-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
The causative agent of tuberculosis, Mycobacterium tuberculosis (Mtb), persists in phagosomes inside infected macrophages. Mtb expresses lipoarabinomannan (LAM), which inhibits fusion of phagosomes with lysosomes as a means for Mtb to evade host defense. LAM is present in the cell envelope, which surrounds Mtb and interfaces with the host, but its localization remains unclear. We show that LprG, an Mtb lipoprotein, binds LAM and controls its distribution in the cell envelope. A mutant strain of Mtb that lacks LprG has less LAM at the surface of the cell envelope. This decreases LAM-mediated inhibition of phagosome-lysosome fusion, thereby impairing an immune evasion mechanism. We propose that LprG facilitates transfer of LAM from the plasma membrane into the cell envelope, enhancing its interaction with the host and ability to regulate host defense. Our results reveal mechanisms that determine bacterial cell envelope function and influence host-pathogen interactions and pathogen evasion of host defense.
Published in the journal: Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion. PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004471
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004471Summary
The causative agent of tuberculosis, Mycobacterium tuberculosis (Mtb), persists in phagosomes inside infected macrophages. Mtb expresses lipoarabinomannan (LAM), which inhibits fusion of phagosomes with lysosomes as a means for Mtb to evade host defense. LAM is present in the cell envelope, which surrounds Mtb and interfaces with the host, but its localization remains unclear. We show that LprG, an Mtb lipoprotein, binds LAM and controls its distribution in the cell envelope. A mutant strain of Mtb that lacks LprG has less LAM at the surface of the cell envelope. This decreases LAM-mediated inhibition of phagosome-lysosome fusion, thereby impairing an immune evasion mechanism. We propose that LprG facilitates transfer of LAM from the plasma membrane into the cell envelope, enhancing its interaction with the host and ability to regulate host defense. Our results reveal mechanisms that determine bacterial cell envelope function and influence host-pathogen interactions and pathogen evasion of host defense.
Introduction
Tuberculosis is the second leading cause of death from an infectious disease worldwide (http://www.who.int/mediacentre/factsheets/fs104/en/index.html). The causative agent, Mycobacterium tuberculosis (Mtb), is an intracellular bacterial pathogen that persists in phagosomes of infected macrophages. Mtb expresses a thick, waxy cell envelope of low permeability that contributes to antibiotic resistance and contains components (e.g. glycolipids, lipoglycans and lipoproteins) that play critical roles in regulating host responses and promoting survival of the pathogen [1]–[9]. Greater understanding of tuberculosis pathogenesis is needed as a foundation for development of drugs or vaccines to prevent or treat tuberculosis. Cell envelope components and the enzymes and transporters that control their synthesis and assembly serve as both determinants of pathogenesis and attractive targets for drug design.
The cell envelope of Mtb is rich in lipids and carbohydrates [1]–[3], [10], including lipoglycans such as lipoarabinomannan (LAM) and lipomannan (LM), and the glycolipid phosphatidyl-myo-inositolmannosides (PIMs). The PIMs include a core acylated glycolipid structure that is incorporated into the more highly glycosylated lipoglycans, LM and LAM, which are synthesized on the on the periplasmic face of the plasma membrane from PIM precursors by the elongation of mannan and arabinan chains [11]. LAM is essential for Mtb survival; the synthesis of LAM and cell envelope arabinans is targeted by the anti-mycobacterial agent ethambutol and DprE1 inhibitors currently under development [12]. LAM inhibits Mtb phagosome-lysosome fusion, providing a mechanism for Mtb evasion of host defense [13]–[18]. This effect is specific to ManLAM, the mannose capped LAM found in slow growing strains that are pathogenic in humans or other host species (e.g. Mtb, M. bovis BCG, M. leprae), and is not produced by phospho-myo-inositol-capped LAM (PI-LAM) found in M. smegmatis and M. fortuitum [13]. Recent studies have identified key steps in the biogenesis of LAM [1], [19], but the mechanisms by which LAM is assembled and organized into the cell envelope are unknown. A model for Mtb cell envelope structure is presented by Kaur et al [1].
Mtb expresses numerous, functionally diverse lipoproteins, many of which are contained in the cell envelope [20]–[22]. Mutation in lipoprotein processing by the disruption of lipoprotein signal peptidase lspA attenuates Mtb virulence [23], causes cell envelope permeability defects and increases sensitivity to antibiotics [24], suggesting potential roles of lipoproteins in maintaining Mtb cell envelope function. A recent proteomic study of the Mtb cell envelope identified LprG among the top 10 most abundant lipoproteins [25]. LprG (Rv1411c) forms an operon with Rv1410c, which encodes a putative efflux pump membrane protein, p55 [26]. Genetic deletion of this operon results in decreased virulence of Mtb in mice, and its deletion in M. smegmatis results in abnormal cell envelope morphology and permeability, suggesting its involvement in cell envelope biogenesis and/or maintenance of cell envelope integrity [26]–[30]. Despite evidence for the importance of LprG to Mtb virulence, its function remains uncertain.
Mtb LprG binds to LAM, LM and PIMs when expressed in M. smegmatis, and acyl moieties of these lipoglycans and glycolipids bind in a hydrophobic binding pocket of LprG [31]. Glycolipid binding was abrogated with a mutated version of LprG (LprG-V91W) with altered hydrophobic binding pocket structure [31]. In the studies reported here, we expressed acylated LprG in the native species, Mtb, to determine the function of LprG in Mtb. Surface plasmon resonance (SPR) binding assays were used to determine the kinetics and affinities for binding of different Mtb lipoglycans and glycolipids to LprG; the results revealed novel interactions between saccharide chains of LAM and LM and a site on LprG outside of the hydrophobic pocket, as well as interactions of the lipoglycan acyl chains with the LprG hydrophobic pocket. Furthermore, an LprG null mutant Mtb (ΔlprG) had reduced levels of surface-exposed LAM and decreased ability to inhibit Mtb phagosome-lysosome fusion, a LAM-dependent effect that is associated with Mtb survival and virulence. We propose that LprG binding to LAM facilitates the transfer of LAM from the plasma membrane (its site of synthesis) to the Mtb outer membrane. This leads to LAM expression at the cell surface, contributing to the ability of Mtb to inhibit phagosome-lysosome fusion and survive in macrophages. Our results highlight the importance of LprG and cell envelope components in the survival and virulence of Mtb and reveal a potentially important target for therapeutic intervention.
Results
LprG functions as a carrier of lipoglycans in Mtb
These studies explored for the first time the lipoglycan/glycolipid binding function of the lipoprotein LprG expressed in Mtb. Previous studies of LprG expressed in a fast-growing saprophytic mycobacterial species, M. smegmatis, had revealed a lipoglycan/glycolipid binding function [31], but the potential roles of LprG in lipoglycan biology in Mtb remained unexplored. The current studies provide a detailed understanding of lipoglycan/glycolipid interactions with LprG and reveal a novel role for LprG in determining the localization of LAM in the cell envelope.
In order to study the association of lipoglycans and glycolipids with LprG in Mtb, the lipoprotein was expressed with a hexahistidine tag and isolated from Mtb lysate by Ni affinity and anion exchange chromatography. These studies used acylated LprG, rather than non-acylated LprG (NA-LprG), which was used in prior studies, as acylation state is likely to affect subcellular localization and intersection with specific glycolipid and lipoglycan species (acylation is linked with translocation across the plasma membrane). SDS-PAGE was used to analyze LprG and any co-purifying molecules. Silver stain and Western blot (Fig. 1A) with anti-hexahistidine antibody showed a single band at ∼25 kDa, corresponding to the approximate molecular weight of LprG. Western blot with CS-35 anti-LAM monoclonal antibody revealed a characteristic diffuse band at ∼37 kDa that was associated with acylated Mtb LprG expressed in both Mtb and M. smegmatis (Fig. 1B). A polyclonal anti-Mtb antibody that detects LAM, LM and PIMs revealed bands at the expected positions for these lipoglycans (∼37 kDa, ∼18 kDa and ∼10 kDa; corresponding to positions of these lipoglycan/glycolipid standards on the gel) in association with Mtb LprG expressed in Mtb and M. smegmatis but not E. coli (Fig. 1C and 1D). The detection of LAM was more intense than the detection of LM and PIMs, and the association of all three lipoglycans/glycolipids was greater when LprG was expressed in Mtb relative to M. smegmatis (Fig. 1B, C). These lipoglycans/glycolipids were not associated with Mtb-expressed acylated LprA, a lipoprotein that is homologous to LprG (hexahistidine-tagged LprA was expressed and purified using the same protocol as for LprG, see Materials and Methods); this control demonstrates the specificity of lipoglycan/glycolipid binding to LprG. These studies establish that acylated LprG binds LAM and LM in Mtb.
Fig. 1. Acylated LprG binds lipoglycans in Mtb. 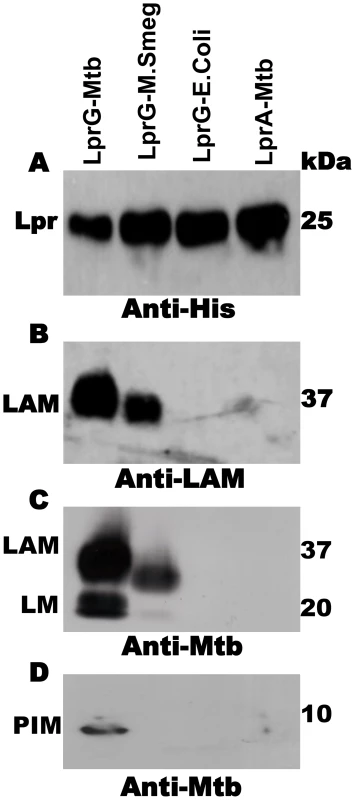
SDS-PAGE analysis of proteins and co-purifying molecules isolated from Mtb H37Ra, M. smegmatis or E. coli. (A) Western blot with monoclonal anti-hexahistidine (anti-His6) to detect LprG or LprA. Mycobacterial components associated with lipoproteins were detected using (B) monoclonal anti-LAM antibody CS-35 or (C and D) rabbit polyclonal anti-Mtb antibody that detects both LAM and LM. Blots are representative of at least three independent experiments. SPR analysis of glycolipid and lipoglycan binding to LprG
While prior studies showed that binding of glycolipids to LprG involved insertion of glycolipid acyl chains into the LprG hydrophobic pocket (blocked by mutation of the hydrophobic pocket in LprG-V91W) [31], we hypothesized that lipoglycans with longer saccharide chains, e.g. LAM, might have additional functional interactions involving their saccharide moieties and other sites on LAM. To obtain insight regarding structures that determine lipoglycan/glycolipid binding to LprG, which may have implications for LprG function, we used SPR to measure the affinity and kinetics of LprG-substrate interactions and the effects of alterations in glycolipid/lipoglycan structures on binding to LprG. In vitro binding assays were performed using non-acylated LprG (NA-LprG) or NA-LprG-V91W, since the non-acylated versions retain glycolipid/lipoglycan binding properties and present technical advantages for expression, purification and use in binding assays (all references to LprG or LprG-V91W in SPR assays refer to the non-acylated versions). Expression of LprG or LprG-V91W in E. coli allowed purification of LprG without the presence of bound substrates (Fig. 1), facilitating subsequent glycolipid and lipoglycan binding assays. LprG and LprG-V91W were immobilized as ligands on sensor chips, and increasing concentrations of substrates were injected. Sensograms representing association and dissociation phases were obtained after subtracting non-specific binding to a blank sensor chip. The results provide the first determination of affinities for binding of different glycolipids and lipoglycans to LprG, allowing us to assess novel contributions of saccharide moieties to lipoglycan binding.
To explore relationships between glycolipid structure and LprG-binding properties, we studied a panel of mycobacterial glycolipids and lipoglycans, including PIM2, PIM6, LM, ManLAM and PI-LAM, all of which share a mannosyl-phosphatidyl-myo-inositol domain with additional specific structural features (Fig. 2A). All of these molecules are expressed by Mtb except for PI-LAM, which is expressed by non-pathogenic mycobacteria. PIM2 (a precursor of PIM6, LM and LAM) has mannose residues at positions 2 and 6 of the myo-inositol ring of PI (Fig. 2A). PIM2 bound to LprG in a dose-dependent manner (Fig. 2B) but with relatively low affinity (KD = 1.09×10−6 M, Table 1). PIM6, LM, ManLAM and PI-LAM all bound to LprG (Fig. 2C–F). Of these glycolipids, LM, which consists of a long linear mannan chain extending from position 6 of the myo-inositol ring, bound to LprG with the highest affinity (KD = 1.58×10−9 M, Fig. 2D, Table 1). PIM6, which consists of a much shorter mannoside motif attached to position 6 of the myo-inositol ring, bound to LprG with the second highest affinity (KD = 4.5×10−8 M, Fig. 2C, Table 1). LAM is generated from LM by the addition of a branched arabinan domain with species-specific terminal caps (ManLAM for Mtb, PI-LAM for non-pathogenic mycobacteria, Fig. 2A). ManLAM and PI-LAM bound to LprG with lower affinities than LM and PIM6 (KD of 1.09×10−7 M for ManLAM and 1.6×10−6 M for PI-LAM, Fig. 2E–F, Table 1). Despite sharing the common acylated core structure that was previously implicated in binding to the hydrophobic pocket of LprG, these glycolipids and lipoglycans displayed distinct kinetics for LprG binding, indicating a previously unknown contribution of polysaccharide structures to LprG.
Fig. 2. SPR analysis of substrate binding to LprG. 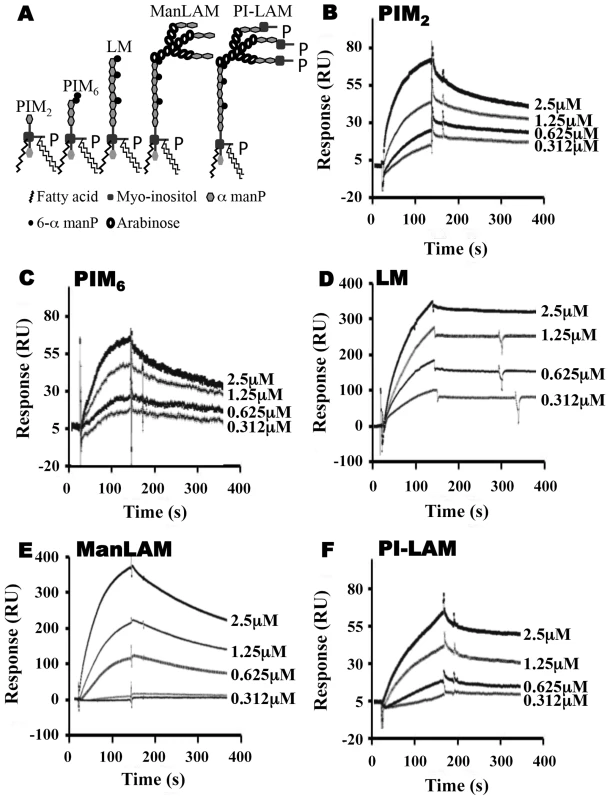
(A) Schematic representation of mycobacterial glycolipids and lipoglycans used in this study. (B–F) Substrate binding to LprG was assessed by SPR. LprG was immobilized on a CM5 sensor chip. Sensograms were obtained by injecting increasing concentrations of (B) PIM2 (C) PIM6 (D) LM (E) ManLAM and (F) PI-LAM. Binding was measured as response units (RU). Binding curves were calculated with BIA evaluation 3.1 software with subtraction of non-specific binding of the substrates to the sensor chip control cells without immobilized LprG. Results are representative of three independent experiments. Tab. 1. Binding kinetics and affinities for interaction of Mtb substrates with LprG and LprG-V91W. 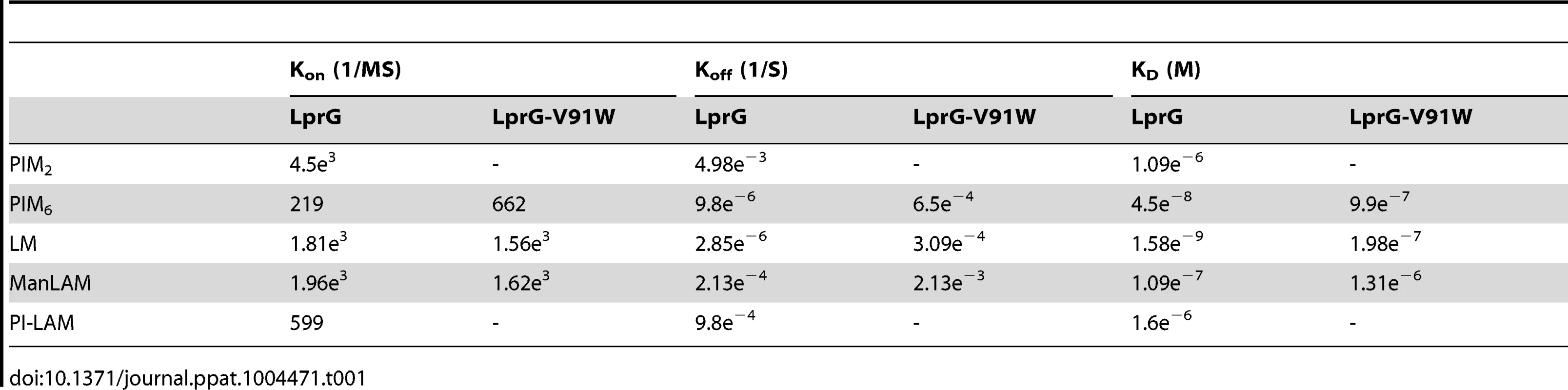
To assess the relative contributions of the hydrophobic binding pocket vs. other sites on LprG, we studied glycolipid and lipoglycan binding to LprG-V91W, which has a mutated pocket that precludes binding of the acyl chains of triacylated glycolipids [31]. PIM2 (Fig. 3A) and PI-LAM failed to bind to LprG-V91W, but PIM6, LM and ManLAM bound to LprG-V91W with an affinity approximately 10 - to 130-fold lower than their affinity for LprG (Fig. 3B–D) (Table 1). The loss of acyl-chain dependent interaction with LprG-V91W increased substrate dissociation rates (Koff), indicating less stable interactions. These results indicate the presence of novel interactions of a subset of glycolipids/lipoglycans with LprG at a site outside of the hydrophobic pocket. Moreover, this suggests that these interactions include glycolipid and lipoglycan structures other than the acyl chains that bind within the hydrophobic pocket of LprG, consistent with the implication of novel contributions of saccharide structures of substrates in interactions with LprG.
Fig. 3. LprG V91W hydrophobic pocket mutation reduces binding of Mtb glycolipids and lipoglycans. 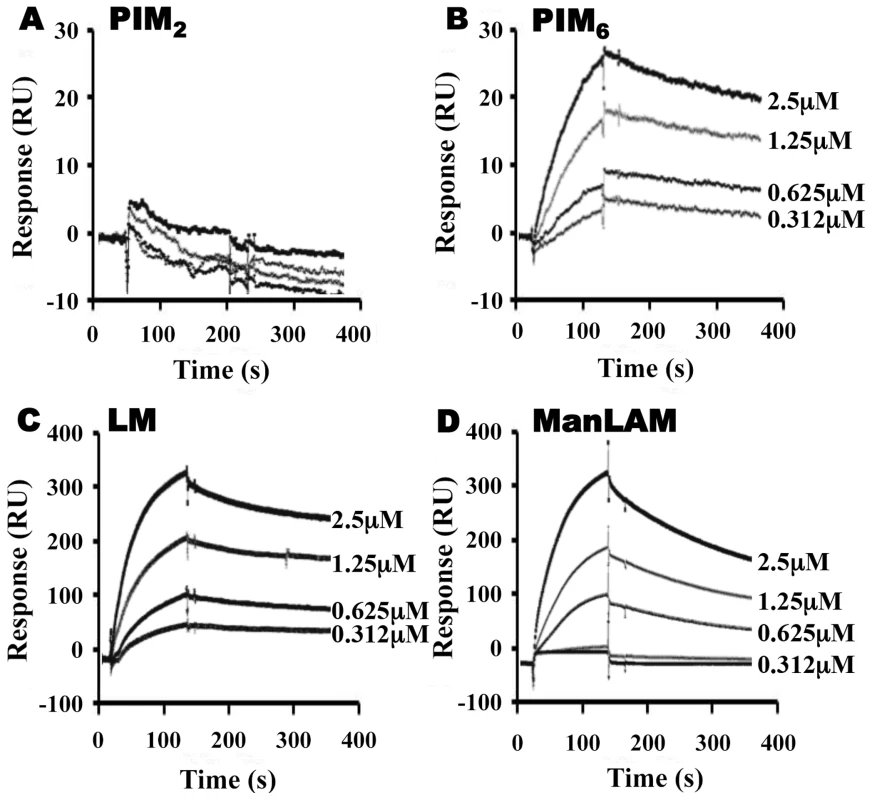
LprG-V91W was immobilized on a CM5 sensor chip, and sensograms were obtained as in Fig. 2 for (A) PIM2 (B) PIM6 (C) LM and (D) ManLAM. Results are from one experiment and representative of three independent experiments. To further analyze the relative contributions of acyl and non-acyl structures of substrates to LprG binding, we subjected LM and ManLAM to alkaline deacylation. Deacylated LM and deacylated ManLAM both bound specifically to LprG (Fig. 4A and B, Table 2), and this binding was similar for LprG and LprG-V91W. This confirms that non-acyl structures of lipoglycans bind to a site on LprG different from the hydrophobic pocket. In support of this conclusion, loss of the acyl chains reduced the affinities of LM and ManLAM for LprG to values similar to the affinities of acylated LM and acylated ManLAM for LprG-V91W. Deacylation had little effect on the association rate for LM or ManLAM binding to LprG or LprG-V91W but resulted in increased dissociation rates with LprG. Overall, these results establish that Mtb lipoglycans and glycolipids bind to LprG via at least two different interactions at different sites on LprG, one being the acyl chain binding in the hydrophobic pocket and the other being interaction of saccharide chains (likely mannose chains) with LprG outside of the hydrophobic pocket. The ability of LprG to discriminate different glycolipids/lipoglycans based on differences in their saccharide structures may allow differential binding and release of different glycolipids/lipoglycans.
Fig. 4. Deacylated ManLAM and LM retain binding to LprG at reduced levels. 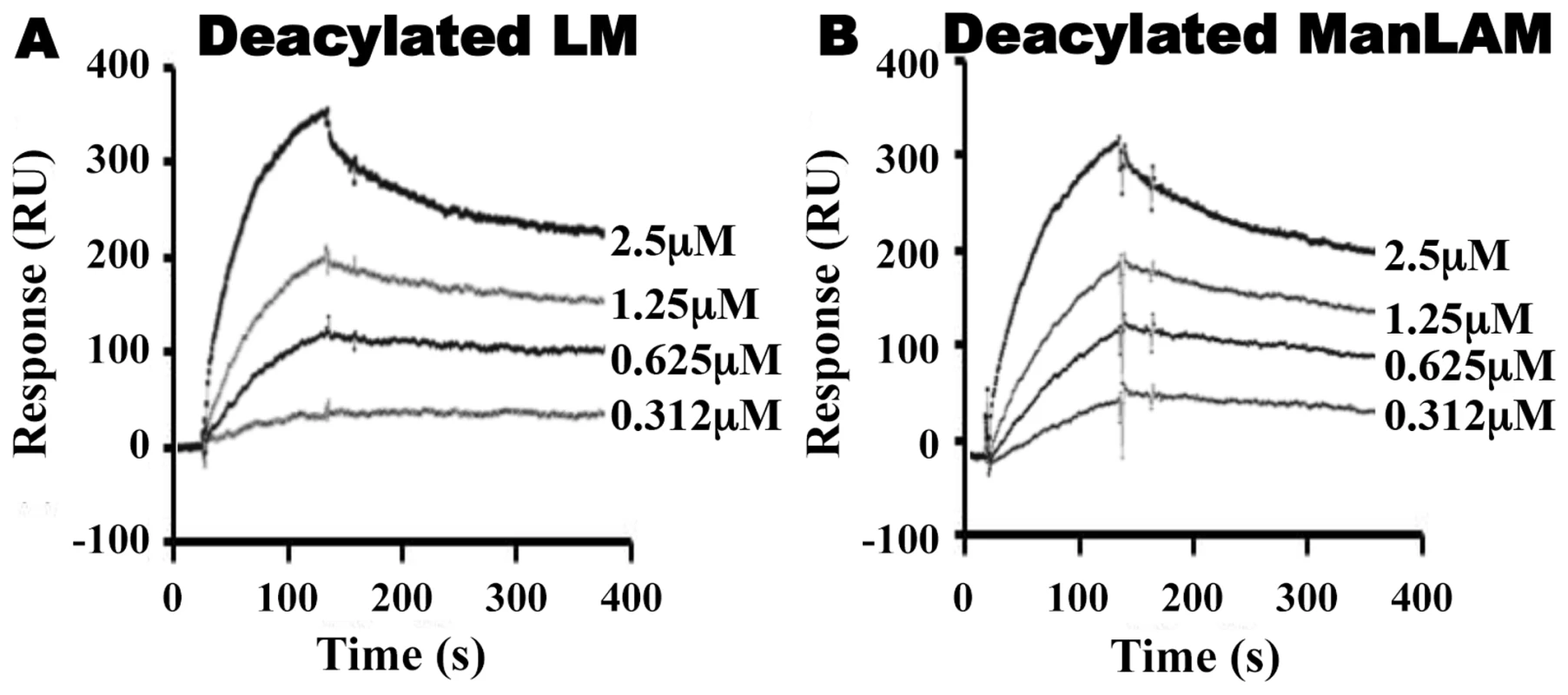
Binding to LprG was assessed as in Fig. 2 for (A) deacylated LM and (B) deacylated ManLAM. Results are from one experiment and representative of at least three independent experiments. Tab. 2. Binding kinetics and affinities for interaction of Mtb deacylated substrates with LprG and LprG-V91W. 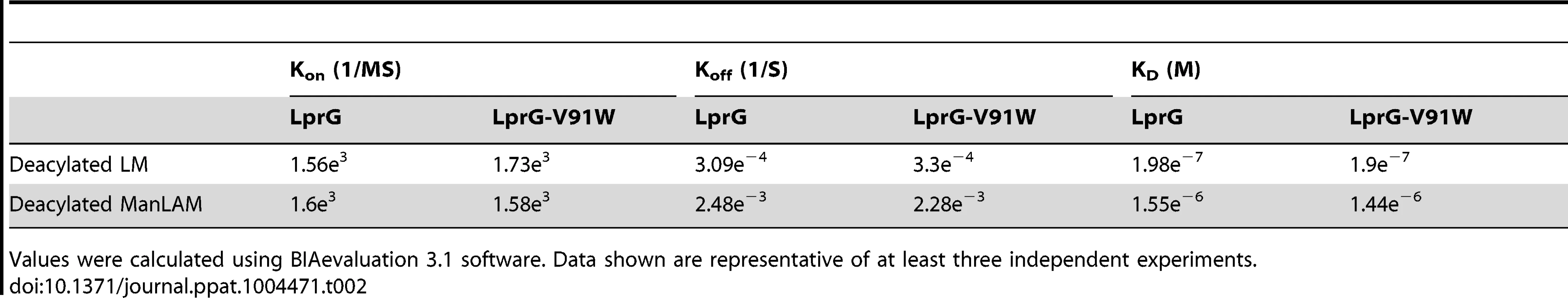
Values were calculated using BIAevaluation 3.1 software. Data shown are representative of at least three independent experiments. Mannose chains contribute to Mtb lipoglycan binding to LprG
To investigate whether mannan components of LAM and LM contribute to lipoglycan binding to LprG, we tested the ability of S. cerevisiae mannan to bind to LprG. S. cerevisiae mannan was selected since its structure is comparable to the polysaccharide components of Mtb lipoglycans, except for the absence of the phosphatidyl-myo-inositol lipid anchor present in Mtb lipoglycans. SPR assays revealed mannan binding to LprG in a dose dependent manner, although with a lower affinity compared to LAM and LM (KD = 8.8×10−5 M, Fig. 5A and Table 3, compare to Table 1). Next, to test whether mannan and Mtb lipoglycans have overlapping binding site(s) on LprG, we assessed their ability to compete for binding to LprG in SPR assays. The potential approach to saturate immobilized LprG with mannan in a first injection and subsequently inject LAM or LM was not feasible due to the rapid dissociation rate of mannan (4.5×10−2 sec−1), which allowed dissociation of pre-bound mannan before injection of LAM or LM could be completed. Accordingly, we first injected the lipoglycan (e.g. LM, 2.5 µM), which bound to LprG (Fig. 5B, “first injection”); a subsequent injection of mannan (2.5 µM; Fig. 5B, “second injection”) did not reveal mannan binding in contrast to the ability of mannan to bind to LprG in the absence of LM (compare Fig. 5A and 5B). Similar results were obtained with LAM and mannan. These results indicate that mannan and Mtb lipoglycans compete for binding to LprG. To confirm this conclusion with a different approach, we used a solid phase binding assay that measured LprG binding to plate-immobilized ManLAM. LprG bound to plate-immobilized ManLAM in a dose-dependent fashion. When LprG was incubated with mannan, however, the binding of LprG to ManLAM was inhibited by the presence of mannan in a dose-dependent manner (Fig. 5C). These data further indicate that mannan and Mtb lipoglycans compete for binding to LprG. Taken together, these results support our other evidence that mannose residues contribute to the binding of Mtb lipoglycans to LprG, in addition to contributions of acyl chain interactions.
Fig. 5. Mannan competition with Mtb lipoglycans for binding to LprG implicates a role for polysaccharide components of lipoglycans in LprG binding. 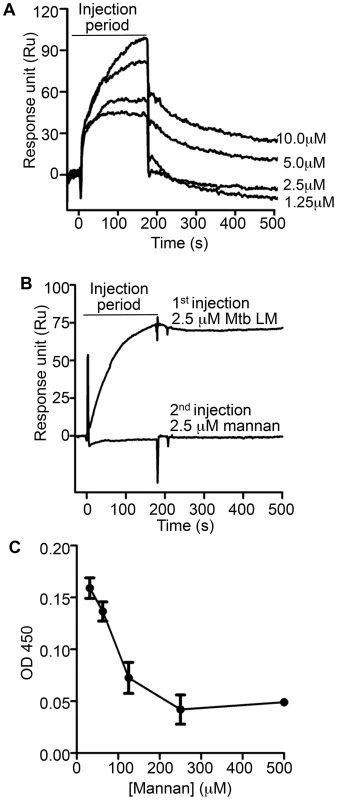
(A) Binding of Mannan to LprG was assessed as in Fig. 2 at the indicated concentrations of mannan. (B) The ability of LM to compete with mannan for binding to LprG was assessed by SPR with sequential injection of LM (“1st injection”) for 3 min followed by buffer for ∼10 min as the instrument prepared for injection of mannan (“2nd injection”). This injection order was chosen due to the fast dissociation rate for mannan. The dissociation rate for LM (shown here) is slower than for LAM, but experiments with LAM provided qualitatively similar results. Results for mannan binding in panels A and B are directly comparable except for the absence (panel A) or presence (panel B) of prior LM injection. Results for panels A and B are representative of two independent experiments. (C) Mannan inhibited LAM binding to LprG in a solid phase competitive binding assay. LprG was incubated with mannan at the indicated concentrations for 30 min and added to wells of plates coated with ManLAM. After incubation and washing, the plates were incubated with anti-LprG antibody followed by an HRP-linked secondary antibody to detect LprG binding to ManLAM (see Materials and Methods). Data are expressed as the means from three independent experiments. Tab. 3. Binding kinetics and affinity for interaction of <i>Saccharomyces cerevisiae</i> mannan with LprG. 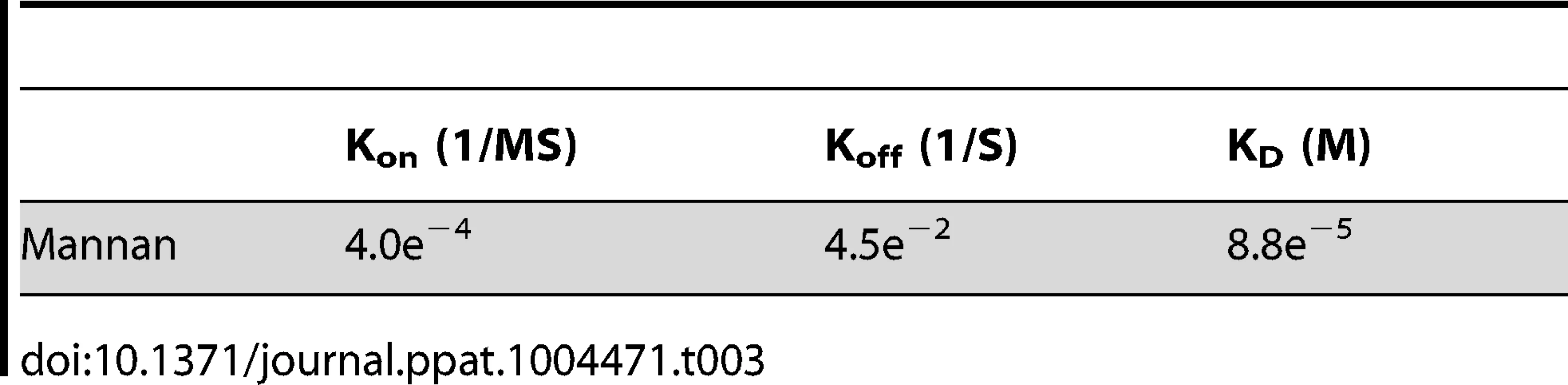
Deletion of lprG results in decreased levels of detectable LAM at the bacterial surface
We hypothesized that LprG may serve a function in the synthesis of LAM (and possibly LM and PIM) or their assembly into the cell envelope (possibly mediating their transfer from anchorage in the plasma membrane to allow localization at sites more peripheral in the cell envelope). This hypothesis suggests that deletion of LprG might alter the expression or localization of these molecules and influence the cell envelope properties of Mtb. Accordingly, we studied the effects of LprG expression on Mtb cell envelope properties using Mtb H37Ra ΔlprG and Mtb H37Rv ΔlprG strains with deletion of the lprG gene, and H37Rv ΔlprG::lprG-Rv1410c, a complemented version of H37Rv ΔlprG expressing the operon encoding LprG and p55.
To assess whether deletion of lprG affects the expression of the glycolipids to which LprG binds, e.g. LAM, we used Western blotting to assess the total bacterial expression of LAM and flow cytometry to detect the expression of LAM at the bacterial cell surface. Whole cell lysates of Mtb H37Ra and H37Ra ΔlprG were analyzed by SDS-PAGE and Western blotting with anti-LprG, which confirmed the absence of LprG in Mtb H37Ra ΔlprG (Fig. S1A). Western blotting with anti-LAM monoclonal antibody CS-35 or anti-Mtb polyclonal antibody (Fig. S1B, C) and detection of glycolipids and lipoglycans with carbohydrate staining (Fig. S1D) showed that Mtb H37Ra and H37Ra ΔlprG had similar total cellular expression of LAM and LM. Assessment of lipoglycan and glycolipid expression in Mtb H37Rv, H37Rv ΔlprG and H37Rv ΔlprG::lprG-Rv1410c strains by SDS-PAGE analysis, thin layer chromatography and periodic acid/Schiff staining revealed comparable amounts of LAM, LM and PIMs in all three strains.
Since deletion of lprG did not affect overall expression of LAM in Mtb, we assessed the hypothesis that LprG may affect the distribution of LAM in Mtb. Specifically, we considered that LprG may affect LAM assembly into the cell envelope, possibly by mediating removal of LAM from anchorage in the plasma membrane to allow its localization to sites more peripheral in the cell envelope. To determine whether the deletion of lprG reduces the amount of LAM detected at the bacterial cell surface, Mtb H37Rv, the knockout strain H37Rv ΔlprG, and the complemented control strain H37Rv ΔlprG::lprG-Rv1410c were stained with rabbit anti-ManLAM anti-serum or control normal rabbit serum and analyzed by flow cytometry. Specific LAM staining was >2-fold higher on Mtb H37Rv relative to Mtb H37Rv ΔlprG, and 1.8-fold higher on Mtb H37Rv ΔlprG::lprG-Rv1410c relative to Mtb H37Rv ΔlprG (Fig. 6). Furthermore, similar results were obtained with Mtb H37Ra (specific LAM staining was 7-fold higher on Mtb H37Ra relative to Mtb H37Ra ΔlprG). In summary, deletion of lprG did not affect total bacterial expression of LAM but did alter LAM distribution to substantially decrease its cell surface exposure.
Fig. 6. Deletion of lprG reduces LAM expression on the Mtb cell surface. 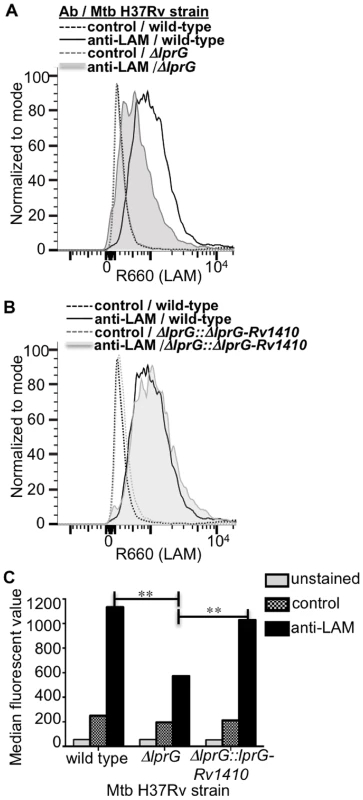
Cultures of H37Rv, H37Rv ΔlprG and H37Rv ΔlprG::lprG-Rv1410c were seeded at a starting density of 0.05 OD600 and harvested after 1 week. Bacteria were stained with rabbit anti-ManLAM antiserum or control normal rabbit serum and analyzed by flow cytometry. (A, B) Histograms showing LAM surface staining of H37Rv, H37Rv ΔlprG and H37Rv ΔlprG::lprG-Rv1410c. (C) Median fluorescence values (MFVs) from panel A. **P<0.001. Specific MFV was defined as MFV with anti-LAM minus MFV with control serum. Specific MFVs were 900 for H37Rv, 360 for H37Rv ΔlprG and 870 for H37Rv ΔlprG::lprG-Rv1410c. Results are representative of two independent experiments. Deletion of lprG causes increased Mtb phagosome-lysosome fusion
Bacterial phagosomes fuse with lysosomes to produce phagolysosomes, which can mediate killing of some bacteria. Inhibition of phagosome-lysosome fusion is one means by which Mtb evades host defenses, and LAM is known to inhibit phagosome-lysosome fusion (i.e. inhibit Mtb phagosome maturation) [13]–[15]. We hypothesized that decreased surface expression of LAM in Mtb ΔlprG would decrease the ability of this Mtb strain to inhibit phagosome maturation. To test this hypothesis, murine bone marrow-derived macrophages were incubated with LysoTracker Red to label lysosomes and then infected with FITC-labeled Mtb H37Rv, H37Rv ΔlprG or H37Rv ΔlprG::lprG-Rv1410c. Mtb phagosome-lysosome fusion was assessed by co-localization of LysoTracker Red with intracellular FITC-Mtb (within DAPI-stained cells) as assessed by fluorescence microscopy (Fig. 7A). After 1 h of infection, Mtb H37Rv demonstrated 50% co-localization with LysoTracker Red, whereas Mtb H37Rv ΔlprG showed 82% co-localization (p = 0.0002). Mtb H37Rv ΔlprG::lprG-Rv1410c showed reversion to the wild-type phenotype with 55% lysosomal co-localization (not significantly different from Mtb H37Rv, p = 0.6181) (Fig. 7B). As a positive control for uninhibited phagosome-lysosome fusion, heat-killed Mtb of all three strains proceeded to near complete phagosome-lysosome fusion (96–97% co-localization of Mtb and lysosomal markers), consistent with prior observations that heat-killed Mtb does not inhibit phagosome maturation [16], [17]. In conclusion, deletion of lprG reduces the availability of LAM for interactions with the host cell machinery that produce inhibition of phagosome-lysosome fusion. These results and the observation that deletion of lprG reduced cell-surface expression of LAM by Mtb both indicate that LprG controls the localization of LAM and suggest that LprG has a role in the insertion or assembly of lipoglycans into the Mtb cell envelope, perhaps by mediating its removal from the plasma membrane to allow localization to more peripheral sites in the cell envelope. The ability of LprG to affect the expression of LAM on the Mtb cell surface has important implications, as this mechanism determines the access of LAM to interaction with host cells.
Fig. 7. Genetic deletion of LprG reduces the ability of Mtb to inhibit phagosome-lysosome fusion. 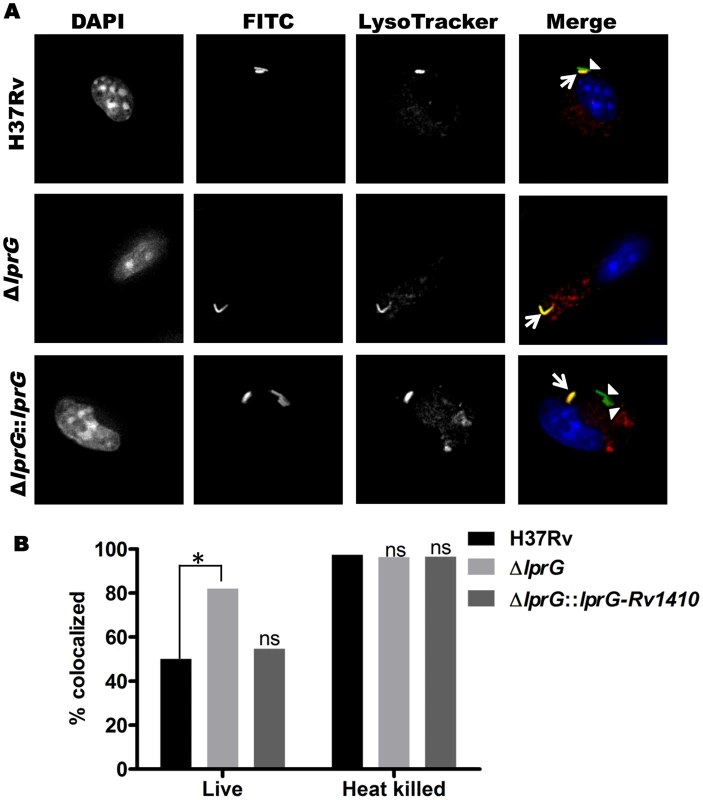
Murine bone marrow-derived macrophages were incubated with LysoTracker Red to label lysosomes and incubated with FITC-labeled Mtb for 60 min (30 min pulse+30 min chase). (A) Representative photomicrographs of macrophages after incubation with Mtb H37Rv, H37Rv ΔlprG and H37Rv ΔlprG::lprG-Rv1410c. Signals are rendered in grey scale in the first three columns, which show signal from a single label. In the fourth column overlay images (“Merge”, in color), Mtb H37Rv ΔlprG shows increased co-localization with lysosomal marker relative to Mtb H37Rv and H37Rv ΔlprG::lprG-Rv1410c. Arrowheads indicate FITC-labeled Mtb contained within phagosomes not colocalized with lysosomes (green); arrows indicate Mtb phagosome-lysosome co-localization (yellow). The p values were calculated using Fisher's exact test. (B) Percent co-localization of Mtb phagosomes with lysosomes (*, p = 0.0002; ns, not significant relative to Mtb H37Rv). Heat-killed bacteria are known to lack the ability to inhibit phagosome-lysosome fusion and were used as a positive control. Results shown here are from one experiment and are representative of at least two independent experiments. Discussion
The studies reported here explore the importance of determinants of cell envelope architecture in host-pathogen interactions and pathogenesis of bacterial infections. In particular, we shed new insight into the role of LprG expression in the pathogenic species, Mtb. Our data reveal novel structure-function relationships that determine glycolipid binding by LprG, including aspects of LAM binding that are specific to the LAM species expressed by Mtb (ManLAM, as opposed to PI-LAM expressed by non-pathogenic mycobacteria). Specifically, we demonstrate that LprG expressed in Mtb binds to ManLAM, a major immunomodulatory molecule of the Mtb cell envelope. Differences in the kinetics and affinities of LprG binding by ManLAM, PI-LAM, LM, PIM2, PIM6 and deacylated versions of LM and LAM reveal novel contributions of the saccharide moieties of these substrates to LprG binding. ManLAM, PI-LAM, LM, PIM2 and PIM6 all share the core acylated PIM structure, allowing their acyl chains to bind in the LprG hydrophobic pocket, as indicated by reductions in binding of all of these species to pocket-mutated LprG V91W relative to LprG (Table 1). The V91W mutation reduced the binding affinity by ∼10–100-fold for ManLAM, LM, and PIM6, and reduced binding to undetectable levels for PIM2 or PI-LAM. However, variation in the saccharide moieties of PIM, LM and LAM also influenced binding to LprG, revealing novel binding interactions of saccharide components at an additional site outside of the hydrophobic pocket (which persist with the LprG V91W pocket mutant). Deacylated ManLAM and deacylated LM were found to bind LprG, albeit with lower affinity than the acylated versions, and their binding was not substantially affected by the V91W pocket mutation. This further establishes a contribution of the substrate saccharide moieties to binding at a site outside of the LprG hydrophobic pocket. PIM2 lacks the longer mannan chain extensions from the PIM core structure that are seen in the other glycolipids, and its lower affinity for LprG binding may reflect lack of mannan interactions with LprG. Variation of the terminal capping of LAM may also affect LprG binding, given the reduced affinity of PI-LAM (which lacks terminal mannose residues) relative to ManLAM.
In summary, these results indicate that binding interactions between Mtb lipoglycans and LprG extend beyond the lipoglycan acyl chains and implicate a role for saccharide chains of Mtb lipoglycans in binding to LprG. First, deacylated Mtb lipoglycans bind to LprG (albeit at lower affinity than acylated lipoglycans). Second, acylated lipoglycans bind to LprG even when the hydrophobic pocket where the acyl chains bind is mutated to prevent their interaction (albeit with lower affinity than to wild-type LprG). Notably, the affinity of binding of the deacylated lipoglycans was not affected by mutation of the hydrophobic pocket (Table 2). Interestingly, in these two situations (acylated lipoglycan binding to pocket-mutated LprG vs. deacylated lipoglycan binding to LprG), whereby different approaches the contribution of acyl chains to LprG binding has been removed and the remaining interactions are implicated to be with saccharide chains, the binding affinities are very similar, consistent with our model (Tables 1 and 2, compare KD for binding of acylated vs. deacylated LAM to LprG vs. LprG-V91W, or compare KD for binding of acylated vs. deacylated LM to LprG vs. LprG-V91W).
The discovery of contributions of lipoglycan polysaccharide moieties to LprG binding suggested that these interactions involve lipoglycan mannan residues, since this property is shared by lipoglycans with mannose saccharides (e.g. LAM) and that lack other saccharide components (e.g. arabinan) found in LAM. Furthermore, we directly tested the contribution of mannan structures to LprG binding by use of S. cerevisiae mannan, which has structural similarity to the mannan chains of Mtb LM and LAM and also binds to LprG, although with a lower affinity. In competition binding assays, LM blocked mannan binding to LprG (Fig. 5B), and mannan inhibited LAM binding to LprG (Fig. 5C). These two findings further support the hypothesis that mannan and the Mtb lipoglycans (LAM, LM) compete for a binding site on LprG and further support the hypothesis that mannan chains of these Mtb lipoglycans contribute to their binding to LprG. In summary, although saccharide interactions are not essential for low affinity glycolipid binding to LprG, mannan chains interact with LprG to enhance substrate-LprG binding. We propose a model for lipoglycan binding to LprG that involves both acyl chain binding in the LprG hydrophobic pocket and mannan chain interactions with LprG outside of the pocket.
In addition to determining the structural basis for LprG-glycolipid interactions, we assessed the functional contributions of LprG using genetic deletion models. Although lprG deletion did not alter the total cellular expression of LAM, LM and PIM by Mtb, our results reveal the striking discovery that LprG controls expression of LAM at the cell surface of Mtb, presumably via its lipoglycan binding function. Thus, LprG influences the spatial organization of LAM within the Mtb cell envelope, perhaps via a role in LAM insertion or assembly into the cell envelope. One possibility is that LprG mediates the removal of lipoglycans from acyl-chain anchorage in the plasma membrane to allow their localization to more peripheral sites in the cell envelope.
Altered localization of LAM in the cell envelope may have significant implications for cell envelope functions and host-pathogen interactions. Deletion of LprG affects general measures of cell envelope integrity (e.g. malachite green decolorization, Congo red binding), consistent with other studies that associated altered cell envelope permeability with deletion of the lprG-Rv1410c operon [28], [30], [32]. Since LAM is implicated in inhibiting the fusion of Mtb phagosomes with lysosomes [13]–[15], we tested the hypothesis that reduced expression of surface-exposed LAM in the Mtb cell envelope would decrease LAM interactions with host cells and limit inhibition of phagosome maturation in Mtb H37Rv ΔlprG. Consistent with our hypothesis, we observed decreased inhibition of Mtb phagosome-lysosome fusion with Mtb H37Rv ΔlprG relative to wild-type Mtb H37Rv. Thus, deletion of lprG reduced the ability of Mtb to inhibit phagosome maturation, consistent with the known role for LAM in inhibiting phagosome maturation and our discovery that deletion of lprG diminishes expression of LAM at the bacterial cell surface, which we surmise diminishes its availability to interact with host cells.
In summary, our results reveal novel structural determinants of the binding of PIMs, LM and LAM to LprG and a novel role for LprG in determining LAM distribution in the cell envelope. While LprG is not necessary for LAM biosynthesis, it controls the expression of LAM at the bacterial cell surface, perhaps via a role in transferring LAM from the plasma membrane for localization in the cell envelope. Thus, LprG determines the accessibility of LAM for interactions with host cells to regulate phagosome-lysosome fusion and other host-pathogen interactions. These findings have important implications for fundamental determinants of host-pathogen interactions during infection with Mtb. LprG clearly influences Mtb phagosome maturation; it is required for optimum inhibition of phagosome maturation by Mtb, and it may thereby contribute to immune evasion by this mechanism. The influence of LprG on LAM localization in the Mtb cell envelope may affect the integrity of the cell envelope and its effectiveness as a permeability barrier, which is critical to Mtb survival within the host. Findings in this study provide novel insight into mechanisms of pathogenesis and host resistance to Mtb infection, and they suggest the possibility that pharmacologic disruption of LprG expression or function might result in a leaky cell envelope (making Mtb less resistant to drugs and antibiotics) and reduced ability of Mtb to evade host defense mechanisms by inhibiting phagosome-lysosome fusion. In conclusion, LprG is a critical determinant of Mtb virulence and host-pathogen interactions during Mtb infection and may be used as a potentially important target for therapeutic intervention.
Materials and Methods
Ethics statement
The Institutional Animal Care and Use Committee of Case Western Reserve University approved all animal studies (protocol 2012–0007). Studies were performed in accordance with recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Mtb strains and preparation of bacterial lysates
Mtb strains H37Rv and H37Ra were obtained from American Type Culture Collection (Manassas, VA). LprG null strains were generated from Mtb H37Rv (H37Rv ΔlprG; N. Banaei) and H37Ra (H37Ra ΔlprG; E.T. Richardson) using a specialized transducing phage targeting 571 bp within the lprG gene locus for homologous recombination with a hygromycin resistance cassette. Mtb H37Rv ΔlprG::lprG-Rv1410c was then generated by complementing Mtb H37Rv ΔlprG with the native Rv1411c/1410c operon expressed off the kanamycin-selective integrating plasmid pMV306. Bacteria were cultured in Middlebrook 7H9 broth (Difco, Lawrence, KS) supplemented with 10% albumin/dextrose/catalase (BD, Franklin Lakes, NJ), 0.05% Tween 80 and 0.2% glycerol plus 100 µg/ml hygromycin B (Invivogen, San Diego, CA) for H37Ra ΔlprG and H37Rv ΔlprG or 50 µg/ml kanamycin (Sigma, St. Louis, MO) for H37Rv ΔlprG::lprG-Rv1410c.
To assess bacterial expression of lipoglycans, lysates were prepared from Mtb H37Ra and H37Ra ΔlprG. Bacteria were grown to mid log phase in 50 ml of Sauton's medium, pelleted, suspended in PBS, and sonicated in an ice bath for 40 min in 10-second cycles at 50 W in a Misonix sonicator (Misonix Inc., Farmingdale, NY). Insoluble debris was pelleted at 2000× g for 5 min, and supernatants were stored at −80°C. Samples were analyzed by SDS-PAGE as described below.
Cloning, expression and purification of hexahistidine-tagged proteins
Constructs for expression of recombinant lipoproteins LprG, LprA, non-acylated LprG (NA-LprG) and non-acylated LprG-V91W (NA-LprG-V91W) were cloned previously [6], [31]. LprG, LprA, NA-LprG and NA-LprG-V91W were expressed in wild-type Mtb H37Ra, M. smegmatis or E. coli and purified as described [6], [31]. For expression in Mtb H37Ra, constructs were digested with NdeI and HindIII (New England Biolabs, Ipswich, MA) and ligated into the shuttle vector pVV16 (provided by J. Belisle, Colorado State University, Fort Collins, CO) under control of the constitutively active hsp60 promoter and in-frame with a C-terminal hexahistidine tag. Mtb was transformed by electroporation with a Gene Pulser (Bio-Rad, Hercules, CA) set at 2.5 kV, 25 µF, and 800 ohms. Mtb was grown with kanamycin selection (50 µg/ml) to late log phase, isolated by centrifugation at 6000 g for 20 min at 4°C, and suspended for 15 min at 37°C in lysis buffer. Bacteria were disrupted by four passages through a French press (2000 psi). Insoluble material was removed by ultracentrifugation at 100,000 g for 1 h at 4°C. The supernatant was incubated for 2–4 h at 4°C with Ni beads (Qiagen, Valencia, CA), which were then washed 3 times with 25 volumes of wash buffer (50 mM sodium phosphate, 1 M NaCl, 20 mM imidazole, 10% glycerol, pH 8.0). Bound protein was dissociated with elution buffer consisting of 50 mM sodium phosphate, 300 mM NaCl, 450 mM imidazole, pH 8.0. Samples were exchanged into 20 mM Tris, pH 8.0 using PD-10 columns (GE Healthcare, Uppsala, Sweden) and further purified by anion-exchange chromatography. Recombinant LprG was bound to HiTrap Q FF columns (GE Healthcare) and eluted by stepwise addition of 50, 150, 200 and 1000 mM NaCl. Recombinant LprG eluted with 50–200 mM NaCl was collected and concentrated using 10-kDa cutoff Centricon units (Amicon, Billerica, MA). Protein yields were determined by BCA protein assay (Pierce, Rockford, IL).
SDS-PAGE and western blots
Protein preparations or bacterial lysates were electrophoresed on 12% or 4–20% Mini-PROTEAN TBX Precast SDS-PAGE gels (BioRad, Hercules, CA). Protein purity was analyzed with Silver Stain Plus (BioRad). For Western blot analysis, materials were transferred to polyvinylidene difluoride membranes (Millipore), which were blocked with 5% milk in PBS supplemented with 0.1% Tween-20 (PBST) for 1 h at room temperature. Membranes were incubated overnight at 4°C with mouse monoclonal anti-hexahistidine antibody (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-Mtb antibody (GENway, Hayward, CA), mouse monoclonal anti-LAM antibody (CS-35) and mouse monoclonal anti-LprG antibody (clone α–Rv1411c, NR-13806, NIH Biodefense and Emerging Infections Research Resources Repository; BEI, Manassas, VA). Blots were washed three times with PBST, incubated for 1–2 h at room temperature with secondary antibodies conjugated to horseradish peroxidase, washed three times with PBST and visualized using a chemiluminescence kit (Pierce). Alternatively, membranes were stained to detect carbohydrate with the Glycoprotein Staining Kit (Pierce, Rockford, IL).
Preparation of deacylated ManLAM and LM
Purified ManLAM and LM from Mtb were deacylated by treatment of 1 mg ManLAM or LM with 0.1 N NaOH for 2 h at 37°C. The reaction mixture was neutralized with 10% acetic acid to pH ∼8.0, and dialyzed against distilled water for one day at room temperature. The sample was dried, loaded onto a Bio-gel P100 column and eluted with 0.1 N acetic acid. Fractions were collected and analyzed by SDS-PAGE to confirm loss of migration due to deacylation. Fractions containing deacylated ManLAM or LM were pooled, dialyzed against distilled water and dried. Sample purity was assessed by gas chromatography-mass spectrometry.
Surface plasmon resonance (SPR) binding assay
SPR binding experiments were performed on a BIAcore 3000 instrument using CM5 (carboxymethylated) sensor chips and HBSN running buffer (10 mM HEPES and 150 mM NaCl, pH 7.4) (GE Healthcare, Uppsala, Sweden). Non-acylated (NA)-LprG was used for SPR studies and purified from E. coli to prevent prior occupancy of LprG binding site(s) by Mtb glycolipids and lipoglycans that might hinder the binding of exogenous purified LprG ligands used in SPR analyses. Our previous studies [31] reveal that NA-LprG retains its ability to bind to Mtb glycolipids and lipoglycans, and a similar spectrum of LprG-associated molecules was found associated with acylated LprG expressed in Mtb (Fig. 1). In addition, it is unlikely that the acyl chains of LprG would interact with residues at or near to the lipoglycan binding pocket, as the N-terminal site of acylation of LprG is ∼50 Å away from the lipoglycan pocket (discussed by Drage et al [31]). Acylated LprG (Fig. 1) and NA-LprG (Figs. 2–5) both bind LAM, LM and PIMs.
NA-LprG and NA-LprG-V91W ligands were immobilized on CM5 sensor chips by amine coupling [33]. A 1∶1 mixture of 0.1 M n-hydroxysuccinimide (NHS) and 0.1 M ethylene diamine (GE Healthcare) was injected to activate carboxyl groups on the CM5 matrix, and 30 ml of a 40 mg/ml solution of ligand in 10 mM acetate buffer pH 4.5 was then injected. Residual NHS-esters were deactivated with 1 M ethanolamine (GE Healthcare). Control flow cells were activated and deactivated in the same manner but without protein ligand. Analytes were ManLAM, LM and PIMs purified from Mtb H37Rv (BEI), PI-LAM purified from M. smegmatis (BEI), and mannan from Saccharomyces cerevisiae (Sigma). The acylation state of PIMs was assessed by thin layer chromatography and mass spectrometry, revealing that the glycolipids were tri-acylated and tetraacylated (other acylation states were not significantly detected). Analytes were diluted in HBSN running buffer and injected at increasing concentrations (0.312 µM to 2.5 µM) at a flow rate of 30 µl/min. At the end of the injection, complexes were allowed to dissociate for 10 min. Chip surfaces were regenerated by 2–3 injections of 20 ml 50–90% ethylene glycol. Injections were separated by an equilibration delay of 30 min with HBSN at a flow rate of 5 ml/min. The final amount of bound analyte, expressed in resonance units (RU), was calculated by subtracting the RU of the control flow cell from the RU of the ligand-conjugated cell. Sensograms were analyzed using BIA evaluation 3.1 software (GE Healthcare). To assess binding competition by SPR, a first injection of LM (2.5 µM for 3 min) was followed by a buffer wash (∼10 min, reflecting the time necessary for the machine to switch to another injection) and then a second injection of mannan from Saccharomyces cerevisiae (2.5 µM for 3 min). This injection order was chosen due to the fast dissociation rate for mannan. Overlaid sensograms of the serial injections (with the signal at the start of each injection normalized) reveal the effect of prior LM-LprG binding on mannan-LprG interactions.
Solid phase competitive binding assay
To assess the ability of mannan to compete with LAM for binding to LprG, solid phase binding assays were performed as described [34] in 96-well plates (Corning Incorporation, NY, USA) coated with ManLAM. ManLAM was added (100 ng in 50 µl of 50% ethanol per well), and plates were air-dried for 1 h at 37°C. Subsequent steps were at room temperature. Plates were washed in wash buffer (10 mM Tris-HCl, pH 7.4, 140 mM NaCl, 1 mM CaCl2, 0.005% Tween 20) to remove unbound ManLAM, blocked with 5% milk in wash buffer for 2 h, washed twice in wash buffer, and incubated overnight with 100 µl of NA-LprG (1 µM) that had been pre-incubated for 30 min with or without mannan (10–500 µM) in wash buffer minus Tween. The plates were washed extensively, incubated with monoclonal anti-LprG antibody (clone α-Rv1411c, NR13806, BEI) for 2 h, washed three times, incubated for 1 h with goat anti-mouse IgG conjugated to horseradish peroxidase (Cell Signaling Technology, Danvers, MA), washed three times and incubated with o-Phenylenediamine dihydrochloride (Sigma) in 0.05 M phosphate-citrate buffer, pH 5.0 (50 µl/well) in the dark for 30 min. To stop the color development reaction, 50 µl of 2N H2SO4 was added. OD450 was determined on a Bio-Rad plate reader.
Flow cytometry
Mtb cultures (Mtb strains H37Rv, H37Rv ΔlprG, H37Rv ΔlprG::lprG-Rv1410c, H37Ra and H37Ra ΔlprG) were seeded to OD600 of 0.05 and harvested after one week. Bacteria were declumped by 10 passages through a 23-gauge needle, incubated in blocking buffer (1% heat inactivated normal rabbit serum, 0.01% Tween 80, PBS) at room temperature for 1 h, and washed twice with FACS buffer (1% BSA, 0.01% Tween 80, PBS). Bacteria were incubated for 1 h with 500 µl of FACS buffer containing rabbit anti-ManLAM antiserum (NR-13821, BEI, Manassas, VA) or control normal rabbit serum (Life Technologies, Grand Island, NY), washed twice with FACS buffer, stained for 1 h with Alexa Fluor 647-anti-rabbit IgG (Life Technologies, Grand Island, NY), washed twice with FACS buffer, fixed with 2% paraformaldehyde for 1 h at room temperature, and suspended in PBS containing 0.01% Tween 80. Anti-LAM labeling of bacteria was assessed with an LSR II flow cytometer (BD Bioscience, San Jose, CA). Data were analyzed using FlowJo software version X (TreeStar, Ashland, OR).
Infection of macrophages and analysis of phagosome-lysosome fusion
Female 8–12 week old C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and housed under specific pathogen-free conditions. Bone marrow-derived macrophages were cultured as described previously [6]. Briefly, bone marrow was flushed from isolated bones in DMEM, and cell suspensions were homogenized and filtered through a 70 µm screen. Bone marrow cells were pelleted and subjected to red blood cell lysis in ACK lysing buffer (Lonza, Walkersville, MD), pelleted again, and cultured in D10F consisting of DMEM (HyClone, Logan, UT) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Carlsbad, CA), 50 µM 2-mercaptoethanol (Bio-Rad, Hercules, CA), 1 mM sodium pyruvate (HyClone), 10 mM HEPES (HyClone), 100 units/ml penicillin, 100 µg/ml streptomycin (HyClone) and 25% LADMAC cell conditioned medium as a source of M-CSF. Cultures were incubated at 37°C in a humidified, 5% CO2 atmosphere. Medium was changed once on day 5 of culture, and differentiated bone marrow-derived macrophages were used on day 7. Macrophages were harvested using 0.25% trypsin/0.02% EDTA (HyClone) and 250,000 cells per well were plated in D10F without penicillin/streptomycin onto autoclaved glass cover slips (Fisher Scientific, Pittsburgh, PA) for in vitro infections and microscopy.
Mtb strains (H37Rv, H37Rv ΔlprG, H37Rv ΔlprG::lprG-Rv1410c) were grown to mid-log phase (OD600 = 0.5 to 0.6), pelleted, suspended in PBS with 0.05% Tween 80, and declumped by vigorous vortexing. Remaining clumps were removed by centrifugation for 15 min at 100 g. Some aliquots of Mtb were heat-killed by incubation at 80°C for 20 min. For fluorescent labeling of bacteria, FITC (Sigma) was added from a stock solution in DMSO to a concentration of 0.2 mg/ml FITC in 1% DMSO for 30 min at 37°C with gentle rotation. Bacteria were washed three times in PBS with 0.05% Tween 80 and suspended in D10F without antibiotics. Bone marrow-derived macrophages (grown on coverslips) were incubated for 30 min with 100 nM LysoTracker Red (Molecular Probes, Carlsbad, CA) in D10F, and FITC-labeled bacteria were added to a multiplicity of infection (MOI) of 10 for 30 min at 37°C. Macrophages were washed and incubated in D10F containing 100 nM LysoTracker Red for 30 min, washed with PBS, and fixed for 1 h in 2% paraformaldehyde. Coverslips were washed three times in PBS and inverted onto ProLong Gold mounting media with DAPI (Molecular Probes). Slides were cured overnight and imaged using a Deltavision RT epifluorescence microscope. Image files were deconvolved using the Softworx software package (Applied Precision, Issaquah, WA). Co-localization was determined by identifying intracellular bacteria (FITC-labeled organisms within DAPI-labeled cells) with overlapping lysosomal marker (LysoTracker Red). Colocalization data were obtained by counting at least six independent fields from each sample, and the results are representative of two independent experiments. Control samples consisting of uninfected/LysoTracker Red-treated or infected/LysoTracker Red-untreated bone marrow-derived macrophages were analyzed to set the lower threshold for FITC and LysoTracker Red positivity, and all infected and LysoTracker-labeled macrophages were analyzed with identical intensity cutoffs.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.01 software (La Jolla, CA). Fisher's exact test or Students two-tailed t-test was used to analyze the statistical significance of differences between groups.
Supporting Information
Zdroje
1. KaurD, GuerinME, SkovierovaH, BrennanPJ, JacksonM (2009) Chapter 2: Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol 69 : 23–78.
2. BrikenV, PorcelliSA, BesraGS, KremerL (2004) Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol Microbiol 53 : 391–403.
3. BrennanPJ, NikaidoH (1995) The envelope of mycobacteria. Annu Rev Biochem 64 : 29–63.
4. TorrellesJB, SchlesingerLS (2010) Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis (Edinb) 90 : 84–93.
5. ShinDM, YukJM, LeeHM, LeeSH, SonJW, et al. (2010) Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signaling. Cell Microbiol 12 : 1648–1665.
6. PecoraND, GehringAJ, CanadayDH, BoomWH, HardingCV (2006) Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol 177 : 422–429.
7. GehringAJ, DobosKM, BelisleJT, HardingCV, BoomWH (2004) Mycobacterium tuberculosis LprG (Rv1411c): A novel TLR-2 ligand that inhibits human macrophage class II MHC antigen processing. J Immunol 173 : 2660–2668.
8. PaiRK, ConveryM, HamiltonTA, BoomWH, HardingCV (2003) Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol 171 : 175–184.
9. NossEH, PaiRK, SellatiTJ, RadolfJD, BelisleJ, et al. (2001) Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19 kD lipoprotein of Mycobacterium tuberculosis. J Immunol 167 : 910–918.
10. BrennanPJ (2003) Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 83 : 91–97.
11. BergS, KaurD, JacksonM, BrennanPJ (2007) The glycosyltransferases of Mycobacterium tuberculosis - roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 17 : 35–56R.
12. MakarovV, ManinaG, MikusovaK, MollmannU, RyabovaO, et al. (2009) Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324 : 801–804.
13. KangPB, AzadAK, TorrellesJB, KaufmanTM, BeharkaA, et al. (2005) The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J Exp Med 202 : 987–999.
14. VergneI, ChuaJ, DereticV (2003) Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med 198 : 653–659.
15. FrattiRA, ChuaJ, VergneI, DereticV (2003) Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci USA 100 : 5437–5442.
16. ClemensDL, HorwitzMA (1995) Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med 181 : 257–270.
17. VergneI, ChuaJ, LeeHH, LucasM, BelisleJ, et al. (2005) Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 102 : 4033–4038.
18. ChuaJ, VergneI, MasterS, DereticV (2004) A tale of two lipids: Mycobacterium tuberculosis phagosome maturation arrest. Curr Opin Microbiol 7 : 71–77.
19. GuerinME, KordulakovaJ, AlzariPM, BrennanPJ, JacksonM (2010) Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J Biol Chem 285 : 33577–33583.
20. ColeST, BroschR, ParkhillJ, GarnierT, ChurcherC, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 : 537–544.
21. SutcliffeIC, HarringtonDJ (2004) Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev 28 : 645–659.
22. RezwanM, GrauT, TschumiA, SanderP (2007) Lipoprotein synthesis in mycobacteria. Microbiology 153 : 652–658.
23. SanderP, RezwanM, WalkerB, RampiniSK, KroppenstedtRM, et al. (2004) Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol Microbiol 52 : 1543–1552.
24. BanaeiN, KincaidEZ, LinSY, DesmondE, JacobsWRJr, et al. (2009) Lipoprotein processing is essential for resistance of Mycobacterium tuberculosis to malachite green. Antimicrob Agents Chemother 53 : 3799–3802.
25. MalenH, PathakS, SoftelandT, de SouzaGA, WikerHG (2010) Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol 10 : 132.
26. BigiF, AlitoA, RomanoMI, ZumarragaM, CaimiK, et al. (2000) The gene encoding P27 lipoprotein and a putative antibiotic-resistance gene form an operon in Mycobacterium tuberculosis and Mycobacterium bovis. Microbiology 146 (Pt 4) 1011–1018.
27. BigiF, GioffreA, KleppL, de la Paz SantangeloM, AlitoA, et al. (2004) The knockout of the lprG-Rv1410 operon produces strong attenuation of Mycobacterium tuberculosis. Microbes Infect 6 : 182–187.
28. BiancoMV, BlancoFC, ImperialeB, ForrelladMA, RochaRV, et al. (2011) Role of P27–P55 operon from Mycobacterium tuberculosis in the resistance to toxic compounds. BMC Infect Dis 11 : 195.
29. BiancoMV, BlancoFC, ForrelladMA, AguilarD, CamposE, et al. (2011) Knockout mutation of p27–p55 operon severely reduces replication of Mycobacterium bovis in a macrophagic cell line and survival in a mouse model of infection. Virulence 2 : 233–237.
30. FarrowMF, RubinEJ (2008) Function of a mycobacterial major facilitator superfamily pump requires a membrane-associated lipoprotein. J Bacteriol 190 : 1783–1791.
31. DrageMG, TsaiHC, PecoraND, ChengTY, AridaAR, et al. (2010) Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nature Struct Mol Biol 17 : 1088–1095.
32. BigiF, EspitiaC, AlitoA, ZumarragaM, RomanoMI, et al. (1997) A novel 27 kDa lipoprotein antigen from Mycobacterium bovis. Microbiology 143 (Pt 11) 3599–3605.
33. JohnssonB, LofasS, LindquistG (1991) Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem 198 : 268–277.
34. SidobreS, NigouJ, PuzoG, RiviereM (2000) Lipoglycans are putative ligands for the human pulmonary surfactant protein A attachment to mycobacteria. Critical role of the lipids for lectin-carbohydrate recognition. J Biol Chem 275 : 2415–2422.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



