-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
The stabilization of the replisome complex is essential in order to achieve highly processive DNA replication and preserve genomic integrity. Conversely, it would also be advantageous for the cell to abrogate replisome functions to prevent inappropriate replication when fork progression is adversely perturbed. However, such mechanisms remain elusive. Here we report that replicative DNA polymerases and helicases, the major components of the replisome, are degraded in concert in the absence of Swi1, a subunit of the replication fork protection complex. In sharp contrast, ORC and PCNA, which are also required for DNA replication, were stably maintained. We demonstrate that this degradation of DNA polymerases and helicases is dependent on the ubiquitin-proteasome system, in which the SCFPof3 ubiquitin ligase is involved. Consistently, we show that Pof3 interacts with DNA polymerase ε. Remarkably, forced accumulation of replisome components leads to abnormal DNA replication and mitotic catastrophes in the absence of Swi1. Swi1 is known to prevent fork collapse at natural replication block sites throughout the genome. Therefore, our results suggest that the cell elicits a program to degrade replisomes upon replication stress in the absence of Swi1. We also suggest that this program prevents inappropriate duplication of the genome, which in turn contributes to the preservation of genomic integrity.
Published in the journal: Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex. PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003213
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003213Summary
The stabilization of the replisome complex is essential in order to achieve highly processive DNA replication and preserve genomic integrity. Conversely, it would also be advantageous for the cell to abrogate replisome functions to prevent inappropriate replication when fork progression is adversely perturbed. However, such mechanisms remain elusive. Here we report that replicative DNA polymerases and helicases, the major components of the replisome, are degraded in concert in the absence of Swi1, a subunit of the replication fork protection complex. In sharp contrast, ORC and PCNA, which are also required for DNA replication, were stably maintained. We demonstrate that this degradation of DNA polymerases and helicases is dependent on the ubiquitin-proteasome system, in which the SCFPof3 ubiquitin ligase is involved. Consistently, we show that Pof3 interacts with DNA polymerase ε. Remarkably, forced accumulation of replisome components leads to abnormal DNA replication and mitotic catastrophes in the absence of Swi1. Swi1 is known to prevent fork collapse at natural replication block sites throughout the genome. Therefore, our results suggest that the cell elicits a program to degrade replisomes upon replication stress in the absence of Swi1. We also suggest that this program prevents inappropriate duplication of the genome, which in turn contributes to the preservation of genomic integrity.
Introduction
Initiation of DNA replication is directed by the formation of the pre-replication complex (pre-RC) at the origin of replication [1]. The pre-RC includes a number of essential replication proteins such as origin recognition complex (ORC), Cdc6, Cdt1, and the mini-chromosome maintenance (MCM) DNA helicase complex. However, to initiate actual DNA synthesis, additional factors are needed to facilitate the unwinding of origins and generation of replication forks. These factors include Cdc45, go-ichi-ni-san (GINS), replication protein A (RPA), proliferating cell nuclear antigen (PCNA), and other accessory factors prior to the loading of DNA polymerases. Together, these factors form the replisome complex at the replication fork [1]. However, how the cell maintains the integrity of the replisome is not well understood.
In response to replication stress, cells activate the DNA replication checkpoint to allow time for DNA repair. Central to this system are protein kinases such as human ATM and ATR, fission yeast Rad3, and budding yeast Mec1 [2]–[6]. These kinases are required for activation of downstream effector kinases by phosphorylation. In the fission yeast Schizosaccharomyces pombe, Rad3 activates Cds1 and Chk1 kinases in response to replication stress or DNA damage, facilitating DNA repair and recombination pathways [2], [4], [7]. Another essential function of the replication checkpoint is to stabilize replication forks by maintaining proper assembly of replisome components and preserving DNA structures during DNA replication problems [8]–[12]. Recent studies found that ancillary factors that are not essential for DNA synthesis but are important for DNA replication accuracy also travel with moving replication forks. Such factors include fission yeast Swi1 and Swi3, which together form the replication fork protection complex (FPC) and are required for efficient activation of the replication checkpoint kinase Cds1 and for the stabilization of replication forks [13]–[16]. In the absence of Swi1 or Swi3, cells accumulate abnormal fork structures that lead to Rad22 (the Rad52 orthologue) DNA repair foci formation and accumulation of recombination structures during S phase [16], [17]. The functions of the Swi1–Swi3 complex appear to be conserved among eukaryotes [14], [15], [18]–[20]. Studies show that Swi1–Swi3 orthologues (Tof1–Csm3 in budding yeast, and Timeless–Tipin in vertebrates) are components of the replisome, are involved in fork stabilization, and regulate the intra-S phase checkpoint [18], [21]–[28]. Furthermore, genetic studies in yeast also suggest that the Swi1–Swi3 FPC has roles in coordinating leading - and lagging-strand DNA synthesis and in coupling DNA polymerase and helicase activities at the replication fork [15], [16], [29]. However, how the FPC protects moving replication forks and coordinates with multiple genome maintenance processes at the replication fork is not well understood.
Replication checkpoint studies have typically used chemical agents to stall replication forks. However, emerging evidence indicates that there are a number of chromosome regions that present obstacles to DNA replication. These include programmed fork blocking sites, DNA-binding proteins such as the transcription machinery, and DNA secondary structures caused by repeat sequences. These sites are considered to be difficult to replicate, causing arrest of replication forks or even fork breakage [30]–[35]. Fork arrest at difficult-to-replicate genome sites can promote both genome instability and stability depending on the circumstances. For example, polar fork pausing at rDNA loci stimulates recombination-dependent rDNA repeat expansion and contraction, which can lead to rDNA instability. On the contrary, this polar fork pausing is also required to coordinate directionality of replication and transcription at rDNA loci, preventing genome instability due to head-on collisions of the replisome and transcriptional machinery [36], [37]. Interestingly, studies found that FPC-related proteins are required for a number of fork arrest events, which are mediated by DNA–protein complexes. These include fork pausing at the rDNA loci, the fission yeast mating-type locus, tRNA loci, and highly transcribed RNA polymerase II genes [14], [29], [38]–[41]. At rDNA loci, loss of FPC causes hyper recombination, leading to contraction of rDNA repeats [17], [40], [42]. Similarly, the high rate of transcription and the presence of DNA-binding factors increase the chances of the replisome colliding with a transcription fork. Indeed, studies in fission yeast revealed that Swi1 is required to prevent DNA damage and hyper recombination activity at these natural obstacles scattered throughout the genome [43]–[45].
In addition to these DNA–protein complex-mediated fork barriers, repeat DNA sequences themselves also cause genome instability in the absence of FPC-related proteins. At these sites, instead of promoting fork stalling, FPC appears to prevent or reduce the rate of fork stalling when the fork encounters DNA secondary structures caused by repeat sequences. Therefore, in the absence of FPC, fork stalling results in elevated levels of ssDNA exposed at the replication fork, which appear to cause genome instability due to expansion and contraction at DNA structure-based impediments [46]–[51]. Thus, the mechanisms of the FPC-dependent fork regulation at repeat regions and at DNA–protein complex-mediated fork barriers are different. However, accumulated evidence indicates that FPC proteins are required for smooth passage of replication forks and for suppression of replication stresses at these natural impediments [14].
Therefore, in this study, we used swi1Δ as a model to understand replication stress response mechanisms. Strikingly, we have found that replicative DNA polymerases and helicases are highly unstable in the absence of Swi1. Our investigation revealed that this degradation is mediated by the ubiquitin-proteasome system, in which the SCFPof3 (Skp1/Cul1/F-box) ubiquitin ligase complex is involved. In the absence of Pof3, swi1Δ cells undergo mitotic catastrophes, suggesting the importance of proteasome-dependent replisome regulation in preserving genomic integrity. Considering that swi1Δ cells accumulate replication stress at difficult-to-replicate regions throughout the genome, our findings suggest that ubiquitin-dependent degradation of replisome components play a critical role in genome duplication in response to replication stresses.
It is widely understood that checkpoint proteins stabilize replication forks and replisomes in response to replication stress. However, our findings suggest an alternative mechanism that cells abrogate replisome functions when the fork encounters obstacles. Therefore, our study proves new mechanistic insights into the understanding of the replication stress response. In addition, although a number of studies have focused on the processes of replication initiation and regulation of fork progression, how the replisome itself is regulated is still largely unknown. Therefore, our findings also fill the knowledge gap in the regulation of replisome components in the DNA replication program.
Results
Replisome components are unstable in swi1Δ cells
Recent studies have shown that fork progression is impaired in the absence of FPC orthologues [24], [27], [28], [38], [52]. We also found a similar defect in S. pombe swi1Δ cells (Figure S1), suggesting that FPC might regulate replisome stability. To test this possibility, we investigated the stability of various replication proteins in cells treated with cycloheximide (CHX), a compound that blocks the synthesis of new proteins and allows for the examination of protein stability. First, we examined the stability of the catalytic subunits of major essential replicative DNA polymerases. For this purpose, we employed S. pombe cells expressing Pol2-FLAG (the catalytic subunit of DNA polymerase ε, required for leading-strand synthesis) [53] and Pol3-FLAG (the catalytic subunit of DNA polymerase δ, required for lagging-strand synthesis) [54] from their genomic loci. Pol2-FLAG showed significant degradation in wild-type cells, whereas, Pol3-FLAG was relatively stable (Figure 1A and 1B). Intriguingly, Pol2 displayed even faster degradation when swi1 was deleted. In addition, Pol3 showed dramatic instability in swi1Δ cells (Figure 1A and 1B). Next, we examined the stability of MCM helicase components. S. pombe cells expressing Mcm2-GFP or Mcm6-GFP from their genomic loci were used, and Mcm4 was detected by the anti-Mcm4 antibody. In wild-type cells, Mcm2-GFP, Mcm4, and Mcm6-GFP were stable and did not undergo significant degradation throughout the CHX treatment (Figure 1C and 1D). In contrast, these helicase subunits were rapidly degraded in swi1Δ cells (Figure 1C and 1D). To determine whether such degradation is specific to certain replication proteins, we also assessed the stability of Orc1 (an ORC subunit), Mrc1 (a mediator of S-phase checkpoints), and PCNA. Although the steady-state levels of Orc1-FLAG and PCNA before the addition of CHX were somewhat lower in swi1Δ cells, their cellular amounts were maintained throughout the 4 h of CHX treatment in both wild-type and swi1Δ cells (Figure 1C). As previously reported [55], Mrc1 was unstable and shows rapid degradation in the presence of CHX, although this degradation was not strengthened by the deletion of swi1 (Figure 1C and 1D). Thus, we concluded that Swi1 is involved in preventing rapid degradation of Pol2 and Pol3, as well as helicase components. Since Swi1 is involved in the suppression of fork collapse at difficult-to-replicate regions in fission yeast [43]–[45], it is possible that chromatin-bound replisome components are susceptible to degradation. Therefore, we fractionated cells into Triton-X-100-soluble (cytosol and nucleoplasm) and Triton-X-100-insoluble (enriched with chromatin - and nuclear matrix-bound proteins) fractions as described in previous studies (Figure 1E) [55], [56]. Tubulin and histone H3 were exclusively fractionated into the Triton-soluble and Triton-insoluble fractions, respectively, indicating that fractionation was successful. Pol2 was mainly fractionated into the Triton-insoluble fraction, while approximately 20% and 30% of Pol3 and Mcm4 were recovered into the Triton-insoluble fraction, respectively. Importantly, degradation of Pol2, Pol3 and Mcm4 was observed in the Triton-insoluble fraction when cells were treated with CHX, suggesting that the chromatin fraction of replisome components undergoes degradation (Figure 1E). Therefore, our results are consistent with the notion that cells promote a fast turnover of replisome components bound to chromatin in response to the accumulation of fork collapse.
Fig. 1. Swi1 prevents degradation of DNA polymerases and helicases. 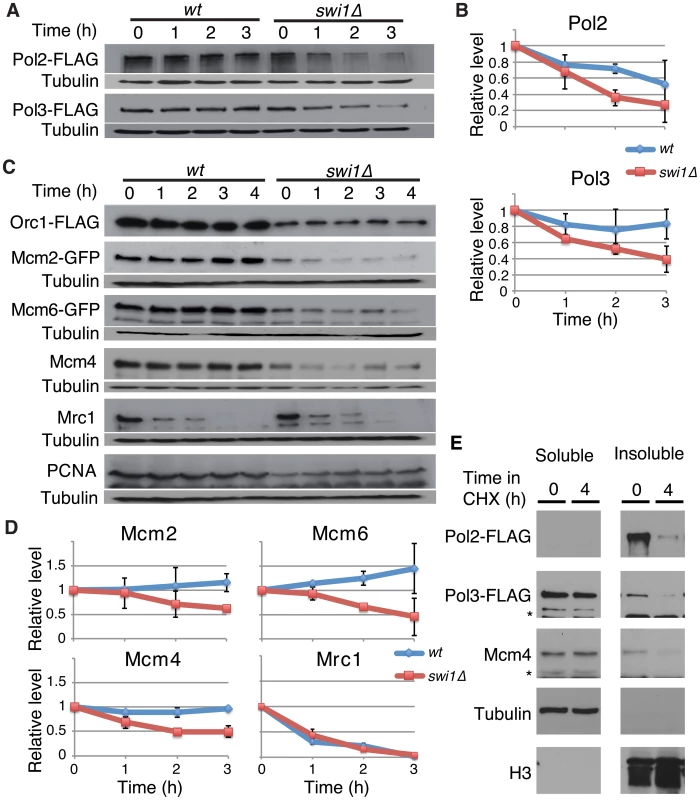
Exponentially growing cells were treated with 0.1 mg/ml CHX at 25°C. (A) Cellular amounts of Pol2-FLAG and Pol3-FLAG were examined from 0 to 3 h of CHX treatment. The anti-FLAG M2 antibody was used to detect Pol2 and Pol3. Western blotting of tubulin was also performed as a loading control. (B) Stability of Pol2-FLAG and Pol3-FLAG shown in A was quantified by NIH ImageJ. Relative intensity of protein bands at 0 h was set to 1 in each experiment. Error bars correspond to standard deviation of three independent experiments. wt, in blue; swi1Δ, in red. (C) Cellular amounts of Mcm2-GFP, Mcm6-GFP, Mcm4, Mrc1, Orc1-FLAG, and PCNA were determined from 0 to 4 h of CHX treatment. Anti-FLAG, anti-GFP, anti-Mcm4, anti-Mrc1, and anti-PCNA antibodies were used for Western blotting. (D) Stability of Mcm2-GFP, Mcm6-GFP, Mcm4, and Mrc1, shown in C, was quantified as described in B. Error bars represent average deviation (n = 2) or standard deviation (n = 3). (E) Replisome components in a chromatin-enriched fraction were degraded in response to CHX. Chromatin-free (Triton-soluble) and chromatin-enriched (Triton-insoluble) fractions were prepared from S. pombe cells treated with CHX for 0 and 4 h. The fractions were analyzed by Western blotting using antibodies to detect the indicated proteins. Swi1 protects the replisome components from proteasome-dependent degradation
To understand the mechanisms of replisome degradation in response to unstable forks in the absence of Swi1, we determined whether the proteasome is responsible for degradation of DNA helicases and polymerases. The mts3-1 temperature-sensitive allele, which has a mutation in a subunit of the 26S proteasome machinery, was used to inactivate the proteasome [57], [58]. It is estimated that proteasome activity of mts3-1 cells is about 50% and 30% of the wild-type enzyme at 25°C and 35°C, respectively [58]. Cells were grown at 25°C or 35°C for 2 h, and then treated with CHX for 2 to 4 h. Strikingly, degradation of Pol2 was substantially inhibited in mts3-1 and swi1Δ mts3-1 cells even at 25°C (Figure 2A and 2C). We observed similar stabilization of Pol3 and Mcm6 in mts3-1 and swi1Δ mts3-1 cells (Figure 2B and 2C). At 35°C, degradation of these replisome components was accelerated both in wild type and swi1Δ cells probably due to increased cell metabolism (Figure 2A and 2B). However, degradation of these replisome components was abolished in mts3-1 and swi1Δ mts3-1 cells at 35°C (Figure 2A and 2B). Thus, our data indicate that Swi1 prevents proteasome-dependent degradation of replisome components.
Fig. 2. Ubiquitin-proteasome-dependent degradation of replisome core components in the absence of Swi1. 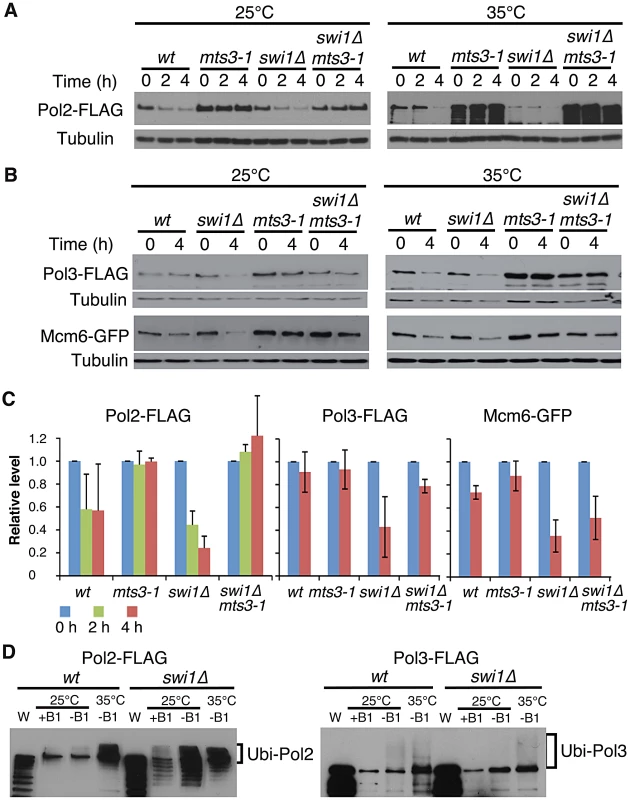
(A) Inactivation of the proteasome stabilizes Pol2. Cells of the indicated genotypes were incubated for 2 h at the indicated temperatures, then treated with CHX for 4 h. Cellular levels of Pol2-FLAG were monitored after 0, 2 and 4 h of CHX treatment. Tubulin was used as a loading control. (B) Cellular levels of Pol3-FLAG and Mcm6-GFP in the indicated cells were determined after 0 and 4 h of CHX treatment as described in A. (C) Stability of Pol2-FLAG, Pol3-FLAG and Mcm6-GFP at 25°C shown in A and B was quantified. Relative intensity of protein bands at 0 h in each cell type was set to 1. Error bars correspond average deviation (n = 2) or standard deviation (n = 3). (D) Pol2 and Pol3 are highly ubiquitinated in the absence of Swi1. 6xHis-Ub peptide was expressed 22 hours in the absence of thiamine (−B1) at 25°C, then cells were placed for 2 hours at 25°C or 35°C. There is some leaking expression of 6xHis–Ub before induction in the presence of thiamin (+B1). Ubiquitinated proteins were purified as described in Materials and Methods. Western blotting of the indicated protein was performed. W, whole cell extract. Representative results of repeat experiments are shown. Ubiquitin moieties (Ub) are conjugated to most of the proteins degraded by the proteasome [59], . Therefore, aforementioned data suggest that replisome core components (polymerases and helicases) are ubiquitinated. To test this possibility and further understand the mechanism of replisome degradation, we investigated whether replisome components were ubiquitinated. Cells harboring FLAG-tagged versions of Pol2 and Pol3 were engineered to express hexahistidine-fused ubiquitin (6xHis–Ub peptide) under the control of the thiamine (B1)-repressible nmt1 promoter. They were first cultured in the presence of thiamin (B1) to repress the nmt1 promoter and then grown in the absence of thiamine for 22 h at 25°C, allowing cells to express 6xHis–Ub peptide. After the 22 h incubation, cultures were divided and further incubated at 25°C or 35°C for 2 h. Ubiquitinated proteins were purified with nickel agarose beads and analyzed by immunoblotting using antibodies against the FLAG-tag (Figure 2B). As shown in Figure 2D, Pol2-FLAG species with slower gel mobility were clearly detected in both wild type and swi1Δ cells, indicating that Pol2 is ubiquitinated. We also observed precipitation of non-ubiquitinated Pol2 with nickel agarose as previously reported for other proteins [61]. In addition, multiple Pol2 bands, which are probably products of degraded Pol2, were detected in swi1Δ cells (Figure 2D), suggesting that Pol2 is more susceptible to degradation in the absence of Swi1. Similarly, ubiquitinated forms of Pol3-FLAG were detected in wild type and swi1Δ cells (Figure 2D). However, with our methods, we were not able to observe ubiquitinated forms of Mcm proteins (data not shown). Considering that Mcm proteins are stabilized in mts3-1 cells (Figure 2A), it is possible that the ubiquitination and degradation processes of Mcm proteins are too rapid to be detected.
Pol2 degradation occurs during S phase and is dependent on SCFPof3
Swi1 and its orthologues are involved in DNA replication, and their defects cause replication stress at difficult-to-replicate genome regions [14], [15], [18], [21], [22], [24], [25], [27], [28], [43]–[45]. Thus, our results suggest that replisome core degradation occurs during S phase in the absence of Swi1. To test this possibility, wild type and swi1Δ cells were synchronized at the G1/S boundary in the presence of 12 mM hydroxyurea (HU) and released into S phase in fresh medium supplemented with CHX. FACS analysis showed that the addition of CHX did not perturb cell cycle progression through S phase after the removal of HU (Figure 3A). There was no significant Pol2 degradation in both wild type and swi1Δ cells in the absence of CHX. In contrast, the level of Pol2-FLAG dramatically dropped between 30 and 45 min after CHX addition in the absence of Swi1 (Figure 3B and 3C), when cells are in S phase (Figure 3A). In contrast, wild-type cells displayed only a mild decrease in the level of Pol2-FLAG (Figure 3B and 3C). We also used the cdc25-22 temperature sensitive allele to synchronize cells at the G2/M boundary at the restrictive temperature (35°C), and cells were released into the cell cycle at permissive temperature (25°C). As determined by the increase of septation index, cells synchronously entered S phase after the release in the absence of CHX (Figure S2A). In this condition, Pol2 levels were maintained throughout the experiments in both cdc25-22 and cdc25-22 swi1Δ cells (Figure S2B and S2C). When cells were treated with CHX, Pol2-FLAG levels gradually decreased in cdc25-22 swi1Δ cells but not in cdc25-22 cells (Figure S2B and S2C). This mild degradation is probably because cells were unable to synchronously progress through S phase in the presence of CHX (Figure S2A), although our data indicate that Pol2-FLAG is unstable in swi1Δ cells. Interestingly, Mcm4 showed rapid degradation as cdc25-22 swi1Δ cells progress through S phase in the absence of CHX (Figure S2B and S2C), indicating that Mcm4 is degraded during replication. Mcm4 degradation in cdc25-22 swi1Δ cells was further exacerbated in the presence of CHX. In contrast, there is no significant Mcm4 degradation in cdc25-22 cells with or without CHX treatment (Figure S2B and S2C). Taken together, we concluded that degradation of replisome core components occurs during DNA replication in the absence of Swi1.
Fig. 3. Pol2 degradation occurs in S phase and is SCFPof3 dependent. 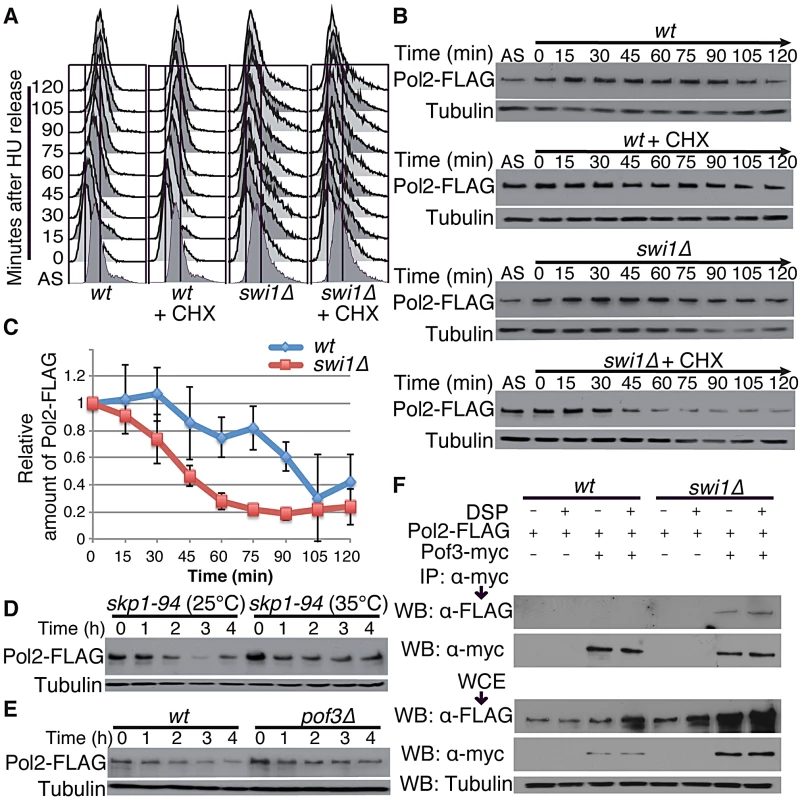
Exponentially growing cells (AS) were synchronized at the G1/S (time zero) boundary in the presence of 12 mM HU for 3 h, and were released into fresh YES medium with or without CHX. Cells were collected and processed for DNA content analysis in A, and for Western blotting in B. (A) Cells were fixed at the indicated times, and DNA contents were analyzed by flow cytometry. (B) Pol2 is degraded during S phase in swi1 mutants. Cellular amounts of Pol2-FLAG were determined at the indicated times. Tubulin levels were also monitored as a loading control. Representative results of repeat experiments are shown. (C) Stability of Pol2-FLAG during the CHX treatment shown in B was quantified as described in Figure 1. For each strain, relative intensity of the Pol2-FLAG band at 0 h was set to 1. (D) Pol2 is stabilized in the skp1-94 mutants. Exponentially growing skp1-94 cells were treated with CHX at 25°C and 35°C. Cellular amounts of Pol2-FLAG were examined from 0 to 4 h of CHX treatment. (E) pof3 deletion stabilizes Pol2. As in D, Pol2-FLAG levels were examined during the CHX treatment of wild-type and pof3Δ cells. (F) Pof3 interacts with Pol2. Cells expressing the indicated fusion proteins (Pol2-FLAG and/or Pof3-Myc) were harvested in the presence or absence of DSP, and protein extracts were prepared. Pof3-Myc was immunoprecipitated, and associated proteins were probed with anti-FLAG antibody. Representative results of repeat experiments are shown. IP, immunoprecipitation; WB, Western blotting; WCE, whole cell extract. Since SCF ubiquitin ligases are often involved in protein degradation during S phase [62], we examined the stability of Pol2 in skp1-94 temperature-sensitive cells, which have a mutation in Skp1, a major component of SCF ubiquitin ligases in S. pombe [63]. Strikingly, Pol2-FLAG was significantly stabilized when skp1-94 cells were incubated at 35°C, indicating the involvement of SCF ubiquitin ligases in Pol2 degradation (Figure 3D; Figure S3B). SCF ligases contain F-box subunits, which are responsible for substrate specificity. Therefore, we examined Pol2 stability in a series of mutants defective for F-box proteins (Figure S3). Among the eleven F-box mutants we tested, we found that Pol2 becomes most stable in the absence of Pof3 (Figure 3E; Figure S3), an F-box protein that has been suggested to be involved in the preservation of genomic integrity [64]. Thus, our data suggest that Pol2 degradation is in part mediated by the SCFPof3 ubiquitin ligase.
To further understand the mechanism of Pol2 degradation, we examined whether Pof3 interacts with Pol2, using immunoprecipitation assays. Cells expressing Pol2-FLAG proteins were engineered to express Pof3-Myc from its genomic locus. As shown in Figure 3F, Pol2-FLAG coprecipitated with Pof3-Myc in the absence of Swi1, indicating that SCFPof3 interacts with Pol2. The Pol2–Pof3 interaction was not detectable in wild-type cells even in the presence of a protein crosslinker dithio-bis succinimidyl propionate (DSP) (Figure 3F). Therefore, our data suggest that SCFPof3–Pol2 interaction is transient in wild-type cells but is enhanced when Pol2 degradation is accelerated in the absence of Swi1.
SCFPof3 is involved in degradation of Mcm4 and Mrc1
SCFPof3 has been shown to interact with fission yeast Mcl1, a DNA polymerase α accessory factor related to budding yeast Ctf4 [64]–. Moreover, in budding yeast, Dia2 (Pof3-related protein) is recruited to the replication fork [67], [68] and is involved in the ubiquitination of Mrc1 [69]. Therefore, SCFPof3-dependent Pol2 degradation suggests that SCFPof3 may also target other replisome components for degradation. We first sought to determine whether Pof3 is also involved in degradation of Mrc1 in S. pombe. As shown in Figure 4, Mrc1 became highly stable in pof3Δ cells under CHX treatment (Figure 4A and 4C). We then examined whether Mcm4 degradation in swi1Δ cells is inhibited by the inactivation of SCFPof3 (Figure 4B and 4D). Intriguingly, Mcm4 was significantly more stable in pof3Δ swi1Δ cells than in swi1Δ cells after CHX treatment. This result suggests that SCFPof3 also targets Mcm4 for proteasome-dependent degradation in response to replication stress provoked by swi1 deletion. Taken together, our results are consistent with the notion that SCFPof3 is involved in degradation of multiple replisome components.
Fig. 4. Pof3-dependent degradation of Mrc1 and Mcm4. 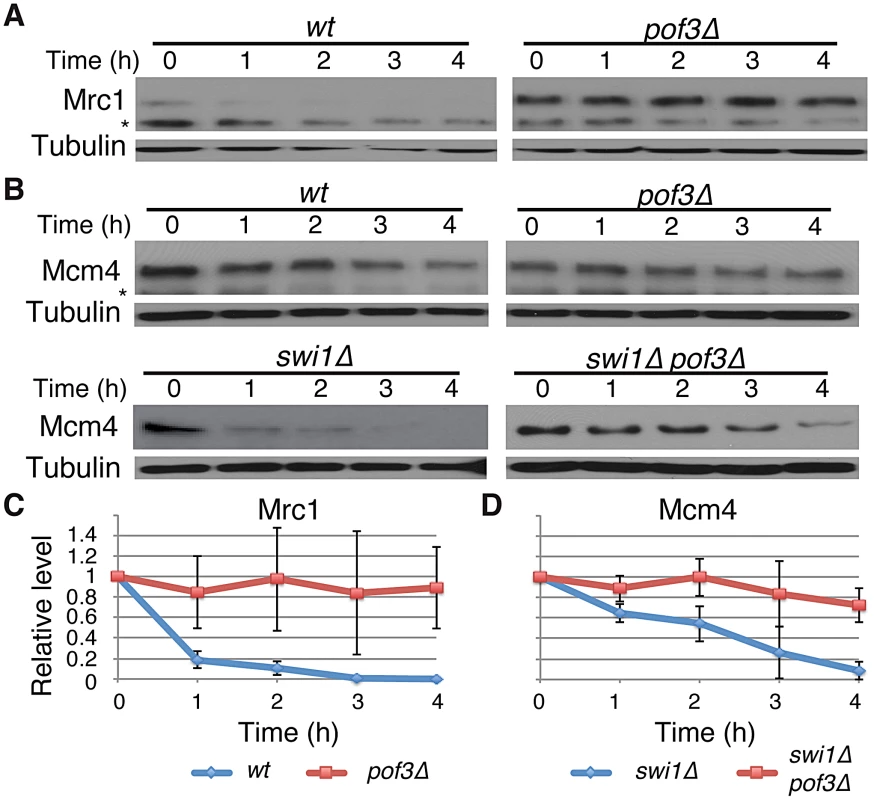
(A) Cellular amount of Mrc1 was determined in wt and pof3Δ cells, from 0 to 4 h of CHX treatment. Western blotting of tubulin was performed as a loading control. (B) As in A, cellular amount of Mcm4 was determined in wt, pof3Δ, swi1Δ, and pof3Δ swi1Δ cells. The asterisks indicate non-specific bands. (C) Stability of Mrc1 during the CHX treatment shown in A was quantified as described in Figure 1. For each strain, relative intensity of the Mrc1 band at 0 h was set to 1. (D) Stability of Mcm4 in swi1Δ and pof3Δ swi1Δ shown in B was quantified as described in C. Samples for Mrc1 or Mcm4 blots were derived from the same experiment and processed in parallel. Representative results of repeat experiments are shown. Replisome degradation prevents mitotic catastrophes in swi1Δ cells
In order to understand the physiological importance of replisome core degradation in the absence of Swi1, we investigated cellular phenotypes of swi1Δ, pof3Δ and swi1Δ pof3Δ double mutant cells. For this purpose, cells were stained with DAPI to visualize nuclear DNA. As shown in Figure 5A and 5B, swi1Δ and pof3Δ cells displayed an increased level of mitotic catastrophes (including chromosome missegregation, aneuploidy, fragmented nuclei, hypercondensed nuclei, “cut” and other aberrant phenotypes, which are shown by arrows) compared to wild-type cells. Importantly, this phenotype was further exacerbated in swi1Δ pof3Δ cells even in the absence of exogenous genotoxic agents (Figure 5A and 5B). We then used HU and camptothecin (CPT) to introduce S phase specific genotoxic stress. HU depletes the dNTP pool and causes an arrest of replication fork progression, while CPT traps topoisomerase I on DNA and induces replication fork breakage. HU or CPT treatment further enhanced the aberrant mitotic phenotypes (Figure 5A and 5B). Consistently, swi1Δ pof3Δ cells were more sensitive to HU and CPT than either single mutant (Figure 5C). In the presence of HU or CPT, swi1Δ cells accumulate DNA damage due to failure in the completion of DNA replication, which causes activation of the DNA damage checkpoint, leading to abnormal cell cycle arrest and a cell elongation phenotype [17], [70]. As expected, HU or CPT treatment caused cell elongation in swi1Δ cells (Figure 5A). However, this elongation phenotype was abolished in swi1Δ pof3Δ cells (Figure 5A), suggesting that the stabilization of replisome components attenuated cell cycle arrest in swi1Δ pof3Δ cells. This attenuation of cell cycle arrest could have caused a growth advantage, leading to the rather weak increase in the HU and CPT sensitivity of swi1Δ pof3Δ cells (Figure 5C), although these cells show strong mitotic catastrophes (Figure 5A).
Fig. 5. Forced accumulation of replisome components in swi1Δ cells causes catastrophic DNA replication and mitotic abnormalities. 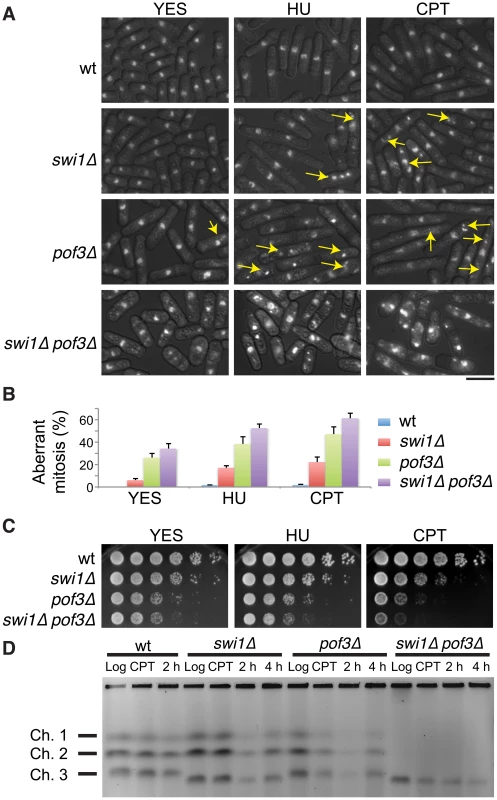
(A) swi1Δ pof3Δ cells have increased levels of mitotic catastrophes. Exponentially growing cells were treated with or without the indicated drugs (12 mM HU or 20 µM CPT for 6 h), fixed in ethanol, and stained with 4′,6-diamidino-2-phenylindole (DAPI). Representative images of observed nuclear phenotypes are shown. Representative mitotic failures are shown by arrows. Arrows were omitted from the images of swi1Δ pof3Δ cells because a large numbers of cells showed mitotic catastrophes. The scale bar represents 10 µM. (B) Quantification of cells with defective mitosis including chromosome missegregation, aneuploidy, cut and other aberrant phenotypes. More than 200 cells were counted for each strain. Error bars correspond to standard deviations obtained from three experiments. (C) DNA damage sensitivity of swi1Δ mutants is increased by pof3 deletion. Five-fold serial dilutions of cells were incubated on YES agar medium supplemented with the indicated drugs (2 mM HU or 1 µM CPT) for 2 to 3 days at 32°C. (D) pof3 deletion exacerbates replication recovery defects of swi1Δ mutants. Exponentially growing cells (Log) were incubated in the presence of 5 µM CPT for 3 h at 30°C (CPT), then washed and returned into fresh medium for 2 h or 4 h (2 h, 4 h). Chromosome samples were examined by PFGE. Representative results of repeat experiments are shown. swi1Δ cells have shorter chromosome III due to hyper recombination at rDNA repeats [42], [70], [101]. Next, we examined the ability of cells to recover DNA replication after CPT-dependent replication fork breakage. Exponentially growing cells (Log) were exposed to a low dose of CPT (5 µM) for 3 h and returned to fresh medium for 2 and 4 h (Figure 5D). Chromosome samples were then analyzed by pulsed-field gel electrophoresis (PFGE), which permits only fully replicated chromosomes to migrate into the gel. In contrast, chromosomes with replication intermediates stay in the well of the gel, allowing us to determine the rate of replication recovery. There was no detectable DNA replication defect in wild-type cells throughout the experiment (Figure 5D, Log, CPT, 2 h), indicating that the low dose of CPT used in this assay did not cause major replication problems in wild-type cells. Although chromosomes from swi1Δ cells migrated into the gel immediately after the CPT exposure (CPT), we observed a marked reduction in chromosome intensity at 2 h after CPT treatment (Figure 5D). This result indicates that the low dose of CPT caused replication problems in swi1Δ cells, which is consistent with previous studies [70]. However, there was a significant recovery at 4 h after the CPT removal due to the completion of DNA synthesis. A similar replication recovery was also observed in pof3Δ cells. In contrast, there was no DNA replication recovery in swi1Δ pof3Δ cells during the course of the experiment (Figure 5D), indicating that these cells experience further difficulties in replication and/or repair of broken replication forks when treated with a replication-stressing agent. Interestingly, we repeatedly observed much less appearance of chromosomes I and II in the gel for swi1Δ pof3Δ cells (Figure 5D; Figure S4), suggesting that these cells experience major problems in DNA replication and chromosome maintenance. Consistently, there was an increased level of mitotic catastrophes in these cells (Figure 5A and 5B). Considering that pof3 deletion stabilizes replisome components (Figure 3E; Figure 4), our results suggest that programmed replisome degradation represents a mechanism to prevent catastrophic DNA replication in response to replication stress caused by swi1 deletion (Figure 6). Similar replication and mitotic phenotypes were observed in swi1Δ mts3-1 cells, which are defective in proteasome functions (Figure S5), strengthening the idea that replisome degradation plays a critical role in the maintenance of genomic integrity.
Fig. 6. Degradation of replisome components prevents genomic instability. 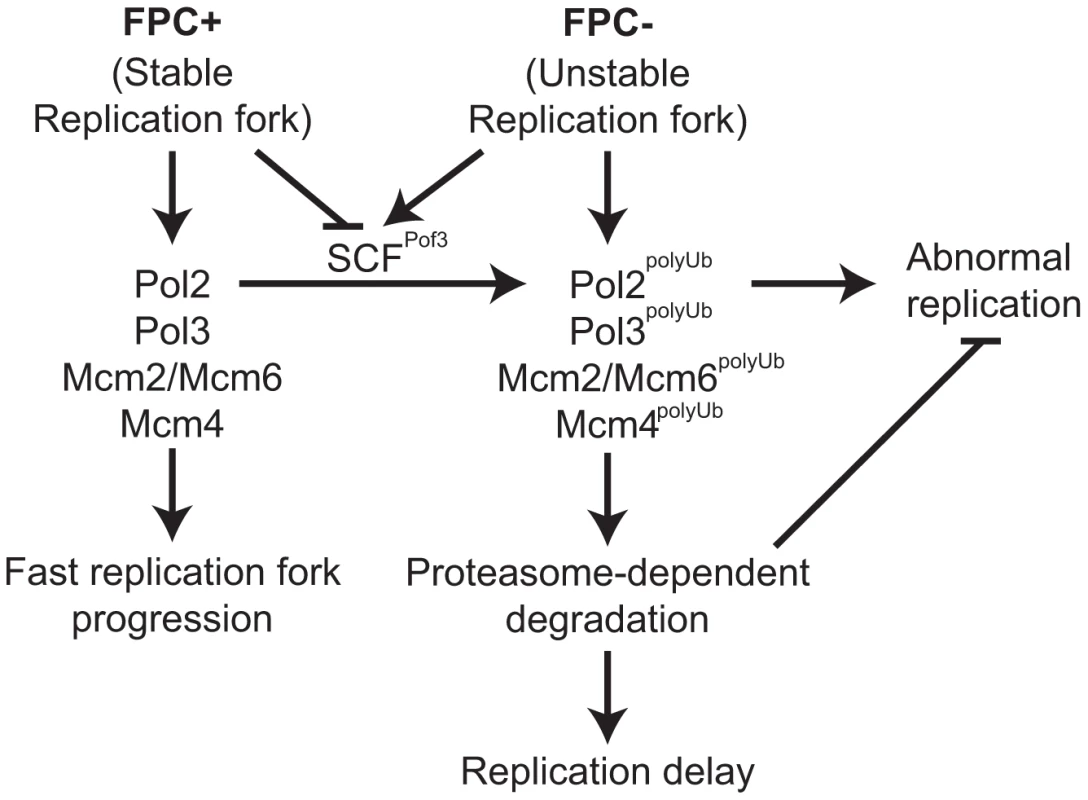
Models for the roles of the FPC in replisome stabilization. The FPC (Swi1-Swi3) stabilizes the replication fork (left; FPC+) and replisome components and promotes an efficient progression of the replication fork. The FPC suppresses replication stress that leads to unstable replication forks (right; FPC−). Under replication stress, SCFPof3 may have access to replisome components and transfer ubiquitin moieties. Ubiquitinated replisome components undergo proteasome-dependent degradation, resulting in delayed replication fork progression. Replisome degradation prevents abnormal DNA replication, thus preserving genomic integrity. Discussion
Accurate transmission of genetic information is one of the major tasks cells need to achieve in order to preserve the species and prevent genetic diseases. Accordingly, eukaryotic cells have developed a variety of genome maintenance mechanisms. In response to DNA damage or replication stress, cells activate checkpoint pathways to coordinate cell cycle arrest with DNA repair activities. It is also known that the replication checkpoint functions to stabilize replication forks by preserving replisome and DNA structures. In this study, we have described an alternative cellular mechanism in response to replication stress. Our studies suggest that cells facilitate proteasome-dependent degradation of replisome components in response to replication stress to preserve genomic integrity.
Proteasome-dependent degradation of replisome components preserves genomic integrity
Swi1 and its orthologues are known to be involved in the stabilization of replication forks to prevent genetic instability during DNA replication. Genetic analyses have suggested that FPC is involved in coordinating leading - and lagging-strand DNA synthesis. It is also suggested that the FPC couples polymerase and helicase activities at stalled forks [14], [15]. Thus, the functions of Swi1 would become even more important to maintain the integrity of the replication fork when it encounters difficult-to-replicate sites or programmed fork pausing sites that are scattered throughout the genome. Consistently, FPC plays a critical role in programmed fork pausing and replication termination events near the mating-type (mat1) locus and at fork pausing sites in rDNA repeats and tRNA loci in yeast [16], [17], [21], [29], [38], [39]. Importantly, recent studies indicated that swi1Δ cells experience fork collapse at these difficult-to-replicate regions [43]–[45]. Therefore, inactivation of Swi1 causes defects in replication fork stabilization at natural impediments, leading to general replication stress at the replication fork.
It is well known that Cdt1 and Cdc6 undergo rapid proteasome-dependent degradation to restrict replication licensing once per cell cycle [71], [72]. However, how replisome degradation contributes to DNA replication process is largely unknown. In this report, we show that DNA polymerases and helicases undergo rapid degradation upon replication stress in the absence of Swi1 (Figure 1). This degradation is dependent on the ubiquitin-proteasome system (Figure 2). In the absence of Swi1, cells experience unstable replication forks that lead to an increased level of replication-dependent DNA damage and hyper-recombination [16], [17], [43]. Such replication stress appears to cause replisome degradation in order to prevent abnormal DNA replication and mitotic catastrophes (Figure 5; Figure S5). These results suggest that replisome degradation functions to maintain genomic integrity during DNA replication in response to replication stress (Figure 6). Similar mechanisms have been described in the transcription-coupled DNA repair (TCR), which is activated by transcription blockage in response to genotoxic agents [73], [74]. In this mechanism, the Cockayne syndrome B protein (budding yeast Rad26) interacts with Def1 to regulate ubiquitination of Rpb1, the large subunit of RNA polymerase II (RNAPII), which results in proteasome-dependent degradation of RNAPII [75],[76]. Ubiquitination of Rbp1 is achieved by the Rsp5/Nedd4 ubiquitin ligase, which promotes DNA-damage induced degradation of RNAPII in budding yeast and human cells [77]–[79]. RNAPII degradation appears to be an alternative mechanism to TCR. Studies indicate that the loss of both TCR and RNAPII degradation pathways renders cells hypersensitive to DNA damage, thus Def1 promotes proteolysis of RNAPII when the lesion cannot be rapidly repaired by TCR [75], [80]–[82]. Therefore, analogous to the DNA damage-induced RNAPII degradation pathway, our present findings suggest that the cell elicits a replisome degradation program when the replication fork is adversely blocked. We speculate that, depending on the degree of replication problems, re-building and re-loading new replisomes might be advantageous to the cell, rather than re-using existing replisome components that are compromised. Therefore, we suggest that replisome degradation is an alternative mechanism to replisome stabilization and prevents DNA synthesis by compromised replisomes.
Roles of Pol2 degradation in replisome dynamics
We also found that Pol2 (Polε) is significantly unstable even in wild-type cells (Figure 1A and 1B), while Pol3 (Polδ) is relatively stable (Figure 1A and 1B). The high rate of Pol2 turnover may suggest that Pol2 needs to be re-loaded during leading-strand synthesis. Since Pol2 is suggested to work continuously on the leading-strand [53], one might think that the high turnover of Pol2 poses a disadvantage to the cells. However, it is possible that the polymerases fall off the chromatin every time the fork arrives at programmed pausing sites or difficult-to-replicate regions. In addition, Pol2 may undergo degradation once it falls off the chromatin. Such a degradation mechanism would also be advantageous for the cell to refresh Pol2 enzymes by efficiently reloading newly synthesized Pol2 at the moving replication fork. On the other hand, the discontinuous nature of Pol3-dependent lagging-strand synthesis would be sufficient to keep Pol3 refreshed at the fork in order to avoid replication-dependent errors or mutations. Another possibility is that this mechanism might simply maintain the coupling of leading - and lagging-strand synthesis. Thus, in addition to the role of replisome degradation in preventing genomic instability described above, polymerase degradation may function to eliminate non-functional replisomes and serve as a mechanism to maintain active DNA polymerases at the replication fork.
Our investigation also revealed that Pol2 and Mcm4 undergo rapid degradation in the presence of CPT (Figure S6), which breaks replication forks. However, in this condition, the Mcm6 level was maintained (Figure S6), although it was highly unstable in swi1Δ cells (Figure 1C and D). It is possible that some replisome components remain stable on the chromatin in the presence of CPT. Interestingly, Trenz et al. reported that polymerases fall off the chromatin in response to CPT, whereas Mcm7 persists [83]. Therefore, swi1 deletion generates a situation distinct from a simple mechanical breakage of the fork caused by DNA damaging agents, where the replisome cannot continue replicating DNA. Importantly, Swi1 functions as an ancillary component of the replisome by interacting with various replisome components, coupling polymerase and helicase activities and coordinating semi-discontinuous DNA synthesis [14], [15]. It is also reported that Swi1 protects replication forks at difficult-to-replicate sites [43]–[45]. Therefore, we suggest that the loss of Swi1 results in unstable replisome structures at the moving replication fork during ongoing DNA synthesis, allowing us to examine replisome degradation pathways during DNA replication.
The FPC–dependent stabilization of replisome components
The FPC moves with the replication fork and interacts with replisome components [16], [18], [21]–[25], [27], [28], [84]–[87]. Surprisingly, Pol2, Pol3, and MCM subunits are rapidly degraded in swi1Δ cells (Figure 1). Consistently, replication fork progression is compromised in FPC deficient cells (Figure S1) [27], [52]. These results suggest that Swi1 prevents degradation of replisome components to maintain efficient progression of replication forks. In wild-type cells, multiple activities required for DNA synthesis are coupled to form a large replisome complex, resulting in efficient progression of the replication fork. However, in the absence of Swi1, DNA replication-related activities are probably uncoupled especially at naturally difficult-to-replicate regions. This uncoupling generates unstable replisome structures, which may expose degradation signals of various replisome components to a ubiquitin ligase(s) associated with the replication fork. Importantly, swi1Δ pof3Δ double mutants showed catastrophic DNA replication and mitosis, suggesting that Pof3-dependent degradation of replisome components prevents genomic instability. However, we cannot exclude the possibility that mitotic catastrophe phenotypes are caused by stabilization of other Pof3 targets. For example, Pof3-dependent proteolysis of Ams2 is responsible for cell cycle-dependent transcriptional activation of core histone genes in S. pombe [88]. Indeed, defects in Ams2 degradation leads to accumulation of histones and alteration of centromere structures [88]. Such dysregulation of histone homeostasis during S phase could also lead to abnormal DNA replication, leading to mitotic problems. However, Dia2, a Pof3-related F-box protein, is associated with the replisome and regulates replication forks in budding yeast. Dia2 is involved in ubiquitination of budding yeast Mrc1, which is a component of the replisome [67]–[69]. Moreover, Tof1 (Swi1 orthologue) collaborates with Dia2 to maintain genomic integrity [89]. These findings suggest that Pof3/Dia2 acts as a part of the replisome. Consistently, we found in fission yeast that SCFPof3 is largely responsible for degradation of some replisome components (Figure 3 and Figure 4). Therefore, Pof3-mediated ubiquitination of replisome components may be prevented by Swi1-dependent replisome stabilization, which may mask potential degradation signals of multiple replisome components (Figure 6). Since many SCF ubiquitin ligases are known to recognize phosphorylated degradation signals (phospho-degrons), it is also possible that replisome components undergo phosphorylation in the absence of Swi1. Therefore, Swi1 might have direct functions in inhibiting SCFPof3 ligase possibly by inhibiting Pof3 or inhibiting potential kinases. In this regard, it is interesting to note that Mrc1 contains a potential phospho-degron, and that the Hsk1 kinase is required for efficient degradation of Mrc1 [55]. Consistently, our present study shows that Pof3 is involved in Mrc1 degradation (Figure 4). Therefore, it is possible that Hsk1-dependent phosphorylation creates Pof3-targeted phospho-degrons on multiple replisome components. However, Mrc1 degradation is independent of replication stress (Figure 1C), raising the possibility that other kinases are responsible for replisome degradation upon replication stress. Further investigation of proteasome-dependent replisome degradation would identify detailed pathways in the regulation of the replisome.
Materials and Methods
General techniques
The methods used for genetic and biochemical analyses of fission yeast have been described previously [90], [91]. Drug sensitivity assays, Western blotting, pulsed-field gel electrophoresis (PFGE) and 4′,6-diamidino-2-phenylindole (DAPI) staining of nuclear DNA were performed as described [70], [92]. Flow cytometry of DNA content has been described [93], [94].
S. pombe strains
S. pombe strains used in this study were constructed using standard techniques [91], and their genotypes and sources are listed in Table S1. swi1Δ (swi1::hphMX6 and swi1::natMX6) and pof3Δ (pof3::ura4MX6) were generated by a two-step PCR method [95], to replace swi1 and pof3 open reading frames with selection marker genes. The two-step PCR method was also used to construct a GFP or 13Myc tag at the C terminus of mcm2, mcm6 and pof3, generating mcm2-GFP:hphMX6 (mcm2-GFP), mcm6-GFP:hphMX6 (mcm6-GFP), and pof3-13Myc:hphMX6 (pof3-13Myc), respectively. Oligonucleotide primers used in the two-step PCR method described above are listed in Table S2. A temperature-sensitive skp1-94 mutation was isolated using error-prone PCR methods [63].
Mutations and epitope-tagged genes have been described for orc1-5FLAG [96], pol2-5FLAG, pol3-5FLAG [97], swi1::kanMX6 [17], cdc45-5FLAG [98], cdc25-22 [99] and mts3-1 [57].
mcm2-GFP, mcm6-GFP, pof3-13MYC, orc1-5FLAG, pol2-5FLAG, pol3-5FLAG, and cdc45-5FLAG cells show normal growth phenotype and were not abnormally sensitive to HU, CPT and MMS, indicating that the tagged version of these proteins are functional.
Cell extract preparation for Western blotting
To examine protein stability, exponentially growing cells were treated with 0.1 mg/ml of cycloheximide (CHX) for the indicated times and collected. Whole-cell extracts were prepared as described [100]. Briefly, cells were washed in STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, and 1 mM NaN3) and lysed by glass beads in lysis buffer U (50 mM Tris-HCl pH 6.8, 2% SDS, 2 mM EDTA, 10% glycerol, and 4 M urea) using a FastPrep Cell disruptor (Qbiogene, Irvine, CA) for 40 seconds at speed 6. Protein extract was clarified by centrifugation at 13,000 rpm in an Eppendorf microcentrifuge 5415R for 10 min at 4°C, and the protein concentration was determined using BCA protein Assay Reagent (Thermo Fisher Scientific, Waltham, MA). Immediately after the protein concentration assay, protein extracts were boiled in the presence of 5% beta-mercaptoethanol and stored at −20°C. For immunoblotting, Myc, GFP, and FLAG fusion proteins were probed with the anti-c-Myc 9E10 antibody (Covance, Princeton, NJ), anti-GFP antibody (Roche, Indianapolis, IN), and anti-FLAG M2 (Sigma-Aldrich) antibody, respectively. The anti-tubulin TAT-1 (gift from Dr. K. Gull), anti-Mcm4 (gift from Drs. S. Kearsey, Z. Lygerou, and H. Nishitani), anti-Mrc1 (gift from Dr. K. Tanaka), and anti-PCNA (gift from Dr. T. Tsurimoto) antibodies were used to detect the corresponding proteins. Quantification of protein bands was performed using NIH ImageJ software.
Fractionation of cells into soluble and chromatin-enriched fractions
Cell fractionation was performed as described elsewhere [55], [56] with modifications. Exponentially growing cells were harvested in 0.01% sodium azide by centrifugation and washed sequentially with STOP buffer, water, and 1.2 M sorbitol, at 4°C. Cells were resuspended in CB1 buffer (50 mM sodium citrate, 40 mM EDTA, 1.2 M sorbitol) and treated with 2.5 mg/ml of Zymolyase for approximately 20 min at 32°C. When cell lysis reached approximately 95%, cell wall digestion by Zymolyase was terminated by adding equal volume of ice-cold CB2 buffer (1.2 M sorbitol, 10 mM Tris-HCl ph7.5), and resulting spheroplasts were washed twice with 1.2 M Sorbitol. Spheroplasts were then incubated in Lysis buffer T (50 mM potassium acetate, 2 mM MgCl2, 20 mM HEPES-KOH pH 7.4, 10 mM EDTA, 1 M Sorbitol, 1% Triton X-100) supplemented with Halt protease inhibitor cocktail (Thermo Fisher Scientific) for 10 min at 4°C. Subsequently, extracts were fractionated into soluble and pellet fractions by centrifugation for 10 min at 4°C. Supernatants (Triton X-100-soluble fraction) were removed, boiled with a one-third volume of 3× SDS-PAGE loading buffer (150 mM Tris-HCl pH 6.8, 6% SDS, 6 mM EDTA, 30% glycerol, 15% beta-mercaptoethanol), and stored at −20°C. The pellets (Triton X-100-insoluble fraction) were washed once with Lysis buffer (without Triton X-100), suspended in Lysis buffer, boiled with a one-third volume of 3× SDS-PAGE loading buffer, and stored at −20°C.
Immunoprecipitation and detection of ubiquitinated proteins
Immunoprecipitation was performed using the anti-myc 9E10 (Covance) antibody with protein G sepharose beads as described [70]. Proteins associated with the anti-myc antibody were analyzed by Western blotting. For detection of ubiquitinated protein, S. pombe cells expressing a hexahistidine-ubiquitin (6xHis-Ub) peptide [61] were lysed in lysis buffer G (6 M guanidine hydrochloride, 100 mM sodium phosphate pH 8.0, and 50 mM Tris-HCl pH 8.0). Hexahistidine-ubiquitinated proteins were purified with Ni-NTA agarose beads (Qiagen, Valencia, CA), eluted in the presence of 4 M urea, and analyzed by Western blotting.
Supporting Information
Zdroje
1. BellSP, DuttaA (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71 : 333–374.
2. BoddyMN, RussellP (2001) DNA replication checkpoint. Curr Biol 11: R953–R956.
3. OsbornAJ, ElledgeSJ, ZouL (2002) Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol 12 : 509–516.
4. NybergKA, MichelsonRJ, PutnamCW, WeinertTA (2002) TOWARD MAINTAINING THE GENOME: DNA Damage and Replication Checkpoints. Annu Rev Genet 36 : 617–656.
5. AbrahamRT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15 : 2177–2196.
6. RouseJ, JacksonSP (2002) Interfaces between the detection, signaling, and repair of DNA damage. Science 297 : 547–551.
7. CarrAM (2002) DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amst) 1 : 983–994.
8. LopesM, Cotta-RamusinoC, PellicioliA, LiberiG, PlevaniP, et al. (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412 : 557–561.
9. PaciottiV, ClericiM, ScottiM, LucchiniG, LongheseMP (2001) Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol 21 : 3913–3925.
10. SogoJM, LopesM, FoianiM (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297 : 599–602.
11. TerceroJA, DiffleyJF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412 : 553–557.
12. TerceroJA, LongheseMP, DiffleyJF (2003) A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11 : 1323–1336.
13. LeeBS, GrewalSI, KlarAJ (2004) Biochemical interactions between proteins and mat1 cis-acting sequences required for imprinting in fission yeast. Mol Cell Biol 24 : 9813–9822.
14. LemanAR, NoguchiE (2012) Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle 11 : 3945–3955.
15. McFarlaneRJ, MianS, DalgaardJZ (2010) The many facets of the Tim-Tipin protein families' roles in chromosome biology. Cell Cycle 9 : 700–705.
16. NoguchiE, NoguchiC, McDonaldWH, YatesJR3rd, RussellP (2004) Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 24 : 8342–8355.
17. NoguchiE, NoguchiC, DuLL, RussellP (2003) Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol 23 : 7861–7874.
18. GotterAL, SuppaC, EmanuelBS (2007) Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol 366 : 36–52.
19. NoguchiE (2010) The DNA Replication Checkpoint and Preserving Genomic Integrity During DNA Synthesis. Nature Education 3 : 46.
20. SabatinosSA (2010) Replication Fork Stalling and the Fork Protection Complex. Nature Education 3 : 40.
21. CalzadaA, HodgsonB, KanemakiM, BuenoA, LabibK (2005) Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 19 : 1905–1919.
22. ChouDM, ElledgeSJ (2006) Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci U S A 103 : 18143–18147.
23. ErricoA, CostanzoV, HuntT (2007) Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc Natl Acad Sci U S A 104 : 14929–14934.
24. KatouY, KanohY, BandoM, NoguchiH, TanakaH, et al. (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 : 1078–1083.
25. LemanAR, NoguchiC, LeeCY, NoguchiE (2010) Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci 123 : 660–670.
26. TanakaH, KubotaY, TsujimuraT, KumanoM, MasaiH, et al. (2009) Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes Cells 14 : 949–963.
27. Unsal-KacmazK, ChastainPD, QuP-P, MinooP, Cordeiro-StoneM, et al. (2007) The Human Tim/Tipin Complex Coordinates an Intra-S Checkpoint Response to UV That Slows Replication Fork Displacement. Mol Cell Biol 27 : 3131–3142.
28. Yoshizawa-SugataN, MasaiH (2007) Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem 282 : 2729–2740.
29. DalgaardJZ, KlarAJ (2000) swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102 : 745–751.
30. AguileraA, Gomez-GonzalezB (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9 : 204–217.
31. AzvolinskyA, DunawayS, TorresJZ, BesslerJB, ZakianVA (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20 : 3104–3116.
32. LemoineFJ, DegtyarevaNP, LobachevK, PetesTD (2005) Chromosomal translocations in yeast induced by low levels of DNA polymerase: A model for chromosome fragile sites. Cell 120 : 587–598.
33. MirkinEV, MirkinSM (2007) Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71 : 13–35.
34. PearsonCE, Nichol EdamuraK, ClearyJD (2005) Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6 : 729–742.
35. RaveendranathanM, ChattopadhyayS, BolonYT, HaworthJ, ClarkeDJ, et al. (2006) Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J 25 : 3627–3639.
36. KobayashiT, HoriuchiT (1996) A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1 : 465–474.
37. TakeuchiY, HoriuchiT, KobayashiT (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17 : 1497–1506.
38. HodgsonB, CalzadaA, LabibK (2007) Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell 18 : 3894–3902.
39. KringsG, BastiaD (2004) swi1 - and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 101 : 14085–14090.
40. MohantyBK, BairwaNK, BastiaD (2006) The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 103 : 897–902.
41. Sanchez-GorostiagaA, Lopez-EstranoC, KrimerDB, SchvartzmanJB, HernandezP (2004) Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol 24 : 398–406.
42. SommarivaE, PellnyTK, KarahanN, KumarS, HubermanJA, et al. (2005) Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol 25 : 2770–2784.
43. PryceDW, RamayahS, JaendlingA, McFarlaneRJ (2009) Recombination at DNA replication fork barriers is not universal and is differentially regulated by Swi1. Proc Natl Acad Sci U S A 106 : 4770–4775.
44. RozenzhakS, Mejia-RamirezE, WilliamsJS, SchafferL, HammondJA, et al. (2010) Rad3ATR decorates critical chromosomal domains with γH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet 6: e1001032 doi:10.1371/journal.pgen.1001032.
45. SabouriN, McDonaldKR, WebbCJ, CristeaIM, ZakianVA (2012) DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev 26 : 581–593.
46. CherngN, ShishkinAA, SchlagerLI, TuckRH, SloanL, et al. (2011) Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc Natl Acad Sci U S A 108 : 2843–2848.
47. LemanAR, DheekolluJ, DengZ, LeeSW, DasMM, et al. (2012) Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle 11 : 2337–2347.
48. LiuG, ChenX, GaoY, LewisT, BarthelemyJ, et al. (2012) Altered replication in human cells promotes DMPK (CTG)(n). (CAG)(n) repeat instability. Mol Cell Biol 32 : 1618–1632.
49. RazidloDF, LahueRS (2008) Mrc1, Tof1 and Csm3 inhibit CAG.CTG repeat instability by at least two mechanisms. DNA Repair (Amst) 7 : 633–640.
50. ShishkinAA, VoineaguI, MateraR, CherngN, ChernetBT, et al. (2009) Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol Cell 35 : 82–92.
51. VoineaguI, NarayananV, LobachevKS, MirkinSM (2008) Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci U S A 105 : 9936–9941.
52. TourriereH, VersiniG, Cordon-PreciadoV, AlabertC, PaseroP (2005) Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19 : 699–706.
53. PursellZF, IsozI, LundstromEB, JohanssonE, KunkelTA (2007) Yeast DNA polymerase ε participates in leading-strand DNA replication. Science 317 : 127–130.
54. Nick McElhinnySA, GordeninDA, StithCM, BurgersPM, KunkelTA (2008) Division of labor at the eukaryotic replication fork. Mol Cell 30 : 137–144.
55. ShimmotoM, MatsumotoS, OdagiriY, NoguchiE, RussellP, et al. (2009) Interactions between Swi1–Swi3, Mrc1 and S phase kinase, Hsk1 may regulate cellular responses to stalled replication forks in fission yeast. Genes Cells 14 : 669–682.
56. KaiM, TanakaH, WangTS (2001) Fission yeast Rad17 associates with chromatin in response to aberrant genomic structures. Mol Cell Biol 21 : 3289–3301.
57. GordonC, McGurkG, WallaceM, HastieND (1996) A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J Biol Chem 271 : 5704–5711.
58. SeegerM, GordonC, FerrellK, DubielW (1996) Characteristics of 26 S proteases from fission yeast mutants, which arrest in mitosis. J Mol Biol 263 : 423–431.
59. FuH, LinYL, FatimababyAS (2010) Proteasomal recognition of ubiquitylated substrates. Trends Plant Sci 15 : 375–386.
60. SchraderEK, HarstadKG, MatouschekA (2009) Targeting proteins for degradation. Nat Chem Biol 5 : 815–822.
61. FramptonJ, IrmischA, GreenCM, NeissA, TrickeyM, et al. (2006) Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell 17 : 2976–2985.
62. NakayamaKI, NakayamaK (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 6 : 369–381.
63. LehmannA, KatayamaS, HarrisonC, DhutS, KitamuraK, et al. (2004) Molecular interactions of fission yeast Skp1 and its role in the DNA damage checkpoint. Genes Cells 9 : 367–382.
64. KatayamaS, KitamuraK, LehmannA, NikaidoO, TodaT (2002) Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol Biol Cell 13 : 211–224.
65. MamnunYM, KatayamaS, TodaT (2006) Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem Biophys Res Commun 350 : 125–130.
66. WilliamsDR, McIntoshJR (2002) mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell 1 : 758–773.
67. KoeppDM, KileAC, SwaminathanS, Rodriguez-RiveraV (2006) The F-box protein Dia2 regulates DNA replication. Mol Biol Cell 17 : 1540–1548.
68. MorohashiH, MaculinsT, LabibK (2009) The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr Biol 19 : 1943–1949.
69. MimuraS, KomataM, KishiT, ShirahigeK, KamuraT (2009) SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J 28 : 3693–3705.
70. RappJB, NoguchiC, DasMM, WongLK, AnsbachAB, et al. (2010) Checkpoint-dependent and -independent roles of Swi3 in replication fork recovery and sister chromatid cohesion in fission yeast. PLoS ONE 5: e13379 doi:10.1371/journal.pone.0013379.
71. HookSS, LinJJ, DuttaA (2007) Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol 19 : 663–671.
72. O'ConnellBC, HarperJW (2007) Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr Opin Cell Biol 19 : 206–214.
73. Hammond-MartelI, YuH, AffarEB (2012) Roles of ubiquitin signaling in transcription regulation. Cell Signal 24 : 410–421.
74. SvejstrupJQ (2007) Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci 32 : 165–171.
75. WoudstraEC, GilbertC, FellowsJ, JansenL, BrouwerJ, et al. (2002) A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415 : 929–933.
76. RatnerJN, BalasubramanianB, CordenJ, WarrenSL, BregmanDB (1998) Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem 273 : 5184–5189.
77. HuibregtseJM, YangJC, BeaudenonSL (1997) The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci U S A 94 : 3656–3661.
78. BeaudenonSL, HuacaniMR, WangG, McDonnellDP, HuibregtseJM (1999) Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol 19 : 6972–6979.
79. AnindyaR, AygunO, SvejstrupJQ (2007) Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell 28 : 386–397.
80. LommelL, BucheliME, SwederKS (2000) Transcription-coupled repair in yeast is independent from ubiquitylation of RNA pol II: implications for Cockayne's syndrome. Proc Natl Acad Sci U S A 97 : 9088–9092.
81. SomeshBP, ReidJ, LiuWF, SogaardTM, Erdjument-BromageH, et al. (2005) Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121 : 913–923.
82. SomeshBP, SigurdssonS, SaekiH, Erdjument-BromageH, TempstP, et al. (2007) Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell 129 : 57–68.
83. TrenzK, SmithE, SmithS, CostanzoV (2006) ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25 : 1764–1774.
84. ErricoA, CosentinoC, RiveraT, LosadaA, SchwobE, et al. (2009) Tipin/Tim1/And1 protein complex promotes Polα chromatin binding and sister chromatid cohesion. EMBO J 28 : 3681–3692.
85. MatsumotoS, OginoK, NoguchiE, RussellP, MasaiH (2005) Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J Biol Chem 280 : 42536–42542.
86. NedelchevaMN, RoguevA, DolapchievLB, ShevchenkoA, TaskovHB, et al. (2005) Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J Mol Biol 347 : 509–521.
87. NumataY, IshiharaS, HasegawaN, NozakiN, IshimiY (2010) Interaction of human MCM2-7 proteins with TIM, TIPIN and Rb. J Biochem 147 : 917–927.
88. TakayamaY, MamnunYM, TrickeyM, DhutS, MasudaF, et al. (2010) Hsk1 - and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell 18 : 385–396.
89. BairwaNK, MohantyBK, StamenovaR, CurcioMJ, BastiaD (2011) The intra-S phase checkpoint protein Tof1 collaborates with the helicase Rrm3 and the F-box protein Dia2 to maintain genome stability in Saccharomyces cerevisiae. J Biol Chem 286 : 2445–2454.
90. MorenoS, KlarA, NurseP (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194 : 795–823.
91. Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E (1993) Experiments with Fission Yeast: A laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
92. NoguchiC, NoguchiE (2007) Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics 175 : 553–566.
93. RhindN, RussellP (1998) The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics 149 : 1729–1737.
94. NoguchiE, ShanahanP, NoguchiC, RussellP (2002) CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr Biol 12 : 599–605.
95. KrawchukMD, WahlsWP (1999) High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15 : 1419–1427.
96. OgawaY, TakahashiT, MasukataH (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol 19 : 7228–7236.
97. MoserBA, SubramanianL, ChangYT, NoguchiC, NoguchiE, et al. (2009) Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J 28 : 810–820.
98. YabuuchiH, YamadaY, UchidaT, SunathvanichkulT, NakagawaT, et al. (2006) Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J 25 : 4663–4674.
99. FantesP (1979) Epistatic gene interactions in the control of division in fission yeast. Nature 279 : 428–430.
100. MasaiH, MiyakeT, AraiK (1995) hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J 14 : 3094–3104.
101. AnsbachAB, NoguchiC, KlansekIW, HeidlebaughM, NakamuraTM, et al. (2008) RFCCtf18 and the Swi1–Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell 19 : 595–607.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



