-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
The dual toxicity/essentiality of copper forces cells to maintain a tightly regulated homeostasis for this metal in all living organisms, from bacteria to humans. Consequently, many genes have previously been reported to participate in copper detoxification in bacteria. Myxococcus xanthus, a prokaryote, encodes many proteins involved in copper homeostasis that are differentially regulated by this metal. A σ factor of the ECF (extracytoplasmic function) family, CorE, has been found to regulate the expression of the multicopper oxidase cuoB, the P1B-type ATPases copA and copB, and a gene encoding a protein with a heavy-metal-associated domain. Characterization of CorE has revealed that it requires copper to bind DNA in vitro. Genes regulated by CorE exhibit a characteristic expression profile, with a peak at 2 h after copper addition. Expression rapidly decreases thereafter to basal levels, although the metal is still present in the medium, indicating that the activity of CorE is modulated by a process of activation and inactivation. The use of monovalent and divalent metals to mimic Cu(I) and Cu(II), respectively, and of additives that favor the formation of the two redox states of this metal, has revealed that CorE is activated by Cu(II) and inactivated by Cu(I). The activation/inactivation properties of CorE reside in a Cys-rich domain located at the C terminus of the protein. Point mutations at these residues have allowed the identification of several Cys involved in the activation and inactivation of CorE. Based on these data, along with comparative genomic studies, a new group of ECF σ factors is proposed, which not only clearly differs mechanistically from the other σ factors so far characterized, but also from other metal regulators.
Published in the journal: CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor. PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002106
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002106Summary
The dual toxicity/essentiality of copper forces cells to maintain a tightly regulated homeostasis for this metal in all living organisms, from bacteria to humans. Consequently, many genes have previously been reported to participate in copper detoxification in bacteria. Myxococcus xanthus, a prokaryote, encodes many proteins involved in copper homeostasis that are differentially regulated by this metal. A σ factor of the ECF (extracytoplasmic function) family, CorE, has been found to regulate the expression of the multicopper oxidase cuoB, the P1B-type ATPases copA and copB, and a gene encoding a protein with a heavy-metal-associated domain. Characterization of CorE has revealed that it requires copper to bind DNA in vitro. Genes regulated by CorE exhibit a characteristic expression profile, with a peak at 2 h after copper addition. Expression rapidly decreases thereafter to basal levels, although the metal is still present in the medium, indicating that the activity of CorE is modulated by a process of activation and inactivation. The use of monovalent and divalent metals to mimic Cu(I) and Cu(II), respectively, and of additives that favor the formation of the two redox states of this metal, has revealed that CorE is activated by Cu(II) and inactivated by Cu(I). The activation/inactivation properties of CorE reside in a Cys-rich domain located at the C terminus of the protein. Point mutations at these residues have allowed the identification of several Cys involved in the activation and inactivation of CorE. Based on these data, along with comparative genomic studies, a new group of ECF σ factors is proposed, which not only clearly differs mechanistically from the other σ factors so far characterized, but also from other metal regulators.
Introduction
Myxococcus xanthus is a soil-dwelling δ-proteobacterium of the group of myxobacteria used as a model to study multicellular behavior and differentiation, because it exhibits a complex developmental cycle triggered by starvation [1]. However, M. xanthus cells not only have to adapt their metabolism and behavior to changing nutritional concentrations, but also to other parameters, such as metals.
Copper is a transition metal that functions as an ideal biological cofactor due to its ability to alternate between the redox states Cu(I) and Cu(II). However, copper also generates reactive oxygen species that cause cell damage [2]. This duality forces organisms to maintain a strict homeostasis for this metal. The most representative examples of the effect of disturbances in copper homeostasis are two inherited human disorders, Wilson disease and Menkes syndrome, which are directly linked to overload and deficiency of this metal, respectively [3].
Copper is required by prokaryotes in trace amounts because it is used as a cofactor by a few proteins. Hence, most bacterial homeostatic mechanisms are devoted to conferring resistance to this metal. The most common mechanisms are copper-transporting P1B-type ATPases, copper chaperones, multicopper oxidases (MCOs), and Cus systems [4]. In bacteria such as Escherichia coli, one of each of these elements is encoded in the genome [4]. In other bacteria, the homeostatic mechanism is even simpler, consisting of two P1B-type ATPases and one chaperone (Synechocystis PCC6803, Enterococcus hirae, and Lactococcus lactis), or one ATPase and one chaperone (Bacillus subtilis) [4], [5]. In contrast, the large M. xanthus genome encodes a large number of paralogous genes to confer copper tolerance: three MCOs, at least two Cus systems, and three P1B-type ATPases, as well as the genes required for the biosynthesis of carotenoids [6]–[9]. This gene redundancy indicates that copper homeostasis in this myxobacterium is more complex than in other prokaryotes. All of these genes have been shown to be differentially regulated [6]–[9], suggesting that this sophisticated network must be finely regulated by specific metal sensors.
One of the signal transduction mechanisms used by bacteria to direct gene expression at the transcriptional level in response to stress signals is represented by alternative σ factors [10]. The largest group of alternative σ factors is the ECF (extracytoplasmic function) family, which corresponds to group 4 of the σ70 proteins [11]. ECF σ factors are small proteins, quite divergent in sequence, that contain only two regions (σ2 and σ4) required for interaction with the RNA polymerase core enzyme and recognition of the promoter [12]. Their ability to promote transcription relies on a protein that is normally cotranscribed with the σ factor, named anti-σ factor. In the absence of external signals, ECF σ factors are sequestered by their cognate anti-σ. After detecting the specific stimulus, the anti-σ factor releases the σ subunit, which can then promote gene expression after recruitment of the core RNA polymerase [13]–[16].
In this report, we identify a novel metal sensor involved in copper homeostasis in M. xanthus named CorE (for copper-regulated ECF σ factor). We demonstrate that CorE requires copper in order to bind to DNA and that its activity is modulated by the redox state of this metal. According to these data, we propose a new group of ECF σ factors, defined by a Cys-rich domain (CRD) located at the C terminus of the protein, which is essential for activation and inactivation of the protein.
Results/Discussion
CorE is an ECF σ factor involved in copper homeostasis
Most M. xanthus genes involved in copper homeostasis are located in the genome in two clusters [8]. In copper region 2, and next to the MCO cuoB, a gene encoding a protein with high similarity to ECF σ factors was found (MXAN_3426), suggesting that it could regulate the expression of genes involved in conferring copper resistance. This σ factor has been designated as CorE. The analysis of the CorE sequence has revealed a domain architecture with the conserved regions σ2 (sigma70_r2, PF04542; E-value of 3.2e-14) and σ4 (sigma70_r4_2, PF08281; E-value of 4.3e-06) typical of this type of σ factors [11], [12].
To determine the role of CorE in copper homeostasis, a strain harboring a corE-lacZ fusion was constructed, and the analysis of this strain revealed that corE was up-regulated by copper (Figure 1A). Additionally, an in-frame deletion mutant (ΔcorE) was also generated, and the phenotypic analysis of this strain confirmed that this regulator conferred copper tolerance (Figure 1B).
Fig. 1. CorE is involved in copper homeostasis. 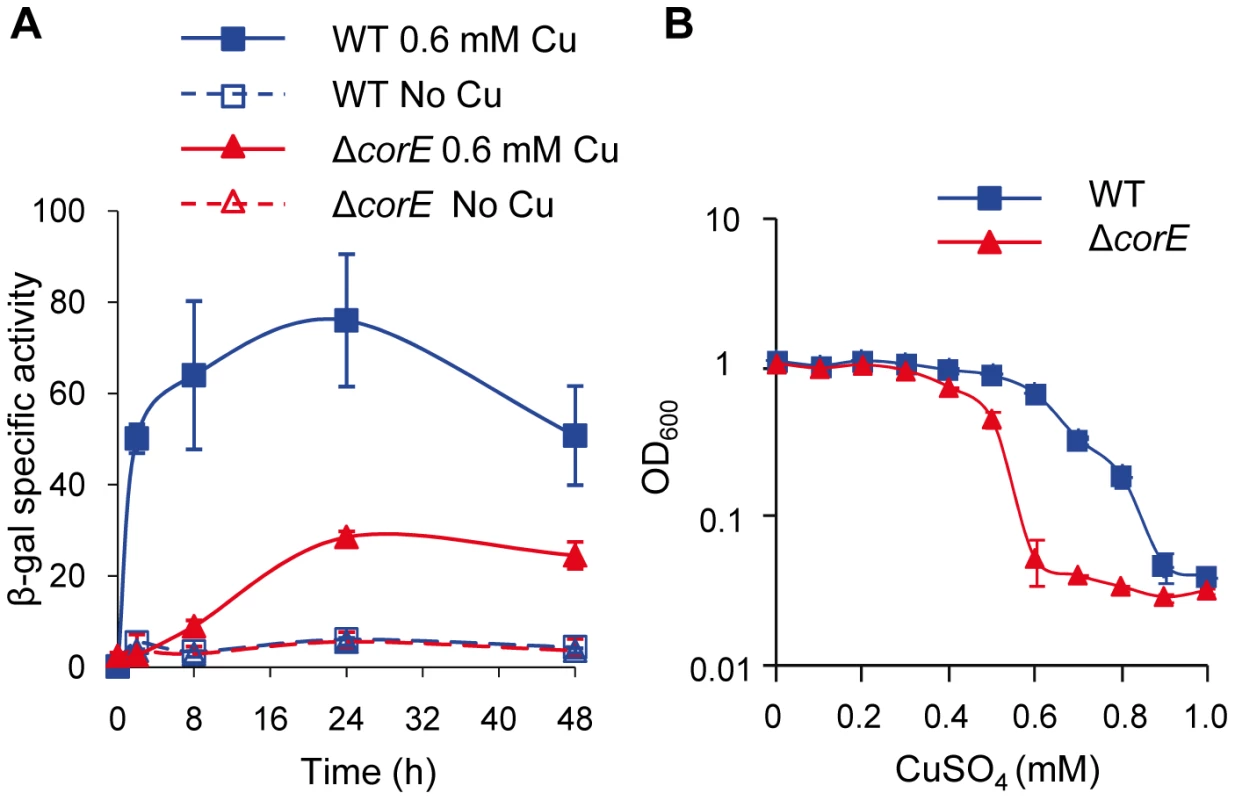
(A) Copper up-regulation of corE. β-gal specific activity was determined in cell extracts of the WT (blue lines) and ΔcorE (red lines) strains (harboring the fusion corE-lacZ) from CTT agar plates containing no copper (open symbols) or 0.6 mM (closed symbols) copper sulfate. (B) Effect of copper on M. xanthus growth. WT (blue line) and ΔcorE (red line) strains were grown in the absence of the metal and diluted to an OD600 of 0.05 into fresh CTT liquid media containing the indicated copper concentrations. The OD600 was then monitored after 24 h of incubation. Error bars indicate standard deviations. Genes regulated by CorE
To identify genes regulated by CorE, plasmids containing fusions between the genes that have so far been involved in copper and/or other metal homeostasis in M. xanthus and lacZ were electroporated into the ΔcorE mutant. When the expression profiles of these genes in the mutant were compared with those exhibited in the wild-type (WT) strain, it was observed that only the MCO cuoB and the P1B-type ATPase copB remained undetectable in the ΔcorE background in the presence of copper (Figure 2A and Figure S1), indicating that they are regulated by this σ factor. Interestingly, these two M. xanthus genes exhibit a characteristic expression profile, with a peak at 2 h after the addition of exogenous copper.
Fig. 2. CorE-dependent genes. 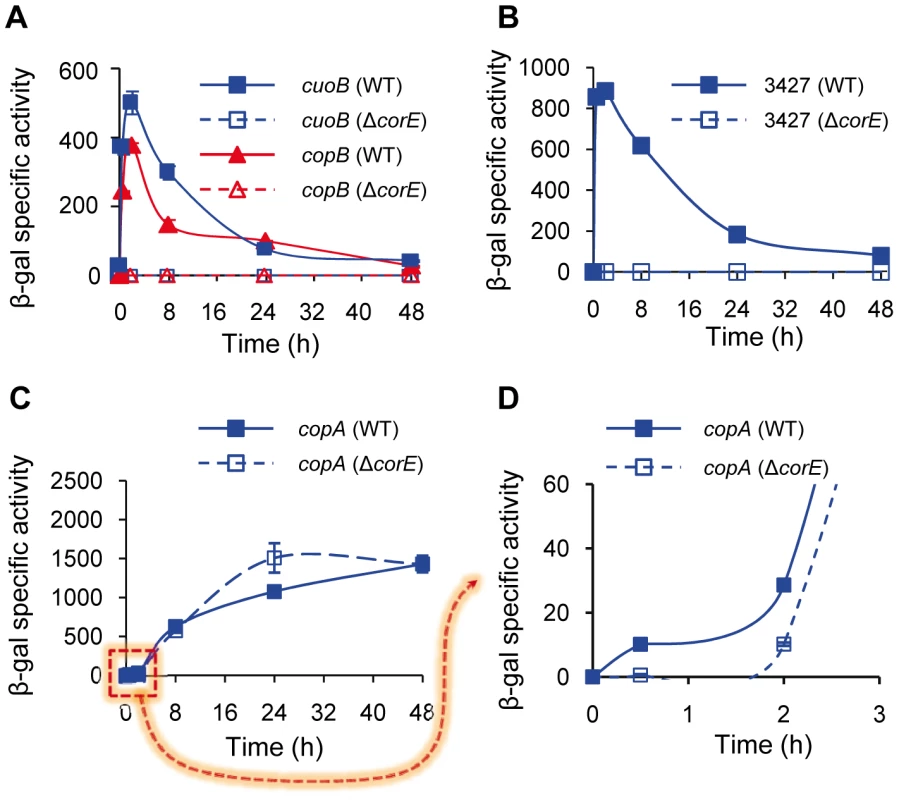
(A) Regulation of cuoB and copB by CorE. Plasmids containing cuoB-lacZ (blue lines) and copB-lacZ (red lines) fusions were introduced into the WT (solid symbols) or the ΔcorE (open symbols) backgrounds, and incubated on CTT agar plates containing 0.6 mM CuSO4. β-gal specific activity was determined in cell extracts harvested at the indicated times. The same approach reported above was followed to study the regulation of MXAN_3427 (B) and copA (C and D) by CorE, although 0.3 mM CuSO4 was used to get an optimal difference in the copA expression levels between the WT and the ΔcorE strains at early times (panels C and D). The dashed arrow from panel C to D indicates that in panel D only the indicated part of panel C is shown. Please note the difference in the scale in each panel, and the different time course of panel D. Error bars indicate standard deviations. As ECF σ factors are usually autoregulated, corE expression was analyzed in the ΔcorE mutant. The results obtained showed that this σ factor is only partially responsible for its own up-regulation by copper, especially in the early stages after metal addition. However, some up-regulation by the metal still remains in the mutant (Figure 1A, red lines), indicating that although cuoB and corE are very close in the genome (Figure S2), their regulation exhibits some differences.
The comparison and analysis of the upstream regions of cuoB and copB has allowed the identification of two similar sequences that could function as the promoter elements recognized by CorE (Figure S3), one located upstream of copB, and the other upstream of a third gene genetically linked to cuoB and corE which encodes an outer membrane efflux protein (MXAN_3424). A manual search for homologous sequences to this putative CorE-binding site in the M. xanthus copper regions 1 and 2 [8] revealed the presence of two other matches, one upstream of the gene for the P1B-type ATPase CopA and the other upstream of the gene identifier MXAN_3427, which encodes a protein with a heavy-metal-associated domain (PF00403, with an E-value of 4.7e-14). To corroborate that these two genes are regulated by CorE, plasmids harboring fusions between these two genes and lacZ were introduced into the WT and ΔcorE backgrounds and β–gal specific activity was determined in these strains. The results obtained revealed that the gene for the heavy-metal-associated protein exhibits an expression profile in the WT strain very similar to those of cuoB and copB after copper supplementation (compare Figure 2A and 2B). Up-regulation by copper was completely eliminated in the ΔcorE mutant, demonstrating that this gene is also part of the CorE regulon. In the case of copA the result was less clear. The expression profile of copA in the WT strain clearly differs from those exhibited by the CorE-regulated genes (compare panel C with panels A and B in Figure 2), and instead of a peak at 2 h, a plateau is reached 24 h after copper addition. Accordingly, the expression profile of copA in the ΔcorE mutant is quite similar to that of the WT strain (Figure 2C). However, when the expression level of copA in these two strains was analyzed with greater precision at short intervals (Figure 2D), it could be observed that the rapid induction of this gene obtained in the WT strain was no longer observed in the mutant. This result suggests that copA is subject to double regulation by CorE and another unidentified copper-dependent regulator. Nevertheless, further work will be required to unambiguously demonstrate that copA is regulated by CorE. Finally, using the consensus sequence of the promoters for these four genes, we tried to determine which other genes could also be under control of CorE. By using the approach described in Materials and Methods, another 13 similar sequences were identified in the M. xanthus genome (Figure S3). However, the fact that only two of them contain the seven invariable residues found in the other promoters, and that none of the proteins encoded by the genes located downstream of these sequences exhibit similarities to other proteins known to be involved in copper handling and trafficking, preventing us from drawing the conclusion that they are indeed regulated by CorE.
As the activation of CorE by copper could be caused either by the general oxidative stress induced by this metal or by the direct binding of the protein to copper in either of its two redox states, cuoB expression in the WT strain was tested in the presence of several concentrations of the oxidants hydrogen peroxide and diamide, and the Cu(II) mimetic divalent metals Cd2+, Ni2+, and Zn2+. Similarly, Ag+ was used to mimic Cu(I). The results obtained revealed that only Cd2+ and Zn2+ could induce cuoB expression (Figure 3A and Figure S4). The fact that Ni2+ does not up-regulate cuoB is not surprising, because the same metals cannot always mimic the copper effect. As an example, the M. xanthus P1B-type ATPase copA has been reported to be induced by copper, Ni2+ and Co2+, but not by Zn2+ [9]. It is notable that the expression levels obtained with Cd2+ and Zn2+ were not only much lower than with copper, but also that the expression profiles were different. In the case of Cd2+, no peak was observed at 2 h; instead, a plateau was reached 24 h after metal supplementation (Figure 3B). Although Zn2+ also yielded a rapid cuoB induction, the peak at 2 h was not as evident as in the case of copper (Figure 3C). cuoB up-regulation by Cd2+ and Zn2+ is also dependent on CorE (Figure 3B and 3C). These data indicate that Cu(II) is the redox state of copper that activates CorE. It should be noted that the Cd2+ and Zn2+ concentrations needed to observe a clear cuoB induction are close to the maxima that M. xanthus cells can tolerate [7], while 0.3 mM copper has almost no effect on myxobacterial growth (Figure 1B). It should also be noted that the addition of metals to the media not only alters the growth rates of the cultures, but also inhibits cell motility, explaining why the morphology of the cell spots is not the same in all of the media tested.
Fig. 3. cuoB is only up-regulated by copper and other divalent metals. 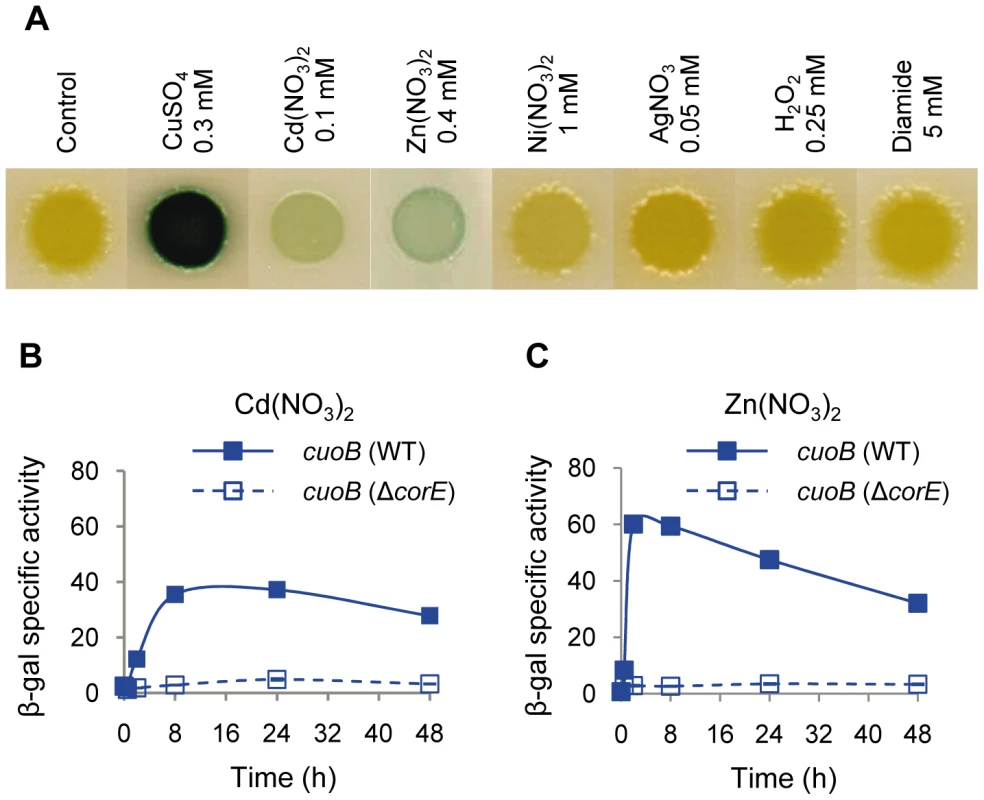
(A) The WT strain harboring the cuoB-lacZ fusion was spotted onto CTT agar plates containing metals or oxidants at the concentrations indicated above each picture. Plates also contained 5-bromo-4-chloro-3-indolyl-β-D-galacto-pyranoside to qualitatively monitor β-gal activity (blue color development). Pictures were taken after 48 h of incubation. (B) Up-regulation of cuoB by Cd2+. The WT (continuous line) and the ΔcorE strains (dashed line) harboring the cuoB-lacZ fusion were incubated on CTT agar plates containing 0.1 mM Cd(NO3)2. β-gal specific activity was determined in cell extracts harvested at the indicated times. (C) Up-regulation of cuoB by Zn2+. The approach followed was the same as the one reported in panel B. The concentration of metal used was 0.4 mM Zn(NO3)2. Error bars indicate standard deviations. Searching for the CorE cognate anti-σ factor
Many ECF σ factors function with a cognate anti-σ which is genetically linked to the σ subunit [11]–[16]. Analyses of the genes located in the proximity of corE revealed that they encode either proteins located in the periplasmic space or in the outer membrane, or that they exhibit striking similarities to well-characterized proteins involved in specific functions, suggesting that no anti-σ factor is cotranscribed with corE. However, the possibility remained that it could be encoded in some other region of the M. xanthus genome. To test for the existence of an anti-σ factor, a strategy was designed consisting of the over-expression of corE [17]. If CorE were present in higher quantities than an unidentified anti-σ factor, it would be released from the antagonistic effect of the anti-σ, and cuoB should be expressed even in the absence of any stimulus. To follow this approach, corE was cloned under control of the oar promoter and introduced into the ΔcorE mutant harboring cuoB-lacZ to facilitate the analysis of cuoB expression. The oar promoter allows genes to be expressed constitutively at high levels [18]. As a control, a corE' cuoB-lacZ strain was also constructed, in which the corE gene was under control of its own promoter (Figure S2 displays the cuoB-lacZ fusions used in this study). Quantitative analyses of cuoB expression in both strains reported no expression of this gene in the absence of copper (Figure 4A), indicating that an excess of CorE was not sufficient to activate the transcription of cuoB. To corroborate that CorE expressed under the oar promoter was functional, copper was added to the media. In this case, up-regulation of cuoB was observed in both strains and with similar expression profiles (Figure 4B). Finally, to confirm that corE was over-expressed when cloned under the oar promoter, we constructed the same two strains described above but introducing a His tag at the N terminus of CorE (hCorE'). Western blot analyses using antibodies against the His tag confirmed that corE was indeed expressed at very high levels in the absence as well as in the presence of copper (Figure 4C and 4D). CorE migrates as a double band, which must correspond to different forms of the protein. Activity of the hCorE' protein was further tested by following cuoB expression. The results obtained indicated that the proteins holding the His tag could promote cuoB transcription in the same manner as the native ones (Figure S5). Although it cannot be completely ruled out that a cognate anti-σ factor for CorE is encoded in the M. xanthus genome, all of these results indicate that CorE functions in a different manner from the one reported for the other characterized ECF σ factors.
Fig. 4. Searching for the CorE anti-σ factor. 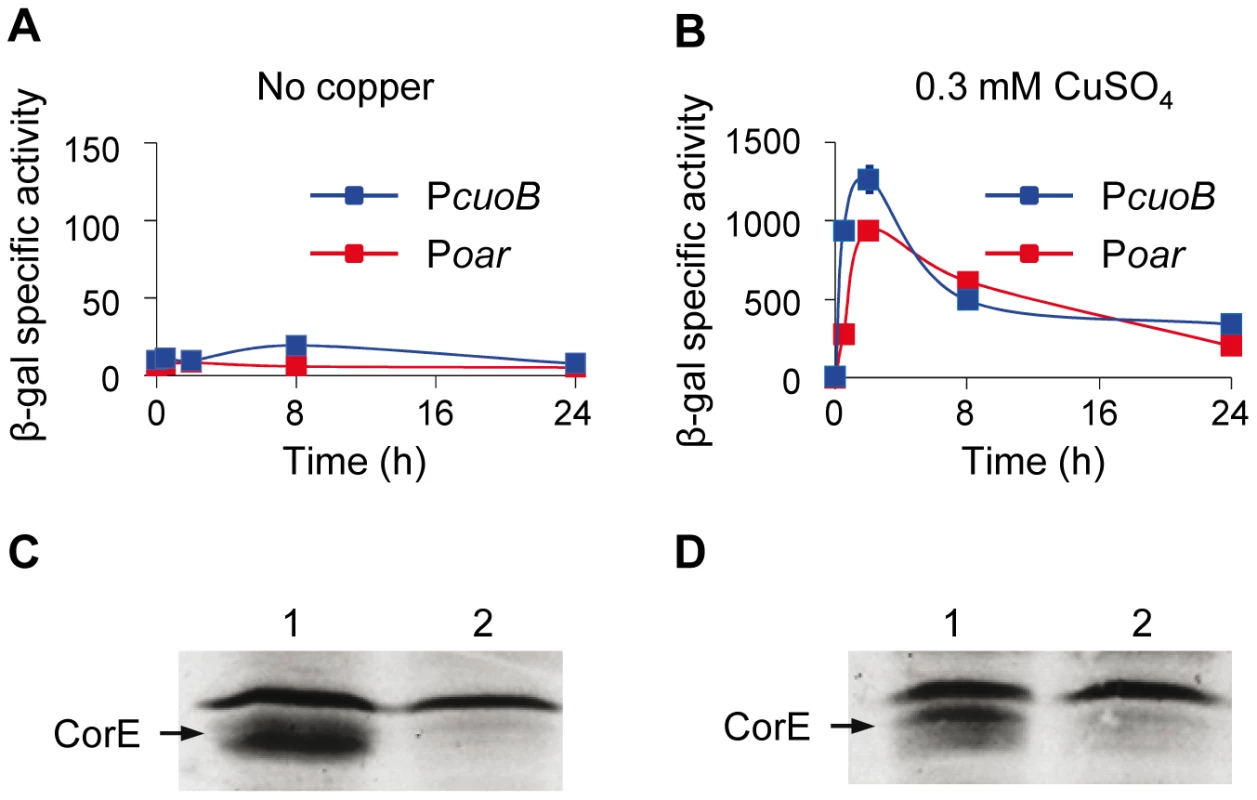
(A and B) Quantification of β-gal activity (cuoB expression) in strains where corE was cloned under the control of the oar promoter (red lines) or its own promoter (blue lines). Activities were determined in the absence of metal (A) or in the presence of 0.3 mM copper (B). Note the difference in the scales of the two panels. Error bars indicate standard deviations. (C and D) Western blot analyses to confirm the over-production of CorE in the absence (C) or in the presence (D) of 0.3 mM copper in the strains harboring the gene corE cloned under the oar promoter (lane 1) or its own promoter (lane 2). Proteins were collected at 2 h after copper addition. The band of equal intensity in all the lanes corresponds to an unidentified M. xanthus protein that reacts with the anti-His tag antibody used in the assay. The intensity of this band, which does not change in the conditions tested, has been used to standardize the amount of protein loaded in each lane. CorE needs copper to bind DNA
The fact that the over-expression of corE did not lead to the induction of cuoB unless copper was added to the medium suggested that CorE might require the binding of copper to promote transcription. Hence, the ability of CorE to bind DNA in vitro was tested by using electrophoretic mobility shift assays. CorE was expressed in E. coli with an N-terminal His tag and purified by affinity chromatography. Additionally, a 265-bp fragment containing the copB promoter was amplified and labeled with 32P to be used as a probe. As shown in Figure 5, an electrophoretic mobility shift was only observed in the reaction mixture containing copper and bathocuproine disulfonic acid (BCS), a specific chelating agent for Cu(I) [19], [20]. These results not only confirm that CorE uses copper as a cofactor, but also suggest that Cu(I) prevents CorE from binding to DNA, and hence, that CorE-Cu(II) is the active form of this σ factor. This is also supported by the fact that only divalent metals can mimic the effect of copper on cuoB up-regulation. No other σ factor has so far been reported to require any metal to bind DNA.
Fig. 5. CorE needs copper and BCS to bind DNA. 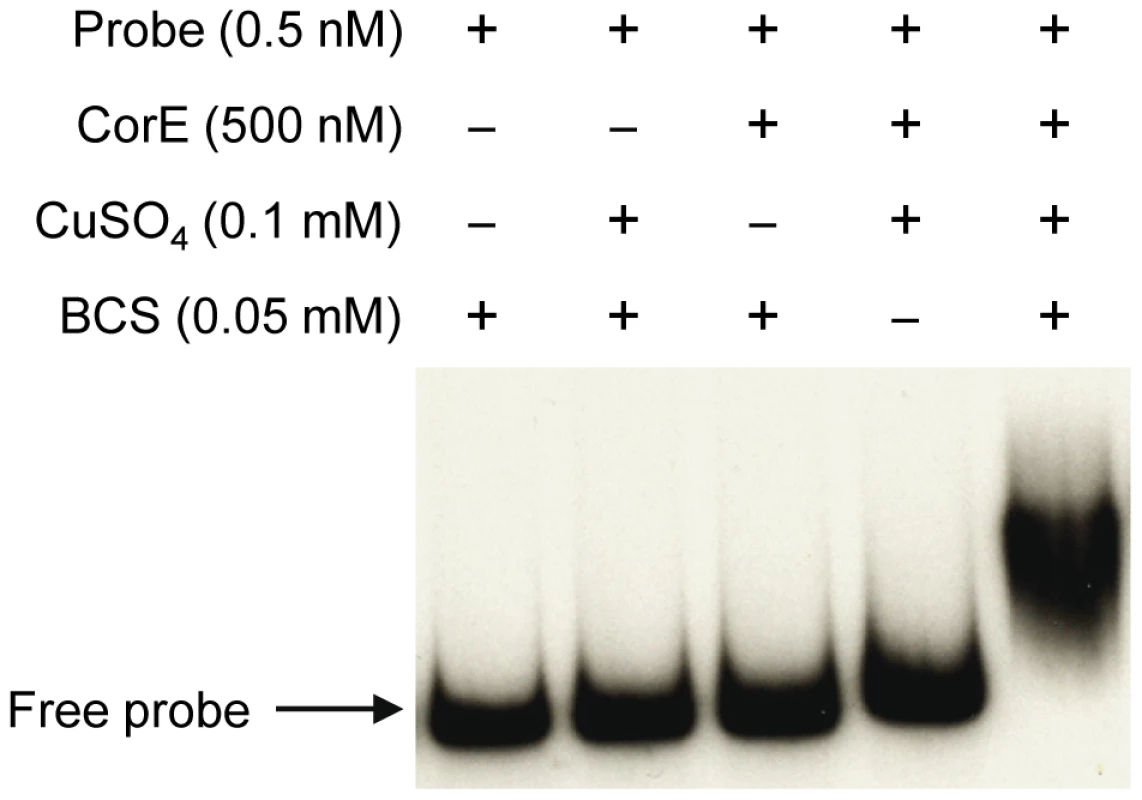
Electrophoretic mobility shift assay with purified hCorE and a radiolabeled DNA fragment containing the copB promoter was carried out with the additives indicated in each lane. Details are given in Materials and Methods. The redox state of copper directs the activation/inactivation of CorE
The expression profiles of the CorE-regulated genes exhibit a peak around 2 h after copper addition (Figure 2A and 2B), in spite of the fact that corE expression is maintained for 48 h (Figure 1A). This observation could be explained by proteolysis of the σ factor. To investigate this option, Western blot analyses were carried out using the hcorE' cuoB-lacZ strain. The data shown in Figure 6A demonstrate that CorE was stable for 24 h after copper addition. Another explanation could be that CorE underwent a cycle of activation/inactivation, whereby the regulator would only be in the active form for a limited period of time. As shown in Figure 5, to obtain binding of CorE to DNA, the reaction mixture must include not only copper, but also a chelating agent for Cu(I). Moreover, only other divalent metals can mimic the copper effect on cuoB up-regulation (Figure 3 and Figure S4). These data suggest that the redox state of copper could be the key in this process. To investigate this possibility, the expression of cuoB was assayed in vivo in conditions that favor the formation of Cu(I) and Cu(II). As shown in Figure 6B and 6C, cuoB up-regulation could only be observed when copper was added to the medium. However, the maximum expression levels were diminished when the reducing agent ascorbate was also included in the medium to favor the formation of Cu(I) (Figure 6C, brown line). Similarly, the addition of Ag+, which mimics Cu(I), also yielded expression levels lower than those obtained with only copper (Figure 6C, green line). In contrast, when copper was added with the Cu(I) chelators BCS or bicinchoninic acid (BCA) [19], [20], cuoB expression was around three times that of the control (Figure 6C, blue versus red and black lines). Moreover, the addition of copper with tetrathiomolybdate (TTM), a chelator of Cu(I) and Cu(II) [21], decreased the up-regulation mediated by this metal to a very basal level (Figure 6C, orange line). In contrast, when these three chelators were tested with Zn2+ as the inducer, the expression levels of cuoB were diminished as the concentrations of all the chelators increased (Figure S6), due to the fact they can also chelate Zn2+, although to a much lesser extent than copper. According to all these data, CorE requires copper for activation, and it only acquires an active conformation in the presence of Cu(II), while the reduced state of the metal leads to an inactive conformation. This notion agrees well with the lack of a peak when up-regulation of cuoB is achieved by Zn2+ and Cd2+, which are metals with only one redox state (Figure 3B and 3C).
Fig. 6. Activation and inactivation of CorE. 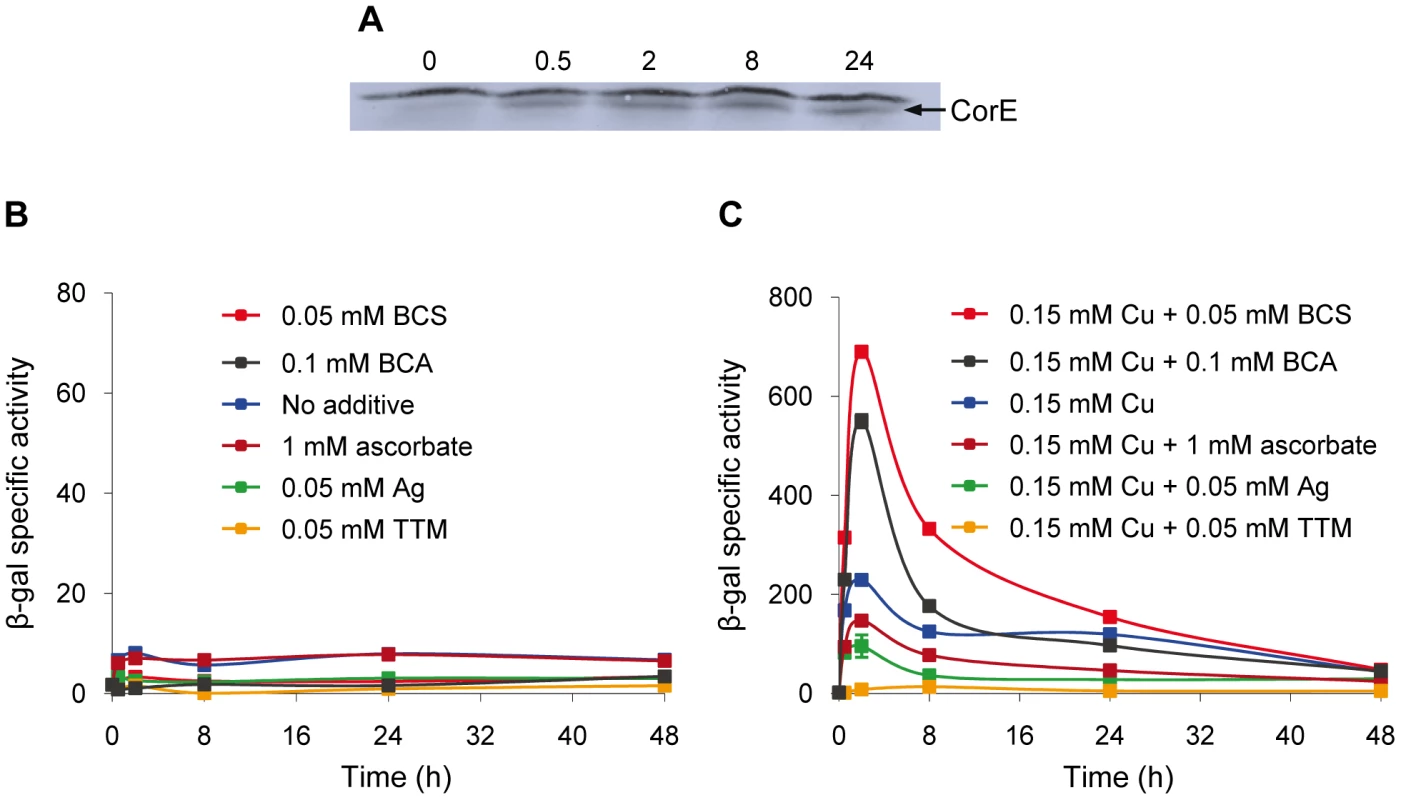
(A) CorE is not degraded. M. xanthus cells harboring the hCorE protein were harvested at the times (h) indicated above each lane after the addition of 0.3 mM copper sulfate and analyzed by Western blot using an anti-His tag antibody. (B) cuoB is not up-regulated by any of the additives indicated in the panel. (C) CorE is activated and inactivated by the redox state of copper. cuoB expression was analyzed in the presence of different additives that modify the redox state of copper, that mimic Cu(I), or that chelate copper in any of its two redox states. For panel B and C, M. xanthus cells harboring the fusion cuoB-lacZ were incubated on CTT agar plates containing the additives indicated. Samples were harvested at different times and β-gal specific activity was determined. Error bars indicate standard deviations. To confirm the results obtained in vivo, the DNA-binding assay was carried out again including Ag+ or TTM in the reaction mixtures. As shown in Figure 7, these two additives overrode the electrophoretic mobility shift achieved by the addition of copper and BCS. It should be reminded that Ag+ mimics Cu(I) and that TTM chelates Cu(II). All of the data presented in this section demonstrate that CorE activity is modulated by the redox state of copper.
Fig. 7. Silver and TTM prevent CorE from binding to DNA. 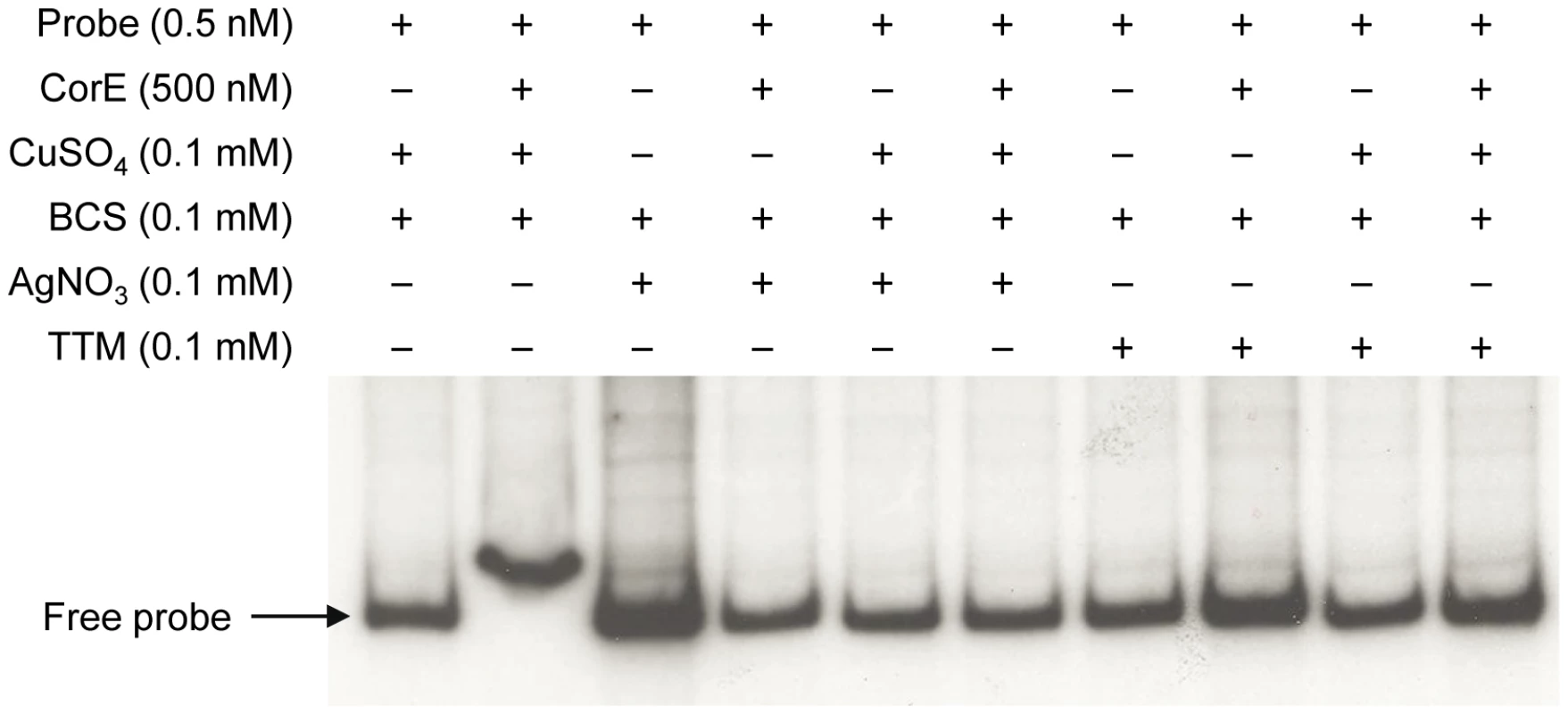
An electrophoretic mobility shift assay was carried out as described in Figure 5, with the additives indicated in each lane. This mechanism of action implies that Cu(II) must be available in the cytosol during the next 2 h after copper supplementation. Although it is assumed that all the copper in the reducing environment of the cytoplasm is present as Cu(I) under normal circumstances [2], [22], it is also expected that the cytoplasm will become more oxidizing in the presence of agents such as copper [23], favoring the formation of Cu(II) until the reducing conditions are restored by the participation of the elements involved in copper detoxification. Furthermore, as free copper in the cells is estimated to be less than one atom per cell [24], it is plausible to speculate that CorE functions with an unidentified Cu(II)-specific metallochaperone, which would ferry the cupric form through the cytoplasm to activate this σ factor. Such an activator working upstream of CorE would explain why the expression levels of cuoB do not increase when corE is over-expressed (Figure 4). However, another possible explanation for this observation could be that CorE aggregates when produced in large amounts.
One paradox is the fact that CorE is activated by Cu(II) and inactivated by Cu(I), while genes under its control encode proteins, such as CopA, CopB, and CuoB, that utilize Cu(I) as a substrate [7], [9]. Nevertheless, this contradiction can be explained by considering two facts: i) Out of the two redox states of copper, Cu(I) is the most toxic form [2]. As CorE-regulated genes represent the first protective barrier against the deleterious effect of copper (please note that these genes are rapidly up-regulated after copper addition, as shown in Figure 2 and Figure S1) and this protein is activated by Cu(II), it is plausible to speculate that copper will initially get into the cytoplasm in the form of Cu(II), activating CorE, and preparing the cells to act on Cu(I) as soon as it appears. At this point, the CorE regulon will be inactivated by the presence of Cu(I) in the cytoplasm. ii) If the presence of copper persists in the environment, M. xanthus cells will obtain protection against the metal by means of at least two other mechanisms (first, by the P1B-type ATPase CopA and the MCO CuoA, and later, by the Cus2 and Cus3 systems), which are sequentially induced after copper addition [7]–[9] (see also Figure 2 and Figure S1).
Although many bacterial transcriptional regulators need metal to bind DNA [25], [26], none of them have been reported to be modulated by the redox state of the metal. Moreover, those that function with copper show selectivity for Cu(I) [24], [27]. Hence, CorE represents a novel type of bacterial copper sensor.
CRD controls the activity of CorE
CorE contains a short C-terminal extension after the σ4 domain consisting of 38 residues named CRD. Six of these residues are Cys. As different arrangements of Cys have been proved to be key elements in several metal-binding proteins [22], [27], [28], we tried to determine whether CRD was involved in the activation/inactivation of CorE mediated by copper. An M. xanthus in-frame deletion mutant was constructed in which most of the CRD region was deleted. This strain, designated as ΔcorECRD, encoded a protein containing the two domains σ2 and σ4 of CorE, but none of the six Cys of CRD. To analyze the activity of CorECRD, the two fusions cuoB-lacZ and copB-lacZ were introduced in this mutant and β-gal activity was assayed in the absence and in the presence of copper. The data obtained revealed that neither cuoB or copB were up-regulated by copper (data not shown), a result identical to that shown in Figure 2A, when the entire corE gene was deleted. These data demonstrate that CRD is essential for the copper-dependent transcription of the genes controlled by CorE.
To determine which Cys are involved in CorE activity, each residue was individually mutated to an Ala by site-directed mutagenesis. The six mutated genes were introduced into the ΔcorE strain harboring the fusion cuoB-lacZ. The effect of the mutations was evaluated by analyzing the expression of cuoB in the absence and in the presence of copper. The results obtained showed different patterns. Mutations C181A and C206A exhibited transcription profiles very similar to those of the WT (Figure 8A), although some small differences regarding the maximum expression levels and timing were observed, indicating that these residues play a minor role in CorE activity.
Fig. 8. Expression of cuoB in strains harboring point mutations in the CRD region of CorE. 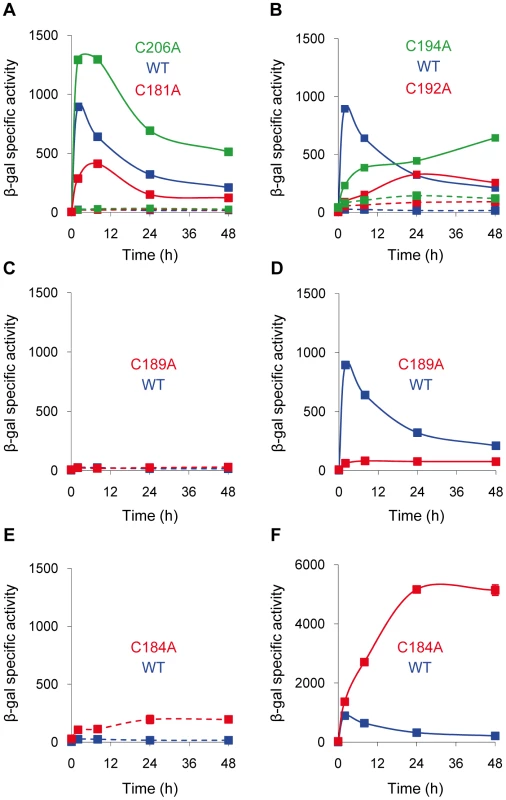
The mutated Cys are indicated in each panel. Cells were incubated on CTT agar plates containing 0.3 mM copper (continuous lines) or without metal (dashed lines), and samples were harvested at different times to determine β-gal specific activity. Note that the scale in panel F is different from that used in the other panels. Error bars indicate standard deviations. More severe effects were obtained with the mutations C192A and C194A. In these mutants, the expression levels of cuoB in the absence of copper were higher than in the WT (Figure 8B, dashed lines), suggesting that both Cys play some role in CorE inactivation. Moreover, although cuoB expression was up-regulated by copper in both mutants, the rapid induction and the peak exhibited by the WT at 2 h were not replicated (Figure 8B, continuous lines). The effect of the mutation C189A was even more drastic, because no expression was observed in the absence of copper and the up-regulation by the metal was almost completely non-existent (Figure 8C and 8D). Accordingly, it can be concluded that these three residues are important in the CorE activation process.
Cys184 was clearly required for CorE inactivation, because mutation C184A yielded a constitutive expression in the absence of copper (Figure 8E) and the addition of copper provoked a rapid up-regulation of cuoB. Interestingly, the expression level did not peak at 2 h, but kept increasing until it reached a plateau at 24 h (Figure 8F).
The effect of each point mutation in cuoB expression was also analyzed in cells grown on media containing copper plus BCS or silver (Figure S7). Mutations C181A and C206, which in the presence of copper yielded expression profiles similar to that of the WT (Figure 8A), also exhibited higher expression levels in the presence of copper plus BCS, and lower levels with copper plus Ag+ (Figure S7). In the case of substitutions C189A, C192A, and C194A, BCS and Ag+ barely affected the expression levels obtained with only copper (Figure S7). However, it should be reminded that these three residues are important for activation, and that cuoB up-regulation by copper was impaired in these mutants (Figure 8B and 8D), suggesting that these three proteins might only have a limited affinity for the metal. Surprisingly, however, the protein with the mutation C184A (the most important residue in CorE inactivation as shown in Figure 8E and 8F), can still be modulated by the two redox states of copper (Figure S7). This result indicates that some other residues must also be involved in the inactivation of CorE. As mentioned above, two of them could be Cys192 and Cys194, because mutations C192A and C194A yield constitutive expression of cuoB. However, it is expected that other residue(s) of CRD might also be required for the proper functioning of the protein (see below).
Taken as a whole, the results demonstrate that at least four of the Cys of CRD form a coordination environment for copper. This domain is able to recognize copper and sense its redox state, allowing the binding of CorE to DNA to activate transcription in those conditions that favor the formation of Cu(II), and inactivating the σ factor in those that favor the formation of Cu(I). However, how exactly CorE distinguishes between Cu(I) and Cu(II) is not easy to predict, because all of the residues identified so far that modulate the activity of CorE are Cys. Although Cys are able to coordinate Cu(I) and Cu(II), they require the presence of other amino acids, such as His, Asp, Glu, or Met to exert this function [22], [29]. Accordingly, it is expected that other residues also participate in the coordination of copper in either of the two redox states. Moreover, thiols are known to allow different types of modifications in an oxidative environment [30]. The exact modification of each individual Cys might also be crucial in the CorE activation/inactivation process. Further genetic, biochemical, and structural studies will be required to elucidate this intriguing question.
The role of CRD in CorE resembles the function of the anti-σ domain present in many anti-σ factors [31]. Anti-σ domains require Zn2+ binding to sequester their cognate σ factor. However, the anti-σ domain and CRD differ in many aspects: i) CRD is an extra portion of the σ factor; ii) elimination of CRD does not activate the σ factor; and iii) CRD senses the redox state of copper to activate or inactivate the σ factor.
ECF σ factors with CRD in other bacteria
BLASTP analyses have allowed the identification of 21 ECF σ factors with CRD, which are distributed in only four phyla. Fourteen belong to Proteobacteria (9 α and 5 δ), four to Acidobacteria, two to Verrucomicrobia, and one to Nitrospira (Figure 9). As in the case of CorE, anti-σ factors are not linked to any of these σ factors.
Fig. 9. ECF σ factors with CRD in other bacteria. 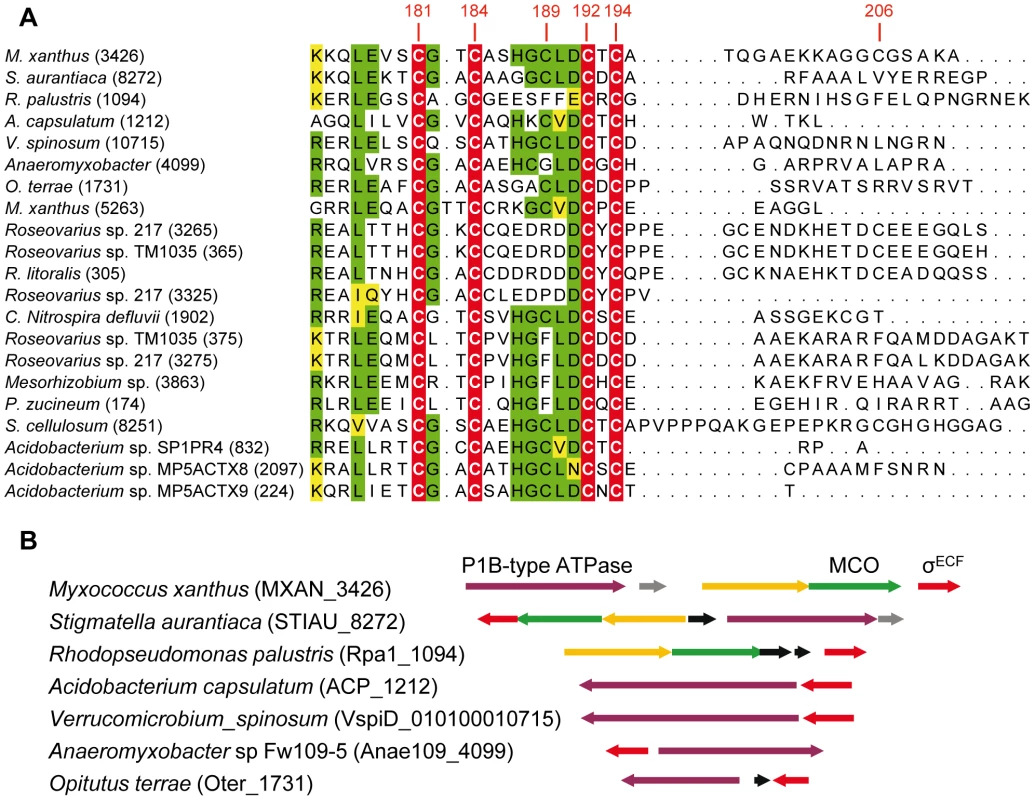
(A) Multiple sequence alignment of the CRD of the 21 ECF σ factors found in prokaryotes. The four invariant Cys are marked on a red background with letters in white. Residues conserved in at least 11 proteins are shown on a green background. The functionally similar amino acids are highlighted in yellow. The number of each gene in the corresponding genome is shown in parenthesis. The codes for the strains, gene identifiers in the genomes (in parentheses), and accession numbers for the proteins are as follows: M. xanthus (MXAN_3426): Myxococcus xanthus DK 1622, YP_631623.1; S. aurantiaca (STIAU_8272): Stigmatella aurantiaca DW4/3-1, ZP_01463028.1; R. palustris (Rpa1_1094): Rhodopseudomonas palustris TIE-1, YP_001990117.1; A. capsulatum (ACP_1212): Acidobacterium capsulatum ATCC 51196, YP_002754310.1; V. spinosum (010100010715): Verrucomicrobium spinosum DSM 4136, ZP_02927111.1; Anaeromyxobacter (Anae109_4009): Anaeromyxobacter sp. Fw109-5, YP_001381261.1; O. terrae (Oter_1731): Opitutus terrae PB90-1, YP_001818615.1; M. xanthus (MXAN_5263): Myxococcus xanthus DK 1622, YP_633415.1; Roseovarius sp. 217 (ROS217_03265): Roseovarius sp. 217, ZP_01038131.1; Roseovarius sp.TM1035 (RTM1035_00365): Roseovarius sp. TM1035, ZP_01881487.1; R. litoralis (RLO149_0305): Roseobacter litoralis Och 149, ZP_02143093.1; Roseovarius sp.217 (ROS217_03325): Roseovarius sp. 217 ZP_01038143.1; C. Nitrospira defluvii (NIDE1902): Candidatus Nitrospira defluvii, YP_003797553.1; Roseovarius sp.TM1035 (RTM1035_00375): Roseovarius sp. TM1035, ZP_01881489.1; Roseovarius sp. 217 (ROS217_03275): Roseovarius sp. 217, ZP_01038133.1; Mesorhizobium (Meso_3863): Mesorhizobium sp. BNC1, YP_676395.1; P. zucineum (PHZ_p0174): Phenylobacterium zucineum HLK1, YP_002128692.1; S. cellulosum (sce8251): Sorangium cellulosum So ce 56,YP_001618901.1; Acidobacterium sp. SP1PR4 (AciPR4DRAFT_0832): Acidobacterium sp. SP1PR4, ZP_07648663.1; Acidobacterium sp.MP5ACTX8 (A9ciX8DRAFT_2097): Acidobacterium sp. MP5ACTX8, ZP_07030792.1; Acidobacterium sp.MP5ACTZ9 (AciX9DRAFT_0224): Acidobacterium sp. MP5ACTX9, ZP_07062224.1. (B) Synteny in the genomic regions where some of the EFC σ factors with CRD are encoded. Genes shown in the same color (except black genes) in the different strains encode proteins with high homology. The alignments of these CRDs have revealed that only 4 Cys (corresponding to residues 181, 184, 192, and 194 in CorE) are absolutely conserved among these σ factors (Figure 9A). Surprisingly, Cys181, whose mutation causes a minor effect on CorE activity, is conserved in all these regulators. In contrast, Cys189, which is the main residue in CorE activation by copper, is only present in 11 σ factors. Interestingly, however, several of the strains with σ factors that conserve this Cys exhibit some synteny in the regions where they are encoded. The surrounding genes encode proteins with high similarities to others known to be implicated in copper handling and trafficking (Figure 9B).
Due to the diversity of the ECF σ factors, Staroń et al. [13] have proposed a classification of this family of regulatory proteins into 44 groups based on sequence similarities and domain architectures. However, CorE did not fit into any of the groups they defined and it was excluded from this classification. The data presented in this report support the notion that a new group should be added to the list, which will include the 21 ECF σ factors that contain CRD.
So far, seven families of metal de-repressors, metal co-repressors, and metal activators are known [25], [26], [32], all of which clearly differ mechanistically from CorE. Hence, elucidation of the exact mode of action of CorE will offer new insights into our current knowledge of metal sensors. Moreover, identification of the factor(s) working upstream of CorE will also help to elucidate how this type of σ factors works and how the trafficking of metals in the bacterial cytoplasm occurs. Finally, characterization of CorE-like proteins identified in other bacteria will also contribute to understanding the role, mechanism of action, and distribution of this novel type of regulators.
Materials and Methods
Plasmids, bacterial strains, and growth conditions
Genotypes of the bacterial strains and plasmids used in this study are listed in Table S1 and Table S2, respectively. M. xanthus was grown in CTT medium at 30°C, supplemented with the additives indicated in each figure, as previously described [6]. E. coli was grown on Luria-Bertani (LB) medium at 37°C [33].
Construction of in-frame deletion mutants and strains harboring lacZ fusions
The methodologies used for obtaining the in-frame deletion mutants and the transcriptional lacZ fusion strains used in this study are the same as previously reported [7]. To generate the corresponding plasmids (listed in Table S2), the desired fragments were amplified by polymerase chain reaction (PCR), using WT chromosomal DNA as a template, the oligonucleotides listed in Table S3 as primers, and the high-fidelity polymerase PrimeSTAR HS (Takara) [33]. PCR products were ligated to vectors pBJ113 and pKY481 [34], [35] to generate in-frame deletion mutants and lacZ fusions, respectively. Plasmids were always introduced into M. xanthus strains by electroporation to obtain integration into the chromosome by homologous recombination. Southern blot analyses were carried out to confirm the proper recombination events. β-gal specific activity in cell extracts obtained by sonication of the strains harboring lacZ fusions was determined as previously described [7], and it is expressed as nmol of o-nitrophenol produced per min and mg of protein. Measurements shown are the averages of data from triplicate experiments.
Over-expression of corE in M. xanthus using the oar promoter
Appropriate oligonucleotide pairs (Table S3) were used to amplify by PCR an 817-bp fragment upstream of the oar gene (MXAN_1450) using M. xanthus chromosomal DNA as a template [33]. Simultaneously, corE was also amplified by PCR. A BamHI site was introduced at the start codon of oar in frame with another BamHI site introduced at the start codon of corE. Both PCR products were cloned in a vector derived from pUC19 in which the ampicillin-resistance gene was substituted by one that encodes resistance to tetracycline (Tetr). The resulting plasmid, pNG06, was introduced by electroporation into an M. xanthus strain with the genotype ΔcorE cuoB-lacZ, and several kanamycin-resistant (Kmr) and Tetr colonies were analyzed by Southern blot to confirm the proper recombination event. β-gal specific activity was determined to quantify cuoB expression. As a control, the plasmid pNG00 was constructed, in which corE was cloned under control of its own promoter. This plasmid was also electroporated into the ΔcorE cuoB-lacZ to restore corE at its original genomic location (see Table S1 and Figure S2). To corroborate that CorE was being over-produced under the constitutive oar promoter, we constructed the same strains described above, but introducing an N-6His tag upstream of CorE, to obtain the strains hcorE' cuoB-lacZ and oar-hcorE' cuoB-lacZ (Table S1). Briefly, the corE gene with an N-6His tag was amplified with appropriate oligonucleotide pairs (Table S3) using pETTOPOCorE plasmid (see below) as a template. The PCR product obtained was cloned under control of the oar promoter and its own promoter as above, obtaining plasmids pNG08 and pNG05, respectively. Plasmids were introduced into the ΔcorE cuoB-lacZ strain, and Kmr Tetr colonies were also analyzed by Southern blot hybridization.
Expression of corE in E. coli and purification of the recombinant protein hCorE
The corE gene was amplified by PCR using the M. xanthus chromosome as a template and the primers CorEcTopoR and CorEcTopoF (Table S3). The PCR product was cloned into pET200/D-TOPO using a Champion pET Directional TOPO Expression Kit supplied by Invitrogen to create pETTOPOCorE. The absence of unwanted mutations in the insert was confirmed by DNA sequencing. As a result, CorE contained an N-6His tag at the N terminus, and this protein has been named hCorE.
The resulting plasmid was used to transform the strain E. coli BL21 Star(DE3). The transformed cells were grown in LB medium containing 25 µg/ml kanamycin at 37°C until the optical density at 600 nm (OD600) reached 0.7 to 0.8. Induction was performed by the addition of 1 mM of isopropyl-β-D-thiogalactopyranoside. Induced cultures were incubated with shaking at 37°C for 6 h. One liter of the cell culture was collected by centrifugation and resuspended in 20 mM Tris-HCl (pH 7.5) containing the protease inhibitors leupeptine and antipain (2 µg/ml each), 10 µg/ml DNAse I, and 5 mM MgCl2. Cells were disrupted in a French pressure cell (at 9000 psi), followed by centrifugation (13000×g for 30 min at 4°C) to remove cell debris. The resulting soluble extract was loaded onto a HisTrapHP column (bed volume 5 ml; GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5) containing 0.5 M NaCl and 30 mM imidazole. Elution was carried out with a linear imidazole gradient (30–250 mM) in the same buffer. Protein fractions were analyzed by using SDS-PAGE. Those fractions containing hCorE were pooled, concentrated by ultrafiltration (cutoff of 10 kDa), and equilibrated to 20 mM Tris-HCl (pH 7.5). The purified hCorE protein content was determined by the Bio-Rad protein assay kit as specified by the manufacturers, using bovine serum albumin as standard. The purity of the samples was higher than 90%.
Western blot analysis
This methodology was used to detect hCorE either in M. xanthus or E. coli extracts. Cells were disrupted by sonication and centrifuged to remove cellular debris. Proteins were separated by SDS-PAGE and transferred onto a membrane of Immobilon-P at 0.8 mA/cm2 for 1.5 hours. hCorE was detected with an anti-His G-AP antibody (Invitrogen), which is conjugated with alkaline phosphatase, using nitro-blue tetrazolium chloride and 5-bromo-4-chloro-3′-indolyphosphate p-toluidine as substrates, following the instructions specified by the manufacturer.
Electrophoretic mobility shift assay
A DNA fragment containing an upstream sequence of the predicted ribosome-binding site of copB was amplified from the M. xanthus genome using primers 3422EMSA265F and 3422EMSA265R (Table S3). After purification, the 265-bp PCR product was labeled with T4 polynucleotide kinase (MBI Fermentas) and [γ-32P]ATP, and purified through a ProbeQuant G-50 Micro Column (GE Healthcare). Binding reactions contained 20 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 0.25 mg/ml of bovine serum albumin, 0.5 mM dithiothreitol, 15% glycerol (v/v), 40 mM KCl, 0.5 nM of labeled DNA (13000 cpm), and a 500-fold molar excess of competitor DNA (polydIdC). When indicated, 500 nM of hCorE protein, 0.1 mM CuSO4, 0.05 or 0.1 mM BCS, 0.1 mM AgNO3, or 0.1 mM TTM were also added to the reaction mixtures. After incubation for 10 min at 30°C, the mixtures were loaded onto a pre-run 5% polyacrilamide gel and run at 100 V for 1 h. The gel was dried under vacuum and exposed to an autoradiography film at −80°C.
Point mutations
Single amino acid substitutions in the CRD of CorE were generated using the QuikChange II site-directed mutagenesis kit (Stratagene). Plasmid pNG00 (containing the WT corE sequence) was used as a template. The primers were designed using the QuikChange Primer Design Program (http://www.genomics.agilent.com/). Oligonucleotides used to generate the six point mutations are listed in Table S3. The presence of the desired mutations in the resulting plasmids pNG181, pNG184, pNG189, pNG192, pNG194, and pNG206 (carrying the mutations C181A, C184A, C189A, C192A, C194A, and C206A, respectively), and the absence of unwanted mutations in other regions of the inserts were confirmed by DNA sequencing. These plasmids were electroporated into M. xanthus JM51EBZY (ΔcorE cuoB-lacZ) to obtain strains SDM181EBZY to SDM206EBZY (Table S1).
Identification of ECF σ factors with CRD and synteny studies
Genes encoding ECF σ factors with a CRD in the prokaryotes were identified by BLASTP analysis of all the genome sequences deposited in the database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and the DOE Joint Genome Institute (http://www.jgi.doe.gov/). All the sequences obtained with an E-value<2e-10 that conserved at least 4 Cys in the C terminus were back-searched against Pfam (http://pfam.sanger.ac.uk/) [36] to unequivocally verify that they matched the σ2 (PF07638) and σ4 (PF08281) regions conserved in all the ECF σ factors [11], [12]. Protein sequence alignments of the 21 σ factors with CRD identified were performed using ClustalX [37]. The graphic representation of the multiple sequence alignment was adjusted and colored manually using the model generated at ESPript.cgi Version 3.06 CGI 3.05 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). For synteny determinations, two upstream and two downstream predicted proteins from CorE were initially aligned with the other 20 predicted proteomes using the BLASTP program (E-value<1e-4). We then manually analyzed the gene organization of positive matches. In the case of conservation, we extended the BLASTP searches to other genes within the same region as already described by Pérez et al. [38].
In silico identification of the CorE-binding site and determination of the genes of the CorE regulon
First, the upstream regions of copB and cuoB were manually analyzed to find the sequence AAC, which is well conserved in the −35 regions of other known ECF σ-factor promoters in M. xanthus and other bacteria [39]. Alignment of the sequences found permitted the identification of two regions that could function as the CorE-binding site (Figure S3). Next, homologous sequences to these ones were manually searched in all the upstream regions of the genes of the copper regions 1 and 2 of the M. xanthus genome [8]. By using this strategy we found, in copper region 2, two other putative CorE-dependent promoters upstream of MXAN_3427 and MXAN_3415 (which encodes the P1B-type ATPase CopA). Experimental approaches demonstrated that the expression of these two genes is dependent on copper and that they are regulated by CorE. The alignment of the four sequences was used to define the CorE-binding motif. The consensus sequence of the −35 region given in the IUPAC code (defined by Nomenclature Committee of the International Union of Biochemistry) was used to analyze the whole M. xanthus genome at the Virtual Footprint server (http://prodoric.tu-bs.de/vfp/) [40] to determine which other genes might be part of the CorE regulon. 754 positive sequences were obtained with a maximum of two mismatches with respect to the defined consensus. All the sequences were manually examined for the proper strand orientation and the conservation of the invariant residues observed in the −35 region. The resulting positive matches were again screened to identify a conserved G in the −10 region, maintaining a distance of 16–18 residues from the AAC of the −35 region. Only 13 positive sequences were finally selected (Figure S3).
Supporting Information
Zdroje
1. WhitworthDE 2008 Myxobacteria. Multicellularity and differentiation Washington DC ASM Press. 520
2. WaldronKJRobinsonNJ 2009 How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol 6 25 35
3. TisatoFMarzanoCPorchiaMPelleiMSantiniC 2010 Copper in diseases and treatments, and copper-based anticancer strategies. Med Res Rev 30 708 49
4. OsmanDCavetJS 2008 Copper homeostasis in bacteria. Adv Appl Microbiol 65 217 247
5. SoliozMAbichtHKMermodMManciniS 2010 Response of Gram-positive bacteria to copper stress. J Biol Inorg Chem 15 3 14
6. Moraleda-MuñozAPérezJFontesMMurilloFJMuñoz-DoradoJ 2005 Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus. Mol Microbiol 56 1159 1168
7. Sánchez-SutilMCGómez-SantosNMoraleda-MuñozAMartinsLOPérezJ 2007 Differential expression of the three multicopper oxidases from Myxococcus xanthus. J Bacteriol 189 4887 4898
8. Moraleda-MuñozAPérezJExtremera-LeónALMuñoz-DoradoJ 2010 Differential regulation of six heavy metal efflux systems in the response of Myxococcus xanthus to copper. Appl Environ Microbiol 76 6069 6076
9. Moraleda-MuñozAPérezJExtremera-LeónALMuñoz-DoradoJ 2010 Expression and physiological role of three Myxococcus xanthus copper-dependent P1B-type ATPases during bacterial growth and development. Appl Environ Microbiol 76 6077 6084
10. Marles-WrightJLewisRJ 2007 Stress responses of bacteria. Curr Opin Struct Biol 17 755 760
11. HelmannJD 2002 The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46 47 110
12. CampbellEAMuzzinOChlenovMSunJLOlsonCA 2002 Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell 9 527 539
13. StarońASofiaHJDietrichSUlrichLELiesegangH 2009 The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol 74 557 581
14. BrooksBEBuchananSK 2008 Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim Biophys Acta 1778 1930 1945
15. RiaivioTLSilhavyTJ 2001 Periplasmic stress and ECF sigma factors. Annu Rev Microbiol 55 591 624
16. HughesKTMatheeK 1998 The anti-sigma factors. Annu Rev Microbiol 52 231 286
17. RodrigueSBrodeurJJacquesPEGervaisALBrzezinskiR 2007 Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J Bacteriol 189 1505 1513
18. Martínez-CañameroMMuñoz-DoradoJFarez-VidalEInouyeMInouyeS 1993 Oar, a 115-kilodalton membrane protein required for development of Myxococcus xanthus. J Bacteriol 175 4756 4763
19. XiaoZDonnellyPSZimmermannMWeddAG 2008 Transfer of copper between bis(thiosemicarbazone) ligands and intracellular copper-binding proteins. Insights into mechanisms of copper uptake and hypoxia selectivity. Inorg Chem 47 4338 4347
20. XiaoZBroseJSchimoSAcklandSMLa FontaineS 2011 Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1 and related proteins: detection probes and affinity standards. J Biol Chem 286 11047 11055
21. AlvarezHMXueYRobinsonCDCanalizo-HernándezMAMarvinRG 2010 Tetrathiomolybdate inhibits copper trafficking proteins through metal cluster formation. Science 327 331 334
22. DavisAVO'HalloranTV 2008 A place for thioether chemistry in cellular copper ion recognition and trafficking. Nat Chem Biol 4 148 151
23. PaulsenCECarrollKS 2010 Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol 5 47 62
24. ChangelaAChenKXueYHolschenJOuttenCE 2003 Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301 1383 1387
25. PenellaMAGiedrocDP 2005 Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometal 18 413 428
26. GiedrocDPArunkumarAI 2007 Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans 29 3107 3120
27. OsmanDCavetJS 2010 Bacterial metal sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep 27 668 680
28. SiluvaiGSMayfieldMNilgesMJDeBeer GeorgeSBlackburnNJ 2010 Anatomy of a red copper center: spectroscopic identification and reactivity of the copper centers of Bacillus subtilis Sco and its Cys-to-Ala variants. J Am Chem Soc 132 5215 5226
29. SiluvaiGSNakanoMMayfieldMBlackburnNJ 2011 The essential role of the Cu(II) state of Sco in the maturation of the CuA center of cytochrome oxidase: evidence from H135Met and H135SeM variants of the Bacillus subtilis Sco. J Biol Inorg Chem 16 285 297
30. CooperCEPatelRPBrookesPSDarley-UsmarVM 2002 Nanotransducers in cellular redox signaling: modification of thiols by reactive oxygen and nitrogen species. Trends Biochem Sci 27 489 492
31. CampbellEAGreenwellRAnthonyJRWangSLimL 2007 A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol Cell 27 793 805
32. WaldronKJRutherfordJCFordDRobinsonNJ 2009 Metalloproteins and metal sensing. Nature 460 823 830
33. SambrookJRussellDW 2001 Molecular cloning: a laboratory manual Cold Spring Harbor NY Cold Spring Harbor Laboratory Press. 2344
34. JulienBKaiserADGarzaA 2000 Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A 97 9098 9103
35. ChoKZusmanDR 1999 AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol Microbiol 34 268 281
36. FinnRDMistryJTateJCoggillJPHegerA 2010 The Pfam protein families database. Nucl Acids Res 38 211 222
37. LarkinMABlackshieldsGBrownNPChennaRMcGettiganPA 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947 2948
38. PérezJCastañeda-GarcíaAJenke-KodamaHMüllerRMuñoz-DoradoJ 2008 Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci U S A 105 15950 15955
39. Elías-ArnanzMFontesMPabmanabhanS 2008 Carotenogenesis in Myxococcus xanthus: a complex regulatory network. WhitworthDE Myxobacteria. Multicellularity and differentiation Washington DC ASM Press 211 225
40. MünchRHillerKGroteAScheerMKleinJ 2005 Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21 4187 4189
41. CrooksGEHonGChandoniaJMBrennerSE 2004 WebLogo: A sequence logo generator. Genome Res 14 1188 1190
Štítky
Genetika Reprodukčná medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



