-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
The conserved DAF-16/FOXO transcription factors and SIR-2.1/SIRT1 deacetylases are critical for diverse biological processes, particularly longevity and stress response; and complex regulation of DAF-16/FOXO by SIR-2.1/SIRT1 is central to appropriate biological outcomes. Caenorhabditis elegans Host Cell Factor 1 (HCF-1) is a longevity determinant previously shown to act as a co-repressor of DAF-16. We report here that HCF-1 represents an integral player in the regulatory loop linking SIR-2.1/SIRT1 and DAF-16/FOXO in both worms and mammals. Genetic analyses showed that hcf-1 acts downstream of sir-2.1 to influence lifespan and oxidative stress response in C. elegans. Gene expression profiling revealed a striking 80% overlap between the DAF-16 target genes responsive to hcf-1 mutation and sir-2.1 overexpression. Subsequent GO-term analyses of HCF-1 and SIR-2.1-coregulated DAF-16 targets suggested that HCF-1 and SIR-2.1 together regulate specific aspects of DAF-16-mediated transcription particularly important for aging and stress responses. Analogous to its role in regulating DAF-16/SIR-2.1 target genes in C. elegans, the mammalian HCF-1 also repressed the expression of several FOXO/SIRT1 target genes. Protein–protein association studies demonstrated that SIR-2.1/SIRT1 and HCF-1 form protein complexes in worms and mammalian cells, highlighting the conservation of their regulatory relationship. Our findings uncover a conserved interaction between the key longevity determinants SIR-2.1/SIRT1 and HCF-1, and they provide new insights into the complex regulation of FOXO proteins.
Published in the journal: The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO. PLoS Genet 7(9): e32767. doi:10.1371/journal.pgen.1002235
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002235Summary
The conserved DAF-16/FOXO transcription factors and SIR-2.1/SIRT1 deacetylases are critical for diverse biological processes, particularly longevity and stress response; and complex regulation of DAF-16/FOXO by SIR-2.1/SIRT1 is central to appropriate biological outcomes. Caenorhabditis elegans Host Cell Factor 1 (HCF-1) is a longevity determinant previously shown to act as a co-repressor of DAF-16. We report here that HCF-1 represents an integral player in the regulatory loop linking SIR-2.1/SIRT1 and DAF-16/FOXO in both worms and mammals. Genetic analyses showed that hcf-1 acts downstream of sir-2.1 to influence lifespan and oxidative stress response in C. elegans. Gene expression profiling revealed a striking 80% overlap between the DAF-16 target genes responsive to hcf-1 mutation and sir-2.1 overexpression. Subsequent GO-term analyses of HCF-1 and SIR-2.1-coregulated DAF-16 targets suggested that HCF-1 and SIR-2.1 together regulate specific aspects of DAF-16-mediated transcription particularly important for aging and stress responses. Analogous to its role in regulating DAF-16/SIR-2.1 target genes in C. elegans, the mammalian HCF-1 also repressed the expression of several FOXO/SIRT1 target genes. Protein–protein association studies demonstrated that SIR-2.1/SIRT1 and HCF-1 form protein complexes in worms and mammalian cells, highlighting the conservation of their regulatory relationship. Our findings uncover a conserved interaction between the key longevity determinants SIR-2.1/SIRT1 and HCF-1, and they provide new insights into the complex regulation of FOXO proteins.
Introduction
The Insulin/Insulin-like Growth Factor-1(IGF-1) signaling (IIS) cascade is one of the most highly conserved and best characterized longevity pathways in eukaryotes. When stimulated, the insulin/IGF-1 like receptors initiate a kinase cascade that leads to the phosphorylation, and cytoplasmic retention of the main downstream effectors, Forkhead box, Class O (FOXO) transcription factors. Reduction in IIS signaling leads to the dephosphorylation of FOXO, allowing nuclear translocation and transcriptional activation of FOXO [1], [2]. The C. elegans FOXO ortholog DAF-16, as well as the Drosophila, mouse, and human FOXO transcription factors are all critical for longevity, metabolism, and stress response [3]–[12], suggesting that the mechanisms underlying FOXOs' ability to affect physiology are highly conserved across species. Indeed, much of our understanding of FOXO regulation comes from studies done on C. elegans DAF-16. When activated, DAF-16 selectively regulates the transcription of a large number of genes which cumulatively act to elevate stress resistance, alter metabolic and developmental responses, improve immunity, and extend lifespan [13]–[16]. To integrate many different environmental stimuli and coordinate proper transcriptional responses, DAF-16 activity must be tightly controlled. DAF-16 activity is known to be regulated by post-translational modifications, nuclear/cytoplasmic translocation and association with transcriptional co-regulators. Although necessary for its activation, translocation of DAF-16 into the nucleus is not sufficient to stimulate its transcriptional activity [17]. Association with additional co-factors is also necessary for nuclear DAF-16 activation [18]–[23]. Little is known about the interplay between DAF-16 and its nuclear regulators and how these multiple factors coordinately act on DAF-16 to ensure proper transcriptional outcomes.
SIR-2.1, the C. elegans homolog of the yeast NAD+-dependent protein deacetylase Sir2p, is an important DAF-16 co-factor. SIR-2.1 is thought to activate DAF-16 in conferring longevity as well as stress resistance [18], [24], [25]. Heat stress stimulates the physical association of SIR-2.1 with DAF-16 via the scaffolding protein 14-3-3, which promotes the transactivation of DAF-16 through an unknown mechanism [18], [25]. Overexpression of Sir2 homologs in worms, yeast and flies extends lifespan [18], [24], [26], [27], emphasizing the evolutionarily conserved role of Sir2 in longevity determination. In mammals, SIRT1 associates with and directly deacetylates FOXO1, 3, and 4 in a stress-dependent manner [28]–[31]. However, the exact mechanism whereby SIR-2.1/SIRT1 affects DAF-16/FOXO activity and whether additional factors are involved in the regulation of DAF-16/FOXO by SIR-2.1/SIRT1 is not well understood.
Host Cell Factor-1 (HCF-1) belongs to a family of highly conserved HCF proteins and acts as a nuclear co-repressor of DAF-16 [21], [32]. Inactivating hcf-1 robustly extends lifespan and confers oxidative stress resistance in a daf-16-dependent manner in C. elegans. In the nucleus, HCF-1 associates with DAF-16 and limits its access to a subset of target gene promoters [21]. C. elegans HCF-1 shares high structural homology with two mammalian counterparts, HCF-1 and HCF-2 [32]. Although mammalian HCF-1 has been studied extensively, HCF-2 functions remain largely unknown. Mammalian HCF-1 was originally identified as a binding partner of the Herpes Simplex Virus VP16 transcription factor [33]. Apart from VP16, HCF-1 has been shown to associate with a number of transcription factors to stimulate or repress their transactivation properties [34]–[39]. HCF-1 is an important regulator of cellular proliferation as it promotes progression through multiple phases of the cell cycle via assembling transcriptional complexes to modulate E2F transcription factor activities [38], [40]. Whether mammalian HCF proteins function as conserved FOXO regulators has yet to be determined.
In this study, we sought to examine whether the two conserved DAF-16/FOXO nuclear regulators, HCF-1 and SIR-2.1/SIRT1, functionally interact in worms and whether this interaction is conserved in mammals. We found that hcf-1 acts downstream of sir-2.1 to regulate daf-16 and thereby modulates lifespan and oxidative stress response in C. elegans. We showed that HCF-1 and SIR-2.1 regulate a common subset of DAF-16 target genes important for ensuring longevity and stress response. Furthermore, we demonstrated that mammalian HCF-1 affects the expression of several SIRT1/FOXO transcriptional targets and physically associates with both FOXO3 and SIRT1. Our findings uncover a new regulatory mechanism between the critical longevity determinants DAF-16/FOXO and SIR-2.1/SIRT1, and implicate an important role of HCF-1 in aging and age-related diseases in diverse organisms.
Results
C. elegans hcf-1 acts downstream of sir-2.1 to modulate longevity and oxidative stress responses
In C. elegans, inactivation of hcf-1 results in a robust lifespan extension, as well as improved survival upon exposure to oxidative stress, in a manner dependent on daf-16. In its role in longevity and stress response, HCF-1 inhibits DAF-16 activity by physically associating with DAF-16 and diminishing DAF-16 localization to a subset of downstream target promoters [21]. In the context of cell cycle progression, mammalian HCF-1 is known to regulate the activities of various transcription factors by promoting the formation of transcriptional regulatory complexes [39], [41]. We reasoned that HCF-1 in C. elegans may function similarly and, in conjunction with other transcriptional regulators, act to fine tune DAF-16 activity. As SIR-2.1 is a well-known, evolutionarily conserved longevity determinant that activates DAF-16 [18], we explored whether HCF-1 and SIR-2.1 functionally interact to regulate DAF-16. As a first step, we examined the putative functional connection between hcf-1 and sir-2.1 in lifespan modulation by performing genetic analyses. We compared the lifespan of hcf-1(pk924) and sir-2.1(ok434) single mutants to that of sir-2.1(ok434) hcf-1(pk924) double mutants. Both hcf-1 and sir-2.1 alleles used in this analysis are putative null mutants [21], [42]. As previously described, hcf-1(pk924) mutant worms displayed a mean lifespan >20% longer than that of wild type and the hcf-1(pk924) long-lived phenotype was fully suppressed by daf-16(mgDf47) mutation (Figure 1A and [21]). sir-2.1(ok434) mutants exhibited lifespan similar to that of wild-type worms and always substantially shorter than that of hcf-1(pk924) (Figure 1A; Table S1A). We found that all four independent lines of the double mutants exhibited lifespans similar to that of hcf-1(pk924) single mutant worms (Figure 1A, Table S1A), suggesting that sir-2.1 is not required for hcf-1(pk924) mutation to extend lifespan. Our genetic data suggest two possibilities: one is that hcf-1 and sir-2.1 may work independently and that sir-2.1 inactivation does not affect hcf-1(pk924) mutant longevity. On the other hand, since the lifespan of the double mutant is similar to that of hcf-1(pk924) single mutant, hcf-1 may act downstream of sir-2.1. To distinguish between these two possibilities, we examined the effect of overexpressing sir-2.1 in worms harboring the hcf-1 mutation. In C. elegans, overexpressing sir-2.1 confers a lifespan extension phenotype that is dependent on daf-16 [18], [24]. We reasoned that if hcf-1 and sir-2.1 work independently, then combining hcf-1 inactivation with sir-2.1 overexpression should further increase lifespan. By contrast, if hcf-1 and sir-2.1 work in the same pathway, and hcf-1 is genetically downstream of sir-2.1, then overexpression of sir-2.1 should not cause further lifespan extension in hcf-1(pk924) mutants. To examine this, we utilized the long-lived, low-copy sir-2.1 overexpressor strain NL3909 pkIs1642 [unc-119 sir-2.1] (pkIs1642[sir-2.1(O/E)]) [18], [43] to generate hcf-1(pk924);pkIs1642[sir-2.1(O/E)] strains. As a control, we outcrossed the pkIs1642 strain and showed that it continues to extend lifespan compared to its transgenic control NL3908 pkIs1641 [unc-119] (pkIs1641[sir-2.1(wt)]) under our assaying conditions (Figure S2A; Table S1G). Furthermore, we knocked-down sir-2.1 in the pkIs1642 strain to show that the lifespan increase is indeed dependent on sir-2.1 (Figure S2B–S2D; Table S1H). hcf-1(pk924) and pkIs1642[sir-2.1(O/E)] worms lived longer than N2 wild type or pkIs1641[sir-2.1(wt)] transgenic controls by 28% and 17%, respectively (Figure 1B; Table S1B, S1G). Interestingly, the hcf-1(pk924);pkIs1642[sir-2.1(O/E)]) worms exhibited a lifespan very similar to, or in some cases shorter than, that of hcf-1(pk924) mutants (Figure 1B; Table S1B). However, in none of the hcf-1(pk924);pkIs1642[sir-2.1(O/E)]) isolates generated did we observe a lifespan longer than that of hcf-1(pk924) mutants (Table S1B). These data support the hypothesis that hcf-1 acts in the same genetic pathway as sir-2.1. Considering our previous findings that hcf-1 can robustly extend the lifespans of long-lived insulin signaling and germline proliferation mutants [21], our current observation that overexpression of sir-2.1 cannot further enhance longevity in worms lacking hcf-1 indicates that the genetic interaction between hcf-1(-) and sir-2.1(O/E) is specific.
Fig. 1. hcf-1 acts downstream of sir-2.1 to modulate lifespan and oxidative stress response. 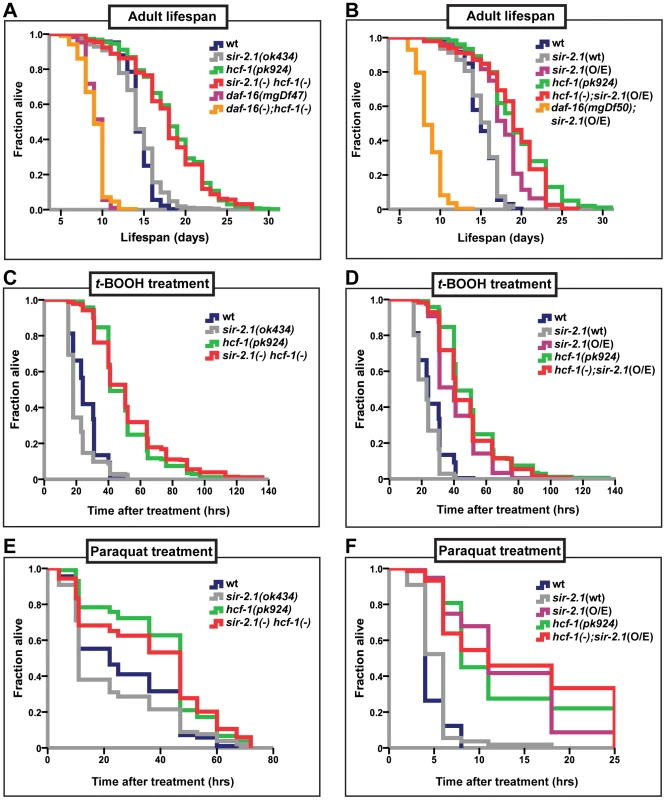
(A–B) Lifespans of synchronized adult populations of indicated genotypes. (A) Data pooled from four independent experiments are plotted. One of four sir-2.1(ok434) hcf-1(pk924) lines is shown. (See Table S1A). (B) Pooled data from three independent experiments are displayed. One of five hcf-1(pk924);pkIs1642[sir-2.1(O/E)] lines is shown (See Table S1B). (C–F) Oxidative stress response of adult worms. (C–D) Day one adult worms were exposed to 6 mM t-BOOH on plates and their survival monitored through time. The survival curves represent pooled data from two independent experiments. (E–F) Day two adult worms were exposed to 150 mM (E) or 200 mM (F) paraquat in M9 buffer and their survival monitored through time. Survival curves are generated using pooled data from two independent experiments (E) or data from one of two representative experiments (F). See Table S1A–S1F for statistics and Figure S1C–S1F for linear mixed model analysis plots. All lifespan and stress experiments were carried out at 25°C. Quantitative data and statistical analyses are displayed in Table S1A–S1D. In addition to their lifespan effects, both HCF-1 and SIR-2.1 regulate the ability of DAF-16 to respond to a variety of environmental stress cues. Adult hcf-1(pk924) mutant worms are resistant to oxidative - and heavy metal-stress [21]. Likewise, sir-2.1 overexpression is protective against exposure to oxidative as well as heat stress, while sir-2.1 mutation increases sensitivity to oxidative, heat, and UV-induced environmental insults [18], [42]. To further investigate the genetic relationship between hcf-1 and sir-2.1, we analyzed the response of sir-2.1(ok434) hcf-1(pk924) double mutants and hcf-1(pk924);pkIs1642[sir-2.1(O/E)]) worms to treatment with two oxidative-stress inducing agents, paraquat and tert-Butyl hydroperoxide (t-BOOH). Paraquat induces cellular damage by elevating intracellular superoxide levels [44], and t-BOOH damages cellular lipids and proteins through peroxidation [45]. Under the paraquat or t-BOOH conditions where sir-2.1(ok434) mutants were sensitive and hcf-1(pk924) worms resistant to the treatments, sir-2.1(ok434) hcf-1(pk924) worms survived the paraquat or t-BOOH exposure as well as hcf-1(pk924) single mutants did, and were significantly more resistant than N2 or sir-2.1(ok434) worms (Figure 1C, 1E; Figure S1A, S1C; Table S1C, S1E). Furthermore, overexpressing sir-2.1 in hcf-1(pk924) mutants did not further enhance the paraquat or t-BOOH-resistance of hcf-1(pk924) worms (Figure 1D, 1F; Figure S1B, S1D; Table S1D, S1F). Overall, our observations are consistent with a model in which hcf-1 acts downstream of sir-2.1 to modulate longevity and oxidative stress responses in C. elegans.
14-3-3 proteins are required for lifespan extension in worms carrying the hcf-1 mutation
In C. elegans, 14-3-3 proteins are required for lifespan extension and stress resistance conferred by extra copies of sir-2.1, as well as for facilitating the association of SIR-2.1 and DAF-16 [18], [25]. Our findings that hcf-1 and sir-2.1 act together to regulate daf-16 raise the possibility that hcf-1 may also functionally interact with 14-3-3. To address this question, we examined the genetic relationship between hcf-1 and 14-3-3 in lifespan. The 14-3-3 homologs in C. elegans are encoded by two highly similar genes ftt-2 and par-5, which share ∼80% sequence identity [46]. RNAi constructs targeting the coding sequences of ftt-2 and par-5 are not specific and will knockdown both genes, whereas RNAi constructs targeting the 3′ UTR of each are gene-specific (Figure S4A and [47]). We found that knocking down either ftt-2 or par-5 alone did not substantially reduce hcf-1(pk924) lifespan, yet simultaneously diminishing the function of both genes through the non-specific RNAi completely abrogated the longevity effect of hcf-1 inactivation (Figure 2A, 2B; Table S2A, S2B). The RNAi data are corroborated by our findings that a null mutation of ftt-2, n4426, was only able to slightly decrease the lifespan of hcf-1 mutants (Figure S3D; Table S2D). Therefore, we conclude that both 14-3-3 genes are necessary for the longevity increase conferred by hcf-1 mutation and likely act downstream of hcf-1.
Fig. 2. 14-3-3 are required for lifespan extension conferred by hcf-1(pk924) mutation. 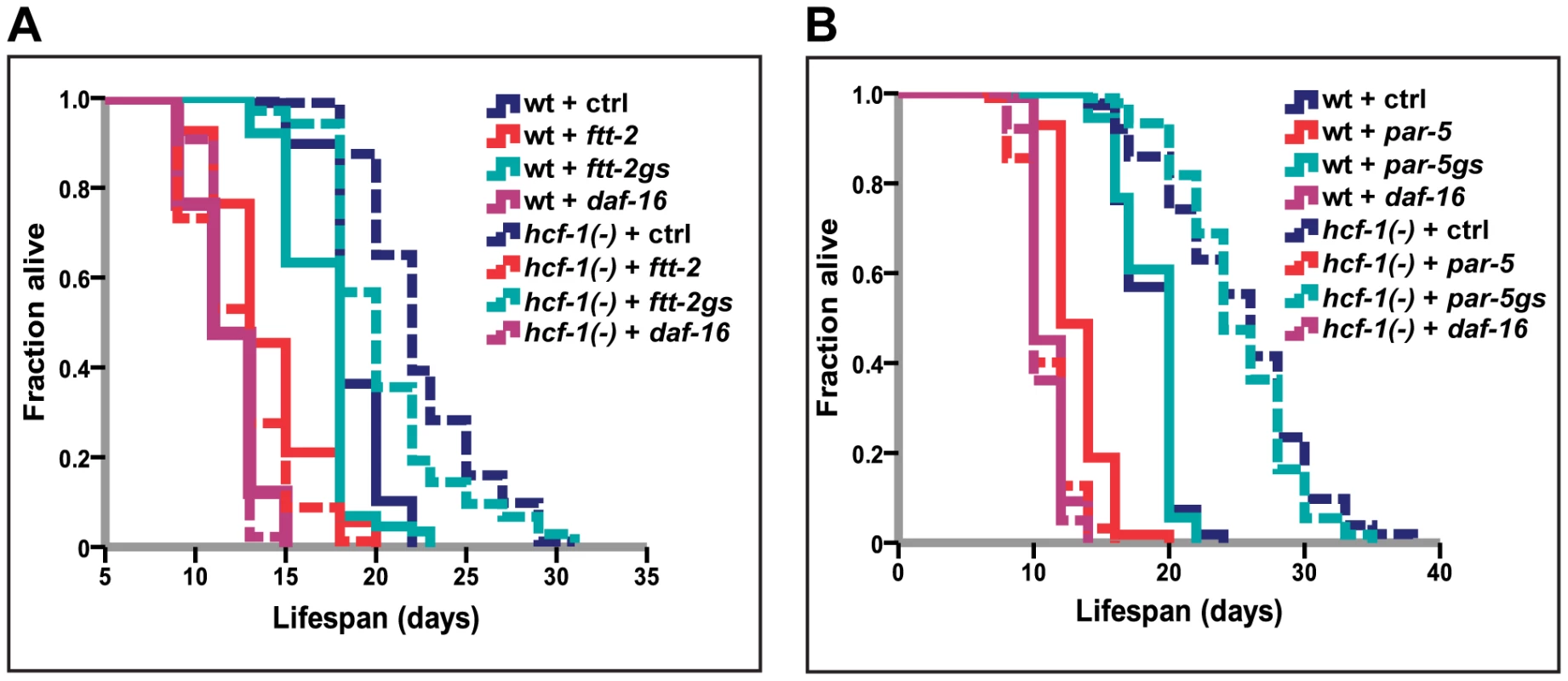
(A–B) Worms were grown on vector, daf-16, ftt-2 (Ahringer - contains multiple stretches of identical sequences to par-5), par-5 (Ahringer - contains overlapping sequences with ftt-2) [47], ftt-2gs (gene specific RNAi targeting 3′ UTR of ftt-2), or par-5gs (gene-specific RNAi targeting 3′ UTR of par-5) [47] from egglay until the end of life. The lifespan experiments were carried out at 20°C. Quantitative data and statistical analyses are included in Table S2A, S2B. hcf-1 and sir-2.1 co-regulate a specific subset of DAF-16 transcriptional targets important for longevity, cellular detoxification, and fatty acid/lipid/amino acid metabolism
DAF-16 responds to different upstream stimuli by selectively activating and repressing groups of target genes, and hence ensuring appropriate responses to specific signals [14]–[16]. We previously proposed that C. elegans HCF-1 acts as a specificity factor for DAF-16 and negatively regulates DAF-16 on a select set of its target genes [21]. Similarly, C. elegans SIR-2.1 is thought to promote DAF-16 regulation of a subset of transcriptional targets [18]. As our genetic data suggest that hcf-1 and sir-2.1 act in the same genetic pathway to modulate longevity in a daf-16-dependent manner, we hypothesized that hcf-1 inactivation and sir-2.1 overexpression would have similar effects on DAF-16-mediated transcription. To test this hypothesis, we compared the daf-16-dependent global transcriptional changes occurring in the long-lived hcf-1(pk924) mutant to those occurring in the long-lived sir-2.1 overexpressor strain.
We identified the genes whose expression was changed in hcf-1(pk924) mutants in a daf-16-dependent manner by comparing the expression profiles of synchronized hcf-1(pk924) mutants to those of daf-16(mgDf47);hcf-1(pk924) double mutants using Agilent C. elegans gene expression microarrays. In addition, to pinpoint the genes that are responsive to the hcf-1(pk924) mutation, instead of those that show expression changes simply due to daf-16 deletion, we focused on genes that showed a similar trend of expression change both in the hcf-1(pk924) vs. N2 and hcf-1(pk924) vs. daf-16(mgDf47);hcf-1(pk924) comparisons (henceforth referred to as hcf-1(-) profile) (Data are available at NCBI Gene Expression Omnibus, accession number GSE30725). Likewise, the genes which were differentially regulated by DAF-16 in response to sir-2.1 overexpression were identified by comparing the strains pkIs1642[sir-2.1(O/E)] to daf-16(mgDf50);pkIs1642[sir-2.1(O/E)] and pkIs1642 [sir-2.1(O/E)] to its transgenic control pkIs1641[sir-2.1(wt)] (henceforth referred to as sir-2.1(O/E) profile). To identify the genes that show consistent and significant expression changes across the independent biological replicates of hcf-1(-) or sir-2.1(O/E), we used Significance Analysis of Microarrays (SAM) [48] with a stringent criteria of expected false discovery rate (FDR) set at 0%. SAM analysis identified 1,032 significantly affected genes in hcf-1(-) and 1,042 genes in sir-2.1(O/E) (Figure 3A; Table S3). Next, we compared the two datasets to determine the extent of overlap. Strikingly, we found 866 genes (473 upregulated and 390 downregulated) whose expression changed similarly in hcf-1(-) and sir-2.1(O/E) profiles, suggesting that the vast majority (>80%) of the genes regulated by DAF-16 in response to hcf-1 inactivation or sir-2.1 activation are shared (Figure 3B). Of the genes that were expressed in a dissimilar manner between hcf-1(-) and sir-2.1(O/E) profiles, ∼10% displayed an opposite expression change and ∼10% were unique to either hcf-1(-) or sir-2.1(O/E) (Figure 3A, 3B). The finding that the transcriptional outcomes conferred by DAF-16 in response to hcf-1 mutation or sir-2.1 overexpression are largely similar corroborates our genetic data suggesting that SIR-2.1 and HCF-1 act in the same pathway to regulate DAF-16.
Fig. 3. hcf-1 inactivation and sir-2.1 overexpression similarly affect a specific subset of daf-16 downstream target genes. 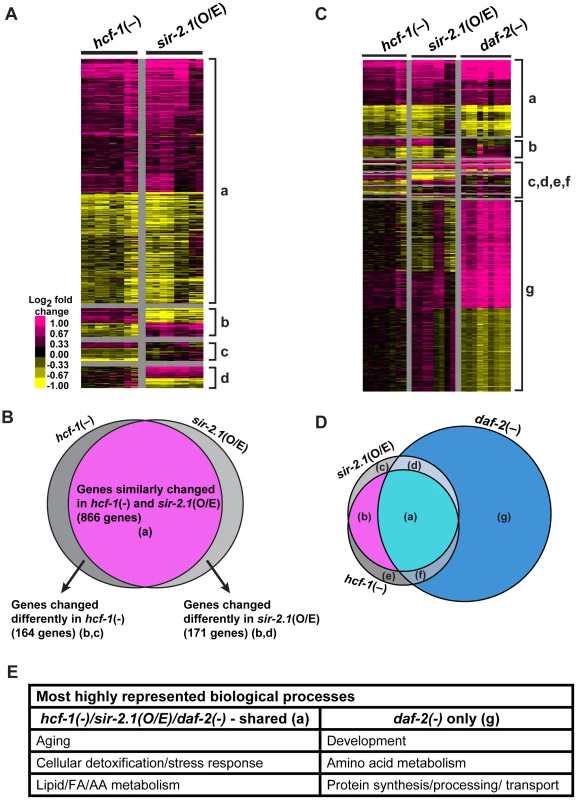
(A–D) Heat maps representing the expression patterns of differentially expressed genes identified by Significance Analysis of Microarrays (SAM) and Venn diagrams showing the overlap between different datasets. For heat maps, each column represents a biological replicate and each row is a gene. Pink = upregulated, Yellow = downregulated, Black = not changed. (A) Heat maps comparing hcf-1(-) and sir-2.1(O/E) arrays. Gene clusters are categorized as: (a) = Genes similarly changed in hcf-1(-) and sir-2.1(O/E) (866), (b) = genes oppositely changed in hcf-1(-) and sir-2.1(O/E) (98), (c) = genes uniquely changed in hcf-1(-) (66), (d) = genes uniquely changed in sir-2.1(O/E) (73). (B) Venn diagram summarizing overlap in (A). (C) Heat maps comparing hcf-1(-), sir-2.1(O/E), and daf-2(-) arrays. Genes are clustered as (a) = similarly expressed in all 3 profiles (693), (b) = similar in only hcf-1(-) and sir-2.1(O/E) (173), (c) = uniquely changed in sir-2.1(O/E) (130), (d) = similar in only sir-2.1(O/E) and daf-2(-) (26), (e) = uniquely changed in hcf-1(-) (140), (f) = similar in only hcf-1(-) and daf-2(-) (46), (g) = uniquely changed in daf-2(-) (1750) (See also Table S3). (D) Venn diagram summarizing overlaps in (C). (E) Most highly enriched GO terms (See also Table S4A, S4B) are summarized based on general biological process. In addition to being regulated by SIR-2.1 and HCF-1, DAF-16 activity is also controlled by the insulin/IGF-1 signaling (IIS) pathway. In response to reduced IIS, DAF-16 translocates into the nucleus and regulates the expression of a large number of genes that together contribute to the diverse functions of IIS, including the regulation of development, metabolism, stress response, and longevity [14]–[16]. To determine how the hcf-1 - and sir-2.1-responsive DAF-16 - target genes compare with the IIS-responsive DAF-16 targets, we further compared the hcf-1(-) and sir-2.1(O/E) profiles to that of the daf-2(-) profile (microarray data from daf-2(e1370) vs. daf-16(mgDf50);daf-2(e1370) [49]). Interestingly, expression of the majority of the shared hcf-1(-)/sir-2.1(O/E)-regulated genes (693/866 = 80%) were also changed in daf-2(-) in the same direction, yet this represented only a fraction of all daf-2(-)-induced changes (693/2515 = 28%) (Figure 3C, 3D). This indicates that, among a large number of potential DAF-16 targets, hcf-1 and sir-2.1 converge to co-regulate a distinct subset of these genes. Our findings from the microarray comparisons support the model that HCF-1 and SIR-2.1 antagonize each other to control a particular aspect of the DAF-16-regulated transcriptional program.
To examine the biological processes that can be carried out by genes affected by hcf-1(-) and sir-2.1(O/E), we queried their Gene Ontology (GO) terms using Database for Annotation, Visualization, and Integrated Discovery (DAVID) [50]. We focused on the GO term categories most significantly enriched in our dataset compared to the C. elegans genome. Our analyses revealed that for the DAF-16 target genes co-regulated by HCF-1/SIR-2.1, GO terms for aging, cellular detoxification (in particular phase 1 & 2 detoxification) and stress response were highly overrepresented among both the upregulated and downregulated genes (Figure 3E; Table S4) [51], [52]. To test whether the DAF-16 targets that are co-regulated by HCF-1/SIR-2.1/DAF-2 might participate in biological functions distinct from the targets uniquely regulated by DAF-2 (and not affected by HCF-1/SIR-2.1), we compared the GO terms represented in the hcf-1(-)/sir-2.1(O/E)-shared genes to those in daf-2(-). Among the genes induced by DAF-16, the most prominent functional categories represented in the hcf-1(-)/sir-2.1(O/E)/daf-2(-)-overlapping set were very similar to those in the hcf-1(-)/sir-2.1(O/E)-co-regulated set (i.e. aging, detoxification, stress response) (Figure 3E; Table S4). By contrast, the DAF-16 target genes that are uniquely upregulated in daf-2(-) are enriched for GO categories for developmental, metabolic (amino acid anabolism/catabolism) and cellular ion transport processes (Figure 3E; Table S4A). Among the genes repressed by DAF-16, the hcf-1(-), sir-2.1(O/E) and daf-2(-) overlapping set is also enriched with GO terms in aging and stress responses, as well as a new category in fatty acid/lipid/amino acid metabolic processes. Interestingly, the daf-2(-)-specific downregulated genes are highly enriched for GO terms in protein biosynthesis, protein degradation, unfolded protein response, protein homeostasis, development and cell division (Figure 3E; Table S4B). Thus, our results suggest that in response to hcf-1 inactivation and sir-2.1 overexpression, DAF-16 specifically induces longevity assurance genes to combat toxic cellular insults/stressors and extend lifespan without strongly affecting developmental, and protein homeostasis pathways.
DAF-16 directly binds a consensus DAF-16 binding element (DBE) to regulate the expression of many downstream target genes [53], [54]. To further investigate how the HCF-1/SIR-2.1-coregulated vs. the IIS-specific DAF-16 target genes might be regulated, we analyzed the 1.5 kb upstream promoter sequences of genes in each group to identify any transcription factor binding sites and regulatory elements that are overrepresented. We submitted the upstream sequences of all genes in hcf-1/sir-2.1-coregulated or daf-2-specific categories to two de novo motif finding algorithms, BioProspector [55] and Regulatory Sequence Analysis Tools (RSAT) [56] and focused on the top highest-scoring motifs from each algorithm. These analyses revealed four common motifs enriched in the promoters of DAF-16 targets, regardless of their responsiveness to HCF-1 & SIR-2.1 (Table S4C), suggesting that DAF-16 likely collaborates with additional yet-to-be identified co-factors in gene regulation.
We were particularly interested in the motifs that are uniquely enriched in the different groups of genes analyzed. The most notable motif highly enriched in the hcf-1/sir-2.1/daf-2-overlapping group, but not in the daf-2-unique group, was the DAF-16-associated element (DAE) (CTTATCA or TGATAAG), previously discovered as a sequence overrepresented in the promoters of DAF-16-regulated genes [16], [54] and shown to be directly bound by DAF-16 in in vitro gel shift assays [54] (Table S4C). Interestingly, the DAE represents a GATA-factor binding motif that is highly enriched in promoters of genes whose expression show age-dependent changes and whose transcription is controlled by C. elegans GATA-factor homologs elt-3, elt-5, and elt-6 [57]. We further compared the expression profiles of hcf-1(-) and sir-2.1(O/E) to that of aging worms [57], and found that 23% of genes that show age-dependent changes were also represented in our hcf-1/sir-2.1 co-regulated set (p-value<2.2e-16 as determined by Chi2 analysis). The large representation of genes that show age-dependent expression changes in the hcf-1/sir-2.1 group correlates well with our observation that HCF-1 and SIR-2.1 together regulate aging - and stress response-specific DAF-16 downstream targets (Figure 3E). Results from the motif analysis also suggest that HCF-1 and SIR-2.1 likely engage additional transcriptional partners, such as GATA factors, in their regulation of DAF-16.
HCF-1 forms a protein complex with SIR-2.1 and 14-3-3 proteins in C. elegans
Our genetic and microarray analyses suggest that SIR-2.1 likely antagonizes HCF-1 to regulate DAF-16 activity. To elucidate the molecular mechanism by which SIR-2.1 may inhibit HCF-1, we first tested whether HCF-1 expression or stability is affected by SIR-2.1. We found that the mRNA and protein levels of HCF-1 did not significantly differ in strains lacking or overexpressing sir-2.1 (data not shown). Since both SIR-2.1 and HCF-1 are known to form a protein complex with DAF-16 in C. elegans [18], [21], we next examined whether SIR-2.1 may also physically associate with HCF-1. We performed co-immunoprecipitation (co-IP) experiments using an affinity-purified anti-HCF-1 antibody and immunoprecipitated HCF-1 from lysates of geIn3[sir-2.1(O/E)], worms overexpressing SIR-2.1 to a greater extent than the pkIs1642[sir-2.1(O/E)] strain we used for lifespan analysis, hcf-1(pk924);geIn3[sir-2.1(O/E)], worms overexpressing SIR-2.1 but lacking hcf-1, and sir-2-1(ok434), worms lacking sir-2.1. SIR-2.1 was co-immunoprecipitated with HCF-1 only in the geIn3[sir-2.1(O/E)] lysate (Figure 4A, left panel). A similar complex formation was also detected in reciprocal co-immunoprecipitation experiments (Figure 4A, right panel).
Fig. 4. C. elegans HCF-1 physically interacts with SIR-2.1 and 14-3-3 proteins. 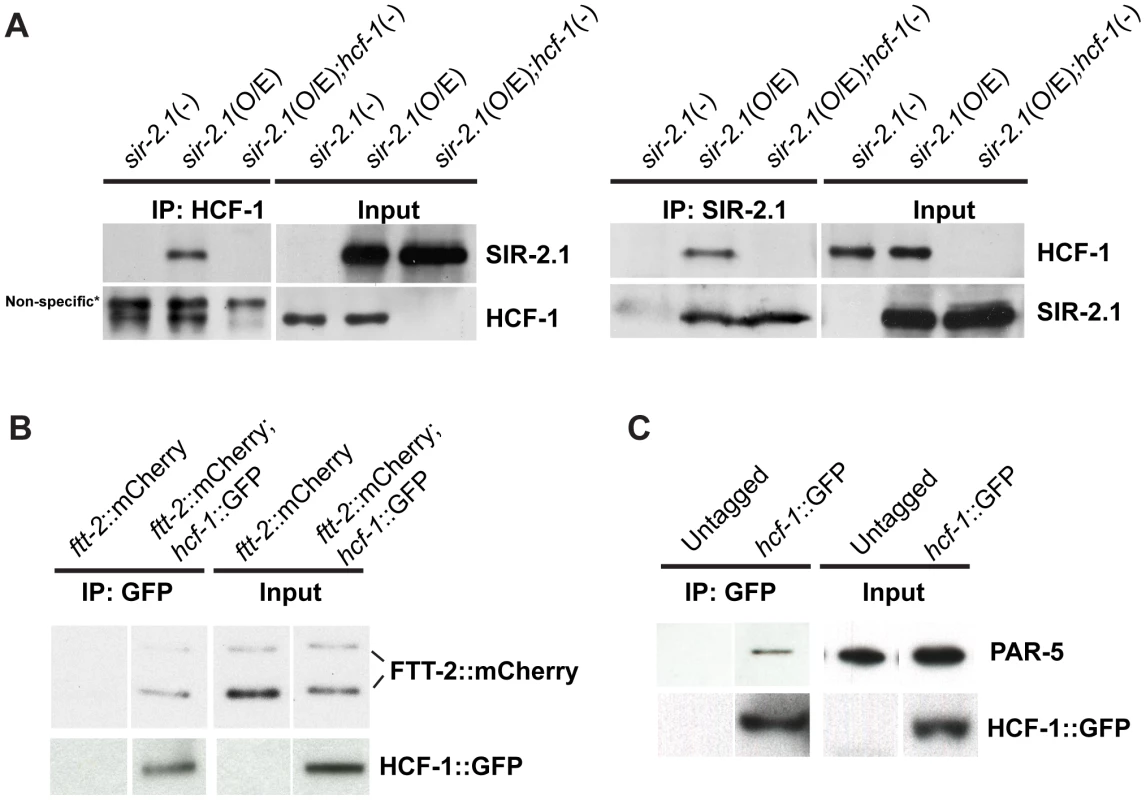
(A) Lysates from sir-2.1(-) (sir-2.1(ok434)), sir-2.1(O/E) (geIn3[sir-2.1(O/E)]), and sir-2.1(O/E);hcf-1(-) (hcf-1(ok559);geIn3[sir-2.1(O/E)]) worms were either immunoprecipitated using anti-HCF-1 antibody (left panel) or anti-SIR-2.1 antibody (right panel). The immunoprecipitated protein complexes were subsequently immunoblotted using anti-HCF-1, or anti-SIR-2.1 antibodies. (B) Lysates from ftt-2::mCherry or hcf-1::GFP;ftt-2::mCherry were immunoprecipitated with an anti-GFP antibody and blotted with anti-mCherry or anti-GFP antibodies. (C) Wild-type or HCF-1::GFP expressing worm lysates were immunoprecipitated using anti-GFP antibodies and blotted with anti-PAR-5 or anti-GFP antibody. Since 14-3-3 proteins are proposed to bridge the physical interactions between SIR-2.1 and DAF-16, especially under stress conditions [18], [25], and our genetic data revealed that 14-3-3 likely function downstream of HCF-1 in longevity modulation, we tested a possible physical association of HCF-1 with 14-3-3 proteins. We immunoprecipitated GFP-fused HCF-1 using anti-GFP antibodies from hcf-1::gfp;ftt-2::mCherry or hcf-1::gfp strains and blotted with anti-mCherry or anti-PAR-5 antibodies to monitor mCherry-tagged FTT-2 and endogenous PAR-5 respectively. HCF-1 was able to form a protein complex with either FTT-2 or PAR-5 (Figure 4B, 4C). Consistent with the co-IP results, a search for HCF-1 binding partners using immunoprecipitation of HCF-1::GFP followed by mass spectrometrical analysis of co-purifying proteins identified the two 14-3-3 proteins FTT-2 and PAR-5 (Figure S4B). Interestingly, sequence analysis (by scansite.mit.edu) predicts that HCF-1 contains a highly significant consensus 14-3-3 binding site, suggesting HCF-1 may directly bind 14-3-3. Taken together, our data reveal that HCF-1 is a new component in the regulatory network involving SIR-2.1, 14-3-3, and DAF-16.
Mammalian HCF-1 and HCF-2 regulate the expression of FOXO target genes
C. elegans HCF-1 belongs to a highly conserved family of proteins [38], [58], [59]. In mammals, two homologs of HCF-1 are present: HCF-1 and HCF-2 [32], [60]. Mammalian HCF-1 plays a role in transcriptional regulation and cell cycle progression, whereas the functions of HCF-2 remain unknown. SIRT1, the mammalian homolog of SIR-2.1, is known to interact with and deacetylate the DAF-16 homologs FOXO1, FOXO3, and FOXO4 and in doing so affects FOXO transcriptional activity [28], [30]. Given that HCF-1, DAF-16 and SIR-2.1 are highly conserved between C. elegans and mammals, we tested whether mammalian homologs of HCF-1 could affect the transcription of FOXO - and SIRT1 - co-regulated target genes. Since mammalian HCF-1 is required for proper cell cycle progression, we employed a transient knockdown approach by transfecting siRNA duplexes targeting the HCF-1 gene into INS-1 rat insulinoma cells. We used two different HCF-1 siRNA duplexes to control for specificity, and found that HCF-1 knockdown did not substantially affect the expression of HCF-2 mRNA as assessed by reverse transcription-quantitative PCR (RT-qPCR) (Figure S5). We examined the expression of Bim, a proapoptotic factor, Gadd45a, which is involved in DNA damage repair, IGFBP1, an insulin-like growth factor-binding protein, and p27, a cyclin-dependent kinase inhibitor. These represent FOXO target genes which are affected by SIRT1 deacetylation of FOXO [28], [30]. Depletion of HCF-1 resulted in a significant increase in the levels of Bim, Gadd45a, and IGFBP1 transcripts, but did not affect p27 expression (Figure 5A). Consistent results were obtained with the two different HCF-1-targeting siRNA duplexes. We next tested whether HCF-2 could also affect FOXO target gene expression. Similar to HCF-1 knockdown, cells treated with HCF-2 siRNA exhibited increased expression of Gadd45a and no change in p27. However, unlike the case with HCF-1, cells depleted of HCF-2 did not show any significant changes in Bim, or IGFBP1 transcripts (Figure 5B). Our data reveal that HCF proteins negatively regulate the expression of a subset of FOXO and SIRT1 transcriptional target genes. Furthermore, HCF-1 appears to play a more substantial role in regulating FOXO target genes relative to HCF-2. The observation that HCF-1 and HCF-2 have specific effects on a subset of FOXO targets tested is also consistent with our findings in C. elegans suggesting HCF-1 to be a specificity factor for DAF-16/FOXO.
Fig. 5. Mammalian HCF-1 and HCF-2 regulate the expression of FOXO target genes. 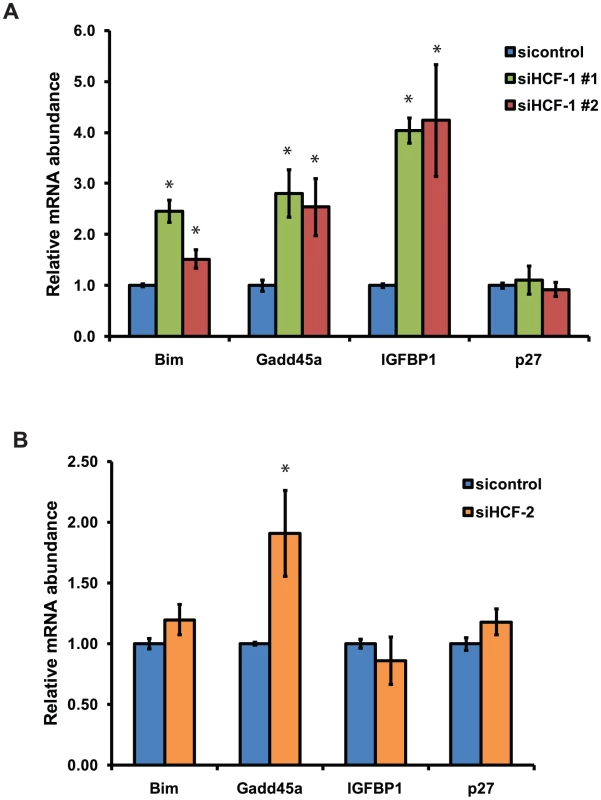
(A) INS-1 cells treated with HCF-1 or control siRNA. (B) INS-1 cells treated with HCF-2 or control siRNA. mRNA levels of Bim, Gadd45a, p27,and IGFBP1 were quantified using RT-qPCR and normalized to the level of β-actin. The mean normalized RNA level for each gene in sicontrol treated cells was set to 1. The data represented are pooled from three independent experiments and are represented as mean +/− SEM. * denotes a p-value<0.05 relative to sicontrol. Mammalian HCF-1 and HCF-2 physically associate with FOXO3 and SIRT1
In C. elegans, HCF-1 is able to physically associate with both DAF-16 and SIR-2.1 (Figure 4A and [21]). We therefore hypothesized that mammalian HCF proteins will also participate in protein complexes with FOXO3 and SIRT1. To examine the physical interactions between these proteins, we transfected HEK293T cells with plasmids encoding either Flag-tagged FOXO3 or Flag-tagged SIRT1. We then performed co-immunoprecipitation experiments with these cell lysates by using Flag-antibody conjugated agarose beads. Both FOXO3 and SIRT1 were found to interact with the endogenous mammalian HCF-1 protein (Figure 6A, 6B; Figure S6A). We also tested whether the closely related HCF-2 protein could also physically interact with FOXO3 and SIRT1. Since antibodies capable of detecting endogenous HCF-2 are not available, we performed co-immunoprecipitation experiments using overexpressed Flag-FOXO3, Flag-SIRT1, and HA-tagged HCF-2. We found that HCF-2 was also present in a protein complex with FOXO3 and SIRT1 when overexpressed (Figure S6B), similar to HCF-1. These results indicate that the physical associations between HCF-1, DAF-16 and SIR-2.1 are highly conserved between C. elegans and mammals.
Fig. 6. Mammalian HCF-1 physically associates with FOXO3 and SIRT1. 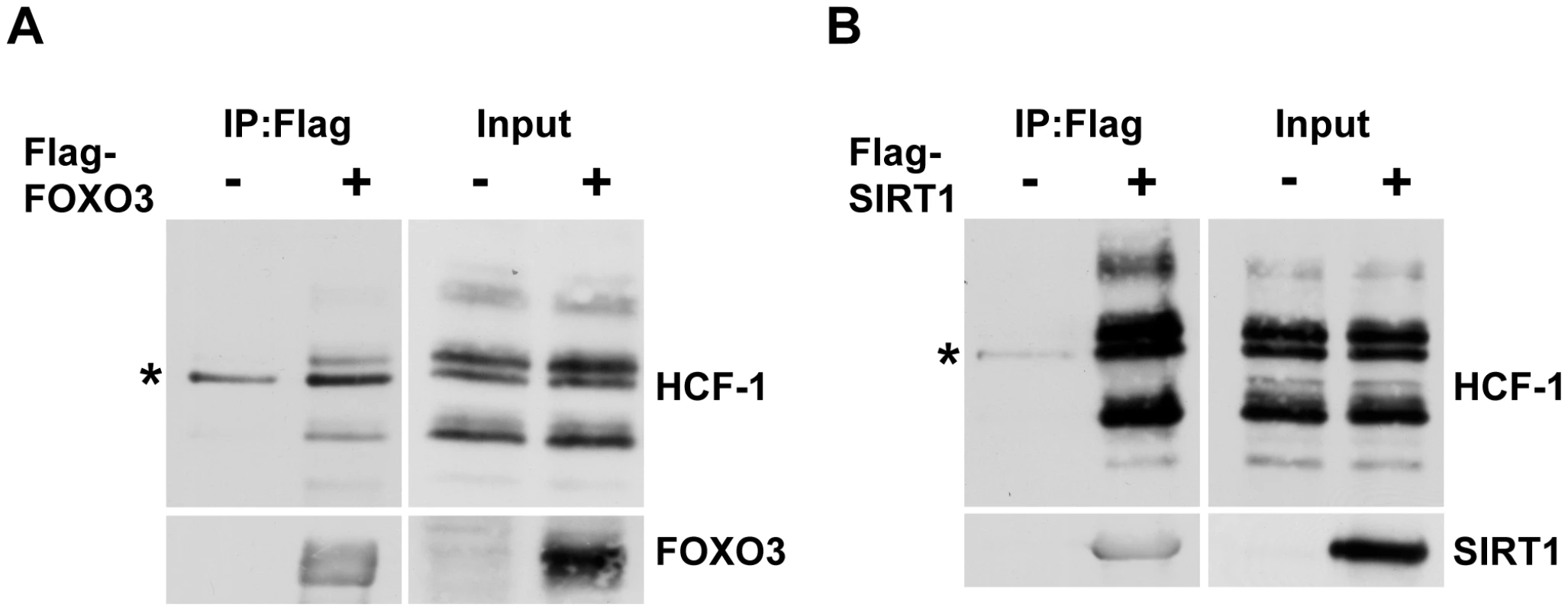
(A, B) HEK293T cells were transfected with plasmids encoding Flag-FOXO3 (A) or Flag-SIRT1 (B). Cell lysates were collected 48 hours later and incubated with anti-Flag-conjugated agarose beads. Immunoprecipitated protein complexes were analyzed by western blot using anti-HCF-1, anti-FOXO3 or anti-SIRT1 antibodies. HCF-1 is known to be proteolytically processed and is detected as multiple bands on SDS-PAGE [76]. * denotes a non-specific band. Discussion
The highly conserved FOXO transcription factors are master regulators of diverse biological processes [61] and as such, their transcriptional activities are tightly controlled [18]–[23]. Although a number of different transcriptional co-factors of DAF-16/FOXO have been identified, little is known about how they functionally interact to fine-tune DAF-16/FOXO activity, and in particular, how they may collaborate to affect DAF-16-mediated lifespan extension. In this study, we identified the DAF-16 nuclear co-repressor HCF-1 as an integral component of the regulatory network involving SIR-2.1/SIRT1, 14-3-3, and DAF-16/FOXO with major consequences to both organismal aging and stress response. Our data indicate that in C. elegans, HCF-1 likely functions downstream of SIR-2.1, and upstream of 14-3-3, to regulate a distinct subset of DAF-16 target genes to affect longevity and oxidative stress response. This regulatory pathway is highly conserved, as mammalian HCF proteins also impact the expression of SIRT1/FOXO co-regulated transcriptional targets, and HCF proteins participate in protein complex formation with SIR-2.1/SIRT1, 14-3-3, and DAF-16/FOXO in worms and in mammals (Figure 7).
Fig. 7. Conserved regulation of DAF-16/FOXO by HCF-1 and SIR-2.1/SIRT1. 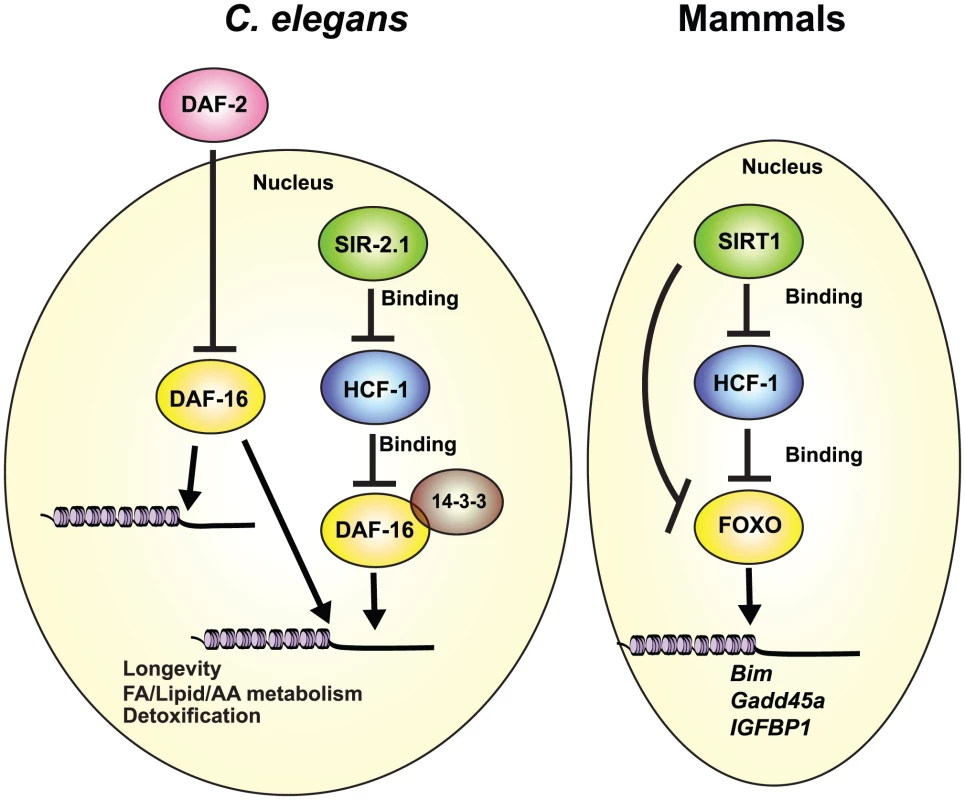
We propose that C. elegans HCF-1 and SIR-2.1 coordinate to fine-tune the transcriptional activity of DAF-16 on a distinct subset of potential target genes. DAF-16 target genes responsive to the hcf-1/sir-2.1 pathway largely overlap with a small subset of IIS-regulated genes, and are specialized in longevity determination, cellular defense, and lipid/fatty acid/amino acid homeostasis. HCF-1 likely represses DAF-16 by forming a complex with SIR-2.1 and 14-3-3, and antagonizing their abilities to stimulate DAF-16. This functional relationship is highly conserved as mammalian HCF proteins also repress the expression of multiple FOXO/SIRT1 target genes and reside in protein complexes with SIRT1 and FOXO3. Our results highlight HCF proteins to be key components of the regulatory network linking SIR-2.1/SIRT1 and DAF-16/FOXO in diverse organisms. Our expression profiling studies indicate that the set of DAF-16 target genes co-regulated by sir-2.1, hcf-1, and daf-2 (area “a” of Figure 3D) is enriched for previously identified longevity-associated genes (annotated as “aging” in GO), whereas the IIS-specific targets (area “g” of Figure 3D) are not. This is somewhat unexpected as the hcf-1 mutant and sir-2.1 overexpressor strains exhibit lifespan extension phenotypes that are much milder than that of the daf-2 mutant. Interestingly, this correlates well with the degree of expression change observed for many of the shared DAF-16 target genes, as they often exhibit more robust expression changes in the daf-2(-) profile compared to the sir-2.1(O/E) or hcf-1(-) profiles. An implication from this observation is that the co-regulated gene set is particularly important for longevity determination, and may thus contain additional targets important for prolonged lifespan that are not currently known to affect aging.
Our previous genetic findings indicated that reduced insulin signaling synergizes with inactivation of hcf-1 to affect longevity and DAF-16-mediated gene regulation [21]. We interpreted those results to suggest that IIS and hcf-1 likely act independently to regulate DAF-16/FOXO. However, a caveat of that interpretation is that the daf-2 mutant we examined was not a null mutant, and formally, loss of hcf-1 can further decrease IIS signaling to further increase lifespan. Similarly, the genetic relationship between the insulin signaling pathway and sir-2.1 has been unclear due to several conflicting reports [18], [24]. In the current study, a comparison of the DAF-16-regulated gene expression changes in response to either daf-2 mutation, hcf-1 inactivation, or sir-2.1 overexpression indicates that a large majority of the HCF-1/SIR-2.1 co-regulated DAF-16 target genes are similarly regulated by reduced IIS. It is possible that upon downregulation of IIS, the majority of DAF-16 migrates into the nucleus but is still subject to regulation by nuclear co-factors. Under this scenario, SIR-2.1 and HCF-1 may be acting as additional “gate keepers” to control DAF-16 activation in the face of reduced IIS. In addition, we saw that the insulin/IGF-1-like peptide, ins-7, which was shown to act as a daf-2 agonist [16], was significantly repressed by hcf-1 inactivation and sir-2.1 overexpression (Table S3). Thus, a possible feedback mechanism in which hcf-1 inactivation or sir-2.1 activation leads to further inhibition of IIS may also explain the genetic results observed with reduced IIS and hcf-1 inactivation or sir-2.1 overexpression.
Our motif analyses revealed additional factors that are likely involved in the regulation of DAF-16 by HCF-1 and SIR-2.1 in C. elegans, in particular the aging-related GATA-factor homologs (ELT-3, -5, -6) known to bind the DAE element, a consensus motif enriched in many of the HCF-1/SIR-2.1 co-regulated genes [57]. Of note, the DAE sequence also shares close resemblance to the mammalian transcription factor Evi1 binding site. Although the C. elegans Evi1 homolog, egl-43, has been shown to be involved in early development [62], a function in longevity and stress response has not been reported. Future functional analysis of HCF-1/SIR-2.1 and ELT-3, -5, -6, and EGL-43 will likely yield new insights into additional layers of DAF-16 regulation.
Considering the high conservation of DAF-16/FOXO-related pathways, it is not surprising that the regulatory relationship among HCF-1, SIR-2.1 and DAF-16 we uncovered in worms turns out to be conserved in mammals. Our findings in mammalian cells are nevertheless very exciting as they implicate the HCF proteins to be key components linking FOXO and SIRT1, two critical master regulators of physiology in mammals. Our results indicate that while both mammalian HCF-1 and HCF-2 are able to interact with SIRT1 and FOXO3, HCF-1 has a greater effect on FOXO target gene expression. Interestingly, while both mammalian HCF-1 and HCF-2 as well as C. elegans HCF-1 are able to support the formation of the Herpes Simplex Virus VP16-transcriptional complex, only mammalian and C. elegans HCF-1 are able to promote VP16 transcriptional activity [32]. Thus, it appears that the evolutionarily conserved functions of HCF proteins are retained in mammalian HCF-1. Alternatively, HCF-1 and HCF-2 likely have tissue-specific functions and are regulated differently under different cellular contexts.
While our data indicate that parallel regulatory mechanisms are shared between C. elegans and mammalian HCF-1, they also suggest the modes of regulation between HCF-1, SIRT1, and FOXO in mammals are likely more complex than what is observed in C. elegans. We note that in the case of the mammalian FOXO target genes Bim and IGFBP1, HCF-1 and SIRT1 appear to affect FOXO target gene expression in a similar manner (Figure 5A and [28], [30]), and thus would appear to act in concert rather than antagonistically. On the other hand, HCF-1 and SIRT1 appear to have antagonistic effects on the FOXO target gene Gadd45a. It is important to keep in mind that in mammals, SIRT1 regulation of FOXO transcription factors is complex; in some instances SIRT1 acts as a repressor and in other cases as an activator of FOXO [28], [30], while in C. elegans the predominant role of SIR-2.1 is as an activator of DAF-16. It is likely that in mammals, the interplay between SIRT1 and HCF-1 results in collaborative as well as antagonistic effects on FOXO transcriptional activity in a gene - and context-dependent manner. Future genome-wide studies examining the effects of HCF-1 on FOXO/SIRT1-regulated gene expression will provide further insights into the relationship between HCF-1 and SIRT1.
We found that HCF-1 physically associates with DAF-16/FOXO and SIR-2.1/SIRT1 in both worms and mammals. Previous studies in C. elegans indicate that 14-3-3 proteins act as bridging molecules that bring SIR-2.1 and DAF-16 into a protein complex in the nucleus [18], [25]. Interestingly, our data suggest 14-3-3 proteins also physically associate with HCF-1. This raises the question of how these different molecules coordinately interact to affect each other's activities. An intriguing model may be that HCF-1 normally binds 14-3-3/DAF-16 and dampens the ability of DAF-16 to activate its target genes; upon appropriate upstream signals, SIR-2.1 ejects HCF-1 off the complex and induces full activation of DAF-16. Whether 14-3-3 proteins are also involved in the regulation of FOXO by SIRT1 and HCF in mammals remain to be investigated. In addition, SIRT1 is known to regulate FOXO transcriptional activity by directly deacetylating FOXO proteins and the FOXO co-activator PGC1α in mammals [63]–[65]. SIRT1 may affect multiple FOXO responses by deacetylating FOXO and specific FOXO co-regulators to achieve activation and/or repression of the appropriate target genes. Future investigation into whether SIRT1 also regulates HCF-1 via deacetylation and whether deacetylation will disrupt protein complexes involving SIRT1/HCF-1/FOXO will provide new insights into the functional interactions among these key longevity determinants.
In conclusion, our findings establish a novel link between two evolutionarily conserved DAF-16/FOXO regulators. This study expands our understanding of the complex role that nuclear factors play in determining the specificity of DAF-16/FOXO activity. These results further implicate HCF-1 as a novel factor that may affect mammalian aging and age-related pathologies through interactions with SIRT1 and FOXO.
Materials and Methods
C. elegans strains
All strain stocks were kept at 16°C and grown under standard growth conditions [66]. The strains used are: Wild type N2, hcf-1(pk924), daf-16(mgDf47);hcf-1(pk924) [21], IU372.1 sir-2.1(ok434) (7 times outcrossed in our lab), NL3908 pkIs1641 [unc-119], NL3909 pkIs1642 [unc-119 sir-2.1] [18], IU91.1 pkIs1641 [unc-119] (1X outcrossed in our lab), IU94 pkIs1642 [unc-119 sir-2.1](1X outcrossed in our lab), geIn3[sir-2.1 rol-6(su1006)] [24] (1X outcrossed in our lab), ftt-2(n4426) [18] (3X outcrossed in our lab), rwIs23 [hcf-1(pk924);Phcf-1::GFP unc-119], GR1680 rwIs23[Phcf-1::GFP; unc-119]; IsB[pCR270(Pftt-2::ftt-2:: Spep-TEV-mCherry::ftt-2-3′UTR; Cb_unc-119)], rwIs9[Phcf-1::hcf-1::GFP Pmec-7::RFP]. Standard genetic methods were utilized to construct the following strains: sir-2.1(ok434) hcf-1(pk924), hcf-1(pk924);pkIs1642[sir-2.1(O/E)], hcf-1(ok559);geIn3[sir-2.1 rol-6(su1006)], ftt-2(n4426);hcf-1(pk924). daf-16(mgDf50); pkIs1642[sir-2.1(O/E)] was a gift from M. Viswanathan and L. Guarente at MIT [43].
Lifespan analysis
All lifespan assays were performed at 25°C, unless otherwise noted, on Nematode Growth Media (NGM) plates seeded with E. coli OP50 or RNAi bacteria. For experiments using OP50, bacteria was grown overnight at 37°C, OD measured after growth and concentrated to OD 7.5 (5X OP50) or used directly, at OD 1.5 (1X). 35 mm NGM plates were seeded with 150 uL of OP50 for egglay plates and dried at room temperature. Plates that would be used for transferring worms throughout the lifespan assay were prepared by adding FUDR to OP50 culture to a final concentration of 50 ug/mL per plate, seeding 150 uL/plate, drying at room temperature, and storing at 4°C until use. For RNAi experiments, HT115 bacteria containing vectors expressing dsRNA were grown at 37°C in LB with 100 ug/mL carbenicillin and 15 ug/mL tetracycline to OD 0.8, induced with 4 mM IPTG for 4 hrs at 37°C, and either concentrated to OD 7.5 and seeded, or seeded at OD 1.5 (1X). RNAi plates were also induced with 4 mM IPTG before use. Well-fed gravid adult worms were allowed to lay eggs at room temperature and the progeny were grown at 25°C until young adult/early gravid adult stage. The synchronized adults were transferred to fresh FUDR-containing plates at Day 0, 2, and 4 of adulthood. For lifespan assays carried out at 20°C, worms were incubated at 25°C for the first three days of adulthood to reduce vulva protrusion defects. The adult worms were scored every other day and worms that did not move when gently prodded by a platinum wire pick were recorded as dead. Worms that bagged, crawled onto the wall of the plate, or had a large protruding vulva were censored on the day of the event. All survival data were analyzed using Kaplan-Meier statistics (SPSS software) to generate statistical values and survival curves. p-values were calculated using the log-rank test. Kaplan-Meier log rank test was employed to determine whether independent experiments displayed statistically similar trends using a cutoff of p-value>0.05. Based on these criteria, data from independent experiments were pooled whenever possible to increase statistical power.
Stress assays
Paraquat
50–60 synchronized worms were grown on three 60 mm NGM/OP50 plates (per strain) at 25°C until day two of adulthood, either directly transferred or washed off the plates with M9 buffer and dispensed into three wells of a 24-well culture plate, and paraquat (Sigma) added to 150 mM or 200 mM final concentration. Plates were kept covered by aluminum foil to prevent excessive light from degrading paraquat, and rocked on a shaker at 25°C. Survival was scored at the indicated time points after paraquat exposure.
tert-Butyl hydroperoxide
Synchronized worms were grown on OP50 plates until day one of adulthood and transferred onto plates containing 6 mM tert-Butyl hydroperoxide (t-BOOH) (Sigma). Survival was scored at indicated time points after treatment.
Kaplan Meier analysis and Log-rank statistics (SPSS software) were used to generate survival curves, calculate mean survival, and compute statistics. Log-rank test was also employed to determine whether independent experiments displayed statistically similar trends using a cutoff of p-value>0.05. Based on these criteria, data from independent experiments were pooled whenever possible to increase statistical power. The mean variation in survival of each strain as compared to either wild-type or pkIs1641[sir-2.1(wt)] was calculated and further analyzed by Linear Mixed model analysis [67] to obtain averaged mean variations relative to control from two or three independent experiments. hcf-1(-) and sir-2.1(-) hcf-1(-) or sir-2.1(O/E) and hcf-1(-);sir-2.1(O/E) were entered as “fixed effect” and experiments as “random effect”. Linear Mixed model analysis allows statistical evaluation of differences between various treatments (mutants) by taking into account the experimental variation.
RNA isolation and microarray preparation
For hcf-1(-) microarrays, total RNA was purified from synchronized L4 or young adult(YA) worms. Worms were synchronized by allowing hypochlorite-treated eggs to hatch in M9 buffer for 20 hrs at 16°C, and plating 500 L1 stage worms onto each of 5–6 10 mm NGM plates seeded with 3X OP50 bacteria. 6 biological replicates of hcf-1(-)/daf-16(-);hcf-1(-), two replicates of hcf-1(-)/N2 were prepared. The synchronized populations were grown to L4 or YA stage at 25°C and harvested by washing off the plates with M9 buffer and freezing the worm pellet in liquid nitrogen. Total RNA was isolated using Tri-reagent (Molecular Research Center, Inc.) [68] and purified with the RNeasy kit (Qiagen). cRNA synthesis/amplification, Cy3/Cy5 dye labeling, and hybridization onto Agilent 4X44K C. elegans oligonucleotide microarrays were performed as previously described [49]. Half the arrays were dye-flip replicates in each comparison.
Details on sir-2.1(O/E) microarrays will be published elsewhere (Rogers*, Jan*, Ashraf, and Murphy, in preparation). daf-2(-) microarray data were published in [49].
Microarray analysis
hcf-1(-) microarrays
Hybridized microarray slides were washed according to Agilent instructions , and images were scanned using an Axon Instruments GenePix 4000B scanner, reading at wavelengths of 635 nm and 532 nm (Axon Instruments, http://www.axon.com) [69]. The arrays were scanned at three different PMT settings to capture spots with low and high signal, and later combined to create a single dataset. The image data were uploaded onto the Princeton University MicroArray database (PUMA [http://puma.princeton.edu]). Log2 transformed fold change data were acquired after normalizing, filtering for array and spot quality, collapsing replicate spots to a mean value on PUMA.
Data for sir-2.1(O/E) and daf-2(-) arrays were similarly normalized and processed on PUMA.
SAM analysis
Log2 transformed fold change data with no cutoff were submitted to SAM [48]. One class analysis was used to identify genes significantly and consistently changed in each database. Two-class unpaired analysis was used to identify genes similarly and divergently changed between different datasets. Genes found to be significantly changed at 0% FDR in only hcf-1(-), sir-2.1(O/E), or daf-2(-) using one-class analysis, and similarly and divergently changed between different datasets using two-class unpaired analysis were combined and sorted based on the SAM output to generate heat maps using Treeview [70].
Gene Ontology classification
Worm Base IDs (WBID) of genes identified in SAM were pasted into the Functional annotation clustering tool in DAVID (http://david.abcc.ncifcrf.gov/) for gene annotation enrichment analysis [50], [71]. Functional annotation clustering was performed with the default criteria and enrichment score for each annotation cluster was determined.
Upstream regulatory motif analysis
1.5 kb upstream sequences were submitted to BioProspector (http://ai.stanford.edu/~xsliu/BioProspector/) [55] and RSAT (http://rsat.ulb.ac.be/rsat/) [56] to identify overrepresented cis-regulatory elements. An oligonucleotide length of 8 bp was specified for both algorithms. The highest scoring (most significantly enriched) 10 motifs from BioProspector and 5 motifs from RSAT were obtained. As BioProspector returns the same sequences multiple times, only unique motifs were reported. Motifs were displayed in WebLogo (weblogo.berkeley.edu) [72]. The matrices associated with each motif were submitted to the TomTom motif comparison tool (http://meme.sdsc.edu/meme/cgi-bin/tomtom.cgi) [73] to compare against a database of known transcription factor binding sites (Transfac).
Immunoprecipitation and mass spectrometry (C. elegans)
Immunoprecipitation was performed as described [21]. For HCF-1/SIR-2.1 co-IPs, mixed stage worms were grown on plates, harvested, and sonicated in IP lysis buffer (50 mM HEPES pH 7.5, 1 mM EDTA, 150 mM NaCl, 10% Glycerol, 0.1% Triton X-100, 1 mM sodium fluoride, 2.5 mM sodium orthovanadate, 1 mM PMSF, and Complete (EDTA-free) protease inhibitor cocktail) and lysates cleared by centrifugation. Lysates were incubated with either affinity purified guinea-pig anti-HCF-1 antibody [21] or rabbit anti-SIR-2.1 antibody (Novus Biologicals) at 4°C overnight. Immunocomplexes were incubated with trysacryl protein A-agarose beads (Pierce) at 4°C for four hours, washed four times with IP lysis buffer, and eluted by boiling in SDS sample buffer. Eluted protein complexes were analyzed by western blotting using the anti-HCF-1, anti-SIR-2.1, or anti-actin (Chemicon, clone C4) antibodies.
For mass spectrometry and 14-3-3 co-IPs, GFP-tagged HCF-1 was purified from mixed stage C. elegans, using a previously reported method [74] with slight modifications. In short, worms were grown in liquid culture as mixed stages to a density of 4000 worms/mL. Worms were washed into lysis buffer (50 mM HEPES at pH 7.4, 1 mM EGTA, 1 mM MgCl2, 150 mM KCl, 10% (v/v) glycerol, protease and phosphatase inhibitors), drop-frozen in liquid nitrogen, and ground using a mortar and pestle. Resulting powder was thawed and NP-40 was added to 0.05% (v/v). Immunoprecipitations were conducted on a 20,000 g supernatant of this extract, using monoclonal mouse-anti-GFP antibody (Invitrogen) coupled to Protein A resin (Biorad). Immunoprecipitated proteins were eluted using 100 mM glycine at pH 2.6. For co-IPs, eluted protein complexes were analyzed by western blotting using anti-mCherry (Ruvkun Lab, MGH Boston) or rabbit anti-PAR-5 (a kind gift from K.J. Kemphues, Cornell University) antibodies. For mass-spectrometrical analysis, immunoprecipitated proteins were eluted using 100 mM glycine at pH 2.6. Eluted proteins were visualized by silver-stained SDS-PAGE and identified by mass spectrometry. For the latter, samples were digested using trypsin and the resulting peptides were separated via nano-capillary liquid chromatography and identified by online tandem mass spectrometry (LTQ-XL, Thermo). Mass spectra were searched against the current wormpep database using Sequest (Thermo) and DTASelect [75].
As a negative-control for the mass-spectrometrical analysis, an identical purification was conducted using C. elegans expressing only untagged endogenous HCF-1. IP and negative-control were compared using Contrast [75].
Plasmids, shRNA, and siRNA for mammalian cells
Flag-FOXO3 and Flag-SIRT1 were obtained from Addgene and have been described previously [28]. The plasmids encoding HA-HCF-1 and HA-HCF-2 were generated by cloning the human HCF-1 and HCF-2 cDNA into the vector pCMV-HA (Clontech). The plasmid encoding the short-hairpin RNA targeting the human SIRT1 gene was generously provided by W.L. Kraus [8]. The plasmid encoding shRNA targeting the firefly luciferase gene was generously provided by L. Qi (Cornell University). siRNA duplexes directed against rat HCF-1 and HCF-2 were purchased from Dharmacon and targeted the following sequences: siHCF-1 #1 : 5′-GGAAGAGACTGAAGGCAAA-3′; siHCF-1 #2 : 5′-AGAACAACATTCCGAGGTA-3′; siHCF-2 : 5′ - GGGAATGGTTGAATATGGA-3′. Non-targeting control siRNA was also from Dharmacon. Cells were collected 48 hours post-transfection, or treated for an additional 6 hours with nicotinamide (10 mM, Sigma).
Cell culture and transfection
HEK293T were maintained in DMEM containing 4.5 g/L glucose and 10% calf serum and were transfected with the indicated plasmids using calcium phosphate. INS-1 cells were maintained in RPMI-1640 medium containing 11.1 mM glucose, 10% fetal bovine serum, 1 mM pyruvate, 10 mM HEPES, and 50 µM 2-mercaptoethanol. INS-1 cells were transfected with siRNA at a concentration of 10 nM using Lipofectamine RNAiMax (Invitrogen). siRNA transfections were performed twice, 24 hours apart, and cells were collected 24 hours after the second transfection.
Reverse-transcription coupled quantitative PCR (RT-qPCR)
RNA was isolated from mammalian cells using Trizol reagent and was reverse-transcribed using Superscript III First-Strand kit (Invitrogen). cDNAs were analyzed by quantitative-PCR using the SYBR Green system on a Roche LightCycler 480 real time PCR machine and quantified relative to a standard curve. β-actin was used as an internal control. The following primers were used: β-actin forward: 5′ - CTAAGGCCAACCGTGAAAAG-3′; : β-actin reverse: 5′-AACACAGCCTGGATGGCTAC-3′; HCF-1 forward: 5′-GCTGGAAAAGCTCCTGTCAC-3′; HCF-1 reverse: 5′ - CACTCATCTGTGGGTTGCTG-3′; HCF-2 forward: 5′ - TTGAAAGCAGAGCAATGGTG-3′; HCF-2 reverse: 5′ - AGTCGGGTACGTCTGCATTT-3′; Bim forward: 5′ - GCCCCTACCTCCCTACAGAC-3′; Bim reverse: 5′ - CAGGTTCCTCCTGAGACTGC-3′; p27 forward: 5′ - GTGGACCAAATGCCTGACTC-3′; p27 reverse: 5′ - TTCTGTTCTGTTGGCCCTTT-3′; Gadd45a forward: 5′ - GCAGAGCTGTTGCTACTGGA-3′; Gadd45a reverse: 5′ - TGTGATGAATGTGGGTTCGT-3′; IGFBP1 forward: 5′ - CTGCCAAACTGCAACAAGAA-3′; IGFBP1 reverse: 5′ - TTCCCACTCCATGGGTAGAC-3′.
Immunoblotting and immunoprecipitations (mammalian cell culture)
For co-immunoprecipitation experiments, HEK293T cells were transfected with the indicated plasmids. 48 hours after transfection, cells were lysed in lysis buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 2 mM EDTA, 1% TritonX-100, 10 mM NaF, 1 mM sodium orthovanadate, 1 mM PMSF, 10 mM nicotinamide, 1 mM trichostatin A, and Roche complete protease inhibitor cocktail). Cell extracts were incubated with either Flag - or HA-conjugated agarose beads (Sigma) overnight at 4°C. Beads were washed 5 times in lysis buffer and eluted by boiling in SDS sample buffer. Immunoprecipitates were analyzed by western blotting using the following antibodies: anti-HA (Covance), anti-FOXO3 (Upstate), anti-SIRT1 (gift from W.L. Kraus), anti-HCF-1 (Bethyl Labs).
Supporting Information
Zdroje
1. KenyonCJ 2010 The genetics of ageing. Nature 464 504 512
2. BurgeringBMKopsGJ 2002 Cell cycle and death control: long live Forkheads. Trends Biochem Sci 27 352 360
3. LinKDormanJBRodanAKenyonC 1997 daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 1319 1322
4. GiannakouMEGossMJungerMAHafenELeeversSJ 2004 Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305 361
5. HwangboDSGershmanBTuMPPalmerMTatarM 2004 Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429 562 566
6. KappelerLDe Magalhaes FilhoCDupontJLeneuvePCerveraP 2008 Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol 6 e254 doi:10.1371/journal.pbio.0060254
7. YuanRTsaihSWPetkovaSBMarin de EvsikovaCXingS 2009 Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell 8 277 287
8. LiYWangWJCaoHLuJWuC 2009 Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet 18 4897 4904
9. WillcoxBJDonlonTAHeQChenRGroveJS 2008 FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A 105 13987 13992
10. ArdenKC 2008 FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene 27 2345 2350
11. KenyonCChangJGenschERudnerATabtiangR 1993 A C. elegans mutant that lives twice as long as wild type. Nature 366 461 464
12. WangMCO'RourkeEJRuvkunG 2008 Fat metabolism links germline stem cells and longevity in C. elegans. Science 322 957 960
13. Halaschek-WienerJKhattraJSMcKaySPouzyrevAStottJM 2005 Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res 15 603 615
14. LeeSSKennedySTolonenACRuvkunG 2003 DAF-16 target genes that control C. elegans life-span and metabolism. Science 300 644 647
15. McElweeJBubbKThomasJH 2003 Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2 111 121
16. MurphyCTMcCarrollSABargmannCIFraserAKamathRS 2003 Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 277 283
17. LinKHsinHLibinaNKenyonC 2001 Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28 139 145
18. BerdichevskyAViswanathanMHorvitzHRGuarenteL 2006 C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell 125 1165 1177
19. BermanJRKenyonC 2006 Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 124 1055 1068
20. EssersMAde Vries-SmitsLMBarkerNPoldermanPEBurgeringBM 2005 Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science 308 1181 1184
21. LiJEbataADongYRizkiGIwataT 2008 Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol 6 e233 doi:10.1371/journal.pbio.0060233
22. WolffSMaHBurchDMacielGAHunterT 2006 SMK-1, an essential regulator of DAF-16-mediated longevity. Cell 124 1039 1053
23. LehtinenMKYuanZBoagPRYangYVillenJ 2006 A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125 987 1001
24. TissenbaumHAGuarenteL 2001 Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410 227 230
25. WangYOhSWDeplanckeBLuoJWalhoutAJ 2006 C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev 127 741 747
26. KaeberleinMMcVeyMGuarenteL 1999 The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13 2570 2580
27. RoginaBHelfandSL 2004 Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 101 15998 16003
28. BrunetASweeneyLBSturgillJFChuaKFGreerPL 2004 Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303 2011 2015
29. DaitokuHHattaMMatsuzakiHArataniSOhshimaT 2004 Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A 101 10042 10047
30. MottaMCDivechaNLemieuxMKamelCChenD 2004 Mammalian SIRT1 represses forkhead transcription factors. Cell 116 551 563
31. YangYHouHHallerEMNicosiaSVBaiW 2005 Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J 24 1021 1032
32. LeeSHerrW 2001 Stabilization but not the transcriptional activity of herpes simplex virus VP16-induced complexes is evolutionarily conserved among HCF family members. J Virol 75 12402 12411
33. GersterTRoederRG 1988 A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci U S A 85 6347 6351
34. GuntherMLaithierMBrisonO 2000 A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem 210 131 142
35. LuRMisraV 2000 Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res 28 2446 2454
36. LuRYangPO'HarePMisraV 1997 Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol 17 5117 5126
37. PilusoDBilanPCaponeJP 2002 Host cell factor-1 interacts with and antagonizes transactivation by the cell cycle regulatory factor Miz-1. J Biol Chem 277 46799 46808
38. TyagiSChabesALWysockaJHerrW 2007 E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell 27 107 119
39. WysockaJMyersMPLahertyCDEisenmanRNHerrW 2003 Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17 896 911
40. JulienEHerrW 2004 A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell 14 713 725
41. TyagiSHerrW 2009 E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J 28 3185 3195
42. WangYTissenbaumHA 2006 Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev 127 48 56
43. ViswanathanMKimSKBerdichevskyAGuarenteL 2005 A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell 9 605 615
44. HassanHMFridovichI 1979 Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys 196 385 395
45. MathewsWRGuidoDMFisherMAJaeschkeH 1994 Lipid peroxidation as molecular mechanism of liver cell injury during reperfusion after ischemia. Free Radic Biol Med 16 763 770
46. WangWShakesDC 1997 Expression patterns and transcript processing of ftt-1 and ftt-2, two C. elegans 14-3-3 homologues. J Mol Biol 268 619 630
47. LiJTewariMVidalMLeeSS 2007 The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev Biol 301 82 91
48. TusherVGTibshiraniRChuG 2001 Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98 5116 5121
49. ShawWMLuoSLandisJAshrafJMurphyCT 2007 The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr Biol 17 1635 1645
50. DennisGJrShermanBTHosackDAYangJGaoW 2003 DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4 P3
51. McElweeJJSchusterEBlancEPiperMDThomasJH 2007 Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol 8 R132
52. XuCLiCYKongAN 2005 Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res 28 249 268
53. FuruyamaTNakazawaTNakanoIMoriN 2000 Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349 629 634
54. CurranSPWuXRiedelCGRuvkunG 2009 A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature 459 1079 1084
55. LiuXBrutlagDLLiuJS 2001 BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127 138
56. Thomas-ChollierMSandOTuratsinzeJVJankyRDefranceM 2008 RSAT: regulatory sequence analysis tools. Nucleic Acids Res 36 W119 127
57. BudovskayaYVWuKSouthworthLKJiangMTedescoP 2008 An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134 291 303
58. JulienEHerrW 2003 Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J 22 2360 2369
59. WysockaJHerrW 2003 The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci 28 294 304
60. JohnsonKMMahajanSSWilsonAC 1999 Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J Virol 73 3930 3940
61. van der HorstABurgeringBM 2007 Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol 8 440 450
62. RimannIHajnalA 2007 Regulation of anchor cell invasion and uterine cell fates by the egl-43 Evi-1 proto-oncogene in Caenorhabditis elegans. Dev Biol 308 187 195
63. PuigserverPRheeJDonovanJWalkeyCJYoonJC 2003 Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423 550 555
64. RodgersJTLerinCHaasWGygiSPSpiegelmanBM 2005 Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434 113 118
65. NemotoSFergussonMMFinkelT 2005 SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J Biol Chem 280 16456 16460
66. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
67. BreslowNEClaytonDG 1993 Approximate Inference in Generalized Linear Mixed Models. Journal of the American Statistical Association 88 9 25
68. TroemelERChuSWReinkeVLeeSSAusubelFM 2006 p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2 e183 doi:10.1371/journal.pgen.0020183
69. PleissJAWhitworthGBBergkesselMGuthrieC 2007 Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol 5 e90 doi:10.1371/journal.pbio.0050090
70. EisenMBSpellmanPTBrownPOBotsteinD 1998 Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95 14863 14868
71. Huang daWShermanBTLempickiRA 2009 Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 44 57
72. CrooksGEHonGChandoniaJMBrennerSE 2004 WebLogo: a sequence logo generator. Genome Res 14 1188 1190
73. GuptaSStamatoyannopoulosJABaileyTLNobleWS 2007 Quantifying similarity between motifs. Genome Biol 8 R24
74. CheesemanIMNiessenSAndersonSHyndmanFYatesJR3rd 2004 A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18 2255 2268
75. TabbDLMcDonaldWHYatesJR3rd 2002 DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1 21 26
76. KristieTMSharpPA 1993 Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-trans-induction factor (VP16). J Biol Chem 268 6525 6534
Štítky
Genetika Reprodukčná medicína
Článek Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and BrainČlánek Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-StateČlánek A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated LociČlánek Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional PolymorphismsČlánek Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of ChromosomesČlánek The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and LearningČlánek PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian CellsČlánek Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Temporal Trends in Results Availability from Genome-Wide Association Studies
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
- Genetic Variants at Chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 Influence the Risk of Breast Cancer in Men
- Large-Scale Gene-Centric Analysis Identifies Novel Variants for Coronary Artery Disease
- Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function
- Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II
- Conserved Regulation of p53 Network Dosage by MicroRNA–125b Occurs through Evolving miRNA–Target Gene Pairs
- Heterozygous Mutations of Are Associated with an Increased Risk of Isolated Metopic Craniosynostosis in Humans and Mice
- Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at and a Potential Indicator of Esophageal Adenocarcinoma Development
- Cholesterol Metabolism Is Required for Intracellular Hedgehog Signal Transduction
- Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain
- Age-Dependent Recombination Rates in Human Pedigrees
- Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont
- Sex Chromosome Mosaicism and Hybrid Speciation among Tiger Swallowtail Butterflies
- A Negative Feedback Loop That Limits the Ectopic Activation of a Cell Type–Specific Sporulation Sigma Factor of
- Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence
- Mutations in or near the Transmembrane Domain Alter PMEL Amyloid Formation from Functional to Pathogenic
- Inactivation of Alters Melanosome Shape But Has Only a Subtle Effect on Visible Pigmentation
- Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency
- Germline Genetic Variants Disturbing the /LIN28 Double-Negative Feedback Loop Alter Breast Cancer Susceptibility
- Separation of Recombination and SOS Response in RecA Suggests LexA Interaction Sites
- Inference of Relationships in Population Data Using Identity-by-Descent and Identity-by-State
- Misregulation of Scm3p/HJURP Causes Chromosome Instability in and Human Cells
- A Noncoding Point Mutation of Causes Multiple Developmental Malformations and Obesity in Twirler Mice
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection
- Bacterial Communities of Diverse Species: Ecological Context of a Host–Microbe Model System
- A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci
- Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification
- Elevated Proteasome Capacity Extends Replicative Lifespan in
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- MicroRNA Predictors of Longevity in
- An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development
- Atypical AT Skew in Firmicute Genomes Results from Selection and Not from Mutation
- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
- The Abnormal Phenotypes of Cartilage and Bone in Calcium-Sensing Receptor Deficient Mice Are Dependent on the Actions of Calcium, Phosphorus, and PTH
- Cell Type–Specific Transcriptome Analysis Reveals a Major Role for and miR-200b in Mouse Inner Ear Morphogenesis
- Essential Roles of BCCIP in Mouse Embryonic Development and Structural Stability of Chromosomes
- IAP1-Mediated Ubiquitylation Controls Activation of the Initiator Caspase DRONC Independent of Protein Degradation
- VANG-1 and PRKL-1 Cooperate to Negatively Regulate Neurite Formation in
- The Receptor Tyrosine Kinase Alk Controls Neurofibromin Functions in Drosophila Growth and Learning
- Comparative and Functional Genomics of PD630 for Biofuels Development
- Identification of Type 1 Diabetes–Associated DNA Methylation Variable Positions That Precede Disease Diagnosis
- PCNA Ubiquitination Is Important, But Not Essential for Translesion DNA Synthesis in Mammalian Cells
- Genetic Effects at Pleiotropic Loci Are Context-Dependent with Consequences for the Maintenance of Genetic Variation in Populations
- Genome-Wide Association Study Identifies Four Loci Associated with Eruption of Permanent Teeth
- Bmp and Nodal Independently Regulate Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish
- Inter-Allelic Prion Propagation Reveals Conformational Relationships among a Multitude of [] Strains
- Emergence and Modular Evolution of a Novel Motility Machinery in Bacteria
- Histone Methyltransferase MET-2 Shields the Male X Chromosome from Checkpoint Machinery and Mediates Meiotic Sex Chromosome Inactivation
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Retrotransposon-Induced Heterochromatin Spreading in the Mouse Revealed by Insertional Polymorphisms
- The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO
- Genome-Wide Analysis of Heteroduplex DNA in Mismatch Repair–Deficient Yeast Cells Reveals Novel Properties of Meiotic Recombination Pathways
- Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



