-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Natural System of Chromosome Transfer in
The High Pathogenicity Island of Yersinia pseudotuberculosis IP32637 was previously shown to be horizontally transferable as part of a large chromosomal segment. We demonstrate here that at low temperature other chromosomal loci, as well as a non-mobilizable plasmid (pUC4K), are also transferable. This transfer, designated GDT4 (Generalized DNA Transfer at 4°C), required the presence of an IP32637 endogenous plasmid (pGDT4) that carries several mobile genetic elements and a conjugation machinery. We established that cure of this plasmid or inactivation of its sex pilus fully abrogates this process. Analysis of the mobilized pUC4K recovered from transconjugants revealed the insertion of one of the pGDT4–borne ISs, designated ISYps1, at different sites on the transferred plasmid molecules. This IS belongs to the IS6 family, which moves by replicative transposition, and thus could drive the formation of cointegrates between pGDT4 and the host chromosome and could mediate the transfer of chromosomal regions in an Hfr-like manner. In support of this model, we show that a suicide plasmid carrying ISYps1 is able to integrate itself, flanked by ISYps1 copies, at multiple locations into the Escherichia coli chromosome. Furthermore, we demonstrate the formation of RecA-independent cointegrates between the ISYps1-harboring plasmid and an ISYps1-free replicon, leading to the passive transfer of the non-conjugative plasmid. We thus demonstrate here a natural mechanism of horizontal gene exchange, which is less constrained and more powerful than the classical Hfr mechanism, as it only requires the presence of an IS6-type element on a conjugative replicon to drive the horizontal transfer of any large block of plasmid or chromosomal DNA. This natural mechanism of chromosome transfer, which occurs under conditions mimicking those found in the environment, may thus play a significant role in bacterial evolution, pathogenesis, and adaptation to new ecological niches.
Published in the journal: A Natural System of Chromosome Transfer in. PLoS Genet 8(3): e32767. doi:10.1371/journal.pgen.1002529
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002529Summary
The High Pathogenicity Island of Yersinia pseudotuberculosis IP32637 was previously shown to be horizontally transferable as part of a large chromosomal segment. We demonstrate here that at low temperature other chromosomal loci, as well as a non-mobilizable plasmid (pUC4K), are also transferable. This transfer, designated GDT4 (Generalized DNA Transfer at 4°C), required the presence of an IP32637 endogenous plasmid (pGDT4) that carries several mobile genetic elements and a conjugation machinery. We established that cure of this plasmid or inactivation of its sex pilus fully abrogates this process. Analysis of the mobilized pUC4K recovered from transconjugants revealed the insertion of one of the pGDT4–borne ISs, designated ISYps1, at different sites on the transferred plasmid molecules. This IS belongs to the IS6 family, which moves by replicative transposition, and thus could drive the formation of cointegrates between pGDT4 and the host chromosome and could mediate the transfer of chromosomal regions in an Hfr-like manner. In support of this model, we show that a suicide plasmid carrying ISYps1 is able to integrate itself, flanked by ISYps1 copies, at multiple locations into the Escherichia coli chromosome. Furthermore, we demonstrate the formation of RecA-independent cointegrates between the ISYps1-harboring plasmid and an ISYps1-free replicon, leading to the passive transfer of the non-conjugative plasmid. We thus demonstrate here a natural mechanism of horizontal gene exchange, which is less constrained and more powerful than the classical Hfr mechanism, as it only requires the presence of an IS6-type element on a conjugative replicon to drive the horizontal transfer of any large block of plasmid or chromosomal DNA. This natural mechanism of chromosome transfer, which occurs under conditions mimicking those found in the environment, may thus play a significant role in bacterial evolution, pathogenesis, and adaptation to new ecological niches.
Introduction
Horizontal gene transfer (HGT) is a driving force for bacterial evolution, as it allows the dispersion of adaptive loci between closely related and also phylogenetically distant bacterial species. Well-characterized mobile genetic elements such as conjugative plasmids, transposons, Integrative conjugative elements (ICE), pathogenicity islands (PAI), or phages are associated with HGT of specific adaptive functions (antibiotic resistance, virulence, metabolic pathways) and participate to genome plasticity. However, exchanges of chromosomal regions that form the core genome and are not part of the mobile genetic pool are also observed. While their importance in bacterial evolution and speciation is now well established, the underlying mechanisms are often loosely described and remain hypothetical in many cases.
The Gram-negative enteropathogen Yersinia pseudotuberculosis carries a PAI termed High Pathogenicity Island (HPI) [1], which encodes the siderophore yersiniabactin [2]. The fact that this island is mobile within the genome of its host strain [3], and is present and often conserved both in terms of genetic organization and nucleotide sequence in various bacterial genera such as Escherichia coli (various pathotypes), Klebsiella or Citrobacter [4], suggested that it may have retained its ability to be horizontally transmitted to new bacterial hosts. Indeed, we evidenced the transfer of the HPI between natural Y. pseudotuberculosis isolates [3]. This phenomenon was observed only when the bacteria were incubated at low temperature (optimal at 4°C) and in broth, and was more efficient in an iron-poor medium [5]. However, this transfer did not require the integration/excision machinery encoded by the HPI, was RecA-dependent in the recipient strain, and involved not only the HPI but also adjacent sequences encompassing at least 46 kb of chromosomal DNA [3]. Similar results were recently obtained for the HPI of natural Escherichia coli isolates, using a multi locus sequence typing approach. The E. coli HPI was found to have been acquired simultaneously with the chromosomal flanking regions of the donor strains [6], indicating again that the island was transmitted as part of a larger chromosomal region. This phenomenon is not restricted to the HPI and to enterobacteria since it has been recently reported that movement of the Enterococcus faecalis PAI was invariably accompanied by transfer of flanking donor chromosome sequences [7].
The aim of this work was to characterize the mechanisms underlying horizontal chromosomal gene transfer in Y. pseudotuberculosis. We describe here a natural system of conjugative transfer, which may be used by a wide variety of bacterial species for gene exchanges, and which may represent a driving force for bacterial evolution.
Results
Generalized transfer of chromosomal and plasmid DNA in Y. pseudotuberculosis IP32637
Since we did not know whether the lateral transfer process previously observed was limited to the region encompassing the HPI or could involve any portion of the chromosome, two other loci (ureB and or5076) were labeled with a spectinomycin (Spe) and trimethoprim (Tmp) resistance cassette, respectively. These two genes were chosen because, based on the IP32953 sequence, they are predicted to be separated from each other and from the HPI (tagged with a kanamycin (Kan) cassette in the irp2 gene) by at least 1.5 Mb of chromosomal DNA (Figure S1). Moreover, the ureB gene, which is part of the urease locus, and or5076, encoding a putative toxin transporter [8] are not predicted to be involved in DNA transfer. After co-incubation of the donor 637-irp2K-ureBS-5076T and recipient 637ΔHPI-NalR strains (Table 1) under conditions (4 days at 4°C in LB-αα' with shaking) that we previously found to be optimal for HPI transfer [3], recipient strains having acquired either the irp2K (NalR, KanR, RifS), ureBS (NalR, SpeR, RifS) or or5076T (NalR, TmpR, RifS) antibiotic resistances were obtained. Acquisitions of the corresponding tagged loci were checked by PCR (Figure S1). Transfer frequencies were of the same magnitude for the three antibiotic-tagged loci (≈10−8, Figure 1). None of the transconjugants obtained had simultaneously acquired two of the antibiotic-tagged loci, indicating that the sizes of the chromosomal fragments transferred were inferior to 1.5 Mb.
Fig. 1. Transfer frequencies at various temperatures of three distantly located chromosomal loci. 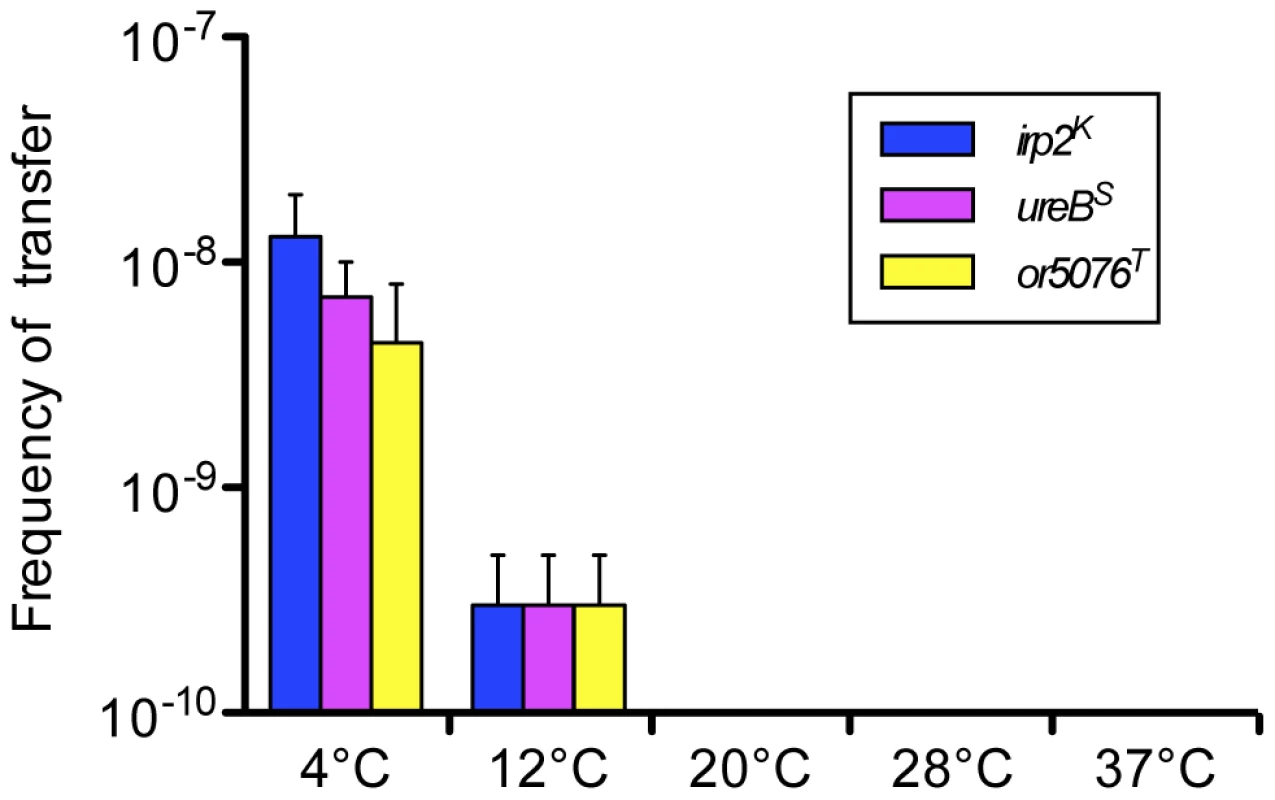
The donor 637-irp2K-ureBS-5076T and recipient 637ΔHPI-NalR strains were co-incubated in LB-αα' with agitation. Transfer frequency was calculated as the number of NalR (or KanR, SpeR, TmpR) and RifS transconjugants per RifR donor cells. Shown are mean values of transfer frequencies (vertical bars) and standard error of the mean (sem, vertical lines) of two independent experiments at each temperature. Mean transfer frequencies (±sem) at 4°C: 1.3(±0.7)×10−8 for irp2K, 0.7(±0.3)×10−8 for ureBS and 0.4(±0.4)×10−8 for or5075T, and at 12°C: 0.03(±0.02)×10−8 for the three loci. Transfer frequencies at temperatures ≥20°C were systematically below the detection limit (10−10). Tab. 1. <i>Y. pseudotuberculosis</i> strains and plasmids used for DNA transfer experiments. 
We previously showed that horizontal transfer of the HPI occurs only at low temperatures [3]. The same temperature dependency was observed for ureBS and or5076T: transfer of each of the three antibiotic-tagged loci was detected only when the donor and recipient strains were co-incubated at temperatures below 20°C (Figure 1), and was more efficient at 4°C than at 12°C (≥13 fold higher), as previously observed for irp2K. Therefore, distantly located chromosomal loci can be transferred with similar efficiencies and temperature regulations.
Whether this transfer mechanism could also mediate horizontal transmission of episomal molecules was addressed by introducing the non-conjugative and non-mobilizable plasmid pUC4K (KanR) into the donor 637-RifR. Using the defined optimal growth conditions, transfer of pUC4K from the 637(pUC4K) to the recipient 637ΔHPI-NalR was obtained and confirmed by PCR with primers 210A/210B (Table S1). This transfer occurred at a frequency of 2.3 (±0.4)×10−7, which is at least 10 times higher than that of chromosomal loci. Therefore, the process of DNA transfer is not limited to chromosomal DNA but can also involve plasmid molecules.
Altogether our results demonstrate the existence of a mechanism that potentially allows transfer of any chromosomal or episomal DNA molecule at low temperature. This mechanism was thus named GDT4 (for Generalized DNA Transfer at 4°C).
IP32637 harbors a plasmid involved in GDT4
The capacity of other Y. pseudotuberculosis strains to mediate GDT4 was studied by tagging the IP32953 and IP32777 strains with both a Kan and Spe cassettes inserted into the irp2 and ureB loci, respectively (Table 1). When these two recombinant strains were used as donors, no IP32637 transconjugants having acquired either irp2K or ureBS were obtained, indicating that GDT4 is not a property common to the entire Y. pseudotuberculosis species.
Strain IP32637 has the peculiarity of harboring an extra high molecular weight (≥100 kb) plasmid [9]. The role of this additional plasmid in chromosomal transfer was assessed by comparing GDT4 in IP32637 and its IP32637c plasmid-cured derivative [9]. Two tagged donor strains, 637c-irp2K and 637c-ureBS (Table 1), were generated and co-incubated with the 953-NalR recipient. No transconjugants were obtained, indicating a role of this plasmid in DNA transfer. The high molecular weight plasmid was thus designated pGDT4.
pGDT4 does not appear to be ubiquitous in the species Y. pseudotuberculosis as the genome sequences of IP32953 and of other Y. pseudotuberculosis strains available in databases did not evidence the presence of this plasmid. To get an insight into the frequency of pGDT4 carriage in this species, a 4 kb HindIII fragment of this episome, designated pGDT4.seq was cloned into pUC18, sequenced, and used to design primers (358A/B) for PCR screening. The analysis of a panel of 39 Y. pseudotuberculosis strains of serotypes I to V (Table S2) for the presence of the pGDT4 sequence identified two isolates (IP32699 and IP30215) that gave a PCR product of the expected size (Table S2). Both strains contained high molecular weight episomes whose HindIII-digestion patterns yielded some restriction fragments with a size similar to those of pGDT4, but the overall profiles of the three episomes were different (data not shown). Therefore, the plasmids found in IP32699 and IP30215 probably share some regions with pGDT4, but they are not identical to this plasmid. Since Yersinia pestis is a recent descent of Y. pseudotuberculosis [10], we also screened by PCR a panel of 51 strains of Y. pestis belonging to the three classical biovars (Antiqua, Medievalis and Orientalis) for the presence of the pGDT4-borne sequence. None of the strains tested yielded an amplification product (Table S2), suggesting the absence of vertical or horizontal transmission of pGDT4 to Y. pestis.
pGDT4 is transferable
Plasmid analysis of transconjugants resulting from the co-incubation of the 637-irp2K-ureBS donor strain with the 637c-NalR recipient revealed that about half of them had acquired pGDT4 together with the chromosomal irp2 (8/20) or ureB (11/20) tagged region, thus indicating that pGDT4 is also transferable.
To further study pGDT4 transfer capacity, the plasmid was labeled by allelic exchange of the pGDT4-4kb segment with a Tmp cassette. When the 637-irp2K-ureBS donor strain carrying the tagged pGDT4T was co-incubated with the 637-NalR recipient, transconjugants harboring pGDT4T were obtained with a frequency approximately 103 times higher than that of chromosomal genes (Figure 2A and Table S3). The transfer frequency of pGDT4T increased 200 fold when 953-NalR instead of 637-NalR was used as a recipient (Figure 2B and Table S3), indicating that properties inherent to the recipient cells may influence their capacity to take up pGDT4. The difference in the ability of the two strains to acquire this plasmid could not be explained by a mechanism of surface exclusion, as the frequency of transfer of pGDT4T to IP32637 harboring or not harboring a resident pGDT4 was similar (Figure 2B and Table S3). Some unidentified intrinsic properties of the recipients such as a difference in their restriction/modification systems may be responsible for this difference. As observed with chromosomal DNA, no transfer of pGDT4T was detected when the bacteria were mated at temperatures ≥28°C (Figure 2C and Table S3). However, in contrast to chromosomal DNA [3], pGDT4T was transferable on nitrocellulose filters, at frequencies similar to those observed in a liquid medium (Table S3), but again, only at low temperature (Figure 2C). Therefore, transfer of pGDT4 is also temperature-dependent, but in contrast to chromosomal genes, it occurs at much higher frequencies, and both in liquid and on solid media.
Fig. 2. Conjugative properties of pGDT4. 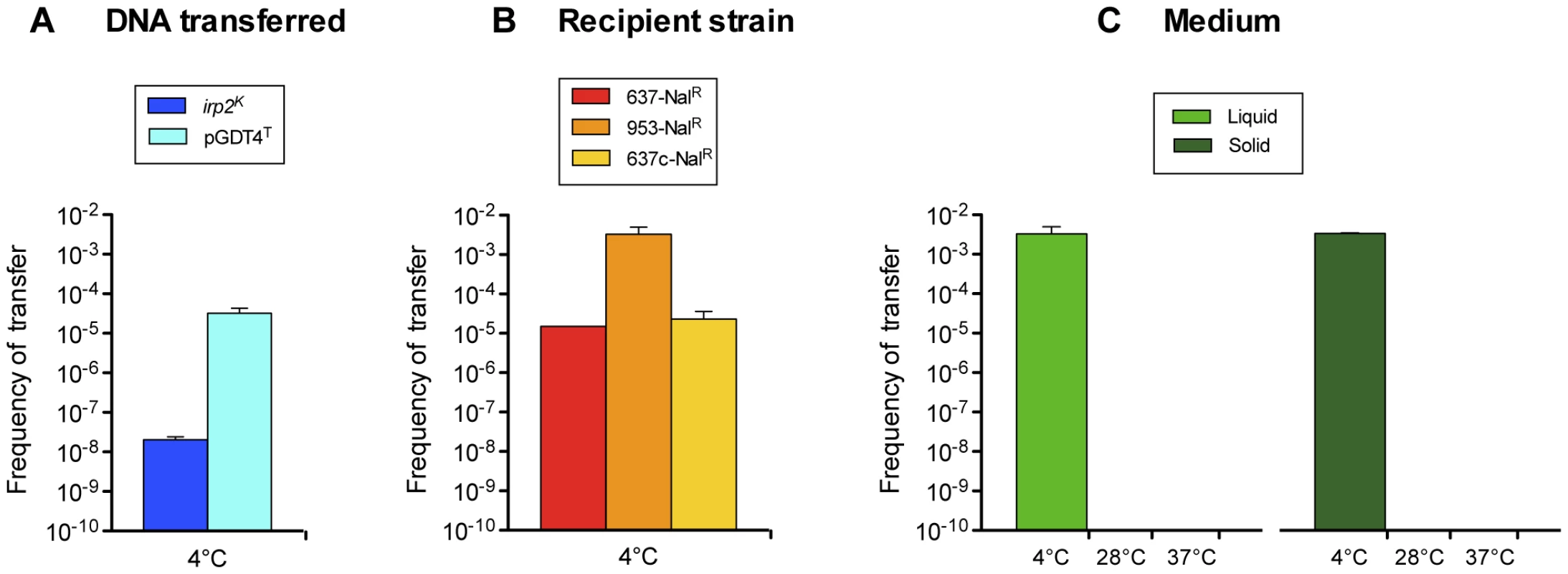
(A) Comparison of the efficiency of transfer of pGDT4 and the irp2K chromosomal locus. The donor 637-irp2K-ureBS(pGDT4T) and the recipient 637-NalR were co-incubated at 4°C in LB-αα' and independent experiments were performed 4 to 5 times. (B) Efficiency of transfer of pGDT4 to various recipient strains. The donor 637-irp2K-ureBS(pGDT4T) was co-incubated at 4°C with either 637-NalR, 953-NalR or 637c-NalR. The results of two independent experiments were combined. (C) Efficiency of transfer of pGDT4 at various temperatures in liquid and on solid media. The experiments were performed twice independently. Shown are mean values of transfer frequencies (vertical bars) and standard error of the mean (sem, vertical lines). The detection limit was 10−10. Since the presence of pGDT4 is required for transfer, we wondered whether its presence could confer GDT4 properties to a strain that is naturally unable to mediate chromosomal transfer. For this purpose, a 953-ureBS transconjugant that had acquired pGDT4 simultaneously with chromosomal genes was used as donor and co-incubated with a 637ΔHPI-RifR recipient. While the parental 953-NalR strain was unable to transfer chromosomal DNA, the 953-ureBS(pGDT4) transconjugant gained the capacity to retransfer the acquired ureBS locus, though with a frequency 10 times lower (10−9) than that observed during the first transfer. These results further point at pGDT4 as a key element in the mechanism of chromosomal transfer.
Sequence analysis of pGDT4
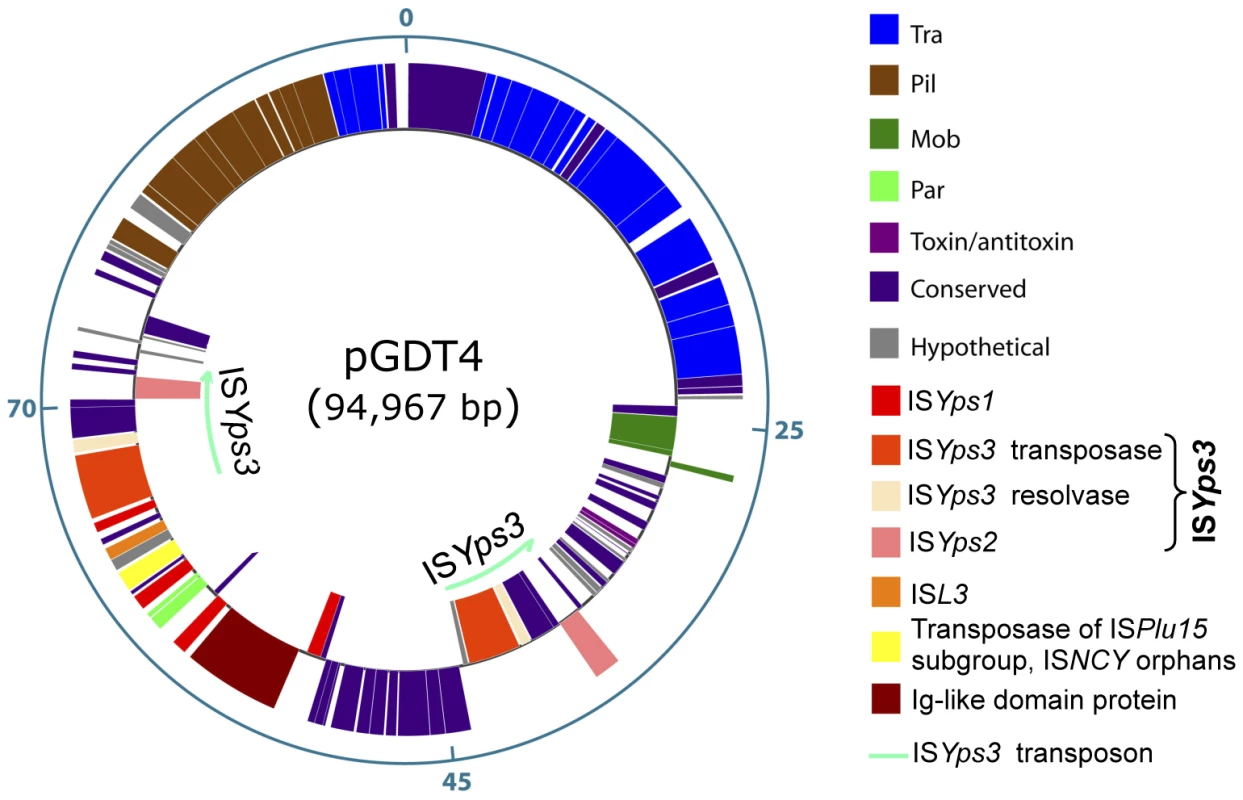
To determine whether pGDT4 could encode its own transfer machinery, the plasmid was sequenced (EMBL accession number FM178282). The schematic map of the 94,967 bp circular plasmid molecule is shown on Figure 3. Of the 102 predicted coding sequences (cds) identified on pGDT4, 74 had homologs in databases (Table S4). Four major functional groups of genes were delineated on pGDT4:
-
A DNA fragment of ≈44 kb (pGDT4_0086-0024) carried genes predicted to be involved in conjugative transfer. Overall, these genes showed the closest similarity with those of the 153 kb conjugative plasmid pADAP of Serratia entomophila [11] that encode proteins involved in mating pair formation (MPF), DNA transfer and post conjugative replication. The pGDT4 MPF belongs to the MPFI class, according to the most recent classification, for which R64 is the paradigm [12]. Interestingly, we found that in pGDT4, traX and traY are fused in a single traXY gene, while they are separated but adjacent in the related pADAP MPF operon. This is the first report of such organization, which seems to be functional as transfer of pGDT4 is occurring at reasonable rates. The classification of conjugative plasmids based on their encoded relaxase has recently emerged as powerful and meaningful in terms of plasmid physiology [13], [14]. The pGDT4 MobA relaxase is likely encoded by pGDT4_0022. This putative relaxase and its closest homolog, the protein pADAP_128 from pADAP, both fall in the MOBP13 class [14]. Proteins encoded by its neighboring genes, pGDT4_0023 and 0024, likely correspond to the relaxosomal accessory proteins MobB and MobC that act together with MobA in this family. Usually, in this type of mobABC backbone, the oriT is located between mobC (pGDT4_0024) and mobB (pGDT4_0023), as demonstrated for instance in pADAP [11].
-
The second group of genes gathered loci involved in plasmid stabilization and replication such as anti-restriction and toxin-antitoxin genes (pGDT4_0028-0031), the parFG operon (pGDT4_0062/0063, Table S4), encoding a partition system of type I (Walker type ATPase), and a replication initiation protein of the IncFII_RepA family (pGDT4_0081, Table S4). This replication protein is highly related to the RepA protein encoded by the Yersinia enterocolitica plasmid pYVe8081 [15], but this plasmid is otherwise unrelated to pGDT4 and has, for example, a partition system from a different family (SopAB).
-
The third group corresponded to mobile genetic elements, mostly transposases and insertion sequences (IS), which are commonly encountered on plasmids. pGDT4 carries one ISL3 transposon, one uncharacterized IS (ISNCY) with a transposase of the ISPlu15 subgroup belonging to the ISNCY orphans in the IS repository ISfinder, four copies (three complete and one truncated) of a new IS6 family member (ISYps1), and two copies of the transposon ISYps3 (Figure 3 and Table S4). The latter is a 7.3 kb Tn3-like transposon flanked by 34 bp inverted repeats and contains the ISYps3 transposase and resolvase, as well as ISYps2, an IS110 family member. None of the IS present on the Y. pseudotuberculosis IP32953 chromosome [8] were present on pGDT4 and conversely, no sequences homologous to the pGDT4-borne IS: ISYps1 (primer pair 727A/B), ISNCY (729A/B), ISYps2 (730A/B), ISL3 (731A/B) and ISYps3 (732A/B) were detected by PCR in the IP32637 genome (data not shown).
-
The remaining genes included a large proportion of putative coding sequences of unknown functions, and a few loci with predictable functions, of which some shared homology with Y. pseudotuberculosis chromosomal genes (Table S4). The most remarkable example was the putative product of pGDT4_0059, which shared 41% amino acid identities with YPTB3789, a chromosomal Ig-like domain protein [16].
pGDT4–mediated generalized DNA transfer
Since pGDT4 carries a large set of genes predicted to be involved in conjugative transfer, we wondered whether this pGDT4-specific mobility function mediates GDT4. To investigate this potential role, the Mpf function was inactivated by allelic exchange of a large portion of the pil region (from pilL to pilV) of pGDT4 with a Tmp cassette in 637(pUC4K). After co-incubation of the resulting strain 637(pUC4K, pGDT4Δpil) with the 953-NalR recipient, no transconjugants having acquired pUC4K were obtained, indicating that the pilus-encoding region of pGDT4 is required for generalized DNA transfer.
As the Mpf region is predicted to encode a conjugative machinery, GDT4 most likely occurs by a mechanism of conjugation. To rule out other possible mechanisms of transfer, DNAse was added to the medium during the co-incubation period. The transfer frequency of the irp2 locus from the donor 637-irp2K-ureBS to the recipient 637ΔHPI-NalR was not affected, arguing against an acquisition of naked DNA through a transformation process. Cell-free filtrates of the supernatant of the donor strain incubated with the recipient strain did not allow DNA transfer, suggesting the absence of transferable DNA released from the bacteria but protected from the action of a DNAse (inside phage particles or membrane vesicles). These results argue against a transfer of DNA by transformation or transduction and further point at conjugation as the most likely mechanism. However, since this conjugative process was observed in liquid medium under agitation, we wondered whether a strong shaking of the culture would disrupt the pilus-mediated interactions between bacterial cells, and therefore decrease the transfer frequency. Surprisingly, when we increased the agitation of the medium containing the donor and recipient cells to 130 rpm (which was vigorous under our experimental conditions), the frequency of transfer of the irp2 locus was not affected (0.9×10−8). Electron microscopy analysis of IP32637 cells grown under conditions optimal for GDT4 did not reveal any pilus structures on the bacterial surface. In contrast, tightly aggregated bacilli that seemed to be connected by “bridges” were observed (Figure 4).
Fig. 4. Electron microscopy of IP32637 grown at 4°C in LB with agitation. 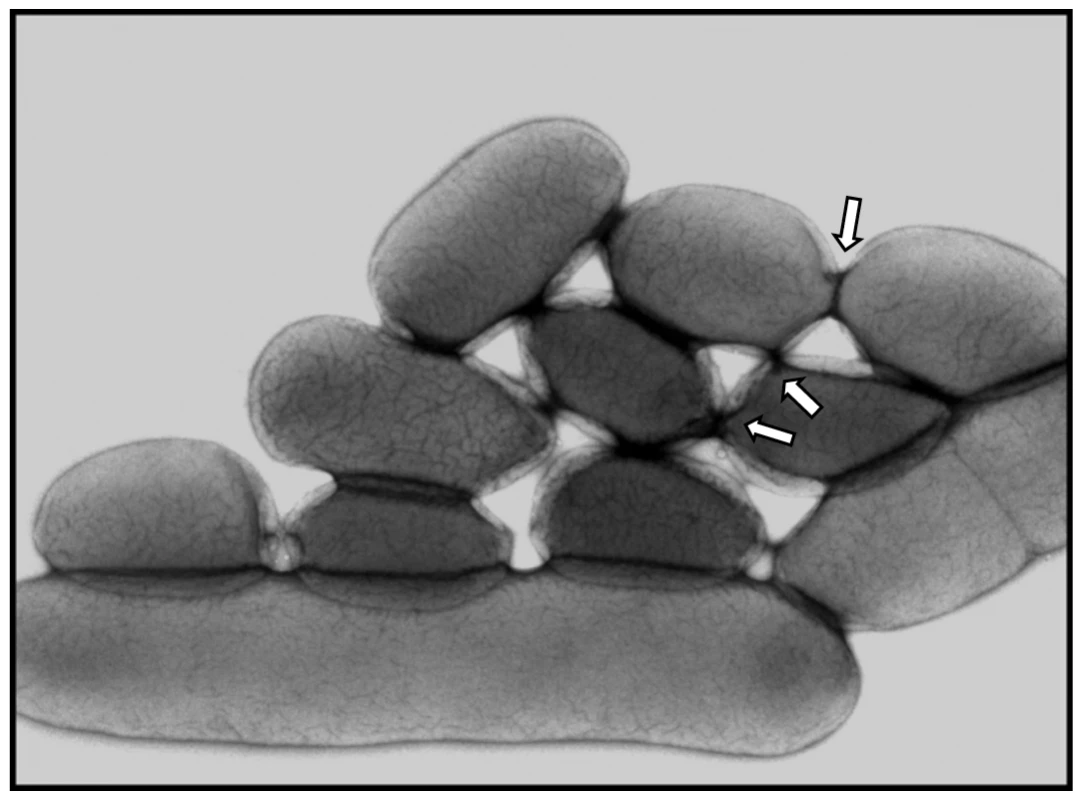
White arrows point at bridge-like structures. IS–mediated DNA mobilization
We noted that after pUC4K transfer, the plasmid sizes of pUC4K in 10 different transconjugants were variable. This was confirmed after digestion of the 10 plasmids with NdeI, an enzyme that has a single restriction site in pUC4K. Three plasmids (rpUC4K-1 to -3) had the expected pUC4K size, while the seven others (rpUC4K-4 to -10) had a size superior to that of the original molecule (data not shown), indicating that various types of rearrangements had occurred during plasmid transfer. Remarkably, a search for the potential transposition of pGDT4-borne IS (ISYps1, ISL3, ISYps2, ISNCY or ISYps3) on rpUC4K molecules by PCR (primers described in Table S1) showed that all seven larger size recombinant plasmids (rpUC4K-4 to -10) harbored ISYps1.
rpUC4K-5 to -9 had a size compatible with the acquisition of a single ISYps1 copy. Digestion with XhoI, an enzyme that cuts once in pUC4K and once in ISYps1, yielded two restriction fragments, thus confirming the presence of a single ISYps1 copy. However, two distinct restriction profiles were observed, one for rpUC4K-5 and -8 and one for rpUC4K-6, -7 and -9 (data not shown), indicating the occurrence of different genetic rearrangements. Sequencing of the regions encompassing the ISYps1 insertion site in one recombinant plasmid of each group (rpUC4K-5 and rpUC4K-6) demonstrated that the IS was inserted at two different sites, approximately 100 bp apart, and in opposite orientation (Figure 5a and 5b). The insertion generated an 8 bp duplication of the target sequence: AAAATAGG in rpUC4K-5 and TATTTGAA in rpUC4K-6.
Fig. 5. Schematic representation of seven recombinant pUC4K molecules recovered from transconjugants. 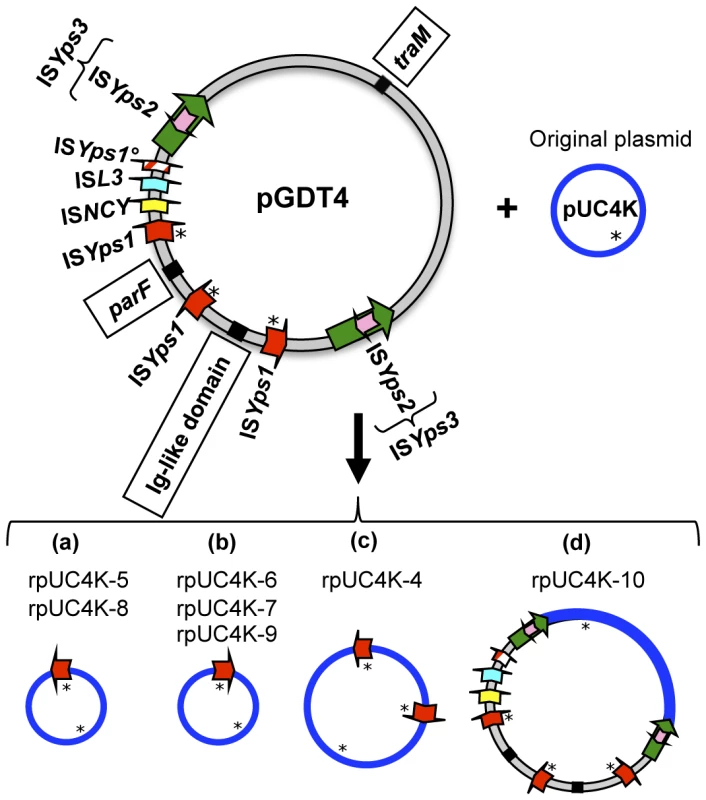
Ten transconjugants resulting from the co incubation of 637(pUC4K) and 637-NalR were analyzed. The donor plasmid pGDT4 harbors two copies of ISYps1 in direct orientation and one copy in opposite direction, two copies of ISYps3 transposon including ISYps3 and ISYps2 transposases, one ISL3 and one ISNCY. ISYps1° is a truncated copy of ISYps1. Stars indicate XhoI restriction sites. Genes used to search by PCR for the presence of inter-ISYps1 regions are boxed. rpUC4K-4 had a size superior to that of the above five plasmids. XhoI digestion revealed the presence of two ISYps1 copies on this plasmid (Figure 5c). To determine whether the region located between these two ISYps1 copies on rpUC4K-4 corresponded to a portion of pGDT4, a PCR amplification of three pGDT4 genes (the Ig-like domain, parF and traM), each located between two different ISYps1 copies on pGDT4 (Figure 5) was performed. No positive signal was detected, suggesting that the region located between the two ISYps1 is a duplicated portion of pUC4K (Figure 5c).
The rpUC4K-10 plasmid was different from all others since the PCR analysis showed that it carries all five pGDT4-borne IS, as well as the Ig-like domain and parF genes (but not traM, Figure 5d). rpUC4K-10 has thus most likely acquired the entire pGDT4 sequence located between the two ISYps3 transposons (Figure 5).
Altogether, these results show that most pUC4K transfers generated a variety of genetic modifications that were systematically accompanied by the transposition of the pGDT4-borne ISYps1 element.
Replicative transposition of ISYps1 and formation of cointegrates
ISYps1 belongs to the IS6 family, known to transpose through replicative transposition. This mode of transposition gives rise exclusively to replicon fusions (cointegrates), in which the donor and target replicons are separated by two IS copies in direct orientation. The cointegrate can be subsequently resolved by recombination between the two IS copies [17]. To determine whether ISYps1 transposes through this mechanism, this IS was cloned into the suicide mobilizable vector pSW23T and introduced into a replication-permissive E. coli strain, yielding ω7249(pSWYps1.1) (Table 1). After mating of this donor strain with ω4826, a non-replication permissive recA- recipient, ω4826::pSWYps1.1 transconjugants resulting from pSWYps1.1 integration into the recipient chromosome were obtained with a frequency of 8.5(±0.4)×10−6 (which corresponds to the frequencies of both conjugation and transposition, Table S5). Since the frequency of conjugation under these conditions was found to be 3.4(±0.9)×10−3 (Table S5), the transposition frequency of ISYps1 is thus approximately 2×10−3. The genomic DNA of eight independent ω4826::pSWYps1.1 colonies were digested with HindIII (which cuts once in pSW23T and not in ISYps1), and hybridized with an ISYps1 probe. All eight clones harbored, as expected, two integrated copies of ISYps1 (Figure S2). Of note, all clones exhibited different hybridization profiles.
To further determine whether association between ISYps1 and a conjugative plasmid allows cointegrate transfer, the IS was cloned on a non-mobilizable vector (pSW23) and introduced into an E. coli strain carrying the conjugative plasmid R388 (Table 1). After mating of the resulting pi3(R388, pSWYps1.2) donor strain with the ω4826 recipient that cannot sustain pSWYps1.2 replication, ω4826(R388::pSWYps1.2) transconjugants were independently selected on Cm (pSWYps1.2 tag) and Tmp (R388 tag) MH agar plates. CmR clones were found at a frequency of 9(±3)×10−5 (Table S5) and were all TmpR, while in the absence of an ISYps1 carried on the pSW23, no CmR transconjugants were obtained (Table S5). This further demonstrates that Yps1 drives the formation of cointegrates that can be subsequently transferred by conjugation. Under these conditions, R388 was transferred at a frequency of 2.2(±0.7)×10−1 (Table S5), indicating that transfer of these cointegrates occurs at high frequencies (≈2×10−5). To further characterize these events, the plasmid profiles of five independent R388::pSWYps1.2 cointegrates were analyzed after restriction with MfeI (10 sites in R388, one in pSWYps1.2). All five pSWYps1.2 insertions were in different locations on R388 (data not shown). The two MfeI junction fragments from one of these R388::pSWYps1.2 cointegrates were cloned into the EcoRI site of pUC18 and the precise cointegrate location was determined by sequencing. Transposition of pSWYps1.2 occurred in the orf5 cassette of the R388 integron [18] and led, as for the two pUC4K insertion events analyzed above, to an 8 bp duplication. The duplicated sequence (GATCCGAG) was different from the other two, further indicating the absence of a specific integration site.
Our results thus demonstrate that ISYps1 is able to transpose into a variety of insertion sites by replicative transposition through cointegrate formation, mediating the transfer of potentially any piece of non-mobilizable DNA molecule.
Discussion
We have evidenced a mechanism of HGT that convey the conjugative transfer or virtually any piece of chromosomal or plasmid DNA in a natural isolate of Y. pseudotuberculosis. This mechanism shares some characteristics with those previously described, but has several novel and unique properties.
GDT4 is not observed at temperatures ≥20°C and its efficiency increases as the temperature decreases. Although some conjugative plasmids have been previously shown to be self-transferable at 14°C but not at 37°C [19], [20], to our knowledge no plasmid able to conjugate at 4°C has ever been described. Temperature-dependent plasmid transfer is primarily mediated by H-NS and Hha proteins, which can be both plasmid and/or chromosome encoded [20]. The pGDT4 sequence did not reveal any gene encoding such proteins, but it is known that chromosomally encoded Hha and YmoA (equivalent to H–NS) act as thermoregulators in Y. enterocolitica [21]. These proteins (also encoded by the Y. pseudotuberculosis genome [8]) may thus modulate pGDT4 transfer at cold temperatures. Interestingly, H-NS is an integral part of bacterial stress response pathways and its function is known to be sensitive to changes in environmental conditions such as temperature [22], [23]. A cold stress could thus be a signal for the bacteria to transfer their genetic material by GDT4. The low temperature and an iron-poor liquid environment may also induce changes in the bacterial membrane structure, as observed for the closely related organism Y. pestis, in which the transcription patterns of various genes encoding components of the bacterial membrane were modified during iron starvation [24] or growth at 10°C [25]. These modifications might facilitate the formation of pores through which chromosomal DNA could translocate. Indeed, large cell aggregates in which bacteria appeared to be connected by bridges were observed. Similar tight bacterial contacts, designated conjugative junctions [26] or conjugational junctions [27] have been observed during RP4 or F-mediated mating of E. coli, respectively. However, these physical properties do not seem to be pGDT4-mediated, as we also observed large bacterial aggregates and possibly intercellular channels with IP32953, a strain that does not harbor this plasmid (data not shown). These bacterial aggregates have some similarities with biofilms in which bacteria are also closely connected. Biofilm formation occurs under natural conditions in a variety of bacterial species [28], including Y. pseudotuberculosis [29]. It could thus be hypothesized that GDT4 may take place between bacteria residing within biofilms in their natural ecological niches. This observation also suggests that acquisition of new functions, including virulence factors, by Y. pseudotuberculosis takes place in the environment rather than in a mammalian host.
Another characteristic feature of GDT4 is that transfer of chromosomal DNA occurs only in a liquid medium. Like other conjugative processes, GDT4 requires a pilus-like mating system and a mating channel to occur, as demonstrated by the fact that inactivation of the pGDT4-borne pilus apparatus abolished this mechanism. While some plasmids transfer better on plates, some others encoding long flexible pili allow DNA transfer efficiencies of the same magnitude in liquid and on solid media [30], [31], and this applied to pGDT4. In contrast, the absence of transfer of chromosomal DNA on agar was unexpected. Also unexpected was the fact that the efficiency of transfer in broth was not affected by a strong agitation, as opposed to recent findings showing that a vigorous shaking negatively affected the transfer of several conjugative plasmids, including the F' plasmid that encodes long and flexible pili [32]. Actually, growth in a liquid medium at low temperature could create the conditions optimal for GDT4. Indeed, this environment might be more favorable for the formation of tight bacterial aggregates and inter-cellular bridges through which long stretches of chromosomal DNA could transit.
pGDT4 also triggered the conjugative transfer of the non-mobilizable plasmid pUC4K. Similarly, transfer of the non mobilizable plasmid pBR325 by an RP4::miniMu mobilizing plasmid was previously observed [33], but the mechanism underlying this genetic transfer was not characterized. cis-mobilization of non-mobilizable plasmid DNA can occur after integration of a conjugative plasmid into the genetic element to be transferred. Integration arises either by homologous recombination between identical elements, often two copies of the same IS located on each DNA molecule (as for the Hfr formation in E. coli [34]), or through the formation of cointegrates mediated by specific transposons or ISs [35]. Integration of pGDT4 into pUC4K could not occur via homologous recombination as the two replicons do not share any common IS or identical DNA sequences. However, pGDT4 carries ISYps1, an IS which is predicted to belong to the IS6 family (http://www-is.biotoul.fr/is.html). ISYps1 is the second IS of the IS6-type identified in the genus Yersinia [36]. Members of this family have the capacity to create cointegrates by replicon fusion in the absence of a homologous IS on the target DNA [37]. Furthermore Tn3, which also moves by replicative transposition, has been found to mediate cis-mobilization of non mobilizable plasmids by this mechanism [35]. The fact that several rpUC4K plasmids obtained after pGDT4-mediated transfer carried a copy of ISYps1 argues for a role of this IS in pGDT4 integration into its target. We have demonstrated that ISYps1 is indeed transposing through replicative transposition. ISYps1 has a low specificity of recognition of the target sequence, as attested by our observation that the three insertion sites sequenced (two on pUC4K and one on R388) were different. This model also predicts the formation of cointegrates carrying two copies of the IS element, each flanking the sequence of the donor plasmid. We do have observed the formation of cointegrates between R388 and pSWYps1 flanked by the expected ISYps1 copies. A resolution step, which occurs through homologous recombination between the two IS copies, is then required to separate the donor and target replicons, leaving a single IS copy in the target and restoring the donor plasmid. The rpUC4K-5 to -9 molecules that were found to carry one ISYps1 copy are most likely the results of such a resolution event. The presence on one recombinant plasmid (rpUC4K-10) of a portion of pGDT4 carrying ISYps1, ISYps2, ISYps3, ISNCY and ISL3 indicates that additional, complex rearrangements involving the Tn3-like transposon ISYps3 can also occur. Finally, the existence in some transconjugants of pUC4K plasmids with a size identical to that of the original molecule could be the result of the resolution of cointegrates containing pUC4K concatemers.
GDT4 thus represents a remarkable illustration and validation of the model of Tn3-mediated transmission of non conjugative plasmids proposed by Crisona et al. in the 1980's [35]. Following this model, the first step is the integration of pGDT4 into its target DNA by ISYps1-mediated replicon fusion during plasmid replication (Figure 6). As mentioned above, this generates a cointegrate which carries the two replicons separated on each side by an ISYps1 copy. This cointegrate then uses the conjugative machinery encoded by pGDT4 to promote its transfer to the recipient strain. The final step is the resolution of the cointegrate by homologous recombination between two ISYps1 copies or any other duplicated sequence present in the cointegrate. Since pGDT4 carries several ISYps1, the resolved molecules have different sizes and DNA composition.
Fig. 6. Model proposed for pUC4K mobilization based on the transposition mechanism of the IS6 family. 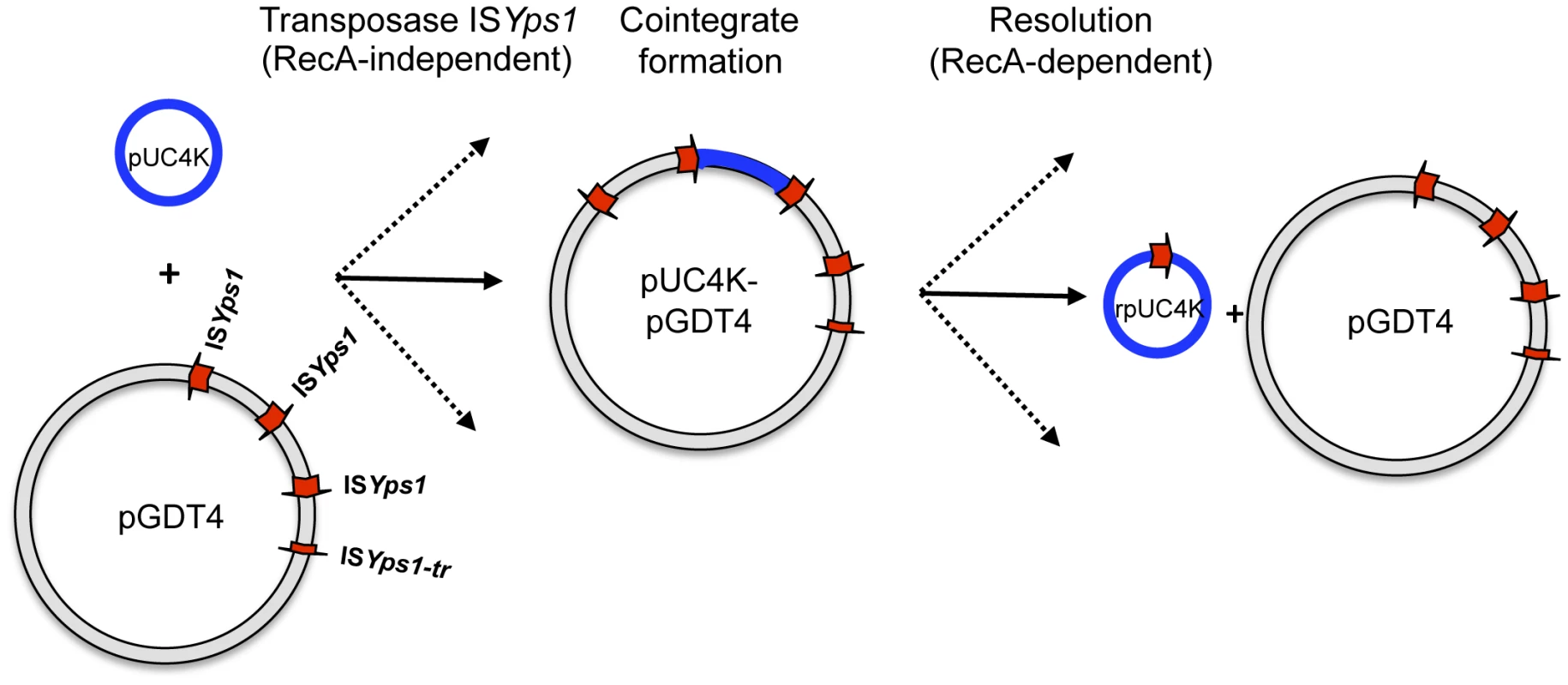
Transposase-mediated replicon fusion of the two plasmid molecules generates a cointegrate carrying an additional copy of ISYps1 in the same orientation. Although only one type of cointegrate is represented here, different types of cointegrates mediated by each ISYps1 copy can be generated (indicated by dashed arrows). RecA-dependent homologous recombination between any two copies of ISYps1 present on the cointegrate will either regenerate the donor plasmid, leaving a single IS copy in the target pUC4K or create a rpUC4K containing a portion of pGDT4. Figure adapted from Mahillon J. and Chandler M. [17]. GDT4 is also able to mediate the translocation of chromosomal DNA, most likely by integration into the bacterial chromosome and transfer in a Hfr-like manner. The Hfr mechanism is one of the earliest and best described examples of chromosomal transfer and is mediated by the F plasmid of E. coli [38]–[40]. F integrates stably into the E. coli chromosome through homologous recombination between IS copies present on both the F plasmid and the bacterial chromosome [38]–[42] to create Hfr strains, with transfer origins located at different chromosomal loci [43], [44]. In contrast to the classical Hfr mechanism, integration of pGDT4 into the chromosome probably occurs, as in pUC4K, via the ISYps1-mediated replication fusion mechanism. At least three pieces of evidence support this hypothesis: (i) no IS element is shared by pGDT4 and the IP32637 chromosome, in contrast to what is expected for the Hfr mechanism, (ii) in Y. pseudotuberculosis, three distantly located chromosomal loci (irp2K, ureBS and or5076T) were transferred with similar frequencies, and (iii) in all eight E. coli transconjugants analyzed, pSWYps1 was inserted at different sites on the chromosome. ISYps1 thus appears to have a very low specificity of recognition, allowing its insertion at multiple sites on bacterial plasmids and chromosomes. After mobilization of the chromosomal fragment adjacent to the pGDT4 integration site and transfer to a recipient strain, following the Hfr-type transfer model, homologous recombination between the incoming DNA and the chromosome is expected to take place, leaving no trace of pGDT4 in the chromosome of the transconjugant. Our previous observation that RecA activity is necessary in the recipient, but not in the donor strain for chromosomal transfer [3], and the results of the present study showing that pGDT4 is absent from some transconjugants that have acquired chromosomal genes further support this model of horizontal transfer. Our study thus validates the model proposed by Willets et al. in the 1980's for the mobilization of the E. coli chromosome via the formation of a cointegrate with the R68.45 plasmid during IS21 transposition [45]. Such cointegrate formations were widely used at that time to establish the genetic map of various bacterial species (see for instance [46], [47]). Most importantly our results show, without the need for heterologous plasmids like RP4 or R68.45, that this type of chromosomal conjugative transfer may occur under natural conditions in wild type bacterial pathogens carrying endogenous plasmids.
The capacity of wild type bacteria to naturally transfer large pieces of chromosomal DNA following the typical Hfr mechanism of homologous recombination between identical IS copies on the chromosome and the plasmids has been documented in a variety of bacteria, including extremophiles [48], Gram-positive cocci [49], and actinomycetes [50]. What we describe here is certainly a less constrained and more powerful mechanism, as it only requires the presence of an IS of the IS6 family on a conjugative replicon to generate cointegrates able to drive the horizontal transfer of any piece of DNA (chromosomal or episomal). It is remarkable that a high density of IS is commonly observed on plasmids. For instance the Shigella plasmid pWR100 carries 93 copies of complete or truncated IS belonging to 21 different types [51]. Thus, more than being IS depository, this location may reflect the broad selective advantage brought by plasmid/IS associations as a chromosomal transfer device. Such a ‘genetic symbiosis’, offers a means for the natural transfer of large blocks of genes conferring new metabolic properties or virulence functions. According to our model, GDT4 does not leave any signature in the recipient genome in most instances, and therefore its contribution to the numerous horizontal gene exchanges that shape bacterial genomes can hardly be quantified. However, according to the ISfinder database, approximately 5% of the known IS belong to the IS6 and Tn3 families, which use a replicative transposition mechanism. As they are found in all bacterial and archaeal phyla, the mechanism we describe here might be responsible for a substantial fraction of gene exchanges occurring among bacterial species.
Remarkably, this mechanism of DNA transfer was optimal when the bacteria were grown under conditions (low temperatures, iron poor medium, biofilm-like bacterial aggregates) that might be close to those met by these microorganisms in their normal ecological niches. This natural GDT mechanism may thus play a significant role in bacterial evolution, genetic polymorphism, pathogenesis and adaptation to new environmental conditions.
Methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table 1 and Table S1. Wild type strains were taken from the collection of the Yersinia Research Unit (Institut Pasteur). Bacteria were grown in LB (Luria Bertani) or MH (Mueller Hinton) medium for 24 h at 28°C (Yersinia) or 37°C (E. coli) with agitation, or for 48 h on LB or MH agar plates. When necessary, kanamycin (Kan: 100 µg ml−1), rifampicin (Rif: 100 µg ml−1), nalidixic acid (Nal: 25 µg ml−1), spectinomycin (Spe: 50 µg ml−1), tetracycline (Tc: 15 µg ml−1), chloramphenicol (Cm: 25 µg ml−1), trimethoprim (Tmp: 20 µg ml−1), thymidine (dT: 0.3 mM) or the iron chelator αα'-dipyridyl (0.2 mM, Sigma) were added to the medium.
Mutagenesis of chromosomal or plasmid genes
Spe (aadA) or Tmp (dfr) non-polar cassettes were PCR-amplified using primers described in Table S1, and pSW25 [52] or pGP704N-dfr [3] as templates, respectively. All allelic exchanges of chromosomal or plasmid genes by an antibiotic resistance cassette were done following the LFHR-PCR procedure [53]. The Spe and Tmp cassettes were introduced into the chromosomal ureB and or5076 genes, respectively, using primers that amplify upstream and downstream fragments of ureB and or5076, as shown on Figure S1 and Table S1. To label pGDT4, the plasmid was digested with HindIII and a 4 kb fragment (pGDT4-4kb) was purified and cloned into pUC18. Approximately 600 bp of each extremity of the cloned fragment were sequenced. These sequences were then used to design primers (358A/B and 359A/B, Table S1) that served for allelic exchange between the Tmp cassette and the target region of pGDT4 in strain 637-irp2K-ureBS. Correct insertion of the Tmp cassette was confirmed by PCR using primer pair 358A/359B. Mutagenesis of the pil region was done by replacing the pGDT4 region extending from pilL (pGDT4_0086) to pilV (pGDT4_0097) by a Tmp cassette, using primer pairs 773A/B and 774A/B (Table S1). The various antibiotic-tagged derivatives cured of pKOBEG-sacB were selected on sucrose plates.
Growth conditions for DNA transfer and determination of the transfer frequency
Optimal conditions for chromosomal DNA transfer in Y. pseudotuberculosis have been previously described [3]. Briefly, the donor strain (usually RifR) harboring chromosomal loci labeled with antibiotic cassettes and the recipient strain (usually NalR) were grown overnight in LB at 28°C with agitation. Equal amounts (5×106) of donor and recipient cells were mixed in 25 ml of LB-αα' and grown at 4°C with mild rotary agitation (80 rpm) for 4 days. Donor and recipient bacteria were quantified on Rif and Nal plates, respectively, and transconjugants were selected on Nal plates containing the appropriate antibiotic. To ensure that the colonies were not spontaneous NalR mutants of the RifR recipient strain, the Rif susceptibility of the transconjugants was systematically checked. For every single DNA transfer experiment, 10 to 20 transconjugant colonies were analyzed by PCR for the acquisition of the corresponding antibiotic-tagged locus with primer pairs 233B/166, 92A/322B and 348B/346A (Table S1) as indicated on Figure S1. When the transfer of the irp2K locus was analyzed, the acquisition of the entire HPI by the recipient strain was further checked with primer pairs A10/144A and A9/143B (Figure S1 and Table S1). The frequency of DNA transfer was determined as the number of NalR (or KanR, SpeR, TmpR) RifS transconjugants per RifR donor cells. To determine whether free DNA molecules in the medium could mediate GDT4, the donor bacteria 637-irp2K-ureBS and the recipient 637ΔHPI-NalR were co-incubated in the presence of 100 U/ml of DNAse in the culture medium. The activity of the DNAse under these conditions was checked by adding 1 ug/ml of bacterial DNA to the culture medium and by observing that the added DNA was degraded.
Transfer of pGDT4T
Transfer of pGDT4T was studied after incubation of the donor (637-irp2K-ureBS(pGDT4T)) and various NalR recipient cells for four days at 4, 28 or 37°C in liquid or solid media. On solid medium, 2×108 donor and recipient cells were mixed on a 0.45 µm nitrocellulose filter (Millipore) and at the end of the incubation period, the bacterial mixture was suspended in 1 ml of MH. Donor and recipient cells were quantified on MH-Rif and MH-Nal plates, respectively. Transconjugants having acquired pGDT4T were identified as NalR/TmpR/RifS colonies. In each transfer experiment, 10 transconjugants were analyzed by PCR for the presence of pGDT4T with primer pair 358A/346B (Table S1). Finally, the pGDT4T transfer frequency was calculated as the number of NalR/TmpR/RifS transconjugants per donor cells.
Sequencing of pGDT4
One transconjugant resulting from the co-incubation of the 637-irp2K-ureBS donor strain with the 637c-NalR recipient was used to obtain a plasmid extract which contained only pGDT4. Sequencing was performed using the whole genome shotgun strategy [54]. A 2–3 kb insert library was generated by random mechanical shearing of pGDT4 DNA and cloning into pcDNA-2.1 (Invitrogen). Recombinant plasmids were used as templates for cycle sequencing reactions consisting in 35 cycles (96°C for 30 s; 50°C for 15 s; 60°C for 4 min) in a thermocycler, using the Big dye terminator kit (V3.1, Applied Biosystems). Samples were precipitated and loaded onto a 96-lane capillary automatic 3700 DNA sequencer (Applied Biosystems). In an initial step, 1000 sequences from the library were assembled into 5 contigs using the Phred/Phrap/Consed software [55], [56] (8-fold sequence coverage). Consed was used to predict links between contigs. PCR products amplified from the pGDT4 template were used to fill gaps and to re-sequence low quality regions using primers designed by Consed. Physical gaps were closed using combinatorial PCR. The correctness of the assembly was confirmed by ensuring that the deduced restriction map was identical to the one obtained experimentally. The traX and traY genes fusion into a single traXY gene was checked by re-sequencing this locus on the original pGDT4 DNA preparation.
ISYps1, ISYps2 and ISYps3 designation were attributed by the ISfinder database (http://www-is.biotoul.fr/). The nucleotide sequence of pGDT4 has been submitted to the EMBL database under accession number FM178282. Details and properties of the different ISYps characterized in this work are accessible through the ISFinder web site.
Electron microscopy
Bacteria were negatively stained with 2% uranyl acetate onto glow discharged copper grids. The samples were observed in a Jeol 1200EXII and/or a JEM 1010 (Jeol) equipped with a Keenview camera (Eloise) at 80-kV accelerating voltage. Images were recorded with an Analysis Pro Software version 3.1 (Eloise).
ISYps1 transposition assay
The sequence corresponding to the ISYps1 copy carried on rpUC4K-6, flanked by its 113 bp upstream and 138 bp downstream regions, was amplified using primers 1039/1040 and cloned as an EcoRI-BamHI insert into the suicide mobilizable vector pSW23T [52], giving rise to pSWYps1.1. This plasmid was then introduced into E. coli ω7249 [57], a strain allowing pSW23T replication and conjugative transfer. Conjugation between this donor strain and E. coli ω4826 was performed as previously described [57], The frequency of conjugation–transposition frequency was calculated as the number of CmR transconjugants (ω4826::pSWYps1.1) per total number of recipients (TcR). The conjugation frequency was established in parallel by conjugation from the same donor ω7249(pSWYps1.1) to a ω4826 pir+ derivative (obtained through transformation with plasmid pSU38Δpir which expresses pir [52]). The frequency of illegitimate recombination of the pSW23T which can lead to CmR transconjugants was established by conjugation between donor ω7249(pSW23T) and ω4826, and found to be 4.6(±1.7)×10−8. Genomic DNA from 8 independent ω4826::pSWYps1.1 colonies were extracted using QIAGEN Genomic Tips and buffer set, digested with HindIII, and hybridized with a probe internal to ISYps1 (generated by PCR amplification with primers 1041/1044 and labeled with α-32P dCTP, using the Random Primed labeling kit (Roche)).
R388::pSWYps1.2 cointegrate formation assay
The EcoRI-BamHI fragment carrying ISYps1 was transferred from pSWYps1.1 to the non-mobilizable version of pSW23 [52], giving rise to pSWYps1.2. This plasmid was then introduced into the E. coli pi3 pir+ strain that harbors the IncW conjugative plasmid R388, which does not carry any IS (GenBank BR000038), giving rise to pi3(R388, pSWYps1.2). Conjugation of this donor strain with ω4826 yielded ω4826(R388::pSWYps1.2). The frequency of cointegrate formation after mating was calculated as the number of CmR transconjugants per total number of recipients harboring R388 (TmpR). The ability of R388 to form transferable cointegrates with pSW23 in the absence of ISYps1 was assessed in the same conditions by replacing pSWYps1.2 by pSW23 in the pi3(R388) donor, and found to be inferior to 10−9.
Supporting Information
Zdroje
1. CarnielEGuilvoutIPrenticeM 1996 Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol 178 6743 6751
2. HeesemannJHantkeKVockeTSakenERakinA 1993 Virulence of Yersinia enterocolitica Is Closely Associated with Siderophore Production, Expression of an Iron-Repressible Outer Membrane Polypeptide of 65000 Da and Pesticin Sensitivity. Mol Microbiol 8 397 408
3. LesicBCarnielE 2005 Horizontal transfer of the high-pathogenicity island of Yersinia pseudotuberculosis. J Bacteriol 187 3352 3358
4. LesicBCarnielE 2004 The High-Pathogenicity Island: a broad-host-range pathogenicity island. CarnielEHinnebuschJ Yersinia: Molecular and Cellular Biology: Horizon Bioscience 285 306
5. LesicBBachSGhigoJMDobrindtUHackerJ 2004 Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol Microbiol 52 1337 1348
6. SchubertSDarluPClermontOWieserAMagistroG 2009 Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog 5 e1000257 doi:10.1371/journal.ppat.1000257
7. MansonJMHancockLEGilmoreMS 2010 Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci USA 107 12269 12274
8. ChainPSCarnielELarimerFWLamerdinJStoutlandPO 2004 Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 101 13826 13831
9. SimonetMMazighDBercheP 1984 Growth of Yersinia pseudotuberculosis in mouse spleen despite loss of a virulence plasmid of mol. wt 47×106. J Med Microbiol 18 371 375
10. AchtmanMZurthKMorelliCTorreaGGuiyouleA 1999 Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 96 14043 14048
11. HurstMRBecherSAO'CallaghanM 2011 Nucleotide sequence of the Serratia entomophila plasmid pADAP and the Serratia proteamaculans pU143 plasmid virulence associated region. Plasmid 65 32 41
12. SmillieCGarcillan-BarciaMPFranciaMVRochaEPde la CruzF 2010 Mobility of plasmids. Microbiol Mol Biol Rev 74 434 452
13. FranciaMVVarsakiAGarcillan-BarciaMPLatorreADrainasC 2004 A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev 28 79 100
14. Garcillan-BarciaMPFranciaMVde la CruzF 2009 The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33 657 687
15. SnellingsNJPopekMLindlerLE 2001 Complete DNA sequence of Yersinia enterocolitica serotype 0∶8 low-calcium-response plasmid reveals a new virulence plasmid-associated replicon. Infect Immun 69 4627 4638
16. ChainPSCarnielELarimerFWLamerdinJStoutlandPO 2004 Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 101 13826 13831
17. MahillonJChandlerM 1998 Insertion sequences. Microbiol Mol Biol Rev 62 725 774
18. SundstromLRadstromPSwedbergGSkoldO 1988 Site-specific recombination promotes linkage between trimethoprim - and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet 213 191 201
19. MaherDTaylorDE 1993 Host range and transfer efficiency of incompatibility group HI plasmids. Can J Microbiol 39 581 587
20. FornsNBanosRCBalsalobreCJuarezAMadridC 2005 Temperature-dependent conjugative transfer of R27: role of chromosome - and plasmid-encoded Hha and H-NS proteins. J Bacteriol 187 3950 3959
21. CornelisGRSluitersCDelorIGeibDKanigaK 1991 ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol 5 1023 1034
22. OnoSGoldbergMDOlssonTEspositoDHintonJC 2005 H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem J 391 203 213
23. TendengCBertinPN 2003 H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol 11 511 518
24. ZhouDSQinLHanYPQiuJFChenZL 2006 Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol Lett 258 9 17
25. HanYPZhouDSPangXZhangLSongYJ 2005 DNA microarray analysis of the heat - and cold-shock stimulons in Yersinia pestis. Microbes Infect 7 335 348
26. SamuelsALLankaEDaviesJE 2000 Conjugative junctions in RP4-mediated mating of Escherichia coli. J Bacteriol 182 2709 2715
27. DurrenbergerMBVilligerWBachiT 1991 Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol 107 146 156
28. BrandlMT 2006 Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol 44 367 392
29. JoshuaGWKarlyshevAVSmithMPIsherwoodKETitballRW 2003 A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149 3221 3229
30. BradleyDE 1980 Morphological and serological relationships of conjugative pili. Plasmid 4 155 169
31. BradleyDETaylorDECohenDR 1980 Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol 143 1466 1470
32. ZhongXKrolJETopEMKroneSM 2010 Accounting for mating pair formation in plasmid population dynamics. J Theor Biol 262 711 719
33. TopEMergeayMSpringaelDVerstraeteW 1990 Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol 56 2471 2479
34. UmedaMOhtsuboE 1989 Mapping of insertion elements IS1, IS2 and IS3 on the Escherichia coli K-12 chromosome. Role of the insertion elements in formation of Hfrs and F' factors and in rearrangement of bacterial chromosomes. J Mol Biol 208 601 614
35. CrisonaNJNowakJANagaishiHClarkAJ 1980 Transposon-mediated conjugational transmission of nonconjugative plasmids. J Bacteriol 142 701 713
36. StrauchEHoffmannBHeinsGAppelB 2000 Isolation of a new insertion element of Yersinia intermedia closely related to remnants of mobile genetic elements present on Yersinia plasmids harboring the Yop virulon. FEMS Microbiol Lett 193 37 44
37. OhtsuboEZenilmanMOhtsuboH 1980 Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci USA 77 750 754
38. WollmanELJacobFHayesW 1956 Conjugation and genetic recombination in Escherichia coli K-12. Cold Spring Harb Symp Quant Biol 21 141 162
39. HayesW 1953 The mechanism of genetic recombination in Escherichia coli. Cold Spring Harb Symp Quant Biol 18 75 93
40. LederbergJTatumEL 1946 Gene recombination in Escherichia coli. Nature 158 558
41. JacobFWollmanE 1961 Sexuality and the Genetics of Bacteria New York Academic Press
42. CurtissR3rdStallionsDR 1969 Probability of F integration and frequency of stable Hfr donors in F+ populations of Escherichia coli K-12. Genetics 63 27 38
43. MatneyTSGoldschmidtEPErwinNSScroggsRA 1964 A preliminary map of genomic sites for F-attachment in Escherichia coli K12. Biochem Biophys Res Commun 17 278 281
44. CurtissRMacrinaFFalkinhamI 1974 Escherichia coli-An overview; KingRC New York Plenum Press 115 135
45. WillettsNSCrowtherCHollowayBW 1981 The insertion sequence IS21 of R68.45 and the molecular basis for mobilization of the bacterial chromosome. Plasmid 6 30 52
46. HaasDHollowayBW 1978 Chromosome mobilization by the R plasmid R68.45: a tool in Pseudomonas genetics. Mol Gen Genet 158 229 237
47. Van GijsegemFToussaintA 1982 Chromosome transfer and R-prime formation by an RP4::mini-Mu derivative in Escherichia coli, Salmonella typhimurium, Klebsiella pneumoniae, and Proteus mirabilis. Plasmid 7 30 44
48. Ramirez-ArcosSFernandez-HerreroLAMarinIBerenguerJ 1998 Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J Bacteriol 180 3137 3143
49. GassonMJGodonJJPillidgeCJEatonTJJuryK 1995 Characterization of conjugation in Lactococcus lactis. Int Dairy J 5 757 762
50. PettisGSCohenSN 1994 Transfer of the plJ101 plasmid in Streptomyces lividans requires a cis-acting function dispensable for chromosomal gene transfer. Mol Microbiol 13 955 964
51. BuchrieserCGlaserPRusniokCNedjariHD'HautevilleH 2000 The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 38 760 771
52. DemarreGGueroutA-MMatsumoto-MashimoCRowe-MagnusDAMarlièreDAMazelD 2005 A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156 245 255
53. DerbiseALesicBDacheuxDGhigoJMCarnielE 2003 A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol Immunol 38 113 116
54. FleischmannRDAdamsMDWhiteOClaytonRAKirknessEF 1995 Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269 496 512
55. GordonDAbajianCGreenP 1998 Consed: a graphical tool for sequence finishing. Genome Res 8 195 202
56. EwingBGreenP 1998 Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8 186 194
57. BabicAGueroutAMMazelD 2008 Construction of an improved RP4 (RK2)-based conjugative system. Res Microbiol 159 545 549
Štítky
Genetika Reprodukčná medicína
Článek Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1Článek Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / TranscriptionČlánek Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding DomainČlánek Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin ComplexesČlánek An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood ObesityČlánek Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAsČlánek Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
- Genomic Analysis of the Hydrocarbon-Producing, Cellulolytic, Endophytic Fungus
- Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders
- Akirin Links Twist-Regulated Transcription with the Brahma Chromatin Remodeling Complex during Embryogenesis
- Too Much Cleavage of Cyclin E Promotes Breast Tumorigenesis
- Imprinted Genes … and the Number Is?
- Genetic Architecture of Highly Complex Chemical Resistance Traits across Four Yeast Strains
- Exploring the Complexity of the HIV-1 Fitness Landscape
- MNS1 Is Essential for Spermiogenesis and Motile Ciliary Functions in Mice
- A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Pathogenicity Islands SPI-1 and SPI-2
- Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution
- Variation in Modifies Risk of Neonatal Intestinal Obstruction in Cystic Fibrosis
- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Critical Evaluation of Imprinted Gene Expression by RNA–Seq: A New Perspective
- A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans
- Mouse Genetics Suggests Cell-Context Dependency for Myc-Regulated Metabolic Enzymes during Tumorigenesis
- Transcriptional Control in Cardiac Progenitors: Tbx1 Interacts with the BAF Chromatin Remodeling Complex and Regulates
- Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators
- APOBEC3G-Induced Hypermutation of Human Immunodeficiency Virus Type-1 Is Typically a Discrete “All or Nothing” Phenomenon
- Interpreting Meta-Analyses of Genome-Wide Association Studies
- Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line
- -Dependent Chemosensory Functions Contribute to Courtship Behavior in
- Diverse Forms of Splicing Are Part of an Evolving Autoregulatory Circuit
- Phenotypic Plasticity of the Drosophila Transcriptome
- Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1
- Precocious Metamorphosis in the Juvenile Hormone–Deficient Mutant of the Silkworm,
- Igf1r Signaling Is Indispensable for Preimplantation Development and Is Activated via a Novel Function of E-cadherin
- Accurate Prediction of Inducible Transcription Factor Binding Intensities In Vivo
- Mitochondrial Oxidative Stress Alters a Pathway in Strongly Resembling That of Bile Acid Biosynthesis and Secretion in Vertebrates
- Mammalian Neurogenesis Requires Treacle-Plk1 for Precise Control of Spindle Orientation, Mitotic Progression, and Maintenance of Neural Progenitor Cells
- Tcf7 Is an Important Regulator of the Switch of Self-Renewal and Differentiation in a Multipotential Hematopoietic Cell Line
- REST–Mediated Recruitment of Polycomb Repressor Complexes in Mammalian Cells
- Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / Transcription
- Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages
- A Genome-Wide Association Study Identifies Variants Underlying the Shade Avoidance Response
- -by- Regulatory Divergence Causes the Asymmetric Lethal Effects of an Ancestral Hybrid Incompatibility Gene
- Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
- A Natural System of Chromosome Transfer in
- Cell Size and the Initiation of DNA Replication in Bacteria
- Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
- Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin Complexes
- Temporal Transcriptional Profiling of Somatic and Germ Cells Reveals Biased Lineage Priming of Sexual Fate in the Fetal Mouse Gonad
- Rapid Analysis of Genome Rearrangements by Multiplex Ligation–Dependent Probe Amplification
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- The Atypical Calpains: Evolutionary Analyses and Roles in Cellular Degeneration
- The Silkworm Coming of Age—Early
- Development of a Panel of Genome-Wide Ancestry Informative Markers to Study Admixture Throughout the Americas
- Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency
- The Min System and Nucleoid Occlusion Are Not Required for Identifying the Division Site in but Ensure Its Efficient Utilization
- Neurobeachin, a Regulator of Synaptic Protein Targeting, Is Associated with Body Fat Mass and Feeding Behavior in Mice and Body-Mass Index in Humans
- Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin
- Gene Reactivation by 5-Aza-2′-Deoxycytidine–Induced Demethylation Requires SRCAP–Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions
- The miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
- Genetic Basis of Hidden Phenotypic Variation Revealed by Increased Translational Readthrough in Yeast
- An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood Obesity
- Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
- Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals
- Polycomb-Like 3 Promotes Polycomb Repressive Complex 2 Binding to CpG Islands and Embryonic Stem Cell Self-Renewal
- Insulin/IGF-1 and Hypoxia Signaling Act in Concert to Regulate Iron Homeostasis in
- EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development
- Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
- Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in
- A Machine Learning Approach for Identifying Novel Cell Type–Specific Transcriptional Regulators of Myogenesis
- Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
- Nos2 Inactivation Promotes the Development of Medulloblastoma in Mice by Deregulation of Gap43–Dependent Granule Cell Precursor Migration
- Intracranial Aneurysm Risk Locus 5q23.2 Is Associated with Elevated Systolic Blood Pressure
- Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits
- A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35.3, 7p14.3, and 13q14.1
- DNA Damage in Nijmegen Breakage Syndrome Cells Leads to PARP Hyperactivation and Increased Oxidative Stress
- DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination
- Genetic Analysis of Floral Symmetry in Van Gogh's Sunflowers Reveals Independent Recruitment of Genes in the Asteraceae
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Promoter Nucleosome Organization Shapes the Evolution of Gene Expression
- The Nucleoside Diphosphate Kinase Gene Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance
- The Ciliogenic Transcription Factor RFX3 Regulates Early Midline Distribution of Guidepost Neurons Required for Corpus Callosum Development
- Phosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
- A Genome-Wide Scan of Ashkenazi Jewish Crohn's Disease Suggests Novel Susceptibility Loci
- Parkinson's Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria
- LMW-E/CDK2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated b-Raf-ERK1/2-mTOR Pathway in Breast Cancer Patients
- Mapping the Hsp90 Genetic Interaction Network in Reveals Environmental Contingency and Rewired Circuitry
- Autoregulation of the Noncoding RNA Gene
- The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of Pro-Inflammatory Cytokines
- Spo0A∼P Imposes a Temporal Gate for the Bimodal Expression of Competence in
- Antagonistic Regulation of Apoptosis and Differentiation by the Cut Transcription Factor Represents a Tumor-Suppressing Mechanism in
- A Downstream CpG Island Controls Transcript Initiation and Elongation and the Methylation State of the Imprinted Macro ncRNA Promoter
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy