-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
Most Cryptococccus neoformans genes are interrupted by introns, and alternative splicing occurs very often. In this study, we examined the influence of introns on C. neoformans gene expression. For most tested genes, elimination of introns greatly reduces mRNA accumulation. Strikingly, the number and the position of introns modulate the gene expression level in a cumulative manner. A screen for mutant strains able to express functionally an intronless allele revealed that the nuclear poly(A) binding protein Pab2 modulates intron-dependent regulation of gene expression in C. neoformans. PAB2 deletion partially restored accumulation of intronless mRNA. In addition, our results demonstrated that the essential nucleases Rrp44p and Xrn2p are implicated in the degradation of mRNA transcribed from an intronless allele in C. neoformans. Double mutant constructions and over-expression experiments suggested that Pab2p and Xrn2p could act in the same pathway whereas Rrp44p appears to act independently. Finally, deletion of the RRP6 or the CID14 gene, encoding the nuclear exosome nuclease and the TRAMP complex associated poly(A) polymerase, respectively, has no effect on intronless allele expression.
Published in the journal: Introns Regulate Gene Expression in in a Pab2p Dependent Pathway. PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003686
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003686Summary
Most Cryptococccus neoformans genes are interrupted by introns, and alternative splicing occurs very often. In this study, we examined the influence of introns on C. neoformans gene expression. For most tested genes, elimination of introns greatly reduces mRNA accumulation. Strikingly, the number and the position of introns modulate the gene expression level in a cumulative manner. A screen for mutant strains able to express functionally an intronless allele revealed that the nuclear poly(A) binding protein Pab2 modulates intron-dependent regulation of gene expression in C. neoformans. PAB2 deletion partially restored accumulation of intronless mRNA. In addition, our results demonstrated that the essential nucleases Rrp44p and Xrn2p are implicated in the degradation of mRNA transcribed from an intronless allele in C. neoformans. Double mutant constructions and over-expression experiments suggested that Pab2p and Xrn2p could act in the same pathway whereas Rrp44p appears to act independently. Finally, deletion of the RRP6 or the CID14 gene, encoding the nuclear exosome nuclease and the TRAMP complex associated poly(A) polymerase, respectively, has no effect on intronless allele expression.
Introduction
Introns, discovered in 1977, are genomic sequences that are removed from the corresponding RNA transcripts of genes [1]. First considered just as elements to be removed for correct gene expression, it has since become obvious that they participate in many aspects of gene regulation. Actually, the presence of introns and their splicing by the RNA-protein complex named spliceosome [2] affect gene expression by different means [3] including transcription, polyadenylation, mRNA export, mRNA localisation, translation efficiency and the rate of mRNA decay (see [4] for review). Most eukaryotic genes contain introns although the proportion of genes containing introns is highly variable between organisms. For example, whereas 92% and 78% of the genes in human and plant genomes contain introns, respectively, [5], [6] introns are found in only 5% of the genes in the yeast Saccharomyces cerevisiae [7]. Furthermore, the influence of introns on gene expression differs from one organism to another and from one gene to another.
In mammals, the expression of most of the genes is reduced in the absence of splicing but the effect of introns on gene expression is generally modest [8]. In contrast, the expression of some genes like the β-globin gene or the purine nucleoside phosphorylase gene has been shown to be highly intron-dependent [9], [10]. Introns act mainly at a post-transcriptional level and their absence reduces nuclear and cytoplasmic mRNA accumulation, alters efficient mRNA 3′end formation and consequently reduces nuclear mRNA export [8], [11], [12]. Introns seem also to regulate mRNA translation efficiency [8], [11], [12]. Similarly in plants most mutations can be complemented by cDNA sequences suggesting that most genes do not require introns for expression. For a few genes however, IME (intron-mediated enhancement) of gene expression has been demonstrated [13]. IME has been shown to act at a post transcriptional level and to be, at least for some genes, independent of splicing per se [14], [15]. More recently, IME has been shown to regulate 3′UTR formation and, to a lesser extent, translation [15]. However, in both plants and mammals, the pathway by which mRNAs transcribed from intronless alleles are degraded has not been described [16].
In fungi, the information is even more sparse, with most data coming from studies on S. cerevisiae, in which introns are rare and generally not necessary for gene expression [17], [18]. In a few examples however, introns have been shown to be necessary for gene expression, controlling the export of mRNA from the nucleus [19], [20]. More recently, introns have been shown to be key modulators of ribosomal protein gene expression in the baker's yeast [21]. In the other hemiascomycete yeasts in which the percentage of intron containing genes goes from 2.4% in Candida glabrata to 14.5% in Yarrowia lipolytica [22], introns do not seem to be necessary for gene expression although no specific studies have been reported. Similarly, in Schizosaccharomyces pombe in which 47% of the genes contain introns [23], these are generally not necessary for gene expression [24]. In filamentous fungi like Aspergillus nidulans or Neurospora crassa, cDNA sequences have been widely used for the production of heterologous or homologous proteins [25]–[27] suggesting only a moderate influence of introns on gene expression. In two cases however, one in Podospora anserina and one in Trichoderma viride, introns were reported to be necessary for gene expression but the mechanisms by which this regulation occurs have not been studied [28], [29]. Similarly in basidiomycetes, although intron density is generally higher than in ascomycetes [30], only a few cases of alteration of gene expression by the elimination or the addition of introns have been described [31]–[35]. In Schizophyllum commune, the addition of one intron in a GFP reporter gene has been shown to increase gene expression by altering mRNA accumulation rather than the level of transcription although no further description of the mechanisms by which this regulation occurs has been reported [31].
Cryptococcus neoformans is a capsular basidiomycete yeast mainly studied because it is responsible for opportunistic infections in patients presenting a cellular immune deficiency (mainly AIDS patients) that are fatal if left untreated [36]. The presence of an antiphagocytic polysaccharide capsule and the production of the antioxidant melanin are its two major virulence factors [37], [38]. The genome (20 Mb) sequences of five strains, two of serotype D, one of serotype A, and two of serotype B are now complete [39], [40]. The sequences of the 14 chromosomes of the serotype D strains were annotated using 21000 cDNA sequences isolated from a normalized library. Of the 6574 predicted genes, 80% had confirmed transcripts associated with them. Interestingly, C. neoformans genes are intron-rich and more than 98% of them have been reported to contain introns. Thus, C. neoformans has probably the intron-richest annotated genome described to date. These introns (5 on average per gene) are very small in size (67 bp) whereas exons have a size (250 bp) close to the human ones [40], [41].). Alternative splicing has been reported to be very common in C. neoformans and intron retention represents its most common manifestation [40], [42]. Finally, the fact that the proteome of C. neoformans contains numbers of proteins sharing sequence similarities with known metazoan SR proteins ([43]; Janbon unpublished data) as well as the identification of a DEAD-box helicase as a central regulator of multiple virulence factors [44] suggest that intron-dependent regulation of gene expression might play a major role in C. neoformans biology and virulence.
In this article, we have addressed the importance of introns for gene expression in C. neoformans. We have shown that introns are necessary for mRNA accumulation for some genes but not for others. We also demonstrated that introns can play a positive or a negative role in this process. Finally, we showed that the nuclear poly(A) binding protein Pab2 and the exosome nuclease Rrp44p are implicated in this intron-dependent regulation of gene expression in C. neoformans. Our results also suggested that Xrn2p might act in the same pathway as Pab2p.
Results
Evidence for intron retention at the CAS3 locus
We previously reported that the CAS3 gene contains 12 introns, all of them but the last one (intron 12) being located within the CDS [45]. We performed RACE experiments and noticed that among the five 5′end cDNAs sequenced, two were copies of RNA molecules not spliced in the intron 1 whereas the intron 2 was spliced. In order to identify the different types of CAS3 mRNA molecules present in the cell we sequenced a large number of full length cDNAs. Poly(A) RNA molecules were purified from C. neoformans var. neoformans cells growing in YPD and used for RT-PCR experiments. After separation by gel electrophoresis and purification, 3 pools of 15 full length cDNA molecules were cloned and sequenced (see Material and Methods). As presented in Figure 1A, a large diversity of CAS3 RNA molecules was identified, ranging from completely spliced molecules to completely unspliced ones. Although these experiments were not quantitative, the pattern of splicing observed revealed that some introns were more rarely spliced than others. Introns 1 and 12 were spliced in only 51% and 18% of the 45 sequenced molecules, respectively. RNA-Seq data alignment pattern analysis confirmed that all the introns from this gene can display a certain level of intron retention (Janbon, unpublished data). With the obvious exception of intron 12 which lies in the 3′UTR, all introns of this gene contain at least one in frame stop codon suggesting that none of these intron-containing mRNA molecules could encode a protein.
Fig. 1. Influence of introns on gene expression. 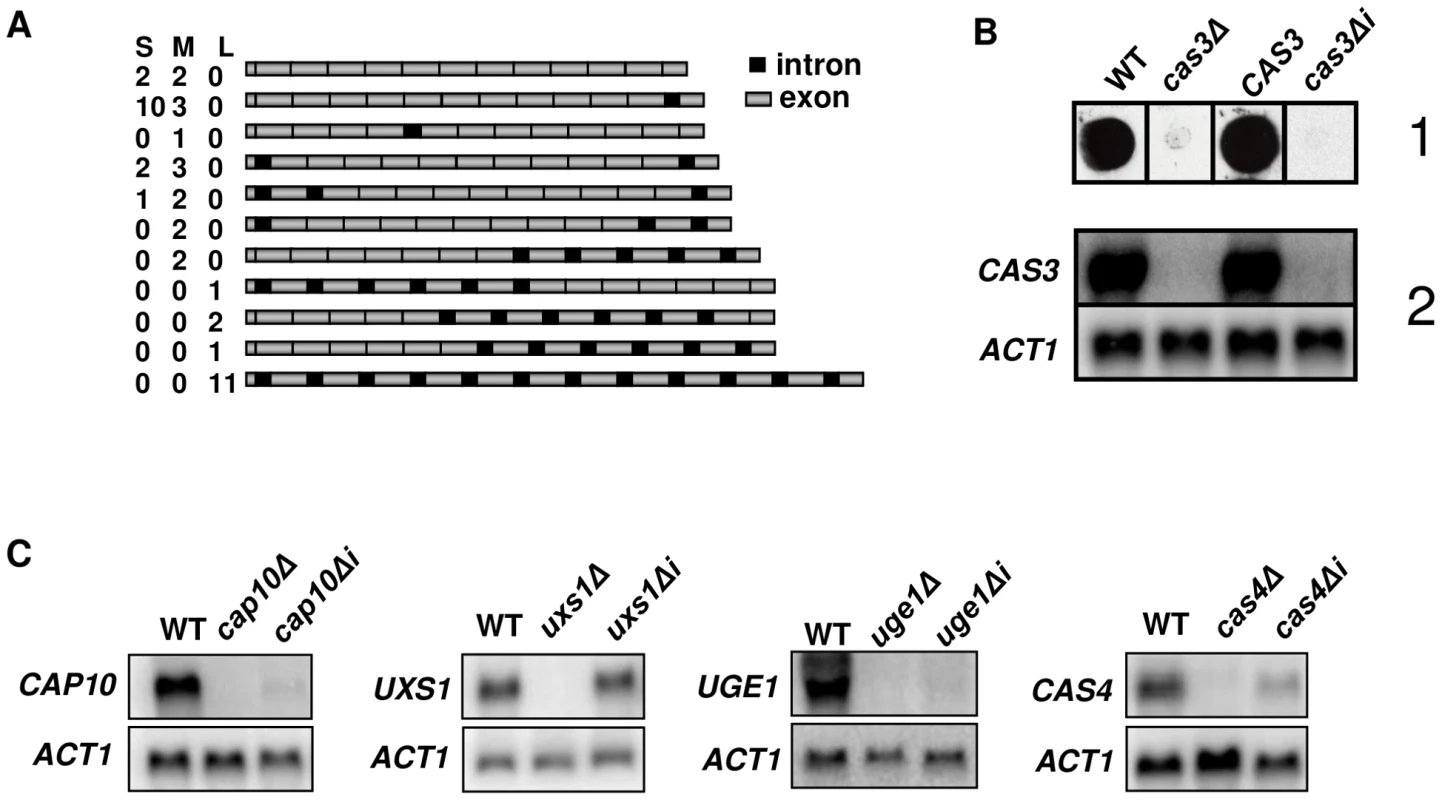
A. Schematic representation of the different cDNA molecules sequenced illustrating intron retention in CAS3 mRNA. CAS3 specific cDNA molecules were RT-PCR-amplified from purified mRNA, separated in three pools according to their sizes (Small, Medium and Large) and cloned. Fifteen clones were sequenced per pool and the presence of introns was analyzed. The numbers of clones obtained for each isoform in each pool are indicated. Introns and exons are not at real scale. B. cas3Δi is a non-functional allele. When the cas3Δ allele was replaced by the intronless allele, there were very little CAS3 mRNAs and no complementation of the capsule phenotype. (1) Antibody reactivity of the GXM-specific Mab CRND-8 with the mutant strains. 104 cells were spotted on a nitrocellulose membrane and probed with CRND-8. (2) Northern blot experiment results showing very low mRNA level of the cas3Δi allele. C. Intron-dependent regulation of mRNA accumulation is gene specific. Two independently obtained Δi strains were tested here giving identical results (data not shown). As a control, we checked that replacing the cas3Δ allele by the wild type gene (CAS3) using the same procedure restored the antibody reactivity and the level of mRNA accumulation. Introns are necessary for CAS3 expression
Evidence for intron retention at the CAS3 locus suggested that at least part of the regulation of this transcript was dependent on introns. So as to analyse the influence of intronic sequences on CAS3 expression we replaced the CAS3 wild type allele by a version without introns (cas3Δi). The co-transformation procedure used here allowed a complete allele replacement at its original locus without any further modification of the local genomic landscape (see Material and Methods). The main phenotype associated with the deletion of CAS3 is a modification of the capsule structure that can be revealed using anti-capsule monoclonal antibodies [46]. As shown in Figure 1B, the capsule structure of the strain bearing cas3Δi was similar to the one in which the gene had been deleted (cas3Δ) suggesting that the intronless allele was not functional. Moreover, Northern blot experiments showed that very little CAS3 specific RNA was present in the cas3Δi strain (Figure 1B). Introns are thus necessary for CAS3 expression.
To verify whether the importance of introns on gene expression was a general feature in C. neoformans, we cloned cDNAs from the genes UXS1, CAP10, UGE1 and CAS4 under the control of their own promoter. These constructs were used to transform the corresponding deletion mutant strains. We then compared mRNA levels of intronless and wild type alleles by Northern blot analysis. As presented in Figure 1C, the influence of introns on mRNA level was gene-dependent. Some genes, like CAP10, UGE1 and to a lesser extend CAS4 were highly intron-dependent whereas others like UXS1 did not depend on the presence of such sequences to be expressed.
CAS3 introns act at a post-transcriptional level
The intron-dependent regulation of mRNA accumulation could act at different levels. Thus, the absence of RNA in the cas3Δi strains could be due to the absence or a very low level of transcription or/and to a decrease of the stability of the corresponding RNA leading to a complete or nearly complete degradation of it. To answer this question, we performed nuclear run-on experiments, thus measuring the frequency of transcription initiation of the different alleles largely independently of the effects of RNA stability [47]. The ratio of the CAS3 specific signal versus the ACT1 specific was not altered by the absence of introns (1.26±0.25) when compared to the wild-type allele (1.29±0.47) (Figure S1). These results suggested that the absence of introns does not alter the transcriptional activity of the gene but rather greatly alters the stability of RNA molecules transcribed from the cas3Δi allele.
Introns influence mRNA accumulation in C. neoformans positively and negatively
So as to better analyse the influence of the introns on gene expression, we constructed a series of alleles bearing different numbers of introns at different positions. These alleles were integrated at the wild-type locus following the same procedure used previously. The level of mRNA was then measured by Northern analysis and confirmed by RT-qPCR (Figure 2). As previously shown, in the absence of introns, very few transcripts can be detected (less than 1% of the wild-type). Surprisingly, the presence of one intron was not enough to restore any expression of the gene as demonstrated by the analysis done with the strains NE293, NE295 and NE300 where the introns 12, 1, or 11 were present, respectively. Even with 2 introns, the expression of the gene remained barely detectable (see strains NE294, NE457 and NE449). The intron 12 and to a lesser extent the intron 1 appeared to play a negative regulatory role in CAS3 expression. Indeed, in strains NE298 and NE299 in which the CAS3 allele lacks the intron 1 and 12, respectively, the expression of this gene went up 2–3 fold. The negative role of the intron 12 in CAS3 gene expression was confirmed by comparing the expression of the CAS3 alleles from the strains NE454 (without intron 12) and NE453 (with intron 12). By comparison the absence of intron 2 influenced poorly the level of expression of the gene (strain NE456). The presence of the other introns regulated CAS3 expression in a positive way. In fact, excepting the regulation by introns 1 and 12, the more introns were present in CAS3 the better the gene was expressed. The identity of the introns did not seem to be important. Indeed, deletion of introns 2 to 6 (strain NE296) altered CAS3 mRNA level to the same degree as the deletion of introns 7 to 11 (NE453). Accordingly, the negative effect of the intron 12 appeared to be more dependent on its position than on its sequence as the re-positioning of the intron 2 at the intron 12 position in the cas3Δi12 allele results in a wild type expression of this intron swapped allele (127% of mRNA accumulation as compared to the wild type) (Figure S2).
Fig. 2. Intron number and position influence mRNA level. 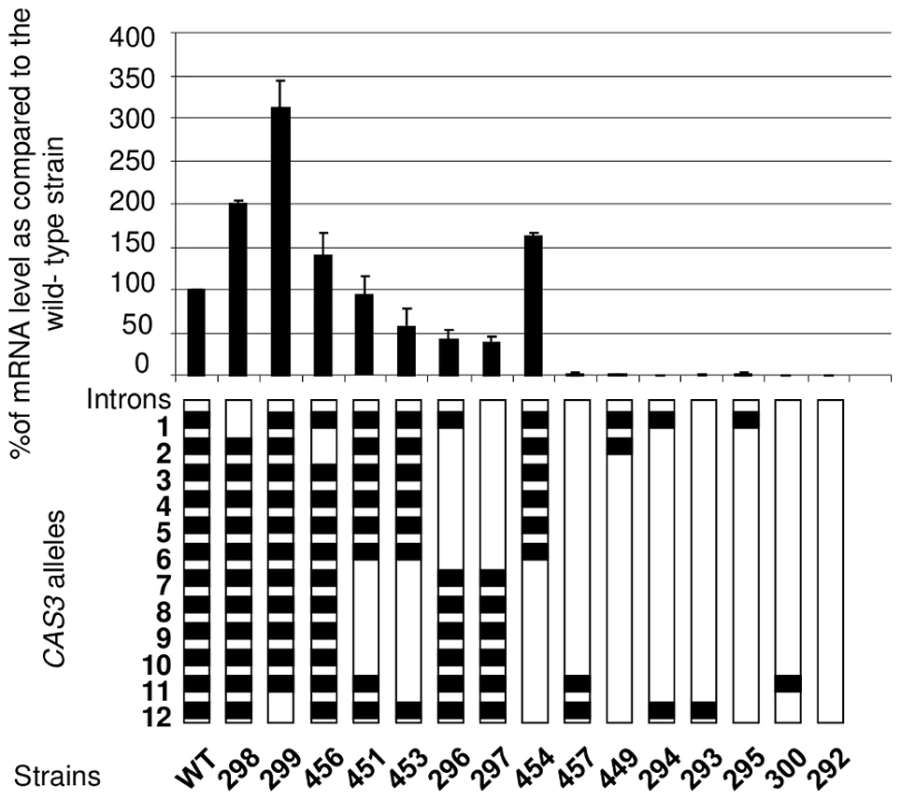
Different CAS3 alleles bearing different numbers and positions of introns were introduced at the original CAS3 locus and the level of CAS3 mRNA was measured by RT qPCR. The reported values are the means ± SD of three independent experiments. Finally, dot blot assays using an anti-capsule antibody were performed to see whether CAS3 mRNA levels correlated with the phenotype of the corresponding strains. Results shown in Figure S3 demonstrated that a level of expression of CAS3 of at least 37% (Figure 2) of the wild-type mRNA level is associated with a wild-type capsule phenotype.
Pab2p regulates intronless allele expression
We aimed to identify elements involved in the degradation of the RNA molecule transcribed from the intronless allele cas3Δi. We constructed and screened an insertional library of C. neoformans mutants using a dot blot assay and the anti-capsule monoclonal antibody CRND-8. More than 5000 mutant strains were tested and fourteen strains were identified as having a low but detectable reactivity with this antibody (data not shown). Northern blot analysis confirmed that all of them expressed the cas3Δi allele at a low level (data not shown). Analysis of the position of the insertion site in the first mutant strain studied, revealed that it was within the gene CNB04570 coding for a protein of 210 amino acids sharing 49% and 32% of amino acid sequence identity with the nuclear poly (A) binding protein of S. pombe [48] and the human one, respectively [49]. Like its human and fission yeast counterparts, the C. neoformans Pab2p sequence presented a single RNA binding domain and an arginine-rich C-terminal domain. However, the poly-alanine domain present in the human protein in which mutations associated with the genetic disease named Oculopharyngeal muscular dystrophy (OPMD) have been identified [49], was absent in both fungal proteins.
We deleted PAB2 using a nourseothricin marker (see Material and Methods). As previously reported in S. pombe, the pab2Δ mutants grow less well at 15°C as compared to the wild-type strains. In C. neoformans, we also found that this mutation results in an alteration of the growth rate at 30°C and 37°C (Figure 3A) and an increased sensitivity to SDS 0.01% as compared to the wild-type. We also studied the classically associated virulence phenotypes and found no evidence of modification of the capsule size or structure and no alteration of the urease production (data not shown). In contrast, we observed a small but reproducible reduction in melanin production (Figure 3A).
Fig. 3. Growth phenotypes associated with PAB2 (panel A), RRP6 (panel B) and RRP44 (panel C) mutations. 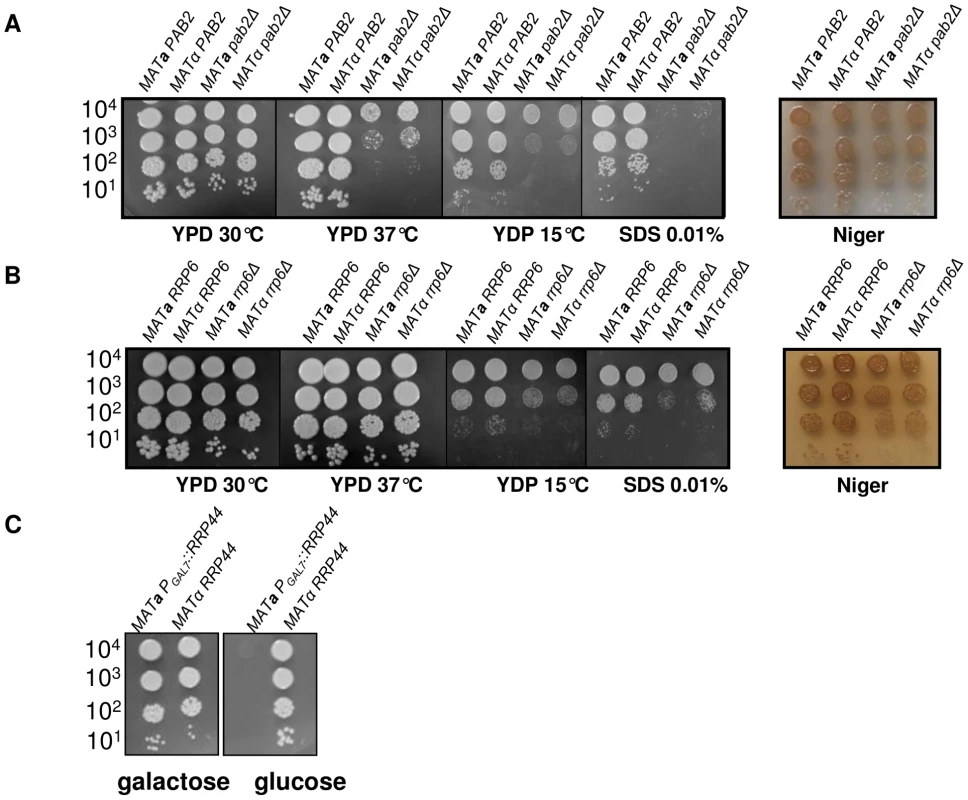
Serial dilutions of cells were spotted on different media. Pictures were taken after three days of incubation. A pab2Δ cas3Δi strain was constructed by selecting adapted progenies after crossing the single mutant strains. Analysis of the expression of this intronless allele in a pab2Δ genetic background confirmed that this protein regulates cas3Δi expression. Indeed, as shown in Figure 4A, whereas PAB2 deletion did not increase the expression of the CAS3 wild type allele, it restored the expression of the intronless allele cas3Δi up to 12% of the wild type (Figure 4A, central panel, grey bars). We also confirmed by ELISA using another anti-capsule monoclonal antibody (Mab 302) that the level of mRNA accumulation in these strains correlated with the phenotype of the corresponding strains (Figure 4A, right panel).
Fig. 4. Pab2p is a nuclear poly(A) binding protein necessary for intron-dependent regulation of CAS3 expression. 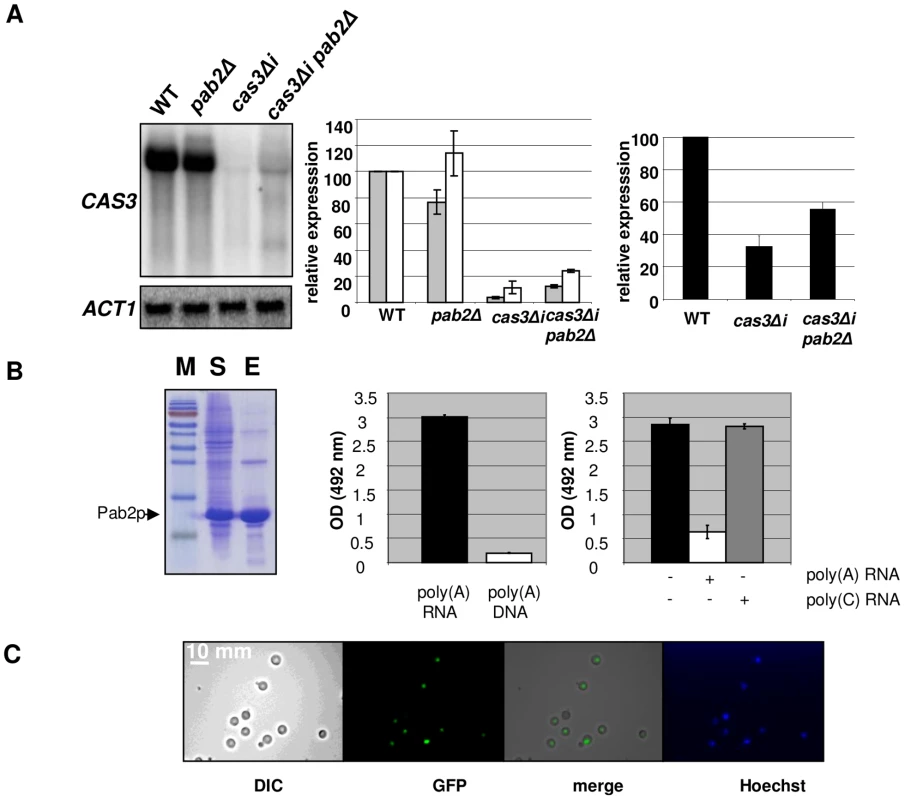
A. Left panel. Northern blot experiment results showing the effect of PAB2 deletion on CAS3 mRNA accumulation. Central panel. Quantification of CAS3 mRNA accumulation in the different genetic backgrounds and normalized to ACT1 mRNA levels. These quantifications were done using RNA extracted from intact cells (grey bars) or from nuclei enriched fractions (white bars). The reported values are the means ± SD of three independent experiments. Right panel. Complementation of the capsule structure phenotypes as measured using the peroxidase-linked anti-capsule Mab 302. B. Left panel. Purification of a recombinant His-tagged-Pab2p in E. coli. 10% SDS/PAGE gel stained with Coomassie blue of E. coli lysate soluble supernatant (lane S), purified protein after affinity Column purification (lane E). Central panel. Pab2p binding assays. Affinity of a recombinant His-tagged Pab2p produced in E. coli to poly(A) was tested in a 96 well-plate format. Each well was coated with a poly(A) 30-mer oligonucleotide RNA. After incubation and washes, the quantity of bound proteins was estimated using an anti-His peroxidase linked monoclonal antibody. The affinity of Pab2p to poly(A) RNA and to poly(A) DNA were compared. Right panel. Competition assays in which 10 µM of unlabeled poly(A) or poly(C) (RNA) were added to the protein solution. C. Cells expressing the GFP-tagged version of the Pab2 protein were grown on glucose at 30°C and examined by epifluorescence or under bright field. Hoechst staining was used as control. We noticed the presence of two additional bands present in Northern blots for the pab2Δ cas3Δi mutant strain (Figure 4A, left panel, lane 4). These bands are probably products of partial degradation of the transcript or the result of partial transcription of the gene. Indeed, hybridizing with oriented RNA probes or with probes specific for the 5′ or 3′ends of the gene demonstrated that these additional bands correspond to the 5′end of the sense transcripts (Figure S4).
We also purified RNA from nuclei isolated using a similar protocol as the one used for the run-on experiments (see Material and Methods) and compared the accumulation of RNA obtained with this nuclei enriched fraction with the ones obtained with RNA extracted from intact cells. As shown in Figure 4A (middle panel), we observed a more pronounced accumulation of the cas3Δi mRNA in the nuclei enriched fraction (white bars), this more pronounced accumulation being exacerbated by a pab2Δ mutation. In good agreement with the localisation experiment data (see below), these results also suggested a nuclear role for Pab2p in the control of intronless allele expression.
Finally, we checked that Pab2p could also modulate the expression of intronless alleles of other genes by constructing a pab2Δ cap10Δi double mutant strain. As shown in Figure S5, the PAB2 deletion also restored the cap10Δi mRNA level close to the wild type level confirming the role of Pab2p in the control of intronless allele expression.
PAB2 encodes a nuclear poly (A) binding protein
To functionally characterise the biophysical properties of Pab2p, we expressed an N-terminal 6XHis-tagged version in E. coli and purified the recombinant protein (see Material and Methods). We tested the affinity of this recombinant Pab2p towards a 30-mer poly(A) RNA coated in a 96-well plate well (see Material and Methods). We found that Pab2p recognized poly(A) oligonucleotides in a dose dependent way (data not shown) and that this recognition was specific to RNA as the same protein presented very little affinity to a 30-mer poly(A) DNA (Figure 4B). Similarly to what has been observed in S. pombe [48], competition assays suggested also that the binding was specific to poly(A) sequences as a poly(C) RNA sequence was not able to compete the binding of Pab2p to poly(A) (Figure 4B).
Next we constructed a GFP::PAB2 allele to localize Pab2p within the cell. We transformed a pab2Δ cas3Δi strain and checked that the transformant selected grew as well as the wild-type strain at all temperatures tested. The functionality of the GFP::Pab2p fusion was confirmed by Northern blot which demonstrated that the fusion protein decreased expression of the allele cas3Δi (data not shown). Examination, by fluorescence microscopy showed a pattern of Pab2p of fluorescence consistent with nuclear localisation (Figure 4C). These results suggested strongly that Pab2p is a nuclear poly(A) binding protein.
The exosome controls cas3Δi expression
Pab2p has been recently shown to interact with the two nucleases of the exosome (i.e. Rrp44p and Rrp6p) to control the synthesis of snoRNAs and the expression of meiotic genes [50]–[52]. It was thus very tempting to hypothesize that this multi-protein complex could regulate the expression of cas3Δi by degrading the RNA transcribed from this allele.
We identified the RRP6 (gene CNC03940) and the RRP44 (gene CND00800) homologues in the genome of C. neoformans and constructed corresponding mutant strains. As in the model yeasts S. cerevisiae and S. pombe, RRP6 was not essential and we were able to delete this gene in C. neoformans. The phenotypes associated with the RRP6 deletion were compared with the ones associated with the PAB2 deletion. As presented in Figures 3A and 3B, pab2Δ and rrp6Δ strains presented a similar growth defect at 30°C. However, in contrast to what we observed with the pab2Δ mutant strains, the rrp6Δ mutants growth defect is not exacerbated when the cells are incubated at 15°C or 37°C and no hyper-sensitivity to SDS 0.01% was observed. The size and structure of the capsule and the urease production were not affected by the deletion of the RRP6 gene (not shown). Interestingly, we observed the same slight defect in melanin production in pab2Δ and rrp6Δ mutant strains when the cells were grown on Niger medium (Figure 3B). Successive unsuccessful attempts to delete RRP44 suggested that this gene is essential in C. neoformans as it is in S. cerevisiae and S. pombe [53], [54]. We thus expressed this gene under the control of the GAL7 promoter which has been shown to be strictly regulated by the presence of galactose in the medium and can be used as a regulatable promoter in promoter swap experiments [55]. On galactose, these cells displayed no specific phenotype although RRP44 was clearly over-expressed (Figure 5B and 5C) whereas on glucose, the PGAL7::RRP44 strains failed to grow, confirming that this gene is essential in C. neoformans (Figure 3C).
Fig. 5. Regulation of cas3Δi mRNA accumulation by the exosome. 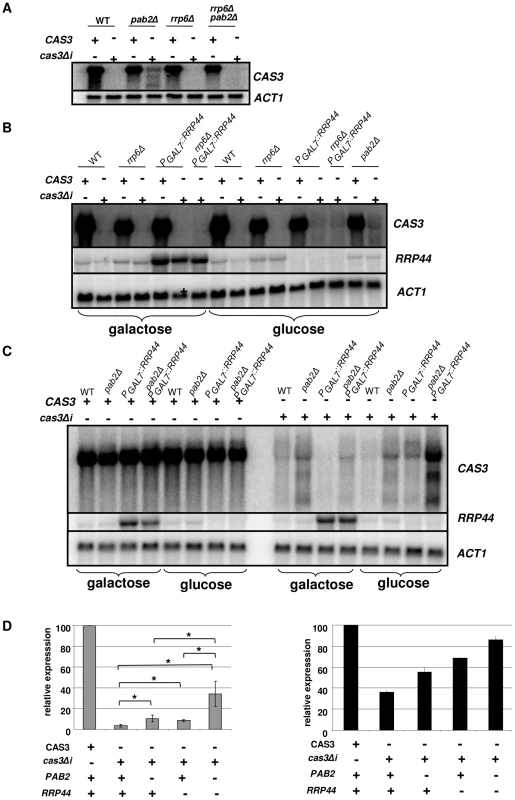
A. Rrp6p does not regulate cas3Δi mRNA accumulation. RNA was extracted from cells growing in YPD (5·107 cells/mL) and analyzed by Northern blot. B. Rrp44p regulates cas3Δi mRNA accumulation. Cells from different mutant strains grown overnight in galactose were washed and transferred to glucose for 6 h before RNA extraction. C. Synergic effect of rrp44 and pab2Δ mutations. Cells from different mutant strains grown overnight in galactose were washed and transferred to glucose for 10 h before RNA extraction D. Left panel. Quantification of CAS3 mRNA accumulation in the different genetic backgrounds. The measurements were normalized to ACT1 mRNA levels. The reported values are the means ± SD of three independent experiments. (* means p<0.05 after student test). Right panel. Complementation of the capsule structure phenotype as measured using the peroxidase-linked anti-capsule Mab 302. To identify the nuclease that regulates expression of intronless CAS3, the cas3Δi allele was next introduced into the rrp6Δ and rrp44 mutant strains. Moreover, we constructed all possible double mutant strains (rrp6Δ pab2Δ, rrp6Δ PGAL7::RRP44 and pab2Δ PGAL7::RRP44) and we introduced the cas3Δi allele in all of them. Deletion of RRP6 did not restore even partially the expression of cas3Δi (Figure 5A), suggesting that Rrp6p is not the nuclease degrading the RNA transcribed from the cas3Δi allele. Surprisingly, RRP6 deletion in a pab2Δ background resulted in reversion to a complete absence of expression of the cas3Δi allele.
We next tested the influence of Rrp44p on the control of cas3Δi expression. To do so, we grew the different strains under the non-restrictive condition (galactose) and then transferred them to the restrictive condition (glucose). Preliminary experiments had shown that as early as 2 hours after the transfer of the cells to glucose medium no RRP44 specific mRNA could be detected by Northern blot analysis when this gene was expressed under the control of the GAL7 promoter (not shown). We compared mRNA levels of CAS3 and cas3Δi under non-restrictive conditions and after 10 hours under restrictive growth conditions in the different genetic backgrounds. Whereas CAS3 mRNA levels were similar in all mutant strains tested, incubation of PGAL7::RRP44 cells under restrictive conditions (glucose) restored the expression of the intronless allele cas3Δi up to 9% of the wild type (Figures 5B and 5D). Similar results were obtained after shorter (6 h) incubation times. This mRNA level was not increased in the rrp6Δ PGAL7::RRP44 double mutant confirming that Rrp6p is not implicated in this regulation. Interestingly, RRP44 appeared to be up-regulated in the absence of RRP6 suggesting a potential explanation for the absence of cas3Δi mRNA in the rrp6Δ pab2Δ double mutant (Figure 5A). The analysis of the double mutant pab2Δ PGAL7::RRP44 revealed a synergic effect of these mutations. As shown in Figures 5C and 5D, the double mutant strains expressed the intronless allele up to 34% of the wild type. Accordingly, the level of mRNA correlated with the phenotypes of the corresponding strains (Figure 5D). These results strongly suggested that Rrp44p and thus the exosome participates in the degradation of mRNA transcribed from the intronless allele cas3Δi. They also demonstrated that the exosome is acting mainly independently of Pab2p suggesting the existence of a least two pathways regulating intronless expression in C. neoformans.
Several RNA species including snRNA, snoRNA, tRNA and rRNA are targeted to degradation by the exosome following polyadenylation by the TRAMP complex [56]. We thus addressed whether this nuclear complex has a role in the regulation of cas3Δi expression. Cid14p has been shown to represent the catalytic subunit responsible for the TRAMP complex poly(A) polymerase activity in S. pombe [57]. We deleted the single homologous gene (CNK02250) in the C. neoformans genome. Neither the cid14Δ strains nor the cid14Δ pab2Δ double mutant strains had any growth phenotype at any temperature tested (30°C, 37°C, 15°C) (not shown). Moreover, no alteration of CAS3 or cas3Δi mRNA levels could be observed (Figure S6). Thus, Cid14p and the TRAMP complex do not seem to be implicated in the regulation of the expression of intronless alleles in C. neoformans.
Finally, as Pab2p has been previously implicated in poly(A) tail length control [58], we performed poly(A)-tests to examine the length of the poly(A) tail in the wild type or in the absence of Pab2p or Cid14p (see Material and Methods). The consequences of these gene deletions on the poly(A) tail length of 10 C. neoformans genes including CAS3 were tested. However, we observed no reproducible modification of length of the poly(A) tails in any of the mutants tested (Figure S7, data not shown).
Depletion of XRN2 is compensated by RRP44 and vice versa
In keeping with a primarily nuclear degradation of cas3Δi (as supported by the nuclear localisation of Pab2p and an enrichment in nuclear fractions) and given the results obtained for the two exosomal nucleases (Rrp6, Rrp44p), the major nuclear 5′→3′ exonuclease Xrn2p/Rat1p appeared to be the most promising candidate for Pab2p-assisted degradation of intronless mRNA. This idea was further encouraged by the finding that deletion of PAB2 not only stabilises full-length cas3Δi but also truncated fragments corresponding to the 5′ end of the sense transcript thus suggesting the involvement of a 5′→3′ exonuclease. In addition, Xrn2p and Rat1p have been shown to be involved in the degradation of unspliced transcripts in human and yeast [59], [60].
Given that XRN2 is likely essential in C. neoformans, we placed the gene (CNF01810) under the control of the GAL7 promoter, similar to the strategy applied for RRP44. As expected, cells failed to grow under restrictive conditions (glucose) showing that XRN2 is indeed essential for C. neoformans viability. However, depletion of XRN2 did not lead to any stabilisation of the cas3Δi transcript neither in a wildtype nor in a pab2Δ context (Figure 6A). Instead a slight decrease of cas3Δi mRNA was observed upon XRN2 depletion in pab2Δ strains (Figure 6A). Accordingly, when we compared the levels of RRP44 expression in wildtype and xrn2 mutant cells, we found an increase in RRP44 expression upon XRN2 depletion (Figure 6B). Likewise expression of XRN2 is elevated in cells depleted for RRP44 (Figure 6B), which might partially explain the rather minor stabilisation of cas3Δi transcripts in rrp44 cells.
Fig. 6. Xrn2p degrades cas3Δi mRNA in a Pab2p-dependent fashion. 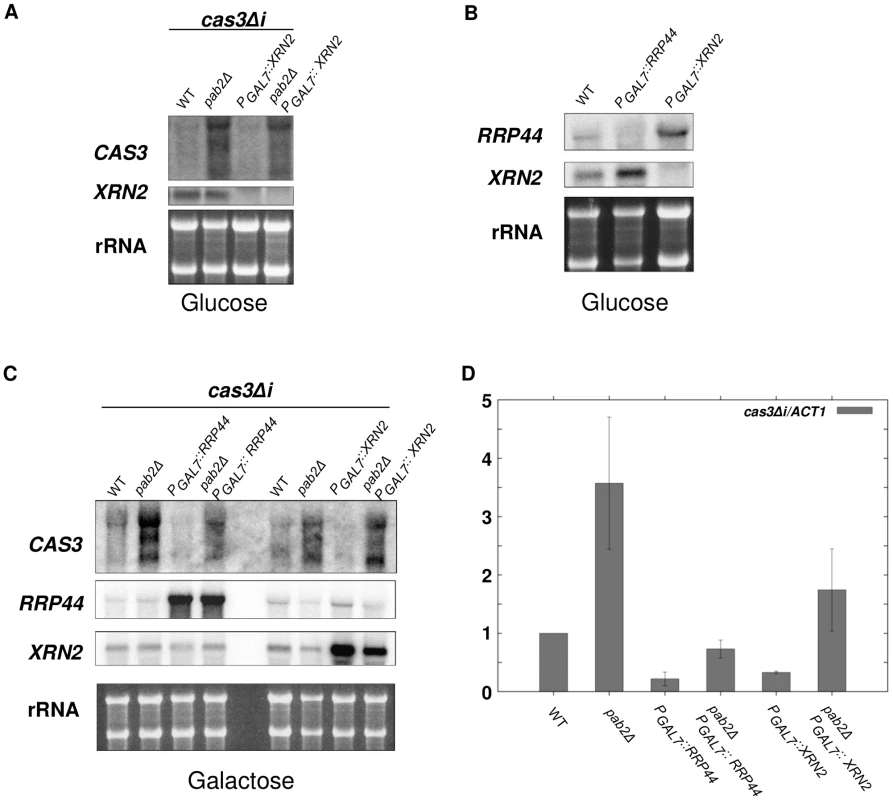
A. Simple depletion of XRN2 does not lead to cas3Δi mRNA stabilisation. Cells from different mutant strains grown overnight in galactose were washed and transferred to glucose for 10 h. RNA was extracted and 5 µg were loaded on a denaturing electrophoresis agarose gel, transferred on a nylon membrane and probed with CAS3 and XRN2 specific probes. B. Depletion of XRN2 leads to increased expression of RRP44 and vice versa. Cells from different mutant strains grown overnight in galactose were washed and transferred to glucose for 10 h. RNA was extracted and 5 µg were loaded on a denaturing electrophoresis agarose gel, transferred on a nylon membrane and probed with RRP44 and XRN2 specific probes. C. and D. The degradation of cas3Δi mRNA by Rrp44p is largely independent of Pab2p whereas that by Xrn2p rather depends on Pab2p. RNA was extracted from cells growing in YPG (5·107 cells/mL). 5 µg were loaded on a denaturing electrophoresis agarose gel, transferred on a nylon membrane and probed with CAS3, RRP44 and XRN2 specific probes C. RNA was treated with DnaseI and 1 µg were used to synthesise cDNA. Each quantitative PCR run was assayed in triplicate. For qPCR analyses primers were chosen that amplify 131 bp of the 5′ part of the cas3Δi transcript ensuring the capture of all isoforms of cas3Δi mRNA stabilised upon PAB2 deletion. The reported values are the means ± SD of three independent experiments. To circumvent this compensatory effect and thus be able to evaluate the role of Xrn2p, we compared cas3Δi mRNA levels when overexpressing either RRP44 or XRN2 in wildtype and pab2Δ backgrounds. To this end, wildtype, pab2Δ, rrp44, xrn2, pab2Δ rrp44 and pab2Δ xrn2 strains were grown in inducing conditions (galactose) and the levels of cas3Δi expression were measured by Northern analysis. Note that upregulation of the intronless allele was reproducibly observed when the strains were grown in galactose (see Figure 5C). Overexpression of RRP44 or XRN2 in a PAB2 wildtype context led to the nearly complete degradation of cas3Δi mRNA preserved by growth in galactose suggesting that both nucleases are implicated in the degradation of these mRNA molecules (Figure 6C). On the other hand, overexpression of RRP6 did not lead to any destabilisation of the cas3Δi transcript thus confirming that Rrp6p has no central role in this regulation (data not shown).
In the absence of Pab2p, RRP44 overexpression led to a strong decrease of cas3Δi mRNA accumulation and to nearly complete elimination of the two additional bands observed by Northern blot in a pab2Δ single mutant context (Figure 6C). In contrast, XRN2 overexpression in a pab2Δ strain led to a very moderate or no decrease of cas3Δi mRNA accumulation. These results were confirmed by RT-qPCR (Figure 6D) although the effect of Xrn2p is probably overestimated in this assay due to the choice of primers specific for the transcript's 5′ end. In conclusion, these overexpression experiments confirm on the one hand that the action of Rrp44p is mainly independent of Pab2p and on the other hand suggest a rather Pab2p-dependent role for Xrn2p in the degradation of the intronless mRNA.
Discussion
The role of introns on gene expression has been the focus of a large number of studies during the last decades [4]. Most of these studies are coming either from mammals or plants or have been performed using the intron-poor micro-organism S. cerevisiae as a model. In these organisms, the replacement of a wild-type gene by an intronless allele generally has a modest effect on gene expression suggesting that introns are more a source of protein diversity or/and regulation of gene expression than a sine qua non condition for a gene to be expressed [8], [18], [21], [61]. In contrast, expression of some genes like the human β-globin or the plant ERECTA genes is highly dependent on the presence of introns [10], [15]. Most of the intron-dependent regulation occurs at a post-transcriptional level although the different steps necessary for the production of a mature mRNA, including transcription and splicing are mutually dependant. [62]. In these cases the presence of introns in a transcript can affect 3′end formation, mRNA export from the nucleus and mRNA stability [8], [63]. However, the pathway(s) by which mRNA molecules transcribed from intronless alleles are recognized and degraded remain unknown [64].
The intron density of the pathogenic yeast C. neoformans is probably the highest yet known for an organism having a completely annotated genome. In fact, a recent re-annotation of the C. neoformans var. grubii genome based on RNA-Seq data showed that 99% of the expressed genes have at least one intron (Janbon, unpublished data). Moreover, 11.5% and 4.1% of genes in C. neoformans have been shown to have 5′ and 3′ UTR introns, respectively [65]. It has also been previously published that alternative splicing is very common in C. neoformans [40], [42]. More recently, a link between transposon pre-mRNA splicing and RNAi dependent degradation has been demonstrated in C. neoformans [66]. Altogether, these data suggested a central role for intron metabolism in the biology and the virulence of C. neoformans. In this study, using the gene CAS3 as a model transcript, we showed that alternative splicing can affect all introns from a single gene although their spliceability appeared to be intron-dependent. We also demonstrated that introns are necessary for the CAS3 gene expression in C. neoformans. Three other tested genes have the same intron-dependence of gene expression whereas another one (UXS1) can be expressed without introns. This insensitivity to the lack of introns does not appear to depend on the number of introns. Indeed CAP10 has only 3 introns, UGE1 only 4 whereas CAS4 and UXS1 have 9 and 7 introns, respectively. Moreover, it probably does not depend on the presence of an intron in the UTR as CAS3 is the only one of the presently studied genes to possess such an intron. It has to be noted that the fact that intronless bacterial antibiotic resistance genes are commonly used for mutant construction in C. neoformans does not contradict this observation. Indeed, all these genes are expressed under the control of the ACT1 promoter, in which an intron is present [67]–[69].
Our results demonstrated that most introns play a positive role on mRNA accumulation and that the absence of introns does not alter the level of transcription as measured by run-on transcription assay. These results are similar to what has been observed in mammals, in the fungus S. commune or for IME in plants in which the regulation of gene expression by introns acts mainly at a post-transcriptional level [8], [31], [61]. In contrast to what has been observed in most cases however, one intron is not enough to restore gene expression. Even with two introns the mRNA level remained below 3% of the wild-type. Most introns played a positive role on gene expression and their action seemed to be more cumulative than specific as previously reported for the ERECTA gene in A. thaliana [15]. The two most external introns (1 and 12) played a negative role on CAS3 mRNA accumulation. Run-on experiments suggested no transcription rate alteration associated with the deletion of either one of these introns (data not shown) suggesting also a post-transcriptional regulatory mechanism. More investigations are obviously needed to understand the role of these introns on mRNA accumulation.
The absence of introns results in an important reduction of mRNA accumulation. Deletion of the PAB2 gene partially stabilized mRNA transcribed from the intronless allele. In contrast, the analysis of the cid14Δ strains suggests no apparent role for the TRAMP complex in this regulation [52]. Pab2p has been shown to interact physically with the two nucleases (Rrp6p and Rrp44p/Dis3p) from the exosome in S. pombe [50]. In C. neoformans, although the deletion of RRP6 encoding the nuclear exosome nuclease has no effect on the accumulation of mRNA transcribed from cas3Δi, the analysis of the level of cas3Δi mRNA in a RRP44 conditional mutant under a restrictive condition strongly implicates this multiprotein complex in this regulation. The analysis of the double mutant strain pab2Δ rrp44 demonstrated a synergic effect of the two mutations suggesting that these two proteins could act in two independent pathways. [70], [71]. As suggested in other Pab2p dependent pathways described to date, Pab2p could be a facilitator for the degradation of cas3Δi mRNA through recruitment of another nuclease. Our double mutant strains analysis and overexpression experiments suggest that the nuclear 5′→3′ exonuclease Xrn2p might represent a good candidate. This model could explain the synergic effect of the pab2Δ rrp44 double mutation. Thus, in the single rrp44 mutant strain, XRN2 is over expressed and can partially compensate the effect of Rrp44p depletion whereas in the absence of Pab2p, XRN2 over-expression would have much less effect on the degradation of cas3Δi transcripts. The fact that Xrn2p/Rat1p has been previously shown to be involved in the degradation of unspliced mRNA in human and yeast [59], [60] sustains this model although no genetic interaction between Xrn2p and Pab2p has been reported to date.
Very recently, Pab2p has been shown to be involved in three different RNA processing and degradation pathways in S. pombe. Thus, together with the exosomal nucleases Rrp6p and independently of the TRAMP complex it controls polyadenylation and synthesis of snoRNAs [50], meiotic gene expression in the Mmi1-dependent pathway [51], [52], [72] and targets ribosomal pre-mRNA RPL30-2 [73]. Similarly, in the meiotic gene expression and pre-mRNA RPL30-2 regulation, a synergic effect was observed when RRP44 and PAB2 were mutated suggesting here also that the effect of Rrp44p could be mainly independent of Pab2p although the other elements involved in this Rrp44p-dependent pathway remain to be identified.
The role of Pab2p in the intronless gene expression regulation remains mysterious. In S. pombe, Pab2p is recruited to the nascent mRNA before 3′ end formation and polyadenylation and controls the length of poly(A) of only a subset of RNAs [48], [50], [71]. It also physically interacts both with the exosome nucleases and the poly(A) polymerase Pla1p [52], [71]. In the absence of Pab2p, some cas3Δi mRNA molecules are exported from the nucleus and translated although most of them are still degraded. The subcellular localisation of Pab2p together with the analysis of the accumulation of the mRNA transcribed from the intronless allele in the nucleus, suggested strongly a nuclear role for this protein although Pab2p has been shown to be able to shuttle to the cytoplasm in S. pombe and in Drosophila [70], [71]. The kinetic of degradation of intronless mRNA in C. neoformans might be the result of a disequilibrium between mRNA export from the nucleus and degradation. Thus, when not enough introns are present the altered dosage of mRNA binding proteins would result in an extended retention time of mRNA in the nucleus giving time to the nucleases to degrade them. The absence of Pab2p would slow down this degradation giving time to some mRNA molecules to be exported in a “take the money and run” strategy [74] (Figure 7).
Fig. 7. Model for intron-dependent gene expression regulation in C. neoformans. 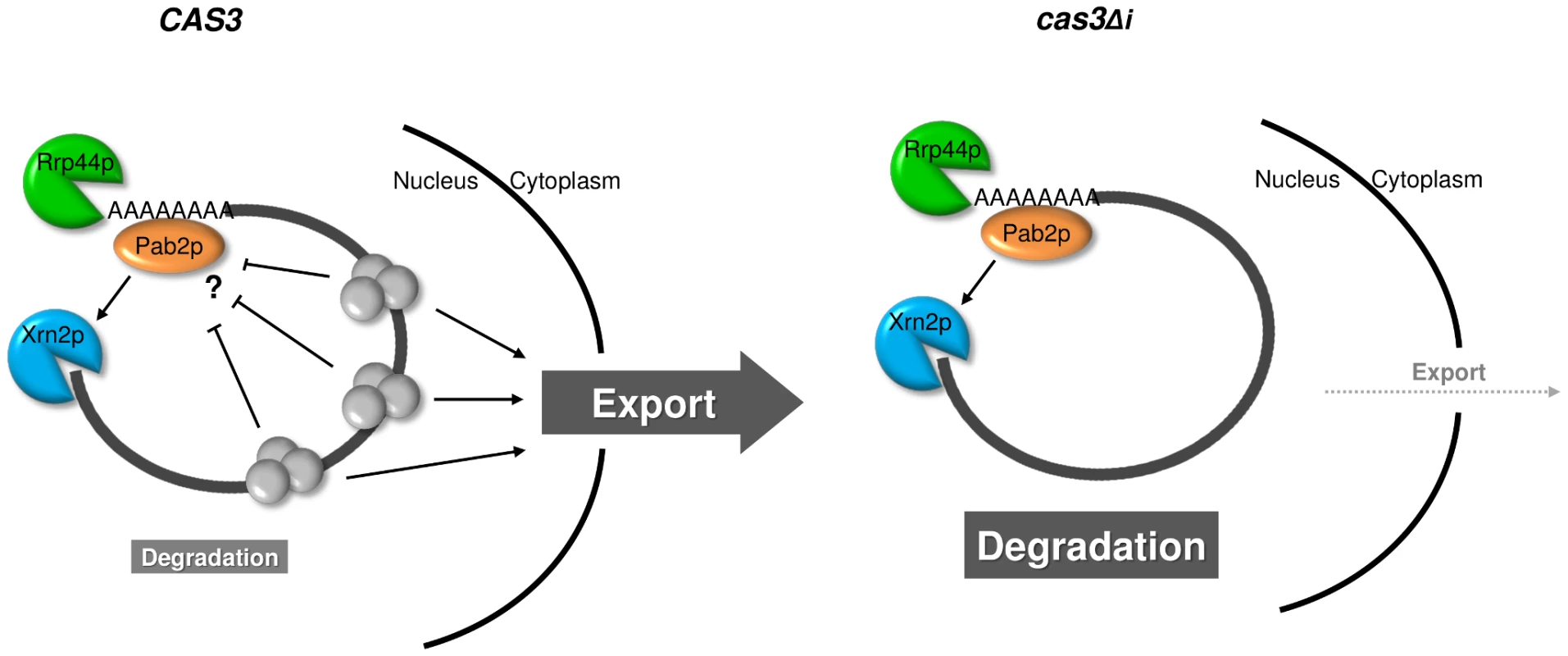
In a wild type context (CAS3), protein complexes (EJC) are deposited on the mRNA upon splicing and facilitate mRNA export. The presence of these complexes might also protect mRNA from degradation. In the intronless context (cas3Δi), the absence of splicing impedes EJC deposition and the mRNA is inefficiently exported giving more time to Rrp44p and Xrn2p/Pab2p to degrade it. In terms of evolution, the comparison of S. cerevisiae and C. neoformans provides a fascinating example of opposite evolutionary choices. Whereas S. cerevisiae has lost almost all its introns and has largely simplified its RNA metabolism (i.e. loss of RNAi pathway, only one SR protein, absence of EJC-like complex…), C. neoformans has conserved and maybe increased its intron number and appears to have a very complex RNA metabolism. The selective pressure that has maintained introns in one organism and has eliminated them in another one is unknown. It has to be noted that C. neoformans is not a unique example among basidiomycete fungi. Thus, genes from Coprinus cinereus and Phanerochaete chrysosporium have an intron density close to that of C. neoformans [30]. In two other pathogens, Ustilago maydis for the plants and Malassezia sp. for human for example, the number of genes with introns is small [75], [76]. This specificity might be related to the fact that C. neoformans is an opportunistic human pathogen living in the environment. As such the diversity of signals to which it can be exposed in the human body or in soil for example is huge. Indeed, this organism has to cope with a large number of different stresses and probably needs a very flexible metabolism. It is tempting to hypothesize that its complex RNA metabolism provides a mechanism to achieve such flexibility.
Materials and Methods
Strains and culture conditions
C. neoformans strains used in this study all originated from the serotype D strain JEC21 [77] and are listed in Table S1. The strains were routinely cultured on YPD medium at 30°C [78]. Synthetic dextrose (SD) was prepared as described [78]. The capsule sizes were estimated after 24 h of growth in capsule-inducing medium at 30°C as previously described [79]. Melanin and urease production were assessed after spotting 105 cells of each strain on Niger or Christensen agar medium, respectively [69], [80]; the plates were read after 48 h of incubation at 30°C. The bacterial strain Escherichia coli XL1-blue (Stratagene) was used for the propagation of all plasmids.
RNA extraction and Northern blot analysis
Cells were routinely harvested after being grown up to 5·107 cells/mL in YPD. RNA was extracted with TRIZOL Reagent (Invitrogen) following the manufacturer's instructions. Total RNA (5 µg) was separated by denaturing agarose gel electrophoresis and transferred onto Hybond-N+ membrane (Amersham) and probed with [32P]dCTP-radiolabelled DNA fragments. The banding pattern was quantified with a Typhoon 9200 imager and Image Quantifier 5.2 software (Molecular dynamics).
CAS3 cDNA analysis
Total RNA was extracted from JEC21 cells growing on YPD. mRNA was purified using Oligotex Direct mRNA Mini Kit (Qiagen) following the manufacturer's instructions. SMARTer RACE cDNA Amplification Kit (Clontech) was used to synthesize the cDNA. CAS3 cDNA was PCR amplified using the primers CAS3a and CAS3AR (see Table S2) and was analysed by agarose gel electrophoresis. The presence of a smeary pattern on the gel suggested the presence of different types of cDNA molecules. The amplified fragments were gel purified in three different pools of sizes and 15 cDNA molecules from each pool were cloned in a pGEMT plasmid and sequenced.
Insertional mutagenesis
An insertional mutant library was constructed in a NE292 (MATa cas3Δi ura5) background using the Agrobacterium tumefaciens strain EHA105 transformed with the plasmid pPZP-NEO1 as previously described [81]. A total of 5796 colonies were transferred from the transformation plates to 96-well plate wells containing 75 µL of capsule inducing medium [79] supplemented with adenine (20 mg/L) and uracil (5 mg/L). The mutants were then tested with the anti-capsule monoclonal antibody CRND-8 [82] as previously described [83]. Positive mutant strains were isolated and tested a second time using the same strategy. Total RNA was extracted and the presence of CAS3 mRNA was analysed by Northern blot.
Recombinant protein production
PAB2 cDNAs were amplified by PCR and inserted into the pQ-30 E. coli expression vector (Qiagen). The E. coli BL21 transformant strains were grown in 50 mL of YT containing ampicillin (50 µg/ml) and kanamycin (30 µg/ml) to an OD600 of 0.5; gene expression was induced by adding 1 mM of IPTG and incubation for 4 hours at 37°C. The cells were then disrupted by sonication and centrifuged at 3000·g. The supernatant was recovered and the recombinant proteins were purified by affinity chromatography on a Ni-NTA column (Qiagen) following the manufacturer's procedures. The protein solution was adjusted to 20% (w/v) glycerol (final concentration 140 µg/mL) and stored in aliquots at −80°C.
Poly(A) binding assays
These experiments were conducted in 96-well Streptavidin coated plates (Nunc). For each sample and concentration to be tested one well was washed three times with 200 µl of washing buffer (Tris 25 mM, Nacl 150 mM, pH 7.2, BSA 1% wt/vol, Tween 20 0.05% wt/vol). Each well was then incubated for 2 h at room temperature and under agitation (700 rpm) with 100 µl of washing buffer containing 0.1 µM of poly(A) 30-mers oligonucleotide 5′ biothinylated. Unbound oligonucleotides were then eliminated through three washes with 200 µL of washing buffer. Each well was then incubated with 100 µl of recombinant Pab2p solution (0.70 mg/mL) for 30 min at room temperature under agitation (700 rpm). After three washes with 200 µL of washing buffer, the quantity of poly(A)-bound protein was estimated using an anti-His peroxidase linked monoclonal antibody (Qiagen) and OPD (O-phenylenediamine dihydrochloride) (Sigma) following the manufacturer's procedures. After 10 min of incubation at room temperature, the colorimetric reaction was stopped by the addition of H2SO4 4% (v/v) and the optic density was measured at 492 nm. For the competition assays, 10 µM of unlabeled poly(A) or poly(C) (Sigma) was added to the protein solution.
Pab2p subcellular localization with fluorescent protein fusion
To localize the Pab2 protein, the PAB2 gene under the control of its own promoter was joined in-frame to a sequence encoding the GFP protein at its N-terminal end. Primers used for amplification are listed in Table S2. A pab2Δ strain was transformed with a plasmid containing the URA5 marker and the Pab2-fluorescent protein fusion by biolistic delivery [84]. Transformants were grown on minimum medium and analyzed for fluorescence.
Intron allele construction
The pBluescript (Stratagene) based plasmid pNE247 contained a 4067 bp DNA fragment containing the complete C. neoformans CAS3 gene PCR amplified using the primers CAS3F and CAS3R (see Table S2) and cloned at the NotI site. This plasmid was used to construct all the CAS3 alleles presented in this study. For the intronless allele, the cDNA from CAS3 was amplified, cloned and sequenced (see above). A completely spliced molecule was digested with SphI and PstI and cloned at the SphI-PstI site of pNE247, thus replacing the wild type gene by an intronless version under the control of its own promoter. 5 µg of the resulting plasmid pNE254 were NotI digested and mixed with 1 µg of the URA5 containing plasmid pNE10 [79] digested with NotI. This DNA solution was used to transform the strain NE128 (MATa cas3Δ:ADE2 ura5) [46] by biolistic transformation. The transformants were selected on a minimum medium containing adenine at 20 mg/L. After three days at 30°C, the transformation plates were transferred to room temperature and one week after transformation some colonies developed a pink phenotype suggesting that the ADE2 gene had been lost and thus that the cas3Δ::ADE2 allele has been replaced by the cas3Δi allele. The pink colonies were then cultured in liquid YPD so as to loose the unstable pNE10 plasmid [79]. Ura− strains were selected on FOA. The absence of the cas3Δ::ADE2 allele and the correct integration of the intronless allele were confirmed by PCR. The absence of additional integrations in the genome was confirmed by Southern blot. Two independent mutant strains were selected and stored at −80°C. Similar strategies were used to construct the other alleles and the other mutant strains.
Nuclear run-on assay and nuclear fraction preparation
For nuclei purification, 500 mL of culture (OD600 = 3) were harvested by centrifugation. Spheroplasts were prepared as previously described [85] and re-suspended in 3 mL of lysing buffer (Pipes 10 mM pH 6.9, sucrose 0.5 M, CaCl2 5 mM, MgSO4 5 mM, DTT 1 mM) containing a complete set of antiproteases (Roche). The spheroplasts were then mechanically disrupted and the intracellular organelles were separated from cellular debris and unbroken cells by centrifugation. Nuclei were purified by differential ultracentifugation (1 hour, 161 000·g, 4°C) through a separation buffer (Pipes 10 mM, sucrose 2.1 M CaCl2 5 mM, MgSO4 5 mM, DTT 1 mM) containing a complete set of antiproteases (Roche). Nuclei were then washed twice with conservation buffer (TrisHCl 50 mM pH 8.3, glycerol 40%, MgCl2 5 mM, EDTA 0.1 mM pH 8), re-suspended in 500 µl of conservation buffer and stored in aliquots at −80°C. For each run on experiment 100 µL of nuclei suspension was used following a protocol previously described [47]. The radioactive transcripts produced were used to hybridize a serial dilution of DNA spotted on a nylon membrane. The plasmid pNE428 containing the 1558 bp fully spliced JEC21 CAS3 cDNA amplified with the primers CAS3a and CAS3AR and cloned in the pGEMT plasmid (Clontech) was used as CAS3 specific DNA. The plasmid pNE435 containing the DNA 519 bp DNA fragment PCR from JEC21 genomic DNA using the primers ACT1F and ACT1R cloned in pGEMT was used as ACT1 specific DNA. The plasmid pGEMT alone was used as negative control. The intensity of the signal was quantified with a Typhoon 9200 imager and Image Quantifier 5.2 software (Molecular dynamics). Each experiment was repeated twice using two independent nuclei preparations. The same protocol of nuclei preparation was used to isolate the nuclear RNA fraction. Electrophoretic analysis of these RNA samples showed a clear decrease in the rRNA proportion confirming the quality of our preparation (data not shown).
Gene disruption
The genes described in this report have been deleted by biolistic transforming a serotype D strain using a disruption cassette constructed by overlapping PCR as previously described [45]. The primer sequences used are given in Table S2. The transformants were then screened for homologous integration as previously described [46]. The plasmid, pNAT used to amplify the NAT selective marker was kindly provided by Dr Jennifer Lodge (Saint Louis University School of Medicine). The plasmid pPZP-NEO1 used to amplify the NEO selective marker was kindly provided by Dr Joe Heitman (Duke University). Multiple mutant strains were obtained through crosses of single mutant strains on V8 medium as previously described [45]. Progenies were selected on minimum medium to which different amino acids were added. Their genotypes were determined by PCR. The mating types of the strains were determined by testing them on V8 medium in the presence of tester strains of known mating type.
Promoter swap
A four way overlap PCR gene deletion was used to generate the promoter-specific exchange cassettes of RRP44 and XRN2, which included a nourseothricin and a neomycin cassette, respectively. The primers used in these experiments are listed in Table S2. The GAL7 promoter was used as the inducible promoter [55]. The 694 bp upstream the RRP44 ATG and the 601 bp upstream the XRN2 ATG were replaced by the 556 bp present upstream of the GAL7 gene. Transformants were screened for homologous integration as previously described [86].
Measure of polyadenylation
The ePAT and TVN-PAT reactions were performed using 1 µg of input RNA as previously described [87]. The sequences of the primers used for PCR amplification are listed in Table S2. The cDNA was column-purified using NucleoSpin Gel and PCR Clean-up columns (MACHEREY-NAGEL). Specific PCR products were analysed by 2% high resolution agarose gel (Ultra pure 1000; Life Technologies) pre-stained with sybr safe (Life Technologies) and imaged against a 100 bp ladder (New England Biolabs) using an LAS 3000 imager and multigauge software (Fujifilm).
RT-qPCR
Total RNA was subjected to an initial DNaseI (Roche) treatment to eliminate contaminating genomic DNA. 1 µg of the DNaseI treated RNA was then reverse transcribed using the kit QuantiTect Reverse Transcription (Qiagen) following the manufacturer's instructions.
Quantitative PCR assays were performed according to Bio-Rad manufacturer's instructions using 96-well optical plates (Thermo Scientific) and an iCycler iQ (170–8740, Biorad).
Each run was assayed in triplicate in a total volume of 25 µL containing the 5 µL cDNA template at an appropriate dilution, 1× Absolute qPCR SYBR Green Fluorescein (Thermo Scientific) and 320 nM of each primer. The primers used are listed in Table S2. PCR conditions were: 95°C/15 min for one cycle, 95°C/30 s for 40 cycles. Amplification of one single specific target DNA was checked with a melting curve analysis (+0.5°C ramping for 10 s from 55°C to 95°C). The Ct values obtained in triplicate were averaged and normalised to that of the housekeeping gene ACT1 using standard curves. To verify the absence of genomic DNA contamination, negative controls in which reverse transcriptase was omitted were used. Three independent biological replicates were performed.
Supporting Information
Zdroje
1. SambrookJ (1977) Adenovirus amazes at Cold Sring Harbour. Nature 268 : 101–104.
2. WahlMC, WillCL, LührmannR (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136 : 701–718.
3. MooreMJ, SilverPA (2008) Global analysis of mRNA splicing. RNA 14 : 197–203.
4. Le HirH, NottA, MooreMJ (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sciences 28 : 215–220.
5. International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431 : 931–945.
6. HaasBJ, WortmanJR, RonningCM, HannickLI, et al. (2005) Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol 22 : 3–7.
7. JuneauK, PalmC, MirandaM, DavisRW (2007) High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci USA 104 : 1522–1527.
8. LuS, CullenBR (2003) Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9 : 618–630.
9. JonssonJJ, GoresmanMD, WilsonN, McIvorRS (1992) Intron requirement for expression of the human purine nucleoside phosphorylase gene. Nucleic Acids Res 20 : 3191–3198.
10. BuchmanAR, BergP (1988) Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol 8 : 4395–4405.
11. RyuWS, MertzJE (1989) Simian virus 40 late transcripts lacking exisable intervening sequences are defective in both stability in the nucleus and transport to the cystoplasm. J Virol 63 : 4386–4394.
12. ValenciaP, DiasAPRR (2008) Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci USA 105 : 3386–3391.
13. MascarenhasD, MettlerIJ, PierceDA, LoweHW (1990) Intronmediated enhancement of heterologous gene expression in maize. Plant Mol Biol 15 : 913–920.
14. RoseAB, BeliakoffJA (2000) Intron-mediated enhancement of gene expression independent of unique intron sequences and splicing. Plant Physiology 122 : 535–542.
15. KarveR, LiuW, WilletSG, TorriiKU, et al. (2011) The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA 17 : 1907–1921.
16. WangHF, FengL, NiuDK (2007) Relation between mRNA stability and intron presence. Biochem Biophys Res Com 354 : 203–208.
17. ParenteauJ, DurandM, VéronneauS, LacombeAA, et al. (2008) Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol Biol Cell 19 : 1932–1945.
18. JuneauK, MirandaM, HillenmeyerME, NislowC, et al. (2006) Introns regulate RNA and protein abundance in yeast. Genetics 174 : 511–508.
19. GalyV, GadalO, Fromont-RacineM, RomanoA, et al. (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116 : 63–73.
20. Cuenca-BonoB, Garcia-MolineroV, Pascual-GarciaP, DopazoH, et al. (2011) SUS1 introns are required for efficient mRNA nuclear export in yeast. Nucleic Acids Res 39 : 8599–8611.
21. ParenteauJ, DurandM, MorinG, GagnonJ, et al. (2011) Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147 : 320–331.
22. NeuvégliseC, MarckC, GaillardinC (2011) The intronome of budding yeasts. C R Biol 334 : 662–670.
23. WoodV, GwilliamR, RajandreamMA, LyneM, et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415 : 871–880.
24. Giga-HamaY, KumagaiH (1999) Expression system for foreign genes using the fission yeast Schizosaccharomyces pombe. Biotechnol Appl Biochem 30 : 235–244.
25. LubertozziD, KeaslingJD (2009) Developing Aspergillus as a host for heterologous expression. Biotech Advances 27 : 53–75.
26. ShellyJ, Rasmussen-WilsonJS, PalasVJ (1997) Expression of a Plant Protein by Neurospora crassa. Appl Environ Microbiol 63 : 3488–3493.
27. AllgaierS, WeilandN, HamadI, KempkenF (2009) Expression of ribonuclease A and ribonuclease N1 in the filamentous fungus Neurospora crassa. Appl Microbiol Biotechnol 85 : 1041–1049.
28. Dequard-ChablatM, RötigA (1997) Homologous and heterologous expression of a ribosomal protein gene in Podospora anserina requires an intron. Mol Gen Genet 253 : 546–552.
29. XuJ, GongZZ (2003) Intron requirement for AFP gene expression in Trichoderma viride. Microbiology 149 : 3093–3097.
30. StajichJE, DietrichFS, RoySW (2007) Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol 8: R223.
31. LugonesLG, ScholtmeijerK, KlootwijkR, WesselsJGH (1999) Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol Microbiol 32 : 681–689.
32. YamazakiT, OkajimaY, KawashimaH, TsukamotoA, et al. (2006) Intron-dependent accumulation of mRNA in Coriolus hirsutus of lignin peroxidase gene the product of which is involved in conversion/degradation of polychlorinated aromatic hydrocarbons. Biosci Biotechnol Biochem 1293–1299.
33. KilaruS, CollinsCM, HartleyAJ, BurnsC, et al. (2009) Investigating dominant selection markers for Coprinopsis cinerea: a carboxin resistance system and re-evaluation of hygromycin and phleomycin resistance vectors. Curr Genet 55 : 543–550.
34. BurnsC, GregoryKE, KirbyM, CheungMK, et al. (2005) Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol 42 : 191–199.
35. MaB, MayfieldMB, GoldMH (2001) The green fluorescent protein genefunctions as a reporter of gene expression in Phanerochaete chrysosporium. Appl Environ Microbiol 67 : 948–955.
36. Casadevall A, Perfect JR (1998) Cryptococcus neoformans. Washington, D.C.: American Society for Microbiology Press.
37. IdnurmA, BahnYS, NielsenK, LinX, et al. (2005) Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol 3 : 753–764.
38. JanbonG (2004) Cryptococcus neoformans capsule biosynthesis and regulation. FEMS Yeast Res 4/8 : 765–771.
39. D'SouzaCA, KronstadJW, TaylorG, WarrenR, et al. (2011) Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2: e00342–10.
40. LoftusB, FungE, RoncagliaP, RowleyD, et al. (2005) The genome and transcriptome of Cryptococcus neoformans, a basidiomycetous fungal pathogen of humans. Science 307 : 1321–1324.
41. KupferDM, DrabenstotSD, BuchananKL, LaiH, et al. (2004) Introns and splicing elements of five diverse fungi. Eukaryot Cell 3 : 1088–1100.
42. McGuireAM, PearsonMD, NeafseyDE, GalaganJE (2008) Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol 9: R50.
43. WarneckeT, J.LP, HurstLD (2008) Finding exonic islands in a sea of non-coding sequence: splicing related constraints on protein composition and evolution are common in intron-rich genomes. Genome Biol 9: R29.
44. PanepintoJ, LiuL, RamosJ, ZhuX, et al. (2005) The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J Clin Invest 115 : 632–641.
45. MoyrandF, ChangYC, HimmelreichU, Kwon-ChungKJ, et al. (2004) Cas3p belongs to a seven member family of capsule structure designer proteins. Eukaryot Cell 3 : 1513–1524.
46. MoyrandF, JanbonG (2004) UGD1 encoding the Cryptococcus neoformans UDP-glucose dehydrogenase is essential for growth at 37°C and for capsule biosynthesis. Eukaryot Cell 3 : 1601–1608.
47. SmaleST (2009) Nuclear Run-On Assay. Cold Spring Harb Protoc doi:10.1101/pdb.prot5329
48. PerreaultA, LemieuxC, BachandF (2007) Regulation of the nuclear poly(A) binding protein by arginine methylation in fission yeast. J Biol Chem 282 : 7552–7562.
49. BraisB, BouchardJP, XieYG, RochefortDL, et al. (1998) Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet 18 : 164–167.
50. LemayJF, D'AmoursA, LemieuxC, LacknerDH, et al. (2010) The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell 37 : 34–45.
51. St-AndréO, LemieuxC, PerreaultA, LacknerDH, et al. (2010) Negative regulation of meiotic gene expression by the nuclear poly(A)-binding protein in fission yeast. J Biol Chem 285 : 27859–27868.
52. YamanakaS, YamashitaA, HarigayaY, IwataR, et al. (2010) Importance of polyadenylation in the selective elimination of meiotic mRNA in growing S. pombe cells. EMBO J 29 : 2173–2181.
53. KinoshitaN, GoeblM, YanagidaM (1991) The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol Cell Biol 11 : 5939–5947.
54. MitchellP, PetfalskiE, ShevchenkoA, MannM, et al. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91 : 457–466.
55. WickesBL, EdmanJC (1995) The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol 16 : 1099–1109.
56. AndersonJT, WangX (2009) Nuclear surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol 44 : 16–24.
57. WinTZ, DraperS, ReadRL, PearceJ, et al. (2006) Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol 26 : 1710–1721.
58. KühnU, WahleE (2004) Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678 : 67–84.
59. Bousquet-AntonelliC, PresuttiC, TollerveyD (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102 : 765–775.
60. DavidsonL, KerrA, WestS (2012) Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J 31 : 2566–2578.
61. Rose AB (2008) Intron-mediated regulation of gene expression, in Reddy A.S.N. and Golovkin M., Eds. Nuclear pre-mRNA processing in plants. Berlin Heidelberg: Springer-Verlag. p. 277–290.
62. MooreMJ, ProudfootNJ (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136 : 688–700.
63. ZhaoC, HamiltonT (2007) Introns regulate the rate of unstable mRNA decay. J Biol Chem 282 : 20230–20237.
64. HouseleyJ, TollerveyD (2009) The many pathways of RNA degradation. Cell 136 : 763–776.
65. RoyWS, PennyD, NeafseyDE (2007) Evolutiannory concervation of UTR intron boundaries in Cryptococcus. Mol Biol Evol 24 : 1140–1148.
66. DumesicPA, NatarajanP, ChenC, DrinnenbergIA, et al. (2013) Stalled Spliceosomes Are a Signal for RNAi-Mediated Genome Defense. Cell 152 : 957–968.
67. McDadeHC, CoxGM (2001) A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol 39 : 151–154.
68. CoxGM, ToffalettiDL, PerfectJR (1996) Dominant selection system for use in Cryptococcus neoformans. J Med Vet Mycol 34 : 385–391.
69. WaltonFJ, IdnurmA, HeitmanJ (2005) Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol 57 : 1381–1396.
70. BenoitB, MitouG, ChartierA, TemmeC, et al. (2005) An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev Cell 9 : 511–522.
71. LemieuxC, BachandF (2009) Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic Acids Res 37 : 3418–3430.
72. ChenHM, FutcherB, LeatherwoodJ (2011) The fission yeast RNA binding protein Mmi1 regulates meiotic genes controlling intron specific splicing and polyadenylation coupled RNA turnover. PloS One 6: e26804.
73. LemieuxC, MargueratS, LafontaineJ, BarbezierN, et al. (2011) A pre-RNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell 44 : 108–119.
74. LibriD (2010) Nuclear poly(a)-binding proteins and nuclear degradation: take the mRNA and run? Mol Cell 37 : 3–5.
75. XuJ, SaundersCW, HuP, GrantRA, et al. (2007) Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci USA 104 : 18730–18735.
76. KämperJ, KahmannR, BölkerM, MaLJ, et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444 : 97–101.
77. Kwon-ChungKJ, BennettJE, RhodesJC (1982) Taxonomic studies of Fillobasidiella species and their anamorphs. Antonie van Leeuwenhoek 48 : 25–38.
78. Sherman F (1992), Getting started with yeast. In: Guthrie C and Fink GR, Eds. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press. p. 3–21.
79. JanbonG, HimmelreichU, MoyrandF, ImprovisiL, et al. (2001) Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol Microbiol 42 : 453–469.
80. RobertsGD, HorstmeierCD, LandGA, FoxworthJH (1978) Rapid urea broth test for yeasts. J Clin Microbiol 7 : 584–588.
81. IdnurnA, ReedyJL, NussbaumJC, HeitmanJ (2004) Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell 3 : 420–429.
82. IkedaR, NishimuraS, NishikawaA, ShinodaT (1996) Production of agglutinating monoclonal antibody against antigen 8 specific for Cryptococcus neoformans serotype D. Clin Diagn Lab Immunol 3 : 89–92.
83. MoyrandF, KlaprothB, HimmelreichU, DromerF, et al. (2002) Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol Microbiol 45 : 837–849.
84. ToffalettiDL, RudeTH, JohnstonSA, DurackDT, et al. (1993) Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol 175 : 1405–1411.
85. VarmaA, Kwon-ChungKJ (1991) Rapid method to extract DNA from Cryptococcus neoformans. J Clin Microbiol 29 : 810–812.
86. MoyrandF, FontaineT, JanbonG (2007) Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol 64 : 771–781.
87. JänickeA, VancuylenbergJ, BoagPR, TravenA, et al. (2012) ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA 18 : 1289–1295.
Štítky
Genetika Reprodukčná medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



