-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca
The Caenorhabditis elegans spermatheca is a myoepithelial tube that stores sperm and undergoes cycles of stretching and constriction as oocytes enter, are fertilized, and exit into the uterus. FLN-1/filamin, a stretch-sensitive structural and signaling scaffold, and PLC-1/phospholipase C-ε, an enzyme that generates the second messenger IP3, are required for embryos to exit normally after fertilization. Using GCaMP, a genetically encoded calcium indicator, we show that entry of an oocyte into the spermatheca initiates a distinctive series of IP3-dependent calcium oscillations that propagate across the tissue via gap junctions and lead to constriction of the spermatheca. PLC-1 is required for the calcium release mechanism triggered by oocyte entry, and FLN-1 is required for timely initiation of the calcium oscillations. INX-12, a gap junction subunit, coordinates propagation of the calcium transients across the spermatheca. Gain-of-function mutations in ITR-1/IP3R, an IP3-dependent calcium channel, and loss-of-function mutations in LFE-2, a negative regulator of IP3 signaling, increase calcium release and suppress the exit defect in filamin-deficient animals. We further demonstrate that a regulatory cassette consisting of MEL-11/myosin phosphatase and NMY-1/non-muscle myosin is required for coordinated contraction of the spermatheca. In summary, this study answers long-standing questions concerning calcium signaling dynamics in the C. elegans spermatheca and suggests FLN-1 is needed in response to oocyte entry to trigger calcium release and coordinated contraction of the spermathecal tissue.
Published in the journal: Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca. PLoS Genet 9(5): e32767. doi:10.1371/journal.pgen.1003510
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003510Summary
The Caenorhabditis elegans spermatheca is a myoepithelial tube that stores sperm and undergoes cycles of stretching and constriction as oocytes enter, are fertilized, and exit into the uterus. FLN-1/filamin, a stretch-sensitive structural and signaling scaffold, and PLC-1/phospholipase C-ε, an enzyme that generates the second messenger IP3, are required for embryos to exit normally after fertilization. Using GCaMP, a genetically encoded calcium indicator, we show that entry of an oocyte into the spermatheca initiates a distinctive series of IP3-dependent calcium oscillations that propagate across the tissue via gap junctions and lead to constriction of the spermatheca. PLC-1 is required for the calcium release mechanism triggered by oocyte entry, and FLN-1 is required for timely initiation of the calcium oscillations. INX-12, a gap junction subunit, coordinates propagation of the calcium transients across the spermatheca. Gain-of-function mutations in ITR-1/IP3R, an IP3-dependent calcium channel, and loss-of-function mutations in LFE-2, a negative regulator of IP3 signaling, increase calcium release and suppress the exit defect in filamin-deficient animals. We further demonstrate that a regulatory cassette consisting of MEL-11/myosin phosphatase and NMY-1/non-muscle myosin is required for coordinated contraction of the spermatheca. In summary, this study answers long-standing questions concerning calcium signaling dynamics in the C. elegans spermatheca and suggests FLN-1 is needed in response to oocyte entry to trigger calcium release and coordinated contraction of the spermathecal tissue.
Introduction
Mechanotransduction, the conversion of physical forces into biochemical signals, is a critical component of cell signaling [1], [2]. Force sensation is essential during organism development and guides cell migration, differentiation, and morphogenesis [3], [4]. Mechanotransduction is essential for normal physiological functioning of the cardiovascular, musculoskeletal, and digestive systems; for example, blood vessel diameter decreases in response to pressure increases to maintain consistent blood volume flow [5], [6]. Cytoskeletal proteins are interconnected in a dynamic, cell-wide structure, and are optimally positioned to sense and transduce changing mechanical parameters [7]. In addition to acting as primary mechanosensors, cytoskeletal proteins are required to anchor and organize stretch-activated membrane ion channels [1].
Filamins are large cytoskeletal scaffolding proteins that consist of an N-terminal actin-binding domain (ABD), followed by a species - and isoform-dependent number of immunoglobulin-like repeats (IgFLN) [8], [9]. Human filamins contain 24 IgFLN domains, and mutations are associated with a broad spectrum of human diseases, including periventricular heterotopia (FLNA), boomerang dysplasia (FLNB), and various myopathies (FLNC) [10]–[13]. Filamin knockout mice show a similar spectrum of phenotypes [14]–[16]. The Drosophila filamin cheerio is composed of 20 IgFLN domains and is important for formation and function of ring canals and follicle cell motility during oogenesis [17]–[19].
Filamins organize the actin cytoskeleton into an elastic three-dimensional network that is responsible for maintaining cell structure and resisting mechanical forces [20], [21]. Experimental evidence suggests that filamin may act as a force-sensitive molecular scaffold [8], [9], [22]. Filamin contains cryptic binding sites that are obscured by adjacent domains [23], [24]. Atomic force microscopy data reveal that filamin molecules unfold when stretched and refold when tension is relieved [25]. Stretching of the filamin rod domain exposes binding sites for integrins and other proteins and leads to strengthening of focal adhesions [23], [26], and a force-dependent filamin-integrin interaction has been observed in cell culture [3]. In addition to acting as a direct mechanosensor, filamin also acts to connect stretch-activated ion channels, such as polycystins, with the cytoskeleton [6].
Our previous work focused on the initial characterization of a well-conserved C. elegans filamin ortholog FLN-1 [27], [28]. FLN-1 consists of an N-terminal ABD followed by 20 IgFLN domains, and is expressed in the spermatheca and uterus, where it colocalizes with F-actin [27], [28]. The allele fln-1(tm545) deletes a portion of the ABD, and a frameshift disrupts translation of all 20 of the IgFLN domains. Therefore, tm545 is a likely strong hypomorph or a null allele [27]. The most striking phenotype of the fln-1(tm545) animals is the failure of fertilized embryos to exit from the spermatheca [27]. The spermatheca, as part of the C. elegans gonad, serves as the site of sperm storage and fertilization [29]. It consists of three distinct regions: the distal constriction, a central accordion-like bag, and the spermatheca-uterine (sp-ut) valve [30]–[33]. The spermatheca, along with a large portion of the oviduct, is enveloped by a single layer of myoepithelial cells [33]. As oocytes are ovulated over the lifetime of the animal, the spermatheca undergoes numerous cycles of stretching, constriction, and relaxation, making it an ideal system to study the role of FLN-1 in cell response to stretch and coordination of cell signaling in an intact tissue.
During each ovulatory cycle, the most proximal oocyte is ovulated into the spermatheca, fertilized, and then released into the uterus [29]. Oocyte entry depends on complex signaling between the oocytes, the sperm, and the sheath cells. LIN-3/EGF is secreted by the proximal oocyte and acts on the proximal sheath cells [34]. In the sheath, LET-23/EGF receptor is predicted to activate PLC-3/phospholipase C-γ, which generates inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG) [34], [35]. The IP3 signal is negatively regulated by IPP-5/inositol 5′-phosphatase and LFE-2/IP3 3′-kinase [34], [36]. ITR-1/IP3 receptor releases calcium from the endoplasmic reticulum when stimulated by IP3 [34]. Calcium likely controls the sheath cell myosin contraction by regulating troponin and tropomyosin [37]. The contraction of the proximal sheath cells propels the oocyte into the spermatheca, where fertilization immediately occurs [29].
After fertilization, directional constriction of the spermatheca propels the embryo into the uterus [29]. The molecular mechanism responsible for initiating and regulating spermathecal constriction is poorly understood, but PLC-1/phospholipase C-ε [38] is required for this process. Loss of PLC-1 results in trapping of embryos in the spermatheca [38]. PLC-1, like PLC-3, generates IP3 and DAG from PIP2; however, PLC-ε enzymes are regulated by small GTPases, while PLC-γ enzymes are regulated by receptor tyrosine kinases [39]–[41]. The specific usage of PLC-3 and PLC-1 in the sheath and the spermatheca suggests that the pathways may be differentially regulated. An increase in cytosolic IP3 likely activates ITR-1/IP3 receptor (IP3R), a tetrameric complex in the endoplasmic reticulum membrane [34], [42]. IP3 binding to IP3R is insufficient for full activation, which requires concomitant calcium binding [42], [43]. Activation of one IP3R in turn activates neighboring IP3Rs by elevating the local calcium concentration [42]. Although intermediate calcium concentration stimulates IP3R, high calcium concentration has an inhibitory effect [42]. This biphasic response of IP3R to calcium can create regenerating calcium waves in the presence of constant IP3 levels [42].
We propose that the release of calcium by the IP3R in the spermathecal cells induces constriction of the spermatheca by activating myosin contraction. In contrast to the sheath cells, the spermatheca does not appear to express a muscle myosin [37]. Smooth muscle regulatory components LET-502/Rho-activated kinase(ROCK) and MEL-11/myosin light chain phosphatase subunit are required for spermathecal function [44], suggesting that the spermathecal myosin belongs to the non-muscle myosin class [45], [46]. NMY-1/non-muscle myosin II is expressed in the spermatheca [39], and here we show NMY-1 is required for spermathecal constriction.
In this study we use GCaMP, a genetically-encoded calcium indicator, to show that oocyte entry stretches the spermathecal cells and triggers coordinated pulses of calcium transients. The calcium transients originate in the distal constriction and appear to propagate proximally across the spermathecal bag. FLN-1 and PLC-1 are required to trigger calcium signaling, and ITR-1 is required downstream of PLC-1 to produce the calcium oscillations. The signal is coordinated across the tissue via gap junctions to produce a directional wave. The directional wave of calcium results in contraction of the actomyosin network and expulsion of the embryo into the uterus. Given the modular protein structure, sub-cellular localization, genetic interaction data, and known mechanosensory functions of filamin, we postulate that FLN-1 is required to convert physical information about the presence of the oocyte into a calcium signal that controls the directional constriction of the spermatheca.
Results
fln-1 interacts genetically with regulators of calcium signaling in the spermatheca
Previous studies have revealed an important role for phosphatidylinositol signaling during ovulation and spermathecal transit in C. elegans, and demonstrated that PLC-1/phospholipase C-ε is required for spermathecal transit [34], [35], [38]. PLC-1 generates IP3, which is thought to trigger calcium release from the endoplasmic reticulum via the IP3 receptor ITR-1 [34], [38]. We have shown previously that fln-1(tm545) and plc-1(rx1) single and double mutants show a very similar phenotype, in which embryos are retained in the spermatheca [27]. Because PLC-1 and FLN-1 are both required in the spermatheca for transit of fertilized oocytes, we explored the possibility that FLN-1 functions in the IP3 signaling pathway to regulate spermathecal function.
Strong itr-1 loss-of-function alleles result in ovulation entry defects, complicating observation of spermathecal transit. To circumvent this problem, we used itr-1 gain-of-function alleles [34], [35]. The itr-1 gain-of-function alleles are located in or near the IP3-binding domain of ITR-1 and are thought to increase the affinity of ITR-1 for IP3 [34], [47], [48]. Using brood size assays, we tested five putative itr-1 gain-of-function alleles for suppression of the fln-1(tm545) brood size defect [27]. itr-1 alleles sy327gf, sy328gf, and sy290gf suppressed the brood size defect (Figure 1), while sy291gf and sy331gf did not (unpublished data).
Fig. 1. fln-1(tm545) brood size defect is suppressed by increased IP3 signaling. 
Brood sizes of fln-1(tm545) animals carrying itr-1(gf) and lfe-2(lf) mutations. The genotypes are indicated along the y-axis. Points represent individual animals, the bar indicates the mean, and the error bar is the standard deviation. Student's t-test was used to compare the genotypes, and p values are indicated with *** for p = 0.0001. We chose to focus on the itr-1(sy290gf) allele for subsequent experiments because it affects a well-characterized residue critical for IP3 binding [49]. Biochemical characterization suggests that the sy290 recombinant protein has a two-fold increase in IP3 binding affinity [48]. The itr-1(sy290gf) animals do not exhibit overt ovulation or spermathecal transit defects; however, the sheath cell contractions are more frequent in these animals and they show a reduced brood size (173±17 SD, n = 6) compared to wildtype animals (301±26 SD, n = 14) [35]. To control for possible effects of the marker phenotypes we used itr-1(sy290gf) strains marked with dpy-20(e1282) (dumpy) or unc-24(e138) (kinker). No brood size differences were observed between the differentially marked strains. Similarly, the brood size of fln-1(tm545) unc-24(e138) (20±4 SD, n = 12) animals is not significantly (p = 0.08, Student's t-test) different from fln-1(tm545) animals. fln-1(tm545) itr-1(sy290) unc-24(e138) and fln-1(tm545) itr-1(sy290) dpy-20(e1282) animals have a significantly (p<0.0001, Student's t-test) higher average brood size (46±22 SD, n = 18, and 46±12 SD, n = 15, respectively) than fln-1(tm545) animals (Figure 1).
LFE-2 negatively regulates IP3 signaling by phosphorylating IP3 into inositol 1,3,4,5-tetrakisphosphate (IP4) [34]. Therefore, lfe-2(sy326), a loss-of-function allele, is predicted to have longer duration or intensity of IP3 signals [34]. Like itr-1(sy290gf), lfe-2(sy326) does not result in obvious ovulation or spermathecal transit defects, but does exhibit a reduced brood size (174±90 SD, n = 6) compared to wildtype animals (301±26 SD, n = 14). We speculated that increased IP3 levels in the lfe-2(sy326) background would also suppress the fln-1(tm545) spermathecal transit defect. We found that, indeed, fln-1(tm545); lfe-2(sy326lf) animals have a significantly (p<0.0001, Student's t-test) increased average brood size (43±27, n = 32) compared to fln-1(tm545) animals (Figure 1).
Characteristic calcium oscillations control spermathecal transit
Because we observed a genetic interaction between fln-1 and components of the phosphatidylinositol signaling pathway, we next examined whether calcium signaling plays a role during spermathecal transit. It has been hypothesized that IP3R-regulated calcium release results in sheath cell contractions, however, IP3R-regulated calcium release has not been observed directly in the sheath or the spermatheca. To monitor calcium levels during spermathecal transit we used GCaMPv3 (GFP-Calmodulin-M13 Peptide, version 3), a genetically-encoded calcium indicator [50]. GCaMP has been previously used in C. elegans neurons and hypodermal cells to image calcium transients [50], [51].
We created transgenic nematodes expressing GCaMP under the control of the fln-1 promoter (xbIs1101[fln-1p::GCaMP]), imaged immobilized animals using wide-field epifluorescence with standardized acquisition parameters, and quantified the GCaMP signal by calculating mean pixel intensity (pixel intensity/area) and normalizing to the initial fluorescence (Figure 2A). Importantly, animals expressing GCaMP exhibit wildtype oocyte entry, ovulation transit times, and normal brood sizes (282±51 SD, n = 6; compare Video S1 and Video S2). Animals expressing regular calcium-insensitive fln-1p::GFP were used as controls to determine whether spermathecal shape changes or photobleaching would affect the GCaMP signal (Figure 2C, Video S2). No alteration in GFP fluorescence signal due to spermathecal shape changes was observed, and photobleaching was not detected over many hours of imaging (n = 6) (Figure 2C, Video S2). As an additional control for possible effects of changing cell shapes on measured intensities, we expressed tdTomato and GCaMP in the spermatheca and performed ratiometric imaging. Because the tdTomato signal remained constant throughout the duration of imaging, normalization of GCaMP to tdTomato signal did not result in any significant differences in signal compared to non-ratiometric imaging (n = 3) (Figure S1).
Fig. 2. Oocyte entry into the spermatheca triggers calcium oscillations. 
(A–B′) A representative first ovulation in xbIs1101[fln-1p::GCaMP] (n = 26) transgenic animals. (A) A plot of GCaMP mean pixel intensity normalized to the pre-ovulation mean pixel intensity is shown, along with a (A′) corresponding kymogram at the same time scale. The top and bottom of the kymogram represent the distal and proximal spermatheca, respectively. Darker regions indicate higher calcium levels. Oocyte entry begins at 30 seconds and is complete by 60 seconds, and the zygote exit is marked by a magenta arrowhead in (A) and is visible in (A′) as a reduction in the size of the spermatheca. (A′) The leftward tilt of dark stripes is indicative of calcium transient propagation from the distal spermatheca. (B–B′) A detailed view of several calcium transients is shown as a (B) trace and (B′) kymogram from the region indicated by the blue line in (A). (B) GCaMP fluorescence intensity was quantified in 1 µm strips (distal is black, and proximal is gray), 50 µm apart as indicated in (B′). The mean peak-to-peak transit time across the spermatheca is 8±1 seconds and is indicated on the trace in (B). (C) A representative first ovulation in xbEx0811[fln-1p::GFP] (n = 6) transgenic animals. The fln-1p::GFP signal does not change during oocyte entry or spermathecal transit. (C′) A corresponding kymogram of fln-1p::GFP spermathecal transit with oocyte entry and sp-ut valve dilation visible as dark horizontal stripes. Time-lapse imaging of GCaMP reveals that oocyte entry into the spermatheca initiates a characteristic and reproducible sequence of calcium oscillations (n = 26) (Figure 2, Video S1). Out of necessity we focused our analysis of calcium signaling on the first ovulation; however, in wild type animals, generally similar calcium transients are observed in subsequent ovulations (Figure S2). Oocyte entry into the spermatheca consistently triggers a single pulse of calcium in the sp-ut valve (Figure 2A′, Video S1). The single pulse of calcium in the sp-ut valve may serve to constrict the valve to prevent premature exit of the oocyte. Neither fertilization nor egg shell formation are required to initiate spermathecal calcium signaling (Figure S3). Following complete entry of the oocyte into the spermatheca, the calcium transients increase in intensity as the oocyte progresses through the spermatheca (Figure 2A and B, Video S1). Embryo exit is concomitant with the strongest calcium pulses, suggesting that the calcium pulses trigger spermathecal constriction (Figure 2A). The final pulse of calcium coincides with constriction of the sp-ut valve following embryo exit (Figure 2A′).
Quantitative analysis of the time-lapse image sequences shows that the calcium transients appear to initiate in the distal spermatheca and propagate proximally (Figure 2A′–2B′). To determine if the calcium transients are directional we measured the fluorescence intensity in the distal and proximal spermatheca (Figure 2B). Calcium pulses are first detected in the distal spermatheca, and then in the proximal spermatheca several seconds later (Figure 2A′–2B′). The calcium transients occur in the direction of oocyte movement, suggesting that directional calcium pulses may control spermathecal constriction. The distal spermathecal cells may act as a pacemaker to trigger and synchronize calcium release in other spermathecal cells.
We predicted that the observed distal to proximal spread of the calcium signal would require cell-cell communication and tissue level coordination. To test this idea, we used RNAi to sequentially deplete the 25 gap junction subunits [52], [53], and determined that loss of the innexin INX-12 results in spermathecal transit defects. In inx-12(RNAi) animals, oocytes enter the spermatheca normally, but variably change direction several times before returning into the ovary or proceeding into the uterus (Figure 3, Video S3). Calcium imaging of the inx-12(RNAi) (n = 4) animals revealed that each spermathecal cell is capable of producing calcium pulses, but that the resulting calcium waves are asynchronous and non-directional (Figure 3A′, Video S3). The random calcium pulses likely result in the observed uncoordinated spermathecal constriction. These results suggest that a small molecule, such as calcium or IP3, propagates through the spermatheca via gap junctions to produce synchronous and directional calcium transients.
Fig. 3. Gap junction subunit INX-12 is required for spermathecal coordination. 
(A) A representative calcium signal trace observed during the first ovulation in inx-12(RNAi) (n = 4) animals imaged with xbIs1101[fln-1p::GCaMP]. The oocyte fails to exit into the uterus and changes direction thrice before returning into the ovary. The arrows indicate direction of movement with blue arrows indicating movement towards the uterus. (A′) Arrowheads in the corresponding kymogram indicate bursts of calcium and correspond to the contractions that stop the oocyte from being refluxed into the ovary. In the kymogram, darker values indicate higher calcium levels. Top is distal, bottom is proximal, and the kymogram is at the same time scale as the calcium trace shown above it. FLN-1, PLC-1, and ITR-1 are essential for normal calcium signaling in the spermatheca
To investigate whether FLN-1 is required for normal calcium signaling during spermathecal transit, we introduced the fln-1p::GCaMP transgene into fln-1(tm545) animals. Although the initial entry pulse of calcium within the sp-ut valve occurs normally (Figure 4A and 4A′, Video S4), filamin-deficient animals fail to initiate calcium transients in the spermatheca itself, with few calcium signals observed during the time normally required for oocyte transit (n = 19) (Figure 4A, Video S4). Abnormal and highly variable transients are observed in fln-1(tm545) animals approximately 15 minutes after oocyte entry—well after the time a wildtype zygote would have exited (Figure 4A). These delayed calcium pulses fail to produce significant constriction of the spermatheca (Figure 4A′, Video S4), suggesting that the contractile mechanism may be compromised in filamin-deficient animals. These data suggest that filamin is required for timely initiation of calcium signaling and for the contractile mechanism, but not for the calcium release mechanism per se.
Fig. 4. FLN-1 and PLC-1 are required for different aspects of calcium signaling. 
Representative calcium signal traces observed during the first ovulation in (A) fln-1(tm545) (n = 19), (B) plc-1(rx1) (n = 5), and (C) fln-1(tm545); plc-1(rx1) (n = 3) animals imaged using xbIs1101[fln-1p::GCaMP]. Corresponding kymograms (A′–C′) are also shown. In the kymograms, darker values indicate higher calcium levels. Top is distal, bottom is proximal, and each kymogram is at the same time scale as the calcium trace shown above it. Homology and genetic interaction data suggest that hydrolysis of PIP2 by PLC-1 generates IP3 in the spermatheca, triggering intracellular calcium release via the IP3R [38], [54]. Consistent with this idea, plc-1(rx1) animals fail to produce calcium signals following oocyte entry into the spermatheca (n = 5) (Figure 4B, Video S5). plc-1(rx1) animals also do not produce the initial entry pulse of calcium in the sp-ut valve. Likewise, a temperature-sensitive reduction-of-function allele of itr-1(sa73ts) results in abnormal calcium signaling at the semi-permissive temperature of 20°C [55]–[57]. Phenotypes observed range from mild perturbations (Figure 5A) to grossly abnormal calcium transients (n = 6) (Figure 5B). The grossly abnormal calcium signaling is of lower intensity, and results in trapping of embryos within the spermatheca. Additionally, itr-1(sa73ts) animals do not produce the initial pulse of calcium in the sp-ut valve during oocyte entry, the timing between pulses is longer, and the calcium release is restricted to the distal spermatheca (Figure 5). These results suggest that the observed calcium transients in the spermatheca require IP3-regulated release of calcium from internal stores, and are consistent with previous findings in the C. elegans intestine [55], [56].
Fig. 5. itr-1(sa73ts) disrupts calcium signaling at the semi-permissive temperature. 
Representative calcium signaling traces in itr-1(sa73ts) animals at the semi-permissive temperature of 20°C imaged with xbIs1101[fln-1p::GCaMP]. (A) At the non-permissive temperature, 66% (n = 6) of the itr-1(sa73ts) successfully propelled oocytes from the spermatheca into the uterus. (B) The remaining ovulations resulted in embryos trapped within the spermatheca. Corresponding kymograms (A′ and B′) are also shown. In the kymograms, darker values indicate higher calcium levels. Top is distal, bottom is proximal, and each kymogram is at the same time scale as the calcium trace shown above it. In both groups the calcium pulse intensity was markedly reduced compared to wildtype animals. In all cases the itr-1(sa73ts) ovulations resulted in a portion of the oocyte being pinched off during ovulation. Unlike fln-1(tm545) single mutant animals, which eventually initiate abnormal calcium pulses, fln-1(tm545); plc-1(rx1) double mutant animals behave like plc-1(rx1) single mutants and fail to produce any calcium transients (n = 3) (Figure 4C, Video S6). This suggests that the delayed calcium pulses seen in fln-1(tm545) animals are generated via the canonical phosphatidylinositol signaling pathway. Although FLN-1 is required for timely initiation of calcium pulses upon oocyte entry, it appears that calcium signaling can eventually be activated by a parallel, filamin-independent pathway.
Because gain-of-function mutations that sensitize ITR-1 to IP3 suppress the fln-1(tm545) brood size defects (Figure 1), we next determined the effect of these mutations on calcium signaling in the spermatheca. We speculated that increased sensitivity of ITR-1 to IP3 would trigger increased calcium release. itr-1(sy290gf) only has a moderate effect on GCaMP intensity in the wildtype background (n = 5) (Figure S4A), which is consistent with the lack of a strong phenotype. We do not detect overt changes in itr-1(sy290gf) calcium dynamics, such as increased propagation speed nor increased frequency of calcium release; however, it is possible that there are higher frequency changes not captured by our imaging parameters. Surprisingly, fln-1(tm545) itr-1(sy290gf) double mutants have markedly increased calcium signaling immediately following embryo entry (n = 3) (Figure 6A, Video S7), and display a novel partial exit phenotype with the embryo held in place by a partially closed sp-ut valve (Figure 6A′). Sp-ut valve constriction around the zygote during eggshell formation results in bow tie-shaped embryos (Video S7). lfe-2(sy326) animals, like itr-1(sy290gf), have marginally increased intensity of calcium signaling in the wildtype background (n = 4) (Figure S4B). However, the calcium transients in lfe-2(sy326) animals are variable from animal to animal, which is consistent with our brood size data (Figure 1), and may reflect incomplete penetrance of the sy326 allele. Similar to the results with itr-1(sy290gf), fln-1(tm545); lfe-2(sy326) animals exhibit increased calcium signaling compared to fln-1(tm545) alone (n = 3) (Figure 6B, Video S8), and partial exit of embryos from the spermatheca (Figure 6B′, Video S8). Importantly, these observations indicate that the fln-1(tm545) spermatheca and valve may be structurally capable of constriction if sufficient calcium is present.
Fig. 6. Increased IP3 signaling suppresses the fln-1(tm545) calcium signaling defect. 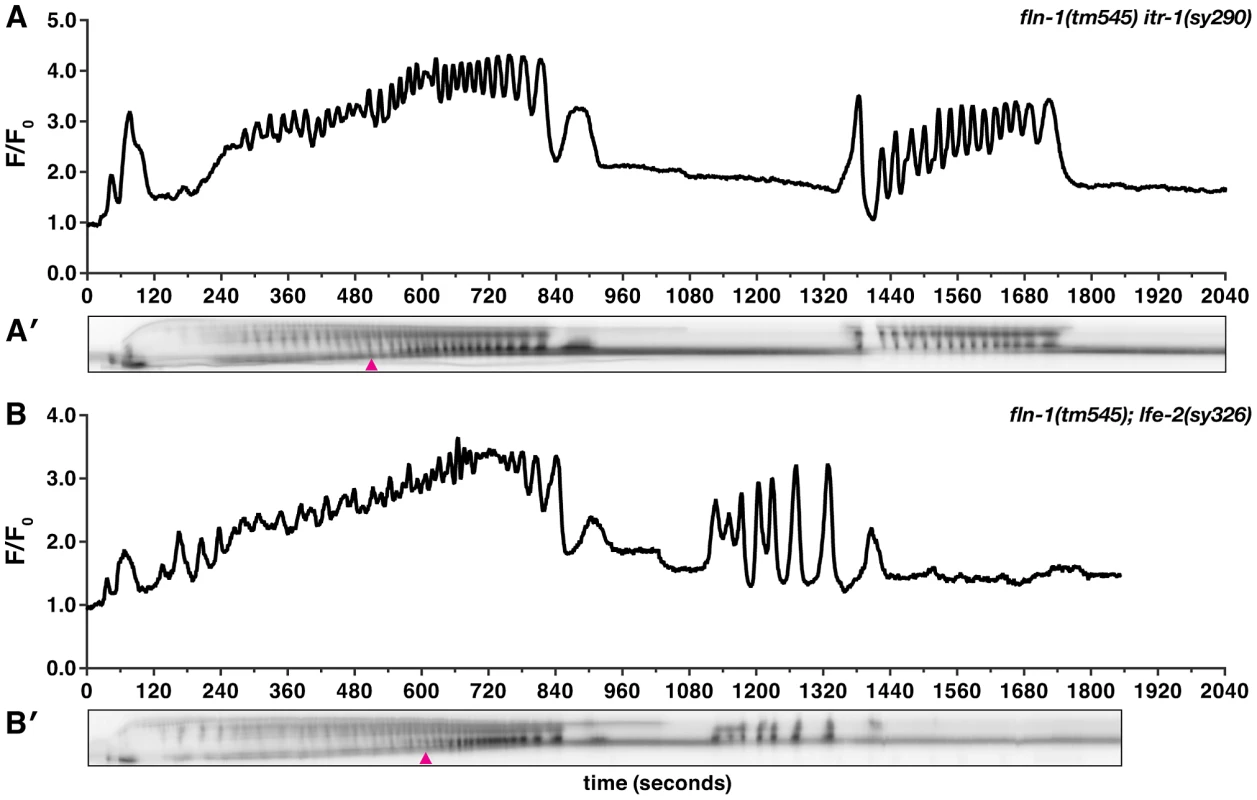
Representative calcium signal traces observed during the first ovulation in (A) fln-1(tm545) itr-1(sy290gf) (n = 3) and (B) fln-1(tm545); lfe-2(sy326) (n = 3) animals imaged with xbIs1101[fln-1p::GCaMP]. The corresponding kymograms (A′ and B′) are shown below the plots. Arrowheads indicate the beginning of partial exit of the embryo from the spermatheca. In the kymograms, darker values indicate higher calcium levels. Top is distal, bottom is proximal, and each kymogram is at the same time scale as the calcium trace shown above it. NMY-1/non-muscle myosin functions in the spermatheca
Cell contractility is generated by the actomyosin cytoskeleton, which consists of myosin, myosin regulatory proteins, and F-actin. Two non-muscle myosin regulatory proteins, Rho-activated kinase LET-502 and a subunit of myosin light chain phosphatase MEL-11, are known to be required for normal spermathecal function [44]. The non-muscle myosin NMY-1, redundantly expressed with NMY-2 during embryonic elongation, is strongly expressed in the spermatheca [45], [46]. NMY-1, MEL-11, and LET-502 have been extensively studied in the context of embryonic elongation where they are required to fine-tune contractility of the hypodermal cells [44], [45], [58]–[61]. We speculated that this contractile module also functions in the spermatheca and is responsible for constriction of the spermatheca.
Similar to the phenotype seen in fln-1(tm545) animals, depletion of nmy-1 by RNAi results in a poorly contractile spermatheca and an sp-ut valve that fails to constrict and completely expel the embryo (Figure 7). Embryos are pushed out of nmy-1(RNAi) spermathecae through a relaxed sp-ut valve due to back pressure from newly ovulated oocytes. In contrast, in mel-11(sb56) animals, oocytes fail to enter into a hyper-constricted spermatheca [44]. We speculated that mel-11(RNAi) might produce a weaker phenotype than the sb56 allele. Indeed, mel-11(RNAi) animals are able to ovulate, allowing observation of the spermathecal transit process (Video S9). Depletion of mel-11 in the spermatheca results in hyper-constriction of the spermatheca around the zygote, rupture of the spermatheca, and escape of the zygote into the body cavity (Figure 8A, Video S9). The distal constriction and the sp-ut valve also appear to hyper-constrict, forcing the embryo in this unusual direction. The mel-11 spermathecal rupture phenotype is strongly suppressed in the fln-1 (Figure 8B) and plc-1 (Figure 8C) backgrounds, consistent with the idea that FLN-1 and PLC-1 are required for spermathecal contractility.
Fig. 7. NMY-1 is required for spermathecal contractility. 
The spermatheca appears compact before the first ovulation (A, C, E), however, the sp-ut valve is constricted only in (A′) wildtype animals but not in (C′) fln-1(tm545) and (E′) nmy-1(RNAi) animals. (B) The wildtype spermatheca and (B′) sp-ut valve fully expel the embryo and re-constrict following every ovulation. In contrast, (D) fln-1(tm545) and (F) nmy-1(RNAi) spermathecae retain embryos, and the sp-ut valves (D′ and F′, respectively) remain open. The spermatheca is labeled with ezIs2[fkh-6p::GFP] (images merged with DIC) and the sp-ut valve is labeled with xbEx1019[tag-312p::GFP]. Arrowheads indicate the sp-ut valve. Vertical lines next to the sp-ut valves indicate the width of the opening. The scale bar indicates 25 µm. Fig. 8. MEL-11 modulates spermathecal contractility. 
(A) mel-11(RNAi) results in hyper-constriction and rupture of the spermatheca. The asterisk indicates an escaped embryo and arrowheads indicate cell fragments in the pseudocoelom. mel-11(RNAi) in the (B) fln-1(tm545) or (C) plc-1(rx1) genetic background does not result in spermathecal rupture. The spermatheca is labeled with ezIs2[fkh-6p::GFP] and the fluorescent image is merged with DIC. The scale bar indicates 25 µm. Discussion
Our previous work described the C. elegans filamin orthologs, and established that FLN-1 is required for function of the spermatheca, a smooth muscle-like tissue in the C. elegans gonad [27], [28]. Filamin-deficient spermathecae are unable to constrict, and as a result trap fertilized embryos [27]. In this study, we show that oocyte entry into the spermatheca triggers calcium oscillations that are likely instructive for spermathecal constriction. We find that FLN-1, an actin-binding protein and a known mechanosensitive scaffold, is required to trigger timely IP3-dependent calcium release in response to oocyte entry. We identify a gap junction subunit, INX-12, required for signal propagation across the spermatheca and the non-muscle myosin, NMY-1, required for spermathecal constriction. Our working hypothesis is that filamin is required in the spermatheca to transduce the physical presence of an oocyte into a biochemical signal, thereby triggering constriction of the spermatheca and expulsion of the embryo (Figure 9).
Fig. 9. Proposed genetic control of calcium signaling in the spermatheca. 
The diagram represents a working model of the spermathecal calcium signaling pathway. Green lines indicate interactions that promote cell contraction, and magenta lines indicate those which inhibit contraction. Dashed lines indicate possible regulatory interactions. We propose FLN-1 acts upstream or in parallel to PLC-1 to initiate calcium release via ITR-1. FLN-1 also plays a downstream role in contraction and maintenance of the F-actin cytoskeleton. Constriction of the spermatheca in response to stretch is reminiscent of myogenic response seen in vascular smooth muscle cells, where increased intraluminal pressure in blood vessels results in vasoconstriction [1], [62]. The myogenic response is triggered via mechanically-sensitive ion channels that stimulate IP3-dependent calcium release. Filamin is required for cytoskeletal anchoring of an inhibitory polycystin channel subunit [6]. Filamin also interacts with other transmembrane channels, such as cystic fibrosis transmembrane regulator (CFTR) [63], Ca(v)1.4 subunit of a voltage-gated L-type channel [64], pacemaker channel HCN1 [65], G protein-coupled calcium-sensing receptor [66], and potassium channels Kir2.1 [67] and Kv4.2 [68]. Filamin therefore appears to be required for normal channel function, and may mechanically couple the channels to the cytoskeleton, as well as controlling their localization [9], [69].
Our calcium signaling and genetic interaction data are consistent with the possibility that FLN-1 might act as a top-level component in the spermatheca signal transduction pathway (Figure 9). However, the molecular details of how filamin might initiate this cascade are unknown. One possibility is that filamin may be a necessary adaptor between stretch-gated ion channels and the cytoskeleton. Strain on the cytoskeleton would then be communicated to the ion channels, allowing a brief influx of calcium ions which could activate PLC-1 either directly via its EF hand domain [70] or indirectly through a calcium/calmodulin-activated protein. In addition to connecting the stretch-gated channels to the cytoskeleton, filamin might also scaffold downstream effectors required to sense or amplify ion channel opening.
Another possibility is that filamin is a direct mechanosensor, and that stretch of the spermathecal cells by oocyte entry directly stretches the filamin molecule, thereby revealing cryptic binding sites. Evidence from biophysical experiments with purified components [22], [24], and study of the mechanosensory role of filamin in the context of focal adhesions in cultured cells support this idea [9], [71]–[73]. Stretch-activated binding sites could localize a RhoGEF, such as Trio [74], to the cortex and increase the level of Rho-GTP, which has been shown to activate PLC-ε [75], [76]. Because PLC-ε also contains a GEF domain, activation of PLC-1 may lead to prolonged activation and increased levels of Rho-GTP in addition to IP3 and DAG [77]–[79]. Activation of ROCK and calcium release would both act to promote contraction. Other downstream pathways could also be activated by DAG and calcium, such as Protein Kinase C (PKC) [80].
Filamin may perform a partially separable signaling and structural role in the spermatheca. We have shown previously that loss of filamin results in a progressive disorganization of filamentous actin in the spermatheca (Figure 9) [27]. F-actin organization is relatively normal initially; however, filamin is required to maintain cytoskeletal structure as the spermatheca is repeatedly stretched by incoming oocytes [27]. Because our calcium measurements are made during the first ovulation, before the actin cytoskeleton becomes grossly abnormal, and because calcium signaling can be strongly disrupted, for example, by loss of PLC-1, in spermathecae with intact actin cytoskeletons, we suspect that the calcium signaling defects in filamin-deficient animals are not simply a consequence of cytoskeletal defects, but we cannot entirely exclude this possibility.
While our results suggest FLN-1 is needed to trigger normal calcium release in the spermatheca, FLN-1 may be acting in parallel with PLC-1 to activate calcium release via ITR-1. We show itr-1(sy290gf) and lfe-2(sy326) ameliorate the effects of fln-1(tm545), suggesting that they function downstream of fln-1. Consistent with this result, over-expression of LFE-2 under a heat-shock promoter results in an exit defect similar to fln-1(tm545) [34]. The brood size defect of plc-1(rx1) animals is not suppressed by itr-1(sy290gf) nor lfe-2(sy326), presumably due to complete absence of IP3 [38], [54]. This suggests that fln-1(tm545) animals possess sufficient IP3 to activate the sensitized ITR-1 receptor. Therefore a key role of FLN-1 might be to regulate some aspect of ITR-1 response to IP3, ultimately resulting in calcium release and spermathecal constriction.
Although IP3 appears to be needed to initiate calcium signaling, we do not know whether IP3 levels oscillate, or whether IP3 simply triggers the first calcium transient. Increased IP3 levels due to inactivation of LFE-2, an IP3 kinase, or a hypersensitive IP3 receptor do not cause abnormal calcium oscillations in the wildtype background, suggesting that precise control of IP3 level is not required. Once triggered, IP3R may be capable of generating self-sustaining calcium oscillations [42], [43], [81], [82]. IP3 binding to IP3R stimulates the release of calcium into the cytosol, which initially stimulates IP3R, but becomes inhibitory at high concentrations (Figure 9) [42], [43]. Endoplasmic reticulum Ca2+-ATPase pumps then remove calcium from the cytosol, returning calcium levels to the stimulatory range. This repeating cycle results in oscillating calcium levels. Sensitivity of the IP3R to IP3 and calcium may be modulated by accessory proteins [43], which may explain the increasing amplitude of calcium release in the spermatheca. With each round of calcium release, ITR-1 may become more activated, leading to larger pulses of calcium. The calcium oscillations may be terminated by an extrinsic signal or a threshold effect of calcium and IP3.
Our observations suggest that calcium pulses are initiated in the distal spermatheca and spread proximally. Although loss of the gap junction subunit INX-12 disrupts the synchronous transients, all spermathecal cells appear to be capable of producing stochastic calcium pulses cell-autonomously. Gap junctions generally permit passive diffusion of small solutes, such as calcium and IP3. Interestingly, the diffusion of calcium within a cell is limited to small domains by the buffering effects of calcium-binding proteins [82], [83], while the diffusion rate of IP3 is much greater, allowing it to act as a global messenger [84]. Given these diffusion rates it seems likely that IP3 is primarily responsible for synchronization of calcium signaling in the spermatheca.
We observed that strengthening and synchronization of the calcium signal coincides with a steady, coordinated contraction of the spermathecal tissue and expulsion of the fertilized embryo. Spermathecal contraction requires the non-muscle myosin NMY-1. Smooth muscle cell contraction is regulated by phosphorylation of the regulatory myosin light chain (rMLC) by MLC kinase (MLCK) [85]. The C. elegans genome lacks an obvious MLCK homolog, suggesting that rMLC phosphorylation is regulated by other kinases, such as ROCK (Figure 9) [86]. In C. elegans loss of LET-502/ROCK leads to loss of contractility of the spermatheca [44], [58], [87]. Contractility is negatively regulated by dephosphorylation of the rMLC by MLCP, and in C. elegans loss of MEL-11, an MLCP subunit, leads to hyper-constriction of the spermatheca. Understanding how this contractile module is regulated by calcium signals is an exciting future area of study.
In summary, in this study we demonstrate that oocyte entry leads to dynamic calcium signaling and coordinated contraction of the tissue, ultimately leading to expulsion of the fertilized embryo. We further demonstrate that FLN-1, PLC-1, ITR-1 and the gap junction component INX-12 are required for normal calcium signaling in the spermatheca. These results establish filamin as a regulator of calcium signaling, and suggest disruption of filamin may result in defective cell response to stretch. This is important because human filamin mutations result in severe myopathies, cardiovascular, and neurological conditions, but it is unclear how loss of filamin results in these diverse pathologies. Our study shows that filamin, in addition to being a cytoskeletal protein, may modulate calcium signaling pathways in spermathecal and other mechanically-sensitive cells. In addition to providing mechanistic insight into how the spermatheca functions, this study helps to establish the spermatheca as an in vivo model system for the study of how cells coordinate tissue-level responses to mechanical input.
Materials and Methods
C. elegans strains and culture
C. elegans strains were cultured on NGM agar plates with OP50 Escherichia coli at 20°C. Nematode observations and manipulations were performed at 20°C unless otherwise noted. For a complete list of strains used in this study please see Table S1. Standard genetic techniques were used to manipulate C. elegans genotypes. Point mutations were tracked with marker alleles. unc-24(e138), a weak kinker allele, was used to follow the itr-1 gain-of-function alleles. dpy-20(e1282), a dumpy allele, was also used to follow itr-1(sy290gf) to control for any marker phenotypes. unc-57(ad592), another weak kinker allele was used to follow lfe-2(sy326). Deletion and insertion alleles were genotyped using polymerase chain reaction (PCR).
RNA interference
RNA interference was performed by feeding animals dsRNA-expressing HT115 DE3 E. coli as described [27], [88]. The RNAi bacteria were seeded onto NGM plates supplemented with carbenicillin and isopropyl β-D-1-thiogalactopyranoside (IPTG). Eggs were obtained from gravid hermaphrodites using alkaline hypochlorite solution and placed on the RNAi plates. RNAi targeting constructs for fln-1 [27], nmy-1, and plc-1 were constructed by PCR amplification of wildtype cDNA using engineered restriction sites, and subsequently cloned into pPD129.36 (Fire Vector Kit). Primer sequences used for plasmid construction are shown in Table S2. mel-11 RNAi targeting construct was isolated from an open reading frame RNAi library (Open Biosystems; Huntsville, AL, USA). Empty pPD129.36 vector was used as a negative control in RNAi experiments.
Gap junction subunit RNAi screen
We used RNA interference to individually deplete the 25 predicted gap junction genes [52], [53] in xbIs1101[fln-1p::GCaMP] animals. RNAi experiments were performed as described above. RNAi targeting constructs were obtained from an open reading frame RNAi library (Open Biosystems; Huntsville, AL, USA) or constructed by PCR amplification of wildtype cDNA and subsequent cloning into pPD129.36 (Fire Vector Kit). Primer sequences are provided in Table S2. We used a low-magnification screen to identify animals with distended spermathecae or embryos present in the ovary as indicators of abnormal spermathecal function. We excluded genes that grossly affected animal development or gonad morphology. Primary screen hits were selected for detailed calcium imaging as described below.
Brood size assays
We define brood size as the number of hatched progeny. The total number of hatchlings was determined by segregating L4 animals to individual, freshly seeded plates. Progeny were counted and aspirated beginning two days after the initial transfer, and continuing for two days after end of egg laying. Brood sizes are reported as the mean ± standard deviation. Two-tailed, unpaired t-tests were used to test for statistical significance between relevant genotypes. Statistical analyses were performed using GraphPad Prism 5.
Construction of fln-1p::GCaMP transgenic animals
GCaMP3 was obtained from Addgene plasmid 22692 [50]. PCR was used to amplify GCaMP with primers IK189 and IK190 (Table S2) containing engineered restriction endonuclease sites XbaI and XmaI. The XbaI-XmaI fragment was cloned downstream of the fln-1 promoter in pUN85 [27] to generate pUN107. pUN107 contains the fln-1 promoter, GCaMP, and the fln-1 3′ UTR. Transgenic animals were created by microinjecting a DNA solution containing 40 ng/µL of pUN107 and 100 ng/µL of pRF4 (rol-6 marker). Progeny displaying the roller phenotype and green fluorescence in the spermatheca were segregated to establish transgenic lines. A strain (UN1037) with low levels of GCaMP expression and moderate transmission frequency was integrated using UV irradiation to generate strain UN1101 xbIs1101[fln-1p::GCaMP] II. UN1101 was outcrossed ten times to create strain UN1108. Standard genetic crosses were used to introduce xbIs1101 into various genetic backgrounds.
Time-lapse microscopy
Acquisition
Animals were anesthetized with 0.01% tetramisole and 0.1% tricaine in M9 buffer [89], mounted on 2% agarose pads and imaged using a 60′ oil immersion objective with a Nikon Eclipse 80i microscope equipped for wide-field epifluorescence and differential interference contrast (DIC). The approximate transverse cross-section of the spermatheca was selected as the focal plane. Fluorescence excitation was provided by a Nikon Intensilight C-HGFI 130W mercury lamp. Neutral density filters were used to reduce illumination intensity to 3.1% (1/32). Fluorescence excitation light was shuttered with a SmartShutter (Sutter Instruments; Novato, CA, USA), and controlled via the camera TTL signal. A SPOT RT3 cooled charge-coupled device camera (Diagnostic Instruments; Sterling Heights, MI, USA) was used to capture images at 1 frame per second using SPOT Advanced 5 (Diagnostic Instruments). For GCaMP imaging the camera exposure time, gain, and binning were set to 75 ms, 8, and 1×1, respectively. All light was redirected to the camera, and the DIC analyzer was removed from the light path. The same microscopy setup was used for all experiments. Room temperature was maintained between 20 and 23°C.
Image processing
Images were acquired at 1600×1200 pixels and saved as 8-bit tagged image file format (TIFF) files. All image processing was done using ImageJ (National Institutes of Health; Bethesda, MD, USA; http://rsb.info.nih.gov/ij/). The GCaMP signal was measured by defining a region of interest around the spermatheca with a standard size of 800×400 pixels (100 µm×50 µm). The mean pixel intensity (total pixel intensity/area) for each frame was then calculated using a custom ImageJ macro. We expressed the GCaMP signal (F) as a ratio with the baseline fluorescence (F0). The baseline fluorescence was calculated as the mean fluorescence of 30 frames preceding the ovulation. Data analysis and plotting were performed with GraphPad Prism 5. Kymograms were generated using a custom ImageJ macro to determine the mean fluorescence intensity of each column of pixels and represent it as a single pixel in the kymogram. Videos were exported as Audio Video Interleave (AVI) files from ImageJ and subsequently encoded as QuickTime (Apple; Cupertino, CA, USA) H.264 movies and scaled down to 400×200 pixels.
Supporting Information
Zdroje
1. OrrAW, HelmkeBP, BlackmanBR, SchwartzMA (2006) Mechanisms of mechanotransduction. Dev Cell 10 : 11–20 doi:10.1016/j.devcel.2005.12.006.
2. JaaloukDE, LammerdingJ (2009) Mechanotransduction gone awry. Nat Rev Mol Cell Biol 10 : 63–73 doi:10.1038/nrm2597.
3. GehlerS, BaldassarreM, LadY, LeightJL, WozniakMA, et al. (2009) Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol Biol Cell 20 : 3224–3238 doi:10.1091/mbc.E08-12-1186.
4. ZhangH, LandmannF, ZahreddineH, RodriguezD, KochM, et al. (2011) A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature 471 : 99–103 doi:10.1038/nature09765.
5. YinJ, KueblerWM (2010) Mechanotransduction by TRP channels: general concepts and specific role in the vasculature. Cell Biochem Biophys 56 : 1–18 doi:10.1007/s12013-009-9067-2.
6. Sharif-NaeiniR, FolgeringJHA, BichetD, DupratF, LauritzenI, et al. (2009) Polycystin-1 and -2 dosage regulates pressure sensing. Cell 139 : 587–596 doi:10.1016/j.cell.2009.08.045.
7. DuFortCC, PaszekMJ, WeaverVM (2011) Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol 12 : 308–319 doi:10.1038/nrm3112.
8. StosselTP, CondeelisJ, CooleyL, HartwigJH, NoegelA, et al. (2001) Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol 2 : 138–145 doi:10.1038/35052082.
9. NakamuraF, OsbornTM, HarteminkCA, HartwigJH, StosselTP (2007) Structural basis of filamin A functions. J Cell Biol 179 : 1011–1025 doi:10.1083/jcb.200707073.
10. FoxJW, LampertiED, EkşioğluYZ, HongSE, FengY, et al. (1998) Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21 : 1315–1325.
11. TsutsumiS, MaekawaA, ObataM, MorganT, RobertsonS, et al. (2012) A Case of Boomerang Dysplasia with a Novel Causative Mutation in Filamin B: Identification of Typical Imaging Findings on Ultrasonography and 3D-CT Imaging. Fetal Diagn Ther doi:10.1159/000335687.
12. GuergueltchevaV, PeetersK, BaetsJ, Ceuterick-de GrooteC, MartinJJ, et al. (2011) Distal myopathy with upper limb predominance caused by filamin C haploinsufficiency. Neurology 77 : 2105–2114 doi:10.1212/WNL.0b013e31823dc51e.
13. VattemiG, NeriM, PifferS, VicartP, GualandiF, et al. (2011) Clinical, morphological and genetic studies in a cohort of 21 patients with myofibrillar myopathy. Acta Myol 30 : 121–126.
14. ZhouX, TianF, SandzénJ, CaoR, FlabergE, et al. (2007) Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc Natl Acad Sci USA 104 : 3919–3924 doi:10.1073/pnas.0608360104.
15. Farrington-RockC, KirilovaV, Dillard-TelmL, BorowskyAD, ChalkS, et al. (2008) Disruption of the Flnb gene in mice phenocopies the human disease spondylocarpotarsal synostosis syndrome. Hum Mol Genet 17 : 631–641 doi:10.1093/hmg/ddm188.
16. DalkilicI, SchiendaJ, ThompsonTG, KunkelLM (2006) Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol 26 : 6522–6534 doi:10.1128/MCB.00243-06.
17. SokolNS, CooleyL (1999) Drosophila filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr Biol 9 : 1221–1230.
18. LiMG, SerrM, EdwardsK, LudmannS, YamamotoD, et al. (1999) Filamin is required for ring canal assembly and actin organization during Drosophila oogenesis. J Cell Biol 146 : 1061–1074.
19. SokolNS, CooleyL (2003) Drosophila filamin is required for follicle cell motility during oogenesis. Developmental Biology 260 : 260–272.
20. SchmollerKM, LielegO, BauschAR (2009) Structural and viscoelastic properties of actin/filamin networks: cross-linked versus bundled networks. Biophys J 97 : 83–89 doi:10.1016/j.bpj.2009.04.040.
21. NakamuraF, OsbornE, JanmeyPA, StosselTP (2002) Comparison of filamin A-induced cross-linking and Arp2/3 complex-mediated branching on the mechanics of actin filaments. J Biol Chem 277 : 9148–9154 doi:10.1074/jbc.M111297200.
22. EhrlicherAJ, NakamuraF, HartwigJH, WeitzDA, StosselTP (2011) Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478 : 260–263 doi:10.1038/nature10430.
23. LadY, KiemaT, JiangP, PentikäinenOT, ColesCH, et al. (2007) Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J 26 : 3993–4004 doi:10.1038/sj.emboj.7601827.
24. IthychandaSS, QinJ (2011) Evidence for multisite ligand binding and stretching of filamin by integrin and migfilin. Biochemistry 50 : 4229–4231 doi:10.1021/bi2003229.
25. FuruikeS, ItoT, YamazakiM (2001) Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. FEBS Lett 498 : 72–75.
26. PentikäinenU, YlänneJ (2009) The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. J Mol Biol 393 : 644–657 doi:10.1016/j.jmb.2009.08.035.
27. KovacevicI, CramEJ (2010) FLN-1/filamin is required for maintenance of actin and exit of fertilized oocytes from the spermatheca in C. elegans. Dev Biol 347 : 247–257 doi:10.1016/j.ydbio.2010.08.005.
28. DeMasoCR, KovacevicI, UzunA, CramEJ (2011) Structural and Functional Evaluation of C. elegans Filamins FLN-1 and FLN-2. PLoS ONE 6: e22428 doi:10.1371/journal.pone.0022428.t004.
29. McCarterJ, BartlettB, DangT, SchedlT (1999) On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol 205 : 111–128 doi:10.1006/dbio.1998.9109.
30. Lints R, Hall D (2009) Reproductive system, somatic gonad. WormAtlas. Available: http://www.wormatlas.org/hermaphrodite/somatic%20gonad/Somframeset.html. Accessed 21 June 2012.
31. McCarterJ, BartlettB, DangT, SchedlT (1997) Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol 181 : 121–143 doi:10.1006/dbio.1996.8429.
32. HubbardEJ, GreensteinD (2000) The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn 218 : 2–22 doi:10.1002/(SICI)1097-0177(200005)218 : 1<2::AID-DVDY2>3.0.CO;2-W.
33. StromeS (1986) Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol 103 : 2241 doi:10.1083/jcb.103.6.2241.
34. ClandininT (1998) Inositol Trisphosphate Mediates a RAS-Independent Response to LET-23 Receptor Tyrosine Kinase Activation in C. elegans. Cell 92 : 523–533 doi:10.1016/S0092-8674(00)80945-9.
35. YinX, GowerNJD, BaylisHA, StrangeK (2004) Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol Biol Cell 15 : 3938–3949 doi:10.1091/mbc.E04-03-0198.
36. Kim BuiY, SternbergPW (2002) Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol Biol Cell 13 : 1641–1651 doi:10.1091/mbc.02-01-0008.
37. OnoK, OnoS (2004) Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol Biol Cell 15 : 2782–2793 doi:10.1091/mbc.E04-03-0179.
38. KariyaK-I, Kim BuiY, GaoX, SternbergPW, KataokaT (2004) Phospholipase Cepsilon regulates ovulation in Caenorhabditis elegans. Dev Biol 274 : 201–210 doi:10.1016/j.ydbio.2004.06.024.
39. ShibatohgeM, KariyaKI, LiaoY, HuCD, WatariY, et al. (1998) Identification of PLC210, a Caenorhabditis elegans phospholipase C, as a putative effector of Ras. J Biol Chem 273 : 6218–6222.
40. KelleyGG, ReksSE, OndrakoJM, SmrckaAV (2001) Phospholipase C(epsilon): a novel Ras effector. EMBO J 20 : 743–754 doi:10.1093/emboj/20.4.743.
41. RheeSG (2001) Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem 70 : 281–312 doi:10.1146/annurev.biochem.70.1.281.
42. TaylorCW, ToveySC (2010) IP3 Receptors: Toward Understanding Their Activation. Cold Spring Harbor Perspectives in Biology 2: a004010–a004010 doi:10.1101/cshperspect.a004010.
43. FoskettJK, WhiteC, CheungKH, MakDOD (2007) Inositol Trisphosphate Receptor Ca2+ Release Channels. Physiological Reviews 87 : 593–658 doi:10.1152/physrev.00035.2006.
44. WissmannA, InglesJ, MainsPE (1999) The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol 209 : 111–127 doi:10.1006/dbio.1999.9242.
45. PieknyAJ (2003) The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 function as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogenesis. Development 130 : 5695–5704 doi:10.1242/dev.00807.
46. Hunt-NewburyR, ViveirosR, JohnsenR, MahA, AnastasD, et al. (2007) High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5: e237 doi:10.1371/journal.pbio.0050237.
47. BaylisHA, Vázquez-ManriqueRP (2011) Genetic analysis of IP3 and calcium signalling pathways in C. elegans. BBA - General Subjects 1–16 doi:10.1016/j.bbagen.2011.11.009.
48. WalkerDS, GowerNJD, LyS, BradleyGL, BaylisHA (2002) Regulated disruption of inositol 1,4,5-trisphosphate signaling in Caenorhabditis elegans reveals new functions in feeding and embryogenesis. Mol Biol Cell 13 : 1329–1337 doi:10.1091/mbc.01-08-0422.
49. YoshikawaF, MoritaM, MonkawaT, MichikawaT, FuruichiT, et al. (1996) Mutational analysis of the ligand binding site of the inositol 1,4,5-trisphosphate receptor. J Biol Chem 271 : 18277–18284.
50. TianL, HiresSA, MaoT, HuberD, ChiappeME, et al. (2009) Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6 : 875–881 doi:10.1038/nmeth.1398.
51. XuS, ChisholmAD (2011) A Gαq-Ca2+ Signaling Pathway Promotes Actin-Mediated Epidermal Wound Closure in C. elegans. Current Biology 21 : 1960–1967 doi:10.1016/j.cub.2011.10.050.
52. AltunZF, ChenB, WangZ-W, HallDH (2009) High resolution map of Caenorhabditis elegans gap junction proteins. Dev Dyn 238 : 1936–1950 doi:10.1002/dvdy.22025.
53. StarichT, SheehanM, JadrichJ, ShawJ (2001) Innexins in C. elegans. Cell Commun Adhes 8 : 311–314.
54. Vázquez-ManriqueRP, NagyAI, LeggJC, BalesOAM, LyS, et al. (2008) Phospholipase C-epsilon regulates epidermal morphogenesis in Caenorhabditis elegans. PLoS Genet 4: e1000043 doi:10.1371/journal.pgen.1000043.
55. EspeltMV, EstevezAY, YinX, StrangeK (2005) Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. The Journal of General Physiology 126 : 379–392 doi:10.1085/jgp.200509355.
56. Dal SantoP, LoganMA, ChisholmAD, JorgensenEM (1999) The inositol trisphosphate receptor regulates a 50-second behavioral rhythm in C. elegans. Cell 98 : 757–767.
57. IwasakiK, LiuDW, ThomasJH (1995) Genes that control a temperature-compensated ultradian clock in Caenorhabditis elegans. Proc Natl Acad Sci USA 92 : 10317–10321.
58. PieknyAJ, MainsPE (2002) Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J Cell Sci 115 : 2271–2282.
59. PieknyAJ, WissmannA, MainsPE (2000) Embryonic morphogenesis in Caenorhabditis elegans integrates the activity of LET-502 Rho-binding kinase, MEL-11 myosin phosphatase, DAF-2 insulin receptor and FEM-2 PP2c phosphatase. Genetics 156 : 1671–1689.
60. GallyC, WisslerF, ZahreddineH, QuintinS, LandmannF, et al. (2009) Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development 136 : 3109–3119 doi:10.1242/dev.039412.
61. DiogonM, WisslerF, QuintinS, NagamatsuY, SookhareeaS, et al. (2007) The RhoGAP RGA-2 and LET-502/ROCK achieve a balance of actomyosin-dependent forces in C. elegans epidermis to control morphogenesis. Development 134 : 2469–2479 doi:10.1242/dev.005074.
62. VoetsT, NiliusB (2009) TRPCs, GPCRs and the Bayliss effect. EMBO J 28 : 4–5 doi:10.1038/emboj.2008.261.
63. PlayfordMP, NurminenE, PentikainenOT, MilgramSL, HartwigJH, et al. (2010) Cystic Fibrosis Transmembrane Conductance Regulator Interacts with Multiple Immunoglobulin Domains of Filamin A. Journal of Biological Chemistry 285 : 17156–17165 doi:10.1074/jbc.M109.080523.
64. LodhaN, BonfieldS, OrtonNC, DoeringCJ, McRoryJE, et al. (2010) Congenital stationary night blindness in mice - a tale of two Cacna1f mutants. Adv Exp Med Biol 664 : 549–558 doi:10.1007/978-1-4419-1399-9_63.
65. GravanteB (2004) Interaction of the Pacemaker Channel HCN1 with Filamin A. Journal of Biological Chemistry 279 : 43847–43853 doi:10.1074/jbc.M401598200.
66. ZhangM, BreitwieserGE (2005) High affinity interaction with filamin A protects against calcium-sensing receptor degradation. J Biol Chem 280 : 11140–11146 doi:10.1074/jbc.M412242200.
67. SampsonLJ (2003) Direct Interaction between the Actin-binding Protein Filamin-A and the Inwardly Rectifying Potassium Channel, Kir2.1. Journal of Biological Chemistry 278 : 41988–41997 doi:10.1074/jbc.M307479200.
68. PetreccaK, MillerDM, ShrierA (2000) Localization and enhanced current density of the Kv4.2 potassium channel by interaction with the actin-binding protein filamin. Journal of Neuroscience 20 : 8736–8744.
69. van der FlierA, SonnenbergA (2001) Structural and functional aspects of filamins. Biochim Biophys Acta 1538 : 99–117.
70. SeifertJP (2004) RhoA Activates Purified Phospholipase C-ε by a Guanine Nucleotide-dependent Mechanism. Journal of Biological Chemistry 279 : 47992–47997 doi:10.1074/jbc.M407111200.
71. KainulainenT, PenderA, D'AddarioM, FengY, LekicP, et al. (2002) Cell death and mechanoprotection by filamin a in connective tissues after challenge by applied tensile forces. J Biol Chem 277 : 21998–22009 doi:10.1074/jbc.M200715200.
72. GlogauerM, AroraP, ChouD, JanmeyPA, DowneyGP, et al. (1998) The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem 273 : 1689–1698.
73. ShifrinY, AroraP, OhtaY, CalderwoodD, McCullochC (2009) The Role of FilGAP-Filamin A Interactions in Mechanoprotection. Mol Biol Cell doi:10.1091/mbc.E08-08-0872.
74. BellangerJM, AstierC, SardetC, OhtaY, StosselTP, et al. (2000) The Rac1 - and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol 2 : 888–892 doi:10.1038/35046533.
75. WingMR, SnyderJT, SondekJ, HardenTK (2003) Direct activation of phospholipase C-epsilon by Rho. J Biol Chem 278 : 41253–41258 doi:10.1074/jbc.M306904200.
76. SongC, SatohT, EdamatsuH, WuD, TadanoM, et al. (2002) Differential roles of Ras and Rap1 in growth factor-dependent activation of phospholipase C epsilon. Oncogene 21 : 8105–8113 doi:10.1038/sj.onc.1206003.
77. SeifertJP, ZhouY, HicksSN, SondekJ, HardenTK (2008) Dual activation of phospholipase C-epsilon by Rho and Ras GTPases. J Biol Chem 283 : 29690–29698 doi:10.1074/jbc.M805038200.
78. BunneyTD, KatanM (2006) Phospholipase C epsilon: linking second messengers and small GTPases. Trends Cell Biol 16 : 640–648 doi:10.1016/j.tcb.2006.10.007.
79. LopezI, MakEC, DingJ, HammHE, LomasneyJW (2001) A novel bifunctional phospholipase c that is regulated by Galpha 12 and stimulates the Ras/mitogen-activated protein kinase pathway. J Biol Chem 276 : 2758–2765 doi:10.1074/jbc.M008119200.
80. LippP, ReitherG (2011) Protein Kinase C: The “Masters” of Calcium and Lipid. Cold Spring Harbor Perspectives in Biology 3: a004556–a004556 doi:10.1101/cshperspect.a004556.
81. WatrasJ, EhrlichBE (1991) Bell-shaped calcium-response curves of lns(l,4,5)P3 - and calcium-gated channels from endoplasmic reticulum of cerebellum.
82. JafriMS, KeizerJ (1995) On the roles of Ca2+ diffusion, Ca2+ buffers, and the endoplasmic reticulum in IP3-induced Ca2+ waves. Biophys J 69 : 2139–2153 doi:10.1016/S0006-3495(95)80088-3.
83. WagnerJ, KeizerJ (1994) Effects of rapid buffers on Ca2+ diffusion and Ca2+ oscillations. Biophys J 67 : 447–456 doi:10.1016/S0006-3495(94)80500-4.
84. BerridgeMJ, TaylorCW (1988) Inositol Trisphosphate and Calcium Signaling. Cold Spring Harbor Symposia on Quantitative Biology 53 : 927–933 doi:10.1101/SQB.1988.053.01.107.
85. SomlyoAP, SomlyoAV (2000) Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J Physiol (Lond) 522 Pt 2 : 177–185.
86. BatchelderEL, Thomas-VirnigCL, HardinJD, WhiteJG (2007) Cytokinesis is not controlled by calmodulin or myosin light chain kinase in the Caenorhabditis elegans early embryo. FEBS Lett 581 : 4337–4341.
87. WissmannA, InglesJ, McGheeJD, MainsPE (1997) Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes & Development 11 : 409–422 doi:10.1101/gad.11.4.409.
88. CramEJ, ShangH, SchwarzbauerJE (2006) A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci 119 : 4811–4818 doi:10.1242/jcs.03274.
89. KirbyC, KuschM, KemphuesK (1990) Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol 142 : 203–215.
Štítky
Genetika Reprodukčná medicína
Článek Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific RecombinationČlánek Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding YeastČlánek High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple IsolatesČlánek Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance inČlánek Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Functional Elements Are Embedded in Structurally Constrained Sequences
- RNA–Mediated Epigenetic Heredity Requires the Cytosine Methyltransferase Dnmt2
- Loss of Expression and Promoter Methylation of SLIT2 Are Associated with Sessile Serrated Adenoma Formation
- Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific Recombination
- Human Genetics in Rheumatoid Arthritis Guides a High-Throughput Drug Screen of the CD40 Signaling Pathway
- Genome-Wide Analysis in German Shepherd Dogs Reveals Association of a Locus on CFA 27 with Atopic Dermatitis
- Liver X Receptors Protect from Development of Prostatic Intra-Epithelial Neoplasia in Mice
- Chromosomal Organization and Segregation in
- A Statistical Framework for Joint eQTL Analysis in Multiple Tissues
- Cell Polarity and Patterning by PIN Trafficking through Early Endosomal Compartments in
- Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast
- High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Isolates
- Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance in
- A Compendium of Nucleosome and Transcript Profiles Reveals Determinants of Chromatin Architecture and Transcription
- Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation
- Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca
- The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts
- Imputation-Based Meta-Analysis of Severe Malaria in Three African Populations
- Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
- Clathrin and AP2 Are Required for Phagocytic Receptor-Mediated Apoptotic Cell Clearance in
- Encodes CDF Transporters That Excrete Zinc from Intestinal Cells of and Act in a Parallel Negative Feedback Circuit That Promotes Homeostasis
- Global Properties and Functional Complexity of Human Gene Regulatory Variation
- DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in and Other
- Side Effects: Substantial Non-Neutral Evolution Flanking Regulatory Sites
- From Paramutation to Paradigm
- From Mouse to Human: Evolutionary Genomics Analysis of Human Orthologs of Essential Genes
- Distinct Translational Control in CD4 T Cell Subsets
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements
- Identification of Meiotic Cyclins Reveals Functional Diversification among Plant Cyclin Genes
- EGL-13/SoxD Specifies Distinct O and CO Sensory Neuron Fates in
- Congruence of Additive and Non-Additive Effects on Gene Expression Estimated from Pedigree and SNP Data
- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression
- Analysis of the Genetic Basis of Disease in the Context of Worldwide Human Relationships and Migration
- Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses
- Strong Purifying Selection at Synonymous Sites in
- , a Susceptibility Gene for Type 1 and Type 2 Diabetes, Modulates Pancreatic Beta Cell Apoptosis via Regulation of a Splice Variant of the BH3-Only Protein
- Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Meiosis
- The Secretory Pathway Calcium ATPase PMR-1/SPCA1 Has Essential Roles in Cell Migration during Embryonic Development
- The Genomic Signature of Crop-Wild Introgression in Maize
- CDK4 T172 Phosphorylation Is Central in a CDK7-Dependent Bidirectional CDK4/CDK2 Interplay Mediated by p21 Phosphorylation at the Restriction Point
- Genome-Wide Identification of Regulatory RNAs in the Human Pathogen
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



