-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
Most plant and animal microRNAs (miRNAs) are transcribed by RNA polymerase II. We previously discovered miRNA–like small RNAs (milRNAs) in the filamentous fungus Neurospora crassa and uncovered at least four different pathways for milRNA production. To understand the evolutionary origin of milRNAs, we determined the roles of polymerases II and III (Pol II and Pol III) in milRNA transcription. Our results show that Pol III is responsible for the transcription of the major milRNAs produced in this organism. The inhibition of Pol III activity by an inhibitor or by gene silencing abolishes the production of most abundant milRNAs and pri–milRNAs. In addition, Pol III associates with these milRNA producing loci. Even though silencing of Pol II does not affect the synthesis of the most abundant milRNAs, Pol II or both Pol II and Pol III are associated with some milRNA–producing loci, suggesting a regulatory interaction between the two polymerases for some milRNA transcription. Furthermore, we show that one of the Pol III–transcribed milRNAs is derived from a tRNA precursor, and its biogenesis requires RNase Z, which cleaves the tRNA moiety to generate pre–milRNA. Our study identifies the transcriptional machinery responsible for the synthesis of fungal milRNAs and sheds light on the evolutionary origin of eukaryotic small RNAs.
Published in the journal: Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III. PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003227
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003227Summary
Most plant and animal microRNAs (miRNAs) are transcribed by RNA polymerase II. We previously discovered miRNA–like small RNAs (milRNAs) in the filamentous fungus Neurospora crassa and uncovered at least four different pathways for milRNA production. To understand the evolutionary origin of milRNAs, we determined the roles of polymerases II and III (Pol II and Pol III) in milRNA transcription. Our results show that Pol III is responsible for the transcription of the major milRNAs produced in this organism. The inhibition of Pol III activity by an inhibitor or by gene silencing abolishes the production of most abundant milRNAs and pri–milRNAs. In addition, Pol III associates with these milRNA producing loci. Even though silencing of Pol II does not affect the synthesis of the most abundant milRNAs, Pol II or both Pol II and Pol III are associated with some milRNA–producing loci, suggesting a regulatory interaction between the two polymerases for some milRNA transcription. Furthermore, we show that one of the Pol III–transcribed milRNAs is derived from a tRNA precursor, and its biogenesis requires RNase Z, which cleaves the tRNA moiety to generate pre–milRNA. Our study identifies the transcriptional machinery responsible for the synthesis of fungal milRNAs and sheds light on the evolutionary origin of eukaryotic small RNAs.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that regulate protein expression levels post-transcriptionally in a wide variety of cellular processes. miRNAs arise from discrete genomic loci and are processed by Dicer-like enzymes from stem-loop RNA precursors [1], [2]. Since their initial discovery in Caenorhabditis elegans, miRNAs have been found in animals, plants and algae [3]–[8]. Most animal and plant miRNAs are transcribed by RNA polymerase II (Pol II) [9]–[12]. Pol II-mediated transcription allows spatial and temporal regulation of the expression of miRNAs. In eukaryotic organisms, RNA polymerase III (Pol III) is the polymerase responsible for the transcription of tRNAs, 5S ribosomal RNA and some small nuclear RNAs, which are all short non-coding RNAs [13], [14].
Compared to transcriptional regulation of Pol II, the regulation of Pol III-mediated transcription is less complex, thus its role in transcribing housekeeping RNAs. Despite their different transcriptomes, Pol II and Pol III are evolutionarily related; the two, share some catalytic subunits, and many of their specific subunits are homologous to each other. In addition, the polymerases share transcription factors and promoter elements to a certain degree, which allows crosstalk between the two enzymes [15]–[17]. Recently, a small number of mammalian and viral miRNAs have been shown to be transcribed by Pol III [12], [18]–[21]. Most of these miRNAs derive from tRNA precursors or are transcribed from regions with repetitive elements.
We previously discovered the first fungal miRNA-like small RNAs (milRNAs) in the filamentous fungus Neurospora crassa [22]. Like plant and animal miRNAs, the Neurospora milRNAs arise from transcripts with stem-loop structures and are mostly dependent on Dicer for production. In addition, the vast majority of milRNA sequences correspond to one arm of the hairpin of the precursor (the milRNA arm) and have a strong preference for U at the 5′end. Interestingly, there are at least four different milRNA production mechanisms that use distinct combinations of factors, including Dicers, QDE-2 and the exonuclease QIP [23]. Studies of the Neurospora milRNAs shed light on the diversity and evolutionary origins of eukaryotic miRNAs.
In this study, we examined the role of Pol II and Pol III in milRNA biogenesis. Our results showed that Pol III is responsible for most milRNAs production in this fungus. On the other hand, Pol II was found to be associated with some milR loci, suggesting a collaboration between the two polymerases in milRNAs production. Furthermore, milR-4 milRNA was derived from a tRNA precursor and requires RNase Z for production.
Results
milRNA production reduced by a Pol III–specific inhibitor
We previously identified ∼25 putative milR loci by analyzing the QDE-2-associated small RNAs [22]. Among these milRs, milR-1, -2, -3 and -4 were experimentally confirmed due to their relative high expression levels. For other miRNA genes, even though they were identified by deep sequencing the QDE-2 associated small RNAs, they could not be detected by Northern blot analysis due to their low abundance. It is worth noting that the milR-1, -2, -3 and -4 genes account for the vast majority (92%) of all milRNAs produced in Neurospora. Therefore, we mostly focused our analyses on these four major milRNAs.
milR-1 is the most abundant milRNA-producing locus in the Neurospora genome. It is localized in the intergenic region between two predicted Neurospora genes [22]. The primary-milRNA (pri-milRNA) transcripts produced from milR-1 are ∼170 nucleotides (nt), much shorter than most of the known Pol II transcripts in Neurospora. Sequence analyses of milR-1 and other experimentally confirmed milR genes (milR-2, milR-3, and mil-4) that produce abundant milRNAs revealed that these four all share a stretch of T residues immediately downstream of the milRNA sequences (Figure 1A and Figure 2). The T cluster is a signature sequence motif for the transcription terminator of Pol III [13], [24], raising the possibility that the pri-milRNAs that generate these milRs are transcribed by Pol III.
Fig. 1. Pol III is required for the production of milRs. 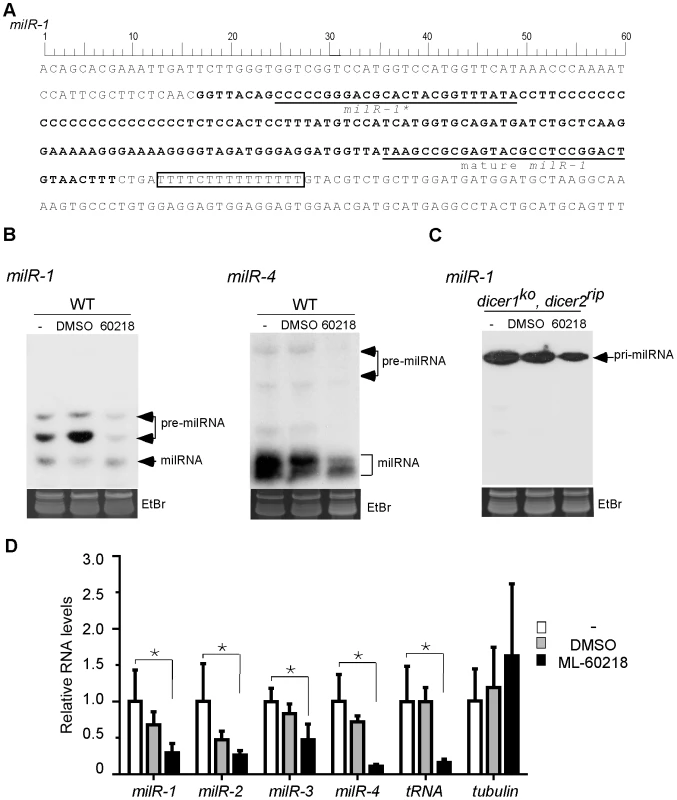
A. DNA sequence of milR-1 locus. B. Northern blot showing the level of small RNAs in indicated strains after indicated treatments. ML-60218 is Pol III specific inhibitor and DMSO is used as the control. The cultures were treated with ML-60218 before they were collected. The ethidium bromide-stained gel in the bottom panel shows equal loading of RNA samples. C. Northern blot result showing the levels of pri-milR-1 in dicer double knockout strain in the presence of the inhibitor. D. qRT-PCR analyses showing that the production of pri-milRNAs for milR-1, milR-2, milR-3 and milR-4 was inhibited by ML-60218. rRNA level was used as the loading control in the qRT-PCR analysis. β-tubulin and a Pol III-transcribed tRNA was served as the negative and positive control, respectively. WT indicates the wild-type strain. The asterisks indicate P value<0.05. Error bars indicate S.D. Fig. 2. DNA sequences of milR-1-4. 
The sequences of milRNA, milRNA* and the poly T sequences are indicated. TATA-like elements are underlined with single dashed lines. Putative A-boxes and B-boxes are underlined with double and triple dashed lines, respectively. The solid triangles in the milR-1 sequence indicate the nucleotides mutated in our promoter analysis. To examine the involvement of Pol III in milRNA production, we treated Neurospora cultures with ML-60218, a Pol III specific inhibitor [25]. Small RNA analysis by northern blot showed that the levels of both forms of pre-milR-1 were dramatically reduced in the presence of the inhibitor (Figure 1B). Similarly, ML-60218 also inhibited the production of pre-milR-4 and mature milR-4. To determine whether the inhibition of milR-1 production is due to the inhibition of the pri-milR-1 production, we examined the levels of pri-milR-1 in a dicer double mutant, in which the pri-milRNAs accumulate to a high level. As shown in Figure 1C, ML-60218 also inhibited the production of pri-milR-1. Quantitative RT-PCR analyses showed that levels of tRNA, pri-milRNA-1, -2, -3 and -4 were reduced in the presence of the inhibitor in the wild-type strain (Figure 1D). In contrast, the levels of β-tubulin transcript were not affected by treatment of cultures with the inhibitor, indicating specific inhibition of Pol III. These results suggest that these milRNAs might be transcribed by Pol III.
Pol III is required for transcription of milR-1-4 and associates with many milR loci
rpc5 is an essential Neurospora gene that encodes the Neurospora homolog of RPC34, a Pol III-specific subunit in Saccharomyces cerevisiae. To obtain genetic evidence for the involvement of Pol III in milR transcription, we generated Neurospora strains (dsrpc5) in which the expression of rpc5 (NCU00727) can be inducibly silenced by an rpc5-specific dsRNA in the presence of quinic acid (QA) inducer. In the absence of QA, race tube assays showed that the dsrpc5 strain grew similarly to the wild-type strain (Figure 3A). The addition of QA to the growth media, however, resulted in a dramatic reduction of growth rate of the dsrpc5 strain and the induction of rpc5-specific siRNA (Figure 3A and 3B), indicating the specific silencing of rpc5 expression.
Fig. 3. Pol III knockdown results in the reduction of milRNA expression. 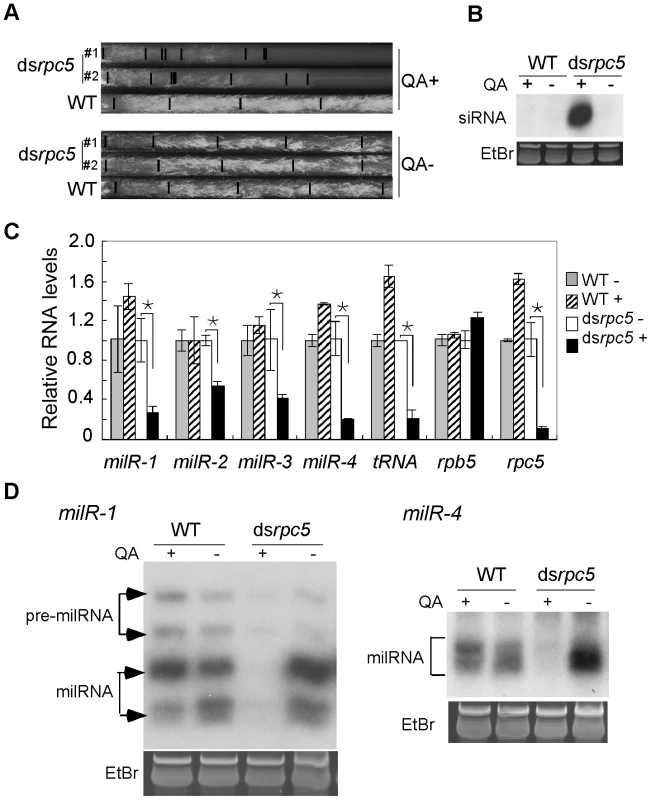
A. Race tube assays showing the growth rates of the indicated strains in the presence or absence of QA. B. Northern blot analysis showing the levels of rpc5-specific siRNA in the indicated strains. C. qRT-PCR analysis showing the reduction of pri-milR levels when rpc5 was silenced. rRNA level was used as the loading control for qRT-PCR. A Pol III-transcribed tRNA was served as a positive control. D. Northern blot showing the levels of milR-1 and milR-4 small RNAs. qRT-PCR analyses showed that the silencing of rpc5 by QA in the dsrpc5 strain resulted in significant reduction of pri-milRNA levels for the four experimentally confirmed milRs that produce vast majority of milRNAs in Neurospora (Figure 3C). In contrast, QA had no inhibitory effects on the pri-milRNA levels in the wild-type strain. Northern blot analyses showed that, as expected, the levels of mature milR-1 and milR-4 were also reduced when rpc5 was silenced in the dsrpc5 strain (Figure 3D). These data indicate that Pol III is required for the transcription of these milRNAs.
To examine whether the involvement of Pol III in milR transcription is direct, we created a Neurospora strain in which a Pol III-specific subunit, RPC7 (NCU07656), is c-Myc tagged, and performed chromatin immunoprecipitation (ChIP) assays using a monoclonal c-Myc antibody for ten of the milRNA-producing loci (milR-1 - milR-10). As shown in Figure 4, a strong RPC7-DNA binding was detected at a selected tRNA locus. In contrast, only background DNA binding was detected at the β-tubulin locus, indicating the specificity for Pol III-DNA binding in our ChIP assays. Significant RPC7-DNA binding by Pol III was detected in the milR-1, milR-2, milR-3 and milR-4 loci. For milR-7 and milR-10 loci, moderate binding was observed. For milR-5, milR-6, milR-8 and milR-9 loci, however, slightly higher than background binding was observed. These results suggest that Pol III is directly involved in the transcription of many of the milRs in Neurospora.
Fig. 4. Pol III specifically binds to milR loci. 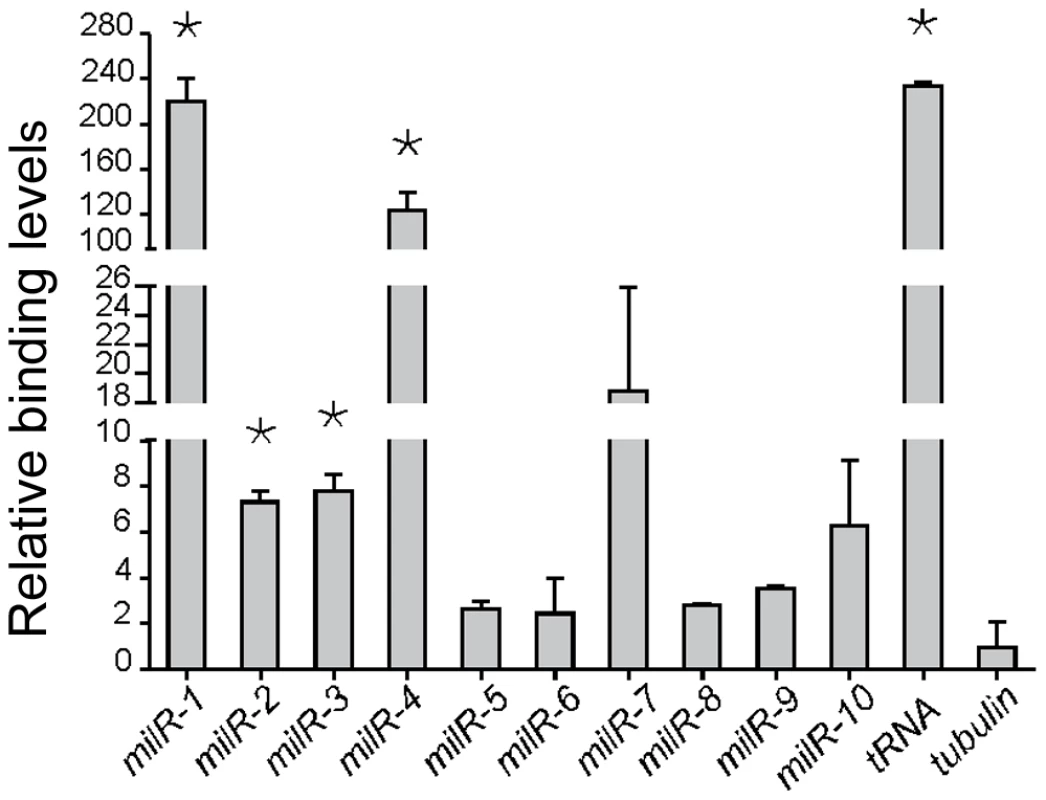
ChIP assay results showing the binding of Pol III to the milR loci. A tRNA and the β-tubulin gene was used as the negative and positive control, respectively. The asterisk indicate P value<0.05. Error bars indicate S.D. Pol II is involved in the transcription of a set of less abundant milRs
The vast majority of animal and plant miRNAs are transcribed by Pol II. To determine the involvement of Pol II in milRNA production in Neurospora, we created a dsrpb5 strain in which rpb5 (NCU02661), the gene encoding for a subunit of Pol II homologous to RPB7 in yeast [26], can be inducibly silenced by addition of QA to the culture media. As shown in Figure 5A and 5B, the addition of QA resulted in significant growth inhibition and the production of rpb5-specific siRNA in the dsrpb5 strain but not in the wild-type strain, indicating the specific silencing of the Pol II subunit in the dsrpb5 strain. As shown in Figure 5C and 5D, the reduced levels of rpb5 and β-tubulin in the presence of QA suggest the silencing of pol II-mediated transcriptin. As shown in Figure 5E, the silencing of rpb5 had no significant effects on the production of the mature milRNAs from the milR-1 and milR-4 genes. Similar results were also obtained by silencing another Pol II-specific subunit RPB6 NCU09378 (data not shown). These results suggest that Pol II is not required for the transcription of these abundant milRs.
Fig. 5. The involvement of Pol II in milRNA production. 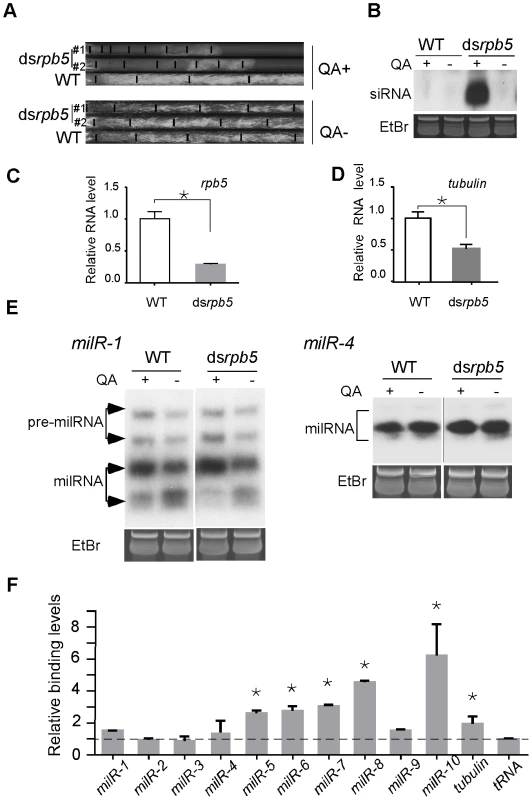
A. Race tube assays showing the growth rates of the indicated strains in the presence or absence of QA. B. Northern blot analysis showing the levels of rpb5-specific siRNA in the indicated strains. C and D. qRT-PCR analysis results showing the reduction of rpb5 and β-tubulin level in dsrpb5 strain in the presence of QA. The asterisks indicate P value<0.05. Error bars indicate S.D. E. Northern blot analysis showing the levels of milR-1 and milR-4 small RNAs. F. ChIP assay using c-Myc antibody showing the binding of Pol II to milR loci in the Myc-RPB6 strain. A tRNA and the β-tubulin gene was served as the negative control and positive control, respectively. The observed weak binding between the Pol III-specific subunit RPC7 and several of the milR loci in ChIP assays raises the possibility that Pol II may be involved in the transcription of these low abundance milRs. To test this, we created a Myc-tagged RPB6 strain and performed ChIP assays using the c-Myc antibody to examine the association of Pol II with these milR loci. For milR-5, milR-6 and milR-8 loci, significant binding was observed, suggesting that Pol II is involved in transcription of these milRNAs. Interestingly, high levels of RPB6-DNA binding were also observed in the milR-7 and milR-10 loci, where Pol III also associates (Figure 5F). In contrast, only background RPB6-DNA binding was observed at the milR-1, milR-2, milR-3 and milR-4 loci that produce the vast majority of milRNAs, further supporting the hypothesis that the transcription of these highly abundant milRNAs is solely mediated by Pol III. These results suggest that even though Pol III plays a major role in transcribing the abundant milRs, Pol II is also involved in the transcription of some of milRs.
To examine the involvement of pol III in the transcription of these low abundance milRs, we performed RNA sequencing using poly(A)-containing RNA of a wild-type strain. As shown in Figure 6, even though milR-1, -2, -3 and -4 loci are responsible for 92% of all Neurospora milRNAs, there was no or only background RNA reads, further supporting the conclusion that they are transcribed by Pol III. On the other hand, poly(A)-containing RNA reads were seen in the milR-5, -6, -7, -8 and -9 loci, suggesting that Pol II is involved in the transcription of these low abundance milRs.
Fig. 6. RNA sequencing of poly(A) RNA results showing the presence/absence of Pol II transcripts in the selected milR loci. 
Viewing window was set as 2000 nt. The vertical line in each panel indicates the location of the indicated milR gene. milR-1's transcription is mediated by a non-conventional Pol III promoter
The milR-1 locus has no predicted Pol III-specific A box or B box promoter elements [13], [27]. Instead, two putative TATA-like elements reside both inside and outside of the pri-milR-1 region (Figure 2). It is known that Pol III can recognize TATA elements for transcription initiation of some genes [13]. To examine the role of these elements in milR-1 transcription, two milR-1 mutant constructs, (mut1 and mut2), in which one of the TATA-like elements was mutated by point mutations, were created and transformed into a milR-1 knock-out strain. Northern blot analysis showed that milR-1 production was dramatically reduced in the mut1 and mut2 strains (Figure 7A). qRT-PCR analysis further showed that the pri-milR-1 levels were also dramatically reduced in the mutants (Figure 7B), suggesting that both of these elements are important for the milR-1 transcription. To determine the role of these elements in Pol III recruitment, the Myc-RPC7 construct was transformed into the wild-type, milR-1KO and mut2 strains. ChIP assay using the c-Myc antibody was performed to compare the binding of Myc-RPC7 at the milR-1 locus in these strains. A wild-type strain without the Myc-RPC7 construct was used as a control. As shown in Figure 7C, the RPC7 binding at the milR-1 locus was dramatically reduced in the mut2 strain, indicating that this TATA-like element is essential for the recruitment of Pol III to milR-1. These results suggest that milR-1 uses a non-conventional Pol III promoter for its transcription.
Fig. 7. milR-1 transcription is mediated by a non-conventional Pol III promoter. 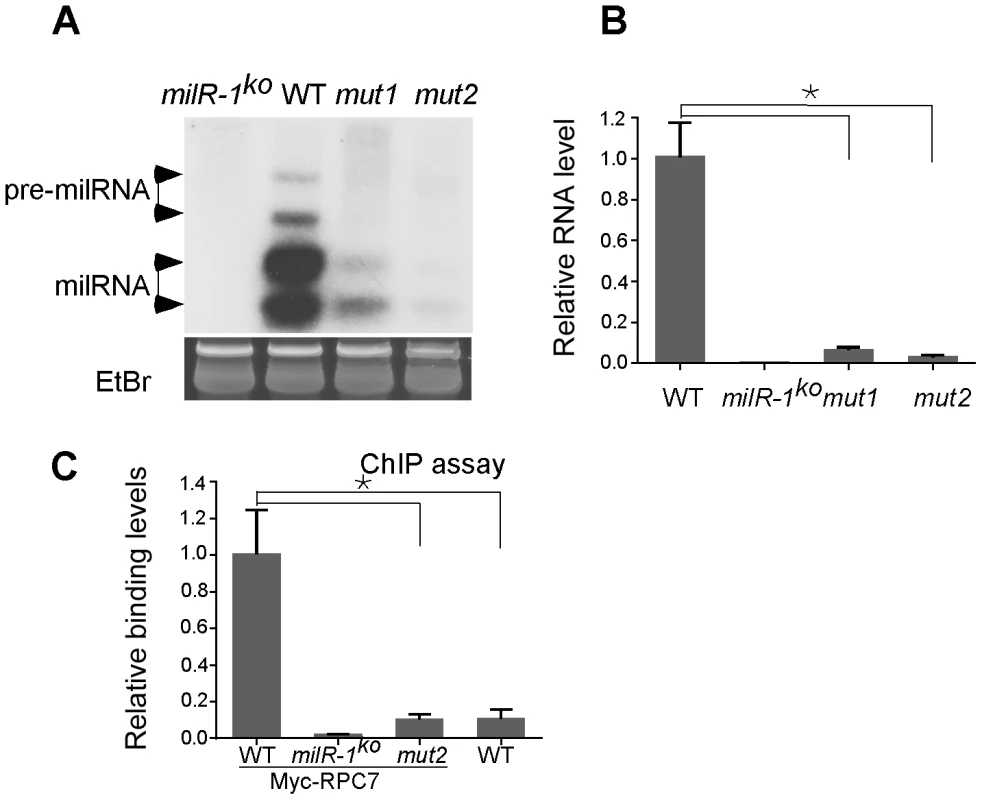
A. Northern blot analysis showing the levels of milR-1 in the indicated strains. B. qRT-PCR analysis showing the reduction of pri-milR-1 levels in the mut1 and mut2 strains. WT and milR-1ko served as the positive and negative control, respectively. C. ChIP assays using the c-Myc antibody showing the reduced binding of Myc-RPC7 at the milR-1 locus with the mutated TATA-like element. The asterisk indicates P value<0.05. Error bars indicate S.D. milR-4 derives from a tRNA–containing transcript and requires RNase Z for production
In eukaryotic organisms, Pol III is the RNA polymerase that transcribes tRNA, 5S ribosomal RNA and other small nuclear RNAs. The major role of Pol III in milR transcription suggests an evolutionary link between milRNAs and these small non-coding RNAs. Although most milRs are found in intergenic regions and are not adjacent to these small RNA genes, the mature milR-4 is located between the predicted genes for two alanine tRNAs and the milRNA* strand is downstream of the second tRNA (Figure 8A), suggesting that milR-4 may derive from RNA transcripts that contain these two tRNAs. To examine this, we performed RT-PCR analysis using a pair of primers that flank the two tRNAAla regions. As shown in Figure 8B, an RT-PCR specific fragment of the predicted size was observed, indicating that milR-4 is indeed co-transcribed with the two tRNAs.
Fig. 8. RNase Z is required for milR-4 processing. 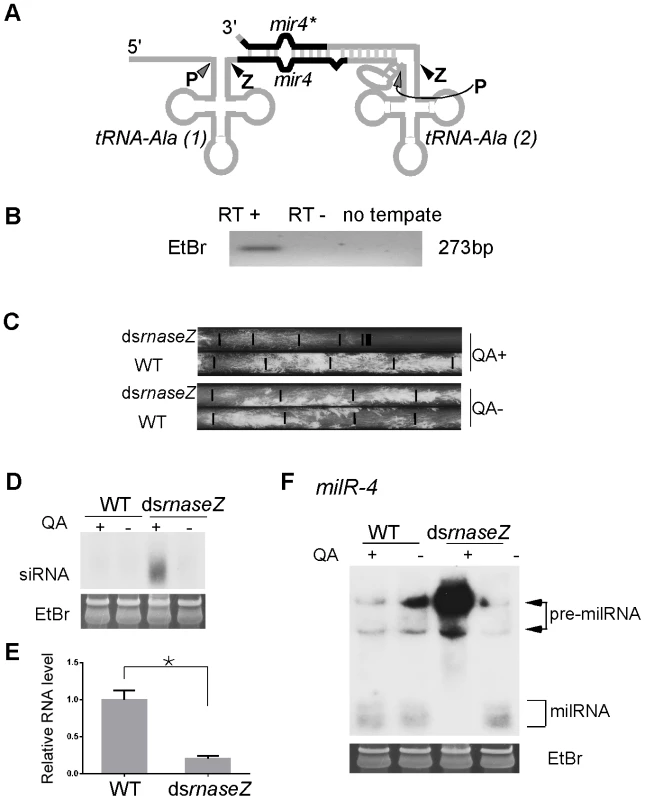
A. A Diagram showing the predicted secondary structure of pri-milR-4. B. RT-PCR analysis, which used a pair of primers indicated in A, showing the presence of transcript that spanning the two alanine tRNAs and milR-4. C. Race tube assays showing the growth rates of the indicated strains in the presence or absence of QA. D. Northern blot analysis showing the levels of rnaseZ-specific siRNA in the indicated strains. E. qRT-PCR analysis showing the reduction of rnaseZ mRNA level in the dsrnaseZ strain in the presence of QA. The asterisk indicates P value<0.05. Error bars indicate S.D. F. Northern blot analysis showing the levels of milR-4 milRNAs in the indicated strains in the presence or absence of QA. After the transcription by Pol III, eukaryotic tRNA precursors are processed at the 5′ and 3′ ends to remove the extraneous sequences, resulting in the generation of functional tRNAs [28]–[30]. The 3′ end sequences of pre-tRNAs are cleaved by the endonuclease RNase Z or other nucleases to generate mature 3′ends of tRNAs. We have previously shown that the production of milR-4 milRNA is partially dependent on Dicer [22]. The predicted RNA secondary structure of tRNA-milR-4 precursor suggests that Dicer should cut the stem region at the 3′ end of milRNA precursor and that another nuclease generates the 5′ end (Figure 8A). Because of predicted processing by RNase Z downstream of the first tRNA, we reasoned that RNase Z is required for the maturation of milR-4. Because RNase Z is likely essential for Neurospora cell growth, we created a strain (dsrnaseZ) in which the expression of the gene that encodes the Neurospora homolog of RNase Z (NCU00232) can be inducibly silenced by dsRNA. The addition of QA resulted in the production of rnaseZ-specific siRNA and dramatic inhibition of cell growth in the race tube assay in the dsrnaseZ strain but not in the wild-type strain (Figure 8C and Figure 8D). qRT-PCR further showed that rnaseZ level was down-regulated in the presence of QA (Figure 8E). In contrast, the growth rates of both strains were comparable in the absence of QA, indicating that the silencing of RNase Z is inducible and specific. As shown by the northern blot analysis result in Figure 8F, the silencing of rnaseZ expression by QA almost completely abolished the production of mature milRNA and resulted in the accumulation of high molecular weight pre-milRNA species. This result indicates that the nuclease RNase Z is required for the maturation of milR-4; it processes the 3′ end of the upstream tRNAAla in the transcript containing milR-4 to generate the 5′ end of the milRNAs.
Discussion
Our results demonstrated that RNA polymerase II and III are both involved in the transcription of the milRNAs in Neurospora. Since milR-1-4 account for the vast majority of milRNAs in Neurospora [22], Pol III is a major RNA polymerase for miRNA production in this organism. We showed that both the treatment of Neurospora by a Pol III-specific inhibitor and the silencing of a Pol III subunit by RNAi resulted in the near abolishment of the production of milR-1 and milR-4. In addition, the levels of pri-miRNAs of milR-1, -2, -3 and -4 were also reduced upon inhibition of Pol III. Furthermore, we showed that a Pol III-specific subunit is specifically associated with milR loci 1, 2, 3 and 4, indicating that Pol III is directly involved in mediating milR transcription.
The prominent role of Pol III in Neurospora milRNA transcription is in contrast with its minor role in miRNA production in animals and plants [9]–[12]. It was recently shown that Pol III is responsible for the transcription of a few animal miRNAs, mostly those derived from tRNA precursors and regions of repetitive elements [12], [18]–[20], [31]. Even though most of the Neurospora milR genes are located in intergenic regions with no repetitive elements, we showed that milR-4, one of the most abundant Neurospora milRNAs, is derived from a region that also encodes tRNAs. Moreover, the maturation of milR-4 requires RNase Z. Our results suggest that RNase Z cleaves the pre-tRNA to generate the 5′ end of pre-milR-4. It is likely that the pre-milR-4 is further processed by Dicer. The production mechanism of milR-4, therefore, is very similar to the biogenesis pathway of the murine γ-herpesvirus 68 encoded miRNAs, which are also cleaved by RNase Z from a tRNA moiety to generate pre-miRNA [19]. Therefore, our results indicate that the tRNA and RNase Z-mediated miRNA production mechanism is conserved from fungi to mammals.
Despite the lack of a significant role for Pol II in the transcription of the most abundant milRNAs, our ChIP assays showed that Pol II was associated with several milR loci at which Pol III binding is low. Poly (A) RNA sequencing results suggest that Pol II transcription is involved in the production of these low abundance milRNAs. Also interestingly, specific Pol III binding was also observed at the milR-7 locus, which also has poly (A) transcripts, suggesting that both polymerases may contribute to the transcription of these milRNAs. It has been previously suggested that these Pol II and III can cooperate to enhance the transcription in mammalian cells [16], [32].
miRNAs function in a wide variety of cellular and developmental processes in animal and plants [1], [33]. The use of Pol II allows the expression of miRNAs to be spatially and temporally regulated. Pol III is normally used for the transcription of small housekeeping non-coding RNAs, including tRNAs, 5S ribosomal RNA and snRNAs. Although the Neurospora milRNAs, like animal miRNAs, can silence endogenous targets with mismatches in vivo, their physiological functions are still unclear [22]. The fact that most Neurospora milRNAs are transcribed by Pol III suggests that expression of these Neurospora small RNAs is not as highly regulated as the production of animal and plant miRNAs. On the other hand, the binding of Pol II alone and the binding of both Pol II and Pol III to some of the milR loci suggest that the transcription of milRNAs may be flexible. Such flexibility may be explained by the fact that both polymerases share some of the same subunits and can recognize similar promoter elements, which allows regulatory interactions between the two enzymes [14]. Alternatively, it is possible that when fungal milRNAs first evolved, they were initially Pol III products. When a milRNA adopted a specific function that had to be highly regulated, interactions between Pol II and Pol III may have resulted in its transcription by Pol II. Future experiments that reveal the functions of different fungal milRNAs should shed light on the biogenesis and evolutionary origins of eukaryotic miRNAs.
Materials and Methods
Strains and growth conditions
The wild-type strain used in this study is FGSC 4200(a). The his-3 locus targeting transformation vector qa.pDE3dBH was used to construct double strand RNA knockdown and Myc-tagged expression vectors. The QA-inducible knockdown strains for dsrpc5, dsrpb5, dsrnaseZ and the QA-inducible Myc-tagged expression strains harboring qaMyc-RPB6 and qaMyc-RPC7 were introduced into the his-3 locus of a wild-type strain (301-6, his-3) as described previously [34]. milR-1ko was generated by gene replacement to delete the entire milR-1 locus with the hygromycin resistance gene (hph), as previously described [35]. Single nucleotide mutation for milR-1 promoter mutants, mut1 and mut2, was performed using one-step site-directed and site-saturation mutagenesis method [36]. The mutation vector was also transformed into Neurospora by his-3 targeting. Primers for milR-1 promoter mutations are: mut1F, CGGGACGCACTACGGTTTAgACCTTCCCCCC, mut1R, GGGGGGAAGGTcTAAACCGTAGTGCGTCCCG, mut2F, GGTCCATGGTTCATgAACCCAAAATCCATTCGC, and mut2R, GCGAATGGATTTTGGGTTcATGAACCATGGACC.
The dcldko strain was generated previously [37]. Liquid medium of 1× Vogel's, 0.1% glucose, and 0.17% arginine cultures with or without QA (0.01 M, pH 5.8), was used for liquid cultures. Race tube tests were carried out as previously described [34].
Chromatin immunoprecipitation assay
Anti-c-Myc antibody was used to perform immunoprecipitation. The WT extract was used as control. qPCR was carried out to quantify the immunoprecipitated DNA. The relative DNA binding levels of ChIP assays were determined by comparing the relative enrichment of the ChIPPed DNA between the Myc-RPC7/RPB6 strain and a wild-type strain (lacking the Myc-tagged protein). rDNA levels in the ChIPPed DNA was used to normalize the loading in different samples.
RNA polymerase III inhibitor treatment
ML-60218 (Calbiochem, catalog no. 557403) was used as an inhibitor of RNA polymerase III transcription [25]. ML-60218 was dissolved in DMSO at a concentration of 20 mg/ml. The final concentration used in tissue treatment was 20 mg/L. Culture medium containing an equal volume of DMSO or medium without DMSO was used as controls.
Conidia were germinated and grown in petri dishes containing liquid medium for one and a half days until even mycelia mats were formed. Discs were cut from the mats using a 7-mm cork borer and were shaken for 4 hrs in liquid medium before the addition of ML-60218. Tissue was treated at room temperature for 2 hours in the dark (to prevent light degradation of the inhibitor) before harvest.
Northern blot, RT–PCR, and qPCR assays
Total RNA and small RNA was prepared as previously described [37], [38]. Small RNA was enriched from total RNA using 5% polyethylene glycol (MW8000) and 500 mM NaCl [39]. RNA concentration was measured by NanoDrop (Thermo Scientific NanoDrop ND-2000 1-position Spectrophotometer). Small RNA was separated by electrophoresis through 16% polyacrylamide, 7 M urea, 0.5× tris-borate EDTA (TBE) gels. Equal amounts of small RNA (20 µg) were loaded in each lane. Chemical cross-linking was used to fix small RNAs to Hybond-NX membrane (GE Healthcare). For siRNA hybridization, a single-stranded RNA was transcribed from a DNA template in antisense orientation in the presence of 32P-labeled uridine triphosphate (PerkinElmer) using MAXIscript T7 kit (Ambion). Then the labeled RNA was treated with TURBO DNase (Ambion) and hydrolyzed at 60°C for 2–3 hr to an average size of 50 nt probe using fresh alkalic solution (80 mM sodium bicarbonate and 120 mM sodium carbonate). The hydrolyzed probe was then added to ULTRAhyb buffer (Ambion) to perform hybridization at 42°C overnight. After hybridization, the membrane was washed three times with 2× SSC and 0.1% SDS buffer for 15 min at 42°C before it was exposed to X-ray film. For miRNA hybridization, probe was labeled with IDT DNA StarFire miRNA Detection Kit in the presence of 32P-labeled deoxyadenosine triphosphate (PerkinElmer). ULTRAhyb-Oligo Buffer (Ambion) was used for hybridization. Other conditions are the same with siRNA hybridization.
qRT-PCR was as previously described [37]. Reverse transcription of total RNA was carried out by using SuperScript II Reverse Transcriptase (RT) (Invitrogen) with random primers. qPCR of the cDNA was performed using iTaq SYBR Green Supermix with ROX (BIO-RAD). The qPCR primer sequences are listed in Table 1. The primer sequences used to verify the pri-milR4 transcript were F1, 5′CTCAGTTGGTAGAGCGTTCG-3′, and R1, 5′-GAGGACCCATCTCACCAGAACA-3′.
Tab. 1. qPCR primer sequences. 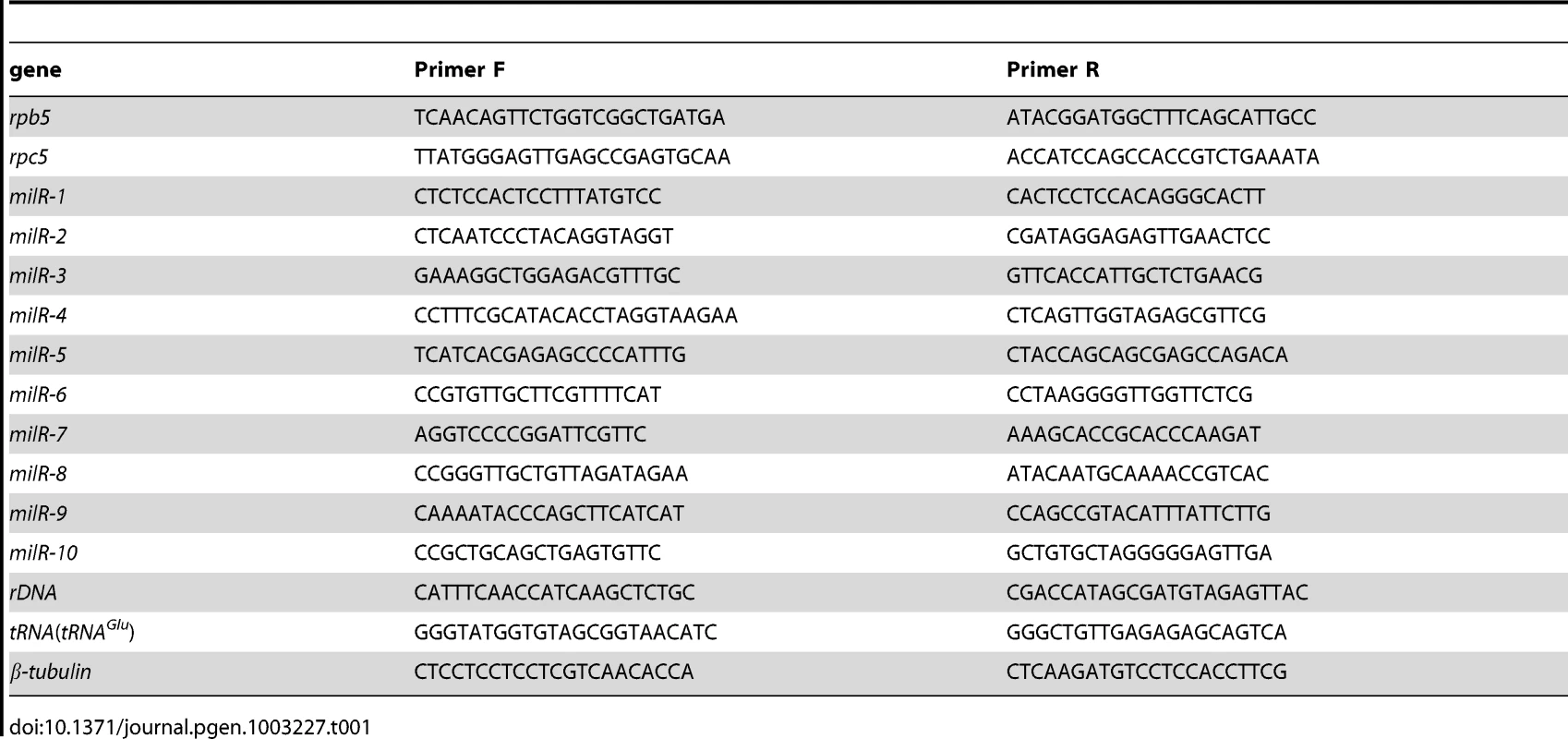
Strand-specific poly(A) mRNA sequencing
Three independent wild-type cultures grown in liquid media were pooled for RNA isolation for sequencing. RNA sequencing and data analyses were performed by Beijing Genomic Institute in Shenzhen, China. Strand specific sequencing was carried out following the method introduced by Dmitri Parkhomchuk [40]. mRNAs that purified from total RNA were sheared and reverse-transcribed into first-strand cDNAs. dTTP was substituted by dUTP during the synthesis of the second-strand cDNAs. After cDNA fragmentation and adaptor ligation, the dUTP containing second strand population were removed by Uracil-N-Glycosylase. The resulting first strand cDNA population were sequenced by Illumina HiSeq 2000. The cleaned reads were assembled and mapped to the reference Neurospora genome sequence (http://www.broadinstitute.org/annotation/genome/neurospora/GenomeDescriptions.html#Neurospora_crassa_OR74A) using SOAP (Short Oligonucleotide Alignment Program) [41], tophat [42], [43] and cufflinks [44]–[46]. The strand-specific results were displayed and analyzed by IGV (Integrative Genomics Viewer) software [47].
Zdroje
1. BartelDP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 : 281–297.
2. AmbrosV, BartelB, BartelDP, BurgeCB, CarringtonJC, et al. (2003) A uniform system for microRNA annotation. RNA 9 : 277–279.
3. LeeRC, FeinbaumRL, AmbrosV (1993) The C. elegans heterochromatic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 : 843–854.
4. Lagos-QuintanaM, RauhutR, LendeckelW, TuschlT (2001) Identification of novel genes coding for small expressed RNAs. Science 294 : 853–858.
5. LeeRC, AmbrosV (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294 : 862–864.
6. LlaveC, KasschauKD, RectorMA, CarringtonJC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14 : 1605–1619.
7. MolnarA, SchwachF, StudholmeDJ, ThuenemannEC, BaulcombeDC (2007) miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447 : 1126–1129.
8. ZhaoT, LiG, MiS, LiS, HannonGJ, et al. (2007) A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 21 : 1190–1203.
9. LeeY, KimM, HanJ, YeomKH, LeeS, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23 : 4051–4060.
10. CaiX, HagedornCH, CullenBR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10 : 1957–1966.
11. KimVN, HanJ, SiomiMC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10 : 126–139.
12. YangJS, LaiEC (2011) Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 43 : 892–903.
13. DieciG, FiorinoG, CastelnuovoM, TeichmannM, PaganoA (2007) The expanding RNA polymerase III transcriptome. Trends Genet 23 : 614–622.
14. WhiteRJ (2011) Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet 12 : 459–463.
15. OlerAJ, AllaRK, RobertsDN, WongA, HollenhorstPC, et al. (2010) Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17 : 620–628.
16. RahaD, WangZ, MoqtaderiZ, WuL, ZhongG, et al. (2010) Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A 107 : 3639–3644.
17. OrioliA, PascaliC, PaganoA, TeichmannM, DieciG (2012) RNA polymerase III transcription control elements: themes and variations. Gene 493 : 185–194.
18. HausseckerD, HuangY, LauA, ParameswaranP, FireAZ, et al. (2010) Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16 : 673–695.
19. BogerdHP, KarnowskiHW, CaiX, ShinJ, PohlersM, et al. (2010) A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell 37 : 135–142.
20. DiebelKW, SmithAL, van DykLF (2010) Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 16 : 170–185.
21. KincaidRP, BurkeJM, SullivanCS (2012) RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci U S A 109 : 3077–3082.
22. LeeHC, LiL, GuW, XueZ, CrosthwaiteSK, et al. (2010) Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell 38 : 803–814.
23. XueZ, YuanH, GuoJ, LiuY (2012) Reconstitution of an Argonaute-Dependent Small RNA Biogenesis Pathway Reveals a Handover Mechanism Involving the RNA Exosome and the Exonuclease QIP. Mol Cell
24. BragliaP, PercudaniR, DieciG (2005) Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem 280 : 19551–19562.
25. WuL, PanJ, ThoroddsenV, WysongDR, BlackmanRK, et al. (2003) Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell 2 : 256–264.
26. WernerM, ThuriauxP, SoutourinaJ (2009) Structure-function analysis of RNA polymerases I and III. Curr Opin Struct Biol 19 : 740–745.
27. SchrammL, HernandezN (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev 16 : 2593–2620.
28. HartmannRK, GossringerM, SpathB, FischerS, MarchfelderA (2009) The making of tRNAs and more - RNase P and tRNase Z. Prog Mol Biol Transl Sci 85 : 319–368.
29. PhizickyEM, HopperAK (2010) tRNA biology charges to the front. Genes Dev 24 : 1832–1860.
30. WalkerSC, EngelkeDR (2006) Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol 41 : 77–102.
31. BorchertGM, LanierW, DavidsonBL (2006) RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13 : 1097–1101.
32. ListermanI, BledauAS, GrishinaI, NeugebauerKM (2007) Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet 3: e212 doi:10.1371/journal.pgen.0030212.
33. GhildiyalM, ZamorePD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10 : 94–108.
34. ChengP, HeQ, HeQ, WangL, LiuY (2005) Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev 19 : 234–241.
35. MaitiM, LeeHC, LiuY (2007) QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev 21 : 590–600.
36. ZhengL, BaumannU, ReymondJL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115.
37. ChoudharyS, LeeHC, MaitiM, HeQ, ChengP, et al. (2007) A double-stranded-RNA response program important for RNA interference efficiency. Mol Cell Biol 27 : 3995–4005.
38. AronsonBD, JohnsonKA, LorosJJ, DunlapJC (1994) Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263 : 1578–1584.
39. CatalanottoC, AzzalinG, MacinoG, CogoniC (2002) Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev 16 : 790–795.
40. ParkhomchukD, BorodinaT, AmstislavskiyV, BanaruM, HallenL, et al. (2009) Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123.
41. LiR, LiY, KristiansenK, WangJ (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24 : 713–714.
42. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
43. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
44. TrapnellC, WilliamsBA, PerteaG, MortazaviA, KwanG, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28 : 511–515.
45. RobertsA, TrapnellC, DonagheyJ, RinnJL, PachterL (2011) Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 12: R22.
46. RobertsA, PimentelH, TrapnellC, PachterL (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27 : 2325–2329.
47. ThorvaldsdottirH, RobinsonJT, MesirovJP (2012) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform
Štítky
Genetika Reprodukčná medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



