-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
The protein kinase Mec1 (ATR ortholog) and its partner Ddc2 (ATRIP ortholog) play a key role in DNA damage checkpoint responses in budding yeast. Previous studies have established the model in which Ddc1, a subunit of the checkpoint clamp, and Dpb11, related to TopBP1, activate Mec1 directly and control DNA damage checkpoint responses at G1 and G2/M. In this study, we show that Ddc2 contributes to Mec1 activation through a Ddc1 - or Dpb11-independent mechanism. The catalytic activity of Mec1 increases after DNA damage in a Ddc2-dependent manner. In contrast, Mec1 activation occurs even in the absence of Ddc1 and Dpb11 function at G2/M. Ddc2 recruits Mec1 to sites of DNA damage. To dissect the role of Ddc2 in Mec1 activation, we isolated and characterized a separation-of-function mutation in DDC2, called ddc2-S4. The ddc2-S4 mutation does not affect Mec1 recruitment but diminishes Mec1 activation. Mec1 phosphorylates histone H2A in response to DNA damage. The ddc2-S4 mutation decreases phosphorylation of histone H2A more significantly than the absence of Ddc1 and Dpb11 function does. Our results suggest that Ddc2 plays a critical role in Mec1 activation as well as Mec1 localization at sites of DNA damage.
Published in the journal: Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism. PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004136
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004136Summary
The protein kinase Mec1 (ATR ortholog) and its partner Ddc2 (ATRIP ortholog) play a key role in DNA damage checkpoint responses in budding yeast. Previous studies have established the model in which Ddc1, a subunit of the checkpoint clamp, and Dpb11, related to TopBP1, activate Mec1 directly and control DNA damage checkpoint responses at G1 and G2/M. In this study, we show that Ddc2 contributes to Mec1 activation through a Ddc1 - or Dpb11-independent mechanism. The catalytic activity of Mec1 increases after DNA damage in a Ddc2-dependent manner. In contrast, Mec1 activation occurs even in the absence of Ddc1 and Dpb11 function at G2/M. Ddc2 recruits Mec1 to sites of DNA damage. To dissect the role of Ddc2 in Mec1 activation, we isolated and characterized a separation-of-function mutation in DDC2, called ddc2-S4. The ddc2-S4 mutation does not affect Mec1 recruitment but diminishes Mec1 activation. Mec1 phosphorylates histone H2A in response to DNA damage. The ddc2-S4 mutation decreases phosphorylation of histone H2A more significantly than the absence of Ddc1 and Dpb11 function does. Our results suggest that Ddc2 plays a critical role in Mec1 activation as well as Mec1 localization at sites of DNA damage.
Introduction
The DNA damage response pathways coordinate DNA repair, replication and cell-cycle progression to maintain genome integrity [1], [2]. Initiation of DNA damage responses requires two evolutionarily conserved phosphoinositide 3-kinase (PI3K)-related protein kinases: ATM and ATR. While ATM responds primarily to DNA double-strand breaks (DSBs), ATR recognizes various types of DNA lesions with single-stranded DNA (ssDNA) [2], [3]. In the budding yeast Saccharomyces cerevisiae, Mec1, the ATR ortholog, plays a critical role in the DNA damage response throughout the cell-cycle [1], [4]. Mec1 forms a stable complex with Ddc2 (ATRIP in human) [5]–[7], which recruits Mec1 to sites of DNA damage by interacting with replication protein A (RPA)-coated ssDNA [8]–[11]. Once recruited to DNA lesions, Mec1 phosphorylates the C-terminal tail of histone H2A [12]. Phosphorylated histone H2A creates a DNA damage mark similar to phosphorylated histone H2AX (γH2AX) in human [4], [13]. Phosphorylation of histone H2A, together with constitutive methylation of histone H3, promotes the recruitment of the checkpoint mediator, Rad9 [14], [15]. In turn, Mec1 phosphorylates Rad9 at sites of DNA damage [16], [17]. Phosphorylated Rad9 creates a docking site for the effector kinase, Rad53 (Chk2 in human) [18]–[20]. The Rad9-Rad53 interaction promotes Rad53 autophosphorylation and allows Mec1 to phosphorylate Rad53, leading to hyperphosphorylation and activation of Rad53 [20]–[23].
To execute full activation of Rad53, Mec1 collaborates with the checkpoint clamp complex and its loader as well [4]. The checkpoint clamp complex, structurally related to PCNA, consists of Ddc1, Mec3 and Rad17 (Rad9, Hus1 and Rad1 in human) [24], [25]. The checkpoint clamp loader is composed of Rad24 (Rad17 in human) and the four small RFC subunits [25]–[27]. The Rad24-RFC complex recognizes and loads the Ddc1-Mec3-Rad17 complex at junctions between ssDNA and double-stranded DNA on partial duplex DNA [25], [28]. Co-accumulation of Mec1 and Ddc1 at sites of DNA damage is essential for checkpoint activation [29], [30]. Supporting this view, tethering of both Ddc1 and Ddc2 on chromatin is sufficient for Rad53 phosphorylation even in the absence of DNA damage [31]. Once loaded at sites of DNA damage, the Ddc1-Mec3-Rad17 complex stimulates the association of Dpb11 (TopBP1 in human) with Mec1 [32]. Ddc1 and Dpb11 play a crucial role in DNA damage checkpoints at G1 and G2/M [33]–[36]. In vitro reconstitution studies have shown that Ddc1 or Dpb11 increases the catalytic activity of Mec1 [34], [35], [37]–[40]. These observations have established the model in which Ddc1 and Dpb11 govern the checkpoint pathway at G1 and G2/M by directly activating Mec1. In parallel with Ddc1 and Dpb11, Dna2 controls DNA damage and replication checkpoints specifically in S-phase by directly activating Mec1 [41]. Interestingly, all Ddc1, Dpb11 and Dna2 proteins utilize the unstructured domains with aromatic amino acid residues (Trp or Tyr) to increase the catalytic activity of Mec1 [34], [35], [41]. Moreover, the activation domain of Ddc1 is functionally replaceable with that of Dna2 [41]. Thus, Ddc1, Dpb11 and Dna2 appear to activate Mec1 through a similar mechanism.
In this study we show that Ddc2 stimulates Mec1 activity by a different mechanism than Ddc1 and Dpb11 at G2/M. Dpb11 possesses the Mec1 activation domain at the C-terminus. We used a strain expressing a C-terminal truncation mutation of Dpb11 encoded by the dpb11-1 mutant allele and the deletion of DDC1, and found that Mec1 activation occurs after DNA damage even in ddc1Δ dpb11-1 mutants at G2/M. However, Mec1 activation was fully dependent on Ddc2. Ddc2 plays an essential role in the recruitment of Mec1 to DNA lesions. To dissect the role of Ddc2 in Mec1 activation, we screened mutations in DDC2, which cause defects in DNA damage response but do not affect Mec1 recruitment. We characterized one of the separation-of-function mutations, ddc2-S4. The ddc2-S4 mutation, unlike the ddc1Δ dpb11-1 mutation, abolished damage-induced Mec1 activation. Cells carrying the ddc2-S4 mutation showed more significant defects in damage-induced phosphorylation of histone H2A than ddc1Δ dpb11-1 mutants at G2/M. These results are consistent with the model in which Ddc2 controls Mec1 activation by a different mechanism than Ddc1 or Dpb11.
Results
Requirement of Ddc2 for damage-induced Mec1 activation
We examined whether Mec1 catalytic activity is increased after DNA damage. Activation of the Mec1 signaling pathway is controlled in a cell-cycle dependent manner [34]–[36]. We thus monitored Mec1 kinase activity in G2/M-arrested cells (Fig. 1A). Cells expressing HA-tagged Mec1 (Mec1-HA) protein were arrested with nocodazole and then treated with methylmethane sulfonate (MMS). As a negative control cells expressing the kinase negative version of Mec1-HA (Mec1-KN-HA) were examined. HA-Mec1 proteins were immunoprecipitated from cell extracts with anti-HA antibodies, and immunoprecipitates were subjected to an in vitro kinase assay. As a substrate we used the GST-fusion protein with the C-terminus of Rad53 (GST-Rad53) [42]. Phosphorylation of the Rad53 C-terminus is critical for Rad53 activation [23]. Phosphorylation of GST-Rad53 was detected with immunoprecipitates from untreated cells, but the level increased after MMS treatment. No phosphorylation was detected with Mec1-KN-HA, indicating that the observed phosphorylation depends on Mec1 kinase activity. Mec1 was activated two-fold after exposure to 0.05% MMS for 1 hr (Fig. 1B). Similar activation was detected even if cells were treated with phleomycin (data not shown).
Fig. 1. Regulation of protein kinase Mec1 after DNA damage. 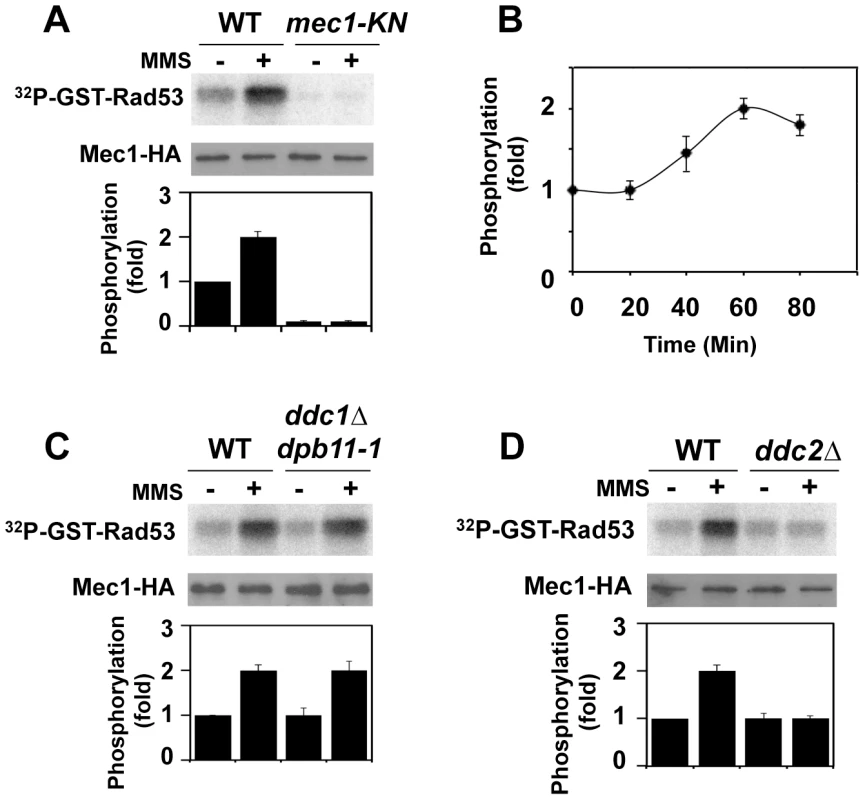
(A) Mec1 activation after MMS treatment. Cells expressing Mec1-HA (KSC1635) or Mec1-KN-HA (KSC1645) were cultured at 25°C and incubated with nocodazole to arrest at G2/M. Cells were then either mock treated (−) or treated (+) with 0.05% MMS for 1 hr. Immunoprecipitated Mec1-HA was subjected to the in vitro kinase assay using GST-Rad53, a recombinant protein purified from E. coli, as a substrate. Incorporation of 32P into GST-Rad53 was monitored by phosphorimager. The amounts of Mec1 or Mec1-KN in the immunoprecipitates were determined by immunoblotting with anti-HA antibody. Phosphorylation of GST-Rad53 was normalized to that observed with immunoprecipitated mock treated wild-type Mec1. Relative phosphorylation is determined from three independent experiments. (B) Mec1 activation kinase after incubation with MMS for various lengths of time. Cells were treated with MMS for indicated lengths of time and were subjected to the in vitro kinase assay as in A. (C) Effect of ddc1Δ dpb11-1 mutation on Mec1 activation. Wild-type (KSC1333) and ddc1Δ dpb11-1 (KSC3130) cells expressing Mec1-HA protein were subjected to the in vitro kinase assay as in A. (D) Effect of ddc2Δ mutation on Mec1 activation. Wild-type (KSC1635) and ddc2Δ (KSC1636) cells expressing HA-Mec1 protein were subjected to the in vitro kinase assay as in A. The unstructured C-terminal tails of Ddc1 and Dpb11 increase Mec1 kinase activity and control the Mec1-Rad53 checkpoint pathway at G2/M [34], [35], [38]. We next addressed whether Mec1 is activated after DNA damage in a Ddc1 - or Dpb11-dependent manner (Fig. 1C). Because DPB11 is essential for cell proliferation, we used the C-terminal truncation mutation in DPB11, dpb11-1, which confers defects in checkpoint activation at G2/M [34], [35], [43]. We examined the effect of ddc1Δ dpb11-1 mutation on Mec1 kinase activity before and after MMS treatment (Fig. 1C). DNA flow cytometry analysis confirmed that ddc1Δ dpb11-1 mutants were largely arrested with G2/M DNA contents after nocodazole treatment (Fig. S1). MMS-induced Mec1 activation was observed in ddc1Δ dpb11-1 mutants, indicating that Mec1 activation occurs after DNA damage independently of Ddc1 and Dpb11. Our assay was not able to detect Ddc1 - or Dpb11-dependent Mec1 activation, but this observation does not contradict the idea that Ddc1 or Dpb11 directly activates Mec1 [34], [35], [38] (see below). Mec1 forms a stable complex with Ddc2 [5]–[7]. We examined the effect of ddc2Δ mutation on Mec1 activity before and after DNA damage (Fig. 1D). DDC2 is essential for cell proliferation [5]–[7]. Since the lethality of ddc2 disruption is suppressed by sml1 mutation [5]–[7], we examined the effect of the ddc2Δ mutation in a sml1Δ background. The ddc2Δ mutation did not impair basal kinase activity of Mec1, but abolished increase in catalytic activity after MMS treatment.
Identification of Ddc2 domain that is involved in Mec1-Ddc2 interaction or DNA damage responses
One explanation for the above results is that Ddc2 mediates Mec1 activation independently of Ddc1 or Dpb11. However, accumulation of Mec1 at sites of DNA damage might be sufficient for Mec1 activation, since Ddc2 plays a key role in recruiting Mec1 to DNA lesions [8], [9], [11]. We therefore sought separation-of-function mutation(s) in DDC2, which confer defects in damage-induced Mec1 activation but not in Mec1 recruitment. To narrow the target area for mutagenesis, we first identified domain(s) in Ddc2 that are necessary for DNA damage response or Mec1-Ddc2 interaction in a modified two-hybrid system (Fig. 2). Mec1-Ddc2 interaction was monitored using the binding domain (BD)-MEC1 fusion and the activation domain (AD)-DDC2 fusion [7]. To assess DNA damage response function, we introduced a ddc2Δ mutation in the two-hybrid tester strain. Cells carrying a ddc2Δ mutation are sensitive to DNA damaging agents including hydroxyurea (HU) or MMS. The full-length AD-DDC2 construct supported both Mec1-Ddc2 interaction and cellular response to HU or MMS treatment (Fig. 2). The C-terminal truncation mutation, ddc2-ΔC, abolishes Mec1-Ddc2 interaction, thereby disrupting Ddc2 function [7]. The modified two-hybrid assay depicted the ddc2-ΔC mutation accurately (Fig. 2). We further explored whether the N-terminus of Ddc2 is required for Mec1-Ddc2 interaction or DNA damage tolerance. We found that the N-terminus of Ddc2 is dispensable for Ddc2 functions; both the ddc2-ΔN1 and ddc2-ΔN2 constructs supported Mec1-Ddc2 interaction and DNA damage tolerance in the modified two-hybrid system (Fig. 2). However, the ddc2-ΔN3 mutation disrupted Mec1-Ddc2 interaction and thereby impaired proper DNA damage responses (Fig. 2). Thus, the middle and C-terminal regions of Ddc2 (amino acid 141–747) are essential for Mec1-Ddc2 complex formation and proper DNA damage responses.
Fig. 2. Identification of the Ddc2 region required for Mec1 interaction and DNA damage response using the modified two-hybrid assay. 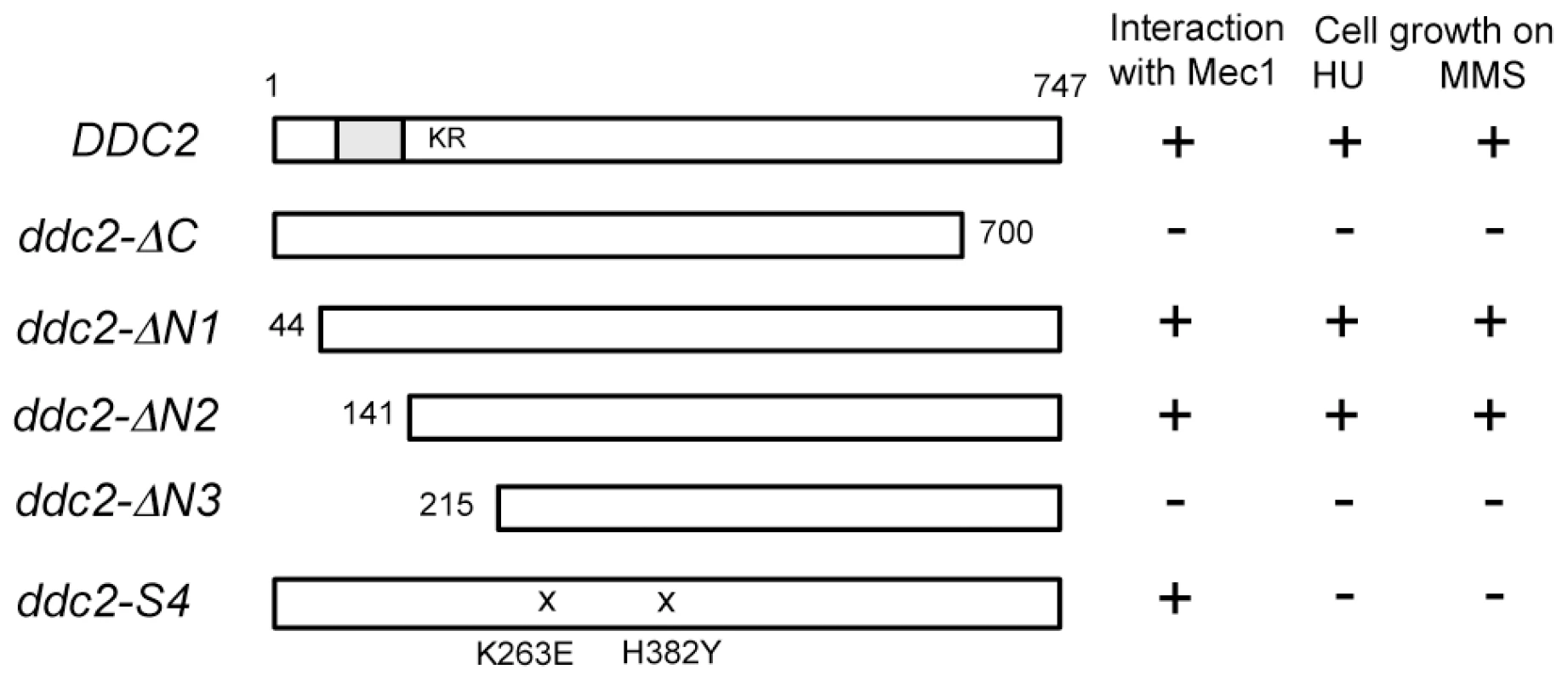
The two-hybrid tester ddc2Δ strain (KSC2077) carrying pBD-MEC1(2-2368) was transformed with pAD-DDC2, pAD-DDC2-ΔC, pAD-DDC2-ΔN1, pAD-DDC2-ΔN2, pAD-DDC2-ΔN3 or pAD-DDC2-S4. The coiled-coil domain (amino acid 68–138) is indicated as a gray bar and the KKRK sequence (amino acid 177–180) is shown as KR. The ddc2-S4 mutation carries two substitution mutations (K263E and H382Y). Transformants were streaked on an SD-Ura-Leu-His plate containing 1 mM AT, YEPD plate containing 1 mg/ml HU or 0.005% MMS. Interaction with Mec1 and DNA damage sensitivity were assessed by cell proliferation on the respective plates. Isolation of the separation-of-function ddc2-S4 mutation by using two-hybrid systems
The above information prompted us to screen separation-of-function mutations within the amino residues (141–747) in the middle and C-terminal regions of Ddc2. We combined two different two-hybrid systems to isolate ddc2 separation-of-function mutations that impair DNA damage responses but do not affect Mec1 localization to DNA damage sites. Proper Mec1 localization depends on Mec1-Ddc2 complex formation [11]. In the first screening, we adopted the functional two-hybrid assay as described above (see Fig. 2). After in vitro mutagenesis in DDC2 of the AD-DDC2 construct, we searched for clones that maintain Mec1-Ddc2 interaction but fail to rescue the ddc2Δ mutation. The Mec1-Ddc2 complex accumulates at sites of DNA damage by interacting with RPA [9]–[11]. RPA consists of three subunits: Rfa1, Rfa2 and Rfa3 in budding yeast [44]. We have shown that the Mec1-Ddc2 complex interacts with Rfa1 or Rfa2 in a two-hybrid system [11]. In the second screening, we further selected clones that retain the interaction with Rfa1 in a two-hybrid system. After the second screening each mutant allele was fused to its own promoter and introduced into its own chromosome locus. One mutation, named ddc2-S4, conferred noteworthy sensitivity to DNA damaging agents (Fig. 3A and 3B). We therefore characterized the ddc2-S4 mutation further. The ddc2-S4 allele contains two substitution mutations; K263E and H382Y, both of which are required to exhibit full DNA damage sensitivity (data not shown).
Fig. 3. DNA damage response and proliferation of ddc2-S4 mutants. 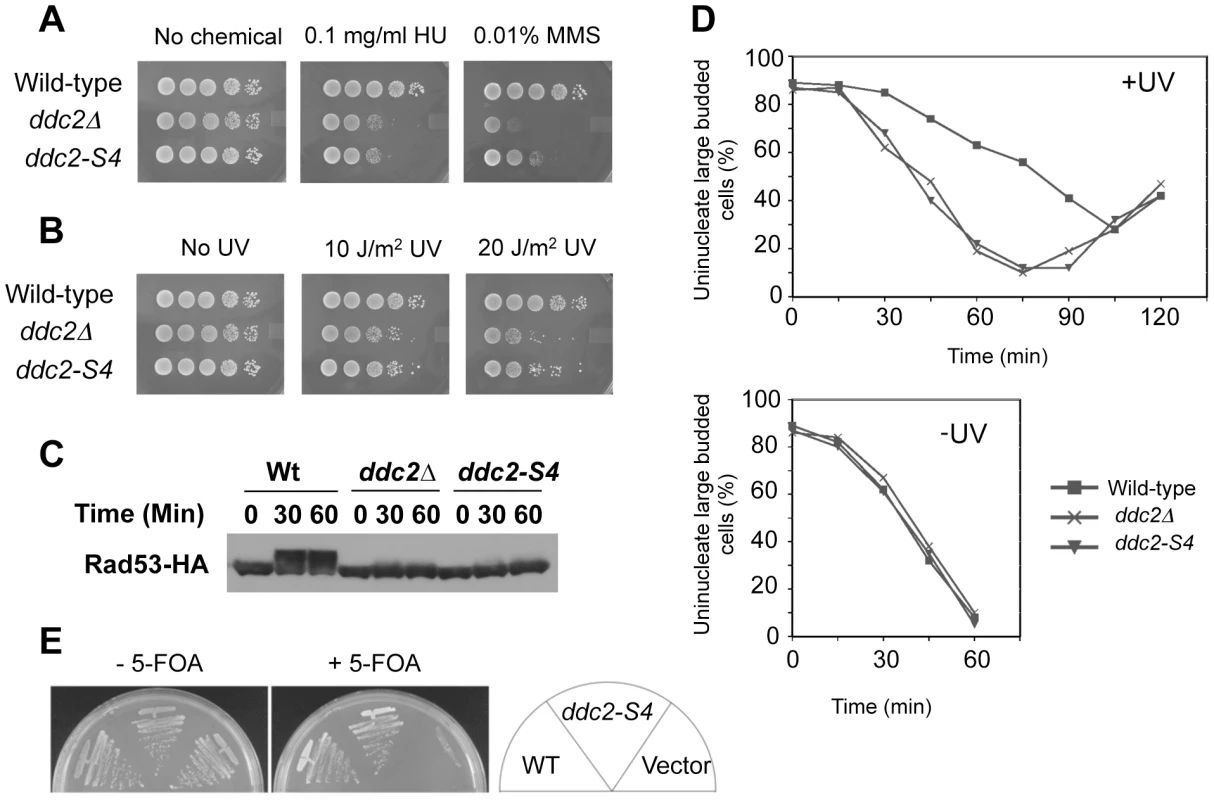
(A, B) DNA damage sensitivity of ddc2-S4 mutants. Serial dilutions of cultures were spotted on YEPD medium without or with MMS or HU (A). Cultures spotted on YEPD medium were irradiated with UV light (B). Plates were incubated at 30°C for two-three days. Strains used were wild-type (KSC1178), ddc2Δ (KSC1234) and ddc2-S4 (KSC3153). (C) Effect of ddc2-S4 on DNA damage checkpoint signaling. Wild-type (KSC1178), ddc2Δ (KSC1234), and ddc2-S4 (KSC3153) cells expressing Rad53-HA were grown to log-phase and exposed to MMS (0.05%) at 30°C for the indicated length of time. Cells were harvested and subjected to immunoblotting analysis with anti-HA antibodies. (D) G2/M-phase DNA damage checkpoint in ddc2-S4 mutants. Wild-type (KSC1178), ddc2Δ (KSC1234) or ddc2-S4 (KSC3153) cells were arrested with nocodazole and irradiated or not irradiated with UV (50 J/m2). At the indicated times after release of UV-irradiated (+UV) and unirradiated (−UV) cultures from nocodazole, the percentage of uninucleate large budded cells was scored by DAPI staining. (E) Effect of ddc2-S4 mutation on cell proliferation in the presence of SML1. ddc2Δ mutants carrying the URA3-marked YCp-DDC2 plasmid (KSC3308) were transformed with YCpT-DDC2, YCpT-DDC2-S4 or the control vector. Transformants were streaked and grown on plates containing medium with or without 5-fluoroorotic acid (5-FOA) at 30°C. Only cells that have lost the URA3-marker plasmid can proliferate in the presence of 5-FOA [62]. We examined the effect of ddc2-S4 mutation on checkpoint signaling in response to DNA damage (Fig. 3C). Checkpoint activation is correlated to the phosphorylation status of Rad53, a kinase that acts downstream of Mec1 [4]. Wild-type, ddc2Δ or ddc2-S4 cells expressing HA-tagged Rad53 protein (Rad53-HA) were incubated with MMS, and subjected to immunoblotting analysis with anti-HA antibody. Rad53 was phosphorylated in wild-type cells after MMS treatment while no apparent phosphorylation was detected in ddc2Δ mutants. As found in ddc2Δ mutants, Rad53 phosphorylation was markedly decreased in ddc2-S4 mutants. We examined whether ddc2-S4 mutants are defective in DNA damage checkpoints. We first examined the G2/M-phase DNA damage checkpoint by monitoring mitotic division following UV irradiation (Fig. 3D). When cell cultures were released from nocodazole arrest following UV irradiation, wild-type cells exhibited delayed nuclear division while ddc2-S4 cells underwent mitosis similar to ddc2Δ cells. Similarly, ddc2-S4 mutant cells progressed faster than wild-type cells through the G1/S transition and S phase following DNA damage (Fig. S2 and Fig. S3). DDC2 is indispensable for cell proliferation [5]–[7] but the ddc2-S4 mutation does not perturb the essential function of Ddc2. Cells carrying the ddc2-S4 mutation proliferated as well as those carrying the wild-type DDC2 gene (Fig. 3E).
Effect of ddc2-S4 mutation on Mec1-Ddc2 complex formation and Mec1 localization to sites of DNA damage
We investigated the effect of ddc2-S4 mutation on Mec1 localization to DNA damage sites. As discussed above, Mec1 localization depends on Mec1-Ddc2 complex formation [11]. We first examined the effect of ddc2-S4 mutation on Mec1-Ddc2 interaction (Fig. 4A). Extracts were prepared from cells expressing Mec1-HA and Ddc2-myc or Ddc2-S4-myc and analyzed by immunoprecipitation with anti-HA antibodies. The immunoprecipitates were then probed with antibodies against the HA and myc epitopes. Co-precipitation of Ddc2 with Mec1 was detected only in extracts from cells expressing both Mec1-HA and Ddc2-myc. Ddc2 and Ddc2-S4 were similarly precipitated with Mec1, indicating that the ddc2-S4 mutation does not impair Mec1-Ddc2 complex formation.
Fig. 4. Effect of ddc2-S4 on Mec1-Ddc2 interaction and Mec1 localization. 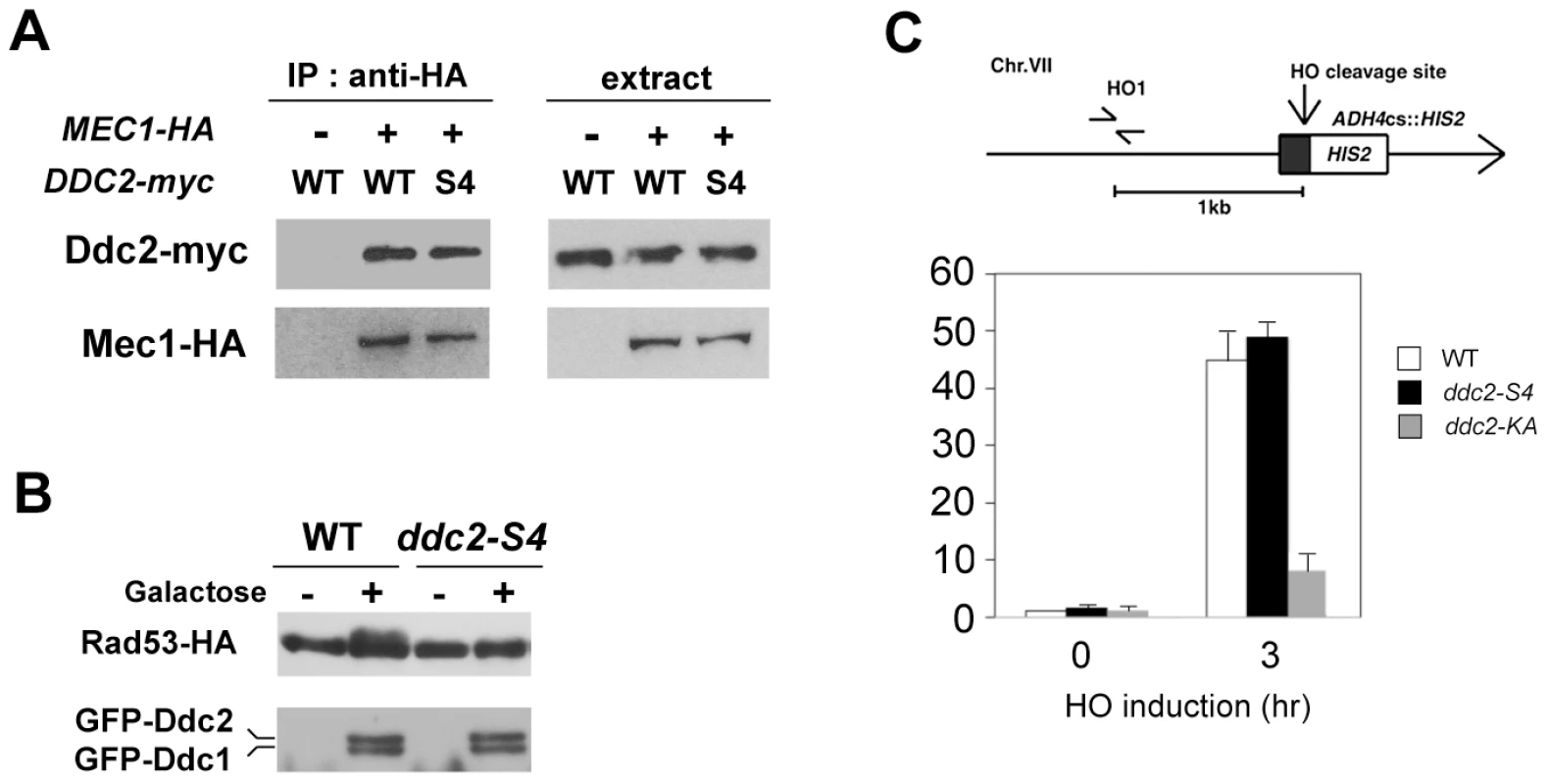
(A) Effect of ddc2-S4 mutation on Mec1-Ddc2 interaction. MEC1-HA ddc2Δ (KSC1340) or ddc2Δ (KSC1234) cells were transformed with YCp-DDC2-myc or YCp-DDC2-S4-myc. Extracts prepared from cells were subjected to immunoprecipitation with anti-HA antibodies. Immunoprecipitates and whole extracts were analyzed by immunoblotting with anti-HA or anti-myc antibodies. (B) Effect of ddc2-S4 or ddc2-KA mutation on Mec1 localization to an HO-induced DSB. Wild-type (KSC1635), ddc2-S4 (KSC2158) or ddc2-KA (KSC2159) cells expressing Mec1-HA were transformed with the YCpA-GAL-HO plasmid. Transformed cells were grown in sucrose and treated with nocodazole. After arrest at G2/M, the culture was incubated with galactose for 3 hr to induce HO expression, while half of the culture was maintained in sucrose to repress HO expression. (Top) The strains contain an HO cleavage site, marked with HIS2, at the ADH4 locus on chromosome (Chr.) VII. The HO1 primer pair amplifies a region 1 kb away from the HO cleavage site. An arrow represents the telomere. (Bottom) Cells were subjected to chromatin immunoprecipitation with anti-HA antibodies. Association of Mec1 with an HO-induced DSB was analyzed by real-time PCR. Relative enrichment was determined from three independent experiments. (C) Effect of ddc2-S4 on Rad53 phosphorylation after co-localization of Ddc1-LacI and Ddc2-LacI to a LacO array. Cells containing the LacO256 array and RAD53-HA with the combination of DDC1-LacI-GFP and DDC2-LacI-GFP (CBY88) or DDC2-S4-LacI-GFP (KSC2419) under the control of GAL promoter were grown in sucrose and arrested in nocodazole for 2 hr. Galactose was pulsed for 2 hr, and then cells were further incubated with glucose. Samples were collected at 2 hr after addition of glucose and subjected to immunoblotting analysis with anti-HA or anti-GFP antibodies. We next compared Mec1 association with HO-induced DSBs in wild-type and ddc2-S4 mutants by chromatin immunoprecipitation (ChIP) assay (Fig. 4B). In budding yeast, HO endonuclease introduces a sequence-specific DSB. We used an experimental system in which cells contain a single HO cleavage site at the ADH4 locus, and HO is expressed from the GAL-HO plasmid after incubation with galactose [11]. Cells expressing Mec1-HA were transformed with the GAL-HO plasmid. Transformed cells were grown initially in sucrose to repress HO expression, and then transferred to medium containing nocodazole to arrest at G2/M. After arrest, galactose was added to induce HO expression. Cells were then subjected to the ChIP assay. Mec1 associated with HO-induced DSBs in ddc2-S4 mutants as efficiently as in wild-type cells. These results show that the ddc2-S4 mutation does not impair Mec1 localization to sites of DNA damage. We previously showed that the substitution mutation (K177A, K178A) within the KKRK sequence (amino acids 177–180), ddc2-KA, confers DNA damage sensitivity similar to the ddc2Δ mutation but does not affect Mec1-Ddc2 complex formation [7]. We wondered whether the ddc2-KA mutation behaves like the ddc2-S4 mutation. However, association of Mec1 with DSBs was significantly decreased in ddc2-KA cells compared with that in wild-type cells (Fig. 4B). Thus, the basic amino acid residues at the middle region of Ddc2 are required for efficient Mec1 localization to sites of DNA damage.
Tethering of multiple Ddc1 and Ddc2 proteins on chromatin stimulates Mec1-mediated checkpoint activation in the absence of DNA damage [31]. In this system an array of lac operator repeats (LacO256) was integrated into the genome and Ddc1 and Ddc2 were fused to lac repressor (LacI). Therefore, Ddc1 - or Ddc2-LacI can be co-localized on chromatin through DNA binding activity of LacI. In turn, Ddc2-LacI can recruit Mec1 to the LacO repeat. We used this artificial tethering system to determine whether the ddc2-S4 mutation is defective in activation of the Mec1-dependent signaling. Co-expression of Ddc1-LacI and Ddc2-LacI promoted Rad53 phosphorylation as found previously (Fig. 4C). By contrast, Rad53 phosphorylation was significantly decreased in cells expressing Ddc1-LacI and Ddc2-S4-LacI (Fig. 4C). These results are consistent with the idea that the ddc2-S4 mutation causes defects in checkpoint activation but not in Mec1 localization.
Effect of ddc2-S4 mutation on Mec1 activation after DNA damage
We examined the effect of ddc2-S4 mutation on Mec1 kinase activity (Fig. 5A). Wild-type and ddc2-S4 cells expressing Mec1-HA protein were arrested with nocodazole at G2/M and then treated with MMS. The ddc2-S4 mutation did not affect basal Mec1 kinase activity but ablated the increase in kinase activity after MMS treatment.
Fig. 5. Effect of ddc2-S4 on Mec1 activation after DNA damage. 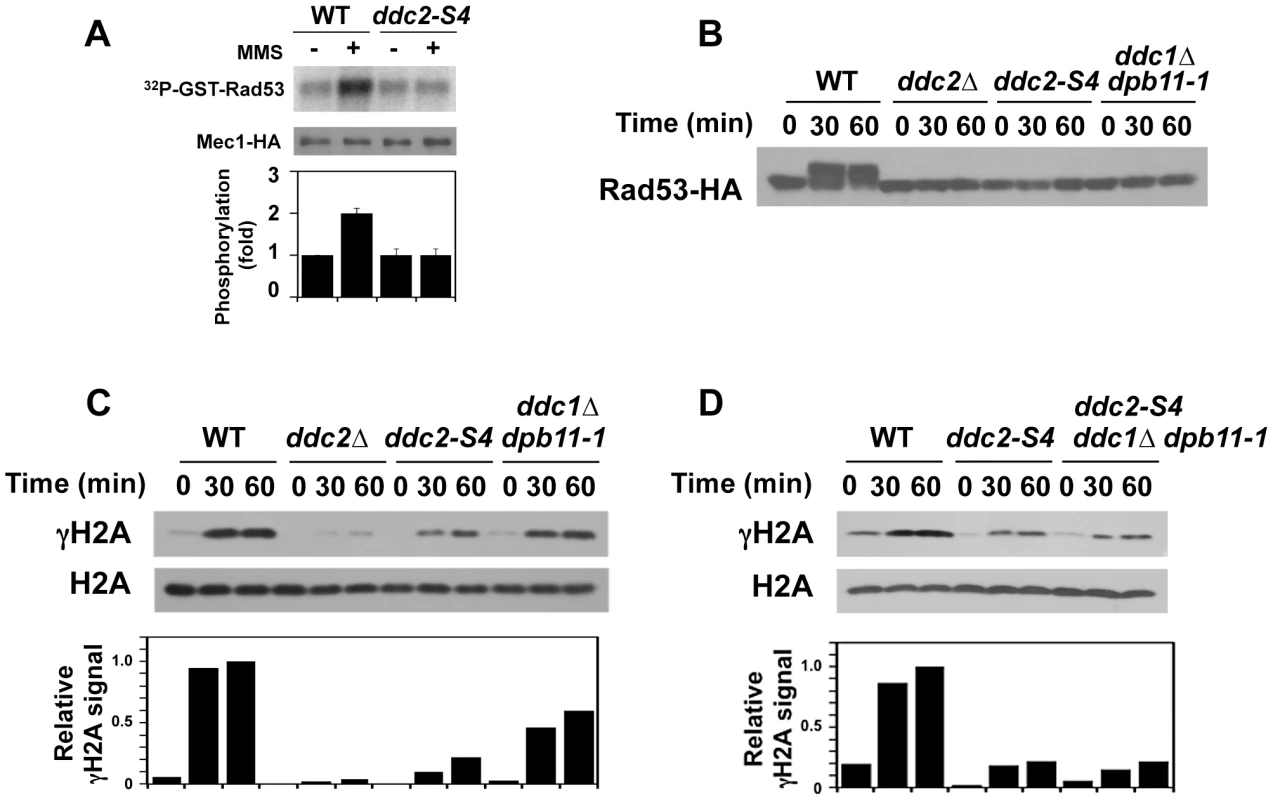
(A) Effect of ddc2-S4 mutation on Mec1 activation. Wild-type (KSC1635) and ddc2-S4 (KSC2158) cells expressing Mec1-HA protein were subjected to the in vitro kinase assay as in Fig. 1. (B) Effect of ddc2-S4 or ddc1Δ dpb11-1 mutation on Rad53 phosphorylation at G2/M. Wild-type (KSC1178), ddc2Δ (KSC1234), ddc2-S4 (KSC3153) or ddc1Δ dpb11-1 (KSC3190) cells expressing Rad53-HA were grown at 25°C and arrested with nocodazole at G2/M. Cells were then exposed to 0.05% MMS. Cells were collected at the indicated time point, and extracts were subjected to immunoblotting analysis with anti-HA antibodies. (C) Effect of ddc2-S4 or ddc1Δ dpb11-1 mutation on histone H2A phosphorylation at G2/M. The same extracts used in B were analyzed by immunoblotting with anti-phospho S129 (γH2A) or anti-H2A antibodies. (D) Histone H2A phosphorylation in ddc2-S4 single and ddc2-S4 ddc1Δ dpb11-1 triple mutants at G2/M. Wild-type (KSC1178), ddc2-S4 (KSC3153) or ddc2-S4 ddc1Δ dpb11-1 (KSC3244) cells were grown at 25°C and arrested with nocodazole at G2/M. Cells were then exposed to 0.05% MMS. Cells were collected at the indicated time point, and extracts were analyzed as in C. As shown above, Mec1 activation occurs even in ddc1Δ dpb11-1 mutants at G2/M (see Fig. 1B). To further dissect Mec1 activation mechanisms, we monitored damage-induced phosphorylation of H2A and Rad53 in wild-type, ddc2Δ, ddc2-S4 and ddc1Δ dpb11-1 mutant cells at G2/M (Fig. 5B and 5C). We note that the sml1Δ mutation does not affect the cell proliferation of ddc1Δ dpb11-1 mutant cells (Fig. S4). Cells expressing Rad53-HA were arrested at G2/M and then treated with MMS. Extracts from cells were subjected to immunoblotting analysis with anti-HA, anti-phospho H2A or anti-H2A antibodies. Rad53 was phosphorylated in wild-type cells after MMS treatment while phosphorylation was markedly decreased in ddc2-S4 mutants as found in ddc2Δ or ddc1Δ dpb11-1 mutants. Thus, ddc2-S4 mutants are as defective in Rad53 phosphorylation as ddc2Δ or ddc1Δ dpb11-1 mutants at G2/M. Histone H2A is phosphorylated after MMS treatment and its phosphorylation largely depends on Mec1 function [12]. Consistently, histone H2A was robustly phosphorylated in wild-type cells after MMS treatment but only faint phosphorylation was observed in ddc2Δ mutants. Phosphorylation was reduced in ddc1Δ dpb11-1 mutants compared with that in wild-type cells [36], supporting the current view that Ddc1 and Dpb11 activate Mec1 [34], [35]. As discussed above, Mec1 localizes to sites of DNA damage in ddc2-S4 mutants (see Fig. 4B) but not in ddc2Δ mutants [11]. Accordingly, histone H2A was phosphorylated more extensively after MMS treatment in ddc2-S4 mutants than in ddc2Δmutants. However, the level of H2A phosphorylation in ddc2-S4 mutants was much lower than that in ddc1Δ dpb11-1 mutants. Ddc1 or Dpb11 could activate Mec1 at sites of DNA damage in ddc2-S4 mutants. We therefore examined the effect of ddc1Δ dpb11-1 mutation on H2A phosphorylation in ddc2-S4 mutants after MMS treatment (Fig. 5D). The ddc2-S4 ddc1Δ dpb11-1 triple mutation did not further decrease H2A phosphorylation compared to the ddc2-S4 single mutation. Thus, the ddc2-S4 mutation causes more significant defects in Mec1 activation than the ddc1Δ dpb11-1 mutation. These results support the idea that Ddc2 mediates Mec1 activation by a different mechanism than Ddc1 or Dpb11.
Discussion
Previous studies have established that Ddc1 and Dpb11 directly activate Mec1 and play a key role in checkpoint activation at G1 and G2/M [34]–[36], [38]–[40]. In this study, we provided evidence suggesting that Ddc2 promotes Mec1 activation after DNA damage through a Ddc1 - or Dpb11-independent mechanism at G2/M. Damage-induced Mec1 activation occurs even in ddc1Δ dpb11-1 double mutants, but the ddc2Δ mutation abolishes Mec1 activation. Ddc2 recruits Mec1 to sites of DNA damage [8]–[11]. To gain more insight into the mechanism, we isolated and characterized the ddc2-S4 mutation that confers defects in Mec1 activation but does not affect Mec1 recruitment. While Rad53 phosphorylation is similarly decreased in ddc2-S4 and ddc1Δ dpb11-1 mutants at G2/M, ddc2-S4 mutants are more defective in phosphorylation of histone H2A than ddc1Δ dpb11-1 mutants. These findings are consistent with the view that Ddc2 plays a pivotal role in not only Mec1 recruitment but also Mec1 activation after DNA damage.
We have shown that the middle and C-terminal regions of Ddc2 is critical for DNA damage recognition, Mec1-Ddc1 interaction and Mec1 activation. The results from the modified two-hybrid assay indicated that the middle and C-terminal regions are required for Mec1-Ddc2 interaction and proper DNA damage responses. Characterization of the ddc2-S4 mutation, consisting of two mutations (K263E and H382Y), revealed that these regions are involved in Mec1 activation as well. Similarly, the middle and C-terminal regions of human and Xenopus ATRIP are involved in DNA damage recognition, ATR-ATRIP complex formation and checkpoint activation [45]–[49]. Thus, although not structurally conserved, the middle and C-terminal regions of ATRIP family proteins share crucial functions. The middle region of ATRIP or Ddc2 also mediates TopBP1 or Dpb11 interaction [38], [49]. It is not determined, however, whether Ddc1 or Dna2 activates Mec1 by interacting with the middle region of Ddc2. The N-terminal region consists of the checkpoint protein recruitment domain (CRD) and the coiled-coil (CC) domain [46], [50]–[52]. Previous studies have shown that the ATRIP/Ddc2 CRD mediates efficient interaction of ATR/Mec1 with RPA [52], while other studies have found that the N-terminus of ATRIP is important but not essential for the interaction with RPA-coated ssDNA [46], [48]. The CRD deletion mutation in DDC2 was found to impede Mec1 localization and sensitize cells to DNA damaging agents [52]. We found that the N-terminal region of Ddc2 is dispensable for proper DNA damage responses; N-terminal truncated Ddc2-ΔN2 (141–747) protein, expressed from its own promoter, also restored DNA damage resistance to ddc2Δ mutants (data not shown). Our results, however, do not exclude the possibility that the N-terminus of Ddc2 contributes to Mec1 localization to sites of DNA damage. Alternatively, we found that the ddc2-KA mutation at the middle region confers defects in Mec1 localization to DNA lesions although the ddc2-KA mutation does not disrupt Mec1-Ddc2 interaction [7]. Thus, multiple regions in ATRIP/Ddc2 appear to control ATR/Mec1 localization to sites of DNA damage. Studies using human cells identified that the CC domain is important for the cellular response to DNA damage or replication stress [50], [51]. We showed that the CC domain of Ddc2 is dispensable for DNA damage responses. In Xenopus, the CC domain of ATRIP is dispensable for Chk1 activation as well [46]. Thus, the CC domain of ATRIP family proteins is structurally similar but not functionally conserved.
We have provided evidence indicating that Ddc2 promotes Mec1 activation through a Ddc1 - or Dpb11-independent mechanism. Previous studies have established that Ddc1 and Dpb11 have crucial functions in G1 - and G2/M-DNA damage checkpoints by increasing the catalytic activity of Mec1 [34]–[36], [38], [39]. Thus, several different mechanisms increase the catalytic activity of Mec1 in response to DNA damage. Our in vitro kinase assay using immunoprecipitated Mec1 protein was not able to detect Ddc1 - or Dpb11-dependent Mec1 activation. One explanation is that Mec1 does not interact stably with Ddc1 or Dpb11 after immunoprecipitation. Previous studies show that the addition of purified Dpb11 protein increases the catalytic activity of immunoprecipitated Mec1 [38], [40]. Co-immunoprecipitation of Ddc1 with Mec1 has not been reported so far.
Activation of the downstream kinase Rad53 requires its interaction with Rad9 at sites of DNA damage [20], [21]. Mec1 phosphorylates histone H2A to promote Rad9 localization to sites of DNA damage [12], [14], [15]. Histone H2A is phosphorylated more strongly in ddc1Δ dpb11-1 mutants than in ddc2-S4 mutants whereas Rad53 phosphorylation is similarly defective in ddc1Δ dpb11-1 and ddc2-S4 mutants. These observations are consistent with the current model in which Ddc1 and Dpb11 are also involved in checkpoint activation events that operate downstream of Mec1 activation. Previous studies have shown that the Ddc1-Mec3-Rad17 complex contributes to Rad9 recruitment [53] and Dpb11 binds Rad9 to positively regulate Rad53 activation [40], [54]. Introduction of the ddc1Δ dpb11-1 double mutation does not further decrease histone H2A phosphorylation in ddc2-S4 mutants. One likely explanation is that Ddc2 and Ddc1/Dpb11 stimulate Mec1 sequentially; that is, Ddc2-dependent activation precedes Ddc1 - and Dpb11-dependent activation although we cannot exclude the possibility that Ddc2-S4 protein is defective for the interaction with Ddc1 and Dpb11. Recent studies show that human ATR undergoes auto-phosphorylation at damage sites and thereby promotes ATR-TopBP1 interaction [55]. Notably, ATR autophosphorylation depends on ATRIP but not on TopBP1 function [55]. As Ddc2 mediates Mec1 activation in a Ddc1 - or Dpb11-independent manner, ATRIP might stimulate ATR independently of TopBP1 at sites of DNA damage in human cells.
Tethering of Ddc2 and Ddc1 protein on chromosomal DNA leads to Rad53 phosphorylation in the absence of DNA damage [31], but the ddc2-S4 mutation was found to abolish Rad53 phosphorylation. Co-localization of Mec1 and Mrc1 can trigger checkpoint activation in the absence of DNA damage as well, but this Mec1 - and Mrc1-mediated checkpoint activation is independent of both Ddc1 and Dpb11 function [56]. Given that the ddc2-S4 mutation is defective in Mec1 activation, these results are consistent with the view that the congregation of Mec1-Ddc2 complexes turns on the Ddc2 function that increases the catalytic activity of Mec1. Since the tethering system triggers checkpoint responses in the absence of DNA damage [31] [56], the interaction of the Mec1-Ddc2 complex with RPA might be dispensable for Mec1 activation. However, tethered Mec1-Ddc2 protein could contact with chromosomal DNA. Studies using Xenopus extracts have suggested that DNA substrates with specific structures can activate ATR-ATRIP in the absence of RPA [46]. Further experiments will be aimed at elucidating the more precise biochemical properties of the Mec1-Ddc2 complex in the presence or absence of RPA and DNA.
Materials and Methods
Plasmids and strains
DNA fragments containing the DDC2 gene were amplified by the primer pair (KS437 and KS1603, KS437 and KS1604, or KS437 and KS1605), digested with BamHI and SalI, and cloned into pGAD-C1 [57], generating pGAD-DDC2-ΔN1, pGAD-DDC2-ΔN2 or pGAD-DDC2-ΔN3, respectively. The pGAD-DDC2-ΔC plasmid was constructed as follows. The ddc2-ΔC (pie1-ΔC) mutation [7] was amplified by PCR, digested with BamHI and SalI and cloned into pGAD-C1, generating pGAD-DDC2-ΔC. PIE1 is an alias of the DDC2 gene [7]. The RFA1 gene was amplified by PCR using the primer pair (KS882 and KS883), digested with BamHI and SalI and cloned into pGBD-C1 [57], creating pGBD-RFA1. To generate YCp-DDC2-myc, an EcoRI-SalI fragment of YCpT-DDC2-myc [7] was cloned in YCplac33 [58]. The pGBD-MEC1(2-2368), pGAD-DDC2, and YCpT-RAD53-HA plasmids were described before [7], [11]. The isolated ddc2 mutations were transferred from pGAD-DDC2 to YCp-DDC2-myc as follows. Each mutation was amplified by PCR using the oligonucleotide pair KS1603 and KSX001. The resulting PCR fragments were treated with XbaI and SalI, cloned into XbaI-SalI-digested YCp-DDC2-myc. YCp-DDC2-S4-myc is a version containing the ddc2-S4 mutation. The SphI-SpeI fragment from the ddc2-S4 allele was introduced into pJAM150 [31], YCp-DDC2 [7] or YCpT-DDC2 [7], resulting in pGFP-DDC2-S4-LacI, YCp-DDC2-S4 or YCpT-DDC2-S4, respectively. The GFP-DDC2-S4-LacI cassette was introduced into the DDC2 locus after digestion with NheI. The ddc2-S4 or ddc2-KA (pie1-KA) mutation [7] was fused to the URA3 marker by PCR using the KS2047 and KS2048 oligonucleotides [59] and integrated into the DDC2 locus. The ddc1Δ mutation has been described [24]. Deletion of DDC2 or SML1 was generated by a PCR-based method [60], [61]. The dpb11-1 mutation [43] was obtained from Dr. H. Araki (National Institute of Genetics, Mishima, Japan). All of the strains and oligonucleotides used in this study are listed in Tables 1 and 2, respectively.
Tab. 1. Strains used in this study. 
All the KSC strains, except for KSC2077 or KSC2419, are isogenic and derived from KSC006 (MATa ade1 his2 leu2 trp1 ura3) [7]. KSC2419 or KSC2077is derived from CBY51 or PJ69-4A, respectively. PJ69-4A has been described [7]. CBY51 and CBY88 have been obtained from David Toczyski [31]. MATa-inc is a mutation of the HO cleavage site [10]. Tab. 2. List of oligonucleotides used in this study. 
Two-hybrid screening
DNA fragments encompassing the middle and C-terminal regions of Ddc2 (amino acid141 to 747) were amplified by error prone PCR in the presence of 0.2 mM MnCl2. The resulting PCR products were introduced with XbaI-SalI digested pGAD-DDC2 into the two-hybrid tester ddc2Δ sml1Δ strain (KSC2077) transformed with pBD-MEC1(2-2368). Transformants were plated on selection medium containing 1 mM 3-aminotriazole (AT) and allowed to grow for 3 days. Colonies on AT were then replica-plated on medium containing 1 mg/ml of HU. Plasmids were recovered from the colonies that did not grow on medium containing HU and retested. Out of ∼10,000 transformants, ten plasmids were found to support proliferation on medium containing AT but not in the presence of HU. Those plasmids were further introduced into the PJ69-4A strain [57] carrying pBD-RFA1. Eight plasmids were found to support Ddc2-Rfa1 interaction. The ddc2-S4 mutation expressed from the own promoter still conferred notable sensitivity to HU and MMS.
Mec1 kinase assay
Cells were grown at 25°C and arrested with nocodazole for 3 hr. Cells were then treated with 0.05% MMS for different length of times. One hundred OD600 cells expressing HA-tagged Mec1 protein were harvested and disrupted in the lysis buffer [20 mM HEPES-KOH pH 7.5, 100 mM NaCl, 0.1% TritonX-100, 1 mM EDTA pH 8.0] containing 15 mM PNPP, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 1 mM sodium orthovanadate, 1 mM PMSF by bead beating with Multi-beads shocker (Yasui Kikai, Osaka, Japan) at 4°C. HA-tagged Mec1 was immunoprecipitated as described previously [61]. Kinase reactions were initiated by addition of 1 µg of gluthatione S-transferase (GST)-Rad53 to immunoprecipitates in 40 µl of the reaction buffer (20 mM Hepes-KOH [pH 7.5], 10 mM MgCl2, 4 mM MnCl2, 50 µM ATP) containing 5 µCi [γ-32P] ATP (3,000 Ci/mmol). After 10 min of incubation at 30°C, the reaction was terminated by the addition of 10 µl of 5× loading dye [61]. The reaction mixtures were separated on SDS-polyacrylamide gels, and phosphorylation was quantified with a phosphorimager system (Typhoon 8600, GE Healthcare).
Checkpoint activation by tethering of Ddc1 and Ddc2 proteins
Cells were treated with some modifications according to the method described previously [31]. LacO256 cells carrying Rad53-HA, GFP-Ddc1-LacI and GFP-Ddc2-LacI or GFP-Ddc2-S4-LacI were grown in 2% sucrose and 0.05% glucose. Cells were arrested by the incubation with nocodazole (final concentration 15 µg/ml) for 2 hr. Galactose (final concentration 2%) was added to one half culture to induce transcription of the LacI fusions for 2 hr. Cells were then incubated with glucose (final concentration 2%) to shut off transcription for 2 hr. After incubation with glucose, cells were harvested for immunoblotting analysis. All the cultures were incubated at 30°C.
Other methods
Chromatin immunoprecipitation assay was carried out as described [11], [61]. The DNA damage sensitivity assay was determined as described [61]. Immunoblotting was described previously [61]. UV and MMS synchrony experiments were carried out as described previously [7], [27]. Plasmid shuffle with 5-fluoroorotic acid (5-FOA) was performed as described previously [62], [63]. Rabbit anti-GFP antibody (ab290) and anti-phospho histone H2A antibody (ab15083) were purchased from Abcam (Cambridge, MA). Rabbit anti-H2A antibody (39235) was obtained from Active Motif (Carlsbad, CA).
Supporting Information
Zdroje
1. ElledgeSJ (1996) Cell cycle checkpoints: preventing an identity crisis. Science 274 : 1664–1672.
2. HarperJW, ElledgeSJ (2007) The DNA damage response: ten years after. Mol Cell 28 : 739–745.
3. CimprichKA, CortezD (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9 : 616–627.
4. HarrisonJC, HaberJE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40 : 209–235.
5. PaciottiV, ClericiM, LucchiniG, LongheseMP (2000) The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev 14 : 2046–2059.
6. RouseJ, JacksonSP (2000) LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J 19 : 5793–5800.
7. WakayamaT, KondoT, AndoS, MatsumotoK, SugimotoK (2001) Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol 21 : 755–764.
8. RouseJ, JacksonSP (2002) Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell 9 : 857–869.
9. ZouL, ElledgeSJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300 : 1542–1548.
10. NakadaD, HiranoY, SugimotoK (2004) Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol Cell Biol 24 : 10016–10025.
11. NakadaD, HiranoY, TanakaY, SugimotoK (2005) Role of the C terminus of mec1 checkpoint kinase in its localization to sites of DNA damage. Mol Biol Cell 16 : 5227–5235.
12. DownsJA, LowndesNF, JacksonSP (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408 : 1001–1004.
13. van AttikumH, GasserSM (2005) The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol 6 : 757–765.
14. GiannattasioM, LazzaroF, PlevaniP, Muzi-FalconiM (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280 : 9879–9886.
15. TohGW, O'ShaughnessyAM, JimenoS, DobbieIM, GrenonM, et al. (2006) Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 5 : 693–703.
16. EmiliA (1998) MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell 2 : 183–189.
17. VialardJE, GilbertCS, GreenCM, LowndesNF (1998) The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J 17 : 5679–5688.
18. SunZ, HsiaoJ, FayDS, SternDF (1998) Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281 : 272–274.
19. DurocherD, HenckelJ, FershtAR, JacksonSP (1999) The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4 : 387–394.
20. SchwartzMF, DuongJK, SunZ, MorrowJS, PradhanD, et al. (2002) Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell 9 : 1055–1065.
21. GilbertCS, GreenCM, LowndesNF (2001) Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell 8 : 129–136.
22. SmolkaMB, AlbuquerqueCP, ChenSH, SchmidtKH, WeiXX, et al. (2005) Dynamic changes in protein-protein interaction and protein phosphorylation probed with amine-reactive isotope tag. Mol Cell Proteomics 4 : 1358–1369.
23. SweeneyFD, YangF, ChiA, ShabanowitzJ, HuntDF, et al. (2005) Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 15 : 1364–1375.
24. KondoT, MatsumotoK, SugimotoK (1999) Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol 19 : 1136–1143.
25. MajkaJ, BurgersPM (2003) Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA 100 : 2249–2254.
26. GreenCM, Erdjument-BromageH, TempstP, LowndesNF (2000) A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol 10 : 39–42.
27. NaikiT, ShimomuraT, KondoT, MatsumotoK, SugimotoK (2000) Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol 20 : 5888–5896.
28. MajkaJ, BinzSK, WoldMS, BurgersPM (2006) Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem 281 : 27855–27861.
29. KondoT, WakayamaT, NaikiT, MatsumotoK, SugimotoK (2001) Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 5543 : 867–870.
30. MeloJA, CohenJ, ToczyskiDP (2001) Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev 21 : 2809–2821.
31. BonillaCY, MeloJA, ToczyskiDP (2008) Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell 30 : 267–276.
32. PudduF, GranataM, Di NolaL, BalestriniA, PiergiovanniG, et al. (2008) Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 28 : 4782–4793.
33. LongheseMP, PaciottiV, FraschiniR, ZaccariniR, PlevaniP, et al. (1997) The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J 16 : 5216–5226.
34. Navadgi-PatilVM, BurgersPM (2009) The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell 36 : 743–753.
35. Navadgi-PatilVM, KumarS, BurgersPM (2011) The unstructured C-terminal tail of yeast Dpb11 (human TopBP1) protein is dispensable for DNA replication and the S phase checkpoint but required for the G2/M checkpoint. J Biol Chem 286 : 40999–41007.
36. PudduF, PiergiovanniG, PlevaniP, Muzi-FalconiM (2011) Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase epsilon. PLoS Genet 7: e1002022.
37. MajkaJ, Niedziela-MajkaA, BurgersPM (2006) The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24 : 891–901.
38. MordesDA, NamEA, CortezD (2008) Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A 105 : 18730–18734.
39. Navadgi-PatilVM, BurgersPM (2008) Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283 : 35853–35859.
40. PfanderB, DiffleyJF (2011) Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J 30 : 4897–4907.
41. KumarS, BurgersPM (2013) Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev 27 : 313–321.
42. NakadaD, ShimomuraT, MatsumotoK, SugimotoK (2003) The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res 31 : 1715–1724.
43. ArakiH, LeemSH, PhongdaraA, SuginoA (1995) Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci U S A 92 : 11791–11795.
44. WoldMS (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66 : 61–92.
45. CortezD, GuntukuS, QinJ, ElledgeSJ (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294 : 1713–1716.
46. KimSM, KumagaiA, LeeJ, DunphyWG (2005) Phosphorylation of Chk1 by ATM - and Rad3-related (ATR) in Xenopus egg extracts requires binding of ATRIP to ATR but not the stable DNA-binding or coiled-coil domains of ATRIP. J Biol Chem 280 : 38355–38364.
47. BallHL, MyersJS, CortezD (2005) ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 16 : 2372–2381.
48. NamikiY, ZouL (2006) ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci U S A 103 : 580–585.
49. MordesDA, GlickGG, ZhaoR, CortezD (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22 : 1478–1489.
50. ItakuraE, TakaiKK, UmedaK, KimuraM, OhsumiM, et al. (2004) Amino-terminal domain of ATRIP contributes to intranuclear relocation of the ATR-ATRIP complex following DNA damage. FEBS Lett 577 : 289–293.
51. BallHL, CortezD (2005) ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem 280 : 31390–31396.
52. BallHL, EhrhardtMR, MordesDA, GlickGG, ChazinWJ, et al. (2007) Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol 27 : 3367–3377.
53. NaikiT, WakayamaT, NakadaD, MatsumotoK, SugimotoK (2004) Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism. Mol Cell Biol 24 : 3277–3285.
54. GranataM, LazzaroF, NovarinaD, PanigadaD, PudduF, et al. (2010) Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet 6: e1001047.
55. LiuS, ShiotaniB, LahiriM, MarechalA, TseA, et al. (2011) ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell 43 : 192–202.
56. BerensTJ, ToczyskiDP (2012) Colocalization of Mec1 and Mrc1 is sufficient for Rad53 phosphorylation in vivo. Mol Biol Cell 23 : 1058–1067.
57. JamesP, HalladayJ, CraigEA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 : 1425–1436.
58. GietzRD, SuginoA (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74 : 527–534.
59. ReidRJ, LisbyM, RothsteinR (2002) Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol 350 : 258–277.
60. WachA, BrachatA, PohlmannR, PhilippsenP (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 13 : 1793–1808.
61. FukunagaK, KwonY, SungP, SugimotoK (2011) Activation of Protein Kinase Tel1 through Recognition of Protein-Bound DNA Ends. Mol Cell Biol 31 : 1959–1971.
62. BoekeJD, TrueheartJ, NatsoulisG, FinkGR (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154 : 164–175.
63. FukunagaK, HiranoY, SugimotoK (2012) Subtelomere-binding protein Tbf1 and telomere-binding protein Rap1 collaborate to inhibit localization of the Mre11 complex to DNA ends in budding yeast. Mol Biol Cell 23 : 347–359.
Štítky
Genetika Reprodukčná medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



