-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Association of Tat with Promoters of PTEN and PP2A Subunits Is Key to Transcriptional Activation of Apoptotic Pathways in HIV-Infected CD4+ T Cells
Apoptosis in HIV-1-infected CD4+ primary T cells is triggered by the alteration of the PI3K and p53 pathways, which converge on the FOXO3a transcriptional activator. Tat alone can cause activation of FOXO3a and of its proapoptotic target genes. To understand how Tat affects this pathway, we carried out ChIP-Chip experiments with Tat. Tat associates with the promoters of PTEN and two PP2A subunit genes, but not with the FOXO3a promoter. PTEN and PP2A encode phosphatases, whose levels and activity are increased when Tat is expressed. They counteract phosphorylation of Akt1 and FOXO3a, and so activate transcriptional activity of FOXO3a. FOXO3a promotes increased transcription of Egr-1, which can further stimulate the transcription of PTEN, thereby reinforcing the pathway that leads to FOXO3a transcriptional activation. RNAi experiments support the role of PTEN and PP2A in the initiation of the Tat-mediated cascade, which is critical to apoptosis. The increased accumulation of PTEN and PP2A subunit mRNAs during Tat expression is more likely to be the result of increased transcription initiation and not relief of promoter-proximal pausing of RNAPII. The Tat-PTEN and -PP2A promoter interactions provide a mechanistic explanation of Tat-mediated apoptosis in CD4+ T cells.
Published in the journal: Association of Tat with Promoters of PTEN and PP2A Subunits Is Key to Transcriptional Activation of Apoptotic Pathways in HIV-Infected CD4+ T Cells. PLoS Pathog 6(9): e32767. doi:10.1371/journal.ppat.1001103
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001103Summary
Apoptosis in HIV-1-infected CD4+ primary T cells is triggered by the alteration of the PI3K and p53 pathways, which converge on the FOXO3a transcriptional activator. Tat alone can cause activation of FOXO3a and of its proapoptotic target genes. To understand how Tat affects this pathway, we carried out ChIP-Chip experiments with Tat. Tat associates with the promoters of PTEN and two PP2A subunit genes, but not with the FOXO3a promoter. PTEN and PP2A encode phosphatases, whose levels and activity are increased when Tat is expressed. They counteract phosphorylation of Akt1 and FOXO3a, and so activate transcriptional activity of FOXO3a. FOXO3a promotes increased transcription of Egr-1, which can further stimulate the transcription of PTEN, thereby reinforcing the pathway that leads to FOXO3a transcriptional activation. RNAi experiments support the role of PTEN and PP2A in the initiation of the Tat-mediated cascade, which is critical to apoptosis. The increased accumulation of PTEN and PP2A subunit mRNAs during Tat expression is more likely to be the result of increased transcription initiation and not relief of promoter-proximal pausing of RNAPII. The Tat-PTEN and -PP2A promoter interactions provide a mechanistic explanation of Tat-mediated apoptosis in CD4+ T cells.
Introduction
HIV-1-infected CD4+ primary T cells progress to the G0 phase of the cell cycle and to cell death [1]. Apoptosis in these cells is triggered by the alteration of transcriptional pathways that converge on the Forkhead box O3 (FOXO3a) transcriptional activator. The induction of FOXO3a target genes, such as Bcl-2-like 11 (BCL2L11 or Bim), TNF-related apoptosis-inducing ligand (TRAIL) and Fas ligand (FasL or CD95L), activates apoptotic intrinsic (via Bim) and extrinsic pathways [2], [3], indicating that HIV infection leads to apoptosis by the engagement of multiple apoptotic pathways. The induction of phosphatase and tensin homolog (PTEN) and FOXO3a was observed in cells that express only the Tat protein, suggesting that Tat may be a key player in the activation of these pathways.
PTEN reduces the phosphorylation of Akt1 and expression of PTEN is transcriptionally regulated by the Early Growth Response Protein 1 (Egr-1) [4], [5], [6]. Egr-1 is expressed at higher levels in HIV-infected T cells [1]. Increased expression of PTEN reduces serine/threonine protein kinase pAkt1 levels, which cause reduced phosphorylation of FOXO3a. Unphosphorylated FOXO3a translocates to the nucleus and becomes transcriptionally active [7].
Transcription of HIV genes from the HIV long terminal repeat (LTR) is strictly dependent on Tat, which interacts with the Positive Transcription Elongation Factor b (P-TEFb) and histone acetyltransferases [8]. The interaction with P-TEFb occurs at the trans-activation-responsive (TAR) element of the nascent RNA and mediates the relief of RNA polymerase II (RNAPII) pausing that occurs at TAR. Tat transcriptional activity is also dependent on lysine acetylation mediated by nuclear histone acetyltransferases p300/CBP (E1A binding protein p300/CREB binding protein) and PCAF (P300/CBP-associated factor). The p300/CBP complex is a transcriptional coactivator of Egr-1 [9], [10], [11], [12]. Tat may enhance the transcriptional activity of p300/CBP by increasing the histone acetyl transferase (HAT) activity on the PTEN promoter, as for histone H4 and the HIV LTR [13]. Inhibition of Sirtuin 1 (SIRT1) deacetylase activity by Tat [14], might also increase transcription of PTEN. Tat can be found in patients' serum [15], [16] and can cross the cell membrane to enter cells [17]. Tat could thus play a role in the apoptosis of uninfected cells by activating the PTEN-FOXO3a pathway after entry. The survival of memory CD4+ T cells correlates with the phosphorylated levels of FOXO3a. The levels of phospho-FOXO3a are reduced in HIV-infected individuals and are higher in elite controllers, who control viral replication to undetectable viremia in the absence of therapy [18], [19]. Activation of the PTEN-FOXO3a pathway via the Tat protein could be the mechanism by which apoptosis is triggered in HIV - infected and non-infected cells and explain the significant decline of the CD4+ T cell memory population in HIV-1-infected individuals [1].
Here we show that the Tat protein triggers apoptosis by altering the Akt-FOXO3a-Egr-1 pathway via its interaction with the promoters of two phosphatases, PTEN and Protein phosphatase 2 (PP2A).
Results
Tat-mediated cellular modulation of gene expression in Jurkat T cells
We reported that HIV-1 Tat-induced FOXO3a is a key mediator of apoptosis in HIV-1-infected primary human CD4 T lymphocytes [1]. To gain insight into the molecular mechanism by which Tat protein affects the PTEN-FOXO3a-Egr-1 signaling pathway, we investigated the impact of the HIV-1 Tat protein on the regulation of FOXO3a in Jurkat T cells. Jurkat T cells are susceptible to adenovirus infection due to high surface levels of the Coxsackie adenovirus receptor (CAR) [20]. Because retroviral transduction of primary CD4+ lymphocytes is inefficient, the use of adenovirus-mediated tat gene transfer in Jurkat T cells is a more amenable model for mechanistic studies. We investigated whether Tat expression in Jurkat cells resulted in modulatory effects on expression of proapoptotic genes, similar to those we observed in primary CD4+ T cells infected with HIVΔ2GFP, HIVΔ3GFP, and wild-type viruses [1]. Jurkat T cells were infected with an Ad-Tat vector expressing Tat in combination with the transactivator Ad-tTA, required to induce Tat expression, or Ad-tTA alone as a control. We conducted real-time RT-PCR using RNA obtained from Jurkat T cells infected with Ad-TatSF2 (a 101 amino acid wild type Tat protein from HIVSF2), Ad-TatSF2K28A,K50A, a mutant that does not associate with p300 [21]–[22]), Ad-TatSF2C25,30,35S, a mutant that does not interact with P-TEFb [23], and Ad-TatSF2G48-R57A, a mutant lacking the nuclear localization signal (NLS), in which the key residues of the NLS are substituted by alanines [24]. RNA was isolated from infected cells 24 48, and 72 hours post-infection at different MOI. Real-time RT-PCR was carried out with primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for normalization. Tat mRNA expression and Tat protein level accumulation was comparable for all cultures examined (Figure 1A and 1B). Intracellular staining for Tat on samples obtained 24 and 48 hrs after Ad-TatSF2 infection ranged between 40 and 60 percent positive cells (data not shown) and was found both in the cytoplasm and in the nucleus in the case of the wild type and the mutants excepts for TatSF2G48-R57A, with was detected virtually exclusively in the cytoplasm (Figure 1C). Levels of Tat expression obtained with different vectors were evaluated by flow cytometry. The Mean Fluorescence Intensity (MFI) for Tat expression after infection with Ad-Tat, eGFP-Tat (a retroviral vector expressing Tat) and HIV-Flag Tat (a HIV infectious virus in which Tat has been tagged with the FLAG epitope) is reported in Figure 1D. Tat MFI was approximately the same after retroviral expression vector or HIV infection and two-fold higher after Ad-TatSF2 infection (Figure 1D). Rates of early and late apoptosis were measured by evaluating the number of Annexin V+/7AAD - and Annexin V+/7AAD+ cells. A significant (p<0.05) increase in apoptosis was observed in Jurkat cells infected with Ad-TatSF2 (Figure 1E) compared to control or Tat mutants.
Fig. 1. Tat and Tat mutants expression and apoptosis in Jurkat T cells. 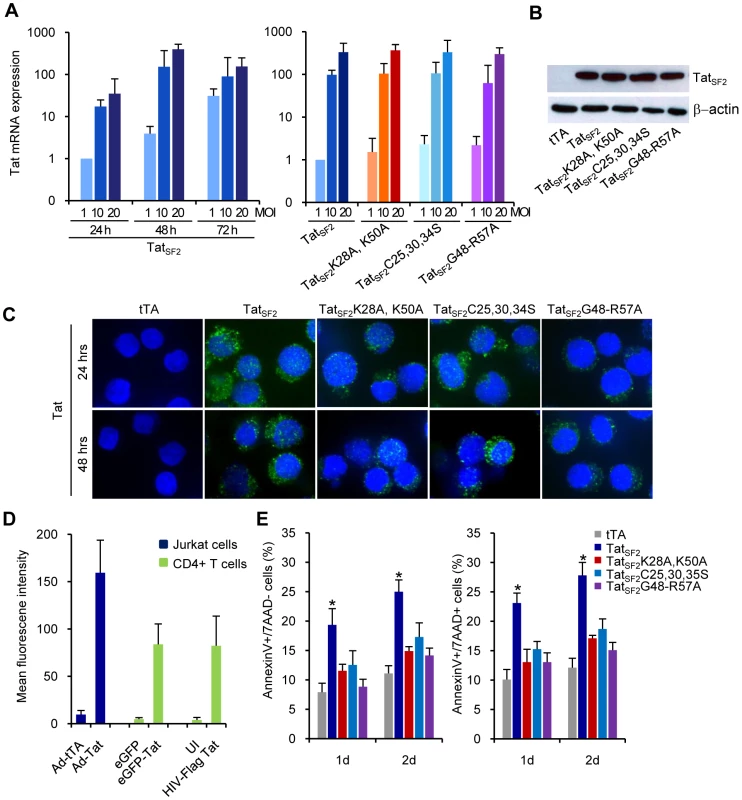
A. mRNA levels of Tat in Jurkat T cells expressing TatSF2 at 24, 48, and 72 hrs after infection at MOI of 1, 10, and 20 (first panel) or wt and Tat mutants at 48 hrs after infection at MOI of 1, 10, and 20 (second panel), analyzed by qRT-PCR. Results are normalized to GAPDH and reported as fold induction relative to Ad-Tat samples infected at MOI of 1. The means ± SEM derived from three independent experiments are reported. B. Western blot analysis of Tat expression in Jurkat cells. C. Detection of wild type and mutant Tat in Jurkat cells. Nuclei are counterstained with DRAG5. D. Tat protein mean fluorescence intensity (MFI) of three independent flow cytometric analyses of Jurkat cells, after infection with different Tat expressing viruses. E. Apoptosis in Tat-expressing Jurkat T cells. Levels of early and late apoptosis are reported as percentage of Jurkat T cells that stain for Annexin V only (left panel) or Annexin V and 7AAD (right panel). The means ± SEM of three experiments are shown. *, p<0.05 when TatSF2 is compared to tTA control. We found that genes up-regulated in primary CD4+ T cells infected with HIVΔ2GFP and HIVΔ3GFP or in HeLa cells infected with Ad-Tat [1] were similarly modulated in Jurkat T cells infected with Ad-TatSF2 (Figure 2A). The Tat mutants had substantially less effect on the expression of these genes. FOXO3a, Egr-1, and PTEN, and TRAIL, critical to apoptosis in primary CD4+ T cells [1], were also induced by Tat expression in Jurkat cells. We found increased accumulation of FOXO3a, Egr-1, and TRAIL as assessed cytofluorimetrically (Figure 2B). The pattern of protein expression was similar when Jurkat cells were infected with adenoviruses expressing two different Tat alleles, TatSF2 and TatHXB2 (a 86 amino acid Tat protein, which misses 15 residues at the carboxyl-terminus) and was not significantly altered after infection with the mutants. The subcellular localization of FOXO3a was assessed by immunofluorescence microscopy using antibodies against FOXO3a and pFOXO3a (Figure 2C and 2D). In the Ad-tTA control, the majority of FOXO3a was phosphorylated (inactive form) and distributed in the cytoplasm (Figure 2D, first column). In contrast, there was significantly less cytoplasmic pFOXO3a in cells infected with Ad-TatSF2 (Figure 2D, second column). We observed different results using anti-FOXO3a antibody. FOXO3a was mostly in the cytoplasm when cells were infected with Ad-tTA alone (Figure 2C, first column). However, Tat expression increased the amount and the translocation of FOXO3a from the cytoplasm to the nucleus (active form), indicating that FOXO3a was no longer phosphorylated (Figure 2C, second column). The intensity of nuclear FOXO fluorescence was higher in cells infected with Ad-TatSF2 than in those infected with Ad-tTA alone. Cells infected with the Tat mutants showed a patter similar to those infected with Ad-tTA. HIV-1 Tat thus increases intracellular levels of FOXO3a, as suggested by RT-PCR for the FOXO3a RNA.
Fig. 2. Tat-mediated cellular gene modulation in Jurkat T cells. 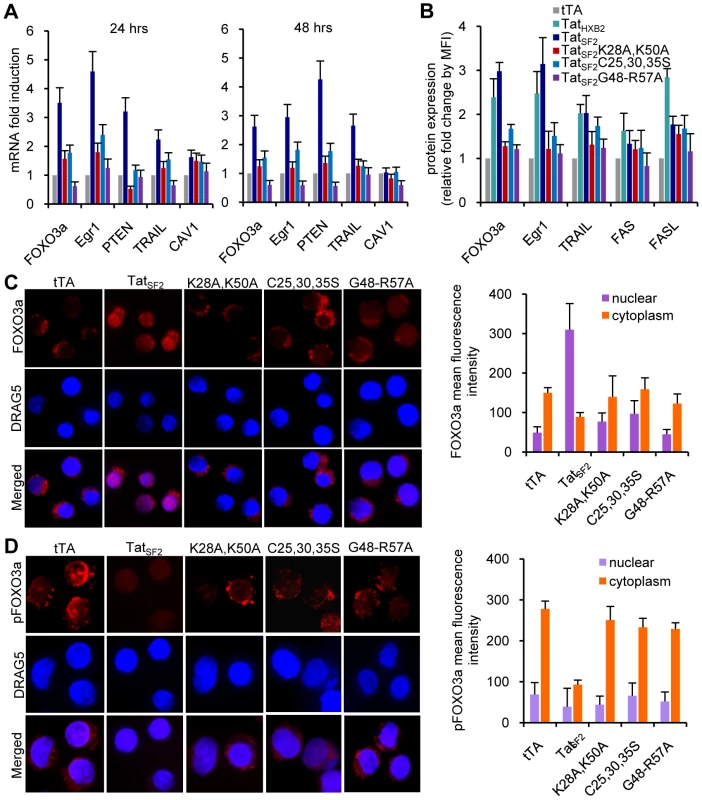
(A) mRNA levels of selected cellular genes 24 and 48 hours after infection with adenoviral vectors expressing wild type Tat and Tat mutants. B. Protein levels of cellular genes analyzed by flow cytometry analysis. Results are reported as fold increase of mean fluorescence intensity (MFI) relative to the tTA control. C. HIV-1 Tat increases FOXO3a nuclear localization in Jurkat T cells. Jurkat T cells infected with Ad-tTA or Ad-tTA+Ad-TatSF2 were analyzed by confocal microscopy 24 hrs after infection. Cells were stained with DRAG5 to visualize the nucleus (blue), and FOXO3a cellular localization (red) was detected using antibodies against FOXO3a (C) or p-FOXO3a (D). A quantitative analysis of nuclear and cytoplasmic FOXO3a or p-FOXO3a fluorescence intensity is provided at the right of each panel. These data confirm the role of Tat in the activation of FOXO3a and in the induction of its target genes involved in apoptosis in Jurkat cells and validate its use to investigate the mechanisms by which HIV-1 Tat affects the PTEN-FOXO3a pathway in primary CD4+ T cells.
Tat associates with the PPP2R1B and PPP2R5E promoters to increase PPP2R1B and PPP2R5E RNA and protein levels as well as PP2A activity in Jurkat cells
How does Tat modulate cellular gene expression during HIV-1 mediated apoptosis? We used ChIP coupled with promoter DNA microarray analysis (ChIP-Chip) [25] to identify genes whose promoters may associate with Tat. We infected Jurkat cells for 6 hours with an adenovirus expressing FLAG-tagged TatSF2 or the mutant Ad-TatSF2G48-R57A lacking the NLS as a negative control. An anti-FLAG antibody suitable for use in ChIP experiments was used because of its lack of background [26], [27]. Two independent experiments were analyzed. We selected a stringent P value threshold of 0.001 to identify genes bound by Tat (Figure 3A). In cells expressing TatSF2, we identified 450 promoters that were occupied by Tat (P<0.001). Control experiments in cells expressing the negative control TatSF2G48-R57A showed only 12 positive signals, which is an acceptable background for ChIP-Chip experiments, supporting the specificity of the results observed with Ad-TatSF2. The 450 genes whose promoters associate with TatSF2 encode proteins that affect many cellular processes, including transcriptional regulation, apoptosis, cell cycle, and immune response. They are listed in Supplemental Table S1, with the P value and the fold enrichment of their association to the promoter compared to input DNA.
Fig. 3. Tat associates with the PPP2R1B and PPP2R5E promoter and increases protein levels of PPP2R1B and PPP2R5E and PP2A activity in Jurkat cells. 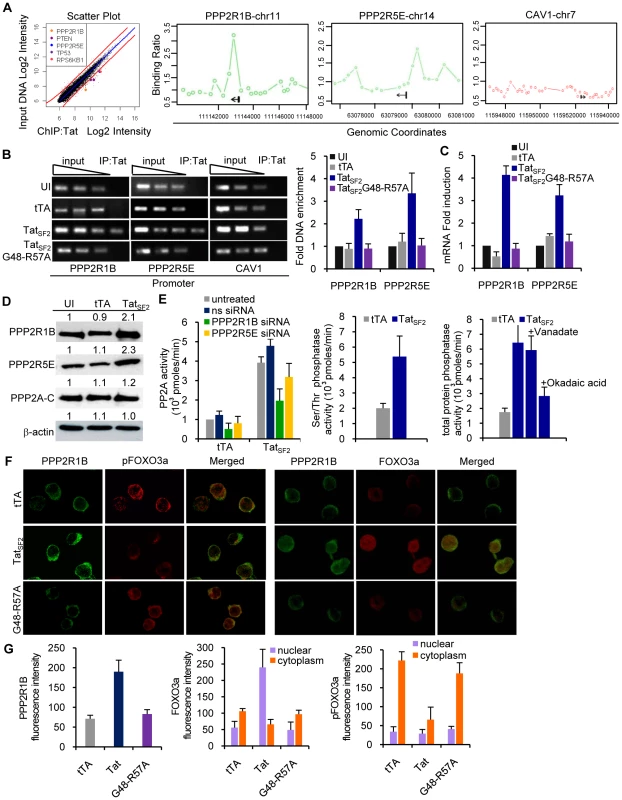
A. Genes enriched in the Tat-immunoprecipitated DNA and associated with the PI3K pathway show a hybridization intensity with a P value lower than 0.001 (hybridization intensities with higher P values fall within the red lines). PPP2R1B, PPP2R5E, and Caveolin 1 (CAV1, negative control) promoter enrichment ratio (ChIP versus total input DNA in ChIP-on-Chip analysis) in Jurkat cells expressing TatSF2. B. ChIP analysis of the PPP2R1B and PPP2R5E promoters in Jurkat T cells expressing TatSF2. DNA from input (90, 30, 10 ng of DNA) and immunoprecipitated samples (3 ng of DNA) was amplified by standard PCR (P2 set of primers, see Supplemental Table S2) and run on 2% agarose gel (second and third panels). One representative experiment is shown in the 3 left panels. In the right panel, the average fold enrichment of a certain promoter in the immunoprecipitated DNA relative to input DNA ± SEM from three independent qPCR experiments is reported. All cycle threshold (Ct) values obtained with 10 ng of immunoprecipitated DNAs were compared with the Ct value obtained with 10 ng of the corresponding input DNA. C. mRNA levels of PPP2R1B and PPP2R5E in Jurkat T cells expressing TatSF2 or the mutant TatSF2G48-R57A, analyzed by qRT-PCR. Results are normalized to GAPDH and reported as fold induction relative to uninfected samples. The means ± SEM of three experiments are shown. D. Western blot analysis of PPP2R1B, PPP2R5E, and the catalytic subunit PP2A-C. Fold-increase compared to the uninfected control (UI) is indicated above the band. E. PP2A enzyme activity in lysates from Jurkat cells expressing TatSF2 alone or in the presence of siRNAs (left panel); serine/threonine phosphatase activity (middle panel), and total phosphatase activity (right panel) in lysates from Jurkat cells infected with Adeno-TatSF2 or with the Adeno-tTA control. Inhibition of tyrosine phosphatases and serine/threonine phosphatases was carried out by incubation of the lysate with sodium vanadate (1 mM) or okadaic acid (0.25 mM) in the phosphatase assay buffer. F. Tat increases PP2A expression and FOXO3a nuclear localization in Jurkat T cells. Jurkat T cells expressing tTA alone, TatSF2+tTA, or TatSF2G48-R57A +tTA were stained with antibodies against PPP2R1B (first and forth columns of panels, green), pFOXO3a (second column, red), and FOXO3a (forth column, red) and analyzed by confocal microscopy. Merged images are shown in the panels in the third and sixth columns. G. Quantitative analysis of the fluorescence intensity of cytoplasmic PPP2R1B, and of cytoplasmic and nuclear FOXO3a and pFOXO3a in Jurkat T cells expressing or tTA alone, TatSF2+tTA, or TatSF2G48-R57A +tTA. PPP2R1B and FOXO3a fluorescence was expressed as total intensity per cell (pixels above threshold x fluorescence intensity). Bars indicate the mean ± SEM of triplicate assays from two separate experiments. At least 100 cells were counted for each condition. To identify the cellular functions affected by the Tat protein, we compared the list of genes occupied by Tat with the biological pathways annotated by the Ingenuity Pathway Analysis (IPA). We found that Tat target genes are significantly associated with promoters of genes that are part of a few pathways (Table 1). A subset of these genes belongs to the PI3K signaling pathway (p = 3.39E-03) (Figure 3A). Among these genes were PTEN, a gene that encodes a truncated, non-functional phosphatase in Jurkat cells [28], [29], and PPP2R1B and PPP2R5E, regulatory subunits of protein phosphatase 2A (PP2A). PPP2R1B (PR65β) is a regulatory subunit A β isoform, tightly associated with the PP2A catalytic subunit C, to form a scaffold onto which the appropriate B subunit can bind. PPP2R5E (B56ε) is a member of the B56 regulatory subunit ε isoform involved in multiple signaling pathways [30], [31], [32], [33], [34], [35], [36], [37]. PP2A affects the phosphorylation status of Akt1 and FOXO3a [38], [39], [40], [41], [42]. High levels of PP2A correlate with reduced phosphorylation of FOXO3a and consequently increase its transcriptional activity [43], [44]. The ChIP-Chip results of Tat binding to the promoters of PPP2R1B and PPP2R5E were further validated by conventional ChIP, performed using site-specific primers on chromatin precipitated from uninfected cells and cells expressing TatSF2 or tTA alone. Primers were designed near the site represented by the oligonucleotides on the promoter arrays that provided a positive signal and were used for qPCR amplification of the corresponding sequences present in the immunoprecipitation-captured chromatin. When the immunoprecipitated DNA samples were evaluated by qPCR, PPP2R1B and PPP2R5E promoter sequences were enriched in IP-captured chromatin from cells expressing TatSF2 compared to the control cells (Figure 3B). To determine whether association of Tat with the PPP2R1B and PPP2R5E promoters affects gene expression, we carried out qRT-PCR with RNA from Tat-expressing cells. mRNA levels of PPP2R1B and PPP2R5E were elevated in cells that express TatSF2, compared to untreated cells or cells that express tTA alone (Figure 3C).
Tab. 1. Analysis of promoters found associated with HIV-Tat in jurkat cells. 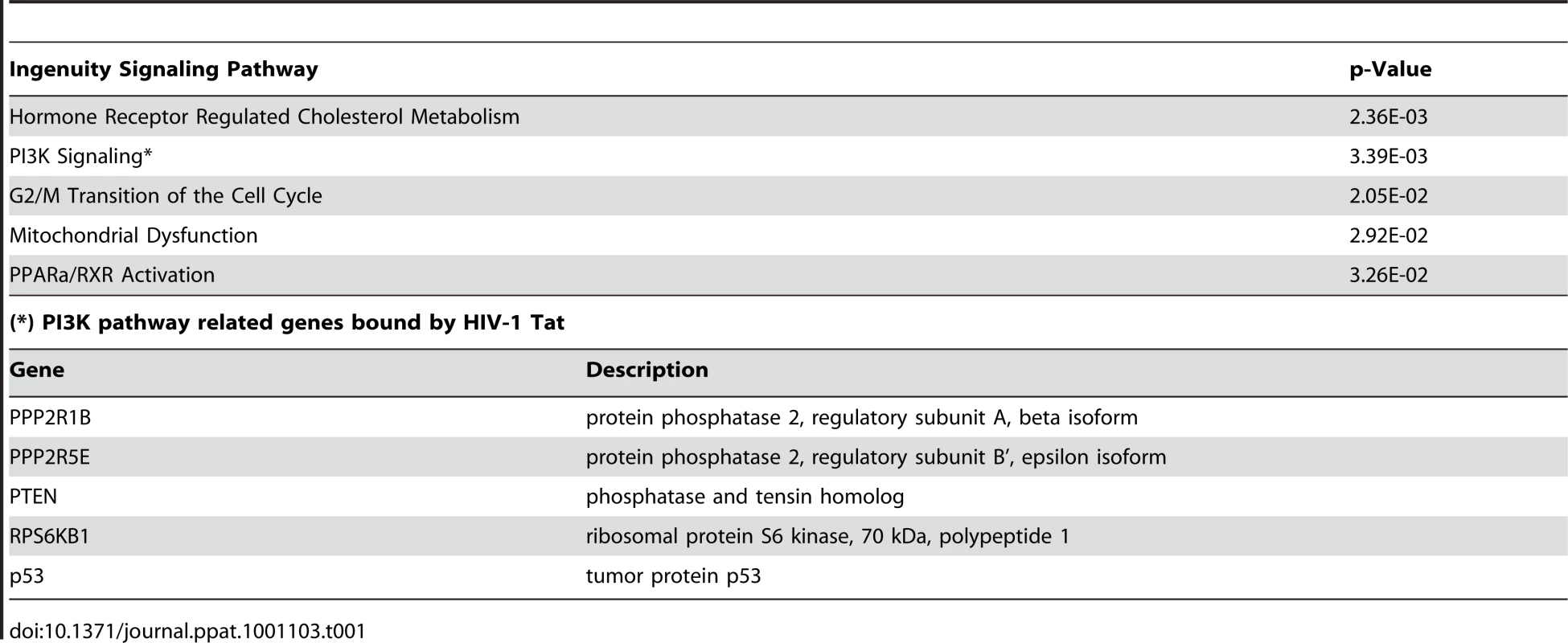
These results confirmed the association of Tat with the PPP2R1B and PPP2R5E promoters identified by the ChIP-Chip analysis and support a role for this association in increased gene expression of PPP2R1B and PPP2R5E seen in Tat expressing Jurkat cells.
We further evaluated the effects of Tat on the protein levels of PPP2R1B and PPP2R5E by immunoblot. PP2A is a ubiquitous enzyme with pleiotropic functions. PP2A is a heterotrimer that consists of a catalytic C subunit, a regulatory A subunit, and a variable regulatory B subunit. Regulation is accomplished mainly by members of a family of regulatory subunits, which determine the substrate specificity. The protein content of PP2A-C was not altered in cells infected with Ad-TatSF2 or Ad-tTA (Figure 3D). In contrast, an approximately two-fold increase in the amount of PPP2R1B and PPP2R5E protein was observed in cells infected with Ad-TatSF2 (Figure 3D).
Does the increase in PPP2R1B and PPP2R5E protein levels correlate with increased PP2A activity and serine/threonine phosphatase activity? We carried out an immunocomplex protein phosphatase assay and a malachite green assay on lysates from cells infected with Ad-TatSF2 or Ad-tTA alone, and treated with a non specific siRNA, a PPP2R1B siRNA, or a PPP2R5E siRNA (Figure 3E, left panel). Cells that express TatSF2 showed an approximately 2-fold increase in PP2A activity compared to cells that were infected with Ad-tTA only or were treated with PP2A subunit specific siRNAs, and a similar two-fold increase in total serine/threonine phosphatase activity (Figure 3E, middle panel), linking increased PP2A activity to the increase in PPP2R1B and PPP2R5E. When we measured total phosphatase activity, we found an approximately 4-fold increase in cells infected with Ad-TatSF2+Ad-tTA compared to the Ad-tTA alone control. Total phosphatase activity in cells that express Tat was reduced to approximately two-fold when cells were treated with okadaic acid, an inhibitor of PP2A, but not when treated with sodium vanadate, a generic inhibitor of tyrosine phosphatases (Figure 3E, right panel). The increase in PPP2R1B and PPP2R5E protein induced by Tat is therefore the critical determinant of the increased serine/threonine phosphatase activity observed in Tat-expressing cells.
We examined the cellular localization of PP2A and FOXO3a in the presence or absence of Tat. Jurkat T cells were infected for 24 hours with Ad-TatSF2 + Ad-tTA or Ad-tTA alone. The subcellular localization of FOXO3a and PPP2R1B was assessed by immunofluorescence microscopy using antibodies against pFOXO3a, FOXO3a, and PPP2R1B (Figure 3F). Cells infected with Ad-TatSF2 revealed stronger fluorescence intensity of PPP2R1B compared to those infected with Ad-tTA alone. This observation confirmed the effect of Tat on the PPP2R1B protein levels detected by Western blot analysis (Figure 3D). Tat expression was associated with increased amounts of nuclear FOXO3a (Figure 3F and 3G). In contrast, FOXO3a was detected predominantly in the cytoplasm in cells infected with Ad-tTA alone or the Tat mutant (Figure 3F, 3G). These observations further confirm that HIV-1 Tat increases PP2A expression and FOXO3a nuclear translocation.
siRNA-mediated PP2A knockdown reduces Tat-induced apoptosis in Jurkat cells
We next investigated the contribution of PP2A to Tat-induced apoptosis using siRNA-mediated knockdown of the PP2A subunits. Jurkat cells were transfected with short interfering RNA (siRNA) targeting PPP2R1B, PPP2R5E or nonspecific siRNA and then infected with Ad-tTA or Ad-TatSF2+Ad-tTA. mRNA levels of PPP2R1B and PPP2R5E were reduced by the PPP2R1B and PPP2R5E-specific siRNAs, respectively, but not by the control siRNA 24 hrs after infection (Figure 4A). The siRNAs targeting the PPP2R1B and PPP255E transcripts also reduced the corresponding protein levels (Figure 4C). Control siRNA had no effect. Levels of both PP2A subunits and of FOXO3a increased after Ad-TatSF2 infection (Figure 3D and 4C). Reduction of PP2A subunits induced by the siRNAs resulted in reduced FOXO3a mRNA and protein (Figure 4A and 4C). Transfection of PP2A subunit siRNAs reduced expression of some of the FOXO3a target genes upregulated after Ad-TatSF2 infection (Figure 4B). Furthermore, the reduction of PPP2R1B and PPP2R5E subunits also resulted in an increase of phosphorylated Akt1 and phosphorylated FOXO3a, supporting the role of the PP2A subunits in the accumulation of transcriptionally active, non phosphorylated FOXO3a and of its target genes (Figure 4C). We evaluated the role of the inhibition of PP2A on the expression of Egr-1, GADD45A and TRAIL, three FOXO3a target genes in Tat-expressing cells [1]. Treatment with siRNAs that target PPP2R1B and/or PPP2R5E reduced expression of Egr-1, GADD45A and TRAIL. The PPP2R1B siRNA had a more pronounced effect on the levels of FOXO3a and pFOXO3a proteins and of Egr-1 mRNA than did treatment of cells with PPP2R5E siRNA. (Figure 4A, 4B, and 4C).
Fig. 4. siRNA-mediated knockdown of PP2A reduces Tat-induced apoptosis in Jurkat T cells. 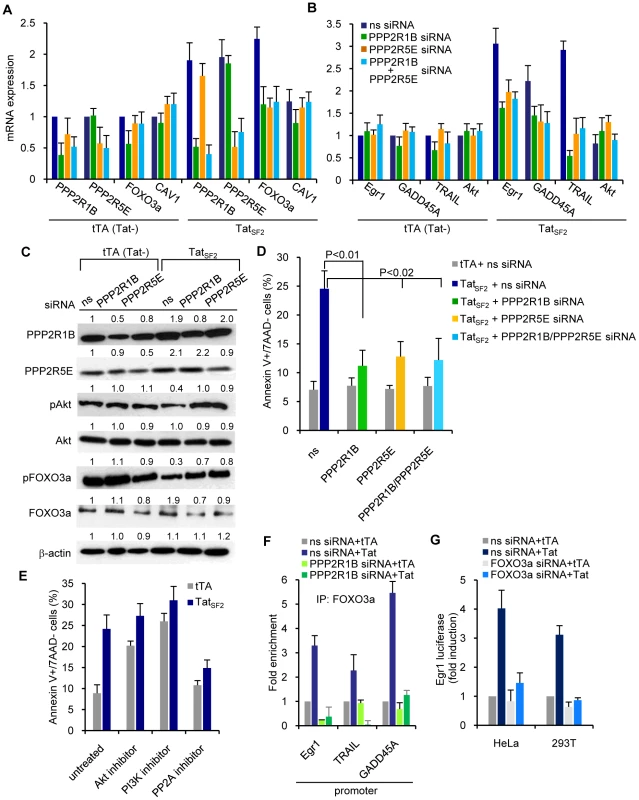
mRNA expression levels of (A) PPP2R1B and PPP2R5E and FOXO3a and (B) Egr1, GADD45A, and TRAIL [1] in Jurkat T cells expressing TatSF2+tTA or tTA alone. Results are normalized to GAPDH and reported as fold induction relative to tTA expressing cells treated with ns siRNA. C. Western blot analysis of cell lysates treated with PPP2R1B and/or PPP2R5E siRNA or ns siRNA. D. Levels of apoptosis in the same cells 48 h after siRNA transduction, measured by staining for Annexin V and 7AAD. The means ± SEM of three independent experiments are reported. E. Levels of apoptosis in Jurkat cells infected with Ad-tTA or Ad-Tat and treated with an Akt inhibitor (50 µM Akt1-1/2), a PI3K inhibitor (10 nM LY294002), or a PP2A inhibitor (100 nM okadaic acid), measured by staining for Annexin V and 7AAD. F. ChIP analysis of the promoters of three FOXO3a target genes. ChIP was carried out with an anti-FOXO3a antibody in lysates of Jurkat T cells treated with ns siRNA or PPP2R1B siRNA and expressing TatSF2 or TatSF2K28A,K50A. Recovered DNA was analyzed by qPCR using primers specific for the Egr1, TRAIL, and GADD45A promoters. The average fold enrichment relative to tTA control from two independent experiments is reported. G. Luciferase activity of lysates from cells expressing tTA or TatSF2 and transfected with an Egr1-luciferase reporter vector. Firely luciferase activity was normalized to Renilla luciferase activity and results are reported as fold induction relative to cells treated with ns siRNA in the presence of tTA. As we previously linked the Tat-mediated increase of FOXO3a to apoptosis, we evaluated the impact of PP2A subunit siRNAs on Tat-induced apoptosis. siRNAs that target PPP2R1B and/or PPP2R5E in Jurkat cells after Ad-TatSF2 infection caused a statistically significant decrease in apoptosis (Figure 4D). The same result was observed with okadaic acid, a PP2A inhibitor, but not with Akt1-1/2 or LY294002, an Akt and a PI3K inhibitor (Figure 4E). These experiments support a direct role of the PP2A subunits, but not of the above kinases, in Tat-mediated apoptosis.
The effect of siRNA-mediated inhibition of PPP2R1B in cells that do or do not express Tat was also evaluated by FOXO3a ChIP and amplification of promoters usually targeted by FOXO3a. Cells were transfected with PPP2R1B siRNA or control siRNA and then infected with Ad-TatSF2. Egr-1, TRAIL, and GADD45A promoters were amplified by PCR using the DNA recovered after FOXO3a ChIP. As expected, the amounts of promoter sequence DNA detected by PCR were increased when the anti-FOXO3a immunoprecipitations of chromatin lysates were performed using Tat-expressing cells compared to cells infected with tTA alone and transfected with ns siRNA (Figure 4F) In contrast, PPP2R1B siRNA treatment was associated with reduced recovery of the same promoter DNAs. Taken together, these data indicate that Tat-induced PP2A subunits can affect transcriptional upregulation of FOXO3a-dependent genes by increasing the amount of transcriptionally active FOXO3a and its binding to the promoters of target genes.
To further evaluate whether the detection of FOXO3a at the Egr-1 promoter plays a role in the transcriptional regulation of Egr-1, we used a dual luciferase reporter assay. HeLa cells were transfected with an Egr-1-luciferase vector with or without FOXO3a siRNA for 24 hours, followed by infection with Ad-TatSF2. TatSF2 expression increased activity of the Egr-1 promoter by approximately four-fold compared to the control (Figure 4G). In contrast, transfection with FOXO3a siRNA reduced Tat-induced Egr-1 promoter activity in HeLa cells (Figure 4G). Similar results were observed when the experiment was carried out with 293T cells transfected with FOXO3a siRNA. Treatment with FOXO3a siRNA reduced expression of both Egr-1 and PTEN mRNA in HIV-infected HeLa cells [1]. Thus Tat can indirectly stimulate Egr-1 promoter activity by increasing the levels of transcriptionally active FOXO3a, which in turn can associate with the Egr-1 promoter. Increased trascription of Egr-1 further stimulated PTEN gene expression.
Tat association with the PTEN promoter
PTEN mRNA is upregulated in HIV-1 infected primary CD4+ T lymphocytes and in Tat-expressing Jurkat T cells [1]. PTEN is a key regulator of the PI3K/Akt1 pathway and controls the phosphorylation status of Akt1 and, indirectly, FOXO3a [45], [46]. High levels of PTEN reduce Akt1 phosphorylation with consequent reduced phosphorylation of FOXO3a. Non-phosphorylated FOXO3a translocates into the nucleus where it is transcriptionally active and increases the expression of FOXO3a transcription-dependent genes involved in either the extrinsic or the intrinsic apoptosis pathway. Furthermore, FOXO3a can also stimulate its own transcription [47]. TatSF2 association with the PTEN promoter was detected by the ChIP-Chip analysis (Figure 3A, 5A left panel) and by conventional ChIP analysis (Figure 5A, right panels) in Jurkat cells. Cells were infected with an Ad-TatSF2 with or without a FLAG tag fused to the tat gene and PCR was used to detect the PTEN promoter in chromatin immunoprecipitated with an anti-FLAG antibody. An enriched PTEN promoter DNA fragment was detected in cells expressing FLAG-TatSF2 but not in cells expressing TatSF2 without FLAG (Figure 5A, right panels). No PTEN amplification was found either in DNA from cells expressing tTA alone or from cells expressing the mutant TatSF2G48-R57A that lacks the NLS. Therefore Tat expression is associated with the PTEN promoter and with an increase in its transcription activity.
Fig. 5. Tat association with the PTEN promoter. 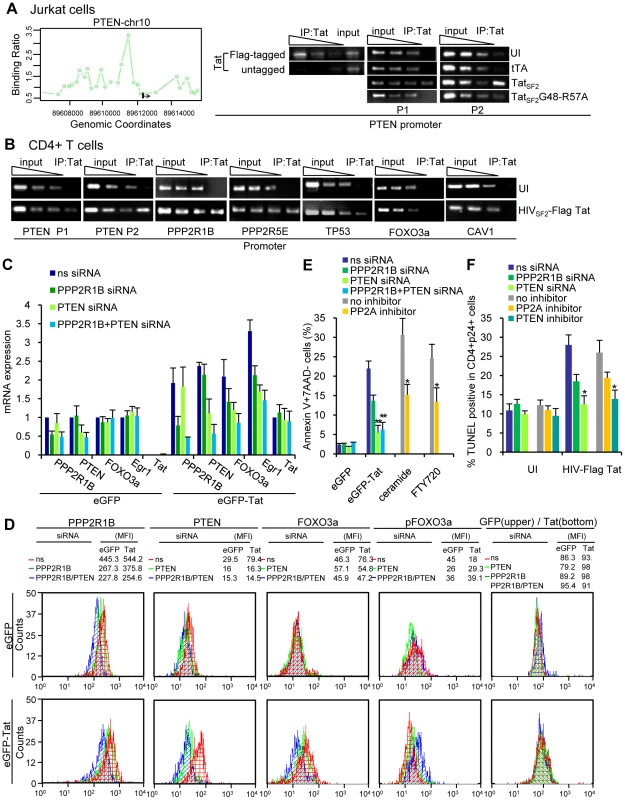
A. PTEN promoter enrichment ratio (ChIP DNA versus total genomic DNA in ChIP-Chip analysis) in Jurkat cells expressing TatSF2 (first panel). ChIP analysis of the PTEN promoter in Jurkat T cells: two fold dilutions, starting from 5 ng, of DNA immunoprecipitated with anti-Flag antibody from samples expressing TatSF2 with or without FLAG or 10 ng of input DNA, used as positive control, were amplified by PCR (first gel). In the second and third gel the signals obtained by PCR carried out with input DNA (90, 30, 10 ng of DNA) or with 3 ng of DNA extracted from immunoprecipitated samples are shown. TatSF2G48-R57A was used as an additional negative control. P1 and P2 indicate 2 different sets of primers (Supplemental Table S3). B. ChIP analysis of the different cellular promoters in primary CD4+ T cells infected with a replication competent HIV expressing a flagged Tat. The signals obtained by standard PCR carried out with input DNA (90, 30, 10 ng of DNA) or with 1–5 ng of DNA extracted from immunoprecipitated samples are shown. The results obtained with two sets of primers, P1 and P2, are shown for the PTEN promoter. C. siRNA-mediated knockdown of PPP2R1B and PTEN reduces PPP2R1B, FOXO3a and FOXO3a-dependent gene expression in CD4+primary T cells infected with eGFP or eGFP-Tat retroviruses. RT-PCR results are normalized to GAPDH and reported as fold induction relative to cells treated with ns siRNA. Two independent experiments are reported. D. Protein expression analysis of PPP2R1B, PTEN, FOXO3a, or pFOXO3a by flow cytometry in CD4+ T cells treated with PPP2R1B and/or PTEN siRNAs and infected with a retrovirus expressing GFP only or Tat and eGFP. Results are reported as MFI. E. Level of apoptosis 48 h after Tat expression in CD4+ T-cells transduced with ns siRNA or PPP2R1B and/or PTEN siRNAs, or in CD4+ T-cells exposed to no inhibitor or 100 nM okadaic acid (a PP2A inhibitor) and then treated with PP2A enhancers (ceramide or FTY720). The mean ±SEM of two independent experiments is reported. (*) indicates p<0.05, (**) indicates p = 0.001. F. Level of apoptosis 48 h after HIV infection in CD4+ T-cells transduced with ns siRNA, PPP2R1B and/or PTEN siRNAs, untreated, treated with 100 nM okadaic acid (a PP2A inhibitor) or 10 nM bpV(HOpic) (a PTEN inhibitor). The mean ±SEM of two independent experiments is reported, (*) indicates p<0.05. The PTEN coding sequence is mutated and the corresponding protein is inactive in Jurkat cells due to the frame-shift mutations in both PTEN alleles, which result in the truncation of PTEN within the C-terminal C2 domain and the rapid degradation of the truncated protein [29]. Therefore we would not expect that the lack of phosphorylation of FOXO3a in Jurkat is dependent on PTEN but only on PP2A, even though an increase of PTEN RNA is observed in these cells. However, association of Tat with the PTEN promoter may be relevant in other cell types where this protein is functional. PTEN mRNA expression is increased in primary CD4+ T cells infected with HIV [1] and in these cells PTEN is functional. Indeed, when the association was investigated by conventional ChIP in primary CD4+ T cells infected with a HIV virus in which the tat gene was tagged with a FLAG epitope (Figure 5B), or a Tat expressing retrovirus (not shown), Tat could be found associated with the PTEN, PP2R1B, PPP2R5E, and TP53 promoters as detected by ChIP-Chip analysis in Jukat cells (Figure 5B). As in Jurkat cells, Tat did not associate with the FOXO3a promoter, excluding a prominent role for Tat in the direct transcriptional activation of FOXO3a observed in HIV-infected and Tat-expressing cells [1], or CAV1 promoter, used as a negative control.
To evaluate the role of PTEN in FOXO3a phosphorylation in primary CD4+ T cells, we investigated the effects of inhibition of PP2A and PTEN on FOXO3a and its target genes. We transfected primary CD4+ T cells with siRNA for PPP2R1B and/or PTEN for 24 hours and then infected them for 48 hours with a retrovirus expressing Tat and eGFP or eGFP alone as a control (Figure 5C). This treatment reduced expression of PPP2R1B and PTEN to approximately 60% of its original level. Reduction of PPP2R1B and PTEN expression resulted in reduced FOXO3a and Egr-1 mRNAs and protein expression and in an increase of p-FOXO3a (Figure 5C and 5D). Treatment with PTEN siRNA or the combination of PTEN and PPP2R1B siRNAs showed a significant reduction of apoptosis (p<0.001) in Tat expressing primary CD4+ T cells (Figure 5E). Treatment with PPP2R1B siRNA alone reduced apoptosis to approximately 46% (non statistically significant). This partial result was not surprising, considering that the siRNA treatment did not result in a complete knockdown of PPP2R1B and that PTEN remained active under these conditions. No synergy was apparent when PTEN and PPP2R1B siRNAs were introduced together, compared to transfection of PTEN siRNA alone. Tat-mediated PTEN activation may play a more significant role than PP2A in FOXO3a transcriptional activation in primary CD4+ T cells compared to Jurkat cells. Primary CD4+ T cells and Jurkat cells show concordant responses. PTEN, in addition to PP2A, is critical for Tat-mediated apoptosis of primary CD4+ T cells. When cells are exposed to a PP2A activator (ceramide or FTY720) apoptosis is induced (Figure 5E, grey bar). Subsequent treatment of these cells with okadaic acid (a PP2A inhibitor) showed a significant reduction of apoptosis (p<0.05) (Figure 5E, yellow bar). Level of apoptosis 48 hr after HIV infection in CD4+ T-cells transduced with PPP2R1B and/or PTEN siRNAs, or treated with a PP2A inhibitor or a PTEN inhibitor were significantly reduced compared to HIV infected cells treated with ns siRNA or not treated with any compound (Figure 5F, p<0.05).
Effect of exogenous Tat on the regulation of the PI3K-PTEN-Akt pathway in primary CD4+ T cells
Exogenous Tat can be taken up by cells in cultures and transactivates HIV-1 LTR, if present in the cells, or induce apoptosis when the cultured cells are CD4+ T [17], [48]. Because of these observations, the possibility has been raised that HIV infected cells could release Tat, which has been detected in the serum of infected individuals [15], [16], and affect uninfected cells. To test the effect of exogenous Tat on the apoptosis of primary CD4+ T cells, recombinant Tat was added to the cultures of PBMC (Figure 6). Levels of early apoptosis were measured by evaluating the number of Annexin V+/7AAD - cells. An increase in apoptosis was observed in CD4+ T cells exposed to recombinant Tat in a dose - and time - dependent manner compared to the untreated control culture (Figure 6A). We also evaluated whether the exposure to recombinant Tat induced apoptosis by altering the PI3K-PTEN-Akt pathway (Figure 6B). PBMC were pretreated with inhibitors of PTEN (10 nM bpV(HOpic)), PP2A (100 nM okadaic acid), Akt (50 µM Akt1-1/2), and PI3K (10 nM LY294002) for 30 min before exposure to 5 µg/ml of recombinant Tat for 24 hr. Treatment with either PTEN or PP2A inhibitors showed a significant reduction of apoptosis in CD4+ T cells exposed to recombinant Tat (Figure 6B). In contrast, treatment with either the Akt or PI3K inhibitor in the absence of Tat increased apoptosis of CD4+ T cells, confirming the critical role of PI3K-Akt pathway in regulating cell growth and survival. Inhibition of Akt or PI3K in Tat expressing cells did not further increase the amount of apoptosis observed with Tat expression alone of CD4+ T cells, excluding the possibility of the involvement of other pathways in Tat-mediated apoptosis. These experiments support the concept that uninfected CD4+ T cells can undergo apoptosis when exposed to Tat released by HIV infected cells and that Tat induces apoptosis in a similar manner, whether expressed endogenously or present exogenously.
Fig. 6. Apoptosis and PI3K pathway modulation in primary CD4+ T cells exposed to exogenous Tat. 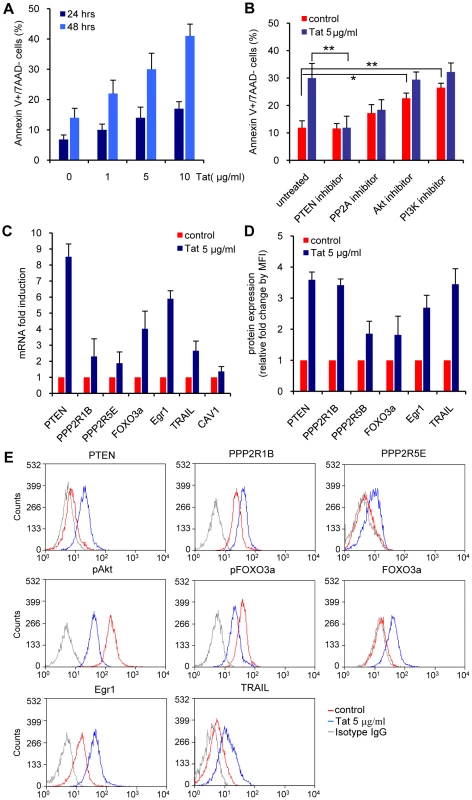
A. Levels of apoptosis 24 and 48 hours after exposure to different Tat concentrations in the medium. B. Levels of apoptosis 24 and 48 hours after exposure to 5 µg/ml of Tat and inhibitors of PTEN (10 nM bpV(HOpic)), PP2A (100 nM okadaic acid), Akt (50 µM Akt1-1/2), and PI3K (10 nM LY294002). The means ± SEM of three experiments are shown. (*) indicates p<0.05, (**) indicates p = 0.001. C. RNA expression levels of different genes part of the PI3K pathway in primary CD4+ T cells exposed to exogenous Tat. D. Corresponding protein levels for the same genes that are part of the PI3K pathway in primary CD4+ T cells 24 hours after exposure to exogenous Tat. RT-PCR results are normalized to GAPDH and reported as fold induction relative to cells untreated control. The average and SEM of three independent experiments is reported. E. One representative flow cytometric protein analysis related to the data reported in D. Results are reported as fold increase of mean fluorescence intensity (MFI) relative to the untreated control. We next validated if exposure to exogenous recombinant Tat modulates the same cellular genes that were found up-regulated in CD4+ T cells expressing Tat endogenously (Figure 6C and 6D). PTEN, PPP2R1B, PPP2R5E, FOXO3a, Egr-1, and TRAIL RNAs were induced by exposure to recombinant Tat (Figure 6C). We also found a corresponding increased accumulation of PTEN, PPP2R1B, PPP2R5E, FOXO3a, Egr-1, and TRAIL proteins in the same cells (Figure 6D and 6E). These data confirm that exposure to exogenous Tat modulates the PI3K-PTEN-Akt pathway in primary CD4+ T cells in the same fashion as in CD4+ T cells expressing Tat endogenously.
PTEN, PPP2R1B and PPP2R5E promoter sequences confer Tat-stimulated transcription on reporter gene
To investigate whether the PTEN, PPP2R1B and PPP2R5E promoter sequences bound by Tat would confer Tat-dependent stimulation of transcription on a reporter gene, ∼1000 bases of the PTEN (-1015 to-1), PPP2R1B (-1074 to-1), or PPP2R5E (-1000 to -1) promoters located 5′ of the transcriptional start site were introduced upstream of the luciferase gene. With all three promoters, the presence of wild type Tat increased luciferase activity relative to that obtained in the absence of Tat or TatSF2G48-R57A (Figure 7B). Luciferase activity was intermediate when measured in cells expressing TatSF2C25,30,35S or TatSF2K28A,K50A (Figure 7B). These results confirm that the transcriptional activity of these promoters can be stimulated by Tat, and suggest that both Tat cofactor-binding domains are necessary for full activity.
Fig. 7. Analysis of transcription initiation and elongation at the PTEN, PP2R1B and PPP2R5E promoters. 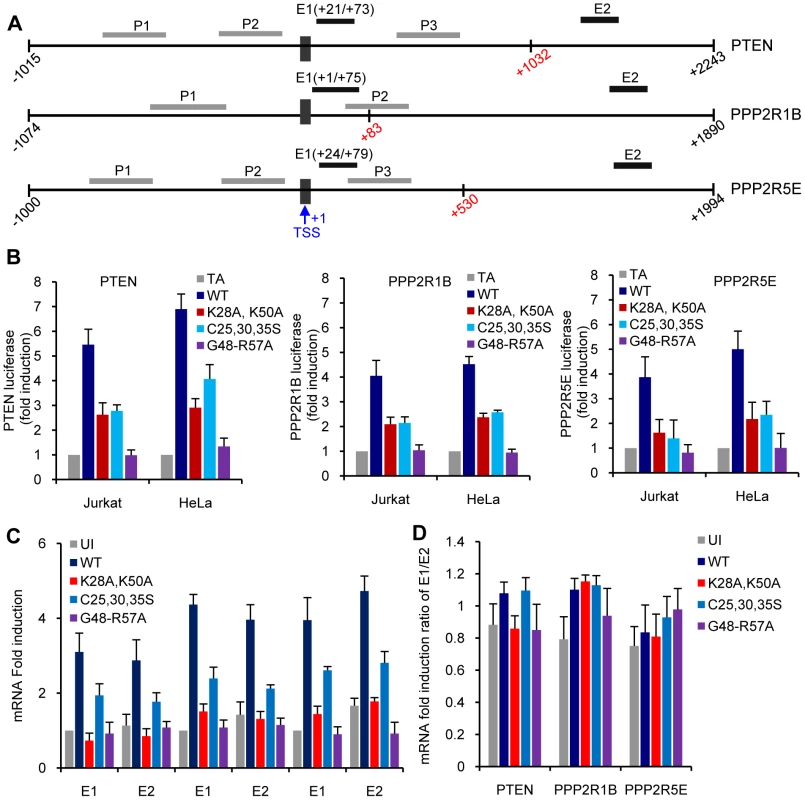
A. Schematic illustration of the location of amplified DNA (P1, P2, and P3) and RNA (E1, E2) fragments in the linear sequence of PTEN, PP2R1B and PPP2R5E genes. The transcriptional start site (TSS) is marked with a black box. Numbers in red mark the first nucleotide of the initiation codon. The promoter fragments used in B. start at the beginning of the individual lines and go to nucleotide −1. B. Luciferase activity in cells expressing Tat or Tat mutants and a luciferase gene under the control of PTEN (−1015 to−1), PPP2R1B (−1074 to−1), or PPP2R5E (−1000 to −1) promoters. C. Detection of PTEN, PP2R1B and PPP2R5E mRNA transcripts using primers in close proximity of (E1) or distant from (E2) the transcription start site. D. mRNA fold induction ratios between E1 and E2. When initiating transcription at the HIV promoter RNA polymerase II generates short RNA species and then pauses at the TAR element of the RNA transcript. Tat recruitment of P-TEFb at TAR releases paused polymerase and allows for accumulation of full length HIV mRNAs. [8], [9], [10], [11], [12]. We searched for evidence that RNAP polymerase II may pause in newly initiated PTEN, PPP2R1B and PPP2R5E transcripts by using RT-PCR to examine the steady-state levels of RNA species within the first 100 bases from the transcriptional start site (E1) and within the coding region of these genes (E2) (Figure 7C). The results did not reveal significant differences in the ratio of accumulation of short compared to long mRNAs in the presence or absence of Tat (Figure 7C, right panel). These results suggest that the increase in PTEN, PPP2R1B and PPP2R5E levels stimulated by Tat is unlikely to be caused by the release of RNA PolII that pauses after generating a short transcript, as it occurs during HIV transcription. Nonetheless, it is possible that Tat does facilitate pause release at the initiation start site or at a site within the first 80 nucleotides of the RNA transcript of PTEN, PPP2R1B and PPP2R5E genes. The lower amounts of Luciferance activity detected when transcription is dependent from the PTEN, PPP2R1B, or PPP2R5E promoters and of accumulated PTEN, PPP2R1B, or PPP2R5E RNA transcripts detected in the presence of TatSF2C25,30,35S or TatSF2K28A,K50A compared to wild type Tat support a role for Tat in recruiting factors that affect the processivity of PolII at the transcription start site of these promoters.
In summary, Tat-mediated activation of apoptotic pathways in T cells starts with the association of Tat with promoters of phosphatase-encoding genes such as PTEN and PP2A. Increased transcription of phosphatase genes leads to reduced phosphorylation of Akt1 and of FOXO3a, its nuclear translocation, followed by the transcriptional activation of its own promoter and of FOXO3a target genes that affect of both the intrinsic and extrinsic apoptotic pathways. FOXO3a increases the transcription of Egr-1, one of its target genes, which further stimulates the transcription of PTEN, thereby reinforcing the pathway that leads to FOXO3a transcriptional activation (Figure 8).
Fig. 8. Tat-mediated alteration of apoptotic pathways regulated by FOXO3a. 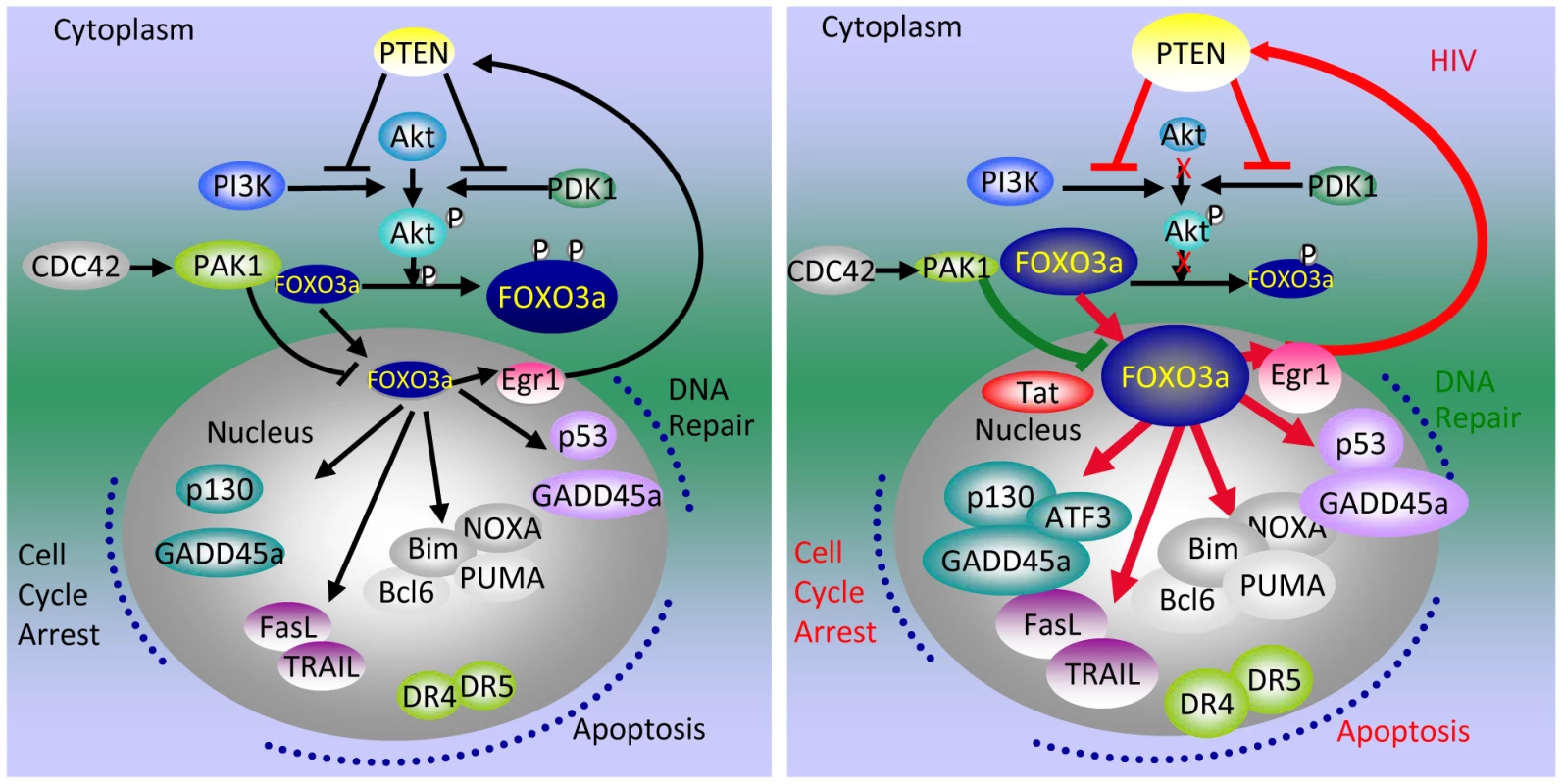
Red lines indicate the association of the protein in the circle at the beginning of the line with the promoter of the factor at the end of the line and its increased transcription. Red arrows connect proteins whose level is increased by the factor indicated at the beginning of the line. The green line that connects PAK1 to FOXO3a indicates reduced levels of PAK1. A red X marks a step that is significantly reduced. Discussion
Association of Tat with the PTEN and PP2A regulatory subunit promoters in T cells is the Tat-mediated event that leads to the activation of FOXO3a and its target genes, many of which are proapoptotic. Inhibition of these phosphatases leads to reduce apoptosis in T cells expressing Tat. A role for PP2A and PP1 in Tat-mediated regulation of HIV transcription was previously reported, but these reports did not investigate the role of Tat-phosphatase interactions in activation of cellular gene expression [49], [50], [51]. A previous investigation proposed that apoptosis in HIV infected CD4+ T cells results from the microtubule perturbation induced by the increased expression of Bim [52]. This mechanism is consistent with our observations, as Bim upregulation was detected during HIV infection in CD4+ T cells and in Tat expressing Jurkat cells [1]. This modulation resulted from the Tat mediated upregulation of trascriptionally active FOXO3a, which regulates Bim gene expression.
We failed to detect association of Tat with the FOXO3a or the Egr-1 promoters. However the level of expression for these genes falls when siRNA for PTEN and/or PP2A are introduced into Tat-expressing cells and partially inhibit expression of these genes. Transcriptionally active FOXO3a, which is also known to stimulate its own transcription [47], is thus linked to the activity of the phosphatases rather than to a closer interaction of Tat with the FOXO3a promoter. However, inhibition of SIRT-1 by Tat [14] might also increase the activity of FOXO3a, as SIRT-1 deacetylase activity can repress the activity of FOXO3a [53]. The detection of FOXO3a at the Egr-1 promoter indicates that Egr-1 is also a transcriptional target of FOXO3a and its increased transcription levels are likely to be directly dependent on the increase of transcriptionally active FOXO3a. The increased levels of Egr-1, which regulates transcription of PTEN [4], [5], [6] and is present at the PTEN promoter as shown by ChIP/PCR analysis, may further elevate the levels of PTEN, and so reinforce the circuit that leads to FOXO3a-mediated transcription.
Inhibition of expression of PTEN and PP2A in primary CD4+ T cells and of PP2A in Jurkat cells in the course of Tat expression reduced transcriptionally active FOXO3a and afforded protection from Tat-induced apoptosis (Figure 5 and 6). This confirms their role in apoptosis, as seen also for FOXO3a and Egr-1 inhibition [1]. Because we failed to detect a direct association of Tat with the promoters of these genes, these experiments point at PTEN or, in its absence, to PP2A, as a Tat target and initiatior of the apoptotic cascade. Inhibition of PPP2R1B did not reach statistical significance in reducing apoptosis in primary CD4+ T-cells, while statistical significance was attained when a siRNA against PTEN was used. It is possible that the activity of PTEN by itself is sufficient to initiate the apoptotic cascade and therefore inhibition of the PP2a subunit alone is not sufficient to drastically reduce apoptosis. Perhaps PTEN expression is more efficiently attenuated by siRNA compared to PPP2R1B, or PTEN governs the regulation of FOXO3a in these cells.
In regulating transcription from the HIV LTR, Tat's major role is in relieving RNAPII pausing at the TAR element by recruiting P-TEFb [8]. Because PTEN and PP2A transcription occurs in the absence of Tat in normal cells, we did not expect that Tat would significantly increase the transcription of these genes by relieving RNAPII pausing at the RNA level. Our data argue that the levels of transcripts that include the first 80 nucleotides occur at the same level as transcripts that are extended through the body of the gene (Figure 7C), but do not exclude the likely possibility that much shorter and unstable transcripts are produced by RNA polymerase II pausing. Because RNA polymerase II pausing now appears to occur very near the start site at all genes [54], it seems likely that Tat binding to these genes increases pause release and RNA polymerase II processivity by recruiting P-TEFb. Furthermore, other transcription factors that bind P-TEFb, such a c-Myc, increase transcript levels by using this mechanism [54]. In addition, the ability of Tat to increase transcription from PTEN and PP2A subunit promoters may depend on Tat facilitating P-TEFb transition form the inactive to the active form [55], [56]. Transcription of luciferance form PTEN and PP2A subunit promoter sequences located upstream of the transcription start site was not as significantly enhanced when a Tat mutant that does not bind P-TEFb was cotransfected with the reporter construct. The same was true when a Tat mutant that does not bind p300/was used in a similar experiment (Figure 7). It has been previously shown that Tat competes with HEXIM1, an inhibitor of P-TEFb activity [57], to increase the active pool of P-TEFb [58]. Tat therefore may act as a factor that increases RNAPII processivity by facilitating the transition of P-TEFb to its transcriptionally active form. Investigation of the details of this step will be the focus of future investigation.
Tat was found associated with 450 promoters during its expression in T cells. We do not know what specifically directs Tat to a subset of all promoters or if Tat associates with all of these promoters also in the context of HIV infection. Our current models are 1) the 450 promoters share one or more transcription factors that bind and recruit Tat to these promoters, and 2) Tat-bound P-TEFb is preferentially recruited to the 450 promoters by various transcription factors. These models will be investigated in future studies.
Levels of phosphorylated FOXO3a correlate with the survival of memory CD4+ T cells pFOXO3a is reduced in HIV-infected individuals and is higher in untreated, infected subjects with undetectable viremia [19]. These data support the in vivo relevance of our data, and suggest that regulating FOXO3a transcriptional activity may provide a target to control CD4+ T-cell apoptosis in vivo. Inhibition of PTEN and PP2A could in principle be beneficial to prevent HIV-mediated T cell apoptosis. However the role of inactive PTEN in many types of cancer is a serious impediment to this approach [6], [45]. Instead, inhibition of Tat may be a viable option to reduce HIV-induced apoptosis, especially in non-infected cells. Efforts aimed at Tat inhibitor discovery have met with limited success, because the primary endpoint was inhibition to reduce viremia. This is probably a poor endpoint because the role of Tat activity in HIV transcription is somewhat redundant and can be replaced by other factors. However, Tat remains a valuable drug target to reduce its deleterious effects on T cell survival. Antibodies against Tat, developed in vaccinated or infected patients, or drugs blocking Tat activity could neutralize Tat released from infected cells and prevent the protein to enter non-infected cells or to block its activity inside uninfected cells and prevent apoptosis of this population. While it is not known how much of the CD4+ T cell death in uninfected cells is due to this mechanism, it would be important to investigate in an experimental setting if these strategies offer some benefit.
Materials and Methods
Plasmids and transfection
Vectors for production of retroviruses (pCMMP-eGFP, pMMP-Tat1b-IGFP, pHDM-G/VSV-G envelope, and pMD.MLVgp) were generously provided by Dr. Jeng-Shin Lee from the Harvard Gene Therapy Initiative. Recombinant adenoviruses: Ad-tTA, Ad-TatSF2, Ad-FLAG-TatSF2, Ad-FLAG-TatSF2G48-R57A, Ad-FLAG-TatSF2C25,30,35S, and Ad-FLAG-TatSF2K28A,K50A were constructed according to previously published procedures [59]. The Tat coding region was cloned into the vector pAd-TRE-MCS1. Tat is under a tetracycline-inducible promoter and co-infection with Ad-tTA, which expresses the tetracycline responsive transactivator, is required. DNA transfection in 293T cells was carried out by calcium phosphate precipitation. Supernatants containing viruses were prepared as described before [1].
Cell lines, primary cells, treatments, Abs, and reagents
Human PBMCs were obtained from healthy donors from the Children's Hospital Boston blood bank. The purity of CD4+ lymphocytes isolated using a CD4+ T cell negative selection kit (Miltenyi Biotec) was more than 95%. All antibodies and siRNAs were previously described [1], except the human anti-PPP2R1B, anti-PPP2R5E antibodies (Santa Cruz Biotechnology), the PPP2R1B, PPP2R5E, siRNAs (Santa Cruz Biotechnology), and the PTEN siGENOME SMARTpool siRNA (Dharmacon).
Transduction of T lymphocytes, infection of cell lines and siRNA treatments
T cells were exposed to the virus-containing supernatants of different vesicular stomatitis virus-G protein (VSV-G)-pseudotyped vectors [enhanced GFP (eGFP) or Tat-GFP] at multiplicity of infection (MOI) of 5 supplemented with 4 µg/ml protamine sulfate for 2 h. Jurkat T cells were exposed to adenoviruses at MOI of 20. Forty-eight hours after transduction T cells were sorted and GFP-positive cells were collected for PCR analysis of the RNA. All siRNAs were transfected into exponentially growing cells or electroporated at a final concentration of 3 µg into stimulated CD4+ T lymphocytes as previously described [1]. For luciferase reporter assay, HeLa and 293T cells were transiently transfected with nonspecific control or FOXO3a siRNA for 24 h and then infected with adenoviruses expressing tTA or TatSF2 for 24 h. Cell lysates were used for luciferase reporter assay according to manufacturer's instructions.
RNA isolation and quantitative real-time RT-PCR
The isolated RNA (100 ng) was reverse transcribed using an iScript cDNA kit (Bio-Rad) followed by amplification of HIV-1 Tat or cellular genes using SYBR Green SuperMix with ROX kit (Bio-Rad) and primers specific for the genes of interest and GAPDH as a control. Supplemental Table S2 reports the list of primers used in the amplification.
Immunofluorescence staining and flow cytometry
Isolated CD4+ T lymphocytes (1×106 cells) were prepared for intracellular staining as described before [1]. For intracellular staining, antibodies against PPP2R1B, pFOXO3a, and FOXO3a, were directly labeled with APC-Cy7, APC, or PE-Cy5 Tandem Conjugation Kit (Innova Biosciences). Flow cytometric acquisition was performed on MoFlow (Dako). Cell analysis was performed on the gated live-cell populations using Summit software (Dako). For apoptosis staining, CD4+ T lymphocytes or Jurkat cells were stained with Annexin V-APE and 7AAD and analyzed by flow cytometry as previously described.
Confocal microscopy analysis
Ad-tTA or Ad-TatSF2 infected cells were fixed and incubated first with a mouse monoclonal anti-PPP2R1B (Santa Cruz Biotechnology), or a rabbit polyclonal anti-FOXO3a, or anti-phospho-FOXO3a antibodies (Cell Signaling Biotechnology), and then with a FITC-conjugated goat anti-mouse IgG or TR-red - conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology). Cells were stained with DRAG5 to visualize the nucleus (blue). The coverslips were mounted using ProLong Gold antifade reagent with DAPI (Molecular probes).
Chromatin immunoprecipitation (ChIP) and DNA microarray analysis (ChIP-Chip)
ChIP was performed as previously described [60]. Briefly, 5×107 cells were infected with adenoviruses, subsequently cross-linked, and sonicated to yield an average DNA fragment of 500 bps. The sheared chromatin was incubated with protein G magnetic beads (Invitrogen) coupled to anti-FLAG (Sigma M2), anti-p300 polyclonal, anti-FOXO3a polyclonal, and anti-Egr-1 polyclonal antibodies (Santa Cruz Biotechnology). These antibodies have been previously used in ChIP and do not have significant background [61], [62], [63]. Amplified DNA was labeled and purified using Bioprime random primer labeling kits (Invitrogen, immunoenriched DNA was labeled with Cy5 fluorophore, whole cell extract DNA was labeled with Cy3 fluorophore). Labeled DNA was mixed and hybridized to Agilent Human Promoter ChIP-on-chip Microarray Set for 20 hours at 40°C. The microarrays (Design ID - 014706 and 014707) have probes for about 17,000 human promoters. Arrays were then washed using standard Agilent protocols and scanned using an Agilent DNA microarray scanner BA. Scans were manually examined for abnormal features and intensities were extracted for each spot automatically using Agilent feature extraction software. We calculated the log of the ratio of median normalized intensity in the IP-enriched channel to median normalized intensity in the genomic DNA channel for each probe and used a whole chip error model [64] to calculate confidence values for each spot on each array (single probe p-value). This error model functions by converting the intensity information in both channels to an X score which is dependent on both the absolute value of intensities and background noise in each channel. The X scores for an array are assumed to be normally distributed which allows for calculation of a p-value for the enrichment ratio seen at each feature. To determine bound regions in the datasets, we calculated the average X score of the 60-mer and its two immediate neighbors. If a feature was flagged as abnormal during scanning, we assumed it gave a neutral contribution to the average X score. Similarly, if an adjacent feature was beyond a reasonable distance from the probe (based on the maximum size of labeled DNA fragments hybridized to the array), we assumed it gave a neutral contribution to the average X score. This set of averaged values gave us a new distribution that was subsequently used to calculate p-values of average X (probe set p-values). If the probe set p-value was less than 0.001, the three probes were marked as potentially bound. In addition, the three probes in a probe set must each have single probe p-values<0.05 or the center probe in the probe set has a single probe p-value<0.01 and one of the flanking probes has a single point p-value<0.1. Association with promoters was identified if the bound regions were 8 kb upstream or 2 kb downstream of the Transcription Start Site [64]. For conventional ChIP analysis the purified DNA was quantified by qPCR. The primers used in qPCR are described in Supplemental Table S3. For the qPCR reaction, 10 ng of immunoprecipitated material was used, and for whole cell extract DNA samples (or input) a range of DNA amounts (10–90 ng of DNA) were used. PCR products obtained by standard PCR for qualitative analysis were visualized on agarose gel with ethidium bromide.
Protein phosphatase and protein phosphatase 2A (PP2A) activity
Protein phosphatase activity was measured using a commercially available Ser/Thr phosphatase assay kit (Upstate Biotechnology). Cell lysates were used to measure phosphatase activity using p-nitrophenyl phosphate (pNPP) or free phosphate released by the phosphatase from a specific peptide substrate KRpTIRR using the malachite green system. For protein phosphatase 2A (PP2A), total cellular protein lysate was incubated with protein A-agarose slurry with anti-PP2A-C (2 µg/ml, Upstate Biotechnology). A commercially available PP2A immunoprecipitation phosphatase assay kit (Upstate Biotechnology) was used to measure phosphate release as an index of phosphatase activity.
Treatments with inhibitors
PBMC (1×106 cells) were infected with HIV-Flag Tat (viral amount corresponding to 20 ng of p24) for 6 hours and then resuspended with fresh medium in the presence or absence of PP2A inhibitor (100 nM okadaic acid) or PTEN inhibitor (10 nM bpV(HOpic). A second dose of the inhibitors was added to the cultures after 48 h. The percentage of TUNEL+ or Annexin V+ cells was determined by fluorescence-activated cell sorter analysis on day 3 or 5. Jurkat T cells were exposed to adenoviruses at MOI of 20 in the presence or absence of inhibitors. After 2 h, cells were washed and fresh media was replaced. Level of apoptosis was expressed as the percentage of Annexin V+7AAD - 24 h after transduction. PBMC were incubated without or with PP2A inhibitor (100 nM okadaic acid) for 1 h and then treated with two PP2A enhancers, sphingolipid ceramide N-Deacyclase (Calbiochem, cat. #567704) (10 µM) or FTY720 (EMD, Gibbstown, NJ) (10 nM), for 48 h. Level of apoptosis was expressed as the percentage of Annexin V+CD4+. Recombinant TatHIV-Bal was added at the indicated concentrations into the cultures of PBMC in the presence or absence of PTEN, PP2A, Akt, and PI3K inhibitors (10 nM bpV(HOpic, 100 nM okadaic acid, 50 µM Akt1-1/2, 10 nM LY294002, respectively), which were added 30 minutes before the addition of Tat.
Luciferase assays
HeLa and 293T cells were transfected with nonspecific siRNA or siRNA targeting FOXO3a with SureFECT transfection (SABiosciences). Luciferase activities were assessed using the dual-luciferase reporter assay system (Promega, Madison, WI). Firefly luciferase activity was normalized to the activity of the Renilla luciferase control.
Immunoblots
Total proteins were separated by SDS-PAGE, transferred onto the nitrocellulose membrane, and incubated with Abs raised against FOXO3a, phospho-FOXO3a (Ser318), PTEN, phospho-Akt1 (Ser473), PPP2R1B, PPP2R5E, and PPP2C (Santa Cruz Biotechnology). The secondary Ab was detected with Pierce ECL substrate. The blots were exposed to Hyperfilm, and the signals were quantified by scanning densitometry with Molecular Analyst 1.5 software (Biorad). To account for any differences in loading, target band densitometries were divided by actin densitometry obtained in the same lane. These corrected densitometries were normalized to controls in each experiment.
Statistical analysis
A two-tailed, two-sample Student t test was used to calculate p-values for differences in means between groups. The data are expressed as means ± SEM. For statistical inference, a p-value of <0.05 was considered significant.
Supporting Information
Zdroje
1. DabrowskaA
KimN
AldoviniA
2008
Tat-Induced Foxo3a Is A Key Mediator Of Apoptosis In Hiv-1-Infected Human Cd4+ T Lymphocytes.
J Immunol
181
8460
8477
2. ModurV
NagarajanR
EversBm
MilbrandtJ
2002
Foxo Proteins Regulate Tumor Necrosis Factor-Related Apoptosis Inducing Ligand Expression. Implications For Pten Mutation In Prostate Cancer.
J Biol Chem
277
47928
47937
3. SuntersA
Fernandez De MattosS
StahlM
BrosensJj
ZoumpoulidouG
2003
Foxo3a Transcriptional Regulation Of Bim Controls Apoptosis In Paclitaxel-Treated Breast Cancer Cell Lines.
J Biol Chem
278
49795
49805
4. BaronV
AdamsonEd
CalogeroA
RagonaG
MercolaD
2006
The Transcription Factor Egr1 Is A Direct Regulator Of Multiple Tumor Suppressors Including Tgfbeta1, Pten, P53, And Fibronectin.
Cancer Gene Ther
13
115
124
5. VirolleT
AdamsonEd
BaronV
BirleD
MercolaD
2001
The Egr-1 Transcription Factor Directly Activates Pten During Irradiation-Induced Signalling.
Nat Cell Biol
3
1124
1128
6. SalmenaL
CarracedoA
PandolfiPp
2008
Tenets Of Pten Tumor Suppression.
Cell
133
403
414
7. BrunetA
BonniA
ZigmondMj
LinMz
JuoP
1999
Akt Promotes Cell Survival By Phosphorylating And Inhibiting A Forkhead Transcription Factor.
Cell
96
857
868
8. GarberMe
JonesKa
1999
Hiv-1 Tat: Coping With Negative Elongation Factors.
Curr Opin Immunol
11
460
465
9. HottigerMo
NabelGj
1998
Interaction Of Human Immunodeficiency Virus Type 1 Tat With The Transcriptional Coactivators P300 And Creb Binding Protein.
J Virol
72
8252
8256
10. MarzioG
TyagiM
GutierrezMi
GiaccaM
1998
Hiv-1 Tat Transactivator Recruits P300 And Creb-Binding Protein Histone Acetyltransferases To The Viral Promoter.
Proc Natl Acad Sci U S A
95
13519
13524
11. OttM
SchnolzerM
GarnicaJ
FischleW
EmilianiS
1999
Acetylation Of The Hiv-1 Tat Protein By P300 Is Important For Its Transcriptional Activity.
Curr Biol
9
1489
1492
12. SilvermanEs
DuJ
WilliamsAj
WadgaonkarR
DrazenJm
1998
Camp-Response-Element-Binding-Protein-Binding Protein (Cbp) And P300 Are Transcriptional Co-Activators Of Early Growth Response Factor-1 (Egr-1).
Biochem J
336
Pt 1
183
189
13. DengL
WangD
De La FuenteC
WangL
LiH
2001
Enhancement Of The P300 Hat Activity By Hiv-1 Tat On Chromatin Dna.
Virology
289
312
326
14. KwonHs
BrentMm
GetachewR
JayakumarP
ChenLf
2008
Human Immunodeficiency Virus Type 1 Tat Protein Inhibits The Sirt1 Deacetylase And Induces T Cell Hyperactivation.
Cell Host Microbe
3
158
167
15. WestendorpMo
FrankR
OchsenbauerC
StrickerK
DheinJ
1995
Sensitization Of T Cells To Cd95-Mediated Apoptosis By Hiv-1 Tat And Gp120.
Nature
375
497
500
16. HudsonL
LiuJ
NathA
JonesM
RaghavanR
2000
Detection Of The Human Immunodeficiency Virus Regulatory Protein Tat In Cns Tissues.
J Neurovirol
6
145
155
17. FrankelAd
PaboCo
1988
Cellular Uptake Of The Tat Protein From Human Immunodeficiency Virus.
Cell
55
1189
1193
18. CuiM
HuangY
ZhaoY
ZhengJ
2008
Transcription Factor Foxo3a Mediates Apoptosis In Hiv-1-Infected Macrophages.
J Immunol
180
898
906
19. Van GrevenyngheJ
ProcopioFa
HeZ
ChomontN
RiouC
2008
Transcription Factor Foxo3a Controls The Persistence Of Memory Cd4(+) T Cells During Hiv Infection.
Nat Med
14
266
274
20. McneesAl
MahrJa
OrnellesD
GoodingLr
2004
Postinternalization Inhibition Of Adenovirus Gene Expression And Infectious Virus Production In Human T-Cell Lines.
J Virol
78
6955
6966
21. WongK
SharmaA
AwasthiS
MatlockEf
RogersL
2005
Hiv-1 Tat Interactions With P300 And Pcaf Transcriptional Coactivators Inhibit Histone Acetylation And Neurotrophin Signaling Through Creb.
J Biol Chem
280
9390
9399
22. BresV
TagamiH
PeloponeseJm
LoretE
JeangKt
2002
Differential Acetylation Of Tat Coordinates Its Interaction With The Co-Activators Cyclin T1 And Pcaf.
Embo J
21
6811
6819
23. GarciaJa
HarrichD
PearsonL
MitsuyasuR
GaynorRb
1988
Functional Domains Required For Tat-Induced Transcriptional Activation Of The Hiv-1 Long Terminal Repeat.
Embo J
7
3143
3147
24. EfthymiadisA
BriggsLj
JansDa
1998
The Hiv-1 Tat Nuclear Localization Sequence Confers Novel Nuclear Import Properties.
J Biol Chem
273
1623
1628
25. RenB
RobertF
WyrickJj
AparicioO
JenningsEg
2000
Genome-Wide Location And Function Of Dna Binding Proteins.
Science
290
2306
2309
26. MarsonA
KretschmerK
FramptonGm
JacobsenEs
PolanskyJk
2007
Foxp3 Occupancy And Regulation Of Key Target Genes During T-Cell Stimulation.
Nature
445
931
935
27. ZhangX
GuoC
ChenY
ShulhaHp
SchnetzMp
2008
Epitope Tagging Of Endogenous Proteins For Genome-Wide Chip-Chip Studies.
Nat Methods
5
163
165
28. GeorgescuMm
KirschKh
AkagiT
ShishidoT
HanafusaH
1999
The Tumor-Suppressor Activity Of Pten Is Regulated By Its Carboxyl-Terminal Region.
Proc Natl Acad Sci U S A
96
10182
10187
29. ShanX
CzarMj
BunnellSc
LiuP
LiuY
2000
Deficiency Of Pten In Jurkat T Cells Causes Constitutive Localization Of Itk To The Plasma Membrane And Hyperresponsiveness To Cd3 Stimulation.
Mol Cell Biol
20
6945
6957
30. WangSs
EsplinEd
LiJl
HuangL
GazdarA
1998
Alterations Of The Ppp2r1b Gene In Human Lung And Colon Cancer.
Science
282
284
287
31. ColellaS
OhgakiH
RuedigerR
YangF
NakamuraM
2001
Reduced Expression Of The Aalpha Subunit Of Protein Phosphatase 2a In Human Gliomas In The Absence Of Mutations In The Aalpha And Abeta Subunit Genes.
Int J Cancer
93
798
804
32. JanssensV
GorisJ
2001
Protein Phosphatase 2a: A Highly Regulated Family Of Serine/Threonine Phosphatases Implicated In Cell Growth And Signalling.
Biochem J
353
417
439
33. SilversteinAm
BarrowCa
DavisAj
MumbyMc
2002
Actions Of Pp2a On The Map Kinase Pathway And Apoptosis Are Mediated By Distinct Regulatory Subunits.
Proc Natl Acad Sci U S A
99
4221
4226
34. YangJ
WuJ
TanC
KleinPs
2003
Pp2a:B56epsilon Is Required For Wnt/Beta-Catenin Signaling During Embryonic Development.
Development
130
5569
5578
35. LetourneuxC
RocherG
PorteuF
2006
B56-Containing Pp2a Dephosphorylate Erk And Their Activity Is Controlled By The Early Gene Iex-1 And Erk.
Embo J
25
727
738
36. MoorheadGb
Trinkle-MulcahyL
Ulke-LemeeA
2007
Emerging Roles Of Nuclear Protein Phosphatases.
Nat Rev Mol Cell Biol
8
234
244
37. JinZ
ShiJ
SarafA
MeiW
ZhuGz
2009
The 48-Kda Alternative Translation Isoform Of Pp2a:B56epsilon Is Required For Wnt Signaling During Midbrain-Hindbrain Boundary Formation.
J Biol Chem
284
7190
7200
38. SatoS
FujitaN
TsuruoT
2000
Modulation Of Akt Kinase Activity By Binding To Hsp90.
Proc Natl Acad Sci U S A
97
10832
10837
39. ResjoS
GoranssonO
HarndahlL
ZolnierowiczS
ManganielloV
2002
Protein Phosphatase 2a Is The Main Phosphatase Involved In The Regulation Of Protein Kinase B In Rat Adipocytes.
Cell Signal
14
231
238
40. TrotmanLc
AlimontiA
ScaglioniPp
KoutcherJa
Cordon-CardoC
2006
Identification Of A Tumour Suppressor Network Opposing Nuclear Akt Function.
Nature
441
523
527
41. YanL
LavinVa
MoserLr
CuiQ
KaniesC
2008
Pp2a Regulates The Pro-Apoptotic Activity Of Foxo1.
J Biol Chem
283
7411
7420
42. BertoliC
CopettiT
LamEw
DemarchiF
SchneiderC
2009
Calpain Small-1 Modulates Akt/Foxo3a Signaling And Apoptosis Through Pp2a.
Oncogene
28
721
733
43. YinKj
HsuCy
HuXy
ChenH
ChenSw
2006
Protein Phosphatase 2a Regulates Bim Expression Via The Akt/Fkhrl1 Signaling Pathway In Amyloid-Beta Peptide-Induced Cerebrovascular Endothelial Cell Death.
J Neurosci
26
2290
2299
44. BarreyroFj
KobayashiS
BronkSf
WerneburgNw
MalhiH
2007
Transcriptional Regulation Of Bim By Foxo3a Mediates Hepatocyte Lipoapoptosis.
J Biol Chem
282
27141
27154
45. DubrovskaA
KimS
SalamoneRj
WalkerJr
MairaSm
2009
The Role Of Pten/Akt/Pi3k Signaling In The Maintenance And Viability Of Prostate Cancer Stem-Like Cell Populations.
Proc Natl Acad Sci U S A
106
268
273
46. LeiH
QuelleFw
2009
Foxo Transcription Factors Enforce Cell Cycle Checkpoints And Promote Survival Of Hematopoietic Cells After Dna Damage.
Mol Cancer Res
7
1294
1303
47. EssaghirA
DifN
MarbehantCy
CofferPj
DemoulinJb
2009
The Transcription Of Foxo Genes Is Stimulated By Foxo3 And Repressed By Growth Factors.
J Biol Chem
284
10334
10342
48. LiCj
FriedmanDj
WangC
MetelevV
PardeeAb
1995
Induction Of Apoptosis In Uninfected Lymphocytes By Hiv-1 Tat Protein.
Science
268
429
431
49. NekhaiS
JerebtsovaM
JacksonA
SoutherlandW
2007
Regulation Of Hiv-1 Transcription By Protein Phosphatase 1.
Curr Hiv Res
5
3
9
50. BharuchaDc
ZhouM
NekhaiS
BradyJn
ShuklaRr
2002
A Protein Phosphatase From Human T Cells Augments Tat Transactivation Of The Human Immunodeficiency Virus Type 1 Long-Terminal Repeat.
Virology
296
6
16
51. RuedigerR
BrewisN
OhstK
WalterG
1997
Increasing The Ratio Of Pp2a Core Enzyme To Holoenzyme Inhibits Tat-Stimulated Hiv-1 Transcription And Virus Production.
Virology
238
432
443
52. ChenD
WangM
ZhouS
ZhouQ
2002
Hiv-1 Tat Targets Microtubules To Induce Apoptosis, A Process Promoted By The Pro-Apoptotic Bcl-2 Relative Bim.
Embo J
21
6801
6810
53. MottaMc
DivechaN
LemieuxM
KamelC
ChenD
2004
Mammalian Sirt1 Represses Forkhead Transcription Factors.
Cell
116
551
563
54. RahlPb
LinCy
SeilaAc
FlynnRa
MccuineS
C-Myc Regulates Transcriptional Pause Release.
Cell
141
432
445
55. ZhouQ
YikJh
2006
The Yin And Yang Of P-Tefb Regulation: Implications For Human Immunodeficiency Virus Gene Expression And Global Control Of Cell Growth And Differentiation.
Microbiol Mol Biol Rev
70
646
659
56. YangZ
YikJh
ChenR
HeN
JangMk
2005
Recruitment Of P-Tefb For Stimulation Of Transcriptional Elongation By The Bromodomain Protein Brd4.
Mol Cell
19
535
545
57. YikJh
ChenR
NishimuraR
JenningsJl
LinkAj
2003
Inhibition Of P-Tefb (Cdk9/Cyclin T) Kinase And Rna Polymerase Ii Transcription By The Coordinated Actions Of Hexim1 And 7sk Snrna.
Mol Cell
12
971
982
58. BarboricM
YikJh
CzudnochowskiN
YangZ
ChenR
2007
Tat Competes With Hexim1 To Increase The Active Pool Of P-Tefb For Hiv-1 Transcription.
Nucleic Acids Res
35
2003
2012
59. ChartierC
DegryseE
GantzerM
DieterleA
PaviraniA
1996
Efficient Generation Of Recombinant Adenovirus Vectors By Homologous Recombination In Escherichia Coli.
J Virol
70
4805
4810
60. LeeTi
JohnstoneSe
YoungRa
2006
Chromatin Immunoprecipitation And Microarray-Based Analysis Of Protein Location.
Nat Protoc
1
729
748
61. SmithJl
FreebernWj
CollinsI
De SierviA
MontanoI
2004
Kinetic Profiles Of P300 Occupancy In Vivo Predict Common Features Of Promoter Structure And Coactivator Recruitment.
Proc Natl Acad Sci U S A
101
11554
11559
62. EmerlingBm
WeinbergF
LiuJl
MakTw
ChandelNs
2008
Pten Regulates P300-Dependent Hypoxia-Inducible Factor 1 Transcriptional Activity Through Forkhead Transcription Factor 3a (Foxo3a).
Proc Natl Acad Sci U S A
105
2622
2627
63. YuJ
De BelleI
LiangH
AdamsonEd
2004
Coactivating Factors P300 And Cbp Are Transcriptionally Crossregulated By Egr1 In Prostate Cells, Leading To Divergent Responses.
Mol Cell
15
83
94
64. HughesJd
EstepPw
TavazoieS
ChurchGm
2000
Computational Identification Of Cis-Regulatory Elements Associated With Groups Of Functionally Related Genes In Saccharomyces Cerevisiae.
J Mol Biol
296
1205
1214
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent AutoimmunityČlánek Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on VirulenceČlánek Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral LoadČlánek A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 9- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Azole Drugs Are Imported By Facilitated Diffusion in and Other Pathogenic Fungi
- Two Genes on A/J Chromosome 18 Are Associated with Susceptibility to Infection by Combined Microarray and QTL Analyses
- Impact of Simian Immunodeficiency Virus Infection on Chimpanzee Population Dynamics
- Breaking the Stereotype: Virulence Factor–Mediated Protection of Host Cells in Bacterial Pathogenesis
- The Canine Papillomavirus and Gamma HPV E7 Proteins Use an Alternative Domain to Bind and Destabilize the Retinoblastoma Protein
- Rescue of HIV-1 Release by Targeting Widely Divergent NEDD4-Type Ubiquitin Ligases and Isolated Catalytic HECT Domains to Gag
- Steric Shielding of Surface Epitopes and Impaired Immune Recognition Induced by the Ebola Virus Glycoprotein
- Dynamics of the Multiplicity of Cellular Infection in a Plant Virus
- HLA Class I Binding of HBZ Determines Outcome in HTLV-1 Infection
- Pathogenic Bacteria Target NEDD8-Conjugated Cullins to Hijack Host-Cell Signaling Pathways
- The HA and NS Genes of Human H5N1 Influenza A Virus Contribute to High Virulence in Ferrets
- SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent Autoimmunity
- Cyclin-Dependent Kinase Activity Controls the Onset of the HCMV Lytic Cycle
- The N-Terminal Domain of the Arenavirus L Protein Is an RNA Endonuclease Essential in mRNA Transcription
- Generation of Neutralizing Antibodies and Divergence of SIVmac239 in Cynomolgus Macaques Following Short-Term Early Antiretroviral Therapy
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- Intracellular Proton Conductance of the Hepatitis C Virus p7 Protein and Its Contribution to Infectious Virus Production
- The Transcriptome of the Human Pathogen at Single-Nucleotide Resolution
- The Epidermal Growth Factor Receptor (EGFR) Promotes Uptake of Influenza A Viruses (IAV) into Host Cells
- Surface Co-Expression of Two Different PfEMP1 Antigens on Single -Infected Erythrocytes Facilitates Binding to ICAM1 and PECAM1
- Sequestration and Tissue Accumulation of Human Malaria Parasites: Can We Learn Anything from Rodent Models of Malaria?
- Phylogenomics of Ligand-Gated Ion Channels Predicts Monepantel Effect
- Generation of Covalently Closed Circular DNA of Hepatitis B Viruses via Intracellular Recycling Is Regulated in a Virus Specific Manner
- CpG-Methylation Regulates a Class of Epstein-Barr Virus Promoters
- Molecular and Evolutionary Bases of Within-Patient Genotypic and Phenotypic Diversity in Extraintestinal Infections
- A Bistable Switch and Anatomical Site Control Virulence Gene Expression in the Intestine
- Are Members of the Fungal Genus (a) Commensals; (b) Opportunists; (c) Pathogens; or (d) All of the Above?
- Structures of Receptor Complexes of a North American H7N2 Influenza Hemagglutinin with a Loop Deletion in the Receptor Binding Site
- Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral Load
- The Coevolution of Virulence: Tolerance in Perspective
- Involvement of the Cytokine MIF in the Snail Host Immune Response to the Parasite
- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
- High Content Phenotypic Cell-Based Visual Screen Identifies Acyltrehalose-Containing Glycolipids Involved in Phagosome Remodeling
- A Novel Small Molecule Inhibitor of Hepatitis C Virus Entry
- The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Diarrhea
- RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral Cap-Dependent Transcription
- Pathogen Specific, IRF3-Dependent Signaling and Innate Resistance to Human Kidney Infection
- Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Formation of Mobile Chromatin-Associated Nuclear Foci Containing HIV-1 Vpr and VPRBP Is Critical for the Induction of G2 Cell Cycle Arrest
- Association of Tat with Promoters of PTEN and PP2A Subunits Is Key to Transcriptional Activation of Apoptotic Pathways in HIV-Infected CD4+ T Cells
- Metal Hyperaccumulation Armors Plants against Disease
- Cyclin-Dependent Kinase-Like Function Is Shared by the Beta- and Gamma- Subset of the Conserved Herpesvirus Protein Kinases
- Role of Acetyl-Phosphate in Activation of the Rrp2-RpoN-RpoS Pathway in
- Ebolavirus Is Internalized into Host Cells Macropinocytosis in a Viral Glycoprotein-Dependent Manner
- A Novel Family of IMC Proteins Displays a Hierarchical Organization and Functions in Coordinating Parasite Division
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- The Coevolution of Virulence: Tolerance in Perspective
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



