-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
In contrast to most RNA viruses, influenza viruses replicate their genome in the nucleus of infected cells. As a result, newly-synthesized vRNA genomes, in the form of viral ribonucleoprotein complexes (vRNPs), must be exported to the cytoplasm for productive infection. To characterize the composition of vRNP export complexes and their interplay with the nucleus of infected cells, we affinity-purified tagged vRNPs from biochemically fractionated infected nuclei. After treatment of infected cells with leptomycin B, a potent inhibitor of Crm1-mediated export, we isolated vRNP export complexes which, unexpectedly, were tethered to the host-cell chromatin with very high affinity. At late time points of infection, the cellular export receptor Crm1 also accumulated at the same regions of the chromatin as vRNPs, which led to a decrease in the export of other nuclear Crm1 substrates from the nucleus. Interestingly, chromatin targeting of vRNP export complexes brought them into association with Rcc1, the Ran guanine exchange factor responsible for generating RanGTP and driving Crm1-dependent nuclear export. Thus, influenza viruses gain preferential access to newly-generated host cell export machinery by targeting vRNP export complexes at the sites of Ran regeneration.
Published in the journal: Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting. PLoS Pathog 7(9): e32767. doi:10.1371/journal.ppat.1002187
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002187Summary
In contrast to most RNA viruses, influenza viruses replicate their genome in the nucleus of infected cells. As a result, newly-synthesized vRNA genomes, in the form of viral ribonucleoprotein complexes (vRNPs), must be exported to the cytoplasm for productive infection. To characterize the composition of vRNP export complexes and their interplay with the nucleus of infected cells, we affinity-purified tagged vRNPs from biochemically fractionated infected nuclei. After treatment of infected cells with leptomycin B, a potent inhibitor of Crm1-mediated export, we isolated vRNP export complexes which, unexpectedly, were tethered to the host-cell chromatin with very high affinity. At late time points of infection, the cellular export receptor Crm1 also accumulated at the same regions of the chromatin as vRNPs, which led to a decrease in the export of other nuclear Crm1 substrates from the nucleus. Interestingly, chromatin targeting of vRNP export complexes brought them into association with Rcc1, the Ran guanine exchange factor responsible for generating RanGTP and driving Crm1-dependent nuclear export. Thus, influenza viruses gain preferential access to newly-generated host cell export machinery by targeting vRNP export complexes at the sites of Ran regeneration.
Introduction
Influenza viruses are nearly unique among RNA viruses, in that they perform all of their viral RNA synthesis in the nucleus of infected cells. While this exceptional attribute provides some advantages to the virus, such as access to capped cellular pre-mRNAs and the host splicing machinery, it also presents the challenge of importing and exporting the viral genome during early and late infection, respectively. Like other negative-strand RNA viruses, the influenza virus genome is encapsidated by the nucleoprotein NP, and is associated with the trimeric viral polymerase complex consisting of the PA, PB1, and PB2 proteins. This complex, known as the viral ribonucleoprotein complex (vRNP), is the minimal infectious unit that is exported from the nucleus at late time points of infection.
The nuclear export of influenza A vRNPs has been well-studied, yet many details remain unclear. First reports implicated both the viral matrix protein M1 as well as the viral nuclear export protein NEP as crucial co-factors [1]–[3]; however, the requirement for each of these proteins has subsequently been questioned [4], [5]. vRNP export was shown to be dependent on the cellular export receptor Crm1, and accordingly cytoplasmic accumulation of vRNPs can be blocked by leptomycin B [5], [6], a potent inhibitor of Crm1 [7]. However, both NP and NEP bind Crm1 and can be exported [2], [5], [8], and thus it is unclear which protein actually drives vRNP export. The current “daisy-chain” model of vRNP nuclear export postulates that M1 binds directly to vRNPs, while NEP acts as a bridge between M1 and Crm1 to facilitate translocation [9]. Despite evidence of binary interactions between each of these components, a fully-formed vRNP export complex has not been isolated from infected cells.
Crm1-dependent nuclear export is driven by a gradient of RanGTP:RanGDP between the nucleus and the cytoplasm. Crm1 association with its export cargo occurs cooperatively in a multi-protein complex containing RanGTP and other factors [10], [11], [12]. This Crm1-RanGTP-cargo complex is escorted through the nuclear pore complex to the cytoplasm [13], where RanGTP is hydrolyzed and the cargo complex dissociates [14]. After nuclear re-import of RanGDP, further export cycles require the regeneration of RanGTP, which is facilitated by the Ran guanine exchange factor Rcc1, a process chromatin-bound during the exchange process [11]. Complex formation of Ran, Crm1 and Rcc1 was shown in biochemical experiments to be facilitated by the chaperone protein RanBP3; however, this has not been confirmed in living cells [12].
Our goal in this work was to investigate both the composition of influenza vRNP export complexes and their interplay with the host cell nucleus by taking advantage of our previously-established affinity purification of vRNPs from infected cells [15]. Using a subnuclear fractionation, we identified putative vRNP export complexes which, surprisingly, were tightly bound to the nuclear substructure. Interestingly, targeting of vRNPs to these chromatin-bound nuclear domains retained Crm1 at these same domains, and thus allowed the virus to usurp the cellular export machinery. The mechanism of this usurpation appears to occur via “snatching” of recycled Crm1-RanGTP complexes after their nucleotide exchange by Rcc1 on chromatin. Thus, influenza virus gains preferential access to host cell export machinery by locating vRNPs proximal to the sites of RanGTP regeneration.
Results
vRNPs accumulate on chromatin upon nuclear export inhibition
To identify the components of a vRNP export complex, we performed vRNP purifications from infected cells at a late time point after infection. Based on previous reports that vRNPs within the nucleus can be either soluble or associated with chromatin [16]–[19], we postulated that subnuclear fractionation before purification might segregate vRNPs by their respective functions and thus better facilitate purification of homogenous vRNP export complexes. We therefore separated cells into cytoplasmic (“cyt”), soluble nucleoplasmic (“nuc”), 150 mM NaCl-extractable chromatin (“ch150”), and 500 mM NaCl-extractable chromatin (“ch500”) fractions (Fig. 1A). Prior to salt extraction, chromatin was digested with a nuclease. Successful fractionation was confirmed by Western blot detection of subcellular marker proteins such as tubulin (cytoplasm), HMGB1 (cytoplasm and nucleoplasm), Pol II or histone H3 (chromatin) (Fig. 1B). In agreement with previous reports [20], [21], differential salt extraction separated chromatin into fractions containing higher or lower levels of markers of active cellular transcription (Pol II, TFIIP, Fig. 1B lanes 5–8), indicative of an approximate functional distinction between the ch150 and ch500 fractions. Interestingly, we repeatedly observed a depletion of Pol II in the ch150 fraction of infected cells, which likely represents the well-described degradation of Pol II during infection. [22], [23]
Fig. 1. Purification of vRNPs after subcellular fractionation. 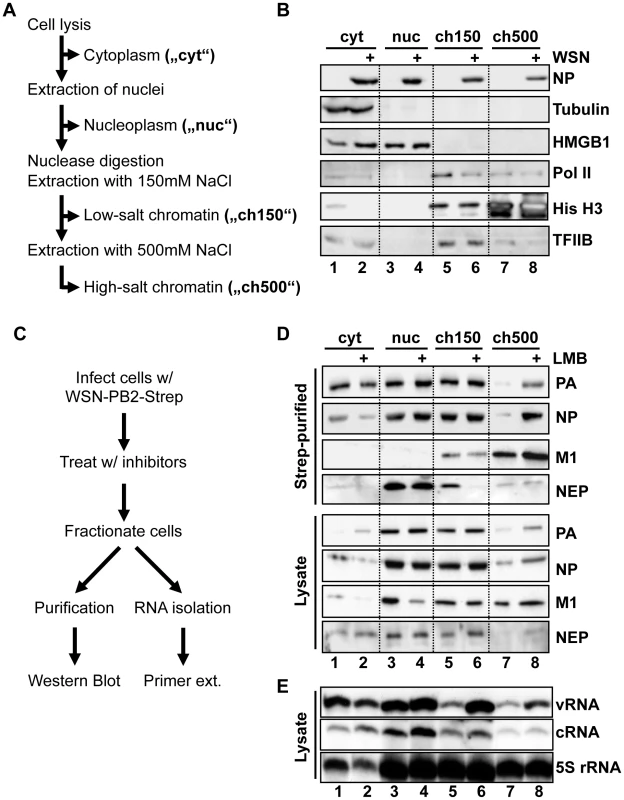
(A) Schematic diagram of subcellular fractionation as described in Materials and Methods. (B) 1×109 HeLa cells were infected with WSN at an MOI of 3 for 9 h or mock-infected. Cells were fractionated as described in (A), and equal amounts of protein from each fraction were analyzed by Western blot analysis for NP or cellular marker proteins. (C) Schematic workflow of Strep purification after LMB treatment. (D) 1×109 HeLa cells were infected with rWSN-Strep at an MOI of 3 for 9 h. At 3hpi, cells were treated with 10 nM LMB or 0.2% EtOH vehicle. Fractionation and Strep-purification were performed as described in (C), and lysates and eluates were analyzed by Western blot for indicated proteins. (E) Cells were infected, treated, and fractionated as in (D), except that 50 U/ml DNase I was used as a nuclease, and total RNA was extracted from each fraction. RNA was analyzed by primer extension using probes specific for segment 6 v/cRNA and 5S rRNA. To obtain a high yield of intact vRNPs from infected cells, we used a recombinant WSN strain encoding a C-terminally Strep-tagged PB2 (rWSN-PB2-Strep). The recombinant virus was generated as previously described [15], [24], and grew to similar titers as an untagged recombinant WSN (rWSN, Fig. S1A). Performing Strep-purification using lysates from rWSN-PB2-Strep-infected cells yields a specific purification of vRNPs with high efficiency (Fig. S1B+C). By infecting cells with rWSN-PB2-Strep followed by fractionation and Strep-purification, we were able to obtain high yields of vRNPs from all 4 fractions assayed (Figs. S1D + E). To differentiate between soluble trimeric polymerase complexes and fully-formed vRNPs, we assayed by Western blot the amounts of co-purified PA, as a marker for the trimeric polymerase complex, as well as co-purified NP as a marker for vRNA/cRNA-containing vRNPs. Silver staining of purified vRNPs from all fractions confirmed that PA and PB2-Strep were always purified in equimolar amounts (data not shown), indicating that PB2 is always integrated into a polymerase complex in our protocol (monomeric, purified PB2-Strep was not detected, data not shown). Despite digestion of DNA and RNA using Benzonase prior to chromatin extraction, the ratio of co-purified NP was not reduced (Fig. S1F), suggesting that the vRNP structure remains intact during this fractionation procedure. Thus, Strep-purification after subnuclear fractionation yielded biochemically equivalent vRNPs from all fractions examined.
In order to enrich vRNP export complexes in the nucleus, we performed the fractionation/purification experiments in HeLa cells with or without the Crm1 inhibitor leptomycin B (LMB), which can potently inhibit the export of influenza vRNPs [7](Fig. 1C). Analysis of vRNP accumulation by Western blot showed an expected decrease of fully-formed vRNPs in the cytoplasm after LMB treatment (Fig. 1D, lanes 1–2, “NP”). Surprisingly, a corresponding increase in nuclear vRNPs after LMB treatment was only found in the chromatin fraction extracted with 500 mM NaCl (Fig. 1D lanes 7–8 “ch500”). The vRNPs derived from this fraction also contained M1, a necessary vRNP export co-factor. Small amounts of NEP were also associated with these complexes, which were enhanced after LMB treatment. Interestingly, NEP also associated with nucleoplasm-derived vRNPs; however, these vRNPs contained no detectable M1 and did not accumulate in the nucleus after LMB treatment, suggesting a different role of NEP in these complexes (see Discussion). These experiments were also performed in A549 cells, yielding similar results (data not shown).
To verify that the complexes enriched in the ch500 fraction after LMB treatment represent vRNP export complexes and not vRNP replication intermediates, we analyzed the amounts of vRNA and cRNA in each fraction using primer extension analysis. vRNA was slightly depleted in the cytoplasm and enriched in both the ch150 and ch500 fractions after LMB treatment (Fig. 1E), while no significant differences in cRNA accumulation were observed. Thus, vRNA, but not cRNA, accumulates on chromatin after treatment with LMB. We observed discrepancies between PA/NP accumulation in Fig. 1D and vRNA accumulation in Fig. 1E, which is likely due to the need to use DNase I, instead of Benzonase, as the nuclease in the latter experiment, which leads to a altered chromatin extraction profile (data not shown). However, in both conditions vRNPs and their components associated with chromatin when vRNP export was inhibited.
Since nuclear export is presumed to be a very dynamic process [25], we were surprised that vRNP export complexes were tightly associated with host cell chromatin and sought a confirmation of this finding using a separate method. We postulated that, if vRNP export complexes are indeed formed on less-soluble chromatin, the major components of these complexes should colocalize in an immunofluorescence analysis (IFA) in cells from which soluble proteins and salt-extractable chromatin has been removed, and the presence of these proteins should be enriched after LMB treatment. To test this hypothesis, we adapted a previously-described in situ extraction for influenza virus-infected cells on coverslips [17]. In a first step, all soluble proteins from the cytoplasm and nucleus were extracted, leaving chromatin-bound material intact (“Total chromatin”, Fig. 2A). This chromatin was then digested with DNase I, and extracted with 250 mM NaCl to reveal the less-soluble dense chromatin (which we so named due to the observation that histones associated with dense packing are not soluble at this concentration [26]), and its associated proteins (Fig. 2A, “Dense chromatin”). We used this extraction process to analyze the localizations of PA, NP, and NEP by IFA in infected cells with or without LMB treatment. In unfractionated cells, NP was retained in the nucleus upon LMB treatment as expected (Fig. 2B, “Whole cells”), whereas when soluble proteins were removed to reveal all chromatin-bound proteins, we observed prominent NP nuclear rings as previously described (Fig. 2B, “Total chromatin”, [27], [28]). However, we detected no difference in staining intensity between LMB-treated and untreated cells, and surprisingly, little colocalization between NP and PA. In contrast, after extraction to reveal dense chromatin-bound material, a strong colocalization of NP and PA was seen in a reticular pattern (Fig. 2B, “Dense chromatin”), which also overlapped with the localization of histone H2B (Fig. S2). This appearance of antigens previously not detectable was surprising, but is a well-known phenomenon likely caused by the exposure of epitopes previously covered by other proteins [29]. As expected, an increase in staining intensity of both PA and NP was observed after LMB treatment on the dense chromatin, but not on total chromatin or whole cells (Fig. 2B).
Fig. 2. Association of vRNP export complexes with subchromatin structure. 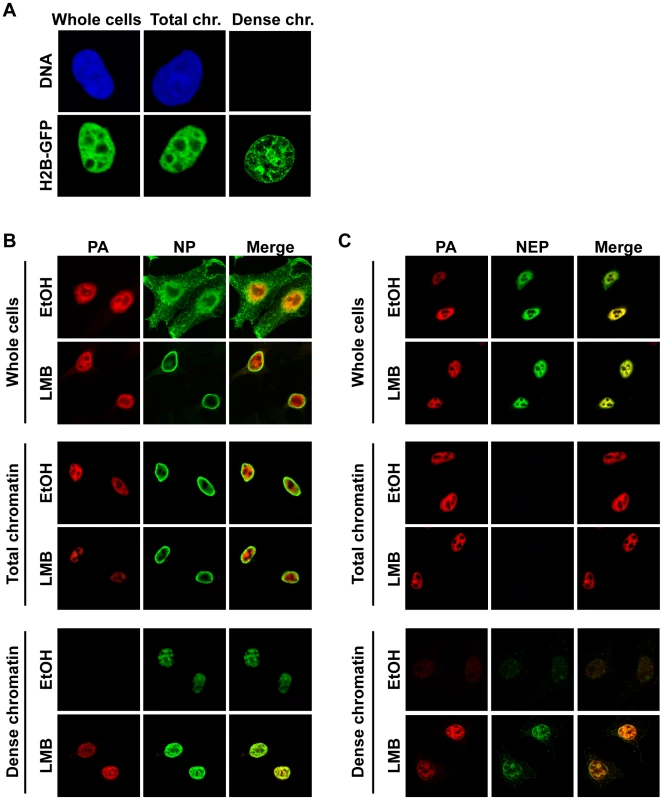
(A) The fractionation protocol was adapted to cells grown on coverslips. Cells were fixed after different fractionation steps to reveal the localization of unextracted proteins. HeLa cells were transfected with histone H2B-GFP or stained with ToPro3 after fixation to demonstrate extraction efficiency. Whole cells = no extraction, Total chr. = detergent extraction (all soluble proteins removed), Dense chr. = low-salt-extractable chromatin removed. (B) HeLa cells were grown on coverslips, infected with WSN at an MOI of 3 for 9 h, and treated with 10 nM LMB or 0.2% EtOH at 3 hpi. Cells were fractionated on the coverslips, fixed after fractionated, and stained for immunofluorescence using antibodies against PA and NP. Optical sections were obtained by confocal microscopy, and detector settings remained constant for each protein between all samples. (C) As in (B), except using antibodies against PA and NEP. Staining of fractionated cells for NEP revealed a similar distribution as PA in unfractionated cells (Fig. 2C, “Whole cells”), but no NEP was detected after soluble proteins were removed (Fig. 2C, “Total chromatin”). When dense chromatin was exposed, no NEP was detectable in control cells, but a faint staining was observed in LMB-treated cells, which colocalized with PA (Fig. 2C, “Dense chromatin”). Similar PA, NP, and NEP distributions after fractionation were observed when MDCK or A549 cells were infected, or when HeLa cells were infected with influenza A strain SC35M (data not shown). Taken together, these results suggest that colocalization of vRNP export complex components can only be visualized when both soluble proteins and soluble chromatin-bound material is removed from the cell. Furthermore, after LMB treatment, vRNP protein components are enriched only on this dense chromatin fraction.
Since LMB targets the general cellular export receptor Crm1, treatment of infected cells may have unspecific effects on vRNP distribution in the nucleus. We therefore generated a recombinant WSN virus (WSN-NEP20/21) containing two previously-described mutations (Q20L, L21C) [30] in the nuclear export sequence (NES) of NEP. This mutant virus can synthesize vRNPs at the same rate as wt (Fig. 3A) but has a delayed vRNP nuclear export [30] (Fig. 3C, left panels). Fractionation of WSN-NEP20/21-infected cells at 10 hpi revealed an accumulation of PA, NP and NEP in the ch500 fraction compared to wt WSN (Fig. 3B). Correspondingly, colocalization and enrichment of PA, NP and NEP on the dense chromatin in WSN-NEP20/21-infected cells were observed by IFA (Fig. 3C and 3D, right panels). Thus, vRNPs accumulate in this fraction during infection when export is impaired through a suboptimal NES.
Fig. 3. Accumulation of vRNPs on chromatin during infection with WSN-NEP20/21. 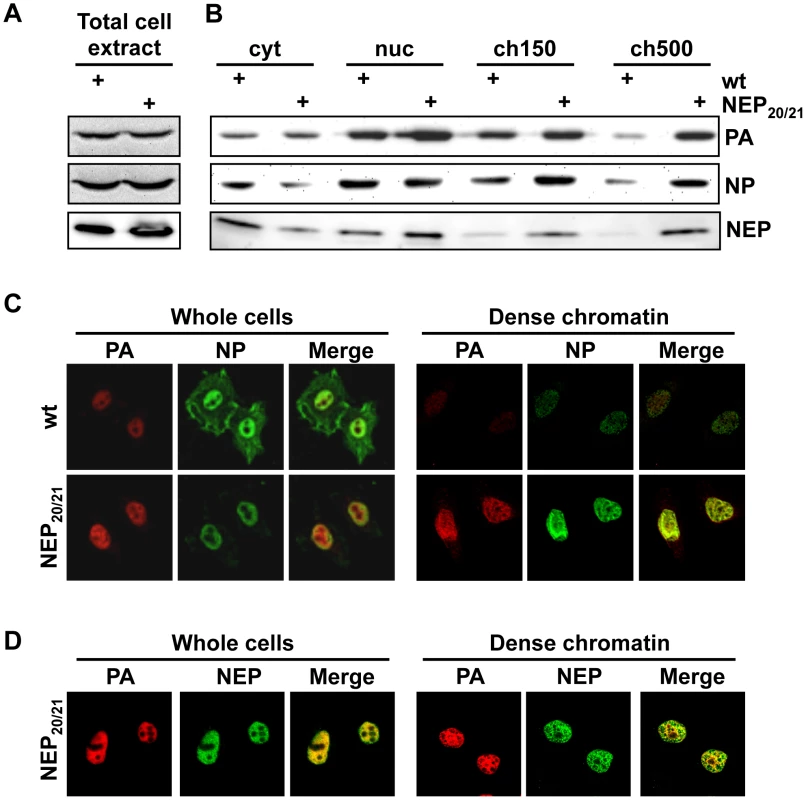
(A) HeLa cells were infected with WSN or WSN-NEP20/21 at an MOI of 3 for 7 h. Whole-cell lysates were analyzed by Western blot for accumulation of PA or NP. (B) 1×109 HeLa cells were infected with WSN or WSN-NEP20/21 at an MOI of 3 for 9 h, then fractionated as in Fig. 1B. Fractionated lysates were analyzed by Western blot for PA, NP or NEP accumulation. (C) HeLa cells on coverslips were infected with WSN or WSN-NEP20/21 at an MOI of 3 for 9 h, then fractionated as described in Fig. 2B and stained for IFA using antibodies against PA and NP. (D) HeLa cells on coverslips were infected with WSN-NEP20/21 at an MOI of 3 for 9 h, then fractionated as described in Fig. 2A and stained for IFA using antibodies against PA and NEP. To confirm the importance of high-affinity chromatin association for vRNP export, we purified vRNPs from fractionated cells after treatment with a second chemical inhibitor with a different mechanism. Among the previously-described inhibitors of influenza vRNP nuclear export, prostaglandin A (PGA) was shown to block export by inducing overexpression of Hsp70, which in turn prevents the association of M1 with vRNPs and therefore the assembly of vRNP export complexes [31]. When we performed fractionation/purification experiments after PGA treatment, we observed a decrease in vRNPs in the ch500 fraction (Figs. 4A and B, lanes 7+8). This decrease corresponded to a lack of detectable NP or PA on the dense chromatin layer by IFA after PGA treatment (Fig. 4C, lower panels and data not shown). Despite the decrease in vRNPs in the ch500 fraction, no decrease in the total amounts of PA or NP was observed in PGA-treated cells (Fig. S3), as previously reported. Concomitantly, a large decrease in the amount of chromatin-bound M1 was detected in PGA-treated cells both by Western blot (Fig. 4B) and IFA (Fig. 4C). These results suggest that Hsp70 overexpression prevents the association of vRNP-M1 complexes with chromatin, thereby leading to a lack of vRNP export.
Fig. 4. Depletion of chromatin-bound vRNPs after PGA treatment. 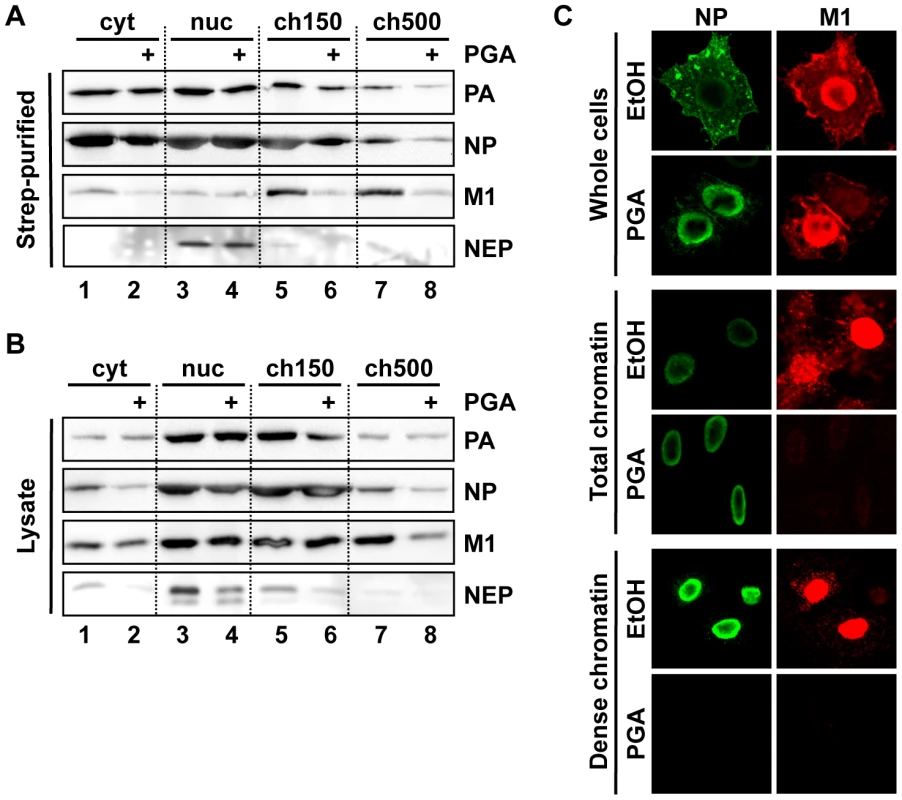
(A) Cells were infected and purified as in Fig. 3A, except that treatment was performed using 20 µg/ml PGA at 3 hpi. Eluted complexes were analyzed by Western blot. (B) Lysates from (A) were analyzed by Western blot. (C) HeLa cells grown on coverslips were infected as in Fig. 4B, treated at 3 hpi with 20 µg/ml PGA, fractionated at 9 hpi as in Fig. 2A, and analyzed by immunofluorescence using antibodies specific for NP and M1. Note that the detector sensitivity was increased for the dense chromatin panels to demonstrate the absence of NP and M1 after PGA treatment. Crm1 is relocalized to chromatin-bound sites of vRNP export complexes
Our data suggested that tight association of influenza vRNPs with chromatin is a prerequisite for their subsequent export, which has not been described for exported cellular proteins. To test whether vRNP chromatin association affects the host cell export machinery, we analyzed the distribution of Crm1 in the nucleus of infected cells. Biochemical fractionation of virus-infected cells at 9 hpi revealed an accumulation of Crm1 in the ch500 fraction, which could be reversed upon LMB treatment (Fig. 5A). Similarly, IFA showed a pronounced relocalization of Crm1 from the nuclear periphery to the interior of the nucleus, which was reversible after LMB or PGA treatment (Fig. 5B). This relocalization was also observed in infected A549 and MDCK cells (data not shown). Although Crm1 co-localized only partially with influenza NP in whole cells (Fig. 5B, middle panels), there was significant co-localization of Crm1-HA with NEP (Fig. 5C). This relocalization of Crm1 upon viral infection correlated with an increase of Crm1 at the sites of NP distribution on the dense chromatin (Fig. 5D), which was also reversed upon LMB or PGA treatment. Interestingly, in WSN-wt-infected cells, redistribution of Crm1 was only observed in cells containing cytoplasmic NP, implying that Crm1 relocalization and active vRNP export are tightly linked (Fig. 5E, upper panels). In contrast, we observed many WSN-NEP20/21-infected cells displaying Crm1 relocalization without cytoplasmic NP staining (Fig. 5E, lower panels), suggesting that Crm1 relocalization is not strictly dependent on robust vRNP export. Thus, Crm1 relocalizes to the sites on dense chromatin where vRNP export complexes also accumulate.
Fig. 5. Relocalization of Crm1 to dense chromatin after influenza virus infection. 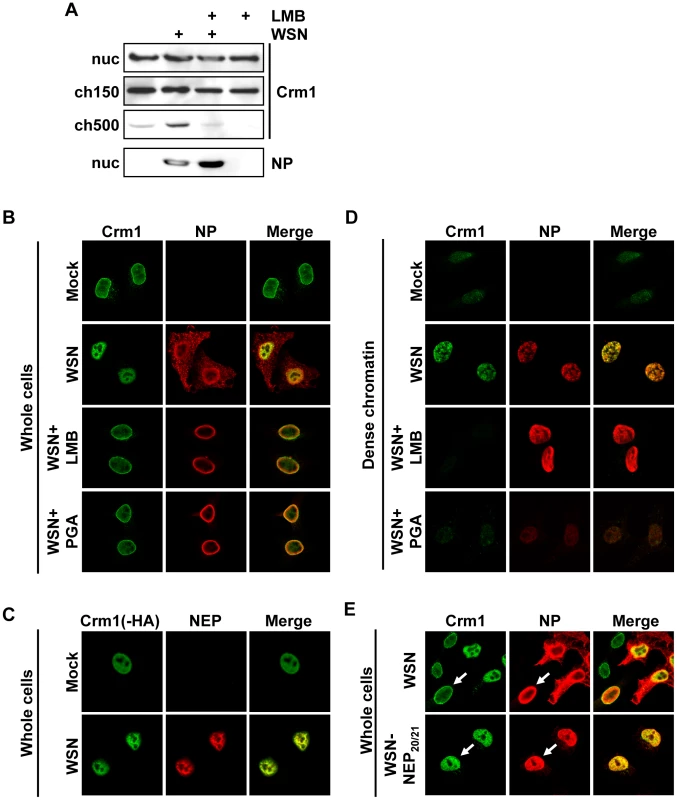
(A) 1×109 HeLa cells were infected with WSN at an MOI of 3 or mock-infected. At 3hpi, cells were treated with LMB or EtOH, and at 9hpi cells were fractionated. Lysates from the ch500 fraction were analyzed by Western blot for Crm1 or NP. (B) HeLa cells grown on coverslips were infected with WSN at an MOI of 3 or mock-infected. Cells were treated with EtOH, LMB or PGA at 3hpi, fixed at 9hpi, and analyzed by immunofluorescence using antibodies against NP and Crm1. (C) HeLa cells were transfected with Crm1-HA, infected with WSN at an MOI of 3 or mock-infected for 9 h, and analyzed by IFA using antibodies against NEP or HA. (D) Cells were infected and treated as in (B), then fractionated to reveal dense chromatin before fixation and immunofluorescence. (E) HeLa cells were infected with WSN or WSN-NEP20/21 for 9 h, and analyzed by IFA using antibodies against NP or Crm1. Export of a non-viral Crm1 substrate is impaired in influenza virus-infected cells
Based on the ability of influenza vRNPs to sequester at least a portion of cellular Crm1 on the chromatin, we postulated that the resulting depletion of free Crm1 might impair the export of cellular Crm1 substrates. To test this hypothesis, we measured the export efficiency of a non-viral Crm1 substrate in influenza virus-infected cells compared to uninfected cells. Due to the targeting of many cellular Crm1 substrates to the nucleus during influenza virus infection, we used GFP containing an HIV Rev-derived NES (GFP-NES) as a marker for Crm1-mediated export. Low-level GFP expression in HeLa cells led to a predominantly nuclear localization, similar to previous findings [32] (Fig. 6A). In contrast, GFP-NES localized almost exclusively in the cytoplasm (Fig. 6B, left panel), and thus provided a suitable measure of an actively-exported Crm1 substrate, as has been previously reported [33]. Upon influenza virus infection, retention of GFP-NES in the nucleus was observed after 10 h of infection (during vRNP export) but not 4 h (Fig. 6B + C), which was also observed in A549 cells (data not shown). Interestingly, while the majority of virus-infected cells contained GFP-NES both in the cytoplasm and the nucleus, treatment of GFP-NES-transfected cells with LMB led to a predominantly nuclear accumulation (Fig. 6B+C). These results suggest that influenza virus infection can impair, but not shut off, the export of non-viral Crm1 substrates. To test whether this impairment of cellular export was dependent on chromatin accumulation of influenza vRNPs, we treated infected cells with PGA, which blocks chromatin association but not total nuclear accumulation of vRNPs (Fig. 4A). Although some GFP-NES was retained in mock-infected cells upon PGA treatment (Fig. 6D), no difference in nuclear retention was observed between mock-infected and influenza virus-infected cells. Thus, impairment of cellular export by influenza virus is not only dependent on accumulation of nuclear vRNPs, but also on their association with the ch500 fraction.
Fig. 6. Influenza infection impairs nuclear export of a Crm1-dependent protein. 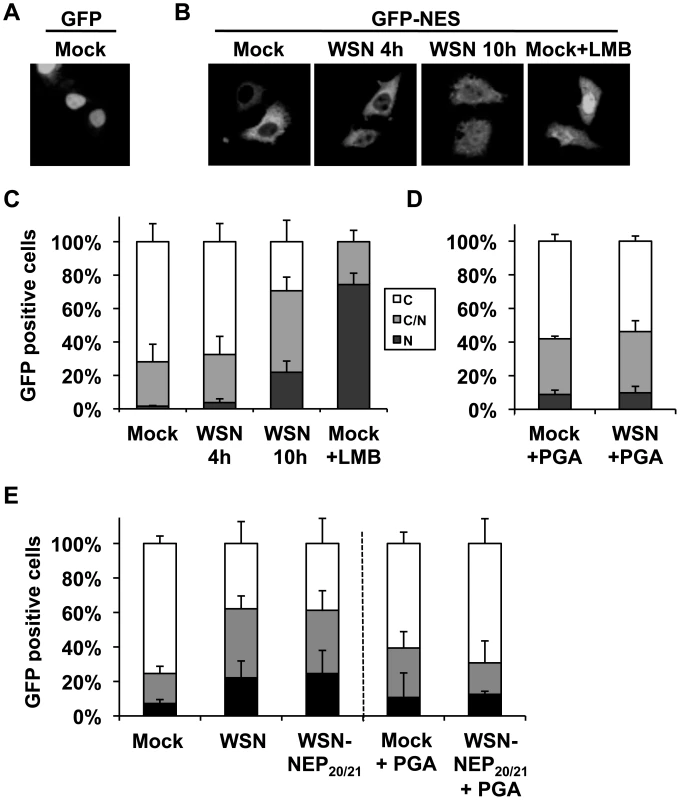
(A) HeLa cells grown on coverslips were transfected with GFP, mock infected, and fixed. (B) HeLa cells grown on coverslips were transfected with GFP-NES and either infected with WSN at an MOI of 3, mock-infected, or treated with LMB. At the times indicated, cells were fixed, and GFP was detected by confocal microscopy. (C) Cells from (B) were scored for the percentage of GFP-expressing cells which displayed either predominantly cytoplasmic (C), approximately equal cytoplasmic and nuclear (C/N), or predominantly nuclear (N) GFP distribution. Values are the average of three independent assays. (D) HeLa cells grown on coverslips were transfected with GFP-NES and either infected or mock-infected at an MOI of 3 for 10 h, and treated with PGA at 3hpi. At the times indicated, cells were fixed, and GFP localization was scored as in (C). (E) HeLa cells transfected with GFP-NES were infected with WSN or WSN-NEP20/21 at an MOI of 3 for 9 h, and some treated with PGA at 3hpi. GFP localization was scored as in (C). As shown in Fig. 5E, infection with the mutant WSN-NEP20/21 could induce Crm1 relocalization to an equivalent extent as wt WSN, despite reduced vRNP export. In GFP-NES transfected cells, WSN-NEP20/21 inhibited GFP-NES nuclear export equally as WSN (Fig. 6E). Treatment of these cells with PGA similarly abrogated any impairment in GFP-NES export (Fig. 6E, right side). Thus, impairment of cellular Crm1-dependent export by influenza virus requires vRNP association with chromatin, but not robust vRNP export.
vRNPs associate with Rcc1 on chromatin
Crm1-dependent nuclear export is driven by association of Crm1 with the small GTPase Ran in its GTP-bound state. Exchange of RanGDP for RanGTP is a chromatin-bound process [11] which is catalyzed by the guanine exchange factor Rcc1, during which a putative Crm1-Ran-Rcc1 complex can transiently form [12]. We were therefore interested whether chromatin-targeting of influenza vRNP export complexes might bring them into proximity with the sites of RanGTP regeneration. First, we performed a biochemical fractionation of uninfected or influenza virus-infected cells to determine the distribution of Rcc1 on chromatin in our fractionation protocol. Interestingly, the highest Rcc1 concentration was found in chromatin extracted with 500 mM NaCl (ch500, Fig. 7A), corresponding to the fraction containing vRNP export complexes. Next, we tested whether Rcc1 and vRNP export complexes can associate on chromatin. Immunoprecipitation of chromatin-extracted, infected cells using Rcc1-specific antibodies co-purified NP, PA, and M1, as well as histone H3 (Fig. 7B). Neither NEP nor Crm1 could be detected in the immunopurified complexes (data not shown). In summary, Rcc1 and components of vRNP export complexes can form a complex, although it is unclear whether this interaction is direct, or indirect via nucleosomes.
Fig. 7. Association of Rcc1 with vRNPs and Crm1. 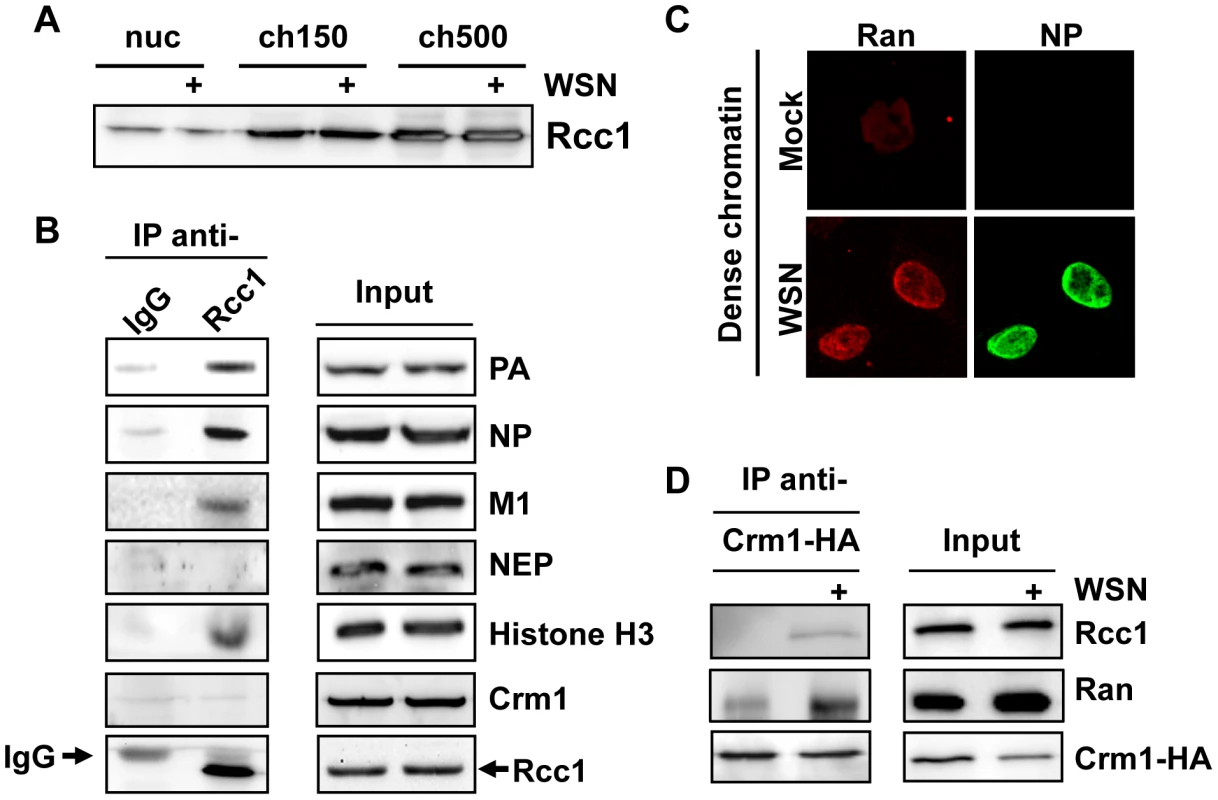
(A) 1×109 HeLa cells were infected with WSN at an MOI of 3 or mock-infected for 9 h. Cells were fractionated, and nuclear lysates were analyzed for distribution of Rcc1. (B) 1×109 HeLa cells were infected with WSN at an MOI of 3 for 9 h. Nuclease-digested lysates were incubated with anti-Rcc1 antibodies or pre-immune serum (IgG) and precipitated with Protein G agarose. Immunoprecipitates and input lysates were analyzed by Western blot for the indicated proteins. (C) HeLa cells grown on coverslips were infected with WSN at an MOI of 3 or mock-infected. Whole cells were fixed at 9hpi, and analyzed by IFA using antibodies against NP and Ran. (D) 2×108 HeLa cells were transfected with an expression plasmid encoding HA-tagged Crm1. 16 h post-transfection, cells were infected with WSN at an MOI of 3 or mock-infected for 9 h. Cells were fractionated, and chromatin lysates were immunoprecipitated using monoclonal HA antibodies. Lysates and precipitates were normalized for Crm1 amounts (to accurately judge amounts of co-precipitated Ran and Rcc1) and analyzed by Western blot for HA-tagged Crm1 or endogenous Rcc1. Based on our finding that Crm1 can be retained on chromatin during influenza virus infection (Fig. 5D), we tested whether infection causes an increase in formation or stability of Crm1-Ran-Rcc1 complexes on chromatin. Virus infection indeed led to accumulation of Ran on dense chromatin (Fig. 7C), partially colocalizing with NP. We then immunoprecipitated HA-tagged Crm1 from uninfected or infected cells after chromatin extraction and measured the amount of bound Ran or Rcc1 by Western blot. At 9 hpi, more Ran and Rcc1 was co-immunoprecipitated by Crm1-HA than in uninfected cells (Fig. 7D). Corresponding to our previous observations, no viral proteins could be detected in the Crm1-HA-bound complexes (data not shown).
Discussion
Unlike most negative-strand RNA viruses, influenza viruses require a functional nuclear export system for nucleocytoplasmic transport of its vRNPs for their incorporation into progeny virions at late time points of infection. Here, we demonstrate that exported vRNPs are assembled on a chromatin fraction which can only be extracted at high salt concentration. Inhibition of Crm1-mediated nuclear export leads to an accumulation of vRNPs containing M1 and NEP on this chromatin fraction, while sequestration of M1 and vRNPs by Hsp70 overexpression prevents their association with this chromatin fraction, thus leading to a lack of vRNP export. Accumulation of vRNPs on the chromatin immediately prior to their export leads to retention of the cellular export proteins Crm1 and Ran at these chromatin-bound sites of vRNP export complex assembly. This relocalization of Crm1 results in a decrease in the nuclear export of other Crm1 substrates. Intriguingly, these sites of vRNP export complex assembly are physically associated with the Ran nucleotide exchange factor Rcc1, and influenza virus infection leads to an increase in the association between Rcc1, Ran, and Crm1.
Several lines of evidence confirm that the vRNPs purified from 500 mM NaCl-extractable chromatin are genuine vRNP export complex precursors. First, these complexes contain both M1 and NEP (Fig. 1D). Although the amount of co-purified NEP was relatively low, it is unclear whether a large amount of the protein is required for export [5]; conversely, vRNPs from the ch500 fraction contained the highest concentration of M1, which is a major driver of vRNP nuclear export [1], [4]. Second, only these complexes are enriched after vRNP nuclear export is blocked either through inhibition of cellular export (Fig. 1D) or mutation of the NES required for vRNP export (WSN-NEP20/21, Fig. 3A + B). Third, while IFA of unfractionated cells revealed a large discrepancy between the distributions of PA and NP in the nucleus (Fig. 2B), extracting all proteins except those bound to chromatin with high affinity showed a colocalization between PA, NP, M1, and NEP, all involved in vRNP export (Figs. 2B + C and data not shown). The exposure of previously-hidden epitopes through extraction has been previously described [29], and likely explains the differences in NP distribution observed between unextracted and chromatin-extracted cells on coverslips. Confirming the biochemical fractionation results, the accumulation of PA, NP, and NEP on dense chromatin also increased after LMB treatment, while no difference was observed on total chromatin or in unextracted cells (Fig. 2B + C). Thus, the vRNP export complex appears to assemble on dense chromatin, from which it translocates to the cytoplasm in a process most likely too transient to capture using our steady-state methods.
One open question in our data is where in the nucleus vRNP export complexes are synthesized. Our finding that PGA treatment reduces the association of vRNPs with chromatin (Fig. 4) suggests that vRNPs are synthesized elsewhere in the nucleus and subsequently associate with the ch500 fraction after synthesis in an M1-dependent manner. Interestingly, previous studies using in vitro transcription assays demonstrated that some portion of vRNA can be synthesized on “nuclear matrix”-like, high-salt insensitive chromatin [18], [34], suggesting the possibility that both vRNP synthesis and export complex assembly are chromatin-bound.
Interestingly, we also purified vRNPs containing NEP from the nucleoplasm (Figs. 1D, 4A). However, these complexes contained no detectable M1, and were not enriched after LMB treatment. Recently, several reports have described a role for NEP in viral RNA replication, including evidence that NEP by itself can increase cRNA and vRNA synthesis in a vRNP reconstitution assay [35], [36]. We therefore speculate that these NEP-containing vRNPs are not exported from the nucleus, but may be involved in the synthesis of full-length viral genomic RNA or cRNA.
Both Crm1 and Ran are very dynamic proteins, which constantly “scan” the nucleus for potential export substrates via diffusion through the soluble region of the nucleus [37], [38]. Although little is known about the nuclear locations at which Crm1 associates with its cargo, many Crm1 substrates, such as nuclear receptors, are very mobile proteins which associate transiently with actively transcribing chromatin [39]. In contrast, RanGTP regeneration by Rcc1 has been shown to be a tightly chromatin-bound process [38], [40]. Our fractionation data suggest that Rcc1 associates with a dense, less transcriptionally-active chromatin fraction, raising the possibility that RanGTP regeneration and Crm1 cargo recognition occur at biochemically and spatially distinct domains in uninfected cells. This separation would provide an unexploited “niche” for influenza vRNPs to access Crm1-RanGTP complexes before they diffuse to other sites of the nucleus. While we cannot determine whether there is a direct contact between Rcc1 and vRNPs (Fig. 7B), both complexes appear to be located on the same nucleosomes, and the ability of influenza virus infection to stabilize the Rcc1-Ran-Crm1 complex (Fig. 7D) suggests that chromatin docking of vRNP export complexes may directly elicit an alteration of Rcc1. The lack of co-precipitated Crm1 or NEP with Rcc1-vRNP complexes (Fig. 7B) is likely due to the very transient nature of both Crm1 and NEP interactions (unpublished observations). Moreover, there is a possibility that different transient complexes between vRNPs, Crm1, and Rcc1 are assembled sequentially on nucleosomes, which could contribute to the asymmetry between Crm1 and Rcc1 immunoprecipitation data.
Based on our findings, we propose that influenza vRNPs gain preferential access to newly-regenerated host cell export machinery through specific chromatin targeting, which confers a physical proximity to the sites of Ran nucleotide exchange (see model in Fig. 8). During late time points of infection, vRNPs associate transiently with dense chromatin, and assemble with M1 and NEP at these sites (Fig. 1D, Fig. 2B+C). This targeting is likely mediated by M1 itself, since M1 can bind to chromatin with high affinity [19], [41], and is sequestered from chromatin after Hsp70 overexpression (Fig. 4B). These chromatin-bound sites of vRNP export complex assembly are associated either directly or via nucleosomes with Rcc1, on which a transient Crm1-Ran-Rcc1 complex is formed in order to regenerate RanGTP from RanGDP [12]. We postulate that the proximity of these components allows influenza vRNPs to “snatch” newly-generated Crm1-RanGTP complexes before they can diffuse to cellular NES-containing substrates at distal nuclear sites (Fig. 8).
Fig. 8. Model of vRNP export. 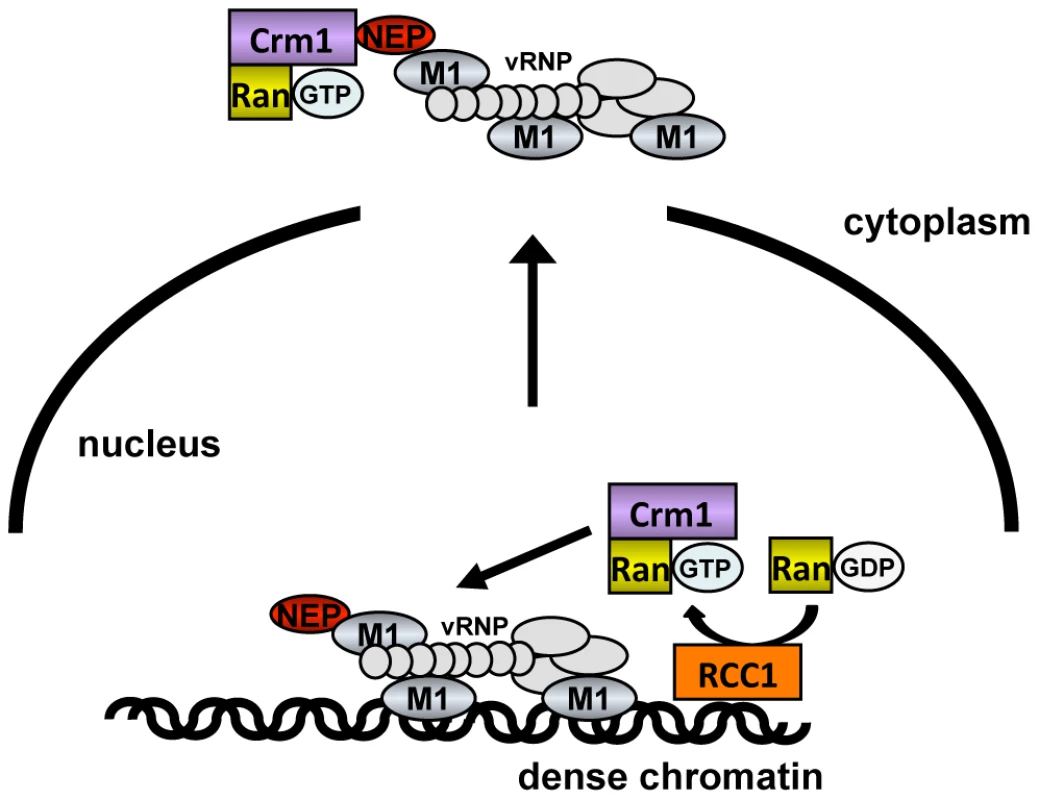
vRNPs (light gray) complexed with M1 (blue) and NEP (red) associate with dense chromatin in proximity to Rcc1 (orange). During or immediately after generation of RanGTP (purple), the proximity of vRNPs to Rcc1 may allow vRNPs access to a RanGTP-Crm1 complex before it can diffuse to other sites in the nucleus. This sequestering of Crm1/RanGTP on the chromatin is a likely cause of GFP-NES retention in the nucleus of infected cells (Fig. 6). Although GFP-NES is not a cellular Crm1 substrate, it is generally accepted as a marker for Crm1 activity [42], and was expressed at low levels in our assay. By retaining a portion of Crm1 on the chromatin, influenza virus infection may reduce the pool of free, soluble Crm1 available to facilitate the export of other proteins more distant from Rcc1. Unlike other RNA viruses [43], [44], influenza virus infection impaired but did not completely block nuclear export, thus keeping intact a cellular system required for viral propagation. Impairment of cellular export seems to be a specific consequence of vRNP association with chromatin: accumulation of vRNPs in the nucleus without chromatin association did not affect export (Fig. 6C, PGA), while vRNP association with chromatin in the absence of vRNP nuclear export led to a decrease in cellular export (Fig. 6D, WSN-NEP20/21). Based on these findings, we postulate that influenza vRNPs gain preferential access to Crm1 in the nucleus, to the detriment of cellular export substrates.
In summary, we show here that influenza viruses use a unique chromatin-targeting strategy to facilitate vRNP export. The chromatin association of vRNPs leads to a retention of Crm1 and Ran, which in turn impairs the ability of other Crm1 substrates to be exported. Thus, we postulate that influenza vRNPs gain an advantage in nuclear export over cellular substrates through chromatin association and subsequent Crm1 “snatching.” While other examples of host-cell hijacking by viruses involve higher binding affinities or shutting off of cellular processes, influenza viruses appear to exploit an unused locational niche in gaining preferential access to the cellular export machinery.
Materials and Methods
Ethics statement
All animal experiments were performed in compliance with the German guidelines according to § 8a Animal Welfare Act of 18. May 2006 (BGBI.I, p. 1206), European Union guidelines 86/609/EWG of 24.11.1986 and according to the European Agreement of 18.3.1996 for protection of animal trials and other for scientific purposes used vertebrates of 11.12.1990 (BGBI. II S. 1486) and NIH/OLAW Animal Welfare Assurance #A5755-01″. The animal welfare committees of the Robert Koch Institute and the Institute of Biotechnology of Vilnius University approved all animal experiments.
Antibodies
Hybridomas producing monoclonal antibodies (MAbs) against PA (A/WSN/33) were generated essentially as described by ([45]). Eight-week-old female BALB/c mice (obtained from a breeding colony at the Institute for Innovative Medicine, Vilnius, Lithuania) were immunized at days 0, 28 and 56 by a subcutaneous injection of 50 µg of recombinant purified PA [46]. The specificity of MAbs was tested by an indirect ELISA. Twenty five hybridomas giving initial ELISA reactivities (OD450) against full-length recombinant PA OD450 >2.0 were identified and subcloned. After subcloning, 22 stable hybridoma cell lines producing MAbs of IgG isotype IgG1 subtype were obtained. The MAb produced by clone 1E7, used in this study, showed high reactive against recombinant full-length C9 terminally hexahistidine-tagged Flu A PA both by ELISA and Western blot (data not shown). The MAb 1E7 did not cross-react using purified C-terminally His-tagged Flu B PA and lysates of either non-transformed yeast cells.
Influenza-specific anti-NP antibodies have been previously described [47], while polyclonal anti-NEP antibody was generated by immunizing a rabbit with recombinant glutathione-S-transferase fused to NEP derived from the influenza A/PR/8/34 virus.
All other antibodies were purchased: anti-M1 (ab22396), anti-histone H3 (ab1791), anti-HMGB1 (ab18256), and anti-Rcc1 (ab54600) from Abcam; anti-Crm1 (sc-5595) from Santa Cruz Biotechnology; anti-TFIIB from Euromedex; anti-Pol II (8WG16) from Covance; anti-tubulin from Sigma-Aldrich; and anti-Ran (610341) from BD Transduction Laboratories. All secondary antibodies were purchased from Jackson Immunoresearch.
Cell lines and chemicals
HeLa, A549 and MDCK cells were cultured in DMEM containing penicillin and streptomycin.
Prostaglandin A (PGA) was purchased from Sigma and stored at a stock concentration of 5 mg/ml in ethanol. Leptomycin B was purchased from Enzo Biochemicals and stored at a stock concentration of 5 µg/ml in ethanol.
Virus strains and infections
Recombinant viruses rWSN-PB2-Strep and rWSN (untagged) were generated as previously described [24]. Viruses were diluted in phosphate-buffered saline (PBS) containing 0.3% bovine serum albumin (BSA), and cells were infected for 1 h at 37°C. Virus titers from supernatants were performed on MDCK cells using standard plaque assay protocol. WSN-NEP20/21 was generated by site-directed mutagenesis based on the sequence in ([30]) in the plasmid pPOLI-NS [48].
Plasmids
Plasmid pCA-GFP-NES was generated from pCA-Flag-GFP [46] using a reverse primer containing the HIV Rev NES sequence (LQLPPLERLTLD) preceded by a linker ecoding three glycine residues and followed by a stop codon and a BglII restriction site. PCR was performed using this primer and a GFP-specific forward primer, and the product was digested and ligated into the digested vector.
Subcellular fractionation
109 HeLa cells were washed and pelleted in PBS. The cell pellet was resuspended in 10 ml sucrose buffer (10 mM HEPES pH 7.9, 10 mM KCl, 2 mM Mg acetate, 3 mM CaCl2, 340 mM sucrose, 1 mM DTT, 1 mM PMSF), and cells were incubated 10 min. on ice. NP-40 was then added to a final concentration of 0.5%, followed by vortexing for 15 sec and centrifugation for 10 min at 4°C at 3,500 g. Supernatant was saved as the cytoplasmic fraction, and the nuclear pellet was washed once with 5 ml sucrose buffer. The nuclear pellet was then resuspended in 1.5 ml nucleoplasm extraction buffer (50 mM HEPES pH 7.9, 150 mM potassium acetate, 1.5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 1 mM PMSF), transferred to an all-glass 4 ml Dounce homogenizer with a tight-fitting pestle, and homogenized with 20 strokes. Homogenate was then incubated 20 min at 4°C on a rotating wheel, followed by centrifugation for 10 min at 4°C at 14,000 rpm in a microcentrifuge. The supernatant was saved as the nucleoplasmic fraction, and the pellet was resuspended in 1.5 ml nuclease incubation buffer (50 mM HEPES pH 7.9, 10 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 1 mM PMSF) containing 100 U/ml Benzonase nuclease (Novagen). Chromatin was digested for 10′ at 37°C, followed by addition of NaCl to 150 mM and further incubation 20 min on ice. Digested lysate was then centrifuged 10 min at 4°C at 14,000 rpm, and supernatant was saved as the low-salt chromatin fraction. The pellet was resuspended in 1.5 ml chromatin extraction buffer (50 mM HEPES pH 7.9, 500 mM NaCl, 1.5 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF) and incubated 20 min on ice, followed by centrifugation as above. The supernatant was saved as the high-salt chromatin fraction.
Strep-tag purification
100 µl Strep-Tactin Sepharose (IBA) was washed with 30 volumes strep wash buffer (20 mM HEPES pH 7.9, 150 mM NaCl, 1 mM EDTA) and added to fractionated lysates. Lysates were incubated with beads for 1 h at 4°C on a rotating wheel. Beads were collected by centrifugation, washed three times with strep wash buffer (containing 500 mM NaCl for high-salt chromatin samples), and eluted with 100 µl strep wash buffer containing 2.5 mM desthiobiotin (IBA). Eluates were diluted 1∶1 with 2x Lämmli buffer for further analysis.
Primer extension
For primer extension, cells were first fractionated as above, except that Benzonase nuclease was replaced with 20 U/ml RNase-free DNase I (Fermentas). RNA was then extracted from each fraction using a Nucleospin RNA II Kit (Macherey-Nagel). Primer extension was performed using WSN NA-specific primers as previously described [35].
In situ fractionation and immunofluorescence
HeLa cells grown on coverslips were washed once with cold PBS, then incubated with 250 µl/well CSK (50 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM PMSF) containing 0.5% Triton X-100 for 3 min. on ice, followed by a brief wash with CSK. Cells were then incubated with 250 µl DB (50 mM PIPES pH 6.8, 50 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM CaCl2, 1 mM PMSF) containing 20 U/ml RNase-free DNase I (Fermentas) for 10 min. at 37°C. This buffer was removed, and cells were incubated in DB containing 250 mM NaCl for 3 min. on ice. Experiments were performed in triplicate, and one coverslip each was fixed before the extraction, after detergent extraction, and after chromatin extraction. Cells were fixed 10′ with 3% paraformaldehyde, followed by standard immunofluorescence. DNA was stained using ToPro3 (Invitrogen). For GFP-NES experiments, HeLa cells were on coverslips in 24-well plates were transfected with 200 ng/well pCA-GFP-NES. After infection, coverslips were mounted on slides directly after fixation. Optical sections were obtained using a Leica TCS confocal microscope.
Immunoprecipitations
HeLa cells were transfected with a plasmid encoding HA-tagged Crm1 ([49], kindly provided by R. Kehlenbach, Göttingen, Germany) using Nanofectin (PAA). 24 h after transfection, cells were infected for 9 h. Fractionation was then performed as above, except that high-salt chromatin extraction was performed using 300 mM NaCl instead of 500 mM to prevent disruption of Crm1 complexes. High salt chromatin lysates were incubated with anti-HA agarose (Sigma-Aldrich) for 1 h at 4°C on a rotating wheel, then washed three times with strep wash buffer containing 300 mM NaCl. Bound complexes were eluted using Lämmli buffer and analyzed by Western blot.
For anti-Rcc1 immunoprecipitation, cells were infected 9 h, then lysed with lysis buffer (50 mM Tris pH 7.5, 250 mM NaCl, 2 mM MgCl2, 0.5% NP-40, 1 mM PMSF, 1 mM DTT) containing 100 U Benzonase per ml 20 min. on ice. Lysates were incubated with anti-Rcc1 (Santa Cruz) and precipitated using Protein G agarose (Pierce). Beads were washed 3 times with lysis buffer without NP-40 and eluted with Lämmli buffer.
Supporting Information
Zdroje
1. MartinKHeleniusA 1991 Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67 117 130
2. NeumannGCastrucciMRKawaokaY 1997 Nuclear import and export of influenza virus nucleoprotein. J Virol 71 9690 9700
3. O'NeillRETalonJPaleseP 1998 The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J 17 288 296
4. BuiMWillsEGHeleniusAWhittakerGR 2000 Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J Virol 74 1781 1786
5. EltonDSimpson-HolleyMArcherKMedcalfLHallamR 2001 Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J Virol 75 408 419
6. WatanabeKTakizawaNKatohMHoshidaKKobayashiN 2001 Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res 77 31 42
7. KudoNMatsumoriNTaokaHFujiwaraDSchreinerEP 1999 Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A 96 9112 9117
8. BuiMMyersJEWhittakerGR 2002 Nucleo-cytoplasmic localization of influenza virus nucleoprotein depends on cell density and phosphorylation. Virus Res 84 37 44
9. AkarsuHBurmeisterWPPetosaCPetitIMullerCW 2003 Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J 22 4646 4655
10. PetosaCSchoehnGAskjaerPBauerUMoulinM 2004 Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell 16 761 775
11. NemergutMEMizzenCAStukenbergTAllisCDMacaraIG 2001 Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292 1540 1543
12. NemergutMELindsayMEBrownawellAMMacaraIG 2002 Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J Biol Chem 277 17385 17388
13. KutayUGuttingerS 2005 Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15 121 124
14. PembertonLFPaschalBM 2005 Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6 187 198
15. Rameix-WeltiMATomoiuADos Santos AfonsoEvan der WerfSNaffakhN 2009 Avian Influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J Virol 83 1320 1331
16. LoucaidesEMvon KirchbachJCFoegleinASharpsJFodorE 2009 Nuclear dynamics of influenza A virus ribonucleoproteins revealed by live-cell imaging studies. Virology 394 154 163
17. EngelhardtOGSmithMFodorE 2005 Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J Virol 79 5812 5818
18. TakizawaNWatanabeKNounoKKobayashiNNagataK 2006 Association of functional influenza viral proteins and RNAs with nuclear chromatin and sub-chromatin structure. Microbes Infect 8 823 833
19. Garcia-RoblesIAkarsuHMullerCWRuigrokRWBaudinF 2005 Interaction of influenza virus proteins with nucleosomes. Virology 332 329 336
20. HenikoffSHenikoffJGSakaiALoebGBAhmadK 2009 Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19 460 469
21. RochaEDavieJRvan HoldeKEWeintraubH 1984 Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J Biol Chem 259 8558 8563
22. VreedeFTChanAYSharpsJFodorE 2010 Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 396 125 134
23. RodriguezAPerez-GonzalezANietoA 2007 Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J Virol 81 5315 5324
24. Dos Santos AfonsoEEscriouNLeclercqIvan der WerfSNaffakhN 2005 The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341 34 46
25. MorASulimanSBen-YishayRYungerSBrodyY 2010 Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol 12 543 552
26. MirskyAEBurdickCJDavidsonEHLittauVC 1968 The role of lysine-rich histone in the maintenance of chromatin structure in metaphase chromosomes. Proc Natl Acad Sci U S A 61 592 597
27. MaKRoyAMWhittakerGR 2001 Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology 282 215 220
28. EltonDAmorimMJMedcalfLDigardP 2005 ‘Genome gating’; polarized intranuclear trafficking of influenza virus RNPs. Biol Lett 1 113 117
29. NickersonJAKrockmalnicGWanKMTurnerCDPenmanS 1992 A normally masked nuclear matrix antigen that appears at mitosis on cytoskeleton filaments adjoining chromosomes, centrioles, and midbodies. J Cell Biol 116 977 987
30. Iwatsuki-HorimotoKHorimotoTFujiiYKawaokaY 2004 Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J Virol 78 10149 10155
31. HirayamaEAtagiHHirakiAKimJ 2004 Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex. J Virol 78 1263 1270
32. SeibelNMEljouniJNalaskowskiMMHampeW 2007 Nuclear localization of enhanced green fluorescent protein homomultimers. Anal Biochem 368 95 99
33. XylourgidisNRothPSabriNTsarouhasVSamakovlisC 2006 The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila. J Cell Sci 119 4409 4419
34. Lopez-TurisoJAMartinezCTanakaTOrtinJ 1990 The synthesis of influenza virus negative-strand RNA takes place in insoluble complexes present in the nuclear matrix fraction. Virus Res 16 325 337
35. RobbNCSmithMVreedeFTFodorE 2009 NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J Gen Virol 90 1398 1407
36. PerezJTVarbleASachidanandamRZlatevIManoharanM 2010 Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A 107 11525 11530
37. DaelemansDCostesSVLockettSPavlakisGN 2005 Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol Cell Biol 25 728 739
38. HutchinsJRMooreWJClarkePR 2009 Dynamic localisation of Ran GTPase during the cell cycle. BMC Cell Biol 10 66
39. ElbiCWalkerDARomeroGSullivanWPToftDO 2004 Molecular chaperones function as steroid receptor nuclear mobility factors. Proc Natl Acad Sci U S A 101 2876 2881
40. CushmanIStenoienDMooreMS 2004 The dynamic association of RCC1 with chromatin is modulated by Ran-dependent nuclear transport. Mol Biol Cell 15 245 255
41. ZhirnovOPKlenkHD 1997 Histones as a target for influenza virus matrix protein M1. Virology 235 302 310
42. SabriNRothPXylourgidisNSadeghifarFAdlerJ 2007 Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol 178 557 565
43. PetersenJMHerLSDahlbergJE 2001 Multiple vesiculoviral matrix proteins inhibit both nuclear export and import. Proc Natl Acad Sci U S A 98 8590 8595
44. LidskyPVHatoSBardinaMVAminevAGPalmenbergAC 2006 Nucleocytoplasmic traffic disorder induced by cardioviruses. J Virol 80 2705 2717
45. KohlerGMilsteinC 1975 Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256 495 497
46. WunderlichKMayerDRanadheeraCHollerASManzB 2009 Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication. PLoS One 4 e7517
47. MayerDMolawiKMartinez-SobridoLGhanemAThomasS 2007 Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J Proteome Res 6 672 682
48. FodorEDevenishLEngelhardtOGPalesePBrownleeGG 1999 Rescue of influenza A virus from recombinant DNA. J Virol 73 9679 9682
49. HilliardMFrohnertCSpillnerCMarconeSNathA 2010 The anti-inflammatory prostaglandin 15-deoxy-delta(12,14)-PGJ2 inhibits CRM1-dependent nuclear protein export. J Biol Chem 285 22202 22210
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage EgressČlánek A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene ExpressionČlánek An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes byČlánek Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine ScrapieČlánek Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 9- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity
- Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
- Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development
- Nrf2, a PPARγ Alternative Pathway to Promote CD36 Expression on Inflammatory Macrophages: Implication for Malaria
- Robust Antigen Specific Th17 T Cell Response to Group A Streptococcus Is Dependent on IL-6 and Intranasal Route of Infection
- Targeting of a Chlamydial Protease Impedes Intracellular Bacterial Growth
- The Protease Cruzain Mediates Immune Evasion
- High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism
- Plague and Climate: Scales Matter
- Exhausted CD8 T Cells Downregulate the IL-18 Receptor and Become Unresponsive to Inflammatory Cytokines and Bacterial Co-infections
- Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles
- Murine Gamma-herpesvirus Immortalization of Fetal Liver-Derived B Cells Requires both the Viral Cyclin D Homolog and Latency-Associated Nuclear Antigen
- Rapid and Efficient Clearance of Blood-borne Virus by Liver Sinusoidal Endothelium
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene Expression
- Strain Specific Resistance to Murine Scrapie Associated with a Naturally Occurring Human Prion Protein Polymorphism at Residue 171
- Development of a Transformation System for : Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector
- Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
- Host Phylogeny Determines Viral Persistence and Replication in Novel Hosts
- BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity
- Expression of the RAE-1 Family of Stimulatory NK-Cell Ligands Requires Activation of the PI3K Pathway during Viral Infection and Transformation
- Structure of the Vesicular Stomatitis Virus N-P Complex
- HSV Infection Induces Production of ROS, which Potentiate Signaling from Pattern Recognition Receptors: Role for S-glutathionylation of TRAF3 and 6
- The Human Papillomavirus E6 Oncogene Represses a Cell Adhesion Pathway and Disrupts Focal Adhesion through Degradation of TAp63β upon Transformation
- Analysis of Behavior and Trafficking of Dendritic Cells within the Brain during Toxoplasmic Encephalitis
- Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance
- Multidrug Resistant 2009 A/H1N1 Influenza Clinical Isolate with a Neuraminidase I223R Mutation Retains Its Virulence and Transmissibility in Ferrets
- Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1
- Step-Wise Loss of Bacterial Flagellar Torsion Confers Progressive Phagocytic Evasion
- Complex Recombination Patterns Arising during Geminivirus Coinfections Preserve and Demarcate Biologically Important Intra-Genome Interaction Networks
- An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by
- Non-Lytic, Actin-Based Exit of Intracellular Parasites from Intestinal Cells
- The Fecal Viral Flora of Wild Rodents
- The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
- Interferon Regulatory Factor-1 (IRF-1) Shapes Both Innate and CD8 T Cell Immune Responses against West Nile Virus Infection
- A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
- Evaluating the Sensitivity of to Biotin Deprivation Using Regulated Gene Expression
- The Motility of a Human Parasite, , Is Regulated by a Novel Lysine Methyltransferase
- Phosphodiesterase-4 Inhibition Alters Gene Expression and Improves Isoniazid – Mediated Clearance of in Rabbit Lungs
- Restoration of IFNγR Subunit Assembly, IFNγ Signaling and Parasite Clearance in Infected Macrophages: Role of Membrane Cholesterol
- Protease ROM1 Is Important for Proper Formation of the Parasitophorous Vacuole
- The Regulated Secretory Pathway in CD4 T cells Contributes to Human Immunodeficiency Virus Type-1 Cell-to-Cell Spread at the Virological Synapse
- Rerouting of Host Lipids by Bacteria: Are You CERTain You Need a Vesicle?
- Transmission Characteristics of the 2009 H1N1 Influenza Pandemic: Comparison of 8 Southern Hemisphere Countries
- Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine Scrapie
- Sequential Bottlenecks Drive Viral Evolution in Early Acute Hepatitis C Virus Infection
- Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen
- Genomic and Proteomic Analyses of the Fungus Provide Insights into Nematode-Trap Formation
- Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
- Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
- Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate
- Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
- c-di-AMP Is a New Second Messenger in with a Role in Controlling Cell Size and Envelope Stress
- Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System
- APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells
- Impairment of Immunoproteasome Function by β5i/LMP7 Subunit Deficiency Results in Severe Enterovirus Myocarditis
- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Tri6 Is a Global Transcription Regulator in the Phytopathogen
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- The Next Opportunity in Anti-Malaria Drug Discovery: The Liver Stage
- Significant Effects of Antiretroviral Therapy on Global Gene Expression in Brain Tissues of Patients with HIV-1-Associated Neurocognitive Disorders
- Inhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
- A Novel Metal Transporter Mediating Manganese Export (MntX) Regulates the Mn to Fe Intracellular Ratio and Virulence
- Rhoptry Kinase ROP16 Activates STAT3 and STAT6 Resulting in Cytokine Inhibition and Arginase-1-Dependent Growth Control
- Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms
- Secretion of Genome-Free Hepatitis B Virus – Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
- Membrane Remodeling by the Double-Barrel Scaffolding Protein of Poxvirus
- A Diverse Population of Molecular Type VGIII in Southern Californian HIV/AIDS Patients
- Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A Virus Protease-Polymerase Processing Intermediate, 3CD
- Quantitative Analyses Reveal Calcium-dependent Phosphorylation Sites and Identifies a Novel Component of the Invasion Motor Complex
- Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes
- Old World Arenaviruses Enter the Host Cell via the Multivesicular Body and Depend on the Endosomal Sorting Complex Required for Transport
- Exploits a Unique Repertoire of Type IV Secretion System Components for Pilus Assembly at the Bacteria-Host Cell Interface
- Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



