-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Recombinogenic Conditions Influence Partner Choice in Spontaneous Mitotic Recombination
Mammalian common fragile sites are loci of frequent chromosome breakage and putative recombination hotspots. Here, we utilized Replication Slow Zones (RSZs), a budding yeast homolog of the mammalian common fragile sites, to examine recombination activities at these loci. We found that rates of URA3 inactivation of a hisG-URA3-hisG reporter at RSZ and non-RSZ loci were comparable under all conditions tested, including those that specifically promote chromosome breakage at RSZs (hydroxyurea [HU], mec1Δ sml1Δ, and high temperature), and those that suppress it (sml1Δ and rrm3Δ). These observations indicate that RSZs are not recombination hotspots and that chromosome fragility and recombination activity can be uncoupled. Results confirmed recombinogenic effects of HU, mec1Δ sml1Δ, and rrm3Δ and identified temperature as a regulator of mitotic recombination. We also found that these conditions altered the nature of recombination outcomes, leading to a significant increase in the frequency of URA3 inactivation via loss of heterozygosity (LOH), the type of genetic alteration involved in cancer development. Further analyses revealed that the increase was likely due to down regulation of intrachromatid and intersister (IC/IS) bias in mitotic recombination, and that RSZs exhibited greater sensitivity to HU dependent loss of IC/IS bias than non RSZ loci. These observations suggest that recombinogenic conditions contribute to genome rearrangements not only by increasing the overall recombination activity, but also by altering the nature of recombination outcomes by their effects on recombination partner choice. Similarly, fragile sites may contribute to cancer more frequently than non-fragile loci due their enhanced sensitivity to certain conditions that down-regulate the IC/IS bias rather than intrinsically higher rates of recombination.
Published in the journal: Recombinogenic Conditions Influence Partner Choice in Spontaneous Mitotic Recombination. PLoS Genet 9(11): e32767. doi:10.1371/journal.pgen.1003931
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003931Summary
Mammalian common fragile sites are loci of frequent chromosome breakage and putative recombination hotspots. Here, we utilized Replication Slow Zones (RSZs), a budding yeast homolog of the mammalian common fragile sites, to examine recombination activities at these loci. We found that rates of URA3 inactivation of a hisG-URA3-hisG reporter at RSZ and non-RSZ loci were comparable under all conditions tested, including those that specifically promote chromosome breakage at RSZs (hydroxyurea [HU], mec1Δ sml1Δ, and high temperature), and those that suppress it (sml1Δ and rrm3Δ). These observations indicate that RSZs are not recombination hotspots and that chromosome fragility and recombination activity can be uncoupled. Results confirmed recombinogenic effects of HU, mec1Δ sml1Δ, and rrm3Δ and identified temperature as a regulator of mitotic recombination. We also found that these conditions altered the nature of recombination outcomes, leading to a significant increase in the frequency of URA3 inactivation via loss of heterozygosity (LOH), the type of genetic alteration involved in cancer development. Further analyses revealed that the increase was likely due to down regulation of intrachromatid and intersister (IC/IS) bias in mitotic recombination, and that RSZs exhibited greater sensitivity to HU dependent loss of IC/IS bias than non RSZ loci. These observations suggest that recombinogenic conditions contribute to genome rearrangements not only by increasing the overall recombination activity, but also by altering the nature of recombination outcomes by their effects on recombination partner choice. Similarly, fragile sites may contribute to cancer more frequently than non-fragile loci due their enhanced sensitivity to certain conditions that down-regulate the IC/IS bias rather than intrinsically higher rates of recombination.
Introduction
Accidental DNA double strand breaks (DSBs) arise during unperturbed proliferation. Such “endogenous” or “spontaneous” chromosome breakage does not occur randomly throughout the genome, but at specific loci, often referred to as fragile sites. Fragile sites have been observed in organisms ranging from bacteria to mammals, suggesting that they might be a ubiquitous feature of the genome [1], [2], [3], [4]. Evidence points to the existence of multiple types of fragile sites that are distinguishable from one another based on its structure, function, and/or genetic requirement(s) for its stability [5], [6], [7].
Mammalian fragile sites are one of the most extensively studied naturally occurring breakage prone regions of the genome. They are classified as either “rare” or “common”, depending on their incidence among general population [4]. Rare fragile sites are found in less than 5% of the population. In most cases, rare fragile sites are tri-nucleotide repeats, expansion of which has been linked to conditions such as Fragile X-syndrome and Huntington disease [8]. In contrast, common fragile sites are present in all individuals and can be found on every chromosome, indicating that they are a normal component of the chromosome. Common fragile sites extend over large regions of the genome, from several hundred kilobases (kb) to over 1 megabase (Mb) with breaks or gaps occurring throughout these regions. There is no sequence determinant that defines common fragile sites [4], [9].
Studies have implicated a link between common fragile sites, genome instability, and cancer [9]. For instance, some fragile sites have been shown to be loci of frequent chromosome deletions, translocations, and/or viral genome integration [10], [11] as well as oncogenic chromosomal rearrangements (e.g. [12]). Combining these with the observations that some fragile sites in model organisms exhibit elevated rates of recombination, it has been proposed that mammalian common fragile sites are recombination hotspots and that increased recombination activities at these loci contribute to cancer.
Replication Slow Zone (RSZ) is a type of fragile site in budding yeast and a putative homolog of the mammalian common fragile sites. It was identified as loci of preferred chromosome breakage following inactivation of Mec1, the budding yeast homolog of ATR, where high levels of replication dependent single stranded DNA (ssDNA), a precursor to DSBs, accumulate [3], [13]. Like its mammalian counterpart, RSZs are relatively large (∼10 kb), and appear to be a normal component of the chromosome. Other similarities between the two include; (i) timing of their replication during normal S-phase, which occurs late, (ii) their sensitivity to mild replication stress and inactivation of the ATR/ATM family kinases, and (iii) the lack of a defining sequence determinant(s) [3], [14], [15], [16]. The mammalian ATR/ATM and their budding yeast homologs Mec1/Tel1 are conserved signal transduction proteins best known for their roles in S-phase and DNA damage checkpoint responses [17]. In addition, they also play essential roles in a number of fundamental DNA and chromosomal processes including genome duplication, meiotic recombination, and DNA repair (e.g. [3], [18], [19]).
Here, we utilized RSZ as a model to test the proposal that mammalian common fragile sites are recombination hotspots. Unexpectedly, we found that recombination rates at RSZ and non RSZ loci were comparable under all conditions tested, indicating that RSZs are not recombination hotspots. Based on these and other observations, we propose a model whereby regulation of the nature of the recombination outcome(s), irrespective of the overall recombination activity, may play a key role in controlling genome rearrangements.
Results
Experimental system
RSZ
During normal S phase, replication forks slow down as they enter the ∼10 kb regions located between active replication origins, referred to as Replication Slow Zones (RSZs). The mechanism and/or significance of the fork slowing down remains unknown, but is mechanistically linked to fragility at these loci [3]. In addition to prolonged fork stalling, chromosome breakage at RSZs requires the onset of Topoisomerase II and condensin mediated mitotic chromosome condensation/compaction [20]. Inactivation of Mec1, high temperature, and/or exposure to a modest concentration of hydroxyurea (HU) promotes RSZ breakage [21]. HU is an inhibitor of the ribonucleotide reductase (RNR) complex and causes a dose dependent reduction in intracellular dNTP levels. Consistent with the notion that RSZ breakage is promoted by insufficient dNTP pools, the breakage is suppressed by sml1Δ, a mutation shown to increase intracellular dNTP levels [22]. Evidence indicates that sml1Δ suppresses RSZ breakage by bypassing the fork slowing that normally occurs at these loci [3].
Recombination tester construct
A URA3 cassette flanked by short hisG direct repeats (hisG-URA3-hisG) was used [23] (Figure 1A; Figure S1). The rate of URA3 inactivation, inferred from the acquisition of resistance to 5FOA (hereupon referred to as 5FOAR), was estimated based on the methods of the median [24] (Figure S2; Figure 2). The nature of genetic alteration associated with URA3 inactivation in each 5FOAR colony was determined by Southern Blot analysis (Figure S3; Figure 3A). Possible mechanisms of URA3 inactivation in a diploid carrying a single copy of the tester construct are summarized in Figure 1C, excluding base pair alternations and small structural changes.
Fig. 1. Experimental system. 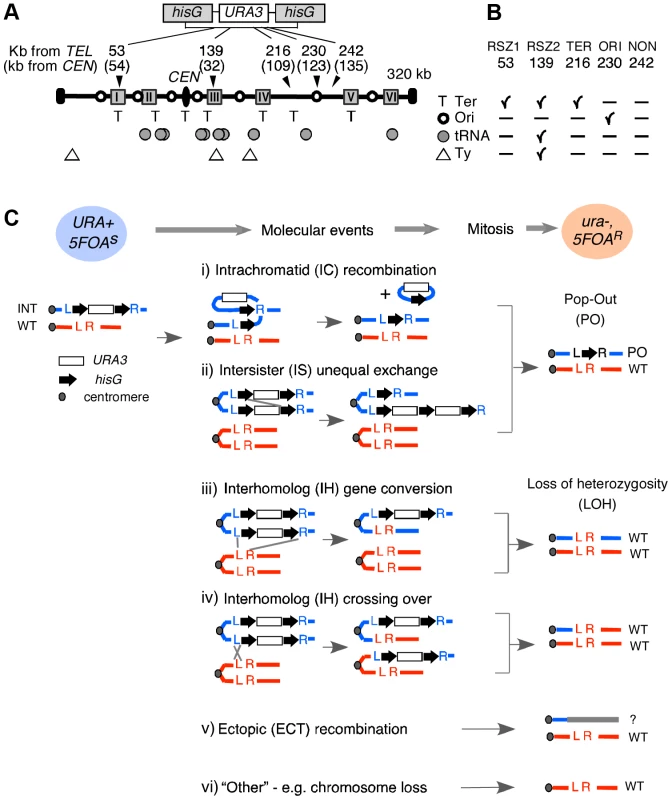
A. Recombination activity at five different loci in chromosome III (ChrIII) was assessed by monitoring URA3 inactivation in hisG-URA3-hisG reporter introduced at each of the five indicated site. The “kb from TEL” and “kb from CEN” are distances from the left telomere and the centromere (CEN) in kb, respectively. Grey boxes represent the six RSZs referred to as I through IV in ChrIII [3]. T: replication fork termination site; Open circle: active replication origin; Grey circle: tRNA gene; Triangle: Ty element. B. Summary of notable features at each locus examined. C. Mechanisms of URA3 inactivation. Heterozygous diploid strains carrying a single copy of hisG-URA3-hisG (blue circle; URA+) were grown under specified conditions and selected for 5FOA resistance. “Molecular events” summarize possible mechanisms of URA3 inactivation. Co-segregation of ura3 chromatids at mitosis would lead to a 5FOAR colony (pink circle) that no longer carries the hisG-URA3-hisG allele. The resulting diploid would carry either the “pop-out” and a single copy of WT (i, ii), two copies of WT (iii, iv), or just one copy of WT (v, vi). L and R; the immediate upstream and downstream sequences of each insertion locus utilized for targeted introduction of the tester construct via homologous recombination (Figure S1). INT: the allele containing the tester construct. Fig. 2. Rate of URA3 inactivation in YPD and in HU. 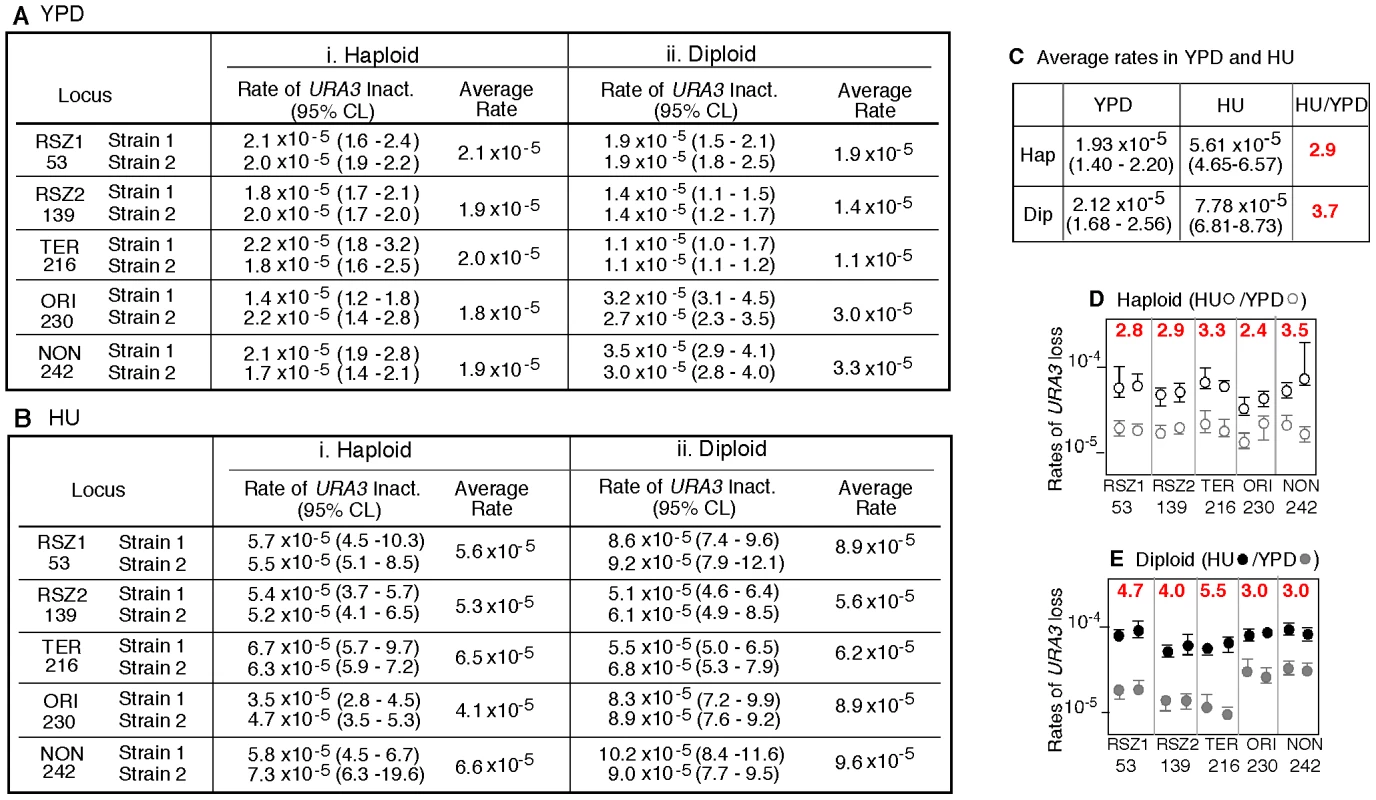
A, B. Rate of URA3 inactivation at the indicated locus in WT haploid- or diploid- strains grown in YPD (A) or in 10 mM HU (B). 95% Confidence Limits (CLs) for each value were calculated as previously described [47]. For each locus, rate measurements from two independently derived strains were obtained by the methods of median (Figure S2) [24]. C. The average recombination rate (and 95% CLs) of the haploid- and diploid- strains analyzed in A and B. “HU/YPD”: The effects of HU over YPD on recombination rate was expressed as the ratio between the two average rates. D, E. Effects of HU on rate of URA3 inactivation at each locus. Graphic representation of the data presented in A and B. Black and grey circles correspond to rate measurements in HU and YPD, respectively. Capped lines indicate 95% CLs. The red number at the top of each box is the ratio between the average rate values in HU and YPD at the indicated locus; the extent increase conferred by HU was statistically significant at every locus (Chi square test, p<0.05). Fig. 3. Intrachromatid and intersister (IC/IS) bias in URA3 inactivation. 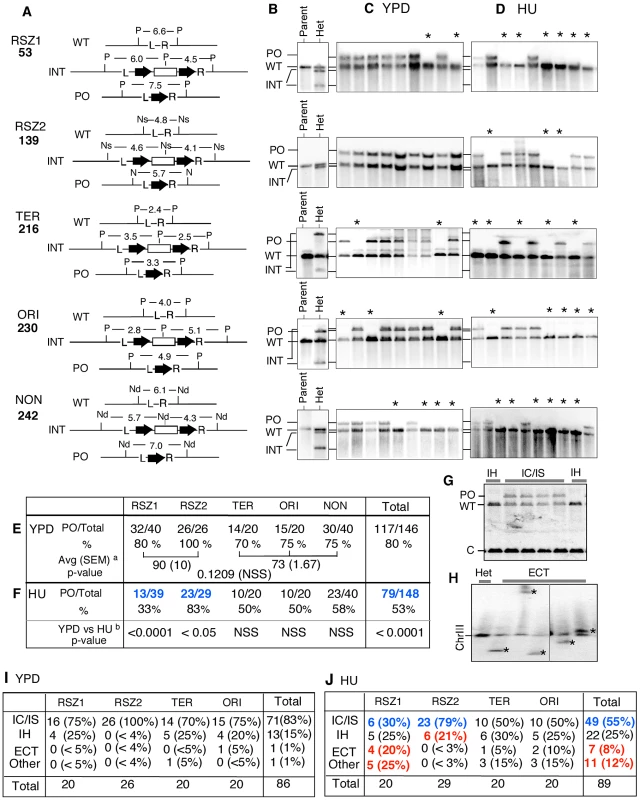
A. For each locus examined, Southern Blot analysis was utilized to monitor the presence of three relevant alleles; (i) WT, (ii) the allele containing the hisG-URA3-hisG reporter integrated, hereupon referred to as “INT” for “integrant”, and (iii) the “pop-out” or “PO” allele, indicative of an intrachromatid or intersister (IC/IS) recombination or recombination related event (Figure 1C). Genomic DNA samples were digested by the indicated restriction enzymes and Southern Blot analysis was performed using the L and the R sequences as a probe (Figure S3). The numbers represent expected sizes of WT, INT, and PO fragments in kb. P: Pst1, Ns: Nsi1, Nd: Nde1. hisG and URA3 sequences are represented by a block-arrow and open rectangle, respectively. B. Representative Southern Blot images of the “Parent” strain (ura3) carrying two copies of WT allele and a heterozygote or “Het”, derived from the parent carrying one WT and one INT allele at the specified locus. Indicated are the migrating positions for WT, INT, and PO fragments described in panel A. C, D. Representative images of Southern Blot analyses on 5FOAR colonies that arose in YPD or HU. All 5FOAR samples examined had lost the INT-fragments, suggesting that URA3 loss in each case was likely to be mediated by one of the mechanisms described in Figure 1B rather than a base pair substitution or small deletion or insertion. * Samples that have lost the INT fragment, but also lacking the diagnostic PO fragment, suggestive of a non IC/IS mediated URA3 inactivation event (See text). E, F. Fraction of 5FOAR colonies at each locus that had undergone URA3 inactivation via an IC/IS mediated event. For each locus, the number of 5FOAR samples exhibiting the diagnostic PO fragment and the total number analyzed are shown. a The average PO fraction of the two RSZs and the three non-RSZ loci. SEM: standard error of the mean. Unpaired student's t test was performed to determine whether the difference between the two averages was statistically significant. b Fischer's exact test was utilized to determine whether the extent of reduction in PO fraction conferred by HU at each locus was statistically significant. Numbers in blue indicate statistically significant reduction. NSS: Not Statistically Significant. G. A representative image of Southern Blot analyses showing 5FOAR colonies that had undergone URA3 inactivation via IH recombination. The occurrence of IH mediated events (either gene conversion or crossing over; Figure 1B) was confirmed by the lack of PO band and a two fold increase in the signal associated with the WT band relative to an IC/IS sample. To assess the WT copy number, the amount of radioactive signal associated with the WT-fragment in each sample was quantified and normalized to the signal found in a control band (“C”) in the same lane (Materials and Methods). H. A representative image of Southern Blot image of a pulse field gel (PFG), where the occurrence of ectopic events (ECT) in each sample was inferred by the presence of a novel chromosome size fragment that hybridized with hisG probe, a part of the hisG-URA3-hisG construct (*). I, J. Frequencies of IC/IS, IH, ECT, and other mechanisms of URA3 inactivation in 5FOAR colonies. Numbers in blue and red in panel J denote statistically significant decrease and increase conferred by HU compared to YPD, respectively. URA3 inactivation via either intrachromatid or intersister (IC/IS) recombination or recombination related process such template switching would result in the allele referred to as “pop-out” or “PO”, where the URA3 and one of the two hisG repeats “pop-out”, leaving only a single copy of the hisG (Figure 1Ci and ii). URA3 inactivation via interhomolog (IH) gene conversion or crossing over would lead to a loss of heterozygosity (LOH), where the WT allele replaces the reporter construct: the resulting 5FOAR colony would now carry two WT copies (Figure 1Ciii, iv). Ectopic recombination resulting in a translocation or chromosome loss would also lead to a LOH; however, unlike the IH-mediated LOH, a 5FOAR colony resulting from the latter is expected to carry only a single copy of WT allele (Figure 1Cv, vi).
In haploid strains, URA3 inactivation via IH recombination cannot occur, and chromosome loss would be lethal. As such, the majority of URA3 loss events in haploids is expected to be via IC/IS mediated events.
Loci analysed
The reporter was integrated into two RSZ and three non RSZ loci in chromosome III (ChrIII) via homologous recombination (Materials and Methods). Notable features associated with each locus include replication origins, replication termination zones, Ty elements, tRNA sequences, and the distance from the centromere (Figure 1A and 1B). 53 kb and 139 kb correspond to the approximate midpoints of an RSZ, each comprising about 10 kb wide. All six RSZs published to date, including the 53 kb and 139 kb, are found at or near a replication termination zone (Figure 1A) [3]. In addition, the RSZ at 139 kb is near a Ty element and several tRNA sequences. The three non RSZ loci chosen were 216 kb, 230 kb, and 242 kb, corresponding to a replication termination zone, a replication origin, and a ‘no feature’ locus, respectively (Figure 1A, B). Hereupon, the five loci are referred to as RSZ1, RSZ2, TER, ORI, and NON.
Tester strains
Two independent haploid strains carrying a single copy of the tester construct at one of the five loci were obtained by transformation, followed by sporulation and selection (Materials and Methods). These haploid strains were used to construct corresponding diploid strains in a WT background as well as both haploid and diploid strains in an sml1Δ, mec1Δ sml1Δ, or rrm3Δ, backgrounds. Control experiments confirmed that the presence of the construct did not impact fitness of the strains examined nor the RSZ-specific chromosome fragility following thermal inactivation of Mec1 (Figure S4).
RSZs are not recombination hotspots during unchallenged proliferation
The observations that stalled - and collapsed - replication forks can promote recombination [6], [7], [25], [26] and that RSZs are loci of slowed replication fork progression during normal S-phase [3], suggest that RSZs might be recombination hotspots during normal proliferation. (NB: In the current context, an arrested fork that can ultimately resume replication without intervention is referred to as a “stalled fork” whereas that requires an active fork-restart process is referred to as a “collapsed fork”). To test this, we compared the rate of URA3 inactivation at the RSZ and non RSZ loci under a standard yeast growth condition (2% glucose at 30°C, hereupon referred to as “YPD”). For each locus, the rate was estimated by the method of the median from two independently derived strains, and the average of the two was used for the locus-to-locus comparison (Figure S2) [24].
In haploids, the locus specific recombination rates varied very little, from 1.8×10−5 per cell generation at ORI to 2.1×10−5 at RSZ1 (Figure 2Ai). In diploids, the variation was greater, about 3 fold, and ranged from 1.1×10−5 per cell generation at TER to 3.3×10−5 at NON (Figure 2Aii). Importantly, the rate of URA3 inactivation at the two RSZs was not higher than the three non RSZ loci in either haploids or diploids. We conclude that RSZs are not spontaneous recombination hotspots, defined as loci of intrinsically higher recombination activities.
RSZs are not recombination hotspots in HU
Chromosome breakage at RSZs does not occur during normal proliferation, but is promoted by a modest level of HU, high temperature, and/or inactivation of Mec1 [3], [21]. Thus, it is possible that the putative recombination hotspot activity associated with RSZs might also require these conditions. To test this, we examined the effect of HU. The same ten haploid and ten diploid strains analyzed above were subjected to a transient (18 hour) exposure to 10 mM HU before 5FOA selection. We found that the exposure lead to a statistically significant increase in rate of URA3 inactivation at every locus (Figure 2D). Importantly, the rates at the two RSZs were not any higher than the three non RSZ loci in either haploids or diploids (Figure 2B). We conclude that RSZs are not recombination hotspots even under a condition that promotes RSZ specific chromosome breakage.
Intrachromatid and intersister (IC/IS) bias in mitotic recombination
Next, we examined the nature of genetic alternations associated with URA3 inactivation in diploids. To this end, we utilized Southern Blot analysis that enabled us to monitor the presence of the following three alleles (Figure 3A, Figure S3); (i) WT, (ii) the allele containing the hisG-URA3-hisG reporter integrated, hereupon referred to as “INT” for “integrant”, and (iii) the “pop-out” or “PO” allele, indicative of an intrachromatid or intersister (IC/IS) recombination or recombination related event (Figure 1C). As expected, the parent ura3 diploid strain exhibited a single band diagnostic of the WT allele at each of the five loci examined; in contrast, each of the URA3 heterozygotes derived from the parent exhibited an additional band(s), corresponding to the INT allele (Figure 3B, Figure S3).
We found that all 5FOAR colonies examined had lost the INT fragment, suggesting that URA3 inactivation in every case involved a relatively large structural change(s) at the integration locus (Figure 3C; data not shown). Overall, ∼80% (117/146) of the samples exhibited the diagnostic PO band, indicating that on average, URA3 inactivation was four times more likely to occur via an IC/IS-mediated event than all other mechanisms combined (Figure 3E “Total”). The latter is consistent with previous reports on strong IC/IS-bias in mitotic recombination (e.g. [27]). Locus specific PO fraction ranged from 70% (14/20) at TER to 100% (26/26) at RSZ2, suggesting that the extent of IC/IS bias might be influenced by local environment (Figure 3E). The average PO fraction for the two RSZs was higher than the three non-RSZ loci (90% versus 73%; Figure 3E); however the difference was not statistically significant (p = 0.1209). We conclude that recombination activity at RSZs during standard growth condition is comparable to non RSZ loci with regard to both the rate of recombination and the extent of the IC/IS bias.
HU down-regulates IC/IS-bias in mitotic recombination
The same Southern Blot analysis was performed on 5FOAR colonies that arose in the presence of HU. Similarly to the YPD samples, all 148 HU 5FOAR samples had lost the diagnostic INT fragment (Figure 3D; data not shown). Overall, 53% (79/148) of the HU 5FOAR colonies carried the PO allele, a significant reduction from the 80% (117/148) observed in YPD (Figure 3F “Total”; Chi square test, p<0.0001).
The negative effect of HU on IC/IS bias was observed at every locus; however, the only statistically significant reduction was at the two RSZs (Fisher's exact test; Figure 3F). At RSZ1, the PO fraction was reduced from 80%in YPD to 33% in HU, indicating that the 4∶1 bias toward IC/IS mediated URA3 inactivation in YPD was completely lost in HU, where the majority of URA3 inactivation occurred via non IC/IS mediated events. RSZ2 was notable because the extent of IC/IS bias in YPD appeared to be unusually strong (26/26; Figure 3E). In HU, we found that six of 29 had undergone a non IC/IS mechanism of URA3 loss, suggesting that the negative effect of HU might be irrespective of the intrinsic robustness of the IC/IS-bias.
HU promotes URA3 inactivation via interhomolog (IH), ectopic (ECT), and other mechanisms
In the current analysis, a 5FOAR colony exhibiting only the WT band (e.g. Figure 3CD, samples denoted by an “*”) was inferred to have undergone URA3 inactivation via a non IC/IS mediated event, such as an IH gene conversion/crossover, ectopic recombination (ECT), or chromosome loss (Figure 1C). As mentioned above, it is possible to distinguish an IH event from the others by the virtue of the fact that a 5FOAR colony that arose via an IH recombination event would carry two copies of WT allele, while the rest carries just one (Figure 1C).
Indeed, quantitative analysis of the WT fragments in Southern Blot images revealed that some of the non IC/IS samples had a two-fold greater signal associated with the WT band relative to an IC/IS sample that carried a copy of the WT and the PO allele each (Figure 3G, compare WT band intensity in “IH” and “IC/IS” lanes); these samples were inferred to have undergone URA3 inactivation via IH-recombination. We were also able to confirm the occurrence of an ectopic (ECT) event by the presence of a novel chromosome sized fragment containing the hisG sequence on a Southern Blot of a pulse field gel (PFG) (e.g. Figure 3F), suggestive of a chromosome translocation event. Applying these criteria, we were able to infer that the majority of 5FOAR colonies that arose in YPD (85/86) or in HU (78/89) had undergone URA3 inactivation via one of these three (IC/IS, IH, or ECT) mechanisms (Figure 3I and J). The rest, 1/86 in YPD and 11/89 in HU, was classified as “Other”, which would include mechanisms such as chromosome loss.
Of the 86 YPD 5FOAR colonies analyzed, about 25%, or 15, had undergone URA3 loss via a non IC/IS event(s). Among them, all but two were mediated by IH recombination. The remaining two corresponded to an ECT and an Other event each (Figure 3I, “Total”). In HU, a reduction in the fraction of IC/IS event was accompanied by a significant increase in the fractions of ECT and Other, where they rose to 8% (7/89) and 12% (11/89) from about 1% in YPD (Figure 3J, “Total”). The only significant increase in IH fraction conferred by HU was at RSZ2, where it rose from 0/26 in YPD to 6/29 in HU (Figure 3I and J).
sml1Δ promotes IC/IS bias in mitotic recombination
To test whether the effect of HU on the IC/IS bias might have been due to reduced dNTP levels, we examined the effect of sml1Δ. Sml1 is a negative regulator of RNR and its deletion leads to a ∼2.5 fold increase in dNTP levels [22]. We found that the mutation conferred a modest, but statistically significant increase in the overall PO fraction from 80% (117/146) in YPD to 89% (89/100) in sml1Δ (Chi-square analysis, p<0.05; Figure 4F, “Total”). The observed effects of HU and sml1Δ suggest a positive correlation between dNTP levels and the extent of IC/IS bias, and implicate dNTP availability in regulation of recombination outcomes and genome rearrangements.
Fig. 4. sml1Δ regulation of mitotic recombination. 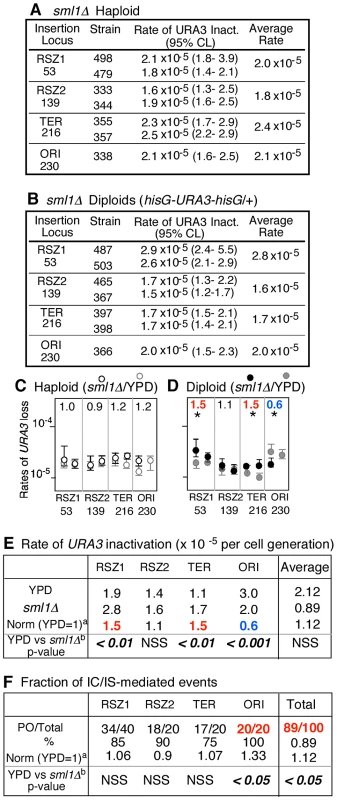
A, B. Rates of URA3 inactivation at the indicated locus in sml1Δ haploid and diploid strains grown in YPD. 95% Confidence Limits (CLs) for each value were calculated as previously described [47]. For each locus except for ORI, rate measurements from two independently derived strains were obtained by methods of median (Figure S2) [24]. C, D. Effects of sml1Δ on rate of URA3 inactivation. Black and grey circles correspond to rate measurements in sml1Δ (panels A and B) and WT (Figure 2A), respectively. Capped lines indicate 95% CLs. The number at the top of each box is the ratio between average rate values in sml1Δ and WT at each locus. * denotes a statistically significant change (Chi square test, p<0.05). Numbers in red and blue denote statistically significant increase or decrease, respectively. E. Locus specific effects of sml1Δ on rate of URA3 inactivation. a The average rate at the indicated locus in sml1Δ was normalized to the corresponding value in WT (Figure 2). Numbers in red and blue denote statistically significant increase or decrease, respectively (Chi square test, p<0.05). NSS: Not Statistically Significant. F. Effect of sml1Δ on IC/IS bias. Fraction of 5FOAR colonies that had undergone URA3 inactivation via an IC/IS mediated event a The fraction in sml1Δ at each locus was normalized to the corresponding value in WT (Figure 3E). b Statistical analysis (Fischer's exact test) was performed on the effects of sml1Δ. The number in red denotes statistically significant increase. The results also revealed that the effect of sml1Δ on IC/IS bias might be locus dependent; while the overall effect was an increase, the mutation actually lead to a modest decrease in the bias at RSZ2 from 100% (26/26) in WT to 90% (18/20) in sml1Δ (Figure 3E; Figure 4F). Similarly, we found that the effect of sml1Δ on recombination rate was locus specific; while the mutation lead to an increase at RSZ1, RSZ2, and TER, but it reduced the rate at ORI (Figure 4D, E). Overall, the effect of sml1Δ on ORI differed from the rest in that it was the only locus where the mutation conferred a statistically significant reduction in recombination rate and a significant increase in the IC/IS-bias. It would be require analysis of additional origin sequences to confirm whether the observed effect might be origin specific.
No significant effect of sml1Δ was observed on the rate of URA3 inactivation in haploids (Figure 4A, C).
Negative correlation between recombination activity and IC/IS bias in URA3 inactivation
If chromosome breakage is mechanistically linked to recombination activity, then HU, a condition that promotes RSZ specific chromosome breakage, should also confer an RSZ specific increase in the rate of URA3 inactivation. Unexpectedly however, we found that HU increased the rate at both RSZ and non RSZ loci, indicating that the breakage was not linked to recombination activity. To confirm this further, we examined the effects of additional conditions shown to regulate chromosome breakage at RSZs. Specifically, we chose high temperature and mec1Δ sml1Δ, the two conditions shown to promote the breakage, and rrm3Δ, a mutation shown to suppress it [21]. Rrm3 encodes a DNA helicase involved in replication fork progression through ∼1000 discrete fork pause sites in the budding yeast genome, whose inactivation leads to fork stalling at these loci [28], [29]. The rrm3Δ mediated fork stalling triggers Mec1/Tel1 dependent S phase checkpoint activation and Sml1 removal. The latter in turn, promotes fork progression through RSZ and prevents RSZ breakage even in the absence of Mec1 function [21], [30]. We limited our analyses to diploids only, where the effect on IC/IS bias can be assessed in addition to the rate of URA3 inactivation.
Results showed that high temperature (37°C), rrm3Δ, and mec1Δ sml1Δ increased the average rate of URA3 inactivation by ∼2.5 fold over WT at 30°C (Figure 5A, “Average”). Low temperature (23°C), on the other hand, lead to ∼30% reduction. These observations confirmed previous reports on recombinogenic effects of rrm3Δ and mec1Δ sml1Δ. Furthermore, they revealed a positive correlation between temperature and recombination rate, suggesting that temperature itself might regulate endogenous recombination activity. Importantly, the observed effect was not specific to RSZs, providing further support for the notion that chromosome fragility and recombination activity at RSZs are not mechanistically linked.
Fig. 5. Regulation of endogenous mitotic recombination. 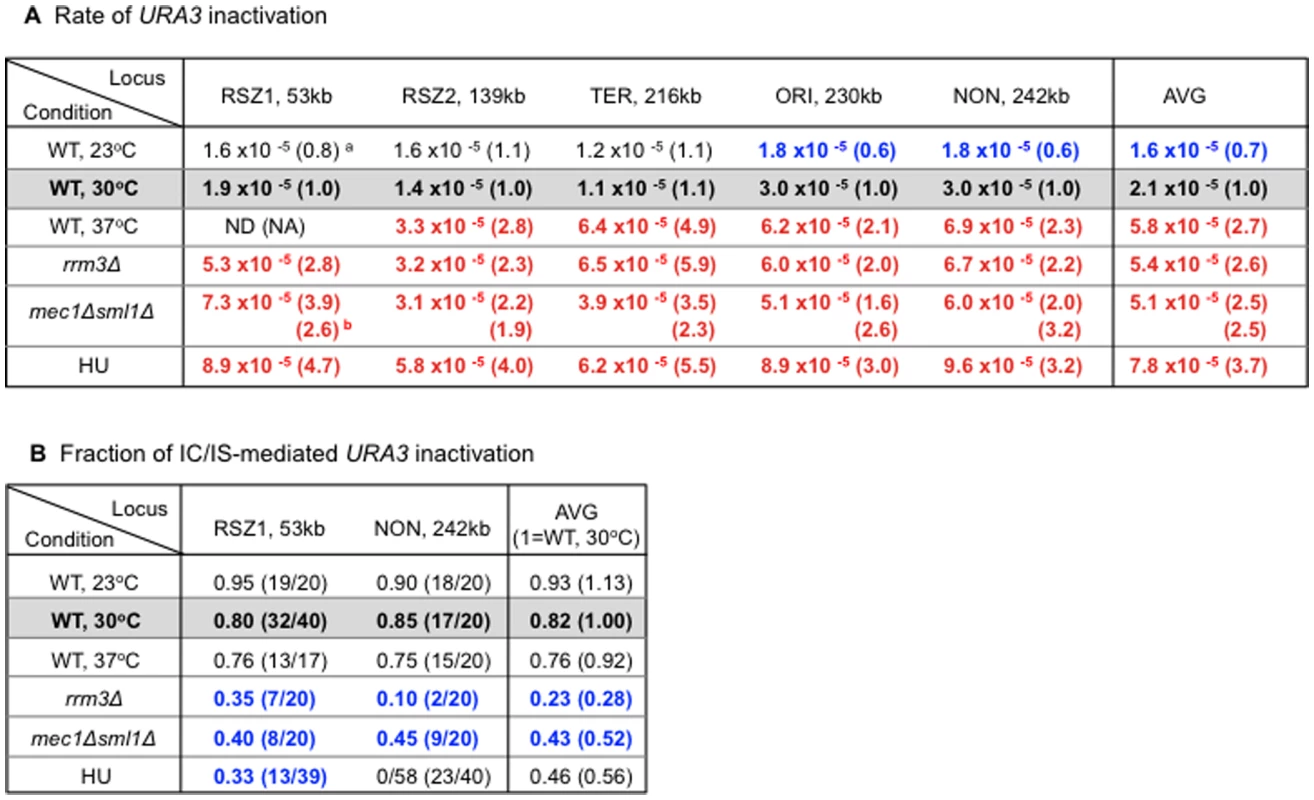
A. Regulation of recombination rate. Rate of URA3 inactivation at each locus in different genetic backgrounds and growth conditions. The numbers represent average rate value at each locus based on two independent rate measurements (Materials and Methods). a Each value is normalized to the corresponding WT value from YPD 30°C shown in blue box. Numbers in red and blue denote statistically significant increase or decrease, respectively. b For the mec1Δ sml1Δ measurements, normalization was also performed using the corresponding values in a sml1Δ background. B. Regulation of IC/IS-bias. Fraction of 5FOAR colonies exhibiting the PO fragment indicative URA3 inactivation via IC/IS-recombination at RSZ1 and NON. Blue numbers denote statistically significant decrease compared to the WT 30°C values. a Fisher's exact test was performed to assess whether the differences between the PO fractions at RSZ1 and NON observed in rrm3Δ and HU were statistically significant. P values for the rrm3Δ and HU values were 0.064 and 0.018, respectively, indicating that only the differences in HU was significant. Similarly to HU, the three recombinogenic conditions, 37°C, rrm3Δ, and mec1Δ sml1Δ lead to a reduction in the overall PO fraction (Figure 5B, “Average”). In contrast, low temperature (23°C), the only condition that decreased the rate of recombination, lead to a modest increase in the fraction. Together, these observations suggest an association between increased recombination activity and loss of IC/IS bias. Finally, results also showed that the effects of different conditions on IC/IS bias can be either RSZ specific, as in the case of HU, or general, as in the case of temperature, rrm3Δ, and mec1Δ sml1Δ (Figure 5B legend).
Discussion
RSZ is a homolog of the mammalian common fragile sites, noted for its sensitivity to dNTP depletion and inactivation of Mec1 and Tel1, the budding yeast ATR and ATM, respectively. Here, we utilized RSZs to address the proposal that mammalian fragile sites are recombination hotspots and that increased recombination activity at these loci contributes to cancer development. Our analyses revealed that; (i) RSZs are not recombination hotspots, (ii) recombinogenic conditions (e.g. rrm3Δ, mec1Δ, HU, and high temperature) down-regulate IC/IS-bias in mitotic recombination, and (iii) RSZs exhibit greater sensitivity to HU dependent loss of IC/IS bias. Below, we discuss each of these findings further.
RSZs are not recombination hotspots
We found that rates of URA3 inactivation at RSZs were comparable to non-RSZ loci under all tested conditions. Based on this, we conclude that RSZs are not recombination hotspots, defined as loci of increased recombination activity. A key assumption is that the URA3 inactivation event monitored in the current study is a readout for recombination activities at the locus, and not for an indirect effect(s) of a more distant element(s). For example, studies of LOH in diploids indicate that most LOH events occur by a crossover between the heterozygous loci, and that the frequency of these events increases as a function of distance from centromere (e.g. [31]). In addition, Ty elements are recombination hotspots that might affect recombination activities downstream of the loci (e.g. [32]). Notably however, the frequency of the LOH events monitored in the current study was independent of the distance from CENIII (Figure 3I, J). Similarly, the rates of overall URA3 inactivation at the loci downstream (the three non RSZ loci) and upstream (the two RSZ loci) of a Ty element were comparable (Figure 2B, 4B, and 5A). Furthermore, the approach employed in the current study – i.e. integration of a reporter construct at a specific locus - is a widely utilized means of assessing local recombination activities (e.g. [33]). Together, these considerations strongly suggest that the assay utilized in the current study monitors local recombination activity.
It was surprising that RSZs did not have intrinsically higher rates of recombination than non RSZs. However, the current observation is actually consistent with the fact that RSZ breakage occurs during prometaphase in the context of topoisomerase II - and condensin - dependent chromosome condensation [3], [20], while most spontaneous mitotic recombination occurs during S phase in the context of stalled - and collapsed-replication forks [5], [7], [26]. The apparent temporal separation and differential genetic requirements suggest that chromosome breakage and recombination at RSZs might each entail a process that is independently regulated. However, it is also possible that the lack of correlation is a feature specific to RSZ (and mammalian fragile sites, by extension) and not a general feature of a fragile site.
Regulation of endogenous mitotic recombination rate
Our results confirmed an earlier observation that the locus-to-locus variation in spontaneous mitotic recombination rate is relatively modest, in contrast to the variation in meiotic recombination rates, which can be several orders of magnitude [34], [35]. This difference is likely due, at least in part, to the nature of DNA structure that leads to recombination in each case: stalled - or collapsed-replication forks for mitotic recombination and developmentally programmed DSBs for meiotic recombination [26], [36]. Formation of meiotic DSBs are regulated at multiple levels, including targeted localization of Spo11, the enzyme that catalyzes the breakage, to the hotspot regions of the genome [37]. This in turn ensures that meiotic DSBs do not occur uniformly throughout the genome, but preferentially at DSB hotspots. In contrast, the occurrence of stalled - or collapsed-forks leading to spontaneous mitotic recombination does not appear to be regulated per se; rather, it appears to be a unintended consequence of a replication fork encountering a locus that is difficult to replicate, for example, due to either unusual DNA or chromatin structures or damaged DNA [6], [7], [25], [26], [38].
Previous studies have shown that genes encoding for proteins involved in processes such as replication fork progression, stalled fork integrity, and fork restart impact endogenous mitotic recombination rates (e.g. [39], [40]). The recombinogenic effects of HU or rrm3Δ observed in current study are likely due to increased incidences (either in the frequency and/or the duration) of fork stalling stemming from depletion of dNTP pools or loss of a replisome associated helicase activity, respectively [28], [41]. The effects of mec1Δ sml1Δ, on the other hand, is unlikely due to increased fork stalling because replication forks proceed faster in an sml1Δ background compared to WT [3]. Given that Mec1 is required for stability of stalled forks and is a key regulator of homologous recombination and recombination related processes involved in replication fork restart [18], [42], the recomginogenic effects of mec1Δ sml1Δ are likely to stem from a defect(s) in processes that occur after fork stalling. The mechanism(s) by which temperature might affect endogenous recombination activity remains unknown. Notably however, there has been a precedent in meiotic recombination, where temperature appears to play an important regulatory role(s) [43].
Regulation of IC/IS bias in mitotic recombination
On average, URA3 inactivation under the standard growth condition was four times more likely to occur via an IC/IS mediated event than all other mechanisms combined. We found that HU, high temperature, rrm3Δ, and mec1Δ sml1Δ abolished this bias while low temperature and sml1Δ enhanced it. These observations suggest that partner choice in mitotic recombination might be subjected to regulation, for example by factors like dNTP availability and temperature. During meiotic recombiantion, a significant fraction of meiotic DSBs is developmentally programmed to be repaired with an IH bias, using an intact homolog as a repair template, rather than a sister chromatid. Evidence indicates that such IH bias in meiotic recombination is mediated, at least in part, by expression of several meiosis specific chromosomal proteins that fundamentally alter meiotic chromatin structure; this in turn, favours physical interaction between the homologs while minimizing that between sister chromatids, overcoming the IC/IS bias intrinsic during mitotic recombination [44], [45], [46]. The latter implicates chromatin structure, notably the status of sister chromatid cohesion, in mitotic IC/IS bias.
Fragile sites and cancer
We utilized RSZ, a model for mammalian common fragile sites, to address the proposal that the fragile sites contribute to cancer due to increased recombination activity at the loci. The evidence presented here, however, suggests an alternative mechanism, which implicates the nature of recombination outcomes, rather than the overall recombination rate. The only RSZ specific recombination activity revealed in the current study was its greater sensitivity to HU induced loss of IC/IS bias. Although the sample size is limiting (i.e. one insertion in each of two RSZs), an implication would be that, depending on the nature of stress, recombination events at RSZs (and at mammalian common fragile sites, by extension), might be more likely to lead to a LOH and translocation, the type of alterations shown to contribute to cancer. Taken together, current observations provide a fresh insight into the ways in which fragile sites and other recombinogenic conditions may contribute to genome rearrangements.
Materials and Methods
Yeast strains
Relevant genotypes of the strains utilized in current study are summarized in Table S1. All URA3 strains were generated by standard yeast genetics procedures including transformation, mating, sporulation, and specific phenotype selections. A wild type haploid strain NKY291, was transformed with each of the five DNA fragments containing the hisG-URA3-hisG cassette flanked by ∼500 bp upstream (L) and ∼500 bp downstream (R) genomic sequences of the targeted loci. The fragments were prepared from integration plasmids (Figure S1) by NotI digestion and gel purification. Correct integration of hisG-URA3-hisG at each locus among randomly selected URA transformants was confirmed by Southern Blot analysis (Figure S3). Two independent transformants of each locus were mated with NKY292, a WT haploid strain of the opposite mating type to generate heterozygous diploid strains, from which URA3 haploids were re-derived. These haploid strains (JDCY 463, 465, 230, 233, 239, 243, 232, 233, 235, 237) were used for all subsequent strain construction.
Rate of URA3 inactivation
The rate of URA3 inactivation was determined by the method of median [24]. For each measurement, 15 colonies of comparable size (1.5–2 mm in diameter) freshly grown on YPD plates, were individually suspended in 5 mls of YPD or YPD+10 mM HU and cultured for 18 hours at 30°C (Figure S2A). Samples were then diluted in water and plated on YPD or 5FOA plates to measure the number of total viable cells or those that had undergone a URA3 inactivation event, respectively. To ensure that only the URA3 inactivation events that occurred during the 18 hour of unselected growth in liquid medium were included for the analyses, only the 5FOARcolonies of comparable size (1.5–2 mm) were counted following a three day incubation at 30°C. Statistical analyses on the rates of URA3 inactivation were assessed as described [47].
Molecular characterization of URA3-loss in 5FOAR recombinants
For each condition examined, a total of 20 or more independent 5FOAR colonies from each locus were subjected to molecular analysis (Figure S2B). Genomic DNA from each colony was restricted using the appropriate restriction enzyme (Figure S3; Figure 3) and subjected to Southern Blot analysis. For each locus, the L and R fragments used for plasmid construction (Figure S1) were used as probes. As a control for quantifying the relative signals associated with different diagnostic fragments (e.g. Figure 3E), a PCR amplified Mec1 fragment corresponding to nucleotide numbers 5539 to 7027 in the OFR was included as a probe (Table S2). Each restriction enzyme used for Southern analysis cleaves endogenous MEC1 sequence to generate a novel sized fragment that hybridizes to the MEC1 probe. To confirm the occurrence of ectopic recombination events, the candidate 5FOAR colonies were analyzed by Pulse Field Gel electrophoresis (PFGE) and Southern Blot analyses using hisG as a probe. For PFGE, Chromosome sized genomic DNA samples were prepared in low melting point agarous plugs as previously described [48]. Electrophoresis condition optimized for resolution around ChrIII was performed as described [49].
Supporting Information
Zdroje
1. Hill T (1996) Features of the Chromosome Terminus Region. In: Neidhardt F, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington DC: ASM Press. pp. 1602–1615.
2. DalgaardJ, KlarA (2000) swi1 and swi3 perform Imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102 : 745–751.
3. ChaRS, KlecknerN (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297 : 602–606.
4. Sutherland G, Hecht F (1985) Fragile sites on human chromosomes. New York: Oxford University Press.
5. RothsteinR, MichelB, GangloffS (2000) Replication fork pausing and recombination or “gimme a break”. Genes Dev 14 : 1–10.
6. MirkinEV, MirkinSM (2007) Replication fork stalling at natural impediments. Micro Mol Biol Rev 71 : 13–35.
7. LambertS, CarrAM (2013) Impediments to replication fork movement: stabilisation, reactivation and genome instability. Chromosoma 122 : 33–45.
8. BrouwerJ, WillemsenR, OostraB (2009) Microsatellite repeat instability and neurological disease. Bioessays 31 : 71–83.
9. DurkinS, GloverT (2007) Chromosome fragile sites. Ann Rev Gen 41 : 169–192.
10. GloverT, SteinC (1988) Chromosome breakage and recombination at fragile sites. Am J Hum Genet 43 : 265–273.
11. RassoolF, McKeithanT, NeillyM, Van MelleE, EspinaR, et al. (1991) Preferential integration of marker DNA into the chromosomal fragile site at 3p14: An approach to cloning fragile sites. Proc Nat Acad Sci 88 : 6657–6661.
12. PopescuN (2003) Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett 192 : 1–17.
13. FengW, Di RienziS, RaghuramanM, BrewerB (2011) Replication stress-induced chromosome breakage is correlated with replication fork progression and is preceded by single-stranded DNA formation. G3 1 : 327–335.
14. LairdC, JaffeE, KarpenG, LambM, NelsonR (1987) Fragile sites in human chromosomes as regions of late-replicating DNA. Trends in Genetics 3 : 274–281.
15. CasperA, NghiemP, ArltM, GloverT (2002) ATR Regulates Fragile Site Stability. Cell 111 : 779–789.
16. Ozer-GalaiE, SchwartzM, RahatA, KeremB (2008) Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene 27 : 2109–2117.
17. HarperJ, ElledgeS (2007) The DNA Damage Response: Ten Years After. Mol Cell 28 : 739–745.
18. CarballoJA, ChaRS (2007) Meiotic roles of Mec1, a budding yeast homolog of mammalian ATR/ATM. Chromosome Res 15 : 539–550.
19. SomyajitK, BasavarajuS, ScullyR, NagarajuG (2013) ATM - and ATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol Cell Biol 33 : 1830–1844.
20. HashashN, JohnsonA, ChaR (2012) Topoisomerase II - and condensin-dependent breakage of MEC1ATR-sensitive fragile sites occurs independently of spindle tension, anaphase, or cytokinesis. PLoS Genet 8: e1002978.
21. HashashN, JohnsonAL, ChaRS (2011) Regulation of fragile sites expression in budding yeast by MEC1, RRM3 and hydroxyurea. J Cell Sci 124 : 181–185.
22. ZhaoX, MullerEGD, RothsteinR (1998) A suppressor of two essential checkpoint genes identifies novel protein that negatively affects dNTP pools. Mol Cell 2 : 329–340.
23. AlaniE, CaoL, KlecknerN (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116 : 541–545.
24. LeaDE, CoulsonCA (1948) The distribution of the numbers of mutants in bacterial populations. J Genet 49 : 264–284.
25. BierneH, MichelB (1994) When replication forks stop. Mol Microbiol 13 : 17–23.
26. CarrA, PaekA, WeinertT (2011) DNA replication: failures and inverted fusions. Semin Cell Dev Biol 22 : 866–874.
27. KadykLC, HartwellLH (1992) Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132 : 387–402.
28. IvessaA, LenzmeierB, BesslerJ, GoudsouzianL, SchnakenbergS, et al. (2003) The Saccharomyces cerevisiae Helicase Rrm3p Facilitates Replication Past Nonhistone Protein-DNA Complexes. Mol Cell 12 : 1525–1536.
29. TorresJ, SchnakenbergS, ZakianV (2004) Sacchromyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells require the intra-S-phase checkpoint and fork restart activities. Mol Cell Biol 24 : 3198–3212.
30. ChengX, QinY, IvessaA (2009) Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p. Mol Genet Genomics 281 : 631–645.
31. BarberaMA, PetesTD (2006) Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci 103 : 12819–12824.
32. MieczkowskiP, LemoineF, PetesT (2006) Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair 5 : 1010–1020.
33. SchwachaA, KlecknerN (1994) Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76 : 51–63.
34. LichtenM, HaberJE (1989) Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123 : 261–268.
35. ManceraE, BourgonR, BrozziA, HuberW, SteinmetzLM (2008) High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454 : 479–485.
36. SzostakJW, Orr-WeaverTL, RothsteinRJ, StahlFW (1983) The double-strand-break repair model for recombination. Cell 33 : 25–35.
37. KugouK, FukudaT, YamadaS, ItoM, SasanumaH, et al. (2009) Rec8 guides canonical Spo11 distribution along yeast metioic chromosomes. Mol Biol Cell 20 : 3064–3076.
38. LambertS, MizunoK, BlaisonneauJ, MartineauS, ChanetR, et al. (2010) Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell 346–359.
39. MyungK, ChenC, KolodnerR (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411 : 1073–1076.
40. AdmireA, ShanksL, DanzlN, WangM, WeierU, et al. (2006) Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint contols in yeast. Genes Dev 20.
41. TimsonJ (1975) Hydroxyurea. Mut Res 32 : 115–132.
42. FriedelA, PikeB, GasserS (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21 : 237–244.
43. BornerGV, KlecknerN, HunterN (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 : 29–45.
44. SchwachaA, KlecknerN (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90 : 1123–1135.
45. NiuH, WanL, BaumgartnerB, SchaeferD, LoidlJ, et al. (2005) Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell 16 : 5804–5818.
46. CarballoJA, JohnsonAL, SedgwickSG, ChaRS (2008) Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132 : 758–770.
47. WierdlM, GreeneC, DattaA, Jinks-RobertsonS, PetesT (1996) Destablization of simple repetitive DNA sequences by transcription in yeast. Genetics 143 : 713–721.
48. BordeV, WuT-C, LichtenM (1999) Use of a recombination-reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol 19 : 4832–4842.
49. HiraokaM, WatanabeK, UmezuK, MakiH (2000) Spontaneous Loss of Heterozygosity in Diploid Saccharomyces serecvisiae Cells. Genetics 156 : 1531–1548.
Štítky
Genetika Reprodukčná medicína
Článek Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis inČlánek Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Molecular Recognition by a Polymorphic Cell Surface Receptor Governs Cooperative Behaviors in Bacteria
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- Ribosome Synthesis and MAPK Activity Modulate Ionizing Radiation-Induced Germ Cell Apoptosis in
- Retrotransposon Silencing During Embryogenesis: Cuts in LINE
- Roles of XRCC2, RAD51B and RAD51D in RAD51-Independent SSA Recombination
- Parallel Evolution of Chordate Regulatory Code for Development
- A Genetic Approach to the Recruitment of PRC2 at the Locus
- Deletion of the Murine Cytochrome P450 Locus by Fused BAC-Mediated Recombination Identifies a Role for in the Pulmonary Vascular Response to Hypoxia
- Elevated Mutagenesis Does Not Explain the Increased Frequency of Antibiotic Resistant Mutants in Starved Aging Colonies
- Deletion of an X-Inactivation Boundary Disrupts Adjacent Gene Silencing
- Interplay between Active Chromatin Marks and RNA-Directed DNA Methylation in
- Recombinogenic Conditions Influence Partner Choice in Spontaneous Mitotic Recombination
- Crosstalk between NSL Histone Acetyltransferase and MLL/SET Complexes: NSL Complex Functions in Promoting Histone H3K4 Di-Methylation Activity by MLL/SET Complexes
- A New Role for the GARP Complex in MicroRNA-Mediated Gene Regulation
- RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Loss of DNMT1o Disrupts Imprinted X Chromosome Inactivation and Accentuates Placental Defects in Females
- Inhibition of the Smc5/6 Complex during Meiosis Perturbs Joint Molecule Formation and Resolution without Significantly Changing Crossover or Non-crossover Levels
- Disruption of Lipid Metabolism Genes Causes Tissue Overgrowth Associated with Altered Developmental Signaling
- Translation Initiation Factors eIF3 and HCR1 Control Translation Termination and Stop Codon Read-Through in Yeast Cells
- Recruitment of TREX to the Transcription Machinery by Its Direct Binding to the Phospho-CTD of RNA Polymerase II
- MYB97, MYB101 and MYB120 Function as Male Factors That Control Pollen Tube-Synergid Interaction in Fertilization
- Oct4 Is Required ∼E7.5 for Proliferation in the Primitive Streak
- Contrasted Patterns of Crossover and Non-crossover at Meiotic Recombination Hotspots
- Transposable Prophage Mu Is Organized as a Stable Chromosomal Domain of
- Ash1l Methylates Lys36 of Histone H3 Independently of Transcriptional Elongation to Counteract Polycomb Silencing
- Fine-Mapping the Genetic Association of the Major Histocompatibility Complex in Multiple Sclerosis: HLA and Non-HLA Effects
- Genomic Mechanisms Accounting for the Adaptation to Parasitism in Nematode-Trapping Fungi
- Decoding a Signature-Based Model of Transcription Cofactor Recruitment Dictated by Cardinal Cis-Regulatory Elements in Proximal Promoter Regions
- Removal of Misincorporated Ribonucleotides from Prokaryotic Genomes: An Unexpected Role for Nucleotide Excision Repair
- Fission Yeast Shelterin Regulates DNA Polymerases and Rad3 Kinase to Limit Telomere Extension
- Activin Signaling Targeted by Insulin/dFOXO Regulates Aging and Muscle Proteostasis in
- Activin-Like Kinase 2 Functions in Peri-implantation Uterine Signaling in Mice and Humans
- Demographic Divergence History of Pied Flycatcher and Collared Flycatcher Inferred from Whole-Genome Re-sequencing Data
- Recurrent Tissue-Specific mtDNA Mutations Are Common in Humans
- The Histone Variant His2Av is Required for Adult Stem Cell Maintenance in the Testis
- The Maternal-to-Zygotic Transition Targets Actin to Promote Robustness during Morphogenesis
- Reconstructing the Population Genetic History of the Caribbean
- and Are Required for Growth under Iron-Limiting Conditions
- Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment
- Neuron-Specific Feeding RNAi in and Its Use in a Screen for Essential Genes Required for GABA Neuron Function
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
- Mouse BAZ1A (ACF1) Is Dispensable for Double-Strand Break Repair but Is Essential for Averting Improper Gene Expression during Spermatogenesis
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis
- Pathways-Driven Sparse Regression Identifies Pathways and Genes Associated with High-Density Lipoprotein Cholesterol in Two Asian Cohorts
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- and Are Required for Growth under Iron-Limiting Conditions
- Genetic and Functional Studies Implicate Synaptic Overgrowth and Ring Gland cAMP/PKA Signaling Defects in the Neurofibromatosis-1 Growth Deficiency
- The Light Skin Allele of in South Asians and Europeans Shares Identity by Descent
- RNA∶DNA Hybrids Initiate Quasi-Palindrome-Associated Mutations in Highly Transcribed Yeast DNA
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



