-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
Insulators can block the action of enhancers on promoters and the spreading of repressive chromatin, as well as facilitating specific enhancer-promoter interactions. However, recent studies have called into question whether the activities ascribed to insulators in model transgene assays actually reflect their functions in the genome. The Drosophila even skipped (eve) gene is a Polycomb (Pc) domain with a Pc-group response element (PRE) at one end, flanked by an insulator, an arrangement also seen in other genes. Here, we show that this insulator has three major functions. It blocks the spreading of the eve Pc domain, preventing repression of the adjacent gene, TER94. It prevents activation of TER94 by eve regulatory DNA. It also facilitates normal eve expression. When Homie is deleted in the context of a large transgene that mimics both eve and TER94 regulation, TER94 is repressed. This repression depends on the eve PRE. Ubiquitous TER94 expression is “replaced” by expression in an eve pattern when Homie is deleted, and this effect is reversed when the PRE is also removed. Repression of TER94 is attributable to spreading of the eve Pc domain into the TER94 locus, accompanied by an increase in histone H3 trimethylation at lysine 27. Other PREs can functionally replace the eve PRE, and other insulators can block PRE-dependent repression in this context. The full activity of the eve promoter is also dependent on Homie, and other insulators can promote normal eve enhancer-promoter communication. Our data suggest that this is not due to preventing promoter competition, but is likely the result of the insulator organizing a chromosomal conformation favorable to normal enhancer-promoter interactions. Thus, insulator activities in a native context include enhancer blocking and enhancer-promoter facilitation, as well as preventing the spread of repressive chromatin.
Published in the journal: The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading. PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003883
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003883Summary
Insulators can block the action of enhancers on promoters and the spreading of repressive chromatin, as well as facilitating specific enhancer-promoter interactions. However, recent studies have called into question whether the activities ascribed to insulators in model transgene assays actually reflect their functions in the genome. The Drosophila even skipped (eve) gene is a Polycomb (Pc) domain with a Pc-group response element (PRE) at one end, flanked by an insulator, an arrangement also seen in other genes. Here, we show that this insulator has three major functions. It blocks the spreading of the eve Pc domain, preventing repression of the adjacent gene, TER94. It prevents activation of TER94 by eve regulatory DNA. It also facilitates normal eve expression. When Homie is deleted in the context of a large transgene that mimics both eve and TER94 regulation, TER94 is repressed. This repression depends on the eve PRE. Ubiquitous TER94 expression is “replaced” by expression in an eve pattern when Homie is deleted, and this effect is reversed when the PRE is also removed. Repression of TER94 is attributable to spreading of the eve Pc domain into the TER94 locus, accompanied by an increase in histone H3 trimethylation at lysine 27. Other PREs can functionally replace the eve PRE, and other insulators can block PRE-dependent repression in this context. The full activity of the eve promoter is also dependent on Homie, and other insulators can promote normal eve enhancer-promoter communication. Our data suggest that this is not due to preventing promoter competition, but is likely the result of the insulator organizing a chromosomal conformation favorable to normal enhancer-promoter interactions. Thus, insulator activities in a native context include enhancer blocking and enhancer-promoter facilitation, as well as preventing the spread of repressive chromatin.
Introduction
A variety of regulatory elements have evolved in higher eukaryotes to regulate gene expression. Cis-regulatory modules (CRMs, or enhancers) are bound by DNA-binding transcription factors that coordinately recruit coactivators and corepressors. Enhancers communicate with basal promoters at least in part through a looping out of intervening DNA, allowing them to act over large distances along a chromosome, or even in trans, with a promoter on another chromosome [1]–[4]. Enhancer activities are regulated by the chromatin environment, which is “managed” by both the enhancers themselves and other DNA elements such as Polycomb-group response elements (PREs) [5]–[8]. Further coordination of these activities is provided by elements such as insulators that affect chromosomal organization and conformation. Insulators harbor activities that can limit the range of action of enhancers and repressive chromatin, as well as facilitate long-range enhancer-promoter communication, depending on context [9]–[12].
Insulators typically show “barrier” function that prevents the spread of heterochromatin, as well as enhancer blocking activity, in model transgene assays [9]–[12]. Pairs of insulators can interact with each other to generate chromosomal loops between them. This has been postulated to create distinct functional domains that somehow prevent enhancer-promoter cross-talk between domains.
Repressive chromatin structures include heterochromatin and Polycomb (Pc) chromatin, which constitutes a form of epigenetic transcriptional memory, stabilizing developmental fate choices, among other functions. Pc chromatin is maintained through the recruitment of Pc-group (PcG) gene products to PREs [5]–[8]. PREs can extend their influence outward to produce Polycomb domains that encompass multiple regulatory regions within a gene or a gene complex [13]–[17]. PREs can also synergize with each other in trans [18], and in some cases facilitate long-range enhancer-promoter communication [19]. Both Pc domains and mammalian X-inactivation involve the histone modification H3K27me3, catalyzed by Pc-repressive complex 2 (PRC2) [20]–[22].
The functions of PREs and insulators have been studied within Drosophila Hox genes [23]–[25]. There, functional chromatin domains are flanked by insulators, so that all the enhancers and PREs within a domain are coordinately regulated. Enhancers acting early in development (“initiators”) are spatially regulated to determine whether a domain will be active or not throughout the rest of development. They do this by inactivating PREs, so that where initiators are active, later-acting enhancers can also be active. The main effect of deleting insulators in this context is to extend the influence of initiators to inactivate PREs in the adjacent domain, which allows its later-acting enhancers to be inappropriately active. However, phenotypic details suggest that in some cells, repressive chromatin may spread instead [26].
Genome-wide chromatin immunoprecipitation (ChIP) analysis of the locations of insulator binding proteins show a wide range of binding patterns [27]–[40]. In Drosophila, insulator proteins include dCTCF (CCCTC-binding factor), Mod(mdg4)67.2, Su(Hw) (Suppressor of hairy wing), CP190 (centrosomal protein 190), BEAF32 (boundary element-associated factor 32), and Zw5 (Zeste-white-5). Recent genome-wide studies also implicate the mitotic spindle protein Chromator [41] and the nuclear lamina [30], [37] in insulator function. In mammals, CTCF is associated with most known insulators [10], [12], [42]–[44]. CTCF functions in the regulation of β-globin [45], [46] and the imprinted Igf2 and H19 loci [47]–[49]. Based on recent genome-wide studies, it has been suggested that insulator proteins bind at many sites that do not function as predicted by model transgene assays [34], [36], [38]. Transgenic dissection in a native context can help to determine their normal functions.
The even skipped (eve) locus is a well-defined Pc domain based on genome-wide analysis [13]–[17], and is regulated by PcG genes [50]–[54]. An insulator flanks its well-characterized regulatory region, which includes the eve PRE at its 3′ end [51], [55]. Thus, this insulator is in a position to separate both positive and negative eve regulatory elements from the constitutively expressed neighboring gene TER94, and/or to prevent ectopic activation of eve by TER94 enhancers. This insulator was shown to have 3 distinct activities in model transgene assays. In addition to enhancer blocking, it causes homing of P-element transgenes to the endogenous eve neighborhood, for which it was nicknamed Homie (Homing insulator at eve). Furthermore, from within a several megabase region flanking endogenous eve, it causes long-range interactions of transgenic promoters with endogenous eve enhancers [55]. Genome-wide analysis showed that most known insulator proteins bind to the Homie region [27], [33].
Homie shares properties with other insulators based on model transgene assays and, like many other putative insulators, is situated close to both a transcription start site (TSS) and a PRE. Thus, understanding Homie's function in its native context can illuminate many of the mysteries that surround this enigmatic group of regulatory elements. In order to investigate its native function, we constructed a transgenic eve-TER94 locus that mimics the normal regulation of both genes. Using this artificial locus, we show that Homie functions as a PRE blocker to protect TER94 from repression due to spreading of the eve Pc domain. Heterologous insulators and PREs can substitute for Homie and the eve PRE, suggesting that limiting the range of PRE action is an important function of insulators generally. Homie also prevents the eve enhancers from activating TER94 in specific tissues. Furthermore, Homie facilitates normal eve expression by augmenting communication between the eve promoter and its 3′ enhancers, likely through a chromosomal looping mechanism.
Results
Insulators are generally considered to have two major functions. First, they can shield promoters from the effects of distal enhancers. Second, they can block the spread of repressive chromatin. Here, we investigate the roles that these activities play in the normal functions of an insulator (Homie) located between the ubiquitously expressed TER94 gene and the highly patterned eve gene. We find that blocking the spread of repressive Polycomb chromatin by Homie is critical for normal TER94 promoter expression. In addition to exhibiting the canonical insulator activities in a near-native context, we find that Homie facilitates certain aspects of normal eve expression, and we present a model for how this occurs.
Homie Shields the TER94 Promoter from eve PRE Activity
In order to analyze the function of the eve 3′ insulator Homie, we employed a pseudo-locus that contains all the regulatory DNA necessary for normal expression of both eve [56]–[59] and the 3′ adjacent gene TER94 [60]–[62]. This transgene extends from −6.4 to +11.3 kb relative to the eve TSS, from the 5′-most enhancer of eve to the 3rd exon of TER94. In addition to all of the eve enhancers, this region contains a characterized PRE [51] located just upstream (on the eve side) of Homie [55]. On the other side of Homie is the TER94 promoter and TSS, which are sufficient for ubiquitous expression, augmented by enhancers in the TER94 introns (data not shown). The eve coding region was replaced with lacZ coding DNA, and the 3rd exon of TER94 was fused with the EGFP coding region (Figure 1A). In this study, we make repeated use of a version of recombinase-mediated cassette exchange (RMCE) [63] that allows modified transgenes to be inserted in either orientation at pre-defined chromosomal landing sites. All aspects of transgene expression were consistent for both orientations and at multiple landing sites, with a few minor exceptions (as noted below).
Fig. 1. Homie shields the TER94 promoter from eve PRE activity. 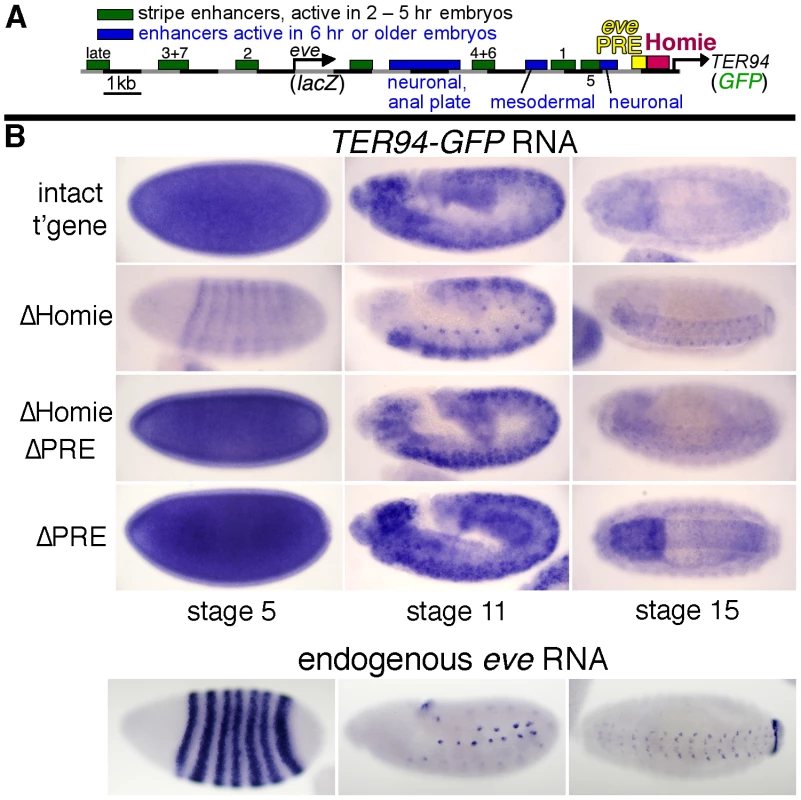
A: map of the eve-TER94 transgene. The 3rd exon of the TER94 protein coding region is fused to that of GFP, while the eve coding region is replaced by that of lacZ. Otherwise, the transgene consists of the entire genomic region from −6.4 kb to +11.3 kb relative to the eve TSS, and includes all of the eve enhancers, plus TER94 enhancers located within its first two introns. The locations of the major eve PRE (“eve PRE”), 3′ insulator (“Homie”), early embryonic stripe enhancers (numbered), and late embryonic enhancers (labeled) are shown as colored boxes. B: embryonic expression, at the indicated stages, of GFP RNA, driven by the TER94 promoter, from the transgene shown in A (top row “intact t'gene”), or the same transgene modified by deletion of either the insulator alone (2nd row “ΔHomie”), the PRE alone (bottom row “ΔPRE”), or both (3rd row “ΔHomie ΔPRE”), each inserted at attP landing site 95E5, visualized by whole-mount in situ hybridization. Note that the normal, ubiquitous expression (mimicking TER94) is changed to resemble the eve pattern (shown for comparison at the bottom) by deletion of Homie, while further deletion of the PRE restores TER94-like expression. Deletion of the PRE alone does not noticeably affect the embryonic pattern. In embryos, TER94 RNA is present ubiquitously at early blastoderm, and begins to fade around stage 10. Most of this RNA is maternally derived, but there is a ubiquitous zygotic contribution as well (see below). At stage 10 and later, strong expression is also observed throughout the brain and central nervous system (CNS) [55]. TER94-GFP expression from our transgene simulates endogenous TER94 expression (Figure 1B, “intact t'gene”). Although the level of expression varies somewhat with chromosomal location, the relative behavior of modified transgenes was consistent at each chromosomal location (compare Figure 1B and Figure S1).
Deletion of Homie caused a severe loss of early, ubiquitous expression driven by the TER94 promoter (Figure 1B “ΔHomie”, Figure S1). When the eve PRE was deleted in addition to Homie, the ubiquitous expression in embryos returned (Figure 1B “ΔHomie ΔPRE”, Figure S1). Deletion of the eve PRE alone did not affect the expression pattern (Figure 1B “ΔPRE”). These results show that the loss of ubiquitous expression from the TER94 promoter caused by deletion of Homie depends on the presence of the PRE. So, one function of Homie is to protect TER94 from PRE-dependent repression.
We also note that when Homie is removed, expression in an eve-like pattern is seen (Figure 1B “ΔHomie”, Figure S1A). This indicates that without Homie, eve enhancers can access the TER94 promoter. We investigate this effect further below.
Homie Blocks eve PRE Activity in Ovaries
Early, ubiquitous expression of TER94 comes from maternally deposited RNA, based on its early appearance and the fact that TER94 is expressed strongly in developing oocytes [60]–[62]. This was confirmed by staining for transgene expression in the absence of a maternal contribution, which is much weaker at early stages than the maternally derived signal (Figure S2 “intact t'gene”; compare to Figure 1B, Figure S1). Since TER94-GFP RNA is deposited maternally, we examined expression in ovaries. TER94 mRNA is present in both the germline, including nurse cells, and somatic epithelial follicle cells [60]–[62]. No eve expression in ovaries has been reported. In our transgenic lines, strong TER94-GFP expression was seen at all stages of oogenesis (Figure 2 “intact t'gene”, Figure S4) in both germline and somatic epithelial cells (Figure S3 “intact t'gene”). However, the level depended to some extent on chromosomal location (compare Figure 2 and Figure S4). In each case, expression was severely repressed when Homie was deleted (Figure 2 “ΔHomie”, Figure S4). As was seen in embryos, it was restored when the PRE was also deleted (Figure 2 “ΔHomie ΔPRE”, Figure S4). These data confirm that in ovaries, Homie is required for TER94 promoter activity, due to its blocking of PRE-dependent repression.
Fig. 2. Homie blocks PRE action in ovaries. 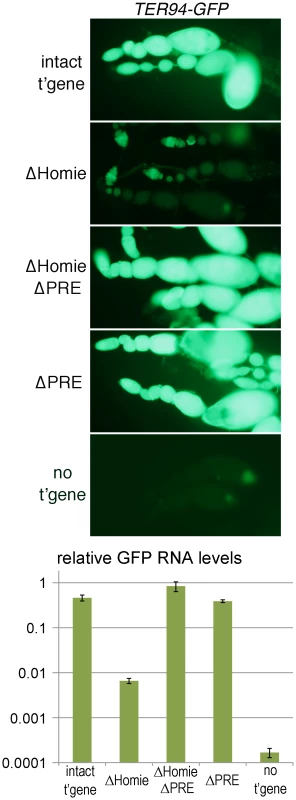
Fluorescence and GFP RNA levels in ovarioles from fly lines carrying the indicated transgenic reporters (described in Figure 1), or no transgene (“no t'gene”). Note the strong fluorescence from TER94 promoter-driven GFP with the intact transgene (“intact t'gene”) at all stages of oogenesis (which proceeds from left to right within each string of ovarioles), and that this is lost when Homie is deleted (“ΔHomie”). Remarkably, strong GFP expression is restored when both Homie and the PRE are deleted (“ΔHomie ΔPRE”), indicating that in the absence of Homie, the PRE is responsible for repression of TER94-GFP. Strong expression is also seen when only the PRE is removed (“ΔPRE”). The graph at the bottom shows, on a log scale, the results of quantitation in triplicate (averages with standard deviations) of GFP RNA from ovaries of lines carrying the indicated transgenes (see Materials and Methods). Note that GFP RNA levels decrease more than 50-fold when Homie is deleted, and are restored by additional deletion of the PRE. The eve PRE Is Redundant in Embryos
Since most of the ubiquitous TER94-GFP RNA seen in early embryos is maternally derived, we tested whether zygotic expression from a paternally-derived transgene is affected by Homie deletion. In this assay, non-transgene-carrying (yw) female flies are crossed with transgene-carrying males, so that there is no maternal GFP RNA in the progeny. Two chromosomal locations were analyzed. In both cases, GFP expression was reduced when Homie was deleted (Figure S2).
Because of the relatively low level of expression, we quantified GFP RNA using RT-PCR. Embryos from three timed collections were analyzed: 2–3 hr. (stages 5–6) and 4–6 hr. (stages 9–11) after egg deposition, and stages 13–15. The effect of Homie deletion paralleled those described above for both ovaries and embryos, in that expression was repressed. However, unlike in ovaries, when both Homie and the PRE were deleted, TER94-GFP expression remained repressed at all stages examined (Figure S2 and data not shown). This is consistent with the idea, confirmed below, that another PRE in the eve locus substitutes in embryos (but not in ovaries) for the eve 3′ PRE. In fact, the eve promoter-proximal region has PRE-like properties [51] (see Discussion).
Other Drosophila Insulators Block eve PRE Action
Are the functions of Homie seen in our assays unique, or are they shared among insulators? In order to test this, we replaced Homie with other known insulators. As a negative control, a >500 bp stretch of λ phage DNA was tested. It had no effect on repression of the TER94 promoter by the eve PRE (Figure 3A “ΔHomie”). In contrast, other characterized Drosophila insulators can substitute for Homie to block repression. gypsy (Figure 3A, B), Fab-7, scs' (Figure 3A), and Fab-8 (Figure 3B) each prevented TER94 promoter repression. Although in one orientation, scs did not work (Figure 3A, “+ scs”), it did work in the opposite orientation (Figure 3A, “+ scs(inv)”). Fab-8 and gypsy showed a minor directionality in their effectiveness (not shown). Restoration of GFP expression is somewhat weaker for scs' and scs(inv) than for gypsy and Fab-7, indicating that they only partially block eve PRE action. Despite differences in efficiency, blocking of PRE action in this context is a shared property of insulators.
Fig. 3. Other Drosophila insulators block eve PRE action, and other PREs are blocked by Homie. 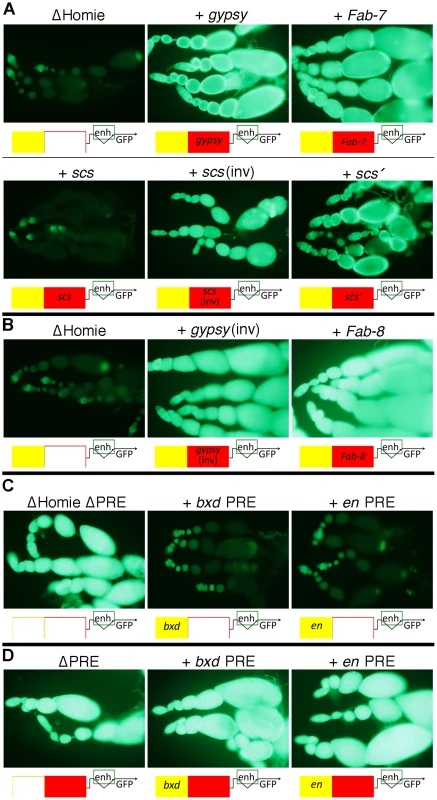
A: fluorescence from transgenic TER94-GFP in the context of the transgene diagrammed in Figure 1A at the 95E5 landing site, with Homie replaced by either λ phage DNA (“ΔHomie”) or the indicated insulator. Diagrams below each panel show the arrangement of regulatory elements affecting GFP expression: yellow-filled boxes are PREs, either from eve (unlabeled), en, or the bxd region of Ubx, as indicated; red-filled boxes are insulators, either Homie (unlabeled), or as labeled. Note that each of these insulators restore TER94 promoter activity, although scs does so when inserted in one orientation (“scs(inv)”) but not the other (“scs”) relative to the TER94-GFP promoter. Note also that restoration of GFP expression is somewhat weaker for scs' and scs (even in the “inverted” orientation) than for gypsy and Fab-7, indicating that they only partially block eve PRE action. B: same as in A, except that the transgenes were inserted in opposite orientation at the same landing site relative to that in A, with either the gypsy insulator or the Fab-8 insulator. Note that eve PRE action is blocked in both cases, causing strong fluorescence. C: fluorescence from transgenic TER94-GFP in the context of the transgene diagrammed in Figure 1A, with Homie deleted, and with either the eve PRE deleted (“ΔHomie ΔPRE”), or replaced by the bxd PRE (“+ bxd PRE”) or the en PRE (“+ en PRE”). Note that each of these PREs repress TER94-GFP to a similar degree as does the eve PRE in the absence of an insulator (compare to “ΔHomie” panels in A and B, where the eve PRE is present). D: fluorescence from transgenic TER94-GFP in the context of the transgene diagrammed in Figure 1A, with the eve PRE either removed (“ΔPRE”), or replaced by either the bxd PRE (“+ bxd PRE”) or the en PRE (“+ en PRE”). Note that neither of these PREs is able to repress TER94-GFP when Homie is present, showing that Homie blocks the repressive action of these heterologous PREs. Heterologous PREs Repress TER94 in the Absence of an Insulator, and Homie Blocks this Repression
In order to test whether the repression of TER94 by the eve PRE is due to some unusual property associated with this PRE, we replaced it with other known PREs. We tested both the bxd PRE and an en PRE for the ability to substitute for the eve PRE in ovaries, in the context of a Homie-deleted transgene. In both cases, repression was seen at a comparable level to that seen with the eve PRE (Figure 3C, compare to Figure 3A,B “ΔHomie”), indicating that TER94 repression is due to a property shared by PREs.
We also tested whether Homie can prevent repression by heterologous PREs. To so this, we replaced the eve PRE with either the bxd PRE or the en PRE. In both cases, Homie blocked their action on the TER94 promoter, and the resulting GFP expression was like that of the wild-type transgene (Figure 3D). This shows that Homie can block repression by a variety of PREs. Taken together, these results suggest that insulators block PRE-dependent repression generally. Thus, the commonly occurring arrangement of PREs flanked on one side by insulators [31] is likely to function to provide a sharp transition in chromatin structure.
Homie Prevents Spreading of the eve Polycomb Domain
The eve locus is a Pc domain, associated with both Polycomb and the characteristic histone modification H3K27me3 [13]–[17]. We asked whether the repression of TER94-GFP in the ΔHomie transgene is accompanied by spreading of this Pc domain over the TER94-GFP promoter. Indeed, in ovaries, we found that H3K27me3 was increased in the TER94-GFP region when Homie was deleted (Figure 4A,B). Additionally removing the PRE reversed this effect almost completely (Figure 4A,B), indicating that spreading of H3K27me3 depends on the eve PRE. Thus, when TER94-GFP is repressed, H3K27me3 is increased, and when this repression is reversed, H3K27me3 levels return to normal. This suggests that Pc domain spreading is likely to be responsible for the repression.
Fig. 4. Homie blocks spreading of the eve Pc/H3K27me3 domain into the adjacent gene, TER94. 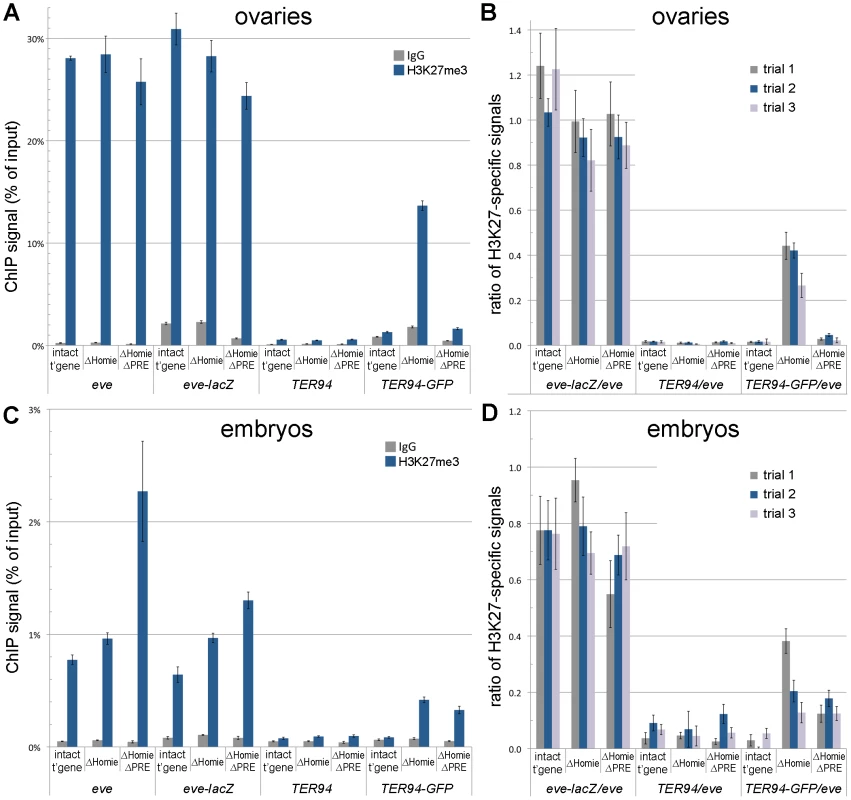
ChIP assays were used to quantify the association of H3K27me3 with the endogenous eve and TER94 coding regions, and with the transgenic eve-lacZ and TER94-GFP coding regions. A, B: ovaries were dissected and subjected to ChIP analysis, as described in Materials and Methods. In A is graphed the averages and standard deviations from triplicate PCR assays using primer pairs specific to each of the 4 coding region indicated along the bottom, and for the transgenic lines indicated (“intact t'gene” carries the entire transgene diagrammed in Figure 1A; “ΔHomie” is the same transgene with Homie deleted; “ΔHomie ΔPRE” has both Homie and the PRE deleted). Either H3K27me3-specific antibodies or non-specific IgG were used to precipitate cross-linked chromatin, as indicated in the inset key. In B is graphed the same data (“trial 2” in the inset key), and two other trials (“trial 1” and “trial 3”) using independent chromatin preparations (see Materials and Methods), normalized to the H3K27me3-specific signal from the eve coding region for each transgenic line. C, D: embryos (2–20 hr. after egg deposition) were collected and subjected to ChIP analysis, as in A and B above. Note that the specific signals from the transgenic eve-lacZ coding region do not change significantly when Homie is deleted, while the transgenic TER94-GFP coding region shows an increase in H3K27 trimethylation when Homie is deleted, correlating with its repression. In embryos, as in ovaries, H3K27me3 spreads into the TER94-GFP region when Homie is deleted (Figure 4C,D). However, in contrast to ovaries, additionally removing the PRE does not reverse the effect (Figure 4C,D), suggesting that there is redundancy between this PRE and other PREs in embryos. This redundant activity may be provided by the eve upstream promoter region [51], or by uncharacterized PREs within the eve locus. Again, recalling that the eve PRE is redundant in embryos for repression of TER94-GFP in the absence of Homie (Figure S2), there is a striking correlation between spreading of the Pc domain and repression of the TER94 promoter.
Homie Shields the TER94 Promoter from eve Enhancers
Intriguingly, when Homie is deleted, the loss of ubiquitous TER94-GFP expression is accompanied by weak expression in an eve pattern (Figure 1B “ΔHomie”, Figure S1A). With the intact transgene, early stripe expression of TER94-GFP driven by eve enhancers might be obscured by early ubiquitous expression, so we cannot rule out that eve enhancers are working on the TER94 promoter at early stages. In fact, eve-like stripe expression from the transgenic TER94 promoter is seen at one chromosomal landing site when the intact transgene is heterozygous and paternally derived, so that there is no maternal contribution (Figure S2B “intact t'gene”). However, eve-like mesodermal, CNS, and anal plate ring (APR) expression seems clearly to be caused by deletion of Homie (Figure 1B “ΔHomie”, stages 11 and 15; Figure S1A “ΔHomie”, stage 13), because with the intact transgene, ubiquitous expression in these tissues is low, yet no such eve-like expression is seen. Furthermore, these later-stage aspects of eve expression are not seen with a paternally derived, intact transgene (Figure S2). Therefore, the data suggest that one of Homie's functions is to prevent interaction between the TER94 promoter and eve enhancers. Accompanying the recovered ubiquitous expression when the PRE is also deleted, expression in an eve pattern is lost (Figure 1B “ΔHomie ΔPRE”, stages 11 and 15; Figure S1A “ΔHomie ΔPRE”, stage 13). This loss of mesodermal, CNS, and APR expression of TER94-GFP caused by additional deletion of the PRE indicates that the PRE not only represses ubiquitous TER94 promoter activity, but also facilitates communication between the eve enhancers and the TER94 promoter in the absence of Homie (see Discussion).
Homie Facilitates eve 3′ Enhancer Action on the eve Promoter
We then tested whether Homie affects eve promoter activity. To do this, we monitored transgenic lacZ expression, which is driven by the eve promoter (Figure 5, Figure S5A,B). When Homie is removed, there is a reduction in expression driven by enhancers located 3′ of the eve coding region. Interestingly, these are the eve enhancers located between Homie and the eve TSS. Comparing “ΔHomie” with the intact transgene at stage 5 (Figure 5 left column), we see that stripes 1, 4, 5, and 6 are weakened relative to stripes 2, 3, and 7. A similar reduction of expression is seen at later stages, where mesodermal, CNS, and APR expression are weakened by deletion of Homie (Figure 5 middle and right columns). This effect is seen at all transgene landing sites tested (Figure 5A, Figure S5A,B), although it varies in strength with the direction of transgene insertion (data not shown). Despite these differences, we consistently see significant disruptions of normal eve expression when Homie is removed.
Fig. 5. Homie facilitates eve 3′ enhancer action on the eve promoter through a mechanism that does not involve the TER94 promoter. 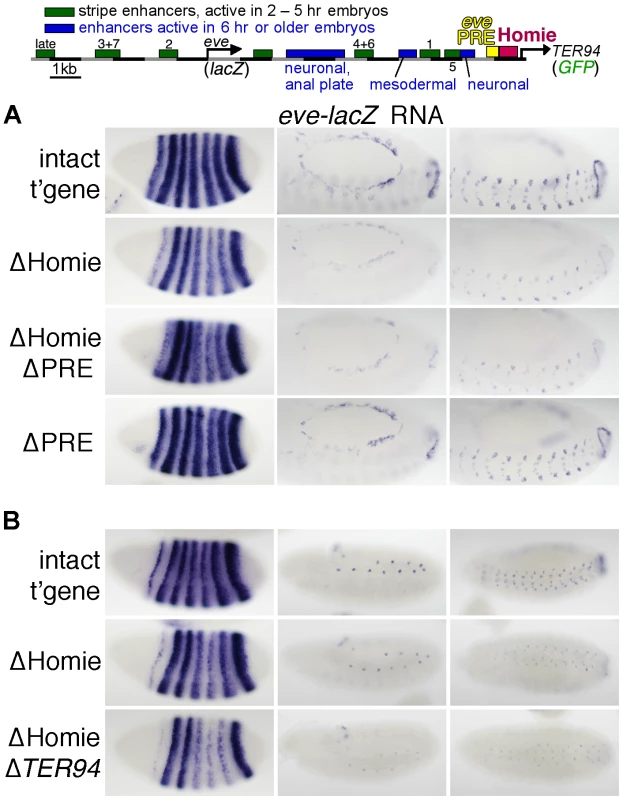
Expression of lacZ RNA driven by the eve promoter from the transgene diagrammed at the top (described in Figure 1A) and its derivatives was monitored by in situ hybridization. A: Representative embryos are shown at either stage 5 (left column) or stage 13 (middle and right columns, which show two different orientations and focal planes at higher magnification). Note that when Homie is deleted (“ΔHomie”), stripes 1, 4, 5, and 6 are weakened relative to stripes 2, 3, and 7 (left column), while all aspects of expression at later stages, in the mesoderm, CNS, and APR, are also weakened (middle and right columns). These weakened expression elements are all driven by enhancers located between the eve and TER94 promoters. These effects are also seen when both Homie and the PRE are deleted (“ΔHomie ΔPRE”), but not when the PRE alone is deleted (“ΔPRE”). B: Representative embryos are shown at 3 embryonic stages (stages 5, 11, and 13, in the left, middle, and right columns, respectively). In ΔHomie ΔTER94, the entire TER94-GFP gene including the TER94 promoter was removed. Note that the weakened activity of 3′ enhancers seen for ΔHomie is also seen for ΔHomie ΔTER94, indicating that competition with the TER94 promoter is not causing the reduced eve promoter activity. It seemed possible that the effects of removing Homie on eve promoter activity were caused by the relief of enhancer blocking, which then might allow eve enhancers access to the TER94 promoter. The resulting promoter competition might reduce eve expression. Alternatively, removing Homie might cause the loss of a chromosome conformation that favors eve enhancer-promoter interactions. This possibility is suggested by the ability of Homie to promote the activation by endogenous eve enhancers of a transgenic eve promoter located up to several megabases away [55]. To distinguish between these possibilities, we performed two sets of experiments. First, we tested whether expression from the TER94 promoter occurs in a pattern that matches the loss of expression from the eve promoter when Homie is deleted. We found that this is not the case. Rather, TER94 expression in an eve striped pattern does not show a difference among the stripes (Figure 1B ΔHomie, compare to Figure 5). Second, we directly tested the promoter competition hypothesis by deleting the TER94 promoter in addition to Homie. Removing the potentially competing promoter did not restore normal eve expression (Figure 5B). While we cannot rule out competition with endogenous promoters, promoter competition seems unlikely to be the primary cause of the eve pattern disruptions that result from removal of Homie. The facilitation of eve promoter activity by Homie may therefore be due to its ability to organize specific chromosomal loops, possibly with the eve promoter (see Discussion).
Interestingly, heterologous insulators are able to restore normal eve enhancer-promoter interactions to different degrees (Figure S5C and data not shown), roughly in parallel to their abilities to restore PRE blocking (Figure 3A,B). For example, gypsy restores normal eve promoter activity, while scs does not (Figure S5C). The abilities of heterologous insulators to perform this function may be due to interactions between them and a region of the eve locus that normally interacts with Homie.
Discussion
A Transgenic eve-TER94 Locus to Assess PRE and Insulator Activity
The eve 3′ insulator, Homie, was shown previously to have three activities: P-element transgene homing, enhancer blocking, and facilitation of long-range enhancer-promoter communication between endogenous eve enhancers and a transgenic promoter [55]. We sought to address how these activities relate to Homie's normal function. Both eve and TER94 are essential genes, and eve is highly dose-dependent, making it problematic to manipulate the endogenous locus. Therefore, we constructed a transgene that contains these genes in their normal configuration. Both the eve and TER94 coding regions were replaced with reporter genes to monitor promoter activity. This transgene simulates the expression pattern of both genes, when inserted at several different chromosomal sites. We used this system to manipulate both Homie and the nearby PRE, to assess their normal functions.
Homie Protects the TER94 Promoter from Spreading of PRE-dependent Repressive Chromatin
A major finding of this study is that Homie is required to prevent PRE-dependent repression of the TER94 promoter. Removal of Homie causes a near-complete loss of the normally ubiquitous TER94 promoter activity. Although Homie is close to the TER94 promoter, its removal does not affect the promoter directly. Rather, removing Homie allows eve enhancers to drive the TER94 promoter in an eve pattern (Figure 1). Furthermore, additional removal of the nearby PRE restores ubiquitous expression. This restoration is complete in some instances (e.g., Figures 1, 2), although it is incomplete in others (e.g., with a paternally-derived transgene in embryos, Figure S2). A simple explanation for the lack of complete restoration in some circumstances is that PRE activity varies in different tissues, and the eve 3′ PRE is partially redundant with other PREs at some times in development.
Ubiquitous expression of TER94 in early embryos, as well as some of the later ubiquitous CNS expression, is due to maternally loaded RNA. Consistent with this, expression in ovaries is robust, and, like early embryonic expression, is strongly repressed without Homie (Figures 2, S4). Accompanying repression in both ovaries and embryos, trimethylation of H3K27 at TER94-GFP is strongly increased when Homie is removed (Figure 4). Thus, without Homie, the eve Pc domain spreads into the adjacent gene, apparently shutting down expression.
Homie is bound in vivo by most known insulator binding proteins, including Su(Hw), CP190, Mod(mdg4)67.2, BEAF32, CTCF, and GAF [27], [33]. In a previous study, depletion of CTCF by RNAi in a cultured cell line caused a reduction in H3K27me3 levels throughout the eve locus [36]. The authors suggested that depleting CTCF altered the activity of insulators flanking eve, which led to a decrease in H3K27me3. In contrast, we found that deletion of Homie did not cause a significant reduction in H3K27me3 levels in the eve-lacZ region of our pseudo-locus, either in embryos or in ovaries (Figure 4). There could be several possible reasons for this discrepancy, including the cell types assayed, and indirect effects of depleting CTCF.
Tissue Specificity and Redundancy of PRE Activity
With removal of Homie, the spreading of H3K27me3 in ovaries is reversed by deletion of the PRE (Figure 4A,B). However, in embryos, this spreading is only partially reversed (Figure 4C,D). A simple explanation for this is that additional PRE activity within the eve locus comes into play in embryos. Consistent with this, the eve promoter-proximal region has PRE-like properties. It causes pairing-sensitive silencing of mini-white in transgenes that carry it [51], a property associated with most known PREs. Furthermore, it has consensus binding sites for several PRE-associated DNA binding proteins [8], [51], and it shares with the eve 3′ PRE the ability to support positive epigenetic maintenance of enhancer activity from embryos to larvae within eve-positive neurons [51].
Perhaps the clearest evidence for redundant PRE activity within the eve locus is that the level of H3K27me3 at the eve-lacZ coding region is not significantly reduced when the 3′ PRE is deleted. This is true in both embryos and ovaries. In contrast, spreading of the Pc domain into TER94 in ovaries requires the 3′ PRE (Figure 4). Our data are consistent with the idea that PREs are the nucleation point for spreading of the H3K27me3 mark, and that PRE activity is regulated, so that PREs are differentially active in different tissues.
Furthermore, because there may be a dynamic balance between active and repressive chromatin, maintaining a boundary between them may have different requirements at different chromosomal locations, and at different times in development. Insulators that are not required to maintain a boundary in one cell type may be required for that function in other cells, or at specific times in development, as previously suggested [34]. One reason for such differences may be regulated PRE activity.
In some cases, spreading of repressive chromatin can be stopped by an active promoter [11], [64]. In the case of the TER94 promoter, although it is robustly expressed, particularly in ovaries, this is not sufficient to stop the spreading of H3K27me3 in the absence of an insulator. This contrasts with the suggestion from recent genome-wide studies in both cultured cells and Drosophila that insulator protein function is generally not required to prevent spreading of H3K27me3 into active genes, or to maintain most normal gene expression [34], [36], [38]. Because many insulator proteins bind to overlapping sets of sites, it is likely that there is considerable redundancy in their function. Thus, knocking out any one of them may not reveal the full function of a majority of their binding sites.
Enhancer Action from Within a Pc Domain
It is intriguing that the eve locus is a Pc domain with well-defined boundaries that flank its extensive regulatory regions. Within chromosomal domains of the Drosophila bithorax complex (BX-C), active enhancers prevent the establishment of repressive Pc-dependent chromatin in early embryos. Conversely, in tissues where such repressive chromatin has been established, such as in parts of the CNS and imaginal discs, later-acting enhancers are repressed [25]. Do similar mechanisms operate within the eve locus? Extensive dissection of eve regulatory DNA has not identified enhancers that can drive expression outside the normal eve pattern, arguing against such a close analogy with the BX-C. However, in PcG mutants, eve is ectopically expressed throughout the late-stage embryonic CNS [50], [54], showing that PcG genes do negatively regulate eve, as they do the Hox complexes.
In our previous studies of eve PRE activity, we found that in a transgenic context, both the 3′ PRE and the PRE-like eve promoter region could facilitate positive maintenance of an eve CNS enhancer from embryonic to larval stages, as well as prevent ectopic expression in cells that normally do not express eve [51]. Unlike maintenance elements [65] in the BX-C, the eve 3′ PRE was found to require the DNA binding PcG protein Pleiohomeotic rather than Trithorax-group members for positive maintenance [51]. In this study, we also see evidence of a positive effect of the eve 3′ PRE on enhancer activity. In this case, it facilitates TER94-GFP expression in an eve pattern when Homie is removed (Figure 1B: eve-like mesodermal and CNS expression are seen when Homie is removed, but are not seen when both Homie and the PRE are removed). One possible explanation for this is that eve enhancers have evolved to function within a Pc domain, and they may be better able to activate the TER94 promoter when the Pc domain spreads over it. In this view, PREs facilitate both the on state and the off state, yet the chromatin may be differently modified in the two cases. This model is similar to the “integration model” proposed for how heterochromatin can have a positive effect on the expression of genes that normally reside within it [66].
Homie's Activity Relative to that of Other Drosophila and Mammalian Insulators
Homie sits adjacent to the eve 3′ PRE, an arrangement that is reminiscent of some boundaries in the BX-C. The mammalian homologs of eve, evx1 and evx2, are located at the 3′ end of the HOX-A and HOX-D clusters, respectively, suggesting that the ancestral eve locus was part of a Hox cluster [67]. Consistent with conservation of the eve insulator-PRE relationship, recent studies identified an enhancer-blocking activity between evx2 and Hoxd13 [68], and a PRE in the HOX-D cluster [69]. The presence of a PRE near an insulator, with a promoter on the other side, may indicate a functionally important boundary between active and repressive chromatin domains.
Previous studies showed that within the BX-C, neither scs nor gypsy could functionally replace Fab-7 [70], indicating that there are different classes or strengths of insulators. In these cases, the primary effects of insulator deletion was ectopic activation, due to early acting enhancers (“initiators”) “turning off” PREs throughout a chromatin domain delineated by insulators [26], [71]. In our system, the major effect of insulator deletion is the spreading of the eve Pc domain, reminiscent of the shielding of transgenic reporter genes from repressive effects at some insertion sites [18], [72]–[75]. Despite the differences in normal function, BX-C insulators can replace Homie in our assay, indicating some degree of universality in insulator function as a PRE blocker. However, our assays did reveal differences in effectiveness in carrying out this function. Specifically, scs' showed slightly weaker activity than either gypsy, Fab-7, or Fab-8, while the activity of scs was highly orientation-dependent (Figure 3).
Homie Blocks Enhancer-promoter Cross-talk, While Facilitating Enhancers that Lie Between It and the eve Promoter
Deletion of Homie results in expression of TER94-GFP in an eve pattern. In fact, the eve early embryonic stripe enhancers may access the TER94 promoter even when Homie is present, because with a paternal-only transgene, we sometimes see eve-like stripe expression from TER94-GFP (Figure S2B “intact t'gene”). However, at later stages of embryogenesis, we do not see eve-like expression in either the mesoderm, CNS, or APR unless Homie is deleted. Therefore, one of Homie's functions is to prevent communication between the TER94 promoter and eve enhancers.
Deletion of Homie, but not deletion of the PRE, also reduced eve-lacZ expression driven by the eve 3′ enhancers (Figures 5, S5). We considered the possibility that because the TER94 promoter has access to eve enhancers in the absence of Homie, the resulting promoter competition might reduce eve promoter activity. However, in ΔHomie lines where we see TER94 expressed in eve stripes, there is no apparent bias in expression toward the 3′ enhancers (Figures 1B, S1A), arguing against this possibility. Furthermore, at later embryonic stages, eve promoter activity is reduced when both Homie and the PRE are removed (in mesoderm, CNS, and APR, which are all the tissues where eve is expressed at these stages, Figure 5A), but this is not accompanied by TER94-GFP expression in an eve pattern (Figures 1B, S1A). Finally, when the TER94 promoter is removed along with Homie, pattern disruptions persist (Figure 5B). While we cannot rule out competition with other promoters in the genome, these lines of evidence together suggest that promoter competition is unlikely to be responsible for this effect.
A second possible explanation for the reduction in eve 3′ enhancer-promoter communication when Homie is deleted is that a 3-dimensional (3-D) conformation that allows the eve promoter to better access the 3′ enhancers is stabilized by the presence of Homie. One possible conformation is a loop between the eve promoter region and Homie (Figure 6). Although we have not tested this directly, evidence consistent with this model is that activation of promoters, including the eve promoter, by downstream Gal4 binding sites can be facilitated by heterologous insulators in a model transgene assay [76]. This possible pairing of Homie with the eve promoter region would result in a loop that would bring the 3′ enhancers in closer proximity to the promoter. Such a model is similar to that proposed for the 3-D organization of regulatory regions upstream of the Abd-B gene [25]. If such loops are anchored to large clusters of insulator proteins, perhaps within insulator bodies, this may serve as a 3-D barrier that separates distinct chromatin domains, and occludes interactions between regulatory elements located on opposite sides of the insulator. At the same time, otherwise distant elements can be brought closer together, facilitating specific enhancer-promoter contacts, particularly if those elements are brought to the same side of the 3-D barrier.
Fig. 6. Model of the effects of Homie deletion on chromosome conformation and chromatin structure. 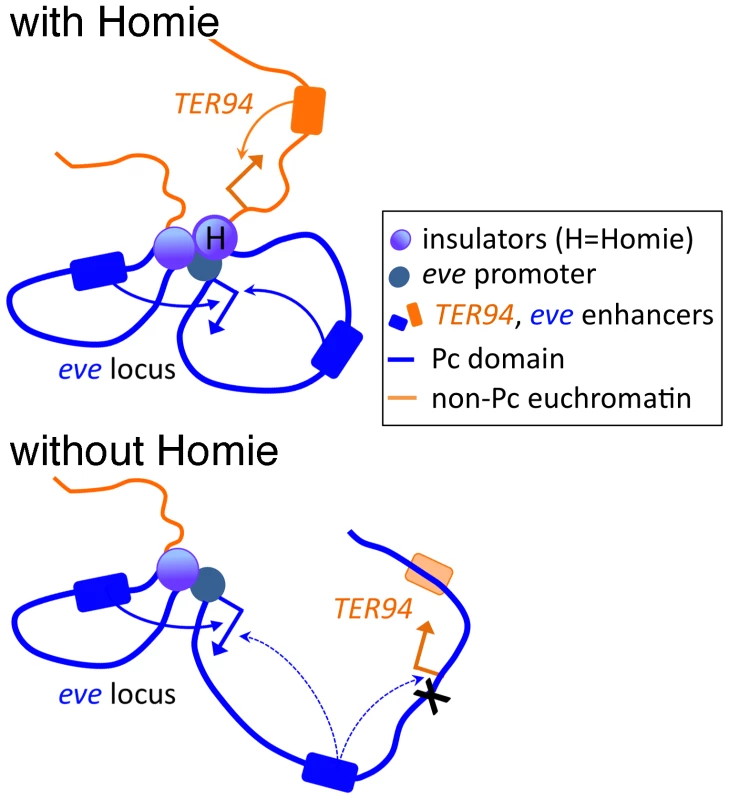
At the top, Homie is present, and separates the eve Pc domain (blue) from the TER94 locus, which is constitutively in active chromatin (orange). The eve enhancers both 5′ and 3′ of the eve start site efficiently activate the eve promoter, and the TER94 enhancers activate the TER94 promoter. Below, when Homie is removed, Pc-dependent chromatin spreads into the TER94 locus, preventing its activation by TER94 enhancers, but allowing eve enhancers to activate it. At the same time, eve 3′ enhancers interact with the eve promoter less efficiently, due to a change in chromosome conformation. Implications for Other Insulators and PREs throughout the Genome
The activities of Homie and the eve PRE are largely interchangeable with those of other insulators and PREs, respectively, in our assay system. Previous studies showed that Homie and the eve PRE have the canonical properties of insulators and PREs when tested in other contexts [51], [55]. Thus, our results are likely to be applicable to many such elements throughout the genome. In particular, a common function of insulators is likely to be to limit the action of PRE-dependent repressive chromatin.
Genome-wide studies using RNAi to knock down specific insulator proteins suggested that insulators may not typically be required in their normal context either to block enhancer-promoter cross-talk or to prevent the spread of repressive chromatin [34], [36]. Our results suggest that Homie is critically important in its normal context for just such activities, functionally separating the loci on either side. Importantly, other insulators function in place of Homie. This suggests that the activities of insulators defined in model transgene assays do in fact correspond to their normal functions. In particular, as with Homie and the TER94 promoter, the tendency of insulator proteins to cluster just upstream of promoters suggests that one of their typical functions is to shield basal promoters from the effects of upstream CRMs, especially PREs. Further, our finding that insulators facilitate enhancer-promoter communication in this context suggests that their ability to organize chromosomal conformations that augment appropriate transcription is also likely to be a common mode of endogenous insulator function.
Materials and Methods
Plasmid Construction and Transgene Production
The eve-TER94 locus construct (“intact t'gene” in figures) was created as follows (detailed sequence coordinates are given in Figure S6). DNA from −6.4 kb to +166 bp relative to the eve TSS was fused to the lacZ coding region. The 3′ end of the lacZ coding region was fused to DNA from +1.3 to +11.4 kb, which includes the eve poly-A signal, and extends into the 3rd exon of TER94. This was joined with the EGFP coding region, followed by the poly-A signal of α–tubulin. The entire construct was placed between two inverted attB sequences [63], [77]. The following deletions were then made in this construct: from +8.4 to +9.2 kb for ΔPRE, from +8.4 to +9.7 kb for ΔHomie ΔPRE, and from +9.2 to +9.7 kb for ΔHomie. To test promoter competition between eve and TER94, DNA from −7.4 to +8.6 kb relative to the eve TSS was used, with the eve coding region replaced by that of lacZ, as described above. This construct does not contain the TER94 promoter.
Replacements of Homie with either heterologous insulators or phage λ DNA were created using the ΔHomie construct, and adding DNA fragments corresponding to gypsy [78], Fab-7 [79]–[82], Fab-8 [83], [84], scs [85], [86], scs' [85], [86], or λ DNA (see Figure S6 for details). For testing repression activity of heterologous PREs, either the engrailed 181PRE [87] or the bxd PRE [88] were inserted into the ΔHomie ΔPRE construct at the site of deletion. For testing Homie activity against these PREs, either the en PRE or the bxd PRE were inserted into the ΔPRE construct at the site of deletion.
All transgenic lines were made using φC31 recombinase-mediated cassette exchange (RMCE) [63]. Three alternative attP target sites were used, at cytological locations 95E5, 74A2, and 30B5. The direction of each insertion was determined by PCR. Both directions were analyzed if obtained. Some variations with insertion site were found, as described in Results.
Analysis of Gene Expression in Embryos and Ovaries
Embryos were collected at time points described in figure legends, and subjected to in situ hybridization using DIG-labeled anti-sense RNA probes against either lacZ or GFP. Expression patterns were visualized by alkaline phosphatase-conjugated anti-DIG with BCIP and NBT as substrates (Roche Applied Science).
GFP expression was detected by fluorescence microscopy in ovaries dissected from 1–2 day-old females. In some cases, expression was also detected using anti-GFP antibody staining (Roche Applied Science), analyzed by confocal microscopy (Zeiss) of material in DAPI-containing mounting medium.
Chromatin Immunoprecipitation
Ovaries were dissected from 2–3 day-old females. Fifty ovaries were cross-linked in 1.8% formaldehyde in PBS for 10 min. After sonication so as to produce a peak near 500 bp in the DNA fragment size distribution, isolated chromatin was immunoprecipitated with anti-H3K27me3 (EMD Millipore), and with rabbit IgG (Jackson ImmunoResearch) as a negative control. Precipitated chromatin samples were collected using ProteinG magnetic beads (EMD Millipore). Immunoprecipitated DNA samples were dissolved in 20–50 µl TE, and 1 µl was used for each PCR reaction. Either duplicate or triplicate samples were analyzed by real-time PCR (Life Technologies, StepOnePlus), using SYBR Green Master Mix with ROX dye (Roche Applied Science). Data were analyzed with StepOne software (Life Technologies), using the standard curve method. Standard deviations were calculated using Excel software (Microsoft). Embryo ChIP analysis was described previously [51], except that results were quantified by real-time PCR, as described above for ovary analysis.
Specific ChIP signals were determined by subtracting the average non-specific IgG signal from the average α-H3K27me3 signal, with standard deviations combined by adding. Errors bars for specific signals relative to that of endogenous eve were determined by adding the relative errors in quadrature; that is, by taking the sum of the squares of the relative standard deviations (the standard deviations divided by their respective averages) to give the square of the relative standard deviation of the ratio.
The following primers were used: TCCAGTCCGGATAACTCCTTGAAC and TGTAGAACTCCTTCTCCAAGCGAC for the endogenous eve coding region, TGAAGCCACCGCGTGGTATTCTTA and TTTGGACATGATCTCCGGTCCGTT for the endogenous TER94 coding region, GCTGTGCCGAAATGGTCCATCAAA and TACTGACGAAACGCCTGCCAGTAT for the transgenic eve-lacZ coding region, and GGGCACAAGCTGGAGTACAACTACAA and TGGCGGATCTTGAAGTTCACCTTG for the transgenic TER94-GFP coding region.
RT-PCR
Total RNA was purified from either five pairs of ovaries from 2–3 day-old females or 10–20 µl of dechorionated embryos for each data point, using an RNA purification kit (Roche Applied Science). RNA was eluted in 50–100 µl elution buffer and stored at −80°C. cDNA was synthesized using the Transcriptor first strand cDNA synthesis kit (Roche Applied Science), and quantified by real-time PCR as described above. A constitutively expressed RNA, RpL32 (a.k.a. RP49), was used to normalize GFP RNA levels. The primers listed above for TER94-GFP were used for GFP, and AAGCCCAAGGGTATCGACAACAGA and TGCACCAGGAACTTCTTGAATCCG were used for RpL32.
Supporting Information
Zdroje
1. DuncanIW (2002) Transvection effects in Drosophila. Annu Rev Genet 36 : 521–556.
2. KennisonJA, SouthworthJW (2002) Transvection in Drosophila. Adv Genet 46 : 399–420.
3. PirrottaV (1999) Transvection and chromosomal trans-interaction effects. Biochimica et Biophysica Acta 1424: M1–8.
4. WilliamsA, SpilianakisCG, FlavellRA (2010) Interchromosomal association and gene regulation in trans. Trends Genet 26 : 188–197.
5. SchuettengruberB, CavalliG (2009) Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136 : 3531–3542.
6. SchwartzYB, PirrottaV (2008) Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20 : 266–273.
7. SimonJA, KingstonRE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nature Reviews Molecular Cell Biology 10 : 697–708.
8. KassisJA, BrownJL (2013) Polycomb group response elements in Drosophila and vertebrates. Adv Genet 81 : 83–118.
9. BusheyAM, DormanER, CorcesVG (2008) Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32 : 1–9.
10. GhirlandoR, GilesK, GowherH, XiaoT, XuZ, et al. (2012) Chromatin domains, insulators, and the regulation of gene expression. Biochimica et Biophysica Acta 1819 : 644–651.
11. RaabJR, KamakakaRT (2010) Insulators and promoters: closer than we think. Nature Reviews Genetics 11 : 439–446.
12. YangJ, CorcesVG (2012) Insulators, long-range interactions, and genome function. Curr Opin Genet Dev 22 : 86–92.
13. KharchenkoPV, AlekseyenkoAA, SchwartzYB, MinodaA, RiddleNC, et al. (2011) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471 : 480–485.
14. SchuettengruberB, GanapathiM, LeblancB, PortosoM, JaschekR, et al. (2009) Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13.
15. SchwartzYB, KahnTG, NixDA, LiXY, BourgonR, et al. (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38 : 700–705.
16. SchwartzYB, KahnTG, StenbergP, OhnoK, BourgonR, et al. (2010) Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805.
17. TolhuisB, de WitE, MuijrersI, TeunissenH, TalhoutW, et al. (2006) Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster.[erratum appears in Nat Genet. 2006 Jul;38(7):850]. Nat Genet 38 : 694–699.
18. SigristCJ, PirrottaV (1997) Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147 : 209–221.
19. DevidoSK, KwonD, BrownJL, KassisJA (2008) The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development 135 : 669–676.
20. JeonY, LeeJT (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146 : 119–133.
21. LeeJT (2012) Epigenetic regulation by long noncoding RNAs. Science 338 : 1435–1439.
22. ZhaoJ, SunBK, ErwinJA, SongJ-J, LeeJT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322 : 750–756.
23. MaedaRK, KarchF (2006) The ABC of the BX-C: the bithorax complex explained. Development 133 : 1413–1422.
24. MaedaRK, KarchF (2007) Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev 17 : 394–399.
25. MaedaRK, KarchF (2009) The bithorax complex of Drosophila an exceptional Hox cluster. Current Topics in Developmental Biology 88 : 1–33.
26. IampietroC, GummallaM, MuteroA, KarchF, MaedaRK (2010) Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet 6: e1001260.
27. CelnikerSE, DillonLAL, GersteinMB, GunsalusKC, HenikoffS, et al. (2009) Unlocking the secrets of the genome. Nature 459 : 927–930.
28. BartkuhnM, StraubT, HeroldM, HerrmannM, RathkeC, et al. (2009) Active promoters and insulators are marked by the centrosomal protein 190. EMBO J 28 : 877–888.
29. BusheyAM, RamosE, CorcesVG (2009) Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23 : 1338–1350.
30. FilionGJ, van BemmelJG, BraunschweigU, TalhoutW, KindJ, et al. (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143 : 212–224.
31. HolohanEE, KwongC, AdryanB, BartkuhnM, HeroldM, et al. (2007) CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3: e112.
32. NegreN, BrownCD, MaL, BristowCA, MillerSW, et al. (2011) A cis-regulatory map of the Drosophila genome. Nature 471 : 527–531.
33. NegreN, BrownCD, ShahPK, KheradpourP, MorrisonCA, et al. (2010) A Comprehensive Map of Insulator Elements for the Drosophila Genome. PLoS Genet 6: e1000814.
34. SchwartzYB, Linder-BassoD, KharchenkoPV, TolstorukovMY, KimM, et al. (2012) Nature and function of insulator protein binding sites in the Drosophila genome.[Erratum appears in Genome Res. 2013 Feb;23(2):409]. Genome Res 22 : 2188–2198.
35. SmithST, WickramasingheP, OlsonA, LoukinovD, LinL, et al. (2009) Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol 328 : 518–528.
36. Van BortleK, RamosE, TakenakaN, YangJ, WahiJE, et al. (2012) Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res 22 : 2176–2187.
37. van BemmelJG, PagieL, BraunschweigU, BrugmanW, MeulemanW, et al. (2010) The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE 5: e15013.
38. SoshnevAA, HeB, BaxleyRM, JiangN, HartCM, et al. (2012) Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res 40 : 5415–5431.
39. WoodAM, Van BortleK, RamosE, TakenakaN, RohrbaughM, et al. (2011) Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell 44 : 29–38.
40. JiangN, EmberlyE, CuvierO, HartCM (2009) Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Molecular & Cellular Biology 29 : 3556–3568.
41. SextonT, YaffeE, KenigsbergE, BantigniesF, LeblancB, et al. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148 : 458–472.
42. HeroldM, BartkuhnM, RenkawitzR (2012) CTCF: insights into insulator function during development. Development 139 : 1045–1057.
43. HouC, CorcesVG (2012) Throwing transcription for a loop: expression of the genome in the 3D nucleus. Chromosoma 121 : 107–116.
44. KimA, DeanA (2012) Chromatin loop formation in the -globin locus and its role in globin gene transcription. Molecules & Cells 34 : 1–5.
45. BellAC, WestAG, FelsenfeldG (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98 : 387–396.
46. HouC, ZhaoH, TanimotoK, DeanA (2008) CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proceedings of the National Academy of Sciences of the United States of America 105 : 20398–20403.
47. BellAC, FelsenfeldG (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene.[see comment]. Nature 405 : 482–485.
48. HarkAT, SchoenherrCJ, KatzDJ, IngramRS, LevorseJM, et al. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus.[see comment]. Nature 405 : 486–489.
49. KanduriC, PantV, LoukinovD, PugachevaE, QiCF, et al. (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10 : 853–856.
50. DuraJM, InghamP (1988) Tissue - and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos by the polyhomeotic gene. Development 103 : 733–741.
51. FujiokaM, YusibovaGL, ZhouJ, JaynesJB (2008) The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development 135 : 4131–4139.
52. KimSN, ShimHP, JeonB-N, ChoiW-I, HurM-W, et al. (2011) The pleiohomeotic functions as a negative regulator of Drosophila even-skipped gene during embryogenesis. Molecules & Cells 32 : 549–554.
53. McKeonJ, SladeE, SinclairDA, ChengN, CoulingM, et al. (1994) Mutations in some Polycomb group genes of Drosophila interfere with regulation of segmentation genes. Molecular & General Genetics 244 : 474–483.
54. SmouseD, GoodmanC, MahowaldA, PerrimonN (1988) polyhomeotic: a gene required for the embryonic development of axon pathways in the central nervous system of Drosophila. Genes Dev 2 : 830–842.
55. FujiokaM, WuX, JaynesJB (2009) A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development 136 : 3077–3087.
56. FujiokaM, Emi-SarkerY, YusibovaGL, GotoT, JaynesJB (1999) Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126 : 2527–2538.
57. GotoT, MacdonaldP, ManiatisT (1989) Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57 : 413–422.
58. HardingK, HoeyT, WarriorR, LevineM (1989) Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. The EMBO Journal 8 : 1205–1212.
59. SackersonC, FujiokaM, GotoT (1999) The even-skipped locus is contained in a 16-kb chromatin domain. Dev Biol 211 : 39–52.
60. LeonA, McKearinD (1999) Identification of TER94, an AAA ATPase protein, as a Bam-dependent component of the Drosophila fusome. Molecular Biology of the Cell 10 : 3825–3834.
61. PinterM, JekelyG, SzepesiRJ, FarkasA, TheopoldU, et al. (1998) TER94, a Drosophila homolog of the membrane fusion protein CDC48/p97, is accumulated in nonproliferating cells: in the reproductive organs and in the brain of the imago. Insect Biochemistry & Molecular Biology 28 : 91–98.
62. RudenDM, SollarsV, WangX, MoriD, AltermanM, et al. (2000) Membrane fusion proteins are required for oskar mRNA localization in the Drosophila egg chamber. Dev Biol 218 : 314–325.
63. BatemanJR, LeeAM, WuCT (2006) Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173 : 769–777.
64. RaabJR, ChiuJ, ZhuJ, KatzmanS, KurukutiS, et al. (2012) Human tRNA genes function as chromatin insulators. EMBO J 31 : 330–350.
65. BrockHW, van LohuizenM (2001) The Polycomb group–no longer an exclusive club? Curr Opin Genet Dev 11 : 175–181.
66. YasuharaJC, WakimotoBT (2006) Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet 22 : 330–338.
67. Garcia-FernandezJ (2005) The genesis and evolution of homeobox gene clusters. Nature Reviews Genetics 6 : 881–892.
68. VasanthiD, AnantM, SrivastavaS, MishraRK (2010) A functionally conserved boundary element from the mouse HoxD locus requires GAGA factor in Drosophila. Development 137 : 4239–4247.
69. WooCJ, KharchenkoPV, DaheronL, ParkPJ, KingstonRE (2010) A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140 : 99–110.
70. HoggaI, MihalyJ, BargesS, KarchF (2001) Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol Cell 8 : 1145–1151.
71. IampietroC, CleardF, GyurkovicsH, MaedaRK, KarchF (2008) Boundary swapping in the Drosophila Bithorax complex. Development 135 : 3983–3987.
72. CometI, SchuettengruberB, SextonT, CavalliG (2011) A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proceedings of the National Academy of Sciences of the United States of America 108 : 2294–2299.
73. ErokhinM, ParshikovA, GeorgievP, ChetverinaD (2010) E(y)2/Sus1 is required for blocking PRE silencing by the Wari insulator in Drosophila melanogaster. Chromosoma 119 : 243–253.
74. KahnTG, SchwartzYB, DellinoGI, PirrottaV (2006) Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem 281 : 29064–29075.
75. MallinDR, MyungJS, PattonJS, GeyerPK (1998) Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148 : 331–339.
76. ErokhinM, DavydovaA, KyrchanovaO, ParshikovA, GeorgievP, et al. (2011) Insulators form gene loops by interacting with promoters in Drosophila. Development 138 : 4097–4106.
77. GrothAC, FishM, NusseR, CalosMP (2004) Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166 : 1775–1782.
78. GeyerPK, CorcesVG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6 : 1865–1873.
79. GyurkovicsH, GauszJ, KummerJ, KarchF (1990) A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J 9 : 2579–2585.
80. HagstromK, MullerM, SchedlP (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev 10 : 3202–3215.
81. KarchF, GalloniM, SiposL, GauszJ, GyurkovicsH, et al. (1994) Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res 22 : 3138–3146.
82. MihalyJ, HoggaI, GauszJ, GyurkovicsH, KarchF (1997) In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124 : 1809–1820.
83. BargesS, MihalyJ, GalloniM, HagstromK, MullerM, et al. (2000) The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127 : 779–790.
84. ZhouJ, AsheH, BurksC, LevineM (1999) Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development 126 : 3057–3065.
85. BlantonJ, GasznerM, SchedlP (2003) Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev 17 : 664–675.
86. KellumR, SchedlP (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell 64 : 941–950.
87. AmericoJ, WhiteleyM, BrownJL, FujiokaM, JaynesJB, et al. (2002) A complex array of DNA-binding proteins required for pairing-sensitive silencing by a polycomb group response element from the Drosophila engrailed gene. Genetics 160 : 1561–1571.
88. FritschC, BrownJL, KassisJA, MullerJ (1999) The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126 : 3905–3913.
Štítky
Genetika Reprodukčná medicína
Článek Defending Sperm FunctionČlánek How to Choose the Right MateČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase inČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



