-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Distinct Pathogenesis and Host Responses during Infection of by and
The genetically tractable model host Caenorhabditis elegans provides a valuable tool to dissect host-microbe interactions in vivo. Pseudomonas aeruginosa and Staphylococcus aureus utilize virulence factors involved in human disease to infect and kill C. elegans. Despite much progress, virtually nothing is known regarding the cytopathology of infection and the proximate causes of nematode death. Using light and electron microscopy, we found that P. aeruginosa infection entails intestinal distention, accumulation of an unidentified extracellular matrix and P. aeruginosa-synthesized outer membrane vesicles in the gut lumen and on the apical surface of intestinal cells, the appearance of abnormal autophagosomes inside intestinal cells, and P. aeruginosa intracellular invasion of C. elegans. Importantly, heat-killed P. aeruginosa fails to elicit a significant host response, suggesting that the C. elegans response to P. aeruginosa is activated either by heat-labile signals or pathogen-induced damage. In contrast, S. aureus infection causes enterocyte effacement, intestinal epithelium destruction, and complete degradation of internal organs. S. aureus activates a strong transcriptional response in C. elegans intestinal epithelial cells, which aids host survival during infection and shares elements with human innate responses. The C. elegans genes induced in response to S. aureus are mostly distinct from those induced by P. aeruginosa. In contrast to P. aeruginosa, heat-killed S. aureus activates a similar response as live S. aureus, which appears to be independent of the single C. elegans Toll-Like Receptor (TLR) protein. These data suggest that the host response to S. aureus is possibly mediated by pathogen-associated molecular patterns (PAMPs). Because our data suggest that neither the P. aeruginosa nor the S. aureus–triggered response requires canonical TLR signaling, they imply the existence of unidentified mechanisms for pathogen detection in C. elegans, with potentially conserved roles also in mammals.
Published in the journal: Distinct Pathogenesis and Host Responses during Infection of by and. PLoS Pathog 6(7): e32767. doi:10.1371/journal.ppat.1000982
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000982Summary
The genetically tractable model host Caenorhabditis elegans provides a valuable tool to dissect host-microbe interactions in vivo. Pseudomonas aeruginosa and Staphylococcus aureus utilize virulence factors involved in human disease to infect and kill C. elegans. Despite much progress, virtually nothing is known regarding the cytopathology of infection and the proximate causes of nematode death. Using light and electron microscopy, we found that P. aeruginosa infection entails intestinal distention, accumulation of an unidentified extracellular matrix and P. aeruginosa-synthesized outer membrane vesicles in the gut lumen and on the apical surface of intestinal cells, the appearance of abnormal autophagosomes inside intestinal cells, and P. aeruginosa intracellular invasion of C. elegans. Importantly, heat-killed P. aeruginosa fails to elicit a significant host response, suggesting that the C. elegans response to P. aeruginosa is activated either by heat-labile signals or pathogen-induced damage. In contrast, S. aureus infection causes enterocyte effacement, intestinal epithelium destruction, and complete degradation of internal organs. S. aureus activates a strong transcriptional response in C. elegans intestinal epithelial cells, which aids host survival during infection and shares elements with human innate responses. The C. elegans genes induced in response to S. aureus are mostly distinct from those induced by P. aeruginosa. In contrast to P. aeruginosa, heat-killed S. aureus activates a similar response as live S. aureus, which appears to be independent of the single C. elegans Toll-Like Receptor (TLR) protein. These data suggest that the host response to S. aureus is possibly mediated by pathogen-associated molecular patterns (PAMPs). Because our data suggest that neither the P. aeruginosa nor the S. aureus–triggered response requires canonical TLR signaling, they imply the existence of unidentified mechanisms for pathogen detection in C. elegans, with potentially conserved roles also in mammals.
Introduction
The study of host-microbe interactions seeks to understand the symbiotic relationships between hosts and microbiota, and their perversion during infectious disease. Essential steps are the identification of bacterial virulence mechanisms and of host defense pathways. In mammalian hosts, Nod-like receptors (NLRs), Toll-like receptors (TLRs), and NF-κB play important roles in the intestinal epithelium, a critical interface of contact between host and microbiota [1], [2], [3]. However, how these signaling pathways function in the context of the whole organism is poorly understood, and potentially novel pathways may yet be uncovered. Likewise, the critical initial stages of infection, before the onset of overt pathogenesis, are poorly defined.
Genetically tractable invertebrate model systems have aided efforts to identify evolutionarily conserved components of the innate immune system [4]. For example, studies using Drosophila melanogaster showed the central importance of the Toll and IMD signaling pathways for the regulation of Relish-family (NF-κB) transcription factors [3], [5]. Likewise, studies using Caenorhabditis elegans revealed the involvement of evolutionarily conserved p38 MAPK, insulin, TGF-β, and β-catenin signaling pathways [6], [7], [8]. In addition to being a genetically tractable model system, C. elegans is a transparent bacterivore, which allows the direct, real-time observation of infection and gene expression reporters in vivo. These qualities make it a useful model host for the study of infection and host defense in the context of the whole organism [9]. C. elegans is particularly useful for studying intestinal epithelial innate defenses, because it has only 20 such cells that are not shed (as are mammalian intestinal epithelia) and are non-renewable [9], [10], allowing the study of defense functions in vivo without potentially confounding cell proliferation and tissue repair. Furthermore, the unique biology of C. elegans allows researchers to focus entirely on epithelial innate defense because it lacks a circulatory system, macrophage-like professional immune phagocytes, and antibody-based adaptive immunity [11].
On the bacterial side, it is important to elucidate the virulence mechanisms that defeat host defenses and establish infection. Pathogenic bacteria are thought to have experienced stepwise additions of virulence factors, as they evolved to survive different host antimicrobial responses and to colonize new niches [12]. Our studies using C. elegans as a model host may thus interrogate early steps in the evolution of bacteria as pathogens, and their interactions with prototypical metazoan epithelial cells. Here we focus on two paradigmatic human pathogenic bacteria of great medical importance that represent two broad categories of evolutionarily distant microbes, the Gram-negative Pseudomonas aeruginosa and Gram-positive Staphylococcus aureus. P. aeruginosa causes systemic acute infections in patients with weakened immune systems [13] and establishes chronic infections in the lungs of cystic fibrosis patients [14]. P. aeruginosa can also infect a wide variety of plants, metazoans, and single-celled eukaryotes [15]. S. aureus is a Gram-positive bacterium that can cause severe diseases in many animal species [16], [17]. In recent years, patients lacking classical risk factors have suffered increasing rates of infection by virulent antibiotic-resistant strains [18]. Human colonization by S. aureus is widespread: 30% of the population carries S. aureus in the microflora of epithelia in the nasopharynx, skin, and intestine [19]. S. aureus can cause severe skin infections, osteomyelitis, endocarditis, food poisoning, pneumonia, and flesh-eating disease [20], for which it deploys an impressive armamentarium of virulence factors, including cytolysins that cause the destruction of host immune cells and tissues [21], [22]. Despite great progress in their identification, the exact contribution of each virulence strategy to disease in vivo is poorly understood. The genetic makeup of the host is suspected to determine susceptibility to infection, but the genetic determinants of susceptibility are unknown [20]. Mice lacking adaptive immunity survive intravenous S. aureus infection as well as wild-type animals, suggesting that innate immunity is the main clearing mechanism for S. aureus infection in mammals, but the exact mechanism is unclear [23]. New approaches are needed to understand the molecular basis of innate host defenses against P. aeruginosa and S. aureus infection.
To this end, our laboratory has developed C. elegans-P. aeruginosa [24], [25] and C. elegans-S. aureus [17], [26] model systems to facilitate the study of the role of innate host defenses in conferring resistance to bacterial infections and to identify host signaling pathways relevant to defense [7], [27], [28], [29]. These infection models recapitulate key aspects of P. aeruginosa or S. aureus disease in mammals (see below), including the requirement of virulence factors necessary for mammalian infection, and have been used to identify novel P. aeruginosa and S. aureus virulence factors [17], [30], [31], [32]. Despite great progress in the dissection of C. elegans host defense signaling pathways since the initial description of the system in 1999 [9], [24], [33], [34], little information has been available on the cellular basis of bacterial pathogenesis and nematode killing.
In this study, we focused on the interactions between C. elegans intestinal cells —as prototypical metazoan epithelial cells, and as the first line of defense against intestinal infection—and P. aeruginosa or S. aureus. We investigated the cytopathologies that occur during infection, which suggest distinct mechanisms of virulence used by each bacterial species in vivo. With P. aeruginosa, we found that initial intestinal distention, putative outer membrane vesicle (OMV) production, and extracellular matrix accumulation on the intestinal cell brush border are followed by host autophagic abnormalities, intracellular invasion, and penetration of the epithelial barrier. Similarly, previous studies found that P. aeruginosa forms biofilms in the lungs of infected patients, where OMV production is also evident [35], [36]. In contrast, faster accumulation of S. aureus in the C. elegans intestine resulted in enterocyte effacement and loss of intestinal cell volume, followed by intestinal epithelial cell lysis and bacterial invasion of the rest of the body, with complete degradation of host internal tissues. Likewise, previous studies showed intestinal colonization by S. aureus, enterocyte effacement in rabbits and neonates, and toxin-mediated cell lysis both in vitro and in vivo [37], [38], [39], [40], [41], [42], [43].
We also evaluated the differential impact of these distinct pathogenic processes on host gene transcription. We previously defined the host transcriptional response to P. aeruginosa infection [44]. To understand if and how the host responds to different virulence mechanisms by employing distinct transcriptional host responses, here we defined the host response to S. aureus. The two responses show minimal overlap; the response to S. aureus apparently involves host damage - and TLR-independent recognition of microbial molecules, potentially pathogen-associated molecular patterns (PAMPs), whereas C. elegans may sense P. aeruginosa-derived heat-labile signals or pathogen-elicited damage. Using functional genomics, we identified host factors critical for host defense against S. aureus, some of which are analogous to human innate defense factors. These observations advance our knowledge of bacterial pathogenesis in C. elegans, and show that the C. elegans infection model illuminates evolutionarily conserved mechanisms of bacterial pathogenesis and epithelial host defense.
Results
P. aeruginosa infection: intestinal distention, extracellular material accumulation, intracellular invasion, outer membrane vesicles, and abnormal autophagy
To determine the cytopathology of P. aeruginosa colonization of the C. elegans intestine, we used transmission electron microscopy (TEM) of the intestinal epithelium (Figure 1A) to evaluate signs of pathogenesis at early times (8 h) or at later times (24 h and 48 h) of infection. At 8 h of infection, we found gross intestinal distention but little bacterial accumulation. Instead, we observed unidentified electron-dense extracellular material accumulating on the apical surface of the brush border (Figure 1C). In addition to coating the brush border, the electron-dense material surrounded bacterial cells that appeared intact and formed clumps in the intestinal lumen. Also surrounding the bacteria and in contact with the extracellular material, we found abundant accumulation of putative outer membrane vesicles (OMVs) (Figure S1A, B). P. aeruginosa OMVs have been shown to act as a virulence factor and toxin delivery mechanism [45]. We did not observe intestinal distention, OMVs, or matrix accumulation in E. coli-fed control animals (Figure 1B).
Fig. 1. P. aeruginosa causes intestinal distention, extracellular material accumulation, intracellular invasion, and abnormal autophagy. 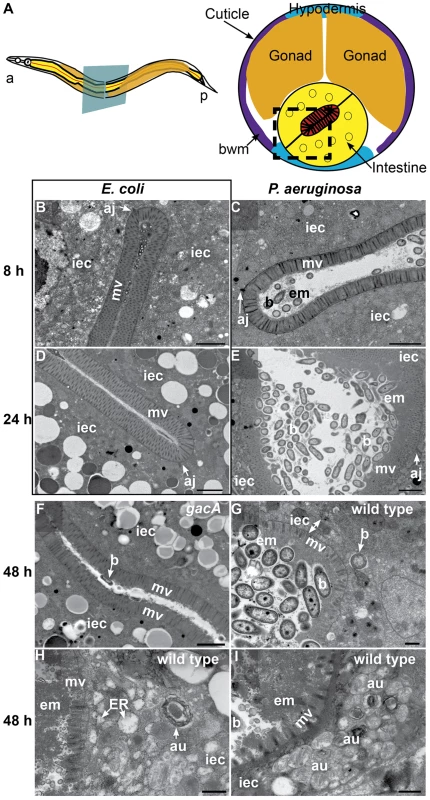
A. Schematic representation of C. elegans body plan and plane of section (Left) and midbody transversal section indicating major organs (Right). Square highlights area of focus in TEM micrographs. B–I. TEM micrographs of transversal midbody sections of animals feeding on non-pathogenic E. coli OP50(B, D), virulent P. aeruginosa PA14(C, E, G, H, I), or attenuated gacA P. aeruginosa PA14 (F) for 8 h (B, C), 24 h (D, E), or 48 h (F–I). Scale bar in B–F, 2 µm; in G–I, 0.5 µm. iec, intestinal epithelial cell; mv, microvilli; b, bacterial cell; bwm, body wall muscle; aj, apical junctions; em, extracellular material; au, arrested autophagosomes, ER, expanded endoplasmic reticulum. At 24 h of infection, the intestine became further distended, with noticeably more bacterial cells accumulating in the lumen in the form of clumps of cells surrounded by extracellular matrix (Figure 1E). There was a thick layer of matrix material coating the microvilli, which were present and of approximately normal length (i.e., ∼1 µm). In contrast, E. coli-fed animals lacked these signs of pathogenesis, exhibiting non-distended intestinal lumina and intestinal epithelial cells filled with lipid droplets and other gut granules characteristic of healthy animals (Figure 1D).
At 48 h of infection, pathogenesis advanced further, resulting in higher levels of bacterial accumulation in the grossly distended intestinal lumen (Figure 1G). The bacterial cells were mostly not in direct contact with the microvillar surface, but separated from it by a thick layer of extracellular material. At this time, there was widespread shortening of the microvilli and intracellular invasion by the bacteria (Figure 1G). Intracellular invasion was observed in 21% of cross sections (N = 14), only after 48 h infection. In some cases, we found bacterial cells at distal sites beyond the intestine, suggesting that P. aeruginosa can penetrate the intestinal cells and invade other tissues (Figure S1C). In addition to these phenotypes, we observed an increased number of autophagosomes, readily identifiable by their multi-membranous structure (Figure 1H and S2). Indeed, most autophagosomes appeared to be either early autophagosomes (Figure 1H) or aberrant multivesicular autophagosomes (Figure 1I). In contrast, mutant gacA P. aeruginosa, lacking the master virulence regulator GacA and therefore attenuated for C. elegans killing [25], caused much lower levels of autophagosome accumulation (Figure S2), pathogenesis (Figure 1F), and OMVs (Figure S1D) by 48 h. Intracellular PA14 gacA was not observed, even after 72 h (N = 15). We observed less dense matrix accumulation in gacA mutant-infected animals than with wild type P. aeruginosa, and did not observe microvillar shortening, intracellular invasion, or severe luminal distention. At all times during the infection by both wild-type and gacA P. aeruginosa, we only observed what appeared to be live P. aeruginosa cells, in contrast to S. aureus as described below.
S. aureus infection: anal deformation, intestinal distention, enterocyte effacement, and cell lysis
Interestingly, the cytopathology of S. aureus infection in the C. elegans intestine is markedly different from a P. aeruginosa infection. First, using GFP-labeled S. aureus, we observed rapid accumulation of bacteria in the intestine 4 h after infection initiation, whereas P. aeruginosa did not start accumulating until 8 after initial exposure (Figure S4A, B). At 4 h, S. aureus accumulated in the anterior and posterior ends of the intestine, and the rectum (Figure S3A, C), with less accumulation in the mid section of the intestinal lumen, where the bacteria appeared to be adhering to the apical surface of intestinal cells (Figure S3B). S. aureus accumulated further over the course of the following 4 h (Figure S4B). The infected animals moved slowly, were smaller (Figure S4C, D), and appeared to produce fewer eggs, than healthy animals.
In addition to the intestinal distention and accumulation phenotypes, we observed a marked deformation of the anal region with S. aureus (Figure S4C) but not with P. aeruginosa (not shown) or non-pathogenic E. coli (Figure S4D). This deformed anal region (Dar) phenotype [46] appeared 4–8 h after initiation of infection and required live S. aureus (Figure S4E). Interestingly, this Dar phenotype was dependent on bar-1/β-catenin and mpk-1/extracellular signal regulated kinase (ERK) (Figures S4F, G, J, K, and L), which is also required for the Dar response to Microbacterium nematophilum [47]. We previously showed that bar-1/β-catenin and its downstream target gene egl-5/HOX exhibit a defective intestinal response to S. aureus [7]. Unexpectedly, mutants defective in egl-5/HOX exhibited a wild-type Dar response (Figure S4H, L), despite having an altered intestinal host response to S. aureus [7] and a defective anal swelling response to M. nematophilum infection [48]. Similarly, pmk-1/p38 MAPK mutants exhibited a slightly less noticeable, but equally frequent, Dar phenotype following S. aureus infection (Figure S4I, L), consistent with our previous observation that pmk-1 mutants are only subtly more susceptible to S. aureus-mediated killing [7]. These data suggest that the Dar phenotype may be a defensive host swelling response to pathogen-mediated host damage, since it requires an active host response to live bacteria.
To investigate the cytopathology of S. aureus infection, we performed TEM of S. aureus-infected animals, focusing on the intestinal epithelium (Figure 2A). After 12 h of infection, we found a striking decrease in the length of the microvilli compared to animals feeding on non-pathogenic E. coli (Figures 2B–E, 3B–C). We also observed significant plasma membrane “blebbing” from the apical surface of intestinal cells (Figures 2D–E, 3C, S6A–B). The intestinal lumina of S. aureus-infected animals were markedly distended, consistent with our previous observations using light microscopy [17]. Distention was apparently a consequence of severe volume loss of the intestinal epithelial cells, with concomitant accumulation of bacterial cells in the enlarged luminal space (Figure 2B, D). In marked contrast to P. aeruginosa, an average 34% of S. aureus cells in the lumen (9–63%, N = 8 cross sections) were lysed at 12 h of infection (Figure 2E); these appeared similar to published TEM micrographs showing S. aureus cells killed with antimicrobial peptides in vitro [49], suggesting that the C. elegans intestine may produce bactericidal factors active against S. aureus.
Fig. 2. Intestinal distention, enterocyte effacement, and bacterial lysis at 12 h of S. aureus infection in C. elegans. 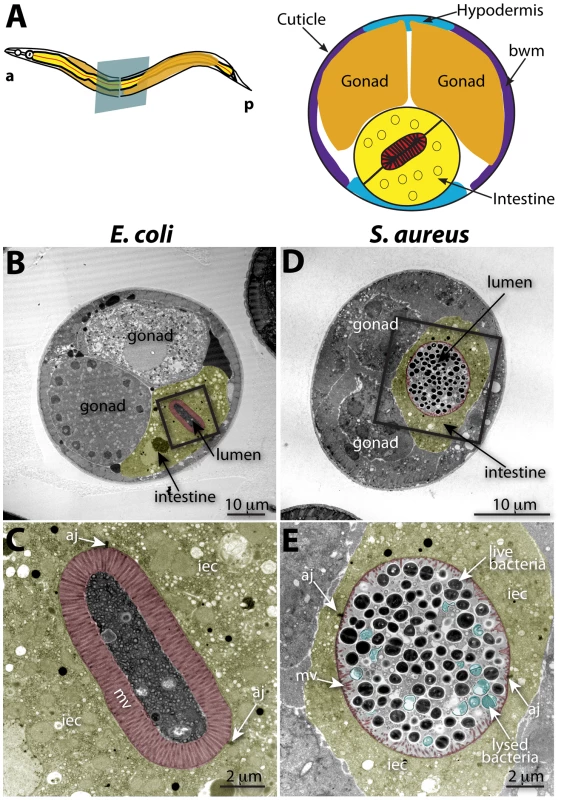
A. Schematic representation of C. elegans body plan and plane of section (Left) and midbody transversal section indicating major organs (Right). B. Midbody transversal section of a control animal fed non-pathogenic E. coli. Square indicates area magnified in C. C. Higher magnification showing healthy microvilli (mv), intestinal epithelial cells (iec), apical junctions (aj), bacteria and bacterial debris in the intestinal lumen. D. Midbody transversal section of an animal infected with S. aureus for 12 h. Square indicates area magnified in E. E. Higher magnification showing short or absent microvilli, shrunken intestinal epithelial cells, apical junctions, and live and lysed bacteria in the distented intestinal lumen. Microvilli are false-colored red from the terminal web to the apical membrane, and intestinal epithelial cells are highlighted in yellow. Lysed bacteria in E. are false-colored blue. Fig. 3. S. aureus causes intestinal cell effacement and lysis in C. elegans. 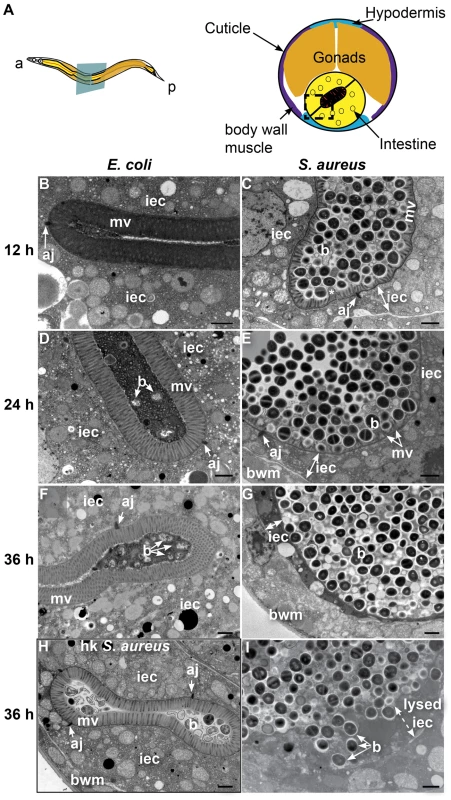
A. Schematic representation of C. elegans body plan and plane of section (Left) and midbody transversal section indicating major organs (Right). Square highlights area of focus in TEM micrographs. B–I. TEM micrographs of transversal midbody sections of animals feeding on heat-killed non-pathogenic E. coli OP50 (B, D, F), virulent S. aureus NCTC8325 (C, E, G, I), or heat-killed S. aureus (H) for 12 h (B, C), 24 h (D, E), or 36 h (F–I). Scale bar, 1 µm. iec, intestinal epithelial cell; mv, microvilli; b, bacterial cell; bwm, body wall muscle; aj, apical junctions, asterisk in C indicates membrane blebbing. In contrast to animals feeding on E. coli, at 24 h of infection the microvilli were almost completely destroyed (Figure 3D, E) and at 36 h were completely absent, in what is termed “enterocyte effacement” (Figure 3F, G, I). Further, at 36 h we observed a reduction of intestinal cell volume (see thin sliver in Figure 3G) and intestinal cell lysis (Figure 3I). Also at 36 h, we observed a few dead animals, the organs of which were completely degraded except for the collagenous cuticle and an unidentified circular internal structure (Figure S5). Heat-killed S. aureus did not cause intestinal distention, microvillar effacement, intestinal cell lysis, or death (Figure 3H). These data show that S. aureus causes membrane and cytoskeletal rearrangements, as well as enterocyte effacement and destruction, possibly by secreted membrane-active bacterial toxins such as cytolysins or other pore-forming toxins (PFTs). Hemolysins α, β, and γ are known S. aureus cytolysins. However, they appeared to not be required for pathogenesis and killing, as a S. aureus strain lacking all three hemolysins exhibited similar kinetics of C. elegans killing as the isogenic wild type (Figure S7). Similarly, the α-hemolysin Δhla mutant was as capable of causing enterocyte effacement, intestinal distention, membrane blebbing, and intestinal cell lysis as wild type (Figure S6C–F). These results indicate that virulence factors other than the hemolysins are responsible for the observed intestinal cell lysis.
S. aureus infection triggers an antimicrobial and detoxifying transcriptional host response
Because host defense from, and digestion of, ingested bacteria are necessarily linked in bacterivorous animals such as C. elegans, the distinction between innate immune responses and digestive responses is blurred. Previous studies have investigated the long-term effects of ingestion of pathogenic bacteria, defining a common necrotic host response that is triggered by several pathogens after 24 h of infection [50]. To investigate gene expression changes more likely to be elicited directly by S. aureus detection, we evaluated gene expression at an earlier infection time, namely 8 h. We previously reported that P. aeruginosa induces a potent transcriptional host response early during infection of C. elegans, which significantly contributed to our understanding of C. elegans defense from P. aeruginosa infection [44]. To determine whether C. elegans mounts a similar host response to S. aureus infection, we performed whole-genome transcriptional profiling of animals infected with S. aureus for 8 h, relative to animals feeding on non-pathogenic E. coli. We found 186 transcripts that increased at least two-fold in abundance and 198 that decreased at least two-fold after infection (Table S1). Focusing on 46 genes up-regulated 4-fold or higher as a smaller sample, we found that the majority had potential xenobiotic detoxification or antimicrobial activities, consistent with their involvement in a protective host response (Table 1). In this group, a number of genes of unknown function appeared to encode short secreted polypeptides that may possess antimicrobial activities (Table 1).
Tab. 1. C. elegans genes induced 4-fold or higher after 8 h infection with S. aureus. 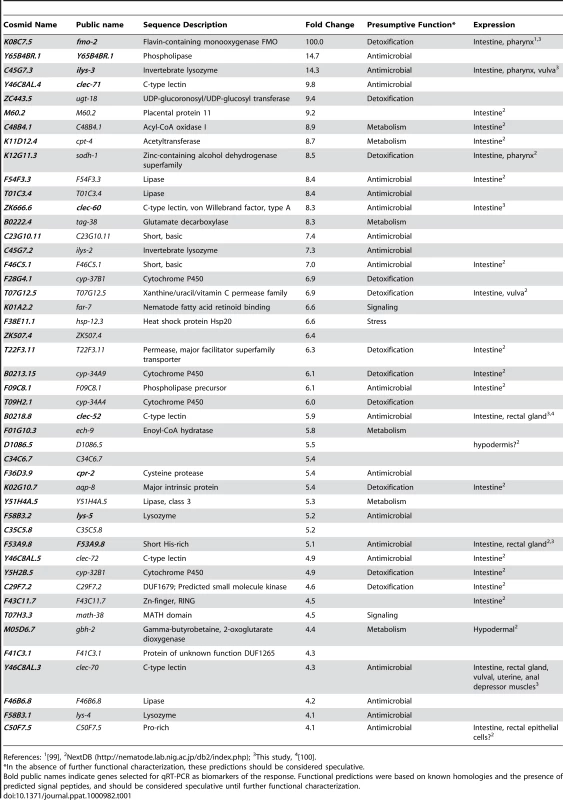
References: 1[99], To identify potential physiological roles of this host response, we used two complementary methods to study the over-representation of gene ontology (GO) classes (see Materials and Methods). These analyses revealed up-regulation of detoxifying and antimicrobial responses and down-regulation of growth-related metabolic pathways and extracellular structural components (Table S7). Significance of representation analysis revealed that the most significantly induced gene class contains sugar-binding proteins including C-type lectins (CTLs, N = 15, p = 1.2E-5), which could act as signaling receptors, opsonizing agents, or direct antimicrobial effectors [51], [52], [53], [54]. The most significantly repressed GO classes contain genes encoding structural constituents of the cuticle (e.g. collagens; N = 47, p = 4.4E-25), phosphate and inorganic anion transport (N = 47, p = 4.5E-25 and p = 5.8E-21), basement membrane components (N = 4, p = 1E-6), and lipid transporters (N = 5, p = 6E-6). A second approach (see Text S1) expanded these observations to include additional metabolic enzymes and transporters (Table S2). These analyses highlight the potential role of CTLs as a major immune effector strategy used by C. elegans during infection by S. aureus, and the significant metabolic component of the host response.
Limited overlap with other stress responses
Because we observed cell membrane rearrangements suggestive of the activity of membrane-active toxins (Figures 2E, 3C, E, G, I, S6), we hypothesized that part of the host response to S. aureus may also be triggered by PFTs. Indeed, we found that 22 of 422 probe sets up-regulated during exposure of worms to the Bacillus thuringiensis PFT Cry5B [55] were also induced during infection with S. aureus (Table S3), significantly higher than the 4 probe sets expected by chance alone. These data suggest that the overlapping set of genes shared by the responses to S. aureus and Cry5B may constitute a host response triggered by intestinal cell membrane disruption.
Because we observed evidence of intestinal destruction and nutritional deprivation in animals infected with S. aureus, we hypothesized that infected animals might be starving. Previous work identified 18 genes whose expression changed during starvation [56]. In contrast, only one out of 9 previously identified fasting-induced genes (acs-2) and 4 out of 9 fasting-repressed genes (lbp-8, acdh-1, fat-7, and F08A8.2) were affected by S. aureus infection. Furthermore, the fasting-induced gene hacd-1 [56] was repressed during infection. These data suggest that the early transcriptional host response to S. aureus infection is minimally impacted by the starvation response.
S. aureus infection triggers at least two waves of transcriptional response in the intestinal epithelium
To validate the microarray experiments, we measured transcript levels for ten selected “biomarker” genes by qRT-PCR over a time course of infection. These ten genes, used as models of the larger host response, were chosen to represent different up-regulation levels and functional annotations (Table 2). All ten genes tested were induced in response to infection (Figure 4A, B). A subset of these biomarkers, fmo-2 (FMO), ilys-3 (lysozyme), cpr-2 (protease), Y65B4BR.1 (lipase), exc-5 (GEF), and F53A9.8 (putative antimicrobial peptide), were already induced 10-fold or higher by the first time-point at 4 h (Figure 4A). A second subset, lys-5 (lysozyme), clec-52, clec-60, and clec-71 (CTLs), were only modestly induced by 4 h and exhibited a further increase by 12 h (Figure 4B). Thus, time-resolved gene expression analysis revealed the existence of at least two kinetic groups, defined by their expression levels at 4 h. We also measured transcript levels for 8 genes predicted to be repressed upon infection, confirming reduced expression for 7 of them (Figure 4C). Together, these results confirm the predictive value of the genome-wide profiling.
Fig. 4. The C. elegans host response to S. aureus infection is comprised of two kinetic groups. 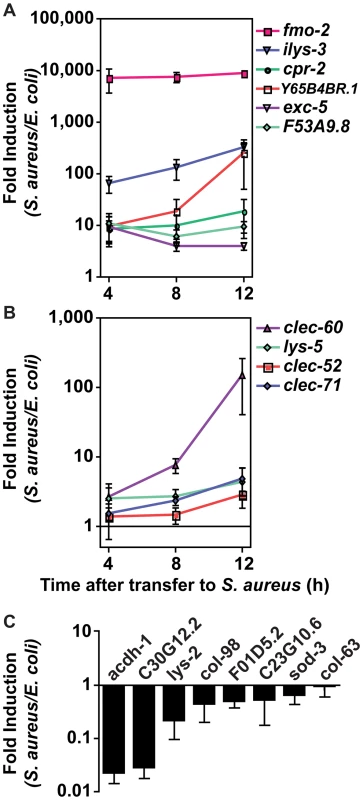
A, B. qRT-PCR analysis of genes predicted to be up-regulated by microarray analysis. Transcript levels were measured in synchronized young adult wild-type animals feeding on heat-killed E. coli OP50 or infected with S. aureus NCTC8325 for 4, 8, and 12 h. Data are the means of three biological replicates, each replicate measured in duplicate and normalized to a control gene, expressed as the ratio of the corresponding S. aureus-induced levels and the basal E. coli levels. A. Immediate-early-induced genes were highly induced by 4 h of infection. B. Later induction of early response genes. C. qRT-PCR analysis of genes predicted to be repressed by microarray analysis. Transcript levels were measured as in A, after 8 h infection with S. aureus. Data are the means of three biological replicates, each replicate measured in duplicate and normalized to a control gene, expressed as the ratio of the corresponding S. aureus-induced levels and the basal E. coli levels. Error bars are SEM. Tab. 2. Selection of 10 biomarker genes to measure the host response to S. aureus. 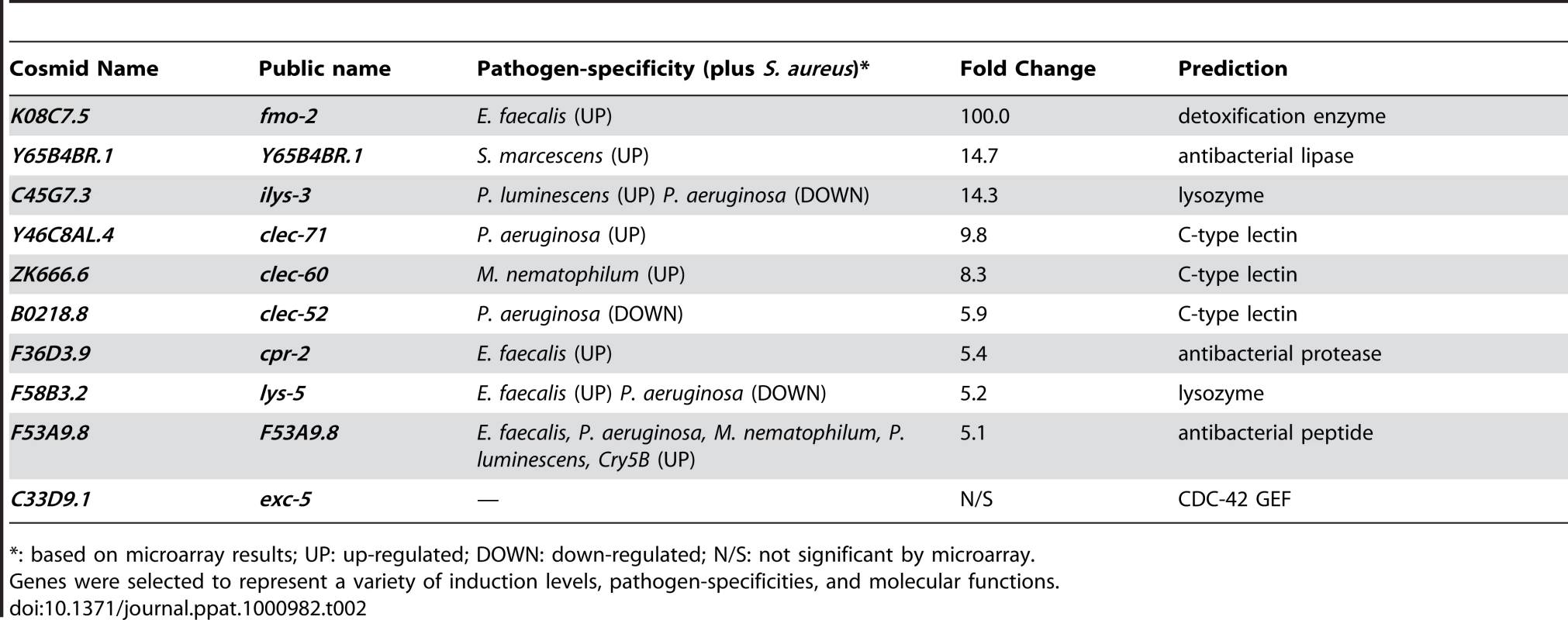
*: based on microarray results; UP: up-regulated; DOWN: down-regulated; N/S: not significant by microarray. To elucidate the spatial pattern of the host response, we used transgenic animals carrying transcriptional reporters in which the promoters for 5 of the 10 biomarker genes (clec-52, clec-60, F53A9.8, fmo-2, and ilys-3), as well as clec-70, an additional CTL gene up-regulated by S. aureus and important for host defense (see below), were fused to GFP. We infected these transgenic animals with S. aureus and compared the intensity and pattern of GFP expression with control animals feeding on E. coli. All the genes tested were expressed at low basal levels in the intestines of the latter (Figure 5, left panels). After infection with S. aureus, all of the GFP reporters were induced in the intestinal epithelial cells (Figure 5, right panels). ilys-3, F53A9.8, clec-52, clec-60, and clec-70 were all expressed more strongly in the posterior end of the intestine than the anterior. ilys-3, fmo-2, and clec-70 were also induced in the pharynx. Although promoter-GFP fusions like these lack potentially important endogenous regulatory 3′ UTR and intronic sequences, these data are consistent with endogenous RNA localization data from ongoing genome-wide in situ hybridization studies of animals feeding on non-pathogenic E. coli (NextDB, http://nematode.lab.nig.ac.jp/db2/index.php, and Table 1). Thus, the C. elegans transcriptional host response to S. aureus is primarily localized in the intestinal epithelial cells and, in some cases, additional sites (Figure S8A–H).
Fig. 5. The C. elegans host response to S. aureus is induced in the intestinal epithelial cells. 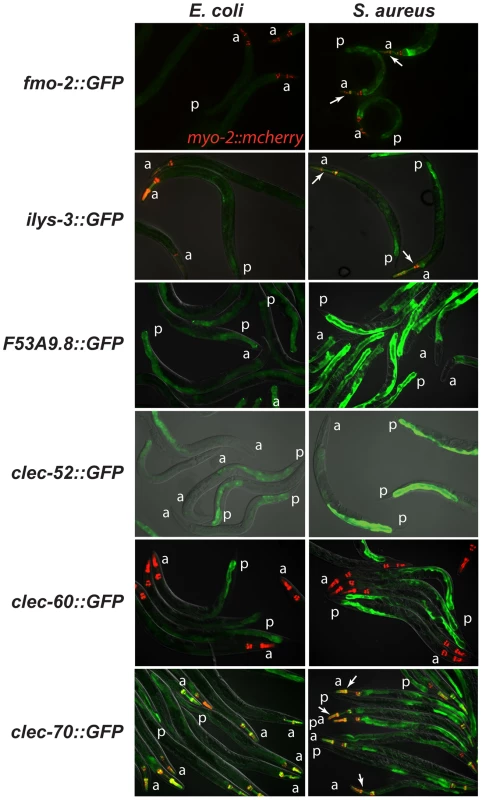
Transcriptional reporters using upstream sequences to fmo-2, ilys-3, F53A9.8, clec-52, clec-60, and clec-70 fused to GFP were induced in the intestinal epithelial cells after 24 h of S. aureus NCTC8325 infection (left panels) compared to parallel E. coli OP50-fed controls (right panels). Despite strong induction of fmo-2 and ilys-3 as measured by qRT-PCR, the corresponding GFP reporters exhibited low levels of expression; this could be due to their low basal expression on E. coli (not shown). Red, myo-2::NLS::cherry coinjection marker expressed in the pharynx in the head. A fold induction of 1 indicates no induction. a, anterior end; p, posterior end. Arrows indicate pharyngeal expression. ilys-3 was also expressed in the pharynx, in an unidentified cell superimposed on the pharynx, and in unidentified cells, possibly epithelial cells, in the vulva (Fig. S6A,B). clec-70 was also expressed in unknown cells in the pharynx (Fig. S6C) and in the uterine muscle (Fig. S6D), but only in one transgenic line of three. F53A9.8 was also expressed in a group of cells surrounding the rectum, possibly the rectal gland cells that secrete molecules into the rectal lumen (Fig. S6G, H). Distinct modes of detection of S. aureus and P. aeruginosa infection
Up-regulation of the 10 biomarker genes could be a result of cell damage caused by S. aureus, or of microbial detection independent of the inflicted damage. To discriminate between these two scenarios, we measured gene induction during exposure to live S. aureus, which causes pathogenesis, or heat-killed S. aureus, which does not (Figures 3G–I and S4C, E). Unexpectedly, all 10 biomarker genes were induced at least equally well on heat-killed S. aureus as on live S. aureus (Figure 6A). This result suggested that the ten biomarker genes form part of a host response against microbe-derived molecules, possibly pathogen-associated molecular patterns (PAMPs) [3], and not of a damage response.
Fig. 6. Potential PAMP-mediated, TLR-independent sensing of S. aureus. 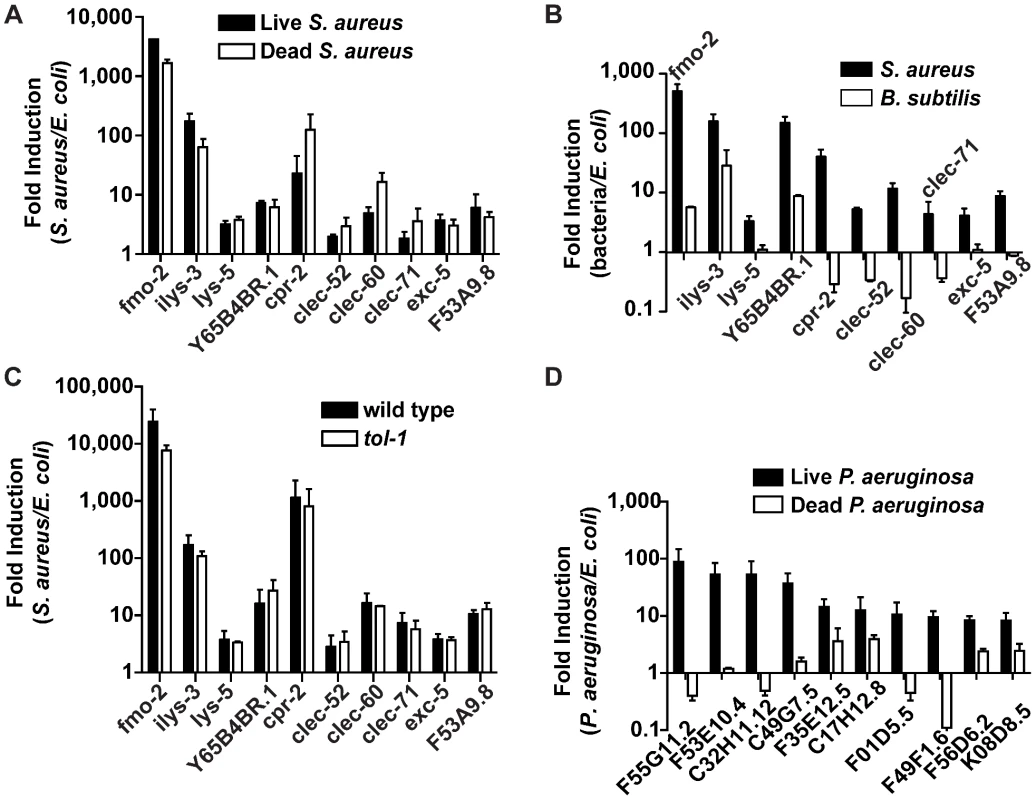
A. qRT-PCR analysis of induced genes in animals feeding on heat-killed S. aureus. Transcript levels were measured in synchronized young adult wild-type animals infected with live S. aureus NCTC8325 or feeding on dead S. aureus or E. coli OP50 for 8 h. B. qRT-PCR analysis of induced genes in animals feeding on B. subtilis. Transcript levels were measured in synchronized young adult wild-type animals feeding on B. subtilis PY79 or heat-killed E. coli for 8 h. C. qRT-PCR analysis of induced genes in tol-1 mutant animals. Transcript levels were measured in synchronized young adult animals feeding on heat-killed E. coli or infected with S. aureus for 8 h. D. qRT-PCR analysis of genes induced by live or heat-killed P. aeruginosa. Transcript levels were measured in synchronized young adult animals feeding on heat-killed E. coli or P. aeruginosa, or infected with live P. aeruginosa PA14 for 4 h. In all cases, data are the means of two biological replicates, each replicate measured in duplicate and normalized to a control gene, expressed as the ratio of the corresponding bacteria-induced levels and the basal E. coli levels. Error bars are SEM. Bacterial cell-wall components, such as LPS and flagellin in Gram-negative bacteria and peptidoglycan in both Gram-negatives and Gram-positives, are common PAMPs in many systems [57]. To test whether Gram-positive cell wall components in general were able to trigger the same S. aureus-induced response, we assayed gene induction in animals feeding on non-pathogenic Gram-positive bacterium Bacillus subtilis compared with E. coli-fed controls. Remarkably, of the ten biomarker genes, only fmo-2, ilys-3, and Y65B4BR.1 were induced by B. subtilis, albeit at much lower levels than with S. aureus (Figure 6B). The remaining 7 genes either were not induced or were repressed by B. subtilis. These results suggest that PAMPs other than shared Gram-positive cell wall molecules may be molecular triggers for the S. aureus-induced host response in C. elegans (see Discussion).
How is S. aureus detected? In fruit flies and mammals, Toll-like receptors (TLR) are involved in PAMP detection. C. elegans has a single gene encoding a TLR, tol-1, which has been shown to be important for avoidance responses to Serratia marcescens and for full induction of antimicrobial peptide abf-2 in response to Salmonella enterica [58], [59]. However, tol-1 was not required for the induction of any of the 10 S. aureus–induced biomarker genes (Figure 6C). Furthermore, tol-1 mutants were not more susceptible to S. aureus-mediated killing than wild type (Figure S10). One caveat is that the tol-1(nr2033) allele used, a deletion that eliminates the cytoplasmic TIR domain necessary for signaling [60], is viable, thus considered a partial loss of function for viability but a null allele for immune signaling [58]. These results show that tol-1 is most likely not required for the C. elegans host response to S. aureus and suggest that alternative mechanisms may exist for PAMP detection.
Since mpk-1/ERK is important for the rectal epithelial cell swelling response to M. nematophilum [47] and for the swelling response to S. aureus (Figure S4K, L), we wondered whether mpk-1 might also be important for the intestinal response to S. aureus. However, mpk-1 animals exhibited no measurable defect in the induction of the biomarker genes relative to E. coli-fed controls (Figure S9). Therefore, mpk-1 is dispensable for at least part of the intestinal transcriptional response to S. aureus, but not the anal swelling response.
In contrast to S. aureus, which either alive or dead triggered the induction of 10 biomarker genes (Figure 6A), heat-killed P. aeruginosa did not trigger the induction of 10 P. aeruginosa-induced biomarker genes (Figure 6D). Together, these data are consistent with the idea that C. elegans may recognize S. aureus infection mainly via TLR-independent PAMP detection, whereas it may recognize P. aeruginosa infection via detection of either Damage-Associated Molecular Patterns (DAMPs), or unidentified heat-labile PAMPs.
P. aeruginosa and S. aureus trigger partially overlapping host responses
Host responses to pathogenic attack in plants and animals are remarkably pathogen-specific. In C. elegans, pathogen-specific gene induction has been observed at late times of infection (i.e., 24 h), when damage and necrosis are apparent [50]. To determine whether pathogen-elicited gene induction at earlier times (i.e., 8 h) is also pathogen-specific, we compared the host response to S. aureus with our previously published study of the early host response to P. aeruginosa [44]. Of 186 genes induced by S. aureus and 259 genes induced by P. aeruginosa, 44 genes were induced by both pathogens, which is more than expected by chance (Figure 7A, Table 3). We validated the results of the microarray comparison using qRT-PCR; four genes predicted to be specifically induced by P. aeruginosa were either unaffected or repressed by S. aureus (Figure 7C), and four genes predicted to be induced by S. aureus were either unaffected or repressed by P. aeruginosa (Figure 7D). In contrast, four of five predicted overlap genes showed increased expression with both S. aureus and P. aeruginosa (Figure 7E). This suggests that the early host response to pathogen attack involves activation of a “pan-pathogen” response against a broad spectrum of pathogens, as well as a more tailored response that is optimized for the defense against the specific class of pathogens that is causing the infection.
Fig. 7. The C. elegans host response is comprised of pathogen-specific and -shared components. 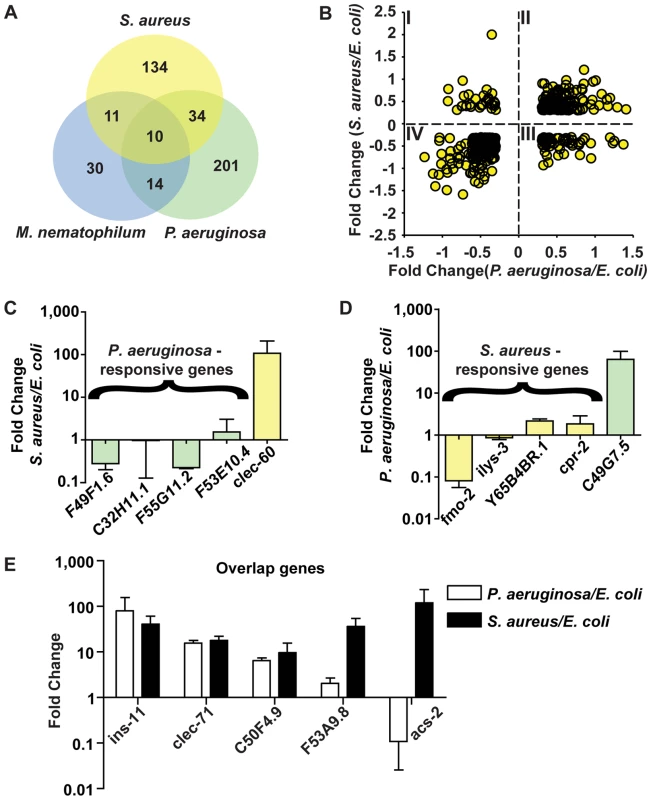
A. Comparison of genes induced 2-fold or higher (p≤0.01) upon infection with S. aureus for 8 h, P. aeruginosa for 8 h, and M. nematophilum for 6 h. Gene identities are presented in Table 3. B. Quadrant analysis of genes with expression changes both during S. aureus and P. aeruginosa infection. X axis, Fold Change for each gene during P. aeruginosa infection (log10 values). Y axis, Fold Change for each gene during S. aureus infection (log10 values). C. qRT-PCR analysis of P. aeruginosa–induced genes during S. aureus infection. D. qRT-PCR analysis of S. aureus–induced genes during P. aeruginosa infection. Transcript levels were measured in synchronized young adult wild-type animals feeding on non-pathogenic E. coli OP50 or infected with pathogen for 8 h (S. aureus NCTC8325) or 4 h (P. aeruginosa PA14). Results are the average of three biological replicates, each replicate measured in duplicate and normalized to a control gene, expressed as the ratio of the corresponding P. aeruginosa -induced levels and the basal E. coli levels. E. qRT-PCR analysis of overlap genes during S. aureus or P. aeruginosa infection. Transcript levels were measured in synchronized young adult wild-type animals feeding on non-pathogenic E. coli or infected for 8 h or 4 h, respectively (on P. aeruginosa, these genes were predicted to change by 4 h as well as 8 h). Results are the average of three biological replicates, each replicate measured in duplicate and normalized to a control gene, expressed as the ratio of the corresponding pathogen-induced levels and the basal E. coli levels. Previous microarray analysis wrongly predicted acs-2 to be induced by P. aeruginosa, the only example of this kind that we have found. Tab. 3. Pathogen specificity of the response to infection. 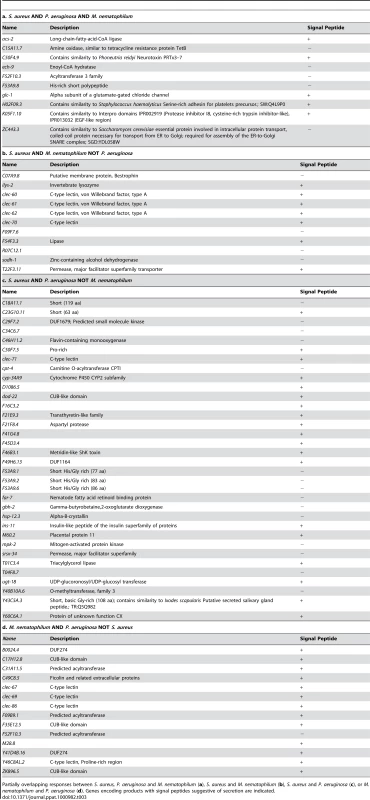
Partially overlapping responses between S. aureus, P. aeruginosa and M. nematophilum (a), S. aureus and M. nematophilum (b), S. aureus and P. aeruginosa (c), or M. nematophilum and P. aeruginosa (d). Genes encoding products with signal peptides suggestive of secretion are indicated. To further test the hypothesis of the existence of pathogen-shared and -specific components of the host response, we compared the genes differentially affected by S. aureus infection with previously published profiling of C. elegans infected with M. nematophilum [61]. The comparison of microarray studies independently performed in these separate laboratories may underestimate overlapping gene sets due to the use of different infection, RNA extraction, and data processing methods. Nonetheless, we found a relatively high degree of overlap between the responses to S. aureus and M. nematophilum (21 genes out of 186); 10 genes were induced by S. aureus, M. nematophilum, and P. aeruginosa (Figure 7A, Table 3). In contrast, 44 (68%) out of 65 genes induced by M. nematophilum were not induced by S. aureus. These data further support the existence of a core, shared response and a specific, tailored response.
GO annotations of genes affected by S. aureus or P. aeruginosa also exhibit a degree of specificity. Whereas the S. aureus-induced host response includes many sugar binding proteins, the P. aeruginosa-induced response is not characterized by any particular over-represented GO annotation (Table S7). Additionally, whereas the repressed response to S. aureus consists of many transporters and cuticle components, the repressed response to P. aeruginosa is mostly represented by metabolic pathways (Table S7). Interestingly, both repressed responses included basement membrane genes (N = 5, p = 1.4E-6 for P. aeruginosa), suggesting host growth suppression by both types of infection.
To investigate whether there were correlated or anti-correlated components of the responses to S. aureus and P. aeruginosa, we focused on genes whose expression changed both during infection with S. aureus and infection with P. aeruginosa. We broke down the two responses by plotting genes whose expression changed more than 2-fold with S. aureus (Y axis in Figure 7B) and with P. aeruginosa (X axis in Figure 7B). Genes whose expression changed only during infection with one of the two pathogens were thus not included. This method defined four quadrants: I) Genes induced by S. aureus and repressed by P. aeruginosa; II) Genes induced by S. aureus and P. aeruginosa; III) Genes repressed by S. aureus and induced by P. aeruginosa; and IV) Genes repressed by S. aureus and P. aeruginosa (Figure 7B). NextBio biogroup representation analysis (see Materials and Methods) on each quadrant failed to detect any over-represented biogroup in Quadrants I and III. However, in Quadrant II (genes up-regulated by both pathogens) several biogroups were over-represented, e.g. genes involved in detoxification, iron sequestration, and energy generation (Table S2a, b). Likewise, biogroups over-represented in Quadrant IV (down-regulated by both pathogens) included transporters, cuticle components, and fatty acid (FA) β-oxidation (Table S2a, b). The common repression of FA β-oxidation is not consistent with a starvation response, where β-oxidation is induced [56]. Furthermore, only 3 of 9 fasting repressed genes (lbp-8, acdh-1, and F08A8.2) [56] were repressed during P. aeruginosa infection [44]. Similarly to S. aureus, the fasting-repressed gene hacd-1 was induced during P. aeruginosa infection [44]. Together, these observations show the distinct nature of the host response to distinct pathogens, and highlights metabolic components of early host responses to infection.
Because overlapping gene expression changes measured by qRT-PCR do not necessarily imply the involvement of the same tissues during infection with different pathogens, we further investigated the expression of clec-60::GFP (up-regulated by S. aureus and M. nematophilum, Figure S11A, B, C, D) and F53A9.8::GFP (up-regulated by S. aureus, M. nematophilum, and P. aeruginosa, Figure S11E, F, G, H). clec-60::GFP was induced by M. nematophilum and by S. aureus in the intestine (Figure S11C, D), and down-regulated by P. aeruginosa (Figure S11B) compared with non-pathogenic E. coli (Figure S11A). Likewise, F53A9.8::GFP was induced by all three pathogens in the intestine (Figure S11F, G, H) compared with E. coli controls (Figure S11E). These data suggest that components of the host response are induced in the intestine by distinct pathogens.
Genes induced by S. aureus influence host survival
To determine whether S. aureus-induced genes have protective functions in host defense, we performed whole-animal RNAi knockdown of 42 of the 46 most highly up-regulated genes (Table 1), and identified 6 genes [tag-38 (Glutamate decarboxylase/sphingosine phosphate lyase), sodh-1 (sorbitol dehydrogenase), cyp-37B1 (Cytochrome P450), F43C11.7 (F-box containing protein), math-38 (MATH domain-containing signaling protein), and clec-70 (secreted CTL)] whose decreased expression caused enhanced susceptibility to killing by S. aureus (Figure 8A,B, C), but not P. aeruginosa (Figure 8G). We also identified one gene,Y51H4A.5 (a putative intracellular lipase) whose decreased expression caused slightly enhanced resistance to S. aureus killing, suggesting that its expression is detrimental to the host, or that it functions as a repressor of host defense (Figure 8B). Importantly, the lifespan of animals continuously fed dsRNA-expressing, non-pathogenic E. coli was near wild type, except for F43C11.7, which actually caused increased lifespan (Figure 8D). These data show that S. aureus-induced genes have important functions in pathogen-specific host defense, and that the enhanced susceptibility to S. aureus mediated by RNAi knockdown is not due to a non-specific decrease in viability. Although the molecular identities of these genes offer clues to their potential functions, the elucidation of their exact mechanisms of action will require further study.
Fig. 8. Host response genes are biologically relevant to host survival during S. aureus infection. 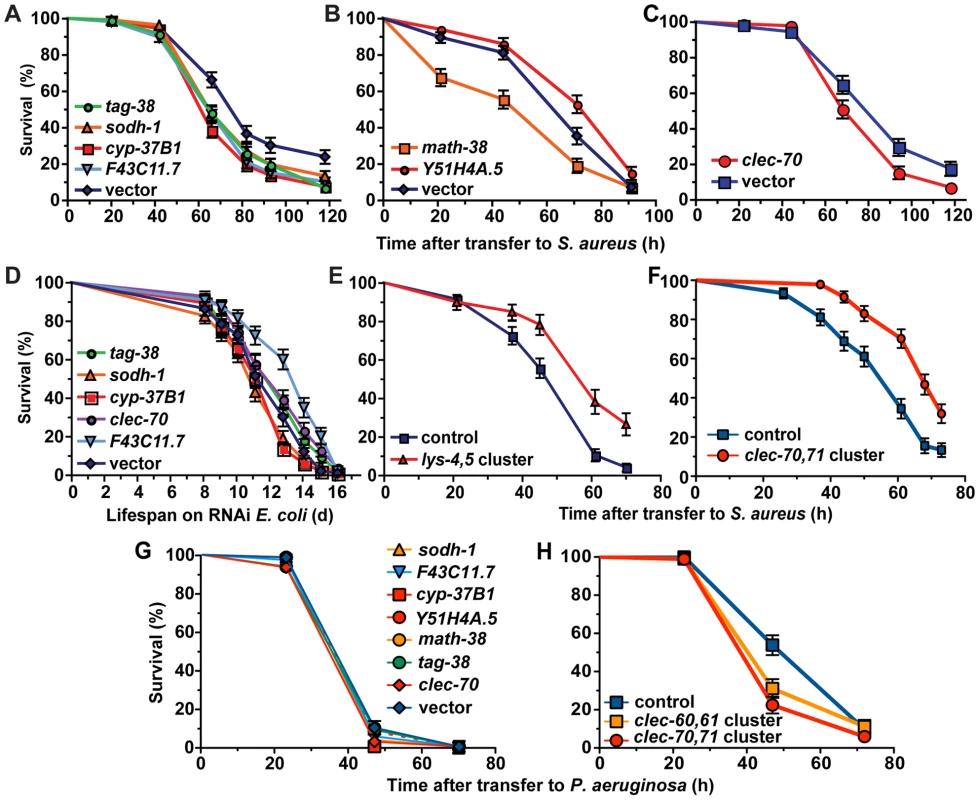
A, B, C. After RNAi knockdown of S. aureus-induced genes, animals were transferred to S. aureus NCTC8325 infection plates. Survival statistics: vector in A (MS = 82 h; N = 125), tag-38 (MS = 66 h, N = 142, p = 0.0002), sodh-1 (MS = 66 h; N = 142; p = 0.009), cyp-37B1 (MS = 66 h; N = 145; p<0.0001), F43C11.7 (MS = 66 h; N = 147; p = 0.0003), vector in B (MS = 71 h; LT50 = 67.6 h, N = 99/8), math-38 (LT50 = 48.6 h; N = 92/7; p = 0.0012), Y51H4A.5 (MS = 91 h; N = 100; p = 0.0122); vector in C (LT50 = 68.1 h; N = 91), clec-70 (LT50 = 67.9 h; N = 98; p = 0.0145). D. Lifespan of E. coli-fed animals. Lifespan statistics: vector (MS = 12.8 d; N = 89), tag-38 (MS = 12.8 d, N = 93, p = 0.1708), sodh-1 (MS = 11.1 d; N = 104; p = 0.1887), cyp-37B1 (MS = 12.8 d; N = 95; p = 0.1473), clec-70 (MS = 12.8 d; N = 99; p = 0.0354), F43C11.7 (MS = 14.08 d; N = 88; p<0.0001). E, F. Overexpression of host response genes enhances host survival during S. aureus infection. E. Animals carrying lys-4,5 cluster transgenes survived longer (LT50 = 54.1 h; N = 60; p<0.0001) during S. aureus infection than control animals bearing transgenes composed of coinjection marker and clec-60::GFP promoter fusion (LT50 = 40.6 h; N = 91/4). F. Animals carrying an integrated array containing a cluster of clec-70,71 (MS = 68 h; N = 94; p<0.0001) were more resistant to S. aureus mediated killing than control animals bearing rol-6 coinjection marker alone (MS = 61 h; N = 90). G. RNAi knockdown of S. aureus induced genes did not result in enhanced susceptibility to P. aeruginosa. Survival statistics: vector (MS = 47 h, N = 80/9); sodh-1 (MS = 47 h, N = 84/8, p = 0.2476); F43C11.7 (MS = 47 h, N = 83/9, p = 0.2592); cyp-37B1 (MS = 47 h, N = 92/11, p = 0.0201); Y51H4A.5 (MS = 47 h, N = 83/5, p = 0.6712); math-38 (MS = 47 h, N = 85/13, p = 0.9919); tag-38 (MS = 47 h, N = 90/10, p = 0.3373); clec-70 (MS = 47 h, N = 84/8, p = 0.0184). H. Overexpression of S. aureus induced C-type lectin genes resulted in mild enhanced susceptibility to P. aeruginosa. clec-60::GFP,myo-2::mCherry control (MS = 72 h; N = 98/20), clec-60,61,myo-2::mCherry (MS = 47 h; N = 94/16; p = 0.0408), clec-70,71, myo-2::mCherry (MS = 47 h; N = 92/12; p = 0.0002). Potential immune effectors identified in our analysis include antimicrobial peptides, lysozymes (enzymes that degrade peptidoglycan in the bacterial cell wall), and CTLs [62]. To test whether lysozymes or CTLs induced by S. aureus can confer resistance to S. aureus-mediated killing when expressed to higher levels than in the wild type, we constructed transgenic C. elegans carrying multiple copies of lys-4 and lys-5, clec-60 and clec-61, or clec-70 and clec-71 (each is a pair of genes that are adjacent to each other in the genome). Transgenic animals carrying the lys-4,5 or clec-60,61 clusters survived significantly longer than transgenic control animals (Figure 8E; Figure S12). Transgenic animals carrying the clec-70,71 cluster extrachromosomally did not exhibit enhanced resistance (not shown); however, three independent spontaneous integrant lines did (Figure 8F). Interestingly, strains with multiple copies of either cluster of C-type lectin genes exhibited enhanced susceptibility to P. aeruginosa-mediated killing (Figure 8H). Collectively, the data suggest that pathogen-induced intestinal expression of epithelial detoxifying and antimicrobial proteins is an important and pathogen-specific mechanism of C. elegans host defense against S. aureus infection.
Discussion
Among the bacteria that cause intestinal infections in C. elegans, the best-studied is P. aeruginosa, a Gram-negative human pathogen. Despite many advances in understanding P. aeruginosa-C. elegans interactions, little was known about the morphological and cell biological consequences of infection in vivo. This report provides unprecedented high-resolution description of bacterial intestinal infections of clinical relevance in C. elegans, with emphasis on the comparative cytopathology of infection by P. aeruginosa and S. aureus. P. aeruginosa and S. aureus cause markedly different symptoms in C. elegans.
During infection with P. aeruginosa, we observed marked intestinal distention, extracellular matrix accumulation in the intestinal lumen, extracellular material accumulation on the apical surface, enlargement of the rough endoplasmic reticulum (RER), and abnormal autophagy in the host intestinal cells. The intestinal distention is likely a result of loss of cytoplasmic volume of the intestinal cells; the identity of the extracellular material is currently unknown. One possibility is that it is a biofilm matrix produced by P. aeruginosa, perhaps as a defensive mechanism. Alternatively, it could be produced by the host in response to P. aeruginosa, but not S. aureus. Further study is required to elucidate the origin of this substance. The cause of abnormal autophagy is also unknown; however, the aberrant structures we observed have also been described in unc-52 mutant animals, which are defective in the early steps of autophagosome assembly [63]. Thus, it is possible that P. aeruginosa infection causes autophagy arrest. It is tempting to speculate that this may represent a virulence mechanism deployed by P. aeruginosa to evade autophagic clearance of intracellular bacteria, an important host defense mechanism in the intestinal cells of C. elegans [64] and humans [65]. The intracellular invasion we observed provides a rationale for the benefit to P. aeruginosa of inhibiting host autophagy. gacA mutant P. aeruginosa was defective in inducing these phenotypes, indicating that GacA orchestrates virulence mechanisms related to host cell disruption in C. elegans. The molecular identity of these virulence mechanisms remains unknown; however, the observed putative OMVs may provide a mechanism for delivery of bacterial virulence factors to C. elegans intestinal cells. Perhaps the reliance on OMVs for virulence may explain why P. aeruginosa defective in type III secretion, an alternative mechanism of virulence factor delivery to host cells, are not defective in nematode killing [66]. OMV production is induced in P. aeruginosa by cellular stress [67]; therefore, the abundant OMV production we observed may indicate that P. aeruginosa perceives the intestinal lumen as a stressful environment, presumably as a result of defensive host factors secreted by the intestinal cells.
The molecular identity of such putative intestinal defense factors remains unknown. In previous work, we defined the early transcriptional host response to P. aeruginosa [68]. Among the genes that are up-regulated by P. aeruginosa infection, several encode putative antimicrobial factors such as ShK toxins. Whatever the molecular identity of the antibacterial factors induced during P. aeruginosa infection, heat-killed P. aeruginosa did not induce a set of 10 biomarkers of the response. This observation is consistent with our previous studies suggesting that P. aeruginosa virulence may be required, at least partially, for response induction [44], [69]. These results are consistent with several interpretations. It is possible that detection of virulent P. aeruginosa is mediated by recognition of the damage inflicted on the host cells, e.g., via DAMP perception [70], or by recognition of PAMPs in the context of host damage, in what has come to be known as a pattern of pathogenesis [71]. Alternatively, PAMPs released only by live bacteria (PAMP-per vitae, or PAMP-PV) may be specifically recognized by C. elegans as a trigger for the response [71]. A third option is the release of PAMP-post mortem (PAMP-PM) upon heat inactivation of P. aeruginosa, which in turn could dampen the host response. PAMP-PM detection has been proposed to be a mechanism used by mammals to limit inflammatory damage to the host once the infection has been controlled [71]. Finally, detection of P. aeruginosa could be mediated by heat-labile signals that were destroyed during heat-inactivation in our experiments. Although further work is required to conclusively show which of these scenarios is correct, we have found that P. aeruginosa strains with increasing levels of virulence cause increasing levels of induction of C. elegans host response gene irg-1 [69], suggesting that the extent of host cell damage determines the magnitude of the host response to the hypothetical P. aeruginosa-produced PAMPs or DAMPs that are detected.
During S. aureus infection, we observed rapid intestinal colonization, swelling of the anus, and effacement and destruction of intestinal epithelial cells, providing important mechanistic information about S. aureus-mediated pathogenesis of host epithelial cells in vivo. The molecular mechanism of cell destruction remains unknown; here we show that it is hemolysin-independent and is abrogated by heat inactivation of S. aureus. One explanation for this observation is that host tissue damage may be caused by the active pathogen, and not an unbridled host response. Alternatively, unknown heat-labile S. aureus toxins may cause cell lysis, as can occur in human cells [72]; further study is required to distinguish between these possibilities. Previous experiments using fertile animals showed that α-hemolysin Δhla mutant S. aureus were defective in C. elegans killing [17]. To our surprise, the Δhla Δhlb, Δhla Δhlg, or Δhla Δhlb Δhlg hemolysin mutants did not exhibit any killing defect in our assay using sterile animals (Figure S7). Thus, it is possible that α-hemolysin-mediated killing of C. elegans requires internal hatching of eggs retained inside the mother as a result of stress.
Eventually, the pathogenic process results in internal tissue degradation and nematode death. These steps recapitulate key features of S. aureus infection in mammals both in vivo and in vitro, e.g. enterocyte effacement during intestinal infection, and cell lysis [37], [38], [39], [40], [41], [42], [43]. Thus, we propose that the C. elegans-S. aureus model has significant relevance to the study of conserved virulence mechanisms used by S. aureus to evade host epithelial defenses and attack host epithelial cells in general.
Transcriptional profiling showed that S. aureus infection elicits changes in expression of a minor fraction of the total genome of ∼22,000 genes, indicating high specificity. This early response does not require live bacteria, suggesting that it involves detection of S. aureus per se (perhaps through PAMP perception) as opposed to indirectly through host cell damage (through DAMPs) [70]. Whatever the relevant PAMPs are, it is clear that they are not shared between S. aureus and B. subtilis, also a Gram-positive bacterium. For example, the peptidoglycan differs greatly between these two species [73], and could potentially be differentially sensed by C. elegans. Other possibilities include differential detection of surface-expressed lipoteichoic acids or differentially expressed surface proteins. Further work is required to elucidate the nature of such signal(s).
Our results provide insight into the cellular biological effects of pathogenic infection on the epithelial barrier in vivo, as well as the early defense mechanisms deployed by C. elegans to fend off attack. The affected genes represent evolutionarily conserved categories relevant to human innate immunity. To gain insight into evolutionarily conserved effector mechanisms of host defense, we compared the genes up-regulated in C. elegans with previously published data using human neutrophils, which are important effector cells in human innate immunity [74]. When grouped by molecular function, we found some overlapping functional classes during the C. elegans and human neutrophil responses to S. aureus infection (Table S4). These classes included detoxification factors [e.g. transporters, UDP-glucuronosyltransferases (UGTs), cytochrome P450s, GSTs, and flavin-containing monooxygenases (FMOs), [75]], antimicrobial effectors (e.g. CTLs, peptidases, and proteases), galectins [76], and signaling components including EGF-like domain containing proteins, Cdc42 guanyl nucleotide exchange factors (GEFs), F-box proteins, mitogen-activated protein kinases (MAPKs), and Leucine-rich repeat domain (LRR) containing proteins. This observation shows that the C. elegans host response to infection shares important components with the human cellular host response, and suggests that human innate responses have ancient components that are conserved across phylogeny. Indeed, it is thought that modern vertebrate innate immunity represents an accretion of ancient invertebrate innate defenses [77].
Comparative genomics identified pathogen-specific as well as pathogen-shared components of the host response. This observation, consistent with similar ones recently made by others using different approaches [50], [78], [79], illustrates how diverse pathogens affect distinct aspects of host physiology as reflected in the distinct nature of the host responses. We also found overlap in gene expression patterns among the responses to three different pathogens, S. aureus (intestinal infection, Gram-positive), M. nematophilum (cuticular infection, Gram-positive), and P. aeruginosa (intestinal infection, Gram-negative), defining a core induced response that involves intracellular detoxification, iron sequestration, and sugar binding. In addition to a common set of up-regulated genes, we observed a repressed core response common to S. aureus and P. aeruginosa that involves anion transport, growth-related genes, lipid - and alcohol-metabolic genes, and acyl-CoA dehydrogenases. The fact that metabolic regulation is a major component of the C. elegans host response to bacterial pathogens provides a rationale to investigate metabolic changes that occur in higher organisms as a result of infection, particularly in innate defense tissues such as epithelia. Recent reports suggest a significant metabolic component during the host response in mammals as well [80], [81], [82].
The non-overlapping responses to S. aureus and P. aeruginosa may reflect the different virulence strategies of the two pathogens and/or may be a consequence of the distinct molecular composition of Gram-positive and -negative cell walls. Further studies are required to dissect the relative contribution of each factor, including the survey of additional Gram-positive and -negative infections in C. elegans. In a first step along those lines, we found that the Gram-positive non-pathogenic bacterium B. subtilis does not induce the same set of 10 biomarkers as S. aureus. Additionally, of 531 genes up-regulated by the Gram-positive pathogen E. faecalis [50], only 15 were shared with the response to S. aureus (Table S5a). The same report characterized the C. elegans late host response (i.e., 24 h) to four additional pathogens, identifying a set of shared genes that defined a pathogen-shared necrotic response [50]. During S. aureus infection we observed up-regulation of none of 16 up-regulated shared late response genes, and down-regulation of only three of six down-regulated shared response genes (Table S5c), suggesting that the host response evolves significantly over time.
Mammalian intestinal epithelial cells directly sense and respond to bacterial stimulation [2], by inducing the expression of antimicrobial genes such as CTLs [53]. Similarly, most of the early C. elegans response to S. aureus occurs in the epithelial cells of the intestine. In Drosophila, mice, and humans, TLRs are important receptors that drive the activation of signaling cascades downstream of microbial stimulation. In C. elegans, however, loss of function of the sole TLR does not result in a defective immune response to S. aureus. Furthermore, C. elegans does not have an NF-kB homolog nor an inflammasome, raising the possibility that in mammals as in C. elegans, at least a portion of the immune response to S. aureus may be regulated independently of the TLR/NF-kB signaling axis [6], [51], [83]. Indeed, we previously reported that β-catenin and HOX genes are required for perception of pathogenic attack by S. aureus to drive the expression of epithelial host response genes [7]. Significantly, we also found that β-catenin and HOX proteins modulated NF-κB signaling in a human epithelial cell line during TLR2 stimulation, illustrating that previously unknown human innate immunity pathways can be identified using C. elegans [7].
We identified 6 host factors out of 42 tested whose lowered expression caused enhanced susceptibility to S. aureus. This corresponds to a 14% hit rate, which was much greater than expected; we had assumed that there would be significant functional redundancy among C. elegans immune effectors. Moreover, RNAi typically exhibits incomplete penetrance and expressivity. In addition to these 6 genes, knockdown of Y51H4A.5 (lipase) caused mild resistance to S. aureus mediated killing, suggesting that Y51H4A.5 acts to limit survival as a negative regulator of the host response or by harming the host instead of protecting it. RNAi of F43C11.7 caused enhanced susceptibility to S. aureus, yet extended lifespan on non-pathogenic E. coli. This is an example of genes that have opposite effects on host defense and lifespan regulation, indicating that these related processes are genetically separable [84]. Increased expression of host defense genes provides a mechanistic explanation of C. elegans defenses, as we found that animals carrying multiple copies of three genomic clusters of lysozymes or CTLs exhibited enhanced resistance to S. aureus. Lysozymes are well-known, evolutionarily ancient antibacterial effector molecules that degrade peptidoglycan and are also produced in human intestinal epithelial cells [1]. Recent studies have shown that some vertebrate CTLs, including human HIP/PAP, have direct bactericidal activity [53]. clec-70 and clec-71 share similar domain architectures with HIP, and clec-60 and clec-61 share similarities with arthropod receptor CTLs, suggesting that they may function respectively as antimicrobials or receptors in the C. elegans host response [85], [86], [87]. Collectively, these observations show that the large number of pathogen-response genes contribute cumulative, incremental defense functions to host survival.
It is interesting that elements of the C. elegans immune response enhance host survival during infection with S. aureus (a human pathogen), supporting the notion that pathogen detection and response, as well as mechanisms of bacterial pathogenesis, share conserved features among distantly related hosts or microbes, respectively. It has been proposed that pathogens have experienced stepwise additions of virulence factors, as they evolved to survive different host antimicrobial responses, and to colonize new niches [12]. Our studies of the C. elegans-S. aureus system may thus probe an early step in the evolution of S. aureus as a pathogen and its interaction with prototypical metazoan epithelial cells. In humans, unknown host and bacterial factors determine whether S. aureus will become an innocuous member of the normal microbiota, or whether it will switch to a more virulent state and become a serious pathogen [23]. In this light, studies of the C. elegans intestinal epithelial response to S. aureus provide a unique starting point to identify previously unknown signaling pathways and molecular mechanisms of host immune response to bacterial virulence. Understanding how S. aureus disrupts host defense and causes host damage and death is critical to identifying new therapeutic targets to treat infectious disease.
Materials and Methods
Strains
C. elegans was grown on nematode-growth media (NGM) plates seeded with E. coli OP50-1 at 15–20°C according to standard procedures [88]. C. elegans strains used in this study are detailed in Table S6a. Bacterial strains are detailed in Table S6b.
Electron microscopy
Wild type N2 Bristol animals were synchronized by hypochlorite treatment and L1 arrest and incubated on NGM plates seeded with E. coli OP50-1. Late L4 animals were collected and plated on 15 cm TSA plates seeded with live S. aureus NCTC8325 or heat-killed NCTC8325, and parallel NGM plates seeded with OP50-1. After 12, 24, and 36 h incubation at 25°C, animals were collected and incubated in fixation buffer (2.5% glutaraldehyde, 1.0% paraformaldehyde in 0.05 M sodium cacodylate buffer, pH 7.4 plus 3.0% sucrose). During the initiation of fixation, animals were cut in half with a surgical blade in a drop of fixative under a dissecting microscope, fixed overnight at 4°C, rinsed in 0.1 M cacodylate buffer, post-fixed in 1.0% osmium tetroxide 0.1 M cacodylate buffer, rinsed in buffer and water, and stained en bloc in 2% aqueous uranyl acetate. After rinsing in water, animals were embedded in 2% agarose in phosphate buffer saline, dehydrated through a graded series of ethanol washes to 100%, then 100% propylene oxide, and finally 1∶1 propylene oxide:EPON overnight. Blocks were infiltrated in 100% EPON and then embedded in fresh EPON overnight at 60°C. Thin sections were cut on a Reichert Ultracut E ultramicrotome and collected on formvar-coated gold grids. Sections were post-stained with uranyl acetate and lead citrate and viewed using a JEOL 1011 transmission electron microscope at 80 kV with an AMT digital imaging system (Advanced Microscopy Techniques, Danvers, MA). For each observation, whenever possible at least 10 cross-sections were evaluated, and representative images were chosen.
Microarray analysis
C. elegans growth and infection
fer-15(b26)ts;fem-1(hc17) animals were synchronized by hypochlorite treatment and L1 arrest [88]. Arrested L1 larvae were placed onto NGM plates seeded with OP50 and grown at the restrictive temperature (25°C) in order to obtain sterile adults. Young adult animals were transferred to slow-killing plates (NGM agar containing 0.35% peptone), seeded with OP50, or tryptic soy agar plates (TSA, see below) seeded with RN6390. Animals were harvested at 8 h after transfer. Three independent replicates of each treatment were isolated.
RNA isolation
Total RNA was extracted using TRI Reagent (Molecular Research Center, http://www.mrcgene.com) according to the manufacturer's instructions, followed by purification on RNeasy columns (Qiagen, http://www1.qiagen.com).
Microarray target preparation and hybridization for Affymetrix GeneChips
RNA samples were prepared and hybridized to Affymetrix full-genome GeneChips for C. elegans at the Harvard Medical School Biopolymer Facility, according to instructions from Affymetrix (http://www.affymetrix.com). Briefly, 5 µg of total RNA was reverse transcribed using an oligo dT-T7 primer and Superscript II reverse transcriptase, followed by second-strand cDNA synthesis. The double-stranded cDNA was then purified using a DNA purification kit (Qiagen), and used as the template for in vitro transcription using T7 RNA polymerase and biotinylated nucleotides. The resulting cRNA was fragmented and hybridized onto C. elegans Affymetrix GeneChips as previously described [44].
Microarray analysis for S. aureus studies
Affymetrix .cel files were uploaded into the Resolver Gene Expression Data Analysis System, version 5.1 (Rosetta Inpharmatics, http://www.rii.com) at the Harvard Center for Genomic Research for analysis. For each condition, three replicate microarrays were normalized and analyzed using the Resolver intensity error model for single color chips [89]. The two conditions were then compared in Resolver to determine fold change for each probe set and a p-value, using a modified t test. Genes with a 2-fold or greater fold change and a p-value <0.01 were considered differentially expressed. Differentially expressed probe sets were compared for S. aureus infection (this study), P. aeruginosa infection [44], and M. nematophilum infection [90] using Resolver.
Significance of over-representation analysis
Analysis of over-representation of GO annotation categories was performed using FuncAssociate (http://llama.med.harvard.edu/cgi/func/funcassociate). GO Bioset analysis was performed using NEXTBIO (www.nextbio.com) [91]. For greater inclusivity, gene expression changes greater than 1 were included in the NEXTBIO analysis, as suggested by NEXTBIO.
Infection and lifespan assays
All assays were conducted at 25°C, 65% relative humidity. Animals were scored as alive or dead by gentle prodding with a platinum wire. Kaplan-Meier statistical analyses were performed using the software Prism (GraphPad, http://www.graphpad.com). Survival data were compared as described using the log-rank test. Data are represented as median survival (MS) or lethal time – 50 (LT50) when MS values were skewed by small number of timepoints, N (number of deaths/censored), and p value. A p-value <0.05 was considered significantly different from control.
S. aureus killing assays
Assays were performed as described [17]. Briefly, NCTC8325 (or mutant derivatives, as noted) was grown overnight in tryptic soy broth (TSB, BD, Sparks, MD) with 10 µg/ml nalidixic acid (Sigma). 5–10 µl of overnight cultures diluted 1∶5 in fresh TSB were seeded on 35 mm tryptic soy agar (TSA, BD, Sparks, MD) plates with 10 µg/ml nalidixic acid. For accumulation experiments using GFP-S. aureus, plates were supplemented with 10 µg/ml chloramphenicol (Sigma) for plasmid maintenance. A total of 25–35 L4 stage hermaphrodites were transferred to each of three replicate plates per strain. Animals that died as a consequence of a bursting vulva or crawled off the agar were censored. Experiments were performed at least twice. For heat-killing of S. aureus, fresh overnight cultures of NCTC8325 were concentrated 10-fold and incubated at 95°C for 45 min. Following heat treatment, no live cells could be detected by plating undiluted cultures on TSA plates.
P. aeruginosa slow-killing assays
Briefly, PA14 was cultured in Luria broth (LB), seeded on slow-killing plates and incubated first for 24 h at 37°C and then for 24 h at 25°C. A total of 25–35 L4 stage hermaphrodites were transferred to each of three replicate plates per strain. Experiments were performed at least twice.
M. nematophilum infection assays
Assays were performed as described [92]. Briefly, OP50 and CBX102 were cultured overnight in LB, mixed at 1∶10 ratio of CBX102:OP50 and plated on NGM. A total of 25–35 L4 stage hermaphrodites were transferred to each of three replicate plates per strain. Assays were conducted at 25°C. For Dar quantification, animals were analyzed directly on infection plates, in triplicate.
Lifespan assays
Briefly, RNAi plates seeded with dsRNA-expressing E. coli HT115 were used. Approximately 100 synchronized eri-1(mg366) L1 larvae were added to each of 3 plates, and incubated for 24 h at 15°C followed by 24 h at 25°C, which causes animal sterility. 35–50 late L4 stage animals were added to each of 3 fresh RNAi plates seeded with dsRNA-expressing E. coli HT115, and incubated at 25°C. Lifespan is defined as the time elapsed from when animals were put on plates to when they were scored as dead. Experiments were performed at least twice. Animals that died of a bursting vulva or crawled off the agar were censored.
Quantitative RT-PCR (qRT-PCR) analysis
Animals were treated essentially as described for killing assays described above, with the following modifications. For S. aureus infection assays, infected samples were compared to parallel samples feeding on E. coli OP50, heat-killed by 30 min incubation at 95°C, plated on the same TSA medium. All strains compared were grown in parallel. Total RNA was extracted using TRI Reagent, and reverse transcribed using the Superscript III kit (Invitrogen). cDNA was subjected to qRT-PCR analysis using SYBR green detection (BIO-RAD SYBR Green supermix) on iCycler (Bio-Rad, http://www.bio-rad.com) and RealPlus (Eppendorf, Germany) machines. Primers for qRT-PCR were designed using Primer3Plus (Massachusetts Institute of Technology, http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi), checked for specificity against the C. elegans genome and tested for efficiency with a dilution series of template. Primer sequences are available upon request. All Ct values are normalized against the control gene snb-1, which did not vary under conditions being tested. Fold change was calculated using the Pfaffl method [93]. We found some variability in gene induction levels from experiment to experiment. The source of this variation has not been conclusively ascertained; however, we suspect it may derive from differences between batches of agar plates used for infection assays. Importantly, all experiments were repeated at least twice (biological replicates) and were internally controlled. Additionally, despite numerical variability in fold induction, all results were internally consistent.
GFP fusions
PCR primers to amplify 1665 bp of sequence upstream of the clec-60 start site, 3505 bp upstream of the clec-70 start site, 1774 bp upstream of the fmo-2 start site, and 913 bp upstream of the ilys-3 start site were designed using the online PCR primer design tool provided by the British Columbia Genome Sciences Center (http://elegans.bcgsc.bc.ca/promoter_primers/index.html). Splicing by overlapping extension PCR (SOE-PCR) was used as described [94] to generate promoter-GFP fusion PCR fragments, which were transformed at 3 ng/ µl into wild type animals by microinjection with 40 ng/ µl of a myo-2::NLS::mCherry construct as coinjection marker used to identify transgenic animals (courtesy of J. Kaplan, Massachusetts General Hospital). Primer sequences are available upon request.
GFP fusion induction
L4 animals carrying extrachromosomal arrays were transferred from NGM plates seeded with OP50-1 to S. aureus, P. aeruginosa, or M. nematophilum killing plates essentially as described above. After incubation, animals were mounted on glass slides with 2% agarose pads, anesthetized with 30 mM NaN3, and immediately used for imaging. Exposure times were set for the most highly expressed condition and kept constant throughout each experiment.
RNAi knockdown
RNAi screen
Enhanced RNAi eri-1(mg366) mutants were propagated at 15°C. RNAi of selected genes was carried out in triplicate using bacterial feeding RNAi [95]. Synchronized L1 animals were transferred to RNAi plates, incubated at 15°C for 25 h and then 25°C for 24 h to induce sterility, and then transferred to NCTC8325-seeded killing assays. The screen was performed once, and positive clones that exhibited altered survival on S. aureus were tested at least once more. RNAi clones were obtained from the Ahringer laboratory (except clec-70 RNAi, which was made by recombining pDONR201.clec-70 from the ORFeome library [96] with pDEST.L4440 [97]. Sequences of positive RNAi clones were confirmed.
cdc-25.1 RNAi
To sterilize worms previous to use in killing assays, cdc-25.1 RNAi was carried out by feeding L4 animals for 24 h at 15°C.
PMN expression analysis
Gene expression microarray data files of S. aureus infection were obtained from Gene Expression Omnibus (GEO accession: GSE2405) and analyzed. The samples were derived from human polymorphonuclear leukocytes (PMNs) from three healthy donors using a separate HU133A GeneChip (Affymetrix) for each donor [74]. We examined the data from PMNs that were either uninfected or infected with live S. aureus for 9 hours. The dataset was MAS5.0-normalized and filtered by excluding probe sets with 100% ‘absent’ calls (MAS5.0 algorithm) across all samples. FDR analysis (q-value<0.005) with 1000 permutations using significance analysis of microarrays [98] was performed to identify genes that were differentially induced in S. aureus-infected PMNs versus uninfected controls.
Epifluorescence microscopy
Images were acquired using a Zeiss AXIO Imager Z1 microscope with an Zeiss AxioCam HRm camera and Axiovision 4.6 (Zeiss) software. Image cropping and minimal manipulation were performed using Photoshop (Adobe).
Gene accession information
Gene Public Name, Gene WormBase ID, Source GenBank ID, Gene CGC Name; bar-1, WBGene00000238, U46673, bar-1; col-63, WBGene00000639, Z81143, col-63; col-98, WBGene00000673, Z81503, col-98; cpr-2, WBGene00000782, Z81531, cpr-2; egl-5, WBGene00001174, L15201, egl-5; exc-5, WBGene00001366, Z68159, exc-5; fmo-2, WBGene00001477, Z70286, fmo-2; ins-11, WBGene00002094, U41279, ins-11; lys-2, WBGene00003091, AL021479, lys-2; lys-5, WBGene00003094, Z73427, lys-5; mpk-1, WBGene00003401, Z46937, mpk-1; pmk-1, WBGene00004055, U58752, pmk-1; sod-3, WBGene00004932, U42844, sod-3; tol-1, WBGene00006593, AF348166, tol-1; unc-32, WBGene00006768, Z11115, unc-32; C32H11.1, WBGene00007864, NM_070062.2; C50F4.9, WBGene00008234, Z70750; F01D5.2, WBGene00008493, Z81493; acs-2, WBGene00009221, Z81071, acs-2; F55G11.2, WBGene00010123, Z82272; clec-60, WBGene00014046, Z49132, clec-60; clec-61, WBGene00014047, Z49132, clec-61; clec-52, WBGene00015052, U58752, clec-52; C23G10.6, WBGene00016013, U39851; C30G12.2, WBGene00016274, U21319; ilys-3, WBGene00016670, AF067611, ilys-3; C49G7.5, WBGene00016783, AF016418; acdh-1, WBGene00016943, AC006625, acdh-1; F49F1.6, WBGene00018646, AF100656; F53A9.8, WBGene00018731, U23523; F53E10.4, WBGene00018760, U88177; clec-70, WBGene00021581, AC024785, clec-70; clec-71, WBGene00021582, AC024785, clec-71; Y65B4BR.1, WBGene00022040, AC024847.
Supporting Information
Zdroje
1. Liévin-Le MoalV
ServinAL
2006 The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev 19 315 337
2. VaishnavaS
BehrendtCL
IsmailAS
EckmannL
HooperLV
2008 Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 105 20858 20863
3. AkiraS
UematsuS
TakeuchiO
2006 Pathogen recognition and innate immunity. Cell 124 783 801
4. HoffmannJA
KafatosFC
JanewayCA
EzekowitzRA
1999 Phylogenetic perspectives in innate immunity. Science 284 1313 1318
5. LemaitreB
HoffmannJA
2007 The host defense of Drosophila melanogaster. Annu Rev Immunol 25 697 743
6. KurzCL
EwbankJJ
2003 Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet 4 380 390
7. IrazoquiJE
NgA
XavierRJ
AusubelFM
2008 Role for beta-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial-pathogen interactions. Proc Natl Acad Sci U S A 105 17469 17474
8. ZugastiO
EwbankJJ
2009 Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-beta signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol 10 249 256
9. IrazoquiJE
UrbachJM
AusubelFM
2010 Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol 10 47 58
10. McGheeJD
2007 The C. elegans intestine. WormBook 1 36
11. SifriCD
BegunJ
AusubelFM
2005 The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol 13 119 127
12. WaterfieldNR
WrenBW
Ffrench-ConstantRH
2004 Invertebrates as a source of emerging human pathogens. Nat Rev Micro 2 833 841
13. LyczakJB
CannonCL
PierGB
2000 Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2 1051 1060
14. LyczakJB
CannonCL
PierGB
2002 Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15 194 222
15. SifriCD
AusubelFM
2004 Use of simple non-vertebrate hosts to model mammalian pathogenesis.
CossartP
BoquetP
NormarkS
RappuoliR
Cellular Microbiology. 2 ed Washington, D.C. ASM Press 543 563
16. CunyC
FriedrichA
KozytskaS
LayerF
NübelU
2009 Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int J Med Microbiol
17. SifriCD
BegunJ
AusubelFM
CalderwoodSB
2003 Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun 71 2208 2217
18. BoucherHW
CoreyGR
2008 Epidemiology of methicillin-resistant Staphylococcus aureus. CLIN INFECT DIS 46 Suppl 5 S344 349
19. GrahamPL
LinSX
LarsonEL
2006 A U.S. population-based survey of Staphylococcus aureus colonization. Annals of Internal Medicine 144 318 325
20. GordonRJ
LowyFD
2008 Pathogenesis of methicillin-resistant Staphylococcus aureus infection. CLIN INFECT DIS 46 Suppl 5 S350 359
21. DiepBA
OttoM
2008 The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol 16 361 369
22. NizetV
2007 Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J Allergy Clin Immunol 120 13 22
23. von Köckritz-BlickwedeM
RohdeM
OehmckeS
MillerLS
CheungAL
2008 Immunological mechanisms underlying the genetic predisposition to severe Staphylococcus aureus infection in the mouse model. Am J Pathol 173 1657 1668
24. TanMW
Mahajan-MiklosS
AusubelFM
1999 Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA 96 715 720
25. TanMW
RahmeLG
SternbergJA
TompkinsRG
AusubelFM
1999 Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA 96 2408 2413
26. GarsinDA
SifriCD
MylonakisE
QinX
SinghKV
2001 A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci USA 98 10892 10897
27. PowellJR
KimDH
AusubelFM
2009 The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc Natl Acad Sci U S A 106 2782 2787
28. GarsinDA
VillanuevaJM
BegunJ
KimDH
SifriCD
2003 Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300 1921
29. KimDH
FeinbaumR
AlloingG
EmersonFE
GarsinDA
2002 A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297 623 626
30. BegunJ
SifriCD
GoldmanS
CalderwoodSB
AusubelFM
2005 Staphylococcus aureus virulence factors identified by using a high-throughput Caenorhabditis elegans-killing model. Infect Immun 73 872 877
31. BaeT
BangerAK
WallaceA
GlassEM
AslundF
2004 Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci USA 101 12312 12317
32. SkaarEP
HumayunM
BaeT
DeBordKL
SchneewindO
2004 Iron-source preference of Staphylococcus aureus infections. Science 305 1626 1628
33. Mahajan-MiklosS
TanMW
RahmeLG
AusubelFM
1999 Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96 47 56
34. DarbyC
CosmaCL
ThomasJH
ManoilC
1999 Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci USA 96 15202 15207
35. KuehnMJ
KestyNC
2005 Bacterial outer membrane vesicles and the host-pathogen interaction. Genes & Development 19 2645 2655
36. LamJ
ChanR
LamK
CostertonJW
1980 Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun 28 546 556
37. NaikS
SmithF
HoJ
CroftNM
DomizioP
2008 Staphylococcal enterotoxins G and I, a cause of severe but reversible neonatal enteropathy. Clin Gastroenterol Hepatol 6 251 254
38. KotlerDP
SandkovskyU
SchlievertPM
SordilloEM
2007 Toxic shock-like syndrome associated with staphylococcal enterocolitis in an HIV-infected man. CLIN INFECT DIS 44 e121 123
39. AmaralMM
CoelhoLR
FloresRP
SouzaRR
Silva-CarvalhoMC
2005 The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant Staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. J Infect Dis 192 801 810
40. KamarasJ
MurrellWG
2001 Intestinal epithelial damage in sids babies and its similarity to that caused by bacterial toxins in the rabbit. Pathology 33 197 203
41. KamarasJ
MurrellWG
2001 The effect of bacterial enterotoxins implicated in SIDS on the rabbit intestine. Pathology 33 187 196
42. da SilvaMCA
ZahmJ-M
GrasD
BajoletO
AbelyM
2004 Dynamic interaction between airway epithelial cells and Staphylococcus aureus. Am J Physiol Lung Cell Mol Physiol 287 L543 551
43. ZetolaN
FrancisJS
NuermbergerEL
BishaiWR
2005 Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis 5 275 286
44. TroemelER
ChuSW
ReinkeV
LeeSS
AusubelFM
2006 p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2 e183
45. Mashburn-WarrenL
McLeanRJC
WhiteleyM
2008 Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology 6 214 219
46. HodgkinJ
KuwabaraPE
CorneliussenB
2000 A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol 10 1615 1618
47. NicholasHR
HodgkinJ
2004 The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr Biol 14 1256 1261
48. NicholasHR
HodgkinJ
2009 The C. elegans Hox gene egl-5 is required for correct development of the hermaphrodite hindgut and for the response to rectal infection by Microbacterium nematophilum. Dev Biol 329 16 24
49. ShimodaM
OhkiK
ShimamotoY
KohashiO
1995 Morphology of defensin-treated Staphylococcus aureus. Infect Immun 63 2886 2891
50. WongD
BazopoulouD
PujolN
TavernarakisN
EwbankJJ
2007 Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol 8 R194
51. GeijtenbeekTBH
GringhuisSI
2009 Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol 9 465 479
52. YuY
YuY
HuangH
FengK
PanM
2007 A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J Immunol 179 8425 8434
53. CashHL
WhithamCV
BehrendtCL
HooperLV
2006 Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313 1126 1130
54. KabelitzD
MedzhitovR
2007 Innate immunity—cross-talk with adaptive immunity through pattern recognition receptors and cytokines. Curr Opin Immunol 19 1 3
55. HuffmanDL
AbramiL
SasikR
CorbeilJ
van der GootFG
2004 Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci USA 101 10995 11000
56. Van GilstMR
HadjivassiliouH
YamamotoKR
2005 A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci USA 102 13496 13501
57. VanceRE
IsbergRR
PortnoyDA
2009 Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host & Microbe 6 10 21
58. PujolN
LinkEM
LiuLX
KurzCL
AlloingG
2001 A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol 11 809 821
59. TenorJL
AballayA
2008 A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep 9 103 109
60. TakedaK
KaishoT
AkiraS
2003 Toll-like receptors. Annu Rev Immunol 21 335 376
61. O'RourkeD
BabanD
DemidovaM
MottR
HodgkinJ
2006 Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res 16 1005 1016
62. SchulenburgH
HoeppnerMP
WeinerJ
Bornberg-BauerE
2008 Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213 237 250
63. SigmondT
FehérJ
BaksaA
PástiG
PálfiaZ
2008 Qualitative and quantitative characterization of autophagy in Caenorhabditis elegans by electron microscopy. Meth Enzymol 451 467 491
64. JiaK
ThomasC
AkbarM
SunQ
Adams-HuetB
2009 Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci USA 106 14564 14569
65. DereticV
2009 Multiple regulatory and effector roles of autophagy in immunity. Curr Opin Immunol 21 53 62
66. WarehamDW
PapakonstantinopoulouA
CurtisMA
2005 The Pseudomonas aeruginosa PA14 type III secretion system is expressed but not essential to virulence in the Caenorhabditis elegans-P. aeruginosa pathogenicity model. FEMS Microbiol Lett 242 209 216
67. McBroomAJ
KuehnMJ
2007 Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol 63 545 558
68. TroemelE
ChuS
ReinkeV
LeeS
AusubelFM
2006 p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2 e183
69. EstesKA
DunbarTL
PowellJR
AusubelFM
TroemelER
2010 bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc Natl Acad Sci USA 107 2153 2158
70. MedzhitovR
2009 Approaching the asymptote: 20 years later. Immunity 30 766 775
71. VanceR
IsbergR
PortnoyD
2009 Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host & Microbe 6 10 21
72. Fernandes-AlnemriT
WuJ
YuJ-W
DattaP
MillerB
2007 The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death and Differentiation 14 1590 1604
73. VollmerW
BlanotD
de PedroMA
2008 Peptidoglycan structure and architecture. FEMS Microbiol Rev 32 149 167
74. BorjessonDL
KobayashiSD
WhitneyAR
VoyichJM
ArgueCM
2005 Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J Immunol 174 6364 6372
75. GemsD
McElweeJJ
2005 Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech Ageing Dev 126 381 387
76. DannSM
EckmannL
2007 Innate immune defenses in the intestinal tract. Curr Opin Gastroenterol 23 115 120
77. SalzetM
2001 Vertebrate innate immunity resembles a mosaic of invertebrate immune responses. Trends Immunol 22 285 288
78. AlperS
LawsR
LackfordB
BoydWA
DunlapP
2008 Identification of innate immunity genes and pathways using a comparative genomics approach. Proc Natl Acad Sci USA 105 7016 7021
79. MuirRE
TanMW
2008 Virulence of Leucobacter chromiireducens subsp. solipictus to Caenorhabditis elegans: characterization of a novel host-pathogen interaction. Appl Environ Microbiol 74 4185 4198
80. PearceEL
WalshMC
CejasPJ
HarmsGM
ShenH
2009 Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460 103 107
81. AnagnostouSH
ShepherdPR
2008 Glucose induces an autocrine activation of the Wnt/beta-catenin pathway in macrophage cell lines. Biochem J 416 211 218
82. BensingerSJ
TontonozP
2008 Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454 470 477
83. FroyO
2005 Regulation of mammalian defensin expression by Toll-like receptor-dependent and independent signalling pathways. Cell Microbiol 7 1387 1397
84. EvansEA
ChenWC
TanM-W
2008 The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell 7 879 893
85. WatanabeA
MiyazawaS
KitamiM
TabunokiH
UedaK
2006 Characterization of a novel C-type lectin, Bombyx mori multibinding protein, from the B. mori hemolymph: mechanism of wide-range microorganism recognition and role in immunity. J Immunol 177 4594 4604
86. YuXQ
GanH
KanostMR
1999 Immulectin, an inducible C-type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidase. Insect Biochem Mol Biol 29 585 597
87. YuX-Q
KanostMR
2004 Immulectin-2, a pattern recognition receptor that stimulates hemocyte encapsulation and melanization in the tobacco hornworm, Manduca sexta. Dev Comp Immunol 28 891 900
88. PowellJR
AusubelFM
2008 Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol Biol 415 403 427
89. WengL
DaiH
ZhanY
HeY
StepaniantsSB
2006 Rosetta error model for gene expression analysis. Bioinformatics 22 1111 1121
90. O'rourkeD
BabanD
DemidovaM
MottR
HodgkinJ
2006 Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Research 16 1005 1016
91. GrewalA
LambertP
StocktonJ
2007 Analysis of expression data: an overview. Current protocols in human genetics/editorial board, Jonathan L Haines [et al] Chapter 11: Unit11.14
92. Gravato-NobreMJ
NicholasHR
NijlandR
O'rourkeD
WhittingtonDE
2005 Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 171 1033 1045
93. PfafflMW
2001 A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45
94. HobertO
2002 PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques 32 728 730
95. TimmonsL
CourtDL
FireA
2001 Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103 112
96. ReboulJ
VaglioP
RualJ-F
LameschP
MartinezM
2003 C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet 34 35 41
97. RualJF
CeronJ
KorethJ
HaoT
NicotAS
2004 Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14 2162 2168
98. TusherVG
TibshiraniR
ChuG
2001 Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98 5116 5121
99. PetalcorinMIR
JoshuaGW
AgapowP-M
DolphinCT
2005 The fmo genes of Caenorhabditis elegans and C. briggsae: characterisation, gene expression and comparative genomic analysis. Gene 346 83 96
100. PauliF
LiuY
KimYA
ChenP-J
KimSK
2006 Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 133 287 295
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System inČlánek A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 7- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The Mouse Resistance Protein Irgm1 (LRG-47): A Regulator or an Effector of Pathogen Defense?
- Leprosy and the Adaptation of Human Toll-Like Receptor 1
- Intergenomic Arms Races: Detection of a Nuclear Rescue Gene of Male-Killing in a Ladybird
- The Role of Chemokines during Viral Infection of the CNS
- Bottlenecks and the Maintenance of Minor Genotypes during the Life Cycle of
- DNA Damage Triggers Genetic Exchange in
- The Role of Coupled Positive Feedback in the Expression of the SPI1 Type Three Secretion System in
- Uropathogenic Modulates Immune Responses and Its Curli Fimbriae Interact with the Antimicrobial Peptide LL-37
- Biogenesis of the Inner Membrane Complex Is Dependent on Vesicular Transport by the Alveolate Specific GTPase Rab11B
- A Spatio-Temporal Analysis of Matrix Protein and Nucleocapsid Trafficking during Vesicular Stomatitis Virus Uncoating
- Hepatitis B Virus Polymerase Blocks Pattern Recognition Receptor Signaling via Interaction with DDX3: Implications for Immune Evasion
- Quasispecies Theory and the Behavior of RNA Viruses
- Bid Regulates the Pathogenesis of Neurotropic Reovirus
- Distinct Roles for Dectin-1 and TLR4 in the Pathogenesis of Keratitis
- Unexpected Inheritance: Multiple Integrations of Ancient Bornavirus and Ebolavirus/Marburgvirus Sequences in Vertebrate Genomes
- Balanced Nuclear and Cytoplasmic Activities of EDS1 Are Required for a Complete Plant Innate Immune Response
- Adaptation of Hepatitis C Virus to Mouse CD81 Permits Infection of Mouse Cells in the Absence of Human Entry Factors
- An Outer Membrane Receptor of Involved in Zinc Acquisition with Vaccine Potential
- Inositol Hexakisphosphate-Induced Autoprocessing of Large Bacterial Protein Toxins
- Plus- and Minus-End Directed Microtubule Motors Bind Simultaneously to Herpes Simplex Virus Capsids Using Different Inner Tegument Structures
- Distinct Pathogenesis and Host Responses during Infection of by and
- Can Bacteria Evolve Resistance to Quorum Sensing Disruption?
- RNA Virus Replication Complexes
- PPARγ and LXR Signaling Inhibit Dendritic Cell-Mediated HIV-1 Capture and -Infection
- The Virulence Protein SopD2 Regulates Membrane Dynamics of -Containing Vacuoles
- Genome-Wide Mutagenesis Reveals That ORF7 Is a Novel VZV Skin-Tropic Factor
- Adaptive Evolution of Includes Retroviral Insertion and Positive Selection at Two Clusters of Residues Flanking the Substrate Groove
- A Systems Immunology Approach to Plasmacytoid Dendritic Cell Function in Cytopathic Virus Infections
- Transduction of Human T Cells with a Novel T-Cell Receptor Confers Anti-HCV Reactivity
- Identification of GBV-D, a Novel GB-like Flavivirus from Old World Frugivorous Bats () in Bangladesh
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- HIV gp41 Engages gC1qR on CD4+ T Cells to Induce the Expression of an NK Ligand through the PIP3/H2O2 Pathway
- Oxidation of Helix-3 Methionines Precedes the Formation of PK Resistant PrP
- Protection from the 2009 H1N1 Pandemic Influenza by an Antibody from Combinatorial Survivor-Based Libraries
- Murine Gamma-Herpesvirus 68 Hijacks MAVS and IKKβ to Initiate Lytic Replication
- Viral Protein Inhibits RISC Activity by Argonaute Binding through Conserved WG/GW Motifs
- TOPO3α Influences Antigenic Variation by Monitoring Expression-Site-Associated Switching in
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- The Meningococcal Vaccine Candidate Neisserial Surface Protein A (NspA) Binds to Factor H and Enhances Meningococcal Resistance to Complement
- Network Modeling Reveals Prevalent Negative Regulatory Relationships between Signaling Sectors in Arabidopsis Immune Signaling
- Endothelial Galectin-1 Binds to Specific Glycans on Nipah Virus Fusion Protein and Inhibits Maturation, Mobility, and Function to Block Syncytia Formation
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
- Integration Preferences of Wildtype AAV-2 for Consensus Rep-Binding Sites at Numerous Loci in the Human Genome
- Epigenetic Analysis of KSHV Latent and Lytic Genomes
- Vaccinia Virus–Encoded Ribonucleotide Reductase Subunits Are Differentially Required for Replication and Pathogenesis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- RNA Virus Replication Complexes
- Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1
- Functional Genetic Diversity among Complex Clinical Isolates: Delineation of Conserved Core and Lineage-Specific Transcriptomes during Intracellular Survival
- Extreme CD8 T Cell Requirements for Anti-Malarial Liver-Stage Immunity following Immunization with Radiation Attenuated Sporozoites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



