-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
Ubiquitylation targets proteins for proteasome-mediated degradation and plays important roles in many biological processes including apoptosis. However, non-proteolytic functions of ubiquitylation are also known. In Drosophila, the inhibitor of apoptosis protein 1 (DIAP1) is known to ubiquitylate the initiator caspase DRONC in vitro. Because DRONC protein accumulates in diap1 mutant cells that are kept alive by caspase inhibition (“undead” cells), it is thought that DIAP1-mediated ubiquitylation causes proteasomal degradation of DRONC, protecting cells from apoptosis. However, contrary to this model, we show here that DIAP1-mediated ubiquitylation does not trigger proteasomal degradation of full-length DRONC, but serves a non-proteolytic function. Our data suggest that DIAP1-mediated ubiquitylation blocks processing and activation of DRONC. Interestingly, while full-length DRONC is not subject to DIAP1-induced degradation, once it is processed and activated it has reduced protein stability. Finally, we show that DRONC protein accumulates in “undead” cells due to increased transcription of dronc in these cells. These data refine current models of caspase regulation by IAPs.
Published in the journal: IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation. PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002261
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002261Summary
Ubiquitylation targets proteins for proteasome-mediated degradation and plays important roles in many biological processes including apoptosis. However, non-proteolytic functions of ubiquitylation are also known. In Drosophila, the inhibitor of apoptosis protein 1 (DIAP1) is known to ubiquitylate the initiator caspase DRONC in vitro. Because DRONC protein accumulates in diap1 mutant cells that are kept alive by caspase inhibition (“undead” cells), it is thought that DIAP1-mediated ubiquitylation causes proteasomal degradation of DRONC, protecting cells from apoptosis. However, contrary to this model, we show here that DIAP1-mediated ubiquitylation does not trigger proteasomal degradation of full-length DRONC, but serves a non-proteolytic function. Our data suggest that DIAP1-mediated ubiquitylation blocks processing and activation of DRONC. Interestingly, while full-length DRONC is not subject to DIAP1-induced degradation, once it is processed and activated it has reduced protein stability. Finally, we show that DRONC protein accumulates in “undead” cells due to increased transcription of dronc in these cells. These data refine current models of caspase regulation by IAPs.
Introduction
Ubiquitylation describes the covalent attachment of ubiquitin, a 76 amino acid polypeptide, to proteins which occurs as a multi-step process (reviewed in [1], [2]). E1-activating enzymes activate ubiquitin and transfer it to E2-conjugating enzymes. E3-ubiquitin ligases mediate the conjugation of ubiquitin from the E2 to the target protein. Repeated ubiquitylation cycles lead to the formation of polyubiquitin chains attached on target proteins. Polyubiquitylated proteins are delivered to the 26S proteasome for degradation. However, non-proteolytic roles of ubiquitylation have also been described (reviewed in [3], [4]). From E1 to E3, there is increasing complexity. For example, the Drosophila genome encodes only one E1 enzyme, termed UBA1, which is required for all ubiquitin-dependent reactions in the cell [5]. In contrast, there are hundreds of E3-ubiquitin ligases which are needed to confer substrate specificity.
Programmed cell death or apoptosis is an essential physiological process for normal development and maintenance of tissue homeostasis in both vertebrates and invertebrates (reviewed in [6]). A highly specialized class of proteases, termed caspases, are central components of the apoptotic pathway (reviewed in [7]). The full-length form (zymogen) of caspases is catalytically inactive and consists of a prodomain, a large and a small subunit. Activation of caspases occurs through dimerization and proteolytic cleavage, separating the large and small subunits. Based on the length of the prodomain, caspases are divided into initiator (also known as apical or upstream) and effector (also known as executioner or downstream) caspases [7]. The long prodomains of initiator caspases harbor regulatory motifs such as the caspase activation and recruitment domain (CARD) in CASPASE-9. Through homotypic CARD/CARD interactions with the adapter protein APAF-1, CASPASE-9 is recruited into the apoptosome, a large multi-subunit complex, where it dimerizes and auto-processes leading to its activation [8], [9]. Activated CASPASE-9 cleaves and activates effector caspases (CASPASE-3, -6, and –7), which are characterized by short prodomains. Effector caspases execute the cell death process by cleaving a large number of cellular proteins [10].
In Drosophila, the initiator caspase DRONC and the effector caspases DrICE and DCP-1 are essential for apoptosis [11]–[18]. Like human CASPASE-9, DRONC carries a CARD motif in its prodomain [19]. Consistently, DRONC interacts with ARK, the APAF-1 ortholog in Drosophila (also known as DARK, HAC-1 or D-APAF-1) [20]–[22] for recruitment into an apoptosome-like complex which is required for DRONC activation [20], [23]–[31]. After recruitment into the ARK apoptosome, DRONC cleaves and activates the effector caspases DrICE and DCP-1 [25], [31]–[34].
Caspases are subject to negative regulation by inhibitor of apoptosis proteins (IAPs) (reviewed in [35], [36]). For example, DRONC is negatively regulated by Drosophila IAP1 (DIAP1) [37], [38]. diap1 mutations cause a dramatic cell death phenotype, in which nearly every mutant cell is apoptotic, suggesting an essential genetic role of diap1 for cellular survival [39]–[41]. DIAP1 is characterized by two tandem repeats known as the Baculovirus IAP Repeat (BIR), and one C-terminally located RING domain [42]. The BIR domains are required for binding to caspases [37], [38], [43]. The RING domain provides DIAP1 with E3-ubiquitin ligase activity, required for ubiquitylation of target proteins [35], [36]. Importantly, the BIR domains can bind to caspases independently of the RING domain [37], [43].
Usually, IAPs bind to and inhibit activated, i.e. processed caspases, including CASPASE-3, CASPASE-7 and CASPASE-9 as well as the Drosophila caspases DrICE and DCP-1 (reviewed in [35], [36]). However, a notable exception to this rule is DRONC. DIAP1 binds to the prodomain of full-length DRONC [37], [38], [43]. This unusual behavior suggests an important mechanism for the control of DRONC activation. Indeed, it has been shown that the RING domain of DIAP1 ubiquitylates full-length DRONC in vitro [38], [44]. It has also been proposed that DIAP1 ubiquitylates auto-processed DRONC [33]. These ubiquitylation events are critical for the control of apoptosis, as homozygous diap1 mutants which lack a functional RING domain (diap1ΔRING) are highly apoptotic [41]. Because the BIR domains are intact in diap1ΔRING mutants, binding of DIAP1 to DRONC is not sufficient for inhibition of DRONC under physiological conditions, and ubiquitylation is the critical event for DRONC inhibition.
Although the importance of DIAP1-mediated ubiquitylation of DRONC is well established, it is still unclear how this ubiquitylation event leads to inactivation of DRONC and of caspases in general. Because DRONC protein accumulates in diap1 mutant cells that are kept alive by expression of the effector caspase inhibitor P35, generating so-called ‘undead’ cells, it has been proposed that DIAP1-mediated ubiquitylation triggers proteasomal degradation of full-length DRONC in living cells, thus protecting them from apoptosis [33], [38], [45], [46]. However, degradation of full-length DRONC in living cells has never been observed and non-degradative models have also been proposed [44]. Furthermore, ubiquitylation of mammalian CASPASE-3 and CASPASE-7 has been demonstrated in vitro [47]–[49]. However, evidence for proteasome-dependent degradation of these caspases in vivo, i.e. in the context of a living animal, is lacking. In fact, a non-degradative mechanism has been demonstrated for the effector caspase DrICE in Drosophila [50].
Here, we further characterize the role of ubiquitylation for the control of DRONC activation. Consistent with a previous report [44], we find that ubiquitylation of DRONC by DIAP1 is critical for inhibition of DRONC's pro-apoptotic activity. Using loss and gain of diap1 function, we provide genetic evidence that DIAP1-mediated ubiquitylation of full-length DRONC regulates this initiator caspase through a non-degradative mechanism. We find that the conjugation of ubiquitin suppresses DRONC processing and activation. Interestingly, once DRONC is processed and activated, it has reduced protein stability. Finally, we show that dronc transcripts accumulate in P35-expressing ‘undead’ cells, accounting for increased DRONC protein levels in these cells. These data refine the current model of caspase regulation by IAPs.
Results
Overexpression of DIAP1 fails to suppress apoptosis of Uba1 mutant cells
It has previously been shown that complete loss of ubiquitylation due to mutations of the E1 enzyme Uba1 causes apoptosis in eye imaginal discs as detected by an antibody that recognizes cleaved, i.e. activated, CASPASE-3 (CAS3*) [5], [51], [52] (see also Figure 1A). Because ubiquitylation of DRONC does not occur in Uba1 mutants, we hypothesized that inappropriate activation of DRONC accounts for the apoptotic phenotype of Uba1 mutants. To test this possibility, we targeted dronc by RNA interference (RNAi) in Uba1 mutant cells in eye imaginal discs using the MARCM system and labeled for apoptosis using CAS3* antibody. In this system, Uba1 mutant cells expressing dronc RNAi are positively marked by GFP. Consistent with our hypothesis, knock-down of dronc strongly reduces apoptosis in Uba1 mutant clones (Figure 1B). Furthermore, we tested clones doubly mutant for Uba1 and ark, the Drosophila ortholog of APAF-1 that is required for DRONC activation (see Introduction). Apoptosis induced in Uba1 mutant clones is strongly suppressed if ark function is removed (Figure S1). These observations suggest that the apoptotic phenotype in Uba1 clones is caused by inappropriate activation of DRONC, presumably due to lack of ubiquitylation.
Fig. 1. Apoptosis in Uba1 mutant clones is dependent on DRONC and cannot be inhibited by expression of DIAP1. 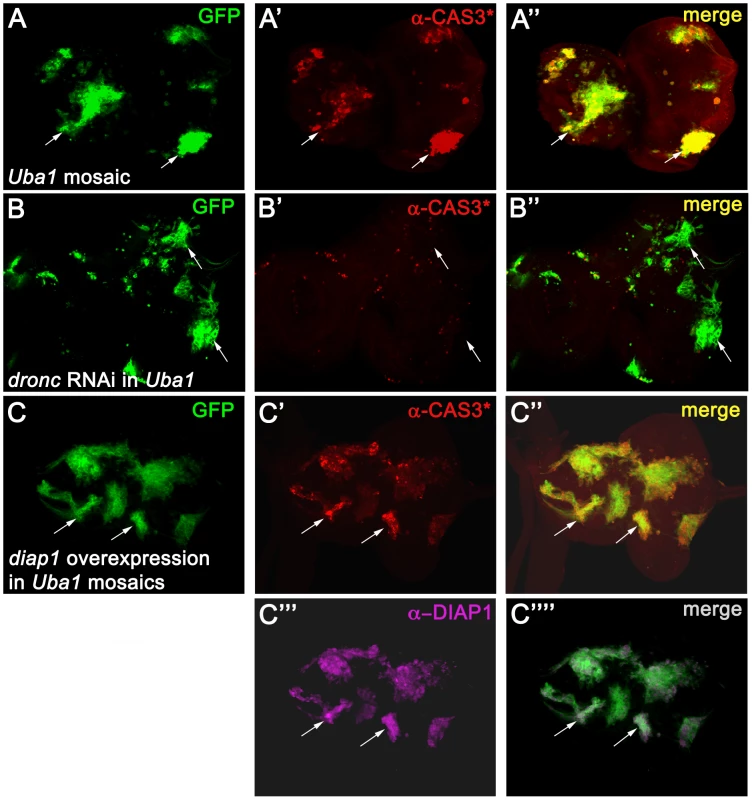
Shown are eye-antennal imaginal discs from third instar larvae. Posterior is to the right. In each panel, arrows highlight two representative clones. (A) Uba1 mosaic eye-antennal discs labeled for cleaved CASPASE-3 (α-CAS3*) antibody (red). These discs were incubated at 30°C 12 hours before dissection (see Material and Methods). Presence of GFP marks the location of Uba1 clones (see arrow). (B) TUNEL labeling of Uba1 mosaic eye-antennal imaginal discs expressing an RNAi transgene targeting dronc (UAS-droncIR (inverted repeat)) using the MARCM technique (see Material and Methods). Clones are positively marked by GFP. TUNEL-positive cell death is largely blocked by dronc knockdown (B′ and B″). (C) Strong overexpression of diap1 in Uba1 clones (magenta in C′″) fails to rescue the apoptotic phenotype, as visualized by CAS3* labeling (red in C′). Uba1 clones are marked by GFP due to the MARCM technique. Please note that diap1 is so strongly overexpressed in the clones that we had to adjust the settings in such a way that endogenous DIAP1 in wild-type tissue is below the detection limit (C′″). Genotypes: (A) hs-FLP UAS-GFP; FRT42D Uba1D6/FRT42D tub-Gal80; tub-GAL4. (B) hs-FLP UAS-GFP; FRT42D Uba1D6/FRT42D tub-Gal80; tub-GAL4/UAS-droncIR. (C) hs-FLP UAS-GFP/UAS-diap1; FRT42D Uba1D6/FRT42D tub-Gal80; tub-GAL4. However, the protein levels of DIAP1 are increased in Uba1 mutant clones [5], [52]. There are two possibilities to explain the apoptotic phenotype in Uba1 mutants despite increased DIAP1 levels. First, the DIAP1 levels may not be sufficiently increased to inhibit DRONC. Alternatively, binding of DIAP1 to DRONC alone may not be sufficient for inhibition of DRONC; instead, ubiquitylation by DIAP1 is required to block DRONC activation, as previously suggested [44]. To distinguish between these two possibilities, we strongly overexpressed diap1 in Uba1 mutant clones in eye discs using the MARCM system and imaged for apoptosis by CAS3* labeling. Surprisingly, despite massive expression of diap1 (>20 fold over wild-type levels; Figure 1C′″), apoptosis still proceeds in Uba1 mutant clones (Figure 1C′), even though expression of the same transgene can block strong apoptotic phenotypes in several apoptotic paradigms (Figure S2). Apparently, overexpression of DIAP1 is not sufficient to inhibit DRONC and to protect Uba1 mutant cells from apoptosis. Because DIAP1 is the key regulator of DRONC and because DRONC is required for the apoptotic phenotype of Uba1 mutant cells, as evidenced by knock-down of dronc (Figure 1B), our data provide genetic evidence that binding of DIAP1 is not sufficient for DRONC inhibition in Uba1 mutant cells.
Consistent with this view, it has previously been shown that DIAP1 does ubiquitylate full-length DRONC in vitro [33], [38], [44]. We tested whether DIAP1 can also ubiquitylate DRONC in vivo. Because the available DRONC antibodies failed to immunoprecipitate endogenous DRONC, we transfected DRONC-V5 along with DIAP1+ or DIAP1ΔRING mutants (CΔ6, lacking the last six C-terminal residues, and F437A changing a critical Phe residue in the RING domain to Ala [53]) and His-tagged Ubiquitin into Drosophila S2 cells. Ubiquitylated proteins were affinity purified under denaturing conditions using Ni columns. The eluates were subsequently examined by immunoblotting with anti-V5 antibodies to detect ubiquitylated forms of DRONC. Under these conditions, DIAP1+ readily ubiquitylates full-length DRONC in S2 cells (Figure 2), whereas the RING mutants DIAP1CΔ6 and DIAP1F437A were significantly impaired in their ability to ubiquitylate DRONC (Figure 2). These results indicate that DIAP1 ubiquitylates full-length DRONC in a RING-dependent manner in cultured cells.
Fig. 2. DIAP1 ubiquitylates DRONC in S2 cells. 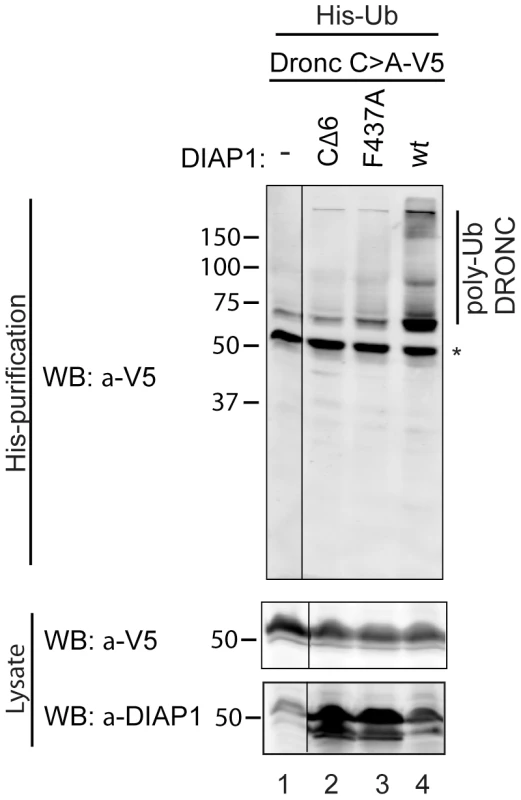
DRONC C>A–V5 was coexpressed with His-Ub and the indicated DIAP1 constructs in S2 cells. Ubiquitylated proteins were purified and analyzed by immunoblot for ubiquitylated DRONC with V5 antibodies. Co-expression of DIAP1wt leads to higher molecular weight modification of DRONC, while the RING-ligase inactive mutants (CΔ6, F437A) cannot ubiquitylate DRONC. * marks non-modified DRONC that is due to unspecific DRONC:matrix association. Overexpression of DIAP1 does not induce degradation of DRONC
Because DIAP1 readily ubiquitylates DRONC, it has been postulated that DIAP1-mediated ubiquitylation leads to proteasomal degradation of DRONC [33], [38], [45]. However, this has never been rigorously tested in vivo. Therefore, we examined, whether overexpression of diap1 in wild-type animals can influence DRONC protein levels in vivo. To this end, we generated clones overexpressing diap1 (marked by absence of GFP) in eye discs, and analyzed the protein abundance of DRONC. Interestingly, despite high expression of diap1 (Figure 3A′″), the levels of DRONC remained unchanged and were not influenced by DIAP1 (Figure 3A′). The anti-DRONC antibody used in this assay is specific for DRONC (Figure S3). Importantly, the diap1 transgene used produces a functional DIAP1 protein that is able to inhibit apoptosis in several paradigms (Figure S2). Therefore, these data suggest that overexpressed DIAP1 does not target DRONC for degradation in living cells.
Fig. 3. Overexpression of diap1 does not trigger degradation of DRONC. 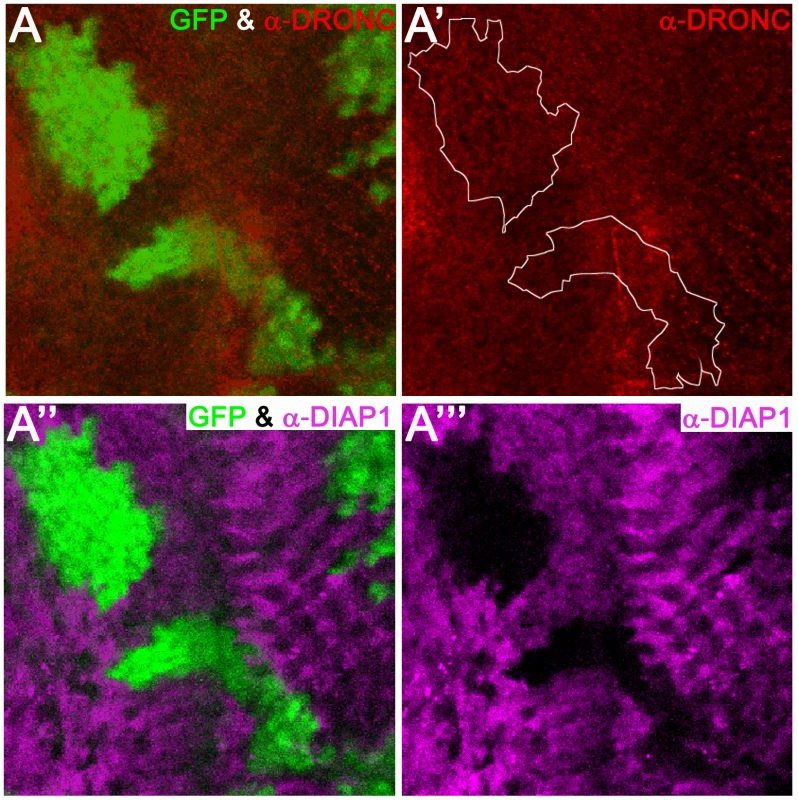
Shown is an eye imaginal disc from a third instar larva. Posterior is to the right. diap1-overexpressing clones are marked by absence of GFP and can be detected using anti-DIAP1 antibodies in magenta (A′″). The boundary between diap1-expressing clones and normal tissue is indicated by a white stippled line in (A′). DRONC levels are unchanged (A′). (A) and (A″) are merged images. Genotype: UAS-diap1/hs-FLP; tub>GFP>GAL4. REAPER-induced loss of DIAP1 does not increase DRONC protein levels
Because of the surprising observation that overexpressed DIAP1 does not cause degradation of DRONC, we tested whether removal of DIAP1 changes DRONC protein levels. Expression of the IAP antagonist reaper (rpr) induces DIAP1 degradation and apoptosis [54]–[58]. Therefore, we examined whether RPR-induced degradation of DIAP1 changes DRONC protein levels. If DIAP1 targets DRONC for degradation, we would expect that DRONC protein levels would accumulate in response to rpr expression. Expression of rpr in eye imaginal discs posterior to the morphogenetic furrow (MF) using the GMR promoter (GMR-rpr) is well suited for this analysis. The MF is a dynamic structure that initiates at the posterior edge of the eye disc and moves towards the anterior during 3rd instar larval stage [59], [60] (Figure 4A, arrow). Expression of rpr by GMR is induced in all cells posterior to the MF [61] (red in Figure 4A). Therefore, GMR-rpr eye discs provide a continuum of all developmental stages in which cells close to the MF have only recently induced rpr expression, while cells towards the posterior edge of the disc have been exposed to rpr progressively longer. Therefore, if DRONC accumulates during any of these stages, we should be able to detect it. In wild-type eye discs, DRONC protein is homogenously distributed throughout the disc. Only in the MF, higher levels of DRONC are detectable (arrowhead in Figure 4B″). This high expression of DRONC in the MF serves as an orientation mark. DIAP1 protein levels are low anterior to the MF, but increase in the MF (arrowhead) and posterior to it in wild-type discs (Figure 4B′). In GMR-rpr eye discs, overall DIAP1 levels are reduced in the rpr-expressing domain posterior to the MF (Figure 4C′), but particularly strongly reduced in the CAS3*-positive area (Figure 4C′, D′, arrow) consistent with previous reports [54]–[58]. However, accumulation of DRONC is not observed (Figure 4C″, D″). In contrast, it appears that DRONC levels are also reduced. They are still high in the MF (Figure 4C″, arrowhead), but drop immediately thereafter.
Fig. 4. Loss of DIAP1 in GMR-rpr eye discs does not alter DRONC protein levels. 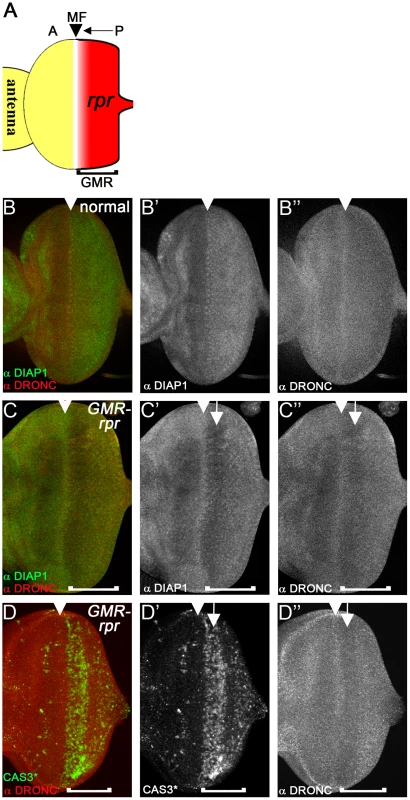
(A) Schematic illustration of the GMR-reaper (GMR-rpr) eye imaginal disc from 3rd instar larvae. The morphogenetic furrow (MF, arrowhead) initiates at the posterior (P) edge of the disc and moves towards the anterior (A) (arrow). The GMR enhancer is active posterior to the MF (bracket) and thus expresses rpr posterior to the MF (red area). (B-B″) Eye disc showing normal protein distribution of DIAP1 (B′) and DRONC (B″). Both DIAP1 and DRONC levels are increased in the MF (arrowhead). (B) is the merged image of DIAP1 and DRONC labeling. (C–C″) Eye discs expressing two copies of GMR-rpr eye disc labeled for DIAP1 (C′) and DRONC (C″). Arrowheads mark the MF. DIAP1 levels are markedly reduced posterior to the MF (C′, arrow). Surprisingly, DRONC protein levels are also reduced (C″, arrow). The brackets indicate the extent of GMR expression. (D–D″) 2×GMR-rpr eye disc labeled for cleaved CASPASE 3 (CAS3*) (D′) and DRONC (D″). DRONC protein levels are reduced in the CAS3*-positive area (arrow). Arrowheads mark the MF. The brackets indicate the extent of GMR expression. We also related DRONC levels to caspase activation. In the MF, where CAS3* activity is not detectable, DRONC is still high (Figure 4D′, D″; arrowhead), but in the CAS3*-positive area, DRONC levels are reduced (Figure 4D′, D″; arrow). These data indicate that loss of DIAP1 does not cause accumulation of DRONC protein implying that DIAP1 does not induce degradation of DRONC. In contrast, it appears that DIAP1 stabilizes DRONC at least under these conditions (see Discussion).
“Undead” diap1 mutant cells induce transcription of dronc
Finally, we analyzed DRONC protein levels in diap1ΔRING mutants which cannot ubiquitylate DRONC [44]. It has previously been shown that clones of the RING mutant diap122-8s accumulate DRONC protein [45], [46] implying that ubiquitylation by the RING domain of DIAP1 causes degradation of DRONC. We repeated these experiments and indeed confirmed that DRONC levels are increased in diap122-8s mutant clones (Figure S4). Thus, these results appear inconsistent with the data presented in Figure 3 and Figure 4 in which manipulating DIAP1 levels did not provide evidence for DIAP1-mediated degradataion of DRONC. However, one caveat with the diap122-8s experiment was the use of the caspase inhibitor P35 which kept diap122-8s mutant cells in an ‘undead’ condition [45]. It has been pointed out that the ‘undead’ state may change the properties of the affected cells (reviewed by [62]) and in fact abnormal induction of transcription in ‘undead’ cells has been reported [45], [63]–[66]. Thus, to explain the conflicting results between the diap122-8s data [45] and our data shown here, we hypothesized that p35-expressing ‘undead’ diap122-8s clones induce dronc transcription, leading to accumulation of DRONC protein. To test this hypothesis, we used a transcriptional lacZ reporter containing 1.33 kb of regulatory genomic sequences upstream of the transcriptional start site of the dronc gene fused to lacZ (dronc1.33-lacZ) [67], [68]. Compared to controls (Figure 5A, 5A′) and consistent with the hypothesis, dronc1.33-lacZ reporter activity is increased in p35-expressing ‘undead’ diap122-8s cells in wing imaginal discs and matches the increased DRONC protein pattern (Figure 5B′-5B′″). We also produced ‘undead’ cells in eye imaginal discs by co-expression of the IAP-antagonist hid and the caspase inhibitor p35 in the dorsal half of the eye disc using a dorsal eye- (DE-) GAL4 driver (Figure 5C). Similar to wing discs, dronc reporter activity is increased in ‘undead’ cells in the dorsal half of the eye (Figure 5D). Expression of p35 alone does not trigger transcription of dronc (Figure 5E) suggesting it is not the mere presence of P35 which causes dronc transcription, but the ‘undead’ nature of the affected cells.
Fig. 5. “Undead” diap1 mutant cells trigger transcription of dronc. 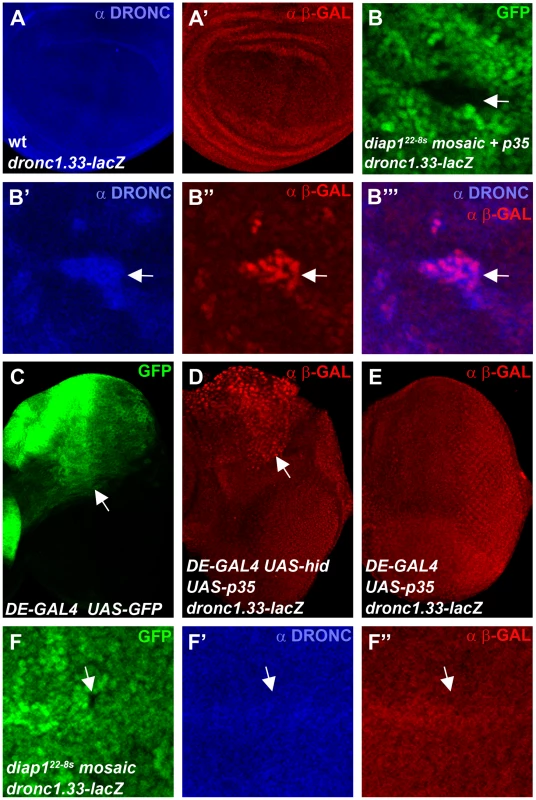
Shown are 3rd instar larval wing (A,B,F) and eye imaginal discs (C,D,E) labeled for DRONC protein levels (blue) and dronc transcriptional activity (red) using the dronc1.33-lacZ reporter (ß-GAL labeling). (A,A′) Co-labeling for DRONC protein (A) and dronc reporter activity (A′) of a wild-type wing disc expressing the dronc1.33-lacZ transgene. (B-B′″) A diap122-8s mosaic wing disc expressing p35 under nub-GAL4 control in a dronc1.33-lacZ background. A mutant clone in the wing pouch is highlighted by an arrow in the GFP-only channel (B). DRONC protein (B′) and ß-GAL immunoreactivity as readout of dronc1.33-lacZ activity (B″) are increased in the same cells and overlap (B′″). Please note that the dronc1.33-lacZ reporter produces nuclear ß-GAL, while DRONC protein appears cytoplasmic. (C) GFP expression in the eye imaginal disc indicates the dorsal expression domain (arrow) of the dorsal eye (DE)-GAL4 driver [95]. (D) Increased dronc reporter activity in the dorsal half of the eye imaginal disc (arrow) in undead cells obtained by co-expression of hid and p35 using DE-GAL4. (E) Expression of p35 alone by DE-GAL4 does not induce dronc reporter activity. (F-F″) A diap122-8s mosaic wing disc in a dronc1.33-lacZ background which does not express p35. diap122-8s mutant clones are marked by the absence of GFP (F). An arrow points to a representative diap122-8s clone in the wing pouch. In the same position, neither DRONC protein (F′) nor dronc reporter activity (F″) are increased. Note, that this clone is present in the wing pouch which has the capacity to upregulate DRONC and dronc transcription in the ‘undead’, p35-expressing condition (see panel B″). Genotypes: (A) dronc1.33-lacZ/+. (B) ubx-FLP; nub-GAL4 UAS-p35/dronc1.33-lacZ; diap122-8s FRT80/ubi-GFP FRT80. (C) DE-GAL4 UAS-GFP/+. (D) UAS-p35 UAS-hid/dronc1.33-lacZ; DE-GAL4. (E) UAS-p35/dronc1.33-lacZ; DE-GAL4. (F) ubx-FLP; nub-GAL4/dronc1.33-lacZ; diap122-8s FRT80/ubi-GFP FRT80. These observations may explain why DRONC protein accumulates in ‘undead’ diap122-8s mutant cells, but they still do not rule out the possibility that DRONC protein accumulates in diap122-8s mutants due to lack of ubiquitylation and thus degradation. To clarify this issue we examined dronc1.33-lacZ and DRONC levels in diap122-8s mutant clones without simultaneous p35 expression. Without the inhibition of apoptosis by P35, diap122-8s clones die rapidly. Nevertheless, we were able to recover wing discs which contained small diap122-8s mutant clones. In these clones, neither dronc1.33-lacZ nor DRONC levels are detectably increased (Figure 5F). Notably, these clones are located in the wing pouch in which we observed accumulation of dronc reporter activity and DRONC protein under ‘undead’ conditions (Figure 5B″). Thus, the ‘undead’ condition of p35-expressing diap122-8s mutant cells causes accumulation of DRONC protein due to induction of dronc transcription, explaining the observations of Ryoo et al. (2004) [45]. In the absence of p35 expression, transcription of dronc and accumulation of DRONC protein are not observed, providing additional evidence that ubiquitylation of DRONC by the RING domain of DIAP1 does not trigger degradation of DRONC.
Ubiquitylation controls processing and thus activation of DRONC in vivo
Our in vivo analysis implies that DIAP1-mediated ubiquitylation does not trigger proteasomal degradation of DRONC. To identify the role of ubiquitylation for regulation of DRONC activity, we analyzed the fate of DRONC protein in RING mutants of diap1. Of note, these mutants retain their ability to bind to DRONC, because DRONC binding is not mediated by the RING domain, but by the BIR2 domain [37], [38], [43]. The RING mutant diap133-1s is especially suitable for this analysis because a premature stop codon results in deletion of the entire RING domain but leaves the BIR domains intact [44] (Figure 6A), thus abrogating its E3 activity, but not caspase binding. Importantly, diap133-1s is characterized by a strong apoptotic phenotype, suggesting inappropriate caspase activation [41], [45]. Indeed, we showed previously that diap1ΔRING mutant phenotypes are dependent upon DRONC, because dronc mutants suppress diap1ΔRING phenotypes [11]. Therefore, ubiquitylation of DRONC by DIAP1 is critical to maintain cell survival.
Fig. 6. Ubiquitylation controls processing of DRONC. 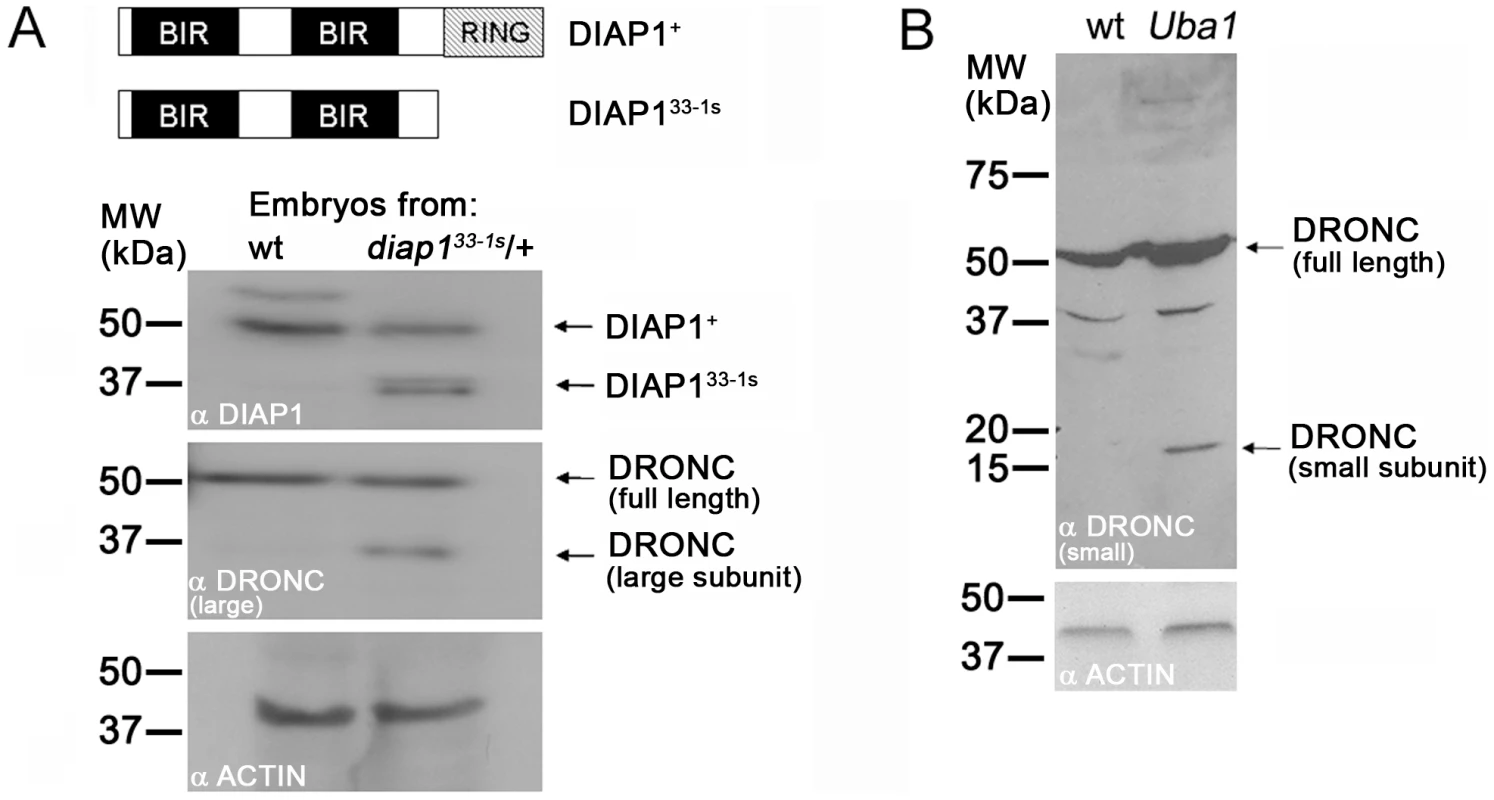
(A) Top: schematic outline of the domain structure of DIAP1+ (wild-type) and RING-deleted DIAP133-1s. Not drawn to scale. Immunoblots of embryonic extracts of stage 6–9 wild-type (wt) and heterozygous diap133-1s mutants were probed with anti-DIAP1 (upper panel) and anti-DRONC antibodies (middle panel). The RING-depleted diap133-1s allele produces a stable protein (DIAP133-1s) that is detectable by its faster electrophoretic mobility (upper panel). In RING-depleted diap133-1s embryos a significant portion of processed DRONC is present (middle panel) which likely accounts for the apoptotic phenotype of diap133-1s embryos [41]. These extracts were obtained from a cross of heterozygous males and females. Thus, only one quarter of the embryos is homozygous mutant for diap133-1s; yet, processed DRONC is easily detectable. The anti-DRONC antibody is specific for the large subunit of DRONC. Lower panel: loading control. (B) Extracts of imaginal discs from wild-type (wt) and mosaic Uba1 imaginal discs were analyzed by immunoblotting using an antibody raised against the small subunit of DRONC. Clones of the temperature sensitive allele Uba1D6 were induced at the permissive temperature in first larval instar and then shifted to the non-permissive temperature (30°C) during third larval instar 12 hours before dissection (see Material and Methods). This treatment ensures that approximately 50% of the mosaic disc is mutant for Uba1. Although only 50% of the disc tissue is mutant for Uba1, processed DRONC is easily detectable. Lower panel: loading control. We examined the cause of the diap133-1s apoptotic phenotype. First, as a control, we determined whether the diap133-1s gene produces a stable protein in vivo. We chose to analyze stage 6–9 embryos, because normal developmental cell death starts at stage 11 [69]; therefore, stage 6–9 diap133-1s mutant embryos allow analysis of DIAP1 in the absence of upstream apoptotic signals. In immunoblots of embryonic extracts obtained from a cross of heterozygous diap133-1s males and females, the DIAP133-1s protein is easily distinguished from wild-type DIAP1+ protein due to its faster electrophoretic mobility (Figure 6A, top panel). The presence of the DIAP133-1s protein suggests that the apoptotic phenotype in diap133-1s mutant embryos is not caused by instability of the mutant protein. Interestingly, the protein levels of DIAP1+ and RING-deleted DIAP133-1s are similar (Figure 6A, top panel) suggesting that loss of the RING domain does not influence the protein stability of DIAP1 in the absence of pro-apoptotic signals.
Next, we analyzed DRONC protein in extracts from diap133-1s mutant embryos. Consistent with the data in Figure 4 and Figure 5, we do not detect a significant increase in the protein levels of DRONC in these embryos (Figure 6A, middle panel). However, a significant amount of DRONC is present in a processed form in extracts of stage 6–9 diap133-1s mutant embryos which is absent in control extracts from wild-type embryos (Figure 6A, middle panel). Therefore, DRONC processing, and thus activation, occurs in RING-depleted diap133-1s mutant embryos despite the fact that the BIR domains of DIAP1 are unaffected. The processed form of DRONC in this mutant of MW ∼36 kDa corresponds to the major apoptotic form of DRONC composed of the large subunit and the prodomain of DRONC [70]. This finding, and the one presented in Figure 1, confirms that binding of DIAP1 to DRONC is not sufficient to prevent processing and activation of DRONC. Instead, the RING domain is required to control DRONC processing. Because the RING domain contains an E3-ubiquitin ligase activity [45], [55]–[58] and because ubiquitylation of full-length DRONC does not trigger proteasomal degradation (Figure 3, Figure 4, and Figure 5), we conclude that ubiquitylation of DRONC by the RING domain of DIAP1 is necessary to inhibit DRONC processing and thus activation.
To further characterize the role of ubiquitylation in the regulation of DRONC processing, we performed an immunoblot analysis with extracts from wild-type and Uba1 mosaic imaginal discs, which, under our experimental conditions, are about half mutant for Uba1 and half wild-type. Immunoblot analysis demonstrated that a significant amount of DRONC is processed in Uba1 mosaic discs (Figure 6B). Thus, these data further support the notion that ubiquitylation of full-length DRONC is necessary for inhibition of DRONC processing.
Discussion
In this paper, we provide three take-home messages. First, we provide genetic evidence that binding of DIAP1 to DRONC is not sufficient for inhibition of DRONC. Instead, ubiquitylation of DRONC controls its apoptotic activity, consistent with the apoptotic phenotype of diap1ΔRING mutants, that retain caspase binding abilities. Second, DIAP1-mediated ubiquitylation of full-length DRONC does not lead to its proteasomal degradation; rather, ubiquitylation directly controls processing and activation of DRONC. Interestingly, processed and active DRONC shows reduced protein stability. Third, ‘undead’ cells accumulate dronc transcripts.
Binding of DIAP1 is not sufficient for Dronc inhibition
Based on biochemical studies in vitro and overexpression studies in cultured cells, many of cancerous origin, it was initially proposed that binding of IAPs to caspases through their BIR domains is sufficient to inhibit caspases [71]–[80]. However, when ubiquitylation of caspases by IAPs was described [38], [44], [47], [48], it was unclear what role ubiquitylation would play for control of caspase activity, especially since for none of them, ubiquitin-mediated degradation has been observed (see below). Because the RING domain is also required for auto-ubiquitylation of DIAP1 [54]–[58], mutations of the RING domain would be expected to increase DIAP1 protein levels and protect cells from apoptosis. However, the opposite phenotype, massive apoptosis, was observed [41]. Nevertheless, despite the biochemical studies showing that the BIR domains of DIAP1 are sufficient for interaction with DRONC [37], [38], [43], one could argue that DIAP1ΔRING mutants have lost the ability to interact with DRONC in vivo. While we cannot exclude this possibility due to the inability of our antibodies to immunoprecipitate endogenous proteins, another experiment supports the notion that ubiquitylation is necessary for DRONC inhibition: when wild-type diap1 was strongly overexpressed in an ubiquitylation-deficient Uba1 mutant background, DRONC-dependent apoptosis was not inhibited (Figure 1C), suggesting that binding of DIAP1 is not sufficient for inhibition of DRONC. Instead, ubiquitylation is critical for inhibition of DRONC activity.
DIAP1 does not control protein levels of full-length DRONC
The current model holds that DIAP1-mediated ubiquitylation leads to proteasomal degradation of full-length DRONC in living cells [33], [38], [45]. However, our data do not support this model in vivo. This model is based on biochemical ubiquitylation studies without in vivo validation and does not take into account that ubiquitylation often serves non-proteolytic functions [1], [3], [4]. Both overexpression and loss of diap1 does not cause a detectable alteration of the protein levels of DRONC (Figure 3, Figure 4, Figure 5), arguing against a degradation model. The only example where DRONC accumulation has been observed is in P35-expressing ‘undead’ diap1ΔRING mutant cells [45], [46], and we showed here that the ‘undead’ nature of these cells causes transcriptional induction of dronc (Figure 5). Together, these observations argue against a degradation model of full-length DRONC and favor a non-traditional (non-proteolytic) role of ubiquitylation regarding control of DRONC activity. Similarly, DIAP1-mediated ubiquitylation of the effector caspase DrICE inactivates this effector caspase through a non-degradative mechanism [50].
Interestingly, in GMR-rpr eye imaginal discs, DRONC protein levels appear to be reduced in apoptotic cells compared to living cells (Figure 4C″, 4D″). Due to the apoptotic activity of REAPER, DRONC is present in its processed and activated form. Reduced protein stability of DRONC has also been reported after incorporation into the ARK apoptosome where it is processed and activated [46]. Combined, these observations suggest that while DIAP1-mediated ubiquitylation of full-length DRONC does not trigger its degradation, processed and activated DRONC has reduced protein stability and may indeed be degraded. It is unclear whether degradation of activated DRONC is mediated by DIAP1, as proposed previously [33]. In GMR-rpr eye imaginal discs, reduced DRONC levels correlate with a reduction of DIAP1 protein (Figure 4C′, 4D′). This correlation indicates that DIAP1 may actually stabilize DRONC protein, at least in part. Alternatively, because DRONC and DIAP1 form a complex [37], REAPER-induced degradation of DIAP1 may result in co-degradation of complexed DRONC in the process. Further studies are needed to determine the cause of decreased DRONC stability in apoptotic cells.
In addition to Drosophila DRONC, mammalian CASPASE-3 and CASPASE-7 have been reported to be ubiquitylated in vitro [47], [48]. However, proteasome-mediated degradation of these caspases in vivo has not been reported. Although a decrease of CASPASE-3 levels has been noted upon overexpression of XIAP, this was shown for an artificial CASPASE-3 mutant, in which the order of the subunits was reversed and the Cys residue in the active site changed to Ser [47]. This catalytically inactive CASPASE-3 mutant is not proteolytically processed [47]. Therefore, physiological in vivo evidence for IAP-mediated degradation of mammalian caspases is lacking.
Moreover, the phenotype of a RING-deleted XIAP mutant mouse is consistent with our data [49]. The XIAPΔRING mutant, which was generated by a knock-in approach in the endogenous XIAP gene, is characterized by increased caspase activity [49]. Intriguingly, the protein levels of CASPASE-3, CASPASE-7 and CASPASE-9 did not significantly change in the XIAPΔRING mutant despite the fact that ubiquitylation of CASPASE-3 was reduced. However, processing of these caspases was easily detectable in XIAPΔRING mutants [49]. These data are very similar to the ones presented here for diap133-1s (Figure 6), and together strongly suggest that non-proteolytic ubiquitylation controls caspase processing and activity in both vertebrates and invertebrates.
Non-proteolytic roles of ubiquitylation have been described in recent years and are involved in many processes including signal transduction, endocytosis, DNA repair, and histone activity (reviewed in [1], [3], [4]). Two types of ubiquitylation lead to non-proteolytic functions. Monoubiquitylation is involved in endocytosis, DNA repair and histone activity. In fact, mammalian CASPASE-3 and CASPASE-7 have been found to be monoubiquitylated in vitro [48]. However, it is unclear whether DRONC is monoubiquitylated by DIAP1. The presence of high molecular-weight ubiquitin conjugates in vitro (Figure 2) suggests that DRONC may be polyubiquitylated, at least under the experimental conditions [38], [44].
Polyubiquitylation serves both proteolytic and non-proteolytic functions depending on the Lysine (K) residue used for polyubiquitin chain formation. In general, if polyubiquitylation occurs via K48, the target protein is subject to proteasome-mediated degradation. If it occurs on a different Lys residue, such as K63, non-proteolytic functions are induced [1], [3], [4]. The best studied examples of both K48 - and K63-linked polyubiquitylation are in the NF-κB pathway (reviewed in [3], [81]). While K48-polyubiquitylation leads to proteasomal degradation, K63-linked polyubiquitin chains act as scaffolds to assemble protein complexes required for NF-κB activation [3], [81]. It is unknown what type of polyubiquitin chain is attached to DRONC, but it is possible that it is not K48-linked. Interestingly, in this context it has been shown that auto-ubiquitylation of DIAP1 preferentially involves K63-linked polyubiquitin chains [82]. By analogy, DIAP1 may ubiquitylate DRONC through formation of K63-linked polyubiquitin chains. This will be an interesting avenue to explore in future experiments.
Conjugated monoubiquitin and polyubiquitin chains can serve as docking sites for factors containing ubiquitin-binding domains (UBD) [2], [4], [83]. The UBD-containing factors interpret the ubiquitylation status of the target protein, and trigger the appropriate response. For example, K48-linked polyubiquitin chains are recognized by Rad23 and Drk2 which deliver them to the proteasome [2]. Other forms of ubiquitin conjugates are recognized by different UBD-containing factors which control the activity of the ubiquitylated protein. Therefore, it is possible that an as yet unknown UBD-containing protein binds to ubiquitylated DRONC and controls its activity. For example, such an interaction could block the recruitment of ubiquitylated DRONC into the ARK apoptosome. Another possibility is that ubiquitylation could block dimerization of DRONC, which is required for activation of DRONC [34].
“Undead” cells trigger dronc transcription
‘Undead’ cells can be obtained by expression of the effector caspase inhibitor P35 [84]. In these cells, apoptosis has been induced, but cannot be executed due to effector caspase inhibition. Nevertheless, the initiator caspase DRONC is active in ‘undead’ cells and can promote non-apoptotic processes [51]. Recent work has suggested that ‘undead’ cells may alter their cellular behavior. For example, ‘undead’ cells change their size and shape, and have some migratory abilities to invade neighboring tissue [62]. They are also able to promote proliferation of neighboring cells causing hyperplastic overgrowth [15], [45], [63]–[66] (reviewed by [85], [86]). Altered transcription of the TGF-ß/BMP member decapentaplegic (dpp), the Wnt-homolog wingless (wg), and the p53 ortholog dp53 has also been reported in ‘undead’ cells [45], [64]–[66]. Intriguingly, while dpp and wg are usually not expressed in the same cells [87], ‘undead’ cells co-express them ectopically, clearly indicating an altered transcriptional program.
As part of this altered transcriptional program, we showed that ‘undead’ cells also stimulate transcription of the initiator caspase dronc (Figure 5). Interestingly, p35 expression in normal cells does not induce dronc transcription suggesting that it is not the mere presence of P35 that induces dronc transcription, but instead the ‘undead’ condition of the affected cells. This transcriptional induction of dronc provides an explanation why DRONC protein levels are increased in ‘undead’ cells. It may also help to explain another observation regarding ‘undead’ cells. It has been demonstrated that although these cells are unable to die, they maintain the apoptotic machinery indefinitely [62], [88]. Therefore, as part of this maintenance program, ‘undead’ cells stimulate dronc transcription. This is likely not restricted for dronc. Martin et al. (2009) [62] also showed that DrICE protein levels remain high in ‘undead’ cells which may also be caused by increased drICE transcription. It is also possible that the induction of dp53 by ‘undead’ cells [66] is part of this maintenance program, because we have shown that Dp53 induces expression of hid and rpr [89] and a positive feedback loop between dp53, hid and dronc exists in ‘undead’ cells [66]. This may all occur at a transcriptional level. The mechanism by which ‘undead’ cells stimulate expression of dpp, wg, dp53, dronc and potentially drICE are currently unknown and are avenues for future research.
Material and Methods
Drosophila genetics
Fly crosses were conducted using standard procedures at 25°C. The following mutants and transgenes were used: Uba1D6 [5]; arkG8 [26]; diap122-8s and diap133-1s [44]; vps25N55 [90]; droncI29 [11]; UAS-droncIR (dronc inverted repeats) [91]; GMR-rpr [92]; dronc1.33-lacZ [67], [68], ubx-FLP [93], nub-GAL4 [94], DE- (dorsal eye-) GAL4 [95], and UAS-hid [96]. nub-FLP is nub-GAL4 UAS-FLP. UAS-p35 and UAS-FLP were obtained from Bloomington. Uba1D6 is a temperature sensitive allele which at 25°C is a hypomorphic allele, but at 30°C is a null allele [5]. In the experiments in Figure 1, Figure 6B, and Figure S1, Uba1D6 and Uba1D6 arkG8 mosaic larvae were incubated at 25°C; 12 hours before dissection the temperature was shifted to 30°C. This treatment allows recovery of Uba1D6 null mutant clones, which otherwise are cell lethal.
Generation of mutant clones and expression of transgenes
Mutant clones were induced in eye-antennal imaginal discs using the FLP/FRT mitotic recombination system [97] using ey-FLP [98]. In this case, clones are marked by loss of GFP. Expression of diap1 and dronc RNAi in Uba1D6 clones (Figure 1) was induced from UAS-diap1 or UAS-droncIR transgenes using the MARCM system [99]. Here, clones are positively marked by GFP. For induction of diap1-expressing clones in Figure 3, the UAS-diap1 transgene was crossed to hs-FLP; tub<GFP<GAL4 (< = FRT). Clones are marked by the absence of GFP. MARCM clones and diap1-overexpressing clones were induced in first instar larvae by heat-shock for one hour in a 37°C water bath. Expression of UAS-p35 in diap122-8s mosaic discs was accomplished by nub-GAL4.
Immunohistochemistry
Eye-antennal imaginal discs from third instar larvae were dissected using standard protocols and labeled with antibodies raised against the following antigens: DIAP1 (a kind gift of Hermann Steller and Hyung Don Ryoo); cleaved CASPASE-3 (CAS3*) (Cell Signaling Technology) and anti-ß-GAL (Promega). The DRONC antibody used for immunofluorescence was raised against the C-terminus of DRONC in guinea pigs [44]. This antibody is specific for DRONC (Figure S3). Cy3 - and Cy-5 fluorescently-conjugated secondary antibodies were obtained from Jackson ImmunoResearch. In each experiment, multiple clones in 10–20 eye and wing imaginal discs were analyzed, unless otherwise noted. Images were captured using an Olympus Optical FV500 confocal microscope.
Ubiquitylation assays
Schneider S2 cells were co-transfected with pMT-DRONC C>A V5, pAcDIAP1 (wt or CΔ6, F437A, respectively, described in [50]) and pAc His-HA-Ub at equal ratios. Expression of DRONC was induced overnight with 350 µM CuSO4. Cells were lysed under denaturing conditions and ubiquitylated proteins were purified using Ni2+-NTA agarose beads (QIAGEN). Immunoblot analysis was performed with α-V5 (Serotec) and α-DIAP1 antibodies [37], [43].
Immunoblot analysis
For the immunoblots in Figure 6A, embryos were collected, decorionated and snap frozen in liquid nitrogen. Embryos were sonicated in Laemmli SDS loading buffer while being frozen. The equivalent of 30 lysed embryos was loaded per lane. Immunoblots were done using standard procedures. The anti-DRONC antibody used in Figure 6A is a peptide antibody raised against the large subunit of DRONC. The anti-DRONC antibody used in Figure 6B was raised against the C-terminus of DRONC in guinea pigs.
Supporting Information
Zdroje
1. WelchmanRLGordonCMayerRJ 2005 Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6 599 609
2. HickeLSchubertHLHillCP 2005 Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6 610 621
3. ChenZJSunLJ 2009 Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33 275 286
4. MukhopadhyayDRiezmanH 2007 Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315 201 205
5. LeeTVDingTChenZRajendranVScherrH 2008 The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development 135 43 52
6. DegterevAYuanJ 2008 Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol 9 378 390
7. KumarS 2007 Caspase function in programmed cell death. Cell Death Differ 14 32 43
8. BaoQShiY 2007 Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ 14 56 65
9. RiedlSJSalvesenGS 2007 The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol 8 405 413
10. TimmerJCSalvesenGS 2007 Caspase substrates. Cell Death Differ 14 66 72
11. XuDLiYArcaroMLackeyMBergmannA 2005 The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development 132 2125 2134
12. XuDWangYWilleckeRChenZDingT 2006 The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ 13 1697 1706
13. ChewSKAkdemirFChenPLuWJMillsK 2004 The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell 7 897 907
14. DaishTJMillsKKumarS 2004 Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell 7 909 915
15. KondoSSenoo-MatsudaNHiromiYMiuraM 2006 DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol 26 7258 7268
16. WaldhuberMEmotoKPetritschC 2005 The Drosophila caspase DRONC is required for metamorphosis and cell death in response to irradiation and developmental signals. Mech Dev 122 914 927
17. KilpatrickZECakourosDKumarS 2005 Ecdysone-mediated up-regulation of the effector caspase DRICE is required for hormone-dependent apoptosis in Drosophila cells. J Biol Chem 280 11981 11986
18. MuroIBerryDLHuhJRChenCHHuangH 2006 The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 133 3305 3315
19. DorstynLColussiPAQuinnLMRichardsonHKumarS 1999 DRONC, an ecdysone-inducible Drosophila caspase. Proc Natl Acad Sci U S A 96 4307 4312
20. KanukaHSawamotoKInoharaNMatsunoKOkanoH 1999 Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell 4 757 769
21. RodriguezAOliverHZouHChenPWangX 1999 Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol 1 272 279
22. ZhouLSongZTittelJStellerH 1999 HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol Cell 4 745 755
23. QuinnLMDorstynLMillsKColussiPAChenP 2000 An essential role for the caspase dronc in developmentally programmed cell death in Drosophila. J Biol Chem 275 40416 40424
24. DorstynLReadSCakourosDHuhJRHayBA 2002 The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J Cell Biol 156 1089 1098
25. DorstynLKumarS 2008 A biochemical analysis of the activation of the Drosophila caspase DRONC. Cell Death Differ 15 461 470
26. SrivastavaMScherrHLackeyMXuDChenZ 2007 ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ 14 92 102
27. MillsKDaishTHarveyKFPflegerCMHariharanIK 2006 The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death. J Cell Biol 172 809 815
28. AkdemirFFarkasRChenPJuhaszGMedved'ovaL 2006 Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development 133 1457 1465
29. MendesCSAramaEBrownSScherrHSrivastavaM 2006 Cytochrome c–d regulates developmental apoptosis in the Drosophila retina. EMBO Rep 7 933 939
30. YuXWangLAcehanDWangXAkeyCW 2006 Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol 355 577 589
31. YuanSYuXTopfMDorstynLKumarS 2011 Structure of the Drosophila apoptosome at 6.9 a resolution. Structure 19 128 140
32. HawkinsCJYooSJPetersonEPWangSLVernooySY 2000 The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem 275 27084 27093
33. MuroIHayBAClemRJ 2002 The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 277 49644 49650
34. SnipasSJDragMStennickeHRSalvesenGS 2008 Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death Differ 15 938 945
35. O'RiordanMXBaulerLDScottFLDuckettCS 2008 Inhibitor of apoptosis proteins in eukaryotic evolution and development: a model of thematic conservation. Dev Cell 15 497 508
36. VauxDLSilkeJ 2005 IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol 6 287 297
37. MeierPSilkeJLeeversSJEvanGI 2000 The Drosophila caspase DRONC is regulated by DIAP1. Embo J 19 598 611
38. ChaiJYanNHuhJRWuJWLiW 2003 Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol 10 892 898
39. WangSLHawkinsCJYooSJMullerHAHayBA 1999 The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98 453 463
40. GoyalLMcCallKAgapiteJHartwiegEStellerH 2000 Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J 19 589 597
41. LisiSMazzonIWhiteK 2000 Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154 669 678
42. HayBAWassarmanDARubinGM 1995 Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83 1253 1262
43. ZachariouATenevTGoyalLAgapiteJStellerH 2003 IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. Embo J 22 6642 6652
44. WilsonRGoyalLDitzelMZachariouABakerDA 2002 The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol 4 445 450
45. RyooHDGorencTStellerH 2004 Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell 7 491 501
46. ShapiroPJHsuHHJungHRobbinsESRyooHD 2008 Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol 10 1440 1446
47. SuzukiYNakabayashiYTakahashiR 2001 Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A 98 8662 8667
48. HuangHJoazeiroCABonfocoEKamadaSLeversonJD 2000 The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J Biol Chem 275 26661 26664
49. SchileAJGarcia-FernandezMStellerH 2008 Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev 22 2256 2266
50. DitzelMBroemerMTenevTBolducCLeeTV 2008 Inactivation of Effector Caspases through non-degradative Polyubiquitylation. Molecular Cell in press
51. FanYBergmannA 2010 The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ 17 534 539
52. PflegerCMHarveyKFYanHHariharanIK 2007 Mutation of the Gene Encoding the Ubiquitin Activating Enzyme Uba1 Causes Tissue Overgrowth in Drosophila. Fly 1 95 105
53. SilkeJKratinaTChuDEkertPGDayCL 2005 Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci U S A 102 16182 16187
54. RyooHDBergmannAGonenHCiechanoverAStellerH 2002 Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 4 432 438
55. HaysRWicklineLCaganR 2002 Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat Cell Biol 4 425 431
56. HolleyCLOlsonMRColon-RamosDAKornbluthS 2002 Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol 4 439 444
57. WingJPSchreaderBAYokokuraTWangYAndrewsPS 2002 Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat Cell Biol 4 451 456
58. YooSJHuhJRMuroIYuHWangL 2002 Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 4 416 424
59. WolffTReadyDF 1991 The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113 841 850
60. CaganRLReadyDF 1989 The emergence of order in the Drosophila pupal retina. Dev Biol 136 346 362
61. EllisMCO'NeillEMRubinGM 1993 Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 119 855 865
62. MartinFAPerez-GarijoAMorataG 2009 Apoptosis in Drosophila: compensatory proliferation and undead cells. Int J Dev Biol 53 1341 1347
63. HuhJRGuoMHayBA 2004 Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol 14 1262 1266
64. Perez-GarijoAMartinFAMorataG 2004 Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development 131 5591 5598
65. Perez-GarijoAShlevkovEMorataG 2009 The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 136 1169 1177
66. WellsBSYoshidaEJohnstonLA 2006 Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 16 1606 1615
67. DaishTJCakourosDKumarS 2003 Distinct promoter regions regulate spatial and temporal expression of the Drosophila caspase dronc. Cell Death Differ 10 1348 1356
68. CakourosDDaishTJKumarS 2004 Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc, regulating its expression in specific tissues. J Cell Biol 165 631 640
69. AbramsJMWhiteKFesslerLIStellerH 1993 Programmed cell death during Drosophila embryogenesis. Development 117 29 43
70. MuroIMonserKClemRJ 2004 Mechanism of Dronc activation in Drosophila cells. J Cell Sci 117 5035 5041
71. DeverauxQLTakahashiRSalvesenGSReedJC 1997 X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388 300 304
72. RoyNDeverauxQLTakahashiRSalvesenGSReedJC 1997 The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J 16 6914 6925
73. TakahashiRDeverauxQTammIWelshKAssa-MuntN 1998 A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem 273 7787 7790
74. SunCCaiMGunasekeraAHMeadowsRPWangH 1999 NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature 401 818 822
75. SunCCaiMMeadowsRPXuNGunasekeraAH 2000 NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP. J Biol Chem 275 33777 33781
76. SrinivasulaSMHegdeRSalehADattaPShiozakiE 2001 A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410 112 116
77. ShiozakiENChaiJRigottiDJRiedlSJLiP 2003 Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell 11 519 527
78. RiedlSJRenatusMSchwarzenbacherRZhouQSunC 2001 Structural basis for the inhibition of caspase-3 by XIAP. Cell 104 791 800
79. ChaiJShiozakiESrinivasulaSMWuQDattaP 2001 Structural basis of caspase-7 inhibition by XIAP. Cell 104 769 780
80. SilkeJHawkinsCJEkertPGChewJDayCL 2002 The anti-apoptotic activity of XIAP is retained upon mutation of both the caspase 3 - and caspase 9-interacting sites. J Cell Biol 157 115 124
81. WertzIEDixitVM 2010 Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol 2 a003350
82. Herman-BachinskyYRyooHDCiechanoverAGonenH 2007 Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ 14 861 871
83. AdhikariAXuMChenZJ 2007 Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26 3214 3226
84. HayBAWolffTRubinGM 1994 Expression of baculovirus P35 prevents cell death in Drosophila. Development 120 2121 2129
85. BergmannAStellerH 2010 Apoptosis, stem cells, and tissue regeneration. Sci Signal 3 re8
86. FanYBergmannA 2008 Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends Cell Biol 18 467 473
87. TabataT 2001 Genetics of morphogen gradients. Nat Rev Genet 2 620 630
88. YuSYYooSJYangLZapataCSrinivasanA 2002 A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development 129 3269 3278
89. FanYLeeTVXuDChenZLamblinAF 2010 Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ 17 912 921
90. HerzHMChenZScherrHLackeyMBolducC 2006 vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development 133 1871 1880
91. LeulierFRibeiroPSPalmerETenevTTakahashiK 2006 Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ 13 1663 1674
92. WhiteKTahaogluEStellerH 1996 Cell killing by the Drosophila gene reaper. Science 271 805 807
93. EmeryGHuttererABerdnikDMayerBWirtz-PeitzF 2005 Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 122 763 773
94. BrandAHPerrimonN 1993 Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401 415
95. MorrisonCMHalderG 2010 Characterization of a dorsal-eye Gal4 Line in Drosophila. Genesis 48 3 7
96. ZhouLSchnitzlerAAgapiteJSchwartzLMStellerH 1997 Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc Natl Acad Sci U S A 94 5131 5136
97. XuTRubinGM 1993 Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117 1223 1237
98. NewsomeTPAslingBDicksonBJ 2000 Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127 851 860
99. LeeTLuoL 2001 Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24 251 254
100. GretherMEAbramsJMAgapiteJWhiteKStellerH 1995 The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 9 1694 1708
Štítky
Genetika Reprodukčná medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



