-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
Genetic variation in the major histocompatibility complex (MHC) affects CD4∶CD8 lineage commitment and MHC expression. However, the contribution of specific genes in this gene-dense region has not yet been resolved. Nor has it been established whether the same genes regulate MHC expression and T cell selection. Here, we assessed the impact of natural genetic variation on MHC expression and CD4∶CD8 lineage commitment using two genetic models in the rat. First, we mapped Quantitative Trait Loci (QTLs) associated with variation in MHC class I and II protein expression and the CD4∶CD8 T cell ratio in outbred Heterogeneous Stock rats. We identified 10 QTLs across the genome and found that QTLs for the individual traits colocalized within a region spanning the MHC. To identify the genes underlying these overlapping QTLs, we generated a large panel of MHC-recombinant congenic strains, and refined the QTLs to two adjacent intervals of ∼0.25 Mb in the MHC-I and II regions, respectively. An interaction between these intervals affected MHC class I expression as well as negative selection and lineage commitment of CD8 single-positive (SP) thymocytes. We mapped this effect to the transporter associated with antigen processing 2 (Tap2) in the MHC-II region and the classical MHC class I gene(s) (RT1-A) in the MHC-I region. This interaction was revealed by a recombination between RT1-A and Tap2, which occurred in 0.2% of the rats. Variants of Tap2 have previously been shown to influence the antigenicity of MHC class I molecules by altering the MHC class I ligandome. Our results show that a restricted peptide repertoire on MHC class I molecules leads to reduced negative selection of CD8SP cells. To our knowledge, this is the first study showing how a recombination between natural alleles of genes in the MHC influences lineage commitment of T cells.
Published in the journal: Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat. PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004151
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004151Summary
Genetic variation in the major histocompatibility complex (MHC) affects CD4∶CD8 lineage commitment and MHC expression. However, the contribution of specific genes in this gene-dense region has not yet been resolved. Nor has it been established whether the same genes regulate MHC expression and T cell selection. Here, we assessed the impact of natural genetic variation on MHC expression and CD4∶CD8 lineage commitment using two genetic models in the rat. First, we mapped Quantitative Trait Loci (QTLs) associated with variation in MHC class I and II protein expression and the CD4∶CD8 T cell ratio in outbred Heterogeneous Stock rats. We identified 10 QTLs across the genome and found that QTLs for the individual traits colocalized within a region spanning the MHC. To identify the genes underlying these overlapping QTLs, we generated a large panel of MHC-recombinant congenic strains, and refined the QTLs to two adjacent intervals of ∼0.25 Mb in the MHC-I and II regions, respectively. An interaction between these intervals affected MHC class I expression as well as negative selection and lineage commitment of CD8 single-positive (SP) thymocytes. We mapped this effect to the transporter associated with antigen processing 2 (Tap2) in the MHC-II region and the classical MHC class I gene(s) (RT1-A) in the MHC-I region. This interaction was revealed by a recombination between RT1-A and Tap2, which occurred in 0.2% of the rats. Variants of Tap2 have previously been shown to influence the antigenicity of MHC class I molecules by altering the MHC class I ligandome. Our results show that a restricted peptide repertoire on MHC class I molecules leads to reduced negative selection of CD8SP cells. To our knowledge, this is the first study showing how a recombination between natural alleles of genes in the MHC influences lineage commitment of T cells.
Introduction
Major histocompatibility complex (MHC) genes have been identified in all vertebrate species [1]. The 3.6 Mb human leukocyte antigen (HLA) was one of the first MHC to be sequenced, and revealed a region with extraordinary complexity [2]. The region contains ∼260 genes that are clustered in sub-regions denoted MHC-I, MHC-II and MHC-III [3]. Genes in the MHC were early recognized for their extreme sequence diversity and association with autoimmune and inflammatory conditions (reviewed in [4]). However, these associations have been difficult to delineate since nearly 40% of the MHC genes have immune-related functions [2]. The interpretation of association data is further complicated by the extensive linkage disequilibrium (LD) across the region [5]. While the LD structure [6], [7] and genetic variation [3], [8] of the HLA in humans is rather well investigated, similar detailed analysis for the MHC in other species is needed.
The first complete sequence of the rat MHC (RT1) on chromosome 20 was derived from the Brown Norway (BN) strain (RT1n) and released in 2004 [9]. The BN genome sequence, which is also the rat reference sequence (RefSeq), was published shortly thereafter [10]. Several inbred rat strains have since then been resequenced, including the Spontaneously Hypertensive Rat (SHR, RT1k) [11], and more recently, the DA (RT1av1), F344 (RT1lv1) [12] and a panel of additional strains [13]. The genetic organization of the MHC is similar in rats and humans, with the exception of a proximal classical MHC class Ia region (RT1-A) [14] and a larger number of telomeric, non-classical, class Ib genes (RT1-CEM) [15] in the rat. MHC class Ib molecules are structurally similar to class Ia molecules but the corresponding class Ib genes are less polymorphic, expressed at lower levels and several are pseudogenes [16], [17]. The number of functional class Ia genes (RT1-A1, RT1-A2, RT1-A3) in the rat varies between one and three in standard inbred strains [18], [19]. These genes are highly polymorphic and densely expressed on the surface of virtually all nucleated cells where they engage in presentation of intracellular antigens to cytotoxic CD8 T cells. The class Ia genes are surrounded by highly conserved framework genes, which are found in the MHC of all mammals, as well as several antigen processing and transportation genes [9]. To this latter group of genes belong the γ-interferon-inducible proteasome subunit beta type 8 (Psmb8) and Psmb9, and Tap1 and Tap2, which are all located in the MHC-II region. An additional gene in this group, Tap binding protein (Tapbp), is located centromeric to the class Ia genes in the MHC-I region. These genes show limited allelic variation in humans and mice [20], [21]. In the rat, by contrast, allelic variants of Tap2 fall into two groups, Tap2A and Tap2B, which encode two functionally distinct allotypic forms of Tap2 [22], [23]. Consequently, the two variants of TAP transporters encoded by Tap1 and Tap2A or Tap2B are denoted TAP-A and TAP-B, respectively [19]. Analyses of inbred rat strains have revealed a significant degree of co-evolution between alleles in Tap2 and the class Ia loci [19] and based on these studies RT1-A molecules have been classified as either TAP-A or TAP-B linked [19], [24]. Livingstone and colleagues have shown that if linkage is lost between RT1-Aa (the DA allele of class Ia) and the Tap2A allele, as in the event of a recombination, the antigenicity of the class Ia molecule is altered [25]. This phenomenon, known as class-I modification (cim), has been explained by the peptide selectivity of the different TAP isoforms [26], [27] and more specifically by the inability of the TAP-B transporter to translocate peptides with C-terminal arginine residues, which are required for optimal loading of RT1-Aa molecules [28]. This was followed by studies showing that cim leads to abnormally slow assembly and reduced extracellular class I expression of RT1-Aa molecules [22], [29]. By contrast, no such effects have been described for TAP-B-linked RT1-A molecules when associated with the promiscuous TAP-A transporter. Within the MHC-II region are four additional antigen processing genes encoding the α and β subunits of RT1-DM and RT1-DO. These non-classical class II molecules are responsible for editing the peptide cargo of the classical class II molecules before these are translocated to the cell surface to present peptides to helper CD4 T cells.
Expression of MHC class I and II molecules on antigen presenting cells is required for T cell selection in the thymus [30], [31] and for the survival of mature T cell subsets in the periphery [32], [33]. The commitment of thymocytes to CD4 or CD8 lineage is determined mainly by the MHC peptide repertoire and the antigen specificity of the T cell receptor (TCR). The variation in CD4 and CD8 T cell numbers is associated with genes in the MHC in humans [34], mice [35] and rats [36]. However, it is not known whether the levels of MHC expression and/or polymorphisms in other genes than the classical MHC class I and II genes, such as the aforementioned antigen processing genes, contribute to this variation.
Here, we assessed the impact of the genetic variation on class I and II expression and T cell selection using two genetic rat models: the NIH-Heterogenous Stock (HS) and a panel of MHC-recombinant congenic strains. The NIH-HS is descended from eight inbred strains through 60 generations of outbreeding [37]. The greater genetic diversity in the HS compared to a conventional F2 cross allows mapping of more QTLs [38], which was exploited here to identify genomic regions (QTLs) that contribute both to MHC expression and the variation in CD4∶CD8 T cell ratio. Recombinant congenic strains (RCS) are powerful tools to resolve complex QTLs in regions with extensive LD. In RCS, haplotype blocks that are the result of rare recombinations can be preserved, which allows investigation of multiple phenotypes using genetically identical individuals [39], [40]. We describe a new panel of rat RCS, in which MHC segments from four inbred strains have been inserted on the background of the DA strain.
In the rat HS we identified a region spanning 4.1–9.7 Mb on chromosome 20 that contributed to both the variation in MHC expression and CD4∶CD8 T cell ratio. Mapping using RCS identified two intervals, each ∼0.25 Mb wide, in the MHC-I and II region that contributed to the phenotypic variation, suggesting that the HS QTL might result from two linked QTLs. First, we showed that the variation in class I expression correlated with alleles of Tap2, which is in line with previously reported data [25]. Second, we identified a novel type of class-I modification that we termed inverse cim, which reduced the expression of TAP-B linked RT1-A molecules if associated with TAP-A. Finally, we showed that polymorphisms in Tap2 significantly contribute to CD8 lineage commitment, likely by altering the class-I peptide repertoire on antigen presenting cells in the thymus.
Results
QTLs for CD4∶CD8 T cell ratio and MHC expression overlap within the MHC region
T cell selection and the maintenance of the peripheral T cell pool rely on the interaction between peptide-MHC complexes and T cell receptors. The variation in the relative proportion of peripheral CD4 and CD8 T cells has been associated with the MHC [34]–[36]. In order to investigate if there is a shared genetic regulation of MHC expression and T cell numbers, we first identified genome-wide QTLs that controlled the CD4∶CD8 T cell ratio and MHC class I and II expression in the rat NIH-HS [41]. We measured the number of circulating CD4 and CD8 T cells and extracellular MHC class I and II expression by flow cytometry in more than 2000 HS rats, of which 1407 were genotyped at 265,551 SNPs. Each HS rat chromosome was reconstructed as a mosaic of the founder genomes, and QTLs were mapped using two methods that take the different levels of relatedness existing in the rat HS into account (see Materials and Methods). We report QTLs detected at a false discovery rate of 10%, which corresponds to a negative logP threshold of 4.2 for the three measures studied here.
The mean ratio of circulating CD4 vs. CD8 T cells in the HS rats was 2.76 (range 1.0–13.8), which is concordant with data previously obtained in the inbred founder strains [42]. Variation in this phenotype was explained mainly by QTLs on chromosomes 9 and 20 (Fig. 1). The intervals most significantly associated with this phenotype were located at 4.20 Mb at the proximal end of chromosome 9 (−logP = 22.1), and at 4.78 Mb in the MHC region on chromosome 20 (−logP = 36.5). The effect sizes of these QTLs were estimated to be 14.4% and 14.6% respectively (upper bound; Fig. 1C). These QTLs also contributed to variation in absolute numbers of CD4 and CD8 T cells (data not shown, genome scans are accessible on the WTCHG website: http://mus.well.ox.ac.uk/gscandb/rat).
Fig. 1. Genetic mapping in heterogeneous stock (HS) rats. 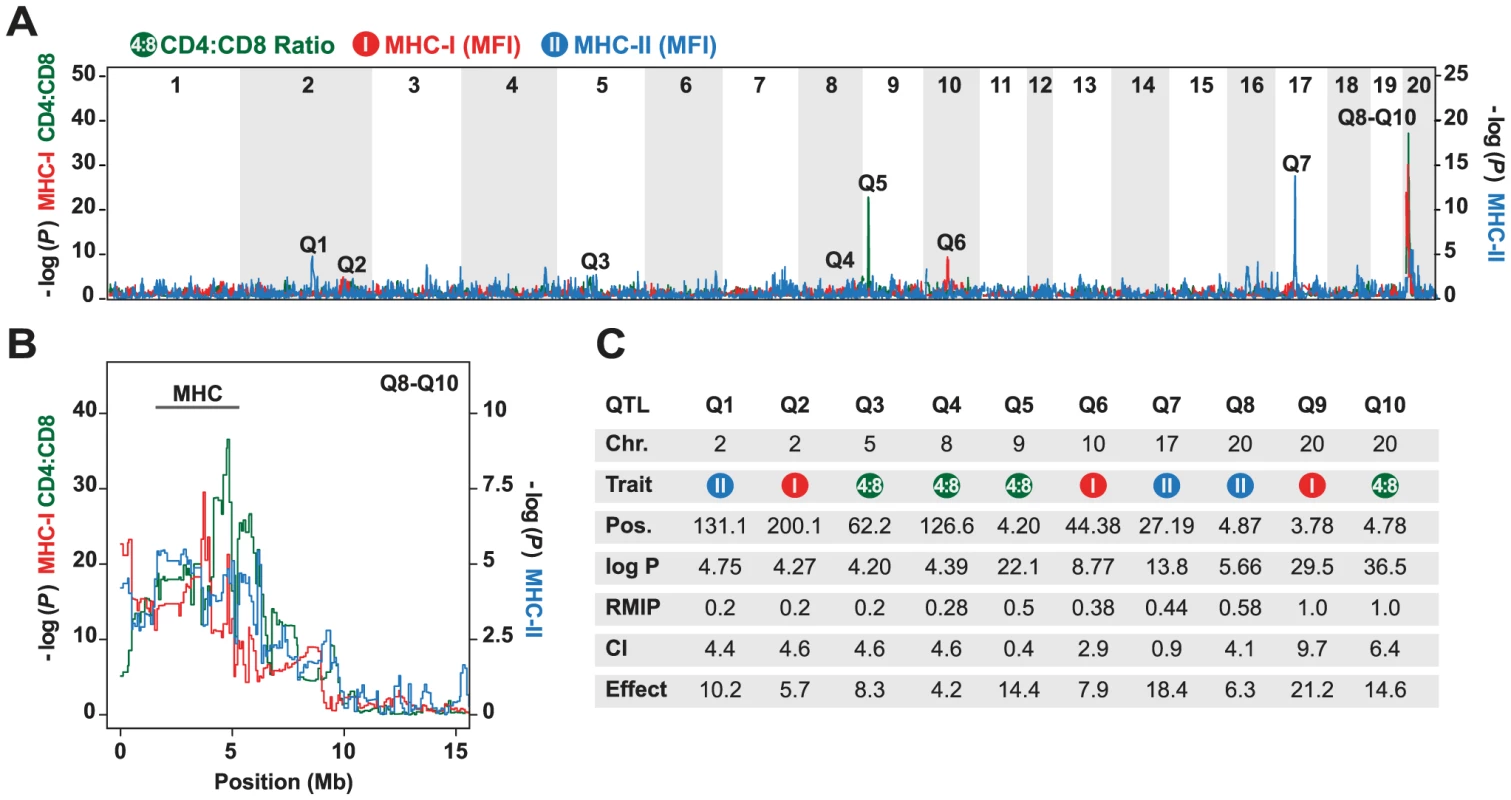
(A) Genome scans of the CD4∶CD8 ratio (green), and the surface expression of MHC class I (MHC-I, red) and MHC class II (MHC-II, blue). The vertical axis shows the negative log P value for the 20 autosomes. Note that the left and right axis show −log P values for different phenotypes. Q1–Q10 indicate significant QTLs. (B) Close-up for Q8–Q10 on chromosome 20 showing the approximate location of the MHC region. (C) Position (in megabases, Mb) of peak marker (Pos.), confidence intervals (CI) in Mb, and effect sizes (%) of the peak markers in the QTLs shown in (A). The RMIP value is a measure of the probability that the loci are correctly identified (max = 1.0). The surface expression of MHC class I was determined on granulocytes using a widely reactive MHC class I antibody to Ia and Ib molecules. QTLs for this phenotype were found on chromosome 2 (−logP = 4.27), 10 (−logP = 8.77) and 20 (−logP = 29.5), of which only the latter coincided with a QTL for CD4∶CD8 T cell ratio. Three regions were associated with MHC class II (RT1-B) expression on B cells. These were located on chromosome 2 (−logP = 4.75), 17 (−logP = 13.8) and 20 (−logP = 5.66). The most significantly associated marker on chromosome 20 was located at 4.87 Mb in the MHC region. Because the median 90% confidence interval for the position of the QTLs mapped in the rat HS is 4.5 Mb, this QTL overlapped with the QTL identified for CD4∶CD8 T cell ratio on this chromosome.
In summary, using a genome-wide approach, we showed that CD4∶CD8 T cell ratio and MHC expression might be regulated by a common QTL or a set of closely linked QTLs in the rat MHC region. However, the majority of identified QTLs for these traits did not co-localize, suggesting that variation in MHC expression is not a strong determinant of the CD4∶CD8 T cell ratio.
Generation of MHC recombinant congenic strains
We next aimed to isolate the intervals associated with the phenotypic variation in the MHC, since the mapping in the HS rats did not distinguish whether a single gene with pleiotropic action or different MHC genes controlled MHC class I and II expression and the CD4∶CD8 T cell ratio.
We therefore established MHC congenic strains with RT1f, RT1i, RT1u and RT1h haplotypes on the homozygous DA (RT1av1) background that showed phenotypic variation in CD4∶CD8 T cell ratio and class I and II expression (Table 1). We produced a panel of more than 40 RCS with segments in the MHC-I, -II and -III regions (selected strains are shown in Figure 2). We identified 70 recombinations within a 2 Mb genomic region (3.4–5.4 Mb) (Fig. 3) and mapped the recombination breakpoints using 67 SNP and short-tandem repeat (STR) markers (Table S1 and Table S2) to intervals of 2–270 kb. Three recombination hotspots were identified near DMb, Btnl2 and Lta (Fig. 3), which are also located close to recombination hotspots in the human HLA [6], [8], [43], [44]. We did not identify any recombination events in the MHC-II region between DMb and Btnl2, despite analyzing ∼29,000 meiotic events. Extensive LD was also observed between Kifc1 and Coll11A2 in the MHC-I region where only a single recombination near the Kifc1 gene was identified among the ∼16,000 analyzed meiotic events. Thus, recombinations in the RT1 between the MHC-I and -II regions occur relatively frequently (0.2%), while the recombination activity within each of these regions is extremely low. The extensive LD between DMb and Btnl2 as well as between Kifc1 and Coll11A2 therefore gave rise to two haplotypes that could be isolated as congenic segments in the RCS panel.
Fig. 2. Physical map of the rat MHC region. 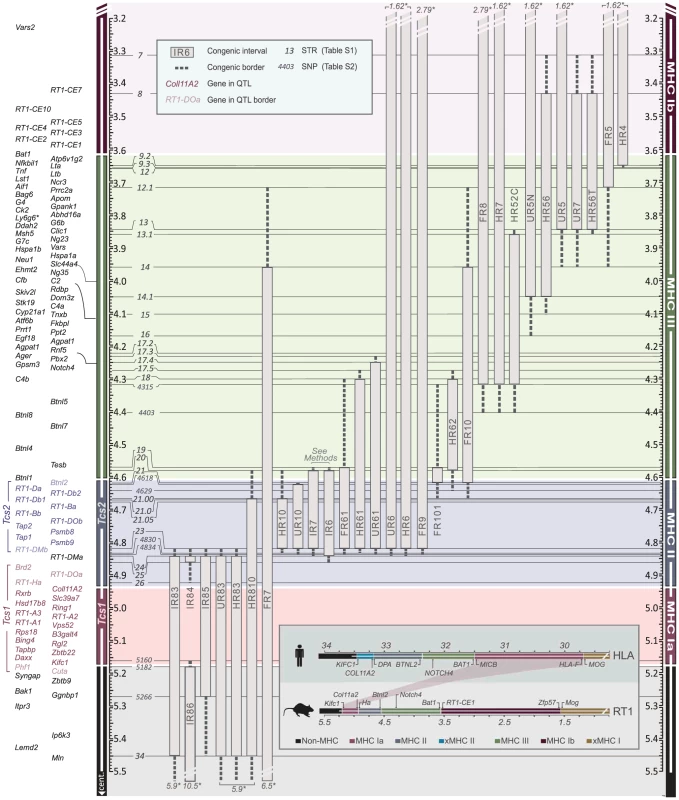
The map was constructed according to the NCBI build 3.4 genome assembly. Genes (left) are depicted according to scale (positions in Mb), except for positions indicated with crotchets. The gross organization of MHC-Ia, II, III and Ib -regions are adopted from Hurt et al. [9]. Recombinant congenic strains are shown as gray vertical bars with dashed lines representing congenic borders (intervals of unknown genotype). Markers, short tandem repeats (STRs) and single nucleotide polymorphisms (SNPs), are shown in italic numbers. Numbers with asterisks at the top and bottom of the figure represent the position of the closest negative (DA) marker. Genes in Tcs1 are red and in Tcs2 blue (see box for definitions). Inset shows the organization of the human (HLA) and rat (RT1) MHC regions. Non-recombinant congenic strains, which have fragments spanning the entire MHC region, are not shown. xMHC-II, extended MHC class II region; xMHC-I, extended MHC class I region. Fig. 3. Recombination hotspots and haplotype blocks in the rat MHC. 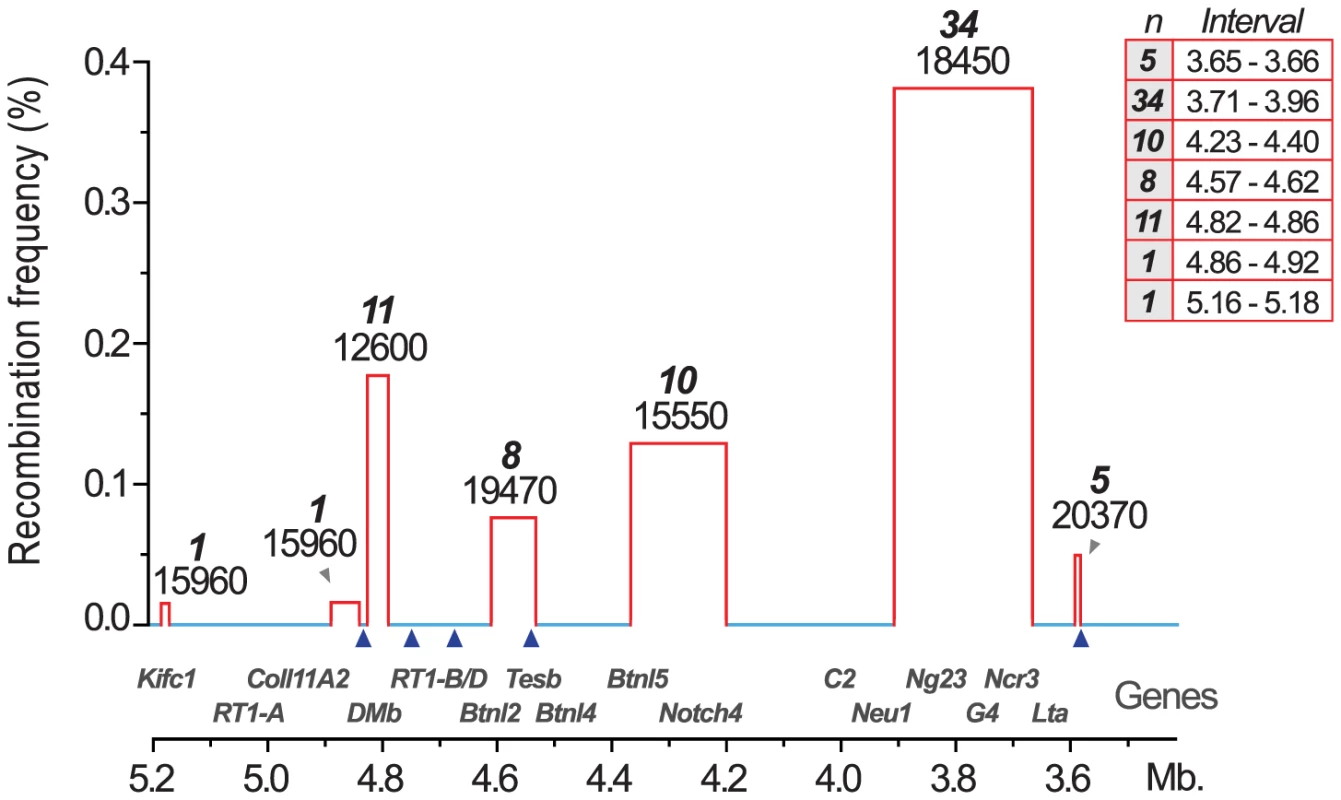
Recombination activity was assessed over 2(3.4–5.4 Mb). Blue lines represent cold regions (haplotype blocks) with low recombination activity. Regions with recombinations are depicted as red bars. The width of the bars, and the table inset (right), represent recombination intervals. Numbers above bars in bold face represent the observed numbers of recombinations within the interval with the number of analyzed meiotic events shown below. The height of the bars indicate individuals with recombinations (in %). Blue triangles represent recombination hotspots in humans and are adopted from Cullen et al. [6]. Tab. 1. CD4∶CD8 T cell ratio and extracellular MHC class I and II expression in spleens of congenic and recombinant strains. 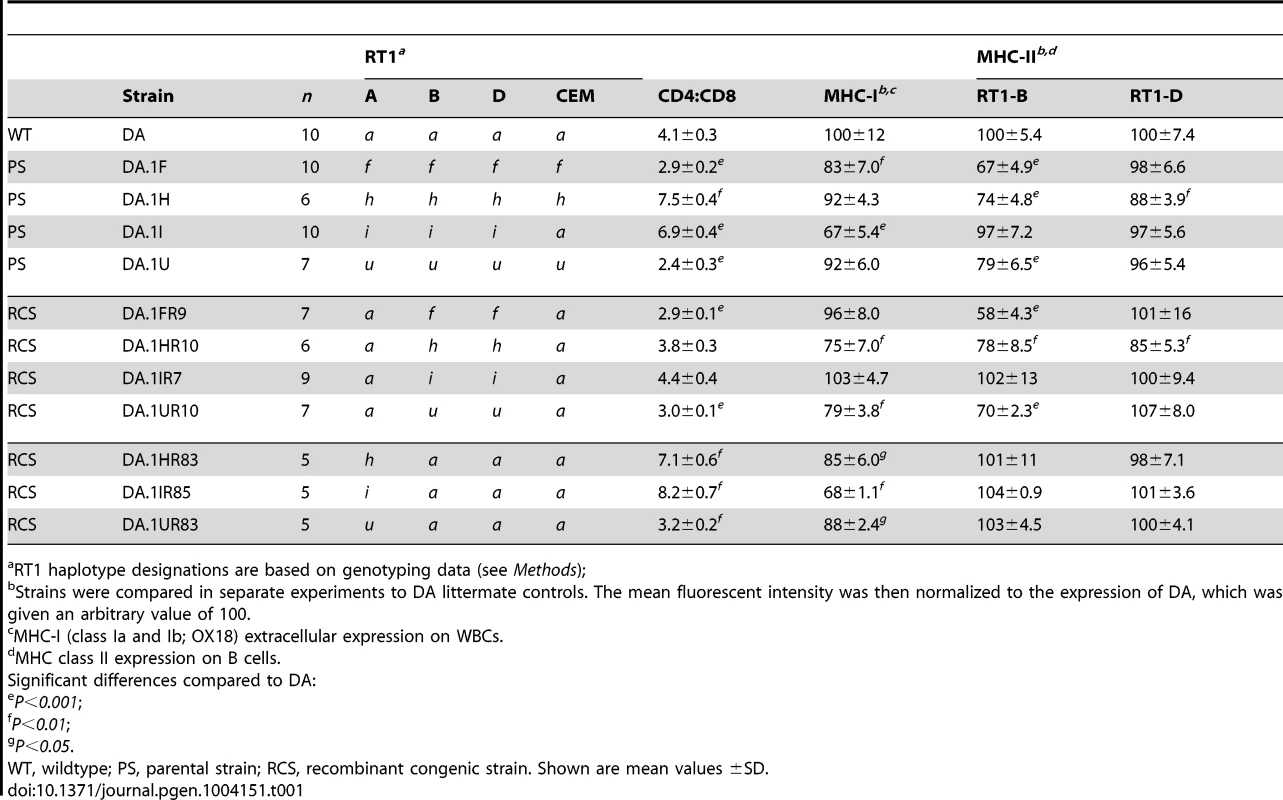
RT1 haplotype designations are based on genotyping data (see Methods); Two linked QTLs in RT1 regulate CD4∶CD8 T cell ratio and MHC expression
The RCS panel allowed the identification of two intervals in RT1 associated with the variation in CD4∶CD8 T cell ratio and MHC expression (Table 1). The first interval, which we named T cell selection QTL 1 (Tcs1), was identified within the MHC-I region (Fig. 2) using DA.1IR85, DA.1HR83 and DA.1UR83 (we refer to these strains as Tcs1-congenic strains). This QTL, which regulated class I cell surface expression and the CD4∶CD8 T cell ratio, was determined to 0.282 Mb (min-max: 0.242–0.323 Mb) based on the average size of the intervals of unknown genotype (congenic borders) in phenotype-negative DA.1IR86 and DA.1IR84 rats (Fig. 2). The second QTL, denoted T cell selection QTL 2 (Tcs2), was mapped to the MHC-II region using the Tcs2-congenic strains DA.1UR10, DA.1HR10 and DA.1FR9 (Fig. 2). Since DA.1FR61 (Fig. 2) was not yet available at the time of the investigation, we used DA.1FR10 and DA.1FR8 (both phenotype-negative) to exclude genes in the MHC-III and RT1-CEM region of DA.1FR9 (Fig. 2). Recombination events in DA.1UR10 within Btnl2 (between intron 5 and 6) and in DA.1HR10 within RT1-DMb (between intron 2 and 5) constituted the telomeric and centromeric boundaries of Tcs2, which was determined to 0.206 Mb (min-max: 0.197–0.214 Mb) (Fig. 2). This QTL regulated the cell surface expression of class I and II as well as the CD4∶CD8 T cell ratio.
Tcs1 and Tcs2 together explained all variation in the CD4∶CD8 T cell ratio and extracellular MHC expression associated with RT1 in the RCS. This suggests that these two linked haplotypes are also responsible for the phenotypic variation mapped to chromosome 20 in the HS. While Tcs2 alone controlled MHC class II expression, both QTLs, Tcs1 and Tcs2, controlled cell surface expression of MHC class I and the variation in CD4∶CD8 T cell ratio. The effect of the MHC-II region on MHC class I expression has previously been ascribed to allelic variants of Tap2 [25]. Since the other genes in Tcs2 are in strong LD with Tap2, we next compared the genetic variation of these genes in order to identify their individual contribution.
Genes encoding proteins in the MHC class I and II pathways show variable degrees of sequence conservation
Two QTLs, Tcs1 in the MHC-I region and Tcs2 in the MHC-II region, were associated with the variation in CD4∶CD8 T cell ratio and MHC expression. In order to identify the causative genes in Tcs1 and Tcs2 associated with these traits, we determined the genetic variation of genes encoding proteins in the MHC class I and II pathways (all genes in Tcs2, Tapbp in Tcs1) by direct sequencing. These analyses of congenic (RT1f,h,i,u), wild-type (RT1a) and RefSeq (RT1n) haplotypes revealed 367 exonic SNPs (synonymous and nonsynonymous), 15 indels (insertions/deletions) and 168 amino acid (aa) substitutions in the corresponding proteins (Table 2).
Tab. 2. Sequence variants and allele distribution of genes in six rat MHC haplotypes. 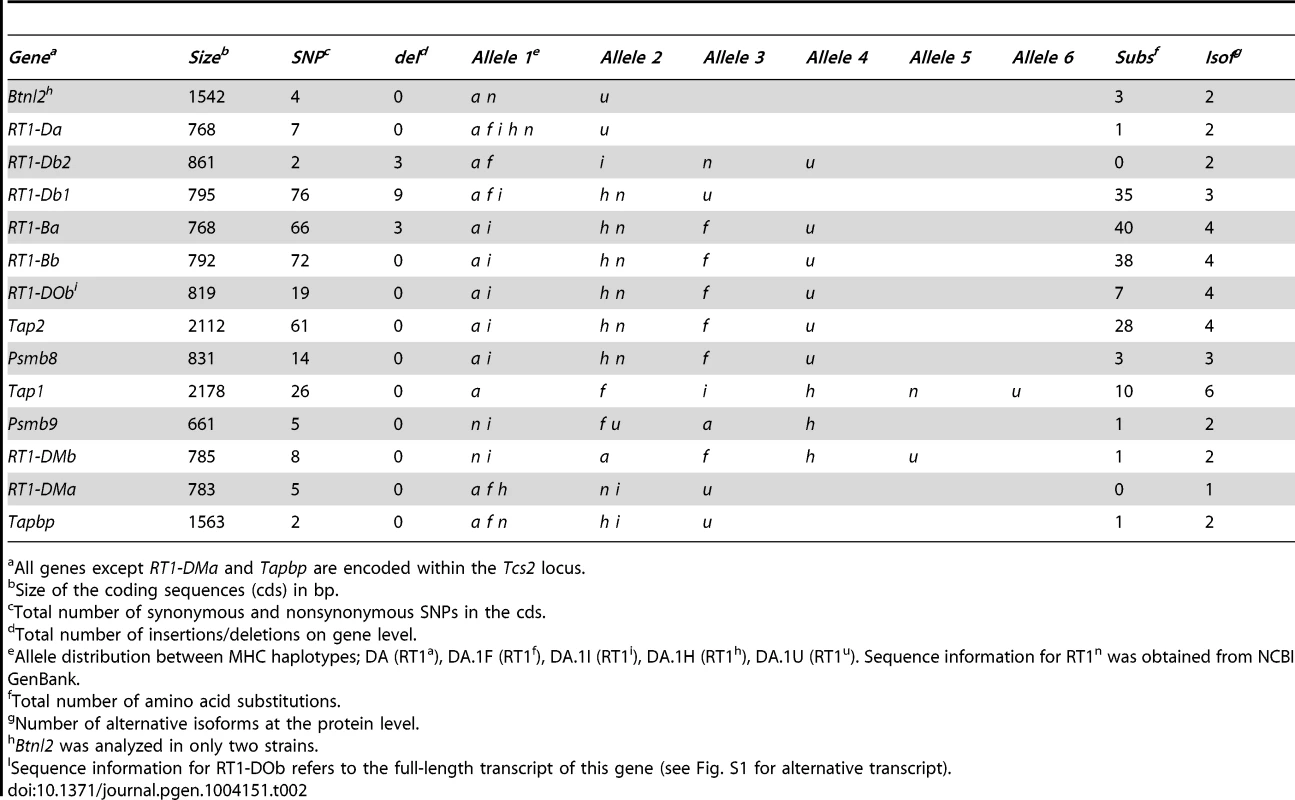
All genes except RT1-DMa and Tapbp are encoded within the Tcs2 locus. The genes associated with the MHC class I pathway (Tap1, Tap2, Psmb8, Psmb9 and Tapbp) showed variable degrees of sequence conservation (Fig. 4). The most conserved gene was Tapbp, with only a single nonsynonymous SNP (nsSNP) coding for R165H in the corresponding protein (alternative allele in the RT1u haplotype, Table 2). Psmb9 contained one nsSNP in a region coding for the propeptide while three nsSNPs were found in Psmb8 (Fig. 4), although not in positions predicted to influence immunoproteasome assembly or the chymotrypsin-like activity of the corresponding protein [45]. By contrast, both Tap1 and Tap2 showed extreme sequence diversity. We identified six allelic variants of Tap1, including unique alleles for the RT1i and RT1h haplotypes, and four allelic variants of Tap2 (Fig. 4). Three of the Tap2 alleles have not been sequenced previously: Tap2h, Tap2f and Tap2i, of which the latter was found to be identical to Tap2a (Table 2). Our sequence analysis confirms previous categorization of these alleles using restriction endonucleases [23] as Tap2A (RT1f and RT1i) and Tap2B (RT1h), respectively. This categorization of Tap2 alleles is based on nsSNPs coding for the amino acids 217, 218, 262, 265, 266 (asterisked in Fig. 4) in the Tap2 polypeptide, which determine the peptide specificity of the TAP complex [46], [47].
Fig. 4. Coding variants in T cell selection QTL 2 (Tcs2). 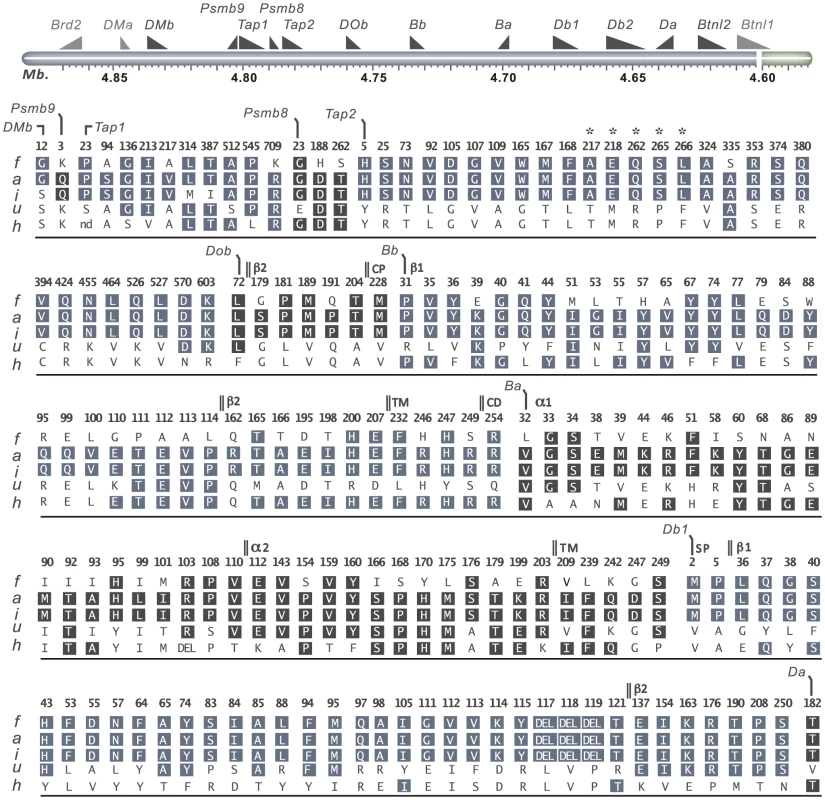
Nonsynonymous- and structural variants in Tcs2 were determined by Sanger sequencing of DA.1F (f), DA (a), DA.1I (i), DA.1U (u) and DA.1H (h) (see also Table 2). Amino acid substitutions are indicated in standard single letter codes and insertions/deletions as DEL. Gene annotations are from UniProt; protein domains depicted in DOb are derived from the human homolog. Letters in boxes depict residues with background allele (DA) with numbers above indicating amino acid positions in the translated cds. On top is a schematic illustration of the genes in the region according to the 3.4 genome assembly (genes outside the QTL are shown in gray). Annotations used: TM, transmembrane domain; CD, cytoplasmic domain; CP, connecting peptide; β1, beta 1 domain; β2, beta 2 domain; α1, alpha 1 domain; α2, alpha 2 domain. *Residues associated with class-I modification (cim). Variable sequence diversity was also observed for the genes in the MHC class II pathway (DOb, DMa and DMb). The Tcs1 and Tcs2 intervals excluded the DMa gene as well as the only nsSNP in DMb (corresponding to aa position 12) (Fig. 4). The DOb gene was found to be more polymorphic in the rat than previously shown for mice and humans [48], with a total of seven aa substitutions in the corresponding protein (Fig. 4). Two transcripts of DOb, which differed in length by 77 bp due to a deletion in exon 3, were identified in both congenic strains and DA (Fig. S1 and Fig. S2).
The rat expresses two classical MHC class II loci, RT1-B and RT1-D. Three MHC class II genes, RT1-Ba, -Bb and -Db1 were highly polymorphic across the six haplotypes studied here, whereas RT1-Da was largely conserved with only a single nsSNP (Fig. 4). The existence of a second putative β-locus of RT1-D has been suggested [9]. Our data show that this gene, RT1-Db2, is expressed and that it is monomorphic at the protein level in DA and MHC-congenic strains (data not shown). We could further show by mass spectrometry that RT1-Db2 forms an αβ-heterodimer with RT1-Da and confirm at the protein level that RT1-Db2 has a 22 residue extended cytoplasmic tail, which does not exist in RT1-Db1 (Fig. S3).
Taken together, among the genes in the MHC class I pathway, only Tap2 had an allele distribution that correlated with the variation in MHC class I expression (Fig. 4, Table 1). The effect of different Tap2 alleles on MHC class I expression has previously been established [25], [29], and we therefore suggest Tap2 as the only causative gene in Tcs2 for this phenotype. In addition, these data exclude functional polymorphisms in Tapbp as responsible for the Tcs1 QTL in MHC-I. Hence, the variations in MHC class I expression and the CD4∶CD8 T cell ratio associated with this QTL are likely to be due to polymorphisms in the RT1-A genes and/or the number of functional protein-coding RT1-A genes per MHC-I haplotype.
Expression of class I proteins in Tcs1-congenic rats
The number of RT1-A genes in standard inbred rat strains varies from one in the RT1lv1 and RT1a haplotypes to three in the RT1n,o,d,m haplotypes [14], [18]. It has therefore been suggested that the number of RT1-A genes influences the selection of CD8 T cells [36]. The expression of RT1-A at the protein level has previously been characterized using allotypic antibodies [18], [49]–[52], while the phenotypes we mapped in the HS and RCS were determined using the widely reactive MHC class I antibody OX18, which does not discriminate between different isoforms of RT1-A. In order to assess the number of functional protein-coding RT1-A genes in DA.1IR85 and to confirm the expression of RT1-Aa, A1h and A2h, and Au as the only class Ia molecules in DA and the Tcs1-congenic strains, we analyzed trypsin-digested cell lysates of IFN-γ stimulated splenocytes from these strains by mass spectrometry. The comparison of DA and the Tcs1-congenic strains also allowed the identification and discrimination of classical Ia and non-classical Ib proteins, since these strains encode different RT1-A genes but the same RT1-CEM genes.
We identified between 5 and 17 peptides (Mascot score >20) per strain that matched rat class Ia and Ib entries in public databases. In DA, 11 of 17 identified peptides matched the RT1-Aa molecule described by Rada et al. [53], while four peptides matched the UniProt entry HA11_RAT (Table 3, Fig. S4). HA11_RAT is the UniProt entry of the class Ib gene RT1-EC2 (RGD), which was isolated from a DA cDNA library as clone 3.6 [53] and predicted to be the rat homologue of mouse H2-Q10 [54]. All four peptides identified in DA.1UR83 were derived from a single class Ia molecule (Table 3, Fig. S5), which is consistent with previous observations that the RT1a and the RT1u haplotypes express only one class Ia gene each (RT1-Aa and RT1-Au, respectively) [53], [55]. In DA.1IR85 and DA.1HR83, 12 and 13 peptides, respectively, allowed the discrimination of two different RT1-A isoforms (A1 and A2) in each strain (Table 3, Fig. S6 and S7). In DA.1HR83 we also identified one peptide unique to clone 3.6 and two peptides that were shared between all identified class I molecules (Fig. S7). None of the analyzed samples contained peptides unique to an A3 isoform, which has been cloned in rats with haplotype RT1o/d/m [18] and RT1n [14].
Tab. 3. MHC class I proteins expressed in splenocytes from DA and Tcs1-congenic strains. 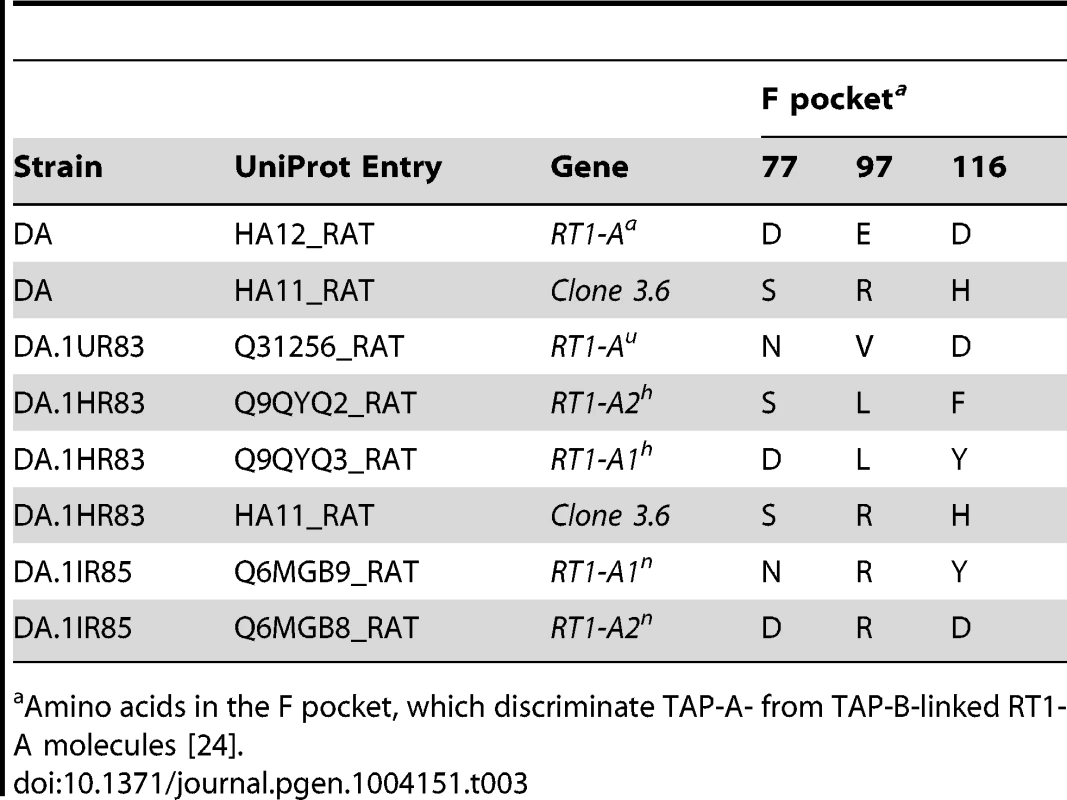
Amino acids in the F pocket, which discriminate TAP-A- from TAP-B-linked RT1-A molecules [24]. Taken together, our results show that DA.1IR85 and DA.1HR83 express two functional protein-coding RT1-A genes, A1n/A2n and A1h/A2h, respectively. We did not identify an A3 isoform in DA.1IR85, which is unexpected since the RT1-A region in DA.1IR85 is supposedly derived from the BN strain (RT1n) [56]. In DA and DA.1UR83 we only identified one RT1-A molecule per haplotype, which is in line with previous studies [53], [55].
A novel type of class-I modification influences class I expression
Having established the number of protein-coding RT1-A genes in DA.1IR85 and confirmed the protein expression of RT1-Aa, A1h and A2h, and Au, we continued assessing the impact of TAP-A and TAP-B on the expression levels of MHC class I molecules in Tcs1 and Tcs2-congenic strains.
Mapping in the RCS identified two adjacent QTLs for class I expression, Tcs1 and Tcs2. Tcs2 aligned with the principles of the classical cim phenomenon [25]. Tcs1, by contrast, was unexpected since the RT1-A genes in the Tcs1-congenic strains are associated with Tap2A, which encodes the promiscuous TAP-A transporter. We therefore hypothesized that this QTL was due to a novel type of class I modification in which the cell surface expression of TAP-B-linked RT1-A molecules (RT1-An, RT1-Au and RT1-Ah) was reduced by the presence of TAP-A. We termed this TAP-A-mediated class-I modification inverse cim. To test this hypothesis, we analyzed the intra - and extracellular expression of class I in the Tcs1-congenic strains (Tap2A), DA (Tap2A) and in two non-recombinant parental strains, DA.1H and DA.1U (both Tap2B).
The expression of class I was determined using the OX18 antibody that binds both A1 and A2 isoforms [57]. However, since OX18 in addition recognizes class Ib molecules [58], which may differ in numbers between the non-recombinant strains and DA, we also used the class Ia specific antibody F16-4-4 [59]. This antibody, on the other hand, reacts with a polymorphic determinant (Fig. S8 and [52]) and was therefore only used to compare the class I expression between strains with the same RT1-A haplotypes.
We first analyzed the extracellular levels of class I on spleen cells expressing CD68, a marker of macrophages and dendritic cells in the rat [60]. Consistent with the leukocyte data shown in Table 1, CD68+ cells in DA.1HR83 and DA.1HR10 showed reduced surface levels of class I compared to DA and DA.1H when stained with OX18 (Fig. 5A) as well as with F16-4-4 (Fig. 5B). The lower levels of class Ia in DA.1HR83 (RT1-Ah, TAP-A) compared to DA.1H (RT1-Ah, TAP-B) suggest that RT1-Ah requires TAP-B for optimal export to the cell surface, which would be consistent with the inverse cim phenomenon.
Fig. 5. Regulation of class I expression by classical and inverse cim. 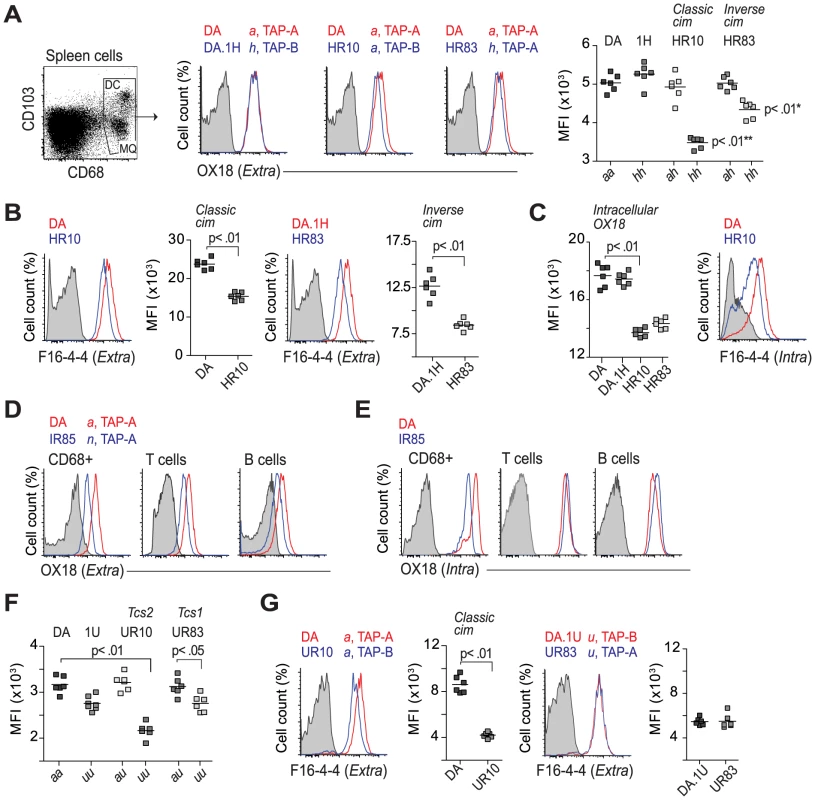
(A) CD68+ cells were stained on the cell surface with OX18 (anti-class Ia and Ib). Histograms show representative samples from DA, DA.1H and DA.1H derived strains with different alleles of RT1-A and Tap2 as stated on top. Data from all individuals are shown in scatterplot (far right); * significant compared to DA.1H (1H); ** significant compared to DA. (B) CD68+ cells stained with a class Ia specific antibody (F16-4-4). (C) T cells from animals shown in (A) stained intracellularly with OX18 (scatterplot) and F16-4-4 (histogram). (D–E) Subsets of leukocytes from DA and DA.1IR85 spleen stained extracellularly (D) and intracellularly (E) with OX18. Data are representative of 6 individuals per group. (F) Surface expression of MHC class I (OX18) on CD68+ cells from DA and DA.1U congenic strains. (G) CD68+ cells (same as in F) stained with F16-4-4. Vertical lines in scatterplots show mean values. Representative results of at least two independent experiments are shown. The association of RT1-Aa to TAP-B has been shown to cause a relative retention of class I molecules in the ER [25], [29], implying that the level of class I should be higher intracellularly in DA.1HR10, and possibly also in DA.1HR83. However, we found that the intracellular levels of class I in these strains were also reduced compared to DA and DA.1H (Fig. 5C). These observations were also confirmed using F16-4-4 in DA.1HR10 (Fig. 5C), suggesting that both classical and inverse cim reduce the class I expression on the cell surface as well as inside the cell. DA.1IR85 showed extremely low extracellular levels of class I staining on all analyzed subsets of leukocytes as well as intracellularly in CD68+ cells (P<0.01; Fig. 5D, E). The intracellular levels of class I in lymphocytes (T and B cells), by contrast, did not differ between DA and DA.1IR85 (Fig. 5E), suggesting that at least one of the two RT1-A molecules in DA.1IR85 is not efficiently transported to the cell surface. It is difficult to fully evaluate the TAP restriction of RT1-An in DA.1IR85 since the parental strain, DA.1I, is itself a recombinant strain that expresses the Tap2A allele (Fig. 4). However, the class Ia genes in DA.1IR85, RT1-A1n and RT1-A2n, are naturally associated with Tap2B in BN, which does not show reduced class I expression compared to ACI (RT1-Aa) [42]. This suggests that TAP-A is not an optimal transporter of peptides for RT1-A1n and/or RT1-A2n and that these molecules in DA.1IR85, as well as in DA.1I, are affected by the inverse cim.
As expected, the class I expression in DA.1IR7 and DA.1FR9 (RT1-Aa, TAP-A), did not differ from DA (Table 1). Moreover, the Tap2B allele in DA.1UR10 has previously been shown to encode an inefficient transporter of peptides for RT1-Aa [61], which explains the low surface expression of class I in this strain (Fig. 5F,G). Leukocytes from the parental congenic strain DA.1U showed a trend towards lower levels of class I on the cell surface compared to DA (Table 1). This reduction in class I expression was statistically significant on CD68+ cells in DA.1U as well as in DA.1UR83 (Fig. 5F). Hence, the reduced expression of class I in DA.1UR83, which did not differ significantly from DA.1U (Fig. 5F,G), cannot be ascribed to polymorphisms in the Tap2 gene and suggest that the expression of RT1-Au is regulated at the transcriptional level.
Our data support a novel type of cim in which RT1-Ah and probably also RT1-An require TAP-B for optimal expression at the cell surface. However, we also found evidence for a cim-independent regulation of extracellular class I levels for RT1-Au, suggesting that regulation also takes place at the gene level.
Weak correlation between transcription and extracellular expression of class I
The variation in MHC class I protein expression between DA.1H and DA.1HR83 revealed that RT1-A1h and/or A2h were reduced in the context of TAP-A. However, whether the expression of MHC class I proteins in addition was influenced by a variation in RT1-A gene expression in the Tcs1 congenic strain was still unclear.
We therefore determined the expression levels in the spleen of the six protein-coding RT1-A genes shown in Table 3. We designed allele-specific primers based on published RT1-A sequences to avoid off-target amplification of RT1-CEM genes (Fig. 6A). Each target was amplified with 2–4 different primer sets and the levels of product were averaged on the gene level and compared to the expression of beta-2-microglobulin (B2M, Fig. 6B) and to three reference genes (Fig. 6C). We observed the highest transcript levels for RT1-A1n in DA.1IR85, which was ∼5-fold higher than RT1-A2n in this strain and ∼3-fold higher than RT1-Aa in DA. Hence, the high levels of intracellular MHC class I proteins in DA.1IR85 T and B cells (Fig. 5E) correlated with a high expression of RT1-A1n on the gene level. By contrast, such correlation could not be found for RT1-A2h in DA.1HR83 and RT1-Au in DA.1UR83, which had comparable expression levels to RT1-Aa in DA despite their significant reduction in protein expression (Fig. 5).
Fig. 6. Transcriptional regulation of MHC class I genes. 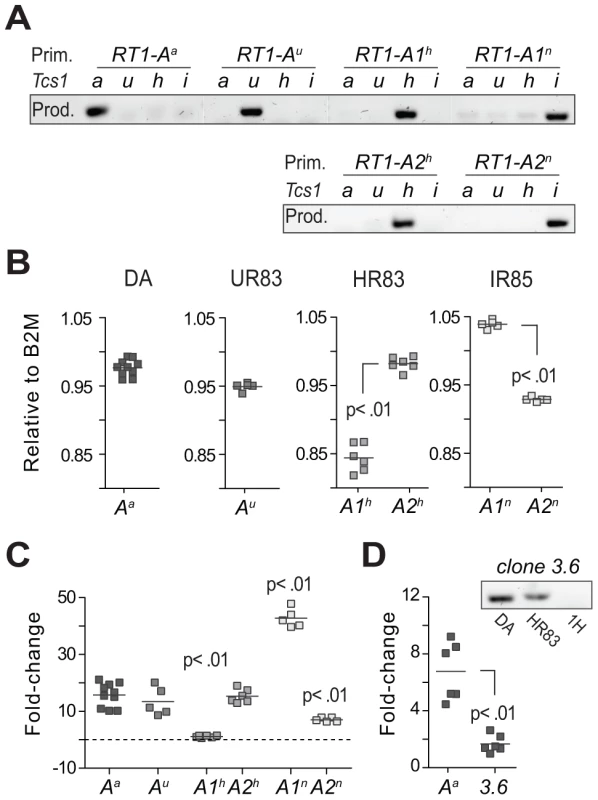
(A) Allele-specific primers (Prim.) used for quantitative RT-PCR showed minimal amplification of other RT1-A alleles and were therefore considered class Ia-specific (i.e. not cross-reacting to class Ib genes); PCR products (Prod.) are shown for DA (a), DA.1UR83 (u), DA.1HR83 (h) and DA.1IR85 (i). (B) Expression in spleen of RT1-A genes in DA and Tcs1-congenic strains relative to the expression of beta-2-microglobuline (B2M). (C) Variation (fold-change) in RT1-A gene expression between different congenic strains. Data show the mean expression of 2–4 different primer sets per target gene after normalization to 3 reference genes (Table S3). Significant differences compared to RT1-Aa. (D) Expression of RT1-Aa (class Ia) and clone 3.6 (class Ib) in spleen from DA rats. The amplification of a product in DA.1HR83 but not in DA.1H (1H) indicates that the primers for clone 3.6 are not cross-reacting to the RT1-Aa gene in DA (adjacent figure). We also compared the expression of class Ia and Ib genes in DA and found ∼6-fold higher levels of RT1-Aa compared to clone 3.6 (Fig. 6D), which was the only class Ib gene that was identified at the protein level (Table 3). Finally, we assessed the impact of Tcs1 on genome-wide transcription by exon-microarray. This showed that all genes that were differentially expressed between DA and DA.1IR83 (which has a slightly larger congenic fragment compared to DA.1IR85) were located within the Tcs1 region (Fig. S9). Hence, the phenotypic variation associated with Tcs1 is unlikely to be due to trans-acting factors outside of the QTL.
Taken together, the extracellular expression of class I proteins correlated poorly with the expression at the gene level. These data therefore support the conclusion that class I expression on the cell surface is largely regulated by polymorphisms affecting the peptide binding pocket of the RT1-A molecules and the TAP transporter.
Regulation of MHC expression by dendritic cells is similar in the thymus and spleen
The genetic variation in Tap2 influenced the extracellular expression of class I molecules in the spleen. Next, we determined the expression of MHC on thymic DCs, which control the egress of T cells by eliminating self-reactive cells during negative selection [62].
Comparing the class I expression at the protein level between thymic DCs (Fig. 7A) and CD68+ cells in the spleen (Fig. 5) showed that the variation between the strains was similar in both organs. This suggests that cim affects a broad repertoire of cells in different tissues. Likewise, the variation in class II expression on DCs in Tcs2-congenic strains was similar in the spleen (Fig. 7B) and in the thymus (Fig. 7C), as well as between DCs and B cells in the spleen (Table 1). Thus, we established that the genetic variation in Tap2 also affects the extracellular class I expression on antigen presenting cells in the thymus. We next aimed to determine if Tap2 had a pleiotropic effect and was also responsible for the variation in CD4∶CD8 lineage commitment.
Fig. 7. Tcs1 and Tcs2-congenic strains show similar variation in extracellular MHC expression in thymus and spleen. 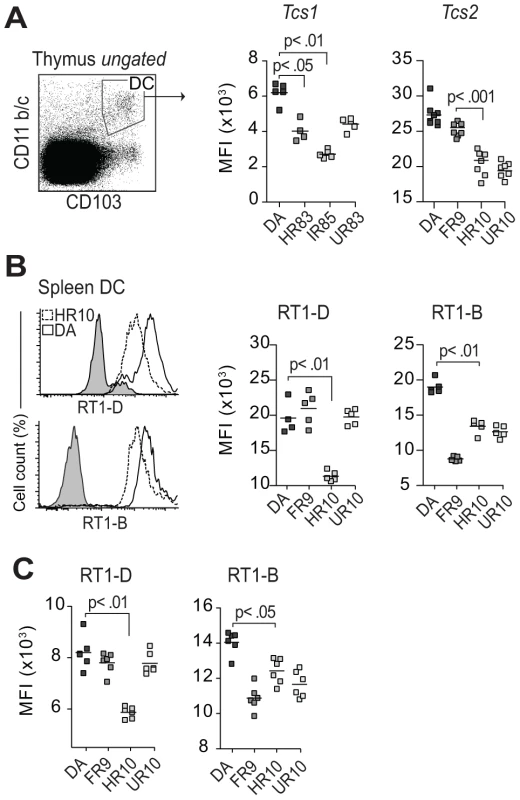
(A) Thymic conventional DCs (CD103+, CD11b/c+) stained extracellularly for class I (OX18). Scatterplots show results from two different experiments with Tcs1 and Tcs2-congenic strains. The variation between the strains is comparable to data shown in Figure 5 for CD68+ cells. (B–C) Splenic DCs (CD103+, CD68+, CD11b/c+) (B) and thymic DCs (C) stained extracellularly for RT1-D (OX17) and RT1-B (OX6). Histograms show representative samples from DA (solid lines) and DA.1HR10 (dashed lines). Classical cim affects lineage commitment and negative selection of CD8 T cells
Classical and inverse cim influenced class I expression on thymic DCs. We hypothesized that the changes associated with cim in class I expression and/or in the quality of the peptide repertoire would affect thymic selection and thereby contribute to the variation in peripheral CD4 and CD8 T cell numbers.
The different maturation stages of thymocytes are defined by the expression of various surface markers, which differ between rats, mice and humans [63], [64]. In the rat, double negative (DN) cells are defined according to the expression of CD45RC and CD2 [65] (see also Figure S10). We first determined the progression from the DN to the double positive (DP) stage in 6.5 week-old Tcs1 - and Tcs2-congenic rats. We found no variation in the total number of DN cells (Fig. 8A, shown for Tcs2-congenic strains), while the frequency of early thymic precursors (CD45RC+, CD2lo) was reduced in DA.1FR9 compared to all other strains (Fig. 8B, shown for DA vs. DA.1FR9). Moreover, all strains showed similar frequencies of immature CD8α single positive (ISP) cells (data not shown), whereas DA.1UR83 showed a greater proportion of cells at late DN stage (corresponding to DN4 in the mouse), in which all cells express TCRβ on the cell surface (Fig. 8C, shown for DA vs. DA.1UR83).
Fig. 8. Class-I modification reduces negative selection of CD8 cells. 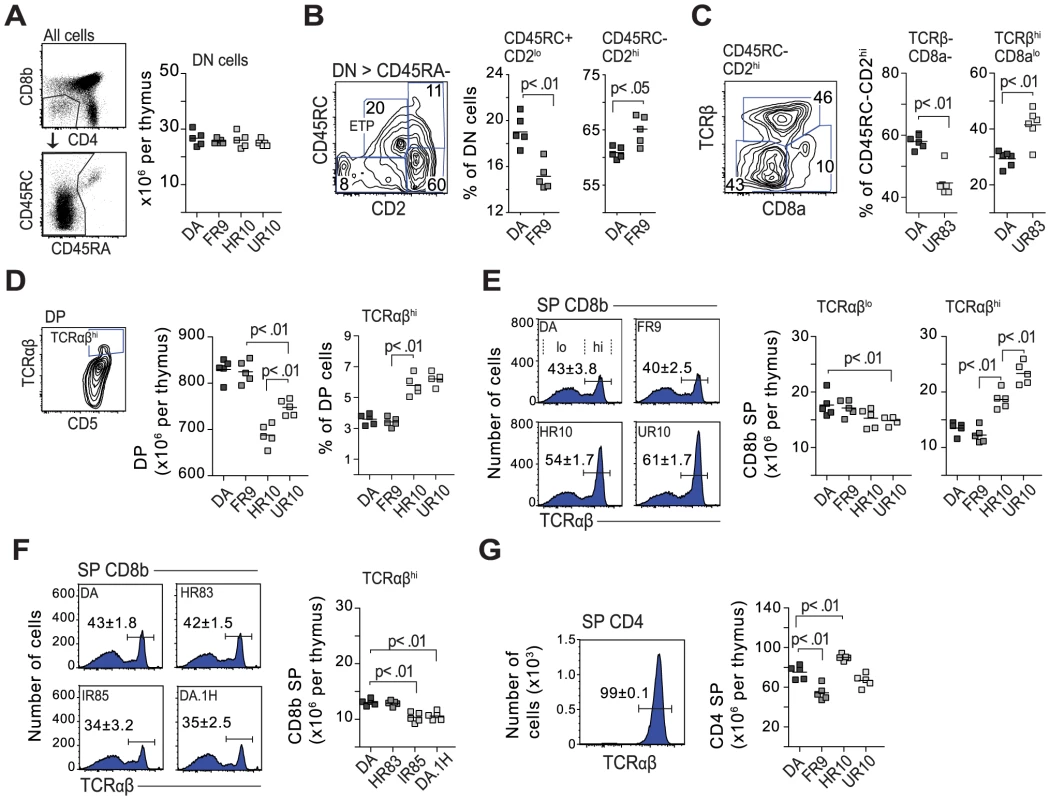
(A) The QTLs in Tcs1 and Tcs2 did not affect the total number of double negative cells (DN; CD4−, CD8b−). B cells (CD45RA+) were excluded from the DN gate. (B) DN cell maturation is defined by CD45RC and CD2 (see Fig. S10). DN cells in DA.1FR9 showed a lower frequency of early thymic precursors (ETP) compared to DA. Counter plot shows gating strategy with numbers indicating percent (%) of parent population (stated above plot). (C) Counter plot shows CD45RC−, CD2hi DN cells from DA.1UR83, and scatter plots the frequency of TCRβ− and TCRβ+ (DN4 in mouse) cells in DA and DA.1UR83. (D) Thymi from Tcs2-congenic strains with TAP-B (HR10 and UR10) contained fewer double positive (DP) cells but more cells (in %) with high TCR expression. (E) TAP-B strains (HR10 and UR10) showed higher frequencies (histograms) and total numbers (scatter plots) of CD8 single positive (SP) cells with high TCR expression. Numbers (%) in histograms represent mean-values ±SD of cells with high TCR expression (gated, n = 5). (F) Frequencies (histograms; n = 5) and total numbers (scatter plot) of CD8SP cells with high TCR expression in DA.1H (RT1-Ah, TAP-B) and in strains with low levels of surface MHC class I (HR83 [RT1-Ah, TAP-A] and IR85 [RT1-An, TAP-A]). (G) Virtually all CD4SP cells express high levels of TCR. Histogram shows expression of TCR on CD4SP thymocytes in DA (n = 5). Scatterplot shows total number of CD4SP cells per thymus in Tcs2-congenic strains. Positive selection takes place in the thymic cortex where DP cells expressing intermediate levels of TCR interact with MHC expressing stromal cells. DP cells that express a functional TCRα chain, which can pair with TCRβ and recognize self-MHC, are positively selected. We determined the total number of DP cells in the thymus and found lower numbers in the strains expressing RT1-Aa and TAP-B (DA.1HR10, DA.1UR10) (Fig. 8D). In addition, DP cells in these strains showed a greater proportion of mature cells, which express high levels of TCR and CD5 (Fig. 8D) [64]. Such a reduction of DP cells was not observed in other strains with low expression of class I, such as DA.1IR85 (RT1-An, TAP-A) (data not shown), suggesting that it is not the lower expression of class I that influences the number of DP cells in DA.1HR10 and DA.1UR10 but rather the quality of the class-I peptide repertoire.
Next, we determined how genes in the MHC-I and II regions influence lineage commitment and negative selection. TCR cross-linking of DP cells in vitro has been shown to generate CD8SP cells in the rat and CD4SP cells in the mouse [66], [67]. As shown in Figure 8E, ∼60% of CD8SP cells in DA express low or intermediate levels of TCR, which is in marked contrast to CD8SP cells in the mouse where essentially all are TCRhi [68]. However, both the frequency and the total number of CD8SP TCRhi cells varied substantially among the strains: the majority of CD8SP cells in strains with TAP-A expressed low TCR levels whereas most CD8SP cells in DA.1HR10 and DA.1UR10 (TAP-B) were TCRhi (Fig. 8E and 8F). A similar increase in TCRhi expressing CD8SP cells was not observed in DA.1H (RT1-Ah, TAP-B) (Fig. 8F), indicating that the increase of CD8SP cells in DA.1HR10 (RT1-Aa, TAP-B) is cim-dependent. Neither was it observed in Tcs1-congenic strains with low surface expression of class I (DA.1HR83, DA.1IR85; Fig. 8F), which suggests that it is the quality of the peptide repertoire and not the lower expression of class I that is responsible for the increase of CD8SP TCRhi cells in DA.1HR10 and DA.1UR10.
In contrast to CD8SP cells, virtually all CD4SP cells were TCRhi (Fig. 8G). All Tcs1-congenic strains had similar numbers of CD4SP cells (data not shown), while we observed an increase of total CD4SP cells in DA.1HR10 and a decrease in the same subset in DA.1FR9 compared to DA and DA.1UR10 (Fig. 8G). The variation in the number of CD4SP cells probably reflects the genetic variation in the classical MHC class II genes, in particular in the RT1-B genes since the RT1-D genes are conserved between DA and DA.1FR9 (Fig. 4). It is further unlikely that the variation in CD4SP cells is cim-dependent since only DA.1HR10, and not DA.1UR10, showed an increase in CD4SP cells.
These data suggest that classical cim reduces negative selection of class I restricted thymocytes, probably by limiting the complexity of available peptides in the ER. In addition, cim may influence positive selection as shown by the reduced number of DP cells in DA.1HR10 and DA.1UR10.
Classical cim influences the number of peripheral CD8 T cells
Class-I modification was associated with an increased number of CD8SP TCRhi cells in the thymus. However, it was unclear whether this also led to an increase of CD8 T cells in the periphery. Thus, we assessed the number of CD8 recent thymic emigrants (RTEs), which in the rat can be distinguished by their expression of CD90 (Thy-1) and CD45RC [69], [70], in the spleen of 6.5 week-old Tcs2-congenic rats. The total number of CD8 RTEs was found to be significantly increased in DA.1HR10 and DA.1UR10 (RT1-Aa, TAP-B) compared to DA and DA.1FR9 (RT1-Aa, TAP-A). Furthermore, the number of CD8 RTEs in the spleen correlated significantly with the number of CD8SP TCRhi cells in the thymus (R2 = 0.83; P<0.001), indicating that CD8SP TCRhi cells constitute a mature subset of thymocytes. CD8SP cells with low or intermediate TCR expression, by contrast, showed a weak inverse correlation with CD8 RTEs in the spleen (data not shown). Similarly to the CD8 RTEs, the CD4 RTEs in the spleen strongly correlated with CD4SP cells in the thymus (Fig. 9B). Differences in CD4 and CD8 RTEs were less pronounced in older rats (13–14 weeks of age), which also showed less, and slightly different, variations in total CD4 and CD8 T cell numbers compared to younger rats (Fig. S11). This emphasizes that other mechanisms, such as clonal expansion, may be more important than thymic output for maintaining the proportions of CD4 and CD8 T cells in older rats.
Fig. 9. Classical cim influences the number of CD8 recent thymic emigrants. 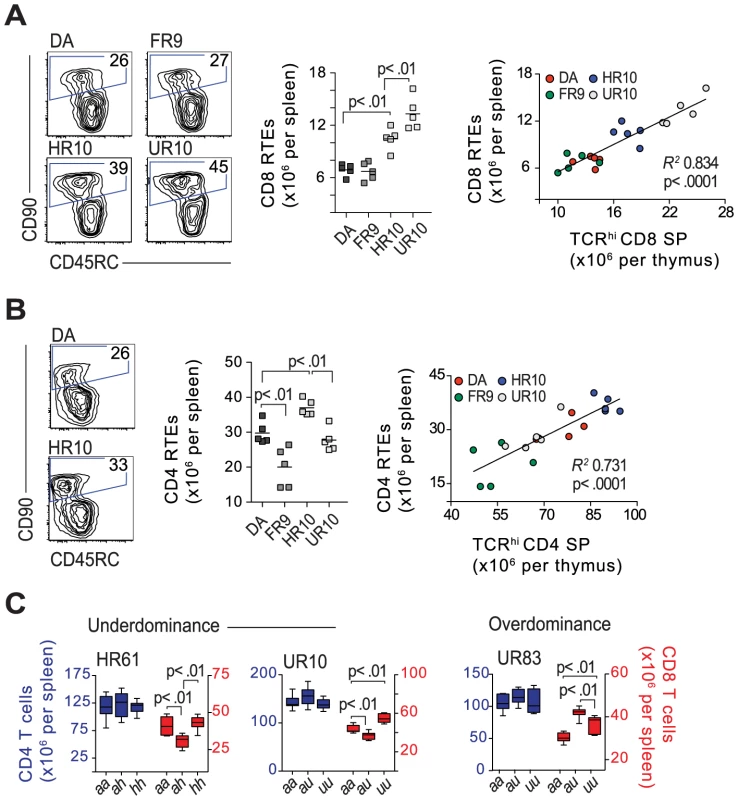
(A) Recent thymic emigrants (RTEs) express CD90 and low levels of CD45RC. Counterplots show CD8 RTEs (gated) in the spleen of 6.5-week old congenic and DA rats (representative samples, numbers in gates show percent of CD8 T cells). Scatterplot shows total number of CD8 RTEs per spleen, which correlates to the absolute number of CD8SP TCRhi cells in the thymi from the same animals (far right). (B) The corresponding staining (as shown in A) for CD4 RTEs, including total numbers (scatterplot) and correlation to CD4SP TCRhi cells in the thymus. (C) Absolute numbers of CD4 (blue) and CD8 (red) T cells in the spleen of 13-week old rats. Strains and genotypes are shown above and below graphs, respectively; n = 5–7 per group. We further compared CD8 T cell numbers between heterozygous and homozygous DA.1HR61 (Fig. 2) and DA.1UR10 rats (both Tap2B). Since the Tap2B allele is recessive [29], it was expected that rats with heterozygous Tap2 alleles had fewer CD8 T cells in the spleen compared to their Tap2B homozygous littermates (Fig. 9C). However, they had also fewer CD8 T cells than their DA littermates, a phenomenon known as underdominance, which has not previously been reported for T cell selection. The impact of heterozygosity was the opposite in DA.1UR83 (Tap2A), in which heterozygous rats had more CD8 T cells than their homozygous littermates (so called overdominance) (Fig. 9C). Hence, these phenomena were not directly related to cim but seem to be specific for the CD8 lineage, and may reflect the default CD8 lineage choice that has been reported in the rat [66], [67].
Taken together, we show that cim reduces negative selection of CD8 cells, increases the thymic output of CD8 T cells and thereby significantly contributes to the peripheral CD8 T cell repertoire.
Discussion
We show that two linked haplotypes in the MHC-I and II regions control MHC expression and T cell selection in rats. A recombination between these haplotypes, which broke the evolutionary conserved linkage between RT1-A and Tap2, affected negative selection and lineage commitment of CD8 cells. This effect was found to be associated with the Tap2B allele and dependent on the co-expression of RT1-Aa. The same combination of alleles has previously been shown to reduce the expression and alter the antigenicity of the RT1-Aa molecule, a phenomenon known as class-I modification (cim) [61]. We found in addition that the combination RT1-Ah and Tap2A, encoding for the promiscuous TAP-A transporter, also reduced the extracellular expression of class I, which we term inverse cim. Thus, our results show how natural polymorphisms in Tap2 modify class I expression and alter T cell selection in rats.
Following the identification of Tap2 as a trans-acting modifier of RT1-Aa antigenicity [25], it has been thoroughly investigated how different TAP transporters are associated with changes in RT1-A protein expression and the RT1-A peptide repertoire. These studies suggest that TAP-A and TAP-B are equally efficient in transporting peptides with hydrophobic C-terminal residues, while TAP-B has weak affinity for peptides with basic C-terminal residues [18], [22], [26]–[28]. These differences were exploited here to assess how an altered spectrum of class I peptides influences T cell selection in animals with natural variations in the TCR and MHC loci.
Thymic DCs in the corticomedullary junction present antigens to positively selected thymocytes ([71], reviewed in [72]). Thymocytes with high affinity for self-peptide:MHC complexes are eliminated through negative selection. However, if the peptide repertoire on the DCs is limited, as in DA.1HR10 and DA.1UR10 (RT1-Aa, TAP-B), fewer thymocytes will express TCRs with high affinity for peptide:MHC complexes and fewer cells will therefore undergo negative selection. We showed in addition that less negative selection in rats with RT1-Aa and TAP-B allotypes correlated with an increased number of CD8 RTEs in the periphery. Thus, it seems possible that cim may lead to an escape of autoreactive CD8 T cells. Preliminary results from our laboratory, however, show that CD8 cells from DA.1HR10 and DA.1UR10 produce lower levels of pro-inflammatory cytokines compared to strains expressing RT1-Aa and TAP-A (Tuncel and Haag, unpublished data).
Although the major effect of cim appears to be during negative selection, it also influenced the number of DP cells. This finding is consistent with studies in fetal thymus organ cultures in which a complex mixture of class-I peptides has been shown to increase positive selection [68], [73]. Hence, it seems reasonable that fewer thymocytes will be positively selected in strains, such as DA.1HR10 and DA.1UR10, where the class I peptide repertoire is limited by the restrictive TAP-B transporter. In contrast to negative selection, positive selection is largely dependent on cortical thymic epithelial cells (cTECs). Interestingly, in the mouse it has been shown that thymic DCs express 10× higher levels of MHC class I than cTECs [74]. This latter subset is still poorly characterized in the rat and we have therefore not been able to determine the expression of class I on these cells. However, it seems likely that different allelic variants of Tap2 would also affect the expression of class I on epithelial cells. The increased number of DP thymocytes should therefore be further investigated considering the lower expression of MHC class I described on cTECs in the mouse and the fact that the processing of antigens in these cells is different due to a thymoproteasome specific subunit encoded by PSMB11 [75], [76].
Cim did not affect TCR selection at the early DN stage. All strains, regardless of Tap2 genotype, had similar number of DN cells and normal transition from early thymic precursors to more mature TCRβ+ cells (DN4). By contrast, the frequency of early thymic progenitor cells was reduced in DA.1FR9, which is congenic for the Tcs2 interval as well as the entire MHC-III region and parts of the non-classical MHC class Ib (RT1-CEM) region. A potential candidate in the MHC-III region, which could influence the regulation of DN cells in this strain, is Notch4. Notch4 is expressed in the thymus [77] and has been shown to influence lymphoid progenitor fate [78] while another Notch family member, Notch1, has been shown to determine early commitment to T cell lineage [79]. We have recently obtained a new RCS, DA.1FR61 (Fig. 2), which excludes Notch4 and future studies on this strain will help delineate whether Notch-4 is involved in the regulation of DN cells.
We did not find evidence for a correlation between class I expression levels and T cell selection. All Tcs1-congenic strains showed low levels of extracellular class I expression (compared to DA) but varied widely in their CD8 cell numbers. Damoiseaux et al. suggested that the expression of two RT1-A genes in the BN strain, compared to one in LEW, was responsible for the increased negative selection of CD8 cells in the LEW.1N strain [36]. This reduction of CD8 cells is probably associated with the RT1-A locus (RT1-An) in LEW.1N and thus the same phenotype as we mapped in DA.1IR85. We determined the expression of the two RT1-A genes (A1n and A2n) in DA.1IR85 and although both genes were highly expressed, the level of class I protein at the cell surface was low compared to DA. However, the low surface level of class I in DA.1IR85 is probably dependent on the presence of TAP-A in this strain, while RT1-An in LEW.1N is associated with TAP-B. Thus, it is unlikely that the reduction of CD8 cells in DA.1IR85 and LEW.1N [36] is associated with the levels of class I expression on the cell surface. However, that CD8 selection would be influenced by the number of protein-coding class I genes, as proposed by Damoiseaux et al. [36], remains an attractive hypothesis. It is further interesting to note that also DA.1HR83 expresses two RT1-A genes and has low numbers of CD8 cells, while DA and DA.1UR83, which have higher numbers of CD8 cells, only express a single RT1-A gene each. However, whether it is the number of expressed class I genes or the increased diversity of class I peptides that promote clonal deletion of CD8 cells remains to be tested in more strains.
We further noted that QTLs for the expression of class I (Q2 on RNO2, Q6 on RNO10) and class II (Q1 on RNO2, Q7 on RNO17) did not co-localize with QTLs for the CD4∶CD8 T cell ratio in the HS rats. Thus, Tap2 remains, at least to our knowledge, the only naturally selected non-classical MHC gene that has been found to be associated with both variations in MHC surface expression and CD4∶CD8 lineage commitment. In addition to the MHC, a second major QTL controlling CD4∶CD8 T cell variation was identified within a narrow region on chromosome 9. A gene at this locus, Satb1, is predominantly expressed in the thymus [80]. Reduced expression of Satb1 at the protein level has been shown to be associated with a reduction in the frequency of CD8SP cells in the thymus as well as in the periphery [81]. Variation in CD8 T cells in the HS rats, however, did not correlate with the allelic distribution of Satb1, nor did the gene expression of Satb1 in the thymus of 136 HS rats (data not shown).
A strong correlation similar to that we showed for Tap2 and CD8 selection could not be found for any gene in Tcs2 and CD4 selection. Apart from the extensive diversity we observed in RT1-Db1, Ba and Bb, several non-synonymous SNPs were in addition identified in RT1-DOb. This gene encodes one subunit of the non-classical class II molecule DO. The human homologue, HLA-DO, which has been shown to be expressed in the thymus [82], competes with the peptide editor HLA-DM for the binding of MHC class II molecules [83], [84]. Hence, polymorphisms that alter the affinity of DO for MHC class II may also affect the peptide repertoire of the MHC class II molecule and, thus, the selection of T cells. We found in addition evidence on transcript and protein level for a second RT1-Db locus (Db2), which is probably the rat ortholog of murine Eb2 [85]. Similar to the mouse gene, Db2 appears to be highly conserved among inbred rat strains. The genetic interaction between allotypes of TAP and RT1-A for the selection of CD8 T cells raises the question whether a similar interaction exists between the two highly polymorphic class II molecules (RT1-B and RT1-D) and alleles of RT1-DOb for the selection of CD4 T cells. Such interaction could be one explanation for the strong LD between these genes.
It has been a long-standing question why the rat has two TAP variants [24], in particular since the restrictive TAP-B transporter does not seem to offer advantages over the promiscuous TAP-A transporter. The redundancy of the TAP-B transporter has also been shown experimentally in the PVG.R23 recombinant strain in which RT1-Au, which is typically found associated with TAP-B, is expressed together with TAP-A [61]. In this strain, the association with TAP-A did not influence the expression of class I, which is consistent with our results in DA.1UR83. It was therefore somewhat surprising that certain other RT1-A allotypes appear to be dependent on the association with TAP-B for optimal expression on the cell surface. We termed this phenomenon inverse cim and showed that the extracellular expression of RT1-Ah is reduced when the molecule is associated with TAP-A compared to TAP-B. The same seems to be true for the RT1-An molecule(s) in DA.1IR85, although the linkage to TAP-A in the parental strain made it difficult to further address this question. One of the antibodies, F16-4-4, used to determine the inverse cim for the RT1-A molecules in DA.1HR83 has been shown to bind the A1h but not the A2h isoform (Anne France Le Rolle, personal communication). Thus, the inverse cim reduces the expression of A1h while it remains to be investigated whether it also reduces the expression of A2h.
The expression of RT1-A at the gene level did not correlate well with the protein expression on the cell surface and did not explain the low levels of extracellular class I in DA.1HR83 and DA.1IR85. In DA.1HR83, the expression of A2 was ∼15-fold higher compared to A1, which is inconsistent with the idea that if any allele is expressed at the A1 locus, this allele will function as the principle class Ia molecule [18]. However, since both isoforms were detected at the protein level, it remains possible that A1 indeed is the dominant class Ia molecule in DA.1HR83, despite being expressed at a lower level. By contrast, RT1-A1n showed a 5-fold increased expression over RT1-A2n in DA.1IR85, and both isoforms were readily detected at the protein level. There was no evidence at the protein level for an A3 isoform, which may suggest that DA.1IR85 has not acquired all functional class Ia genes from BN. The class Ia molecules in DA.1IR85 further differed from DA.1HR83 by a high intracellular expression in lymphocytes, while a similar difference was not observed for myeloid cells. It should be noted that the OX18 antibody used for the intracellular staining of MHC class I also recognizes MHC class I in the absence of β2-microglobulin. Hence, the high intracellular levels of MHC class I in DA.1IR85 lymphocytes do not necessarily need to reflect an increase in mature peptide-loaded molecules. An interesting possibility is that the differences in intracellular class I levels between leukocyte lineages is regulated at the transcript level, which could suggest that the expression of A1 and A2 in DA.1IR85 is lineage specific. This has indeed been shown previously in rhesus macaques where specific class Ia transcripts have been associated with either myeloid or lymphoid lineages [86].
Co-evolution of genes in the class I and Tap loci is certainly not an isolated phenomenon in the rat, although the class I genes are separated from the TAP genes by the class II region in the majority of mammals. The class I molecules in both humans and mice have evolved to accommodate peptides that are supported by their associated TAP transporters, while the Tap1 and Tap2 genes themselves have remained functionally monomorphic. By contrast, functional alleles of Tap2 that have co-evolved specifically with different alleles of class I have been described in the chicken [87]. The Tap genes in the chicken are directly flanked by class I genes [88], which probably have preserved the linkage. In the rat, by contrast, the Tap2 and class I loci are separated by a ∼250 kb interval, which is relatively susceptible to recombinations (Fig. 3). On the contrary, the recombination activity between Tap2 and the class II genes is extremely low as shown in this study. This may suggest that alleles of Tap2 have been conserved in the rat in cis-configuration with alleles in the RT1-B and RT1-D loci. This linkage may not have been maintained in species that lack functional polymorphisms in the Tap genes, such as in the mouse [89]–[91] and in humans [92]. Highly conserved MHC-II haplotypes are also evident in many inbred and partially outbred rat strains [13], with the only exception to our knowledge being the WRC strain that has a recombination in the RT1-B locus [93]. In addition, several related inbred strains, which have been derived from outbred stocks, have recombinations between Tap2 and RT1-A, e.g. LN (Tap2B) vs. LL (Tap2A), and FHL (Tap2B) vs. FHH (Tap2A) [13]. With an increasing number of genome sequences available, and the possibility to obtain sequence information from wild rats, it might be possible to investigate whether the strong linkage between Tap2 and the class II genes is associated with certain haplotypes and bears an evolutionary advantage.
In summary, we mapped two QTLs associated with variations in CD4∶CD8 lineage commitment and MHC expression to the MHC-I and MHC-II region in the rat. A recombination between these two regions modifies class I expression by breaking the linkage between co-evolved RT1-A and Tap2 alleles, which previously has been described as class-I modification (cim). We demonstrate a novel type of cim and also show that certain combinations of RT1-A and Tap2 alleles are not affected by cim. Furthermore, we show that cim had a pronounced effect on thymic selection. Cim did not influence DN stages, but decreased the number of DP thymocytes. Most importantly, cim rescued CD8 T cells from negative selection and thereby increased the number of CD8 T cell in the periphery.
Materials and Methods
Ethics statement
All experiments involving animals were approved by the local ethics committees at Karolinska Institutet, Stockholm, or by the Spanish legislation on “Protection of Animals Used for Experimental and Other Scientific Purposes” and the European Communities Council Directive (86/609/EEC) on this subject.
Animals
NIH-HS
The origin of the strains and the outbreeding regime that was used to create and maintain the NIH-HS have been described elsewhere [42]. Briefly, the colony was founded from 8 inbred progenitor strains: BN/SsN (RT1n), MR/N (RT1d), BUF/N (RT1b), M520/N (RT1b), WN/N (RT1l), ACI/N (RT1a), WKY/N (RT1l), and F344/N (RT1lv1) [37]. Animals were housed in open polycarbonate (Makrolon) cages at a temperature of 22°C±2°C and with 12 h light-dark cycle and fed standard rodent chow and tap water ad libitum.
Congenic strains
Inbred DA/Ztm rats were obtained from the Zentralinstitut für Versuchstierzucht (Hannover, Germany) and DA/OlaHsd from Harlan Europe (Horst, the Netherlands). Rats were maintained in a barrier facility by sister-brother mating and were specific pathogen free according to the current FELASA guidelines. Animals were kept in a climate-controlled environment with 14 h light/10 h dark cycles, in individually ventilated microisolator-cages (Allentown Inc. NJ, USA) containing wood shavings and fed standard rodent chow and microfiltered water ad libitum. Congenic strains were originally established on DA/Ztm background (N>20) and thereafter further backcrossed (N>5) to DA/OlaHsd. The RCS were produced by crossing F1 hybrid rats. The derivation of DA.1FR9, DA.1FR10, DA.1FR8 and DA.1FR5 from congenic DA.1F (DA.LEW-RT1f) has been described previously [94]. RCS with MHC haplotype RT1u were derived from DA.1U, which was generated by introgression of the corresponding E3/ZtmRhd fragment on chromosome 20. RCS with MHC haplotypes RT1i and RT1h were derived from DA.1I and DA.1H, respectively (both established at the Zentralinstitut fur Versuchstierzucht). The RT1i haplotype originates from the now extinct BI (formerly called B3) strain [95] and the RT1h haplotype from the KHW strain. DA.1I contains a derivative MHC haplotype (RT1-AiBaDa); however, several STRs and SNPs in the MHC-II region and, to a lesser extent, in the MHC-III and Ib regions are polymorphic between DA.1I and DA. RT1h is a standard haplotype [56] but most genetic variants in the MHC-II region are shared with RT1n (this study). Several genes in the MHC-Ia region (including RT1-A1 and RT1-A2 [sequences from GenBank]) and the MHC-III region, however, are unique for RT1h (Tuncel and Yau, unpublished data). Additional information about strains can be found on our website http://www.inflam.mbb.ki.se/rat/MHC. The QTLs described have identification numbers 7175096 (Tcs1) and 7175099 (Tcs2) in the Rat Genome Database (RGD) (http://rgd.mcw.edu/rgdweb/search/qtls.html).
Genotyping
NIH-HS
Information on polymorphic SNPs was provided through the STAR consortium that identified SNPs and haplotypes in the rat to assist complex trait analysis as part of the EURATRANS consortium. DNA was extracted from liver biopsies using a standard proteinase K protocol. Genotyping was performed using a custom-designed high density Affymetrix SNP genotyping array (RATDIV), which is based on sequence information from 13 inbred rat strains. The array interrogates 803,485 SNPs of which 265,551 polymorphic high quality SNPs were chosen for the reconstruction of the HS chromosomes as a mosaic of the founder haplotypes as described below.
Congenic strains
Genomic DNA was extracted from biopsies as has been described previously [94]. PCR primers for short tandem repeats (STRs) and SNPs were retrieved from the rat genome sequence 3.4/rn4 2004 assembly and designed using PrimerSelect 8.1.3 (Lasergene, DNAstar Inc., WI, USA). Forward primers were fluorescently labelled with 6-FAM, HEX, VIC, PET or NED (MWG Biotech, Riskov, Denmark). STRs were amplified with PCR according to standard protocols, diluted in HPLC water, combined with the size standard Liz-600 in 10 µl HiDi formamide (both from Applied Biosystems, CA, USA) and analyzed on a 48-capillary 3730 DNA analyzer (Applied Biosystems). SNPs were genotyped by Sanger sequencing as described below.
Exon sequencing
RNA was extracted from spleen using the RNeasy Mini kit (Qiagen, Ballerup, Denmark) and treated with DNase I (Roche, Mannhein, Germany). Complementary DNA (cDNA) was synthesized with iScript (Bio-Rad, CA, USA). Genomic DNA (gDNA) was isolated from spleen by proteinase K digestion (AquaPure Genomic DNA kit, Bio-Rad). Primer sequences were obtained from the RefSeq 3.4 genome assembly. All sequences were obtained through conventional capillary sequencing except Btnl2, which was sequenced on the SOLiD platform (Applied Biosystems). For capillary sequencing, products were amplified in a 25 µl PCR reaction containing 0.5 µg DNA, 2 µl 2.5 mM dNTP (New England Biolabs), 1 µl 50 mM MgCl2, 0.1 µl Platinum Taq (Invitrogen, CA, USA) and 0.5 µl of each 10 µM primer. The products were purified using the Millipore's Montage Cleanup Kit and diluted in HPLC water or were isolated by agarose digestion after gel electrophoresis. Sequencing was performed with BigDye Terminator v3.1 according to instructions (Applied Biosystems) using 0.4 µM of single primers. Products were purified by ethanol precipitation, resuspended in 10 µl HiDi formamide and analyzed on a 48-capillary 3730 DNA analyzer. Nucleotide sequences have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession numbers KC222882–KC222951.
Flow cytometry
Analysis of PBMCs from NIH-HS rats has been described recently [41]. Briefly, phenotyping were performed in 8 cohorts of 230–270 individuals over 3 years. Rats had been subjected to behavioral tests (week 8–10) prior to blood sampling (week 13), but besides from a wound-healing test (puncture of the external ear) in week 7 and a glucose tolerance test (week 11), no invasive procedures had been performed. Twenty µl blood was stained for 20 min in duplicates with saturating concentrations of fluorescently-labeled monoclonal antibodies (MAbs, see below). After erythrolysis, cells were fixed for 20 min at RT in a 2% phosphate-buffered formaldehyde solution and then washed twice in PBS before acquisition.
Congenic strains
For analysis of MHC expression on DCs, tissues were cut into ∼1 mm3 cubes and incubated with 2 ml digestion buffer containing 2.5 mg/ml collagenase IV (Sigma-Aldrich, MO, USA), 0.2 mg/ml bovine pancreas DNase I (Roche Applied Science), 2% fetal calf serum (FCS, Gibco Laboratories, MA, USA) in Hank's Balanced Salt Solution (HBSS) (Sigma-Aldrich) for 20 min at 37°C. Undigested material was disrupted by pipetting, 2 ml fresh digestion buffer was added and the incubation was continued for 10–15 min. The digestion was stopped by adding EDTA (Merck) to a finale concentration of 20 mM. After incubation for 5 min in EDTA on an orbital shaker (100 rpm) at RT, the suspensions were filtered through 40 µm cell strainers (BD Falcon), and washed twice in ice-cold FACS buffer (calcium - and magnesium-free PBS-D supplemented with 1% FCS, 10 mM EDTA and 0.02% NaN3). For thymocyte and lymphocyte analyses, single-cell suspensions were prepared without collagenase/DNase I treatment, filtered through 40 µm cell strainers, washed twice in cold EDTA-FACS buffer and resuspended in the same buffer and counted on a Sysmex KX-21N. 106 cells/well were added in duplicates to 96-well v-bottom polypropylene plates (BD Falcon) and stained with saturating concentrations of MAbs (30 µl finale staining volume). The following Alexa Fluor 488, FITC, PE, Pe-Cy5, PerCP-Cy5.5, APC, APC-Cy7, Pe-Cy7, Alexa Fluor 648 and biotin conjugated antibodies were used: CD4 (OX35), CD8a (OX8), CD8b (341), CD45RA (OX22), anti-granulcoytes (His48), RT1-B (OX6), RT1-D (OX17), MHC class I (OX18) were purchased from BD Pharmingen (CA, USA); CD45 (OX1), CD45RA (OX33), CD90 (OX7), CD4 (W3/25), CD11b/c (OX42), CD103 (OX62) and αβ-TCR (R73) were purchased from BioLegend (CA, USA); CD68 (ED1) and RT1-A (F16-4-4, conjugated in-house with Alexa Fluor 647 using the APEX labeling kit, Invitrogen) were obtained from AbD Serotec (Düsseldorf, Germany). CD2 (OX34) and CD5 (OX19) were produced in-house. To stain for β2-microglobulin (using clone TLD-3H12B, BD), as an alternative to OX18, was evaluated but did not result in reproducible data. R73 (αβ-TCR) was used to gate TCRβ+ cells in the DN population (Fig. 8). LIVE/Dead Violet (Invitrogen) was included in all stains to exclude necrotic cells. For the intracellular staining of class I, unlabeled OX18 or F16-4-4 was used to block extracellular epitopes and the cells were thereafter incubated in BD Cytofix/Cytoperm for 20 min at RT. Cells were then washed twice in BD PermWash and stained intracellularly with labelled OX18 and F16-4-4. A SORP BD LSRII Analytic Flow Cytometer or a FACSCalibur (BD) were used for acquisition and the data was analyzed with FlowJo (Tree Star Inc., OR, USA). Significant differences between groups were analyzed using a non-parametric test (Mann-Whitney U).
Quantitative RT-PCR
RNA was extracted from 2×106 non-stimulated spleen cells using the RNeasy Mini kit (Qiagen) and treated with DNase I (Roche). RNA concentration was determined spectrophotometrically on a NanoDrop ND-1000 (NanoDrop Technologies, DE, USA), diluted to 60 ng/µl and reverse transcribed to cDNA using High Capacity cDNA Reverse Transcription (Applied Biosystems). Quantitative real-time PCR (qPCR) was performed on an ABI 7900HT (Applied Biosystems) using SYBR Green (Applied Biosystems) and a two-step PCR protocol (95°C for 10 min followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec). Allele-specific primers (Table S3) for RT1-Aa, RT1-Au, RT1-A1n, RT1-A2n, RT1-A1h, RT1-A2h and clone 3.6 were designed using NCBI/Primer-BLAST with sequences retrieved from public databases. The performances of all primers were evaluated prior to use to ensure that all PCR reactions were performed at comparable efficiencies. In the final experiments, each target was amplified using 2–4 different primer pairs and the cycle threshold (Ct) values were then averaged at the gene level. Average Ct-values were compared to beta-2-microglobuline or normalized to the geometric mean of the reference genes Arbp, Hprt-1 and Mdh-1 (Table S3), whereafter fold-change variations were determined using the relative quantification method (ΔΔCt).
Microarray
Array hybridization
Thymi and inguinal lymph nodes (iLN) were removed from DA.1IR83 rats and non-congenic littermates (n = 6) immediately post mortem. To minimize the risk of blood contamination in the thymi, the aorta was first incised below the chest to allow blood to drain into the abdominal cavity. Whole organs were excised, instantly frozen in liquid nitrogen and stored at −70°C until prepared. RNA extraction and array hybridization were performed as previously described [96]. Briefly, total RNA was extracted using TRIzol reagent, and further purified and DNase I treated using an RNeasy Mini kit (Qiagen) and RNase-Free DNase Set (Qiagen), according to the manufacturer protocols. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). A total of 1 µg RNA was used for array hybridization against Affymetrix GeneChip Rat Exon 1.0 ST Arrays in accordance with the recommendations of the manufacturer (Affymetrix).
Data analysis
CEL intensity files were produced using GeneChip Operating Software version 1.4 (Affymetrix) and quality tested using the Affymetrix Expression Console. Detection of differential expression was performed at the gene level using the Partek Genomics Suite 6.4 (Partek Incorporated, MO, USA) and summarized at the gene level using a One-Step Tukey's Biweight Algorithm.
Mass spectrometry
For the detection of class I derived peptides, single cell suspensions were prepared from spleens in PBS after lysis of erythrocytes. Cells were washed and taken up in 5% FCS supplemented DMEM-medium. 30 million cells were incubated for 7 hours at 37°C with 10 ng/ml IFN-γ, cells were washed in PBS and subsequently lysed in PBS containing 1.2% (w/v) CHAPS (GE Healthcare Life Sciences) and protease inhibitors (Complete, Roche Applied Science). RT1-D was immunoprecipitated from spleen cell-lysate using the monoclonal antibody OX-17. Proteins were reduced using 5 mM dithiothreitol (DTT) in 50 mM NH4CO3 for 10 min at 90°C and alkylated with 10 mM iodoacetamide (IAA) for 30 min at room temperature in the dark. Protein was precipitated overnight at −20°C in 95% acetone, precipitate was collected by centrifugation and washed with 80% acetone, 10% methanol, 0.2% acetic acid and incubated for 30 min at −20°C. Protein was pelleted by centrifugation and dissolved in 20 µl DMSO, diluted to 50 mM NH4CO3 and 30% DMSO with a final trypsin∶protein ratio of 1∶20 for in-solution digestion overnight at 37°C. Prior to MS analysis, the peptide mixture was dried, reconstituted in 5% formic acid, and cleaned using ZipTipC18 (Millipore) [97]. LC-MS/MS analyses were performed on an Easy-nLC system (Thermo Scientific, Bremen, Germany) directly on-line coupled to a hybrid QExcative Orbitrap mass spectrometer (Thermo Scientific). 1 µg of each sample was injected from a cooled auto sampler onto the LC column, peptide separation was performed on a 10 cm long fused silica tip column (SilicaTips, New Objective Inc., MA, USA) packed in-house with 3 µm C18-AQ ReproSil-Pur (Dr. Maisch GmbH, Ammerbuch, Germany). The chromatographic separation was performed using an acetonitrile (ACN)/water solvent system containing 0.2% formic acid with the following gradient set up: 5–35% ACN in 90 min, 35–95% ACN in 5 min and 95% ACN for 5 min all at a flow rate of 0.3 µl/min. The MS acquisition method consisted of one survey scan ranging from m/z 300 to m/z 1650 acquired in the FT-Orbitrap with a resolution of R = 70,000 at m/z 200, followed by ten consecutive data-dependent MS/MS scans from the top ten precursor ions with a charge state ≥2. The MS/MS scans were fragmented using HCD and acquired with a resolution of 17,500 at m/z 200. The instrument was calibrated externally according to the manufacturer's instructions and all samples were acquired using internal lock mass calibration on m/z 429.088735 and 445.120025. Mass lists were extracted from the raw data using the in-house written software Raw2MGF, and searched using the Mascot search engine (Matrix Science Ltd., London, UK) against a database consisting of the IPI rat database (v. 3.82), all accessions matching rat in the SwissProt database (downloaded 2011.04.05), as well as a protein FASTA file containing translated Sanger-sequenced regions of congenic MHC-II haplotypes (Fig. 4). The following parameters were used: tryptic digestion with maximum 2 miscleavages; carbamidomethylation of cysteines as a fixed modification; oxidation of methionine and glutamine to pyroglutamate conversions at peptide N-termini as variable modifications; precursor and fragment tolerance set to 10 ppm and 0.1 Da respectively.
QTL mapping and statistical analysis
Genetic analysis was performed using HAPPY (http://www.well.ox.ac.uk/happy/) to calculate the probabilities of descent from the eight HS founders as described previously in [98]. To account for relatedness, the Efficient Mixed-Model Association eXpedited (EMMAX) method was used [41]. The negative logP threshold necessary to achieve a false discovery rate (FDR) of 10% across all traits was estimated by simulation to ∼4.2. To account for the complex family structure in the HS, we alternatively used Bagphenotype [99], where each QTL is scored by its Resample-based Model Inclusion Probability (RMIP). Confidence intervals were calculated by simulation of phenotypes, each arising from a single QTL, in addition to a correlated genetic random effect and uncorrelated errors. All the genome scans and annotated QTLs are publicly available from the website http://mus.well.ox.ac.uk/gscandb/rat.
Supporting Information
Zdroje
1. FlajnikMF, KasaharaM (2001) Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15 : 351–362.
2. Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing (1999) Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature 401 : 921–923 doi:10.1038/44853
3. TrowsdaleJ, KnightJC (2013) Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet 14 : 301–323 doi:10.1146/annurev-genom-091212-153455
4. FernandoMMA, StevensCR, WalshEC, De JagerPL, GoyetteP, et al. (2008) Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet 4: e1000024 doi:10.1371/journal.pgen.1000024
5. GabrielSB, SchaffnerSF, NguyenH, MooreJM, RoyJ, et al. (2002) The structure of haplotype blocks in the human genome. Science 296 : 2225–2229 doi:10.1126/science.1069424
6. CullenM, PerfettoSP, KlitzW, NelsonG, CarringtonM (2002) High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet 71 : 759–776 doi:10.1086/342973
7. JeffreysAJ, NeumannR (2002) Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet 31 : 267–271 doi:10.1038/ng910
8. TraherneJA, HortonR, RobertsAN, MirettiMM, HurlesME, et al. (2006) Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet 2: e9 doi:10.1371/journal.pgen.0020009
9. HurtP (2004) The Genomic Sequence and Comparative Analysis of the Rat Major Histocompatibility Complex. Genome Research 14 : 631–639 doi:10.1101/gr.1987704
10. GibbsRA, WeinstockGM, MetzkerML, MuznyDM, SodergrenEJ, et al. (2004) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428 : 493–521 doi:10.1038/nature02426
11. AtanurSS, BirolI, GuryevV, HirstM, HummelO, et al. (2010) The genome sequence of the spontaneously hypertensive rat: Analysis and functional significance. Genome Research 20 : 791–803 doi:10.1101/gr.103499.109
12. GuoX, BrennerM, ZhangX, LaragioneT, TaiS, et al. (2013) Whole-genome sequences of DA and F344 rats with different susceptibilities to arthritis, autoimmunity, inflammation and cancer. Genetics 194 : 1017–1028 doi:10.1534/genetics.113.153049
13. AtanurSS, DiazAG, MaratouK, SarkisA, RotivalM, et al. (2013) Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell 154 : 691–703 doi:10.1016/j.cell.2013.06.040
14. WalterL, GüntherE (2000) Physical mapping and evolution of the centromeric class I gene-containing region of the rat MHC. Immunogenetics 51 : 829–837.
15. IoanniduS, WalterL, DresselR, GüntherE (2001) Physical map and expression profile of genes of the telomeric class I gene region of the rat MHC. J Immunol 166 : 3957–3965.
16. StrongRK, HolmesMA, LiP, BraunL, LeeN, et al. (2003) HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem 278 : 5082–5090 doi:10.1074/jbc.M208268200
17. HowcroftTK, SingerDS (2003) Expression of nonclassical MHC class Ib genes: comparison of regulatory elements. Immunol Res 27 : 1–30 doi:10.1385/IR:27 : 1:1
18. González-MuñozAL, Le RolleA-F, BrunH, HedrichHJ, WedekindD, et al. (2003) A novel instance of class I modification (cim) affecting two of three rat class I RT1-A molecules within one MHC haplotype. J Immunol 171 : 274–284.
19. JolyE, Le RolleAF, GonzélezAL, MehlingB, StevensJ, et al. (1998) Co-evolution of rat TAP transporters and MHC class I RT1-A molecules. Current Biology 8 : 169–180 Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=9443915&retmode=ref&cmd=prlinks.
20. BahramS, ArnoldD, BresnahanM, StromingerJL, SpiesT (1991) Two putative subunits of a peptide pump encoded in the human major histocompatibility complex class II region. Proc Natl Acad Sci USA 88 : 10094–10098.
21. PowisSH, MockridgeI, KellyA, KerrLA, GlynneR, et al. (1992) Polymorphism in a second ABC transporter gene located within the class II region of the human major histocompatibility complex. Proc Natl Acad Sci USA 89 : 1463–1467.
22. PowisSJ, DeversonEV, CoadwellWJ, CiruelaA, HuskissonNS, et al. (1992) Effect of polymorphism of an MHC-linked transporter on the peptides assembled in a class I molecule. Nature 357 : 211–215 doi:10.1038/357211a0
23. The distribution of Tap2 alleles among laboratory rat RT1 haplotypes (1994) The distribution of Tap2 alleles among laboratory rat RT1 haplotypes. 40 : 45–53 Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=8206525&retmode=ref&cmd=prlinks.
24. JolyE, ButcherGW (1998) Why are there two rat TAPs? Immunol Today 19 : 580–585 doi:10.1016/S0167-5699(98)01348-6
25. LivingstoneAM, PowisSJ, DiamondAG, ButcherGW, HowardJC (1989) A trans-acting major histocompatibility complex-linked gene whose alleles determine gain and loss changes in the antigenic structure of a classical class I molecule. J Exp Med 170 : 777–795.
26. MomburgF, RoelseJ, HowardJC, ButcherGW, HammerlingGJ, et al. (1994) Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature 367 : 648–651 doi:10.1038/367648a0
27. HeemelsMT, SchumacherTN, WonigeitK, PloeghHL (1993) Peptide translocation by variants of the transporter associated with antigen processing. Science 262 : 2059–2063.
28. PowisSJ, YoungLL, JolyE, BarkerPJ, RichardsonL, et al. (1996) The rat cim effect: TAP allele-dependent changes in a class I MHC anchor motif and evidence against C-terminal trimming of peptides in the ER. Immunity 4 : 159–165.
29. PowisSJ, HowardJC, ButcherGW (1991) The major histocompatibility complex class II-linked cim locus controls the kinetics of intracellular transport of a classical class I molecule. J Exp Med 173 : 913–921.
30. CosgroveD, GrayD, DierichA, KaufmanJ, LemeurM, et al. (1991) Mice lacking MHC class II molecules. Cell 66 : 1051–1066.
31. ZijlstraM, BixM, SimisterNE, LoringJM, RauletDH, et al. (1990) Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature 344 : 742–746 doi:10.1038/344742a0
32. ViretC, WongFS, JanewayCA (1999) Designing and maintaining the mature TCR repertoire: the continuum of self-peptide∶self-MHC complex recognition. Immunity 10 : 559–568.
33. KirbergJ, BernsA, Boehmer vonH (1997) Peripheral T cell survival requires continual ligation of the T cell receptor to major histocompatibility complex-encoded molecules. J Exp Med 186 : 1269–1275.
34. FerreiraMAR, ManginoM, BrummeCJ, ZhaoZZ, MedlandSE, et al. (2010) Quantitative trait loci for CD4∶CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet 86 : 88–92 doi:10.1016/j.ajhg.2009.12.008
35. YalcinB, NicodJ, BhomraA, DavidsonS, CleakJ, et al. (2010) Commercially available outbred mice for genome-wide association studies. PLoS Genet 6 doi:10.1371/journal.pgen.1001085
36. DamoiseauxJG, CautainB, BernardI, MasM, van Breda VriesmanPJ, et al. (1999) A dominant role for the thymus and MHC genes in determining the peripheral CD4/CD8 T cell ratio in the rat. J Immunol 163 : 2983–2989.
37. HansenC, SpuhlerK (1984) Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res 8 : 477–479.
38. TalbotCJ, NicodA, ChernySS, FulkerDW, CollinsAC, et al. (1999) High-resolution mapping of quantitative trait loci in outbred mice. Nat Genet 21 : 305–308 doi:10.1038/6825
39. YazbekSN, BuchnerDA, GeisingerJM, BurrageLC, SpiezioSH, et al. (2011) Deep congenic analysis identifies many strong, context-dependent QTLs, one of which, Slc35b4, regulates obesity and glucose homeostasis. Genome Research 21 : 1065–1073 doi:10.1101/gr.120741.111
40. TanIKL, MackinL, WangN, PapenfussAT, ElsoCM, et al. (2010) A recombination hotspot leads to sequence variability within a novel gene (AK005651) and contributes to type 1 diabetes susceptibility. Genome Research 20 : 1629–1638 doi:10.1101/gr.101881.109
41. Rat Genome Sequencing and Mapping Consortium (2013) BaudA, HermsenR, GuryevV, StridhP, et al. (2013) Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat Genet 45 : 767–775 doi:10.1038/ng.2644
42. JohannessonM, Lopez-AumatellR, StridhP, DiezM, TuncelJ, et al. (2009) A resource for the simultaneous high-resolution mapping of multiple quantitative trait loci in rats: the NIH heterogeneous stock. Genome Research 19 : 150–158 doi:10.1101/gr.081497.108
43. JeffreysAJ, KauppiL, NeumannR (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29 : 217–222 doi:10.1038/ng1001-217
44. MyersS, BottoloL, FreemanC, McVeanG, DonnellyP (2005) A fine-scale map of recombination rates and hotspots across the human genome. Science 310 : 321–324 doi:10.1126/science.1117196
45. AgarwalAK, XingC, DeMartinoGN, MizrachiD, HernandezMD, et al. (2010) PSMB8 encoding the β5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am J Hum Genet 87 : 866–872 doi:10.1016/j.ajhg.2010.10.031
46. DeversonEV, LeongL, SeeligA, CoadwellWJ, TredgettEM, et al. (1998) Functional analysis by site-directed mutagenesis of the complex polymorphism in rat transporter associated with antigen processing. J Immunol 160 : 2767–2779.
47. MomburgF, ArmandolaEA, PostM, HammerlingGJ (1996) Residues in TAP2 peptide transporters controlling substrate specificity. J Immunol 156 : 1756–1763.
48. JonssonAK, RaskL (1989) Human class II DNA and DOB genes display low sequence variability. Immunogenetics 29 : 411–413.
49. ButcherGW (1987) A list of monoclonal antibodies specific for alloantigens of the rat. J Immunogenet 14 : 163–176.
50. DiamondAG, LarkinsAP, WrightB, EllisST, ButcherGW, et al. (1984) The alloantigenic organization of RT1Aa, a class I major histocompatibility complex molecule of the rat. Eur J Immunol 14 : 405–412 doi:10.1002/eji.1830140505
51. HowardJC, ButcherGW, GalfreG, MilsteinC, MilsteinCP (1979) Monoclonal Antibodies as Tools to Analyze the Serological and Genetic Complexities of Major Transplantation Antigens. Immunol Rev 47 : 139–174 doi:10.1111/j.1600-065X.1979.tb00292.x
52. JolyE, LeongL, CoadwellWJ, ClarksonC, ButcherGW (1996) The rat MHC haplotype RT1c expresses two classical class I molecules. J Immunol 157 : 1551–1558.
53. RadaC, LorenziR, PowisSJ, van den BogaerdeJ, ParhamP, et al. (1990) Concerted evolution of class I genes in the major histocompatibility complex of murine rodents. Proc Natl Acad Sci USA 87 : 2167–2171.
54. CosmanD, KressM, KhouryG, JayG (1982) Tissue-specific expression of an unusual H-2 (class I)-related gene. Proc Natl Acad Sci USA 79 : 4947–4951.
55. JolyE, ClarksonC, HowardJC, ButcherGW (1995) Isolation of a functional cDNA encoding the RT1.Au MHC class I heavy chain by a novel PCR-based method. Immunogenetics 41 : 326–328.
56. HedrichHJ, AdamsM, ScienceICFLA (1990) Genetic monitoring of inbred strains of rats. Gustav Fischer 1.
57. FukumotoT, Robert McMasterW, WilliamsAF (1982) Mouse monoclonal antibodies against rat major histocompatibility antigens. Two Ia antigens and expression of Ia and class I antigens in rat thymus. Eur J Immunol 12 : 237–243 doi:10.1002/eji.1830120313
58. SpencerSC, FabreJW (1987) Identification in rat liver and serum of water-soluble class I MHC molecules possibly homologous to the murine Q10 gene product. Journal of Experimental Medicine 165 : 1595–1608 Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=3585249&retmode=ref&cmd=prlinks.
59. SpencerSC, FabreJW (1987) Water-soluble form of RT1.A class I MHC molecules in the kidney and liver of the rat. Immunogenetics 25 : 91–98.
60. DijkstraCD, DöppEA, JolingP, KraalG (1985) The heterogeneity of mononuclear phagocytes in lymphoid organs: distinct macrophage subpopulations in the rat recognized by monoclonal antibodies ED1, ED2 and ED3. Immunology 54 : 589–599.
61. LivingstoneAM, PowisSJ, GüntherE, CramerDV, HowardJC, et al. (1991) Cim: an MHC class II-linked allelism affecting the antigenicity of a classical class I molecule for T lymphocytes. Immunogenetics 34 : 157–163.
62. SpeiserDE, LeesRK, HengartnerH, ZinkernagelRM, MacDonaldHR (1989) Positive and negative selection of T cell receptor V beta domains controlled by distinct cell populations in the thymus. J Exp Med 170 : 2165–2170.
63. BommhardtU, BeyerM, HünigT, ReichardtHM (2004) Molecular and cellular mechanisms of T cell development. Cell Mol Life Sci 61 : 263–280 doi:10.1007/s00018-003-3224-3
64. HünigT, Torres-NagelN, MehlingB, ParkHJ, HerrmannT (2001) Thymic development and repertoire selection: the rat perspective. Immunol Rev 184 : 7–19.
65. LawDA, SpruytLL, PatersonDJ, WilliamsAF (1989) Subsets of thymopoietic rat thymocytes defined by expression of the CD2 antigen and the MRC OX-22 determinant of the leukocyte-common antigen CD45. Eur J Immunol 19 : 2289–2295 doi:10.1002/eji.1830191217
66. MitnachtR, BischofA, Torres-NagelN, HünigT (1998) Opposite CD4/CD8 lineage decisions of CD4+8+ mouse and rat thymocytes to equivalent triggering signals: correlation with thymic expression of a truncated CD8 alpha chain in mice but not rats. J Immunol 160 : 700–707.
67. HünigT, MitnachtR (1991) T cell receptor-mediated selection of functional rat CD8 T cells from defined immature thymocyte precursors in short-term suspension culture. J Exp Med 173 : 561–568.
68. HogquistKA, GavinMA, BevanMJ (1993) Positive selection of CD8+ T cells induced by major histocompatibility complex binding peptides in fetal thymic organ culture. J Exp Med 177 : 1469–1473.
69. YangCP, BellEB (1992) Functional maturation of recent thymic emigrants in the periphery: development of alloreactivity correlates with the cyclic expression of CD45RC isoforms. Eur J Immunol 22 : 2261–2269 doi:10.1002/eji.1830220913
70. HosseinzadehH, GoldschneiderI (1993) Recent thymic emigrants in the rat express a unique antigenic phenotype and undergo post-thymic maturation in peripheral lymphoid tissues. J Immunol 150 : 1670–1679.
71. VolkmannA, ZalT, StockingerB (1997) Antigen-presenting cells in the thymus that can negatively select MHC class II-restricted T cells recognizing a circulating self antigen. J Immunol 158 : 693–706.
72. KleinL, HinterbergerM, Rohrscheidt vonJ, AichingerM (2011) Autonomous versus dendritic cell-dependent contributions of medullary thymic epithelial cells to central tolerance. Trends Immunol 32 : 188–193 doi:10.1016/j.it.2011.03.002
73. Ashton-RickardtPG, Van KaerL, SchumacherTN, PloeghHL, TonegawaS (1993) Peptide contributes to the specificity of positive selection of CD8+ T cells in the thymus. Cell 73 : 1041–1049.
74. DelaneyJR, SykulevY, EisenHN, TonegawaS (1998) Differences in the level of expression of class I major histocompatibility complex proteins on thymic epithelial and dendritic cells influence the decision of immature thymocytes between positive and negative selection. Proc Natl Acad Sci USA 95 : 5235–5240.
75. MurataS, SasakiK, KishimotoT, NiwaS-I, HayashiH, et al. (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316 : 1349–1353 doi:10.1126/science.1141915
76. XingY, JamesonSC, HogquistKA (2013) Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proc Natl Acad Sci USA 110 : 6979–6984 doi:10.1073/pnas.1222244110
77. KanetaM, OsawaM, SudoK, NakauchiH, FarrAG, et al. (2000) A role for pref-1 and HES-1 in thymocyte development. J Immunol 164 : 256–264.
78. VercauterenSM, SutherlandHJ (2004) Constitutively active Notch4 promotes early human hematopoietic progenitor cell maintenance while inhibiting differentiation and causes lymphoid abnormalities in vivo. Blood 104 : 2315–2322 doi:10.1182/blood-2004-01-0204
79. PuiJC, AllmanD, XuL, DeRoccoS, KarnellFG, et al. (1999) Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11 : 299–308.
80. DickinsonLA, JohT, KohwiY, Kohwi-ShigematsuT (1992) A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70 : 631–645.
81. NieH, MaikaSD, TuckerPW, GottliebPD (2005) A role for SATB1, a nuclear matrix association region-binding protein, in the development of CD8SP thymocytes and peripheral T lymphocytes. J Immunol 174 : 4745–4752.
82. DouekDC, AltmannDM (1997) HLA-DO is an intracellular class II molecule with distinctive thymic expression. Int Immunol 9 : 355–364.
83. GuceAI, MortimerSE, YoonT, PainterCA, JiangW, et al. (2013) HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat Struct Mol Biol 20 : 90–98 doi:10.1038/nsmb.2460
84. YoonT, MacmillanH, MortimerSE, JiangW, RinderknechtCH, et al. (2012) Mapping the HLA-DO/HLA-DM complex by FRET and mutagenesis. Proc Natl Acad Sci USA 109 : 11276–11281 doi:10.1073/pnas.1113966109
85. BraunsteinNS, GermainRN (1986) The mouse E beta 2 gene: a class II MHC beta gene with limited intraspecies polymorphism and an unusual pattern of transcription. EMBO J 5 : 2469–2476.
86. GreeneJM, WisemanRW, LankSM, BimberBN, KarlJA, et al. (2011) Differential MHC class I expression in distinct leukocyte subsets. BMC Immunol 12 : 39 doi:10.1186/1471-2172-12-39
87. WalkerBA, HuntLG, SowaAK, SkjødtK, GöbelTW, et al. (2011) The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc Natl Acad Sci USA 108 : 8396–8401 doi:10.1073/pnas.1019496108
88. KaufmanJ, MilneS, GöbelTW, WalkerBA, JacobJP, et al. (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401 : 923–925 doi:10.1038/44856
89. BaudatF, de MassyB (2007) Cis - and trans-acting elements regulate the mouse Psmb9 meiotic recombination hotspot. PLoS Genet 3: e100 doi:10.1371/journal.pgen.0030100
90. GuillonH, de MassyB (2002) An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat Genet 32 : 296–299 doi:10.1038/ng990
91. QinJ, RichardsonLL, JasinM, HandelMA, ArnheimN (2004) Mouse strains with an active H2-Ea meiotic recombination hot spot exhibit increased levels of H2-Ea-specific DNA breaks in testicular germ cells. Mol Cell Biol 24 : 1655–1666.
92. CullenM, NobleJ, ErlichH, ThorpeK, BeckS, et al. (1997) Characterization of recombination in the HLA class II region. Am J Hum Genet 60 : 397–407.
93. LobelSA, CramerDV (1981) Demonstration of a new genetic locus in the major histocompatibility system of the rat. Immunogenetics 13 : 465–473.
94. TuncelJ, HaagS, CarlsenS, YauACY, LuS, et al. (2012) Class II major histocompatibility complex-associated response to type XI collagen regulates the development of chronic arthritis in rats. Arthritis Rheum 64 : 2537–2547 doi:10.1002/art.34461
95. GillTJ, KunzHW (1979) Gene complex controlling growth and fertility linked to the major histocompatibility complex in the rat. The American Journal of Pathology 96 : 185–206.
96. GillettA, MaratouK, FewingsC, HarrisRA, JagodicM, et al. (2009) Alternative splicing and transcriptome profiling of experimental autoimmune encephalomyelitis using genome-wide exon arrays. PLoS ONE 4: e7773 doi:10.1371/journal.pone.0007773
97. YtterbergAJ, PeltierJ-B, van WijkKJ (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140 : 984–997 doi:10.1104/pp.105.076083
98. MottR, TalbotCJ, TurriMG, CollinsAC, FlintJ (2000) A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA 97 : 12649–12654 doi:10.1073/pnas.230304397
99. ValdarW, HolmesCC, MottR, FlintJ (2009) Mapping in structured populations by resample model averaging. Genetics 182 : 1263–1277 doi:10.1534/genetics.109.100727
Štítky
Genetika Reprodukčná medicína
Článek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



