-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24
Elucidation of the biological role of linker histone (H1) and heterochromatin protein 1 (HP1) in mammals has been difficult owing to the existence of a least 11 distinct H1 and three HP1 subtypes in mice. Caenorhabditis elegans possesses two HP1 homologues (HPL-1 and HPL-2) and eight H1 variants. Remarkably, one of eight H1 variants, HIS-24, is important for C. elegans development. Therefore we decided to analyse in parallel the transcriptional profiles of HIS-24, HPL-1/-2 deficient animals, and their phenotype, since hpl-1, hpl-2, and his-24 deficient nematodes are viable. Global transcriptional analysis of the double and triple mutants revealed that HPL proteins and HIS-24 play gene-specific roles, rather than a general repressive function. We showed that HIS-24 acts synergistically with HPL to allow normal reproduction, somatic gonad development, and vulval cell fate decision. Furthermore, the hpl-2; his-24 double mutant animals displayed abnormal development of the male tail and ectopic expression of C. elegans HOM-C/Hox genes (egl-5 and mab-5), which are involved in the developmental patterning of male mating structures. We found that HPL-2 and the methylated form of HIS-24 specifically interact with the histone H3 K27 region in the trimethylated state, and HIS-24 associates with the egl-5 and mab-5 genes. Our results establish the interplay between HPL-1/-2 and HIS-24 proteins in the regulation of positional identity in C. elegans males.
Published in the journal: Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24. PLoS Genet 8(9): e32767. doi:10.1371/journal.pgen.1002940
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002940Summary
Elucidation of the biological role of linker histone (H1) and heterochromatin protein 1 (HP1) in mammals has been difficult owing to the existence of a least 11 distinct H1 and three HP1 subtypes in mice. Caenorhabditis elegans possesses two HP1 homologues (HPL-1 and HPL-2) and eight H1 variants. Remarkably, one of eight H1 variants, HIS-24, is important for C. elegans development. Therefore we decided to analyse in parallel the transcriptional profiles of HIS-24, HPL-1/-2 deficient animals, and their phenotype, since hpl-1, hpl-2, and his-24 deficient nematodes are viable. Global transcriptional analysis of the double and triple mutants revealed that HPL proteins and HIS-24 play gene-specific roles, rather than a general repressive function. We showed that HIS-24 acts synergistically with HPL to allow normal reproduction, somatic gonad development, and vulval cell fate decision. Furthermore, the hpl-2; his-24 double mutant animals displayed abnormal development of the male tail and ectopic expression of C. elegans HOM-C/Hox genes (egl-5 and mab-5), which are involved in the developmental patterning of male mating structures. We found that HPL-2 and the methylated form of HIS-24 specifically interact with the histone H3 K27 region in the trimethylated state, and HIS-24 associates with the egl-5 and mab-5 genes. Our results establish the interplay between HPL-1/-2 and HIS-24 proteins in the regulation of positional identity in C. elegans males.
Introduction
Linker histone H1 and heterochromatin protein HP1 are involved in numerous processes ranging from stabilizing heterochromatin condensation to the regulation of gene expression [1]–[5]. As has been reported, a methylation mark on vertebrate histone H1 is specifically recognized by the chromodomain of HP1. However, the exact biological role of HP1 binding to linker histone has not been determined [6].
The functions of HP1 and H1 proteins are mainly dependent on the cell type in which particular variants are expressed. Although the number of H1 (11) and HP1 variants (3) presents difficulties in studying the effect of H1 and HP1 depletion in mice, some data has emerged [3], [7]–[10]. For example, loss of HP1β results in defective development of neuromuscular junctions and the cerebral cortex [10], whereas depletion of three of eleven H1 genes causes lethality connected with a very broad range of defects in mice [11]–[12]. In ES cells, the lack of three somatic H1 variants leads to changes in nucleosome spacing and local chromatin compaction, and this is correlated with decreased levels of H3K27 trimethylation [11]. Additionally, H1 is necessary to establish and maintain the DNA methylation pattern in a subset of genes including the reproductive homeobox (Rhox) gene cluster [13].
C. elegans possesses eight linker histone variants and two HP1 homologues, HPL-1 and HPL-2 [14]–[16]. Mutation of hpl-2 results in defective vulval and germline development at elevated temperatures [15]–[17]. hpl-1, in contrast to hpl-2, does not have visible effects on C. elegans development at different temperatures, however, hpl-1 acts redundantly with hpl-2 to control larval development, somatic gonad development and vulval cell fate determination [17]. Our previous study revealed that HPL-1 recognizes the linker histone variant HIS-24 when it is mono-methylated at lysine 14 (HIS-24K14me1), similar to the situation in vertebrates [16]. Additionally, we showed that HIS-24 interacts with H3K27me3 [18]. The H3K27me3 modification correlates with a repressive chromatin state that inhibits expression of many developmentally regulated genes. This is consistent with studies of Hox loci demonstrating that enrichment of H3K27me3 recruits the binding of Polycomb group proteins (PcG) [19].
The Hox genes encode conserved homeodomain-containing transcription factors that control the positional identities of cells along the anterior–posterior axis [20]–[21]. The expression pattern of Hox genes appears to be regulated by two evolutionarily conserved PcG complexes, the ESC/E(Z) complex and the PRC1 complex. Both have been identified in flies and mammals and are linked to modulation of repressive chromatin structures [21]. The C. elegans Hox cluster consisting of lin-39, ceh-13, mab-5 and egl-5 (orthologs of Drosophila Scr, labial, ftz and Abd-B, respectively) is quite degenerated in comparison to Hox clusters in other species [22] but, as in mammals, is also globally repressed by Polycomb group (PcG) proteins [20], [23]. Mutations in mes-2 and mes-6, which encode the C. elegans ESC/E(Z) complex, result in ectopic expression of Hox genes [23]. A similar phenotype has also been observed in the absence of sop-2 or sor-1 genes. SOP-2 and SOR-1 form another C. elegans PcG-like complex which shares many structural and functional properties with the Drosophila PRC1, and is involved in the global repression of Hox gene expression. Loss of sop-2 and sor-1 results in gross homeotic transformations [24]–[25].
To elucidate the function of H1 and HP1 related proteins in C. elegans, we decided to generate double and triple mutants, since hpl-1, hpl-2 and his-24 deficient nematodes are viable, and since HIS-24K14me1 is recognized by HPL-1 [16]–[17], [26]. We performed global transcriptional analyses of single, double and triple mutant animals, and we found that HPL-1/-2 and HIS-24 regulate a relatively small number of genes. We provide evidence that the methylated form of HIS-24 (HIS-24K14me1) and HPL-2 are involved in the regulation of mab-5 and egl-5 expression by binding to H3K27me3, although HIS-24K14me1 does not interact with HPL-2 [16]. Furthermore, we observed that HIS-24 and HPL-2 act in parallel pathway as MES (PcG) proteins, and loss of their activity causes defects of male tail structures. Overall, our data suggest a common and dual role for C. elegans H1 and HP1, functioning both as chromatin architectural proteins and at the same time as modifiers of a small subset of genes. Furthermore, we provide the first direct evidence for redundant functions of H1 and HP1 in metazoan development.
Results
HP1 and HIS-24 are not global repressors of transcriptional activity in C. elegans
C. elegans contains two related HP1 proteins (HPL) and eight linker histone variants [14]–[15]. Only one of the eight linker histone variants, HIS-24 is important for germline development, with its absence resulting in reduced fertility and de-repression of extrachromosomal transgenic arrays in the germline [14]. As we previously reported, the absence of HIS-24 did not affect protein levels of the other histone variants, in contrast to the mammalian H1 subtypes which are sufficient to compensate for the loss of a single linker histone [7], [16]. Furthermore, we showed that C. elegans heterochromatin protein 1 variant, HPL-1 recognizes and binds the methylated form of HIS-24 [16]. Given the physical interaction of HPL-1 with HIS-24 mono-methylated at lysine 14 and their role in chromatin silencing and germline developmental processes [15]–[17], we decided to study HPL and HIS-24 function in transcriptional regulation in C. elegans. It was of great interest to determine how the HPL subtypes (HPL-1 and HPL-2) and HIS-24 affect gene expression. To determine the contribution of HIS-24 and HPL-1/-2 to the control of gene transcription, we compared the gene-expression profiles of single null mutations in the hpl-1, his-24 and hpl-2 as well as profiles of hpl-1his-24, and hpl-2; his-24 double, and hpl-2; hpl-1his-24 triple mutant animals in L4 larval stages grown at 21°C. We decided to use L4 larval stages because HIS-24 is the most abundant linker histone H1 variant at this stage according to mass spectrometry-based protein expression data (Figure 1).
Fig. 1. C. elegans H1 proteomics. 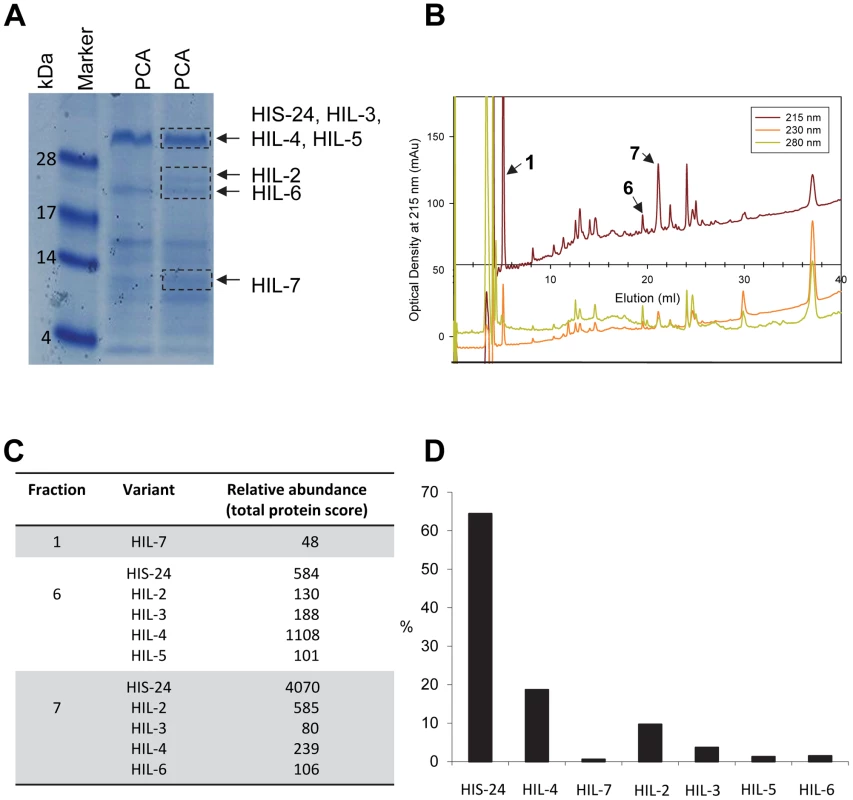
(A) L4 larval stage of wild type animals were extracted with 5% PCA. The PCA extract was chromatographed on C18 column. (B) Fractions 1, 6, and 7 containing H1 were analyzed. (C) Absolute protein abundances of linker histone variants were calculated from spectral counts. The table shows the relative abundance of the variants on the basis on their protein scores (Mascot). The total protein score corresponds directly to the protein abundance, because the histones have similar masses. HIL-1 (H1.X) was found only in an HCl extract (data not shown). (D) Abundance of C. elegans H1 variants in percent (%). By microarray we observed very few changes in the gene expression profiles of either single, double, or triple mutants when compared with wild type animals at L4 larval stages. Among the 16,278 target probe sets assayed, we identified only modest changes in expression of just a small number of genes (Figure 2A–2H, Table 1). The majority of genes exhibiting changes were upregulated (6.5%) in the absence of the three heterochromatin components HIS-24, HPL-1 and HPL-2, in contrast to 3.7% downregulated genes from a total of 16,278 genes analyzed (FDR<0.05) suggesting that HPL-1/-2 and HIS-24 are not global repressors of transcriptional activity (Table 1). The deletion of both hpl-1 and hpl-2 genes caused up-regulation of 4.5% genes and downregulation of 2.1% of a total 16,278 genes when compared to wild type (WT) animals.
Fig. 2. HPL and HIS-24 are not global repressors of transcriptional activity. 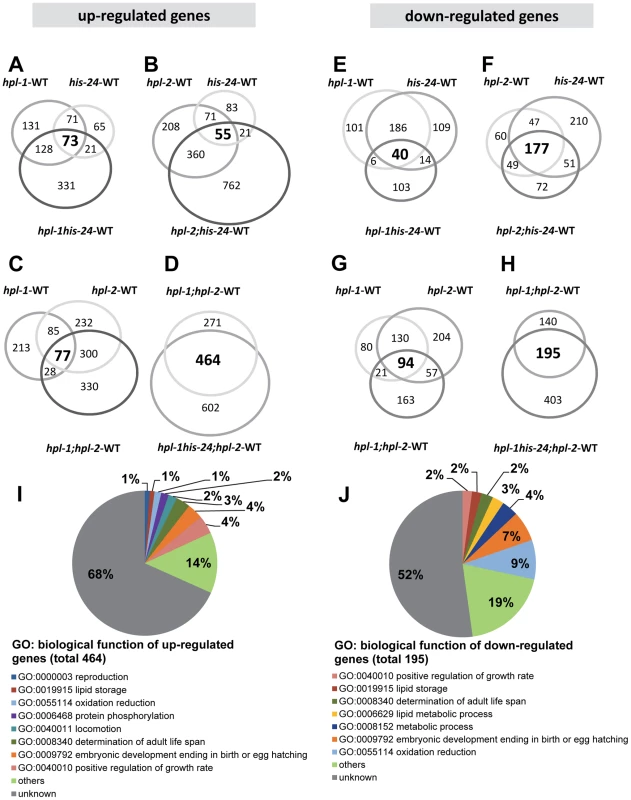
(A–H) Venn diagrams on the basis of whole genome microarray (16278 genes) showing the extent of overlap among genes that are associated with HIS-24 and HPLs activity. Values for fold change are the average from 2 independent biological replicates (FDR<0.05; |log2-fold change|>1.5). WT-wild type; significant overlapping among the candidate lists - p-value<0.000001 for all pair-wise comparisons by hypergeometric tests. (I, J) Functional clusters of HPL-1, HPL-2 and HIS-24 co-regulated genes in C. elegans. The molecular functions and the percentage of the total for each group are indicated. Gene Ontology (GO) terms taken from WormBase (http://www.wormbase.org). Tab. 1. Summary of agilent microarray analysis of gene expression in mutant animals versus wild type (16, 278 genes analysed). 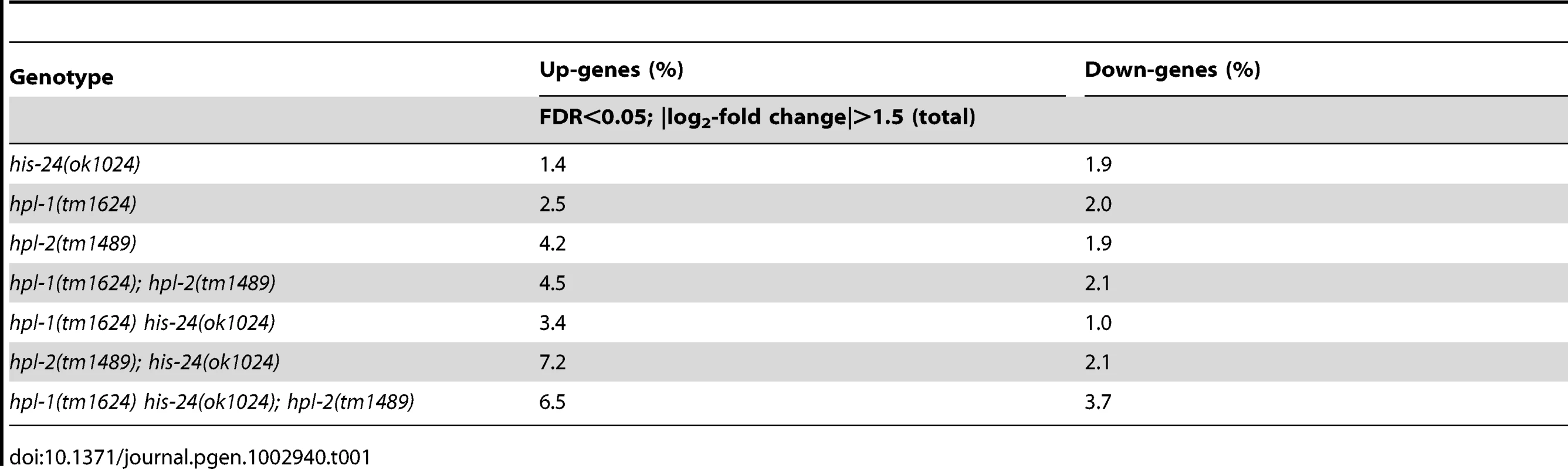
As previously reported, HPL-2 binds to HIS-24K14me1 through its association with HPL-1, and the heterochromatin proteins HPL-1 and HPL-2 play redundant roles in C. elegans development [16]–[17]. Considering these observations we compared transcriptional profiles between hpl-2 (tm1489); hpl-1(tm1624) double mutants and hpl-2 (tm1489); hpl-1(tm1624) his-24(ok1024) triple mutant animals. We found that 464 up-regulated (2.9% of 16,278) and 195 down-regulated (1.2% of 16,278) genes were commonly affected (FDR<0.05; p-value<0.000001 for all pair-wise comparisons by hypergeometric tests) (Figure 2, Table S1). Among the 464 up-regulated genes we identified some significantly enriched in GO terms associated with growth regulation (Fisher exact test (FET) P = 4×10−6), determination of adult life span (FET P = 2×10−6), locomotion (FET P = 0,003), protein phosphorylation (FET P = 0,04), reproduction (FET P = 0,05) and lipid storage (FET P = 0,05). The 195 genes that are down-regulated are enriched in GO terms associated with oxidation reduction (FET P = 0,003), embryonic development (FET P = 0,002) and metabolic process (FET P = 0,04). We identified common response proteins including heat shock proteins (HSP-12.3, -12.6, -16.2 and -17), enzymes (cytochromes) of the P450 family involved in protection against toxins (CYP-13A12, CYP-33C4, CYP-33D3, CYP-34A2, CYP-34A4 and CYP-34A9), metabolic enzymes such as the fatty acid-coenzyme A (CoA) synthetase ACS-1 and the fatty acid/retinol binding proteins FAR-5, -7 (Table S1). Furthermore, we observed the induction of oxidative stress proteins such as glutathione S-transferases (GST) and genes commonly associated with increased stress resistance – for example, the mitochondrial sod-3 superoxide dismutase gene (Table S1).
In conclusion, deletion of the different HPL variants and HIS-24 caused an alteration in the expression of a limited number of genes, different in each HPL variant and HIS-24. Most of the genes are affected by a single HPL variant and HIS-24, supporting the theory that HPL isoforms or HIS-24 play specific roles in gene expression. Nonetheless, a proportion of genes are altered by more than one HPL variant as well as HIS-24, suggesting redundant roles for HIS-24 and HPL variants, and for HPL-1/-2 may also exist.
HIS-24 acts synergistically with HPL proteins to allow normal reproduction, somatic gonad development, and vulval cell fate decisions
In parallel to microarray analysis we investigated the biological role of HIS-24 and HPL proteins in C. elegans. For morphological defects we scored hpl-1(tm1624) his-24(ok1024), and hpl-2(tm1489); his-24(ok1024) double mutants as well as hpl-2(tm1489); hpl-1(tm1624) his-24(ok1024) triple mutant animals. In particular, we focused on germline nuclei morphology, hermaphrodite vulval development and the somatic patterning of the male tail since these tissues are known to be affected by mutations in chromatin factors, and HPL-2 influences vulval cell fate specification in the synMuv (synthetic multivulva) pathway [14]–[15], [27]. We found that the deletion of hpl-2(tm1489) together with his-24(ok1024) results in synergistic non-lethal defects of vulval cell fate specification (everted vulva, multivulva) and sterility at 21°C, and at 25°C (Table 2). While the observed phenotypic effects at 21°C were minor in contrast to the situation at 25°C, it is tempting to speculate that the effects can be also modulated through unknown mechanisms, environmental cues (temperature), which in itself may also lead to significant side-effects. Additionally, decreased brood sizes were observed in hpl-2(tm1489); his-24(ok1024) double and hpl-2(tm1489); hpl-1(tm1624) his-24(ok1024) triple mutant animals grown at 21°C (Figure 3). The brood size of the hpl-2(tm1489); his-24(ok1024) was strongly decreased by 35% of wild type worms, and was further decreased to about 50% in the hpl-2(tm1489); hpl-1(tm1624) his-24(ok1024) triple mutant animals (Figure 3). These results were consistent with our microarray data analysis that revealed differential expression of genes involved in the embryonic development or reproduction (Table S1). Furthermore, we observed several defects in the morphology of the somatic gonad of hpl-1(tm1624) his-24(ok1024) double mutant animals grown at 21°C. In wild type, single mutant and hpl-2(tm1489); his-24(ok1024) double mutant the gonad arms form an U-shaped structure (Figure 4A–4D). In contrast, in the double mutant hpl-1(tm1624) his-24(ok1024) 25% of gonad arms (161 of 642) form a loop (Figure 4E). These results suggest that both proteins HIS-24 and HPL-1 are involved in the somatic gonad development whereas HIS-24 and HPL-2 influence vulva cell fate specification and reproduction (Table 2).
Fig. 3. HIS-24 act synergistically with the hpl-genes to control brood size at 21°C. 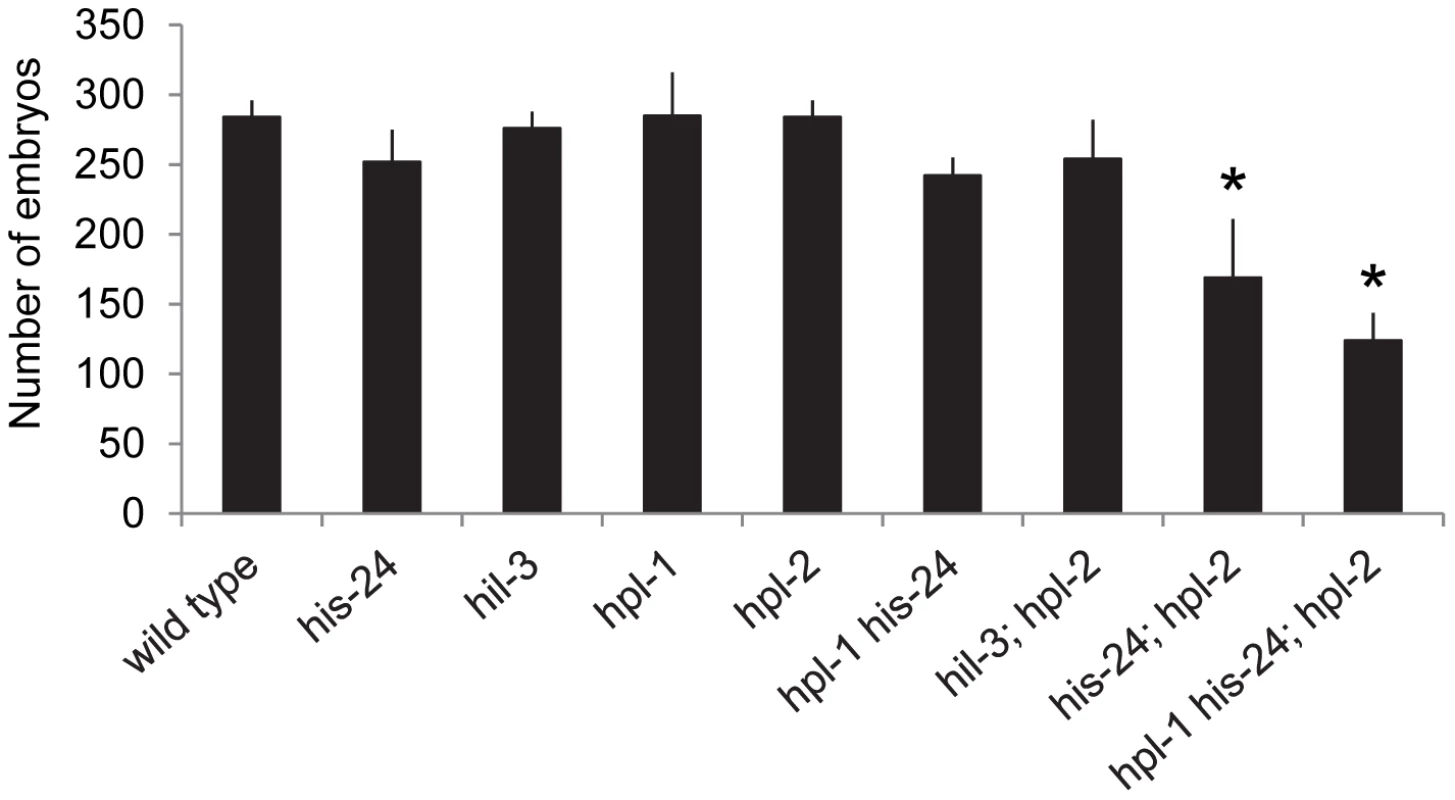
Number of embryos (± SEM), 20 animals scored: * p<0.0001, vs. wild type. Fig. 4. HIS-24 and HPL-1/-2 depletion results in abnormal somatic gonad development and reduction of chromatin compaction. 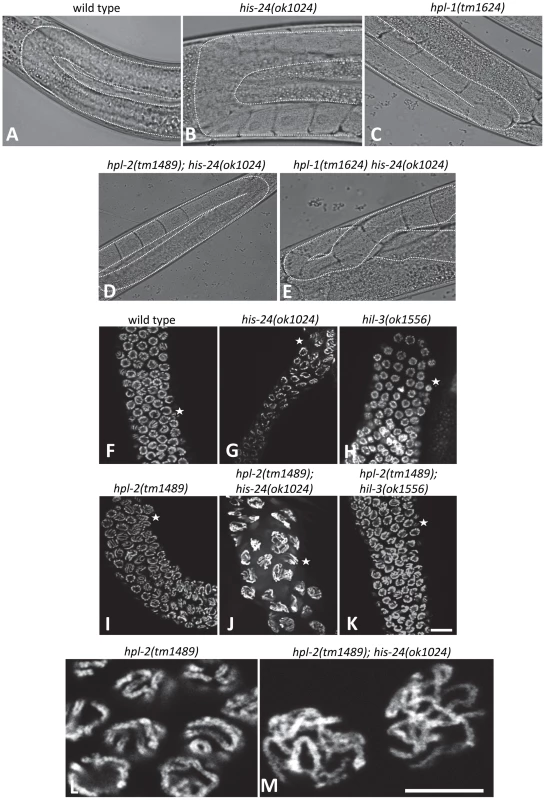
(A–D) Normal U-shaped form of gonad arm of wild type, his-24 or hpl-1 single, and hpl-2; his-24 double mutants. (E) hpl-1his-24 mutant animal demonstrates aberrant loop form of gonad arm. (F–K) DAPI stained gonad arms of his-24, hil-3 or hpl-2 single and hpl-2; his-24 or hpl-2; hil-3 double mutants. Stars point to pachytene stage germ nuclei. Scale bar: 10 µm. (L, M) Morphology of pachytene stage nuclei of the germ line of hpl-2 mutant animals compared to the nuclei of hpl-2; his-24 double mutants. Scale bar: 7.5 µm. Tab. 2. Genetic interaction of hpl-1, hpl-2, and his-24. 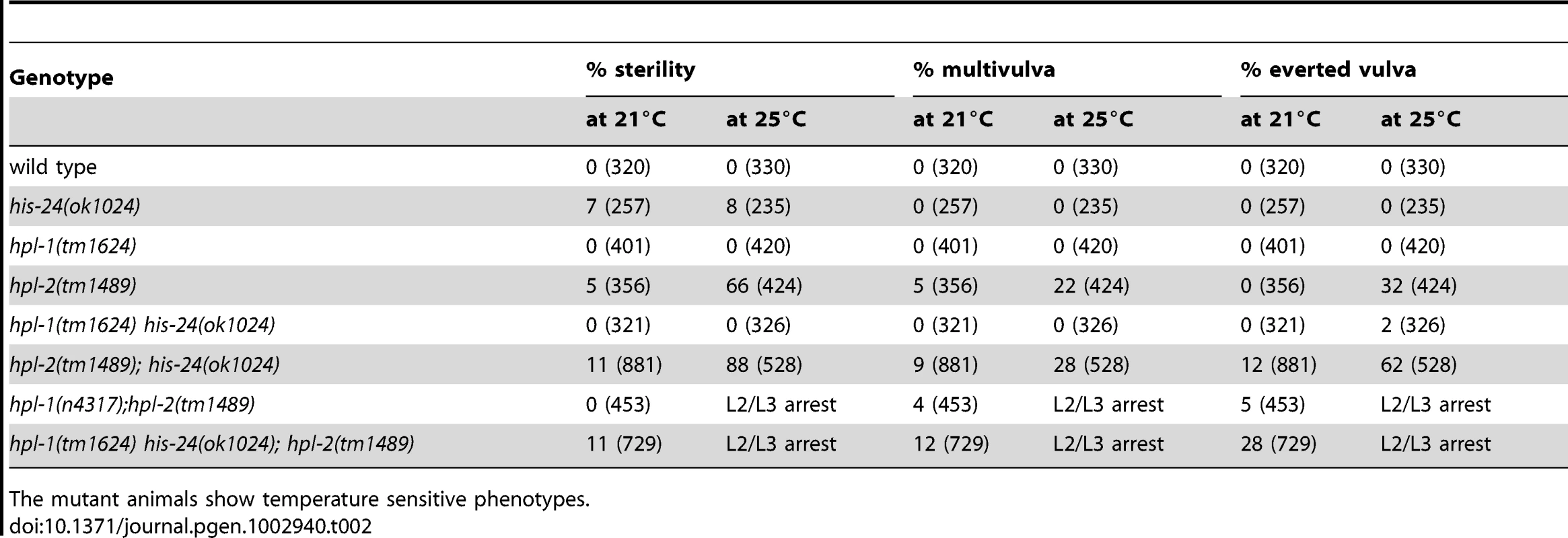
The mutant animals show temperature sensitive phenotypes. HIS-24 and HPL-2 are required for chromatin compaction
Since HPL-2 and HIS-24 are required for germline development and for the chromatin based germline-specific silencing mechanism [14]–[15], [26], we asked whether they influence the structure of nuclei. In-depth analysis revealed that the germline nuclei of hpl-2(tm1489); his-24(ok1024) double mutants differ in size and morphology when compared to single mutants or to wild type worms grown at 21°C (Figure 4F–4I, 4L). The observed chromatin of 86% of gonad arms (36 of 42) had a more open, relaxed structure suggesting that HIS-24 and HPL-2 play a function in chromatin condensation in the germline (Figure 4J, 4M). To assess the specific requirements for HIS-24 among the H1 isoforms, we also tested hpl-2(tm1489);hil-3(ok1556) double mutant strain to determine if the observed changes in the chromatin compaction is linker histone variant specific (Figure 4K). As shown, loss of hpl-2 and linker histone variant hil-3 did not cause defects in chromatin compaction in contrast to hpl-2; his-24 strain. In addition, we also did not observe involvement of HPL-2 and HIL-3 on brood size (Figure 3).
To determine if the loss of HIS-24 and HPL proteins also influence chromatin histone modifications as well as core histone H3 level, we performed western blot analysis of mutant animals. No gross changes were observed in the methylation and core histone H3 levels using antibodies directed against H3K9me3, H3K27me3, and H3 (Figure S1). In addition, we did not detect changes in chromatin modification marks on a cellular level by immunofluorescence (data not shown) indicating that the observed effects of chromatin compaction are not correlated with alterations of histone modifications in hpl-2(tm1489); his-24(ok1024) double mutant animals.
Mutations in his-24 and hpl-2 cause male tail defects
Loss of hpl-1, -2 and his-24 function results in changes of transcriptional regulation of genes encoding nuclear hormone receptor family genes (nhr-60, nhr-156), transcription factors (miz-1, zip-3, zip-8, madf-2), homeobox ceh-82 and homeodomain lim-7 genes (Table S1). Moreover, hpl-2 regulates lin-39 Hox gene expression in vulval precursor cells (VPCs) [27]. Therefore we tested whether hpl-1, -2 and his-24 genes are involved in the regulation of Hox gene expression during the somatic patterning of the male tail.
The wild type male tail possesses nine pairs of bilateral sensory rays that function in locating and mating with hermaphrodites. Normally, the posterior hypodermal blast cells V5 and V6 produce six pairs of rays (ray 1 to ray 6), while the blast cell T gives rise to the three rays (rays 7–9) [23]–[25]. We found that mutations in both his-24 and hpl-2 (37%, 51 of 73 males with defected rays) as well as in his-24, hpl-1 and hpl-2 (83%, 76 of 107 males with defected rays) cause abnormalities in patterning of blast cells V that result in fused and atypical (under-developed) rays, while the single and hpl-1; hpl-2 and hpl-1 his-24 double mutations have normal development of rays (Table S2, Figure 5A–5E, 5G). Although hpl-1 mutation alone or in combination with his-24 or hpl-2 had no visible effect on the male tail at 21°C (Figure 5C, 5G–5H), it appeared to be partially redundant in combination with hpl-2 and his-24 double mutations. As Figure 5J and Table S2 show, the number of under-developed rays is significantly increased (up to 42%, 39 of 107 males) in the hpl-2(tm1489); hpl-1(tm1624) his-24(ok1024) triple mutant compared to the hpl-2(tm1489); his-24(ok1024) double mutant males (13%, 18 of 73 males) (Figure 5I). This synergism suggests that hpl-1 only in combination with his-24 and hpl-2 plays functions in the patterning of the male tail. We also tested hil-3; hpl-2 double mutant animals for the mail tail phenotype. We did not observe any defects in the patterning of the male tail of hil-3; hpl-2 double mutant animals in contrast to hpl-2; his-24 animals suggesting that HIS-24 (in combination with HPL-2) specifically affects the patterning of the mating structures in C. elegans (Figure 5F–5I).
Fig. 5. his-24 and hpl-2 genes are involved in proper postembryonic development of male-specific structures. 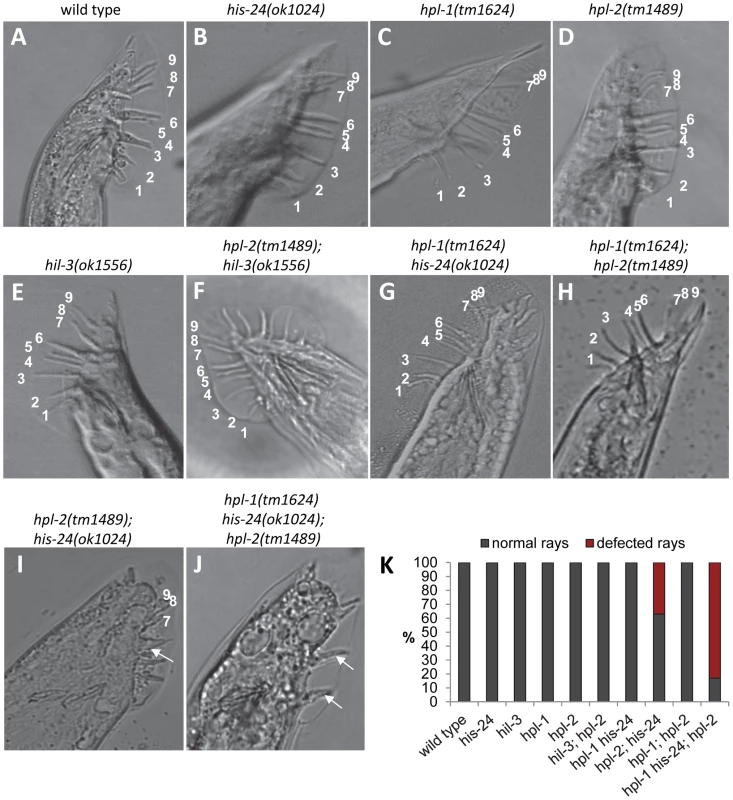
(A–F, I–J) All nine rays are formed normally in his-24, hil-3, hpl-1 or hpl-2 single mutants and in hpl-1his-24 as well as in hpl-1; hpl-2 and hpl-2; hil-3 double mutant males. (G, H) Abnormal male tails of hpl-2; his-24 and hpl-2; hpl-1his-24 mutant animals. Arrows point to ray fusions. (K) Quantification of ray defects associated with single, hpl-2; his-24 double and hpl-1his-24; hpl-2 triple mutations. The animals were growing on him-14 (high incidence of males) feeding plates at 21°C. HIS-24 and HPL-2 are required for inhibiting the ectopic expression of mab-5 and egl-5 Hox genes
In agreement with previous observations we analyzed the ability of his-24, hpl-1 and hpl-2 genes to regulate mab-5 and egl-5 expression [28]–[29]. Interestingly, these two Hox genes are required for V ray development [23] and mab-5 was slightly up-regulated in our microarray analysis of hpl-2(tm1489); hpl-1(tm1624) his-24(ok1024) mutant animals (data not shown).
We compared expression of egl-5::gfp and mab-5::gfp reporter genes in wild type animals and in combination with his-24(ok1024), hpl-1(tm1624) and hpl-2(tm1489) background mutations (Figure 6). We observed that mab-5::gfp reporter is ectopically expressed in approximately 30% of early L3 stage of hpl-2(tm1489); his-24(ok1024) double mutant males scored (n = 100) (Figure 6A, 6B). Altered expression of this reporter was also detected in adult males. Similarly, about 80% of L3 stage of hpl-2(tm1489); his-24(ok1024) double mutant males (n = 100) displayed ectopic expression of EGL-5::GFP protein in two daughters of ray precursors anterior to R4, R5 and R6 sublineages (Figure 6C–6E). We did not observe any significant enhancement of the ectopic expression of mab-5 and egl-5 Hox genes in hpl-1 depleted hpl-2(tm1489); his-24(ok1024) double mutant animals. For the crossing with mab-5::gfp transgenic strain we did not use triple mutant animals due to hpl-1 his-24; hpl-2 phenotype (sterile worms, worms with everted vulva or multivulva; Table 2).
Fig. 6. mab-5 and egl-5 Hox genes are ectopically expressed in hpl-2; his-24 mutants. 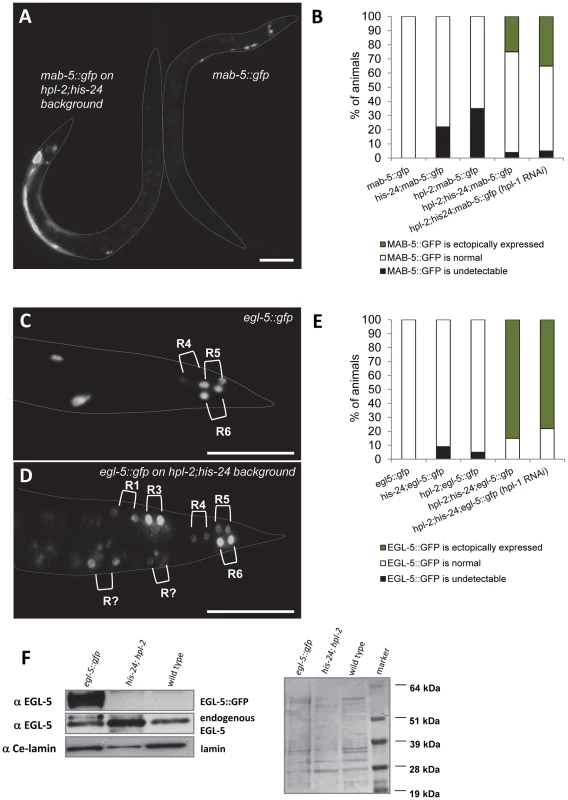
(A) A hpl-2; his-24 early L3 mutant male ectopically expresses mab-5::gfp in hypodermal syncytium cell ((hyp7); animal on the left side). Expression of mab-5::gfp in a wild type early L3 male marks very few cells at the posterior (animal on the right side). Scale bar: 25 µm. (B) Quantification of progeny of single and double hpl-2; his-24 mutant hermaphrodites with males carrying mab-5::gfp reporter versus wild type males. (C, D) In a wild type L3 male, expression of egl-5::gfp is limited to the daughters of the ray precursor cells R4, R5 and R6 which give rise to rays 3–6. In hpl-2; his-24 L3 mutant male the reporter is expressed in additional ray sublineages. Scale bar: 25 µm. (E) Quantification of progeny of single and double hpl-2; his-24 mutant hermaphrodites with males carrying egl-5::gfp reporter versus wild type males. (F) Western blot of protein extracts (150 males) from C. elegans wild type, hpl-2; his-24 double mutant and egl-5 transgenic strain probed with antibody raised against EGL-5. Anti-EGL-5 recognized the EGL-5::GFP fusion protein and endogenous EGL-5 in egl-5::gfp transgenic strain. Protein loading was confirmed by probing with an anti-Ce-lamin antibody and western blot stained with Ponceau S. To verify HPL-1 depletion directly and to examine the extent of HPL-1 knockdown we tested the hpl-1 depleted hpl-2; his-24 mutant animals for presence of HPL-1 on the western blot. We found that hpl-1RNAi strongly reduces HPL-1 level compared to the controls (Figure S2).
Since mutations in hpl-2 and his-24 affect transgene expression in C. elegans [14]–[15] we assessed the expression level of the endogenous EGL-5 in hpl-2; his-24 double mutant males. Western blot of hpl-2; his-24 double mutant males probed with EGL-5 antibody revealed an increased level of endogenous EGL-5 protein of predicted size (26 kDa) compared to EGL-5 level of wild type C. elegans and egl-5::gfp transgenic line (Figure 6F) [29]. Altogether, these results suggest that HIS-24 and HPL-2 silence the Hox gene cluster, either by general repression of the transcriptional activity, or through a specific biochemical and structural function in Hox gene silencing.
HIS-24 binds to egl-5 and mab-5 loci
Since HIS-24 and HPL-2 are required for inhibiting the ectopic expression of mab-5 and egl-5 Hox genes, we tested if HIS-24 and HPL-2 bind directly to their promoters in vivo and therefore regulate egl-5 and mab-5 transcription. The primer sets used for quantitative ChIP-PCR (qChIP-PCR) analysis were directed to the promoters, introns and 3′UTR regions of mab-5 and egl-5 genes. Remarkably, mab-5 and egl-5 are tightly clustered on chromosome III, suggesting that chromatin structure coordinately regulates the expression of these genes (Figure 7A). qChIP-PCR analysis revealed that HIS-24 is indeed associated with the promoters and introns of mab-5 and egl-5 genes (Figure 7B). In contrast, we did not see any HIS-24 binding to 3′UTR regions (Figure 7B). However they are occupied by H3 (Figure 7D). As shown, the anti-HIS-24 antibody binds with higher affinity to egl-5 and mab-5 genes than the anti-HIL-4 antibody, which is cross-reactive to C. elegans linker histone variants [14] (Figure 7C). Next, to verify the specificity of the HIS-24 binding to Hox genes, we tested the HIS-24 binding to mab-5 gene ectopically expressed in sor-1 background mutation. As previously reported, SOR-1 (together with SOP-2) shares many structural and functional properties with the PRC1 complex, and is involved in the global repression of egl-5 or mab-5 Hox gene expression [25]. As shown, we detected a significantly decreased level of HIS-24 at this region compared to the situation in wild type animals, implicating that HIS-24 enables mab-5 transcriptional repression, thereby influencing its expression (Figure 7D). Additionally, we observed lower levels of histone H3 occupancy at the mab-5 promoter in sor-1 background mutation than in wild type animals, suggesting that the difference in H3 levels could be due to the nucleosome free region that forms at high levels of expression (Figure 7D). In addition, mab-5 promoter and intron regions in the his-24 mutant animals showed decreased enrichment of the histone H3 than in wild type animals, suggesting that binding of H3 and HIS-24 can be positively correlated at regulatory regions. In comparison, the H3 changes at 3′UTR region of mab-5 in sor-1 and his-24 background mutation were relatively mild than in wild type animals (Figure 7D).
Fig. 7. HIS-24 associates with promoters of mab-5 and egl-5 genes. 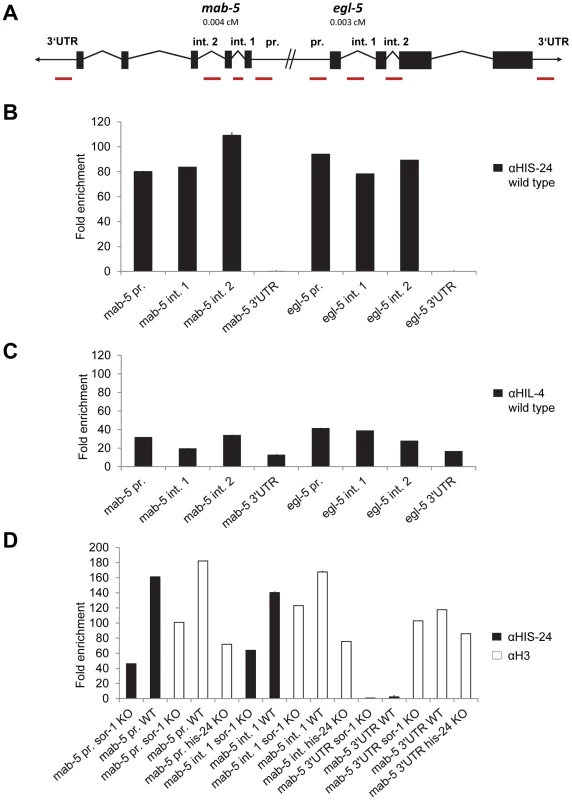
(A) mab-5 and egl-5 genes are tightly clustered on chromosome III. Primer sets for qChIP-PCR are directed to the mab-5 and egl-5 promoters, introns and 3′UTR (red bars). (B) qChIP-PCR assay determining HIS-24 occupancy at the mab-5, and egl-5 genes. The results were normalized to binding by anti-HIS-24 antibody in his-24 mutant strains and performed in triplicates. Error bars indicate ±SD (see Table S3). (C) qChIP using anti-HIL-4 antibody and total protein isolated from wild type worms. (D) Decreased level of HIS-24 enrichment at mab-5 using anti-HIS-24 antibody and mab-5::gfp transgenic strain in sor-1 background mutation (KO) as well as wild type (WT) animals. H3 occupancy at mab-5 loci is affected in sor-1 and his-24 background. In sor-1 and his-24 background mab-5 is ectopically expressed in contrast to repressed mab-5 in wild type animals. (C, D) All results were normalized to binding by control IgG antibody and performed in triplicates. Error bars indicate ±SD (see Table S3). Unfortunately, we have failed so far to detect HPL-2 at this region using direct ChIP approach.
HIS-24 and HPL-2 act in parallel pathway as MES-2/-3
Hox genes are transcriptionally repressed by the evolutionally conserved Polycomb group (PcG) proteins through the H3K27me3 mark in a lineage specific fashion [30]–[31]. In Drosophila, a member of the Polycomb group (PcG), the H3K27 histone methyltransferase E(Z) has been identified as a stable repressor of Hox genes [32]. In C. elegans, orthologs of the PcG chromatin repressors E(Z) and ESC, namely MES-2 and MES-6 influence expression of Hox genes and male tail development [23]. Since Polycomb group (PcG) proteins (MES-2/3/6 complex) are involved in the repression of Hox genes, we performed genetic epistasis analysis of mes-2 - and mes-3-depleted triple mutant animals [23], [33]. Interestingly, hpl-2(tm1489); his-24(ok1024) double as well as hpl-2(tm1489); hpl-1(tm1624) his-24(ok1024) triple mutant males on mes-2 or mes-3 feeding plates showed an increased number of ectopic rays (∼2-fold) and defective rays in comparison to mes-3 - or mes-2 - depleted double mutant males (Figure 8A, 8C; Table S2). As shown, loss of HPL and HIS-24 together with depletion of mes-2 or mes-3 resulted in additive defects implying that HPL and HIS-24 act in parallel pathway as MES-2 or MES-3.
Fig. 8. MES-2 and MES-3 enhances the number of defected rays in animals lacking hpl-2 and his-24. 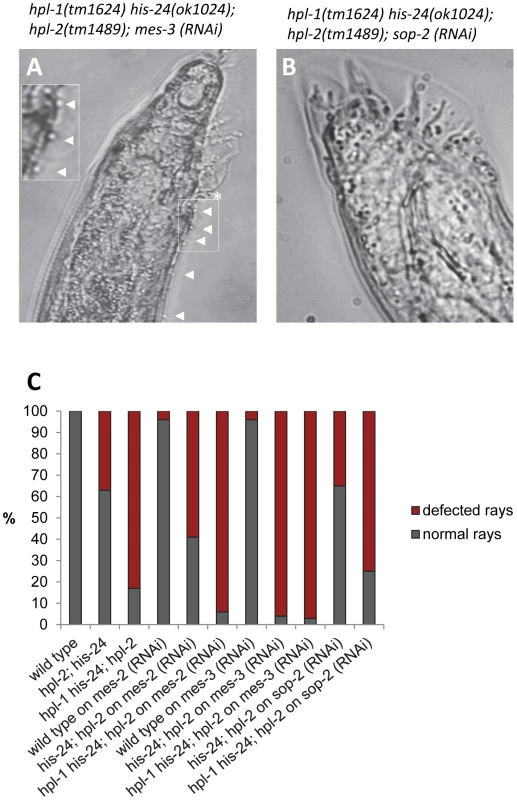
(A) RNA interference (RNAi) of mes-3 leads to enhancement of hpl-2; hpl-1his-24 mutant male phenotype in contrast to (B) sop-2-depleted triple mutant animals. Ectopic rays are indicated with arrowheads and under-developed rays by a star. (C) Quantification of ray defects associated with hpl-2; his-24 double and hpl-2; hpl-1his-24 triple mutations. The animals were growing on him-14 and mes-2 or mes-3 as well as sop-2 feeding plates at 21°C. sop-2 deletion is epistatic to hpl-2; his-24 double mutation. We also phenotyped hpl-2(tm1489); his-24(ok1024) double and hpl-2(tm1489); hpl-1(tm1624) his-24(ok1024) triple mutant males on sop-2 feeding plates (Figure 8B, 8C, Table S2). As previously reported, SOP-2 forms a novel PcG-like complex that may function analogously to PRC1 in C. elegans and regulates expression of Hox genes [25]. Homologs of SOR-1 and SOP-2 are not found in other organisms, including even the very closely related C. briggsae suggesting a C. elegans specific mechanism on an essential global gene regulatory system [25]. Remarkably, we did not observe any influence of SOP-2 depletion in the hpl-2; his-24 double and hpl-2; hpl-1 his-24 triple mutant background suggesting that sop-2 appears to be epistatic to hpl-2; his-24 deletion.
HIS-24K14me1 and HPL-2 bind H3K27me3 chromatin mark
Recently, we have reported that HIS-24 specifically interacts with H3K27 trimethylated and H3K27 unmodified peptides [18]. While HPL-1 and HPL-2 were able to pull down native HIS-24K14me1, and HPL-2 failed to bind either modified or unmodified HIS-24 peptides in vitro, we asked whether HPL-2 and HIS-24K14me1 repress the transcription of egl-5 and mab-5 genes by binding to H3K27me3 [16], [18].
By peptide pull down assay (PD) we observed that HIS-24K14me1 interacts preferentially with H3K27me3 peptide when compared to the unmodified, mono - or di-methylated H3K27 peptides, and conversely, native H3K27me3 binds only the methylated form of HIS-24 peptide (Figure 9A). Furthermore, we found strong preference of HPL-2 for the trimethylated form of H3K27, as well as for H3K27me2 and H3K9me2/3 as previously reported (Figure 9A) [16]. No interaction was observed between H3K9me0/1 or H3K27me0/1. We confirmed the results obtained from peptide pull down (PD) by an immunoprecipitation (IP) approach using antibodies raised against different chromatin modification marks and lysates of wild type animals (Figure 9B). Additionally, we were able to pull down native H3K27me3 using a GFP-antibody directed against GFP-tagged HPL-2 and HIS-24 (Figure 9C). As a control we used GFP expressed protein under the his-24 promoter to demonstrate the specificity of HPL-2 and HIS-24 binding to H3K27me3 (Figure 9C). To confirm that HPL-2 and HIS-24 indeed display H3K27me3 binding, we expressed HPL-2 and HIS-24 in E. coli. We did not detect the interaction of HPL-2 with H3K27me3 in contrast to HIS-24, suggesting that additional factors (transcription factors, RNAi machinery, post-translational modifications of HPL-2) are involved in the mediation of HPL-2 binding to H3K27me3 (Figure 9D, 9F). In the case of HIS-24 we detected strong preference for H3K27me peptides apart from H3K27me1 (Figure 9D). The differences in the binding to H3K27me3 between bacterially expressed HIS-24 and native HIS-24 can be explained by the fact that bacterially expressed proteins are not methylated and only the methylated form of HIS-24 binds specifically the H3K27me3. Finally, to exclude that the binding of HPL-2 to H3K27me3 takes place via interaction with the C. elegans HIS-24, we repeated the pull downs using extracts obtained from his-24(ok1024) mutant animals (Figure 9E). We detected a preference of HPL-2 for H3K27me3 independently of HIS-24 however this binding was reduced compared to binding of HPL-2 to H3K27me3 in the presence of HIS-24 (Figure 9B, 9E).
Fig. 9. HIS-24K14me1 and HPL-2 bind to the H3K27me3 chromatin mark. 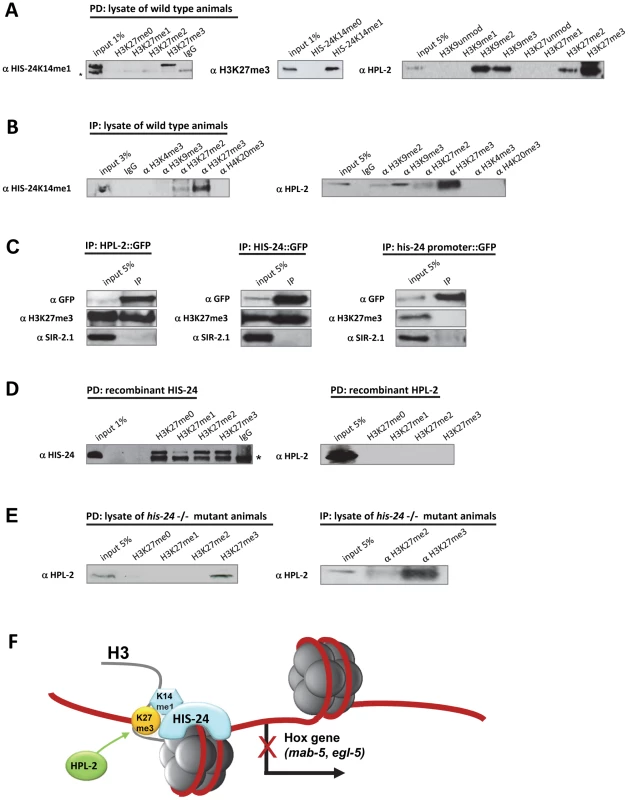
(A, B) HIS-24K14me1 protein specifically recognizes H3K27me3 chromatin mark and HPL-2 binds H3K27me2/me3, and H3K9me2/me3 in peptide pulldown assay (PD) and immunoprecipitation experiment (IP). Non-specific bands are indicated by a star. (C) HPL-2::GFP and HIS-24::GFP pull down H3K27me3. In contrast, GFP under his-24 promoter did not precipitate H3K27me3. In the bottom panel, SIR-2.1 was used as a negative control [18]. (D) Recombinant expressed HIS-24 binds H3K27me0/2/3 in contrast to HPL-2. (E) Deficiency of HIS-24 does not influence the binding of HPL-2 to H3K27me3. (F) Simplified model of HIS-24K14me1 and HPL-2 regulation of Hox gene expression. HIS-24K14me1 does not interact with HPL-2. HIS-24K14me1 rescues the developmental patterning of the male tail
To assess whether the methylated form of HIS-24 has a causal role in the observed changes of the male tail morphology, we generated his-24::gfp and his-24K14A::gfp transgenic worms in the hpl-2(tm1489); his-24(ok1024) mutant background. We observed that the restoration of HIS-24 levels by expression of HIS-24::GFP rescued the male phenotype and the fused/missing rays were down nearly to zero in the transgenic line (Figure 10). Importantly, the nonmethylatable HIS-24K14A::GFP mutant failed to rescue the wild type rays development in hpl-2; his-24 animals, suggesting that HIS-24 methylation at lysine 14 is necessary to regulate male tail development (Figure 10B, 10D, 10E). These results also imply that, at 21°C, hpl-2 and his-24 play a redundant role in the regulation of positional identity in the C. elegans males. Importantly, the analysis of transgene expression at the cellular level by immunostaining and immunoblotting of the rescued hpl-2(tm1489); his-24(ok1024) animals verified that the exogenous HIS-24K14A::GFP mutated form was expressed at a level comparable to that in animals carrying HIS-24::GFP wild type form (Figure 10C).
Fig. 10. HIS-24::GFP expression in hpl-2;his-24 double mutant animals restores normal male tail development. 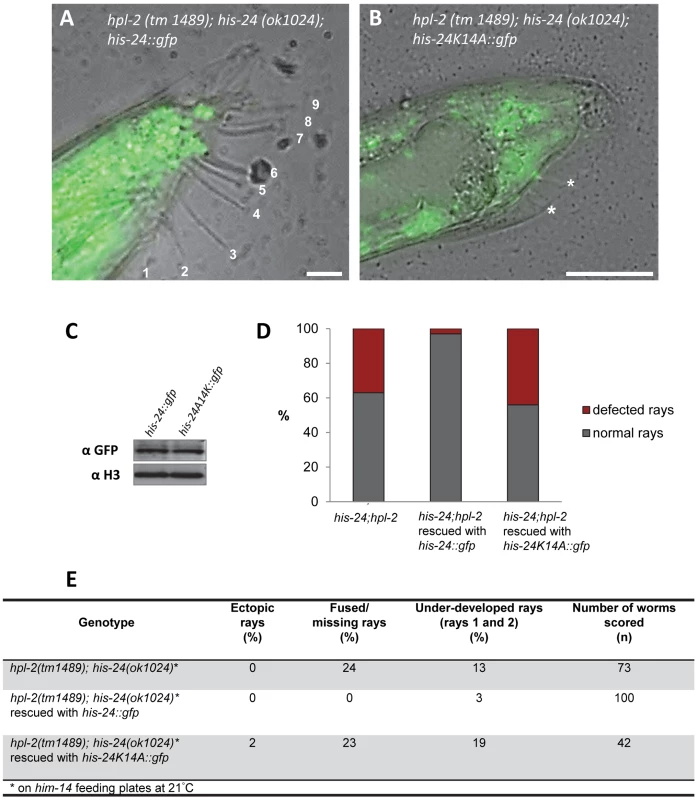
(A–C) The exogenous, mutated form of his-24K14A::gfp is expressed at the level comparable to those in animals carrying his-24::gfp wild type form. (A) Scale bar 10 µm. (B) Scale bar 25 µm. Stars point to under-developed rays. (D) Quantification of ray defects (E) associated with his-24K14A::gfp and his-24::gfp in hpl-2; his-24 background mutation. The animals were growing on him-14 feeding plates at 21°C. Discussion
HP1 and H1 are heterochromatin components that are believed to be associated with global repression of transcriptional activity 4–5. Surprisingly, our microarray analysis showed that H1 and HP1 play more dynamic and gene-specific roles in the roundworm C. elegans. They grossly affect only a few genes and can have an overlapping function in the same or parallel pathways where they regulate common target genes.
In particular, we found that HIS-24 and HPL-2 can regulate a shared target, the Hox genes. Although, the C. elegans homeobox genes (egl-5, mab-5) are silenced by mechanisms involving H3K27 trimethylation, we showed that the methylated form of HIS-24 and HPL-2 can also serve as essential protein components in establishing and/or maintaining the repressive chromatin state at the selected Hox genes, presumably through their binding to H3K27me3 (Figure 9F).
Effect of HIS-24 and HPL on gene expression profile and chromatin organization
Our microarray analyses support a role of H1 and HP1 in specific gene regulation, rather than a general repressive function [34]–[36]. Despite global changes in chromatin compaction and synergism of HIS-24 and HPL in aspects of many developmental processes we observed very few and slight changes in gene expression profile of mutants when compared with wild type animals. We detected a set of shared up - and down-regulated genes by HIS-24 and HPL suggesting that redundant roles for HIS-24 and HPL also exist. The relatively small number of regulated genes in observed triple mutant animals may indicate that HPL proteins and HIS-24 serve to fine-tune the regulation of key genes during development or differentiation. This model can be explained by the fact that the sequential arrangement of the linker histone HIS-24 and HPL-2 on the chromatin fibre might influence higher-order chromatin structure and effect nucleosome positioning, and stability [36]. It is possible that the different HPL subtypes and HIS-24 confer subtle differences in the properties of the chromatin fiber which allow for quantitative modulation of gene expression [34], [35]. Although the changes in gene transcription are subtle, we think that even 1.5-fold differences in expression can contribute to the marked phenotypic consequences we observed.
Model of transcriptional regulation of egl-5 and mab-5 by HPL-2 and HIS-24K14me1
We demonstrated that HIS-24K14me1, together with HPL-2, has a causal role in transcriptional silencing of egl-5 and mab-5. We propose that HPL-2 and HIS-24K14me1 may serve as essential protein components in establishing and/or maintaining the repressive chromatin state at the selected Hox genes through their interactions with H3K27me3. While we did not observe any phenotypic effects on male tail development either in hpl-2; hpl-1 nor in hpl-1 his-24 background, we speculate that HPL-2 acts redundantly with HIS-24K14me1 to regulate the positional identity in the C. elegans males. Loss of the two heterochromatin components, HIS-24K14me1 and HPL-2, causes significant changes in chromatin structure affecting Hox gene expression in C. elegans. However, since no interaction of HPL-2 and HIS-24K14me1 was observed in immunoprecipitation experiments, it is possible that HPL-2 together with HIS-24K14me1 might be a part of the same protein group involved in the regulation of Hox gene expression. The high degree of redundancy between his-24 and hpl-2 in Hox gene regulation might indicate that these two proteins are the only readers acting in parallel to perform the same role in translating the effects of histone H3K27 trimethylation. However, since we have failed so far to detect HPL-2 at the Hox gene region using direct ChIP approach, it is possible that the mechanisms by which HPL-2 regulates mab-5 and egl-5 might be indirect, involve intermediate factors (RNAi machinery, transcription factors) and depend on an architectural level in the cellular context.
HIS-24K14me1 and HPL-2 as a part of the PcG silencing complex
In mammals, H1 regulates Hox gene activation by promoting DNA demethylation [13]. Although C. elegans does not possess methylated DNA, we speculate that H1 can still influence Hox gene regulation and, together with HPL-2, regulate Hox gene expression as a part of the PcG silencing complex. The interaction of HPL-2 and HIS-24K14me1 with H3K27me3 can regulate the Hox gene in parallel pathway as MES-2 or MES-3, and can be directed to specific parts of the genome. Notably C. elegans HP1/HPL-2 does not follow the H3K9me2/me3 code [37]–[41] but it is sufficient to recognize, and to bind H3K27me2/me3. Remarkably, HIS-24 is required for optimal HPL-2 binding to H3K27me3 in vivo.
Interestingly, some PcG proteins containing a chromodomain similar to that found in C. elegans HPL-2 and mammalian HP1s have been shown to bind H3K27me3 [30], [42].
Overall, these and our previous results implicate that HPL and HIS-24 share some common functions even though there are differences among these proteins [16]–[17], [26]. We conclude that a combination of the H3K27me3 methylation mark, HPL-2 and HIS-24K14me1 could be a major factor in the establishment of stable patterns of selected homeotic gene expression.
Methods
Strains
Nematodes strains were cultured and genetically manipulated as previously described [43]. The Bristol strain (N2) was used as wild type. The following strains, obtained from the Caenorhabditis Genetics Center (CGC), were used in this study: his-24(ok1024)X, hil-3(ok1556)X (both strains outcrossed 1×), hpl-1(tm1624)X (outcrossed 4×), hpl-2(tm1489)III (outcrossed 4×). Transgenic strain (transcriptional reporter) expressing GFP under the control of the his-24 promoter was kindly provided by BC C. elegans Gene Expression Consortium, Canada. The double mutants hpl-1(tm1624)X his-24(ok1024)X, hpl-2(tm1489)III; his-24(ok1024)X, hpl-2(tm1489)III; hil-3(ok1556)X and the triple mutant strain hpl-2(tm1489)III; hpl-1(tm1624)X his-24(ok1024)X were generated by crossing. his-24::gfp (stable integrated EC602 strain [26]) and his-24K14A::gfp transgenic strains were crossed with the hpl-2(tm1489)III; his-24(ok1024)X. The generation of his-24K14A::gfp transgenic strain was previously described [16].
For the reporter gene analysis following transgenic strains: EM599 [egl-5::gfp; him-5(e1490)V; lin-15B(n765)X; bxIs13], OP27 [unc-119(ed3)III; wgIs27], OP54 [unc-119(ed3)III; wgIs54] and HZ111 [mab-5::gfp; muIs16 II; sor-1(bp1)/qC1 dpy-19(e1259) glp-1(q339)III; him-5(e1490)V], kindly provided by CCG, were used.
The brood size was scored as previously described [14].
All C. elegans strains were maintained at 15°C or at 21°C, unless otherwise specified.
Protein extraction, purification, and identification
C. elegans H1 extraction was performed as previously described [16].
Immunofluorescence analysis
Worms from wild type strain and the mutant worms were fixed and stained as previously described [26]. Gonads of worms were stained with fluorescent dye 4′,6′-diamidino-2-phenylindole (DAPI) diluted 1∶1000. The slides were mounted with Vectashield Mounting Medium and analyzed by using Leica DMI 6000 microscope.
Microarray analysis
Microarray analysis from two biological replicates was performed as previously described [16], [44]. In brief, 80 to 100 animals in L4 larval stage raised at 21°C were used. The gene expression fold change was calculated from the duplicate microarray data. The fold change cut-off was 1.5 from 2 biological replicates.
Analysis of ray phenotypes
Abnormalities of rays were identified in single, double and triple mutant males in comparison to the wild type worms. Animals were transferred on agar pads (2% agarose) and examined with differential interference contrast (DIC), using Leica DMI 6000 microscope. The number of rays, their position in relation to the anterior-posterior body axis and their shape served as basics of the analysis. Rays which were found outside of their normal formation region were defined as ectopic.
RNA interference (RNAi) experiments
RNAi feeding experiment was performed in 50 mm NGM feeding plates (NGM plates with 100 µg/ml ampicillin, 1 mM IPTG). him-14 (RNAi) (CGC, USA), hpl-1 (RNAi), mes-2 (RNAi) and mes-3 (RNAi) bacterial clones (Sanger Institute, UK) were grown overnight at 37°C in LB medium with 100 mg/ml ampicillin and were spotted onto 50 mm NGM plates. Mixed stage L3 and L4 mutant larval worms were transferred onto feeding plates and incubated at 21°C through several generations. Males were examined on the agar pads using Leica DMI 6000 microscope. Male progeny were scored for the presence of ectopic, under-developed rays and/or ray fusions.
Analysis of EGL-5::GFP and MAB-5::GFP expression in the single, double, and triple mutant strains
L3 stage and adult animals from each line were mounted on the agar pads and examined under Leica DMI 6000 microscope. Males were scored for the presence of ectopic EGL-5::GFP or MAB-5::GFP expression.
Chromatin immunoprecipitation
ChIP was performed as previously described [45] with several modifications. Mixed stage L4 and adult worms were homogenized in ice-cold FA lysis buffer (50 mM HEPES/KOH pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate; 150 mM NaCl) with complete protease inhibitor cocktail (Roche) and 1% Triton X-100 using liquid nitrogen. Worm lysate was sonicated with a Branson Digital Sonifier using following settings: 30% amplitude for 3 min total. Protein concentration of the extract was determined by the Coomassie Plus (Bradford) Protein Assay. Worm extract was incubated with the following antibodies: anti-H3 (Abcam 1791), anti-H3K27me2 (Upstate 07-322), anti-H3K27me3 (kindly provided by T. Jenuwein), anti-H3K9me2 (Abcam 1220), anti-H3K4me3 (Abcam 1012), anti-GFP (Roche) and anti-HIS-24. Proteins were immunoprecipitated using G-agarose beads (Pierce). mab-5 and egl-5 genes were detected by qPCR using iCycler iQ™ Multi - Color real time PCR detection system (Bio-Rad). Primer sequences are available on request.
Peptide pull down analysis
Peptide pull downs were performed as previously described [46]. 10 µg of each biotinylated peptide was coupled to streptavidin - agarose beads (Pierce). For peptide binding experiments following peptides were used: H3 mono-, di - or trimethylated at K9, H3 mono-, di - or trimethylated at K27, H3 unmethylated at K27, HIS-24 monomethylated at K14 and HIS-24 unmethylated at K14. Peptides were generated by Squarix (Germany). Worm extracts were incubated for 2 h at 4°C with the beads (constant rotation). Beads were washed six times with PD 150 buffer (20 mM Hepes pH 7.9, 150 mM KCl, 0.2% Triton-X 100, complete protease inhibitor cocktail, 20% glycerol). Bounded proteins were separated on gradient NuPAGE SDS gel (4–12%).
Western and dot blot
C. elegans lysates were prepared and analyzed by western blot as previously described [16], [18].
Immunoprecipitation
Mixed populations of L4 worms carrying the hpl-2::gfp transgene or wild type worms were homogenized [47]. About 1.5 mg of total precleared protein was incubated with following antibodies GFP-TrapR –A beads (Chromotek, Germany), anti - H3 (Abcam 1791), anti-H3K27me2 (Upstate 07-322), anti-H3K27me3 (kindly provided by T. Jenuwein), anti-H3K9me2 (Abcam 1220), anti-H3K4me3 (Abcam 1012), anti-H3K9me3 (Abcam 8898) or anti-H4K20me3 (Abcam 9053) at 4°C overnight. Next, the complexes were washed six times with PD150 buffer for 5 minutes at 4°C (20 mM Hepes, pH 7.9; 150 mM KCl, 0.2% Triton X-100, 1× Protease Inhibitor (Roche), 20% glycerol). Finally, the immunoprecipitated proteins were resolved on NuPAGE SDS gradient gel (4–12%) and western blotted with antibodies against H3K27me3 (1∶20 000 dilution), HPL-2 (1∶2000 dilution; kindly provided by F. Palladino), HIS-24K14me1 (1∶10000 dilution) and GFP (Roche; 1∶20000 dilution).
Expression of recombinant HPL-2 protein
The pGEX HPL-2a plasmid (kindly provided by F. Palladino) and HIS-24 pet3a plasmid were expressed in E. coli BL21(DE3) and the recombinant proteins were used for the peptide pull down assay.
Accession numbers
The microarray data can be found in the Gene Expression Omnibus (GEO) of NCBI through accession number GSE33339.
Supporting Information
Zdroje
1. ShenX, GorovskyMA (1996) Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86 : 475–83.
2. HellauerK, SirardE, TurcotteB (2001) Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem 276 : 13587–92.
3. GilbertN, BoyleS, SutherlandH, de Las HerasJ, AllanJ, et al. (2003) Formation of facultative heterochromatin in the absence of HP1. EMBO J 22 : 5540–5550.
4. LaybournPJ, KadonagaJT (1991) Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254 : 238–245.
5. VerschurePJ, van der KraanI, de LeeuwW, van der VlagJ, CarpenterAE, et al. (2005) In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol 25 : 4552–64.
6. DaujatS, ZeisslerU, WaldmannT, HappelN, SchneiderR (2005) HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem 280 : 38090–5.
7. SirotkinAM, EdelmannW, ChengG, Klein-SzantoA, KucherlapatiR, et al. (1995) Mice develop normally without the H10 linker histone. Proc Natl Acad Sci USA 92 : 6434–6438.
8. SanchoM, DianiE, BeatoM, JordanA (2008) Depletion of Human Histone H1 Variants Uncovers Specific Roles in Gene Expression and Cell Growth. PLoS Genet 4 (10) e1000227.
9. FilesiI, CardinaleA, van der SarS, CowellIG, SinghPB, et al. (2002) Loss of heterochromatin protein 1 (HP1) chromodomain function in mammalian cells by intracellular antibodies causes cell death. J Cell Sci 115 : 1803–1813.
10. AucottR, BullwinkelJ, YuY, ShiW, BillurM, et al. (2008) HP1-beta is required for development of the cerebral neocortex and neuromuscular junctions. J Cell Biol 183 : 597–606.
11. FanY, NikitinaT, ZhaoJ, FleuryTJ, BouhassiraEE, et al. (2005) Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123 : 1199–212.
12. FanY, NikitinaT, Morin-KensickiEM, ZhaoJ, MagnusonTR, et al. (2003) H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol 23 : 4559–72.
13. MacleanJA, BettegowdaA, KimBJ, LouCH, YangSM, et al. (2011) The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone H1 and DNA methylation. Mol Cell Biol 31 : 1275–87.
14. JedrusikMA, SchulzeE (2001) Single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development 128 : 1069–80.
15. CouteauF, GuerryF, MullerF, PalladinoF (2002) A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep 3 : 235–41.
16. StudenckaM, KonzerA, MoneronG, WenzelD, OpitzL, et al. (2011) Novel roles of C. elegans heterochromatin protein HP1 and linker histone in the regulation of innate immune gene expression. Mol Cell Biol 32 : 251–65.
17. SchottS, CousthamV, SimonetT, BedetC, PalladinoF (2006) Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev Biol 298 : 176–187.
18. WirthM, PaapF, FischleW, WenzelD, AgafonovDE, et al. (2009) HIS-24 linker histone and SIR-2.1 deacetylase induce H3K27me3 in the Caenorhabditis elegans germ line. Mol Cell Biol 29 : 3700–9.
19. MargueronR, ReinbergD (2011) The Polycomb complex PRC2 and its mark in life. Nature 469 : 343–9.
20. GellonG, McGinnisW (1998) Shaping animal body plans in development and evolution by modulation of Hox expression patterns. BioEssays 20 : 116–125.
21. LevineSS, WeissA, Erdjument-BromageH, ShaoZ, TempstP, et al. (2002) The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol 22 : 6070–8.
22. AboobakerAA, BlaxterML (2003) Hox Gene Loss during Dynamic Evolution of the Nematode Cluster. Curr Biol 13 : 37–40.
23. RossJM, ZarkowerD (2003) Polycomb group regulation of Hox gene expression in C. elegans. Dev Cell 4 : 891–901.
24. ZhangH, AzevedoRB, LintsR, DoyleC, TengY, et al. (2003) Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev Cell 4 : 903–15.
25. ZhangT, SunY, TianE, DengH, ZhangY, et al. (2006) RNA-binding proteins SOP-2 and SOR-1 form a novel PcG-like complex in C. elegans. Development 133 : 1023–33.
26. JedrusikMA, SchulzeE (2007) Linker histone HIS-24 (H1.1) cytoplasmic retention promotes germ line development and influences histone H3 methylation in Caenorhabditis elegans. Mol Cell Biol 27 : 2229–39.
27. SchottS, RamosF, CousthamV, PalladinoF (2009) HPL-2/HP1 prevents inappropriate vulval induction in Caenorhabditis elegans by acting in both HYP7 and vulval precursor cells. Genetics 181 : 797–801.
28. SalserSJ, KenyonC (1996) A C. elegans Hox gene switches on, off, on and off again to regulate proliferation, differentiation and morphogenesis. Development 122 : 1651–1661.
29. FerreiraHB, ZhangY, ZhaoC, EmmonsSW (1999) Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev Biol 207 : 215–228.
30. CaoR, WangL, WangH, XiaL, Erdjument-BromageH, et al. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 : 1039–1043.
31. CaoR, ZhangY (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14 : 155–164.
32. MüllerJ, HartCM, FrancisNJ, VargasML, SenguptaA, et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111 : 197–208.
33. HoldemanR, NehrtS, StromeS (1998) MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development 125 : 2457–2467.
34. AlamiR, FanY, PackS, SonbuchnerTM, BesseA, et al. (2003) Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci U S A 100 : 5920–5.
35. SanchoM, DianiE, BeatoM, JordanA (2008) Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genetics 4 (10) e1000227.
36. KoopR, Di CroceL, BeatoM (2003) Histone H1 enhances synergestic activation of the MMTV promoter in chromatin. EMBO Journal 22 : 588–599.
37. DialynasGK, TerjungS, BrownJP, AucottRL, Baron-LuhrB, et al. (2007) Plasticity of HP1 proteins in mammalian cells. JSC 120 : 3415–3424.
38. LiY, KirschmannDA, WallrathLL (2002) Does heterochromatin protein 1 always follow code? Proc Natl Acad Sci USA 99 : 16462–16469.
39. SinghPB, GeorgatosSD (2002) HP1: facts, open questions, and speculation. J Struct Biol 140 : 10–16.
40. FlueckC, BartfaiR, VolzJ, NiederwieserI, Salcedo-AmayaAM, et al. (2009) Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog 5 (9) e1000569.
41. NielsenPR, NietlispachD, MottHR, CallaghanJ, BannisterA, et al. (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416 : 103–107.
42. CzerminB, MelfiR, McCabeD, SeitzV, ImhofA, et al. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 : 185–196.
43. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
44. OpitzL, Salinas-RiesterG, GradeM, JungK, JoP, et al. (2010) Impact of RNA degradation on gene expression profiling. BMC Medical Genomics 3 : 36.
45. ErcanS, GiresiPG, WhittleCM, ZhangX, GreenRD, et al. (2007) X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet 39 : 403–8.
46. WysockaJ (2006) Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods 40 : 339–43.
47. CheesemanIM, DesaiA (2005) A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 266, pl1.
Štítky
Genetika Reprodukčná medicína
Článek Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese SubjectsČlánek Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in RiceČlánek Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System inČlánek An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural CrestČlánek A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of BacteriaČlánek A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Heterozygous Mutations in DNA Repair Genes and Hereditary Breast Cancer: A Question of Power
- GWAS of Diabetic Nephropathy: Is the GENIE out of the Bottle?
- The Conflict within and the Escalating War between the Sex Chromosomes
- Proteome-Wide Analysis of Disease-Associated SNPs That Show Allele-Specific Transcription Factor Binding
- Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes and as Potential Breast Cancer Susceptibility Alleles
- A Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in
- Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese Subjects
- Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in Rice
- Co-Evolution of Mitochondrial tRNA Import and Codon Usage Determines Translational Efficiency in the Green Alga
- SIRT6/7 Homolog SIR-2.4 Promotes DAF-16 Relocalization and Function during Stress
- CNV Formation in Mouse Embryonic Stem Cells Occurs in the Absence of Xrcc4-Dependent Nonhomologous End Joining
- Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System in
- Citrullination of Histone H3 Interferes with HP1-Mediated Transcriptional Repression
- Variation in Genes Related to Cochlear Biology Is Strongly Associated with Adult-Onset Deafness in Border Collies
- The Long Non-Coding RNA Affects Chromatin Conformation and Expression of , but Does Not Regulate Its Imprinting in the Developing Heart
- Rif2 Promotes a Telomere Fold-Back Structure through Rpd3L Recruitment in Budding Yeast
- Is a Metastasis Susceptibility Gene That Suppresses Metastasis by Modifying Tumor Interaction with the Cell-Mediated Immunity
- The p38/MK2-Driven Exchange between Tristetraprolin and HuR Regulates AU–Rich Element–Dependent Translation
- Rare Copy Number Variants Contribute to Congenital Left-Sided Heart Disease
- A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse
- An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural Crest
- Characterization of Inducible Models of Tay-Sachs and Related Disease
- Hominoid-Specific Protein-Coding Genes Originating from Long Non-Coding RNAs
- Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24
- Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Virulence
- Convergence of the Transcriptional Responses to Heat Shock and Singlet Oxygen Stresses
- Genomics of Adaptation during Experimental Evolution of the Opportunistic Pathogen
- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Vsx2 Controls Eye Organogenesis and Retinal Progenitor Identity Via Homeodomain and Non-Homeodomain Residues Required for High Affinity DNA Binding
- The Long Path from QTL to Gene
- TCF7L2 Modulates Glucose Homeostasis by Regulating CREB- and FoxO1-Dependent Transcriptional Pathway in the Liver
- The Non-Flagellar Type III Secretion System Evolved from the Bacterial Flagellum and Diversified into Host-Cell Adapted Systems
- Complex Chromosomal Rearrangements Mediated by Break-Induced Replication Involve Structure-Selective Endonucleases
- Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-A in Fission Yeast
- A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of Bacteria
- Determinants of Human Adipose Tissue Gene Expression: Impact of Diet, Sex, Metabolic Status, and Genetic Regulation
- Genome-Wide Association Studies Identify Heavy Metal ATPase3 as the Primary Determinant of Natural Variation in Leaf Cadmium in
- Tethering of the Conserved piggyBac Transposase Fusion Protein CSB-PGBD3 to Chromosomal AP-1 Proteins Regulates Expression of Nearby Genes in Humans
- A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- H4K20me1 Contributes to Downregulation of X-Linked Genes for Dosage Compensation
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Coevolution within and between Regulatory Loci Can Preserve Promoter Function Despite Evolutionary Rate Acceleration
- New Susceptibility Loci Associated with Kidney Disease in Type 1 Diabetes
- SWI/SNF-Like Chromatin Remodeling Factor Fun30 Supports Point Centromere Function in
- A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
- Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription
- Genetic Modifiers of Chromatin Acetylation Antagonize the Reprogramming of Epi-Polymorphisms
- UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development
- A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



