-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene
In plants, expression of certain imprinted genes is restricted to embryo nourishing tissue, the endosperm. Since these genes are silenced by epigenetic mechanisms during vegetative growth, it has been assumed that they have no role in this phase of the plant life cycle. Here, we report on heat-mediated release of epigenetic silencing and ectopic activation of the Arabidopsis thaliana endosperm-imprinted gene SDC. The stress induced activation of SDC involves epigenetic regulation but not the canonical heat-shock perception and signaling, and it seems to be required for efficient growth recovery after the stress. Our results exemplify a potential concealed role of an imprinted gene in plant responses to environmental challenges.
Published in the journal: Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene. PLoS Genet 10(11): e32767. doi:10.1371/journal.pgen.1004806
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004806Summary
In plants, expression of certain imprinted genes is restricted to embryo nourishing tissue, the endosperm. Since these genes are silenced by epigenetic mechanisms during vegetative growth, it has been assumed that they have no role in this phase of the plant life cycle. Here, we report on heat-mediated release of epigenetic silencing and ectopic activation of the Arabidopsis thaliana endosperm-imprinted gene SDC. The stress induced activation of SDC involves epigenetic regulation but not the canonical heat-shock perception and signaling, and it seems to be required for efficient growth recovery after the stress. Our results exemplify a potential concealed role of an imprinted gene in plant responses to environmental challenges.
Introduction
It has been long recognized that transcriptional gene silencing (TGS) in plants is associated mainly with increased levels of DNA methylation [1], [2]. DNA methylation is found in cytosines (C) residing in CG, CHG and CHH sequence contexts (where H stands for A, T or C). Methyltransferase 1 (MET1) perpetuates CG methylation patterns during DNA replication. Cytosine methylation in CHG and CHH sequences is mediated by Chromomethylase 3 (CMT3) and Chromomethylase 2 (CMT2), respectively [3]–[6]. Cytosine methylation in asymmetric CHH sequences cannot be maintained in a replicative manner and the RNA-dependent DNA methylation (RdDM) pathway leads to their methylation de novo through sequence-specific targeting with small interfering RNAs, and thus the mitotic persistence of TGS [7]. De novo DNA methylation occurs in all sequence contexts and is mainly catalyzed by Domains Rearranged Methyltransferase 2 (DRM2) [3], [8].
TGS is involved in the epigenetic suppression of invading DNA, such as that of pathogens but also of endogenous transposons, which threaten genome stability by their mutational capacity and deleterious regulatory effects on neighboring genes [9], [10]. However, TGS is also involved in genomic imprinting, i.e. allele-specific expression dependent on the parent-of-origin. The expression of some imprinted genes in plants is restricted to seed endosperm and is associated with silencing during somatic growth [11]. Such strict developmental regulation of imprinted gene expression is critical for seed and plant development. In Arabidopsis thaliana, aberrant expression of imprinted genes such as Medea (MEA) and Fertilization Independent Seed 2 (FIS2) has strong phenotypic consequences that lead to seed abortion [12]. Ectopic expression of imprinted genes during vegetative growth may also have phenotypic consequences. For example, the imprinted A. thaliana gene SDC is epigenetically silenced in somatic tissues due to DNA methylation targeted by the RdDM pathway to tandem-repeats within its promoter. This locus is highly activated in particular combinations of TGS mutants such as drm1/drm2/cmt3 and ddm1/drd1, which results in leaf curling and plant dwarfism [5], [13].
Although epigenetic mechanisms can suppress transcription at ambient temperatures, it was reported recently that transcriptional activation can occur transiently during prolonged exposure to heat [14]–[17]. The degree of activation was proportional to the duration of the stress and was associated with decreased nucleosome occupancy and resulting chromatin decondensation. Importantly, chromatin assembly factors restored silencing within 48 h after heat stress [14]. Here, we report on heat stress-mediated ectopic activation of the imprinted SDC gene in vegetative tissues. The stress-triggered transcriptional response of SDC occurred particularly in young developing leaves and the kinetics of re-silencing could be entrained by repeated heat stress cycles. We provide evidence for a physiological role of this unexpected regulation of an imprinted gene during recovery from heat stress.
Results
To analyze the heat-mediated release of TGS, we used a transgenic line carrying a silent 35S::GUS construct, referred to as L5-GUS [2]. The transcription of this transgene is repressed by DNA methylation of the promoter, and its silencing is released in several epigenetic mutants such as mom1, ddm1 and met1 [2], [14]. L5-GUS A. thaliana seedlings were subjected to an acclimation treatment consisting of a varying number of diurnal heat cycles as entrainment. Each cycle comprised 12 h at 37°C in the light and 12 h at 21°C in the dark. This experimental design with elevated temperature associated with light periods closely models natural growth conditions, when plants experience high temperatures mostly during the day. The entrainment was followed by a recovery period of 3 days at 21°C with a 12 h/12 h light/dark cycle. After recovery, an additional heat cycle (second stress) was applied to a subset of the entrained seedlings (Figure 1A). As expected, GUS transcription was released upon heat stress but resulted in similar transcript levels between each heat cycle and the second stress. However, GUS mRNA levels during recovery showed a stepwise increase proportional to the number of heat stress cycles (Figure 1B). This result suggests either transcriptional memory related to the previous heat-induced release of silencing or merely the physiological consequence of a higher perceived stress dose. To distinguish between these possibilities, we examined the transcriptional regulation of typical heat stress-responsive genes after the entrainment. These loci did not show an L5-GUS-related pattern of mRNA accumulation, during either activation or recovery (Figure S1), indicating that the transcriptional consequences of entrainment were specific to the epigenetically regulated L5-GUS transgene.
Fig. 1. Transcriptional memory of the heat-induced SDC activation state. 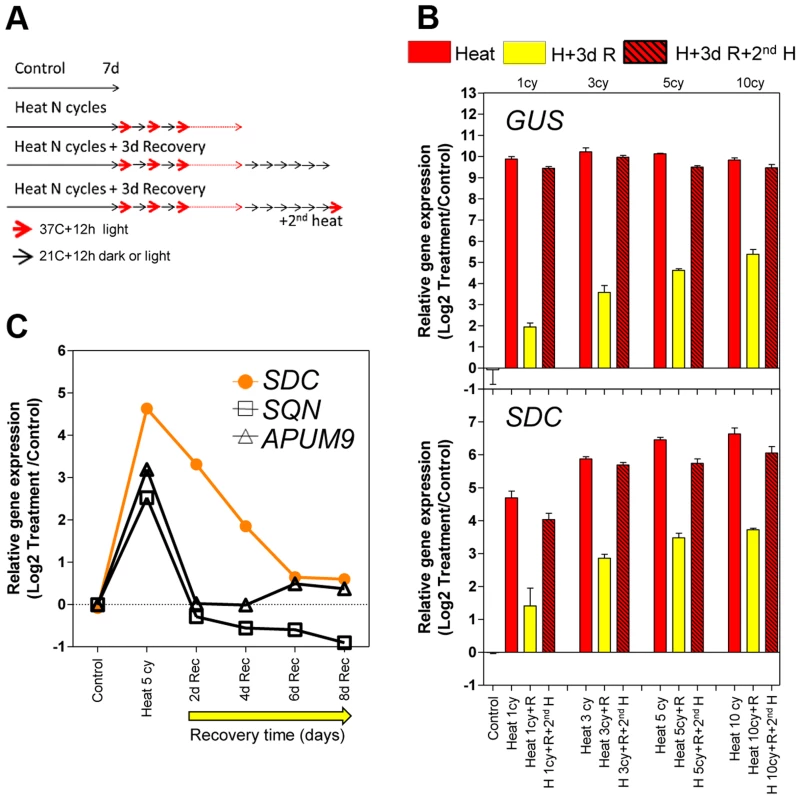
A, Design of the heat-entrainment experiment. Seven-day-old A. thaliana seedlings were subjected to standard conditions, heat cycle entrainment (day 37°C and night 21°C, 12/12 h), heat cycle entrainment +3 days recovery at 21°C, and heat cycle entrainment +3 days recovery +2nd heat treatment (day 37°C,12 h). Heat entrainment was performed for 1, 3, 5 or 10 cycles. B, Transcript levels of L5-GUS and SDC loci during recovery from heat-induced release of silencing. Bars represent means ± SE as a log2 ratio with the non-treated control condition (i.e., control = 0); replicated samples were pooled from 40–60 whole seedlings. C, Kinetics of transcript level re-silencing of the partially silent genes during the recovery phase following a 5 heat-cycle entrainment. Dots represent mean gene expression as a log2 ratio with the non-treated control condition (i.e., control = 0); replicated samples were pooled from 40–60 whole seedlings. In a search for protein-coding genes displaying responses similar to the L5-GUS transgene, we examined a subset of loci known to be transcriptionally suppressed by epigenetic modification of their promoters but activated in epigenetic mutants or under heat stress [16], [18]–[20]. We tested the heat stress entrainment of the genes SDC (AT2G17690), SQN (AT2G15790), and APUM9 (AT1G35730). Of these three candidates, only SDC showed a response pattern similar to L5-GUS (Figure 1B and S1). We hypothesized that for both L5-GUS and SDC, the positive correlation between elevated transcript levels during recovery and the number of heat stress cycles reflects an altered speed of re-silencing as a consequence of the entrainment (Figure S2). To test this possibility, we focused on SDC re-silencing kinetics. Indeed, after entrainment by 5 heat cycles, SDC transcripts displayed significantly slower re-silencing kinetics than SQN and APUM9 (Figure 1C). In this experiment, we also assayed some heat-induced transposable elements from different families. We observed cases of fast and slow re-silencing, suggesting that both patterns are possible in TGS targets (Figure S2).
The SDC promoter contains tandem-repeats targeted by the TGS machinery and it is possible that this particular promoter structure contributes to the observed transcriptional entrainment. To address this, we constructed a vector containing the SDC promoter linked to the luciferase reporter (-1200PromSDC::LUC+) and transformed A. thaliana Col-0 wild type and the drm2-2/cmt3-11 double mutant (referred to as dc), which is deficient in RdDM and CMT3-mediated DNA methylation responsible for SDC silencing [13]. In dc transgenic plants under control conditions, a strong luciferase signal was recorded from -1200PromSDC::LUC+ throughout entire seedlings, implying that the SDC promoter does not require heat for activation (Figure 2A). In the Col-0 transgenic plants, -1200PromSDC::LUC+ was transcriptionally suppressed but remained responsive to activation by heat stress (Figure 2A), demonstrating that DNA methyltransferases targeted the -1200PromSDC::LUC+ transgene and the promoter of the endogenous SDC gene in a similar way. Closer examination of the luciferase signals showed them to be highest in young true leaves, lower in cotyledons, and absent from roots (Figure 2A). To determine whether the transcriptional regulation of the -1200PromSDC::LUC+ transgene indeed reflects the heat-induced activation and developmental regulation of the SDC gene, we compared their relative transcript levels in various tissues of seedlings subjected to heat stress. The levels and tissue distribution of mRNA were very similar for both transgenic and endogenous loci, with highest heat induction in young leaves and the lowest in roots. Moreover, they clearly differed from the expression patterns of typical heat-responsive genes, which are induced ubiquitously throughout all seedlings tissues (Figure 2B and S3A). After entrainment for 5 heat-cycles, the 1st and 2nd leaves of -1200PromSDC::LUC+ Col-0 plants showed high transgenic transcript levels and high luciferase signals, decreasing during the recovery phase with kinetics similar to that observed previously for SDC (Figure S3B). Interestingly, leaves 3 to 5 developed during the 4 days of recovery and also showed luciferase signals (Figure 2C). Furthermore, when older plants were subjected to heat stress, marked luciferase signals were found mostly in developing leaves 5 and 6 but were largely absent from older leaves (1 to 4) developed before stress application (Figure S3C). This indicated that not fully expanded young leaves, or possibly even their primordia in the apical meristem, respond predominantly to heat stress by activation of the SDC promoter. Moreover, the slow re-silencing kinetics of endogenous SDC and the luciferase signals from -1200PromSDC::LUC+ Col-0 plants suggest that the acquired active state is maintained during the maturation of leaves when recovering from stress.
Fig. 2. Tissue-specific heat-induced release of SDC gene silencing. 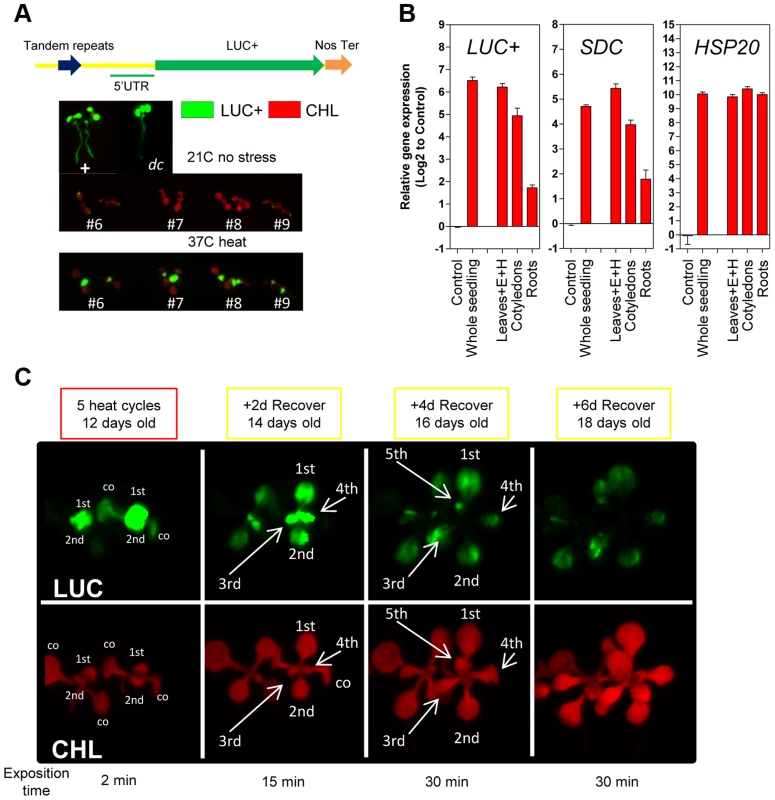
A, In vivo luciferase activity in transgenic seedlings. Top: Positive control UBQ3::LUC+ and transgenic dc mutant (-1200SDCProm::LUC+). Bottom: #6, #7, #8 and #9 represent independent Col-0 transgenic lines (-1200SDCProm::LUC+) under standard conditions or heat stress. B, Transcript levels of LUC+, SDC and HSP20 in different tissues of transgenic Col-0 seedlings (-1200SDCProm::LUC+) after a 5 heat cycle entrainment. Bars represent means ± SE as a log2 ratio with the non-treated control condition (i.e., control = 0); replicated samples were pooled from 20–30 seedlings (E: epicotyl, H: hypocotyl). C, In vivo luciferase activity in 7-days-old Col-0 transgenic seedlings (-1200SDCProm::LUC+) after an entrainment of 5 heat-cycles and varying recovery times. LUC: luciferase signal, CHL: chlorophyll signal. Arrows depict specific leaves; co: cotyledon, and 1st, 2nd, 3th, 4th, 5th represent developing true leaves. We determined that the number of tandem repeats within the SDC promoter in a subset of A. thaliana accessions is typically seven or eight. Therefore, we compared the kinetics of heat-induced SDC activation and re-silencing in these two categories by applying 5 heat cycles and 3 days of recovery. Across all accessions tested, the SDC gene was silent under control conditions and activated by heat stress, suggesting an evolutionary conservation of the heat-induced transcriptional response (Figure S4). However, the relative transcript levels induced by heat stress and their persistence during recovery varied significantly between accessions (Figure S4). The observed differences in SDC regulation could not be attributed to the different number of repeats, demonstrating that a slight variation in the genetic constitution of the promoter does not determine differences in the kinetics of heat-induced release of SDC silencing.
Next, we compared the pattern of transcriptional responses of SDC to typical heat-responsive loci. The thermal threshold of their activation was tested with daily increases in the ambient temperature of growing seedlings (between 28°C and 36°C, Figure 3A). Heat-responsive genes were already activated transcriptionally when plants were moved from 21°C to 28°C, and transcript levels further increased stepwise with increasing temperature (Figure 3A and Figure S5A). However, the SDC locus displayed a distinct thermal threshold for activation within a window of 2°C, from 32°C to 34°C (Figure 3A). A further experiment using a single-step change in temperature yielded similar results, demonstrating that the narrow thermal threshold was independent of the temperature applied on the previous day (Figure 3B and Figure S5B). In a heat time-course (76 h at a constant 37°C), expression of the typical heat-responsive genes peaked rapidly 3 h after the start of the treatment and remained at high levels relative to the control conditions. However, the accumulation of SDC transcripts showed no peak but developed in proportion to the length of the heat stress (Figure 3C and S5C), similar to the responses of other epigenetically regulated loci [14], [16], [17]. Notably, SDC activation occurred only when the heat stress reached the thermal threshold, unlike the heat-responsive genes (Figure 3D and Figure S5D). Overall, similar activation patterns to SDC were observed for the luciferase transcript from -1200PromSDC::LUC+ Col-0 transgene (Figure S5A and C). Taken together, these data support the notion that the particular transcriptional regulation of SDC takes place independently of canonical heat-shock perception and signaling.
Fig. 3. Transcriptional patterns of SDC expression under heat compared to a typical heat-shock gene. 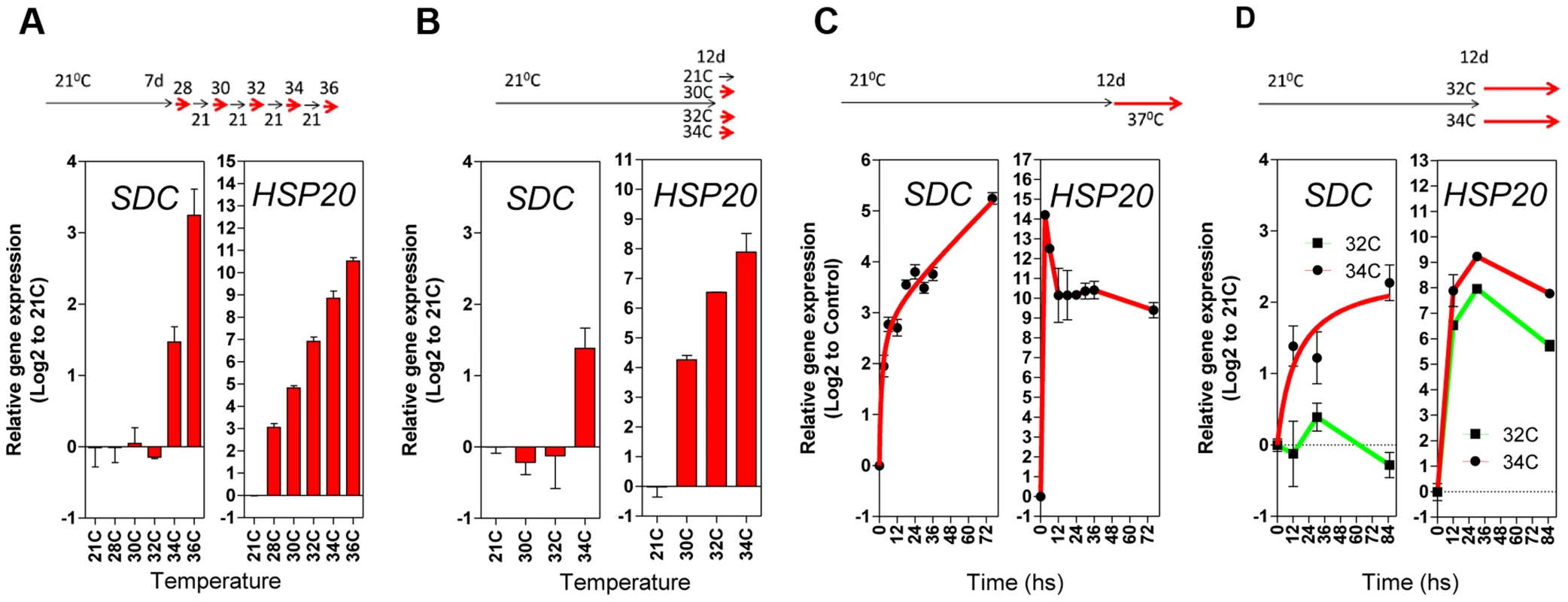
A and B, Thermal threshold of transcriptional activation for SDC and HSP20. A, Seedlings were grown under standard conditions for 7 days and then subjected to daily increases in temperature. B, Seedlings were grown under standard conditions for 12 days and then subjected to a one-step increase in temperature for 12 h. C and D, Heat time-course for transcript levels of SDC and HSP20. C, Seedlings were grown under standard condition for 7 days and then subjected to constant heat for at least 74 h. D, Seedlings were grown under standard condition for 12 days and then subjected to a one-step increase in temperature to a constant 32°C or 34°C. Bars and dots represent the means ± SE as a log2 ratio with the non-treated control conditions (i.e., control = 0); replicated samples were pooled from 40–60 seedlings. In all cases, the experimental design is shown at the top. To assess whether compromised heat stress tolerance contributes to SDC regulation, we tested the heat stress hypersensitive mutant hot1-3, which is impaired in the Heat Shock Protein 101 (HSP101) [21]. Heat-induced SDC transcription, re-silencing and thermal-threshold patterns in hot1-3 were identical to that in wild-type plants (Figure 4A and B). Moreover, experiments performed with this mutant should be indicative of the relative level of heat stress perceived by plants under our experimental conditions. Transcriptional regulation of typical heat-responsive genes was not altered in hot1-3, consistent with the heat stress levels applied in our experimental conditions being relatively low (Figure S6).
Fig. 4. Epigenetic regulation of heat-induced SDC release from silencing. 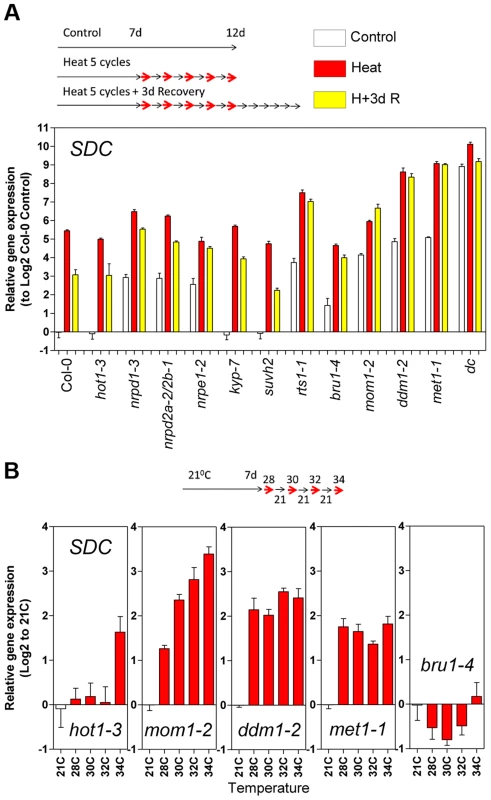
A, Relative levels of SDC transcripts in A. thaliana mutants under control conditions, after a 5 heat cycle entrainment and a 5 heat cycle entrainment +3 days recovery. Bars represent means ± SE as a log2 ratio with the non-treated wild-type Col-0 control conditions (i.e., Col-0 control = 0); replicated samples were pooled from 40–60 whole seedlings. B, Thermal threshold of transcriptional activation of SDC in epigenetic mutants. Bars represent means ± SE as a log2 ratio with the non-treated control conditions (i.e., control = 0); replicated samples were pooled from 40–60 seedlings. Since transcription of SDC is suppressed during vegetative growth by DNA methylation and possibly other epigenetic mechanisms, we examined SDC transcriptional heat-stress responses, re-silencing kinetics, and thermal threshold properties in mutants impaired in various aspects of TGS. The transcriptional heat-stress responses of SDC observed in these epigenetic mutants could be divided into three categories: a) not different to the wild type (kyp-7 and suvh2); b) almost complete release of SDC silencing and thus loss of additional transcriptional activation induced by heat (dc); c) partial release of SDC silencing under control conditions but heat-stress induction maintained (nrpd1-3, nrpd2a-2/2b-1, nrpe1-2, rts1-1, bru1-4, mom1-2, ddm1-2 and met1-1) (Figure 4A). In the latter category, re-silencing of SDC to control levels was largely impaired in some mutants, especially in mom1-2, ddm1-2 and met1-1 (Figure 4A). Importantly, typical heat-responsive genes displayed unaltered transcriptional responses across all the mutants tested (Figure S6), indicating that canonical heat-shock signaling is not influenced by the epigenetic mechanisms examined here. We further tested whether the specific thermal threshold for SDC activation is affected by the mutations in epigenetic regulation. The threshold was clearly disturbed in these mutants and was moved towards lower (mom1-2, ddm1-2 and met1-1) or higher temperatures (bru1-4) (Figure 4B). Because MOM1 and BRU1 influence the stability of particular chromatin states at target loci [18], [20], [22] but their mutated alleles do not affect DNA methylation within the SDC promoter [23], their effect on the thermal threshold is consistent with the chromatin state per se being a candidate for the threshold regulation. We examined DNA methylation and a set of histone modifications within the SDC locus in control and heat-stressed plants but found no evidence for major stress-induced alterations in these epigenetic marks (Figure S7 and S8). Therefore other as yet undefined chromatin properties may determine the narrow temperature range of the transcriptional activation of the SDC gene.
SDC is an imprinted locus, with maternal allele activation during endosperm development and otherwise silent during the entire vegetative growth [24], [25]. To test whether additional endosperm-imprinted genes may be subjected to stress-triggered transcriptional activation, we examined 93 genes for SDC-like transcriptional activation triggered by environmental stress. These 93, selected from 114 previously confirmed endosperm-imprinted genes [24], were represented on the Affymetrix GeneChip ATH1 used in experiments with plants subjected to various stress conditions [26]. A number of stresses including cold, osmotic stress, salinity, wounding, oxidative stress, UV-B irradiation and heat were able to induce transcription of many of these genes during vegetative growth (Figure S9). Although these results suggest that they may contribute to the stress responses and possibly stress tolerance, this hypothesis requires further experimental support, comparable to the study on the SDC gene described below.
The sdc mutant displays neither seed nor somatic developmental abnormalities [13], [25] and, thus, it's physiological or developmental role remains unknown. However, epigenetic suppression of SDC seems to be required for proper plant development. DNA methylation mutants such as dc display abnormal phenotypes during vegetative growth, which can be attributed to high ectopic activity of SDC, given that the phenotype is suppressed in the sdc/dc mutant [13]. Therefore, we tested the effect of SDC induction in vegetative tissues under elevated temperatures, comparing the heat stress responses of wild type to the sdc, dc and sdc/dc mutants. Under our standard heat-entrainment/recovery conditions, transcripts levels of typical heat-responsive genes did not change in any of these mutants (Figure S10), indicating unaltered heat perception and signaling. As a consequence, we did not expect any disturbance of heat shock-induced acquired-thermotolerance [27], so we performed a non-lethal long-term heat stress experiment of wild type and sdc, dc or sdc/dc. Seven-day-old seedlings received 15 entrainment heat-cycles followed by 3 days of recovery and were then grown in soil for a further 15 days under standard conditions, before harvesting and determination of their total aerial fresh weight. The sdc and sdc/dc mutants showed significantly reduced biomass than the corresponding controls, wild-type and dc respectively (Figure 5A, left). Absence of a functional SDC gene accounted on average for approximately 30% of biomass deficit (Figure 5A, right), suggesting a role for the SDC protein in the response to long-term heat. The growth of sdc/dc was more affected by non-lethal heat stress than sdc or dc separately, suggesting that mutations leading to depletion of CHG and CHH DNA methylation may not behave epistatic to a mutation of SDC under specific heat stress treatments. To test this hypothesis in an independent experimental setup, we examined the survival of wild type, sdc, dc, and sdc/dc under moderately high temperatures that resulted in 50% lethality of the wild type [27]. Seedling survival was scored after growth consecutively at 21°C, 35°C, and then 21°C, each for 7 days. The wild-type, sdc and dc lines showed survival rates of approximately 50% but the survival of sdc/dc was significantly lower at 13% (Figure 5B). These results are consistent with at least two parallel pathways contributing to recovery from moderately high temperatures; the first mediated by SDC activity and the second involving epigenetic regulation of CHG/CHH methylation, potentially influencing the transcriptome.
Fig. 5. Physiological and transcriptomic data for sdc, dc and sdc/dc mutants. 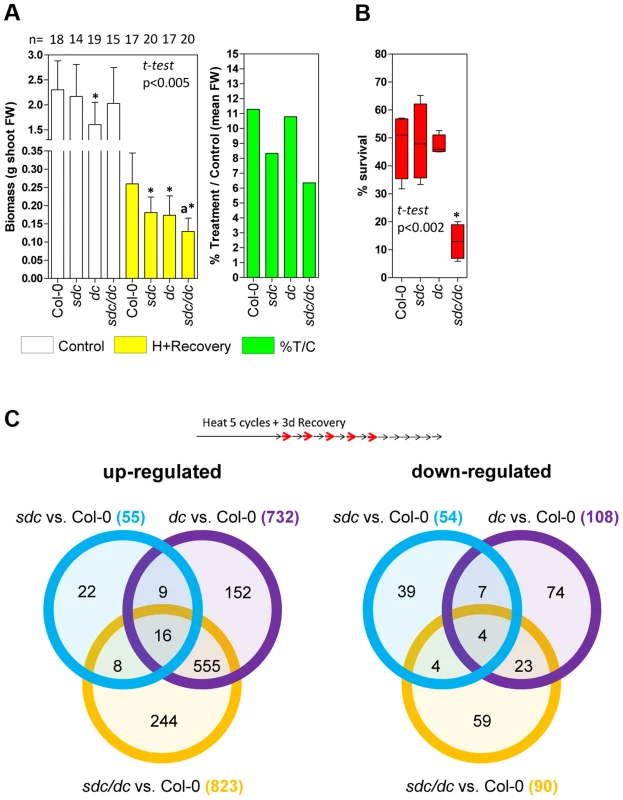
A, Left: Physiological tolerance measured as final aerial biomass. Bars represent means ± SD of shoot fresh weigh (FW) of controls and plants treated for 15 heat cycles plus 18 days of recovery under standard conditions. Numbers at the top represent the independent plants tested.* means statistically significant to the corresponding wild type Col-0 control, and a means statistically significant to the dc heat + recovery plants (P<0.005, Student t-test). Right: Relative mean aerial biomass as percentage of controls obtained from the data shown on the left, denoting a decrease of at least 30% in relative biomass after treatment of genetic backgrounds harboring the sdc mutation. B, Survival to moderately high temperatures, expressed as percentage of seedling survival after 7 days of continuous heat treatment at 35°C from four independent biologically replicated pools of 30–50 seedlings. Data are depicted as box plots. * means statistically significant to wild type Col-0 (P<0.002, Student t-test). C, Venn diagrams comparing differentially regulated genomic features in the sdc, dc and sdc/dc mutants. Transcriptome analysis was performed in duplicated biological samples after a 5 heat-cycle entrainment +3 days of recovery (experimental design shown at the top). Venn diagrams show up- or down-regulated genomic features for each mutant compared with the wild-type. Within the diagrams, the numbers represent differentially regulated genomic features either unique or shared, whereas the legend brackets show the total number of up- or down-regulated genomic features. As a consequence, we compared the transcriptomes of wild type, sdc, dc and sdc/dc during recovery from heat stress entrainment (Table S1). Compared with the wild type, a total of 109, 840 and 913 loci were differentially regulated in sdc, dc and sdc/dc, respectively (Figure 5C). Selected genes found to be altered in the transcriptome analysis were validated in an independent experiment, using qRT-PCR (Figure S11). The 109 genes differentially regulated in sdc are involved in a wide range of cellular processes (Figure S12A). Of these, 68 were represented on the Affymetrix GeneChip ATH1 microarray used earlier for expression profiling that revealed genes differentially expressed during a prolonged heat treatment that led to release of TGS [14]. Out of these 68 loci, transcript levels of 27 (ca. 40%) changed under long-term constant heat [14] (Table S1), supporting the notion that SDC is involved in the regulation of a sub-set of responses to heat stress. Surprisingly, one-third of the genes with altered transcript levels in sdc were altered similarly in dc (Figure S12B). This raises the possibility that SDC controls a sub-set of genes regulated by CHG and CHH methylation. To determine whether this regulation is mediated directly by changes in DNA methylation, we examined available DNA methylation data and observed that none of the loci are subjected to DNA methylation, either in wild-type or in epigenetic mutants [23]. Therefore, the influence on their transcription in both sdc and dc mutants seems to be indirectly linked to DNA methylation.
Although approximately two-thirds of the loci in dc or sdc/dc that differed from wild type overlapped and encoded mostly transposons activated in the dc mutant, approximately one-third (297) displayed transcriptional changes linked specifically to sdc/dc and not shared with either sdc or dc (Figure 5C). The sdc/dc miss-regulated transcripts observed during heat stress recovery are potentially linked to the higher heat sensitivity of the sdc/dc mutant. Indeed, 163 of these 297 transcripts were represented on the Affymetrix GeneChip ATH1 microarray used for profiling after long-term heat as described before [14], and 48 of them (ca. 30%) were altered under these condition (Table S1). This is again consistent with the involvement of SDC in heat-stress responses, acting together but only partially redundantly with activities involved in the maintenance of CHG and CHH methylation. Taken together, the gene expression profiling data and the observed altered recovery of sdc and sdc/dc mutants point towards the physiological significance of SDC during plant vegetative growth in adverse environmental conditions.
SDC is an F-Box protein putatively involved in ubiquitin-mediated degradation of target proteins by the proteasome [13] but its substrate(s) are unknown. Our attempt to recover potential target interactors using high-throughput tandem-affinity-purification/mass-spectrometry with a TAP-tag fusion [28] with SDC was unsuccessful. However, since the activities of the RdDM pathway and CMT3 that influence SDC expression are restricted to the nucleus, we used a vector containing the ubiquitin promoter linked to the coding sequence of SDC fused to the GFP reporter (UBQ10::SDC-GFP) in transient transformation assays and obtained clear evidence for the nuclear localization of the SDC-GFP signal (Figure S13).
Discussion
Epigenetic regulation, typically involving modification of histones and/or remodeling of chromatin, has been implicated previously in plant responses to biotic and abiotic stress [29]. Components of the RdDM pathway and histone deacethylase activity seem to support the survival of plants subjected to lethal heat [30]. Moreover, a heat-sensitive mutant has been isolated in which the heat-induced release of heterochromatic silencing is attenuated [17]. However, the physiological significance of TGS disturbance under long-term heat remained largely unknown, and was suggested previously to be merely a consequence of the thermal disruption of protein-DNA and/or protein-protein interactions [14]. The results presented here suggest that one potential role of silencing release may be the transient expression of epigenetically suppressed loci that encode genes whose activities contribute to stress tolerance. We provide the example of the epigenetically silenced and imprinted gene SDC, with a physiological role in responses to long-term heat stress. Our findings suggest that the silencing of SDC in vegetative tissues was concealing its involvement in stress responses, when transcriptional reactivation occurs following exposure to heat stress. Interestingly, this appears to take place independently of canonical heat-stress signaling pathways.
The expression of a subset of imprinted loci is restricted to the endosperm and although some imprinted genes are active in seed development and maturation, many of them have as yet no ascribed roles in seeds [11]. The results presented here for SDC provide an example that their activity may be revealed under particular growth conditions and that their functions could be executed beyond the tissue of parent-of-origin expression. Furthermore, it may be possible that such concealed activities exist for other imprinted genes, under different stress conditions. For example, it was reported previously that pathogens, UV, cold, and freezing treatments may also transiently disturb epigenetic silencing [15], [31], [32]. In line with this, stress-induced change in DNA methylation has been proposed to impart regulatory control over defense genes that become activated by pathogen attack [32]. Our analyses of published data demonstrated that some endosperm-imprinted genes can be activated by various environmental stresses. Thus, it is plausible that imprinted or epigenetically suppressed loci may exert their activities during vegetative growth upon trigger-specific destabilization of TGS. However, the interactions of particular stress(es)/gene(s) require detailed studies of individual examples.
It was shown previously that heat-induced release of silencing occurs across all plant tissues [16]. However, in the case of SDC, TGS destabilization seemed to occur mostly in young, expanding leaves. Together with the decreased shoot biomass observed with the sdc mutant following heat stress, these results implicate this gene in the expansion/maturation of leaves of plants exposed to high temperatures. This reinforces the current concept that epigenetic silencing may bring about new and unexpected plasticity to gene regulation and plant phenotypes [33]. As a remarkable example, we also observed that SDC activation occurred only above a certain window of absolute temperature, resembling a thermal-sensing mechanism [34]. The epigenetic machinery may, therefore, mediate transcriptional control of certain stress responses in a threshold fashion, as defined previously [35]. However, it is not currently clear whether the SDC locus itself senses absolute temperature through its dynamic epigenetic landscape.
We have shown that the kinetics of SDC re-silencing following variable repeated heat stress is tailored to the previous entrainment, thus displaying transcriptional memory that is especially apparent in the recovery phase. Such a convoluted transcriptional regulation appears also to rely on post-stress epigenetic resetting, since SDC transcripts after heat treatment did not recover to the control levels in epigenetic mutants like mom1-2, ddm1-2 and met1-1. This implies that epigenetic regulation in plants stores previous stress experiences during vegetative development. In connection to this, somatic transcriptional memory was demonstrated recently in plants subjected to osmotic stress and this was attributed to dynamic changes in histone modification [36], [37]. Major stress-induced changes in several common histone marks were not observed within the SDC locus. Also, since the heat-mediated activation of SDC gene occurs in mutants impaired in siRNA biogenesis, a regulatory involvement of siRNAs is unlikely. However, it remains possible that other not tested histone modifications or physical properties of chromatin are responsible for the memory phenomenon. Previously, it have been demonstrated that the transient release of epigenetic suppression under heat stress coincides with decreased nucleosome occupancy, and that chromatin remodelling or assembly factors are a requirement for the fast restoration of silencing [14], [38]. Although these mechanisms may contribute to the regulation of SDC, publically available data suggest that nucleosome density in its promoter area is already low without heat stress [39].
The SDC protein is present in cell nuclei and belongs to the F-Box protein family, which mediates ubiquitin-tagged degradation of proteins and are among the fastest evolving gene families in plants [40]. It is therefore possible that SDC targets a yet unknown nuclear protein for degradation. The A. thaliana gene Upward Curly Leaf 1 (UCL1), which encodes a protein very similar to SDC, has been shown to target Curly Leave (CLF) [41]. CLF is a histone-methyl-transferase of the polycomb-repressive-complex-2 (PRC2), which is involved in various aspects of sporophyte development [42]. Despite an intensive search, we failed to reveal a SDC substrate but CLF or CLF-related proteins remain as potential candidates. Regardless of the actual target protein(s), our transcriptome analysis pointed towards the involvement of SDC in the transcriptional regulation of a sub-set of genes responding to long-term heat.
We propose a silencing/de-silencing loop model illustrating the thermal control of SDC expression (Figure 6). In this model, heat-induced destabilization of the suppressive chromatin allows the transcriptional machinery to access the SDC promoter, but this occurs only above a particular window of absolute temperature. Moreover, the level of transcriptional activation depends on the severity and duration of the heat stress. Expression of SDC leading to synthesis and nuclear translocation of the SDC protein subjects certain nuclear protein(s) to ubiquitin-mediated degradation. Following termination of the heat stress, slow SDC re-silencing allows a temporal extension of SDC activity. All these regulatory mechanisms occur independently and in parallel to canonical heat-shock perception and signaling, but rely on epigenetic properties. It is likely that these arise through the targeting of TGS to the tandem-repeats residing in the SDC promoter. We propose that two steps, the emergence of these repeats and the subsequent epigenetic control, led to rapid evolution of a novel type of environmentally regulated transcriptional output.
Fig. 6. Hypothesized silence/de-silence loop model showing the different steps of transcriptional epigenetic control in the heat-induced expression of the <i>SDC</i> gene. 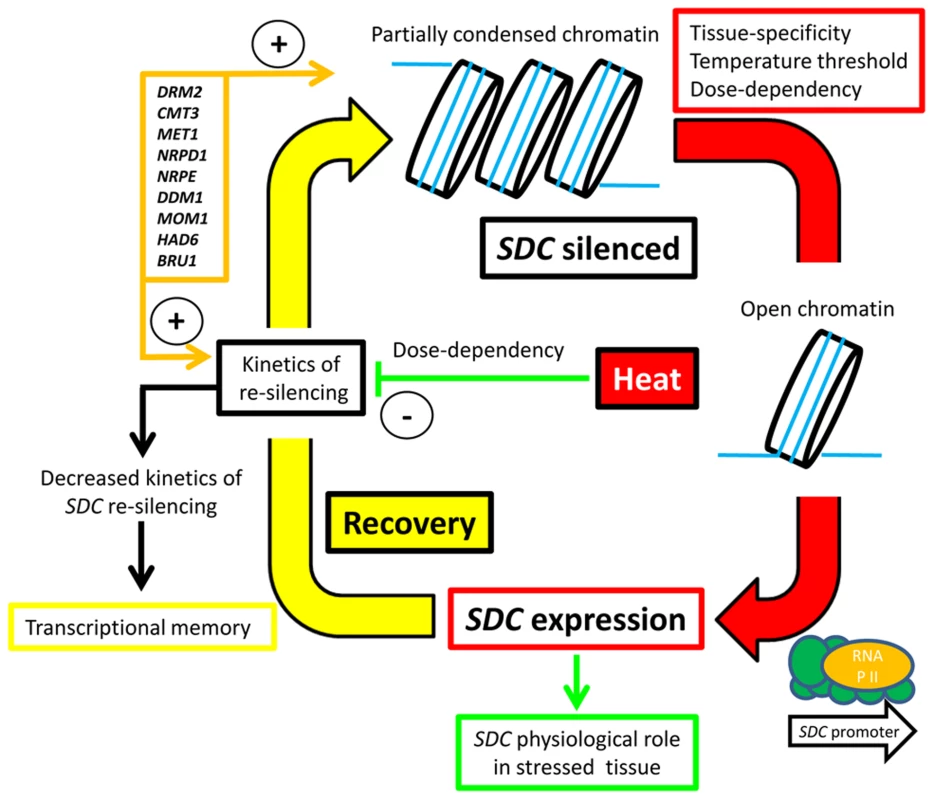
Materials and Methods
Plant material, growth and experimental conditions
All Arabidopsis thaliana Col-0 mutants used in this study have been described and characterized previously: sdc, dc (drm2-2/cmt3-11) and sdc/dc [13], met1-1 [43], mom1-2 [18], ddm1-2 [1], nrpd1-3 and nrpd2a-2/2b-1 [44], nrpe1-2 [45], kyp-7 [4], suvh2 [46], bru1-4 [23], rts1-1 (HAD6, Aufsatz et al. [47]), and hot1-3 [21]. As control wild type, we used the Col-0 line N22681 (The Nottingham Arabidopsis Stock Centre, NASC). The L5-GUS silenced transgenic is the b5b line of Morel et al. [2], and the LUC+ positive control was the UBQ3::LUC+ line (named LUC26) from Yokthongwattana et al. [20]. The different A.thaliana accessions are available at ABRC (http://abrc.osu.edu/) and NASC (http://arabidopsis.info/).
Seeds were surfaced sterilized and sown in sealed petri dishes with 0.5× MS medium containing 1% sucrose, 0.8% agar, and 0.05% MES at pH 5.7. Stratification was applied for 3 days at 4°C. Seedlings were grown for 7 or 12 days at 21°C in a CU-22L growth chamber (Percival) with a 12 h/12 h (day/night) light cycle (termed “standard conditions”), and then subjected to changes in temperature only during the light period (with the exception of the heat time-course and the survival under moderately high temperatures experiments). The experimental design for each particular experiment is shown at the top of the corresponding graphs in the Results section. In all cases, the light cycle was maintained. Heat always means 37°C unless otherwise stated. Long-term heat-entrainment experiments consisted of a varying number of heat cycles (day at 37°C, night at 21°C, 12/12 h), followed by 3 days of recovery at 21°C and a second stress treatment of 37°C for 12 h. For the non-lethal long-term heat experiment, seedlings were subjected to 15 days of heat cycles followed by 3 days of recovery. Plants were then transplanted to soil for a further 15 days of recovery in a growth room at 21°C, after which total above-ground fresh weight was determined. For survival under moderately high temperatures, seedlings were growth in vitro consecutively at 21°C, 35°C, and then 21°C, each for 7 days [27].
Gene expression analysis
Total RNA was isolated using the RNeasy Plant Mini kit (Invitrogen). cDNA synthesis and quantitative real-time RT-PCR analysis were performed as described previously [48] using the geometric mean of three housekeeping genes for normalization. In short, 5 ug of total RNA was treated with TURBO DNA-free kit (Ambion) and first-strand cDNA was synthesized with an oligo dT primer using the SuperScript III First-Strand Synthesis System (Invitrogen). Real-time PCR was performed with the Power SYBR Green PCR Master Mix (Applied Biosystems) in a final reaction volume of 10 µl and with a 1/10 dilution of the cDNA. Cycling and dissociation curves were analyzed in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Primer design, reaction parameters and analysis of expression data were performed as described previously [49], [50]. We used the geometric mean of three housekeeping genes for normalization; these were UBQ10 (AT4G05320), SAND (AT2G28390) and PDF2 (AT1G13320). However, only SAND and PDF2 were used as housekeeping genes for comparisons across A. thaliana accessions. Typical heat-responsive genes were selected from those showing high transcriptional induction under long-term heat [14]. A list of the primers used is available in Table S2.
Whole transcriptome analysis was performed by the Functional Genomics Center of ETH University (Zurich, Switzerland) using a HiSeq 2000/2500 (Illumina) platform to perform unstranded RNA-seq from purified poly-A RNA obtained from duplicated biological replicates. For annotation, mapping of reads was carried out using gene models from the TAIR10 genome assembly (http://www.arabidopsis.org/). Statistical analysis was performed with the edgeR Bioconductor package and the false-discovery-rate (FDR) was computed with the Benjamini-Hochberg algorithm. A genomic feature was considered differentially changed in a mutant versus wild type comparison when the Benjamini-Hochberg's FDR was <0.1 and the log2 fold change was >1 or <−1. Raw GeneChip Arabidopsis ATH1 Genome Array (Affymetrix) data were analyzed with the RobiNA software [51] and the probesets were considered differentially changed by the heat treatment using the above parameters. The non-redundant functional categories of the differentially changed features were assessed with the MapMan software [52]. Mean-normalized expression data of confirmed endosperm-imprinted genes in seedlings under stress was taken from Kilian et al. [26].
Vector design and transgenic plants
For cloning purposes, PCR was performed using the Phusion High-Fidelity DNA Polymerase (NEB) and blunt-end products were cloned using the CloneJET PCR Cloning Kit (Thermo Scientific). A fragment of the SDC promoter (1198 bp upstream of the ORF) was PCR amplified from genomic DNA, cloned, sequenced, and re-cloned into a pGPTVII-bar-MCS (multi-cloning-site) plasmid using the BamHI/XhoI sites [53], producing the pGPTVII-bar-1200PromSDC vector. LUC+ was PCR amplified from genomic DNA of a UBQ3::LUC+ line [20], cloned, sequenced, and re-cloned into the previous construct using the XhoI/XmaI sites, thus producing the -1200PromSDC::LUC+ construct. The ORF of SDC without a stop codon was PCR amplified from genomic DNA, cloned, sequenced, and re-cloned into the pGPTVII-bar-UBQ10-GFP5 using the BamHI/XhoI sites in frame with the GFP5, giving the UBQ10::SDC-GFP construct. All original pGPTVII binary plasmids were kindly provided by Dr. Rainer Waadt (University of California SD, USA). A list of the primers used is available in Table ST2. The Agrobacterium tumefaciens pGV3101 strain was used to transform A. thaliana using the standard floral-dip method. Transgenic lines were selected in vitro for resistance to BASTA (dl-phosphinothricin, Duchefa).
Other methods
In vivo measurements of luciferase activity were performed by spraying the treated transgenic seedlings with luciferin (Biosynth, 1 mM in water). After 5 min in the dark, images were captured with a CDD ORCA2 C4742-98 digital camera (Hamamatsu) and then analyzed with Wasabi Imaging software. Luciferase activity was detected without a filter, whereas a 632.8 nm filter and blue light was used to detect the chlorophyll signal.
DNA methylation was analyzed by cloning and sequencing of PCR products from bisulfite-treated genomic DNA, from whole young seedlings subjected to entrainment by 5 heat cycles along the corresponding controls. Genomic DNA was isolated by standard CTAB buffer and further fenol-chloroform extractions and precipitation. Bisulfite treatment was performed with the Epitect Bisulfite Kit (Quiagen). PCR products were amplified with Taq polymerase (Promega) using a touch-down PCR strategy and cloned with the pGEM-T Easy Vector System I (Promega). Primer design and analysis of sequences with differentially methylated cytosines were performed with Kismeth and CyMATE [54], [55]. Samples used to assess histone modifications in the SDC locus were kindly provided by Dr. Herve Gaubert (University of Cambridge, UK). Chromatin immunoprecipitation was performed in tissue from whole young seedlings following a protocol adapted from Gendrel et al. [56] and Nelson et al. [57]. A list of the primers used to test these samples is available in Table ST2.
For cellular localization of the SDC-GFP fusion protein, 4-week-old Nicotiana benthamiana plants were infiltrated with A. tumefaciens carrying the corresponding constructs according to Schütze et al. [58]. After 3 days, pieces of leaves were mounted and the GFP signal from transiently transformed epidermal cells photographed with a confocal LSM 700 laser scanning microscope (Zeiss) housed by the UNIGE Bioimaging Core Facilities (http://www.unige.ch/medecine/bioimaging/index.html).
Supporting Information
Zdroje
1. VongsA, KakutaniT, MartienssenRA, RichardsEJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260 : 1926–1928.
2. MorelJB, MourrainP, BeclinC, VaucheretH (2000) DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol 10 : 1591–1594.
3. CaoX, AufsatzW, ZilbermanD, MetteMF, HuangMS, et al. (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13 : 2212–2217.
4. MathieuO, ReindersJ, CaikovskiM, SmathajittC, PaszkowskiJ (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130 : 851–862.
5. ZemachA, KimMY, HsiehPH, Coleman-DerrD, Eshed-WilliamsL, et al. (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153 : 193–205.
6. CaoX, JacobsenSE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. PNAS 99 : 16491–16498.
7. PikaardCS, HaagJR, ReamT, WierzbickiAT (2008) Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 13 : 390–397.
8. CaoX, JacobsenSE (2002) Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12 : 1138–1144.
9. LischD (2009) Epigenetic regulation of transposable elements in plants. Ann Rev Plant Biol 60 : 43–66.
10. RajaP, SanvilleBC, BuchmannRC, BisaroDM (2008) Viral genome methylation as an epigenetic defense against geminiviruses. J Virology 82 : 8997–9007.
11. KohlerC, WolffP, SpillaneC (2012) Epigenetic mechanisms underlying genomic imprinting in plants. Ann Rev Plant Biol 63 : 331–352.
12. RaissigMT, BarouxC, GrossniklausU (2011) Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23 : 16–26.
13. HendersonIR, JacobsenSE (2008) Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes & Dev 22 : 1597–1606.
14. PecinkaA, DinhHQ, BaubecT, RosaM, LettnerN, et al. (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22 : 3118–3129.
15. Lang-MladekC, PopovaO, KiokK, BerlingerM, RakicB, et al. (2010) Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant 3 : 594–602.
16. Tittel-ElmerM, BucherE, BrogerL, MathieuO, PaszkowskiJ, et al. (2010) Stress-induced activation of heterochromatic transcription. PLoS Genet 6: e1001175.
17. WangLC, WuJR, ChangWL, YehCH, KeYT, et al. (2013) Arabidopsis HIT4 encodes a novel chromocentre-localized protein involved in the heat reactivation of transcriptionally silent loci and is essential for heat tolerance in plants. J Exp Bot 64 : 1689–1701.
18. HabuY, MathieuO, TariqM, ProbstAV, SmathajittC, et al. (2006) Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep 7 : 1279–1284.
19. ListerR, O'MalleyRC, Tonti-FilippiniJ, GregoryBD, BerryCC, et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 : 523–536.
20. YokthongwattanaC, BucherE, CaikovskiM, VaillantI, NicoletJ, et al. (2010) MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J 29 : 340–351.
21. HongSW, VierlingE (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27 : 25–35.
22. OhnoY, NarangajavanaJ, YamamotoA, HattoriT, KagayaY, et al. (2011) Ectopic gene expression and organogenesis in Arabidopsis mutants missing BRU1 required for genome maintenance. Genetics 189 : 83–95.
23. StroudH, GreenbergMV, FengS, BernatavichuteYV, JacobsenSE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152 : 352–364.
24. HsiehTF, ShinJ, UzawaR, SilvaP, CohenS, et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. PNAS 108 : 1755–1762.
25. VuTM, NakamuraM, CalarcoJP, SusakiD, LimPQ, et al. (2013) RNA-directed DNA methylation regulates parental genomic imprinting at several loci in Arabidopsis. Development 140 : 2953–2960.
26. KilianJ, WhiteheadD, HorakJ, WankeD, WeinlS, et al. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 : 347–363.
27. YehCH, KaplinskyNJ, HuC, CharngYY (2012) Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci 195 : 10–23.
28. Van LeeneJ, WittersE, InzeD, De JaegerG (2008) Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci 13 : 517–520.
29. GutzatR, Mittelsten ScheidO (2012) Epigenetic responses to stress: triple defense? Curr Op Plant Biol 15 : 568–573.
30. PopovaOV, DinhHQ, AufsatzW, JonakC (2013) The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant 6 : 396–410.
31. HuY, ZhangL, HeS, HuangM, TanJ, et al. (2012) Cold stress selectively unsilences tandem repeats in heterochromatin associated with accumulation of H3K9ac. Plant Cell Environ 35 : 2130–2142.
32. DowenRH, PelizzolaM, SchmitzRJ, ListerR, DowenJM, et al. (2012) Widespread dynamic DNA methylation in response to biotic stress. PNAS 109: E2183–2191.
33. CortijoS, WardenaarR, Colomé-TatchéM, GillyA, EtcheverryM, et al. (2014) Mapping the Epigenetic Basis of Complex Traits. Science 343 : 1145–1148.
34. McClungCR, DavisSJ (2010) Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Current Biol CB 20: R1086–1092.
35. SanchezDH, LippoldF, RedestigH, HannahMA, ErbanA, et al. (2008) Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J 53 : 973–987.
36. DingY, FrommM, AvramovaZ (2012) Multiple exposures to drought 'train' transcriptional responses in Arabidopsis. Nat Comm 3 : 740.
37. SaniE, HerzykP, PerrellaG, ColotV, AmtmannA (2013) Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 14: R59.
38. IwasakiM, PaszkowskiJ (2014) Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. PNAS 111 : 8547–8552.
39. ChodavarapuRK, FengS, BernatavichuteYV, ChenPY, StroudH, et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466 : 388–392.
40. HuaZ, PoolJE, SchmitzRJ, SchultzMD, ShiuSH, et al. (2013) Epigenomic programming contributes to the genomic drift evolution of the F-Box protein superfamily in Arabidopsis. PNAS 110 : 16927–16932.
41. JeongCW, RohH, DangTV, ChoiYD, FischerRL, et al. (2011) An E3 ligase complex regulates SET-domain polycomb group protein activity in Arabidopsis thaliana. PNAS 108 : 8036–8041.
42. BemerM, GrossniklausU (2012) Dynamic regulation of Polycomb group activity during plant development. Curr Op Plant Biol 15 : 523–529.
43. KankelMW, RamseyDE, StokesTL, FlowersSK, HaagJR, et al. (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163 : 1109–1122.
44. OnoderaY, HaagJR, ReamT, Costa NunesP, PontesO, et al. (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 : 613–622.
45. PontierD, YahubyanG, VegaD, BulskiA, Saez-VasquezJ, et al. (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & Dev 19 : 2030–2040.
46. NaumannK, FischerA, HofmannI, KraussV, PhalkeS, et al. (2005) Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J 24 : 1418–1429.
47. AufsatzW, MetteMF, van der WindenJ, MatzkeM, MatzkeAJ (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 21 : 6832–6841.
48. LippoldF, SanchezDH, MusialakM, SchlerethA, ScheibleW-R, et al. (2009) AtMyb41 Regulates Transcriptional and Metabolic Responses to Osmotic Stress in Arabidopsis. Plant Physiol 149 : 1761–1772.
49. CzechowskiT, BariRP, StittM, ScheibleWR, UdvardiMK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root - and shoot-specific genes. Plant J 38 : 366–379.
50. CzechowskiT, StittM, AltmannT, UdvardiMK, ScheibleWR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 : 5–17.
51. LohseM, BolgerAM, NagelA, FernieAR, LunnJE, et al. (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40: W622–627.
52. UsadelB, NagelA, ThimmO, RedestigH, BlaesingOE, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138 : 1195–1204.
53. WalterM, ChabanC, SchutzeK, BatisticO, WeckermannK, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 : 428–438.
54. HetzlJ, FoersterAM, RaidlG, Mittelsten ScheidO (2007) CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51 : 526–536.
55. GruntmanE, QiY, SlotkinRK, RoederT, MartienssenRA, et al. (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9 : 371.
56. GendrelAV, LippmanZ, YordanC, ColotV, MartienssenRA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 : 1871–1873.
57. NelsonJD, DenisenkoO, SovaP, BomsztykK (2006) Fast chromatin immunoprecipitation assay. Nucleic Acids Res 34: e2.
58. Schütze K, Harter K, Chaban C (2009) Bimolecular Fluorescence Complementation (BiFC) to Study Protein-protein Interactions in Living Plant Cells. In: Pfannschmidt T, editor. Plant Signal Transduction: Humana Press. pp.189–202.
Štítky
Genetika Reprodukčná medicína
Článek The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural CompetenceČlánek Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the TestisČlánek The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I ofČlánek GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and AnnotationČlánek Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant BiomassČlánek p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide SecretionČlánek The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of MitophagyČlánek Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of SenescenceČlánek ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Establishing a Multidisciplinary Context for Modeling 3D Facial Shape from DNA
- RNA Processing Factors Swd2.2 and Sen1 Antagonize RNA Pol III-Dependent Transcription and the Localization of Condensin at Pol III Genes
- Inversion of the Chromosomal Region between Two Mating Type Loci Switches the Mating Type in
- A Thermolabile Aldolase A Mutant Causes Fever-Induced Recurrent Rhabdomyolysis without Hemolytic Anemia
- The Role of Regulatory Evolution in Maize Domestication
- Stress Granule-Defective Mutants Deregulate Stress Responsive Transcripts
- 24-Hour Rhythms of DNA Methylation and Their Relation with Rhythms of RNA Expression in the Human Dorsolateral Prefrontal Cortex
- Pseudoautosomal Region 1 Length Polymorphism in the Human Population
- Fungal Communication Requires the MAK-2 Pathway Elements STE-20 and RAS-2, the NRC-1 Adapter STE-50 and the MAP Kinase Scaffold HAM-5
- The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural Competence
- The Protein -glucosyltransferase Rumi Modifies Eyes Shut to Promote Rhabdomere Separation in
- The Talin Head Domain Reinforces Integrin-Mediated Adhesion by Promoting Adhesion Complex Stability and Clustering
- Quantitative Genetics of CTCF Binding Reveal Local Sequence Effects and Different Modes of X-Chromosome Association
- Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the Testis
- Genetic Analysis of a Novel Tubulin Mutation That Redirects Synaptic Vesicle Targeting and Causes Neurite Degeneration in
- A Systems Genetics Approach Identifies , , and as Novel Aggressive Prostate Cancer Susceptibility Genes
- Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in
- Approximation to the Distribution of Fitness Effects across Functional Categories in Human Segregating Polymorphisms
- The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I of
- SAS-1 Is a C2 Domain Protein Critical for Centriole Integrity in
- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and Annotation
- Let's Face It—Complex Traits Are Just Not That Simple
- Glutamate Receptor Gene , Coffee, and Parkinson Disease
- The Red Queen Model of Recombination Hotspots Evolution in the Light of Archaic and Modern Human Genomes
- The Ethics of Our Inquiry: An Interview with Hank Greely
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
- Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-response
- Lack of Replication of the -by-Coffee Interaction in Parkinson Disease
- Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity
- A Germline Polymorphism of Thymine DNA Glycosylase Induces Genomic Instability and Cellular Transformation
- Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene
- ATPase-Independent Type-III Protein Secretion in
- p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide Secretion
- The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of Mitophagy
- Evolution of DNA Methylation Patterns in the Brassicaceae is Driven by Differences in Genome Organization
- Regulation of mRNA Abundance by Polypyrimidine Tract-Binding Protein-Controlled Alternate 5′ Splice Site Choice
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of Senescence
- A Functional Portrait of Med7 and the Mediator Complex in
- Systematic Analysis of the Role of RNA-Binding Proteins in the Regulation of RNA Stability
- ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
- Genomic Evidence of Rapid and Stable Adaptive Oscillations over Seasonal Time Scales in Drosophila
- Genome-Wide Associations between Genetic and Epigenetic Variation Influence mRNA Expression and Insulin Secretion in Human Pancreatic Islets
- HAM-5 Functions As a MAP Kinase Scaffold during Cell Fusion in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



