-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
For most organisms, chromosome segregation during meiosis relies on deliberate induction of DNA double-strand breaks (DSBs) and repair of a subset of these DSBs as inter-homolog crossovers (COs). However, timing and levels of DSB formation must be tightly controlled to avoid jeopardizing genome integrity. Here we identify the DSB-2 protein, which is required for efficient DSB formation during C. elegans meiosis but is dispensable for later steps of meiotic recombination. DSB-2 localizes to chromatin during the time of DSB formation, and its disappearance coincides with a decline in RAD-51 foci marking early recombination intermediates and precedes appearance of COSA-1 foci marking CO-designated sites. These and other data suggest that DSB-2 and its paralog DSB-1 promote competence for DSB formation. Further, immunofluorescence analyses of wild-type gonads and various meiotic mutants reveal that association of DSB-2 with chromatin is coordinated with multiple distinct aspects of the meiotic program, including the phosphorylation state of nuclear envelope protein SUN-1 and dependence on RAD-50 to load the RAD-51 recombinase at DSB sites. Moreover, association of DSB-2 with chromatin is prolonged in mutants impaired for either DSB formation or formation of downstream CO intermediates. These and other data suggest that association of DSB-2 with chromatin is an indicator of competence for DSB formation, and that cells respond to a deficit of CO-competent recombination intermediates by prolonging the DSB-competent state. In the context of this model, we propose that formation of sufficient CO-competent intermediates engages a negative feedback response that leads to cessation of DSB formation as part of a major coordinated transition in meiotic prophase progression. The proposed negative feedback regulation of DSB formation simultaneously (1) ensures that sufficient DSBs are made to guarantee CO formation and (2) prevents excessive DSB levels that could have deleterious effects.
Published in the journal: The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance. PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003674
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003674Summary
For most organisms, chromosome segregation during meiosis relies on deliberate induction of DNA double-strand breaks (DSBs) and repair of a subset of these DSBs as inter-homolog crossovers (COs). However, timing and levels of DSB formation must be tightly controlled to avoid jeopardizing genome integrity. Here we identify the DSB-2 protein, which is required for efficient DSB formation during C. elegans meiosis but is dispensable for later steps of meiotic recombination. DSB-2 localizes to chromatin during the time of DSB formation, and its disappearance coincides with a decline in RAD-51 foci marking early recombination intermediates and precedes appearance of COSA-1 foci marking CO-designated sites. These and other data suggest that DSB-2 and its paralog DSB-1 promote competence for DSB formation. Further, immunofluorescence analyses of wild-type gonads and various meiotic mutants reveal that association of DSB-2 with chromatin is coordinated with multiple distinct aspects of the meiotic program, including the phosphorylation state of nuclear envelope protein SUN-1 and dependence on RAD-50 to load the RAD-51 recombinase at DSB sites. Moreover, association of DSB-2 with chromatin is prolonged in mutants impaired for either DSB formation or formation of downstream CO intermediates. These and other data suggest that association of DSB-2 with chromatin is an indicator of competence for DSB formation, and that cells respond to a deficit of CO-competent recombination intermediates by prolonging the DSB-competent state. In the context of this model, we propose that formation of sufficient CO-competent intermediates engages a negative feedback response that leads to cessation of DSB formation as part of a major coordinated transition in meiotic prophase progression. The proposed negative feedback regulation of DSB formation simultaneously (1) ensures that sufficient DSBs are made to guarantee CO formation and (2) prevents excessive DSB levels that could have deleterious effects.
Introduction
For most diploid organisms, the formation of haploid gametes relies on crossover (CO) recombination between homologous chromosomes for accurate chromosome segregation. Recombination is initiated during meiotic prophase by the programmed induction of DNA double strand breaks (DSBs), catalyzed by the evolutionarily conserved topoisomerase-like protein Spo11 [1]. A subset of these DSBs are repaired by a specialized meiotic DSB repair pathway that uses the homolog as a recombination partner and generates intermediates that can be resolved as COs. This specialized repair is completed during the pachytene stage of meiotic prophase, in the context of meiosis-specific chromosome organization in which homologs are paired and connected along their axes by a structure known as the synaptonemal complex (SC). By the last stage of meiotic prophase (diakinesis), the SC has disassembled, and chromosomes have further condensed and reorganized to reveal CO-dependent structures called chiasmata, which connect homologous chromosomes and allow them to orient and segregate to opposite poles at the meiosis I division [2].
DSB formation must be tightly regulated to ensure successful meiosis: cells must both turn on DSB formation to achieve inter-homolog COs, but also turn off DSB formation to allow repair and subsequent chromosome re-organization in preparation for the meiotic divisions. Thus, DSB formation and repair must be coordinated with other aspects of meiotic chromosome dynamics. In addition, cells must make enough DSBs to guarantee one CO per chromosome pair, but too many DSBs could lead to unrepaired DNA damage and compromise genomic integrity.
While Spo11 catalyzes DSB formation, little is known about how Spo11 activity is regulated and how the timing and number of DSBs are controlled. Several proteins besides Spo11 are required for meiotic DSB formation in various systems, although their mode(s) of action are not well understood [3], [4], [5]. The highly conserved Rad50/Mre11 complex is required for DSB formation in some systems but not in others, and even in an organism where it is normally required (C. elegans), Spo11-dependent DSBs can form independently of Rad50/Mre11 in some contexts [6], [7]. Further, many of the known DSB-promoting proteins are not well conserved at the sequence level, showing rapid divergence even among closely related species [4]. In C. elegans, the chromatin-associated proteins HIM-17, XND-1, and HIM-5 have been implicated in promoting normal levels and/or timing of DSB formation, particularly on the X chromosomes [8], [9], [10]. These proteins localize to chromatin throughout the germ line and are proposed to exert their effects by modulating the chromatin environment to affect accessibility of the DSB machinery. However, the localization of these proteins is not limited to the time of DSB formation, suggesting that other factors must control when the DSB machinery is active.
In the current work, we identify the C. elegans DSB-2 protein (encoded by dsb-2, member of new gene class dsb for DNA double-strand break factor) as a novel factor required specifically to promote the DSB step of meiotic recombination. We show that DSB-2 localizes to chromatin in meiotic prophase germ cells, and that the timing of its appearance and disappearance corresponds to the time window during which DSBs are formed. These and other data implicate DSB-2 in regulating the timing of competence for DSB formation by SPO-11. Further, we find that the presence of DSB-2 on chromatin is regulated coordinately with multiple distinct aspects of the meiotic program, including specialized meiotic DSB repair features and the phosphorylation state of nuclear envelope protein SUN-1. Thus, we propose that disappearance of DSB-2 reflects loss of competence for DSB formation, which occurs as part of a major coordinated transition in meiotic prophase progression. Moreover, our data suggest the existence of a regulatory network wherein germ cells can detect the presence or absence of downstream CO-eligible recombination intermediates. In the context of this model, successful formation of monitored intermediates would trigger removal of DSB-2 (and other factors) from chromatin and consequent shut-down of DSB formation, whereas a deficit of relevant intermediates would elicit a delay in DSB-2 removal (and in other aspects of meiotic progression). We propose that the negative feedback property inherent in such a regulatory network provides a means to ensure that sufficient DSBs are made to guarantee CO formation, while at the same time protecting the chromosomes against formation of excessive levels of DSBs that could jeopardize genomic integrity.
Results
Identification of dsb-2, a novel gene required for robust chiasma formation
The dsb-2(me96) allele was isolated following EMS mutagenesis in a screen for meiotic abnormalities visible in oocytes at diakinesis, the last stage of meiotic prophase (see Materials and Methods). Whereas WT oocyte nuclei consistently exhibit 6 pairs of homologous chromosomes attached by chiasmata (bivalents), oocyte nuclei in the dsb-2(me96) mutant exhibit a variable number of unattached (achiasmate) chromosomes (univalents), indicating a defect in chiasma formation (Figure 1A) resulting from an underlying defect in CO formation (below).
Fig. 1. dsb-2 mutant worms exhibit a defect in CO/chiasma formation that worsens with age. 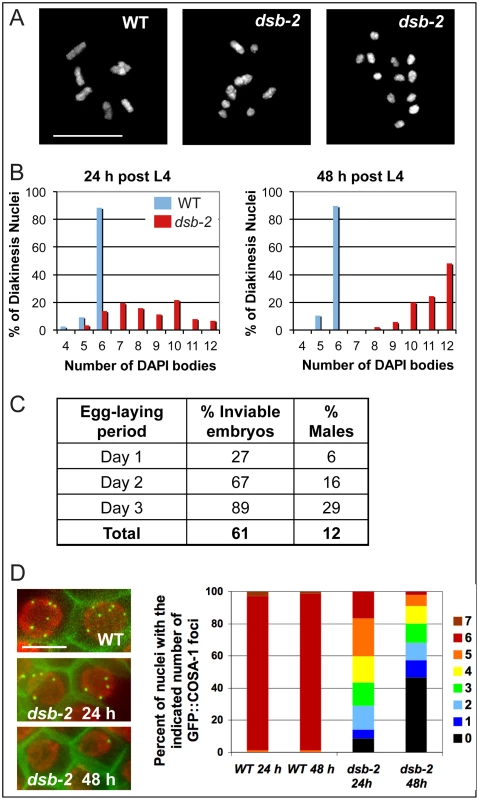
(A) Each panel shows the full complement of chromosomes in a single diakinesis-stage oocyte, stained by DAPI. Left: WT nucleus with six DAPI bodies corresponding to six pairs of homologs connected by chiasmata (bivalents). Middle: dsb-2(me96) nucleus with eight DAPI bodies (4 bivalents and 4 univalents), indicating that only four pairs of homologs have formed chiasmata. Right: dsb-2(me96) nucleus with 12 DAPI bodies (all univalents), indicating absence of chiasmata between all homolog pairs. Scale bar, 10 µm. (B) Graphs showing frequencies of diakinesis-stage nuclei with the indicated number of DAPI bodies in dsb-2(me96) and WT hermaphrodites fixed and stained at 1 day and 2 days post L4. (C) Table showing frequency of inviable embryos and frequency of males (among surviving progeny) from eggs laid by dsb-2(me96) rol-1(e91) hermaphrodites (where rol-1 is a marker that does not affect meiosis) during the indicated time interval after the L4 stage. Inviable embryos that do not hatch are indicative of autosomal mis-segregation, while male progeny indicate X-chromosome mis-segregation. For comparison, wild-type hermaphrodites produce less than 1% inviable embryos and approximately 0.2% males during their entire reproductive lives. (D) Left: images of GFP::COSA-1 foci in late pachytene nuclei of live anesthetized worms, with chromatin visualized by mCherry::H2B and plasma membranes marked by GFP::PH. Each WT nucleus has 6 GFP::COSA-1 foci, corresponding to the single CO site on each homolog pair; reduced numbers of GFP::COSA-1 foci in the dsb-2(me97) nuclei reflect reduced CO formation. Scale bar, 5 µm. Right: Graph showing frequencies of nuclei with indicated numbers of GFP::COSA-1 foci in late pachytene nuclei of worms examined at 24 or 48 h post L4, revealing worsening of the CO deficit with age in dsb-2(me97) mutant worms. The me96 mutation was mapped to a 126 kb interval on chromosome II, and RNAi against F26H11.6 (a gene in the candidate interval) phenocopied the me96 mutant (Materials and Methods). Sequencing revealed a T-to-A transversion in F26H11.6 in the me96 mutant; as this mutation results in an early stop at codon 14 (TTA = >TAA) of F26H11.6 (280 codons total), me96 is presumed to be a null allele. A second, independently-isolated, dsb-2 allele (me97) contains a premature stop at codon 168 in the same gene, further confirming the identity of F26H11.6 as dsb-2.
Homology searches revealed an F26H11.6 paralog in C. elegans (F08G5.1), and genes encoding orthologs of both proteins were found in other Caenorhabditis species (briggsae, remanei and japonica) but were not identified in other organisms (Figures S1 and S2). Based on independent analyses implicating both genes in meiotic double-strand break formation [11], F08G5.1 and F26H11.6 were designated as dsb-1 and dsb-2, respectively. Multiple sequence alignment of this highly diverged protein family shows two readily alignable regions corresponding to residues 1–103 and 195–251 of the C. elegans F26H11.6/DSB-2 protein, each containing several conserved sites (Figure S1). These two regions are connected by a variable segment that in each protein contains an S/T Q cluster domain [12], a feature suggesting that these proteins are potential targets for phosphorylation by the ATM/ATR family of protein kinases.
Chiasma/crossover formation defects in dsb-2 worms aggravate with age
The number of achiasmate chromosomes detected in diakinesis-stage oocytes of dsb-2 mutant hermaphrodites increases with maternal age, indicating a worsening of phenotype over time (Figure 1B). Whereas adult dsb-2(me96) hermaphrodites fixed one day after the L4 larval stage (24 hours post-L4) had an average of 8.5 DAPI-stained bodies at diakinesis (reflecting a mixture of bivalents and univalents), 48 hour post-L4 hermaphrodites had an average of 11.1 DAPI bodies (indicating that nearly all chromosome pairs lacked chiasmata). Further, as lack of chiasmata connecting homologs results in mis-segregation of chromosomes, both the frequency of inviable embryos (reflecting autosomal aneuploidy) and the frequency of males (XO, reflecting X chromosome mis-segregation) produced by dsb-2(me96) hermaphrodites likewise increased with maternal age (Figure 1C): frequencies rose from 27% dead embryos and 6% males on day 1 of egg-laying to 89% dead embryos and 29% males on day 3.
Age dependence of the dsb-2 mutant phenotype was also observed for the dsb-2(me97) allele, using GFP::COSA-1 as a cytological marker of crossover (CO) sites (Figure 1D). During wild-type meiosis, GFP::COSA-1 localizes to 6 foci per nucleus during the late pachytene and diplotene stages, marking the single CO/emerging chiasma on each homolog pair [13]. Whereas 6 GFP::COSA-1 foci were consistently observed in late pachytene nuclei of control worms regardless of maternal age, the number of GFP::COSA-1 foci was substantially reduced in dsb-2(me97) worms at 24 hours post-L4 and further declined by 48 hours post-L4 .
The age effect in dsb-2 mutants is not caused by persistence of maternal gene product in the germ line, as it was observed in homozygous mutant worms derived from either heterozygous parents or homozygous mutant parents (where no maternal product should be present). In addition, the age effect is evident at both standard (20°C), and elevated (25°C) growth temperatures.
Together, our data indicate that the function of DSB-2 is required throughout reproductive life to generate normal levels of COs and chiasmata, and becomes increasingly important for meiotic success in germ cell nuclei that enter the meiotic program at progressively later times. This implies that changes must occur as the worms age that render crossing over and chiasma formation increasingly sensitive to the loss of DSB-2 protein.
DSB-2 promotes meiotic DSB formation
Successful chiasma formation requires pairing of homologous chromosomes, assembly of the synaptonemal complex (SC), and CO recombination between the homologs. Homolog pairing and SC assembly are not dependent on initiation or progression of recombination during C. elegans meiosis [14], facilitating investigation of potential involvement of DSB-2 in these events. To this end, we conducted immunofluorescence analyses on germ lines dissected from dsb-2 worms at 48 hours post-L4, when the CO/chiasma deficit is severe. Several lines of evidence indicate that the lack of chiasmata in dsb-2 mutants is due to a defect in the initiation of meiotic recombination.
First, dsb-2 mutant worms are proficient for pairing of the X chromosomes, as immunofluorescence of pachytene nuclei showed a single focus of HIM-8, a protein that binds a specific region of the X chromosome known as the pairing center [15], [16] (Figure 2A). Second, dsb-2 mutants are proficient for assembly of the SC, as immunostaining revealed proper loading of HIM-3 (an SC lateral element component) and SYP-1 (an SC central region component) [17], [18] along the lengths of aligned homologs (Figure 2B). Proficiency for pairing and synapsis suggests that dsb-2 mutants are deficient in the process of meiotic recombination per se.
Fig. 2. DSB-2 is required for meiotic DSB formation. 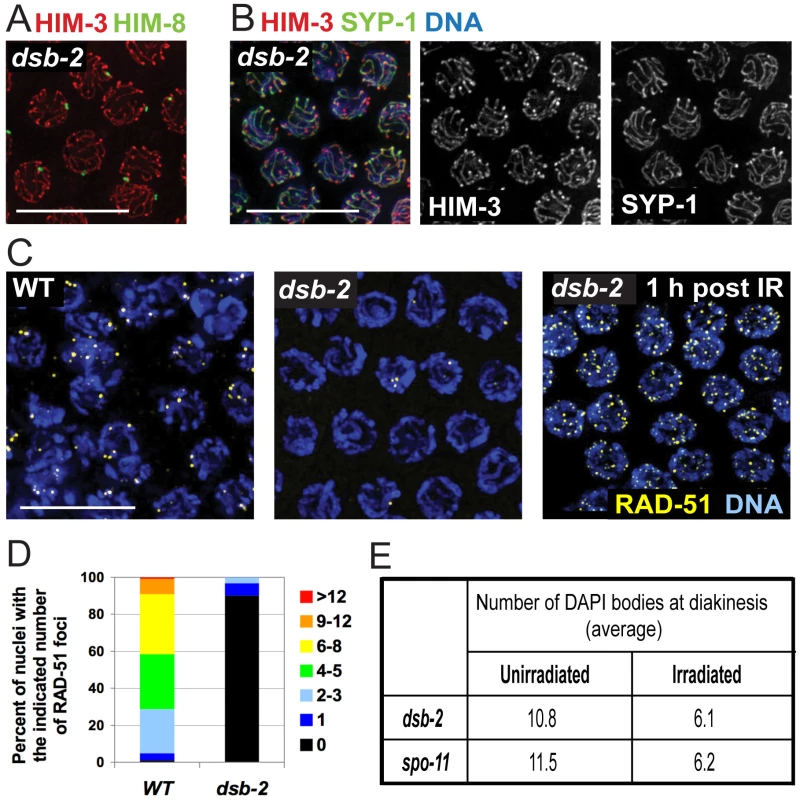
(A) Homolog pairing assayed by immunofluorescence of X-chromosome pairing center (X-PC) binding protein HIM-8 in dsb-2(me96) pachytene nuclei. A single HIM-8 focus is observed in each nucleus, indicating successful pairing at the X-PC. HIM-3 marks chromosome axes. Scale bar, 10 µm. (B) Synapsis assayed by immunofluorescence in dsb-2(me96) pachytene nuclei. Axis protein HIM-3 and SC central region protein SYP-1 co-localize in continuous stretches between chromosome pairs, indicating successful synapsis. Scale bar, 10 µm. (C) Immunolocalization of RAD-51 in early pachytene nuclei. RAD-51 foci mark DSBs and are greatly reduced or absent in untreated dsb-2(me96) nuclei compared to WT. Following irradiation (IR), RAD-51 foci are abundant in dsb-2(me96) nuclei, indicating that the dsb-2 mutant retains that capacity to load RAD-51 at induced DSBs. Scale bar, 10 µm. (D) Quantitation of reduced RAD-51 foci in the dsb-2(me96) mutant. RAD-51 foci were scored in 8 contiguous rows of pachytene nuclei from the region of the germ line where foci were most abundant in wild type (see Materials and Methods). (E) Table showing average numbers of DAPI bodies in diakinesis-stage oocytes in dsb-2(me96) and spo-11 mutant worms with and without irradiation, showing restoration of chiasma formation by IR-induced DSBs. Worms were exposed to 1 kRad of gamma-irradiation at 36 hours post L4, and irradiated and age-matched controls were fixed and stained with DAPI 18 hours post irradiation. As in Figure 1A and B, the number of DAPI bodies reflects success or failure of chiasma formation: 12 indicates lack of chiasmata for all homolog pairs, and 6 indicates successful chiasma for all homolog pairs. This assay tends to underestimate the incidence of achiasmate chromosomes, as some lie too close together to be resolved. Meiotic recombination is initiated by formation of DNA double-strand breaks (DSBs) by the SPO-11 protein [14], [19], followed by processing of these DSBs to enable loading of the DNA-strand exchange protein RAD-51, which can be detected as foci from zygotene to mid-pachytene stages in WT germlines [20], [21]. dsb-2 germ lines display greatly reduced levels of RAD-51 foci, with most nuclei having no foci (Figure 2C,D), suggesting either that fewer DSBs are made or that loading of RAD-51 is impaired. However, the dsb-2 mutant is proficient for loading of RAD-51 when DSBs are induced by gamma-irradiation, as seen by the presence of RAD-51 foci in germline nuclei fixed 1 hour post-irradiation (Figure 2C).
Furthermore, irradiation bypasses the requirement for DSB-2 and restores chiasma formation (Figure 2E). It was previously shown that in C. elegans, providing DSBs by irradiation rescues chiasma formation in the spo-11 mutant, which lacks the enzyme responsible for making programmed DSBs [14], [22]. The same effect was seen upon irradiation of dsb-2 mutant worms, demonstrating that the chiasma defect in dsb-2 worms is a result of a defect in SPO-11-induced DSB formation. In both dsb-2 worms and age-matched spo-11 worms, 1 kRad of irradiation resulted in efficient restoration of chiasmata in diakinesis-stage oocytes examined 18 hours post-irradiation (Figure 2E). Thus DSB-2 is a novel protein required for robust meiotic DSB formation.
DSB-2 localizes to chromatin during the presumed timing of DSB formation
Immunofluorescence experiments using an antibody against the DSB-2 protein (Materials and Methods) showed that DSB-2 localizes to chromatin in germ cell nuclei from meiotic entry to mid-pachytene (Figure 3A). DSB-2 staining is first detected in the transition zone (TZ; corresponds to leptotene and zygotene stages, when pairing and SC assembly occur), and is strongest overall in early pachytene, where it localizes to chromatin in an uneven pattern, showing a few bright patches per nucleus as well as fainter stretches/foci associated with most of the chromatin. Towards mid-pachytene, the bright patches diminish and the chromatin signal fades and then disappears from most nuclei. However, in a subset of nuclei in the mid/late pachytene region, DSB-2 staining becomes brighter, with bright stretches/foci along most of the chromatin; a few of these “outlier” brightly-staining nuclei are present in later pachytene and likely represent nuclei destined for apoptosis (see Discussion).
Fig. 3. DSB-2 localizes to chromatin in early meiotic prophase nuclei, concurrent with DSB marker and SUN-1 Ser8-phosphorylation. 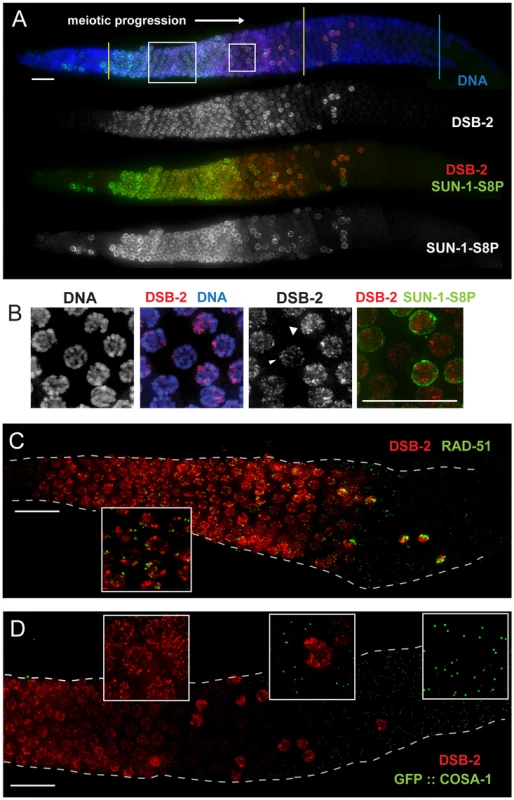
(A) Immunofluorescence image of a WT hermaphrodite gonad (from distal tip to end of pachytene) stained with DAPI and antibodies that recognize DSB-2 and SUN-1 S8P. In this and all subsequent figures, the distal tip (which contains mitotically cycling germ cells) is at the left, and meiotic progression is from left to right. DSB-2 first appears on chromatin at meiotic onset in transition zone nuclei (corresponding to the leptotene/zygotene stages of meiotic prophase) and disappears around mid-pachytene stage of meiotic prophase, with a few outlier nuclei retaining DSB-2 in later pachytene. Here and in subsequent figures, yellow lines demarcate the “DSB-2-positive” zone and a cyan line marks the end of the pachytene zone. Co-staining shows correlation between DSB-2-positive and SUN-1 S8P-positive meiotic nuclei. (Note: SUN-1 S8P is also present in a few pre-meiotic nuclei; [23]). A close-up of the large box is shown in Figure 5C. Scale bar, 15 µm. (B) Close-up of nuclei outlined by the small box in (A). DSB-2 localizes to a few bright patches/foci, as well as fainter stretches/foci along the entire chromatin (see also Fig. 5C). As nuclei reach mid-pachytene, the DSB-2 signal becomes fainter (narrow arrowhead), however in some nuclei signal gets brighter along most of chromatin (broad arrowhead). (C) Immunofluorescence image of a WT hermaphrodite gonad from entry into meiotic prophase to mid-to-late pachytene, stained with antibodies that recognize DSB-2 and RAD-51. RAD-51 foci (marking processed DSBs) appear in nuclei shortly after DSB-2 staining appears on chromatin upon meiotic entry, and the RAD-51 foci disappear shortly after DSB-2 is no longer present on chromatin in mid-pachytene nuclei. Inset shows that RAD-51 foci mostly do not co-localize with concentrated DSB-2. DSB-2-bright outlier nuclei in late pachytene contain high levels of RAD-51 foci. Scale bar, 15 µm. (D) Immunofluorescence image of the early mid-pachytene to late pachytene region of a WT hermaphrodite gonad expressing GFP::COSA-1 (strain AV630), stained with antibodies that recognize DSB-2 and GFP. COSA-1 foci marking designated CO sites appear in nuclei only after the removal of DSB-2 from chromatin; DSB-2-bright outlier nuclei in the late pachytene region lack COSA-1 foci, even when COSA-1 foci are present in neighboring nuclei. Close-ups are shown in insets. Scale bar, 15 µm. Apart from the outlier nuclei, the “DSB-2-positive” region of the germ line corresponds to nuclei at the stages in which DSB formation is presumed to occur [8], [21]. Indeed, co-immunostaining experiments showed that RAD-51 foci (marking DSB-dependent recombination intermediates) appear in nuclei shortly after DSB-2 staining appears on chromatin upon meiotic entry, and the RAD-51 foci disappear shortly after DSB-2 is no longer present on chromatin in mid-pachytene nuclei (Figure 3C). Previous work has demonstrated that germ cell nuclei at later stages of meiotic prophase are proficient to load RAD-51 when DSBs are introduced by irradiation [6]. Thus, the disappearance of RAD-51 foci in the endogenous case likely indicates that DSBs are no longer being formed and that existing DSBs have progressed to subsequent stages of repair. Indeed, we observe that COSA-1 foci marking designated CO intermediates appear in nuclei only after the removal of DSB-2 from chromatin (Figure 3D). In addition, the “outlier” bright-staining DSB-2 nuclei in late pachytene contain high levels of RAD-51 foci and lack COSA-1 foci, suggesting these nuclei are arrested in their progression and may be triggering a checkpoint response. Thus, the close correspondence between the zone where DSB-2 localizes on chromatin and the zone where RAD-51 foci are detected is not only consistent with the demonstrated role for DSB-2 in promoting DSB formation, but further suggests that loss of DSB-2 coincides with loss of competence for DSB formation and progression to a subsequent stage of DSB repair.
Relationship of DSB-2 to other factors promoting DSB formation
We used immunofluorescence analyses to investigate the relationships between DSB-2 and other meiotic factors that act at the DSB formation step. Figure 4 shows the relationship between DSB-2 and its paralog DSB-1, which was independently implicated in DSB formation [11]. Nuclear localization of DSB-1 and DSB-2 is detected in the same region of the gonad, and their staining patterns on chromatin have a similar appearance (Figure 4A). However, the relative intensity patterns of the two proteins differ during meiotic progression. Within the gonad, DSB-1 signal is detected on nuclei slightly before DSB-2 and has a stronger intensity early on, which then declines as nuclei progress through pachytene (except for the outlier nuclei); DSB-2 signal is weaker early on and peaks in intensity later than DSB-1 before eventually declining. Both proteins disappear from nuclei at the same time, and both localize to the same outlier nuclei. Within each nucleus, the intensity patterns on chromatin are also different, such that the DSB-1 and DSB-2 signals partially overlap but do not match each other (Fig. 4A inset).
Fig. 4. Relationship between DSB-2 and paralog DSB-1. 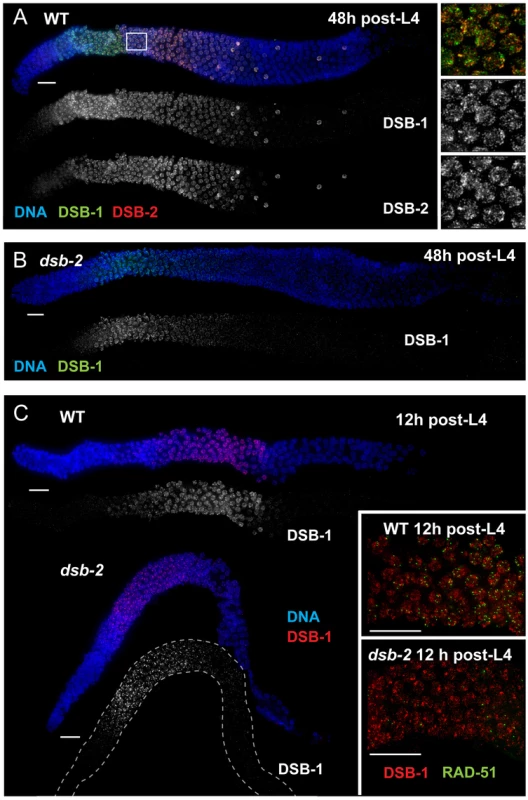
(A) Immunofluorescence image of a WT hermaphrodite gonad (from distal tip to end of pachytene), stained with DAPI and antibodies that recognize DSB-2 and DSB-1. DSB-2 and DSB-1 are detected in a highly correlated subset of germline nuclei (within a region spanning from meiotic prophase onset through mid-pachytene), although the relative intensities of the DSB-2 and DSB-1 signals vary during prophase progression (see text). Inset: close-up of the field of early pachytene nuclei outlined in (A) showing that although DSB-1 and DSB-2 are present in the same nuclei, the DSB-1 and DSB-2 signals on chromatin sometimes overlap but mostly do not match each other. Scale bar, 15 µm. (B, C) DSB-1 immunolocalization in the dsb-2(me96) mutant. Zoomed out images show that DSB-1 staining is fairly similar to wild-type control in a 12 h post-L4 dsb-2 mutant gonad (C), but DSB-1 staining is reduced (relative to wild-type) in the pachytene region of a 48 h post-L4 dsb-2 mutant gonad (B). Insets in C show fields of pachytene nuclei illustrating that RAD-51 foci are already reduced in the dsb-2 mutant at 12 h post L4. Scale bars, 15 µm. Whereas DSB-2 localization is abolished in dsb-1 mutant germ lines [11], some DSB-1 protein is present on chromatin in the dsb-2 mutant (Figure 4B,C). DSB-1 staining in dsb-2 young adult germ lines (12 hours post-L4) appears comparable to age-matched wild-type controls despite that fact that RAD-51 foci are already substantially diminished by this stage; this indicates that the presence of DSB-1 on chromatin is not sufficient to promote efficient DSB formation in the absence of DSB-2. Further, the association of residual DSB-1 protein in the dsb-2 mutant appears to change with age, as DSB-1 staining in older dsb-2 germ lines (48 hours post-L4) is typically fainter and declines and disappears sooner than in WT. Together these data suggest that DSB-2 may be required to augment the DSB-promoting activity of DSB-1, possibly by affecting the nature of its association with chromatin, and that the reliance on DSB-2 for this augmentation becomes more acute with increasing age.
We further showed that DSB-2 localizes to chromatin independently of DSB formation, indicating that DSB-2 localization is not a consequence of DSB formation. Specifically, in spo-11 mutants, which lack endogenous DSBs, DSB-2 is detected on chromosomes in transition zone and pachytene nuclei, and the overall appearance of the staining within nuclei is similar to that in WT nuclei. However, DSB-2 association with chromatin extends further into late pachytene, suggesting that endogenous DSB formation affects timing of DSB-2 removal (Figure 5A; see below).
Fig. 5. DSB-2 and SUN-1 S8P persist when DSB formation is defective. 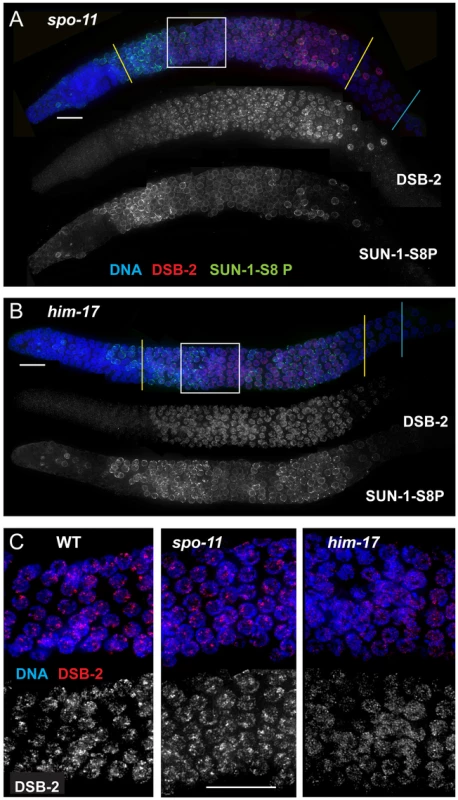
(A) and (B) Immunofluorescence images of gonads from the distal pre-meiotic region to end of pachytene, stained with DAPI and antibodies that recognize DSB-2 and SUN-1 S8P. The zone of DSB-2 and SUN-1 S8P-positive nuclei is extended in both spo-11 (A) and him-17 (B) mutants, which are defective in DSB formation. (C) Close-up images of fields of nuclei in early pachytene, as outlined in Figure 3A and (A), (B) above. WT as well as spo-11 nuclei show bright patches of DSB-2 staining, whereas him-17 nuclei do not. Scale bar, 15 µm. Finally, we assessed DSB-2 localization in germ lines lacking HIM-17, a THAP-domain containing protein that associates with germline chromatin and is required for normal levels of meiotic DSB formation [8]. In him-17 mutant germ lines, DSB-2 is detected on chromatin in nuclei from transition zone to late pachytene (Figure 5B), but the DSB-2 signal has an altered appearance within the nuclei: the bright patches characteristic of DSB-2 localization in WT germ cells are not observed, and DSB-2 instead displays only the fainter, more uniform distribution (Figure 5C). Thus, improper localization of DSB-2 may contribute to the observed defect in DSB formation in him-17 mutants. Taken together, these data suggest that association of DSB-2 and DSB-1 with chromatin is required to regulate competence for DSB formation by SPO-11.
DSB-2 localization to chromatin is CHK-2 dependent and correlates with Ser-8 phosphorylation of nuclear envelope protein SUN-1
The distribution of DSB-2 positive nuclei within the germ line is similar to that reported for nuclei exhibiting phosphorylation of serine-8 of nuclear envelope (NE) protein SUN-1[23]. SUN-1 is a part of a conserved protein complex that spans the NE and mediates attachment of the chromosomes to the cytoskeletal motility machinery [24], [25]. Although the SUN-1 protein is present throughout the germ line, SUN-1 S8P is detected only in a subset of nuclei during meiotic prophase [23]: SUN-1 S8P appears abruptly at the onset of meiotic prophase, with TZ nuclei exhibiting both bright SUN-1 S8P patches, corresponding to the chromosome attachment points that mediate chromosome movement, and a diffuse SUN-1 S8P staining throughout the NE; in early pachytene, the patches dissipate (except for one), but the diffuse NE staining persists, weakening until it disappears around mid-pachytene; however, a few outlier nuclei maintain SUN-1 S8P staining in later pachytene (Figure 3A and B) [26].
Co-staining experiments revealed that DSB-2 and SUN-1 S8P tend to be detected in the same nuclei (Figure 3A). The relative intensity patterns are different, with SUN-1 S8P exhibiting a much stronger signal in the TZ, and showing generally weaker signal towards mid-pachytene when compared with DSB-2 (Figure 3A). Most outlier nuclei are bright for both marks, but some are bright only for one of the marks and weak for the other. Nevertheless, the correlation is striking, suggesting that these two features (presence of DSB-2 on chromatin and of SUN-1 S8P on the NE) may be co-regulated.
In support of this hypothesis, we found that DSB-2 localization depends on the CHK-2 protein kinase. CHK-2 was previously shown to be required for several early prophase events including DSB formation, homolog pairing and synapsis, reorganization of chromosomes within the nucleus, chromosome movement, and associated phosphorylation of SUN-1 [23], [24], [27], [28]. We found that both DSB-2 staining and SUN-1 S8P (in early meiotic prophase) were severely reduced or absent in chk-2 mutant gonads (Figure 6B), indicating that CHK-2 represents a common regulator of these two distinct features of the meiotic program. Although chromatin-associated DSB-2 staining was not observed by immunofluorescence, Western blot analysis indicated that the DSB-2 protein is expressed in the chk-2 mutant (Figure 6B).
Fig. 6. DSB-2 and SUN-1 S8P are coordinately regulated by common upstream regulator CHK-2. 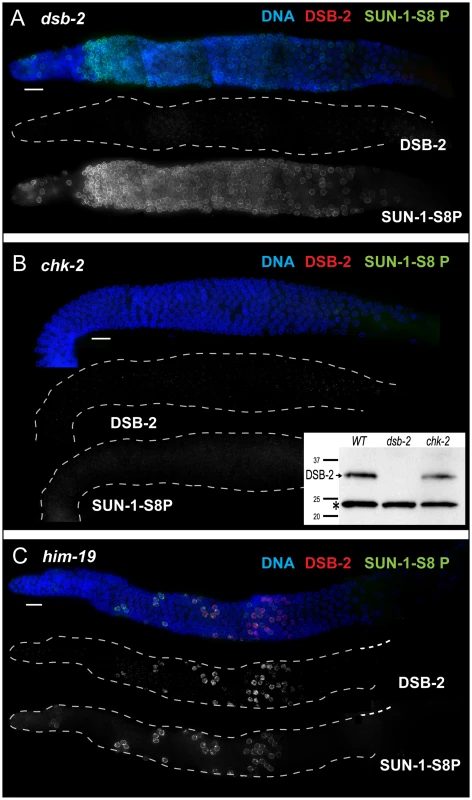
Immunofluorescence images of gonads of indicated genotypes from the distal pre-meiotic region to end of pachytene, stained with DAPI and antibodies that recognize DSB-2 and SUN-1 S8P. Scale bar, 15 µm. (A) SUN-1 S8P is detected at the NE in meiotic prophase nuclei in the dsb-2 mutant germ line, indicating that although these features are coordinated during wild-type meiosis, acquisition of meiotic SUN-1 S8P does not depend on DSB-2. However, the SUN-1 S8P zone is extended in the dsb-2 mutant, indicating that the timing of its removal is affected. DSB-2 staining is absent from chromatin, indicating antibody specificity. (B) Main panel: Immunofluorescence images showing that localization of DSB-2 on chromatin and SUN-1 S8P staining at the NE are both severely reduced in the chk-2 mutant in the indicated meiotic region. Note: SUN-1 S8P signal remains present on some pre-meiotic nuclei and on late diakinesis oocytes in chk-2 mutants (data not shown; [23]). Inset: Western blot of whole-worm protein lysates from the indicated genotypes (60 worms per lane) stained with anti-DSB-2 antibodies. The arrow indicates the DSB-2 protein (32 kD), which is absent in the dsb-2 mutant but is still present in the chk-2 mutant; the asterisk indicates a non-specific band that serves as a loading control. (C) The presence of DSB-2 on chromatin and SUN-1 S8P at the NE are correlated in the him-19 mutant, in which only a small subset of nuclei are positive for these marks. Whereas localization of DSB-2 on chromatin and Ser-8 phosphorylation of SUN-1 at the NE in meiotic prophase nuclei tend to be correlated, they do not depend on each other. SUN-1 S8P immunostaining is present on meiotic prophase nuclei in dsb-2 mutant worms, and the zone of SUN-1 S8P-positive nuclei is extended into later pachytene (Figure 6A, see below). Conversely, DSB-2 is able to load on chromatin in nuclei in sun-1(gk199) null mutant germ lines despite severe defects in germline organization and abnormal chromosome morphology (data not shown). Thus, these two features appear to be independent downstream readouts of CHK-2 activity in meiosis. Together, our data suggest that CHK-2 coordinates the meiotic program by acting as a common upstream regulator of two parallel pathways, thereby linking competence for DSB formation (mediated through DSB-2) with chromosome and NE dynamics (mediated through SUN-1 S8P).
The correlation between DSB-2 and SUN-1 S8P was also tested in him-19 mutants, which show an age-dependent pleiotropic phenotype that includes multiple defects (in DSB formation, chromosome clustering and movement in TZ, pairing and synapsis) that are hypothesized to result from mis-regulation of CHK-2 activity [29]. In 2-day old him-19 worms, SUN-1 S8P is missing from most of the TZ and early pachytene regions, but is present on a few scattered nuclei [23] that are also positive for DSB-2 (Figure 6C), consistent with these two features being controlled by common factors including CHK-2.
DSB-2 and SUN-1 S8P persist when CO recombination is impaired
The removal of DSB-2 and SUN-1 S8P at mid-pachytene during WT meiosis, concurrent with the timing of disappearance of RAD-51 foci, led us to hypothesize the existence of a coordinated regulatory mechanism that simultaneously shuts down competence for DSB formation and changes other properties of the nucleus as it enters another stage of meiotic progression. In spo-11 and him-17 mutants, the zone of DSB-2 and SUN-1 S8P marked nuclei was extended beyond what was seen in WT (Figure 5A and B, Figure 7); extension of the SUN-1 S8P-positive zone in the spo-11 mutant was also reported by Woglar et al.[26]. In addition, in dsb-2 mutants, the zone of SUN-1 S8P staining was also prolonged (Figures 6A, 7). All of these mutants have defective DSB formation, and thus lack or have a deficit of downstream recombination intermediates and COs. We hypothesized that the deficit of appropriate recombination intermediates prolonged the zone of nuclei marked by DSB-2 and SUN-1 S8P. To test this hypothesis, we analyzed DSB-2 and SUN-1 S8P staining in several classes of meiotic mutants.
Fig. 7. Quantitation of the DSB-2 positive zone in WT and meiotic mutants. 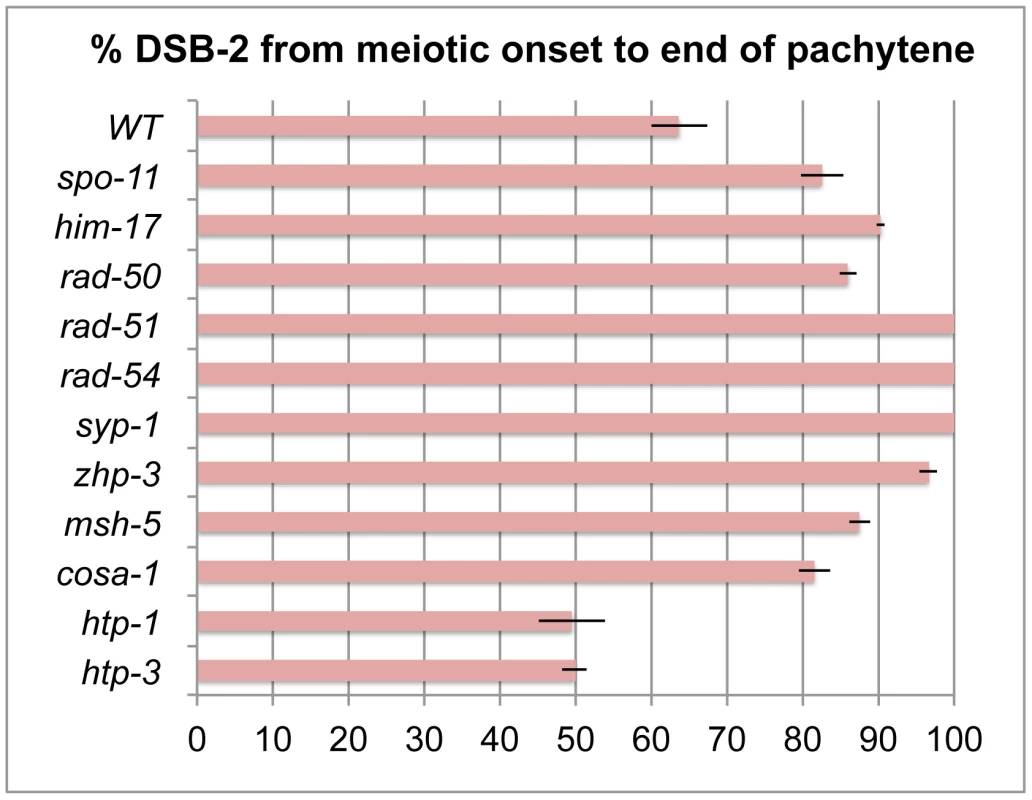
Bar graph showing the extent of the region of DSB-2 positive nuclei in germ lines of indicated genotypes. The presence/absence of DSB-2 signals was assessed in the portion of the germ line extending from the onset of DSB-2 staining to the end of the pachytene region. The extent of the DSB-2 positive zone was defined as the percentage of continuous rows of nuclei in which all or most nuclei exhibited DSB-2 staining out of total rows of nuclei in the scored region. Representative germ lines were imaged and scored: 5 for WT and 3 for each of the meiotic mutants. Error bars show standard deviation. We tested mutants lacking proteins involved in early steps of DSB processing and repair: the rad50 mutant, which lacks the RAD-50 protein that has been implicated in meiotic DSB formation, DSB resection and RAD51 loading [6], [30]; the rad51 mutant, which lacks the RAD-51 recombinase that catalyzes strand exchange [20]; and the rad54 mutant, in which unloading of RAD-51 and progression of DSB repair are disrupted [31]. We found that in all of these mutants, DSB-2 and SUN-1 S8P staining are extended over most of the pachytene region (which also tends to be smaller than in WT gonads) (Figures 8, 7). This prolonged staining in mutants defective in DSB formation, processing, and repair suggests that such mutants lack the signals that would normally trigger removal of DSB-2 and SUN-1 S8P.
Fig. 8. DSB-2 and SUN-1 S8P persist in mutants defective for DSB repair. 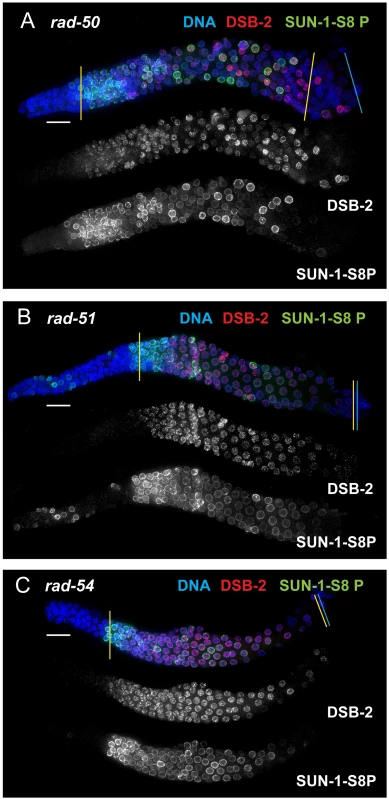
Immunofluorescence images of gonads of indicated genotypes from the distal pre-meiotic region to end of pachytene, stained with DAPI and antibodies that recognize DSB-2 and SUN-1 S8P. The zone of DSB-2 and SUN-1 S8P-positive nuclei is extended in all three mutants depicted: (A) rad-50, which is defective in both DSB formation and DSB repair; (B) and (C) rad-51 and rad-54, which are defective in DSB repair. In the rad-50 mutant germ line, pachytene nuclei have variable staining intensities, with some bright DSB-2 and/or SUN-1 S8P-positive nuclei scattered over the entire pachytene zone; this likely reflects the fact that although the rad-50 mutant lacks SPO-11-dependent DSBs, many nuclei enter meiotic prophase with existing DNA damage resulting from failure to repair lesions arising during DNA replication [6]. DSB-2 staining persists until the end of the pachytene region of the rad-51 and rad-54 mutant gonads, which are also shorter than the gonads of wild-type controls. Scale bar, 15 µm. We next assessed zhp-3, msh-5, and cosa-1 mutants, which have a specific defect in CO formation. These mutants are proficient for homolog pairing and synapsis and can initiate and repair DSBs, but not as COs [13], [21], [22], [32]. All of these mutants showed an extended zone of DSB-2 and SUN-1 S8P staining (Figure 9 B, C, D), thus suggesting that lack of the CO-eligible recombination intermediates that depend on ZHP-3, MSH-5 and COSA-1 will prolong DSB-2 localization to chromatin and phosphorylation of SUN-1 S8.
Fig. 9. DSB-2 and SUN-1 S8P persist when CO formation is impaired. 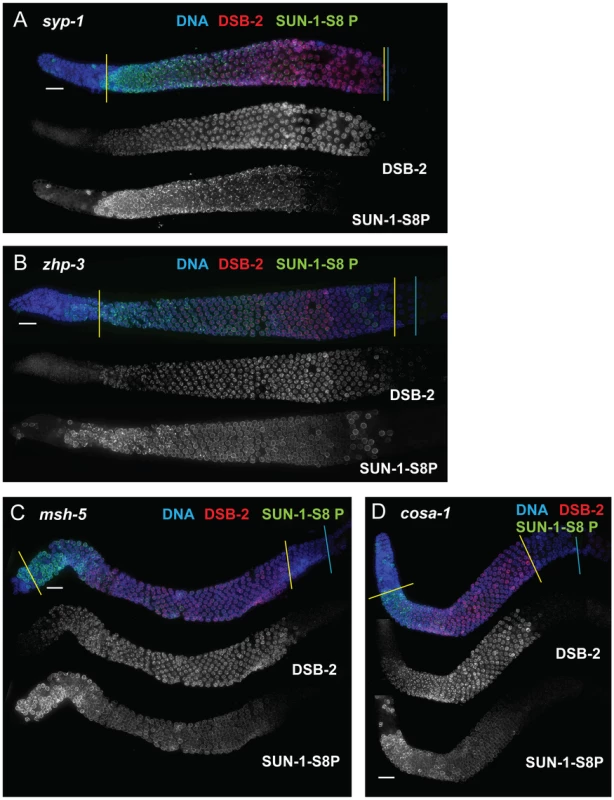
Immunofluorescence images of gonads of indicated genotypes from the distal pre-meiotic region to end of pachytene, stained with DAPI and antibodies that recognize DSB-2 and SUN-1 S8P. The zone of DSB-2 and SUN-1 S8P-positive nuclei is extended in: (A) the syp-1 mutant, which is defective for SC formation and for formation of interhomolog COs; and (B, C and D) the zhp-3, msh-5 and cosa-1 mutants, respectively, which are proficient for synapsis and DNA repair but are defective in conversion of DSBs to COs. Scale bar, 15 µm. Finally, we tested whether meiosis-specific chromosome structures are required to mediate the persistence of DSB-2 and SUN-1 S8P when CO-eligible inter-homolog recombination intermediates are reduced or lacking. We first examined the syp-1 mutant, which loads chromosome axis proteins but lacks a key structural component of the central region of the synaptonemal complex, and thus cannot establish synapsis between homologs [18]. In this mutant, DSB-dependent RAD-51 foci form and persist at elevated levels before disappearing at the very end of pachytene, and COs do not form [18], [21]; in addition, chromosome clustering, chromosome movement and SUN-1 phosphorylation are all greatly prolonged [18], [26], [28], [33]. We found that DSB-2 and SUN-1 S8P staining were both extended to the end of the pachytene region in the syp-1 mutant (Figure 9A). Thus, lack of SYP proteins leads to both lack of inter-homolog COs and prolonged DSB-2 and SUN-1 S8P staining.
In contrast, lack of HORMA domain chromosome axis proteins HTP-1 or HTP-3 does not lead to extended DSB-2 or SUN-1 S8P staining in the respective mutant gonads, despite a lack or severe deficit of inter-homolog COs (Figure 10). htp-1 mutants are defective in pairing of autosomes and assemble SCs between nonhomologous chromosomes, and they exhibit reduced RAD-51 foci reflecting reduced DSB formation and/or altered kinetics of repair [34], [35]; htp-3 mutants are defective in pairing and SC formation for all chromosomes and appear to lack DSBs [36], [37]. We find that despite the deficit or lack of COs in the htp-1 and htp-3 mutants, the zone of DSB-2 and SUN-1 S8P-positive nuclei was not extended (Figures 10, 7). This finding suggests that HTP-1 and HTP-3, or features of axis organization that are dependent on these proteins, are needed for DSB-2 and SUN-1 S8P to persist when CO recombination intermediates are absent.
Fig. 10. DSB-2 and SUN-1 S8P persistence requires axis proteins HTP-1 and HTP-3. 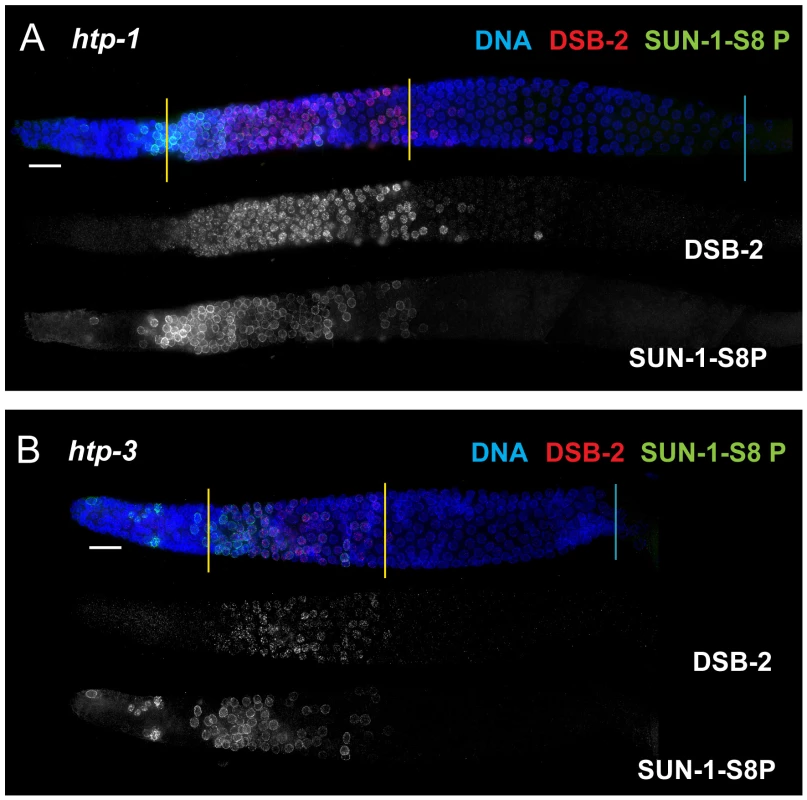
(A and B) Immunofluorescence images of gonads of indicated genotypes from the distal pre-meiotic region to end of pachytene, stained with DAPI and antibodies that recognize DSB-2 and SUN-1 S8P. The zone of DSB-2 and SUN-1 S8P-positive nuclei is not extended in the htp-1 and htp-3 mutants, which lack major components of the meiotic chromosome axes, despite the fact that these mutants are impaired in formation of interhomolog COs. DSB-2 marked nuclei require RAD-50 for formation of RAD-51 foci after irradiation
In addition to acquiring and subsequently losing competence to form DSBs during meiotic prophase progression, C. elegans germ cells also switch on, then subsequently switch off, a specialized meiotic mode of DSB repair [6], [13], [38], [39]. Whereas switching on this meiotic DSB repair mode enables formation of inter-homolog intermediates capable of yielding COs, switching off this repair mode is proposed to facilitate repair of any remaining DSBs in order to guarantee restoration of genome integrity prior to cell division. One notable feature of this specialized meiotic DSB repair mode is a requirement for RAD-50 to load RAD-51 on DSBs induced by gamma-irradiation: whereas essentially all germ cells in wild-type gonads rapidly acquire RAD-51 foci following gamma-irradiation, formation of irradiation-induced RAD-51 foci is strongly inhibited in a specific subset of rad-50 mutant germ cells, from meiotic prophase onset until after the transition to late pachytene [6]. Thus, dependence on RAD-50 for RAD-51 loading at DSBs provides a means to visualize germ cells in which the meiotic DSB repair mode is engaged.
We used this feature to test the hypothesis that the presence of DSB-2 on chromatin correlates with engagement of the meiotic mode of DSB repair. By co-staining for DSB-2 and RAD-51 following irradiation of rad-50 mutant gonads, we found a striking correspondence between the nuclei in which DSB-2 was present on chromatin and the nuclei in which RAD-51 loading was inhibited (Figure 11A). Further, we similarly observed strong correspondence between the presence of DSB-2 and inhibition of RAD-51 loading in htp-1; rad-50 double mutant gonads, in which both features are restricted to a smaller region of the germ line than in the rad-50 single mutant [6]; Figure 11B). Moreover, in both rad-50 and htp-1; rad-50 gonads, nuclei exhibited this inverse correlation between DSB-2 and RAD-51 staining even when neighboring nuclei were in a different mode. In the context of a model in which association of DSB-2 with chromatin is a marker for a DSB-competent state, these results suggest that competence for DSB formation and utilization of the meiotic DSB repair mode are coordinately turned on and shut off, and that coordination of these processes occurs at the level of individual nuclei.
Fig. 11. DSB-2 marked nuclei require RAD-50 for formation of RAD-51 foci after irradiation. 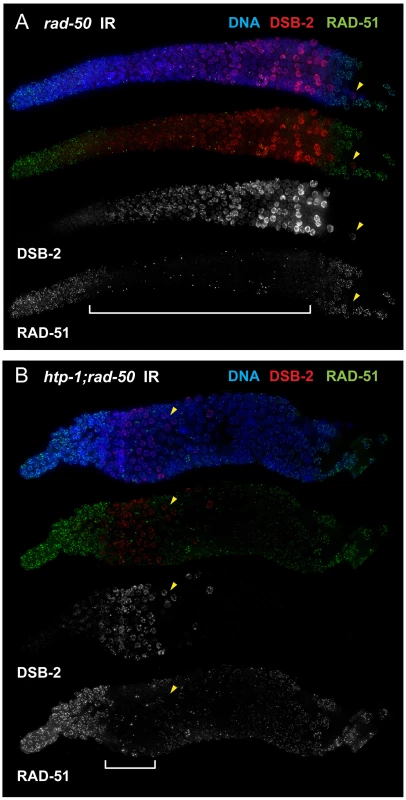
Immunofluorescence images of rad-50 (A) and htp-1; rad-50 (B) mutant gonads from the distal pre-meiotic region to end of pachytene, stained with DAPI and antibodies that recognize DSB-2 and RAD-51. Worms were fixed and stained 1 hour after exposure to 5 kRad of gamma-irradiation. A reciprocal relationship is observed between DSB-2 and RAD-51 immunolocalization: in nuclei where DSB-2 signal is detected on chromatin, formation of irradiation-induced RAD-51 foci is inhibited, and in nuclei where IR-induced RAD-51 foci are present, DSB-2 is absent. The zone of DSB-2 staining/RAD-51 inhibition is indicated by brackets. (Occasional bright RAD-51 foci in the “inhibited” zone are thought to represent pre-existing DNA damage acquired during mitotic cell cycles in mutants lacking RAD-50, as they are both irradiation- and SPO11-independent [6].) Arrowheads point to examples of nuclei that retain DSB-2 staining/RAD-51 inhibition in a region of the germ line where their neighbors do not. Scale bar, 15 µm. While the zone of DSB-2 staining/RAD-51 inhibition in the irradiated rad-50 single mutant extends from meiotic prophase entry to late pachytene, the zone of DSB-2 staining/RAD-51 inhibition is limited to a smaller domain from meiotic entry to the early pachytene region in the irradiated htp-1; rad-50 double mutant. Discussion
DSB-2 as a regulator of DSB competence
In this work, we identify DSB-2 as a protein that is required for efficient meiotic DSB formation and that localizes to chromatin during the stages of meiotic prophase when DSBs are thought to form. DSB-2 localizes to chromatin independently of SPO-11 (and thus of DSB formation) and is restricted to the region of the gonad where RAD-51 foci mark processed DSBs (from TZ to mid-pachytene). Further, the fact that exogenous DSBs induced by irradiation rescue the chiasma defect in dsb-2 mutant germ cells indicates that the downstream DNA processing and CO formation machinery are functional in the mutant. Moreover, the timing of disappearance of DSB-2 coincides with the cessation of DSB formation (implied by the disappearance of RAD-51 foci), suggesting a model in which removal of DSB-2 (and presumably other factors) results in shutting down of DSB formation. Based on these data, we propose that DSB-2 regulates competence for SPO-11-dependent DSB formation during C. elegans meiosis.
Several properties distinguish DSB-2 from other previously identified chromatin-associated proteins (HIM-17, XND-1 and HIM-5) that influence DSB formation in C. elegans. Whereas HIM-17, XND-1 and HIM-5 proteins localize to chromatin in nuclei throughout the germ line [8], [9], [10], the presence of DSB-2 on chromatin correlates with the timing of DSB formation. Further, while him-17 and xnd-1 mutants display pleiotropic phenotypes indicating that HIM-17 and XND-1 have additional roles regulating germ line proliferation and/or organization [9], [40], dsb-2 mutants are specifically defective in meiotic DSB formation. In addition, whereas XND-1 and HIM-5 affect DSB formation predominantly on the X chromosomes, DSB-2 is required for efficient DSB formation on all chromosomes. Together these data suggest that DSB-2 has a more direct role in promoting DSB formation than do HIM-17, XND-1 or HIM-5.
We interpret the region of the germ line where nuclei are positive for DSB-2 localization to represent the zone in which nuclei are competent to undergo DSB formation. Consistent with this interpretation, in meiotic mutants in which the DSB-2-positive zone is extended (and that are capable of making DSBs and loading RAD-51), RAD-51 foci are higher in number and persist beyond mid-pachytene [20], [21], [31], [32]. In principle, persistence of RAD-51 foci could be due to excess/prolonged DSB formation, delayed RAD-51 removal, or both. Thus, caution is warranted when using such mutants to estimate numbers of DSBs. We suggest that in mutants with an extended DSB-2 positive zone (in which the DSB machinery is functional) germ cells may continue to make additional DSBs for a prolonged period, whether or not they are ultimately competent to repair them.
How might DSB-2 control DSB competence? Given its broad yet uneven localization on chromatin, it might act by altering chromatin structure to create an environment that is permissive for the activity of SPO-11 and the DSB machinery. It might also act directly upon SPO-11 and the DSB machinery, by recruiting and/or activating it at certain locations depending upon the underlying chromatin structure. It is intriguing that DSB-2 localizes to a few bright patches/foci in addition to its broader chromatin staining. The fact that these bright patches are absent in him-17 mutants, which are defective in DSB formation, suggests that the patches may have functional significance.
Evidence for feedback regulation coordinating multiple distinct aspects of the meiotic program
Immunofluorescence analyses of DSB-2 in both wild type and meiotic mutants were highly informative regarding how DSB formation is coordinated with multiple distinct aspects of the meiotic program. We found that presence of DSB-2 on chromosomes and the presence of SUN-1 S8P are highly correlated, despite the fact that neither feature is required for the other. Further, we identified CHK-2 as a common upstream regulator of these two features, and we suggest that CHK-2 links acquisition of competence for DSB formation (promoted by DSB-2) with nuclear/chromosomal processes required for successful pairing and synapsis of homologous chromosomes (mediated by SUN-1 at the NE). Moreover, the correlated removal of both DSB-2 and SUN-1 S8P at mid-pachytene, at the same time that RAD-51 foci disappear, further suggests the existence of coordinated regulatory mechanisms that shut down competence for DSB formation and change other properties of the nucleus as germ cells transition to a later stage of meiotic progression.
As seen in multiple experimental systems, DSB formation is restricted to a specific time window in early prophase, indicating that cells must have a means to shut down the meiotic DSB machinery [3]. However, little is known about what controls this transition. Recent evidence from Drosphila, mice and budding yeast suggests that ATM, a protein kinase involved in DNA damage response, may play a role in limiting meiotic DSB formation [41], [42], [43]. It was suggested that ATM is activated by meiotic DSBs and inhibits further DSB formation at the local level by triggering a negative feedback loop. Based on the current work, we propose that additional negative feedback regulation operates at the nucleus-wide level to mediate shutdown of DSB formation during C. elegans meiosis.
Our evidence that germ cells have the capacity to monitor and respond to the presence or absence of DSB-dependent CO-eligible recombination intermediates is based on the analysis of DSB-2 localization in various meiotic mutants. We found that DSB-2 persists in mutants with defects in DSB formation (spo-11, him-17, rad-50), in mutants with defects in early steps of DSB processing (rad-50, rad-51, rad-54), as well as in mutants that can make DSBs but repair them by pathways that do not yield inter-homolog COs (zhp-3, msh-5, cosa-1). Although we cannot exclude the possibility that different defects in these mutants elicit the same response, the parsimonious explanation is that DSB-2 persistence reflects a response to the common deficit shared by all of these mutants, i.e., the inability to generate CO recombination intermediates. Thus, we infer that CO-eligible recombination intermediates are required for removal of DSB-2 with WT timing. We propose a model in which the appearance of CO-eligible recombination intermediates results in a signal (or quenching of an inhibitory signal) that is necessary to trigger the shutdown of DSB formation, in part by removal of DSB-2 (Figure 12). We suggest that this change occurs at the nucleus-wide level when cells sense that sufficient CO-eligible intermediates have been formed to guarantee one CO per chromosome pair. Once this requirement is met, cells are permitted to enter a different state of meiotic progression; if this condition is not met, cells experience a delay in this transition. This type of coupling can be viewed as analogous to checkpoint mechanisms that make cell cycle progression contingent upon fulfillment of a requirement to complete a monitored event. However, it is also appropriate to consider such a coupling as reflecting operation of a negative feedback circuit wherein the formation of threshold levels of a downstream product (i.e. CO-eligible recombination intermediates) feeds back to inhibit an earlier step in the pathway (i.e. DSB formation). Thus, we envision a regulatory network governing DSB formation that involves negative feedback operating on (at least) two levels, one that inhibits DSB formation locally (in a region where a DSB has already formed [41], [42], [43]), and one that inhibits DSB formation nucleus-wide once sufficient CO-eligible recombination intermediates are established. This regulatory network would ensure that sufficient DSBs are made to guarantee that every chromosome pair undergoes a CO [13], [31], [39], while protecting against excessive DSB levels or local concentration of DSBs that could have deleterious effects.
Fig. 12. Model for regulatory network coordinating DSB competence with other aspects of meiotic progression. 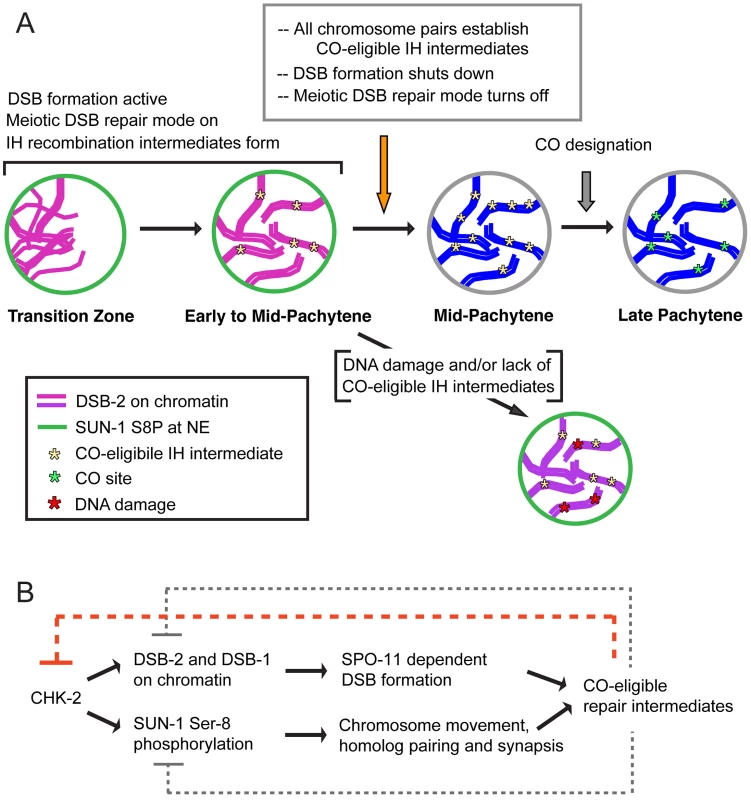
(A) Diagram depicting progression of germ cells through meiotic prophase. DSB-2 localization to chromatin (magenta) and SUN-1 S8P (green) at the nuclear envelope (NE) indicate a cellular state in which chromatin is permissive for DSB formation by SPO-11. Once a germ cell senses that sufficient CO-eligible inter-homolog (IH) intermediates have been formed to guarantee one CO per chromosome pair, the cell responds by shutting down both DSB formation and several specialized features of the meiotic mode of DNA repair (access to the homolog as a repair partner, dependence on RAD-50 for RAD-51 loading). This coordinate transition corresponds to removal of DSB-2 from chromatin and loss of SUN-1 S8P, and enables progression to CO designation and maturation of CO sites. In some nuclei, persistent DNA damage and/or lack of CO-eligible IH intermediates can lead to increased DSB-2 on chromatin (purple) and persistence of SUN-1 S8P at the NE, features that may mark germ cells destined for apoptosis. (B) The CHK-2 protein kinase acts as a common upstream regulator of both DSB-2 localization on chromatin and SUN-1 S8P at the NE, which function in parallel to promote competence for DSB formation (DSB-2) and homologous chromosome synapsis (SUN-1 S8P). Both of these processes are essential for formation of inter-homolog COs. The emergence of sufficient CO-eligible repair intermediates leads to negative feedback regulation by which DSB-2 and SUN-1 S8P are removed, shutting down DSB formation and leading to a change in nucleus state and properties. This coordinated negative feedback regulation may occur through common regulator CHK-2. We further propose that multiple aspects of the meiotic recombination program undergo a coordinated transition that in wild type germ cells is marked by disappearance of DSB-2 and SUN-1 S8P (Figure 12). We proposed in a previous study that access to the homologous chromosome as a repair partner is shut down once sufficient CO-eligible recombination intermediates are formed [39]. We suggested that this transition occurs around mid-pachytene in WT germ lines, and we showed that inter-homolog access is prolonged in msh-5 mutants [39]. In light of the current results, an attractive possibility is that the appearance of sufficient CO-eligible recombination intermediates simultaneously signals both shut-down of DSB formation and shut down of inter-homolog access for DSB repair. Moreover, we found that another specialized aspect of the meiotic DSB repair program, namely the dependence on RAD-50 for rapid loading of RAD-51 on IR-induced DSBs, is restricted to nuclei positive for DSB-2. This finding further strengthens the case that cessation of programmed DSB formation is coordinated with a major transition in the mode of DSB repair.
It is notable that in mutants defective for HORMA domain axis proteins HTP-1 and HTP-3, the DSB-2/SUN-1 S8P - positive zone is not extended despite the absence of CO-eligible recombination intermediates on most or all chromosomes. This finding raises the possibility that this family of proteins, which was previously implicated in the operation of checkpoint-like coupling mechanisms that coordinate early prophase chromosome movement, homolog recognition and SC assembly [34], may also be required for operation of checkpoint-like mechanisms that make later events in meiotic progression contingent upon the formation of CO-eligible recombination intermediates.
We speculate that the regulatory network that coordinates this meiotic transition (i.e. the shutdown of DSB formation and accompanying changes) likely involves the activities of one or more protein kinases. As the CHK-2 protein kinase is required to promote the acquisition of both DSB-2 and SUN-1 S8P, it is likely that the disappearance of DSB-2 and SUN-1 S8P requires inactivation of CHK-2, suggesting that CHK-2 may be a key target of feedback regulation. Further, DSB-2 contains several potential phosphorylation sites both for CHK-2 and for the ATM/ATR protein kinases [12], [44]. Future work will investigate the significance of these for DSB-2 function and regulation.
The fact that DSB-2 and SUN-1 S8P are coordinately removed in wild-type meiosis (and coordinately prolonged in mutants) implies that the NE also responds to signaling from CO-eligible recombination intermediates. Our findings confirm and extend the recent report of Woglar et al., who similarly showed that the SUN-1 phosphorylation is prolonged in spo-11 and rad-51 mutants and concluded that establishment of CO intermediates is necessary for exit from early pachytene (as defined by loss of phospho-SUN-1) [26]. The change in SUN-1 phosphorylation status at this transition may be indicative of global changes in properties of the nucleus that occur as it enters a different stage of meiotic progression; e.g., the fluidity of the nuclear membrane, which is modified upon entry into meiotic prophase [28], may revert to a more constrained state similar to that of non-meiotic germ cells. Such a change would be analogous to the observed reversion to the non-meiotic mode of DSB repair that occurs at this same transition.
DSB-2 and SUN-1 may function in activation of the DNA damage checkpoint and apoptosis signaling
While DSB-2 and SUN-1 S8P immunofluorescence signals become dimmer and disappear from most nuclei by the time they reach the mid-pachytene region of the germ line, a few “outlier” nuclei show bright DSB-2 and SUN-1 S8P staining later in the pachytene region. Sometimes the chromatin in these nuclei has a clustered organization reminiscent of zygotene or early pachytene stages, but in contrast to earlier nuclei, these outlier nuclei have brighter DSB-2 staining covering most of the chromatin as well as high levels of RAD-51 foci. This difference suggests that these nuclei are arrested in their progression and may have triggered a checkpoint response. This response could be due to failure to make appropriate CO-eligible recombination intermediates and/or to the presence of excess or persistent DNA breaks. These processes may be inter-related: if the failure to make CO-eligible recombination intermediates keeps DSB formation active, this could increase the chance of accumulating levels of DNA damage that challenge the capacity for repair. Accumulation of high levels of DSB-2 and SUN-1 S8P may indicate that these nuclei are triggering the recombination/DNA damage checkpoint and will be targeted for future apoptosis. While these outlier nuclei may be destined for apoptosis, however, they likely have not yet engaged the cell death program, as outlier nuclei are still detected in mutants lacking the pro-apoptotic factors CED-3 or CED-4 [11], [26].
Meiotic recombination robustness in a changing germline environment
An intriguing aspect of the dsb-2 mutant phenotype is that the defect in meiotic recombination worsens with age. This implies that the DSB-1 protein retains some residual DSB-promoting activity in the absence of its paralog, but also indicates that the requirement for DSB-2 becomes more acute in older germ cells. Interestingly, CO distribution has also been found to differ between young and old WT C. elegans oocytes [45]. This suggests that meiotic recombination processes such as DSB formation and CO distribution are sensitive to changes in the germline environment that occur as worms age. However, the ability to achieve accurate and reliable meiosis in the context of a changing environment is advantageous for the reproductive success of the organism. The C. elegans reproductive system has substantial plasticity in this regard, as the duration of progression through meiotic prophase varies markedly with both sex and age and can be modulated dramatically in the female germ line by the availability of sperm [46]. The operation of feedback networks such as that demonstrated here provides a means to regulate and coordinate key events and transitions in a manner that buffers the system against a varying environment, thereby promoting reproductive success.
Materials and Methods
C. elegans strains
Strains were maintained at 20°C under standard conditions. Experiments were performed at 20°C unless otherwise noted. Strains used in this study:
AV334 unc-119 III; ruIs32 [Ppie-1::GFP-his-11; unc-119(+)] III mnT12 (IV;X)
AZ212 unc-119 III; ruIs32 [Ppie-1::GFP-his-11; unc-119(+)] III
AV477 dsb-2(me96) II 4X outcrossed
AV501 rol-1(e91) dsb-2(me96) II
AV511 rol-1(e91) dsb-2(me96) unc-52(e998) II
AV539 rol-1(e91) dsb-2(me96)/mnC1 [dpy-10(e128) unc-52(e444)] II
AV727 meIs8[unc-119(+) pie-1promoter::gfp::cosa-1] II ;; ItIs37[unc-119(+)pie-1::mcherry::histoneH2B]; ltIs38[pAA1;pie-1 promoter::GFP::PH::unc-119(+)]
AV758 dsb-2(me97) meIs8[unc-119(+) pie-1promoter::gfp::cosa-1] II ;; ItIs37[unc-119(+)pie-1::mcherry::histoneH2B]; ltIs38[pAA1;pie-1 promoter::GFP::PH::unc-119(+)]
AV630 meIs8[unc-119(+) pie-1promoter::gfp::cosa-1] II
AV645 spo-11(ok79)/nT1 IV; +/nT1[qIs51] V
AV146 chk-2(me64) rol-9(sc148)/unc-57(e369) rol-9(sc148) V
AV660 chk-2(me64) rol-9(sc148)/sC4(s2172)[dpy-21(e428)] V
VC292 +/nT1 IV; sun-1(gk199)/nT1 V
VC255 +/nT1 IV, him-17(ok424)/nT1 V
AV158 +/nT1 IV; rad-50(ok197)/nT1 [unc-?(n754) let-? qIs50] V
TG9 dpy-13(e184) rad-51(lg8701) IV/nT1[let-?(m435)] (IV;V)
VC531 rad-54 and tag-157(ok615) I/hT2[gli-4(e937) let(9782) qIs48] I; III
AV449 zhp-3(me95)/hT2 [bli-4(e937) let-? (q782) qIs48] I
AV603 msh-5(me23)/nT1 IV; +/nT1[qIs51] V
AV596 cosa-1(tm3298)/qC1[qIs26] III
AV307 +/nT1 IV; syp-1(me17)/nT1 V
AV393 htp-1(gk174) IV/nT1[unc-?(n754) let-? qIs50] (IV;V)
TY4986 htp-3(y428) ccIs4251 I/hT2[bli-4(e937)let-?(q782) qIs48] (I,III).
AV473 +/nT1 IV; rad-50(ok197)/nT1[qIs51] V
AV443 htp-1(gk174)/nT1[ unc-?(n754) let-? qIs50] IV; rad-50 (ok197)/nT1 [qIs51] V
Bristol (N2) wild type
CB4856 Hawaiian wild type
Isolation, mapping and molecular identification of dsb-2 mutations
The dsb-2(me96) allele was isolated in a genetic screen for meiotic mutants exhibiting defects in chiasma formation or chromosome organization in diakinesis-stage oocytes, conducted in collaboration with M. Hayashi [47]. The AV334 strain used for this screen, which allows visualization of chromosomes using a germline-expressed GFP::histone H2B fusion protein, also contains a fusion of chromosomes IV and X. Parental (P0) L4 hermaphrodites were treated with ethyl methanosulfonate (EMS) as in [48] and were plated individually. F1 progeny were picked to individual plates to produce progeny, and pools of F2 progeny worms from each F1 plate were mounted on multi-well slides in anesthetic (0.1% tricaine and 0.01% tetramisole in M9 buffer) and their germ lines were visualized for meiotic defects. Two mutations affecting meiotic recombination, me95 and me96, were identified based on the presence of univalents at diakinesis in a the subset of F2s (from independent F1s) and were recovered by plating of siblings; repeated outcrossing (with N2) and selecting for the mutant phenotype removed the chromosome fusion as well as the GFP::H2B transgene. Mapping, complementation testing and sequencing revealed that me95 is an allele of zhp-3, containing a C-to-T transition that results in a premature stop at codon 348 of the predicted 387 amino acid coding sequence of K02B12.8a. Mapping of the me96 mutation (below) indicated that it identified a new component of the meiotic machinery.
Initial SNP mapping based on the methods of [49] and [50] placed me96 near genetic map position 20 on the right side of chromosome II. To select for informative COs near this region, rol-1 me96 unc-52 worms were crossed with CB4856 males, and Rol non-Unc F2 progeny were selected and genotyped for SNP pkP2117 at genetic map position 17.9 to select for COs occurring between this marker and unc-52 (genetic map position 23). Informative recombinants were assessed for me96 phenotype and typed using additional SNP markers in the region, narrowing down the position of me96 to a 165 kb interval (between SNP markers uCE2-2315 and uCE2-2332) comprising 36 candidate genes. RNAi of candidate genes was performed using bacterial clones from the RNAi feeding library [51], [52] as in [53]. Worms used were AZ212 worms, which contain GFP::histone, as this genotype was shown to be more sensitive to RNAi [54]. RNAi against candidate gene F26H11.6 (at 15°C, but not at 20°C) phenocopied the me96 mutation, eliciting a mixture of bivalents and univalents at diakinesis. Sequencing identified a T-to-A transversion generating an early stop at codon 14 (TTA = >TAA) of the predicted F26H11.6 coding sequence (280 codons total) in the me96 mutant.
A second dsb-2 allele (me97) was isolated independently in a screen for mutants with altered numbers of GFP::COSA-1 foci (which mark CO sites in late pachytene); me97 fails to complement me96 and contains a premature stop at codon 168, further confirming the identity of F26H11.6 as the dsb-2 gene. Except where otherwise noted, all analyses were conducted using the me96 allele.
Assessment of inviable embryos and males
L4 hermaphrodites were picked to individual plates, allowed to lay eggs, and transferred to fresh plates every 24 hours for three days. Hermaphrodites start laying eggs after they transition from L4 to adult, and lay most eggs in the first three days. Inviable embryos that do not hatch are indicative of autosomal mis-segregation, while male progeny indicate X-chromosome mis-segregation. Eggs from eight dsb-2(me96); rol-1(e91) hermaphrodites (where rol-1 is a marker with no meiotic defects) were counted. The number of eggs counted for each day is: 573 (day 1), 879 (day 2) and 488 (day 3), with an average of 243 eggs per hermaphrodite over the three day interval.
Cytological methods
Numbers of DNA bodies present in diakinesis oocytes were assessed in intact adult hermaphrodites of indicated age, fixed in ethanol and stained with 4′,6-diamidino-2-phenylindole (DAPI) as in [40]. This method underestimates the frequency of achiasmate chromosomes, as some univalents lie too close to each other to be resolved unambiguously. Numbers of nuclei scored were: 88 and 106 for dsb-2 worms, 1 day and 2 day post L4 respectively; 43 and 57 for WT worms, 1 day and 2 day post L4 respectively.
Numbers of GFP::COSA-1 foci in late pachytene nuclei of live anesthetized worms (0.1% tricaine and 0.01% tetramisole in M9 buffer ) were quantified by taking 3D image stacks on a DeltaVision microscope. GFP foci were counted in the last five rows of pachytene nuclei; only nuclei completely contained within the stack were scored, and nuclei with features indicative of apoptosis (compact and bright mCherry::histoneH2B signal) were excluded. 24 h control data were from fixed immunofluorescence images [13]. Numbers of nuclei scored: dsb-2 24 h, n = 127; dsb-2 48 h, n = 101; WT 24 h, n = 76; WT 48 h, n = 78. (Note: Whereas most nuclei in a mutant that lacks DSBs and COs have zero COSA-1 foci, 20% have one or two foci presumably reflecting non-specific aggregation of CO proteins when a suitable substrate is absent [13]. Thus, a subset of dsb-2 nuclei with one or two COSA-1 foci may similarly lack COs, especially at 48 h post L4 where nuclei with zero foci are frequent.)
Immunofluorescence was conducted as in [55] with minor modifications. Unless otherwise noted, all experiments were performed at 40–48 hours post L4. Worms were cut at the vulva to dissect the gonads (in egg buffer with 0.1% Tween-20) and fixed with 1% paraformaldehyde (in egg buffer) for 5 minutes. Slides (Superfost Plus) were covered with a coverslip and frozen in liquid nitrogen. The coverslip was removed, and slides were immersed in cold (−20°C) methanol for 1 minute. Slides were washed three times for 8–10 minutes in phosphate-buffered saline containing 0.1% Tween-20 (PBST) and then blocked for one hour with 0.5% bovine serum albumin (BSA) diluted in PBST. Primary antibody solution was added (50 µl) on top of the dissected gonads and covered with a parafilm square. Slides were incubated overnight in a humid chamber at room temperature, then washed three times for 8–10 minutes in PBST. Secondary antibody solution was added (50 µl) and slides were incubated with parafilm cover for 2 hours at room temperature in the dark. Slides were washed three times with PBST and incubated for 5 minutes with 2 µg/ml DAPI solution in the dark, followed by two more washes. Slides were mounted with Vectashield and the coverslip was sealed with nail polish.
The following primary antibodies were used at the indicated dilutions in PBST with 0.5% BSA: guinea pig anti-HIM-8 (1∶500) [16], rabbit anti-HIM-3 (1∶200) [17], guinea pig anti-SYP-1 (1∶200) [18], rabbit anti-RAD-51 (1∶500) [21] , guinea pig anti-SUN1 S8P (1∶1000) [23], rabbit anti-DSB-2 (1∶5000), rat anti-RAD-51 (1∶250), guinea pig anti-DSB-1 (1∶500) [11].
An affinity-purified rabbit polyclonal antibody against DSB-2 was generated by SDIX (Newark, DE) using the C-terminal 100 amino acids of F26H11.6 as the immunogen. Specificity of the antibody was demonstrated both by the lack of chromatin staining in immunofluorescence analysis of dsb-2 mutant gonads (Figure 6A) and by Western blot analysis (Figure 6B).
Rat anti-RAD-51 antibody was generated using a His-tagged fusion protein expressed from plasmid pET28a containing the entire RAD-51S coding sequence [56]; immunizations and bleeds were performed by SDIX. Rat anti -RAD-51 was affinity purified against membrane-bound protein as described in [57] with the following modifications: nitrocellulose membrane was blocked in 5% milk in 1×TBST; and, eluates containing rat anti -RAD-51 were further purified by dialysis with 12–14 kDa dialysis tubing (Spectrum) in 1×TBST for 1 hour and overnight at 4°C. Specificity was demonstrated by showing that rat anti-RAD-51 foci colocalize with rabbit anti-RAD-51 foci [21] by immunofluorescence and that these recombination-dependent foci are eliminated in spo-11(me44) gonads.
All secondary antibodies were Alexa Fluor goat from Invitrogen used at 1∶200 dilution in PBST with 0.5% BSA.
Immunofluorescence images were acquired using the DeltaVision microscopy system (Applied Precision) and deconvolved using softWoRx software. Images shown are maximum-intensity projections of Z-stacks acquired at 0.3 µm intervals.
Quantitation of RAD-51 foci
For each wild-type germ line evaluated, RAD-51 foci were quantified in 8 contiguous rows of pachytene nuclei from the region where foci were most abundant. The average distance (in rows of nuclei) between the position of this peak and the end of the transition zone was calculated for wild-type germ lines, and this distance was used to define the corresponding regions to be scored in dsb-2(me96) mutant germ lines (in which the abundance of RAD-51 foci was low throughout). Quantitation was carried out on deconvolved 3D image stacks using SoftWoRx software; only nuclei that were completely contained with in the image stack were scored. Occasional atypical nuclei with condensed, bright DAPI signals were excluded. Numbers of nuclei scored: WT, n = 335; dsb-2, n = 196.
Western blot analysis
For each genotype, sixty adult worms (24 hours post-L4) were picked into M9+0.05% Tween 20, washed gently three times, then lysed by resuspension in 2× Laemmli Sample Buffer (Bio-Rad). Gel electrophoresis was performed on a 4–15% Criteriot TGX gradient gel (Bio-Rad), followed by transfer of proteins to a PVDF membrane. Blots were probed with rabbit anti-DSB-2 (1∶1000 in 5% milk) for 2 hours, followed by HRP-conjugated secondary antibody and detection by ECL.
Rescue of chiasma formation by gamma-irradiation
Worms were exposed to 1 kRad (10Gy) of gamma-irradiation using a Cs-137 source. The 1 kRad dose was chosen based on its sufficiency to restore chiasmata to 95% of chromosome pairs in affected nuclei of the spo-11(ok79) mutant [6]. Worms were irradiated at 36 hours post L4, and the number of DNA bodies at diakinesis was assessed in worms fixed at 18 hours post-irradiation, for both dsb-2 and age-matched spo-11 mutants. The dsb-2 worms also carried the rol-1 marker, which does not affect meiosis. This assay tends to underestimate the incidence of achiasmate chromosomes, as some lie too close together to be resolved. Numbers of nuclei scored were: 71 and 76 for dsb-2 worms, untreated and irradiated respectively; 76 and 45 for spo-11 worms, untreated and irradiated respectively.
Assessment of RAD-51 foci following gamma-irradiation
Worms were exposed to 5 kRad (50Gy) of gamma-irradiation using a Cs-137 source. Formation of RAD-51 foci was assessed by immunofluorescence in gonads dissected and fixed 1 hour after irradiation. Germ lines from rad-50 and htp-1; rad-50 mutants were irradiated at 24 hours post-L4, and stained with DAPI, RAD-51 antibody and DSB-2 antibody. Germ lines from dsb-2 mutants were irradiated at 48 hours post-L4 and stained with DAPI and RAD-51 antibody.
Supporting Information
Zdroje
1. KeeneyS (2001) Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52 : 1–53.
2. PageSL, HawleyRS (2003) Chromosome choreography: the meiotic ballet. Science 301 : 785–789.
3. KeeneyS (2008) Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn Stab 2 : 81–123.
4. KumarR, BourbonHM, de MassyB (2010) Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 24 : 1266–1280.
5. LakeCM, NielsenRJ, HawleyRS (2011) The Drosophila zinc finger protein trade embargo is required for double strand break formation in meiosis. PLoS Genet 7: e1002005.
6. HayashiM, ChinGM, VilleneuveAM (2007) C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet 3: e191.
7. BaudrimontA, PenknerA, WoglarA, MamnunYM, HulekM, et al. (2011) A new thermosensitive smc-3 allele reveals involvement of cohesin in homologous recombination in C. elegans. PLoS One 6: e24799.
8. ReddyKC, VilleneuveAM (2004) C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118 : 439–452.
9. WagnerCR, KuerversL, BaillieDL, YanowitzJL (2010) xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467 : 839–843.
10. MeneelyPM, McGovernOL, HeinisFI, YanowitzJL (2012) Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190 : 1251–1266.
11. StamperEL, RodenbuschSE, RosuS, AhringerJ, VilleneuveAM, et al. (2013) Identification of DSB-1, a protein required for initiation of meiotic recombination in C. elegans, illuminates a checkpoint that promotes crossover assurance. PLoS Genet In press.
12. TravenA, HeierhorstJ (2005) SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays 27 : 397–407.
13. YokooR, ZawadzkiKA, NabeshimaK, DrakeM, ArurS, et al. (2012) COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149 : 75–87.
14. DernburgAF, McDonaldK, MoulderG, BarsteadR, DresserM, et al. (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94 : 387–398.
15. MacQueenAJ, PhillipsCM, BhallaN, WeiserP, VilleneuveAM, et al. (2005) Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123 : 1037–1050.
16. PhillipsCM, WongC, BhallaN, CarltonPM, WeiserP, et al. (2005) HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123 : 1051–1063.
17. ZetkaMC, KawasakiI, StromeS, MullerF (1999) Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13 : 2258–2270.
18. MacQueenAJ, ColaiacovoMP, McDonaldK, VilleneuveAM (2002) Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev 16 : 2428–2442.
19. KeeneyS, GirouxCN, KlecknerN (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 : 375–384.
20. AlpiA, PasierbekP, GartnerA, LoidlJ (2003) Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112 : 6–16.
21. ColaiacovoMP, MacQueenAJ, Martinez-PerezE, McDonaldK, AdamoA, et al. (2003) Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell 5 : 463–474.
22. KellyKO, DernburgAF, StanfieldGM, VilleneuveAM (2000) Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156 : 617–630.
23. PenknerAM, FridkinA, GloggnitzerJ, BaudrimontA, MachacekT, et al. (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139 : 920–933.
24. PenknerA, TangL, NovatchkovaM, LadurnerM, FridkinA, et al. (2007) The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell 12 : 873–885.
25. SatoA, IsaacB, PhillipsCM, RilloR, CarltonPM, et al. (2009) Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139 : 907–919.
26. WoglarA, DaryabeigiA, AdamoA, HabacherC, MachacekT, et al. (2013) Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet 9: e1003335.
27. MacQueenAJ, VilleneuveAM (2001) Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev 15 : 1674–1687.
28. WynneDJ, RogO, CarltonPM, DernburgAF (2012) Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol 196 : 47–64.
29. TangL, MachacekT, MamnunYM, PenknerA, GloggnitzerJ, et al. (2010) Mutations in Caenorhabditis elegans him-19 show meiotic defects that worsen with age. Mol Biol Cell 21 : 885–896.
30. ChinGM, VilleneuveAM (2001) C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev 15 : 522–534.
31. MetsDG, MeyerBJ (2009) Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139 : 73–86.
32. JantschV, PasierbekP, MuellerMM, SchweizerD, JantschM, et al. (2004) Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol 24 : 7998–8006.
33. BaudrimontA, PenknerA, WoglarA, MachacekT, WegrostekC, et al. (2010) Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet 6: e1001219.
34. Martinez-PerezE, VilleneuveAM (2005) HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev 19 : 2727–2743.
35. CouteauF, ZetkaM (2005) HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev 19 : 2744–2756.
36. GoodyerW, KaitnaS, CouteauF, WardJD, BoultonSJ, et al. (2008) HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell 14 : 263–274.
37. SeversonAF, LingL, van ZuylenV, MeyerBJ (2009) The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev 23 : 1763–1778.
38. SmolikovS, EizingerA, HurlburtA, RogersE, VilleneuveAM, et al. (2007) Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 176 : 2027–2033.
39. RosuS, LibudaDE, VilleneuveAM (2011) Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334 : 1286–1289.
40. BesslerJB, ReddyKC, HayashiM, HodgkinJ, VilleneuveAM (2007) A role for Caenorhabditis elegans chromatin-associated protein HIM-17 in the proliferation vs. meiotic entry decision. Genetics 175 : 2029–2037.
41. JoyceEF, PedersenM, TiongS, White-BrownSK, PaulA, et al. (2011) Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J Cell Biol 195 : 359–367.
42. ZhangL, KimKP, KlecknerNE, StorlazziA (2011) Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc Natl Acad Sci U S A 108 : 20036–20041.
43. LangeJ, PanJ, ColeF, ThelenMP, JasinM, et al. (2011) ATM controls meiotic double-strand-break formation. Nature 479 : 237–240.
44. AhnJ, UristM, PrivesC (2004) The Chk2 protein kinase. DNA Repair (Amst) 3 : 1039–1047.
45. LimJG, StineRR, YanowitzJL (2008) Domain-specific regulation of recombination in Caenorhabditis elegans in response to temperature, age and sex. Genetics 180 : 715–726.
46. Jaramillo-LambertA, EllefsonM, VilleneuveAM, EngebrechtJ (2007) Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol 308 : 206–221.
47. Rosu S (2012) Regulation of meiotic recombination: DNA double-strand break formation and repair in C. elegans [PhD thesis]. Stanford (California): Department of Genetics, Stanford University. Available: http://purl.stanford.edu/qz167mc2859.
48. VilleneuveAM (1994) A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136 : 887–902.
49. WicksSR, YehRT, GishWR, WaterstonRH, PlasterkRH (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet 28 : 160–164.
50. DavisMW, HammarlundM, HarrachT, HullettP, OlsenS, et al. (2005) Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6 : 118.
51. FraserAG, KamathRS, ZipperlenP, Martinez-CamposM, SohrmannM, et al. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408 : 325–330.
52. KamathRS, AhringerJ (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30 : 313–321.
53. WignallSM, VilleneuveAM (2009) Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol 11 : 839–844.
54. HayashiM, Mlynarczyk-EvansS, VilleneuveAM (2010) The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186 : 45–58.
55. NabeshimaK, VilleneuveAM, HillersKJ (2004) Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168 : 1275–1292.
56. RinaldoC, EderleS, RoccoV, La VolpeA (1998) The Caenorhabditis elegans RAD51 homolog is transcribed into two alternative mRNAs potentially encoding proteins of different sizes. Mol Gen Genet 260 : 289–294.
57. KoelleMR, HorvitzHR (1996) EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84 : 115–125.
58. NotredameC, HigginsDG, HeringaJ (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 : 205–217.
59. KiontkeK, FitchDH (2005) The phylogenetic relationships of Caenorhabditis and other rhabditids. WormBook 1–11.
Štítky
Genetika Reprodukčná medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



