-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
Transcriptome analyses indicate that a core 10%–15% of the yeast genome is modulated by a variety of different stresses. However, not all the induced genes undergo translation, and null mutants of many induced genes do not show elevated sensitivity to the particular stress. Elucidation of the RNA lifecycle reveals accumulation of non-translating mRNAs in cytoplasmic granules, P-bodies, and stress granules for future regulation. P-bodies contain enzymes for mRNA degradation; under stress conditions mRNAs may be transferred to stress granules for storage and return to translation. Protein degradation by the ubiquitin-proteasome system is elevated by stress; and here we analyzed the steady state levels, decay, and subcellular localization of the mRNA of the gene encoding the F-box protein, UFO1, that is induced by stress. Using the MS2L mRNA reporter system UFO1 mRNA was observed in granules that colocalized with P-bodies and stress granules. These P-bodies stored diverse mRNAs. Granules of two mRNAs transported prior to translation, ASH1-MS2L and OXA1-MS2L, docked with P-bodies. HSP12 mRNA that gave rise to highly elevated protein levels was not observed in granules under these stress conditions. ecd3, pat1 double mutants that are defective in P-body formation were sensitive to mRNAs expressed ectopically from strong promoters. These highly expressed mRNAs showed elevated translation compared with wild-type cells, and the viability of the mutants was strongly reduced. ecd3, pat1 mutants also exhibited increased sensitivity to different stresses. Our interpretation is that sequestration of highly expressed mRNAs in P-bodies is essential for viability. Storage of mRNAs for future regulation may contribute to the discrepancy between the steady state levels of many stress-induced mRNAs and their proteins. Sorting of mRNAs for future translation or decay by individual cells could generate potentially different phenotypes in a genetically identical population and enhance its ability to withstand stress.
Published in the journal: Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability. PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002527
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002527Summary
Transcriptome analyses indicate that a core 10%–15% of the yeast genome is modulated by a variety of different stresses. However, not all the induced genes undergo translation, and null mutants of many induced genes do not show elevated sensitivity to the particular stress. Elucidation of the RNA lifecycle reveals accumulation of non-translating mRNAs in cytoplasmic granules, P-bodies, and stress granules for future regulation. P-bodies contain enzymes for mRNA degradation; under stress conditions mRNAs may be transferred to stress granules for storage and return to translation. Protein degradation by the ubiquitin-proteasome system is elevated by stress; and here we analyzed the steady state levels, decay, and subcellular localization of the mRNA of the gene encoding the F-box protein, UFO1, that is induced by stress. Using the MS2L mRNA reporter system UFO1 mRNA was observed in granules that colocalized with P-bodies and stress granules. These P-bodies stored diverse mRNAs. Granules of two mRNAs transported prior to translation, ASH1-MS2L and OXA1-MS2L, docked with P-bodies. HSP12 mRNA that gave rise to highly elevated protein levels was not observed in granules under these stress conditions. ecd3, pat1 double mutants that are defective in P-body formation were sensitive to mRNAs expressed ectopically from strong promoters. These highly expressed mRNAs showed elevated translation compared with wild-type cells, and the viability of the mutants was strongly reduced. ecd3, pat1 mutants also exhibited increased sensitivity to different stresses. Our interpretation is that sequestration of highly expressed mRNAs in P-bodies is essential for viability. Storage of mRNAs for future regulation may contribute to the discrepancy between the steady state levels of many stress-induced mRNAs and their proteins. Sorting of mRNAs for future translation or decay by individual cells could generate potentially different phenotypes in a genetically identical population and enhance its ability to withstand stress.
Introduction
High throughput yeast microarray studies indicate that the mRNA abundance of a common core of 10–15% of the yeast genome is modulated by a variety of different environmental challenges such as DNA damage, heat, oxidative, osmotic, heavy metal, and salt stress [1]. This response, known as the environmental stress response (ESR) represents a network of interlinked functions that preserves cell integrity [2], [3]. A hallmark of the ESR is a down-regulation of protein synthesis genes and an up-regulation of genes that encode chaperones and genes involved in protein degradation [4].
The ubiquitin-proteasome system is the major pathway for regulated protein degradation in the cell. In most cases proteins targeted to the proteasome are covalently linked to chains of ubiquitin by a cascade of E1, ubiquitin activating - and E2 ubiquitin conjugating enzymes, and an E3 ubiquitin ligase [5]. The large family of Skp1-Cdc53-F-box protein (SCF) ubiquitin ligase complexes regulates growth and cell cycle progression in all eukaryotes [6]. In yeast about seventeen different F-box proteins recruit degradation substrates to the SCF complex. Of all the F-box proteins, transcription only of UFO1 is highly induced by DNA damage and arsenate stress (four - and sixfold respectively [7], [8]. UFO1 may function in maintenance of genome stability as in the absence of Pif1 helicase ufo1Δ mutants show a 74-fold increase in gross chromosomal rearrangements [9]. This role is consistent with the key function of SCFUfo1 in degradation of the mating switch Ho endonuclease [10]–[13], and the translesion DNA polymerase, Rad30 [14]. However, despite the robust induction of transcription of UFO1 mRNA in response to stress, ufo1Δ mutants do not display enhanced sensitivity to arsenate [8] or UV [15] compared with their isogenic wild types. Functional profiling in yeast shows that this is a widespread phenomenon as deletion mutants of many genes highly induced by a particular stress do not exhibit enhanced sensitivity to the specific stress (reviewed by [16]).
The dynamics of the transcriptome in response to changing conditions is mainly determined by the balance between transcription and mRNA decay and in many instances functionally related genes show a negative correlation between transcription and decay [17], [18]. Genome-wide proteomics revealed that only ca. 70% of the steady state protein levels can be attributed to mRNA abundance, indicative of translational regulation [19]–[23]. Furthermore the lifecycle of mRNA molecules is complex and involves dynamic changes in subcellular localization to distinct cytoplasmic bodies. These include processing bodies (PBs) that are rich in mRNA decay enzymes such as mRNA decapping enzymes (the Dcp1/Dcp2 heterodimer and its activator, Dhh1), Xrn1 5′-3′ exonuclease, and repressors of translation [24]–[27]. The protein composition of PBs suggests that they are centers of mRNA degradation [28]–[30]. Under stress conditions mRNAs are also present in Stress Granules (SGs) that form when initiation of translation is impaired. SGs contain primarily translation initiation factors and may therefore serve as storage centers from which the mRNAs can be returned to the polysomes for translation [27], [29], [31]–[34]. Despite their apparently distinct roles several lines of evidence suggest a functional relationship between PBs and SGs. First, genetic studies show that formation of yeast SGs depends on biogenesis of PBs [27]. Second, PBs and SGs are often colocalized suggesting that mRNAs may be transferred from one body to the other [27]. Third, yeast deficient for the decapping activators, Pat1 and Dhh1, are blocked in both repression of translation and in PB formation [30], [35]. Finally, recent studies link transcription with both mRNA nuclear export, decay, and translation: two subunits of the RNA Pol II holoenzyme transcription complex, Rpb4 and Rpb7, escort the mRNA transcripts from the nucleus to the cytoplasm where they physically interact with subunits of the PBs [36], [37]; these subunits are important for coordination of mRNA synthesis with decay [18]. Additional PB-associated proteins, yeast Dhh1 and its Xenopus ortholog, shuttle between the nucleus and cytoplasm [38], [39]. Thus the discrepancy between transcript and protein level could be attributable to the complex regulatory mechanisms of the mRNA lifecycle.
Here we report analysis of the lifecycle of UFO1 mRNA whose transcription is highly induced by several kinds of stress, yet the null mutant shows no enhanced sensitivity to these stresses. We compared UFO1 mRNA lifecycle with that of the heat shock gene, HSP12 that represents a different paradigm of regulation. HSP12 is regulated by the ESR and encodes a membrane protein important for preserving membrane organization under stress conditions [40]. Induction of HSP12 mRNA is slow [41], but the protein attains a high cellular level after stress. We measured induction of UFO1 and HSP12 mRNAs and the stability of their mRNAs after stress by quantitative real time PCR (qRT-PCR). We also compared their steady state protein levels in response to different stresses. Furthermore to dissect the UFO1 mRNA lifecycle we followed the mRNA molecules in single cells by tagging genomic UFO1 with bacteriophage MS2L sequence for detection with the viral capsid protein fused to GFP [42], [43]. As further reference mRNAs we used MFA2 mRNA that is constitutively expressed at a high level from a strong promoter [24], [44], the low copy mRNAs of ASH1 and OXA1 that are localized to the bud [42] and the mitochondrion [43], respectively, prior to their translation. We visualized the subcellular localization of these mRNA molecules in single cells that expressed fluorescent subunits of the PBs and SGs [27], [31], [45] and found that the highly expressed UFO1 and MFA2 mRNAs enter PBs. Genetic analysis using mutants unable to form PBs showed that cell viability is strongly reduced when UFO1, MFA2 or HSP12 genes are expressed at a high level from the GAL promoter. Our interpretation is that sequestration in PBs plays a major role in preserving cell viability. We suggest that the ability to store highly abundant mRNAs in PBs for future regulation is a key facet of the stress response allowing individual cells to sort mRNAs for decay or translation. This mechanism has the potential to facilitate acquisition of a variety of different phenotypes in a genetically identical population enhancing its ability to withstand the stress.
Results
Expression of UFO1 mRNA in cell populations in response to stress
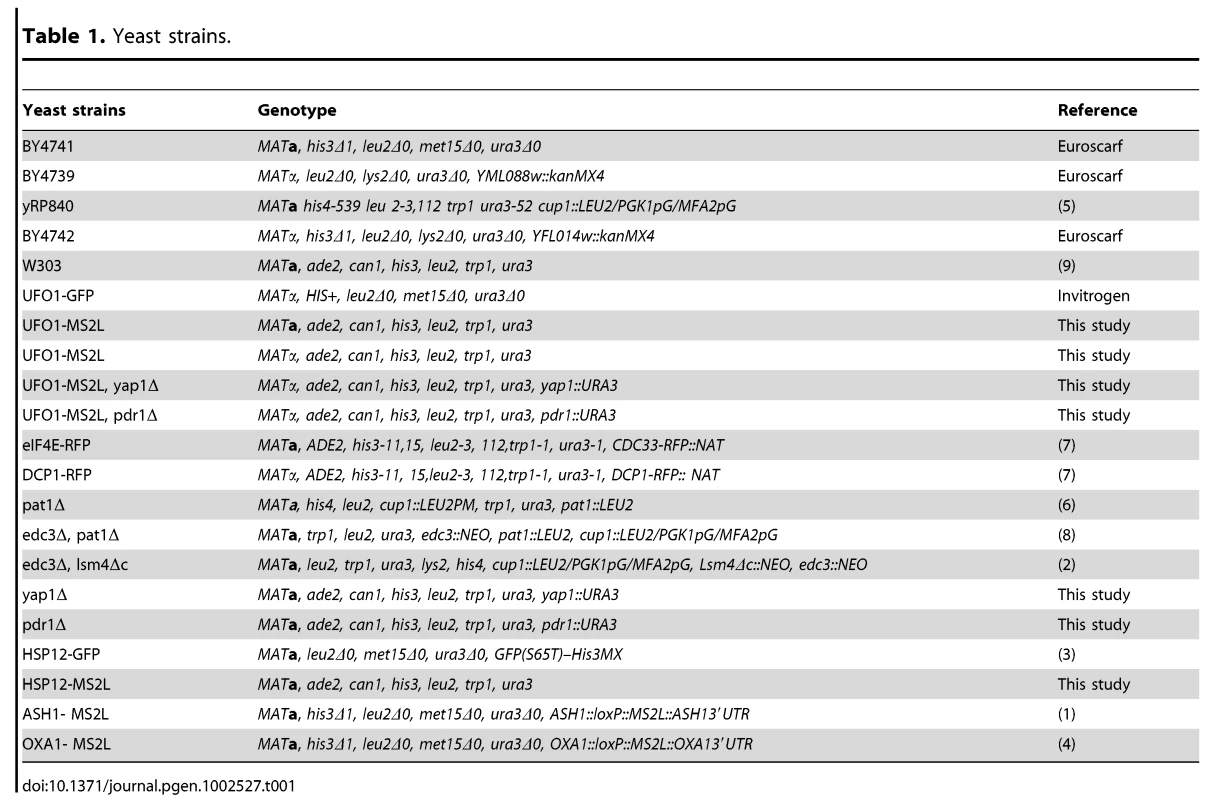
We followed UFO1 expression using qRT-PCR on mRNA extracted from cells at different times after treatment with arsenate, H2O2, or UV irradiation. UFO1 is under repression during normal growth conditions [46] and treatment with arsenate led to a fourfold increase in the level of UFO1 mRNA after 15 minutes that stayed high for at least one hour during which the cells were assayed. Treatment with H2O2 gave a threefold increase in mRNA level after 15 minutes followed by a decrease back to the basal level of the untreated control from the 30 minute time point. After UV irradiation the UFO1 transcript level increased fourfold after 15 minutes and remained high for one hour (Figure 1A). The mRNA abundance is dependent upon the relative rates of transcription and decay, and when we examined the decay of UFO1 mRNA in response to the above stresses we observed stabilization of UFO1 mRNA in response to UV. The half-life of UFO1 mRNA was ca. 7 minutes in untreated cells and in cells treated with arsenate or H2O2, but extended to almost 30 minutes after UV irradiation (Figure 1B). Ufo1 protein levels are very low in untreated cells [47], however, by using 10-fold the amount of cells we could observe the protein after arsenate, H2O2, or UV stress (Figure 1C). GFPUfo1 protein was stabilized in cells treated with arsenate or H2O2, but has the same half-life after UV irradiation as in the untreated control cells (Figure S1). Thus the Ufo1 protein could reflect accumulation during the prolonged arsenate or H2O2 treatments, but in contrast in UV-irradiated cells could be newly translated from the stabilized UFO1 mRNA. The UFO1 promoter has sequence elements similar to those regulated by the Yap1 oxidative stress - and Pdr1 pleiotropic drug response transcription factors. Untreated mutant cells showed a reduced basal level of UFO1 transcription in yap1Δ and pdr1Δ mutants compared with wild type cells (Figure 1D) and there was no induction of UFO1 transcription in response to arsenate, H2O2, and UV stresses indicating that induction of UFO1 mRNA is Yap1 - and Pdr1-dependent (Figure 1E).
Fig. 1. Transcription of UFO1 in response to arsenate, H2O2, and UV. 
A. Wild type cells were grown in SC 2% glucose medium overnight, diluted to A600 = 0.1, regrown to A600 = 0.5 and treated with 1 mM arsenate, 8.8 mM H2O2, or irradiated with 40 mJ/cm2 UV. Aliquots were collected at the indicated times and analyzed by qRT-PCR. mRNA levels were normalized to ACT1 and to time 0 (untreated log cells). B. pGAL-GFP-UFO1 was expressed in ufo1Δ mutants by overnight induction with 2% galactose. Next morning cells were diluted to A600 = 0.1, regrown to A600 = 0.5 then untreated, or stressed with 1 mM arsenate or 8.8 mM H2O2 for 30 minutes, or irradiated with 40 mJ/cm2 UV. The cells were washed and transferred to SC with 4% glucose. Samples were collected immediately after addition of glucose and at the times indicated and analyzed by qRT-PCR. mRNA levels were normalized to ACT1 and to time 0 (untreated log cells). C. Western blot analysis of Ufo1GFP protein produced from the tagged genomic UFO1-GFP gene. Anti-GFP antibodies were used to detect Ufo1GFP and anti-α-tubulin antibodies to detect α-tubulin that serves as a loading control. D. UFO1 mRNA levels in untreated wild type, yap1Δ or pdr1Δ mutants. mRNA levels were normalized to ACT1 and to w.t. mRNA levels. E. yap1Δ or pdr1Δ mutants grown, treated and analyzed as in Figure 1A. mRNA levels were normalized to ACT1 and to time 0 (untreated log cells). Detection of UFO1 mRNA in single cells in response to stress
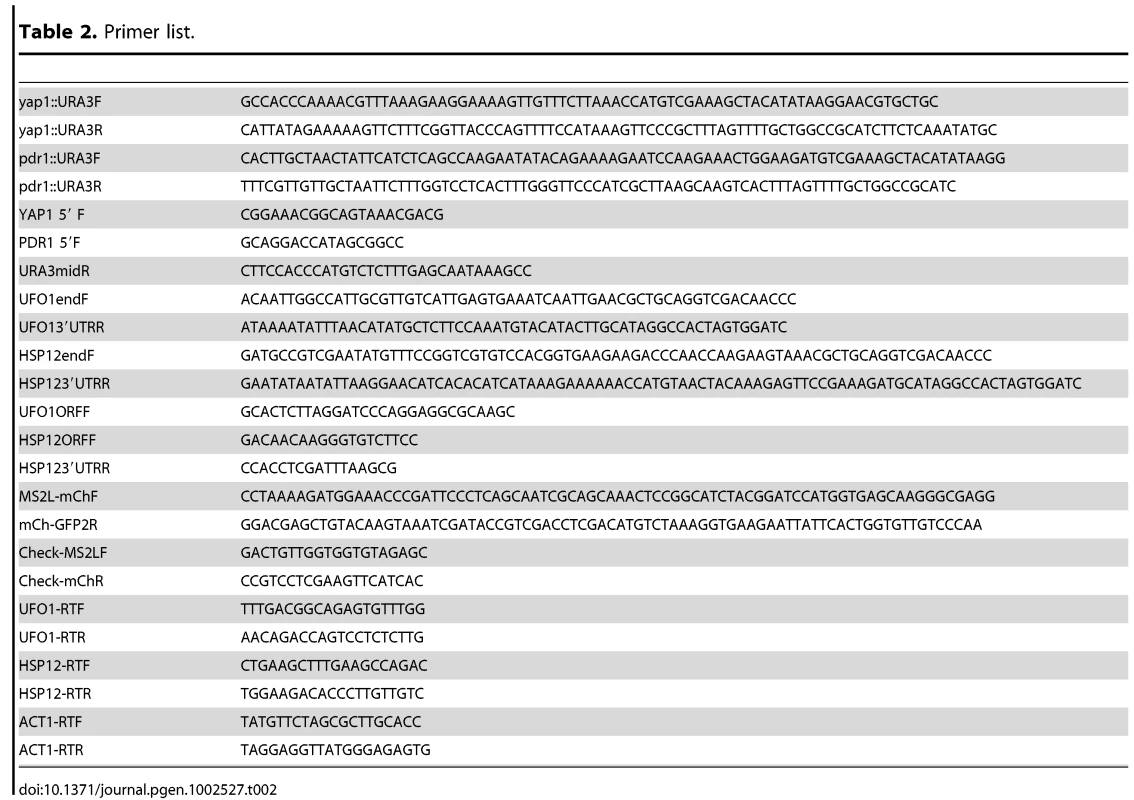
Within a population of cells of identical genotype the response of individual cells to stress can differ [48], [49]. Therefore to test the response of single cells we tagged the UFO1 ORF with MS2L DNA and expressed the coat protein (CP) that binds the MS2L RNA as a GFP-fusion protein [43]. To ensure the specificity of detection of UFO1 mRNA by the CPGFP fusion protein we induced CP expression in logarithmic cells with SC glucose medium lacking methionine for one hour. Subsequently, both wild type control and UFO1-MS2L cells were treated with arsenate, H2O2, or UV irradiation and observed after 30 minutes by confocal microscopy. In the control cells the CPGFP protein was visible diffusely throughout the cytoplasm both before and after each stress treatment whereas in response to arsenate, H2O2, and UV the UFO1-MS2L cells showed pronounced cytoplasmic granules ranging in number from zero to five (Figure 2A). Cessation of the treatment by incubation in fresh medium led to their disappearance (Figure 2B). No granules were observed after 30 minutes in treated UFO1-MS2L, yap1Δ or UFO1-MS2L, pdr1Δ mutant cells (Figure 2C) similar to the qRT-PCR results (Figure 1B).
Fig. 2. Stress-induced granules appear only in UFO1-MS2L stressed cells. 
A. Wild type and UFO1-MS2L cells transformed with pCP-MS2L-GFPx3 at A600 = 0.5, were transferred to SC 2% glucose without methionine for 1 hour to induce the CPGFP. Cells were untreated or exposed to 1 mM arsenate, 8.8 mM H2O2, or UV-irradiated with 40 mJ/cm2. Aliquots were collected 30 minutes after each treatment. Fluorescent granules appear only in cells with the tagged UFO1-MS2L gene. B. UFO1-MS2L cells expressing pCP-MS2L-GFPx3 grown as above, treated with 1 mM arsenate for 30 minutes (stress), and transferred to fresh SC glucose medium. Stress recovery 30′ and 60′ indicate time after transfer to fresh SC. C. UFO1-MS2L, yap1Δ or UFO1-MS2L, pdr1Δ cells grown as above and untreated (Utrd), exposed to 1 mM arsenate, 8.8 mM H2O2, or UV-irradiated with 40 mJ/cm2. Time course of granule appearance after stress
Accumulation of the granules was gradual and not all the cells in the population responded within the same time frame and to the same extent. We therefore quantified 150–200 cells for each treatment by defining three different classes of granules per cell: zero, 1–2 (few), or >3 (multiple). In response to arsenate after 30 minutes we observed an increase in the number of cells with a few or multiple granules followed by an increase in cells with a few granules over the 90-minutes of the experiment (Figure 3A). H2O2 led to an increase in cells with a few and with multiple granules after 30 minutes. By 90 minutes there was a slight increase in cells without granules and a slight decrease in the number of cells with 1–2 granules; the relative number of cells with multiple granules was unaltered (Figure 3B). After UV-irradiation UFO1-MS2L-CPGFP granules were visible after15 minutes; this included cells with a few granules, but mostly with multiple granules. The relative number of cells with a few granules increased during the first hour and then decreased; the number of cells with multiple granules decreased from 30 minutes post-irradiation until the end of the experiment (Figure 3C).
Fig. 3. Time course of granule appearance in UFO1-MS2L cells after stress. 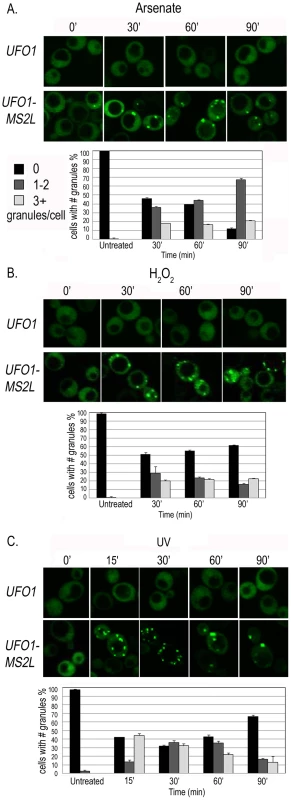
Cells treated with A. 1 mM arsenate, B. 8.8 mM H2O2, or C. 40 mJ/cm2 UV, and visualized by confocal microscopy at the times indicated. Histograms show number of cells with different numbers of granules per cell at the times indicated (n = >150 cells). Characterization of the UFO1 mRNA granules
To determine whether the granules that appeared after stress treatment of UFO1-MS2L cells correspond to PBs or SGs we tested for colocalization of the UFO1 mRNA with the PB marker protein, Dcp1RFP, and with the SG marker protein, eIF4ERFP, by mating UFO1-MS2L cells with PB - or SG-tagged strains [31]. Neither UFO1-MS2L granules nor PBs or SGs were visible in untreated cells growing in glucose medium (Figure 4A); cells in SC medium without glucose showed a single red fluorescent PB or SG, however, there was no induction of UFO1 mRNA in response to glucose deprivation (Figure 4B) and [31]. After 30 minutes of arsenate or H2O2 treatment of the glucose-deprived cells, we observed induction of UFO1 mRNA granules and these colocalized with both the PB and SG marker proteins (Figure 4C and 4D, respectively). Indeed control cells that coexpressed the PB marker protein, Dhh1GFP, and the SG marker protein, eIF4ERFP, showed overlap between the two types of granules after arsenate treatment (Figure 4E).
Fig. 4. UFO1-MS2L mRNAs induced by stress colocalize with subunits of PBs and SGs. 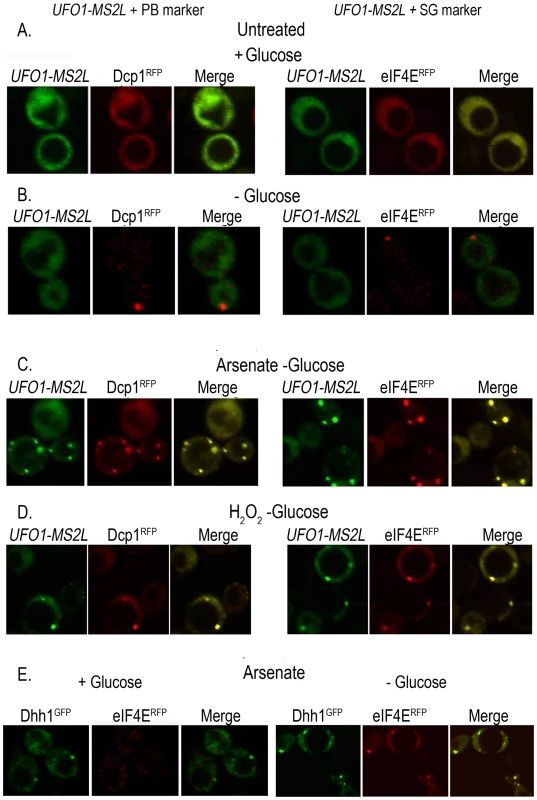
A. UFO1-MS2L cells at A600 = 0.5 that produce PB marker protein, Dcp1RFP, or the SG marker, eIF4ERFP, were transferred to fresh SC 2% glucose medium without methionine for 1 hour. B. Cells as in A, deprived for glucose for 30 minutes. C. Cells as in B, treated for 30 minutes with 1 mM arsenate, or D. 8.8 mM H2O2. E. Cells at A600 = 0.5 that produce both the PB marker protein, Dhh1GFP, and eIF4ERFP, treated with 1 mM arsenate for 30 minutes in the presence or the absence of glucose. Two distinct highly abundant mRNAs, UFO1 and MFA2, colocalize to the same PBs
The colocalization of stress-induced UFO1 mRNA with proteins associated with PBs and SGs together with the overlap of these bodies with one another after stress (Figure 4) suggest that these granules may house highly expressed mRNAs. To determine whether more than one mRNA species is present in the same granule we used a second well-characterized mRNA tagging system in which U1A binding sites are inserted into the 3′-UTR of the mRNA of interest and coexpressed with the U1AGFP RNA-binding protein [44]. Wild type cells were transformed with the plasmid for producing U1AGFP protein alone, or cotransformed with pMFA2-U1A and the above plasmid. Expression of MFA2 is regulated by the strong constitutive GPD (glyceraldehyde-3-phosphate dehydrogenase) promoter [24], [44] and reaches a high steady state level. As both the U1A and the CP proteins are fused to GFP, we changed the marker of the CPGFP protein to mCherry so that we could detect each mRNA species separately in the same cells. Control untreated or arsenate stressed cells that expressed only U1AGFP protein showed no granule formation. Untreated cotransformants of pMFA2-U1A and UFO1-MS2L showed multiple MFA2-U1A granules, but no granules attributable to UFO1-MS2L mRNA. After arsenate treatment UFO1-MS2L mRNA granules bound by red CPmCherry were observed and these colocalized with the MFA2-U1A granules. This result indicates that multiple mRNA species are found in the same PB (Figure 5).
Fig. 5. UFO1-MS2L and MFA2-U1A mRNAs are sequestered in the same PBs after arsenate stress. 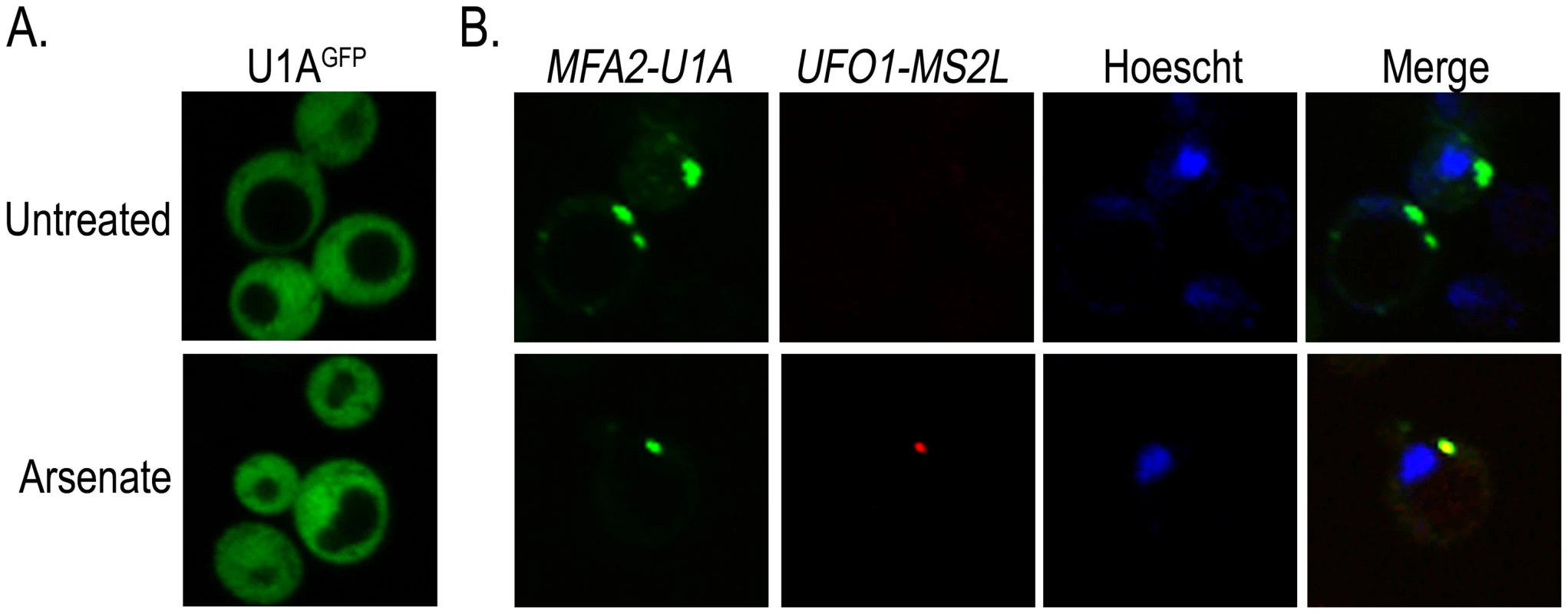
A. Control wild type cells at A600 = 0.5 producing U1AGFP untreated or treated with 1 mM arsenate for 30 minutes. B. wild type UFO1-MS2L cells at A600 = 0.5 expressing pMFA2-U1A, with their respective RNA-binding proteins, CPmCherry and U1AGFP, untreated or treated with 1 mM arsenate and stained with Hoechst 33342 at a final concentration of 2.5 µg/mL for 30 minutes. Low-abundance mRNAs ASH1-MS2L and OXA1-MS2L partially colocalize with PBs and SGs
To determine whether all mRNA granules correspond to PBs we assayed two further mRNAs that are specifically localized prior to their translation: ASH1-MS2L mRNA that is transported from the mother cell to the bud [42], and OXA1-MS2L mRNA that is localized to the mitochondria [43]. The PB marker Edc3mCherry was produced in cells that expressed ASH1-MS2L or OXA1-MS2L together with pCP-MS2L-GFPx3 for detection of their mRNAs. The cells were treated with the above stresses and imaged after 30 minutes with the confocal microscope. Both ASH1-MS2L and OXA1-MS2L mRNAs showed a similar response to the stress treatments. A small number of untreated cells showed at most a few Edc3mCherry-marked PBs, however, most cells lacked CPGFP granules of either ASH1-MS2L or OXA1-MS2L mRNAs. After 30 minutes with arsenate, H2O2, or glucose deprivation, we observed CPGFP granules corresponding to ASH1-MS2L (Figure 6A) or OXA1-MS2L (Figure 6B) mRNAs. These granules did not fully colocalize with PBs as did UFO1 mRNA, however, they were often docked, partially colocalized (overlapping), or appeared in the same cells with the PB marker protein Edc3mCherry, but at a distinct location in the same cell (Figure 6C).
Fig. 6. ASH1 and OXA1 mRNA granules interact with PBs. 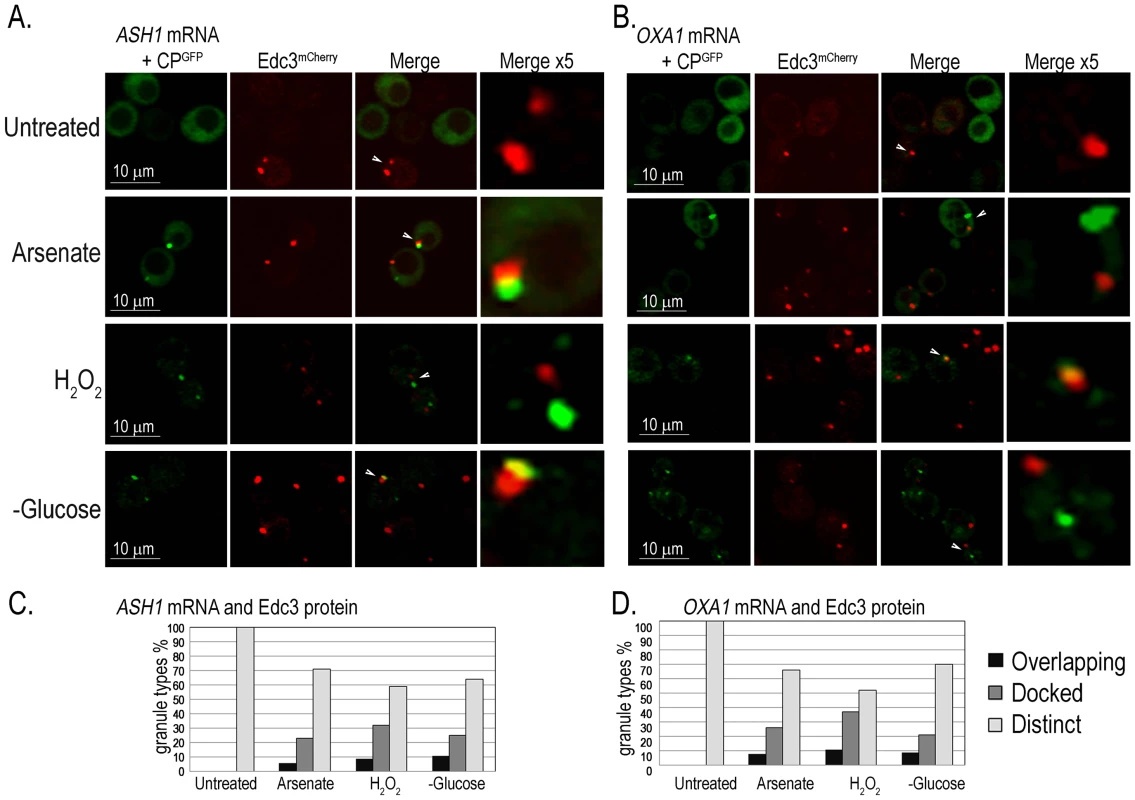
A. ASH1-MS2L cells at A600 = 0.5 with pCP-MS2L-GFPx3 and the PB marker, Edc3mCherry, either untreated, treated with 1 mM arsenate or with 8.8 mM H2O2 for 30 minutes or transferred to SC without glucose for 30 minutes. Merge ×5 represents 5 times enlargement of selected granules indicated with white arrows in the whole cells. B. OXA1-MS2L cells treated as in A., and visualized by confocal microscopy. C. Histograms of ASH1-MS2L cells or D. OXA1-MS2L cells, showing percentages of overlapping, docked, or distinct granule types in a population of cells untreated, treated with 1 mM arsenate or with 8.8 mM H2O2 for 30 minutes or stressed in SC without glucose (n = >100 cells). Highly expressed Hsp12 protein is encoded by a mRNA not observed in mRNA granules
Both whole-genome microarray experiments [7], [8] and our qRT-PCR data (Figure 1) indicate that UFO1 steady state mRNA levels are elevated in response to stress. However, it is only by using 10-fold the amount of cells compared with our standard protocols that we are able to detect Ufo1GFP protein after any of the stresses applied. We therefore examined the fate of the mRNA of HSP12, the protein of which is highly expressed in response to stress [40]. We treated cells with arsenate, H2O2, UV, glucose deprivation, 37°C or NaCl and analyzed induction of the protein by Western blotting. Hsp12GFP protein was induced after all the treatments, particularly after 37°C (Figure 7A and Figure S2). To examine mRNA localization we fused the MS2L tag to the HSP12 ORF and treated the cells with the same stresses. In contrast to UFO1-MS2L, we did not detect granules in HSP12-MS2L cells and the CPGFP signal remained diffuse throughout the cytoplasm (Figure S3 and [50]). The basal level of HSP12 mRNA in untreated cells was higher than that of UFO1 (Figure 7B). however, the relative increase of HSP12 steady state mRNA level after stress was lower than the high induced levels of UFO1 mRNA in these samples (cf. Figure 7C and Figure 1A). The elevation of HSP12 mRNA level results from increased transcription as HSP12 mRNA is slightly destablized after arsenate, whereas after H2O2, and UV treatments the stability is similar to the untreated cells with a half-life of ca. 10 minutes in both untreated and stressed cells (Figure 7D). There was no stabilization of the HSP12 mRNA after UV irradiation as observed for UFO1 mRNA (Figure 1B).
Fig. 7. Induction of Hsp12 protein and mRNA, and mRNA decay after stress. 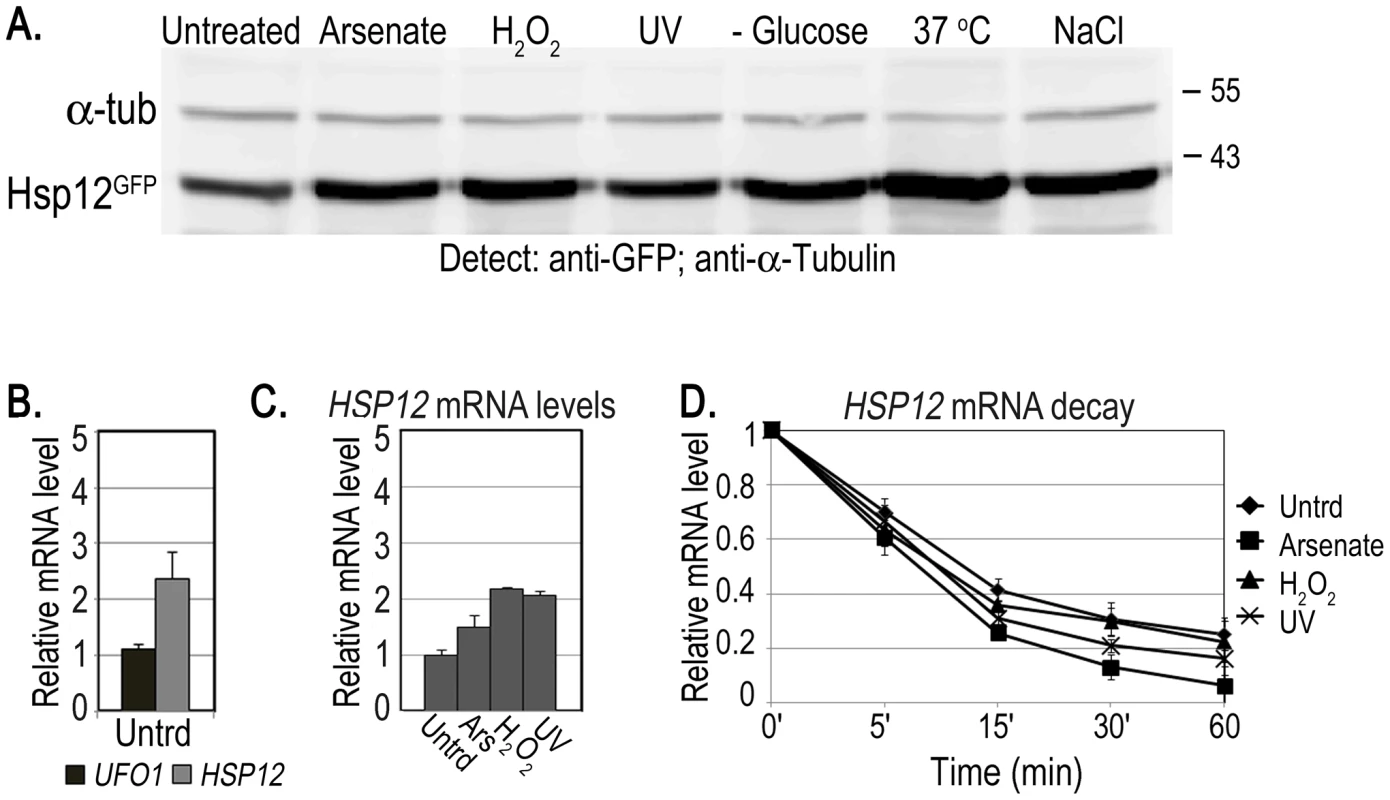
A. WB of protein produced from genomic HSP12-GFP in response to stress. B. UFO1 and HSP12 mRNA levels in untreated cells analyzed by qRT-PCR. mRNA levels were normalized to ACT1. C. Induction of HSP12 mRNA by stress. Wild type cells at A600 = 0.5, treated with 1 mM arsenate, 8.8 mM H2O2, irradiated with 40 mJ/cm2 UV or shifted from 30°C to 37°C for 40 minutes. Aliquots were collected at the times indicated and analyzed by qRT-PCR. D. HSP12 mRNA decay. pGAL-HSP12 was expressed in hsp12Δ mutants by overnight induction with 2% galactose. Next morning cells at A600 = 0.5 were untreated, or stressed with 1 mM arsenate or 8.8 mM H2O2 for 30 minutes, or irradiated with 40 mJ/cm2 UV. The cells were washed and transferred to SC medium with 4% glucose. Samples were collected immediately after addition of glucose and at the times indicated and analyzed by qRT-PCR. mRNA levels were normalized to ACT1 and to time 0 (untreated log cells). High-level expression of mRNA affects the viability of mutants unable to form PBs
To determine what role sequestration of UFO1 and of MFA2 mRNAs in PBs may have we examined the viability of cells defective in the formation of PBs under two different conditions: (a) exposure to arsenate, H2O2, UV, or glucose deprivation, and (b) high level expression of UFO1 or HSP12 from the GAL promoter, and of MFA2 from the strong GPD promoter. We used a single pat1Δ mutant defective in a mRNA decapping enzyme and two different double mutants: edc3Δ, pat1Δ mutants defective in both PB and SG formation, and edc3Δ, lsm4Δc mutants that have reduced PB and SG formation. (Edc3 enhances mRNA decapping [51] and Lsm4 is a subunit of a heptomeric complex involved in mRNA decay [27]). The edc3Δ and lsm4Δ single mutants are able to form PBs under conditions of glucose deprivation, however, in the absence of Edc3, there is a requirement for the C-terminal prionlike domain of Lsm4 for PB formation [52]. To visualize granule formation isogenic wild type, pat1Δ, edc3Δ, pat1Δ, and edc3Δ, lsm4Δc cells were transformed to express the PB marker, Dcp2mCherry, and the SG marker protein, Pab1GFP from pRP1658. There were no granules in untreated cells in the wild type or in the single pat1Δ mutant, PBs appeared in response to arsenate, H2O2, and UV, and both PBs and SGs appeared after glucose deprivation. No granules were visible in edc3Δ, pat1Δ mutants consistent with [27]; in response to glucose deprivation the edc3Δ, lsm4Δc mutants showed very faint PBs and SGs (Figure 8A). Arsenate, H2O2, and UV treatments caused a decrease in viability of the edc3Δ, pat1Δ mutants but not of the w.t., pat1Δ, or edc3Δ, lsm4Δc mutants (Figure 8B). Induction of the YCpGAL control vector on galactose plates did not affect viability of any of the mutants. In contrast, induction of high levels of UFO1 or HSP12 mRNA from the GAL1 promoter led to a strong decrease in the viability of edc3Δ, pat1Δ but not of the pat1Δ, or edc3Δ, lsm4Δc mutants compared with wild type or with the noninduced cells growing on glucose plates. MFA2 is expressed from the constitutive GPD promoter and affected viability of these mutants on both glucose and galactose plates (Figure 8C). To determine whether the differential susceptibility of the edc3Δ, pat1Δ mutants may be the result of non-regulated translation of the UFO1, HSP12 or MFA2 mRNAs we compared the level of protein in wild type and mutant cells when these three genes were expressed from the GAL promoter. Western blot analysis of wild type and edc3Δ, pat1Δ mutants growing under noninducing (glucose) or inducing (galactose) conditions indicated enhanced translation of each mRNA in the edc3Δ, pat1Δ mutants (Figure 8D and Figure S4). There was no difference in the rate of decay of UFO1 or of HSP12 mRNAs between wild type and edc3Δ, pat1Δ mutants (Figure 8E).
Fig. 8. Ectopic high level gene expression affects viability of mutants unable to form PBs and SGs. 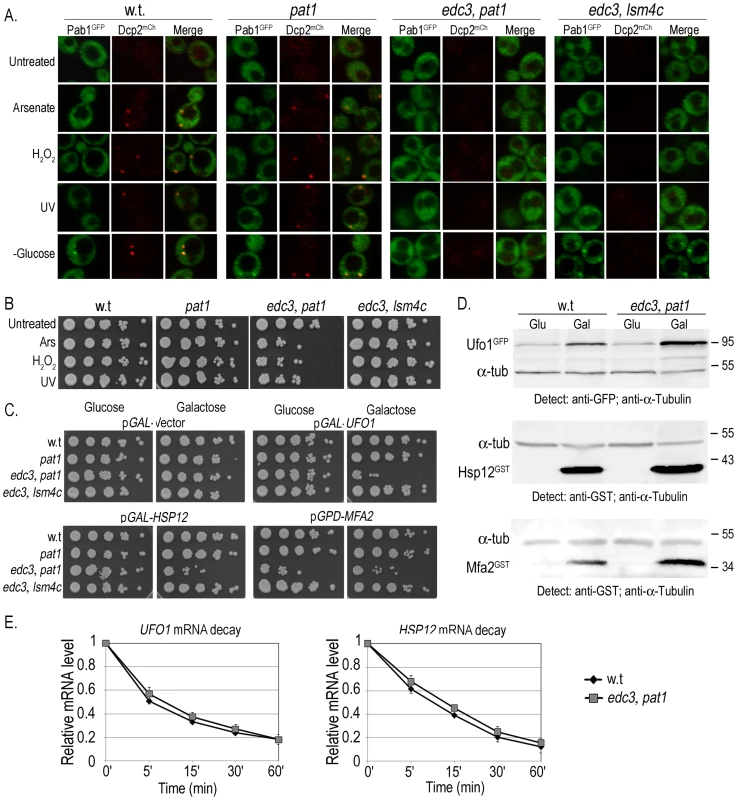
A. Visualization of wild type, pat1Δ, edc3Δ, pat1Δ or edc3Δ, lsm4Δc cells expressing the PB marker Dcp2mCherry and the SG marker Pab1GFP untreated or exposed for 30 minutes to 1 mM arsenate or 8.8 mM H2O2, irradiated with 40 mJ/cm2 UV, or incubated in SC medium without glucose for 30 minutes. B. Viability of wild type, pat1Δ; edc3Δ, pat1Δ or edc3Δ, lsm4Δc cells untreated (Unt) or treated with 1 mM arsenate, 8.8 mM H2O2 or irradiated with 40 mJ/cm2 UV analyzed by the spot test viability assay. C. Wild type, pat1Δ; edc3Δ, pat1Δ or edc3Δ, lsm4Δc cells expressing empty pGAL-vector (YCp), pGAL-GFP-UFO1, pGAL-HSP12 or MFA2-U1A (pRP1193) were grown in SC medium with 2% glucose or induced in 2% galactose medium overnight, diluted and regrown in the same media to A600 = 0.5 for spot test analysis on SC plates with 2% glucose or 2% galactose, respectively. D. WB analysis of w.t or edc3Δ, pat1Δ cells expressing UFO1, HSP12 or MFA2 from the GAL promoter. Glu (noninducing conditions) and Gal (inducing). The intensities of each protein band were normalized to the α-tubulin loading control using ImageJ [79]. E. Comparison of UFO1 and HSP12 mRNA decay. pGAL-GFP-UFO1 or pGAL-HSP12 was expressed in w.t or edc3Δ, pat1Δ cells by overnight induction with 2% galactose. Next morning cells at A600 = 0.5 were washed and transferred to SC medium with 4% glucose. Samples were collected immediately after addition of glucose and at the times indicated and analyzed by qRT-PCR. mRNA levels were normalized to ACT1 and to time 0 (untreated log cells). Discussion
We analyzed the functional consequence of accumulation of high levels of mRNA in yeast cells under different stress conditions and identified stations in the lifecycle of mRNAs that could explain the discrepancy between transcript abundance and protein level. Our data suggest that the ability to store highly abundant mRNAs in PBs for future regulation is a key facet of the stress response as it allows individual cells to sort mRNAs for decay or translation. A prototype of this kind of regulation is induction of UFO1 mRNA under stress conditions. Our qRT-PCR experiments show that steady state levels of UFO1 are elevated in cells stressed by arsenate, H2O2, or UV. This observation is compatible with microarray data [8], [17], [53]. The Yap1 and Pdr1 dependence of UFO1 transcription suggests that UFO1 is part of the early stress response that includes genes that encode several heat shock proteins [54]. Transcriptome studies have shown that production and degradation of mRNA are often coordinated [18]. However, in our experiments even though the steady state UFO1 mRNA level was elevated after each stress, the mRNA lifecycle was different for UV, an acute stress, compared with arsenate and H2O2 treatments. Arsenate did not affect UFO1 mRNA stability, and after H2O2 there was a very slight destabilization of the mRNA. In contrast UV irradiation led to an elevation of the steady state UFO1 mRNA level that remained high due to stabilization of the mRNA. In contrast to UFO1 mRNA there was no marked change in the stability of HSP12 mRNA after arsenate, H2O2, or UV. A general stabilization of mRNAs expressed from the GAL promoter in response to UV was reported by [55] who did not observe mRNA stabilization after starvation, heat or osmotic stress. UV irradiation did not lead to disassembly of polysomes, however, the mRNAs were no longer associated with the polysomes but accumulated in cytoplasmic granules without losing their polyA tails suggesting they were not designated for decay [55].
By increasing the yeast extract concentration tenfold we were able for the first time to detect Ufo1 protein expressed from genomic UFO1 after arsenate, H2O2, and UV. The protein steady state level was very low in untreated cells (1% that of α-tubulin) but after these stresses showed a 10-fold elevation. Examination of the Ufo1 protein half-life after arsenate and H2O2 showed that the protein was stabilized thus the higher level can be attributed to lack of proteasomal degradation. In contrast UV irradiation did not affect the half-life of Ufo1 protein and in light of Galliard's observation that polysomes are not disassembled after UV irradiation, our interpretation is that the elevated steady state level of Ufo1 protein is due to renewed translation of the stabilized UFO1 mRNA. This is compatible with the proposed role for Ufo1 in maintenance of genome stability [9] and may be indicative of a remodeling of the genome during recovery. These steady state Ufo1 protein values are comparable to results of a study in which we expressed the bacterial reporter genes, luxA and luxB, in yeast from the UFO1 promoter [46]. Luciferase activity was elevated 10-fold in response to 40 mJ/cm2 UV whereas arsenate or H2O2 led to a threefold elevation of enzyme activity. Direct measurement of UFO1 transcription here by qRT-PCR shows that all three stress treatments lead to very similar elevation of transcription suggesting that luciferase activity may have been affected by prolonged treatment with arsenate or H2O2. HSP12 represents a different paradigm of regulation in that the basic steady state mRNA level was higher in unstressed cells and fold-elevation of steady state level was less than for UFO1. However, the salient difference was in the cellular amounts of each protein in untreated cells: steady state Hsp12 protein levels are two orders of magnitude those of Ufo1 (Figure S3) and [47]. This may be due to differences in translatability of HSP12 mRNA that encodes a protein of 107 amino acids compared with UFO1 whose gene product has 668 residues [56]. Ribosome profiling has shown that translation efficiency can differ 100-fold between different genes with shorter genes having a higher ribosome density [22]. Nuclear export is an important regulatory step for molecular chaperones [57]; in addition the 5′-untranslated region (5′-UTR) plays an important role in determining translation efficiency of HSP12 mRNA in Aspergillus oryzae [58], [59] and Arabidopsis [60].
Cells with the UFO1-MS2L reporter showed granules after stress; these granules correspond to UFO1-MS2L mRNA molecules bound to the fluorescent capsid protein, CPGFP. Granules were only visible when the genomic UFO1 ORF was fused to MS2L excluding the possibility they were comprised of aggregated CPGFP. Moreover, after cessation of arsenate stress, the granules disappeared; this could be indicative of decay or return to the polysomes. There was a similar decrease of cells with granules after H2O2 treatment that could be due to cellular responses to reactive oxygen species [3]. UFO1-MS2L granules were not visible in UFO1-MS2L, yap1Δ or pdr1Δ mutants that lack the transcription factors shown by our qRT-PCR results to be essential for induction of UFO1 by stress. Therefore the granules are indeed UFO1-MS2L mRNA bound to CPGFP. Individual cells showed considerable variation both in their response time and in their number of granules as observed in many other systems. Genes involved in the stress response are expressed with a high level of cell-to-cell variation, stochastic noise attributed to epigenetic factors [61]–[64]. This is considered to enhance the ability of the population to survive adverse conditions by enabling them to sample multiple phenotypes [16], [62].
Colocalization of the UFO1-MS2L mRNA granules with the PB and SG marker proteins, Dcp1GFP and eIF4ERFP, respectively, indicated that the UFO1-MS2L mRNA is sequestered in PBs and SGs. SGs were only visible in the absence of glucose [31] and this necessitated incubation of the cells in SC medium without glucose prior to stress treatment in these experiments. The mRNAs of two highly expressed genes - UFO1-MS2L induced by arsenate stress, and MFA2-U1A constitutively expressed from the strong GPD promoter - colocalized to the same PBs indicating that these granules house multiple copies of diverse mRNAs. Using protein markers for PBs and SGs we found that these two bodies colocalize after stress consistent with a role for PBs as a sorting station for future regulation of mRNAs for decay or storage [28], [65].
Our data indicate clear differences in the lifecycles of UFO1 and HSP12 mRNAs. UFO1 transcription was elevated three - to fourfold in response to arsenate, H2O2, or UV and the UFO1 mRNA was present in PBs and SGs. In contrast, HSP12 mRNA levels that were double those of UFO1 mRNA in untreated cells were elevated at most twofold after these stresses. Furthermore under these conditions in which we and [40] observed strong induction of the Hsp12 protein we did not observe granules corrresponding to HSP12-MS2L mRNA. The half-life of UFO1 mRNA was not affected by arsenate and H2O2 stresses, but the mRNA was stabilized after UV irradiation; none of these stress treatments affected the stability of HSP12 mRNA. On the protein level we could only observe Ufo1GFP protein in cells with genomic UFO1-GFP after stress by taking 10-fold the number of cells for analysis; in contrast genomic Hsp12GFP protein was easily detectable in untreated cells and was elevated between two - and fivefold in cells treated with H2O2, 37°C, and NaCl using our standard experimental protocols.
Our genetic analysis suggests that sequestration of highly expressed mRNAs in PBs is an important mechanism for survival. In edc3Δ, pat1Δ mutants that are unable to form PBs we observe elevated protein levels compared with the isogenic wild type cells. mRNA decay rates are not affected in these mutants and our interpretation is that the mRNAs lose their regulation and enter the polysome fraction. Here again we observe a difference between the PB-associated UFO1 and MFA2 mRNAs and HSP12 mRNA: UFO1 and MFA2 mRNAs show a six - to eightfold elevation of steady state protein levels, respectively, in the edc3Δ, pat1Δ mutants compared with Hsp12 that shows a threefold elevation of protein level compared with wild type cells. Overexpression of all three genes affects the viability of the edc3Δ, pat1Δ mutants. Pat1 and Dhh1 function in the coordination of translation and PB formation acting as repressors of translation and enhancers of PB formation [30], [35]. The nonregulated elevated translation we observe in the edc3Δ, pat1Δ mutants may exert its effect on viability through depletion of translation factors required for maintenance of basic essential cell functions. Fold-elevation of HSP12 mRNA levels in response to stress is lower than that of UFO1 and the high levels of HSP12 mRNA produced from the GAL promoter may create a requirement for their sequestration in PBs to maintain the balance between storage, decay, and translation. In addition the high level of HSP12 mRNA could lead to a depletion of essential HSP-gene specific regulatory factors and affect biosynthesis of molecular chaperones crucial for withstanding the stress [66], [67].
It is not clear whether the lifecycle of every mRNA species involves a sojourn in PBs. Transcripts of housekeeping genes could go straight to the polysomes [18], [28], or a certain fraction of transcripts, depending on the environmental conditions, could be imprinted with the RNA polymerase II associated Rpb4-Rpb7 heterodimer for sojourn and future sorting in PBs [18]. This heterodimer escorts mRNA transcripts from the nucleus to the cytoplasm and physically interacts with the PB proteins, Pat1 and Lsm2, prior to being reimported into the nucleus [36], [68]. Besides nuclear export [69], Rpb4 and Rpb7 regulate other stages of the mRNA lifecycle such as exit from PBs [70], and 5′ to 3′ and 3′ to 5′ decay [36], [68]. Moreover even though ASH1-MS2L and OXA1-MS2L CPGFP granules did not show the full colocalization observed for UFO1 and MFA2 mRNAs we did observe docking of their granules with PBs, similar to the dynamic docking interaction reported for dendritically localized mRNAs in Drosophila neurons [71]. Transport granules share some factors with PBs and this may allow reciprocal transfer of mRNA and proteins between them [28].
We present a diagram that summarizes putative alternative lifecycles for mRNA based on the sample of genes studied here (Figure 9). The ability to store UFO1 and other mRNAs in PBs and SGs provides a mechanism for individual cells to regulate their future sorting into pathways for decay or translation. Under normal growth conditions mRNAs could be translated immediately (a), or could pass through PBs that could lead to delayed translation (b); or direction of the mRNA for decay (c). Under stress conditions pre-existing mRNAs undergo enhanced translation as we propose for HSP12 mRNA (α), or are retracted from the polysomes (β) for future sorting for storage (γ) or decay (δ). Furthermore, after stress mRNAs can shuttle between PBs and SGs (γ) from where they can return to translation (ε). Both extrinsic conditions such as strength and duration of the stress, and the intrinsic metabolic state of a particular cell would influence the outcome [62]–[64]. This regulatory mechanism would result in a population of cells each with the potential to express a unique subset of mRNAs and to acquire a different phenotype thus increasing the ability of the population of genetically identical cells to withstand the stress.
Fig. 9. Pathways for mRNA under normal and stress conditions. 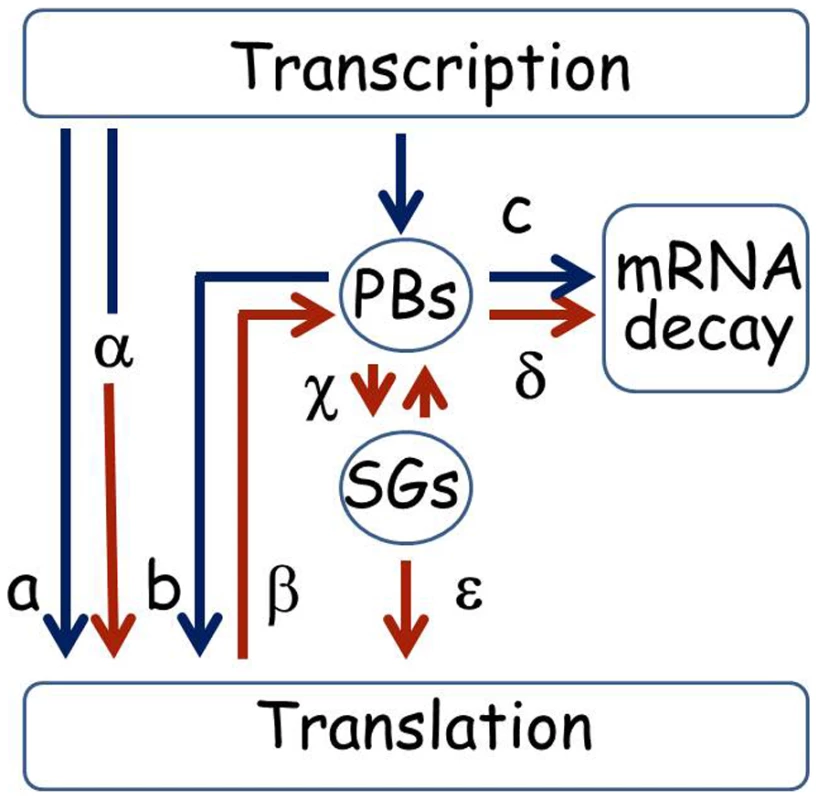
Under normal growth conditions after transcription a mRNA could go directly to the polysomes for translation (a), or the mRNAs could be escorted by subunits of the RNA polymerase complex to the PBs and sorted either for the polysomes for translation (b), or the mRNA could undergo decay in the PBs (c). Under stress conditions pre-existing mRNAs could undergo enhanced translation as we propose for HSP12 mRNA (α), or be retracted from the polysomes (β) for future sorting for storage or decay (γ or δ). Furthermore, after stress mRNAs can shuttle between PBs and SGs (γ) from where they can return to translation (ε). mRNA lifecycles under normal growth conditions have blue arrows, stress conditions are indicated with brown arrows. A more comprehensive description of the mRNA lifecycle, particularly of mRNA decay, can be found in [80]. Materials and Methods
Yeast strains
Strain genotypes are listed in Table 1. Strain yMK1366 has genomic DCP1 fused to RFP as a marker for PBs, and strain yMK1307 has genomic eIF4E fused to RFP as a marker for SGs [31]. Strain yRP1600 is pat1Δ; strain yRP1752 has double deletions of edc3Δ, pat1Δ [72];and strain yRP2338 has double deletions, edc3Δ, lsm4Δc [52]. yap1Δ or pdr1Δ deletion strains were constructed by homologous recombination [73] with a PCR fragment that encodes an auxotrophic URA3 marker gene amplified from YCpGAL [74] flanked with 40 bps for targeting, using the primer pairs yap1::URA3F/R to delete YAP1, and pdr1::URA3F/R to delete PDR1 (Table 2). The deletions were confirmed using the primer pair YAP15′F for yap1Δ, and PDR15′F for pdr1Δ with the reverse primer URA3midR complementary to the middle of the URA3 gene.
Fusion of genomic UFO1 and heat-shock gene, HSP12, to the MS2L cassette
To the UFO1 or HSP12 ORFs we fused a PCR-amplified cassette comprising 12 MS2L-CP binding sites and the Schizosaccharomyces pombe his5+ selectable marker between two copies of the lambda phage loxP sequence from plasmid pLoxHis5MS2L [43]. Forty nts of homology to UFO1 or HSP12 were introduced into each primer for site-specific integration of the PCR cassette. The entire loxP::Sphis5+::loxP::MS2L PCR cassette was integrated after the STOP codon of UFO1 using the primers UFO1endF and UFO13′UTRR, and at the end of the HSP12 ORF using primers HSP12end F and HSP123′UTRR. UFO1 cassette integration was confirmed by colony PCR amplification using the primer pair, UFO1ORFF/UFO13′UTRR and of HSP12 with the primer pair, HSP12ORFF/HSP12 3′UTRR. Subsequently the S. pombe his5+ marker was excised with Cre recombinase by transforming positive colonies with plasmid pSH47 that encodes Cre recombinase under the GAL1 promoter [43]. Transformants were isolated on glucose plates and transferred to galactose medium for excision of the Sphis5+ gene. This yielded histidine auxotrophic colonies in which the MS2L sequence was separated from the UFO1 - or HSP12 ORF by LoxP followed immediately by the UFO1 or HSP12 3′-UTR. Genomic integration of the MS2L cassette downstream of the UFO1 - and HSP12 ORF was verified by sequencing the PCR fragment amplified with the primers used to check cassette integration.
Plasmids
pDHH1-GFP [45] and pRP1574 that encodes EDC3-mCherry [27] were used as markers of PBs. pRP1658 that encodes both PAB1-GFP and DCP2-mCherry was used to provide markers for PBs and SGs [44]. pRP1187 encoding U1AGFP [44] was used to detect MFA2 mRNA expressed from pRP1193 [24], both are regulated by the GPD constitutive promoter. pCP-MS2L-GFPx3 [43] and pCP-MS2L-mCherry (below) were used to detect mRNAs with MS2L binding loops; pGAL-GFP-UFO1 [13], pGAL-HSP12 [75] and pBM123 (YCpGAL) [74] were used in growth analysis studies. pGAL-GFP-UFO1 [13], pGAL-HSP12 [75] and pGAL-MFA2 [75] were used in WB analysis in Figure 8D.
pCP-MS2L-mCherry plasmid construction: pCP-MS2L-mCherry was constructed by homologous recombination in vivo by replacing the first GFP ORF in pCP-MS2L-GFPx3 with a PCR product that encodes mCherry. The primer pair MS2L-mChF/mCh-GFP2R was designed with homologous ends to MS2L and to the third copy of GFP. The reverse primer has a STOP codon after the mCherry ORF so that only mCherry will be expressed fused to the CP. The mCherry PCR product together with pCP-MS2L-GFPx3 were transformed into wild type yeast for recombination in vivo. pCP-MS2L-mCherry was extracted from yeast and amplified in E. coli for verification of replacement of GFP with mCherry by PCR and sequencing using primer pair Check-MS2LF/Check-mChR. The pCP-MS2L-mCherry plasmid was then transformed into UFO1-MS2L yeast and treated with arsenate for confirmation of a red signal. Subsequently plasmids pRP1193 [24] and pRP1187 [44] for expression and visualization of MFA2-U1A mRNA were cotransformed into the cells for treatment and confocal microscopy.
Growth conditions
Yeast cultures were grown overnight at 30°C in a rotary thermoshaker at 120 rpm in synthetic minimal medium (SC) supplemented with the appropriate amino acids and carbon source [76]. Next morning cultures were diluted to A600 = 0.1, regrown to early exponential phase, A600 = 0.5, and treated as detailed below.
Arsenate and H2O2
For arsenate treatment cells at A600 = 0.5 in SC glucose medium were incubated with 1 mM Na2HAsO4; for H2O2 we diluted the 30% stock solution 1∶1000 to a final concentration of 8.8 mM. Aliquots were collected by brief centrifugation at the times indicated in the figures.
UV irradiation
Cells at A600 = 0.5 were irradiated with 40 mJ/cm2 UV on a transilluminator and aliquots were collected as above.
Glucose deprivation
Cells at A600 = 0.5 were collected by brief centrifugation, washed in fresh SC medium without glucose and incubated at 30°C for 30 minutes in fresh SC medium without glucose. Cells were collected by brief centrifugation and visualized by confocal microscopy.
Osmotic stress
Cells at A600 = 0.5 were incubated in 0.5 M NaCl for 30 minutes prior to visualization by confocal microscopy.
Heat shock
Cells at A600 = 0.5 were shifted from 30°C to 37°C for 40 minutes prior to visualization by confocal microscopy.
Growth analysis
For over-expression experiments YCpGAL [74], pGAL - GFP-UFO1 [13], pRP1193 encoding MFA2-U1A [24] or pGAL-HSP12 [75] were transformed into wild type or mutant yeast. Cells were grown overnight in SC with 2% glucose or induced with 2% galactose, diluted to A600 = 0.1 in the appropriate media, and regrown to A600 = 0.5. At this stage viability was assayed using a spot test with 5 µl drops of 10-fold serial dilutions plated on the appropriate medium. Plates were incubated at 30°C for 3 days and scanned.
Detection of UFO1-MS2L or HSP12-MS2L mRNA
UFO1-MS2L or HSP12-MS2L cells were transformed with pCP-MS2L-GFPx3 that encodes the MS2L-CP fused to three tandem repeats of GFP or with pCP-MS2L-mCherry (above), both expressed from the inducible MET25 promoter [42]. Transformed cells at A600 = 0.5 were incubated in SC medium with 2% glucose but lacking methionine for 1 hour prior to treatment with stress agents as above and visualized by confocal microscopy.
Quantitative real-time PCR (qRT–PCR)
Total RNA was extracted using a MasterPure Yeast RNA Purification kit (Epicentre) according to the manufacturer's protocol. The amount of total RNA extracted from the cells was measured by absorbance at 260 nm. First-strand cDNA was synthesized from 1 µg total RNA using a blend of RNA primers (random hexamers and anchored oligodT 3∶1 (v/v)) and Verso Enzyme mix following the manufacturer's instructions (Thermo Scientific Verso cDNA Kit). A final concentration of 20 ng/µl of synthesized cDNA was used as template for the PCR reaction. qRT-PCR was performed using Thermo-Start DNA Polymerase, UFO1 primers (UFO1-RTF and UFO1-RTR), HSP12 primers (HSP12-RTF and HSP12-RTR), or ACT1 primers (ACT1-RTF and ACT1-RTR) and an ABsolute Blue QPCR SYBR Green ROX Mix, as follows: denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, and extension at 72°C for 30 sec, for 30 cycles. Appropriate non-RT and non-template controls were included in each PCR reaction, and dissociation analysis was performed at the end of each run to confirm the specificity of the reaction. Primer efficiency for UFO1, HSP12 and ACT1 mRNAs was determined (Figure S5). Data were analyzed using the comparative Ct method: [delta][delta]Ct = [delta]Ct,sample−[delta]Ct,reference. Here [delta]CT,sample is the Ct value for each treated sample normalized to the endogenous housekeeping gene Actin and [delta]Ct,reference is the Ct value for the calibrator (untreated cells) also normalized to Actin (Figure 1). A simple [delta]CT calculation (sample normalized to Actin) was used in Figure 7B.
mRNA decay
pGAL-GFP-UFO1 or pGAL-HSP12 were expressed in ufo1Δ or hsp12Δ mutants, respectively, by overnight induction with 2% galactose. Next morning cells were diluted to A600 = 0.1, and regrown to A600 = 0.5, then treated with the indicated stresses. The cells were washed and transferred to SC with 4% glucose. Samples were collected immediately after glucose addition and at the times indicated and analyzed by qRT-PCR.
Coexpression of MS2L-tagged mRNAs with markers of PB or SGs
Cells expressing DCP1-RFP or eIF4E-RFP were mated with the UFO1-MS2L strain and the diploids were transformed with pMS2L-CP-GFPx3. ASH1-MS2L or OXA1-MS2L cells were transformed with the PB marker plasmid pRP1574 [27] that encodes EDC3-mCherry. All strains were treated with stress as described in the relevant figures and visualized by confocal microscopy. To study coexpression of PBs and SGs yeast strains eIF4E-RFP [31] transformed with pDHH1-GFP [45] or wild type transformed with pRP1658 [27] were treated as indicated in the relevant figures and visualized by confocal microscopy.
Microscopy
All images were acquired with an Olympus FV1000 laser-scanning confocal microscope using the ×60 objective lens. The fluorescence was excited with 543 nm for the red fluorescent markers and 488 nm for GFP. For the coexpression experiments sequential screening was used to avoid overlapping. Images are representative of three independent experiments. For the time course experiments images of untreated cells and those exposed to stress (1 mM arsenate, 8.8 mM H2O2, or 40 mJ/cm2 UV) were subjected to quantitative analysis by defining 3 different classes of cells with zero, 1–2, or 3 or more granules. 150–200 cells were assayed for each treatment at each time point. Hoescht staining was used for cell nuclei.
Induction of protein expression in yeast
For expression from the GAL1 promoter, yeast cells were grown overnight in 2% galactose medium. GFP-tagged protein was observed using a Nikon fluorescence microscope fitted with GFP-specific filter set: dichromic 505 nm, excitation 450–490 nm, emission (low pass) 515 nm (Nikon).
Western blot analysis
For Western blot analysis cells were grown overnight in 2% glucose medium, or for induction from the GAL promoter with 2% galactose. Next morning the cells were diluted to A600 = 0.1 and regrown to A600 = 0.5 for TCA precipitation [77] and Western blotting [78].
Supporting Information
Zdroje
1. GaschAWerner-WashburneM 2002 The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics 2 181 192
2. GaschAPHuangMMetznerSBotsteinDElledgeSJ 2001 Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12 2987 3003
3. TempleMDPerroneGGDawesIW 2005 Complex cellular responses to reactive oxygen species. Trends Cell Biol 15 319 326
4. GaschAPSpellmanPTKaoCMCarmel-HarelOEisenMB 2000 Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11 4241 4257
5. HershkoACiechanoverA 1998 The ubiquitin system. Annu Rev Biochem 67 425 479
6. PetroskiMDDeshaiesRJ 2005 Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 9 20
7. JelinskySASamsonLD 1999 Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci U S A 96 1486 1491
8. HaugenACKelleyRCollinsJBTuckerCJDengC 2004 Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol 5 R95
9. SmithSHwangJYBanerjeeSMajeedAGuptaA 2004 Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101 9039 9044
10. KaplunLIvantsivYKornitzerDRavehD 2000 Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc Natl Acad Sci U S A 97 10077 10082
11. KaplunLIvantsivYBakhratARavehD 2003 DNA damage response-mediated degradation of Ho endonuclease via the ubiquitin system involves its nuclear export. J Biol Chem 278 48727 48734
12. KaplunLTzirkinRBakhratAShabekNIvantsivY 2005 The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol Cell Biol 25 5355 5362
13. IvantsivYKaplunLTzirkin-GoldinRShabekNRavehD 2006 Turnover of SCFUfo1 complexes requires the UbL-UbA motif protein, Ddi1. Mol Cell Biol 26 1579 1588
14. SkonecznaAMcIntyreJSkonecznyMPolicinskaZSledziewska-GojskaE 2007 Polymerase eta Is a Short-lived, Proteasomally Degraded Protein that Is Temporarily Stabilized Following UV Irradiation in Saccharomyces cerevisiae. J Mol Biol 366 1074 1086
15. KaplunL 2002 Ph.D. thesis, Ben Gurion University of the Negev
16. López-MauryLMargueratSBählerJ 2008 Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 9 583 593
17. ShalemODahanOLevoMMartinezMFurmanI 2008 Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol 4 223
18. ShalemOGroismanBChoderMDahanOPilpelY 2011 Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet 9 e1002273 doi:10.1371/journal.pgen.1002273
19. AravaYBoasFEBrownPOHerschlagD 2005 Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Research 33 2421 2432
20. Beyer AHJNasheuerHPWilhelmT 2004 Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol Cell Proteomics 3 1083 1092
21. LuPVogelCWangRYaoXMarcotteE 2007 Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25 117 124
22. IngoliaNTGhaemmaghamiSNewmanJRSWeissmanJS 2009 Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 324 218 223
23. TullerTCarmiAVestsigianKNavonSDorfanY 2010 An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation. Cell and Molec Life Sci 141 344 354
24. BrenguesMTeixeiraDParkerR 2005 Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310 486 489
25. TeixeiraDParkerR 2007 Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell 18 2274 2287
26. FranksTMLykke-AndersenJ 2008 The control of mRNA decapping and P-body formation. Mol Cell 32 605 615
27. BuchanJRMuhlradDParkerR 2008 P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183 441 455
28. ParkerRShethU 2007 P bodies and the control of mRNA translation and degradation. Mol Cell Biol 25 635 646
29. BalagopalVParkerR 2009 Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 21 403 408
30. CarrollJSMunchelSEWeisK 2011 The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol 194 527 537
31. HoyleNPCastelliLMCampbellSGHolmesLEAsheMP 2007 Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol 179 65 74
32. HoyleNPAsheMP 2008 Subcellular localization of mRNA and factors involved in translation initiation. Biochem Soc Trans 36 648 652
33. EulalioABehm-AnsmantIIzaurraldeE 2007 P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8 9 22
34. DamgaardCKLykke-AndersenJ 2011 Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev 25 2057 2068
35. CollerJParkerR 2005 General translational repression by activators of mRNA decapping. Cell 122 875 886
36. LotanRGoler-BaronVDuekLHaimovichGChoderM 2007 The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J Cell Biol 178 1133 1143
37. Goler-BaronVSelitrennikMBarkaiOHaimovichGLotanR 2008 Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev 22 2022 2027
38. SmillieDASommervilleJ 2002 RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J Cell Sci 115 395 407
39. FischerNWeisK 2002 The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J 21 2788 2797
40. SadehAMovshovichNVolokhMGheberLAharoniA 2011 Fine-Tuning of the Msn2/4-Mediated Yeast Stress Responses as Revealed by Systematic Deletion of Msn2/4 Partners. Mol Biol Cell 22 3127 3138
41. ArandaAQuerolAdel OlmoM 2002 Correlation between acetaldehyde and ethanol resistance and expression of HSP genes in yeast strains isolated during the biological aging of sherry wines. Arch Microbiol 177 304 312
42. BeachDLBloomK 2001 ASH1 mRNA localization in three acts. Mol Biol Cell 12 2567 2577
43. HaimLZiporGAronovSGerstJE 2007 A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 4 409 412
44. TeixeiraDShethUValencia-SanchezMABrenguesMParkerR 2005 Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11 371 382
45. ShethUParkerR 2003 Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300 805 808
46. BakhratAEltzovEFinkelsteinYMarksRRavehD 2011 UV and arsenate toxicity: a specific and sensitive yeast bioluminescence assay. Cell Biol Toxicol 27 227 236
47. GhaemmaghamiSHuhWKBowerKHowsonRWBelleA 2003 Global analysis of protein expression in yeast. Nature 425 737 741
48. RajAPeskinCSTranchinaDVargasDYTyagiS 2006 Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biol 4 e309 doi:10.1371/journal.pbio.0040309
49. RaserJMO'SheaEK 2004 Control of stochasticity in eukaryotic gene expression. Science 304 1811 1814
50. LavutA 2011 Multi-level Regulation Of The Yeast F-box Protein, Ufo1, In Response To Stress. PhD thesis, Ben Gurion University of the Negev, Beersheba, Israel (submitted)
51. KshirsagarMParkerR 2004 Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166 729 739
52. DeckerCJTeixeiraDParkerR 2007 Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179 437 449
53. JelinskySAEstepPChurchGMSamsonLD 2000 Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol 20 8157 8167
54. Lucau-DanilaALelandaisGKozovskaZTantyVDelaveauT 2005 Early expression of yeast genes affected by chemical stress. Mol Cell Biol 25 1860 1868
55. GaillardHAguileraA 2008 A novel class of mRNA-containing cytoplasmic granules are produced in response to UV-irradiation. Mol Biol Cell 19 4980 4992
56. GingoldHPilpelY 2011 Determinants of translation efficiency and accuracy. Mol Syst Biol 7 481
57. SaavedraCTungKSAmbergDCHopperAKColeCN 1996 Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev 10 1608 1620
58. KodaAMinetokiTOzekiKHirotsuneM 2004 Translation efficiency mediated by the 5′ untranslated region greatly affects protein production in Aspergillus oryzae. Appl Microbiol Biotechnol 66 291 296
59. KodaABogakiTMinetokiTHirotsuneM 2006 5′ Untranslated region of the Hsp12 gene contributes to efficient translation in Aspergillus oryzae. Appl Microbiol Biotechnol 70 333 336
60. DansakoTKatoKSatohJSekineMYoshidaK 2003 5′ Untranslated region of the HSP18.2 gene contributes to efficient translation in plant cells. J Biosci Bioeng 95 52 58
61. Bar-EvenAPaulssonJMaheshriNCarmiMO'SheaE 2006 Noise in protein expression scales with natural protein abundance. Nat Genet 38 636 643
62. MaheshriNO'SheaEARBBS 2007 Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct 36 413 434
63. ChoiJKKimY-K 2009 Implications of the nucleosome code in regulatory variation, adaptation and evolution. Epigenetics 4 291 295
64. WhitelawNCChongSWhitelawE 2010 Tuning in to noise: epigenetics and intangible variation. Dev Cell 19 649 650
65. AdeliK 2011 Translational Control Mechanisms in Metabolic Regulation: Critical Role of RNA binding proteins, miRNAs, and Cytoplasmic RNA granules. Am J Physiol Endocrinol Metab Epub ahead of print
66. MorimotoRI 1998 Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12 3788 3796
67. ErkinaTYErkineAM 2006 Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Mol Cell Biol 26 7587 7600
68. LotanRBar-OnVGHarel-SharvitLDuekLMelamedD 2005 The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19 3004 3016
69. FaragoMNahariTHammelCColeCNChoderM 2003 Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol Biol Cell 14 2744 2755
70. Harel-SharvitLEldadNHaimovichGBarkaiODuekL 2010 RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143 552 563
71. ZeitelhoferMKarraDMacchiPTolinoMThomasS 2008 Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci 28 7555 7562
72. KshirsagarMParkerR 2004 Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166 729 739
73. LongtineMSMcKenzieA3rdDemariniDJShahNGWachA 1998 Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953 961
74. JohnstonMDavisR 1984 Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol 4 1440 1448
75. ZhuHBilginMBanghamRHallDCasamayorA 2001 Global analysis of protein activities using proteome chips. Science 14 2101 2105
76. AdamsAGottschlingDEKaiserCAStearnsT 1997 Methods in Yeast Genetics CSHL Press, NY
77. CoxJSChapmanREWalterP 1997 The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell 8 1805 1814
78. Baranes-BacharKKhalailaIIvantsivYLavutAVoloshinO 2008 New interacting partners of the F-box protein Ufo1 of yeast. Yeast 25 733 743
79. RasbandW 1997–2011 ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA
80. HarigayaYParkerR 2010 No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA 1 132 141
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy