-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A New Role for the GARP Complex in MicroRNA-Mediated Gene Regulation
Many core components of the microRNA pathway have been elucidated and knowledge of their mechanisms of action actively progresses. In contrast, factors with modulatory roles on the pathway are just starting to become known and understood. Using a genetic screen in Caenorhabditis elegans, we identify a component of the GARP (Golgi Associated Retrograde Protein) complex, vps-52, as a novel genetic interactor of the microRNA pathway. The loss of vps-52 in distinct sensitized genetic backgrounds induces the enhancement of defective microRNA-mediated gene silencing. It synergizes with the core microRNA components, alg-1 Argonaute and ain-1 (GW182), in enhancing seam cell defects and exacerbates the gene silencing defects of the let-7 family and lsy-6 microRNAs in the regulation of seam cell, vulva and ASEL neuron development. Underpinning the observed genetic interactions, we found that VPS-52 impinges on the abundance of the GW182 proteins as well as the levels of microRNAs including the let-7 family. Altogether, we demonstrate that GARP complex fulfills a positive modulatory role on microRNA function and postulate that acting through GARP, vps-52 participates in a membrane-related process of the microRNA pathway.
Published in the journal: A New Role for the GARP Complex in MicroRNA-Mediated Gene Regulation. PLoS Genet 9(11): e32767. doi:10.1371/journal.pgen.1003961
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003961Summary
Many core components of the microRNA pathway have been elucidated and knowledge of their mechanisms of action actively progresses. In contrast, factors with modulatory roles on the pathway are just starting to become known and understood. Using a genetic screen in Caenorhabditis elegans, we identify a component of the GARP (Golgi Associated Retrograde Protein) complex, vps-52, as a novel genetic interactor of the microRNA pathway. The loss of vps-52 in distinct sensitized genetic backgrounds induces the enhancement of defective microRNA-mediated gene silencing. It synergizes with the core microRNA components, alg-1 Argonaute and ain-1 (GW182), in enhancing seam cell defects and exacerbates the gene silencing defects of the let-7 family and lsy-6 microRNAs in the regulation of seam cell, vulva and ASEL neuron development. Underpinning the observed genetic interactions, we found that VPS-52 impinges on the abundance of the GW182 proteins as well as the levels of microRNAs including the let-7 family. Altogether, we demonstrate that GARP complex fulfills a positive modulatory role on microRNA function and postulate that acting through GARP, vps-52 participates in a membrane-related process of the microRNA pathway.
Introduction
The microRNA (miRNA) pathway is a gene regulatory system that uses small non-coding RNAs to target messenger RNAs (mRNAs) for post-transcriptional regulation. In the canonical form of miRNA biogenesis, miRNA-containing transcripts are processed through sequential cleavage operated by the Drosha and Dicer enzymes into mature miRNA species (21–23 nucleotides long) that associate with an Argonaute protein (reviewed in [1]). In its effector phase, the miRNA-loaded Argonaute, as part of the core miRNA-induced silencing complex (miRISC), regulates target mRNAs through binding sites in their 3′UTRs. The most detailed repressive effector function of this complex is mediated by its association to GW182 proteins (reviewed in [2]). The miRISC-mediated effector phase of target regulation, may involve the repression of multiple target molecules by each single miRNA through the process of ‘miRNA recycling’, and this mRNA regulation affects miRNA stability [3]–[7]. Finally, all miRISC components would be subjected to degradation, nucleases have been shown to degrade miRNAs (reviewed in [8]), and autophagy mediates the degradation of Dicer, Argonaute [9], [10] and GW182 [11].
In the nematode Caenorhabditis elegans (C. elegans), the miRNA pathway comprises over 120 miRNAs [12], two GW182 homologues (ain-1 and ain-2) [13], [14], the Argonautes alg-1 and alg-2 (both referred to as alg-1/2) [15] and single genes for Dicer (dcr-1) [15], [16] and Drosha (drsh-1) [17]. In worms, as in other animals, the miRNA pathway is essential for development and reproduction. Animals mutant for dcr-1 or drsh-1 genes are sterile [15]–[17], while at the Argonaute level, the loss of both alg-1/2 results in embryonic arrest [15], [18]. In contrast, single mutants of alg-1 or alg-2 display differentially penetrant post-embryonic, somatic and germ line defects [19], [20]. As exemplified here, the existence of these two gene paralogs, with specialized and partially redundant functions provides an opportunity to study the miRNA pathway in a sensitized genetic condition where miRNA activity is reduced albeit not completely abolished, by screening for genetic enhancers of the partial loss-of-miRNA condition.
In the present study, we identify the vps-52 gene, encoding a component of the GARP complex, as a genetic interactor of the miRNA-specific alg-1 Argonaute, and establish that this complex fulfills a positive modulatory role in regulating the activity of miRNAs. The loss of vps-52 in distinct sensitized genetic backgrounds induces the reiteration of the miRNA-controlled proliferative seam cell division program, enhances the let-7-related lethal phenotype, exacerbates the abnormal vulval development associated to the lessened miRNA-regulation of the let-60 gene, as well as augments the defective expression of a reporter of the lsy-6 miRNA activity in the ASEL neuron. Our phenotypic analyses thus suggest a broad role for GARP in miRNA function. Underpinning these GARP effects, we found decreased abundance of miRNAs and the GW182 proteins. Based on our data, we propose that the GARP complex operation facilitates a transition of miRISC occurring at endomembranes.
Results
The gene vps-52 is a genetic interactor of the microRNA pathway Argonaute alg-1
In order to identify new components and modulators of the miRNA pathway, we conducted a forward genetic screen for interactors of the alg-1/2 Argonautes, based on a design that allows the recovery of gene enhancers, including synthetic lethal gene pairs [21]. In brief, we subjected to mutagenesis worms carrying a partially inheritable extrachromosomal array containing a functional GFP-tagged alg-2 gene expressed in the alg-2(ok304) mutant (referred to as alg-2 mutant) background. F2 clones were scored and selected as candidate interactors if their progeny was uniformly transgenic (i.e. there was no segregation of viable worms lacking the extrachromosomal array in the population), indicative of a possible genetic interaction of an unknown mutated factor, in the transgenic setting, with the alg-2(ok304) background. Upon removal of the screening background (array and alg-2 mutation), a strain with increased growth and fertility defects in alg-1/2(RNAi) was selected, mapped and mutations identified by whole-genome sequencing (further details in the Materials and Methods section). We transgenically rescued the growth and fertility defects of the mutant strain, confirming the identity of the genetic interactor as vps-52. In addition to the single allele vps-52(qbc4) retrieved from the screen (Figure 1A), an available deletion allele, vps-52(ok853) was studied and found to display similar phenotypes (Figure 2). As expected, vps-52 behaved genetically as an enhancer, but interestingly double mutants of vps-52 with either alg-2 or alg-1 were obtained as viable strains, and the loss of vps-52 induced a visible phenotypic enhancement in combination with the loss-of-function alg-1(gk214) mutant (referred to as alg-1(0)), but not with alg-2(ok304) as reported below. The mutant strain initially isolated from the screen does not sustain the segregation of viable animals without the extrachromosomal array, which could be accounted for by array-mediated overexpression effects of the GFP::ALG-2 fusion protein or additional secondary background mutations; its segregation was not further investigated.
Fig. 1. Features and mutants of the vps-52 gene. 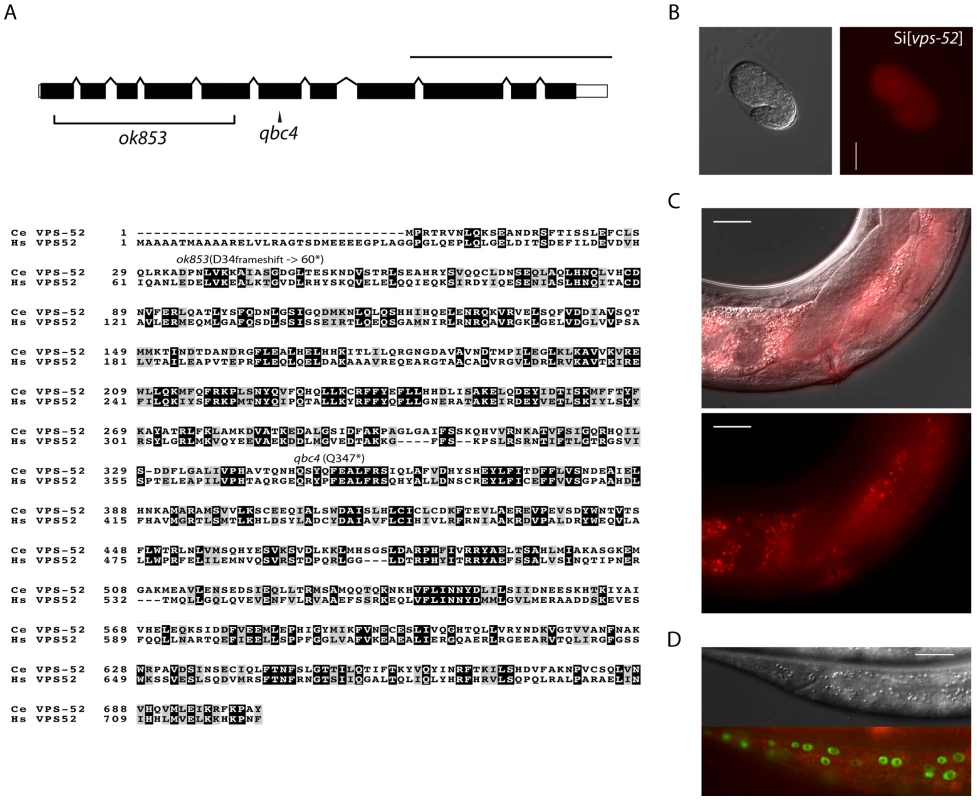
A) (Top) Schematic representation of the intron-exon structure of the C. elegans vps-52 gene. The two studied alleles are indicated (the allele qbc4 is a non-sense mutation and the allele ok853 is a frameshift deletion). VPS-52 contains predicted coiled coils stretches, but no distinct protein domains have been identified. (Bottom) vps-52 is conserved from worms to humans. ClustalW alignment (edited with Boxshade program) of human (Hs) and C. elegans (Ce) VPS-52 orthologs, the proteins share a 56% of sequence similarity. The amino acid changes introduced by the two alleles are indicated. B–D) Expression of a single copy insertion of Pvps-52::vps-52::mCherry::vps-52 3′UTR (referred to as Si[vps-52]). B) The expression of VPS-52 in early embryos. Micrographs of Nomarski (left panel) and mCherry fluorescence (right panel). Scale bar measures 20 µm. C–D) The vps-52 gene is widely expressed in somatic tissues. C) VPS-52 expression in the vulva and somatic gonad. Top panel: micrograph of merged Nomarski with mCherry fluorescence pictures. Bottom panel: micrograph of mCherry fluorescence. The VPS-52 protein localizes to the cytoplasm within punctated intracellular foci. Scale bar measures 20 µm. D) Expression of VPS-52::mCherry in the hypodermal cells. Micrographs of Nomarski (top) and merged GFP and mCherry fluorescence (bottom). Tagged VPS-52 (in red) is expressed in the cytoplasm around the seam cells nuclei (in green, GFP tagged). Scale bar measures 20 µm. Fig. 2. Seam cell-related phenotypes of the GARP mutants. 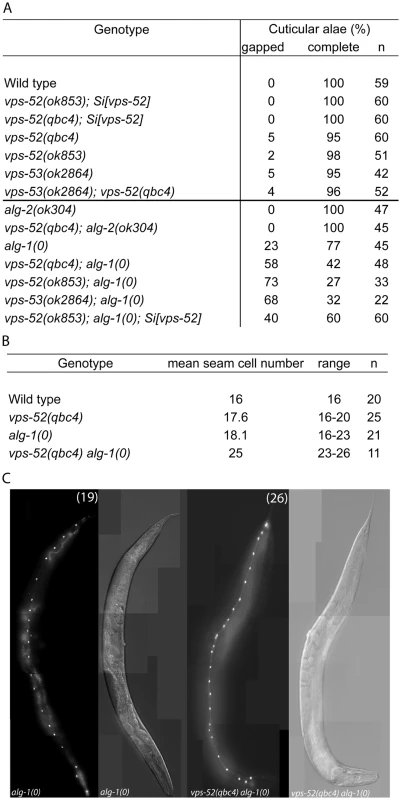
A) Percentage of young adult worms with defective cuticular alae. Young adult hermaphrodites from synchronized populations were mounted and analyzed under Nomarski optics. The continuity of the cuticular alae was scored and categorized as gapped (one or more interruptions) or complete (no gaps present). Strains above the horizontal black line were scored at 20°C, those below that line were grown and scored at 15°C. B) Average and range of seam cells number at adulthood. The indicated strains were crossed with a strain expressing GFP in the seam cells (scm::GFP) and the number of seam cells scored in young adult hermaphrodites grown at 15°C. The mean value and range covered by the individual counts are indicated. C) Representative average of the seam cells at adulthood. The indicated mutants were crossed with a strain expressing GFP in the seam cells (scm::GFP) and scored. The pictures correspond to GFP and DIC fluorescence, the number of seam cell nuclei is indicated within parentheses. The number of animals scored (n) is indicated (A–B). The vps-52 gene encodes a conserved structural protein (Figure 1A), that functions in the traffic of vesicles to the trans-Golgi network (TGN). It forms part of a conserved TGN-localized multimeric complex, known as the GARP (Golgi Associated Retrograde Protein) complex [22], [23], which comprises in C. elegans the genes vps-51, vps-52, vps-53 and vps-54 [24]. Expressing a functional fluorescently tagged VPS-52 from a single copy genomic insert controlled by endogenous gene regulatory elements (referred to as Si[vps-52]), we detected widespread VPS-52 expression in cytoplasmic puncta of many somatic tissues from early embryos on (Figure 1B–D), consistent with a previous report [24]. In particular, VPS-52 is expressed in temporal continuity at all larval stages in the vulval and seam cells (Figure S1).
GARP mutants enhance the seam cell defects of the animals lacking the Argonaute alg-1
To address whether the phenotypes of vps-52 mutants reflect an impairment of the GARP complex activity, we included in our study a strain defective for another subunit of this complex, the vps-53(ok2864) mutant. To analyze the function of vps-52 and vps-53 in the miRNA pathway, we first studied the development of seam cells. These lateral rows of hypodermal cells have a postembryonic developmental program, consisting of patterned rounds of division during each larval stage (L1 to L4), ended by terminal differentiation encompassing exit from the cell cycle, cell fusion and production of a cuticular structure (named alae) at the transition to adult. The seam cell developmental program is controlled at different larval stages by the miRNA lin-4 [25] and those of the let-7 family (miR-48, miR-84, miR-241 and let-7) [26], [27] and their targets lin-14, lin-28, hbl-1, daf-12 and lin-41 [26]–[33]. The repetition of the symmetrical seam cell division program that normally occurs once at the L2 stage is a frequently observed defect in mutants of core components of the miRNA pathway, such as alg-1/2, dcr-1 and ain-1/2 [13], [15]. Similarly to these mutants, other pathway modulators and components also display distinctive seam cells defects [34]–[36]. Discontinuities in the cuticular alae (in particular, gaps) arise from inappropriate terminal differentiation, and are an indicator of possible alterations in the development of seam cells. We analyzed the vps-52 and vps-53 mutants and noticed mild penetrant defects on the alae structures (Figure 2A) that were not exacerbated in the vps-52; vps-53 double mutant, demonstrating epistasis consistent with the affiliation of both gene products to a common complex.
We then proceeded to analyze the defects of the vps-52 and vps-53 mutants (referred to as GARP mutants) in the absence of functional alg-1/2 Argonautes. Given that defects of the alg-1 null mutant are less penetrant at lower temperatures (data not shown), we conducted the following scoring of alae and seam cell counts under more beneficial condition (15°C) to allow for better phenotypic enhancement. No enhancement of alae defects was observed in vps-52(qbc4); alg-2 double mutant animals (Figure 2A). However, combining either vps-52 or vps-53 with the alg-1 null mutant caused a very prominent increase in alae defects, which were partially rescued by transgenic vps-52 expression (Figure 2A). The enhancement of the alg-1(0) alae defects was correlated with an increase in the number of seam cells. While single GARP mutants showed minor deviations from the wild-type lineage (16 seam cells at adulthood), the mean number of seam cells in alg-1 mutant at 15°C (18 cells), reached 25 in vps-52(qbc4) alg-1(0) (Figure 2B–C). This increase in seam cells was not observed in the L1 larval stage (data not shown), indicating that it likely results from the reiteration of the L2 stage proliferative division of the seam cells. We then addressed whether the effects of vps-52 mutant on the seam cell phenotype of alg-1(0) worms were recapitulated with related mutants of vesicle trafficking processes. We tested the effect of disrupted Golgi trafficking by impairing the action of the worm small GTPase rab-6.2, whose gene product physically interacts with the GARP complex [24]. While single mutants of the putative null rab-6.2(ok2254) did not display any gapped alae, the exposure of rab-6.2(ok2254) to alg-1(RNAi) caused a strong interruption in the continuity of alae (Table S1). Moreover, this disruption was much stronger than that obtained for vps-52(qbc4) assayed under the same experimental conditions (100% vs 41% gapped alae; Table S1). We conclude that the loss of vps-52 or vps-53 function does not prominently affect seam cell development, but effectively synergizes in the absence of ALG-1 to induce reiteration of seam cell division program. Our observations of similar synergy for the mutant of rab-6.2, implicate the Golgi-associated function of these genes and miRNA-controlled development of the seam cells.
The loss of GARP enhances the defects of miRNA mutants in a target-dependent manner
The let-7 family members miR-48, miR-84 and miR-241 redundantly regulate the expression of their target gene hbl-1 during the L2-to-L3 larval stage transition [26], [28]. Abolishing completely the function of these three miRNAs causes seam cells to reiterate their L2 developmental program [26]. Loss of singleton or pairs of these genes leads to seam cell defects of varied penetrance, thereby constituting useful sensitized genetic backgrounds where the action of pathway modulators can be unveiled, as previously exemplified for the nhl-2 modulator [36]. It was therefore possible that the altered seam cell development observed in alg-1 mutant and enhanced by loss of vps-52 resulted from impaired miR-48, miR-84 and miR-241 miRNAs function. Consequently, we evaluated whether GARP mutants would alter seam cell development in the absence of miR-48, with the mir-48(n4097) mutant allele (referred to as mir-48(0)). Although loss of this miRNA did not induce seam cells defects on its own, the concomitant loss of vps-52 did lead to increase gapped or absent alae (Table 1). We next investigated the genetic interaction of vps-52 with hbl-1, a main target of the let-7 miRNA family that controls developmental timing at the L2 stage. Reduced hbl-1 function results in the precocious terminal differentiation of the seam cells at the third larval molt evidenced by the production of alae [28], [36]. The combination of the reduced-function allele, hbl-1(ve18), with a vps-52 mutant resulted in the partial suppression of the hbl-1(ve18) precocious phenotype (Table 1 and Figure S2). Similarly, suppression of this hbl-1 mutant phenotype has been reported upon concomitant loss of the let-7 miRNA family [36]. Altogether, these results suggest that loss of vps-52 diminishes the activity of let-7 family miRNAs. This effect likely underpins the enhanced defects in the developmental program of the seam cells observed with the alg-1(0) and mir-48(0) mutants.
Tab. 1. Alae defects for the vps-52, hbl-1 and mir-48 mutant animals. 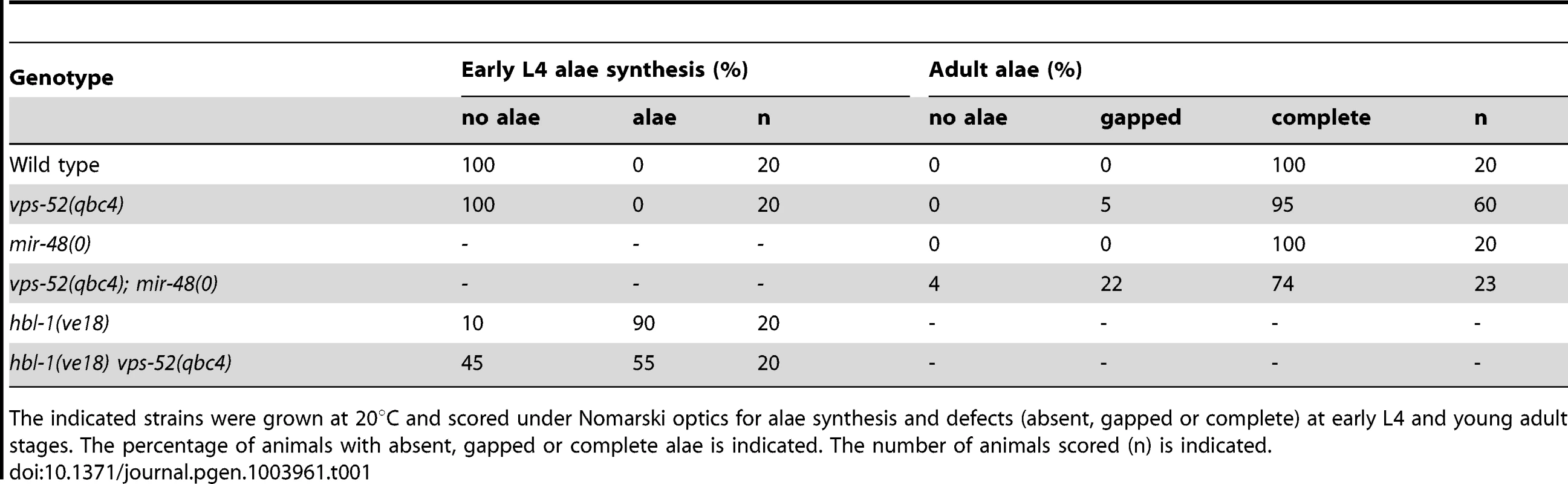
The indicated strains were grown at 20°C and scored under Nomarski optics for alae synthesis and defects (absent, gapped or complete) at early L4 and young adult stages. The percentage of animals with absent, gapped or complete alae is indicated. The number of animals scored (n) is indicated. We further extended our analysis to the let-7 miRNA, which regulates developmental programs at the L4 to adult transition. The complete loss of the let-7 miRNA gene produces a highly penetrant phenotype of bursting through the vulva [27] that is classically used in phenotypic assessments of miRNA functions (e.g. [37]). In this respect, we did not observe any bursting phenotype in the GARP mutants (Table 2). In addition, the bursting of GARP mutants in combination with alg-1(0) was not overtly different from that of single alg-1 mutant (Table 2). We then investigated if there was any effect on sensitized let-7 genetic backgrounds. We used, to that aim, the hypomorphic let-7 mutation, the n2853 allele, that carries a point mutation in the miRNA seed region, which leads to reduced mature let-7 miRNA level [27], [38] and to temperature-sensitive reduction in the activity of this miRNA [27]. Remarkably, the GARP mutants strongly enhanced the let-7(n2853) bursting phenotype at permissive temperature (Table 2). The penetrance of the defect prevented the propagation of double vps-52(qbc4) let7(n2853) mutants to useful populations, but, upon combination with the transgenic vps-52 gene, a rescued viable strain could be obtained (Table 2). The let-7 miRNA represses expression of lin-41, one of its major target genes; and reducing lin-41 function suppresses the bursting phenotype of let-7 mutants [27], [32], [39]. We therefore decided to verify whether the lin-41 impairment could suppress the penetrant bursting of the double vps-52(qbc4) let-7(n2853) mutant. The combination of the lin-41(ma104) hypomorph with vps-52(qbc4) let-7(n2853) resulted in a viable triple mutant and the complete loss of bursting (Table 2). Thus, the effect of vps-52 on the sensitized let-7 background is dependent on its target, lin-41.
Tab. 2. Lethal vulva bursting phenotype observed in different mutant backgrounds. 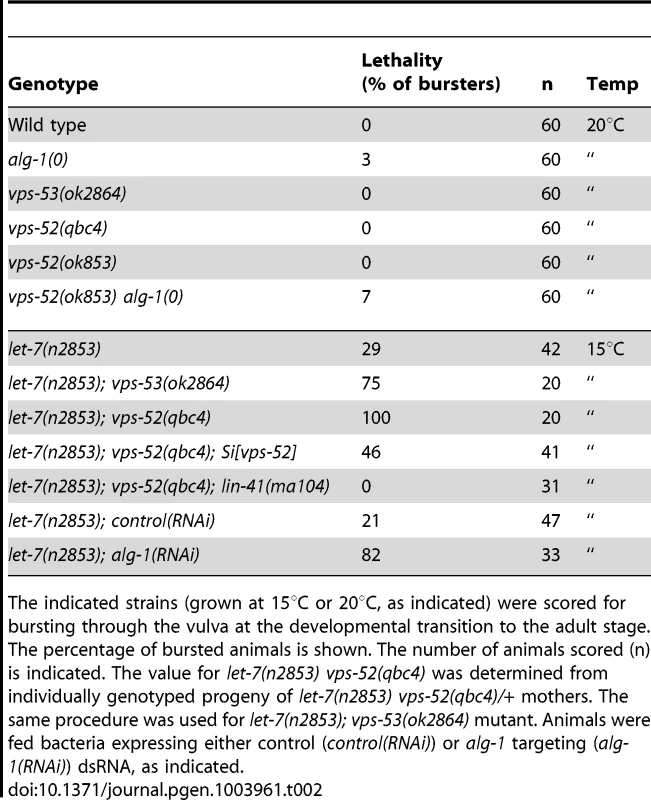
The indicated strains (grown at 15°C or 20°C, as indicated) were scored for bursting through the vulva at the developmental transition to the adult stage. The percentage of bursted animals is shown. The number of animals scored (n) is indicated. The value for let-7(n2853) vps-52(qbc4) was determined from individually genotyped progeny of let-7(n2853) vps-52(qbc4)/+ mothers. The same procedure was used for let-7(n2853); vps-53(ok2864) mutant. Animals were fed bacteria expressing either control (control(RNAi)) or alg-1 targeting (alg-1(RNAi)) dsRNA, as indicated. We next expanded our study to an additional target of the let-7 miRNA family through the investigation of the regulation of vulva development by the miRNA-targeted let-60 gene. During vulva development, an inductive signal from the somatic gonad promotes the vulval cell fates among a set of vulva precursor cells (VPCs). A restricted subset of VPCs adopts vulval fates and produces the egg-laying organ, while the remaining VPCs acquire a non-vulval epidermal fate [40]. The response to the inductive gonadal signal in the VPCs, requires the action of the let-60 gene (RAS homolog). Increased let-60 activity results in ectopic adoption of the vulval fate by VPCs and supernumerary vulva-like structures (known as the Muv phenotype) [41], [42]. The let-7 family miRNAs participate in this developmental process through the regulation of their target gene let-60 [36], [43]. The miRNA-mediated regulation of this gene can modulate the penetrance of the Muv phenotype in let-60 gain-of-function mutants [36]. Similar to wild type worms, GARP mutants did not display the Muv phenotype (Figure 3A). A missense gain-of-function mutation, let-60(n1046) produces a weakly penetrant Muv phenotype in heterozygous condition (Figure 3A). The concomitant impairment of vps-52 in let-60(n1046)/+ mutants exacerbated the percentage of defective Muv animals (4% vs 32%; Figure 3A). This result suggests that vps-52 gene function contributes to the negative regulation of let-60 activity during vulva development, likely through the regulation of the let-7 family miRNAs.
Fig. 3. Effects of vps-52 on the miRNA-mediated regulation of vulva and ASEL neuron development. 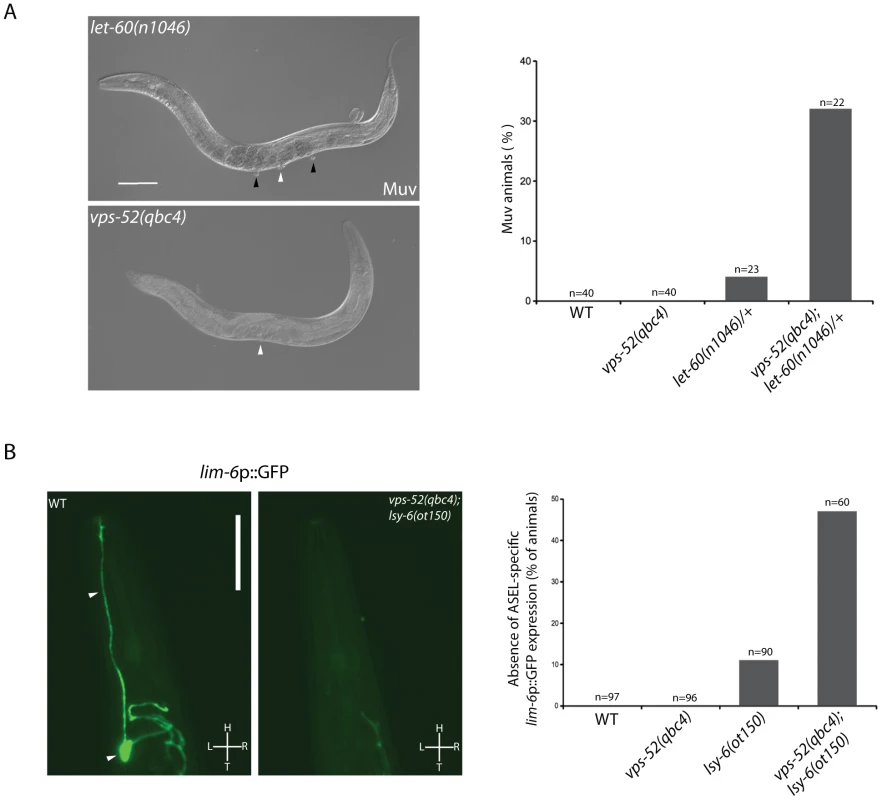
A) Analysis of the let-60-mediated Muv phenotype. Left panels: Nomarski pictures of adult hermaphrodites of the indicated genotypes. A single vulva (white arrowhead) is observed in the GARP mutant. In let-60(n1046) mutants multiple vulval-like structures (black arrowheads, Muv phenotype) are observed in addition to vulva (white arrowhead). Scale bar measures 100 µm. Right panel: Percentage of adult worms of the indicated genotypes with the Muv phenotype. B) Analysis of the lsy-6-mediated ASEL-specific gene expression. Left Panels: GFP fluorescence pictures of the lim-6p::GFP reporter. In wild type (WT) animals, gene silencing mediated by the lsy-6 miRNA leads to GFP expression in the ASEL neuron (arrowheads). Impairment of the miRNA activity in vps-52(qbc4); lsy-6(ot150) mutant blocks reporter expression. Scale bar measures 50 µm. L: Left, R: Right, H: Head, T: Tail. Right panel: Percentage of adult worms of the indicated genotypes that failed to express the lim-6p::GFP reporter in ASEL. The strains were grown and scored at 15°C. The number of animals scored (n) is indicated. In order to address if GARP fulfills a restricted or broad modulatory activity on miRNA function, we then studied the regulation of ASEL neuron development by the lsy-6 miRNA. The lsy-6 miRNA promotes the adoption of a unique cell fate by the ASEL chemosensory neuron. This results in a functional right-left asymmetry between it and its counterpart, the ASER neuron [44]. In the ASEL neuron, the reduced expression of the lsy-6 targeted gene cog-1 leads to a subsequent gene regulatory cascade that activates ASEL-specific gene expression. In particular, the lsy-6-mediated silencing of cog-1 expression leads to transcriptional derepression of lim-6 [44]. Thus, a transcriptional fluorescent reporter of lim-6 (lim-6p::GFP) serves as an indicator of achieved lsy-6-mediated cog-1 silencing, when its expression is switched on in ASEL [44]. In wild type and vps-52(qbc4) animals, this reporter was expressed in the ASEL neuron of every worm (Figure 3B). In contrast, complete absence of lsy-6 activity causes the total loss of expression of the reporter from the ASEL neuron [44]. While the hypomorphic lsy-6(ot150) mutation induced a mild penetrant loss of expression of this reporter (Figure 3B) [45], the loss of vps-52 in lsy-6(ot150) mutants augmented the absence of reporter expression in ASEL (11% vs 47%; Figure 3B). This result suggests that vps-52 gene function facilitates the activity of the lsy-6 miRNA in silencing cog-1 expression that subsequently contributes to the establishment of ASEL-specific gene expression.
In summary, our phenotypic analysis indicates that vps-52 and vps-53 mutants display weakly penetrant miRNA-related defects, but synergize with the alg-1 and miRNA mutants in enhancing developmental defects in a miRNA target-dependent manner. In addition, the loss of vps-52 suppresses the precocious phenotypes of a reduced function hbl-1 mutant, increases the misregulation of the miRNA-targeted gene let-60 during vulva development, as well as enhances the defective gene silencing activity of an hypomorphic lsy-6 mutant in the ASEL neuron. These collective results, obtained in the context of distinct miRNA-dependent phenotypes in sensitized genetic backgrounds, suggest therefore, a positive and general role for the GARP complex in miRNA function.
The loss of GARP leads to reduced abundance of the GW182 protein and miRNAs
In order to gain insights into the steps at which vps-52 regulate miRNA activity, we investigated the physical association of VPS-52 with components of the microRNA pathway in C. elegans. Using immunoprecipitation, we did not observe physical interaction between VPS-52 and the miRISC components AIN-1 or ALG-1 (data not shown). We next investigated if the GARP mutations affected the abundance of key miRNA pathway proteins in C. elegans. While we did not detect any effect on the DCR-1 or ALG-1/2 protein levels (Figure S3), the abundance of the GW182 AIN-1 protein was reduced in both vps-52 and vps-52 alg-1 mutants (Figure 4A) and this without a corresponding decrease in the expression at the mRNA level of the ain-1 gene (Figure S3B). In order to determine if this effect of vps-52 was specific to ain-1, we addressed genetically the effect of loss of vps-52 in ain-1 function. Interestingly, vps-52(qbc4) and the loss-of-function ain-1(ku322) mutant synergized, as their combination induced a strong enhancement of seam cell defects (Figure 4B). Thus, vps-52 may also modulate miRNA activity in parallel to ain-1. It is likely that the ain-2 gene, which encodes a second GW182 protein that provides redundant miRISC function with AIN-1 [13], is also affected by the loss of vps-52. We therefore examined the effect of vps-52 on a AIN-2::GFP translational reporter [13] to overcome the lack of specific antibodies. We observed that the knockdown of alg-1 in the vps-52(qbc4) mutant induced a moderate decrease of AIN-2::GFP abundance notably in vulva cells (Figure S4). The moderate effect detected with this transgenic GW182 reporter may be due to its inability to recapitulate the dynamics and expression level of the endogenous AIN-2 protein. Altogether these results support that the GARP complex may impinge on the abundance of the GW182 protein.
Fig. 4. Effects of vps-52 on the GW182 protein and GW182 mutant animals. 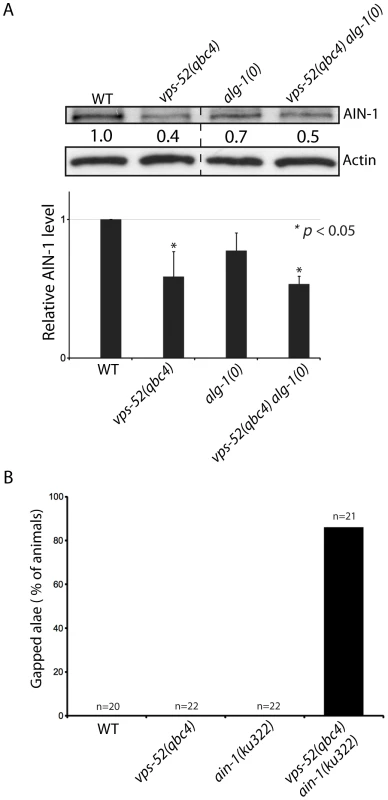
A) Abundance of the GW182 protein, AIN-1. (Top) Populations of the indicated genotyped were synchronized to the adult stage and Western blot performed on these samples. Actin served as loading control. (Bottom) Quantification of the AIN-1 signal compared with the level found in wild type worms (WT: 1). The error bars represent standard deviation from three independent experiments. p values were obtained using a two-sided Student's test. B) Synergy between vps-52 and ain-1 mutants. The indicated strains were grown at 15°C and the continuity of the cuticular alae analyzed in young adult animals. The number of animals scored (n) is indicated. Next, we determined the effects of the alteration of vps-52 on miRNA levels. Considering that several of the observed genetic interactions are congruent with reduced activity of the let-7 family miRNAs, we monitored their abundance. While in the single vps-52 mutant, we observed a significant decrease of the let-7 family miRNA members miR-48 and miR-241, their levels were further reduced in vps-52 alg-1(0) double mutant (Figure 5). Moreover, this reduced miR-48 and miR-241 abundance was rescued to the levels found in the single alg-1(0) mutant upon expression of a vps-52 rescue transgene (Figure 5). The level of primary and precursors miRNA molecules remained intact or was mildly altered (Figures S5A and 5), supporting that the decreased miRNA abundance is not likely due to diminished transcription or biogenesis of these miRNAs. The diminished miRNA abundance induced by the loss of vps-52 was not restricted only to these two let-7 family miRNAs. In vps-52 mutant, the abundance of three other miRNAs tested was also significantly reduced as observed for miR-48 and miR-241 (Figure S5C). This result supports that the loss of vps-52 impinges on a broad or general manner on miRNAs. We conclude that the GARP-mediated modulation of miRNA function, observed upon loss of GARP in alg-1(0) and multiple other mutants of miRNAs and their targets, is likely underpinned by the lowered abundance of the miRISC component GW182 as well as reduced miRNA abundance.
Fig. 5. Synergistic effects of vps-52 on the abundance of mature let-7 family miRNAs. 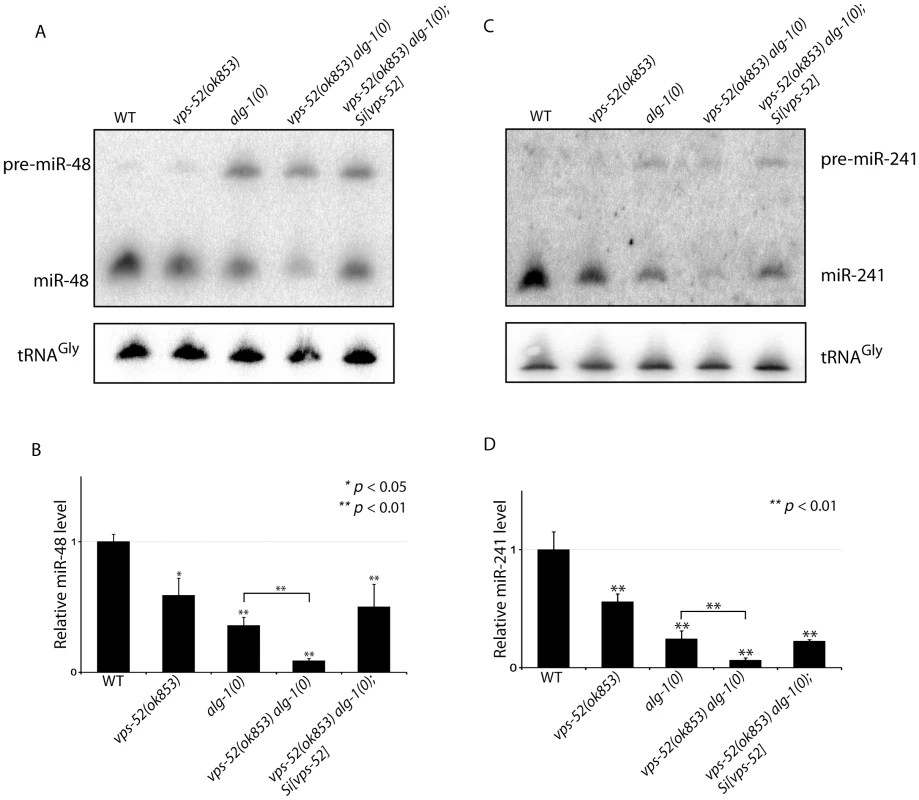
(A, C) The precursor and mature forms of miR-48 (A) and miR-241 (C) were investigated by Northern blotting in mid-L3 synchronized animals. The tRNAGly was used as control RNA. (B, D) The abundance of the miR-48 (B) and miR-241 (D). miRNAs level was measured by quantitative real-time PCR (TaqMan assay) in mid-L3 synchronized mutant animals and compared with the level found in wild type worms (WT: 1). The small nucleolar RNAs sn2841 was used as the normalization control. The error bars represent the standard deviation of three independent experiments and p values were obtained using a two-sided Student t-test. Discussion
Employing a genetic screen for alg-1/2 Argonaute interactors, we have identified the gene vps-52, encoding a component of the GARP complex, as a genetic enhancer of the miRNA pathway activity in C. elegans. The vps-52 mutants displayed weakly penetrant miRNA defects, and correspondingly mild molecular alterations of the pathway. However, upon vps-52 loss, a positive role was uncovered by the induction of increased defects in sensitized mutant backgrounds for Argonautes, miRNAs and their targets. The phenotypes and interactions we observed for vps-52, establish it as a modulator of miRNA activity rather than a core pathway component, which is incidentally the type of factors expect to be retrieved from a modifier genetic screen.
We encountered similar phenotypes for the mutants of vps-52 and vps-53, and both synergized with the alg-1 and let-7 mutants. Given that this two genes encode components of the GARP complex, it is likely that the observed regulation of miRNA activity corresponds to the impairment of a GARP complex function, rather than other putative additional role(s) of these genes. The GARP complex is involved in the tethering of endosome-derived vesicles reaching the TGN, in a recycling pathway known as ‘retrograde transport’ (reviewed in [46]). This pathway allows the recycling of proteins such as the cation-independent mannose 6-phosphate receptors in mammalian cells, and its impairment in both mammals and yeast leads to protein missorting [22], [47]. Consequently, we hypothesize that the mechanism underlying the observed regulation of miRNA activity is related to the known function of the GARP complex in tethering endosome-derived vesicles at the TGN and the consequences of it [46]. Noteworthy, a different sensitized let-7 background was previously employed in a genome-wide screen for regulators of the miRNA pathway [37]. Among the top hits identified were components of the COG (Conserved Oligomeric Golgi) complex, a vesicle tethering complex [48], exemplifying again the link between Golgi trafficking functions and miRNA activity.
Numerous studies have now reported on several endomembrane-related aspects of the miRNA pathway in diverse organisms: 1) the presence of Argonaute at the Golgi of certain cultured cells [49], [50]; 2) the co-fractionation of pathway components with Multivesicular Bodies (MVBs) and the negative effects of disrupting components of this compartment [4]; 3) the association of Argonautes and Dicer with the endoplasmic reticulum [51], [52]; 4) the miRNA regulatory effects of the BLOC-3 complex [53]; 5) those of disrupting the isoprenoid producing enzymes of the mevalonate pathway [54], [55] and; 6) the selective autophagic degradation of Dicer and Argonaute [9], [10] and C. elegans AIN-1 (GW182) protein [11]. Although all these findings may be underpinned by different mechanisms, they highlight the importance of membrane-regulated aspects on the function of the miRNA pathway. The impairment of autophagy leads to suppression of defective miRNA-mediated gene silencing in C. elegans and affects the abundance of AIN-1 [11]. Thus, it is conceivable that lysosomal mediated pathways, such as autophagy, underpin the lessened GW182 abundance we observed upon the loss of GARP.
An interesting possibility regarding the membrane association of the miRNA pathway is that it relates to the sorting and secretion of miRNAs in MVB-derived exosomes. Indeed, circulating miRNAs have been detected in diverse body fluids and proposed to be involved in some form of intercellular communication (reviewed in [56]). Similarly, if miRNA secretion were eventually used to alter gene expression in other cells, it should be expected to alter the cell autonomy of miRNA action to certain extent. In C. elegans this aspect has been only studied for lin-4 in the seam cells, where this miRNA functions cell autonomously [57]. Nonetheless, the sorting of the miRNA and pathway components in exosomes, their putative secretion and role in intercellular communication in C. elegans and other organisms are research topics that require further exploration.
An alternative and non-mutually exclusive possibility with that of miRNA secretion, is that the membrane association of the miRNA pathway could be part of a process that facilitates certain transitions occurring during the course of miRNA action [5]. This process being facilitating rather than necessarily required, its absence would only impair, but not completely abolish, miRNA-mediated gene regulation. It can be envisioned that the miRISC components would be associated with endomembranes, such as endosomal vesicles, to facilitate a transition of the miRISC, including, possibly, the recycling of miRISC components, its assembly or disassembly. In this context, impairing the GARP complex function in retrieving endosomal vesicles carrying miRISC components or other factors required for proper miRISC activity would have two foreseeable consequences: i) missorting of miRISC components, such as the GW182 proteins and; ii) a ‘block’ of the miRISC at membranes. As a result, the affected complexes would be disallowed from engaging in further repression of target mRNAs, and also from participating in the accumulation of new miRNAs. Although a role for GW182 in regulating miRNA stability has been recently proposed in mammalian cells [58], other reports indicated that the abundance of miRNAs is not affected by the absence of the GW182 proteins [13], [59], suggesting that miRNA abundance and GW182 accumulation can be uncoupled. Consistently with these last reports, we did not find a decrease in miRNA abundance upon the loss of AIN-1 (Figure S5B–C).
In addition to the processes of miRNA biogenesis and core effector functions, the understanding of subsequent phase(s) of microRNA activity will likely unveil the existence of new components that facilitate miRNA activity and regulate its recycling and turnover. The present trends of discoveries suggest that these facets may be, at least in part, dependent on processes occurring at the interface of endomembranes. Future studies will be required to establish the mechanism by which GARP regulates miRNA activity. Similarly, further research will be helpful to better understand the mechanisms by which membrane-based processes regulate the function of miRNAs.
Materials and Methods
Culture conditions and general methods
Worms were cultured in standard conditions [60]. All experiments were performed at 20°C unless otherwise noted. The strains used in the present study were outcrossed four times before analysis (detailed on the strains can be found in Text S1). The RNAi by feeding was performed on nematode growth media (NGM) plates containing 1 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) after overnight induction (25°C) of the bacterial culture. The alg-1/2(RNAi) was performed as described in [15]. The alg-1(RNAi) was performed with a construct targeting the alg-1-specific N-terminal gene region [35].
Genetic screen and mutant identification
The genetic screen followed a design to isolate enhancers of the queried gene (including synthetic lethal interactors) previously used in C. elegans [21], [61]. EMS was used to mutagenize alg-2(ok304) worms carrying an extrachromosomal array containing GFP::alg-2 copies (strain MJS11). From a pool of 1,000 mutagenized fluorescent F1, F2 clones where the non-integrated array becomes necessary for worm survival, evidenced by a homogeneous population of GFP expressing animals were kept and further investigated. Next, single mutant strains (with no transgenic array or alg-2(ok304)) were obtained and tested as follows. Upon outcrossing with wild type N2 males, a random set of F2 worms (chosen to be GFP negative, without the alg-2(ok304) deletion), was fed with alg-1/2(RNAi) and the mendelian segregation of a discernible miRNA-related phenotype (gapped alae or bursting) in the RNAi condition was assessed. One single mutant strain displaying increased defects upon alg-1/2(RNAi) with the expected segregation frequency was selected for further characterization.
Using alg-1/2(RNAi) the mutant locus was SNP mapped to the X chromosome in the genetic interval (−2.9, −0.76 cM). Mutations inside the interval were unveiled by whole genome sequencing (in collaboration with Dr Don Moerman and the British Columbia Cancer Agency). Only a single nonsense mutation inside the interval (in the F08C6.3 gene) was found. A transgenic strain carrying the wild-type vps-52 gene, rescued all the visible defects of the mutant strain. To note, the obtained vps-52(qbc4) mutant is sensitive to both germline and somatic RNAi (data not shown).
Plasmids
The Mos transposase plasmid (pJL44) and co-injection markers (pGH8, pCFJ90, pCFJ104) were used following the MosSCI method [62]. To generate the vps-52 rescue plasmid MSp166 (Pvps-52::vps-52::mCherry::vps-52 3′UTR), two genomic regions comprising the whole vps-52 gene were amplified, introducing a NotI site adjacent to the stop codon and terminal restriction digestion sites (AvrII and BsiWI). Upon NotI ligation, the resulting gene fragment was introduced into double digested (AvrII, BsiWI) pCFJ151 plasmid and verified by sequencing. Using the introduced C-terminal NotI site, a mCherry NotI cassette was ligated to produce MSp166. The oligonucleotides used for plasmid construction are listed in Text S1.
Transgenic strains
A single copy transgenic line containing the promoter, coding sequence and both 5′ and 3′ untranslated regions of the vps-52 gene was obtained as follows. Mutant unc-119(ed9) worms (strain EG4322) were injected with a mix of the MSp166 plasmid, Mos transposase and marker plasmids following the MosSCI method [62]. The obtained transgenic lines were processed according to the heat-shock protocol of the method. The integrity of the single copy insert was tested by PCR and sequencing. The obtained line (carrying the qbcSi01 transgene) was then used for crosses.
Western blotting
Synchronized worm populations of the desired stage were disrupted and dissolved in Laemmli buffer. Protein bands in the immunoblots were visualized using the western lighting plus ECL kit (Perkin-Elmer). Antibodies were used at the following dilutions. Actin-HRP (1∶10,000); rabbit anti-ALG-1 (1∶5,000); rat anti-AIN-1 (1∶10,000) [13]; rabbit anti-ALG-1/2 (1∶2,000) [63]; rabbit anti-DCR-1 (1∶5,000) [64].
Real-time quantitative PCR
Total RNA from synchronized worm populations was prepared using Tri-Reagent (Sigma-Aldrich). Reverse transcription was performed with the high capacity cDNA reverse transcription kit (Life technologies). A 7900HT PCR system was used for quantitative real-time PCR. SYBR Green I (Invitrogen) was used to monitor pri-miRNA and mRNA levels. TaqMan small RNA assays (Life technologies) were used to measure miRNA levels (miR-48/miR-241, let-7) following the manufacturer protocol. The sn2841 (small nucleolar RNA) Taqman assay was used as control. The oligonucleotides used for qPCR are listed in Text S1.
Northern blotting
Total RNA was separated by gel electrophoresis, transferred into a Genescreen plus membrane (Perkin-Elmer) and crosslinked using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) (Sigma) as described in [65]. DNA probes radiolabeled with the Starfire system (IDT) were hybridized to the membrane. After washing, the membrane was exposed to an image plate and scanned with the FLA-5100 phosphoimager. Image quantification was done using the ImageGauge 4.1 (Fujifilm) software. The oligonucleotide probes are listed in Text S1.
Supporting Information
Zdroje
1. KrolJ, LoedigeI, FilipowiczW (2010) The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics 11 : 597–610.
2. FabianMR, SonenbergN (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature structural & molecular biology 19 : 586–593.
3. BaccariniA, ChauhanH, GardnerTJ, JayaprakashAD, SachidanandamR, et al. (2011) Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Current biology 21 : 369–376.
4. GibbingsDJ, CiaudoC, ErhardtM, VoinnetO (2009) Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11 : 1143–1149.
5. GibbingsD, VoinnetO (2010) Control of RNA silencing and localization by endolysosomes. Trends Cell Biol 20 : 491–501.
6. AmeresSL, HorwichMD, HungJH, XuJ, GhildiyalM, et al. (2010) Target RNA-directed trimming and tailing of small silencing RNAs. Science 328 : 1534–1539.
7. ChatterjeeS, FaslerM, BussingI, GrosshansH (2011) Target-mediated protection of endogenous microRNAs in C. elegans. Developmental Cell 20 : 388–396.
8. RueggerS, GrosshansH (2012) MicroRNA turnover: when, how, and why. Trends in biochemical sciences 37 : 436–446.
9. GibbingsD, MostowyS, JayF, SchwabY, CossartP, et al. (2012) Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nature cell biology 14 : 1314–1321.
10. DerrienB, BaumbergerN, SchepetilnikovM, ViottiC, De CilliaJ, et al. (2012) Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proceedings of the National Academy of Sciences of the United States of America 109 : 15942–15946.
11. ZhangP, ZhangH (2013) Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO reports 14 : 568–576.
12. WarfMB, JohnsonWE, BassBL (2011) Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA 17 : 563–577.
13. ZhangL, DingL, CheungTH, DongMQ, ChenJ, et al. (2007) Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Molecular Cell 28 : 598–613.
14. DingL, SpencerA, MoritaK, HanM (2005) The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Molecular Cell 19 : 437–447.
15. GrishokA, PasquinelliAE, ConteD, LiN, ParrishS, et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106 : 23–34.
16. KnightSW, BassBL (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293 : 2269–2271.
17. DenliAM, TopsBB, PlasterkRH, KettingRF, HannonGJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432 : 231–235.
18. Vasquez-RifoA, JannotG, ArmisenJ, LabouesseM, BukhariSI, et al. (2012) Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One 7: e33750.
19. BukhariSI, Vasquez-RifoA, GagnéD, PaquetER, ZetkaM, et al. (2012) The microRNA pathway controls germ cell proliferation and differentiation in C. elegans. Cell Res 22 : 1034–1045.
20. BouaskerS, SimardMJ (2012) The slicing activity of miRNA-specific Argonautes is essential for the miRNA pathway in C. elegans. Nucleic acids research 40 : 10452–10462.
21. FayDS, KeenanS, HanM (2002) fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes & development 16 : 503–517.
22. ConibearE, StevensTH (2000) Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell 11 : 305–323.
23. LiewenH, Meinhold-HeerleinI, OliveiraV, SchwarzenbacherR, LuoG, et al. (2005) Characterization of the human GARP (Golgi associated retrograde protein) complex. Experimental cell research 306 : 24–34.
24. LuoL, HannemannM, KoenigS, HegermannJ, AilionM, et al. (2011) The Caenorhabditis elegans GARP complex contains the conserved Vps51 subunit and is required to maintain lysosomal morphology. Mol Biol Cell 22 : 2564–2578.
25. AmbrosV, HorvitzHR (1984) Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226 : 409–416.
26. AbbottAL, Alvarez-SaavedraE, MiskaEA, LauNC, BartelDP, et al. (2005) The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Developmental Cell 9 : 403–414.
27. ReinhartBJ, SlackFJ, BassonM, PasquinelliAE, BettingerJC, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 : 901–906.
28. AbrahanteJE, DaulAL, LiM, VolkML, TennessenJM, et al. (2003) The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Developmental Cell 4 : 625–637.
29. LeeRC, FeinbaumRL, AmbrosV (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 : 843–854.
30. WightmanB, HaI, RuvkunG (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 : 855–862.
31. MossEG, LeeRC, AmbrosV (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88 : 637–646.
32. SlackFJ, BassonM, LiuZ, AmbrosV, HorvitzHR, et al. (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Molecular Cell 5 : 659–669.
33. GrosshansH, JohnsonT, ReinertKL, GersteinM, SlackFJ (2005) The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Developmental Cell 8 : 321–330.
34. BussingI, YangJS, LaiEC, GrosshansH (2010) The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. The EMBO journal 29 : 1830–1839.
35. JannotG, BajanS, GiguèreNJ, BouaskerS, BanvilleIH, et al. (2011) The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO reports 12 : 581–586.
36. HammellCM, LubinI, BoagPR, BlackwellTK, AmbrosV (2009) nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136 : 926–938.
37. ParryDH, XuJ, RuvkunG (2007) A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol 17 : 2013–2022.
38. BaggaS, BrachtJ, HunterS, MassirerK, HoltzJ, et al. (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122 : 553–563.
39. VellaMC, ChoiEY, LinSY, ReinertK, SlackFJ (2004) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev 18 : 132–137.
40. SternbergPW, HorvitzHR (1986) Pattern formation during vulval development in C. elegans. Cell 44 : 761–772.
41. BeitelGJ, ClarkSG, HorvitzHR (1990) Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348 : 503–509.
42. HanM, AroianRV, SternbergPW (1990) The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics 126 : 899–913.
43. JohnsonSM, GrosshansH, ShingaraJ, ByromM, JarvisR, et al. (2005) RAS is regulated by the let-7 microRNA family. Cell 120 : 635–647.
44. JohnstonRJ, HobertO (2003) A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 : 845–849.
45. SarinS, O'MearaMM, FlowersEB, AntonioC, PooleRJ, et al. (2007) Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176 : 2109–2130.
46. BonifacinoJS, HierroA (2011) Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol 21 : 159–167.
47. Perez-VictoriaFJ, MardonesGA, BonifacinoJS (2008) Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol Biol Cell 19 : 2350–2362.
48. MillerVJ, UngarD (2012) Re'COG'nition at the Golgi. Traffic 13 : 891–897.
49. CikalukDE, TahbazN, HendricksLC, DiMattiaGE, HansenD, et al. (1999) GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell 10 : 3357–3372.
50. TahbazN, CarmichaelJB, HobmanTC (2001) GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. The Journal of biological chemistry 276 : 43294–43299.
51. LiS, LiuL, ZhuangX, YuY, LiuX, et al. (2013) MicroRNAs Inhibit the Translation of Target mRNAs on the Endoplasmic Reticulum in Arabidopsis. Cell 153 : 562–574.
52. StalderL, HeusermannW, SokolL, TrojerD, WirzJ, et al. (2013) The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. The EMBO journal 32 : 1115–1127.
53. LeeYS, PressmanS, AndressAP, KimK, WhiteJL, et al. (2009) Silencing by small RNAs is linked to endosomal trafficking. Nature cell biology 11 : 1150–1156.
54. ShiZ, RuvkunG (2012) The mevalonate pathway regulates microRNA activity in Caenorhabditis elegans. Proc Natl Acad Sci U S A 109 : 4568–4573.
55. BrodersenP, Sakvarelidze-AchardL, SchallerH, KhafifM, SchottG, et al. (2012) Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc Natl Acad Sci U S A 109 : 1778–1783.
56. ChenX, LiangH, ZhangJ, ZenK, ZhangCY (2012) Secreted microRNAs: a new form of intercellular communication. Trends in cell biology 22 : 125–132.
57. ZhangH, FireAZ (2010) Cell autonomous specification of temporal identity by Caenorhabditis elegans microRNA lin-4. Developmental biology 344 : 603–610.
58. YaoB, LaLB, ChenYC, ChangLJ, ChanEK (2012) Defining a new role of GW182 in maintaining miRNA stability. EMBO reports 13 : 1102–1108.
59. JiangZ, YuN, KuangP, ChenM, ShaoF, et al. (2012) Trinucleotide repeat containing 6a (Tnrc6a)-mediated microRNA function is required for development of yolk sac endoderm. J Biol Chem 287 : 5979–5987.
60. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
61. BosséGD, RueggerS, OwMC, Vasquez-RifoA, RondeauEL, et al. (2013) The Decapping Scavenger Enzyme DCS-1 Controls MicroRNA Levels in Caenorhabditis elegans. Molecular Cell 50 : 281–287.
62. Frokjaer-JensenC, DavisMW, HopkinsCE, NewmanBJ, ThummelJM, et al. (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nature genetics 40 : 1375–1383.
63. WuE, ThiviergeC, FlamandM, MathonnetG, VashishtAA, et al. (2010) Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Molecular Cell 40 : 558–570.
64. DuchaineTF, WohlschlegelJA, KennedyS, BeiY, ConteDJr, et al. (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124 : 343–354.
65. DrinnenbergIA, WeinbergDE, XieKT, MowerJP, WolfeKH, et al. (2009) RNAi in budding yeast. Science 326 : 544–550.
Štítky
Genetika Reprodukčná medicína
Článek Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis inČlánek Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Molecular Recognition by a Polymorphic Cell Surface Receptor Governs Cooperative Behaviors in Bacteria
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis in
- Retrotransposon Silencing During Embryogenesis: Cuts in LINE
- Roles of XRCC2, RAD51B and RAD51D in RAD51-Independent SSA Recombination
- Parallel Evolution of Chordate Regulatory Code for Development
- A Genetic Approach to the Recruitment of PRC2 at the Locus
- Deletion of the Murine Cytochrome P450 Locus by Fused BAC-Mediated Recombination Identifies a Role for in the Pulmonary Vascular Response to Hypoxia
- Elevated Mutagenesis Does Not Explain the Increased Frequency of Antibiotic Resistant Mutants in Starved Aging Colonies
- Deletion of an X-Inactivation Boundary Disrupts Adjacent Gene Silencing
- Interplay between Active Chromatin Marks and RNA-Directed DNA Methylation in
- Recombinogenic Conditions Influence Partner Choice in Spontaneous Mitotic Recombination
- Crosstalk between NSL Histone Acetyltransferase and MLL/SET Complexes: NSL Complex Functions in Promoting Histone H3K4 Di-Methylation Activity by MLL/SET Complexes
- A New Role for the GARP Complex in MicroRNA-Mediated Gene Regulation
- RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Loss of DNMT1o Disrupts Imprinted X Chromosome Inactivation and Accentuates Placental Defects in Females
- Inhibition of the Smc5/6 Complex during Meiosis Perturbs Joint Molecule Formation and Resolution without Significantly Changing Crossover or Non-crossover Levels
- Disruption of Lipid Metabolism Genes Causes Tissue Overgrowth Associated with Altered Developmental Signaling
- Translation Initiation Factors eIF3 and HCR1 Control Translation Termination and Stop Codon Read-Through in Yeast Cells
- Recruitment of TREX to the Transcription Machinery by Its Direct Binding to the Phospho-CTD of RNA Polymerase II
- MYB97, MYB101 and MYB120 Function as Male Factors That Control Pollen Tube-Synergid Interaction in Fertilization
- Oct4 Is Required ∼E7.5 for Proliferation in the Primitive Streak
- Contrasted Patterns of Crossover and Non-crossover at Meiotic Recombination Hotspots
- Transposable Prophage Mu Is Organized as a Stable Chromosomal Domain of
- Ash1l Methylates Lys36 of Histone H3 Independently of Transcriptional Elongation to Counteract Polycomb Silencing
- Fine-Mapping the Genetic Association of the Major Histocompatibility Complex in Multiple Sclerosis: HLA and Non-HLA Effects
- Genomic Mechanisms Accounting for the Adaptation to Parasitism in Nematode-Trapping Fungi
- Decoding a Signature-Based Model of Transcription Cofactor Recruitment Dictated by Cardinal Cis-Regulatory Elements in Proximal Promoter Regions
- Removal of Misincorporated Ribonucleotides from Prokaryotic Genomes: An Unexpected Role for Nucleotide Excision Repair
- Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
- Activin Signaling Targeted by Insulin/dFOXO Regulates Aging and Muscle Proteostasis in
- Activin-Like Kinase 2 Functions in Peri-implantation Uterine Signaling in Mice and Humans
- Demographic Divergence History of Pied Flycatcher and Collared Flycatcher Inferred from Whole-Genome Re-sequencing Data
- Recurrent Tissue-Specific mtDNA Mutations Are Common in Humans
- The Histone Variant His2Av is Required for Adult Stem Cell Maintenance in the Testis
- The Maternal-to-Zygotic Transition Targets Actin to Promote Robustness during Morphogenesis
- Reconstructing the Population Genetic History of the Caribbean
- and Are Required for Growth under Iron-Limiting Conditions
- Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment
- Neuron-Specific Feeding RNAi in and Its Use in a Screen for Essential Genes Required for GABA Neuron Function
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
- Mouse BAZ1A (ACF1) Is Dispensable for Double-Strand Break Repair but Is Essential for Averting Improper Gene Expression during Spermatogenesis
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis
- Pathways-Driven Sparse Regression Identifies Pathways and Genes Associated with High-Density Lipoprotein Cholesterol in Two Asian Cohorts
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- and Are Required for Growth under Iron-Limiting Conditions
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



