5′ UTR regions near the Shine-Dalgarno (SD) sequence of both the hilE and sopA mRNAs and inhibits the expression of these two proteins. While it is generally believed that binding of the SD region by sRNA results in translation repression, the exact mechanism of how IsrM reduces the expression of HilE and SopA proteins is currently unknown. Equally elusive is whether the IsrM-mediated reduction is mediated by bacterial proteins. The Sm-like RNA-binding protein Hfq, which has been found to be potentially associated with more than 40 Salmonella sRNAs [25], acts as a RNA chaperone that modulates the intracellular stability of many small non-coding RNAs and their annealing with target mRNAs for translation repression or stimulation [17], [43], [44]. It is unclear whether IsrM RNA is associated with or bound to Hfq, based on recent co-immunoprecipitation experiments [23], [25]. The SopA mRNA was found to be enriched moderately by co-immunoprecipitation with anti-Hfq antibodies while the HilE mRNA was not detected in these experiments [25]. Further studies are needed to characterize the molecular mechanism of the regulation mediated by IsrM.
It is generally believed that the amounts of various Salmonella SPI-1 proteins expressed in vivo are in a delicate balance as there are hierarchical transports of different effectors during Salmonella entry and extensively ordered synergistic and antagonist relationships between these effectors following their delivery into host cells [3], [45], [46]. It is conceivable that the ability of the bacteria to establish successful infection and cause pathogenesis in specific tissues may be significantly influenced by the balance of the amounts of these factors during infection. IsrM was found to be expressed in vitro under conditions resembling those during the initial infection in the gastrointestinal tract. Moreover, IsrM was found to be differentially expressed in vivo, with expression levels higher in the ileum than that in the spleen. Thus, controlled expression of IsrM is likely a mechanism Salmonella uses to achieve both global and specific regulation of the expression of SPI-1 proteins in vivo at the appropriate time and place in the host. It would be interesting to determine the expression patterns of IsrM as well as other sRNAs and their potential target mRNAs in different tissues and at different stages of infection in vivo, and study how their expression affects Salmonella infection. Further characterization of these sRNAs should provide significant insights into their exact roles in Salmonella infection and pathogenesis.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol for all animal experiments was approved by the Animal Care and Use Committee of the University of California-Berkeley (Protocol #R240 and #R276). All efforts were made to minimize suffering.
Generation of Salmonella mutants and constructs
Table 1 lists the bacterial strains while Table 2 and 3 list the plasmid constructs and the primers used in the study, respectively. Salmonella strain ΔIsrM was derived from S. typhimurium strain ST14028s by deleting isrM with the λ Red recombinase method [47], following the procedures described previously [48]. Briefly, primers P5ΔIsrM and P3ΔIsrM (Table 3) were used to amplify the kanamycin resistance gene sequence in plamsid pKD4. The resulting PCR products were electrophorated into ST14028s carrying plasmid pKD46. Mutants undergoing homologous recombination were selected from electroporated ST14028s and confirmed by PCR using primers P5IsrM and P3IsrM. Once the deletion mutation was confirmed, the mutation was introduced into fresh culture of ST14028s by transduction using phage P22, and P22-free colonies were selected. The non-polar strain ΔIsrM was constructed using plasmid pCP20 [47], selected for its sensitivity to kanamycin, and further confirmed using PCR with the primers P5IsrM and P3IsrM (Table 3) [12], [13], [48].
Tab. 2. Plasmid constructs used in the study. 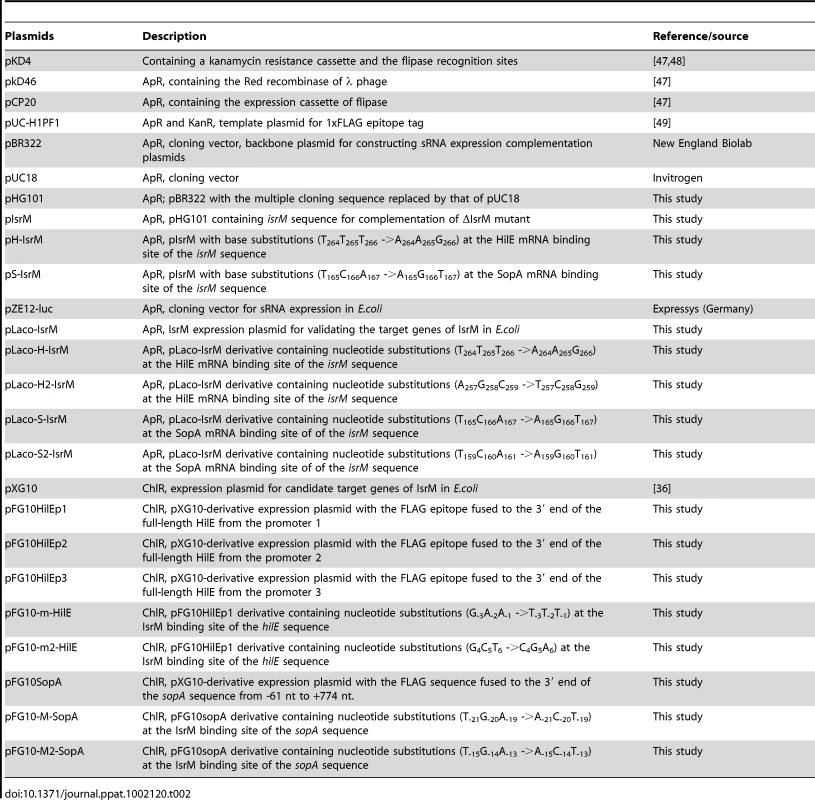
Tab. 3. Primers used in the study. 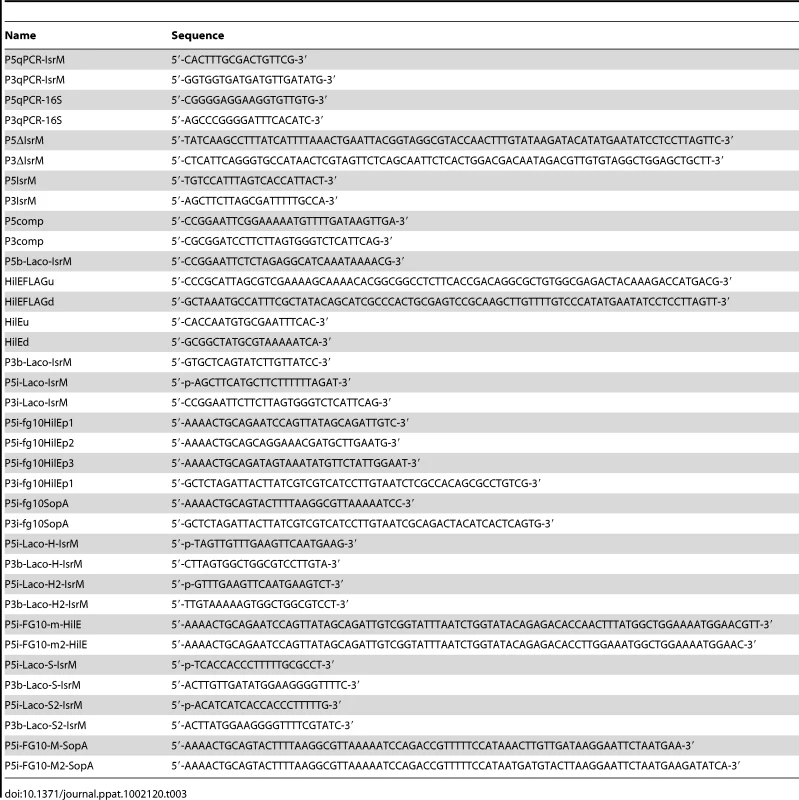
Mutant T-hilE was constructed using the λ Red recombinase method [47], following the procedures as described previously [12], [13], [48]. A pair of primers was designed to amplify the FLAG epitope coding sequence and kanamycin resistance gene using pUC-H1PF1 as the template [49]. The resulting PCR products were electroporated into Salmonella ST14028s strain carrying plasmid pKD46. The non-polar strain T-hilE was constructed using plasmid pCP20 [47], selected for its sensitivity to kanamycin, and further confirmed using PCR and subsequent DNA sequencing. To generate Salmonella strains bearing different tagged SPI-1 proteins and the isrM-deletion mutation, Salmonella SPI-1 protein-tagged non-polar strains T-invJ, T-sipA, T-sipB, T-sipC, T-sipD, T-sopA, T-sopB, T-sopE2, T-spaO, T-sptP, which were previously constructed in our laboratory [12], [13], were transduced with phage P22 carrying the isrM-deletion mutation, following the procedures described previously [12], [13]. Similar procedures were used to generate mutant ΔM-T-hilE. The regions for the tagged ORFs in all the generated tagged strains were sequenced to confirm that no other mutations were present in these regions [12], [13], [48].
To generate complementation plasmids to express the wild type and mutant IsrM RNA under the control of its native promoter, plasmid pBR322 was used as the backbone to generate construct pHG101. Construct pIsrM was generated by inserting to pHG101 with the IsrM sequence generated by PCR with primers P5comp and P3comp (Table 3) that annealed to a 20-nt sequence located at 208 nucleotides upstream from the transcription initiating site and 51 nucleotides downstream from the transcription terminator of IsrM, respectively.
In vitro growth analysis of Salmonella
Growth analysis of bacteria in LB broth was carried out by first inoculating a single colony from a fresh plate in 2 ml LB broth and culturing at 37°C with shaking at 250 RPM overnight for at least 15 hours [12]. Thirty microliters of the overnight culture were then inoculated into 3 ml fresh LB broth and cultured at 37°C and 250 RPM. At time points of 0, 2, 4, 6, 8, 10, 12, 14, 16, and 24 hours after inoculation, 100 microliters of bacterial culture were collected and used for analysis by serial dilution in sterile 96-well plates, and then plated on LB agar plates to determine their CFU/ml. Each sample was analyzed in triplicates and the analysis was repeated at least three times. The average value of CFU/ml was used to generate the growth curve [12].
Expression of IsrM sRNA in vitro
To study the effect of different culture conditions on the IsrM expression in vitro, 20 µl of overnight bacterial cultures were inoculated into 1 ml of LB broth and shaken at 250 RPM and 37°C for 4 hours. The bacterial cultures were centrifuged at 5,000xg for 5 minutes. To study the effect of pH, the pelleted bacteria were re-suspended in 1 ml fresh LB broth (control, pH7.0) or 1 ml LB broth at pH 3.0, 4.4, 5.4, 6.4, 7.2, or 8.4, respectively, shaken at 250 RPM and 37°C for an additional 3 hours, and then collected. To study the effect of osmolarity, the pelleted bacteria were re-suspended in 1 ml NaCl-free LB broth supplemented with 0, 42.5, 85, 160, 340, and 680 mM sodium chloride, respectively, shaken at 250 RPM and 37°C for an additional 3 hours, and were collected. Regular LB broth, which contained 170 mM NaCl, was used as the control. To study the effect of oxygen limitation, the pelleted bacteria were re-suspended in 1.5 ml fresh LB broth. One group of bacteria was shaken at 250 RPM and 37°C for an additional 3 hours with good aeration (control) while another group of bacteria was transferred into 1.5 ml microcentrifuge tubes with their covers closed tightly and wrapped with parafilm, and incubated at 37°C without shaking for an additional 3 hours. Bacteria were pelleted by centrifugation (2 min, 13,000 rpm, 4°C), and RNA samples were isolated with the Trizol method (Invitrogen, Carlsbad, CA), treated with DNase I (TURBO DNA-free, Ambion, Austin, TX), and used for quantitative RT-PCR analysis, following the procedures described previously [13], [48], [50]. The analyses were repeated three times and the average of three experiments are presented. For Northern blot analysis, the Salmonella RNA samples were separated in 2% agarose gels that contained formaldehyde, transferred to nitrocellulose membranes, hybridized with the [32P]-radiolabeled DNA probes that contained the DNA sequence coding for Salmonella IsrM and 5S RNA, and analyzed with a STORM840 Phosphorimager [49].
Validation of IsrM targets in E.coli
The TargetRNA and RNAhybrid programs were used to predict the target mRNAs of IsrM against the entire genome of Salmonella typhimurium ST14028s [33], [34], and the RNAup software (http://rna.tbi.univie.ac.at/cgi-bin/RNAup.cgi) was used to analyze the potential interaction of IsrM to the hilE and sopA mRNAs. Constructs to validate the targets of IsrM in E.coli were generated following a modified procedure described previously [36]. To generate construct pLaco-IsrM expressing IsrM, a 2.2 kb DNA fragment, which contained a PLlacO promoter (from the position -1), an ampicillin resistance cassette, a ColE1 replicon, and a strong rrnB terminator, was amplified from the high-copy plasmid, pZE12luc (EXPRESSYS, Ruelzheim, Germany), with primers P5b-Laco-IsrM and P3b-Laco-IsrM. This DNA fragment was used as the backbone to ligate with the DNA fragment (generated in PCR with primers P5i-Laco-IsrM and P3i-Laco-IsrM) containing the entire IsrM coding sequence and its transcription terminator sequence, generating construct pLaco-IsrM.
To generate constructs expressing HilE and SopA proteins in E.coli, the low-copy plasmid, pXG10 (a gift from Dr. J. Vogel, Germany) [36], was used as the backbone to insert the hilE and sopA sequence for expression of 1×FLAG fusion proteins. The entire coding sequence of hilE was included in the FLAG-fusion protein. As hilE has three initiating sites of transcription, three plasmid constructs (pFG10HilEp1, pFG10HilEp2, and pFG10HilEp3) were generated to express the HilE-FLAG fusion protein. Since the transcription initiation site of the sopA mRNA has not definitively been determined, construct pFG10SopA was generated to contain the sequence between the position 61 nt upstream the translation initiation codon and the position coding for the 258th amino acid residue of SopA, a protein of 782 amino acids [51]. Constructs expressing mutated isrM and its target genes carrying different mutations (e.g. pLaco-H-IsrM, pFG10-m-HilE, pLaco-S-IsrM, and pFG10-M-SopA) were generated by a PCR-based mutagenesis approach, using primers listed in Table 3.
Expression of tagged proteins in bacteria
To study the expression of tagged proteins in E.coli, the plasmids for expressing HilE - and SopA-FLAG proteins (e.g. pFG10HilEp1, pFG10HilEp2, pFG10HilEp3, and pFG10SopA) were co-transformed into E coli Top10 with pLaco-IsrM and its derivatives, which contained the wild type and mutated isrM sequence, respectively. The transformants expressed HilE - and SopA-FLAG proteins in the presence of 20 µg/ml tetracycline and expressed isrM and its related RNA in the presence of 1 mM IPTG. Plasmid pZE12lu (Table 2), which served as the backbone of RNA expression plasmids for target validation, was used as the control plasmid.
To determine the effect of the wild type and mutated IsrM RNA on the expression of HilE and SopA, 60 µl of overnight culture of E.coli Top10 that contained both an expression plasmid for IsrM and an expression plasmid for SopA - or HilE-FLAG protein was inoculated in 3 ml of LB broth containing ampicillin (100 µg/ml) and chloramphenicol (20 µg/ml), and shaken at 37°C, 250 RPM for 4 hours. Tetracycline and IPTG were added into bacterial cultures to a final concentration of 20 µg/ml and 1 mM, respectively, and shaken at 37°C, 250 RPM for an additional six hours. Bacterial pellets were collected and protein samples were isolated. Western blot analysis was performed to determine the levels of the tagged proteins with the expression of GroEL as the internal control [13], [48], [50].
To study protein expression in Salmonella, the SPI-1 protein-tagged strains carrying the deletion mutation of isrM were inoculated in 2 ml of LB broth and grew at 37°C and 250 RPM overnight. Sixty microliters of overnight bacterial culture was inoculated in 3 ml of LB broth and grew at 37°C and 250 RPM for 6 hours. Protein samples were prepared and Western blot analysis was performed using the expression of DnaK as the internal control, following the procedure described previously [13], [48], [50]. The analyses were repeated three times.
Western blot analyses
Polypeptides from bacterial lysates were separated on SDS-containing 10-12% polyacrylamide gels, transferred to nitrocellulose membranes, and reacted with anti-mouse IgG conjugated with alkaline phosphatase in addition to the antibodies against DnaK protein (StressGen, Victoria, BC, Canada), GroEL protein (Sigma, St Louis, MO), or the FLAG sequence (Sigma, St Louis, MO), following the procedure described previously [13]. The membranes were subsequently stained with a chemiluminescent substrate with the aid of a Western chemiluminescent substrate kit (GE Healthcare, Waukesha, WI) and quantified with a STORM840 phosphorimager. Normalization of samples was also carried out by loading total proteins extracted from the same CFU (e.g. 5×107 CFU) of bacteria in each lane. Quantitation was performed in the linear range of protein detection.
Invasion and intracellular replication assays
Assays for bacterial invasion in HeLa cells and intracellular survival/replication in J774 macrophages were performed as described previously [37], [49], [50]. Briefly, Salmonella was added to HeLa cells at multiplicities of infection (MOI) of 5 : 1 to 10 : 1, and intracellular bacteria were quantified after 1 h of incubation for invasion, which was followed by incubation in the presence of 50 µg/ml of gentamicin to kill extracellular bacteria. The invasiveness of Salmonella was measured by determining the ratio of intracellular bacteria, which was calculated as follows: (number of intracellular bacteria/number of input bacteria) ×100 [49], [50].
To study intracellular growth of Salmonella in macrophages, mouse J774 macrophages were infected with Salmonella for 1 h at an MOI of 10, washed with phosphate-buffered saline (PBS), and then incubated in fresh medium containing 50 µg/ml of gentamicin for 1 hour. A set of cells was washed and lysed to quantify the intracellular bacteria and used as the zero-time (0-h) sample. Additional sets of cells were harvested after an additional 8 h of incubation to quantify intracellular Salmonella (8-h samples). The growth of Salmonella inside J774 cells was measured by determining the increase in the number of intracellular Salmonella, which was calculated by dividing the number of intracellular bacteria in the 8-h sample by that in the zero-time sample [49], [50]. All analyses were repeated three times.
Animal studies
Female BALB/c and SCID mice (6–8 weeks old) were obtained from Jackson Laboratory (Bar Harbor, ME). Overnight bacterial cultures were serially diluted to suitable colony forming unit (CFU)/ml in phosphate-buffered saline (PBS) for infection. To assess the virulence of the Salmonella strains, mice (5 animals per group) were either inoculated intragastrically with 5×106 CFU per BALB/c mouse and 1×103 CFU per SCID mouse or intraperitoneally with 1×102 CFU per BALB/c mouse and 1×101 CFU per SCID mouse. The survival of the infected animals was monitored and the survival rate was recorded.
For IsrM sRNA expression and organ colonization experiments, mice (5 animals per group) were inoculated intraperitoneally with 1×105 or 1×107 CFU per BALB/c mouse or 1×102 or 1×104 CFU per SCID mouse of the bacterial strains, and were euthanized at 7 days or 18 hours after inoculation, respectively. Mice (5 animals per group) were also inoculated intragastrically with 1×105 or 1×108 CFU per BALB/c mouse or 1×102 or 1×104 CFU per SCID mouse of the bacterial strains and were euthanized at 7 days or 18 hours after inoculation, respectively. Spleen was collected from infected mice, and ileum was dissected from the small intestine. All organs were homogenized in cold PBS, and bacterial CFU was determined by plating of serially diluted organ homogenates [13], [48], [50]. The analyses were repeated three times.
To determine the level of IsrM RNA, the homogenates of the spleen and ileum samples were centrifuged at 9,000xg at 4°C for 10 minutes [13], [48], [50]. Pellets were incubated in lysis buffer (120 mM NaCl, 4 mM MgCl2, 20 mM Tris/HCl, pH 7.5, 1% Triton-X100) at 4°C for 1 hour, and released bacteria were collected by centrifugation at 18,000xg for 10 minutes. Harvested bacteria were then resuspended in PBS, and adjusted for the bacterial CFU. RNA samples were prepared from isolated Salmonella and analyzed by qRT-PCR to determine the expression of IsrM RNA [13], [48], [50]. The analyses were repeated three times.
Supporting Information
Zdroje
1. OlsenSJMacKinnonLCGouldingJSBeanNHSlutskerL 2000 Surveillance for foodborne-disease outbreaks–United States, 1993–1997. MMWR CDC Surveill Summ 49 1 62
2. SchmidtHHenselM 2004 Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 17 14 56
3. GalanJEWolf-WatzH 2006 Protein delivery into eukaryotic cells by type III secretion machines. Nature 444 567 573
4. AbrahamsGLHenselM 2006 Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol 8 728 737
5. GalanJE 2001 Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol 17 53 86
6. WatermanSRHoldenDW 2003 Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol 5 501 511
7. HueckCJ 1998 Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62 379 433
8. EllermeierJRSlauchJM 2007 Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol 10 24 29
9. GiacomodonatoMNUzzauSBacciuDCaccuriRSarnackiSH 2007 SipA, SopA, SopB, SopD and SopE2 effector proteins of Salmonella enterica serovar Typhimurium are synthesized at late stages of infection in mice. Microbiology 153 1221 1228
10. PfeiferCGMarcusSLSteele-MortimerOKnodlerLAFinlayBB 1999 Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun 67 5690 5698
11. Steele-MortimerOBrumellJHKnodlerLAMeresseSLopezA 2002 The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol 4 43 54
12. GongHSuJBaiYMiaoLKimK 2009 Characterization of the expression of Salmonella Type III secretion system factor PrgI, SipA, SipB, SopE2, SpaO, and SptP in cultures and in mice. BMC Microbiol 9 73
13. GongHVuGPBaiYYangELiuF 2010 Differential expression of Salmonella type III secretion system factors InvJ, PrgJ, SipC, SipD, SopA and SopB in cultures and in mice. Microbiology 156 116 127
14. HannonGJRivasFVMurchisonEPSteitzJA 2006 The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol 71 551 564
15. StorzG 2002 An expanding universe of noncoding RNAs. Science 296 1260 1263
16. Toledo-AranaARepoilaFCossartP 2007 Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol 10 182 188
17. PapenfortKVogelJ 2010 Regulatory RNA in bacterial pathogens. Cell Host Microbe 8 116 127
18. StorzGOpdykeJAWassarmanKM 2006 Regulating bacterial transcription with small RNAs. Cold Spring Harb Symp Quant Biol 71 269 273
19. VogelJ 2009 A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol 71 1 11
20. StorzGOpdykeJAZhangA 2004 Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol 7 140 144
21. BabitzkePRomeoT 2007 CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10 156 163
22. WassarmanKM 2007 6S RNA: a small RNA regulator of transcription. Curr Opin Microbiol 10 164 168
23. Padalon-BrauchGHershbergRElgrably-WeissMBaruchKRosenshineI 2008 Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res 36 1913 1927
24. PfeifferVSittkaATomerRTedinKBrinkmannV 2007 A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 66 1174 1191
25. SittkaALucchiniSPapenfortKSharmaCMRolleK 2008 Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4 e1000163
26. ArricauNHermantDWaxinHEcobichonCDuffeyPS 1998 The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol Microbiol 29 835 850
27. GalanJECurtissR3rd 1990 Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun 58 1879 1885
28. TarteraCMetcalfES 1993 Osmolarity and growth phase overlap in regulation of Salmonella typhi adherence to and invasion of human intestinal cells. Infect Immun 61 3084 3089
29. JonesBDFalkowS 1994 Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect Immun 62 3745 3752
30. LeeCAFalkowS 1990 The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci U S A 87 4304 4308
31. CummingsJHPomareEWBranchWJNaylorCPMacfarlaneGT 1987 Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28 1221 1227
32. Van ImmerseelFDe BuckJDe SmetIPasmansFHaesebrouckF 2004 Interactions of butyric acid - and acetic acid-treated Salmonella with chicken primary cecal epithelial cells in vitro. Avian Dis 48 384 391
33. KrugerJRehmsmeierM 2006 RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34 W451 454
34. TjadenBGoodwinSSOpdykeJAGuillierMFuDX 2006 Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res 34 2791 2802
35. BaxterMAFahlenTFWilsonRLJonesBD 2003 HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect Immun 71 1295 1305
36. UrbanJHVogelJ 2007 Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35 1018 1037
37. BuchmeierNAHeffronF 1989 Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun 57 1 7
38. EllermeierCDEllermeierJRSlauchJM 2005 HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57 691 705
39. ZhangYHigashideWMMcCormickBAChenJZhouD 2006 The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol 62 786 793
40. DiaoJZhangYHuibregtseJMZhouDChenJ 2008 Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol 15 65 70
41. HicksSWGalánJE 2010 Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol 13 41 46
42. RaffatelluMWilsonRPChessaDAndrews-PolymenisHTranQT 2005 SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect Immun 73 146 154
43. BrennanRGLinkTM 2007 Hfq structure, function and ligand binding. Curr Opin Microbiol 10 125 133
44. Valentin-HansenPEriksenMUdesenC 2004 The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol 51 1525 1533
45. CainRJHaywardRDKoronakisV 2008 Deciphering interplay between Salmonella invasion effectors. PLoS Pathog 4 e1000037
46. WinnenBSchlumbergerMCSturmASchupbachKSiebenmannS 2008 Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS ONE 3 e2178
47. DatsenkoKAWannerBL 2000 One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 6640 6645
48. LuSKilloranPBFangFCRileyLW 2002 The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun 70 451 461
49. SuJGongHLaiJMainALuS 2009 The potassium transporter Trk and external potassium modulate Salmonella enterica protein secretion and virulence. Infect Immun 77 667 675
50. LuSMangesARXuYFangFCRileyLW 1999 Analysis of virulence of clinical isolates of Salmonella enteritidis in vivo and in vitro. Infect Immun 67 5651 5657
51. McClellandMSandersonKESpiethJCliftonSWLatreilleP 2001 Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413 852 856
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage EgressČlánek A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene ExpressionČlánek An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes byČlánek Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine ScrapieČlánek Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 9- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity
- Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
- Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development
- Nrf2, a PPARγ Alternative Pathway to Promote CD36 Expression on Inflammatory Macrophages: Implication for Malaria
- Robust Antigen Specific Th17 T Cell Response to Group A Streptococcus Is Dependent on IL-6 and Intranasal Route of Infection
- Targeting of a Chlamydial Protease Impedes Intracellular Bacterial Growth
- The Protease Cruzain Mediates Immune Evasion
- High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism
- Plague and Climate: Scales Matter
- Exhausted CD8 T Cells Downregulate the IL-18 Receptor and Become Unresponsive to Inflammatory Cytokines and Bacterial Co-infections
- Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles
- Murine Gamma-herpesvirus Immortalization of Fetal Liver-Derived B Cells Requires both the Viral Cyclin D Homolog and Latency-Associated Nuclear Antigen
- Rapid and Efficient Clearance of Blood-borne Virus by Liver Sinusoidal Endothelium
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene Expression
- Strain Specific Resistance to Murine Scrapie Associated with a Naturally Occurring Human Prion Protein Polymorphism at Residue 171
- Development of a Transformation System for : Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector
- Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
- Host Phylogeny Determines Viral Persistence and Replication in Novel Hosts
- BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity
- Expression of the RAE-1 Family of Stimulatory NK-Cell Ligands Requires Activation of the PI3K Pathway during Viral Infection and Transformation
- Structure of the Vesicular Stomatitis Virus N-P Complex
- HSV Infection Induces Production of ROS, which Potentiate Signaling from Pattern Recognition Receptors: Role for S-glutathionylation of TRAF3 and 6
- The Human Papillomavirus E6 Oncogene Represses a Cell Adhesion Pathway and Disrupts Focal Adhesion through Degradation of TAp63β upon Transformation
- Analysis of Behavior and Trafficking of Dendritic Cells within the Brain during Toxoplasmic Encephalitis
- Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance
- Multidrug Resistant 2009 A/H1N1 Influenza Clinical Isolate with a Neuraminidase I223R Mutation Retains Its Virulence and Transmissibility in Ferrets
- Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1
- Step-Wise Loss of Bacterial Flagellar Torsion Confers Progressive Phagocytic Evasion
- Complex Recombination Patterns Arising during Geminivirus Coinfections Preserve and Demarcate Biologically Important Intra-Genome Interaction Networks
- An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by
- Non-Lytic, Actin-Based Exit of Intracellular Parasites from Intestinal Cells
- The Fecal Viral Flora of Wild Rodents
- The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
- Interferon Regulatory Factor-1 (IRF-1) Shapes Both Innate and CD8 T Cell Immune Responses against West Nile Virus Infection
- A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
- Evaluating the Sensitivity of to Biotin Deprivation Using Regulated Gene Expression
- The Motility of a Human Parasite, , Is Regulated by a Novel Lysine Methyltransferase
- Phosphodiesterase-4 Inhibition Alters Gene Expression and Improves Isoniazid – Mediated Clearance of in Rabbit Lungs
- Restoration of IFNγR Subunit Assembly, IFNγ Signaling and Parasite Clearance in Infected Macrophages: Role of Membrane Cholesterol
- Protease ROM1 Is Important for Proper Formation of the Parasitophorous Vacuole
- The Regulated Secretory Pathway in CD4 T cells Contributes to Human Immunodeficiency Virus Type-1 Cell-to-Cell Spread at the Virological Synapse
- Rerouting of Host Lipids by Bacteria: Are You CERTain You Need a Vesicle?
- Transmission Characteristics of the 2009 H1N1 Influenza Pandemic: Comparison of 8 Southern Hemisphere Countries
- Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine Scrapie
- Sequential Bottlenecks Drive Viral Evolution in Early Acute Hepatitis C Virus Infection
- Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen
- Genomic and Proteomic Analyses of the Fungus Provide Insights into Nematode-Trap Formation
- Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
- Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
- Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate
- Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
- c-di-AMP Is a New Second Messenger in with a Role in Controlling Cell Size and Envelope Stress
- Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System
- APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells
- Impairment of Immunoproteasome Function by β5i/LMP7 Subunit Deficiency Results in Severe Enterovirus Myocarditis
- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Tri6 Is a Global Transcription Regulator in the Phytopathogen
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- The Next Opportunity in Anti-Malaria Drug Discovery: The Liver Stage
- Significant Effects of Antiretroviral Therapy on Global Gene Expression in Brain Tissues of Patients with HIV-1-Associated Neurocognitive Disorders
- Inhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
- A Novel Metal Transporter Mediating Manganese Export (MntX) Regulates the Mn to Fe Intracellular Ratio and Virulence
- Rhoptry Kinase ROP16 Activates STAT3 and STAT6 Resulting in Cytokine Inhibition and Arginase-1-Dependent Growth Control
- Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms
- Secretion of Genome-Free Hepatitis B Virus – Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
- Membrane Remodeling by the Double-Barrel Scaffolding Protein of Poxvirus
- A Diverse Population of Molecular Type VGIII in Southern Californian HIV/AIDS Patients
- Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A Virus Protease-Polymerase Processing Intermediate, 3CD
- Quantitative Analyses Reveal Calcium-dependent Phosphorylation Sites and Identifies a Novel Component of the Invasion Motor Complex
- Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes
- Old World Arenaviruses Enter the Host Cell via the Multivesicular Body and Depend on the Endosomal Sorting Complex Required for Transport
- Exploits a Unique Repertoire of Type IV Secretion System Components for Pilus Assembly at the Bacteria-Host Cell Interface
- Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



