-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse
Intragenomic conflicts arise when a genetic element favours its own transmission to the detriment of others. Conflicts over sex chromosome transmission are expected to have influenced genome structure, gene regulation, and speciation. In the mouse, the existence of an intragenomic conflict between X - and Y-linked multicopy genes has long been suggested but never demonstrated. The Y-encoded multicopy gene Sly has been shown to have a predominant role in the epigenetic repression of post meiotic sex chromatin (PMSC) and, as such, represses X and Y genes, among which are its X-linked homologs Slx and Slxl1. Here, we produced mice that are deficient for both Sly and Slx/Slxl1 and observed that Slx/Slxl1 has an opposite role to that of Sly, in that it stimulates XY gene expression in spermatids. Slx/Slxl1 deficiency rescues the sperm differentiation defects and near sterility caused by Sly deficiency and vice versa. Slx/Slxl1 deficiency also causes a sex ratio distortion towards the production of male offspring that is corrected by Sly deficiency. All in all, our data show that Slx/Slxl1 and Sly have antagonistic effects during sperm differentiation and are involved in a postmeiotic intragenomic conflict that causes segregation distortion and male sterility. This is undoubtedly what drove the massive gene amplification on the mouse X and Y chromosomes. It may also be at the basis of cases of F1 male hybrid sterility where the balance between Slx/Slxl1 and Sly copy number, and therefore expression, is disrupted. To the best of our knowledge, our work is the first demonstration of a competition occurring between X and Y related genes in mammals. It also provides a biological basis for the concept that intragenomic conflict is an important evolutionary force which impacts on gene expression, genome structure, and speciation.
Published in the journal: A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse. PLoS Genet 8(9): e32767. doi:10.1371/journal.pgen.1002900
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002900Summary
Intragenomic conflicts arise when a genetic element favours its own transmission to the detriment of others. Conflicts over sex chromosome transmission are expected to have influenced genome structure, gene regulation, and speciation. In the mouse, the existence of an intragenomic conflict between X - and Y-linked multicopy genes has long been suggested but never demonstrated. The Y-encoded multicopy gene Sly has been shown to have a predominant role in the epigenetic repression of post meiotic sex chromatin (PMSC) and, as such, represses X and Y genes, among which are its X-linked homologs Slx and Slxl1. Here, we produced mice that are deficient for both Sly and Slx/Slxl1 and observed that Slx/Slxl1 has an opposite role to that of Sly, in that it stimulates XY gene expression in spermatids. Slx/Slxl1 deficiency rescues the sperm differentiation defects and near sterility caused by Sly deficiency and vice versa. Slx/Slxl1 deficiency also causes a sex ratio distortion towards the production of male offspring that is corrected by Sly deficiency. All in all, our data show that Slx/Slxl1 and Sly have antagonistic effects during sperm differentiation and are involved in a postmeiotic intragenomic conflict that causes segregation distortion and male sterility. This is undoubtedly what drove the massive gene amplification on the mouse X and Y chromosomes. It may also be at the basis of cases of F1 male hybrid sterility where the balance between Slx/Slxl1 and Sly copy number, and therefore expression, is disrupted. To the best of our knowledge, our work is the first demonstration of a competition occurring between X and Y related genes in mammals. It also provides a biological basis for the concept that intragenomic conflict is an important evolutionary force which impacts on gene expression, genome structure, and speciation.
Introduction
Transmission distorters (TDs), also known as segregation distorters or meiotic drivers, are genetic elements that are transmitted to the next generation with a higher frequency than the expected 1∶1 Mendelian inheritance ratio. TDs have the tendency to accumulate in low recombination regions where tight linkage allows cooperation between TDs and responder genes to evolve, as seen in the mouse t-complex [1] [for recent reviews see [2], [3]]. The non-recombining region of the heteromorphic sex chromosomes is the largest genomic example of recombination suppression [4], with the consequent potential for TDs to arise and distort the population sex ratio. Theory predicts that an unlinked suppressor of the sex ratio distortion (whether autosomal or on the other sex chromosome) would rapidly be selected for to restore the Fisherian 1∶1 sex ratio [5]. A subsequent evolutionary arms race between the distorter and its suppressor may follow and lead to repeated bouts of amplification of the genes involved in this intragenomic conflict [6]. In Drosophila, the X - and Y-encoded multicopy genes Stellate and Suppressor of Stellate are believed to illustrate the genomic conflict theory since deletions of Su(Ste) locus lead to a derepression of Stellate associated with a distorted sex ratio towards an excess of females; but to date it remains unclear whether or not Stellate is a transmission distorter [7], [8]. Intragenomic conflicts over sex chromosome transmission are predicted to have influenced genome structure, gene expression and speciation [2], [3]. Several cases of sex chromosome transmission distortion have been reported in the literature but they mostly concern Drosophila species [2], [9]–[14] and remain poorly characterized in mammals. Sex ratio segregation distortion may be more frequent than observed as the distortion is often masked by the presence of a suppressor in wild-type (WT) populations [2], [9]–[15].
In the mouse, the existence of an intragenomic conflict between X - and Y-linked genes has long been suggested: males with a partial deletion of the male specific region of the Y long arm (MSYq) produce offspring with a sex ratio skewed towards females [16], suggesting that MSYq encodes a factor(s) suppressing sex ratio distortion. MSYq consists of multicopy gene families, present in ∼60 to 100 copies [17]–[24], many of which possess X-linked multicopy homologous genes [20], [25], [26]. This has been considered a manifestation of a conflict between an X-encoded TD and a Y-encoded suppressor that remain to be identified [16], [20], [26], [27].
We have previously shown that the MSYq-encoded multicopy gene Sly (Sycp3-like Y-linked) represses the postmeiotic expression of X and Y genes [19]. Sly-deficient males – also known as shSLY males since they carry a short hairpin RNA-expressing transgene which triggers the specific degradation of Sly transcripts by RNA interference – present a remarkable up-regulation of sex chromosome genes in postmeiotic germ cells (spermatids) associated with a loss of repressive epigenetic marks, such as trimethylated histone H3 (H3K9me3) and CBX1 [19]. SLY therefore limits sex chromosome expression via the recruitment/maintenance of repressive epigenetic marks to post meiotic sex chromatin (PMSC) and has been proposed to associate with the sex chromosomes through its Cor1 domain – a domain thought to mediate chromatin interactions (Conserved Domain Database from the National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=147120).
Interestingly, Slx and Slxl1, two multicopy X-linked genes related to Sly [25] have been co-amplified with Sly during the evolution of the mouse genome [18], [20] and are among the genes that are up-regulated when Sly expression is reduced/absent [19]. Using a strategy of transgenically-delivered short hairpin RNA similar to the one previously used to disrupt the function of Sly, we have recently produced Slx/Slxl1-deficient mice (also known as shSLX mice). This study has shown that Slx/Slxl1 are indispensable for normal sperm differentiation, and that Slx/Slxl1 deficiency leads to the deregulation of a number of autosomal genes [28]. Moreover, both SLY and SLXL1 proteins have now been shown to interact with the acrosomal protein DKKL1 [21], [29].
In the present study we show that SLX/SLXL1 and SLY proteins have antagonistic effects on gene expression for both the sex chromosomal genes deregulated in shSLY and the set of autosomal genes deregulated in shSLX, and furthermore have antagonistic effects on offspring sex ratio. Our data demonstrate that Slx/Slxl1 and Sly are involved in a postmeiotic intragenomic conflict; we propose this phenomenon has had a strong impact on the structure and epigenetic regulation of the sex chromosomes, and may also have influenced the evolution of hybrid sterility in the mouse lineage.
Results
In Sly-deficient mice, SLX/SLXL1 proteins relocate to the nuclear sites vacated by SLY proteins
In normal males, SLX/SLXL1 proteins are located in the cytoplasmic compartment of spermatids [28], whereas SLY is additionally detected in the spermatid nucleus where it has been shown to colocalize with the X and Y chromosomes [19]. When performing immunofluorescence detection of SLX/SLXL1 proteins on spermatids devoid of SLY protein (i.e. on shSLY testicular sections), we observed an augmented SLX/SLXL1 signal in the cytoplasm compared to controls (WT) – confirming up-regulation at the protein level – and some signal in shSLY round spermatid nuclei that was not visible in WT (Figure 1A). The presence of SLX/SLXL1 proteins in shSLY spermatid nuclei was confirmed by Western blot analyses of nuclear fractions (Figure 1B). We then investigated in more detail the nuclear localization of SLX/SLXL1 in the context of Sly deficiency. The vast majority of shSLY spermatid nuclei showed a strong SLX/SLXL1 signal (280/369, 76%) (Figure 1C and Figure S1). This signal colocalized with the postmeiotic sex chromatin (PMSC, i.e. the X or the Y chromosome since spermatids are haploid) in 96.5% of round spermatids (82/85; 32/34 for X-bearing and 50/51 for Y-bearing spermatids). In comparison, 84% of WT round spermatid nuclei (265/316) did not have any SLX/SLXL1 signal. The nuclear SLX/SLXL1 signal observed in the remaining ∼16% of WT round spermatid was very weak when compared to the nuclear signal in shSLY round spermatids but appeared to colocalize with the PMSC in the majority of the cases (Figure S1).
Fig. 1. SLX/SLXL1 proteins behave similarly to SLY in its absence. 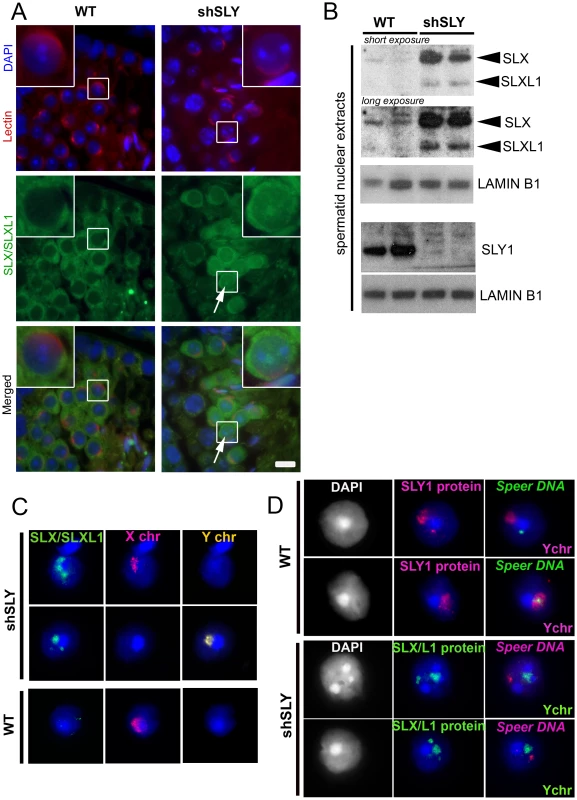
A) Immunofluorescence detection of SLX/SLXL1 protein (green) in wild-type (WT) and Sly-deficient (shSLY) testicular sections. DAPI (blue) was used to stain nuclei and lectin-PNA (red) was used to stain acrosomes in order to determine tubular stage. The inset represents a 3× magnification. Pictures were taken using the same image capture parameters. Scale bar indicates 10 µm. B) Western blot detection of SLX/SLXL1 proteins in nuclear extracts from shSLY and WT round spermatids. SLY1 antibody was used on the same extracts to confirm the absence of SLY protein in the shSLY nuclear fraction. Sly gene encodes two alternative splice variants (Sly1 and Sly2) which are predicted to be translated into a long and a short protein isoform (SLY1 and SLY2), but only SLY1 proteins have been detected so far and it remains unclear whether Sly2 transcripts are translated [21]. LAMIN-B1 detection was used as a loading control. C) Immunofluorescence detection of SLX/SLXL1 protein (green) in shSLY and WT round spermatid nuclei. DAPI (blue) was used to stain nuclei. X and Y chromosome painting were performed sequentially. A strong SLX/SLXL1 signal was observed in the majority of shSLY spermatid nuclei (76%). This signal colocalized with either sex chromosome in 96.5% of the cases. No signal could be detected in the majority of WT round spermatid nuclei (84%). D) Immunofluorescence detection of SLY1 (pink) or SLX/SLXL1 (green) protein in WT or shSLY round spermatids. Hybridization with a DNA probe detecting Speer gene cluster was subsequently performed, followed by Y chromosome painting. DAPI (white or blue) was used to stain nuclei. SLY1 protein colocalized with Speer gene cluster in 78.5% of WT spermatids while SLX/SLXL1 proteins colocalized with Speer gene cluster in 73% of shSLY spermatids. In addition to colocalizing with the PMSC, foci of SLX/SLXL1 proteins were observed outside the sex chromatin, reminiscent of the SLY signal present in the nucleus of WT spermatids [19]. We have since established that these ‘ectopic’ SLY sites include a ∼14 Mb cluster of 7 Speer genes on chromosome 5 that are up-regulated in shSLY spermatids. As a result, SLY immunofluorescence followed by fluorescent hybridization of a Speer DNA probe (DNA FISH) showed that, in the majority of WT round spermatids (107/136, 78.5%), SLY protein colocalized with the Speer DNA FISH signal (Figure 1D). We next looked at SLX/SLXL1 proteins in Sly-deficient round spermatids and observed that they colocalized with the Speer gene cluster in 73% of the cases (130/178) (Figure 1D). Thus, in the absence of SLY, SLX/SLXL1 proteins colocalize with the sex chromatin and with the autosomal Speer gene cluster, mimicking the pattern observed for SLY protein in WT spermatids.
Transgenic delivery of shSLX and shSLY short hairpin RNAs leads to a dramatic reduction in Slx/Slxl1 and Sly RNA and protein levels
We then wondered if the localization of SLX/SLXL1 proteins to the PMSC in the absence of SLY also affects postmeiotic sex chromosome gene expression. To address this question, we generated males that were deficient for SLX/SLXL1 and SLY proteins: we produced males carrying shSLY (Sly specific short hairpin RNA) transgene [19] together with one or two shSLX (Slx/Slxl1 specific short hairpin RNA) transgenes, shSLX1 and/or shSLX2 [28]. Firstly, we checked the efficiency of Slx/Slxl1 and Sly knockdowns in round spermatids from males carrying shSLX1 and shSLY transgenes (hereafter named shSLX1shSLY males). The reduction in Slx/Slxl1 transcript level was similar in shSLX1shSLY males and in shSLX1 siblings, while Sly knockdown was even stronger in shSLX1shSLY males compared to shSLY siblings (Figure 2A and 2B). Sly transcript quantification included both alternative splice variants (Sly1 and Sly2) [21] which were knocked-down with the same efficiency [19]. No SLY1 protein could be detected in shSLY or in shSLX1shSLY tissues (Figure 2D–2E). To date it remains unclear whether Sly2 transcripts are translated since anti-SLY1 antibody cannot detect SLY2 protein [21]. The discrepancy between transcript and protein levels is likely due to the presence of non-coding Sly transcripts, as previously observed [19]. Reduction in SLX and SLXL1 proteins was similar in shSLX1shSLY males and in shSLX1 siblings (Figure 2C). We also produced shSLX1/2shSLY males that carry the two shSLX transgenes along with the shSLY transgene. As expected, shSLX1/2shSLY males showed a very efficient knockdown of Slx and Slxl1 (Figure S2A); Sly knockdown in these males was similar to that in shSLX1shSLY males (Figure S2B). Thus, the combination of shSLX and shSLY transgenes gives an efficient knockdown of Slx/Slxl1 and Sly genes; the resulting transgenic males are therefore deficient for Slx/Slxl1 and Sly transcripts and proteins (hereafter named Slx/y-deficient males).
Fig. 2. The combination of shSLX and shSLY transgenes produces an efficient knockdown of Slx/Slxl1 and Sly genes. 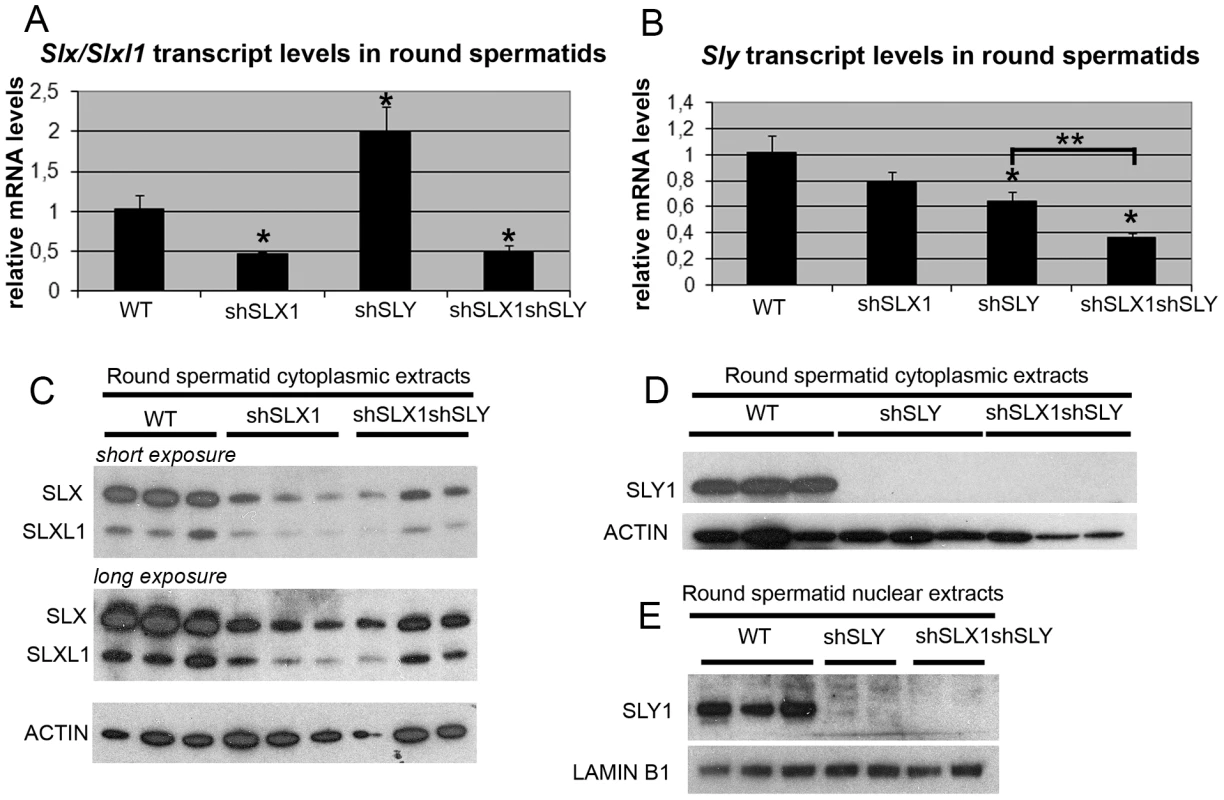
A–B) Real time PCR quantification of Slx/Slxl1 (Slx-all primers) (A) and Sly (Sly1 and Sly2 variants) (B) transcript levels in WT, shSLX1, shSLY and shSLX1shSLY round spermatids. The y-axis indicates the level of expression compared to WT after normalization with Acrv1 (2ΔΔCt ± standard errors). The reduction in Slx/Slxl1 transcript level was similar in shSLX1shSLY males and in shSLX1 siblings. As observed before [19], Slx/Slxl1 transcript level was found increased in shSLY males. One asterisk indicates significant difference from WT (p<0.05; t test on ΔΔCt values). Sly knockdown was even stronger in shSLX1shSLY males compared to shSLY siblings [two asterisks indicate significant difference between shSLX1shSLY and shSLY (p = 0.02; t test on ΔΔCt values)]. C–E) Western blot detection of SLY1, SLX and SLXL1 proteins in nuclear and cytoplasmic fractions from WT, shSLY, shSLX1 and shSLX1shSLY round spermatids. LAMIN B1 and ACTIN detection were used as loading controls for nuclear and cytoplasmic fractions, respectively. No SLY1 protein could be detected in shSLY or in shSLX1shSLY samples. In Sly-deficient spermatids, SLX/SLXL1 proteins increase sex chromosome gene expression associated with a reduction of H3K9me3 marks on PMSC
We then performed microarray transcriptome analyses on Slx/y-deficient purified round spermatids and compared these results to those obtained from Sly-deficient and from WT round spermatids (Figure 3 and Figure S3). The up-regulation of X - and Y-encoded spermatid transcripts was significantly less pronounced in Slx/y-deficient males than in Sly-deficient males (Figure 3A–3C). Specifically, 222 genes showed a greater than 1.5 fold-increase in Sly-deficient spermatids relative to WT, and 196 of them were corrected to some degree by the additional Slx/Slxl1 deficiency (i.e. in Slx/y-deficient spermatids). As a Y-encoded gene, Sly itself is affected by Slx/Slxl1 knockdown and thus is expressed at a lower level in Slx/y-deficient males than in Sly-deficient males (Figure 2B and Figure S3). The microarray findings were confirmed for several representative X and Y genes by real time PCR (Figure 3B and Figure S2C). These opposite effects of Sly and Slx/Slxl1 deficiency show that, in the absence of SLY protein, SLX/SLXL1 proteins localize to PMSC where they increase sex chromosome gene expression; when both SLX/SLXL1 and SLY proteins are reduced/absent in PMSC (in Slx/y-deficient males), the level of X - and Y - encoded transcripts is closer to the WT value. It is worth noting that while Slx/Slxl1 deficiency significantly reduces the up-regulation of XY genes induced by Sly deficiency, it does not bring expression all the way back down to WT levels. This may indicate that Slx/Slxl1 knockdown is not sufficient to fully compensate for the effect of Sly deficiency; alternatively it may be that in the WT MF1 laboratory strain, the combined effect of the presence of both SLX/SLXL1 and SLY is a net reduction of XY expression level, thus leading to a net increase when both genes are deficient.
Fig. 3. SLX/SLXL1 and SLY have opposite effects on gene expression and on the recruitment/maintenance of H3K9me3 on the sex chromatin. 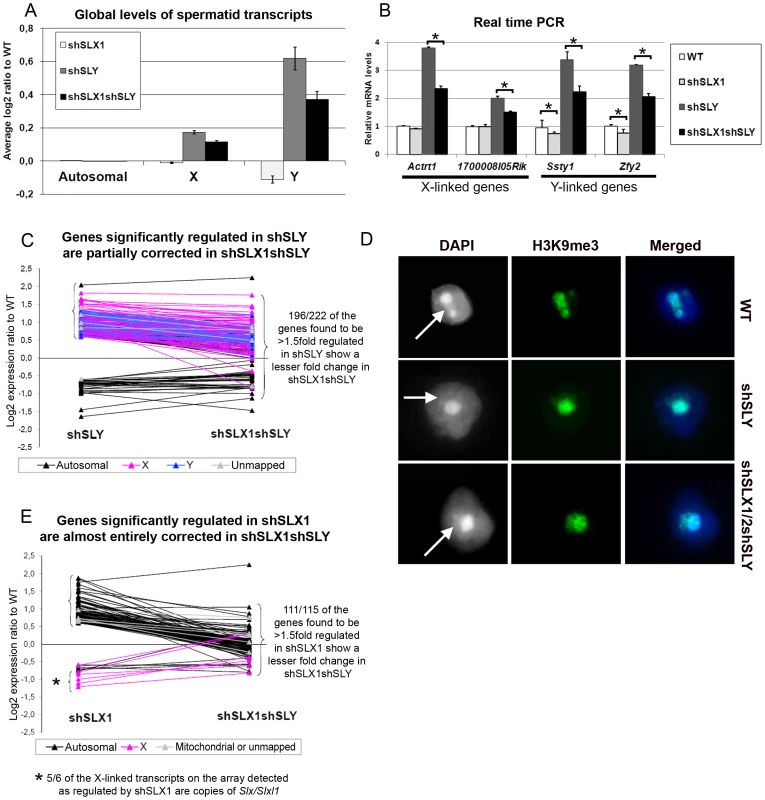
A) Representation of the microarray results obtained for Slx/Slxl1-deficient (shSLX1), Sly-deficient (shSLY) and Slx/y-deficient (shSLX1shSLY) compared to WT spermatids. B) Real time PCR quantification showed that transcript levels of X-encoded (Actrt1 and 1700008I05Rik) and of Y-encoded genes (Ssty1 and Zfy2) were lower in shSLX1shSLY than in shSLY spermatids. Transcript levels were also lower in shSLX1 spermatids compared to WT spermatids. The y-axis indicates the level of expression compared to WT after normalization with Acrv1 (2ΔΔCt ± standard errors). One asterisk indicates significant difference from corresponding shSLY or WT value (t test on ΔΔCt values; p<0.05). C) Graphic representation of the expression ratio relative to WT for the 222 genes showing greater than 1.5 fold-change in shSLY. Most genes affected by shSLY are sex-linked and up-regulated [19]. The majority of them (196/222) were corrected to some degree by addition of the shSLX transgene (in shSLX1shSLY). For 46 of these genes, the difference between shSLY and shSLX1shSLY was itself statistically significant. D) Immunofluorescence detection of H3K9me3 (green) in WT, shSLY and shSLX1/2shSLY round spermatid nuclei. DAPI (white or blue) was used to stain nuclei. The round DAPI-dense structure is the chromocenter. The less DAPI-dense structure at the periphery of the chromocenter is the postmeiotic sex chromatin (PMSC) and is indicated by an arrow. Pictures were taken using the same image capture parameters. Note the decreased H3K9me3 signal on the sex chromatin of Sly-deficient spermatids; this is almost completely restored by Slx/Slxl1 deficiency (in shSLX1/2shSLY spermatids). See also Figure S4. E) Graphic representation of the expression ratio relative to WT for the 115 genes showing greater than 1.5 fold-change in shSLX1. Most genes affected by shSLX1 are autosomal and up-regulated [28]. Note that almost all of them (111/115) have a lower fold change in shSLX1shSLY than in shSLX1. For 91 of these genes, the difference between shSLX1 and shSLX1shSLY was itself statistically significant. The up-regulation of X - and Y-encoded spermatid genes in Sly-deficient spermatids has been shown to be concurrent with a diminution of the repressive epigenetic marks (such as H3K9me3) normally associated with PMSC [19]. We therefore decided to study these repressive marks in Slx/y-deficient spermatids, and observed that H3K9me3 staining on PMSC (as compared to H3K9me3 chromocenter staining) was significantly higher (p = 0.00003) in Slx/y-deficient spermatids than in Sly-deficient spermatids (average staining intensity: 0.59 and 0.51 respectively), and closer to but significantly different from the WT value (average staining intensity in WT: 0.65, p = 0.003) (Figure 3D and Figure S4 for quantification). Therefore Slx/Slxl1 deficiency partially compensates the loss of H3K9me3 marks induced by Sly deficiency. These results correlate with the global effect of Slx/Slxl1 transcript knockdown on sex chromosome expression and suggest that SLX/SLXL1 and SLY proteins compete in spermatids for access to PMSC where they have activator and repressive effects respectively, at the whole-chromosome level.
We then compared the transcriptomes of WT, Slx/Slxl1-deficient and Slx/y-deficient spermatids. This revealed a 10% reduction in Y transcription in Slx/Slxl1-deficient spermatids compared to WT that was not seen in an earlier study [28] (Figure 3A–3B). This reduction is congruent with our observation of some SLX/SLXL1 proteins in a small number of WT spermatid nuclei (Figure 1B and Figure S1); this small fraction of SLX/SLXL1 proteins most likely increases sex chromosome gene expression in the nucleus of WT spermatids, while the loss of these proteins leads to a slight reduction of XY expression in Slx/Slxl1-deficient spermatids. A faint reduction of expression was observed for some X genes (for instance Actrt1, see Figure 3B) but this did not significantly differ from the WT value.
Sly knockdown corrects the gene deregulation induced by Slx/Slxl1 deficiency
We have previously shown that Slx/Slxl1 deficiency leads to delay in spermatid elongation and sperm release, associated with the deregulation (principally the up-regulation) of 115 genes, the majority of which are located on the autosomes. Given that SLX/SLXL1 proteins are almost entirely cytoplasmic in wild type, we proposed that these transcriptional changes were a manifestation of “cytoplasmic” defects, rather than a direct effect of SLX/SLXL1 proteins on autosomal gene expression; for instance, an as yet unidentified cytoplasmic partner of SLX/SLXL1 could mediate the transcriptional changes that are necessary for normal spermatid elongation, or it may be that the transcriptional changes seen reflect an altered cellular proportion of different step spermatids in shSLX [28]. In the present study, we compared microarray results from Slx/Slxl1-deficient and Slx/y-deficient spermatids and, surprisingly, observed that most of the genes deregulated by Slx/Slxl1 deficiency were less affected in Slx/y-deficient spermatids (111/115 genes, Figure 3E and Figure S5). Therefore, Sly knockdown corrects the deregulation of autosomal genes induced by Slx/Slxl1 (with autosomal gene expression values close to WT levels in Slx/y-deficient spermatids; Figure 3E and Figure S5). These results show that SLX/Y proteins have opposite regulatory effects on autosomal gene expression as well as on sex chromosome gene expression.
Slx/y-deficient males have better reproductive parameters and overall fertility than males that are deficient for either Slx/Slxl1 or for Sly
Our microarray results demonstrate that the deregulation of sex chromosome-linked or autosomal genes observed in Sly-deficient or in Slx/Slxl1-deficient spermatids respectively, is corrected in Slx/y-deficient spermatids; we therefore compared the reproductive parameters of Slx/y-deficient males with those from males that are singly deficient for either Slx/Slxl1 or for Sly. Firstly, Slx/y-deficient males had significantly improved sperm numbers (Table 1). This was particularly striking for the comparison between shSLX1/2 and shSLX1/2shSLY males: shSLX1/2 males had dramatically reduced spermatozoa numbers but the addition of shSLY transgene to this genotype increased the number of sperm produced ∼50-fold (Table 1). Low sperm count in shSLX males was attributed to the apoptosis of delayed elongating spermatids [28]. We therefore analyzed spermatid elongation delay and apoptosis in Slx/y-deficient males and in their Slx/Slxl1-deficient siblings. Remarkably, while Slx/Slxl1-deficient males presented a high number of delayed and apoptotic elongating spermatids, Slx/y-deficient models did not significantly differ from WT (Figure 4A–4B). The spermatozoa morphology of Slx/y-deficient males was also much improved compared to that of Slx/Slxl1 - or Sly-deficient males (Figure 4C and Figure S6). Finally, we compared the fertility of Slx/y-deficient males with Slx/Slxl1 - or Sly-deficient siblings: Slx/y-deficient males had overall better fertility than males that are deficient for either Slx/Slxl1 or Sly, with reproductive parameters close to WT values (Table 1). Strikingly, the addition of Sly deficiency was able to reverse the sterility observed in Slx/Slxl1 deficient-males (line shSLX1/2) (Table 1). All in all, males that were deficient for both Slx/Slxl1 and Sly had considerably better reproductive parameters than males that were deficient for Slx/Slxl1 or Sly alone.
Fig. 4. Slx/y-deficient males have fewer spermiogenesis defects than males that are deficient for either Sly or Slx/Slxl1 alone. 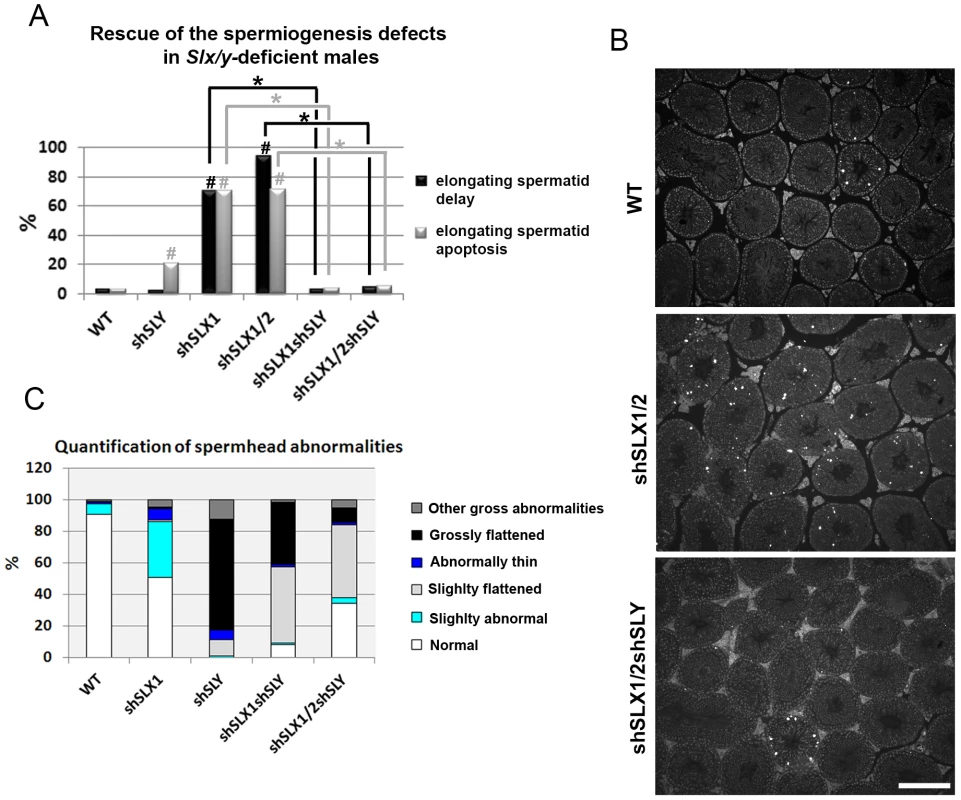
A) Bar graph representing the percentage of tubules containing apoptotic elongating spermatids measured by TUNEL assay (in grey) and the percentage of tubules containing delayed elongating spermatids (in black). Hatch symbol (#) indicates significant difference from WT (ANOVA, p<0.0001). One asterisk indicates significant difference between values obtained for Slx/y-deficient males (shSLX1shSLY or shSLX1/2shSLY) and Slx/Slxl1-deficient siblings (shSLX1 or shSLX1/2) (ANOVA, p<0.00001). B) Representative black and white TUNEL pictures of WT, shSLX1/2 and shSLX1/2shSLY testicular sections. Note the presence of apoptotic cells (TUNEL+ cells in white) in most shSLX1/2 seminiferous tubules, while in WT and in shSLX1/2shSLY they are largely restricted to stage XII tubules (and correspond to metaphasic cells undergoing normal apoptosis). Scale bar represents 200 µm. C) Bar graph representing the percentage of sperm head abnormalities in Slx/Slxl1-deficient, Sly-deficient and Slx/y-deficient males (see also Figure S6). The percentage of total abnormal sperm heads is significantly lower in Slx/y-deficient males (shSLX1shSLY and shSLX1/2shSLY) than in Sly-deficient males (shSLY). In particular, the number of grossly flattened spermheads – which are specifically observed in Sly-deficient males – is reduced (ANOVA, p<0.001). ShSLX1/2shSLY males also show a significant decrease in this percentage compared to shSLX1shSLY males (ANOVA, p<0.03). The number of slightly abnormal sperm heads – which are specifically observed in Slx/Slxl1-deficient males – is also reduced in Slx/y-deficient males compared to Slx/Slxl1-deficient males (ANOVA, p<0.0008). For this category of sperm abnormalities, Slx/y-deficient males did not significantly differ from WT. Tab. 1. Analysis of the reproductive parameters of Slx/y-deficient mice compared to controls. 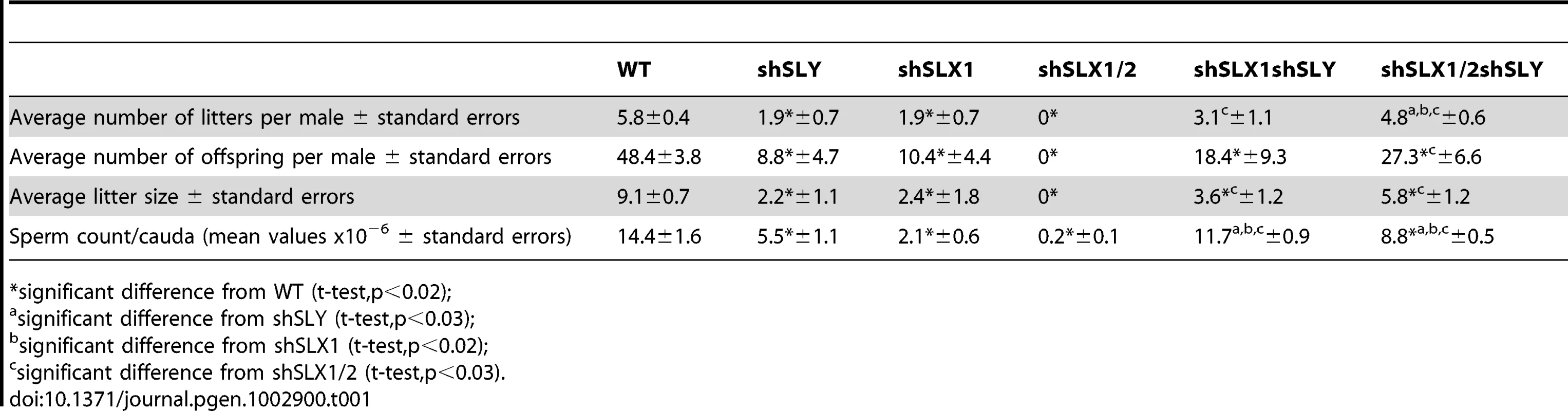
significant difference from WT (t-test,p<0.02); These analyses show that Sly deficiency almost completely rescues the defects and gene deregulation induced by Slx/Slxl1 deficiency, while Slx/Slxl1 knockdown only partially rescues those subsequent to Sly deficiency. This may be due to a different knockdown efficiency: indeed, no SLY1 protein can be detected in Slx/y-deficient samples while some SLX/SLXL1 proteins remain (Figure 2C–2E).
Slx/Slxl1 deficiency causes a sex ratio distortion in favor of males that is restored by Sly deficiency
We previously reported a tendency of an excess of females in the progeny of Sly-deficient males (7.7% excess of females, Chi-square p = 0.0569) [19]. While analyzing the fertility of our transgenic lines, we observed that Slx/Slxl1-deficient males (i.e. shSLX1) yielded an offspring sex ratio of 40% (74/187) female progeny, compared to a ratio of 51% (234/461) in WT siblings. This represents a statistically significant sex ratio distortion of 11% in favour of male offspring (Chi-square p = 0.006). Importantly, a normal sex ratio was restored by the addition of Sly deficiency: Slx/y-deficient males produced an offspring sex ratio of 50% (103/208) female progeny that did not differ from WT and was also significantly different from the offspring sex ratio of shSLX1 males (Chi-square p = 0.03). These data show that both Slx/Slxl1 and Sly affect the transmission of X - and Y-bearing gametes, Slx/Slxl1 favouring X transmission while Sly favours Y transmission.
Discussion
SLX/SLXL1 and SLY proteins have antagonistic effects on the expression of two distinct sets of genes
In recent years, we have identified the transcriptional consequences of Sly and Slx/Slxl1 deficiency, and related these to the observed phenotypes in terms of spermatid development, sperm morphology and offspring sex ratio [19], [28]. Remarkably, we now show that in dual shRNA knockdown models where both genes are deficient, the transcriptional and phenotypic consequences of the individual knockdown are dramatically ameliorated, correcting the X/Y/Speer up-regulation and sperm shape abnormalities seen in Sly-deficient spermatids; the autosomal gene up-regulation, spermatid elongation delay and apoptosis, and sperm shape abnormalities seen in Slx/Slxl1-deficient spermatids; and improving fertility in both cases.
Strikingly, however, two different and almost entirely non-overlapping sets of genes are affected by the mutual antagonism of SLX/SLXL1 and SLY. In this discussion, we refer to “Group 1” genes as the set of X/Y/Speer genes up-regulated in Sly-deficient spermatids and (partially) corrected in the dual knockdown, and “Group 2” genes as the set of metabolism-related autosomal genes up-regulated in Slx/Slxl1-deficient spermatids and (almost fully) corrected in the dual knockdown.
Group 1 genes: Nuclear consequences of antagonism between SLX/SLXL1 and SLY
Sly regulates the epigenetic repression of post meiotic sex chromatin (PMSC) and a few specific autosomal genes such as the Speer cluster. In the nucleus, SLY appears to act via the recruitment/maintenance of the repressive heterochromatin marks CBX1 and H3K9me3, which consequently limits the expression of X and Y genes in spermatids, among which are its X-linked homologs Slx and Slxl1 [19]. Here, we show that, in the absence of SLY, SLX/SLXL1 proteins relocate to the nuclear sites (both sex-linked and autosomal) vacated by SLY proteins. It is unlikely that SLX/SLXL1 nuclear localization in Sly-deficient spermatids is solely a consequence of increased SLX/SLXL1 protein abundance, since there is no clear enrichment in nuclear SLX/SLXL1 proteins in spermatids of transgenic mice overexpressing SLX or SLXL1 (our unpublished preliminary data).
Moreover, in the double transgenic model (Slx/y-deficient males) where SLX/SLXL1 family members are also reduced/absent, XY gene expression, Speer expression and the intensity of H3K9me3 marks on the sex chromatin are closer to normal values. This indicates that SLX and/or SLXL1 have consequences both for transcriptional activity and for histone modification when present on sex chromatin, and that these are directly opposed to the effects of SLY. We therefore propose that SLX/SLXL1 and SLY proteins compete for access to nuclear sites in spermatids, where they act as positive and negative transcriptional regulators respectively. We cannot at this point say precisely where the competition occurs: it may be directly at the level of chromatin binding within the nucleus, or SLX/SLXL1 and SLY may compete for access to factors affecting nuclear import. We note that SLX and SLXL1 proteins lack nuclear localization signals (NLS) while SLY NLS is mutated/truncated [22], [25]; as such they probably depend on other interacting factors to enter the nucleus. It also remains possible that the competition is mediated indirectly: for example, SLY could affect SLX/SLXL1 intracellular localization via regulating the expression of a third factor controlling SLX/SLXL1 access to the nuclear sites.
Group 2 genes: Cytoplasmic consequences of antagonism between SLX/SLXL1 and SLY
Slx/Slxl1 deficiency has been shown to increase the level of ∼100 autosomal transcripts which code for proteins of the cytoskeleton and the extracellular matrix, or are implicated in various cytoplasmic processes (i.e. energy production, lipid metabolism, ubiquitin-mediated degradation, etc.) [28]. These transcriptional effects are corrected in Slx/y-deficient males, suggesting that these changes may also be manifestations of the same nuclear/chromatin regulatory antagonism exhibited by Group 1 genes, perhaps via relocation of repressive factors from sex chromatin to autosomal locations and vice versa. There are, however, three significant objections to this interpretation. Firstly, as noted previously, in WT spermatids SLX/SLXL1 are predominantly cytoplasmic proteins, and the levels in the nucleus are almost undetectable: it is hard therefore to see i) how Slx/Slxl1 knockdown could directly induce widespread transcriptional changes, ii) what would then be the function of the abundant SLX/SLXL1 proteins in the cytoplasm. Secondly, this interpretation would require not only that SLX/SLXL1 act simultaneously as transcriptional activators of Group 1 genes and as transcriptional repressors of Group 2 genes, but that SLY has the reverse effect in both cases: it is challenging to imagine a mechanism that could explain this. Thirdly, if both Group 1 and Group 2 gene effects are a manifestation of the changing balance of SLX/Y proteins in the nucleus and/or of a relocation of repressive factors from the sex chromosomes to autosomes, then both groups of genes would be expected to change together. This is not the case: Group 1 genes are affected in shSLY but not in shSLX, and Group 2 genes vice versa.
For this reason, we favour our existing interpretation that Group 2 gene deregulation is a manifestation of the spermiogenesis defects occasioned by cytoplasmic Slx/Slxl1 deficiency (i.e. spermatid elongation delay and apoptosis, reduced sperm count, abnormal head to tail connections of the spermatozoa and male infertility) [28], and is not a direct effect of SLX/SLXL1 proteins on autosomal gene transcription. Given that the (cytoplasmic) spermiogenesis defects are corrected in the dual mutant, it stands to reason that the secondary expression changes follow the same pattern. We therefore propose that, in addition to the nuclear effects on Group 1 genes, SLY protein has a cytoplasmic role, opposing that of SLX/SLXL1. SLY proteins have been shown to be present in both the spermatid nucleus and cytoplasm [19], [21]. Intriguingly, a recent report indicates that the acrosomal (cytoplasmic) protein DKKL1, which we previously identified as a binding partner of SLY1 [21], also interacts with SLXL1 [29]. We have performed additional experiments and observed that all SLX/Y family members (i.e. SLY1, SLY2, SLX and SLXL1) can interact with DKKL1 (Figure S7). Therefore, SLX/SLXL1 and SLY proteins could compete for interaction with (a) common partner(s) in the cytoplasm, and this competition could be at the basis of the opposite effects of SLX/SLXL1 and SLY on spermiogenesis and autosomal gene expression. A combined model proposing how SLX/SLXL1 and SLY proteins have antagonistic effects in both the spermatid nucleus and cytoplasm is presented in Figure 5.
Fig. 5. Model presenting how SLX/SLXL1 and SLY proteins have antagonistic effects in the spermatid nucleus and cytoplasm. 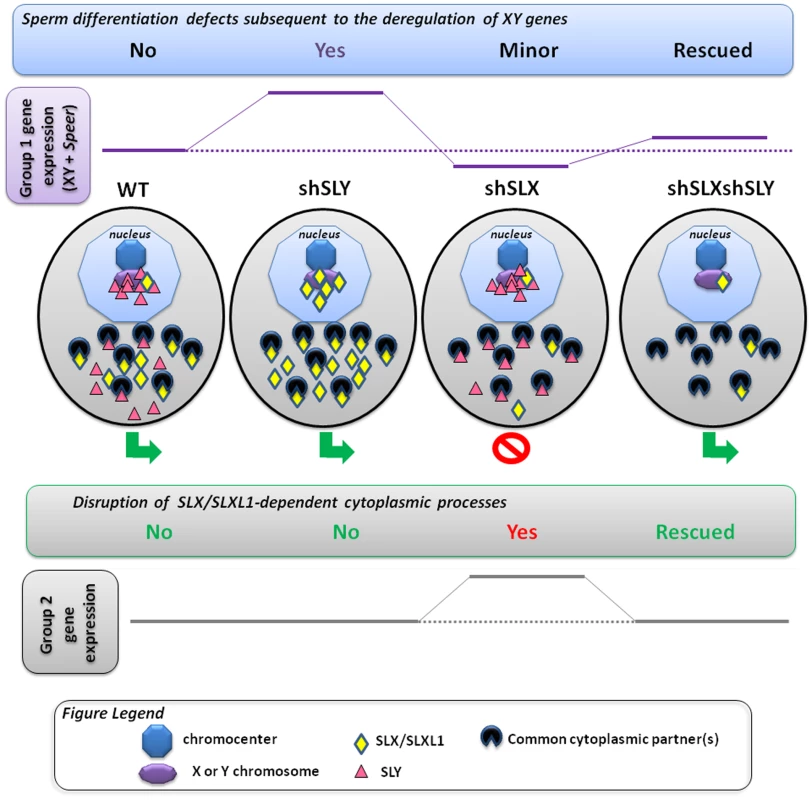
In the spermatids, SLX/SLXL1 (yellow lozenges) and SLY (pink triangles) proteins have antagonistic effects i) in the nucleus, on the expression of XY genes and of a few autosomal genes, such as Speer (group 1 genes); ii) in the cytoplasm, on the regulation of metabolic processes which secondarily causes a deregulation of ∼100 autosomal genes (group 2 genes). i) In WT, SLY proteins are located in both the nucleus and cytoplasm, while SLX/SLXL1 proteins are almost exclusively in the cytoplasm. The nuclear fraction of SLY proteins colocalizes with the sex chromosomes and the autosomal Speer gene cluster, and represses their expression. A very small fraction of SLX/SLXL1 proteins also appears to colocalize with the sex chromatin. In Sly-deficient spermatids (shSLY), SLX/SLXL1 proteins relocate to the nuclear sites (both sex-linked and autosomal) vacated by SLY proteins; however, SLX/SLXL1 proteins have an opposite effect to that of SLY, and activate XY gene expression. This is associated with a reduction in the repressive epigenetic mark H3K9me3 on the sex chromatin (purple octagon), and produces sperm differentiation defects such as spermhead abnormalities, shedding delay, motility defects and subsequent male infertility. In Slx/Slxl1-deficient spermatids (shSLX), the absence of SLX/SLXL1 nuclear proteins has only minor effect on gene regulation, since it does not change SLY localization profile. There is only a slight reduction in XY transcription, congruent with the idea that SLX/SLXL1 is a transcription activator sharing the targets of SLY when present in the nucleus. In the double knock-down (shSLXshSLY), Slx/Slxl1 deficiency almost fully abrogates the effects of Sly knockdown: in shSLXshSLY spermatids, group 1 gene expression and repressive epigenetic marks are close to WT values. This is correlated with a rescue of SLY-dependent sperm differentiation defects. In sum, in the nucleus, the experimental observations indicate that SLX/SLXL1 competes with SLY at the level of sex chromatin regulation: SLY acts as a repressor while SLX/SLXL1 acts as a positive regulator. ii) Slx/Slxl1 deficiency induces various spermiogenic defects (such as spermatid elongation delay and apoptosis, reduced sperm count, abnormal head to tail connections of the spermatozoa and subsequent male infertility) associated with an up-regulation of ∼100 autosomal genes which code for proteins of the cytoskeleton, the extracellular matrix, or implicated in various metabolic processes (i.e. group 2 genes). Since SLX/SLXL1 proteins are predominantly cytoplasmic in WT spermatids, we propose that this gene deregulation is a manifestation of the spermiogenesis defects occasioned by Slx/Slxl1 deficiency, and not a direct effect of SLX/SLXL1 proteins on autosomal gene transcription. In the case of Sly deficiency, group 2 gene expression is unaffected; however, in the double knock-down, Sly deficiency corrects SLX/SLXL1-dependent phenotypes which abrogates the subsequent group 2 gene up-regulation. This means that SLY protein has a cytoplasmic role, opposing that of SLX/SLXL1. This antagonism could be mediated via interaction with (a) common partner(s) in the cytoplasm; the absence of competition between SLX/SLXL1 and SLY proteins in the dual knockdown model would explain the absence of defects. In sum, SLX/SLXL1 and SLY proteins apparently compete in the cytoplasm for the regulation of spermiogenic processes. The functional role of SLX/SLXL1 could be to prevent the access of SLY to cytoplasmic proteins that are necessary for spermiogenesis. We recognize that under our preferred model, it is difficult to explain the directionality of the expression changes seen in shSLX relative to WT, which was predominantly up-regulation of autosomal genes with comparatively few down-regulated genes [28]. A potential explanation for this lies in the spermatid developmental delay resulting in delayed spermatid elongation in shSLX. This could potentially skew the round spermatid population in shSLX testes towards earlier stages, i.e. proportionally more step 1 spermatids and fewer step 7–8 spermatids. Since there is a progressive transcriptional shutdown throughout spermatid development as chromatin is repackaged in preparation for nuclear condensation, this would thus manifest in shSLX as a selective up-regulation of those genes expressed specifically in early stage round spermatids (which in turn is plausible given the annotated functional categories for these Group 2 genes). Testing this interpretation will require further experiments on fractionated, staged sub-populations of round spermatids.
The mouse X and Y chromosomes are involved in an intragenomic conflict that is regulated by Slx/Slxl1 and Sly
Irrespective of the precise molecular mechanism(s) underlying the antagonistic effects of SLX/SLXL1 and SLY, our results demonstrate that both genes have an effect on offspring sex ratio. In particular, comparing shSLX (where Sly is still present) to the dual knockdown, there is a significant excess of males; and when comparing shSLY (where Slx/Slxl1 are still present) to the dual mutant, there is a trend towards excess of females. Thus, the net effect of these genes on inheritance is for X-linked family members to favour X chromosome transmission, and Y-linked members to favour Y chromosome transmission, constituting a prima facie genomic conflict. Such a conflict was first postulated in the 1990s following observations that male mice with a partial deletion of the Y long arm produce an excess of female offspring, however supporting evidence has not been forthcoming until recently [16], [20], [26], [27]. The present study demonstrates that such a conflict exists between the sex chromosome-linked Sycp3-related genes. An intragenomic conflict is often not visible under normal conditions (i.e. in a WT population) [11], [13] and here the positive effect of Slx/Slxl1 on sex chromosome transcription was uncovered by the production of mice that are deficient for both Sly and Slx/Slxl1; similarly, the effects of Slx/Slxl1 deficiency are also corrected in the dual mutant, although the molecular mechanisms involved are less clear.
Can sex ratio distortion be directly attributed to Slx/Slxl1 and Sly?
Under the distorter/responder model exemplified by the t complex [1], both Slx/Slxl1 and Sly are transmission distorters in that changes in their expression levels lead to a distortion of the sex ratio. However, it is unlikely that they are directly responsible for mediating the transmission skew (i.e. responder genes). Indeed, the physiological mechanism of the skew in the present model is an asymmetry in fertilizing ability between X and Y sperm [30]. This implies an underlying molecular/functional asymmetry, namely the presence of a responder gene product which is not evenly shared between X and Y sperm. Both of the known mammalian examples of transmission ratio distortion depend on non-sharing of gene products (both transcript and protein) between sister spermatids: Spam1 in the case of Rb(6.16) and Rb(6.15) translocation heterozygotes, and TcrSmok in the case of driving t haplotypes [31]–[33]. We note that SLX/Y proteins appear to be similarly expressed in X - and Y-bearing spermatids. It therefore seems likely that the distortion in Yq deleted mice and in shSLXshSLY transgenic models is mediated by an as yet unidentified sex-linked gene or gene(s) (i.e. the responder), for which Slx/Slxl1 and Sly are competing regulators via their global effects on sex chromatin expression. Among the deregulated genes, a few appear as promising candidates, such as the X-encoded homolog of Tcp11, which is one of the genes involved in the t-complex transmission distortion, albeit as a distorter rather than a responder [34], and Alkbh7, since another Alkbh gene has recently been found to cause sex ratio distortion [35].
However, there may be several linked genes involved, at least one of which is likely to evade transcript sharing. In view of this possibility, it is worth noting that both regulators of the conflict have a global effect on sex chromatin; this is an efficient way to control multiple sex chromosome-linked distorters and/or responders simultaneously. The ease of identifying the responder(s) will depend on how directly SLX/Y regulate them and how many there are. Finally, it is possible that autosomal factors also contribute to the regulation of sex-linked transmission distortion. We note that historically, Slx appeared on the X before Sly appeared on the Y, and its distorting effect on sex ratio may have subsequently been countered by a combination of Sly-mediated repression and other autosomal genes being selected to favour a balanced sex ratio [20].
The intragenomic conflict in which Slx/Slxl1 and Sly are involved has influenced the structure of the mouse sex chromosomes
In the mouse lineage, there has been a remarkable amplification of spermatid-expressed sex chromosome genes (all of which fall into Group 1 identified above), and which has had a dramatic influence on the structure of the mouse sex chromosomes. This expansion occurred subsequent to the appearance of Sly, but was not accompanied by a matching increase in XY transcript levels [20]. It is therefore very likely that essential sex-linked spermatid-expressed genes have become amplified in order to maintain a steady expression in the face of the enhancement of Sly-mediated repression and in a sense constitute a “collateral damage” arising from the conflict between Sly and Slx/Slxl1 that we unravel here. Interestingly, the Speer gene cluster is one of the autosomal gene families that have experienced the largest rodent-specific expansions [36] and is also repressed by SLY. Slx/Slxl1 and Sly competition may therefore have led to the amplification of reproductive genes outside the sex chromosomes as well as on them.
The intragenomic conflict in which Slx/Slxl1 and Sly are involved may have played an important role in mouse speciation
F1 hybrid sterile males produced by asymmetric crosses between M. m. musculus and M. m. domesticus display sperm differentiation defects and wide-spread overexpression of X-encoded spermiogenic genes [37]. Intriguingly, this only occurs in males with a M. m. musculus X chromosome and M. m. domesticus Y and autosomal chromosomes [38]. These males have an excess of Slx/Slxl1 copies compared to Sly copies, since the M. m. domesticus X and Y chromosomes carry ∼40 to 60 copies of Slx/Slxl1 and Sly, while XY encoded Sycp3-related genes have been more amplified in M. m. musculus, with >100 of Slx/Slxl1 and Sly on the X and Y [18], [20]. Our data show that a balance between Slx/Slxl1 and Sly expression exists in wild-type populations and that disruption of this balance can cause male infertility. In light of these data, we propose that deficiency in the number of Sly copies compared to Slx/Slxl1 copies contributes to F1 male hybrid sterility (see Figure 6) in some of these crosses. This would explain the observed over-expression of X-encoded spermiogenic genes observed in some F1 hybrid males [37] and subsequent sperm differentiation defects and infertility. The observation that F1 males born from the reciprocal cross domesticus x musculus are reproductively normal [39] does not necessarily challenge this model. These males have an excess of Sly copies compared to Slx/Slxl1 copies and, according to our model, could be considered as Slx/Slxl1-deficient mice and thus display some spermiogenic defects. This however depends critically on the mechanism of the antagonistic effects of SLY and SLX/SLXL1 in the cytoplasm, and on the threshold of copy number imbalance required to trigger abnormal spermatogenesis and/or sex ratio skewing. Given that autosomal genes will be selected to maintain a balanced sex ratio, the Slx/Sly conflict may well be “buffered” to some extent by epistatic interactions with autosomal genes.
Fig. 6. Model comparing Slx:Sly copy number imbalance in natural and laboratory mouse strains to Slx:Sly gene expression imbalance in shRNA knockdown models. 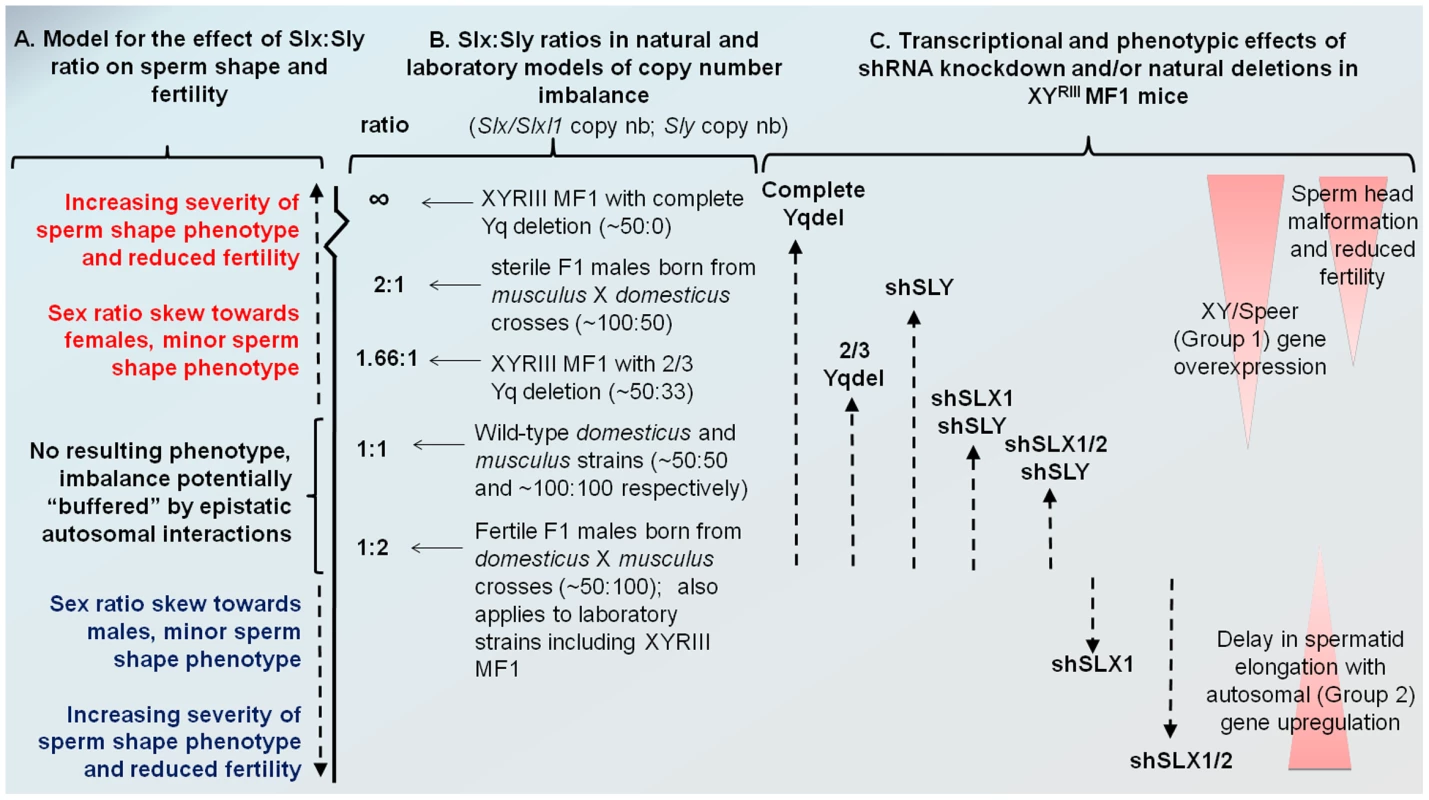
A. A model for how Slx/Slxl1:Sly imbalance affects sperm shape, offspring sex ratio and fertility. B. Approximate copy number ratio of Slx/Slxl1 and Sly in the reciprocal crosses studied by Good et al. [37] based on an estimate of ∼100 copies of each gene in musculus and ∼50 in domesticus, in the WT laboratory strain MF1 YRIII which has a domesticus X and autosomes but a musculus-derived Y [40]–[42], and in the two natural mutants from the same background studied by us and others [18], [20]. C. The relative magnitude of Group 1 and Group 2 transcriptional responses seen in the various shRNA/deletion models on the MF1 YRIII background. The double and triple shRNA models show a partial Group 1 response, but no Group 2 response. Importantly, in this model, the shSLX1 and shSLX1/2 phenotypes are expected to fall outside the range of variation seen in the natural mutant and reciprocal cross males, since they are on a background which has already a deficiency in Slx/Slxl1 copy number compared to Sly (50∶100). We emphasise that the effects of Slx/Slxl1:Sly imbalance are only one contributor to hybrid sterility: sperm shape and testis size QTLs on the musculus X map to distinct locations and show different interactions with the domesticus autosomes and Y chromosome [50]. We have observed that mice with a partial knockdown of Slx/Slxl1 (shSLX1 or shSLX2) have comparatively minor spermiogenic defects compared to mice with a severe knock-down (shSLX1/2) [28]. We also note that laboratory strain X chromosomes (including MF1 mice which were used in the present study) are predominantly derived from a domesticus background [40], [41], yet are paired in these strains with a musculus Y chromosome YRIII [42]. Thus laboratory strains are intrinsically comparable to the reciprocal cross. Our shSLX models therefore involve skewing the balance of SLX/SLXL1 and SLY even further, to pathogenic effect (see Figure 6). In this light it is intriguing that WT MF1 males have lower XY gene transcription than Slx/y deficient males: might this reflect the fact that laboratory strains are inherently “overdosed” for Sly relative to Slx/Slxl1 by virtue of their hybrid origin?
Male hybrid sterility is a complex trait involving several X-linked loci (as demonstrated by the mapping of several quantitative trait loci – QTL – on the X chromosome [38], [43], [44]) as well as autosomal factors [45], [46]). It is worth noting that among the four non-overlapping X-chromosome-linked QTL associated with abnormal spermheads and hybrid sterility, one encompasses Slx (0–37.1 Mb), the other, Slxl1 (47.9–81.8 Mb) [38]. Interestingly, it has been shown that one of the autosomal loci linked to hybrid sterility, Prdm9, encodes a histone H3 lysine 4 methyltransferase involved in the silencing of the sex chromosomes during meiosis (Meiotic Sex Chromosome Inactivation). It therefore epigenetically represses multiple X-chromosome loci, some of which part of the hybrid sterility gene network, and epistatic interactions between Prdm9 and multiple X and autosomal loci have been shown to cause asymmetric hybrid male sterility associated with a disruption of MSCI and thus a de-repression of the X chromosome [43], [46]. However, Prdm9 does not appear to be involved in the X-chromosome up-regulation and sterility observed in F1 hybrid males studied by Good et al. [37].
Taken together, the genetic basis of reproductive isolation in mice is complex, and disruption of the transcriptional regulation of the X seems to contribute to the evolution of hybrid male sterility. The antagonistic effects of Slx/Slxl1 and Sly at the transcriptional and phenotypic level, in particular the effects on postmeiotic XY gene regulation, may therefore be among the important elements contributing to the evolution of hybrid sterility between mouse species. The production of F1 males with a transgene-derived increased Sly expression or with a knockdown of Slx/Slxl1 expression should help address this question.
In conclusion, we have demonstrated that the mouse X and Y chromosomes are involved in an intragenomic conflict that is regulated by the multicopy genes Slx/Slxl1 and Sly. SLX/SLXL1 and SLY proteins compete during sperm differentiation, and notably have opposite effects on the regulation of sex chromosome gene expression. Disruption of Slx/y balance causes sex ratio distortion, sperm differentiation defects and male infertility. To the best of our knowledge, our work is the first characterization of a conflict over sex chromosome transmission in mammals and provides further evidence to support the hypothesis that intragenomic conflicts can have major consequences on gene regulation, genome structure and speciation.
Materials and Methods
Generation and breeding of transgenic mice
shSLY (aka sh367), shSLX1 and shSLX1/2 males were produced and maintained as described before [19], [28]. To produce shSLX1shSLY and shSLX1/2shSLY double transgenic mice, shSLX1 females were mated to shSLY or to shSLYshSLX2 transgenic males. Double transgenic females were then mated to MF1 XYRIII males (see [19]) to maintain the stock, since shSLY males are subfertile and give progeny only rarely. Two-month-old males single or double transgenic for sh367 (shSLY), shSLX1 or shSLX1/2 transgenes, as well as their non-transgenic siblings (WT) were processed for all the analyses presented here. Animal procedures were in accordance with the United Kingdom Animal Scientific Procedures Act 1986 and were subject to local ethical review.
Elutriation of spermatids
Fractions enriched in round spermatids (>90%) were obtained from the above described transgenic and control (WT) males as described previously [19]. Each sample has been purified from a pool of testes obtained from 2 to 5 males.
Transfection
The coding sequence of mouse Dkkl1 and Slx cDNA were amplified by PCR and cloned into a C-terminal Myc-tagged pCMV vector; the coding sequence of mouse Slx, Slxl1, Sly1 and Sly2 cDNA were amplified by PCR and cloned into a N-terminal Flag tagged pCMV vector using EcoRI and NotI restriction sites (see Table S1 for a full list of primers). Co-transfections of HEK293 or COS cells were performed in 6-well plates using 1.5 µg of each DNA and 5 µl of Lifofectamine (Invitrogen) following the manufacturer's instructions. Proteins were extracted 24 hours post transfection in 200 µl of Lysis buffer (25 mM NaCl, 10 mM Tris-HCl, 5 mM EDTA, 0.1%NP-40) and immunoprecipitated as described below.
Protein analyses
Nuclear and cytoplasmic protein extracts were obtained as follow. The powder obtained from two adult testes crushed on dry ice was homogenized in a glass pestle with 1 mL of lysis buffer (0.6 M Sucrose, 10 mM Hepes pH 7.7, 25 mM KCl, 2 mM EDTA, 0.5 mM EGTA and protease inhibitors). After the addition of 0.2% NP40, the lysate was centrifuged for 15 minutes at 800 g. The supernatant corresponded to the cytoplasmic fraction. The pellet was washed twice with 1 mL of lysis buffer and then resuspended in 100 µl of nuclear protein extraction buffer (20 mM Hepes pH 7.7, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol and protease inhibitors) plus 10 µl of 4 M NaCl. After 30 minutes of homogenization at 4°C, the samples were centrifuged for 30 minutes at 11000 g; the supernatant corresponded to the nuclear protein extract. A pellet of ∼1×107 round spermatids was extracted following the same protocol using 250 µl of lysis buffer and 50 µl of nuclear protein extraction buffer. Whole testicular protein extraction was performed as described previously [19]. For immunoprecipitation experiments, proteins extracted from transfected cells were first pre-cleared with protein A/G sepharose for 1 hour at 4°C. They were then incubated overnight with Protein G - or Protein A - sepharose which had been covalently bound to MYC (Santa Cruz Biotechnology) or FLAG (Sigma) antibody beforehand (see [21] for a detailed protocol). Western blot experiments were performed as described previously [19]. Membranes were incubated overnight with anti-SLX/SLXL1 [28] diluted at 1/3000, anti-SLY1 [21] at 1/3000, anti–β-actin (Sigma) at 1/50000, or anti-LAMIN B1 (Santa Cruz Biotechnology) at 1/1000, anti-FLAG (Sigma) at 1/1000, or anti-MYC (Santa Cruz Biotechnology) at 1/1000. Detection by chemiluminescence was carried out after incubation with the corresponding secondary antibody coupled to peroxidase, as described by the manufacturer (Millipore).
Immunofluorescence and TUNEL
Immunofluorescence experiments were performed on testis material fixed in 4% buffered paraformaldehyde and sectioned as described before [25]. DAPI (4′,6-diamidino-2-phenylindole) was used to stain nuclei (Vectashield DAPI, Vectorlab). Alexa Fluor 594-conjugated peanut agglutinin lectin (Invitrogen), which stains the developing acrosome of spermatids, was used to stage the testis tubules. For the analysis of apoptotic elongating spermatids and delayed elongating spermatids, approximately 150 tubules were counted per individual (4 to 6 individuals per genotype). The percentage of tubules containing apoptotic elongating spermatids was determined on testis sections fluorescently stained using an in situ cell death detection kit (TUNEL, terminal deoxynucleotidyltransferase dUTP nick end labeling) as described by the manufacturer (Roche Diagnostics, Indianapolis, IN). The percentage of tubules containing delayed elongating spermatids (i.e. stage I to VIII tubules containing elongating spermatids) was assessed on testis sections fluorescently stained by H4K12Ac antibody (Millipore, Bedford, MA), a known marker of stage 9–12 elongating spermatids.
Antibody detection, chromosome painting, and DNA–FISH on surface-spread testicular cells
Antibody detection was performed on surface-spread testicular cells following a protocol described previously [19] adapted from Barlow et al. [47]. Incubation with the primary antibody (anti-SLY1 [17], anti-SLX/SLXL1 [28] or anti-H3K9me3 [Upstate] diluted 1/100) was carried out over-night in a humid chamber at 37°C. DNA-FISH, then chromosome painting were performed after antibody detection as described previously [48]. Speer DNA-FISH was carried out using mouse BACs RP23-212A20 and RP24-310N20 (CHORI). As a control for specificity (see Figure S1), SLX/SLXL1 antibody was preabsorbed with 8 mg of SLX immunogenic peptide or with 8 mg of a noncompeting peptide (SLY peptide). For the quantification of H3K9me3 signal over the PMSC, the chromocenter domain was defined using the corresponding black and white DAPI picture. Then, H3K9me3 signal outside this chromocenter domain was measured and normalized to that of H3K9me3 signal over the chromocenter for each cell (100 cells per genotype), using Metamorph and ImageJ (See Figure S4). Slides corresponding to 3 individuals per genotype were coded and randomized before the analysis; the analysis was therefore carried out blind as to genotype.
Analysis of sperm head morphology
For the quantification of spermhead abnormalities, sperm collected from the initial caput epididymis were suspended in phosphate-buffered saline. The suspension was smeared on slides (two slides per individual) and fixed in 3∶1 methanol∶acetic acid. The slides were then dipped in 0.4% Photoflo for 2 min, air dried and stained on a plate heated at 60°C with one drop of 50% silver nitrate mixed with one drop of 2% gelatin (Sigma). The slides were coded and randomized. Sperm scoring was carried out ‘blind’ as to genotype (4 to 6 individuals per genotype) and 100 sperm per slide were classified into 6 categories on the basis of the type and severity of abnormalities observed, using criteria described by Yamauchi et al. [49] and in Figure S6. In the text and figures, spermheads from category N were termed “normal”; category 1S, “slightly abnormal”; category 2S, “slightly flattened”; category 3G, “abnormally thin”; category 4G, “grossly flattened” and categories 5G to 8G were pooled and named “other gross abnormalities” (cf. Figure S6).
Fertility testing and sex ratio of the offspring
To assess fertility and obtain sex ratio data from the offspring, five males of each genotype were mated with MF1 WT females over a period of six months.
Real-time PCR and microarray analyses
Real-time Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and microarray analyses were performed as previously described on RNA extracted from 2-month old testis or from round spermatids obtained after elutriation [19] (cf. Table S1 for a list of the primers used in this study). Real-time RT-PCR experiments were performed in parallel for all the genotypes described in this study, with between 3 to 5 individuals per genotype. For the microarray analysis, three shSLX1, three shSLY, three shSLX1shSLY and four wild type spermatid batches were analyzed (Illumina BeadChip, mouse whole-genome array, v2). These data thus include and extend our previously-reported results for shSLX1 round spermatids and for shSLY round spermatids in previous analyses [19], [28], which collectively used two shSLY, two shSLX1 and four WT spermatid batches. Data normalization and calculation of FDR-adjusted p values was carried out in BeadStudio (Illumina) as previously described [19], [28]. The full data set has been uploaded to GEO, accession number GSE39109.
Statistical analysis
For comparisons of the incidence of sperm head abnormalities, differences between genotypes were assessed by ANOVA after angular transformation of percentages, using the General Linear Models ANOVA provided by NCSS statistical data analysis software. The same test was applied to the frequency of abnormal head-tail connections, TUNEL positive elongating spermatids, delayed elongating spermatids (assessed by H4K12Ac staining) and H3K9me3 quantification. Student's t test was used to compare the data obtained for fecundity, sperm number and real-time PCR (performed on the ΔCt values). A Chi-square test was used for sex ratio data. Microarray results were analyzed as described in Figure S3 and Figure S5.
Supporting Information
Zdroje
1. LyonMF (1984) Transmission ratio distortion in mouse t-haplotypes is due to multiple distorter genes acting on a responder locus. Cell 37 : 621–628.
2. MeiklejohnCD, TaoY (2009) Genetic conflict and sex chromosome evolution. Trends Ecol Evol 25 : 215–223.
3. WerrenJH (2011) Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci U S A 108 Suppl 2 : 10863–10870.
4. HurstLD, PomiankowskiA (1991) Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics 128 : 841–858.
5. HamiltonWD (1967) Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science 156 : 477–488.
6. PartridgeL, HurstLD (1998) Sex and conflict. Science 281 : 2003–2008.
7. HurstLD (1996) Further evidence consistent with Stellate's involvement in meiotic drive. Genetics 142 : 641–643.
8. BelloniM, TrittoP, BozzettiMP, PalumboG, RobbinsLG (2002) Does Stellate cause meiotic drive in Drosophila melanogaster? Genetics 161 : 1551–1559.
9. FaulhaberSH (1967) An abnormal sex ratio in Drosophila simulans. Genetics 56 : 189–213.
10. CazemajorM, LandreC, Montchamp-MoreauC (1997) The sex-ratio trait in Drosophila simulans: genetic analysis of distortion and suppression. Genetics 147 : 635–642.
11. TaoY, HartlDL, LaurieCC (2001) Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A 98 : 13183–13188.
12. TaoY, AraripeL, KinganSB, KeY, XiaoH, et al. (2007) A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol 5: e293 doi:10.1371/journal.pbio.0050293.
13. PhadnisN, OrrHA (2009) A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323 : 376–379.
14. JaenikeJ (2001) Sex chromosome meiotic drive. Annu Rev Ecol Syst 32 : 25–49.
15. SandlerL, NovitskiE (1957) Meiotic drive as an evolutionary force. Am Nat 41 : 105–110.
16. ConwaySJ, MahadevaiahSK, DarlingSM, CapelB, RattiganÁM, et al. (1994) Y353/B: a candidate multiple-copy spermiogenesis gene on the mouse Y chromosome. Mammalian Genome 5 : 203–210.
17. Alfoldi JE (2008) Sequence of the Mouse Y Chromosome [Thesis (Ph. D.)]. Cambridge: Massachusetts Institute of Technology. 93 p.
18. ScavettaRJ, TautzD (2010) Copy number changes of CNV regions in intersubspecific crosses of the house mouse. Mol Biol Evol 27 : 1845–1856.
19. CocquetJ, EllisPJ, YamauchiY, MahadevaiahSK, AffaraNA, et al. (2009) The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 7: e1000244 doi:10.1371/journal.pbio.1000244.
20. EllisPJ, BaconJ, AffaraNA (2011) Association of Sly with sex-linked gene amplification during mouse evolution: a side effect of genomic conflict in spermatids? Hum Mol Genet 20 : 3010–3021.
21. ReynardLN, CocquetJ, BurgoynePS (2009) The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol Reprod 81 : 250–257.
22. EllisPJ, FergusonL, ClementeEJ, AffaraNA (2007) Bidirectional transcription of a novel chimeric gene mapping to mouse chromosome Yq. BMC Evol Biol 7 : 171.
23. TouréA, GrigorievV, MahadevaiahSK, RattiganA, OjarikreOA, et al. (2004) A protein encoded by a member of the multicopy Ssty gene family located on the long arm of the mouse Y chromosome is expressed during sperm development. Genomics 83 : 140–147.
24. ToureA, ClementeEJ, EllisP, MahadevaiahSK, OjarikreOA, et al. (2005) Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol 6: R102.
25. ReynardLN, TurnerJM, CocquetJ, MahadevaiahSK, ToureA, et al. (2007) Expression analysis of the mouse multi-copy X-linked gene Xlr-related, meiosis-regulated (Xmr), reveals that Xmr encodes a spermatid-expressed cytoplasmic protein, SLX/XMR. Biol Reprod 77 : 329–335.
26. EllisPJI, ClementeEJ, BallP, ToureA, FergusonL, et al. (2005) Deletions on mouse Yq lead to upregulation of multiple X - and Y-linked transcripts in spermatids. Human Molecular Genetics 14 : 2705–2715.
27. EllisPJ, AffaraNA (2006) Spermatogenesis and sex chromosome gene content: an evolutionary perspective. Hum Fertil (Camb) 9 : 1–7.
28. CocquetJ, EllisPJ, YamauchiY, RielJM, KaracsTP, et al. (2010) Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol Biol Cell 21 : 3497–3505.
29. ZhuangXJ, HouXJ, LiaoSY, WangXX, CookeHJ, et al. (2011) SLXL1, a novel acrosomal protein, interacts with DKKL1 and is involved in fertilization in mice. PLoS ONE 6: e20866 doi:10.1371/journal.pone.0020866.
30. WardMA, BurgoynePS (2006) The Effects of Deletions of the Mouse Y Chromosome Long Arm on Sperm Function - Intracytoplasmic Sperm Injection (ICSI) - Based Analysis. Biol Reprod 74 : 652–658.
31. Martin-DeLeonPA, ZhangH, MoralesCR, ZhaoY, RulonM, et al. (2005) Spam1-associated transmission ratio distortion in mice: elucidating the mechanism. Reprod Biol Endocrinol 3 : 32.
32. VeronN, BauerH, WeisseAY, LuderG, WerberM, et al. (2009) Retention of gene products in syncytial spermatids promotes non-Mendelian inheritance as revealed by the t complex responder. Genes Dev 23 : 2705–2710.
33. EllisPJ, YuY, ZhangS (2011) Transcriptional dynamics of the sex chromosomes and the search for offspring sex specific antigens in sperm. Reproduction 142 : 609–619.
34. FraserLR, HosseiniR, HanyalogouA, TalmorA, DudleyRK (1997) TCP-11, the product of a mouse t-complex gene, plays a role in stimulation of capacitation and inhibition of the spontaneous acrosome reaction. Mol Reprod Dev 48 : 375–382.
35. NordstrandLM, SvardJ, LarsenE, NilsenA, OuglandR, et al. (2010) Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS ONE 5: e13827 doi:10.1371/journal.pone.0013827.
36. ChurchDM, GoodstadtL, HillierLW, ZodyMC, GoldsteinS, et al. (2009) Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi:10.1371/journal.pbio.1000112.
37. GoodJM, GigerT, DeanMD, NachmanMW (2010) Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet 6: e1001148 doi:10.1371/journal.pgen.1001148.
38. GoodJM, DeanMD, NachmanMW (2008) A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179 : 2213–2228.
39. GoodJM, HandelMA, NachmanMW (2008) Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution 62 : 50–65.
40. YalcinB, NicodJ, BhomraA, DavidsonS, CleakJ, et al. (2010) Commercially available outbred mice for genome-wide association studies. PLoS Genet 6: e1001085 doi:10.1371/journal.pgen.1001085.
41. YangH, WangJR, DidionJP, BuusRJ, BellTA, et al. (2011) Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43 : 648–655.
42. MardonG, MosherR, DistecheCM, NishiokaY, McLarenA, et al. (1989) Duplication, deletion, and polymorphism in the sex-determining region of the mouse Y chromosome. Science 243 : 78–80.
43. StorchovaR, GregorovaS, BuckiovaD, KyselovaV, DivinaP, et al. (2004) Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome 15 : 515–524.
44. OkaA, MitaA, TakadaY, KosekiH, ShiroishiT (2010) Reproductive isolation in hybrid mice due to spermatogenesis defects at three meiotic stages. Genetics 186 : 339–351.
45. ForejtJ (1996) Hybrid sterility in the mouse. Trends in Genetics 12 : 412–417.
46. MiholaO, TrachtulecZ, VlcekC, SchimentiJC, ForejtJ (2009) A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323 : 373–375.
47. BarlowAL, BensonFE, WestSC, Hult‚nMA (1997) Distribution of the RAD51 recombinase in human and mouse spermatocytes. EMBO Journal 16 : 5207–5215.
48. TurnerJM, MahadevaiahSK, EllisPJ, MitchellMJ, BurgoynePS (2006) Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell 10 : 521–529.
49. YamauchiY, RielJM, WongSJ, OjarikreOA, BurgoynePS, et al. (2009) Live offspring from mice lacking the Y chromosome long arm gene complement. Biol Reprod 81 : 353–361.
50. CampbellP, GoodJM, DeanMD, TuckerPK, NachmanMW (2012) The Contribution of the Y Chromosome to Hybrid Male Sterility in House Mice. Genetics
Štítky
Genetika Reprodukčná medicína
Článek Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese SubjectsČlánek Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in RiceČlánek Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System inČlánek An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural CrestČlánek A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of BacteriaČlánek A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Heterozygous Mutations in DNA Repair Genes and Hereditary Breast Cancer: A Question of Power
- GWAS of Diabetic Nephropathy: Is the GENIE out of the Bottle?
- The Conflict within and the Escalating War between the Sex Chromosomes
- Proteome-Wide Analysis of Disease-Associated SNPs That Show Allele-Specific Transcription Factor Binding
- Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes and as Potential Breast Cancer Susceptibility Alleles
- A Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in
- Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese Subjects
- Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in Rice
- Co-Evolution of Mitochondrial tRNA Import and Codon Usage Determines Translational Efficiency in the Green Alga
- SIRT6/7 Homolog SIR-2.4 Promotes DAF-16 Relocalization and Function during Stress
- CNV Formation in Mouse Embryonic Stem Cells Occurs in the Absence of Xrcc4-Dependent Nonhomologous End Joining
- Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System in
- Citrullination of Histone H3 Interferes with HP1-Mediated Transcriptional Repression
- Variation in Genes Related to Cochlear Biology Is Strongly Associated with Adult-Onset Deafness in Border Collies
- The Long Non-Coding RNA Affects Chromatin Conformation and Expression of , but Does Not Regulate Its Imprinting in the Developing Heart
- Rif2 Promotes a Telomere Fold-Back Structure through Rpd3L Recruitment in Budding Yeast
- Is a Metastasis Susceptibility Gene That Suppresses Metastasis by Modifying Tumor Interaction with the Cell-Mediated Immunity
- The p38/MK2-Driven Exchange between Tristetraprolin and HuR Regulates AU–Rich Element–Dependent Translation
- Rare Copy Number Variants Contribute to Congenital Left-Sided Heart Disease
- A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse
- An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural Crest
- Characterization of Inducible Models of Tay-Sachs and Related Disease
- Hominoid-Specific Protein-Coding Genes Originating from Long Non-Coding RNAs
- Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24
- Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Virulence
- Convergence of the Transcriptional Responses to Heat Shock and Singlet Oxygen Stresses
- Genomics of Adaptation during Experimental Evolution of the Opportunistic Pathogen
- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Vsx2 Controls Eye Organogenesis and Retinal Progenitor Identity Via Homeodomain and Non-Homeodomain Residues Required for High Affinity DNA Binding
- The Long Path from QTL to Gene
- TCF7L2 Modulates Glucose Homeostasis by Regulating CREB- and FoxO1-Dependent Transcriptional Pathway in the Liver
- The Non-Flagellar Type III Secretion System Evolved from the Bacterial Flagellum and Diversified into Host-Cell Adapted Systems
- Complex Chromosomal Rearrangements Mediated by Break-Induced Replication Involve Structure-Selective Endonucleases
- Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-A in Fission Yeast
- A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of Bacteria
- Determinants of Human Adipose Tissue Gene Expression: Impact of Diet, Sex, Metabolic Status, and Genetic Regulation
- Genome-Wide Association Studies Identify Heavy Metal ATPase3 as the Primary Determinant of Natural Variation in Leaf Cadmium in
- Tethering of the Conserved piggyBac Transposase Fusion Protein CSB-PGBD3 to Chromosomal AP-1 Proteins Regulates Expression of Nearby Genes in Humans
- A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- H4K20me1 Contributes to Downregulation of X-Linked Genes for Dosage Compensation
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Coevolution within and between Regulatory Loci Can Preserve Promoter Function Despite Evolutionary Rate Acceleration
- New Susceptibility Loci Associated with Kidney Disease in Type 1 Diabetes
- SWI/SNF-Like Chromatin Remodeling Factor Fun30 Supports Point Centromere Function in
- A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
- Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription
- Genetic Modifiers of Chromatin Acetylation Antagonize the Reprogramming of Epi-Polymorphisms
- UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development
- A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



