-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Comprehensive Methylome Characterization of and at Single-Base Resolution
In the bacterial world, methylation is most commonly associated with restriction-modification systems that provide a defense mechanism against invading foreign genomes. In addition, it is known that methylation plays functionally important roles, including timing of DNA replication, chromosome partitioning, DNA repair, and regulation of gene expression. However, full DNA methylome analyses are scarce due to a lack of a simple methodology for rapid and sensitive detection of common epigenetic marks (ie N6-methyladenine (6 mA) and N4-methylcytosine (4 mC)), in these organisms. Here, we use Single-Molecule Real-Time (SMRT) sequencing to determine the methylomes of two related human pathogen species, Mycoplasma genitalium G-37 and Mycoplasma pneumoniae M129, with single-base resolution. Our analysis identified two new methylation motifs not previously described in bacteria: a widespread 6 mA methylation motif common to both bacteria (5′-CTAT-3′), as well as a more complex Type I m6A sequence motif in M. pneumoniae (5′-GAN7TAY-3′/3′-CTN7ATR-5′). We identify the methyltransferase responsible for the common motif and suggest the one involved in M. pneumoniae only. Analysis of the distribution of methylation sites across the genome of M. pneumoniae suggests a potential role for methylation in regulating the cell cycle, as well as in regulation of gene expression. To our knowledge, this is one of the first direct methylome profiling studies with single-base resolution from a bacterial organism.
Published in the journal: Comprehensive Methylome Characterization of and at Single-Base Resolution. PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003191
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003191Summary
In the bacterial world, methylation is most commonly associated with restriction-modification systems that provide a defense mechanism against invading foreign genomes. In addition, it is known that methylation plays functionally important roles, including timing of DNA replication, chromosome partitioning, DNA repair, and regulation of gene expression. However, full DNA methylome analyses are scarce due to a lack of a simple methodology for rapid and sensitive detection of common epigenetic marks (ie N6-methyladenine (6 mA) and N4-methylcytosine (4 mC)), in these organisms. Here, we use Single-Molecule Real-Time (SMRT) sequencing to determine the methylomes of two related human pathogen species, Mycoplasma genitalium G-37 and Mycoplasma pneumoniae M129, with single-base resolution. Our analysis identified two new methylation motifs not previously described in bacteria: a widespread 6 mA methylation motif common to both bacteria (5′-CTAT-3′), as well as a more complex Type I m6A sequence motif in M. pneumoniae (5′-GAN7TAY-3′/3′-CTN7ATR-5′). We identify the methyltransferase responsible for the common motif and suggest the one involved in M. pneumoniae only. Analysis of the distribution of methylation sites across the genome of M. pneumoniae suggests a potential role for methylation in regulating the cell cycle, as well as in regulation of gene expression. To our knowledge, this is one of the first direct methylome profiling studies with single-base resolution from a bacterial organism.
Introduction
Among a few documented mechanisms, methylation of specific DNA sequences by DNA methyltransferases provides one way by which epigenetic inheritance can be orchestrated [1]. For instance, in many eukaryotes, methylated cytosine residues at 5′-CG-3′ (CpG) sequences are recognized by methyl-CpG binding proteins that usually repress the transcription of local DNA regions [2]–[5]. In the bacterial world, methylation is most commonly associated with restriction-modification (R-M) systems that provide a defense mechanism against invading foreign genomes [6]. In addition, it is known that a variety of enzymes capable of methylating DNA at adenine [7] and cytosine [8], [9] play functionally important roles, including timing of DNA replication, chromosome partitioning, DNA repair, transposition and conjugal transfer of plasmids, and regulation of gene expression [7], [10]–[16]. Phenomena involving inheritance of DNA methylation patterns are also known in bacteria. These systems use DNA methylation patterns to pass on information regarding the phenotypic expression state of the mother cell to the daughter cells. Methylation can alter the DNA structure and affect the binding of regulatory protein(s) to its DNA target site, thereby controlling gene expression [17], [18]. Notably, most adhesion genes in Escherichia coli are regulated by DNA methylation patterns [19], [20]. Little is known about how widespread heritable epigenetic control is in the bacterial world or the roles that epigenetic regulatory systems play in bacterial biology, including pathogenesis. For instance, it has been shown that DNA methylation in Streptococcus mutans up-regulates the expression of virulence factors like gbpC and bacteriocins [21]. It has also been shown that in E. coli, the expression of the Type IV secretion gene cluster is regulated by a non-stochastic epigenetic switch that depends on methylation of the Fur binding box [22].
In some gram-positive and gram-negative species that have been studied, adenine methylation plays a critical role in regulating chromosome replication. Adenine is generally methylated by members of the Dam family of methyltransferases, such as Dam in E. coli and DpnII in Streptococcus pneumoniae, that recognize the sequence motif 5′-GATC-3′ [23]. In these bacteria, the protein SeqA binds to hemi-methylated DNA target sites (5′-GATC-3′) clustered at the origin of replication (oriC) and sequesters the origin from replication initiation. SeqA also binds to hemi-methylated 5′-GATC-3′ sites in the dnaA promoter, blocking the synthesis of DnaA protein, which is necessary for replication initiation [24]–[27]. All of these events use the hemi-methylated state of newly replicated DNA as a signal. This hemi-methylated DNA is generated by semi-conservative replication of a fully methylated DNA molecule. Because of the transient nature of the hemi-methylation state, none of these phenomena are heritable. However, this mechanism is not universal, and other bacteria, like Bacillus subtilis, lack the Dam methyltransferase and SeqA proteins that E. coli employs to repress (sequester) its oriC during replication [28].
While there are many studies demonstrating the potential roles of methylation in epigenetic control of bacteria, the number of studies is significantly smaller than those for eukaryotes. This dearth of studies on bacterial epigenetics is partly due to a lack of a simple methodology that would allow rapid and sensitive detection of common epigenetic markers, such as N6-methyladenine (6 mA) and N4-methylcytosine (4 mC), in these organisms. Through bisulfite treatment, 5-methylcytosine (5 mC) was the only base modification detectable with efficiency and sensitivity suitable for genome wide epigenetic studies [29], [30]. Recently, Single-Molecule, Real-Time (SMRT) sequencing was described to provide the capability of directly detecting different base modifications beyond the canonical A, C, G, and T bases, in addition to yielding the sequence information [31]. The technique has been successfully demonstrated to identify methyltransferase specificities on plasmids [32].
Here, we use SMRT sequencing to comprehensively determine the methylomes of two mycoplasma species, Mycoplasma genitalium and Mycoplasma pneumoniae, with single-base and -strand resolution. M. pneumoniae and M. genitalium are closely related human pathogens that cause atypical pneumonia and non-gonococcal urethritis, respectively [33], [34]. These bacteria are members of the Mollicutes class characterized by the lack of a cell wall and by their reduced genomes with a low GC content. The genome sizes of M. pneumoniae and M. genitalium are 816 kb and 580 kb, respectively [35], [36]. M. genitalium is widely considered to have the smallest genome of any bacteria that can be grown in a test tube in the absence of host cells [37]. Our analysis identified a widespread 6 mA methylation sequence motif common to both bacteria (5′-CTAT-3′, with m6A in italics), as well as a more complex Type I m6A sequence motif in M. pneumoniae (5′-GAN7TAY-3′/3′-CTN7ATR-5′). Analysis of the chromosome distribution pattern of the first motif in M. pneumoniae suggests that methylation is involved in regulating cell division. To our knowledge, this work is one of the first comprehensive methylome analysis of bacteria.
Results
Putative restriction modification systems in M. pneumoniae and M. genitalium
We analyzed the genomes of M. pneumoniae and M. genitalium for all the putative methyltransferase genes using comparative sequence analysis and our previous functional assignment [38]. In the M. pneumoniae genome, we identified different putative Type I and Type II restriction modification systems. Type I involves a complex consisting of three polypeptides: R (restriction), M (modification), and S (specificity). The resulting complex can both cleave and methylate DNA. The S subunit determines the specificity of both restriction and methylation [39]. M. pneumoniae Type I system includes a methyltransferase (mpn342), a DNA specific recognition protein that brings the methyltransferase to the target DNA (HdsS, mpn343), and a restriction enzyme that cleaves unmethylated DNA (HdsR, mpn345). The restriction protein HdsR gene contains three frameshift mutations which likely make it inactive (additional protein fragments could be coded by mpn346 and mpn347). There are also some isolated genes encoding duplicated copies of the specificity determining subunit HdsS (mpn089, mpn289, mpn290, mpn365, mpn507, mpn615, and mpn638). In the Type II, methyltransferase and endonuclease are typically encoded as two separate proteins and act independently [39]. In M. pneumoniae, Type II systems could consist of the methyltransferase protein (HsdM, mpn107, mpn108 or mpn111) and the restriction enzyme (HsdR, mpn109 or mpn110). Additionally, a putative uncharacterized methyltransferase (mte1; mpn198), annotated as an EcoRI-like methylase in Uniprot and not associated with any R-M system, was identified. EcoRI restriction/modification system (R/M) is a Type II system that has been well characterized in vivo and in vitro [40], [41]. M. genitalium has an orthologous of mpn198 (mg184) and only one of the Type II-specificity determining subunits HdsS, mpn638 (mg438) (Table 1).
Tab. 1. Levels of RNA and protein for different proteins of M-R systems of M. pneumoniae. 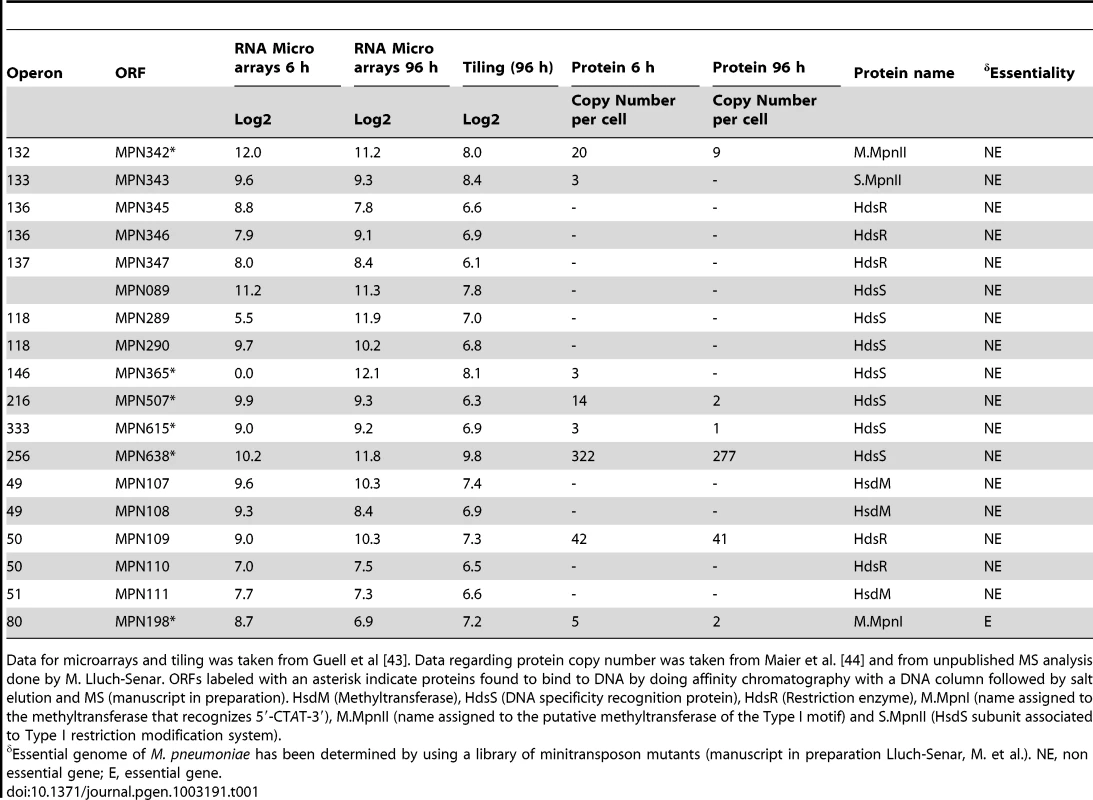
Data for microarrays and tiling was taken from Guell et al [43]. Data regarding protein copy number was taken from Maier et al. [44] and from unpublished MS analysis done by M. Lluch-Senar. ORFs labeled with an asterisk indicate proteins found to bind to DNA by doing affinity chromatography with a DNA column followed by salt elution and MS (manuscript in preparation). HsdM (Methyltransferase), HdsS (DNA specificity recognition protein), HdsR (Restriction enzyme), M.MpnI (name assigned to the methyltransferase that recognizes 5′-CTAT-3′), M.MpnII (name assigned to the putative methyltransferase of the Type I motif) and S.MpnII (HsdS subunit associated to Type I restriction modification system). We looked at the transcript and protein levels for the putative genes involved in methylation systems by using information of the transcriptome [42], [43] and proteome [44] of M. pneumoniae (Table 1). Although we could detect transcripts in the tiling array for all genes, albeit at very low level for many of them, we could identify in multiple MS experiments unique peptides for only six of them: mpn109, mpn198, mpn342, mpn343, mpn615, and mpn638 (Table 1). Of these, mpn198, mpn342, mpn615 and mpn638 were found to bind DNA by doing affinity chromatography with a DNA column followed by salt elution and MS analysis (manuscript in preparation). Only mpn198 (mte1; EcoRI-like) and mpn342 (Type I) are putative DNA adenine methyltransferases.
Methylome characterization of M. pneumoniae and M. genitalium by SMRT sequencing
Identification of methylated bases in M. pneumoniae and M. genitalium genomes was performed by SMRT sequencing at exponential (6 h) and stationary phases (96 h). Figure 1A shows the results of the genome-wide base modification detection analysis for the M. pneumoniae genome in stationary phase. The inner and outer most tracks in the Circos plot are the modification values (Qmod) of polymerase kinetics for the reverse and forward strands of the genome relative to an unmodified WGA (whole genome amplification) control. Qmod is the −10log(Pvalue) from a t-test and described in further details in the Materials and Methods section. The plot shows many significant peaks which correspond to methylated template positions. Figure 1B shows examples of the IPD (interpulse duration) ratios of a representative genomic section, highlighting both the base and strand resolutions of the technique. The statistically significant peaks, which were defined as Qmod >100 (Figure 1C; see Methods), were clustered as a function of sequence context to determine the recognition motifs of the methyltransferases responsible for the observed signals. The clustering results for M. pneumoniae identified >99.9% of all detected genomic positions as falling into two distinct sequence motifs: 5′-CTAT-3′ and 5′-GAN7TAY-3′/3′-CTN7ATR-5′ (Y = T or C and R = A or G, with m6A in italics). The first motif is found in both bacteria and is methylated on only one of the two DNA strands. In the second motif, the first adenines in the plus and minus strands are methylated (Figure 1B). The stretch of degenerate bases that separates the two recognition elements in the motif is characteristic of Type I methyltransferase signatures (Figure 1C) [45]. Despite the fact that the second sequence motif appears 1825 times per strand in M. genitalium (Table 2), there was no instance where it was detected as methylated. In contrast, this motif appears 1681 times in the genome of M. pneumoniae and 1678 are methylated (99,8%, Table 2). Approximately 1–2% of the assigned peaks were secondary peaks of the primary detected m6A and treated as redundant information for the tabulation in Table 2 [31].
Fig. 1. Methylome determination of M. pneumoniae by SMRT sequencing. 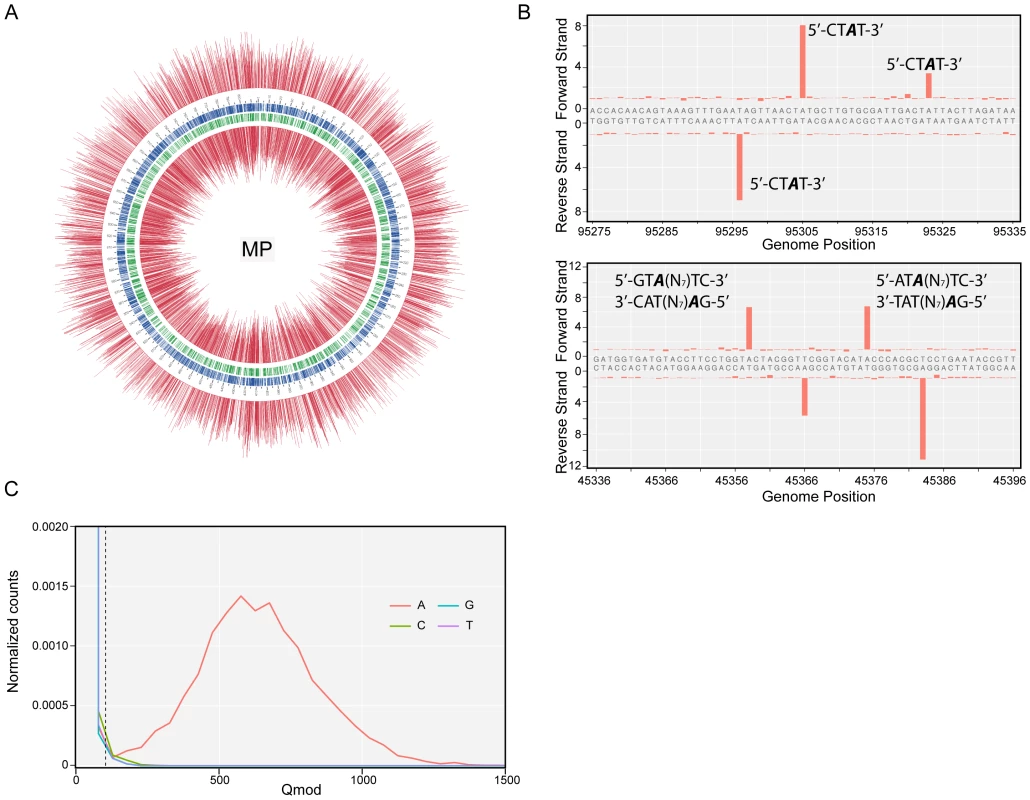
(A) Circos plot of kinetic variation across the genome. Red tracks represent the Qmod (−10log(Pvalue) from t-test) values of the forward (outer track) and reverse (inner track) strands. Blue and green tracks represent the location of the 5′-CTAT-3′ and Type I motifs discovered by filtering on Qmod values as shown in (C). (B) Example of IPD ratio plots of the two discovered motifs for a section of the genome. The top plot shows 3 instances of 5′-CTAT-3′, two of them are asymmetrically methylated. The bottom plot shows 2 Type I examples, where each one is fully methylated; that is each Type I recognition site is methylated on both strands. (C) Qmod distribution showing the filtering threshold of 100 used (black dash line) for determining modified positions. Tab. 2. Summary of discovered methylation motifs in M. genitalium and M.pneumoniae. 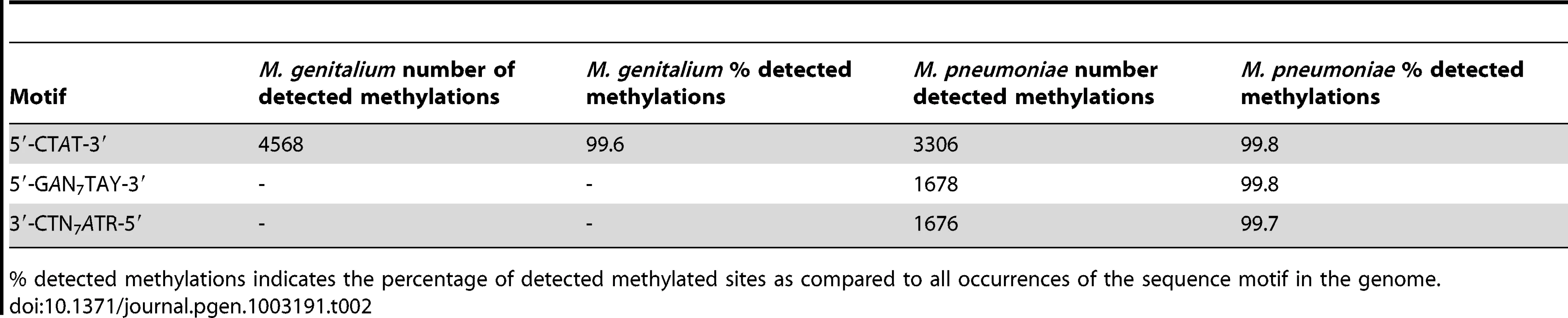
% detected methylations indicates the percentage of detected methylated sites as compared to all occurrences of the sequence motif in the genome. Analysis of two biological replicates of M. pneumoniae grown for 96 hours showed a reproducibility of 99.88% in the assignment of methylated positions.
Validation of methylation motifs and assignment to specific methyltransferase genes
Putative Type II independent methyltransferases (HsdM) (mpn198, mpn107, and mpn108) without an associated DNA recognition partner (HsdS), considered as possible candidates for the methylation of 5′-CTAT-3′ motif, were cloned into pRSS vector and then transformed into a methyltransferase-free E. coli ER2796 (DB24) [46] (Table S9b) following procedures described previously [32]. Mpn111 was discarded because it is a duplication of mpn108.
After cloning, the different plasmids were isolated and analyzed by SMRT sequencing. Of the three putative single proteins with methyltransferase activity, only mpn198 was capable of modifying the 5′-CTAT-3′ sequence. Interestingly, this is the only one of this group of methyltransferases that was found to be expressed by mass spectroscopy (MS) analyses (Table 1). As expected, no methyltransferase was identified by this approach for the Type I 5′-GAN7TAY-3′/3′ - CTN7ATR-5′motif, since Type I motifs also require the DNA recognition protein HsdS [45]. These results agree with the finding that both mycoplasma species are methylated at the same motif (5′-CTAT-3′) and share a common methyltransferase, namely, mpn198 in M. pneumoniae and mg184 in M. genitalium.
The fact that our MS analysis in M. pneumoniae detected protein expression only for DNA methylases MPN198 and MPN343, together with the lack of a mpn343 ortologue and the absence of the 5′-GAN7TAY-3′/3′-CTN7ATR-5′ methylated motif in M. genitalium, suggest that MPN343 could be responsible for the methylation of the 5′-GAN7TAY-3′/3′-CTN7ATR-5′ motif. These results validated the motifs observed for M. genitalium and M. pneumoniae and identified them as the recognition sequences of previously unassigned methyltransferases.
The new identified methyltransferases have been submitted in the REBASE and re-named using the standard nomenclature (mpn198: M.MpnI, mpn342: M.MpnII, mpn343: S.MpnII, mg184: M.MgeI). M indicates methyltransferase; S refers to the specificity subunit for Type I system; Mpn indicates M. pneumoniae and Mge indicates M. genitalium.
Genome-wide methylome analysis
We next focused on M. pneumoniae to study the role of methylation in regulating gene expression and DNA replication, since the transcriptome and proteome data are currently available for it [42], [43].
To study the putative role of methylation in DNA replication, we analyzed the density distribution of the 5′-CTAT-3′ methylation motifs in a sliding window of 1 kb along the M. pneumoniae genome (Figure 2A). The mean number of 5′-CTAT-3′motifs per 1 kb window is two (±1.6 standard deviation). Regions with more than five 5′-CTAT-3′ motifs (Pvalue<0.01) were considered to be “hot spots of methylation” for 5′-CTAT-3′ (Table S2b). A functional enrichment analysis of all the genes in M. pneumoniae present at the 5′-CTAT-3′ hotspots showed two functional categories of clusters of orthologous groups (COGs) over-represented: defense mechanisms (Pvalue = 0.025) and genes coding for membrane proteins or lipoproteins (Pvalue = 9×10−4) (Table S4a). Of the hot spots, there are three regions that have more than 10 motifs/kb. Interestingly, these regions are symmetrically distributed around the first kb of the genome (Figure 2B). This region of the genome comprises an intergenic region of 687 bp with three non-coding RNAs (MPNs200, MPNs201, and MPNs381) that frame eight repetitive 5′-TATTA-3′ sequences (identified as DnaA boxes based on Chip-seq analysis; Yus et al manuscript in preparation; Figure 2C [47]). There are three 5′-CTAT-3′ methylation motifs, two of them in overlapping and opposite strands of the region with the putative DnaA boxes suggesting that DNA methylation, although different from E. coli, could play a role in DNA replication. The other two regions are located at approximately 105 kb to the left and right from the putative origin of replication (Figure 2B). Search of common motifs in these two methylation hot spots revealed a common motif of 14 bp (5′-GATAG/ACCAAGG/AAGC-3′) (Figure 2D). This motif is found at opposite strands in the two regions, but only the left side region contains the 5′-CTAT-3′ sequence overlapping.
Fig. 2. Genome-wide distribution of 5′-CT 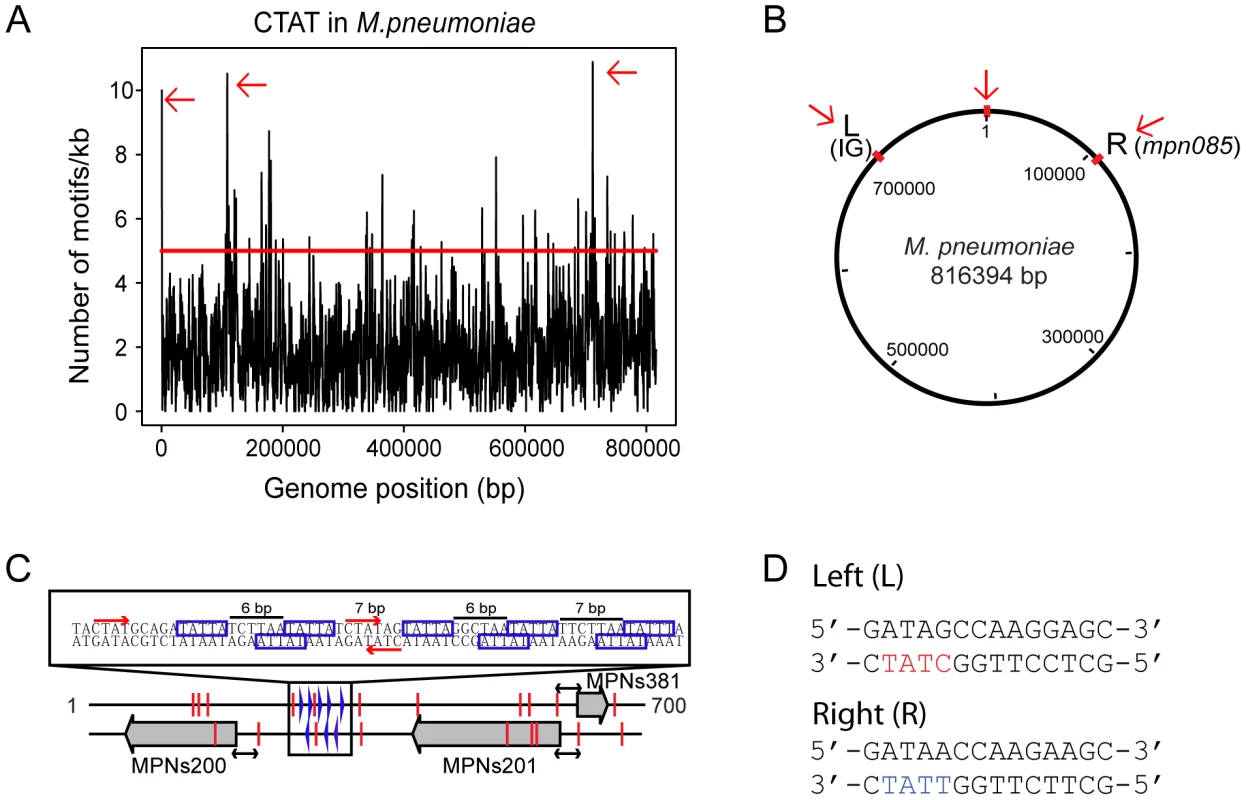
AT-3′ motif. (A) Hot spots of methylation for the 5′-CTAT-3′ motif. The graph represents the number of motifs per 1 kb window. The red line indicates the threshold (5). Regions with more than five 5′-CTAT-3′ motifs are considered “hot spots of methylation”. Red arrows indicate the most enriched regions. (B) Circular representation of the M. pneumoniae genome. Red marks indicate genome locations of the three main enriched regions of methylation. (C) Methylation in the putative origin of replication of M. pneumoniae. Blue boxes indicate putative DnaA boxes. Red arrows and lines indicate methylation sites. Black arrows indicated a common distance (24 bp) from methylation sites to the TSSs of the three MPNs. (D) Motif sequences of two putative cell division “check points” (L, left and R, right). Noteworthy only in L motif contains the recognition motif 5′-CTAT-3′ (showed in red letters) on the complementary strand. In contrast, the R motif contains 5′-TTAT-3′ (showed in blue letters) on the complementary strand instead. The three large grey arrows are the three non-coding RNAs (MPNs200, MPNs201 and MPNs381). We also analyzed the genome-wide distribution of the Type I motif. The average distribution for the 5′-GAN7TAY-3′/3′ - CTN7ATR-5′motif in 1 kb is 1 motif/kb (±1.1 standard deviation), and hot spot regions were considered to be those with more than 3 motifs within 1 kb (Pvalue<0.01). Most of the genes that overlap with these hotspots are of unknown function with a Pvalue of 0.04 (Table S4b). There are four 1 kb regions in the genome that have more than five instances of 5′-GAN7TAY-3′/3′-CTN7ATR-5′methylation (Figure 3A, and Table S2a). Interestingly, this highly methylated region with the most motifs (6 in 582 bp), is within mpn140, the first gene of the cytadherence operon that contains one of the main virulence factors of M. pneumoniae (Figure 3B). These motifs are located just upstream of the transcriptional start site (TSS) of an antisense transcript (MPNs383) that could be involved in regulating the expression of mpn140 (Figure 3C). The other three enriched regions correspond to mpn684 (that encodes a conserved hypothetical protein), mpn357 (DNA ligase), and mpn358 (conserved hypothetical protein) and, surprisingly, to the region containing mpn342 (M.MpnII) and mpn343 (S.MpnII). As mentioned above, M.MpnII is the putative methyltransferase responsible for 5′-GAN7TAY-3′/3′-CTN7ATR-5′ methylation.
Fig. 3. Genome-wide distribution of 5′-GAN7TAY-3′/3′-CTN7ATR-5′ motif. 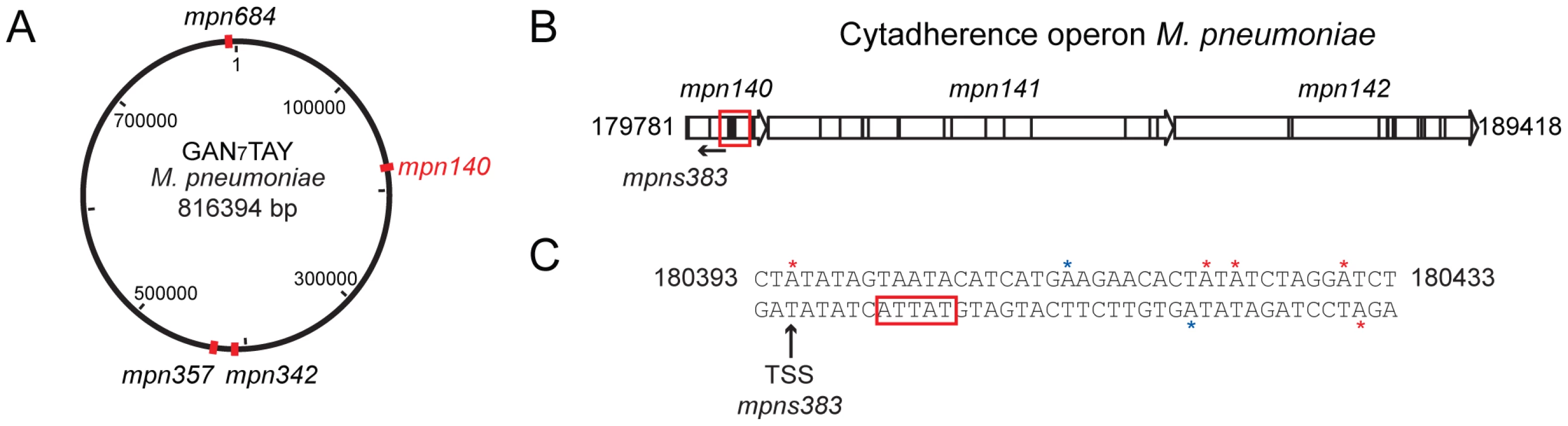
(A) Map of the genome of M. pneumoniae representing the enriched regions for the 5′-GAN7TAY-3′/3′-CTN7ATR-5′ motif. (B) Schematic representation of the cytadherence operon. The red square indicates the main “hot spot” of methylation for the Type I motif. (C) Upstream sequence of MPNs383. Red stars indicate the methylation by M.MpnI and blue stars indicate methylation by M.MpnII. The black arrow shows the transcriptional start site (TSS) of the MPNs383 and the red box the promoter sequence of this antisense RNA. Analysis of unmethylated motifs
The genome-wide access to methylation information allows for the interrogation of genomic locations which match the methyltransferases sequence targets, but are kept in an unmethylated state by the bacterium. The results in Table 3 show 5′-CTAT-3′ and 5′-GAN7TAY-3′/3′-CTN7ATR-5′ sites that are always unmethylated, two examples are shown in Figure 4. Only one unmethylated 5′-CTAT-3′ site was identified (genome position: 466475). This motif is overlapping with the stop codon of the mpn390 gene that codifies for the dihydrolipoamide dehydrogenase (PdhD). This gene together with mpn391 (PdhC, dihydrolipoamide acetyltransferase) constitute an operon involved in pyruvate metabolism. Also, three 5′-GAN7TAY-3′/3′-CTN7ATR-5′ unmethylated sites were detected. One is located in an intergenic region and the other two sites are located inside mpn493 (UlaD, 3-keto-L-gulonate-6-phosphate decarboxylase) involved in ascorbate and aldarate metabolism and mpn503 (cytadherence protein) (Table 3). We hypothesize that these unmethylated sites indicate the presence of an interacting protein or a DNA structure that is protecting from methylation along the different phases of growth.
Fig. 4. Unmethylated sites. 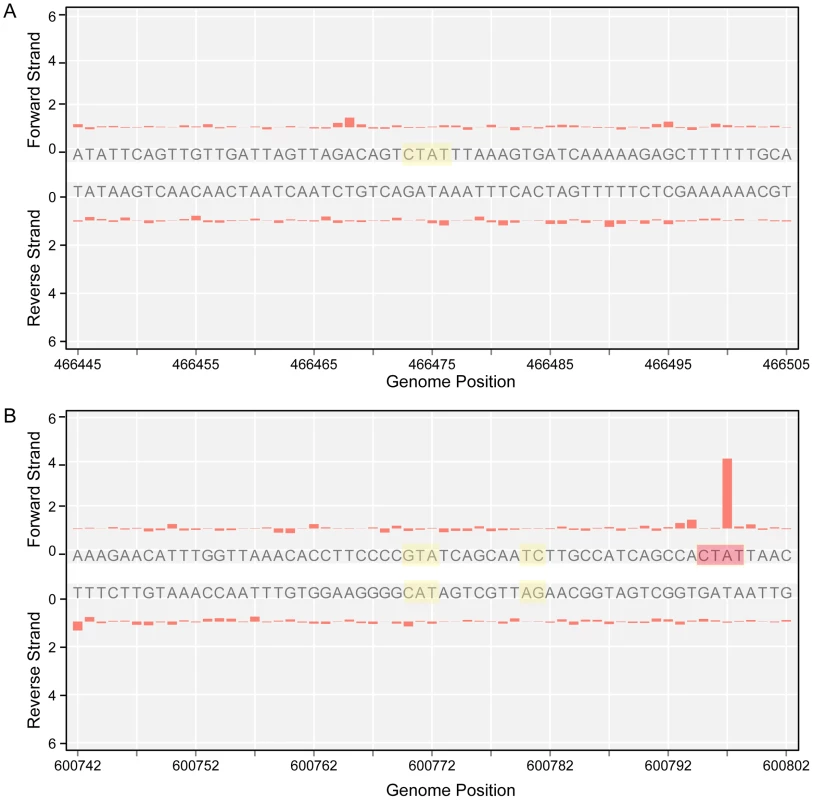
A) IPD ratio plot of a 5′-CTAT-3′ site not detected as methylated (shadowed in yellow). B) IPD ratio plot of an unmethylated 5′-GAN7TAY-3′/3′-CTN7ATR-5′ motif (shadowed in yellow). For comparison of signal intensity, a methylated 5′-CTAT-3′ is also shown in the bottom plot (shadowed in red). Tab. 3. Instances of genomic positions in <i>M. pneumoniae</i> consistently detected as lacking methylation across all three samples sequenced. 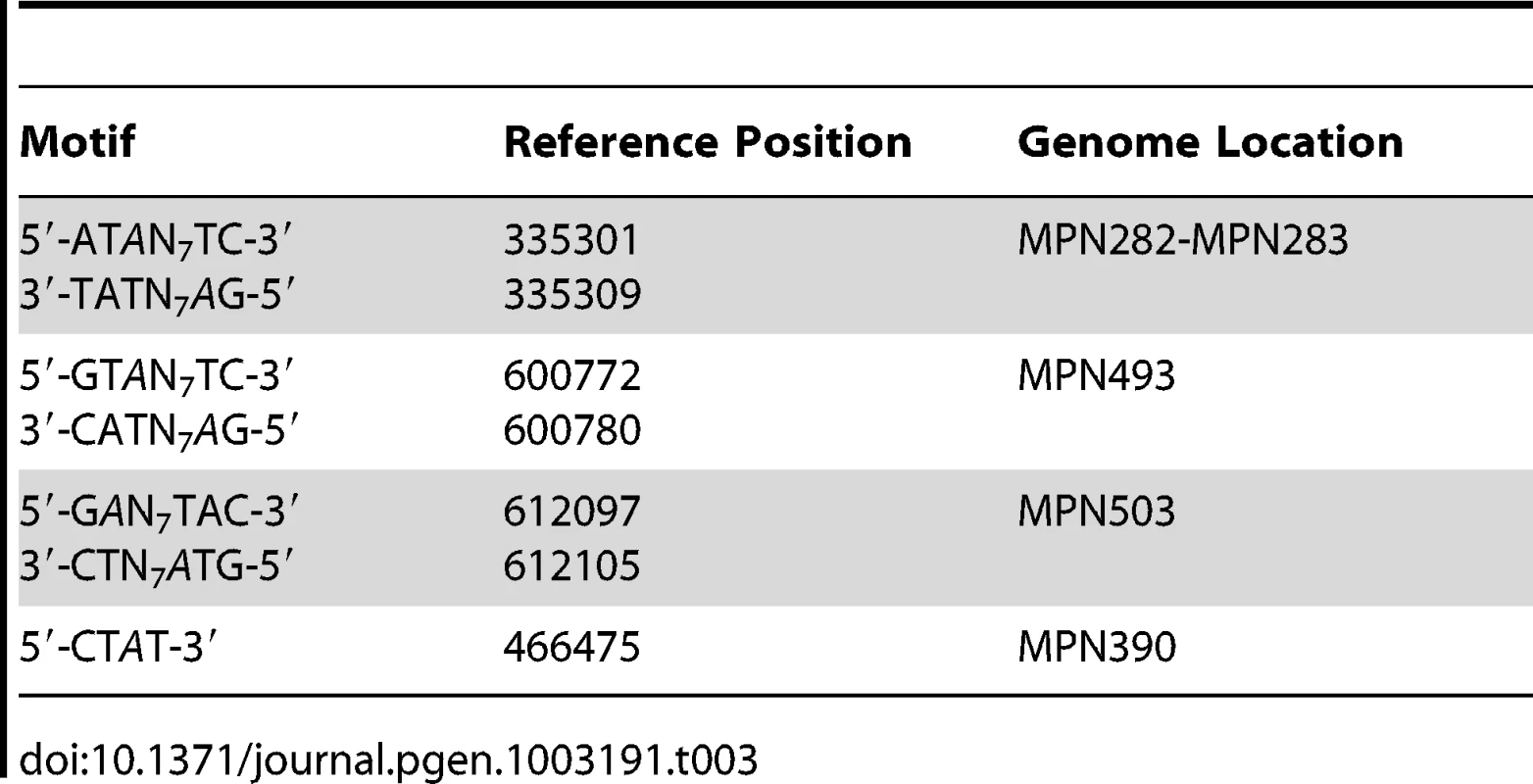
Functional correlations of the M. pneumoniae methylome
Recent identification of TSSs in M. pneumoniae [42] allowed us to study methylation patterns in promoter regions. We analyzed the regions comprising 40 bp upstream from the TSS (e.g. the promoter region) for 663 transcripts with TSS assigned and found 197 that were methylated in the promoter region (Table S5), with a total of 162 5′-CTAT-3′ and 74 5′-GAN7TAY-3′/3′-CTN7ATR-5′ motifs (located on both strands at the context site). Of these 197 transcripts, 103 are for non-coding RNAs (MPNs) and 89 correspond to ORFs. Fisher's exact test shows that there is a strong enrichment in methylation of MPNs promoters, with a Pvalue of 8.98×10−11. No functional enrichment is found for genes or MPNs (considering coding genes that overlap) methylated at the promoter regions (Table S6a).
Figure 5 shows the distribution in promoter regions of the distances from the methylation site (located upstream) to the TSS. Both motifs show that the highest frequency of methylation is at positions near the TSS and the Pribnow box (∼10–12 bases) (Pvalue of 0.03 for the 5′-CTAT-3′ motif, and of 0.005 for the Type I motif). These results could suggest the methylation has a potential role in transcription by affecting interaction of the sigma70, or of specific transcription factors, with the promoter.
Fig. 5. Histogram of distances of methylation motifs to TSS in the promoter regions. 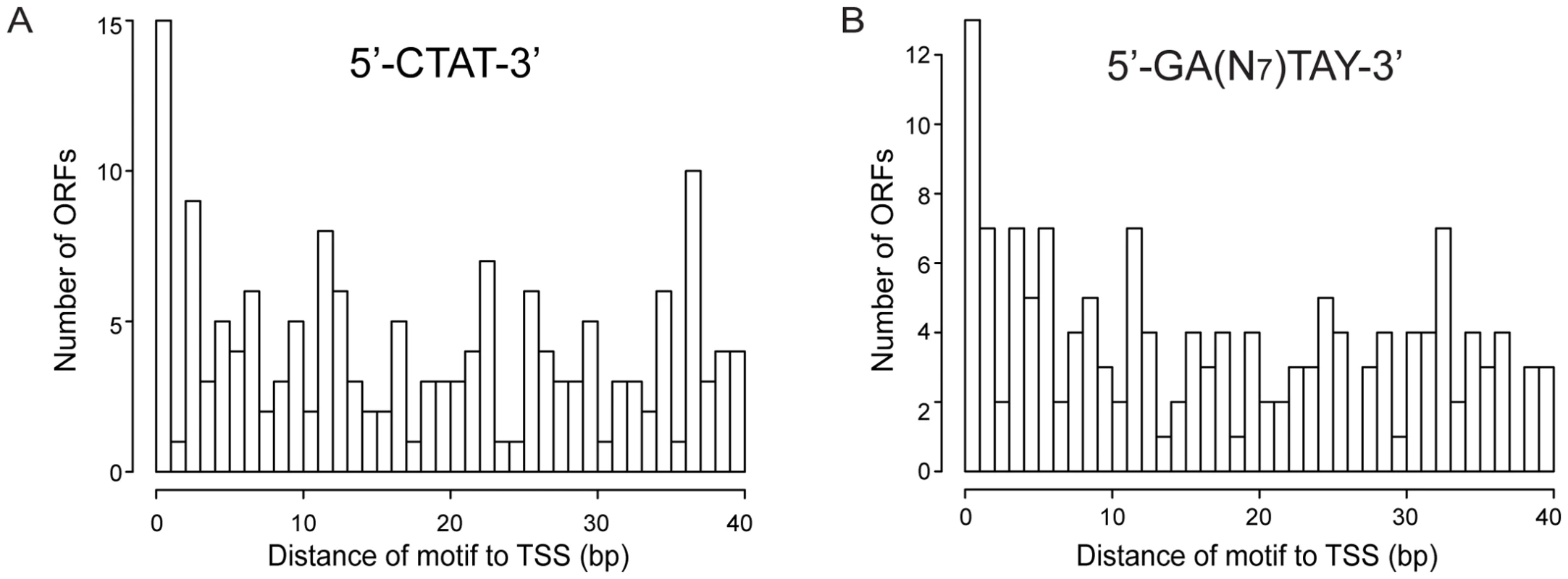
(A) Distances for the 5′-CTAT-3′ motif to TSS. (B) Distances for the 5′-GAN7TAY-3′/3′-CTN7ATR-5′ motif to TSS. We have also investigated the methylation pattern of 5′UTR regions encompassing the DNA sequences between the TSS and the translational start codon longer than 40 bp (long 5′UTR). Ninety two of 154 ORFs that have long 5′UTR regions showed methylation (Table S7). COG analysis of genes showing methylation in long 5′UTRs (Table S6b) revealed that genes involved in defense mechanism were three times more represented, with a Pvalue of 0.02. Interestingly, mpn342 gene (M.MpnII) has a 56 bp 5′UTR with two5′-GAN7TAY-3′/3′-CTN7ATR-5′ motifs, with 11 bp distance between the TSS and the motifs. As mentioned above, this gene could be responsible for methylating the 5′-GAN7TAY-3′/3′-CTN7ATR-5′motif, suggesting an autoregulatory gene expression mechanism.
Changes in methylation status as a function of growth phase
Although the majority of the 5′-CTAT-3′ sites were methylated in both exponential (6 h) and stationary (96 h) phases, using the conservative Qmod threshold of 100, a few sites were identified as having significantly different Qmod values which would suggest a change in methylation fraction at the given sites. Figure 6 illustrates the decrease in the 5′CTAT-3′ Qmod distributions from stationary to exponential growth samples, while the 5′-GAN7TAY-3′/3′-CTN7ATR-5′ Qmod distributions remain unchanged. This drop in the Qmod values points to a potential decrease in the methylation fraction at some 5′-CTAT-3′ sites at exponential growth as compared to stationary phase. To address this question of methylation changes at any given 5′-CTAT-3′ site between the growth phases at 6 h vs 96 h, we performed a direct comparison analysis between M. pneumoniae 6 h and 96 h. From this analysis, there are 35 5′-CTAT-3′ sites that were unmethylated at 6 h but became methylated by 96 h (Qmod≥60), indicating a change in methylation status between exponential and stationary phases of growth (Table S3). Twenty-five of the 35 methylation motifs are inside genes coding for membrane proteins, one in a 5′UTR, and the rest in intergenic regions. Analyzing the transcriptome for these 25 genes at 6 h and 96 h showed that their expression levels did not significantly change (Table S3), suggesting that this change in methylation state inside the genes is not related to the regulation of gene expression at different phases of growth. It was also observed that the fraction of methylation increased from 6 h to 96 h but not vice versa, further suggesting that the methylation in these regions are dependent on the phase of growth. It is noteworthy that M.MpnI reaches its maximal level of expression at exponential growth [39]. No general increase or decrease in gene expression was found associated with methylation. However, some specific cases, such as MPNs111, displayed an increase in promoter methylation with a significant decrease in transcript levels (fold change log2 = 2.93) (Table S8).
Fig. 6. Qmod distributions. 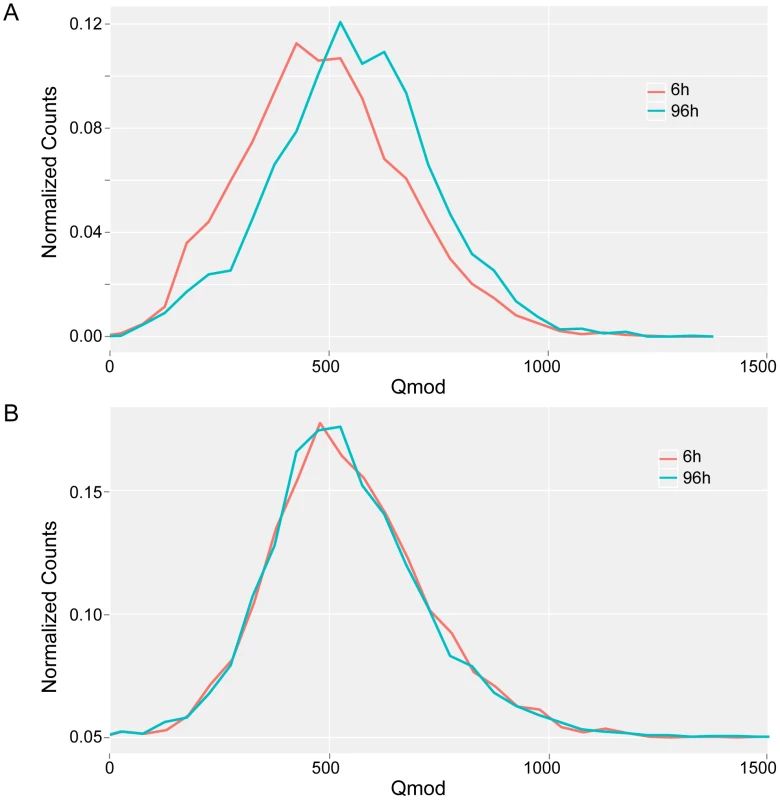
Qmod distributions of 5′-CTAT-3′ (A) and 5′-GAN7TAY-3′/3′-CTN7ATR-5′ motifs (B) for M. pneumoniae genome at exponential (6 h, red line) and stationary (96 h, blue line) phases of growth. Discussion
Previous analysis of DNA methylation in several mycoplasma species by HPLC revealed the presence of 6 mA in all of them, and of 5 mC in Mycoplasma hyorhinis [48]. Further studies performed in Mycoplasma arthritidis, to increase the efficiency of transformation, revealed methylated cytosine residues at 5′-AGCT-3′ and 5′-GCGC-3′ sites [49], [50]. Our current bioinformatic analysis in M. pneumoniae and M. genitalium did not find any evidence for 5 mC and only detected 6 mA. The study of proteome data (Table S1), together with a comparative analysis of gene conservation between these two species, suggest that there is an adenine methyltransferase (M.MpnI in M. pneumoniae, and M.MgI in M. genitalium) common to both genomes, and a putative Type I system in M. pneumoniae (mpn342 for HsdM (M.MpnII), mpn343 for HdsS (S.MpnII), and mpn345 for HdsR). It also revealed other putative methyltransferases in M. pneumoniae, and parts of the Type I system identified at the genome level, but these were not detected by proteome analysis of extracts from the bacteria exposed to different stresses or along the growth curve [44], or from SDS gels. These results suggest that there are two functional methylation systems in M. pneumoniae, and one in M. genitalium.
We employed SMRT sequencing to test these hypotheses by comprehensively characterizing the methylomes of M. pneumoniae and M. genitalium. The unique capability of SMRT sequencing to have both base and DNA strand specificities in base modification detection enable whole microbial methylome profiling with unprecedented resolution. We identified an asymmetric adenine methylation motif common to both bacteria, 5′-CTAT-3′, and a Type I motif with methylated adenines in both strands (5′-GAN7TAY-3′/3′-CTN7ATR-5′) found only in M. pneumoniae. The role of M.MpnI in the methylation of the 5′-CTAT-3′ motif was experimentally validated by expressing the methyltransferase in an E. coli strain devoid of methyltransferases [32].
The 5′-CTAT-3′motif was found enriched at the putative origin of replication (ORI) in M. pneumoniae as well as at two sites ∼100 kbs distant on both sides of the ORI which could be putative replication checkpoints, like the ψ sites described in B. subtilis [51]. The presence of two methylated 5′-CTAT-3′sites on the top and bottom strands at the mid-position of the putative DNA boxes at the ORI suggests a role for methylation in regulating DNA replication by M.MpnI. This hypothesis is reinforced by the fact that we did not find a restriction enzyme associated to this gene like in a classical EcoRI Type II system, similar to Dam methyltransferase in E. coli. The oriC of E. coli also contains an enriched region of methylated motifs (5′-GATC-3′). SeqA preferentially binds to clusters of two or more hemimethylated 5′-GATC-3′sites, delaying re-methylation and preventing binding of DnaA, which controls the initiation of DNA replication [52], [53]. No orthologous to E. coli SeqA protein has been identified in M. pneumoniae. However, a fundamental difference is found between the Dam system of E. coli and the M.MpnI methyltransferase of M. pneumoniae: in M. pneumoniae, only the one strand harboring the motif at any given genomic position is methylated, while in E. coli, both strands of the 5′-GATC-3′ motif can be methylated. Thus, it is not expected that M. pneumoniae will use a similar system with SeqA as E. coli to control DNA duplication. In fact, the M pneumoniae firmicute relative B. subtilis also lacks seqA and dam orthologous but contains several other proteins, like Spo0, that regulate oriC [54], [55]. Interestingly, analysis of transcript levels along the growth curve shows that M.MpnI correlates with genes involved in transcription like mpn515 (rpoC) and mpn516 (rpoB), DNA duplication (mpn003 [gyrB] and mpn004 [gyrA]) and growth (ribosomal proteins like mpn538, mpn539, and mpn540) (Figure S1). This suggests a coordination between expression of M.MpnI and other genes involved in cell division and growth. Additionally, M.MpnI is the only methyltransferase that is essential for M. pneumoniae growth reinforcing its key role in cell cycle regulation.
Analysis of COG categories for ORFs located in regions enriched for 5′-CTAT-3′ showed that these are involved in virulence, similar to previously described adhesion genes regulated by DNA methylation in E. coli [19], [20]. We also found genes in M. pneumoniae methylated at their promoter or 5′UTR regions that have orthologous known to be regulated by methylation in other bacteria, such as trpS [56] and the SOS regulon [57] in E. coli, and ClpB in Streptococus mutans. However, no relationship between methylation and transcription levels was observed when we studied the correlation between M.MpnI and ORFs with methylation in their regulatory sequences. Nonetheless, this apparent lack of correlation may be due to the lack of synchrony in the bacterial population, which may therefore exhibit different phenotypic properties. The high number of antisense RNAs that show methylation in promoter regions could imply that in the absence of regulatory proteins, methylation could serve as a mechanism to regulate the expression of the antisense strand and, consequently, any overlapping genes.
In most active R-M systems, all sites recognized by the restriction enzyme are protected by methylation in order to prevent the microbe's own defense mechanism from damage to its genome. However, there are incidences in which a protein protects certain sites from restriction digestion or methylation. For example, a 5′-GATC-3′sequence within the regulatory region of the car operon in E. coli was found to be protected from Dam methylation [58]. Indeed, CarP and IHF were shown to bind in this regulatory region and protect the 5′-GATC-3′ site from methylation [59].We have detected unmethylated 5′-GAN7TAY-3′/3′-CTN7ATR-5′ and 5′-CTAT-3′ sites, which could indicate that there is a protein interacting with these regions. A comparative study of the transcriptome at 6 h and 96 h in M. pneumoniae did not reveal any difference in transcription of genes containing unmethylated motifs when they are compared with the rest of the genes in the genome. Thus, these regions could be interaction sites for DNA-binding proteins that protect the DNA from methylation; in this case, methylation could play a role in transcription when the interacting protein is not occupying the region [60], [61]. However, interaction of structural elements that determine the structure of chromosome cannot be ruled out. Studies of protein occupancy could help to reveal why these regions are protected from methylation.
Conclusion
In conclusion, using SMRT DNA sequencing, we were able to directly observe and analyze with single-base and strand resolution the genome-wide methylomes of M. genitalium and M. pneumoniae. The two strains share an analogous methlytransferase that targets the sequence 5′-CTAT-3. M. pneumoniae additionally has a Type I methyltransferase with a 5′-GAN7TAY-3′/5′-CTN7ATR-3′ specificity. Together, these 2 motifs correspond to more than 99.9% of all sites directly detected by SMRT sequencing as modified. While ongoing work involving methyltransferase knock-out and over-expression studies are underway to help establish the relationship, this work demonstrates the unique capability of SMRT sequencing to directly sequence and profile the methylome of a whole microbial genome, allowing for unprecedented progress towards understanding the role of epigenomics in the world of prokaryotes.
Materials and Methods
Bacterial strains and growth conditions
Escherichia coli TOP 10 strain (Invitrogen) and E. coli ER2796 (DB24) [46] deficient in methyltransferases, also called DB24 (New England Biolabs), were grown at 37°C in LB broth or LB agar plates containing 100 µgml−1 ampicillin. The M. genitalium G-37 WT and M. pneumoniae M129 strains were grown in SP-4 and Hayflick media, respectively [62] at 37°C under 5% CO2 in tissue culture flasks (TPP). Cells were grown for 96 h for the stationary phase of growth. Alternatively, after 96 h of growth, the media was removed and replaced by fresh media, and the cells were scraped and re-grown for 6 h (exponential phase of growth).
Genomic DNA of M. genitalium and M. pneumoniae was isolated using the Illustra bacteria genomic Prep Mini Spin Kit (GE Healthcare). Plasmid DNA was obtained using the QIAprep Spin Miniprep Kit (Qiagen). All primers and plasmids used in this work are summarized in Table S9a and S9b. PCR products and digested fragments from agarose gels were purified using the QIAquick PCR purification Kit (Qiagen).
Sequencing library preparation and SMRT sequencing
Genomic and plasmid samples of M. genitalium and M. pneumoniae were prepared for SMRT sequencing following standard SMRTbell template preparation protocols for base modification detection on the PacBio RS [63]. In brief, each genomic sample was used to construct two SMRTbell template libraries: a ∼500 bp randomly sheared insert library of native genomic DNA, and a whole-genome-amplified (WGA) library of the same insert size to remove any existing base modifications in the genomic DNA. The WGA sample served as a control. SMRT sequencing was performed using C2 chemistry. At 2–4 SMRTCells each, all samples achieved ∼500× average sequencing coverage across the genome.
SMRT sequencing analysis
The principle of base modification detection using SMRT sequencing by synthesis was detailed in previous publications [31], [32]. The technique relies on the sensitivity of the polymerase kinetics to the DNA template structure as DNA synthesis is recorded in real time. It was observed that the time between base incorporations, or interpulse duration (IPD), is on average longer when the nucleotide incorporation occurs opposite of a methylated base in the DNA template, as compared to an incorporation opposite of a canonical base.
In previous studies, the analysis involved computing the ratio of the mean IPD of the native sample to the mean IPD of the WGA control sample for every reference template position, and setting a threshold to call certain template positions as methylated. The data analysis implemented here uses a t-test with a log-normal distribution model for the IPDs and associated Pvalue at every position for identifying the methylated sites. The null hypothesis in this analysis is that the IPDs from the native and WGA samples are part of the same population, and the alternate hypothesis is that the native set of IPDs stems from a population with larger IPDs, namely from incorporations opposite of a methylated rather than canonical template base. A threshold value of 100 for the log-transformed Pvalue from the t-test (called Qmod = −10log(Pvalue)) at each reference position was used for assigning the given position as methylated. The value of 100 was chosen based on the Qmod distribution observed in the data, where there was a clear bimodal distribution arising from unmodified background and modified positions. Furthermore, a Qmod≥100 corresponds to better than the Bonferroni corrected Pvalue of 0.0001 for the 816 kb genome.
To detect relative changes of the methylation status between samples grown for different time periods, the two native samples were directly compared against each other, rather than against a WGA control sample, thus highlighting the methylome difference between those samples. This analysis is performed after whole methylome analysis of the genome of interest. Hence, all sites of the discovered motifs were used as the n independent test sites giving a Bonferroni corrected Pvalue of better than 0.01 (0.0067) at Qmod≥60. Plots were made using Circos [64].
Both modes of analysis were carried out using SMRT Portal (http://www.smrtcommunity.com/SMRT-Analysis/Software/SMRT-Portal), while sequence motif cluster analysis was done using Pacific Biosciences's Motif Finder (http://www.smrtcommunity.com/CodeShare_Project?id=a1q70000000GtatAAC). Data sets containing kinetic values for each reference position and DNA strand are available at http://www.pacbiodevnet.com/Share/Datasets/Senar-et-al.
Molecular cloning
M. pneumoniae mpn107 gene was obtained by PCR using genomic DNA as template and specific primers (Table S9a). 5′-end oligonucleotides incorporated a PstI site followed by the sequence 5′-TTAAGG-3′ (to terminate translation of the lac α-peptide reading frame of the pRSS plasmid vector and to reinitiate translation of the cloned methyltransferse (MTase) genes, followed by an eight nucleotide spacer sequence 5′-TTAATCAT-3′ and sequences complementary to the 5′-end of the relevant MTase coding sequence. 3′-end oligonucleotides were complementary to the 3′-end of the MTase coding sequences, including translation termination codons and a BamHI restriction site. Since the TGA codon encodes tryptophan in Mycoplasma but an opal stop codon in E. coli, the mpn198 and mpn108 genes having several opal codons were codon-transformed and synthesized by GeneScript. After PCR amplification, the different genes were cloned into a PstI-BamHI digested pRSS vector. The resulting vectors were termed pRSS107, pRSS198, and pRSS108 (Table S9b).
Confirmation of methylation motifs by SMRT sequencing
The vectors described above were used to transform the E. coli deficient in methyltransferases ER2796 strain (kindly provided by R. Roberts, NEB). The plasmid DNA of every transformed strain was analyzed by SMRT sequencing as described previously [32].
Transcriptome data
Transcriptional start sites of the M.pneumoniae transcriptome have been described recently [42]. This information was used to define the 5′-UTR (RNA sequences from transcriptional start site to translational start codon). Transcription levels of M. pneumoniae genes at 6 h and 96 h were previously determined by tiling and ultrasequencing [43]. These data were used to study the relation between methylation and transcription in M. pneumoniae (Table S1).
Supporting Information
Zdroje
1. CasadesusJ, D'AriR (2002) Memory in bacteria and phage. Bioessays 24 : 512–518.
2. JorgensenHF, BirdA (2002) MeCP2 and other methyl-CpG binding proteins. Ment Retard Dev Disabil Res Rev 8 : 87–93.
3. KloseRJ, BirdAP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31 : 89–97.
4. LewisJD, MeehanRR, HenzelWJ, Maurer-FogyI, JeppesenP, et al. (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69 : 905–914.
5. NanX, CrossS, BirdA (1998) Gene silencing by methyl-CpG-binding proteins. Novartis Found Symp 214 : 6–16 discussion 16–21, 46–50.
6. RobertsRJ, VinczeT, PosfaiJ, MacelisD (2010) REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 38: D234–236.
7. Lobner-OlesenA, SkovgaardO, MarinusMG (2005) Dam methylation: coordinating cellular processes. Curr Opin Microbiol 8 : 154–160.
8. MarinusMG, MorrisNR (1973) Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol 114 : 1143–1150.
9. MayMS, HattmanS (1975) Analysis of bacteriophage deoxyribonucleic acid sequences methylated by host - and R-factor-controlled enzymes. J Bacteriol 123 : 768–770.
10. BarrasF, MarinusMG (1989) The great GATC: DNA methylation in E. coli. Trends Genet 5 : 139–143.
11. ModrichP (1989) Methyl-directed DNA mismatch correction. J Biol Chem 264 : 6597–6600.
12. PalmerBR, MarinusMG (1994) The dam and dcm strains of Escherichia coli–a review. Gene 143 : 1–12.
13. WionD, CasadesusJ (2006) N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4 : 183–192.
14. CasadesusJ, LowD (2006) Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70 : 830–856.
15. SohanpalBK, El-LabanyS, LahootiM, PlumbridgeJA, BlomfieldIC (2004) Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc Natl Acad Sci U S A 101 : 16322–16327.
16. BlynLB, BraatenBA, LowDA (1990) Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J 9 : 4045–4054.
17. PolaczekP, KwanK, CampbellJL (1998) GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA-protein recognition. Mol Gen Genet 258 : 488–493.
18. PolaczekP, KwanK, LiberiesDA, CampbellJL (1997) Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol Microbiol 26 : 261–275.
19. HerndayA, BraatenB, LowD (2004) The intricate workings of a bacterial epigenetic switch. Adv Exp Med Biol 547 : 83–89.
20. HerndayA, KrabbeM, BraatenB, LowD (2002) Self-perpetuating epigenetic pili switches in bacteria. Proc Natl Acad Sci U S A 99 Suppl 4 : 16470–16476.
21. BanasJA, BiswasS, ZhuM (2011) DNA Methylation Affects Virulence Gene Expression in Streptococcus mutans. Appl Environ Microbiol 77 : 7236–7242.
22. BrunetYR, BernardCS, GavioliM, LloubesR, CascalesE (2011) An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet 7: e1002205 doi:10.1371/journal.pgen.1002205.
23. MannarelliBM, BalganeshTS, GreenbergB, SpringhornSS, LacksSA (1985) Nucleotide sequence of the Dpn II DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci U S A 82 : 4468–4472.
24. TaghbaloutA, LandoulsiA, KernR, YamazoeM, HiragaS, et al. (2000) Competition between the replication initiator DnaA and the sequestration factor SeqA for binding to the hemimethylated chromosomal origin of E. coli in vitro. Genes Cells 5 : 873–884.
25. MolinaF, SkarstadK (2004) Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol Microbiol 52 : 1597–1612.
26. KangS, LeeH, HanJS, HwangDS (1999) Interaction of SeqA and Dam methylase on the hemimethylated origin of Escherichia coli chromosomal DNA replication. J Biol Chem 274 : 11463–11468.
27. CampbellJL, KlecknerN (1990) E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62 : 967–979.
28. KaguniJM (2006) DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol 60 : 351–375.
29. CokusSJ, FengS, ZhangX, ChenZ, MerrimanB, et al. (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 : 215–219.
30. ListerR, O'MalleyRC, Tonti-FilippiniJ, GregoryBD, BerryCC, et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 : 523–536.
31. FlusbergBA, WebsterDR, LeeJH, TraversKJ, OlivaresEC, et al. (2010) Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7 : 461–465.
32. ClarkTA, MurrayIA, MorganRD, KislyukAO, SpittleKE, et al. (2012) Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res 40: e29.
33. ChinerE, Signes-CostaJ, AndreuAL, AndreuL (2003) [Mycoplasma pneumoniae pneumonia: and uncommon cause of adult respiratory distress syndrome]. An Med Interna 20 : 597–598.
34. JensenJS (2004) Mycoplasma genitalium: the aetiological agent of urethritis and other sexually transmitted diseases. J Eur Acad Dermatol Venereol 18 : 1–11.
35. DandekarT, HuynenM, RegulaJT, UeberleB, ZimmermannCU, et al. (2000) Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res 28 : 3278–3288.
36. PetersonSN, HuPC, BottKF, HutchisonCA3rd (1993) A survey of the Mycoplasma genitalium genome by using random sequencing. J Bacteriol 175 : 7918–7930.
37. FraserCM, GocayneJD, WhiteO, AdamsMD, ClaytonRA, et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science 270 : 397–403.
38. YusE, MaierT, MichalodimitrakisK, van NoortV, YamadaT, et al. (2009) Impact of genome reduction on bacterial metabolism and its regulation. Science 326 : 1263–1268.
39. WilsonGG, MurrayNE (1991) Restriction and modification systems. Annu Rev Genet 25 : 585–627.
40. SmithDW, CrowderSW, ReichNO (1992) In vivo specificity of EcoRI DNA methyltransferase. Nucleic Acids Res 20 : 6091–6096.
41. ReichNO, OlsenC, OstiF, MurphyJ (1992) In vitro specificity of EcoRI DNA methyltransferase. J Biol Chem 267 : 15802–15807.
42. YusE, GuellM, VivancosAP, ChenWH, Lluch-SenarM, et al. (2012) Transcription start site associated RNAs in bacteria. Mol Syst Biol 8 : 585.
43. GuellM, van NoortV, YusE, ChenWH, Leigh-BellJ, et al. (2009) Transcriptome complexity in a genome-reduced bacterium. Science 326 : 1268–1271.
44. MaierT, GuellM, SerranoL (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett 583 : 3966–3973.
45. MurrayNE (2000) Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev 64 : 412–434.
46. KongH, LinLF, PorterN, StickelS, ByrdD, et al. (2000) Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res 28 : 3216–3223.
47. MottML, BergerJM (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5 : 343–354.
48. RazinA, RazinS (1980) Methylated bases in mycoplasmal DNA. Nucleic Acids Res 8 : 1383–1390.
49. VoelkerLL, DybvigK (1996) Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J Bacteriol 178 : 6078–6081.
50. LuoW, TuAH, CaoZ, YuH, DybvigK (2009) Identification of an isoschizomer of the HhaI DNA methyltransferase in Mycoplasma arthritidis. FEMS Microbiol Lett 290 : 195–198.
51. GautamA, BastiaD (2001) A replication terminus located at or near a replication checkpoint of Bacillus subtilis functions independently of stringent control. J Biol Chem 276 : 8771–8777.
52. SkarstadK, TorheimN, WoldS, LurzR, MesserW, et al. (2001) The Escherichia coli SeqA protein binds specifically to two sites in fully and hemimethylated oriC and has the capacity to inhibit DNA replication and affect chromosome topology. Biochimie 83 : 49–51.
53. SkarstadK, LuederG, LurzR, SpeckC, MesserW (2000) The Escherichia coli SeqA protein binds specifically and co-operatively to two sites in hemimethylated and fully methylated oriC. Mol Microbiol 36 : 1319–1326.
54. HiragaS, IchinoseC, OnogiT, NikiH, YamazoeM (2000) Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells 5 : 327–341.
55. Castilla-LlorenteV, Munoz-EspinD, VillarL, SalasM, MeijerWJ (2006) Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J 25 : 3890–3899.
56. Marinus MG (1996) Methylation of DNA; al. Ne, editor. Washington, D.C.: ASM Press. 782–791 p.
57. Lobner-OlesenA, MarinusMG, HansenFG (2003) Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc Natl Acad Sci U S A 100 : 4672–4677.
58. WangMX, ChurchGM (1992) A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature 360 : 606–610.
59. CharlierD, GigotD, HuysveldN, RooversM, PierardA, et al. (1995) Pyrimidine regulation of the Escherichia coli and Salmonella typhimurium carAB operons: CarP and integration host factor (IHF) modulate the methylation status of a GATC site present in the control region. J Mol Biol 250 : 383–391.
60. WallechaA, MunsterV, CorrentiJ, ChanT, van der WoudeM (2002) Dam - and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J Bacteriol 184 : 3338–3347.
61. CorrentiJ, MunsterV, ChanT, WoudeM (2002) Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol Microbiol 44 : 521–532.
62. TullyJG, RoseDL, WhitcombRF, WenzelRP (1979) Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J Infect Dis 139 : 478–482.
63. TraversKJ, ChinCS, RankDR, EidJS, TurnerSW (2010) A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res 38: e159.
64. KrzywinskiM, ScheinJ, BirolI, ConnorsJ, GascoyneR, et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19 : 1639–1645.
Štítky
Genetika Reprodukčná medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



