-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
The coordination of cell proliferation and cell fate determination is critical during development but the mechanisms through which this is accomplished are unclear. We present evidence that the Snail-related transcription factor CES-1 of Caenorhabditis elegans coordinates these processes in a specific cell lineage. CES-1 can cause loss of cell polarity in the NSM neuroblast. By repressing the transcription of the BH3-only gene egl-1, CES-1 can also suppress apoptosis in the daughters of the NSM neuroblasts. We now demonstrate that CES-1 also affects cell cycle progression in this lineage. Specifically, we found that CES-1 can repress the transcription of the cdc-25.2 gene, which encodes a Cdc25-like phosphatase, thereby enhancing the block in NSM neuroblast division caused by the partial loss of cya-1, which encodes Cyclin A. Our results indicate that CDC-25.2 and CYA-1 control specific cell divisions and that the over-expression of the ces-1 gene leads to incorrect regulation of this functional ‘module’. Finally, we provide evidence that dnj-11 MIDA1 not only regulate CES-1 activity in the context of cell polarity and apoptosis but also in the context of cell cycle progression. In mammals, the over-expression of Snail-related genes has been implicated in tumorigenesis. Our findings support the notion that the oncogenic potential of Snail-related transcription factors lies in their capability to, simultaneously, affect cell cycle progression, cell polarity and apoptosis and, hence, the coordination of cell proliferation and cell fate determination.
Published in the journal: Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail. PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003884
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003884Summary
The coordination of cell proliferation and cell fate determination is critical during development but the mechanisms through which this is accomplished are unclear. We present evidence that the Snail-related transcription factor CES-1 of Caenorhabditis elegans coordinates these processes in a specific cell lineage. CES-1 can cause loss of cell polarity in the NSM neuroblast. By repressing the transcription of the BH3-only gene egl-1, CES-1 can also suppress apoptosis in the daughters of the NSM neuroblasts. We now demonstrate that CES-1 also affects cell cycle progression in this lineage. Specifically, we found that CES-1 can repress the transcription of the cdc-25.2 gene, which encodes a Cdc25-like phosphatase, thereby enhancing the block in NSM neuroblast division caused by the partial loss of cya-1, which encodes Cyclin A. Our results indicate that CDC-25.2 and CYA-1 control specific cell divisions and that the over-expression of the ces-1 gene leads to incorrect regulation of this functional ‘module’. Finally, we provide evidence that dnj-11 MIDA1 not only regulate CES-1 activity in the context of cell polarity and apoptosis but also in the context of cell cycle progression. In mammals, the over-expression of Snail-related genes has been implicated in tumorigenesis. Our findings support the notion that the oncogenic potential of Snail-related transcription factors lies in their capability to, simultaneously, affect cell cycle progression, cell polarity and apoptosis and, hence, the coordination of cell proliferation and cell fate determination.
Introduction
Members of the Snail superfamily of zinc-finger transcription factors are essential during development and their deregulation has been implicated in various malignancies including tumorigenesis [1]–[4]. One of the best known functions of Snail-related proteins is their role in the induction of epithelial-mesenchymal transitions (EMTs) [1], [2], [4], [5]. EMTs are fundamentally important for normal development and, in particular, for processes such as mesoderm formation, gastrulation and neural tube formation. EMTs are also important for tumorigenesis since they are responsible for the invasive behavior of certain types of tumor cells [1], [2], [5]. Hallmarks of EMTs are the loss of apico-basal polarity and adhesive properties, which is critical for the ability of epithelial cells to become migratory. Snail-related proteins contribute to these cellular changes by repressing the transcription of genes that encode factors required for apico-basal polarity and cell adhesion, such as Crumbs and E-cadherin, respectively [6]–[8].
Snail-related proteins have additional cellular functions that can operate independently of the induction of EMT. In Drosophila melanogaster, for example, the Snail family members Snail, Worniu and Escargot are important for both the cell polarity of neuroblasts and the ability of these cells to divide [9], [10]. Snail, Worniu and Escargot are required for the polarity of embryonic neuroblasts because they promote the expression of the gene inscuteable, which encodes an adaptor protein that, by forming a physical link between the proteins Par3 and Pins, is thought to connect cell polarity to spindle position [10]–[12]. In the case of the division of neuroblasts, Snail, Worniu and Escargot are thought to enhance cell cycle progression by promoting the expression of the gene cdc25string, which encodes a Cdc25 phosphatase homolog required for the removal of inhibitory phosphates on Cyclin-dependent kinases (CDKs) and, hence, CDK activation [10], [13], [14]. However, whether the effect of Snail, Worniu and Escargot on cdc25string is direct or indirect remains to be determined. In mammals, Snail-related proteins have also been shown to regulate cell proliferation [4]. Specifically, a reduced rate of cell proliferation is observed in cultured epithelial cells transfected with Snail1 (formerly referred to as ‘Snail’) and in regions of the mouse embryo that express endogenous Snail1 [15]. The inhibitory effect of Snail1 expression on cell proliferation is due to the ability of the Snail1 protein to directly repress the transcription of the cyclin D2 gene, which is required for the G1 to S phase transition [15]. In the same study, an inverse correlation was also found between Snail1 expression and apoptosis in the mouse embryo, suggesting that Snail1 can repress apoptosis. Additional evidence that Snail-related transcription factors can repress apoptosis in mammals comes from studies on radiation-induced apoptosis in hematopoietic precursor cells. Snail2 (formerly referred to as ‘Slug’) was found to block apoptosis by repressing the transcription of the pro-apoptotic BH3-only gene puma [16].
The ability of Snail-related transcription factors to block apoptosis was initially discovered in Caenorhabditis elegans and during the analysis of the NSM (NSM, neuro-secretory motoneuron) lineages (Two bilaterally symmetric NSM lineages exist, the left and right NSM lineage). About 410 min after the 1st division of the embryo (referred throughout the manuscript as “1st round of division”), the two NSM neuroblasts (which are generated about 280 min after the 1st division) divide asymmetrically along the ventral-lateral dorsal-medial axis to each generate two daughter cells of different sizes and different cell fates, the larger NSM, which survives and differentiates into a serotonergic neuro-secretory motorneuron, and the smaller NSM sister cell, which undergoes apoptosis and forms a cell corpse about 30 min after the completion of the NSM neuroblast division [17], [18]. A dominant gain-of-function (gf) mutation of the ces-1 (ces, cell-death specification) gene, which encodes a Snail-related protein, was found to block the death of the NSM sister cells and the I2 sister cells [19], [20]. Otherwise, ces-1(n703gf) animals are indistinguishable from wild-type animals at least at the dissecting microscope level. This ces-1 gf mutation affects a regulatory region of the ces-1 locus, which is likely to results in the over-expression of the ces-1 gene in specific lineages, including the NSM lineage [20]. In ces-1 gf mutants, the CES-1 protein blocks the death of the NSM sister cells by binding to a cis-acting element of the BH3-only gene egl-1 (egl, egg-laying defective), thereby preventing the HLH-2/HLH-3 - (HLH, basic helix-loop-helix transcription factor) dependent activation of egl-1 transcription [21]. Interestingly, the ces-1 gf mutation also affects the cell polarity of the NSM neuroblast. Specifically, in ces-1 gf mutant animals, the NSM neuroblast divides symmetrically along randomly selected axes rather than dividing asymmetrically along the ventral-lateral dorsal-medial axis [17]. The same polarity defect is observed in animals that lack a functional ces-2 or dnj-11 (dnj, DnaJ domain) gene, which encode a HLF-like bZIP transcription and a MIDA1-like chaperone, respectively, and which act upstream of ces-1 to repress ces-1 transcription in the NSM neuroblast [17], [19], [20], [22]. Furthermore, in a wild-type background, expression from a functional Pces-1ces-1::yfp construct is detected only in the larger NSM daughter that is destined to survive (the NSM); however, in a ces-2 or dnj-11 mutant background expression of this construct is detected in the NSM neuroblast as well as both daughter cells [17]. Based on these findings it has been proposed that in the NSM neuroblast, CES-2 and DNJ-11 maintain ces-1 expression below a certain level and that this is important for the establishment and/or maintenance of NSM neuroblast polarity and the ability of the NSM neuroblast to divide asymmetrically. After the NSM neuroblast divides, CES-1 protein is restricted to the NSM, where it acts as a direct repressor of egl-1 transcription and hence, apoptosis. In ces-2, dnj-11 or ces-1 gf mutant animals, the level of CES-1 protein in the NSM neuroblast is elevated and this leads to the symmetric, random division of the NSM neuroblast. This results in the formation of two daughters of similar sizes, both of which contain CES-1 protein and, consequently, survive [17], [23].
We now demonstrate that CES-1 has an additional function in the NSM lineage. Specifically, we present evidence that CES-1 can also regulate cell cycle progression in the NSM neuroblast by functionally interacting with core components of the cell cycle machinery. By simultaneously controlling cell cycle progression, cell polarity and apoptosis, the Snail-related transcription factor CES-1 plays a crucial role in the coordination of cell proliferation and cell fate specification in the NSM lineage.
Results
In ces-1(n703gf); bc416 animals, the division of the NSM neuroblast is blocked
Wild-type larvae carrying the Ptph-1his-24::gfp reporter (tph, tryptophane hydroxylase; his, histone structural gene), which is specifically expressed in serotonergic neurons (and labels the nuclei of these neurons) [24], have two GFP-positive neurons in the anterior pharynx, the left and right NSM (Figure 1A, +/+). In animals carrying a gf mutation of ces-1, n703, the NSM neuroblast divides symmetrically, resulting in two daughter cells of similar sizes, both of which survive [17], [19]. Therefore, ces-1(n703gf) larvae carrying the Ptph-1his-24::gfp reporter have four GFP-positive neurons in the head region, the left and right NSM and the left and right ‘undead’ NSM sister cell (Figure 1A, ces-1(n703gf)). To identify targets of the CES-1 protein involved in the asymmetric division of the NSM neuroblast, we performed a ces-1(n703gf) suppressor screen using the Ptph-1his-24::gfp reporter as a tool. Specifically, we screened mutagenized ces-1(n703gf) animals for mutations that cause a reduction in the number of GFP-positive NSMs and undead NSM sister cells (i.e. less than four GFP-positive cells in the anterior pharynx). Using this approach, we isolated the mutation bc416. At 15°C, 100% of ces-1(n703gf) larvae have four GFP-positive cells. In contrast, only 5% of ces-1(n703gf) larvae homozygous for bc416 (ces-1(n703gf); bc416) have four GFP-positive cells (Table 1). The bc416 mutation is recessive and does not show maternal rescue (data not shown; Table S1).
Fig. 1. ces-1(n703gf); cya-1(bc416) affects the division of the NSM neuroblast. 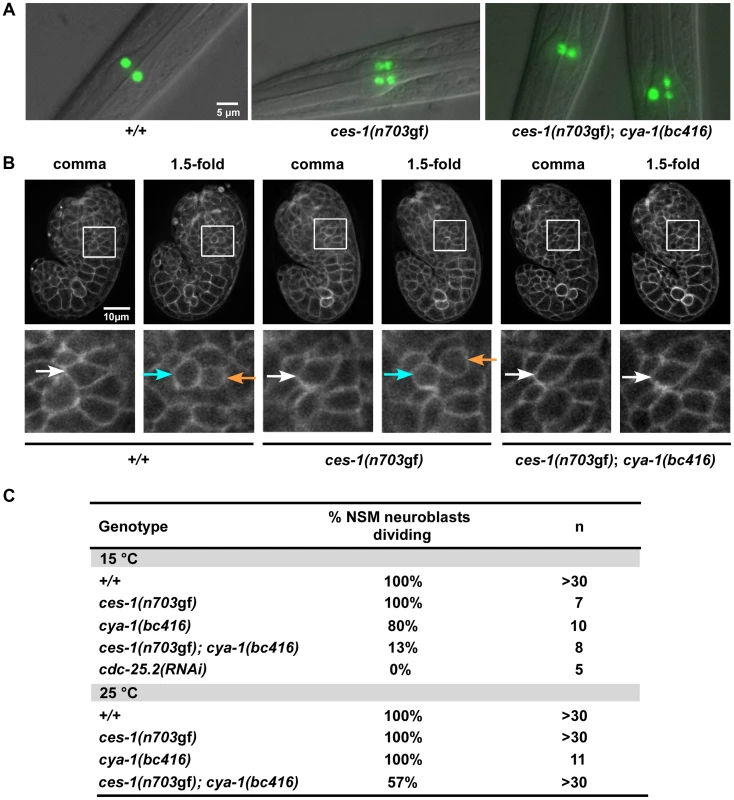
(A) The presence of NSMs, undead NSM sister cells, and non-dividing NSM neuroblasts was analyzed in L3, L4 larvae using the reporter Ptph-1his-24::gfp (bcIs66). All strains analyzed were homozygous for bcIs66. Epifluorescence images overlaid with DIC. (B) The NSM neuroblast division was analyzed in embryos using the reporter Ppie-1mCherry::phPLC1δ (ltIs44) or Ppie-1gfp::phPLC1δ (ltIs38). Epifluorescence images were taken before (‘comma’) and after the NSM neuroblast division (‘1.5-fold’). In the case of ces-1(n703gf); cya-1(bc416), the NSM neuroblasts had not divided at the time the analysis had to be terminated due to the beginning of muscle twitching (around the 2-fold stage). White arrows point to the NSM neuroblasts, blue and orange arrows point to the NSMs and NSM sister cells, respectively. All strains analyzed were homozygous for ltIs44 and bcIs66, except ces-1(n703gf), which was homozygous for ltIs38 and bcIs66. (C) Quantification of the percentage of NSM neuroblasts dividing in wild-type, ces-1(n703gf), ces-1(n703gf); cya-1(bc416), cya-1(bc416) and cdc-25.2(RNAi) embryos. cdc-25.2(RNAi) was performed by injection. n indicates the number of NSM neuroblasts analyzed. Tab. 1. ces-1(n703gf); cya-1(bc416) affects the number of ‘NSM-like’ cells. 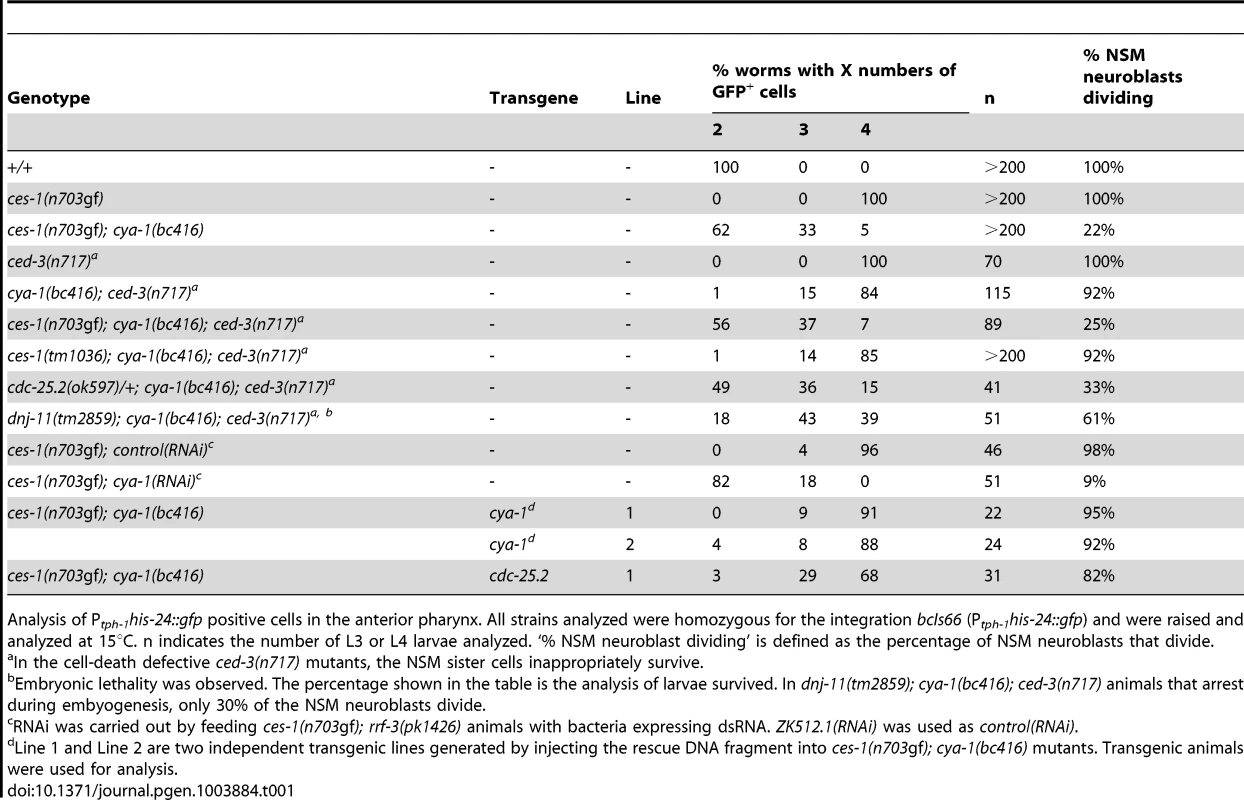
Analysis of Ptph-1his-24::gfp positive cells in the anterior pharynx. All strains analyzed were homozygous for the integration bcIs66 (Ptph-1his-24::gfp) and were raised and analyzed at 15°C. n indicates the number of L3 or L4 larvae analyzed. ‘% NSM neuroblast dividing’ is defined as the percentage of NSM neuroblasts that divide. At 15°C, 33% of ces-1(n703gf); bc416 larvae have three GFP-positive cells (Table 1). Interestingly, based on Ptph-1his-24::gfp labeling, in these animals, one nucleus is larger than the other two nuclei (Figure 1A, ces-1(n703gf); bc416). This phenomenon is observed at a high frequency. Based on this observation, we hypothesized that instead of suppressing the inappropriate survival of NSM sister cells in ces-1(n703gf) animals, the bc416 mutation might affect the division of the NSM neuroblasts. To test this, we directly analyzed the division of the NSM neuroblasts in ces-1(n703gf); bc416 embryos. To that end, we used a transgene that expresses a plasma membrane-targeted mCherry fusion protein as a tool [25]. We identified the NSM neuroblasts based on their positions during the comma stage of embryogenesis and tracked their fates until the 2-fold stage, which is the stage during which in wild-type animals, the NSM neuroblasts complete their division and the NSM sister cells undergo apoptosis [17], [18]. We found that at 15°C, only 13% of the NSM neuroblasts divide in ces-1(n703gf); bc416 embryos (Figure 1B, C). To rule out the possibility that the division of the NSM neuroblasts in ces-1(n703gf); bc416 animals is delayed rather than blocked, we scored ces-1(n703gf); bc416 animals in the background of the ced-3 loss-of-function mutation n717, which causes a general block in apoptosis [26]. If the NSM neuroblasts divided in late embryos or in larvae and the resulting NSM sister cells underwent apoptosis, using Ptph-1his-24::gfp as a tool, we should observed an increased number of animals with four GFP-positive cells in the ced-3(n717) background. However, we found that the percentage of animals with two, three, or four GFP-positive cells is not affected by ced-3(n717) (Table 1, ces-1(n703gf); bc416; ced-3(n717)). Therefore, the reduction in GFP-positive cells observed in ces-1(n703gf); bc416 larvae is the result of a failure of the NSM neuroblasts to divide. For this reason, we are presenting the data acquired in larvae using Ptph-1his-24::gfp not only in the form of ‘% animals with two, three, or four GFP-positive cells’ but also as ‘% NSM neuroblasts dividing’, which is defined as the percentage of the NSM neuroblasts that divide (Table 1).
The NSM neuroblast division is blocked between S phase and M phase
To further examine the cell cycle defect in ces-1(n703gf); bc416 animals, we determined the relative DNA content in non-dividing NSM neuroblasts. We labeled DNA in ces-1(n703gf); bc416 animals with the fluorescent dye DAPI and measured fluorescence intensity in non-dividing NSM neuroblasts [27], [28], in NSMs and in undead NSM sister cells. We found that in ces-1(n703gf); bc416 mutants with three GFP-positive cells, the average DNA content of the cells with the larger nuclei (presumably the non-dividing NSM neuroblasts) is 1.8 times greater than the average DNA content of the cells with the smaller nuclei (presumably the NSMs and undead NSM sister cells) (Figure S1). Furthermore, in ces-1(n703gf); bc416 animals with two GFP-positive cells, the average DNA content of both cells is about 2-fold higher than that of control pharyngeal muscle cells (data not shown). Taken together, these observations suggest that in ces-1(n703gf); bc416 animals, the NSM neuroblasts complete DNA replication but fail to undergo mitosis. Hence, in this mutant background, the NSM neuroblasts arrest between S phase and M phase. Interestingly, just like NSMs, the non-dividing, tetraploid NSM neuroblasts express the Ptph-1his-24::gfp reporter during larval stages, which suggests that they differentiate into serotonergic neurons.
ces-1(n703gf); bc416 causes embryonic lethality and sterility
Besides exhibiting a defect in NSM neuroblast division, ces-1(n703gf); bc416 animals have additional defects. When raised at 15°C or 25°C, 8% or 76% of ces-1(n703gf); bc416 animals, respectively, exhibit an embryonic lethal (Emb) phenotype and arrest at the elongation stage during embryogenesis (Figure 2A, B). Arrested embryos have multiple defects in hypodermal morphogenesis (Figure 2B). Since the Emb phenotype is temperature sensitive, we performed temperature-shift experiments to define the temperature-sensitive period (TSP) of ces-1(n703gf); bc416 animals. Embryos were shifted from 25°C to 15°C or vice versa at different stages during embryonic development, and viability was assessed 24 h to 48 h later. As shown in Figure S2, the TSP of ces-1(n703gf); bc416 animals lies between the 50-cell stage and the comma stage of embryogenesis. Therefore, at least in the ces-1(n703gf) mutant background, the gene defined by the bc416 mutation is essential and its activity required for embryonic development between the 50-cell stage and the comma stage. Furthermore, animals that escape embryonic lethality and hatch also display morphological abnormalities (Figure 2C). Finally, when raised at 25°C, about 30% of ces-1(n703gf); bc416 animals that develop into adults are sterile (Ste phenotype) (data not shown) and the average brood size of the fertile ces-1(n703gf); bc416 adults is smaller than that of wild-type adults (Table S2).
Fig. 2. ces-1(n703gf); cya-1(bc416) causes temperature-sensitive embryonic lethality. 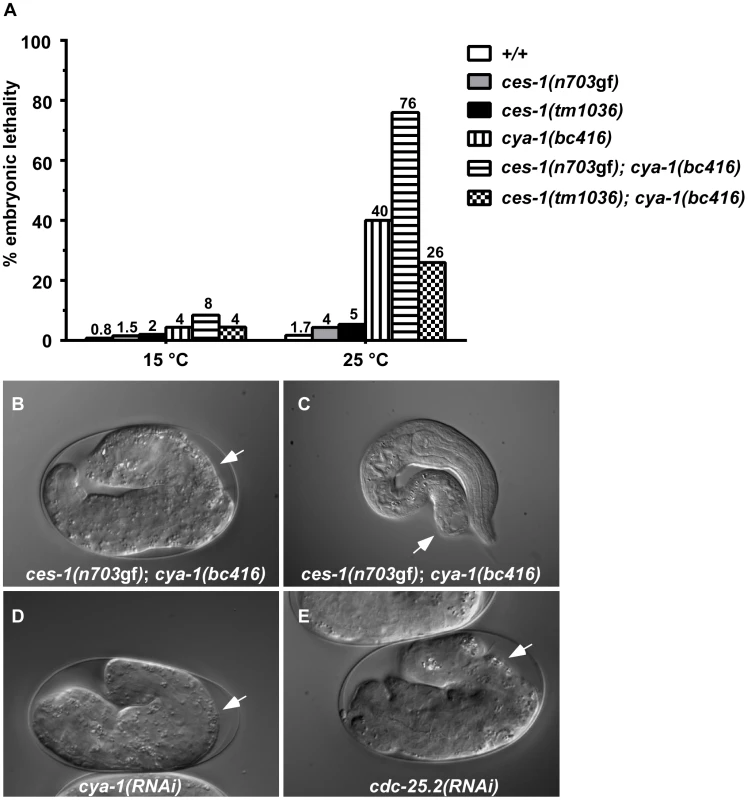
(A) The percentages of embryonic lethality at 15°C and 25°C. The numbers above the bars represent the percentage of embryonic lethality. For each genotype, around 1000 embryos were scored. DIC images of embryos arrested during the elongation stage of embryogenesis (B, D, E) or during the first larval stage (L1) (C) when grown at 25°C are shown. White arrows point to abnormalities in the hypodermis. All strains analyzed were homozygous for bcIs66. RNAi was performed by injection. ces-1(n703gf); bc416 mutants display cell division defects in the ABarp, C, and E lineages
Since a highly penetrant Emb phenotype was observed in ces-1(n703gf); bc416 animals raised at 25°C, we investigated whether cell divisions other than the divisions of the NSM neuroblasts are affected in these animals. A systematic analysis of all cell lineages using 4D lineage analysis [29] showed that cell division defects are not restricted to the NSM lineage. We found that at 25°C, the ABarp, C and E lineages are also affected in ces-1(n703gf); bc416 animals (Figure 3). All other lineages were not affected. ABarp is a major hypodermal precursor and the C founder cell generates additional posterior and dorsal hypodermal cells [18]. In the ABarp lineage, most cell divisions that give rise to ventrolateral ectoblasts (V1 to V6) are blocked (Figure 3). Furthermore, in the C lineage, many cell divisions that generate the embryonic large hypodermal syncytium (hyp7) fail to occur (Figure 3). The defects in the ABarp and C lineage most likely cause or contribute to the observed hypodermal abnormalities (Figure 2B, C). A failure in the formation of the hypodermis has previously been shown to cause embryonic lethality [30]. In addition, the phenotype of arrested ces-1(n703gf); bc416 embryos is similar to the phenotype of mutants with hypodermal defects [30]. Therefore, the cell division defects in the ABarp and C lineages observed in ces-1(n703gf); bc416 animals most probably cause the Emb phenotype exhibited by these animals. In addition, we identified variable defects in the E lineage. Specifically, some cell divisions of the 7th round of division during C. elegans embryogenesis do not occur in the E lineage (Figure 3). Based on these observations, we conclude that in the ces-1(n703gf) background and at 25°C, bc416 affects the divisions of cells other than the NSM neuroblasts. (The defects caused by bc416 in an otherwise wild-type background will be discussed below.)
Fig. 3. ces-1(n703gf); cya-1(bc416) blocks cell divisions in the ABarp, C and E lineages. 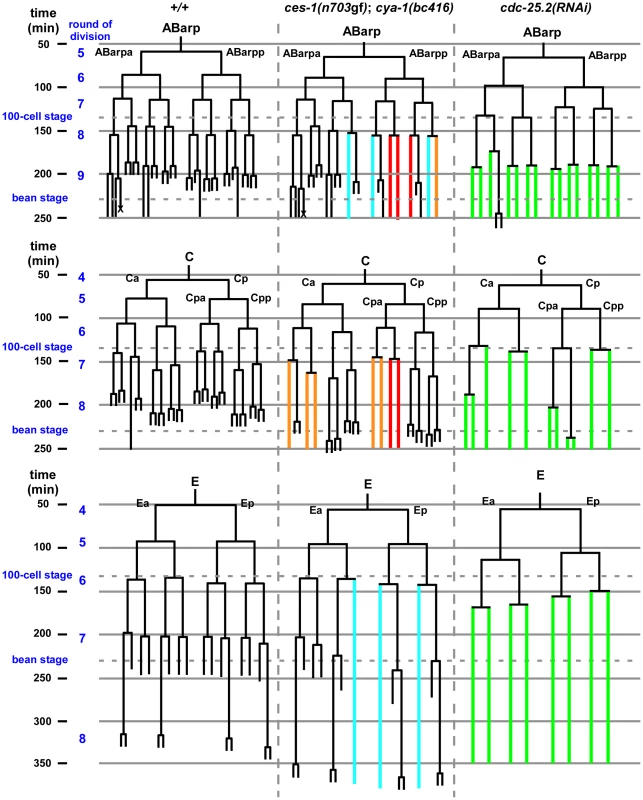
All strains analyzed were homozygous for bcIs66. Lineage analyses were performed for two (wild-type, +/+), three (ces-1(n703gf); cya-1(bc416)) and three (cdc-25.2(RNAi)) embryos raised at 25°C. The ABarp, C and E lineages are shown. Vertical axis indicates approximate time in min after the 1st round of embryonic division, in which P0 divides into AB and P1. In the case of ces-1(n703gf); cya-1(bc416), cell division defects observed in three out of three embryos are depicted in red, defects found in two out of three embryos are depicted in blue, and defects found in one out of three embryos are depicted in orange. In the case of cdc-25.2(RNAi), RNAi was carried out by injection. Since there is some variability of the RNAi effect, the lineage shown here was derived from the embryo with the strongest phenotype (cell division defects observed in this embryo are depicted in green), and the lineages from the other two cdc-25.2(RNAi) embryos are shown in Figure S6. The severe cell division defects in the ABarp, C and E lineages were seen in all three cdc-25.2(RNAi) embryos. The cell death in the ABarp lineage is labeled with the cross. The defects in the C lineage and ABarp lineage result in a defect in the formation of the hypodermis (the mitoses that generate hyp7, hyp5, hyp11, H0, H1, H2, V1, V2, V4, and V6 fail to occur). bc416 is a mutation in the C. elegans Cyclin A homolog, cya-1
The bc416 mutation was mapped genetically to a 900 kb region (between SNPs F22B7 : 15755 and ZK1098 : 19075) on LGIII using linkage analysis, three-factor mapping and SNP mapping (Figure S3A). In addition, we used Illumina deep sequencing technology to sequence the entire genome of ces-1(n703gf); bc416 animals. In the F22B7 : 15755 - ZK1098 : 19075 region, we found a G to A transition at the conserved 5′ splice-donor site of the first intron of the gene cya-1 (ZK507.6), which encodes one of two C. elegans Cyclin A homologs, CYA-1 (Figure S3B) [31].
A 4.3 kb genomic DNA fragment that contains the entire coding region of cya-1 rescues the NSM neuroblast division defect (Table 1; ces-1(n703gf); bc416 plus ‘cya-1’ transgene) and the Emb phenotype observed in ces-1(n703gf); bc416 animals (data not shown). Similar to bc416, partially reducing cya-1 function by RNA-mediated interference (RNAi) blocks 90% of the NSM neuroblast divisions in the ces-1(n703gf) mutant background (Table 1; ces-1(n703gf); cya-1(RNAi)). In addition, cya-1(RNAi) leads to embryonic lethality and the terminal phenotype of arrested embryos is similar to the terminal phenotype of arrested ces-1(n703gf); bc416 embryos (Figure 2D). (For the cya-1 RNAi experiment, sequences of exons 4 and 5 of cya-1 were used. Sequence alignments reveal that these two exons are highly homologous [≥60%] to cya-2, the second C. elegans cyclin A gene. For this reason, it is possible that cya-1(RNAi) also causes a decrease in cya-2 function.) Finally, a null mutation of the cya-1 gene, he153, which causes embryonic lethality, fails to complement bc416 in the ces-1(n703gf) mutant background (data not shown; S. van der Heuvel, personal communication). In conclusion, the gene defined by bc416 is identical to the cya-1 gene.
Since the accuracy of the 5′ splice-donor site is important for the recognition and removal of introns, we determined whether the bc416 mutation influences the splicing of the primary cya-1 transcript. Using reverse transcriptase PCR (RT-PCR), we found that, at both 15°C and 25°C, bc416 affects the splicing of the cya-1 gene and results in aberrantly spliced messages, in which parts of the first intron are retained (Figure S3D). The translation of these aberrant messages is predicted to result in the synthesis of a truncated, non-functional CYA-1 protein that includes only the first 12 amino acids of the full-length protein. Using quantitative real-time PCR (qPCR), next, we determined the level of correctly spliced, wild-type cya-1 transcript. We found that at both 15°C and 25°C, compared to wild-type (cya-1(+/+)) animals, the level of correctly spliced cya-1 transcript is reduced by about 50% in cya-1(bc416) animals (Figure S3E). These results suggest that bc416 affects the pre-mRNA splicing of the cya-1 gene, resulting in a reduction of correctly spliced mRNA and hence, presumably, full-length CYA-1 protein. Therefore, bc416 most likely represents a partial loss-of-function (lf) mutation of cya-1.
ces-1(n703gf) enhances the phenotypes caused by cya-1(bc416)
While performing cya-1 RNAi experiments, we noticed that cya-1 RNAi by injection results in a more penetrant Emb phenotype when performed in the ces-1(n703gf) background (64% embryonic lethality at 25°C) compared to the wild-type background (40% embryonic lethality at 25°C). To test whether the defects observed in ces-1(n703gf); cya-1(bc416) animals are dependent on the presence of the ces-1(n703gf) mutation, we analyzed bc416 in an otherwise wild-type background. Using the plasma membrane-targeted mCherry fusion protein as a tool, we found that at 15°C, 80% of the NSM neuroblasts divide in cya-1(bc416) embryos (Figure 1C). For comparison, only 13% of the NSM neuroblasts divide in ces-1(n703gf); cya-1(bc416) embryos. Similarly, using the Ptph-1his-24::gfp reporter as a tool, we found that 92% of NSM neuroblasts divide in cya-1(bc416) animals in the ced-3 mutant background (Table 1, cya-1(bc416); ced-3(n717)). For comparison, in ces-1(n703gf); cya-1(bc416) animals in the ced-3 mutant background only 25% of the NSM neuroblasts divide. Finally, we found that at 25°C, 40% of cya-1(bc416) animals arrest at the elongation stage during embryogenesis and therefore exhibit an Emb phenotype (Figure 2A). For comparison, 76% of ces-1(n703gf); cya-1(bc416) animals exhibited an Emb phenotype when raised at 25°C. Temperature-shift experiments revealed that the TSP of cya-1(bc416) animals lies between the 50-cell stage and the comma stage of embryogenesis and therefore is identical to the TSP of ces-1(n703gf); cya-1(bc416) animals (Figure S2). Lineage analysis revealed that cya-1(bc416) embryos have cell division defects in the ABarp, C and E lineages (Figure S4). However, cell division defects in the ABarp and C lineages were only observed in one out of three cya-1(bc416) embryos analyzed. For comparison, cell division defects in the ABarp and C lineages were observed in all three ces-1(n703gf); cya-1(bc416) embryos analyzed. Lineage analysis also revealed that the ces-1(n703gf) embryos have no cell division defects in the ABarp, C or E lineages (Figure S4). Therefore, ces-1(n703gf) increases the penetrance of the cell division defects caused by cya-1(bc416), especially in the ABarp and C lineages. This is consistent with the more penetrant Emb phenotype observed in ces-1(n703gf); cya-1(bc416) animals (76% embryonic lethality at 25°C, Figure 2). These findings demonstrate that ces-1(n703gf) enhances the NSM neuroblast division defect and the Emb phenotype caused by cya-1(bc416). Interestingly, ces-1(n703gf) does not enhance the defect in brood size caused by cya-1(bc416). When raised at 25°C, the brood size of both cya-1(bc416) animals and ces-1(n703gf); cya-1(bc416) animals is reduced by about 2.5-fold when compared to the brood size of wild-type animals (Table S2).
ces-1 over-expression reduces the relative expression level of cdc-25.2
Snail-related transcription factors are thought to predominantly act as repressors of transcription [1]. To determine the mechanism through which ces-1(n703gf) enhances cya-1(bc416), we therefore analyzed the expression of candidate target genes. The Drosophila melanogaster Snail family has been implicated in the control of the expression of the gene cdc25string, which encodes the D. melanogaster ortholog of Cdc25 [10], [13], [14]. For this reason, we analyzed the level of expression of the four C. elegans cdc25 homologs (cdc-25.1, cdc-25.2, cdc-25.3 and cdc-25.4) [32] in wild-type animals and in animals over-expressing the ces-1 gene using qPCR. To that end, using a heat-inducible promoter, ces-1 expression was induced for 1 h in embryos and embryos were collected after a 1.5 h recovery period. This induction scheme resulted in a 3-fold increase in the relative expression level of ces-1 (Figure 4). Using this experimental set-up, we found that the relative expression levels of cdc-25.1 and cdc-25.4 are not significantly changed in embryos over-expressing ces-1. In contrast, the relative expression level of cdc-25.2 is significantly decreased, indicating that ces-1 over-expression can repress the expression of cdc-25.2 (Figure 4). We also found that the relative expression level of cdc-25.3 is significantly increased. Since CES-1 is thought to predominantly act as repressor of transcription and since it has been suggested that mammalian Cdc25A and Cdc25B may compensate for the loss of Cdc25C [33], [34], we analyzed whether the increase in the relative expression level of cdc-25.3 in embryos over-expressing ces-1 is an indirect effect that is triggered by decreased cdc-25.2 expression. We found that the relative expression level of cdc-25.3 is not increased in embryos in which cdc-25.2 function is knocked-down by RNAi (Figure S5). This suggests that, independently of decreasing cdc-25.2 expression, ces-1 over-expression increases cdc-25.3 expression. Finally, the relative expression level of cya-1 expression is not significantly changed in embryos over-expressing ces-1 (Figure 4). In summary, our data indicate that CES-1 directly or indirectly represses the expression of cdc-25.2.
Fig. 4. cdc-25.2 expression is down-regulated by ces-1 over-expression. 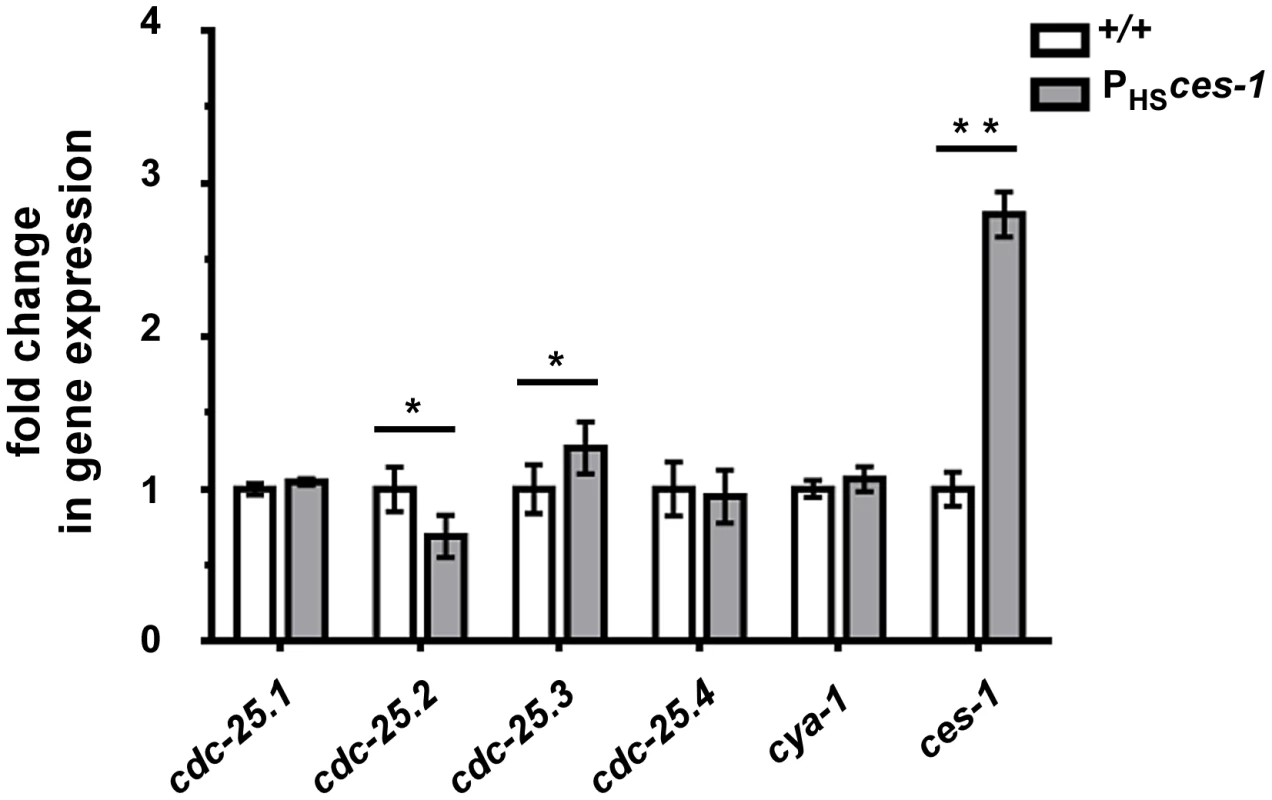
Transgenic animals carrying an extra-chromosomal array of ces-1 heat-shock plasmids and coinjection marker were used as the sample group (PHSces-1), while transgenic animals carrying an extra-chromosomal array of only coinjection marker were used as control (+/+). Relative expression levels of cdc-25 genes and cya-1 gene in control animals (+/+) and animals over-expressing ces-1 (PHSces-1) were determined by real-time PCR (qPCR). Data are represented as fold change relative to control. Data shown are the means ± SEM from four independent repeats. Paired t-test was used to determine significance. The level of cdc-25.2 in PHSces-1 is significantly lower than in control. The level of cdc-25.3 in PHSces-1 is significantly higher than in control. The levels of cdc-25.1, cdc-25.4, cya-1 are not significantly changed in response to ces-1 over-expression. *p<0.05, **p<0.01 significantly different from the control. CES-1 binds to the cdc-25.2 locus in vivo
To determine whether CES-1 directly controls the transcription of the cdc-25.2 gene, we analyzed CES-1 ChIP-seq (ChIP-seq, chromatin immunoprecipitation with massively parallel DNA sequencing) data acquired by the modENCODE Project (http://www.modencode.org/) [35], [36] (M. Snyder, S. Kim, T. Kawli, personal communication). This data was acquired using, as starting material, embryos that harbor multiple copies of an engineered, stably integrated ces-1 fosmid, which expresses a ces-1::gfp transgene under the endogenous ces-1 promoter [37], [38]. (We have previously shown that a Pces-1ces-1::yfp transgene can rescue the ces-1 loss-of-function mutant phenotype and, hence, is generating a fusion protein that is functional [17]. ) The ChIP-seq data obtained indicate that CES-1 binds to a 1.7 kb region that is located 4.8 kb to 6.5 kb upstream of the predicted transcriptional start site of cdc-25.2 (Figure 5) (Integrated Genome Browser [39]). In contrast, we did not identify peaks indicative of CES-1 binding sites in the immediate regions 5′ or 3′ of the predicted cdc-25.1, cdc-25.3 or cdc-25.4 transcription units, nor within their introns (Figure 5). Based on these findings we conclude that cdc-25.2 most likely is a direct target of CES-1 and, hence, that the effect of ces-1 over-expression on the relative expression level of cdc-25.2 is a direct effect.
Fig. 5. CES-1 binds to an upstream region of the cdc-25.2 locus. 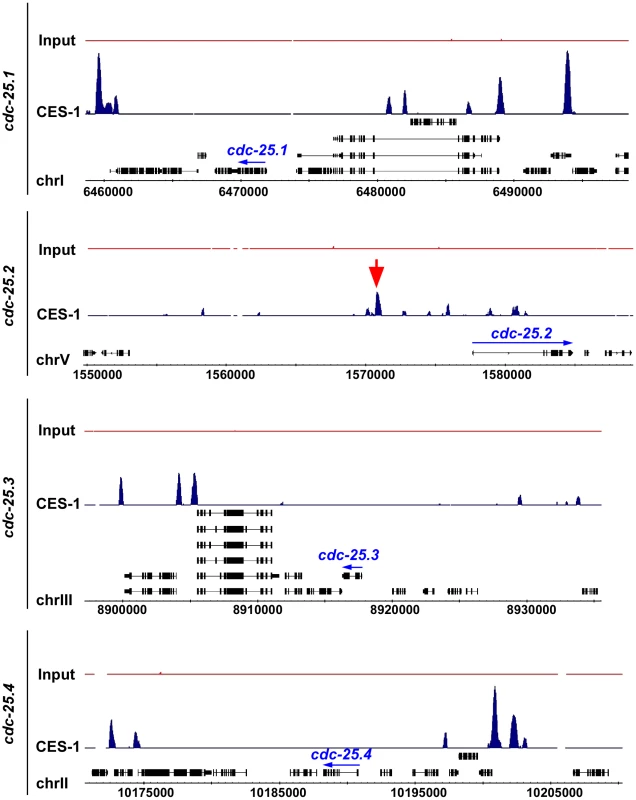
The genome-wide binding sites of the CES-1 protein were identified using ChIP-seq. Shown are the distributions of CES-1-bound regions around the genomic loci of the four cdc-25 genes, whose transcription units are indicated by blue arrows. The black boxes correspond to the gene exons. The red arrow points to the CES-1-bound region upstream of cdc-25.2. Data was visualized using Integrated Genome Browser based on genome WS190 of C. elegans [39]. cdc-25.2 acts downstream of ces-1 to promote cell cycle progression
The over-expression of ces-1 results in a reduction of the relative expression level of cdc-25.2 by about 30% (Figure 4). To determine whether a decrease in cdc-25.2 dosage by 50% is sufficient to enhance the NSM neuroblast division defect caused by cya-1(bc416), we analyzed cya-1(bc416) animals heterozygous for cdc-25.2(ok597), a deletion of the cdc-25.2 gene that removes 2.7 kb of the cdc-25.2 locus, including four of its six exons [40]. We found that cdc-25.2(ok597)/+; cya-1(bc416) animals exhibit a NSM neuroblast division defect similar to the defect observed in ces-1(n703gf); cya-1(bc416) animals. Specifically, at 15°C, 25% and 33% of the NSM neuroblasts divide in ces-1(n703gf); cya-1(bc416) or cdc-25.2(ok597)/+; cya-1(bc416) animals, respectively (in the ced-3 mutant background) (Table 1). Conversely, we tested whether the transgenic expression of the cdc-25.2 transcription unit under the control of the endogenous cdc-25.2 promoter can rescue the NSM neuroblast division defect observed in ces-1(n703gf); cya-1(bc416) animals. We found that the expression of cdc-25.2 significantly reduces the NSM neuroblast division defect in ces-1(n703gf); cya-1(bc416) animals. Specifically, at 15°C, the expression of cdc-25.2 increases the percentage of NSM neuroblasts dividing from 22% to 82% (Table 1; ces-1(n703gf); cya-1(bc416) plus ‘cdc-25.2’ transgene). Together, these findings support the notion that ces-1(n703gf) enhances the NSM neuroblast division defect caused by cya-1(bc416) by decreasing cdc-25.2 expression.
ces-1, cdc-25.2 and cya-1 act together to control cell cycle progression in specific lineages
Next, we analyzed the phenotypes caused by the downregulation of the four cdc-25 genes. We found that the downregulation by RNAi of cdc-25.1 or cdc-25.2 results in embryonic lethality. In contrast, the downregulation by RNAi of cdc-25.3 or cdc-25.4 does not cause any obvious abnormalities, which is consistent with previous observations [32]. While cdc-25.1(RNAi) embryos arrest during early embryonic stages (as early as the 4-cell stage) (data not shown), cdc-25.2(RNAi) embryos arrest at the elongation stage during embryogenesis (Figure 2E, cdc-25.2(RNAi)). Using lineage analyses, we determined the phenotype of cdc-25.2(RNAi) embryos in more detail. We found that the inactivation of cdc-25.2 causes an increase in cell cycle length in all lineages (Table S4). For example, in wild-type animals, the average time between the 6th and 7th and between the 7th and 8th round of division in the ABala lineage is 28 min and 36 min, respectively. In cdc-25.2(RNAi) animals, the average time is 36 min and 52 min, respectively. In general, many cell divisions of the last three rounds of division during embryonic development are blocked (Figure 3 and Figure S6). For example, the divisions of the NSM neuroblasts are blocked in cdc-25.2(RNAi) embryos (Figure 1C). In addition, the ABarp lineage is particularly sensitive to reduced levels of cdc-25.2 function. Most of the cell divisions during the 9th round of division are blocked in the ABarp lineage (Table S3, Figure 3 and Figure S6). For comparison, in the case of other AB descendants, only half or less than half of the cell divisions during the 9th round of division are blocked (Table S3). We also observed severe cell division defects in the C and E lineages in cdc-25.2(RNAi) animals (Figure 3 and Figure S6). Interestingly, the cell lineages that exhibit increased sensitivity to reduced levels of cdc-25.2 activity are identical to the cell lineages that are affected in ces-1(n703gf); cya-1(bc416) animals. In summary, these findings support the notion that, in a specific set of lineages, ces-1, cdc-25.2 and cya-1 act together to control cell cycle progression. These lineages and cells include the ABarp, C and E lineages as well as the NSM neuroblasts.
Finally, to determine whether the loss of ces-1 function affects the phenotype caused by cya-1(bc416), we analyzed the NSM neuroblast division in animals homozygous for cya-1(bc416) and the ces-1 deletion allele tm1036. tm1036 is a 1.3 kb deletion that removes exons 2, 3 and 4 of the ces-1 transcription unit and that is predicted to result in the synthesis of a truncated protein lacking two of the five zinc-finger domains of the CES-1 protein [20]. ces-1(tm1036) animals are indistinguishable from wild-type animals at the dissection microscope level, and in an otherwise wild-type background, the loss of ces-1 function causes no obvious phenotype in the NSM lineage [17], [19], [20]. We found that, in the ced-3 mutant background, ces-1(tm1036) does not suppress the NSM neuroblast division defect caused by cya-1(bc416) (Table 1). However, ces-1(tm1036) reduces the embryonic lethality caused by cya-1(bc416). While 40% of cya-1(bc416) animals exhibit an Emb phenotype, only 26% of ces-1(tm1036); cya-1(bc416) animals exhibit an Emb phenotype (Figure 2A). Based on these observations, we suggest that ces-1 may play a role in the control of cell cycle progression at least in certain cell lineages.
Genetic interactions between cya-1 and dnj-11
Like ces-1(n703gf), the loss of dnj-11 function causes symmetric NSM neuroblast division and inappropriate NSM sister cell survival [17], [19]. Therefore, we determined whether the loss of dnj-11 function also enhances the NSM neuroblast division defect caused by cya-1(bc416). We found that animals homozygous for cya-1(bc416) and dnj-11(tm2859), a deletion allele of dnj-11 that removes 614 base pairs of the coding region [17], exhibit a partially penetrant Emb phenotype (data not shown). Whereas in viable larvae 61% of the NSM neuroblasts had divided, only 30% of the NSM neuroblasts divide in dnj-11(tm2859); cya-1(bc416) animals that arrest during embyogenesis (at 15°C, Table 1). For comparison, 92% of NSM neuroblasts divide in cya-1(bc416) animals raised at 15°C. These findings demonstrate that, like ces-1(n703gf), the loss of dnj-11 function enhances the NSM neuroblast division defect caused by cya-1(bc416). Based on these findings, we conclude that dnj-11 regulates ces-1 function also in the context of cell cycle progression.
Discussion
CES-1 has previously been shown to cause loss of cell polarity in the NSM neuroblast and to suppress apoptosis in its daughter cells [17], [19]. Through a combination of genetic and cell biological studies, we now show that CES-1 also affects cell cycle progression in the NSM neuroblast (See model, Figure 6). In one and the same cell lineage and within a short period of time (<150 min), CES-1 therefore impacts on at least three processes (cell cycle progression, cell polarity and apoptosis) that are fundamentally important to normal development. We speculate that it is through their ability to impact on these processes in a spatially and temporally coordinated manner that Snail-related transcription factors play a crucial role in normal development and tumorigenesis.
Fig. 6. ces-1 Snail represents a functional link between cell cycle progression, cell polarity and apoptosis in the NSM lineage. 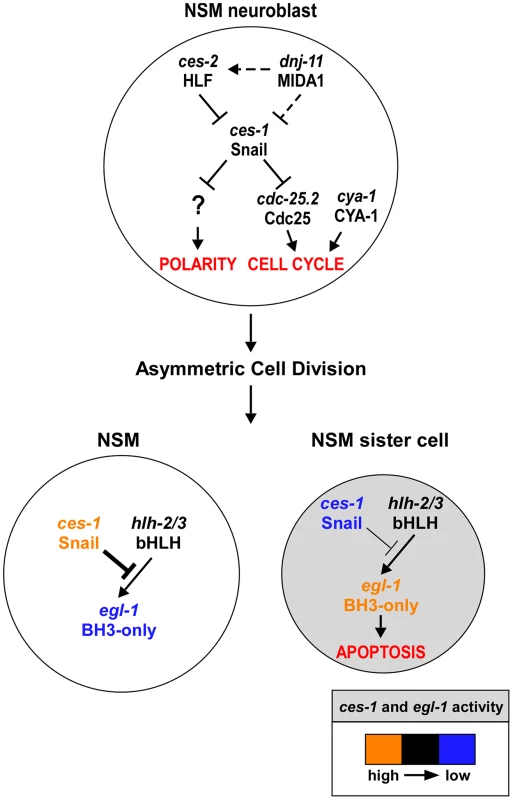
Genetic model of ces-1 Snail functions in the NSM neuroblast (top), the NSM and the NSM sister cell (bottom). In the NSM neuroblast, ces-1 function is negatively regulated by the genes dnj-11 MIDA1 and ces-2 bZIP. ces-1 affects cell cycle progression in the NSM neuroblast by negatively regulating cdc-25.2 Cdc25. ces-1 also affects the polarity of the NSM neuroblast. However, to date, it is unclear through what mechanism. After the asymmetric division of the NSM neuroblast, the level of ces-1 activity is high in the larger NSM (left) and low in the smaller NSM sister cell (right). The activity of ces-1 in the NSM is sufficient to block the function of hlh-2/3 bHLH, thereby resulting in a level of egl-1 BH3-only activity that is too low to induce apoptosis. Conversely, in the NSM sister cell, the activity of ces-1 is not sufficient to block the function of hlh-2/3, thereby resulting in a level of egl-1 activity that is high enough to induce apoptosis. See text for details and molecular interpretations. The role of C. elegans Cyclin A in cell cycle progression
We have isolated and characterized a hypomorphic allele of the cya-1 gene, one of two C. elegans cyclin A genes [31]. This cya-1 mutation, bc416, presumably results in a reduction in the level of CYA-1 protein thereby causing cell division defects in specific lineages (ABarp, C, E and NSM lineages) and partially penetrant embryonic lethality. Given that animals homozygous for a cya-1 deletion allele are not viable (S. van der Heuvel, personal communication), we conclude that cya-1 is essential for embryogenesis. We also present evidence that cya-1(bc416) causes a block in cell cycle progression between S phase and M phase, which is consistent with the proposed function of the CYA-1 protein, as predicted based on the function of Cyclin A in other organisms [31], [41]. Based on the function of Cyclin A in other organisms, we also speculate that it is the C. elegans CDKs CDK-1 and/or CDK-2 that CYA-1 binds to and activates [31], [42]. Furthermore, we provide evidence that in the ABarp, C, E and NSM lineages, cya-1 acts with cdc-25.2 to cause CDK activation and, hence, cell cycle progression.
Interestingly, in cya-1(bc416) animals, at 15°C around 10% of the NSM neuroblast divisions are blocked, while no block in cell division was observed in the ABarp, C or E lineage (Table 1 and Figure 1C). However, at 25°C, a cell division defect was not observed for the NSM neuroblasts, while cell division defects were observed in the ABarp, C and E lineages in around 30% of the cya-1(bc416) embryos (Figure 1C and Figure S4). Therefore, the cell division defect in the NSM lineage is more severe at 15°C, whereas the cell division defects in the ABarp, C and E lineages are more severe at 25°C. We found that compared to wild-type animals, the reduction in the level of correctly spliced cya-1 transcript is similar at both 15°C and 25°C in cya-1(bc416) animals (Figure S3E). This suggests that the splicing defect is not temperature sensitive. Why different cell lineages (NSM neuroblast, ABarp, C and E lineages) respond differently to a reduction in cya-1 function and why their responses differ in their temperature sensitivity is unclear. Lineage or tissue-specific control of cell cycle progression has previously been observed in C. elegans [42]–[46]. One determining factor could be cell cycle length. Since cell cycle length is influenced by temperature and diverges greatly in different lineages, this behavior might reflect different CDK-activity thresholds for different lineages and/or differential regulation of CDKs within specific lineages at different temperatures [42]–[46].
The role of the dnj-11 MIDA1, ces-2 HLF, ces-1 Snail pathway in cell cycle progression
We propose that the dnj-11 MIDA1, ces-2 HLF, ces-1 Snail pathway, which has previously been shown to control asymmetric cell division and apoptosis in the NSM lineage [17], [19], [20], also controls cell cycle progression in this lineage (Figure 6). Specifically, we demonstrate that the dnj-11 loss-of-function mutation tm2859 or the ces-1 gain-of-function mutation n703 enhances a defect in NSM neuroblast division caused by cya-1(bc416). Furthermore, we have uncovered the molecular mechanism through which this pathway controls cell cycle progression in this lineage. We provide evidence in support of the notion that the Snail-related transcriptional repressor CES-1 directly represses the transcription of the cdc-25.2 gene thereby decreasing the level of CDC-25.2. While in an otherwise wild-type background, this does not lead to a block in NSM neuroblast division, it does cause a block in NSM neuroblast division in a cya-1(bc416) mutant background, in which the level of CYA-1 presumably is reduced. The observation that the loss of dnj-11 function or the ces-1 gain-of-function mutation n703 are synthetic lethal with cya-1(bc416) furthermore suggests that the dnj-11 MIDA1, ces-2 HLF, ces-1 Snail pathway may act to control cya-1 - and cdc-25.2-dependent cell cycle progression in lineages other than the NSM lineage.
Unlike dnj-11(tm2859) and ces-1(n703gf), the ces-1 deletion allele tm1036 did not affect the NSM neuroblast division defect caused by cya-1(bc416). This finding suggests that ces-1 may not have a physiological role in cell cycle progression in the NSM neuroblast. Alternatively, the function of ces-1 in cell cycle progression in the NSM neuroblast may be redundant with that of another gene or genes. Interestingly, the functions of the D. melanogaster Snail-related genes snail, escargot and worniu in cell cycle progression and polarity in embryonic neuroblasts are redundant and defects are only observed in animals in which all three genes are inactivated [9], [10]. Apart from ces-1, the C. elegans genome contains at least two additional genes that encode Snail-related transcription factors, scrt-1 and K02D7.2 (http://www.wormbase.org) [1]. Therefore, we speculate that the functions of ces-1 in cell cycle progression, polarity and apoptosis in the NSM lineage are redundant with the functions in these processes of scrt-1 and K02D7.2. Finally, the observation that ces-1(n703gf) enhances but ces-1(tm1036) partially suppresses the embryonic lethality caused by cya-1(bc416) supports the notion that ces-1 has a physiological role in cell cycle progression in some cell lineages, such as the ABarp and C lineages.
The role of Snail-related transcription factors in the regulation of cell cycle progression
Members of the Snail superfamily have previously been shown to affect cell cycle progression. Mammalian Snail1 has been shown to block cell cycle progression in cultured epithelial cells or in mouse embryos, and this effect appears to be mediated through the direct repression of cyclin D2 transcription [15]. In contrast, over-expression of the Snail1 gene in mouse epidermis causes hyperproliferation [47]. In D. melanogaster, the Snail-related proteins Snail, Escargot and Worniu have been shown to promote cell cycle progression in embryonic neuroblasts in part by, directly or indirectly, promoting cdc25string expression [10]. Cdc25string is a critical regulator of M phase during D. melanogaster development, whose activity is regulated at the transcriptional level [13], [14]. In support of the model that cdc25string acts as an integrator of signals that regulate cell division during D. melanogaster development, the cdc25string locus is subject to complex transcriptional regulation. Interestingly, it has been shown that in D. melanogaster larval neuroblasts, the level of Worniu has to be precisely regulated as well. A low level of Worniu in larval neuroblasts leads to a delay in cell cycle progression and premature differentiation, whereas an elevated level of Worniu results in cell cycle arrest due to increased Prospero expression [48]. These findings suggest that the roles of Snail-related proteins in cell cycle progression are complex and might be cell - or tissue-type specific.
However, so far, no Snail-related transcription factor has been implicated in the cdc25-mediated block of cell cycle progression. Here we have identified a new mechanism through which Snail-related proteins can block cell cycle progression. Specifically, we present evidence that, by binding to a region 4.8 kb to 6.5 kb upstream of the cdc-25.2 transcription unit, CES-1 most likely directly represses cdc-25.2 transcription thereby causing a block in cell cycle progression in a sensitized background (the cya-1(bc416) background). Interestingly, the region 4.8 kb to 6.5 kb upstream of the cdc-25.2 transcription unit is at least partially conserved in other Caenorhabditis species such as Caenorhabditis remanei or Caenorhabditis briggsae (UCSC genome browser http://genome.ucsc.edu/ [49]). In analogy to D. melanogaster cdc25string, this finding suggests that the transcriptional regulation of cdc-25.2 might be an important aspect of the developmental control of cell division in C. elegans, a notion that is supported by the observation that the cdc-25.2 transcription unit is flanked by extensive intergenic regions that are conserved (UCSC genome browser [49]). As mentioned above, while D. melanogaster Snail, Escargot und Worniu promote cell cycle progression and cdc25string expression, C. elegans CES-1 blocks cell cycle progression and represses cdc-25.2 expression. Interestingly, we found that besides repressing cdc-25.2 transcription, the over-expression of ces-1 directly or indirectly increases the relative level of cdc-25.3, which encodes another member of the Cdc25 phosphatase family of C. elegans. Hence, depending on the cell lineage and cellular context, members of the Snail superfamily may enhance or repress the expression of Cdc25 phosphatases.
The role of the Snail-related transcription factors in tumorigenesis
The over-expression of Snail-related transcription factors has been implicated in the formation and progression of metastatic cancers, in part due to the ability of Snail-related transcription factors to induce EMTs [1]–[4], [50]. Their potency as proto-oncogenes is thought to lie in their capability to cause loss of cell polarity and adhesive functions on the one hand and acquisition of migratory properties on the other. We argue that their ability to, within the same cell lineage, also block cell cycle progression and apoptosis is similarly important for the formation of metastases. Cells undergoing EMT have high invasive potential and are primarily found at the margins of tumors. Whether EMT has to be accompanied by a reduction in proliferation is a question still under debate. It has previously been shown that increased expression of the gene encoding the transcription factor YB-1 (Y-box binding protein), which is frequently observed in human cancers and which results in increased Snail1 expression, induces EMT accompanied by enhanced metastatic potential and reduced cellular proliferation [51], [52]. Here we demonstrate that the ces-1 gf mutation not only affects cell polarity and apoptosis in the NSM lineage but also enhances the defect in cell cycle progression caused by a partial cya-1 loss-of-function mutation. These findings support the notion that a block in cell cycle progression and, hence, cell proliferation may be important for EMT. A block in cell cycle progression could, for example, provide the time necessary for cytoskeletal reorganizations or cell polarity transitions. Finally, Snail-related transcription factors have recently been implicated in the acquisition and maintenance of the stem cell fate in mammals [4], [53]–[55]. We speculate that it is the ability of Snail-like transcription factors to coordinately influence cell cycle progression, cell polarity and apoptosis that allows specific cells to adopt and maintain the cancer stem cell fate.
Models of metastatic cancers have mainly focused on the analysis of the starting and end points of the cellular transformations that cells undergo during the formation of metastases (i.e. the epithelial and mesenchymal phenotype) [5]. There is a need for in vivo models that allow the analysis of intermediate stages of this process. We suggest that the over-expression of the Snail-related gene ces-1 in C. elegans (i.e. the ces-1 gf phenotype) may serve as such a model at least for certain stages of this process. The ability to combine systems biology approaches (such as ChIP-seq analyses) with cell biological and genetic dissection at single cell resolution will allow us to further dissect the complex role of Snail-like transcription factors during normal development and tumorigenesis.
Materials and Methods
Strains and genetics
C. elegans strains were maintained and cultured as described [56]. Bristol N2 was used as the wild-type strain, unless noted otherwise. CB4856 (Hawaii) was used for SNP mapping. Mutations and transgenes used in this study are listed below and are described by Riddle et al. unless noted otherwise [57]: LGI: ces-1(n703gf), ces-1(tm1036) (National BioResource Project) (3 times backcrossed). LGII: rrf-3(pk1426) [58]. LGIII: dpy-17(e164), cya-1(bc416) (this study) (5 times backcrossed), bcIs66 (Ptph-1his-24::gfp) (this study), unc-69(e587), ltIs38 (Ppie-1gfp::phPLC1δ) [25]. LGIV: ced-3(n717), dnj-11(tm2859) [17]. LGV: ltIs44 (Ppie-1mCherry::phPLC1δ) [25], cdc-25.2(ok597) [40]. LGX: lin-15(n765ts).
Molecular analysis
The plasmids pBC1153 (cdc-25.2) and pBC1282 (cdc-25.3) used for in vitro transcription of double-strand (ds) RNA were generated by cloning PCR fragments containing exons of the targeted genes into the EcoRV site of pBluescript II KS+. The plasmid pBC1098 (cya-1), which was used for RNAi by feeding as well as in vitro transcription of dsRNA, was generated by cloning a PCR fragment containing genomic DNA of the cya-1 locus into the NcoI and XmaI sites of pPD129.36 [59]. The plasmids pMM#47 (PHSces-1; pPD49.78 based) and pMM#48 (PHSces-1; pPD49.83 based) were generated using a full-length ces-1 cDNA (R.H. Horvitz and M.M. Metzstein, personal communication). For rescue experiments, DNA fragments containing the gene of interests (including regulatory regions) were amplified by PCR (NEB LongAmp Taq) and purified. The sequences of oligonucleotides used for PCR are provided in Table S5.
RNA interference
RNAi by feeding was performed as described by Fire and co-workers [59] using 6 mM IPTG. For RNAi experiments by microinjection [60], pBC1153 (cdc-25.2) and pBC1282 (cdc-25.3) were used as templates and oligonucleotides 5′-ttgtaaaacgacggccag-3′ and 5′-catgattacgccaagcgc-3′ as primers to generate PCR products containing at their ends, either the T3 or T7 promoter. pBC1098 (cya-1) was used as template and oligonucleotides 5′ - taatacgactcactataggg-3′ as primer to generate PCR products containing T7 promoter at both ends. These PCR products were used to synthesize dsRNA in vitro using T3 and T7 polymerase (Ambion). RNAi was performed by microinjection of dsRNA into young adults. Injected animals were incubated at 25°C for at least 20 h and the phenotype of their progeny was determined.
Transgenic animals
Germline transformation was performed as described [61]. For rescue experiments, ces-1(n703gf); cya-1(bc416) animals were injected with purified PCR products (0.5–6 ng/µl) using pRF4 (100 ng/µl) as coinjection marker, which confers a dominant Rol phenotype. For ces-1 over-expression experiment, pMM#47 (5 ng/µl) and pMM#48 (5 ng/µl) were injected into lin-15(n765ts) animals using pL15EK (80 ng/µl), which rescues the Muv phenotype caused by lin-15(n765ts), as coinjection marker.
Phenotypic analyses
The NSMs and the surviving NSM sister cells were identified in L3 or L4 larvae carrying the Ptph-1his-24::gfp reporter using fluorescence microscopy essentially as described for Ptph-1gfp [21]. The division of the NSM neuroblast was analyzed in embryos using a plasma membrane-targeted GFP fusion protein (Ppie-1gfp::phPLC1δ) or mCherry fusion protein (Ppie-1mCherry::phPLC1δ) as described [17], [25]. Embryos were imaged using 4D microscopy and cell lineage analysis was performed using a Zeiss Imager microscope and SIMIBioCell software (Simi Reality Motion Systems GmbH, Unterschleissheim, Germany) as described [29].
DAPI staining and DNA content quantification
L4 larvae were fixed and stained with DAPI as described [62], with the exception that slides were mounted in 1.0 µg/ml DAPI in PBS diluted 1∶1 with VectaShield (Vector Laboratories). Fluorescence intensities were measured using Metamorph software. Fluorescence intensities were normalized by comparing the intensities of the nuclei of interest with the intensities of nuclei of neighboring, pharyngeal muscle nuclei. The GFP signal from the Ptph-1his-24::gfp reporter was still observed after fixation and DAPI staining, and was used to identify the NSMs, NSM sister cells and non-dividing NSM neuroblasts.
RNA preparation and cDNA synthesis
Embryos were collected and frozen at −80°C in TRIzol (Invitrogen). Frozen embryo pellets were disrupted using a 7-ml tight Dounce tissue grinder (Fisher Scientific) and total RNA was prepared using the RNeasy Mini Kit (Qiagen). The first strand cDNA synthesis reaction was performed using the SuperScript III system (Invitrogen). For cDNA synthesis, an oligo (dT) primer was used.
ces-1 over-expression experiment
Transgenic animals carrying an extra-chromosomal array of pMM#47, pMM#48 and the coinjection marker pL15EK were used as the sample group. Early stage embryos were isolated by bleaching synchronized hermaphrodites that contain four to six embryos. Isolated embryos were allowed to develop on large NGM plates at 20°C for 70 min. ces-1 expression was induced by heat-shocking the embryos at 32°C for 1 h. After a 1.5 h recovery period at 20°C embryos were collected. RNA extraction and cDNA synthesis were performed as described above. Transgenic animals carrying an extra-chromosomal array of only coinjection marker pL15EK were used as control and were treated the same way.
Quantitative real-time PCR (qPCR)
Fast SYBR green master mix (Applied Biosystems) was used to amplify cDNA templates by real-time PCR. Each sample was performed in triplicate on a Biorad CFX96 real-time PCR machine. The sequences of the primers used are provided in Table S5. The ‘housekeeping’ gene act-1 served as endogenous control [63]. Results were analyzed using the relative standard curve method. To produce the standard curve for each target sequence including act-1, first, 5-fold dilution series of standard N2 cDNA were prepared and subjected to real-time PCR to determine Ct values for each dilution. Second, average Ct values were plotted versus the logarithm of the concentration (base 5) of the template. To determine the amounts of the target sequences in the starting samples, average Ct values for each sample were compared to the standard curve. To normalize the samples using act-1, the result of a particular target sequence was divided by the act-1 control result.
ChIP-seq (chromatin immunoprecipitation with massively parallel DNA sequencing)
A ces-1::gfp fosmid reporter was generated as described [37], [38]. Briefly, using recombineering, a GFP::3×FLAG tag was inserted in-frame to the C-terminus of the ces-1 transcription unit in a fosmid that contains the entire locus of ces-1. This fosmid reporter was integrated into the worm genome using the method of bombardment, which produces transgenic animals with low-copy integrated arrays. The in vivo binding sites of the CES-1::GFP::3×FLAG fusion protein synthesized in these animals were then determined using the method of ChIP-seq as described [64].
Supporting Information
Zdroje
1. NietoMA (2002) The Snail superfamily of zinc-finger transcription factors. Nature reviews Molecular cell biology 3 : 155–166.
2. Barrallo-GimenoA, NietoMA (2005) The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132 : 3151–3161.
3. CobaledaC, Perez-CaroM, Vicente-DuenasC, Sanchez-GarciaI (2007) Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annual review of genetics 41 : 41–61.
4. PeinadoH, OlmedaD, CanoA (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature reviews Cancer 7 : 415–428.
5. NietoMA (2011) The ins and outs of the epithelial to mesenchymal transition in health and disease. Annual review of cell and developmental biology 27 : 347–376.
6. WhitemanEL, LiuCJ, FearonER, MargolisB (2008) The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene 27 : 3875–3879.
7. CanoA, Perez-MorenoMA, RodrigoI, LocascioA, BlancoMJ, et al. (2000) The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature cell biology 2 : 76–83.
8. BatlleE, SanchoE, FranciC, DominguezD, MonfarM, et al. (2000) The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature cell biology 2 : 84–89.
9. CaiY, ChiaW, YangX (2001) A family of Snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. The EMBO journal 20 : 1704–1714.
10. AshrafSI, IpYT (2001) The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development 128 : 4757–4767.
11. KnoblichJA (2010) Asymmetric cell division: recent developments and their implications for tumour biology. Nature reviews Molecular cell biology 11 : 849–860.
12. BetschingerJ, KnoblichJA (2004) Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Current biology : CB 14: R674–685.
13. LehmanDA, PattersonB, JohnstonLA, BalzerT, BrittonJS, et al. (1999) Cis-regulatory elements of the mitotic regulator, string/Cdc25. Development 126 : 1793–1803.
14. EdgarBA, LehmanDA, O'FarrellPH (1994) Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development 120 : 3131–3143.
15. VegaS, MoralesAV, OcanaOH, ValdesF, FabregatI, et al. (2004) Snail blocks the cell cycle and confers resistance to cell death. Genes & development 18 : 1131–1143.
16. WuWS, HeinrichsS, XuD, GarrisonSP, ZambettiGP, et al. (2005) Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123 : 641–653.
17. HatzoldJ, ConradtB (2008) Control of apoptosis by asymmetric cell division. Plos Biology 6: e84.
18. SulstonJE, SchierenbergE, WhiteJG, ThomsonJN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental biology 100 : 64–119.
19. EllisRE, HorvitzHR (1991) Two C. elegans genes control the programmed deaths of specific cells in the pharynx. Development 112 : 591–603.
20. MetzsteinMM, HorvitzHR (1999) The C. elegans cell death specification gene ces-1 encodes a Snail family zinc finger protein. Molecular cell 4 : 309–319.
21. ThellmannM, HatzoldJ, ConradtB (2003) The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development 130 : 4057–4071.
22. MetzsteinMM, HengartnerMO, TsungN, EllisRE, HorvitzHR (1996) Transcriptional regulator of programmed cell death encoded by Caenorhabditis elegans gene ces-2. Nature 382 : 545–547.
23. NehmeR, ConradtB (2008) egl-1: a key activator of apoptotic cell death in C. elegans. Oncogene 27 Suppl 1: S30–40.
24. SzeJY, ZhangS, LiJ, RuvkunG (2002) The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development 129 : 3901–3911.
25. AudhyaA, HyndmanF, McLeodIX, MaddoxAS, YatesJR (2005) A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. The Journal of cell biology 171 : 267–279.
26. EllisHM, HorvitzHR (1986) Genetic control of programmed cell death in the nematode C. elegans. Cell 44 : 817–829.
27. YanowitzJ, FireA (2005) Cyclin D involvement demarcates a late transition in C. elegans embryogenesis. Developmental biology 279 : 244–251.
28. FayDS, HanM (2000) Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development 127 : 4049–4060.
29. SchnabelR, HutterH, MoermanD, SchnabelH (1997) Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Developmental biology 184 : 234–265.
30. ZipperlenP, FraserAG, KamathRS, Martinez-CamposM, AhringerJ (2001) Roles for 147 embryonic lethal genes on C.elegans chromosome I identified by RNA interference and video microscopy. The EMBO journal 20 : 3984–3992.
31. BoxemM (2006) Cyclin-dependent kinases in C. elegans. Cell division 1 : 6.
32. AshcroftNR, SraykoM, KosinskiME, MainsPE, GoldenA (1999) RNA-Mediated interference of a cdc25 homolog in Caenorhabditis elegans results in defects in the embryonic cortical membrane, meiosis, and mitosis. Developmental biology 206 : 15–32.
33. ChenMS, HurovJ, WhiteLS, Woodford-ThomasT, Piwnica-WormsH (2001) Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Molecular and cellular biology 21 : 3853–3861.
34. FergusonAM, WhiteLS, DonovanPJ, Piwnica-WormsH (2005) Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Molecular and cellular biology 25 : 2853–2860.
35. GersteinMB, LuZJ, Van NostrandEL, ChengC, ArshinoffBI, et al. (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330 : 1775–1787.
36. CelnikerSE, DillonLA, GersteinMB, GunsalusKC, HenikoffS, et al. (2009) Unlocking the secrets of the genome. Nature 459 : 927–930.
37. SarovM, MurrayJI, SchanzeK, PozniakovskiA, NiuW, et al. (2012) A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150 : 855–866.
38. SarovM, SchneiderS, PozniakovskiA, RoguevA, ErnstS, et al. (2006) A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nature methods 3 : 839–844.
39. NicolJW, HeltGA, BlanchardSGJr, RajaA, LoraineAE (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25 : 2730–2731.
40. KimJ, KawasakiI, ShimYH (2010) cdc-25.2, a C. elegans ortholog of cdc25, is required to promote oocyte maturation. Journal of cell science 123 : 993–1000.
41. BudirahardjaY, GonczyP (2009) Coupling the cell cycle to development. Development 136 : 2861–2872.
42. van den HeuvelS (2005) Cell-cycle regulation. WormBook : the online review of C. elegans biology 1–16.
43. SegrefA, CabelloJ, ClucasC, SchnabelR, JohnstoneIL (2010) Fate specification and tissue-specific cell cycle control of the Caenorhabditis elegans intestine. Molecular biology of the cell 21 : 725–738.
44. KirienkoNV, ManiK, FayDS (2010) Cancer models in Caenorhabditis elegans. Developmental dynamics : an official publication of the American Association of Anatomists 239 : 1413–1448.
45. KosticI, RoyR (2002) Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans. Development 129 : 2155–2165.
46. AshcroftN, GoldenA (2002) CDC-25.1 regulates germline proliferation in Caenorhabditis elegans. Genesis 33 : 1–7.
47. JamoraC, LeeP, KocieniewskiP, AzharM, HosokawaR, et al. (2005) A signaling pathway involving TGF-beta2 and Snail in hair follicle morphogenesis. Plos Biology 3: e11.
48. LaiSL, MillerMR, RobinsonKJ, DoeCQ (2012) The Snail family member Worniu is continuously required in neuroblasts to prevent Elav-induced premature differentiation. Developmental cell 23 : 849–857.
49. KentWJ, SugnetCW, FureyTS, RoskinKM, PringleTH, et al. (2002) The human genome browser at UCSC. Genome research 12 : 996–1006.
50. Moreno-BuenoG, PortilloF, CanoA (2008) Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 27 : 6958–6969.
51. EvdokimovaV, TognonC, NgT, SorensenPH (2009) Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell cycle 8 : 2901–2906.
52. EvdokimovaV, TognonC, NgT, RuzanovP, MelnykN, et al. (2009) Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer cell 15 : 402–415.
53. GuoW, KeckesovaZ, DonaherJL, ShibueT, TischlerV, et al. (2012) Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148 : 1015–1028.
54. JordanNV, JohnsonGL, AbellAN (2011) Tracking the intermediate stages of epithelial-mesenchymal transition in epithelial stem cells and cancer. Cell cycle 10 : 2865–2873.
55. ManiSA, GuoW, LiaoMJ, EatonEN, AyyananA, et al. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133 : 704–715.
56. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
57. Riddle DL, Blumenthal T, Meyer B, Priess JR, editors(1997) C. elegans II: Cold Spring Harbor: Cold Spring Harobr Laboratory Press.
58. SimmerF, TijstermanM, ParrishS, KoushikaSP, NonetML, et al. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Current biology 12 : 1317–1319.
59. TimmonsL, FireA (1998) Specific interference by ingested dsRNA. Nature 395 : 854.
60. FireA, XuS, MontgomeryMK, KostasSA, DriverSE, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 : 806–811.
61. MelloC, FireA (1995) DNA transformation. Methods in cell biology 48 : 451–482.
62. StromeS, WoodWB (1982) Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 79 : 1558–1562.
63. KimJ, LeeAR, KawasakiI, StromeS, ShimYH (2009) A mutation of cdc-25.1 causes defects in germ cells but not in somatic tissues in C. elegans. Molecules and Cells 28 : 43–48.
64. ZhongM, NiuW, LuZJ, SarovM, MurrayJI, et al. (2010) Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS genetics 6: e1000848.
Štítky
Genetika Reprodukčná medicína
Článek Defending Sperm FunctionČlánek How to Choose the Right MateČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase inČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



