-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
Mini-chromosome maintenance (MCM) 2–9 proteins are related helicases. The first six, MCM2–7, are essential for DNA replication in all eukaryotes. In contrast, MCM8 is not always conserved in eukaryotes but is present in Arabidopsis thaliana. MCM8 is required for 95% of meiotic crossovers (COs) in Drosophila and is essential for meiosis completion in mouse, prompting us to study this gene in Arabidopsis meiosis. Three allelic Atmcm8 mutants showed a limited level of chromosome fragmentation at meiosis. This defect was dependent on programmed meiotic double-strand break (DSB) formation, revealing a role for AtMCM8 in meiotic DSB repair. In contrast, CO formation was not affected, as shown both genetically and cytologically. The Atmcm8 DSB repair defect was greatly amplified in the absence of the DMC1 recombinase or in mutants affected in DMC1 dynamics (sds, asy1). The Atmcm8 fragmentation defect was also amplified in plants heterozygous for a mutation in either recombinase, DMC1 or RAD51. Finally, in the context of absence of homologous chromosomes (i.e. haploid), mutation of AtMCM8 also provoked a low level of chromosome fragmentation. This fragmentation was amplified by the absence of DMC1 showing that both MCM8 and DMC1 can promote repair on the sister chromatid in Arabidopsis haploids. Altogether, this establishes a role for AtMCM8 in meiotic DSB repair, in parallel to DMC1. We propose that MCM8 is involved with RAD51 in a backup pathway that repairs meiotic DSB without giving CO when the major pathway, which relies on DMC1, fails.
Published in the journal: MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in. PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003165
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003165Summary
Mini-chromosome maintenance (MCM) 2–9 proteins are related helicases. The first six, MCM2–7, are essential for DNA replication in all eukaryotes. In contrast, MCM8 is not always conserved in eukaryotes but is present in Arabidopsis thaliana. MCM8 is required for 95% of meiotic crossovers (COs) in Drosophila and is essential for meiosis completion in mouse, prompting us to study this gene in Arabidopsis meiosis. Three allelic Atmcm8 mutants showed a limited level of chromosome fragmentation at meiosis. This defect was dependent on programmed meiotic double-strand break (DSB) formation, revealing a role for AtMCM8 in meiotic DSB repair. In contrast, CO formation was not affected, as shown both genetically and cytologically. The Atmcm8 DSB repair defect was greatly amplified in the absence of the DMC1 recombinase or in mutants affected in DMC1 dynamics (sds, asy1). The Atmcm8 fragmentation defect was also amplified in plants heterozygous for a mutation in either recombinase, DMC1 or RAD51. Finally, in the context of absence of homologous chromosomes (i.e. haploid), mutation of AtMCM8 also provoked a low level of chromosome fragmentation. This fragmentation was amplified by the absence of DMC1 showing that both MCM8 and DMC1 can promote repair on the sister chromatid in Arabidopsis haploids. Altogether, this establishes a role for AtMCM8 in meiotic DSB repair, in parallel to DMC1. We propose that MCM8 is involved with RAD51 in a backup pathway that repairs meiotic DSB without giving CO when the major pathway, which relies on DMC1, fails.
Introduction
Meiosis is a process that occurs in the germlines of sexually reproducing organisms. Two successive rounds of chromosome segregation (meiosis I and II) follow a single round of DNA replication (S phase). The resulting four cells each contain half the genetic content of the pre-meiotic mother cell. The genetic complement of these gametes is a mosaic of the paternal and maternal DNA due to meiotic recombination that occurs between S phase and the first meiotic division [1].
Meiotic recombination begins with programmed DSBs that are dependent on SPO11 and multiple cofactors, including PRD1 in plants [2], [3]. DSBs are subsequently resected to yield 3′ overhangs that invade the homologous chromosome. At this step, two recombinases co-operate to achieve efficient strand exchange with the homolog, RAD51 and DMC1 [4]. RAD51 is a recombinase involved both at mitosis and meiosis while DMC1 is specific to meiosis. Importantly, it has been recently shown in S. cerevisiae that only the strand exchange activity of DMC1, and not of RAD51, is required for meiotic crossover formation [5]. RAD51 appears thus to be an accessory factor of DMC1 for meiotic homologous crossover formation, but may also serve as a backup to repair breaks when DMC1 fails [5]. In Arabidopsis thaliana, RAD51 is indispensable for repair of meiotic DSBs as shown by the extensive meiotic chromosome fragmentation which occurs at meiosis in Atrad51 mutants [6], [7]. AtDMC1 is required for CO formation but not meiotic DSB repair. Indeed, in Atdmc1 mutant, meiotic DSBs are repaired in a AtRAD51-dependent manner which does not promote chromosome pairing and does not yield COs between homologs, likely using the sister chromatid as a template [7], [8]. In addition, consistent with a role of RAD51 in helping DMC1 in wild type, the number of DMC1 foci is severely decreased in a Atrad51 mutant [7], [9], while RAD51 foci are unaffected in Atdmc1 [9]. Thus two meiotic functions of RAD51 emerge, helping DMC1 to promote COs and promoting DSB repair on the sister without DMC1.
Two other Arabidopsis mutants, sds and asy1, have phenotypes reminiscent of Atdmc1, repairing breaks using AtRAD51 but exhibiting major homologous chromosome pairing defects and making no or few COs [10]–[12]. Both sds and asy1 show localization defects of AtDMC1 but not of AtRAD51, suggesting that they work with DMC1 to promote interhomolog recombination [12], [13]. Based on its amino acid sequence, SDS is a cyclin-like protein and ASY1 is a HORMA domain protein making it the likely functional homologue of S. cerevisiae Hop1.
DSB repair events form intermediates that are resolved as either crossovers (COs) or non-crossovers (NCOs) (gene conversion). COs are required for accurate segregation of chromosomes during meiosis I and can arise from at least two independent pathways known as class I and class II COs. These two pathways coexist in budding yeast, mammals and Arabidopsis [1], [14]–[17]. Class I COs are subject to a phenomenon known as interference, whereby the occurrence of a CO significantly reduces the probability of a CO occurring at an adjacent locus, in a distance dependent manner. This pathway is dependent on the ZMM proteins (defined as ZIP1, ZIP2/SHOC1, ZIP3, ZIP4, MSH4, MSH5, MER3) and, in most eukaryotes, is responsible for the majority of COs during meiosis. Class II COs, that do not display interference, require MUS81 [1], [14]–[17].
Here we addressed the meiotic function of MCM8. MCM8 is a member of the eight MCM family proteins (MCM2–9), that all share a well conserved helicase domain. Together MCM2–7, as a hexamer, form a well characterized DNA helicase, which is essential for replication in all eukaryotes [18]. In contrast, MCM8–9 is not present in all eukaryotes [19], being notably missing in S. cerevisiae, S. pombe and C. elegans, but existing in vertebrates and plants. A study in Xenopus showed that MCM8 functions during DNA replication at the elongation stage but it is not required for replication licensing. The Xenopus MCM8 protein is the only MCM8 representative for which helicase activity has been demonstrated in vitro [20]. MCM8 is also involved in, but not essential for the assembly of the pre-replicative complex in human [21]. Very recently, MCM8 and MCM9 has been shown to be involved in homologous recombination-mediated DNA repair in mouse and chicken somatic cells [22], [23]. MCM8 has also been shown to be involved in meiosis. In the fruit fly (Drosophila melanogaster), in which MCM9 has not been identified, MCM8 (also known as REC) is required for 95% of meiotic COs. In contrast to COs, the frequency of NCOs increases in the absence of Dmrec [24]. Finally, a very recent study pointed out a role for MCM8, but not MCM9, in meiotic recombination in mouse [22]. Indeed meiocytes in the mouse mcm8 mutant accumulate DMC1 foci, display synapsis defects and go into apoptosis, consistent with a defect in meiotic DSB repair. The meiotic function of MCM8 has been analyzed only in Drosophila and mouse, with contrasting conclusions. This raises the question of the conservation of this function in eukaryotes. The aim of the present study was to further explore the meiotic function of MCM8 by deciphering its role in the model plant Arabidopsis.
Results
Identification of the AtMCM8 gene and Atmcm8 mutations
Phylogenetic analyses of the MCM family [19], [24], showed that the Arabidopsis genome contains one clear homolog for each MCM2–9, At3g09660 being the MCM8 homolog. We sequenced the At3g09660 CDS using RT-PCR on mRNA from Arabidopsis inflorescences. Because of some differences in splicing sites, the At3g09660 CDS slightly differed from the predicted sequence found in the genebank (NM_111800), measured 2,406 bp and contained 17 exons (Figure 1) (genebank BankIt1577803 MCM8 KC109786). We nonetheless confirmed by reciprocal BLAST analysis and multiple protein alignment that At3g09660 encodes the Arabidopsis MCM8 homolog (Figure S1 and [24]).
Fig. 1. AtMCM8 gene structure. 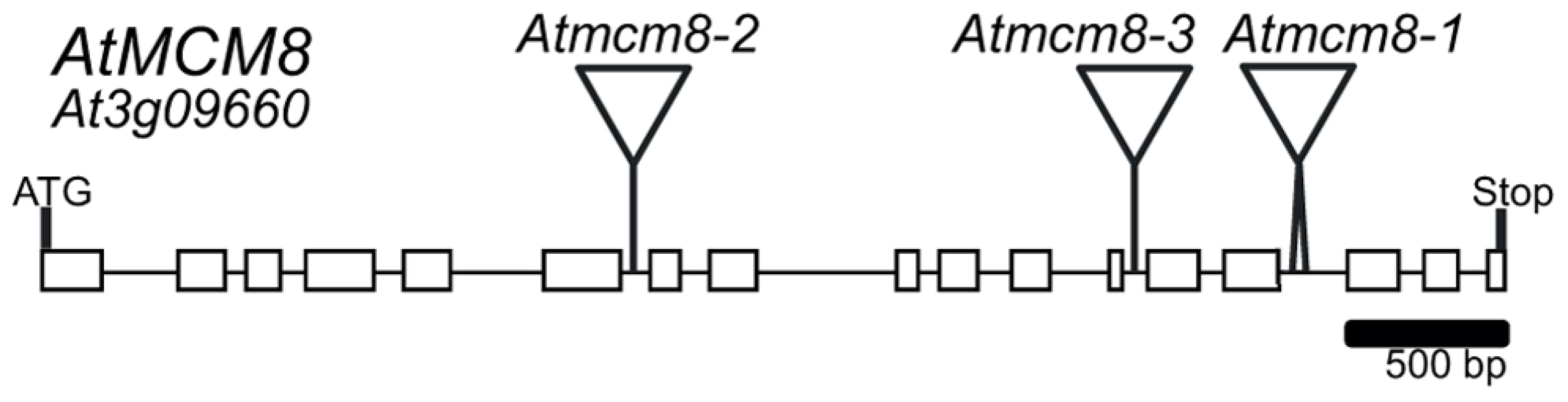
Exons are represented as black boxes and T-DNA insertions in Atmcm8-1, Atmcm8-2 and Atmcm8-3 alleles are indicated by triangles. We identified three T-DNA insertions from the public collections within the AtMCM8 gene: Atmcm8-1, Atmcm8-2 and Atmcm8-3 (Figure 1). Plants homozygous for the insertions showed normal vegetative growth but reduced fertility as shown by Alexander staining of pollen (Figure 2). This phenotype (and others described below) was detected only in homozygotes of each mutant. Moreover seed counts showed that Atmcm8-1 has significantly less seeds than wild type (44.8±5.2 (n = 41) compared to 52.4±5.8 (n = 77), Z test p<10−13). Allelism tests showed that the meiotic defects observed (see below) were due to the insertions in Atmcm8.
Fig. 2. Alexander staining. 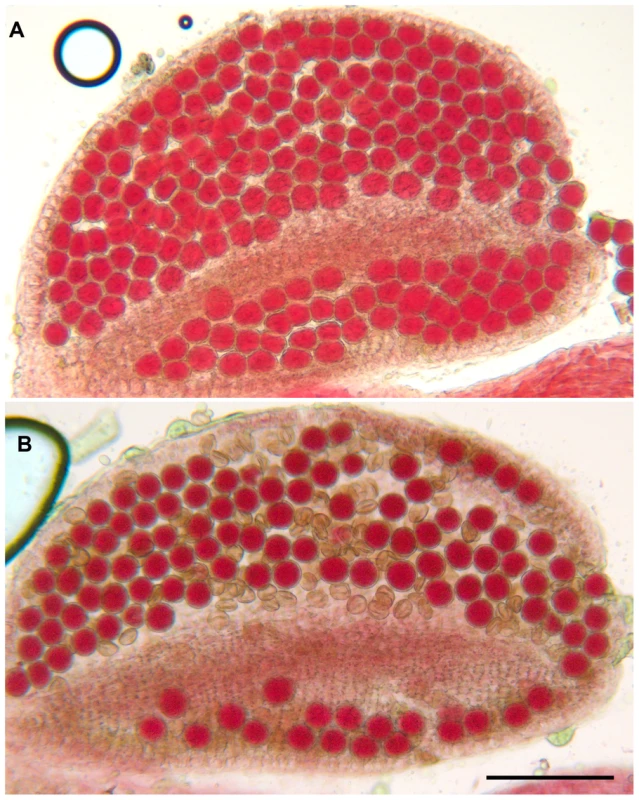
(A) A wild type anther containing pollen grains that are all viable, as indicated by their red staining and round shape. (B) An Atmcm8-1 anther containing viable and dead pollen grains as indicated by their abnormal shapes and green coloration. Bar, 100 µm. Atmcm8 chromosomes fragment during meiosis
To investigate if this reduction in fertility was linked to a meiotic defect, we analyzed meiotic progression by DAPI (4′,6-diamidino-2-phenylindole) staining of meiotic chromosome spreads in all three mutant alleles. In wild type meiosis (Figure 3A–3E), chromosomes condense at leptotene. Then, synapsis is initiated at zygotene until its completion in pachytene when the two homologous chromosomes are connected along their entire length by a proteinous structure called the synaptonemal complex [25] (Figure 3A and Figure 4A). Desynapsis occurs at diplotene and further condensation of the chromosomes occurs. Five bivalents continue to condense and become visible at diakinesis. At metaphase I, the five bivalents align on the metaphase I plate (Figure 3B). At anaphase I homologous chromosomes segregate to opposite poles (Figure 3C). At telophase I the two groups of five recombinant chromosomes begin to decondense. At prometaphase II chromosomes recondense and align on the two metaphase II plates (Figure 3D). At anaphase II each of the ten chromosomes segregate their two sister chromatids to opposite poles resulting in four balanced groups of five chromatids (Figure 3E).
Fig. 3. Male meiosis in wild-type and in Atmcm8. 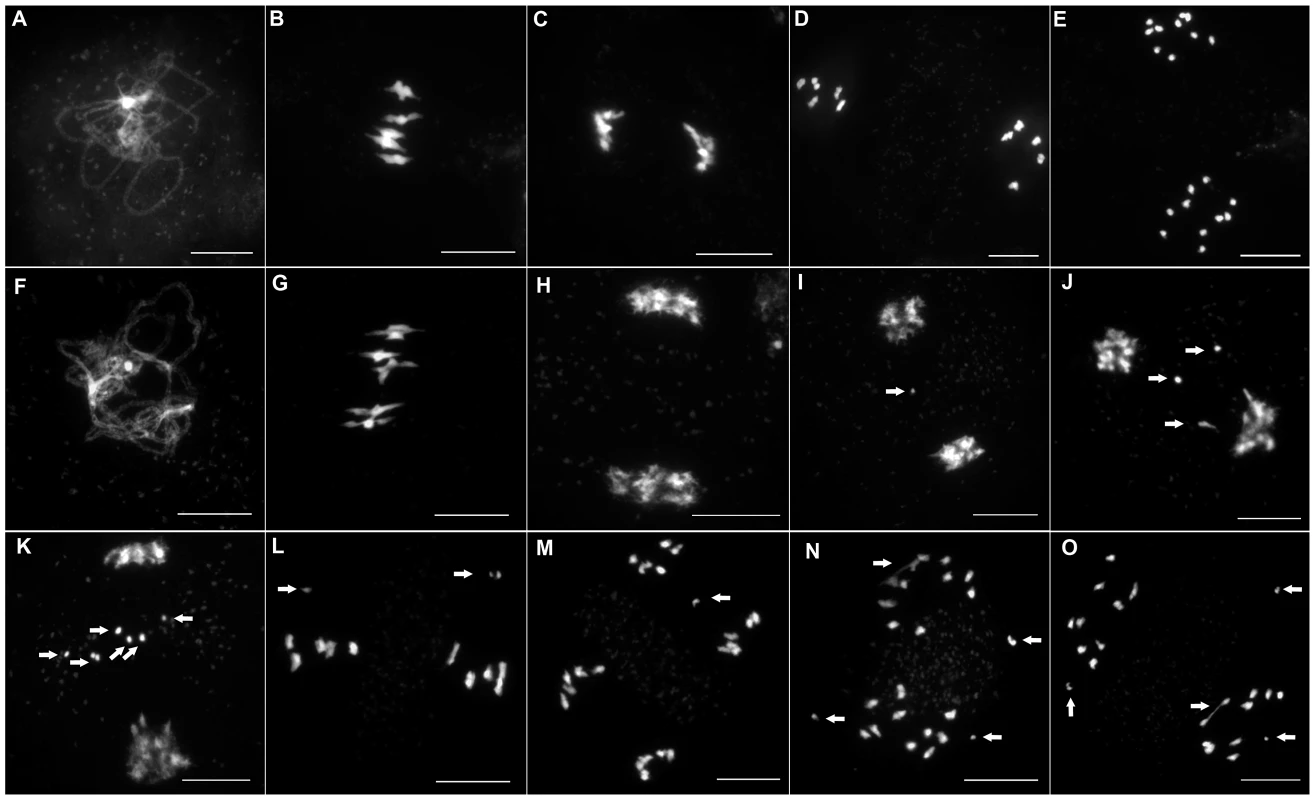
Male meiosis is shown (A–E) in wild type and (F–O) in Atmcm8. Chromosome spreads at (A and F) pachytene, (B and G) metaphase I, (C and H–K) end of anaphase I, (D and L) metaphase II, (E and M–O) anaphase II, using DAPI staining. Fragments and chromosome bridges are indicated with arrows. Bar, 10 µm. Fig. 4. Coimmunolocalization of ASY1 and ZYP1. 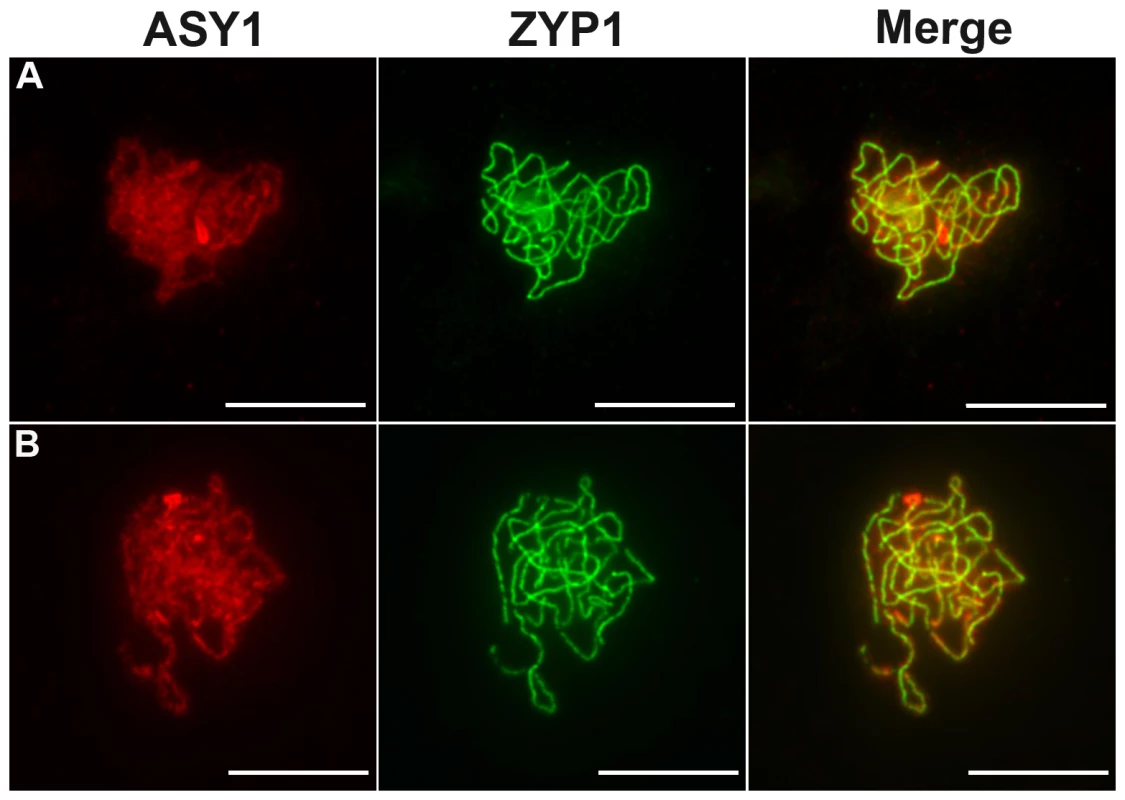
ASY1 (red), ZYP1 (green) are shown as well as the overlay of both signals (merge) at pachytene in (A) wild type and in (B) Atmcm8 mutant. In both wild type and mutant the polymerization of the synaptonemal complex, revealed by ZYP1, is completed at pachytene. The ASY1 signal is largely depleted from the chromosomes as the synaptonemal complex forms. Bar, 10 µm. In all three Atmcm8 alleles, meiosis appeared to progress normally from leptotene through to pachytene (Figure 3F) where chromosomes condensed, aligned and fully synapsed like wild type. The completion of synapsis in Atmcm8 was confirmed by immunolabelling meiotic chromosomes with antibodies against ASY1 and AtZYP1 (Figure 4B), that are components of the axial elements and of the transverse filament of the synaptonemal complex, respectively [26], [27]. Chromosomes desynapsed normally during diplotene and we observed five bivalents as condensation progressed during diakinesis, revealing the presence of chiasmata (the cytological manifestation of CO). At metaphase I, five bivalents were systematically observed in all mutant alleles, showing that at least one CO is formed per pair of homologous chromosomes (Figure 3G). Anaphase I proceeded, however chromosome fragmentation was observed in all three Atmcm8 alleles (Figure 3H–3K), with 1 to 10 chromosome fragments detected in 60 to 80% of the cells (Figure 5). Chromosomes aligned on the metaphase II plate, with fragments dispersed throughout the cell (Figure 3L). Anaphase II proceeded but additional chromosome fragments appeared (Figure 3M–3O). This fragmentation persists at telophase II. We also observed fragmentation in female meiosis showing that Atmcm8 mutation also affects female meiosis (data not shown).
Fig. 5. Quantification of chromosome fragmentation levels. 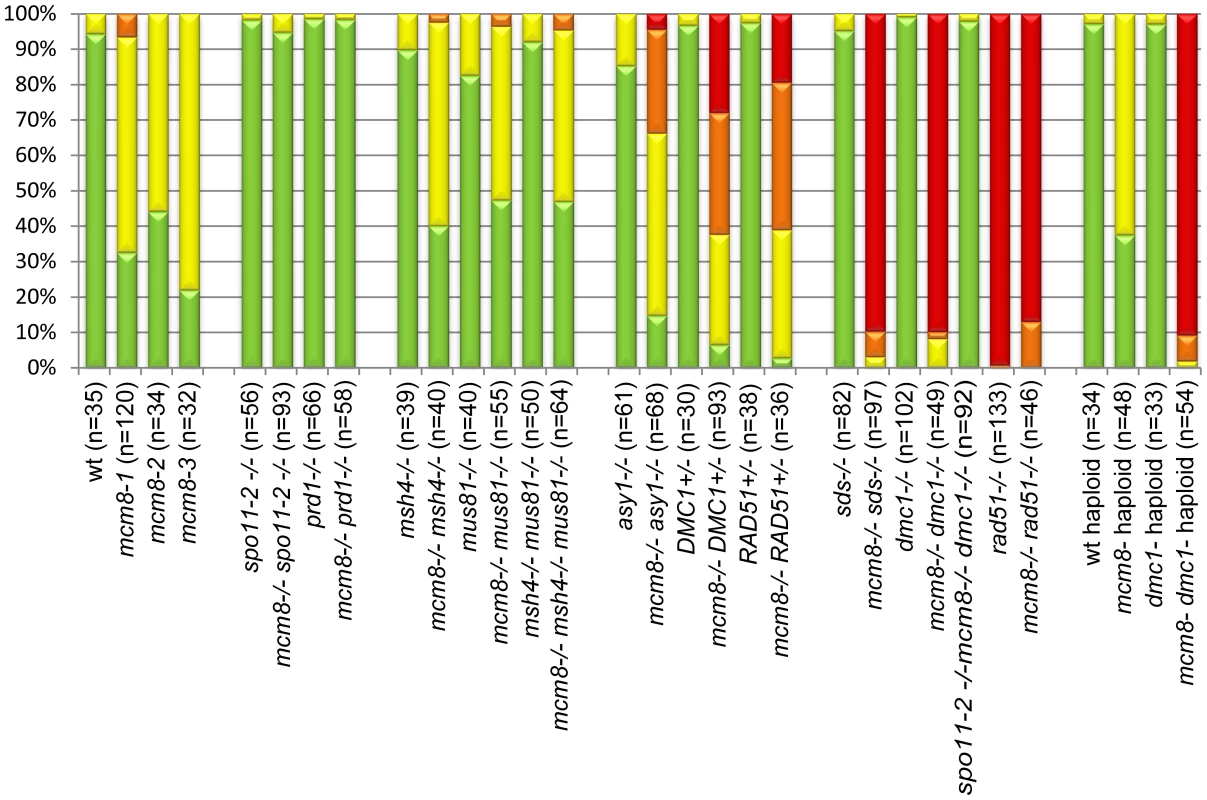
For each genotype, a number (indicated in brackets) of late anaphase I/telophase I cells were observed after chromosome spreads and DAPI staining. DAPI-stained bodies observed above the expected number of chromosomes were counted as fragments, and cells were classified has having 0 (green), 1–5 (yellow), 6–10 (orange), or more than 10 fragments (red). The percentage of each class is shown. Chromosome fragmentation in Atmcm8 is dependent on meiotic DSB formation
In Atspo11-2 and Atprd1, no meiotic DSBs are formed and therefore recombination does not occur [3], [28]. Thus at metaphase I, ten univalents are observed and segregate randomly (Figure 6A–6B and 6E–6F). To test whether the chromosome fragmentation seen in Atmcm8 mutants are dependent on DSB formation or not, we introduced the Atspo11-2 and Atprd1 mutations independently into Atmcm8. At meiosis, we observed ten univalents at metaphase I in the Atmcm8/Atspo11-2 or Atmcm8/Atprd1 and, importantly, the chromosome fragmentation was abolished (Figure 6C–6D and 6G–6H, Figure 5). Therefore, the fragmentation defect of Atmcm8 is dependent on AtSPO11-2 and AtPRD1. Thus, AtMCM8 is required for efficient repair of the DSBs that initiate meiotic recombination.
Fig. 6. Epistasis tests between Atmcm8 and two mutants affected in DSB formation. 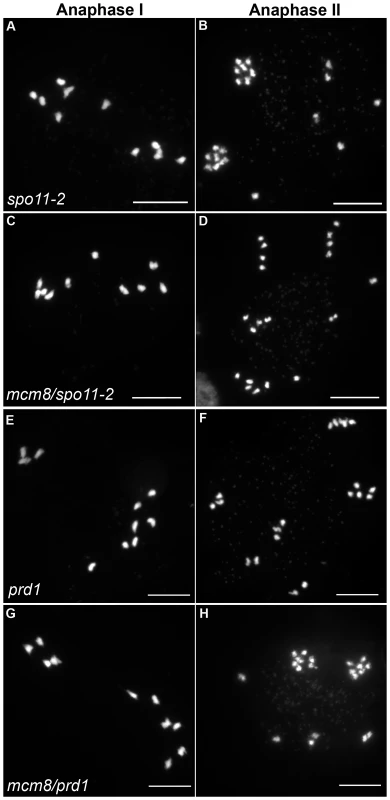
Meiotic spreads with (A–B) Atspo11-2, (C–D) Atmcm8/Atspo11-2, (E–F) prd1, (G–H) Atmcm8/Atspo11-2 using DAPI staining at anaphase I and anaphase II. Bar, 10 µm. Atmcm8 does not affect CO frequency
We then tested if the Atmcm8 fragmentation phenotype is dependent on the presence of any of the known pathways of CO formation, using epistasis tests. We used Atmsh4 and Atzip4 that are both required for class I CO formation and Atmus81 that is required for class II CO formation. In the Atmcm8/Atmsh4, Atmcm8/Atzip4, Atmcm8/Atmus81 double mutants and the Atmcm8/Atmsh4/Atmus81 triple mutant, we still observed a chromosome fragmentation defect as in the Atmcm8 single mutant (Figure 5 and Figure 7, data not shown for Atmcm8/Atzip4). Thus the Atmcm8 fragmentation phenotype is independent of MSH4, ZIP4 and MUS81.
Fig. 7. Epistasis tests between Atmcm8 and mutants affected in crossover formation. 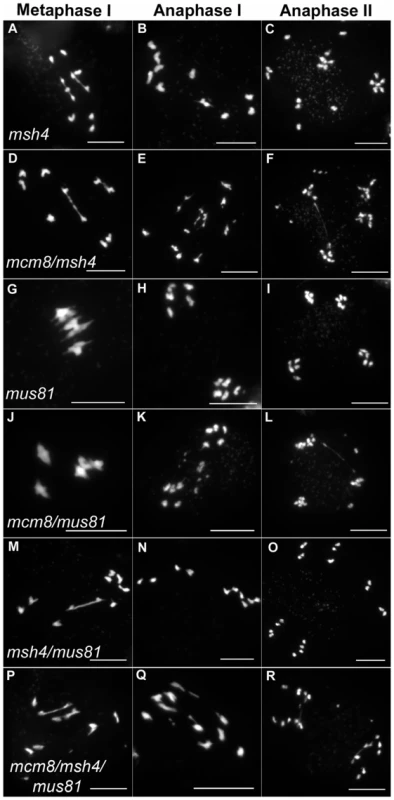
Meiotic spreads with (A–C) Atmsh4, (D–F) Atmcm8/Atmsh4, (G–I) Atmus81, (J–L) Atmcm8/Atmus81, (M–O) Atmsh4/Atmus81, (P–R) Atmcm8/Atmsh4/Atmus81 using DAPI staining at metaphase I, anaphase I and anaphase II. Bar, 10 µm. In Atmcm8 and Atmcm8/Atmus81 we invariably observed five bivalents at metaphase I, suggesting that the formation of class I COs, which account for most of the CO in wild type, is not grossly affected by the Atmcm8 mutation. This was further supported by counts of AtMLH1 foci, a marker of class I COs at late prophase of meiosis I [29], [30] (Figure S2), that revealed no significant differences between wild type (10.1±1.4 per cell; n = 81) and the Atmcm8 mutant (10.3±1.9; n = 86 (Z p = 0.55)). In Atmcm8/Atmsh4 (Figure 6), the residual number of bivalents at metaphase I was unchanged compared to the single Atmsh4 mutant (1.5±1; n = 91 vs 1.3±1.1; n = 91 (Z p = 0.94)), strongly suggesting that class II CO formation is not affected neither by Atmcm8 mutation. We then measured recombination frequency and crossover interference genetically in Atmcm8. This was achieved using tetrad analysis (Fluorescent-Tagged Lines, FTL) which is a visual pollen assay allowing the measurement of multiple COs simultaneously with access to all four chromatids from the same meiosis [31]. Two different sets of adjacent intervals on chromosome 5 have been analyzed, (I5aI5b and I5cI5d), representing four intervals in total. We did not detect any difference in recombination frequency between the Atmcm8 and wild type for any of these intervals (Table 1, Genetic Distance), consistent with the cytological data. Also, interference, that affects the distribution of crossovers, was unchanged compared to wild type for both sets of adjacent intervals (Table 1, Interference Ratio). Taken together these data suggest that AtMCM8 is not involved in CO formation. This contrasts from the observation that the absence of MCM8 reduces COs frequency by 95% in Drosophila [24].
Tab. 1. Genetic distances and interference in Atmcm8 using FTLs. 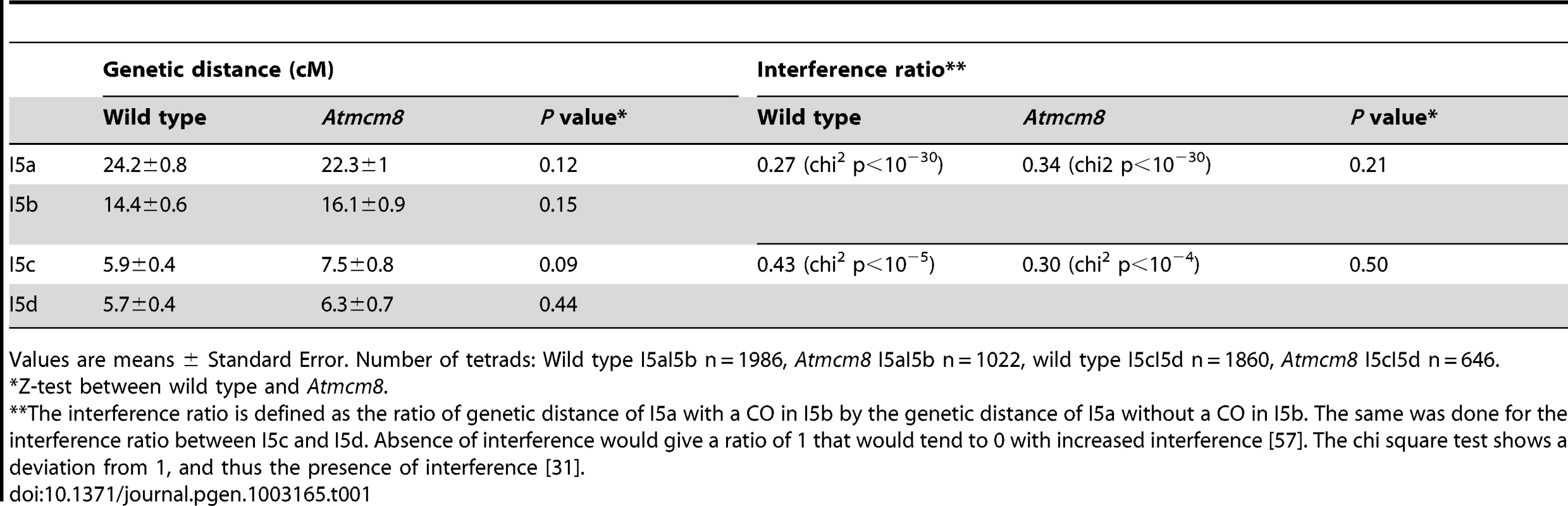
Values are means ± Standard Error. Number of tetrads: Wild type I5aI5b n = 1986, Atmcm8 I5aI5b n = 1022, wild type I5cI5d n = 1860, Atmcm8 I5cI5d n = 646. Mei9/Rad1 is another gene required for the formation of more than 90% of the COs in Drosophila [32]. Given the major difference in MCM8 function between Arabidopsis and Drosophila, we tested the role of AtRAD1 [33]–[35] in crossover formation in Arabidopsis. Cytological analysis showed that the single Atrad1 mutant has no obvious defect in CO formation. We then analyzed if AtRAD1 has a minor effect. To achieve this, we constructed a shoc1/Atrad1 double mutant and a Atmus81/shoc1/Atrad1 triple mutant to be able to detect a weak reduction in CO formation, in a sensitive context where there are no class I and class II COs. However, this triple mutant was not different from Atmus81/shoc1 (0.99±0.84 (n = 74) versus 1.15±1.28 (n = 75), Z p = 0.36) and neither was shoc1/Atrad1 different from shoc1 (1.47±1.07 (n = 51) versus 1.56±0.86 (n = 32), Z p = 0.67). These genes, MCM8 and MEI9/RAD1, are essential for CO formation in Drosophila but not in Arabidopsis showing divergent functions. However, contrary to RAD1, MCM8 has conserved a meiotic function in Arabidopsis.
The Atmcm8 DSB repair defect is amplified by DMC1 mutation
DMC1 is involved at the strand invasion stage of meiotic recombination and Atdmc1 mutants fail to synapse and to make COs (Figure 8A–8B, 8G–8H). However, DSBs are repaired in Atdmc1, in an AtRAD51-dependent manner, without CO formation, suggesting that the DSBs are repaired on sister chromatids in these mutants [8], [12]. In the Atmcm8/Atdmc1 double mutant, from metaphase I to the end of the meiosis we observed extensive chromosome fragmentation in all cells, which was much more intense than in the single Atmcm8 mutant (compare Figure 8C–8D to Figure 3I–3K and see quantification in Figure 5). Consistently, the Atmcm8/Atdmc1 double mutant was completely sterile whereas Atmcm8 has moderate fertility reduction and Atdmc1 produce some residual seeds [8], [12] (Table 2). Mutating SPO11-2 in this Atmcm8/Atdmc1 double mutant abolished the chromosome fragmentation (Figure 8E–8F, Figure 5), demonstrating that MCM8 and DMC1 act in parallel pathways of meiotic DSB repair.
Fig. 8. Epistasis tests between Atmcm8 and Atdmc1. 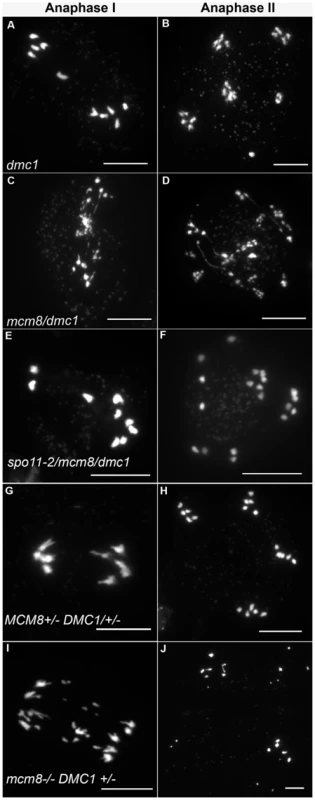
Meiotic spreads with (A–B) Atdmc1−/−, (C–D) Atmcm8−/−/Atdmc1−/−, (E–F) Atmcm8−/−/Atdmc1−/−/Atspo11−/−, (G–H) Atmcm8+/−/AtDMC1+/−, (I–J) Atmcm8−/−/AtDMC1+/−, using DAPI staining at anaphase I and anaphase II. Bar, 10 µm. Tab. 2. Seed per fruit and fragmentation levels in different combinations of double mutants. 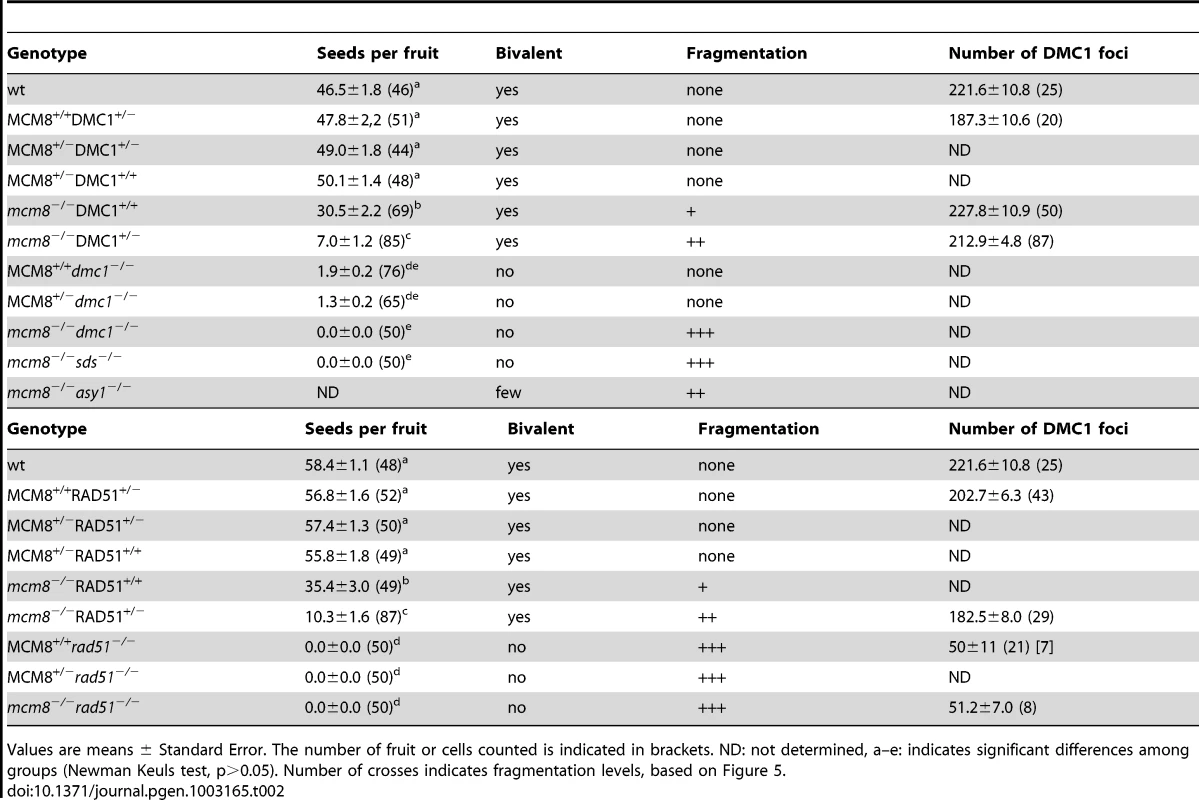
Values are means ± Standard Error. The number of fruit or cells counted is indicated in brackets. ND: not determined, a–e: indicates significant differences among groups (Newman Keuls test, p>0.05). Number of crosses indicates fragmentation levels, based on Figure 5. Furthermore in the Atmcm8 mutant context, we observed a more drastic meiotic chromosome fragmentation in plants heterozygous for DMC1 (Atmcm8−/−AtDMC1+/−) than wild type for DMC1 (Atmcm8−/−AtDMC1+/+) (compare Figure 8I–8J to Figure 3I–3K, quantification on Figure 5), accompanied by a strong reduction of fertility (Table 2). However, the fragmentation observed in Atmcm8−/−AtDMC1+/− was less dramatic than in the double mutant (Atmcm8−/−Atdmc1−/−) (Figure 5), which is also supported by the fertility levels (Table 2). This is despite the AtDMC1 mutation being recessive (in an AtMCM8+/+ or AtMCM8+/− context). Thus, in the absence of Atmcm8, the mutation of one of the two copies of DMC1 was enough to enhance fragmentation, which is even more drastic when both DMC1 alleles are disrupted.
The Atmcm8 DSB repair defect is amplified by mutation of ASY1, SDS, or one copy of RAD51
Therefore we tested the relationship of AtMCM8 with ASY1 and SDS, two proteins that are required for normal DMC1 localization [3], [13]. In the sds and asy1 single mutants, COs are greatly reduced (Figure 9E–9F) [10], [11]. In the Atmcm8/asy1 and Atmcm8/sds double mutant, we observed chromosome fragmentation from anaphase I onwards, which was much greater than that seen in the Atmcm8−/− single mutant (compare Figure 9G–9H with Figure 3I–3K, quantification on Figure 5). Thus, mutation of SDS or ASY1 amplified the fragmentation phenotype of Atmcm8. Finally, both the single Atrad51 mutant and the double Atmcm8/Atrad51 mutant show intense chromosome fragmentation (Figure 10). Interestingly, while AtRAD51+/− does not show chromosome fragmentation, Atmcm8−/−/AtRAD51+/− showed more chromosome fragmentation that Atmcm8 (Figure 10, Figure 5). Thus, in the absence of Atmcm8, the mutation of one of the two copies of RAD51 was enough to enhance fragmentation.
Fig. 9. Epistasis tests between Atmcm8 and sds or asy1. 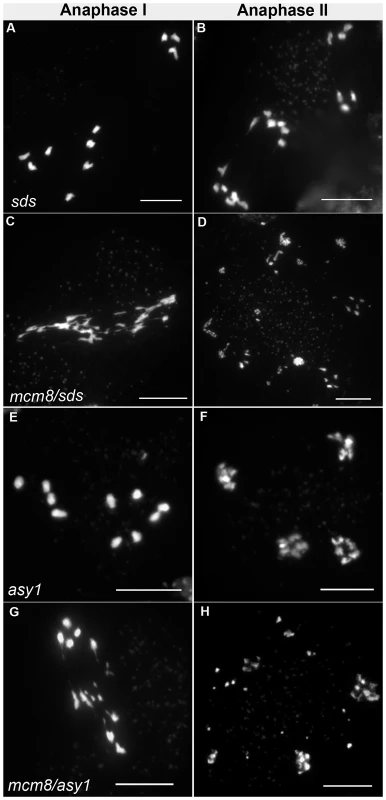
Meiotic spreads with (A–B) sds−/−, (C–D) Atmcm8−/−/sds−/−, (E–F) asy1−/−, (G–H) Atmcm8−/−/asy1−/, using DAPI staining at anaphase I and anaphase II. Bar, 10 µm. Fig. 10. Epistasis tests between Atmcm8 and Atrad51. 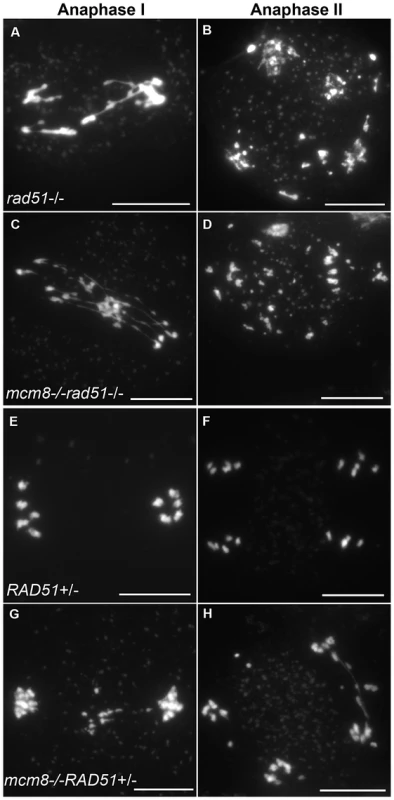
Meiotic spreads with (A–B) Atrad51−/−, (C–D) Atmcm8−/−/Atrad51−/−, (E–F) Atrad51+/−, (G–H) Atmcm8−/−/Atrad51+/−, using DAPI staining at anaphase I and anaphase II. Bar, 10 µm. AtDMC1 foci number is unaffected in Atmcm8
Given the relationship between DMC1 functional gene copy number and the degree of Atmcm8-dependent fragmentation, we looked at DMC1 behavior in Atmcm8. No significant difference in DMC1 foci shape or number was observed in Atmcm8−/− compared to wild type (Table 2). Similarly, we did not detect any differences in number or shape of DMC1 foci in Atmcm8−/−AtDMC1+/− or Atmcm8−/−AtRAD51+/− compared to either wild type or Atmcm8−/− (Figure S3, Table 2). In the Atmcm8 Atrad51 double mutant, we observed a marked decrease of DMC1 foci number, which was however similar to what was previously observed in a single Atrad51 mutant [7] (Table 2). It is intriguing that Atmcm8−/−AtDMC1+/− and Atmcm8−/−AtRAD51+/− exhibit a more drastic meiotic defect than Atmcm8−/−AtDMC1+/+, while DMC1 foci number and shape appear similar. However, it is possible that immunolocalization fails to detect subtle differences in DMC1 protein quantity or dynamics.
In the absence of homologous chromosomes, DSBs fail to be repaired in the absence of both MCM8 and DMC1
Next we explored the functional relationship between MCM8 and DMC1, in haploid plants, where homologous chromosomes are not present. Thus, the only template available for meiotic DSB repair is the sister chromatid. Meiotic chromosome spreads, in a wild-type haploid, showed that the five chromosomes were intact and segregated randomly at anaphase I [36] (Figure 11A–11B), suggesting that DSBs are efficiently repaired. The haploid Atmcm8 mutant had a limited fragmentation defect (Figure 11C–11D), similar to the defect in the diploid Atmcm8 mutant (Figure 5 for quantification). The Atdmc1 haploid had no fragmentation (Figure 11C–11F). In clear contrast, in the double Atmcm8/Atdmc1 haploid, we observed extensive meiotic chromosome fragmentation (Figure 11G–11H, see Figure 5 for quantification). This shows that in a haploid context, DSB repair is efficient in wild type and Atdmc1, only slightly affected in Atmcm8, but ineffective in the Atmcm8/Atdmc1 double mutant. This suggests that in the absence of a homologous template, AtMCM8 and AtDMC1 catalyze DSB repair on the sister chromatid in a redundant manner.
Fig. 11. Atmcm8 haploids during anaphase I and anaphase II. 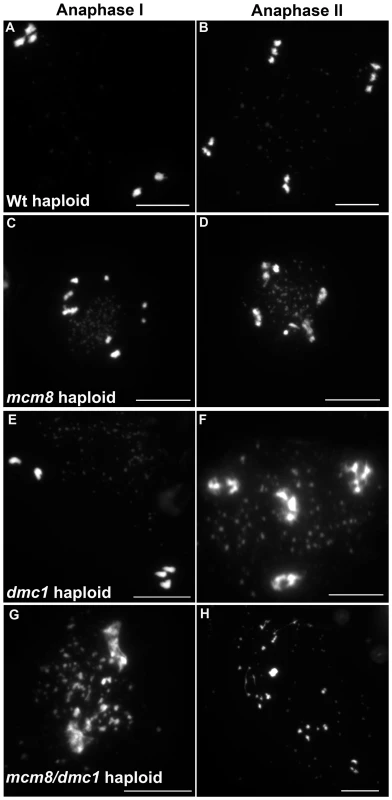
Meiotic spreads with (A–B) wild type haploid, (C–D) Atmcm8 haploid, (E–F) Atdmc1 haploid, (G–H) Atmcm8/Atdmc1 haploid using DAPI staining. Bar, 10 µm. Discussion
Here AtMCM8 was shown to be involved in meiotic DSB repair but not CO formation. This study thus revealed a pathway for DNA DSB repair that does not yield COs. This pathway depends on AtMCM8 and acts in parallel to the AtDMC1 pathway from which COs originate.
AtMCM8 is required for efficient meiotic DSB repair but not for CO formation
Arabidopsis MCM8 is required for effective meiotic DSB repair as all Atmcm8 mutant alleles had a clear, albeit limited, chromosome fragmentation defect at meiosis. The fragmentation is dependent on meiotic DSB formation as it disappears when AtSPO11-2 or AtPRD1 is absent. However, in contrast to Drosophila rec (mcm8) mutants, genetic and cytological data strongly support that CO formation is not affected by AtMCM8 mutation: (1) In the absence of AtMSH4 or AtZIP4 (class I COs) or AtMUS81 (class II COs) fragmentation still occurred and the number of bivalents was unchanged. (2) MLH1 foci numbers, a marker of class I COs, were unchanged in Atmcm8. (3) The genetic analysis using FTLs revealed no difference in terms of genetic distance and the strength of interference. These data showed that AtMCM8 acts in a pathway which repairs a subset of meiotic DSB and does not lead to CO formation.
Two pathways for DSB repair: one dependent on AtMCM8 and one on AtDMC1
A striking finding was that AtMCM8 becomes crucial when the DMC1 pathway was affected. Indeed, we observed a drastic amplification of the Atmcm8 mutant chromosome fragmentation defect when one of the two allelic copies of DMC1 was mutated, which was even more drastic when both DMC1 copies were mutated. This extensive fragmentation defect reflects a failure of DSB repair, as it is abolished by SPO11-2 mutation. Further, this extensive fragmentation was consistently confirmed in the absence of AtMCM8 and SDS, or AtMCM8 and ASY1. SDS and ASY1 are essential for AtDMC1 loading/stability [12], [37]. Extensive fragmentation was also observed when one copy of RAD51 was mutated in the Atmcm8 mutant. A function of RAD51 as a cofactor of DMC1 has been recently identified in yeast [5], and consistently DMC1 foci number is drastically reduced in the Arabidopsis rad51 mutant [7], [9]. We thus propose that two pathways of DSB repair coexist, one dependent on AtMCM8 and the other one on AtDMC1. In the absence of AtDMC1, efficient DSB repair occurs without CO formation. This repair depends on AtRAD51 [7], [8], [12] and on AtMCM8 (this study). Such RAD51-mediated, DMC1-independent, repair also exists in S. cerevisiae but is normally inhibited by RAD51 regulators [38]–[42]. Consequently, we suggest that, in the Atdmc1 context, AtMCM8 and AtRAD51 can co-operate to repair DSBs using the sister as a template. In addition to this function, AtRAD51 is required for the AtDMC1-dependent pathway (possibly as an accessory factor for the DMC1 strand-exchange activity as shown in yeast [5]) as repair is completely defective in the single Atrad51 mutant [6], like in the double Atmcm8/Atdmc1 mutant.
The fact that the fragmentation defect is limited in the single Atmcm8 mutant, suggests that the AtMCM8/AtRAD51 pathway would be essential for a limited number of events in wild type, when DMC1 fails. The repair events promoted by AtMCM8 are likely not intended to become a CO, as CO formation was not affected in Atmcm8, leaving sister chromatid repair or NCOs as the only other known possibilities. The absence of synapsis in Atdmc1 [7], [8], in which the AtMCM8/AtRAD51 pathway must be active, favors the hypothesis of sister chromatid repair. In contrast, the DMC1 pathway promotes CO formation. However, DMC1 foci in wild type, outnumber COs by approximately 25 to 1 [7], [43]. This suggests that repair of many DSBs catalyzed by DMC1 do not become CO, but NCO (that involve the homologous chromosome) or sister chromatid exchange (SCE). In Arabidopsis, the genome-wide frequency of NCOs and SCEs is currently unknown. We favor the hypothesis that DMC1 promotes NCOs, as DMC1 promotes synapsis. However, it should be noted that DMC1 is also able to promote SCE, notably in the haploid mcm8 context. Indeed, only the simultaneous mutation of AtDMC1 and AtMCM8 in haploids led to extensive chromosome fragmentation. The capacity of DMC1 to promote inter-sister repair was previously shown in other mutant background in both Arabidopsis [9] and yeast [44].
In summary we suggest that two pathways of DSB repair exist in wild type meiosis: The first pathway relies on the strand exchange activity of DMC1, and is also promoted by ASY1, SDS and RAD51 as a co-factor of DMC1 [5]. This pathway generates the COs, but also NCOs and SCEs in a ratio that remains to be determined. The second pathway of the model, which may be viewed as a backup pathway in case of failure of DMC1, relies on the strand exchange activity of RAD51 and the helicase activity of MCM8, and uses the sister chromatid as a template.
MCM8 function varies among eukaryotes
The function of MCM8 appears to differ markedly in Arabidopsis and in Drosophila. Interestingly, DMC1 and MCM8 appear to be partially redundant in Arabidopsis while the Drosophila genome seems devoid of a DMC1 homolog [45]. Thus CO formation in Drosophila appears to rely on a RAD51/MCM8 pathway, which has only a minor role in wild type meiotic DSB repair in Arabidopsis. The CO pathways appear to differ considerably in the two species, mainly using ZMMs in Arabidopsis but not RAD1, and the reverse in Drosophila, i.e. RAD1 but not ZMMs (that are absent from the Drosophila genome). Drosophila appears to be unique, as in distant species like S. cerevisiae, mammals and C. elegans CO formation depends mainly on ZMM. Adding to the complexity, MCM8 exists in mammals but not in S. cerevisiae and C. elegans [19], [24]. In mouse, MCM8 mutation leads to a meiotic arrest, with defects in homologous synapsis and over-accumulation of DMC1 foci before apoptosis, suggestive of defects in DSB repair [22]. We would like to suggest that these defects may compatible with MCM8 being required for a backup pathway in the case of failure of DMC1 to repair breaks, like in Arabidopsis. The lack of the backup pathway may lead to the accumulation of DMC1 foci, and a failure to repair a subset of breaks, triggering apoptosis (it is noteworthy that DSB repair defects do not trigger meiotic arrest or apoptosis in Arabidopsis). This illustrates the variety of mechanisms that arose in the course of evolution to fulfill the conserved outcome of meiotic DSB repair and CO formation.
In conclusion, our data reveals the meiotic function of MCM8 in Arabidopsis. Cytological and genetic analyses showed that AtMCM8 is involved in DSB repair but it is not a determinant for CO formation. This study identified a new pathway of meiotic DSB repair independent of AtDMC1.
Materials and Methods
Plant material
A. thaliana accession Columbia (Col-0) was the wild type reference. Atmcm8-1 (Salk_032764, N532764), Atmcm8-2 (Salk_104007, N604007) Atmcm8-3 (Salk_099327, N599327) were obtained from the collection of T-DNA mutants at the Salk Institute Genomic Analysis Laboratory (SIGnAL, http://signal.salk.edu/cgi-bin/tdnaexpress) [46] via NASC (http://nasc.nott.ac.uk/). Other mutants used in this study were Atspo11-2 (Gabi_749C12, N359272) [47], Atprd1 (Salk_024703, N524703) [3], Atdmc1-3 (Sail_170_F08, N871769) [48], Atrad51 (Atrad51-1) [6], asy1-4 (Salk_046272, N546272), sds-2 (Sail_129_F09, N806294) [12], Atzip4-2 (Salk_068052, N568052) [43], Atmsh4 (Salk_136296, N636296) [49], mus81-2 (Salk_107515, N607515), mus81-3 (Salk_002761, N502761) [50], [51], and shoc1-1 (Salk_057589, N557589). rad1-1 (uvh1-1) has a EMS (ethyl methanesulfonate) mutation [33], [34] and was provided by C. White.
Growth conditions
Plants were cultivated in greenhouse or growth chamber with a 16 h/day and 8 h/night photoperiod, at 20°C and 70% humidity.
Genetic analysis
Allelism tests were performed by crossing Atmcm8-1+/− with Atmcm8-2+/− and selecting F1 plants hemizygous for both alleles and likewise for Atmcm8-2+/− with Atmcm8-3+/−. Double mutants were obtained by crossing heterozygous plants for each mutation and selfing the double heterozygous F1 plants. Atmcm8/Atmhs4/Atmus81 triple mutant was identified by crossing Atmcm8/Atmsh4 double heterozygous with Atmus81 single mutant. As Atmsh4 and Atmus81 are linked, a plant heterozygous for Atmcm8/Atmsh4 was self-fertilized and homozygous for Atmus81 to identify the triple mutant in the offspring. Haploid Atmcm8 and Atmcm8/Atdmc1 were obtained by crossing a heterozygous plant for Atmcm8 or Atmcm8/Atdmc1 mutations as male and the GEM line as female [36], [52]. In F1, haploid plants of the desired genotype were selected.
Oligonucleotides for PCR genotyping
Plants of interest were selected by PCR genotyping using diagnostic primer sets. The three AtMCM8 insertions were genotyped by PCR using following primer combinations to amplify genomic DNA flanking the T-DNA insertions. Atmcm8-1: left borders (LB) with LBsalk2 (5′-GCTTTCTTCCCTTCCTTTCTC-3′)/N532764L (5′-AGCGCCATTAGCAAAATGTC-3′) or with LBsalk2/N532764U (5′-GCAGCTTCATTCTGCAAGTG-3′). Wild type allele with N532764U/N532764L. Atmcm8-2 LB with LBsalk2/N604007L (5′ - TCACTACAGCAACGGTGAGC -3′), right border (RB) with RBsalk1 (5′-TCA GAG CAG CCG ATT GTC-3′)/N604007U (5′-GCTGATGGAAGACCTTGTGG-3′). Wild type allele with N604007U/N604007L. Atmcm8-3 LB with LBsalk2/N599327L (5′-TGGTGTGGAATCAGCAGATG-3′) or with Lbsalk2/N599327U (5′-TGTGTCTCTGTTGCAAAGGC-3′). Wild type allele with N599327U/N599327L. T-DNA right and left borders were analyzed by sequencing PCR products. AtSPO11-2 wild type allele was amplified using primers 749C12U (5′-GAGCGAGAATTTTTGGTTGG-3′) and 749C12L (5′ - CCACAAGGTCAATTCTTCAAC-3′) and mutant allele using N524703L and LBgabi1 (5′-CCCATTTGGACGTGAATGTAGACAC-3′). AtPRD1 wild type allele was amplified using primers N524703U (5′-AAGTCTGCCCATGGTCACGATTCTCTCTG-3′) and N524703L (5′-GCCTGCTCAAAGGGTCCAGC-3′) and mutant allele using N524703L and LbSalk2. AtDMC1 wild type allele was amplified using primers N871769U (5′ - TTTTTAATTGTTTACAGAGGAAATCAG-3′) and N871769L (5′-TCCACTCGGAATAAAGCAATG-3′) and mutant allele using N871769L and Lb3sail (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′). AtRAD51 wild type allele was amplified using primers RAD51-1U (5′-ATGCCAAGGTTGACAAGATTG-3′) and RAD51-1L (5′ - CTCCCCTTCCAGAGAAATCTG -3′) and mutant allele using RAD51-1U and LBgabi1 (5′-CCCATTTGGACGTGAATGTAGACAC-3′). We amplified SDS wild type allele using primers N806294U (5′-CTGCTCCCTGATTACAAGCAG-3′) and N806294L (5′-CTTAACGCATTCAGGCAACTC-3′) and mutant allele using N806294U and Lb3sail. AtMSH4 wild type allele was amplified using primers N636296U (5′-CTTCTTGCAGGTTGTGTTTG-3′) and N636296L (5′-GCCAGCTGTTTTTGTTGTC-3′) and mutant allele using N636296L and LbSalk2. AtMUS81A wild type allele was amplified for Salk_107515 using primers N607515U (5′-CATGCTGACAGTTGAAGGTC-3′) and N607515L (5′-CCTCAAACGTTTCTCCAAAT-3′) and mutant allele using N607515L and LbSalk2. AtMUS81A wild type allele was amplified for Salk_002176 using primers N502176U (5′-CACATACGTTTTTGGTTCCC-3′) and N502176L (5′-AGTGTCCAAGTCCTGCTTTC-3′) and mutant allele using N607515L and LbSalk2. AtZIP4 wild type allele was amplified using primers N568052U (5′-TCCTTCCCACACCTTGACCC-3′) and N568052L (5′-GACTGCTGGAGCAGAAACT-3′) and mutant allele using N568052L and LbSalk2. ASY1 wild type allele was amplified using primers N546272U (5′-TCTATGTTTGTTACGCGTTAATCAG-3′) and N546272L (5′-AGGTGGCTCGTAATCTGGTGGCTGC-3′) and mutant allele using N546272L and LbSalk2. SHOC1 wild type allele was amplified using primers N557589U (5′-TTACCGGAGTTTGAAAACCG-3′) and N557589L (5′-GGCAAAGACTTGAAGGCATC-3′) and mutant allele using N557589L and LbSalk2. AtRAD1 was amplified using primers o629 (5′-CTGGTGAAGAACATTTGGTAG-3′) and o630 (5′-CTCTTATGGCTGCTGCGTCTTC-3′). Polymorphism between wild type and mutant alleles was revealed with Dde1 digestion.
Fluorescent tagged lines
FTL lines were obtained from G.P. Copenhaver. For this study, we used two couple of adjacent intervals: I5aI5b and I5cI5d [31]. The procedure to create plants of interest and to collect data was described in [31], [53]. Statical analysis was performed as described in [31].
Cytology, immunolocalization, and antibodies
Alexander staining for pollen viability was performed as described [54]. The protocol described by [55] was used to observe the female meiosis and the protocol described by [29] for male meiotic spreads. Immunolocalization of AtMLH1 was made as described by [29]. Immunolocalization of AtZYP1 and AtDMC1 was performed according to [56] with the modifications described in [43]. The anti-ASY1 polyclonal [56] and anti-ZYP1 polyclonal [49] antibodies were used at a dilution of 1∶250. The anti-MLH1 antibody [29] was used at a dilution of 1∶200. The anti-DMC1 antibody was described in [43] and the purified serum was used at 1∶20.
Microscopy
For male meiotic spreads, observations were made with a Leica DM RXA2 epifluorescence microscope using an oil PL APO 100X/1.40 objective (Leica). Photographs were taken using a CoolSNAP HQ (Roper Scientific) camera driven by Open-LAB 4.0.4 software (Improvision). For immunocytology and FTLs analyzes, observations were made using a Zeiss Axio Imager2 microscope. We analyzed FTLs using the automatic slide-scanner function of the ZEISS AxioObserver DIC FISH Apotome and its workbench. Photographs were taken using an AxioCam MRm (Zeiss) camera driven by Open-LAB 4.0.4 software AxioVision 4.8. All pictures were processed with AdobePhotoshop 7.0 (Adobe Systems Inc.).
Supporting Information
Zdroje
1. CromieGa, SmithGR (2007) Branching out: meiotic recombination and its regulation. Trends in cell biology 17 : 448–455 doi:10.1016/j.tcb.2007.07.007.
2. KeeneyS (2008) Spo11 and the formation of DNA double-strand breaks in meiosis. Recombination and meiosis 81–123 doi:10.1007/7050.
3. De MuytA, VezonD, GendrotG, GalloisJ-L, StevensR, et al. (2007) AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. The EMBO journal 26 : 4126–4137 doi:10.1038/sj.emboj.7601815.
4. KagawaW, KurumizakaH (2010) From meiosis to postmeiotic events: uncovering the molecular roles of the meiosis-specific recombinase Dmc1. The FEBS journal 277 : 590–598 doi:10.1111/j.1742-4658.2009.07503.x.
5. CloudV, ChanY-L, GrubbJ, BudkeB, BishopDK (2012) Rad51 Is an Accessory Factor for Dmc1-Mediated Joint Molecule Formation During Meiosis. Science 337 : 1222–1225 doi:10.1126/science.1219379.
6. LiW, ChenC, Markmann-MulischU, TimofejevaL, SchmelzerE, et al. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proceedings of the National Academy of Sciences of the United States of America 101 : 10596–10601 doi:10.1073/pnas.0404110101.
7. VignardJ, SiwiecT, ChelyshevaL, VrielynckN, GonordF, et al. (2007) The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet 3: e176 doi:10.1371/journal.pgen.0030176.
8. CouteauF, BelzileF, HorlowC, GrandjeanO, VezonD, et al. (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. The Plant cell 11 : 1623–1634.
9. Kurzbauer M-T, Uanschou C, Chen D, Schlögelhofer P (2012) The Recombinases DMC1 and RAD51 Are Functionally and Spatially Separated during Meiosis in Arabidopsis. The Plant cell: 1–14. doi:10.1105/tpc.112.098459.
10. CarylAPP, ArmstrongSJ, JonesGH, FranklinFCH (2000) A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109 : 62–71.
11. AzumiY, LiuD, ZhaoD, LiW, WangG, et al. (2002) Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. The EMBO journal 21 : 3081–3095 doi:10.1093/emboj/cdf285.
12. De MuytA, PereiraL, VezonD, ChelyshevaL, GendrotG, et al. (2009) A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 5: e1000654 doi:10.1371/journal.pgen.1000654.
13. Sanchez-MoranE, SantosJL, JonesGH, FranklinFCH (2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes & development 21 : 2220–2233 doi:10.1101/gad.439007.
14. OsmanK, HigginsJD, Sanchez-MoranE, ArmstrongSJ, FranklinFCH (2011) Pathways to meiotic recombination in Arabidopsis thaliana. The New phytologist 190 : 523–544 doi:10.1111/j.1469-8137.2011.03665.x.
15. HarrisonCJ, AlveyE, HendersonIR (2010) Meiosis in flowering plants and other green organisms. Journal of experimental botany 61 : 2863–2875 doi:10.1093/jxb/erq191.
16. LynnA, SoucekR, BörnerGV (2007) ZMM proteins during meiosis: crossover artists at work. Chromosome Research 15 : 591–605 doi:10.1007/s10577-007-1150-1.
17. MézardC, VignardJ, DrouaudJ, MercierR (2007) The road to crossovers: plants have their say. Trends in genetics: TIG 23 : 91–99 doi:10.1016/j.tig.2006.12.007.
18. MaioranoD, LutzmannM, MéchaliM (2006) MCM proteins and DNA replication. Current opinion in cell biology 18 : 130–136 doi:10.1016/j.ceb.2006.02.006.
19. LiuY, RichardsTa, AvesSJ (2009) Ancient diversification of eukaryotic MCM DNA replication proteins. BMC evolutionary biology 9 : 60 doi:10.1186/1471-2148-9-60.
20. MaioranoD, CuvierO, DanisE, MéchaliM (2005) MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell 120 : 315–328 doi:10.1016/j.cell.2004.12.010.
21. VolkeningM, HoffmannI (2005) Involvement of Human MCM8 in Prereplication Complex Assembly by Recruiting hcdc6 to Chromatin Involvement of Human MCM8 in Prereplication Complex Assembly by Recruiting hcdc6 to Chromatin. 25 doi:10.1128/MCB.25.4.1560.
22. LutzmannM, GreyC, TraverS, GanierO, Maya-MendozaA, et al. (2012) MCM8 - and MCM9-Deficient Mice Reveal Gametogenesis Defects and Genome Instability Due to Impaired Homologous Recombination. Molecular cell 47 : 523–534 doi:10.1016/j.molcel.2012.05.048.
23. NishimuraK, IshiaiM, HorikawaK, FukagawaT, TakataM, et al. (2012) Mcm8 and Mcm9 Form a Complex that Functions in Homologous Recombination Repair Induced by DNA Interstrand Crosslinks. Molecular cell 47 : 511–522 doi:10.1016/j.molcel.2012.05.047.
24. BlantonHL, RadfordSJ, McMahanS, KearneyHM, IbrahimJG, et al. (2005) REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet 1: e40 doi:10.1371/journal.pgen.0010040.
25. PageSL, HawleyRS (2004) The genetics and molecular biology of the synaptonemal complex. Annual review of cell and developmental biology 20 : 525–558 doi:10.1146/annurev.cellbio.19.111301.155141.
26. ArmstrongSJ, CarylAPP, JonesGH, FranklinFCH (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. Journal of Cell Science 115 : 3645–3655 doi:10.1242/jcs.00048.
27. HigginsJD, Sanchez-MoranE, ArmstrongSJ, JonesGH, FranklinFCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes & development 19 : 2488–2500 doi:10.1101/gad.354705.
28. StaceyNJ, KuromoriT, AzumiY, RobertsG, BreuerC, et al. (2006) Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. The Plant Journal 48 : 206–216 doi:10.1111/j.1365-313X.2006.02867.x.
29. ChelyshevaL, GrandontL, VrielynckN, le GuinS, MercierR, et al. (2010) An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenetic and genome research 129 : 143–153 doi:10.1159/000314096.
30. JacksonN, Sanchez-MoranE, BucklingE, ArmstrongSJ, JonesGH, et al. (2006) Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. The EMBO journal 25 : 1315–1323 doi:10.1038/sj.emboj.7600992.
31. BerchowitzLE, CopenhaverGP (2008) Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nature protocols 3 : 41–50 doi:10.1038/nprot.2007.491.
32. SekelskyJJ, McKimKS, ChinGM, HawleyRS (1995) The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141 : 619–627.
33. LiuZ, HossainGS, Islas-OsunaMA, MitchellDL, MountDW (2000) Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. The Plant journal: for cell and molecular biology 21 : 519–528.
34. GallegoF, FleckO, LiA, WyrzykowskaJ, TinlandB (2000) AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. The Plant journal: for cell and molecular biology 21 : 507–518.
35. DubestS, GallegoME, WhiteCI (2002) Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO reports 3 : 1049–1054 doi:10.1093/embo-reports/kvf211.
36. RaviM, ChanSWL (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464 : 615–618 doi:10.1038/nature08842.
37. Sanchez-moranE, SantosJL, JonesGH, FranklinFCH (2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes & Development 1 : 2220–2233 doi:10.1101/gad.439007.2002.
38. SheridanS, BishopDK (2006) Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes & development 20 : 1685–1691 doi:10.1101/gad.1447606.
39. TsubouchiH, RoederGS (2006) Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes & development 20 : 1766–1775 doi:10.1101/gad.1422506.
40. NiuH, WanL, BusyginaV, KwonY, AllenJa, et al. (2009) Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Molecular cell 36 : 393–404 doi:10.1016/j.molcel.2009.09.029.
41. ShinoharaA, GasiorS, OgawaT, KlecknerN, BishopDK (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes to cells: devoted to molecular & cellular mechanisms 2 : 615–629.
42. NiuH, LiX, JobE, ParkC, MoazedD, et al. (2007) Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Molecular and cellular biology 27 : 5456–5467 doi:10.1128/MCB.00416-07.
43. ChelyshevaL, GendrotG, VezonD, DoutriauxM-P, MercierR, et al. (2007) Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3: e83 doi:10.1371/journal.pgen.0030083.
44. Schwachaa, KlecknerN (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90 : 1123–1135.
45. YoudsJL, BoultonSJ (2011) The choice in meiosis - defining the factors that influence crossover or non-crossover formation. Journal of cell science 124 : 501–513 doi:10.1242/jcs.074427.
46. AlonsoJM, StepanovaAN, LeisseTJ, KimCJ, ChenH, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (New York, NY) 301 : 653–657 doi:10.1126/science.1086391.
47. HartungF, Wurz-WildersinnR, FuchsJ, SchubertI, SuerS, et al. (2007) The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. The Plant cell 19 : 3090–3099 doi:10.1105/tpc.107.054817.
48. PradilloM, LópezE, LinaceroR, RomeroC, CuñadoN, et al. (2012) Together yes, but not coupled: new insights into the roles of RAD51 and DMC1 in plant meiotic recombination. The Plant journal: for cell and molecular biology 69 : 921–933 doi:10.1111/j.1365-313X.2011.04845.x.
49. Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes & Development: 2557–2570. doi:10.1101/gad.317504.eukaryote.
50. HigginsJD, BucklingEF, FranklinFCH, JonesGH (2008) Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. The Plant Journal 54 : 152–162 doi:10.1111/j.1365-313X.2008.03403.x.
51. BerchowitzLE, FrancisKE, BeyAL, CopenhaverGP (2007) The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3: e132 doi:10.1371/journal.pgen.0030132.
52. MarimuthuMPa, JolivetS, RaviM, PereiraL, DavdaJN, et al. (2011) Synthetic clonal reproduction through seeds. Science 331 : 876 doi:10.1126/science.1199682.
53. MacaisneN, VignardJ, MercierR (2011) SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. Journal of cell science 124 : 2687–2691 doi:10.1242/jcs.088229.
54. AlexanderM (1969) Differential staining of aborted and nonaborted pollen. Biotechnic & Histochemistry 44 : 117–122.
55. MotamayorJ, VezonD, BajonC (2000) Switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sexual Plant Reproduction 12 : 209–218.
56. ArmstrongSJ (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. Journal of Cell Science 115 : 3645–3655 doi:10.1242/jcs.00048.
57. MalkovaA, SwansonJ, GermanM, McCuskerJH, HousworthEa, et al. (2004) Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics 168 : 49–63 doi:10.1534/genetics.104.027961.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



