-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
Pseudomonas entomophila is an entomopathogenic bacterium that infects and kills Drosophila. P. entomophila pathogenicity is linked to its ability to cause irreversible damages to the Drosophila gut, preventing epithelium renewal and repair. Here we report the identification of a novel pore-forming toxin (PFT), Monalysin, which contributes to the virulence of P. entomophila against Drosophila. Our data show that Monalysin requires N-terminal cleavage to become fully active, forms oligomers in vitro, and induces pore-formation in artificial lipid membranes. The prediction of the secondary structure of the membrane-spanning domain indicates that Monalysin is a PFT of the ß-type. The expression of Monalysin is regulated by both the GacS/GacA two-component system and the Pvf regulator, two signaling systems that control P. entomophila pathogenicity. In addition, AprA, a metallo-protease secreted by P. entomophila, can induce the rapid cleavage of pro-Monalysin into its active form. Reduced cell death is observed upon infection with a mutant deficient in Monalysin production showing that Monalysin plays a role in P. entomophila ability to induce intestinal cell damages, which is consistent with its activity as a PFT. Our study together with the well-established action of Bacillus thuringiensis Cry toxins suggests that production of PFTs is a common strategy of entomopathogens to disrupt insect gut homeostasis.
Published in the journal: Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality. PLoS Pathog 7(9): e32767. doi:10.1371/journal.ppat.1002259
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002259Summary
Pseudomonas entomophila is an entomopathogenic bacterium that infects and kills Drosophila. P. entomophila pathogenicity is linked to its ability to cause irreversible damages to the Drosophila gut, preventing epithelium renewal and repair. Here we report the identification of a novel pore-forming toxin (PFT), Monalysin, which contributes to the virulence of P. entomophila against Drosophila. Our data show that Monalysin requires N-terminal cleavage to become fully active, forms oligomers in vitro, and induces pore-formation in artificial lipid membranes. The prediction of the secondary structure of the membrane-spanning domain indicates that Monalysin is a PFT of the ß-type. The expression of Monalysin is regulated by both the GacS/GacA two-component system and the Pvf regulator, two signaling systems that control P. entomophila pathogenicity. In addition, AprA, a metallo-protease secreted by P. entomophila, can induce the rapid cleavage of pro-Monalysin into its active form. Reduced cell death is observed upon infection with a mutant deficient in Monalysin production showing that Monalysin plays a role in P. entomophila ability to induce intestinal cell damages, which is consistent with its activity as a PFT. Our study together with the well-established action of Bacillus thuringiensis Cry toxins suggests that production of PFTs is a common strategy of entomopathogens to disrupt insect gut homeostasis.
Introduction
The intestinal epithelium has a role in defining the barrier between the host and the external environment [1]. This barrier protects the host against invasion and systemic dissemination of both pathogenic and commensal microorganisms. Both resistance and tolerance mechanisms contribute to maintain the gut integrity from the assault of infectious bacteria [2]. Resistance mechanisms involve the activation of various local immune responses that directly target pathogens. In contrast, tolerance mechanisms involve the activation of repair and stress pathways that quickly seal damages caused by infectious agents. Pathogenic bacteria have the capacity to overcome gut defenses and impede the return to homeostasis [3]. To study how pathogenic bacteria disrupt gut homeostasis, we chose to investigate the interactions between Drosophila and a newly identified entomopathogen, Pseudomonas entomophila. P. entomophila is closely related to the saprophytic soil bacterium Pseudomonas putida [4], [5]. It was originally isolated from a fly sampled in Guadeloupe and subsequently shown to be lethal to Drosophila larvae and adults after ingestion. P. entomophila can also effectively kill members of other insect orders (e.g. Bombyx mori, Anopheles gambiae, Galleria mellonella). After ingestion, P. entomophila is able to persist in the Drosophila gut. It induces the expression of antimicrobial peptide genes via the Imd pathway, both locally in the intestinal epithelium and systemically in the fat body, an organ analog to the mammalian liver [4]. It was shown that P. entomophila virulence is under the control of two global regulatory systems: the well known GacS/GacA two component system, and a second system involving a secreted secondary metabolite synthesized by the pvf gene products [4], [6]. The Gac system also controls the production of a secreted protease, AprA, which is important for P. entomophila to counteract the local immune response of Drosophila [7].
Recent studies revealed that upon bacterial infection, homeostasis in the gut is restored only when bacterial clearance is coordinated with the repair of infection-induced damage through epithelium renewal [8]–[10]. Epithelium renewal of the Drosophila gut is stimulated by the release of the secreted ligand Upd3 from damaged enterocytes, which then activates the JAK/STAT pathway in intestinal stem cells to promote both their division and differentiation, establishing a homeostatic regulatory loop [8], [9]. In contrast to infection with non-lethal bacteria, P. entomophila infection inflicts strong damage to its host without triggering an epithelial renewal [8], [11]. This suggests that the damages inflicted by P. entomophila are too severe to be repaired. How damages are inflicted however remains unknown. One hypothesis was that P. entomophila produces cytotoxic factors that damage the intestinal epithelium.
In this study, we identified a secreted protein that plays an important role in the damage inflicted by P. entomophila to the Drosophila gut. We showed that this protein is a pore-forming toxin (PFT) that we called Monalysin. Our work indicates that production of PFTs is a strategy used by entomopathogenic bacteria to disrupt gut homeostasis.
Results
Identification of a secreted protein involved in P. entomophila pathogenicity
We previously showed that P. entomophila secretes large amount of the metalloprotease, AprA, which can degrade antimicrobial peptides [7]. The production of this protease is regulated by the GacS-GasA system, known to control secondary metabolite production, protein secretion, and virulence determinants in γ-proteobacteria [12]. To identify additional factors responsible for P. entomophila virulence, we analyzed the culture supernatant of the wild-type bacterium and a gacA mutant by SDS-PAGE (Figure 1A). Bands corresponding to major secreted proteins in the wild type strain, but not in the gacA mutant were submitted to analysis by mass spectrometry. This allowed us to confirm that one of the major bands corresponds to the 51 kDa AprA. Three bands contained Hcp, Vgr and Rhs, proteins known to be secreted by the Type VI Secretion System (T6SS). T6SS are bacterial needle-like structure involved in the injection of effectors into the cytoplasm of eukaryotic but also prokaryotic cells [13]. We also identified a band with an apparent molecular weight of 30 kDa, containing a protein encoded by the uncharacterized gene pseen3174 that we named Monalysin.
Fig. 1. PSEEN3174 encodes a secreted protein, Monalysin, required for P. entomophila virulence. 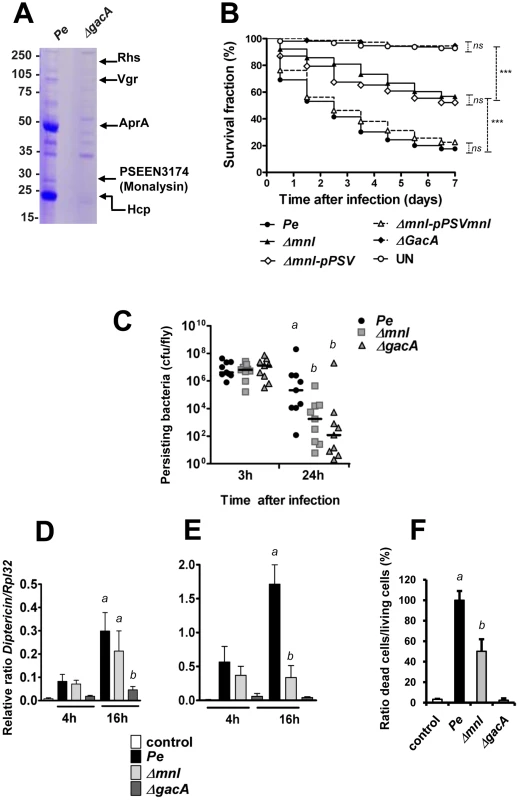
(A) SDS-PAGE analysis of culture supernatants from wild type P. entomophila and the ΔgacA derivative. Proteins extracted from culture supernatants were loaded on a SDS-PAGE and stained with coomassie blue. The nature of proteins identified by MALDI-TOF analysis of tryptic fragments is shown on the right. (B) Survival analysis of wild-type Oregon adult flies following infection by feeding with the P. entomophila wild-type strain (Pe), the gacA-deficient strain (ΔgacA), the mnl deficient strain (Δmnl) the mnl-deficient strain carrying a plasmid expressing a wild-type copy of the monalysin gene (Δmnl-pPSVmnl), or carrying the plasmid pPSV35 without any insert (Δmnl-pPSV). UN: unchallenged. The Kaplan-Meier log rank test was used to determine statistical significance. Dashed brackets represent the significance between the different infections (***: p<0.001, ns: not significant). (C) Bacterial persistence in wild-type Oregon flies as the number of colony-forming-unit (cfu) per fly. After infection by the P. entomophila wild-type strain (Pe), the gacA-deficient strain (ΔgacA), the mnl deficient strain (Δmnl), the number of cfu per fly was determined at the indicated time point. (D and E )Time-course analysis of Diptericin expression measured by RT-qPCR in (D) guts (local) or (E) whole flies (corresponding to the systemic fat body expression). (F) Cell death quantification using acridine orange staining. Results represent the percentage of dead cells (acridine orange positive nuclei) in the midguts of flies infected for 16 hours with the indicated bacterial strains. Results represent the average of four independent experiments. Statistical analysis was performed using a Wilcoxon test, and different letters indicate significantly different values (P<0.05). In order to investigate the role of these secreted proteins, we made a T6SS mutant (affecting the ORF pseen0535) and a monalysin (mnl) mutant (Δmnl), and tested their virulence in Drosophila. No difference could be observed between the wild type strain and the T6SS mutant. Interestingly, the mnl mutant presented a decreased pathogenicity. Indeed, survival analysis of Drosophila adults after oral infection with the wild-type strain, the gacA mutant, and the mnl mutant showed that only 40% of the flies infected with the mnl mutant succumbed within 3 days, while 70% of the flies died after infection with wild-type P. entomophila (Figure 1B and Figure S6). As previously shown [4], a gacA deficient mutant did not show any pathogenicity using this assay. The attenuated virulence of the mnl mutant was fully rescued by complementation with a wild-type copy of the monalysin gene.
A mutant deficient in Monalysin production is affected in its abilities to induce cell damage in the Drosophila gut
It was previously shown that P. entomophila virulence towards Drosophila is associated to its ability to persist in the gut and the transcription of antibacterial peptide genes both locally and systematically [4]. In order to better characterize the role of the monalysin gene in the infectious process, we next compared the ability of the mnl mutant (Δmnl) to persist to that of the wild type strain or a gacA mutant. Flies were infected by feeding and bacterial loads were quantified at two time points (Figure 1C). While bacterial loads were indistinguishable after 3 hrs, persistence of the mnl mutant and the gacA mutant were significantly decreased when compared to wild type bacteria [4]. We then compared the activation of the Imd pathway after infection by the wild type, the gacA, and the mnl mutant. We used reverse transcriptase quantitative PCR (RT-qPCR) to measure the expression of the Diptericin gene, a target of the Imd pathway, specifically in the gut (local response) or in whole flies (reflecting mostly the systemic expression of Diptericin by the fat body) (Figure 1D and E). As previously shown [7], Diptericin expression increased already 4 h after infection by P. entomophila and even more after 16 h, both in the gut and the fat body, an increase that was not observed for the gacA mutant [4]. The mnl mutant leads to an increase in Diptericin expression in the gut similar to that observed for the wild-type bacterium (Figure 1D). However, while Diptericin expression increased to wild-type levels in the fat body 4 h after infection, no further increase was observed (16 h) in flies infected with the mnl mutant (Figure 1E).
We next investigated the contribution of Monalysin in the damage caused by P. entomophila to the Drosophila gut. We first monitor the induction of cell death upon bacterial ingestion using an acridine orange staining. A high number of dead cells were detected in guts from flies infected by wild type P. entomophila, but not in guts from flies infected by a gacA mutant as previously reported [6]. Interestingly, a reduced level of cell death was observed in the mnl mutant (Figure 1F and S7). Oral infection with P. entomophila resulted in a decrease of the adherens junction marker Cadherin-GFP (Figure 2A) and to morphologically altered guts, with regions devoid of enterocytes indicative of a disruption of tissue integrity (see the lack of nuclear DAPI staining due to the loss of cell in Figure 2A3). Interestingly, gut collected 16 hrs after oral infection with gacA and mnl mutants did not show any Cadherin-GFP signal decreases or a rupture of the gut integrity (Figure 2A4 to 2A6). Previous studies showed that ingestion of P. entomophila activates both JAK-STAT and the Jun N-terminal kinase (JNK) pathway in the Drosophila gut [8] that participate in the repair and stress responses, respectively [8], [14]–[16]. The activation of both pathways can be monitored by measuring by RT-qPCR the expression of puckered (puc) (a direct downstream target of JNK signaling) or upd3 (a target of JAK-STAT signaling) and Socs36E (a target of JAK-STAT signaling that encodes a negative regulator of this pathway). Figure 2B, 2C and 2D shows that the mnl mutant was less efficient than wild type P. entomophila to activate the JNK and JAK-STAT pathways, yet more efficient than a gacA mutant. Consistent with the RT-qPCR analysis, expression of the upd3-GFP reporter gene (upd3-Gal4, UAS-GFP) was strongly induced in the gut of flies orally infected with a sub-lethal dose of P. entomophila but not the mnl mutant (Figure 2E). Altogether, these data show that even though the mnl mutant retains some ability to cause intestinal damage, this ability is strongly diminished compared to wild type P. entomophila. This suggested a specific role of the Monalysin protein in P. entomophila cytotoxicity towards Drosophila.
Fig. 2. Monalysin contributes to P. entomophila-inflicted damage to the Drosophila gut. 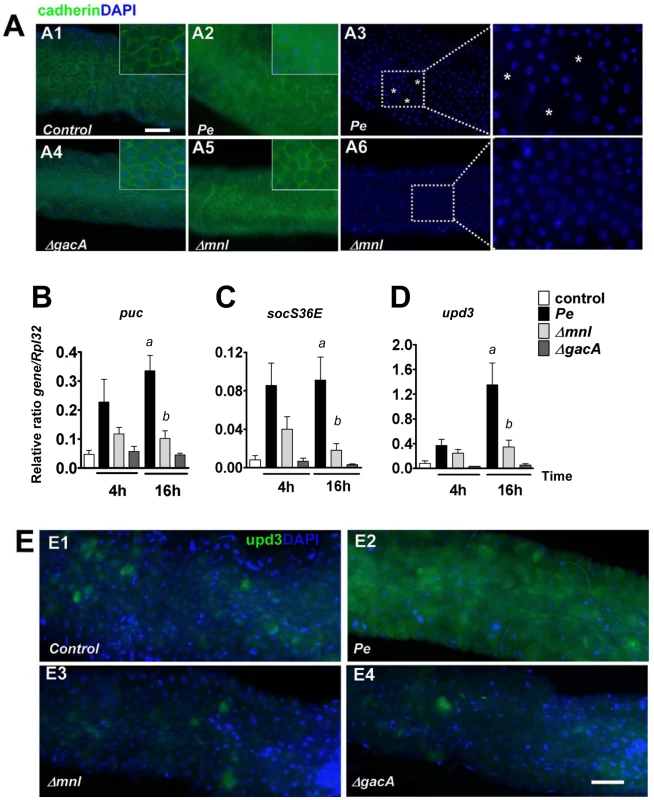
(A) Expression of the marker of adherens junction Cadherin-GFP 16 hours after infection with lethal doses of the indicated bacteria. Ingestion of wild type P. entomophila disrupts the pattern of Cadherin-GFP. A3 and A6 show DAPI of A2 and A5 respectively. The symbol (*) marks regions where DAPI staining is absent. (B–D) Analysis of puckered (puc), unpaired3 (upd3), and socs36E expression measured by RT-qPCR in guts of infected flies. Statistical analysis was performed using a Wilcoxon test and letters indicate significantly different values (P<0.05). (E) Expression of the upd3-Gal4, UAS-GFP reporter in (E1) unchallenged flies or( E2–E4) 4 hours after infections with a sublethal dose of bacteria (OD600 = 10). In contrast to the wild type P. entomophila strain (E2), the Δmnl (E3) and the ΔgacA (E4) strains were unable to elicit upd3-GFP expression. Scale bars represent 50 µm. Monalysin is a secreted cytotoxic protein
In order to characterize the activity of the secreted protein encoded by monalysin, we produced and purified a his-tag version of it in E. coli. Ingestion of the recombinant protein at high dose had no impact on fly survival. However, the Monalysin protein was highly toxic when directly injected in the body cavity (Figure 3A). This dose-dependent lethal activity suggests that Monalysin function as a bacterial toxin. Results shown in Figures 3B, 3C and Table S2 indicate that Monalysin has a strong cytotoxicity towards S2 cells (derived from Drosophila embryonic hemocytes) and SF9 cells (from the Lepidoptera Spodoptera frugiperda). Moreover, Monalysin treated S2 cells showed DNA fragmentation and condensation that are sign of apoptosis (Figure 3D and E). Along the same line, Figure 4A shows that the recombinant toxin rapidly induced hemolysis in a dose dependant manner as measured by the loss of sample turbidity. In addition we found that two mammalian culture cell lines – Hela and RPE1 – were also sensitive to Monalysin (Figure 4B and Table S2). Altogether these observations show that Monalysin is a secreted cytotoxic factor of P. entomophila with a broad range of activity.
Fig. 3. Monalysin encodes a cytotoxic protein secreted by P. entomophila. 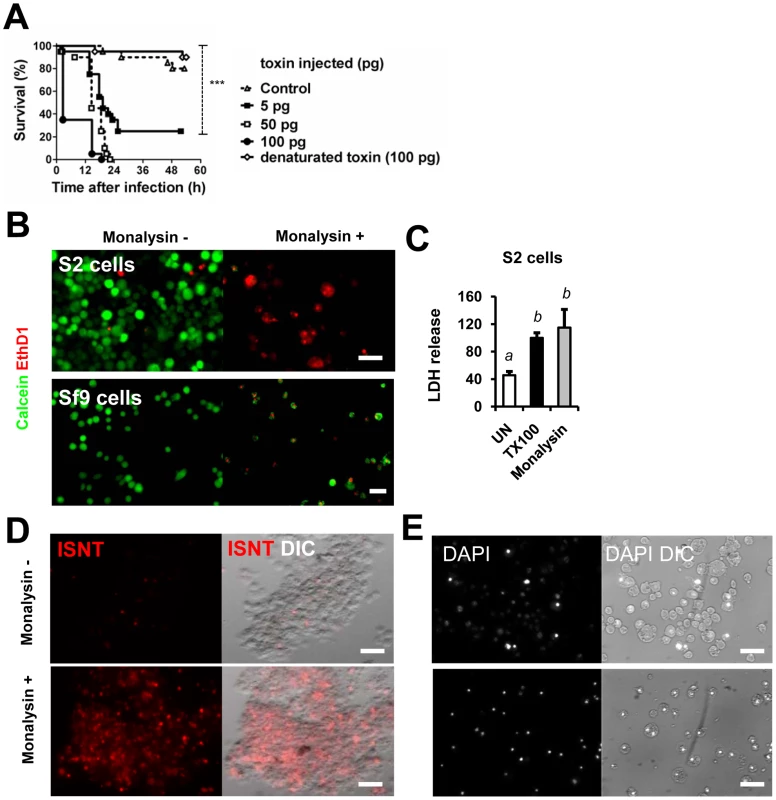
(A) Survival analysis of wild-type Oregon adult flies after injection of various quantities of Monalysin or heat-inactivated (denaturated) Monalysin. (B and C) Cytotoxic effect of Monalysin on insect culture cell line S2. (B) Drosophila S2 cells and Spodoptera frugiperda Sf9 cells were treated with Monalysin (Final concentration = 100nM) and stained with a live-dead viability reagent. Living cells are stained in green with Calcein while dead cells are stained in red with Ethidium homodimer 1 (EthD1, red). (C) The loss of viability was quantified by measuring the release of lactate dehydrogenase (LDH) from S2 cells. (D) DNA fragmentation in S2 cells was monitored by ISNT (in situ nick translation). (E) Chromatin condensation on untreated and Monalysin treated S2 cells was examined by DAPI staining. Phase-contrast and fluorescence views of the same microscopic fields are shown. (−) untreated cells, (+) = cells treated with Monalysin 100nM. Fig. 4. Monalysin hemolytic activity and cytotoxicity towards mammalian cells. 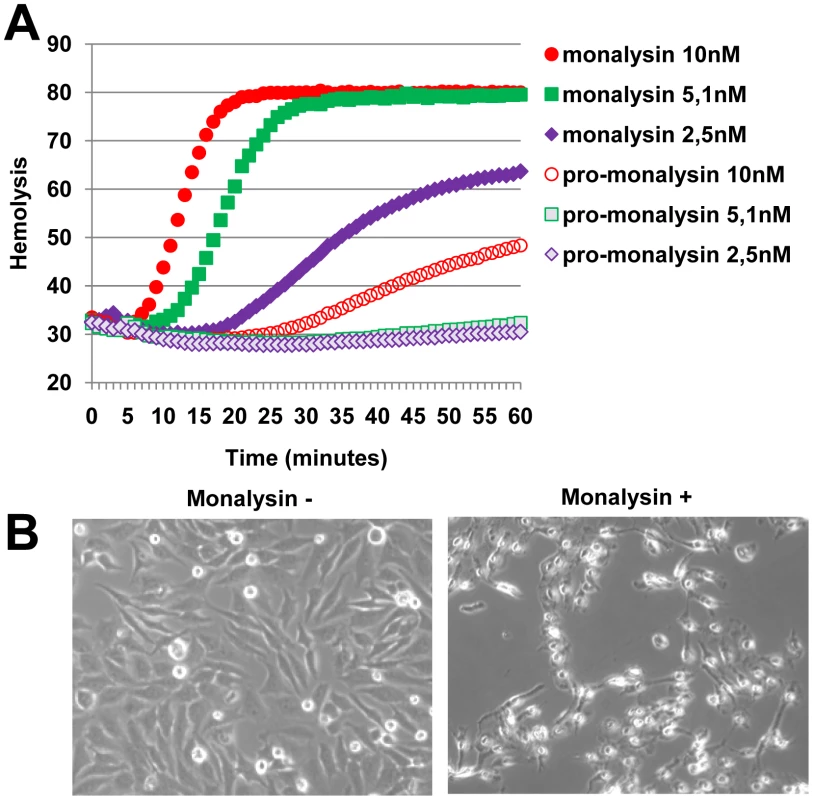
(A) Hemolytic activity was measured with pro and matured Monalysin incubated with red blood cells. The mature Monalysin was obtained by limited trypsinolysis of a fresh extract of recombinant Pro-Monalysin. (B) Phase contrast microscopy of Hela cells untreated or treated with Monalysin 100 nM for 24h. Cells were shrinking and displayed irreversible loss of adherence. Regulation and processing of Monalysin
P. entomophila virulence is controlled by several regulatory systems: i) the two component system GacS/GacA that functions at the post-transcriptional level, ii) a secreted signaling molecule produced by the pvf genes, and iii) AlgR that is known to control alginate production in other bacteria [5], [6], [7]. To determine which of these mechanisms regulate Monalysin production, crude cell extracts or filtered supernatants from wild-type and mutant P. entomophila were analyzed using a specific antiserum (Figure 5A, Figure S5). Monalysin was undetectable in both cells and medium of P. entomophila lacking the two-component system GacS/GacA and the pvf signaling molecule.
Fig. 5. Regulation and processing of Monalysin. 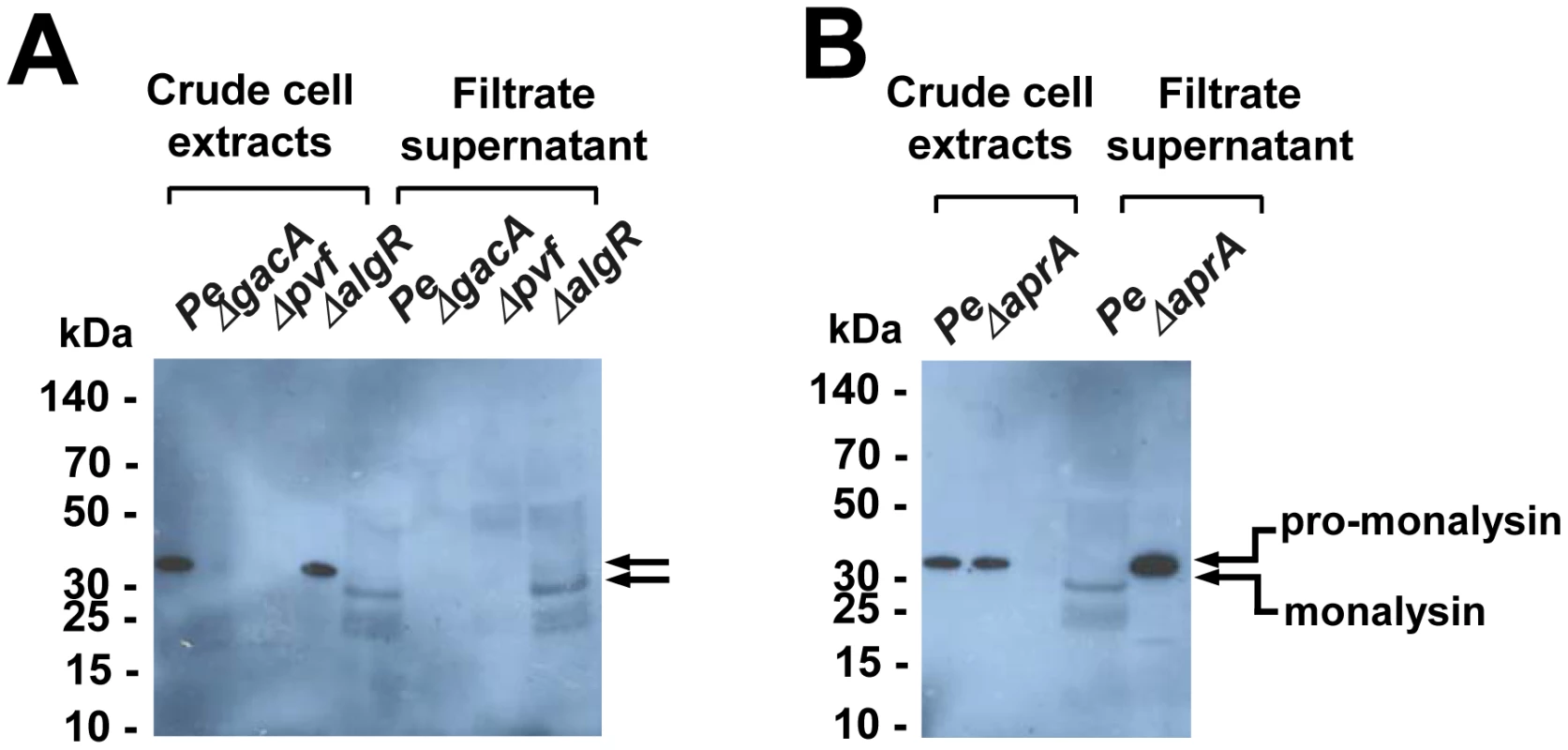
(A) Western-blot analysis of proteins from crude cell extracts or filtrate supernatants shows that Monalysin was not produced in gacA and pvf mutants, but was produced in the algR mutant. (B) Western-blot analysis of bacterial crude cell extracts and filtrated supernatants of Pe wt and ΔaprA shows that pro-monalysin is not processed in the supernatant of an AprA mutant. The stronger signal in the AprA mutant lane is due to the fact that the serum recognized better the pro-monalysin than the monalysin (see Figure S5). Interestingly, this analysis revealed that the supernatant of P. entomophila contained a shorter form of Monalysin when compared to the form detected in cell extracts. A N-terminal Edman sequencing of the shorter form found in culture supernatant (extracted from SDS gels) was performed, which revealed that the size shift was due to a cleavage taking place before Asparagine 34 (indicated in Figure 6A). Many toxins require a proteolytic activation, which can be performed by proteases produced by the bacterium itself or by enzymes of the host digestive tract [17]. Interestingly P. entomophila secretes large amounts of the metallo-protease AprA. To test whether AprA could be responsible for maturation of pro-Monalysin to Monalysin, we analyzed the supernatants of AprA-deficient and wild-type P. entomophila by Western blotting. Figure 5B shows that pro-Monalysin was found in supernatant derived from the aprA mutant while the mature form predominates in supernatant from wild-type P. entomophila.
Fig. 6. Monalysin is a ß pore-forming toxin. 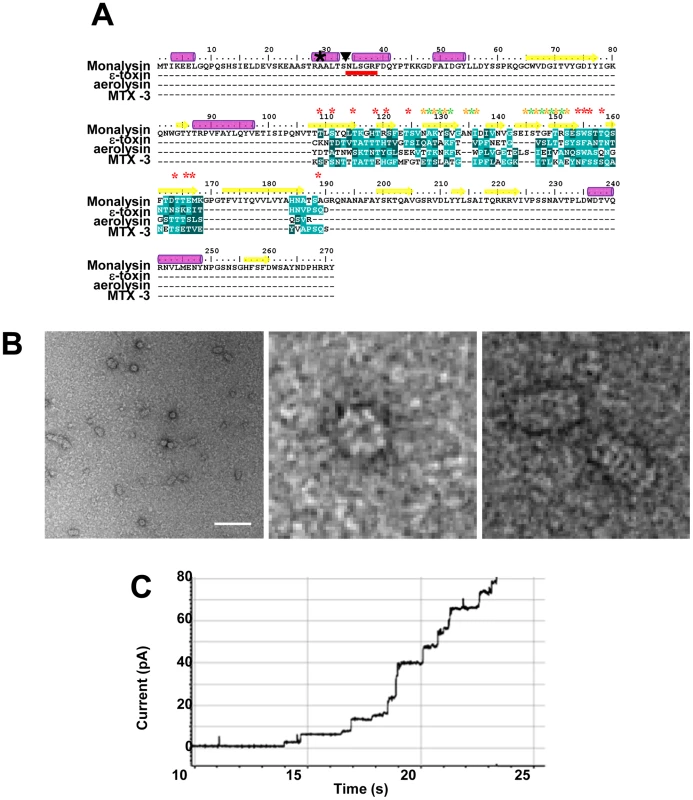
(A) Protein sequence analysis of Monalysin reveals an internal domain with amphipathic patches flanked by serine- and threonine-rich sequences that shares similarities with the membrane-spanning domain of ß-PFT (ε-toxin from Clostridium perfringiens, aerolysin from Aeromonas hydrophila and MTX-3 from Bacillus sphaericus). The multiple sequence alignment reveals the presence in P. entomophila Monalysin of putative membrane exposed residues (Yellow stars), solvent-exposed residues (green stars), and serine and threonine residues (red stars). A black star (★) shows the first amino acid detected by MALDI-TOF analysis of tryptic fragment of the recombinant Monalysin. The N-terminal residues of the mature Monalysin, identified by Edman sequencing, present in P. entomophila supernatant are underlined in red; a triangle (▾) indicates the potential cleavage site of pro-monalysin deduced from the N-terminal Edman sequencing. Purple cylinders indicate predicted α-helixes and yellow arrows indicate predicted β-sheets. (B) Scanning Electron micrographs show that Monalysin forms circular-like structures (top view) and barrel-like aggregates (side view). Scale bar represents 100 nm. (C) Monalysin (5 µg.ml) is able to form pores in a planar lipid bilayer. Collectively, our data indicate that Monalysin production is controlled by the global regulatory systems Gac and Pvf and that its N-terminus is cleaved by AprA upon secretion into the extracellular medium.
Monalysin is a novel ß-type pore-forming toxin
The Monalysin amino acid sequence does not show any homology to other sequences using P Blast, except for two uncharacterized orthologs found in Pseudomonas putida F1 strain (Figure S1). Neither the P. entomophila nor the P. putida gene products displayed any obvious protein domains. Nevertheless, the use of the HHpred software (Homology detection & structure prediction by HMM-HMM comparison) revealed the presence of an internal region with alternating polar and hydrophobic residues flanked by stretch of serine - and threonine residues, a hallmark of the membrane-spanning region of ß-barrel pore-forming toxins (Figure 6A). PFTs can be classified according to the secondary structure of their membrane-spanning region as α - and ß-PFTs. Far-UV circular dichroism analysis of Monalysin revealed a spectrum typical of structured proteins (Figure S2). The content of α-helixes and β-sheets was estimated to be 13% and 40%, respectively in agreement with the secondary structure prediction obtained with the program JPRED giving 17% of α-helixes and 35% of β-sheets as indicated in Figure 6A. This program also indicated that the putative membrane-spanning region of Monalysin was formed of a ß-sheet. This sequence analysis suggests that Monalysin is related to PFT of the ß-type.
ß-PFTs are synthesized as soluble proteins and have the ability to multimerize into circular polymers at high concentration, a step that for certain toxins, such as Aerolysin, requires proteolytic activation [18]. We next investigated whether Monalysin shared these properties with PFTs. SDS-PAGE analysis of a fresh recombinant Monalysin solution revealed a major band at the expected size (30 kDa) as well as several high molecular weight bands corresponding to oligomers that were resistant to SDS (see below). Interestingly, a shorter form of the protein was observed upon storage of samples at 4°C (Figure S3A). This together with the observation that Monalysin is matured by AprA indicates the existence of a protease sensitive site in the N-terminus part of Monalysin (Figure 5B). The cleavage of the recombinant pro-Monalysin into its shorter form could also be induced by a limited trypsinolysis (Figure S3B). This processed form has a molecular weight of 26.5 kDa as determined by MALDI-TOF analysis, as opposed to 30.2 kDa for the full-length pro-toxin. Interestingly, Monalysin had stronger hemolytic activity than pro-monalysin (Figure 4A), indicating that the removal of the N-terminal fragment constitutes a maturation step that enhances the cytotoxic activity. Processing of pro-monalysin to its mature form was accompanied by an increase and change in the higher order SDS-resistant complexes (Figure S3B).
While multiple size oligomers were observed by SDS-PAGE, a single species was observed by native PAGE analysis of Monalysin (Figure S3C). Multi-Angle Light Scattering analysis (MALS/UV/RI) confirmed the presence of a single species with a molecular mass of 546 kDa and hydrodynamic radius of 7.5 nm hence a diameter of 15 nm, which would correspond to about 18 monomers (Figure S4). This was further confirmed by electron microscopy of negatively stained recombinant Monalysin which showed circular like (top view) and barrel like structures (side view) similar to that observed with other ß-PFT (Figure 6B).
Sequence analysis of Monalysin, its ability to form ring like high order structures combined with its hemolytic activity strongly indicate that the toxin is a PFT. To address this issue directly, we analyzed its ability to form channels in planar lipid bilayers, an extremely sensitive electrophysiological method that enables the study of single-channel events. Addition of Monalysin led to a stepwise increase in membrane current, reflecting the formation of pores (Figure 6C). Collectively, our data show that P. entomophila Monalysin is a bona fide pore-forming toxin of the ß type.
Discussion
Many bacterial pathogens, both Gram-positive and Gram-negative, produce PFTs that contribute to their virulence [17]. Here we report the identification of a novel PFT that contributes to the virulence of P. entomophila against Drosophila. Our data show that Monalysin requires N-terminal cleavage to become fully active, forms oligomers in vitro, and induces pore formation in artificial lipid membranes. The prediction of the secondary structure of the membrane-spanning domain indicates that Monalysin is a PFT of the ß-type. Outside of this domain, Monalysin does not show any homology to any other PFT and appears rather different from previously identified insecticidal PFTs such as B. thuringiensis Cry toxins. Nevertheless, Monalysin has two homologs in the closely related P. putida F1 strain. These proteins could participate to the interaction of some Pseudomonas species with eukaryotic cells, defining a new family of PFTs.
We previously showed that P. entomophila virulence is multi-factorial and regulated by multiple signaling modules. Taking advantage of the genetic amenability of both the host and the pathogen, we aimed to identify P. entomophila and Drosophila pathways and effectors involved in the infectious process. Using this integrated approach, we previously proposed a role for the AprA metalloprotease in protection against antimicrobial peptides [7]. We now identify a second virulence factor, the ß-PFT Monalysin. Like AprA, a mnl mutant is affected in several, but not all, aspects of P. entomophila virulence. This attenuated virulence of the mnl mutant is clearly shown by survival analysis, which monitors the global outcome of infection. Our study indicates that Monalysin significantly contributes to the damage inflicted to intestinal cells by the bacterium, which is fully consistent with its activity as a pore-forming toxin. Supporting this notion, we observe that a mnl-deficient mutant induced less cell damage and a lower level of stress and repair pathway activity. The mnl mutant still induced a local immune response but the systemic immune response is drastically attenuated. This is also consistent with a role of Monalysin as a cytotoxin since activation of a systemic immune response is probably linked to damage of intestinal tract rendering possible the translocation of peptidoglycan, the bacteria elicitor activating the Imd pathway, from the lumen to the hemolymph compartment.
We also show that Monalysin production is regulated both by the GacS/GacA two component-system and the pvf genes. However, a mnl mutant still causes higher levels of stress and damage to the intestinal epithelium than gacA or pvf mutants. This indicates that these signaling modules regulate additional virulence factors contributing to P. entomophila cytotoxicity. Alternatively, it is possible that the overall cytotoxicity is caused by a synergy between the metalloprotease AprA and the PFT Monalysin, both of them being regulated by GacA. Along this line, we observed that AprA promotes the rapid cleavage of the pro-Monalysin into its active form. Since Monalysin can also be processed by trypsin, it is likely that AprA is not essential for PFT function of Monalysin in the Drosophila gut, as this toxin could also be processed by host enzymes. Both Monalysin and AprA are expressed in the algR mutant that affects a transcriptional regulator involved in alginate production as well as genes that are often associated to virulence (ie: pili biosynthesis, cyanide production…) [19]. The observation that an algR mutant is still avirulent (Vodovar 2005), although expressing both Monalysin and AprA, indicates the existence of additional virulence factors. Future studies should investigate at which level Pvf and GacS/GacA affect Monalysin production as well as identify other potential virulence factors regulated by the Pvf, Gac or AlgR.
Recent studies have shown that cells respond to PFTs by inducing repair and stress signal-transduction pathways to repair damage. Studies in C. elegans and mammalian cells have revealed a role for the P38 pathway, the unfolded protein response, and hypoxia in cellular resistance to the action of PFT [20]–[22]. The reduced expression of JAK-STAT and JNK pathway activities in guts infected with the mnl mutant indicate that Drosophila epithelial cells respond to PFT by activating stress and repair pathways. Thus, the P. entomophila/Drosophila interaction provides an interesting model to dissect the host response to PFTs in a natural infectious context.
Insects are potential reservoirs for microbes and are ideal vectors for their transmission due to their motility and their capacity to live in bacteria-rich environments [23]. This is exemplified by fruit flies that live in rotting fruits and are capable of transmitting phytopathogenic bacteria [24]. Insects are notably resistant to microbial infection allowing them to colonize these microbe-rich environments. This is largely due to the existence of very efficient physical barriers that block entry of microbes in the body cavity. As an illustration, injection of less than 10 cells of P. aeruginosa or Serratia marcescens in the body cavity rapidly kills flies, while high doses of these bacteria have only modest effects on survival when ingested [25]. In contrast to mammals, the gut of insects is lined with a chitinous matrix, the peritrophic matrix [26], that blocks the direct interaction between bacteria and epithelia cells and prevents the use of virulence devices such as type III and VI secretion systems that allow the injection of virulence factors directly into target cells. Rare bacterial species such as Photorhabdus luminescens can bypass this physical barrier since there are transported by symbiotic nematodes that can pierce the insect cuticle [27]. Other entomopathogens that enter through the oral route have to escape the local immune response and breach the gut barrier [23]. Despite the characterization of several virulence factors in few species, the mechanisms by which enteric pathogens kill insects remain poorly understood. This paper together with the well-characterized action of Bacillus thuringiensis cytotoxin Cry suggests that PFTs efficiently promote bacterial colonization of the insect gut [28]–[30]. This heavy artillery strategy does not require a direct contact between bacteria and host cells since PFTs can cross the pores of the peritrophic matrix and reach intestinal cells. PFTs can induce gut damage and rupture of intestinal homeostasis that in fine will lead to a weakening of the gut barrier and an inhibition of gut peristaltism promoting bacteria persistence. Gut damage and food uptake blockage are two symptoms of insect pathogenesis and could reflect the action of PFTs [23]. It would be interesting to know if other entomopathogens such as Serratia marcescens and Serratia entomophila also used PFT to colonize their insect host. In conclusion, this and other studies using different bacteria species contribute to uncovering strategies used by entomopathogens to breach insect barriers. A better knowledge of these strategies could also open the route to new methods of insect pests control.
Materials and Methods
Bacterial strains, media and antibiotics
P. entomophila L48 [4] was grown in LB for all experiments. P. entomophila mutated for the gacA, aprA, algR, and the pvf gene are described elsewhere [4], [5], [6], [7], [11]. The mnl deletion construct was generated by amplifying flanking regions of the monalysin gene (pseen3174 or mnl) by PCR. The resulting PCR product was cloned into the plasmid pEXG2. This plasmid was then used to create the strain Δ3174 (alternatively Δmnl), containing a deletion of the gene pseen3174. Complementation construct were made by cloning into the plasmid pPSV35 of PCR-amplified DNA fragments from P. entomophila containing the mutated genes. Pseudomonas Isolation agar (PIA, Difco) was used for selection after conjugations and persistence experiments. When E. coli was grown, antibiotics were used when necessary at the following concentrations: G418, 25 µg/ml and tetracycline, 5 µg/ml. When P. entomophila was grown, antibiotics were used when necessary at the following concentrations: gentamicin, 50 µg/ml for liquid cultures and 150 µg/ml for solid media, tetracycline 40 µg/ml and rifampicin, 30 µg/ml. The bacterial strains used in this study and the culture conditions are presented in Table S1. All primer sequences are available upon request. Insertion constructs were generated as previously described [6], [11].
Sequence analysis
DNA sequence searches and analysis were performed using the Pseudomonas genome database (www.pseudomonas.com). The monalysin gene (ORF PSEEN3174) corresponds to the accession number YP_608728.1 . Monalysin putative orthologs in Pseudomonas putida Pput_1063 and Pput_1064 correspond to the accessions numbers YP_001266408.1 , YP_001266409.1 respectively. The ORF PSEEN0535 involved in the production of the type VI secretion system corresponds to the accession number YP_606298.1 Monalysin amino-acids sequence analysis was performed using the HHpred software (Homology detection & structure prediction by HMM-HMM comparison http://toolkit.tuebingen.mpg.de/hhpred).
Fly stocks and infection assays
Oregon R flies were used as a standard wild-type strain and were maintained at 25°C. Adherens junctions were visualized using ubi-DE-cadherin-GFP flies [14], [31]. Upd3 expression in unchallenged gut and following infection, was monitored using upd3-Gal4, UAS-GFP flies (Buchon et al., 2009). Fly natural infections were carried out at 29°C on 4 - to 8 - day-old adult females as previously described. All the infections, except when specified, were carried out with bacterial preparation adjusted to an OD600 = 100 which correspond to 6.5E10 colony forming units per ml [11]. Monalysin was injected in the body cavity of fly using a Nanodrop microinjector (Nanoject). Virulence assays were performed at least three times in triplicate.
Reverse transcriptase quantitative PCR analysis
Total RNA was extracted from whole flies (5 for each assay) or from dissected guts without Malpighian tubules (14 for each assay) using TRIzol (Invitrogen). RT-qPCR was performed using SYBR Green I (Roche) on a Lightcycler 2.0 (Roche) as previously described [32]. Data represent ratio of the amount of mRNA detected normalized to the amount of the control rpl32 mRNA. Experiments were performed at least three times independently. Averages of more than three experiments are shown.
Cell cultures, treatments, cytotoxicity assays and live imaging
The macrophage-like lineage S2 cells derived from D. melanogaster embryos where grown in Schneider's medium (Invitrogen). The Sf9 cells (Invitrogen) derived from Spodoptera frugiperda (Lepidoptera) pupal ovarian tissue were cultured in complete TNM-FH (Invitrogen). The mammalian cell lines Hela and the Retinal Pigmented Epithelial (RPE1) were grown in a humidified incubator with 5% CO2 at 37°C. Hela cells were cultured in MEM media supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, 1% glutamine and 1% NEAA (Gibco). RPE1 cells were cultured in DMEM media supplemented with 10% fetal calf serum, 1% penicillin-streptomycin and 1% glutamine (Gibco). Cell viability was observed using the LIVE/DEAD Viability/Cytotoxicity Assay Kit (Invitrogen) according to the provider instruction. Briefly cells are simultaneously labeled with calcein AM that reveals intracellular esterase activity in live cells and ethidium homodimer (EthD-1) that reveals plasma membrane damages. LDH release from damaged cells was measured following the instructions of the CytoTox-One Homogeneous Membrane integrity Assay kit (Promega). In Situ Nick Translation was performed to detect fragmented DNA in nuclei. In situ DNA synthesis was performed by a DNA polymerase I (150 units/ml) (Takara) in the presence of a dNTP mix in which dUTP is tetramethylrhodamine-conjugated (Roche). The reaction was carried out for 90 min at room temperature. Live imaging and immunofluorescence were performed as previously described [8]. After treatment, cells were recovered, fixed with 4% PFA and permeabilized with 0.3% Triton X-100. Dead cells were detected using acridine orange staining (Invitrogen). Dead cells quantification was performed as follows: 16 hours after infection, guts were dissected and stained with acridine orange and DAPI. Pictures were taken using a fluorescent microscope. From these pictures, groups of 100 hundred DAPI stained nuclei were randomly defined and the number of acridine orange positive nuclei (ie dead cells) was determined. Three parcels per guts were analyzed. The results are the mean of four independent experiments. Nuclei were stained by DAPI (Sigma). All the images were performed using a Zeiss Axioimager Z1.
Monalysin expression, purification, and analysis
All cloning steps were performed as described earlier [33]. The sequence of Monalysin (from residue 1 to 271, access number pseen3174) was PCR-amplified from genomic DNA (isolated from P. entomophila) and cloned into pDONR201 (Invitrogen). The ORF was then subcloned into the pETG-20A E. coli (a generous gift from Dr A. Geerlof, EMBL) destination vector to generate a constructs encoding Monalysin with an N-terminal fusion composed of the thioredoxin (TRX) protein, followed by a 6xHis-tag and a Tobacco Etch Virus (TEV) protease cleavage site. The construct was sequenced verified. The production and purification were performed as described earlier [34]. Briefly, the pETG-20A-Monalysin was transformed into Rosetta (DE3) pLysS E. coli cells (Novagen). An overnight LB pre-culture (with 100 mg mL-1 ampicillin and 34 mg mL-1 chloramphenicol) was used to inoculate large cultures in ZYP-5052 auto-induction media [35] supplemented with the same antibiotics and incubated with vigorous shaking (250 rpm) at 37°C during 4 h. At this stage, the temperature was decreased to 17°C, and the cultures were allowed to grow for an additional 18 h with vigorous shaking (250 rpm). After 18 h, cells were harvested by centrifugation (4000g for 10 min) and the pellet was homogenized and frozen in lysis buffer (50 mM Tris-HCl; 500 mM NaCl; 0.5 mM lysozyme; 10 mM imidazole and 1 mM phenylmethylsulfonyl fluoride (PMSF), pH 8). The cell pellets were thawed and lysed with a sonicator after the addition of DNase I at 20 mg mL−1 and 1 mM MgSO4. The pellet and soluble fraction were separated by centrifugation (30 min at 16,000g) of an early stationary phase culture. The supernatants were filtered through a 0.22 µm filter and were concentrated 50-fold by using 5 kDa cutoff Centricon membranes (Biorad). The cell pellets were washed in PBS, resuspended in PBS containing protease inhibitors and lysed by sonication. The soluble fraction was purified by immobilized metal ion affinity chromatography using a 5 mL HisTrap crude (GE Healthcare) Ni2+-chelating column equilibrated in buffer A (500 mM NaCl; 50 mM Tris–HCl; 10 mM imidazole; pH 8). After the loading of the soluble fraction and a column wash (buffer A with 50 mM Imidazole), the protein was eluted with buffer A supplemented with 250 mM imidazole. The eluted fraction was desalted in buffer A (Hiprep 25/10 Desalting column, GE) and the protein concentration of the TRX-His6-TEV-Monalysin determined. After a 4°C overnight cleavage of the protein with 1∶20 w:w His-TEV protease, the TRX-His6-TEV and the His-TEV were separated from the pure Pro-Monalysin by collecting the Flow Through (FT) of a second Nickel purification. The final purification and the characterization of the oligomeric state of the monalysin were achieved by the separation of the FT on a size exclusion chromatography (HiLoad 16/60 Superdex 200 prep grade, GE), equilibrated in Tris 10mM, NaCl 500mM pH8. The pure Pro-Monalysin was used for the functional characterizations. For the MultiAngle Light Scattering analysis, size exclusion chromatography was carried out on an Alliance 2695 HPLC system (Waters) using a Silica Gel KW804 column (Shodex) equilibrated in 10 mM Tris and 150 mM NaCl at pH 7.5 at a flow of 0.5 ml/min. Detection was performed using a triple-angle light scattering detector (Mini-DAWN TREOS, Wyatt Technology), a quasi-elastic light scattering instrument (Dynapro, Wyatt Technology), and a differential refractometer (OptilabrEX, Wyatt Technology). Proteins were analyzed by SDS-PAGE. Native PAGE was performed to determine the oligomeric state of Monalysin. To generate Monalysin, pro-monalysin samples were submitted to limited trypsinolysis by adding trypsin (1∶100 w:w). The reaction was stopped by using a trypsin-chymotrypsin inhibitor (Invitrogen). Pro-monalysin and Monalysin were detected by Western-blot using a specific serum that recognized better the pro-monalysin than the Monalysin (see Figure S5).
Production of the antibody anti-monalysin
The Guinea pig antibody anti-Monalysin was provided by Eurogentec.
Circular dichroism, molecular weight and hydrodynamic radius determination
Far-UV Circular Dichroism (CD) spectra (Figure S2) were recorded with a JASCO J-810 spectropolarimeter (JASCO Corporation) equipped with a Peltier temperature control and using 1-mm thick quartz cells. The molecular weight of recombinant pro-monalysin and Monalysin was determined by MALDI-TOF/TOF. Molecular weight and hydrodynamic radius determination was performed by the ASTRA V software (Wyatt Technology). Proteins were loaded at a final concentration of 0.02 mM.
Edman sequencing
After SDS-PAGE electrophoresis and Coomassie blue staining, protein bands were excised. Proteins were extracted from gel and blotted onto polyvinylidene difluoride membranes with the ProSob system (Applied Biosystems). The N-terminal sequences of proteins were determined by automated Edman degradation by introducing the blots into a Procise P494 automated protein sequencer (Applied Biosystems). The sequences obtained were compared to sequences in public protein sequence databases.
Planar lipid bilayer
Planar lipid bilayer experiments were performed as previously described [36]. The bilayer was formed by painting a solution of 50% PC (egg lecithin) / 50% DOPE (w:w) in n -decane (40 mg ml −1) on an aperture (d = 150 µm, pretreated with the same solution) in a delrin cuvette separating two chambers, each containing 1 ml of 1 M NaCl, 5 mM CaCl2 10 mM HEPES, pH 7 and agar bridge connection (1 M KCl) to Ag/AgCl electrodes (Warner Instrument Corp. Hamden, CT). Monalysin was added to the cis chamber at room temperature.
Statistical analysis
Survival assays have been performed at least three times in triplicate. The Kaplan-Meier log rank test was used to determine statistical significance. Dashed brackets represent the significance between the different infections (*: p<0.05, **: p<0.01, ***: p<0.001, ns = not significant). RT-qPCR analysis and cell death quantification using acridine orange staining are averages of at least 4 independent experiments. Error bars indicate standard errors. Statistical analysis was performed using a Wilcoxon test, and letters indicate significantly different values (P<0.05).
Supporting Information
Zdroje
1. SansonettiPJ 2004 War and peace at mucosal surfaces. Nat Rev Immunol 4 953 964
2. SchneiderDSAyresJS 2008 Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol 8 889 895
3. HooperLVMacphersonAJ 2010 Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10 159 169
4. VodovarNVinalsMLiehlPBassetADegrouardJ 2005 Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci U S A 102 11414 11419
5. VodovarNVallenetDCruveillerSRouyZBarbeV 2006 Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol 24 673 679
6. Vallet-GelyIOpotaOBonifaceANovikovALemaitreB 2010 A secondary metabolite acting as a signalling molecule controls Pseudomonas entomophila virulence. Cell Microbiol 12 1666 1679
7. LiehlPBlightMVodovarNBoccardFLemaitreB 2006 Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog 2 e56
8. BuchonNBroderickNAChakrabartiSLemaitreB 2009 Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev 23 2333 2344
9. JiangHPatelPHKohlmaierAGrenleyMOMcEwenDG 2009 Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137 1343 1355
10. CroninSJNehmeNTLimmerSLiegeoisSPospisilikJA 2009 Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325 340 343
11. Vallet-GelyINovikovAAugustoLLiehlPBolbachG 2010 Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl Environ Microbiol 76 910 921
12. LapougeKSchubertMAllainFHHaasD 2008 Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67 241 253
13. PukatzkiSMcAuleySBMiyataST 2009 The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol 12 11 17
14. BuchonNBroderickNAKuraishiTLemaitreB 2010 Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol 8 152
15. AmcheslavskyAJiangJIpYT 2009 Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4 49 61
16. JiangHGrenleyMOBravoMJBlumhagenRZEdgarBA 2011 EGFR/Ras/MAPK Signaling Mediates Adult Midgut Epithelial Homeostasis and Regeneration in Drosophila. Cell Stem Cell 8 84 95
17. GonzalezMRBischofbergerMPernotLvan der GootFGFrecheB 2008 Bacterial pore-forming toxins: the (w)hole story. Cell Mol Life Sci 65 493 507
18. BischofbergerMGonzalezMRvan der GootFG 2009 Membrane injury by pore-forming proteins. Curr Opin Cell Biol 21 589 595
19. LizewskiSELundbergDSSchurrMJ 2002 The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect Immun 70 6083 6093
20. BellierAChenCSKaoCYCinarHNAroianRV 2009 Hypoxia and the hypoxic response pathway protect against pore-forming toxins in C. elegans. PLoS Pathog 5 e1000689
21. BischofLJKaoCYLosFCGonzalezMRShenZ 2008 Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathog 4 e1000176
22. WeiJZHaleKCartaLPlatzerEWongC 2003 Bacillus thuringiensis crystal proteins that target nematodes. Proc Natl Acad Sci U S A 100 2760 2765
23. Vallet-GelyILemaitreBBoccardF 2008 Bacterial strategies to overcome insect defences. Nat Rev Microbiol 6 302 313
24. BassetATzouPLemaitreBBoccardF 2003 A single gene that promotes interactions of a phytopathogenic bacterium with its insect vector, Drosophila melanogaster. EMBO Rep 4 205 209
25. NehmeNTLiegeoisSKeleBGiammarinaroPPradelE 2007 A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog 3 e173
26. HegedusDErlandsonMGillottCToprakU 2009 New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol 54 285 302
27. WaterfieldNRCicheTClarkeD 2009 Photorhabdus and a host of hosts. Annu Rev Microbiol 63 557 574
28. BravoAGillSSSoberonM 2007 Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49 423 435
29. BravoALikitvivatanavongSGillSSSoberonM 2011 Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem Mol Biol 41 423 431
30. SoberonMFernandezLEPerezCGillSSBravoA 2007 Mode of action of mosquitocidal Bacillus thuringiensis toxins. Toxicon 49 597 600
31. MaedaKTakemuraMUmemoriMAdachi-YamadaT 2008 E-cadherin prolongs the moment for interaction between intestinal stem cell and its progenitor cell to ensure Notch signaling in adult Drosophila midgut. Genes Cells 13 1219 1227
32. RomeoYLemaitreB 2008 Drosophila immunity : methods for monitoring the activity of toll and imd signaling pathways. Methods Mol Biol 415 379 394
33. VincentelliRBignonCGruezACanaanSSulzenbacherG 2003 Medium-scale structural genomics: strategies for protein expression and crystallization. Acc Chem Res 36 165 172
34. GraslundSNordlundPWeigeltJHallbergBMBrayJ 2008 Protein production and purification. Nat Methods 5 135 146
35. StudierFW 2005 Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41 207 234
36. IacovacheIPaumardPScheibHLesieurCSakaiN 2006 A rivet model for channel formation by aerolysin-like pore-forming toxins. EMBO J 25 457 466
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage EgressČlánek A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene ExpressionČlánek An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes byČlánek Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine ScrapieČlánek Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 9- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity
- Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
- Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development
- Nrf2, a PPARγ Alternative Pathway to Promote CD36 Expression on Inflammatory Macrophages: Implication for Malaria
- Robust Antigen Specific Th17 T Cell Response to Group A Streptococcus Is Dependent on IL-6 and Intranasal Route of Infection
- Targeting of a Chlamydial Protease Impedes Intracellular Bacterial Growth
- The Protease Cruzain Mediates Immune Evasion
- High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism
- Plague and Climate: Scales Matter
- Exhausted CD8 T Cells Downregulate the IL-18 Receptor and Become Unresponsive to Inflammatory Cytokines and Bacterial Co-infections
- Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles
- Murine Gamma-herpesvirus Immortalization of Fetal Liver-Derived B Cells Requires both the Viral Cyclin D Homolog and Latency-Associated Nuclear Antigen
- Rapid and Efficient Clearance of Blood-borne Virus by Liver Sinusoidal Endothelium
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene Expression
- Strain Specific Resistance to Murine Scrapie Associated with a Naturally Occurring Human Prion Protein Polymorphism at Residue 171
- Development of a Transformation System for : Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector
- Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
- Host Phylogeny Determines Viral Persistence and Replication in Novel Hosts
- BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity
- Expression of the RAE-1 Family of Stimulatory NK-Cell Ligands Requires Activation of the PI3K Pathway during Viral Infection and Transformation
- Structure of the Vesicular Stomatitis Virus N-P Complex
- HSV Infection Induces Production of ROS, which Potentiate Signaling from Pattern Recognition Receptors: Role for S-glutathionylation of TRAF3 and 6
- The Human Papillomavirus E6 Oncogene Represses a Cell Adhesion Pathway and Disrupts Focal Adhesion through Degradation of TAp63β upon Transformation
- Analysis of Behavior and Trafficking of Dendritic Cells within the Brain during Toxoplasmic Encephalitis
- Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance
- Multidrug Resistant 2009 A/H1N1 Influenza Clinical Isolate with a Neuraminidase I223R Mutation Retains Its Virulence and Transmissibility in Ferrets
- Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1
- Step-Wise Loss of Bacterial Flagellar Torsion Confers Progressive Phagocytic Evasion
- Complex Recombination Patterns Arising during Geminivirus Coinfections Preserve and Demarcate Biologically Important Intra-Genome Interaction Networks
- An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by
- Non-Lytic, Actin-Based Exit of Intracellular Parasites from Intestinal Cells
- The Fecal Viral Flora of Wild Rodents
- The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
- Interferon Regulatory Factor-1 (IRF-1) Shapes Both Innate and CD8 T Cell Immune Responses against West Nile Virus Infection
- A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
- Evaluating the Sensitivity of to Biotin Deprivation Using Regulated Gene Expression
- The Motility of a Human Parasite, , Is Regulated by a Novel Lysine Methyltransferase
- Phosphodiesterase-4 Inhibition Alters Gene Expression and Improves Isoniazid – Mediated Clearance of in Rabbit Lungs
- Restoration of IFNγR Subunit Assembly, IFNγ Signaling and Parasite Clearance in Infected Macrophages: Role of Membrane Cholesterol
- Protease ROM1 Is Important for Proper Formation of the Parasitophorous Vacuole
- The Regulated Secretory Pathway in CD4 T cells Contributes to Human Immunodeficiency Virus Type-1 Cell-to-Cell Spread at the Virological Synapse
- Rerouting of Host Lipids by Bacteria: Are You CERTain You Need a Vesicle?
- Transmission Characteristics of the 2009 H1N1 Influenza Pandemic: Comparison of 8 Southern Hemisphere Countries
- Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine Scrapie
- Sequential Bottlenecks Drive Viral Evolution in Early Acute Hepatitis C Virus Infection
- Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen
- Genomic and Proteomic Analyses of the Fungus Provide Insights into Nematode-Trap Formation
- Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
- Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
- Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate
- Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
- c-di-AMP Is a New Second Messenger in with a Role in Controlling Cell Size and Envelope Stress
- Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System
- APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells
- Impairment of Immunoproteasome Function by β5i/LMP7 Subunit Deficiency Results in Severe Enterovirus Myocarditis
- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Tri6 Is a Global Transcription Regulator in the Phytopathogen
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- The Next Opportunity in Anti-Malaria Drug Discovery: The Liver Stage
- Significant Effects of Antiretroviral Therapy on Global Gene Expression in Brain Tissues of Patients with HIV-1-Associated Neurocognitive Disorders
- Inhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
- A Novel Metal Transporter Mediating Manganese Export (MntX) Regulates the Mn to Fe Intracellular Ratio and Virulence
- Rhoptry Kinase ROP16 Activates STAT3 and STAT6 Resulting in Cytokine Inhibition and Arginase-1-Dependent Growth Control
- Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms
- Secretion of Genome-Free Hepatitis B Virus – Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
- Membrane Remodeling by the Double-Barrel Scaffolding Protein of Poxvirus
- A Diverse Population of Molecular Type VGIII in Southern Californian HIV/AIDS Patients
- Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A Virus Protease-Polymerase Processing Intermediate, 3CD
- Quantitative Analyses Reveal Calcium-dependent Phosphorylation Sites and Identifies a Novel Component of the Invasion Motor Complex
- Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes
- Old World Arenaviruses Enter the Host Cell via the Multivesicular Body and Depend on the Endosomal Sorting Complex Required for Transport
- Exploits a Unique Repertoire of Type IV Secretion System Components for Pilus Assembly at the Bacteria-Host Cell Interface
- Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



