-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
The telomere-ending binding protein complex CST (Cdc13-Stn1-Ten1) mediates critical functions in both telomere protection and replication. We devised a co-expression and affinity purification strategy for isolating large quantities of the complete Candida glabrata CST complex. The complex was found to exhibit a 2∶4∶2 or 2∶6∶2 stoichiometry as judged by the ratio of the subunits and the native size of the complex. Stn1, but not Ten1 alone, can directly and stably interact with Cdc13. In gel mobility shift assays, both Cdc13 and CST manifested high-affinity and sequence-specific binding to the cognate telomeric repeats. Single molecule FRET-based analysis indicates that Cdc13 and CST can bind and unfold higher order G-tail structures. The protein and the complex can also interact with non-telomeric DNA in the absence of high-affinity target sites. Comparison of the DNA–protein complexes formed by Cdc13 and CST suggests that the latter can occupy a longer DNA target site and that Stn1 and Ten1 may contact DNA directly in the full CST–DNA assembly. Both Stn1 and Ten1 can be cross-linked to photo-reactive telomeric DNA. Mutating residues on the putative DNA–binding surface of Candida albicans Stn1 OB fold domain caused a reduction in its crosslinking efficiency in vitro and engendered long and heterogeneous telomeres in vivo, indicating that the DNA–binding activity of Stn1 is required for telomere protection. Our data provide insights on the assembly and mechanisms of CST, and our robust reconstitution system will facilitate future biochemical analysis of this important complex.
Published in the journal: The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures. PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003145
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003145Summary
The telomere-ending binding protein complex CST (Cdc13-Stn1-Ten1) mediates critical functions in both telomere protection and replication. We devised a co-expression and affinity purification strategy for isolating large quantities of the complete Candida glabrata CST complex. The complex was found to exhibit a 2∶4∶2 or 2∶6∶2 stoichiometry as judged by the ratio of the subunits and the native size of the complex. Stn1, but not Ten1 alone, can directly and stably interact with Cdc13. In gel mobility shift assays, both Cdc13 and CST manifested high-affinity and sequence-specific binding to the cognate telomeric repeats. Single molecule FRET-based analysis indicates that Cdc13 and CST can bind and unfold higher order G-tail structures. The protein and the complex can also interact with non-telomeric DNA in the absence of high-affinity target sites. Comparison of the DNA–protein complexes formed by Cdc13 and CST suggests that the latter can occupy a longer DNA target site and that Stn1 and Ten1 may contact DNA directly in the full CST–DNA assembly. Both Stn1 and Ten1 can be cross-linked to photo-reactive telomeric DNA. Mutating residues on the putative DNA–binding surface of Candida albicans Stn1 OB fold domain caused a reduction in its crosslinking efficiency in vitro and engendered long and heterogeneous telomeres in vivo, indicating that the DNA–binding activity of Stn1 is required for telomere protection. Our data provide insights on the assembly and mechanisms of CST, and our robust reconstitution system will facilitate future biochemical analysis of this important complex.
Introduction
The special nucleoprotein structures located at the ends of linear eukaryotic chromosomes, known as telomeres, are critical for chromosome stability; they protect the terminal DNAs from degradation, end-to-end fusion, and other abnormal transactions [1]–[3]. In most organisms, telomeric DNA consists of short G-rich repeats that terminate in 3′ overhangs referred to as G-tails. The G-tails of budding yeast are bound by CST (Cdc13-Stn1-Ten1), a complex that is critical for both telomere protection and replication [4]–[7]. Genetic and structural analyses suggest that CST exhibits many feature of the RPA complex, which constitutes the major single-strand DNA-binding complex in eukaryotes [5], [6], [8]. Interestingly, even though CST proteins were initially discerned only in budding yeast, recent studies have uncovered Stn1 and Ten1 homologues in fission yeast, as well as CST-like complexes in plants and mammals, indicating that this complex is widely conserved [9]–[11]. Notably, a Stn1 homologue named Verrocchio is now known to be critical for telomere stability in Drosophila, an organism that utilizes retrotransposons rather than short repeats to cap chromosome ends [12], [13]. CST components thus appear to be universally present and involved in telomere regulation. In support of this idea, the human CST complex was recently shown to regulate telomerase activity, telomere replication, and telomere C-strand synthesis [14]–[17].
Among all the CST complexes, the structures and mechanisms of the S. cerevisiae subunits are the most extensively characterized. ScCdc13 is a large multifunctional protein that contains probably four OB fold domains and that binds to nucleic acid as well as protein targets. The first, third and last OB fold domains of Cdc13 (named OB1, DBD and OB4) are known to interact with Pol1 (the catalytic subunit of Pol α), G-tail, and Stn1, respectively [18], [19]. The OB1 domain also mediates dimerization, which in turn creates an extended surface for Pol1 binding [18]. Moreover, the recruitment domain (RD) of Cdc13, located between OB1 and OB2, binds to the telomerase subunit Est1 to promote the recruitment of telomerase to chromosome ends and the activation of telomerase [20]–[22]. In comparison to ScCdc13, fewer interaction partners have been identified for ScStn1 and ScTen1. ScStn1 and ScTen1 together form a stable subcomplex and each protein has been reported to bind telomere G-strand with moderate to low affinity [5], [23]. ScStn1 is also known to interact with Pol12, a regulatory subunit of the Pol α complex [24]. The multiplicity of interactions between CST and Pol α supports a role for CST in regulating telomere C-strand synthesis, which is presumed to be mediated by pol α [7].
Analyses of CST homologues or analogues in diverse organisms have revealed intriguing structural and mechanistic variations in these critical telomere capping proteins. For instance, CTC1s (the largest subunit of the mammalian and plant complex), despite having multiple OB fold domains, exhibit little sequence similarity to Cdc13 and possess no autonomous DNA-binding activity [10]. Unlike ScCdc13, the mammalian CST complex manifests distinct recognition properties for short and long single stranded (ss) DNA; it binds preferentially to short oligonucleotides (<20 nts) with G-strand repeats, but indiscriminately to long oligonucleotides (>50 nts) with a wide variety of sequences [10], [14], [25]. Other instances of striking variation are found in many Candida species, where the Cdc13 orthologues are quite small and consist of just the DBD and OB4 domains [6], [26]. Notwithstanding the absence of the OB1 domain, these orthologues nevertheless form dimers through an alternative interface involving specialized loops in their OB4 regions [26]. Moreover, in contrast to ScCdc13, dimerization of Candida Cdc13s is required for high affinity and sequence-specific recognition of telomeric DNA [26]. These observations raise fascinating questions concerning the mechanistic diversity and evolutionary plasticity of the CST complex.
Not withstanding considerable knowledge on the structure and function of fungal CST subunits, studies of the complex has been hampered by an inability to reconstitute and isolate adequate quantities of the full complex for detailed biochemical investigations. Thus, the precise assembly mechanisms of the complex (e.g., how the subunits interact with one another) remain undefined. Whether the incorporation of the Stn1 and Ten1 subunit alters the DNA-binding property of Cdc13 is likewise unclear. To address such deficiencies, we systematically screened CST homologues for co-expression and complex assembly in E. coli. After considerable trials, we developed a strategy for isolating large quantities of the Candida glabrata CST complex, which was shown to have an unusual stoichiometry. Both Cdc13 and the CST complex were found to recognize G-tails with high affinity and sequence-specificity, and to be capable of unfolding higher order G-tail structures. Additional studies suggest that Stn1 and Ten1 can contact DNA directly in the context of the full CST-DNA assembly. Mutating residues on a hypothesized DNA-binding surface of Candida albicans Stn1 OB fold domain caused a reduction in its DNA-binding in vitro (as measured by a photo-crosslinking assay) and engendered long and heterogeneous telomeres in vivo, supporting the physiologic significance of DNA-binding by Stn1.
Results
The C. glabrata CST complex has an unusual stoichiometry
To reconstitute the CST complex encoded by the C. glabrata genome, we co-expressed all three subunits as fusion proteins in E. coli (Figure 1A). The CDC13, STN1 and TEN1 gene were fused to the FLAG, HIS6, and GST tag, respectively to allow sequential affinity purification of the complex. The Cdc13 and Stn1 fusion proteins also contained a SUMO tag, which improved their expression level and solubility. Unless explained otherwise, the fusion proteins will henceforth be referred to as Cdc13, Stn1 and Ten1 to simplify the discussion. Both Ten1 and Cdc13 were recovered from the initial Ni-NTA column, indicating that they can both associate with Stn1 (Figure 1B, lane 4 and 5). The much higher concentrations of Stn1 and Ten1 in comparison to Cdc13 in these fractions are consistent with the expression levels of these proteins (data not shown). As expected, Cdc13 was further enriched after purification on the M2 (anti-FLAG) resin. More importantly, both Stn1 and Ten1 were again recovered, at concentrations that were either equal to Cdc13 (Ten1), or higher than Cdc13 (Stn1) (Figure 1B, lane 7 and 8). Notably, the binding of Stn1 and Ten1 to M2-agarose could not be detected in the absence of Cdc13 (Figure 1B, lane 6). To confirm the formation of the ternary complex, we subjected the M2-derived fractions to Glutathione-Sepharose chromatography and once more recovered all three proteins (Figure 1C). The protein yield from the Glutathione column was low and the composition of the Glutathione fractions was similar to the M2 fractions. Hence, we carried out all subsequent analysis of CST using the M2 fractions.
Fig. 1. Purification and characterization of the C. glabrata CST complex. 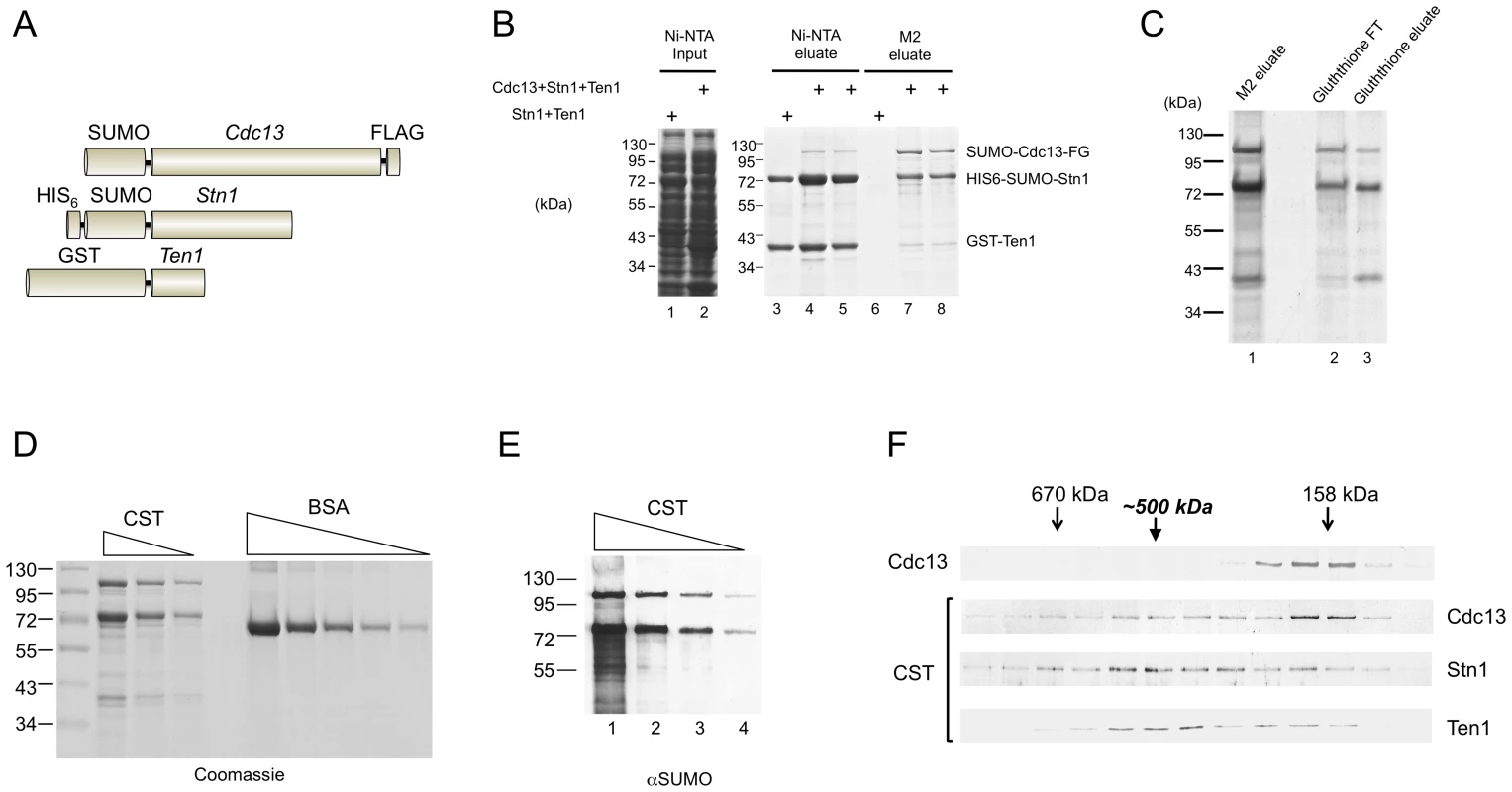
(A) The fusion proteins co-expressed in E. coli are schematically illustrated. (B) Extracts were isolated from E. coli strains co-expressing Stn1 and Ten1 or all three subunits, and subjected sequentially to Ni-NTA and M2 affinity chromatography. The extracts and eluates from the Ni-NTA and M2 resins were analyzed by SDS-PAGE and Coomassie staining. (C) The M2 eluate containing the CST complex from part B was subjected to Glutathione-Sepharose affinity chromatography. The flow through (FT) and eluate fractions as well as the starting material were analyzed by SDS-PAGE and Coomassie staining. (D) Different amounts of the purified CST complex (10 µl, 5 µl and 2.5 µl) and bovine serum albumin (5 µg, 2.5 µg, 1.25 µg, 0.63 µg, and 0.32 µg) were analyzed by SDS-PAGE and Coomassie staining. (E) Serial three fold dilutions of the purified CST complex were subjected to Western analysis using antibodies directed against the SUMO tag, which is present in both the Cdc13 and Stn1 fusion proteins. (F) Purified SUMO-Cdc13 and the CST complex were separately fractionated on 15–30% glycerol gradients. The distributions of the proteins in the fractions were analyzed by Western using antibodies directed against SUMO (the Cdc13 and Stn1 fusions) and GST (the Ten1 fusion protein). The results for free Cdc13 and the CST complex are displayed in the top panel and the bottom three panels, respectively. The apparently higher levels of Stn1 in the CST complex prompted us to carry out a more detailed analysis of the stoichiometry of the complex. First, we quantified the levels of individual proteins by comparing their Coomassie staining intensities to BSA standards (Figure 1D). Assuming that the intensities of Cdc13, Stn1 and Ten1 are proportional to their molecular weights, we obtained a ratio of roughly 1∶2.5∶1 for these subunits in the complex. Second, we estimated the relative amounts of Cdc13 and Stn1 by Western analysis using antibodies directed against the SUMO tag that is present in both fusion proteins. This method again yielded a Cdc13: Stn1 ratio of 1∶2.5 (Figure 1E). In particular, the Western signal of Stn1 in a 3-fold diluted CST sample was slightly less than that of Cdc13 in the undiluted sample (Figure 1E, compare each pair of neighboring samples, e.g., lane 3 & 4). We analyzed multiple CST preparations and found the Stn1: Cdc13 ratio to be always between 2∶1 and 3∶1, and the Ten1: Cdc13 ratio to be nearly 1∶1. We were unable to discriminate firmly between the 2∶1 or 3∶1 stoichiometry for Stn1: Cdc13. It is possible that some heterogeneity exists in the composition of the complex. Alternatively, the staining intensity of Stn1 may not be strictly proportional to its size, thus causing errors in the calculation of molar ratios.
Because Cdc13 is known to dimerize [18], [26], our estimated ratio for the subunits suggests that the complex may have a 2∶4∶2 or 2∶6∶2 stoichiometry, hence a native size of ∼500–600 kDa. Consistent with this notion, glycerol gradient sedimentation analysis of purified CST revealed a peak for each subunit at ∼500 kDa (Figure 1F, the lower 3 panels). Even though the distribution of all three proteins in the gradient are broad, plots of the relative levels of each protein across the gradient are consistent with co-sedimentation around 500 kDa (Figure S1A). A lighter peak for Cdc13 was also observed, implying partial disassembly of the complex during the gradient run. The position of this peak (∼160 kDa) was precisely that predicted for a Cdc13 dimer and was identical to the sedimentation position of purified Cdc13 alone (Figure 1F, the top 2 panels). We also analyzed the Stn1: Cdc13 ratio in the 500 kDa gradient fraction using anti-SUMO antibodies and again obtained a ratio of ∼2.5∶1 (Figure S1B). We conclude that the CST complex has a stoichiometry of 2∶4∶2 or 2∶6∶2.
Stn1, but not Ten1, can independently form a stable complex with Cdc13
To begin to analyze the assembly mechanisms of the CST complex, we separately co-expressed Stn1 and Cdc13, as well as Ten1 and Cdc13, and then used the same affinity purification strategy to assess complex formation. Interestingly, Cdc13 could be recovered in the Ni-NTA fraction that contained highly purified Stn1, but was not detectable in the Glutathione-Sepharose fraction that contained purified Ten1 (Figure 2A, lane 3 and 4). Conversely, Stn1 but not Ten1 could be recovered in the M2 fractions that were highly enriched for Cdc13 (Figure 2A, lane 5 and 6). These results imply that Stn1 but not Ten1 directly interacts with Cdc13. Because Stn1 was at least 20 fold more abundant than Cdc13 in the extract (data not shown), one would expect the majority of Cdc13 to be associated with Stn1, but not vice versa. Consistent with this expectation, Stn1 was ∼2–3 times more abundant than Cdc13 in the M2 fraction, but was present at far greater excess over Cdc13 in the Ni-NTA fraction (Figure 2A, lane 3 and 5). As further controls, we expressed Stn1 or Ten1 alone in the absence of Cdc13 and subjected the extracts to M2 affinity chromatography (Figure 2B and 2C). Little of either protein was found in the eluate, thus ruling out non-specific binding of Stn1 to the M2 resin. Altogether, these experiments support a model of CST assembly that entails a direct interaction between Cdc13 and Stn1, but not between Cdc13 and Ten1.
Fig. 2. Stn1, but not Ten1, independently binds to Cdc13. 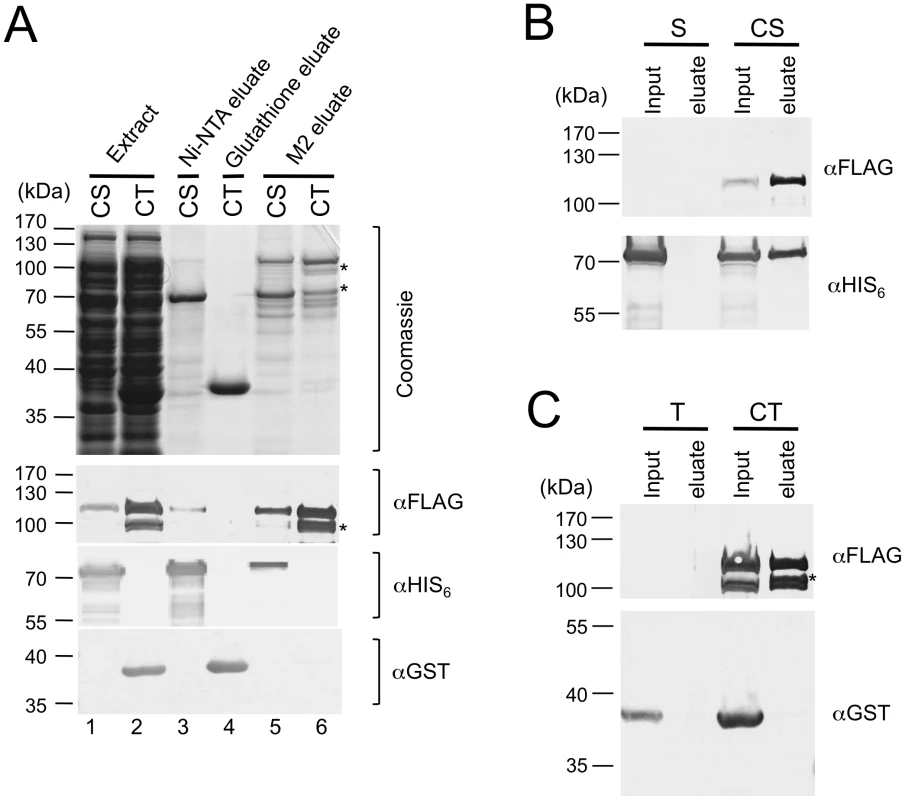
(A) E. coli extracts prepared from strains co-expressing SUMO-Cdc13-FG and HIS6-SUMO-Stn1 (CS) or SUMO-Cdc13-FG and GST-Ten1 (CT) were subjected to Ni-NTA, Glutathione-Sepharose and M2 affinity purification as indicated. The extracts and eluates were analyzed by SDS-PAGE and Coomassie staining (top), as well as Western using antibodies directed against the FLAG tag, the HIS6 tag and the GST tag of Cdc13, Stn1 and Ten1 fusion protein, respectively (bottom). Significant amounts of Cdc13 proteolytic fragments (*) can be detected in the CT eluate from the M2 resin. (B) E. coli extracts prepared from strains expressing HIS6-SUMO-Stn1 alone (S) or HIS6-SUMO-Stn1 and SUMO-Cdc13-FG (CS) were subjected to M2 affinity purification. The Cdc13 and Stn1 fusion proteins in the input and eluate samples were detected by Western using antibodies directed against the FLAG and HIS6 tag, respectively. (C) E. coli extracts prepared from strains expressing GST-Ten1 alone (T) or GST-Ten1 and SUMO-Cdc13-FG (CT) were subjected to M2 affinity purification. The Cdc13 and Ten1 fusion proteins in the input and eluate samples were detected by Western using antibodies directed against the FLAG and GST tag, respectively. Characterization of the DNA–binding properties of C. glabrata Cdc13 and CST complex
We next attempted to characterize the DNA-binding properties of individual subunits or sub-complexes as well as the full CST assembly in order to assess the contributions of these subunits. Among the subunits and sub-complexes, we were able to obtain substantial amounts of Cdc13, Ten1 and the Stn1-Ten1 (ST) sub-complex for gel mobility shift analysis. Neither Ten1 nor ST could form a detectable complex with C. glabrata telomere oligonucleotides in a variety of buffer conditions in these assays (data not shown). In contrast, Cdc13 and CST each bound telomere oligoes with a Kd of ∼10–20 nM (Figure 3A). The Cdc13-DNA and CST-DNA complexes were relatively stable with a ½ life of ∼10 min (Figure 3B and data not shown). Binding was also highly sequence-specific. In competition assays, more than 200 fold higher concentrations of a random unlabeled oligo (R1) as well as a heterologous telomere oligo (LeTELX2, corresponding to 2 copies of the L. elongisporus telomere repeat) were needed to achieve the same degree of inhibition as the C. glabrata telomere oligo (Figure S2). Interestingly, an oligo based on the C. lusitaniae telomere repeat (ClTELX2) exhibited moderate affinity for the C. glabrata Cdc13 and CST; just 20-fold higher concentration of this oligo competed as well as the C. glabrata telomere repeat (Figure S2).
Fig. 3. DNA–protein complexes generated by Cdc13 and CST. 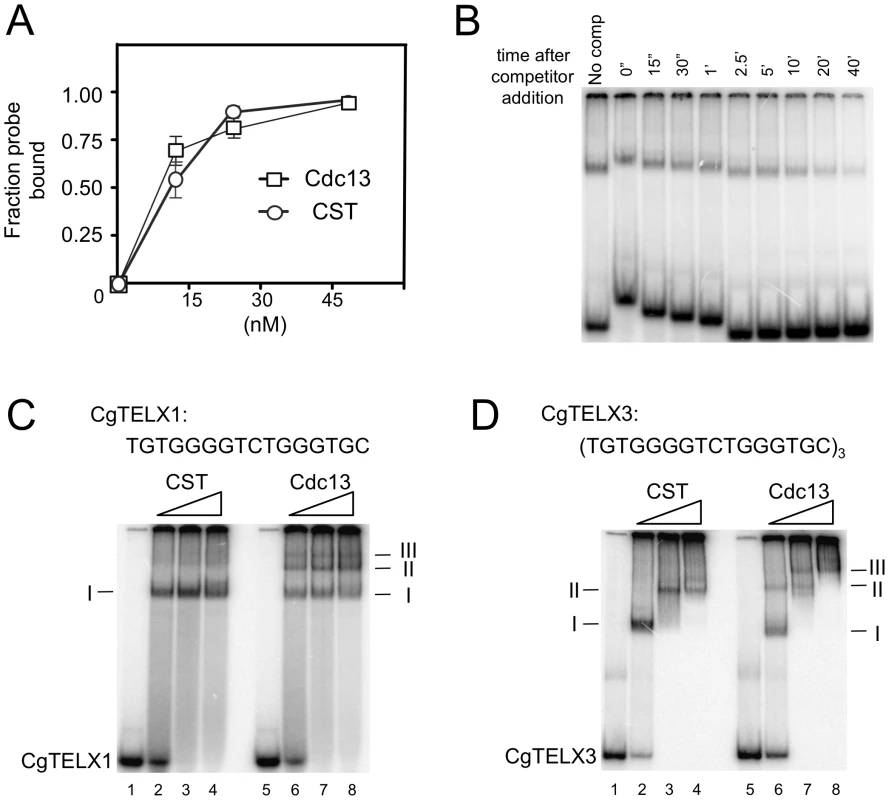
(A) The apparent Kd of Cdc13 and CST for the CgTELX1 probe (TGTGGGGTCTGGGTGC) was estimated in gel shift assays using different concentrations of the protein or protein complex. Data (average ± standard deviation) are from three independent experiments. (B) The stability of the CST-DNA complex was estimated in a competition experiment. CST (10 nM) was allowed to form complexes with labeled CgTELX1 (7.5 nM) for 20 min. One-hundred fold excess unlabeled CgTELX1 was then added, and the mixture applied to a native gel at various time points following the addition of the competitor. The non-uniform mobility of the free probe and the complex was due to the fact that samples were applied at different times. The fractions of labeled complexes remaining at various time points are determined and used to calculate t½. The experiments were repeated three times and the estimated t½ ranges from 5 to 13 min. (C) Increasing concentrations of CST and Cdc13 (15 nM, 30 nM and 60 nM) were incubated with P32-labeled CgTELX1 oligo (7.5 nM) and the resulting complexes analyzed by electrophoresis through a native gel. Complexes of decreasing mobility are designated by I, II, etc. (D) Same as part C except that P32-labeled CgTELX3 oligo ([TGTGGGGTCTGGGTGC]3) was used as the probe. We then attempted to determine the lengths of DNA occupied by Cdc13 and CST through titration analysis. We reasoned that a fast migrating complex should be converted to a slower complex at higher protein concentrations if the former contains sufficient naked DNA to capture additional proteins. Conversely, if the DNA in a complex is fully occupied, then adding more proteins should not alter its mobility (unless the added proteins can form oligomers with the proteins in the complex). Characterization of the DNA-protein species formed by Cdc13 and CST on a 16-nt (one repeat) probe suggests that both can fully occupy this DNA target (Figure 3C). In the case of CST, only one predominant gel shift band could be detected, and this band was not converted to other forms even at a protein concentration that was twice than necessary to retard all of the free probe. For Cdc13, even though three gel shift bands could be observed, the fastest migrating complex (I) was not converted into other forms even at much higher protein concentrations. This finding suggests that while multiple Cdc13 dimers may be able to land on a 16-nt DNA (Figure 3C, complex II and III), a significant fraction of the probe was fully occupied by just one Cdc13 dimer.
Interestingly, characterization of the complexes formed on a 48-nt probe suggests different occupancy sizes for Cdc13 and CST (Figure 3D). For Cdc13, three complexes could be observed, and the highest mobility species (complex I) was progressively converted to lower mobility species (complex II and III) with increasing Cdc13 concentrations. By contrast, only two species of CST-DNA complex could be detected over the same range of protein concentrations. These observations suggest that the 48-nt probe can be fully occupied by three Cdc13 dimers, but just two CST complexes. Hence the CST complex appears to occupy a longer stretch of G-tail than the Cdc13 dimer. However, the difference is evidently subtle and requires additional studies (e.g., footprinting) for confirmation.
Binding of CST to telomere G-strand alters DNA conformation
G-tails have the propensity to form salt-dependent higher order structures (e.g., G-quadruplex) [27], [28]. Cdc13 and other G-tail binding proteins have been reported to bind or modulate such higher order structures [27], [29], [30], but the activity of the complete CST complex in this regard has not been characterized. We used a single-molecule FRET strategy [31], [32], which has been used to study the dynamics of various proteins on ssDNA [33], to characterize the interaction between C. glabrata telomeric DNA and either Cdc13 or CST. The standard DNA construct contains 48 nt of G-tail DNA ([TGTGGGGTCTGGGTGC]3) and harbors fluorescent dyes (a donor-acceptor FRET pair) near its 5′ and 3′ ends (Table 1, Figure 4A, and Figure S3A). In the presence of low concentrations of salt (e.g., 10 mM K+), the majority of DNA probes appear to be unfolded as indicated by the low FRET efficiencies (∼0.1 to 0.2) (Figure S3B). FRET efficiencies increased steadily in the presence of progressively higher salt concentrations (Figure S3B), consistent with the formation of salt-dependent higher order DNA structures with reduced distance between the fluorescent donor and acceptor [34]. Evidence for higher order G-tail structures can also be gleaned from comparison to the FRET signals generated from a 32-nt G-tail and (dT)32 (Figure S4B and S4D). FRET efficiencies for the 32-nt G-tail (∼0.7) was substantially higher than that for (dT)32 (∼0.35) despite their identical lengths, suggesting that the G-tail is folded into a compact conformation rather than forming a random coil. Additional evidence suggesting “folded G-tails” came from FRET experiments that utilized Li+, which is known to be less efficient than Na+ and K+ in stabilizing G-quadruplex [35]. Indeed, the single molecule FRET-time traces in 100 mM Li+ exhibited greater dynamics than in 100 mM Na+, indicating that the folded state is more short-lived in Li+ (Figure S4C). Moreover, the peaks of the FRET histogram were consistently lower in Li+ than in the same concentrations of K+ (e.g., 0.15 in 20 mM Li+ and 0.25 in 20 mM K+) (Figure S3B and S3D). However, it is important to stress that folded DNA structures other than G-quadruplex are also compatible with our data.
Fig. 4. Cdc13 and CST can bind and unfold higher order G-tail structure. 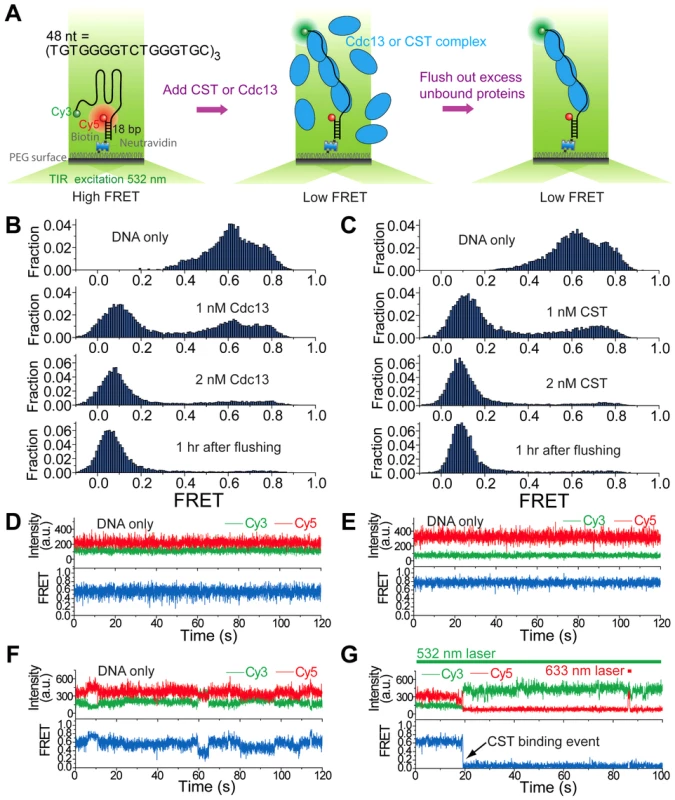
(A) A schematic depiction of reaction steps for single molecule FRET experiments. (B) Single molecule FRET efficiency histograms in the presence of the indicated concentrations of Cdc13. (C) Single molecule FRET efficiency histograms in the presence of the indicated concentrations of CST. (D–F) Representative single-molecule FRET-time traces of the 48-nt G-tail construct in 3 mM MgCl2 and 100 mM NaCl before adding any protein. A 532 nm laser was used for excitation. (G) A representative single-molecule FRET-time trace of the 48-nt G-tail construct in 3 mM MgCl2 and 100 mM NaCl immediately after adding 1 nM CST, showing a CST binding event at ∼19 sec (the black arrow). A 532 nm laser was used for Cy3 excitation throughout the entire course of data acquisition. A 633 nm laser was briefly turned on for direct Cy5 excitation at ∼86 sec in order to confirm that Cy5 is still active rather than photobleached. Tab. 1. Oligonucleotides used in the single-molecule FRET experiments. 
Binding of Cdc13 and CST to the 48-nt G-tail construct was analyzed in solution containing 3 mM MgCl2 and 100 mM NaCl. Prior to protein addition, the G-tail was evidently folded and yielded a broad FRET distribution in which at least three high FRET states (∼0.4, ∼0.6 and ∼0.8) can be discerned (Figure 4B and 4C, top panels), similar to the FRET distribution obtained in 500 mM K+ (Figure S3B). This implies the existence of several folded conformations for the 48-nt G-tail, which is expected for a G-tail containing six repeats of the ‘GGG’ motif. In single-molecule FRET-time traces, the G-tail molecules show two heterogeneous behaviors: Some G-tails maintain their particular folded conformation for a long duration (Figure 4D and 4E), whereas others show fluctuations between different conformations (Figure 4F). This heterogeneity in the conformational dynamics of single molecules was previously observed for the human telomeric G-tail [34]. The introduction of either Cdc13 and CST resulted in the disappearance of the three high FRET species (∼0.4, ∼0.6 and ∼0.8) and a corresponding increase in low FRET (∼0.1) molecules, indicating that both Cdc13 and CST can bind and disrupt the higher order structures associated with high FRET efficiencies (Figure 4B and 4C, second and third panels). The CST binding event can be observed in single-molecule FRET-time traces as an abrupt decrease in FRET (Figure 4G). Similar shifts from the high FRET to low FRET states were observed when we added CST to two other DNA constructs with G-tail lengths of 16 or 32 nt (Figure S4A and S4B). Notably, we observed an inverse relation between the lengths of G-tail and the FRET efficiencies of protein-bound DNA; the peak FRET efficiencies were ∼0.35, 0.11, and 0.09 for the 16 nt, 32 nt and 48 nt constructs, respectively. This correlation suggests that the CST-bound DNAs adopt an extended conformation.
Similar to the results obtained in the gel mobility shift assays, we found very stable binding of Cdc13 and CST to the G-tail in the FRET experiments. For the 32 nt and 48 nt G-tail constructs, we were unable to detect the reappearance of high FRET DNA molecules even an hour after flushing the system with a protein-free buffer (Figure 4B and 4C, Figure S4B). For the 16 nt G-tail construct, about half of the complexes appear to disassemble 30 min after the application of protein-free buffer (Figure S4A). Thus, maximal stability of the CST-DNA and Cdc13-DNA complex appear to require more than one telomere repeat unit.
Binding of Cdc13 and CST to non-telomeric DNA
During the course of the single molecule FRET analysis, we found that Cdc13 and CST can also alter the FRET of a poly-dT control construct. For example, in one experiment, the addition of 100 nM CST to fluorescent (dT)58 (in which the Cy5 and Cy3 fluorophores are ∼16 nt apart) resulted in a shift of FRET from ∼0.6 to ∼0.45 for a significant fraction of the molecules (Figure S5). Importantly, CST binding to this oligonucleotide is evidently unstable; the number of DNA molecules with ∼0.45 FRET efficiency was greatly reduced just 2 minutes after the system was flushed with the binding buffer (Figure S5B and S5C). This observation suggests that Cdc13 and CST may have the ability to bind transiently to non-telomeric DNA, and prompted us to re-investigate this issue using a variety of non-telomeric oligonucleotides in gel mobility shift assays. Indeed, we found that Cdc13 can alter the mobility of the (dT)58 as well as several other non-telomeric DNA (Figure 5A and 5B). Notably, the complexes formed on the non-telomeric oligonucleotides did not migrate as discreet species, but rather as broad smears that trail upward from the position of the free probe (Figure 5B and 5C). Such behavior can be explained by gradual dissociation of the complexes during gel electrophoresis, and is hence quite consistent with the FRET findings. Similar binding to non-telomeric DNAs was also observed for the CST complex (Figure 5C and data not shown). These results provide biochemical support for the notion that the CST complex may have the ability to act on non-telomeric DNA in vivo.
Fig. 5. Cdc13 and CST form unstable complexes with non-telomeric DNA. 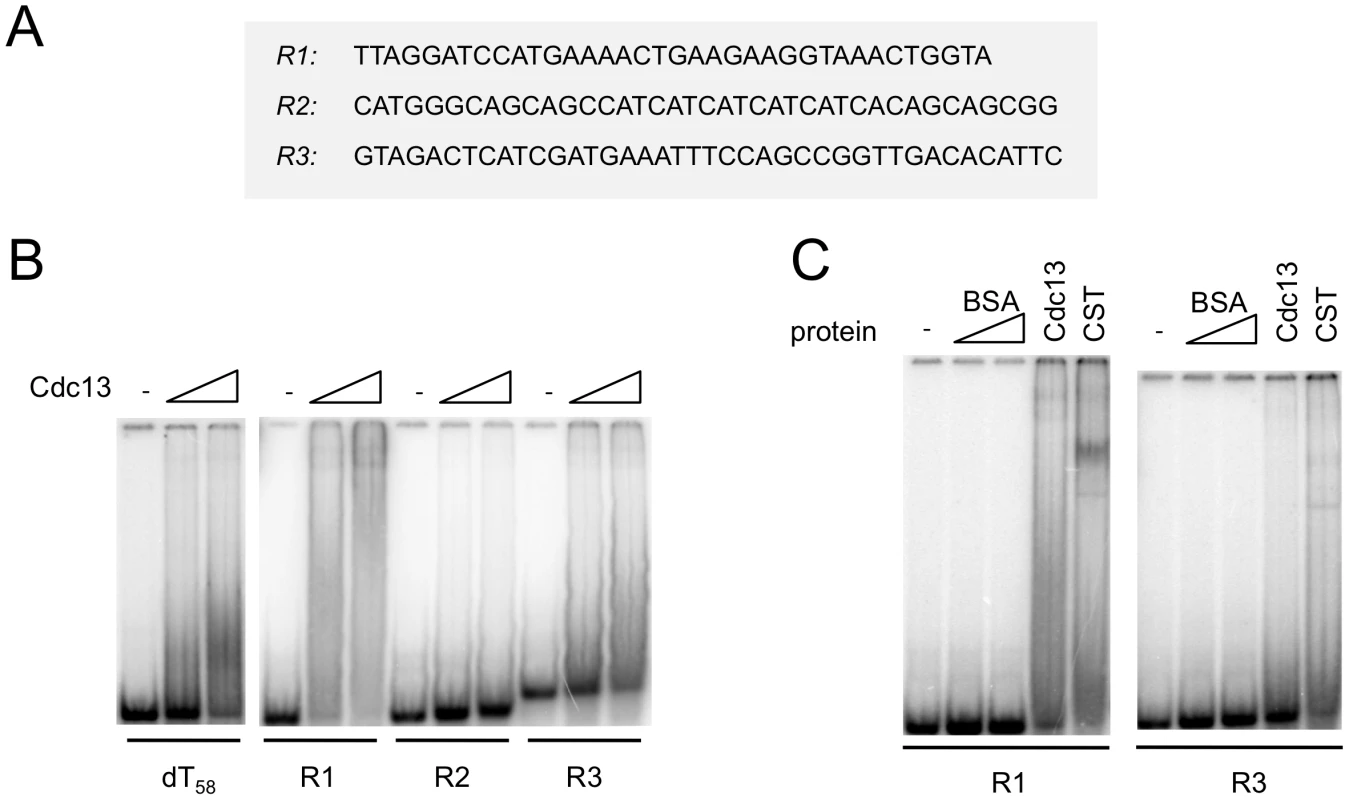
(A) The sequences of the non-telomeric oligonucleotides used in the gel mobility shift assays are displayed. (B) Increasing concentrations of purified Cdc13 (0 nM, 40 nM and 80 nM) were incubated with the indicated oligonucleotide probes (7.5 nM) and the resulting complexes analyzed by gel electrophoresis and PhosphorImager scanning. (C) Labeled R1 and R3 probes (7.5 nM) were incubated with BSA (80 and 240 nM), Cdc13 (40 nM), and CST (40 nM), and the resulting complexes analyzed by gel electrophoresis and PhosphorImager scanning. CgStn1 and CgTen1 have weak intrinsic DNA–binding activities
The potential difference between the occupancy size of the Cdc13 dimer and CST complex suggests that Stn1 and Ten1 may contact DNA directly in the context of the full CST-DNA assembly. To address this possibility, we first carried out crosslinking studies using purified CST and a telomere oligonucleotide containing an Iodo-dU substitution. Even though putative Stn1-DNA adducts could be detected, the signals for such adducts were quite weak in comparison to the Cdc13-DNA products, making it difficult to draw clear conclusions (data not shown). We therefore repeated the crosslinking assays using just the Stn1-Ten1 subcomplex. Because the N-terminal OB fold of Stn1 is presumed to be the DNA-binding domain, we also tested a Stn1N-Ten1 complex. Analysis of labeled products indicates that both Stn1 and Ten1 can be readily crosslinked to DNA (Figure 6A and 6B). The efficiency of Stn1-DNA or Stn1N-DNA adduct formation was consistently about 3 fold higher than Ten1-DNA. Furthermore, Ten1 alone was capable of direct binding to DNA, and the efficiency of Ten1-DNA crosslinking was not stimulated by its association with Stn1 (Figure 6A and 6C). Interestingly, moving the Iodo-dU substitution in the substrate from the 5′ to the 3′ end did not significantly change the crosslinking efficiency of either protein, suggesting that the proteins may not be positioned in a specific manner in relation to the DNA (Figure 6B and 6C, compare signals generated by IdU-1 and IdU-5). Moreover, in competition assays, a non-telomeric oligonucleotide inhibited the crosslinking efficiency to the same extent as the telomere oligo (Figure 6D). Thus, in the absence of Cdc13, Stn1 and Ten1 appear to bind DNA weakly in a non-sequence specific manner.
Fig. 6. Crosslinking of C. glabrata Stn1 and Ten1 to photo-reactive telomere oligonucleotides. 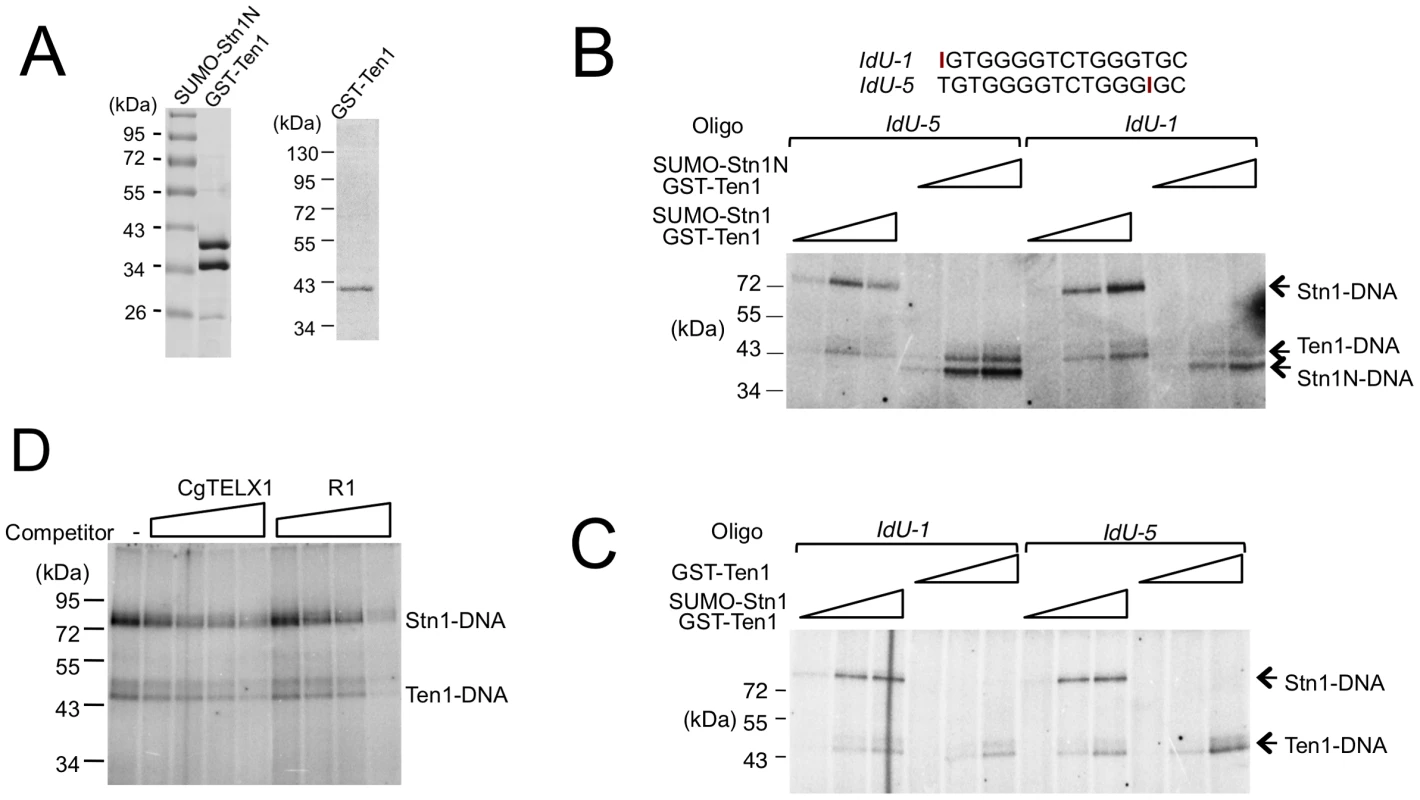
(A) Purified protein complex containing SUMO-Stn1N and GST-Ten1 fusion, as well as GST-Ten1 alone, was analyzed by SDS-PAGE and Coomassie staining. (B) Increasing concentrations of the indicated Stn1-Ten1 complexes (6.6 nM, 20 nM, 60 nM) were crosslinked to labeled IdU-1 or IdU-5 oligonucleotides and the covalent products detected by SDS-PAGE and PhosphorImager scanning. (C) Increasing concentrations of the Stn1-Ten1 complex or GST-Ten1 alone (6.6 nM, 20 nM, 60 nM) were crosslinked to labeled IdU-1 or IdU-5 oligonucleotides and the covalent products detected by SDS-PAGE and PhosphorImager scanning. (D) Purified CgStn1-Ten1 complex (240 nM) was crosslinked to labeled IdU-1 oligonucleotide (16 nM) in the presence of increasing concentrations of the indicated competitor oligonucleotides (125 nM, 500 nM, 2 µM and 8 µM). The DNA–binding activity of C. albicans Stn1 is required for telomere protection
The weak and non-sequence specific DNA binding activities of CgStn1 and Ten1 are consistent with a domain-swapping study, in which the OB fold of ScStn1 was shown to function in place of the DNA-binding domain of ScRpa2 [5]. To investigate the relevance of this activity in telomere protection, we mutated potential DNA-binding residues in C. albicans Stn1 and tested the effects of mutations on telomere length regulation. Inspection of a homology model of CaStn1 revealed two basic residues (K98 in L12 and K170 in L45) that are well positioned to contact DNA directly (Figure 7A). We mutated these residues separately or together to either Ala or Glu, and tested the ability of the resulting alleles to complement the stn1 null mutant. As described in a previous report, the telomeres of the null mutant are substantially longer and more heterogeneous than the parental strain (Figure 7B, lane 3–4) [6]. Re-integrating a tagged but otherwise wild type STN1 gene (STN1-GSCP) suppressed the telomere defects (Figure 7B, lane 2) [6]. Interestingly, the Ala substitution mutants (K98A, K170A and K98A/K170A) exhibited little or no defects in telomere regulation; discreet telomere fragments can still be detected in the mutants and only the K98A/K170A combination mutant manifested a slight increase in telomere lengths (Figure 7B, lane 5–10). In contrast, all of the Glu mutants exhibited significant increases in their telomere lengths and heterogeneity, with the combination mutant (K98E/K170E) manifesting the most severe defect (Figure 7B, lane 11–16). The dysfunction of the Stn1 mutants cannot be attributed to diminished protein levels (Figure 7C and data not shown). In addition, we found that K98E/K170E mutant exhibited normal binding to C. albicans Cdc13 in a co-expression/co-immunoprecipitation analysis (Figure S6). Thus, we surmise that the negative charges introduced by the Glu mutations probably rendered Stn1 defective in telomere protection by disrupting the direct contacts between Stn1 and DNA.
Fig. 7. The effects of C. albicans Stn1 mutations on telomere lengths and Stn1–DNA crosslinking. 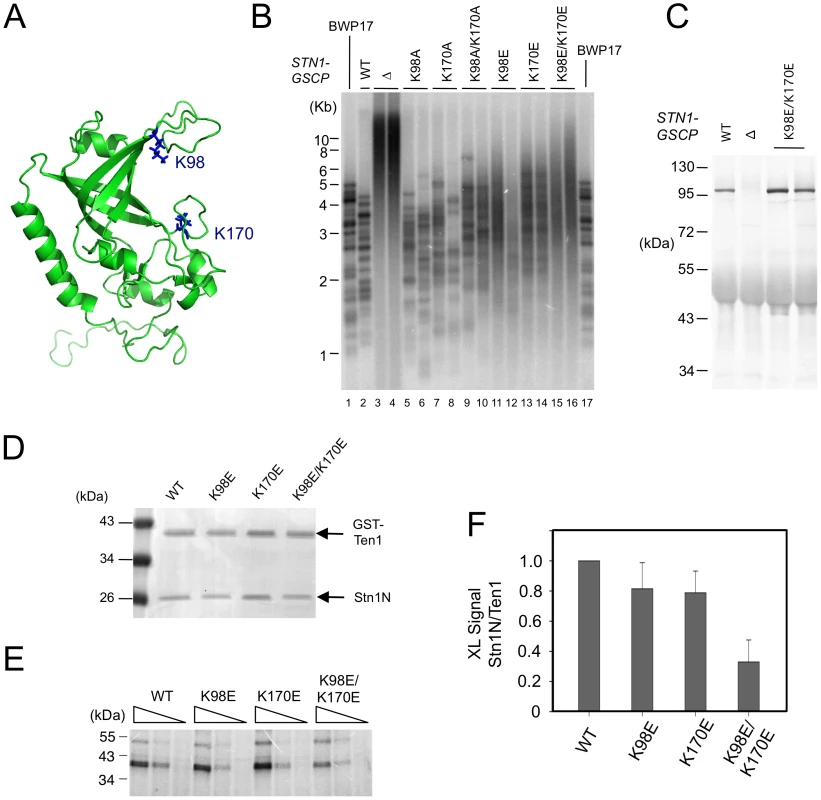
(A) A homology model of C. albicans Stn1N is displayed with two putative DNA-binding residues highlighted in blue. (B) The distributions of telomere restriction fragments in the indicated strains were analyzed by Southern using a probe that corresponds to two copies of the C. albicans telomere repeat unit. (C) The levels of CaStn1-GSCP proteins in extracts derived from the indicated strains were analyzed by Western using antibodies against protein A. (D) Purified complexes containing C. albicans GST-Ten1 and the indicated Stn1N (wild type or mutant) were analyzed by SDS-PAGE and Coomassie staining. (E) Different concentrations of the complexes shown in D (300 nM, 100 nM, 33 nM) were crosslinked to a C. albicans telomere oligonucleotide containing a single Iodo-dU substitution. The covalent products were detected by SDS-PAGE and PhosphorImager scanning. (F) The levels of crosslinked products for each Stn1N protein were normalized against that for the corresponding GST-Ten1 protein and plotted. Data (average ± standard deviation) are derived from three independent experiments. To confirm that the Stn1 mutants are indeed defective in DNA-binding, we purified and analyzed C. albicans ST complexes containing either wild type or mutated CaStn1N. Because the His6-SUMO-CaStn1N and GST-CaTen1 fusion proteins expressed in E. coli exhibited very similar mobility in SDS-PAGE, we removed the His6-SUMO tag before subjecting the complex to the photo-crosslinking assay (Figure 7D). In accordance with the telomere length defects described earlier, the K98E and K170E single mutants each exhibited a mild reduction in crosslinking efficiency, whereas the double mutant manifested a more severe, 3 fold reduction (Figure 7E and 7F). The correlation between the in vivo and in vitro phenotypes suggests that the DNA-binding activity of CaStn1 is required for full telomere protection.
Discussion
The chief significance of the current work resides in the discovery of an unanticipated subunit arrangements for the CST complex and the functional characterization of the DNA-binding properties of individual subunits and complexes. The mechanistic and evolutionary implications of these findings are discussed below.
CST subunit arrangement is different from the RPA complex
Not withstanding the strong similarities between Stn1-Ten1 and Rpa2-Rpa3, previous studies have revealed two major differences between Cdc13 and Rpa1. First, the available high-resolution structures of Cdc13 OB fold domains do not resemble those of Rpa1 [18], [26], [36]. Second, homo-dimerization appears to be a common feature of Cdc13, but not of Rpa1. The current study discloses additional differences between the CST and RPA complex with respect to subunit stoichiometry and inter-subunit interactions. Specifically, RPA has a well-characterized 1∶1∶1 stoichiometry that is established by a trio of interacting α helices, one from each of the subunits [37]. CST, on the other hand, are probably assembled through separate Cdc13-Stn1 and Stn1-Ten1 interactions (this work and [6]). The higher copy number of Stn1 additionally suggests the existence of multiple Stn1-binding sites on a single Cdc13 molecule. Clearly more structural studies are necessary to delineate the molecular basis of CST assembly, but existing data already points a different organizing principle for CST from RPA. The higher Stn1 copy number also raises the intriguing possibility of functional specialization, i.e., different Stn1 in the same complex may bind to different targets (e.g., Pol α and DNA) to mediate distinct functions. Of the three CST components, Stn1 is clearly the best conserved at the level of amino acid sequence [5], suggesting greater evolutionary constraint that could be due to a large number of binding targets.
Stn1 has a weak but functionally significant DNA–binding activity
Previous investigations of DNA-binding by Stn1 and Ten1 have yielded disparate results. Whereas the S. cerevisiae Stn1 and Ten1 were shown individually to bind G-rich telomeric DNA with moderate to low affinity, the C. tropicalis Stn1-Ten1 failed to form detectable complexes with the corresponding telomere repeats [5], [6]. The current report provides a potential explanation for this discrepancy. Even though the C. glabrata Stn1-Ten1 complex or Ten1 protein did not show evidence of interaction with telomeric DNA in native gels, they can clearly make direct contacts to DNA as judged by the photo-crosslinking analysis. Moreover, our discovery of comparable DNA-crosslinking by C. albicans Stn1-Ten1 strongly supports the notion that DNA-binding is a conserved (and hence functionally important) property of these proteins. This notion was presaged by an earlier analysis of the Drosophila orthologue Verrocchio; a quadruple substitution on the putative DNA-binding surface of this protein caused telomere dysfunction [12]. In the current study, we found a good correlation between the DNA-binding defects of Stn1 mutants and telomere dysfunction, thus providing further support for the physiologic significance of this activity. However, we cannot exclude the possibility that the DNA-binding mutants are deficient in some other aspects of Stn1 function. The notion of direct-DNA contacts by Stn1 and Ten1 extends further the parallel between the RPA and CST complex in terms of the mechanisms of DNA-binding. Even though the two smaller subunits of each complex have very low intrinsic affinity for single-stranded DNAs, they probably do touch DNAs directly in the context of the full assembly ([38] and this work).
The CST complex can unfold higher order G-tail structures
Higher order G-tail structures can inhibit the action of telomerase and other nucleic acid enzymes, and the propensity of C. glabrata G-tails for G-quadruplex formation is well documented [27], [28]. Previous investigations suggest that one function of G-tail binding proteins such as Pot1 and Cdc13 is to unfold such structures to facilitate important telomere transactions [27], [29], [30]. However, all such earlier studies utilized individual G-tail binding proteins rather than the more physiologically relevant complexes. Here we show that the C. glabrata CST complex can indeed unfold higher order G-tail structures as judged by single-molecule FRET analysis. The near complete loss of DNA FRET signals upon protein binding, together with the inverse relationship between FRET efficiencies and G-tail lengths, suggests that the G-tail may adopt a relatively extended conformation after the loading of CST complex. In this respect, the yeast G-tail coated by CST may be structurally distinct from the mammalian G-tail coated by the corresponding binding complex known as POT1-TPP1, which appears to form a compact and potentially more ordered structure [39]. Yeast telomere G-tails are known to undergo S-phase specific lengthening due to the resection of the complementary C-strand [40], [41]. The long G-tails generated during the S-phase presumably have a strong propensity to form folded structures that may pose a challenge for nucleic acid enzymes. Indeed, helicases such as Pif1 and Sgs1 have been proposed to help the cell unwind problematic G-quadruplex structures [42], [43]. The significance of the G-tail unfolding activity of the CST complex in relation to quadruplex-unwinding helicases, and whether there is coordination between these activities are important issues worthy of further investigation.
C. glabrata Cdc13 and CST can bind non-telomeric single-stranded DNA with moderate affinity
One of our surprising observations is the ability of Cdc13 and CST to form unstable complexes with non-telomeric DNA. Even though in competition assays, the protein and the complex exhibited at least 200-fold preference for the cognate telomere repeat, they are able to bind with moderate affinity to some non-telomeric DNAs of similar sizes in the absence of the preferred target sequence. This observation recalls analysis of other OB fold proteins such as the ciliate TEBP, which was found to be capable of using different molecular contacts to bind non-cognate target DNAs [44]. The recognition mechanisms of C. glabrata CST for telomeric and non-telomeric DNAs are likely to be distinct given the dramatic differences in target sequences and in the stability of the complexes. It is tempting to speculate that the non-telomeric DNA-binding activity of the C. glabrata CST complex may allow it to mediate non-telomeric functions. Hints that yeast CST may mediate non-telomeric functions came from the observation that over-expression of Stn1 engenders HU sensitivity and checkpoint defects [45]. Evidence for non-telomeric activities of the mammalian complex is even stronger. For example, there is incomplete co-localization of mammalian CST and telomere DNA in the nucleus, and knockdown of CTC1 results in DNA-damage foci both at and away from telomeres [10], [11]. In addition, the human disease(s) caused by CTC1 mutations (known as Coats plus or CRMCC) has a phenotypic spectrum that is distinct from the prototypical telomere diseases such as dyskeratosis congenita [46], [47]. Finally, the human CST complex was recently implicated in promoting replication re-start after replication stress [17]. Along this line, it is worth noting that the mammalian CST complex has been shown to bind long ssDNA of non-telomere sequences [10], [14]. Our discovery of a non-telomeric DNA binding activity of the CgCST complex suggests a greater degree of mechanistic conservation of the yeast and mammalian complex than previously realized.
Materials and Methods
Construction and growth of Candida strains
The C. albicans laboratory strain BWP17 (ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG) and the stn1 null derivative have been described previously [6], [48]. To analyze the functions of CaStn1 DNA-binding mutants in vivo, the relevant mutations were introduced into the integrating plasmid pGEM-URA3-STN1-GSCP (containing an epitope tagged and functional STN1 allele and the URA3 selectable marker) by QuikChange mutagenesis [6]. The resulting plasmids were linearized by Hpa I digestion and then used to transform the C. albicans stn1 null mutant. Correct integration of the linearized DNA in the transformants was confirmed by Southern analysis.
Telomere analyses
The telomere length analysis was performed as previously described [49].
Expression and purification of CST subunits and complexes
The following plasmids were constructed in order to express C. glabrata CST subunits and complexes: pSMT3-CgCDC13-FG, encoding a HIS6-SUMO-CgCdc13-FLAG fusion protein; pACYC1-DUET-CgCDC13-FG, encoding a SUMO-CgCdc13-FLAG fusion; pSMT3-CgSTN1, encoding a HIS6-SUMO-CgStn1 fusion; and pGEX-6P1-CgTEN1, encoding a GST-Ten1 fusion. Individual plasmids or combinations thereof were transformed into BL21(DE3), and the strains were cultured in LB and induced for protein expression with IPTG as described earlier [26]. Following induction, extracts were prepared and the expressed protein or protein complexes purified by appropriate combinations of affinity chromatography as follows: HIS6-SUMO-CgCdc13-FG, Ni-NTA-agarose and M2-agarose columns; the CST complex, Ni-NTA-agarose and M2-agarose columns; the Stn1-Ten1 (ST) sub-complex, Ni-NTA-agarose and glutathione-Sepharose columns. The Ni-NTA and Glutathione-Sepharose chromatography procedures were performed as previously described. For purification on the M2 resin, the starting extracts or fractions were adjusted to the composition of the FLAG binding buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, 0.1% NP-40, and 10% Glycerol) and incubated with 1/10 to 1/20 vol. of M2-Agarose resin at 4°C on a rotator for 3 h. The resin was then washed 5 times with the FLAG binding buffer, and eluted with FLAG binding buffer containing 0.2 mg/ml 3XFLAG peptide.
For the analysis of C. albicans Stn1N-Ten1 complex, PCR fragments encoding CaStn1N (amino acid 1–239) and CaTen1 were inserted into the pSMT3 and pGEX-4T2 vectors to allow for the expression of HIS6-SUMO-CaStn1N and GST-CaTen1 fusion proteins, respectively. The complex was synthesized in E. coli and isolated by tandem affinity chromatography as described above for the C. glabrata Stn1-Ten1 complex. The roles of the K98 and K170 residues of Stn1 in DNA-binding were tested by mutating the corresponding codons in pSMT3-CaSTN1N to either Ala or Glu. Mutant complexes were made and analyzed using the same method as that described for the wild type complex.
Gel electrophoretic mobility shift analysis
Binding reactions contained 10 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM spermidine, 1 mM DTT, 10% Glycerol, 7.5 nM oligonucleotide probe (∼80,000 c.p.m), and the indicated concentrations of purified proteins in 20 µl total volume. Following incubation at 22°C for 20 m, the reaction mixtures were electrophoresed through a 5% nondenaturing polyacrylamide gel (acrylamide/bis-acrylamide 44 : 1) to resolve the free probe from the DNA-protein complex. Binding activity was analyzed using a Typhoon PhosphorImager and the ImageQuant software (GE Healthcare).
UV crosslinking assay
Crosslinking reactions were conducted in 12 µl mixtures that included 50 mM Hepes-NaOH, pH 7.5, 10% glycerol, 30 mM K-acetate, 2 mM Mg-acetate, 1 mM DTT, 2 nM 5′ P32-labeled oligonucleotide containing a single Iodo-dU substitution (∼50,000 c.p.m.), and indicated concentrations of proteins. The binding between the proteins and the oligonucleotide was allowed to proceed at 22°C for 10 min, followed by 20 min of UV irradiation (Model UVM-57, UVP Inc.) on ice. The conjugates were separated in SDS-PAGE (10–11%) and detected by PhosphorImager analysis.
Single-molecule FRET experiments
The oligonucleotides used in the FRET experiments are listed in Table 1. The amine-modified thymine (/iAmMC6T/) shown in the sequence enables the oligonucleotides to be labeled with the monofunctional NHS ester form of Cy3 dyes (GE Healthcare)./Cy5/represents the Cy5 dye that was inserted directly to the DNA backbone using phosphoramidite chemistry. The partial duplex DNA substrates (18 bp dsDNA) with single-stranded tails carrying fluorescence dyes were generated by annealing ∼5 µM of biotinylated strand and ∼7 µM of the longer strand in 10 mM Tris-HCl (pH 8.0) and 50 mM NaCl, followed by slow cooling from 90°C to room temperature for ∼2 hours.
All single-molecule measurements were performed at 22±1°C. 50–100 pM of the annealed DNA substrates were immobilized on a quartz slide surface which is coated with polyethyleneglycol (mPEG-SC, Laysan Bio) in order to eliminate nonspecific surface adsorption of proteins [50], [51]. The immobilization was mediated by biotin-Neutravidin binding between biotinylated DNA, Neutravidin (Pierce), and biotinylated polymer (Bio-PEG-SC, Laysan Bio). Next, an imaging buffer containing 20 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 100 mM NaCl, 1 mM DTT, 0.1 mg/ml BSA, 2% (v/v) glycerol, 0.5% (w/v) D-glucose, 165 U/ml glucose oxidase, 2170 U/ml catalase, 3 mM Trolox and with desired CST or Cdc13 concentrations was directly added and incubated with the surface-tethered DNA substrates for 5 min before data acquisition. A total internal reflection fluorescence (TIRF) microscope previously described [32] was used for data acquisition. Finally, the same imaging buffer (but with no protein) was used to flush out the excess unbound protein from solution for another round of data acquisition. Single-molecule FRET-time traces was recorded with a time resolution of 30 ms and the FRET histograms were generated by averaging for 300 ms. To study the monovalent ion induced G-tail structures for the 48-nt three telomere repeats, desired concentration of KCl was used to replace 3 mM MgCl2 and 100 mM NaCl in the imaging buffer.
Homology modeling
The homology model of CaStn1N was calculated using the I-Tasser server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) with 3KF8.pdb (the structure file for CtStn1N) specified as the template. The resulting model was visualized and analyzed in Pymol (http://pymol.sourceforge.net/index.html).
Supporting Information
Zdroje
1. JainD, CooperJP (2011) Telomeric strategies: means to an end. Annu Rev Genet 44 : 243–269.
2. de LangeT (2009) How telomeres solve the end-protection problem. Science 326 : 948–952.
3. O'SullivanRJ, KarlsederJ (2010) Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11 : 171–181.
4. Lundblad V (2006) Budding yeast telomeres. In: de Lange T, Lundblad V, Blackburn E, editors. Telomeres and Telomerase. 2nd ed: Cold Spring Harbor Laboratory Press. pp. 345–386.
5. GaoH, CervantesRB, MandellEK, OteroJH, LundbladV (2007) RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14 : 208–214.
6. SunJ, YuEY, YangY, ConferLA, SunSH, et al. (2009) Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev 23 : 2900–2914.
7. Giraud-PanisMJ, TeixeiraMT, GeliV, GilsonE (2010) CST meets shelterin to keep telomeres in check. Mol Cell 39 : 665–676.
8. GelinasAD, PaschiniM, ReyesFE, HerouxA, BateyRT, et al. (2009) Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci U S A 106 : 19298–19303.
9. MartinV, DuLL, RozenzhakS, RussellP (2007) Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci U S A 104 : 14038–14043.
10. MiyakeY, NakamuraM, NabetaniA, ShimamuraS, TamuraM, et al. (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36 : 193–206.
11. SurovtsevaYV, ChurikovD, BoltzKA, SongX, LambJC, et al. (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36 : 207–218.
12. RaffaGD, RaimondoD, SorinoC, CugusiS, CenciG, et al. (2010) Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev 24 : 1596–1601.
13. PardueML, DebaryshePG (2011) Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A 108 : 20317–20324.
14. ChenLY, RedonS, LingnerJ (2012) The human CST complex is a terminator of telomerase activity. Nature 488 : 540–544.
15. WuP, TakaiH, de LangeT (2012) Telomeric 3′ Overhangs Derive from Resection by Exo1 and Apollo and Fill-In by POT1b-Associated CST. Cell 150 : 39–52.
16. HuangC, DaiX, ChaiW (2012) Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res doi: 10.1038/cr.2012.132. [Epub ahead of print].
17. StewartJA, WangF, ChaikenMF, KasbekC, ChastainPD2nd, et al. (2012) Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J 31 : 3537–3549.
18. SunJ, YangY, WanK, MaoN, YuTY, et al. (2011) Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res 21 : 258–274.
19. Mitton-FryRM, AndersonEM, TheobaldDL, GlustromLW, WuttkeDS (2004) Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol 338 : 241–255.
20. PennockE, BuckleyK, LundbladV (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: p387–396.
21. ChanA, BouleJB, ZakianVA (2008) Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet 4: e1000236 doi:10.1371/journal.pgen.1000236.
22. WuY, ZakianVA (2011) The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci U S A 108 : 20362–20369.
23. PetreacaRC, ChiuHC, EckelhoeferHA, ChuangC, XuL, et al. (2006) Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol 8 : 748–755.
24. GrossiS, PuglisiA, DmitrievPV, LopesM, ShoreD (2004) Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev 18 : 992–1006.
25. NakaokaH, NishiyamaA, SaitoM, IshikawaF (2011) Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 287 : 619–627.
26. YuEY, SunJ, LeiM, LueNF (2012) Analyses of Candida Cdc13 orthologues revealed a novel OB fold dimer arrangement, dimerization-assisted DNA binding, and substantial structural differences between Cdc13 and RPA70. Mol Cell Biol 32 : 186–198.
27. WangH, NoraGJ, GhodkeH, OpreskoPL (2011) Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J Biol Chem 286 : 7479–7489.
28. TranPL, MergnyJL, AlbertiP (2011) Stability of telomeric G-quadruplexes. Nucleic Acids Res 39 : 3282–3294.
29. LinYC, ShihJW, HsuCL, LinJJ (2001) Binding and partial denaturing of G-quartet DNA by Cdc13p of Saccharomyces cerevisiae. J Biol Chem 276 : 47671–47674.
30. ZaugAJ, PodellER, CechTR (2005) Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A 102 : 10864–10869.
31. HaT, EnderleT, OgletreeDF, ChemlaDS, SelvinPR, et al. (1996) Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A 93 : 6264–6268.
32. RoyR, HohngS, HaT (2008) A practical guide to single-molecule FRET. Nat Methods 5 : 507–516.
33. HaT, KozlovAG, LohmanTM (2012) Single-molecule views of protein movement on single-stranded DNA. Annu Rev Biophys 41 : 295–319.
34. LeeJY, OkumusB, KimDS, HaT (2005) Extreme conformational diversity in human telomeric DNA. Proc Natl Acad Sci U S A 102 : 18938–18943.
35. LeeJY, YoonJ, KihmHW, KimDS (2008) Structural diversity and extreme stability of unimolecular Oxytricha nova telomeric G-quadruplex. Biochemistry 47 : 3389–3396.
36. Mitton-FryR, AndersonE, HughesT, LundbladV, WuttkeD (2002) Conserved structure for single-stranded telomeric DNA recognition. Science 296 : 145–147.
37. BochkarevaE, KorolevS, Lees-MillerSP, BochkarevA (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 21 : 1855–1863.
38. SalasTR, PetrusevaI, LavrikO, SaintomeC (2009) Evidence for direct contact between the RPA3 subunit of the human replication protein A and single-stranded DNA. Nucleic Acids Res 37 : 38–46.
39. TaylorDJ, PodellER, TaatjesDJ, CechTR (2011) Multiple POT1-TPP1 proteins coat and compact long telomeric single-stranded DNA. J Mol Biol 410 : 10–17.
40. LarriveeM, Le BelC, WellingerR (2004) The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18 : 1391–1396.
41. BonettiD, MartinaM, ClericiM, LucchiniG, LongheseMP (2009) Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35 : 70–81.
42. AzamM, LeeJY, AbrahamV, ChanouxR, SchoenlyKA, et al. (2006) Evidence that the S.cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res 34 : 506–516.
43. PaeschkeK, CapraJA, ZakianVA (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145 : 678–691.
44. TheobaldDL, SchultzSC (2003) Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J 22 : 4314–4324.
45. GasparyanHJ, XuL, PetreacaRC, RexAE, SmallVY, et al. (2009) Yeast telomere capping protein Stn1 overrides DNA replication control through the S phase checkpoint. Proc Natl Acad Sci U S A 106 : 2206–2211.
46. AndersonBH, KasherPR, MayerJ, SzynkiewiczM, JenkinsonEM, et al. (2012) Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 44 : 338–342.
47. PolviA, LinnankiviT, KivelaT, HervaR, KeatingJP, et al. (2012) Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet 90 : 540–549.
48. EnloeB, DiamondA, MitchellA (2000) A single-transformation gene function test in diploid Candida albicans. J Bacteriol 182 : 5730–5736.
49. YuEY, YenWF, Steinberg-NeifachO, LueNF (2010) Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol Cell Biol 30 : 1254–1268.
50. HaT, RasnikI, ChengW, BabcockHP, GaussGH, et al. (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419 : 638–641.
51. ZhouR, KozlovAG, RoyR, ZhangJ, KorolevS, et al. SSB functions as a sliding platform that migrates on DNA via reptation. Cell 146 : 222–232.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



