-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
An increasing body of literature from genome-wide association studies and human whole-genome sequencing highlights the identification of large numbers of candidate regulatory variants of potential therapeutic interest in numerous diseases. Our relatively poor understanding of the functions of non-coding genomic sequence, and the slow and laborious process of experimental validation of the functional significance of human regulatory variants, limits our ability to fully benefit from this information in our efforts to comprehend human disease. Humanized mouse models (HuMMs), in which human genes are introduced into the mouse, suggest an approach to this problem. In the past, HuMMs have been used successfully to study human disease variants; e.g., the complex genetic condition arising from Down syndrome, common monogenic disorders such as Huntington disease and β-thalassemia, and cancer susceptibility genes such as BRCA1. In this commentary, we highlight a novel method for high-throughput single-copy site-specific generation of HuMMs entitled High-throughput Human Genes on the X Chromosome (HuGX). This method can be applied to most human genes for which a bacterial artificial chromosome (BAC) construct can be derived and a mouse-null allele exists. This strategy comprises (1) the use of recombineering technology to create a human variant–harbouring BAC, (2) knock-in of this BAC into the mouse genome using Hprt docking technology, and (3) allele comparison by interspecies complementation. We demonstrate the throughput of the HuGX method by generating a series of seven different alleles for the human NR2E1 gene at Hprt. In future challenges, we consider the current limitations of experimental approaches and call for a concerted effort by the genetics community, for both human and mouse, to solve the challenge of the functional analysis of human regulatory variation.
Published in the journal: Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing. PLoS Genet 8(3): e32767. doi:10.1371/journal.pgen.1002544
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1002544Summary
An increasing body of literature from genome-wide association studies and human whole-genome sequencing highlights the identification of large numbers of candidate regulatory variants of potential therapeutic interest in numerous diseases. Our relatively poor understanding of the functions of non-coding genomic sequence, and the slow and laborious process of experimental validation of the functional significance of human regulatory variants, limits our ability to fully benefit from this information in our efforts to comprehend human disease. Humanized mouse models (HuMMs), in which human genes are introduced into the mouse, suggest an approach to this problem. In the past, HuMMs have been used successfully to study human disease variants; e.g., the complex genetic condition arising from Down syndrome, common monogenic disorders such as Huntington disease and β-thalassemia, and cancer susceptibility genes such as BRCA1. In this commentary, we highlight a novel method for high-throughput single-copy site-specific generation of HuMMs entitled High-throughput Human Genes on the X Chromosome (HuGX). This method can be applied to most human genes for which a bacterial artificial chromosome (BAC) construct can be derived and a mouse-null allele exists. This strategy comprises (1) the use of recombineering technology to create a human variant–harbouring BAC, (2) knock-in of this BAC into the mouse genome using Hprt docking technology, and (3) allele comparison by interspecies complementation. We demonstrate the throughput of the HuGX method by generating a series of seven different alleles for the human NR2E1 gene at Hprt. In future challenges, we consider the current limitations of experimental approaches and call for a concerted effort by the genetics community, for both human and mouse, to solve the challenge of the functional analysis of human regulatory variation.
Introduction
A decade ago, the Human Genome Project published its first human DNA sequence draft, followed shortly by the full version in 2003 [1]–[3]. This project and the SNP Consortium and the International HapMap Project have provided geneticists with invaluable tools for their research on human populations [4], [5]. Their activities have resulted in an exponential growth of PubMed entries related to genome-wide association studies (GWASs) plus human whole-genome sequencing (HWGS) over the past decade (Figure 1, white bars). The increasing numbers of studies cumulated at 2,649 entries in 2010; these studies mainly focused on understanding the genetic variants affecting the development of diseases and disorders in humans. For obvious reasons, protein-coding variants have been the most extensively studied so far. However, an increasing body of literature from GWASs and candidate gene association studies also highlights the identification of candidate regulatory variants of potential therapeutic interest in numerous diseases [6]–[14]. Furthermore, with the cost of HWGS being driven down by cheaper sequencing technologies, we envision a continuing large increase in the identification of candidate regulatory variants. In general, the biological role of variants found in putative regulatory regions is harder to predict than that for protein-coding variants, in part because of our poor understanding of the functions of non-coding genomic sequence, and the slow and laborious process of experimental validation of the functional significance of human regulatory variants. In this commentary, we will review current efforts at modelling human variation in mouse and highlight a novel method for high-throughput generation of humanized mouse models (HuMMs) entitled High-throughput Human Genes on the X Chromosome (HuGX, pronounced “hugs”).
Fig. 1. The literature is increasing more slowly for humanized mouse models than for GWASs and HWGS or novel mouse models. 
Interrogation of the PubMed literature database (http://www.ncbi.nlm.nih.gov/pubmed) reveals a faster growing body of literature related to GWASs and HWGS (white bars) or novel mouse models (grey bars) than to HuMMs (black bars). Interrogation of the database was done using the online search option from EndNote (http://www.endnote.com/). Individual numbers of entries for the search terms “genome wide association studies” and “human whole genome sequencing” were added together for the figure. Search terms for novel mouse models were “novel knockout mouse”, “novel knockin mouse”, and “novel knock-in mouse”. The entries for the search term “humanized mouse models” were not restricted to genetic mouse models but included xenograft mouse models as well. Search terms were interrogated in “all fields” per year. Typical Humanized Mouse Models Are Powerful but Not Ideal for Regulatory Variants
It is always important to remember that mice are not “little humans”, and that species-specific differences limit the value of any model organism. Nevertheless, throughout history, the laboratory mouse has been the human disease model of choice for geneticists, in part because of the rapid breeding rate of mice, which led to the generation of a wide variety of inbred and spontaneous-mutation-harbouring strains. Contributing to the mouse as a model was the advancement in embryonic technologies, allowing the engineering of the mouse genome and resulting in the generation of transgenic random-insertion, knock-out, and knock-in mouse models. Furthermore, the laboratory mouse genome sequence was released in 2002 and demonstrated that 99% of mouse genes have human homologues, strengthening the importance of mouse models in probing human biology and disease [15]–[17]. This importance has been reflected by a continually growing literature describing novel mouse models over the past decade (Figure 1, grey bars). However, in contrast to coding regions, human–mouse comparative genomic analysis demonstrated a lower level of conservation in putative regulatory regions of the genome [15]. This finding strengthened a hypothesis posed more than 25 years ago suggesting that regulatory regions may play a crucial role in underlying species differences and human-specific biology and disease [18]. It also raises a problem for mouse modelling when a strictly mouse-genome-based approach is used to validate human candidate regulatory variants, since the equivalent DNA sequence and/or epigenomic environment may not be present.
HuMMs, in which human genes are introduced into the mouse, suggest an approach to this problem. Surprisingly, the number of entries in the literature for HuMMs is very modest when compared to the two previous categories (Figure 1, black bars). Many of the HuMM entries are not genetic per se but are related to immunity studies—using human cells or tissues engrafted in nude mice—and thus are unrelated to the data generated by GWASs and HWGS. Nevertheless, there are numerous examples of successful genetic HuMMs.
A HuMM approach was used to study the complex genetic condition arising from Down syndrome, also known as trisomy 21. This syndrome results from an altered dosage of wild-type (WT) genes on human Chromosome 21, a phenomenon that can be mimicked by generating trans-species aneuploid mice carrying a human chromosome [19]. In this example, the mouse strain generated contained an estimated 92% of all known human Chromosome 21 genes, and a large-scale analysis demonstrated that 81% of human Chromosome 21 genes were expressed in mouse tissues [19], [20]. Additional investigation, using a set of conserved and well-characterized transcription factors responsible for hepatocyte development and function, revealed that genetic sequence rather than interspecies differences in epigenetic machinery or cellular environment is largely responsible for directing transcriptional programs [21]. These results demonstrated that human gene regulation is generally conserved in mice, strengthening the argument that HuMMs can be a good approach for understanding the role of candidate regulatory variants in disease development.
Other examples of successful HuMMs to study the role of genetic mutations are found in common monogenic disorders such as Huntington disease and β-thalassemia, as well as cancer susceptibility genes such as BRCA1 [22]–[25]. All of these WT human genes in HuMMs successfully rescued the embryonic lethal phenotype from the mouse gene knock-out animals, thereby providing valuable information regarding the human gene function by demonstrating an interspecies complementation of the human gene in the mouse null background. This complementation was due not only to the similarity of the genes in terms of protein function, but also to the identical tissue expression distribution of the human gene [22]–[25]. This was surprising considering the low percentage of identity between human and mouse for some of these genes in both the regulatory and coding sequences [25].
These results were invaluable, as they demonstrated that HuMMs can be used to study the biological role of mutant forms of these human genes. In the case of Huntington disease, this line of investigation has led to the generation of several human yeast artificial chromosome (YAC)–harbouring strains to study the biological implication of expanded glutamine repeats in Huntington disease development [26]–[28]. Advancements in site-specific bacterial artificial chromosome (BAC) mutagenesis techniques supported the shift to generation of BAC-based mutation-harbouring mouse models [29]–[32]. These included the generation of HuMMs harbouring codon-specific mutations for β-thalassemia and the BRCA1 cancer susceptibility gene. These HuMMs provided information regarding the biological implication of such mutations and their potential underlying role in human health [33], [34]. However, the approaches used to generate these HuMMs, which were suitable when protein-coding variants were being tested, encountered serious limitations in probing the role of human candidate regulatory variants.
In general, HuMM generation has used microinjection of DNA into zygotic pronuclei [35]–[37]. This technique is widely used in the field of mammalian genetics, but is not without limitations. For one, it requires extensive characterization of the different founder lines to control for variability in gene expression, a phenomenon due in part to the influence of the genomic environment at the site of insertion (i.e., position effect) and the number of copies often found tandemly inserted (i.e., copy effect) [38]–[41]. The transgene can potentially lead to disruption of endogenous gene function and repeat-induced gene silencing, two factors that must be taken into account when generating mice by random-insertion pronuclear injection [42]. Since each strain is unique, reproducibility between the different mouse strains becomes a major limiting factor when using random insertion as a mean to generate HuMMs. This lack of reproducibility is less than ideal for any comparison between transgenes in different mouse strains, but is particularly concerning when probing for candidate regulatory variant differences. The ideal method would control for both the site of insertion and the copy numbers inserted in the genome.
Excellent Techniques Exist for Single-Copy Non-Random Docking in the Mouse Genome
One type of approach, which allows single-copy insertion in the genome, includes the use of retroviruses and transposon activity [43]–[45]. Although quite successful, this approach has limitations as it does not provide controls for the site of insertion in the genome, leading to variability in expression due to genomic environment, as well as potential disruption of endogenous genes. Another potentially powerful approach, called recombinase-mediated genomic replacement, allows the cre-based insertion of a human gene at the site of, and replacing, the endogenous mouse gene [46]. This approach provides stringent control over the genomic environment surrounding the insertion site. However, the method simultaneously creates two inseparable genetic events in the same gene: (1) heterozygosity at the mouse locus and (2) addition of the human gene. Thus, the human gene can be studied only on the heterozygous mutant mouse background. Other limitations include the fact that the replacement is a low-frequency event, and the “gene by gene” approach will restrict throughput. Another novel approach was described recently using pronuclear injection coupled to integrase activity to achieve single-copy site-specific insertion in the mouse genome [47]. This approach used ϕC31-integrase-mediated recombination activity between attB sites from recombinant DNA with attP sites previously inserted at a specific locus in the mouse genome. Although also quite promising, this approach yielded up to 40% site-specific integration at best, and was only tested on small construct plasmids, another limitation, since many genes require large constructs [47].
Traditionally, two mouse genes have been used as genomic docking sites: the autosomal Rosa26 (reverse orientation splice acceptor 26) and X chromosome Hprt (hypoxanthine guanine phosphoribosyl transferase) [48], [49]. The Rosa26 locus has most often been used to dock constructs when strong ubiquitous expression is required [50]–[54]. Plasmid-size docking is readily achieved; however, large BAC insertions have not been reported. Also, insertion at the Rosa26 locus typically results in disruption of the gene, which in turn may lead to mild phenotypic consequences [55]. Use of the Hprt docking site has also been widely reported in the literature, and despite the wide expression of Hprt itself, this locus is more often chosen for tissue - or cell-type-specific expression of the targeted construct [56]–[58]. This locus readily accepts plasmid-size constructs but also large (>200 kb) BAC constructs [48], [56]. In the past, docking has been done in such a way that it disrupts the Hprt gene, resulting in mice with a mild phenotype [59]–[61]. However, this disruption is now typically avoided by a strategy that uses embryonic stem cells (ESCs) that already carry a spontaneous deletion of the 5′ end of the Hprt gene [62]. In this strategy, docking involves construct insertion 5′ of Hprt and repairing the expression of the Hprt gene itself [56], [57], [63]. This repair of Hprt enables direct selection of high-frequency correctly targeted ESC clones [63].
GWASs and HWGS Require High-Throughput Humanized Mouse Models
Huge strides have been made bringing high throughput to mouse functional genomics. One such stride is simple and highly efficient BAC recombineering in Escherichia coli [31], [64], [65]. This technology provides researchers with limitless possibilities for DNA modification via homologous recombination in E. coli. It employs the BAC-adapted strain harbouring a defective lambda prophage that allows the recombination genes exo, bet, and gam to be expressed under the control of a temperature-sensitive λ cI-repressor [31], [64]–[66]. DNA modification possibilities include insertion of exogenous DNA fragments in the endogenous BAC DNA, size-specific DNA deletion, single-base-pair-specific DNA alteration, and BAC fusion (i.e., recombining overlapping BAC constructs into a single, larger BAC) [30], [31], [66], [67]. Hence, generation of any variant-harbouring allele in a high-throughput manner can be easily achieved using this technology. Such approaches are already being adopted by large-scale mouse knock-out programmes such as the International Knockout Mouse Consortium [68].
Another stride is the generation of important resources by the currently ongoing large-scale mouse projects [58], [68], [69]. For example, the International Knockout Mouse Consortium is generating ESC-targeted mutations in all protein-coding genes [68]. This resource will have many impacts, but specific to this discussion, it enables complementation approaches to be undertaken for most human genes. To date, this group has generated 16,878 targeted alleles in germline-competent C57BL/6N ESCs (http://www.knockoutmouse.org) [68], [70]–[72].
Finally, C57BL/6 is the most widely used inbred mouse strain and one of the best characterized [68]. The increasing use of ESCs derived from this strain, especially by large-scale projects, will greatly reduce the need for backcrossing by projects using this mouse strain, thus increasing the throughput of most projects [68].
Even with these game-changing strides, HuMM generation will never achieve the throughput of array and sequence technologies. Thus, variants identified by GWASs and HWGS will always need to be extensively pre-screened as strong candidate regulatory variants and suitable for cross-species analysis before entering a HuMM project pipeline.
HuGX for High-Throughput Assaying of Human Candidate Regulatory Variants
Here we present an approach, HuGX, aimed at understanding the role of candidate human regulatory variants in the development of human diseases and disorders. The strategy comprises (1) the use of the BAC-adapted recombineering technology to create a human-variant-harbouring BAC, (2) knock-in of this BAC into the mouse genome using Hprt docking technology, and (3) allele comparison by interspecies complementation. This approach can be applied to human genes for which an expressing BAC construct can be derived, which can complement at least a component of a mouse phenotype.
The first step is to find a suitable BAC for “your favourite gene” (YFG). This BAC should be computationally analyzed to determine the likelihood that it includes the entire coding sequence as well as 5′ and 3′ regulatory sequences. The GENSAT project, having generated >1,000 mouse BAC random-insertion transgenics, reports ∼85% endogenous-like expression for genes ≤100 kb [69], [73]. Since ∼86% of human genes are ≤100 kb (Ensemble assembly, February 2009, GRCh37/hg19), we estimate there will be suitable BACs for ∼75% of them. In addition, recombineering approaches are available to fuse two BACs, isolate only the 5′ region, or delete unwanted sequences (e.g., neighbouring genes) as needed [67]. Alternatively, the recombineering technology can be applied to a human P1 artificial chromosome should YFG be small [30], [66]. Our approach highlights the use of the RPCI-11 Human Male BAC Library (http://bacpac.chori.org/hmale11.htm), which was built in the pBACe3.6 vector (Figure 2A). The backbone of this BAC vector contains a SacB gene that can be used as a targeting site for the first retrofitting step, adding the HPRT homology regions from plasmids pJDH8A/246b or the pEMS1306 series [56], [58], [74]. This BAC construct can then be used as the substrate for subsequent retrofitting steps, to add “your favourite regulatory variant” (YFRV), a DNA insertion, deletion, or single-base-pair alteration as needed. These retrofitting steps can be carried out in a high-throughput manner to rapidly develop an allele series of different variants to be tested. Since, both the WT-YFG and YFRV-YFG BAC constructs contain the homology regions that allow proper targeting at the Hprt locus, each can be electroporated into ESCs and selected in hypoxanthine-aminopterin-thymidine (HAT) medium, and homologous recombinant clones can be identified and microinjected into mouse embryos (Figure 2A and 2B) [63]. Male chimeras are bred to generate germline females that carry a site-specific single-copy WT-YFG BAC, or YFRV-YFG BAC, on their X chromosome. Assuming the genetic background is suitable, genetic complementation can be tested immediately by performing two generations of mating (Figure 2C). The resulting animals will carry a single copy of the human WT-YFG, or YFRV-YFG BAC, on the Yfg mouse null background (Yfg−/−, HprtWT-YFG/Y or Yfg−/−, HprtYFRV-YFG/Y). Animals studied on the null background will be males, thus avoiding X inactivation [56], [75]. Using this HuGX strategy, the phenotype of the Yfg−/−, HprtYFRV-YFG/Y animals can be directly compared to that of the Yfg−/−, HprtWT-YFG/Y animals. Regardless of whether interspecies complementation is complete or partial, any differences can be attributed to the function of the human variant.
Fig. 2. Strategy for high-throughput human genes on the X chromosome (HuGX). 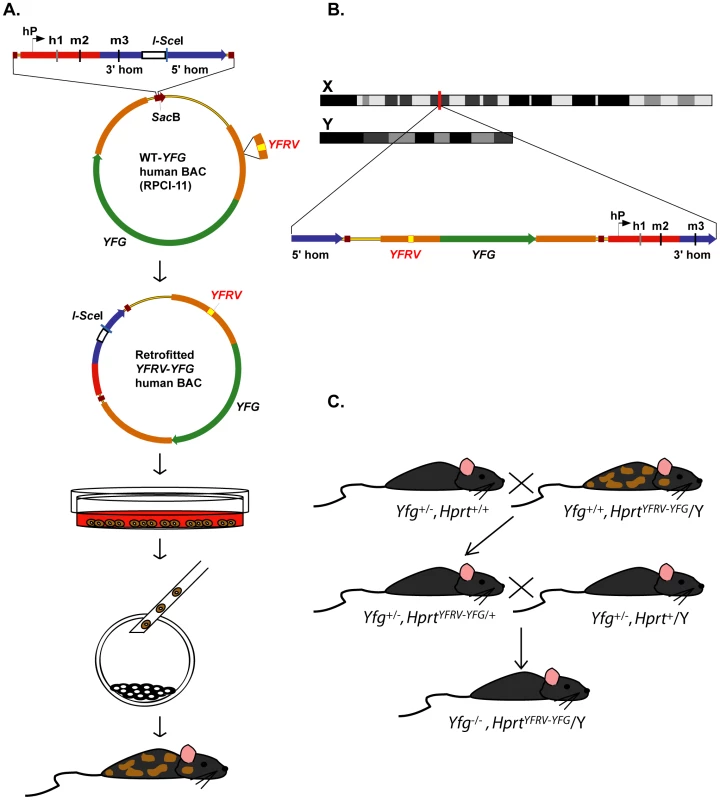
(A) Flow diagram representing the major steps of the HuGX strategy, which builds on previous methods [56], [74]. Starting with a human BAC carrying your favourite gene (YFG) from the RPCI-11 library, for example, two retrofitting steps are employed: (1) addition of the HPRT homology regions for recombination (WT-YFG) and (2) introduction of your favourite regulatory variant (YFRV) into YFG (YFRV-YFG). In this example the resulting BAC YFRV-YFG is linearized, typically using I-SceI, and electroporated into ESCs. 129P2/OlaHsd, B6129F1 hybrid, and C57BL/6NTac ESCs are all available carrying the 36-kb (Hprtb-m3) deletion used for docking. Selection of homologous recombinant clones is performed using hypoxanthine-aminopterin-thymidine medium, and clones carrying correctly targeted complete-BAC inserts are injected into blastocysts to generate chimeras. Schematic, not to scale. (B) Details of knock-in 5′ of the Hprt locus on X chromosome. The linearized BAC construct is introduced into the Hprtb-m3 deletion by electroporation. Hprt gene expression is restored by the presence of the human HPRT promoter (hP), first exon (h1), and second mouse exon (m2). Mouse homology arms (blue); Hprt coding regions (red); vector backbone (narrow yellow line); SacB gene from BAC vector backbone (brown); 5′ and 3′ untranslated regions of YFG (orange); YFRV (yellow); coding region of YFG (green); hP (black arrow); h1 (grey); m2 and m3 (black). Schematic, not to scale. (C) Breeding strategy to achieve complementation. Assuming the genetic background is suitable, male chimeras can immediately be bred to females heterozygous for a null allele at the mouse copy of Yfg (Yfg+/−) to generate germline females heterozygous for Yfg (Yfg+/−) and HprtYFRV-YFG/+. On the other hand, assuming the challenging situation in which no heterozygous phenotype exists to complement, these females will be mated with a Yfg+/−male, resulting in males for study carrying a single copy of the human retrofitted HprtYFRV-YFG and the mouse null (Yfg−/−) gene. We have recently used this approach to generate a directly comparable allele series for our favourite gene, nuclear receptor 2E1 (NR2E1). This gene encodes an orphan nuclear receptor (also known as TLX) that is highly conserved between human and mouse, and has an important role in the maintenance of the neural/progenitor stem cell populations of both the forebrain and retina [76]–[80]. Nr2e1-null mice have brain and eye abnormalities such as hyperactivity, extreme aggressive behaviours, and blindness [81]–[83]. These phenotypes can be rescued by human NR2E1 under its endogenous promoter, thereby demonstrating the functional equivalence of the human and mouse genes in mouse [81], [84]. Recently, positive association results between NR2E1 and bipolar disorder have been reported, along with an increase in detection of rare variants in patients [85]. The objective of our approach was to generate seven human alleles knocked in to mouse, including one harbouring a human WT BAC, an ∼2-kb regulatory deletion, four single-base-pair candidate regulatory variants, and one two-base-pair candidate regulatory variant (Figure 3A). Recombineering and targeting of these constructs at the Hprt locus was performed. As shown by others and our own data (Figure 3A), it is important to molecularly characterize the integrity of the BAC insertions [56]. Nevertheless, the low number of clones that needed to be picked per construct, and the high percentage of correctly targeted clones, which varied from 13% to 73% with an average of 48%, highlights the ease of this strategy and its applicability to high throughput.
Fig. 3. High-throughput generation of regulatory allele series. 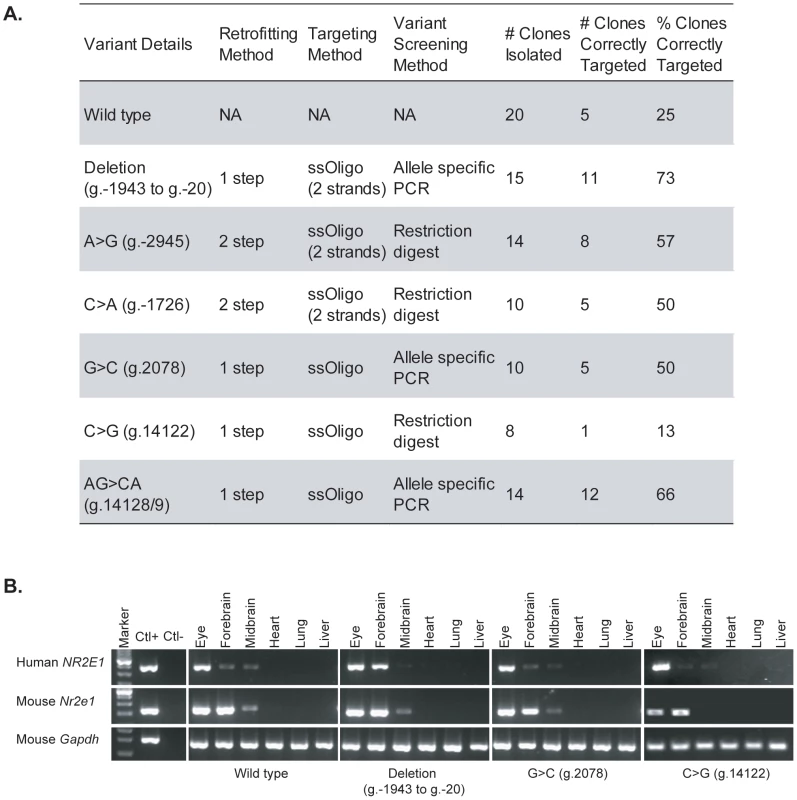
(A) Human BAC RP11-144P8 was retrofitted seven times to generate the different regulatory variants (column 1). The method of retrofitting (column 2), targeting (column 3), and variant screening (column 4) is presented for each variant. Also given are the number of ESC clones isolated after electroporation (column 5), the number of correctly targeted clones after PCR validation using assays an average of 6 kb, and a maximum of 11 kb, apart (column 6), and the resulting percentage of correctly targeted clones (column 7). (B) Species-specific reverse transcriptase PCR demonstrates transcription from the human BAC in germline animals from four of the strains generated by the high-throughput approach. One-step reverse transcription PCR reactions were performed using oligonucleotides specific for human NR2E1, mouse Nr2e1, and mouse Gapdh. The results show, as expected, expression of the human NR2E1 gene in adult eye, forebrain, and midbrain, but not in adult lung, heart, and liver. Marker, 100-bp ladder; positive control (Ctl+), human RNA for human NR2E1 assay and mouse RNA for mouse Nr2e1 and Gapdh assays; negative control (Ctl−), human RNA for mouse Nr2e1 and Gapdh assays and mouse RNA for human NR2E1 assay. Mouse strains were generated from these different constructs, and species-specific reverse transcription PCR (RT-PCR) assays on different tissue samples were performed for four of these mouse strains (WT, Deletion [g.−1943 to g.−20], G>C [g.2078], and C>G [g.14122]) (Figure 3B). These assays demonstrated expression of the human NR2E1 BACs in the eyes, forebrain, and midbrain of adult mice, and the absence of expression in the adult heart, lung, and liver (Figure 3B). These results, when compared to the mouse endogenous Nr2e1 expression pattern, suggest endogenous-like tissue-specific expression of the human NR2E1 BACs in the mouse strains. Backcrossing to the appropriate background and subsequent crossing to the Nr2e1-null background will allow us to evaluate the importance of these variants in the development of diseases and disorders.
Overall, generation of these seven strains has demonstrated that six to nine months is necessary to generate a single HuGX mouse model. Since the components of the HuGX methodology are scalable, and applicable to a large-scale parallel approach, this strategy is suitable for high-throughput mouse model generation to study the relevance of candidate mutations.
Challenges for the Future
The exponential growth of data in the literature coming from GWASs and HWGS requires novel high-throughput approaches to test the biological importance of the large numbers of variants being identified, particularly candidate regulatory variants. In considering experimental approaches, three challenges face our field. The first is a consideration of the balance between construct flexibility and size. Small plasmid-based constructs lead in flexibility, especially with the option of DNA synthesis, allowing the efficient generation of any desired sequence [86]. But plasmids will almost certainly fail to capture the genomic context of the regulatory variant, especially factors such as the chromatin structure. BACs are often gene-sized (holding ∼200 kb) and are relatively easily manipulated by recombineering, and so are the construct of choice for many large projects, e.g., the GENSAT project and the International Knockout Mouse Consortium [30], [68], [69]. However, some human genes can span more than one megabase (e.g., dystrophin) [87]–[89]. YACs can accommodate this size of genomic DNA, and site-specific mutagenesis can be readily performed using the homologous recombination system of the yeast [90]. However, site-specific docking of YAC constructs is beyond our current abilities and makes YACs presently unsuitable for high-throughput single-copy approaches. The second challenge is a consideration of docking sites and technology. The Hprt locus provides a reliable and highly efficient docking site for BAC insertion into the mouse genome. The position of this locus on the X chromosome can be an advantage, i.e., all female offspring of a carrier male are carriers, but also a disadvantage, i.e., X inactivation in females results in mosaic expression in heterozygotes; thus, in neither sex can you obtain two functional copies of the human gene. Although the Hprt locus has been used to dock the largest fragments yet into the genome, up to 200 kb, size is still a limiting factor for certain human genes [56]. Hence, the generation of an alternative autosomal docking site that does not disrupt a gene, and allows insertion of large DNA fragments, would be ideal in the near future. The third challenge is a consideration of the value of stem cells and in vitro differentiation to assay candidate regulatory variant function. Mouse ESCs, as already described, can be derived to carry a matched pair of human alleles that differ only by the variant. Assuming an appropriate differentiation protocol [91], [92], differences in expression in almost any cell type could be detected, and this would add to the overall understanding of the variant. However, generation of such in vitro data alone would presumably be less successful in leading to an understanding of human disease, than when accompanied by information on the in vivo phenotype of mice derived from these same cells. A species-relevant, powerful in vitro assay can be envisaged for the near future when it would be possible to derive a matched pair of human-induced pluripotent stem cells, differing only by the variant. However, it would still remain necessary to undertake an in vivo analysis using HuMM or HuGX mice to comprehensively study the variant.
We conclude by calling for a concerted effort by the genetics community, those studying human and mouse, to move forward to solve the challenge of functional analysis of human regulatory variation in human disease and disorders.
Ethics Statement
All mice were maintained in the pathogen-free Centre for Molecular Medicine and Therapeutics animal facility on a 6 am–6 pm light cycle, 20±2°C, with 50%±5% relative humidity, and had food and water ad libitum. All procedures involving animals were in accordance with the Canadian Council on Animal Care and University of British Columbia Animal Care Committee (Protocol# A07-0435).
Zdroje
1. LanderESLintonLMBirrenBNusbaumCZodyMC 2001 Initial sequencing and analysis of the human genome. Nature 409 860 921
2. VenterJCAdamsMDMyersEWLiPWMuralRJ 2001 The sequence of the human genome. Science 291 1304 1351
3. CollinsFSGreenEDGuttmacherAEGuyerMS 2003 A vision for the future of genomics research. Nature 422 835 847
4. SachidanandamRWeissmanDSchmidtSCKakolJMSteinLD 2001 A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409 928 933
5. FrazerKABallingerDGCoxDRHindsDAStuveLL 2007 A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851 861
6. OnoSEzuraYEmiMFujitaYTakadaD 2003 A promoter SNP (−1323T>C) in G-substrate gene (GSBS) correlates with hypercholesterolemia. J Hum Genet 48 447 450
7. SugataniJYamakawaKYoshinariKMachidaTTakagiH 2002 Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Biophys Res Commun 292 492 497
8. BosmaPJChowdhuryJRBakkerCGantlaSde BoerA 1995 The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. New Engl J Med 333 1171 1175
9. NakamuraSKugiyamaKSugiyamaSMiyamotoSKoideS 2002 Polymorphism in the 5′-flanking region of human glutamate-cysteine ligase modifier subunit gene is associated with myocardial infarction. Circulation 105 2968 2973
10. MarzecJMChristieJDReddySPJedlickaAEVuongH 2007 Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 21 2237 2246
11. JinnaiNSakagamiTSekigawaTKakiharaMNakajimaT 2004 Polymorphisms in the prostaglandin E2 receptor subtype 2 gene confer susceptibility to aspirin-intolerant asthma: a candidate gene approach. Hum Mol Genet 13 3203 3217
12. SpeliotesEKWillerCJBerndtSIMondaKLThorleifssonG 2010 Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42 937 948
13. DuboisPCTrynkaGFrankeLHuntKARomanosJ 2010 Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 42 295 302
14. AnttilaVStefanssonHKallelaMTodtUTerwindtGM 2010 Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet 42 869 873
15. WaterstonRHLindblad-TohKBirneyERogersJAbrilJF 2002 Initial sequencing and comparative analysis of the mouse genome. Nature 420 520 562
16. RossantJMcKerlieC 2001 Mouse-based phenogenomics for modelling human disease. Trends Mol Med 7 502 507
17. PaigenK 1995 A miracle enough: the power of mice. Nat Med 1 215 220
18. KingMCWilsonAC 1975 Evolution at two levels in humans and chimpanzees. Science 188 107 116
19. O'DohertyARufSMulliganCHildrethVErringtonML 2005 An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309 2033 2037
20. ReynoldsLEWatsonARBakerMJonesTAD'AmicoG 2010 Tumour angiogenesis is reduced in the Tc1 mouse model of Down's syndrome. Nature 465 813 817
21. WilsonMDBarbosa-MoraisNLSchmidtDConboyCMVanesL 2008 Species-specific transcription in mice carrying human chromosome 21. Science 322 434 438
22. HodgsonJGSmithDJMcCutcheonKKoideHBNishiyamaK 1996 Human huntingtin derived from YAC transgenes compensates for loss of murine huntingtin by rescue of the embryonic lethal phenotype. Hum Mol Genet 5 1875 1885
23. VadolasJWardanHBosmansMZaibakFJamsaiD 2005 Transgene copy number-dependent rescue of murine beta-globin knockout mice carrying a 183 kb human beta-globin BAC genomic fragment. Biochim Biophys Acta 1728 150 162
24. LaneTFLinCBrownMASolomonELederP 2000 Gene replacement with the human BRCA1 locus: tissue specific expression and rescue of embryonic lethality in mice. Oncogene 19 4085 4090
25. ChandlerJHohensteinPSwingDATessarolloLSharanSK 2001 Human BRCA1 gene rescues the embryonic lethality of Brca1 mutant mice. Genesis 29 72 77
26. HodgsonJGAgopyanNGutekunstCALeavittBRLePianeF 1999 A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 23 181 192
27. SlowEJvan RaamsdonkJRogersDColemanSHGrahamRK 2003 Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet 12 1555 1567
28. KuhnAGoldsteinDRHodgesAStrandADSengstagT 2007 Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet 16 1845 1861
29. YuYBradleyA 2001 Engineering chromosomal rearrangements in mice. Nat Rev Genet 2 780 790
30. CopelandNGJenkinsNACourtDL 2001 Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2 769 779
31. SwaminathanSEllisHMWatersLSYuDLeeEC 2001 Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis 29 14 21
32. NarayananKWilliamsonRZhangYStewartAFIoannouPA 1999 Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther 6 442 447
33. YangYSwaminathanSMartinBKSharanSK 2003 Aberrant splicing induced by missense mutations in BRCA1: clues from a humanized mouse model. Hum Mol Genet 12 2121 2131
34. JamsaiDZaibakFKhongniumWVadolasJVoullaireL 2005 A humanized mouse model for a common beta0-thalassemia mutation. Genomics 85 453 461
35. GordonJWScangosGAPlotkinDJBarbosaJARuddleFH 1980 Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci U S A 77 7380 7384
36. GordonJWRuddleFH 1981 Integration and stable germ line transmission of genes injected into mouse pronuclei. Science 214 1244 1246
37. BrinsterRLChenHYTrumbauerMSenearAWWarrenR 1981 Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell 27 223 231
38. MilotEStrouboulisJTrimbornTWijgerdeMde BoerE 1996 Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87 105 114
39. PedramMSprungCNGaoQLoAWReynoldsGE 2006 Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol 26 1865 1878
40. GaoQReynoldsGEInnesLPedramMJonesE 2007 Telomeric transgenes are silenced in adult mouse tissues and embryo fibroblasts but are expressed in embryonic stem cells. Stem Cells 25 3085 3092
41. WilliamsAHarkerNKtistakiEVeiga-FernandesHRoderickK 2008 Position effect variegation and imprinting of transgenes in lymphocytes. Nucleic Acids Res 36 2320 2329
42. GarrickDFieringSMartinDIWhitelawE 1998 Repeat-induced gene silencing in mammals. Nat Genet 18 56 59
43. LoisCHongEJPeaseSBrownEJBaltimoreD 2002 Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295 868 872
44. DingSWuXLiGHanMZhuangY 2005 Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122 473 483
45. MatesLChuahMKBelayEJerchowBManojN 2009 Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41 753 761
46. WallaceHAMarques-KrancFRichardsonMLuna-CrespoFSharpeJA 2007 Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128 197 209
47. TasicBHippenmeyerSWangCGamboaMZongH 2011 From the cover: site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc Natl Acad Sci U S A 108 7902 7907
48. DoetschmanTGreggRGMaedaNHooperMLMeltonDW 1987 Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330 576 578
49. FriedrichGSorianoP 1991 Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev 5 1513 1523
50. ZambrowiczBPImamotoAFieringSHerzenbergLAKerrWG 1997 Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A 94 3789 3794
51. MaoXFujiwaraYOrkinSH 1999 Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A 96 5037 5042
52. AbeTKiyonariHShioiGInoueKNakaoK 2011 Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis 49 579 590
53. SorianoP 1999 Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21 70 71
54. MadisenLZwingmanTASunkinSMOhSWZariwalaHA 2010 A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13 133 140
55. KohlheppRLHeggeLFMoserAR 2001 The ROSA26 LacZ-neo(R) insertion confers resistance to mammary tumors in Apc(Min/+) mice. Mamm Genome 12 606 611
56. HeaneyJDRettewANBronsonSK 2004 Tissue-specific expression of a BAC transgene targeted to the Hprt locus in mouse embryonic stem cells. Genomics 83 1072 1082
57. YurchenkoEFriedmanHHayVPetersonAPiccirilloCA 2007 Ubiquitous expression of mRFP-1 in vivo by site-directed transgenesis. Transgenic Res 16 29 40
58. Portales-CasamarESwansonDJLiuLde LeeuwCNBanksKG 2010 A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc Natl Acad Sci U S A 107 16589 16594
59. JinnahHAGageFHFriedmannT 1991 Amphetamine-induced behavioral phenotype in a hypoxanthine-guanine phosphoribosyltransferase-deficient mouse model of Lesch-Nyhan syndrome. Behav Neurosci 105 1004 1012
60. FingerSHeavensRPSirinathsinghjiDJKuehnMRDunnettSB 1988 Behavioral and neurochemical evaluation of a transgenic mouse model of Lesch-Nyhan syndrome. J Neurol Sci 86 203 213
61. DunnettSBSirinathsinghjiDJHeavensRRogersDCKuehnMR 1989 Monoamine deficiency in a transgenic (Hprt-) mouse model of Lesch-Nyhan syndrome. Brain Res 501 401 406
62. HooperMHardyKHandysideAHunterSMonkM 1987 HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326 292 295
63. BronsonSKPlaehnEGKluckmanKDHagamanJRMaedaN 1996 Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci U S A 93 9067 9072
64. YuDEllisHMLeeECJenkinsNACopelandNG 2000 An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97 5978 5983
65. WarmingSCostantinoNCourtDLJenkinsNACopelandNG 2005 Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33 e36
66. SharanSKThomasonLCKuznetsovSGCourtDL 2009 Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4 206 223
67. SopherBLLa SpadaAR 2006 Efficient recombination-based methods for bacterial artificial chromosome fusion and mutagenesis. Gene 371 136 143
68. SkarnesWCRosenBWestAPKoutsourakisMBushellW 2011 A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474 337 342
69. GongSZhengCDoughtyMLLososKDidkovskyN 2003 A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425 917 925
70. GertsensteinMNutterLMReidTPereiraMStanfordWL 2010 Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS ONE 5 e11260 doi:10.1371/journal.pone.0011260
71. PettittSJLiangQRairdanXYMoranJLProsserHM 2009 Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods 6 493 495
72. PoueymirouWTAuerbachWFrendeweyDHickeyJFEscaravageJM 2007 F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol 25 91 99
73. GongSKusLHeintzN 2010 Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nat Protoc 5 1678 1696
74. YangGSBanksKGBonaguroRJWilsonGDreoliniL 2009 Next generation tools for high-throughput promoter and expression analysis employing single-copy knock-ins at the Hprt1 locus. Genomics 93 196 204
75. LiskayRMEvansRJ 1980 Inactive X chromosome DNA does not function in DNA-mediated cell transformation for the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A 77 4895 4898
76. JacksonAPanayiotidisPForoniL 1998 The human homologue of the Drosophila tailless gene (TLX): characterization and mapping to a region of common deletion in human lymphoid leukemia on chromosome 6q21. Genomics 50 34 43
77. MiyawakiTUemuraADezawaMYuRTIdeC 2004 Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci 24 8124 8134
78. ZhangCLZouYYuRTGageFHEvansRM 2006 Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev 20 1308 1320
79. RoyKKuznickiKWuQSunZBockD 2004 The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci 24 8333 8345
80. LiWSunGYangSQuQNakashimaK 2008 Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol Endocrinol 22 56 64
81. AbrahamsBSKwokMCTrinhEBudaghzadehSHossainSM 2005 Pathological aggression in “fierce” mice corrected by human nuclear receptor 2E1. J Neurosci 25 6263 6270
82. YuRTChiangMYTanabeTKobayashiMYasudaK 2000 The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci U S A 97 2621 2625
83. RoyKThielsEMonaghanAP 2002 Loss of the tailless gene affects forebrain development and emotional behavior. Physiol Behav 77 595 600
84. SchmouthJFBanksKGMathelierAGregory-EvansCYCastellarinM 2012 Retina restored and brain abnormalities ameliorated by single-copy knock in of human NR2E1 in null mice. Mol Cell Biol E-pub ahead of print
85. KumarRAMcGheeKALeachSBonaguroRMacleanA 2008 Initial association of NR2E1 with bipolar disorder and identification of candidate mutations in bipolar disorder, schizophrenia, and aggression through resequencing. Am J Med Genet B Neuropsychiatr Genet 147B 880 889
86. KhakshoorOKoolET 2011 Chemistry of nucleic acids: impacts in multiple fields. Chem Commun 47 7018 7024
87. IoannouPAAmemiyaCTGarnesJKroiselPMShizuyaH 1994 A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet 6 84 89
88. ShizuyaHBirrenBKimUJMancinoVSlepakT 1992 Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U S A 89 8794 8797
89. den DunnenJTBakkerEvan OmmenGJPearsonPL 1989 The DMD gene analysed by field inversion gel electrophoresis. Br Med Bull 45 644 658
90. DuffKHuxleyC 1996 Targeting mutations to YACs by homologous recombination. Methods Mol Biol 54 187 198
91. GaspardNBouschetTHerpoelANaeijeGvan den AmeeleJ 2009 Generation of cortical neurons from mouse embryonic stem cells. Nat Protoc 4 1454 1463
92. BarberiTKlivenyiPCalingasanNYLeeHKawamataH 2003 Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol 21 1200 1207
Štítky
Genetika Reprodukčná medicína
Článek Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1Článek Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / TranscriptionČlánek Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding DomainČlánek Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin ComplexesČlánek An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood ObesityČlánek Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAsČlánek Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
- Genomic Analysis of the Hydrocarbon-Producing, Cellulolytic, Endophytic Fungus
- Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders
- Akirin Links Twist-Regulated Transcription with the Brahma Chromatin Remodeling Complex during Embryogenesis
- Too Much Cleavage of Cyclin E Promotes Breast Tumorigenesis
- Imprinted Genes … and the Number Is?
- Genetic Architecture of Highly Complex Chemical Resistance Traits across Four Yeast Strains
- Exploring the Complexity of the HIV-1 Fitness Landscape
- MNS1 Is Essential for Spermiogenesis and Motile Ciliary Functions in Mice
- A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Pathogenicity Islands SPI-1 and SPI-2
- Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution
- Variation in Modifies Risk of Neonatal Intestinal Obstruction in Cystic Fibrosis
- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Critical Evaluation of Imprinted Gene Expression by RNA–Seq: A New Perspective
- A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans
- Mouse Genetics Suggests Cell-Context Dependency for Myc-Regulated Metabolic Enzymes during Tumorigenesis
- Transcriptional Control in Cardiac Progenitors: Tbx1 Interacts with the BAF Chromatin Remodeling Complex and Regulates
- Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators
- APOBEC3G-Induced Hypermutation of Human Immunodeficiency Virus Type-1 Is Typically a Discrete “All or Nothing” Phenomenon
- Interpreting Meta-Analyses of Genome-Wide Association Studies
- Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line
- -Dependent Chemosensory Functions Contribute to Courtship Behavior in
- Diverse Forms of Splicing Are Part of an Evolving Autoregulatory Circuit
- Phenotypic Plasticity of the Drosophila Transcriptome
- Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1
- Precocious Metamorphosis in the Juvenile Hormone–Deficient Mutant of the Silkworm,
- Igf1r Signaling Is Indispensable for Preimplantation Development and Is Activated via a Novel Function of E-cadherin
- Accurate Prediction of Inducible Transcription Factor Binding Intensities In Vivo
- Mitochondrial Oxidative Stress Alters a Pathway in Strongly Resembling That of Bile Acid Biosynthesis and Secretion in Vertebrates
- Mammalian Neurogenesis Requires Treacle-Plk1 for Precise Control of Spindle Orientation, Mitotic Progression, and Maintenance of Neural Progenitor Cells
- Tcf7 Is an Important Regulator of the Switch of Self-Renewal and Differentiation in a Multipotential Hematopoietic Cell Line
- REST–Mediated Recruitment of Polycomb Repressor Complexes in Mammalian Cells
- Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / Transcription
- Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages
- A Genome-Wide Association Study Identifies Variants Underlying the Shade Avoidance Response
- -by- Regulatory Divergence Causes the Asymmetric Lethal Effects of an Ancestral Hybrid Incompatibility Gene
- Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
- A Natural System of Chromosome Transfer in
- Cell Size and the Initiation of DNA Replication in Bacteria
- Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
- Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin Complexes
- Temporal Transcriptional Profiling of Somatic and Germ Cells Reveals Biased Lineage Priming of Sexual Fate in the Fetal Mouse Gonad
- Rapid Analysis of Genome Rearrangements by Multiplex Ligation–Dependent Probe Amplification
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- The Atypical Calpains: Evolutionary Analyses and Roles in Cellular Degeneration
- The Silkworm Coming of Age—Early
- Development of a Panel of Genome-Wide Ancestry Informative Markers to Study Admixture Throughout the Americas
- Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency
- The Min System and Nucleoid Occlusion Are Not Required for Identifying the Division Site in but Ensure Its Efficient Utilization
- Neurobeachin, a Regulator of Synaptic Protein Targeting, Is Associated with Body Fat Mass and Feeding Behavior in Mice and Body-Mass Index in Humans
- Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin
- Gene Reactivation by 5-Aza-2′-Deoxycytidine–Induced Demethylation Requires SRCAP–Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions
- The miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
- Genetic Basis of Hidden Phenotypic Variation Revealed by Increased Translational Readthrough in Yeast
- An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood Obesity
- Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
- Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals
- Polycomb-Like 3 Promotes Polycomb Repressive Complex 2 Binding to CpG Islands and Embryonic Stem Cell Self-Renewal
- Insulin/IGF-1 and Hypoxia Signaling Act in Concert to Regulate Iron Homeostasis in
- EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development
- Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
- Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in
- A Machine Learning Approach for Identifying Novel Cell Type–Specific Transcriptional Regulators of Myogenesis
- Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
- Nos2 Inactivation Promotes the Development of Medulloblastoma in Mice by Deregulation of Gap43–Dependent Granule Cell Precursor Migration
- Intracranial Aneurysm Risk Locus 5q23.2 Is Associated with Elevated Systolic Blood Pressure
- Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits
- A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35.3, 7p14.3, and 13q14.1
- DNA Damage in Nijmegen Breakage Syndrome Cells Leads to PARP Hyperactivation and Increased Oxidative Stress
- DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination
- Genetic Analysis of Floral Symmetry in Van Gogh's Sunflowers Reveals Independent Recruitment of Genes in the Asteraceae
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Promoter Nucleosome Organization Shapes the Evolution of Gene Expression
- The Nucleoside Diphosphate Kinase Gene Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance
- The Ciliogenic Transcription Factor RFX3 Regulates Early Midline Distribution of Guidepost Neurons Required for Corpus Callosum Development
- Phosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
- A Genome-Wide Scan of Ashkenazi Jewish Crohn's Disease Suggests Novel Susceptibility Loci
- Parkinson's Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria
- LMW-E/CDK2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated b-Raf-ERK1/2-mTOR Pathway in Breast Cancer Patients
- Mapping the Hsp90 Genetic Interaction Network in Reveals Environmental Contingency and Rewired Circuitry
- Autoregulation of the Noncoding RNA Gene
- The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of Pro-Inflammatory Cytokines
- Spo0A∼P Imposes a Temporal Gate for the Bimodal Expression of Competence in
- Antagonistic Regulation of Apoptosis and Differentiation by the Cut Transcription Factor Represents a Tumor-Suppressing Mechanism in
- A Downstream CpG Island Controls Transcript Initiation and Elongation and the Methylation State of the Imprinted Macro ncRNA Promoter
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



