-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Genetic Association Study Identifies as a Risk Gene for Idiopathic Dilated Cardiomyopathy
Dilated cardiomyopathy (DCM) is a structural heart disease with strong genetic background. Monogenic forms of DCM are observed in families with mutations located mostly in genes encoding structural and sarcomeric proteins. However, strong evidence suggests that genetic factors also affect the susceptibility to idiopathic DCM. To identify risk alleles for non-familial forms of DCM, we carried out a case-control association study, genotyping 664 DCM cases and 1,874 population-based healthy controls from Germany using a 50K human cardiovascular disease bead chip covering more than 2,000 genes pre-selected for cardiovascular relevance. After quality control, 30,920 single nucleotide polymorphisms (SNP) were tested for association with the disease by logistic regression adjusted for gender, and results were genomic-control corrected. The analysis revealed a significant association between a SNP in HSPB7 gene (rs1739843, minor allele frequency 39%) and idiopathic DCM (p = 1.06×10−6, OR = 0.67 [95% CI 0.57–0.79] for the minor allele T). Three more SNPs showed p < 2.21×10−5. De novo genotyping of these four SNPs was done in three independent case-control studies of idiopathic DCM. Association between SNP rs1739843 and DCM was significant in all replication samples: Germany (n = 564, n = 981 controls, p = 2.07×10−3, OR = 0.79 [95% CI 0.67–0.92]), France 1 (n = 433 cases, n = 395 controls, p = 3.73×10−3, OR = 0.74 [95% CI 0.60–0.91]), and France 2 (n = 249 cases, n = 380 controls, p = 2.26×10−4, OR = 0.63 [95% CI 0.50–0.81]). The combined analysis of all four studies including a total of n = 1,910 cases and n = 3,630 controls showed highly significant evidence for association between rs1739843 and idiopathic DCM (p = 5.28×10−13, OR = 0.72 [95% CI 0.65–0.78]). None of the other three SNPs showed significant results in the replication stage.
This finding of the HSPB7 gene from a genetic search for idiopathic DCM using a large SNP panel underscores the influence of common polymorphisms on DCM susceptibility.
Published in the journal: Genetic Association Study Identifies as a Risk Gene for Idiopathic Dilated Cardiomyopathy. PLoS Genet 6(10): e32767. doi:10.1371/journal.pgen.1001167
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001167Summary
Dilated cardiomyopathy (DCM) is a structural heart disease with strong genetic background. Monogenic forms of DCM are observed in families with mutations located mostly in genes encoding structural and sarcomeric proteins. However, strong evidence suggests that genetic factors also affect the susceptibility to idiopathic DCM. To identify risk alleles for non-familial forms of DCM, we carried out a case-control association study, genotyping 664 DCM cases and 1,874 population-based healthy controls from Germany using a 50K human cardiovascular disease bead chip covering more than 2,000 genes pre-selected for cardiovascular relevance. After quality control, 30,920 single nucleotide polymorphisms (SNP) were tested for association with the disease by logistic regression adjusted for gender, and results were genomic-control corrected. The analysis revealed a significant association between a SNP in HSPB7 gene (rs1739843, minor allele frequency 39%) and idiopathic DCM (p = 1.06×10−6, OR = 0.67 [95% CI 0.57–0.79] for the minor allele T). Three more SNPs showed p < 2.21×10−5. De novo genotyping of these four SNPs was done in three independent case-control studies of idiopathic DCM. Association between SNP rs1739843 and DCM was significant in all replication samples: Germany (n = 564, n = 981 controls, p = 2.07×10−3, OR = 0.79 [95% CI 0.67–0.92]), France 1 (n = 433 cases, n = 395 controls, p = 3.73×10−3, OR = 0.74 [95% CI 0.60–0.91]), and France 2 (n = 249 cases, n = 380 controls, p = 2.26×10−4, OR = 0.63 [95% CI 0.50–0.81]). The combined analysis of all four studies including a total of n = 1,910 cases and n = 3,630 controls showed highly significant evidence for association between rs1739843 and idiopathic DCM (p = 5.28×10−13, OR = 0.72 [95% CI 0.65–0.78]). None of the other three SNPs showed significant results in the replication stage.
This finding of the HSPB7 gene from a genetic search for idiopathic DCM using a large SNP panel underscores the influence of common polymorphisms on DCM susceptibility.Introduction
Dilated cardiomyopathy (DCM) is a common form of heart muscle disease with a prevalence of 1∶2,500 in the general population. It represents a major cause of cardiovascular morbidity and mortality and is characterized by systolic dysfunction as well as dilation and impaired contraction of the ventricles, often leading to chronic heart failure and eventually requiring cardiac transplantation [1]. In about 35% of cases DCM is a familial disease [2]. However, in the sporadic form of DCM, i. e. after exclusion of affected family members and all detectable causes (also called idiopathic DCM), a genetic component is discussed, but can thus far not be assigned to single gene defects. Knowledge of genetic risk factors for both, familial and non-familial forms of DCM is important to initiate treatment prior to symptomatic onset of the disease, to delay its occurrence or possibly halt its progression. To date, only a few common susceptibility alleles for sporadic DCM were identified from candidate-gene approaches, but could not be confirmed in replication samples [2], [3], this being a common problem of single gene based analyses [4]. In contrast, unbiased genome-wide association studies (GWAS) allow the identification of genetic risk factors even outside of known genes, but higher power is needed to compensate for multiple testing [5]. No comprehensive GWAS was performed to date on sporadic form of DCM.
The cardiovascular gene-centric 50K single nucleotide polymorphism (SNP) ITMAT-Broad-CARe (IBC) array represents an established compromise between GWAS and hypothesis-driven candidate gene approach by analyzing polymorphisms in more than 2,000 genes known or predicted to be involved in cardiovascular phenotypes [6].
In this study, we conducted a screening based on the cardiovascular 50K SNP array with three independent replication studies to reveal insight in genetic contribution to idiopathic DCM. The four samples from Germany and France included 1,910 sporadic DCM cases and 3,630 healthy controls individuals. We identified a common intronic variant in HSPB7, encoding a cardiovascular small heat shock protein, to be associated with sporadic form of DCM.
Results
Screening stage
In our screening case-control sample, DCM cases were more likely men, were slightly younger and less frequently smokers, had a lower BMI and a higher prevalence of hypertension, hypercholesterolemia as well as type 2 diabetes (Table 1).
Tab. 1. Characteristics of DCM cases and controls used for initial screening. 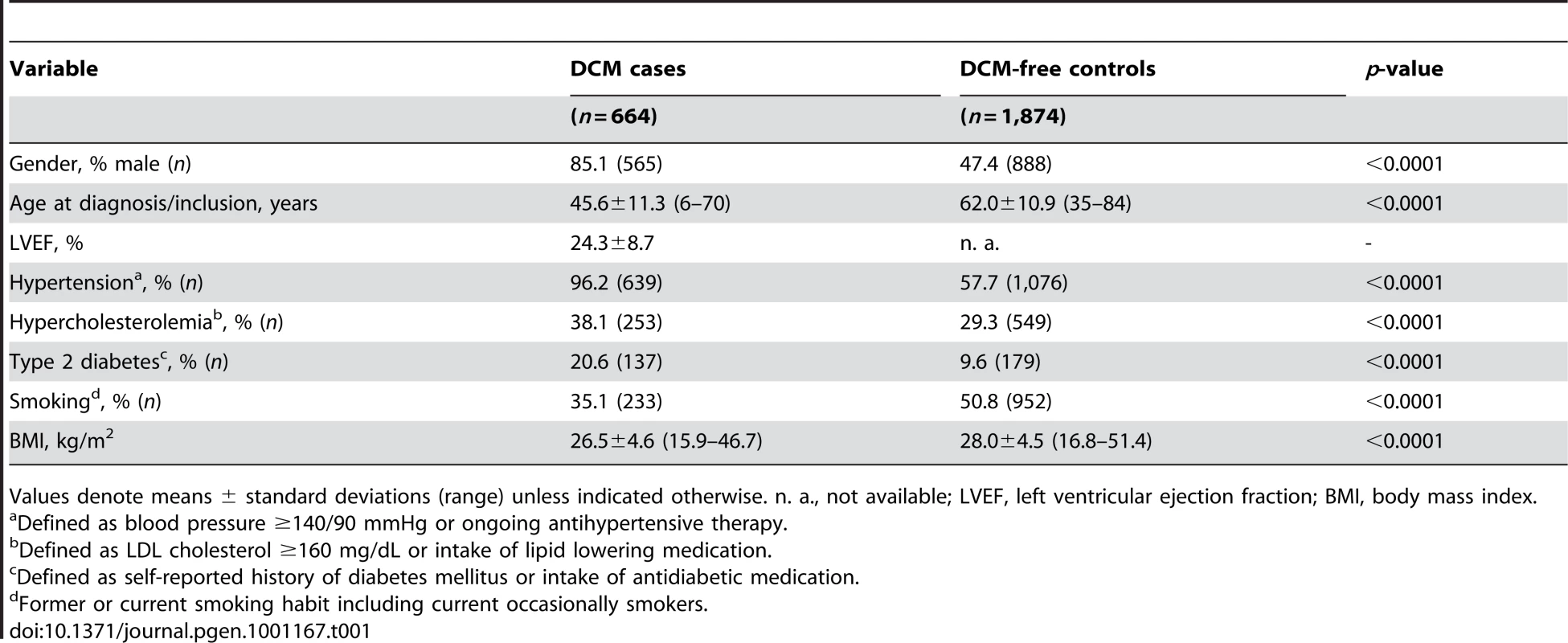
Values denote means ± standard deviations (range) unless indicated otherwise. n. a., not available; LVEF, left ventricular ejection fraction; BMI, body mass index. After quality control, 30,920 SNPs were available for analysis with 23,307 independent markers (defined as SNPs with pairwise r2<0.8 based on linkage disequilibrium (LD) in the control group). Therefore, we set a significance threshold to 0.05/23,307 = 2.15×10−6 to account for the multiple testing. In association analyses of this stage 1 study applying logistic regression adjusted for gender, four SNPs, namely rs1739843 (HSPB7, intron 2), rs11701453 (RUNX1, intron 1), rs7597774 (ADD2, intron 1) and rs2229714 (RPS6KA1, 3′ untranslated region) showed a p-value below this threshold (3.16*10−8, 1.65*10−7, 2.05*10−7, and 1.51*10−6, respectively). Results were similar when additionally adjusting for age (e.g. for rs1739843 p = 2.40*10−8). None of the four polymorphisms showed deviation from Hardy-Weinberg equilibrium. The lowest p-value for association with DCM was observed for a SNP located in HSPB7 intron 2 (rs1739843) leading to a protective effect of the minor allele (OR = 0.67 [95% CI 0.58–0.77]). Analysis of the region around the SNP rs1739843 using HapMap data (release #22) revealed the presence of six genes and 27 polymorphisms in LD with the lead SNP (r2-value>0.5) (Figure 1A). Nine of these SNPs were present on the cardiovascular 50K array after quality control and were located in HSPB7 gene as well as two genes downstream, CLCNKA and CLCNKB (Figure 1B; Table S1).
Fig. 1. Linkage disequilibrium (LD) structure of HSPB7 genomic region and association results. 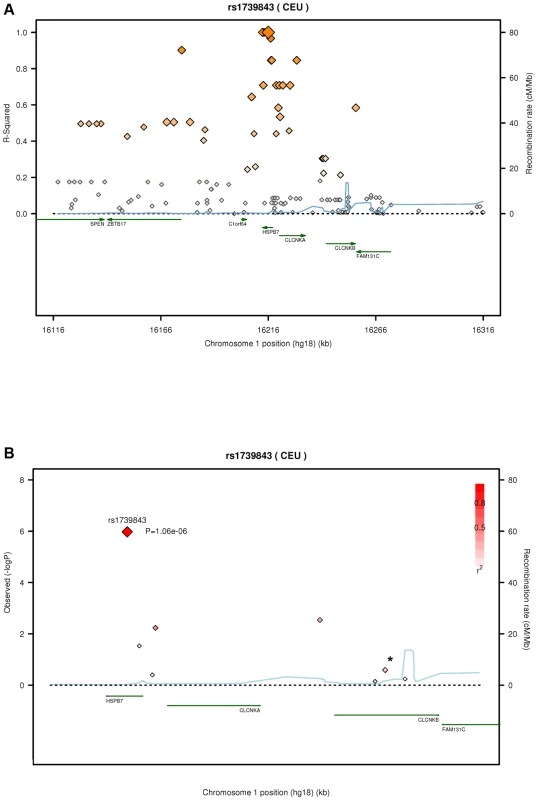
(A) LD measurement (r2) of HapMap data on CEU samples (release #22) in relation to rs1739843. On each side of the SNP, 100 kb were analyzed and plotted (n = 138 SNPs). (B) SNPs (n = 9, Table S1; *, two SNPs) in HSPB7 gene region on the 50K gene-centric human CVD bead chip after quality control and λ-corrected association results in 664 DCM cases and 1,874 controls. Plots were generated by using the SNAP tool [33]. In this sample, the genomic inflation factor λ was 1.285 for the highest 90% of the 30,920 observed p-values. When correcting rs1739843 for this λ factor, the p-value was 1.06*10−6 and OR = 0.67 [95% CI 0.57–0.79] (Table 2).
Tab. 2. Association of SNPs showing p-values < 2×10−6 in the initial screening sample and follow-up in three independent replication samples analyzed by logistic regression adjusted for gender. 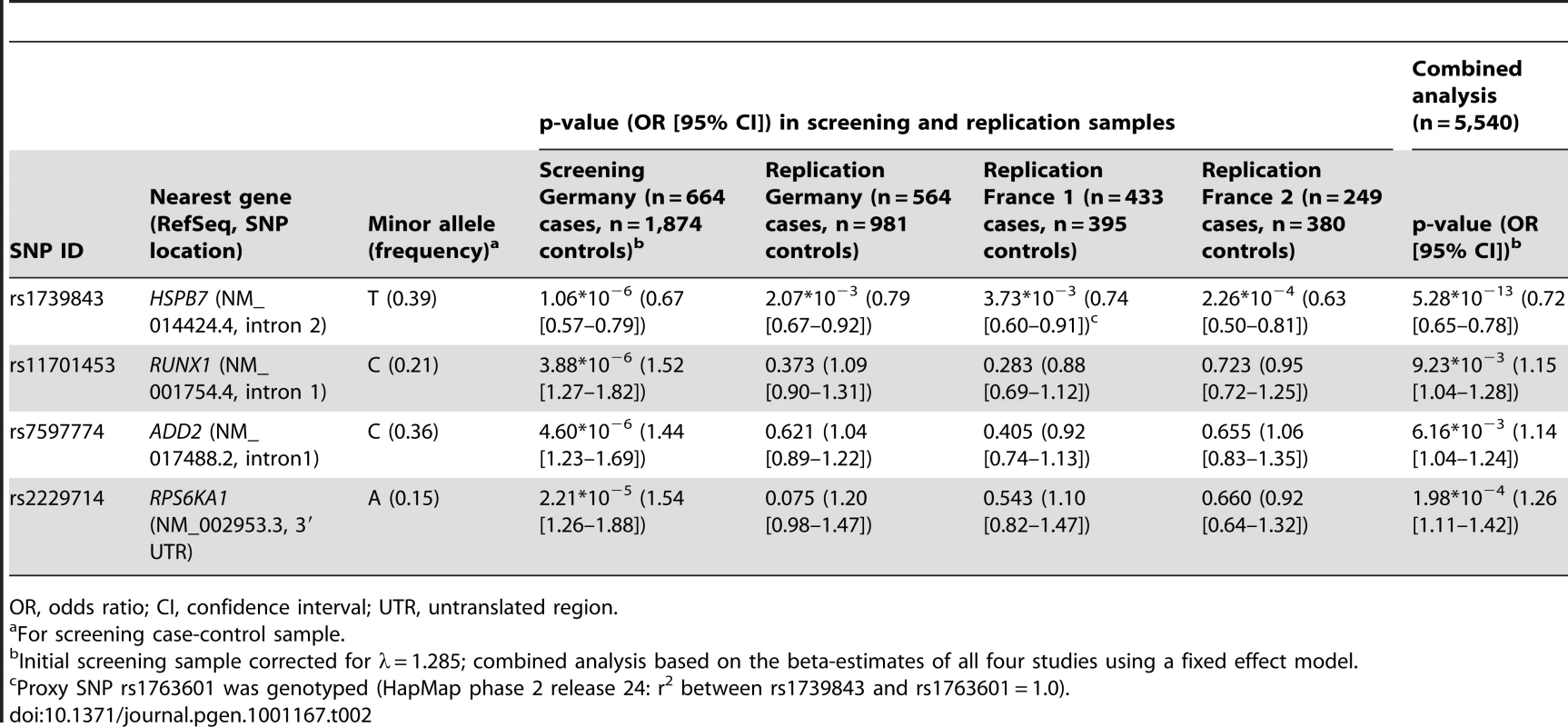
OR, odds ratio; CI, confidence interval; UTR, untranslated region. Replication
The four SNPs with uncorrected p<2.15×10−6 in the initial scan (rs1739843, rs11701453, rs7597774 and rs2229714) were analyzed using logistic regression adjusted for gender in three independent replication samples. First, n = 564 additional German DCM patients and n = 981 controls were genotyped for the four SNPs. Marker rs1739843 showed strong association with DCM (p = 2.07*10−3, OR = 0.79 [95% CI 0.67–0.92]). Conversely, for rs2229714 (p = 0.075, OR = 1.20 [95% CI 0.98–1.47]), rs11701453 (p = 0.373, OR = 1.09 [95% CI 0.90–1.31]) and rs7597774 (p = 0.621, OR = 1.04 [95% CI 0.89–1.22]) the initial association results were not replicated. Second, a French replication sample (France 1) consisted of n = 433 cases and n = 395 controls. Only rs1739843 showed association with DCM after adjustment for gender (p = 3.73*10−3, OR = 0.74 [95% CI 0.60–0.91]). For the other SNPs, no significant association was seen in this sample. Third, in an independent second French replication sample (France 2), again only rs1739843 showed association with DCM after adjustment for gender (p = 2.26*10−4, OR = 0.63 [95% CI 0.50–0.81]). Replication results are summarized in Table 2. None of the four polymorphisms showed deviation from Hardy-Weinberg equilibrium in any replication samples.
In a combined analysis of the screening step, corrected for the λ factor of 1.285, and the three follow-up studies (n = 5,540), the SNP rs1739843 reached a p-value of 5.28*10−13 (OR = 0.72 [95% CI 0.65–0.78]) for association with idiopathic DCM (Table 2, Figure 2). There was no between-study heterogeneity for this effect (I2 = 6.9%, p = 0.36).
Fig. 2. Forest plot for rs1739843 in initial screening sample (λ-corrected) and three replication samples (Germany, France 1, and France 2), together with results from the combined analysis. 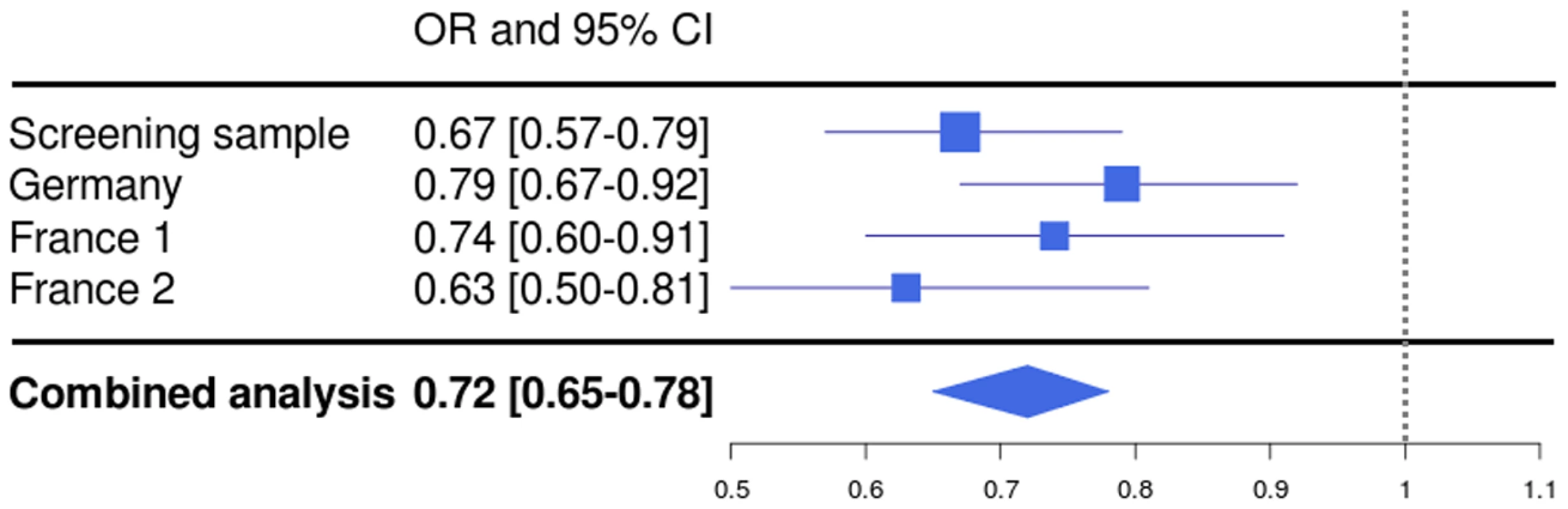
Resequencing
To reveal potential causal variants, the coding region of HSPB7 was resequenced in a total of 48 DCM patients. We detected three known synonymous variants (rs945416, rs732286 and rs1739840). The synonymous variants rs945416 (position 19, serine) and rs732286 (position 33, alanine) are in high LD with rs1739843 (r2 = 0.96, HapMap CEU data release #24). SNP rs1739840 (position 117, threonine) is not available in HapMap. In the initial sample of 664 DCM patients, all three synonymous polymorphisms are in perfect LD to each other and to rs1739843 as shown by genotyping. Neither missense nor splice site de novo mutations were identified by sequencing. Synonymous SNP rs11807575, as well as non-synonymous variants rs77021870 and rs74626772 were listed in databases, but not found to be polymorphic in our sample.
Analysis of DCM candidate genes
Since the design of the 50K human gene-centric bead chip (IBC array) aims at a large-scale gene-based approach, we screened candidate genes which are known for or potentially involved in susceptibility to DCM in our initial screening sample utilizing the information on 30,920 SNPs. We established a list of previously reported genes for DCM by searching PubMed and OMIM databases (http://www.ncbi.nlm.nih.gov/) for “CARDIOMYOPATHY, DILATED” and “GENETIC”. A total of 315 SNPs including 234 independent SNPs (defined as SNPs with pairwise r2<0.8 based on LD in the control group) were located in or near (+/−10kb) the chosen candidate genes, representing 1.01% of array content. DCM association results for these SNPs were obtained from our screening study on 664 cases and 1,874 controls (Table 3, more details in Table S2). On a single candidate gene level, polymorphisms in or near ABCC9, DES, MYH6 and TPM1 showed nominal significance after Bonferroni correction for the number of SNPs tested in gene regions (p = 0.010, p = 0.022, p = 0.005 and p = 0.018, respectively). However, none of these SNPs remained significant after correction for the 234 independent markers tested in this DCM candidate gene approach. Our study was powered to detect moderate to large effects (e. g. for OR>1.3 and MAF = 30% or OR>1.5 and MAF = 20% or OR>1.7 and MAF = 10%, the power was 56%, 96% and 97% for two-sided p<0.05/234 = 2.14*10−4, respectively).
Tab. 3. Candidate gene approach on DCM causing or susceptibility genes. 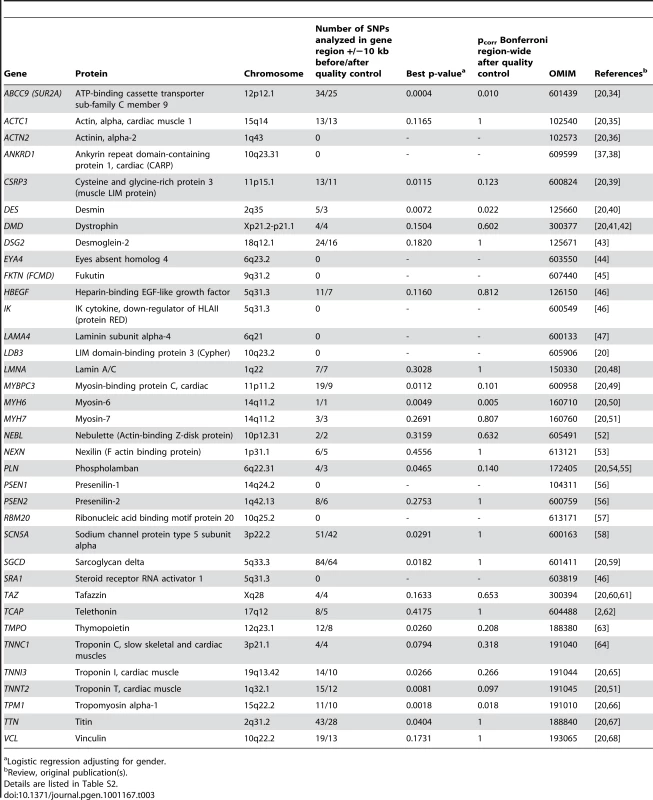
Logistic regression adjusting for gender. Discussion
In the present case-control study, we evaluated the relationship of common SNPs with sporadic DCM using a large-scale screening approach. Our comprehensive strategy set out to analyze the human gene-centric 50K bead chip (IBC array), which focuses on loci with a potential functional link to cardiovascular disease (CVD) and covers more than 45,000 SNPs from about 2,000 genes [6].
Our study identified a polymorphism (rs1739843) in intron 2 of the HSPB7 gene being associated with susceptibility to DCM in a German case-control sample with three replication steps. Recently, Cappola et al. reported an association between rs1739843 and both, ischemic and non-ischemic heart failure, applying the same gene-centric 50K bead chip [7]. They found a protective effect of the minor allele, which is in conformity with our results on DCM. As DCM is a potential preliminary stage for non-ischemic heart failure, these independent findings point to a possible common pathophysiologic cascade. However, a second association signal for heart failure located in the FRMD4B region (rs6787362, minor allele frequency (MAF) 10.4%) identified by Cappola et al. [7] could not be detected in our DCM case-control sample (p = 0.64). Our study had a power of 99% to find a nominal association between DCM and rs6787362 with p<0.05 and an OR = 0.67.
The finding on HSPB7 is also in-line with a previously reported large-scale re-sequencing approach in four biologically relevant cardiac signaling genes, which detected HSPB7 sequence diversity in sporadic cardiomyopathy [8]. Our data together with the results from Cappola et al. [7] and Matkovich et al. [8], substantiate the importance of rs1739843 or related polymorphisms in the HSPB7 locus for DCM and heart failure and possibly underscore a common genetic basis for these related phenotypes.
Matkovich et al. further report that none of the detected HSPB7 gene variants altered amino acid sequence [8], which is also consistent with the fact that we found neither missense nor splice site mutations in the HSPB7 sequence. Therefore, the biological mechanism explaining the association between the polymorphism rs1739843 and DCM risk remains still unclear. The three detected synonymous variants (rs945416, rs732286 and rs1739840) are in high LD with each other as well as with our lead SNP rs1739843 and lie on one LD block. Therefore, it could be hypothesized that these SNPs represent causal risk factors for DCM, as described for the P-glycoprotein encoding gene MCP1 and affected drug and inhibitor interactions [9]. Synonymous SNPs lead to changes in codon usage and may cause functional implications by conformational changes in protein structure due to translation efficiency. Alternatively, a de novo splice site could be created by a SNP or other (unmapped) polymorphisms outside the HSPB7 coding region may alter its gene expression. Clearly, functional studies would be required to prove these hypotheses.
Besides the HSPB7 gene, where the lead SNP is located, also five genes (CLCNKA, CLCNKB, C1orf64, ZBTB17 and SPEN) lying on the same LD block may potentially be responsible for the association with DCM. CLCNKA and CLCNKB encode for two members of the family of voltage-gated chloride channels. These proteins are predominantly expressed in the kidney and participate in renal salt reabsorption [10]. The function of C1orf64 is currently unknown. ZBTB17, also known as MIZ-1, encodes a zinc finger protein involved in the regulation of c-myc [11]. SPEN (RBM15C or MINT) encodes a conserved transcriptional repressor that controls the expression of regulators in diverse signaling pathways [12], [13].
HSPB7, encoding the small heat shock protein cvHsp (also known as HspB7), is the functionally most plausible candidate gene in this genomic region. It is known to be expressed in cardiovascular and insulin-sensitive tissues [14]. In general, the expression and activation of heat shock proteins is influenced by elevated temperatures as well as ischemia, hypoxia and acute cellular stress [15], [16]. In the aging skeletal muscle increase of cvHsp protein content was observed [17]. cvHsp was shown to be constitutively localized under non-stressful conditions to nuclear splicing speckles and may influence mRNA processing [18]. Recent data suggest co-localization between cvHsp and α-B-crystallin in the z-band of cardiac tissue and interaction with other small heat shock proteins [19]. However, further investigations like genomic fine-mapping and subgroup analyses in the context of cardiomyopathies are needed.
Genetic analyses in familial forms of DCM led to the identification of risk loci showing X-linked, autosomal dominant or autosomal recessive patterns of inheritance [2], [20], [21]. Some of the DCM causing genes or plausible candidate genes were also covered by the 50K bead chip, wherefore we specifically tested those SNPs lying in risk gene regions (10 kb upstream and downstream, respectively). In these analyses, no significant association with any of the gene variants was found, indicating that in sporadic cases of DCM probably other pathways are involved than in familial DCM. However, less frequent variants may have been missed due to insufficient power of our screening sample. Furthermore, the distinction between familial and sporadic forms of DCM is, to a certain degree, somewhat arbitrary. Screening of family members is rarely done in clinical routine, but when carried out on a systematic basis, up to 7% of previously healthy first-degree relatives have reduced left ventricular function or dilation without presence of cardiac symptoms [22]. Therefore, it might be anticipated that genetic testing could help to identify individuals at risk in familial DCM but also in families of patients affected by so-called idiopathic forms of the disease.
Already known genetic factors account for only a fraction of DCM heritability [20]. Given a 1.5-fold increased risk of DCM among heterozygous subjects in our screening sample (48% in the general population-based KORA study) and a 2.25 times increased risk among homozygous subjects (34% in KORA), 49% of DCM cases would be attributable to the SNP rs1739843 (or correlated polymorphisms) with 19% attributable to heterozygous and 30% to homozygous carriers, respectively. Therefore, the genetic component seems to comprise a large proportion for this disease. However, with the prevalence of the idiopathic form of the disease being about 1∶2,700 [23], a genetic screening of the general population would include four cases out of 10,000 screened persons and two of these would have the disease due to this SNP. Therefore, the great potential of this variant might rather be screening of high risk populations, or this pathway indicates potential drug targets. Further investigations should aim (1) to identify additional variants underlying DCM susceptibility with otherwise unknown etiology and (2) to analyze potential influence of these common alleles as modifiers for familial forms of DCM. Taken together for both, modifiers of familial forms and susceptibility alleles in idiopathic DCM, knowledge of genetic background will support preventive medical measures in the future.
Some limitations of our study should be mentioned. First, we conducted a large-scale SNP analysis focused on genes potentially involved in cardiovascular traits. Therefore, on the one hand we were able to detect associations between DCM and polymorphisms only in these pre-selected genes. On the other hand, the 50K human CVD bead chip allows comprehensive gene-based analysis with more than 2,000 well covered loci. Second, our sample size only allowed to detect moderate to large effects (e. g. for OR>1.3 and MAF = 30% or OR>1.5 and MAF = 20% or OR>1.7 and MAF = 10%, the power was 19%, 75% and 80% for p<2.15*10−6, respectively). Therefore, we may have overlooked real association signals in our screening step. Third, there could be some population stratification in our initial screen sample. However, the observed λ could also be caused - in part - by underlying association due to the analysis of pre-selected loci known or suggested to be involved in cardiovascular phenotypes. The fact that the association between rs1739843 in HSPB7 and idiopathic DCM was replicated in three independent samples strongly enhances the confidence in our results.
Materials and Methods
Ethics statement
The ethics committees of the participating study centers approved the study protocol and all participants gave their written informed consent. The study was in accordance with the principles of the current version of the Declaration of Helsinki.
Case-control samples and phenotyping
Cases for the initial German screening study were recruited from the German Heart Institute (Berlin), and controls were from a population-based German KORA study (follow-up survey F3, Augsburg) [24]. Phenotypic details are summarized in Table 1. Controls (n = 1,874) had no medical history for coronary artery disease (CAD), myocardial infarction or DCM; mean age was 62±11 years and slightly more women (n = 986) than men (n = 888) were present in the control group. Inclusion criteria for DCM cases were the following: reduced systolic function (left ventricular ejection fraction (LVEF) <45%) without angiographically assessed evidence of major CAD, significant valvular heart disease (>grade 2, i. e. such as mitral or aortic regurgitation), hypertensive heart disease, congenital heart disease, myocarditis (by endomyocardial biopsy, when available) or other secondary forms of heart failure. Patients with a positive family history were also excluded from this study. In DCM cases (n = 664), mean LVEF was 24±3% and mean age of disease diagnosis was 46±11 years.
For the first replication step, additional German DCM cases (mean age 53±13 years; n = 564, n = 440 men, n = 124 women) were recruited from different German study centers: Berlin, n = 64; Lübeck, n = 96 (Angio-Lueb); Marburg, n = 61 (EUROGENE); Münster, n = 101 (EUROGENE); Regensburg, n = 150 (EUROGENE); Regensburg, n = 92 (GoKard). Independent German KORA controls from surveys S1 and S2 (n = 981, n = 539 men, n = 442 women) had a mean age of 52±10 years [24]. Inclusion and exclusion criteria were identical to the initial case-control sample.
A second replication study (France 1) was recruited in France (CARDIGENE) [25], [26]. The French cases were of white European origin (all born in France, from parents born in France or neighboring countries) with a diagnosis of DCM, i. e. enlarged left ventricle end-diastolic volume/diameter >140 ml/m2 on ventriculography or >34 mm/m2 on echocardiography and LVEF ≤40% confirmed over a six-month period, in the absence of causal factors such as CAD or sustained hypertension, intrinsic valvular disease, documented myocarditis, congenital malformation, insulin-dependent diabetes. Only apparently sporadic DCM cases without additional (first degree) relative with DCM were included (but 8% were in fact with familial form after careful cardiac examination in relatives). Recruitment was performed in ten hospitals in six regions in France (Lille, Lyon, Nancy, Nantes, Paris-Ile de France, Strasbourg) from September 1994 to February 1996. A total of 433 patients (229 had undergone a cardiac transplantation) were included (n = 345 men, n = 88 women). Mean age of patients was 45±11 years, mean LVEF was 23±7% and mean end-diastolic volume was 195±67 ml/m2. Controls (n = 395) were age - and gender-matched (n = 310 men, n = 85 women).
The third replication sample was also of French origin (France 2). Inclusion criteria were identical to the France 1 sample. A total of 249 patients from EUROGENE and PHRC were included (n = 198 men, n = 51 women). Mean age of patients at diagnosis was 51±10 years. Controls (n = 380) were free of medical history for CAD, myocardial infarction or DCM and mean age was 46±11 years (n = 301 men, n = 79 women).
Genotyping
Initial genotyping was carried out using the 50K gene-centric human CVD bead chip version 1 (IBC v1 array) (Illumina, San Diego, CA, USA) [6] following the manufacturer's protocol. Data were analyzed (calling and sample clustering) and exported employing BeadStudio analysis software (Illumina). From the initial 45,707 SNPs, those markers with low call rates (<95%) or low frequency (MAF<1%) were excluded. Minimal call rate per individual was 90%. We used identity-by-descent methods to exclude unknown first-degree relation of participants.
Replication samples were taken from human CVD bead chip data or genotyped with 5′ exonuclease TaqMan technology (Applied Biosystems, Foster City, CA, USA) as previously described [27]. A by-design assay for rs1739843 was used with primer sequences 5′-CTCTGCCATCACCATCTCACA-3′ and 5′-GGCAGAGGGAGCCTGAG-3′ and probe sequences 5′-VIC-AGGGTGGGAGGTGACAG-NFQ-3′ and 5′-FAM-AGGGTGGGAGATGACAG-NFQ-3′ (site of rs1739843 is underlined; fluorescence dyes VIC and FAM on 5′ end and non-fluorescence quencher (NFQ) on 3′ end are indicated). All other assays were obtained pre-designed directly from Applied Biosystems. Detailed information on assays used in France 2 sample are available at http://genecanvas.ecgene.net/infusions/genecanvas/Polymorphisms/PolymorphismsList.php.
SNP rs1739843 was re-genotyped using the by-design TaqMan assay in initial case sample (n = 664) to check for discrepancies between human CVD bead chip and TaqMan genotypes. A >99.8% concordance of genotypes was found. For all genotyped samples a call rate >97% for each SNP assay was reached.
Resequencing
Polymerase chain reaction (PCR) primer were generated using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) [28] to cover the coding parts of the three HSPB7 exons (GenBank accession No. NM_14424.4). The primer sequences and PCR amplification products are listed in Table S3. Included intronic regions were 267 bp for 5′ end of intron 1, 156 bp for 3′ end of intron 1, 136 bp for 5′ end of intron 2, and 89 bp for 3′ end of intron 2, respectively. PCR cycling conditions consisted of an initial denaturation at 95°C for 9 min, followed by 40 cycles with denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 30 s, with a final elongation step at 72°C for 7 min.
After PCR amplification, primers and dNTPs were removed using ExoSAP-IT (USB Europe, Staufen, Germany) following the manufacturer's instructions. The purified PCR products were directly sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit Version 3.1 on the ABI 3730 (Applied Biosystems, Foster City, CA, USA).
Statistical analyses
For initial screening and replication analyses, logistic regression adjusted for gender was used. P-values, odds ratios (OR) and their 95% confidence intervals (CI) were reported. The inflation factor λ was computed in the 50K initial screening analysis for logistic regression analysis assuming a χ2 distribution with two degrees of freedom of the minus two-times logep measures (90% highest p-values). The p-values and CI from initial screening analysis were genomic-control corrected using this λ factor via standard errors (standard error[corrected] = sqrt(λ)*standard error) and beta estimates (95%CI beta[corrected] = beta±1.96*standard error[corrected]). Deviation from Hardy-Weinberg equilibrium was calculated with an exact test [29]. Statistical and association analyses were performed using JMP 7.0.2 (SAS Institute Inc, Cary, NC, USA) and PLINK v1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/) [30], respectively. Power analysis was carried out using Quanto 1.2.4 (http://hydra.usc.edu/gxe/). We combined the initial scan results corrected for λ with the replication studies' results using a fixed effect model. Annotation of association results on a genome level was performed with WGAViewer software (http://people.genome.duke.edu/~dg48/WGAViewer/) [31]. LD patterns were calculated using HapMap releases #22 and #24 (http://www.hapmap.org/) [32].
Supporting Information
Zdroje
1. MaronBJ
TowbinJA
ThieneG
AntzelevitchC
CorradoD
2006 Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 113 1807 1816
2. RichardP
VillardE
CharronP
IsnardR
2006 The Genetic Bases of Cardiomyopathies. Journal of the American College of Cardiology 48 A79 A89
3. RampersaudE
KinnamonDD
HamiltonK
KhuriS
HershbergerRE
2010 Common Susceptibility Variants Examined for Association with Dilated Cardiomyopathy. Ann Hum Genet 74 110 116
4. IoannidisJP
2005 Why most published research findings are false. PLoS Med 2 e124 doi:10.1371/journal.pmed.0020124
5. PearsonTA
ManolioTA
2008 How to interpret a genome-wide association study. JAMA 299 1335 1344
6. KeatingBJ
TischfieldS
MurraySS
BhangaleT
PriceTS
2008 Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE 3 e3583 doi:10.1371/journal.pone.0003583
7. CappolaTP
LiM
HeJ
KyB
GilmoreJ
2010 Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet 3 147 154
8. MatkovichSJ
Van BoovenDJ
HindesA
KangMY
DruleyTE
2010 Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing HSPB7 polymorphisms associated with disease. J Clin Invest 120 280 289
9. Kimchi-SarfatyC
OhJM
KimIW
SaunaZE
CalcagnoAM
2007 A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315 525 528
10. UchidaS
2000 In vivo role of CLC chloride channels in the kidney. Am J Physiol Renal Physiol 279 F802 F808
11. IkegakiN
GotohT
KungB
RicebergJS
KimDY
2007 De novo identification of MIZ-1 (ZBTB17) encoding a MYC-interacting zinc-finger protein as a new favorable neuroblastoma gene. Clin Cancer Res 13 6001 6009
12. NewberryEP
LatifiT
TowlerDA
1999 The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry 38 10678 10690
13. LiJ
WangJ
YangX
LiJ
QinH
2006 The Spen homolog Msx2-interacting nuclear target protein interacts with the E2 ubiquitin-conjugating enzyme UbcH8. Mol Cell Biochem 288 151 157
14. KriefS
FaivreJF
RobertP
LeDB
Brument-LarignonN
1999 Identification and characterization of cvHsp. A novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J Biol Chem 274 36592 36600
15. LindquistS
CraigEA
1988 The heat-shock proteins. Annu Rev Genet 22 631 677
16. AnckarJ
SistonenL
2007 Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv Exp Med Biol 594 78 88
17. DoranP
GannonJ
O'ConnellK
OhlendieckK
2007 Aging skeletal muscle shows a drastic increase in the small heat shock proteins alphaB-crystallin/HspB5 and cvHsp/HspB7. Eur J Cell Biol 86 629 640
18. VosMJ
KanonB
KampingaHH
2009 HSPB7 is a SC35 speckle resident small heat shock protein. Biochim Biophys Acta 1793 1343 1353
19. BrodehlA
MartinI
GawlowskiT
StorkI
GummertJ
2010 The small heat shock proteins cvHSP and alpha-B-Crystallin are colocalized and interact in human myocardial tissue. Clin Res Cardiol 99 P467
20. BurkettEL
HershbergerRE
2005 Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol 45 969 981
21. OsterzielKJ
HassfeldS
GeierC
PerrotA
2005 [Familial dilated cardiomyopathy]. Herz 30 529 534
22. MichelsVV
OlsonTM
MillerFA
BallmanKV
RosalesAG
2003 Frequency of development of idiopathic dilated cardiomyopathy among relatives of patients with idiopathic dilated cardiomyopathy. Am J Cardiol 91 1389 1392
23. CoddMB
SugrueDD
GershBJ
MeltonLJIII
1989 Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in Olmsted County, Minnesota, 1975–1984. Circulation 80 564 572
24. WichmannHE
GiegerC
IlligT
2005 KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 67 Suppl 1 S26 S30
25. TessonF
CharronP
PeuchmaurdM
NicaudV
CambienF
1999 Characterization of a unique genetic variant in the beta1-adrenoceptor gene and evaluation of its role in idiopathic dilated cardiomyopathy. CARDIGENE Group. J Mol Cell Cardiol 31 1025 1032
26. CharronP
TessonF
PoirierO
NicaudV
PeuchmaurdM
1999 Identification of a genetic risk factor for idiopathic dilated cardiomyopathy. Involvement of a polymorphism in the endothelin receptor type A gene. CARDIGENE group. Eur Heart J 20 1587 1591
27. StarkK
ReinhardW
NeureutherK
WiedmannS
SedlacekK
2008 Association of common polymorphisms in GLUT9 gene with gout but not with coronary artery disease in a large case-control study. PLoS ONE 3 e1948 doi:10.1371/journal.pone.0001948
28. UntergasserA
NijveenH
RaoX
BisselingT
GeurtsR
2007 Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35 W71 W74
29. WiggintonJE
CutlerDJ
AbecasisGR
2005 A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 76 887 893
30. PurcellS
NealeB
Todd-BrownK
ThomasL
FerreiraMA
2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
31. GeD
ZhangK
NeedAC
MartinO
FellayJ
2008 WGAViewer: software for genomic annotation of whole genome association studies. Genome Res 18 640 643
32. FrazerKA
BallingerDG
CoxDR
HindsDA
StuveLL
2007 A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851 861
33. JohnsonAD
HandsakerRE
PulitSL
NizzariMM
O'DonnellCJ
2008 SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24 2938 2939
34. BienengraeberM
OlsonTM
SelivanovVA
KathmannEC
O'CochlainF
2004 ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet 36 382 387
35. OlsonTM
MichelsVV
ThibodeauSN
TaiYS
KeatingMT
1998 Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 280 750 752
36. MohapatraB
JimenezS
LinJH
BowlesKR
CovelerKJ
2003 Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab 80 207 215
37. Duboscq-BidotL
CharronP
RuppertV
FauchierL
RichterA
2009 Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur Heart J 30 2128 2136
38. MoulikM
VattaM
WittSH
ArolaAM
MurphyRT
2009 ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol 54 325 333
39. KnollR
HoshijimaM
HoffmanHM
PersonV
Lorenzen-SchmidtI
2002 The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111 943 955
40. LiD
TapscoftT
GonzalezO
BurchPE
QuinonesMA
1999 Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation 100 461 464
41. MuntoniF
CauM
GanauA
CongiuR
ArvediG
1993 Brief report: deletion of the dystrophin muscle-promoter region associated with X-linked dilated cardiomyopathy. N Engl J Med 329 921 925
42. TowbinJA
HejtmancikJF
BrinkP
GelbB
ZhuXM
1993 X-linked dilated cardiomyopathy. Molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation 87 1854 1865
43. PoschMG
PoschMJ
GeierC
ErdmannB
MuellerW
2008 A missense variant in desmoglein-2 predisposes to dilated cardiomyopathy. Mol Genet Metab 95 74 80
44. SchönbergerJ
WangL
ShinJT
KimSD
DepreuxFF
2005 Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet 37 418 422
45. ArimuraT
HayashiYK
MurakamiT
OyaY
FunabeS
2009 Mutational analysis of fukutin gene in dilated cardiomyopathy and hypertrophic cardiomyopathy. Circ J 73 158 161
46. FriedrichsF
ZugckC
RauchGJ
IvandicB
WeichenhanD
2009 HBEGF, SRA1, and IK: Three cosegregating genes as determinants of cardiomyopathy. Genome Res 19 395 403
47. SylviusN
TessonF
GayetC
CharronP
BenaicheA
2001 A new locus for autosomal dominant dilated cardiomyopathy identified on chromosome 6q12-q16. Am J Hum Genet 68 241 246
48. FatkinD
MacRaeC
SasakiT
WolffMR
PorcuM
1999 Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 341 1715 1724
49. DaehmlowS
ErdmannJ
KnueppelT
GilleC
FroemmelC
2002 Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem Biophys Res Commun 298 116 120
50. CarnielE
TaylorMR
SinagraG
DiLA
KuL
2005 Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation 112 54 59
51. KamisagoM
SharmaSD
DePalmaSR
SolomonS
SharmaP
2000 Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med 343 1688 1696
52. ArimuraT
NakamuraT
HiroiS
SatohM
TakahashiM
2000 Characterization of the human nebulette gene: a polymorphism in an actin-binding motif is associated with nonfamilial idiopathic dilated cardiomyopathy. Hum Genet 107 440 451
53. HasselD
DahmeT
ErdmannJ
MederB
HugeA
2009 Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med 15 1281 1288
54. HaghighiK
KolokathisF
PaterL
LynchRA
AsahiM
2003 Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest 111 869 876
55. SchmittJP
KamisagoM
AsahiM
LiGH
AhmadF
2003 Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299 1410 1413
56. LiD
ParksSB
KushnerJD
NaumanD
BurgessD
2006 Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am J Hum Genet 79 1030 1039
57. BrauchKM
KarstML
HerronKJ
deAM
PellikkaPA
2009 Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol 54 930 941
58. OlsonTM
MichelsVV
BallewJD
ReynaSP
KarstML
2005 Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA 293 447 454
59. TsubataS
BowlesKR
VattaM
ZintzC
TitusJ
2000 Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest 106 655 662
60. BioneS
D'AdamoP
MaestriniE
GedeonAK
BolhuisPA
1996 A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet 12 385 389
61. D'AdamoP
FassoneL
GedeonA
JanssenEA
BioneS
1997 The X-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am J Hum Genet 61 862 867
62. HayashiT
ArimuraT
Itoh-SatohM
UedaK
HohdaS
2004 Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol 44 2192 2201
63. TaylorMR
SlavovD
GajewskiA
VlcekS
KuL
2005 Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Hum Mutat 26 566 574
64. MogensenJ
MurphyRT
ShawT
BahlA
RedwoodC
2004 Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol 44 2033 2040
65. MurphyRT
MogensenJ
ShawA
KuboT
HughesS
2004 Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet 363 371 372
66. OlsonTM
KishimotoNY
WhitbyFG
MichelsVV
2001 Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol 33 723 732
67. GerullB
GramlichM
AthertonJ
McNabbM
TrombitasK
2002 Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet 30 201 204
68. OlsonTM
IllenbergerS
KishimotoNY
HuttelmaierS
KeatingMT
2002 Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation 105 431 437
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2010 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Common Genetic Variants and Modification of Penetrance of -Associated Breast Cancer
- FSHD: A Repeat Contraction Disease Finally Ready to Expand (Our Understanding of Its Pathogenesis)
- Genome-Wide Identification of Targets and Function of Individual MicroRNAs in Mouse Embryonic Stem Cells
- Allele-Specific Down-Regulation of Expression Induced by Retinoids Contributes to Climate Adaptations
- The Meiotic Recombination Checkpoint Suppresses NHK-1 Kinase to Prevent Reorganisation of the Oocyte Nucleus in
- Actin Depolymerizing Factors Cofilin1 and Destrin Are Required for Ureteric Bud Branching Morphogenesis
- DSIF and RNA Polymerase II CTD Phosphorylation Coordinate the Recruitment of Rpd3S to Actively Transcribed Genes
- Continuous Requirement for the Clr4 Complex But Not RNAi for Centromeric Heterochromatin Assembly in Fission Yeast Harboring a Disrupted RITS Complex
- Genome-Wide Association Study of Blood Pressure Extremes Identifies Variant near Associated with Hypertension
- The Cytosine Methyltransferase DRM2 Requires Intact UBA Domains and a Catalytically Mutated Paralog DRM3 during RNA–Directed DNA Methylation in
- β-Actin and γ-Actin Are Each Dispensable for Auditory Hair Cell Development But Required for Stereocilia Maintenance
- Genetic Association Study Identifies as a Risk Gene for Idiopathic Dilated Cardiomyopathy
- Evidence for a Xer/ System for Chromosome Resolution in Archaea
- Four Novel Loci (19q13, 6q24, 12q24, and 5q14) Influence the Microcirculation
- Lifespan Extension by Preserving Proliferative Homeostasis in
- Ancient and Recent Adaptive Evolution of Primate Non-Homologous End Joining Genes
- Loss of the p53/p63 Regulated Desmosomal Protein Perp Promotes Tumorigenesis
- Altering a Histone H3K4 Methylation Pathway in Glomerular Podocytes Promotes a Chronic Disease Phenotype
- Characterization of LINE-1 Ribonucleoprotein Particles
- Conserved Genes Act as Modifiers of Invertebrate SMN Loss of Function Defects
- Alternative Splicing at a NAGNAG Acceptor Site as a Novel Phenotype Modifier
- Tight Regulation of the Gene of the KplE1 Prophage: A New Paradigm for Integrase Gene Regulation
- Conjugative DNA Transfer Induces the Bacterial SOS Response and Promotes Antibiotic Resistance Development through Integron Activation
- Nasty Viruses, Costly Plasmids, Population Dynamics, and the Conditions for Establishing and Maintaining CRISPR-Mediated Adaptive Immunity in Bacteria
- Stress-Induced Activation of Heterochromatic Transcription
- H3K27me3 Profiling of the Endosperm Implies Exclusion of Polycomb Group Protein Targeting by DNA Methylation
- Simultaneous Disruption of Two DNA Polymerases, Polη and Polζ, in Avian DT40 Cells Unmasks the Role of Polη in Cellular Response to Various DNA Lesions
- Characterising and Predicting Haploinsufficiency in the Human Genome
- Dual Functions of ASCIZ in the DNA Base Damage Response and Pulmonary Organogenesis
- Pervasive Cryptic Epistasis in Molecular Evolution
- Transition from Positive to Neutral in Mutation Fixation along with Continuing Rising Fitness in Thermal Adaptive Evolution
- Comprehensive Analysis Reveals Dynamic and Evolutionary Plasticity of Rab GTPases and Membrane Traffic in
- Regulates Tissue-Specific Mitochondrial DNA Segregation
- Role for the Mammalian Swi5-Sfr1 Complex in DNA Strand Break Repair through Homologous Recombination
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genome-Wide Identification of Targets and Function of Individual MicroRNAs in Mouse Embryonic Stem Cells
- Common Genetic Variants and Modification of Penetrance of -Associated Breast Cancer
- Allele-Specific Down-Regulation of Expression Induced by Retinoids Contributes to Climate Adaptations
- Simultaneous Disruption of Two DNA Polymerases, Polη and Polζ, in Avian DT40 Cells Unmasks the Role of Polη in Cellular Response to Various DNA Lesions
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



