-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in
In regenerative tissues, the successive differentiation of stem cell lineage is well controlled and coordinated with proper cell proliferation at each differentiation stage. Disruption of the control mechanism can lead to tumor growth or tissue degeneration. The germline stem cell lineage of Drosophila spermatogenesis provides an ideal research model to unravel the genetic network coordinating proliferation and differentiation. In a genetic screen, we identified a male-sterile mutant whose germ cells are under-differentiated and overproliferating. The responsible gene encodes an RNA-binding protein whose target belongs to a tumor suppressor family. We demonstrate that this and two other RNA-binding proteins form a physical and functional unit to ensure the proper differentiation and accurate proliferation of germline stem cell lineage.
Published in the journal: Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in. PLoS Genet 10(11): e32767. doi:10.1371/journal.pgen.1004797
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004797Summary
In regenerative tissues, the successive differentiation of stem cell lineage is well controlled and coordinated with proper cell proliferation at each differentiation stage. Disruption of the control mechanism can lead to tumor growth or tissue degeneration. The germline stem cell lineage of Drosophila spermatogenesis provides an ideal research model to unravel the genetic network coordinating proliferation and differentiation. In a genetic screen, we identified a male-sterile mutant whose germ cells are under-differentiated and overproliferating. The responsible gene encodes an RNA-binding protein whose target belongs to a tumor suppressor family. We demonstrate that this and two other RNA-binding proteins form a physical and functional unit to ensure the proper differentiation and accurate proliferation of germline stem cell lineage.
Introduction
Adult stem cells divide to replenish differentiated, dead, or damaged cells in regenerative tissues. To produce sufficient number of differentiated progeny for tissue homeostasis and to avoid the accumulation of oncogenic mutations derived from frequent cell divisions, stem cell daughters undergo multiple rounds of transit-amplifying (TA) divisions prior to terminal differentiation [1], [2], [3], [4]. However, failure to stop TA divisions and enter programmed differentiation may contribute to tumorigenesis in adult stem cell lineages [2], [5], [6], [7].
Drosophila spermatogenesis is a highly stereotyped and accessible system to study the control mechanisms of accurate TA divisions in adult stem cell lineage. At the apical tip of testis, germline stem cells (GSCs) and somatic cyst stem cells form a rosette surrounding the hub (Figure 1A). GSC divides asymmetrically to generate a daughter cell adjacent to the hub remaining as a GSC while the other one away from the hub differentiating as a gonialblast (GB). As the founder giving rise to a clonal production of gametes, GB in turn undergoes four rounds of TA divisions to form a cluster of 16 interconnected spermatogonial cells which develop in synchrony thereafter. After the four TA divisions, spermatogonia switch to the meiotic/spermatocyte program and increase 25-fold in cell size [8], [9], [10]. The dramatic differences in morphology, the availability of molecular markers to distinguish germ cells at different stages, and especially the accurate number of TA divisions make Drosophila spermatogonial proliferation a perfect model to look for deviations upon genetic manipulations.
Fig. 1. tut restricts spermatogonial proliferation cell-autonomously. 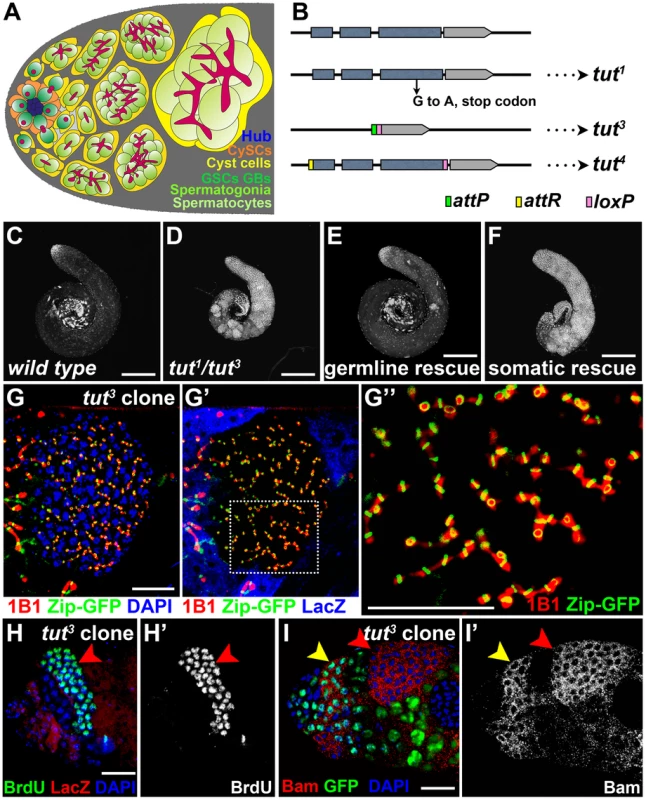
(A) Schematic illustration of the cellular architecture at the apical part of Drosophila testis. CySC: cyst stem cell; GSC: germline stem cell; GB: gonialblast. Note the morphological differences of fusome (red) in different cells: dot in GSC and GB, or branched network in spermatogonia and spermatocytes. (B) Molecular information of tut alleles. tut3, a null allele, was generated by replacing tut genomic region with attP and loxP elements. tut4 is a weak allele, with tut coding region flanked by attR (before start codon) and loxP (after stop codon). (C–F) Low magnification images showing the testes of w1118 (C), tut1/3 (D), bam-Gal4/Y; UAS-GFP-tut/+; tut1/3 (E), and tj-Gal4/UAS-GFP-tut; tut1/3 (F) stained with DNA dye DAPI. (G–I) Immunofluorescence images of the apical part of the testes containing tut mutant clones. tut3 clones were marked by the absence of LacZ (G′, H) or GFP (I). (G) A clone of over-proliferating germ cells with branched fusome (1B1) running through ring canal (Zip-GFP). G″ shows the high magnification view of the boxed region in G′. (H–H′) A testis stained for clone marker LacZ and S phase marker BrdU. Red arrowhead points to the tut3 mutant clone with all cells entering S phase in synchrony. (I–I′) A testis containing tut3 mutant clones was stained for Bam, GFP, and DNA (DAPI). Bam was expressed both in wild-type spermatogonia (yellow arrowhead) and tut3 mutant cells (red arrowhead). Scale bars: 200 µm (C–F) and 25 µm (G–I). See also Figure S1. In the past decades, many intrinsic and extrinsic factors regulating TA divisions have been found in Drosophila spermatogenesis [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25]. Among them, bag of marbles (bam) is at the center of the picture not only because it was the first player identified in this process, also because in germ cells Bam protein accumulation signals the stop of TA division and/or the start of further differentiation [14], [26], [27]. Ectopic expression of Bam protein in GSCs leads to the premature differentiation of all stem cells [28], [29], [30].
Benign gonial cell neoplasm (Bgcn) is an ‘intimate’ partner of Bam, given that they have the same mutant phenotype (i.e., spermatogonial over-amplification) [14] and they are present in the same protein complex to confer translational repression in both male and female germ cells [15], [31], [32], [33]. In cultured Drosophila S2 cells, Bam and Bgcn repress the expression of a reporter coupled with the 3′ untranslated region (3′UTR) of DE-Cadherin [33]. In female germline, Bam-Bgcn complex antagonizes Nanos (Nos) expression via nos 3′UTR [34]. While in male germ cells, this complex binds mei-P26 3′UTR directly to repress Mei-P26, whose initial expression in early TA cells is required for Bam accumulation [15]. Thus, a negative feedback loop is formed between Mei-P26 and Bam to ensure proper accumulation of Bam and accurate TA divisions. However, overexpression of Mei-P26 in late TA cells did not resemble bam or bgcn mutant phenotype [15]. Identification of more genes involved in this process will unravel the regulatory network governing the switch from TA division to meiotic differentiation.
From a mutagenesis screen combined with germline clonal analysis, we isolated a mutant showing spermatogonial over-proliferation, a phenotype similar to that of mei-P26, bam, or bgcn. This mutation disrupts CG32364 which we named tumorous testis (tut). We found by genetic and biochemical methods that Tut, Bam, and Bgcn act in a complex to accurately coordinate TA division and differentiation of germline stem cell daughters.
Results
Identification of tut as an Intrinsic Factor Restricting Transit-amplification
To search for more factors regulating the proliferation and/or differentiation of fly germline stem cell lineage, we performed a large scale EMS screen, and obtained a male sterile mutant line exhibiting germ cell overgrowth. We mapped the gene responsible for this phenotype to CG32364, and designated it as tumorous testis (tut). tut encodes a protein containing a putative RNA recognition motif (RRM). The EMS-induced point mutation in tut1 generates a premature stop codon (Figure 1B and S2D). The tut mRNA transcribed from tut1 genome is much lower than that from wild-type (Figure S1B). We also generated a null allele, tut3 (Figure 1B), by homologous recombination-based gene targeting (Materials and Methods). tut1, tut3 and tut1/3 mutant testes all exhibited the same phenotypes including the failure to exit TA division and the spatial/temporal pattern of molecular markers (See below). Compared to the wild-type (Figure 1C), tut mutant testis was filled with early germ cells brightly stained by the DNA dye (Figure 1D), but lacked late germ cells such as spermatocytes (compare the spermatocyte marker Hrb98DE-GFP [35], [36] in Figure S1C and D) or spermatids (compare Dj-GFP [55], [36] in Figure S1E and F). tut mutant testis could be completely rescued by the expression of tut cDNA in germ cells (Figure 1E), but not in somatic cells (Figure 1F), indicating that tut functions in germ cells. Consistently, the over-proliferating germ cells were observed only when tut was knocked down in germ cells (Figure S1G–H), but not in somatic cells (Figure S1I).
To determine the cell stage at which tut mutant germ cells were arrested, we chose several well-characterized molecular markers for early germ cells including GSC, GB, and spermatogonia [14], [37], [38]. The over-proliferating germ cells in tut mutant clones possessed all the characteristics of TA spermatogonia, such as branched fusome passing through ring canal (Figure 1G), synchronized cell division (Figure 1H), Bam protein expression (Figure 1I), and bam transcription revealed by bamP-GFP (Figure S1J–K). Additionally, no expansion of GSC and GB was detected in the tut mutant testes by the commonly used markers for these cell types (Figure S1L–O). Thus, tut mutant germ cells arrested at spermatogonial TA stage and over-proliferated. Taken together, we conclude that tut is intrinsically required in germ cells to ensure proper transit amplification of spermatogonia.
Tut Is Required for the Translational Repression of mei-P26 via Its 3′UTR
tut mutant phenotype is similar to bam, bgcn, or mei-P26 mutant. Bam-Bgcn complex has been shown to regulate mei-P26 expression by binding to its 3′UTR [15]. Since Tut protein contains a predicted RRM, we wonder if Tut binds mei-P26 3′UTR directly. We found a longer isoform of mei-P26 3′UTR in tut, bam, and bgcn mutant testes (Figure S2A–B), which also existed at low abundance in wild-type testes (Figure S2C). To examine the interaction between Tut and mei-P26 3′UTR, we performed a series of RNA immunoprecipitations and quantified mei-P26 3′UTR by realtime PCR. Interestingly, Tut protein binds the longer isoform of mei-P26 3′UTR more efficiently than the shorter one that has been reported to interact with Bam (Figure 2A) [15].
Fig. 2. Tut is required to repress mei-P26 expression via mei-P26 3′UTR. 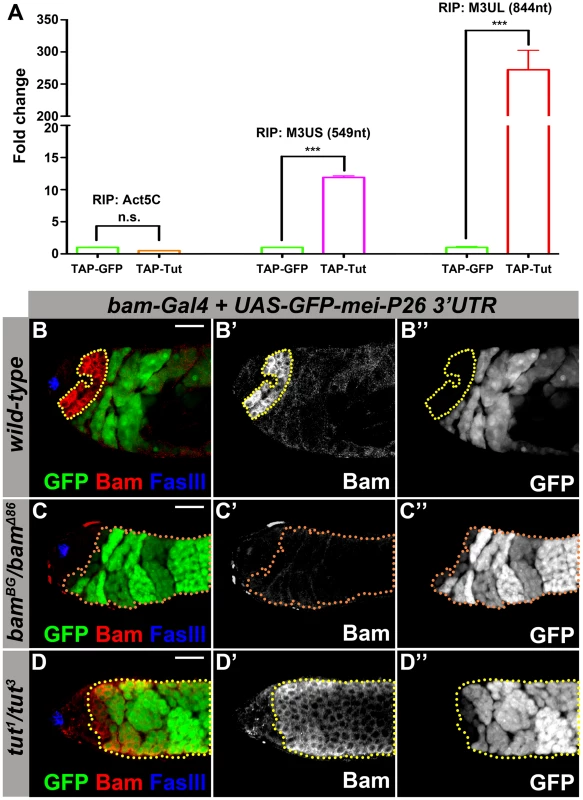
(A) Physical interaction of Tut protein and mei-P26 3′UTR detected by RNA immunoprecipitation (RIP). TAP-Tut or TAP-GFP and mei-P26 3′UTR were co-expressed in S2 cells. IgG beads were used to enrich TAP-Tut or TAP-GFP followed by TEV digestion to release Tut or GFP and bound RNA. RNA was extracted and reverse transcribed. The quantity of Actin5C mRNA, short (M3US) or long (M3UL) isoform of mei-P26 3′UTR was determined by real-time PCR. The Y axis represents the ratio of RIP/Input, which was normalized to 1 for TAP-GFP. Error bar indicates SD. ***, p<0.0001 in t test. n.s., not significant. (B–B″) Genotype: bam-Gal4/Y; UAS-GFP-mei-P26 3′UTR (2k)/+. A genomic region of 2 kb in length downstream of mei-P26 stop codon was selected to cover both short and long isoforms (see Figure S2A). Yellow dots outline Bam-expressing spermatogonia. GFP was repressed in most Bam-expressing spermatogonia. (C–C″) Genotype: bam-Gal4/Y; UAS-GFP-mei-P26 3′UTR (2k)/+; bamBG/bamΔ86. Orange dots outline GFP-positive spermatogonia. (D–D″) Genotype: bam-Gal4/Y; UAS-GFP-mei-P26 3′UTR (2k)/+; tut1/tut3. Yellow dotted outline indicates Bam-expressing spermatogonia. GFP was de-repressed in tut mutant even in the presence of Bam. Scale bars: 25 µm. See also Figure S2-S3. Yeast 3-hybrid is an easy and efficient assay to detect the physical interaction between protein and RNA [39]. Consistently, Tut binds the long isoform of mei-P26 3′UTR at high stringent conditions and binds also the short isoform of mei-P26 3′UTR at low stringent conditions (Figure S2E–F). However, we could not detect any interaction between Bam and long mei-P26 3′UTR (Figure S2E–F). Deletion of the RRM domain abolished the association between Tut and mei-P26 3′UTR, further supporting that Tut functions as an RBP and binds to mei-P26 3′UTR (Figure S2F).
To test whether Mei-P26 protein level is changed in tut mutant germ cells, we generated an antibody against Mei-P26. We confirmed the specificity of this antibody by immunostaining mei-P26 mutant testis (Figure S3H). Mei-P26 protein was detectable at low level in wild-type spermatogonia (Figure S3G) and was up-regulated in tut mutant (Figure S3I).
To test whether tut is required via mei-P26 3′UTR for Bam-Bgcn complex-mediated repression of mei-P26, we generated a reporter containing GFP coding sequence and mei-P26 3′UTR region (2 kb downstream of the stop codon), from which both long and short isoforms were detected when the reporter was expressed (detected by 3′RACE specific for the reporter). The reporter expression driven by bam-Gal4 was repressed in ∼80% of Bam-positive cysts in the presence of mei-P26 3′UTR (Figure 2B, n = 60), consistent with the reported pattern using a shorter mei-P26 3′UTR in a similar construct [15]. As expected, in the absence of Bam, the GFP reporter was de-repressed in spermatogonial TA cells (Figure 2C and S3B). In tut mutant testis, though Bam and Bgcn were expressed, the GFP reporter was nonetheless de-repressed (Figure 2D and Figure S3A,D), indicating that Tut is essential for the translational repression mediated by mei-P26 3′UTR. Similarly, the GFP reporter was also de-repressed in bgcn mutant testis (Figure S3C,J), suggesting that Tut, Bam, and Bgcn may act together on mei-P26 regulation as well as in the development of GSC lineage.
Genetic and Physical Interactions between Tut and Bam in Regulating Spermatogonial TA Division
To examine the expression pattern of Tut during spermatogenesis, we first tried to raise an antibody against Tut but failed after several attempts using different strategies. We then sought to add a tag into tut locus by genomic engineering, but the extra sequences introduced into tut locus affected tut function, generating the weak allele tut4 instead (Figure 1B and Figure 3G). Then we tagged a genomic fragment containing tut regulatory sequences (Figure S4A) that was sufficient to rescue tut mutant phenotype (Figure 3A–B). The expression pattern of this genomic construct resembled that of Bam protein (Figure 3C–D), though very weak expression was detected in GSCs (Figure 3C′).
Fig. 3. tut interacts with bam both genetically and physically. 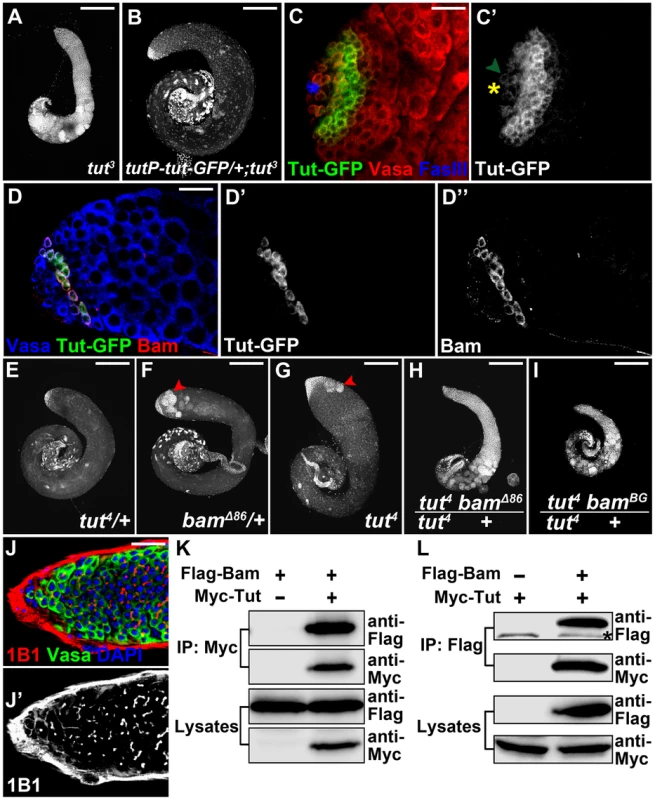
(A–B) Low magnification view of tut3 (A) and tutP-tut-GFP/+; tut3 (B) testes stained with DAPI. (C&C′) tutP-tut-GFP/+; tut3 testis stained for GFP, germline marker Vasa, and hub cell marker FasIII. Yellow asterisk indicates hub and hereafter. Green arrowhead points to a GSC expressing weak Tut-GFP signal. (D–D″) Immunofluorescence images of tutP-tut-GFP/+; tut3 testis. Tut-GFP and Bam were both expressed in spermatogonia. (E–I) tut4/+ (E), bamΔ86/+ (F), tut4 (G), tut4, bamΔ86/+ (H), and tut4, bamBG/+ (I) testes stained with DAPI. Red arrowheads point to over-proliferational cysts. (J&J′) Immunofluorescence images of tut4, bamBG/+ testis. Note the branched fusome. (K–L) Bam and Tut coimmunoprecipitated from S2 cells expressing tagged proteins. Flag-Bam and Myc-Tut were over-expressed in S2 cells and the cell lysates were used for anti-Myc (K) or anti-Flag (L) immunoprecipitation. Western analysis with corresponding antibodies was performed to detect the presence of Flag-Bam and Myc-Tut. Asterisk indicates a nonspecific band. Scale bars: 25 µm (A, B, E–I); 200 µm (C,D,J). See also Figure S4. We next tested whether or not tut functions in the same pathway as that of bam by genetic assays. tut4 is a weak allele and its heterozygous testes were indistinguishable from the wild-type (Figure 3E). bamΔ86 is a null allele whose heterozygous testes showed ∼60% ‘tumor’ rate (Figure 3F and Figure S4D). Comparing the severity of spermatogonial accumulation, we found that tut4 homozygotes contained mostly spermatocytes mixed with a few spermatogonial tumors (Figure 3G); whereas disrupting a copy of bam in tut4 background blocked the germline development at spermatogonial stage (Figure 3H–J; 100%, n>50). The genetic interaction between tut and bam was confirmed by different alleles of both genes (Figure S4B–D).
Expression of either tut or bam did not rescue each other's mutant phenotype (Figure S4E–F). This prompted us to ask whether Tut and Bam act in the same protein complex. We first tried yeast 2-hybrid assay and found that Tut and Bam could form a complex (Figure S4G). Then we co-expressed Tut and Bam in cultured Drosophila S2 cells followed by co-immunoprecipitation assay. Myc-Tut and Flag-Bam co-immunoprecipitated with each other in both ways (Figure 3K–L). As in the testis, Tut and Bam protein expressed in S2 cells were localized in the cytoplasm (Figure S4H). Taken together, it is likely that Tut and Bam function in the same protein complex to regulate spermatogonial TA division.
Tut, Bam, and Bgcn Are Present in the Same Protein Complex
Bgcn and Bam have been demonstrated to form a complex in both female and male germline [15], [32] (Figure S5D). Although genetic interaction between bam and bgcn in germ cell development has been revealed in fly females [40], their interaction in male germline was not known due to the lack of weak allele of bam or bgcn. We generated a weak allele of bgcn named bgcn2. Like tut4 (Figure 3G), bgcn2 testis exhibited mild over-proliferation phenotype (Figure S5E). Removing one copy of bam dramatically enhanced bgcn2 mutant phenotype (Figure S5F).
Given the close relationship between Tut and Bam as well as Bam and Bgcn, plus the concurrence of the three proteins in spermatogonia (Figure 3D, 4A, and S5H), it is conceivable that three of them form a functional unit in spermatogonial TA cells. We then tested the genetic relationship between tut and bgcn. The double heterozygotes of tut4 and bgcn (20093 = null; QS2 = C-terminal truncation) did not show any spermatogonial tumor growth (Figure 4B–C; 100%, n>50). Further disrupting the other copy of tut made the majority of germ cells to keep dividing but unable to differentiate beyond spermatogonial stage (Figure 4D–E and S5G; 100%, n>50; also see Figure 3G for tut4 homozygous phenotype). Consistently, removing one copy of bgcn dramatically enhanced tut knockdown phenotype (Figure S5A–C). These data suggest that tut functions with bgcn in controlling spermatogonial TA proliferation.
Fig. 4. Tut, Bam, and Bgcn form a protein complex. 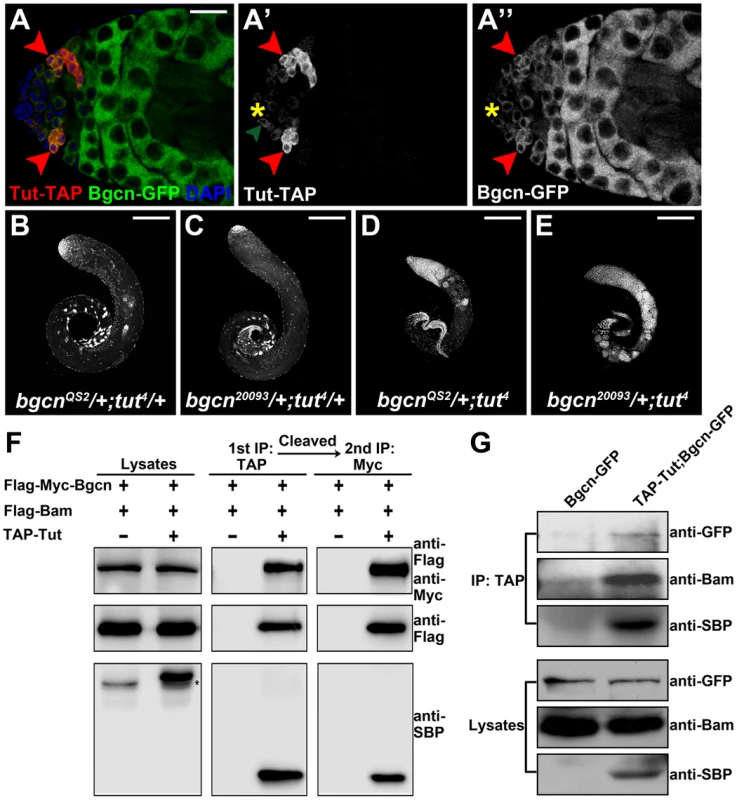
(A–A″) Immunofluorescence images of tutP-tut-TAP/+; bgcnP-bgcn-GFP/+ testis. Red arrowheads point to spermatogonial cells expressing both Tut-TAP and Bgcn-GFP. Green arrowhead points to a germline stem cell expressing weak Tut-TAP. (B-E) Genetic interactions between tut and bgcn. bgcnQS2/+; tut4/+ (A), bgcn20093/+; tut4/+ (B), bgcnQS2/+; tut4 (C), and bgcn20093/+; tut4 (D) testes were stained with DAPI. (F) S2 cells were transfected with different combinations of DNA constructs as indicated. Lysates from transfected S2 cells were used in a two-step immunoprecipitation method employing IgG and anti-Myc beads successively. Western analyses with anti-SBP (streptavidin binding protein), anti-Flag, and anti-Myc were performed to detect the presence of TAP-Tut, Flag-Bam, and Flag-Myc-Bgcn, respectively. TAP tag contains two TEV cleavage sites joining Protein G and SBP. After TEV digestion, the size of TAP-Tut changed from 48.35 kD to 32.86 kD. * indicates a nonspecific band. (G) Testes extracts of bamP-bgcnGFP/+ or tutP-tutTAP/Y; tutP-tutTAP/+;bamP-bgcnGFP/+ were immunoprecipitated with IgG beads. Tut-TAP was detected by anti-SBP on Western blot. Scale bars: 25 µm (A); 200 µm (B–E). See also Figure S5. To determine if Tut, Bam, and Bgcn are present in the same protein complex, we carried out the two-step co-immunoprecipitation assay [34], [41], [42] by co-expressing TAP-Tut, Flag-Bam, and Flag-Myc-Bgcn in S2 cells. After two rounds of successive immunoprecipitations, Bam and Bgcn were still present in Tut complex (Figure 4F), suggesting that these three proteins form a trimeric complex rather than exclusive heterodimers such as Bam/Tut, Bam/Bgcn, or Tut/Bgcn. To examine the existence of this complex in vivo, we used the extracts of fly testes to do the co-immunoprecipitation and again, demonstrate that these three factors are physically associated with each other (Figure 4G).
Although Tut and Bgcn formed a complex in the presence of Bam (Figure 5A–B, left panels), we failed to detect physical interaction between Tut and Bgcn in the absence of Bam in co-immunoprecipitation (Figure 5A–B, right panels) or in yeast 2-hybrid assays (Figure S6A). These observations raised the possibility that Bam brings Tut and Bgcn together to form a complex. Bgcn has been reported to interact with Bam C-terminus [32]. To map which region of Bam associates with Tut, we expressed different fragments of Bam in yeast and tested the interacting activity by yeast 2-hybrid assay. The Fragment containing N-terminal 100 amino acids of Bam was both necessary and sufficient to bind Tut (Figure S6B). We confirmed this interaction by co-immunoprecipitation using S2 cells (Figure 5C–F). These data suggest that Bam recruits Tut and Bgcn proteins to form a complex via its N-terminus and C-terminus, respectively.
Fig. 5. Tut and Bgcn are recruited by Bam to form a complex. 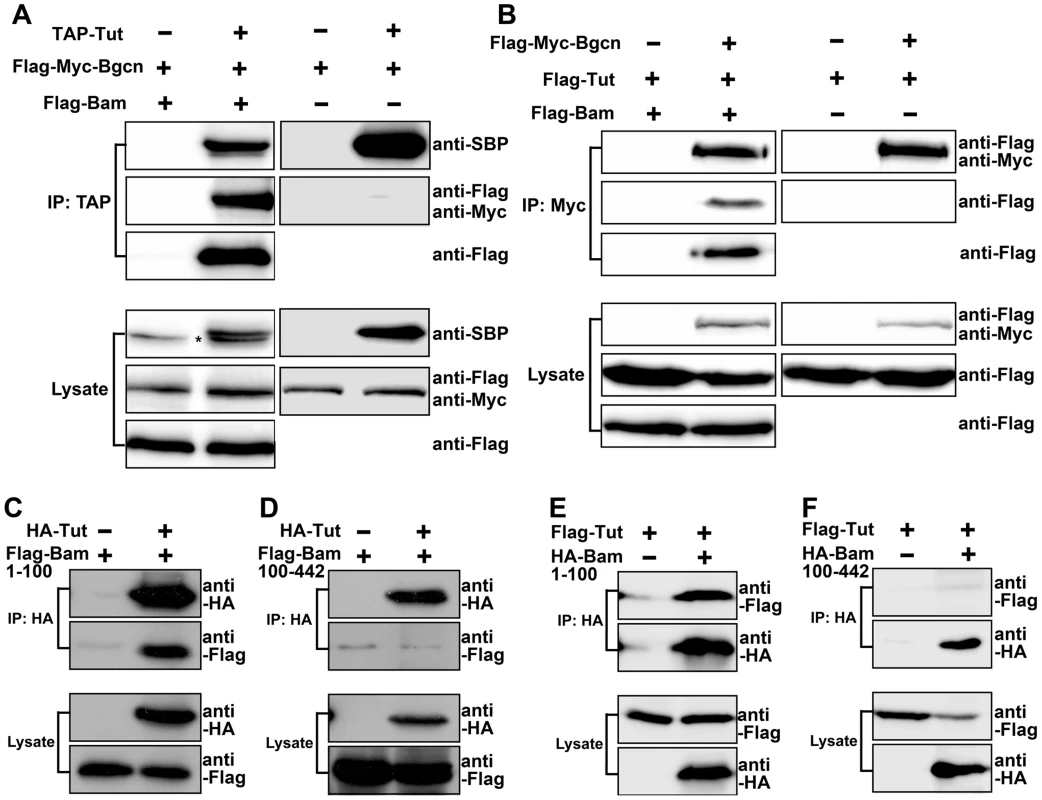
(A–B) S2 cells were transfected with the combinations of DNA constructs as indicated. Lysates from transfected S2 cells were immunoprecipitated with IgG (A) or anti-Myc (B) beads. Western blots were used to analyze the presence of TAP-, Flag-, or Myc-tagged proteins. Interaction between Tut and Bgcn was not detected in the absence of Bam. Asterisk in (A) indicates non-specific bands. (C–F) S2 cells were transfected with different combinations of DNA constructs as indicated. Lysates from transfected S2 cells were immunoprecipitated with anti-HA beads. Western blots were used to analyze the presence of Flag- or HA-tagged proteins. See also Figure S6. Given that all three proteins are RNA binding proteins (This study and [15]), we wonder if the formation of Tut-Bam-Bgcn complex is RNA-dependent. We found that the formation of this complex in S2 cells was not disrupted by the treatment of RNaseA (Figure S6C). Thus, Tut, Bam, and Bgcn form a protein complex in an RNA-independent manner.
Tut Is Required for Bam to Drive Germline Stem Cell Differentiation
Bam has been shown to promote differentiation when over-expressed in GSCs [29], [30] or in TA spermatogonia [26]. We wondered if Tut is required for Bam pro-differentiation function, and compared the consequences of Bam over-expression in GSCs (by the combination of nos-Gal4 and UASp-bam-GFP) in tut mutant v.s. wild-type background. As expected, ectopic expression of Bam in GSCs eliminated all germ cells (Figure 6A–B) [30]. However, in tut mutant background, Bam over-expression in GSCs just resembled tut mutant phenotype (Figure 6C–D), suggesting that bam requires tut to promote GSC differentiation.
Fig. 6. Tut is required for Bam to drive germline stem cell differentiation. 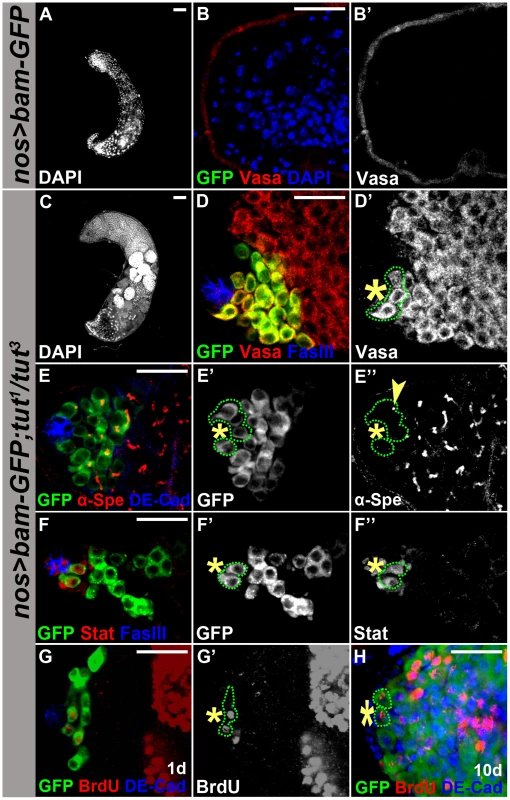
(A–B&B′) Genotype: UASp-bam-GFP/+; nos-Gal4/+. Forced expression of Bam in GSCs eliminated all germ cells. (C–D&D′) Genotype: UASp-bam-GFP/+; tut1 nos-Gal4/tut3. Germ cells were present in tut mutant background. Green dots outline the hub-adjacent germ cells expressing Bam-GFP. (E–E″) Immunofluorescence images of UASp-bam-GFP/+; tut1 nos-Gal4/tut3 testis. Green dots outline hub-adjacent germ cells expressing Bam-GFP and yellow arrowhead points to a dot-shape spectrosome. (F–F″) Immunofluorescence images of UASp-bam-GFP/+; tut1 nos-Gal4/tut3 testis. Green dots outline the hub-adjacent germ cells expressing Stat92E and Bam-GFP. (G&G′) 1 day old UASp-bam-GFP/+;tut1 nos-Gal4/tut3 testis labeled with BrdU for 1 hour. Green dotted-line highlights three hub-adjacent germ cells positive for Bam-GFP and two of them incorporated BrdU. (H) 10 day old UASp-bam-GFP/+;tut1 nos-Gal4/tut3 testis labeled with BrdU for 1 hour. Green dotted-line indicates two hub-adjacent germ cells positive for Bam-GFP and BrdU. Scale bars: 50 µm (A,C); 25 µm (B, D–H). See also Figure S7. In wild-type testis, GSCs are the first tier of germ cells surrounding the hub where Bam is repressed [8], [9]. When Bam-GFP was over-expressed in tut mutant background, it was present in the germ cells next to the hub (Figure 6D). To determine whether this tier of Bam-GFP positive germ cells were indeed GSCs, we immunostained these cells for fusome marker alpha-spectrin and GSC marker Stat92E. Fusome morphology changes from spherical in GSCs/GBs to branched in spermatogonia/spermatocytes (Figure 1A). We found that these hub-adjacent Bam-GFP-positive cells contained dot-shaped fusome (Figure 6E, yellow arrowhead) and expressed Stat92E (Figure 6F). Moreover, these Bam-GFP positive GSC-like cells maintained cell division, which was revealed by BrdU incorporation (Figure 6G), and such dividing activity was detected even 10 days after eclosure (Figure 6H). Thus, we demonstrate that Tut is required for Bam to drive GSC differentiation. Similarly, Bam also failed to drive GSC differentiation in bgcn mutant background (Figure S7), further supporting that Tut, Bam, and Bgcn form a functional unit to promote differentiation in GSC lineage.
Discussion
In the development of regenerative tissues, the successive differentiation of stem cell lineage is well controlled and coordinated with proper cell proliferation at each differentiation stage. This is clearly exemplified in Drosophila spermatogenesis which provides a nice research system for us to address related questions. RBPs play major roles in germline development but the molecular mechanisms how they exert their function remain largely unclear.
Tut, Bam, and Bgcn Act in the Same Complex to Regulate Spermatogonial TA Division in Drosophila
From a large-scale genetic screen, we identified tut as an intrinsic factor restricting spermatogonial proliferation. Using cultured S2 cells or fly testes, we demonstrated that Tut, Bam, and Bgcn formed a protein complex (Figure 4F–G). Mutations disrupting any of the three components block differentiation and lead to severe spermatogonial over-proliferation (Figure 1D and [14]). Furthermore, either Tut or Bgcn is required for Bam to drive GSC differentiation in testis (Figure 6 and Figure S7), suggesting that they could function as a complex in GSCs as well. Given the genetic and physical associations observed, we conclude that Tut, Bam, and Bgcn form a complex in spermatogonia to ensure precise TA divisions.
TA divisions are very sensitive to bam dosage and Bam protein level is under intricate control [26], [43]. Why is Bam protein level so critical for the timely transition from TA divisions to meiotic differentiation? In the Tut-Bam-Bgcn complex, Tut and Bgcn do not interact with each other unless Bam protein is present (Figure 5 and S6A–B). Bam acts as a nexus to bring Tut and Bgcn together, in the manner that the N-terminus of Bam interacts with Tut and the C-terminus with Bgcn. Bam expression is dynamic, first detected in 2∼4-cell of TA spermatogonia and peaked at 8-cell stage. Thus, Bam level determines the quantity of Tut-Bam-Bgcn complex. This may explain why Bam protein level serves as an ‘index’ for the spermatogonia to respond as when to stop TA divisions and start differentiation.
Regarding how mei-P26, the downstream target of the complex, is regulated, there is a discrepancy between the study by Insco et al. [15] and our current one. They found that Bam bound to the segments contained in the 549 nt of mei-P26 3′UTR, which we designated as the short form (Figure S2A–C). However, we could not detect any interaction between Bam and mei-P26 3′UTR long form by Y3H (Figure S2E–F). This could be due to the different assays we employed. It has been demonstrated that correct RNA folding is essential for protein-RNA interaction in Y3H [39]. Nonetheless, we both demonstrated that Bam and Bgcn are present in a complex required for the repression of mei-P26.
Different Expression Patterns of Tut, Bam, Bgcn, and Mei-P26 Are Associated with Different States of Germline Differentiation
Based on the previous and our current findings of the Tut-Bam-Bgcn complex and its target Mei-P26, we propose a model describing how the dynamic expression patterns of these proteins are associated with germline differentiation (Figure S8). Under normal conditions, Bgcn is present in all stages of spermatogenic cells whereas the other 3 proteins are not (Figure S8A). Tut is very weakly expressed in GSC, GB, or early TA cells, thus Mei-P26 cannot be completely repressed. Because Mei-P26 promotes Bam expression [15] and Bam is required for the full expression of Tut (Figure S3E–F), Tut accumulates and peaks in the late TA cells in which Tut, Bam, and Bgcn can form a complex bound on the 3′UTR of mei-P26 to repress its expression (Figure S8A). The shorter version of mei-P26 3′UTR is much more predominant than the longer one under this circumstance (Figure S2B–C). At the end of TA stage when Tut is degraded and Bam decreases, Mei-P26 gets derepressed and the germ cells enter the meiotic cycle.
In the mutants of tut, bam, or bgcn, the protein complex cannot be formed on the 3′UTR of mei-P26 to repress its expression, and mei-P26 3′UTR exists as a longer isoform (Figure S8B). However, derepression of Mei-P26 in TA cells is not sufficient to block the differentiation towards spermatocytes because overexpression of Mei-P26 in late TA cells did not phenocopy tut, bam, or bgcn mutant (this study and [15]). Even if Tut-Bam-Bgcn complex is the ‘master switch’ for TA cell transition to meiosis, there should be more downstream targets than just mei-P26 mRNA.
Bam is normally not expressed in GSCs whereas Tut and Bgcn are present, but ectopic expression of Bam in GSCs leads to GSC premature differentiation and eventually GSC loss ([29], [30] and Figure 6A–B), indicating that Bam could exert its ‘pro-differentiation’ function in GSCs. However, in the absence of Tut or Bgcn, ectopic Bam cannot drive GSC to differentiate, further suggesting the coordinated action of the three proteins in germ cell differentiation (Figure S8C).
Tut May Be a Male-Specific Component of the Translational Repression Complex in Germline
RBPs play central roles in germline development across species [44], [45]. Bam-Bgcn complex may act as part of the translational machinery but execute different functions in female and male germline by binding to different RNA targets with different partners. In female germline, Sxl binds to nos 3′UTR directly and associates with Bam-Bgcn complex to repress nos translation [46], [47]. But Sxl is not expressed in testis [46]. Although tut mutant spermatogonia fail to differentiate and over-proliferate, tut3 females are fully fertile and their ovarioles were indistinguishable from the wildtype by immunostaining (Figure S1P–Q). Thus, Tut may represent a male-specific partner of Bam-Bgcn complex.
It is plausible that Tut-Bam-Bgcn complex functions as part of the translational repression machinery to inhibit target mRNA translation in fly male germline. First, Tut also binds to mei-P26 3′UTR, though the preferred isoform of binding is different from Bam or Bgcn (Figure 2A, S2E–F). Secondly, Tut, Bam, and Bgcn are all expressed in spermatogonia (Figure S5H), and their mutant testes exhibit the same phenotype (this study and [14]). Thirdly, Tut, Bam, and Bgcn form a protein complex in Drosophila testes and S2 cells (Figure 4F–G). Fourthly, Bam protein binds the translation initiation factor eIF4A directly, and removing one copy of eIF4A partially suppresses the phenotype of bam mutants in both male and female systems [26], [33]. We speculate that Tut-Bam-Bgcn complex binds mei-P26 3′UTR to repress the translation of mei-P26 mRNA in TA cells. Noticeably, Dnd1, the putative homolog of Tut in Zebrafish, has been reported to protect mRNA from miRNA-mediated repression by binding to the 3′UTR in germline [48]. Furthermore, Tut-Bam-Bgcn complex is likely to target additional mRNAs in spermatogonia. Characterizing more of these mRNAs will further elucidate the molecular mechanisms how Tut-Bam-Bgcn complex promotes differentiation in GSC lineage.
Materials and Methods
Drosophila Strains and Husbandry
The fly strains used: bamΔ86 [49], bamBG [50], bamBW [40], bam-Gal4 [51], bamP-GFP [51], UASp-bam-GFP [51], bamP-bam-HA;bgcnP-bgcn-GFP [32], bgcnQS2 [40], bgcn20093 [52], UAS-dcr2 (gifts from T.Tabata), Zip-GFP [53], [54], dj-GFP [36], [55], Hrb98DE-GFP [35], [36]. nos-Gal4 and UAS-Flp were ordered from Bloomington Drosophila Stock Center; UAS-tutRNAi (v26044) was ordered from Vienna Drosophila RNAi Center. bgcn2 was generated in our lab and contains the deletion of TGACG in the 2nd intron of the gene.
Fly stocks were maintained under standard culture conditions and all flies were dissected 0–2 days after eclosure unless otherwise indicated. For RNAi experiments, flies were cultured at 25°C for 6 days and transferred to 29°C for another 6 days before dissection. For germline clonal analysis, flies were heat-shocked in 37°C water bath for 1 hour at late pupal stage and dissected 4–5 days after clone induction. tut4 homozygous and bam heterozygous phenotype varies at different temperatures, age, or nutritions. For tut4, bgcn2, and bam/+ related experiments, flies were cultured at 24°C, fed with fresh yeast daily, and dissected within 12 hours after eclosure.
EMS Mutagenesis
Isogenized flies bearing FRT and UAS-Flp were fed with EMS overnight. The progeny of EMS-treated flies were crossed to flies carrying FRT-GFP and nos-Gal4. Their male offspring were dissected and stained with DAPI [19], [56], [57], [58]. tut1 was one of the mutants with germline over-growth phenotype, and was mapped by deficiency screen and candidate gene sequencing.
tut Alleles
tut1 bears a point mutation (3L: G8203128A) that creates a new stop codon. Wild-type Tut protein is 230 amino acids in length, and tut1 is expected to produce only the N-terminal 174 amino acids. tut3 and tut4 alleles were generated by genomic engineering [59] using 3.1 kb upstream from start codon and 3.1 kb downstream from stop codon flanking sequences. Genomic coding region of tut was replaced with attP and loxP, generating the null allele tut3. tut genomic region was introduced back into the tut locus of in tut3 background via attP-attB incorporation. However, after this manipulation, 91 bp (attR and vector sequence) sequences were inserted upstream of tut start codon and 68 bp (loxP and vector sequence) downstream of stop codon, generating the weak allele tut4.
tut-Related Transgenic Flies
The w1118 and p51D stocks were chosen as the hosts for P-element and attB-attP mediated transgenesis, respectively [60], [61]. tutP-tut-GFP contains 1.5 kb sequence upstream of tut start codon, tut genomic region (introns included and stop codon removed) tagged with GFP at its C-terminus, and 2.2 kb sequence downstream of tut stop codon. tutP-tut-TAP and tutP-GFP contain the same regulatory sequences as tutP-tut-GFP [41], [62], [63]. UAS-GFP-mei-P26 3′UTR was generated by cloning GFP coding sequence and mei-P26 3′UTR (2 kb downstream of stop codon) to replace the SV40 element in pUAST vector.
Immunofluorescence
Fly testes were prepared and immunostained as previously described [18]. The following antibodies were used: 1B1 (1∶50, DSHB, 1B1), mouse anti α-Spectrin (1∶50, DSHB, 3A9), rabbit anti-pH3 (1∶1000, Upstate, 06-570), rabbit anti-Bam (1∶2000) [64], mouse anti-BrdU (1∶200, BD), mouse anti-FasIII (1∶200, DSHB, 7G10), rabbit anti-GFP (1∶2000, Invitrogen, A6455), rat anti-GFP (1∶200, MBL, D153-3), rabbit anti-LacZ (1∶50000, Cappel), mouse anti-SBP (1∶200, Santa Cruz, sc-101595), rabbit anti-Stat92E (1∶5000) [25], rabbit anti-Mei-P26 (1∶4000; against (KLH)-SFDGSEHQNRLSAVFIEC-OH) rabbit anti-Vasa (1∶8000; against (KLH)-MSDDWDDEPIVDTRGARC-OH), guinea pig anti-Vasa (1∶4000; against 6xHis-Vasa produced in E. coli), rat anti-Vasa (1∶50, DSHB).
Cell Culture, Immunoprecipitation, and Western Analysis
S2 cells were cultured in SFM serum free medium (Gibco, 10902). Transfection was performed using Cellfectin Reagent (Invitrogen, 10362-100) according to the manufacturer's instructions. An act-Gal4 construct was co-transfected with pUAST expression vectors for all transfection experiments except for pAFMW-bgcn. 48 hours after transfection, cells were lysed in Default Lysis Buffer (50 mM Tris pH 7.5, 5% glycerol, 0.2% IGEPAL, 1 mM DTT, 1.5 mM MgCl2, 125 mM NaCl, 25 mM NaF, 1 mM Na3VO4, proteinase inhibitor cocktail, 1 mM PMSF) for 30 minutes on ice. Then the supernatants were incubated with corresponding beads for 4 hours at 4°C. The beads were washed 4 times with washing buffer (50 mM Tris pH 7.5, 5% glycerol, 0.2% IGEPAL, 1.5 mM MgCl2, 125 mM NaCl, 25 mM NaF, 1 mM Na3VO4), followed by Western analyses. To assess the RNA dependence of protein-protein interaction, S2 cell lysates were incubated with 0.5 µg/uL RNaseA for 30 min at room temperature [65]. For co-immunoprecipitation with testis extracts, 300–500 pairs of testes from freshly eclosed flies were lysed in Default Lysis Buffer.
The beads used in co-immunoprecipitation: IgG-beads (Sigma, A2909), anti-Flag-beads (Sigma, A2220), anti-Myc-beads (Sigma, A7470), and anti-GFP-beads (MBL, D153-9). The primary antibodies used in Western analyses: mouse anti-Flag (1∶2000, Sigma, F1804), mouse anti-GFP (1∶2000, Santa Cruz, sc9996), mouse anti-HA (1∶5 000, MBL, M180-3), mouse anti-Myc (1∶2000, Santa Cruz, sc40), mouse anti-SBP (1∶2000, Santa Cruz, sc-101595), rabbit anti-Vasa (1∶8000), mouse anti-Bam (1∶10000, gift from D. Chen).
Two-Step Co-immunoprecipitation
Two-step co-immunoprecipitation was performed according to the procedures described previously [34], [41], [42]. After first immunoprecipitation, IgG-beads were incubated in TEV cleavage buffer (10 mM Tris pH 7.5, 100 mM NaCl, 0.1% IGEPAL, 0.5 mM EDTA) with TEV protease (Invitrogen, 12575-015) for 2 hours at 16°C with shaking. TEV eluate was subjected to the second immunoprecipitation by incubating with anti-Myc-beads.
RNA Immunoprecipitation (RIP)
RNase inhibitor (Takara 2313A) was used for all RIP-related experiments. mei-P26 3′UTR (549 bp and 844 bp from stop codon) was cloned into pUAS-GFP vector between GFP stop codon and SV40 3′UTR. S2 cells transfected with TAP-Tut and mei-P26 3′UTR or TAP-GFP and mei-P26 3′UTR were lysed in polysome lysis buffer according to [66]. IgG beads were used to enrich TAP-Tut or TAP-GFP. Tut or GFP and their bound RNA were released by TEV digestion. RNA from the digested elutes as well as from 5% cell lysis (input, used for normalization) was extracted, treated with DNaseI (Takara 2270A), reverse transcribed with primer mixture (100 nM each of 5′-CGTTGATAGGGGACTATACA, 5′-TTTGTTGCATTTTGTTTATC, 5′-TCAAGTCGCATTCAACGCAT, 5′-TTTTTTTAGTAGTAGCGCTAATTG) complementary to mei-P26 3′UTR, and quantified by real-time PCR with primers 5′-TCTTGGCAAGGAGTCAACAC and 5′-CTGTCGATGAGGCAAATGTT. Oligo-dT primer was used for reverse transcription to examine the actin5C mRNA bound to TAP-Tut or TAP-GFP.
Yeast 2-Hybrid Assay
Yeasts were cultured on SD/-Ade/-His/-Leu/-Trp medium supplemented with Aureobasidin A and X-α-Gal (QDO/X/A) to test protein-protein interactions or on SD/-Leu/-Trp medium (DDO) to confirm the transformation of testing plasmid DNA.
Yeast 3-Hybrid Assay
Yeasts were cultured on SD/-His/-Leu/-Ura medium supplemented with X-β-Gal (TDO/X) to test protein-RNA interactions or on SD/-Leu/-Ura medium (DDO) to confirm the transformation of testing plasmid DNA [39].
3′RACE (Rapid Amplification of cDNA Ends)
Total RNA was extracted using TRIzol (Ambion, 15596-018) from 10 pairs of w1118 testes and 3′RACE was performed by following the manufacturer's instructions (TAKARA, 6106). Outer primer (5′-TCCGAGGGCTATGTGGTTAC-3′) and inner primer (5′-GTTCTAGTCCTGAACACCCT-3′) were used to amplify mei-P26 3′UTR. PCR products were loaded into 2% argarose gel and electrophoresed at 100 V for 1.5 h on ice.
Supporting Information
Zdroje
1. AlonsoL, FuchsE (2003) Stem cells in the skin: waste not, Wnt not. Genes & Development 17 : 1189–1200.
2. ClarkeMF, FullerM (2006) Stem cells and cancer: Two faces of eve. Cell 124 : 1111–1115.
3. KondoM, WagersAJ, ManzMG, ProhaskaSS, SchererDC, et al. (2003) Biology of hematopoietic stem cells and progenitors: Implications for clinical application. Annual Review of Immunology 21 : 759–806.
4. SchuijersJ, CleversH (2012) Adult mammalian stem cells: the role of Wnt, Lgr5 and R-spondins. Embo Journal 31 : 2685–2696.
5. JamiesonCHM, AillesLE, DyllaSJ, MuijtjensM, JonesC, et al. (2004) Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. New England Journal of Medicine 351 : 657–667.
6. KrivtsovAV, TwomeyD, FengZ, StubbsMC, WangY, et al. (2006) Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442 : 818–822.
7. VisvaderJE (2011) Cells of origin in cancer. Nature 469 : 314–322.
8. de CuevasM, MatunisEL (2011) The stem cell niche: lessons from the Drosophila testis. Development 138 : 2861–2869.
9. FullerMT (1998) Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Seminars in Cell & Developmental Biology 9 : 433–444.
10. SpradlingA, FullerMT, BraunRE, YoshidaS (2011) Germline stem cells. Cold Spring Harbor perspectives in biology 3: a002642–a002642.
11. ChangY-J, PiH, HsiehC-C, FullerMT (2013) Smurf-mediated differential proteolysis generates dynamic BMP signaling in germline stem cells during Drosophila testis development. Developmental Biology 383 : 106–120.
12. DaviesEL, LimJGY, JooWJ, TamCH, FullerMT (2013) The transcriptional regulator lola is required for stem cell maintenance and germ cell differentiation in the Drosophila testis. Developmental Biology 373 : 310–321.
13. EunSH, StoiberPM, WrightHJ, McMurdieKE, ChoiCH, et al. (2013) MicroRNAs downregulate Bag of marbles to ensure proper terminal differentiation in the Drosophila male germline. Development 140 : 23–30.
14. GonczyP, MatunisE, DiNardoS (1997) bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development 124 : 4361–4371.
15. Insco MeganL, Bailey AlexisS, KimJ, Olivares GonzaloH, Wapinski OrlyL, et al. (2012) A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell stem cell 11 : 689–700.
16. JotiP, Ghosh-RoyA, RayK (2011) Dynein light chain 1 functions in somatic cyst cells regulate spermatogonial divisions in Drosophila. Scientific Reports 1.
17. KigerAA, White-CooperH, FullerMT (2000) Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature 407 : 750–754.
18. LiC-Y, GuoZ, WangZ (2007) TGF beta receptor saxophone non-autonomously regulates germline proliferation in a Smox/dSmad2-dependent manner in Drosophila testis. Developmental Biology 309 : 70–77.
19. MatunisE, TranJ, GonczyP, CaldwellK, DiNardoS (1997) punt and schnurri regulate a somatically derived signal that restricts proliferation of committed progenitors in the germline. Development 124 : 4383–4391.
20. ParrottBB, ChiangYT, HudsonA, SarkarA, GuichetA, et al. (2011) Nucleoporin98-96 Function Is Required for Transit Amplification Divisions in the Germ Line of Drosophila melanogaster. Plos One 6.
21. SarkarA, ParikhN, HearnSA, FullerMT, TazukeSI, et al. (2007) Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Current Biology 17 : 1253–1258.
22. SchulzC, WoodCG, JonesDL, TazukeSI, FullerMT (2002) Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development 129 : 4523–4534.
23. SunS, ZhaoS, WangZ (2008) Genes of Hippo signaling network act unconventionally in the control of germline proliferation in Drosophila. Developmental Dynamics 237 : 270–275.
24. TranJ, BrennerTJ, DiNardoS (2000) Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature 407 : 754–757.
25. ZhaoSW, ChenD, GengQ, WangZH (2013) The highly conserved LAMMER/CLK2 protein kinases prevent germ cell overproliferation in Drosophila. Developmental Biology 376 : 163–170.
26. InscoML, LeonA, TamCH, McKearinDM, FullerMT (2009) Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proceedings of the National Academy of Sciences of the United States of America 106 : 22311–22316.
27. McKearinDM, SpradlingAC (1990) bag-of-marbles - a Drosophila gene required to initiate both male and female gametogenesis. Genes & Development 4 : 2242–2251.
28. OhlsteinB, McKearinD (1997) Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 124 : 3651–3662.
29. ShengXR, BrawleyCM, MatunisEL (2009) Dedifferentiating Spermatogonia Outcompete Somatic Stem Cells for Niche Occupancy in the Drosophila Testis. Cell Stem Cell 5 : 191–203.
30. ShivdasaniAA, InghamPW (2003) Regulation of stem cell maintenance and transit amplifying cell proliferation by TGF-beta signaling in Drosophila spermatogenesis. Current Biology 13 : 2065–2072.
31. KimJY, LeeYC, KimC (2010) Direct inhibition of Pumilo activity by Bam and Bgcn in Drosophila germ line stem cell differentiation. Journal of Biological Chemistry 285 : 4741–4746.
32. LiY, MinorNT, ParkJK, McKearinDM, MainesJZ (2009) Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proceedings of the National Academy of Sciences of the United States of America 106 : 9304–9309.
33. ShenR, WengC, YuJ, XieT (2009) eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proceedings of the National Academy of Sciences of the United States of America 106 : 11623–11628.
34. LiQ, LinS, WangX, LianG, LuZ, et al. (2009) Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nature Cell Biology 11 : 1128–U1184.
35. MorinX, DanemanR, ZavortinkM, ChiaW (2001) A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 98 : 15050–15055.
36. SarkarS, LakhotiaSC (2005) The Hsp60C gene in the 25F cytogenetic region in Drosophila melanogaster is essential for tracheal development and fertility. Journal of Genetics 84 : 265–281.
37. KawaseE, WongMD, DingBC, XieT (2004) Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 131 : 1365–1375.
38. PekJW, LimAK, KaiT (2009) Drosophila Maelstrom Ensures Proper Germline Stem Cell Lineage Differentiation by Repressing microRNA-7. Developmental Cell 17 : 417–424.
39. BernsteinDS, ButerN, StumpfC, WickensM (2002) Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26 : 123–141.
40. OhlsteinB, LavoieCA, VefO, GateffE, McKearinDM (2000) The Drosophila cystoblast differentiation factor, benign genial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics 155 : 1809–1819.
41. KyriakakisP, TippingM, AbedL, VeraksaA (2008) Tandem affinity purification in Drosophila - The advantages of the GS-TAP system. Fly 2 : 229–235.
42. XiaL, JiaS, HuangS, WangH, ZhuY, et al. (2010) The Fused/Smurf Complex Controls the Fate of Drosophila Germline Stem Cells by Generating a Gradient BMP Response. Cell 143 : 978–990.
43. MonkAC, SiddallNA, VolkT, FraserB, QuinnLM, et al. (2010) HOW is required for stem cell maintenance in the Drosophila testis and for the onset of transit-amplifying divisions. Cell Stem Cell 6 : 348–360.
44. SadaA, SuzukiA, SuzukiH, SagaY (2009) The RNA-Binding Protein NANOS2 Is Required to Maintain Murine Spermatogonial Stem Cells. Science 325 : 139–1398.
45. SaffmanEE, LaskoP (1999) Germline development in vertebrates and invertebrates. Cellular and Molecular Life Sciences 55 : 1141–1163.
46. ChauJ, KulnaneLS, SalzHK (2012) Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proceedings of the National Academy of Sciences of the United States of America 109 : 9465–9470.
47. LiY, ZhangQ, Carreira-RosarioA, MainesJZ, McKearinDM, et al. (2013) Mei-P26 Cooperates with Bam, Bgcn and Sxl to Promote Early Germline Development in the Drosophila Ovary. Plos One 8.
48. KeddeM, StrasserMJ, BoldajipourB, Oude VrielinkJA, SlanchevK, et al. (2007) RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131 : 1273–1286.
49. McKearinD, OhlsteinB (1995) A role for the Drosophila Bag-of-marbles protein in the differentiation of cystoblasts from germline stem-cells. Development 121 : 2937–2947.
50. ChenDH, McKearinD (2005) Gene circuitry controlling a stem cell niche. Current Biology 15 : 179–184.
51. ChenDH, McKearinDM (2003) A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130 : 1159–1170.
52. JinZ, KirillyD, WengC, KawaseE, SongX, et al. (2008) Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell 2 : 39–49.
53. BuszczakM, PaternoS, LighthouseD, BachmanJ, PlanckJ, et al. (2007) The Carnegie protein trap library: A versatile tool for Drosophila developmental studies. Genetics 175 : 1505–1531.
54. EikenesAH, BrechA, StenmarkH, HaglundK (2013) Spatiotemporal control of Cindr at ring canals during incomplete cytokinesis in the Drosophila male germline. Developmental Biology 377 : 9–20.
55. SantelA, WinhauerT, BlumerN, RenkawitzPohlR (1997) The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterized by an unusual domain of a repetitive amino acid motif. Mechanisms of Development 64 : 19–30.
56. BrandAH, PerrimonN (1993) Targeted gene-expression as a means of altering cell fates and generating dominant phenotypes. Development 118 : 401–415.
57. Van DorenM, WilliamsonAL, LehmannR (1998) Regulation of zygotic gene expression in Drosophila primordial germ cells. Current Biology 8 : 243–246.
58. XuT, RubinGM (1993) Analysis of genetic mosaics in developing and adult drosophila tissues. Development 117 : 1223–1237.
59. HuangJ, ZhouW, DongW, WatsonAM, HongY (2009) Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proceedings of the National Academy of Sciences of the United States of America 106 : 8284–8289.
60. BischofJ, MaedaRK, HedigerM, KarchF, BaslerK (2007) An optimized transgenesis system for Drosophila using germ-line-specific phi C31 integrases. Proceedings of the National Academy of Sciences of the United States of America 104 : 3312–3317.
61. RubinGM, SpradlingAC (1982) Genetic-transformation of drosophila with transposable element vectors. Science 218 : 348–353.
62. BuerckstuemmerT, BennettKL, PreradovicA, SchutzeG, HantschelO, et al. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nature Methods 3 : 1013–1019.
63. VeraksaA, BauerA, Artavanis-TsakonasS (2005) Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Developmental Dynamics 232 : 827–834.
64. ChenD, WangQ, HuangH, XiaL, JiangX, et al. (2009) Effete-mediated degradation of Cyclin A is essential for the maintenance of germline stem cells in Drosophila. Development 136 : 4133–4142.
65. LiuJD, Valencia-SanchezMA, HannonGJ, ParkerR (2005) MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature Cell Biology 7 : 719–U118.
66. PeritzT, ZengF, KannanayakalTJ, KilkK, EiriksdottirE, et al. (2006) Immunoprecipitation of mRNA-protein complexes. Nat Protoc 1 : 577–580.
Štítky
Genetika Reprodukčná medicína
Článek The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural CompetenceČlánek Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the TestisČlánek The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I ofČlánek GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and AnnotationČlánek Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant BiomassČlánek Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant GeneČlánek p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide SecretionČlánek The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of MitophagyČlánek Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of SenescenceČlánek ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Establishing a Multidisciplinary Context for Modeling 3D Facial Shape from DNA
- RNA Processing Factors Swd2.2 and Sen1 Antagonize RNA Pol III-Dependent Transcription and the Localization of Condensin at Pol III Genes
- Inversion of the Chromosomal Region between Two Mating Type Loci Switches the Mating Type in
- A Thermolabile Aldolase A Mutant Causes Fever-Induced Recurrent Rhabdomyolysis without Hemolytic Anemia
- The Role of Regulatory Evolution in Maize Domestication
- Stress Granule-Defective Mutants Deregulate Stress Responsive Transcripts
- 24-Hour Rhythms of DNA Methylation and Their Relation with Rhythms of RNA Expression in the Human Dorsolateral Prefrontal Cortex
- Pseudoautosomal Region 1 Length Polymorphism in the Human Population
- Fungal Communication Requires the MAK-2 Pathway Elements STE-20 and RAS-2, the NRC-1 Adapter STE-50 and the MAP Kinase Scaffold HAM-5
- The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural Competence
- The Protein -glucosyltransferase Rumi Modifies Eyes Shut to Promote Rhabdomere Separation in
- The Talin Head Domain Reinforces Integrin-Mediated Adhesion by Promoting Adhesion Complex Stability and Clustering
- Quantitative Genetics of CTCF Binding Reveal Local Sequence Effects and Different Modes of X-Chromosome Association
- Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the Testis
- Genetic Analysis of a Novel Tubulin Mutation That Redirects Synaptic Vesicle Targeting and Causes Neurite Degeneration in
- A Systems Genetics Approach Identifies , , and as Novel Aggressive Prostate Cancer Susceptibility Genes
- Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in
- Approximation to the Distribution of Fitness Effects across Functional Categories in Human Segregating Polymorphisms
- The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I of
- SAS-1 Is a C2 Domain Protein Critical for Centriole Integrity in
- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and Annotation
- Let's Face It—Complex Traits Are Just Not That Simple
- Glutamate Receptor Gene , Coffee, and Parkinson Disease
- The Red Queen Model of Recombination Hotspots Evolution in the Light of Archaic and Modern Human Genomes
- The Ethics of Our Inquiry: An Interview with Hank Greely
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
- Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-response
- Lack of Replication of the -by-Coffee Interaction in Parkinson Disease
- Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity
- A Germline Polymorphism of Thymine DNA Glycosylase Induces Genomic Instability and Cellular Transformation
- Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene
- ATPase-Independent Type-III Protein Secretion in
- p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide Secretion
- The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of Mitophagy
- Evolution of DNA Methylation Patterns in the Brassicaceae is Driven by Differences in Genome Organization
- Regulation of mRNA Abundance by Polypyrimidine Tract-Binding Protein-Controlled Alternate 5′ Splice Site Choice
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of Senescence
- A Functional Portrait of Med7 and the Mediator Complex in
- Systematic Analysis of the Role of RNA-Binding Proteins in the Regulation of RNA Stability
- ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
- Genomic Evidence of Rapid and Stable Adaptive Oscillations over Seasonal Time Scales in Drosophila
- Genome-Wide Associations between Genetic and Epigenetic Variation Influence mRNA Expression and Insulin Secretion in Human Pancreatic Islets
- HAM-5 Functions As a MAP Kinase Scaffold during Cell Fusion in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



