-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Induces T-Cell Lymphoma and Systemic Inflammation
Human T-cell leukemia virus type 1 (HTLV-1) is the causal agent of a neoplastic disease of CD4+ T cells, adult T-cell leukemia (ATL), and inflammatory diseases including HTLV-1 associated myelopathy/tropical spastic paraparesis, dermatitis, and inflammatory lung diseases. ATL cells, which constitutively express CD25, resemble CD25+CD4+ regulatory T cells (Treg). Approximately 60% of ATL cases indeed harbor leukemic cells that express FoxP3, a key transcription factor for Treg cells. HTLV-1 encodes an antisense transcript, HTLV-1 bZIP factor (HBZ), which is expressed in all ATL cases. In this study, we show that transgenic expression of HBZ in CD4+ T cells induced T-cell lymphomas and systemic inflammation in mice, resembling diseases observed in HTLV-1 infected individuals. In HBZ-transgenic mice, CD4+Foxp3+ Treg cells and effector/memory CD4+ T cells increased in vivo. As a mechanism of increased Treg cells, HBZ expression directly induced Foxp3 gene transcription in T cells. The increased CD4+Foxp3+ Treg cells in HBZ transgenic mice were functionally impaired while their proliferation was enhanced. HBZ could physically interact with Foxp3 and NFAT, thereby impairing the suppressive function of Treg cells. Thus, the expression of HBZ in CD4+ T cells is a key mechanism of HTLV-1-induced neoplastic and inflammatory diseases.
Published in the journal: Induces T-Cell Lymphoma and Systemic Inflammation. PLoS Pathog 7(2): e32767. doi:10.1371/journal.ppat.1001274
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001274Summary
Human T-cell leukemia virus type 1 (HTLV-1) is the causal agent of a neoplastic disease of CD4+ T cells, adult T-cell leukemia (ATL), and inflammatory diseases including HTLV-1 associated myelopathy/tropical spastic paraparesis, dermatitis, and inflammatory lung diseases. ATL cells, which constitutively express CD25, resemble CD25+CD4+ regulatory T cells (Treg). Approximately 60% of ATL cases indeed harbor leukemic cells that express FoxP3, a key transcription factor for Treg cells. HTLV-1 encodes an antisense transcript, HTLV-1 bZIP factor (HBZ), which is expressed in all ATL cases. In this study, we show that transgenic expression of HBZ in CD4+ T cells induced T-cell lymphomas and systemic inflammation in mice, resembling diseases observed in HTLV-1 infected individuals. In HBZ-transgenic mice, CD4+Foxp3+ Treg cells and effector/memory CD4+ T cells increased in vivo. As a mechanism of increased Treg cells, HBZ expression directly induced Foxp3 gene transcription in T cells. The increased CD4+Foxp3+ Treg cells in HBZ transgenic mice were functionally impaired while their proliferation was enhanced. HBZ could physically interact with Foxp3 and NFAT, thereby impairing the suppressive function of Treg cells. Thus, the expression of HBZ in CD4+ T cells is a key mechanism of HTLV-1-induced neoplastic and inflammatory diseases.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) was the first human retrovirus associated with human diseases including adult T-cell leukemia (ATL) [1], [2] and HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP)[3], [4]. One of the virological attributes of HTLV-1 is that it transmits mainly by cell-to-cell contact [5], [6]. Therefore, HTLV-1 induces the proliferation of infected CD4+ T cells to increase further transmission [7]. HTLV-1 encodes several regulatory and accessory genes in the pX region located between the env gene and the 3′ LTR [7], [8]. Among the viral genes, tax possesses in vitro transforming activity and can induce cancers in transgenic (Tg) animals via its pleiotropic actions [9], [10]. Yet the expression of Tax is frequently disrupted in ATL [7]. In contrast, the HTLV-1 bZIP factor (HBZ) gene, which is encoded in the minus strand of the HTLV-1 genome [11], [12], is transcribed in all ATL cases [13]. Recently, it has been reported that APOBEC3G generates nonsense mutations in all HTLV-1 genes except HBZ [14], suggesting that the HBZ gene is indispensable for the growth and/or survival of ATL cells and HTLV-1 infected cells. The HBZ gene product promotes the proliferation of ATL cells [13], [15]. Further, HBZ mRNA expression in HAM/TSP patients was well correlated with disease severity [16]. These findings suggest that HBZ has a critical role in the development of ATL and HAM/TSP.
It has been shown that ATL cells functionally and phenotypically resemble Foxp3+ CD25+CD4+ regulatory T (Treg) cells, which control immune responses against self - and non-self-antigen [17]. ATL cells constitutively express CD25 and scarcely produce interleukin-2 (IL-2)[18], [19]. Furthermore, two thirds of ATL cases harbor leukemic cells expressing FoxP3 [20], [21], a key transcription factor for the generation and function of Treg cells [22], [23], [24]. In HTLV-1 carriers, HTLV-1 provirus is detected mainly in CD4+ effector/memory T cells and Treg cells [25], [26], [27]. Thus, HTLV-1 favors Treg cells and effector/memory T cells in vivo, and transforms them. However, how HTLV-1 targets these T cell subpopulations remains to be elucidated.
In this study, we show that transgenic expression of HBZ increases Foxp3+ Treg cells and effector/memory T cells, leading to development of T-cell lymphomas and systemic inflammatory diseases. In addition, the suppressive function of Treg cells is severely impaired in HBZ transgenic mice. At the molecular level, we show that HBZ interacts with Foxp3 and NFAT, interrupting the function of each molecule, and leading to the deregulation of Foxp3-mediated transcriptional control of the genes associated with Treg functions. These results indicate that HBZ plays a critical role in neoplastic and inflammatory diseases arising from HTLV-1 infection.
Results
HBZ transgenic mice spontaneously develop inflammatory lesions in the skin and lung
Since HTLV-1 mainly infects CD4+ T cells in vivo, we generated Tg mice expressing the HBZ gene under the control of the murine CD4-specific promoter/enhancer/silencer (Figure S1) [13]. We analyzed the HBZ transgenes (Figure S1) and their expression in the three lines generated. HBZ gene expression was specifically detected in CD4+ T cells (Figure 1A). HBZ protein was also detected in these transgenic mice (Figure 1B). The level of HBZ gene transcripts in line 12 was the most abundant but similar to that of endogenous expression of the HBZ gene in ATL cell lines (Figure 1C). Therefore, unless specifically described, we used line 12 in this study. Notably, the majority of HBZ-Tg mice developed skin lesions by 18 weeks of age, in contrast with no disease in non-transgenic littermates (non-Tg mice) (Figure 1, D and E). Histological analyses revealed infiltration of CD3+CD4+ T cells into the dermis and epidermis, and also the alveolar septa of the lung (Figure 1, F, G and S2), whereas no obvious evidence of inflammation in other tissues, including liver, kidney, muscle, heart, stomach, spinal cord, intestines and brain. Since massive infiltration of lymphocytes in the skin and lung was observed in line 9 and 12, but not in line 2, level of HBZ expression is likely associated with these phenotypes. Thus, HBZ-Tg mice spontaneously developed dermatitis and alveolitis. Similar lesions have been observed in HTLV-1 carriers, especially in those harboring large numbers of infected cells [28], [29].
Fig. 1. HBZ-Tg mice spontaneously develop inflammatory diseases in skin and lung. 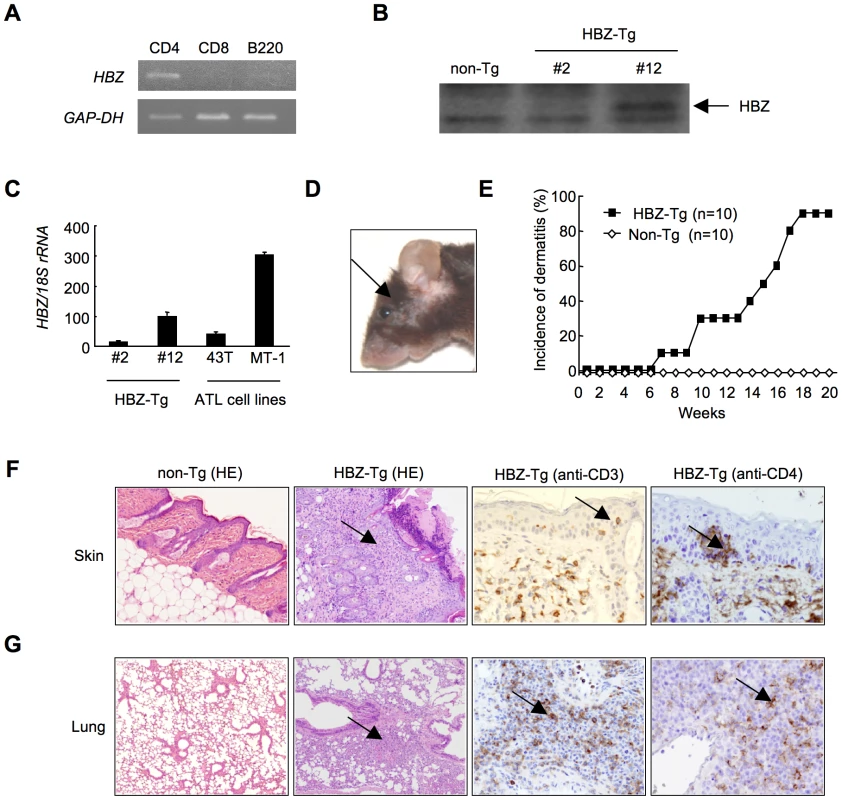
(A) Cell-type specific transcription of the transgene in line 12 was confirmed by RT-PCR in each sorted cell population. (B) The expression of HBZ protein in CD4+ splenocytes was confirmed by Western blotting. (C) Transcripts of the HBZ gene in CD4+ splenocyte of HBZ-Tg mice or ATL cell lines were quantified by real time PCR. ATL-43T and MT-1 are derived from ATL cells. (D) An HBZ-Tg mouse with typical skin symptom (Arrow indicates skin lesion). (E) The incidence of dermatitis in HBZ-Tg (line 12) and non-Tg mice. (F and G) Histological findings of the skin and the lung in HBZ-Tg mice. Lymphocytes massively infiltrated the dermis and epidermis (F) and the alveolar septum (G) (Arrows present infiltration of lymphocytes). Infiltration of CD3+, CD4+ T cells into these tissues was shown by immunohistochemistry compared with non-Tg mice as control. HBZ-Tg mice develop T-cell lymphoma after a long latent period
To study the growth-promoting activity of the HBZ gene, we assessed the proliferation of CD4+ T cells in HBZ-Tg mice by incorporation of bromodeoxyuridine (BrdU), and found that the proliferation was three fold-higher than in non-Tg mice, whereas the proliferation of CD8+ T cells or B cells was not altered (Figure 2A, Table S1A). Transgenic expression of HBZ enhances the in vivo proliferation of mouse T cells, as ectopic expression of HBZ enhances the proliferation of human T cells [13], [15]. It is known that HTLV-1 transforms CD4+ T cells after a long latent period in a fraction of asymptomatic carriers [7]. Analogous to the development of ATL in humans, 14 of 37 (37.8%) HBZ-Tg mice of all three-founder lines developed T-cell lymphomas after 16 months, in contrast with 2 of 27 non-Tg mice (7.4%) (P<0.001 by the logrank test) (Figure 2B). In some transgenic mice, lymphoma cells infiltrated various organs including the lung, bone marrow, spleen and liver (Figure 2C). All of the lymphomas in HBZ-Tg mice were CD3+ and CD4+ by immunohistochemical analyses when examined before the mice became moribund (Figure 2D). Lymphoma cells also expressed αβT cell receptors on their surfaces (Figure S3). Monoclonal proliferation of these lymphoma cells was shown by single strand conformation polymorphism in Vγ2-Jγ1 junction region of T cell receptor γchain gene (Figure S4). Notably, the primary lymphoma cells expressed Foxp3 at various intensities in the majority of cases (Figure 2E, Table 1), exhibiting a similar FoxP3 staining pattern to that in lymph nodes in human ATL cases (Figure S5). Thus, the T-cell lymphomas in HBZ-Tg mice phenotypically resemble ATL, suggesting that HBZ promotes proliferation of CD4+ T cells and predisposes expressing cells to transform in due course.
Fig. 2. HBZ-Tg mice develop T-cell lymphoma after a long latent period. 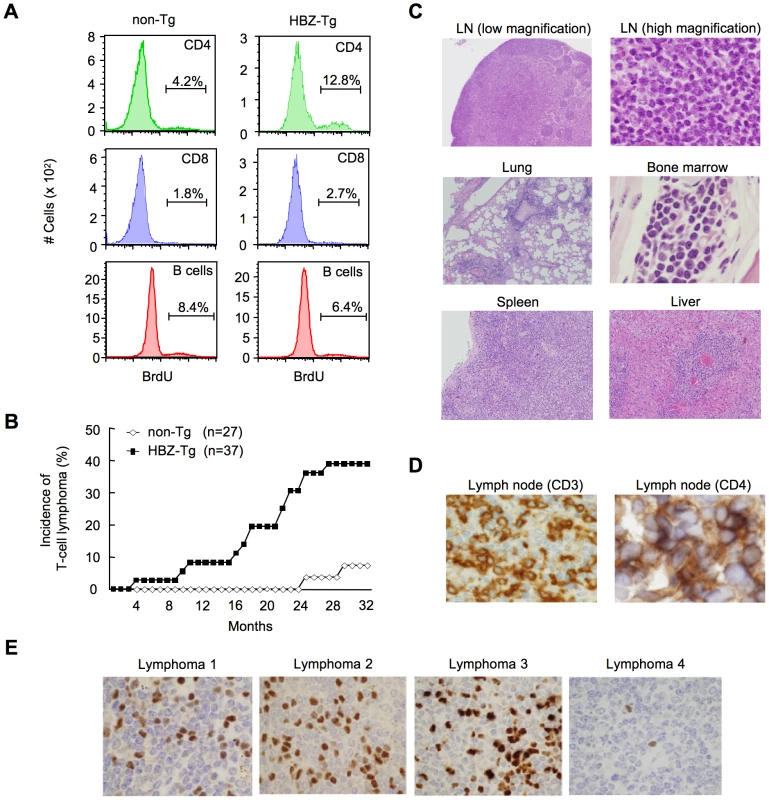
(A) BrdU was injected into mice twice a day for three days, and splenocytes were stained with antibodies to BrdU, CD4, CD8, and B220. (B) Incidence of T-cell lymphoma in HBZ-Tg mice was statistically significant compared with that in non-Tg mice (P<0.001 by the logrank test). (C) Pleomorphic lymphoma in the cervical lymph node in a representative HBZ-Tg mouse. Infiltrations of lymphoma cells into lung, bone marrow, spleen and liver are also shown. (D) Expression of CD3 and CD4 in lymphoma cells was shown by immunohistochemical staining. (E) Immunohistochemical staining for Foxp3 in primary lymphomas of HBZ-Tg mice. Tab. 1. Characteristics of lymphomas in HBZ-Tg and non-Tg littermates. 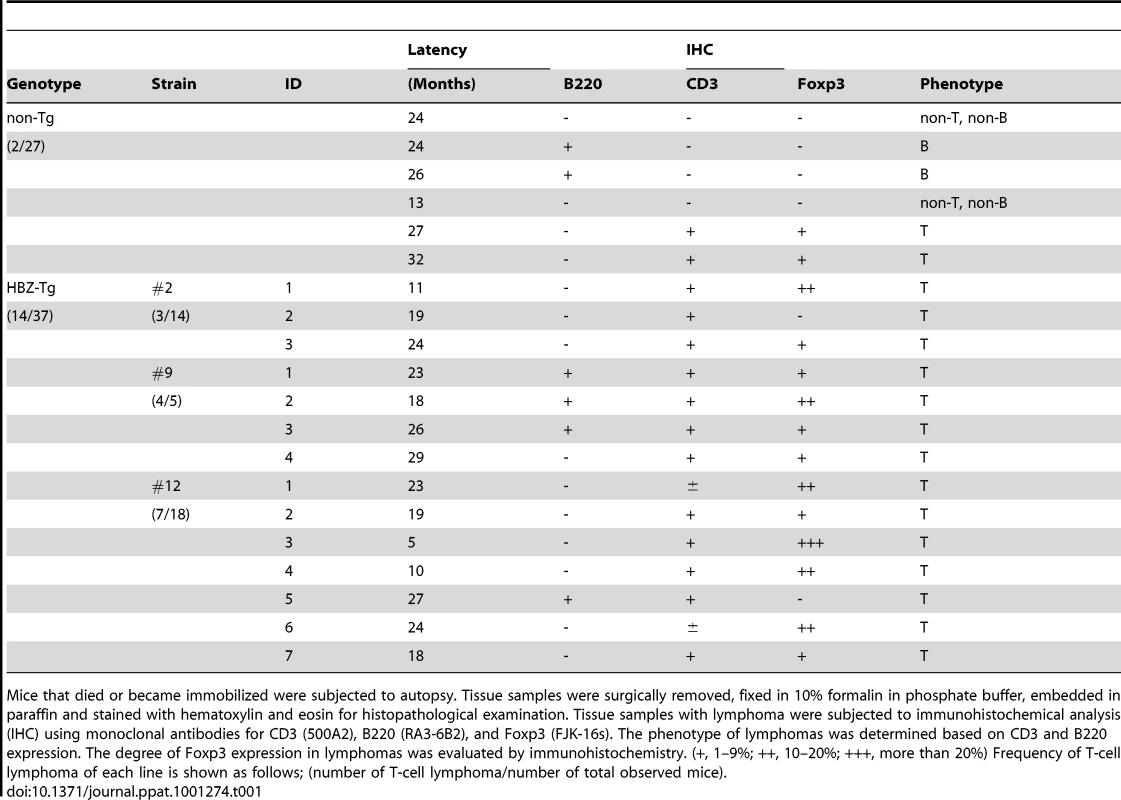
Mice that died or became immobilized were subjected to autopsy. Tissue samples were surgically removed, fixed in 10% formalin in phosphate buffer, embedded in paraffin and stained with hematoxylin and eosin for histopathological examination. Tissue samples with lymphoma were subjected to immunohistochemical analysis (IHC) using monoclonal antibodies for CD3 (500A2), B220 (RA3-6B2), and Foxp3 (FJK-16s). The phenotype of lymphomas was determined based on CD3 and B220 expression. The degree of Foxp3 expression in lymphomas was evaluated by immunohistochemistry. (+, 1–9%; ++, 10–20%; +++, more than 20%) Frequency of T-cell lymphoma of each line is shown as follows; (number of T-cell lymphoma/number of total observed mice). Increased effector/memory and regulatory CD4+ T cells in HBZ-Tg mice
To study the cellular basis of the lymphomagenesis and inflammation in HBZ-Tg mice, we analyzed the phenotype and function of T cells, especially Treg cells, in 3-month-old HBZ-Tg line 12 mice before their pathological manifestations. CD44high CD62Llow effector/memory CD4+ T cells increased in HBZ-Tg mice (Figure 3A). CD4 single positive T cells also increased in the thymus (Figure S6). Further, not only the ratio but also the absolute number of Foxp3+ T cells was markedly increased in HBZ-Tg mice compared with non-Tg mice, while the numbers of Foxp3− T cells were equivalent (Figure 3, B and C). Increased Treg cells were also observed in thymus, lymph node and peripheral blood mononuclear cells (Figure 3D and Figure S7). We also observed the increased Treg cells and effector/memory T cells in the HBZ-Tg line 2 (Figure S8), which showed quite lower expression of HBZ than line 12 (Figure 1C). The proportion of Treg cells in skin and lung was rather low compared with that in spleen (Figure 3B and S2), indicating that Foxp3− T cells are predominant in the infiltrating T cells.
Fig. 3. Transgenic expression of HBZ in CD4+ T cells increases Foxp3+ Treg cells with impaired suppressive function. 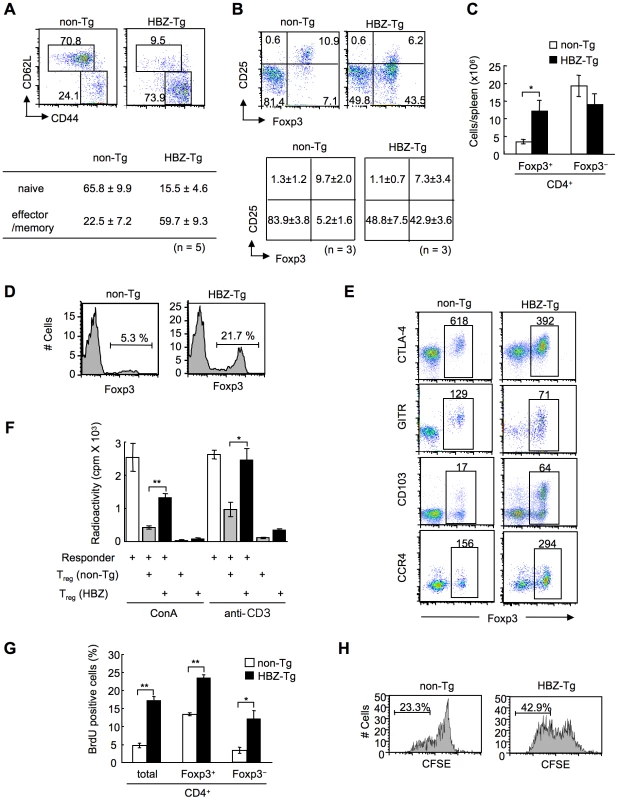
(A and B) Mouse splenocytes were stained with the indicated antibodies, and analyzed by flow cytometry. Representative dot plots gated on the CD4+ population are shown. For these experiments, HBZ-Tg mice without any symptoms were used. Tables show the mean ± SD (n = 5 for A, n = 3 for B). (C) The absolute number of Foxp3+ or Foxp3− CD4+ T cells in HBZ-Tg and non-Tg mice. The results shown are the mean ± SD (n = 3). (D) Flow cytometric analysis for the Foxp3 expression in CD4 single positive thymocytes. Representative dot plots gated on the CD4 single positive population are shown from three independent analyses. (E) Flow cytometric analyses of CD4+ T cells for Treg related molecules. Numbers in dot plots indicate mean fluorescence intensity (MFI) of each molecule in the rectangular gates. (F) Suppressive activity of Treg cells from HBZ- or non-Tg mice on T-cell proliferation. Sorted Foxp3+ T cells were cultured with CD4+CD25− cells of non-Tg mice as responder cells for 72 h with ConA or soluble anti-CD3 antibody and x-irradiated antigen presenting cells (APCs), and [3H] thymidine incorporation during the last 6 hours was measured. Results are means ± SD for triplicate cultures. (G) In vivo BrdU incorporation in total CD4+, Foxp3+CD4+, or Foxp3−CD4+ T cells. The results shown are the mean ± SD (n = 3). (H) Sorted Foxp3+ cells were labeled with CFSE and cultured with anti-CD3 antibody and x-irradiated APCs. After 96 hours, the cells were stained with anti-Foxp3, and CFSE dilution was analyzed for Foxp3+ cells. *, P<0.01; **, P<0.001 by two-tailed Student t-test. This result indicates that transgenic expression of HBZ induces systemic inflammation despite an increase in Foxp3+ Treg cells. It has been reported that IL-2 is critical in the homeostasis of Treg cells [30]. To study mechanisms by which HBZ expression increases Treg cells, we analyzed IL-2 production in the CD4+ T cells of HBZ-Tg mice after stimulation by PMA and ionomycin. IL-2 production was not augmented in either the Foxp3+ or Foxp3− populations from HBZ-Tg mice (Figure S9), indicating that the increase in the number of Treg cells was not due to enhanced IL-2 production.
Previous studies showed that Tax is a critical viral protein for the pathogenesis of HTLV-1. Therefore, we generated Tax transgenic (tax-Tg) mice using the same promoter/enhancer/silencer. In the tax-Tg mice, we did not observe increased effector/memory T cells or Treg cells (Figure S10). Thus, this increase in effector/memory T cells and Treg cells was specific to HBZ and not associated with similar transgenic expression of tax in this transgenic model system.
We next analyzed the phenotype and function of the increased Foxp3+ Treg cells in HBZ-Tg mice. CD4+Foxp3+ T cells of HBZ-Tg mice expressed Treg-associated molecules, such as cytotoxic T-lymphocyte associated antigen-4 (CTLA-4), glucocorticoid-induced TNF receptor family-related-protein (GITR), CD103, and CD25 [31]; yet the expression levels of CTLA-4, GITR and CD25 were lower than those of Foxp3+ T cells in non-Tg mice (Figure 3, B and E, Table S1B). In contrast, both Foxp3+ and Foxp3− CD4+ T cells of HBZ-Tg mice expressed CCR4 and CD103 at higher levels than those in non-Tg mice, suggesting that this might contribute to the migration and infiltration of HBZ-Tg CD4+ T cells into the skin (Figure 1F) [32], [33]. Further, it is of note that the in vitro suppressive function of HBZ-Tg Treg cells was severely impaired. When CD4+GITRhigh T cells, which were >90% Foxp3+ [23], from HBZ-Tg or non-Tg mice were co-cultured with CD4+CD25− T cells from wild-type mice and stimulated with Con A or anti-CD3 antibody, HBZ-Tg Treg cells were much less suppressive (Figure 3F). These results indicate that HBZ expression increases functionally impaired Treg cells.
Next, we assessed the proliferation of CD4+ T cells in HBZ-Tg mice. BrdU incorporation of Foxp3+ as well as Foxp3−CD4+ T cells from HBZ-Tg mice was also significantly higher than those in non-Tg mice (Figure 3G). In general, proliferation of Treg cells in response to mitogenic stimulation is suppressed in vitro. However, Foxp3+ T cells from HBZ-Tg mice proliferated more vigorously in vitro in response to anti-CD3 antibody than did non-Tg Foxp3+ T cells (Figure 3H). Thus, transgenic expression of HBZ in CD4+ T cells induces the expansion of Foxp3+ Treg cells, yet impairs their suppressive function.
HBZ directly induces Foxp3 expression in a CD4+ T-cell intrinsic manner
To study whether HBZ increases Foxp3+ Treg cells in a cell intrinsic manner, we expressed HBZ in naive CD4+ T cells in vitro using a retrovirus vector (Figure 4A). Interestingly, HBZ induced Foxp3 expression in 16.8% of HBZ expressing T cells, which is a similar enhancement to that due to TGF-β treatment (14.8%). The expression was markedly augmented in HBZ expressing T cells treated with TGF-β (72.2%) (Figure 4B). A reporter assay using the enhancer and promoter of the Foxp3 gene [34] demonstrated that HBZ induced transcription of the Foxp3 gene (Figure 4C), which was enhanced in the presence of TGF-β. Thus, HBZ-induced Foxp3 expression could be a mechanism for the increase of Foxp3+ T cells in HBZ-Tg mice.
Fig. 4. HBZ directly induces Foxp3 expression in CD4+ T cells. 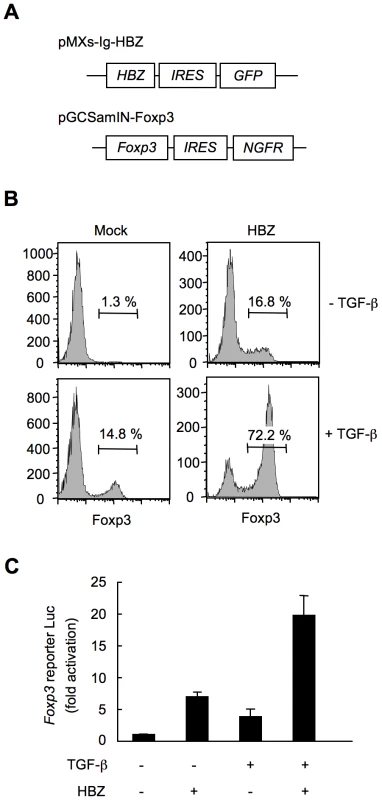
(A) Schematic diagrams of retrovirus vectors used in this study. (B) Mouse CD4+CD25− T cells transduced with retrovirus vector encoding HBZ or empty vector with or without TGF-β were stained with anti-Foxp3 antibody and analyzed by flow cytometry. (C) To study the effect of HBZ on promoter activity of the Foxp3 gene, EL4 cells were transfected with Foxp3 reporter plasmid and/or HBZ expressing plasmid. Representative data shown are firefly luciferase activities normalized to those of renilla luciferase (mean ± SD). HBZ physically interacts with Foxp3
Previous studies have shown that Foxp3 controls Treg function by cooperating with transcription factors including NFAT [35] and AML-1/Runx1[36]. Impaired interactions of Foxp3 with these factors not only alter the suppressive function of Treg cells but also suppress the expression of Treg associated molecules, such as CD25, CTLA-4, and GITR [23], [35], [36], [37], which is similar to the phenotype observed in HBZ-Tg mice (Figure 3, B and E). These findings prompted us to assess the possibility that HBZ might be involved in Foxp3-dependent transcriptional regulation. To address this, we first examined direct interaction among HBZ, NFAT and Foxp3. Immunoprecipitation experiments showed that HBZ physically interacted with both NFAT and Foxp3 (Figure 5A). Moreover, to study the interaction of endogenous HBZ and Foxp3, we performed immunoprecipitation using ATL-43T, a Foxp3-expressing ATL cell line. An anti-HBZ antibody co-precipitated endogenous Foxp3 in the ATL-43T cells, demonstrating that the interaction occurs not only in an enforced over-expressed state but also under physiological conditions (Figure 5B). It has been previously reported that human FoxP3 protein migrates as a doublet, which coincides with this result [38]. Analyses using HBZ deletion mutants showed that the central domain of HBZ interacted with Foxp3 (Figure 5C). Experiments with Foxp3 deletion mutants revealed that HBZ interacted with the forkhead (FH) domain of Foxp3 (Figure 5D). It has been reported that the region between the forkhead domain and the leucine zipper domain of Foxp3 interacted with AML-1 [36]. HBZ did not inhibit the binding between Foxp3 and AML-1 nor the suppressive effect of Foxp3 on AML-1-mediated transcription from the IL-2 gene promoter (Figure S11), indicating that HBZ does not influence Foxp3/AML1 mediated gene regulation.
Fig. 5. HBZ physically interacts with Foxp3 and NFAT. 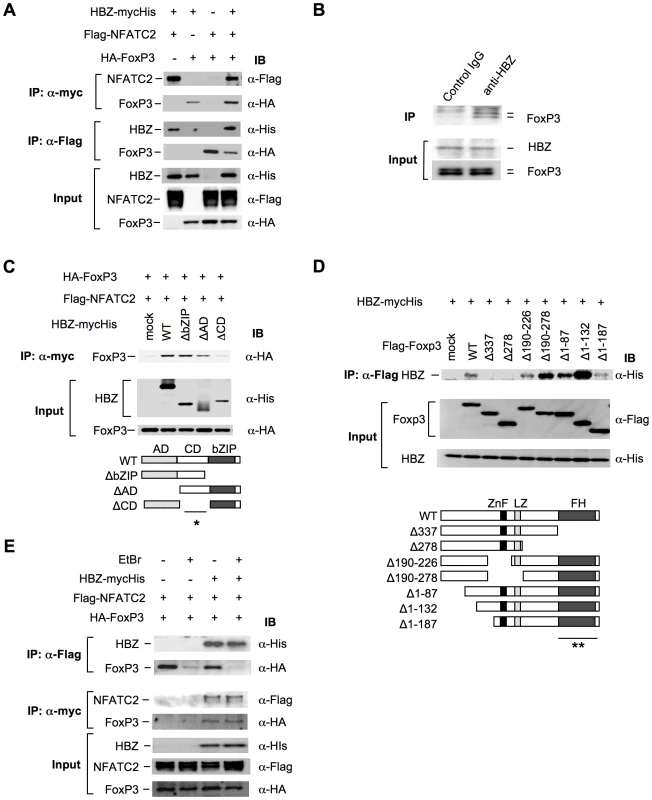
(A) The expression vectors of the indicated proteins were co-transfected into 293FT cells, and their interactions were analyzed by immunoprecipitation (IP). (B) Nuclear extract of ATL-43T cells was subjected to IP with anti-HBZ antibody or control IgG, and detected by anti-FoxP3 antibody. (C and D) The interactions of HBZ and Foxp3 were analyzed by IP using HBZ mutants (C) or Foxp3 mutants (D). A schematic diagram of Foxp3 mutants is shown. ZnF, zinc finger; LZ, leucine zipper; FH, forkhead domain. Asterisks (* or **) show responsible region for each molecular interaction. (E) The interactions among HBZ, Foxp3 and NFATc2 were analyzed with or without EtBr. To study whether HBZ independently interacts with Foxp3 and NFAT or, alternatively, if these molecules form a ternary complex, we studied the effect of the DNA intercalator ethidium bromide (EtBr) on their interactions. As shown in Figure 5E, the interactions of HBZ with Foxp3 or NFAT were not affected by EtBr while the interaction between NFAT and Foxp3 was diminished by EtBr as reported previously [35]. These findings suggest that the interactions of HBZ with NFAT and Foxp3 are independent of DNA while the interaction between NFAT and Foxp3 requires the presence of DNA.
HBZ inhibits Foxp3-mediated CTLA-4 and GITR expression in CD4+ T cells in vitro
In HBZ-Tg mice, the expression of Treg-associated molecules including CTLA-4, GITR and CD25 was suppressed when compared with their expression in Treg cells from non-Tg mice (Figure 3B and E). This finding may account for the impaired function of Treg cells since these molecules, in particular CTLA-4, play a critical role in Treg-mediated suppression [39]. To further study the effect of HBZ on the expression of Treg-associated molecules, we transduced HBZ along with Foxp3 into naive CD4+ T cells in vitro using retrovirus vectors (Figure 4A). HBZ expression suppressed Foxp3-induced GITR and CTLA-4 expression whereas it did not inhibit CD25 expression (Figure 6A). Expression of HBZ alone increased CD25 expression (Figure 6A), which might obscure the suppressive effect of HBZ under these conditions. Suppression of GITR and CTLA-4 expression required both the activation and the central domains of HBZ (Figure 6, B and C), which correspond to the binding sites of HBZ to Foxp3 (Figure 5C) and NFAT (Figure S12). Since both Foxp3 and NFAT are critical for Treg function [35], it is likely that HBZ suppresses the expression of GITR and CTLA-4 by interacting with Foxp3 and NFAT and thereby interfering with their transcriptional regulation in Treg cells. To examine suppressive effect of HBZ on expression of GITR, CTLA-4 and CD25, we isolated Treg cells from wild type mice and expressed HBZ using retroviral vectors. As shown in Figure 6D, HBZ suppressed endogenous expression of CD25, GITR and CTLA-4 in Treg cells, confirming that HBZ is responsible for suppressed expression of these molecules.
Fig. 6. HBZ inhibites Foxp3-mediated CTLA-4 and GITR expression in vitro. 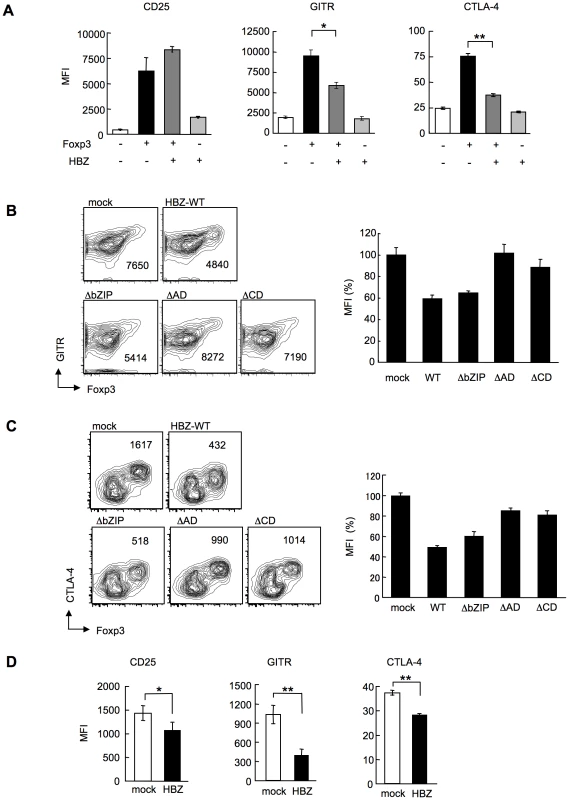
(A) Mouse CD4+CD25− T cells co-transduced with the retroviral vectors were stained with the indicated antibodies. Mean fluorescence intensity (MFI) of CD25, GITR, and CTLA-4 in GFP/NGFR double-positive cells are shown as mean ± SD. for triplicate culture. *, P<0.01; **, P<0.001 by two-tailed Student t-test. (B and C) CD4+CD25− T cells transduced with the pMXs-Ig vector encoding wild-type or mutant HBZ, and pGCSamIN-Foxp3 vector were stained with anti-GITR (B) or anti-CTLA-4 (C) antibody in addition to anti-NGFR antibody, and then analyzed by flow cytometry. Left, numbers in density plots indicate MFI of GITR (B) or CTLA-4 (C) in GFP/NGFR double-positive cells. Representative data from three independent experiments are shown. Right, relative MFI of wild type or mutated HBZ compared to mock transduced cells was shown as mean ± SD (n = 3). (D) HBZ transduction in Foxp3+ Treg cells inhibited the endogenous expression of Treg associated molecules. Mean fluorescence intensity (MFI) of CD25, GITR, and CTLA-4 in CD4+Foxp3+NGFR+ cells are shown as mean ± SD. for triplicate culture. *, P<0.05; **, P<0.01 by two-tailed Student t-test. Discussion
HTLV-1 targets CD4+ T cells; cell central to immune regulation. In contrast to human immunodeficiency virus, which destroys CD4+ T cells, HTLV-1 increases its copy number by inducing clonal proliferation of infected cells in vivo [40], [41]. Since HTLV-1 spreads mainly by cell-to-cell transmission [5], increased number of infected cells facilitates transmission of HTLV-1 to new cells. Recent studies showed that glucose transporter 1, heparan sulfate proteoglycans and neuropilin-1 are important for the entry of HTLV-1[42], [43], [44], consistent with the finding that this virus can infect a variety of cell types [45], [46]. However, HTLV-1 provirus is detected mainly in the regulatory and effector/memory CD4+ T cells of HTLV-1 carriers (Figure S13) [25], [26], [27], which indicates that HTLV-1 favors these specific subpopulations of CD4+ T cells. These findings suggest that HTLV-1 induces proliferation and/or facilitates survival of the regulatory and effector/memory CD4+ T cells. The mechanism(s) by which HTLV-1 targets Treg cells, however, remained unclear until now. In this study, we showed that HBZ could enhance transcription of the Foxp3 gene, and also promote proliferation of Foxp3+CD4+ T cells in transgenic mice, indicating that HBZ enhances both the generation and proliferation of Foxp3+ T cells. Impaired Foxp3 function is associated with proliferation of Treg cells [37], so the HBZ-mediated Treg dysfunction may also contribute to Treg proliferation in addition to direct growth proliferation by the HBZ transcript [13]. Another possible explanation is that Treg cells might be more susceptible to HTLV-1 infection, since Treg cells proliferate vigorously in vivo presumably by recognizing self-antigen and commensal microbes [47]. With these strategies, HTLV-1 likely targets this specific T-cell population as its host, which might be beneficial for their survival.
As mechanisms of the HBZ-mediated effect on Foxp3 functions, we demonstrated that HBZ physically interacted with Foxp3 and impaired its function in vitro. HBZ lacking the Foxp3-binding region showed a slight inhibitory effect on Foxp3 function, indicating that direct interaction between HBZ and Foxp3 is, at least in part, responsible for suppression. The results of immunoprecipitation analyses using Foxp3 mutants showed that the forkhead domain of Foxp3 was responsible for the molecular interaction between HBZ and Foxp3. Since the forkhead domain is the DNA-binding domain of Foxp3 [17], HBZ might inhibit the transcriptional function of Foxp3 by interfering with the DNA binding activity. Foxp3 play a key role in the function and homeostasis of Treg cells [22], [23], [24], indicating that HBZ-mediated dysfunction of Foxp3 contributes to impaired Treg function in HBZ-Tg mice. This impaired Treg function allows non-regulatory T cells to become hyper-reactive to commensal microbes and self-antigens, provoking enhanced proliferation of non-regulatory T cells and T cell-mediated autoimmune/inflammatory disease. These data collectively suggest that the viral protein HBZ hijacks the transcriptional machinery of host Treg cells leading to inflammatory disorders in the host. Conversely, Tax, another HTLV-1 protein, has been reported to suppress FoxP3 expression in human T cells in vitro [48]. Therefore, it is likely that both viral proteins target Foxp3 albeit with apparently different effects. Considering that HBZ is consistently expressed while Tax expression is sporadic, Tax might control excess expression of Foxp3 in HTLV-1 infected cells.
In this study, we demonstrated that the characteristics of CD4+ T cells in HBZ-Tg mice resemble those of human ATL cells or HTLV-1 infected cells in carriers. First, the frequency of Foxp3 positive cells in T-cell lymphomas was similar in HBZ-Tg mice and in ATL [20]. Second, the suppressive function of Foxp3+ T cells was impaired in both ATL and HBZ-Tg mice [49]. Third, CD4+ T cells in HBZ-Tg mice, HTLV-1-infected cells in carriers, and ATL cells possess similar effector/memory and regulatory phenotypes [25], [27]. As shown in this study, transgenic mice expressing Tax under the same promoter as the HBZ-Tg mice did not show any changes in the number of Foxp3+ Treg cells or effector/memory T cells. These data suggest that HBZ, rather than Tax, is responsible for conferring the specific phenotype of HTLV-1 infected cells and ATL cells.
It has been reported that tax transgenic animals develop tumors [50], [51], [52]. In these reports, Tax induced tumors, the type of which depends on the promoter used. However, irrespective of the possible oncogenic activity of Tax, leukemic cells in ATL patients frequently lose Tax expression [7], whereas HBZ expression has been detected in all ATL cases studied so far [13]. We reported that the HBZ gene transcript itself has growth-promoting activity in vitro [13]. Taken together, our results suggest that HBZ is responsible for the specific phenotype, function and proliferation of HTLV-1-infected CD4+ T cells and ATL cells, and that HBZ plays important roles for the oncogenic activity of HTLV-1 in addition to Tax. Further, the long latent period before the onset of T-cell lymphomas in HBZ-Tg mice suggests that additional genetic and/or epigenetic alterations in CD4+ T cells are necessary for the development of T-cell lymphomas in HBZ-Tg mice as well as for ATL.
In conclusion, the HBZ-mediated dysregulation of Treg function and proliferation that we report here provides novel insights into the interaction between the host and the virus and may be exploited to treat and prevent HTLV-1-induced diseases.
Materials and Methods
Ethics statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Board of Kyoto University (E921). All patients provided written informed consent for the collection of samples and subsequent analysis. Animal experimentation was performed in strict accordance with the Japanese animal welfare bodies (Law No. 105 dated 19 October 1973 modified on 2 June 2006), and the Regulation on Animal Experimentation at Kyoto University. The protocol was approved by the Institutional Animal Research Committee of Kyoto University (Permit Number: D09-3). All efforts were made to minimize suffering.
Mice and cell cultures
C57BL/6J mice were purchased from CLEA Japan. The HBZ cDNA was cloned into the SalI site of the H/M/T-CD4 vector, which was designed for restricted expression of a transgene in CD4+ cells. The purified fragment containing the HBZ transgene was microinjected into C57BL/6J F1 fertilized eggs. Transgenic founders were screened for the integration of transgenes in their genomic DNA by PCR and mated with C57BL/6J mice to generate transgenic progeny [13], [15]. All HBZ-Tg mice were heterozygotes for the transgene. The phenotype of HBZ-Tg mice was stable in the different generations. They express the spliced HBZ gene under the control of the CD4-specific promoter/enhancer/silencer. All mice were used at 10-16 weeks of age unless specifically described.
The human embryonic kidney cell line, 293FT, was cultured in DMEM containing 10% FCS and G418 (500 µg/ml). The 293FT cell line is derived from the 293F cell line and stably expresses the SV40 large T antigen. 293FT cell line was purchased from Invitrogen. The packaging cell line, Plat-E (kindly provided by T. Kitamura, Tokyo University) was cultured in DMEM supplemented with 10% FCS containing 10 µg/ml blasticidin and 1 µg/ml puromycin. ATL-43T(−) (kindly provided by M. Maeda, Kyoto University) and MT-1 cells were derived from ATL cells, and cultured in RPMI containing 10% FCS and antibiotics (penicillin and streptomycin). A mouse T-cell lymphoma line, EL4 cells, were cultured with RPMI1640 containing 10% FCS, antibiotics, and 50 µM 2-mercaptoethanol (2-ME; Invitrogen).
Plasmids
In order to construct the vectors expressing tagged spliced HBZ and its mutants, their coding sequences were amplified by PCR, and cloned into the expression vector, pcDNA 3.1(−)/myc-His (Invitrogen). A cDNA clone that contains NFATc2 coding sequence was kindly provided by Kazusa DNA Research Institute. To construct the FLAG-tagged NFATc2 expression vector, its coding region was cloned into pCMV-Tag2 (Stratagene). pCMV-HA (Clontech) was used to generate HA-tagged Foxp3 expression vectors. The vectors expressing Flag-tagged Foxp3 mutants were also used for immunoprecipitation.
Antibodies and reagents
The following antibodies were used for immunoprecipitation and Western blotting: mouse anti-Flag (clone M2; Sigma, Saint Louis, MO), mouse anti-c-myc (clone 9E10; Sigma), mouse anti-HA (clone HA-7; Sigma), rabbit anti-His polyclonal antibody (MBL), rabbit anti-FOXP3 (polyclonal antibody; Abcam), and rabbit anti-HBZ polyclonal antisera [15].
The following antibodies were purchased from BD PharMingen; purified monoclonal antibody (mAb) for mouse CD4 (RM4-5), CD8α (53-6.7), CD25 (PC61), CD44 (IM7), CD103 (M290), and IL-2 (JES6-5H4). Purified monoclonal antibodies for mouse GITR (DTA-1), CTLA-4 (UC10-4B9), CD62L (MEL-14), TCRβ (H57-597), TCRγδ (eBioGL3) and Foxp3 (FJK-16s) or human FoxP3 (236A/E7) were purchased from eBioscience. Anti-mouse CCR4 antibody (polyclonal antibody; Capralogics) and FITC-labeled anti-goat IgG antibody (Santa Cruz Biotechnology) were used for the detection of mouse CCR4. The following reagents were used for cell culture: anti-CD3ε antibody (145-2C11; R&D systems), Con A (Sigma), PMA (Sigma), and ionomycin (Sigma).
Synthesis of cDNA and semiquantitative RT-PCR
cDNAs were synthesized from 1 µg total RNA of purified mouse CD4+ T cells by a reverse transcriptase SuperScript III and random primers according to the manufacturer's instructions (Invitrogen). Spliced HBZ and GAPDH transcripts were quantified using RT-PCR. The primers used were as follows: sHBZ gene: 5′-TAAACTTACCTAGACGGCGG-3′ (sense), 5′-CTGCCGATCACGATGCGTTT -3′ (antisense); GAPDH gene: 5′-GTGGAGA TTGTTGCCATCAACG -3′ (sense) and 5′-AGAGGGGCCATCCACAGTCTT-3′ (antisense). PCR was performed in a PC-808 (Astec) under the following conditions: HBZ: 2 minutes at 95°C, followed by 26 cycles of 30 seconds at 95°C, 30 seconds at 59°C and 60 seconds at 72°C; GAPDH: 3 minutes at 95°C, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 61°C and 30 seconds at 72°C.
Quantitative RT-PCR
To quantify the expression level of HBZ, a TaqMan probe and primers for HBZ were designed. The sequences of primers and probe for HBZ were as follows; HBZ primers; 5′-GGACGCAGTTCAGGAGGCAC-3′ (sense) and 5′-CCTCCAAGGATAATAGCCCG-3′ (antisense); HBZ probe; 5′-CCTGTGCCATGCCCGGAGGACCTGC-3′. We used the TaqMan Gene expression Assay for 18S rRNA (Applied Biosystems) as an internal control. Relative expression level of HBZ or IL-2 was calculated with the delta delta Ct method.
Retroviral constructs and transduction
For retroviral gene transduction experiments, spliced HBZ cDNA was cloned into a retroviral vector, pMXs-Ig (a gift from T. Kitamura), to generate pMXs-Ig-HBZ. pGCSamIN (kindly provided from M. Onodera) and pGCSamIN-Foxp3 were used as previously described. Transfection of the packaging cell line, Plat-E, was performed as described. For retroviral transduction, CD25−CD4+ cells were enriched by a CD4 enrichment kit (BD Pharmingen) and were activated by 0.5 µg/ml anti-CD3 Ab and 50 U/ml rIL-2 in the presence of T-cell-depleted and x-irradiated (20Gy) C57BL/6J splenocytes as APCs in 12 well plates. After 16 hours, activated T cells were transduced with viral supernatant and 4 µg/ml polybrene, and centrifuged at 3,000 rpm for 60 min. Cells were cultured in medium supplemented with 50 U/ml rIL-2. Activation of naïve T cells by anti-CD3 antibody influenced expression of these molecules. Therefore, we analyzed their expression after influence by activation was lost [35]. Two days later, Foxp3-mediated CTLA-4 expression was detected by a flow cytometry, and five days later, expression of GITR or CD25 was analyzed. After two days, we stimulated the transduced cells with 50 ng/ml PMA and 1 µg/ml ionomycin in the presence of protein transport inhibitor (BD PharMingen) for 6 hours, and then analyzed intracellular IL-2 expression using intracellular cytokine staining kits (BD Pharmingen) according to the manufacturer's instructions.
To elucidate the effect of HBZ on endogenous expression of Treg associated molecules, we transduced HBZ into CD4+Foxp3+ cells purified from mouse splenocytes. Three days after transduction, the expression levels of Treg associated molecules were evaluated by a flow cytometry.
Preparation of splenocytes, flow cytometric analyses, cell sorting, and assays of regulatory T cells
Cell suspensions were prepared from murine spleens by forcing the organs through a nylon mesh, and splenic erythrocytes were eliminated with NH4Cl. Proliferation of murine cells was measured by 3H-thymidine uptake after 3 days of incubation in RPMI1640 medium supplemented with 10% FCS and 50 µM 2-ME. Flow cytometric analyses and cell sorting were carried out using a FACS CantoII or FACS Aria with Diva Software (BD Pharmingen) and the data was analyzed by FlowJo software (Treestar). For cell surface staining, 106 cells were incubated with mAbs for 30 min at 4°C, and then analyzed. For intracellular staining, we used a mouse Foxp3 staining kit according to its protocol (eBioscience). To sort Foxp3+ cells, suspended splenocytes were stained with mAb for CD4 and GITR, and the CD4+GITRhigh fraction was sorted by FACS Aria. Purity of the sorted population was always >90% by re-analysis of Foxp3 staining. For the ex vivo proliferation assay of Foxp3+ cells, carboxy-fluorescein diacetate, succinimidyl ester (CFSE)(Molecular Probe) was used according to the manufacturer's instructions. Foxp3+ T cells (2×104/well) were stimulated with anti-CD3 antibody (4 µg/ml) in round-bottomed 96-well plates in the presence of x-irradiated splenocytes as antigen presenting cells (APC; 5×104/well) for 96 hours. Then, cells were permeabilized, and stained with anti-Foxp3. CFSE dilution was analyzed by flow cytometry. To evaluate the suppressive activity of Foxp3+ T cells sorted from HBZ-Tg or non-Tg mice, Foxp3+ T cells (2×104/well) were cultured with CD25−CD4+ cells (2×104/well) and APCs (5×104/well) from wild-type mice for 72 h in the presence of soluble anti-CD3 (4 µg/ml) or Con A (1 µg/ml), and then [3H] thymidine incorporation was measured.
BrdU staining
In vivo proliferation was measured by BrdU incorporation. BrdU (Nacalai Tesque) was dissolved in PBS (3 µg/ml), and then 200 µl was injected intraperitoneally into HBZ-Tg and non-transgenic mice twice a day for three days as reported previously [53]. BrdU incorporation in CD4+, CD8+, or B220+ splenocytes was detected using FITC BrdU Flow Kits (BD Pharmingen) according to the manufacturer's instructions. Flow cytometric analyses were performed on a FACS CantoII with Diva Software (BD Pharmingen).
Foxp3 reporter assay
We constructed Foxp3 promoter and enhancer reporter plasmids as the previous report [34]. A murine T-cell line, EL4 cells (1×107), were transiently cotransfected by electroporation with the following plasmid DNAs: Foxp3 reporter plasmid, Renilla luciferase control vector (pRL-TK), and HBZ expression vector (pME18SneoHBZ). Cells were cultured with or without TGF-β (2 ng/ml). Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). Relative luciferase activities were calculated as the ratio of firefly and Renilla luciferase activities. The luciferase values are shown as relative values. Values represent means plus standard deviations (error bars) (n = 3).
Histological analyses
The study of clinical samples was approved by the local research ethics committee of the appropriate hospital. Tissue samples were fixed in 10% formalin in phosphate buffer and then embedded in paraffin. Haematoxylin and eosin (H&E) staining was performed according to standard procedures. Images were captured using a Provis AX80 microscope (Olympus) equipped with OLYMPUS DP70 digital camera, and detected using a DP manager system (Olympus).
For analysis of tumors, mice that became immobilized were sacrificed and subjected to autopsy. Tissue samples were surgically removed and fixed in 10% formalin in phosphate buffer and embedded in paraffin. Sections were stained with H&E for histopathologic examination. After we obtained informed consent, tissue samples from patients who were diagnosed as lymphoma-type ATL were analyzed by immunohistochemical methods to determine FoxP3 expression. Monoclonal antibodies for CD3ε(500A2; BD Pharmingen), B220 (RA3-6B2; BD Pharmingen), and Foxp3 (FJK-16s; eBioscience) were used for immunohistochemistry. We judged CD3+B220+ cases to be T-cell lymphomas since some activated T cells and T cells of the lpr/lpr mutant mouse expressed B220 [54], [55].
PCR/single stranded conformation polymorphism (SSCP) analysis
To investigate clonality of lymphoma cells observed in HBZ-Tg mice, lymphoma tissue samples of HBZ-Tg were analyzed for the clonality of T-cell receptor (TCR) γ locus using PCR-SSCP analysis of the TCR γ-gene. Genomic DNA was subjected to PCR amplification using primers for the Vγ2 gene and the Jγ1. The primers used were as follows: Vγ2 : 5′-ACCAAGAGATGAGACTGCACAA-3′ (sense), Jγ1 : 5′-GCGTCTGATCCTCAAAATAACTTCC-3′ (antisense); PCR was performed in a PC-808 (Astec) under the following conditions: 3 minutes at 95°C, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 55°C and 30 seconds at 72°C. We used EL-4 as a positive control and splenic DNA from young non-Tg or HBZ-Tg mice as negative control. PCR products were run on a 6% polyacrylamide gel and visualized by staining with DNA Silver Staining Kit (GE Healthcare).
Coimmunoprecipitation assay and immunoblotting
Expression vectors for the relevant genes were transiently cotransfected into 293FT cells using the TransIT-LT1 reagent (Mirus Bio). 24 hours later, transfected cells were stimulated with 50 ng/ml PMA and 1 µg/ml ionomycin for another 6 hours. Coimmunoprecipitation assays were performed using the Nuclear Complex Co-IP Kit (Active motif). Briefly, the nuclear extracts of transfected cells were prepared in the presence or absence of ethidium bromide (10 µg/ml). They were precleared with Protein G Sepharose 4 Fast Flow (GE Healthcare), and their supernatants were incubated with anti-myc tag (clone 9E10, Sigma) or anti-Flag tag (M2, Sigma) antibody overnight at 4°C. The immunocomplexes were precipitated with Protein G Sepharose 4 Fast Flow, fractionated in SDS-PAGE, and transferred to PVDF membranes. HBZ-myc-His was detected with horseradish peroxidase (HRP)-conjugated anti-His tag (MBL) antibody. HRP-conjugated anti-Flag tag (Sigma) and anti-HA tag (Sigma) antibodies were used to detect Flag-tagged and HA-tagged proteins, respectively. To detect endogenous interaction between HBZ and FoxP3, immunoprecipitation was performed using an ATL cell line, ATL-43T(-), as described above with anti-HBZ antisera and anti-FOXP3 antibody (Abcam). To examine the expression of HBZ in transgenic mice, CD4+ splenocytes from wild type or HBZ-Tg mice were enriched by a mouse CD4 T lymphocyte enrichment set (Pharmingen). Whole cell extracts were prepared with the lysis buffer (50 mM Tris-HCL, PH 7.5, 150 mM NaCl, 1% NP-40), and analyzed by western blotting with anti-HBZ antisera.
Flow cytometric analysis for HTLV-1 carrier cells
A previous report demonstrated that Tax expression could not be detected in freshly isolated PBMC from HTLV-1 infected carriers but could be detected when they were cultivated ex vivo for 12 hours [56]. We cultured PBMCs from asymptomatic HTLV-1 carriers for 12 hours and stained with monoclonal antibodies against FoxP3 or Tax (MI-73), and then analyzed by flow cytometry.
Statistical analysis
For in vitro experiments, multiple data comparisons were performed using Student's unpaired t-test. Statistical differences in the incidence of T-cell lymphoma were analyzed using a logrank test.
Supporting Information
Zdroje
1. TakatsukiK
2005
Discovery of adult T-cell leukemia.
Retrovirology
2
16
2. GalloRC
2005
The discovery of the first human retrovirus: HTLV-1 and HTLV-2.
Retrovirology
2
17
3. GessainA
BarinF
VernantJC
GoutO
MaursL
1985
Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis.
Lancet
2
407
410
4. OsameM
UsukuK
IzumoS
IjichiN
AmitaniH
1986
HTLV-I associated myelopathy, a new clinical entity.
Lancet
1
1031
1032
5. IgakuraT
StinchcombeJC
GoonPK
TaylorGP
WeberJN
2003
Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton.
Science
299
1713
1716
6. Pais-CorreiaAM
SachseM
GuadagniniS
RobbiatiV
LasserreR
2010
Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses.
Nat Med
16
83
89
7. MatsuokaM
JeangKT
2007
Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation.
Nat Rev Cancer
7
270
280
8. JournoC
DouceronE
MahieuxR
2009
HTLV gene regulation: because size matters, transcription is not enough.
Future Microbiol
4
425
440
9. GrassmannR
AboudM
JeangKT
2005
Molecular mechanisms of cellular transformation by HTLV-1 Tax.
Oncogene
24
5976
5985
10. LairmoreMD
SilvermanL
RatnerL
2005
Animal models for human T-lymphotropic virus type 1 (HTLV-1) infection and transformation.
Oncogene
24
6005
6015
11. LaroccaD
ChaoLA
SetoMH
BrunckTK
1989
Human T-cell leukemia virus minus strand transcription in infected T-cells.
Biochem Biophys Res Commun
163
1006
1013
12. GaudrayG
GachonF
BasbousJ
Biard-PiechaczykM
DevauxC
2002
The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription.
J Virol
76
12813
12822
13. SatouY
YasunagaJ
YoshidaM
MatsuokaM
2006
HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells.
Proc Natl Acad Sci U S A
103
720
725
14. FanJ
MaG
NosakaK
TanabeJ
SatouY
2010
APOBEC3G generates nonsense mutations in human T-cell leukemia virus type 1 proviral genomes in vivo.
J Virol
84
7278
7287
15. ArnoldJ
ZimmermanB
LiM
LairmoreMD
GreenPL
2008
Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation.
Blood
112
3788
3797
16. SaitoM
MatsuzakiT
SatouY
YasunagaJ
SaitoK
2009
In vivo expression of the HBZ gene of HTLV-1 correlates with proviral load, inflammatory markers and disease severity in HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP).
Retrovirology
6
19
17. SakaguchiS
YamaguchiT
NomuraT
OnoM
2008
Regulatory T cells and immune tolerance.
Cell
133
775
787
18. HattoriT
UchiyamaT
ToibanaT
TakatsukiK
UchinoH
1981
Surface phenotype of Japanese adult T-cell leukemia cells characterized by monoclonal antibodies.
Blood
58
645
647
19. UchiyamaT
HoriT
TsudoM
WanoY
UmadomeH
1985
Interleukin-2 receptor (Tac antigen) expressed on adult T cell leukemia cells.
J Clin Invest
76
446
453
20. KarubeK
OhshimaK
TsuchiyaT
YamaguchiT
KawanoR
2004
Expression of FoxP3, a key molecule in CD4CD25 regulatory T cells, in adult T-cell leukaemia/lymphoma cells.
Br J Haematol
126
81
84
21. ToulzaF
NosakaK
TakiguchiM
PagliucaT
MitsuyaH
2009
FoxP3+ regulatory T cells are distinct from leukemia cells in HTLV-1-associated adult T-cell leukemia.
Int J Cancer
125
2375
2382
22. FontenotJD
GavinMA
RudenskyAY
2003
Foxp3 programs the development and function of CD4+CD25+ regulatory T cells.
Nat Immunol
4
330
336
23. HoriS
NomuraT
SakaguchiS
2003
Control of regulatory T cell development by the transcription factor Foxp3.
Science
299
1057
1061
24. KhattriR
CoxT
YasaykoSA
RamsdellF
2003
An essential role for Scurfin in CD4+CD25+ T regulatory cells.
Nat Immunol
4
337
342
25. YasunagaJ
SakaiT
NosakaK
EtohK
TamiyaS
2001
Impaired production of naive T lymphocytes in human T-cell leukemia virus type I-infected individuals: its implications in the immunodeficient state.
Blood
97
3177
3183
26. ToulzaF
HeapsA
TanakaY
TaylorGP
BanghamCR
2008
High frequency of CD4+FoxP3+ cells in HTLV-1 infection: inverse correlation with HTLV-1-specific CTL response.
Blood
111
5047
5053
27. RichardsonJH
EdwardsAJ
CruickshankJK
RudgeP
DalgleishAG
1990
In vivo cellular tropism of human T-cell leukemia virus type 1.
J Virol
64
5682
5687
28. SugimotoM
NakashimaH
WatanabeS
UyamaE
TanakaF
1987
T-lymphocyte alveolitis in HTLV-I-associated myelopathy.
Lancet
2
1220
29. BittencourtAL
de Oliveira MdeF
2010
Cutaneous manifestations associated with HTLV-1 infection.
Int J Dermatol
49
1099
1110
30. FontenotJD
RasmussenJP
GavinMA
RudenskyAY
2005
A function for interleukin 2 in Foxp3-expressing regulatory T cells.
Nat Immunol
6
1142
1151
31. SakaguchiS
2005
Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self.
Nat Immunol
6
345
352
32. LehmannJ
HuehnJ
de la RosaM
MaszynaF
KretschmerU
2002
Expression of the integrin alpha Ebeta 7 identifies unique subsets of CD25+ as well as CD25 - regulatory T cells.
Proc Natl Acad Sci U S A
99
13031
13036
33. SatherBD
TreutingP
PerdueN
MiazgowiczM
FontenotJD
2007
Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease.
J Exp Med
204
1335
1347
34. ToneY
FuruuchiK
KojimaY
TykocinskiML
GreeneMI
2008
Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer.
Nat Immunol
9
194
202
35. WuY
BordeM
HeissmeyerV
FeuererM
LapanAD
2006
FOXP3 controls regulatory T cell function through cooperation with NFAT.
Cell
126
375
387
36. OnoM
YaguchiH
OhkuraN
KitabayashiI
NagamuraY
2007
Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1.
Nature
446
685
689
37. ChaeWJ
HenegariuO
LeeSK
BothwellAL
2006
The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells.
Proc Natl Acad Sci U S A
103
9631
9636
38. WalkerMR
KasprowiczDJ
GersukVH
BenardA
Van LandeghenM
2003
Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25 - T cells.
J Clin Invest
112
1437
1443
39. WingK
OnishiY
Prieto-MartinP
YamaguchiT
MiyaraM
2008
CTLA-4 control over Foxp3+ regulatory T cell function.
Science
322
271
275
40. WattelE
VartanianJP
PannetierC
Wain-HobsonS
1995
Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy.
J Virol
69
2863
2868
41. EtohK
TamiyaS
YamaguchiK
OkayamaA
TsubouchiH
1997
Persistent clonal proliferation of human T-lymphotropic virus type I-infected cells in vivo.
Cancer Res
57
4862
4867
42. ManelN
KimFJ
KinetS
TaylorN
SitbonM
2003
The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV.
Cell
115
449
459
43. JonesKS
Petrow-SadowskiC
BertoletteDC
HuangY
RuscettiFW
2005
Heparan sulfate proteoglycans mediate attachment and entry of human T-cell leukemia virus type 1 virions into CD4+ T cells.
J Virol
79
12692
12702
44. LambertS
BouttierM
VassyR
SeigneuretM
Petrow-SadowskiC
2009
HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165.
Blood
113
5176
5185
45. KoyanagiY
ItoyamaY
NakamuraN
TakamatsuK
KiraJ
1993
In vivo infection of human T-cell leukemia virus type I in non-T cells.
Virology
196
25
33
46. JonesKS
Petrow-SadowskiC
HuangYK
BertoletteDC
RuscettiFW
2008
Cell-free HTLV-1 infects dendritic cells leading to transmission and transformation of CD4(+) T cells.
Nat Med
14
429
436
47. Vukmanovic-StejicM
ZhangY
CookJE
FletcherJM
McQuaidA
2006
Human CD4+ CD25hi Foxp3+ regulatory T cells are derived by rapid turnover of memory populations in vivo.
J Clin Invest
116
2423
2433
48. YamanoY
TakenouchiN
LiHC
TomaruU
YaoK
2005
Virus-induced dysfunction of CD4+CD25+ T cells in patients with HTLV-I-associated neuroimmunological disease.
J Clin Invest
115
1361
1368
49. ShimauchiT
KabashimaK
TokuraY
2008
Adult T-cell leukemia/lymphoma cells from blood and skin tumors express cytotoxic T lymphocyte-associated antigen-4 and Foxp3 but lack suppressor activity toward autologous CD8+ T cells.
Cancer Sci
99
98
106
50. NerenbergM
HinrichsSH
ReynoldsRK
KhouryG
JayG
1987
The tat gene of human T-lymphotropic virus type 1 induces mesenchymal tumors in transgenic mice.
Science
237
1324
1329
51. GrossmanWJ
KimataJT
WongFH
ZutterM
LeyTJ
1995
Development of leukemia in mice transgenic for the tax gene of human T-cell leukemia virus type I.
Proc Natl Acad Sci U S A
92
1057
1061
52. HasegawaH
SawaH
LewisMJ
OrbaY
SheehyN
2006
Thymus-derived leukemia-lymphoma in mice transgenic for the Tax gene of human T-lymphotropic virus type I.
Nat Med
12
466
472
53. von BoehmerH
HafenK
1993
The life span of naive alpha/beta T cells in secondary lymphoid organs.
J Exp Med
177
891
896
54. AsanoT
TomookaS
SerushagoBA
HimenoK
NomotoK
1988
A new T cell subset expressing B220 and CD4 in lpr mice: defects in the response to mitogens and in the production of IL-2.
Clin Exp Immunol
74
36
40
55. IshimotoY
Tomiyama-MiyajiC
WatanabeH
YokoyamaH
EbeK
2004
Age-dependent variation in the proportion and number of intestinal lymphocyte subsets, especially natural killer T cells, double-positive CD4+ CD8+ cells and B220+ T cells, in mice.
Immunology
113
371
377
56. HanonE
HallS
TaylorGP
SaitoM
DavisR
2000
Abundant tax protein expression in CD4+ T cells infected with human T-cell lymphotropic virus type I (HTLV-I) is prevented by cytotoxic T lymphocytes.
Blood
95
1386
1392
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance inČlánek The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding ModuleČlánek A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 2- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- A Fresh Look at the Origin of , the Most Malignant Malaria Agent
- In Situ Photodegradation of Incorporated Polyanion Does Not Alter Prion Infectivity
- Highly Efficient Protein Misfolding Cyclic Amplification
- Positive Signature-Tagged Mutagenesis in : Tracking Patho-Adaptive Mutations Promoting Airways Chronic Infection
- Charge-Surrounded Pockets and Electrostatic Interactions with Small Ions Modulate the Activity of Retroviral Fusion Proteins
- Whole-Body Analysis of a Viral Infection: Vascular Endothelium is a Primary Target of Infectious Hematopoietic Necrosis Virus in Zebrafish Larvae
- Inhibition of Nox2 Oxidase Activity Ameliorates Influenza A Virus-Induced Lung Inflammation
- STAT2 Mediates Innate Immunity to Dengue Virus in the Absence of STAT1 via the Type I Interferon Receptor
- Uropathogenic P and Type 1 Fimbriae Act in Synergy in a Living Host to Facilitate Renal Colonization Leading to Nephron Obstruction
- Elite Suppressors Harbor Low Levels of Integrated HIV DNA and High Levels of 2-LTR Circular HIV DNA Compared to HIV+ Patients On and Off HAART
- DC-SIGN Mediated Sphingomyelinase-Activation and Ceramide Generation Is Essential for Enhancement of Viral Uptake in Dendritic Cells
- Short-Lived IFN-γ Effector Responses, but Long-Lived IL-10 Memory Responses, to Malaria in an Area of Low Malaria Endemicity
- Induces T-Cell Lymphoma and Systemic Inflammation
- The C-Terminus of RON2 Provides the Crucial Link between AMA1 and the Host-Associated Invasion Complex
- Critical Role of the Virus-Encoded MicroRNA-155 Ortholog in the Induction of Marek's Disease Lymphomas
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Atypical/Nor98 Scrapie Infectivity in Sheep Peripheral Tissues
- Innate Sensing of HIV-Infected Cells
- BosR (BB0647) Controls the RpoN-RpoS Regulatory Pathway and Virulence Expression in by a Novel DNA-Binding Mechanism
- Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance in
- Expression of Genes Involves Exchange of the Histone Variant H2A.Z at the Promoter
- The RON2-AMA1 Interaction is a Critical Step in Moving Junction-Dependent Invasion by Apicomplexan Parasites
- Epstein-Barr Virus Nuclear Antigen 3C Facilitates G1-S Transition by Stabilizing and Enhancing the Function of Cyclin D1
- Transcription and Translation Products of the Cytolysin Gene on the Mobile Genetic Element SCC Regulate Virulence
- Phosphatidylinositol 3-Monophosphate Is Involved in Apicoplast Biogenesis
- The Rubella Virus Capsid Is an Anti-Apoptotic Protein that Attenuates the Pore-Forming Ability of Bax
- Episomal Viral cDNAs Identify a Reservoir That Fuels Viral Rebound after Treatment Interruption and That Contributes to Treatment Failure
- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Relationship between Functional Profile of HIV-1 Specific CD8 T Cells and Epitope Variability with the Selection of Escape Mutants in Acute HIV-1 Infection
- The Genotype of Early-Transmitting HIV gp120s Promotes αβ –Reactivity, Revealing αβ/CD4 T cells As Key Targets in Mucosal Transmission
- Small Molecule Inhibitors of RnpA Alter Cellular mRNA Turnover, Exhibit Antimicrobial Activity, and Attenuate Pathogenesis
- The bZIP Transcription Factor MoAP1 Mediates the Oxidative Stress Response and Is Critical for Pathogenicity of the Rice Blast Fungus
- Entrapment of Viral Capsids in Nuclear PML Cages Is an Intrinsic Antiviral Host Defense against Varicella-Zoster Virus
- NS2 Protein of Hepatitis C Virus Interacts with Structural and Non-Structural Proteins towards Virus Assembly
- Measles Outbreak in Africa—Is There a Link to the HIV-1 Epidemic?
- New Models of Microsporidiosis: Infections in Zebrafish, , and Honey Bee
- The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding Module
- A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- Secreted Bacterial Effectors That Inhibit Host Protein Synthesis Are Critical for Induction of the Innate Immune Response to Virulent
- Genital Tract Sequestration of SIV following Acute Infection
- Functional Coupling between HIV-1 Integrase and the SWI/SNF Chromatin Remodeling Complex for Efficient Integration into Stable Nucleosomes
- DNA Damage and Reactive Nitrogen Species are Barriers to Colonization of the Infant Mouse Intestine
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
- Targeted Disruption of : Invasion of Erythrocytes by Using an Alternative Py235 Erythrocyte Binding Protein
- Trivalent Adenovirus Type 5 HIV Recombinant Vaccine Primes for Modest Cytotoxic Capacity That Is Greatest in Humans with Protective HLA Class I Alleles
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



