-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
Seed development in flowering plants is initiated after a double fertilization event with two sperm cells fertilizing two female gametes, the egg cell and the central cell, leading to the formation of embryo and endosperm, respectively. In most species the endosperm is a polyploid tissue inheriting two maternal genomes and one paternal genome. As a consequence of this particular genomic configuration the endosperm is a dosage sensitive tissue, and changes in the ratio of maternal to paternal contributions strongly impact on endosperm development. The FERTILIZATION INDEPENDENT SEED (FIS) Polycomb Repressive Complex 2 (PRC2) is essential for endosperm development; however, the underlying forces that led to the evolution of the FIS-PRC2 remained unknown. Here, we show that the functional requirement of the FIS-PRC2 can be bypassed by increasing the ratio of maternal to paternal genomes in the endosperm, suggesting that the main functional requirement of the FIS-PRC2 is to balance parental genome contributions and to reduce genetic conflict. We furthermore reveal that the AGAMOUS LIKE (AGL) gene AGL62 acts as a dosage-sensitive seed size regulator and that reduced expression of AGL62 might be responsible for reduced size of seeds with increased maternal genome dosage.
Published in the journal: Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development. PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003163
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003163Summary
Seed development in flowering plants is initiated after a double fertilization event with two sperm cells fertilizing two female gametes, the egg cell and the central cell, leading to the formation of embryo and endosperm, respectively. In most species the endosperm is a polyploid tissue inheriting two maternal genomes and one paternal genome. As a consequence of this particular genomic configuration the endosperm is a dosage sensitive tissue, and changes in the ratio of maternal to paternal contributions strongly impact on endosperm development. The FERTILIZATION INDEPENDENT SEED (FIS) Polycomb Repressive Complex 2 (PRC2) is essential for endosperm development; however, the underlying forces that led to the evolution of the FIS-PRC2 remained unknown. Here, we show that the functional requirement of the FIS-PRC2 can be bypassed by increasing the ratio of maternal to paternal genomes in the endosperm, suggesting that the main functional requirement of the FIS-PRC2 is to balance parental genome contributions and to reduce genetic conflict. We furthermore reveal that the AGAMOUS LIKE (AGL) gene AGL62 acts as a dosage-sensitive seed size regulator and that reduced expression of AGL62 might be responsible for reduced size of seeds with increased maternal genome dosage.
Introduction
Seed development in flowering plants is initiated by double fertilization of the female gametophyte. Within the female gametophyte there are two distinct gametic cells that have divergent fates after fertilization. The haploid egg cell will give rise to the diploid embryo, while the homodiploid central cell will form the triploid endosperm [1]. The endosperm supports embryo growth by delivering nutrients acquired from the mother plant [2]. As most angiosperms, the endosperm of Arabidopsis thaliana follows the nuclear-type of development where an initial syncytial phase of free nuclear divisions without cytokinesis is followed by cellularization [3]. At the eighth mitotic cycle cellularization of the syncytial endosperm is initiated in the micropylar domain around the embryo, coinciding with the early heart stage of embryo development [4], [5]. The timing of endosperm cellularization correlates with final seed size. Precocious endosperm cellularization results in small seeds, while delayed endosperm cellularization causes the formation of enlarged seeds [6], [7]. Timing of endosperm cellularization can be manipulated by interploidy hybridizations, which have opposite effects on endosperm cellularization and seed size dependent on the direction of the increased parental genome contribution. Increased maternal genome contribution (4n×2n, corresponds to maternal excess hybridization) causes precocious endosperm cellularization and the formation of small seeds. Conversely, increased paternal genome dosage (2n×4n, corresponds to paternal excess hybridization) results in delayed or complete failure of endosperm cellularization, causing seed abortion with an accession-dependent frequency [6], [8], [9]. Developmental defects caused by interploidy hybridizations with increased paternal genome contribution are associated with deregulation of genes that are directly or indirectly controlled by the FERTILIZATION INDEPENDENT SEED (FIS) Polycomb Repressive Complex 2 (PRC2), implicating that developmental aberrations in response to interploidy crosses are largely caused by deregulated FIS-PRC2 target genes [9]. PRC2 is a chromatin-modifying complex that ensures mitotically stable repression of specific target genes by applying trimethylation marks at lysine 27 of histone H3 (H3K27me3) [10], [11]. In plants, several PRC2 subunits are encoded by small gene families that form specific complexes with distinct functions during plant development [10]. The FIS-PRC2 is comprised of the subunits MEDEA (MEA), FERTILIZATION INDEPENDENT SEED2 (FIS2), FERTILIZATION INDEPENDENT ENDOSPERM (FIE) and MULTICOPY SUPPRESSOR OF IRA1 (MSI1) [10]. The FIS-PRC2 complex plays a pivotal role in suppressing initiation of endosperm and seed development in the absence of fertilization [12]–[15]. After fertilization, loss of FIS function causes endosperm overproliferation and cellularization failure, ultimately leading to seed abortion [12], [16], [17]. The phenomenon of decreased seed size in response to maternal excess interploidy hybridizations is known since long [6]; however, the underlying molecular mechanism for this phenomenon remains unknown. A recent study revealed increased expression of the FIS-PRC2 subunit FIS2 in response to maternal excess hybridizations [18], possibly linking increased FIS-PRC2 activity with decreased seed size. Other recent work proposed increased levels of 24-nt small interfering RNAs (p4-siRNAs) to cause decreased seed size by decreasing expression of AGAMOUS-LIKE (AGL) MADS-box transcription factor encoding genes in the endosperm [19]. In this study we tested the role of FIS-PRC2 as well as p4-siRNAs in mediating maternal excess interploidy effects. Surprisingly, our study revealed that neither changed levels of FIS-PRC2 nor p4-siRNAs are likely to be involved in mediating effects caused by maternal excess interploidy hybridizations. Instead, our results strongly suggest that reduced AGL gene expression as a consequence of reduced paternal genome dosage causes decreased seed size and we reveal that AGL62 acts as a dosage sensitive seed size regulator. We furthermore show that FIS-PRC2 function can be bypassed in maternal excess triploid seeds. Loss of FIS-PRC2 causes the formation of enlarged viable triploid seeds containing a cellularized endosperm and a developed embryo, contrasting the strict requirement of FIS-PRC2 function in diploid seeds. Development of viable fis triploid seeds is connected with normalized expression of AGL genes, suggesting that reduced AGL gene expression as a consequence of reduced paternal genome dosage allows bypassing the need of the FIS-PRC2 complex.
Results
Increased Maternal Genome Dosage Caused by the osd1 Mutation Phenocopies Maternal Excess Interploidy Hybridizations
To investigate the effect of increased maternal genome dosage on embryo and endosperm development, we made use of the meiotic omission of second division 1 (osd1) mutant that forms unreduced diploid male and female gametes at high frequency, whereas the ploidy of the parental plant remains unchanged [20]. Pollinating the osd1-1 mutant (introgressed into the Col accession) with wild-type pollen allowed us to mimic maternal excess interploidy crosses (4n×2n) without changing the ploidy of the maternal plant. Pollination of an osd1 plant with wild-type pollen resulted in 91.5% triploid seeds and 8.5% diploid seeds (n = 1921; Table S1), in close agreement with previously published results [20]. Triploid seeds derived from an osd1×2n cross were significantly smaller and lighter than diploid wild-type seeds (Figure 1A, 1B, p<0.001). Segregating diploid seeds from the osd1×2n cross were significantly bigger than wild-type seeds (p<0.001) (Figure 1A, 1B), maybe because the reduced seed size of the triploid seeds allows diploid sibling seeds to acquire more resources. Alternatively it is possible that loss of OSD1 affects seed size not only by altering ploidy of the endosperm but also by an unknown mechanism in the diploid maternal tissue. Tetraploid seeds derived from self-fertilized osd1 mutants were only slightly larger than wild-type seeds, contrasting the formation of considerably enlarged seeds by self-fertilized tetraploid plants (Col accession, Figure 1B; p<0.001). This finding suggests that increased size of seeds derived from tetraploid plants is largely caused by maternal sporophytic effects. Triploid seeds derived from 4n×2n interploidy crosses were smaller and lighter compared to tetraploid seeds (p<0.001), but of similar size and weight as wild-type diploid seeds (Figure 1B), contrasting previous data revealing decreased size of maternal excess triploid seeds in Landsberg erecta and C24 accession backgrounds [6]. Similar to triploid seeds derived from 4n×2n interploidy crosses [6], triploid seeds derived from osd1×2n crosses had a reduced endosperm proliferation rate (Figure 1C) and an early onset of endosperm cellularization (Figure 1D), correlating with decreased seed size compared to the self-fertilized maternal parents. Whereas endosperm cellularization in wild-type diploid and tetraploid seeds started at 6 DAP and was completed at 7–8 DAP, endosperm cellularization in triploid seeds derived from osd1×2n as well as 4n×2n crosses started two days earlier at 4 DAP and was completed at 5 DAP (Figure 1D, Figure S1). Precocious endosperm cellularization has previously been connected with small seed size [6], [7], [21], [22], implicating the early onset of endosperm cellularization in triploid seeds as the main cause for reduced seed size. We did not detect reproducible developmental differences of triploid embryos compared to diploid embryos, making it rather unlikely that alteration in triploid embryo development were causally responsible for decreased seed size.
Fig. 1. Seeds Derived from Crosses osd1×2n Mimic the Effect of Maternal Excess Interploidy Hybridizations. 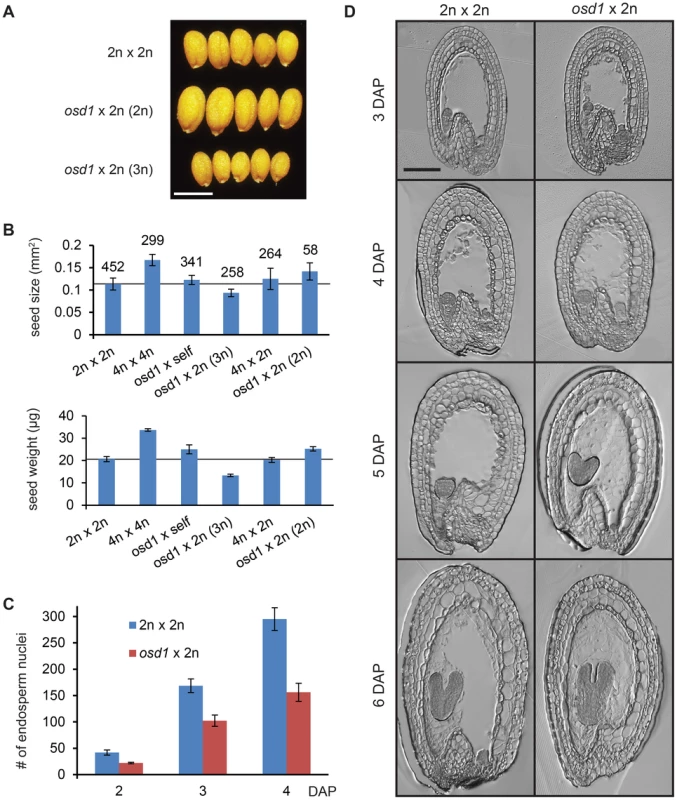
(A) Seeds from crosses of Col 2n×2n (upper panel) and osd1×2n (diploid and triploid seeds, middle and lower panel, respectively. Ploidy refers to ploidy of the embryo). Bar = 0.5 mm. (B) Average seed size (upper panel) and seed weight (lower panel) of different crosses. Numbers on top of bars correspond to number of analyzed seeds. For seed weight the average of 100 seeds was calculated in triplicates. Error bars indicate SD. The reference line corresponds to wild-type seed size and weight. (C) Number of endosperm nuclei in Col 2n×2n and osd1×2n crosses. n = 5. Error bars indicate SD. (D) Sections of seeds from Col 2n×2n and osd1×2n crosses. Bar = 100 µm. Together, we conclude that increased maternal genome contribution inherited through female gametes is sufficient to cause reduced seed size, establishing the osd1 mutant as a suitable tool to investigate the effect of maternal excess interploidy hybridizations. While also the dyad mutant forms unreduced female gametes and small-sized triploid seeds [23], the very low frequency of viable seed formation (ranging from one to ten viable seeds per dyad plant) does not allow detailed investigations of the effect of increased maternal ploidy on seed development using this mutant.
FIS-PRC2 Target Genes Are Deregulated in Response to Maternal Excess Interploidy Hybridizations
Paternal excess interploidy hybridizations cause similar seed developmental defects as mutants lacking FIS-PRC2 function. This is reflected by strikingly similar sets of deregulated genes [9], suggesting failure of FIS-PRC2 function in response to increased paternal genome dosage. As maternal and paternal genome excess cause reciprocal phenotypes [6], we addressed the question whether also maternal excess hybridizations cause global deregulation of FIS-PRC2 target genes. Transcriptome profiling of seeds derived from osd1×2n crosses at 6 DAP identified 342 and 510 genes as significantly up - and down-regulated, respectively (Signal Log Ratio (SLR)>1, or SLR<−1, p<0.05; Figure S2, Tables S2 and S3) that significantly overlapped with previously identified genes deregulated in 4n×2n interploidy hybridizations [24] (Table S4). While the overlap of deregulated genes was significant, there was also a high number of non-overlapping genes that are likely a consequence of different tissue types and accession backgrounds used to generate both datasets. Whereas transcriptome data of 4n×2n hybridizations were generated from entire siliques of C24 plants, osd1×2n transcriptional profiles were specifically generated from seeds of Columbia plants. Both, up - and down-regulated genes are significantly enriched for the PRC2 hallmark H3K27me3 (Figure 2, Table S2), suggesting a role of FIS-PRC2 in response to maternal excess interploidy hybridizations. We tested whether the genes that are deregulated in interploidy crosses are deregulated also in seeds lacking FIS2 function. Down-regulated genes were significantly enriched for genes affected by loss of FIS2 function, whereas no enrichment was detected for up-regulated genes (Figure 2). Together, the enrichment for H3K27me3 and the overlap with FIS2-responsive genes suggests that the FIS-PRC2 might be involved in mediating the seed phenotype in response to maternal excess hybridizations.
Fig. 2. FIS-PRC2 Target Genes Are Deregulated in Response to Maternal Excess Interploidy Hybridizations. 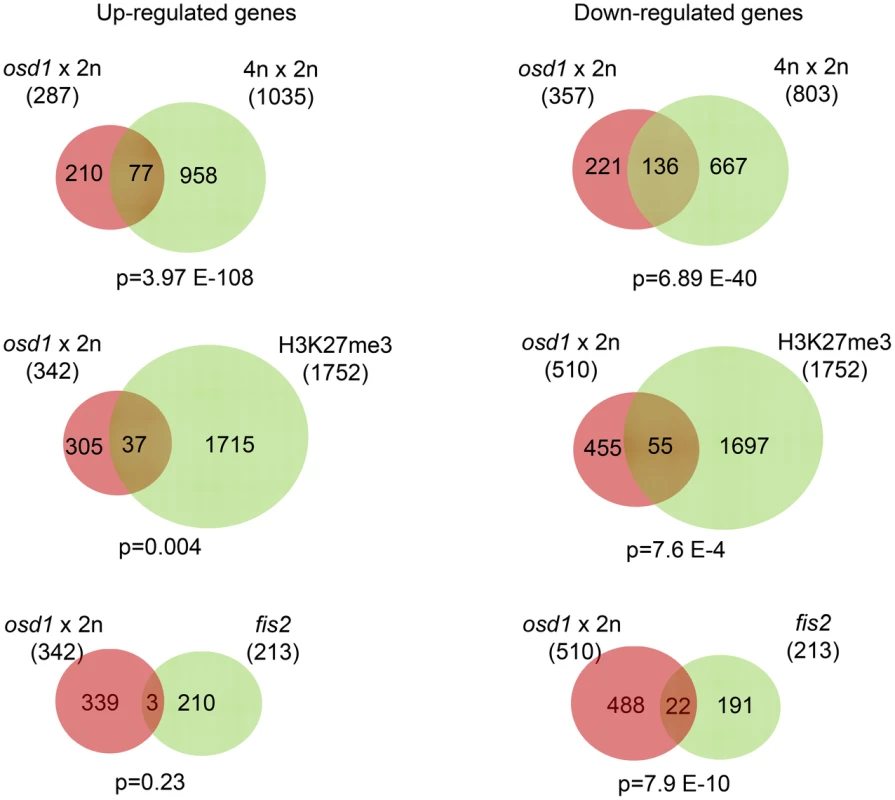
Venn diagrams showing overlap of genes being deregulated in seeds derived from osd1×2n crosses with genes deregulated in seeds of 4n×2n crosses [24] (upper panel); overlap of deregulated genes in seeds derived from osd1×2n crosses with H3K27me3 target genes in the endosperm [49] (middle panel), and genes that are upregulated in fis2 mutant seeds [9] (lower panel). In the upper panel only deregulated genes that are present on the ATH1 array have been used to calculate the overlap. Decreased Size of Triploid Seeds Persists when FIS-PRC2 Dosage Is Reduced
We wished to further test the idea whether decreased expression of FIS2-responsive genes in triploid maternal excess seeds was mediated by increased FIS activity. FIS-PRC2 components FIS2 and MEA are regulated by genomic imprinting and exclusively expressed from the maternally inherited alleles [25]–[27]. Therefore, it was possible that increased maternal genome dosage could cause increased expression of FIS2 and MEA that might in turn be responsible for increased FIS activity. We tested this hypothesis by measuring mRNA levels of FIS2 and MEA in triploid osd1 seeds and triploid seeds derived from 4n×2n crosses. Levels of MEA mRNA were only increased in 4n×2n derived seeds compared to wild-type seeds at 3 DAP, whereas no increase but rather a decrease was detected in triploid osd1 seeds (Figure 3 and Figure S3). In contrast, FIS2 mRNA levels were strongly increased in triploid osd1 seeds and slightly increased in triploid seeds derived from 4n×2n crosses at 2 and 3 DAP (Figure 3 and Figure S3) in agreement with previously published data [18].
Fig. 3. Expression Level of FIS2 but Not of MEA Is Increased in osd1×2n Crosses. 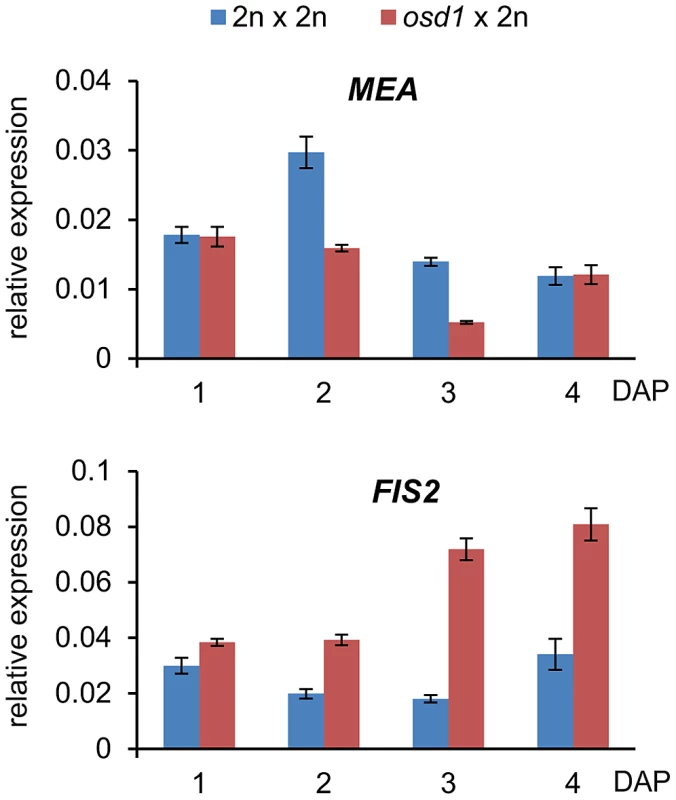
Quantitative RT-PCR analysis of MEA and FIS2 expression in seeds derived from osd1×2n crosses at 1–4 days after pollination (DAP). Error bars indicate s.e.m. Increased FIS2 mRNA levels might cause increased FIS-PRC2 activity that in turn could be causally responsible for phenotypic abnormalities of triploid seeds. To test this hypothesis, we generated mea/MEA; osd1/osd1 and fis2/FIS2 osd1/osd1 double mutants (Figure S4). MEA and FIS2 are unlinked to the centromere, therefore, mutant and wild-type alleles of both genes will frequently recombine. Consequently, most central cells formed in theses double mutants will be duplex for the mea or fis2 mutation (mea/mea/MEA/MEA or fis2/fis2/FIS2/FIS2), with a small fraction of central cells being tetraplex for mea or fis2 (mea/mea/mea/mea or fis2/fis2/fis2/fis2). We pollinated these double mutants with wild-type pollen and analyzed the ploidy and genotype of the resulting progeny. Based on this analysis we infer that about 8% of central cells are nulliplex for MEA (tetraplex for mea), whereas 10% are nulliplex for FIS2 (tetraplex for fis2) (Table S5). If increased FIS activity was causally responsible for reduced triploid seed size, we expected that reducing the copy number of active MEA or FIS2 alleles should result in the formation of enlarged triploid seeds. More than 70% of seeds derived from pollination of mea/MEA; osd1/osd1 and fis2/FIS2; osd1/osd1 with wild-type pollen had only two active maternally inherited wild-type MEA or FIS2 alleles in the pentaploid endosperm, respectively (meaM/meaM/MEAM/MEAM/MEAP; fis2M/fis2M/FIS2M/FIS2M/FIS2P superscribed M and P correspond to maternal and paternal alleles, Table S5). These seeds were viable and the seed size distribution of the majority of seeds was almost identical to triploid osd1 seeds (Figure 4A and Figure S5). Based on these results we conclude that increased expression of FIS2 is unlikely to cause decreased size of triploid seeds.
Fig. 4. Maternal Excess Triploid mea and fis2 Seeds Are Strongly Enlarged. 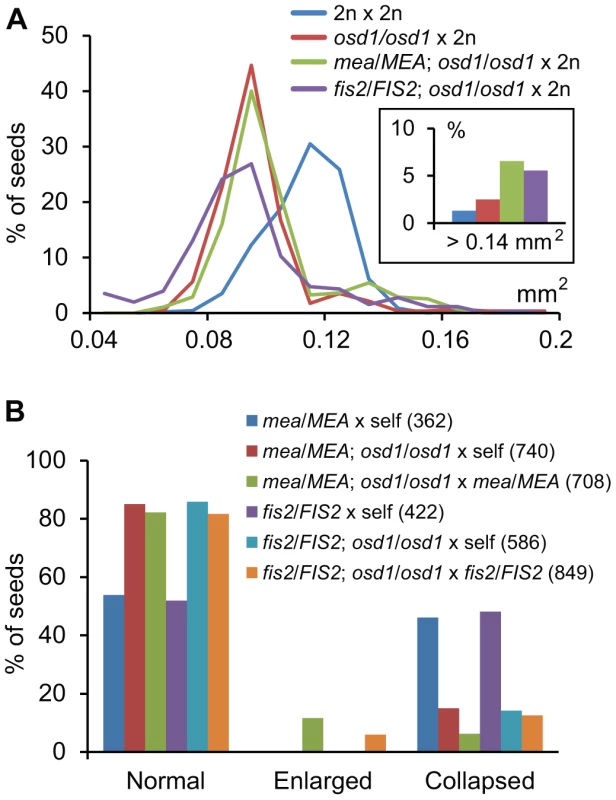
(A) Distribution of seed sizes from different crosses. A minimum of 250 seeds was analyzed for each cross. Inset shows percentage of seeds larger than 0.14 mm2. (B) Percentages of normal, enlarged and collapsed seeds from different crosses. Numbers in parenthesis correspond to numbers of analyzed seeds. Maternal Genome Excess Rescues Lethality of fis Mutant Seeds
We noted that a small fraction of seeds derived from pollination of mea/MEA; osd1/osd1 and fis2/FIS2; osd1/osd1 with wild-type pollen were strongly enlarged compared to triploid osd1 seeds (Figure 4A, Figure 5, Figure S6). Similarly, we detected about 10% and 5% of enlarged seeds in the progeny of the cross mea/MEA; osd1/osd1×mea/MEA and fis2/FIS2; osd1/osd1×fis2/FIS2, respectively (Figure 4B) that were not detected in the progeny of self-fertilized mea/MEA and fis2/FIS2 mutants (Figure 4B). About 4.3% of seeds derived from crosses mea/MEA; osd1/osd1×mea/MEA are expected to be diploid mea seeds (Tables S1 and S5), corresponding closely to the observed 5% of collapsed seeds that largely failed to germinate (only 1 out of 35 tested collapsed seeds germinated). We analyzed ploidy and genotype of enlarged seeds derived from crosses mea/MEA; osd1/osd1×mea/MEA. Genotyping revealed them being either duplex or triplex for the mea mutation (meaM/meaM/MEAP; meaM/meaM/meaP; Figure S7), revealing that triploid seeds can bypass the requirement of MEA function. Among collapsed fis2 seeds we found that 25% (n = 76) of those seeds were able to germinate. Similar to enlarged mea seeds, ploidy analysis and genotyping revealed that these seeds were triploid seeds duplex or triplex for the fis2 mutation (fis2M/fis2M/FIS2P; fis2M/fis2M/fis2P; Figure 4B, Figure S7), adding strong support to the view that triploid seeds can bypass the requirement of FIS-PRC2 function. In contrast, self-fertilized mea/MEA; osd1/osd1 and fis2/FIS2; osd1/osd1 mutants did not form enlarged seeds and none of the collapsed seeds was able to germinate, indicating that bypass of FIS function depends on an increased ratio of maternal to paternal genomes, rather than an absolute increase of maternal genome dosage.
Fig. 5. Endosperm Cellularization Is Restored in Triploid mea Seeds. 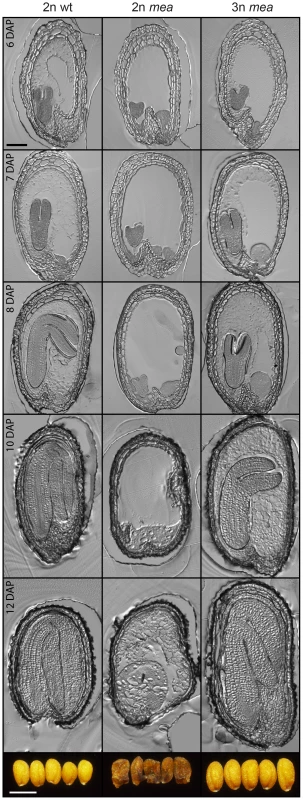
Sections of seeds from 2n×2n (left panels), mea/MEA×2n (middle panels) and mea/MEA; osd1/osd1×2n (right panels) crosses at 6–12 days after pollination (DAP). Bar = 100 µm. Images of mature seeds are shown in bottom panels. Bar = 0.5 mm. Lethality of fis mutant seeds is associated with a failure of endosperm cellularization [28], [29]. We asked the question whether rescue of mea and fis2 mutant triploid seeds would be associated with a restoration of endosperm cellularization. Indeed, we found that in contrast to diploid mea and fis2 seeds, endosperm cellularization of triploid duplex or triplex mea and fis2 seeds was initiated and completed, albeit it occurred delayed compared to wild-type seeds. Whereas cellularization of wild-type seeds was largely progressed at 6 DAP, it was only initiated in triploid mea and fis2 seeds at this time and was completed only at 10 DAP (Figure 5 and Figure S6).
Together, we conclude that relative increase of maternal to paternal genome dosage allows bypass of FIS function and restores endosperm cellularization in fis mutant seeds.
Expression of AGL MADS Box Genes Is Normalized in Triploid mea and fis2 Seeds
Endosperm cellularization is negatively regulated by the MADS-box transcription factor AGL62, and complete loss of AGL62 causes precocious endosperm cellularization after few mitotic divisions [30]. We addressed the question whether precocious endosperm cellularization in triploid seeds correlates with decreased expression levels of AGL62. In agreement with this notion, expression of AGL62 was reduced in triploid osd1 seeds at timepoints before cellularization at 5 DAP (Figure 6), similar to decreased expression of AGL62 in 4n×2n interploidy hybridizations [19] (Figure S8). Yeast two-hybrid interaction studies revealed that AGL62 interacts directly with AGL transcription factors such as the PEG PHERES1 (PHE1), the MEG AGL36, and AGL90. AGL62 also interacts indirectly with the MEG AGL28 and AGL40 that both directly interact with PHE1 [31], [32]. We tested whether expression of the genes encoding direct and indirect interaction partners of AGL62 was altered in triploid seeds. Similar to the reduced expression of AGL62, expression of all tested AGL genes was strongly reduced in triploid osd1 seeds as well as triploid seeds derived from 4n×2n crosses (Figure 6 and Figure S8).
Fig. 6. Expression of AGL MADS Box Genes Is Decreased in osd1×2n Crosses. 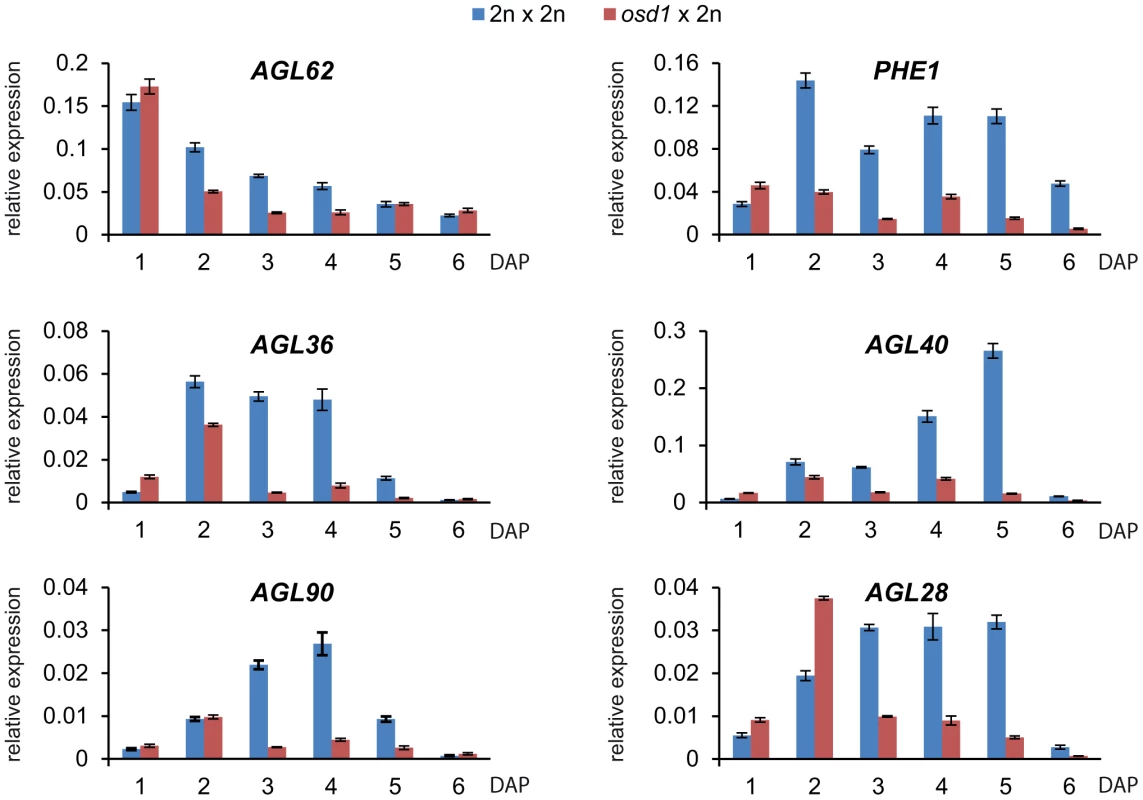
Quantitative RT-PCR analysis of AGL62, PHE1, AGL90, AGL36, AGL40 and AGL28. Error bars indicate s.e.m. Reduced maternal genome dosage of AGL62 can suppress fis2 seed abortion [29] likely by initiating endosperm cellularization. We therefore addressed the question whether restoration of endosperm cellularization in triploid mea and fis2 seeds is accompanied by normalized expression of AGL genes. Because of the extreme size difference, triploid and diploid mea and fis2 seeds can easily be distinguished from other seeds at 8 DAP and manually isolated. Expression of six AGL genes implicated in endosperm cellularization was measured, and all tested genes had reduced transcript levels in triploid mea and fis2 seeds (Figure 7), correlating with the initiation of endosperm cellularization (Figure 5). Together, we conclude that decreased AGL gene expression in triploid maternal excess seeds alleviates the need for FIS-PRC2 function.
Fig. 7. Expression of AGL MADS Box Genes Is Normalized in Triploid mea and fis2 Seeds. 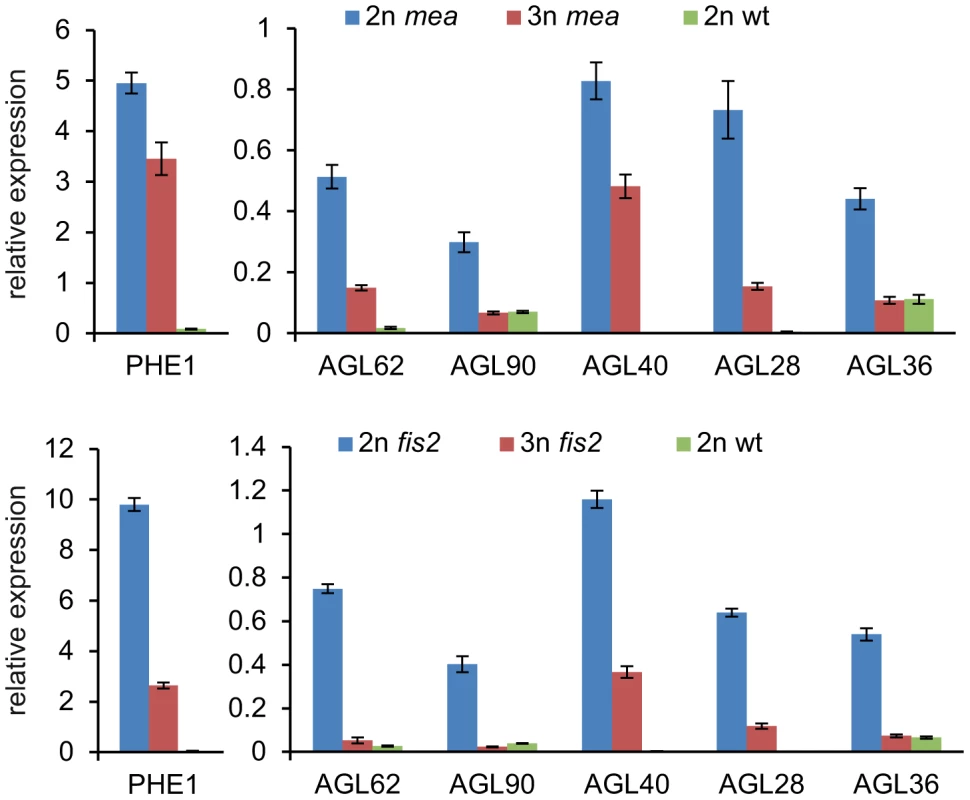
Quantitative RT-PCR analysis of AGL62, PHE1, AGL90, AGL40, AGL28 and AGL36 at 8 DAP in mea and fis2 seeds (upper and lower panel, respectively). Enlarged seeds with a fis phenotype were selected as 3n mea and fis2 seeds from mea/MEA; osd1/osd1×2n and fis2/FIS2; osd1/osd1×2n crosses, respectively. Diploid mea seeds are a mixture of wild-type and mea seeds. Error bars indicate s.e.m. Reduced Size of Triploid Seeds Is Not Dependent on the p4-siRNA Pathway
Previous work proposed a possible connection of increased levels of p4-siRNAs small interfering RNAs (siRNAs) in seeds derived from 4n×2n hybridizations and decreased expression levels of AGL genes [19]. Biosynthesis of p4-siRNAs is dependent on RNA polymerase IV (PolIV) encoded by NRPD1a [33], [34]. To test the requirement of p4 - siRNAs for dampening expression of AGL genes, we generated nrpd1a/nrpd1a; osd1/osd1 double mutants and pollinated double homozygous mutants with wild-type pollen. In seeds resulting from this cross at 3 DAP expression of AGL62, PHE1, AGL28 and AGL40 was increased compared to wild-type triploid seeds, but remained significantly below wild-type expression levels (p<0.005; Figure 8A). In contrast, expression levels of AGL36 and AGL90 remained unchanged in triploid seeds lacking maternal NRPD1a function (Figure 8A). Maternal loss of NRPD1a in diploid seeds caused decreased expression levels of all tested AGL genes except for AGL40, which remained expressed at wild-type levels (Figure 8A). We also analyzed the size of seeds derived from osd1 nrpd1a×wild-type hybridizations and found that loss of NRPD1 was not connected with increased size of triploid maternal excess seeds (Figure 8B). Together, our results reveal that maternal loss of NRPD1a affects expression of only a subset of AGL genes and has no impact on seed size, strongly arguing against a causal role of p4-siRNAs in regulating triploid seed size.
Fig. 8. Maternal Loss of NRPD1a Does Not Restore Wild-Type Levels of AGL Expression and Wild-Type Seed Size. 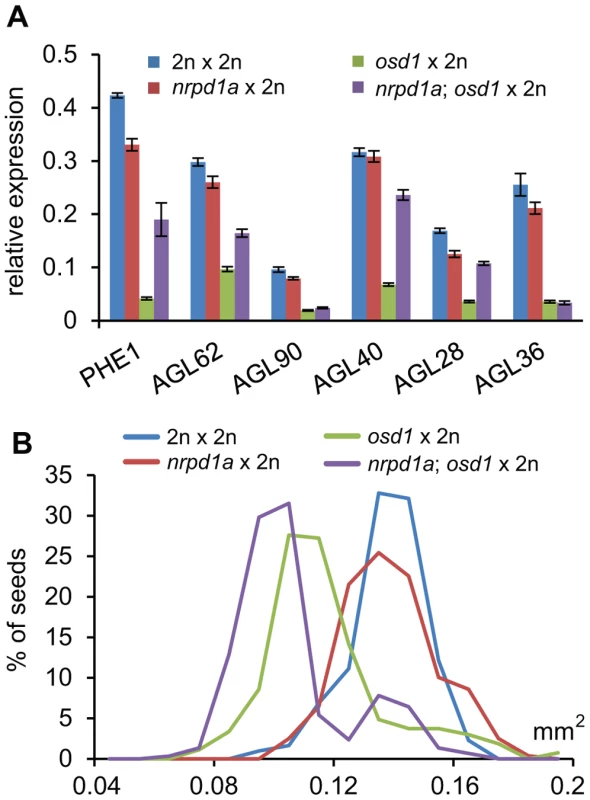
(A) Quantitative RT-PCR analysis of PHE1, AGL62, AGL90, AGL40, AGL28 and AGL36 in seeds derived from crosses 2n×2n, nrpd1a/nrpd1a×2n, osd1/osd1×2n, and nrpd1a/nrpd1a; osd1/osd1×2n at 3 DAP. Error bars indicate s.e.m. (B) Distribution of seed sizes from different crosses. A minimum of 250 seeds was analyzed for each cross. Reduced Dosage of AGL62 Causes Reduced Seed Size
Finally, we addressed the question whether AGL62 acts as a dosage dependent seed size regulator. Therefore, we generated agl62/AGL62; osd1/osd1 double mutants and fertilized them with wild-type pollen. The majority of triploid seeds was found to be simplex for the agl62 mutation and about 17% to be duplex, corresponding to allele frequencies in the pentaploid endosperm of agl62M/agl62M/AGL62M/AGL62M/AGL62P and agl62M/asgl62M/agl62M/agl62M/AGL62P respectively, Table S6). We analyzed the size of triploid seeds being simplex or duplex for the agl62 mutation and observed an AGL62 dosage-dependent decrease in size, with seeds having five functional AGL62 alleles in the endosperm being larger than seeds with only three or one functional AGL62 allele (Figure 9A, 9B). In contrast, heterozygous diploid agl62/AGL62 seeds were similar in size to wild-type seeds (Figure 9A), revealing a differential response of diploid and triploid seeds to reduced AGL62 dosage. If reduced dosage of AGL62 is responsible for decreased size of maternal excess triploid seeds, we expected to find an enrichment of MADS-box binding motifs in those genes that are down-regulated in triploid maternal excess seeds. We tested for the presence of CArG-box motifs of the SRE type (CC(A/T)6GG) as well as of the MEF2-type (CTA(A/T)4TAG), as both have been reported to be bound by plant MADS-box proteins [35], [36]. Among down-regulated genes both motifs were significantly enriched, with 54 (10.6%; Table S2) genes containing a MEF-type motif within 1000 bp upstream of the transcriptional start site, which was significantly higher compared to the genome-wide presence of this motif (6.9%, p<0.001). A similar number of genes contain a SRF-type motif in their promoter region (55 genes, 10.7%, Table S2), which is slightly, but significantly higher compared to the genome-wide frequency of 8.1% (p<0.01). Most strikingly, genes containing a MEF2-type motif in their promoter region were predominantly expressed in the chalazal region of the endosperm (17%; Figure S9), consistent with a preferential expression of AGL62 interacting AGLs in this region of the endosperm [37]. Together, we conclude that AGL62 is a dosage sensitive regulator of triploid seed size, strongly suggesting that reduced expression of AGL62 causes decreased size of triploid maternal excess seeds.
Fig. 9. Size of Triploid Seeds Is Dependent on the Dosage of AGL62. 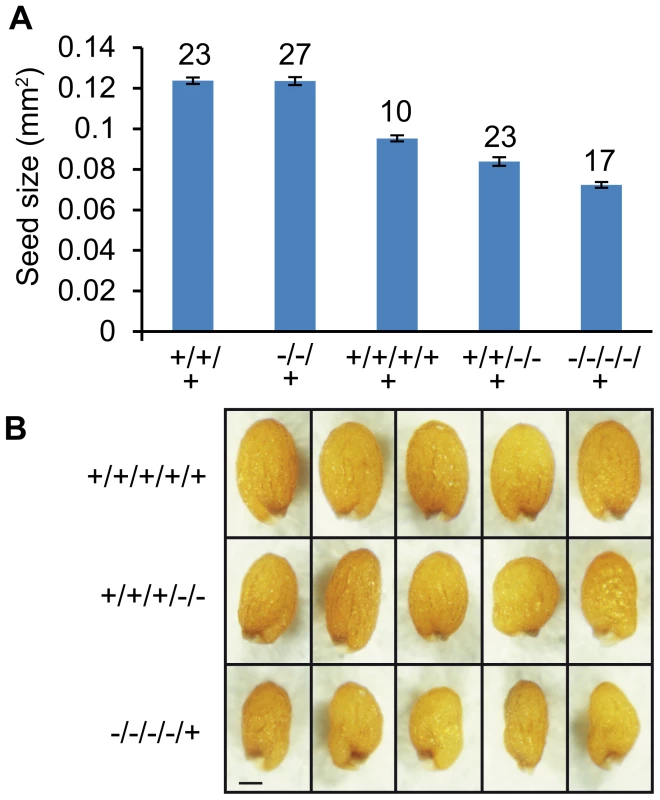
(A) Size of seeds derived from crosses agl62/AGL62; osd1/osd1×wild type and diploid seeds from crosses agl62/AGL62×wild type. Average seed size of different genotypes is shown. Wild-type (+) and mutant (−) allele frequencies of AGL62 in the endosperm are shown below the x axis. Numbers on top of bars refer to numbers of analyzed seeds. Error bars indicate s.e.m. (B) Images of triploid seeds from the three categories shown in (A). Bar = 0.1 mm. Discussion
In this study we report the following new discoveries: (1) The functional requirement of the FIS-PRC2 can be bypassed by increasing the ratio of maternal to paternal genomes. (2) Bypass of FIS-PRC2 function is connected with decreased expression of AGL62 and interacting AGLs. (3) Decreased seed size of maternal excess triploid seeds is neither mediated by increased activity of FIS-PRC2 and nor by increased levels of p4-siRNAs. (4) Decreased size of maternal excess triploid seeds is likely a consequence of decreased expression of paternally expressed genes. (5) AGL62 is a dosage-sensitive seed size regulator.
The FIS-PRC2 complex is essential for viable seed development; however, the underlying forces that lead to the evolution of the FIS-PRC2 remained obscure. Our study demonstrates that maternal excess triploid seeds can bypass FIS-PRC2 function, suggesting that FIS-PRC2 is mainly required to balance expression of maternally and paternally expressed dosage sensitive genes in the endosperm. Consistent with previous work revealing that fis mutant seed abortion can be partially suppressed by maternal loss of AGL62 [29], we show that normalized fis triploid seed development is connected with normalized AGL62 gene expression. Based on yeast-two-hybrid data AGL62 interacts with several AGLs [31], with at least two of them, PHE1 and AGL36 are regulated by genomic imprinting [38], [39]. Therefore, altering the parental genome dosage is expected to impair the balance of AGL gene expression. This view is supported by the fact that increased paternal genome dosage causes strongly increased expression of PHE1, AGL62, and AGL36 [9], [24], [32], whereas increased maternal genome dosage exerts the converse effect [24] and data shown in this study). Type II MADS-box proteins are well known to form multimeric complexes [40] and changing the expression level of individual members of these complexes strongly impacts on plant development [41]–[43]. It can thus be assumed that unbalanced expression changes of AGL genes in the endosperm will similarly impair functional AGL complex formation and strongly affect endosperm development.
Our study furthermore revealed that decreased seed size in maternal excess interploidy seeds is neither connected with increased FIS-PRC2 function, nor with increased p4-siRNA levels. Instead, we propose that reduced expression of paternally expressed genes causes decreased seed size in maternal excess crosses and conversely, that increased expression of paternally expressed genes causes increased seed size in fis mutants. This model predicts that decreasing paternal genome dosage should reduce seed size in fis mutants and potentially even partially suppress seed abortion. This prediction is confirmed by the data presented in this study. FIS-PRC2 function can also be bypassed in fis seeds that form a sexual embryo and an asexual endosperm [44]. These surviving fis2 seeds with diploid endosperm remain smaller than wild-type seeds. Our results show that another class of surviving fis2 seeds, triploid maternal excess fis mutant seeds with a pentaploid endosperm, is larger than wild-type seeds. These two classes of surviving fis2 seeds have two major differences that could explain the very different seed size: First, fis2 seeds with pentaploid endosperm have four, whereas fis2 seeds with diploid endosperm have only two maternal genomes. Second, fis2 seeds with pentaploid endosperm but not those with diploid endosperm contain a paternal genome. Because increased maternal genome dosage usually causes decreased rather than increased seed size, the different maternal genome dosage in the two classes of surviving fis2 seeds is unlikely causing the observed differences in seed size. Instead, we conclude that the presence of a paternal genome is the main determinant for seed size in fis2 mutant seeds. Consequently, the FIS-PRC2 regulates paternally contributed seed size regulators that cause endosperm abnormalities and seed abortion upon overexpression. Our work revealed that AGL62 is a dosage-dependent seed size regulator, suggesting that either activation or function of AGL62 depends on a paternally contributed factor. AGL62 physically interacts with the paternally expressed PHE1 [31], raising the possibility that decreased expression of PHE1 and possibly other proteins reduces the number of functional AGL62 complexes.
Recent data implicate a link between increased levels of PolIV-dependent maternal p4-siRNAs and decreased size of maternal excess seeds [19]. It has been suggested that maternal p4-siRNAs target AGL genes and that increased siRNA levels in triploid maternal excess seeds causes decreased AGL transcript levels [19]. The results shown in this study reveal that maternal loss of NRPD1a in triploid seeds only affects expression of a subset of AGL genes with expression of none of the tested AGL genes being restored to wild-type levels. Maternal loss of NRPD1a in triploid seeds did neither affect size of triploid seeds. Therefore, the connection between maternal p4-siRNAs and regulation of AGL genes in response to interploidy hybridizations requires further investigations.
Many theoretical considerations argue that endosperms with higher levels of maternal ploidy and reduced levels of interparental genomic conflict have adaptive benefits and should be evolutionary favored [45]–[47]. In agreement with that view, transitions from triploid to higher endosperm ploidy occurred frequently by changing the mode of female gametophyte formation [47]. Based on the data presented in this study we propose that the FIS-PRC2 is needed to counteract excessive parental conflict. Therefore, reduced genetic conflict as a consequence of higher levels of maternal ploidy in the endosperm can bypass the need of the FIS-PRC2. We speculate that the FIS-PRC2 will be of less importance in species forming endosperms with higher maternal ploidy.
Materials and Methods
Plant Material and Growth Conditions
Plants were grown in a growth chamber at 60% humidity and daily cycles of 16 h light at 21°C and 8 h darkness at 18°C. Arabidopsis thaliana mutants mea-8 [48], fis2-5 [49], agl62-2 [30], and nrpd1a-3 [34] are in the Columbia accession. The osd1-1 mutant [20] was kindly provided by Raphael Mercier. The mutant was originally identified in the Nossen background and subsequently introgressed into Columbia by repeated backcrossing over five generations. Tetraploid Columbia plants were kindly provided by Ortrun Mittelsten Scheid. For crosses, designated female partners were emasculated, and the pistils hand-pollinated two day after emasculation.
RNA Extraction and qPCR Analysis
For analysis of crosses in Figure 7, siliques were opened and a minimum of 50 seeds were harvested into RNA later (Sigma, Buchs, Switzerland). For analysis in all other Figures, three siliques were harvested for each timepoint and frozen in liquid nitrogen.
Glass beads (1.25–1.55 mm) were added, and the samples were ground in a Silamat S5 (IvoclarVivadent, Ellwangen, Germany). RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Residual DNA was removed using the Qiagen RNase-free DNase Set and cDNA was synthesized using the Fermentas First strand cDNA synthesis kit (Fermentas, Burlington, Canada) according to the manufacturer's instruction.
Quantitative RT-PCR was performed using an iQ5 Real-Time PCR Detection System (BioRad, Hercules, USA) and Maxima SYBR green qPCR master mix (Fermentas, Burlington, Canada) according to the manufacturer's instruction. Quantitative RT-PCR was performed with three replicates using primers as indicated in Table S7, and results were analyzed as described [50]. Expression of PP2A and ACTIN11 did not change in triploid versus diploid seeds (data not shown), and both genes were used as reference genes with similar results (expression normalized to PP2A is shown).
For quantification of agl62 alleles in triploid seeds, DNA was isolated from seedlings and the number of mutant alleles was determined by quantitative PCR using three different primer pairs (Table S7) for each mutant (one pair that specifically amplifies the T-DNA allele, one pair that amplifies only the wild-type allele and one pair that amplifies both the T-DNA and wild-type allele equally). To distinguish between simplex and duplex mutants, the ratio of T-DNA to wild-type alleles was calculated (Figure S10).
Microscopy
Tissue sections and clearing analysis were performed as previously described [51]. Pictures were taken using a Leica DMI 4000B microscope and Leica DFC360 FX camera and processed using Adobe Photoshop CS5.
Seed Size Analysis
Seeds were arranged on glass slides and pictures were taken using a Leica Z16apoA microscope. Images were converted to black and white using the “threshold” function in Adobe Photoshop CS5. Seed size was measured in ImageJ (http://rsbweb.nih.gov/ij/) using the “Analyze Particles” function. Seed size analysis shown in Figure 9A and Figure S5 was done from individual seeds that were later on germinated to determine genotype and ploidy.
Flow Cytometry
Ploidy levels were measured by flow cytometry with a CyFlow Ploidy Analyzer (Partec, Münster, Germany). For seed ploidy analysis, seeds were allowed to germinate and 10 day old seedlings were analyzed. Plant tissue was chopped with a razor blade in CyStain extraction buffer (Partec), filtered through a 30-µm CellTrics filter (Partec) into a sample tube, and stained with CyStain Staining buffer (Partec).
Microarray Analysis
Samples, array design, and hybridizations
The transcriptional profile of wild-type and fis2 seeds at 6 DAP has been previously published [9]. Reference transcript profiles during seed development were taken from [52]. To generate transcript profiles of osd1×2n crosses and 2n×2n crosses, seeds of at least 20 siliques per sample and three independent biological replicates were harvested into 40 µl RNAlater (Sigma, Buchs, Switzerland) at 6DAP. RNA extraction and labeling was performed as previously described [9]. AGRONOMICS1 microarrays (Affymetrix, Santa Clara, CA) were hybridized as previously described [53]. Analysis was based upon annotations compiled in TAIR9. Data were deposited into the ArrayExpress database (Accession number E-MTAB-1061).
Bioinformatic analysis
All data processing was essentially performed as previously described [9].
Probe sets were called significantly differentially expressed when q<0.05. To enrich for biologically relevant changes, only probe sets with FCreal>2 or FCreal<−2 were selected. Data for H3K27me3 target loci were from [49]. The significance of enrichment was estimated based on the hypergeometric test. Analysis of tissue-specificity of differentially expressed genes was performed using MeV4 (http://www.tm4.org/mev/).
Supporting Information
Zdroje
1. DrewsGN, YadegariR (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36 : 99–124.
2. IngramGC (2010) Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247 : 195–214.
3. CostaLM, Gutierrez-MarcosJF, DickinsonHG (2004) More than a yolk: the short life and complex times of the plant endosperm. Trends Plant Sci 9 : 507–514.
4. BrownRC, LemmonBE, NguyenH, OlsenO-A (1999) Development of the endosperm in Arabidopsis thaliana. Sex Plant Reprod 12 : 32–42.
5. Boisnard-LorigC, Colon-CarmonaA, BauchM, HodgeS, DoernerP, et al. (2001) YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 : 495–509.
6. ScottRJ, SpielmanM, BaileyJ, DickinsonHG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 : 3329–3341.
7. GarciaD, SaingeryV, ChambrierP, MayerU, JürgensG, et al. (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol 131 : 1661–1670.
8. DilkesBP, SpielmanM, WeizbauerR, WatsonB, Burkart-WacoD, et al. (2008) The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol 6: e308 doi:10.1371/journal.pbio.0060308.
9. ErilovaA, BrownfieldL, ExnerV, RosaM, TwellD, et al. (2009) Imprinting of the Polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet 5: e1000663 doi:10.1371/journal.pgen.1000663.
10. HennigL, DerkachevaM (2009) Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet 25 : 414–423.
11. BeiselC, ParoR (2011) Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet 12 : 123–135.
12. ChaudhuryAM, MingL, MillerC, CraigS, DennisES, et al. (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94 : 4223–4228.
13. OhadN, MargossianL, HsuY-C, WilliamsC, FischerR (1996) A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA 93 : 5319–5324.
14. KöhlerC, HennigL, BouveretR, GheyselinckJ, GrossniklausU, et al. (2003) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22 : 4804–4814.
15. GuittonAE, PageDR, ChambrierP, LionnetC, FaureJE, et al. (2004) Identification of new members of FERTILIZATION INDEPENDENT SEED Polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131 : 2971–2981.
16. KiyosueT, OhadN, YadegariR, HannonM, DinnenyJ, et al. (1999) Control of fertilization-independent endosperm development by the MEDEA Polycomb gene in Arabidopsis. Proc Natl Acad Sci USA 96 : 4186–4191.
17. SorensenMB, ChaudhuryAM, RobertH, BancharelE, BergerF (2001) Polycomb group genes control pattern formation in plant seed. Curr Biol 11 : 277–281.
18. JullienPE, BergerF (2010) Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet 6: e1000885 doi:10.1371/journal.pgen.1000885.
19. LuJ, ZhangC, BaulcombeDC, ChenZJ (2012) Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci USA 109 : 5529–5534.
20. d'ErfurthI, JolivetS, FrogerN, CatriceO, NovatchkovaM, et al. (2009) Turning meiosis into mitosis. PLoS Biol 7: e1000124 doi:10.1371/journal.pbio.1000124.
21. GarciaD, Fitz GeraldJN, BergerF (2005) Maternal Control of Integument Cell Elongation and Zygotic Control of Endosperm Growth Are Coordinated to Determine Seed Size in Arabidopsis. Plant Cell 17 : 52–60.
22. LuoM, DennisES, BergerF, PeacockWJ, ChaudhuryA (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci U S A 102 : 17531–17536.
23. RaviM, MarimuthuMP, SiddiqiI (2008) Gamete formation without meiosis in Arabidopsis. Nature 451 : 1121–1124.
24. TiwariS, SpielmanM, SchulzR, OakeyRJ, KelseyG, et al. (2010) Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol 10 : 72.
25. KinoshitaT, YadegariR, HaradaJJ, GoldbergRB, FischerRL (1999) Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11 : 1945–1952.
26. Vielle-CalzadaJP, ThomasJ, SpillaneC, ColuccioA, HoeppnerMA, et al. (1999) Maintenance of genomic imprinting at the Arabidopsis MEDEA locus requires zygotic DDM1 activity. Genes Dev 13 : 2971–2982.
27. LuoM, BilodeauP, DennisES, PeacockWJ, ChaudhuryA (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97 : 10637–10642.
28. van de RheeMD, LemmersR, BolJF (1993) Analysis of regulatory elements involved in stress-induced and organ-specific expression of tobacco acidic and basic b-1,3-glucanase genes. Plant Molecular Biology 21 : 451–461.
29. HehenbergerE, KradolferD, KöhlerC (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139 : 2031–2039.
30. KangIH, SteffenJG, PortereikoMF, LloydA, DrewsGN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20 : 635–647.
31. de FolterS, ImminkRG, KiefferM, ParenicovaL, HenzSR, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17 : 1424–1433.
32. WolffP, WeinhoferI, SeguinJ, RoszakP, BeiselC, et al. (2011) High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm. PLoS Genet 7: e1002126 doi:10.1371/journal.pgen.1002126.
33. OnoderaY, HaagJR, ReamT, NunesPC, PontesO, et al. (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 : 613–622.
34. HerrAJ, JensenMB, DalmayT, BaulcombeDC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308 : 118–120.
35. de FolterS, AngenentGC (2006) trans meets cis in MADS science. Trends Plant Sci 11 : 224–231.
36. VerelstW, SaedlerH, MunsterT (2007) MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol 143 : 447–460.
37. WaliaH, JosefssonC, DilkesB, KirkbrideR, HaradaJ, et al. (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19 : 1128–1132.
38. KöhlerC, PageDR, GagliardiniV, GrossniklausU (2005) The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37 : 28–30.
39. ShirzadiR, AndersenED, BjerkanKN, GloeckleBM, HeeseM, et al. (2011) Genome-Wide Transcript Profiling of Endosperm without Paternal Contribution Identifies Parent-of-Origin-Dependent Regulation of AGAMOUS-LIKE36. PLoS Genet 7: e1001303 doi:10.1371/journal.pgen.1001303.
40. ImminkRG, TonacoIA, de FolterS, ShchennikovaA, van DijkAD, et al. (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24.
41. FavaroR, PinyopichA, BattagliaR, KooikerM, BorghiL, et al. (2003) Mads-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15 : 2603–2611.
42. PelazS, DittaGS, BaumannE, WismanE, YanofskyMF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 : 200–203.
43. HonmaT, GotoK (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 : 525–529.
44. NowackMK, GriniPE, JakobyMJ, LafosM, KonczC, et al. (2006) A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat Genet 38 : 63–67.
45. HaigD (1987) Kin conflict in seed plants. Trends Ecol Evol 2 : 337–340.
46. HaigD, WestobyM (1989) Parent specific gene expression and the triploid endosperm. Am Nature 134 : 147–155.
47. FriedmanW, MadridE, WilliamsJ (2008) Origin of the fittest and survival of the fittest: relating female gametophyte development to endosperm genetics. Int J Plant Sci 169 : 79–92.
48. NgoQA, MooreJM, BaskarR, GrossniklausU, SundaresanV (2007) Arabidopsis GLAUCE promotes fertilization-independent endosperm development and expression of paternally inherited alleles. Development 134 : 4107–4117.
49. WeinhoferI, HehenbergerE, RoszakP, HennigL, KöhlerC (2010) H3K27me3 profiling of the endosperm implies exclusion of Polycomb group protein targeting by DNA methylation. PLoS Genet 6: e1001152 doi:10.1371/journal.pgen.1001152.
50. SimonP (2003) Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19 : 1439–1440.
51. RoszakP, KöhlerC (2011) Polycomb group proteins are required to couple seed coat initiation to fertilization. Proc Natl Acad Sci USA 20826–20831.
52. LeBH, ChengC, BuiAQ, WagmaisterJA, HenryKF, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107 : 8063–8070.
53. RehrauerH, AquinoC, GruissemW, HenzS, HilsonP, et al. (2010) AGRONOMICS1 - A new resource for Arabidopsis transcriptome profiling. Plant Physiol 152 : 487–499.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



