-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
The DAF-16/FoxO transcription factor controls growth, metabolism and aging in Caenorhabditis elegans. The large number of genes that it regulates has been an obstacle to understanding its function. However, recent analysis of transcript and chromatin profiling implies that DAF-16 regulates relatively few genes directly, and that many of these encode other regulatory proteins. We have investigated the regulation by DAF-16 of genes encoding the AMP-activated protein kinase (AMPK), which has α, β and γ subunits. C. elegans has 5 genes encoding putative AMP-binding regulatory γ subunits, aakg-1-5. aakg-4 and aakg-5 are closely related, atypical isoforms, with orthologs throughout the Chromadorea class of nematodes. We report that ∼75% of total γ subunit mRNA encodes these 2 divergent isoforms, which lack consensus AMP-binding residues, suggesting AMP-independent kinase activity. DAF-16 directly activates expression of aakg-4, reduction of which suppresses longevity in daf-2 insulin/IGF-1 receptor mutants. This implies that an increase in the activity of AMPK containing the AAKG-4 γ subunit caused by direct activation by DAF-16 slows aging in daf-2 mutants. Knock down of aakg-4 expression caused a transient decrease in activation of expression in multiple DAF-16 target genes. This, taken together with previous evidence that AMPK promotes DAF-16 activity, implies the action of these two metabolic regulators in a positive feedback loop that accelerates the induction of DAF-16 target gene expression. The AMPK β subunit, aakb-1, also proved to be up-regulated by DAF-16, but had no effect on lifespan. These findings reveal key features of the architecture of the gene-regulatory network centered on DAF-16, and raise the possibility that activation of AMP-independent AMPK in nutritionally replete daf-2 mutant adults slows aging in C. elegans. Evidence of activation of AMPK subunits in mammals suggests that such FoxO-AMPK interactions may be evolutionarily conserved.
Published in the journal: DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in. PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004109
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004109Summary
The DAF-16/FoxO transcription factor controls growth, metabolism and aging in Caenorhabditis elegans. The large number of genes that it regulates has been an obstacle to understanding its function. However, recent analysis of transcript and chromatin profiling implies that DAF-16 regulates relatively few genes directly, and that many of these encode other regulatory proteins. We have investigated the regulation by DAF-16 of genes encoding the AMP-activated protein kinase (AMPK), which has α, β and γ subunits. C. elegans has 5 genes encoding putative AMP-binding regulatory γ subunits, aakg-1-5. aakg-4 and aakg-5 are closely related, atypical isoforms, with orthologs throughout the Chromadorea class of nematodes. We report that ∼75% of total γ subunit mRNA encodes these 2 divergent isoforms, which lack consensus AMP-binding residues, suggesting AMP-independent kinase activity. DAF-16 directly activates expression of aakg-4, reduction of which suppresses longevity in daf-2 insulin/IGF-1 receptor mutants. This implies that an increase in the activity of AMPK containing the AAKG-4 γ subunit caused by direct activation by DAF-16 slows aging in daf-2 mutants. Knock down of aakg-4 expression caused a transient decrease in activation of expression in multiple DAF-16 target genes. This, taken together with previous evidence that AMPK promotes DAF-16 activity, implies the action of these two metabolic regulators in a positive feedback loop that accelerates the induction of DAF-16 target gene expression. The AMPK β subunit, aakb-1, also proved to be up-regulated by DAF-16, but had no effect on lifespan. These findings reveal key features of the architecture of the gene-regulatory network centered on DAF-16, and raise the possibility that activation of AMP-independent AMPK in nutritionally replete daf-2 mutant adults slows aging in C. elegans. Evidence of activation of AMPK subunits in mammals suggests that such FoxO-AMPK interactions may be evolutionarily conserved.
Introduction
In Caenorhabditis elegans, it has long been known that reduction of insulin/IGF-1 signaling (IIS) increases lifespan [1]–[3], reviewed in [4]. However, the mechanisms by which IIS controls aging, and the nature of the aging process itself, remain unclear [5]. One protein that is required for daf-2 mutants to promote longevity is the FoxO transcription factor DAF-16 [2], [6], [7]. This suggests that DAF-16 regulates expression of terminal effectors of aging. Over the last decade a number of studies have characterized the set of genes regulated by IIS and DAF-16, e.g. [8]–[15]. Although there are many suggestions for how IIS and DAF-16 control aging [9], [16]–[21] how they actually do has proved difficult to determine, particularly because, directly or indirectly, DAF-16 regulates a very large number of other genes.
One approach to investigate DAF-16 function is to define the topology of the gene regulatory network within which it acts. To this end we recently used new chromatin profiling data, cross-referenced to mRNA profile data, to identify with high confidence 65 genes subject to direct transcriptional activation by DAF-16 [22]. The identity of genes in this set gave rise to a new view of DAF-16 action. Rather than acting to regulate a range of somatic maintenance proteins (e.g. detoxification enzymes and chaperonins), DAF-16 targets are involved primarily in signaling and gene regulation, e.g. kinases, phosphatases and transcription factors. This suggests that DAF-16 functions as a central node within a gene regulatory sub-network. Predicted direct DAF-16 target genes include a number of major regulators of metabolism and aging including the AMP-activated protein kinase (AMPK) and, perhaps, DAF-16 itself.
AMPK acts as a fuel gauge within cells: when the AMP∶ATP ratio rises as energy availability drops, activation of AMPK increases catabolism and reduces biosynthesis [23]. Thus, this enzyme helps to coordinate energy availability with rates of biosynthesis and growth. AMPK is heterotrimeric and is formed of an α catalytic subunit and β and γ regulatory subunits. In C. elegans there are multiple isoforms of each subunit, aak-1 and aak-2 (α), aakb-1 and aakb-2 (β), and aakg-1-5 (γ) (WormBase version WS238). Mammalian AMPK is activated by binding of AMP or ADP to the γ subunit [23], [24].
AMPK inhibits aging in C. elegans: The increased lifespan seen in mutants with reduced IIS (e.g. daf-2) or TOR (e.g. rsks-1) pathways, as well as those with mutations affecting mitochondrial function (e.g. isp-1) are aak-2 (i.e. AMPK) dependent [25]–[27] as is the longevity incurred by a specific form of dietary restriction [28]. Moreover, over-expression or activation of AMPK genes can modestly increase lifespan [25], [28], [29]. Evidence suggests that AMPK may also protect against aging in other species. In Drosophila activation of AMPK by modulating enzymes involved in AMP biosynthesis extends lifespan [30] as does a gain-of-function mutation in lkb1 (an upstream activator of AMPK) [31], although metformin-induced activation of AMPK did not increase lifespan [32]. In mice, mutation of ribosomal S6 kinase (S6K1) increases AMPK activity and slows aging [27], and AMPK activation in response to either AICAR, dietary restriction or physical stimuli declines with age in both rats and mice; reviewed in [33].
That daf-2 longevity requires aak-2, the catalytic subunit of AMPK, may reflect regulation of AMPK by IIS, but the nature of such regulation remains poorly defined [25]. One possibility is that in IIS mutants, DAF-16 promotes longevity by stimulating expression of genes encoding AMPK. Chromatin and mRNA profile data suggest that DAF-16 binds to and activates expression of aakg-4 and aakg-5, and mRNA profile data suggests that DAF-16 also activates expression of aak-2 and aakb-1 [22]. Notably, DAF-16 is also phospho-activated by AMPK [28], as is mammalian FoxO3 [34]. This raises the possibility that DAF-16 and AMPK are mutual activators, acting in a positive feedback loop. In this study we have carefully examined the role of DAF-16/FoxO in the regulation of genes encoding AMPK α, β, and γ subunits. We report that the atypical aakg-4 subunit is a direct target of DAF-16, leading to increased aakg-4 gene expression. Moreover, reducing aakg-4 expression attenuates daf-2 mutant longevity, implying that this regulation contributes to longevity. Effects of aakg-4 and daf-2 on expression of DAF-16 target genes support the existence of an AMPK-DAF-16 positive feedback loop promoting diapause and longevity. In addition, the daf-16 gene itself contains strong DAF-16 binding sites [22], suggesting a possible second positive feedback loop. However, we could find no further evidence that DAF-16/FoxO regulates its own expression.
Results
Atypical AMPK γ isoforms in C. elegans
Our initial mRNA and chromatin profiling analysis suggested that two AMPK γ subunits, aakg-4 and aakg-5, are direct targets of, and transcriptionally activated by DAF-16 [22]. Binding of AMP to the AMPK γ subunit allosterically activates the enzyme heterotrimer and facilitates phosphorylation of the α subunit by the upstream kinase PAR-4/LKB-1 [23]. C. elegans is unusual in possessing five AMPK γ isoforms, compared to one in Drosophila [35] and three in humans [36]. Relatively little is known about the function of the five C. elegans AMPK γ isoforms but over-expression of a constitutively active form of aakg-2 (R81Q) can modestly increase worm lifespan [28]. To better understand the function of the five AMPK γ subunits we examined their respective sequences and expression levels.
In terms of sequence similarity to mammalian AMPK γ isoforms, aakg-1 is the most similar, followed by aakg-2 and aakg-3 (Figure 1A and Figures S1 and S2). The remaining isoforms, aakg-4 and aakg-5, are more divergent and form a distinct clade, suggesting that they resulted from a duplication of an ancestral atypical AMPK γ isoform. The sequence alignment of the C. elegans γ isoforms with the human AMPK γ1 (PRKAG1) isoform highlights conserved regions corresponding to the mammalian AMPK cystathione-β-synthase (CBS) domains. These domains mediate AMP/ADP and ATP binding to the enzyme allowing AMPK to respond to changes in the energy status of the cell. Interestingly, there is much less CBS domain conservation in aakg-4 and aakg-5 (Table S1) and some key residues required for interaction with AMP/ADP/ATP are missing: residues Arg-70 (human PRKAG1 numbering); His-150/Arg-151; and His-298/Arg-299 (Figure 1B) [37]. Their importance is highlighted by the fact that mutation of some of these residues in the heart-specific human AMPK γ2 isoform (R302Q, H383R, R384T, R531G/Q) have been identified in humans suffering from Wolff-Parkinson-White syndrome with cardiac hypertrophy and glycogen storage disease [38]–[41]. Studies have shown that these mutations lead to impaired activation by AMP/ADP [24], [42], [43]. Thus, AMPK complexes harboring any of these mutations no longer respond to stimuli that alter the AMP/ADP∶ATP ratio [44]. This suggests that C. elegans AMPK complexes containing AAKG-4 or AAKG-5 might render it AMP/ADP insensitive.
Fig. 1. Characterization of five AMPK γ subunits in C. elegans. 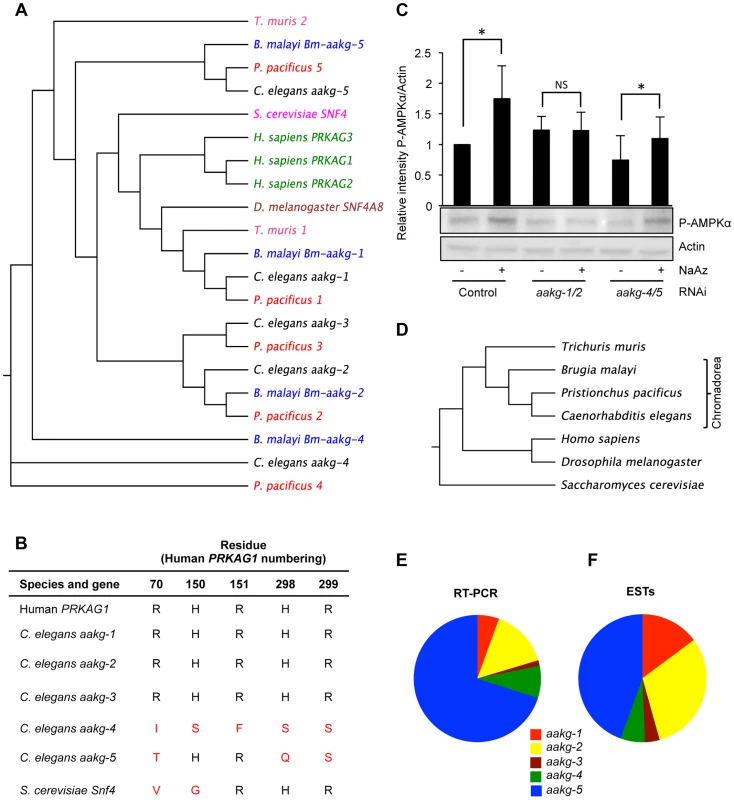
A) Phylogenetic relationship between AMPK γ orthologs in selected animal species. B) Critical AMP binding residues are missing in yeast Snf4 and C. elegans aakg-4 and aakg-5 (shown in red). An alignment is shown of the human AMPK γ1 (PRAKG1), C. elegans γ isoforms (aakg-1-5), and budding yeast (Saccharomyces cerevisiae) γ, Snf4. Sequences were aligned using the ClusterW multiple alignment application. More broadly, the AMP binding cystathione-β-synthase (CBS) domains [37] are less well conserved in aakg-4 and aakg-5 (Table S1). C) Evidence that Chromadorian AMPK (containing AAKG-4 or AAKG-5) does not respond to increased AMP levels. Western blot analysis shows levels of phosphorylated AMPKα in wild type (N2) worms in response to 1 mM sodium azide. The assumption was made that aakg-3 makes little contribution to overall AMPK activity as it is expressed at very low levels (Figures 1E and 1F). Quantification shows means from 4 independent trials. Error bars, standard error. *p<0.05 vs control for that RNAi treatment. One representative blot is shown. D) Phylogenetic relationship between animal species in 1A. E, F) Relative abundance of mRNA of C. elegans AMPK γ isoforms based on E) quantitative qRT-PCR and F) numbers of ESTs (WormBase version WS237). To probe whether AMPK containing AAKG-4 or AAKG-5 might be AMP/ADP insensitive, we compared worms subjected to simultaneous aakg-4/5(RNAi), in which one would expect all AMPK to be AMP/ADP sensitive, with aakg-1/2(RNAi) worms in which one would expect most AMPK to be AMP/ADP insensitive. To test AMPK activity, we measured levels of AMPKα phosphorylation, either in control, nutritionally replete animals, or in ATP-depleted animals (treated with the respiratory inhibitor sodium azide). As predicted, RNAi control animals treated with azide showed increased AMPK activity (Figure 1C). Our expectation was that this increase was mediated by AMPK containing AAKG-1 or AAKG-2. Consistent with this, azide increased AMPK activity in aakg-4/5(RNAi) worms but not aakg-1/2(RNAi) worms (Figure 1C). In nutritionally replete animals, presumably with lower AMP levels, one would expect that most AMPK activity was from enzyme containing AAKG-4 or AAKG-5. Consistent with this, we observed a slight decrease in AMPK activity in response to aakg-4/5(RNAi) but not aakg-1/2(RNAi) (Figure 1C). These results support the hypothesis that the activity of AMPK containing AAKG-4 or AAKG-5 is constitutively active, and not AMP dependent. However, to be certain of this, further studies would be needed, e.g. of the properties of reconstituted C. elegans AMPK in vitro using recombinant proteins.
Evolution of Chromadorean AMPK γ isoforms
To investigate the evolution of the five C. elegans aakg subunits, and the extent of the existence of atypical AAKG isoforms in other species, we identified and analyzed AAKG protein sequences from a range of nematode species. Orthologs of aakg-4 and aakg-5 were found in the nematodes Pristionchus pacificus (order: Rhabditida) and Brugia malayi (order: Spirurida) (Figures 1A, and 1D), demonstrating that these two isoforms are widespread at least among the large Chromadorea class of nematodes. However, orthologs of AAKG-4 and -5 are not clearly identifiable among either the more distantly related nematodes of the class Enoplea (e.g. Trichuris muris), or among platyhelminth species (Figures S1 and S3). This phylogenetic analysis suggests that duplications separating aakg-4 from aakg-5 and aakg-1 from aakg-2,-3 occurred before the common ancestor of the Rhabditida and Spirurida (Figure 1D). The final duplication separating aakg-2 from aakg-3 appears to have happened later, prior to the ancestor of the genera Caenorhabditis and Pristionchus as most of the Rhabditid species contain five aakg paralogs (Figure S2), but Spirurid nematodes such as B. malayi possess an ortholog of aakg-2 but not aakg-3 (Figure S1).
Notably, all orthologs of C. elegans aakg-4 and -5 also lack key AMP-binding residues (Tables S2 and S3), implying that the single ancestor of these two genes also lacked these residues. Some AAKG isoforms in the more distantly related Enoplea nematodes also lack key AMP binding residues (Figure 1A, Figure S1 and Table S3), raising the possibility that atypical AAKG isoforms might have existed in the common ancestor of the Enoplea and the Chromadorea.
In order to gain some indication of the relative importance of the five AMPK γ isoforms in C. elegans, we measured levels of their respective mRNAs in wild-type adult hermaphrodites. The most abundant mRNA was that of the atypical isoform aakg-5 (70.1% of total aakg mRNA) (Figure 1E), while the other atypical isoform aakg-4 accounted for 8.2% of aakg mRNA (Figure 1E). A count of the number of ESTs for each isoform listed on WormBase confirmed that aakg-5 is the most abundant mRNA (Figure 1F). Thus, the two atypical isoforms account for more than 3/4 of the total aakg mRNA. This implies that that much of the AMPK trimer in adult hermaphrodites contains either AAKG-4 or AAKG-5. For convenience, we suggest the term Chromadorean AMP kinase to describe this unusual form of this major metabolic regulator.
IIS regulation of atypical AMPK isoforms
Next we examined regulation by IIS and DAF-16 of genes encoding AMPK γ subunits. A previous transcript profile analysis showed 6.6-fold and 3.5-fold (both p<0.001) increases in aakg-4 and aakg-5 mRNA, respectively, in glp-4(bn2); daf-2(m577) relative to daf-16(mgDf50) glp-4; daf-2 young adult hermaphrodites [8]. Further data mining showed similar results for aakg-4 in an additional two profiles [15], [45] and for aakg-5 in one profile [15] (Table S4). For verification, we used qRT-PCR to compare mRNA levels in daf-2(e1370) and daf-16; daf-2 young adults. This confirmed that aakg-4 mRNA levels are elevated in daf-2 relative to daf-16; daf-2, but for aakg-5 this could not be confirmed (Figure 2A, 2B and Table S4).
Fig. 2. aakg-4 transcription is directly regulated by DAF-16/FoxO. 
A, B) aakg-4 but not aakg-5 mRNA levels are increased in daf-2 animals in a daf-16 dependent manner. ♦p<0.05, compared to N2, *p<0.05, compared to daf-2. Mean values from 4 trials. Error bars, standard deviation. C) DAF-16 binds to the promoter of aakg-4 but not aakg-5. aakg-4 peak 2 is positioned within 1 Kb of the transcriptional start site and contains two DAF-16 binding elements (Figure S5). One representative experiment is shown which contained 3 immuno-precipitation replicates from the same chromatin preparation (error bars show the standard deviation between them). The horizontal dotted line shows the % input from a negative control region (3′ of gene R11A5.4) that does not bind DAF-16 [22]. Statistical analysis of additional trials is presented in Table S5. A western blot showing the specificity of the DAF-16 antibody is shown in Figure S4. D) Confocal microscopy shows Paakg-4::gfp to be broadly expressed in C. elegans. Images show Paakg-4::gfp expression pattern in 1 day old hermaphrodites. (i) Whole worm expression pattern. (ii) Paakg-4::gfp is seen in tail sensory organs: phasmid sheath (PhaSh), socket cells (PhaSc) and neurons (PhaN), as well as in epithelial rectal cells (RectE), anal-depressor muscle (ADM), pre-anal ganglion rectal neurons (RectN), body wall muscles (BWM), posterior intestine (Int) and dorsal cord neuronal processes (DC). (iii) Paakg-4::gfp is expressed in vulval (VlvM), uterine muscles (UtM) and ventral cord processes (VC). (iv) Head expression mostly localizes to the ring ganglia (RingN) plus 6 pharyngeal neurons (PhxN). (v) It is also seen in amphid sensory organs including sheath (AmphSh), socket cells (AmphSc) and neurons (AmphN). Table S10 compares expression of Paakg-4::gfp with other AMPK subunits. E) Quantification of GFP fluorescence in worms expressing the Paakg-4::gfp reporter shows that fluorescence increased in daf-2 animals dependent on daf-16. The same was also true for a second set of strains generated from a different extrachromosomal array. Means from 3 independent trials shown. Error bars, standard error. Animals contained the wuEx256 transgene array. ♦p<0.01 compared to N2, *p<0.001 compared to daf-2. F) gfp mRNA was increased in daf-2 animals in a daf-16-dependent fashion. ♦p<0.05 compared to N2, *p<0.01 compared to daf-2. Means from 3 independent trials shown. Error bars, standard deviation. Prior to transgene quantification animals were maintained at 15°C and then shifted to 25°C for 24 hr at the L4 stage. Next we verified binding of DAF-16 to the promoters of aakg-4 and aakg-5, comparing daf-2(e1370) and daf-16; daf-2 adults using chromatin immunoprecipitation (ChIP) with a DAF-16-specific antibody (Santa Cruz) (Figure S4) and PCR. Our initial chromatin profiling experiment [22] had identified two DAF-16 binding peaks associated with aakg-4 and one with aakg-5 (Figure S5) and we examined each individually. We confirmed DAF-16 binding in the aakg-4 peak situated within 1 Kb of the transcriptional start site (peak 2) (Figure 2C and Table S5). However, we did not consistently observe DAF-16 binding at the first aakg-4 peak or that of aakg-5 (Figure 2C and Table S5). Thus, it seems that aakg-4 is a bona fide DAF-16 target gene and DAF-16 binding to the aakg-5 promoter is either non-existent or condition dependent.
To examine further the expression of aakg-4 and aakg-5, and their regulation by IIS and DAF-16, we created transgenic lines containing Paakg-4::gfp and Paakg-5::gfp transcriptional reporters. Paakg-4::gfp expression was observed in many tissues, particularly the intestine, body wall muscle, vulval muscles and several neurons in the head and tail including amphids and phasmids, respectively (Figure 2D). A broadly similar pattern of expression was seen in all larval stages and adults (data not shown). Paakg-5::gfp was also widely expressed (Figure S6A) and although expression of the two isoforms overlapped to an extent, there were also clear differences. For instance, aakg-5 was not expressed in amphid neurons nor aakg-4 in the pharynx (Figure 2D and Figure S6A).
To further assess the impact of IIS on aakg-4 and aakg-5 expression, transgene arrays were crossed into daf-2(e1370) and daf-16; daf-2 backgrounds and GFP fluorescence measured. Consistent with aakg-4 mRNA data, Paakg-4::gfp expression was increased 1.6 fold by daf-2, and this increase was daf-16 dependent (Figure 2E). No changes were detected in the distribution of expression (data not shown).
It was recently shown that mutation of daf-2 globally reduces protein levels by inhibiting protein synthesis [46], [47] raising the concern that GFP levels in daf-2; Paakg::gfp worms might be affected by non-specific effects of daf-2 on protein synthesis. We therefore introduced an additional control, using qRT-PCR to measure effects of IIS on gfp mRNA levels. In Paakg-4::gfp transgenic worms this revealed a greater, 6-fold induction in expression of gfp mRNA by daf-2 (Figure 2F). In contrast, no effect of IIS was detected on the level of Paakg-5::gfp GFP fluorescence, gfp mRNA or on distribution of expression (Figures S6B, S6C and data not shown).
We also examined the aakg-4 promoter sequence, which proved to contain two predicted DAF-16-binding elements (DBE) [48] (POSSUM scores 9.18 and 10.1), one of which is canonical and within 1 Kb of the translational start site (Figure S5). This is within the DAF-16-binding site previously identified by chromatin profiling using DamID and confirmed by ChIP PCR [22] (Figure 2C). One canonical DBE (POSSUM score 9.06) is also present in the same region of the C. briggsae aakg-4 promoter, suggesting conservation of regulation by IIS/FoxO, but not in C. remanei. DBEs were not detected in the aakg-5 promoter of any of these species. These findings strongly imply that DAF-16 binds to the promoter of aakg-4 and activates its expression.
aak-2, aakb-1 and IIS/DAF-16 regulation
We also checked our previous whole genome mRNA profiling study comparing glp-4; daf-2(m577) relative to daf-16; glp-4; daf-2 young adult hermaphrodites [8] for changes in expression of other AMPK subunits. aak-1 (α1 subunit) and aakb-2 (β2) mRNA levels were not different between the two strains, but aak-2 (α2) and aakb-1 (β1) were both elevated in daf-2 vs daf-16; daf-2, showing 2.28-fold and 7.51-fold (both p<0.001) higher levels, respectively, in daf-2. aakb-1 activation by DAF-16 was also implied by two other mRNA profile studies [15], [45], although not by a third study [49]. aakb-1 mRNA levels were also higher in daf-2(e1370) relative to N2 in individually profiled 19 day old worms [50] (Table S4). Changes in aak-2 mRNA levels were not seen in any of these other studies. For verification, we then used qRT-PCR to compare aak-2 and aakb-1 mRNA levels in 1 day old daf-2(e1370) and daf-16; daf-2 adults. This confirmed a statistically significant increase for aakb-1, but also suggested a possible increase in aak-2 mRNA levels (Figure S7 and Table S4). These results demonstrate that DAF-16 robustly increases aakb-1 expression, and suggest that it might also activate aak-2 expression. We also compared expression of Paak-2::gfp and Paakb-1::gfp transcriptional reporters [51] (Figures S8A and S9A) in wild type, daf-2 and daf-16; daf-2 backgrounds. For neither reporter was any effect of IIS on expression detected, either in terms of level (Figures S8B and S9B) or distribution (data not shown). However, in the case of Paak-2::gfp levels of gfp mRNA were higher in daf-2 relative to daf-16; daf-2 (Figures S8C), and possibly also for Paakb-1::gfp (Figures S9C). Thus, there appears to be a modest daf-16-dependent increase in both aak-2 and aakb-1 expression in daf-2 mutants, but one which is, perhaps, condition dependent and near the borderline of detectability.
The evidence of up-regulation of aakb-1 expression by DAF-16 suggests that this AMPK subunit might play a role in daf-2 mutant longevity (the Age phenotype). To test this the deletion mutation aakb-1(tm2658) was combined with daf-2(m577), and lifespan measured at 25°C. aakb-1 proved to shorten lifespan in daf-2(m577) and wild type worms to a similar degree (12% and 11%, respectively; mean of 4 trials) (Figure S10 and Table S6). To estimate whether the effect of aakb-1 differed between wild type and daf-2 backgrounds we used Cox proportional hazard analysis (CPHA) on combined data for all trials, but no difference was detected (p = 0.25). This suggests that activation of aakb-1 expression does not contribute to daf-2 Age.
aakg-4 contributes to the daf-2 Age phenotype
To assess whether aakg-4 contributes to daf-2 Age, we obtained the mutation aakg-4(tm5539). This deletes 405 bp from the gene, in a region encoding portions of both CBS3 and CBS4, and is therefore predicted to be a null. aakg-4 mutants appeared healthy and showed normal fertility (Figure S11). aakg-4 reduced lifespan in a daf-2(m577) background (−24.4%, combined data for 4 trials), but not in a daf-2(+) background (Figure 3A, Table 1 and Table S7). Statistical analysis confirmed that aakg-4 effects on lifespan were greater in a daf-2 background (CPHA p<0.0001), implying that aakg-4 contributes to daf-2 Age. To confirm this, we abrogated aakg-4 expression using RNAi, and again observed a greater reduction in lifespan in daf-2 than in wild-type worms (9 trials, CPHA p = 0.0024) (Figure 3B, Table 1 and Table S8).
Fig. 3. aakg-4 mutation or aakg-4(RNAi) shorten the lifespan of daf-2(m577) animals. 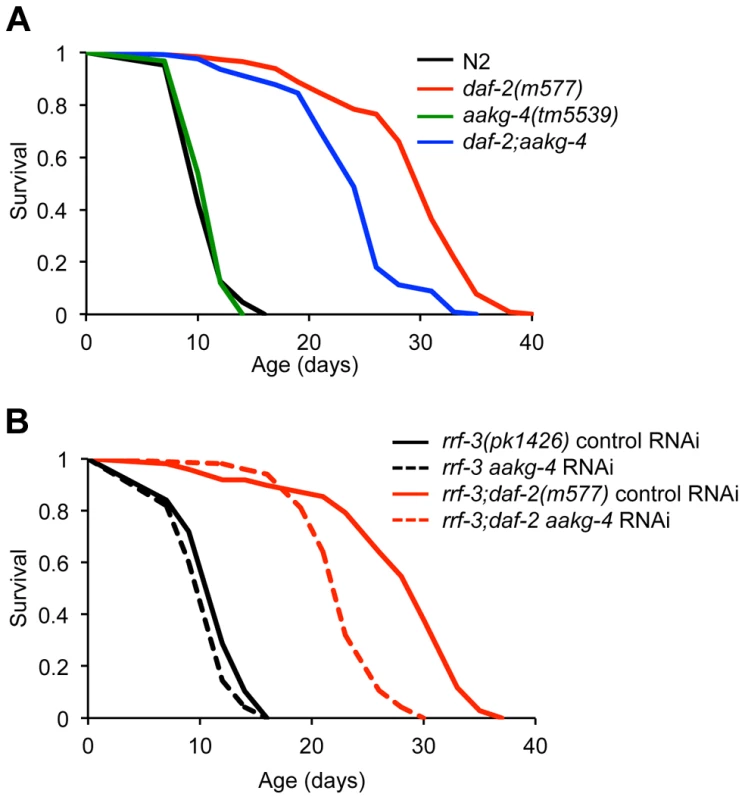
A and B) One representative experiment is shown in each case, corresponding to A) Trial 1 in Table S7 and B) Trial 5 in Table S8. Lifespan measurements carried out at 25°C. Tab. 1. aakg-4(tm5539) and aakg-4 RNAi shortens daf-2 lifespan. 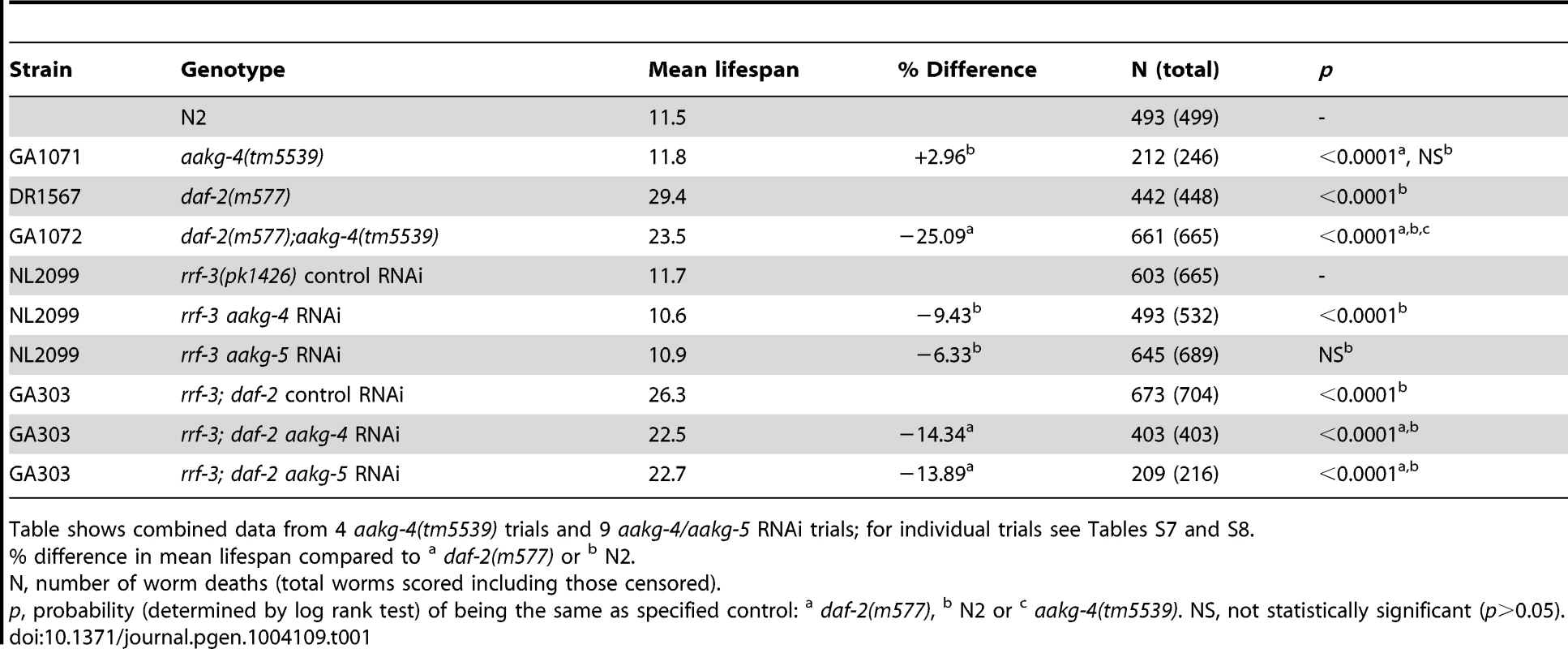
Table shows combined data from 4 aakg-4(tm5539) trials and 9 aakg-4/aakg-5 RNAi trials; for individual trials see Tables S7 and S8. To test whether aakg-5 also contributed to daf-2 Age we used aakg-5(RNAi). Like aakg-4(RNAi), this too markedly reduced lifespan in daf-2 but not N2 (4 out of 9 trials, CPHA, p = 0.0005) (Figure S12 and Table S8). We also tested effects of RNAi of aakg-1, aakg-2 and aakg-3 but none consistently suppressed daf-2 Age (Table S9).
aakg-4 and aakg-5 form a distinct clade within the C. elegans AMPK γ subunits (Figure 1) and both contribute to daf-2 Age. This raises the possibility is that these two genes are partially redundant in function. To probe this we tested whether aakg-5(RNAi) could further reduce daf-2 Age in daf-2;aakg-4 worms, but it could not (Figure S12 and Table S8), suggesting that aakg-4 and aakg-5 are not functionally redundant.
Taken together, these findings imply that in daf-2 mutants, activation of DAF-16 leads to increased aakg-4 expression, which in turn contributes to the daf-2 Age phenotype. aakg-5 may also be important for daf-2 Age but is not directly regulated by DAF-16. The absence of key residues important for interacting with AMP in AAKG-4 and our data implying that AAKG-4 and AAKG-5 activity may be unresponsive to AMP/ADP (Figures 1B and 1C) suggests the possibility that daf-2 mutants have elevated expression of an AMP-independent AMPK complex (Chromadorean AMPK).
Evidence of a positive feedback loop involving DAF-16 and AMPK
We have identified daf-16-dependent up-regulation of expression of aakg-4, and possibly also aak-2 and aakb-1, in daf-2 mutants. It was shown previously that AMPK directly phosphorylates and activates DAF-16 [28]. Taken together, this suggests the presence of a positive feedback loop, where DAF-16 increases AMPK activity by promoting expression of genes encoding AMPK subunits, and this in turn activates DAF-16 activity. If such a positive feedback loop exists, then aakg-4 should promote DAF-16 activity. To test this we used expression of DAF-16 target genes as a readout of DAF-16 activity.
Expression of the well-characterized DAF-16 target gene sod-3 is induced by reduced IIS and has also been shown to be regulated by aak-2 [28]. sod-3 expression can be induced by shifting rrf-3; daf-2(m577) worms from 15°C to 25°C (the non-permissive temperature for this temperature-sensitive daf-2 allele) (Figure S13). The rrf-3 mutation sensitizes the animals to RNAi [52]. A temperature shift does not induce sod-3 expression in either rrf-3 or rrf-3; daf-16; daf-2 animals (Figure S13). Notably, if aakg-4(RNAi) was initiated at the same time as the temperature shift there was a significant reduction in sod-3 expression after 24 hr in animals treated with aakg-4(RNAi) (Figure 4A and 4B). Moreover, this reduction was transient, and absent at later time points (48 hr or 72 hr).
Fig. 4. aakg-4 RNAi alters dynamics of DAF-16 target gene expression. 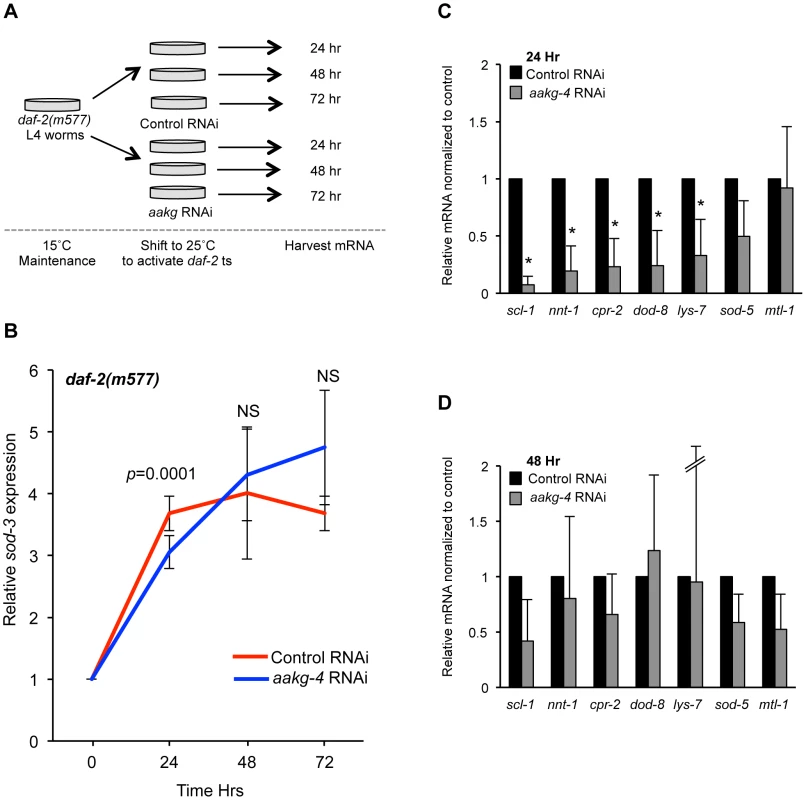
A) Experimental design: L4 daf-2(m577) animals were treated with either control (L4440) or aakg-4 RNAi and shifted to 25°C prior to RNA extraction. B) The induction of sod-3 mRNA expression is reduced after 24 hr by aakg-4(RNAi) but this reduction was not seen after 48 or 72 hr. The level of sod-3 at 0 hr is that of adults that have not been shifted to 25°C (i.e. maintained at 15°C). C, D) The expression of other DAF-16 target genes was also reduced after 24 hr by aakg-4(RNAi) but this reduction was not seen after 48 hr. Error bars, standard deviation. *p<0.05 compared to daf-2 animals fed control RNAi. We then looked at the effect of aakg-4(RNAi) in rrf-3; daf-2(m577) worms on expression of seven other known DAF-16 target genes [53], [54] 24 hr or 48 hr after temperature shift/RNAi and found that 5 out of 7 also showed reduced expression compared with control RNAi after 24 hr but, again, not 48 hr (Figure 4C and 4D). Next, we tested the effects of aakg-1, aakg-2 and aakg-5 RNAi on sod-3 expression but could detect no significant effect (Figure S14), implying that DAF-16 activity is specifically dependent on aakg-4. Finally, we tested whether aakg(RNAi) in daf-2 mutant animals reduced nuclear localization of DAF-16::GFP, but only marginal effects were detected (Figure S15). This is consistent with a previous report that phosphorylation of DAF-16 by AMPK does not increase nuclear localization [28].
These results imply that AAKG-4 can increase DAF-16 activity, consistent with the existence of a positive feedback loop involving Chromadorean AMPK (Figure 5A). That reduced induction of DAF-16 target gene expression is seen after 24 hr but not 48 hr suggests that the function of the positive feedback loop is to accelerate DAF-16 target gene induction, rather than to alter their endpoint expression levels (Figure 5B).
Fig. 5. DAF-16 and AMPK form a positive feedback loop. 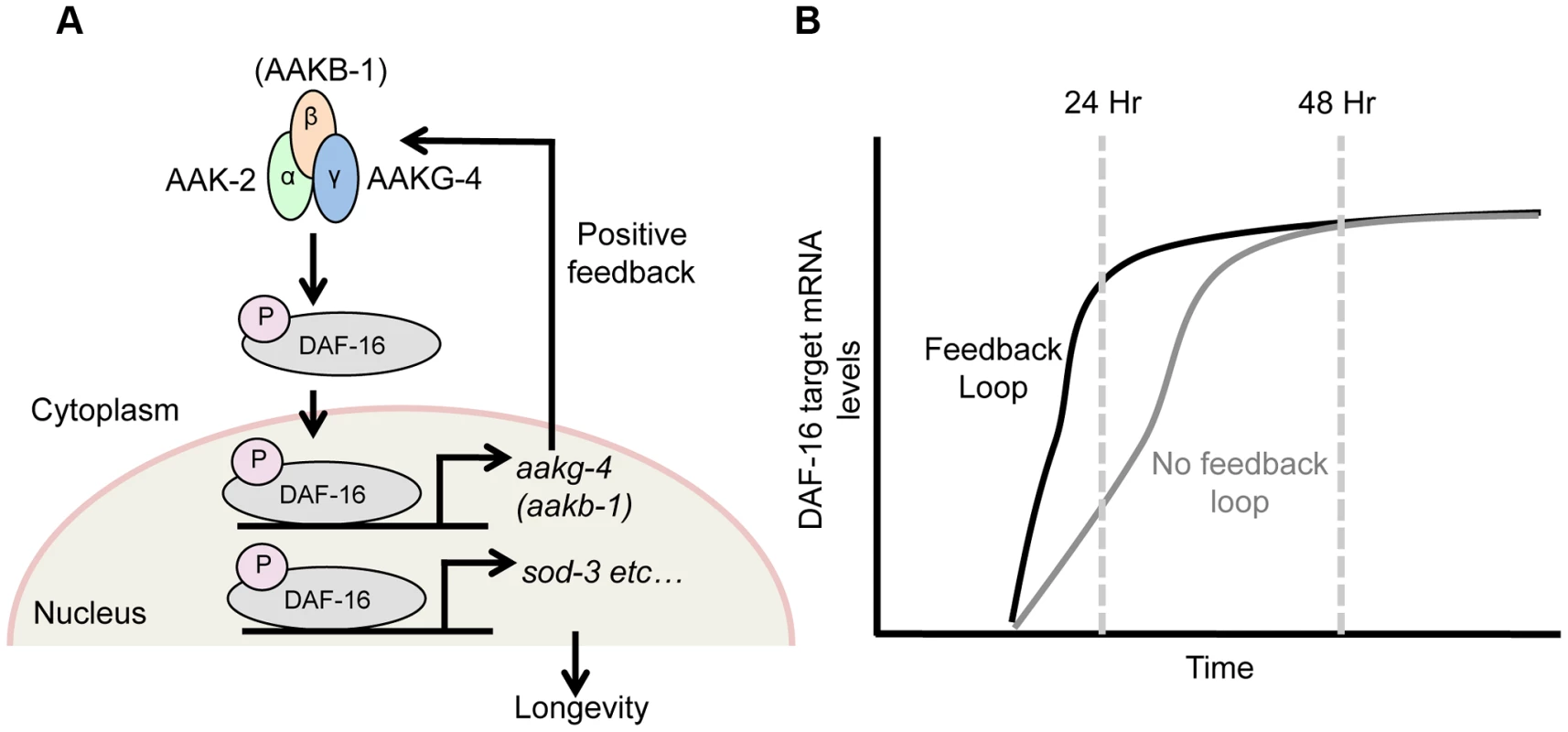
Schematic model suggested by combining findings presented here, and elsewhere [28]. A) AMPK phospho-activates DAF-16 [28] and directly controls the transcription of 2 AMPK subunits, aakg-4 and possibly aakb-1 (this study). This in turn further increases AMPK levels, which further activate DAF-16 and target gene expression, and so on, creating a positive feedback loop. This broadly affects expression of DAF-16 target genes (e.g. sod-3), including those that determine aging rate (though note that sod-3 itself does not affect aging [77], [78]. B) This feedback loop appears to speed up DAF-16-mediated induction of gene expression in response to reduced IIS, rather than altering final expression level. daf-16 does not activate its own expression
Chromatin profiling data [22] implies that DAF-16 binds strongly to its own promoter, suggesting the possible action of DAF-16 in a second positive feedback loop involving activation of its own transcription. To test whether daf-16 activates its own transcription, we compared daf-16 mRNA levels in daf-2(e1370) mutants in the presence of the daf-16(m26) point mutation. No difference was detected, implying that daf-16 does not activate its own expression (Figure S16A). This conclusion was further supported by studies of Pdaf-16a::gfp reporters (Figures S16B–F).
Discussion
In this study, we used existing mRNA and chromatin profiling data as a starting point for careful testing of predicted gene regulatory relationships involving DAF-16. This has revealed the presence of a positive feedback loop involving DAF-16 and AMPK, where DAF-16 activation of an atypical AMPK γ subunit plays a key role.
aakg-4 as an effector of DAF-16/FoxO
Our results show that DAF-16 acts directly on the promoter of aakg-4 to activate its transcription. The observation that daf-2 mutants have elevated aakg-4 levels suggested that this may be a potential mechanism that mediates the lifespan extension seen in these worms. This is confirmed by our experiments showing that mutation or RNAi of aakg-4 shortened the lifespan of daf-2 mutants. Taken together, this implies that AMPK complexes containing the AAKG-4 γ subunit are functionally important downstream of IIS. One possibility is that when IIS is reduced, AAKG-4 replaces other AMPK γ isoforms thereby increasing its capacity to inhibit aging.
Microarray analysis suggests that expression of aak-2 and aakb-1 is also promoted by DAF-16, i.e. that there is activation of α, β and γ AMPK subunits. Further analysis of regulation of these 2 genes, e.g. using qRT-PCR and GFP reporter analysis, confirmed this for aakb-1 (Figure S7B and Figure S9C). However, such activation appears to contribute little to daf-2 Age (Figure S10). By contrast, aak-2 is either not regulated by DAF-16, or subject to regulation that is relatively weak and at the limits of detectability and/or condition dependent. This latter result contrasts with the absolute requirement of aak-2 for daf-2 longevity and suggests that although this catalytic subunit is essential for AMPK activity [25], [26], IIS regulates AMPK via the γ subunits.
In summary, the α subunit AAK-2 is important for daf-2 Age [25]–[27] but aak-2 is not robustly up-regulated by DAF-16; the β subunit AAKB-1 is not important for daf-2 Age, but is up-regulated by DAF-16; and the γ subunit AAKG-4 is both important for daf-2 Age and up-regulated directly by DAF-16.
Atypical AMPK γ isoforms lack AMP binding sequences
Given the high degree of protein sequence conservation, it seems highly likely that worm AMPK, like mammalian AMPK, is regulated by AMP/ADP. However, the observation that C. elegans AAKG-4 and AAKG-5 (Figure 1B, Table S2) lack key residues involved in interacting with AMP/ADP in mammals [37] suggests the possibility that AMPK trimers formed with AAKG-4 or AAKG-5 are active in the absence of AMP. Moreover, like AAKG-4 and AAKG-5, budding yeast Snf4 lacks some of the key residues that are important in binding AMP in the human γ1 isoform (e.g. human γ1 R70 and H150) (Figure 1B), and the yeast enzyme is not regulated by AMP. Consistent with the hypothesis that Chromadorean AMPK is AMP independent, we observed that a stress known to increase AMP∶ATP ratio [25] could increase levels of active AMPK in aakg-4/-5(RNAi) worms, predicted to contain only AMP-dependent AMPK, but not in aakg-1/2(RNAi) worms, predicted to contain only Chromadorean AMPK (Figure 1C). It will be interesting to examine further the enzymology of Chromadorean AMPK, e.g. using in vitro analysis of recombinant C. elegans proteins.
One further influence of mutating the key nucleotide-interacting residues is that they also cause an increase in the phosphorylation and basal activity of AMPK [39]–[41], [55]. This means that these mutant complexes are effectively constitutively active. Thus, C. elegans AMPK complexes containing the AAKG-4 or AAKG-5 γ isoforms may not only be unable to respond to changes in the energy status of the cell but they may also have a higher basal activity compared to AAKG-1, AAKG -2 or AAKG-3-containing AMPK, i.e. show constitutive activity. Consistent with this, worms treated with aakg-4/-5(RNAi) but not aakg-2/-3(RNAi) did show slightly reduced pAMPKα levels in unstressed, fully fed worms (Figure 1C). Both isoforms are present in nematodes throughout the large Chromadorea clade, implying an important and non-redundant function for these two atypical AMPK γ isoforms.
Up-regulation of AAKG-4 in daf-2 and the abundant expression of AAKG-5 suggest an interesting speculation: that abundant AMP-independent Chromadorean AMPK in daf-2 mutants permits a high level of AMPK activity in nutritionally replete animals, where AMP/ATP ratios are low. Possibly this AMPK equivalent of having your cake and eating it contributes to the unusual longevity of C. elegans IIS mutants. Consistent with this, daf-2 Age is partially aakg-4 dependent. It will be interesting to examine levels of AMPK activity in daf-2 mutants in future studies.
The presence of an AMP-independent AMPK suggests that fitness among Chromadorean nematodes is enhanced by high levels of AMPK activity under nutritionally replete conditions. In C. elegans this might aid entry into the dauer state while food levels are still relatively high. Moreover, AMPK activity might help maintain diapause in developmentally arrested parasitic larvae that exist in a nutritionally replete state, e.g. in microfilariae of filarial worms such as Onchocerca volvulus, or second stage larvae of ascarids such as Toxocara canis.
Tissue-specific expression of AMPK subunits
The presence of two α, two β and five γ AMPK subunits in C. elegans implies considerable complexity in AMPK function in this organism. Presumably AMPK subunits that are expressed in the same tissues combine to make active trimers. We have used GFP transcriptional reporters to characterize the expression patterns of aakg-4, aakg-5 and aakb-1 and to verify that of aak-2 (Table S10). Both daf-16 and its direct target aakg-4 show a broad and similar distribution of expression, including several tissues critical for regulating adult lifespan, e.g. the amphid neurons and intestine [56], [57]. In adults the expression of aak-2 and aakb-1 appears to be more restricted than that of aakg-4, but all 3 are expressed in the intestine (Table S10). If the C. elegans γ subunits do have distinct tissue specific roles this would mimic the situation in mammals where e.g. human AMPK γ2 (PRKAG2) is particularly important in cardiac function [58].
It seems likely that there is at least some functional redundancy between subunit isoforms. In dauer larvae aak-2 can act with either aakb-1 or aakb-2 and both β subunits need to be removed to fully abrogate the long lifespan of dauer larvae [59]. Such redundancy may explain why aakb-1 mutation alone does not affect daf-2 adult lifespan (Figure S10).
Reduced IIS generates a DAF-16-AMPK positive feedback loop
AMPK activates DAF-16 [28] and DAF-16 increases expression of several AMPK subunits, suggesting the presence of a positive feedback loop (Figure 5A). Gene regulatory feedback loops have previously been well characterized in unicellular organisms such as S. pombe and E. coli [60]. Supporting the existence of such a loop, aakg-4(RNAi) delays induction of the DAF-16 target gene sod-3 (Figure 4B). Such a positive feedback loop could, in principle, serve any of several roles: to increase the stability of the activated status of AMPK and DAF-16; to accelerate the induction of DAF-16 target genes; or to increase the final level of expression of DAF-16 target genes after induction. Our findings provide support for the second but not the third scenario, since blunting of DAF-16 target gene induction by aakg-4(RNAi) was transient i.e. detected after 24 hr but not 48 hr (Figure 5B). Thus, the presence of the positive feedback loop may facilitate rapid metabolic response to a changing nutrient environment. For example, it might reduce response time for larvae to activate the dauer entry program in a deteriorating environment, thereby increasing fitness.
Possible evolutionary conservation of FoxO-AMPK interactions
Many aspects of the IIS-regulated signaling network are conserved in higher organisms, and this may extend to AMPK. In Drosophila, the AMPK γ subunit SNF4Aγ is up-regulated in flies expressing a dominant negative form of the insulin receptor, although binding of dFOXO to the corresponding promoter is not detected in whole flies [61]. We also explored the possibility that similar relationships exist in mammals but data mining did not reveal any FoxO-dependent changes in AMPK γ subunits in mice [34], [62]–[65]. However, this does not rule out FoxO regulation AMPK gene transcription as these data sets only examine a limited number of tissues. Notably, a recent study showed that human FoxO3 and FoxO4 directly activate LKB1 expression in vitro [66]. Given that LKB1 phosphorylation activates AMPK, this suggests that FoxO-AMPK positive feedback loops could be operational in mammals.
Conclusions
In this study we have carefully mapped out part of the gene regulatory network centered on DAF-16. This has revealed a positive feedback loop involving DAF-16 and AMPK, involving an atypical AMPK γ subunit which may act independently of AMP. Thus, AMPK acts an effector as well as an activator of DAF-16 in its promotion of longevity.
Materials and Methods
Strains and strain construction
Worms were maintained at 20°C unless otherwise indicated. The following strains were used: N2, NL2099 rrf-3(pk1426), GA303 rrf-3(pk1426); daf-2(m577), GA1073 rrf-3(pk1426); daf-16(mgDf50); daf-2(m577), GA1061 aakb-1(tm2658) (crossed 3 times into recently thawed cultivar of CGC male N2 stock), GA1069 daf-2(m577); aakb-1(tm2658), GA1071 aakg-4(tm5539) (crossed 3 times into a recently thawed cultivar of CGC male N2 stock), GA1072 daf-2(m577); aakg-4(tm5539), GA1052 sEx10615 [Paak-2::gfp], GA1048 daf-2(e1370) sEx10615 [Paak-2::gfp], GA1050 daf-16(mgDf50); daf-2(e1370) sEx10615 [Paak-2::gfp], GA1051 sEx11830 [Paakb-1::gfp], GA1047 daf-2(e1370) sEx11830 [Paakb-1::gfp], GA1049 daf-16(mgDf50); daf-2(e1370) sEx11830 [Paakb-1::gfp], GA1403 wuEx256 [Paakg-4::gfp], GA1404 daf-2(e1370) wuEx256 [Paakg-4::gfp], GA1405 daf-16(mgDf50); daf-2(e1370) wuEx256 [Paakg-4::gfp], GA1410 wuEx257 [Paakg-4::gfp], GA1411 daf-2(e1370) wuEx257 [Paakg-4::gfp], GA1412 daf-16(mgDf50); daf-2(e1370) wuEx257 [Paakg-4::GFP], GA1400 wuEx251 [Paakg-5::gfp], GA1401 daf-2(e1370) wuEx251 [Paakg-5::gfp], GA1402 daf-16(mgDf50); daf-2(e1370) wuEx251 [Paakg-5::gfp], GA1413 wuEx258 [Paakg-5::gfp], GA1414 daf-2(e1370) wuEx258 [Paakg-5::gfp], GA1415 wuEx258 daf-16(mgDf50); daf-2(e1370) [Paakg-5::gfp], GA1431 wuEx261 [Pdaf-16a(i)::gfp], GA1433 daf-2(e1370) wuEx261 [Pdaf-16a(i)::gfp], GA1435 daf-16(mgDf50); daf-2(e1370) wuEx261 [Pdaf-16a(i)::gfp], GA1419 wuEx260 [Pdaf-16a(i)::gfp], GA1420 daf-2(e1370) wuEx260 [Pdaf-16a(i)::gfp], GA1421 daf-16(mgDf50); daf-2(e1370) wuEx260 [Pdaf-16a(i)::gfp], GA1437 wuEx263 [Pdaf-16a(ii)::gfp], GA1440 wuEx263 [Pdaf-16a(ii)::gfp], GA1442 daf-2(e1370) wuEx263 [Pdaf-16a(ii)::gfp], GA1438 daf-16(mgDf50); daf-2(e1370) wuEx264 [Pdaf-16a(ii)::gfp], GA1441 daf-2(e1370) wuEx264 [Pdaf-16a(ii)::gfp], GA1443 daf-16(mgDf50); daf-2(e1370) wuEx264 [Pdaf-16a(ii)::gfp].
Multiple mutants were created using standard methodologies and the presence of genomic deletions was tested either by screening for characteristic phenotypes or via PCR. In the case of PCR, genotyping, of aakb-1(tm2658), was carried out by lysis of parent animals using proteinase K (Sigma) and subsequent PCR using the following primers. F:gcaatgtgattaaaagttatggg, In:cgatgataatcagcaaaagacg, R:cagggtatactacacatgtacc. Primers for aakg-4(tm5539) were F: agtctctgacacgccgagtt, In: gcacaggcttttagacttcg, R: cgaagtctaaaagcctgtgc.
The promoter::gfp transgenes of daf-16a were created using Gateway cloning (Invitrogen). The primer sequences that define the region used for Pdaf-16a(i) were: F:cctccatcaacaagagcgttc and R:gcaaatgcaacacggagaaaacg; and for the Pdaf-16a(ii): F:catgtctcgtgtgcctctcctttcca and R:taacgtcttcgggaatttcagccaaag. The sequences used to define the daf-16 3′UTR were F:attctcttcattttgtttcccctggtgttgttcg and R:catcatcatacagtcgcaaatatatttggggg. The promoter::gfp fusions for aakg-4 and aakg-5 were made as previously described [67] using the PCR to fuse the promoter of interest to GFP and the unc-54 3′UTR. The sequences that define the promoter used for Paakg-4 were F:aaaagacacactcaatttccataaatatat and R:tgataaatgatgatattttgaggttgtgaaa and for Paakg-5 F:agatcaaaggcttattgtgcatttctattt and R:tctggaaaataaaagcattaaagtgaaaaat. In each case the transgenic strains were created by microinjection of the plasmid (Pdaf-16) or PCR product (Paakg-4 and 5).
aakg-1, aakg-2 and aakg-4 RNAi clones were obtained from the Ahringer library and those for aakg-3 and aakg-5 were constructed de novo. The primer sequences defining the regions used for aakg-3 were: F:attaaatgcggccgctaaggataggagttcgtaagtatcaattag and R:attaaagacgtcgtaatatcctacgatgtgtcaatatgtacg; and for aakg-5 F:attaaagtcgacacacaccagcatccgaacaacgtcgtaact and R:attaaagagctcctaaagattcttcagcttgctgcagacatt. The primers contain NotI, PstI, SalI and SacI restriction sites, respectively, allowing the resulting PCR fragments to be cloned into pL4440.
Alteration of C. elegans AMP levels and western blot analysis
Adult worms were allowed to lay eggs for 12 hr on plates seeded with E. coli HT115 transformed with RNAi plasmids. The resulting progeny were themselves allowed to develop to young adults at 20°C before harvesting in M9 buffer. Worms were then placed either on control plates or plates containing 1 mM sodium azide for 2 hr at 20°C [25]. Sodium azide plates were made by pipetting a stock solution directly onto NGM plates seeded with E. coli OP50 72 hr before use.
Following treatment worms were washed off plates using M9, collected by settling and resuspended in 100 µl Phosphosafe extraction reagent (Millipore) supplemented with a protease inhibitor cocktail (Roche). Protein concentration was determined using BCA (Pierce) and 40 µg protein was separated by SDS-gel electrophoresis. Proteins were subsequently transferred to nitrocellulose membrane and probed using a 1∶1000 dilution of phospho-AMPKα (Thr172) antibody (Cell Signaling). β-actin antibody (Santa Cruz) was used as a loading control. Imaging and quantification of bands was carried out using the ImageQuant LAS4000 imaging system and software (GE Healthcare).
Microscopy
Worms were raised at 15°C, picked at L4 stage, and shifted to 25°C for 24 hr to increase transgenic extrachromosomal array expression and to induce the daf-2 phenotype in the daf-2(m577) 1 day old adults. For each slide, 30–40 worms were mounted in M9+0.2% levamisole on a 2% agarose pad and imaged within the following 30 min. Quantification of GFP expression from transgenic strains was carried out using a Leica DMRXA2 microscope using a GFP filter cube (excitation: 470/40 nm; emission: 525/50 nm), an Orca C10600 digital camera (Hamamatsu) and Volocity image analysis software (Improvision). Low level intestinal autofluorescence was detected in the GFP range in all our strains. For the highly expressed Paakg-4::gfp and Paakg-5::gfp reporters this did not interfere with our analysis but for Paak-2::gfp and Paakb-1::gfp which required longer exposure times we corrected for autofluorescence using GFP measurements from non-transgenic animals or by measuring GFP levels only in the head where autofluorescence is minimal.
Confocal images were acquired using the Zen2009 software driving a LSM-710 confocal station with an inverted Axio Observer microscope (Carl Zeiss Microscopy, Germany). Whole worm images were generated from the Z-projection of 12–18 XY-planes acquired every 3 µm through a 10× Plan-Apochromat 0.45 NA dry objective (Carl Zeiss Microscopy, Germany). High-resolution images were generated from the Z-projection of 45–60 XY-planes acquired every 0.75 µm through a 40× Plan-Apochromat 1.3 NA oil objective (Carl Zeiss Microscopy, Germany). Z-projections were created using the freeware LSM5 image browser (Carl Zeiss Microscopy, Germany). Tiff images were then exported to Photoshop CS4 (Adobe), background was subtracted and contrast adjusted for optimal display.
Quantitative RT-PCR
RNA was isolated from adult worms after transfer of the worms to an unseeded NGM plate to remove E. coli. 50–100 worms were used for each assay. RNA was extracted using Trizol (Sigma) and cDNA synthesized using SuperScript II reverse transcriptase with oligo dT (Invitrogen). qRT-PCR was carried out using Fast SYBR Green Master Mix (Applied Biosystems) and the 7900 HT Fast PCR system (Applied Biosystems). Normalization of transcript quantity was carried out using the geometric mean of three stably expressed reference genes Y45F10D.4, pmp-3, and cdc-42 in order to control for cDNA input, as previously described [68]. Primer sequences for DAF-16 target genes are as previously described [53], [54]. Primer sequences for ChIP PCR are as follows: M01H9.3 F:gcatgtgaccacgtgaattt and R:aacccctctaacactatcca; aakg-4 (peak 1) F:tctcacactcccttcccact and R:gccgtcgtcacaaatactga; aakg-4 (peak 2) F: aaaaagcgagcaaagcaaaa R: ttccacatttgtcgcacttc; and aakg-5 (peak 1 primers 1) F:aggacggactgttttgttgc and R:gctcctcgttttcaatgctt. The relative levels of aakg isoform mRNA were determined using custom made primers designed and synthesized by Primerdesign. These were optimized to amplify each of the 5 subunits at the same rate, thus allowing relative quantification without the need for an internal standard.
Chromatin immunoprecipitation
The protocol for chromatin immunoprecipitation was adapted from a previous report [69]. C. elegans cultures were grown on plates on OP50 at 20°C. The worms were collected, washed 4× in PBS buffer, frozen in liquid nitrogen and pulverized with a mortar and pestle. The resulting powder was then re-suspended in 8 ml PBS containing 1% formaldehyde and incubated for 20 min with gentle mixing at room temperature. Crosslinking was stopped by addition of 400 µl 2.5 M glycine solution and 20 min further incubation at room temperature. After 4 washes in PBS containing EDTA-free protease inhibitor tablets (Complete, Roche), samples were flash frozen and stored at −80°C. After thawing, 2 ml of HLB buffer [50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% (wt/vol) sodium deoxycholate, 1% (vol/vol) Triton X-100, 0.1% (wt/vol) SDS and 1× Complete protease inhibitor] was added and sonication was carried out at 70% intensity for 7 bursts of 30 s in a Vibracell sonicator (Sonics). Protein quantity was estimated by BCA (Pierce) and 2 mg were diluted to 500 µl in HLB buffer.
3×50 µl aliquots were removed at this point. DNA isolated from these samples was subsequently used for input controls. Samples were pre-cleared for 1 hr in prewashed salmon sperm DNA/protein-A agarose beads (Millipore) and then incubated overnight with 10 µl of anti-DAF-16 Ab (Santa Cruz). Samples were then incubated with prewashed salmon sperm DNA/protein-A agarose beads for 2 hr. The beads were then washed twice in WB1 [50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% (wt/vol) sodium deoxycholate, 1% (vol/vol) Triton X-100, 0.1% (wt/vol) SDS and 1× Complete protease inhibitor], twice in WB2 [50 mM HEPES-KOH, pH 7.5, 1 M NaCl, 1 mM EDTA, 1% (wt/vol) sodium deoxycholate, 1% (vol/vol) Triton X-100, 0.1% (wt/vol) SDS and 1× Complete protease inhibitor] and once in WB3 [50 mM Tris-HCl, pH 8, 0.25 mM LiCl, 1 mM EDTA, 0.5% (vol/vol) NP-40 and 0.5% (wt/vol) sodium deoxycholate]. Crosslinking was reversed by addition of proteinase K solution [50 mM Tris-HCl, pH 8, 25 mM EDTA, 1.25% (wt/vol) SDS, 160 µg/ml proteinase K (Qiagen)] and incubation for 2 hr at 45°C and overnight at 65°C. DNA was isolated by applying solution to Qiagen PCR purification columns.
Lifespan analysis
Prior to experiments animals were maintained at the permissive temperature and grown for at least one generation in the presence of food to assure full viability. Lifespan assays were performed essentially as described [70]. Survival plots and p values (log rank test) were determined using JMP software, version 7. Cox proportional hazard p values were determined in R from combined experiments using mixed effects models (Christensen, R. H. B., 2012 http://www.cran.r-project.org/package=ordinal). For lifespans using RNAi worms were grown on bacteria expressing aakg RNAi clones from the L4 stage. Empty pL4440 vector was used as a control.
Bioinformatics
For the majority of species orthologs of the C. elegans genes aakg-1-5 were identified by local alignment searches of the five protein sequences to the gene models in WormBase. Since not all genomes were available in WormBase orthologs for the parasitic helminths (B. xylophilus, E. granulosus, E. multilocularis, T. solium, H. microstoma, S. mansoni and S. japonicum) were derived from GeneDB. Orthologs for G. pallida, H. contortus, O. volvulus, S. ratti and T. muris were derived using data available on the Sanger Institute Scientific Resources website. Since there are no gene model predictions available for O. volvulus and T. muris, we identified contigs that aligned closely to the C. elegans gene sequences of interest using a “protein versus translated DNA” Blast search. We then predicted genes within these contigs using the Eukaryotic GeneMark.hmm [71]. Of the several genes predicted, we kept for further analysis the ones with protein sequences that showed high similarity to the C. elegans aakg genes. Finally, orthologs of P. pacificus were derived from the gene predictions available from www.pristionchus.org.
Multiple sequence alignments of the protein sequences were done using MUSCLE [72]. Trees were constructed with ClustalW2 Phylogeny [73] using the Neighbour-joining method. The species tree was constructed by integrating the taxonomy information available through WormBase, Uniprot Taxonomy [74] and PhyloExplorer [75] (based on data from TreeBASE). All trees were visualized and annotated using EvolView [76].
Supporting Information
Zdroje
1. FriedmanDB, JohnsonTE (1988) Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J Gerontol 43: B102–109.
2. KenyonC, ChangJ, GenschE, RudnerA, TabtiangR (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366 : 461–464.
3. KimuraKD, TissenbaumHA, LiuY, RuvkunG (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 : 942–946.
4. KenyonCJ (2010) The genetics of ageing. Nature 464 : 504–512.
5. GemsD, PartridgeL (2013) Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol 75 : 621–644.
6. LinK, HsinH, LibinaN, KenyonC (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28 : 139–145.
7. OggS, ParadisS, GottliebS, PattersonGI, LeeL, et al. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 : 994–999.
8. McElweeJJ, SchusterE, BlancE, ThomasJH, GemsD (2004) Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem 279 : 44533–44543.
9. MurphyCT, McCarrollSA, BargmannCI, FraserA, KamathRS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 : 277–283.
10. MurphyCT, LeeSJ, KenyonC (2007) Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A 104 : 19046–19050.
11. Halaschek-WienerJ, KhattraJS, McKayS, PouzyrevA, StottJM, et al. (2005) Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res 15 : 603–615.
12. OhSW, MukhopadhyayA, DixitBL, RahaT, GreenMR, et al. (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38 : 251–257.
13. DongMQ, VenableJD, AuN, XuT, ParkSK, et al. (2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317 : 660–663.
14. LeeSJ, MurphyCT, KenyonC (2009) Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab 10 : 379–391.
15. McElweeJ, BubbK, ThomasJH (2003) Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2 : 111–121.
16. VilchezD, MorantteI, LiuZ, DouglasPM, MerkwirthC, et al. (2012) RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489 : 263–268.
17. GemsD, McElweeJJ (2005) Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech Ageing Dev 126 : 381–387.
18. HsuAL, MurphyCT, KenyonC (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 : 1142–1145.
19. CohenE, BieschkeJ, PerciavalleRM, KellyJW, DillinA (2006) Opposing activities protect against age-onset proteotoxicity. Science 313 : 1604–1610.
20. Ben-ZviA, MillerEA, MorimotoRI (2009) Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A 106 : 14914–14919.
21. VanfleterenJR (1993) Oxidative stress and ageing in Caenorhabditis elegans. Biochem J 292(Pt 2): 605–608.
22. SchusterE, McElweeJJ, TulletJM, DoonanR, MatthijssensF, et al. (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol 6 : 399.
23. HardieDG, CarlingD (1997) The AMP-activated protein kinase–fuel gauge of the mammalian cell? Eur J Biochem 246 : 259–273.
24. XiaoB, SandersMJ, UnderwoodE, HeathR, MayerFV, et al. (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472 : 230–233.
25. ApfeldJ, O'ConnorG, McDonaghT, DiStefanoPS, CurtisR (2004) The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev 18 : 3004–3009.
26. CurtisR, O'ConnorG, DiStefanoPS (2006) Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell 5 : 119–126.
27. SelmanC, TulletJM, WieserD, IrvineE, LingardSJ, et al. (2009) Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326 : 140–144.
28. GreerEL, DowlatshahiD, BankoMR, VillenJ, HoangK, et al. (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 17 : 1646–1656.
29. MairW, MorantteI, RodriguesAP, ManningG, MontminyM, et al. (2011) Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature 470 : 404–408.
30. StenesenD, SuhJM, SeoJ, YuK, LeeKS, et al. (2013) Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell Metab 17 : 101–112.
31. FunakoshiM, TsudaM, MuramatsuK, HatsudaH, MorishitaS, et al. (2011) A gain-of-function screen identifies wdb and lkb1 as lifespan-extending genes in Drosophila. Biochem Biophys Res Commun 405 : 667–672.
32. SlackC, FoleyA, PartridgeL (2012) Activation of AMPK by the putative dietary restriction mimetic Metformin is insufficient to extend lifespan in Drosophila. PLoS One 7: e47699.
33. SalminenA, KaarnirantaK (2012) AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res Rev 11 : 230–241.
34. GreerEL, OskouiPR, BankoMR, ManiarJM, GygiMP, et al. (2007) The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 282 : 30107–30119.
35. PanDA, HardieDG (2002) A homologue of AMP-activated protein kinase in Drosophila melanogaster is sensitive to AMP and is activated by ATP depletion. Biochem J 367 : 179–186.
36. HardieDG, CarlingD, GamblinSJ (2011) AMP-activated protein kinase: also regulated by ADP? Trends Biochem Sci 36 : 470–477.
37. XiaoB, HeathR, SaiuP, LeiperFC, LeoneP, et al. (2007) Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 449 : 496–500.
38. AradM, BensonDW, Perez-AtaydeAR, McKennaWJ, SparksEA, et al. (2002) Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J Clin Invest 109 : 357–362.
39. ZouL, ShenM, AradM, HeH, LofgrenB, et al. (2005) N488I mutation of the gamma2-subunit results in bidirectional changes in AMP-activated protein kinase activity. Circ Res 97 : 323–328.
40. LuptakI, ShenM, HeH, HirshmanMF, MusiN, et al. (2007) Aberrant activation of AMP-activated protein kinase remodels metabolic network in favor of cardiac glycogen storage. J Clin Invest 117 : 1432–1439.
41. AhmadF, AradM, MusiN, HeH, WolfC, et al. (2005) Increased alpha2 subunit-associated AMPK activity and PRKAG2 cardiomyopathy. Circulation 112 : 3140–3148.
42. SandersMJ, GrondinPO, HegartyBD, SnowdenMA, CarlingD (2007) Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J 403 : 139–148.
43. DanielT, CarlingD (2002) Functional analysis of mutations in the gamma 2 subunit of AMP-activated protein kinase associated with cardiac hypertrophy and Wolff-Parkinson-White syndrome. J Biol Chem 277 : 51017–51024.
44. HawleySA, RossFA, ChevtzoffC, GreenKA, EvansA, et al. (2010) Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab 11 : 554–565.
45. BudovskayaYV, WuK, SouthworthLK, JiangM, TedescoP, et al. (2008) An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134 : 291–303.
46. DePinaAS, IserWB, ParkSS, MaudsleyS, WilsonMA, et al. (2011) Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol 11 : 11.
47. DepuydtG, XieF, PetyukVA, ShanmugamN, SmoldersA, et al. (2013) Reduced insulin/IGF-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol Cell Proteomics. 12 : 3624–39.
48. FuruyamaT, NakazawaT, NakanoI, MoriN (2000) Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349 : 629–634.
49. TroemelER, ChuSW, ReinkeV, LeeSS, AusubelFM, et al. (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2: e183.
50. GoldenTR, MelovS (2004) Microarray analysis of gene expression with age in individual nematodes. Aging Cell 3 : 111–124.
51. Hunt-NewburyR, ViveirosR, JohnsenR, MahA, AnastasD, et al. (2007) High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5: e237.
52. SimmerF, TijstermanM, ParrishS, KoushikaSP, NonetML, et al. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12 : 1317–1319.
53. KwonES, NarasimhanSD, YenK, TissenbaumHA (2010) A new DAF-16 isoform regulates longevity. Nature 466 : 498–502.
54. ZhangP, JudyM, LeeSJ, KenyonC (2013) Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab 17 : 85–100.
55. HamiltonSR, StapletonD, O'DonnellJBJr, KungJT, DalalSR, et al. (2001) An activating mutation in the gamma1 subunit of the AMP-activated protein kinase. FEBS Lett 500 : 163–168.
56. LibinaN, BermanJR, KenyonC (2003) Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115 : 489–502.
57. AlcedoJ, KenyonC (2004) Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41 : 45–55.
58. GollobMH (2003) Glycogen storage disease as a unifying mechanism of disease in the PRKAG2 cardiac syndrome. Biochem Soc Trans 31 : 228–231.
59. NarbonneP, RoyR (2009) Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457 : 210–214.
60. AlonU (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8 : 450–461.
61. AlicN, AndrewsTD, GiannakouME, PapatheodorouI, SlackC, et al. (2011) Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7 : 502.
62. EijkelenboomA, MokryM, de WitE, SmitsLM, PoldermanPE, et al. (2013) Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol Syst Biol 9 : 638.
63. TothovaZ, KolliparaR, HuntlyBJ, LeeBH, CastrillonDH, et al. (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128 : 325–339.
64. PaikJH, KolliparaR, ChuG, JiH, XiaoY, et al. (2007) FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128 : 309–323.
65. BakkerWJ, van DijkTB, Parren-van AmelsvoortM, KolbusA, YamamotoK, et al. (2007) Differential regulation of Foxo3a target genes in erythropoiesis. Mol Cell Biol 27 : 3839–3854.
66. LutznerN, De-Castro ArceJ, RoslF (2012) Gene expression of the tumour suppressor LKB1 is mediated by Sp1, NF-Y and FOXO transcription factors. PLoS One 7: e32590.
67. HobertO (2002) PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32 : 728–730.
68. HoogewijsD, HouthoofdK, MatthijssensF, VandesompeleJ, VanfleterenJR (2008) Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9 : 9 doi:10.1186/1471-2199-9-9
69. MukhopadhyayA, DeplanckeB, WalhoutAJ, TissenbaumHA (2008) Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc 3 : 698–709.
70. HsinH, KenyonC (1999) Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399 : 362–366.
71. LomsadzeA, Ter-HovhannisyanV, ChernoffYO, BorodovskyM (2005) Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33 : 6494–6506.
72. EdgarRC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 : 113.
73. LarkinMA, BlackshieldsG, BrownNP, ChennaR, McGettiganPA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 : 2947–2948.
74. UniProtC (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–75.
75. RanwezV, ClaironN, DelsucF, PouraliS, AubervalN, et al. (2009) PhyloExplorer: a web server to validate, explore and query phylogenetic trees. BMC Evol Biol 9 : 108.
76. ZhangH, GaoS, LercherMJ, HuS, ChenWH (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40: W569–572.
77. DoonanR, McElweeJJ, MatthijssensF, WalkerGA, HouthoofdK, et al. (2008) Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev 22 : 3236–3241.
78. Van RaamsdonkJM, HekimiS (2012) Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci U S A 109 : 5785–5790.
79. EtchbergerJF, HobertO (2008) Vector-free DNA constructs improve transgene expression in C. elegans. Nat Methods 5 : 3.
80. LeeH, ChoJS, LambacherN, LeeJ, LeeSJ, et al. (2008) The Caenorhabditis elegans AMP-activated protein kinase AAK-2 is phosphorylated by LKB1 and is required for resistance to oxidative stress and for normal motility and foraging behavior. J Biol Chem 283 : 14988–14993.
81. HendersonST, JohnsonTE (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11 : 1975–1980.
82. PatelDS, Garza-GarciaA, NanjiM, McElweeJJ, AckermanD, et al. (2008) Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics 178 : 931–946.
83. TulletJM, HertweckM, AnJH, BakerJ, HwangJY, et al. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132 : 1025–1038.
84. BatemanA (1997) The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci 22 : 12–13.
Štítky
Genetika Reprodukčná medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



