-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
All pancreatic endocrine cell types arise from a common endocrine precursor cell population, yet the molecular mechanisms that establish and maintain the unique gene expression programs of each endocrine cell lineage have remained largely elusive. Such knowledge would improve our ability to correctly program or reprogram cells to adopt specific endocrine fates. Here, we show that the transcription factor Nkx6.1 is both necessary and sufficient to specify insulin-producing beta cells. Heritable expression of Nkx6.1 in endocrine precursors of mice is sufficient to respecify non-beta endocrine precursors towards the beta cell lineage, while endocrine precursor - or beta cell-specific inactivation of Nkx6.1 converts beta cells to alternative endocrine lineages. Remaining insulin+ cells in conditional Nkx6.1 mutants fail to express the beta cell transcription factors Pdx1 and MafA and ectopically express genes found in non-beta endocrine cells. By showing that Nkx6.1 binds to and represses the alpha cell determinant Arx, we identify Arx as a direct target of Nkx6.1. Moreover, we demonstrate that Nkx6.1 and the Arx activator Isl1 regulate Arx transcription antagonistically, thus establishing competition between Isl1 and Nkx6.1 as a critical mechanism for determining alpha versus beta cell identity. Our findings establish Nkx6.1 as a beta cell programming factor and demonstrate that repression of alternative lineage programs is a fundamental principle by which beta cells are specified and maintained. Given the lack of Nkx6.1 expression and aberrant activation of non-beta endocrine hormones in human embryonic stem cell (hESC)–derived insulin+ cells, our study has significant implications for developing cell replacement therapies.
Published in the journal: Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity. PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003274
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003274Summary
All pancreatic endocrine cell types arise from a common endocrine precursor cell population, yet the molecular mechanisms that establish and maintain the unique gene expression programs of each endocrine cell lineage have remained largely elusive. Such knowledge would improve our ability to correctly program or reprogram cells to adopt specific endocrine fates. Here, we show that the transcription factor Nkx6.1 is both necessary and sufficient to specify insulin-producing beta cells. Heritable expression of Nkx6.1 in endocrine precursors of mice is sufficient to respecify non-beta endocrine precursors towards the beta cell lineage, while endocrine precursor - or beta cell-specific inactivation of Nkx6.1 converts beta cells to alternative endocrine lineages. Remaining insulin+ cells in conditional Nkx6.1 mutants fail to express the beta cell transcription factors Pdx1 and MafA and ectopically express genes found in non-beta endocrine cells. By showing that Nkx6.1 binds to and represses the alpha cell determinant Arx, we identify Arx as a direct target of Nkx6.1. Moreover, we demonstrate that Nkx6.1 and the Arx activator Isl1 regulate Arx transcription antagonistically, thus establishing competition between Isl1 and Nkx6.1 as a critical mechanism for determining alpha versus beta cell identity. Our findings establish Nkx6.1 as a beta cell programming factor and demonstrate that repression of alternative lineage programs is a fundamental principle by which beta cells are specified and maintained. Given the lack of Nkx6.1 expression and aberrant activation of non-beta endocrine hormones in human embryonic stem cell (hESC)–derived insulin+ cells, our study has significant implications for developing cell replacement therapies.
Introduction
Innovative strategies for diabetes therapy aim to replace lost insulin-producing beta cells by reprogramming other cell types or by deriving beta cells from pluripotent cells. Ectopic expression of the transcription factors Pdx1, Neurogenin 3 (encoded by the Neurog3 gene; Ngn3), and MafA has been shown to reprogram pancreatic exocrine acinar cells into beta-like cells [1]. Similarly, some success in reprogramming of liver cells into beta cells has been reported after misexpression of Pdx1, Ngn3, MafA, NeuroD, or Nkx6.1 [2]–[6]. Moreover, recent studies have demonstrated that pancreatic endocrine alpha cells can spontaneously convert into beta cells after near complete ablation of beta cells in adult mice [7]. Conversely, loss of beta cell identity and partial conversion of beta cells into other endocrine cell types has recently been identified as an early event marking beta cell failure in diabetes [8]. Thus, substantial plasticity exists between pancreatic cell types, and this plasticity could potentially be exploited to halt diabetes progression or to replenish beta cells in diabetic individuals. However, little is still known about the factors that control this plasticity.
During embryonic development, all endocrine cell types are derived from a common endocrine precursor population marked by the transcription factor Ngn3 [9], [10]. Ngn3 activity is required for the specification of all endocrine cells [11] and the expression of Arx and Pax4, two transcription factors that control endocrine subtype choices downstream of Ngn3. Arx-deficient mice display a loss of alpha cells and concomitant increase in beta and delta cells, while Pax4-deficiency results in the opposite phenotype of reduced beta and delta cells but increased alpha cells [12], [13]. Strikingly, forced expression of Pax4 in endocrine precursors and their differentiated progeny imparts a beta-like cell identity to differentiating precursors, resulting in hyperplastic islets with an excess of beta-like cells at the expense of the other endocrine cell types [14]. However, despite their increased beta cell mass, mice misexpressing Pax4 eventually become diabetic and succumb prematurely, suggesting that sustained expression of Pax4 is not compatible with normal beta cell function. Since Pax4 is normally absent from beta cells and only transiently expressed in endocrine precursors during embryogenesis [15], it is possible that proper beta cell development and maturation requires Pax4 downregulation. Similar to Pax4, misexpression of Pdx1 in endocrine precursors has also been shown to favor a beta cell fate choice over other endocrine cell types [16]. Unlike ectopic Pax4 expression, forced expression of Pdx1 did not reduce the numbers of delta and PP cells, but selectively affected the ratio between beta and alpha cells. Therefore, Pdx1 activity appears to primarily control the alpha versus beta cell fate decision, which is consistent with its expression in both beta and delta cells [13]. Nkx2.2 has recently been identified as a beta cell maintenance factor and stabilizes beta cell fate by repressing the alpha cell fate determinant Arx [17]. While these studies have provided insight into the factors involved in endocrine cell type specification and maintenance, still little is known about how these factors interact to establish and maintain gene expression programs characteristic of each endocrine cell type. In particular, it is unclear which molecular mechanisms operate in beta cell precursors to ensure that alternative endocrine lineage programs are repressed, while beta cell-specific programs are activated. Given the simultaneous initiation of multiple endocrine subtype programs in one cell with current human embryonic stem cell (hESC) differentiation protocols [18], [19], such knowledge is critical for refining these protocols to support the differentiation of mature and functional beta cells in vitro.
In addition to MafA and Mnx1 (also called Hb9) [20]–[22], in the adult pancreas Nkx6.1 is among the few transcription factors exclusively detected in beta cells. During development, Nkx6.1 is first expressed in multipotent pancreatic progenitors, where it specifies an endocrine identity by repressing the pre-acinar transcription factor Ptf1a [23]. At later developmental stages, Nkx6.1 expression persists in common progenitor cells for the ductal and endocrine cell lineages before becoming eventually restricted to the beta cell lineage [24]. Whether or not Nkx6.1 plays a role in beta cell specification and maintenance remains unknown, largely due to the lack of appropriate genetic models to study this question. Excessive early acinar cell specification and reduced numbers of Ngn3+ cells in Nkx6.1 null mutant mice preclude their utility for such studies.
To determine the function of Nkx6.1 in endocrine cell type specification and beta cell maintenance, we generated novel genetic mouse models to conditionally inactivate or misexpress Nkx6.1 after endocrine precursors have been specified. Our studies reveal that Nkx6.1 is both necessary and sufficient to specify the beta cell lineage. Nkx6.1 prevents alpha cell specification in cooperation with Pdx1 by directly repressing Arx through competition with the Arx gene activator Isl1. Furthermore, inactivation of Nkx6.1 in beta cells causes loss of beta cell identity and conversion into delta cells. Our findings identify Nkx6.1 as a beta cell programming factor and uncover a transcriptional network that initiates and maintains beta cell-specific gene expression programs, while repressing programs of alternative endocrine lineages.
Results
Heritable expression of Nkx6.1 in endocrine precursors favors a beta cell fate choice at the expense of other islet cell types
During pancreatic development, Nkx6.1 is expressed in a subset of Ngn3+ cells and is then exclusively maintained in beta cells [25], suggesting that the specification of non-beta endocrine cell types might require Nkx6.1 downregulation. To explore whether expression of Nkx6.1 in all, or at least the majority of, Ngn3+ cells is sufficient to allocate precursors to the beta cell lineage, we heritably expressed Nkx6.1 in Ngn3+ cells, utilizing a mouse line that allows for conditional overexpression of Nkx6.1 after expression of Cre recombinase (Nkx6.1OE mice). In Nkx6.1OE mice, concomitant expression of Nkx6.1 and enhanced green fluorescent protein (eGFP) is induced by Cre recombinase-mediated excision of a lacZ expression cassette flanked by loxP sites. The Nkx6.1OE transgene design is analogous to the Z/EG transgene, in which Cre recombinase induces expression of GFP by recombining loxP sites flanking a lacZ cassette. Therefore, Z/EG mice were used as controls for the Nkx6.1OE strain (Figure 1A, 1B). We induced transgene recombination with Ngn3-Cre and compared the relative contribution of recombined GFP+ cells to each of the five endocrine cell types in Ngn3-Cre;Nkx6.1OE and Ngn3-Cre;Z/EG control mice. Notably, due to mosaic expression of the transgenes, not all hormone+ cells expressed GFP. At postnatal day (P) 2, 55.1±1.7% of the GFP+ cells expressed insulin in Ngn3-Cre;Z/EG control mice, while 86.1±2.5% of the GFP+ cells were insulin+ in Ngn3-Cre;Nkx6.1OE mice (Figure 1C, 1H, 1M; P<0.001), suggesting that Nkx6.1 favors a beta cell fate choice. Consistent with this notion, Nkx6.1-expressing endocrine precursor cells displayed a significantly decreased propensity to differentiate into glucagon+, somatostatin+, pancreatic polypeptide (PP)+, or ghrelin+ cells compared to endocrine precursor cells from control mice (Figure 1D–1G, 1I–1M). Since Nkx6.1 expression did not affect cell replication, as shown by analysis of the proliferation marker Ki67 (Figure 1N), or survival [23], these data indicate that Nkx6.1 promotes beta cell differentiation from endocrine precursors at the expense of alternative endocrine fates.
Fig. 1. Forced Nkx6.1 expression favors the beta cell fate choice. 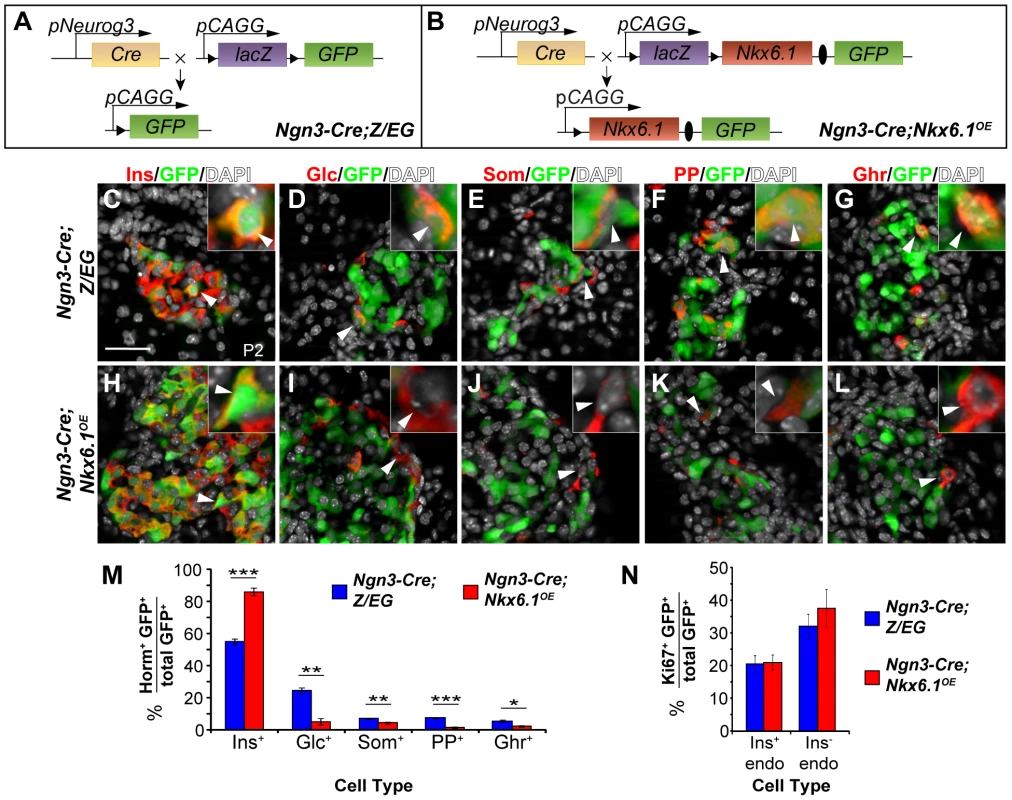
(A, B) Schematic of the transgenes for conditional Nkx6.1 misexpression and cell lineage tracing; Triangles, loxP sites; Ovals, internal ribosomal entry site (IRES). (C–L) Immunofluorescence staining of pancreata from Ngn3-Cre;Z/EG and Ngn3-Cre;Nkx6.1OE mice at postnatal day (P) 2 for GFP together with each of the five endocrine hormones. The insets display higher magnification images. Arrowheads point to GFP+ cells expressing each of the five hormones in Ngn3-Cre;Z/EG mice and insulin, but not glucagon, somatostatin, pancreatic polypeptide, or ghrelin in Ngn3-Cre;Nkx6.1OE mice. Quantification of hormone+GFP+ (M) or Ki67+GFP+ (N) co-positive cells as a percentage of all GFP-expressing cells in pancreata of Nkx6.1f/−;Ngn3-Cre;Z/EG and Ngn3-Cre;Z/EG mice at P2 (n = 4). Forced expression of Nkx6.1 in endocrine precursors favors a beta cell fate choice over all other non-beta endocrine cell fate choices. Horm, hormones; Ins, insulin; Glc, glucagon; Som, somatostatin; PP, pancreatic polypeptide; Ghr, ghrelin; endo, endocrine. Scale bar = 50 µm. Error bars represent S.E.M; *p<0.05, **p<0.01, ***p<0.001. Similar to Nkx6.1, conditional expression of Pax4 in mouse endocrine precursors results in beta cell specification at the expense of all other endocrine cell types [14]. The expression of Pax4 leads to oversized islets and is eventually accompanied by beta cell dysfunction and diabetes. To determine whether transgenic Nkx6.1 expression similarly causes islet and beta cell hyperplasia, we compared islet size in Ngn3-Cre;Nkx6.1OE and control mice at P2 and at 5 months of age. Consistent with our observation that Nkx6.1 overexpression in adult beta cells does not stimulate beta cell expansion [26], Ngn3-Cre;Nkx6.1OE mice displayed normal islet cell mass (Figure S1A, S1B). Furthermore, 5-month-old Ngn3-Cre;Nkx6.1OE mice exhibited normal glucose tolerance (Figure S1C), showing that sustained expression of the Nkx6.1 transgene does not perturb glucose homeostasis.
To next explore the extent of endocrine precursor reprograming and to assess the maturity of beta cells in Ngn3-Cre;Nkx6.1OE mice, we analyzed insulin+ progeny of targeted endocrine precursors for the expression of critical beta cell markers and possible ectopic expression of non-beta endocrine cell markers. As expected, insulin+GFP+ cells in Ngn3-Cre;Nkx6.1OE mice expressed the beta cell marker Pdx1, MafA, and Pax6 at P2, showing that transgenic Nkx6.1 expression in endocrine precursors and their progeny does not impair beta cell maturation (Figure S1D–S1I). Next, to determine whether Nkx6.1 is sufficient to fully repress alternative endocrine lineage programs during endocrine cell differentiation, we analyzed insulin+ cells in Ngn3-Cre;Nkx6.1OE mice for expression of Arx, Brn4, glucagon, and somatostatin. At P2, targeted GFP+ cells in Ngn3-Cre;Nkx6.1OE mice were largely indistinguishable from their counterparts in control mice and rarely displayed coexpression of insulin with any of these non-beta endocrine markers (Figure S1J–S1Q; arrowheads), suggesting that Nkx6.1 is effective in fully establishing a beta cell expression program.
Forced Nkx6.1 expression is not sufficient to reprogram differentiated non-beta endocrine cells into beta cells
We next sought to determine whether Nkx6.1 acts by inducing cell fate conversion during differentiation of Ngn3+ endocrine precursors or by converting already differentiated non-beta endocrine cells. Since we observed residual glucagon+, somatostatin+, pancreatic polypeptide (PP)+, and ghrelin+ cells misexpressing Nkx6.1 at P2 (Figure 1M), we first examined whether these cells convert into beta cells during postnatal life, as observed after Pdx1 expression in Ngn3+ cells [16]. Different from Ngn3-Cre;Pdx1OE mice, we found that targeted non-beta endocrine cells persisted in 5-month-old Ngn3-Cre;Nkx6.1OE mice (Figure S2A–S2H). Notably, the endocrine cell type ratios observed in Ngn3-Cre;Nkx6.1OE mice at P2 are largely maintained at 5 months of age (Figure S2I), suggesting that additional non-beta-to-beta cell fate conversion does not occur postnatally. To directly test whether forced expression of Nkx6.1 in differentiated alpha cells triggers their conversion into beta cells, we induced recombination of the Nkx6.1OE transgene with Glucagon-Cre (Glc-Cre). Consistent with the persistence of targeted non-beta endocrine cells in adult Ngn3-Cre;Nkx6.1OE mice, we failed to observe insulin+ cells expressing GFP (Figure S3). These findings pinpoint Nkx6.1 beta cell programming activity to a period between the Ngn3+ state and activation of hormone gene expression.
Nkx6.1 is required for beta cell specification downstream of Ngn3
Since global loss of Nkx6.1 impairs the generation of Ngn3+ endocrine precursors [23], it has remained unclear whether beta cell development requires Nkx6.1 activity downstream of Ngn3. To investigate a potential requirement for Nkx6.1 in this process, we constructed a conditional mutant allele for Nkx6.1 by flanking exon 2 with loxP sites (Figure S4A). Cre recombinase-mediated deletion of exon 2 eliminates a large portion of the DNA-binding homeodomain and additionally introduces a frameshift, resulting in three premature stop codons in exon 3, which cause termination of translation [27]. Importantly, mice heterozygous or homozygous for the Nkx6.1flox (Nkx6.1f) allele show no abnormalities, suggesting that the floxed allele of Nkx6.1 is fully functional. To verify that Cre-mediated recombination of the Nkx6.1f allele generates a null allele, we intercrossed Nkx6.1f/+;Prm1-Cre and Nkx6.1+/− mice to induce recombination of the Nkx6.1f allele in germ cells (Nkx6.1Δf/− allele). As expected, these Nkx6.1Δf/− mice phenocopied Nkx6.1 germline null mutant mice [28], and died immediately after birth, manifesting paralysis of their upper extremities and asphyxia (Figure S4B). Western blot analysis of Nkx6.1 protein expression in pancreata from Nkx6.1Δf/− embryos at embryonic day (e) 14.5 showed a complete absence of Nkx6.1 (Figure S4C). The pancreas of Nkx6.1Δf/− embryos was of normal size, but displayed a drastic reduction in insulin+ cells at e18.5 (Figure S4D–S4F), phenocopying Nkx6.1−/− mice [29].
To determine whether Nkx6.1 is required for beta cell formation from Ngn3+ precursors, we utilized Ngn3-Cre to simultaneously induce recombination of the Nkx6.1f allele and the Z/EG reporter transgene for stable lineage tracing of all progeny derived from Ngn3+ cells. In Nkx6.1f/−;Ngn3-Cre;Z/EG embryos, Cre recombines the loxP sites in both the Nkx6.1f allele and the Z/EG transgene to produce cells that are deficient for Nkx6.1 and express eGFP (Figure 2A, 2B). At e15.5, when Ngn3 expression peaks [11], Nkx6.1 was detected in a large subset of Ngn3+ and GFP+ cells derived from the Ngn3-expressing domain in Ngn3-Cre;Z/EG control embryos (Figure 2C). In Nkx6.1f/−;Ngn3-Cre;Z/EG mice, GFP+ cells were devoid of Nkx6.1 (Figure 2D, Figure S5B), showing the Ngn3-Cre transgene efficiently deletes Nkx6.1 in Ngn3+ cells and their progeny. Ngn3 was similarly expressed in Nkx6.1f/−;Ngn3-Cre;Z/EG and control embryos (Figure 2C, 2D), demonstrating that loss of Nkx6.1 in endocrine precursors does not affect Ngn3 expression.
Fig. 2. Nkx6.1 is required for beta cell specification downstream of Ngn3. 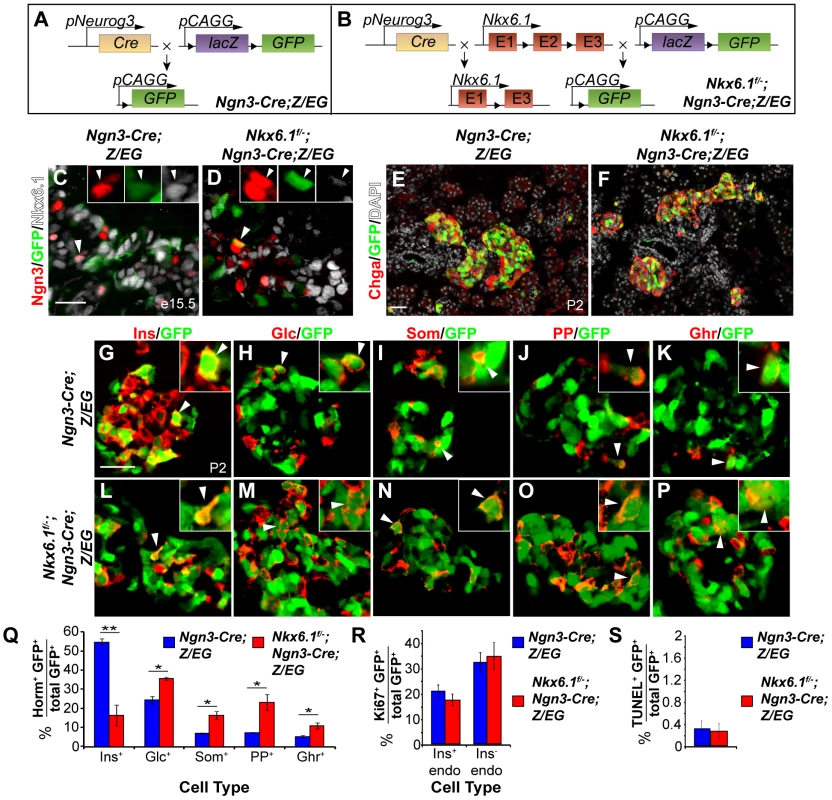
(A, B) Schematic of the alleles and transgenes for Nkx6.1 inactivation and lineage tracing; Triangles, loxP sites. Immunofluorescence staining of pancreata at e15.5 (C, D) or postnatal day (P) 2 (E–P). Recombined, GFP+ cells are restricted to the endocrine compartment (antibody against the pan-endocrine marker Chromogranin A, Chga) in control (E) and Nkx6.1f/−;Ngn3-Cre;Z/EG mice (F). The insets show higher magnifications and arrowheads point to GFP+ cells expressing Ngn3 (C, D) or hormones (G–P). Quantification of hormone+GFP+ (Q), Ki67+GFP+ (R), or TUNEL+GFP+ (S) co-positive cells as a percentage of all GFP-expressing cells in pancreata of Nkx6.1f/−;Ngn3-Cre;Z/EG and Ngn3-Cre;Z/EG mice at P2 (n = 4). Loss of Nkx6.1 in endocrine precursors favors alternative, non-beta endocrine cell fate choices over beta cell fate. Horm, hormones; Ins, insulin; Glc, glucagon; Som, somatostatin; PP, pancreatic polypeptide; Ghr, ghrelin; endo, endocrine. Scale bar = 50 µm. Error bars represent S.E.M; *p<0.05, **p<0.01. Nkx6.1f/−;Ngn3-Cre;Z/EG mice were born at the expected Mendelian frequency, but died within the first few days after birth from dehydration and hyperglycemia; a phenotype indicative of a beta cell defect. To determine whether loss of Nkx6.1 affects the cell fate choice of endocrine precursors, we analyzed the fate of Ngn3+ cells in Ngn3-Cre;Z/EG and Nkx6.1f/−;Ngn3-Cre;Z/EG mice. Based on our previous finding that Nkx6.1 prevents acinar cell fate specification [23], we first examined whether loss of Nkx6.1 in Ngn3+ cells allocates endocrine precursors to the acinar lineage. The ability of Ngn3+ cells to undergo endocrine-to-acinar cell fate conversion has been previously demonstrated in conditions of reduced Ngn3 gene dosage or impaired Notch signaling activity [30], [31]. At P2, we found GFP+ cells to be exclusively restricted to endocrine islets in both Ngn3-Cre;Z/EG and Nkx6.1f/−;Ngn3-Cre;Z/EG mice (Figure 2E, 2F), revealing that Nkx6.1 deletion in Ngn3+ cells does not cause endocrine-to-acinar fate conversion. Thus, unlike multipotent pancreatic progenitors, which adopt an acinar cell identity in the absence of Nkx6.1 activity [23], Ngn3+ endocrine precursors are no longer competent to activate acinar gene expression programs after deletion of Nkx6.1.
Because Nkx6.1-deficient endocrine precursors differentiate into endocrine cells (Figure 2F), we next sought to determine whether loss of Nkx6.1 affects the relative proportion of the different endocrine cell types arising from Ngn3+ cells. To examine the endocrine cell fate choice of Ngn3+ cells, we quantified how many of the recombined Ngn3-expressing cells were allocated to each endocrine cell lineage by co-staining for GFP as a lineage marker of Ngn3-cell progeny together with each of the five hormones individually. At P2, 55.1±1.7% of recombined cells were insulin+ in Ngn3-Cre;Z/EG control mice, while only 16.6±5.4% of recombined cells were insulin+ in Nkx6.1f/−;Ngn3-Cre;Z/EG mice (Figure 2G, 2L, 2Q; P<0.01), suggesting that Nkx6.1-deficient precursors have a lower propensity to differentiate into insulin+ cells. To test whether Nkx6.1-deficient Ngn3+ cells instead adopt non-beta endocrine cell identities, we compared the percentage of Ngn3+ cells that contributed to each non-beta endocrine cell lineage in Ngn3-Cre;Z/EG and Nkx6.1f/−;Ngn3-Cre;Z/EG mice. Nkx6.1f/−;Ngn3-Cre;Z/EG mice at P2 exhibited significantly more glucagon+GFP+ (35.9±0.8% vs. 24.6±2.0%; P<0.05), somatostatin+GFP+ (16.6±2.1% vs. 7.2±0.2%; P<0.05), PP+GFP+ (23.3±4.1% vs. 7.5±0.1%; P<0.025), and ghrelin+GFP+ (11.1±1.5% vs. 5.5±0.7%; P<0.05) cells than Ngn3-Cre;Z/EG control mice (Figure 2H–2K, 2M–2Q). Together, these findings suggest that endocrine precursor cells require Nkx6.1 activity to differentiate into beta cells and that Nkx6.1 prevents precursors from adopting non-beta endocrine fates. To ascertain that the differences in islet cell type composition between Ngn3-Cre;Z/EG and Nkx6.1f/−;Ngn3-Cre;Z/EG mice are indeed the result of preferential precursor cell fate choices and not due to different proliferation or survival rates, we analyzed GFP+ cells in each genotype for their rates of proliferation and apoptosis at P2. Both insulin+GFP+ and insulin−GFP+ endocrine cells displayed similar proliferation and apoptotic rates in Ngn3-Cre;Z/EG and Nkx6.1f/−;Ngn3-Cre;Z/EG mice (Figure 2R, 2S), demonstrating that loss of Nkx6.1 does not affect proliferation or survival. Together, we show that Nkx6.1 controls the fate choice between beta and non-beta endocrine cell lineages in endocrine precursor cells, without favoring any one non-beta endocrine cell type in particular.
Nkx6.1-deficient insulin+ cells are polyhormonal and ectopically express alpha cell markers
To investigate whether Nkx6.1 mediates beta cell specification downstream of Ngn3 by regulating transcription factors necessary for beta cell development, we analyzed expression of the beta cell progenitor markers Pax4, MafB, and Pdx1 in Ngn3-Cre;Z/EG and Nkx6.1f/−;Ngn3-Cre;Z/EG embryos during the peak period of beta cell differentiation at e15.5. Confirming our previous findings in Nkx6.1 null mutant embryos [24], Pax4 expression was not affected in Nkx6.1f/−;Ngn3-Cre;Z/EG embryos (Figure 3A, 3B; 9.6% of GFP+ cells expressed Pax4 in control mice vs. 10.1% in Nkx6.1f/−;Ngn3-Cre;Z/EG mice). In contrast, the marker of newly-born alpha and beta cells, MafB, was absent from the majority of targeted cells in Nkx6.1f/−;Ngn3-Cre;Z/EG embryos (Figure 3C, 3D; 76.2% of GFP+ cells expressed MafB in control mice vs. 20.7% in Nkx6.1f/−;Ngn3-Cre;Z/EG mice), suggesting that MafB, similar to its homolog MafA [20], is controlled by Nkx6.1. Likewise, while a large percentage of Ngn3+ cell progeny were Pdx1+ in control embryos, only a small percentage expressed Pdx1 in Nkx6.1-deficient embryos (Figure 3E, 3F; 81.0% of GFP+ cells expressed Pdx1 in control mice vs. 18.6% of GFP+ cells in Nkx6.1f/−;Ngn3-Cre;Z/EG mice). Thus, Nkx6.1 controls the expression of MafB and Pdx1, but not Pax4 in embryonic beta cell precursors.
Fig. 3. Nkx6.1 controls Pdx1 and MafB expression. 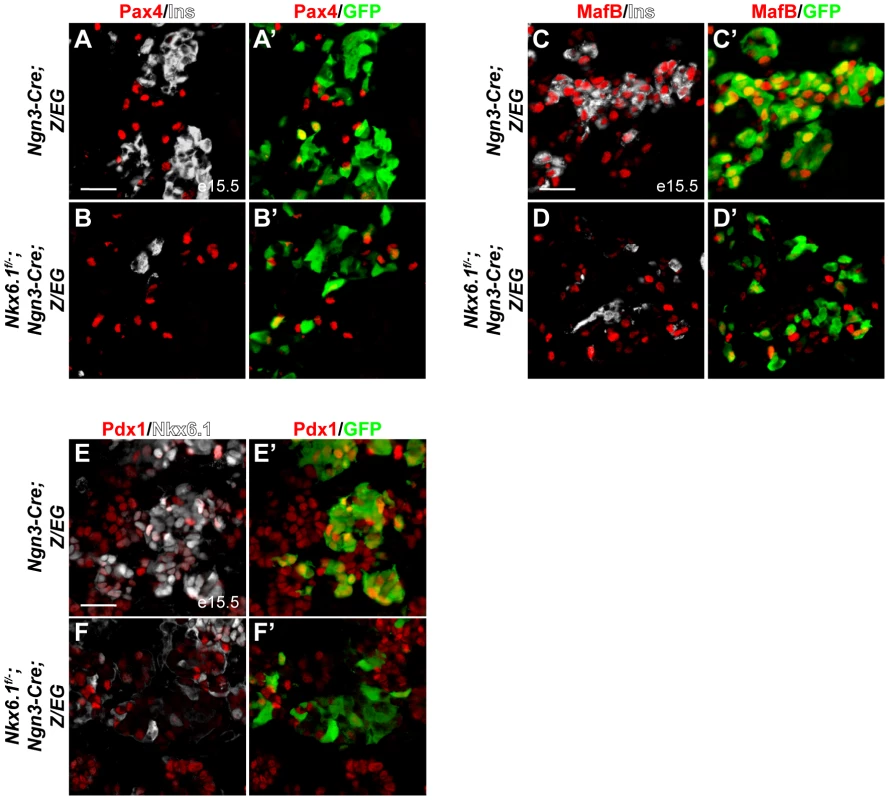
Immunofluorescence staining of pancreata at e15.5 shows no difference in Pax4 expression (A, B) in recombined, GFP+ cells between Nkx6.1f/−;Ngn3-Cre;Z/EG embryos and control Ngn3-Cre;Z/EG embryos, while MafB (C, D) and Pdx1 (E, F) expression is reduced in Nkx6.1-deficient, GFP+ cells compared to control embryos. Ins, insulin. Scale bar = 50 µm. Although reduced in numbers, we still observed targeted GFP+ cells expressing insulin in neonatal Nkx6.1f/−;Ngn3-Cre;Z/EG mice, raising the question of whether these Nkx6.1-deficient insulin+ cells properly differentiate into beta cells. To investigate how lack of Nkx6.1 affects beta cell gene expression programs, we analyzed insulin+ cells in neonatal Nkx6.1f/−;Ngn3-Cre;Z/EG mice for the expression of Pax6, Pdx1, and MafA. Expression of the islet cell marker Pax6 was not affected in Nkx6.1f/−;Ngn3-Cre;Z/EG mice (Figure S5C, S5D; 68.4% of insulin+GFP+ cells expressed Pax6 in control mice vs. 72.7% in Nkx6.1f/−;Ngn3-Cre;Z/EG mice). By contrast, confirming our findings at embryonic stages (Figure 3E, 3F), Pdx1 expression was markedly reduced in Nkx6.1-deficient insulin+ cells at P2 (Figure 4A, 4B; 76.5% of insulin+GFP+ cells expressed Pdx1 in control mice vs. 30.2% in Nkx6.1f/−;Ngn3-Cre;Z/EG mice). Similarly, the mature beta cell marker MafA was absent from insulin+ cells in Nkx6.1f/−;Ngn3-Cre;Z/EG mice (Figure 4C, 4D; 79.3% of insulin+GFP+ cells expressed MafA in control mice vs. 0% in Nkx6.1f/−;Ngn3-Cre;Z/EG mice). This demonstrates that beta cells require Nkx6.1 activity during their differentiation to initiate MafA expression and to maintain high levels of Pdx1. These findings are consistent with the phenotype of Nkx6.1 null mutant mice, in which limited numbers of insulin+ cells lacking MafA are observed [20], [24]. We conclude that insulin expression can still be initiated in the absence of Nkx6.1, but that these insulin+ cells lack key features of normal beta cells.
Fig. 4. Loss of Nkx6.1 in endocrine precursors results in activation of non-beta endocrine genes. 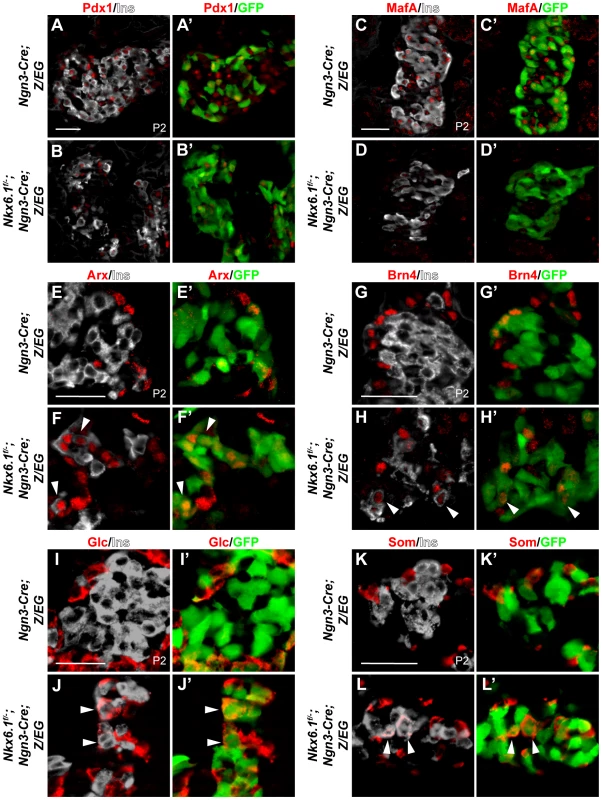
(A–L) Immunofluorescence staining of pancreata from Ngn3-Cre;Z/EG and Nkx6.1f/−;Ngn3-Cre;Z/EG mice at postnatal day (P) 2 shows reduced Pdx1 (A, B), absence of MafA (C, D), and ectopic expression of Arx (E, F), Brn4 (G, H), glucagon (Glc; I, J), and somatostatin (Som; K, L) in Nkx6.1-deficient, recombined, insulin+GFP+ cells. Arrowheads point to insulin+ cells ectopically expressing non-beta endocrine markers. Ins, insulin. Scale bar = 50 µm. The lack of beta cell-specific markers in Nkx6.1-deficient insulin+ cells raised the question of whether these insulin+ cells also carry features of alternative endocrine lineages. To determine whether loss of Nkx6.1 in endocrine precursors results in the activation of mixed endocrine gene expression programs, we analyzed insulin+ cells in Nkx6.1f/−;Ngn3-Cre;Z/EG mice for coexpression of the alpha cell lineage determinants Arx and Brn4. As expected, in Ngn3-Cre;Z/EG control mice virtually no colocalization of Arx and Brn4 with insulin was observed at P2 (Figure 4E, 4G; 0.2% of insulin+GFP+ cells expressed Arx and 0% Brn4 in control mice). In contrast, a subset of recombined insulin+GFP+ cells in Nkx6.1f/−;Ngn3-Cre;Z/EG mice also expressed Arx and Brn4 (Figure 4F, 4H; arrowheads; 26.1% of insulin+GFP+ cells expressed Arx and 26.5% Brn4 in Nkx6.1f/−;Ngn3-Cre;Z/EG mice), showing aberrant activation of alpha cell differentiation genes. Notably, Nkx6.1 deletion or misexpression did not affect Arx expression in the immediate progeny of endocrine precursors at e15.5 (Figure S6A–S6D), pinpointing Nkx6.1-mediated regulation of Arx to a time window between e15.5 and birth.
At P2, loss of Nkx6.1 activity was also associated with aberrant expression of glucagon in insulin+ cells (Figure 4I, 4J; arrowheads in J; 0.5% of insulin+GFP+ cells expressed glucagon in control mice vs. 29.8% in Nkx6.1f/−;Ngn3-Cre;Z/EG mice). Moreover, we found that many of the targeted insulin+ cells ectopically expressed somatostatin in Nkx6.1f/−;Ngn3-Cre;Z/EG mice (Figure 4K, 4L; arrowheads in L; 0% of insulin+GFP+ cells expressed somatostatin in control mice vs. 19.6% in Nkx6.1f/−;Ngn3-Cre;Z/EG mice). These findings demonstrate that Nkx6.1 is critical for repressing alternative endocrine lineage programs and that beta cell-specific programs can only be induced to a limited extent when Nkx6.1 is lost.
Previous studies have shown that the number of glucagon+ cells is increased in Pdx1 heterozygous mutant mice [32], [33]. Furthermore, beta cells lose Nkx6.1 expression upon Pdx1 deletion in beta cells [34]. Combined with our observation that Nkx6.1 maintains Pdx1 expression during beta cell differentiation, these findings raise the possibility that Pdx1 and Nkx6.1 cooperate through a positive feedback loop to establish and maintain beta cell identity and to repress non-beta endocrine lineage programs. To test this idea, we analyzed wild type, Nkx6.1f/+;Ngn3-Cre, Pdx1+/−, and compound heterozygous Nkx6.1f/+;Ngn3-Cre;Pdx1+/− mice for the ectopic expression of non-beta endocrine hormones in insulin+ cells at P2. While we saw no coexpression of somatostatin or PP with insulin in any of the four genotypes (Figure 5A–5D; data not shown), glucagon and insulin co-positive cells were occasionally detected in all genotypes, including wild type mice (Figure 5A–5D, 5I). Quantification of the percentage of insulin+ cells also expressing glucagon revealed significantly more dual hormone-positive cells in compound heterozygous Ngn3-Cre;Nkx6.1f/+;Pdx1+/− mice than in either single heterozygous mutant or in wild type mice (Figure 5A–5D, 5I). As previously reported [32], [33], Pdx1+/− mice displayed an increase in the glucagon to insulin cell ratio that was also seen in compound heterozygous Ngn3-Cre;Nkx6.1f/+;Pdx1+/− but not in Nkx6.1f/+;Ngn3-Cre mice (Figure 5J). Thus, haploinsufficiency for Pdx1 but not Nkx6.1 increases alpha cell numbers, which may reflect a non-cell autonomous effect on alpha cell proliferation, as recently shown in a mouse model of conditional deletion of Pdx1 in beta cells [35]. To determine whether the mixed lineage identity of insulin+glucagon+ cells is associated with the expression of Arx, we co-stained pancreatic sections of mice at P2 from all genotypes for insulin, glucagon, and Arx. The majority of insulin+glucagon+ cells expressed Arx in all genotypes, although occasional insulin+glucagon+Arx− cells were also detected (Figure 5E–5H; arrowheads). The observed increase in cells exhibiting mixed alpha/beta cell identity and Arx expression in Nkx6.1/Pdx1 compound heterozygous mice supports the notion that Pdx1 and Nkx6.1 cooperate in beta cell fate specification by preventing activation of alpha cell-specific gene expression programs during endocrine cell differentiation.
Fig. 5. Nkx6.1 and Pdx1 collectively stabilize beta cell identity. 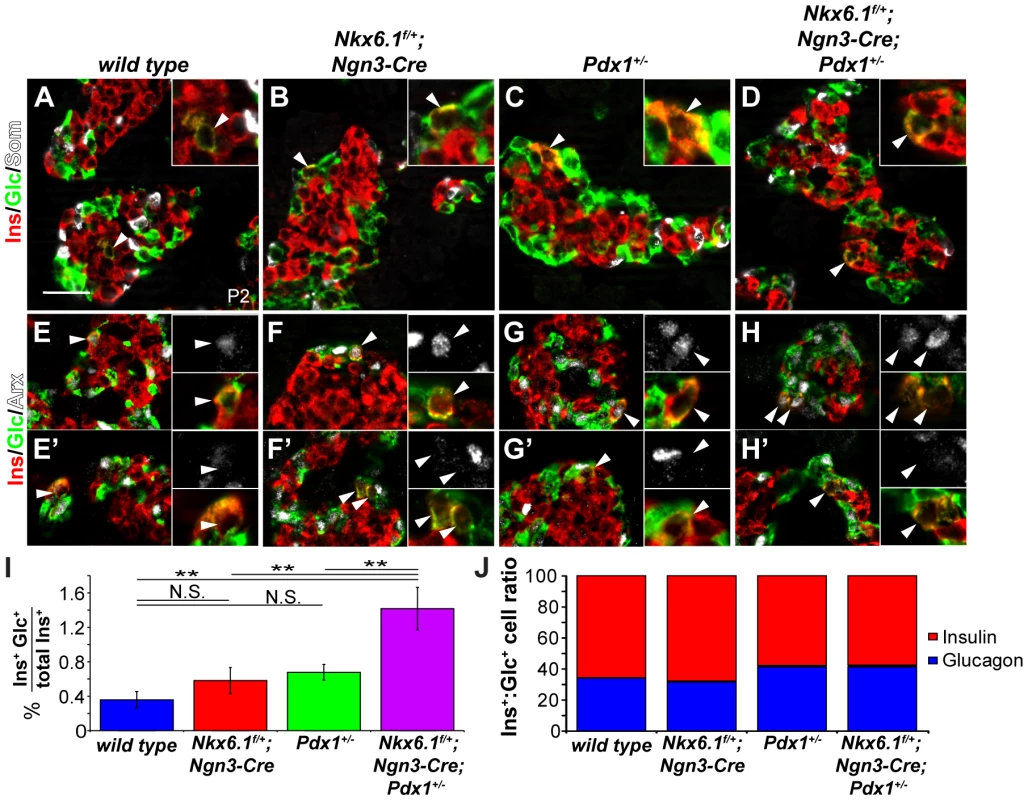
(A–H) Immunofluorescence staining of pancreata from wild type, Nkx6.1f/+;Ngn3-Cre, Pdx1+/−, and Nkx6.1f/+;Ngn3-Cre;Pdx1+/− mice at postnatal day (P) 2 reveals occasional coexpression of insulin with glucagon but not with somatostatin in all genotypes (A–D; arrowheads and insets). Both Arx+ (E–H) and Arx− (E′–H′) insulin+glucagon+ cells (arrowheads and insets) are found in all genotypes. (I) Quantification of the percentage of insulin+ cells co-expressing glucagon at P2 reveals significantly more insulin+glucagon+ cells in Nkx6.1f/+;Ngn3-Cre;Pdx1+/−, Nkx6.1f/+;Ngn3-Cre, and Pdx1+/− mice compared to wild type controls. In addition, Nkx6.1f/+;Ngn3-Cre;Pdx1+/− mice show more insulin+glucagon+ cells than either single heterozygous mutant (n = 3). (J) Quantification of insulin+ and glucagon+ cell numbers in P2 pancreata shows an increase in glucagon+ cells in Nkx6.1f/+;Ngn3-Cre;Pdx1+/− and Pdx1+/− mice compared to Nkx6.1f/+;Ngn3-Cre and wild type mice demonstrating that loss of a single Nkx6.1 allele does not significantly affect alpha cell numbers (n = 3). Ins, insulin; Glc, glucagon; Som, somatostatin. Scale bar = 50 µm. Error bars represent S.E.M; **p<0.01, N.S. = not significant. Nkx6.1 is a direct repressor of Arx and competes with the Arx activator Isl1 at an Arx intronic enhancer
To further explore how Nkx6.1 prevents endocrine precursors from adopting alpha cell identity, we examined the relationship of the Nkx6.1 and Arx expression domains in progeny of Ngn3-expressing cells during development. Consistent with the dependence of Arx expression on Ngn3 [12], Arx was confined to a domain that marks descendants of Ngn3-expressing cells (Figure 6A′″, 6B′″; note, occasional Arx+GFP− cells in A and B can be explained by mosaic expression of the Z/EG and/or Ngn3-Cre transgenes). At e14.5, preceding the onset of the major wave of beta cell differentiation, the majority of Arx+ cells also expressed Nkx6.1 and Arx+Nkx6.1− cells were rare (Figure 6A; arrowheads). By e16.5, however, when large numbers of beta cells arise [36], GFP+ cells seldom coexpressed Nkx6.1 and Arx (Figure 6B; arrowheads). Thus, endocrine precursors initially activate Nkx6.1 and Arx concomitantly, but their expression domains become mutually exclusive during beta cell differentiation. Together with our finding that Nkx6.1 regulates Arx during this time window, the observed expression pattern of Arx and Nkx6.1 raised the possibility that Nkx6.1 functions as a transcriptional repressor of Arx.
Fig. 6. Nkx6.1 and Isl1 function as antagonistic transcriptional regulators of the Arx Re1 enhancer. 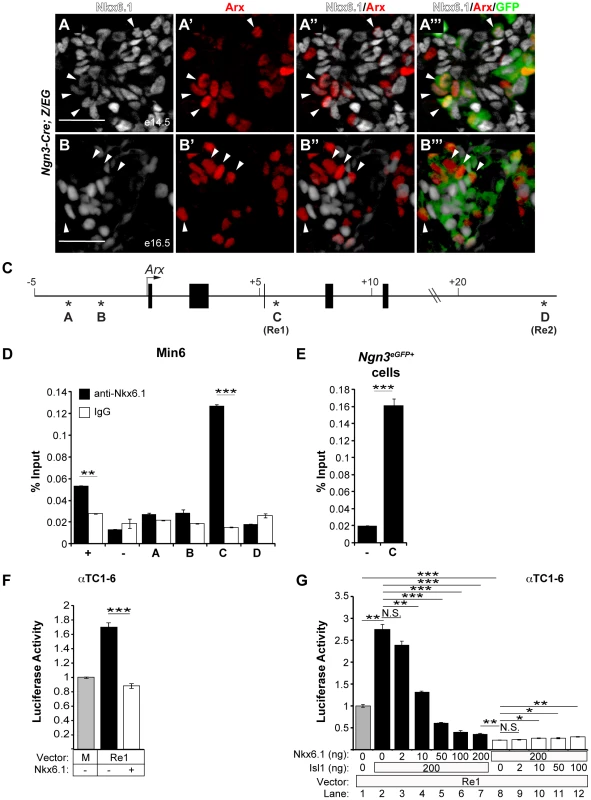
Immunofluorescence staining of pancreata from Ngn3-Cre;Z/EG mice at e14.5 (A) and e16.5 (B) for Nkx6.1, Arx, and GFP shows that the majority of progeny of Ngn3-expressing cells (GFP+) co-express Arx and Nkx6.1 at e14.5 (arrowheads in A), while the Arx+ and Nkx6.1+ domains are distinct at e16.5 (arrowheads in B point to GFP+Arx+Nkx6.1− cells). (C) Schematic of the Arx locus; asterisks indicate phylogenetically-conserved Nkx6.1 binding motifs and numbers indicate the distance from the transcriptional start site. Nkx6.1 binds to site C (Re1 element) in the Arx locus in chromatin from Min6 cells (D) and FACS-sorted GFP+ cells (E) from e15.5 pancreata of Neurog3eGFP embryos analyzed by ChIP with antibodies against Nkx6.1 or control immunoglobulin G (IgG). Mouse glucagon promoter and intergenic primers were used as positive (+) and negative (−) controls, respectively. (F) Co-transfection of αTC1–6 cells with the Arx Re1 enhancer-luciferase construct, the CMV-Renilla expression construct, and with or without the CMV-Nkx6.1 expression construct. Lane one (M) represents basal luciferase expression of the minimal promoter. Luciferase activity was quantified relative to the expression of the minimal promoter. Activity of the Re1 enhancer is repressed by Nkx6.1. (G) Co-transfection of αTC1–6 cells with the Arx Re1 enhancer-luciferase construct, CMV-Renilla, and with different concentration of CMV-Nkx6.1 and CMV-Isl1, as indicated. Nkx6.1 prevents activation of the Arx Re1 enhancer by Isl1 in a dose-dependent manner (lanes 2–7). Luciferase activity was quantified relative to the expression of the Re1 enhancer. Increasing concentrations of Isl1 are not sufficient to restore baseline activity of the Re1 enhancer in the presence of 200 ng of CMV-Nkx6.1 (lanes 8–12). Scale bar = 50 µm. Error bars represent S.E.M; *p<0.05, **p<0.01, ***p<0.001. To explore this hypothesis, we tested whether Nkx6.1 occupies Arx regulatory sequences and is capable of repressing Arx transcription. We identified two conserved Nkx6.1 binding motifs within 5 kb of the 5′ end flanking region from the Arx transcriptional start site (Figure 6C; site A and B) as well as 12 potential Nkx6.1 binding sites located in two previously characterized enhancers within the third intron of the Arx genomic sequence (Figure 6C; site C, Re1) and the 3′ flanking region (Figure 6C; site D, Re2) [37]. Recently, the Arx Re1 and Re2 enhancers have been shown to be required for Isl1-mediated activation of Arx in alpha cells, suggesting that these two enhancers are critical for Arx transcription [37]. Chromatin immunoprecipitation (ChIP) analyses for Nkx6.1 revealed that Nkx6.1 directly and specifically associates with the Arx Re1 enhancer (site C) in the Min6 beta cell line (Figure 6D) and in embryonic endocrine precursors isolated by fluorescence-activated cell sorting (FACS) from Ngn3eGFP/+ embryonic pancreata at e15.5 (Figure 6E). Discordant with a previous report, which reported binding of Nkx6.1 to site B in a beta cell line [17], no association was observed with the other sites (sites A, B, or D) containing Nkx6.1 motifs (Figure 6D). Transfection of the αTC1–6 alpha cell line with an expression plasmid for Nkx6.1 and a luciferase reporter construct containing the Arx Re1 enhancer sequence revealed that Nkx6.1 significantly reduced reporter gene activity (Figure 6F). Confirming previous findings [37], transfection of an expression plasmid for Isl1 activated the Re1 enhancer (Figure 6G). Co-transfection of CMV-Nkx6.1 abolished the ability of Isl1 to activate the Re1 enhancer in a dosage-dependent manner, showing that Nkx6.1 and Isl1 regulate Arx antagonistically through competition at the Re1 enhancer. However, in the presence of Nkx6.1, addition of CMV-Isl1 was not sufficient to revert Nkx6.1-mediated repression (Figure 6G), indicating dominance of Nkx6.1 repressive over Isl1 activator activity. While our experiments show that Nkx6.1 is able to repress the Arx Re1 enhancer in alpha cell lines, Nkx6.1 cannot evoke an alpha-to-beta cell fate change when misexpressed in differentiated alpha cells (Figure S3). Thus, Nkx6.1-dependent Arx repression through the Re1 enhancer appears to be functionally most relevant during endocrine cell type specification, when Nkx6.1 prevents initiation of Arx expression (Figure 4E, 4F).
Nkx6.1 is necessary for maintaining beta cell identity
It has recently been shown that Nkx2.2 is an obligatory repressor of Arx in differentiated beta cells and that the absence of Nkx2.2 repressor activity causes beta-to-alpha cell conversion in mice [17]. Since Nkx2.2 is expressed in both beta and alpha cells, it has been speculated that beta cell-specific repression of Arx might depend on Nkx6.1 [17]. To directly test this hypothesis, we deleted Nkx6.1 selectively in beta cells, using the rat insulin promoter II (RIP)-Cre transgene to recombine the Nkx6.1f allele and the Rosa26 (R26)-YFP reporter allele. As expected, the YFP lineage label was largely confined to beta cells in control Nkx6.1f/+;RIP-Cre;R26-YFP mice at 6 weeks of age (Figure 7A–7E, 7O). In striking contrast, only a few Nkx6.1-deficient, YFP-labeled cells expressed insulin in Nkx6.1f/−;RIP-Cre;R26-YFP mice (Figure 7F, 7G, 7O), suggesting that cells that once activated the insulin promoter no longer expressed insulin. Analysis of YFP expression in conjunction with glucagon, somatostatin, and PP revealed that Nkx6.1-deficient beta cells adopted delta cell identity, but did not convert into alpha or PP cells (Figure 7H–7J, 7O). Thus, loss of Nkx6.1 in differentiated beta cells no longer causes activation of glucagon and PP, as observed after Nkx6.1 inactivation in Ngn3+ endocrine precursors. These findings suggest that Nkx6.1 is necessary to repress delta cell-specific genes in beta cells, but that expression of alpha and PP cell-specific genes are inhibited through an Nkx6.1-independent mechanism once beta cells have formed.
Fig. 7. Loss of Nkx6.1 in beta cells causes beta-to-delta cell conversion. 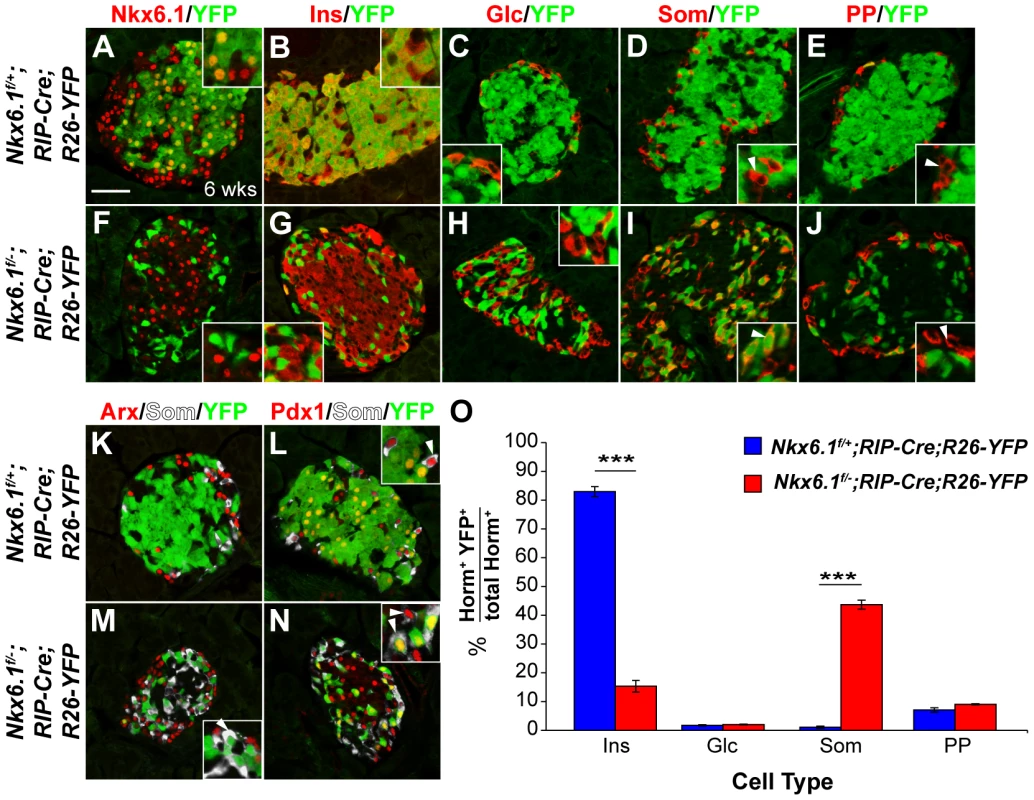
Immunofluorescence staining of pancreata from Nkx6.1f/+;RIP-Cre;R26-YFP and Nkx6.1f/−;RIP-Cre;R26-YFP mice at 6 weeks of age shows Nkx6.1 (A) and insulin (B) expression in YFP+ cells of Nkx6.1f/+;RIP-Cre;R26-YFP control mice, but loss of Nkx6.1 (F) and insulin (G) in YFP+ cells of Nkx6.1f/−;RIP-Cre;R26-YFP mice. The insets display higher magnification images. YFP+ cells do not express glucagon (C, H) and rarely express pancreatic polypeptide (E, J; insets, arrowheads) in either genotype. While YFP+ cells are somatostatin− in Nkx6.1f/+;RIP-Cre;R26-YFP mice (D; insets, arrowheads), YFP-labeled cells are mostly somatostatin+ in Nkx6.1f/−;RIP-Cre;R26-YFP mice (I; insets, arrowheads), suggesting beta-to-delta cell conversion. Arx expression is similar in both genotypes and absent from lineage-labeled YFP+ cells (K, M; inset, arrowhead), showing that loss of Nkx6.1 in beta cells does not activate Arx. Pdx1+somatostatin+ cells are found in both genotypes (L, N; insets, arrowheads), but express YFP only in Nkx6.1f/−;RIP-Cre;R26-YFP mice (L; inset, arrowhead). (O) Quantification of the percentage of hormone+YFP+ cells relative to all hormone+ cells for each islet cell type shows reduced numbers of insulin+YFP+ cells and increased numbers of in somatostatin+YFP+ cells in Nkx6.1f/−;RIP-Cre;R26-YFP mice compared to Nkx6.1f/+;RIP-Cre;R26-YFP mice at 6 weeks (n = 3). Wks, weeks; Ins, insulin; Glc, glucagon; PP, pancreatic polypeptide; Som, somatostatin; Horm, hormones. Scale bar = 50 µm. Error bars represent S.E.M; ***p<0.0001. To further test whether Nkx6.1 deficiency in beta cells could lead to partial activation of an alpha cell gene expression program, we examined YFP+ cells for Arx expression. YFP+ cells rarely expressed Arx in both control and Nkx6.1f/−;RIP-Cre;R26-YFP mice (Figure 7K, 7M), demonstrating that Nkx6.1 is no longer necessary for Arx repression after beta cells have differentiated. Likewise, Pdx1 expression in adult islet cells was Nkx6.1-independent, as somatostatin+ cells that arose from Nkx6.1-deficient insulin-expressing cells were Pdx1+ (Figure 7L, 7N). Our data reveal that gene regulation by Nkx6.1 is highly context-dependent. While Nkx6.1 is necessary for Arx repression and Pdx1 activation in beta cell precursors, both genes are regulated by Nkx6.1-independent mechanisms in mature beta cells.
Discussion
The specification of pancreatic endocrine cell types is governed by the transcription factors Pdx1, Pax4, and Arx [12], [13], [16]. However, as none of these transcription factors control the development of solely one endocrine cell type, the molecular mechanisms that confer lineage specificity have remained largely elusive. Here, we demonstrate that the beta cell-specific transcription factor Nkx6.1 is both necessary and sufficient to specify the beta cell lineage (Figure 8A). We show that Nkx6.1 and Isl1 antagonistically regulate Arx expression in endocrine precursors through a direct transcriptional mechanism (Figure 8B). Thus, our study identifies Nkx6.1 as a critical beta cell programming factor that promotes the beta cell fate choice by simultaneously inducing beta cell genes and repressing non-beta endocrine genes.
Fig. 8. Model of Nkx6.1 function in endocrine precursor cells. 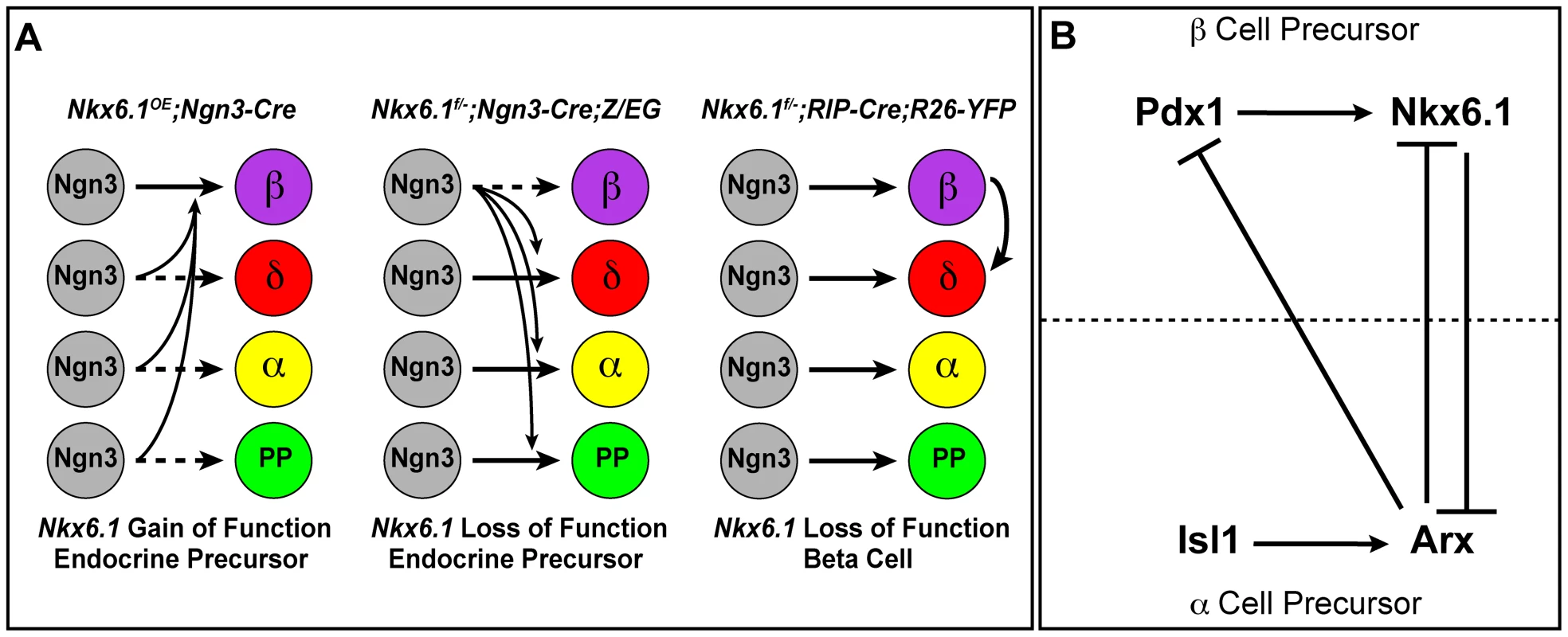
(A) Expression of Nkx6.1 results in allocation of precursors from all non-beta endocrine lineages to the beta cell lineage. Deletion of Nkx6.1 in endocrine precursors has the opposite effect. When Nkx6.1 is deleted in beta cells, beta cells convert into delta cells, but not into alpha or pancreatic polypeptide (PP)-producing cells. (B) Our study suggests that in endocrine precursors, Nkx6.1 and Isl1 compete for repression and activation, respectively, of the alpha cell fate determinant Arx. We also demonstrate that the expression of Pdx1 in endocrine precursors depends on Nkx6.1. In conjunction with previous studies, showing repression of Pdx1 and Nkx6.1 by Arx [39] and activation of Nkx6.1 by Pdx1 [34], our data support a model whereby cross-repression between Arx and Nkx6.1 confers alpha versus beta cell precursor identity. In beta cell precursors, Nkx6.1 expression is reinforced by Pdx1, which is repressed by Arx in alpha cell precursors. Nkx6.1: a master regulator of the beta cell fate choice
We found that forced expression of Nkx6.1 allocates endocrine precursors to the beta cell lineage. While a similar activity has been described for Pdx1 [16], Nkx6.1 and Pdx1 display different kinetics of beta cell programming. As evidenced by cells coexpressing insulin, glucagon, and Arx at birth [16], forced expression of Pdx1 in endocrine precursor cells initially produces cells with mixed alpha and beta cell identity. In contrast to Ngn3-Cre;Pdx1OE mice, we rarely observed cells exhibiting both alpha and beta cell features in neonatal Ngn3-Cre;Nkx6.1OE mice, suggesting that Nkx6.1 plays a critical role early during cell fate specification of endocrine precursors. This notion is consistent with our finding that Nkx6.1 acts as a direct transcriptional repressor of the Arx gene. While Pax4 has also been shown to act as a direct repressor of Arx [38], our observation that Pax4 expression is not affected by Nkx6.1 ablation suggests that Pax4 alone is not sufficient to repress Arx in beta cell precursors. Given the dependence of Nkx6.1 expression on Pax4 activity [15], it is possible that the observed derepression of Arx and glucagon in Pax4 mutant mice [12], [38] is a consequence of Nkx6.1-deficiency.
In addition to repressing alpha cell-specific genes through the regulation of Arx, Nkx6.1 also reallocated delta, PP, and epsilon cell precursors to the beta cell lineage. Likewise, inactivation of Nkx6.1 in endocrine precursors resulted in increased production of cells of all non-beta endocrine cell types and ectopic expression of non-beta cell hormones in insulin+ cells. The phenotype observed upon conditional activation or deletion of Nkx6.1 in endocrine precursors identifies Nkx6.1 as a potent general repressor of non-beta endocrine gene expression programs. While our study establishes Nkx6.1 as a direct repressor of the alpha cell fate determinant Arx, Nkx6.1 likely represses additional cell fate determinants critical for the specification of delta, PP, and epsilon cells. As little is known about the transcription factors mediating non-beta endocrine cell fate choices, identification of additional direct target genes for Nkx6.1 in endocrine cell fate specification will have to await future studies.
Similar to Nkx6.1, forced expression of Pax4 also conferred beta cell identity to precursors of all endocrine cell lineages [14]. However, in contrast to Pax4 misexpressing mice [14], we did not observe oversized islets or diabetes in adult mice misexpressing Nkx6.1. Our observations in Ngn3-Cre;Nkx6.1OE mice are consistent with our previous study, showing that transgenic overexpression of Nkx6.1 in beta cells does not stimulate beta cell proliferation or perturb beta cell function [26]. Thus, despite their shared property as a direct repressor of Arx [38], Nkx6.1 and Pax4 must also have distinct targets in endocrine cells. The lack of adverse effects on beta cell function makes Nkx6.1 an excellent candidate for beta cell programming strategies.
The transcriptional network that specifies beta cells
Unlike Pax4 expression, which we found to be independent of Nkx6.1, maintenance of Pdx1 expression downstream of Ngn3 required Nkx6.1 activity. Previous genetic studies in mice have shown that the expression of Nkx6.1 in beta cells also depends on Pdx1 activity [34], suggesting that these two “pro-beta” transcription factors reinforce each other's expression. Since both Pdx1 and Nkx6.1 control the expression of critical genes for beta cell function, such as MafA and Glut2 ([20], [25], [34]; this study), the positive regulatory loop between Pdx1 and Nkx6.1 might be critical for initiating and stabilizing beta cell-specific gene expression programs during endocrine cell differentiation. The mutual reinforcement in gene expression between these two transcription factors also explains why the combined deletion of one copy of Pdx1 and Nkx6.1 was sufficient to destabilize the beta cell fate choice and to cause ectopic expression of glucagon in insulin+ cells. Nkx6.1 could regulate Pdx1 either directly or indirectly by repressing an inhibitor of Pdx1 expression. Since Nkx6.1 directly represses Arx and Arx has previously been shown to repress both Pdx1 and Nkx6.1 [39], loss of Pdx1 in Nkx6.1-deficient precursors is most likely the consequence of Arx derepression (Figure 8B).
Competence window of beta cell programming
We found that expression of Nkx6.1 did not force all Ngn3+ cells to differentiate into beta cells and that small numbers of alpha, delta, PP, and epsilon cells expressing Nkx6.1 persisted during adulthood. This implies that the competence of precursors to adopt a beta cell fate upon Nkx6.1 activation is limited to a short period during development when cells are still plastic and lineage-specific gene expression programs have not been fully established. Our observation that misexpression of Nkx6.1 in alpha cells failed to induce conversion of alpha into beta cells supports the notion that cells quickly lose the competence to respond to Nkx6.1 repressive cues as they undergo endocrine differentiation.
Our study shows that Nkx6.1 occupies the Arx enhancer in beta cells (Figure 6D), but unlike its inactivation in endocrine precursors, Nkx6.1 inactivation in beta cells does not activate Arx. This finding suggests that Nkx6.1 is only capable of repressing Arx during endocrine cell differentiation. One potential mechanism that could account for Nkx6.1-independent Arx repression in beta cells is DNA methylation, which has recently been shown to occur at the Arx locus [40]. Interestingly, deletion of the DNA methyltransferase Dnmt3a or Dnmt1 in beta cells results in Arx derepression and spontaneous conversion of beta into alpha cells [17], [40], suggesting that DNA methylation alone is sufficient to keep Arx repressed in beta cells. Dnmt3a is recruited to Arx by Nkx2.2 and loss of Nkx2.2 repressor activity causes spontaneous conversion of beta into alpha cells [17], demonstrating that Arx expression in beta cells can be readily induced when Nkx2.2 repressor function is removed. Whereas Nkx2.2 forms a complex with Dnmt3a in the 5′ regulatory region of Arx [17], we show that Nkx6.1 occupies an intronic Arx enhancer, where it competes with Isl1 and prevents Isl1 from activating Arx. Thus, it appears that DNA methylation-dependent repression of Arx is particularly important for keeping Arx repressed in beta cells, whereas Nkx6.1 prevents Arx activation in differentiating beta cell precursors. Overall, the role of DNA methylation in restraining cell plasticity during development is still poorly understood. Knowledge of how islet cell type-specific genes are epigenetically modified as cells differentiate and how this process can be reversed will prove important for devising effective cell programming and reprogramming strategies.
Our study highlights the importance of Nkx6.1 as a beta cell programming factor during endocrine cell differentiation and shows that insulin expression can be initiated independent of Nkx6.1 and Pdx1. Strikingly, the insulin+Nkx6.1−Pdx1− cells observed after conditional inactivation of Nkx6.1 in Ngn3+ cells display a similar molecular profile as insulin+ cells generated in vitro with current hESC differentiation protocols. Like Nkx6.1-deficient insulin+ cells in Nkx6.1f/−;Ngn3-Cre mice, hESC-derived insulin+ cells are polyhormonal and fail to express Nkx6.1, Pdx1, MafA, and critical glucose transporters ([18], [19]; Sander laboratory, unpublished data), which suggests that Nkx6.1 is required for complete beta cell programming in mice and humans. Our studies now pave the way for exploring the effectiveness of Nkx6.1 in (re)-programming strategies to generate functional beta cells for diabetes therapy.
Materials and Methods
Nkx6.1 gene targeting, mice, and glucose tolerance test
CAG-Bgeo,-Nkx6.1,-eGFP (Nkx6.1OE) [23], Nkx6.1+/− [29], Pdx1+/− [41], Neurog3eGFP [42], Prm1-Cre [43], Ngn3-Cre [44], RIP-Cre [45], Rosa26-YFP [46], CAG-Bgeo,-eGFP (Z/EG) [47], and Glc-Cre mice [48] have been previously described. To create the Nkx6.1flox (Nkx6.1f) allele, a targeting vector consisting of two loxP sites inserted into the first and second introns of Nkx6.1 was generated (Figure S4A). The herpes simplex virus-thymidine kinase gene was placed outside of the Nkx6.1 gene homology region for negative selection. After electroporation of 129S6-derived mouse embryonic stem cells, 375 clones survived neomycinR selection. Southern blotting identified 14 clones as correctly targeted. Two clones carrying the Nkx6.1flox-PGK-Neo allele were independently injected in mouse blastocysts, and chimeric mice bred with C57BL/6J mice for germline transmission screening. The FRT-flanked neomycinR gene in intron 2 was subsequently removed by crossing Nkx6.1flox-PGK-Neo mice with ActB-FlpE mice (JAX; more information at http://www.mmrrc.org/strains/29994/029994.html).
Midday on the day of vaginal plug appearance was considered e0.5. Glucose tolerance tests were performed as previously described [26].
Immunohistochemistry, morphometry, and cell quantification
Tissue preparation, immunofluorescence and TUNEL staining were performed as previously described [49]. For detection of nuclear antigens, antigen retrieval was performed in pH 6.0 citrate buffer and sections were permeabilized in 0.15% Triton X-100 in PBS. The following primary antibodies were used at the given dilutions: mouse anti-insulin (Sigma), 1∶5000; guinea pig anti-insulin (Dako), 1∶2000; guinea pig anti-glucagon (Sigma), 1∶2000; rabbit anti-PP (Dako), 1∶2000; goat anti-ghrelin (Santa Cruz), 1∶1000; rabbit anti-somatostatin (Dako), 1∶3000; goat anti-chromogranin A (Santa Cruz), 1∶500; rat anti-GFP (C. Kioussi), 1∶1000; mouse anti-Nkx6.1 (BCBC clone #2023; against C-terminal part of Nkx6.1), 1∶500; guinea pig anti-Ngn3 [24], 1∶2000; rabbit anti-Pax6 (Chemicon), 1∶1000; rabbit anti-Brn4 (M. Rosenfeld), 1∶500; rabbit anti-Ki67 (Lab Vision), 1∶500; rabbit anti-MafB (Bethyl Labs), 1∶1000; rabbit anti-MafA (Bethyl Labs), 1∶1000; rabbit anti-Pax4 (B. Sosa-Pineda), 1∶100; rabbit anti-Arx (P. Collombat), 1∶500; rabbit anti-Arx (K. Morohashi), 1∶250; guinea-pig anti-Pdx1 (C. Wright), 1∶10,000. Staining with antibodies raised in mice was conducted using the M.O.M. Kit (Vector Labs) in conjunction with streptavidin-conjugated secondary antibodies (Jackson ImmunoResearch). When necessary, nuclei were counterstained with Hoechst 33342 (Invitrogen) at 10 µg/ml. Primary antibodies were detected with donkey-raised secondary antibodies conjugated to Cy3, Cy5, DyLight488 (Jackson ImmunoResearch) or Alexa488 (Molecular Probes) at 1∶1500 dilution (1∶500 for Cy5). ApoTome images were captured on a Zeiss Axio Observer Z1 microscope with Zeiss AxioVision 4.8 and figures prepared with Adobe Photoshop/Illustrator CS4. Where necessary, the Cy5 channel was pseudo-colored white. Images were processed in accordance with the Journal of Cell Biology figure manipulation guidelines.
For all morphometric analyses and cell quantifications, a total of 10 sections per mouse from at least three mice per genotype were analyzed. For TUNEL, proliferation, and cell lineage analyses, the number of GFP+Hoechst+marker+ cells was manually counted, divided by the total number of GFP+Hoechst+ (Ngn3-Cre-mediated lineage tracing) or marker+Hoechst+ (RIP-Cre-mediated lineage tracing) cells, and multiplied by 100. For the analysis of Pdx1 and Nkx6.1 single and compound heterozygous mice, sections were stained for insulin, glucagon, and somatatostatin and all marker+ cells (on average 2100 cells per pancreas) were manually counted. Insulin+ cells were analyzed for the expression of glucagon or somatostatin. For quantification of GFP+marker+ cells, on average 500 GFP+ cells from at least five different sections per mouse were counted.
For islet cell mass measurements, images covering an entire pancreas section were tiled using a Zeiss Axio Oberver Z1 microscope with the Zeiss ApoTome module. The hormone+ area and total pancreas area were measured using ImagePro Plus 5.0.1 software (Media Cybernetics) and islet cell mass was calculated as follows: (hormone+ area/total pancreatic area) multiplied by pancreatic weight.
Western blot, ChIP, and reporter gene assays
Western blot analysis using anti-Nkx6.1 (P. Serup) and anti-HDAC1 (Santa Cruz) antibodies was performed as previously described [26].
Based on previously identified motifs [50], [51], a custom positional weight matrix was used to identify putative Nkx6.1 binding sites. ChIP assays were performed as described [52], using Nkx6.1 (P. Serup, 1∶250) or rabbit IgG antisera. Immunoprecipitations were performed on Min6 cells (ATCC) or on 1.5×106 GFP+ cells isolated from 300 pancreata of Neurog3eGFP/+ embryos at e15.5 by fluorescence activated cell sorting. Each ChIP assay was quantified in triplicate by qPCR. The following primer sequences were used: (site A) 5′-CAT CCG GTG ATA CTG GAA GCC C -3′ and 5′-GTC TTT ATC TGA GGG GGG GCT G -3′; (site B) 5′-GCA GAG GGG GGA GGA GGG -3′ and 5′ - CGG CAG GGA AAT CCA CAA AAC -3′; (site C; Re1) 5′ - CCA TTT GAA GGC AAA ATG CT -3′ and 5′ - GTA TGG GCT GCA AAC ACC TT -3′; (site C; Re2) 5′ - TGA AGT GGC TGA ATG AGA GC -3′ and 5′ - AGT TGG AGC GCG TTT TGT AG -3′; glucagon 5′ - AAG CAG ATG AGC AAA GTG AGT G -3′ and 5′ - AGG CTG TTT AGC CTT GCA GAT A -3′; and intergenic control 5′ - CAC TCA GAT CCT GAG CCA CA -3′ and 5′ - GCT CTC TGC CTT CCA CTT TG -3′.
The Isl1 Re1 enhancer and CMV-Isl1 constructs as well as procedures for transient transfections and luciferase assays have been described previously [53]. Unless indicated otherwise, 0.1 µg of CMV-Nkx6.1 was transfected. Luciferase and Renilla expression were measured 48 hours post transfection. For each data point relative luciferase activity was quantified as the total luciferase units divided by the total Renilla units. All reporter gene analyses were performed in triplicate.
Statistical analysis
All values are shown as mean ± standard error of the mean (SEM); p-values were calculated using student's 2-tailed t-test; P<0.05 was considered significant.
Supporting Information
Zdroje
1. ZhouQ, BrownJ, KanarekA, RajagopalJ, MeltonDA (2008) In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455 : 627–632.
2. Gefen-HaleviS, RachmutIH, MolakandovK, BernemanD, MorE, et al. (2010) NKX6.1 promotes PDX-1-induced liver to pancreatic beta-cells reprogramming. Cell Reprogram 12 : 655–664.
3. FerberS, HalkinA, CohenH, BerI, EinavY, et al. (2000) Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med 6 : 568–572.
4. YechoorV, LiuV, PaulA, LeeJ, BurasE, et al. (2009) Gene therapy with neurogenin 3 and betacellulin reverses major metabolic problems in insulin-deficient diabetic mice. Endocrinology 150 : 4863–4873.
5. KojimaH, FujimiyaM, MatsumuraK, YounanP, ImaedaH, et al. (2003) NeuroD-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat Med 9 : 596–603.
6. SongYD, LeeEJ, YasharP, PfaffLE, KimSY, et al. (2007) Islet cell differentiation in liver by combinatorial expression of transcription factors neurogenin-3, BETA2, and RIPE3b1. Biochem Biophys Res Commun 354 : 334–339.
7. ThorelF, NepoteV, AvrilI, KohnoK, DesgrazR, et al. (2010) Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464 : 1149–1154.
8. TalchaiC, XuanS, SusselL, AcciliD (2012) Pancreatic beta cell destruction as a mechanism of diabetic beta cell failure. Cell 150 : 1223–1234.
9. DesgrazR, HerreraPL (2009) Pancreatic neurogenin 3-expressing cells are unipotent islet precursors. Development 136 : 3567–3574.
10. GuG, DubauskaiteJ, MeltonDA (2002) Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129 : 2447–2457.
11. GradwohlG, DierichA, LeMeurM, GuillemotF (2000) neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 97 : 1607–1611.
12. CollombatP, MansouriA, Hecksher-SorensenJ, SerupP, KrullJ, et al. (2003) Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 17 : 2591–2603.
13. Sosa-PinedaB, ChowdhuryK, TorresM, OliverG, GrussP (1997) The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature 386 : 399–402.
14. CollombatP, XuX, RavassardP, Sosa-PinedaB, DussaudS, et al. (2009) The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell 138 : 449–462.
15. WangJ, ElghaziL, ParkerSE, KizilocakH, AsanoM, et al. (2004) The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic beta-cell differentiation. Dev Biol 266 : 178–189.
16. YangYP, ThorelF, BoyerDF, HerreraPL, WrightCV (2011) Context-specific {alpha}-to-{beta}-cell reprogramming by forced Pdx1 expression. Genes Dev 25 : 1680–1685.
17. PapizanJB, SingerRA, TschenSI, DhawanS, FrielJM, et al. (2011) Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev 25 : 2291–2305.
18. D'AmourKA, BangAG, EliazerS, KellyOG, AgulnickAD, et al. (2006) Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 24 : 1392–1401.
19. KroonE, MartinsonLA, KadoyaK, BangAG, KellyOG, et al. (2008) Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol 26 : 443–452.
20. MatsuokaTA, ArtnerI, HendersonE, MeansA, SanderM, et al. (2004) The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A 101 : 2930–2933.
21. LiH, ArberS, JessellTM, EdlundH (1999) Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat Genet 23 : 67–70.
22. HarrisonKA, ThalerJ, PfaffSL, GuH, KehrlJH (1999) Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet 23 : 71–75.
23. SchafferAE, FreudeKK, NelsonSB, SanderM (2010) Nkx6 Transcription Factors and Ptf1a Function as Antagonistic Lineage Determinants in Multipotent Pancreatic Progenitors. Developmental Cell 18 : 1022–1029.
24. HenseleitKD, NelsonSB, KuhlbrodtK, HenningsJC, EricsonJ, et al. (2005) NKX6 transcription factor activity is required for alpha - and beta-cell development in the pancreas. Development 132 : 3139–3149.
25. NelsonSB, SchafferAE, SanderM (2007) The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development 134 : 2491–2500.
26. SchafferAE, YangAJ, ThorelF, HerreraPL, SanderM (2011) Transgenic in vivo overexpression of the transcription factor Nkx6.1 in beta cells does not increase beta cell proliferation, beta cell mass, or improve glucose clearance. Molecular Endocrinology in press.
27. RudnickA, LingTY, OdagiriH, RutterWJ, GermanMS (1994) Pancreatic beta cells express a diverse set of homeobox genes. Proc Natl Acad Sci U S A 91 : 12203–12207.
28. SanderM, PaydarS, EricsonJ, BriscoeJ, BerberE, et al. (2000) Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev 14 : 2134–2139.
29. SanderM, SusselL, ConnersJ, ScheelD, KalamarasJ, et al. (2000) Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development 127 : 5533–5540.
30. WangS, YanJ, AndersonDA, XuY, KanalMC, et al. (2010) Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Developmental Biology 339 : 26–37.
31. Cras-MeneurC, LiL, KopanR, PermuttMA (2009) Presenilins, Notch dose control the fate of pancreatic endocrine progenitors during a narrow developmental window. Genes & Development 23 : 2088–2101.
32. DuttaS, Bonner-WeirS, MontminyM, WrightC (1998) Regulatory factor linked to late-onset diabetes? Nature 392 : 560.
33. JohnsonJD, AhmedNT, LucianiDS, HanZ, TranH, et al. (2003) Increased islet apoptosis in Pdx1+/ − mice. The Journal of Clinical Investigation 111 : 1147–1160.
34. AhlgrenU, JonssonJ, JonssonL, SimuK, EdlundH (1998) beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev 12 : 1763–1768.
35. GannonM, AblesET, CrawfordL, LoweD, OffieldMF, et al. (2008) pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev Biol 314 : 406–417.
36. JohanssonKA, DursunU, JordanN, GuG, BeermannF, et al. (2007) Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell 12 : 457–465.
37. LiuJ, HunterCS, DuA, EdigerB, WalpE, et al. (2011) Islet-1 Regulates Arx Transcription during Pancreatic Islet Alpha-Cell Development. Journal of Biological Chemistry 286 : 15352–15360.
38. CollombatP, Hecksher-SorensenJ, BroccoliV, KrullJ, PonteI, et al. (2005) The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha - and beta-cell lineages in the mouse endocrine pancreas. Development 132 : 2969–2980.
39. CollombatP, Hecksher-SorensenJ, KrullJ, BergerJ, RiedelD, et al. (2007) Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest 117 : 961–970.
40. DhawanS, GeorgiaS, TschenSI, FanG, BhushanA (2011) Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell 20 : 419–429.
41. OffieldMF, JettonTL, LaboskyPA, RayM, SteinRW, et al. (1996) PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122 : 983–995.
42. LeeCS, PerreaultN, BrestelliJE, KaestnerKH (2002) Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes & Development 16 : 1488–1497.
43. MatsumuraH, HasuwaH, InoueN, IkawaM, OkabeM (2004) Lineage-specific cell disruption in living mice by Cre-mediated expression of diphtheria toxin A chain. Biochem Biophys Res Commun 321 : 275–279.
44. SchonhoffSE, Giel-MoloneyM, LeiterAB (2004) Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Developmental Biology 270 : 443–454.
45. PosticC, ShiotaM, NiswenderKD, JettonTL, ChenY, et al. (1999) Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274 : 305–315.
46. SrinivasS (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1 : 4.
47. NovakA, GuoC, YangW, NagyA, LobeCG (2000) Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon cre-mediated excision. genesis 28 : 147–155.
48. HerreraPL (2000) Adult insulin - and glucagon-producing cells differentiate from two independent cell lineages. Development 127 : 2317–2322.
49. SeymourPA, FreudeKK, DuboisCL, ShihH-P, PatelNA, et al. (2008) A dosage-dependent requirement for Sox9 in pancreatic endocrine cell formation. Developmental Biology 323 : 19–30.
50. JorgensenMC, Vestergard PetersenH, EricsonJ, MadsenOD, SerupP (1999) Cloning and DNA-binding properties of the rat pancreatic beta-cell-specific factor Nkx6.1. FEBS Lett 461 : 287–294.
51. MirmiraRG, WatadaH, GermanMS (2000) Beta-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J Biol Chem 275 : 14743–14751.
52. GerrishK, CissellMA, SteinR (2001) The Role of Hepatic Nuclear Factor 1 alpha and PDX-1 in Transcriptional Regulation of the pdx-1 Gene. Journal of Biological Chemistry 276 : 47775–47784.
53. LiuJ, HunterCS, DuA, EdigerB, WalpE, et al. (2011) Islet-1 regulates Arx transcription during pancreatic islet alpha-cell development. J Biol Chem 286 : 15352–15360.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



