-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
Pseudomonas aeruginosa is an important human pathogen that employs a vast arsenal of virulence factors and infects immunocompromised hosts, such as patients in intensive care units, causing pneumonia and other illnesses. Macrophages are cells in the first line of defense against pathogens in the lungs. After pathogen recognition, macrophages release pro-inflammatory cytokines to recruit other immune cells and employ a process known as oxidative burst to kill invading microbes. P. aeruginosa can counteract oxidative stress using antioxidant proteins, such as peroxiredoxins. We show here that LsfA, which belongs to the poorly characterized Prx6 subfamily of peroxiredoxins, is indeed endowed with a thiol-dependent activity that is required for full virulence. In vitro and in vivo infection models confirmed that LsfA peroxidase activity is required for the immunomodulation caused by P. aeruginosa and that its absence allows the host to overcome the infection. This study demonstrates for the first time the involvement of a bacterial Prx6 in virulence.
Published in the journal: Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence. PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004442
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004442Summary
Pseudomonas aeruginosa is an important human pathogen that employs a vast arsenal of virulence factors and infects immunocompromised hosts, such as patients in intensive care units, causing pneumonia and other illnesses. Macrophages are cells in the first line of defense against pathogens in the lungs. After pathogen recognition, macrophages release pro-inflammatory cytokines to recruit other immune cells and employ a process known as oxidative burst to kill invading microbes. P. aeruginosa can counteract oxidative stress using antioxidant proteins, such as peroxiredoxins. We show here that LsfA, which belongs to the poorly characterized Prx6 subfamily of peroxiredoxins, is indeed endowed with a thiol-dependent activity that is required for full virulence. In vitro and in vivo infection models confirmed that LsfA peroxidase activity is required for the immunomodulation caused by P. aeruginosa and that its absence allows the host to overcome the infection. This study demonstrates for the first time the involvement of a bacterial Prx6 in virulence.
Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium that can cause diseases in several hosts [1]. P. aeruginosa acute infections are one of the major problems in immunodeficient subjects, burn victims and mechanical ventilation patients. Pulmonary infections caused by P. aeruginosa, including ventilator-associated pneumonia and chronic pulmonary infection in cystic fibrosis patients, are associated with high mortality rates, and chronic pulmonary infection impairs life quality and life expectancy of the infected individuals [2]–[4]. The high intrinsic resistance of P. aeruginosa to antibiotics adds to the difficulties of treating infections caused by this versatile opportunist [5].
Macrophages are the first line of defense in pulmonary infections and play a major role in the host response to P. aeruginosa infections. Pathogens are recognized by the immune system, which detects pathogen-associated molecular patterns (PAMPs) by the corresponding pattern recognition receptor. The activation of signaling cascades by the binding of PAMPs to Toll-like receptors (TLRs), except for TLR-3, depends on MyD88 and results in the activation of the NF-κB and MAPK pathways [6]; their activation leads to the production of cytokines, including TNF-α, IL-6 and IL-1. The role of TLR-4 and TLR-5 in triggering a protective immunity against P. aeruginosa has been shown in vivo; as mice lacking TLR-4/5 have increased susceptibility to pulmonary infections [7], [8].
After the recognition of bacteria by the TLRs in macrophages, the signaling cascade leads to the generation of reactive oxygen species (ROS) in a process known as the oxidative burst, which depends on NADPH oxidase [9]. The oxidative burst is bactericidal and can cause lipid, protein and DNA lesions, resulting in pathogen clearing. However, to overcome or prevent these lesions, pathogens have developed a complex detoxification system that includes superoxide dismutase, and catalase/peroxidases that have been extensively studied in several pathogens, including P. aeruginosa [10], [11].
Among hydroperoxide-reducing enzymes, the peroxiredoxins (Prxs) are considered cellular sensors due to their abundance and reactivity [12]. Prxs catalyze the reaction ROOH+2e−→ROH+H2O and reduce hydrogen peroxide, peroxynitrite and a wide range of organic hydroperoxide compounds [13]–[15]. Prxs are found in organisms belonging to all Domains of life, indicating their crucial physiological function, but their role in P. aeruginosa virulence remains uncharacterized.
Prxs are a large family of proteins that can be divided into six sub-groups with distinct amino acid sequences, but all contain the thioredoxin fold and the PXXT(S)XXC motif [16]. Among these six-subgroups, Prx enzymes can display 2-Cys Prx or 1-Cys-Prx mechanisms, depending on the number of cysteine residues involved in catalysis [16]. AhpC, a 2-Cys Prx, is involved in the virulence of Helicobacter cinaedi and Staphylococcus aureus [17], [18], but it does not seem to be a virulence determinant for other bacteria that have been analyzed [19]–[21].
The genome of the highly virulent P. aeruginosa strain PA14 contains at least eight genes that encode Prxs, including AhpC and Tpx. Ohr is another Cys-based peroxidase from P. aeruginosa that has been structurally and enzymatically characterized [22], but it is not required for virulence [23].
Among the six Prx sub-groups, Prx6 is the least well studied. The precise physiological roles of the Prx6 sub-group remain unknown, with few reports addressing their kinetics and structural functions; most of these reports were from studies of eukaryotes [24]–[29]. Thus far, all of the Prx6 proteins characterized display the 1-Cys Prx mechanism. Remarkably, although the bacterial Domain contains hundreds of Prx6 representatives [16], no characterization of their roles has been reported.
In the P. aeruginosa genome, only one gene coding for a putative Prx6 is present (PA14_19490 in PA14; PA3450 in PA01). It was named lsfA because its expression is up-regulated, together with a gene cluster coding for an ABC-transport system involved in organic sulfur uptake, in cells grown in low-sulfate medium [30]. However, there is no experimental evidence for the mechanism underlying the function of LsfA in this process. It has been suggested that LsfA up-regulation and AhpC expression in low sulfate conditions may be a response to the oxidative stress caused by the excess levels of reduced flavin nucleotides due to sulfonate utilization [31]. Several transcriptomic analyses have revealed that lsfA expression is up-regulated in other stressful conditions, including in the presence of sodium hypochlorite [32], a product of the macrophage oxidative burst. Proteomic analyses identified LsfA as differentially expressed during other stressful conditions. Three LsfA isoforms are induced by the superoxide-generating drug paraquat [33] and in P. aeruginosa biofilms [34]. In iron starvation conditions, Pseudomonas putida also showed increased levels of the LsfA ortholog protein [35]. Interestingly, indole treatment, which may mimic conditions of iron abundance, decreased lsfA expression as well as virulence-related traits [36]. A more recent report found that the oxidation responsive OxyR activator protein binds to the lsfA promoter region [37]; this observation supports the role of LsfA in the bacterial response to H2O2.
Here, we show that the antioxidant function of the bacterial 1-Cys Prx LsfA is important for P. aeruginosa virulence, both in a macrophage model in vitro and in an acute pneumonia model in vivo. This work reveals the role of this protein as a novel virulence factor that contributes to the P. aeruginosa arsenal against host defenses and allows it address other stresses in various environmental conditions.
Results
LsfA is an active peroxiredoxin and is important for the antioxidant response
A previous sequence alignment revealed that LsfA belongs to the Prx6 subfamily [16], with the Cys45 of LsfA as the putative peroxidasic cysteine (Fig. S1). To determine whether P. aeruginosa LsfA is indeed endowed with thiol-dependent peroxidase activity, the recombinant wild-type (His-LsfA) protein and a mutant protein without the putative catalytic cysteine (His-C45A) were expressed in Escherichia coli and purified by affinity chromatography. As predicted, H2O2 was reduced in the presence of wild-type His-LsfA but not when His-C45A was employed (Fig. 1A), showing that Cys45 is essential for catalysis and confirming that LsfA is an active 1-Cys Prx.
Fig. 1. LsfA confers PA14 resistance to hydrogen peroxide and reduces it in vitro. 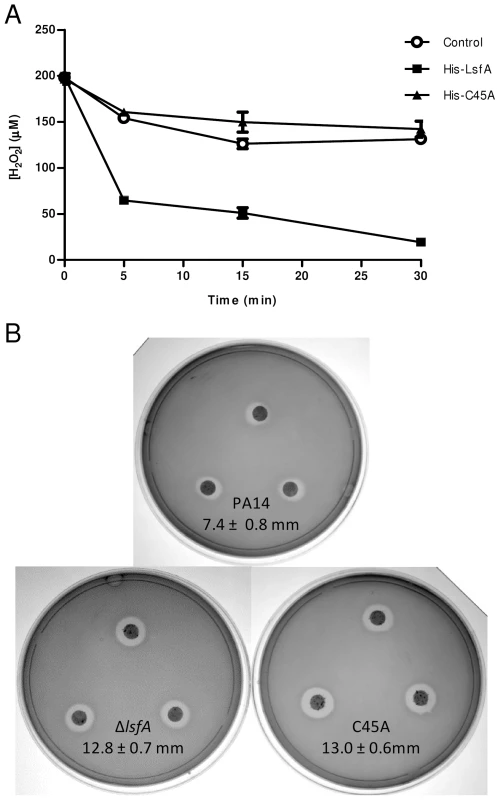
(A) The recombinant proteins His-LsfA and His-C45A were incubated with 200 µM H2O2. The amount of H2O2 remaining in the reaction mixtures was determined by ferric-thiocyanate assays at the indicated time points. In a control assay, H2O2 was incubated with buffer without recombinant proteins. The data are the means ± SD from at least three independent experiments performed in triplicate. (B) Bacteria were grown to an OD600 nm of 1 and spread on LB plates. A 6 mm filter disk containing 2.5% H2O2 was placed on top of the bacterial lawn, and the plates were incubated at 37°C for 16 hours. The data represent 1 of 5 independent experiments and the diameters of the inhibition haloes are shown as the means ± SD (mm). Because 2-Cys Prxs play an important role in bacterial protection against H2O2, tert-butyl hydroperoxide and paraquat [13], [14], we assessed whether LsfA was also important for P. aeruginosa resistance to such oxidants. To test this hypothesis, an in-frame deletion mutant strain (ΔlsfA) and a strain with a point mutation in the catalytic cysteine (C45A) were constructed. Both mutant strains grow like wild-type in minimal medium and in biofilms, excluding any growth defects (Fig. S2). The wild-type, ΔlsfA and C45A strains were tested using disk diffusion halo assays in the presence of oxidants. Larger inhibition haloes due to H2O2 were observed for both mutant strains (12.8±0.7 mm for ΔlsfA, 13.0±0.6 mm for C45A) compared with PA14 (7.4±0.8 mm for PA14) (Fig. 1B), indicating that LsfA is important for oxidative stress resistance in P. aeruginosa and confirming that the C45 catalytic cysteine is essential for LsfA activity. Complementation of the mutant strains with a copy of the lsfA gene in a plasmid restored the wild-type phenotype (Fig. S3), confirming that the larger haloes were due to a lack of LsfA activity. The lsfA mutants did not demonstrate increased sensitivity to paraquat or tert-butyl hydroperoxide (data not shown), which may reflect a compensatory effect of other Prxs and/or Ohr.
LsfA is required for the inhibition of macrophage activation by P. aeruginosa
Because macrophages and neutrophils produce ROS and reactive nitrogen species in response to pathogens, bacterial antioxidant systems are important mechanisms that allow bacteria to overcome the deleterious effects of oxidative lesions and to survive during infection. To assess whether the P. aeruginosa LsfA is related to virulence, an in vitro model of infection in J774 macrophages was used. The macrophages were infected with PA14 and the lsfA mutants, and incubated for 1 hour prior to the gentamicin addition. At regular time points, the number of bacterial cells remaining in the culture supernatants was assessed (Fig. 2A), the macrophages were washed and lysed, and the released bacteria were counted (Fig. 2B). The phagocytosed bacteria counts were similar for all strains and showed a slight increase over time (Fig. 2B), suggesting that all of the strains were internalized by macrophages at the same extent. However, for the lsfA mutants, a 2-fold reduction in extracellular colony-forming units (CFUs) compared with the wild-type PA14 was observed (Fig. 2A). Thus, the peroxidase activity of LsfA contributes to bacterial viability in the presence of macrophages. The number of remaining macrophages at the end of the assay, assessed by a LDH release assay, was similar for all bacterial strains tested, showing that LsfA does not alter cytotoxicity (Fig. S4).
Fig. 2. The lsfA mutants are phagocytosed at the same rate as PA14, but do not survive as well as PA14 in the cultures supernatants. 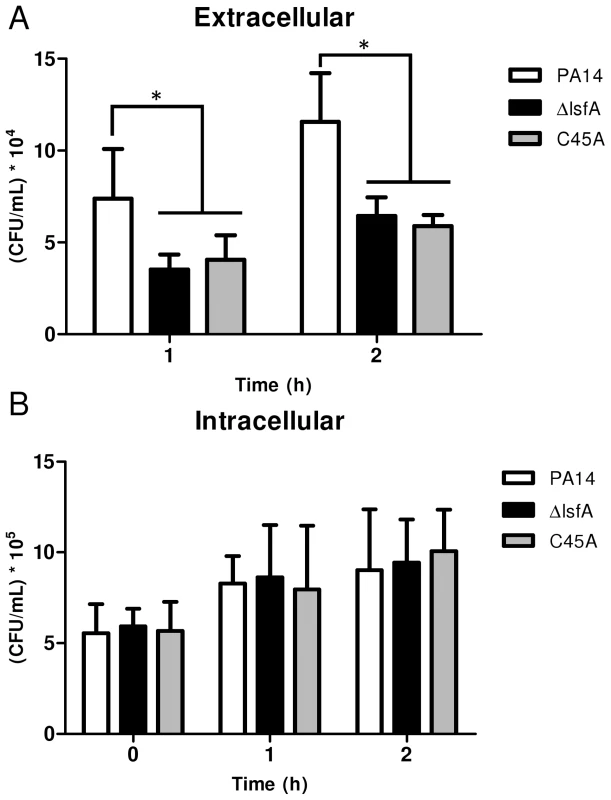
J774 macrophages were incubated with P. aeruginosa PA14 or the ΔlsfA or C45A mutants at an MOI of 10. (A) At the indicated time points, the supernatants were collected and diluted, and the number of CFU was determined. (B) The cells were washed with PBS and R-10 containing 200 µg/mL gentamicin was added to the wells. At the indicated time points, the macrophages were lysed with Triton X-100. The released bacteria were diluted, and number of CFU was determined. The data shown are the means ± SD from at least three independent experiments performed in triplicate. *, p<0.05. The increased survival of PA14 compared with the lsfA mutants may reflect a change in the oxidative status of the macrophages in addition to an improved bacterial response to the oxidative burst. To address this hypothesis, macrophages were incubated with wild-type or mutant strains, and at different times, the cells were washed and incubated with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), a probe that can sense shifts in the cellular redox state. After 3 hours of treatment, an increase in the intracellular oxidative status in macrophages incubated with the lsfA mutant strains was observed, in comparison with macrophages infected with PA14 (Fig. 3A). This result suggests that LsfA participates in the response of P. aeruginosa to the oxidative insult caused by the macrophages, most likely due to its peroxidase activity. Because LsfA affects the macrophage redox status, it can also impact virulence by subverting the host signaling pathways [38]. The mutant for gacA, which encodes a protein that plays a role in P. aeruginosa pathogenicity, has been extensively studied [39] and caused a similar increase in the macrophage oxidative state (Fig. 3A), suggesting that LsfA may indirectly inhibit the oxidative burst.
Fig. 3. LsfA downregulates macrophages oxidative state that protected the bacteria against NADPH oxidase-generated ROS, and inhibits TNF-α production via the MAPK and NF-κB pathways. 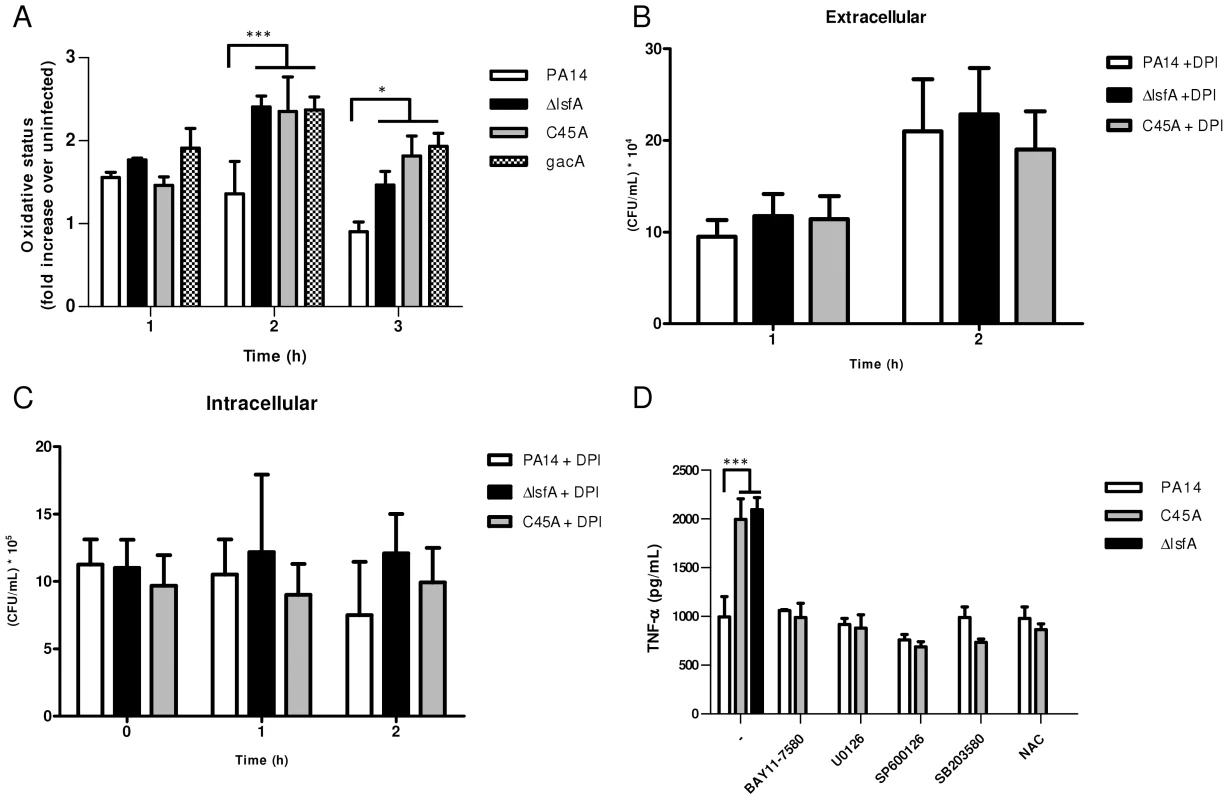
(A) J774 cells were infected with PA14 or the ΔlsfA, C45A, or gacA mutants. After 1, 2 or 3 hours, the macrophages were washed with PBS and incubated for 15 min with H2DCFDA. Fluorescence was analyzed by FACS. (B) Macrophages were treated with DPI prior to infection. The supernatants were collected and diluted at the indicated time points, and the number of CFU was determined. (C) Cells were washed with PBS and R-10 containing 200 µg/mL gentamicin was added to the wells. At the indicated time points, the macrophages were lysed with Triton X-100. The released bacteria were diluted and number of CFU was determined (D) Macrophages were infected with PA14 or the lsfA mutants in the absence (-) or the presence of inhibitors of NF-κB (BAY11-7580), ERK (U0126), JNK (SP600125) and p38 (SB2035-80) or the antioxidant N-acetylcysteine (NAC). After 3 hours of infection, the supernatants were recovered, and TNF-α secretion was determined by ELISA. Data are the means ± SD from three independent experiments performed in triplicate. *, p<0.05; ***, p<0.001. To evaluate the role of ROS generated by NADPH oxidase in the clearance of lsfA mutant strains, a gentamicin exclusion assay was performed again with macrophages, now pre-treated with DPI, a NADPH oxidase inhibitor. The CFU number corresponding to the bacteria that survive inside the DPI-treated macrophages is higher than in untreated ones (Fig. 2B and 3B), suggesting that they are no longer able to kill the bacteria. Moreover, the number of PA14 or the mutant cells under DPI treatment were similar both intra and extracellular, suggesting that LsfA counteracted NADPH oxidase activity during PA14 evasion from the macrophages (Fig. 3B and C). To understand whether P. aeruginosa LsfA modulates macrophage activation, J774 cells were infected with PA14 or the lsfA mutant strains. The supernatants were recovered after 3 hours, and cytokine levels were measured. Macrophages infected with the lsfA mutants secreted more TNF-α than macrophages infected with PA14 (Fig. 3D). This result suggests that the LsfA function decreases macrophage activation and, thus, its oxidative state because the oxidative burst is less pronounced when macrophages are infected with the wild-type strain.
C45A-infected macrophages treated with the thiol-reductant N-acetylcysteine (NAC) showed a reduced oxidative state (Fig. S5) and lower TNF-α production, similar to that of PA14-infected macrophages either in the presence or absence of NAC (Fig. 3D). Both NAC and LsfA may prevent macrophage activation by decreasing the oxidative state of phagocyte and thereby subverting signaling pathways involved in the immunological response.
Indeed, ROS can act as signaling molecules that lead to NF-κB and MAPK activation and increased cytokine production [40]. To determine which signaling pathways are up-regulated when the macrophages are infected with lsfA mutant strains, specific inhibitors of NF-κB or the MAPKs ERK1/2, JNK or p38 were used. In the presence of these inhibitors, no differences in TNF-α production were observed when macrophages were infected with either PA14 or C45A strains (Fig. 3D). These data suggest that LsfA peroxidase activity in PA14 decreases the activation of the NF-κB and MAPK pathways, and this decreased activation is reflected in the lower oxidative state of macrophages.
LsfA is essential for P. aeruginosa full virulence in vivo
Because LsfA is required for virulence in macrophages in vitro, the next step was to ascertain whether this 1-Cys Prx belonging to Prx6 group was also relevant in an acute pneumonia model in mice. Although all mice infected intratracheally (i.t.) with PA14 were dead 48 hours after infection, mice infected with lsfA strains had higher survival rates, with 37.5% of animals still alive after 13 days (Fig. 4A). After 60 days, surviving mice that had previously been infected with ΔlsfA (37.5%) and C45A (25%) seemed healthy, indicating that the infection had resolved or became chronic. In conclusion, the requirement of LsfA activity for P. aeruginosa virulence was confirmed.
Fig. 4. LsfA plays an important role in virulence in an acute pneumonia model. 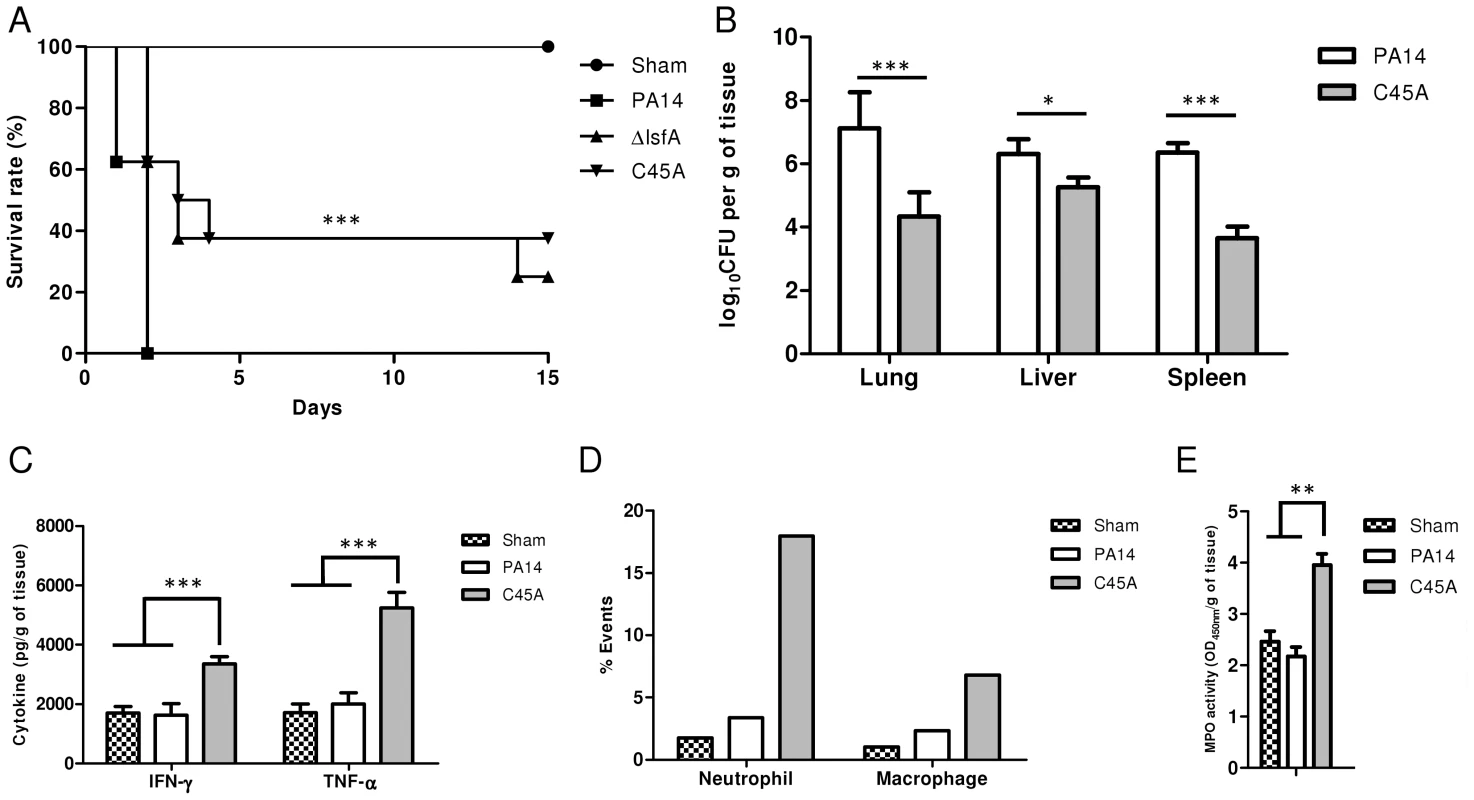
(A) BALB/c mice were infected i.t. with 2×106 bacteria per mouse with wild-type strain PA14 or the ΔlsfA or C45A mutants (n = 8 per group). Control mice were inoculated with PBS (n = 3) and the survival of the mice was followed for the course of the experiment. (B) At 24 hours post-infection, three animals were sacrificed, the organs were macerated, and the bacterial CFU were enumerated. (C) The cytokines in the lungs were determined by ELISA. (D) Mice were infected with PA14 or the C45A mutant. At 24 hours post-infection, the animals were euthanized, and the lungs were macerated. The cell suspensions were labeled for macrophages (F4/80+; CD11b+) or neutrophils (Ly6G/Ly6C+; F4/80−) and analyzed by FACS. (E) Mice were infected with PA14 or the C45A mutant. At 24 hours post-infection, the animals were euthanized, and the lungs were macerated, the cells were lysed by sonication and the suspension was centrifuged. The resulting supernatant was used for MPO activity assay with TMB., and OD was measured at 450 nm. The data are representative of three (B, C) or two (D, E). independent experiments independent experiments *, p<0.05; **, p<0.01; ***, p<0.001. P. aeruginosa introduced i.t. into mice disseminates quickly and affects other organs, leading to death [8]. Bacterial loads were assessed in the primary site of infection as well as in secondary organs (liver and spleen) as an indication of sepsis. Lungs, spleen and liver were recovered 24 hours p.i., and bacterial counts were evaluated. Animals infected with the C45A mutant strain showed a reduced bacterial burden in all organs analyzed compared with PA14-infected animals (Fig. 4B). This result indicates an improved bacterial resolution in mice infected with the C45A strain and demonstrates the relevance of the 1-Cys Prxs to virulence.
To ascertain whether LsfA would impact cytokine production in vivo, thus potentially decreasing the immune system activation and neutrophil recruitment to the infection site, the TNF-α and IFN-γ levels in the lungs of infected animals were determined by ELISA at 24 hours p.i. The wild-type strain induced local immunosuppression, with TNF-α and IFN-γ release similar to that observed in the control mice (Fig. 4C). However, mice infected with C45A released higher levels of these cytokines (Fig. 4C). No differences were observed in the anti-inflammatory cytokine IL-10 in mice infected with either PA14 or the lsfA mutant strain compared to the control (Fig. S6). In addition, C45A-infected mice showed an increased recruitment of neutrophils (Ly6G/Ly6C+; F4/80−) and macrophages (F4/80+; CD11b+) to the lungs compared with the controls (Fig. 4D). To determine if the recruited neutrophils were activated, myeloperoxidase (MPO) activity was present in all treatments, but it was higher when mice were infected with the C45A mutant strain (Fig. 4E), suggesting that thiol peroxidase activity is also involved in neutrophil activation. This set of data confirms that a better immune response that is reflected in the higher survival of the lsfA mutant-infected animals compared to those infected with PA14.
Discussion
In this study, we show for the first time the relevance of the 1-Cys Prx LsfA in bacterial virulence. LsfA, a Prx from P. aeruginosa, is the first protein in the Prx6 sub-group to be connected to pathogenicity. This protein, LsfA, has orthologues in other pathogens, including Burkholderia and Bordetella, suggesting that this host-pathogen interaction may be present in other bacteria. Biochemical analysis demonstrated that LsfA has peroxidase activity that depends on the catalytic cysteine (C45). Inactivating this peroxidase activity makes lsfA mutant cells more sensitive than wild-type cells to hydrogen peroxide but not to organic peroxides, including tert-butyl-hydroperoxide. In addition, LsfA may also be endowed with other enzymatic activity because LsfA displays a conserved lipase motif (GDSWG) that is also present in the Prx6 from humans and mice. Curiously, the lipase motif is not conserved in all 1-Cys Prxs, and a deeper analysis of the evolution and function of 1-Cys Prxs is in progress. However, the lipase motif is not required for virulence in the mice macrophage and lung models we used here, as a point mutation in the catalytic cysteine was sufficient to abrogate the virulence to the same extent as when the entire lsfA coding region was deleted from the P. aeruginosa chromosome. Prx proteins have been implied in several diseases in humans. The 1-Cys Prdx6 has been characterized as a tumor inhibitor because it protects mice and human skin cells against lipid peroxidation [41], and the levels of this protein are lower in papillary thyroid carcinomas than in normal thyroid tissue [42]. Prx6 may also be involved in degenerative neuronal disorders, including Alzheimer's disease and prion diseases [43], [44]. In mice infected i.t. with P. aeruginosa, LsfA directly or indirectly downregulates the host innate immune response, enabling the pathogen to spread and colonize other organs, leading to an acute infection that results in death. In the macrophage infection model, LsfA is required for P. aeruginosa resistance to clearance, with lower TNF-α production in macrophages infected with the PA14 strain compared with macrophages infected with lsfA mutants, suggesting that LsfA plays a role in the PA14 immunomodulatory effect. This immunomodulation seems to be related to the macrophages' oxidative state, which is higher when the macrophages are infected with lsfA mutant strains than when they are infected with the wild-type PA14. We also found that NADPH oxidase activity is required for the clearance of the lsfA mutant, indicating that LsfA is important for PA14 resistance to ROS generated by macrophages. Other P. aeruginosa virulence factors also promote immunomodulation. ExoU, a secreted phospholipase, inhibits caspase-1 activation, which is related to pro-IL-1β maturation [45]. The quinolones HHQ and PQS can also negatively regulate the immune system, reducing NF-κB activation, bacterial clearance and TNF-α and IL-6 production [46]. Nevertheless, this is the first evidence of a Prx exerting an immunomodulatory function, protecting the pathogen against phagocytes, reducing phagocyte activation and leading to increased bacterial virulence. We also found that the oxidation of H2DCFDA was higher in macrophages infected with lsfA mutant strains than in macrophages infected with PA14. In support to our findings, LPS stimulated macrophages, carrying a knockout of the 2-Cys PrxII gene, released more pro-inflammatory cytokines, including TNF-α and IL-6, than wild-type macrophages. This increase in cytokine release is correlated with higher ROS production by the PrxII knockout macrophages, leading to activation of the MAPK and NF-κB pathways [47]. In this study, we show that the lack of a bacterial Prx seems to have the same effect as the lack of PrxII in macrophages, and we speculate that a balance between ROS production and turnover might be necessary to allow the outcome of a host-pathogen interaction to swing from an efficient immune response to virulence, favoring either the host or the bacteria.
In addition to bactericidal activity, ROS can also act as signaling molecules, but the mechanisms involved are poorly understood. Further studies are required to investigate how oxidative stress leads to NF-κB and MAPK activation. However, it appears that bacterial antioxidants play a role in protecting microorganisms from oxidative insult [38] and subvert signaling pathways such as those involved in the immunological response [48].
With the increasing resistance of pathogens to antibiotics, it is crucial to explore new paradigms to develop novel anti-infective drugs, taking advantage of a deeper understanding of bacterial virulence and host defense mechanisms. One aspect that may be explored to achieve this goal is the ROS sensing and production that is employed by both pathogens and hosts. Understanding the role of LsfA in the P. aeruginosa immunomodulatory effect may lead to novel therapeutics to overcome the effects of infection by targeting LsfA itself or by improving the host immune response.
Methods
Bacterial strains, plasmids, oligonucleotides and culture conditions
All of the strains and plasmids used in this study are listed in Supporting Table S1. The P. aeruginosa strains were grown at 37°C in LB broth. The E. coli strains were grown in LB supplemented with 100 µg/mL of ampicillin, 50 µg/mL of kanamycin, or 10 µg/mL of gentamicin, when required. The P. aeruginosa strains were grown in 250 µg/mL of kanamycin, 20 µg/mL nalidixic acid or 30 µg/mL of gentamicin, when required.
To construct the unmarked in-frame deletion of lsfA, primers flanking the upstream and downstream regions of lsfA were designed. Amplicons were cloned into pNPTS138 at the HindIII and EcoRI sites to generate pNPTS138ΔlsfA. The resulting construct was used to introduce the lsfA deletion into the wild-type PA14 genome by homologous recombination [49], resulting in the ΔlsfA mutant, that contains only the first eight N-terminal aminoacids and 31 aminoacids at the LsfA C-terminus. No polar effect is expected, because lsfA coding region is distant 249 bp from the next open reading frame in the PA14 genome and the frame of translation was maintained.
The oligonucleotide-directed mutagenesis of the LsfA Cys45 to Ala was performed using the primer pairs listed in Supporting Table S2, and a two-step procedure was performed as previously described [50]. The resulting amplicon was cloned into pNPTS138 and introduced into the PA14 genome by homologous recombination. Mutant clones were screened by PCR followed by digestion or direct sequencing. Again, no polar effects are anticipated, because only one codon was changed.
To construct the lsfA complementation strains, the lsfA coding region was amplified by PCR using the primer pairs listed in the Supporting Table S2. The resulting amplicon was cloned into pJN105 at the EcoRI and SpeI sites to generate pLsfA. pLsfA and pJN105 plasmids were introduced into the ΔlsfA and the C45A mutants, generating the ΔlsfA/pJN105, ΔlsfA/pLsfA, C45A/pJN105 and C45A/pLsfA strains.
Protein expression and purification
The lsfA or lsfAC45A coding regions were cloned into pProEX-Hta to overexpress His-LsfA or His-LsfAC45A in E. coli BL21. Briefly, E. coli cultures were grown in 250 mL of LB at 37°C until the culture reached an OD600 nm of 0.5. IPTG was added to a final concentration of 0.6 mM, and the cultures were grown at 30°C for 6 h. Cells were harvested by centrifugation and resuspended in 25 mL lysis buffer (20 mM sodium phosphate pH 7.4, 500 mM NaCl, 20 mM imidazole, 1 mM PMSF). Cell suspensions were lysed by ten 15 second sonication cycles in an ice bath. The lysate was centrifuged at 16000 g for 20 min at 4°C. The recombinant proteins His-LsfA and His-LsfAC45A were purified using an Ni-NTA column (Invitrogen) equilibrated with lysis buffer and eluted with an imidazole gradient (20–1000 mM). The eluted fractions were analyzed by SDS-PAGE and pooled, and the buffer was exchanged using a PD-10 column (GE) equilibrated with incomplete reaction buffer (20 mM sodium phosphate pH 7.4, 500 mM NaCl). The proteins were concentrated with Centriprep-Ultracel YM-10.000 MWCO (Millipore). The protein concentration was determined by UV spectroscopy using an extinction coefficient of 33920 M−1 cm−1, calculated as described by Gill and von Hippel [51], and confirmed using the Bradford reagent (Sigma-Aldrich) according to the manufacturer's protocol.
Peroxidase activity determination
The in vitro peroxidase activity was determined using the ferric-thiocyanate assay [15]. Briefly, 10 µM of purified recombinant protein was incubated at 37°C in 100 µL of reaction buffer (20 mM sodium phosphate pH 7.4, 500 mM NaCl, 1 mM DTT, 10 µM DTPA, 10 µM sodium azide) in the presence or absence of 200 µM H2O2. At the times indicated, the reaction was stopped by the addition of 20 µL of 2 M HCl and incubated for 10 min at 37°C. Then, 100 µL of 2.5 M KSCN, 100 µL of 20 mM FeSO4 and 680 µL of H2O were added to the reaction. The absorbance was read at 480 nm. As controls, we used the same conditions in the absence of DTT or without recombinant protein. After the reaction times, the H2O2 concentration was determined by comparison with a standard curve with different H2O2 concentrations (15.625–1000 µM).
Disk diffusion halo assays
Halo inhibition assays were performed as described before with modifications [52]. Cultures of P. aeruginosa PA14 and the ΔlsfA and C45A mutant and complemented strains were grown overnight in LB broth. Cultures were diluted into fresh LB broth to an OD600 nm of 0.1 and grown to an OD600 nm of 1.0. Plate assays were performed by adding 200 µL of a cell culture to 3 mL of 0.7% LB soft agar. The agar suspensions were spread on LB plates. Sterile paper disks (6 mm in diameter) were saturated with 10 µL of 2.5% hydrogen peroxide and placed on the plates, which were incubated for 16 hours at 37°C. For the complementation assays, 30 ug/mL gentamicin (to maintain the plasmids) and 0.2% arabinose (to induce expression from the ara promoter) were added into the LB.
Cell culture
The macrophage cell line J774 was maintained in R-10 (RPMI 1640 supplemented with 2 mM glutamine, 10% fetal bovine serum (FBS) and 40 µg/mL gentamicin) at 37°C in 5% CO2. Macrophages were counted using a Neubauer chamber, and dead cells were excluded by the trypan blue exclusion assay. Macrophages were seeded at 1×105 cells/well (96-well plates) or 2×106 (6-well plates) in R-10 without antibiotic and primed overnight with 10 ng/mL IFN-γ at 37°C in 5% CO2.
In vitro infection experiments
PA14 and the ΔlsfA and C45A mutant strains were grown in LB broth to an OD600 nm of 2.0. The bacteria were diluted in R-10 without antibiotics. Macrophages that had been previously seeded in 96-well plates were infected at a multiplicity of infection (MOI) of 10. After 1 hour of infection, the supernatant was removed and 200 µg/mL of gentamicin was added to cell cultures to kill extracellular bacteria. After 30 minutes the cells were washed once with PBS and fresh media were added. This point was defined as time 0 (t = 0 h). To determine the numbers of extracellular bacteria, the supernatant was collected, serially diluted, the cells were plated and the CFU were enumerated. To access intracellular bacteria, macrophages were lysed with PBS+0.1% Triton X-100, the lysates were serially diluted, and the CFU were determined. Cytotoxicity was measured for samples taken at the indicated times by quantifying the release of lactate dehydrogenase (LDH) using the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega) following the manufacturer's instructions. For NADPH oxidase inhibition, macrophages were treated with 10 µM DPI (diphenylene iodonium) 4 hours before infection and the same steps described above were performed. To quantify TNF-α, macrophages were seeded in six-well plates and infected at an MOI of 10. For inhibition experiments, where indicated, cells were pretreated for 4 hours before infection with an NF-κB inhibitor (10 µM BAY11-7085) or MAPK inhibitors of ERK1/2 (1 µM U0126), p38 (1 µM SB203580), or JNK (1 µM SP600125); alternatively, the cells were treated with 2 mM NAC. At 3 hours post-infection, the supernatants were removed, centrifuged and stored at −20°C. Cytokine quantification was performed by ELISA (R&D systems) following the manufacturer's instructions.
Oxidative state evaluation
The macrophage oxidative state was determined as described previously [53]. Briefly, macrophages were pre-treated with or without 2 mM NAC for 4 hours before infection. Macrophages were infected with PA14 or with the ΔlsfA, C45A or gacA::tn mutant strains at an MOI of 10. After the indicated time in post-infection culture medium, the cells were washed with PBS and then incubated with H2DCFDA (Invitrogen) at a 2.5 µM final concentration for 30 min at 37°C. Cells were washed with warmed PBS (37°C), resuspended in cold PBS containing 1% FBS and analyzed by fluorescence-activated cell sorting (FACS). Unstained controls were treated similarly. For the baseline fluorescence control, macrophages were uninfected but stained according to the above procedure. The mean fluorescence intensity values were calculated by dividing the values of the infected macrophages by those of the uninfected control.
Ethics statement
The animal experiments were performed in agreement with the Ethical Principles in Animal Research adopted by the Conselho Nacional de Controle da Experimentação Animal (CONCEA) and in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Internal Animal Care and Use Committee of the Instituto de Química, Universidade de São Paulo (N°08/2012).
Animals
Female BALB/c mice (8–12 weeks old) were obtained from the in-house animal facility (Biotério de Produção e Experimentação da Faculdade de Ciências Farmacêuticas e do Instituto de Química da Universidade de São Paulo). Mice were kept on a 12/12-h light/dark cycle with free access to food and water and were maintained under specific pathogen-free conditions. All mice were euthanized in CO2 chamber, and every effort was made to minimize suffering.
Animal inoculation with bacteria
PA14 and the ΔlsfA and C45A mutant strains were used for i.t. inoculation as described before [54], with few modifications. Bacteria were grown as described above, harvested by centrifugation at 12000 g for 2 min, washed twice in sterile PBS and resuspended in PBS at a concentration of 2×106 bacteria. The CFU/mL were validated by plating serial dilutions of the suspensions. Each mouse received 60 µL of a bacterial suspension. A ketamine/xylazine mixture was injected i.p. to anesthetize the mice before surgery. A midventral incision was made, and the trachea was exposed. The bacterial suspension was inoculated i.t. Controls were inoculated i.t. with 60 µL sterile PBS.
In vivo CFU determination
At 24 hours after infection, the lungs, spleen and liver were harvested for CFU and cytokine measurements. The tissues were homogenized in 1 mL PBS for the lung and spleen and in 2 mL PBS for the liver. The supernatants were collected, and the CFUs were assayed by serial dilution and plating on LB plates. For cytokine measurements, the lung tissues were homogenized, and the supernatants were centrifuged at 12000 g for 10 min at 4°C. The cytokines TNF-α, IFN-γ and IL-10 were quantified by ELISA (R&D systems), following the manufacturer's instructions.
FACS
At 24 hours after infection, the lungs were harvested, minced and digested with collagenase for 30 min at 37°C. The RBCs were lysed by adding NH4Cl lysing buffer. The cells were resuspended in PBS with 3% FBS and stained with different combinations of conjugated antibodies, including F4/80-PECy5 (BM8), CD11c-FITC (HL3), CD11b-PE (M1/70) and Ly6G/Ly6C-APC (RB6-8C5), followed by incubation for 20 min on ice. Finally, the cells were washed and resuspended for flow cytometry analysis. FlowJo software (Tree Star) was used to analyze the data.
Myeloperoxidase activity assay
The myeloperoxidase activity assay was performed as previously described with a few modifications [55], [56]. The animals were infected as described above, and the lungs were harvested, lysed mechanically in the presence of 50 mM sodium phosphate pH 5.4, 5 mM EDTA and 0.5% cetyltrimethylammonium bromide, ultrasonicated and centrifuged. The supernatant (50 µL) was mixed with an equal volume of 3 mM 3,3′,5,5′-tetramethylbenzidine dihydrochloride (TMB) for 2 minutes. The reaction was stopped by the addition of 25 µL of 2M H2SO4. The optical density (OD) was measured at 450 nm.
Survival
After i.t. infection with wild-type PA14 or the ΔlsfA or C45A strains, the treated (N = 8) and control (N = 3) groups were observed for survival. All deaths reported were from moribund/euthanized mice. Mice with labored or rapid breathing, decreased motility, ruffled or abnormal-looking fur or other obvious signs of distress were considered to be moribund as described before [57].
Statistical analyses
Prism 5 (GraphPad Inc.) was used for all statistical analyses. Kaplan-Meier survival curves were plotted, and significance was calculated using the log-rank test. The data were compared using the one-way or two-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison test.
Supporting Information
Zdroje
1. RahmeLG, StevensEJ, WolfortSF, ShaoJ, TompkinsRG, et al. (1995) Common virulence factors for bacterial pathogenicity in plants and animals. Science 268 : 1899–1902.
2. Crouch BrewerS, WunderinkRG, JonesCB, LeeperKVJr (1996) Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest 109 : 1019–1029.
3. GaynesR, EdwardsJR (2005) Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis 41 : 848–854.
4. WilliamsBJ, DehnbostelJ, BlackwellTS (2010) Pseudomonas aeruginosa: host defence in lung diseases. Respirology 15 : 1037–1056.
5. DrenkardE (2003) Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect 5 : 1213–1219.
6. KawaiT, AkiraS (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunol 11 : 373–384.
7. FeuilletV, MedjaneS, MondorI, DemariaO, PagniPP, et al. (2006) Involvement of Toll-like receptor 5 in the recognition of flagellated bacteria. Proc Natl Acad Sci U S A 103 : 12487–12492.
8. RamphalR, BalloyV, JyotJ, VermaA, Si-TaharM, et al. (2008) Control of Pseudomonas aeruginosa in the lung requires the recognition of either lipopolysaccharide or flagellin. J Immunol 181 : 586–592.
9. PawateS, ShenQ, FanF, BhatNR (2004) Redox regulation of glial inflammatory response to lipopolysaccharide and interferongamma. J Neurosci Res 77 : 540–551.
10. IiyamaK, ChiedaY, LeeJM, KusakabeT, Yasunaga-AokiC, et al. (2007) Effect of superoxide dismutase gene inactivation on virulence of Pseudomonas aeruginosa PAO1 toward the silkworm, Bombyx mori. Appl Environ Microbiol 73 : 1569–1575.
11. LeeJS, HeoYJ, LeeJK, ChoYH (2005) KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infect Immun 73 : 4399–4403.
12. WinterbournCC, HamptonMB (2008) Thiol chemistry and specificity in redox signaling. Free Radic Biol Med 45 : 549–561.
13. BrykR, GriffinP, NathanC (2000) Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407 : 211–215.
14. ChristmanMF, MorganRW, JacobsonFS, AmesBN (1985) Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41 : 753–762.
15. NettoLES, ChaeHZ, KangSW, RheeSG, StadtmanER (1996) Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. TSA possesses thiol peroxidase activity. J Biol Chem 271 : 15315–15321.
16. NelsonKJ, KnutsonST, SoitoL, KlomsiriC, PooleLB, et al. (2011) Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Proteins 79 : 947–964.
17. CharoenlapN, ShenZ, McBeeME, MuthupalaniS, WoganGN, et al. (2012) Alkyl hydroperoxide reductase is required for Helicobacter cinaedi intestinal colonization and survival under oxidative stress in BALB/c and BALB/c interleukin-10−/ − mice. Infect Immun 80 : 921–928.
18. CosgroveK, CouttsG, JonssonIM, TarkowskiA, Kokai-KunJF, et al. (2007) Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol 189 : 1025–1035.
19. RankinS, LiZ, IsbergRR (2002) Macrophage-induced genes of Legionella pneumophila: protection from reactive intermediates and solute imbalance during intracellular growth. Infect Immun 70 : 3637–3648.
20. SpringerB, MasterS, SanderP, ZahrtT, McFaloneM, et al. (2001) Silencing of oxidative stress response in Mycobacterium tuberculosis: expression patterns of ahpC in virulent and avirulent strains and effect of ahpC inactivation. Infect Immun 69 : 5967–5973.
21. TaylorPD, InchleyCJ, GallagherMP (1998) The Salmonella typhimurium AhpC polypeptide is not essential for virulence in BALB/c mice but is recognized as an antigen during infection. Infect Immun 66 : 3208–3217.
22. LesniakJ, BartonWA, NikolovDB (2002) Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J 21 : 6649–6659.
23. AtichartpongkulS, FuangthongM, VattanaviboonP, MongkolsukS (2010) Analyses of the regulatory mechanism and physiological roles of Pseudomonas aeruginosa OhrR, a transcription regulator and a sensor of organic hydroperoxides. J Bacteriol 192 : 2093–2101.
24. LiS, PetersonNA, KimMY, KimCY, HungLW, et al. (2005) Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys peroxiredoxin. J Mol Biol 346 : 1035–1046.
25. LoumayeE, Ferrer-SuetaG, AlvarezB, ReesJF, ClippeA, et al. (2011) Kinetic studies of peroxiredoxin 6 from Arenicola marina: rapid oxidation by hydrogen peroxide and peroxynitrite but lack of reduction by hydrogen sulfide. Arch Biochem Biophys 514 : 1–7.
26. MizohataE, SakaiH, FusatomiE, TeradaT, MurayamaK, et al. (2005) Crystal structure of an archaeal peroxiredoxin from the aerobic hyperthermophilic crenarchaeon Aeropyrum pernix K1. J Mol Biol 354 : 317–329.
27. MonteiroG, HortaBB, PimentaDC, AugustoO, NettoLE (2007) Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci U S A 104 : 4886–4891.
28. PedrajasJR, PadillaCA, McDonaghB, BarcenaJA (2010) Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae. Antioxid Redox Signal 13 : 249–258.
29. SarmaGN, NickelC, RahlfsS, FischerM, BeckerK, et al. (2005) Crystal structure of a novel Plasmodium falciparum 1-Cys peroxiredoxin. J Mol Biol 346 : 1021–1034.
30. HummerjohannJ, KuttelE, QuadroniM, RagallerJ, LeisingerT, et al. (1998) Regulation of the sulfate starvation response in Pseudomonas aeruginosa: role of cysteine biosynthetic intermediates. Microbiology 144 (Pt 5): 1375–1386.
31. KerteszMA (2000) Riding the sulfur cycle–metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol Rev 24 : 135–175.
32. SmallDA, ChangW, ToghrolF, BentleyWE (2007) Comparative global transcription analysis of sodium hypochlorite, peracetic acid, and hydrogen peroxide on Pseudomonas aeruginosa. Appl Microbiol Biotechnol 76 : 1093–1105.
33. HareNJ, ScottNE, ShinEH, ConnollyAM, LarsenMR, et al. (2011) Proteomics of the oxidative stress response induced by hydrogen peroxide and paraquat reveals a novel AhpC-like protein in Pseudomonas aeruginosa. Proteomics 11 : 3056–3069.
34. PatrauchanMA, SarkisovaSA, FranklinMJ (2007) Strain-specific proteome responses of Pseudomonas aeruginosa to biofilm-associated growth and to calcium. Microbiology 153 : 3838–3851.
35. HeimS, FerrerM, HeuerH, RegenhardtD, NimtzM, et al. (2003) Proteome reference map of Pseudomonas putida strain KT2440 for genome expression profiling: distinct responses of KT2440 and Pseudomonas aeruginosa strain PAO1 to iron deprivation and a new form of superoxide dismutase. Environ Microbiol 5 : 1257–1269.
36. LeeJ, AttilaC, CirilloSL, CirilloJD, WoodTK (2009) Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol 2 : 75–90.
37. WeiQ, MinhPN, DotschA, HildebrandF, PanmaneeW, et al. (2012) Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40 : 4320–4333.
38. MelilloAA, BakshiCS, MelendezJA (2010) Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J Biol Chem 285 : 27553–27560.
39. GooderhamWJ, HancockRE (2009) Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33 : 279–294.
40. GwinnMR, VallyathanV (2006) Respiratory burst: role in signal transduction in alveolar macrophages. J Toxicol Environ Health B Crit Rev 9 : 27–39.
41. RolfsF, HuberM, GruberF, BöhmF, PfisterHJ, et al. (2013) Dual role of the antioxidant enzyme peroxiredoxin 6 in skin carcinogenesis. Cancer Res 73 : 3460–3469.
42. NicolussiA, D'InzeoS, MincioneG, BuffoneA, Di MarcantonioMC, et al. (2014) PRDX1 and PRDX6 are repressed in papillary thyroid carcinomas via BRAF V600E-dependent and -independent mechanisms. Int J Oncol 44 : 548–556.
43. SizovaD, CharbautE, DelalandeF, PoirierF, HighAA, et al. (2007) Proteomic analysis of brain tissue from an Alzheimer's disease mouse model by two-dimensional difference gel electrophoresis. Neurobiol Aging 28 : 357–370.
44. WagnerW, ReuterA, HüllerP, LöwerJ, WesslerS (2012) Peroxiredoxin 6 promotes upregulation of the prion protein (PrP) in neuronal cells of prion-infected mice. Cell Commun Signal 10 : 38.
45. SutterwalaFS, MijaresLA, LiL, OguraY, KazmierczakBI, et al. (2007) Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med 204 : 3235–3245.
46. KimK, KimYU, KohBH, HwangSS, KimSH, et al. (2010) HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-kappaB pathway. Immunology 129 : 578–588.
47. YangCS, LeeDS, SongCH, AnSJ, LiS, et al. (2007) Roles of peroxiredoxin II in the regulation of proinflammatory responses to LPS and protection against endotoxin-induced lethal shock. J Exp Med 204 : 583–594.
48. BaxtLA, Garza-MayersAC, GoldbergMB (2013) Bacterial subversion of host innate immune pathways. Science 340 : 697–701.
49. SimonR, PrieferU, PuhlerA (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (N Y) 1 : 784–790.
50. KongW, ChenL, ZhaoJ, ShenT, SuretteMG, et al. (2013) Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol microbiol 88 : 784–797.
51. GillSC, von HippelPH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182 : 319–326.
52. PassalacquaKD, BergmanNH, Herring-PalmerA, HannaP (2006) The superoxide dismutases of Bacillus anthracis do not cooperatively protect against endogenous superoxide stress. J Bacteriol 188 : 3837–3848.
53. WestAP, BrodskyIE, RahnerC, WooDK, Erdjument-BromageH, et al. (2011) TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472 : 476–480.
54. CaiS, BatraS, WakamatsuN, PacherP, JeyaseelanS (2012) NLRC4 inflammasome-mediated production of IL-1beta modulates mucosal immunity in the lung against gram-negative bacterial infection. J Immunol 188 : 5623–5635.
55. Veliz RodriguezT, MoalliF, PolentaruttiN, ParoniM, BonavitaE, et al. (2012) Role of Toll interleukin-1 receptor (IL-1R) 8, a negative regulator of IL-1R/Toll-like receptor signaling, in resistance to acute Pseudomonas aeruginosa lung infection. Infect Immun 80 : 100–109.
56. PulliB, AliM, ForghaniR, SchobS, HsiehKL, et al. (2013) Measuring myeloperoxidase activity in biological samples. PLoS One 8: e67976.
57. KohAY, PriebeGP, RayC, Van RooijenN, PierGB (2009) Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect Immun 77 : 5300–5310.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



